- 1School of Biomedical Engineering, Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, China
- 2Department of Biochemistry, Periyar University, Salem, India
- 3Department of Pharmacognosy, ISF College of Pharmacy, Moga Punjab, India
The surface chemical characteristics of nanomaterials have a substantial impact on the affinity probe used to enrich proteins and peptides for MALDI-MS analysis of a real human sample. Detecting phosphoproteins involved in signalling is always difficult, even with recent developments in mass spectrometry, because protein phosphorylation is often temporary from complicated mixtures. This review summarizes current research on the successful enrichment of various intriguing glycoproteins and glycol peptides using surface affinity materials with distinctive qualities such as low cost, excellent structural stability, diversity, and multifunction. As a consequence, this review will provide a quick overview of the scholars from various backgrounds who are working in this intriguing interdisciplinary field. Label-free cancer biomarkers and other diseases will benefit from future challenges.
Introduction
Phosphorylation is the main post-translational protein adjustment, influencing signal transduction, quality articulation, cell cycle, cytoskeletal guideline, and apoptosis, among other cell flagging pathways (Wang et al., 2014; Müller, 2018; Ramazi and Zahiri, 2021). Phosphoproteomics has been a prominent study issue since it is critical for interpreting biological changes and discovering new techniques to identify and treat sickness (Arrington et al., 2017; Farooq et al., 2022). Enrichment of objective biomolecules is usually required in today’s biological examinations because direct detection of target biomolecules is often difficult due to numerous typical constraints (e.g., low abundance of objective biomolecules, the presence of contaminants) (Angel et al., 2012; Rejeeth et al., 2018). Affinity materials with unique benefits such as reasonable price range, amazing physical consistency, diversity, and functionality have been widely used for enriching objective biomolecules from complicated biological samples based on the peculiar affinity communication systems between the affinity materials and the objective biomolecules (Limo et al., 2018; Zhou and Pang, 2018). In response of the fast development in material science, particularly the development of nanotechnologies, a variety of excellent affinity materials have been developed to enrich different biological molecules (Ray et al., 2010; Tiambeng et al., 2020). Interestingly, in the field of phosphoproteomics, affinity materials for increasing phosphorylated proteins/peptides are getting a lot of attention (Wang et al., 2015) (Riley and Coon, 2016).
Mass spectrometry (MS) with great specificity and faster data processing has been the most dynamic and crucial tool for phosphoproteomics research in recent years (Iwamoto and Shimada, 2018; Macklin et al., 2020). Regardless, due to significant issues such as the low abundance of phosphorylated proteins/peptides resulting from high heterogeneity and low stoichiometry of protein phosphorylation, the sign victory impact of high-overflow parts (e.g., nonphosphorylated atoms, lipids, and salts), and the low ionisation productivity of phosphorylated proteins/peptides in MS examination, direct MS-based phosphoproteomics investigation is not recommended (Goel et al., 2018; Bekker-Jensen et al., 2020). As a result, performing preliminary analysis before MS has become a crucial step in efficiently reducing the complexity of proteome preliminaries and concentrating phosphorylated proteins/peptides to improve MS investigation outcomes (Angel et al., 2012). Enrichment analysis prior to MS has become a vital step in successfully lowering the complexity of proteome trials and concentrating phosphorylated proteins/peptides in order to improve MS analysis outcomes up to this point (Nakayasu et al., 2021). The employment of a linker molecule to render metal ions impotent on a porosity bead, polymer bead, or nanomaterial is one of the most extensively used technologies, and great effort has gone into constructing the IMAC material (Colón and Furukawa, 2020). Linkers like iminodiacetic acid and nitrilotriacetic acid are commonly used to chelate metal ions. The attached metal ions were easily washed away during the sample loading and washing procedure due to the comparatively anaemic interaction, dramatically lowering the enrichment efficiency (Rodzik et al., 2020) (Fang et al., 2017).
Unfortunately, intrinsic difficulties such as high-cost antibodies, limited universal applicability, and biological sample decay hamper these approaches (Morofuji et al., 2016; Zhou and Rossi, 2017). Affinity material-based enrichment approaches, on the other hand, have a number of advantages, including universality, low cost, and minimal impact on the structural integrity of phosphorylated proteins/peptides (Wang et al., 2015; Rejeeth et al., 2018; Zhang et al., 2019; Rejeeth et al., 2022). However, affinity material-based approaches still face nonspecific binding of nonphosphorylated proteins/peptides, which can significantly hamper the selective enrichment of phosphorylated proteins/peptides (Delom and Chevet, 2006; Rainer and Bonn, 2015). Developing high-performance affinity materials remains the most important and broadly used way to improve the enrichment performance of affinity material-based strategies, despite the addition of some auxiliary measures (e.g., chemical treatment of the biological sample, pH directive of loading buffers, and additive addition) (Gabriel et al., 2019; Keeble Anthony et al., 2019).
The present state of affinity materials for the enrichment of phosphorylated proteins and peptides is discussed in this review. The focus will be on the design and creation of affinity materials, as well as the presentation and discussion of relevant enrichment methods. The difficulties and future directions of this discipline will also be highlighted. Because several assessments on specific topics, such as MS technology, enrichment conditions, and enrichment protocols, have already been completed, the details of some related issues, such as MS technology, enrichment conditions, and enrichment protocols, are not presented here.
Affinity-bound nanoparticles: Manufacturing and properties
Biomolecule-conjugated nanoparticles (NPs) have been shown to have promising uses in bioanalysis. The reviewed affinity materials are grouped into three types depending on the species of the affinity site: magnetic-based affinity materials, silica-based affinity matrials and hydrogel-based affinity materials (Scheme 1). In the meantime, the enrichment processes utilised by these affinity materials will be described. In general, the three particles involve the use of various reducing agents, formation management, size preparation, and the discovery of some interesting previous literature reports for medication and gene delivery against various cancers and other ailments.
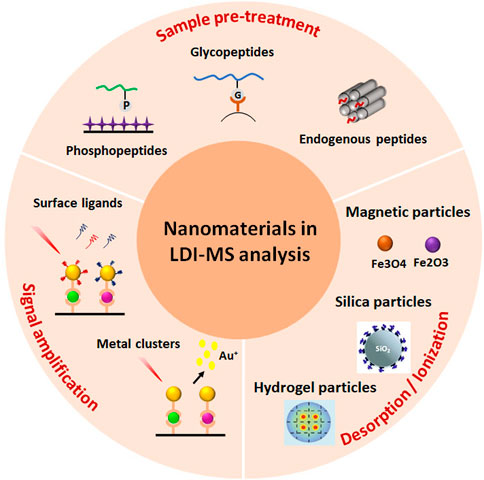
SCHEME 1. Schematic illustration of phosphopeptide enrichment for MS detection using diverse affinity materials.
Magnetic nanoparticles
Magnetic nanoparticles are made up of an inorganic core containing paramagnetic/superparamagnetic structures like magnetite (Fe3O4) or maghemite (Fe2O3), as well as a shielding layer, which is often made up of polymers and gives colloidal stability and solubility (Gao et al., 2009; Wu et al., 2015). Magnetic nanoparticles (MNPs) have been studied for a variety of uses. In order to provide imaging, treatments, and targeting modalities, MNPs must be encased in biocompatible materials, primarily polymeric ligands containing chemical functional groups to conjugate biomolecules and ligands (See Figure 1).
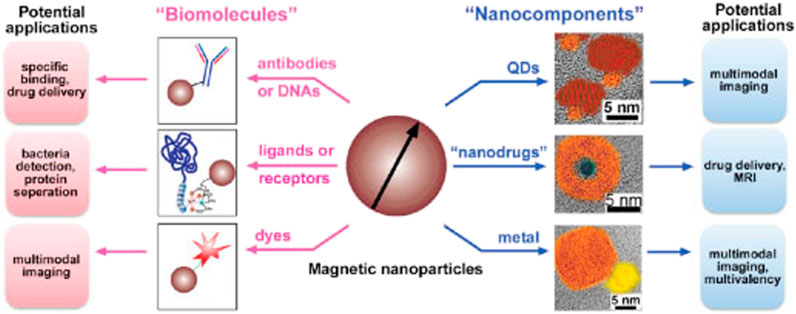
FIGURE 1. The scheme illustrates two strategies to fabricate multifunctional magnetic nanoparticles and their potential applications. Reproduced from ref 30. Copyright 2003 American Chemical Society.
Due to their unique properties and modalities in fields such as imaging, drug administration, magnetic separation, and purification, magnetic particles, particularly superparamagnetic MNPs, have increased in prominence in biomedical research in recent decades (Ali et al., 2021). Superparamagnetic iron oxide nanoparticles (SPIONs) have been used in tumour imaging, stem cell detection, metastatic breast cancer imaging, Alzheimer’s disease detection, lymph node imaging, and many more imaging applications as an intrinsic T2 magnetic resonance imaging (MRI) contrast agent (Cheng et al., 2014; Fatima et al., 2021). Readers can be enticed to look into extensive evaluations in this sector. SPIONs have been used in a variety of applications, including chemotherapeutics, gene transfer, and hyperthermia (Musielak et al., 2019; Suciu et al., 2020).
Magnetic separation has significant potential in biological research because it allows for targeted capture that is selective, sensitive, and regulated (Yildiz, 2016). Magnetic separation, which employs an external magnetic field to trap target molecules with MNPs via affinity interaction, may be more time, labour, and yield efficient than complex and time-consuming chromatographic separations and purifications (Zborowski and Chalmers, 2011; Musielak et al., 2019). To create a design a superparamagnetic-magnetically responsive only in the presence of an external magnetic field-adsorbent or catcher with high magnetization must be used with proper features such as stability, biocompatibility, and having binding units affinity ligands to capture the target of interest with high specificity and selectivity, and a superparamagnetic-magnetically responsive only in the presence of an external magnetic field-adsorbent or catcher with high magnetization must be used with proper features such as stability (Suciu et al., 2020).
The immobilisation of affinity ligands on the surface of particles toward the target proteins is the basis for magnetic particle-based separation of proteins such as antibodies, biomarkers, and enzymes, and a comprehensive list of applications, including homemade and commercial magnetic particles, has been reviewed elsewhere (Mani et al., 2011; Yildiz, 2016). On the other hand, the goal of this article is to discuss recent research in this field. Immobilized metal affinity chromatography (IMAC) has long been a favoured enrichment method in phosphoproteomics (Ndassa et al., 2006). Apart from electronic attraction, IMAC is prepared using metal cations with unoccupied organisation orbitals, and chelation between the immobilised metal cations and the phosphoryl oxygen in phosphorylated proteins/peptides is the primary enrichment mechanism (Balmant et al., 2016). The matrix supports carrying chelating ligands are normally synthesised first, and then the metal cations are immobilised onto the chelating ligands to form IMAC affinity materials. Cu2+, Zn2+, Fe3+, Ga3+, Al3+, Ti4+, and Zr4+ are some of the metal cations that have been employed in IMAC thus far. In traditional IMAC materials, the most frequent metal cations are Fe3+ and Ga3+ (Ruprecht et al., 2015). Antibody-free approaches are promising and need to be developed for real-case applications in serum to address the limitations of antibody-based techniques in terms of robustness, expense, and throughput. In this work, we demonstrated a novel approach using hyaluronic acid (HA)-modified materials/devices for the extraction, detection, and profiling of serum biomarkers via ligand-protein interactions. Figure 2 shown using hyaluronic acid (HA)-modified materials/devices (e.g., Fe3O4@SiO2@HA) (Rejeeth et al., 2018).
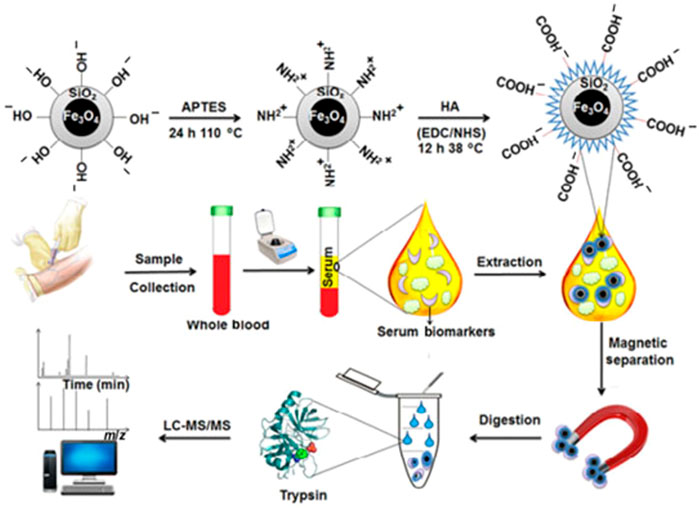
FIGURE 2. Schematic illustration for the preparation of Fe3O4@SiO2@HA particles and their application in the extraction and detection of biomarkers from human serum samples by MS. Reproduced from ref 7. Copyright 2018 Springer.
Due to the unusual coordination specificity of metal (IV)-phosphate chemistry, Zr4+-IMAC and Ti4+-IMAC with phosphate group as chelating ligands displayed greater selectivity for phosphopeptides than regular Fe3+-IMAC beads (Ruprecht et al., 2015). Iminodicitic acid (IDA) and nitrilotriacetic acid (NTA) are two chelating ligands that are frequently employed to immobilise metal cations (e.g., Fe3+ and Ga3+) (Blankespoor et al., 2005). Sample loading and washing could impair the immobilised metal cations since each metal cation can only bind to one NTA or IDA ligand. Figure 3 discusses, scientists are on the lookout for new chelating ligands with greater chelating ability. As a result, novel, extremely effective chelating ligands such as arsenate (AsO32), phosphate (PO32), adenosine triphosphate (ATP), and dopamine have proved successful in immobilising metal cations, particularly Ti4+ and Zr4+ (Yan et al., 2013).
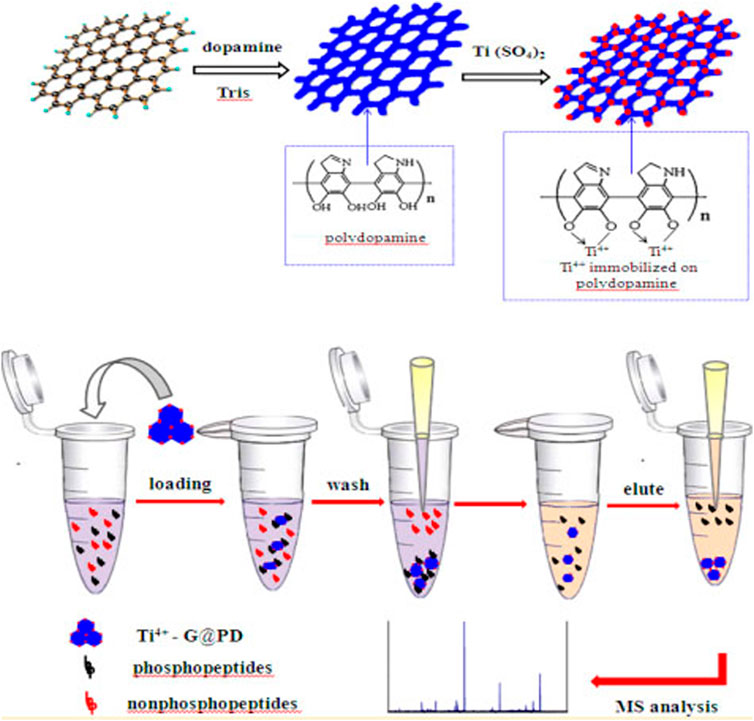
FIGURE 3. The Ti4+-G@PD with the above unique properties was anticipated to have excellent performance for the selective enrichment of phosphopeptides, which would be highly beneficial for mass spectrometric analysis. Reproduced from ref 45. Copyright 2003 American Chemical Society.
As affinity probes, magnetic nanoparticles (Fe3O4@Al2O3) efficiently concentrate polyhistidine (His)-tagged proteins/peptides from complicated materials (Zhang et al., 2013). This method can be used to quickly determine whether or not a sample contains His6-tagged organisms. Cell lysates containing recombinant Shiga-like toxins labelled with His6 were used as samples to demonstrate that this technology may be used to examine exceedingly complex samples (Schmit et al., 2019). The entire analysis procedure takes around 10 min, including on-plate enrichment and enzymatic digestion, followed by MALDI-MS analysis (Rühl et al., 2017). For similar reasons, magnetic nanoparticles (MNP) with amine functionalization are roughly four times smaller than those previously utilised. Researchers compared the enrichment performance of the magnetic nanoparticle approach to a commercially available glycopeptide enrichment kit offered by EMD Millipore using tandem mass tags from Thermo Scientific (Bodnar and Perreault, 2013). The MNP approach is easy to apply and takes less than 10 min, and it enriches sialylatedglycopeptides quantitatively and qualitatively more than a commercially available kit (Bodnar and Perreault, 2013). Another study used magnetic nanoparticles modified with polyethyleneimine (PEI) as a highly selective affinity probe. The particles collected phosphopeptides from a tryptic digest of a protein mixture that contained 0.07% (mole/mole) phosphoproteins, which is the highest selectivity ever observed. In matrix-assisted laser desorption/ionization mass spectrometry analysis, the detection limits of phosphopeptides generated from bovine casein and casein were 5 fmol (Chen et al., 2011). This method was also utilised to boost the amount of phosphopeptides in a non-fat milk protein digest. Using antibodies against target peptides, MNPs could concentrate serum biomarker peptide fragments. MNPs were conjugated to polyclonal antibodies against the tumour necrosis factor (TNF) peptide and utilised to collect TNF from a TNF-spiked blood sample, followed by MS analysis (Dunning and Lame, 2018). These effective applications to single glycoproteins and complete proteome combinations collected from physiological fluids revealed magnetic practical flexibility that is unrivalled.
Silica particles
Two synthesis methods for silica nanoparticles include the Stober process and reverse micro emulsion (Koźlecki et al., 2016). A water in oil (w/o) micro emulsion is formed up of a homogeneous mixture of water, oil, and surfactant molecules (Koźlecki et al., 2016). The Stober method involves hydrolysis and condensation of siloxane precursors (such as tetraethylorthosilicate TEOS) with ethanol and ammonia to produce silica particles (Kim and Kim, 2002). This method has been used to incorporate a variety of organic colour chemicals. They are classified as spherical, mesoporous, or macrospores in biomedical applications (Borzęcka et al., 2022). Spherical silica NPs, on the other hand, carry cargo through encapsulation or conjugation. Mesopores (pore sizes ranging from 2 to 30 nm) and macrospores are the two types of pores found in mesoporous silica NPs (50–100 nm pore size) (Narayan et al., 2018). Physical and chemical adsorption are frequently utilised to disperse active payloads. The release profile of payloads delivered by porous silica NPs is controlled by chemical linkers or silica matrix degradation, whereas the “gatekeeper” strategy of modifying the inner surface of the pores to control the binding affinity with drugs controls the release profile of payloads delivered by porous silica NPs (Jafari et al., 2019).
MO modified mesoporous silica composites with various affinity sites were made using simple techniques that took into account the unique qualities of mesoporous silica (e.g., size exclusion effect, high surface area, easy modifiability, and stable skeleton) as well as MO affinity ability (e.g., solgel) (Mamaeva et al., 2013). These compounds can extract phosphopeptides from complicated biosamples quickly and selectively (e.g., human serum). Using mesoporous silica particles, titanium phosphonate was employed to selectively extract phosphopeptides from complicated peptide and protein combinations (Kim et al., 2010). Using MALDI-TOFMS detection, the limit of detection for phosphopeptides from -casein and standard phosphopeptide spikes in human serum was as low as 1.25 fmol (Karayel et al., 2020). The modified mesoporous silica particles were then utilised to enrich phosphopeptides from the serum of patients with hepatocellular carcinoma and healthy persons, which were subsequently evaluated using MALDI-TOFMS. MALDI-TOF MS/MS results for blood phosphopeptide profiling were also confirmed utilising a combination of isobaric tagging for relative and absolute quantitative labelling (Rauniyar and Yates, 2014). The substantial disparities in serum phosphopeptide profiling between cancer patients and healthy people emphasise the technique’s utility in cancer detection and biomarker development. This approach could be applied to a wide range of biological samples and disorders. Microspheres with a Fe3O4@nSiO2 core and a perpendicularly oriented mesoporous SiO2 shell (Fe3O4@nSiO2@mSiO2) were used to develop a novel peptide enrichment approach (Chen et al., 2010). The magnetic receptivity of the Fe3O4@nSiO2@mSiO2 microspheres is advantageous, as it speeds up and simplifies the enrichment process. The microspheres’ highly organised nanoscale pores (2 nm) and high surface areas were shown to have a good size-exclusion effect for peptide adsorption. A Fe3O4@nSiO2@mSiO2 microsphere was also investigated. The microspheres were used to concentrate endogenous peptides in rat brain extract on a large scale. After enrichment, the sample was examined with an Automated nano-LC-ESI-MS/MS, revealing a total of 60 distinct peptides (Bian et al., 2020). Figure 4 shown the Fe3O4@nSiO2@mSiO2 microspheres are a viable contender for selectively separating and enriching endogenous peptides from difficult biological samples due to their simple and low-cost production as well as the convenient and efficient enrichment procedure (Zhu et al., 2012).
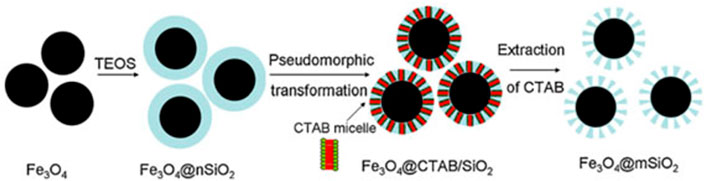
FIGURE 4. Schematic diagram for the preparation of monodisperse Fe3O4@mSiO2 microspheres. Reproduced from ref 63. Copyright 2012 Elsevier.
Phosphate-imprinted mesoporous silica nanoparticles (MSNs) as a sorbent for phosphopeptide selective enrichment and an offline combination with matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) for very efficient protein phosphorylation analysis (Rahi et al., 2016). To make the molecule, a new technology called dual-template docking guided molecular imprinting was applied (DTD-OMI). High phosphopeptide selectivity, interference tolerance, rapid binding equilibrium, and substantial binding capacity were among the attributes of the molecularly imprinted mesoporous material produced, making it an ideal sorbent for selective phosphopeptide enrichment.
Macroporous silica with open big pores outperforms mesoporous silica in terms of MOs loading and subsequent phosphorylated protein/peptide binding to affinity composites. For chromatography, a mechanically stable silica microparticle with macrosized interior pores (1.6 m particles with 100 nm pores) was developed (Mann et al., 2013). Concanavalin A (Con A) and Aleuriaaurantia lectin (AAL) were used to derivatize the material, and the binding capabilities were examined using conventional glycoproteins. Con A was able to bind 75 g of a common glycoprotein in a 50 1 mm column, demonstrating good binding capacity for microaffinity enrichment. In a 50 1 mm column, Con A was able to bind 75 g of a common glycoprotein, exhibiting good binding capacity for microaffinity enrichment (Mann et al., 2013). Importantly, in some mesoporous materials with lengthy channels, the open macroporous structure can allow effective mass transmission while avoiding the so-called “shadow effect.” Figure 5 shown to make Al-MOSF and Ti-MOSF, Al2O3 and TiO2 were placed onto macroporous ordered silica foam, respectively (MOSF) (Wan et al., 2009).
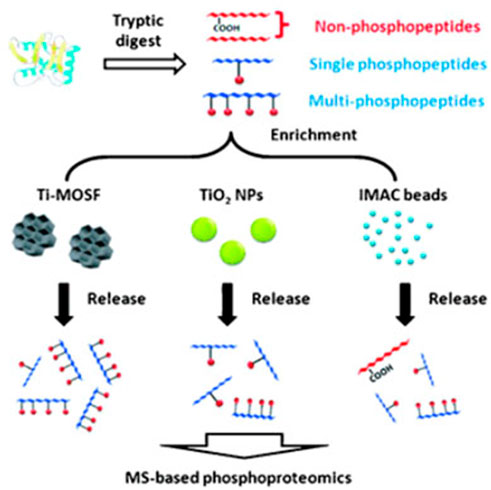
FIGURE 5. Schematic illustration of strategies employed for enriching phosphopeptides. Reproduced from ref 66. Copyright 2009 Wiley.
Hydrogel particles
Using cellulose as a matrix, a new simple synthetic technique for producing magneticcellulose or nanocomposite microspheres was successfully developed. The cellulose matrix’s micro-nano porous architectures were critical in in situ synthesising TiO2 nanoparticles while preserving TiO2’s basic structure and character, enabling for enrichment in a hostile environment (Montoya et al., 2011). Due to their strong capture capability, the novel magnetic cellulose/TiO2 nanocomposite microspheres (MCTiMs) showed high effectiveness for selective extraction of trace phosphopeptides from tryptic digest of alp-casein and human serum samples. With a molar ratio of 1:1000, MCTiMs were used to preferentially concentrate phosphopeptides from alpha-casein and BSA mixtures, which was significantly better than commercial TiO2 nanoparticles (Kupcik et al., 2019). The Figure 6 shown TiO2 nanoparticles implanted in the magnetic cellulose microspheres had high specific surface areas and a high adsorbing nature, resulting in a strong Lewis acid-base interaction, according to our findings. As a result, MCTiMs will be a great choice for phosphopeptide enrichment in complex systems using MS (Tan et al., 2013).
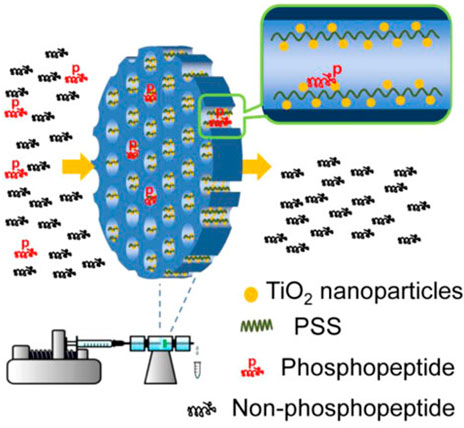
FIGURE 6. Schematic illustration of strategies employed for enriching phosphopeptides. Reproduced from ref 69. Copyright 2013 American Chemical Society.
For the first time, magnetic cellulose/TiO2 nanocomposite microspheres (MCTiMs) were made using a two-step process. In the first stage, magnetic cellulose microspheres (MCMs) were created by microwave heating cellulose/Fe3O4 colloidal micro-droplets efficiently to induce the sol-gel transition (Luo et al., 2009). MCTiMs were generated in the second step by dispersing MCMs in TiO2 precursor solution and then generating TiO2 nanoparticles in situ in MCMs’ micro-nano pores. Meanwhile, the amount of TiO2 in MCTiMs may be controlled by changing the precursor solution feeding volume to make MCTiMs-1, MCTiMs-2, MCTiMs-3, and MCTiMs-5, a series of MCTiMs (Duan et al., 2015). Meanwhile, the amount of TiO2 in MCTiMs may be controlled by changing the precursor solution feeding volume to make MCTiMs-1, MCTiMs-2, MCTiMs-3, and MCTiMs-5, a series of MCTiMs. The growth of a more compact shell layer at a higher level of TBOT resulted in an impediment for TBOT diffusing into the microspheres due to the uncontrolled creation of TiO2 nanoparticles on the surface of microspheres. The successful application of phosphopeptide enrichment necessitates the use of microspheres with excellent magnetic response qualities. Using an external magnetic field, MCTiMs with a sensitive magnetic response can be swiftly removed from peptide solutions, allowing for convenient recycling. A tryptic digest of alpha-casein, a combination of alpha casein and BSA, and fresh human blood samples were used to demonstrate the selectivity of MCTiMs for phosphopeptides. The process and procedure for phosphopeptide enrichment utilising MCTiMs are proposed. The TiO2 nanoparticles were firmly attached in the pores of MCTiMs, and the magnetic cellulose matrix’s pore wall safeguarded their structure and reaction activity. During the incubation process, phosphopeptides diffused rapidly into the magnetic cellulose microspheres, colliding with TiO2 nanoparticles and quickly anchoring on the TiO2 surface via Lewis reaction. Enrichment was successful and effective. Based on Lewis acid-base interaction, TiO2 with a positively charged surface can selectively adsorb phosphorylated species at acidic pH, and TiO2 with a large surface area has a stronger binding ability on phosphopeptides (Tan et al., 2013).
Magnetic celluloseTiO2 was developed by Duan’s research team for selective phosphopeptide enrichment. Magnetic cellulose microspheres were created using Solgel (MCMs). The transition of cellulose Fe3O4 colloidal microdroplets is caused by microwave heating. Then MCMs were distributed in TiO2 precursor solution and in situ manufactured TiO2 nanoparticles within the porous structure of MCMs to make magnetic celluloseTiO2nanocomposite microspheres (MCTiMs) (Long et al., 2017). MCTiMs had their magnetic hysteresis curve reduced, but they still allowed for efficient separation under external magnetic fields. Using the Lewis acid-base method, the phosphopeptides could be efficiently bonded to the nanosphere and separated, as shown in Figure 7.
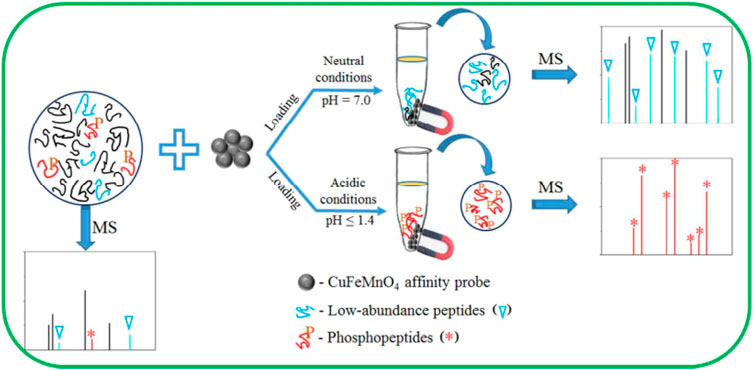
FIGURE 7. Schematic illustration of strategies employed for enriching phosphopeptides. Reproduced from ref 72. Copyright 2017 American Chemical Society.
Conclusion
A wide range of affinity materials for enrichment of phosphorylated proteins/peptides based on diverse enrichment processes have been developed during the last few decades. Despite significant progress, the majority of these materials’ applications are still in their infancy. The bulk of these affinity materials were tested utilising similar or even rigid assessment systems (e.g., digest mixes of standard pure phosphorylated and nonphosphorylated proteins, human serum, and nonfat milk), demanding more extensive and precise testing. It is important to note that proper enrichment conditions, such as pH and solvent composition, are also necessary for effective phosphoproteomics uses of these materials. Furthermore, because different affinity materials might have complementary enrichment capability, a mixture of affinity materials can be used when a comprehensive analysis of exceedingly complicated biosamples is required. Furthermore, the interaction mechanisms and concepts between affinity materials and target phosphorylated molecules are critical for the design of desired efficient affinity materials, though the bulk of these are unknown and require further research.
A substantial amount of research will be committed to the enhancement and production of affinity materials to meet the needs of phosphoproteomics analysis, in tandem with the development of phosphoproteomics analysis. More powerful multifunctional affinity materials are projected to come from the combination of established preparation methods and emerging technologies (e.g., nanotechnology). Understanding the basic microscopic enrichment methods and principles could be another challenge, thus further research will be done in this area to provide theoretical guidance for the design and fabrication of powerful affinity materials.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors are gratefully acknowledged by Dr. D. S. Kothari Postdoctoral Fellowship, No. F.4-2/2006 (BSR)/BL/19-20/0217 from the University Grants Commission (UGC) of India and Periyar University, Salem, India for providing the necessary facilities in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, A., Shah, T., Ullah, R., Zhou, P., Guo, M., Ovais, M., et al. (2021). Review on recent progress in magnetic nanoparticles: Synthesis, characterization, and diverse applications. Front. Chem. 9, 629054. doi:10.3389/fchem.2021.629054
Angel, T. E., Aryal, U. K., Hengel, S. M., Baker, E. S., Kelly, R. T., Robinson, E. W., et al. (2012). Mass spectrometry-based proteomics: Existing capabilities and future directions. Chem. Soc. Rev. 41 (10), 3912–3928. doi:10.1039/c2cs15331a
Arrington, J. V., Hsu, C.-C., Elder, S. G., and Andy Tao, W. (2017). Recent advances in phosphoproteomics and application to neurological diseases. Analyst 142 (23), 4373–4387. doi:10.1039/c7an00985b
Balmant, K. M., Zhang, T., and Chen, S. (2016). Protein phosphorylation and redox modification in stomatal guard cells. Front. Physiol. 7, 26. doi:10.3389/fphys.2016.00026
Bekker-Jensen, D. B., Bernhardt, O. M., Hogrebe, A., Martinez-Val, A., Verbeke, L., Gandhi, T., et al. (2020). Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 11 (1), 787. doi:10.1038/s41467-020-14609-1
Bian, Y., Zheng, R., Bayer, F. P., Wong, C., Chang, Y.-C., Meng, C., et al. (2020). Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS. Nat. Commun. 11 (1), 157. doi:10.1038/s41467-019-13973-x
Blankespoor, R., Limoges, B., Schöllhorn, B., Syssa-Magalé, J.-L., and Yazidi, D. (2005). Dense monolayers of metal-chelating ligands covalently attached to carbon electrodes electrochemically and their useful application in affinity binding of histidine-tagged proteins. Langmuir 21 (8), 3362–3375. doi:10.1021/la047139y
Bodnar, E. D., and Perreault, H. (2013). Qualitative and quantitative assessment on the use of magnetic nanoparticles for glycopeptide enrichment. Anal. Chem. 85 (22), 10895–10903. doi:10.1021/ac402332z
Borzęcka, W., Pereira, P. M. R., Fernandes, R., Trindade, T., Torres, T., and Tomé, J. P. C. (2022). Spherical and rod shaped mesoporous silica nanoparticles for cancer-targeted and photosensitizer delivery in photodynamic therapy. J. Mat. Chem. B 10, 3248–3259. doi:10.1039/d1tb02299g
Chen, C.-T., Wang, L.-Y., and Ho, Y.-P. (2011). Use of polyethylenimine-modified magnetic nanoparticles for highly specific enrichment of phosphopeptides for mass spectrometric analysis. Anal. Bioanal. Chem. 399 (8), 2795–2806. doi:10.1007/s00216-010-4623-6
Chen, H., Liu, S., Yang, H., Mao, Y., Deng, C., Zhang, X., et al. (2010). Selective separation and enrichment of peptides for MS analysis using the microspheres composed of Fe3O4@nSiO2 core and perpendicularly aligned mesoporous SiO2 shell. Proteomics 10 (5), 930–939. doi:10.1002/pmic.200900553
Cheng, K., Yang, M., Zhang, R., Qin, C., Su, X., and Cheng, Z. (2014). Hybrid nanotrimers for dual T1 and T2-weighted magnetic resonance imaging. ACS Nano 8 (10), 9884–9896. doi:10.1021/nn500188y
Colón, Y. J., and Furukawa, S. (2020). Understanding the role of linker flexibility in soft porous coordination polymers. Mol. Syst. Des. Eng. 5 (1), 284–293. doi:10.1039/c9me00117d
Delom, F., and Chevet, E. (2006). Phosphoprotein analysis: From proteins to proteomes. Proteome Sci. 4 (1), 15. doi:10.1186/1477-5956-4-15
Duan, J., He, X., and Zhang, L. (2015). Magnetic cellulose–TiO2 nanocomposite microspheres for highly selective enrichment of phosphopeptides. Chem. Commun. 51 (2), 338–341. doi:10.1039/c4cc08442j
Dunning, C. M., and Lame, M. E. (2018). Development of a hybrid immunoaffinity-LC-MS/MS method for the quantification of active biotherapeutics targeting TNF-α in serum. 720006317. Waters Corporation.
Fang, X., Liu, Y., Jimenez, L., Duan, Y., Adkins, G. B., Qiao, L., et al. (2017). Rapid enrichment and sensitive detection of multiple metal ions enabled by macroporous graphene foam. Anal. Chem. 89 (21), 11758–11764. doi:10.1021/acs.analchem.7b03336
Farooq, A., Bhat, K. A., Mir, R. A., Mahajan, R., Nazir, M., Sharma, V., et al. (2022). Emerging trends in developing biosensor techniques to undertake plant phosphoproteomic analysis. J. Proteomics 253, 104458. doi:10.1016/j.jprot.2021.104458
Fatima, A., Ahmad, M. W., Al Saidi, A. K. A., Choudhury, A., Chang, Y., and Lee, G. H. (2021). Recent advances in gadolinium based contrast agents for bioimaging applications. Nanomater. (Basel) 11 (9), 2449. doi:10.3390/nano11092449
Gabriel, J., Höfner, G., and Wanner, K. T. (2019). A library screening strategy combining the concepts of MS binding assays and affinity selection mass spectrometry. Front. Chem. 7, 665. doi:10.3389/fchem.2019.00665
Gao, J., Gu, H., and Xu, B. (2009). Multifunctional magnetic nanoparticles: Design, synthesis, and biomedical applications. Acc. Chem. Res. 42 (8), 1097–1107. doi:10.1021/ar9000026
Goel, R. K., Paczkowska, M., Reimand, J., Napper, S., and Lukong, K. E. (2018). Phosphoproteomics analysis identifies novel candidate substrates of the nonreceptor tyrosine kinase, src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristoylation sites (SRMS). Mol. Cell. Proteomics 17 (5), 925–947. doi:10.1074/mcp.ra118.000643
Iwamoto, N., and Shimada, T. (2018). Recent advances in mass spectrometry-based approaches for proteomics and biologics: Great contribution for developing therapeutic antibodies. Pharmacol. Ther. 185, 147–154. doi:10.1016/j.pharmthera.2017.12.007
Jafari, S., Derakhshankhah, H., Alaei, L., Fattahi, A., Varnamkhasti, B. S., and Saboury, A. A. (2019). Mesoporous silica nanoparticles for therapeutic/diagnostic applications. Biomed. Pharmacother. 109, 1100–1111. doi:10.1016/j.biopha.2018.10.167
Karayel, Ö., Tonelli, F., Virreira Winter, S., Geyer, P. E., Fan, Y., Sammler, E. M., et al. (2020). Accurate MS-based Rab10 phosphorylation stoichiometry determination as readout for LRRK2 activity in Parkinson's disease. Mol. Cell. Proteomics 19 (9), 1546–1560. doi:10.1074/mcp.ra120.002055
Keeble Anthony, H., Turkki, P., Stokes, S., Khairil Anuar Irsyad, N. A., Rahikainen, R., Hytönen Vesa, P., et al. (2019). Approaching infinite affinity through engineering of peptide–protein interaction. Proc. Natl. Acad. Sci. U. S. A. 116 (52), 26523–26533. doi:10.1073/pnas.1909653116
Kim, J., Kim, B. C., Lopez-Ferrer, D., Petritis, K., and Smith, R. D. (2010). Nanobiocatalysis for protein digestion in proteomic analysis. Proteomics 10 (4), 687–699. doi:10.1002/pmic.200900519
Kim, K. D., and Kim, H. T. (2002). Formation of silica nanoparticles by hydrolysis of TEOS using a mixed semi-batch/batch method. J. Sol-Gel Sci. Technol. 25 (3), 183–189. doi:10.1023/a:1020217105290
Koźlecki, T., Polowczyk, I., Bastrzyk, A., and Sawiński, W. (2016). Improved synthesis of nanosized silica in water-in-oil microemulsions. J. Nanoparticles 2016, 1–9. doi:10.1155/2016/8203260
Kupcik, R., Macak, J. M., Rehulkova, H., Sopha, H., Fabrik, I., Anitha, V. C., et al. (2019). Amorphous TiO(2) nanotubes as a platform for highly selective phosphopeptide enrichment. ACS Omega 4 (7), 12156–12166. doi:10.1021/acsomega.9b00571
Limo, M. J., Sola-Rabada, A., Boix, E., Thota, V., Westcott, Z. C., Puddu, V., et al. (2018). Interactions between metal oxides and biomolecules: From fundamental understanding to applications. Chem. Rev. 118 (22), 11118–11193. doi:10.1021/acs.chemrev.7b00660
Long, X.-Y., Zhang, Z.-J., Li, J.-Y., Sheng, D., and Lian, H.-Z. (2017). Controllable preparation of CuFeMnO4 nanospheres as a novel multifunctional affinity probe for efficient adsorption and selective enrichment of low-abundance peptides and phosphopeptides. Anal. Chem. 89 (19), 10446–10453. doi:10.1021/acs.analchem.7b02476
Luo, X., Liu, S., Zhou, J., and Zhang, L. (2009). In situ synthesis of Fe3O4/cellulose microspheres with magnetic-induced protein delivery. J. Mat. Chem. 19 (21), 3538–3545. doi:10.1039/b900103d
Macklin, A., Khan, S., and Kislinger, T. (2020). Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteomics 17 (1), 17. doi:10.1186/s12014-020-09283-w
Mamaeva, V., Sahlgren, C., and Lindén, M. (2013). Mesoporous silica nanoparticles in medicine—recent advances. Adv. Drug Deliv. Rev. 65 (5), 689–702. doi:10.1016/j.addr.2012.07.018
Mani, V., Chikkaveeraiah, B. V., and Rusling, J. F. (2011). Magnetic particles in ultrasensitive biomarker protein measurements for cancer detection and monitoring. Expert Opin. Med. diagn. 5 (5), 381–391. doi:10.1517/17530059.2011.607161
Mann, B. F., Mann, A. K. P., Skrabalak, S. E., and Novotny, M. V. (2013). Sub 2-μm macroporous silica particles derivatized for enhanced lectin affinity enrichment of glycoproteins. Anal. Chem. 85 (3), 1905–1912. doi:10.1021/ac303274w
Montoya, A., Beltran, L., Casado, P., Rodríguez-Prados, J.-C., and Cutillas, P. R. (2011). Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods 54 (4), 370–378. doi:10.1016/j.ymeth.2011.02.004
Morofuji, N., Ojima, H., Hiraoka, N., Okusaka, T., Esaki, M., Nara, S., et al. (2016). Antibody-based proteomics to identify an apoptosis signature for early recurrence of hepatocellular carcinoma. Clin. Proteomics 13 (1), 28. doi:10.1186/s12014-016-9130-0
Müller, M. M. (2018). Post-translational modifications of protein backbones: Unique functions, mechanisms, and challenges. Biochemistry 57 (2), 177–185. doi:10.1021/acs.biochem.7b00861
Musielak, M., Piotrowski, I., and Suchorska, W. M. (2019). Superparamagnetic iron oxide nanoparticles (SPIONs) as a multifunctional tool in various cancer therapies. Rep. Pract. Oncol. Radiother. 24 (4), 307–314. doi:10.1016/j.rpor.2019.04.002
Nakayasu, E. S., Gritsenko, M., Piehowski, P. D., Gao, Y., Orton, D. J., Schepmoes, A. A., et al. (2021). Tutorial: Best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 16 (8), 3737–3760. doi:10.1038/s41596-021-00566-6
Narayan, R., Nayak, U. Y., Raichur, A. M., and Garg, S. (2018). Mesoporous silica nanoparticles: A comprehensive review on synthesis and recent advances. Pharmaceutics 10 (3), 118. doi:10.3390/pharmaceutics10030118
Ndassa, Y. M., Orsi, C., Marto, J. A., Chen, S., and Ross, M. M. (2006). Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. J. Proteome Res. 5 (10), 2789–2799. doi:10.1021/pr0602803
Rahi, P., Prakash, O., and Shouche, Y. S. (2016). Matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry (MALDI-TOF MS) based microbial identifications: Challenges and scopes for microbial ecologists. Front. Microbiol. 7, 1359. doi:10.3389/fmicb.2016.01359
Rainer, M., and Bonn, G. K. (2015). Enrichment of phosphorylated peptides and proteins by selective precipitation methods. Bioanalysis 7 (2), 243–252. doi:10.4155/bio.14.281
Ramazi, S., and Zahiri, J. (2021). Post-translational modifications in proteins: Resources, tools and prediction methods. Database. 2021, baab012. doi:10.1093/database/baab012
Rauniyar, N., and Yates, J. R. (2014). Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 13 (12), 5293–5309. doi:10.1021/pr500880b
Ray, S., Chandra, H., and Srivastava, S. (2010). Nanotechniques in proteomics: Current status, promises and challenges. Biosens. Bioelectron. 25 (11), 2389–2401. doi:10.1016/j.bios.2010.04.010
Rejeeth, C., Pang, X., Zhang, R., Xu, W., Sun, X., Liu, B., et al. (2018). Extraction, detection, and profiling of serum biomarkers using designed Fe3O4@SiO2@HA core–shell particles. Nano Res. 11 (1), 68–79. doi:10.1007/s12274-017-1591-6
Rejeeth, C., Sharma, A., Kannan, S., Kumar, R. S., Almansour, A. I., Arumugam, N., et al. (2022). Label-free electrochemical detection of the cancer biomarker platelet-derived growth factor receptor in human serum and cancer cells. ACS Biomater. Sci. Eng. 8 (2), 826–833. doi:10.1021/acsbiomaterials.1c01135
Riley, N. M., and Coon, J. J. (2016). Phosphoproteomics in the age of rapid and deep proteome profiling. Anal. Chem. 88 (1), 74–94. doi:10.1021/acs.analchem.5b04123
Rodzik, A., Pomastowski, P., Sagandykova, G. N., and Buszewski, B. (2020). Interactions of whey proteins with metal ions. Int. J. Mol. Sci. 21 (6), 2156. doi:10.3390/ijms21062156
Rühl, M., Golghalyani, V., Barka, G., Bahr, U., and Karas, M. (2017). Enhanced on-plate digestion of proteins using a MALDI-digestion chamber. Int. J. Mass Spectrom. 416, 37–45. doi:10.1016/j.ijms.2016.11.011
Ruprecht, B., Koch, H., Medard, G., Mundt, M., Kuster, B., and Lemeer, S. (2015). Comprehensive and reproducible phosphopeptide enrichment using iron immobilized metal ion affinity chromatography (Fe-IMAC) columns. Mol. Cell. Proteomics 14 (1), 205–215. doi:10.1074/mcp.m114.043109
Schmit, N. E., Neopane, K., and Hantschel, O. (2019). Targeted protein degradation through cytosolic delivery of monobody binders using bacterial toxins. ACS Chem. Biol. 14 (5), 916–924. doi:10.1021/acschembio.9b00113
Suciu, M., Ionescu, C. M., Ciorita, A., Tripon, S. C., Nica, D., Al-Salami, H., et al. (2020). Applications of superparamagnetic iron oxide nanoparticles in drug and therapeutic delivery, and biotechnological advancements. Beilstein J. Nanotechnol. 11, 1092–1109. doi:10.3762/bjnano.11.94
Tan, Y.-J., Sui, D., Wang, W.-H., Kuo, M.-H., Reid, G. E., and Bruening, M. L. (2013). Phosphopeptide enrichment with TiO2-modified membranes and investigation of tau protein phosphorylation. Anal. Chem. 85 (12), 5699–5706. doi:10.1021/ac400198n
Tiambeng, T. N., Roberts, D. S., Brown, K. A., Zhu, Y., Chen, B., Wu, Z., et al. (2020). Nanoproteomics enables proteoform-resolved analysis of low-abundance proteins in human serum. Nat. Commun. 11 (1), 3903. doi:10.1038/s41467-020-17643-1
Wan, J., Qian, K., Qiao, L., Wang, Y., Kong, J., Yang, P., et al. (2009). TiO2-Modified macroporous silica foams for advanced enrichment of multi-phosphorylated peptides. Chem. Eur. J. 15 (11), 2504–2508. doi:10.1002/chem.200802079
Wang, Y.-C., Peterson, S. E., and Loring, J. F. (2014). Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24 (2), 143–160. doi:10.1038/cr.2013.151
Wang, Z.-G., Lv, N., Bi, W.-Z., Zhang, J.-L., and Ni, J.-Z. (2015). Development of the affinity materials for phosphorylated proteins/peptides enrichment in phosphoproteomics analysis. ACS Appl. Mat. Interfaces 7 (16), 8377–8392. doi:10.1021/acsami.5b01254
Wu, W., Wu, Z., Yu, T., Jiang, C., and Kim, W.-S. (2015). Recent progress on magnetic iron oxide nanoparticles: Synthesis, surface functional strategies and biomedical applications. Sci. Technol. Adv. Mat. 16 (2), 023501. doi:10.1088/1468-6996/16/2/023501
Yan, Y., Zheng, Z., Deng, C., Li, Y., Zhang, X., and Yang, P. (2013). Hydrophilic polydopamine-coated graphene for metal ion immobilization as a novel immobilized metal ion affinity chromatography platform for phosphoproteome analysis. Anal. Chem. 85 (18), 8483–8487. doi:10.1021/ac401668e
Yildiz, I. (2016). Applications of magnetic nanoparticles in biomedical separation and purification. Nanotechnol. Rev. 5 (3), 331–340. doi:10.1515/ntrev-2015-0012
Zborowski, M., and Chalmers, J. J. (2011). Rare cell separation and analysis by magnetic sorting. Anal. Chem. 83 (21), 8050–8056. doi:10.1021/ac200550d
Zhang, L., Zhu, X., Jiao, D., Sun, Y., and Sun, H. (2013). Efficient purification of His-tagged protein by superparamagnetic Fe3O4/Au–ANTA–Co2+ nanoparticles. Mater. Sci. Eng. C 33 (4), 1989–1992. doi:10.1016/j.msec.2013.01.011
Zhang, R., Rejeeth, C., Xu, W., Zhu, C., Liu, X., Wan, J., et al. (2019). Label-free electrochemical sensor for CD44 by ligand-protein interaction. Anal. Chem. 91 (11), 7078–7085. doi:10.1021/acs.analchem.8b05966
Zhou, H.-X., and Pang, X. (2018). Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 118 (4), 1691–1741. doi:10.1021/acs.chemrev.7b00305
Zhou, J., and Rossi, J. (2017). Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 16 (3), 181–202. doi:10.1038/nrd.2016.199
Keywords: label-free, nanometerials, MALDI-MS, proteins, peptides
Citation: Rejeeth C and Sharma A (2022) Label-free designed nanomaterials enrichment and separation techniques for phosphoproteomics based on mass spectrometry. Front. Nanotechnol. 4:1047055. doi: 10.3389/fnano.2022.1047055
Received: 17 September 2022; Accepted: 21 November 2022;
Published: 01 December 2022.
Edited by:
Jan M. Macak, University of Pardubice, CzechiaReviewed by:
Vadivalagan Chithravel, China Medical University, TaiwanNipun Babu, University of Nebraska-Lincoln, United States
Copyright © 2022 Rejeeth and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chandrababu Rejeeth, Y3JlamVlQGdtYWlsLmNvbQ==
 Chandrababu Rejeeth
Chandrababu Rejeeth Alok Sharma
Alok Sharma