- 1Department of Food and Nutrition, National Agri-Food Biotechnology Institute, Mohali, India
- 2Department of Biochemistry, Panjab University, Chandigarh, India
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a pandemic has been validated as an extreme clinical calamity and has affected several socio-economic activities globally. Proven transmission of this virus occurs through airborne droplets from an infected person. The recent upsurge in the number of infected individuals has already exceeded the number of intensive care beds available to patients. These extraordinary circumstances have elicited the need for the development of diagnostic tools for the detection of the virus and, hence, prevent the spread of the disease. Early diagnosis and effective immediate treatment can reduce and prevent an increase in the number of cases. Conventional methods of detection such as quantitative real-time polymerase chain reaction and chest computed tomography scans have been used extensively for diagnostic purposes. However, these present several challenges, including prolonged assay requirements, labor-intensive testing, low sensitivity, and unavailability of these resources in remote locations. Such challenges urgently require fast, sensitive, and accurate diagnostic techniques for the timely detection and treatment of coronavirus disease 2019 (COVID-19) infections. Point-of-care biosensors that include paper- and chip-based diagnostic systems are rapid, cost-effective, and user friendly. In this article nanotechnology-based potential biosensors for SARS-CoV-2 diagnosis are discussed with particular emphasis on a lateral flow assay, a surface-enhanced Raman scattering-based biosensor, a localized surface plasmon resonance-based biosensor, Förster resonance energy transfer, an electrochemical biosensor, and artificial intelligence-based biosensors. Several biomolecules, such as nucleic acids, antibodies/enzymes, or aptamers, can serve as potential detection molecules on an appropriate platform, such as graphene oxide, nanoparticles, or quantum dots. An effective biosensor can be developed by using appropriate combinations of nanomaterials and technologies.
Introduction
Coronavirus is a spherical or pleomorphic (Dharmendra and Rishabha, 2020), single-stranded, positive-sense, polyadenylated, 30 kb enveloped RNA virus covered with club-shaped glycoproteins (Kahn and McIntosh, 2005). Coronaviruses are related to outbreaks of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) that emerged in 2002 and 2012, respectively. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the family Coronaviridae. It is a recently emerged coronavirus that is responsible for coronavirus disease 2019 (COVID-19). Owing to its worldwide spread across continents, it was declared a pandemic by the World Health Organization (WHO) (Rodriguez-Morales et al., 2020).
Coronavirus was first reported in 1960, and the recently emerged COVID-19 has been reported as a global threat to public health. Novel SARS-CoV-2 has been the third coronavirus outbreak in humans in the past two decades, after SARS and MERS (Munster et al., 2020). COVID infection is affecting a huge population because of its high reproduction number (R0), which represents the rate of transmission of disease from an infected person to a healthy population in a susceptible society. The value of R0 depends on the type of study model chosen by a research team and its value varies significantly from one research group to another. Wu et al. (2020) gave a range for the R0 value for SARS-CoV-2 of 2.47–2.86. The R0 value was reported to be in the range of 2.0–3.3 by Majumder and Mandl (2020) using an incidence decay and exponential adjustment study model. The R0 value of other β-coronaviruses is reported to be in the range of 2.2–3.6 (Lipsitch et al., 2003), while the R0 value of MERS is estimated to be in the range of 2.0–6.7 (Majumder et al., 2014).
The novel coronavirus was first reported in December 2019 in Wuhan City, Hubei Province, China, and was regarded as a pneumonia of unknown etiology (Harapan et al., 2020). It spread through China and to other countries within 2 months. Because of the seriousness of the disease, WHO recognized COVID-19 as a global health emergency and further declared it as a pandemic (WHO, 2020a). Initially, the coronavirus cases were reported in patients who were working or living around a seafood wholesale market in Hunan. WHO identified the novel coronavirus from nasopharyngeal swabs from patients. Previously, the name of the virus was abbreviated to 2019-nCoV, but it was subsequently termed severe acute respiratory syndrome 2 (SARS-CoV-2) (Gorbalenya et al., 2020) and became known as coronavirus disease 2019 (COVID-19). At present, several research organizations throughout the world are involved in the development of antiviral drugs or vaccines against coronavirus and governments are simultaneously implementing preventive measures to restrict the spread of COVID-19.
Nearly 88% similarity was observed between the phylogeny of SARS-CoV-2 and bat-derived SARS-like coronavirus (bat-SL-CoVZC45 and bat-SL-CoVZXC21) that is found in east central China, but SARS-CoV-2 was found to be genetically dissimilar to MERS-CoV and SARS-CoV (Lu et al., 2020). A comparison of genomic sequences of SARS-CoV, RaTG13, and SARS-CoV-2 demonstrated nearly 96.2% genomic similarity between SARS-CoV-2 and bat-CoV-RaTG13, which was formerly identified in Rhinolophus affinis in Yunnan Province (Zhou et al., 2020). Studies have provided evidence that no further genetic recombination occurred in SARS-CoV-2 from other viruses originating from bats (SARS-CoV, bat-CoV-RaTG13, and SARS-CoVr-CoVs) (Zhou et al., 2020). Collectively, different studies have suggested that the original host of the coronavirus might be bats, with the possibility of an unknown intermediate host. Further research is needed, however, to fill the knowledge gap in understanding the transmission of coronavirus to humans. There are reasons that strongly negate bats being the virus-transmitting agent in humans. For example, close relatives of the virus, i.e., bat-SL-CoVZXC21 and bat-SL-CoVZC45, possess a genome identity of <90% with SARS-CoV-2, suggesting that these viruses are not direct ancestors of SARS-CoV-2.
According to data from May 2020, globally there were 4,170,424 reported infections of COVID-19, with 287,399 deaths, affecting more than 210 countries (WHO Situation Report).
Demographics
Men in the age group of 34–59 are more prone to being infected by SARS-CoV-2 (Bai et al., 2020; Chang et al., 2020; Huang et al., 2020a; Wang D. et al., 2020). Patients with heart disease (e.g., cardiovascular disease), cerebrovascular disease, and diabetes are also more prone to SARS-CoV-2 infection (Chen N. et al., 2020). The highest infection rate occurred in adults older than 60 years who have co-morbidities such as cardiovascular disease, chronic respiratory disease, and cerebrovascular disease. Severe manifestations may also be associated with bacterial or fungal infections. A very few cases of SARS-CoV-2 have been reported in children younger than 15 years (Bai et al., 2020; Chang et al., 2020; Huang et al., 2020a; Wang D. et al., 2020). According to a study of 425 COVID-19 patients in Wuhan, there was no report of infection among children aged 15 years or younger. However, another study suggested that pediatric patients' symptoms vary, with the majority showing minor symptoms with no fever or pneumonia-like symptoms and with a good prognosis (Shen and Yang, 2020). The majority of patients with COVID-19 infection demonstrate respiratory illness of mild to moderate severity and they recover without any medical intervention. In summary, children are less prone to being infected or will have mild symptoms if they are infected (Lee et al., 2020).
Transmission of the virus among humans is through close contact with a person carrying the virus, which is mainly spread through saliva droplets or discharge from the nose. In general, the virus spreads via airborne droplets and replicates in the ciliated epithelium, causing cellular damage and infection. Angiotensin-converting enzyme 2 (ACE-2) is a receptor used by SARS-CoV-2 to enter human cells (Letko and Munster, 2020). Previously, ACE-2 was thought to be expressed by the epithelial cells of blood vessels, the intestine, and kidneys (Hamming et al., 2004). Recent investigations have demonstrated the presence of ACE-2 on the mucosa of the oral cavity, which aids the entry of the virus into the host cell. All coronaviruses encode two types of protein, that is, nucleocapsid protein (N-protein) and spike protein (S-protein). N-protein is present on the outer shell of SARS-CoV-2, enclosing the genetic material of the virus, which helps in virus nucleic acid synthesis and interaction with the virus membrane during assembly of the virus structure. S-protein helps in the attachment of the virus to the cell surface receptor of the host cell and allows entry of the virus particle into the host cell. S-protein present on the SARS-CoV-2 envelope is cleaved into two subunits, S1 and S2, during infection (Kirchdoerfer et al., 2016). SARS-CoV-2 infects the host cell by interacting with the receptor-binding domain (RBD) of the S1 subunit by means of the ACE-2 peptidase domain (Turner et al., 2002; Li et al., 2003; Yan et al., 2020), while the S2 subunit assists with the infusion of the virion particle into the membrane of the host cell. The RBDs of SARS-CoV-2 and SARS-CoV are highly similar with few differences in the amino acid sequence present in the binding domain, which supports the reason for strong binding with ACE-2 (Wang Q. et al., 2020; Yan et al., 2020). The clinical manifestations of SARS-CoV-2 include symptoms such as fever, myalgia, dry cough, fatigue, dyspnea, and chest pain (Zhu N. et al., 2020). Vomiting, abdominal pain, headache, nausea, dizziness, and diarrhea are less common symptoms (Dhama et al., 2020; Huang et al., 2020a).
Morbidity and Mortality
The early stages of coronavirus infection exhibited high mortality and morbidity rates (Velavan and Meyer, 2020). With time COVID-19 spread to Italy and Spain in Europe, and currently the USA has the highest numbers of confirmed cases and deaths. Recent increases in the number of COVID-19 cases have surpassed the numbers of intensive care unit beds, which are available only to critically ill patients. The limited numbers of intensive care beds have progressively contributed to the high mortality rate during the COVID-19 outbreak.
Several studies have suggested that blood plasma from recovered patients can be an effective treatment for MERS and SARS and can reduce the rate of mortality (Mair-Jenkins et al., 2015; Arabi et al., 2016; Mustafa et al., 2018). Figure 1 shows the statistics associated with infection and mortality in different world zones. The numbers of cases and deaths keep on increasing on a daily basis because of the relaxation of lockdown measures and increasing testing per day. Even though the number of cases is increasing at a rapid rate, compared with countries such as Italy, Spain, the UK, and the USA, India's health facilities are not robust and have a poor infrastructure, but the number of mortalities in India is less. Also, the recovery rate in India is higher than in many other nations because of timely diagnosis and treatment. As per the Statista 2020 data on September 4, 2020, India has a mortality rate of 1.74%, which is similar to the mortality rate in Russia and much less than the mortality rate in other countries like Italy (13.01%), the UK (12.2%), Belgium (11.44%), Mexico (10.75%), Spain (5.98%), China (5.25%), and the USA (3.05%) (Best, 2020). The low mortality rate in India is due to a higher proportion of younger people in the population (75% of the Indian population are younger than 45 years) and exposure to other infectious disease might have built a stronger innate immunity in the Indian population.
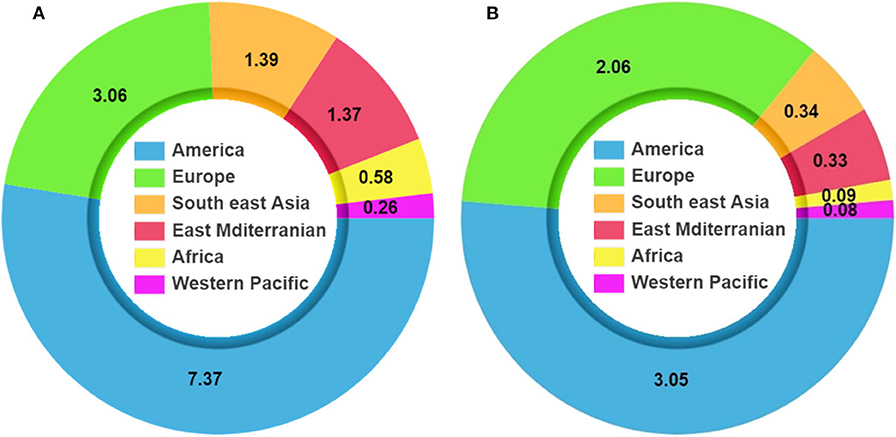
Figure 1. Coronavirus disease 2019 (COVID-19) pandemic statistics. (A) Total number of infections. (B) Mortality due to COVID-19 (Source: WHO data, July 19, 2020).
Diagnosis of SARS-CoV-2
Detection of SARS-CoV-2 is performed by reverse transcription polymerase chain reaction (RT-PCR) or next-generation sequencing (NGS), and computed tomography (CT) scan (Gao, 2020; Wang W. et al., 2020; Xiao et al., 2020; Xie et al., 2020). Presently RT-PCR is recognized as the principal and most effective method for coronavirus detection. Researchers and multiple companies are engaged in developing effective diagnostic kits that include detection of the virus by using nucleic acid RT-PCR and a double-fluorescence PCR method for the simultaneous detection of coronavirus and influenza (Flu A and Flu B) by Jiangsu Shushi Biotechnology Co. Ltd. (Yu et al., 2020). Twenty-two in vitro diagnostic methods were authorized by the US Food and Drug Administration (FDA) under emergency use authorization (EUA), which also includes RT-PCR for the detection of SARS-CoV-2.
Current Methods of Detection
RT-PCR and CT scans are used extensively for diagnostic purposes (Udugama et al., 2020; Waller et al., 2020). In addition, several diagnostic techniques such as clustered regularly interspaced short palindromic repeats (CRISPR)–specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), reverse transcription loop-mediated isothermal amplification (RT-LAMP), enzyme-linked immunosorbent assay (ELISA), and sequencing are under development for enhanced detection of the virus in a minimum amount of time (Carter et al., 2020). However, RT-PCR remains the primary and highly specific mode for diagnosis and quantification of SARS-CoV-2. Commonly used diagnostic technique are represented schematically in Figure 2.
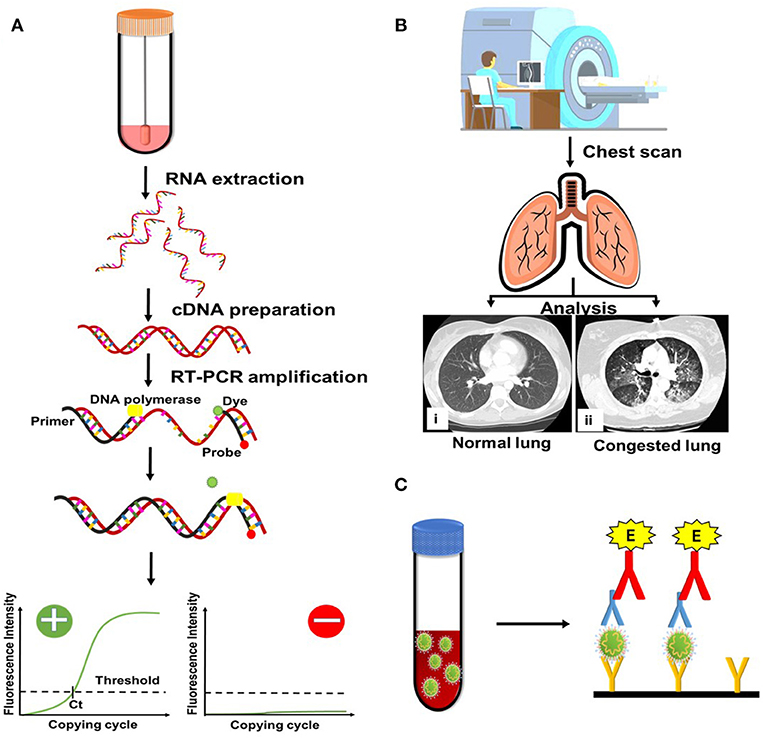
Figure 2. Conventional methods currently being used for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. (A) Reverse transcription polymerase chain reaction (RT-PCR). cDNA, complementary DNA. (B) Computed tomography scan. (C) Enzyme-linked immunosorbent assay.
Real-Time Polymerase Chain Reaction
The accessibility of SARS-CoV-2 genome sequences by means of the Global Initiative on Sharing All Influenza Data (GISAID) has proved beneficial in the development of specific test kits for its detection (Forster et al., 2020). Presently, COVID-19 is diagnosed by detecting the presence of SARS-CoV-2 RNA in oropharyngeal and nasopharyngeal swabs (WHO, 2020b). Since its inception, several RT-PCR-based kits have been developed that involve reverse transcription of SARS-CoV-2 RNA to complementary DNA (cDNA). The genes targeted so far in the detection kits are the E gene (encodes for envelope protein), N gene (encodes for N-protein), and RdRP gene (encodes for RNA-dependent RNA polymerase), where the E gene is used for initial screening, the RdRP gene for confirmation, and the N gene serves as an additional confirmatory assay (Corman V. et al., 2020). The technical limit of detection (LOD) for the RdRP and E genes is 3.8 and 5.2 RNA copies per reaction, respectively, while it is 8.3 for the N gene at the 95% confidence interval (Corman V. et al., 2020). van Kasteren et al. (2020) compared seven RT-PCR kits and found that all were equally efficient with LOD95 variation within a 6-fold range. Each kit included either one of the E, N, or RdRP gene or all of them as a basic detection system, most of which were CE-IVD approved. Chan et al. (2020) reported the performance of an assay based on RT-PCR targeting nucleocapsid (N), RdRP/Helicase (Hel), and spike (S) of SARS-CoV-2 in comparison with RdRp-P2. They observed that COVID-19-RdRp/Hel had more sensitivity than the other two assays as well as RdRP-2 (P ≤ 0.001) in the clinical samples procured from the respiratory tract as well as non-respiratory tract samples. COVID-19-RdRp/Hel was able to detect 15.4% more samples as positive that tested negative with RdRp-P2 and no cross-reactivity was observed with other respiratory coronaviruses (Chan et al., 2020). However, RT-PCR-based detection is also associated with false-negative results, which might be due to the low viral load in patients' throats, improper handling of RNA samples, or lack of adequate internal controls (Di Paolo et al., 2020; Kelly et al., 2020). To improve this limitation of RT-PCR, droplet digital PCR was optimized for the detection of SARS-CoV-2 with a lower LOD than conventional RT-PCR (Suo et al., 2020).
Computed Tomography Scans
Chest CT scans have also been used for the diagnosis and management of COVID-19 (Pascarella et al., 2020; Udugama et al., 2020; Waller et al., 2020). Chest CT scanning for the detection of COVID-19 involves X-ray imaging of a patient's chest at different angles (Meng et al., 2020). Any anomalous features on the CT scan image, as per radiological reports, may be due to COVID-19 infestation. Commonly observed features on a chest scan of a patient with COVID-19 are ground-glass opacification (GGO), consolidations (increase in the opacity of the parenchyma, which results in shrouding of the underlying vessels), crazy-paving pattern (GGO with intralobular and interlobular septal thickening), and linear opacities (Ding et al., 2020; Huang et al., 2020a; Li and Xia, 2020). In a radiological assay by Song F. et al. (2020), the CT findings of 51 patients were analyzed. Nearly 86% of patients exhibited bilateral-type pneumonia affecting both lungs. Patients aged 50 or younger showed more GGO whereas elderly patients had organized pneumonia-like features with enhanced consolidations (Song F. et al., 2020). However, these features may vary in different stages of the disease and various age groups; hence, it is not suggested as a primary detection method for SARS-CoV-2 (Abbasi-Oshaghi et al., 2020). Pan et al. (2020) reported four stages of COVID pneumonia progression from 0 to 26 days, based on CT scans and the degree of lung involvement in patients with COVID-19. During days 0–4 days (stage 1), they observed a low CT score of 2 ± 2 with the appearance of GGO and a slight crazy-paving pattern. As the disease progressed, during days 5–8 days (stage 2), the CT score increased and a crazy-paving pattern was observed that further increased during the late stage at days 9–13 (stage 3) with the appearance of consolidation and a CT score of 7 ± 4. At the later stage of 14 days or more (stage 4), the CT score and crazy-paving pattern started to decrease, along with a reduction in the consolidations (Pan et al., 2020). Use of high-resolution CT could aid in the detection of GGOs in the early stages of infection (Wei et al., 2020). Current techniques used for the detection of SARS-CoV-2 are given in Table 1.
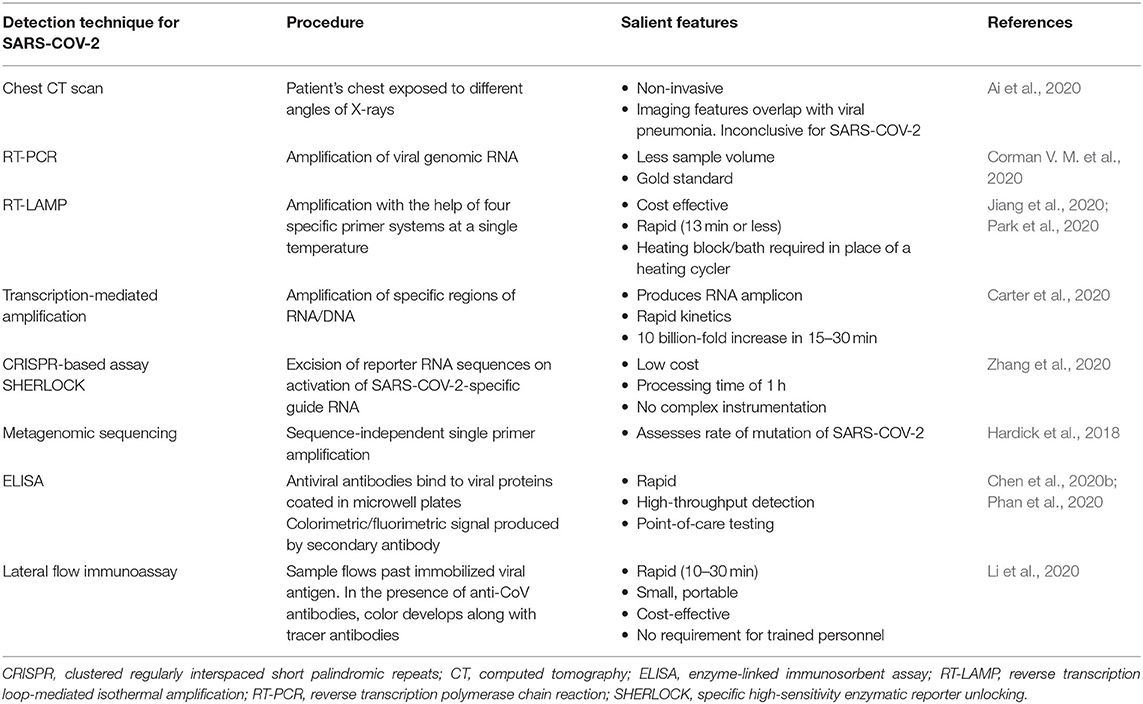
Table 1. Current techniques used for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection.
Other Detection Techniques
Other than RT-PCR and CT scans, various other detection systems have also been designed for the purpose of SARS-CoV-2 diagnosis. Zhang et al. (2020) have developed a technique named SHERLOCK to detect RNA fragments of SARS-CoV-2 with 10–100 copies/μl of the input. The basic principle of SHERLOCK-based diagnosis is CRISPR-based detection. This test can be performed in < 60 min, without requiring specific instruments. They chose two targets, the S gene and Orf1ab gene, from the SARS-CoV-2 genome. To minimize cross-reactivity with other respiratory virus genomes, they also selected specific guide sequences. Another group developed a rapid, low-cost, and accurate test, SARS-CoV-2 DETECTR (~30 min), which is based on a CRISPR/Cas12 lateral flow assay (LFA) (Broughton et al., 2020). An alternative for COVID-19 diagnosis is RT-LAMP. An optimized RT-LAMP-based detection technique has more sensitivity than conventional RT-PCR methods and requires less time. This technique can be utilized for the rapid diagnosis of coronavirus and increases the testing capacity by 2–2.5-fold (Jiang et al., 2020; Park et al., 2020). Chen et al. (2020b) developed a rapid and sensitive lateral flow immunoassay using nanoparticles prepared from lanthanide-doped polystyrene, known as LNPs, for the detection of anti-sSARS-Cov-2 immunoglobulin G (IgG) in serum samples. A specific N phosphoprotein of SARS-CoV-2 was used for the detection of specific antibodies in serum. The detection time for the assay was only 10 min (Chen et al., 2020b). Another study by Phan et al. (2020) utilized an in silico and serological approach to detect specific antibodies of SARS-CoV-2 in serum. In a recent development, the FDA has allowed the use of Illumina's NGS test for COVID-19 as an EUA. By using 98 DNA fragments, which cover nearly 30 kb in the genome of SARS-CoV-2, this technology can run ~3,000 test samples in a single 24 h turnaround. The minimum amount of sample required is 1,000 copies/mL, but the strategy is yet to receive approval from the FDA.
Emerging Tools and Technologies
The quick spread of the SARS-CoV-2 virus with an increased number of infected individuals worldwide makes it an absolute necessity to develop rapid and accurate detection technologies that will help to prevent illness and contain the disease (Green et al., 2019). The specificity and accuracy of RT-PCR-based detection of SARS-CoV-2 is not absolute owing to the genetic diversity and rapid evolution of the virus and technical skills (Ai et al., 2020). In this regard, sample collection, transport, and kit sensitivity are important hallmarks for the accuracy of the test. In developing countries, the problem is more pronounced because of minimal medical facilities and services. Moreover, the extended time required to obtain results makes it futile for public health emergencies such as the COVID-19 pandemic, in which rapid, easy, and accurate detection is required. Meanwhile, CT scan-based diagnosis has a low specificity, as other viral phenomena cannot be distinguished from SARS-CoV-2 by means of the scanned image. Therefore, there is an urgent need for the development of low-cost, efficient, and point-of-care (PoC) devices that are capable of providing fast and reliable detection results. The sensor platform should be simple enough to be used without the requirement for trained specialists for its operation.
Nanotechnology-Based Biosensors for COVID-19
A biosensor is a device that is used to diagnose analytes within liquids, solutions, and body fluids on combination with a biorecognition element and a physical transducer. When an analyte interacts with a biological element, a signal is produced that is converted into a measurable and quantifiable entity by the physical transducer. Physical transducers are mostly optical, electrochemical, and piezoelectric systems, whereas the biorecognition element includes nucleic acids such as DNA or RNA, proteins such as enzymes or antibodies, organic and biological receptors, whole cells, and tissues (Rocchitta et al., 2016). Large quantities of biosensor components can be manufactured according to requirements. Nanomaterials can be selected and used as transductors because of their highly stable nature in various media and their biocompatibility with physiological fluids. In addition, nanomaterials provide surface chemistry, which can be advantageous for the bioconjugation of molecules, as well as high surface energy and a strong amplification effect on signals. Because of their advantages, metallic nanoparticles such as gold and silver nanoparticles, nanomaterials based on carbon (nanotubes and graphene), photonic crystals, nanogels, and microgels are being used for biosensor applications. Nano-carbon-based particles have already been proven to provide a good platform for bacterial detection using various techniques such as electrochemistry and piezoelectric and also for the preparation of microfluidic-based kits (Sharma et al., 2020).
Developments in Nanotechnology-Based COVID-19 Detection
Point-of-Care Testing
PoC testing is defined as testing done using a kit or strip at home or at a place where the patient is being cared for. The most critical component of PoC testing is the biosensor, which is used to perform a biochemical assay to detect the pathogen (Vashist, 2017). The advantages of using PoC testing are: (i) minimal space requirement for testing and storage; (ii) wide-scale analysis; (iii) testing can be performed in a variety of locations; and (iv) flexible in meeting diverse medical needs.
PoC tests enable diagnosis without sending samples to specialized laboratories. An LFA for SARS-CoV-2 detection is one such PoC method that is under development for the diagnosis of COVID-19 (Xiang et al., 2020). An LFA conjugated with nanoparticles makes the device more reliable and sensitive. Lateral flow strips consist of a paper-like membrane, generally made up of nitrocellulose coated with two lines: the first line is the nanoparticle-conjugated antibody, which is known as the test line; the second is to capture antibodies, known as the control line. The sample is deposited on the membrane, also known as the sample pad, and analytes travel across the strip until they reach the first line by capillary action. As the analytes move further through the first test line, antigens in the analyte bind to the conjugate and this complex moves forward through the membrane. When it reaches the second line, it binds with the capture antibodies and a colored line appears. An LFA has shown a clinical sensitivity, specificity, and accuracy of 81, 100, and 86% for IgG and 57, 100, and 69% for immunoglobulin M (IgM), respectively (Xiang et al., 2020). This test detects IgM and IgG with a sensitivity of 82% (Xiang et al., 2020). Nucleic acid testing with DNA or RNA strands or aptamer, which is a single-stranded DNA (ssDNA) or RNA strand, can also be used for LFAs.
A gold nanoparticle-based lateral flow (AuNP-LF) assay for the rapid detection of the antibody IgM generated against SARS-CoV-2 was developed by Huang et al. (2020b). AuNP-LF strips were prepared using an analytical membrane coated with the SARS-CoV-2 N-protein for capturing the sample and an anti-human IgM–AuNP conjugate was used as the detector. AuNP-LF assay applicability was confirmed using blood serum samples from infected patients and uninfected healthy controls. The results obtained were compared with the gold standard RT-PCR method. The sensitivity and specificity of the AuNP-LF assay were found to be 100 and 93.3%, respectively (Huang et al., 2020b). The process of an LFA is explained schematically in Figure 3.
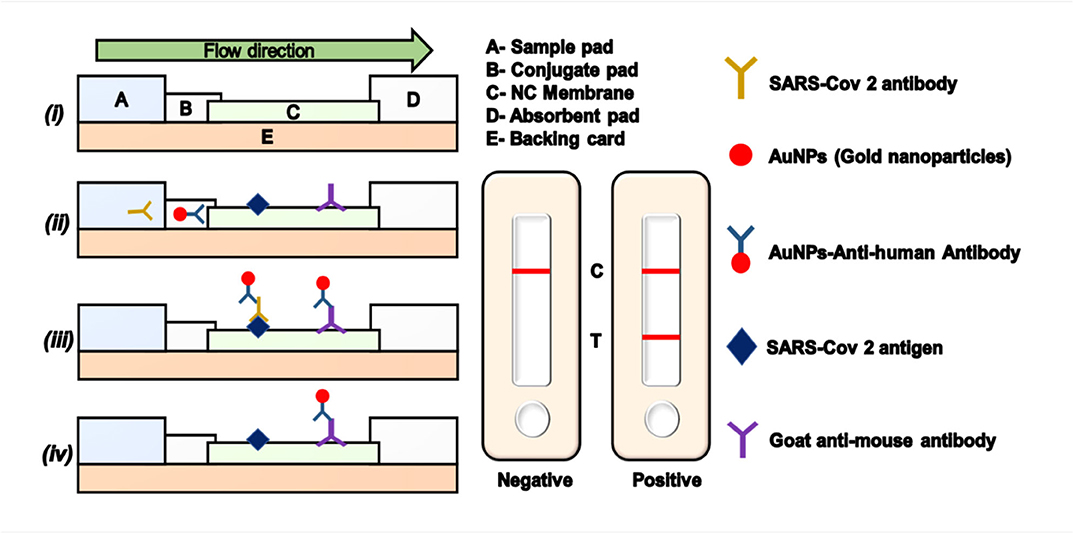
Figure 3. Lateral flow assay (LFA) using gold nanoparticles. (i) Model representation of an LFA strip. (ii) Conjugation of antigen and antibody on the LFA strip. (iii) Appearance of the test and control lines in a positive sample. (iv) Appearance of a single control line in a negative sample. NC, nitrocellulose; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The FDA has approved the BD Veritor System for SARS-CoV-2 rapid PoC detection under an EUA (FDA newsroom, 2020). This was the second antigen-based assay approved by the FDA, with the first being the Sofia 2 SARS Antigen FIA test (Shuren, 2020).
Colorimetric Assay
An easy, fast, and specific “naked-eye” colorimetric SARS-CoV-2 detection test is an urgent requirement at this time. Colorimetric assays exploit the optical properties of AuNPs, and specific antisense oligonucleotides (ASOs) for the N phosphoprotein of SARS-CoV-2 were modified by thiol for its conjugation. Detection was performed within 10 min after RNA isolation. When the target RNA sequence from SARS-CoV-2 was present, thiol-modified ASOs capped the AuNP (ASO) aggregate and a change in surface plasmon resonance was observed, as shown in Figure 4. On adding RNaseH, the RNA strand is cleaved from the RNA–DNA hybrid, which leads to the formation of a visually detectable precipitate facilitated by the agglomeration among AuNPs. The LOD using this method was 0.18 ng/μL of RNA having a SARS-CoV-2 viral load in the presence of MERS-CoV viral RNA (Moitra et al., 2020).
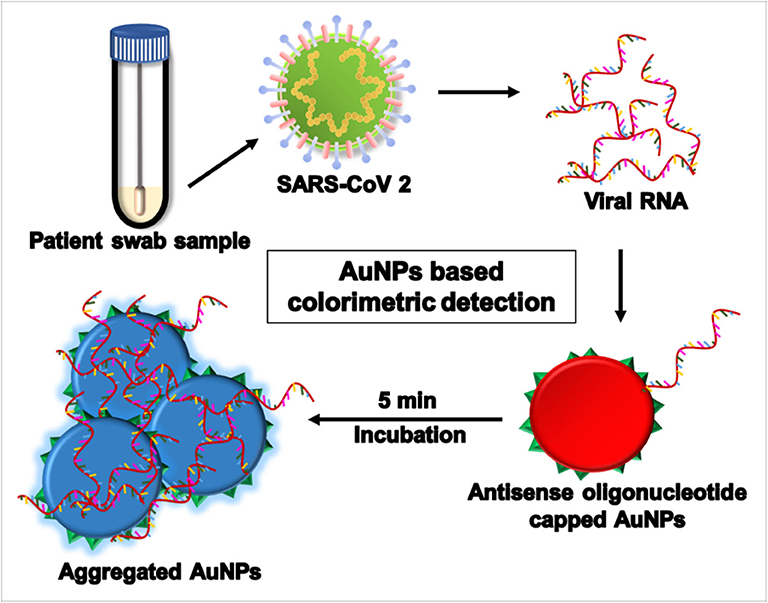
Figure 4. Aggregation assay using gold nanoparticles (AuNPs) for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection in a swab sample.
Microfluidic Devices
Microfluidic devices constitute another approach for use as a PoC test. These are palm-sized chips imprinted with micrometer-sized channels and reaction chambers made from a material such as poly-dimethylsulfoxide, paper, or glass, providing advantages such as miniaturized size and requiring only a small sample volume and a short detection time. The basic principle of these microfluidic chips is that they mix and segregate liquid samples using capillary action and electrokinetic properties (Foudeh et al., 2012). Microfluidic devices along with a smartphone application attachment can detect different antibodies against three sexually transmitted infections. In Rwanda, when tested in 96 patients, the platform showed specificity and sensitivity of 87 and 100%, respectively (Laksanasopin et al., 2015). Because of the ease and reliability of microfluidic devices, they can be modified to detect SARS-CoV-2 RNA or proteins. Different modes of detection using a microfluidic channel are shown in Figure 5.
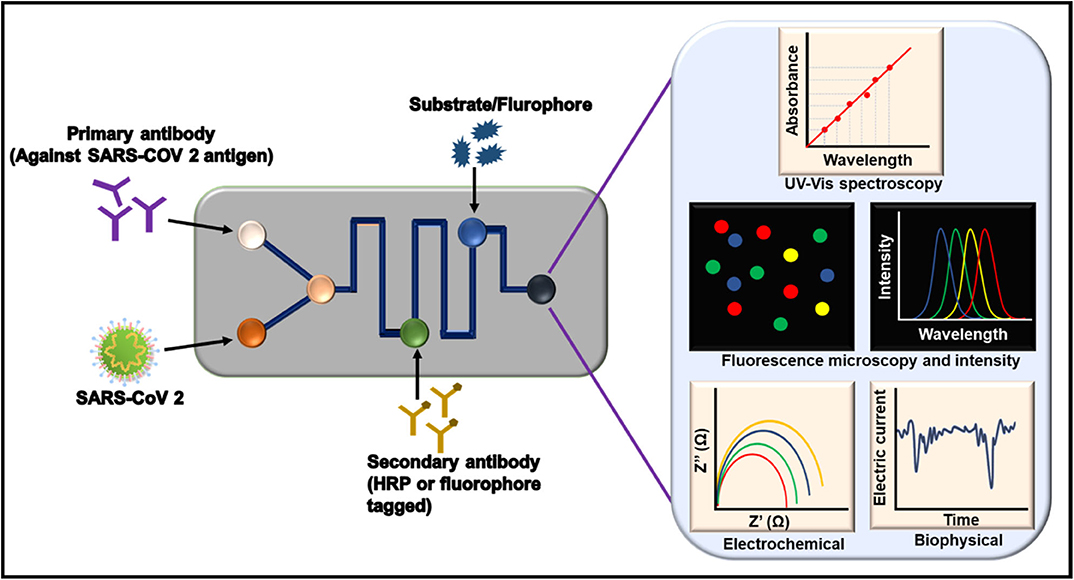
Figure 5. Potential microfluidics techniques for coronavirus detection using antibody against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). HRP, horseradish peroxidase; UV, ultraviolet; Vis, visible.
Magnetic Nanoparticle-Based Separation
Nucleic acid extraction from a clinical sample is the first step in the molecular diagnosis of SARS-CoV-2. This is labor intensive and time consuming. To extract viral RNA, magnetic nanoparticles coated with carboxyl polymer (pcMNPs) were developed to simplify the extraction for the sensitive detection of SARS-CoV-2 RNA (Zhao et al., 2020). Also, pcMNP–RNA complexes can be used for an RT-PCR protocol. pcMNPs have several advantages over a conventional column-based extraction method, such as showing good viral RNA binding performance. They also result in a 10-fold increase in sensitivity. Further, the pcMNP–RNA complexes obtained by the MNP separation method can be conveniently used with various isothermal amplification methods, such as LAMP. Thus, this method can be used for the development of PoC devices (Zhao et al., 2020).
Apart from the emerging technologies discussed above, Table 2 provides a few more nanotechnology-based techniques for detecting viruses. Multiple platforms such as paper-based systems, microfluidics, surface-enhanced Raman scattering, colorimetric systems, smartphone-based systems, and electrochemical sensors are being developed. Such approaches are in their early stages and are currently difficult to use for diagnosing COVID-19.
Detection Based on Biomolecules
A biosensor contains biorecognition elements, which are also known as biomolecules. They are used to detect the target depending upon the type of assay employed. Biomolecules can be antibodies that are traditionally used for ELISAs. Other than antibodies, enzymes and nucleic acids can be used. Nucleic acids are generally used in RT-PCR and LAMPS. Other than the existing biorecognition elements, nowadays, aptamers are gaining popularity. Different assays and detection kits are being developed using these biomolecules. Aptamer-based detection assays comprise optical, electrochemical, impedance, fluorescence, ELISA (also known as enzyme-linked aptamer assays), and other nanoparticle-conjugated assays (Zou et al., 2019). The properties and roles of these biomolecules are explained in the following sections. Moreover, common biomolecules and their roles in different techniques using nanoparticles are summarized in Figure 6.
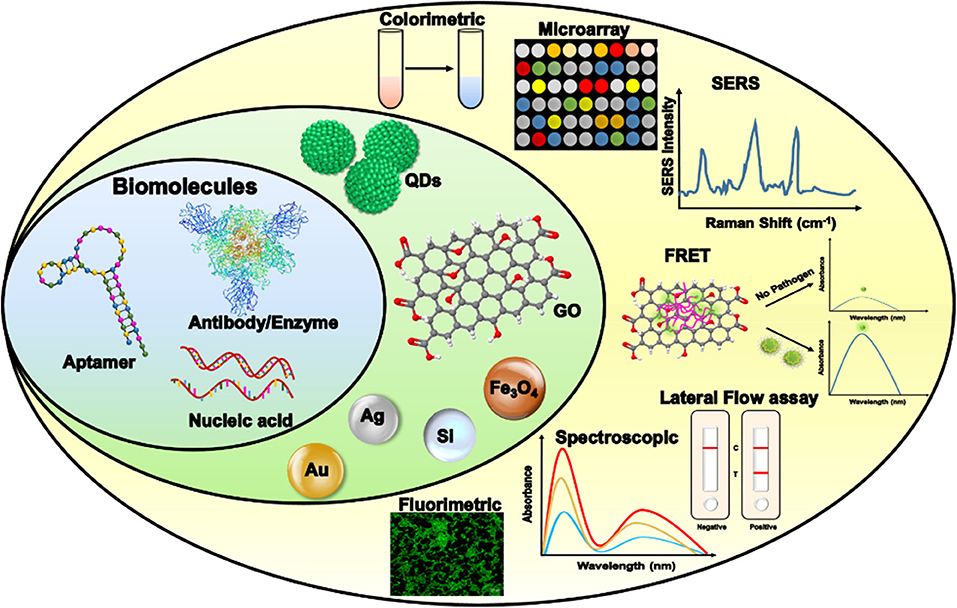
Figure 6. Application of biorecognition elements of a biosensor to develop a sensing platform against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). FRET, Förster resonance energy transfer; GO, graphene oxide; SERS, surface-enhanced Raman spectroscopy; QD, quantum dot.
Nucleic Acid-Based Biosensors
A double functional plasmonic biosensor was developed for the sensitive diagnosis of SARS-CoV-2 nucleic acid. This biosensor is a dual platform that combines the plasmonic photothermal (PPT) effect and localized surface plasmon resonance sensing transduction. The device consists of an integrated chip of two-dimensional gold nano-islands (AuNIs) on which a cDNA receptor for RdRp, ORF1ab, or the E gene have been immobilized by gold–thiol linkage. Detection of SARS-CoV-2 nucleic acid using this method utilizes the principle of nucleic acid hybridization with an LOD of 0.22 pM. The enhanced sensitivity of the device is the result of enhancement in the hybridization kinetics of the complementary strands, which is the result of the local PPT heat-generating capacity of AuNI plasmonic chips. The performance of this biosensor in clinical samples is yet to be analyzed (Qiu et al., 2020). Another assay used an integrated approach combining LAMP amplification, multiplex analysis, and reverse transcription with a nanoparticle-based lateral flow immunoassay biosensor (COVID-19 RT-LAMP-LFB) (Zhu X. et al., 2020). The LOD of this platform was observed to be 12 copies per reaction with no cross-reactivity. ORF1 antibody, two LAMP primer sets, and the N gene of SARS-CoV-2 were amplified simultaneously and detected with streptavidin-coated polymer nanoparticles.
Antibody-Based Biosensors
Spike protein of SARS-CoV-2 was reportedly detected using a novel antibody-based biosensor. Here, SARS-CoV-2 antibody was coated on graphene sheets of a field-effect transistor (FET) using nasopharyngeal swab samples from clinical patients as antigens. The LOD of the developed biosensor in a universal transport medium and phosphate-buffered saline was found to be 100 and 1 fg/mL, respectively. This sensor also detected the virus in culture medium with a detection limit of 1.6 × 101 pfu/mL. The COVID-19 FET sensor can differentiate between infected and healthy individuals with a detection limit of 2.42 × 102 copies/mL (Seo et al., 2020).
IgG and IgM antibodies were detected using an ELISA kit based on recombinant N- and S-protein that was developed and tested on patients. Among 214 patients, 146 (68.2%) and 150 (70.1%) were diagnosed with N-protein-based IgM and IgG ELISAs, respectively, whereas 165 (77.1%) and 159 (74.3%) were diagnosed using spike protein-based ELISAs. The positive rate for both kits was 82.2 and 80.4% for S- and N-protein, respectively (Liu et al., 2020).
Aptamer-Based Biosensors
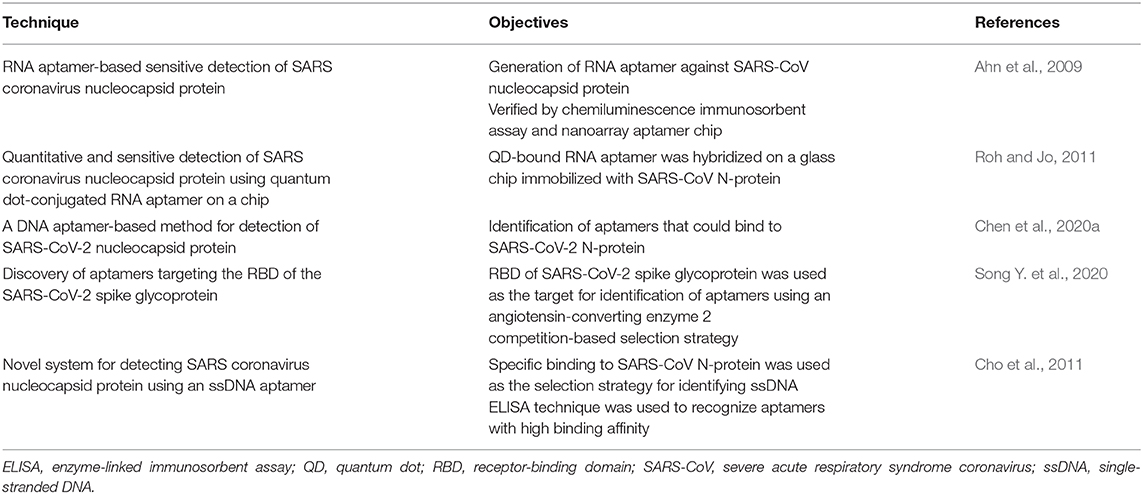
Recently, aptamers have gained popularity in virus diagnosis. These consist of ssRNA or ssDNA that binds conformationally and detects multiple targets with high sensitivity and specificity. Owing to a robust screening method, aptamers are able to detect viral genes, proteins, or any other viral infection markers. By modifying the designed assays, aptamer-based sensors can differentiate between infected and uninfected host cells or between active and inactive viral forms (Wandtke et al., 2015). Because of their properties, aptamer-based detection has considerable advantages over antibodies, including high stability at a vast range of temperatures and conditions, easy synthesis through a systematic evolution of ligands by exponential enrichment (SELEX) method, and easy modification according to the requirements of the assay. Here we describe some diagnostic approaches by aptamers. Very few research articles are published that describe the use of aptamers for detecting coronaviruses (Table 3). One study showed that N-protein of SARS-CoV can be detected in blood serum using an aptamer-based approach. The SELEX method was used to screen RNA aptamer 1 and RNA aptamer 2, which bind specifically to the C-terminal region of SARS-CoV N-protein with a low dissociation constant of 0.81 nM, and 3.35 nM, respectively. Using a short palindromic repeat technique, binding association was determined that confirmed that aptamer 1 has higher binding affinity than aptamer 2 (KD 1.65 nM for aptamer 1 vs. KD 2.1 μM for aptamer 2) and also aptamer binds with higher affinity than antibody. To detect N-protein, an ELISA was designed in which a streptavidin-coated plate was used for aptamer immobilization. After incubating N-protein, polyclonal antibody and a labeled monoclonal antibody were engaged to produce detectable signals. The LOD of this method was 20 pg/mL (420 fM) for N-protein (Ahn et al., 2009), which is similar to that of ELISA using both polyclonal and monoclonal antibodies (He et al., 2005). Further, a nanoarray chip was designed using an antibody, and the fluorescently labeled secondary antibody was proved to be 10 times more sensitive than the chemiluminescence assay, which had a lower detection limit of 42 fM (Ahn et al., 2009).
In another study, an ssDNA aptamer was isolated that specifically binds to N-protein of SARS-CoV with an efficient binding affinity and KD of 4.93 ± 0.30 nM (Cho et al., 2011). SARS-CoV-2 N-protein was immobilized on an ELISA plate, followed by the addition of modified aptamer with biotin at its 5′ terminal. The avidin labeled with horseradish peroxidase was added to produce a colored signal that was quantified using tetramethylbenzidine and measured by recording the absorbance. The above assay provided results that prove that all aptamers could specifically and sensitively detect SARS-CoV-2 N-protein (Chen et al., 2020a).
Recently, a novel aptamer against the RBD (anti-RBD-SARS-CoV-2) was screened using a SELEX and ACE-2 competition-based selection strategy. This development also utilized a machine learning screening algorithm. To measure the enrichment of interacted aptamers with SARS-CoV-2-RBD, histidine-tagged RBD-modified nickel beads were used for flow cytometry. Finally, two aptamers were selected with a high affinity for binding with KD values of 19.9 nM and 5.8 nM (Song Y. et al., 2020).
Other Detection Systems
Since the outbreak of COVID-19, efforts have been made to develop various detection systems that are portable, reliable, sensitive, rapid, and user friendly. One such option is the serology-based test (SBT). This is a blood-based method where the immune response to viral infection is monitored. This test measures antibodies and protein present in the blood of an infected person when the body reacts to SARS-CoV-2 antigens. A reliable SBT is highly recommended as the study of immune response provides significant and important details related to the prevalence of COVID-19. This information would help considerably in determining the immunity level attained within a community. In this regard, out of 33 kits approved by the FDA, one is a rapid SBT for the presence of the antibodies IgM and IgG in blood samples. However, SBTs are less sensitive (86.66%) than the highly sensitive qRT-PCR and LAMP methods (Nguyen et al., 2020).
Conclusion and Future Prospects
The containment of epidemics requires widespread communication, surveillance, data sharing, monitoring of patients, and availability of resources (Smith, 2006). To achieve this purpose, smartphones have been considered very useful because of their properties such as computational power, hardware to improve electronic reporting, PoC testing, preparation of epidemiological databases (Wood et al., 2019), and most importantly connectivity. Pre-existing diagnostic tests can be linked with smartphones, providing real-time information that helps global agencies to incorporate and implement control strategies. Smartphones not only provide risk-free direct contact between patients and clinicians but also sharing of field data with public health databases to control and monitor outbreaks becomes possible and easy. Cameras, flashlights, audio jacks, and other smartphone components can be used in surveillance along with the statistics and databasing of diagnostic experiments (Malekjahani et al., 2019). Use of smartphones with forward-looking infrared radar for thermal scanning and body temperature monitoring due to inflammation for diagnosing fever as a common coronavirus symptom has been developed (Kanazawa et al., 2016).
The availability of modern techniques has contributed toward diagnosing SARS-CoV-2 with high accuracy. These techniques have further increased the daily number of tests globally, providing information related to the severity of the infection. RT-PCR is currently the most widely used technique but it takes 4–6 h to get results. To simplify and increase testing, the use of nanotechnology in PoC testing and microfluidics should be thoroughly used. These technologies should be further developed so that they can be rapidly implemented during unforeseen medical emergencies.
Author Contributions
RG and PS work equally in drafting and writing the whole review. NP was instrumental in making the figures. All the table framing and extra documents file was drafted by SK. RS help in drafting the content and correction of the review. VR and NS were responsible for idea, writing the review, and designing the figure. NS finally corrected and communicated the review. All authors contributed to the article and approved the submitted version.
Funding
The authors would like to acknowledge the Department of Biotechnology, India, grant number BT/PR10353/PFN/20/889/2013, the National Agri-Food Biotechnology Institute, Mohali, India, and the Indian Council of Medical Research, India, for providing financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbasi-Oshaghi, E., Mirzaei, F., Farahani, F., Khodadadi, I., and Tayebinia, H. (2020). Diagnosis and treatment of coronavirus disease 2019 (COVID-19): laboratory, PCR, and chest CT imaging findings. Int. J. Surg. 79, 143–153. doi: 10.1016/j.ijsu.2020.05.018
Ahmed, S. R., Kang, S. W., Oh, S., Lee, J., and Neethirajan, S. (2018). Chiral zirconium quantum dots: a new class of nanocrystals for optical detection of coronavirus. Heliyon 4:e00962. doi: 10.1016/j.heliyon.2018.e00766
Ahn, D. G., Jeon, I. J., Kim, J. D., Song, M. S., Han, S. R., Lee, S. W., et al. (2009). RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 134, 1896–1901. doi: 10.1039/b906788d
Ai, T., Yang, Z., Hou, H., Zhan, C., Chen, C., Lv, W., et al. (2020). Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296:E32–E40. doi: 10.1148/radiol.2020200642
Arabi, Y. M., Hajeer, A. H., Luke, T., Raviprakash, K., Balkhy, H., Johani, S., et al. (2016). Feasibility of using convalescent plasma immunotherapy for MERS-CoV infection, Saudi Arabia. Emerg. Infect. Dis. 22, 1554–1561. doi: 10.3201/eid2209.151164
Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D. Y., Chen, L., et al. (2020). Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 323, 1406–1407. doi: 10.1001/jama.2020.2565
Best (2020). COVID-19 death rates worldwide as of September 4, 2020, by country. from Retrieved from: https://www.statista.com/statistics/1105914/coronavirus-death-rates-worldwide/ (Retrieved September 7, 2020).
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Singh, J., Streithorst, J., et al. (2020). Rapid detection of 2019 novel coronavirus SARSCoV-2 using a CRISPR-based DETECTR lateral flow assay. MedRxiv. 2020.03.06.20032334. doi: 10.1101/2020.03.06.20032334
Carter, L. J., Garner, L. V., Smoot, J. W., Li, Y., Zhou, Q., Saveson, C. J., et al. (2020). Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 6, 591–605. doi: 10.1021/acscentsci.0c00501
Chan, J. F. W., Yip, C. C. Y., To, K. K. W., Tang, T. H. C., Wong, S. C. Y., Leung, K. H., et al. (2020). Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 58:e00310-20. doi: 10.1128/JCM.00
Chang, D., Lin, M., Wei, L., Xie, L., Zhu, G., Dela Cruz, C. S., et al. (2020). Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA 323, 1092–1093. doi: 10.1001/jama.2020.1623
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513. doi: 10.1016/S0140-6736(20)30211-7
Chen, Z., Wu, Q., Chen, J., Ni, X., and Dai, J. (2020a). A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 35, 351–354. doi: 10.1007/s12250-020-00236-z
Chen, Z., Zhang, Z., Zhai, X., Li, Y., Lin, L., Zhao, H., Bian, L., et al. (2020b). Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 92, 7226–7231. doi: 10.1021/acs.analchem.0c00784
Cho, S. J., Woo, H. M., Kim, K. S., Oh, J. W., and Jeong, Y. J. (2011). Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 112, 535–540. doi: 10.1016/j.jbiosc.2011.08.014
Corman, V., Bleicker, T., Brünink, S., Drosten, C., Landt, O., Koopmans, M., et al. (2020). Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RTPCR Wuhan-Virus-Assay-V1991527E5122341D99287a1B17C111902.
Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K. W., et al. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
Dhama, K., Patel, S. K., Pathak, M., Yatoo, M. I., Tiwari, R., Malik, Y. S., et al. (2020). An update on SARS-CoV-2/COVID-19 with particular reference to its clinical pathology, pathogenesis, immunopathology and mitigation strategies. Travel Med. Infect. Dis. 37:101755. doi: 10.1016/j.tmaid.2020.101755
Dharmendra, K. M., and Rishabha, K. S. P. (2020). Corona virus: a review of COVID-19. Eurasian J. Med. Oncology 4, 8–25. doi: 10.14744/ejmo.2020.51418
Di Paolo, M., Iacovelli, A., Olmati, F., Menichini, I., Oliva, A., Carnevalini, M., et al. (2020). False-negative RT-PCR in SARS-CoV-2 disease: experience from an Italian COVID-19 unit. ERJ Open Res. 6, 324–2020. doi: 10.1183/23120541.00324-2020
Ding, X., Xu, J., Zhou, J., and Long, Q. (2020). Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur. J. Radiol. 127:109009. doi: 10.1016/j.ejrad.2020.109009
FDA newsroom (2020). Coronavirus (COVID-19) Update : FDA Issued Emergency Use Authorization for Point of Care Antigen Test Inquiries. 1–2. Retrieved from: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issued-emergency-use-authorization-point-care-antigen-test
Forster, P., Forster, L., Renfrew, C., and Forster, M. (2020). Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U.S.A. 117, 9241–9243. doi: 10.1073/pnas.2004999117
Foudeh, A. M., Fatanat Didar, T., Veres, T., and Tabrizian, M. (2012). Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 12, 3249–3266. doi: 10.1039/c2lc40630f
Gao, Z. C. (2020). [Efficient management of novel coronavirus pneumonia by efficient prevention and control in scientific manner]. Zhonghua Jie He He Hu Xi Za Zhi 43, 163–166. doi: 10.3760/cma.issn.1001-0939.2020.03.002
Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., et al. (2020). Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. Nat. Microbiol. 5, 536–544. doi: 10.1038/s41564-020-0695-z
Green, K., Winter, A., Dickinson, R., Graziadio, S., Wolff, R., Mallett, S., et al. (2019). What Tests Could Potentially be Used for the Screening, Diagnosis and Monitoring of COVID-19 and What are Their Advantages and Disadvantages? The Centre for Evidence Based Medicine. Retrieved from: https://www.cebm.net/covid-19/what-tests-could-potentially-be-used-for-the-screening-diagnosis-and-monitoring-of-covid-19-and-what-are-their-advantages-and-disadvantages/
Hamming, I., Timens, W., Bulthuis, M. L. C., Lely, A. T., Navis, G. J., and van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637. doi: 10.1002/path.1570
Harapan, H., Itoh, N., Yufika, A., Winardi, W., Keam, S., Te, H., et al. (2020). Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health 13, 667–673. doi: 10.1016/j.jiph.2020.03.019
Hardick, J., Metzgar, D., Risen, L., Myers, C., Balansay, M., Malcom, T., et al. (2018). Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagn. Microbiol. Infect. Dis. 91, 245–247. doi: 10.1016/j.diagmicrobio.2018.02.011
He, Q., Du, Q., Lau, S., Manopo, I., Lu, L., Chan, S. W., et al. (2005). Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods 127, 46–53. doi: 10.1016/j.jviromet.2005.03.004
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., et al. (2020a). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5
Huang, C., Wen, T., Shi, F. J., Zeng, X. Y., and Jiao, Y. J. (2020b). Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 5, 12550–12556. doi: 10.1021/acsomega.0c01554
Jiang, M., Pan, W., Arasthfer, A., Fang, W., Ling, L., Fang, H., et al. (2020). Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front. Cell. Infect. Microbiol. 10:331. doi: 10.3389/fcimb.2020.00331
Kahn, J. S., and McIntosh, K. (2005). History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 24:60. doi: 10.1097/01.inf.0000188166.17324.60
Kanazawa, T., Nakagami, G., Goto, T., Noguchi, H., Oe, M., Miyagaki, T., et al. (2016). Use of smartphone attached mobile thermography assessing subclinical inflammation: a pilot study. J. Wound Care 25, 177–180, 182. doi: 10.12968/jowc.2016.25.4.177
Kelly, J. C., Dombrowksi, M., O'Neil-Callahan, M., Kernberg, A. S., Frolova, A. I., and Stout, M. J. (2020). False-negative testing for severe acute respiratory syndrome coronavirus 2: consideration in obstetrical care. Am. J. Obstet. Gynecol. 2(Suppl. 3):100130. doi: 10.1016/j.ajogmf.2020.100130
Kirchdoerfer, R. N., Cottrell, C. A., Wang, N., Pallesen, J., Yassine, H. M., Turner, H. L., et al. (2016). Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121. doi: 10.1038/nature17200
Laksanasopin, T., Guo, T. W., Nayak, S., Sridhara, A. A., Xie, S., Olowookere, O. O., et al. (2015). A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 7:273re1. doi: 10.1126/scitranslmed.aaa0056
Lee, P. I., Hu, Y. L., Chen, P. Y., Huang, Y. C., and Hsueh, P. R. (2020). Are children less susceptible to COVID-19? J. Microbiol. Immunol. Infect. 53, 371–372. doi: 10.1016/j.jmii.2020.02.011
Letko, M., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for lineage B b-coronaviruses, including 2019-nCoV. BioRxiv. 2020.01.22.915660. doi: 10.1101/2020.01.22.915660
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi: 10.1038/nature02145
Li, Y., and Xia, L. (2020). Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 214, 1280–1286. doi: 10.2214/AJR.20.22954
Li, Z., Yi, Y., Luo, X., Xiong, N., Liu, Y., Li, S., et al. (2020). Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 92, 1518–1524. doi: 10.1002/jmv.25727
Lipsitch, M., Cohen, T., Cooper, B., Robins, J. M., Ma, S., James, L., et al. (2003). Transmission dynamics and control of severe acute respiratory syndrome. Science 300, 1966–1970. doi: 10.1126/science.1086616
Liu, W., Liu, L., Kou, G., Zheng, Y., Ding, Y., Ni, W., et al. (2020). Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 58:e00461–20. doi: 10.1128/JCM.00461-20
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Mair-Jenkins, J., Saavedra-Campos, M., Baillie, J. K., Cleary, P., Khaw, F. M., Lim, W. S., et al. (2015). The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 211, 80–90. doi: 10.1093/infdis/jiu396
Majumder, M. S., and Mandl, K. D. (2020). Early transmissibility assessment of a novel coronavirus in Wuhan, China. SSRN. 3524675. doi: 10.2139/ssrn.3524675
Majumder, M. S., Rivers, C., Lofgren, E., and Fisman, D. (2014). Estimation of MERS-coronavirus reproductive number and case fatality rate for the spring 2014 Saudi Arabia outbreak: insights from publicly available data. PLoS Curr. 6:ecurrents.outbreaks.98d2f8f3382d84f390736cd5f5fe133c. doi: 10.1371/currents.outbreaks.98d2f8f3382d84f390736cd5f5fe133c
Malekjahani, A., Sindhwani, S., Syed, A. M., and Chan, W. C. W. (2019). Engineering steps for mobile point-of-care diagnostic devices. Acc. Chem. Res. 52, 2406–2414. doi: 10.1021/acs.accounts.9b00200
Meng, H., Xiong, R., He, R., Lin, W., Hao, B., Zhang, L., et al. (2020). CT imaging and clinical course of asymptomatic cases with COVID-19 pneumonia at admission in Wuhan, China. J. Infect. 81, e33–e39. doi: 10.1016/j.jinf.2020.04.004
Moitra, P., Alafeef, M., Dighe, K., Frieman, M. B., and Pan, D. (2020). Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 14, 7617–7627. doi: 10.1021/acsnano.0c03822
Munster, V. J., Koopmans, M., van Doremalen, N., van Riel, D., and de Wit, E. (2020). A novel coronavirus emerging in China — key questions for impact assessment. N. Engl. J. Med. 382, 692–694. doi: 10.1056/NEJMp2000929
Mustafa, S., Balkhy, H., and Gabere, M. N. (2018). Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J. Infect. Public Health 11, 9–17. doi: 10.1016/j.jiph.2017.08.009
Nguyen, T., Duong Bang, D., and Wolff, A. (2020). 2019 novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines 11:306. doi: 10.3390/mi11030306
Pan, F., Ye, T., Sun, P., Gui, S., Liang, B., Li, L., et al. (2020). Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295, 715–721. doi: 10.1148/radiol.2020200370
Park, G. S., Ku, K., Baek, S. H., Kim, S. J., Kim, S., Il, K.im, B. T., et al. (2020). Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 22, 729–735. doi: 10.1016/j.jmoldx.2020.03.006
Pascarella, G., Strumia, A., Piliego, C., Bruno, F., Del Buono, R., Costa, F., et al. (2020). COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med. 288, 192–206. doi: 10.1111/joim.13091
Phan, I. Q., Subramanian, S., Kim, D., Carter, L., King, N., Anishchenko, I., et al. (2020). In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19. BioRxiv. 2020.05.22.111526. doi: 10.1101/2020.05.22.111526
Qiu, G., Gai, Z., Tao, Y., Schmitt, J., Kullak-Ublick, G. A., and Wang, J. (2020). Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 14, 5268–5277. doi: 10.1021/acsnano.0c02439
Rocchitta, G., Spanu, A., Babudieri, S., Latte, G., Madeddu, G., Galleri, G., et al. (2016). Enzyme biosensors for biomedical applications: strategies for safeguarding analytical performances in biological fluids. Sensors 16:780. doi: 10.3390/s16060780
Rodriguez-Morales, A. J., Bonilla-Aldana, D. K., Balbin-Ramon, G. J., Rabaan, A. A., Sah, R., Paniz-Mondolfi, A., et al. (2020). History is repeating itself: probable zoonotic spillover as the cause of the 2019 novel coronavirus epidemic. Infez. Med. 28, 3–5.
Roh, C., and Jo, S. K. (2011). Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 86, 1475–1479. doi: 10.1002/jctb.2721
Seo, G., Lee, G., Kim, M. J., Baek, S. H., Choi, M., Ku, K. B., et al. (2020). Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal Swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142. doi: 10.1021/acsnano.0c02823
Sharma, A., Sharma, N., Kumari, A., Lee, H. J., Kim, T., and Tripathi, K. M. (2020). Nano-carbon based sensors for bacterial detection and discrimination in clinical diagnosis: a junction between material science and biology. Appl. Mater. Today 18:100467. doi: 10.1016/j.apmt.2019.100467
Shen, K. L., and Yang, Y. H. (2020). Diagnosis and treatment of 2019 novel coronavirus infection in children: a pressing issue. World J. Pediatr. 16, 219–221. doi: 10.1007/s12519-020-00344-6
Shuren, J. E. (2020). Coronavirus (COVID-19) Update: FDA Authorizes First Antigen Test to Help in the Rapid Detection of the Virus that Causes COVID-19 in Patients | FDA. Commissioner of Food and Drugs - Food and Drug Administration Stephen M. Hahn M.D. 10–11.
Smith, R. D. (2006). Responding to global infectious disease outbreaks: lessons from SARS on the role of risk perception, communication and management. Soc. Sci. Med. 63, 3113–3123. doi: 10.1016/j.socscimed.2006.08.004
Song, F., Shi, N., Shan, F., Zhang, Z., Shen, J., Lu, H., et al. (2020). Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 295, 210–217. doi: 10.1148/radiol.2020200274
Song, Y., Song, J., Wei, X., Huang, M., Sun, M., Zhu, L., et al. (2020). Discovery of aptamers targeting receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 92, 9895–9900. doi: 10.1021/acs.analchem.0c01394
Suo, T., Liu, X., Feng, J., Guo, M., Hu, W., Guo, D., et al. (2020). ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. MedRxiv. doi: 10.1101/2020.02.29.20029439
Turner, A. J., Tipnis, S. R., Guy, J. L., Rice, G., and Hooper, N. M. (2002). ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can. J. Physiol. Pharmacol. 80, 346−353. doi: 10.1139/y02-021
Udugama, B., Kadhiresan, P., Kozlowski, H. N., Malekjahani, A., Osborne, M., Li, V. Y. C., et al. (2020). Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14, 3822–3835. doi: 10.1021/acsnano.0c02624
van Kasteren, P. B., van der Veer, B., van den Brink, S., Wijsman, L., de Jonge, J., van den Brandt, A., et al. (2020). Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 128:104412. doi: 10.1016/j.jcv.2020.104412
Vashist, S. K. (2017). Point-of-Care diagnostics: recent advances and trends. Biosensors 7:62. doi: 10.3390/bios7040062
Velavan, T. P., and Meyer, C. G. (2020). The COVID-19 epidemic. Trop. Med. Int. Health 25, 278–280. doi: 10.1111/tmi.13383
Waller, J. V., Kaur, P., Tucker, A., Lin, K. K., Diaz, M. J., Henry, T. S., et al. (2020). Diagnostic tools for coronavirus disease (COVID-19): comparing CT and RT-PCR viral nucleic acid testing. Am. J. Roentgenol. 215, 834–838. doi: 10.2214/AJR.20.23418
Wandtke, T., Wozniak, J., and Kopiński, P. (2015). Aptamers in diagnostics and treatment of viral infections. Viruses 7, 751–780. doi: 10.3390/v7020751
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323, 1061–1069. doi: 10.1001/jama.2020.1585
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e9. doi: 10.1016/j.cell.2020.03.045
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., et al. (2020). Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844. doi: 10.1001/jama.2020.3786
Wei, J., Xu, H., Xiong, J., Shen, Q., Fan, B., Ye, C., et al. (2020). 2019 Novel coronavirus (COVID-19) pneumonia: serial computed tomography findings. Korean J. Radiol. 21, 501–504. doi: 10.3348/kjr.2020.0112
Wen, T., Huang, C., Shi, F. J., Zeng, X. Y., Lu, T., Ding, S. N., et al. (2020). Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst. 145, 5345–5352. doi: 10.1039/D0AN00629G
WHO (2020a). Coronavirus Disease 2019 Situation Report 51 11th March 2020. Geneva: World Health Organization, 2633. doi: 10.1001/jama.2020.2633
WHO (2020b). Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March 2020. Geneva: World Health Organization. Retrieved from: https://apps.who.int/iris/handle/10665/331329
Wood, C. S., Thomas, M. R., Budd, J., Mashamba-Thompson, T. P., Herbst, K., Pillay, D., et al. (2019). Taking connected mobile-health diagnostics of infectious diseases to the field. Nature 566, 467–474. doi: 10.1038/s41586-019-0956-2
Wu, J. T., Leung, K., and Leung, G. M. (2020). Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 395, 689–697. doi: 10.1016/S0140-6736(20)30260-9
Xiang, J., Yan, M., Li, H., Liu, T., Lin, C., Huang, S., et al. (2020). Evaluation of enzyme-linked immunoassay and colloidal gold- immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). MedRxiv. 2020.02.27.20028787. doi: 10.1101/2020.02.27.20028787
Xiao, S. Y., Wu, Y., and Liu, H. (2020). Evolving status of the 2019 novel coronavirus infection: Proposal of conventional serologic assays for disease diagnosis and infection monitoring. J. Med. Virol. 92, 464–467. doi: 10.1002/jmv.25702
Xie, C., Jiang, L., Huang, G., Pu, H., Gong, B., Lin, H., et al. (2020). Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 93, 264–267. doi: 10.1016/j.ijid.2020.02.050
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Yu, F., Du, L., Ojcius, D. M., Pan, C., and Jiang, S. (2020). Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 22, 74–79. doi: 10.1016/j.micinf.2020.01.003
Zhang, F., Abudayyeh, O. O., Gootenberg, J. S., Sciences, C., and Mathers, L. (2020). A Protocol for Detection of COVID-19 Using CRISPR Diagnostics.
Zhao, Z., Cui, H., Song, W., Ru, X., Zhou, W., and Yu, X. (2020). A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. BioRxiv. doi: 10.1101/2020.02.22.961268
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Zhu, X., Wang, X., Han, L., Chen, T., Wang, L., Li, H., et al. (2020). Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosensors Bioelectron. 166:112437. doi: 10.1016/j.bios.2020.112437
Keywords: pandemic, diagnosis, SARS-CoV-2, COVID-19, RT-PCR, nanotechnology, point of care, aptamer
Citation: Gupta R, Sagar P, Priyadarshi N, Kaul S, Sandhir R, Rishi V and Singhal NK (2020) Nanotechnology-Based Approaches for the Detection of SARS-CoV-2. Front. Nanotechnol. 2:589832. doi: 10.3389/fnano.2020.589832
Received: 31 July 2020; Accepted: 10 September 2020;
Published: 18 December 2020.
Edited by:
Ajeet Kaushik, Florida Polytechnic University, United StatesReviewed by:
Kumud Malika Tripathi, Indian Institute of Petroleum and Energy (IIPE), IndiaSneham Tiwari, Florida International University, United States
Copyright © 2020 Gupta, Sagar, Priyadarshi, Kaul, Sandhir, Rishi and Singhal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nitin Kumar Singhal, bml0aW5AbmFiaS5yZXMuaW4=; Vikas Rishi, dmlrYXNyaXNoaUBuYWJpLnJlcy5pbg==
†These authors have contributed equally to this work
 Ritika Gupta
Ritika Gupta Poonam Sagar
Poonam Sagar Nitesh Priyadarshi
Nitesh Priyadarshi Sunaina Kaul
Sunaina Kaul Rajat Sandhir
Rajat Sandhir Vikas Rishi
Vikas Rishi Nitin Kumar Singhal
Nitin Kumar Singhal