
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 24 March 2025
Sec. Brain Disease Mechanisms
Volume 18 - 2025 | https://doi.org/10.3389/fnmol.2025.1460403
Background: Migraine is a paroxysmal headache disorder, which seriously affects the patients’ quality of life. However, the pathogenesis of migraine is not clear yet. Proteomics is an emerging technology for studying small molecules and protein components in biological systems. This study aimed to analyze the serum proteome of migraine patients and healthy controls and identify differentially expressed proteins, which could provide a reference for the study of biomarkers and pathophysiological mechanisms of migraine.
Methods: Fasting venous blood was collected, and serum was separated. Liquid chromatography–mass spectrometry was used to detect the proteome of the two groups, and MaxQuant was used to analyze the protein profile and identify the differentially expressed proteins.
Results: Twenty-seven migraine patients and 20 healthy people matching the age and sex ratio of the migraine group were collected. A total of 27 differentially expressed proteins were identified between migraine and control groups, which were mainly related to immune response, inflammation, glycolysis, lipid metabolism, neurotrophy and development, and so on. Subgroup analysis also identified several differentially expressed proteins between the migraine with aura and the migraine without aura groups and between the ictal and interictal migraine groups. Moreover, the signal pathways that may be related to migraine include the glycolysis/gluconeogenesis pathway and the hypoxia-inducible factor-1 signal pathway. Differentially expressed proteins are mainly distributed in the extracellular area. Related biological processes include complement activation, immunoglobulin receptor binding, and phagocytosis.
Discussion: The research screened out several differentially expressed proteins of migraine patients, which may be potential biomarkers, but it still needs verification in further studies with larger sample sizes. Various proteins related to inflammation, immune response, and energy metabolism are differentially expressed between the migraine group and the control group, suggesting that the pathogenesis of migraine may be related to inflammation, immunity, and energy metabolism disorders. In the future, we can further explore the therapeutic targets of migraine in terms of these biological processes.
Migraine is a paroxysmal headache disorder characterized by recurrent, mostly unilateral, pulsating, moderate-to-severe headaches, often accompanied by nausea, vomiting, photophobia, and phonophobia, which may also have vision, sensory, or other kinds of aura. Patients would suffer from headaches for 4 to 72 h without treatment or with ineffective treatment (Headache Classification Committee of the International Headache Society (IHS), 2018). Migraine is a common disease. The global prevalence of migraine is 14.7%, and the Chinese annual prevalence is 9.3% (Yu et al., 2011). The prevalence in women is two to three times more than men. The highest prevalence occurs between the ages of 25 and 55 years. According to the Global Burden of Disease Study by the World Health Organization in 2021, migraine was the third most disabling disease of neurological disorders in the world, which seriously affected the patients’ quality of life (GBD 2021 Nervous System Disorders Collaborators, 2024).
However, the pathophysiology of migraine is still incompletely understood. There are generally accepted mechanisms that may play a role in the migraine attack, including cortical spreading depression, alteration in thalamocortical circuits, altered brain connectivity, and sensitization of the trigemino-cervical complex (Charles, 2018). The diagnosis of migraine depends on clinical history at present. Although well-accepted diagnostic criteria exist for migraine, it is still a complex disorder that remains both underdiagnosed and misdiagnosed. The lack of appropriate biomarkers is an obstacle to developing more effective diagnostic criteria and treatments (Durham and Papapetropoulos, 2013).
Proteomics studies the expression and function of proteins in a biological system, together with the pathophysiological processes in which they are involved, helping to define more precisely the pathogenesis of diseases. Currently, proteomics is one of the most significant methodologies for the identification of the overall protein content of cells, tissues, or biofluids (Lionetto et al., 2013; Hasin et al., 2017; Hosp and Mann, 2017). Compared to genomics or metabolomics, proteomics provides a new perspective to explore the mechanism and biomarker of migraine. Moreover, proteins are the direct reflection of biological function, and the occurrence and development of many diseases are caused by the abnormal expression or dysfunction of proteins, rather than the genes. Therefore, the proteomic approach could be appropriate to explore promising biomarkers of multifactorial diseases such as migraine. However, at present, few proteomics studies have been developed in an attempt to identify biomarkers and pathophysiological mechanisms associated with migraine. Klychnikov et al. (2010) focused on the cortical synapse proteomics of a transgenic migraine mouse model with mutated CaV2.1 calcium and observed some upregulated proteins in the mutant mice related to neurite outgrowth and actin dynamics, vesicle turnover, and glutamate transporters. Guyuron et al. (2014) compared the protein expression of the zygomaticotemporal branch of the trigeminal nerve in patients with and without migraine headaches and found five significant pathways involved in cytoskeletal organization, myelination of axons, and nervous system development. Bellei et al. (2020, 2021) examined the serum and urinary proteome of females suffering from menstrually related migraine and postmenopause migraine, in the search for potential biomarkers.
This study aimed to analyze the serum proteome of migraine patients in comparison with healthy controls and to search and identify the differentially expressed proteins as potential biomarkers of migraine, which could be informative on the accurate diagnosis and treatment of migraine.
In this study, participants who were diagnosed as migraine without aura or migraine with aura according to the diagnostic criteria of the International Classification of Headache Disorders, third edition (ICHD-3) and had a headache history of more than 1 year were included as the migraine group. Healthy adults who were age-matched and sex-matched with the migraine group and had no headache were included as the control group. Patients and healthy adults with the following conditions were excluded: (1) with severe infection, blood system disease, liver disease, malignant tumor, severe mental disease, or immune system disease, (2) received blood products transfusions in the past 6 months, and (3) with pregnancy and lactation (Figure 1). The baseline demographic data and clinical history were collected through questionnaires. All participants gave their written informed consent for inclusion before they participated in the study. The study was approved by the Medical Ethics Committee of the Second Clinical Hospital of Peking University.
Fasting venous blood (4 mL) was collected in the morning from migraine patients and non-headache controls. Blood samples were collected into 4 mL BD Biosciences gold top and placed on ice. Serum was subsequently subjected to centrifugation at 2,000–3,000×g for 10 min at +4°C. To preserve proteins, serum was added with a protease inhibitor cocktail (Roche.) in a ratio of 1:500 prior to being divided into aliquots and stored at −80°C until proteomic analysis.
The serum proteins were digested according to the manufacturer’s protocol for filter-aided sample preparation (FASP) with minor modifications. The serum (10 μL) was diluted in 8 M urea (90 μL). The mixture was heated at 95°C for 10 min. When the mixture was left to cool to room temperature, tris-(2-carboxyethyl) phosphine (TCEP) was added to the serum solution to a final concentration of 10 mM and heated at 67°C for 10 min. Then, 50 mM iodoacetamide (IAA) was added to the alkylated solution. After the sample was incubated at room temperature for 30 min in the dark, all of the samples were transferred to the Vivacon 500 filtration tubes (Cat No. VNO1HO2, Sartorius Stedim Biotech, UK, MWCO 10 kDa), mixed with 150 μL of 8 M urea in 0.1 M Tris/HCl (pH 8.5), and centrifuged at 14,000 g for 15 min at 20°C. This step was performed two times. Then, the samples were washed three times with 100 μL of 50 mM NH4HCO3. Finally, 6 μg of trypsin (Promega, Madison, WI) in 100 μL of 50 mM NH4HCO3 was added to each filter. The protein-to-enzyme ratio was 100:1. The samples were incubated overnight at 37°C. Released peptides were collected by centrifugation and vacuum-dried.
After redissolving with 0.1% FA, the peptide samples (1 μg) were analyzed by an LC–MS/MS system consisting of UltiMate 3,000 UHPLC (Thermo Scientific) and Q-Exactive HF (Thermo Fisher Scientific). The peptides were loaded on a homemade pre-column (2 cm × 100 μm i.d., 15–20 μm, 300 A, YWG-C18) before being separated on a homemade analytical column (12 cm × 75 μm i.d., 15–20 μm, 300 A, YWG-C18) in the 65°C thermostat at a flow rate of 400 nL/min. For a gradient separation, H2O/FA (99.9:0.1) was used as mobile phase A, whereas ACN/H2O/FA (80:19.9:0.1) was used as mobile phase B. The LC gradient began with 5% phase B for 5 min and ramped to 10% for 3 min, from 10 to 40% for 92 min, and then from 40 to 100% for 10 min; it was held at 100% for 4 min. Finally, the concentration was restored to 5% phase B and held for 6 min before the next run.
MS/MS spectra were obtained in a data-dependent high-energy collision dissociation (HCD) model. The parameters for serum peptide analysis were as follows: (1) full MS: scan range = 350–1,500 m/z; resolution = 60,000; AGC target = 3,000,000; maximum injection time = 20 ms; included charge state = 2–6; dynamic exclusion duration = 30 s; (2) dd-MS2: isolation window = 1.6 m/z; resolution = 15,000; AGC target = 100,000; maximum injection time = 45 ms; loop count = 20; stepped collision energy = 27.
Raw data were analyzed using the MaxQuant software (version 1.5.6.0). MS data were searched against the UniProt Human database, with the following settings: Carbamidomethylation of cysteine was defined as fixed modification and methionine oxidation (+15.99 Da) as a variable modification of the peptides. Trypsin was set as the protease with two missed cleavage sites allowed. Label-free quantification (LFQ) can calibrate the raw intensity values between samples to eliminate experimental batch effects or technical errors between samples and finish data normalization. LFQ was performed in MaxQuant following that the minimum ratio count for LFQ was set to 2 and the match-between-runs option was enabled. Other parameters were set as default. The continuous variables normally distributed were expressed as mean ± standard deviation (SD) and the dichotomous variables as percentages. The false discovery rate was set as less than 0.01. The comparison between means was made using a t-test. Differences were considered significant with a p-value <0.05. Regarding the fold changes of spot abundance, values >1.2 were considered as significant.
All statistical analyses were performed using SPSS 27.0. Values are presented as the mean ± standard deviation for data that were normally distributed or median and interquartile range for data that were not normally distributed for continuous variables and number (%) for categorical variables. For two-group comparison, p-values were derived from the one-way Student t-test to determine the differences between groups with the normally distributed data and Mann–Whitney non-parametric test with other data. For all comparisons, a p-value of <0.05 was considered statistically significant.
A total of 27 migraine patients and 20 healthy controls were enrolled in this study. There was no statistical difference in gender ratio and average age between the two groups. The BMI of the migraine group was significantly lower than that of the control group, and the proportion of allergic diseases and the SAS scale score were significantly higher than those of the control group (Table 1).
There were 27 differentially expressed proteins identified between the migraine and control groups (Table 2). Compared with healthy controls, nine proteins were upregulated, which were involved in neurotrophy, neuroprotection, molecular chaperone, immunity, and inflammation, and 18 were downregulated, which were related to cell motility, glycolysis, lipid metabolism, immunity, neurogenesis, and so on (Figures 2, 3).
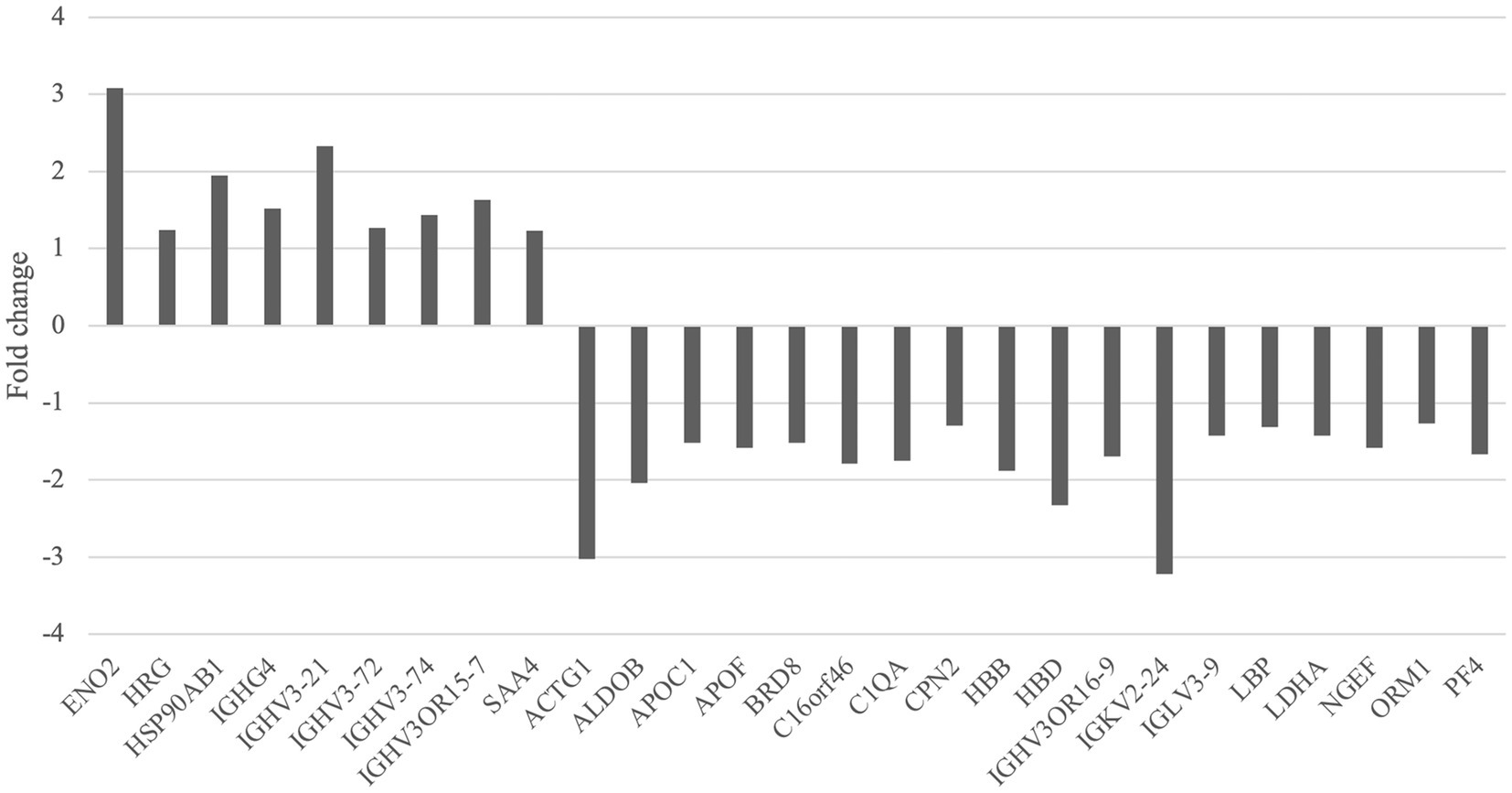
Figure 2. Upregulated and downregulated proteins of the migraine group compared with the control group.

Figure 3. Volcano plot demonstrating differentially expressed proteins of the migraine group compared with the control group.
We performed a search of these 27 proteins in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Table 3). The results of GO and KEGG analysis indicated that the signal pathways that might be related to migraine included the glycolysis/gluconeogenesis pathway and the hypoxia-inducible factor-1 (HIF-1) signal pathway (Figure 4). Differentially expressed proteins were mainly distributed in the extracellular area. Related biological processes included complement activation, immunoglobulin receptor binding, and phagocytosis.
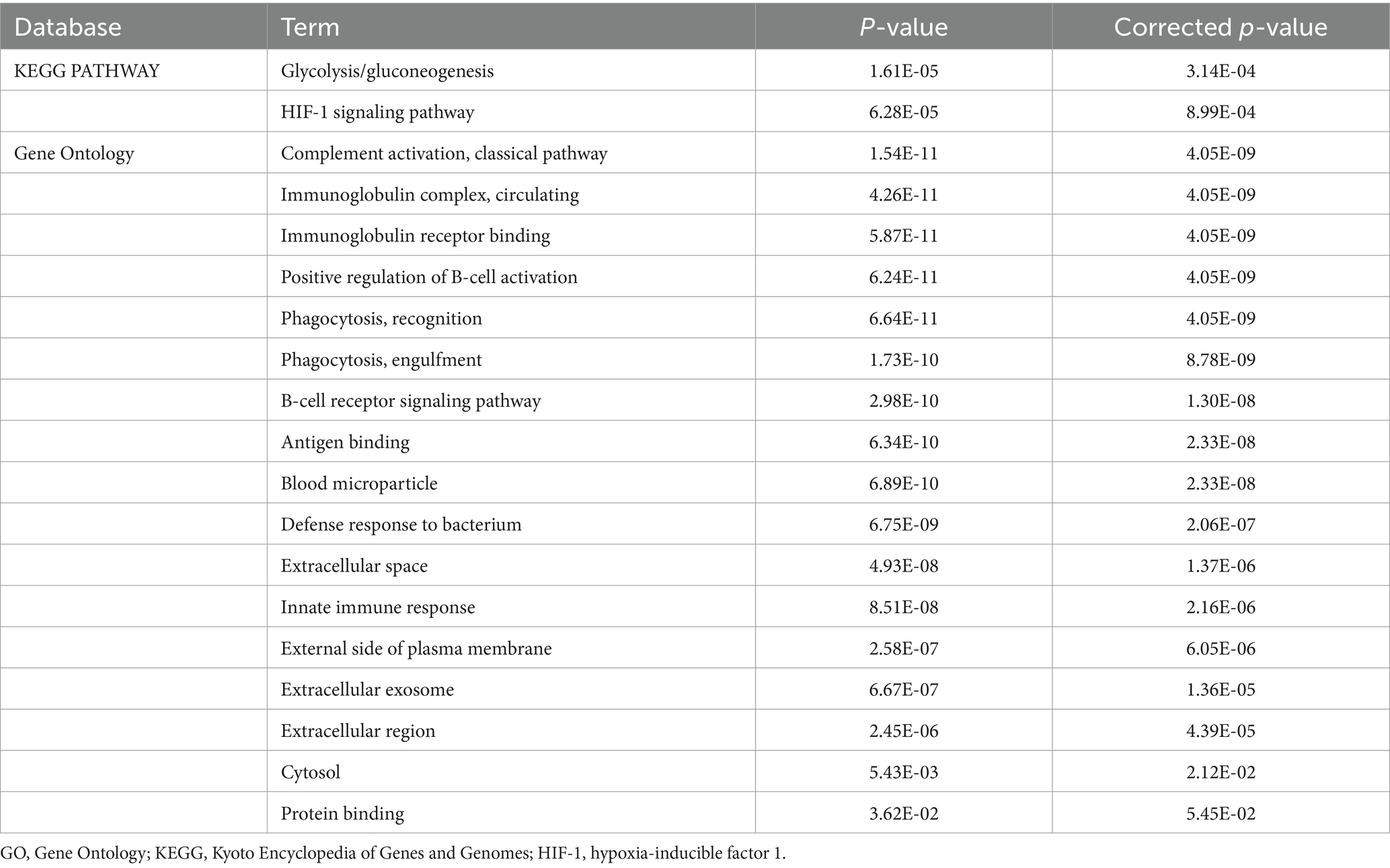
Table 3. GO and KEGG analysis of differentially expressed proteins between the migraine and control groups.
Compared with the migraine without aura (MO) group, migraine with aura (MA) group had eight differentially expressed proteins, of which seven were upregulated and one was downregulated, which were involved with oxygen transport, immunity, blood coagulation, cell adhesion, and lipid metabolism (Table 4).
Compared with the interictal migraine group, there were four differentially expressed proteins in the ictal migraine group, of which two were upregulated and two were downregulated, which were related to inflammation and immunity (Table 5).
Among migraine patients, compared with patients who had no family history of headaches, three proteins were upregulated and four proteins were downregulated in the patients who had a family history of headaches, which were related to embryonic organ development, coagulation, immunity, cell adhesion, and cell movement (Table 6).
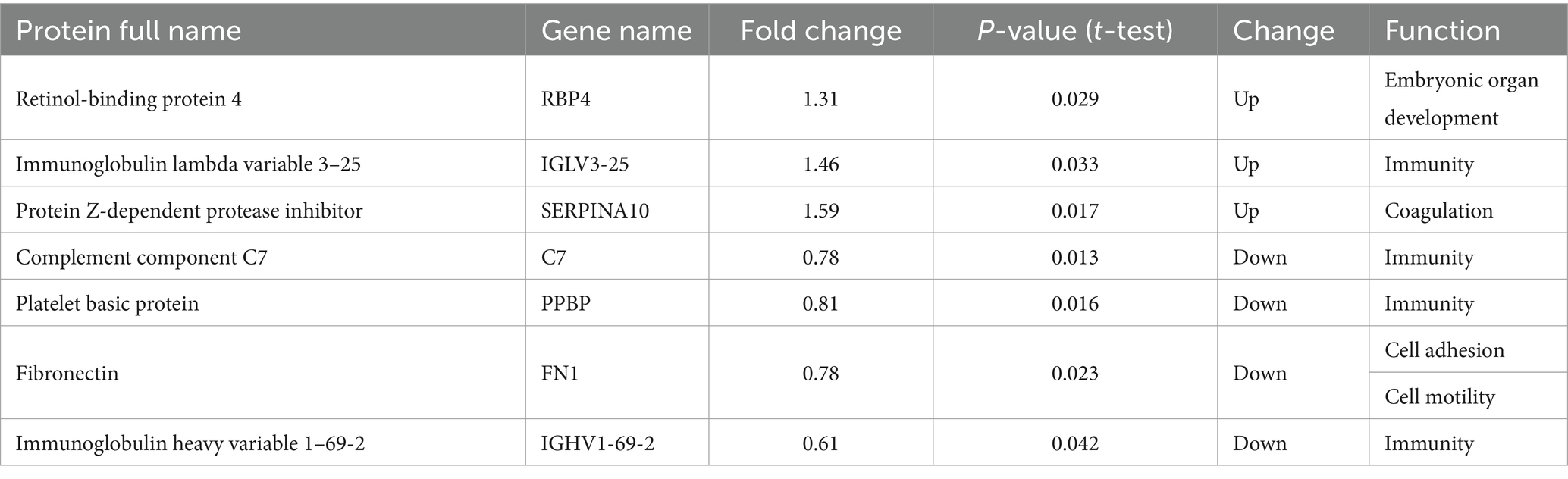
Table 6. Differentially expressed proteins between migraine patients with and without headache family history.
Migraine is a common primary headache in clinical practice, whose clinical manifestations are episodic moderate-to-severe pulsatile headache. According to the Global Burden of Disease Study, migraine is the most disabling neurological disease among the ages 5–19 years and second after stroke among adults aged 20–59 years (Peres et al., 2024). Frequent migraine attacks and chronic migraine have a serious impact on the study, work, and daily life of migraine patients (Deuschl et al., 2020). Currently, it is believed that migraine is a chronic neurovascular disease that involves multiple pathogenesis mechanisms such as genetics, immunity, endocrine, and metabolism. However, looking for suitable biomarkers to assist in the diagnosis and treatment of migraine still remains an urgent issue to be addressed.
In this study, a total of 27 differential proteins between migraine patients and healthy controls were identified by liquid chromatography–mass spectrometry. Among these 27 proteins, eight belong to the component of immunoglobulins, and C1QA, LBP, HRG, ORM1, and SAA4 are also related to inflammation and immunity. The differential proteins between the ictal and the interictal groups are also related to inflammation and immunity, suggesting that neuroinflammation and immune disorders may participate in the pathophysiological mechanism of migraine. Previous migraine proteomics studies had similar findings. Similar to our study, Woldeamanuel et al. (2024) performed untargeted proteomics and found proteins for classifying migraine patients with migraine from healthy controls, including immunoglobulin heavy variable 3–74 (IGHV 3–74). Bellei et al. (2020, 2021) observed that IGKC and CO4A levels were higher in the serum of women with menstrual-related migraine and postmenopausal migraine, which was also found in the urine proteomics study of the same participants. Togha et al. (2022) analyzed proteomics changes of migraine patients compared to controls or their pain-free period and found that the expression levels of several proteins such as haptoglobin, clusterin, and fibrinogen alpha chain showed considerable changes, and most of them were associated with inflammation, oxidative stress, and neuroprotection. The discrepancies in migraine proteomics studies may be related to sample heterogeneity, such as different tissue and disease courses, and different methods of sample preparation and proteomics techniques.
Some evidence supporting the role of immune disorders in the pathogenesis of migraine exists. Studies have found changes in cytokine levels and lymphocyte subsets in migraine patients (Perini et al., 2005; Aydn et al., 2015; Arumugam and Parthasarathy, 2016). In addition, migraine and autoimmune diseases have been found to have common genetic and environmental pathogenic factors. Polymorphisms in genes encoding human leukocyte antigen (HLA) and cytokines are considered risk factors for autoimmune diseases. Some studies have found that these genes are also involved in the pathogenesis of migraine (Rainero et al., 2005; Yilmaz et al., 2010).
γ-Enolase, also known as neuron-specific enolase (NSE), is an enzyme located in the axons and cytoplasm of neurons and plays a key role in glycolysis. Its elevated concentration in serum indicated neuronal damage caused by stroke or brain injury (Teepker et al., 2009). This study found that the serum NSE level in migraine patients increased, which suggested that migraine might be related to the injury of glial cells or neurons in the brain and damage to blood–brain barrier. Previous studies found inconsistent results. Yilmaz (2020) found that the serum NSE level in migraine patients was lower than that in controls, whereas Gönen et al. (2021) observed that the serum NSE level in both paroxysmal and chronic migraine patients was significantly higher than that in the control group. Yilmaz et al. (2011) found that the serum NSE level in MO patients during the ictal period was higher than that in the control group. Other studies have shown that there is no significant difference in serum NSE levels in children between the migraine and control groups (Azapağasi et al., 2012).
Compared with healthy controls, the levels of APOC1 and APOF in migraine group decreased, and the levels of APOL1 in MA patients increased compared with MO patients, indicating that lipid metabolism disorders may be related to the pathogenesis of migraine. Previous studies about lipidomics have found abnormal levels of lipids and apolipoprotein in migraine patients (Ren et al., 2018; Onderwater et al., 2019). Dyslipidemia is known as a vascular risk factor, and the incidence rate of cardiovascular and cerebrovascular diseases in migraine patients has also increased, but the specific relationship between migraine and dyslipidemia is still unclear.
We found two signal pathways based on 27 differentially expressed proteins, including the glycolysis/gluconeogenesis pathway and HIF-1 signal pathway, indicating that the pathogenesis of migraine is related to energy metabolism and oxidative stress. Liu et al. (2022) observed that joint pathway analysis of proteomics and metabolomics revealed significant enrichment of the pentose phosphate and glycolysis/gluconeogenesis pathways, which provided evidence for the influence of energy metabolism on migraine. Previous studies have suggested that migraine is a stress response to brain energy deficiency and oxidative stress. Some researchers used magnetic resonance spectroscopy (MRS) to detect ATP, lactic acid, magnesium, and other substances in the brains of migraine patients. It has been found that oxidative phosphorylation of mitochondria in migraine patients’ brains is impaired, manifested by increased ADP levels, decreased organic phosphate, and free magnesium levels (Gross et al., 2019). These findings suggest that the pathogenesis of migraine may be related to the imbalance of energy and glucose metabolism in the brain.
Compared with the control group, multiple proteins were significantly upregulated or downregulated in the migraine group, and subgroup analysis also showed that there was a differential expression of proteins between the ictal and interictal period of migraine between MO and MA as well as between with and without family history of headache. These proteins may be potential biomarkers of migraine, but further validation experiments are needed. The differential expression of various proteins related to inflammation, immune response, and energy metabolism suggests that the pathogenesis of migraine may be related to inflammation, immunity, and energy metabolism disorders. Specific molecular mechanisms can be researched in the future.
This study has some limitations. At first, the sample size was small. Further study needs to be performed in larger-sized populations. Second, the differentially expressed proteins identified through proteomics need to be further confirmed by enzyme-linked immunosorbent assay (ELISA) or Western blot. Because of the long storage time of original serum samples, new samples need to be collected for further validation. In addition, the signal pathways related to migraine need to be deeply studied. To further validate the involvement of the HIF-1 pathway and inflammation in migraine, we plan to establish a migraine model in mice by repeated injection of nitroglycerin (10 mg/kg, i.p.). Roxadustat, a HIF-1α stabilizer, will be orally administered before or after nitroglycerin injection. Mice will be randomly divided into three groups in the prevention and acute treatment experiments: the normal saline control group (NS), the NTG-induced migraine group (NTG), and the NTG plus roxadustat treatment group (NTG + Rox). Pressure application measurement and tail flick and light-aversive behavior tests will be performed to determine the pressure pain threshold, thermal nociceptive sensitivity, and migraine-related light sensitivity. At the end of experiments, mouse serum samples and brain tissues will be collected to analyze the expression of HIF-1α and proinflammatory, including factors interleukin (IL)-1β and IL-6 and tumor necrosis factor (TNF)-α. The expected results include that compared with the NS group, mice in the NTG group show a significant decrease in both the pressure pain threshold and thermal nociceptive threshold after NTG injection, and prevention or treatment with roxadustat can improve NTG-induced light sensitivity. HIF-1α will be reduced by NTG injection and be enhanced by roxadustat. In addition, roxadustat can improve the neuroinflammation induced by the repeated administration of NTG, as evidenced by the decreased protein and mRNA levels of proinflammatory factors. In a word, the hypothesis of further study is that HIF-1α stabilization can improve migraine-like behaviors and inhibits inflammation in nitroglycerin-injected mice, which may be mediated by the HIF-1α/NF-κB/inflammation pathway, suggesting the potential of HIF-1α activators as therapeutics for migraine.
In conclusion, this study could provide some new insights into the treatment of migraine. We can further explore therapeutic targets of migraine in the fields of inflammation, immunity, and energy metabolism in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving humans were approved by Ethical Review Committee of Peking University People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
YH: Data curation, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YW: Methodology, Software, Validation, Writing – original draft. XZ: Methodology, Software, Supervision, Validation, Writing – original draft. HG: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 81230023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2025.1460403/full#supplementary-material
Arumugam, M., and Parthasarathy, V. (2016). Reduction of CD4(+) CD25(+) regulatory T-cells in migraine: is migraine an autoimmune disorder? J. Neuroimmunol. 290, 54–59. doi: 10.1016/j.jneuroim.2015.11.015
Aydn, M., Demir, C., Arkanolu, A., Bulut, S., and Lhan, N. (2015). Plasma cytokine levels in migraineurs during and outside of attacks. Eur. J. Gen. Med. 12, 307–312. doi: 10.15197/ejgm.01255
Azapağasi, E., Alehan, F., Saygi, S., Bayraktar, N., and Yazici, A. C. (2012). Serum concentrations of neuron-specific enolase in pediatric migraine. Turk. J. Pediatr. 54, 150–155
Bellei, E., Bergamini, S., Rustichelli, C., Monari, E., Dal Porto, M., Fiorini, A., et al. (2021). Urinary proteomics reveals promising biomarkers in menstrually related and post-menopause migraine. J. Clin. Med. 10:1854. doi: 10.3390/jcm10091854
Bellei, E., Rustichelli, C., Bergamini, S., Monari, E., Baraldi, C., Lo Castro, F., et al. (2020). Proteomic serum profile in menstrual-related and post menopause migraine. J. Pharm. Biomed. Anal. 184:113165. doi: 10.1016/j.jpba.2020.113165
Charles, A. (2018). The pathophysiology of migraine: implications for clinical management. Lancet Neurol. 17, 174–182. doi: 10.1016/S1474-4422(17)30435-0
Deuschl, G., Beghi, E., Fazekas, F., Varga, T., Christoforidi, K. A., Sipido, E., et al. (2020). The burden of neurological diseases in Europe: an analysis for the global burden of disease study 2017. Lancet Public Health 5, e551–e567. doi: 10.1016/S2468-2667(20)30190-0
Durham, P., and Papapetropoulos, S. (2013). Biomarkers associated with migraine and their potential role in migraine management. Headache 53, 1262–1277. doi: 10.1111/head.12174
GBD 2021 Nervous System Disorders Collaborators (2024). Global, regional, and national burden of disorders affecting the nervous system, 1990-2021: a systematic analysis for the global burden of disease study 2021. Lancet Neurol. 23, 344–381. doi: 10.1016/S1474-4422(24)00038-3
Gönen, M., Özdoğan, S., Balgetir, F., Demir, C. F., Aytaç, E., and Müngen, B. (2021). S100B and neuron-specific enolase levels in episodic and chronic migraine. Acta Neurol. Scand. 143, 298–302. doi: 10.1111/ane.13365
Gross, E. C., Lisicki, M., Fischer, D., Sándor, P. S., and Schoenen, J. (2019). The metabolic face of migraine – from pathophysiology to treatment. Nat. Rev. Neurol. 15, 627–643. doi: 10.1038/s41582-019-0255-4
Guyuron, B., Yohannes, E., Miller, R., Chim, H., Reed, D., and Chance, M. R. (2014). Electron microscopic and proteomic comparison of terminal branches of the trigeminal nerve in patients with and without migraine headaches. Plast. Reconstr. Surg. 134, 796e–805e. doi: 10.1097/PRS.0000000000000696
Hasin, Y., Seldin, M., and Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol. 18:83. doi: 10.1186/s13059-017-1215-1
Headache Classification Committee of the International Headache Society (IHS) (2018). The international classification of headache disorders, 3rd edition. Cephalalgia 38, 1–211. doi: 10.1177/0333102417738202
Hosp, F., and Mann, M. (2017). A primer on concepts and applications of proteomics in neuroscience. Neuron 96, 558–571. doi: 10.1016/j.neuron.2017.09.025
Klychnikov, O. I., Li, K. W., Sidorov, I. A., Loos, M., Spijker, S., Broos, L. A., et al. (2010). Quantitative cortical synapse proteomics of a transgenic migraine mouse model with mutated ca(V)2.1 calcium channels. Proteomics 10, 2531–2535. doi: 10.1002/pmic.200900733
Lionetto, L., Gentile, G., Bellei, E., Capi, M., Sabato, D., Marsibilio, F., et al. (2013). The omics in migraine. J. Headache Pain 14:55. doi: 10.1186/1129-2377-14-55
Liu, L., Li, W., Wang, L., Gong, P., Lyu, T., Liu, D., et al. (2022). Proteomic and metabolomic profiling of acupuncture for migraine reveals a correlative link via energy metabolism. Front. Neurosci. 16:1013328. doi: 10.3389/fnins.2022.1013328
Onderwater, G. L. J., Ligthart, L., Bot, M., Demirkan, A., Fu, J., van der Kallen, C. J. H., et al. (2019). Large-scale plasma metabolome analysis reveals alterations in HDL metabolism in migraine. Neurology 92, e1899–e1911. doi: 10.1212/WNL.0000000000007313
Peres, M. F. P., Sacco, S., Pozo-Rosich, P., Tassorelli, C., Ahmed, F., Burstein, R., et al. (2024). Migraine is the most disabling neurological disease among children and adolescents, and second after stroke among adults: a call to action. Cephalalgia 44:3331024241267309. doi: 10.1177/03331024241267309
Perini, F., D'Andrea, G., Galloni, E., Pignatelli, F., Billo, G., Alba, S., et al. (2005). Plasma cytokine levels in migraineurs and controls. Headache 45, 926–931. doi: 10.1111/j.1526-4610.2005.05135.x
Rainero, I., Fasano, E., Rubino, E., Rivoiro, C., Valfrè, W., Gallone, S., et al. (2005). Association between migraine and HLA-DRB1 gene polymorphisms. J. Headache Pain 6, 185–187. doi: 10.1007/s10194-005-0180-3
Ren, C., Liu, J., Zhou, J., Liang, H., Wang, Y., Sun, Y., et al. (2018). Lipidomic analysis of serum samples from migraine patients. Lipids Health Dis. 17:22. doi: 10.1186/s12944-018-0665-0
Teepker, M., Munk, K., Mylius, V., Haag, A., Möller, J. C., Oertel, W. H., et al. (2009). Serum concentrations of s100b and NSE in migraine. Headache 49, 245–252. doi: 10.1111/j.1526-4610.2008.01228.x
Togha, M., Rahimi, P., Farajzadeh, A., Ghorbani, Z., Faridi, N., and Zahra Bathaie, S. (2022). Proteomics analysis revealed the presence of inflammatory and oxidative stress markers in the plasma of migraine patients during the pain period. Brain Res. 1797:148100. doi: 10.1016/j.brainres.2022.148100
Woldeamanuel, Y. W., Sanjanwala, B. M., and Cowan, R. P. (2024). Deep and unbiased proteomics, pathway enrichment analysis, and protein-protein interaction of biomarker signatures in migraine. Ther. Adv. Chronic Dis. 15:20406223241274302. doi: 10.1177/20406223241274302
Yilmaz, S. (2020). Serum NO, S100B, NSE concentrations in migraine and their relationship. J. Clin. Neurosci. 82, 32–35. doi: 10.1016/j.jocn.2020.10.046
Yilmaz, N., Karaali, K., Ozdem, S., Turkay, M., Unal, A., and Dora, B. (2011). Elevated S100B and neuron specific enolase levels in patients with migraine-without aura: evidence for neurodegeneration? Cell. Mol. Neurobiol. 31, 579–585. doi: 10.1007/s10571-011-9651-z
Yilmaz, I. A., Ozge, A., Erdal, M. E., Edgünlü, T. G., Cakmak, S. E., and Yalin, O. O. (2010). Cytokine polymorphism in patients with migraine: some suggestive clues of migraine and inflammation. Pain Med. 11, 492–497. doi: 10.1111/j.1526-4637.2009.00791.x
Keywords: ACTG1, ENO2, glycolysis, HIF-1 signaling pathway, migraine, proteomics
Citation: Han Y, Wang Y, Zou X and Guo H (2025) The serum proteomic profile in patients with migraine. Front. Mol. Neurosci. 18:1460403. doi: 10.3389/fnmol.2025.1460403
Received: 06 July 2024; Accepted: 07 March 2025;
Published: 24 March 2025.
Edited by:
Claudia Altamura, Fondazione Policlinico Campus Bio-Medico, ItalyReviewed by:
Parisa Gazerani, Oslo Metropolitan University, NorwayCopyright © 2025 Han, Wang, Zou and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huailian Guo, Z3VvaEBiam11LmVkdS5jbg==; Xiajuan Zou, em91eGpAYmptdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.