- 1Microbiology, College of Science and Engineering, University of Galway, Galway, Ireland
- 2SFI Centre for Medical Devices (CÚRAM), Galway, Ireland
- 3Chemical Engineering, University of Wyoming, Laramie, WY, United States
COVID-19 is a severe acute respiratory disease caused by SARS-CoV-2. From its initial appearance in Wuhan, China in 2019, it developed rapidly into a global pandemic. In addition to vaccines, therapeutic antibodies play an important role in immediately treating susceptible individuals to lessen severity of the disease. In this study, phage display technology was utilised to isolate human scFv antibody fragments that bind the receptor-binding domain (RBD) of SARS-CoV-2 Wuhan-Hu-1 spike protein. Of eight RBD-binding scFvs isolated, two inhibited interaction of RBD with ACE2 protein on VeroE6 cells. Both scFvs also exhibited binding to SARS-CoV-2 Delta variant spike protein but not to Omicron variant spike protein in a Raman spectroscopy immunotest. The study demonstrates the potential of recombinant antibody approaches to rapidly isolate antibody moieties with virus neutralisation potential.
Introduction
COVID-19 disease is caused by the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It first emerged in China in late 2019 and was defined as a pandemic by the World Health Organisation (WHO) in March 2020. It is associated with high rates of morbidity and mortality, and over 608 million infections and 6.5 million deaths have been attributed to it worldwide in September 2022 (World Health Organization, 2022).
SARS-CoV-2 gains entry to host cells by attaching to the host angiotensin-converting enzyme 2 (ACE2) receptor via the virus homotrimeric spike (S) glycoprotein, leading to entry and replication in ACE2-expressing cells including lung, heart, intestine and kidney cells. The spike protein is composed of domains S1, which contains the host receptor-binding domain (RBD) and an N-terminal domain (NTD), and S2, which mediates virus-cell membrane fusion (Letko et al., 2020). RBD is highly dynamic and alternates between a receptor-accessible “up”/“open” and a receptor-inaccessible “down”/“closed” state (Wrapp et al., 2020). Numerous studies of previously infected patients have identified antibodies against spike protein, and RBD in particular, which potently neutralise virus interaction with cells in vitro and prevent disease in animal models (Liu et al., 2020; Ke et al., 2022).
While the rapid development and roll-out of vaccines, at least in the developed world, greatly reduced the mortality associated with SARS-CoV-2, the development of therapeutics is also an important facet to lessen disease severity. This is particularly true in elderly or immunocompromised patients, or individuals with severe pre-existing conditions, in whom breakthrough infections can be life threatening (Beaudoin-Bussières et al., 2020; Jeyanathan et al., 2020; Havlin et al., 2021). Passive immunisation with antibodies against the virus spike protein has potential to neutralise SARS-CoV-2 (Yang et al., 2020) with immediate effect, rather than waiting for a number of weeks for active immunity to accrue from vaccination. A number of groups have reported the isolation of monoclonal antibodies which neutralise SARS-CoV-2 (reviewed in Corti et al., 2021; Hurt and Wheatley, 2021). Antibodies that have progressed to clinical trial stages to date are predominantly RBD-binding antibodies due to their high potency at blocking interaction of RBD with ACE2 (Li et al., 2022). Current details of monoclonal antibodies and antibody cocktails in Phase two or Phase three clinical trials for COVID-19 treatment or prophylaxis are available at https://www.antibodysociety.org/covid-19-biologics-tracker/.
Following FDA EUA approval of the use of convalescent plasma as a treatment for COVID-19 patients (Food and Drug Administration, 2020), studies confirmed the clinical benefit of administering COVID-19 convalescent plasma to hospitalised patients early in the course of disease (Casadevall et al., 2020). This led to U.S. Food and Drug Administration emergency use authorisations for a number of monoclonal antibodies and antibody combinations to reduce mortality in COVID-19 patients (Kumar et al., 2021; www.fda.gov). While a number of antibodies have demonstrated high efficacy in trials, their high administration doses (0.5–1.2 g per antibody per dose; Kumar et al., 2021) place considerable pressure on mammalian cell production pipelines, limiting their widespread use in disease treatment (Yang et al., 2021).
Therapeutic antibodies are traditionally produced via immunisation approaches, or reverse transcriptase-polymerase chain reaction (RT-PCR) amplification from B cells isolated from infected individuals, followed by in vitro screening (Hwang et al., 2022). Considerations of ethical and regulatory requirements, immunogenicity, cost, scale-up and timeliness have led, however, to increasing use of in vitro approaches to isolate (and engineer) instead recombinant antibody fragments with similar or improved properties compared to monoclonal antibodies. Isolation approaches, such as phage display technology, involve identification of binders of a ligand of interest from large, synthetic libraries of antibody fragments, typically of human origin, in a process which mimics antigen selection and affinity maturation (Marks et al., 1991). The approaches typically utilise antibody fragments that retain the variable V regions (encompassing the binding pocket) of full-length immunoglobulin molecules, but contain few or no constant (C) domains that mediate effector functions in vivo–but are unnecessary for virus neutralisation. The single-chain fragment variable (scFv) fragments utilised in this study contain the variable heavy (VH) and variable light (VL) domains of an antibody, joined by a peptide linker (Wronska et al., 2016). These small-sized fragments are easily produced in expression hosts such as Escherichia coli that are considerably cheaper, more robust and produce higher yields than the mammalian cells used to express whole antibodies (Wronska et al., 2016). Additionally, protein engineering to rapidly tune antibody properties such as affinity and specificity is facilitated in E. coli (Kiguchi et al., 2020).
Since its appearance, SARS-CoV-2 continues to undergo significant antigenic drift, resulting in the emergence of several variants of concern with higher virulence and/or reduced neutralisation by therapeutic antibodies (Jeong et al., 2022; Sievers et al., 2022). Safe and effective therapies are therefore needed to combat the transmissibility and disease severity of new SARS-CoV-2 variants. The ability to rapidly isolate antibody fragments using in vitro screening technologies, and to effectively and cheaply scale-up production in recombinant expression platforms, enables a quick pivot to antibodies which neutralise emerging virus variants. This can form part of a core toolset to respond to emerging variants in this and future epidemics, by providing faster responses, easier manufacturability and lower cost therapeutics.
A variety of recombinant antibody fragments have been isolated against SARS-CoV-2 to date, using cloning approaches from antibodies from COVID-19 patients (Ebihara et al., 2021), antibody gene libraries derived from convalescent COVID-19 patients (Bertoglio et al., 2021; Mendoza-Salazar et al., 2022; Minenkova et al., 2022), or by immunising animals such as alpacas (Güttler et al., 2021). Phage display has also been utilised to isolate nanobodies (single-domain antibodies derived from camelids; Muyldermans et al., 2013) with SARS-CoV-2 neutralising ability (Zhao et al., 2022), as well as scFv-Fc fusion proteins against nucleocapsid protein for use in diagnostics (Kim et al., 2021). Numerous studies have also reported the use of phage display to isolate scFvs against RBD but without investigating their virus neutralisation ability (Salem et al., 2022; Parray et al., 2020).
Therefore, the goal of our work was to demonstrate rapid isolation of SARS-CoV-2-binding antibody fragments from a naïve human scFv antibody library and characterisation of their virus neutralisation potential. Phage display was utilised to identify fragments that bind spike protein RBD, followed by screening for inhibition of the RBD-ACE2 interaction and investigation of binding of emerging virus variant spike proteins to determine the potential for cross-reactive neutralising ability. The work demonstrates the potential of recombinant antibody approaches to rapidly develop virus-neutralising antibodies for use in passive immunotherapy.
Materials and methods
Materials
All chemicals were from Sigma-Aldrich unless otherwise specified. E. coli strain HB2151 was used for soluble expression of scFvs. The YamoI human scFv library was from Montarop Yamabhai (Pansri et al., 2009). SARS-CoV-2 proteins and inactivated viruses from BEI Resources (Manasses, Virginia) are listed in the Acknowledgements. The Omicron (B.1.1.529) variant spike trimer was purchased from Sino Biological (cat. #40589-V08H26).
ELISA analysis
Procedures for phage library panning were as described previously (Antoine et al., 2022). After three rounds of panning, eluted phages were rescued and titred in E. coli TG1 cells for RBD-binding assessment by ELISA. Wells of a 96-well microtitre plate (Maxisorb, Nunc) were coated overnight at 4°C with 2 μg/ml of RBD (BEI Resources) in PBS. After 3 washes with PBS, wells were blocked using PBS/4% bovine serum albumin (BSA) for 2 h at room temperature, followed by five washes with PBS. Wells were incubated with 100 µl of scFv-phage or soluble scFvs for 1 h at room temperature, and washed three times with PBS/0.1% Tween-20 and three times with PBS. After 1 h incubation at room temperature with anti-M13 horseradish peroxidase (HRP)-conjugated IgG (0.4 μg/ml in PBS/1% BSA) for scFv-phage analysis, or anti-c-myc HRP-conjugated IgG (Abcam, United Kingdom; diluted 1:250 in PBS/1% BSA) for soluble scFv analysis, the wash step was repeated and 100 µl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added. Reactions were stopped using 100 µl 1 M H2SO4 and absorbances were read at OD450. Half maximal effective concentrations (EC50) were calculated with GraphPad Prism Version 9.3, utilising fitting to a four-parameter logistic curve.
ScFv expression
Plasmid DNA was purified from individual E. coli TG1 clones expressing RBD-binding scFv-phage and used to transform E. coli HB2151 cells. Overnight E. coli cultures were used to inoculate 50 ml of ZY-5052 autoinducing medium containing 100 μg/ml ampicillin to a starting OD600 of 0.05. Supernatants from 50 ml cultures were collected by centrifugation to pellet cells after 48 h induction at 37°C and 225 rpm, followed by purification by immobilised metal affinity chromatography (IMAC) (Antoine et al., 2022).
ScFv purification
For IMAC purification of scFvs, a 1 ml Hitrap column (GE Healthcare, United Kingdom) was loaded with Ni2+ before equilibration with wash buffer (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4) containing 20 mM imidazole. Protein samples were filtered through a 0.4 µm filter, followed by addition of 20 mM imidazole before loading onto the column. The column was washed with 20, 30 and 10 ml wash buffer containing 20, 50 and 70 mM imidazole, respectively, and eluted using wash buffer containing 400 mM imidazole. Eluted fractions were dialysed against PBS, and purified proteins were analysed under denaturing conditions in 12% SDS-PAGE, with staining with InstantBlue Coomassie stain (Expedeon, United Kingdom). Protein concentration was determined using a BCA Assay kit (Thermo Fisher Scientific, United Kingdom).
DNA sequencing and analysis
ScFv diversity was analysed by BstNI restriction analysis of plasmid DNA from RBD-binding clones and electrophoresis on 1.3% agarose gels. ScFv genes were sequenced at Eurofins Genomics (Ebersberg, Germany).
Inhibition of RBD binding to ACE2-expressing cells
Investigation of inhibition of RBD-ACE2 binding was performed using flow cytometry. For each scFv, 3 μg/ml of SARS-CoV-2 Spike RBD Alexa Fluor 647 (Bio-Techne, Abingdon, United Kingdom) was added to varying concentrations of scFv in a 100 µl final volume and incubated for 1 h at room temperature. The RBD-scFv mixture was incubated with 1 × 105 ACE2-expressing Vero E6 cells (Thermo Fisher Scientific) for 1 h at room temperature. Cells incubated with RBD Alexa Fluor 647 alone were used as the negative control. Positive control cells were pre-incubated with 100 µL of 3 μg/ml unlabelled RBD (BEI Resources) for 1 h at room temperature to block RBD binding sites of ACE2 prior to addition of the RBD Alexa Fluor 647. After three washes with 2% fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with fetal calf serum), the fluorescence of positive cells was measured using a BD Accuri C6™ flow cytometer.
Binding of variant spike proteins
ScFvs were evaluated for binding of SARS-CoV-2 variant spike proteins using an immunoassay previously described (Antoine et al., 2022). SERS nanotags, which consist of an scFv conjugated to 60 nm gold nanoparticles, were generated by a layer-by-layer method as described elsewhere (Antoine et al., 2022), while scFvs were conjugated to magnetic particles (Pierce NHS-Activated Magnetic Beads, cat. #88826, Invitrogen) following the manufacturer’s protocol. SERS immunoassays were carried out by combining scFv-conjugated magnetic beads, scFv-conjugated SERS nanotags and 50 pg of target antigen in PBS containing 1% BSA in a final volume of 500 μl, followed by incubation at room temperature with shaking at 200 rpm for 20 min and pelleting of immunocomplexes using an external magnet (Antoine et al., 2022). For single-scFv assays, the same scFv was conjugated to both magnetic beads and SERS nanotags; for two-scFv assays, scFv 10 was conjugated to magnetic beads and scFv D4 to SERS nanotags. Raman spectra of pellets were measured with a Mira DS handheld Raman spectrometer (Metrohm). Spectra were collected using a laser power of 5 (50 mW), integration time 1 s, and raster off, and the peak height was measured at 591 cm−1. Assays were performed in triplicate, and data is represented as the average of those triplicates with standard deviations.
Results
ScFv isolation and sequence analysis
After plating of phage clones isolated from Round 3 of the panning (Antoine et al., 2022), 104 clones were screened for binding to RBD (Figure 1). Of these, 13 positive clones were identified, which yielded eight distinct profiles upon restriction analysis using BstNI (Figure 2). After confirmation by PCR of the presence of a gene of the expected size of scFvs in each clone, DNA sequencing identified the sequence of the eight different scFvs.
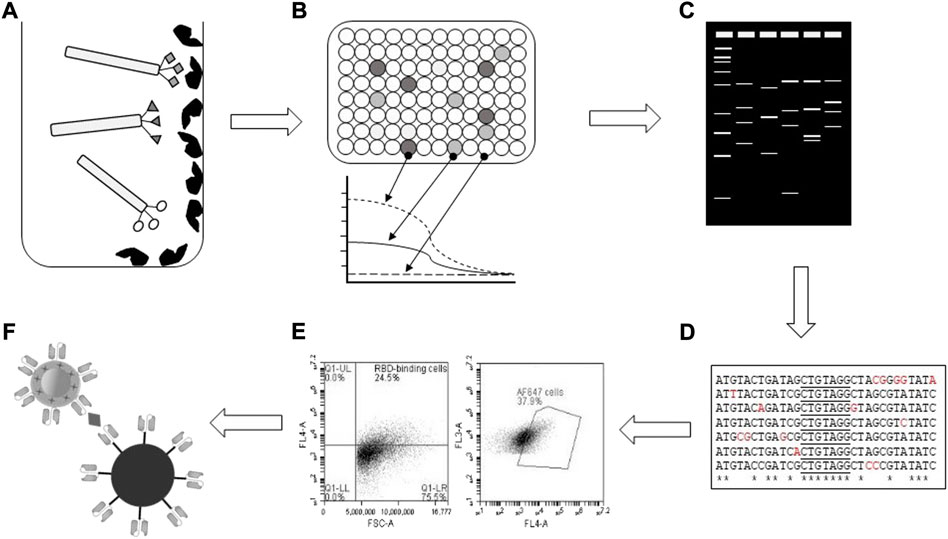
FIGURE 1. Schematic of experimental workflow. (A) RBD-binding bacteriophage-scFvs were selected based on binding to immobilised RBD in vitro, followed by (B) ELISA confirmation of binding and EC50 determinations. (C) Restriction profiling of isolated RBD-binding clones identified unique patterns for (D) DNA sequencing and bioinformatic analysis. (E) Flow cytometry screening identified scFvs which blocked RBD interaction with ACE2-expressing Vero E6 cells, with (F) screening for binding to variant spike proteins carried out by immunocomplex formation and SERS Raman spectroscopy.
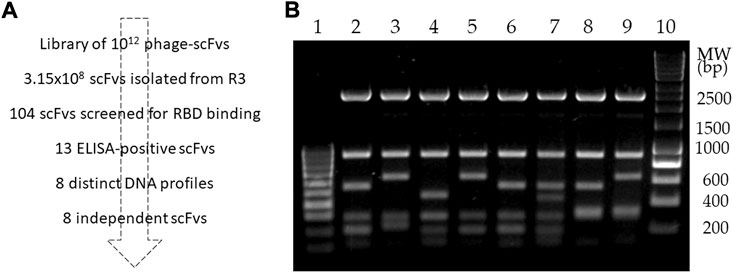
FIGURE 2. Isolation and DNA fingerprinting of RBD-binding scFvs. (A) Workflow showing isolation of RBD-binding scFvs. (B) Restriction profile analysis of eight RBD-binding scFvs isolated from library screening. Lanes: 1, 10: molecular weight markers; 2–9: BstNI-digested DNA from scFvs 2, 3, 10, 37, D4, E10, F11 and G12, respectively.
Recombinant protein expression
The eight RBD-binding scFvs were produced in soluble, non-phage-displayed form in 50 ml culture scale using an optimised autoinduction method utilising accumulation of scFvs in the extracellular medium, with equivalent yields after purification by IMAC ranging from 0.5 to 40 mg per liter of bacterial culture (Table 1). The binding of each purified scFv to RBD was confirmed by ELISA (data not shown) prior to RBD-ACE2 inhibition analysis.
RBD-ACE2 binding inhibition
The ability of the purified scFvs to inhibit binding of RBD to ACE2-positive Vero E6 cells was assessed in a cell-based assay in order to identify potential neutralising molecules. A fluorochrome-conjugated RBD was used in the place of SARS-CoV-2 virus and allowed precise gating of positive cells during the flow cytometry analysis (Supplementary Figure S1). Using a scFv:RBD molar ratio of 100:1, two scFvs were found to inhibit RBD binding to the ACE2-expressing cells by more than 50%: scFv 10 reduced RBD binding to cells by 55%, while scFv D4 inhibited 94% of RBD-cell binding compared to the no-scFv control (Figure 3A).
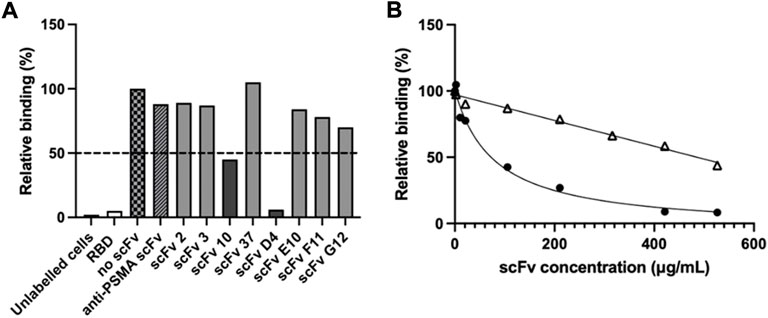
FIGURE 3. Inhibition of RBD binding to ACE-2 cells by scFvs. Flow cytometry was used to determine inhibition of RBD-ACE-2 binding. (A) Alexa Fluor 647-labelled SARS-CoV-2 RBD was pre-incubated with scFv (100:1 scFv:RBD ratio), prior to incubation with ACE-2-expressing Vero E6 cells. Unlabelled cells, cells pre-incubated with unlabelled RBD (“RBD”), cells pre-incubated with Alexa Fluor 647-labelled RBD but no scFv (“no scFv”), cells pre-incubated with Alexa Fluor 647-labelled RBD and an irrelevant “anti-PSMA scFv” were used as controls. Values show single measurements. The dotted line indicates 50% inhibition of RBD-cell binding. (B) IC50 determination of RBD binding to ACE-2 cells with inhibitory scFvs 10 (△) and D4 (●). 80 nM Alexa Fluor 647-labelled SARS-CoV-2 RBD was pre-incubated with scFv 10 or scFv D4 prior to addition to Vero E6 cells. Cells incubated with RBD-Alexa Fluor 647 without scFv were used as a control. Values show single measurements. The IC50 was calculated with GraphPad Prism version 9.3, using fitting to a four-parametric nonlinear regression curve.
To further characterise the neutralisation potential of scFvs D4 and 10, their ability to inhibit RBD binding to ACE2-expressing Vero E6 cells was measured over a concentration range of 2–530 μg/ml scFv in a cell-based flow cytometry assay (Figure 3B). While scFv D4 showed a half-maximal inhibitory concentration (IC50) of RBD binding of 79 μg/ml, signal saturation was not achieved at 530 μg/ml for scFv 10. However, 56% inhibition was measured at 530 μg/ml scFv 10. Meanwhile, half-maximal effective concentrations (EC50) of 11 μg/ml and 280 μg/ml were determined by ELISA for scFvs D4 and 10, respectively, for SARS-CoV-2 RBD (data not shown). The EC50 of scFv D4 for spike protein was also measured and was similar to the EC50 for RBD, at 8 μg/ml.
ScFv binding of variant spike proteins
To determine the potential of the neutralising scFvs raised against Wuhan-Hu-1 SARS-CoV-2 RBD to also block host cell interactions of emerging virus variants, the scFvs were also screened for their binding to Delta and Omicron spike proteins. Both scFvs exhibited binding to spike protein from the Delta variant, albeit with SERS signals reduced by 26% and 14% for scFv D4 and scFv 10, respectively. No binding of the Omicron variant protein was detectable for either scFv, however (Figure 4).
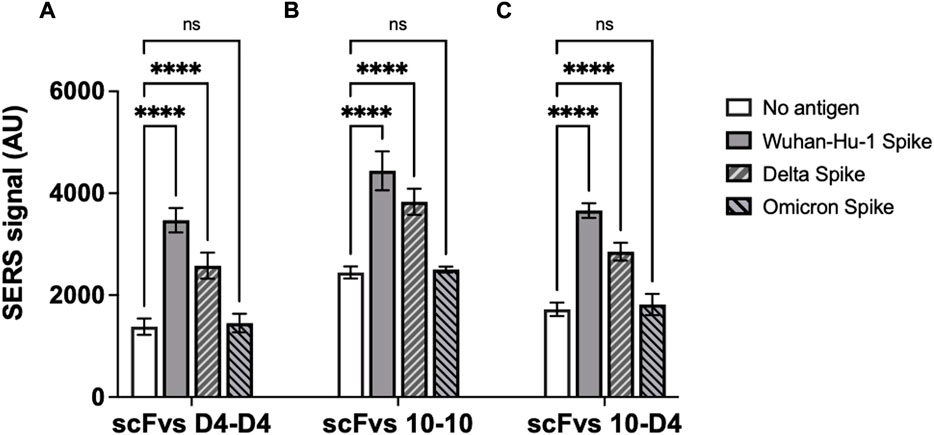
FIGURE 4. ScFv D4 and scFv 10 binding to spike proteins from SARS-CoV-2 Wuhan-Hu-1 virus and Delta and Omicron variants. Panels represent assays carried out with (A) scFv D4 on both magnetic nanoparticles and SERS nanotags, (B) scFv 10 on both particles, and (C) scFv 10 on magnetic nanoparticles with scFv D4 on SERS nanotags. Values represent the average of three replicate wells, and error bars indicate the standard deviation. For statistical analysis, two-way ANOVA followed by Dunnett’s multiple comparisons test was performed: ****: p < 0.0001; ns: not significant.
Discussion
The ongoing COVID-19 pandemic has crystallised the need for rapid isolation of virus-binding moieties, both for use in diagnosis, in order to identify and isolate potentially infectious individuals and thereby reduce spread of the disease, and for treatment, in order to mitigate the severity of the disease in infected and particularly high-risk individuals (Zhu et al., 2020).
Antibodies occupy a central role in the detection and identification of pathogens due to their natural binding specificity and affinity for their targets. In addition, the ability of antibodies to block or outcompete specific binding interactions has led to their use in neutralising viruses and toxins (Cheedarla and Hanna, 2019). As the classical hybridoma technology developed with murine B cells (Köhler and Milstein, 1975) has proven difficult to adapt to the generation of human monoclonal antibodies, however, recombinant approaches have been widely developed to create human antibodies, or derived antibody fragments (Wronska et al., 2016), which can be used in vivo in applications such as drug delivery and neutralisation, and in vitro methodologies like immunodiagnosis (Raeisi et al., 2022; Wang et al., 2022).
While the Fc region of antibodies–which is absent from the scFv fragments utilised in this study–is essential for many in vivo functions such as antibody-dependent cell-mediated cytotoxicity, antibody-dependent cellular phagocytosis and complement activation, recombinant fragments containing only V regions can effectively neutralise viruses and toxins by blocking their interaction with cells. In the case of SARS-CoV-2, Fc effector functions have been demonstrated to not be required for optimal protection against lethal infections in transgenic mice (Noy-Porat et al., 2021). This ability to neutralise SARS-CoV-2 in the absence of antibody C domains is important as it allows the use of E. coli expression systems for production and scale-up of small-sized recombinant fragments that do not require glycosylation, resulting in faster production, higher yields and reduced costs than for traditional monoclonal antibody production–all critical considerations when rapidly responding to an infection. The biophysical properties of antigen specificity and affinity are typically retained in recombinant antibody fragments compared to monoclonal antibodies from mammalian cells–with the added advantage of the ease of affinity maturation of fragments routinely allowing selection of molecules with affinities 10–50 times higher than their parent antibodies (Marks et al., 1992). While scFvs retain the intramolecular disulfide bonds of whole antibody VH and VL regions, they are nevertheless more prone to thermally-induced aggregation (Jager and Plückthun, 1999), which can be reduced using directed evolution or rational design approaches (Kang and Seong, 2020). Similarly, while their smaller size leads to faster clearance than whole antibody molecules in vivo, protein engineering can be used to extend their in vivo half-life and increase their pharmaceutical efficacy (Esquerda-Canals et al., 2019; Seifert and Kontermann, 2022).
E. coli can also have difficulty in expressing some mammalian proteins, leading to their accumulation in inclusion bodies or protein degradation, while it also struggles with multiple disulfide bridges and cannot glycosylate protiens (Rosano and Ceccarelli, 2014). ScFv yields in this study were as high as 40 mg/L in simple batch cultures, however, though yields can typically be further increased by approaches such as process optimisation or molecular chaperone overproduction (Hu et al., 2007), or particularly by adoption of a fed-batch culture system to increase cell density (Gąciarz et al., 2017).
Only two of the eight scFvs, which were isolated from the library based on their RBD binding, prevented RBD from interacting with the ACE2 receptor in a cell-based assay. The RBD domain used in biopanning is a 25-kDa protein containing 223 amino acids but only 17 amino acid residues make contact with (i.e. occur within a distance cut-off of 4 Å of) the ACE2 receptor (Lan et al., 2020). Therefore, it is unsurprising that most of the isolated RBD-binding scFvs did not bind to the domain at a site that impedes its interaction with ACE2.
ScFv D4 was particularly effective at inhibiting the interaction of RBD with ACE2-expressing cells, indicating its binding target is likely to overlap more completely with the ACE2 interaction site than scFv 10. RBD epitopes are categorised into four classes, with Classes 1 and 2 overlapping the ACE2 binding site and thereby constituting the major target for neutralising antibodies (Onodera et al., 2021). Many neutralising molecules have been demonstrated to bind at, or close to, the receptor binding motif of the RBD and inhibit its attachment to the ACE2 protein (Xiang et al., 2020; Hanke et al., 2022), which is necessary to infect host cells, or lock the RBD in its “down” conformation to prevent productive RBD-ACE2 fusion (Schoof et al., 2020).
The differing neutralisation properties of the two RBD-ACE2-blocking scFvs is suggestive of different binding epitopes in RBD. Investigation of the predicted amino acid sequences of the two scFvs revealed only 27% identity and 64% similarity between their VH CDR sequences, compared to 66% and 89%, respectively, across their entire gene sequences (not shown). As the use of multiple neutralising molecules binding different epitopes can be used in combination therapies to mitigate mutagenic escape in newly emerging virus variants (Makdasi et al., 2021), further investigation will be necessary to determine the precise epitopes bound by the two scFvs. This cocktail approach is seen in the emergency use authorisation (EUA) by the U.S. FDA of commercial monoclonal antibody combinations for treatment of SARS-CoV-2 infections (Baum et al., 2020; Bennett et al., 2021). Furthermore, simultaneous targeting of more than one epitope in a diagnostic test could be used to improve detection of virus, including new VOCs, and, importantly, to rapidly distinguish between VOCs in a point-of-care test.
While both of the present scFvs retained binding to spike protein from the Delta variant virus, neither scFv exhibited detectable binding of Omicron spike protein. This contrasts with scFv 3 which was also isolated from the biopanning and bound Alpha, Beta, Delta and Omicron spike proteins with similar affinity (Mohammadi et al., 2022), but failed to inhibit the RBD-ACE2 interaction in this study. The inability of the Wuhan-Hu-1- and Delta-binding scFvs 10 and D4 to detect Omicron spike protein mirrors clinical experience, with the ongoing emergence of virus variants which are resistant to detection and/or neutralisation by antibodies protective against earlier strains (Focosi et al., 2021). The large number of amino acid substitutions in Omicron in particular (37 in spike glycoprotein, compared to approximately 10 in Alpha and Beta variants of concern (VOCs), and 15 mutations in the RBD alone (Ou et al., 2022)) has exacerbated its evasion of immunoprotection from previous infections. Many Class 1 RBD-binding antibodies, with epitopes centred around loop 477–489, are frequently subject to Omicron escape due to mutations in this protein region, though neutralisation towards Omicron may be retained by antibodies which target the highly conserved SD1 region of the spike protein (Hong et al., 2022).
This study outlines an approach to rapidly isolate scFvs against current SARS-CoV-2 virus variants, followed by screening of the scFv panel for binding and neutralisation of newly emerging VOCs, with de novo biopanning against new variant proteins in 3–4 weeks if necessary. This provides a promising strategy to isolate detection and treatment moieties which can be more rapidly tailored to detect or treat new variants in immunocompromised, young or elderly individuals than monoclonal antibody-based reagents.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
PJ, KW, and GW contributed to conception and design of the study. DA, MM, CM, and EW carried out the experimental work. DA, MM, PJ, KW, and GW performed the data analysis. DA and GW wrote the first draft of the manuscript. PJ, KW, and GW acquired funding for the work. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was funded by Health Research Board, Ireland COVID-19 Pandemic Rapid Response Funding Opportunity 2020 (grant no. COV19-2020-081), funds granted from CARES Act COVID Innovation Fund (United States), and by the National Institute of General Medical Sciences of the National Institutes of Health (United States) under award number P20GM103432.
Acknowledgments
The authors thank Shirley Hanley for help with flow cytometry and Dr. Merve Zeden for assistance with statistical analysis. The following reagents were obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein (Stabilised) from SARS-CoV-2, Wuhan-Hu-1 with C-Terminal Histidine and Twin-strep Tags, Recombinant from HEK293 Cells, NR-53589; Spike Glycoprotein (Stabilised) from SARS-CoV-2, AY.2 Lineage (Delta variant) with C-Terminal Histidine and Avi Tags, Recombinant from HEK293 Cells, NR-55711.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2022.1028186/full#supplementary-material
References
Antoine, D., Mohammadi, M., Vitt, M., Dickie, J. M., Jyoti, S. S., Tilbury, M. A., et al. (2022). Rapid, point-of-care scFv-SERS assay for femtogram level detection of SARS-CoV-2. ACS Sens. 7, 866–873. doi:10.1021/acssensors.1c02664
Baum, A., Fulton, B. O., Wloga, E., Copin, R., Pascal, K. E., Russo, V., et al. (2020). Antibody cocktail to SARS- CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018. doi:10.1126/science.abd0831
Beaudoin-Bussières, G., Laumaea, A., Anand, S. P., Prévost, J., Gasser, R., Goyette, G., et al. (2020). Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio 11, e02590–e02620. doi:10.1128/mBio.02590-20
Bennett, R. S., Postnikova, E. N., Liang, J., Gross, R., Mazur, S., Dixit, S., et al. (2021). Scalable, micro-neutralization assay for qualitative assessment of SARS-CoV-2 (COVID-19) virus-neutralizing antibodies in human clinical samples. bioRxiv : the preprint server for biology 2021.03.05.434152. doi:10.1101/2021.03.05.434152
Bertoglio, F., Fühner, V., Ruschig, M., Heine, P. A., Abassi, L., Klünemann, T., et al. (2021). A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations. Cell Rep. 36, 109433. doi:10.1016/j.celrep.2021.109433
Casadevall, A., Joyner, M. J., and Pirofski, L. A. (2020). SARS-CoV-2 viral load and antibody responses: The case for convalescent plasma therapy. J. Clin. Invest. 130, 5112–5114. doi:10.1172/JCI139760
Cheedarla, N., and Hanna, L. E. (2019). “Functional and protective role of neutralizing antibodies (NAbs) against viral infections,” in Recent developments in applied microbiology and biochemistry. Editor V. Buddolla (London. U.K. Academic Press), 83–93.
Corti, D., Purcell, L. A., Snell, G., and Veesler, D. (2021). Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 3086–3108. doi:10.1016/j.cell.2021.05.005
Ebihara, T., Masuda, A., Takahashi, D., Hino, M., Mon, H., Kakino, K., et al. (2021). Production of scFv, Fab, and IgG of CR3022 antibodies against SARS-CoV-2 using silkworm-Baculovirus expression system. Mol. Biotechnol. 63, 1223–1234. doi:10.1007/s12033-021-00373-0
Esquerda-Canals, G., Martí-Clúa, J., and Villegas, S. (2019). Pharmacokinetic parameters and mechanism of action of an efficient anti-Aβ single chain antibody fragment. PloS one 14, e0217793. doi:10.1371/journal.pone.0217793
Focosi, D., Maggi, F., Franchini, M., McConnell, S., and Casadevall, A. (2021). Analysis of immune escape variants from antibody-based therapeutics against COVID-19: A systematic review. Int. J. Mol. Sci. 23, 29. doi:10.3390/ijms23010029
Food and Drug Administration (2020). Emergency use authorization for convalescent plasma as potential promising COVID-19 treatment, another achievement in administration’s fight against pandemic. U.S.A. Available at: https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment (Accessed 07 08, 2022).
Gąciarz, A., Khatri, N. K., Velez-Suberbie, M. L., Saaranen, M. J., Uchida, Y., Keshavarz-Moore, E., et al. (2017). Efficient soluble expression of disulfide bonded proteins in the cytoplasm of Escherichia coli in fed-batch fermentations on chemically defined minimal media. Microb. Cell Fact. 16, 108. doi:10.1186/s12934-017-0721-x
Güttler, T., Aksu, M., Dickmanns, A., Stegmann, K. M., Gregor, K., Rees, R., et al. (2021). Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies. EMBO J. 40, e107985. doi:10.15252/embj.2021107985
Hanke, L., Das, H., Sheward, D. J., Perez Vidakovics, L., Urgard, E., Moliner-Morro, A., et al. (2022). A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo. Nat. Commun. 13, 155. doi:10.1038/s41467-021-27610-z
Havlin, J., Svorcova, M., Dvorackova, E., Lastovicka, J., Lischke, R., Kalina, T., et al. (2021). Immunogenicity of BNT162b2 mRNA COVID-19 vaccine and SARS-CoV-2 infection in lung transplant recipients. J. Heart Lung Transpl. 40, 754–758. doi:10.1016/j.healun.2021.05.004
Hong, Q., Han, W., Li, J., Xu, S., Wang, Y., Xu, C., et al. (2022). Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 604, 546–552. doi:10.1038/s41586-022-04581-9
Hu, X., O'Hara, L., White, S., Magner, E., Kane, M., and Wall, J. G. (2007). Optimisation of production of a domoic acid-binding scFv antibody fragment in Escherichia coli using molecular chaperones and functional immobilisation on a mesoporous silicate support. Protein Expr. Purif. 52, 194–201. doi:10.1016/j.pep.2006.08.009
Hurt, A. C., and Wheatley, A. K. (2021). Neutralizing antibody therapeutics for COVID-19. Viruses 16, 628. doi:10.3390/v13040628
Hwang, Y.-C., Lu, R.-M., Su, S.-C., Chiang, P.-Y., Ko, S.-H., Ke, F.-Y., et al. (2022). Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 29, 1. doi:10.1186/s12929-021-00784-w
Jager, M., and Plückthun, A. (1999). Domain interactions in antibody fv and scFv fragments: Effects on unfolding kinetics and equilibria. FEBS Lett. 462, 307–312. doi:10.1016/s0014-5793(99)01532-x
Jeong, B. S., Cha, J. S., Hwang, I., Kim, U., Adolf-Bryfogle, J., Coventry, B., et al. (2022). Computational design of a neutralizing antibody with picomolar binding affinity for all concerning SARS-CoV-2 variants. MAbs 14, 2021601. doi:10.1080/19420862.2021.2021601
Jeyanathan, M., Afkhami, S., Smaill, F., Miller, M. S., Lichty, B. D., and Xing, Z. (2020). Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 20, 615–632. doi:10.1038/s41577-020-00434-6
Kang, T. H., and Seong, B. L. (2020). Solubility, stability, and avidity of recombinant antibody fragments expressed in microorganisms. Front. Microbiol. 11, 1927. doi:10.3389/fmicb.2020.01927
Ke, Q., Sun, P., Wang, T., Mi, T., Xu, H., Wu, J., et al. (2022). Non-glycosylated SARS-CoV-2 RBD elicited a robust neutralizing antibody response in mice. J. Immunol. Methods 506, 113279. doi:10.1016/j.jim.2022.113279
Kiguchi, Y., Oyama, H., Morita, I., Morikawa, M., Nakano, A., Fujihara, W., et al. (2020). Clonal array profiling of scFv-displaying phages for high-throughput discovery of affinity-matured antibody mutants. Sci. Rep. 10, 14103. doi:10.1038/s41598-020-71037-3
Kim, H. Y., Lee, J. H., Kim, M. J., Park, S. C., Choi, M., Lee, W., et al. (2021). Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelec. 175, 112868. doi:10.1016/j.bios.2020.112868
Köhler, G., and Milstein, C. (1975). Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497. doi:10.1038/256495a0
Kumar, S., Chandele, A., and Sharma, A. (2021). Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog. 17, e1009885. doi:10.1371/journal.ppat.1009885
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi:10.1038/s41586-020-2180-5
Letko, M., Marzi, A., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569. doi:10.1038/s41564-020-0688-y
Li, D., Sempowski, G. D., Saunders, K. O., Acharya, P., and Haynes, B. H. (2022). SARS-CoV-2 neutralizing antibodies for COVID-19 prevention and treatment. Annu. Rev. Med. 73, 1–16. doi:10.1146/annurev-med-042420-113838
Liu, L., Wang, P., Nair, M. S., Yu, J., Rapp, M., Wang, Q., et al. (2020). Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456. doi:10.1038/s41586-020-2571-7
Makdasi, E., Levy, Y., Alcalay, R., Noy-Porat, T., Zahavy, E., Mechaly, A., et al. (2021). Neutralizing monoclonal anti-SARS-CoV-2 antibodies isolated from immunized rabbits define novel vulnerable spike-protein epitope. Viruses 13, 566. doi:10.3390/v13040566
Marks, J. D., Griffiths, A. D., Malmqvist, M., Clackson, T. P., Bye, J. M., and Winter, G. (1992). By-passing immunization: Building high affinity human antibodies by chain shuffling. Nat. Biotechnol. 10, 779–783. doi:10.1038/nbt0792-779
Marks, J. D., Hoogenboom, H. R., Bonnert, T. P., McCafferty, J., Griffiths, A. D., and Winter, G. (1991). By-passing immunization. J. Mol. Biol. 222, 581–597. doi:10.1016/0022-2836(91)90498-u
Mendoza-Salazar, I., Gómez-Castellano, K. M., González-González, E., Gamboa-Suasnavart, R., Rodríguez-Luna, S. D., Santiago-Casas, G., et al. (2022). Anti-SARS-CoV-2 Omicron antibodies isolated from a SARS-CoV-2 Delta semi-immune phage display library. Antibodies 11, 13. doi:10.3390/antib11010013
Minenkova, O., Santapaola, D., Milazzo, F. M., Anastasi, A. M., Battistuzzi, G., Chiapparino, C., et al. (2022). Human inhalable antibody fragments neutralizing SARS-CoV-2 variants for COVID-19 therapy. Mol. Ther. 30, 1979–1993. doi:10.1016/j.ymthe.2022.02.013
Mohammadi, M., Antoine, D., Vitt, M., Dickie, J. M., Jyoti, S. S., Wall, J. G., et al. (2022). A fast, ultrasensitive SERS immunoassay to detect SARS-CoV-2 in saliva. Anal. Chim. Acta X. 1229, 340290. doi:10.1016/j.aca.2022.340290
Muyldermans, S. (2013). Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797. doi:10.1146/annurev-biochem-063011-092449
Noy-Porat, T., Edri, A., Alcalay, R., Makdasi, E., Gur, D., Aftalion, M., et al. (2021). Fc-independent protection from SARS-CoV-2 infection by recombinant human monoclonal antibodies, Antibodies (Basel) 10, 45. doi:10.3390/antib10040045
Onodera, T., Kita, S., Adachi, Y., Moriyama, S., Sato, A., Nomura, T., et al. (2021). A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site. Immunity 54, 2385–2398. e10. doi:10.1016/j.immuni.2021.08.025
Ou, J., Lan, W., Wu, X., Zhao, T., Duan, B., Peipei, Y., et al. (2022). Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Sig. Transduct. Target. Ther. 7, 138. doi:10.1038/s41392-022-00992-2
Pansri, P., Jaruseranee, N., Rangnoi, K., Kristensen, P., and Yamabhai, M. (2009). A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 9, 6. doi:10.1186/1472-6750-9-6
Parray, H. A., Chiranjivi, A. K., Asthana, S., Yadav, N., Shrivastava, T., Mani, S., et al. (2020). Identification of an anti-SARS-CoV-2 receptor-binding domain-directed human monoclonal antibody from a naïve semisynthetic library. J. Biol. Chem. 295, 12814–12821. doi:10.1074/jbc.AC120.014918
Raeisi, H., Safarnejad, M. R., and Sadeghkhani, F. (2022). A new single-chain variable fragment (scFv) antibody provides sensitive and specific detection of citrus tristeza virus. J. Virol. Methods 300, 114412. doi:10.1016/j.jviromet.2021.114412
Rosano, G., and Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 5, 172. doi:10.3389/fmicb.2014.00172
Schoof, M., Faust, B., Saunders, R. A., Sangwan, S., Rezelj, V., Hoppe, N., et al. (2020). An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479. doi:10.1126/science.abe3255
Salem, R., El-Kholy, A. A., Waly, F. R., Ayman, D., Sakr, A., Hussein, M., et al. (2022). Generation and utility of a single-chain fragment variable monoclonal antibody platform against a baculovirus expressed recombinant receptor binding domain of SARS-CoV-2 spike protein. Mol. Immunol. 141, 287–296. doi:10.1016/j.molimm.2021.12.006
Seifert, O., and Kontermann, R. E. (2022). GlycoTAIL and FlexiTAIL as half-life extension modules for recombinant antibody fragments. Molecules 27, 3272. doi:10.3390/molecules27103272
Sievers, B. L., Chakraborty, S., Xue, Y., Gelbart, T., Gonzalez, J. C., Cassidy, A. G., et al. (2022). Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 14, eabn7842. doi:10.1126/scitranslmed.abn7842
Wang, J., Kang, G., Yuan, H., Cao, X., Huang, H., and de Marco, A. (2022). Research progress and applications of multivalent, multispecific and modified nanobodies for disease treatment. Front. Immunol. 12, 838082. doi:10.3389/fimmu.2021.838082
World Health Organization (2022). WHO coronavirus disease (COVID-19) dashboard. Available at: https://covid19.who.int/(Accessed 07 08, 2022).
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi:10.1126/science.abb2507
Wronska, M. A., O'Connor, I. B., Tilbury, M. A., Srivastava, A., and Wall, J. G. (2016). Adding functions to biomaterial surfaces through protein incorporation. Adv. Mat. 28, 5485–5508. doi:10.1002/adma.201504310
Xiang, Y., Nambulli, S., Xiao, Z., Liu, H., Sang, Z., Duprex, W. P., et al. (2020). Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370, 1479–1484. doi:10.1126/science.abe4747
Yang, L., Liu, W., Yu, X., Wu, M., Reichert, J. M., and Ho, M. (2020). COVID-19 antibody therapeutics tracker: A global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib. Ther. 3, 205–212. doi:10.1093/abt/tbaa020
Yang, Z., Wang, Y., Jin, Y., Zhu, Y., Wu, Y., Li, C., et al. (2021). A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 6, 378. doi:10.1038/s41392-021-00810-1
Zhao, D., Liu, L., Liu, X., Zhang, J., Yin, Y., Luan, L., et al. (2022). A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode. J. Nanobiotechnol. 20, 411. doi:10.1186/s12951-022-01619-y
Keywords: SARS-CoV-2, scFv, recombinant antibody, phage display, neutralisation, variant of concern
Citation: Antoine D, Mohammadi M, McDermott CE, Walsh E, Johnson PA, Wawrousek KE and Wall JG (2022) Isolation of SARS-CoV-2-blocking recombinant antibody fragments and characterisation of their binding to variant spike proteins. Front. Nanotechnol. 4:1028186. doi: 10.3389/fnano.2022.1028186
Received: 25 August 2022; Accepted: 26 September 2022;
Published: 12 October 2022.
Edited by:
Linqi Zhang, Tsinghua University, ChinaReviewed by:
Diana Lousa, Universidade Nova de Lisboa, PortugalFang Xin Hu, Suzhou University of Science and Technology, China
Copyright © 2022 Antoine, Mohammadi, McDermott, Walsh, Johnson, Wawrousek and Wall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Gerard Wall, Z2VyYXJkLndhbGxAdW5pdmVyc2l0eW9mZ2Fsd2F5Lmll
†These authors have contributed equally to this work
 Delphine Antoine
Delphine Antoine Moein Mohammadi3
Moein Mohammadi3 J. Gerard Wall
J. Gerard Wall