
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 03 September 2024
Sec. Brain Disease Mechanisms
Volume 17 - 2024 | https://doi.org/10.3389/fnmol.2024.1417961
The progressive degeneration of motor neurons in amyotrophic lateral sclerosis (ALS) is accompanied by the formation of a broad array of cytoplasmic and nuclear neuronal inclusions (protein aggregates) largely containing RNA-binding proteins such as TAR DNA-binding protein 43 (TDP-43) or fused in sarcoma/translocated in liposarcoma (FUS/TLS). This process is driven by a liquid-to-solid phase separation generally from proteins in membrane-less organelles giving rise to pathological biomolecular condensates. The formation of these protein aggregates suggests a fundamental alteration in the mRNA expression or the levels of the proteins involved. Considering the role of the epigenome in gene expression, alterations in DNA methylation, histone modifications, chromatin remodeling, non-coding RNAs, and RNA modifications become highly relevant to understanding how this pathological process takes effect. In this review, we explore the evidence that links epigenetic mechanisms with the formation of protein aggregates in ALS. We propose that a greater understanding of the role of the epigenome and how this inter-relates with the formation of pathological LLPS in ALS will provide an attractive therapeutic target.
Amyotrophic lateral sclerosis (ALS) is characterized by the relentless degeneration of motor neurons subserving both upper and lower motor neuron functions (UMN & LMN, respectively), ultimately giving rise to progressive muscle weakness, spasticity, and atrophy, with lethal respiratory failure within two to five years of the disease onset (Strong et al., 2005; Robberecht and Philips, 2013). The majority of ALS cases are sporadic (sALS) with the remainder being genetic, often in association with family history (fALS). Approximately two out of three individuals with fALS and one out of ten individuals with sALS have a mutation in at least one of the over forty genes associated with the condition. These genes vary in the degree of evidence supporting their association with ALS, with many having limited or weak evidence (http://alsod.iop.kcl.ac.uk) (Ghasemi and Brown, 2018; Masrori and Van Damme, 2020).
While classically ALS is considered to be a pure disorder of the motor neurons, it is now accepted that over 50% of ALS patients have an associated disorder of frontotemporal dysfunction, including either presenting with or developing frontotemporal dementia (FTD) (Hardiman et al., 2017; Strong et al., 2017). The progressive degeneration of motor neurons in ALS is accompanied by the formation of a broad array of nuclear and cytoplasmic inclusions which are driven by liquid-to-solid phase separation (Chakraborty and Zweckstetter, 2023; Shen et al., 2023). Amongst a broad array of proteins associated with these inclusions are RNA-binding proteins (RBPs), of which inclusions containing the TAR DNA-binding protein 43 (TDP-43) are observed in the vast majority of degenerating neurons (Ling et al., 2013).
Considering the complexity of this disease at the molecular level (Goutman et al., 2022), the mechanisms associated with the pathogenesis of ALS are currently a priority area of study. In recent years, it has been suggested that epigenetic mechanisms could contribute to the etiology of ALS (Bennett et al., 2019). A contemporary definition of epigenetics by Cavalli and Heard indicates that it is “the study of molecules and mechanisms that can perpetuate alternative gene activity states in the context of the same DNA sequence” (Cavalli and Heard, 2019). This includes both mitotic and transgenerational inheritance, as well as the long-term maintenance of gene activity or chromatin states in the absence of cell division, with a DNA sequence that is biological system-dependent (Cavalli and Heard, 2019). Nowadays, the consensus is that epigenetics includes DNA methylation, histone modifications, and RNA-based mechanisms, a multilayer process that controls gene expression through altering DNA packing and accessibility and regulates the levels of proteins using RNA machinery (Dulac, 2010; Gibney and Nolan, 2010; Aristizabal et al., 2020; Jiang et al., 2023). The most strongly ALS-associated genes for which there is evidence of a potential association between aggregate formation and alterations in the epigenome are included in Table 1.
These mechanisms mediate the diversified gene expression profiles in a variety of cells and tissues in multicellular organisms. Studying how these mechanisms can be perturbed in neurodegeneration, and most specifically are related to pathological biomolecular condensate formation in ALS, is a critical step in understanding the etiology of the disease beyond those causes mediated by heritable protein-coding gene mutations (Sultan and Day, 2011; Hwang et al., 2017).
Liquid–liquid phase separation (LLPS) segregates proteins and nucleic acids into liquid-like structures characterized as membrane-less organelles (MLOs). These structures play a crucial role in a variety of normal biological processes, such as chromatin organization, genomic stability, DNA damage response and repair, transcription, and signal transduction, and contribute to intracellular spatiotemporal coordination (Hyman et al., 2014; Nesterov et al., 2021; Wang et al., 2021a; Chen et al., 2022b). Additionally, LLPS is critical for the control of cellular functions such as metabolic processing and cellular compartment control (Hyman et al., 2014).
Nuclear bodies (NBs) are MLOs with various nuclear functions. The nucleolus is involved in pre-rRNA transcription, processing, and ribosomal ribonucleoprotein (RNP) assembly; nuclear speckles are associated with storage and assembly of spliceosomal components; nuclear stress bodies regulate transcription and splicing under stress; paraspeckles modulate gene expression by sequestering specific mRNAs and proteins; Promyelocytic leukemia gene product (PML) play key roles in genome stability, DNA repair, control of transcription, and viral defense; and Cajal bodies are involved in the biogenesis, maturation, and recycling of small RNAs (Lamond and Spector, 2003; Handwerger and Gall, 2006; Boulon et al., 2010).
Cytoplasmic MLOs, generally known as RNP granules, are essential in mRNA metabolism and homeostasis. These molecular structures include processing bodies (P-bodies) involved in post-transcriptional gene regulation and mRNA metabolism; stress granules (SGs) that play crucial roles in regulating mRNA metabolism, translation, and stress response pathways; and germ granules that orchestrate RNA regulation, genome integrity, epigenetic regulation, and stress responses in germ cells (Kedersha et al., 2005; Buchan, 2014; Jain et al., 2016; Luo et al., 2018).
In general, the interaction of the intrinsically disordered regions (IDRs) of proteins are responsible for their phase separation. IDRs are usually made up of a limited number of amino acids and/or repetitive sequence elements, referred to as low-complexity domains (LCDs) (Pakravan et al., 2021). Many ALS-related proteins have LCDs, making them prone to forming aggregates through LLPS. In ALS, dysfunctional LLPS can lead to pathological protein aggregation (liquid-to-solid phase transition), contributing to disease progression (Pakravan et al., 2021). As a result, to avoid aggregate formation and protein accumulation, both LLPS and clearance functions must be balanced properly.
In healthy cells, TDP-43 forms various oligomeric states within the nucleus through interactions of its N-terminal domains (Afroz et al., 2017). Under pathological conditions such as observed in ALS, TDP-43 becomes predominantly cytosolic with relative nuclear clearance and forms biomolecular condensates through LLPS. Both its N-terminal and C-terminal domains contribute to aggregation. In solution, LCDs of TDP-43 can self-aggregate with higher concentrations leading to the formation of gel-like structures. ALS-linked mutations in TDP-43 decrease its liquid properties and disrupt its phase-separation behavior (Conicella et al., 2016). Similar to TDP-43, mutations in the LCD of fused in sarcoma/translocated in liposarcoma (FUS/TLS) disrupt the phase-separating properties of the protein (Han et al., 2012). Full-length FUS/TLS has been observed to form droplets in vitro, affecting its LLPS by the RNA-to-protein ratio (Maharana et al., 2018).
In pathological states, MLOs can act as sites for the accumulation of abnormal protein species, contributing to the formation of neuronal inclusions in ALS or frontotemporal dementia (FTD) (Butti and Patten, 2018; Ismail et al., 2021). Neuronal inclusions are common in a wide range of neurodegenerative diseases (Dugger and Dickson, 2017; Wilson 3rd et al., 2023); however, there remains considerable debate as to whether the presence of cellular inclusions is beneficial or toxic for the cell (Lippi and Krisko, 2023). Liquid-to-solid phase transition in pathological conditions has been observed for several RBPs, including the ALS-related proteins TDP-43 (encoded by TARDBP) and FUS/TLS (encoded by FUS) (Carey and Guo, 2022), in a process that can be accelerated by ALS-associated mutations (Patel et al., 2015). These biomolecular condensates can be observed either in the nucleus or the cytoplasm of neurons (Banani et al., 2017).
Growing evidence points to the importance of structural and functional nuclear alterations in the etiology of many neurodegenerative illnesses. The formation of neuronal intranuclear inclusions (NIIs) is a consequence of sequestering essential nuclear factors by mutant proteins or RNAs within the nucleus (Woulfe, 2007). NIIs can trigger a progressive neurodegenerative condition typified by the presence of pathologic eosinophilic hyaline intranuclear inclusions. These inclusions are observed in both the central and peripheral nervous systems as well as in multiple visceral organs (Seilhean et al., 2004; Liu et al., 2022c; Tai et al., 2023). The presence of NIIs and the malfunction of the ubiquitin-proteasome system (UPS) such as elevated levels of ubiquitinated proteins and P62 protein are common pathological traits observed in neuronal intranuclear inclusion disease (NIID) as well as other neurodegenerative conditions. NBs, such as PML, function as sites for protein degradation and are linked to the ubiquitin-proteasome pathway (Mattsson et al., 2001). Intranuclear eosinophilic inclusions displaying ubiquitin, PML gene product, proteasome, and ataxin-3 immunoreactivity have been observed within the hippocampus and motor cortex in ALS (Kakita et al., 1997; Seilhean et al., 2004). TDP-43(+) neuronal inclusions with a distribution and morphology similar to NII and neuronal cytoplasmic inclusions (NCIs) have been observed in brain samples from patients with ALS and a concomitant FTD (Cairns et al., 2007; Seelaar et al., 2007).
NCIs within degenerating motor neurons in association with UMN and LMN loss have long been the neuropathological hallmark of ALS. Classically, such inclusions were characterized based on their morphological appearance and included Bunina bodies which are small, electron dense, eosinophilic cytoplasmic ubiquitin-negative, TDP-43 positive inclusions (Tomonaga et al., 1978; Okamoto et al., 2008; Mori et al., 2014), Lewy body-like hyaline or skein-like inclusions which are ubiquitinated (Leigh et al., 1988; Lowe et al., 1988; Leigh et al., 1991; Okamoto et al., 2011); and neurofilament-rich “hyaline conglomerate inclusions” in degenerating motor neurons (Wada et al., 1999; Strong et al., 2005). The contemporary view of ALS, however, includes not only inclusions consisting of neuronal intermediate filaments but also the far more commonly observed cytoplasmic inclusions containing an array of proteins associated with RNA metabolism (Chou et al., 2018; Alberti and Dormann, 2019; Elbaum-Garfinkle, 2019). Amongst these, neuronal and glial cytoplasmic inclusions immunoreactive for TDP-43 are observed in virtually (approaching 97%) all ALS patients, except superoxide dismutase 1 (SOD1) and FUS/TLS cases (Andersen et al., 1995; Al-Sarraj et al., 2011; Boxer et al., 2011; Couthouis et al., 2011; Deng et al., 2011; Hortobagyi et al., 2011; Couthouis et al., 2012).
Little is known about the effect of NCIs directly on gene expression, but their presence can interfere with a broad range of normal cellular processes, including mRNA metabolism, protein trafficking, synaptic function, and intracellular signaling. Impaired expression of RBPs and their resulting aggregation lead to disruptions in RNA regulation, which manifest as abnormalities in processes such as splicing, polyadenylation, transport, translation, and decay of RNA targets (Kanai et al., 2004; Lagier-Tourenne et al., 2010; Nussbacher et al., 2019). Aggregates can have a broad variety of detrimental consequences in the cell, including interference with protein degradation pathways, disruption of cellular membranes, induction of oxidative stress, and activation of inflammatory responses. These toxic effects can ultimately lead to cellular dysfunction and, in severe cases, cell death (Stefanis, 2012; Prasad et al., 2019; Michalska and Leon, 2020).
While genetic mutations are well-established contributors to ALS pathology, emerging evidence suggests that epigenetic alterations also significantly influence aggregate formation in the disease. DNA methylation, histone modifications, and non-coding RNA-mediated mechanisms are among the key epigenetic processes implicated in ALS aggregate formation.
One of the most studied epigenetic processes is DNA methylation in which a covalent transfer of a methyl group from S-adenosyl methionine (SAM) to carbon-5 of the cytosine pyrimidine ring of DNA occurs to form 5-methylcytosine (5mC). This modification in the DNA effectively inhibits gene transcription when adjacent to promoter regions (Robertson, 2005; Moore et al., 2013). DNA methylation is regulated by a family of DNA methyltransferases (DNMT) and is implicated in the regulation of several cellular processes (Bestor, 2000). Conversely, DNA demethylation is promoted by the ten-eleven translocation family of proteins (TET proteins), which oxidizes 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) (Guo et al., 2013). DNA methylation occurs almost exclusively on a cytosine followed by a guanine nucleotide (CpG dinucleotide). Over 80% of CpG dinucleotides located along the gene are typically subject to methylation; in contrast, CpG islands, regions that are greater than 500 base pairs in size with high CpG content, are hypomethylated (Bird et al., 1985; Esteller, 2005). This pattern changes with age such that CpG islands become hypermethylated and silenced (Robertson, 2005), whereas non-CpG sites are hypomethylated (Egger et al., 2004; Fraga et al., 2007).
Methylation of CpG (mCpG) has a role in transcription and replication by decreasing the speed of RNA/DNA polymerase and DNA helicase while stabilizing the DNA helix and raising its melting temperature in vitro (Rausch et al., 2021). Transcription factor (TF) binding is either facilitated or prevented when DNA methylation occurs at promoters of CpG islands, leading to gene silencing (Kass et al., 1997). TFs that recognize mCpG are classified as methyl-CpG binding proteins (MBPs) (Fournier et al., 2012; Shimbo and Wade, 2016) and are associated with increased chromatin density through LLPS. Specifically, methyl-CpG-binding protein 2 (MeCP2) induces compaction and LLPS of nucleosomal arrays in vitro and further enhances the formation of chromatin condensates by DNA methylation (Wang et al., 2020).
This is of importance given the evidence that DNA methylation is altered in ALS. It has been observed that the level of DNA methylation in the blood DNA of ALS patients with SOD1 mutation is notably higher compared to asymptomatic carriers or family members without SOD1 mutations. Also, a direct relationship between overall DNA methylation levels and the duration of the disease has been found in patients with SOD1 mutations (Coppede et al., 2018). An examination of DNA methylation across the entire genome in the frontal cortex of individuals with sALS has uncovered a notable increase in the methylation levels of genes related to calcium regulation (including those for calcium channels and a sodium-calcium exchanger), neurotransmission (genes involved in glutamate transport at synaptic vesicles), oxidative stress (genes related with repair of oxidative damage, and play a role in neuroinflammation) and mechanisms that are involved in protein aggregation (Morahan et al., 2009).
There is evidence that suggests the methylation status of the DNA is linked to the formation of pathological aggregates in ALS (Table 1) (Chen et al., 2012; Chouliaras et al., 2012). Reductions in methylated DNA in the presence of high expression levels of pathological TDP-43 have been described, suggesting a relationship between TDP-43 proteinopathy and DNA methylation (Appleby-Mallinder et al., 2021). Moreover, the DNA of the autoregulatory region that codifies the TARDBP 3’untranslated regions (3’UTR) is demethylated which is associated with reduced alternative splicing, increased levels of TARDBP mRNA, and in the end, augmenting the levels of cytoplasmic TDP-43 protein in affected neurons in ALS (Figure 1A) (Koike et al., 2021).
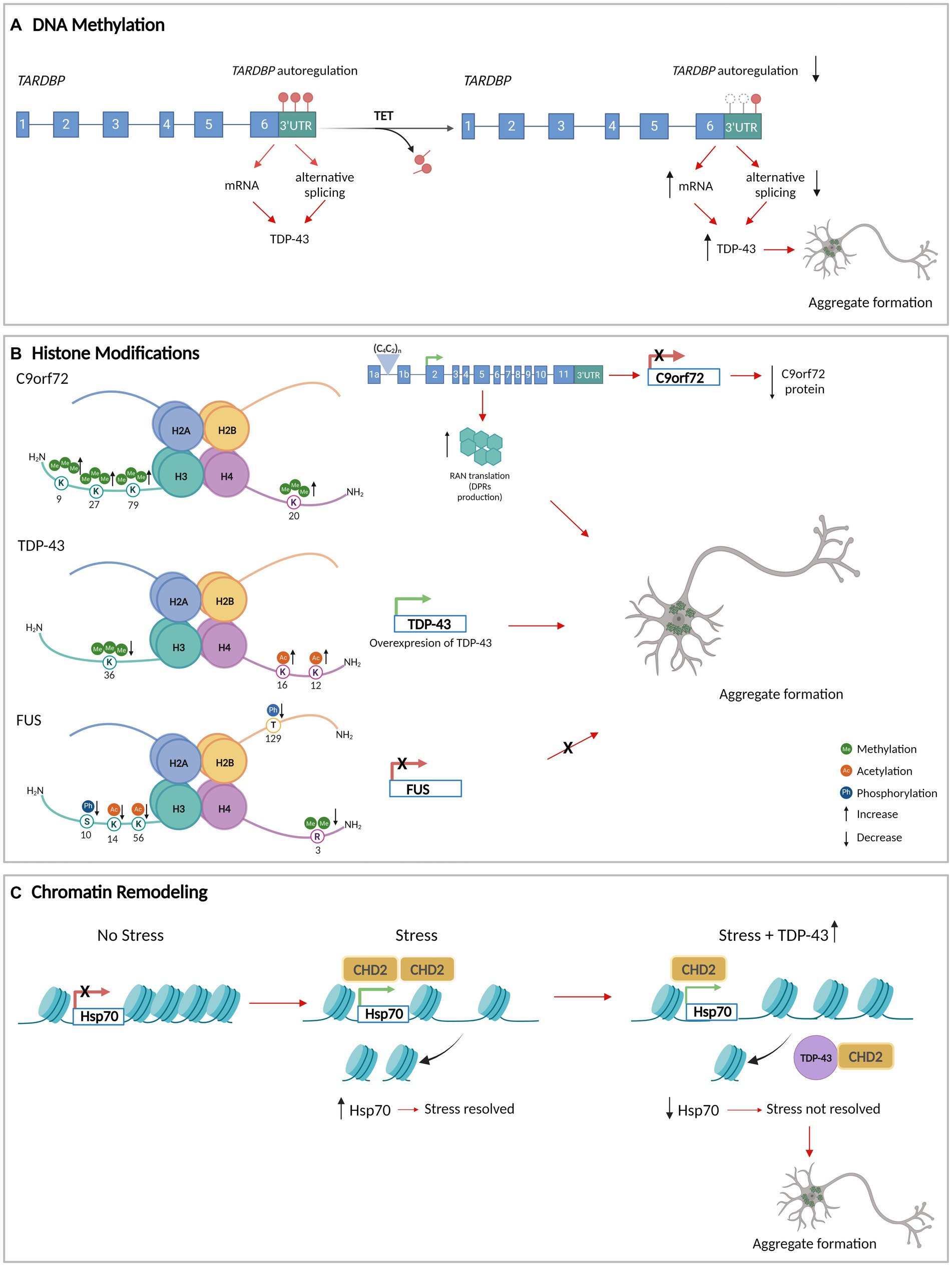
Figure 1. DNA methylation, histone modifications, and chromatin remodeling in aggregate formation in ALS. (A) The methylation of the DNA that encodes TARDBP 3’UTR (red circles) allows its autoregulation and normal levels of TDP-43. TET enzymes demethylate TARDBP 3’UTR DNA (white circles), decreasing the autoregulation and increasing the levels of TDP-43. (B) Histone modifications at the level of the genes that encode C9orf72, TDP-43, and FUS/TLS. The modifications generate an increase in the protein levels of TDP-43 with an increase in aggregate formation. For C9orf72 and FUS/TLS, histone modifications decrease the expression of the gene. In the case of C9orf72, the histones tri-methylation produces C9orf72 repeat expansions leading to repeat-associated non-AUG translation (RAN translation) resulting in dipeptide repeat proteins (DPRs) that accumulate as insoluble aggregates in the cytoplasm of neurons. (C) TDP-43 interacts with CHD2 impeding its recruitment in the chromatin. This results in the impairment of nucleosome clearance from the Hsp70 gene which prevents its activation under stress. Consequently, there is an increase in TDP-43 levels and aggregation formation. Figure created with BioRender.com.
Methylation of the chromosome 9 open reading frame 72 (C9orf72) promoter region has been observed to decrease the gene expression, which may contribute to the reduction of RNA foci and dipeptide repeat aggregates formation (Babic Leko et al., 2019). This suggests that methylation of the C9orf72 promoter could have a neuroprotective role in ALS/FTD and conversely that demethylation could have a pathological effect.
Niclosamide, an anti-helminthic drug that reduces methylation in promoter CpG islands but increments methylation in the gene body region (Jiang et al., 2022) decreases the cytoplasmic accumulation of phosphorylated TDP-43 in the motoneurons of SOD1-G93A and FUS transgenic mice models. This drug also partially prevents the cytoplasmic mislocalization of FUS/TLS in the motoneurons (Milani et al., 2024). These results show that the hypomethylation in promoter CpG islands can decrease protein aggregation.
Other studies have examined site-specific promoter methylation and the expression of FUS/TLS and DNMTs in motor neurons derived from ALS patients with FUS mutations. Reduced FUS mRNA expression and cytoplasmic mislocalization of FUS/TLS leads to a loss of its nuclear function and the formation of toxic aggregates that represent a gain of function (Hartung et al., 2021). Both mechanisms may be partially driven by altered methylation. Higher expression levels of DNMT1, DNMT2, and DNMT3a have been observed in mutant FUS motoneurons (Hartung et al., 2021) as has been an inverse correlation of FUS expression and proximal FUS promoter methylation in ALS cell lines. Despite lower FUS expression levels, mutant FUS motor neurons show significantly more pathological cytoplasmic FUS/TLS aggregates suggesting that FUS methylation and repression act as protective mechanisms to counteract the formation of cytoplasmic aggregates (Hartung et al., 2021).
Finally, it has been observed that the methylation patterns from a specific region down to a single CpG can be identified using fragmentation patterns of circulating free DNA (cfDNA) molecules (Zhou et al., 2022b). A recent study found that methylation patterns on cfDNA from ALS patients accurately predicts ALS status and disease severity (Caggiano et al., 2024). Additionally, ALS cases were distinguished from FTD and other neurological conditions by the model. These findings suggest that cfDNA methylation patterns are potential quantitative biomarkers for ALS.
Nevertheless, while DNA methylation patterns may function as epigenetic indicators in ALS, it is still not clear if DNA methylation is the cause or consequence of protein aggregation and further research is necessary on this topic.
Each nucleosome consists of DNA wrapped around a core of eight histone proteins, including two copies of each of histones H2A, H2B, H3, and H4 (Marino-Ramirez et al., 2005; Woodcock, 2005). The DNA wraps around this histone core forming the nucleosome structure together with histone H1 bound to the linker DNA. DNA segments connect adjacent nucleosomes aiding in the compaction of chromatin and the formation of higher-order chromatin structures (Widom, 1998). This organization of the chromatin helps to regulate gene expression, DNA replication, and DNA repair processes (Luger et al., 1997). The amino terminal tail of histones is exposed and subject to extensive PTMs such as acetylation (lysine residues), phosphorylation (serine or threonine residues), methylation (mono-, di-, and tri-methylation on lysine or arginine residues), SUMOylation (lysine residues), ubiquitination (lysine residues), glycosylation, and ADP-ribosylation (serine/threonine residues). These PTMs are important in gene regulation because they control the accessibility of DNA to the transcriptional machinery (both activation and inactivation) and chromatin remodeling processes without changing the DNA sequence (Becker and Workman, 2013; Stavreva and Hager, 2015; Allis and Jenuwein, 2016). Certain histone modifications, such as histone acetylation, are associated with active gene transcription, whereas methylations are associated with the formation of condensed and transcriptionally repressed chromatin (Heard et al., 2001; Bannister and Kouzarides, 2011).
The group of enzymes responsible for post-translational modification of histones includes histone acetyltransferase (HAT), histone methyltransferase (HMT), histone deacetylase (HDAC) and histone demethylase (HDM) (Hyun et al., 2017; Hwang et al., 2021; Liu et al., 2023a). Several of these enzymes are associated with ALS pathogenesis. For example, the reduction in the levels of cAMP-responsive element-binding protein (CREB)-binding protein (CBP) with HAT activity produces the histone H3 hypoacetylation detected in cholinergic motor neurons from the lumbar spinal cord in an ALS animal model (SOD1-G86R). This alteration has been associated with reduced motor neuron survival (Rouaux et al., 2003; Rouaux et al., 2007).
By acetylating histones H3 and H4, elongator complex protein 3 (ELP3) that has HAT activity directly controls the production of heat shock protein 70 (Hsp70) in yeast (Han et al., 2008). As a result, deficiencies in ELP3 may cause a reduction in Hsp70 transcription, which in turn may lead to motor neuron degeneration (Simpson et al., 2009). It has been shown that the knock-down of Hsp70 does not influence SGs assembly but results in the liquid-to-solid transition in SGs. Furthermore, by preventing the transition to a solid state, Hsp70 can assist in maintaining TDP-43-positive MLOs in a liquid state (Kitamura et al., 2018; Gu et al., 2021). Similarly, a propensity for liquid phase transition was observed with Hsp70 in FUS/TLS pathology. In this case, Hsp70 presence in FUS/TLS-related SGs correlates with a decrease in the transition from liquid to solid state (Li et al., 2022b).
In a variety of neurologic and psychiatric conditions, HDAC inhibitors can improve deficiencies in synaptic plasticity, cognition, and stress-related behaviors (Abel and Zukin, 2008). HDAC2 expression was found to be upregulated in the motor cortex and spinal cord grey matter, specifically in motor neuron nuclei of ALS patients, supporting the idea that the inhibition of HDAC2 has a protective role in the pathogenesis of ALS. The specific mechanism underlying this effect has not been described (Janssen et al., 2010). A selective increase in HDAC 2 levels has also been found in the muscle of a mouse model of muscular dystrophy, while functional and morphological parameters have been observed to improve with HDAC 2 downregulation in the same model (Minetti et al., 2006; Colussi et al., 2008). The neuroprotective role of HDAC reduction has also been observed using ACY-738, a strong HDAC inhibitor, in FUS/TLS transgenic mice. ACY-738 improves the motor phenotype and significantly increases the life span (Rossaert et al., 2019).
The link between histone modifications and aggregate formation has begun to be elucidated. In yeast, the deletion of a member of the HDAC complex known as a Set3 suppresses the toxicity of TDP-43 inclusions (Sanna et al., 2020). In SOD1 mutant cells, a connection between HDAC activity and aggresome formation has been observed through an intracellular structure that sequesters potentially toxic misfolded proteins and facilitates their clearance by autophagy. The percentage of cells containing aggresomes decreases when cells are treated with an HDAC inhibitor, implying a pathological role for HDACs (Corcoran et al., 2004). A study using a screening system for TDP-43 aggregation in mouse neuroblastoma Neuro2a cells analyzing a library of genes associated with fALS/FTD has shown that microtubule-related proteins (MRPs) and RBPs co-aggregate with TDP-43 in the cytosol via different mechanisms, involving microtubules and LLPS, respectively. TDP-43 aggregates induced by MRPs co-localize with markers for aggresomes and rely on HDAC6, further supporting a role for the aggresome formation in to aggregate formation (Watanabe et al., 2020).
The expression of HDAC4, an epigenetic factor responsive to stress, is significantly increased in the skeletal muscle of ALS patients and mice (SOD1-G93A) with denervation (Pigna et al., 2019) and is present in neuronal cytoplasmic inclusions (Federspiel et al., 2019). HDAC6 has been also implicated in protein aggregation through binding to ubiquitinated misfolded proteins (Simoes-Pires et al., 2013). Mutated SOD1 (mtSOD1) associated with ALS can regulate HDAC6 activity, leading to enhanced tubulin acetylation. This process subsequently promotes the aggregation of mtSOD1 in a process facilitated by dynein-mediated retrograde transport along microtubules (Gal et al., 2013).
Histone methylation has also been associated with ALS pathogenesis. Histone 3 (H3) trimethylation at lysine 9 (H3K9me3), 27 (H3K27me3), 79 (H3K79me3), and H4 trimethylation at lysine 20 (H4K20me3) have been shown to reduce C9orf72 gene expression in patients carrying pathological C9orf72 hexanucleotide repeat expansions (Belzil et al., 2013). These histone modifications create RNA foci that may sequester RBPs and serve as foci for C9orf72 dipeptide repeat protein aggregates formation (Ash et al., 2013; Mori et al., 2013) (Figure 1B). This suggests that the upregulation of H3 trimethylation-dependent C9orf72 expression could lead to dipeptide repeat protein aggregates formation.
It has been described that the bromodomain and extraterminal domain (BET) family of proteins that includes four conserved mammalian members of bromodomain-containing proteins (BRDs): BRD2, BRD3, BRD4, and BRDT, regulate the C9orf72 locus in ALS and that BRD inhibitors could have therapeutic potential for this disease (Zeier et al., 2015). These proteins function as epigenetic readers of histone acetylation, recruiting transcriptional regulator complexes to chromatin and binding to acetylated histones (Fujisawa and Filippakopoulos, 2017; Wang et al., 2021b; To et al., 2023).
BET inhibitors have been shown to effectively displace BET proteins from acetylated histones and elements of the transcriptional machinery (Filippakopoulos et al., 2010; Dawson et al., 2011; Delmore et al., 2011). Specifically, in primary cortical neurons from a C9orf72 ALS/FTD transgenic mouse model (C9BAC), treatment with PFI-1 and JQ1, both BRD inhibitors, enhanced the expression of the human mutant C9orf72 gene, increased the accumulation of nuclear RNA foci, and reduced poly (GP)-DPR inclusions, with reduced hippocampal-dependent cognitive impairments (Zeier et al., 2015).
Additionally, a role for a MAPK/MAK/MRK overlapping kinase (MOK), a Ser/Thr kinase in the mitogen-activated protein kinase (MAPK) superfamily, in regulating inflammatory and type-I interferon (IFN) responses in microglia through Brd4-dependent mechanisms has been uncovered (Perez-Cabello et al., 2023). MOK interacts with and colocalizes with cytoplasmic TDP-43 inclusions in microglia when these cells are exposed to external TDP-43 aggregates (Leal-Lasarte et al., 2017). It was found that MOK-mediated immune functions are dysregulated and actively contribute to ALS pathophysiology in a mouse preclinical model.
In yeast models, histone modifications and the expression of ALS-linked RBPs have been correlated. It has been observed that TDP-43wt overexpression is associated with an increase in several histone PTMs, while FUS/TLS overexpression is associated with a decrease in histone modification levels. In the TDP-43 model, hyperacetylation of histone H4 on lysine 12 (H4K12ac) and 16 (H4K16ac) and reduced levels of trimethylation in histone H3 on lysine 36 (H3K36me3) have been observed, both of which result in increased TARDBP transcription (Figure 1B) (Chen et al., 2018). In the FUS/TLS yeast model, a decrease of dimethylation on arginine 3 of histone H4 (H4R3me2) has been observed as has been reduced levels of phosphorylation of H3 on serine 10 (H3S10ph) and H2B on threonine 129 (H2BT129ph), and hypoacetylation of H3 on lysine 14 (H3K14ac) and 56 (H3K56ac). These histone PTM changes collectively decrease FUS transcription (Chen et al., 2018) (Figure 1B). This suggests that hypermethylation of arginine 3, hyperphosphorylation of serine 10 and threonine 129, and hyperacetylation of lysine 14 and lysine 56 could be associated with an increase in FUS/TLS aggregate formation.
The chromatin remodeling complex is constituted by a group of proteins responsible for modulating chromatin architecture and is an essential component of the DNA damage response (Chou et al., 2010). This complex regulates gene expression temporally and spatially by altering transcriptional machinery accessibility and by controlling RNA polymerase-mediated transcription (Flaus et al., 2006; Wilson et al., 2021). The main families of chromatin remodelers include switch/sucrose non-fermenting (SWI/SNF), the imitation switch (ISWI), inositol requiring 80 (INO80), and the chromodomain helicase DNA binding proteins (CHD) (Clapier and Cairns, 2009).
The CHD protein family comprises nine DNA-binding proteins that contain chromo (chromatin organization modifier) and SWI/SNF helicase domains that are highly conserved between different organisms (Woodage et al., 1997). Beyond its role in regulating chromatin accessibility, CHD2 is also involved in DNA damage repair. CHD2 is recruited by poly (ADP-ribose) polymerase 1 (PARP1) and stimulates classical non-homologous DNA end joining (NHEJ), DNA repair (Al-Sarraj et al., 2011; Wang et al., 2013; Ray Chaudhuri and Nussenzweig, 2017). CHD2 triggers the deposition of histone variant H3.3 at sites of damage which facilitates the recruitment of X-ray repair cross-complementing protein 4 (XRCC4) and DNA ligase 4 (LIG4). The inhibition of NHEJ is associated with TDP-43 mislocalization in the cytoplasm, suggesting that persistent DNA damage and impaired DNA repair may further disrupt the nuclear function of TDP-43 in DNA repair, exacerbating nuclear DNA damage and the accumulation of TDP-43 in the cytoplasm (Al-Sarraj et al., 2011).
In normal conditions, CHD2, the homolog of CHD1 in Drosophila melanogaster, interacts with TDP-43 in modulating the stress response (heat shock and oxidative stress). Under these conditions, CHD2 facilitates nucleosome clearance from the Hsp70 transcription start site (TSS) of the gene, allowing its activation. However, through the interaction with CHD2, TDP-43 overexpression led to the inhibition of proper nucleosome clearance from the TSS of Hsp70, thereby impeding its activation and causing a failure in the stress response (Figure 1C) (Berson et al., 2017). In addition, the knockdown of CHD1 in Drosophila melanogaster induces SGs formation under oxidative stress, increasing both the number and size of SGs per cell and the percentage of cells with SGs (Berson et al., 2017). This supports the idea that CHD2 plays an important role in neuroprotection with the decrease of aggregate formation.
Non-coding RNA (ncRNAs) regulate gene expression at multiple levels in both the nucleus and cytosol using a variety of mechanisms. This property renders them critical elements in the fine control of the levels of RNAs and proteins involved in the formation of pathological aggregates. Certain long ncRNAs (lncRNAs) are also architectural in the formation of MLOs that have been involved in aggregate formation after stress. Any sustained alteration in this intricate network of thousands of ncRNAs and their targets that is not compensated for has the potential to lead to aberrant condensates that might evolve into insoluble aggregates through liquid-to-solid phase separation. This section is focused on those ncRNAs subgroups that regulate mRNA expression and levels of proteins of aggregates in ALS including examples of ncRNAs that control the function of these proteins and in doing so, are linked to granule and aggregate formation, maintenance, and removal.
Mature microRNAs (miRNAs) are 20–25 nt single stranded RNAs that regulate the expression of the whole human genome. In canonical biogenesis, miRNAs are formed from long primary sequences (pri-miRNAs) in sequential steps dependant on two ribonucleases, Drosha that processes pri-miRNAs in the nucleus, and Dicer that slices precursor miRNAs (pre-miRNAs) in the cytosol. MiRNAs bind through imperfect pairing with miRNA recognition elements (MREs) usually in 3’UTRs of RNA targets, which they use to guide Argonaute proteins from the RNA-induced silencing complex (RISC) to exert their slicing role and silence specific targets. Even though miRNAs are better known for their silencing effects at the transcript level, they can also activate or repress translation (Vasudevan et al., 2008; Treiber et al., 2019) and activate or repress transcription. The latter occurs through binding to complementary sequences in promoters and enhancers, with or without the participation of other epigenetic factors, interacting with single stranded DNA (miRNA: DNA) or double stranded DNA (miRNA:DNA:DNA) (Campos-Melo et al., 2022; Santovito and Weber, 2022; Hu et al., 2023).
MiRNAs are deeply interconnected with TDP-43 and FUS/TLS in that both RBPs have critical roles in miRNA biogenesis. TDP-43 regulates miRNA processing by interacting with Drosha and Dicer complexes and with selected pri- and pre-miRNAs (Kawahara and Mieda-Sato, 2012), while FUS/TLS facilitates miRNA processing by co-transcriptional recruitment of Drosha on the chromatin and binding to specific nascent pri-miRNA (Morlando et al., 2012). These functions have been linked to the vast downregulation of miRNAs observed in spinal cord in ALS, specifically in motor neurons (Campos-Melo et al., 2013; Emde et al., 2015). Further, the expression of pathogenic ALS-causing mutations of TDP-43 (A315T), FUS/TLS (R495X), and SOD1-G93A or their respective wild-type variants, or an oxidative stressor or endoplasmic reticulum (ER) stress was sufficient to reduce or inhibit Dicer catalytic activity (Emde et al., 2015).
Numerous miRNAs have been reported to modulate the levels of TDP-43, FUS/TLS, and other proteins involved in aggregate formation in ALS or the regulation of this process (Liu et al., 2022a) (Table 1). Many of these miRNAs have been studied in detail in the context of cancer; however, examples of miRNAs that are linked to neurodegeneration, and specifically to ALS, have also been reported (Figure 2A). For instance, miR-27b-3p and miR-181c-5p, two miRNAs that are reduced in ALS, downregulate TDP-43 expression as a part of a negative feedback loop dependent on TDP-43 nuclear localization (Campos-Melo et al., 2013; Hawley et al., 2020). It is important to note that miRNAs have multiple targets. Thus, a single miRNA can alter the levels of different proteins associated with aggregate formation leading to complex effects. This is the case of miR-183-5p, which has been reported to be upregulated in the spinal cord in ALS and downregulated in the hippocampus and cerebellum (Kim et al., 2023). MiR-183-5p is induced after stress and increases neuronal survival by directly targeting programmed cell death 4 (PDCD4) and receptor interacting serine/threonine kinase 3 (RIPK3) transcripts (Li et al., 2020). In addition, miR-183-5p reduces sequestosome-1/ubiquitin-binding protein p62 (SQSTM1/p62) and increases TDP-43 levels in neuronal cells, regulating the formation of SGs and protecting against cytotoxicity (Kim et al., 2023). Other examples of miRNAs with several ALS-linked targets are miR-194 and miR-b2122 in which the former is involved in the regulation of inflammation and the latter a novel miRNA. Both miRNAs regulate TDP-43 and FUS/TLS and show reduced levels in ALS patients (Hawley et al., 2017).
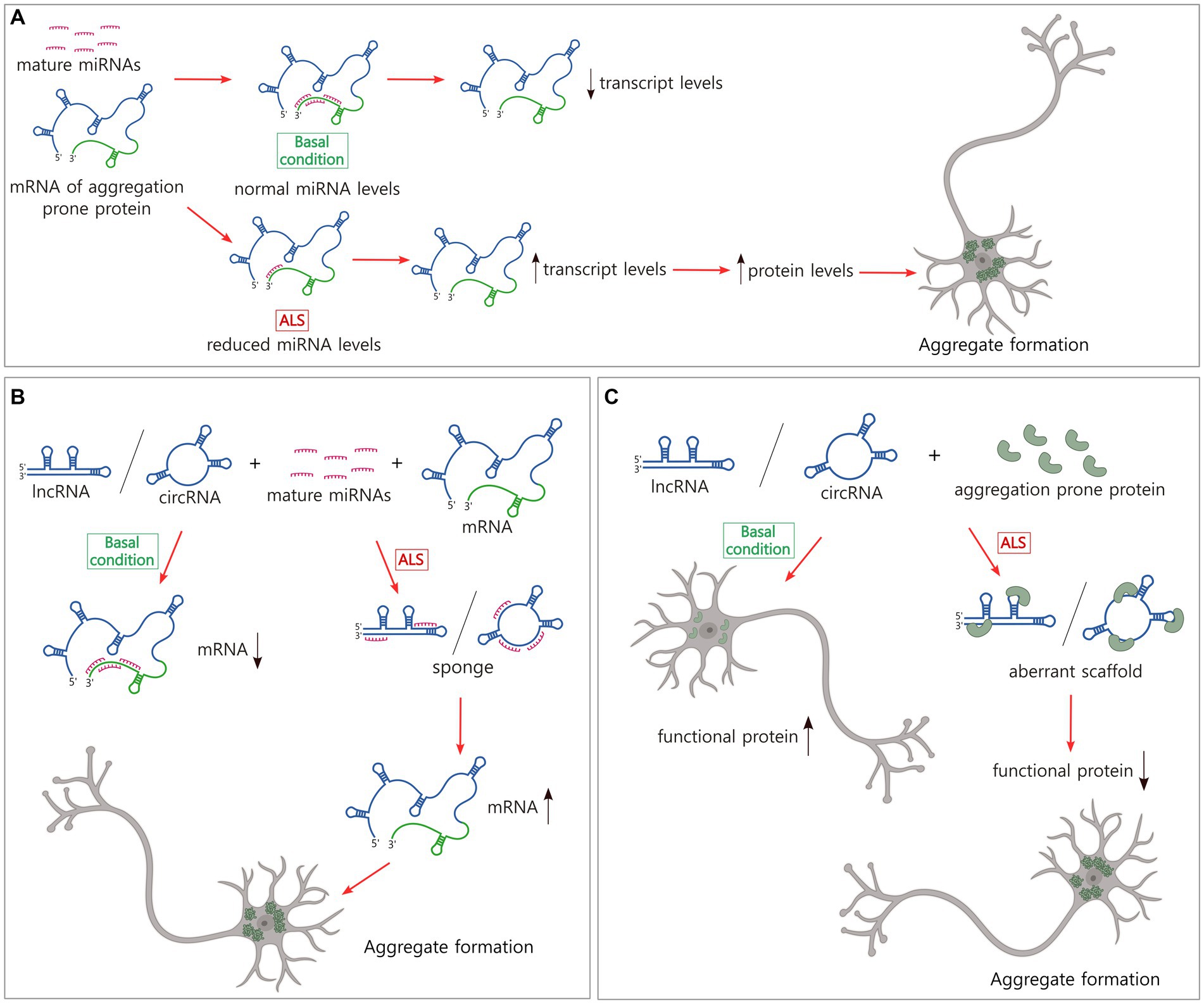
Figure 2. Non-coding RNAs modulate the levels of transcripts and proteins that form pathological inclusions in ALS. (A) The majority of miRNAs downregulate mRNA levels. In ALS, there is a broad reduction of miRNAs that might explain the increase in the levels of certain ALS-related transcripts and proteins and the subsequent aggregate formation. (B) LncRNAs and circRNAs sponge miRNAs which might increase the levels of certain transcripts and proteins, and the pathological aggregation. (C) LncRNAs and circRNAs bind to aggregation prone proteins, inducing aberrant scaffolds and reduction of levels of functional proteins. This loss of balance could generate protein inclusion formation. Figure created with BioRender.com.
One of the most studied miRNAs is miR-206, which is upregulated in blood samples of ALS patients (Toivonen et al., 2014; Dobrowolny et al., 2021; Gomes et al., 2023). MiR-206 is also significantly upregulated in the skeletal muscle of SOD1-G93A transgenic mice in which motor deficits are expressed. MiR-206 −/−; SOD1-G93A mice showed that loss of miR-206 accelerated the progression of motor impairments and decreased survival (Williams et al., 2009). Details of the mechanism include that miR-206 downregulated the translation of HDAC4 in vitro and both have opposite effects on fibroblast growth factor binding protein 1 pathway, promoting and impeding neuromuscular junction innervation after injury, respectively (Williams et al., 2009). This study and the protective role of HDAC inhibitors in animal models (described in the histone modifications section) support the idea that increased levels of miR-206 in the skeletal muscle might have a protective function in ALS. However, an investigation that analyzed muscle samples of a small group of ALS patients observed that HDAC4 levels are increased and occurred exclusively in rapidly progressing ALS, and that only HDAC4 (not miR-206) had a correlation with reinnervation (negative) and disease progression rate (positive) (Bruneteau et al., 2013).
Other studies have explored the link between miR-206 and aggregate formation in neurodegeneration. It has been reported that miR-206 downregulates Fas apoptotic inhibitory molecule (FAIM), a protein implicated in blocking the formation of protein aggregates under stress conditions and whose isoforms are de-regulated in Alzheimer’s disease and dopaminergic neurons (Yu et al., 2008; Carriba et al., 2015; Coccia et al., 2020). In in vitro experiments, FAIM counteracted SOD1-G93A aggregation in cells in filter trap and sedimentation assays (Kaku et al., 2020). Altogether these results suggest that the upregulation of miR-206 and the reduction of HDAC4 might be protective in SOD1-G93A mice. However, in patients, there could be elements in this regulatory network that partially block the beneficial effect of miR-206. It would be interesting to study specific regulatory molecules of miR-206, such as the hepatocyte growth factor and circular_0057558 (circ_0057558) (Choi et al., 2018; Chen et al., 2021b), as well as investigate FAIM levels in ALS patients. It is possible that the upregulation of miR-206 might be increasing the formation of aggregates via FAIM reduction. Finally, the multilayer mechanism of action of miR-206 also included the interaction of miRNA-206 with TDP-43, which limits miRNA activity by disrupting their RISC association (King et al., 2014).
LncRNAs are molecules of ≥200 nt in length that show diverse mechanisms of biogenesis. Many lncRNAs are spliced, 7-methylguanosine capped, and polyadenylated as mRNAs; others are not. LncRNAs are expressed from RNA polymerase I (Pol I) or Pol III promoters or are processed from precursors that come from introns or repetitive elements. LncRNAs can also be antisense with respect to protein coding genes, transcribed from intergenic regions or pseudogenes, or derived from 3’UTR mRNAs (Mercer et al., 2011; Mattick et al., 2023). LncRNAs can be linear or circular (circRNAs), and the majority of the latest generated by back-splicing of coding and non-coding transcripts using the spliceosomal machinery (Liu and Chen, 2022).
Linear lncRNAs (called lncRNAs hereafter) and circRNAs have multiple dynamic functions in the regulation of gene expression but only a limited group have been characterized in neurodegeneration. LncRNAs act as guides and scaffolds, binding transcription factors and chromatin-modifiers, and assembling protein complexes to direct them to specific genomic locations. They can function as molecular decoys, sponging miRNAs inhibiting their binding to target mRNAs or sequestering regulatory proteins and transcription factors, and chromatin modifiers from gene promoters. LncRNAs also have enhancer-like functions, activating gene transcription in cis and circRNAs regulate splicing of protein coding transcripts (Kopp and Mendell, 2018; Liu and Chen, 2022; Mattick et al., 2023). While studies of the regulatory mechanisms utilized by lncRNAs and circRNAs are also abundant in the cancer field, our understanding of their role in ALS is still early. However, given that the functional strategies these molecules use are extensively spread in nature, further examining their role in aggregate formation in ALS is timely (Figures 2B,C).
Currently, a limited number of lncRNA expression profiles exist for ALS tissue. The most representative examples show the dysregulation of this population of ncRNAs in the blood cells of ALS patients (Gagliardi et al., 2018; Yu et al., 2022). Among the lncRNAs that modulate the levels of expression of ALS-linked genes (Table 1) is SOD1-DT, a divergent lncRNA encoded near the locus of SOD1 gene that is transcribed in the opposite direction. SOD1-DT is expressed in neuronal cells and regulates the expression of SOD1 using an unknown mechanism, suggesting a role in ALS (Guerra et al., 2023). Other examples are long intergenic non-protein coding RNA 2231 (linc02231) and lncRNA ANCR that sponge miR-939-5p and miR-140-3p, respectively, and prevent heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) degradation (Wen et al., 2020; Xu et al., 2023), and lncRNA SOX2-OT that sponges miR-146b-5p increasing the levels of hnRNPA2B1 (Zhang and Li, 2019). In the pathogenesis of ALS, it has been observed that lnc-HIBADH-4 levels correlate with disease severity and survival. Lnc-HIBADH-4 is another example of a molecular sponge, but it controls the clearance of protein aggregates in neurons by regulating lysosomal function through the lnc-HIBADH-4/miR-326/cathepsin D pathway (Figure 2B) (Huang et al., 2023).
Nuclear paraspeckle assembly transcript 1 (NEAT1) is a lncRNA that functions as a scaffold of proteins in the formation of nuclear paraspeckles, MLOs that modulate gene expression by sequestering mRNAs from translation and proteins from their gene regulatory functions (Hirose et al., 2014; Jacq et al., 2021; McCluggage and Fox, 2021). At a physiological level, paraspeckles protect the cell in stress conditions and have been involved in different neurodegenerative diseases (An et al., 2018). TDP-43, FUS/TLS, TATA-box binding protein associated factor 15 (TAF15), Ewing’s sarcoma RBP 1 (EWSR1), and HNRNPA1, all proteins that form pathological aggregates in ALS, are recruited in paraspeckles (Naganuma et al., 2012). Then, alterations in NEAT1 levels compromise the abundance and biophysical properties of paraspeckles, the availability of free paraspeckle proteins, and as with other MLOs, might potentially induce the formation of protein aggregates in neurodegeneration (Figure 2C). In fact, it has been observed that in the early phase of ALS, NEAT1_2, one of the two isoforms of NEAT1, is upregulated, binds to TDP-43 and FUS/TLS, and induces paraspeckle formation in motor neurons, modulating the levels and functions of these ALS-associated RBPs (Nishimoto et al., 2013). Reduction of nuclear TDP-43 but not its accumulation in the cytosol or aggregation results in this paraspeckle hyper-assembly (Shelkovnikova et al., 2018). Poly-proline-arginine, the most toxic dipeptide repeat from C9orf72, binds to and upregulates NEAT1, increasing also paraspeckle formation (Suzuki et al., 2019).
The expression and function of circRNAs in ALS is also beginning to be elucidated. Two independent groups have analyzed the levels of circRNAs in skeletal muscle and spinal cord in ALS patients to evaluate their biomarker potential. It has been observed that a group of circRNAs is altered in ALS-relevant tissues, some of them in opposite directions (Aquilina-Reid et al., 2022; Tsitsipatis et al., 2022). Evidence that might link circRNAs with aggregate formation in ALS arises mainly from FUS/TLS studies. FUS/TLS and circRNAs are closely interrelated. FUS/TLS regulates circRNA biogenesis by binding the introns flanking the back-splicing junctions inducing cyclization (Errichelli et al., 2017; He et al., 2023; Liu et al., 2023b). However, circRNAs such as circRNA-0004904 and circRNA_0000285 also modulate FUS/TLS levels (Chen et al., 2019; Dai and Liu, 2021). In a different mechanism, circRNAs recruit/sponge FUS/TLS to participate in mRNA stability in different regulatory axes that involve SGs (Liu et al., 2023b; Liu et al., 2023c).
The intracellular traffic of the multi-exonic circ-Hdgfrp3 has also provided some insights. In wild-type stressed motor neurons, circ-Hdgfrp3 localized to SGs are proposed to serve as a protein scaffold. In motor neurons carrying mutant FUS/TLS, a high proportion of circ-Hdgfrp3 is trapped in cytoplasmic aggregates, from where it’s not efficiently released upon stress removal (D'Ambra et al., 2021). Aggregation of TDP-43 might also be connected to circRNAs function. The overexpression of circTmeff1 triggers skeletal muscle atrophy in vivo and in vitro, and its knockdown partially rescues muscle mass in an atrophy model in mice (Chen et al., 2023). Mechanistically, circTmeff1 directly interacts with TDP-43 and promotes its aggregation in mitochondria (Figure 2C), triggering the release of mitochondrial DNA into the cytosol and the activation of the cyclic GMP-AMP synthase (cGAS)/stimulator of interferon genes (STING) pathway (Chen et al., 2023).
The epitranscriptome describes changes to the transcriptome that do not involve changes in the ribonucleotide sequence but are based on nucleotide chemical modifications (Saletore et al., 2012). There are more than 100 different types of chemical modifications identified for RNA in both coding RNAs and noncoding RNAs during their cycle of life (Roundtree et al., 2017) including N6-methyladenosine (m6A) (Meyer et al., 2012), N1-methyladenosine (m1A) (Safra et al., 2017), and inosine-to-adenine editing (Bazak et al., 2014). Some of them are relevant to ALS.
m6A is the most abundant conserved modification and plays a crucial role in regulating gene expression. This modification has been extensively associated with nuclear splicing, mRNA stability, translation speed, and the subcellular localization of targeted mRNAs (Widagdo and Anggono, 2018). For lncRNAs, m6A regulates splicing and nuclear export (Akhtar et al., 2021). m6A is catalyzed by the Mettl3/Mettl14 methyltransferase complex and can also undergo demethylation by fat mass and obesity-associated protein (FTO) and AlKBH5 (AlkB family homolog 5) demethylases. m6A are recognized by “readers” such as the YT521-B homology (YTH) domain family proteins (YTHDFs), YTH domain-containing proteins (YTHDCs), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and eukaryotic initiation factor 3 (eIF3), potentially influencing mRNA translation, stability, splicing, or localization (Scuteri et al., 2007; Jia et al., 2011; Wang et al., 2016a).
It has been shown that TDP-43 interacts with its own m6A-modified RNA and in doing so downregulates its expression. The extensive RNA hypermethylation in post-mortem spinal cord tissue from sALS patients also suggests a direct association between the regulation of TDP-43 levels in ALS and m6A modification (McMillan et al., 2023). Moreover, an overlap between m6A and cytoplasmic TDP-43 inclusions has been observed in motor neurons (McMillan et al., 2023), and recently, a mechanism by which m1A CAG repeat RNA binds TDP-43 and stimulates its mislocalization in the cytosol and the formation of gel-like aggregates has been described (Sun et al., 2023). Together these studies confirm TDP-43’s capacity to detect methylated RNA and the link between this modification and protein aggregate formation in neurodegeneration.
RNA editing involves post-transcriptional modifications mediated by cytosine and adenosine deaminases, wherein nucleotides are modified, inserted, or deleted, often facilitated by enzymes, sometimes modifying the RNA’s coding potential. Apobec enzyme naturally converts cytosines to uracils, while adenosine deaminases, such as the adenosine deaminases acting on RNA (ADARs) enzyme family, transform adenines into inosines (Tan et al., 2017; Walkley and Li, 2017). These changes can influence RNA translation into proteins, potentially impacting protein folding and aggregation. Furthermore, RNA editing can influence RNA stability and localization, affecting protein aggregation dynamics (Eisenberg and Levanon, 2018).
In sALS motor neurons, there is a concomitant occurrence of TDP-43 aggregation and reduced expression of ADAR2 (Aizawa et al., 2010; Hideyama et al., 2012). The enzyme ADAR2 primarily targets the Q/R site in GluA2 (a subunit of AMPA receptor) pre-mRNA and its deficiency leads to excessive Ca2+ permeability of glutamate-AMPA receptors due to the failure of A-to-I editing (Kawahara et al., 2004; Kawahara and Kwak, 2005). This conversion is crucial for motor neuron survival (Kwak and Kawahara, 2005; Hideyama et al., 2010). The loss of ADAR2 promotes the activation of calpain proteases due to this excess of Ca2+ permeability, which induces cleavage of the TDP-43 C-terminal fragment, generating aggregation-prone N-terminal fragments. Persistent calpain activation results in the gradual expansion of TDP-43 aggregates, contributing to disease progression (Yamashita and Kwak, 2014). The evidence shows that the variation in TDP-43 expression does not affect the expression levels of ADAR2, suggesting that the aggregation or abnormal TDP-43 processing is a consequence of an inefficient GluA2 Q/R site RNA editing in the motor neurons of sALS patients (Yamashita et al., 2012).
Notably, a deficiency of ADAR2 was also observed in the spinal motor neurons of an ALS patient with a FUS/TLS mutation, although it is still unclear if this deficiency is related to the presence of FUS-positive inclusions (Aizawa et al., 2016). In addition, ADAR2 undergoes nucleocytoplasmic mislocalization, leading to abnormal RNA editing in the postmortem tissue of individuals with C9orf72 mutations. A significant proportion of neurons with cytoplasmic accumulation of ADAR2 along with TDP-43 pathology was observed in the spinal cord of C9orf72 ALS/FTD patients (Moore et al., 2019). In this study, the analysis of the whole transcriptome for RNA A-to-I editing changes was evaluated by RNA-seq, indicating that these RNA alterations occur in 1,526 genes, including ALS-related transcripts (Moore et al., 2019).
Currently, the evidence suggests that the formation of protein aggregates and epigenetic modifications can also be related to aging (Lopez-Otin et al., 2023). The alterations of epigenetic modifications affect DNA replication and repair, gene transcription and silencing, cell division, and telomere length maintenance (Gonzalo, 2010). With age, chromatin undergoes various changes, such as structural remodeling, alterations in chromatin architecture, loss of histones, and modifications to histones. For example, reduced global histone acetylation can disrupt metabolic gene expression and homeostasis. The balance of histone acetylation and deacetylation is critical, and any disruption in this balance can lead to issues in stress response and DNA repair mechanisms (Eberharter and Becker, 2002; Lazo-Gomez et al., 2013; Jagaraj et al., 2024).
In the context of ALS pathology, it has been described that aging accelerates DNA methylation in the CpG-island 5′ region which is linked to a more severe ALS disease phenotype, earlier onset, and shorter disease duration in patients with C9orf72 mutations (Zhang et al., 2017).
It makes sense to hypothesize that alterations of the epigenome due to aging could trigger or contribute to a cascade of events that could contribute to the formation of protein aggregates in pathologies such as ALS. However, this is a vastly unexplored field, and more studies are necessary to elucidate this link.
In this review, we presented the current evidence that suggests how epigenetic mechanisms can be related to inclusion formation in ALS, and by extension to other related neurodegenerative diseases. This holds significant therapeutic potential for ALS since targeting specific epigenetic regulators involved in protein aggregation pathways could offer novel strategies for mitigating neuronal dysfunction and slowing disease progression in ALS patients. In the course of studying the evidence of the link between epigenetics and inclusion formation, we realized two main hindrances in the field: first, the definition of epigenetics itself, and second, the causality association between epigenetics and biomolecular condensate formation.
The definition of epigenetics has been a controversial topic since it was first conceptualized (Felsenfeld, 2014). Many definitions have been created since then, depending especially on the field to which the author belongs (Nicoglou and Merlin, 2017). In general, the contemporary definition includes DNA modifications, chromatin regulation, and the possibility of heritability. However, where the definition becomes open to interpretation is concerning the role of RNA in epigenetics. In this work, we used the contemporary definition of Cavalli (Cavalli and Heard, 2019) which includes RNA (ncRNA and RNA modifications). However, it is clear to us that this boundary is diffuse. The RNA field has had an explosive development over the last decade and the question that emerges is if it will be necessary to include all the RNA-related regulatory processes as a separate mechanism with its own definition, leaving only the chromatin-related mechanisms to the epigenetics field.
Despite the evidence showing how epigenetics can affect directly or indirectly the aggregate formation in ALS, we also noticed several examples where only a correlation between both was detected. It becomes quite complex trying to elucidate which one has a causal role. Specifically, it remains unclear in many cases if changes in epigenetics modifications are leading to disease processes or if disease processes lead to changes in the epigenome. In this context, the field seems rather stalled with few mechanistic studies published. We believe that greater efforts are needed in deciphering these causal processes.
As the majority of ALS cases do not have a clear genetic explanation, studying the epigenome becomes highly relevant for understanding the pathogenesis of ALS and the goal of finding new therapeutic targets. This would also help us to understand if feedback loops between aggregate formation and epigenetics alterations exist, a key point in the design of new drugs that could reduce or stop the disease progression.
VN: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. DC-M: Conceptualization, Supervision, Visualization, Writing – review & editing. CD: Conceptualization, Visualization, Writing – review & editing. MS: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Research funding provided by an operating grant from the Canadian Institutes of Health research (grant #201806SOP-411481) and by a generous donation from the Temerty Family Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abel, T., and Zukin, R. (2008). Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 8, 57–64. doi: 10.1016/j.coph.2007.12.002
Afroz, T., Hock, E. M., Ernst, P., Foglieni, C., Jambeau, M., Gilhespy, L., et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8:45. doi: 10.1038/s41467-017-00062-0
Aizawa, H., Hideyama, T., Yamashita, T., Kimura, T., Suzuki, N., Aoki, M., et al. (2016). Deficient RNA-editing enzyme ADAR2 in an amyotrophic lateral sclerosis patient with a FUS(P525L) mutation. J. Clin. Neurosci. 32, 128–129. doi: 10.1016/j.jocn.2015.12.039
Aizawa, H., Sawada, J., Hideyama, T., Yamashita, T., Katayama, T., Hasebe, N., et al. (2010). TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 120, 75–84. doi: 10.1007/s00401-010-0678-x
Akhtar, J., Lugoboni, M., and Junion, G. (2021). M(6)a RNA modification in transcription regulation. Transcription 12, 266–276. doi: 10.1080/21541264.2022.2057177
Alberti, S., and Dormann, D. (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194. doi: 10.1146/annurev-genet-112618-043527
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Al-Sarraj, S., King, A., Troakes, C., Smith, B., Maekawa, S., Bodi, I., et al. (2011). p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 122, 691–702. doi: 10.1007/s00401-011-0911-2
An, H., Williams, N. G., and Shelkovnikova, T. A. (2018). NEAT1 and paraspeckles in neurodegenerative diseases: a missing lnc found? Noncoding RNA Res 3, 243–252. doi: 10.1016/j.ncrna.2018.11.003
Andersen, P. M., Nilsson, P., Ala-Hurula, V., Keranen, M. L., Tarvainen, I., Haltia, T., et al. (1995). Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat. Genet. 10, 61–66. doi: 10.1038/ng0595-61
Appleby-Mallinder, C., Schaber, E., Kirby, J., Shaw, P. J., Cooper-Knock, J., Heath, P. R., et al. (2021). TDP43 proteinopathy is associated with aberrant DNA methylation in human amyotrophic lateral sclerosis. Neuropathol. Appl. Neurobiol. 47, 61–72. doi: 10.1111/nan.12625
Aquilina-Reid, C., Brennan, S., Curry-Hyde, A., Teunisse, G. M. S. C., and Janitz, M. (2022). Circular RNA expression and interaction patterns are perturbed in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 23:14665. doi: 10.3390/ijms232314665
Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H., et al. (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611. doi: 10.1016/j.bbrc.2006.10.093
Aristizabal, M. J., Anreiter, I., Halldorsdottir, T., Odgers, C. L., Mcdade, T. W., Goldenberg, A., et al. (2020). Biological embedding of experience: a primer on epigenetics. Proc. Natl. Acad. Sci. USA 117, 23261–23269. doi: 10.1073/pnas.1820838116
Ash, P. E., Bieniek, K. F., Gendron, T. F., Caulfield, T., Lin, W. L., Dejesus-Hernandez, M., et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. doi: 10.1016/j.neuron.2013.02.004
Ayers, J. I., Mcmahon, B., Gill, S., Lelie, H. L., Fromholt, S., Brown, H., et al. (2017). Relationship between mutant cu/Zn superoxide dismutase 1 maturation and inclusion formation in cell models. J. Neurochem. 140, 140–150. doi: 10.1111/jnc.13864
Babic Leko, M., Zupunski, V., Kirincich, J., Smilovic, D., Hortobagyi, T., Hof, P. R., et al. (2019). Molecular mechanisms of neurodegeneration related to C9orf72 Hexanucleotide repeat expansion. Behav. Neurol. 2019:2909168. doi: 10.1155/2019/2909168
Baek, M., Choe, Y. J., Bannwarth, S., Kim, J., Maitra, S., Dorn, G. W. 2nd, et al. (2021). TDP-43 and PINK1 mediate CHCHD10(S59L) mutation-induced defects in Drosophila and in vitro. Nat. Commun. 12:1924. doi: 10.1038/s41467-021-22145-9
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi: 10.1038/nrm.2017.7
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Bauer, P. O. (2016). Methylation of C9orf72 expansion reduces RNA foci formation and dipeptide-repeat proteins expression in cells. Neurosci. Lett. 612, 204–209. doi: 10.1016/j.neulet.2015.12.018
Bazak, L., Haviv, A., Barak, M., Jacob-Hirsch, J., Deng, P., Zhang, R., et al. (2014). A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376. doi: 10.1101/gr.164749.113
Becker, P. B., and Workman, J. L. (2013). Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5:a017905. doi: 10.1101/cshperspect.a017905
Belzil, V. V., Bauer, P. O., Prudencio, M., Gendron, T. F., Stetler, C. T., Yan, I. K., et al. (2013). Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 126, 895–905. doi: 10.1007/s00401-013-1199-1
Bennett, S. A., Tanaz, R., Cobos, S. N., and Torrente, M. P. (2019). Epigenetics in amyotrophic lateral sclerosis: a role for histone post-translational modifications in neurodegenerative disease. Transl. Res. 204, 19–30. doi: 10.1016/j.trsl.2018.10.002
Berson, A., Sartoris, A., Nativio, R., Van Deerlin, V., Toledo, J. B., Porta, S., et al. (2017). TDP-43 promotes neurodegeneration by impairing chromatin remodeling. Curr. Biol. 27, 3579–3590.e6. doi: 10.1016/j.cub.2017.10.024
Bestor, T. H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402. doi: 10.1093/hmg/9.16.2395
Bi, D., Shi, M., Zheng, D., Hu, Q., Wang, H., Peng, L., et al. (2022a). Mechanism underlying the targeted regulation of the SOD1 3'UTR by the AUF1/Dicer1/miR-155/SOD1 pathway in sodium arsenite-induced liver injury. Ecotoxicol. Environ. Saf. 243:113990. doi: 10.1016/j.ecoenv.2022.113990
Bi, X., Zhang, Y., Yu, Y., Yuan, J., Xu, S., Liu, F., et al. (2022b). MiRNA-339-5p promotes isoproterenol-induced cardiomyocyte hypertrophy by targeting VCP to activate the mTOR signaling. Cell Biol. Int. 46, 288–299. doi: 10.1002/cbin.11731
Bird, A., Taggart, M., Frommer, M., Miller, O. J., and Macleod, D. (1985). A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40, 91–99. doi: 10.1016/0092-8674(85)90312-5
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M., and Lamond, A. I. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. doi: 10.1016/j.molcel.2010.09.024
Boxer, A. L., Mackenzie, I. R., Boeve, B. F., Baker, M., Seeley, W. W., Crook, R., et al. (2011). Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J. Neurol. Neurosurg. Psychiatry 82, 196–203. doi: 10.1136/jnnp.2009.204081
Bruneteau, G., Simonet, T., Bauche, S., Mandjee, N., Malfatti, E., Girard, E., et al. (2013). Muscle histone deacetylase 4 upregulation in amyotrophic lateral sclerosis: potential role in reinnervation ability and disease progression. Brain 136, 2359–2368. doi: 10.1093/brain/awt164
Buchan, J. R. (2014). mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11, 1019–1030. doi: 10.4161/15476286.2014.972208
Buchan, J. R., Kolaitis, R. M., Taylor, J. P., and Parker, R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. doi: 10.1016/j.cell.2013.05.037
Butti, Z., and Patten, S. A. (2018). RNA dysregulation in amyotrophic lateral sclerosis. Front. Genet. 9:712. doi: 10.3389/fgene.2018.00712
Caggiano, C., Morselli, M., Qian, X., Celona, B., Thompson, M., Wani, S., et al. (2024). Tissue informative cell-free DNA methylation sites in amyotrophic lateral sclerosis. medRxiv. doi: 10.1101/2024.04.08.24305503
Cairns, N. J., Neumann, M., Bigio, E. H., Holm, I. E., Troost, D., Hatanpaa, K. J., et al. (2007). TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am. J. Pathol. 171, 227–240. doi: 10.2353/ajpath.2007.070182
Campos-Melo, D., Droppelmann, C. A., He, Z., Volkening, K., and Strong, M. J. (2013). Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain 6:26. doi: 10.1186/1756-6606-6-26
Campos-Melo, D., Hawley, Z. C. E., Mclellan, C., and Strong, M. J. (2022). “Chapter 7 - MicroRNA turnover and nuclear function” in MicroRNA. ed. J. Xiao (Academic Press), 109–140.
Campos-Melo, D., Hawley, Z. C. E., and Strong, M. J. (2018). Dysregulation of human NEFM and NEFH mRNA stability by ALS-linked miRNAs. Mol. Brain 11:43. doi: 10.1186/s13041-018-0386-3
Carey, J. L., and Guo, L. (2022). Liquid-liquid phase separation of TDP-43 and FUS in physiology and pathology of neurodegenerative diseases. Front. Mol. Biosci. 9:826719. doi: 10.3389/fmolb.2022.826719
Carriba, P., Jimenez, S., Navarro, V., Moreno-Gonzalez, I., Barneda-Zahonero, B., Moubarak, R. S., et al. (2015). Amyloid-beta reduces the expression of neuronal FAIM-L, thereby shifting the inflammatory response mediated by TNFalpha from neuronal protection to death. Cell Death Dis. 6:e1639. doi: 10.1038/cddis.2015.6
Cavalli, G., and Heard, E. (2019). Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499. doi: 10.1038/s41586-019-1411-0
Chakraborty, P., and Zweckstetter, M. (2023). Role of aberrant phase separation in pathological protein aggregation. Curr. Opin. Struct. Biol. 82:102678. doi: 10.1016/j.sbi.2023.102678
Chang, R., Chu, Q., Zheng, W., Zhang, L., and Xu, T. (2020b). The Sp1-responsive microRNA-15b negatively regulates Rhabdovirus-triggered innate immune responses in lower vertebrates by targeting TBK1. Front. Immunol. 11:625828. doi: 10.3389/fimmu.2020.625828
Chang, H., Li, Z. B., Wu, J. Y., and Zhang, L. (2020a). Circ-100338 induces angiogenesis after myocardial ischemia-reperfusion injury by sponging miR-200a-3p. Eur. Rev. Med. Pharmacol. Sci. 24, 6323–6332. doi: 10.26355/eurrev_202006_21530
Chen, K., Bennett, S. A., Rana, N., Yousuf, H., Said, M., Taaseen, S., et al. (2018). Neurodegenerative disease Proteinopathies are connected to distinct histone post-translational modification landscapes. ACS Chem. Neurosci. 9, 838–848. doi: 10.1021/acschemneuro.7b00297
Chen, H., Dzitoyeva, S., and Manev, H. (2012). Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor. Neurol. Neurosci. 30, 237–245. doi: 10.3233/RNN-2012-110223
Chen, W., Guo, L., Li, M., Wei, C., Li, S., and Xu, R. (2022a). The pathogenesis of amyotrophic lateral sclerosis: mitochondrial dysfunction, protein misfolding and epigenetics. Brain Res. 1786:147904. doi: 10.1016/j.brainres.2022.147904
Chen, M. L., Hong, C. G., Yue, T., Li, H. M., Duan, R., Hu, W. B., et al. (2021a). Inhibition of miR-331-3p and miR-9-5p ameliorates Alzheimer's disease by enhancing autophagy. Theranostics 11, 2395–2409. doi: 10.7150/thno.47408
Chen, Z., Huai, Y., Mao, W., Wang, X., Ru, K., Qian, A., et al. (2022b). Liquid-liquid phase separation of biomacromolecules and its roles in metabolic diseases. Cells 11:3023 doi: 10.3390/cells11193023
Chen, R. X., Liu, H. L., Yang, L. L., Kang, F. H., Xin, L. P., Huang, L. R., et al. (2019). Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur. Rev. Med. Pharmacol. Sci. 23, 8771–8778. doi: 10.26355/eurrev_201910_19271
Chen, F. R., Sha, S. M., Wang, S. H., Shi, H. T., Dong, L., Liu, D., et al. (2020). RP11-81H3.2 promotes gastric cancer progression through miR-339-HNRNPA1 interaction network. Cancer Med. 9, 2524–2534. doi: 10.1002/cam4.2867
Chen, X., Tan, Q. Q., Tan, X. R., Li, S. J., and Zhang, X. X. (2021b). Circ_0057558 promotes nonalcoholic fatty liver disease by regulating ROCK1/AMPK signaling through targeting miR-206. Cell Death Dis. 12:809. doi: 10.1038/s41419-021-04090-z
Chen, R., Yang, T., Jin, B., Xu, W., Yan, Y., Wood, N., et al. (2023). CircTmeff1 promotes muscle atrophy by interacting with TDP-43 and encoding a novel TMEFF1-339aa protein. Adv. Sci. 10:e2206732. doi: 10.1002/advs.202303215
Choi, W., Lee, J., Lee, J., Ko, K. R., and Kim, S. (2018). Hepatocyte growth factor regulates the miR-206-HDAC4 Cascade to control neurogenic muscle atrophy following surgical denervation in mice. Mol. Ther. Nucleic Acids 12, 568–577. doi: 10.1016/j.omtn.2018.06.013
Chou, D. M., Adamson, B., Dephoure, N. E., Tan, X., Nottke, A. C., Hurov, K. E., et al. (2010). A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 107, 18475–18480. doi: 10.1073/pnas.1012946107
Chou, C. C., Zhang, Y., Umoh, M. E., Vaughan, S. W., Lorenzini, I., Liu, F., et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239. doi: 10.1038/s41593-017-0047-3
Chouliaras, L., Van Den Hove, D. L., Kenis, G., Keitel, S., Hof, P. R., Van Os, J., et al. (2012). Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr. Alzheimer Res. 9, 536–544. doi: 10.2174/156720512800618035
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Coccia, E., Masanas, M., Lopez-Soriano, J., Segura, M. F., Comella, J. X., and Perez-Garcia, M. J. (2020). FAIM is regulated by MiR-206, MiR-1-3p and MiR-133b. Front. Cell Dev. Biol. 8:584606. doi: 10.3389/fcell.2020.584606
Colussi, C., Mozzetta, C., Gurtner, A., Illi, B., Rosati, J., Straino, S., et al. (2008). HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. USA 105, 19183–19187. doi: 10.1073/pnas.0805514105
Conicella, A. E., Zerze, G. H., Mittal, J., and Fawzi, N. L. (2016). ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549. doi: 10.1016/j.str.2016.07.007
Coppede, F., Stoccoro, A., Mosca, L., Gallo, R., Tarlarini, C., Lunetta, C., et al. (2018). Increase in DNA methylation in patients with amyotrophic lateral sclerosis carriers of not fully penetrant SOD1 mutations. Amyotroph Lateral Scler Frontotemporal Degener 19, 93–101. doi: 10.1080/21678421.2017.1367401
Corcoran, L. J., Mitchison, T. J., and Liu, Q. (2004). A novel action of histone deacetylase inhibitors in a protein aggresome disease model. Curr. Biol. 14, 488–492. doi: 10.1016/j.cub.2004.03.003
Couthouis, J., Hart, M. P., Erion, R., King, O. D., Diaz, Z., Nakaya, T., et al. (2012). Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2899–2911. doi: 10.1093/hmg/dds116
Couthouis, J., Hart, M. P., Shorter, J., Dejesus-Hernandez, M., Erion, R., Oristano, R., et al. (2011). A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA 108, 20881–20890. doi: 10.1073/pnas.1109434108
Dai, W., and Liu, X. (2021). Circular RNA 0004904 promotes autophagy and regulates the fused in sarcoma/vascular endothelial growth factor axis in preeclampsia. Int. J. Mol. Med. 47:111. doi: 10.3892/ijmm.2021.4944
D'ambra, E., Santini, T., Vitiello, E., D'uva, S., Silenzi, V., Morlando, M., et al. (2021). Circ-Hdgfrp3 shuttles along neurites and is trapped in aggregates formed by ALS-associated mutant FUS. iScience 24:103504. doi: 10.1016/j.isci.2021.103504
Dawson, M. A., Prinjha, R. K., Dittmann, A., Giotopoulos, G., Bantscheff, M., Chan, W. I., et al. (2011). Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. doi: 10.1038/nature10509
Delmore, J. E., Issa, G. C., Lemieux, M. E., Rahl, P. B., Shi, J., Jacobs, H. M., et al. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. doi: 10.1016/j.cell.2011.08.017
Deng, H. X., Chen, W., Hong, S. T., Boycott, K. M., Gorrie, G. H., Siddique, N., et al. (2011). Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215. doi: 10.1038/nature10353
Deng, H. X., Zhai, H., Bigio, E. H., Yan, J., Fecto, F., Ajroud, K., et al. (2010). FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 67, 739–748. doi: 10.1002/ana.22051
Dini Modigliani, S., Morlando, M., Errichelli, L., Sabatelli, M., and Bozzoni, I. (2014). An ALS-associated mutation in the FUS 3'-UTR disrupts a microRNA-FUS regulatory circuitry. Nat. Commun. 5:4335. doi: 10.1038/ncomms5335
Dobrowolny, G., Martone, J., Lepore, E., Casola, I., Petrucci, A., Inghilleri, M., et al. (2021). A longitudinal study defined circulating microRNAs as reliable biomarkers for disease prognosis and progression in ALS human patients. Cell Death Discov 7:4. doi: 10.1038/s41420-020-00397-6
Droppelmann, C. A., Campos-Melo, D., Moszczynski, A. J., Amzil, H., and Strong, M. J. (2019). TDP-43 aggregation inside micronuclei reveals a potential mechanism for protein inclusion formation in ALS. Sci. Rep. 9:19928. doi: 10.1038/s41598-019-56483-y
Droppelmann, C. A., Wang, J., Campos-Melo, D., Keller, B., Volkening, K., Hegele, R. A., et al. (2013). Detection of a novel frameshift mutation and regions with homozygosis within ARHGEF28 gene in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener 14, 444–451. doi: 10.3109/21678421.2012.758288
Dugger, B. N., and Dickson, D. W. (2017). Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9:9. doi: 10.1101/cshperspect.a028035
Dulac, C. (2010). Brain function and chromatin plasticity. Nature 465, 728–735. doi: 10.1038/nature09231
Eberharter, A., and Becker, P. B. (2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229. doi: 10.1093/embo-reports/kvf053
Egger, G., Liang, G., Aparicio, A., and Jones, P. A. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463. doi: 10.1038/nature02625
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490. doi: 10.1038/s41576-018-0006-1
Elbaum-Garfinkle, S. (2019). Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem. 294, 7160–7168. doi: 10.1074/jbc.REV118.001188
Emde, A., Eitan, C., Liou, L. L., Libby, R. T., Rivkin, N., Magen, I., et al. (2015). Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J. 34, 2633–2651. doi: 10.15252/embj.201490493
Errichelli, L., Dini Modigliani, S., Laneve, P., Colantoni, A., Legnini, I., Capauto, D., et al. (2017). FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 8:14741. doi: 10.1038/ncomms14741
Esteller, M. (2005). DNA methylation and cancer therapy: new developments and expectations. Curr. Opin. Oncol. 17, 55–60. doi: 10.1097/01.cco.0000147383.04709.10
Fang, L., Wang, S. H., Cui, Y. G., and Huang, L. (2021). LINC00941 promotes proliferation and metastasis of pancreatic adenocarcinoma by competitively binding miR-873-3p and thus upregulates ATXN2. Eur. Rev. Med. Pharmacol. Sci. 25, 1861–1868. doi: 10.26355/eurrev_202102_25081
Fang, A., Yuan, Y., Sui, B., Wang, Z., Zhang, Y., Zhou, M., et al. (2023). Inhibition of miR-200b-3p confers broad-spectrum resistance to viral infection by targeting TBK1. MBio 14:e0086723. doi: 10.1128/mbio.00867-23
Federspiel, J. D., Greco, T. M., Lum, K. K., and Cristea, I. M. (2019). Hdac4 interactions in Huntington's disease viewed through the prism of multiomics. Mol. Cell. Proteomics 18, S92–S113. doi: 10.1074/mcp.RA118.001253
Felsenfeld, G. (2014). A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 6:6. doi: 10.1101/cshperspect.a018200
Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W. B., Fedorov, O., et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. doi: 10.1038/nature09504
Flaus, A., Martin, D. M., Barton, G. J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Fournier, A., Sasai, N., Nakao, M., and Defossez, P. A. (2012). The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief. Funct. Genomics 11, 251–264. doi: 10.1093/bfgp/elr040
Fraga, M. F., Agrelo, R., and Esteller, M. (2007). Cross-talk between aging and cancer: the epigenetic language. Ann. N. Y. Acad. Sci. 1100, 60–74. doi: 10.1196/annals.1395.005
Fu, R., Yang, P., Amin, S., and Li, Z. (2020). A novel miR-206/hnRNPA1/PKM2 axis reshapes the Warburg effect to suppress colon cancer growth. Biochem. Biophys. Res. Commun. 531, 465–471. doi: 10.1016/j.bbrc.2020.08.019
Fujisawa, T., and Filippakopoulos, P. (2017). Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246–262. doi: 10.1038/nrm.2016.143
Fujiya, M., Konishi, H., Mohamed Kamel, M. K., Ueno, N., Inaba, Y., Moriichi, K., et al. (2014). microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene 33, 4847–4856. doi: 10.1038/onc.2013.429
Fung, L., Guzman, H., Sevrioukov, E., Idica, A., Park, E., Bochnakian, A., et al. (2019). miR-128 restriction of LINE-1 (L1) Retrotransposition is dependent on targeting hnRNPA1 mRNA. Int. J. Mol. Sci. 20:1955. doi: 10.3390/ijms20081955
Gagliardi, S., Zucca, S., Pandini, C., Diamanti, L., Bordoni, M., Sproviero, D., et al. (2018). Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci. Rep. 8:2378. doi: 10.1038/s41598-018-20679-5
Gal, J., Chen, J., Barnett, K. R., Yang, L., Brumley, E., and Zhu, H. (2013). HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J. Biol. Chem. 288, 15035–15045. doi: 10.1074/jbc.M112.431957
Ghasemi, M., and Brown, R. H. Jr. (2018). Genetics of amyotrophic lateral sclerosis. Cold Spring Harb. Perspect. Med. 8:a024125. doi: 10.1101/cshperspect.a024125
Ghazi-Noori, S., Froud, K. E., Mizielinska, S., Powell, C., Smidak, M., Fernandez De Marco, M., et al. (2012). Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain 135, 819–832. doi: 10.1093/brain/aws006
Gibney, E. R., and Nolan, C. M. (2010). Epigenetics and gene expression, Epigenetics and gene expression. Heredity 105, 4–13. doi: 10.1038/hdy.2010.54
Goljanek-Whysall, K., Soriano-Arroquia, A., Mccormick, R., Chinda, C., and Mcdonagh, B. (2020). miR-181a regulates p62/SQSTM1, parkin, and protein DJ-1 promoting mitochondrial dynamics in skeletal muscle aging. Aging Cell 19:e13140. doi: 10.1111/acel.13140
Gomes, B. C., Peixinho, N., Pisco, R., Gromicho, M., Pronto-Laborinho, A. C., Rueff, J., et al. (2023). Differential expression of miRNAs in amyotrophic lateral sclerosis patients. Mol. Neurobiol. 60, 7104–7117. doi: 10.1007/s12035-023-03520-7
Gonzalo, S. (2010). Epigenetic alterations in aging. J. Appl. Physiol. 109, 586–597. doi: 10.1152/japplphysiol.00238.2010
Goutman, S. A., Hardiman, O., Al-Chalabi, A., Chio, A., Savelieff, M. G., Kiernan, M. C., et al. (2022). Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 21, 465–479. doi: 10.1016/S1474-4422(21)00414-2
Gu, J., Wang, C., Hu, R., Li, Y., Zhang, S., Sun, Y., et al. (2021). Hsp70 chaperones TDP-43 in dynamic, liquid-like phase and prevents it from amyloid aggregation. Cell Res. 31, 1024–1027. doi: 10.1038/s41422-021-00526-5
Guerra, M., Meola, L., Lattante, S., Conte, A., Sabatelli, M., Sette, C., et al. (2023). Characterization of SOD1-DT, a divergent long non-coding RNA in the locus of the SOD1 human gene. Cells 12:2058. doi: 10.3390/cells12162058
Guo, C., Sun, L., Chen, X., and Zhang, D. (2013). Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 8, 2003–2014. doi: 10.3969/j.issn.1673-5374.2013.21.009
Han, T. W., Kato, M., Xie, S., Wu, L. C., Mirzaei, H., Pei, J., et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. doi: 10.1016/j.cell.2012.04.016
Han, Q., Lu, J., Duan, J., Su, D., Hou, X., Li, F., et al. (2008). Gcn5- and Elp3-induced histone H3 acetylation regulates hsp70 gene transcription in yeast. Biochem. J. 409, 779–788. doi: 10.1042/BJ20070578
Handwerger, K. E., and Gall, J. G. (2006). Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16, 19–26. doi: 10.1016/j.tcb.2005.11.005
Hardiman, O., Al-Chalabi, A., Chio, A., Corr, E. M., Logroscino, G., Robberecht, W., et al. (2017). Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3:17071. doi: 10.1038/nrdp.2017.71
Hart, M. P., Brettschneider, J., Lee, V. M., Trojanowski, J. Q., and Gitler, A. D. (2012). Distinct TDP-43 pathology in ALS patients with ataxin 2 intermediate-length polyQ expansions. Acta Neuropathol. 124, 221–230. doi: 10.1007/s00401-012-0985-5
Hartung, T., Rhein, M., Kalmbach, N., Thau-Habermann, N., Naujock, M., Muschen, L., et al. (2021). Methylation and expression of mutant FUS in motor neurons differentiated from induced pluripotent stem cells from ALS patients. Front. Cell Dev. Biol. 9:774751. doi: 10.3389/fcell.2021.774751
Hawley, Z. C. E., Campos-Melo, D., and Strong, M. J. (2017). Novel miR-b2122 regulates several ALS-related RNA-binding proteins. Mol. Brain 10:46. doi: 10.1186/s13041-017-0326-7
Hawley, Z. C. E., Campos-Melo, D., and Strong, M. J. (2020). Evidence of a negative feedback network between TDP-43 and miRNAs dependent on TDP-43 nuclear localization. J. Mol. Biol. 432:166695. doi: 10.1016/j.jmb.2020.10.029
He, T., Zhang, Q., Xu, P., Tao, W., Lin, F., Liu, R., et al. (2023). Extracellular vesicle-circEHD2 promotes the progression of renal cell carcinoma by activating cancer-associated fibroblasts. Mol. Cancer 22:117. doi: 10.1186/s12943-023-01824-9
Heard, E., Rougeulle, C., Arnaud, D., Avner, P., Allis, C. D., and Spector, D. L. (2001). Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107, 727–738. doi: 10.1016/S0092-8674(01)00598-0
Helferich, A. M., Brockmann, S. J., Reinders, J., Deshpande, D., Holzmann, K., Brenner, D., et al. (2018). Dysregulation of a novel miR-1825/TBCB/TUBA4A pathway in sporadic and familial ALS. Cell. Mol. Life Sci. 75, 4301–4319. doi: 10.1007/s00018-018-2873-1
Hewitt, C., Kirby, J., Highley, J. R., Hartley, J. A., Hibberd, R., Hollinger, H. C., et al. (2010). Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 67, 455–461. doi: 10.1001/archneurol.2010.52
Hideyama, T., Yamashita, T., Aizawa, H., Tsuji, S., Kakita, A., Takahashi, H., et al. (2012). Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 45, 1121–1128. doi: 10.1016/j.nbd.2011.12.033
Hideyama, T., Yamashita, T., Suzuki, T., Tsuji, S., Higuchi, M., Seeburg, P. H., et al. (2010). Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 30, 11917–11925. doi: 10.1523/JNEUROSCI.2021-10.2010
Higashi, K., Yamada, Y., Minatoguchi, S., Baba, S., Iwasa, M., Kanamori, H., et al. (2015). MicroRNA-145 repairs infarcted myocardium by accelerating cardiomyocyte autophagy. Am. J. Physiol. Heart Circ. Physiol. 309, H1813–H1826. doi: 10.1152/ajpheart.00709.2014
Hirose, T., Virnicchi, G., Tanigawa, A., Naganuma, T., Li, R., Kimura, H., et al. (2014). NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183. doi: 10.1091/mbc.E13-09-0558
Honda, H., Hamasaki, H., Wakamiya, T., Koyama, S., Suzuki, S. O., Fujii, N., et al. (2015). Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions. Neuropathology 35, 37–43. doi: 10.1111/neup.12153
Hortobagyi, T., Troakes, C., Nishimura, A. L., Vance, C., Van Swieten, J. C., Seelaar, H., et al. (2011). Optineurin inclusions occur in a minority of TDP-43 positive ALS and FTLD-TDP cases and are rarely observed in other neurodegenerative disorders. Acta Neuropathol. 121, 519–527. doi: 10.1007/s00401-011-0813-3
Hu, X., Yin, G., Zhang, Y., Zhu, L., Huang, H., and Lv, K. (2023). Recent advances in the functional explorations of nuclear microRNAs. Front. Immunol. 14:1097491. doi: 10.3389/fimmu.2023.1097491
Huang, T., Wan, X., Alvarez, A. A., James, C. D., Song, X., Yang, Y., et al. (2019). MIR93 (microRNA −93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy 15, 1100–1111. doi: 10.1080/15548627.2019.1569947
Huang, J., Yu, Y., Pang, D., Li, C., Wei, Q., Cheng, Y., et al. (2023). Lnc-HIBADH-4 regulates autophagy-lysosome pathway in amyotrophic lateral sclerosis by targeting Cathepsin D. Mol. Neurobiol. 61, 4768–4782. doi: 10.1007/s12035-023-03835-5
Hwang, J. Y., Aromolaran, K. A., and Zukin, R. S. (2017). The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361. doi: 10.1038/nrn.2017.46
Hwang, J. W., Cho, Y., Bae, G. U., Kim, S. N., and Kim, Y. K. (2021). Protein arginine methyltransferases: promising targets for cancer therapy. Exp. Mol. Med. 53, 788–808. doi: 10.1038/s12276-021-00613-y
Hyman, A. A., Weber, C. A., and Julicher, F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58. doi: 10.1146/annurev-cellbio-100913-013325
Hyun, K., Jeon, J., Park, K., and Kim, J. (2017). Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49:e324. doi: 10.1038/emm.2017.11
Ismail, H., Liu, X., Yang, F., Li, J., Zahid, A., Dou, Z., et al. (2021). Mechanisms and regulation underlying membraneless organelle plasticity control. J. Mol. Cell Biol. 13, 239–258. doi: 10.1093/jmcb/mjab028
Jacq, A., Becquet, D., Guillen, S., Boyer, B., Bello-Goutierrez, M. M., Franc, J. L., et al. (2021). Direct RNA-RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention. RNA Biol. 18, 2016–2027. doi: 10.1080/15476286.2021.1889253
Jagaraj, C. J., Shadfar, S., Kashani, S. A., Saravanabavan, S., Farzana, F., and Atkin, J. D. (2024). Molecular hallmarks of ageing in amyotrophic lateral sclerosis. Cell. Mol. Life Sci. 81:111. doi: 10.1007/s00018-024-05164-9
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. doi: 10.1016/j.cell.2015.12.038
Janssen, C., Schmalbach, S., Boeselt, S., Sarlette, A., Dengler, R., and Petri, S. (2010). Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 69, 573–581. doi: 10.1097/NEN.0b013e3181ddd404
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jian, J., and Xia, L. (2021). miR-1226-3p promotes eNOS expression of pulmonary arterial endothelial cells to mitigate hypertension in rats via targeting Profilin-1. Biomed. Res. Int. 2021:1724722. doi: 10.1155/2021/1724722
Jiang, D., Li, T., Guo, C., Tang, T. S., and Liu, H. (2023). Small molecule modulators of chromatin remodeling: from neurodevelopment to neurodegeneration. Cell Biosci. 13:10. doi: 10.1186/s13578-023-00953-4
Jiang, H., Li, A. M., and Ye, J. (2022). The magic bullet: Niclosamide. Front. Oncologia 12:1004978. doi: 10.3389/fonc.2022.1004978
Johnson, J. O., Pioro, E. P., Boehringer, A., Chia, R., Feit, H., Renton, A. E., et al. (2014). Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666. doi: 10.1038/nn.3688
Kakita, A., Oyanagi, K., Nagai, H., and Takahashi, H. (1997). Eosinophilic intranuclear inclusions in the hippocampal pyramidal neurons of a patient with amyotrophic lateral sclerosis. Acta Neuropathol. 93, 532–536. doi: 10.1007/s004010050649
Kaku, H., Ludlow, A. V., Gutknecht, M. F., and Rothstein, T. L. (2020). FAIM opposes aggregation of mutant SOD1 that typifies some forms of familial amyotrophic lateral sclerosis. Front. Neurosci. 14:110. doi: 10.3389/fnins.2020.00110
Kanai, Y., Dohmae, N., and Hirokawa, N. (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525. doi: 10.1016/j.neuron.2004.07.022
Kass, S. U., Pruss, D., and Wolffe, A. P. (1997). How does DNA methylation repress transcription? Trends Genet. 13, 444–449. doi: 10.1016/S0168-9525(97)01268-7
Kawahara, Y., Ito, K., Sun, H., Aizawa, H., Kanazawa, I., and Kwak, S. (2004). Glutamate receptors: RNA editing and death of motor neurons. Nature 427:801. doi: 10.1038/427801a
Kawahara, Y., and Kwak, S. (2005). Excitotoxicity and ALS: what is unique about the AMPA receptors expressed on spinal motor neurons? Amyotroph. Lateral Scler. Other Motor Neuron Disord. 6, 131–144. doi: 10.1080/14660820510037872
Kawahara, Y., and Mieda-Sato, A. (2012). TDP-43 promotes microRNA biogenesis as a component of the Drosha and dicer complexes. Proc. Natl. Acad. Sci. USA 109, 3347–3352. doi: 10.1073/pnas.1112427109
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. doi: 10.1083/jcb.200502088
Kim, K. Y., Hwang, Y. J., Jung, M. K., Choe, J., Kim, Y., Kim, S., et al. (2014). A multifunctional protein EWS regulates the expression of Drosha and microRNAs. Cell Death Differ. 21, 136–145. doi: 10.1038/cdd.2013.144
Kim, H. C., Zhang, Y., King, P. H., and Lu, L. (2023). MicroRNA-183-5p regulates TAR DNA-binding protein 43 neurotoxicity via SQSTM1/p62 in amyotrophic lateral sclerosis. J. Neurochem. 164, 643–657. doi: 10.1111/jnc.15744
King, I. N., Yartseva, V., Salas, D., Kumar, A., Heidersbach, A., Ando, D. M., et al. (2014). The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J. Biol. Chem. 289, 14263–14271. doi: 10.1074/jbc.M114.561902
Kitamura, A., Iwasaki, N., and Kinjo, M. (2018). Molecular chaperone HSP70 prevents formation of inclusion bodies of the 25-kDa C-terminal fragment of TDP-43 by preventing aggregate accumulation. Cell Stress Chaperones 23, 1177–1183. doi: 10.1007/s12192-018-0930-1
Koike, Y., Sugai, A., Hara, N., Ito, J., Yokoseki, A., Ishihara, T., et al. (2021). Age-related demethylation of the TDP-43 autoregulatory region in the human motor cortex. Commun Biol 4:1107. doi: 10.1038/s42003-021-02621-0
Konno, M., Koseki, J., Kawamoto, K., Nishida, N., Matsui, H., Dewi, D. L., et al. (2015). Embryonic MicroRNA-369 controls metabolic splicing factors and urges cellular reprograming. PLoS One 10:e0132789. doi: 10.1371/journal.pone.0132789
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Koppers, M., Van Blitterswijk, M. M., Vlam, L., Rowicka, P. A., Van Vught, P. W., Groen, E. J., et al. (2012). VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 33, 837.e7–837.e13. doi: 10.1016/j.neurobiolaging.2011.10.006
Korac, J., Schaeffer, V., Kovacevic, I., Clement, A. M., Jungblut, B., Behl, C., et al. (2013). Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126, 580–592. doi: 10.1242/jcs.114926
Kwak, S., and Kawahara, Y. (2005). Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J. Mol. Med. (Berl) 83, 110–120. doi: 10.1007/s00109-004-0599-z
Laffita-Mesa, J. M., Bauer, P. O., Kouri, V., Pena Serrano, L., Roskams, J., Almaguer Gotay, D., et al. (2012). Epigenetics DNA methylation in the core ataxin-2 gene promoter: novel physiological and pathological implications. Hum. Genet. 131, 625–638. doi: 10.1007/s00439-011-1101-y
Lagier-Tourenne, C., Polymenidou, M., and Cleveland, D. W. (2010). TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64. doi: 10.1093/hmg/ddq137
Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612. doi: 10.1038/nrm1172
Lazo-Gomez, R., Ramirez-Jarquin, U. N., Tovar, Y. R. L. B., and Tapia, R. (2013). Histone deacetylases and their role in motor neuron degeneration. Front. Cell. Neurosci. 7:243. doi: 10.3389/fncel.2013.00243
Leal-Lasarte, M. M., Franco, J. M., Labrador-Garrido, A., Pozo, D., and Roodveldt, C. (2017). Extracellular TDP-43 aggregates target MAPK/MAK/MRK overlapping kinase (MOK) and trigger caspase-3/IL-18 signaling in microglia. FASEB J. 31, 2797–2816. doi: 10.1096/fj.201601163R
Lee, D. Y., Deng, Z., Wang, C. H., and Yang, B. B. (2007). MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. USA 104, 20350–20355. doi: 10.1073/pnas.0706901104
Lee, M., Nam, H. Y., Kang, H. B., Lee, W. H., Lee, G. H., Sung, G. J., et al. (2021). Epigenetic regulation of p62/SQSTM1 overcomes the radioresistance of head and neck cancer cells via autophagy-dependent senescence induction. Cell Death Dis. 12:250. doi: 10.1038/s41419-021-03539-5
Leigh, P. N., Anderton, B. H., Dodson, A., Gallo, J. M., Swash, M., and Power, D. M. (1988). Ubiquitin deposits in anterior horn cells in motor neurone disease. Neurosci. Lett. 93, 197–203. doi: 10.1016/0304-3940(88)90081-X
Leigh, P. N., Whitwell, H., Garofalo, O., Buller, J., Swash, M., Martin, J. E., et al. (1991). Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain 114, 775–788.
Li, Z., Cao, Z., Li, N., Wang, L., Fu, C., Huo, R., et al. (2023). M2 macrophage-derived Exosomal lncRNA MIR4435-2HG promotes progression of infantile hemangiomas by targeting HNRNPA1. Int. J. Nanomedicine 18, 5943–5960. doi: 10.2147/IJN.S435132
Li, C., Chen, Y., Chen, X., Wei, Q., Ou, R., Gu, X., et al. (2020). MicroRNA-183-5p is stress-inducible and protects neurons against cell death in amyotrophic lateral sclerosis. J. Cell. Mol. Med. 24, 8614–8622. doi: 10.1111/jcmm.15490
Li, Y., Gu, J., Wang, C., Hu, J., Zhang, S., Liu, C., et al. (2022b). Hsp70 exhibits a liquid-liquid phase separation ability and chaperones condensed FUS against amyloid aggregation. iScience 25:104356. doi: 10.1016/j.isci.2022.104356
Li, C., Lie, H., and Sun, W. (2022a). Inhibitory effect of miR-182-5p on retinal neovascularization by targeting angiogenin and BDNF. Mol. Med. Rep. 25:61. doi: 10.3892/mmr.2021.12577
Li, H., Miao, D., Zhu, Q., Huang, J., Lu, G., and Xu, W. (2018). MicroRNA-17-5p contributes to osteoarthritis progression by binding p62/SQSTM1. Exp. Ther. Med. 15, 1789–1794. doi: 10.3892/etm.2017.5622
Liguori, M., Nuzziello, N., Introna, A., Consiglio, A., Licciulli, F., D'errico, E., et al. (2018). Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 11:288. doi: 10.3389/fnmol.2018.00288
Ling, S. C., Polymenidou, M., and Cleveland, D. W. (2013). Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. doi: 10.1016/j.neuron.2013.07.033
Lippi, A., and Krisko, A. (2023). Protein aggregation: a detrimental symptom or an adaptation mechanism? J. Neurochem. 168, 1426–1441. doi: 10.1111/jnc.15955
Liu, C. X., and Chen, L. L. (2022). Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034. doi: 10.1016/j.cell.2022.04.021
Liu, S., and Feng, P. (2015). MiR-203 determines poor outcome and suppresses tumor growth by targeting TBK1 in osteosarcoma. Cell. Physiol. Biochem. 37, 1956–1966. doi: 10.1159/000438556
Liu, Y., Hei, Y., Shu, Q., Dong, J., Gao, Y., Fu, H., et al. (2012). VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS One 7:e35800. doi: 10.1371/journal.pone.0053178
Liu, Y., Li, H., Liu, X., Wang, B., Yang, H., Wan, B., et al. (2022c). Clinical and mechanism advances of neuronal intranuclear inclusion disease. Front. Aging Neurosci. 14:934725. doi: 10.3389/fnagi.2022.934725
Liu, Y., Liu, Y., He, Y., Zhang, N., Zhang, S., Li, Y., et al. (2023b). Hypoxia-induced FUS-circTBC1D14 stress granules promote autophagy in TNBC. Adv. Sci. 10:e2204988. doi: 10.1002/advs.202204988
Liu, W., Song, H., Xu, J., Guo, Y., Zhang, C., Yao, Y., et al. (2022b). Low shear stress inhibits endothelial mitophagy via caveolin-1/miR-7-5p/SQSTM1 signaling pathway. Atherosclerosis 356, 9–17. doi: 10.1016/j.atherosclerosis.2022.07.014
Liu, H., Wang, Y., Li, X., Zhang, Y. J., Li, J., Zheng, Y. Q., et al. (2013). Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumour Biol. 34, 1713–1722. doi: 10.1007/s13277-013-0708-0
Liu, R., Wu, J., Guo, H., Yao, W., Li, S., Lu, Y., et al. (2023a). Post-translational modifications of histones: mechanisms, biological functions, and therapeutic targets. MedComm 4:e292. doi: 10.1002/mco2.292
Liu, Y., Yang, Y., Xu, C., Liu, J., Chen, J., Li, G., et al. (2023c). Circular RNA circGlis3 protects against islet beta-cell dysfunction and apoptosis in obesity. Nat. Commun. 14:351. doi: 10.1038/s41467-023-35998-z
Liu, J., Zhou, F., Guan, Y., Meng, F., Zhao, Z., Su, Q., et al. (2022a). The biogenesis of miRNAs and their role in the development of amyotrophic lateral sclerosis. Cells 11:572. doi: 10.3390/cells11030572
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2023). Hallmarks of aging: An expanding universe. Cell 186, 243–278. doi: 10.1016/j.cell.2022.11.001
Lowe, J., Lennox, G., Jefferson, D., Morrell, K., Mcquire, D., Gray, T., et al. (1988). A filamentous inclusion body within anterior horn neurones in motor neurone disease defined by immunocytochemical localisation of ubiquitin. Neurosci. Lett. 94, 203–210. doi: 10.1016/0304-3940(88)90296-0
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 389, 251–260. doi: 10.1038/38444
Luo, Y., Na, Z., and Slavoff, S. A. (2018). P-bodies: composition, properties, and functions. Biochemistry 57, 2424–2431. doi: 10.1021/acs.biochem.7b01162
Luo, J., Zheng, J., Hao, W., Zeng, H., Zhang, Z., and Shao, G. (2021). lncRNA PCAT6 facilitates cell proliferation and invasion via regulating the miR-326/hnRNPA2B1 axis in liver cancer. Oncol. Lett. 21:471. doi: 10.3892/ol.2021.12732
Ma, J., Lin, J., Qian, J., Qian, W., Yin, J., Yang, B., et al. (2014). MiR-378 promotes the migration of liver cancer cells by down-regulating Fus expression. Cell. Physiol. Biochem. 34, 2266–2274. doi: 10.1159/000369669
Mackenzie, I. R., and Neumann, M. (2012). FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res. 1462, 40–43. doi: 10.1016/j.brainres.2011.12.010
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. doi: 10.1126/science.aar7366
Marino-Ramirez, L., Kann, M. G., Shoemaker, B. A., and Landsman, D. (2005). Histone structure and nucleosome stability. Expert Rev. Proteomics 2, 719–729. doi: 10.1586/14789450.2.5.719
Maruyama, H., Morino, H., Ito, H., Izumi, Y., Kato, H., Watanabe, Y., et al. (2010). Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226. doi: 10.1038/nature08971
Masrori, P., and Van Damme, P. (2020). Amyotrophic lateral sclerosis: a clinical review. Eur. J. Neurol. 27, 1918–1929. doi: 10.1111/ene.14393
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447. doi: 10.1038/s41580-022-00566-8
Mattsson, K., Pokrovskaja, K., Kiss, C., Klein, G., and Szekely, L. (2001). Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. USA 98, 1012–1017. doi: 10.1073/pnas.98.3.1012
Mccluggage, F., and Fox, A. H. (2021). Paraspeckle nuclear condensates: global sensors of cell stress? BioEssays 43:e2000245. doi: 10.1002/bies.202000245
Mcmillan, M., Gomez, N., Hsieh, C., Bekier, M., Li, X., Miguez, R., et al. (2023). RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol. Cell 83:e217, 219–236.e7. doi: 10.1016/j.molcel.2022.12.019
Meenhuis, A., Van Veelen, P. A., De Looper, H., Van Boxtel, N., Van Den Berge, I. J., Sun, S. M., et al. (2011). MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood 118, 916–925. doi: 10.1182/blood-2011-02-336487
Mercer, T. R., Wilhelm, D., Dinger, M. E., Solda, G., Korbie, D. J., Glazov, E. A., et al. (2011). Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 39, 2393–2403. doi: 10.1093/nar/gkq1158
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Michalska, P., and Leon, R. (2020). When it comes to an end: oxidative stress crosstalk with protein aggregation and Neuroinflammation induce neurodegeneration. Antioxidants 9:740. doi: 10.3390/antiox9080740
Milani, M., Della Valle, I., Rossi, S., Fabbrizio, P., Margotta, C., Nardo, G., et al. (2024). Neuroprotective effects of niclosamide on disease progression via inflammatory pathways modulation in SOD1-G93A and FUS-associated amyotrophic lateral sclerosis models. Neurotherapeutics 21:e00346. doi: 10.1016/j.neurot.2024.e00346
Minetti, G. C., Colussi, C., Adami, R., Serra, C., Mozzetta, C., Parente, V., et al. (2006). Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 12, 1147–1150. doi: 10.1038/nm1479
Moore, S., Alsop, E., Lorenzini, I., Starr, A., Rabichow, B. E., Mendez, E., et al. (2019). ADAR2 mislocalization and widespread RNA editing aberrations in C9orf72-mediated ALS/FTD. Acta Neuropathol. 138, 49–65. doi: 10.1007/s00401-019-01999-w
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Morahan, J. M., Yu, B., Trent, R. J., and Pamphlett, R. (2009). A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 10, 418–429. doi: 10.3109/17482960802635397
Mori, F., Kakita, A., Takahashi, H., and Wakabayashi, K. (2014). Co-localization of Bunina bodies and TDP-43 inclusions in lower motor neurons in amyotrophic lateral sclerosis. Neuropathology 34, 71–76. doi: 10.1111/neup.12044
Mori, K., Weng, S. M., Arzberger, T., May, S., Rentzsch, K., Kremmer, E., et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. doi: 10.1126/science.1232927
Morlando, M., Dini Modigliani, S., Torrelli, G., Rosa, A., Di Carlo, V., Caffarelli, E., et al. (2012). FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 31, 4502–4510. doi: 10.1038/emboj.2012.319
Naganuma, T., Nakagawa, S., Tanigawa, A., Sasaki, Y. F., Goshima, N., and Hirose, T. (2012). Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31, 4020–4034. doi: 10.1038/emboj.2012.251
Nesterov, S. V., Ilyinsky, N. S., and Uversky, V. N. (2021). Liquid-liquid phase separation as a common organizing principle of intracellular space and biomembranes providing dynamic adaptive responses. Biochim. Biophys. Acta, Mol. Cell Res. 1868:119102. doi: 10.1016/j.bbamcr.2021.119102
Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. doi: 10.1126/science.1134108
Ni, C., Zheng, K., Liu, W., Ou, Y., Li, G., and Jin, G. (2022). LINC00483 promotes proliferation and metastasis through the miR-19a-3p/TBK1/MAPK axis in pancreatic ductal adenocarcinoma (PDAC). Ann. Transl. Med. 10:317. doi: 10.21037/atm-22-907
Nicoglou, A., and Merlin, F. (2017). Epigenetics: a way to bridge the gap between biological fields. Stud. Hist. Phil. Biol. Biomed. Sci. 66, 73–82. doi: 10.1016/j.shpsc.2017.10.002
Nishimoto, Y., Nakagawa, S., Hirose, T., Okano, H. J., Takao, M., Shibata, S., et al. (2013). The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 6:31. doi: 10.1186/1756-6606-6-31
Nussbacher, J. K., Tabet, R., Yeo, G. W., and Lagier-Tourenne, C. (2019). Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320. doi: 10.1016/j.neuron.2019.03.014
Oakes, J. A., Davies, M. C., and Collins, M. O. (2017). TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol. Brain 10:5. doi: 10.1186/s13041-017-0287-x
Oates, N., and Pamphlett, R. (2007). An epigenetic analysis of SOD1 and VEGF in ALS. Amyotroph. Lateral Scler. 8, 83–86. doi: 10.1080/17482960601149160
Okamoto, K., Mizuno, Y., and Fujita, Y. (2008). Bunina bodies in amyotrophic lateral sclerosis. Neuropathology 28, 109–115. doi: 10.1111/j.1440-1789.2007.00873.x
Okamoto, Y., Shirakashi, Y., Ihara, M., Urushitani, M., Oono, M., Kawamoto, Y., et al. (2011). Colocalization of 14-3-3 proteins with SOD1 in Lewy body-like hyaline inclusions in familial amyotrophic lateral sclerosis cases and the animal model. PLoS One 6:e20427. doi: 10.1371/journal.pone.0020427
Pakravan, D., Orlando, G., Bercier, V., and Van Den Bosch, L. (2021). Role and therapeutic potential of liquid-liquid phase separation in amyotrophic lateral sclerosis. J. Mol. Cell Biol. 13, 15–28. doi: 10.1093/jmcb/mjaa049
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi: 10.1016/j.cell.2015.07.047
Perez-Cabello, J. A., Silvera-Carrasco, L., Franco, J. M., Capilla-Gonzalez, V., Armaos, A., Gomez-Lima, M., et al. (2023). MAPK/MAK/MRK overlapping kinase (MOK) controls microglial inflammatory/type-I IFN responses via Brd4 and is involved in ALS. Proc. Natl. Acad. Sci. USA 120:e2302143120. doi: 10.1073/pnas.2302143120
Picher-Martel, V., Dutta, K., Phaneuf, D., Sobue, G., and Julien, J. P. (2015). Ubiquilin-2 drives NF-kappaB activity and cytosolic TDP-43 aggregation in neuronal cells. Mol. Brain 8:71. doi: 10.1186/s13041-015-0162-6
Pigna, E., Simonazzi, E., Sanna, K., Bernadzki, K. M., Proszynski, T., Heil, C., et al. (2019). Histone deacetylase 4 protects from denervation and skeletal muscle atrophy in a murine model of amyotrophic lateral sclerosis. EBioMedicine 40, 717–732. doi: 10.1016/j.ebiom.2019.01.038
Prasad, A., Bharathi, V., Sivalingam, V., Girdhar, A., and Patel, B. K. (2019). Molecular mechanisms of TDP-43 Misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 12:25. doi: 10.3389/fnmol.2019.00025
Rausch, C., Zhang, P., Casas-Delucchi, C. S., Daiss, J. L., Engel, C., Coster, G., et al. (2021). Cytosine base modifications regulate DNA duplex stability and metabolism. Nucleic Acids Res. 49, 12870–12894. doi: 10.1093/nar/gkab509
Ray Chaudhuri, A., and Nussenzweig, A. (2017). The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621. doi: 10.1038/nrm.2017.53
Robberecht, W., and Philips, T. (2013). The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 14, 248–264. doi: 10.1038/nrn3430
Robertson, K. D. (2005). DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610. doi: 10.1038/nrg1655
Rodriguez-Aguayo, C., Monroig, P. D. C., Redis, R. S., Bayraktar, E., Almeida, M. I., Ivan, C., et al. (2017). Regulation of hnRNPA1 by microRNAs controls the miR-18a-K-RAS axis in chemotherapy-resistant ovarian cancer. Cell Discov. 3:17029. doi: 10.1038/celldisc.2017.29
Rossaert, E., Pollari, E., Jaspers, T., Van Helleputte, L., Jarpe, M., Van Damme, P., et al. (2019). Restoration of histone acetylation ameliorates disease and metabolic abnormalities in a FUS mouse model. Acta Neuropathol. Commun. 7:107. doi: 10.1186/s40478-019-0750-2
Rouaux, C., Jokic, N., Mbebi, C., Boutillier, S., Loeffler, J. P., and Boutillier, A. L. (2003). Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 22, 6537–6549. doi: 10.1093/emboj/cdg615
Rouaux, C., Panteleeva, I., Rene, F., Gonzalez De Aguilar, J. L., Echaniz-Laguna, A., Dupuis, L., et al. (2007). Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J. Neurosci. 27, 5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017). The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255. doi: 10.1038/nature24456
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S., and Mason, C. E. (2012). The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13:175. doi: 10.1186/gb-2012-13-10-175
Salifu, A. R., Pang, Y., He, Y., Liu, Y., Shen, Y., Xu, X., et al. (2023). MiR-217 targets TBK1 to modulate inflammatory response in grass carp following infection with Aeromonas hydrophila. Dev. Comp. Immunol. 143:104583. doi: 10.1016/j.dci.2022.104583
Sanna, S., Esposito, S., Masala, A., Sini, P., Nieddu, G., Galioto, M., et al. (2020). HDAC1 inhibition ameliorates TDP-43-induced cell death in vitro and in vivo. Cell Death Dis. 11:369. doi: 10.1038/s41419-020-2580-3
Santovito, D., and Weber, C. (2022). Non-canonical features of microRNAs: paradigms emerging from cardiovascular disease. Nat. Rev. Cardiol. 19, 620–638. doi: 10.1038/s41569-022-00680-2
Scuteri, A., Sanna, S., Chen, W. M., Uda, M., Albai, G., Strait, J., et al. (2007). Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3:e115. doi: 10.1371/journal.pgen.0030115
Seelaar, H., Schelhaas, H. J., Azmani, A., Kusters, B., Rosso, S., Majoor-Krakauer, D., et al. (2007). TDP-43 pathology in familial frontotemporal dementia and motor neuron disease without Progranulin mutations. Brain 130, 1375–1385. doi: 10.1093/brain/awm024
Seilhean, D., Takahashi, J., El Hachimi, K. H., Fujigasaki, H., Lebre, A. S., Biancalana, V., et al. (2004). Amyotrophic lateral sclerosis with neuronal intranuclear protein inclusions. Acta Neuropathol. 108, 81–87. doi: 10.1007/s00401-004-0855-x
Shelkovnikova, T. A., Kukharsky, M. S., An, H., Dimasi, P., Alexeeva, S., Shabir, O., et al. (2018). Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 13:30. doi: 10.1186/s13024-018-0263-7
Shen, Y., Chen, A., Wang, W., Shen, Y., Ruggeri, F. S., Aime, S., et al. (2023). The liquid-to-solid transition of FUS is promoted by the condensate surface. Proc. Natl. Acad. Sci. USA 120:e2301366120. doi: 10.1073/pnas.2301366120
Sheng, J., Yu, W., Gao, X., Xu, Z., and Hu, G. F. (2014). Angiogenin stimulates ribosomal RNA transcription by epigenetic activation of the ribosomal DNA promoter. J. Cell. Physiol. 229, 521–529. doi: 10.1002/jcp.24477
Shimbo, T., and Wade, P. A. (2016). Proteins that read DNA methylation. Adv. Exp. Med. Biol. 945, 303–320. doi: 10.1007/978-3-319-43624-1_13
Shu, F., Xiao, H., Li, Q. N., Ren, X. S., Liu, Z. G., Hu, B. W., et al. (2023). Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets. Signal Transduct. Target. Ther. 8:32. doi: 10.1038/s41392-022-01300-8
Simoes-Pires, C., Zwick, V., Nurisso, A., Schenker, E., Carrupt, P. A., and Cuendet, M. (2013). HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol. Neurodegener. 8:7. doi: 10.1186/1750-1326-8-7
Simpson, C. L., Lemmens, R., Miskiewicz, K., Broom, W. J., Hansen, V. K., Van Vught, P. W., et al. (2009). Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481. doi: 10.1093/hmg/ddn375
Smith, B. N., Ticozzi, N., Fallini, C., Gkazi, A. S., Topp, S., Kenna, K. P., et al. (2014). Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 84, 324–331. doi: 10.1016/j.neuron.2014.09.027
Song, B., Zheng, K., Ma, H., Liu, A., Jing, W., Shao, C., et al. (2015). miR-429 determines poor outcome and inhibits pancreatic ductal adenocarcinoma growth by targeting TBK1. Cell. Physiol. Biochem. 35, 1846–1856. doi: 10.1159/000373995
Stavreva, D. A., and Hager, G. L. (2015). Chromatin structure and gene regulation: a dynamic view of enhancer function. Nucleus 6, 442–448. doi: 10.1080/19491034.2015.1107689
Stefanis, L. (2012). Alpha-Synuclein in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2:a009399. doi: 10.1101/cshperspect.a009399
Strong, M. J., Abrahams, S., Goldstein, L. H., Woolley, S., Mclaughlin, P., Snowden, J., et al. (2017). Amyotrophic lateral sclerosis - frontotemporal spectrum disorder (ALS-FTSD): revised diagnostic criteria. Amyotroph Lateral Scler. Frontotemporal. Degener. 18, 153–174. doi: 10.1080/21678421.2016.1267768
Strong, M. J., Kesavapany, S., and Pant, H. C. (2005). The pathobiology of amyotrophic lateral sclerosis: a proteinopathy? J. Neuropathol. Exp. Neurol. 64, 649–664. doi: 10.1097/01.jnen.0000173889.71434.ea
Sultan, F. A., and Day, J. J. (2011). Epigenetic mechanisms in memory and synaptic function. Epigenomics 3, 157–181. doi: 10.2217/epi.11.6
Sun, Y., Dai, H., Dai, X., Yin, J., Cui, Y., Liu, X., et al. (2023). M(1)a in CAG repeat RNA binds to TDP-43 and induces neurodegeneration. Nature 623, 580–587. doi: 10.1038/s41586-023-06701-5
Suzuki, N., Kato, S., Kato, M., Warita, H., Mizuno, H., Kato, M., et al. (2012). FUS/TLS-immunoreactive neuronal and glial cell inclusions increase with disease duration in familial amyotrophic lateral sclerosis with an R521C FUS/TLS mutation. J. Neuropathol. Exp. Neurol. 71, 779–788. doi: 10.1097/NEN.0b013e318264f164
Suzuki, H., Shibagaki, Y., Hattori, S., and Matsuoka, M. (2019). C9-ALS/FTD-linked proline-arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 10:746. doi: 10.1038/s41419-019-1983-5
Svetoni, F., De Paola, E., La Rosa, P., Mercatelli, N., Caporossi, D., Sette, C., et al. (2017). Post-transcriptional regulation of FUS and EWS protein expression by miR-141 during neural differentiation. Hum. Mol. Genet. 26, 2732–2746. doi: 10.1093/hmg/ddx160
Tada, M., Doi, H., Koyano, S., Kubota, S., Fukai, R., Hashiguchi, S., et al. (2018). Matrin 3 is a component of neuronal cytoplasmic inclusions of motor neurons in sporadic amyotrophic lateral sclerosis. Am. J. Pathol. 188, 507–514. doi: 10.1016/j.ajpath.2017.10.007
Tai, H., Wang, A., Zhang, Y., Liu, S., Pan, Y., Li, K., et al. (2023). Clinical features and classification of neuronal Intranuclear inclusion disease. Neurol Genet 9:e200057. doi: 10.1212/NXG.0000000000200057
Tan, M. H., Li, Q., Shanmugam, R., Piskol, R., Kohler, J., Young, A. N., et al. (2017). Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254. doi: 10.1038/nature24041
Thiyagarajan, N., Ferguson, R., Subramanian, V., and Acharya, K. R. (2012). Structural and molecular insights into the mechanism of action of human angiogenin-ALS variants in neurons. Nat. Commun. 3:1121. doi: 10.1038/ncomms2126
Tibshirani, M., Tradewell, M. L., Mattina, K. R., Minotti, S., Yang, W., Zhou, H., et al. (2015). Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1. Hum. Mol. Genet. 24, 773–786. doi: 10.1093/hmg/ddu494
To, K. K. W., Xing, E., Larue, R. C., and Li, P. K. (2023). BET Bromodomain inhibitors: novel design strategies and therapeutic applications. Molecules 28:3043. doi: 10.3390/molecules28073043
Toivonen, J. M., Manzano, R., Olivan, S., Zaragoza, P., Garcia-Redondo, A., and Osta, R. (2014). MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One 9:e89065. doi: 10.1371/journal.pone.0089065
Tomonaga, M., Saito, M., Yoshimura, M., Shimada, H., and Tohgi, H. (1978). Ultrastructure of the Bunina bodies in anterior horn cells of amyotrophic lateral sclerosis. Acta Neuropathol. 42, 81–86. doi: 10.1007/BF00690971
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20. doi: 10.1038/s41580-018-0059-1
Tsitsipatis, D., Mazan-Mamczarz, K., Si, Y., Herman, A. B., Yang, J. H., Guha, A., et al. (2022). Transcriptomic analysis of human ALS skeletal muscle reveals a disease-specific pattern of dysregulated circRNAs. Aging (Albany NY) 14, 9832–9859. doi: 10.18632/aging.204450
Vasudevan, S., Tong, Y., and Steitz, J. A. (2008). Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7, 1545–1549. doi: 10.4161/cc.7.11.6018
Vaz, B., Halder, S., and Ramadan, K. (2013). Role of p97/VCP (Cdc48) in genome stability. Front. Genet. 4:60. doi: 10.3389/fgene.2013.00060
Veerappan, C. S., Sleiman, S., and Coppola, G. (2013). Epigenetics of Alzheimer's disease and frontotemporal dementia. Neurotherapeutics 10, 709–721. doi: 10.1007/s13311-013-0219-0
Wada, M., Uchihara, T., Nakamura, A., and Oyanagi, K. (1999). Bunina bodies in amyotrophic lateral sclerosis on Guam: a histochemical, immunohistochemical and ultrastructural investigation. Acta Neuropathol. 98, 150–156. doi: 10.1007/s004010051063
Walkley, C. R., and Li, J. B. (2017). Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol. 18:205. doi: 10.1186/s13059-017-1347-3
Wang, P., Doxtader, K. A., and Nam, Y. (2016a). Structural basis for cooperative function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, L., Hu, M., Zuo, M. Q., Zhao, J., Wu, D., Huang, L., et al. (2020). Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 30, 393–407. doi: 10.1038/s41422-020-0288-7
Wang, J., Hussain, T., Yue, R., Liao, Y., Li, Q., Yao, J., et al. (2018). MicroRNA-199a inhibits cellular autophagy and downregulates IFN-beta expression by targeting TBK1 in Mycobacterium bovis infected cells. Front. Cell. Infect. Microbiol. 8:238. doi: 10.3389/fcimb.2018.00238
Wang, W. Y., Pan, L., Su, S. C., Quinn, E. J., Sasaki, M., Jimenez, J. C., et al. (2013). Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 16, 1383–1391. doi: 10.1038/nn.3514
Wang, Z., Shi, Z., Zhang, L., Zhang, H., and Zhang, Y. (2019). Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in human hepatocellular carcinoma. Pathol. Res. Pract. 215, 499–505. doi: 10.1016/j.prp.2018.12.012
Wang, N., Wu, R., Tang, D., and Kang, R. (2021b). The BET family in immunity and disease. Signal Transduct. Target. Ther. 6:23. doi: 10.1038/s41392-020-00384-4
Wang, B., Zhang, L., Dai, T., Qin, Z., Lu, H., Zhang, L., et al. (2021a). Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target. Ther. 6:290. doi: 10.1038/s41392-021-00678-1
Wang, Y., Zhang, N., Zhang, L., Li, R., Fu, W., Ma, K., et al. (2016b). Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol. Cell 63, 34–48. doi: 10.1016/j.molcel.2016.05.027
Watanabe, S., Inami, H., Oiwa, K., Murata, Y., Sakai, S., Komine, O., et al. (2020). Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis. 11:909. doi: 10.1038/s41419-020-03116-2
Wen, Z., Lian, L., Ding, H., Hu, Y., Xiao, Z., Xiong, K., et al. (2020). LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 17, 381–394. doi: 10.1080/15476286.2019.1708547
Weng, C., Dong, H., Chen, G., Zhai, Y., Bai, R., Hu, H., et al. (2012). miR-409-3p inhibits HT1080 cell proliferation, vascularization and metastasis by targeting angiogenin. Cancer Lett. 323, 171–179. doi: 10.1016/j.canlet.2012.04.010
Widagdo, J., and Anggono, V. (2018). The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J. Neurochem. 147, 137–152. doi: 10.1111/jnc.14481
Widom, J. (1998). Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27, 285–327. doi: 10.1146/annurev.biophys.27.1.285
Williams, A. H., Valdez, G., Moresi, V., Qi, X., Mcanally, J., Elliott, J. L., et al. (2009). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554. doi: 10.1126/science.1181046
Wilson, D. M. 3rd, Cookson, M. R., Van Den Bosch, L., Zetterberg, H., Holtzman, D. M., and Dewachter, I. (2023). Hallmarks of neurodegenerative diseases. Cell 186, 693–714. doi: 10.1016/j.cell.2022.12.032
Wilson, M. M., Henshall, D. C., Byrne, S. M., and Brennan, G. P. (2021). CHD2-related CNS pathologies. Int. J. Mol. Sci. 22:588. doi: 10.3390/ijms22020588
Woo, J. A., Liu, T., Trotter, C., Fang, C. C., De Narvaez, E., Lepochat, P., et al. (2017). Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat. Commun. 8:15558. doi: 10.1038/ncomms15558
Woodage, T., Basrai, M. A., Baxevanis, A. D., Hieter, P., and Collins, F. S. (1997). Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94, 11472–11477. doi: 10.1073/pnas.94.21.11472
Woodcock, C. L. (2005). A milestone in the odyssey of higher-order chromatin structure. Nat. Struct. Mol. Biol. 12, 639–640. doi: 10.1038/nsmb0805-639
Woulfe, J. M. (2007). Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 33, 2–42. doi: 10.1111/j.1365-2990.2006.00819.x
Wu, C. H., Fallini, C., Ticozzi, N., Keagle, P. J., Sapp, P. C., Piotrowska, K., et al. (2012). Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503. doi: 10.1038/nature11280
Wu, Q., Li, G., Gong, L., Cai, J., Chen, L., Xu, X., et al. (2023). Identification of miR-30c-5p as a tumor suppressor by targeting the m(6) a reader HNRNPA2B1 in ovarian cancer. Cancer Med. 12, 5055–5070. doi: 10.1002/cam4.5246
Xi, Z., Zhang, M., Bruni, A. C., Maletta, R. G., Colao, R., Fratta, P., et al. (2015). The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol. 129, 715–727. doi: 10.1007/s00401-015-1401-8
Xiao, S., Mclean, J., and Robertson, J. (2006). Neuronal intermediate filaments and ALS: a new look at an old question. Biochim. Biophys. Acta 1762, 1001–1012. doi: 10.1016/j.bbadis.2006.09.003
Xu, S., Fang, Y., Chang, L., Bian, Y., Wang, Y., Ding, J., et al. (2023). STAT2-induced linc02231 promotes tumorigenesis and angiogenesis through modulation of hnRNPA1/ANGPTL4 in colorectal cancer. J. Gene Med. 25:e3506. doi: 10.1002/jgm.3506
Yadav, S., Singh, N., Shah, P. P., Rowbotham, D. A., Malik, D., Srivastav, A., et al. (2017). MIR155 regulation of Ubiquilin1 and Ubiquilin2: implications in cellular protection and tumorigenesis. Neoplasia 19, 321–332. doi: 10.1016/j.neo.2017.02.001
Yamashita, T., Hideyama, T., Teramoto, S., and Kwak, S. (2012). The abnormal processing of TDP-43 is not an upstream event of reduced ADAR2 activity in ALS motor neurons. Neurosci. Res. 73, 153–160. doi: 10.1016/j.neures.2012.02.015
Yamashita, T., and Kwak, S. (2014). The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 1584, 28–38. doi: 10.1016/j.brainres.2013.12.011
Yang, Z., Dong, X., Pu, M., Yang, H., Chang, W., Ji, F., et al. (2020). LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer 23, 449–463. doi: 10.1007/s10120-019-01019-6
Yang, C., Rong, R., Li, Y., Cheng, M., and Luo, Y. (2022). Decrease in LINC00963 attenuates the progression of pulmonary arterial hypertension via microRNA-328-3p/profilin 1 axis. J. Clin. Lab. Anal. 36:e24383. doi: 10.1002/jcla.24383
Yeh, L. Y., Liu, C. J., Wong, Y. K., Chang, C., Lin, S. C., and Chang, K. W. (2015). miR-372 inhibits p62 in head and neck squamous cell carcinoma in vitro and in vivo. Oncotarget 6, 6062–6075. doi: 10.18632/oncotarget.3340
Yin, N., Yang, Y., Wang, X., Yang, C., Ma, X., Shaukat, A., et al. (2019). MiR-19a mediates the negative regulation of the NF-kappaB pathway in lipopolysaccharide-induced endometritis by targeting TBK1. Inflamm. Res. 68, 231–240. doi: 10.1007/s00011-019-01213-3
You, L., Zheng, Y., Yang, J., Hou, Q., Wang, L., Zhang, Y., et al. (2022). LncRNA MDRL mitigates atherosclerosis through miR-361/SQSTM1/NLRP3 signaling. Mediat. Inflamm. 2022, 1–13. doi: 10.1155/2022/5463505
Yu, Y., Pang, D., Li, C., Gu, X., Chen, Y., Ou, R., et al. (2022). The expression discrepancy and characteristics of long non-coding RNAs in peripheral blood leukocytes from amyotrophic lateral sclerosis patients. Mol. Neurobiol. 59, 3678–3689. doi: 10.1007/s12035-022-02789-4
Yu, L. Y., Saarma, M., and Arumae, U. (2008). Death receptors and caspases but not mitochondria are activated in the GDNF- or BDNF-deprived dopaminergic neurons. J. Neurosci. 28, 7467–7475. doi: 10.1523/JNEUROSCI.1877-08.2008
Yu, Z., Zhu, M., Shu, D., Zhang, R., Xiang, Z., Jiang, A., et al. (2023). LncRNA PEG11as aggravates cerebral ischemia/reperfusion injury after ischemic stroke through miR-342-5p/PFN1 axis. Life Sci. 313:121276. doi: 10.1016/j.lfs.2022.121276
Zeier, Z., Esanov, R., Belle, K. C., Volmar, C. H., Johnstone, A. L., Halley, P., et al. (2015). Bromodomain inhibitors regulate the C9ORF72 locus in ALS. Exp. Neurol. 271, 241–250. doi: 10.1016/j.expneurol.2015.06.017
Zeng, T., Zhang, S., He, Y., Liu, Z., and Cheng, Q. (2021). MiR-361-5p promotes oxygen-glucose deprivation/re-oxygenation induced neuronal injury by negatively regulating SQSTM1 in vitro. Metab. Brain Dis. 36, 2359–2368. doi: 10.1007/s11011-021-00845-x
Zhang, E., and Li, X. (2019). LncRNA SOX2-OT regulates proliferation and metastasis of nasopharyngeal carcinoma cells through miR-146b-5p/HNRNPA2B1 pathway. J. Cell. Biochem. 120, 16575–16588. doi: 10.1002/jcb.28917
Zhang, C., Nie, P., Zhou, C., Hu, Y., Duan, S., Gu, M., et al. (2021). Oxidative stress-induced mitophagy is suppressed by the miR-106b-93-25 cluster in a protective manner. Cell Death Dis. 12:209. doi: 10.1038/s41419-021-03484-3
Zhang, L., Sun, P., Zhang, Y., Xu, Y., and Sun, Y. (2020). miR-182-5p inhibits the pathogenic Th17 response in experimental autoimmune uveitis mice via suppressing TAF15. Biochem. Biophys. Res. Commun. 529, 784–792. doi: 10.1016/j.bbrc.2020.06.073
Zhang, M., Tartaglia, M. C., Moreno, D., Sato, C., Mckeever, P., Weichert, A., et al. (2017). DNA methylation age-acceleration is associated with disease duration and age at onset in C9orf72 patients. Acta Neuropathol. 134, 271–279. doi: 10.1007/s00401-017-1713-y
Zhang, Y., Zheng, S., Geng, Y., Xue, J., Wang, Z., Xie, X., et al. (2015). MicroRNA profiling of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS One 10:e0122674. doi: 10.1371/journal.pone.0145988
Zhao, Y., Ran, Z., Jiang, Q., Hu, N., Yu, B., Zhu, L., et al. (2019). Vitamin D alleviates rotavirus infection through a Microrna-155-5p mediated regulation of the TBK1/IRF3 signaling pathway in vivo and in vitro. Int. J. Mol. Sci. 20:3562. doi: 10.3390/ijms20143562
Zheng, L., Kang, Y., Zhang, L., and Zou, W. (2020). MiR-133a-5p inhibits androgen receptor (AR)-induced proliferation in prostate cancer cells via targeting FUsed in sarcoma (FUS) and AR. Cancer Biol. Ther. 21, 34–42. doi: 10.1080/15384047.2019.1665393
Zhou, C., Cao, H., Meng, X., and Zhang, Q. (2022a). Lnc-MEG3 inhibits invasion, migration, and epithelial- mesenchymal transition of nasopharyngeal carcinoma cells by regulating sequestosome 1. Head Neck 44, 201–211. doi: 10.1002/hed.26917
Zhou, Q., Kang, G., Jiang, P., Qiao, R., Lam, W. K. J., Yu, S. C. Y., et al. (2022b). Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proc. Natl. Acad. Sci. USA 119:e2209852119. doi: 10.1073/pnas.2209852119
Zhou, B., Wang, Y., Jiang, J., Jiang, H., Song, J., Han, T., et al. (2016). The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life 68, 201–210. doi: 10.1002/iub.1474
Keywords: ALS, epigenetics, protein aggregates, DNA methylation, histone modifications, chromatin remodeling enzymes, non-coding RNAs, RNA modifications
Citation: Noches V, Campos-Melo D, Droppelmann CA and Strong MJ (2024) Epigenetics in the formation of pathological aggregates in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 17:1417961. doi: 10.3389/fnmol.2024.1417961
Received: 15 April 2024; Accepted: 23 August 2024;
Published: 03 September 2024.
Edited by:
Baojin Ding, Louisiana State University Health Shreveport, United StatesReviewed by:
Ashley Watson, University of California, San Diego, United StatesCopyright © 2024 Noches, Campos-Melo, Droppelmann and Strong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Strong, bXN0cm9uZ0B1d28uY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.