- LifeArc, Accelerator Building, Open Innovation Campus, Stevenage, United Kingdom
Targeted protein degradation (TPD) is a rapidly expanding field, with various PROTACs (proteolysis-targeting chimeras) in clinical trials and molecular glues such as immunomodulatory imide drugs (IMiDs) already well established in the treatment of certain blood cancers. Many current approaches are focused on oncology targets, leaving numerous potential applications underexplored. Targeting proteins for degradation offers a novel therapeutic route for targets whose inhibition remains challenging, such as protein aggregates in neurodegenerative diseases. This mini review focuses on the prospect of utilizing TPD for neurodegenerative disease targets, particularly PROTAC and molecular glue formats and opportunities for novel CNS E3 ligases. Some key challenges of utilizing such modalities including molecular design of degrader molecules, drug delivery and blood brain barrier penetrance will be discussed.
Introduction
Targeted protein degradation (TPD) is a new modality with potential to drug poorly tractable targets. PROTAC (proteolysis-targeting chimera) or molecular glue (MG)-driven ternary complex formation with an E3 ligase utilizes cells’ ubiquitin-proteasome system (UPS) to degrade targets. Several such molecules have entered clinical development (Kong and Jones, 2023). Two E3 ligases, Von Hippel–Lindau (VHL) and Cereblon (CRBN), are regularly harnessed for therapeutic TPD approaches; both belong to the Cullin-RING E3 Ligase (CRL) family (Bondeson et al., 2015; Zengerle et al., 2015). Most current activity is in oncology, with indications including central nervous system (CNS)-associated pathologies less explored. Therapeutic development for CNS diseases is challenging due to blood brain barrier (BBB) permeability constraints, and in the druggability of protein aggregates which often characterize neurodegenerative pathologies. Commonly used degrader approaches have the potential to target proteins or aggregated complexes for degradation by the ubiquitin-proteasome (UPS) system (Bekes et al., 2022; Zhu et al., 2024) or autophagy-lysosome machinery (Pei et al., 2021; Ji et al., 2022). Targeted protein degraders act through event-driven pharmacology via non-reversible removal of functional components, and their potency is boosted by a catalytic mode of action (MOA), enabling sub-stoichiometric dosing regimens (Bekes et al., 2022). An extended pharmacokinetic (PK)- pharmacodynamic (PD) disconnect (Mares et al., 2020), can afford a reduction in off-target toxicity. These represent advantages over occupancy driven pharmacology often displayed by small molecule inhibitors. Further, degradation can be driven by ternary complex stability (Bondeson et al., 2018), for which protein–protein interaction (PPI) driven cooperativity is a key factor. As such, ligand-binding site affinity can be lower, potentially useful in targeting proteins without functional sites, such as the protein aggregates observed in neurodegenerative disease (Figure 1).
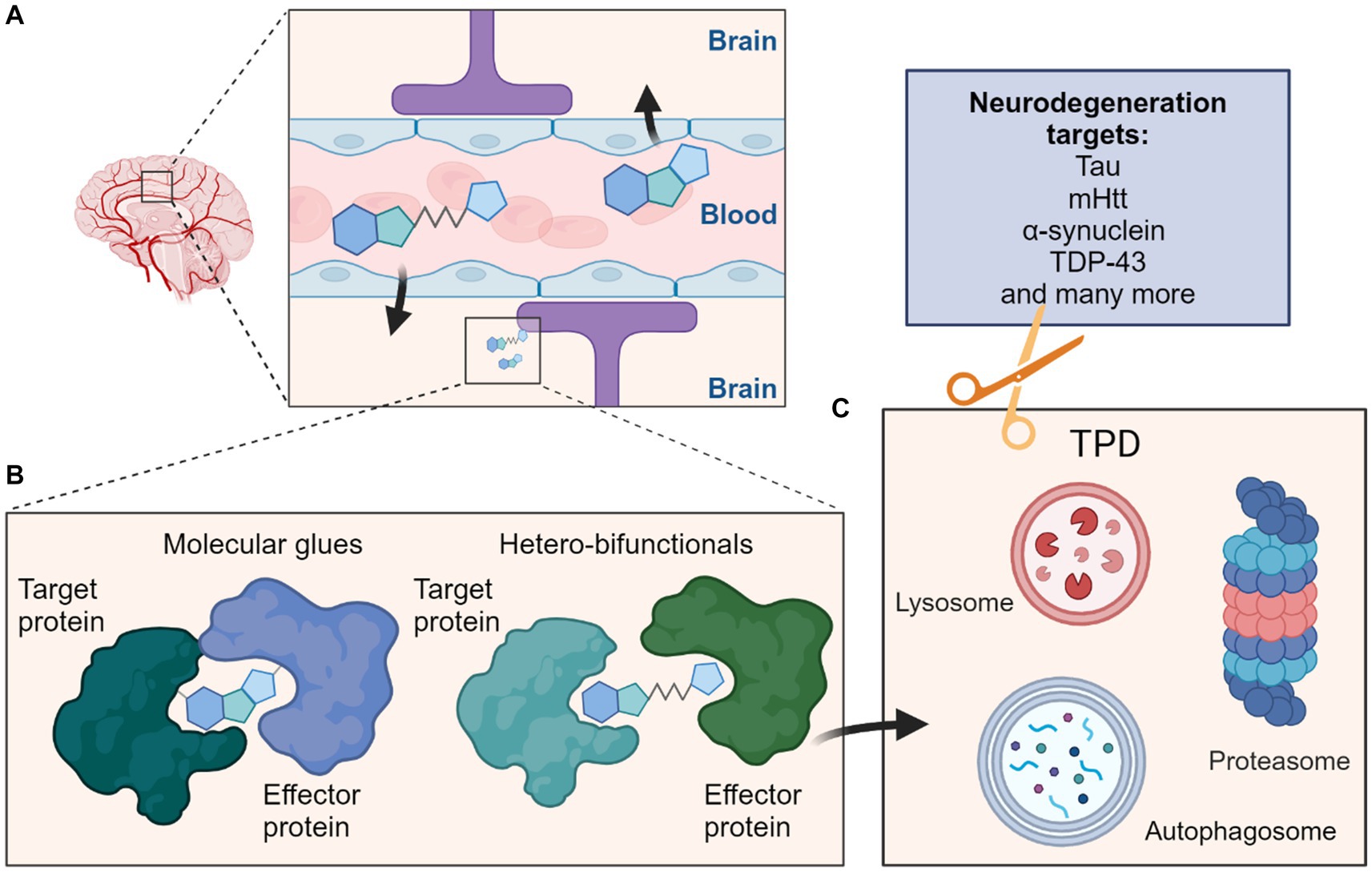
Figure 1. Schematic overview depicting targeted protein degradation approaches for neurodegeneration targets. (A) One major consideration for CNS-targeted ligands is the requirement for sufficient permeation of the blood–brain barrier (BBB); this is most often achieved via appropriate compound design and property metric strategies, although actively co-opting cross-BBB transport mechanisms remains an alternative possibility. (B) Such ligands could be MG typically of lower molecular weight which induce interactions between a target protein and effector protein, or the larger heterobifunctionals which classically contain ligands for both the target protein and effector protein connected by a linker. (C) These molecules can then induce protein degradation of the target protein by targeting them to cellular degradation machineries such as the ubiquitin proteasome system (UPS) or autophagy-lysosome pathways. Many neurodegeneration targets such as proteins prone to aggregation would be amenable to such strategies and their degradation may ameliorate effects of these diseases.
PROTACs in CNS disease
Degradation approaches have been examined in proof-of-concept studies for neurodegenerative diseases (Thomas et al., 2023). Conceptually, protein or aggregates can be degraded if a selective small molecule binder is available. Utilizing degron tagging to induce proximity between a given target and an E3 ligase of interest may validate the approach (Ottis et al., 2017).
Many neurodegenerative disorders feature accumulation of aggregated proteins. Examples include Tauopathies, described in frontotemporal dementia (FTD) and Alzheimer’s disease (AD), and characterized by accumulation of aberrant Tau proteins (Morris et al., 2011; Gotz et al., 2013; Wang et al., 2014). Post-translational modifications of Tau lead to misfolding, mislocalization and oligomerization, resulting in neuronal toxicity (Gotz et al., 2013; Medina, 2018; Congdon et al., 2023). Studies utilizing a KEAP1-dependent peptide PROTAC showed degradation of intracellular Tau (Lu et al., 2018), providing evidence that Tau can be degraded by TPD approaches. Several studies report ternary complex formation and degradation of toxic forms of Tau using PROTACs to recruit the E3 ligases VHL or CRBN (Silva et al., 2019; Wang et al., 2021; Silva et al., 2022). A Tau-selective degrader QC-01-175, showed preferential degradation of aberrant Tau in FTD patient-derived neuronal cell models compared to healthy controls (Silva et al., 2019). A follow-up study optimized the linkers and demonstrated improved degradation of insoluble protein (Silva et al., 2022).
α-Synucleinopathies in Parkinson’s disease (PD) are also a potential degrader target. Abnormal accumulation in neurons leads to the formation of Lewy bodies and neurites, hallmarks of PD (Spillantini et al., 1997, 1998), and α-synuclein may disrupt normal lysosomal function (Bourdenx et al., 2014), suggesting degradation via the UPS as a possible approach. Utilizing an α-synuclein peptide and proteasome-targeting motif resulted in ubiquitination and degradation, attenuating neuronal toxicity (Qu et al., 2020). PROTACs based on three E3 ligase binders and α-synuclein aggregation inhibitor sery384, which binds oligomeric α-synuclein (Wagner et al., 2013; Pena-Diaz et al., 2023), induced degradation of aggregated α-synuclein in an overexpression system using preformed fibrils (Wen et al., 2023). The efficacy of these molecules was low and PROTAC MOA experiments are required to determine the value of this approach.
Mutant huntingtin (mHTT) aggregation is thought to cause neuronal injury and apoptosis in Huntington’s disease (Fiorillo et al., 2021). One approach to targeting mHTT for degradation linked amyloid binding imaging agents (Olsen et al., 2010; Matsumura et al., 2012) to IAP binders and showed degradation of HTT and mHTT in cells (Tomoshige et al., 2017, 2018).
Amyotrophic Lateral Sclerosis (ALS) and FTD show formation of cytoplasmic TAR DNA-binding protein (TDP-43) aggregates (Jo et al., 2020). In healthy cells, TDP-43 is predominantly nuclear and involved in transcriptional and post-transcriptional regulation and pre-mRNA splicing (Jo et al., 2020). A proportion of TDP-43 shuttles between the cytoplasm and nucleus as part of cellular stress responses. In ALS pathology, TDP-43 is found in insoluble cytoplasmic aggregates where it is often hyperphosphorylated, ubiquitinated and fragmented (Neumann et al., 2006; Hasegawa et al., 2008). Loss of nuclear TDP-43 into cytoplasmic aggregates could be a driver of ALS pathology (Ling et al., 2013). PROTACs linking aggregate binders, benzothiazole-aniline derivatives (BTA), to Pomalidomide, degraded overexpressed truncated c-terminal TDP-43, attenuating the reduction in cell viability (Tseng et al., 2023). Both varying expression and subcellular localization of target proteins would offer opportunities for selectivity, giving spatiotemporal control of therapeutic activity.
PROTAC molecular design constraints
Constraining small molecule property design space within thresholds improves toxicity and PKPD outcomes in later development (Lipinski et al., 2001; Wager et al., 2010; Shultz, 2019). CNS-targeted PROTACs are large molecules which also require BBB penetration, and as such occupy several restrictive and complex design spaces. Property considerations are stricter for CNS-targeted ligands requiring passive BBB permeation (Wager et al., 2016). A detailed analysis of CNS metrics is beyond the scope of this review, but consideration of the widely used Pfizer CNS MPO metric (Wager et al., 2010, 2016) favors chemotypes of moderate size and limited polarity (total polar surface area, TPSA, LogD, hydrogen bond donors, HBD), a pattern reproduced in the B3DB compound database with annotated BBB permeation characteristics (Meng et al., 2021).
PROTACs are also large molecules which extend into challenging non-traditional, “beyond rule of 5” (bRo5) property space (Matsson et al., 2016; Volak et al., 2023). There are additional resulting challenges: (i) larger molecules can behave differently especially regarding permeability and transport (Guimaraes et al., 2012; Doak et al., 2014; Matsson et al., 2016; Matsson and Kihlberg, 2017; Pye et al., 2017; Linker et al., 2023); (ii) available chemical space is larger; (iii) large molecules are often more conformationally complex (Rossi Sebastiano et al., 2018; Mobitz, 2023); (iv) fewer molecules from bRo5 space have been studied; (v) those studied molecules cluster in niches (e.g., macrocycles) and represent a less even distribution (Doak et al., 2014). Metric-based approaches nonetheless exist (Degoey et al., 2018; Poongavanam et al., 2018; Mobitz, 2023). Broadly, these suggest that higher LogP values are tolerated; greater ligand complexity could compensate for lipophilicity driven promiscuity (Hann et al., 2001; Doak et al., 2016; Matsson et al., 2016; Egbert et al., 2019). This may compensate for size associated permeability challenges (Pye et al., 2017; Pike et al., 2020). With increasing size, TPSA becomes less representative of ligand polarity, and conformational changes which modulate exposed polarity in response to environment (chameleonic behavior) become accessible (Rossi Sebastiano et al., 2018; Ono et al., 2021; Garcia Jimenez et al., 2023). While the extent to which this behavior is observed in bRo5 chemical matter is debated (Mobitz, 2023), accessible conformations of significantly lower polarity than suggested by TPSA values may be required (Whitty et al., 2016; Mobitz, 2023). Tight restrictions on hydrogen bonding are suggested (Doak et al., 2014; Matsson et al., 2016; Whitty et al., 2016; Mobitz, 2023; Volak et al., 2023).
Analysis of PROTAC chemotypes shows these molecules broadly comply with bRo5 paradigms (Edmondson et al., 2019), although stricter limits on HBD were observed in rat absorption data (Hornberger and Araujo, 2023). A CNS penetrant PROTAC must balance sufficient polarity to provide specific target engagement and solubility while crossing the BBB (Doak et al., 2014). Additionally, increased efflux is common in bRo5 (Doak et al., 2014; Matsson et al., 2016; Cantrill et al., 2020; Cox et al., 2023) and PROTAC development space (Klein et al., 2021). Beyond CNS considerations, the combination of differentiated property space (in particular lipophilicity) and pharmacology results in additional considerations around absorption, distribution, metabolism, and excretion (ADME) testing (Cantrill et al., 2020; Pike et al., 2020; Apprato et al., 2023; Hornberger and Araujo, 2023; Volak et al., 2023).
Lastly, heterobifunctional molecules display behaviors not attributable to a single component, including different target specificity to their POI ligands, whose affinity does not predict ternary complex stability or productivity (Bondeson et al., 2018). Cooperative binding behavior can arise from induced PPI (Song et al., 2021). Given the limitations implied by the combination of CNS permeation, bRo5 space and efflux liabilities, the ‘budget’ of polar contacts and hydrogen bonds available for the POI binder alone is limited (Hughes et al., 2021; Hornberger and Araujo, 2023). The interactions induced in ternary complex formation can generate contact areas equivalent to those in other bRo5 modalities (Doak and Kihlberg, 2017; Bondeson et al., 2018). Considered alongside the importance of conformational masking of polarity in bRo5 space, property-based linker design and rigidification (Troup et al., 2020; Klein et al., 2021; Poongavanam et al., 2022), and holistic consideration of molecular properties will prove vital. The combination of CNS and PROTAC constraints will require greater chameleonicity and cooperativity (Figure 2), and each component to adopt multiple roles to achieve sufficient permeability and binding specificity.
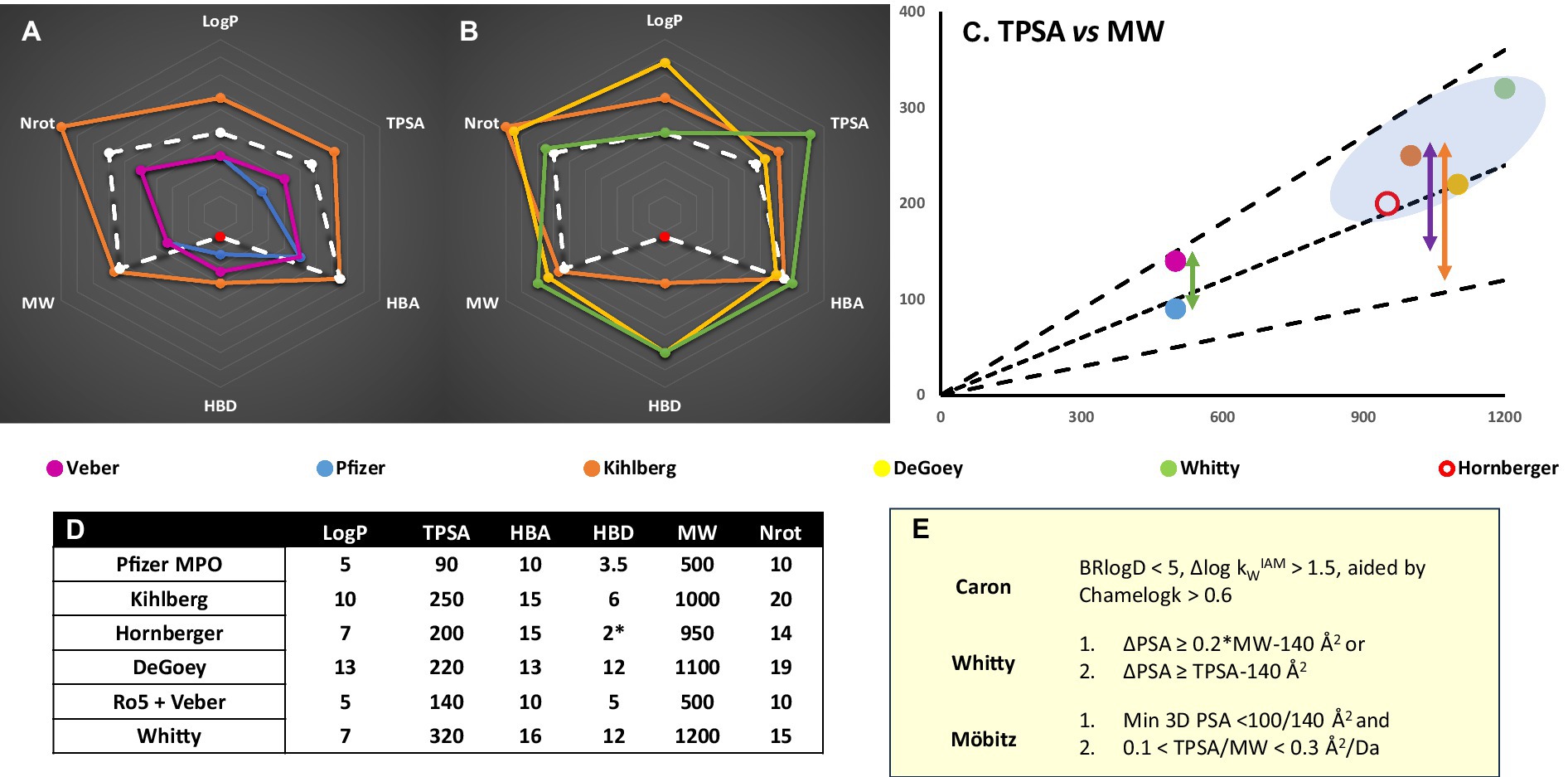
Figure 2. (A) Chemical space representation for CNS (Pfizer MPO) vs Ro5 (Lipinski et al., 2001; Veber et al., 2002) bRo5 (Doak et al., 2014; Villar et al., 2014; Poongavanam et al., 2018) and PROTAC guidelines (Hornberger and Araujo, 2023) for oral absorption. Red data point refers to unsatisfied HBD – intramolecular hydrogen bond pairs not counted. MGD designs would be expected to fall within Ro5/CNS metric space. (B) PROTAC specific values vs different metrics or limitations identified from bRo5 space studies (Degoey et al., 2018). (C) Several TPSA guidelines (E) scaled by MW; (Whitty et al., 2016; Mobitz, 2023) black lines are TPSA = 0.1, 0.2, 0.3*MW, plotted points are the MW and TPSA outer limits of metric sets from (A,B). The green arrow shows the TPSA difference between Ro5 and CNS Ro5 space. The purple arrow represents the TPSA vs minimum/3D PSA difference seemingly required for bRo5 oral absorption. BBB penetration will impose stricter minimum/3D PSA limitations and a correspondingly larger ΔPSA is likely necessary (orange). (D) Numerical values plotted in (A,B) for different metric spaces. (E). Polarity based chameleonicity or ΔPSA/3D TPSA metrics defined in the literature (Whitty et al., 2016; Garcia Jimenez et al., 2023; Mobitz, 2023).
Several CNS penetrant PROTACs have been reported, utilizing VHL (Wang et al., 2021; Liu et al., 2022) and CRBN binders (Mihalic, 2023); oral exposure was demonstrated for each (Liu et al., 2022; Mihalic, 2023). All display low Kp,uu values (Loryan et al., 2022), but nonetheless several demonstrate in vivo activity (Wang et al., 2021; Mihalic, 2023). These observations may reflect exposure requirements depending on potency and event vs occupancy driven pharmacology (Volak et al., 2023). Notably the clinical candidate NX-5948 minimizes size and hydrogen bonding consistent with earlier considerations (Mihalic, 2023). Though development of PROTACs for neurodegenerative diseases will have to meet the challenges of BBB and brain penetrance, there is considerable commercial activity in employing this therapeutic strategy (Kargbo, 2020).
Expanding the repertoire of E3 ligases
Two key E3 ligases have been routinely harnessed for TPD approaches. CRBN ligands dominate PROTACs and MG in clinical stages (Kong and Jones, 2023), mainly due to their size and “drug-like” properties, although the risk of teratogenicity remains a concern (Smith and Mitchell, 2018). Two VHL-based PROTACs, namely BCL-XL degrader DT2216 (Khan et al., 2019) and KRASG12D degrader ASP-3082 (Nagashima et al., 2022) are in clinical trials, and E7820, which glues RBM39 to the DCAF15 ligase complex (Chirnomas et al., 2023; Kong and Jones, 2023). Other E3 ligases such as RNF114 (Spradlin et al., 2019; Luo et al., 2021), KEAP1 (Wei et al., 2021; Du et al., 2022), DCAF11 (Zhang et al., 2021; Xue et al., 2023) and DCAF16 (Zhang et al., 2019) have been harnessed for TPD, but not progressed beyond pre-clinical investigations and their effectiveness in degrading a wide range of targets remains unclear. Several of these E3s are ubiquitously expressed, likely leading to target degradation in multiple tissues increasing toxicity when targeting essential proteins. The use of tissue enriched or selective E3 ligases as PROTAC or molecular glue degrader (MGD) targets has huge potential for diseases restricted to one tissue or organ such as CNS pathologies. One clinical example is DT2116, which degrades BCL-XL. BCL-XL inhibition causes on-target and dose-limiting thrombocytopenia (Kaefer et al., 2014); however, the PROTAC showed significantly lower toxicity due to low VHL expression in platelets (Khan et al., 2019). CNS enriched E3s may allow CNS target degradation while limiting potential toxicity in unwanted tissues. Examples include RNF182 identified as a brain-enriched E3 upregulated in AD patients (Liu et al., 2008). Several E3s of the Kelch family such as ENC1, KBTBD11, KLHL2, KLHL32, KLHL35 and KLHL4 also have preferential brain expression (Ehrlich et al., 2020), where other family members such as KEAP1 have already been utilized in PROTAC-based degraders (Wei et al., 2021). Two TRIM-family E3 Ligases (TRIM9 and TRIM67) are neuronally enriched and regulate neuronal morphological changes (Tanji et al., 2010; Menon et al., 2015; Boyer et al., 2020). A related E3 Ligase TRIM21 shows proximity-induced degradation using the “Trim-away” technique, degrading targets through binding the Fc region of antibodies (Clift et al., 2017). Developments in specific fragment libraries (Whitehurst et al., 2023) and covalent approaches have been investigated for E3 ligand finding (Tao et al., 2022; Belcher et al., 2023; Toriki et al., 2023), but not demonstrated in CNS space. Advances in understanding which E3 ligases could be utilized for TPD purposes (Liu et al., 2023) will support efforts to discover and develop new E3 ligase ligands (Ishida and Ciulli, 2021).
Molecular glue degraders (MGDs) for CNS disease
Most MGDs facilitate degradation via induced protein–protein interactions between an E3 ligase and a target (Rui et al., 2023). Historically, the discovery of MG and their mechanism was retrospective. For example, the severe birth defects caused by thalidomide (Vargesson, 2015) are thought to be caused by degradation of the neosubstrate transcription factor SALL4 via thalidomide binding to the E3 ligase CRBN (Ito et al., 2010) The landscape of neosubstrates depends on ligand structure (Matyskiela et al., 2016; Nowak et al., 2023; Szewczyk et al., 2024), unlocking opportunities for novel target discovery. The aryl sulfonamide Indisulam blocks cell cycle progression (Owa et al., 1999) through degradation of mRNA splicing factor RBM39 mediated by recruitment of the E3 ligase DCAF15 (Han et al., 2017). Indisulam and derivatives do not display high affinity to either target protein; ternary complex formation results in nanomolar affinities (Du et al., 2019). Linking aryl sulfonamides to the BET bromodomain inhibitor JQ1 demonstrated that BRD4 degradation was independent of DCAF15 and driven by stabilizing existing interactions of adjacent bromodomains with the E3 ligase DCAF16 (Hsia et al., 2024). These intramolecular bivalent glues highlight another mechanistic possibility, demonstrating the pharmacological scope of these approaches and the importance of evaluating off-target degradation profiles of novel degraders.
Efforts are underway to rationalize the discovery of MG. One approach is the E3 ligase centric generation of compound libraries, currently focused on CRBN-binding chemotypes (Powell et al., 2020) but which will extend to other E3s upon ligand and neosubstrate discovery. Another is to engineer cell lines to overexpress, eliminate, conformationally trap or inactivate a given E3 ligase; then screen libraries for their ability to degrade targets (Mayor-Ruiz et al., 2019; Hanzl et al., 2023; Ng et al., 2023). However, CNS relevant cell types are often more difficult to manipulate than cancer cell lines. DNA-encoded library (DEL) screening has been adapted to screen for ternary complex formation (Mason et al., 2023) and in combination with disease relevant recombinant aggregates or proteins would be a definite starting point for CNS relevant MG identification. Opportunities for novel MG identification depend on an understanding of interactions between relevant proteins. Techniques such as Bio-ID/Turbo-ID can identify potential E3 ligase substrates and weak protein–protein interactions (Branon et al., 2018; Barroso-Gomila et al., 2023) in disease relevant cell types as a precursor to DEL screening for novel MG discovery. Matching an E3 ligase to a target protein through CRISPR-based degron mapping would further annotate pre-selection (Timms et al., 2023; Zhang et al., 2023) to identify CNS-enriched E3s and target pairings.
Structure-based design of MG remains an enticing but challenging prospect. Modeling or obtaining structural information on ternary complexes is difficult due to their complexity (Casement et al., 2021). Artificial Intelligence (AI) and Machine Learning (ML) approaches have driven advances in protein structure prediction, but their utility in predicting protein–protein complexes remains uncertain (Burke et al., 2023), although improvements in accuracy within classes may be realized as more structural data is acquired.
Glue molecular design considerations
A CNS-targeted MG would generally comprise a small molecule requiring BBB permeability; although differences in binding (Cao et al., 2022; Sasso et al., 2023) and the specifics of event driven pharmacology (Riching et al., 2022) may create distinctions. MG chemotype diversity (e.g., thalidomide vs rapamycin), and scaffold over-representation complicate definition of generic design constraints (Dong et al., 2021; Sasso et al., 2023) although it appears that most non-natural product MG can achieve their function within Ro5 parameter space (Dong et al., 2021; Dewey et al., 2023). Re-positioning existing chemotypes is common in glue development- (Guo et al., 2019; Geiger et al., 2022; Sasso et al., 2023; Toriki et al., 2023), so scaffold specific considerations may be widely applicable.
The most studied MGD class are immunomodulatory imide drugs (IMiD) analogues, which bind to CRBN (Cao et al., 2022) via structures related to biological signaling motifs (Ichikawa et al., 2022) and engage a recurring POI motif represented in numerous targets, largely via hydrophobic groups (Oleinikovas et al., 2024). Recent work on parameter optimization for CRBN ligand specificity (Szewczyk et al., 2024) suggested significant target dependence, but reduction in aromatic carbocycles, introduction of heteroatoms and increased spatial complexity appear beneficial. LogP or TPSA were not determinative, while increasing HBD count was detrimental in several classes. This suggests that CRBN MGD selectivity optimization is not necessarily in tension with CNS property space. However, the determinative power of these MPOs was weak across much of the chemical space surveyed. The observation that CRBN ligands extend the PPI surface (Oleinikovas et al., 2024), and can cause PPI relevant conformational changes (Watson et al., 2022) is consistent with groups apparently distal from the common binding site influencing substrate selectivity (Nowak et al., 2023; Nguyen et al., 2024). In studies of CRBN binders it was observed that more proteins interact than are degraded (Sievers et al., 2018) an effect which may give rise to stronger selectivity in cells (Oleinikovas et al., 2024).
Among other known degrader classes, Cyclin K degraders have been shown to function throughout the Ro5 LogP and MW range, with a hydrophobic contact and ligand modification of the PPI again noted as key considerations in binding and selectivity (Kozicka et al., 2023). Indisulam analogues have less binary affinity and bind in a less conserved site but are involved in extensive interface interactions which modify the resulting PPI surfaces (Faust et al., 2020). From the limited studies to date the influence of conformational and PPI effects such as extending or remodeling surfaces and steric exclusion appear to be considerations for the efficacy and selectivity of glue interactions, which may influence the choices made in glue design (Geiger et al., 2022; Kozicka et al., 2023; Jiang et al., 2024; Oleinikovas et al., 2024).
In practical terms non-natural product MG fall mostly within chemical space for which existing ADME assays are effective. Compound efficiency metrics, however, may need to be re-defined to adequately discriminate between early stage ligands (Jia et al., 2024).
Non E3-ligase driven degradation
Strategies to target proteins or aggregates to the autophagy-lysosome machinery are less well understood than UPS-targeting degraders. For example, microarray-based screening led to the discovery of mHTT–light chain 3 (LC3) binders which induced autophagy-mediated degradation of mHTT (Li et al., 2019). The underlying MOA has since been questioned as it was found that the indolinone LC3 binders covalently bind to the E3 Ligase DCAF11, leading to UPS-driven degradation (Xue et al., 2023). α-Synuclein has been targeted to the lysosomal proteolytic machinery by peptides containing α-synuclein-binding, membrane-penetrating and chaperone-mediated autophagy-targeting motifs (Tong et al., 2023). α-Synuclein has also been degraded by macroautophagy, employing an autophagy-targeting chimera (AUTOTAC) binding both α-Synuclein aggregates and autophagy receptor p62/SQSTM1/Sequestosome-1 (Lee et al., 2023). Strategies to clear extracellular proteins or aggregates include lysosome-targeting chimeras (LYTACs), which are bifunctional molecules binding both a cell-surface lysosome-shuttling receptor and the target of interest (Banik et al., 2020), which could be especially valuable for neurodegenerative diseases characterized by toxic insoluble extracellular aggregates. Some of these techniques may lead to the activation of autophagy having an added benefit, since mutations in genes encoding regulators of the autophagy and UPS machineries, such as p62/SQSTM1, VCP or Ubiquilin-2 are often found in FTD or ALS and are thought to cause downregulation of autophagy/proteasomal functions (Deng et al., 2017). Further, protein aggregates or oligomers may interfere with proteasomal function, resulting in lower PROTAC efficacy. One study describes allosteric impairment of the 20S proteasome substrate gate by soluble oligomers composed of either amyloid-β (Aβ) 1–42, α-synuclein, or mHTT, preventing substrates from entering the 19S pore (Thibaudeau et al., 2018). It is not known whether these oligomers inhibit the proteasome sufficiently to prevent efficient degradation.
Concluding remarks and outlook
Several proof-of-concept studies have been published on the degradation of known neurodegenerative disease targets. Key challenges remain in the discovery of selective small molecules suitable for degradation approaches and their development towards therapeutic applications in CNS space. For heterobifunctional molecules such as PROTACs, the most important aspects center on complex mechanistic and property design considerations. By contrast, MG discovery space particularly requires identification of degraders and the PPI which they enable. For both contexts, novel CNS-enriched E3 ligase enablement will drive progress in this area. Despite these challenges, the event driven pharmacology allowed by degraders may be especially suited to CNS disease targets, allowing for innovative therapeutic developments in an area of significant unmet medical need.
Author contributions
SK: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. AC: Conceptualization, Writing – original draft, Writing – review & editing. KB: Writing – original draft, Writing – review & editing. ZY: Writing – original draft, Writing – review & editing. JL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Figure 1 was created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Apprato, G., Ermondi, G., and Caron, G. (2023). The quest for Oral Protac drugs: evaluating the weaknesses of the screening pipeline. ACS Med. Chem. Lett. 14, 879–883. doi: 10.1021/acsmedchemlett.3c00231
Banik, S. M., Pedram, K., Wisnovsky, S., Ahn, G., Riley, N. M., and Bertozzi, C. R. (2020). Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584, 291–297. doi: 10.1038/s41586-020-2545-9
Barroso-Gomila, O., Merino-Cacho, L., Muratore, V., Perez, C., Taibi, V., Maspero, E., et al. (2023). BioE3 identifies specific substrates of ubiquitin E3 ligases. Nat. Commun. 14:7656. doi: 10.1038/s41467-023-43326-8
Bekes, M., Langley, D. R., and Crews, C. M. (2022). Protac targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200. doi: 10.1038/s41573-021-00371-6
Belcher, B. P., Ward, C. C., and Nomura, D. K. (2023). Ligandability of E3 ligases for targeted protein degradation applications. Biochemistry 62, 588–600. doi: 10.1021/acs.biochem.1c00464
Bondeson, D. P., Mares, A., Smith, I. E., Ko, E., Campos, S., Miah, A. H., et al. (2015). Catalytic in vivo protein knockdown by small-molecule Protacs. Nat. Chem. Biol. 11, 611–617. doi: 10.1038/nchembio.1858
Bondeson, D. P., Smith, B. E., Burslem, G. M., Buhimschi, A. D., Hines, J., Jaime-Figueroa, S., et al. (2018). Lessons in Protac design from selective degradation with a promiscuous warhead. Cell. Chem. Biol. 25, 78–87.e5. doi: 10.1016/j.chembiol.2017.09.010
Bourdenx, M., Bezard, E., and Dehay, B. (2014). Lysosomes and alpha-synuclein form a dangerous duet leading to neuronal cell death. Front. Neuroanat. 8:83. doi: 10.3389/fnana.2014.00083
Boyer, N. P., Mccormick, L. E., Menon, S., Urbina, F. L., and Gupton, S. L. (2020). A pair of E3 ubiquitin ligases compete to regulate filopodial dynamics and axon guidance. J. Cell Biol. 219:e201902088. doi: 10.1083/jcb.201902088
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with Turboid. Nat. Biotechnol. 36, 880–887. doi: 10.1038/nbt.4201
Burke, D. F., Bryant, P., Barrio-Hernandez, I., Memon, D., Pozzati, G., Shenoy, A., et al. (2023). Towards a structurally resolved human protein interaction network. Nat. Struct. Mol. Biol. 30, 216–225. doi: 10.1038/s41594-022-00910-8
Cantrill, C., Chaturvedi, P., Rynn, C., Petrig Schaffland, J., Walter, I., and Wittwer, M. B. (2020). Fundamental aspects of Dmpk optimization of targeted protein degraders. Drug Discov. Today 25, 969–982. doi: 10.1016/j.drudis.2020.03.012
Cao, S., Kang, S., Mao, H., Yao, J., Gu, L., and Zheng, N. (2022). Defining molecular glues with a dual-nanobody cannabidiol sensor. Nat. Commun. 13:815. doi: 10.1038/s41467-022-28507-1
Casement, R., Bond, A., Craigon, C., and Ciulli, A. (2021). Mechanistic and structural features of Protac ternary complexes. Methods Mol. Biol. 2365, 79–113. doi: 10.1007/978-1-0716-1665-9_5
Chirnomas, D., Hornberger, K. R., and Crews, C. M. (2023). Protein degraders enter the clinic - a new approach to cancer therapy. Nat. Rev. Clin. Oncol. 20, 265–278. doi: 10.1038/s41571-023-00736-3
Clift, D., Mcewan, W. A., Labzin, L. I., Konieczny, V., Mogessie, B., James, L. C., et al. (2017). A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706.e18. doi: 10.1016/j.cell.2017.10.033
Congdon, E. E., Ji, C., Tetlow, A. M., Jiang, Y., and Sigurdsson, E. M. (2023). Tau-targeting therapies for Alzheimer disease: current status and future directions. Nat. Rev. Neurol. 19, 715–736. doi: 10.1038/s41582-023-00883-2
Cox, B., Nicolai, J., and Williamson, B. (2023). The role of the efflux transporter, P-glycoprotein, at the blood-brain barrier in drug discovery. Biopharm. Drug Dispos. 44, 113–126. doi: 10.1002/bdd.2331
Degoey, D. A., Chen, H. J., Cox, P. B., and Wendt, M. D. (2018). Beyond the rule of 5: lessons Learned from AbbVie's drugs and compound collection. J. Med. Chem. 61, 2636–2651. doi: 10.1021/acs.jmedchem.7b00717
Deng, Z., Sheehan, P., Chen, S., and Yue, Z. (2017). Is amyotrophic lateral sclerosis/frontotemporal dementia an autophagy disease? Mol. Neurodegener. 12:90. doi: 10.1186/s13024-017-0232-6
Dewey, J. A., Delalande, C., Azizi, S. A., Lu, V., Antonopoulos, D., and Babnigg, G. (2023). Molecular glue discovery: current and future approaches. J. Med. Chem. 66, 9278–9296. doi: 10.1021/acs.jmedchem.3c00449
Doak, B. C., and Kihlberg, J. (2017). Drug discovery beyond the rule of 5 - opportunities and challenges. Expert Opin. Drug Discov. 12, 115–119. doi: 10.1080/17460441.2017.1264385
Doak, B. C., Over, B., Giordanetto, F., and Kihlberg, J. (2014). Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol. 21, 1115–1142. doi: 10.1016/j.chembiol.2014.08.013
Doak, B. C., Zheng, J., Dobritzsch, D., and Kihlberg, J. (2016). How beyond rule of 5 drugs and clinical candidates bind to their targets. J. Med. Chem. 59, 2312–2327. doi: 10.1021/acs.jmedchem.5b01286
Dong, G., Ding, Y., He, S., and Sheng, C. (2021). Molecular glues for targeted protein degradation: from serendipity to rational discovery. J. Med. Chem. 64, 10606–10620. doi: 10.1021/acs.jmedchem.1c00895
Du, G., Jiang, J., Henning, N. J., Safaee, N., Koide, E., Nowak, R. P., et al. (2022). Exploring the target scope of Keap1 E3 ligase-based Protacs. Cell. Chem. Biol. 29, 1470–1481.e31. doi: 10.1016/j.chembiol.2022.08.003
Du, X., Volkov, O. A., Czerwinski, R. M., Tan, H., Huerta, C., Morton, E. R., et al. (2019). Structural basis and kinetic pathway of Rbm39 recruitment to Dcaf15 by a sulfonamide molecular glue E7820. Structure 27, 1625–1633.e3. doi: 10.1016/j.str.2019.10.005
Edmondson, S. D., Yang, B., and Fallan, C. (2019). Proteolysis targeting chimeras (Protacs) in 'beyond rule-of-five' chemical space: recent progress and future challenges. Bioorg. Med. Chem. Lett. 29, 1555–1564. doi: 10.1016/j.bmcl.2019.04.030
Egbert, M., Whitty, A., Keseru, G. M., and Vajda, S. (2019). Why some targets benefit from beyond rule of five drugs. J. Med. Chem. 62, 10005–10025. doi: 10.1021/acs.jmedchem.8b01732
Ehrlich, K. C., Baribault, C., and Ehrlich, M. (2020). Epigenetics of muscle- and brain-specific expression of Klhl family genes. Int. J. Mol. Sci. 21:8394. doi: 10.3390/ijms21218394
Faust, T. B., Yoon, H., Nowak, R. P., Donovan, K. A., Li, Z., Cai, Q., et al. (2020). Structural complementarity facilitates E7820-mediated degradation of Rbm39 by Dcaf15. Nat. Chem. Biol. 16, 7–14. doi: 10.1038/s41589-019-0378-3
Fiorillo, A., Morea, V., Colotti, G., and Ilari, A. (2021). Huntingtin ubiquitination mechanisms and novel possible therapies to decrease the toxic effects of mutated huntingtin. J. Pers. Med. 11:1309. doi: 10.3390/jpm11121309
Garcia Jimenez, D., Vallaro, M., Rossi Sebastiano, M., Apprato, G., D'agostini, G., Rossetti, P., et al. (2023). Chamelogk: A chromatographic Chameleonicity quantifier to design orally bioavailable beyond-rule-of-5 drugs. J. Med. Chem. 66, 10681–10693. doi: 10.1021/acs.jmedchem.3c00823
Geiger, T. M., Schäfer, S. C., Dreizler, J. K., Walz, M., and Hausch, F. (2022). Clues to molecular glues. Curr. Chem. Biol. 2:100018. doi: 10.1016/j.crchbi.2021.100018
Gotz, J., Xia, D., Leinenga, G., Chew, Y. L., and Nicholas, H. (2013). What Renders Tau Toxic. Front. Neurol. 4:72. doi: 10.3389/fneur.2013.00072
Guimaraes, C. R., Mathiowetz, A. M., Shalaeva, M., Goetz, G., and Liras, S. (2012). Use of 3D properties to characterize beyond rule-of-5 property space for passive permeation. J. Chem. Inf. Model. 52, 882–890. doi: 10.1021/ci300010y
Guo, Z., Hong, S. Y., Wang, J., Rehan, S., Liu, W., Peng, H., et al. (2019). Rapamycin-inspired macrocycles with new target specificity. Nat. Chem. 11, 254–263. doi: 10.1038/s41557-018-0187-4
Han, T., Goralski, M., Gaskill, N., Capota, E., Kim, J., Ting, T. C., et al. (2017). Anticancer sulfonamides target splicing by inducing Rbm39 degradation via recruitment to Dcaf15. Science 356:eaal3755. doi: 10.1126/science.aal3755
Hann, M. M., Leach, A. R., and Harper, G. (2001). Molecular complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 41, 856–864. doi: 10.1021/ci000403i
Hanzl, A., Barone, E., Bauer, S., Yue, H., Nowak, R. P., Hahn, E., et al. (2023). E3-specific degrader discovery by dynamic tracing of substrate receptor abundance. J. Am. Chem. Soc. 145, 1176–1184. doi: 10.1021/jacs.2c10784
Hasegawa, M., Arai, T., Nonaka, T., Kametani, F., Yoshida, M., Hashizume, Y., et al. (2008). Phosphorylated Tdp-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70. doi: 10.1002/ana.21425
Hornberger, K. R., and Araujo, E. M. V. (2023). Physicochemical property determinants of Oral absorption for Protac protein degraders. J. Med. Chem. 66, 8281–8287. doi: 10.1021/acs.jmedchem.3c00740
Hsia, O., Hinterndorfer, M., Cowan, A. D., Iso, K., Ishida, T., Sundaramoorthy, R., et al. (2024). Targeted protein degradation via intramolecular bivalent glues. Nature 627, 204–211. doi: 10.1038/s41586-024-07089-6
Hughes, S. J., Testa, A., Thompson, N., and Churcher, I. (2021). The rise and rise of protein degradation: opportunities and challenges ahead. Drug Discov. Today 26, 2889–2897. doi: 10.1016/j.drudis.2021.08.006
Ichikawa, S., Flaxman, H. A., Xu, W., Vallavoju, N., Lloyd, H. C., Wang, B., et al. (2022). The E3 ligase adapter cereblon targets the C-terminal cyclic imide degron. Nature 610, 775–782. doi: 10.1038/s41586-022-05333-5
Ishida, T., and Ciulli, A. (2021). E3 ligase ligands for Protacs: how they were found and how to discover new ones. Slas. Discov. 26, 484–502. doi: 10.1177/2472555220965528
Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Imamura, Y., et al. (2010). Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350. doi: 10.1126/science.1177319
Ji, C. H., Kim, H. Y., Lee, M. J., Heo, A. J., Park, D. Y., Lim, S., et al. (2022). The Autotac chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat. Commun. 13:904. doi: 10.1038/s41467-022-28520-4
Jia, L., Weiss, D., Shields, B., Claus, B., Shanmugasundaram, V., Johnson, S., et al. (2024). Scoring methods in lead optimization of molecular glues. ChemRxiv. doi: 10.26434/chemrxiv-2023-4hn4s-v2
Jiang, W., Jiang, Y., Luo, Y., Qiao, W., and Yang, T. (2024). Facilitating the development of molecular glues: opportunities from serendipity and rational design. Eur. J. Med. Chem. 263:115950. doi: 10.1016/j.ejmech.2023.115950
Jo, M., Lee, S., Jeon, Y. M., Kim, S., Kwon, Y., and Kim, H. J. (2020). The role of Tdp-43 propagation in neurodegenerative diseases: integrating insights from clinical and experimental studies. Exp. Mol. Med. 52, 1652–1662. doi: 10.1038/s12276-020-00513-7
Kaefer, A., Yang, J., Noertersheuser, P., Mensing, S., Humerickhouse, R., Awni, W., et al. (2014). Mechanism-based pharmacokinetic/pharmacodynamic meta-analysis of navitoclax (Abt-263) induced thrombocytopenia. Cancer Chemother. Pharmacol. 74, 593–602. doi: 10.1007/s00280-014-2530-9
Kargbo, R. B. (2020). Protac compounds targeting alpha-Synuclein protein for treating Neurogenerative disorders: Alzheimer's and Parkinson's diseases. ACS Med. Chem. Lett. 11, 1086–1087. doi: 10.1021/acsmedchemlett.0c00192
Khan, S., Zhang, X., Lv, D., Zhang, Q., He, Y., Zhang, P., et al. (2019). A selective Bcl-X(L) Protac degrader achieves safe and potent antitumor activity. Nat. Med. 25, 1938–1947. doi: 10.1038/s41591-019-0668-z
Klein, V. G., Bond, A. G., Craigon, C., Lokey, R. S., and Ciulli, A. (2021). Amide-to-Ester substitution as a strategy for optimizing Protac permeability and cellular activity. J. Med. Chem. 64, 18082–18101. doi: 10.1021/acs.jmedchem.1c01496
Kong, N. R., and Jones, L. H. (2023). Clinical translation of targeted protein degraders. Clin. Pharmacol. Ther. 114, 558–568. doi: 10.1002/cpt.2985
Kozicka, Z., Suchyta, D. J., Focht, V., Kempf, G., Petzold, G., Jentzsch, M., et al. (2023). Design principles for cyclin K molecular glue degraders. Nat. Chem. Biol. 20, 93–102. doi: 10.1038/s41589-023-01409-z
Lee, J., Sung, K. W., Bae, E. J., Yoon, D., Kim, D., Lee, J. S., et al. (2023). Targeted degradation of ⍺-synuclein aggregates in Parkinson's disease using the Autotac technology. Mol. Neurodegener. 18:41. doi: 10.1186/s13024-023-00630-7
Li, Z., Wang, C., Wang, Z., Zhu, C., Li, J., Sha, T., et al. (2019). Allele-selective lowering of mutant Htt protein by Htt-Lc3 linker compounds. Nature 575, 203–209. doi: 10.1038/s41586-019-1722-1
Ling, S. C., Polymenidou, M., and Cleveland, D. W. (2013). Converging mechanisms in Als and Ftd: disrupted Rna and protein homeostasis. Neuron 79, 416–438. doi: 10.1016/j.neuron.2013.07.033
Linker, S. M., Schellhaas, C., Kamenik, A. S., Veldhuizen, M. M., Waibl, F., Roth, H. J., et al. (2023). Lessons for Oral bioavailability: how Conformationally flexible cyclic peptides enter and cross lipid membranes. J. Med. Chem. 66, 2773–2788. doi: 10.1021/acs.jmedchem.2c01837
Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26. doi: 10.1016/S0169-409X(00)00129-0
Liu, X., Kalogeropulou, A. F., Domingos, S., Makukhin, N., Nirujogi, R. S., Singh, F., et al. (2022). Discovery of Xl01126: A potent, fast, cooperative, selective, orally bioavailable, and blood-brain barrier penetrant Protac degrader of leucine-rich repeat kinase 2. J. Am. Chem. Soc. 144, 16930–16952. doi: 10.1021/jacs.2c05499
Liu, Q. Y., Lei, J. X., Sikorska, M., and Liu, R. (2008). A novel brain-enriched E3 ubiquitin ligase Rnf182 is up regulated in the brains of Alzheimer's patients and targets Atp6V0C for degradation. Mol. Neurodegener. 3:4. doi: 10.1186/1750-1326-3-4
Liu, Y., Yang, J., Wang, T., Luo, M., Chen, Y., Chen, C., et al. (2023). Expanding Protactable genome universe of E3 ligases. Nat. Commun. 14:6509. doi: 10.1038/s41467-023-42233-2
Loryan, I., Reichel, A., Feng, B., Bundgaard, C., Shaffer, C., Kalvass, C., et al. (2022). Unbound brain-to-plasma partition coefficient, Kp,uu,brain—a game changing parameter for CNS drug discovery and development. Pharm. Res. 39, 1321–1341. doi: 10.1007/s11095-022-03246-6
Lu, M., Liu, T., Jiao, Q., Ji, J., Tao, M., Liu, Y., et al. (2018). Discovery of a Keap1-dependent peptide Protac to knockdown tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 146, 251–259. doi: 10.1016/j.ejmech.2018.01.063
Luo, M., Spradlin, J. N., Boike, L., Tong, B., Brittain, S. M., Mckenna, J. M., et al. (2021). Chemoproteomics-enabled discovery of covalent Rnf114-based degraders that mimic natural product function. Cell. Chem. Biol. 28, 559–566.e15. doi: 10.1016/j.chembiol.2021.01.005
Mares, A., Miah, A. H., Smith, I. E. D., Rackham, M., Thawani, A. R., Cryan, J., et al. (2020). Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 3:140. doi: 10.1038/s42003-020-0868-6
Mason, J. W., Chow, Y. T., Hudson, L., Tutter, A., Michaud, G., Westphal, M. V., et al. (2023). DNA-encoded library-enabled discovery of proximity-inducing small molecules. Nat. Chem. Biol. 20, 170–179. doi: 10.1038/s41589-023-01458-4
Matsson, P., Doak, B. C., Over, B., and Kihlberg, J. (2016). Cell permeability beyond the rule of 5. Adv. Drug Deliv. Rev. 101, 42–61. doi: 10.1016/j.addr.2016.03.013
Matsson, P., and Kihlberg, J. (2017). How big is too big for cell permeability? J. Med. Chem. 60, 1662–1664. doi: 10.1021/acs.jmedchem.7b00237
Matsumura, K., Ono, M., Kimura, H., Ueda, M., Nakamoto, Y., Togashi, K., et al. (2012). (18)F-labeled phenyldiazenyl benzothiazole for in vivo imaging of neurofibrillary tangles in Alzheimer's disease brains. ACS Med. Chem. Lett. 3, 58–62. doi: 10.1021/ml200230e
Matyskiela, M. E., Lu, G., Ito, T., Pagarigan, B., Lu, C. C., Miller, K., et al. (2016). A novel cereblon modulator recruits Gspt1 to the Crl4(Crbn) ubiquitin ligase. Nature 535, 252–257. doi: 10.1038/nature18611
Mayor-Ruiz, C., Jaeger, M. G., Bauer, S., Brand, M., Sin, C., Hanzl, A., et al. (2019). Plasticity of the Cullin-Ring ligase repertoire shapes sensitivity to ligand-induced protein degradation. Mol. Cell 75, 849–858.e8.
Medina, M. (2018). An overview on the clinical development of tau-based therapeutics. Int. J. Mol. Sci. 19:1160. doi: 10.3390/ijms19041160
Meng, F., Xi, Y., Huang, J., and Ayers, P. W. (2021). A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. Sci. Data 8:289. doi: 10.1038/s41597-021-01069-5
Menon, S., Boyer, N. P., Winkle, C. C., Mcclain, L. M., Hanlin, C. C., Pandey, D., et al. (2015). The E3 ubiquitin ligase Trim9 is a Filopodia off switch required for netrin-dependent axon guidance. Dev. Cell 35, 698–712. doi: 10.1016/j.devcel.2015.11.022
Mihalic, J. (2023) First disclosure of Nx-5948, an Oral targeted degrader of Bruton's tyrosine kinase (Btk) for the treatment of B-cell malignancies. American Chemical Society conference - first time disclosures.
Mobitz, H. (2023). Design principles for balancing lipophilicity and permeability in beyond rule of 5 space. ChemMedChem 19:e202300395. doi: 10.1002/cmdc.202300395
Morris, M., Maeda, S., Vossel, K., and Mucke, L. (2011). The many faces of tau. Neuron 70, 410–426. doi: 10.1016/j.neuron.2011.04.009
Nagashima, T., Inamura, K., Nishizono, Y., Suzuki, A., Tanaka, H., Yoshinari, T., et al. (2022). 85 (Pb075) - Asp3082, a first-in-class novel Kras G12D degrader, exhibits remarkable anti-tumor activity in Kras G12D mutated cancer models. Eur. J. Cancer 174:S30. doi: 10.1016/S0959-8049(22)00881-4
Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., et al. (2006). Ubiquitinated Tdp-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. doi: 10.1126/science.1134108
Ng, A., Offensperger, F., Cisneros, J. A., Scholes, N. S., Malik, M., Villanti, L., et al. (2023). Discovery of molecular glue degraders via isogenic morphological profiling. ACS Chem. Biol. 18, 2464–2473. doi: 10.1021/acschembio.3c00598
Nguyen, T. M., Sreekanth, V., Deb, A., Kokkonda, P., Tiwari, P. K., Donovan, K. A., et al. (2024). Proteolysis-targeting chimeras with reduced off-targets. Nat. Chem. 16, 218–228. doi: 10.1038/s41557-023-01379-8
Nowak, R. P., Che, J., Ferrao, S., Kong, N. R., Liu, H., Zerfas, B. L., et al. (2023). Structural rationalization of Gspt1 and Ikzf1 degradation by thalidomide molecular glue derivatives. Rsc Med. Chem. 14, 501–506. doi: 10.1039/D2MD00347C
Oleinikovas, V., Gainza, P., Ryckmans, T., Fasching, B., and Thoma, N. H. (2024). From thalidomide to rational molecular glue Design for Targeted Protein Degradation. Annu. Rev. Pharmacol. Toxicol. 64, 291–312. doi: 10.1146/annurev-pharmtox-022123-104147
Olsen, J. S., Brown, C., Capule, C. C., Rubinshtein, M., Doran, T. M., Srivastava, R. K., et al. (2010). Amyloid-binding small molecules efficiently block Sevi (semen-derived enhancer of virus infection)- and semen-mediated enhancement of Hiv-1 infection. J. Biol. Chem. 285, 35488–35496. doi: 10.1074/jbc.M110.163659
Ono, S., Naylor, M. R., Townsend, C. E., Okumura, C., Okada, O., Lee, H. W., et al. (2021). Cyclosporin A: conformational complexity and Chameleonicity. J. Chem. Inf. Model. 61, 5601–5613. doi: 10.1021/acs.jcim.1c00771
Ottis, P., Toure, M., Cromm, P. M., Ko, E., Gustafson, J. L., and Crews, C. M. (2017). Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem. Biol. 12, 2570–2578. doi: 10.1021/acschembio.7b00485
Owa, T., Yoshino, H., Okauchi, T., Yoshimatsu, K., Ozawa, Y., Sugi, N. H., et al. (1999). Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J. Med. Chem. 42, 3789–3799. doi: 10.1021/jm9902638
Pei, J., Pan, X., Wang, A., Shuai, W., Bu, F., Tang, P., et al. (2021). Developing potent Lc3-targeting Autac tools for protein degradation with selective autophagy. Chem. Commun. (Camb.) 57, 13194–13197. doi: 10.1039/D1CC04661F
Pena-Diaz, S., Garcia-Pardo, J., and Ventura, S. (2023). Development of small molecules targeting alpha-Synuclein aggregation: A promising strategy to treat Parkinson's disease. Pharmaceutics 15:839. doi: 10.3390/pharmaceutics15030839
Pike, A., Williamson, B., Harlfinger, S., Martin, S., and Mcginnity, D. F. (2020). Optimising proteolysis-targeting chimeras (Protacs) for oral drug delivery: a drug metabolism and pharmacokinetics perspective. Drug Discov. Today 25, 1793–1800. doi: 10.1016/j.drudis.2020.07.013
Poongavanam, V., Atilaw, Y., Siegel, S., Giese, A., Lehmann, L., Meibom, D., et al. (2022). Linker-dependent folding rationalizes Protac cell permeability. J. Med. Chem. 65, 13029–13040. doi: 10.1021/acs.jmedchem.2c00877
Poongavanam, V., Doak, B. C., and Kihlberg, J. (2018). Opportunities and guidelines for discovery of orally absorbed drugs in beyond rule of 5 space. Curr. Opin. Chem. Biol. 44, 23–29. doi: 10.1016/j.cbpa.2018.05.010
Powell, C. E., Du, G., Che, J., He, Z., Donovan, K. A., Yue, H., et al. (2020). Selective degradation of Gspt1 by Cereblon modulators identified via a focused combinatorial library. ACS Chem. Biol. 15, 2722–2730. doi: 10.1021/acschembio.0c00520
Pye, C. R., Hewitt, W. M., Schwochert, J., Haddad, T. D., Townsend, C. E., Etienne, L., et al. (2017). Nonclassical size dependence of permeation defines bounds for passive adsorption of large drug molecules. J. Med. Chem. 60, 1665–1672. doi: 10.1021/acs.jmedchem.6b01483
Qu, J., Ren, X., Xue, F., He, Y., Zhang, R., Zheng, Y., et al. (2020). Specific knockdown of alpha-Synuclein by peptide-directed proteasome degradation rescued its associated neurotoxicity. Cell. Chem. Biol. 27, 751–762.e4. doi: 10.1016/j.chembiol.2020.03.010
Riching, K. M., Caine, E. A., Urh, M., and Daniels, D. L. (2022). The importance of cellular degradation kinetics for understanding mechanisms in targeted protein degradation. Chem. Soc. Rev. 51, 6210–6221. doi: 10.1039/D2CS00339B
Rossi Sebastiano, M., Doak, B. C., Backlund, M., Poongavanam, V., Over, B., Ermondi, G., et al. (2018). Impact of dynamically exposed polarity on permeability and solubility of chameleonic drugs beyond the rule of 5. J. Med. Chem. 61, 4189–4202. doi: 10.1021/acs.jmedchem.8b00347
Rui, H., Ashton, K. S., Min, J., Wang, C., and Potts, P. R. (2023). Protein-protein interfaces in molecular glue-induced ternary complexes: classification, characterization, and prediction. Rsc Chem. Biol. 4, 192–215. doi: 10.1039/D2CB00207H
Sasso, J. M., Tenchov, R., Wang, D., Johnson, L. S., Wang, X., and Zhou, Q. A. (2023). Molecular glues: the adhesive connecting targeted protein degradation to the clinic. Biochemistry 62, 601–623. doi: 10.1021/acs.biochem.2c00245
Shultz, M. D. (2019). Two decades under the influence of the rule of five and the changing properties of approved Oral drugs. J. Med. Chem. 62, 1701–1714. doi: 10.1021/acs.jmedchem.8b00686
Sievers, Q. L., Petzold, G., Bunker, R. D., Renneville, A., Slabicki, M., Liddicoat, B. J., et al. (2018). Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through Crbn. Science 362:eaat0572. doi: 10.1126/science.aat0572
Silva, M. C., Ferguson, F. M., Cai, Q., Donovan, K. A., Nandi, G., Patnaik, D., et al. (2019). Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife 8:e45457. doi: 10.7554/eLife.45457
Silva, M. C., Nandi, G., Donovan, K. A., Cai, Q., Berry, B. C., Nowak, R. P., et al. (2022). Discovery and optimization of tau targeted protein degraders enabled by patient induced pluripotent stem cells-derived neuronal models of Tauopathy. Front. Cell. Neurosci. 16:801179. doi: 10.3389/fncel.2022.801179
Smith, R. L., and Mitchell, S. C. (2018). Thalidomide-type teratogenicity: structure-activity relationships for congeners. Toxicol. Res. (Camb) 7, 1036–1047. doi: 10.1039/c8tx000187a
Song, J. H., Wagner, N. D., Yan, J., Li, J., Huang, R. Y., Balog, A. J., et al. (2021). Native mass spectrometry and gas-phase fragmentation provide rapid and in-depth topological characterization of a Protac ternary complex. Cell. Chem. Biol. 28, 1528–1538.e4. doi: 10.1016/j.chembiol.2021.05.005
Spillantini, M. G., Crowther, R. A., Jakes, R., Hasegawa, M., and Goedert, M. (1998). Alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 95, 6469–6473. doi: 10.1073/pnas.95.11.6469
Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R., and Goedert, M. (1997). Alpha-synuclein in Lewy bodies. Nature 388, 839–840. doi: 10.1038/42166
Spradlin, J. N., Hu, X., Ward, C. C., Brittain, S. M., Jones, M. D., Ou, L., et al. (2019). Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 15, 747–755. doi: 10.1038/s41589-019-0304-8
Szewczyk, S. M., Verma, I., Edwards, J. T., Weiss, D. R., and Chekler, E. L. P. (2024). Trends in Neosubstrate degradation by Cereblon-based molecular glues and the development of novel multiparameter optimization scores. J. Med. Chem. 67, 1327–1335. doi: 10.1021/acs.jmedchem.3c01872
Tanji, K., Kamitani, T., Mori, F., Kakita, A., Takahashi, H., and Wakabayashi, K. (2010). Trim9, a novel brain-specific E3 ubiquitin ligase, is repressed in the brain of Parkinson's disease and dementia with Lewy bodies. Neurobiol. Dis. 38, 210–218. doi: 10.1016/j.nbd.2010.01.007
Tao, Y., Remillard, D., Vinogradova, E. V., Yokoyama, M., Banchenko, S., Schwefel, D., et al. (2022). Targeted protein degradation by electrophilic Protacs that Stereoselectively and site-specifically engage Dcaf1. J. Am. Chem. Soc. 144, 18688–18699. doi: 10.1021/jacs.2c08964
Thibaudeau, T. A., Anderson, R. T., and Smith, D. M. (2018). A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 9:1097. doi: 10.1038/s41467-018-03509-0
Thomas, B. A. I., Lewis, H. L., Jones, D. H., and Ward, S. E. (2023). Central nervous system targeted protein degraders. Biomol. Ther. 13:1164. doi: 10.3390/biom13081164
Timms, R. T., Mena, E. L., Leng, Y., Li, M. Z., Tchasovnikarova, I. A., Koren, I., et al. (2023). Defining E3 ligase-substrate relationships through multiplex Crispr screening. Nat. Cell Biol. 25, 1535–1545. doi: 10.1038/s41556-023-01229-2
Tomoshige, S., Nomura, S., Ohgane, K., Hashimoto, Y., and Ishikawa, M. (2017). Discovery of small molecules that induce the degradation of huntingtin. Angew. Chem. Int. Ed. Engl. 56, 11530–11533. doi: 10.1002/anie.201706529
Tomoshige, S., Nomura, S., Ohgane, K., Hashimoto, Y., and Ishikawa, M. (2018). Degradation of huntingtin mediated by a hybrid molecule composed of Iap antagonist linked to phenyldiazenyl benzothiazole derivative. Bioorg. Med. Chem. Lett. 28, 707–710. doi: 10.1016/j.bmcl.2018.01.012
Tong, Y., Zhu, W., Chen, J., Zhang, W., Xu, F., and Pang, J. (2023). Targeted degradation of alpha-Synuclein by autophagosome-anchoring chimera peptides. J. Med. Chem. 66, 12614–12628. doi: 10.1021/acs.jmedchem.3c01303
Toriki, E. S., Papatzimas, J. W., Nishikawa, K., Dovala, D., Frank, A. O., Hesse, M. J., et al. (2023). Rational chemical Design of Molecular Glue Degraders. Acs Cent. Sci. 9, 915–926. doi: 10.1021/acscentsci.2c01317
Troup, R. I., Fallan, C., and Baud, M. G. J. (2020). Current strategies for the design of Protac linkers: a critical review. Explor Target Antitumor. Ther. 1, 273–312. doi: 10.37349/etat.2020.00018
Tseng, Y. L., Lu, P. C., Lee, C. C., He, R. Y., Huang, Y. A., Tseng, Y. C., et al. (2023). Degradation of neurodegenerative disease-associated Tdp-43 aggregates and oligomers via a proteolysis-targeting chimera. J. Biomed. Sci. 30:27. doi: 10.1186/s12929-023-00921-7
Vargesson, N. (2015). Thalidomide-induced teratogenesis: history and mechanisms. Birth Defects Res. C Embryo Today 105, 140–156. doi: 10.1002/bdrc.21096
Veber, D. F., Johnson, S. R., Cheng, H. Y., Smith, B. R., Ward, K. W., and Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623. doi: 10.1021/jm020017n
Villar, E. A., Beglov, D., Chennamadhavuni, S., Porco, J. A. Jr., Kozakov, D., Vajda, S., et al. (2014). How proteins bind macrocycles. Nat. Chem. Biol. 10, 723–731. doi: 10.1038/nchembio.1584
Volak, L. P., Duevel, H. M., Humphreys, S., Nettleton, D., Phipps, C., Pike, A., et al. (2023). Industry perspective on the pharmacokinetic and absorption, distribution, metabolism, and excretion characterization of heterobifunctional protein degraders. Drug Metab. Dispos. 51, 792–803. doi: 10.1124/dmd.122.001154
Wager, T. T., Hou, X., Verhoest, P. R., and Villalobos, A. (2010). Moving beyond rules: the development of a central nervous system multiparameter optimization (Cns Mpo) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 1, 435–449. doi: 10.1021/cn100008c
Wager, T. T., Hou, X., Verhoest, P. R., and Villalobos, A. (2016). Central nervous system multiparameter optimization desirability: application in drug discovery. ACS Chem. Neurosci. 7, 767–775. doi: 10.1021/acschemneuro.6b00029
Wagner, J., Ryazanov, S., Leonov, A., Levin, J., Shi, S., Schmidt, F., et al. (2013). Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson's disease. Acta Neuropathol. 125, 795–813. doi: 10.1007/s00401-013-1114-9
Wang, J. Z., Gao, X., and Wang, Z. H. (2014). The physiology and pathology of microtubule-associated protein tau. Essays Biochem. 56, 111–123. doi: 10.1042/bse0560111
Wang, W., Zhou, Q., Jiang, T., Li, S., Ye, J., Zheng, J., et al. (2021). A novel small-molecule Protac selectively promotes tau clearance to improve cognitive functions in Alzheimer-like models. Theranostics 11, 5279–5295. doi: 10.7150/thno.55680
Watson, E. R., Novick, S., Matyskiela, M. E., Chamberlain, P. P. A. H. D. L. P., Zhu, J., Tran, E., et al. (2022). Molecular glue CelmoD compounds are regulators of cereblon conformation. Science 378, 549–553. doi: 10.1126/science.add7574
Wei, J., Meng, F., Park, K. S., Yim, H., Velez, J., Kumar, P., et al. (2021). Harnessing the E3 ligase Keap1 for targeted protein degradation. J. Am. Chem. Soc. 143, 15073–15083. doi: 10.1021/jacs.1c04841
Wen, T., Chen, J., Zhang, W., and Pang, J. (2023). Design, synthesis and biological evaluation of alpha-Synuclein proteolysis-targeting chimeras. Molecules 28:4458. doi: 10.3390/molecules28114458
Whitehurst, B. C., Bauer, M. R., Edfeldt, F., Gunnarsson, A., Margreitter, C., Rawlins, P. B., et al. (2023). Design and evaluation of a low hydrogen Bond donor count fragment screening set to aid hit generation of Protacs intended for Oral delivery. J. Med. Chem. 66, 7594–7604. doi: 10.1021/acs.jmedchem.3c00493
Whitty, A., Zhong, M., Viarengo, L., Beglov, D., Hall, D. R., and Vajda, S. (2016). Quantifying the chameleonic properties of macrocycles and other high-molecular-weight drugs. Drug Discov. Today 21, 712–717. doi: 10.1016/j.drudis.2016.02.005
Xue, G., Xie, J., Hinterndorfer, M., Cigler, M., Dotsch, L., Imrichova, H., et al. (2023). Discovery of a drug-like, natural product-inspired Dcaf11 ligand Chemotype. Nat. Commun. 14:7908. doi: 10.1038/s41467-023-43657-6
Zengerle, M., Chan, K. H., and Ciulli, A. (2015). Selective small molecule induced degradation of the bet Bromodomain protein Brd4. ACS Chem. Biol. 10, 1770–1777. doi: 10.1021/acschembio.5b00216
Zhang, X., Crowley, V. M., Wucherpfennig, T. G., Dix, M. M., and Cravatt, B. F. (2019). Electrophilic Protacs that degrade nuclear proteins by engaging Dcaf16. Nat. Chem. Biol. 15, 737–746. doi: 10.1038/s41589-019-0279-5
Zhang, X., Luukkonen, L. M., Eissler, C. L., Crowley, V. M., Yamashita, Y., Schafroth, M. A., et al. (2021). Dcaf11 supports targeted protein degradation by electrophilic proteolysis-targeting chimeras. J. Am. Chem. Soc. 143, 5141–5149. doi: 10.1021/jacs.1c00990
Zhang, Z., Sie, B., Chang, A., Leng, Y., Nardone, C., Timms, R. T., et al. (2023). Elucidation of E3 ubiquitin ligase specificity through proteome-wide internal degron mapping. Mol. Cell 83, 3377–3392.e6. doi: 10.1016/j.molcel.2023.08.022
Keywords: CNS degraders, TPD, PROTAC, molecular glue, UPS, E3 ligase
Citation: Kuemper S, Cairns AG, Birchall K, Yao Z and Large JM (2024) Targeted protein degradation in CNS disorders: a promising route to novel therapeutics? Front. Mol. Neurosci. 17:1370509. doi: 10.3389/fnmol.2024.1370509
Edited by:
Robert Kazimierz Lesniak, Alzheimer's Research UK UCL Drug Discovery Institute, United KingdomReviewed by:
John Harling, GlaxoSmithKline, United KingdomCharles Aaron Wartchow, Novartis, United States
Copyright © 2024 Kuemper, Cairns, Birchall, Yao and Large. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra Kuemper, U2FuZHJhLkt1ZW1wZXJAbGlmZWFyYy5vcmc=; Andrew G. Cairns, QW5kcmV3LkNhaXJuc0BsaWZlYXJjLm9yZw==
 Sandra Kuemper
Sandra Kuemper Andrew G. Cairns
Andrew G. Cairns Kristian Birchall
Kristian Birchall Jonathan M. Large
Jonathan M. Large