
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Nanotechnol., 27 November 2020
Sec. Biomedical Nanotechnology
Volume 2 - 2020 | https://doi.org/10.3389/fnano.2020.574328
This article is part of the Research TopicThe DNA Molecule as an object of Nanotechnology and the Creation of Helical-Structured Metamaterials and MetasurfacesView all 5 articles
Nanostructures specifically responsive to endogenous biomolecules hold great potential in accurate diagnosis and precision therapy of cancers. In the pool of nanostructures with responsiveness to unique triggers, nanomaterials derived from DNA self-assembly have drawn particular attention due to their intrinsic biocompatibility and structural programmability, enabling the selective bioimaging, and site-specific drug delivery in cancer cells and tumor tissues. In this mini review, we summarize the most recent advances in the development of endogenous stimuli-responsive DNA nanostructures featured with precise self-assembly, targeted delivery, and controlled drug release for cancer theranostics. This mini review briefly discusses the diverse dynamic DNA nanostructures aiming at bioimaging and biomedicine, including DNA self-assembling materials, DNA origami structures, DNA hydrogels, etc. We then elaborate the working principles of DNA nanostructures activated by biomarkers (e.g., miRNA, mRNA, and proteins) in tumor cells and microenvironments of tumor tissue (e.g., pH, ATP, and redox gradient). Subsequently, applications of the endogenous stimuli-responsive DNA nanostructures in biological imaging probes for detecting cancer hallmarks as well as intelligent carriers for drug release in vivo are discussed. In the end, we highlight the current challenges of DNA nanotechnology and the further development of this promising research direction.
DNA nanotechnology has made brilliant accomplishments in chemistry, materials, and other fields. In the 1980s, Ned Seeman proposed the concept of DNA nanotechnology by introducing the principle of complementary pairing of DNA bases into the construction of nanostructures (Seeman, 1982; Kallenbach et al., 1983). Later on, the introduction of DNA origami (Rothemund, 2006) heralded the beginning of a new era in DNA nanotechnology, further increasing the size and complexity of self-assembled DNA building blocks. DNA technology allows specific recognition structures to induce the transformation and deconstruction of DNA nanostructures in the presence of triggers (Yu et al., 2019; Kong et al., 2020) to generate sensor signals or effectors. With the structure switch and signal activation, nano-constructs such as DNA sensors and DNA machinery have been elaborately devised, holding enormous potential in biological imaging and biomedical research (Liu S. et al., 2019).
As a global health problem, accurate diagnosis and efficient treatment of cancer is the current research hotspot. Chemotherapy is an important means of cancer treatment, but there are some problems like low drug utilization, less accumulation at the tumor site, and serious toxic and side effects on normal tissues. Therefore, the advances of nanotechnology provide new opportunities for molecular diagnosis and therapy of cancer. Compared with external stimuli, such as light, ultrasound, and magnetic field, that enable remote modulation of nanostructures, endogenous stimuli including pH, ATP, redox, proteins, and RNAs possess closer ties with microenvironment in biological systems (Hu et al., 2020). Thus, the design of endogenous stimuli-responsive nanomaterials activated by tumor biomarkers and microenvironmental features is significant for cancer diagnosis and treatment. In this minireview, we will discuss the latest developments of DNA nanostructures responsive to endogenous stimuli in recent years, and elaborate on their design and working principles (Figure 1). The detection of cancer hallmarks in biological imaging probes and the application of intelligent carriers to release drugs in vivo will also be summarized. In the end, more challenges and further advances of this research direction will be prospected.
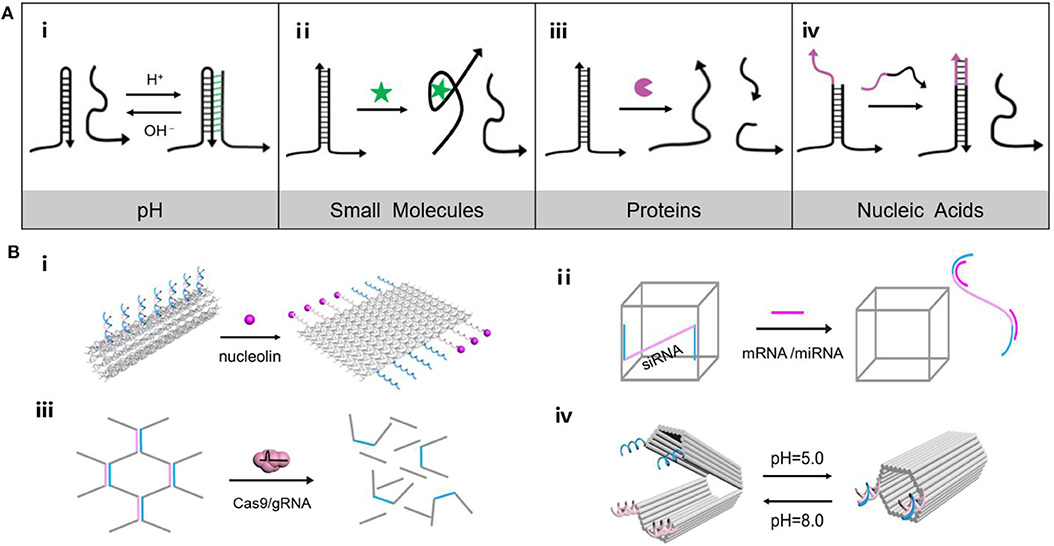
Figure 1. (A) Versatility of endogenous stimulus response principle of DNA nanomaterials. (i) Triplex/duplex DNAs transform along with pH changes, (ii) Aptamer structure combined with small molecules, (iii) Enzymatic cleavage of DNA, (iv) DNA strand displacement reaction. (B) (i) Tube nanocarrier opens in response to the presence of nucleolin, (ii) Oligonucleotides triggered DNA “Nanosuitcases” for siRNA releasing, (iii) DNA hydrogel degrades under the precise excision of Cas9/sgRNA, (iv) A DNA origami nanocapsule can be opened and closed with the help of pH-responsive latches.
pH anomaly is considered a common feature of cancer cells. As the energy acquisition mode of tumor cells are largely dependent on glycolysis, the accumulation of glycolytic metabolites such as lactic acid is a major factor in the change of a tumor's microenvironment (Boedtkjer and Pedersen, 2020; Wang C. C. et al., 2020). Coincidentally, DNA nanostructures such as i-motif structure and triplex/duplex DNAs could transform along with pH changes, which show great potential in construction of stimuli response nanomaterials (Dong et al., 2014; Hu et al., 2017). Gehring et al. (1993) firstly proposed the i-motif structure, in which cytosine-cytosine base pairs are protonated to obtain a tetrameric DNA structure in acidic conditions, and switch into DNA single strands under neutral environments. Following the emergence of the i-motif structure, triplex/duplex DNAs formed by C-G·C+ and T-A·T triplets of Watson Crick (“–”), and Hoogsteen hydrogen bonds (“·”) was found to show responsiveness to pH (Figure 1Ai). Under acidic conditions, the protonated cytosine (C+) tends to bind with the guanine (G) following the Hoogsteen hydrogen bond, while the central G is paired with another C according to the Watson Crick model to form the stable triplex C-G·C+. In neutral conditions, C+ loses a proton to become C, resulting in dissociation of the triplet C-G·C+ into two or single chain. Since then, a number of pH responsive nanodevices were designed. For example, DNA triplex was introduced to build a pH-triggered self-assembly DNA tetrahedron (Liu et al., 2013). The DNA components could obtain triplet C-G·C+, which configured DNA tetrahedron at pH 5.0 and dissociated at pH 8.0, achieving reversible assembling of DNA nanostructure. By employing the foldable i-motifs in key parts of DNA nanodevices, DNA machines were developed including DNA tweezers (Wang et al., 2010a), DNA walkers (Wang et al., 2010b), and DNA pendulums (Qi et al., 2013), which were deformable in response to pH.
Small molecules of biological functions play an essential role in physiological processes such as cell growth, cell proliferation and differentiation. For example, due to the accelerated metabolism, glucose and glutamine are used by tumor cells to produce ATP to promote cell growth. The concentration of ATP is lower in the extracellular environment and higher in the intracellular cytosol (Leist et al., 1997; Gorman et al., 2007). Therefore, the prominent difference in ATP levels is used to design ATP-responsive systems. Since the ATP-binding aptamer can recognize ATP with high binding affinity, the ligand-aptamer complex is able to serve as the actuator of stimuli-responsive DNA nanostructures (Figure 1Ai). Mao's group presented a double-stranded DNA motif with three helical domains (Li et al., 2020). Two of the outer horizontal helix domains were stable, while the central helix domain was an ATP binding aptamer sequence. In the ATP environment, the central domain combined with two ATP molecules and adapted to a stable helical conformation, whereas the central domain and the raised ring form a larger flexible inner ring in the ATP-free conditions. Additionally, ATP-induced DNA conformational variation also created a driven force for downstream cargo release. Liao et al. (2016) constructed a microcapsule consisting of DNA shells crosslinked by anti-ATP aptamers, in which specific recognition of ATP initiated the dissociation of the DNA outer frames.
Tumor cells have a strong reductive environment because of the excessive production of intracellular glutathione (GSH) (Luo et al., 2016). The concentration of GSH in tumor cells is about four times higher than that in normal cells (Yang et al., 2018). Typically, incorporation of disulfide or selenium bonds in the phosphodiester scaffold of DNA nanostructures offers an effective strategy to develop GSH-responsive nanocarriers. Ding's group recently reported DNA origami nanocarriers grafted with the tumor suppressor gene P53 by disulfide bonds (Liu et al., 2018), allowing the GSH induced bond cleavage to liberate therapeutic segments.
Aberrant expression of functional proteins including enzymes, antigens and hormones involves the dysregulation of signal transduction pathways of cancer cells (Mura et al., 2013). In turn, DNA nanostructures with responsiveness to tumor-associated proteins afford predefined nanoplatforms for investigating and combating cancer. In principle, protein-responsive systems can be achieved by identifying tumor-associated proteins via nucleic acid aptamer or by enzymatic reactions in the extracellular environment (Figure 1Ai) (Mu et al., 2018). For example, a hollow tube-shaped DNA nanorobot (Li et al., 2018) was fabricated by rolling up a two-dimensional (2D) origami sheet containing DNA aptamers that bound nucleolin, a protein specifically expressed on tumor-associated endothelial cells (Figure 1Bi). When the robot arrived at tumor-associated endothelial cells, it was triggered by the nucleolin to expose the cargo inside. Guo's group (Lyu et al., 2018) constructed a stimuli-responsive hydrogel system with liposomes. Upon being exposed to the endonuclease enzyme, EcoR I, the opening of the DNA motifs triggers the transformation of the hydrogel to the subsequent cargo release. Besides, Lu's group demonstrated an enzyme-stimulated DNA nanostructure (Zhang et al., 2017). The aptamer-based activatable photoacoustic DNA probe could recognize thrombin to separate the fluorophore and quencher, thus recovering photoacoustic signal for molecular imaging.
Several coding or non-coding nucleic acids including messenger RNA (mRNA) and microRNA (miRNA) directly regulate the progression of tumors. Considering the intrinsic affinity to DNA structures through complementary base pairing, these endogenous nucleic acids have been working as an ideal activator of DNA nanostructure in a wide spectrum of biomedical applications (Figure 1Ai) (Yeung et al., 2005; Keller and Linko, 2020). Survivin mRNA, a biomarker which had a high expression in lung adenocarcinoma, was employed to initiate the hybridization chain reaction (HCR) for the construction of DNA “nano string light” (Ren et al., 2018). Target mRNA triggered the HCR reaction between hairpin DNA H1 and H2, leading to a quick assembling of a DNA “nano string light” with superior cargo transportation efficiency and remarkable amplification outcomes. Besides, a highly-ordered self-assembled 3D DNA nanostructure equipped with electrochemiluminescence emitter -labeled DNA nipper units was constructed. The sensing platform could respond to miRNA to induce DNA nippers opening for signal switching, achieving miRNA analysis in breast cancer cells (Zhang et al., 2018). In another example, DNA “Nanosuitcases” were utilized as the host for loading small interfering RNA (siRNA), capable of releasing siRNA by strand-displacement of intracellular mRNA or miRNA (Figure 1Bi) (Bujold et al., 2016). The stimuli-responsiveness not only enabled site-specific release of siRNA for gene silencing, but also facilitated exhaustion of intracellular mRNA or miRNA for synergistic therapy.
Since differences in the abundance of proteins and nucleic acids, activation level of enzymes, and physiological environment exist between tumor and normal tissues, nanomaterials that respond to abovementioned tumor-related molecules offer a toolbox to achieve selective sensing or imaging of tumor markers and on-demand delivery of therapeutic agents to tumor sites. In the past decades, the dynamic changes of DNA nanostructures caused by the characteristics of the tumor microenvironment have been used to implement signal modulation and cargo transportation, aiming at delineating and regulating the tumor microenvironment to realize the tumor diagnosis and treatment (Table 1).
Biomarker detection and imaging hold promising prospects in revealing physiological and pathological mechanisms in clinical medicine, including the definition of benign and malignant tumors, early pathological information collection, and the design of metabolically relevant imaging targets (Kumar et al., 2019). Benefitting from the programmability and modifiability of DNA, DNA nanostructures permit embedment of activable DNA motifs equipped with signal modules that can be governed by tumor microenvironments (Chen et al., 2015).
HCR reactions offer an amplification tool for outputting the DNA assembly events triggered by target nucleic acids. In Wang's study, miRNA first triggered a catalyzed hairpin assembly of hairpin DNA H1 and H2, which further led to a HCR reaction between hairpin DNA H3 and H4, achieving a dual-enhanced system for selective miRNA detection in cell media (Liu C. X. et al., 2019). Then DNA nanostructures incorporated with the molecular actuator responsive to pH were also designed (Zhou et al., 2019). The formation of i-motif structure led a significant fluorescence resonance energy transfer between Cy3 and Cy5, realizing pH sensing and mapping in the pH range of 5.0–7.4 in living cells. By integrating the above two aspects, a functionalized DNA nanoprobe activated by high intracellular pH was constructed to release DNA substrates for miRNA triggered HCR reaction, achieving both pH sensing and mRNA imaging in living cells (Zhu et al., 2019). Furthermore, Li's study revealed an endogenous multi-stimuli-responsive system (Peng et al., 2018). Equipped with split-i-motif and halved-ATP aptamer, the Cy3-modified tetrahedron could only be combined with Cy5-labled tetrahedron in the milieu of high ATP and low pH, which can map ATP, and pH in cervical cancer cells.
DNA hydrogels, self-assembled by DNA linker of building blocks to obtain a three-dimensional network, have great applications in different fields including biosensing and imaging due to their unique phase transition between gel and sol caused by destruction of DNA cross-linking points. Hu and co-authors (Wang Q. et al., 2020) introduced a protein-responsive DNA hydrogel for sensitive detection of α-fetoprotein (AFP, biomarker of hepatocellular carcinoma). By recognition and combination of linkers and AFP, the encapsulated immunoglobulin G (IgG) was released and later combined with SERS probes and magnetic beads, leading a SERS signal for AFP detection with an ultrasensitive detection limit of 50 pg mL−1. Considering the requirements of convenient operation and visual inspection in practical applications, Liu and co-authors constructed a visual detection strategy based on pH-responsive DNA hydrogel (Cheng et al., 2009). The individual three-armed DNA nanostructure (Y-unit) involving part of i-motif structures underwent self-assembly with other ones to form complete i-motif structure under the acidic environment, which finally extended a three-dimensional network of DNA hydrogel. In this process, the gold nanoparticles dispersed initially into the upper layer were trapped in the hydrogel as “tracer agents” to indicate the pH.
Although various stimuli-responsive DNA nanostructures have received success, visualizing the signals activation initiated by single or few molecules remains challenging (Jiang et al., 2019a). DNA origami structures are highly programmable DNA nanoassemblies with prescribed scaffold shapes and sequence addressability, which can magnify single-molecule recognition events to conformational change of a single DNA nanostructure. Kuzuya et al. (2011) reported versatile DNA origami systems for sensing endogenous stimuli including tumor-related proteins, small molecules like ATP, and even miRNAs. In this design, the functional nanomechanical DNA origami structures were devised as “DNA origami pliers” with two levers linked at a fulcrum, following two ends of a lever labeled with fluorescein and another lever labeled with quencher. By locating aptamers on the DNA origami pliers, the endogenous IgG, ATP, and miRNAs could control the opening and closing of DNA pliers, which finally led to a fluorescence turn-off for targets analyze. Liu's group presented a pair of antiparallel DNA origami filaments that allowed gold nanocrystals-mediate coordinated sliding powered by DNA fuels, achieving visualization of dynamic single molecules by transmission electron microscopy and in situ fluorescence spectroscopy (Urban et al., 2018).
DNA nanostructures are capturing increasing attention owing to its molecular programmability and precise addressability (Yuan et al., 2019). Compared with the traditional drug delivery systems (DDSs), the endogenous stimuli-responsive DDSs have shown more excellent properties, such as superior bioavailability and site-specific drug release. In this regard, DNA nanostructures responsive to endogenous tumor markers point toward a new way of engineering DDSs for cancer therapy.
DNA-involved nanoaggregates have been constructed by combining organic or inorganic molecules with DNA to mediate the spontaneous DNA assembly into various structures. The principle that endogenous stimuli elicit the structural transformation of DNA nanomaterials, including conformational expansion or contraction, assembly or disassembly, aggregation or dissociation, makes DNA an attractive scaffold for high-capacity drug loading and controlled drug delivery (Dai et al., 2017). For example, an example of DNA nanoaggregates for chemotherapy (Mo et al., 2014) was a DNA nanogel assembled from DOX-intercalated DNA scaffolds with the aid of protamine. In the presence of ATP, the scaffold containing ATP-responsive DNA motif and its complementary single-stranded DNA was changed from duplex to tertiary aptamer structure, resulting in the release of the embedded DOX from the duplex. Additionally, as an expected immune checkpoint inhibitor, PD-1 DNA aptamers were selected to construct hydrogels containing sgRNA sequences (Lee et al., 2019). Upon exposure to Cas9/sgRNA systems, the PD-1 DNA aptamer hydrogel lost its gel properties and released the PD-1 aptamer sequence to enhance antitumor immunotherapeutic effects (Figure 1Bi). In the same year, a series of metal-ion-chelated curcumin and DNA nanostructure conjugates were constructed as drug carriers (Vellampatti et al., 2018a,b). Triggered by pH, it is found that the metal curcumin conjugate reaches the maximum release from the DNA complex under acidic conditions. Subsequently, self-catabolic DNAzyme nanosponges were designed by encapsulating pH-responsive ZnO nanoparticles in the DNAzyme substrate scaffold prepared by roll ring polymerization (Wang et al., 2019). Zn2+ ions resolved from ZnO in the acidic microenvironment could catalyze the decomposition of RCA products and generate intracellular destructive reactive oxygen species (ROS). Simultaneously, the collapsed nanoparticles spark the encapsulated chemotherapeutic agents into the cytoplasm.
Different from the DNA nanoaggregates, DNA origami (Zhan et al., 2014; Linko et al., 2015; Ijäs et al., 2018; Jiang et al., 2019b) featured well-shaped nanostructures with structure rigidity and sequence addressability, has been increasingly used for drug delivery. In addition to the common merits of DNA nanomaterials in biological applications, DNA origami nanostructures are capable of positioning multiple desired functional components like therapeutic agents and tumor targeting ligands in a predefined number and pattern throughout the addressable structure (Nummelin et al., 2020). In 2017, an example of reconfigurable DNA origami device that encapsulated enzyme has been reported (Grossi et al., 2017). The locks are designed to be reversibly opened and closed by toehold-mediated DNA strand displacement. Similarly, Ryadnov's group constructed a hollow cube containing a pH responsive lid that controlled the encapsulation of functional proteins (Burns et al., 2018). Later, a cargo-shielding DNA origami nanocapsule controlled by pH has been assembled (Ijäs et al., 2019). The “pH-latch,” which comprised of a single-stranded DNA and a double-stranded DNA, could either form a parallel triplex DNA via Hoogsteen bonding (closed state, low pH) or freely suspended on the capsule (open state, high pH) according to the environmental pH (Figure 1Bi). These nanocapsules can be loaded with a variety of molecular cargo and selectively display them to the special external cue.
So far, quantities of cancer-related endogenous stimuli-responsive DNA nanostructures have been developed and demonstrated in a wide spectrum of application scenarios. Based on these achievements, we reviewed numerous endogenous stimuli-responsive DNA nanostructures with the corresponding actuation mechanisms and applications. Since DNA has good biocompatibility and excellent modifiability, molecular sensing and drug delivery can be implemented by combining DNA nanostructures with signal tags and therapeutic cargoes. Aiming at characteristic tumor markers and tumor microenvironment, DNA nanostructures designed for responding these cancer-associated stimuli offer great opportunities for early cancer diagnosis and localizable drug release in biomedical applications.
Although endogenous stimuli-responsive DNA nanostructures have made remarkable achievements, it still faces many challenges. Firstly, DNA nanostructures are less stable under physiological conditions in vivo. Not only does it require significant ionic strength to maintain a complete morphology, but it is also easily degraded by nucleases (Ramakrishnan et al., 2018; Bila et al., 2019). Many methods were shown to improve the stability of DNA nanostructures, such as using protective coatings (Ponnuswamy et al., 2017) or chemical crosslinking (Cassinelli et al., 2015). More endeavors are anticipated to be devoted to improving the structural stability via outer protection and structural tailoring for biomedical use. Secondly, attention on the endogenous stimuli mentioned is primarily restricted to pH, small molecules, enzymes and nucleic acids. Other molecules indicative of tumor pathological features, including peroxynitrite (Wu et al., 2020), H2S (An et al., 2018), and hormones (Sun and Zhao, 2017), present a larger library of stimuli for creating stimuli-responsive DNA nanostructures, conducive to broadening the applications in bioimaging and biomedicine. Thirdly, due to the complexity of the cancer treatment mechanisms, DNA nanostructures responding to multi-stimuli and including multiple therapeutic components such as proteins, genes, antibodies, photothermal agents and photosensitizers represent the general trend in clinical treatment, aiming at improving the medical precision and augmenting the curative effects. We truly believe that endogenous stimulus-responsive DNA nanostructures would establish an appealing toolbox to boost the development of personalized cancer theranostics.
XH and ZX designed and wrote the review with input from QM, CT, and YT for conceiving, writing, and editing the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (Grant Nos. 21971109 and 21834004), Jiangsu Youth Fund (Grant No. BK20180337), and the Fundamental Research Funds for the Central Universities (Grant No. 14380151). It was also supported by the Program for Innovative Talents and Entrepreneur in Jiangsu Province, China (No. 133181), Shenzhen International Cooperation Research Project, China (No. GJHZ20180930090602235), Shenzhen Science and Technology Innovation Committee (GJHZ20190819151807167), and the Nanjing Science and Technology Innovation Project for Oversea Scholars' Merit Funding, China (No. 133170).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
An, L., Wang, X., Rui, X., Lin, J., Yang, H., Tian, Q., et al. (2018). The in situ sulfidation of Cu2O by endogenous H2S for colon cancer theranostics. Angew. Chem. 57, 15782–15786. doi: 10.1002/anie.201810082
Bila, H., Kurisinkal, E. E., and Bastings, M. M. (2019). Engineering a stable future for DNA-origami as a biomaterial. Biomater. Sci. 7, 532–541. doi: 10.1039/C8BM01249K
Boedtkjer, E., and Pedersen, S. F. (2020). The acidic tumor microenvironment as a driver of cancer. Annu. Rev. Physiol. 82, 103–126. doi: 10.1146/annurev-physiol-021119-034627
Bujold, K. E., Hsu, J. C. C., and Sleiman, H. F. (2016). Optimized DNA “nanosuitcases” for encapsulation and conditional release of siRNA. J. Am. Chem. Soc. 138, 14030–14038. doi: 10.1021/jacs.6b08369
Burns, J. R., Lamarre, B., Pyne, A. L., Noble, J. E., and Ryadnov, M. G. (2018). DNA origami inside-out viruses. ACS Synth. Biol. 7, 767–773. doi: 10.1021/acssynbio.7b00278
Cassinelli, V., Oberleitner, B., Sobotta, J., Nickels, P., Grossi, G., Kempter, S., et al. (2015). One-Step formation of “chain-armor” -stabilized DNA nanostructures. Angew. Chem. Int. Ed. 54, 7795–7798. doi: 10.1002/anie.201500561
Chen, Y. H., Groves, B., Muscat, R. A., and Seelig, G. (2015). DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 10, 748–760. doi: 10.1038/nnano.2015.195
Cheng, E. J., Xing, Y. Z., Chen, P., Yang, Y., Sun, Y. W., Zhou, D. J., et al. (2009). A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. Int. Ed. 48, 7660–7663. doi: 10.1002/anie.200902538
Dai, Z., Leung, H. M., and Lo, P. K. (2017). Stimuli-responsive self-assembled DNA nanomaterials for biomedical applications. Small 13:1602881. doi: 10.1002/smll.201602881
Dong, Y. C., Yang, Z. Q., and Liu, D. S. (2014). DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 47, 1853–1860. doi: 10.1021/ar500073a
Gehring, K., Leroy, J. L., and Guéron, M. (1993). A Tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 363, 561–565. doi: 10.1038/363561a0
Gorman, M. W., Feigl, E. O., and Buffington, C. W. (2007). Human plasma ATP concentration. Clin. Chem. 53, 318–325. doi: 10.1373/clinchem.2006.076364
Grossi, G., Jepsen, M. D. E., Kjems, J., and Andersen, E. S. (2017). Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 8, 1–8. doi: 10.1038/s41467-017-01072-8
Hu, Y., Wang, Y., Yan, J., Wen, N., Xiong, H., Cai, S., et al. (2020). Dynamic DNA assemblies in biomedical applications. Adv. Sci. 7:2000557. doi: 10.1002/advs.202000557
Hu, Y. B., Cecconello, A., Idili, A., Ricci, F., and Willner, I. (2017). Triplex DNA nanostructures: from basic properties to applications. Angew. Chem. Int. Ed. 56, 15210–15233. doi: 10.1002/anie.201701868
Ijäs, H., Hakaste, I., Shen, B., Kostiainen, M. A., and Linko, V. (2019). Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 13, 5959–5967. doi: 10.1021/acsnano.9b01857
Ijäs, H., Nummelin, S., Shen, B., Kostiainen, M. A., and Linko, V. (2018). Dynamic DNA origami devices: from strand-displacement reactions to external-stimuli responsive systems. Int. J. Mol. Sci. 19:2114. doi: 10.3390/ijms19072114
Jiang, Q., Liu, S., Liu, J., Wang, Z., and Ding, B. (2019a). Rationally designed DNA-origami nanomaterials for drug delivery in vivo. Adv. Mater. 31:1804785. doi: 10.1002/adma.201804785
Jiang, Q., Zhao, S., Liu, J. B., Song, L. L., Wang, Z. G., and Ding, B. Q. (2019b). Rationally designed DNA-based nanocarriers. Adv. Drug Deliv. Rev. 147, 2–21. doi: 10.1016/j.addr.2019.02.003
Kallenbach, N. R., Ma, R.-I., and Seeman, N. C. (1983). An immobile nucleic acid junction constructed from oligonucleotides. Nature 305, 829–831. doi: 10.1038/305829a0
Keller, A., and Linko, V. (2020). Challenges and perspectives of DNA nanostructures in biomedicine. Angew. Chem. Int. Ed. 59, 15818–15833. doi: 10.1002/anie.201916390
Kong, G., Zhang, M., Xiong, M., Fu, X., Ke, G., and Zhang, X. B. (2020). DNA nanostructure-based fluorescent probes for cellular sensing. Anal. Methods 12, 1415–1429. doi: 10.1039/D0AY00105H
Kumar, V., Kukkar, D., Hashemi, B., Kim, K. H., and Deep, A. (2019). Advanced functional structure-based sensing and imaging strategies for cancer detection: possibilities, opportunities, challenges, and prospects. Adv. Func. Mater. 29:1807859. doi: 10.1002/adfm.201807859
Kuzuya, A., Sakai, Y., Yamazaki, T., Xu, Y., and Komiyama, M. (2011). Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2:449. doi: 10.1038/ncomms1452
Lee, J., Le, Q. V., Yang, G., and Oh, Y. K. (2019). Cas9-edited immune checkpoint blockade PD-1 DNA polyaptamer hydrogel for cancer immunotherapy. Biomaterials 218:119359. doi: 10.1016/j.biomaterials.2019.119359
Leist, M., Single, B., Castoldi, A. F., Kuhnle, S., and Nicotera, P. (1997). Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J. Exp. Med. 185, 1481–1486. doi: 10.1084/jem.185.8.1481
Li, Q., Liu, L., Mao, D., Yu, Y., Li, W., Zhao, X., et al. (2020). ATP-triggered, allosteric self-assembly of DNA nanostructures. J. Am. Chem. Soc. 142, 665–668. doi: 10.1021/jacs.9b10272
Li, S., Jiang, Q., Liu, S., Zhang, Y., Tian, Y., Song, C., et al. (2018). A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264. doi: 10.1038/nbt.4071
Liao, W. C., Sohn, Y. S., Riutin, M., Cecconello, A., Parak, W. J., Nechushtai, R., et al. (2016). The application of stimuli-responsive VEGF- and ATP-aptamer-based microcapsules for the controlled release of an anticancer drug, and the selective targeted cytotoxicity toward cancer cells. Adv. Funct. Mater. 26, 4262–4273. doi: 10.1002/adfm.201600069
Linko, V., Ora, A., and Kostiainen, M. A. (2015). DNA nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 33, 586–594. doi: 10.1016/j.tibtech.2015.08.001
Liu, C. X., An, Y. Y., Zhang, Y. F., Li, X., Xue, Q. W., and Wang, H. S. (2019). Digital quantitative detection of serum circulating MiRNAs using dual-enhanced magnetobiosensors based on cascaded nucleic acid circuits. Chem. Commun. 55, 13733–13736. doi: 10.1039/C9CC07841J
Liu, J., Song, L., Liu, S., Jiang, Q., Liu, Q., Li, N., et al. (2018). A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 18, 3328–3334. doi: 10.1021/acs.nanolett.7b04812
Liu, S., Jiang, Q., Wang, Y., and Ding, B. (2019). Biomedical applications of DNA-based molecular devices. Adv. Healthc. Mater. 8:1801658. doi: 10.1002/adhm.201801658
Liu, Z. Y., Li, Y. M., Tian, C., and Mao, C. D. (2013). A Smart DNA tetrahedron that isothermally assembles or dissociates in response to the solution pH value changes. Biomacromolecules 14, 1711–1714. doi: 10.1021/bm400426f
Luo, C., Sun, J., Liu, D., Sun, B., Miao, L., Musetti, S., et al. (2016). Self-assembled redox dual-responsive prodrug-nanosystem formed by single thioether-bridged paclitaxel-fatty acid conjugate for cancer chemotherapy. Nano Lett. 16, 5401–5408. doi: 10.1021/acs.nanolett.6b01632
Lyu, D., Chen, S., and Guo, W. (2018). Liposome crosslinked polyacrylamide/DNA hydrogel: a smart controlled-release system for small molecular payloads. Small 14:1704039. doi: 10.1002/smll.201704039
Mo, R., Jiang, T., DiSanto, R., Tai, W., and Gu, Z. (2014). ATP-triggered anticancer drug delivery. Nat. Commun 5:3364. doi: 10.1038/ncomms4364
Mu, J., Lin, J., Huang, P., and Chen, X. (2018). Development of endogenous enzyme-responsive nanomaterials for theranostics. Chem. Soc. Rev. 47, 5554–5573. doi: 10.1039/C7CS00663B
Mura, S., Nicolas, J., and Couvreur, P. (2013). Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 12, 991–1003. doi: 10.1038/nmat3776
Nummelin, S., Shen, B., Piskunen, P., Liu, Q., Kostiainen, M. A., and Linko, V. (2020). Robotic DNA nanostructures. ACS Synth. Biol. 9, 1923–1940. doi: 10.1021/acssynbio.0c00235
Peng, P., Du, Y., Zheng, J., Wang, H. H., and Li, T. (2018). Reconfigurable bioinspired framework nucleic acid nanoplatform dynamically manipulated in living cells for subcellular imaging. Angew. Chem. Int. Ed. 58, 1648–1653. doi: 10.1002/anie.201811117
Ponnuswamy, N., Bastings, M. M., Nathwani, B., Ryu, J. H., Chou, L. Y., Vinther, M., et al. (2017). Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8:15654. doi: 10.1038/ncomms15654
Qi, X. J., Lu, C. H., Liu, X. Q., Shimron, S., Yang, H. H., and Willner, I. (2013). Autonomous control of interfacial electron transfer and the activation of DNA machines by an oscillatory pH system. Nano Lett. 13, 4920–4924. doi: 10.1021/nl402873y
Ramakrishnan, S., Ijäs, H., Linko, V., and Keller, A. (2018). Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 16, 342–349. doi: 10.1016/j.csbj.2018.09.002
Ren, K. W., Xu, Y. F., Liu, Y., Yang, M., and Ju, H. X. (2018). Responsive “nano string light” for highly efficient mrna imaging in living cells via accelerated DNA cascade reaction. ACS Nano 12, 263–271. doi: 10.1021/acsnano.7b06200
Rothemund, P. W. K. (2006). Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302. doi: 10.1038/nature04586
Seeman, N. C. (1982). Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247. doi: 10.1016/0022-5193(82)90002-9
Sun, Q., and Zhao, Z. (2017). Peptide hormones as tumor markers in clinical practice. Enzymes 42, 65–79. doi: 10.1016/bs.enz.2017.09.001
Urban, M. J., Both, S., Zhou, C., Kuzyk, A., Lindfors, K., Weiss, T., et al. (2018). Gold nanocrystal-mediated sliding of doublet DNA origami filaments. Nat. Commun. 9:1454. doi: 10.1038/s41467-018-03882-w
Vellampatti, S., Chandrasekaran, G., Mitta, S. B., Dugasani, S. R., Lakshmanan, V. K., and Park, S. H. (2018a). Bacterial resistance and prostate cancer susceptibility toward metal-ion-doped DNA complexes. ACS Appl. Mater. Interfaces 10, 44290–44300. doi: 10.1021/acsami.8b17013
Vellampatti, S., Chandrasekaran, G., Mitta, S. B., Lakshmanan, V. K., and Park, S. H. (2018b). Metallo-curcumin-conjugated DNA complexes induces preferential prostate cancer cells cytotoxicity and pause growth of bacterial cells. Sci. Rep. 8:14929. doi: 10.1038/s41598-018-33369-z
Wang, C. C., Zhao, P., Yang, G. L., Chen, X. Y., Jiang, Y. Q., Jiang, X. W., et al. (2020). Reconstructing the Intracellular pH microenvironment for enhancing photodynamic therapy. Mater. Horizons 4, 1180–1185. doi: 10.1039/C9MH01824G
Wang, J., Wang, H., Wang, H., He, S., Li, R., Deng, Z., et al. (2019). Nonviolent self-catabolic dnazyme nanosponges for smart anticancer drug delivery. ACS Nano 13, 5852–5863. doi: 10.1021/acsnano.9b01589
Wang, Q., Hu, Y. J., Jiang, N. J., Wang, J. J., Yu, M., and Zhuang, X. M. (2020). Preparation of aptamer responsive DNA functionalized hydrogels for the sensitive detection of α-fetoprotein using SERS method. Bioconjug. Chem. 31, 813–820. doi: 10.1021/acs.bioconjchem.9b00874
Wang, Z. G., Elbaz, J., Remacle, F., Levine, R. D., and Willner, I. (2010a). All-DNA finite-state automata with finite memory. Proc. Natl. Acad. Sci. U.S.A. 107, 21996–22001. doi: 10.1073/pnas.1015858107
Wang, Z. G., Elbaz, J., and Willner, I. (2010b). DNA machines: bipedal walker and stepper. Nano Lett. 11, 304–309. doi: 10.1021/nl104088s
Wu, W., Zhang, C., Rees, T. W., Liao, X., Yan, X., Chen, Y., et al. (2020). A lysosome-targeting iridium(III) Probe with near-infrared emission for the visualization of NO/O2- crosstalk via in vivo peroxynitrite imaging. Anal. Chem. 92, 6003–6009. doi: 10.1021/acs.analchem.0c00259
Yang, Z., Fan, W., Tang, W., Shen, Z., Dai, Y., Song, J., et al. (2018). Near-infrared semiconducting polymer brush and pH/GSH-responsive polyoxometalate cluster hybrid platform for enhanced tumor-specific phototheranostics. Angew. Chem. 57, 14101–14105. doi: 10.1002/anie.201808074
Yeung, M. L., Bennasser, Y., Le, S. Y., and Jeang, K. T. (2005). SiRNA, MiRNA and HIV: promises and challenges. Cell Res. 15, 935–946. doi: 10.1038/sj.cr.7290371
Yu, Y., Jin, B., Li, Y., and Deng, Z. (2019). Stimuli-responsive DNA self-assembly: from principles to applications. Chem. Eur. J. 25, 9785–9798. doi: 10.1002/chem.201900491
Yuan, Y., Gu, Z., Yao, C., Luo, D., and Yang, D. (2019). Nucleic acid–based functional nanomaterials as advanced cancer therapeutics. Small 15:1900172. doi: 10.1002/smll.201900172
Zhan, P., Jiang, Q., Wang, Z. G., Li, N., Yu, H., and Ding, B. (2014). DNA nanostructure-based imaging probes and drug carriers. ChemMedChem 9, 2013–2020. doi: 10.1002/cmdc.201402137
Zhang, J. J., Smaga, L. P., Satyavolu, N. S. R., Chan, J., and Lu, Y. (2017). DNA aptamer-based activatable probes for photoacoustic imaging in living mice. ACS Nano 139, 17225–17228. doi: 10.1021/jacs.7b07913
Zhang, P., Jiang, j., Yuan, R., Zhuo, Y., and Chai, Y. Q. (2018). Highly ordered and field-free 3D DNA nanostructure: the next generation of DNA nanomachine for rapid single-step sensing. J. Am. Chem. Soc, 140, 9361–9364. doi: 10.1021/jacs.8b04648
Zhou, Y. J., Wan, Y. H., Nie, C. P., Zhang, J., Chen, T. T., and Chu, X. (2019). Molecular switching of a self-assembled 3D DNA nanomachine for spatiotemporal pH mapping in living cells. Anal. Chem. 91, 10366–10370. doi: 10.1021/acs.analchem.9b02514
Keywords: nanotechnology, medicines, DNA nanostructures, stimuli-responsive, cancer theranostics
Citation: Hu X, Xu Z, Min Q, Teng C and Tian Y (2020) Endogenous Stimuli-Responsive DNA Nanostructures Toward Cancer Theranostics. Front. Nanotechnol. 2:574328. doi: 10.3389/fnano.2020.574328
Received: 24 June 2020; Accepted: 23 October 2020;
Published: 27 November 2020.
Edited by:
Nagesh Kolishetti, Florida International University, United StatesReviewed by:
Vinoth Kumar Lakshmanan, Sri Ramachandra Institute of Higher Education and Research, IndiaCopyright © 2020 Hu, Xu, Min, Teng and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Tian, eXRpYW5Abmp1LmVkdS5jbg==; Chao Teng, dGVuZ2NoYW9Ac3pwdC5lZHUuY24=; Qianhao Min, bWlucWlhbmhhb0BuanUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.