- Kirori Mal College, University of Delhi, New Delhi, India
Globally, breast cancer is the foremost cause of mortality among women detected with cancer, with 21% diagnosed in India alone. Etoposide loaded gelatin nanoparticles (EGNP) were prepared and its physical characterization (size:150nm±0.241; zeta potential −29.32 mV) was done along with in-vitro studies to assess biotoxicity, intracellular ROS, cell cycle arrest and death caused by EGNPs. We report the molecular pathways induced by EGNP in-vitro, pharmacokinetics, biodistribution and tumor regression in-vivo in Balb/c mice.Gene expression profiling of Bax, Bcl2, p53, Caspase-3, RIPK1, RIPK3 and ß-actin as internal control were done by RT-PCR wherein Etoposide and EGNP treated MCF-7 cells showed higher expressions of apoptotic genes-Bax, p53, caspase-3, lower expression of anti-apoptotic gene-Bcl2 when compared to control. Enhanced expression of necroptosis-RIPK1 were observed, while RIPK3 was insignificant. Since, RIPK1 regulates necroptosis and apoptosis, expression of apoptotic markers confirmed apoptotic molecular mechanisms. Negligible hemolysis of Gelatin nanoparticles (GNP), and EGNP at selected dosages confirmed biocompatibility. In vivo pharmacokinetics and biodistribution were done by 99Tc-labelled nanoparticles indicating increased circulation of EGNPs, allowing accumulation at the tumor site by Enhanced permeability and retention (EPR) phenomena. Tumor regression indicates the efficacy of EGNP by reducing the tumor burden when compared to void GNP and Etop per se, resulting in increased life span. High biocompatibility and bio-efficacy of EGNPs prove their therapeutic potential in cancer treatment.
Introduction
Cancer is a global life-threatening problem, only second to cardiovascular disease. Past few decades have witnessed an exponential rise in incidence of cancer, by about 33%. Breast Cancer is the most frequently occurring cancer worldwide affecting ∼2.5 million females every year and the total cancer cases in India are predicted to be ∼170,000 as per WHO report which is higher by ∼14% amongst all cancers in India (Jain et al., 2020). Breast cancer incidence in both sexes and in age-specific manner has increased by 14% uptil 2018. Globally, there is a 43% increase in cancer incidence as a result of aging (contributing 13%) and burst in population growth (15%) (Fitzmaurice et al., 2017).
Successful cancer therapeutics include alkylating agent, antimetabolites drugs, and naturally occurring alkaloids such as Vinblastine, Vincristine, Etoposide, Irinotecan, Teniposide, Topotecan, Paclitaxel, Docetaxel etc (Shu-Ting et al., 2016). Etoposide (Etop) being a podophyllotoxin derivative and an inhibitor of deoxyribonucleic acid (DNA) topoisomerase II is preferred as it primarily effects the G2 phase in cell cycle (Siddiqui et al., 2015). Etop administration is challenging due to its lipophilic nature and short half-life in serum. It further prevents cancer cell growth and DNA replication by inducing apoptosis in cancer cell. Physiologically, apoptosis is the main mechanism for eliminating unwanted cells (Montecucco et al., 2015). Apoptosis resistance is a hallmark of innumerable cancer cells that limits the success of conventional chemotherapy. Reports further suggest that the response of tumor cells to various chemotherapeutic drug was not confined only to apoptosis but also includes other modes of cell death (Brown and Attardi, 2005) like caspase-independent apoptosis, necroptosis, paraptosis, pyroptosis, and slow cell death, whose morphologic and biochemical characteristics vary from current definitions of the major cell death pathways (Ricci and Zong, 2006). Therefore, targeting non-apoptotic pathway may emerge as an alternative strategy to combat the menace of cancer. The programmed necrosis emerged as a potential backup mechanism to apoptosis that was assessed here. Unlike apoptosis, necroptosis is a caspase-independent death program that may be mediated by receptor interacting protein kinase-3 (RIPK3) (Moriwaki and Chan, 2013). When caspase inhibition occurs, the cellular inhibitor of apoptosis proteins (cIAPs) get depleted, tumor necrosis factor (TNF) receptor-1 then stimulates a cascade that terminates by binding RIPK3 with its upstream molecule-RIPK1 via the RIP homotypic interaction motif (RHIM). Phosphorylation of RIPK1 and RIPK3 stabilizes the complex triggering formation of necrosome. Mixed lineage kinase domain-like (MLKL) a substrate of RIPK3 is recruited by stimulated, RIPK3 (Sun et al., 2012), and gets phosphorylated to form oligomers that gets transported to the intracellular membranes as well as the plasma membrane, which finally ruptures the membrane (Cai et al., 2014; Chen et al., 2014; Dondelinger et al., 2014; Wang et al., 2014).
Toxicogenomics has been considered as a promising tool for measurement of the gene expression pattern elicited by toxicant exposure (Nuwaysir et al., 1999). Toxicity can be reliably indicated by toxicogenomics (Pognan, 2007; Afshari et al., 2011); gene expression profiles at an early stage accurately projected the potential for non-genotoxic carcinogenicity, carcinogenicity and hepato-carcinogenicity (Nie et al., 2006; Ellinger-Ziegelbauer et al., 2008).
Exceptional progress in nanotechnological development towards administering lipophilic drugs offer immense promise to enhance circulation time of drug combined with drug stability to achieve targeted delivery of therapeutic molecule (Elzoghby et al., 2012; Singh et al., 2013). A vital parameter for innovatively engineering a nanoparticle for application both in vitro and in vivo is its biocompatibility in the biological system (Hubbell and Chilkoti, 2012) that can be achieved by modulating various factors like fabrication procedures, careful selection of cross-linkers and surfactants; nevertheless, its the biopolymers that have the most critical role as they contribute the framework in which the drug molecules have to be entrapped, and finally, it is the surface of the polymer that will intermingle with the biological fluids (Vonarbourg et al., 2006; De Jong and Borm, 2008; Jabr-Milane et al., 2008). Therefore, use of natural polymers like chitosan, alginate, and gelatin has aroused excessive attention in nanotechnology as these biopolymers display all the earlier-mentioned characteristics, and additionally, they are water soluble and biodegradable (Tabata, 2009; Cesàro et al., 2012; Singh et al., 2014). Gelatin is a naturally occurring versatile FDA (United States) approved polymer having exceptional properties like biocompatibility, biodegradability with ease of chemical modification attained by the fractional hydrolysis of collagen (Kaul and Amiji, 2004; Magadala and Amiji, 2008; Elzoghby, 2013). Being a polyampholyte, gelatin exhibits both cationic and anionic groups, along with the hydrophobic group in an estimated ratio of 1:1:1, making this polypeptide unique. Gelatin based nanoparticles have been synthesized using various techniques like desolation that uses acetone/alcohol as a desolvating agent for gelatin in aqueous solution for dehydrating the polymer chain resulting in a conformational change (Fuchs et al., 2012). Irrespective of the protocol for fabrication, GNPs have been expansively used for drug/gene delivery carrying cargo that is either hydrophilic or hydrophobic anticancer drugs, vaccine and proteins (Saxena et al., 2005; Zwiorek et al., 2005). Herein, we report the varied adverse effects of Etop that can be reduced by encapsulating it in GNPs (EGNPs). The synthesized GNPs were characterized for its size, zeta potential, surface morphology and encapsulation. We report the bio-efficacy, ROS generation and cell cycle analysis induced by EGNPs in MCF-7 breast cancer cell line and HEK-293 cells and compared these results with etoposide per se. Furthermore, these nanoparticles were evaluated for its ability to regulate different cell death pathways. EGNPs might emerge as a promising drug delivery system for improved therapeutic index of lipophilic Etop as assessed by pharmacokinetics, biodistribution and tumor regression in-vivo in Balb/c mice.
Chemicals Used for Experiments
Gelatin (Bovine, Type B, 175 Bloom) Etoposide, MTT [(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide], Nec-1[Necrostatin-1], zVAD, were all procured from Sigma Chemical Co., (St. Louis, MO, United States). Acetone, Chloroform, Isopropanol, Ethanol, Glutaraldehyde (25% v/v), L-cysteine and Dimethyl sulphoxide (DMSO) were obtained from SRL Pvt Ltd. India. Dulbecco’s modified Eagle’s medium (DMEM), Antibiotic cocktail containing streptomycin and penicillin, Trypsin were procured from HiMedia Pvt Ltd. Fetal bovine serum (FBS) was purchased from GIBCO. Trizol was obtained from Invitrogen. Primers (Forward and reverse) for RIPK-1, RIPK-3, MLKL, Bax, Bcl2, Caspase 9, p53 were procured from Thermo scientific.
Methods
Synthesis of Gelatin Nanoparticles
Gelatin-5% (type-B; 75 bloom) dissolved in warm(∼50°C) distilled water (25ml) was UV-degraded at room temperature (RT) for 12h. The UV-degraded gelatin solution was then titrated with an equal volume of acetone and the precipitate was dissolved in an equal volume of warm deionized water (∼50°C), further pH was adjusted to 2.5. A second step desolvation was carried out and 7% glutaraldehyde was added dropwise. The clear supernatant was discarded after centrifugation at 20,000g for 19min. After washing the pellet, it was suspended in acetone and left overnight under constant stirring. After 24h slightly turbid solution containing nanoparticles was harvested (Saxena et al., 2005; Moin et al., 2018).
Dynamic Laser Scattering and Nanoparticle Tracking Analysis
Dynamic Light Scattering (DLS) and zeta potential measurements were calculated on Malvern Zetasizer Nano ZS equipped with a solid state HeNe laser (= 633nm). All the samples were measured three times at 25°C. The intensity-weighted mean diameter was used as the hydrodynamic size. The particle size distribution was evaluated using polydispersity index (PdI) (Verma et al., 2011).
Nano Sight NS 300 (Nano Sight, Malvern, United Kingdom) was used for the NTA measurement, equipped with a Viton fluoro-elastomer O-ring and a sample chamber with 640nm laser. The chambers were injected with a requisite amount of samples using sterile syringe. The samples were measured with gain adjustments and manual shutter for 40s and data was captured by NTA 2.0 Build 127. The NTA software analyze the size of all the particles to calculate the arithmetic values which corresponds to the SD values and mean size obtained by the software.
Entrapment Efficiency (%)
Etop was entrapped by our earlier published procedure. Etop was solubilized in DMSO under vigorous stirring and bath sonication was added to 5mg/ml of GNPs. The free Etop was entrapped in GNPs. The solution was filtered through a filter [100KDa cut off] to separate free Etop from EGNP. The absorbance of free Etop was read in a spectrophotometer (Cary 60 UV-Vis, Agilent Technologies) (Verma et al., 2005). The EE% was calculated as:
Where, free Drug and total Drug are the sum of total Drug added along with free Drug, respectively.
Bio-Toxicity Assay
Cytotoxicity of EGNP, void gelatin nanoparticles (GNP) and Etop per se was estimated by established/published protocol (Leekha et al., 2016). Briefly, the requisite cells were seeded in ninty six well microplates. Cells were subjected to varied concentrations of GNP, EGNP and Etop per se, and incubated for 24 and 48h at 37oC. After incubation, MTT was added in each well. After 4 h, media was discarded carefully. Added DMSO to solubilize the formazan crystals formed after the reaction, and using a UV-spectrometer absorbance was observed at 570nm.
The % Cytotoxicity was calculated by
Where, Absorbance (control) is the absorbance of control sample and Absorbance (test) is absorbance of test sample.
Quantitation of Intracellular ROS Generation
For investigation of ROS generation, 1 × 106 cells were seeded in petri-plates and kept overnight in RPMI-1640 supplemented with 10% FBS were incubated with Etop, GNP and EGNP. Cells were harvested after treatment, and incubated with 25μM of DCF-DA at 37 °C for 40 min, and the plates were subsequently wrapped in aluminium foil to restrain entering of light. Fluorescence intensity was measured in arbitrary units by means of spectrofluorimeter (Agilent Technologies, United States) having an excitation at 485nm, and an emission at 529nm, respectively, having the slit-width adjusted to 5.0 (Leekha et al., 2016)
Cell Cycle Assessment
MCF-7 cells and HEK-293 cells were cultured in 6-well plates till they reach the logarithmic growth phase, after 24h they were treated with various concentrations of Etop, EGNP and GNPs. Post 18h of treatment, cells were trypsinized and centrifuged for 10min at 1,100rpm in a cooled centrifuge. The harvested cells were washed three times with PBS, and fixed with 70% ethanol for 4h at 4°C. After fixing, the cells were washed again with PBS and then cells were incubated with Propidium Iodide (20μg/ml) and DNase-free RNase. Samples were analyzed at the BD Aria11: Becton and Dickinson, San Jose, CA Flow Cytometer. The percentages of cells in the G1, S, and G2 phases of the cell cycle were calculated using FlowJO software.
Expression of Apoptotic Proteins
Cells (1 × 106) were plated and allowed to adhere overnight. After 24h of the treatment with selected concentration of the Etop, EGNP, Nec1+Etop, Nec1+EGNP, zVAD + Etop and zVAD + EGNP; PBS was used to wash the cells and total RNA were extracted from treated and untreated MCF-7 cells using Trizol reagent (Invitrogen, CA, United States) as per the manufacture’s instruction. The concentration and quality of RNA were quantified using a nanophotometer (Thermo Fisher, United States). cDNA was synthesized through the commercially available kit easy i-script cDNA synthesis (Bio-Rad, USA). Real time PCR (SYBR green) was done on Quant Studio 5 Applied Biosystem PCR, through defined annealing temperature for specific genes to check the expressions of apoptotic and necroptotic genes. The sequences of primer used for Bax, Bcl2, p53, Caspase-3, RIPK1, RIPK3, MLKL and Actin were designed as described in Supplementary Table S3.
Ex vivo Hemolysis
To measure the hemolytic activity, mice whole blood was used and evaluated as per established protocol (Verma et al., 2005). The heparinized blood was washed twice with PBS (pH 7.4). RBC were suspended in PBS at 1:1 ratio. The RBC were incubated with PBS, GNP, Etop per se, and EGNP up to 4h at 37°C. The absorbance of lysed RBC was measured using UV-visible spectrophotometer (Bio-Tek, United States) at 540nm, after incubation of 2 and 4h, and percent hemolysis was evaluated using the following formula
Radiolabeling of Gelatin Nanoparticles
The radiolabeling of Etop per se and EGNP was done with 99mTcO4־ by direct labelling method. Shortly, 100µl of 99mTcO4־ normal saline was added to Etop and EGNP. Then, the pH was adjusted to 6.5 and incubated for 15min at 37°C. The radiolabeled formulation was purified from free 99mTcO4−, by passing through a column [Sephadex G-20] radiocolloids and then eluted with normal saline. The purity of the labelled formulation was verified by Instant thin-layer chromatography (ITLC) by means of silica gel-coated fiber sheets (Gelman Science Inc., Ann Arbor, MI, United States) and the mobile phase was acetone. The Rf value of free 99mTcO4־ was found to be in the range of 0.9–1.0, a well-type γ ray spectrometer (Type CRS 23C, Electronic Corporation of India Ltd. Hyderabad, India) was used for counting the radioactivity.
Biodistribution
Inbred, 4–6weeks old female Balb/c mice weighing around 20–25g were selected and studies were performed as per our standardized previously published protocol (Verma and Sachin, 2008). The animals were anaesthetized followed by injection of 99mTc-labeled EGNP and Etop (1mg/kg body weight) were administrated through the tail vein. As per decided time point, the study groups of four mice Etop per se, and EGNP 99mTc- were used. Post the required time period the animals were sacrificed by cervical dislocation and various tissues (liver, kidney, spleen, heart, lung, intestine, stomach, brain, and blood) were harvested. Using shielded well-type γ-ray spectrometer (Type CRS 23C, Electronic Corporation of India Ltd., Hyderabad, India), the radioactivity was measured.
Gamma Scintigraphic Imaging
100μCi of the 99mTc- dosage formulation of GNP, Etop per se, and EGNP was injected in rabbit through the external ear vein. Single Photon Emission Computerized Tomography (SPECT, LC 75-005, Diacam, Siemens, United States) was conducted after 4 and 24h of administration of formulation.
Tumor Regression
Female Balb/c mice were purchased from Small Animal Facility (SAF), AIIMS Delhi and were maintained at relative humidity of 55 ± 5%, temperature 25 ± 2°C, and normal chow diet (Golden feed). All the experiments performed were approved by Institutional Animal Ethics Committee (IAEC), K.M. College, University of Delhi (Reg No. 1666/GO/Re/S/12/CPCSEA and Protocol no. DU/KR/IAEC/2018/03). Subcutaneous Ehrlich Ascites Carcinoma (EAC) mice model was used to assess tumor regression. EAC cells were maintained in the peritoneum of female Balb/c mice (20–25g) by serial intraperitoneal passage at regular intervals of 7–10days (Verma et al., 2018). To assess tumor inhibition ∼1.5 × 106 cells in 100μl of sterile PBS were injected subcutaneously on the dorsal lower back. Mice were monitored regularly and tumor size was measured every alternate day till it reached a measurable size.
Tumor bearing female Balb/c mice were randomly divided into groups, having five animals per group. Group I was considered the Control (tumor bearing, untreated) receiving Saline, Group II Etop receiving 1mg/kg; Group III GNP receiving 1mg/kg; Group IV EGNP receiving 1mg/kg daily intravenously (i.v) for 14days (7days after the tumor inoculum). All the mice were observed for tumor regression and survival. The tumor volume was measured on alternate days using a vernier calliper.
After 21days, the mice were euthanised and tumors were harvested and weighed. To minimize personal bias, all measurements were done by same examiner.
Statistical Analysis
The results were articulated as the Mean standard deviation. Evaluation amongst groups were evaluated by One-way ANOVA and the respective means were separated by Tukey’s test using the Prism (5.0) software (Prism software Inc. CA, United States). The levels of significance were accepted at ≤0.05 level.
Results and Discussion
Currently, there is an increased demand for biopolymeric nanoparticles as a means for enhanced delivery of lipophilic chemotherapeutic moieties. For the preparation of homogenous nanoparticles under controlled parameters two-step desolvation method was used to synthesize GNPs; any perturbation of pH i.e a slight increase or decrease alters the net negative or positive charges, principally due to the ionization of -COOH and -NH2 groups, respectively. As a cross linker, glutaraldehyde was used to saturate the functional amino groups that were accessible for the interaction of the covalent amino-reactive ligands. Size of GNPs was dependent on the number of cross-links, but the cross-linker was used as minimal as possible in order to prevent toxicity from free aldehyde groups. Additionally, L-cysteine was used to quench the aldehyde groups that might possibly produce the free sulfhydryl groups on the surface of GNPs (Coester et al., 2000). Figure 1 depicts the characterization of GNP where, Fig (A) shows video frame of the morphology of the nanoparticles followed by size by DLS ie. 150nm ±0.241 (1B). The zeta potential (ZP) of GNP was -29.32mV as represented in Figure 1C. Nanoparticles having ZP of > +25mV or < −25mV usually have high degree of stability. Lower dispersions ZP values will often lead to aggregation, coagulation, or flocculation due to Van der Waal’s interparticle attraction (Shnoudeh et al., 2019), Figure 1D indicates the UV spectra of both Etop per se and EGNP. Figure S1 represents the UV spectra of Gelatin polymer and GNPs. The entrapment efficiency of Etop in EGNPs was ∼29% (Figure 1D).
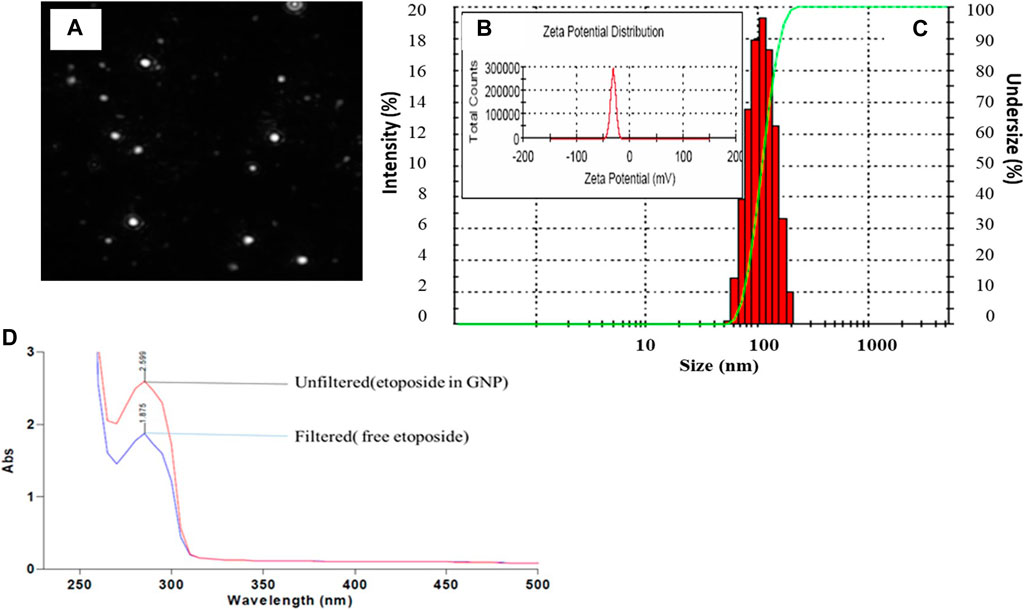
FIGURE 1. Physical Characterization of GNP (A) NTA video frame (A) (B) size of the GNP (C) Zeta potential of GNP = −29.5mV (D) UV Spectra of Etop and EGNP before filtering and after filtering at 257nm. Calculated Entrapment Efficiency = 29%.
The GNPs were stable for 90h at different pH and temperature (Saxena et al., 2005) The nanoparticle sizing is critical, as the biodistribution and pharmacokinetics of the gelatin nanoparticles is largely going to be governed by this factor, with surface charge. The synthesis of Gelatin nanoparticles involves using a desolvation agent to remove low molecular weight fractions, allows the remaining high molecular weight precipitate to form ∼100–300nm particles after chemical crosslinking. Experimental modifications to pH, crosslinker concentration and desolvation agent are routinely employed to obtain desirable nanoparticle sizes (typically less than 200nm), charges (higher charges indicate greater nanoparticle stability) and chemical bonding (Elzoghby, 2013; Hudson and Margaritis, 2014).
The IC 50 was calculated to assess the biotoxicity Etop, GNP and EGNP on MCF-7 and HEK-293 cells by cell viability assay using MTT that reduces the yellow coloured tetrazolium salt to purple coloured formazan crystals. The metabolic activities are proportional to the developed colour owing to the NAD(P)H-dependent cellular oxidoreductase enzymes in living cells (Mosmann, 1983; Berridge et al., 2005). This accurate quantitation regarding proliferation rate or viability of cells is regularly used to assess a linear relationship among the metabolically active cells with respect to the color intensity. Table S4 represents the IC50 of Etop per se was 142.67μg/ml in MCF–7 cells, post 24 h, while in HEK-293 it was 1,337.49μg/ml. GNP had negligible cytotoxicity on both the cell lines. IC50 of EGNP was observed to be lowest i. e 74.14μg/ml indicating maximum anti-cancer efficacy (Figure 2).
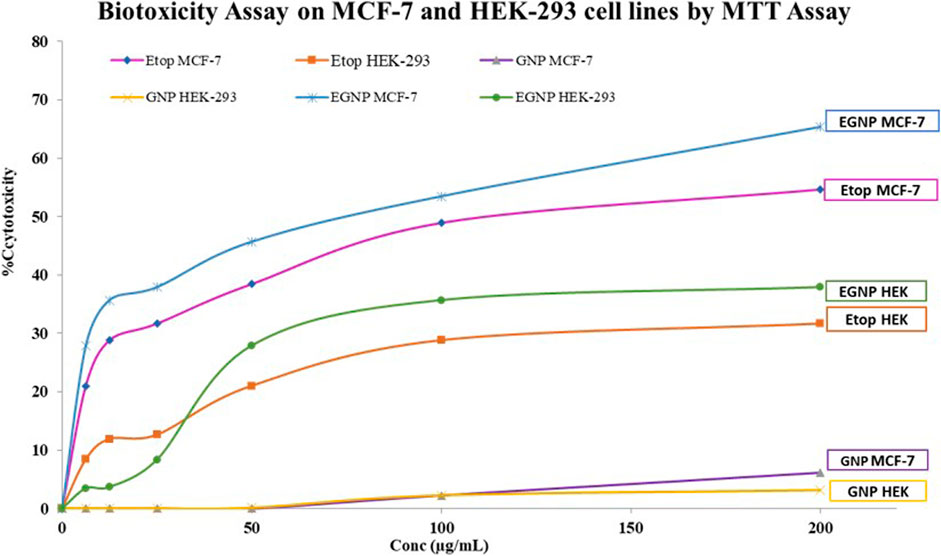
FIGURE 2. Dose dependent cytotoxicity on MCF-7 and HEK-293 of Etop, GNP and EGNP for 24h (A)Etoposide (B)GNP (C) EGNP. Data were expressed in mean ± S.E.M (N = 3).
For further evaluation of mechanistic pathways 100μg/ml dose of Etop per se and equivalent Etop in EGNP was carefully chosen. In our earlier published studies GNPs and EGNPs were seen to localize in cells leading to buildup of oxidative stress and DNA damage (Moin et al., 2018). The mechanical pathways of nanoparticles to induce cellular damage and cell death induction are not yet fully understood. The pathways induced by EGNP in cellular damage and cell death are not completely known but most frequently p53 appears to play a major role in ROS induced DNA damage and apoptotic cellular responses (Shafagh et al., 2015). Increased intensity of DCFDA indicated enhanced ROS generation in MCF-7 when compared to HEK-293 cells (Supplementary Figure S2).
EGNPs triggered enhanced release of intracellular Ca2+ in MCF-7 making them susceptible to Etop at a lower concentration. The twin-functional role of Etop makes it a pro-oxidant owing to its affinity to intracellular thiols in the cells. This redox perturbation possibly contributes to its carcinogenic potential and genotoxicity resulting in antitumor efficacy. In recent years, the paradigm of cell death regulation infers not only apoptosis but other form of programmed or regulated necrosis i.e. necroptosis. Moin et al., 2018, discussed the mechanism of Etop and EGNP in MCF-7 cells hypothesizing the occurrence of necroptosis. We have previously reported the influence of redox paradox of Etop on intracellular biochemical pathways that influenced cell death whereby cells were progressing both towards necroptosis and apoptosis. (Moin et al., 2018).
Herein, we assessed the levels of the major proteins responsible for the formation of Necroptosome i.e. RIPK1, RIPK3 and MLKL in the presence of both an apoptotic inhibitor zVAD and a necroptotic inhibitor Nec-1. The role of RIPK1 and RIPK3 have been expansively explored in infectious diseases. We evaluated the expression of RIPK1, RIPK3 and MLKL along with the expression of apoptotic biomarkers in MCF-7 cell line by Real-time PCR (qPCR). High expression of RIPK1 in EGNP + zVAD confirmed necroptosis, as zVAD is known to block the caspase activity (Figure 3). Expression of RIPK3 in Etop and EGNP were visible, but the expression of EGNP + zVAD was low and upregulation of RIPK3 expression in EGNP + Nec-1 was significant. Although, this indicated the presence of RIPK3, but interestingly there was no evidence of necroptosis. Negligible expression of MLKL led to the understanding that lack of formation of a necroptosome was inhibiting necroptosis as it is essential for triggering necroptosis. EGNP induced high expression of RIPK1 too, so we could hypothesize that high expression of RIPK1 may push the cell towards apoptosis. Also, earlier reports suggest that RIPK1 can regulate not only necroptosis but also apoptotic cell death (Vandenabeele et al., 2013). We further confirmed the expression of apoptotic markers like p53, Bax, Caspase 9 and anti-apoptotic genes Bcl2 (Figure 4). Our findings suggested that Etop and EGNP treated MCF-7 cells showed enhanced expression of apoptotic gene Bax, and lower expression of anti-apoptotic gene i.e. Bcl2, when compared to their individual treatment and control. The mRNA for pro-apoptotic protein Bax was significantly elevated in cells treated with Etop and EGNP whereas the anti-apoptotic Bcl-2 signal was significantly decreased. The Bax/Bcl-2 ratio indicates enhanced apoptotic process (Figure 5). Etop induces apoptosis either via caspase dependent pathways or caspase independent pathways (Degterev et al., 2005). Literature suggests that necroptosis is normally driven by expression of RIPK3, that may be activated following phosphorylation of the serine/threonine kinase RIPK1 (Cho, 2018). RIPK3 possibly stimulates a switch in the cell’s metabolism, that causes an increase in production of mitochondrial ROS that culminates in cell death (Zhang et al., 2009; Vanlangenakker et al., 2011). Further, necroptosis may emerge as an alternative form of cell death that can substitute apoptosis, when caspase activation is blocked. Undoubtedly, necroptosis embodies a significant pathway for increasing sensitivity of tumor cells to anticancer therapies. Moreover, its potentiation may represent a crucial and an important treatment window to eliminate tumor cells, particularly those resistant to apoptosis (Giampietri et al., 2014). Even though, the conventional form of cell death is frequently concealed by other forms of cell death, it recommences as a back-up mechanism, when the other pathways are blocked.
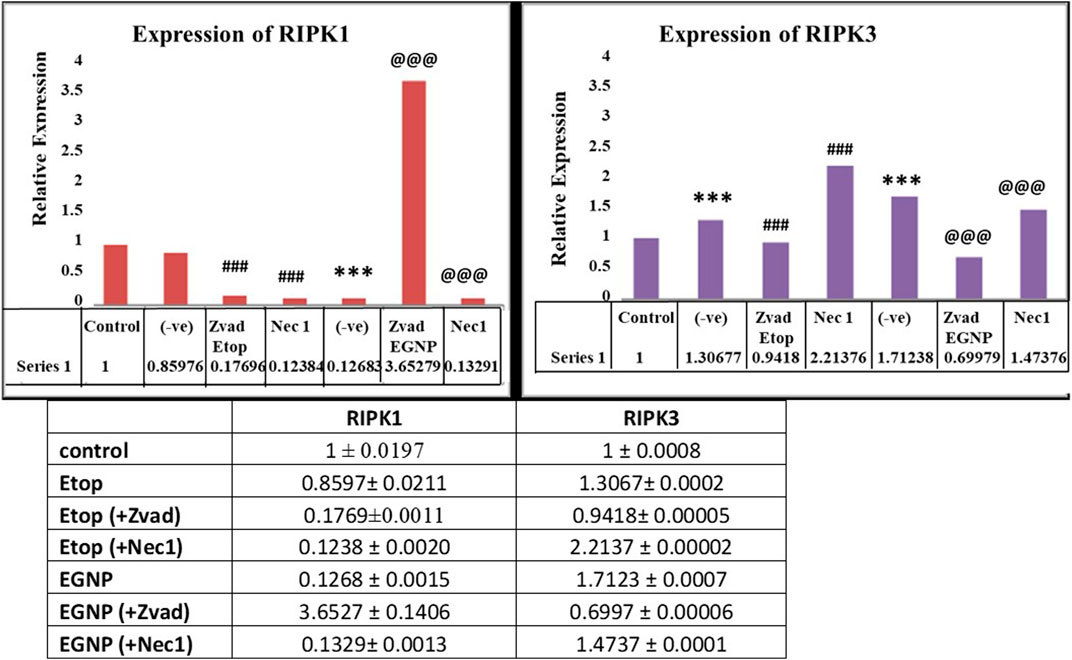
FIGURE 3. Gene expression of RIPK1 and RIPK3 with. Etop, EGNP, zVAD+Etop, zVAD+EGNP, Nec-1+Etop and Nec-1+EGNP treated groups. Expression of GAPDH taken as internal control. Data were expressed in mean ± S.E.M (N = 3). *denotes significant difference between Control vs treated groups. # denotes significant difference between Etop vs Nec1/Zvad. @ denotes significant difference between Etop vs Nec1/Zvad. *p < 0.05.
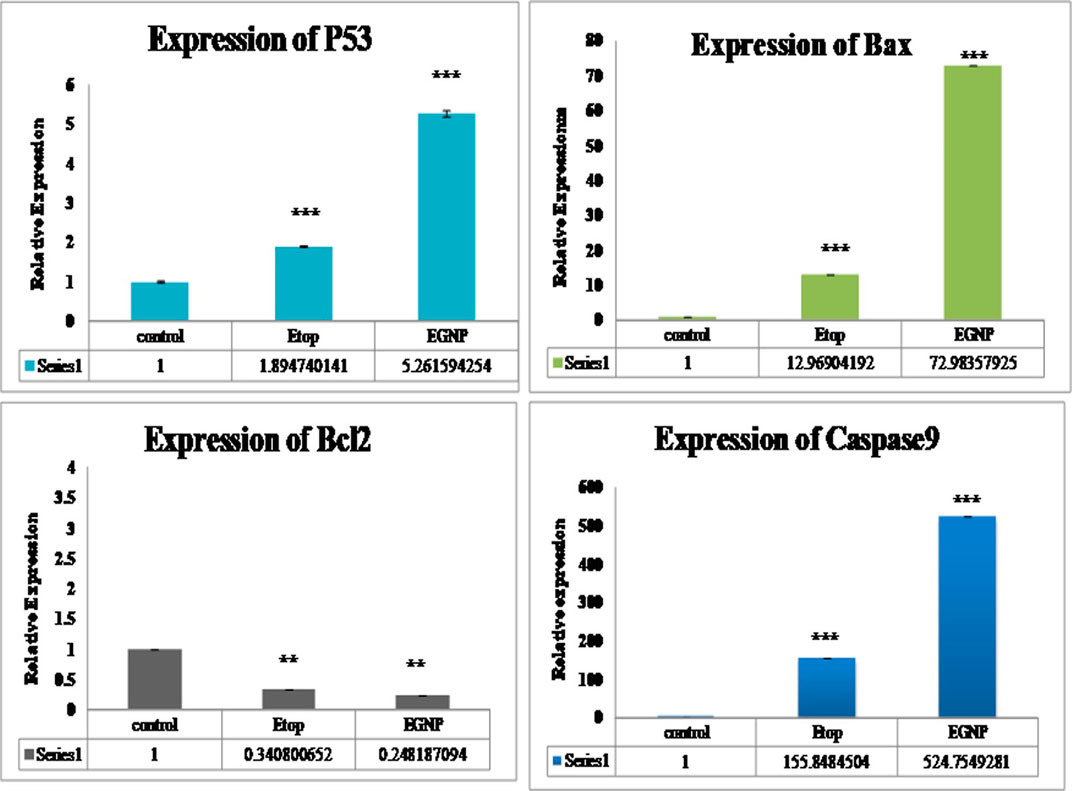
FIGURE 4. Gene expression of p53, Bcl2, Bax, Caspase 9 with Etop, EGNP, treated groups. Expression of Beta-Actin taken as internal control. Data were expressed in mean ± S.E.M (N = 3). *denotes significant difference between Control vs treated groups. *p < 0.05.
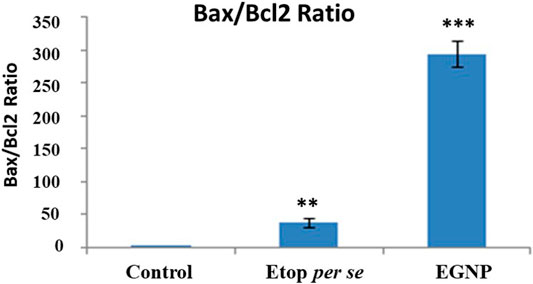
FIGURE 5. Determination of Bax, Bcl-2 and their ratio in control, Etop and EGNP treatment. The mRNA for pro-apoptotic protein Bax was significantly elevated in cells treated Etop and EGNP whereas the anti-apoptotic Bcl-2 signal is significantly decreased. The Bax/Bcl-2 ratio may indicate an enhanced apoptotic process. Data were expressed in mean ± S.E.M (N = 3). *denotes significant difference between Control vs treated groups. *p < 0.05.
Cell cycle arrest after treatment with Etop, GNP and EGNP along with control group on MCF-7 and HEK-293 cells was performed and the percentages of cells in G1, S, and G2/M were calculated using FlowJo software. We observed that MCF-7 cells treated with Etop, GNP and ENGP showed the significant cell cycle perturbation, G1/G0 arrest as compared to the untreated control. However, in case of HEK-293 cells treated with Etop, GNP and ENGP showed non-significant cell cycle perturbation. (Supplementary Figure S3, Supplementary data). Figure 6 shows the hypothesized mechanism of the action of Etoposide/Etoposide formulation.
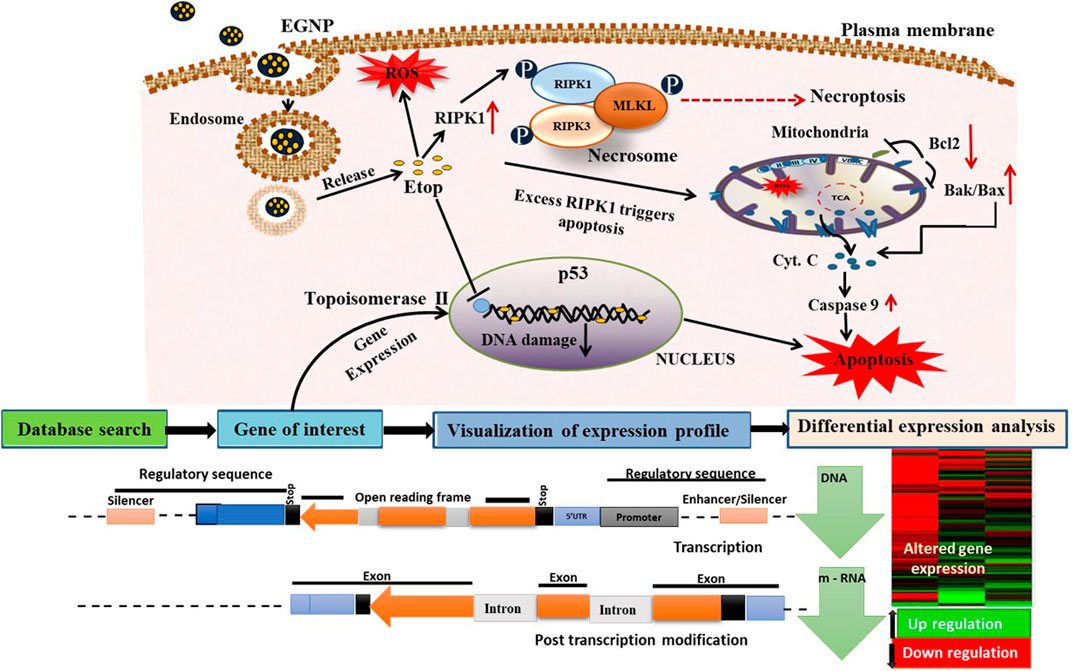
FIGURE 6. Hypothesized mechanism of action of etoposide/etoposide formulation and toxicological approach to study the nanoparticles toxicity. EGNPs enter the cells via endocytosis to form the endosomal complex. Due to the differences in pH, etoposide is released from EGNPs to inhibit topoisomerase ǁ activity. Free etoposide can also activate RIPK1 which further phosphorylates the RIPK3. This then recruits MLKL to form the necrosome that triggers necropoptosis. Over expression of RIPK1 can also stimulate mitochondrial based apoptosis. Our results show enhanced expression of RIPK1 and RIPK3 and decreased expression of MLKL but exhibits necroptosis that is probably because of enhanced RIPK3.
In order to conduct any in vivo study, it was essential to evaluate the biocompatibility of the nanoparticles with red blood cells (RBC) (Dobrovolskaia et al., 2008). Biocompatibility was evaluated by hemolysis assay induced by Etop per se, GNP, EGNP incubated with fresh mice blood for 2 and 4h. Around 4% hemolysis (Supplementary Figure S4, Supplementary data) was detected in all experimental groups proving their biocompatibility (p < 0.001) with red blood cells compared to that of positive control (Triton X) at both the time intervals.
In spite of the anti-neoplastic efficacy of Etop, the adverse effects and pharmacokinetic hindrances such as fever, nausea, bone marrow suppression and hair loss are often limiting its use (Urban et al., 2019). Another significant pharmacokinetic drawback of Etop is its hydrophobicity (Soni and Yadav, 2014) and inconsistent bioavailability ranging from 25–74%, which is considerably high intra- and inter-patient variation, and a half-life of 4–11h (Duong et al., 2019). This short half-life often reduces the duration of exposure of Etop to cancer cells, thereby, minimizing its efficacy. Clinical results have revealed that exposure time coupled with slow release of Etop are decisive factors for maximizing its impact, since its target, i.e., topoisomerase II, will only be expressed in cells that are in mitosis phase (Snehalatha et al., 2008). To overcome the pharmacokinetic deterrents of Etop, FDA approved gelatin has been used as an excipient.
Influence of stannuous chloride on labelling efficiency of Etop, GNPs and EGNPs was assessed. Etop, GNPs were labelled with 99mTc with high labelling efficiency by direct method. Amount of stannous chloride(SnCl2) required to reduce 99mTec is necessary in the labelling process as an enhanced amount results in the formation of radio colloids, that are unwanted. Also, reduced amounts of SnCl2 results in meagre labelling efficiency. The influence of SnCl2 on the labelling efficiency is shown in Supplementary Table S2 (Supplementary Data).
Herein, the optimal amount of SnCl2 required for high labelling efficiency, with a low amount of free and R/H 99mTc, was observed to be 100µg for all preparations. Incubation time in which maximum percentage of labelling occurs was further optimized as 15min post-addition of 99mTc -pertechnetate to the formulation. To optimize the above parameters at each time point, quality control checks were done by TLC using ITLC strips.
In vitro Stability
Radiolabelled preparations were assessed for their stability in the presence of Phosphate buffered saline (PBS) and sera. In vivo physiological micro-environment like pH and temperature was simulated for evaluating the stability of the labelled Etop, GNPs and EGNPs. Stability data in PBS and sera at different time points are represented in Supplementary Table S3. All preparations were stable in upto 24h at different time points and in their respective media, 99mTc- labelled preparations have shown a percentage of radiolabelling of more than 90%. The stability of the labelled complexes in different conditions clearly indicates the utility of the label as a marker for biodistribution studies.
With this assurance, biodistribution and pharmacokinetic studies were conducted in Balb/c mice and rabbit, respectively. Biodistribution of 99mTcGNP and 99mTcEGNP in Balb/c mice indicated maximum accumulation in kidney in terms of % uptake/gm in kidney (13.10 %uptake/gm), liver and blood after 1 h, that subsequently decreased upto 24h (Figures 7A,B). The activity in lung, liver and spleen after first hour of administration was found to be almost same at all time points. 111In and 64Cu Radiolabelling of nanoparticles enables the visualization of the interactions between the nanoparticles and the kidneys by PET and SPECT, requiring only attomolar doses of injected particles (10–18mol l−1). For example, labelling silica nanoparticles with 124I allows imaging of their in vivo transport through the kidneys in patients with cancer (Bujie et al., 2018); The retention in the non-target tissues was comparable for all the probes, with low uptake in most of the assessed organs and higher levels of activity in the kidneys. (Starmans et al., 2013). Half-life (t1/2) in context of GNPs and EGNPs is a useful evaluation to observe the pharmacokinetics and is referred to as the time taken for nanoparticles concentration in the blood to reduce to half its initial value. The elimination rate constant and half life time values indicated prolonged circulation of 99mTcEGNP in the blood owing to a lower elimination rate when compared to 99mTcGNP and 99mTcEtop (Figure 7C). This result when coupled with pharmacokinetics of 99mTcEGNP illustrated the reduced activity of 99mTcEGNP at 4h. Both liver and kidney showed enhanced accumulation of 99mTcEGNP. Figure 5B as indicated by the gamma scintigraphy images in rabbit exhibiting accumulation of 99mTcGNP and 99mTcEGNP in various organs performed post 2 and 24h administration (Figure 5D. The hot spots in rabbit represents the accumulation of 99mTcGNP and 99mTcEGNP that was coupled with the result of bio-distribution. Plasma molecules and particles may perfuse deeper and more evenly into the tumor tissue. Therefore, while designing nanoparticles it is essential to control the size to overcome the criteria described earlier. Consequently, an optimal size of nanoparticles that can deliver adequate cargo of drugs to be dispersed homogeneously in solid tumors is desired to induce maximum therapeutic efficacy that is reported between 10 and 200nm in diameter (Allen and Cullis, 2004).
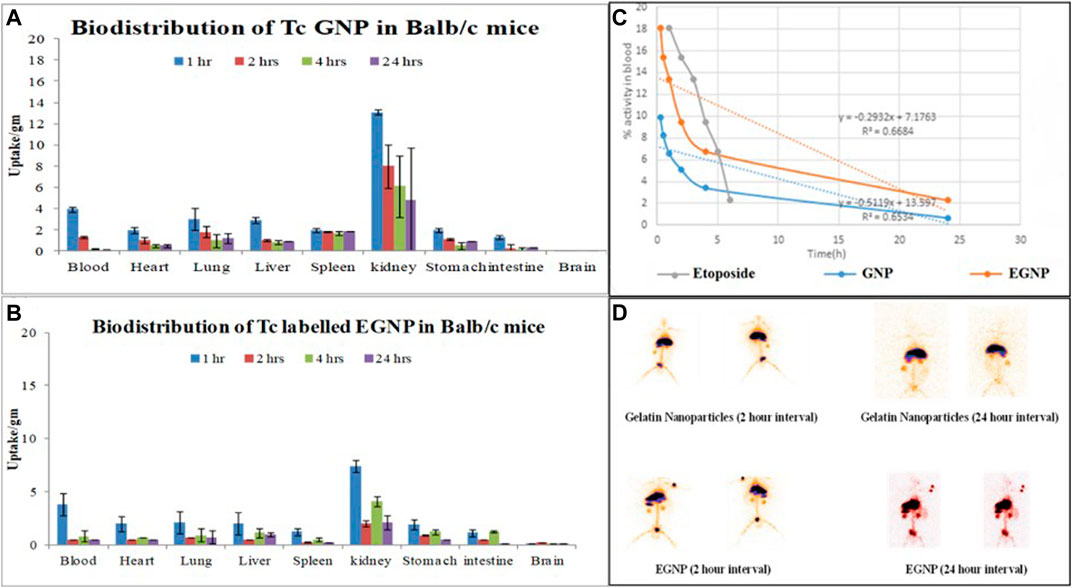
FIGURE 7. Graph illustrate the in vivo bio-distribution of 99mTcGNP in (A,B)99mTcEGNP various organs of Balb/c mice (C) blood pharmacokinetics of intra-venously injected 99mTc-Etop, 99mTcGNP and 99mTcEGNP in rabbit (D)In vivo gamma scintigraphic imaging representing the accumulation of 99mTcGNP and 99mTcEGNP post 2 h and 24 h of injection through the ear vein of rabbit.
EAC is a homogenous tumor showing efficient capacity for ease of transplant, quick proliferation, assured malignancy, originally hyper-diploid, and described due to the absence of tumor-specific transplantation antigen (Othman et al., 2018).
Tumor regression efficacy of synthesized EGNP were evaluated. Our subcutaneous EAC solid tumor model does not exceed 20mm at the largest diameter, which is considered acceptable in standard procedures using mouse models. After 21days, results of tumor regression analysis indicated that our EGNP formulation causes significant reduction in tumor size when compared to Etop per se. Tumor progression was evident in Control and GNP treated mice (Figure 8).
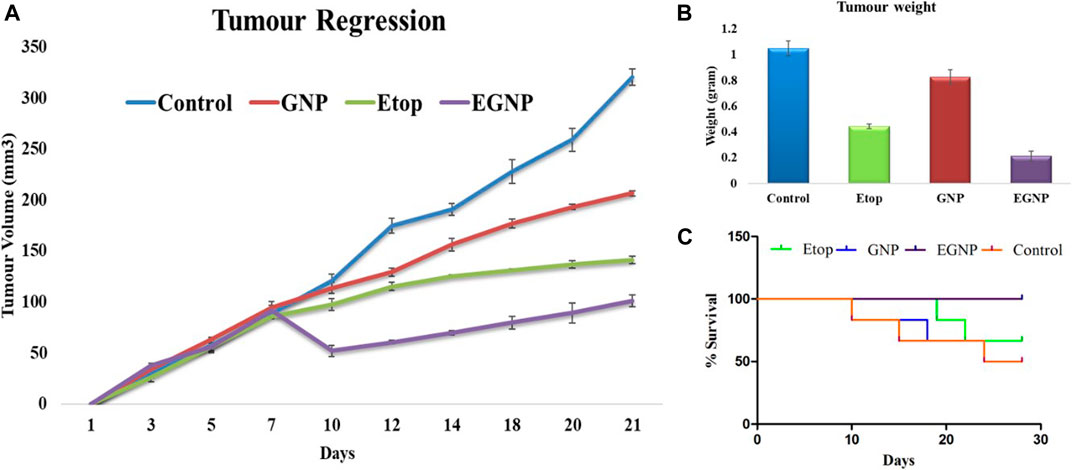
FIGURE 8. In vivo studies in Balb/c mice after administering Etop, GNP and EGNP. (A) Tumour regression study for a period of 21 days. showed EGNP has better anti-tumour efficacy as compared to Etop per se. (B) Tumours were weighed after 21 days. EGNP treatment has been able to reduce the weight and size of tumours effectively in comparison to Etop per se and control. (C) Percent Survival.
Anti-tumor efficacy of EGNP may be contributed to the Enhanced permeability and retention effect (EPR). EPR effect is possible in solid tumors as the drug delivery system can be passively accumulated based on its size and enhanced circulation time in the blood thereby reducing the tumor burden (Allen and Cullis, 2004; Maeda, 2013).
The promising results in the terms of percentage survival (100%) and decreased tumor volume in case of EGNP are accredited to increase in accumulation at the tumor site that causes sustained release of etoposide drug from our smart, pH responsive gelatin nanoparticles resulting in long-time circulation in the body. The steady release of Etop from GNPs could be attributed to lysosomal degradation and acidic tumor environment. Hence, the results proved the therapeutic efficacy of the GNPs as a biocompatible drug delivery carrier for Etop delivery for cancer therapy.
Conclusion
The sustained expansion of nanotechnology entails an inclusive understanding of the probable mechanisms of their toxicity for appropriate safety evaluation and identification of expressed biomarkers. Their impact on biological pathways have begun to emerge as the complex death paradigms that are needed to be explained by activation of different death pathways in a context-dependent manner. We report preparation of GNPs in the size range of ∼150nm with PDI of 0.257; Physical characterization specified a spherical morphology and a size that was in unison with the NTA data. Etop, a highly hydrophobic, low molecular weight drug was entrapped in GNPs. The therapeutic efficacy is only possible by its hydrolysis inside the target cells where Etop per se is released. Increased biotoxicity was observed by EGNPs at a much lower concentration of Etop in MCF-7 cells when compared to HEK-293 cells. Crucially, EGNPs were efficient as it enabled reduction in the concentration of etoposide without negotiating the efficacy of cell mortality. Results of the long term biotoxicity study concluded that the EGNPs formulations were efficient in reducing the viability of MCF-7 cells over a period of three days, whereas pure Etop exerted its maximum efficiency within 24h. Moreover, specific apoptosis inhibitors, such as zVAD, can further shed light on the presence of apoptosis. With respect to caspase inhibition, the serine proteases RIPK1 and RIPK3 could possibly switch apoptosis to necroptosis, a pro-inflammatory and regulated form of cell death, characterized by loss of plasma membrane permeability and release of intracellular contents, such as LDH. Therefore, explicating the nanoparticles induced activation of molecular mechanistic cell death signaling pathways would be crucial for development of novel strategies to abate nanotoxicity. Gene expression profiling is further expected to recognize the various mechanisms that trigger the potential toxicity of drug molecules. Overall, to improve the efficacy of classical cancer drugs, treatment-driven gene expression vagaries can be exploited and their pre-treatment prediction could be of enormous clinical value to evolve a guided cancer therapy regimen.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors. However, the animal study was reviewed and approved by Institutional Animal Ethics Committee, K.M. College, University of Delhi.
Author Contributions
AV has made significant contributions to the conception and design of this work, as well as the drafting, data interpretation, and revision of this manuscript. IM has contributed significantly to the experimental work, data acquisition and analysis of data. LB, ZY, NK, AL, DM, contributed towards paper in data acquisition, formatting, revision and final approval of this manuscript.
Funding
Funding will be done by the Organizers of National Conference on Nano/Bio-Technology 2019, India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
IM acknowledges CSIR for his fellowship. LB and DM gratefully acknowledge UGC for SRFs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2021.624083/full#supplementary-material.
References
Afshari, C. A., Hamadeh, H. K., and Bushel, P. R. (2011). The evolution of bioinformatics in toxicology: advancing toxicogenomics. Toxicol. Sci. 120 (Suppl_1), S225–S237. doi:10.1093/toxsci/kfq373
Allen, T. M., and Cullis, P. R. (2004). Drug delivery systems: entering the mainstream. Science 303 (5665), 1818–1822. doi:10.1126/science.1095833
Berridge, M. V., Herst, P. M., and Tan, A. S. (2005). Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11, 127–152. doi:10.1016/S1387-2656(05)11004-7
Brown, J. M., and Attardi, L. D. (2005). The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer. 5, 231–237. doi:10.1038/nrc1560
Bujie, D., Mengxiao, Y., and Zheng, J. (2018). Transport and interactions of nanoparticles in the kidneys. Nat. Rev. Mater. 3, 358–374. doi:10.1038/s41578-018-0038-3
Cai, Z., Jitkaew, S., Zhao, J., Chiang, H. C., Choksi, S., Liu, J., et al. (2014). Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 16, 55–65. doi:10.1038/ncb2883
Cesàro, A., Bellich, B., and Borgogna, M. (2012). Biophysical functionality in polysaccharides: from Lego-blocks to nano-particles. Eur. Biophys. J. 41 (4), 379–395. doi:10.1007/s00249-011-0753-9
Chen, X., Li, W., Ren, J., Huang, D., He, W. T., Song, Y., et al. (2014). Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 24, 105–121. doi:10.1038/cr.2013.171
Cho, Y. S. (2018). The role of necroptosis in the treatment of diseases. BMB Rep. 51 (5), 219–224. (2018). doi:10.5483/bmbrep.2018.51.5.074
Coester, C., Kreuter, J., von Briesen, H., and Langer, K. (2000). Preparation of avidin-labelled gelatin nanoparticles as carriers for biotinylated peptide nucleic acid (PNA). Int. J. Pharm. 196 (2), 147–149. doi:10.1016/s0378-5173(99)00409-3
De Jong, W. H., and Borm, P. J. (2008). Drug delivery and nanoparticles:applications and hazards. Int. J. Nanomedicine. 3 (2), 133–149. doi:10.2147/ijn.s596
Degterev, A., Huang, Z., Boyce, M., Li, Y., Jagtap, P., Mizushima, N., et al. (2005). Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1 (2), 112–119. doi:10.1038/nchembio711
Dobrovolskaia, M. A., Clogston, J. D., Neun, B. W., Hall, J. B., Patri, A. K., and McNeil, S. E. (2008). Method for analysis of nanoparticle hemolytic properties in vitro. Nano Lett. 8 (8), 2180–2187. doi:10.1021/nl0805615
Dondelinger, Y., Declercq, W., Montessuit, S., Roelandt, R., Goncalves, A., Bruggeman, I., et al. (2014). MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 7, 971–981. doi:10.1016/j.celrep.2014.04.026
Duong, J., Veal, G., Nath, C., Shaw, P., Errington, J., Ladenstein, R., et al. (2019). Population pharmacokinetics of carboplatin, etoposide and melphalan in children: a re-evaluation of paediatric dosing formulas for carboplatin in patients with normal or mild impairment of renal function, Br. J. Clin. Pharmacol. 85, 136–146. doi:10.1111/bcp.13774
Ellinger-Ziegelbauer, H., Gmuender, H., Bandenburg, A., and Ahr, H. J. (2008). Prediction of a carcinogenic potential of rat hepatocarcinogens using toxicogenomics analysis of short-term in vivo studies. Mutat. Res. 637 (1–2), 23–39. doi:10.1016/j.mrfmmm.2007.06.010
Elzoghby, A. O., Samy, W. M., and Elgindy, N. A. (2012). Protein-based nanocarriers as promising drug and gene delivery systems. J. Control. Release. 161 (1), 38–49. doi:10.1016/j.jconrel.2012.04.036
Elzoghby, A. O. (2013). Gelatin-based nanoparticles as drug and gene delivery systems: reviewing three decades of research. J. Control. Release. 172 (3), 1075–1091. doi:10.1016/j.jconrel.2013.09.019
Fitzmaurice, C., Fitzmaurice, C., Allen, C., Barber, R. M., Barregard, L., Bhutta, Z. A., et al. (2017). Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 3 (4), 524–548. doi:10.1001/jamaoncol.2016.5688
Fuchs, S., Klier, J., May, A., Winter, G., Coester, C., and Gehlen, H. (2012). Towards an inhalative invivo application of immunomodulating gelatin nanoparticles in horse-related preformulation studies. J. Microencapsul. 29 (7), 615–625. doi:10.3109/02652048.2012.668962
Giampietri, C., Starace, D., Petrungaro, S., Filippini, A., and Ziparo, E. (2014). Necroptosis: molecular signalling and translational implications. Int. J. Cell Biol. 2014, 490275. doi:10.1155/2014/490275
Henwood, J. M., and Brogden, R. N. (1990). Etoposide. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in combination chemotherapy of cancer. Drugs 39 (3), 438–490. doi:10.2165/00003495-199039030-00008
Hubbell, J. A., and Chilkoti, A. (2012). Chemistry. Nanomaterials for drug delivery. Science 337, 303–305. doi:10.1126/science.1219657
Hudson, D., and Margaritis, A. (2014). Biopolymer nanoparticle production for controlled release of biopharmaceuticals. Crit. Rev. Biotechnol. 34 (2), 161–179. doi:10.3109/07388551.2012.743503
Jabr-Milane, L., van Vlerken, L., Devalapally, H., Shenoy, D., Komareddy, S., Bhavsar, M., et al. (2008). Multi-functional nanocarriers for targeted delivery of drugs and genes. J. Control. Release. 130 (2), 121–128. doi:10.1016/j.jconrel.2008.04.016
Jain, V., Kumar, H., Anod, H. V., Chand, P., Gupta, N. V., Dey, S., et al. (2020). A review of nanotechnology-based approaches for breast cancer and triple-negative breast cancer. J. Control. Release. 326, 628–647. doi:10.1016/j.jconrel.2020.07.003
Kaul, G., and Amiji, M. (2004). Biodistribution and targeting potential of poly(ethylene glycol)-modified gelatin nanoparticles in subcutaneous murine tumor model. J. Drug Target. 12 (9–10), 585–591. doi:10.1080/10611860400013451
Leekha, A., Gurjar, B. S., Tyagi, A., Rizvi, M. A., and Verma, A. K. (2016). Vitamin C in synergism with cisplatin induces cell death in cervical cancer cells through altered redox cycling and p53 upregulation. J. Cancer Res. Clin. Oncol. 142 (12), 2503–2514. doi:10.1007/s00432-016-2235-z
Maeda, H. (2013). The link between infection and cancer: tumor vasculature, free radicals, and drug delivery to tumors via the EPR effect. Cancer Sci. 104 (7), 779–789. doi:10.1111/cas.1215
Magadala, P., and Amiji, M. (2008). Epidermal growth factor receptor-targeted gelatin-based engineered nanocarriers for DNA delivery and transfection in human pancreatic cancer cells. AAPS J. 10 (4), 565–576. doi:10.1208/s12248-008-9065-0
Moin, I., Biswas, L., Mittal, D., Leekha, A., Kumari, N., and Verma, A. K. (2018). Crosstalk of ER stress, mitochondrial membrane potential and ROS determines cell death mechanisms induced by etoposide loaded gelatin nanoparticles in MCF-7 breast cancer cells. J. Nanomed Nanotechnol. 9 (513), 2. doi:10.4172/2157-7439.1000513
Montecucco, A., Zanetta, F., and Biamonti, G. (2015). Molecular mechanisms of etoposide. EXCLI J. 14, 95–108. doi:10.17179/excli2015-561
Moriwaki, K., and Chan, F. K. (2013). RIP3: a molecular switch for necrosis and inflammation. Genes Dev. 27, 1640–1649. doi:10.1101/gad.223321.113
Mosmann, T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. doi:10.1016/0022-1759(83)90303-4
Nie, A. Y., McMillian, M., Parker, J., Leone, A., Bryant, S., Yieh, L., et al. (2006). Predictive toxicogenomics approaches reveal underlying molecular mechanisms of nongenotoxic carcinogenicity. Mol. Carcinog. 45 (12), 914–933. doi:10.1002/mc.20205
Nuwaysir, E. F., Bittner, M., Trent, J., Barrett, J. C., and Afshari, C. A. (1999). Microarrays and toxicology: the advent of toxicogenomics. Mol. Carcinog. 24 (3), 153–159. doi:10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p
Othman, A. I., El-Sherbiny, I. M., ElMissiry, M. A., Ali, D. A., and AbdElhakim, E. (2018). Polyphenon-E encapsulated into chitosan nanoparticles inhibited proliferation and growth of Ehrlich solid tumor in mice. Egypt. J. Basic Appl. Sci. 5 (1), 110–120. doi:10.1016/j.ejbas.2017.10.008
Plowman, J., Dykes, D. J., Hollingshead, M., Simpson-Herren, L., and Alley, M. C. (1997). “Human tumor xenograft models,” in Anticancer drug development guide: preclinical screening, clinical trials, and approval. Editor B Teicher (Totowa, NJ: Humana Press), 101–125.
Pognan, F. (2007). Toxicogenomics applied to predictive and exploratory toxicology for the safety assessment of new chemical entities: a long road with deep potholes, Prog. Drug Res. 64, 219–238. doi:10.1007/978-3-7643-7567-6_9
Ricci, M. S., and Zong, W. X. (2006). Chemotherapeutic approaches for targeting cell death pathways. Oncologist. 11 (4), 342–357. doi:10.1634/theoncologist.11-4-342
Saxena, A., Sachin, K., Bohidar, H., and Verma, A. K. (2005). Effect of molecular weight heterogeneity on drug encapsulation efficiency of gelatin nano-particles. Colloids Surf. B Biointerfaces. 45, 42–48. doi:10.1016/j.colsurfb.2005.07.005
Shafagh, M., Rahmani, F., and Delirezh, N. (2015). CuO nanoparticles induce cytotoxicity and apoptosis in human K562 cancer cell line via mitochondrial pathway, through reactive oxygen species and p53. Iran J. Basic Med. Sci. 18 (10), 993–1000.
Shnoudeh, A. J., Hamad, I., Abdo, R. W., Qadumii, L., Jaber, A. Y., Surchi, H. S., et al. (2019). Synthesis, characterization, and applications of metal nanoparticles. Adv. Pharm. Product. Develop. Res. 30, 527–612. doi:10.1016/b978-0-12-814427-5.00015-9
Shu-Ting, P., Zhi- Ling, L., Zhi-Xu, H., Jia-Xuan, Q., and Shu-Feng, Z. (2016). Molecular mechanisms for tumour resistance to chemotherapy. Clin. Exp. Pharmacol. Physiol. 43, 723–737. doi:10.1111/1440-1681.12581
Siddiqui, M. F., Muqaddas, M., and Sarwar, S. (2015). Biochemical mechanisms of etoposide; upshot of cell death. Int. J. Pharm. Sci. Res. 6 (12), 4920–4939. doi:10.13040/IJPSR.0975-8232.6(12).4920-39
Singh, D., Singh, D., Choi, S. M., and Han, S. S. (2013). Enhanced proliferation and growth of human lung epithelial cells on gelatin microparticle loaded with Ephedra extracts. J. Nanomater. 2013, 909120. doi:10.1155/2013/909120
Singh, D., Han, S. S., and Shin, E. J. (2014). Polysaccharides as nanocarriers for therapeutic applications. J. Biomed. Nanotechnol. 10 (9), 2149–2172. doi:10.1166/jbn.2014.1958
Snehalatha, M., Venugopal, K., Saha, N., Babbar, M.K., and Sharma, R.K. (2008). Etoposide loaded PLGA and PCL nanoparticles II: biodistribution and pharmacokinetics after radiolabeling with Tc-99m. Drug Deliv. 15, 277–287.
Soni, G., and Yadav, K. S. (2014). High encapsulation efficiency of poloxamer-based injectable thermoresponsive hydrogels of etoposide. Pharm. Dev. Technol. 19, 651–661. doi:10.3109/10837450.2013.819014
Starmans, L. W., van Duijnhoven, S. M., Rossin, R., Berben, M., Aime, S., Daemen, M. J., et al. (2013). Evaluation of 111In-labeled EPep and FibPep as tracers for fibrin SPECT imaging. Mol. Pharm. 10 (11), 4309–4321. doi:10.1021/mp400406x
Sun, L., Wang, H., Wang, Z., He, S., Chen, S., Liao, D., et al. (2009). Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 148, 213–227. doi:10.1016/j.cell.2011.11.031
Tabata, Y. (2009). Biomaterial technology for tissue engineering applications. J. R. Soc. Interf. 6 (3), S311–S324. doi:10.1098/rsif.2008.0448.focus
Urban, T. A., Lucena, M., Gaffney, K., Dean, R. M., Gerds, A. T., Hamilton, B. K., et al. (2019). Comparison of the tolerability of busulfan, cyclophosphamide, etoposide (BuCyVP) versus carmustine, etoposide, cytarabine, melphalan (BEAM) for autologous hematopoietic cell transplant (AHCT) in Hodgkin lymphoma, Biol. Blood Marrow Transplant. 25, S277–S278. doi:10.1016/j.bbmt.2018.12.344
Vandenabeele, P., Grootjans, S., Callewaert, N., and Takahashi, N. (2013). Necrostatin-1 blocks both RIPK1 and IDO: consequences for the study of cell death in experimental disease models. Cell Death Differ. 20 (2), 185–187. doi:10.1038/cdd.2012.151
Vanlangenakker, N., Vanden Berghe, T., Bogaert, P., Laukens, B., Zobel, K., Deshayes, K., et al. (2011). cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 18 (4), 656–665. doi:10.1038/cdd.2010.138
Verma, A. K., Pandey, R. P., Chanchal, A., and Sharma, P. (2011). Immuno-potentiating role of 687 encapsulated proteins of infectious diseases in biopolymeric nanoparticles as a potential delivery system. J. Biomed. Nanotechnol. 7 (1), 63–64.
Verma, A. K., and Sachin, K. (2008). Novel hydrophilic drug polymer nano-conjugates of Cisplatin showing long blood retention profile: its release kinetics, cellular uptake and bio-distribution. Curr. Drug Deliv. 5, 120–126. (2008). doi:10.2174/156720108783954806
Verma, A. K., Sachin, K., Saxena, A., and Bohidar, H. B. (2005). Release kinetics from bio-polymeric nanoparticles encapsulating protein synthesis inhibitor-cycloheximide, for possible therapeutic applications. Curr. Pharmaceut. Biotechnol. 6 (2), 121–130.
Vonarbourg, A., Passirani, C., Saulnier, P., and Benoit, J. P. (2006). Parameters influencing the stealthiness of colloidal drug delivery systems. Biomaterials. 27 (24), 4356–4373. doi:10.1016/j.biomaterials.2006.03.039
Wang, H., Sun, L., Su, L., Rizo, J., Liu, L., Wang, L. F., et al. (2014). Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 54, 133–146. doi:10.1016/j.molcel.2014.03.003
Zhang, D. W., Shao, J., Lin, J., Zhang, N., Lu, B. J., Lin, S. C., et al. (2009). RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325 (5938), 332–336. doi:10.1126/science.1172308
Keywords: EGNPs, biodistribution, pharmacokinetics, necroptosis, apoptosis, toxicogenomics, tumor regression
Citation: Moin I, Biswas L, Zafaryab M, Kumari N, Leekha A, Mittal D and Verma AK (2021) In vitro Toxico-genomics of Etoposide Loaded Gelatin Nanoparticles and Its in-vivo Therapeutic Potential: Pharmacokinetics, Biodistribution and Tumor Regression in Ehrlich Ascites Carcinoma (EAC) Mice Model. Front. Nanotechnol. 3:624083. doi: 10.3389/fnano.2021.624083
Received: 30 October 2020; Accepted: 08 February 2021;
Published: 21 April 2021.
Edited by:
Pratima R. Solanki, Jawaharlal Nehru University, IndiaReviewed by:
Vinoth Kumar Lakshmanan, Sri Ramachandra Institute of Higher Education and Research, IndiaPayal Gulati, Jawaharlal Nehru University, India
Copyright © 2021 Moin, Biswas, Zafaryab, Kumari, Leekha, Mittal and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anita Kamra Verma, YWt2ZXJtYUBrbWMuZHUuYWMuaW4=
 Imran Moin
Imran Moin Largee Biswas
Largee Biswas Anita Kamra Verma
Anita Kamra Verma