- 1Department of Translational Medicine – Hand Surgery, Lund University, Malmö, Sweden
- 2Department of Hand Surgery, Skåne University Hospital, Malmö, Sweden
- 3Faculty of Medicine, Department of Clinical Sciences, Mass Spectrometry, Lund University, Lund, Sweden
- 4Unit for Social Epidemiology, Department of Clinical Sciences, Malmö, Lund University, Malmö, Sweden
- 5Department of Biomedical and Clinical Sciences, Linköping University, Linköping, Sweden
Introduction: Proteomic analysis of injured human peripheral nerves, particularly focusing on events occurring in the proximal and distal nerve ends, remains relatively underexplored. This study aimed to investigate the molecular patterns underlying a digital nerve injury, focusing on differences in protein expression between the proximal and distal nerve ends.
Methods: A total of 26 human injured digital nerve samples (24 men; 2 women; median age 47 [30–66] years), harvested during primary nerve repair within 48 h post-injury from proximal and distal nerve ends, were analyzed using mass spectrometry.
Results: A total of 3,914 proteins were identified, with 127 proteins showing significant differences in abundance between the proximal and the distal nerve ends. The downregulation of proteins in the distal nerve end was associated with synaptic transmission, autophagy, neurotransmitter regulation, cell adhesion and migration. Conversely, proteins upregulated in the distal nerve end were implicated in cellular stress response, neuromuscular junction stability and muscle contraction, neuronal excitability and neurotransmitter release, synaptic vesicle recycling and axon guidance and angiogenesis.
Discussion: Investigation of proteins, with functional annotations analysis, in proximal and the distal ends of human injured digital nerves, revealed dynamic cellular responses aimed at promoting tissue degeneration and restoration, while suppressing non-essential processes.
1 Introduction
Peripheral nerve injuries can lead to significant functional impairments, severely affecting patients’ quality of life. Understanding the immediate molecular mechanisms that govern nerve injury response and regeneration is crucial for developing effective therapeutic surgical and pharmacological interventions. Data on up- and down-regulated proteins from experimental studies must also be confirmed in human studies (Krishnan et al., 2024). Following a nerve transection, the proximal nerve end around the site of the lesion and the entire distal nerve end undergoes distinct and extensive biological processes. From the proximal nerve end, the axons attempt to regenerate across the site of injury and into the distal nerve end, where Wallerian degeneration is rapidly initiated to prepare the environment for potential regeneration. Despite the clinical importance of these processes, comprehensive proteomic differences between these nerve ends have not been extensively studied in a well-defined time perspective, nor have they been adequately related to findings in experimental studies.
The peripheral nervous system possesses the ability to undergo self-regeneration following an injury, where the efficiency may depend on the extent and location of the injury, the surgical methods of repair, or reconstruction as well as the timing of the surgical procedure. However, the functional recovery is often slow and incomplete due to its reliance on the synthesis and transport of intracellular substances in the neuron that are crucial for axonal outgrowth, which must occur along the entire distal nerve end to the target. The regeneration process also involves expression of substances by for example Schwann cells (SC) and recruited macrophages (Court et al., 2008; Jessen and Mirsky, 2016; Clements et al., 2017). The regenerative capacity of axons decreases with increasing distance from the injury site, suggesting the presence and impact of crucial signals originating particularly from the Schwann cells (SC) in the distal nerve end. Timing of surgery, related to utilization of up- and downregulated molecules by the injury over time, plays an important role in guiding axonal outgrowth (Danielsen et al., 1983; Huebner and Strittmatter, 2009).
Although individual proteins and pathways involved in nerve regeneration have been investigated in mice and rats (Dahlin, 2023; Krishnan et al., 2024), there is a lack of comprehensive proteomic analysis comparing the protein profiles of proximal and distal nerve ends post-injury in humans, where the proximal nerve end is considered to exhibit molecular alterations. This gap of knowledge limits our understanding of the molecular environment and sophisticated regulatory mechanisms, particularly in the distal nerve end, which are essential for devising targeted therapeutic strategies and for interpreting findings from experimental studies.
From the proximal injured nerve end, neurons immediately after injury, attempt to regrow their axons across the site of injury and down into the distal nerve end to their target tissue, guided by dedifferentiated SC that become activated and proliferate distal to the injury, forming structures known as bands of Büngner (Nocera and Jacob, 2020). Schwann cells undergo a transition from a myelinating state to a growth-supportive state after loss of axonal contact. Changes occurs also in the SC related to the non-myelinated nerve fibers (Saito et al., 2010). In both the neuronal cell body and SC, several signaling pathways are initiated to facilitate the regenerative response (Bolívar et al., 2020), which is supported and facilitated by other key players, such as the crucial macrophages (Zigmond and Echevarria, 2019). The changes in the proximal nerve end usually involve only a limited distance from the site of injury, extending to the nearest node of Ranvier in myelinated nerve fibers, resulting in limited alteration of the proteomic pattern. Thus, understanding the short-term proteomic differences between the proximal and distal nerve in humas is a suitable model to identify proteins that are differentially expressed, potentially revealing novel targets for therapeutic intervention. Proteins uniquely up- or downregulated in either nerve end can serve as biomarkers to monitor the progression of nerve degeneration and regeneration and relate to the effectiveness of treatments. Analyzing the early events, such as within the first days after injury, in the proximal and distal nerve ends is an initial step using proteomics in humans with a well-defined injury.
The distal nerve end is believed to release trophic factors and other signaling molecules that promote axonal regeneration and guide growing axons towards their targets (Lundborg, 1987; Neurotropism and nerve growth, 1987). Furthermore, the distal nerve end may undergo changes over time following injury, such as alterations in gene expression of the key players or composition of extracellular matrix, including the inflammatory response and also the presence of pro-healing macrophages (Zigmond and Echevarria, 2019) with risk of formation of scarring, which affect its ability to support axonal outgrowth over longer distances. As the distance between the proximal and distal nerve ends increases, the concentration of signaling molecules may decrease or become insufficient to sustain robust axonal outgrowth, despite formation of a formed and crucial fibrin matrix with macrophages between the nerve ends. Such a fibrin matrix is influenced by the distance between the nerve ends (Zhao et al., 1993). Additionally, the time frame during which these signals remain active may be limited, further limiting the window of opportunity for successful regeneration (Sulaiman and Gordon, 2013).
In recent years, significant progress has been made in understanding the specific signaling molecules emitted by the injured peripheral nerves with mass spectrometry techniques. Neurotrophic factors, extracellular matrix molecules, and signaling pathways have emerged as key players in promoting axonal outgrowth and survival (Rozenbaum et al., 2018; Vergara et al., 2018; He et al., 2021) Knowledge has been raised from animal experimental studies, using both mice and rats along with in vitro studies (Raivich, 2011; Jessen and Mirsky, 2019; Jessen and Mirsky, 2021). However, studies with focus on proteomic changes at the tip of the injured human proximal and distal nerve ends are limited.
Thus, understanding the differences between the proximal and distal nerve ends after injury is vital for crafting effective strategies to enhance nerve regeneration. While alterations are thought to be limited in the proximal nerve end after nerve injury, the distal end may undergo significant cellular and molecular alterations with the intention to clear the path for the outgrowing axons. The aim of the study is to investigate the molecular changes underlying a human digital nerve injury, focusing on changes in protein expression in the distal nerve end compared to the proximal nerve end of human digital nerves.
2 Materials and methods
2.1 Ethical approval
The study has been approved by the Regional Ethical Review Board in Lund, Sweden (no 311/2016). All study participants provided informed written consent. All procedures were carried out in line with relevant current guidelines and regulations. Prior to surgery an informed consent was retrieved from the donor. The research was conducted in accordance with the principles of the Helsinki Declaration.
2.2 Subjects
Patients with a transected digital nerve injury, where the proximal and the distal digital nerve ends could be visualized, and a primary surgical intervention was performed within 48 h from injury were included in the study. Only patients with a clearcut digital nerve injury that did not require extensive resection were included, while patients undergoing a nerve reconstruction procedure were not included. A short segment of the injured proximal and distal end of the digital nerve was removed, during an acute surgical procedure at the Department of Hand Surgery in Malmö, Sweden, as part of preparing the injured nerve for a direct nerve repair with sutures. Thereby, only nerve injuries with a limited impact on the nerve ends were included. The resected nerve pieces were collected and stored at −80°C until further analysis.
The study included 19 individuals (men n = 24; women n = 2; median age 47 [interquartile range; IQR 30–66]), four individuals had multiple digital nerve injuries leading to a final inclusion of 26 paired digital nerve injuries, each containing a proximal and a distal nerve end harvested during the surgical repair, subsequently leading to the analysis of 52 nerve ends (Figure 1).
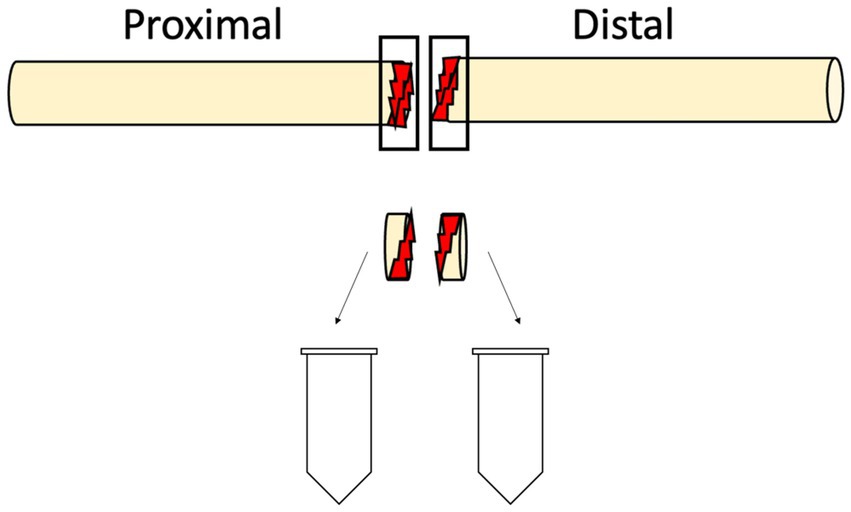
Figure 1. A schematic drawing of the harvesting procedure of nerve tissue from the transected digital nerve performed within 48 h after injury. The injured segment of the proximal and distal end of the digital nerve were collected and stored at −80°C.
2.3 Sample preparation
Samples were prepared as previously described (Frostadottir et al., 2024). Briefly, fresh frozen human digital nerve tissue pieces were homogenized using a Bullet Blender Storm Pro (BT24M, Next Advance, Inc. Troy, NY, United States) first with Ripa buffer, and then followed by 8 M Urea. Enzyme digestion was performed by LysC for 3 h at 37 C followed by trypsination overnight at 37 C.
2.3.1 LC–MS analysis
The samples were analyzed on an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fischer Scientific) coupled with an Ultimate 3,000 RSLCnano system (Thermo Fischer Scientific). Two-column setup was used on the HPLC system and peptides were loaded into an Acclaim PepMap 100 C18 precolumn (75 μm x 2 cm, Thermo Scientific, Waltham, MA) and then separated on an EASY spray column (75 μm x 25 cm, nanoViper, C18, 2 μm, 100 Å) with the flow rate 300 nL/min. The column temperature was set at 45°C. Solvent A (0.1% FA in water) and solvent B (0.1% FA in 80% ACN) were used to create a non-linear gradient to elute the peptides. For the gradient, the percentage of solvent B was maintained at 2% for 4 min, increased from 2 to 25% for 100 min and then increased to 40% for 20 min and then increased to 95% for 1 min and then kept at 95% for another 5 min to wash the column.
The Eclipse was operated in the data-independent acquisition mode. Full MS survey scans from m/z 350–1,450 with a resolution 120,000 were performed in the Orbitrap detector. The automatic gain control (AGC) target was set to 1 × 106 with the maximum injection time of 50 ms. One segment for MS1 was kept constant. Forty-four windows with isolation width of 25 m/z. were acquired for MS2 with the resolution of 30,000. The normalized collision energy for HCD and the AGC target for MS2 were 30 and 1 × 106, respectively. The maximum injection time was set to 54 ms.
2.3.2 Data analysis
A total of 52 DIA runs were loaded in Spectronaut™ (version 18.4, Biognosys AG) and the protein search was performed using the human protein data base downloaded from Uniprot 20240216_ UP5640. For protein identification, an FDR < 0.01 was stablished in Spectronaut both at protein and peptide level. Default settings (BGS factory settings) were used for the search with additional modifications: cysteine carbamidomethylation as a fixed modification, and N-terminal acetylation and methionine oxidation as variable modifications. The precursor quantification was performed at MS2 level, and the peak area of proteotypic peptides for each protein was used for quantification.
2.4 Statistics
Students t-tests were performed to evaluate differential abundances of the proteins between distal and proximal nerve ends. All reported p-values are two-tailed, with p < 0.05 being considered as statistically significant. If an individual had multiple samples by type of nerve end and protein, median value was used.
3 Results
A total of 3,914 proteins were found in our dataset. A summary of these proteins with their related biological functions, cellular components and molecular functions are presented in the Supplementary Figures S1–S3.
3.1 Protein expression profile in the injured digital nerve ends
From the nerves of the 19 individuals analyzed, the proximal nerve end contained 3,846 proteins, while the distal nerve end consistently presented 3,755 proteins. Of those, 3,687 proteins were expressed in both nerve ends (Figure 2).
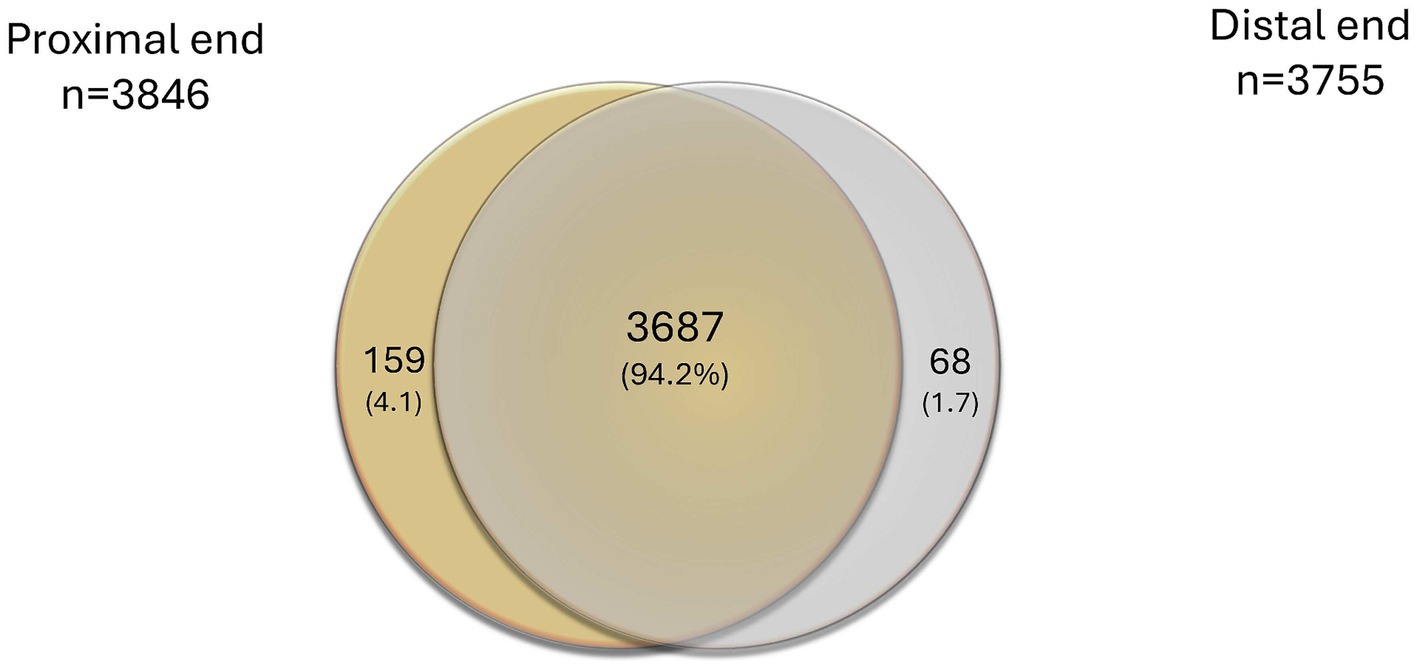
Figure 2. Venn diagram presenting overlaps of all identified proteins in the proximal and the distal nerve ends, alone or combined, expressed as number of proteins and % in brackets.
3.2 The difference in protein abundance between the proximal and distal nerve ends
To identify differentially expressed proteins in the proximal and the distal nerve ends, the relative abundance was compared between the two groups using Student’s t-test. For each group any significant changes with p-value of less than 0.05 were presented as up or downregulated in the distal nerve end compared to the proximal end. There were 127 proteins statistically significant up- or downregulated in the data set (Figure 3).
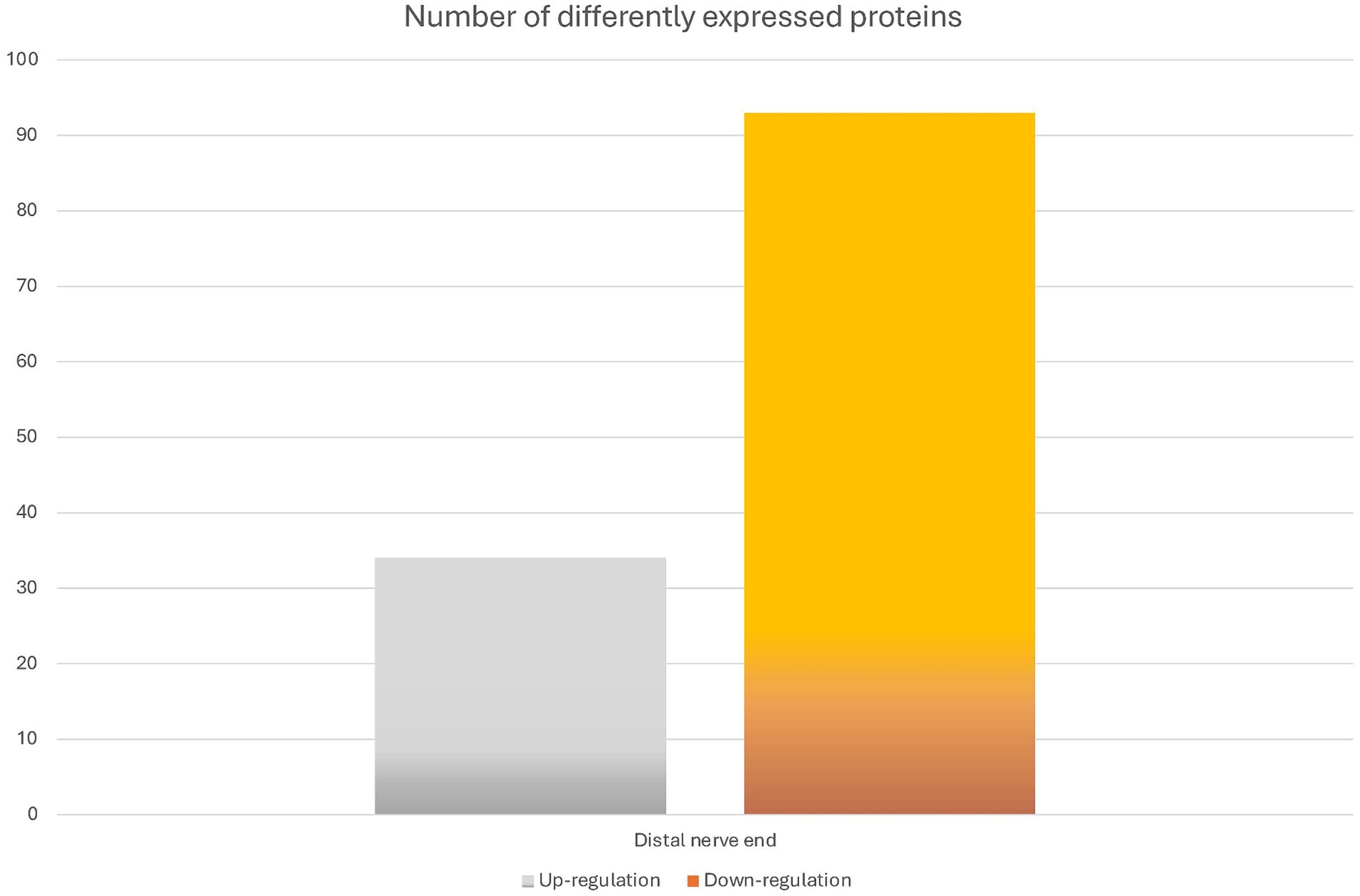
Figure 3. Overview of the differentially expressed proteins between the proximal and distal nerve ends. The bar graph shows the number of identified proteins that were differentially expressed when comparing the samples of the proximal (n = 26 nerve samples) and the distal nerve ends (n = 26 nerve samples). The difference is presented as numbers of proteins significantly either up- or down-regulated in the distal nerve end compared to the proximal nerve end with a p-value of less than 0.05.
3.2.1 Visualizing differential protein expression: volcano plot analysis of distal vs. proximal nerve ends
A Volcano plot analysis, including a total of presently expressed 3,914 proteins between the proximal and distal nerve ends, was performed. Fold changes (x axis) were plotted against the significance -log10 p values (y axis) for each protein highlighting proteins with the largest and most significant differences presented in Figure 4 and in Tables 1, 2. The proteins found with the most significant differences in expression in the distal end compared to the proximal end were cross-referenced with The Human Protein Atlas (HPA) (Karlsson et al., 2021) as well as The Injured Sciatic Nerve Atlas (iSNAT) (Zhao et al., 2022) where C57BL/6 mice were studied.
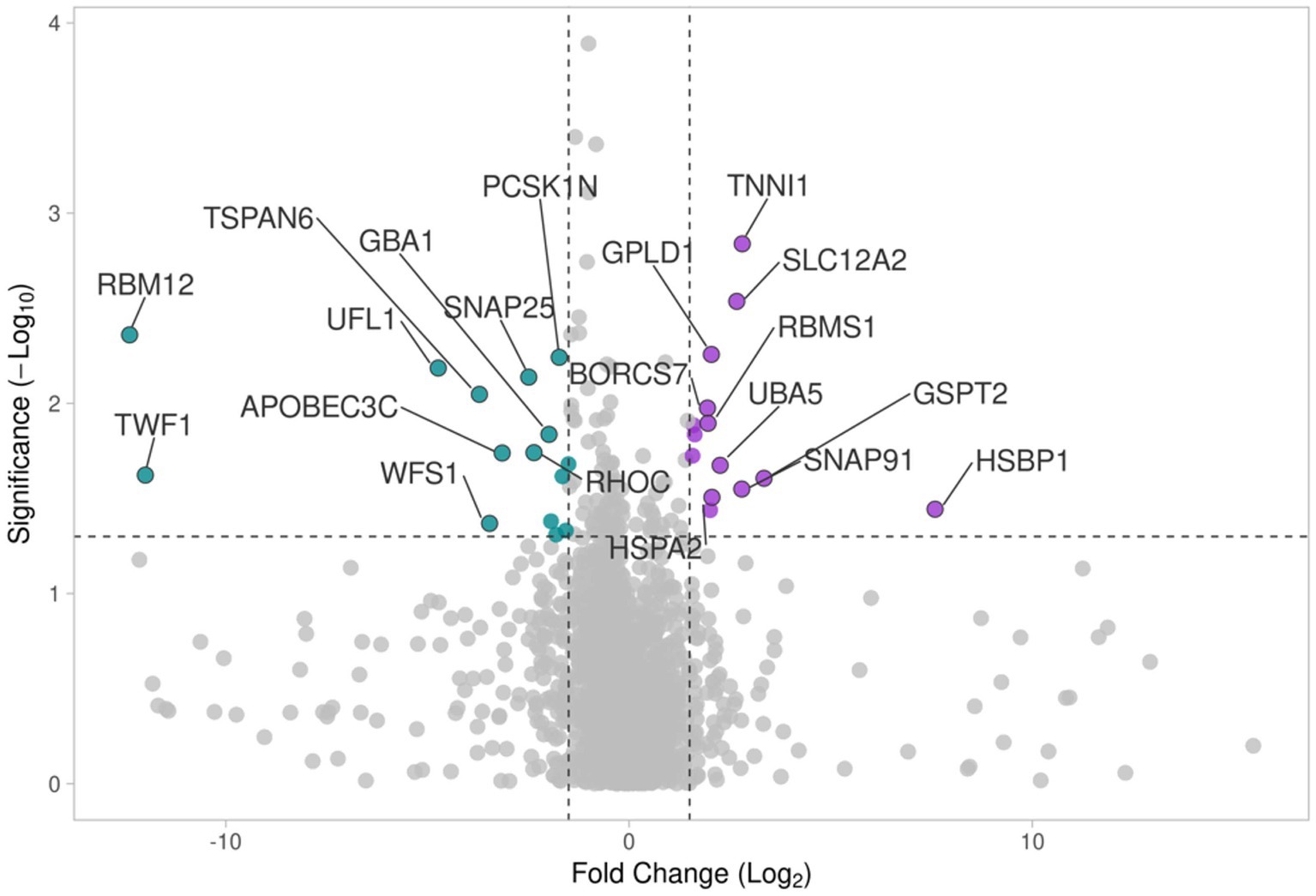
Figure 4. The volcano plot comparing differences in protein expression in the proximal and distal nerve ends shows log2 ratio versus -log10 p value of differentially expressed proteins. Colors show Up-regulated (purple) and down-regulated (blue) proteins.
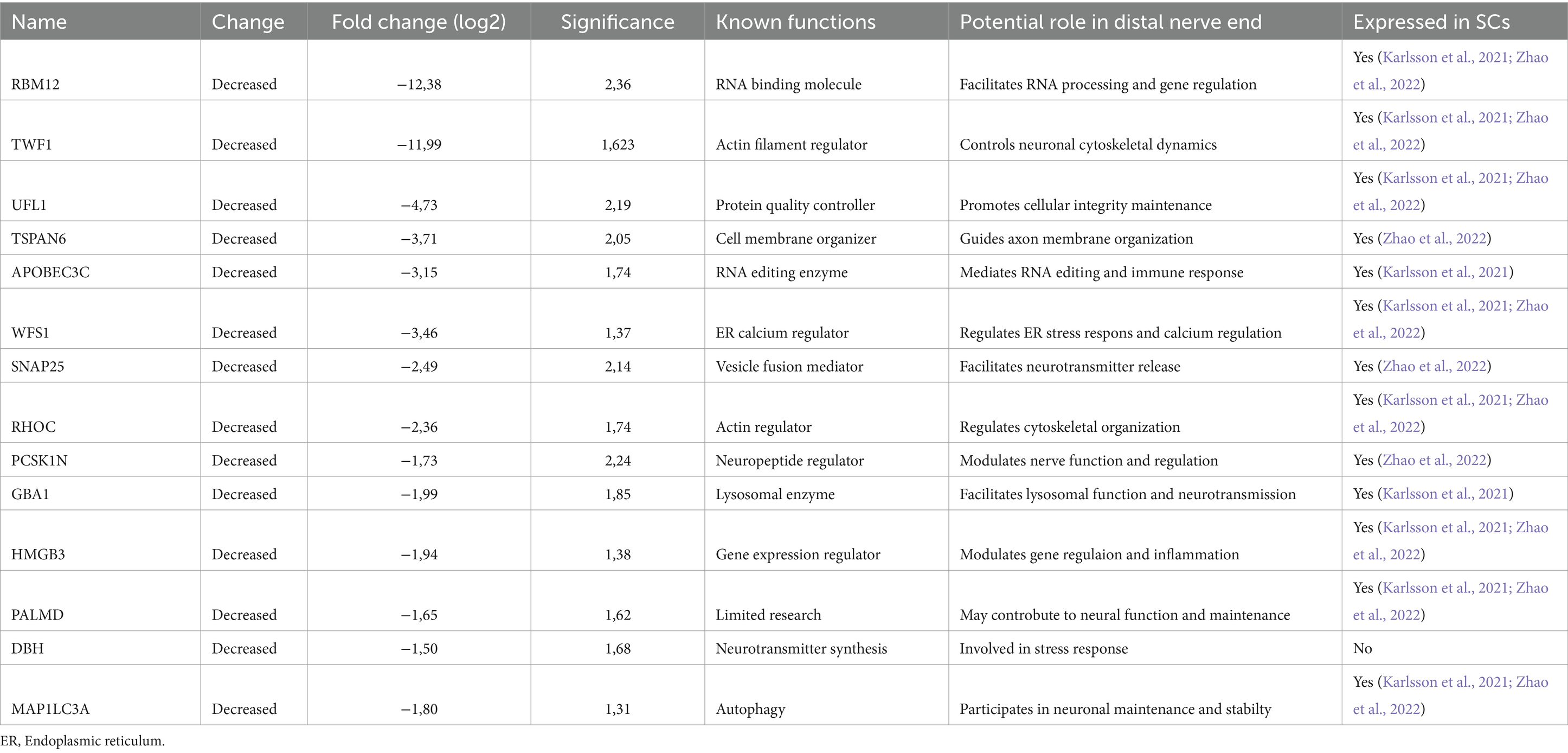
Table 1. Proteins down-regulated in the distal nerve end, after an injury in 26 human injured digital nerves repaired within 48 h post-injury, compared with the proximal nerve end with the most substantial differences in fold change and highest significance (−log10 p value).
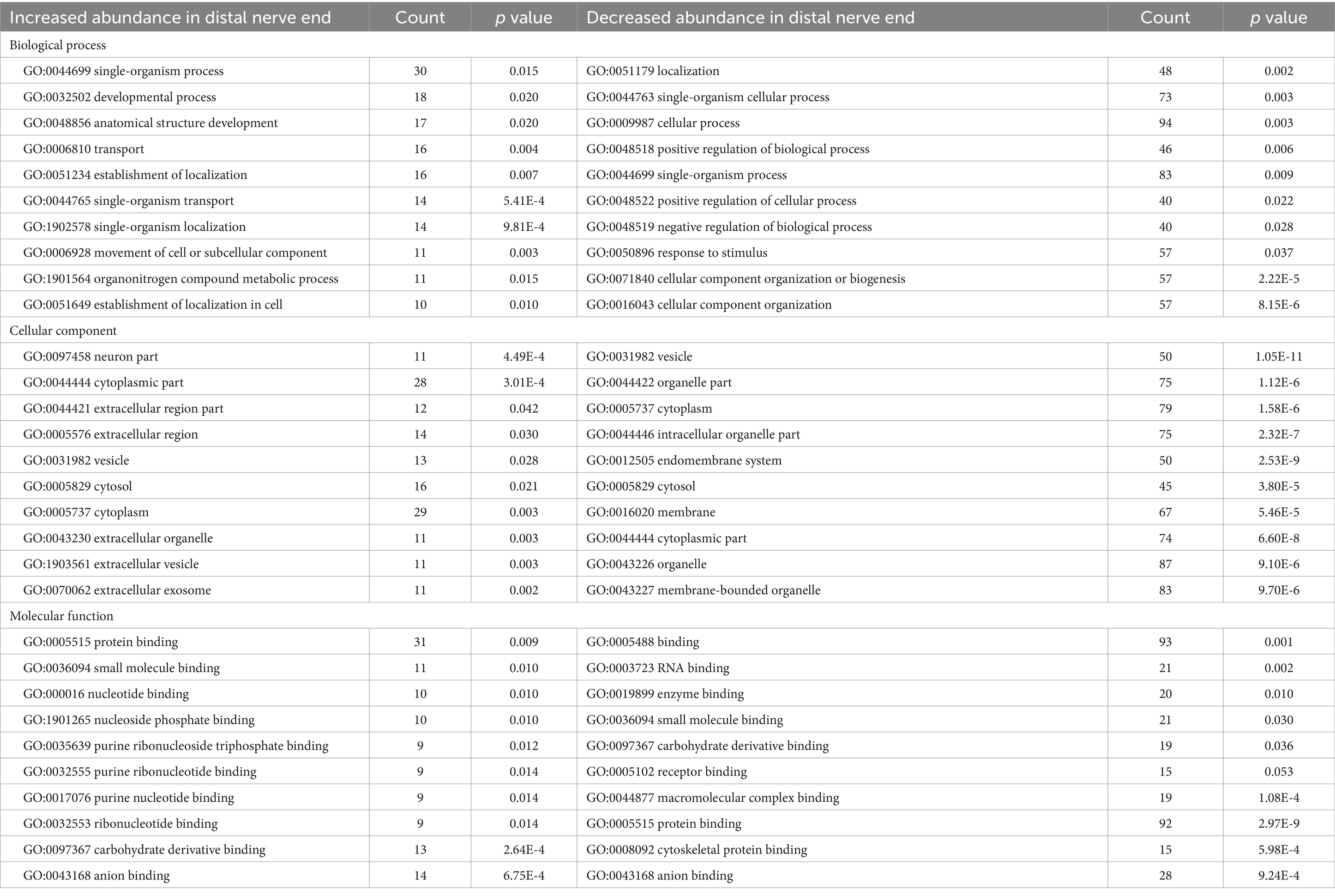
Table 2. The top 10 functional annotations (GO) Pathways for significantly up- and down-regulated proteins in the distal compared to the proximal nerve end after an injury in 26 human injured digital nerves repaired within 48 h post-injury.
3.2.1.1 Proteins up-regulated in the distal nerve end
Proteins up-regulated in the distal nerve end included Heat Shock Factor Binding Protein 1 (HSBP1), Troponin I (TNNI1), Solute Carrier Family 12 Member 2 (SLC12A2), G1 to S phase Transition 2 (GSPT2), Clathrin coat assembly protein AP180 (SNAP91), Glycosylphosphatidylinositol Specific Phospholipase D1 (GPLD1), Ubiquitin Like Modifier Activating Enzyme 5 (UBA5), BLOC-1 Related Complex Subunit 7 (BORCS7), RNA Binding Motif Single Stranded Interacting Protein 1 (RBMS1), Heat Shock Protein Family A (Hsp70) member 2 (HSPA2), Integrin alpha-2 (ITGA2), Ependymin Related 1 (EPDR1), Protein FAM3C (FAM3C), and Epidermal growth factor-like protein 8 (EGFL8). All proteins except SNAP91 were found, when cross-referenced with the HPA and the iSNAT, to be expressed by SCs (Table 3).
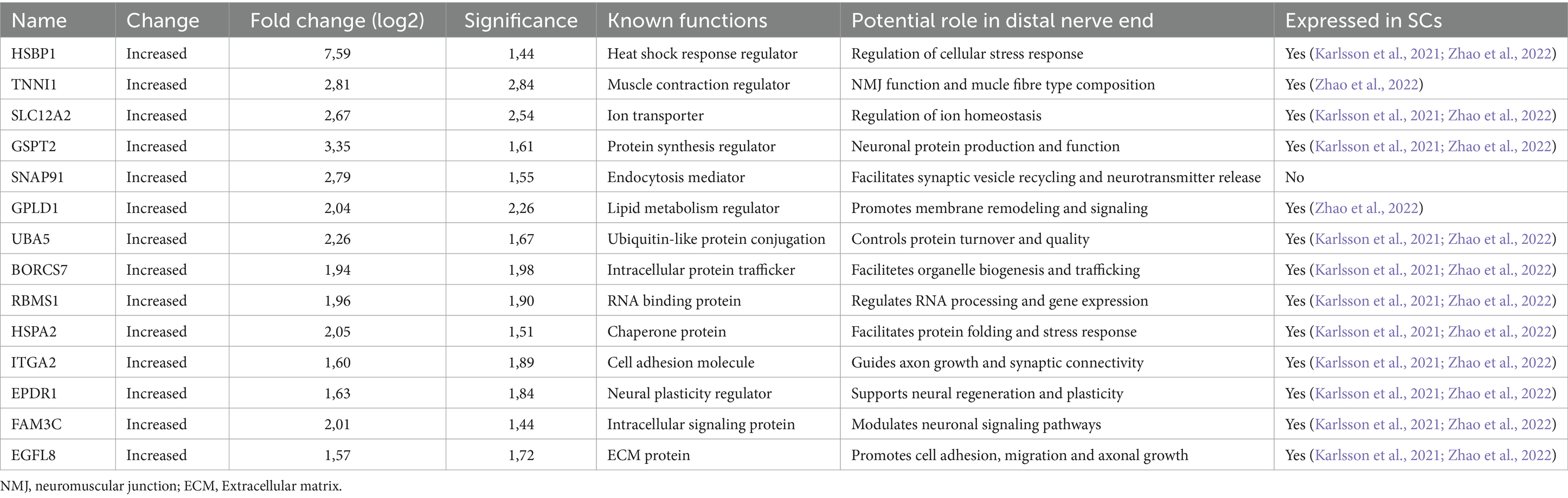
Table 3. Up-regulated proteins in the distal end compared with the proximal nerve end, after an injury in 26 injured digital nerves repaired within 48 h post-injury, with the most substantial differences in expression levels and highest significance (−log10 p value).
3.2.1.2 Proteins down-regulated in the distal nerve end
Among the proteins down-regulated in the distal nerve end of the injured nerve compared to the proximal nerve end were Synaptosome Associated Protein 25 (SNAP25), Microtubule Associated Protein 1 Light Chain 3 Alpha (MAP1LC3A), Wolfram Syndrome 1 (WFS1), Dopamine Beta-Hydroxylase (DBH), Tetraspanin-6 (TSPAN6), RNA Binding Protein 12 (RBM12), Twinfilin Actin Binding Protein 1 (TWF1), E3 UFM1-protein ligase 1 (UFL1), DNA dC- > dU-editing enzyme APOBEC-3C (APOBEC3C), Rho-related GTP-binding protein RhoC (RHOC), Proprotein Convertase Subtilisin/Kexin Type 1 Inhibitor (PCSK1N), Glucosidase Beta Acid 1 (GBA1), High Mobility Group Protein 3 (HMGB3), and Palmdelphin (PALMD). All proteins except DBH were found, when cross-referenced with the HPA and the iSNAT, to be expressed by SCs (Table 2).
3.2.2 Functional profiling of a digital nerve injury and regeneration with DAVID pathway analysis
DAVID pathway analysis (Huang da et al., 2009; Sherman et al., 2022) was performed, using the Uniprot Accession number, to group together genes with similar cell components, biological processes, and molecular functions. Of the 127 proteins, 33 proteins showed to be significantly increased in abundance at the distal nerve end, while 93 proteins were significantly decreased in the distal nerve end compared to the proximal nerve end. Table 3 presents the top 10 functional annotations (GO) pathways for the proteins significantly increased and decreased in the distal nerve end compared to the proximal nerve end.
3.2.3 Functional enrichment analysis for digital nerve injury and regeneration
3.2.3.1 Proteins up-regulated in the distal nerve end
When analyzing the functional annotations of proteins with increased abundance in the distal compared to the proximal nerve ends, we observed a clustering pattern based on shared biological functions and properties. In the cluster presented in Figure 5A, annotations reveal the common themes among Ubiquitin Like Modifier Activating Enzyme 5 (UBA5), Heat Shock Protein Family A (Hsp70) member 2 (HSPA2), Ribosomal Protein S6 Kinase A3 (RPS6KA3,) Protein tyrosine phosphatase non-receptor type 11 (PTPN11), Glycosylphosphatidylinositol Specific Phospholipase D1 (GPLD1), Membrane Metalloendopeptidase (MME), dCTP Pyrophosphatase 1(DCTPP1), Ectonucleoside Triphosphate Diphosphohydrolase 2 (ENTPD2), Atlastin GTPase 1 (ATL1), ADP Ribosylation Factor Like GTPase 8B (ARL8B), G1 to S phase Transition 2 (GSPT2), and Obg like ATPase 1 (OLA1) of nucleotide binding, hydrolase activity and P-loop containing nucleoside triphosphate hydrolase domain (Figure 5A).
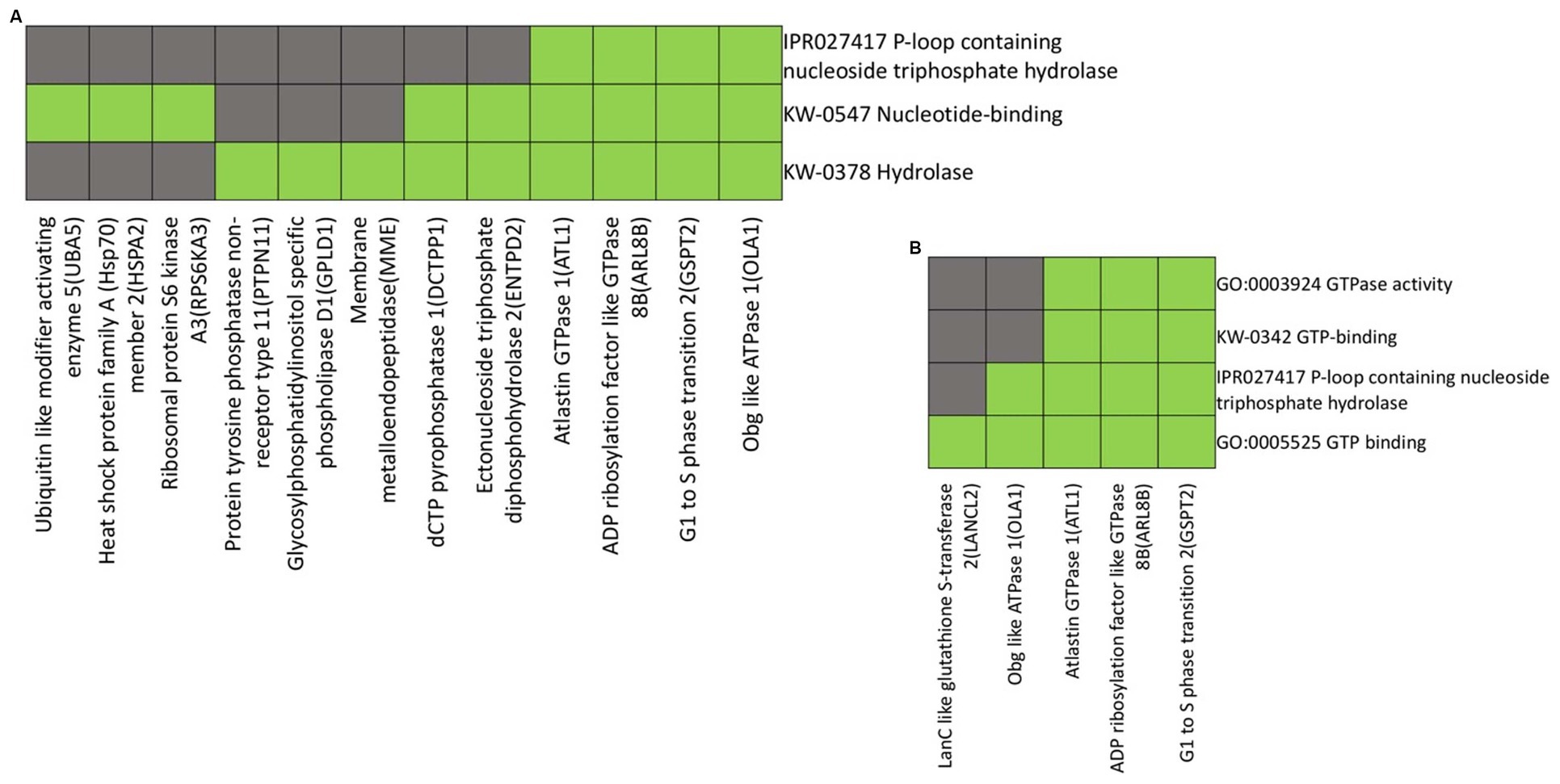
Figure 5. Heatmap presenting gene members and their associated annotation terms showing the gene–gene and term-term relationships within a group. Clusters (A and B) show associations of proteins up-regulated in the distal nerve end compared to the proximal nerve end with the top two highest enrichment score (< 1.0). Corresponding gene-term association positively reported presented in green, while corresponding gene-term association not reported yet is presented in black. The genes in the gene group are presented at the y axis of the heatmap, the annotation terms at the x axis.
The clustering proteins LanC like glutathione S-transferase 2 (LANCL2), Obg like ATPase 1 (OLA1), Atlastin GTPase 1 (ATL1), ADP Ribosylation Factor Like GTPase 8B (ARL8B) and G1 to S phase Transition 2 (GSPT2) share a common functional annotation related to GTPase activity, GTP binding as well as a P-loop containing nucleoside triphosphate hydrolase domain (Figure 5B).
3.2.3.2 Proteins down-regulated in the distal nerve end
The functional annotations and clustering were performed on proteins down-regulated in the distal nerve end compared to the proximal nerve end. The proteins Microtubule Associated Protein 1B (MAP1B), Stathmin 2 (STMN2), Dystrobrevin Binding Protein 1 (DTNBP1), Synaptosome Associated Protein 25 (SNAP25), Platelet Activating Factor Acetylhydrolase 1b Regulatory Subunit 1 (PAFAH1B1), Kinesin Family Member 5C (KIF5C), Kinesin Light Chain 1 (KLC1), Dynamin 2 (DNM2), IQ Motif Containing GTPase Activating Protein 1 (IQGAP1), Microtubule Associated Protein 1A (MAP1A), and Mesencephalic Astrocyte Derived Neurotrophic Factor (MANF) exhibit shared functional annotations related to neuron projection development, neuronal cell body, and growth cones, as indicated by Gene Ontology (GO) terms. (Figure 6A).
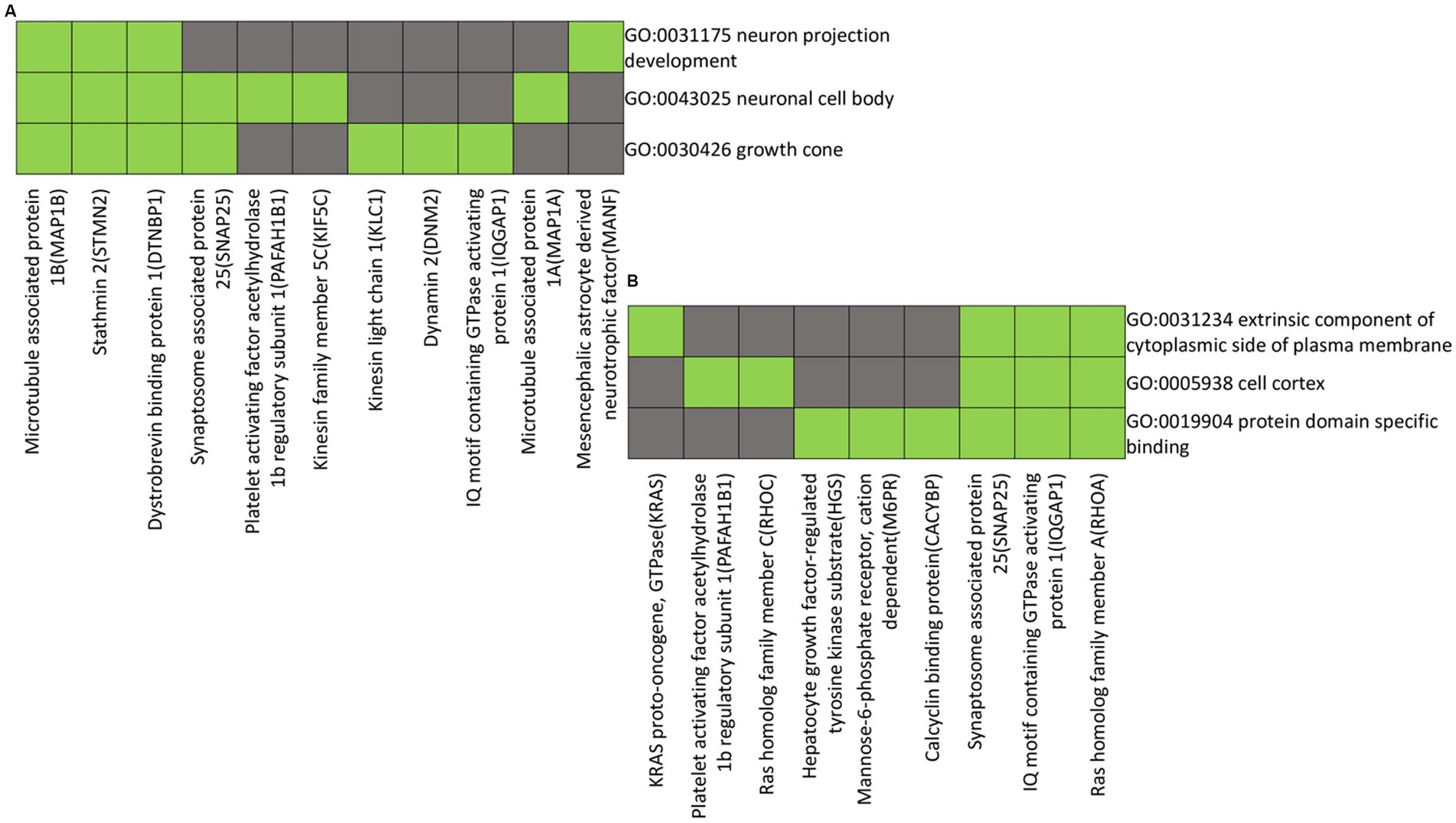
Figure 6. Heatmap presenting gene members and their associated annotation terms showing the gene–gene and term-term relationships within a group. Clusters (A and B) show associations of proteins with down-regulated in the distal compared to the proximal nerve ends with the top two highest enrichment score (< 2.0). Corresponding gene-term association positively reported presented in green, while corresponding gene-term association not reported yet is presented in black. The genes in the gene group are presented at the y axis of the heatmap, the annotation terms at the x axis.
The functional annotations and clustering of proteins KRAS proto-oncogene, GTPase (KRAS), Platelet Activating Factor Acetylhydrolase 1b Regulatory Subunit 1 (PAFAH1B1), Ras Homolog Family Member C (RHOC), Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (HGS), Mannose-6-Phosphate Receptor, Cation Dependent (M6PR), Calcyclin Binding Protein (CACYBP), Synaptosome Associated Protein 25 (SNAP25), IQ Motif Containing GTPase Activating Protein 1 (IQGAP1), and Ras Homolog Family Member A (RHOA) show them to exhibit shared functional annotations related to the extrinsic component of the cytoplasmic side of the plasma membrane, cell cortex, and protein domain-specific binding, as indicated by Gene Ontology (GO) terms (Figure 6B).
4 Discussion
In this study, 3,914 proteins were identified in clear-cut injured digital nerves of the hand within 48 h post-injury. The proteomic pattern, characterized by up- and down-regulated abundances of proteins in the proximal and distal nerve ends, were investigated in connection with surgery before the nerve repair. Comparative analysis revealed 127 proteins with significant differences in regulation, with 28 proteins showing the most substantial difference between the distal and proximal nerve ends. Cross-referencing these findings with single cell RNA sequencing data from The Human Protein Atlas (HPA) (Karlsson et al., 2021) and The Injured Sciatic Nerve Atlas (iSNAT) (Zhao et al., 2022) confirmed the presences of 26 of these proteins in nerve tissue, more importantly the SCs.
During the first 48 h post-injury, the focus on the events taking place in the injured nerve is on degeneration, where axons disintegrate, and myelin sheaths break down, while SCs activate, and macrophages are recruited (Dahlin, 2023).
Our analysis of up-regulated proteins in the distal nerve end identified protein that are expressed in the axons and SCs (Karlsson et al., 2021; Zhao et al., 2022) (Table 1), as well as potentially other related cells in the nerve trunk during the process were our data and interpretation suggests significant roles in cellular processes of these cells during the nerve degeneration. The up-regulation of EGFL8, a protein highly expressed in the axons and SCs (Karlsson et al., 2021), in response to nerve injury could serve as an axon-saving mechanism facilitating early axonal outgrowth and guidance through enhanced cell adhesion, migration, and angiogenesis (Yi and Dahlin, 2010; Weiss et al., 2021) since the sprouting mechanism is initiated early. Integrin Subunit Alpha 2 (ITGA2) may guide axonal regeneration, given its role in cell adhesion and migration (Nieuwenhuis et al., 2018; Tilot et al., 2018) by promoting interactions between extracellular matrix components and guidance signals (Nieuwenhuis et al., 2018). Further, BORCS7, essential in axonal transport of lysosomes (Snouwaert et al., 2018) showed an increased expression in the distal nerve end, indicating enhanced autophagy and lysosomal degradation pathways, aiding cellular clearance processes and potentially contributing to energy conserving mechanism. Axonal transport may also continue in the distal nerve end for some time after nerve injury with accumulation of transported substances distal to the injury (Dahlin et al., 1986; Curtis et al., 1994; Lindwall and Kanje, 2005). The presence of increased SLC12A2 in the distal nerve end suggests enhanced neuronal excitability and neurotransmitter release (Ayka and Şehirli, 2020; Hu et al., 2020), crucial for synaptic plasticity and regeneration. SLC12A2, also known as NKCC1 (Zhang et al., 2021), may regulate ion transport to maintain cellular homeostasis and enhance inter-synaptic cross talk necessary for neuronal function and restoratio (Virtanen et al., 2020). Additionally, elevated HSPA2(HSP70) expression, previously identified in biopsies of the terminal branch of the posterior interosseous nerve in the forearm of type 1 diabetes (Ising et al., 2023), in the distal nerve end acting as a molecular chaperone might assist proper protein folding and degradation (Stetler et al., 2010), promoting neuronal survival and functional recovery. Its function in maintaining protein homeostasis is critical for neuronal survival and regeneration post-nerve injury (Moin et al., 2020). The UBA5 participates in activating the ubiquitin-fold modifier 1 protein (UFM1) (Fujihara and Ikawa, 2016), which regulates cellular stress responses and protein quality control mechanisms. Efficient protein quality control is important for maintaining proteostasis during nerve degeneration, and UBA5 up-regulation may enhance cellular stress responses, facilitating proper protein turnover and promoting neuronal survival.
While HSBP1 direct impact on nerve injury remains uncertain, its role in regulating the heat shock response may indicate the initial response to cellular stress immediately upon injury. It regulates heat shock factors (HSFs) promoting the expression of heat shock proteins like HSP27, crucial for protecting neurons from stress-induced injury and aiding regeneration (Almeida-Souza et al., 2011; Ylikallio et al., 2015). Given the substantial stress experienced by the distal nerve end of the injured nerve due to the nerve fiber injury and the inflammatory response, the up-regulation of HSBP1 could activate protective and survival mechanisms of the neurons and SCs facilitating cellular restoration (Collier and Benesch, 2020). Finally, FAM3C is known to be involved in metabolic regulation and resolving inflammation (Nakano et al., 2022) and may contribute to creating a supportive environment for stable nerve degeneration in the distal nerve end.
For the down-regulated proteins in the distal nerve end compared to the proximal nerve end, the proteins previously identified were found to be expressed in SCs (Zhao et al., 2022) (Table 2). This may suggest these proteins have important roles in the SCs during the nerve injury response, but their reduced abundance in the distal nerve end compared to the proximal end may indicate a functional shift or reduction. However, their alterations over time after the injury have to be clarified. Impaired axonal guidance signals in the distal nerve end may contribute to a decreased expression of TSPAN6 (Hemler, 2001) involved in cell adhesion and migration (Humbert et al., 2022). WFS1 has primarily been associated with Wolfram syndrome, a genetic disorder characterized by optic atrophy and peripheral neuropathy. While its precise involvement in nerve injury is not fully understood, its functions in calcium homeostasis and endoplasmic reticulum stress response (Takei et al., 2006) suggest potential relevance in the degeneration process because of the calcium dependent proteases needed for the degeneration process (George et al., 1995). During nerve injury calcium dysregulation can trigger various cellular processes, including activation of calpain proteases (Metwally et al., 2023), which contributes to cytoskeletal disintegration (Ma et al., 2013). Additionally cellular resources are diverted towards managing endoplasmic reticulum stress, there may be a concurrent decrease in WFS1 expression, potentially contributing to the degeneration process in the distal nerve end after injury. For SNAP25 and PCSK1N, the reduced levels of these proteins in the distal end could imply the breakdown of synaptic vesicle regulation and prohormone processing in the distal segment post-injury. SNAP25 has garnered interest not only for its role in synaptic transmission, but also for its implications in neuronal regeneration and axonal outgrowth post-injury (Osen-Sand et al., 1993; Tao-Cheng et al., 2000; Noor and Zahid, 2016; Batista et al., 2017). For RHOC, lower abundance distally might reflect specific regulatory changes in cellular adhesion and migration processes at the site of injury (Qu et al., 2019). Further HMGB3, a protein involved in DNA binding and repair, shows decreased levels distally, potentially being an indication of reduced cellular proliferation or stress responses in this region (Stros, 2010). The MAP1LC3A was among the first markers identified to associate with autophagosome structures and is the most widely used markers for autophagy (Osen-Sand et al., 1993; Yamamoto and Yue, 2014; Bonam et al., 2020). The role of autophagy in the maintenance of cell homeostasis is well documented (Bonam et al., 2020), showing that autophagy clearance mechanisms can improve the microenvironment and provide basal energy for SC survival (Huang et al., 2016). Further, alterations in autophagic activities have been linked to neuropathic pain after nerve injury (Liao et al., 2022). We lack an explanation for the observed down-regulation in MAP1LC3A, where the findings imply a potential delay in autophagic processes responsible for cellular clearance of damaged components (Osman et al., 2015; Mohseni et al., 2017), findings which have to be clarified in the future in response to nerve injury over time.
The functional annotations and clustering of up-regulated proteins in the distal nerve end suggest their association with nucleotide-binding proteins crucial roles for signaling, energy metabolism, and gene regulation. These proteins, linked to enhanced cellular signaling pathways, are involved in neuronal degeneration and restoration (The UniProt Consortium, 2017). Many of these proteins contain the P-loop nucleoside triphosphate hydrolase domain (IPR027417), indicating roles in ATPase activity (The UniProt Consortium, 2017; Kozlova et al., 2022), essential for energy metabolism, vesicular trafficking, protein folding, and cytoskeletal dynamics. Increase in ATPase activity in the injured distal nerve end supports the energy-demanding processes of nerve restoration and axonal regeneration. This up-regulation suggests an increase in energy dependent processes for nerve degeneration.
Functional annotations and clustering show that proteins down-regulated in the distal nerve end are involved in development and maintenance of sprouts and growth cones, which guide axonal growth. The down-regulation suggest that these processes have not yet begun in the distal nerve end as the nerve repair was performed after the harvest of the nerve pieces and done within 48 h post-injury. Early axonal sprouting and rapid crossing of a repaired gap can occur after nerve repair (Holmquist et al., 1993; Kerns et al., 1993; Dahlin, 2008). Proteins like MAP1B and MAP1A essential for stabilizing microtubules in growth cones and regulating axonal outgrowth, are naturally down-regulated in the distal end before axons sprouting after the nerve repair (Noiges et al., 2002; Sainath and Gallo, 2015). Similarly, STMN2, which aids in microtubule dynamics and sprout formation (Noiges et al., 2002; Krus et al., 2022) is also down-regulated. A delay in autophagy activation, important for clearing damaged cellular components, may contribute to the down-regulation of MAP1LC3A in the distal nerve end.
In the distal nerve end, proteins significantly increased linked to single-organism processes, development, anatomical structure development, and transport indicating a specialized response to nerve injury focused on tissue restoration and regeneration rather than basic cellular maintenance. This upregulation suggests effort towards rebuilding and restructuring the injured nerve tissue integral to the initial stages of nerve degeneration and subsequent restoration process.
Conversely, down-regulated proteins in the distal nerve end are associated with cellular localization, cellular processes and regulatory mechanisms. This shift likely conserves energy and resources, focusing on essential degeneration and restoration. The suppression of certain cellular activities and the reduced response to external stimuli suggest a focus on internal restoration during the degenerative phase, preparing for tissue regeneration.
Up-regulated proteins are linked to neuronal and cytoplasmic elements, indicating active neuron involvement in degeneration. Structural and functional changes in nerve cell, along with active extracellular communication, aids tissue restoration. Vesicular trafficking and secretion processes support intracellular communication (Perlson et al., 2004; Sun and Cavalli, 2010; di Paolo et al., 2021; Wu et al., 2024).
Down-regulated proteins are related to intracellular organelles and the endomembrane system, indicating decreased intracellular trafficking and organelle dynamics. Reduced membrane-bound organelles and cytosolic components reflect decreased cellular activity, conserving energy for essential restoration processes.
Molecular functions of up-regulated proteins include protein binding activities that may increase molecular interactions and signaling, important for cellular regeneration. Conversely, down-regulated proteins show reduced RNA and enzyme binding, diminishing cell signaling and cytoskeletal dynamics, and reflecting cellular quiescence during the degeneration.
The study’s specific focus on human digital nerve injuries repaired within 48 h, provides clinically relevant insights. Understanding the proteomic changes specific to human nerve injuries enhances the translational potential of the findings to future clinical practice. Moreover, the study utilized advanced mass spectrometry techniques for proteomic analysis, ensuring high sensitivity and coverage of protein identification. This integration of proteomic methodologies enhances the reliability and robustness of the study findings. We also corroborated our protein findings with the RNA sequencing databases of HPA and iSNAT to verify their expression in a peripheral nerve of mice (Karlsson et al., 2021; Zhao et al., 2022). However, the study primarily examined events during the degenerative phase of nerve injury within the initial 48 h post-injury, thereby not capturing later-stage changes or long-term outcomes associated with nerve repair and regeneration. Although the study identified proteins associated with various cellular processes and pathways, functional validation through experimental assays was not conducted. Additionally, while the study did not directly address underlying diseases, age, and potentially sex differences, these factors could influence the observed proteomic changes and warrant consideration in future investigations. Another factor to consider is the influence of the injury distance from the soma on protein expression and composition. Protein concentrations in nerve segments can vary based on their distance from the neuronal cell body due to transport dynamics of protein synthesized in the soma. Although the relatively short length of digital nerves means that variations due to distance from the soma are most likely minor, this factor may potentially introduce subtle differences in protein composition, bur the digital nerves in the fingers are located anatomically at a long distance from the related nerve cell bodies in the spinal cord and dorsal root ganglia. These limitations underscore the need for further to validate the functional roles of these proteins in nerve degeneration, regeneration, and repair or reconstruction, including the factor timing of surgery as well as investigations of proteomic gradients, which may be more relevant when addressing injuries to the major nerve trunks in the upper arm and forearm. Moreover, it is important to note that interpretation of proteomic data may be subject to bias and assumptions, potentially influencing the conclusions drawn from the study. These limitations highlight the need for further research to comprehensively understand the molecular mechanisms underlying peripheral nerve injury, degeneration, and regeneration processes, facilitating the development of more effective diagnostic and therapeutic interventions for nerve injuries.
5 Conclusion
Distinct differences in abundance of 127 proteins between the distal and proximal nerve ends were found within 48 h after a human digital nerve injury before nerve repair. The up-regulated proteins in the distal nerve end suggest a specialized response during the degenerative phase aimed at tissue restoration and subsequent regeneration, while down-regulated proteins indicate a shift in cellular priorities. These insights emphasize the complexity of the nerve degeneration and regeneration processes in humans, urging further detailed exploration into the underlying molecular mechanisms, particularly in relation to time after injury and timing of surgery.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, PXD051266.
Ethics statement
The studies involving humans were approved by Regional Ethical Review Board in Lund, Sweden (no 311/2016). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DF: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. CW: Data curation, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. RP: Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. LD: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Swedish Research Council [#2022–01942], the Swedish Diabetes Foundation [#DIA2020-492], the Regional Agreement on Medical Training and Clinical Research [ALF] between Region Skåne and Lund University [#2018-Projekt 0104] and Funds from Skåne University Hospital [#2019–659], and Elly Olsson’s Foundation for scientific research.
Acknowledgments
We thank Lena Stenberg, Lund University for help with handling and storage of the fresh frozen specimens. We would like to thank the Center for Translational Proteomics at Medical Faculty, Lund University for providing facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2024.1425780/full#supplementary-material
References
Almeida-Souza, L., Asselbergh, B., d'Ydewalle, C., Moonens, K., Goethals, S., de Winter, V., et al. (2011). Small heat-shock protein HSPB1 mutants stabilize microtubules in Charcot-Marie-tooth neuropathy. J. Neurosci. 31, 15320–15328. doi: 10.1523/jneurosci.3266-11.2011
Ayka, A., and Şehirli, A. (2020). The role of the SLC transporters protein in the neurodegenerative disorders. Clin Psychopharmacol. Neurosci. 18, 174–187. doi: 10.9758/cpn.2020.18.2.174
Batista, A. F. R., Martínez, J. C., and Hengst, U. (2017). Intra-axonal synthesis of SNAP25 is required for the formation of presynaptic terminals. Cell Rep. 20, 3085–3098. doi: 10.1016/j.celrep.2017.08.097
Bolívar, S., Navarro, X., and Udina, E. (2020). Schwann cell role in selectivity of nerve regeneration. Cells 9:2131. doi: 10.3390/cells9092131
Bonam, S. R., Bayry, J., Tschan, M. P., and Muller, S. (2020). Progress and challenges in the use of MAP1LC3 as a legitimate marker for measuring dynamic autophagy in vivo. Cells 9:1321. doi: 10.3390/cells9051321
Clements, M. P., Byrne, E., Camarillo Guerrero, L. F., Cattin, A. L., Zakka, L., Ashraf, A., et al. (2017). The wound microenvironment reprograms Schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron 96, 98–114.e7. doi: 10.1016/j.neuron.2017.09.008
Collier, M. P., and Benesch, J. L. P. (2020). Small heat-shock proteins and their role in mechanical stress. Cell Stress Chaperones 25, 601–613. doi: 10.1007/s12192-020-01095-z
Court, F. A., Hendriks, W. T., MacGillavry, H. D., Alvarez, J., and van Minnen, J. (2008). Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 28, 11024–11029. doi: 10.1523/jneurosci.2429-08.2008
Curtis, R., Scherer, S. S., Somogyi, R., Adryan, K. M., Ip, N. Y., Zhu, Y., et al. (1994). Retrograde axonal transport of LIF is increased by peripheral nerve injury: correlation with increased LIF expression in distal nerve. Neuron 12, 191–204. doi: 10.1016/0896-6273(94)90163-5
Dahlin, L. B. (2008). Techniques of peripheral nerve repair. Scand. J. Surg. 97, 310–316. doi: 10.1177/145749690809700407
Dahlin, L. B. (2023). The dynamics of nerve degeneration and regeneration in a healthy milieu and in diabetes. Int. J. Mol. Sci. 24:15241. doi: 10.3390/ijms242015241
Dahlin, L. B., Sjöstrand, J., and McLean, W. G. (1986). Graded inhibition of retrograde axonal transport by compression of rabbit vagus nerve. J. Neurol. Sci. 76, 221–230. doi: 10.1016/0022-510x(86)90170-x
Danielsen, N., Dahlin, L. B., Lee, Y. F., and Lundborg, G. (1983). Axonal growth in mesothelial chambers. The role of the distal nerve segment. Scand. J. Plast. Reconstr. Surg. 17, 119–125. doi: 10.3109/02844318309013106
di Paolo, A., Farias, J., Garat, J., Macklin, A., Ignatchenko, V., Kislinger, T., et al. (2021). Rat sciatic nerve Axoplasm proteome is enriched with ribosomal proteins during regeneration processes. J. Proteome Res. 20, 2506–2520. doi: 10.1021/acs.jproteome.0c00980
Frostadottir, D, Welinder, C, Perez, R, and Dahlin, L. B. (2024). Quantitative Mass Spectrometry Analysis of Fresh Frozen Human Sural Nerve: Optimizing Methods for Enhanced Nerve Regeneration Research. J. Proteome Res. (Under revision).
Fujihara, Y., and Ikawa, M. (2016). GPI-AP release in cellular, developmental, and reproductive biology. J. Lipid Res. 57, 538–545. doi: 10.1194/jlr.R063032
George, E. B., Glass, J. D., and Griffin, J. W. (1995). Axotomy-induced axonal degeneration is mediated by calcium influx through ion-specific channels. J. Neurosci. 15, 6445–6452. doi: 10.1523/jneurosci.15-10-06445.1995
He, Q., Yu, F., Cong, M., Ji, Y., Zhang, Q., and Ding, F. (2021). Comparative proteomic analysis of differentially expressed proteins between injured sensory and motor nerves after peripheral nerve transection. J. Proteome Res. 20, 1488–1508. doi: 10.1021/acs.jproteome.0c00639
Hemler, M. E. (2001). Specific tetraspanin functions. J. Cell Biol. 155, 1103–1108. doi: 10.1083/jcb.200108061
Holmquist, B., Kanje, M., Kerns, J. M., and Danielsen, N. (1993). A mathematical model for regeneration rate and initial delay following surgical repair of peripheral nerves. J. Neurosci. Methods 48, 27–33. doi: 10.1016/s0165-0270(05)80004-4
Hu, C., Tao, L., Cao, X., and Chen, L. (2020). The solute carrier transporters and the brain: physiological and pharmacological implications. Asian J. Pharm. Sci. 15, 131–144. doi: 10.1016/j.ajps.2019.09.002
Huang, H. C., Chen, L., Zhang, H. X., Li, S. F., Liu, P., Zhao, T. Y., et al. (2016). Autophagy promotes peripheral nerve regeneration and motor recovery following sciatic nerve crush injury in rats. J. Mol. Neurosci. 58, 416–423. doi: 10.1007/s12031-015-0672-9
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huebner, E. A., and Strittmatter, S. M. (2009). Axon regeneration in the peripheral and central nervous systems. Results Probl. Cell Differ. 48, 339–351. doi: 10.1007/400_2009_19
Humbert, P., Pryjda, T. Z., Pranjic, B., Farrell, A., Fujikura, K., de Matos Simoes, R., et al. (2022). TSPAN6 is a suppressor of Ras-driven cancer. Oncogene 41, 2095–2105. doi: 10.1038/s41388-022-02223-y
Ising, E., Åhrman, E., Thomsen, N. O. B., Åkesson, A., Malmström, J., and Dahlin, L. B. (2023). Quantification of heat shock proteins in the posterior interosseous nerve among subjects with type 1 and type 2 diabetes compared to healthy controls. Front. Neurosci. 17:1227557. doi: 10.3389/fnins.2023.1227557
Jessen, K. R., and Mirsky, R. (2016). The repair Schwann cell and its function in regenerating nerves. J. Physiol. 594, 3521–3531. doi: 10.1113/JP270874
Jessen, K. R., and Mirsky, R. (2019). The success and failure of the Schwann cell response to nerve injury. Front. Cell. Neurosci. 13:33. doi: 10.3389/fncel.2019.00033
Jessen, K. R., and Mirsky, R. (2021). The role of c-Jun and autocrine signaling loops in the control of repair Schwann cells and regeneration. Front. Cell. Neurosci. 15:820216. doi: 10.3389/fncel.2021.820216
Karlsson, M., Zhang, C., Méar, L., Zhong, W., Digre, A., Katona, B., et al. (2021). A single–cell type transcriptomics map of human tissues. Sci. Adv. 7:eabh2169. doi: 10.1126/sciadv.abh2169
Kerns, J. M., Danielsen, N., Holmquist, B., Kanje, M., and Lundborg, G. (1993). The influence of predegeneration on regeneration through peripheral nerve grafts in the rat. Exp. Neurol. 122, 28–36. doi: 10.1006/exnr.1993.1104
Kozlova, M. I., Shalaeva, D. N., Dibrova, D. V., and Mulkidjanian, A. Y. (2022). Common patterns of hydrolysis initiation in P-loop fold nucleoside triphosphatases. Biomol. Ther. 12:1345. doi: 10.3390/biom12101345
Krishnan, A., Verge, V. M. K., and Zochodne, D. W. (2024). Hallmarks of peripheral nerve injury and regeneration. Handb. Clin. Neurol. 201, 1–17. doi: 10.1016/b978-0-323-90108-6.00014-4
Krus, K. L., Strickland, A., Yamada, Y., Devault, L., Schmidt, R. E., Bloom, A. J., et al. (2022). Loss of Stathmin-2, a hallmark of TDP-43-associated ALS, causes motor neuropathy. Cell Rep. 39:111001. doi: 10.1016/j.celrep.2022.111001
Liao, M. F., Lu, K. T., Hsu, J. L., Lee, C. H., Cheng, M. Y., and Ro, L. S. (2022). The role of autophagy and apoptosis in neuropathic pain formation. Int. J. Mol. Sci. 23:2685. doi: 10.3390/ijms23052685
Lindwall, C., and Kanje, M. (2005). Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol. Cell. Neurosci. 29, 269–282. doi: 10.1016/j.mcn.2005.03.002
Lundborg, G. (1987). Nerve regeneration and repair. A review. Acta Orthop. Scand. 58, 145–169. doi: 10.3109/17453678709146461
Ma, M., Ferguson, T. A., Schoch, K. M., Li, J., Qian, Y., Shofer, F. S., et al. (2013). Calpains mediate axonal cytoskeleton disintegration during Wallerian degeneration. Neurobiol. Dis. 56, 34–46. doi: 10.1016/j.nbd.2013.03.009
Metwally, E., Al-Abbadi, H. A., Hussain, T., Murtaza, G., Abdellatif, A. M., and Ahmed, M. F. (2023). Calpain signaling: from biology to therapeutic opportunities in neurodegenerative disorders. Front. Vet. Sci. 10:1235163. doi: 10.3389/fvets.2023.1235163
Mohseni, S., Badii, M., Kylhammar, A., Thomsen, N. O. B., Eriksson, K. F., Malik, R. A., et al. (2017). Longitudinal study of neuropathy, microangiopathy, and autophagy in sural nerve: implications for diabetic neuropathy. Brain Behav. 7:e00763. doi: 10.1002/brb3.763
Moin, A. S. M., Nandakumar, M., Diane, A., Dehbi, M., and Butler, A. E. (2020). The role of heat shock proteins in type 1 diabetes. Front. Immunol. 11:612584. doi: 10.3389/fimmu.2020.612584
Nakano, M., Imamura, R., Sugi, T., and Nishimura, M. (2022). Human FAM3C restores memory-based thermotaxis of Caenorhabditis elegans famp-1/m70.4 loss-of-function mutants. PNAS Nexus. 1:pgac242. doi: 10.1093/pnasnexus/pgac242
Neurotropism and nerve growth (1987). The evolution of a concept. Acta Orthop. Scand. 58, 91–92. doi: 10.3109/17453678709146446
Nieuwenhuis, B., Haenzi, B., Andrews, M. R., Verhaagen, J., and Fawcett, J. W. (2018). Integrins promote axonal regeneration after injury of the nervous system. Biol. Rev. Camb. Philos. Soc. 93, 1339–1362. doi: 10.1111/brv.12398
Nocera, G., and Jacob, C. (2020). Mechanisms of Schwann cell plasticity involved in peripheral nerve repair after injury. Cell. Mol. Life Sci. 77, 3977–3989. doi: 10.1007/s00018-020-03516-9
Noiges, R., Eichinger, R., Kutschera, W., Fischer, I., Németh, Z., Wiche, G., et al. (2002). Microtubule-associated protein 1A (MAP1A) and MAP1B: light chains determine distinct functional properties. J. Neurosci. 22, 2106–2114. doi: 10.1523/jneurosci.22-06-02106.2002
Noor, A., and Zahid, S. (2016). A review of the role of synaptosomal-associated protein 25 (SNAP- 25) in neurological disorders. Int. J. Neurosci. 127, 805–811. doi: 10.1080/00207454.2016.1248240
Osen-Sand, A., Catsicas, M., Staple, J. K., Jones, K. A., Ayala, G., Knowles, J., et al. (1993). Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature 364, 445–448. doi: 10.1038/364445a0
Osman, A. A., Dahlin, L. B., Thomsen, N. O., and Mohseni, S. (2015). Autophagy in the posterior interosseous nerve of patients with type 1 and type 2 diabetes mellitus: an ultrastructural study. Diabetologia 58, 625–632. doi: 10.1007/s00125-014-3477-4
Perlson, E., Medzihradszky, K. F., Darula, Z., Munno, D. W., Syed, N. I., Burlingame, A. L., et al. (2004). Differential proteomics reveals multiple components in retrogradely transported axoplasm after nerve injury. Mol. Cell. Proteomics 3, 510–520. doi: 10.1074/mcp.M400004-MCP200
Qu, L., Pan, C., He, S. M., Lang, B., Gao, G. D., Wang, X. L., et al. (2019). The Ras superfamily of small GTPases in non-neoplastic cerebral diseases. Front. Mol. Neurosci. 12:121. doi: 10.3389/fnmol.2019.00121
Raivich, G. (2011). Transcribing the path to neurological recovery-from early signals through transcription factors to downstream effectors of successful regeneration. Ann. Anat. 193, 248–258. doi: 10.1016/j.aanat.2011.01.010
Rozenbaum, M., Rajman, M., Rishal, I., Koppel, I., Koley, S., Medzihradszky, K. F., et al. (2018). Translatome regulation in neuronal injury and axon regrowth. eNeuro 5, ENEURO.0276–ENEU17.2018. doi: 10.1523/eneuro.0276-17.2018
Sainath, R., and Gallo, G. (2015). Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 359, 267–278. doi: 10.1007/s00441-014-1955-0
Saito, H., Kanje, M., and Dahlin, L. B. (2010). Crossed over repair of the femoral sensory and motor branches influences N-CAM. Neuroreport 21, 841–845. doi: 10.1097/WNR.0b013e32833d40e2
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–w221. doi: 10.1093/nar/gkac194
Snouwaert, J. N., Church, R. J., Jania, L., Nguyen, M. T., Wheeler, M. L., Saintsing, A., et al. (2018). A mutation in the Borcs7 subunit of the lysosome regulatory BORC complex results in motor deficits and dystrophic Axonopathy in mice. Cell Rep. 24, 1254–1265. doi: 10.1016/j.celrep.2018.06.118
Stetler, R. A., Gan, Y., Zhang, W., Liou, A. K., Gao, Y., Cao, G., et al. (2010). Heat shock proteins: cellular and molecular mechanisms in the central nervous system. Prog. Neurobiol. 92, 184–211. doi: 10.1016/j.pneurobio.2010.05.002
Stros, M. (2010). HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta 1799, 101–113. doi: 10.1016/j.bbagrm.2009.09.008
Sulaiman, W., and Gordon, T. (2013). Neurobiology of peripheral nerve injury, regeneration, and functional recovery: from bench top research to bedside application. Ochsner J. 13, 100–108.
Sun, F., and Cavalli, V. (2010). Neuroproteomics approaches to decipher neuronal regeneration and degeneration. Mol. Cell. Proteomics 9, 963–975. doi: 10.1074/mcp.R900003-MCP200
Takei, D., Ishihara, H., Yamaguchi, S., Yamada, T., Tamura, A., Katagiri, H., et al. (2006). WFS1 protein modulates the free ca(2+) concentration in the endoplasmic reticulum. FEBS Lett. 580, 5635–5640. doi: 10.1016/j.febslet.2006.09.007
Tao-Cheng, J. H., Du, J., and McBain, C. J. (2000). Snap-25 is polarized to axons and abundant along the axolemma: an immunogold study of intact neurons. J. Neurocytol. 29, 67–77. doi: 10.1023/a:1007168231323
The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–d169. doi: 10.1093/nar/gkw1099
Tilot, A. K., Kucera, K. S., Vino, A., Asher, J. E., Baron-Cohen, S., and Fisher, S. E. (2018). Rare variants in axonogenesis genes connect three families with sound-color synesthesia. Proc. Natl. Acad. Sci. USA 115, 3168–3173. doi: 10.1073/pnas.1715492115
Vergara, D., Romano, A., Stanca, E., la Pesa, V., Aloisi, L., de Domenico, S., et al. (2018). Proteomic expression profile of injured rat peripheral nerves revealed biological networks and processes associated with nerve regeneration. J. Cell. Physiol. 233, 6207–6223. doi: 10.1002/jcp.26478
Virtanen, M. A., Uvarov, P., Hübner, C. A., and Kaila, K. (2020). NKCC1, an elusive molecular target in brain development: making sense of the existing data. Cells 9:2607. doi: 10.3390/cells9122607
Weiss, T., Taschner-Mandl, S., Janker, L., Bileck, A., Rifatbegovic, F., Kromp, F., et al. (2021). Schwann cell plasticity regulates neuroblastic tumor cell differentiation via epidermal growth factor-like protein 8. Nat. Commun. 12:1624. doi: 10.1038/s41467-021-21859-0
Wu, W., Zhang, J., Chen, Y., Chen, Q., Liu, Q., Zhang, F., et al. (2024). Genes in axonal regeneration. Mol. Neurobiol. doi: 10.1007/s12035-024-04049-z
Yamamoto, A., and Yue, Z. (2014). Autophagy and its Normal and pathogenic states in the brain. Annu. Rev. Neurosci. 37, 55–78. doi: 10.1146/annurev-neuro-071013-014149
Yi, C., and Dahlin, L. B. (2010). Impaired nerve regeneration and Schwann cell activation after repair with tension. Neuroreport 21, 958–962. doi: 10.1097/WNR.0b013e32833e787f
Ylikallio, E., Konovalova, S., Dhungana, Y., Hilander, T., Junna, N., Partanen, J. V., et al. (2015). Truncated HSPB1 causes axonal neuropathy and impairs tolerance to unfolded protein stress. BBA Clin. 3, 233–242. doi: 10.1016/j.bbacli.2015.03.002
Zhang, S., Zhou, J., Zhang, Y., Liu, T., Friedel, P., Zhuo, W., et al. (2021). The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2. Commun. Biol. 4:226. doi: 10.1038/s42003-021-01750-w
Zhao, Q., Dahlin, L. B., Kanje, M., and Lundborg, G. (1993). Repair of the transected rat sciatic nerve: matrix formation within implanted silicone tubes. Restor. Neurol. Neurosci. 5, 197–204. doi: 10.3233/rnn-1993-5304
Zhao, X. F., Huffman, L. D., Hafner, H., Athaiya, M., Finneran, M. C., Kalinski, A. L., et al. (2022). The injured sciatic nerve atlas (iSNAT), insights into the cellular and molecular basis of neural tissue degeneration and regeneration. eLife 11:e80881. doi: 10.7554/eLife.80881
Keywords: digital nerve injury, quantitative mass spectrometry, proteomics, peripheral nerve injury, nerve injury pathways, signal transduction, extracellular matrix
Citation: Frostadottir D, Welinder C, Perez R and Dahlin LB (2024) Quantitative mass spectrometry analysis of the injured proximal and distal human digital nerve ends. Front. Mol. Neurosci. 17:1425780. doi: 10.3389/fnmol.2024.1425780
Edited by:
Richard Zigmond, Case Western Reserve University, United StatesReviewed by:
Roman J. Giger, University of Michigan, United StatesDianna E. Willis, Burke-Cornell Medical Research Institute, United States
Copyright © 2024 Frostadottir, Welinder, Perez and Dahlin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Drifa Frostadottir, ZHJpZmEuZnJvc3RhZG90dGlyQG1lZC5sdS5zZQ==
 Drifa Frostadottir
Drifa Frostadottir Charlotte Welinder3
Charlotte Welinder3 Lars B. Dahlin
Lars B. Dahlin