
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 15 July 2024
Sec. Neuroplasticity and Development
Volume 17 - 2024 | https://doi.org/10.3389/fnmol.2024.1405109
This article is part of the Research TopicMechanisms of Auditory Development, Maintenance, Damage, and ProtectionView all 9 articles
 Binling Xie1,2,3†
Binling Xie1,2,3† Jiaxin Liang1,2,3†
Jiaxin Liang1,2,3† Jifan Jiang1,2†
Jifan Jiang1,2† Ting Zeng1,2
Ting Zeng1,2 Ling Liu1,2
Ling Liu1,2 Dinghua Xie4
Dinghua Xie4 Ganghua Zhu4
Ganghua Zhu4 Lei Xiong1,2,3
Lei Xiong1,2,3 Kanjia Zhang4
Kanjia Zhang4 Dong Liu5
Dong Liu5 Jie Gong5
Jie Gong5 Xiangding Chen3
Xiangding Chen3 Ruosha Lai4*
Ruosha Lai4* Huaping Xie1*
Huaping Xie1*Introduction: myo7aa, the homolog of the human Usher 1B syndrome pathogenic gene, myo7A, plays an important role in stereociliary development and maintenance, therefore, is critical for hearing and balance. However, the molecular mechanisms that myo7aa regulate hearing and balance still need to be studied.
Methods: In this study, we generated two independent zebrafish myo7aa knockout lines using CRISPR/Cas9 technology. To investigate the effects of myo7aa on hearing, YO-PRO-1 staining and startle response assay were used. To gain insight into the specific molecular mechanisms by which myo7aa affects hearing, transcriptome sequencing and bioinformatics analysis were employed.
Results: Our study showed that hair cells of myo7aa-/- zebrafish can not take up YO-PRO-1 fluorescent dye and are insensitive to acoustic stimulation in myo7aa-/- zebrafish compared to wild type. Genes related to the Rho GTPase signaling pathway, such as arhgap33, dab2ip, and arghef40, are significantly down-regulated in myo7aa-/- zebrafish embryos at 3 dpf. GTP and ATP compensation can partially rescue the hair cell defects in myo7aa knockout zebrafish.
Discussion: Our findings suggest that zebrafish myo7aa affects congenital hearing by regulating Rho GTPase signaling, and loss of myo7aa leads to abnormal Rho GTPase signaling and impairs hair cell function. myo7aa, myo7A, arhgap33, dab2ip, arghef40 and myo7aa-/- fonts in the abstract are italicized. -/- is a superscript format.
Deafness and hearing impairment are major global health problems. The ear is an important hearing and balance organ for humans, and the inner ear hair cells are the key sensory cells for auditory conversion.
There are about 15,000 hair cells in the human inner ear, among which there are about 3,000 cochlear hair cells as auditory receptors (Rao et al., 2019). When the hair cell is stimulated, the apical stereociliary deflection occurs, leading to the opening of the mechanical conduction channel at the tip of the stereociliary, K+ influx, and membrane potential changes. The calcium channel at the hair cell synapse opens, and the synaptic vesicle fusing and releasing neurotransmitters to transmit sound signals to the brain (Fettiplace, 2017; Nicolson, 2017). Zebrafish is widely used in the auditory research because of its embryonic transparency, morphological structure and physiological function of inner ear and lateral line hair cells, which are similar to those of human cochlear hair cells (Pickett and Raible, 2019).
More than 160 different MYO7A mutations have been found to cause a variety of human hearing disorders, including Usher 1B syndrome, recessive nonsyndromic deafness (DFNB2), and dominant nonsyndromic deafness (DFNA11) (Ma et al., 2016).
The MYO7A protein in zebrafish is highly homologous to the proteins found in human and mice. All the interaction domains and signaling protein domains of the MYO7A protein are evolutionary conserved. The myo7aa gene in zebrafish is located on chromosome 18 and spans a length of 96,803 bp, with 49 exons. It encodes one MYSc protein domain, four IQ motifs, two MyTH4_B41 tandem domains, and one SH3 protein domain. Mariner zebrafish is the first fish model associated with human hereditary deafness (Ernest et al., 2000). Five alleles of Mariner have been identified by forward genetic screening methods. Mariner zebrafish larvae are characterized by defects in inner ear hair cell bundle, reduced sensitivity to acoustic vibration, and reduced or absent extracellular hair cell potential. These characteristics are similar to the phenotype of shaker-1 mice (Self et al., 1998).
Stereocilia are actin-based cell processes on the apical surface of hair cells in the inner ear, which play a key role in hearing and balance sensation. Stereocilia development and maintenance are tightly regulated, and defects in this process often lead to hearing or balance disorders. The Rho GTPase family consists of about 20 members, including CDC42, RAC1, RhoA, and others, which play important roles in cytoskeleton rearrangement, cell motility, cell polarity, axon guidance, vesicle trafficking, and cell cycle progression (Heasman and Ridley, 2008). RhoA regulates F-actin formation and adhesion through its downstream effectors, such as mDia (Watanabe et al., 1999). The Rho GTPase cell division cycle 42 (CDC42) is located in hair cell stereocilia and is responsible for filopodia formation and actin stress fiber assembly (Du et al., 2021; Dai et al., 2023). Rac1 regulates the interaction between kinocilium and stereocilia via p21-activated kinase (PAK), inducing membrane folding and lamellipodia formation, which are required for cohesion of developing hair bundles. Loss of Rac1 in mouse ear epithelium leads to abnormal cochlear epithelial morphology and reduced number of auditory hair cells. Hair cells also exhibit defects in planar cell polarity and morphogenesis of stereociliary bundles, including bundle fragmentation or deformation (Grimsley-Myers et al., 2009).
In this study, we utilized CRISPR/Cas9 technology to generate zebrafish myo7aa gene knockout lines and examined the role of myo7aa in sensory hair cells. The lack of YO-PRO-1 fluorescence staining and the startle response assay indicated the knockout of myo7aa affected the normal function of the hair cells. Comparative transcriptome analysis showed significant changes in the expression levels of genes involved in endocytosis and exocytosis and Rho GTPase signaling in myo7aa mutant larvae. GTP and ATP compensation can partially rescue the hair cell defect in myo7aa knockout zebrafish. Taken together, our findings suggest that loss of myo7aa leads to aberrant Rho GTPase signaling, which is essential for normal hair cell function.
Tübingen (TU) zebrafish (Laboratory of Animal Nutrition and Human Health, College of Life Science, Hunan Normal University) was used in this study. The embryos were were obtained through natural mating and incubated at 28.5°C. Additionally, the embryos were cultured in E3 water (286.7 mg NaCl, 12.7 mg KCl, 48.3 mg CaCl2·2H2O, 81.7 mg MgSO4·7H2O, 50 μL 0.01% methylene blue per liter of pure water) in Petri dishes until day 5. When developed to desired stages, embryos were collected and fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) overnight at 4°C.
The myo7aa mutant lines were generated with the CRISPR/Cas9 system. Firstly, we obtained the mRNA sequence and amino acid sequence of myo7aa gene from NCBI database and analyzed the structural domain of MYO7AA protein on SMART website.1 Then, We searched the intron and exon sequences of zebrafish myo7aa using the UCSC (UCSC Genome Browser Home) database, and designed two target sites in exon 6, which is located in the coding region of the MYSc structural domain: 5’-GTATACGGGGTCCATCTTAGTGGG-3′ and 5’-GAATAGCAGTTGTCTGCGACGGG-3′. The T7 promoter (TAATACGACTCACTATA) was added at the 5′ end of the target sequence, and the upstream sequence of the sgRNA backbone sequence (GTTTTTAGAGCTAGAAATAG) was added at the 3′ end of the target sequence as the forward priming sequence, and the downstream sequence of the sgRNA backbone as the reverse priming sequence for PCR amplification. The sgRNA was synthesized in vitro using the T7 Transcription Kit (Thermo Fisher Scientific). sgRNA1 and sgRNA2 were purified and recovered using the RNA Purification Kit (Qiagen). Cas9 protein was purchased from Thermo Fisher Scientific. Finally, the two sgRNAs and Cas9 protein were prepared and mixed together (final concentration was 20 μg/μL sgRNA and 500 μg/μL Cas9 protein). Approximately 1 nL of the mixture was injected into wild-type embryos at the single-cell stage of zebrafish embryos. After injection, embryos were incubated at 28.5°C.
A pair of primers for genotyping was designed at both ends of the two target sites: myo7aa-Fwd-GT (TGTTGCAATGTCCTGATGGG) and myo7aa-Rev-GT (ACAGCATATCGTCCCATGGA). For wild-type, the PCR product is 361 bp. The targeted genomic regions were amplified by PCR and subjected to 1.5% agarose gel electrophoresis to determine the success of the knockdown.
Control and injected embryos were randomly selected for validity testing at 36 hpf. The two target sites are 142 bp apart, if both target sites work well, there will be a deletion of approximately 142 bp compared to the control. Once the mutation was confirmed in the injected embryos, the remaining fish were cultured to 45 dpf, then each fish was tail clipped, genomic DNA was extracted for genotyping, and the fish with the ~142 bp deletion were cultured to adulthood as the F0 generation and then crossed to WT. Two independently inherited heterozygous mutant zebrafish lines, the PCR product sizes of line1 and line2 are 247 and 475 bp, respectively, were obtained from the offspring of the same F0 generation mutant. Two PCR products with sizes of 247 bp and 475 bp were purified and sent to Qingke Biotech for Sanger sequencing.
The mRNA sequence of the myo7aa gene was obtained from NCBI, and the primer sequence for anti-sense RNA probe was designed using the primer3 input online primer design software. The primers were then sent to Sangon Biological Company for synthesis. The primers used for the in situ hybridization probes in this study were as follows: myo7aa Fwd (GTATACGGGGTCCATCTTAGT) and myo7aa Rvs (CATCAGATCCTTCAAGTCCAC). The reverse primer added to the T7 core promoter sequence. Zebrafish embryos at the desired stage were fixed in 4% paraformaldehyde (PFA) overnight and then analyzed by WISH as described previously (Chitramuthu and Bennett, 2013). A digoxigenin UTP-labeled antisense RNA probe for myo7aa was generated by an in vitro transcription method using T7 RNA polymerase (Thermo Fisher, Waltham, MA, United States).
Twenty control and mutant embryos that developed to 60 hpf were stained overnight with YO-PRO-1 dye at a dilution of 1:2000 in E3 water. The next day, the embryos were washed with E3 water three times at 5-min intervals. The zebrafish were then anesthetized with 0.4% MS-222 (tricaine) solution, and 1% low-melting-point agarose was added to the laser confocal petri dish. All fluorescence imaging was conducted using a Zeiss LSM 900 laser scanning confocal microscope. Z-stacks were analyzed using ImageJ and brightness and contrast were equally adjusted for all relevant control and experimental images.
Homozygous mutant embryos were screened by YO-PRO-1 staining, and the total RNA was extracted from control and mutant embryos at 3 dpf using trizol reagent. The cDNA was synthesized using a reverse transcription kit (TaKaRa) with random primers. Real-time quantitative PCR reactions (qPCR) were performed on a QuantStudio 3 real-time PCR system (Thermo Fisher Scientific) using a SYBR kit (Vazyme). β-actin was used as the reference gene, and the data were analyzed using the (2-△△Ct) method (t-test, p < 0.05).
Five days after fertilization (dpf) of 20 larvae, the experiment was conducted in a petri dish with a layer of thin embryo culture medium. To provide sound stimuli, a mini-vibrator was attached to the bottom of the Petri dish. The acoustic stimuli used a frequency of 600 Hz and an intensity of 9 dB re.1 ms−2, and were applied for duration of 30 ms (Wei et al., 2022; Wang et al., 2023). For each stimulus level, this was repeated 20 times with a 3-min interval. The behavioral responses of the larvae to sound stimuli were also recorded by an infrared camera over a period of 6 s. Then, the movement trajectory of the larvae was extracted from the recorded film. The average movement distance and peak velocity were analyzed as typical parameters to quantify the startle response of larvae to sound stimuli.
In this experiment, we tested wild-type zebrafish larvae, myo7aa−/−zebrafish larvae, ATP-treated zebrafish larvae, and GTP-treated zebrafish larvae. The concentration of ATP was 3 mM, and the concentration of GTP treatment was 4 mM. The treatments were initiated 2 h prior to the test and remained in either ATP or GTP solution throughout the duration of the test.
Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion) and the Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) was used to evaluate RNA integrity. The samples with RNA Integrity Number (RIN) ≥ 7 were subjected to the subsequent analysis. According to the manufacturer’s instructions, the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) was used to construct libraries. These libraries were then sequenced on the Illumina sequencing platform (HiSeqTM 2,500 or Illumina HiSeq X Ten) to generate 125 bp/150 bp paired-end reads. After the raw data had been processed by Trimmomatic, the cleaned reads were mapped into the GRCz11 reference genome using Hisat2. The FPKM value of each transcript was calculated using Ballgown, and the FPKM of each transcript is calculated using Cufflinks. The differentially expressed transcripts were filtered with p-value <0.05 and | log2 fold change | > 1 as thresholds.
Differential expression analysis between myo7aa knockout group and control group was performed using R (v3.6.1) and RStudio (v4.3.0), package DESeq2 (v1.24.0). Genes with Padj (FDR) < 0.05 were considered as significantly differentially expressed genes. The results of differentially expressed genes were heatmapped using the pheatmapR package. Gene ontology (GO) analysis is a common method for large-scale functional enrichment analysis. GO database can annotate genes, and gene products can be enriched from three aspects: molecular function (MF), biological process (BP) and cellular component (CC) Gene Ontology Consortium (2015). GO biological process analysis was performed using clusterProfiler package (Yu et al., 2012). Item screening criteria was adjusted p-value <0.05 was considered statistically significant, and the p-value correction method was Benjamini Hochberg (BH). The results were visualized by Sangerbox, a bioinformatics analysis website, and the important key genes in the pathways were shown. KEGG and reactome pathway enrichment was performed by KOBAS (v3.0) (Kanehisa and Goto, 2000; Xie et al., 2011; Kanehisa, 2019; Kanehisa et al., 2021), transcript annotations were retrieved using the Bioconductor org.Dr.eg.db package, and ggplot2 (Wickham, 2016) package was used to visualize the correlation analysis results.
Zebrafish myo7aa was the ortholog of the human deafness pathogenic gene MYO7A. In mice and zebrafish, MYO7A is present in sensory hair cells, regions rich in F-actin (Wasfy et al., 2014). It has been reported in the literature that myo7aa is expressed in the maculae of the ellipsoid capsule and the globular capsule in zebrafish embryos developing to 24 hpf and 32 hpf (Schwarzer et al., 2017). However, the spatiotemporal expression pattern of this gene in zebrafish is still not well understood. To investigate the spatiotemporal expression model of this gene, we synthesized a digoxigenin-labeled antisense RNA probe of myo7aa. In situ hybridization results showed that the myo7aa is maternally expressed at the 1-cell stage (Figure 1A), and at the 4-cell stage (Figure 1B). It is ubiquitously expressed at 4 hpf (Figure 1C), 9 hpf (Figure 1D), and 12 hpf (Figure 1E). myo7aa was specifically expressed in the hair cells of the inner ear and retina at 48 hpf (Figure 1F), in the hair cells of inner ear, retina and lateral line at 3 dpf (Figure 1G), and strongly expressed in the hair cells of inner ear, retina and lateral line at 4 dpf and 5 dpf (Figures 1H,I). These results further suggest that zebrafish myo7aa may affect hair cell development and loss of myo7aa may lead to hearing impairment.
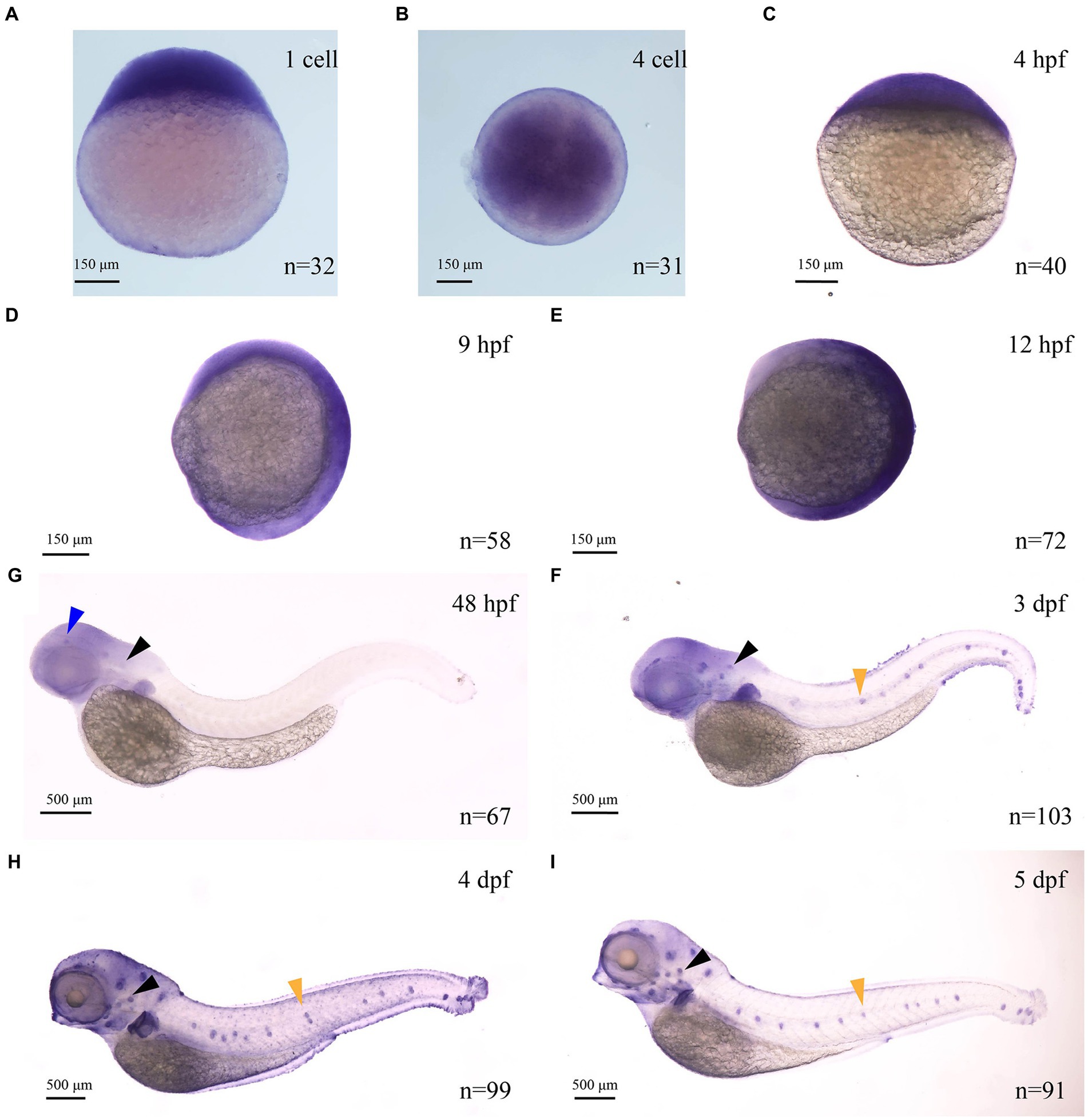
Figure 1. The expression of myo7aa at different stages (A–I), arrowheads indicate zebrafish retina (blue), inner ear (black) and lateral line hair cells (yellow); n: the number of embryos.
To study the effect of the zebrafish myo7aa gene on hair cells, zebrafish myo7aa knockout lines that can be stably inherited by CRISPR/Cas9 technology were generated. The target site was designed in exon 6 of the MYSc domain coding region (Figure 2A), and genotyping primers for detecting wild-type PCR products were designed at both ends with a length of 361 bp (Figure 2B). Two independently inherited heterozygous mutant zebrafish lines, line1 and line2, with 114 bp deletion (Figure 2C) and 104 bp insertion (Figure 2D), were selected from the offspring of the same F0 generation mutant (Figure 2E). Line1 mutant caused a 38-amino acid deletion in the MYSc domain of MYO7AA protein (Figure 2C), and line2 mutant caused a frameshift mutation in the open reading frame of myo7aa, leading to premature termination of protein translation (Figure 2D). The myo7aa homozygous mutant embryos showed no obvious defects in body size during early development (not shown), but the homozygous juveniles of both mutant lines died at 10–12 days.
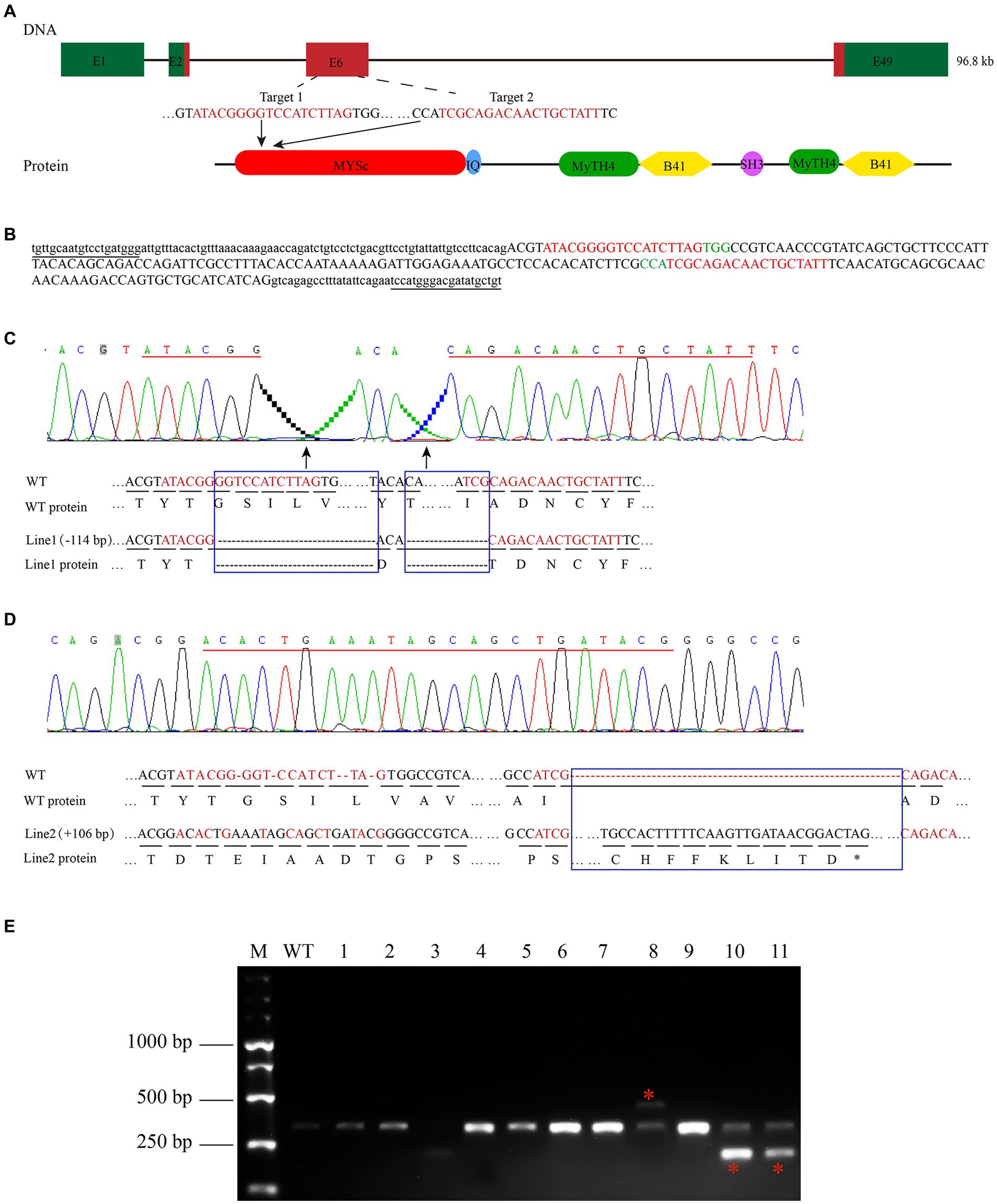
Figure 2. Zebrafish myo7aa gene knockout. (A) Schematic representation of the CRISPR/Cas9 target region in myo7aa gene. Green box represents the 3′UTR and 5′UTR, while the red box indicates exons. (B) Sequence of the CRSIPR/Cas9 target region in the myo7aa gene. Underlined: detection primers; red font: target site sequence; green font: PAM sequence; capital letters: exon sequence; lowercase letters: intron sequences. (C,D) Sequencing peak map, genomic DNA sequence, and amino acid sequence between two independent mutant alleles. Compared with the wild type, the myo7aa line1 mutation had a 114 bp deletion and the line2 mutation had a 106 bp insertion. The sequence of the target site is shown in red. Rectangular boxes indicate deletion or insertion sites in the mutant. *Indicates termination of protein translation. (E) The screening results of the F1 generation adult fish showed the presence of DNA marker, wild type fish, and mutant fish.
Previous studies have shown that myo7aa is expressed in hair cells, which was confirmed by in situ hybridization. To investigate the role of myo7aa in hair cells, YO-PRO-1 staining was used to detect the abnormal function of hair cells. YO-PRO-1 is a cyanine dye that binds to DNA and fluoresces after entering the cell. Labeling of hair cells in the lateral line with YO-PRO-1 can be used as a reliable index to evaluate the viability of hair cells (Santos et al., 2006). At 5 dpf, the wild-type zebrafish larvae exhibited a bright and intact inner ear (Figure 3C) and lateral line hair cell signal (Figure 3A), whereas the myo7aa mutant embryos showed a complete lack of inner ear (Figure 3D) and lateral line hair cell fluorescence signal (Figure 3B). We confirmed the genotypes of YO-PRO-1 stained embryos, and the embryos without fluorescence signal were myo7aa homozygous mutant (Figure 3E). To confirm the effect of myo7aa on zebrafish hearing, we used YO-PRO-1 staining to screen myo7aa homozygous mutant larvae for startle response testing. Vibrators were connected beneath small dishes containing 20 wild-type or myo7aa−/− zebrafish larvae, and the zebrafish larvae were stimulated with an acoustic wave at a frequency of 600 Hz and a pitch of 9 dB re.1 ms−2 for 30 ms. When frightened by sound waves, the average movement distance of wild-type zebrafish larvae was 1.134 mm per 0.3 s, while that of myo7aa−/− larvae was only moved 0.069 mm (Figure 3F; Table 1). Additionally, the reaction speed of myo7aa−/− larvae was much slower than that of the wild-type larvae. The maximum movement speed of the wild type zebrafish larvae was 7.388 mm/s, while the maximum movement speed of the myo7aa−/− larvae was only 0.468 mm/s (Figure 3G; Table 2). These results indicate that myo7aa gene knockout impairs the normal physiological function of hair cells and causes hearing impairment.
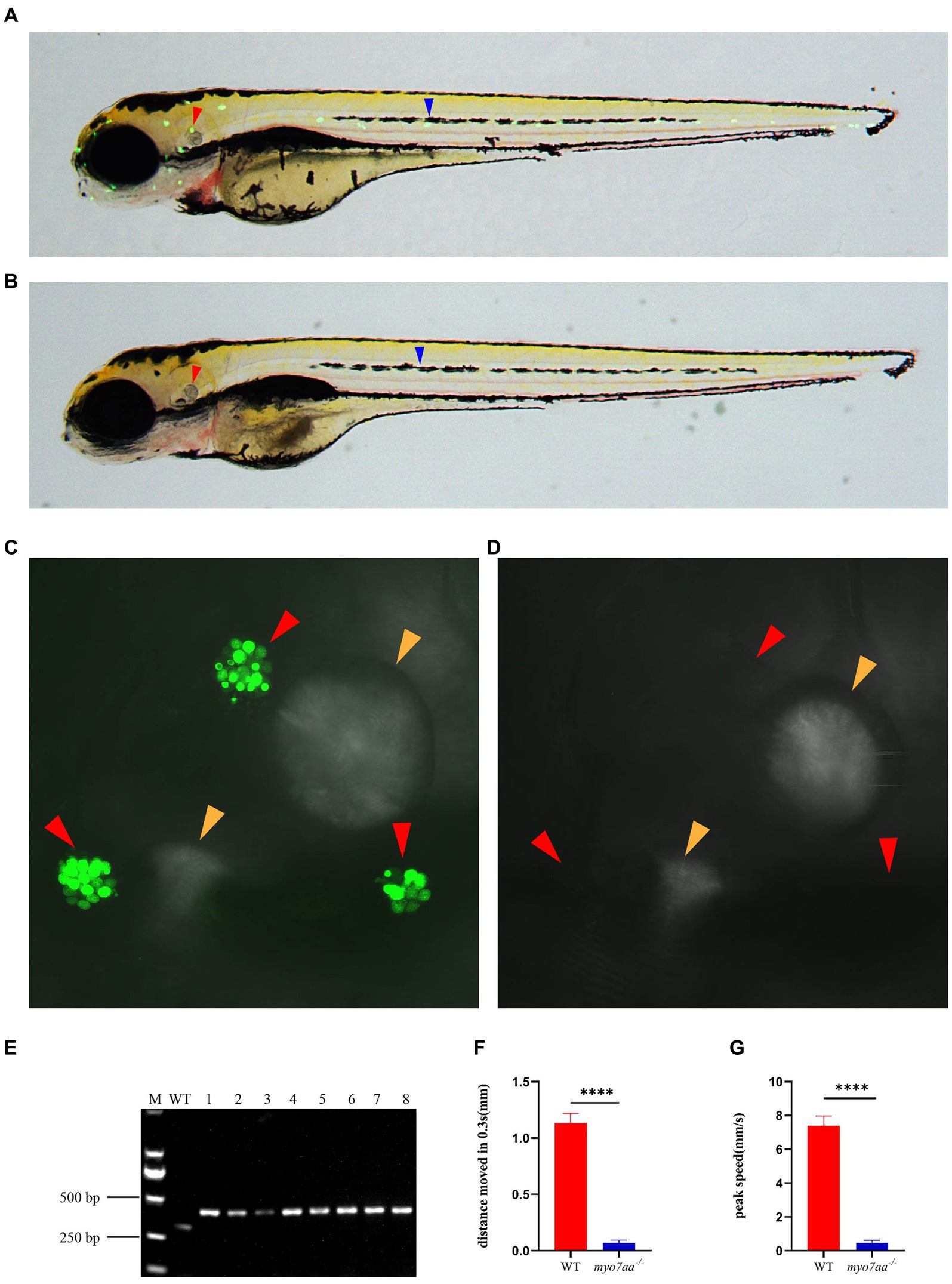
Figure 3. Loss of myo7aa causes hearing impairment. (A,B) YO-PRO-1 fluorescence staining results of wild-type and myo7aa mutant hair cells at 5 dpf, arrowheads indicate zebrafish inner ear (red) and lateral line hair cells (blue); (C,D) YO-PRO-1 fluorescent staining results of ear hair cells in wild-type and myo7aa mutant at 5 dpf, arrowheads indicate zebrafish inner ear hair cells (red) and otoliths (yellow). These results demonstrate the impact of myo7aa loss on the function of hair cells in the ear, leading to hearing impairment. Additionally, the results of genotype identification of embryos shown in (B,D) are presented. (E) Genotyping results of line2. M is DNA marker, WT indicates wild type, 1–8 are embryos shown in (B,D). (F,G) The movement distance (F) and reaction speed (G) of wild type and myo7aa mutant within 0.3 s after acoustic stimulation at 5 dpf are measured; n = 20; mean with SEM; with t-test, **** indicates a p-value <0.0001.
To further analyze the molecular mechanism of deafness caused by myo7aa deletion, we collected myo7aa knockout lines and TU embryos at 3 dpf for RNA-seq. Each group had three replicates, with 50 embryos per replicate. The principal component dimension reduction (PCA) analysis results showed that there was a clear separation between myo7aa−/− mutant and control. The samples in the two groups were clustered together, indicating that the samples within each groups were well grouped and correlated (Figure 4A). The differentially expressed genes (DEGs) results showed that the gene expression profile of myo7aa−/− mutant was significantly changed. A total of 364 DEGs were screened between myo7aa−/− mutant and control, 49 of which were down-regulated and 315 were up-regulated (Figure 4B).
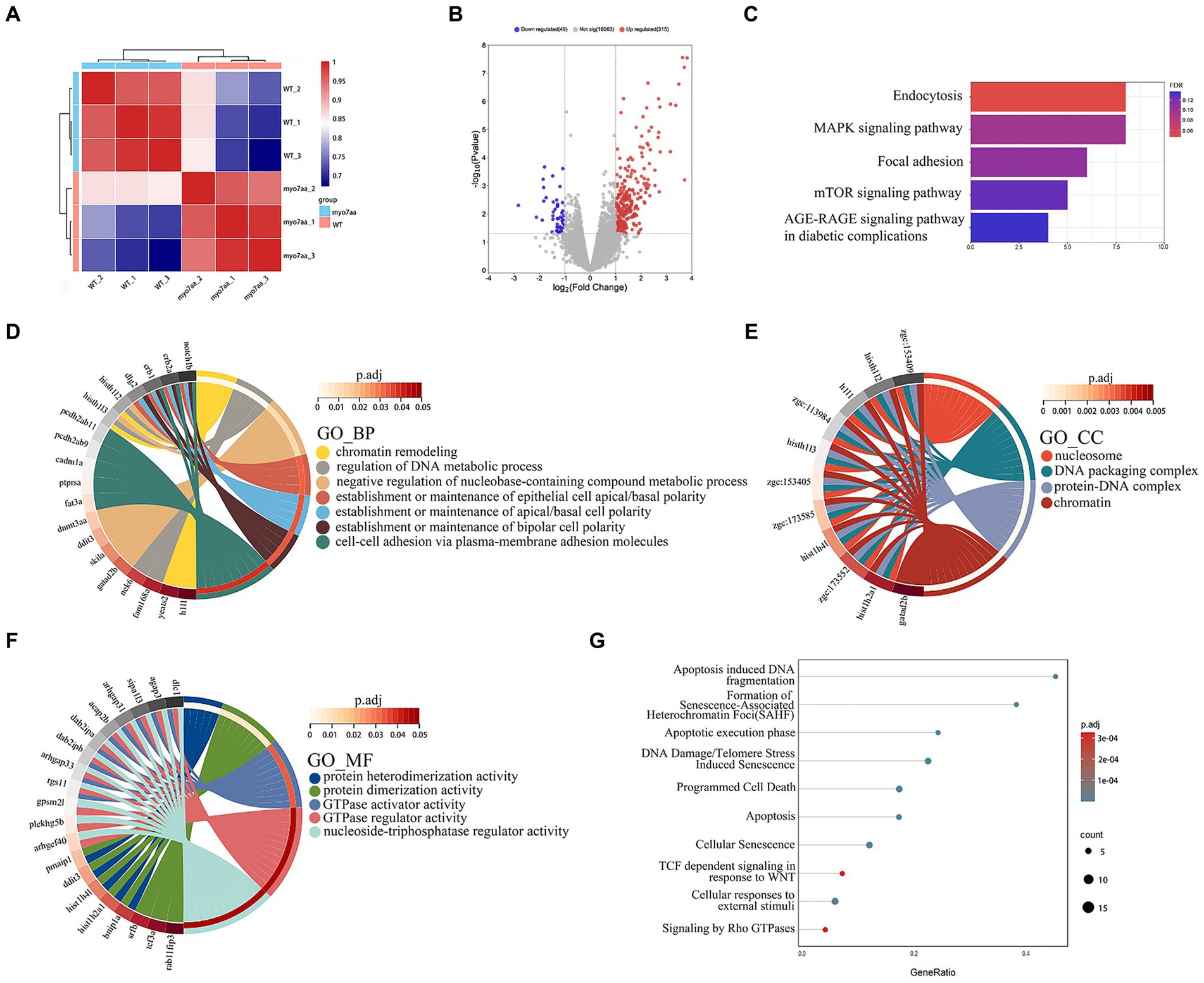
Figure 4. The results of RNA-seq analysis, (A) the Principal Component Analysis (PCA) diagram; (B) the volcano plot of differentially expressed genes. Red dots indicate upregulated genes. Blue dots indicate genes that are down-regulated; (C) the enrichment bar chart of gene function annotation analysis in the KEGG database. The bar length in the chart represents the number of genes. The bar color represents the p corrected value; (D–F) the GO database enrichment analysis results are provided for Biological process (BP), cellular component (CC), and molecular function (MF); (G) the Reactome database enrichment analysis results.
KEGG pathway annotation of DEGs was performed in the KEGG database, and the results of the enrichment analysis showed that all DEGs were significantly different in KEGG pathways (p < 0.05). The DEGs were mainly significantly enriched in the endocytosis and MAPK signaling pathway, and the genes involved in these pathways included grk3, rab11fip3, agap3, etc. (Figure 4C). The gene function annotation of DEGs in GO database showed that DEGs were involved in biological processes such as “regulation of DNA metabolism,” “establishment or maintenance of apical/basal cell polarity,” and “establishment or maintenance of bipolar cell polarity” (Figure 4D). Cellular components such as “nucleosome,” “DNA assembly complex,” “protein-DNA complex” and “nuclear chromatin” (Figure 4E). And molecular functions such as “GTPase-activating protein activity,” “GTPase-regulator activity” and “nucleic acid triphosphate regulator activity” (Figure 4F) were significantly enriched.
Gene function annotation of DEGs in Reactome database showed that the results were closely related to “DNA damage,” “telomere inhibition,” “apoptosis,” “programmed cell necrosis,” “Rho GTPase signaling” and so on (Figure 4G).
In order to further validate the results of transcriptome and bioinformatics analyses (Figure 5A), we collected control and mutant embryos at 3 days after embryonic development, 30 embryos were collected from each sample, with 3 replicates in each group. Total RNA was extracted and reverse transcribed using the PrimeScript™ RT reagent Kit (Takara), and then the qPCR primers for the relevant pathways were synthesized and subjected to qPCR experiments. qPCR validation results showed that Rho GTPase signaling pathway-related differential genes, such as arhgap33, a member of the sorted connexin family with the structural domain of Rho GTPase-activating protein (RhoGAP), and dab2ipb, a member of the Ras GTPase-activating protein family were significantly down-regulated. The guanine nucleotide exchange factor arghef40, which targets RhoA, and dab2ipa, a member of the Ras GTPase-activating protein family, are barely expressed in myo7aa−/− embryos (Figure 5B); endocytosis-associated genes, such as scaffolding protein family member rab11fip3 and kinesin kif5bb, were significantly downregulated (Figure 5C); and MAPK signaling pathway-associated genes, such as mapk8 (mitogen-activated protein kinase 2) and mapk8ip2 (p38 family scaffolding protein) were significantly upregulated (Figure 5D).
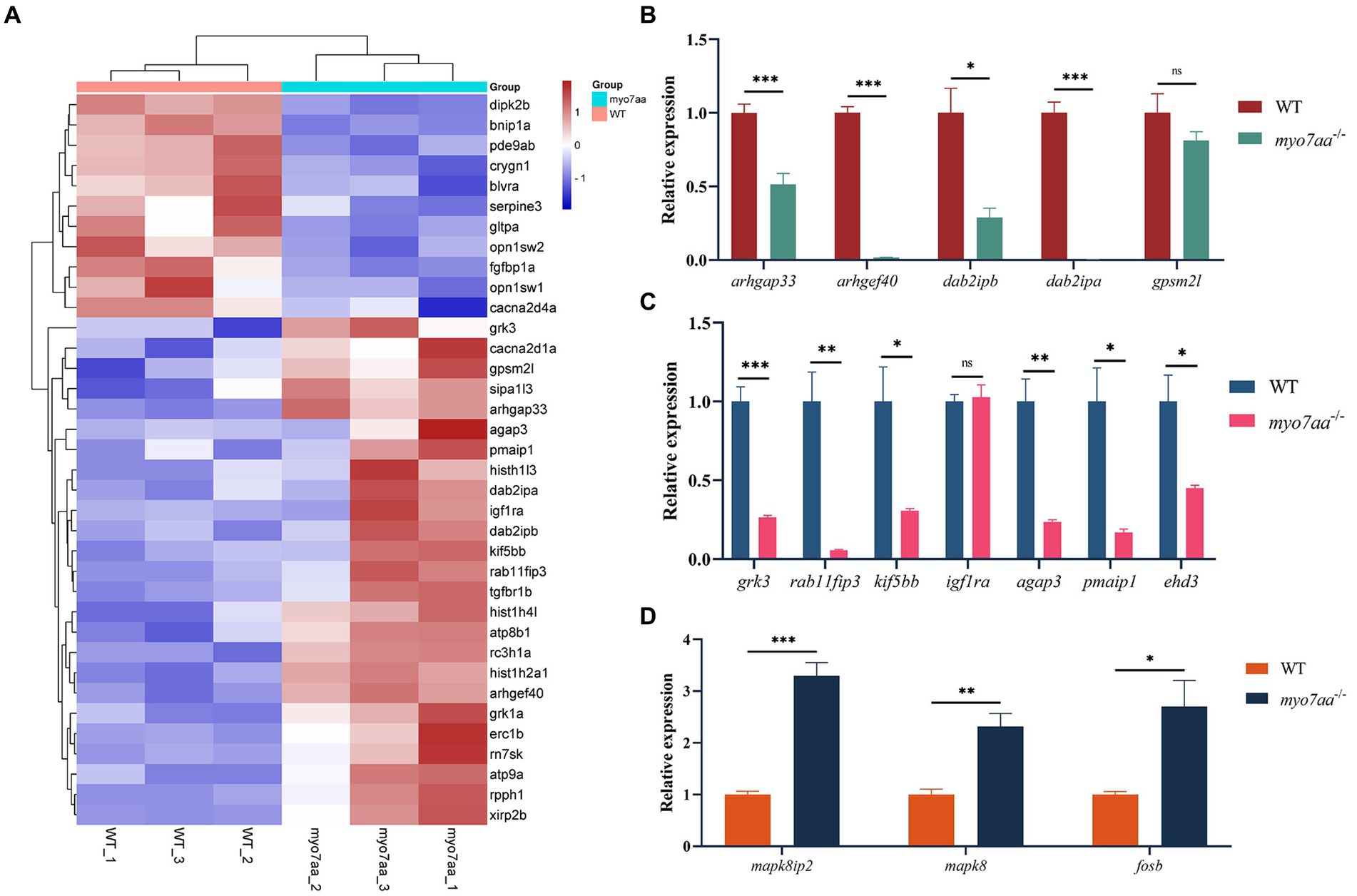
Figure 5. myo7aa regulates G-protein signaling. (A) The gene heatmap is depicted in (A), while (B–D) display the relative expression of genes in the Rho GTPase signaling pathway (B), cellular endocytosis pathway (C), and MAPK signaling pathway (D) between WT and myo7aa−/− at 3 dpf, *p < 0.05, **p < 0.01, ***p < 0.001 (t-test).
These results suggest that a variety of cellular processes, including apoptosis and endocytosis, are affected in myo7aa deficiency. Notably, the disorder of Rho GTPase signaling system in myo7aa−/− zebrafish may lead to abnormal hair cell function and thus hearing damage.
Transcriptome and qPCR results showed that myo7aa deficiency resulted in dysregulation of GTPase activity, disordered GTP metabolism, and impaired cellular endocytosis. We next determined whether GTP compensation could rescue the loss of hair cell function in myo7aa homozygous mutants. After the YO-PRO-1 staining, we conducted GTP compensation experiments on sibling embryos, as previously described. YO-PRO-1 dye can enter inner ear hair cells and lateral line cells in sibling embryos, while the fluorescence signal is completely absent in myo7aa−/− embryos. Next, we added a 4 mM GTP solution to E3 water and incubated it for 6 h. The results showed that the fluorescent signal was still present in the hair cells of sibling embryos (Figures 6A,B). In contrast, the myo7aa−/− group embryos showed partial restoration of the YO-PRO-1 signal in the inner ear and lateral line hair cells (Figures 6C,D). Subsequently, we collected the control and GTP-treated embryos for qPCR Experiment. The results showed that the expression of endocytosis related genes grk3, rab11fip3, and kif5bb was partially restored in myo7aa−/− embryos after GTP treatment (Figure 6E). The expression of arghef40 and dab2ipa genes, encoding guanine nucleotide exchange factor and GTPase activating protein, was also restored (Figure 6F). The startle response experiment after GTP treatment showed that the group treated with GTP was more sensitive to sound stimulation compared to the myo7aa homozygous mutant. Additionally, there was a significantly improvement in both the movement distance and response speed to stimulation (ANOVA). When stimulated by sound waves, the average movement distance of wild-type zebrafish larvae was 1.466 mm per 0.3 s, whereas the myo7aa−/− larvae only moved 0.051 mm, and the GTP-treated larvae moved 0.124 mm (Figure 6G; Table 3). After sound stimulation, the maximum movement speed of wild-type zebrafish larvae was 13.427 mm/s, In contrast, the maximum movement speed of myo7aa−/− larvae was 0.488 mm/s, and the maximum movement speed of GTP-treated larvae was 1.241 mm/s (Figure 6H; Table 4). These results suggest that knockout of myo7aa leads to dysfunction of GTPase signaling, which is caused by disrupted GTPase activity. Additionally, the application of GTP can partially restore hair cell function.
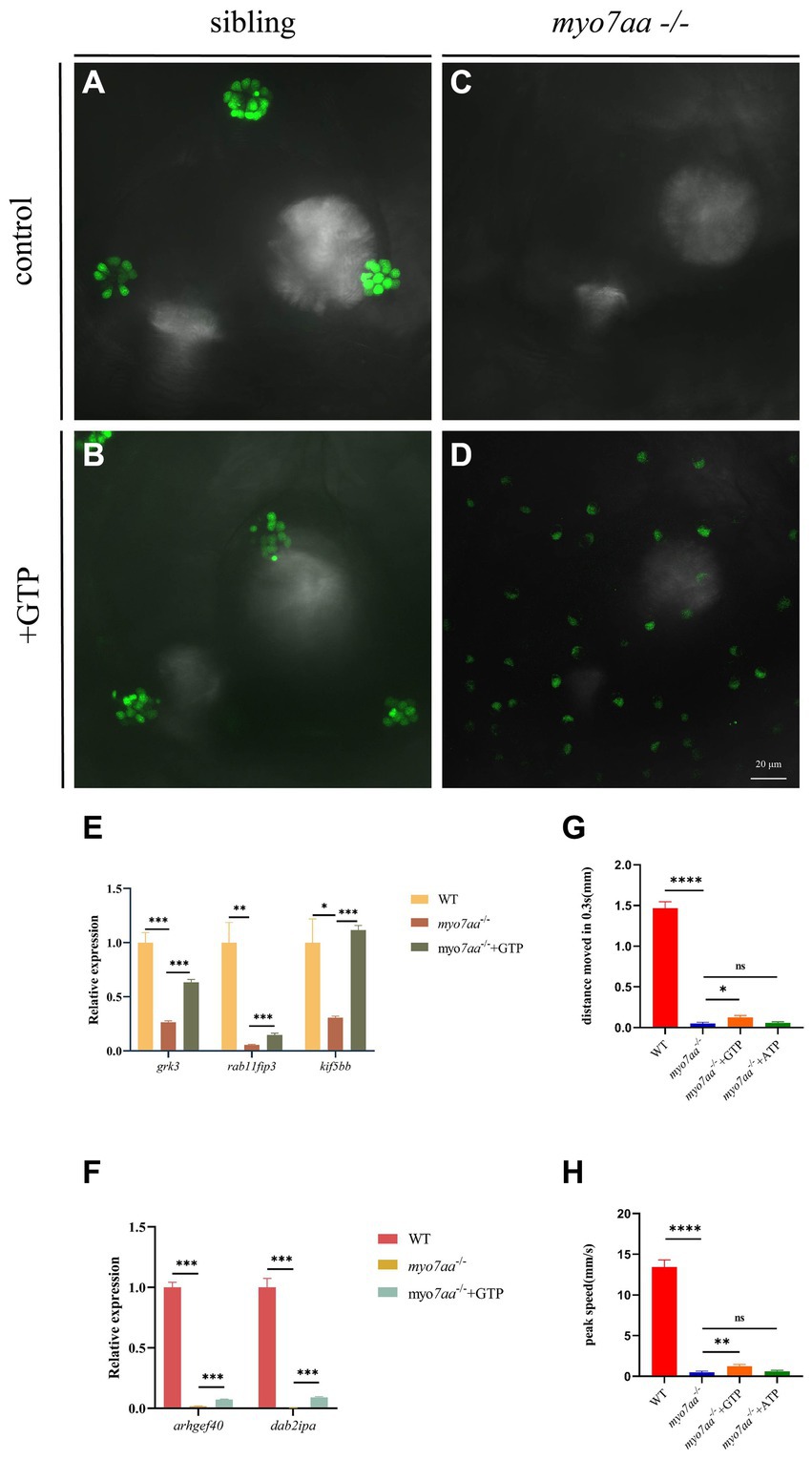
Figure 6. GTP compensation partially restored the hearing impairment caused by myo7aa deficiency. (A–D) YO-PRO-1 staining results of inner ear hair cells were observed in sibling (A), myo7aa−/− (B), GTP-compensated sibling (C), and myo7aa−/− (D) embryos at 5 dpf. (E,F) The relative expression of some genes in the cellular endocytic pathway (E) and G protein signaling pathway (F) is examined between WT, myo7aa−/−, and GTP-compensated myo7aa−/− embryos at 3 dpf is examined. (G,H) The movement distance (G) and reaction speed (H) of wild WT, myo7aa−/−, and GTP-compensated myo7aa−/− embryos are measured within 0.3 s after acoustic stimulation at 5 dpf are measured. n = 20; mean with SEM; with ANOVA, ****p < 0.0001, **p < 0.01, *p < 0.05, nsp > 0.05.
Nucleoside diphosphate kinase (NDK) can transphosphorylate GDP and ATP near the cell membrane and G protein to generate GTP and ADP (Piacentini and Niroomand, 1996). To verify whether ATP affects the G-protein signaling process, ATP compensation experiments were performed after the YO-PRO-1 staining experiment, as described previously. Zebrafish embryos at 5 dpf were treated with 3 mM ATP in combination with YO-PRO-1 and observed under a fluorescence microscope after 6 h. Fluorescence signals were still observed in the inner ear and lateral line hair cells of Sibling embryos (Figures 7A,B), and YO-PRO-1 fluorescence signals were partially recovered in myo7aa−/− embryos (Figures 7C,D). The expression of endocytosis related genes grk3, rab11fip3, kif5bb and pmaip1 was partially restored (Figure 7E), and the mRNA expression level of arghef40 was also restored (Figure 7F). These results indicate that the application of ATP could partially restore the function of hair cells in myo7aa −/−embryos. The startle response experiment conducted after ATP treatment revealed that neither the ATP treatment group nor the myo7aa homozygous mutant showed sensitivity to acoustic stimulation. When subjected to by acoustic stimulation, myo7aa−/− larvae moved a distance of 0.051 mm, while the ATP treatment group moved a distance of 0.057 mm (Figure 6G; Table 3). When stimulated by sound waves, the maximum movement speed of myo7aa−/− and ATP-treated larvae was 0.488 mm/s and 0.591 mm/s, respectively (Figure 6H; Table 4). There was no significant difference in movement distance and reaction speed between the two groups (ANOVA) (see Figure 8).
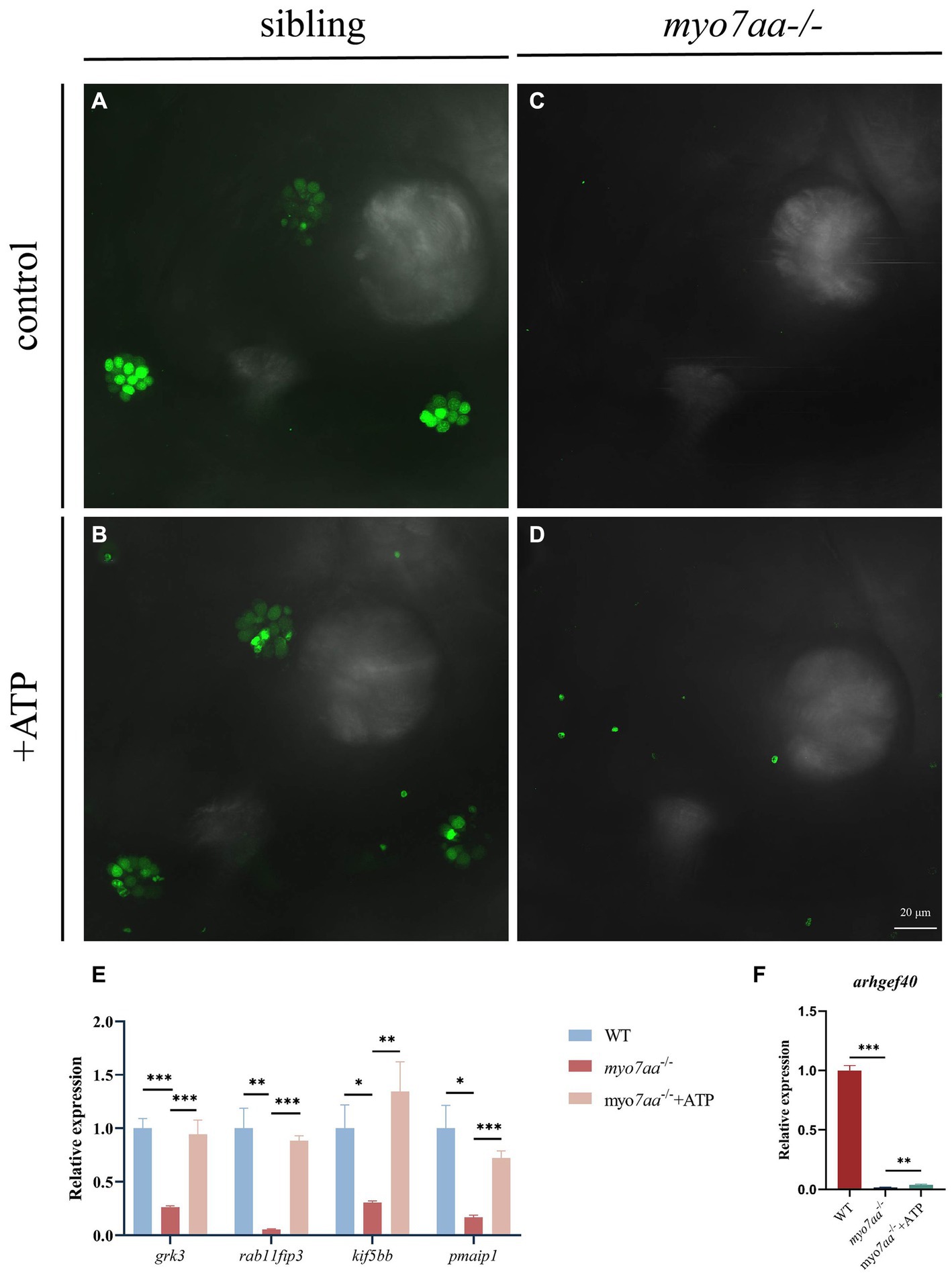
Figure 7. ATP compensation partially restored the hearing defects caused by myo7aa deficiency. (A–D) The YO-PRO-1 staining results of inner ear hair cells were observed in sibling (A), myo7aa−/− (B) and ATP-compensated sibling (C), myo7aa−/− (D) embryos at 5 dpf. (E,F) The relative expression of some genes in the cellular endocytic pathway (E) and G protein signaling pathway (F) was compared between WT, myo7aa−/− and ATP-compensated myo7aa−/− embryos at 3 dpf.
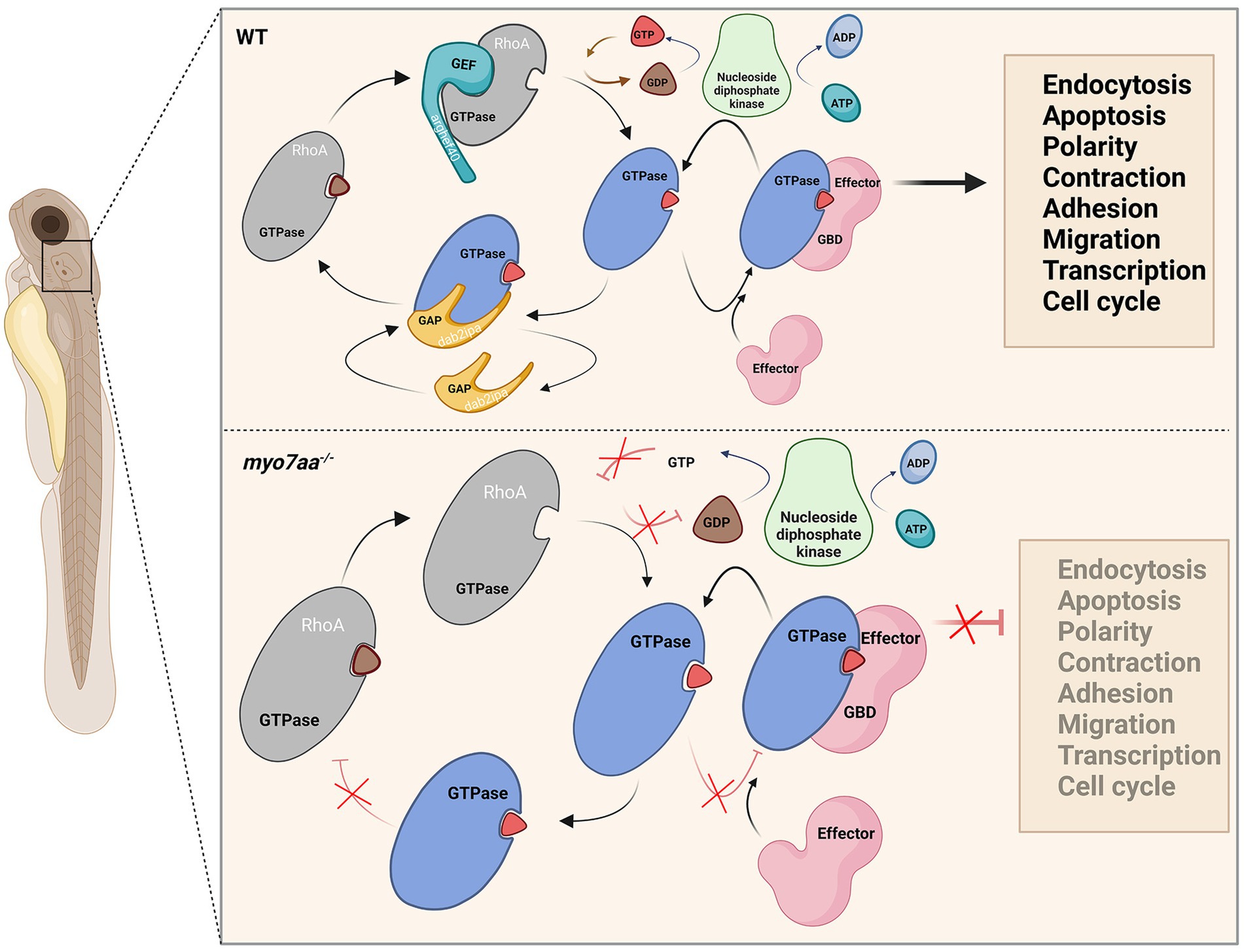
Figure 8. Loss of myo7aa leads to impaired Rho GTPase signaling, resulting in hearing impairment (image created at BioRender.com).
Previous studies have shown that MYO7A is the causative gene for Usher 1B syndrome, recessive nonsyndromic deafness (DFNB2), and dominant nonsyndromic deafness (DFNA11). In myo7aa−/− zebrafish, hair cell function is impaired, but the downstream regulatory network is not well understood. In this study, we generated a myo7aa knockout zebrafish line to investigate the molecular functions of myo7aa in hair cells. To compare the transcriptome data of wild-type and myo7aa knockout zebrafish, We analyzed the differentially expressed genes. We found that these genes were mainly involved in pathways such as “nucleosome assembly,” “protein-DNA complex,” “establishment or maintenance of epithelial cell apex,” “establishment or maintenance of bipolar cell polarity,” “establishment or maintenance of apical/basal cell polarity,” “GTPase-activating protein activity,” “GTPase regulator activity,” “nucleoside triphosphatase regulator activity,” “endocytosis exocytosis,” “autophagy” and other pathways were enriched. The genes involved in these pathways, such as grk3, rab11fip3, and agap3, play crucial roles in the regulation of various biological processes.
Endocytic transport plays a crucial role in the establishment and maintenance of cell polarity. Polarized cells, especially highly polarized cells such as hair cells, rely heavily on the proper organization of their intracellular components to achieve their function (Nakazawa et al., 2016). Subcellular localization of organelles, proteins, and RNA is essential for the compartmental function of cells. Rab11 GTPase has been shown to be a master regulator of endosomal trafficking via recycling, which is required to establish and maintain epithelial polarity (Jing and Prekeris, 2009). Rab11-FIP3 is a member of the Rab11-FIP family. Rab11-FIP is a family of scaffold proteins that play a major role in mediating endocytic recycling in a variety of cells (Horgan et al., 2004, 2007; Inoue et al., 2008). Kinesin superfamily proteins are a class of kinesin proteins. Kif5s has a conserved carved structure consisting of an N-terminal globular head/motor domain and a C-terminal globular tail domain connected to the coiled-rich stem region via a neck domain. The motor domain is responsible for ATP hydrolysis and microtubule binding, allowing the complex to move along microtubules (Campbell and Marlow, 2013). Kif5s has been implicated in a number of transport processes including retrograde transport of vesicles from the Golgi to the endoplasmic reticulum (ER) (Lippincott-Schwartz et al., 1995), anterograde transport of lysosomes to the plasma membrane (Nakata and Hirokawa, 1995), pigment dispersion in melanocytes (Hara et al., 2000), and anterograde axonal transport of organelles, proteins, vesicles, and RNA in neurons (Hirokawa et al., 2010). Eps15 homeodomain-containing protein 3 (EHD3) is an endocytic trafficking regulator (Juszczak and Stankiewicz, 2018) that controls trafficking to and from the endocytic recycling compartment (ERC) to the plasma membrane (Naslavsky et al., 2009; Cabasso et al., 2015) and is required for the stabilization of tubular recycling endosomes (Bahl et al., 2016). ARHGAP33 (also known as SNX26, TCGAP or NOMA-GAP) is a member of the sorting junction protein (SNX) family. Mediating interactions with phosphatidylinositol via the conserved PX domain, SNX proteins normally regulate membrane protein sorting in the Golgi apparatus and endosomes, where they are localized (Liu et al., 2006; Kim et al., 2013; Nakazawa et al., 2016).
igf1r is essential for the early development of zebrafish. igf1ra encodes the insulin-like growth factor 1 receptor, which is expressed in the ear capsule, an inner ear (auditory) and vestibular (balance) organ of zebrafish (Li et al., 2014).
Purine nucleotide GTP is involved in a variety of cellular processes, including RNA and DNA synthesis, G-protein signaling, protein biosynthesis, gluconeogenesis, and tubulin formation (Wolff et al., 2022). GTPase activity is essential for accelerating synaptic vesicle recruitment (Watanabe et al., 1999). Rho GTPase regulates actin dynamics near the cell membrane, and GTP plays an important role in cellular endocytosis (Yamashita et al., 2005; Xu et al., 2008). AGAP3 is a member of a family of proteins containing functional ArfGAP domains and GTpase-like domains (AGAP1-3) with bifunctional enzyme activity (Oku and Huganir, 2013). Solo(arghef40) is a guanine nucleotide exchange factor that targets RhoA and regulates cell morphology by interacting with keratin filaments (Nishimura et al., 2018). DOC-2 / DAB-2 interacting protein (Dab2IP) is a member of the Ras gtpase activating protein (GAP) family (Lee et al., 2012; Qiao et al., 2013). Guanine nucleotide exchange factors (GEFs) can facilitate the exchange of GDP-GTP bound by small guanine nucleotide-binding proteins (G proteins), while GTPase activating proteins (GAPs) facilitate the hydrolysis of GTP by G proteins to GDP (Fujiwara et al., 2018). Together, they regulate the activity of G proteins. Upon binding to GTP, the G protein is activated and can bind to other proteins and trigger downstream signaling targets. Upon binding to GDP, the G protein loses its activity and is unable to bind to its target proteins, resulting in impaired RNA and DNA synthesis, G protein signaling, protein biosynthesis (Jewett et al., 2009), gluconeogenesis, and tubulin formation (Muraoka and Sakai, 1999; Pickett and Raible, 2019). GRK3 (G protein-coupled receptor kinase 3) is highly expressed in sensory epithelial cells and mediates the agonist-dependent phosphorylation and uncoupling of many G protein-coupled receptors (Peppel et al., 1997).
Both GTP and ATP compensation could partially rescue the defects in myo7aa knockout zebrafish. Therefore, we hypothesized that knockout of myo7aa results in decreased expression of GTPase activator protein dab2ip and guanine nucleotide exchange factor arghef40, which prevents G protein from properly binding and activating GTP and leads to Rho GTPase signaling dysfunction. It affects RNA and DNA synthesis, protein biosynthesis, endocytosis and exocytosis, and the establishment or maintenance of cell polarity. Compensating GTP can increase the concentration of GTP in hair cells, accelerate the exchange of GTP and GDP, and lead to the activation of Rho GTPase, so as to transmit signals normally. When ATP is compensated, nucleotide diphosphate kinase catalyzes the regeneration of GTP in GDP due to the presence of ATP, and at the same time converts ATP to ADP (Aamodt and Williams, 1984; Dewulf et al., 2022), making Rho GTPase bound to GTP into an activated state, and G protein signaling can be partially restored (Figure 8). Since ATP affects G-protein signaling by promoting GTP production, ATP compensation did not significantly restore hearing in myo7aa homozygous mutant embryos.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1086823?reviewer=atbodt8tbhj1tqo6bpv6l7n3cq.
The animal study was approved by Biomedical Research Ethics Committee of Hunan Normal University. The study was conducted in accordance with the local legislation and institutional requirements.
BX: Data curation, Investigation, Project administration, Validation, Visualization, Writing – original draft. JL: Data curation, Formal analysis, Software, Visualization, Writing – original draft. JJ: Investigation, Validation, Writing – original draft. TZ: Writing – original draft. LL: Data curation. DX: Funding acquisition, Resources, Writing – review & editing. GZ: Writing – original draft, Resources. LX: Writing – review & editing. KZ: Resources, Writing – original draft. DL: Resources, Writing – review & editing. JG: Resources, Writing – review & editing. XC: Writing – review & editing. RL: Funding acquisition, Investigation, Resources, Writing – review & editing. HX: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Natural Science Foundation of China (82170308), Xiaoxiang Scholar Distinguished Professor Startup Funding for H-pX (053112–3826), Science Foundation of Hunan Provincial Health Commission (202000666); Natural Science Foundation of Hunan Province (2023JJ30753) and Natural Science Foundation of Changsha (kq2208326).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2024.1405109/full#supplementary-material
Aamodt, E. J., and Williams, R. C. (1984). Association of microtubules and neurofilaments in vitro is not mediated by ATP. Biochemistry 23, 6031–6035. doi: 10.1021/bi00320a020
Bahl, K., Xie, S., Spagnol, G., Sorgen, P., Naslavsky, N., and Caplan, S. (2016). EHD3 protein is required for tubular recycling endosome stabilization, and an asparagine-glutamic acid residue pair within its Eps15 homology (EH) domain dictates its selective binding to NPF peptides. J. Biol. Chem. 291, 13465–13478. doi: 10.1074/jbc.M116.716407
Cabasso, O., Pekar, O., and Horowitz, M. (2015). SUMOylation of EHD3 modulates Tubulation of the endocytic recycling compartment. PLoS One 10:e0134053. doi: 10.1371/journal.pone.0134053
Campbell, P. D., and Marlow, F. L. (2013). Temporal and tissue specific gene expression patterns of the zebrafish kinesin-1 heavy chain family, kif5s, during development. Gene Expr. Patterns 13, 271–279. doi: 10.1016/j.gep.2013.05.002
Chitramuthu, B. P., and Bennett, H. P. (2013). High resolution whole mount in situ hybridization within zebrafish embryos to study gene expression and function. J. Vis. Exp. 80:e50644. doi: 10.3791/50644
Dai, Y. B., Gao, X., Liu, D., and Gong, J. (2023). The role of rho GTPase family in cochlear hair cells and hearing. Neural Regen. Res. 18, 2167–2172. doi: 10.4103/1673-5374.369101
Dewulf, J. P., Marie, S., and Nassogne, M. C. (2022). Disorders of purine biosynthesis metabolism. Mol. Genet. Metab. 136, 190–198. doi: 10.1016/j.ymgme.2021.12.016
Du, H., Zhou, H., Sun, Y., Zhai, X., Chen, Z., Wang, Y., et al. (2021). The rho GTPase cell division cycle 42 regulates Stereocilia development in Cochlear hair cells. Front. Cell Dev. Biol. 9:765559. doi: 10.3389/fcell.2021.765559
Ernest, S., Rauch, G. J., Haffter, P., Geisler, R., Petit, C., and Nicolson, T. (2000). Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum. Mol. Genet. 9, 2189–2196. doi: 10.1093/hmg/9.14.2189
Fettiplace, R. (2017). Hair cell transduction, tuning, and synaptic transmission in the mammalian cochlea. Compr. Physiol. 7, 1197–1227. doi: 10.1002/cphy.c160049
Fujiwara, S., Matsui, T. S., Ohashi, K., Deguchi, S., and Mizuno, K. (2018). Solo, a RhoA-targeting guanine nucleotide exchange factor, is critical for hemidesmosome formation and acinar development in epithelial cells. PLoS One 13:e0195124. doi: 10.1371/journal.pone.0195124
Gene Ontology Consortium (2015). Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056. doi: 10.1093/nar/gku1179
Grimsley-Myers, C. M., Sipe, C. W., Géléoc, G. S., and Lu, X. (2009). The small GTPase Rac1 regulates auditory hair cell morphogenesis. J. Neurosci. 29, 15859–15869. doi: 10.1523/jneurosci.3998-09.2009
Hara, M., Yaar, M., Byers, H. R., Goukassian, D., Fine, R. E., Gonsalves, J., et al. (2000). Kinesin participates in melanosomal movement along melanocyte dendrites. J. Invest. Dermatol. 114, 438–443. doi: 10.1046/j.1523-1747.2000.00894.x
Heasman, S. J., and Ridley, A. J. (2008). Mammalian rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701. doi: 10.1038/nrm2476
Hirokawa, N., Niwa, S., and Tanaka, Y. (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638. doi: 10.1016/j.neuron.2010.09.039
Horgan, C. P., Oleksy, A., Zhdanov, A. V., Lall, P. Y., White, I. J., Khan, A. R., et al. (2007). Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic 8, 414–430. doi: 10.1111/j.1600-0854.2007.00543.x
Horgan, C. P., Walsh, M., Zurawski, T. H., and McCaffrey, M. W. (2004). Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem. Biophys. Res. Commun. 319, 83–94. doi: 10.1016/j.bbrc.2004.04.157
Inoue, H., Ha, V. L., Prekeris, R., and Randazzo, P. A. (2008). Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell 19, 4224–4237. doi: 10.1091/mbc.e08-03-0290
Jewett, M. C., Miller, M. L., Chen, Y., and Swartz, J. R. (2009). Continued protein synthesis at low [ATP] and [GTP] enables cell adaptation during energy limitation. J. Bacteriol. 191, 1083–1091. doi: 10.1128/jb.00852-08
Jing, J., and Prekeris, R. (2009). Polarized endocytic transport: the roles of Rab11 and Rab11-FIPs in regulating cell polarity. Histol. Histopathol. 24, 1171–1180. doi: 10.14670/hh-24.1171
Juszczak, G. R., and Stankiewicz, A. M. (2018). Glucocorticoids, genes and brain function. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 82, 136–168. doi: 10.1016/j.pnpbp.2017.11.020
Kanehisa, M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. doi: 10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., and Tanabe, M. (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–d551. doi: 10.1093/nar/gkaa970
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kim, Y., Ha, C. M., and Chang, S. (2013). SNX26, a GTPase-activating protein for Cdc42, interacts with PSD-95 protein and is involved in activity-dependent dendritic spine formation in mature neurons. J. Biol. Chem. 288, 29453–29466. doi: 10.1074/jbc.M113.468801
Lee, G. H., Kim, S. H., Homayouni, R., and D'Arcangelo, G. (2012). Dab2ip regulates neuronal migration and neurite outgrowth in the developing neocortex. PLoS One 7:e46592. doi: 10.1371/journal.pone.0046592
Li, J., Wu, P., Liu, Y., Wang, D., and Cheng, C. H. (2014). Temporal and spatial expression of the four Igf ligands and two Igf type 1 receptors in zebrafish during early embryonic development. Gene Expr. Patterns 15, 104–111. doi: 10.1016/j.gep.2014.05.006
Lippincott-Schwartz, J., Cole, N. B., Marotta, A., Conrad, P. A., and Bloom, G. S. (1995). Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J. Cell Biol. 128, 293–306. doi: 10.1083/jcb.128.3.293
Liu, H., Nakazawa, T., Tezuka, T., and Yamamoto, T. (2006). Physical and functional interaction of Fyn tyrosine kinase with a brain-enriched rho GTPase-activating protein TCGAP. J. Biol. Chem. 281, 23611–23619. doi: 10.1074/jbc.M511205200
Ma, Y., Xiao, Y., Zhang, F., Han, Y., Li, J., Xu, L., et al. (2016). Novel compound heterozygous mutations in MYO7A gene associated with autosomal recessive sensorineural hearing loss in a Chinese family. Int. J. Pediatr. Otorhinolaryngol. 83, 179–185. doi: 10.1016/j.ijporl.2016.01.001
Muraoka, M., and Sakai, H. (1999). Effects of purinenucleotide analogues on microtubule assembly. Cell Struct. Funct. 24, 305–312. doi: 10.1247/csf.24.305
Nakata, T., and Hirokawa, N. (1995). Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 131, 1039–1053. doi: 10.1083/jcb.131.4.1039
Nakazawa, T., Hashimoto, R., Sakoori, K., Sugaya, Y., Tanimura, A., Hashimotodani, Y., et al. (2016). Emerging roles of ARHGAP33 in intracellular trafficking of TrkB and pathophysiology of neuropsychiatric disorders. Nat. Commun. 7:10594. doi: 10.1038/ncomms10594
Naslavsky, N., McKenzie, J., Altan-Bonnet, N., Sheff, D., and Caplan, S. (2009). EHD3 regulates early-endosome-to-Golgi transport and preserves Golgi morphology. J. Cell Sci. 122, 389–400. doi: 10.1242/jcs.037051
Nicolson, T. (2017). The genetics of hair-cell function in zebrafish. J. Neurogenet. 31, 102–112. doi: 10.1080/01677063.2017.1342246
Nishimura, R., Kato, K., Fujiwara, S., Ohashi, K., and Mizuno, K. (2018). Solo and keratin filaments regulate epithelial tubule morphology. Cell Struct. Funct. 43, 95–105. doi: 10.1247/csf.18010
Oku, Y., and Huganir, R. L. (2013). AGAP3 and Arf6 regulate trafficking of AMPA receptors and synaptic plasticity. J. Neurosci. 33, 12586–12598. doi: 10.1523/jneurosci.0341-13.2013
Peppel, K., Boekhoff, I., McDonald, P., Breer, H., Caron, M. G., and Lefkowitz, R. J. (1997). G protein-coupled receptor kinase 3 (GRK3) gene disruption leads to loss of odorant receptor desensitization. J. Biol. Chem. 272, 25425–25428. doi: 10.1074/jbc.272.41.25425
Piacentini, L., and Niroomand, F. (1996). Phosphotransfer reactions as a means of G protein activation. Mol. Cell. Biochem. 157, 59–63. doi: 10.1007/bf00227881
Pickett, S. B., and Raible, D. W. (2019). Water waves to sound waves: using zebrafish to explore hair cell biology. J. Assoc. Res. Otolaryngol. 20, 1–19. doi: 10.1007/s10162-018-00711-1
Qiao, S., Kim, S. H., Heck, D., Goldowitz, D., LeDoux, M. S., and Homayouni, R. (2013). Dab2IP GTPase activating protein regulates dendrite development and synapse number in cerebellum. PLoS One 8:e53635. doi: 10.1371/journal.pone.0053635
Rao, L., Meng, F. L., Fang, R., Cai, C. Y., and Zhao, X. L. (2019). Molecular mechanism of microRNA in regulating cochlear hair cell development. Yi Chuan 41, 994–1008. doi: 10.16288/j.yczz.19-119
Santos, F., MacDonald, G., Rubel, E. W., and Raible, D. W. (2006). Lateral line hair cell maturation is a determinant of aminoglycoside susceptibility in zebrafish (Danio rerio). Hear. Res. 213, 25–33. doi: 10.1016/j.heares.2005.12.009
Schwarzer, S., Spieß, S., Brand, M., and Hans, S. (2017). Dlx3b/4b is required for early-born but not later-forming sensory hair cells during zebrafish inner ear development. Biol Open 6, 1270–1278. doi: 10.1242/bio.026211
Self, T., Mahony, M., Fleming, J., Walsh, J., Brown, S. D., and Steel, K. P. (1998). Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 125, 557–566. doi: 10.1242/dev.125.4.557
Wang, C., Wang, X., Zheng, H., Yao, J., Xiang, Y., and Liu, D. (2023). The ndrg2 gene regulates hair cell morphogenesis and auditory function during zebrafish development. Int. J. Mol. Sci. 24:10002. doi: 10.3390/ijms241210002
Wasfy, M. M., Matsui, J. I., Miller, J., Dowling, J. E., and Perkins, B. D. (2014). Myosin 7aa(−/−) mutant zebrafish show mild photoreceptor degeneration and reduced electroretinographic responses. Exp. Eye Res. 122, 65–76. doi: 10.1016/j.exer.2014.03.007
Watanabe, N., Kato, T., Fujita, A., Ishizaki, T., and Narumiya, S. (1999). Cooperation between mDia1 and ROCK in rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143. doi: 10.1038/11056
Wei, G., Zhang, X., Cai, C., Sheng, J., Xu, M., Wang, C., et al. (2022). Dual-specificity phosphatase 14 regulates zebrafish hair cell formation through activation of p38 signaling pathway. Front. Cell. Neurosci. 16:840143. doi: 10.3389/fncel.2022.840143
Wolff, D. W., Bianchi-Smiraglia, A., and Nikiforov, M. A. (2022). Compartmentalization and regulation of GTP in control of cellular phenotypes. Trends Mol. Med. 28, 758–769. doi: 10.1016/j.molmed.2022.05.012
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, J., McNeil, B., Wu, W., Nees, D., Bai, L., and Wu, L. G. (2008). GTP-independent rapid and slow endocytosis at a central synapse. Nat. Neurosci. 11, 45–53. doi: 10.1038/nn2021
Yamashita, T., Hige, T., and Takahashi, T. (2005). Vesicle endocytosis requires dynamin-dependent GTP hydrolysis at a fast CNS synapse. Science 307, 124–127. doi: 10.1126/science.1103631
Keywords: myo7aa, Rho GTPase signaling, hearing, zebrafish, CRISPR/Cas9
Citation: Xie B, Liang J, Jiang J, Zeng T, Liu L, Xie D, Zhu G, Xiong L, Zhang K, Liu D, Gong J, Chen X, Lai R and Xie H (2024) Zebrafish myo7aa affects congenital hearing by regulating Rho-GTPase signaling. Front. Mol. Neurosci. 17:1405109. doi: 10.3389/fnmol.2024.1405109
Received: 22 March 2024; Accepted: 31 May 2024;
Published: 15 July 2024.
Edited by:
Katsuhiko Tabuchi, Shinshu University, JapanCopyright © 2024 Xie, Liang, Jiang, Zeng, Liu, Xie, Zhu, Xiong, Zhang, Liu, Gong, Chen, Lai and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruosha Lai, bGFpcnVvc2hhQGNzdS5lZHUuY24=; Huaping Xie, aHB4aWVASHVubnUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.