
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 22 January 2024
Sec. Neuroplasticity and Development
Volume 17 - 2024 | https://doi.org/10.3389/fnmol.2024.1334862
 Jennifer M. Achiro1*
Jennifer M. Achiro1* Yang Tao1
Yang Tao1 Fuying Gao2
Fuying Gao2 Chia-Ho Lin3
Chia-Ho Lin3 Marika Watanabe1
Marika Watanabe1 Sylvia Neumann1
Sylvia Neumann1 Giovanni Coppola2
Giovanni Coppola2 Douglas L. Black3
Douglas L. Black3 Kelsey C. Martin1,2*
Kelsey C. Martin1,2*Aging-related memory impairment and pathological memory disorders such as Alzheimer’s disease differ between males and females, and yet little is known about how aging-related changes in the transcriptome and chromatin environment differ between sexes in the hippocampus. To investigate this question, we compared the chromatin accessibility landscape and gene expression/alternative splicing pattern of young adult and aged mouse hippocampus in both males and females using ATAC-seq and RNA-seq. We detected significant aging-dependent changes in the expression of genes involved in immune response and synaptic function and aging-dependent changes in the alternative splicing of myelin sheath genes. We found significant sex-bias in the expression and alternative splicing of hundreds of genes, including aging-dependent female-biased expression of myelin sheath genes and aging-dependent male-biased expression of genes involved in synaptic function. Aging was associated with increased chromatin accessibility in both male and female hippocampus, especially in repetitive elements, and with an increase in LINE-1 transcription. We detected significant sex-bias in chromatin accessibility in both autosomes and the X chromosome, with male-biased accessibility enriched at promoters and CpG-rich regions. Sex differences in gene expression and chromatin accessibility were amplified with aging, findings that may shed light on sex differences in aging-related and pathological memory loss.
Aging is associated with cognitive decline and memory impairment, including spatial and episodic memory deficits (Nyberg et al., 2012). Research has revealed sex differences in memory performance with aging, as well as sex differences in the incidence of Alzheimer’s disease, a condition associated with memory loss and damage to the hippocampal brain region (Gao et al., 1998; Herlitz and Rehnman, 2008; Jack et al., 2015; McCarrey et al., 2016; Beam et al., 2018; Anstey et al., 2021; Davis et al., 2021). The hippocampus is critical for spatial, episodic, and long-term memory formation (Bird and Burgess, 2008), and therefore, understanding how sex differences impact aging in the hippocampus can provide insights into aging- and sex-dependent declines in memory.
Numerous studies have shown that the abundance of cell types in the brain remains largely constant during aging (Stilling et al., 2014; Ximerakis et al., 2019; Hajdarovic et al., 2022; Allen et al., 2023; Hahn et al., 2023); however, increased immune activation, blood–brain barrier breakdown, altered synaptic function, as well as changes in gene expression, alternative splicing, epigenetic marks, and chromatin accessibility have been associated with aging in the brain (Shankar et al., 1998; Morrison and Baxter, 2012; Stilling et al., 2014; Barrientos et al., 2015; Barter and Foster, 2018; Ximerakis et al., 2019; Zhang et al., 2022; Bieri et al., 2023). Regulated chromatin accessibility is critical to the precise control of gene expression patterns (Su et al., 2017), and hippocampus-dependent long-term memory formation requires new gene expression (Bambah-Mukku et al., 2014; Eagle et al., 2016; Marco et al., 2020). However, few studies have examined sex differences in aging-related changes in gene expression in the hippocampus (Mangold et al., 2017; Hadad et al., 2019), and none, to the best of our knowledge, have examined how sex differences affect aging-related alternative splicing and chromatin accessibility in the hippocampus. Understanding how gene regulatory networks and chromatin dynamics associated with aging differ between sexes in the hippocampus may guide sex-dependent therapies for age-related memory disorders.
Male and female C57BL6/J mice, aged 8 weeks and 78 weeks, were purchased from Jackson Laboratories. The use of mice in this study was approved by the UCLA Institutional Animal Care and Use Committee. Mice were housed in groups under a 12:12 h light/dark cycle with food provided ad libitum until they reached the age of 10 weeks (young adult) and 80 weeks (aged). For sample collection, mice were deeply anesthetized with isoflurane and euthanized by cervical dislocation. The brain was quickly removed and cooled for 1 min in ice-cold PBS before the hippocampus from each hemisphere was dissected on ice and processed immediately. For sequencing experiments, one hippocampus was homogenized in Trizol for RNA preparation, and the other hippocampus was homogenized for nuclei preparation for ATAC-seq (see below). For qPCR and Western blot validation, hippocampal tissue was homogenized in Trizol or RIPA buffer, respectively (see below).
RNA was extracted using a Trizol/RNeasy hybrid protocol. In brief, after phase separation, the aqueous phase was mixed with one volume of 70% ethanol and passed through an RNeasy spin column (QIAGEN RNeasy Mini Kit). RNA quality was determined using the high-sensitivity RNA ScreenTape assay (Agilent 5067-5579), and all samples had RIN scores >7.0. RNA-seq libraries for differential expression or splicing analysis were prepared for four biological replicates/conditions, including four young adult males, four aged males, four young adult females, and four aged females per library type. For differential expression analysis, total RNA libraries were prepared using the TruSeq Stranded Total RNA library prep kit with Ribo-Zero (Illumina 20020596). Paired-end 100 bp sequencing to a depth of ~50 million reads per sample was performed using the Illumina HiSeq 2500 system at the UCLA Broad Stem Cell Research Center Sequencing Core. For splicing analysis, mRNA libraries were prepared with poly-A selection using the TruSeq Stranded mRNA library prep kit (Illumina 20020594), and paired-end 75 bp sequencing to a depth of ~50 million reads per sample was performed using the Illumina HiSeq 2500 system at the UCLA Neuroscience Genomics Core.
ATAC-seq sample preparation was performed for eight biological replicates/conditions according to detailed protocols obtained from Hongjun Song’s lab at the University of Pennsylvania (Buenrostro et al., 2015; Su et al., 2017). In brief, one hippocampus was homogenized with a Dounce tissue grinder (Wheaton 357544) in 2 mL HB buffer (1 mM DTT, 0.15 mM spermine, 0.5 mM spermidine, protease inhibitor (Sigma-Aldrich 04693159001), 0.3% IGEPAL-630, 0.25 M sucrose, 25 mM MgCl2, 20 mM Tricine-KOH). The homogenate was filtered through a 40-μm strainer and centrifuged over one volume of cushion buffer (0.5 mM MgCl2, 0.5 mM DTT, protease inhibitor, 0.88 M sucrose) at 2,800g for 10 min in a swinging bucket centrifuge at 4°C. The nuclei pellet was resuspended in 20 μL PBS. Then, 1 μL of the nuclei sample was stained with Hoechst 33342 to calculate nuclei concentration, and 50,000 nuclei per sample were used for downstream library preparation. Libraries were prepared using the Nextera DNA library prep kit (Illumina FC-121-1030), and quality was analyzed using the D1000 ScreenTape Assay (Agilent 5067-5582). Paired-end 50 bp sequencing to a depth of ~40 million reads per sample was performed using the Illumina HiSeq 2500 system at the UCLA Broad Stem Cell Research Center Sequencing Core.
RNA-seq reads were mapped to the mouse genome (mm10) using STAR with the two-pass option (Dobin et al., 2013). Only uniquely mapped reads were used for downstream analysis. Lowly expressed genes were removed by retaining only genes with counts per million >0.1 in at least four samples (17,821 genes). Read counts were normalized via the trimmed mean method before differential expression analysis (DEA) (Robinson and Oshlack, 2010). For DEA, the analysis package edgeR (Robinson et al., 2010) was used, with an FDR <0.05 cutoff. For aging-dependent expression differences, data from female and male samples were analyzed separately. For differential splicing analysis, the analysis package rMATS version 4.0.2 (Shen et al., 2014) was used. Alternative splicing events were included for analysis if events had FDR-adjusted p-value <0.05, total reads ≥50, and skipping/junction reads >20. For gene ontology enrichment analysis, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool was used (Huang et al., 2009; Huang da et al., 2009). RNA-sequencing data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE244506.1
Eight replicates per condition were sequenced for ATAC-seq; however, one young male sample had less than one million reads and was not used in further analyses. Adapter content was trimmed using Scythe,2 and ATAC-seq reads were mapped to the mouse genome (mm10) using Bowtie 2 (Langmead and Salzberg, 2012). Reads in blacklist regions (Dunham et al., 2012; Amemiya et al., 2019) and reads mapped to mitochondria were removed. Peaks were called on each individual sample using MACS3 (with parameter settings—nomodel-f BAMPE and FDR cutoff of 0.05) (Zhang et al., 2008). For each sample, read pairs (fragments) were counted for each peak using featureCount (Liao et al., 2014) (with parameter settings—p—countReadPairs–M—fraction). For quality control, we calculated a transcription start site (TSS) enrichment score using ATACseqQC (Ou et al., 2018) and a FRiP score (fraction of reads/fragments in peaks). One young male and one aged male sample had a TSS enrichment score < 10 and/or a FRiP score < 0.2; therefore, these samples were excluded from further analyses. The remaining samples had an average TSS enrichment score of 20.2 ± 1.1 (mean ± s.e.m.) and an average FRiP score of 0.46 ± 0.03 (mean ± s.e.m.). For each condition, narrow peak sets were merged, and peaks were included in consensus peak sets for each condition if there was at least a 50% overlap with a peak in at least two replicates (consensus sets: 65,088 peaks in young male, 71,719 in aged male, 61,264 in young female, and 68,655 in aged female). A total consensus peak set (92,233 peaks) was generated by merging the consensus peak sets from each condition. Fragments were counted for each total consensus peak in each sample using featureCount as described above, and differential analysis was performed using DESeq2, including sequencing batch as a variable (Love et al., 2014). Batch correction for principal components analysis was performed using limma (Ritchie et al., 2015). Peak annotation, peak histograms, and motif searches were done using HOMER (Heinz et al., 2010). Browser track examples were generated from the UCSC genome browser3 (Kent et al., 2002). For the ATAC-seq peak profile heat map, normalized peak histograms for each peak were generated in HOMER for each sample using 10 bp bins. Then, for each peak, an average ATAC histogram was calculated for each condition and ranked by young male ATAC signal intensity ± 100 bp around the peak center. For ATAC-seq profile plots, normalized peak histograms for each sample were generated in HOMER using 10 bp bins, producing one histogram per sample based on the number of peaks (or gene TSSs) indicated in figures, and then averages and standard error of the means were calculated based on the sample histograms (with n = number of samples, i.e., 6 young male, 7 aged male, 8 young female, and 8 aged female). Genomic locations of full-length intact long interspersed element-1 (LINE-1) regions were obtained using L1Base 2 (Penzkofer et al., 2017). Where indicated in the results, we used smooth-quantile with GC-content normalization (QSmooth-GC) using qsmooth (Hicks et al., 2017; Van den Berge et al., 2022). This study used computational and storage services associated with the Hoffman2 Shared Cluster provided by UCLA Institute for Digital Research and Education’s Research Technology Group. ATAC-sequencing data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE244506.4
Samples for qPCR validation were prepared from separate animals from those used for library preparation. Then, 50 ng of total RNA was reverse transcribed into cDNA using SuperScript III First-Strand Synthesis System (Invitrogen 18080051) with random hexamer primers for four biological replicates of each sex and age group. Technical triplicates were prepared for each primer set using SYBR Green PCR Master Mix (Applied Biosystems 4309155). The following primers were used:
*Gapdh (F: AGGTCGGTGTGAACGGATTTG; R: TGTAGACCATGTAGTTGAGGTCA),
*Tubb3 (F: TAGACCCCAGCGGCAACTAT; R: GTTCCAGGTTCCAAGTCCACC),
*Gfap (F: CCCTGGCTCGTGTGGATTT; R: GACCGATACCACTCCTCTGTC),
*Pcdhb9 (F: ACTGCTCTTGAGAATACCAGAGA; R: AGGACGTGAAAATAAGGGTTGG),
Gpr17 (F: TCACAGCTTACCTGCTTCCC; R: CCGTTCATCTTGTGGCTCTTG),
*Ptpro (F: AACATCCTGCCGTATGACTTTAG; R: GGGACTTCTGTTGTAGGACCATC),
*Npnt (F: GGACAGGTCCGATGTCAGTG; R: CTTCCAGTCGCACATTCATCA),
*Mag (F: CTGCCGCTGTTTTGGATAATGA; R: CATCGGGGAAGTCGAAACGG),
*Mbp (F: GACCATCCAAGAAGACCCCAC; R: GCCATAATGGGTAGTTCTCGTGT),
LINE-1 5’UTR (F: TGAGTGGAACACAACTTCTGC; R: CAGGCAAGCTCTCTTCTTGC),
LINE-1 ORF1 (F: ATGGCGAAAGGCAAACGTAAG; R: ATTTTCGGTTGTGTTGGGGTG),
LINE-1 5’UTR:ORF1 (F: CTGCCTTGCAAGAAGAGAGC; R: AGTGCTGCGTTCTGATGATG).
Gene primers from PrimerBank5 are indicated with a *. Primers for LINE-1 were obtained from previous publications (De Cecco et al., 2013; Van Meter et al., 2014). The rest of the gene primers were custom-designed to span exon-exon junctions. The samples were run on the Bio-Rad CFX Connect real-time PCR detection system. We used the delta–delta Ct method for calculating fold gene expression, with delta Cts calculated relative to Gapdh (for RNA-seq validation) or Tubb3 (for LINE-1) and delta–delta Cts calculated relative to young male samples. Results were tested for normality using the Shapiro–Wilk test. Unpaired t-tests/one-way ANOVAs with Sidak’s multiple comparisons test or Mann–Whitney/Kruskal–Wallis tests with Dunn’s multiple comparison tests were used for normally and non-normally distributed data, respectively.
Western blot samples were prepared from four biological replicates. For each replicate, one hippocampus was homogenized in a RIPA buffer with protease and phosphatase inhibitors and centrifuged at 10,000g for 10 min at 4°C to remove cell debris. Protein concentration was determined using the Pierce BCA protein assay kit (Thermo Fisher 23225). All protein samples were diluted to a protein concentration of 0.5 mg/mL. Then, 4X sample loading buffer (0.2 M Tris–HCl pH 6.5, 4.3 M glycerol, 8.0% (w/v) SDS, 6 mM bromophenol blue, and 0.4 M DTT) was added, and the samples were boiled at 95–100°C for 10 min. The samples were then aliquoted and stored at −20°C. Samples were loaded on NuPAGE 4–12% Bis-Tris protein gels (Invitrogen NP0335BOX) and transferred onto 0.2 μm nitrocellulose membranes. After transfer, blots were blocked with Odyssey TBS blocking buffer (LI-COR 927-50000) for 1 h at room temperature before being incubated in the primary antibody at 4°C overnight. The following primary antibodies were used: anti-MBP (Abcam, ab218011) and anti-TUJ1 (Biolegend, 801201). Blots were incubated in secondary antibodies for 2 h at room temperature. Blots were imaged using the LI-COR Odyssey imaging system and quantified using LI-COR Image Studio software.
To investigate sex differences in aging-related changes in the transcriptome (including alternative splicing) and chromatin accessibility in the hippocampus, we performed RNA-sequencing (RNA-seq) and Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) (Buenrostro et al., 2013, 2015) on the hippocampus of young adult (10 weeks) or aged (80 weeks) mice of both sexes (Figure 1A). Principle component analysis of the RNA-seq data showed that samples were well-separated by sex and age by the first and second principal components, respectively (Figure 1B). Overall, 905 genes were differentially expressed (DE) either by sex or aging, with the largest number of differences occurring with aging irrespective of sex (Figure 1C; Supplementary Table S1). In the hippocampus of females, we found 446 genes that were upregulated and 261 genes downregulated with aging. Significantly fewer aging-associated changes were detected in the hippocampus of males compared to females (chi-square test, p < 0.001, X2 = 108.1), with 206 and 164 genes being upregulated and downregulated in males, respectively, with aging (Figures 1D,E). These results indicate that aging is associated with widespread changes in gene expression, with a larger number of genes undergoing aging-related changes in expression in the hippocampus of females than males.
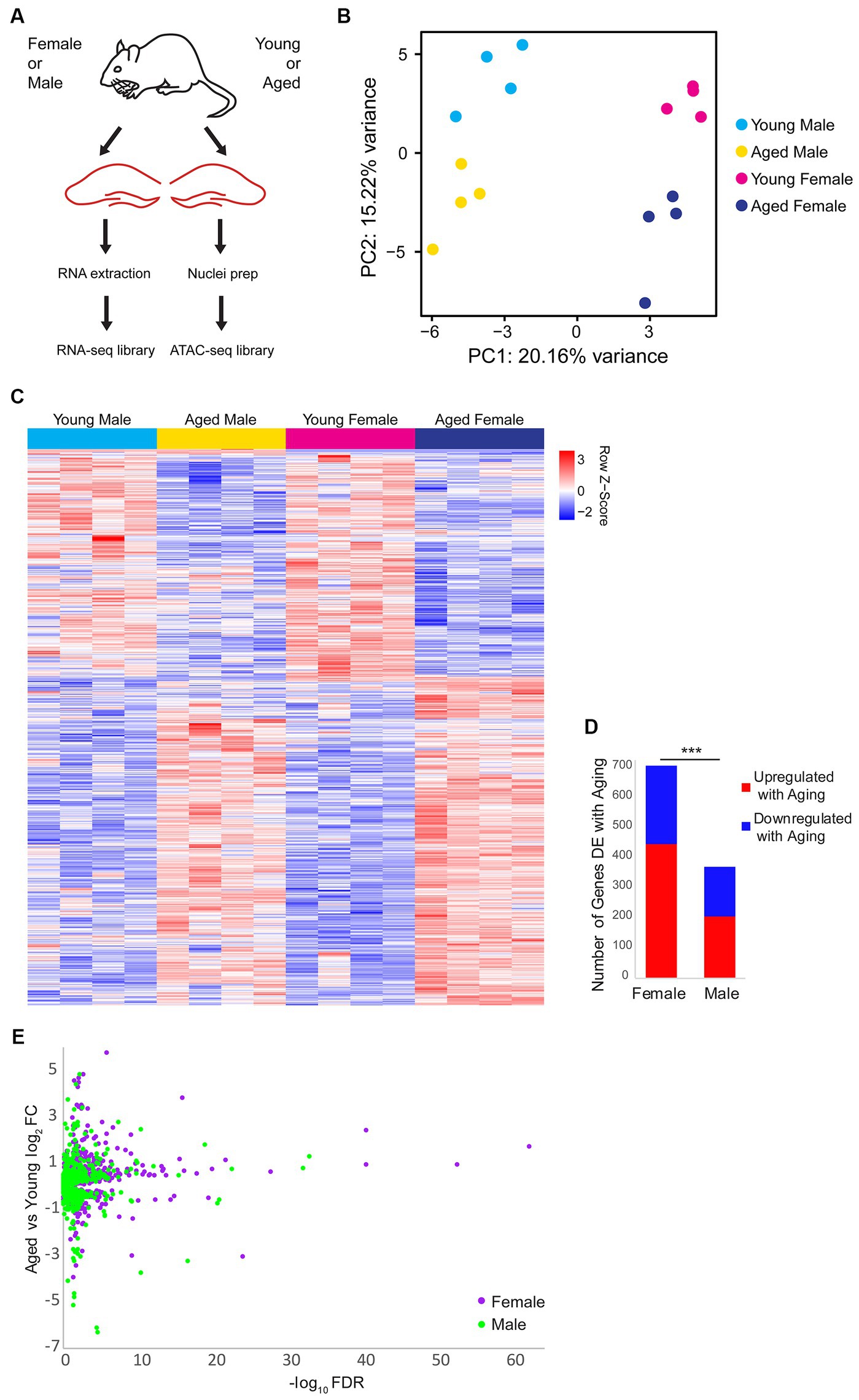
Figure 1. Gene expression changes with aging and sex bias in female and male mouse hippocampus. (A) Illustration of the experimental design. (B) Principle component analysis (PCA) of RNA-seq gene expression data. N = four biological replicates for each group (young adult female, aged female, young adult male, and aged male). (C) Expression heat map of 905 sex-biased or aging-related differentially expressed (DE) genes (FDR < 0.05). Each row represents a gene and each column is a biological replicate. Red row z-scores indicate high relative expression and blue indicates low relative expression (see Supplementary Table S1 for all FPKM and log2FC values). (D) Number of genes that are DE between aged and young animals in females and males (FDR < 0.05). *** indicates p < 0.001 for the chi-square test. (E) Volcano plot of aging-related DE genes in males (green) and females (purple).
First, we focused on sex-independent gene expression changes with aging. We found that 136 genes were upregulated and 64 genes were downregulated with aging in both sexes (Figure 2A). We noted that although these genes were DE with aging in both sexes, for many genes, the aging-related fold change in female hippocampus was larger than in male hippocampus (Figure 2B). We ranked DE genes by lowest FDR in either males or females and found that many of the aging-dependent genes encoded immune response genes, protocadherins, extracellular matrix proteins, and neurodevelopment-related proteins (Figure 2C). Gene ontology analysis revealed that genes upregulated with aging showed enrichment for cell adhesion (GO:0007155, FDR = 5.92E-08) and innate immune response (GO:0045087, FDR = 0.02), whereas genes downregulated with aging showed enrichment for nervous system development (GO:0007399, FDR = 5.38E-04) and neuron migration (GO:0001764, FDR = 0.002; Figure 2D). To validate these findings, we performed qPCR using a separate cohort of animals for a number of aging-dependent genes (Figure 2E). We also analyzed gene expression changes from an existing dataset in a mouse model of Alzheimer’s disease (AD) (Forner et al., 2021). This analysis showed that, similar to our finding with aging, more genes showed AD-related changes in the female hippocampus compared to the male AD hippocampus (Supplementary Figure S1A). Furthermore, we found that ~30% of the aging-related genes we identified also showed AD-related differential expression in the hippocampus, including immune response genes that showed AD-related changes in both young adult and aged AD mice (Supplementary Figure S1B). These results are consistent with previous reports showing similar aging-related changes in the brain, including increases in immune-response genes and decreases in neurodevelopment-related genes (Walter et al., 2011; Uddin and Singh, 2013; Stilling et al., 2014; Barrientos et al., 2015; Carlock et al., 2017; Lanke et al., 2018; Ximerakis et al., 2019; Li M. et al., 2020; Rivera et al., 2021; Hahn et al., 2023).
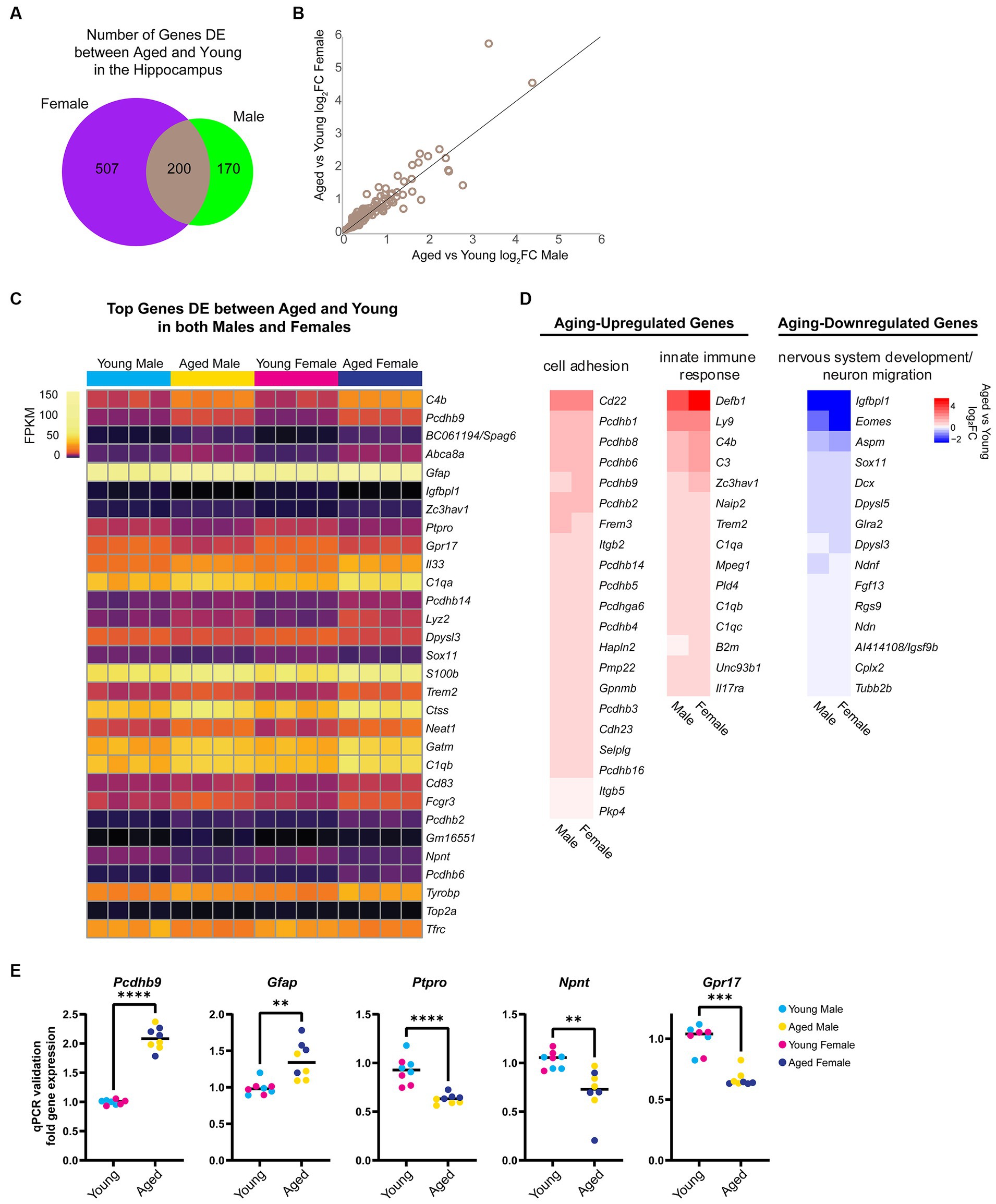
Figure 2. Aging is associated with changes in the expression of cell adhesion, immune response, and nervous system development genes. (A) Venn diagram of number of genes DE (FDR < 0.05) with aging in female and male mouse hippocampus. (B) Scatter plot of aged vs. young adult log2FC in males and females for the 200 genes that showed aging-related differential expression in the hippocampus of both sexes. (C) Expression heat map of top 30 genes (sorted by lowest FDR) DE in both male and female hippocampus with aging. Each row represents a gene and each column is a biological replicate. FPKM for each gene/replicate is represented by color (see Supplementary Table S1 for all FPKM and log2FC values). (D) Fold change heat maps of genes in biological process categories from gene ontology analysis that were upregulated (red) and downregulated (blue) in the hippocampus with aging. (E) qPCR validation for genes Pcdhb9, Gfap, Ptpro, Npnt, and Gpr17 in young and aged male and female hippocampus (n = four each sex/age). Pcdhb9 unpaired t-test p < 0.0001; Gfap unpaired t-test p = 0.001; Ptpro unpaired t-test p < 0.0001; Npnt unpaired t-test p = 0.003; Gpr17 Mann–Whitney test p < 0.001. ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001.
To determine whether and how sex impacts aging-dependent changes in gene expression, we compared gene expression profiles between the hippocampus of young adult males and females and between aged males and females. In the hippocampus of young animals, we identified 17 sex-biased genes. Among these, neuropeptide Y (Npy), a neuropeptide whose expression is correlated with female-biased stress-related memory disorders (Nahvi and Sabban, 2020), showed a 1.3-fold higher expression in the hippocampus of young females compared to young males, but a female-specific downregulation with aging (young female vs. male log2FC = 0.41, FDR < 0.001; average FPKM: young male = 64.34, aged male = 67.38, young female = 88.11, aged female = 71.34). In the hippocampus of aged animals, significantly more genes (42) were DE between sexes (chi-square test, p = 0.002, X2 = 9.8; Figure 3A). Most of the sex-biased genes in the hippocampus of young animals also showed sex bias in the hippocampus of aged animals (Figure 3B). Of the genes with sex-biased expression in both young and aged, all were on sex chromosomes except for Prl (prolactin) and Cplx2 (complexin 2). Complexin 2, a protein involved in synaptic vesicle fusion and whose dysregulated expression is associated with a number of cognitive disorders (Brose, 2008; Hass et al., 2015), was more highly expressed in males compared to females in both age groups, and this gene was also significantly downregulated with aging in both sexes (female vs. male: young log2FC = −0.16, aged log2FC = −0.21; aged vs. young: male log2FC = −0.16, female = −0.21). Of the sex-biased genes located on the X chromosome, all are known to escape X-inactivation in the mouse brain (Deng et al., 2014). Interestingly, we also found that Xist, a female-specific non-coding RNA critical for X-inactivation (Pandya-Jones and Plath, 2016; Galupa and Heard, 2018), was significantly upregulated in the female hippocampus with aging (log2FC = 0.21, FDR = 0.009; average FPKM: young male = 0.06, aged male = 0.04, young female = 27.75, aged female = 32.08). This finding is consistent with a recent report showing increased Xist expression with aging in the mouse hypothalamus (Hajdarovic et al., 2022). Altogether, these results suggest that aging results in more divergence in the hippocampal transcriptome between males and females.
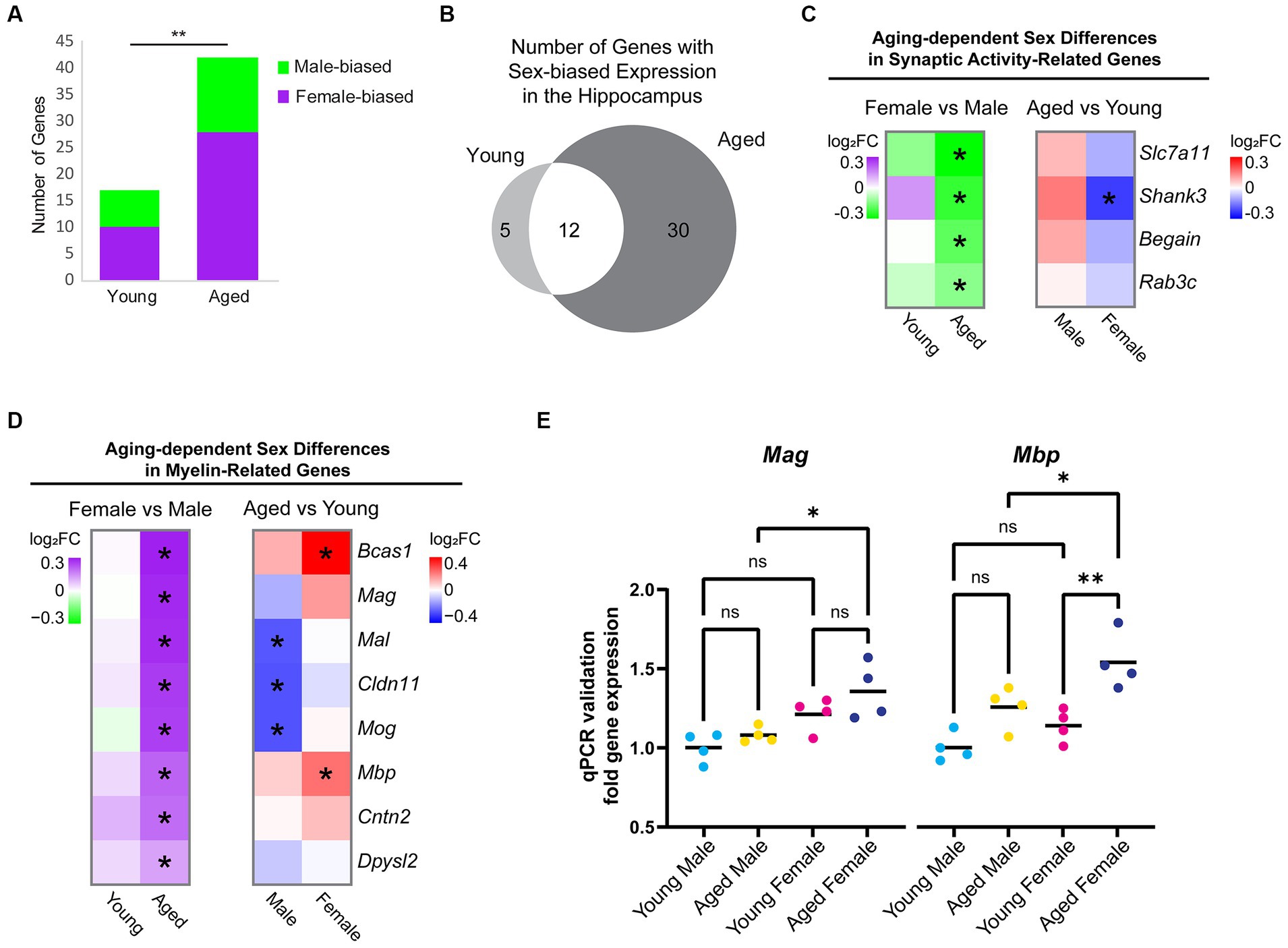
Figure 3. Sex differences in expression of hippocampal genes during aging. (A) Number of genes that were DE between female and male hippocampus (FDR < 0.05) in young adult and aged animals. Purple indicates higher expression in female and green indicates higher expression in male. ** indicates p < 0.01 by chi-square test. (B) Venn diagram of sex-biased DE genes in the hippocampus. (C) Fold change heat maps of synaptic activity-related genes showing higher expression in aged male vs. female hippocampus. Left heat map shows sex-biased differential expression (purple indicates higher in female and green indicates higher in male). Right heat map shows aging-related differential expression (red indicates upregulation with aging and blue indicates downregulation with aging).* indicates significant differential expression (FDR < 0.05). (D) Fold change heat maps [as in (C)] of myelin-related genes showing sex-biased expression. (E) qPCR validation for myelin sheath genes Mag and Mbp in young adult and aged male and female hippocampus (n = four for each condition). Mag aged male vs. aged female ANOVA with Sidak’s multiple comparison test adjusted p = 0.022. Mbp aged male vs. aged female ANOVA with Sidak’s multiple comparison test adjusted p = 0.039; young female vs. aged female adjusted p = 0.004. *indicates p < 0.05, ** indicates p < 0.01, ns = not significant.
To further explore sex differences in gene expression that arise with aging, we focused on the 30 genes with sex-biased expression only in aged animals. Eight genes were male-biased (Slc7a11/xCT, Begain, Rab3c, Shank3, Hist1h4d, D830046C22Rik, Ncl, and Scarf2), the first four of which encode proteins involved in synaptic transmission (Figure 3C) (Deguchi et al., 1998; Schlüter et al., 2004; Kouser et al., 2013; Massie et al., 2015). We found that 22 genes showed female-biased expression in aged hippocampus, including the myelination-related gene, Bcas1, which showed an aging-related increase in expression in female but not male hippocampus, resulting in significantly higher expression in aged females than aged males (log2FC = 0.36, FDR < 0.001; average FPKM: young male = 33.4; aged male = 37.3, young female = 33.8, aged female = 47.9).
Gene ontology analysis of female-biased genes in aged hippocampus revealed a significant enrichment of the term myelin sheath (GO:0043209; FDR < 0.001). We then assessed all genes associated with the GO terms “myelin sheath” and “myelination” (GO:0043209 and GO:0042552) and found that 8 genes (Bcas1, Mag, Mal, Mbp, Cldn11, Cntn2, Mog, and Dpysl2) had sex-biased expression in the hippocampus of aged animals, all of which showed a female bias (Figure 3D). In contrast, other oligodendrocyte-related genes such as Olig1, Olig2, Opalin, Cnp, Plp1, Myrf, Gpr17, and Mobp were not DE between aged female and aged male hippocampus, suggesting that this was not due to a difference in the number of oligodendrocytes or oligodendrocyte precursor cells (OPCs), as has been previously reported (Hahn et al., 2023). Furthermore, we found no sex bias in the expression of myelination-related genes (GO:0043209 and GO:0042552) in young adult animals. The sex bias in myelin genes in aged hippocampus resulted from aging-related changes in both males and females: aging resulted in a significant downregulation of Mog, Cldn11, and Mal in the hippocampus of males but not females, whereas aging resulted in significant upregulation in Bcas1 and Mbp in the hippocampus of females but not males (Figure 3D). We also performed qPCR for two of the myelin sheath genes, Mag and Mbp, using samples from a separate cohort of animals and verified that the expression of both had female-biased expression in aged hippocampus (Figure 3E). This sex-biased expression of myelin sheath genes suggests that the hippocampus of aged females may have less myelin degeneration or more remyelination than aged males.
Alternative splicing patterns have been reported to be altered with aging in the mouse hippocampus and show sex-specific differences in the human brain (Trabzuni et al., 2013; Stilling et al., 2014; Bhadra et al., 2020). To examine sex bias and aging-related changes in alternative splicing, we generated a poly-A selected RNA-library from the hippocampus of young adult and aged male and female mice and queried five types of alternative splicing (AS) events: skipped exon (SE), alternative 5′ splice site (A5SS), alternative 3’splice site (A3SS), mutually exclusive exons (MXE), and retained introns (RI). We identified 591 significant AS events in 452 genes that showed sex bias or aging-related changes (FDR < 0.05, Supplementary Table S2). Aging in the male hippocampus resulted in 171 AS events in 154 genes, significantly more events than that occurred with aging in the female hippocampus (Figure 4A; 125 AS events in 124 genes; chi-square test, X2 = 7.2, p = 0.007).
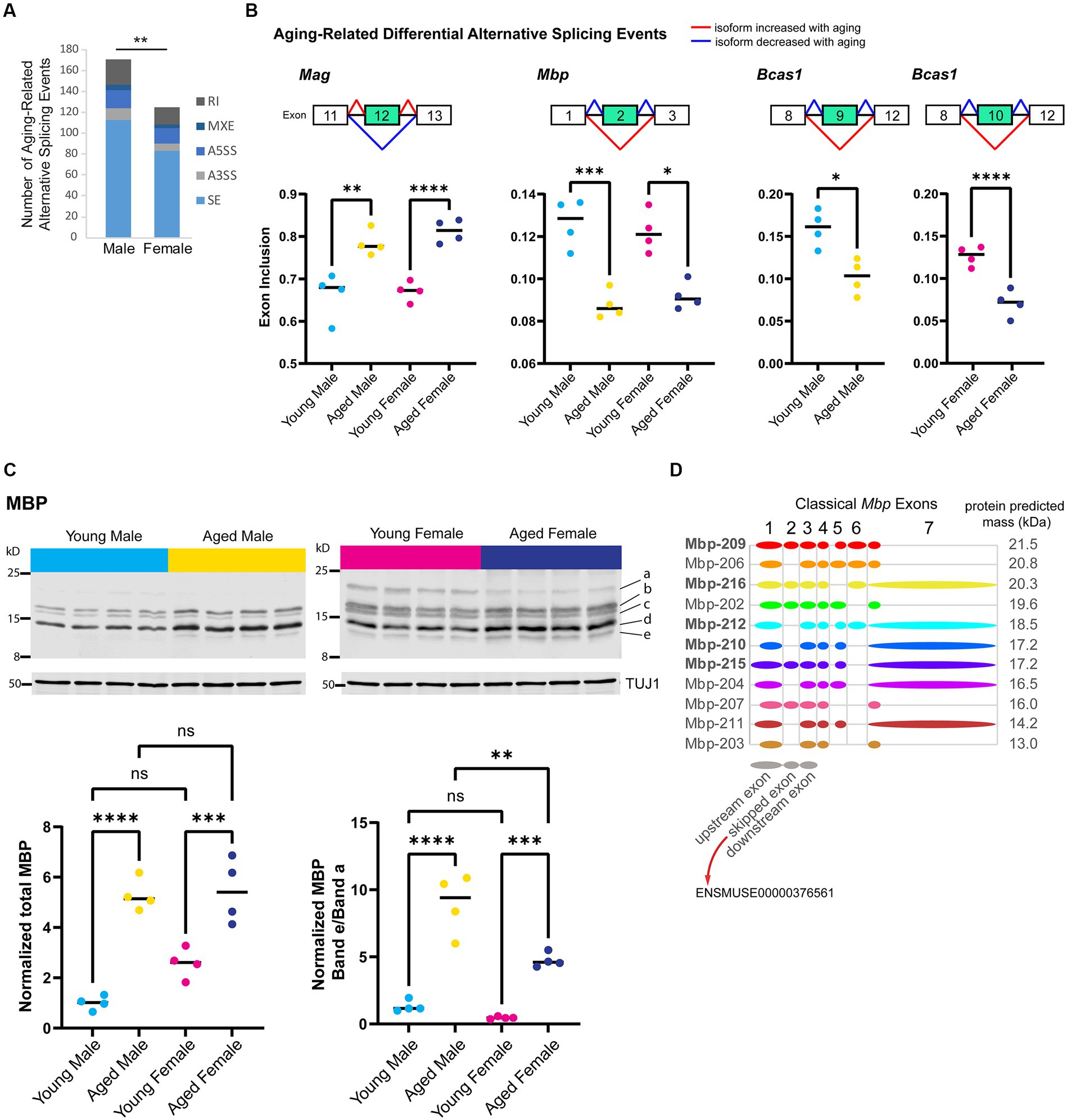
Figure 4. Aging is associated with alternative splicing of myelin sheath-related genes. (A) Number of each type of aging-related AS event in male and female hippocampus. SE = skipped exon, A5SS = alternative 5′ splice site, A3SS = alternative 3’splice site, MXE = mutually exclusive exons, RI = retained intron. (B) Genes that exhibited aging-related alternative splicing events in both male and female hippocampus. Under each gene name is a diagram showing the skipped and flanking exons involved in each alternative splicing event. Below the diagram, plots show exon inclusion values and statistical significance calculated from rMATS (Shen et al., 2014). Mag aging-related inclusion of exon 12 (ENSMUSE00000373997) in males rMATS FDR = 0.001 and females FDR < 0.0001. Mbp aging-related skipping of exon 2 (ENSMUSE00000376561) in males FDR < 0.001 and females FDR = 0.016. Bcas1 aging-related skipping of exons 9–11 (ENSMUSE00000170506, ENSMUSE00000170504, ENSMUSE00001234083) in males FDR = 0.029 and female FDR < 0.0001 (also see Supplementary Figures S2A–C). (C) Western blot analysis for MBP protein isoforms in young adult and aged hippocampus. Top row shows Western blot images for MBP protein and loading control TUJ1 (below) for young adult and aged male and female hippocampus (4 biological replicates each). Letters on the right side indicate the bands that were quantified. Bottom panels show the quantification of total MBP (left plot) and the ratio of the lowest-migrating MBP isoform (e) to the highest-migrating MBP isoform (a). See Supplementary Figures S2D,E for quantification of each band. Total MBP young male vs. aged male ANOVA with Sidak’s multiple comparison test adjusted p < 0.0001; young female vs. aged female adjusted p = 0.001. MBP band e/a young male vs. aged male ANOVA with Sidak’s multiple comparison test adjusted p < 0.0001; young female vs. aged female adjusted p = 0.001; aged male vs. aged female adjusted p = 0.001. (D) Classical (not including Golli) Mbp protein-coding splice variants from Ensembl (Cunningham et al., 2021) release 102 with a diagram of included exons. Diagrams of upstream, skipped, and downstream exons are shown in gray below splice variant diagrams. Bolded transcript names indicate Ensembl’s designation as stable, reviewed, and high-quality transcript annotations. The predicted protein mass from Ensembl for the product of each transcript is indicated to the right of the diagram. ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001, ns = not significant.
First, we focused on sex-independent AS in the hippocampus with aging and identified seven genes (Mag, Bcas1, Mbp, 4732471J01Rik, Huwe1, Ctxn3, and Mthfsl) with skipped exon events that showed the same aging-related AS event in both males and females. The top three most significant aging-related AS events occurred in myelin-related genes: (1) inclusion of exon 12 (ENSMUSE00000373997) in Mag, (2) skipping of exon 2 (ENSMUSE00000376561) in Mbp, and (3) skipping of exons 9–11 (ENSMUSE00000170506, ENSMUSE00000170504, ENSMUSE00001234083) in Bcas1 (Figure 4B; Supplementary Figures S2A–C). As mentioned previously, the overall transcript abundance of myelin sheath genes Mag, Mbp, and Basc1 were female-biased in the aged hippocampus (Figure 3D); however, the proportions of exon inclusion for these genes were similar between the sexes. The aging-related skipping of exon 2 in Mbp was also supported at the protein level: aged animals of both sexes exhibited an overall increase in MBP protein compared with young animals but specifically showed a loss of a higher-migrating MBP species and a gain of a low-migrating species that correspond to Mbp splice variants with the inclusion or skipping of exon 2, respectively (Figures 4C,D; Supplementary Figures S2D,E). These aging-related changes in Mag and Mbp exon skipping are similar to those that have been reported to occur throughout development and aging (Tropak et al., 1988; Fujita et al., 1998; Woodruff and Franklin, 1998; Li et al., 2000; Sugiyama et al., 2002). In addition to Mag and Mbp, Bcas1 is also known to be involved in myelination (Darbelli et al., 2017; Ishimoto et al., 2017); therefore, aging results in changes in the expression of different isoforms of myelin-related genes.
To determine whether and how sex impacts AS, we compared AS profiles of male and female young adult and aged hippocampus. We found 156 sex-biased AS events in 144 genes in young adult hippocampus and 139 sex-biased AS events in 129 genes in aged hippocampus (Figure 5A), including 9 genes with sex-biased AS in both young and aged (Ankrd13c, Ccdc62, Epn2, Lrrc14, Lrrcc1, Mapk12, Pofut2, Rtel1, and Tle1). We ranked the sex-biased AS events by lowest FDR-adjusted p-value in either the young or aged hippocampus and found that within the 30 most significant autosomal AS events (Figure 5B), many occurred in genes encoding nuclear proteins such as the polycomb repressive complex-2 protein EZH2, as well as other genes encoding DNA-binding proteins such as Hmga1, Rorc, Ccdc62, and Zmym5. Furthermore, we detected an A5SS event that showed sex-biased AS in Miat/Gomafu, a lncRNA that regulates splicing and represses transcription through association with the polycomb repressive complex (Barry et al., 2014; Zakutansky and Feng, 2022). One of the top 10 most significant AS events in young hippocampus was an A3SS splicing event in Ift88, a transport protein important for primary cilia maintenance in neurons (Tereshko et al., 2021). Indeed, in young but not aged hippocampus, we found sex-biased AS in a number of genes associated with primary cilia, including Bbs4, Fuz, C2cd3, Rab34, Fam149b, and Cdk10.
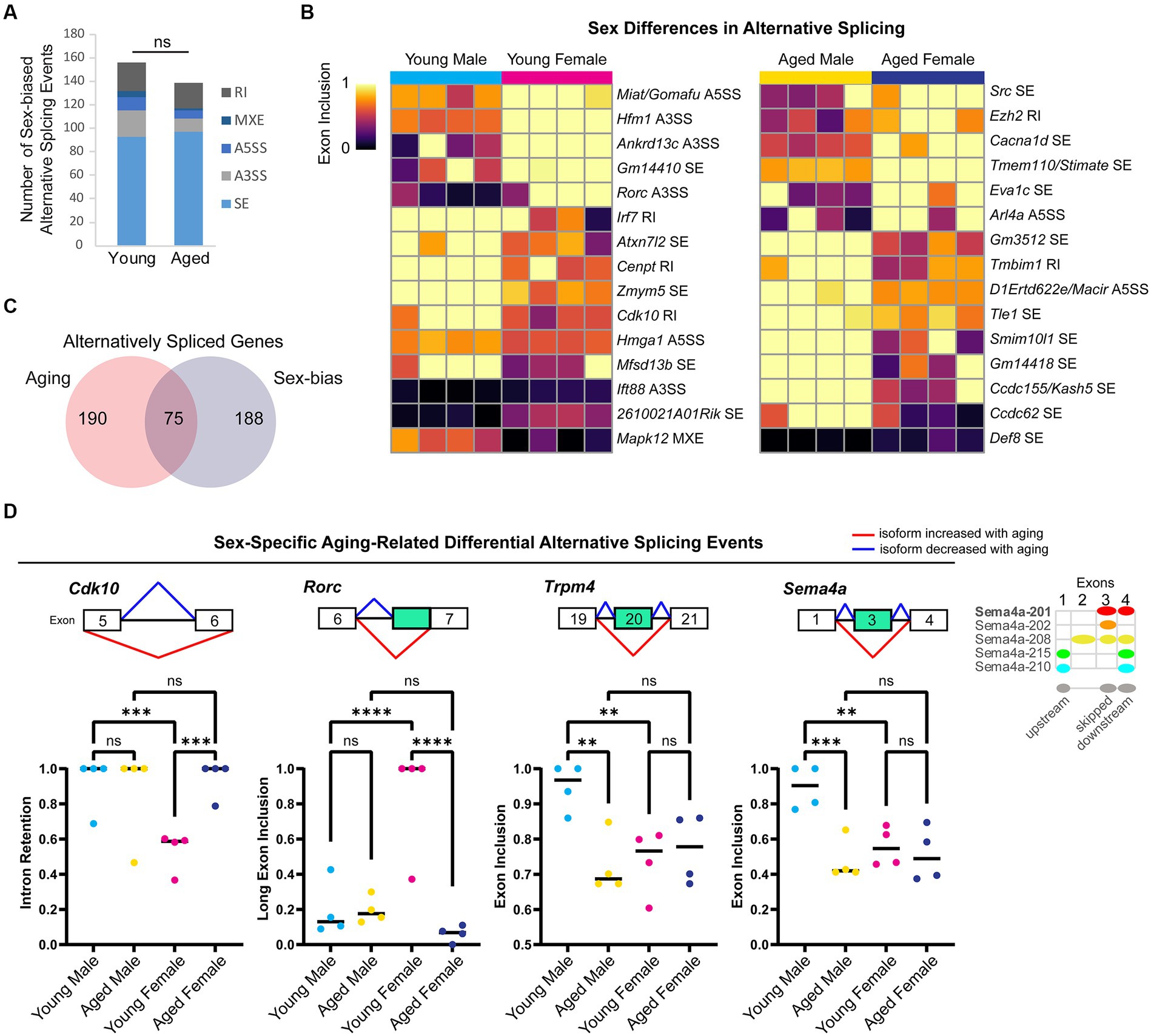
Figure 5. Sex differences in alternative splicing in the hippocampus. (A) Number of each type of sex-biased AS event in young adult and aged hippocampus. SE = skipped exon, A5SS = alternative 5′ splice site, A3SS = alternative 3’splice site, MXE = mutually exclusive exons, RI = retained intron. ns = not significant. (B) Exon inclusion heat map of top most significant sex-biased AS events in young adult or aged hippocampus (by rMATS FDR). Each column represents a biological replicate, and each row represents an AS event labeled on the right. (C) Venn diagram showing the number of genes with aging-related or sex-biased AS events. (D) Example genes with sex-specific aging-related alternative splicing events in the hippocampus. Under each gene name is a diagram showing the skipped and flanking exons involved in each alternative splicing event. Below the diagram, plots show exon inclusion values and statistical significance calculated from rMATS. Cdk10 female bias in exclusion of intron between exons 5 (ENSMUSE00000292260) and 6 (ENSMUSE00000292228) in young hippocampus (FDR < 0.001) and aging-related intron retention in females (FDR < 0.001). Rorc female bias in long exon inclusion of exon 7 (ENSMUSE00000566508) in young hippocampus (FDR < 0.0001) and aging-related short exon usage in females (FDR < 0.0001). Trpm4 male bias in exon inclusion of exon 20 (ENSMUSE00000594904) in young hippocampus (FDR < 0.01) and aging-related exon skipping in males (FDR < 0.01). Sema4a male bias in exon inclusion of exon 3 (ENSMUSE00000832345) in the young hippocampus (FDR < 0.01) and aging-related exon skipping in males (FDR < 0.001). Diagram on the right shows Ensembl protein-coding transcripts for Sema4a with the skipped exon corresponding to the second exon of Sema4a-208. ** indicates p < 0.01, *** indicates p < 0.001, **** indicates p < 0.0001, ns = not significant.
For genes on the X chromosome, we found sex bias in 10 splicing events in nine genes. For example, in the aged hippocampus, the X-chromosome gene Huwe1, encoding an E3 ubiquitin ligase linked to intellectual disability (Moortgat et al., 2018), was associated with an SE event that showed both a change with aging in females and a sex bias in the aged hippocampus. Overall, we found ~30% of genes with sex bias in AS also showed a change in splicing with aging (Figure 5C). Sex-specific aging-related alternative splicing events occurred in many of the genes mentioned above (e.g., Ezh2, Rorc, Ift88, Fuz, and Cdk10) and included isoform ratios that were specific to either young male or young female hippocampus (Figure 5D). Altogether, these results suggest that AS may contribute more to sex differences in the transcriptome than expression and that aging-related changes in AS are often sex-specific.
Synaptic plasticity in the hippocampus is important for long-term memory formation (Bird and Burgess, 2008; Nabavi et al., 2014); therefore, we examined aging-dependent or sex-biased expression and splicing of synapse-related genes. Most of the synapse-related genes showed an aging-related decrease in expression in either male or female hippocampus and encode proteins critical for the excitatory postsynaptic terminal such as DLG4/PSD95, DLG2/PSD93, SYNGAP1, SHANK1, SHANK3, CACNG5, CAMK2A, and RGS14 (Table 1). Additionally, we found aging-related changes in the expression of genes regulating inhibitory neuron synaptic functions, including decreased expression of the inhibitory synaptogenic gene Igsf21 (in females), the inhibitory synaptic adhesion gene Igsf9b (in both sexes), and calcium-binding protein gene Calb1 (in both sexes) and increased expression of the calcium-binding protein gene Pvalb (in females) and inhibitory synaptic vesicle gene Syt2 (in females) (Tanabe et al., 2017; Babaev et al., 2018). Together, these findings support previous reports of changing excitatory and inhibitory synaptic dynamics in the hippocampus with aging (McQuail et al., 2015; Rozycka and Liguz-Lecznar, 2017).
Overall, we found significant AS events in a number of genes important for synaptic function (Table 2). One of the most significant sex-biased AS events in the aged hippocampus was the female-biased inclusion of exon 8a Cacna1d, a gene important for synaptic plasticity that encodes the pore-forming subunit of an L-type voltage-gated Ca2+ channel. Mutations in Cacna1d exon 8a have been implicated in autism spectrum disorder (Pinggera et al., 2015, 2017; Pinggera and Striessnig, 2016), and alternative splicing of exon 8 may contribute to channel voltage dependency differences (Hofer et al., 2021). We also found aging-related AS events in voltage-gated calcium channel genes Cacna1c, Cacnb1, and Cacna1g that may impact calcium channel function (Ernst and Noebels, 2009; Buraei and Yang, 2010; Lipscombe et al., 2013; Hu et al., 2017). We also detected sex-biased splicing (MXE in young, SE in aged) of Mapk12, a gene encoding the neuroprotective MAPK, p38γ, that is involved in regulating the post-synaptic density (Ittner et al., 2016; Asih et al., 2020). These results reveal that many synapse-related genes show sex-biased AS that may affect synaptic function.
Dynamic changes in chromatin accessibility are critical to the precise control of gene expression patterns and have been shown to be altered with aging (Su et al., 2017; Zhang et al., 2022). We used ATAC-seq to probe for chromatin accessibility changes during aging in the hippocampus of male and female mice. The hippocampus of young adult and aged males and females showed a similar number of ATAC peaks, with a similar distribution of genome annotations: ~40% intergenic, 35% introns, and 18% promoter-TSS (Figure 6A; Supplementary Figure S3A). We analyzed a merged set of 92,233 replicated peaks (see Methods section; Figure 6A; Supplementary Figure S3B) and, similar to the results from our RNA-seq analysis, principle component analysis of fragment counts from ATAC-seq regions showed that samples were separated by sex and age by the first and second principle component, respectively (Figure 6B; Supplementary Figure S3C). Overall, 774 genomic regions were differentially accessible either with sex or aging (FDR < 0.05). There were 393 regions with sex-biased chromatin accessibility and 404 regions showing accessibility changes with aging (Figure 6C), including 23 regions that showed changes with both aging and sex. More differentially accessible regions (DARs) exhibited aging-related changes in female compared to male hippocampus (366 vs. 109 regions, chi-square test, X2 = 139.4, p < 0.00001), and this result was consistent even when reduced numbers of female samples were included to match the number of male samples (Supplementary Figure S3D). Approximately 90% of aging-related DARs showed increased chromatin accessibility with aging, with intergenic regions making up the majority of DARs that either opened or closed with aging (Figure 6D). For example, an intergenic region in chromosome 13 was ~10 times more accessible in aged male and female hippocampus compared to the young adult hippocampus (Figure 6E left), whereas an intergenic region in chromosome 8 was ~1.5 times less accessible in aged compared to young adult hippocampus (Figure 6E right). On average, regions that opened with aging showed a 3.4- and 2.3-fold increase in accessibility with aging in males and females, respectively, and those regions that closed with aging showed a 1.8-fold aging-related decrease in accessibility in both males and females (Figure 6F). Altogether, these results indicate that aging leads to increased chromatin accessibility in the hippocampus, especially in non-coding regions.
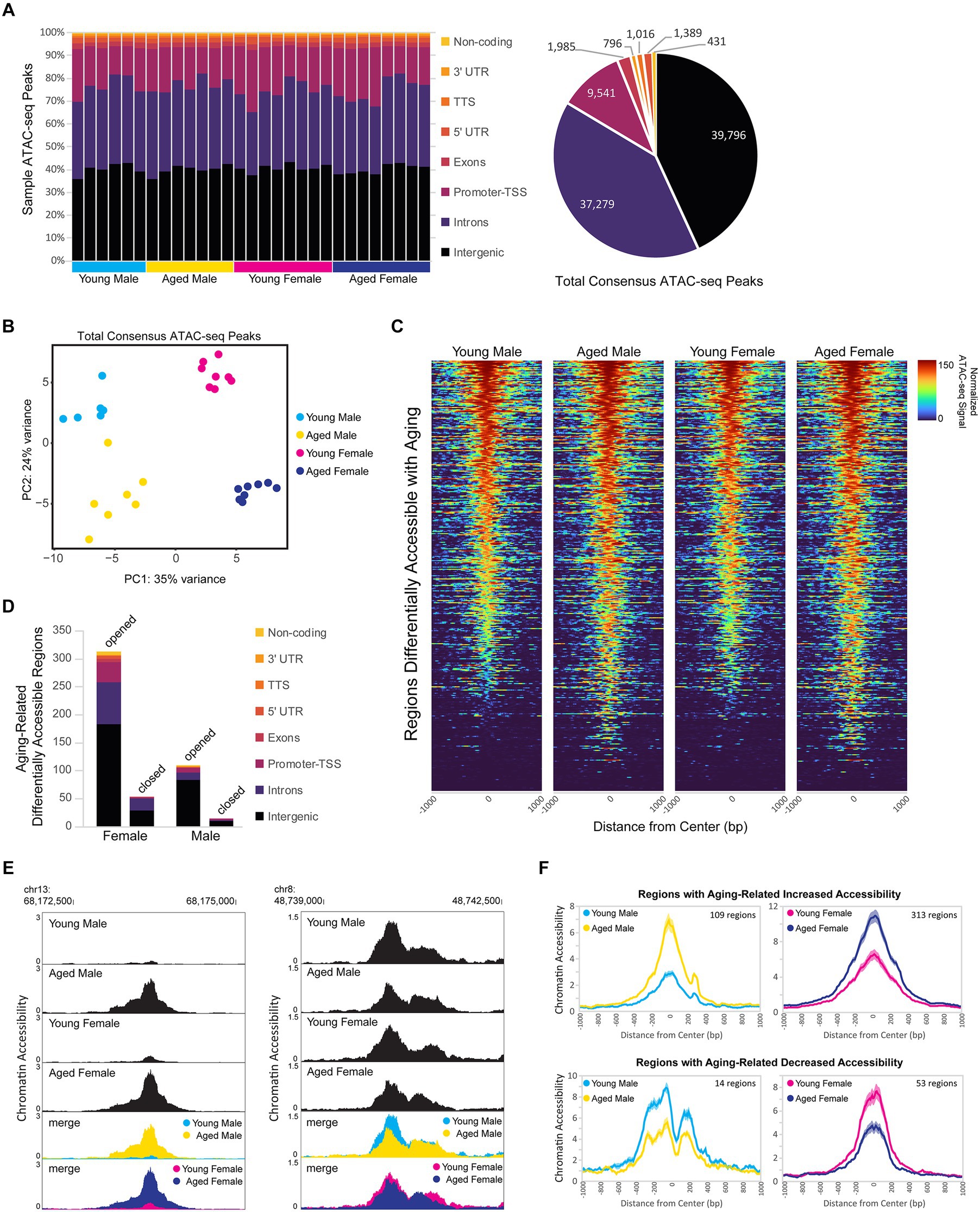
Figure 6. Aging is associated with higher chromatin accessibility, with most of the changes occurring in intergenic regions. (A) Genome annotation of individual sample ATAC-seq peaks, with each column representing a biological replicate (left) and genome annotation for total consensus ATAC-seq peaks (right). (B) Principle component analysis of ATAC-seq fragments in the total consensus peak set. (C) Heat maps of average normalized ATAC-seq signal per condition for aging-related differentially accessible regions (DARs). Normalized ATAC-seq signal/10 bp bin is shown for ± 1,000 bp around the ATAC-seq region center, and regions are sorted by young male ATAC signal in the ± 100 bp around the center. (D) Genome annotation of regions that were differentially accessible (FDR < 0.05) between young adult and aged hippocampus. (E) Browser track examples of regions showing aging-dependent differences in chromatin accessibility. Panel on the left shows the average normalized ATAC-seq signal from young and aged male and female hippocampus in the intergenic-MuRRS-int|LTR|ERV1 region chr13: 68173077-68174082 that was significantly more accessible in aged compared to young males and females (male aged vs. young log2FC = 3.85, FDR < 0.0001; aged vs. young log2FC = 3.00, FDR < 0.0001). Panel on the right shows the average normalized ATAC-seq signal in the intergenic region chr8: 48739982-48741266 that was significantly more accessible in young compared to aged males and females (male aged vs. young log2FC = −0.63, FDR = 0.04; female aged vs. young log2FC = −0.57, FDR =0.02). (F) ATAC-seq profiles for aging-related DARs in either males or females. Solid lines indicate the average of each condition’s normalized histogram (n = 6 young males, 7 aged males, 8 young females, and 8 aged females) with shading indicating s.e.m.
First, we focused on the sex-independent chromatin accessibility changes in the hippocampus with aging. We found that 85 regions showed differential ATAC-seq signals with aging in both sexes (Figure 7A), and 99% of these regions exhibited aging-related increases in chromatin accessibility (84/85 regions opened, 1/85 closed). For the regions that gained accessibility in both sexes, there was ~3.8-fold increase in accessibility with aging (Figure 7B), and motif enrichment analysis showed that these regions were enriched for HOXA- and MEF2-like motifs (Supplementary Figure S4A). We ranked DARs by lowest FDR-adjusted p-value in either males or females and found that within the 30 most significant DARs, 13 were located in retrotransposable element-derived sequences, and all of these regions showed an opening in accessibility with aging (Figure 7C). For example, an intergenic L1MA5A-type LINE-1 region in chromosome 14 showed ~4-fold increase in accessibility in the hippocampus of aged animals of both sexes compared to young animals (Figure 7D). Twenty LINE-1 retrotransposon elements showed an aging-associated increase in accessibility in the hippocampus of both sexes (Supplementary Figure S4B). These regions encode truncated sequences incapable of retrotransposition; however, truncated LINE-1 sequences can be transcribed (Rangwala et al., 2009), leading to DNA damage (Kines et al., 2014) and immune activation (De Cecco et al., 2019; Simon et al., 2019). To test for an aging-related increase in the transcription of LINE-1 elements in the hippocampus, we performed qPCR for the retrotransposon LINE-1 with three different primer sets. We found that with aging, there was an average increase of 32% in LINE-1 transcript abundance in the hippocampus (LINE-1 5’UTR p < 0.001, LINE-1 ORF1 p < 0.01, LINE-1 5’UTR:ORF1 p < 0.05; Figure 7E), in agreement with the finding that LINE-ORF1 protein increased in the frontal cortex with aging (Zhang et al., 2022). Previous studies have found that LINE-1 retrotransposon activity is more common in neurons than other somatic cells and that it increases with aging (Reilly et al., 2013; Van Meter et al., 2014; Zhao et al., 2019; Gorbunova et al., 2021). We examined the chromatin accessibility of full-length intact LINE-1 genomic regions (Penzkofer et al., 2017) and found suppression of chromatin accessibility in these regions in both young adult and aged hippocampus (Supplementary Figure S4C), indicating that full-length LINE-1 regions as a whole remained repressed during aging. These results indicate that there is an opening of chromatin during aging, especially in retrotransposon-derived sequences, with an accompanying increase in LINE-1 transcript abundance, which may contribute to aging-related genome instability (Gorbunova et al., 2021).
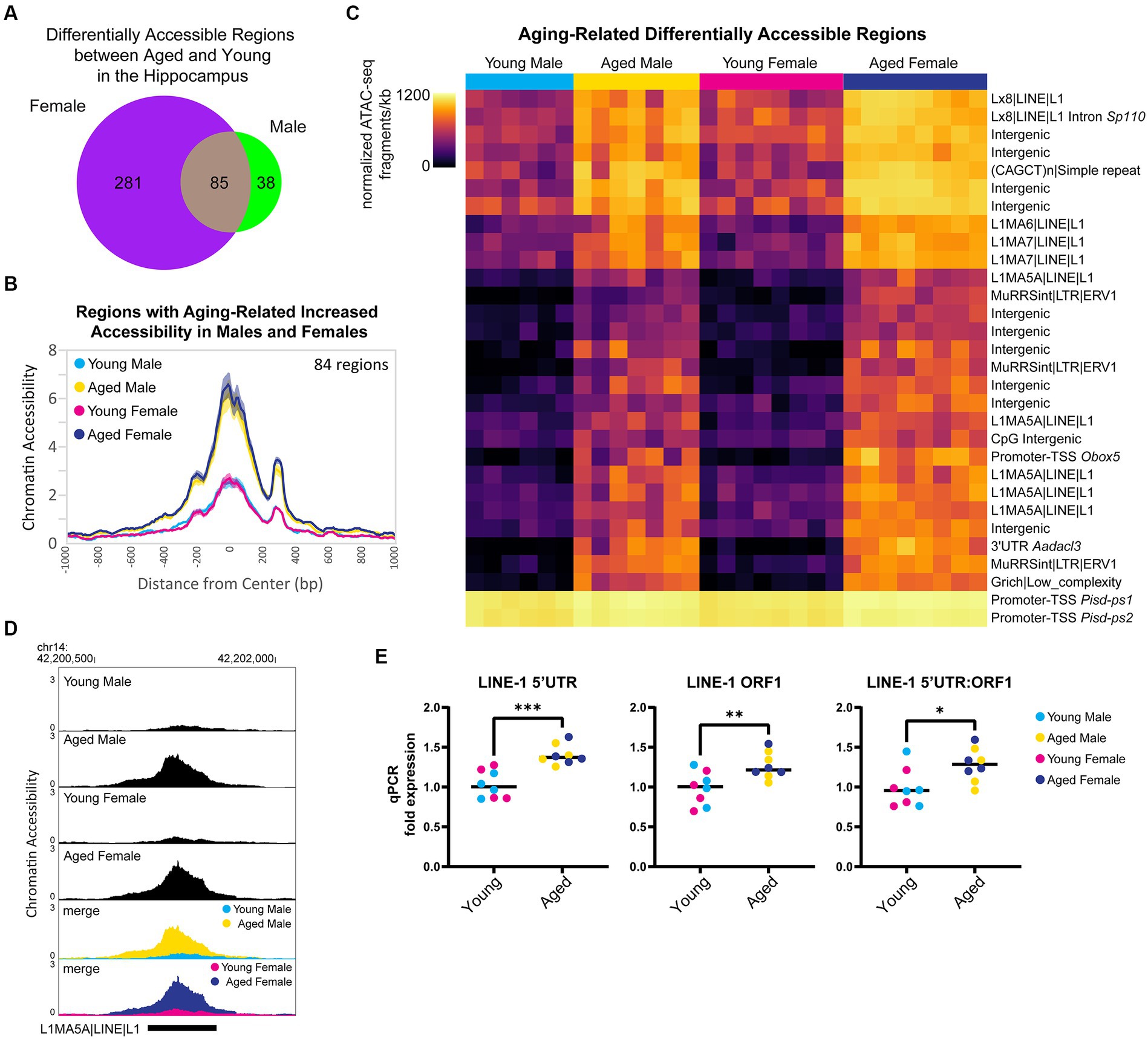
Figure 7. Aging is associated with higher accessibility in repetitive elements and higher expression of retrotransposon transcripts. (A) Venn diagram of the number of DARs (FDR < 0.05) in young adult and aged hippocampus. (B) ATAC-seq profiles for regions that showed increased accessibility with aging in both males and females. Solid lines indicate the average of each condition’s normalized histogram (n = 6 young males, 7 aged males, 8 young females, and 8 aged females) with shading indicating s.e.m. (C) Heat map of normalized total ATAC-seq signal per region for the top 30 aging-related DARs in both male and female hippocampus (top 30 by FDR, clustered by ATAC-seq signal). Each row represents a chromatin region with detailed annotation labeled on the right, and each column is a biological replicate. Normalized ATAC-seq fragments/kb are represented by color (color scale is non-linear, see Supplementary Table S3 for all normalized signal and log2FC values). (D) Browser track example of LINE-1 region showing aging-related increase in chromatin accessibility. Shown are the average normalized ATAC-seq signals from young and aged male and female hippocampus in the intergenic L1MA5A|LINE|L1 region chr14: 42200723-42201552 that was significantly more accessible with aging in males and females (aged vs. young male log2FC = 2.22, FDR < 0.0001; aged vs. young female log2FC = 2.05, FDR < 0.0001). (E) qPCR of LINE-1 transcript expression in young adult and aged hippocampus (n = four age/sex). LINE-1 5’UTR, LINE-1 ORF1, and LINE-1 5’UTR:ORF1 are three different primers for LINE-1 annealing to sequences within the 5’UTR, within ORF1, and spanning the 5’UTR and ORF1, respectively. Unpaired t-tests: LINE-1 5’UTR p < 0.001, LINE-1 ORF1 p = 0.009, LINE-1 5’UTR:ORF1 p = 0.02. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
To assess sex-dependent differences in chromatin accessibility in the hippocampus, we analyzed the DARs between males and females from young adult and aged mice. We identified 29 autosomal regions in young animals and 281 autosomal regions in aged animals that showed sex-biased differential accessibility, with aging associated with more sex differences in autosome chromatin accessibility (chi-square test, young vs. aged X2 = 205.2, p < 0.00001; Figure 8A). Sex bias in chromatin accessibility resulted in ~1.7-fold change in peak ATAC-seq signal in sex-biased DARs (Figure 8B), relatively smaller than those seen with aging-related differential accessibility (Figure 6F). When we ranked sex-biased regions by lowest FDR in either young or aged animals, we found that the majority of the most significant male-biased regions were annotated to promoters, whereas the most significant female-biased regions were intergenic (Figure 8C). Similarly, when we analyzed the genomic ontology for all sex-biased DARs, we found that most regions that were more open in females versus males were annotated to intergenic regions (Figure 8D). In contrast, most male-biased accessible regions were promoter-TSS regions (8/15 in young, 80/157 in aged). For example, an intergenic region (1.5 Mb from nearest gene, Ncam2) was more accessible in aged females compared to aged males (Figure 8E left), and the TSS-promoter region of Stub1 was more accessible in aged males compared to aged females (Figure 8E right). Motif analysis revealed that promoters with male-biased accessibility were enriched for CpG island-associated motifs, including CGCG/GFX and GC-box/SP/KLF motifs (Supplementary Figure S5A). Non-promoter regions with male-biased accessibility were enriched in CTCF, HOXA, ZNF382, and MEF2 motifs whereas non-promoter regions with female-biased accessibility were enriched for CTCF, E-box/NEUROD1, AP-1, and TLX motifs (Supplementary Figure S5B). Most mammalian promoter sequences are GC-rich and contain CpG islands (Yang et al., 2007). Indeed, we found the CpG content of accessible promoter regions in the hippocampus was ~7%, whereas that of accessible non-promoter regions was ~2% (Supplementary Figures S6A,B). However, we found that in the young and aged hippocampus, the CpG content in male-biased non-promoter regions was higher than average (~4%; Kruskal–Wallis tests with Dunn’s correction, p < 0.05; Supplementary Figure S6B). To test if the sex differences in accessibility for promoter and CpG-rich regions could be due to GC bias, we used smooth-quantile with GC-content normalization (Hicks et al., 2017; Van den Berge et al., 2022), which can correct for GC-content effects in ATAC-seq datasets. With this normalization technique, we found similar male bias in accessibility at promoters (QSmooth-GC; Supplementary Figure S6C) and CpG-rich regions (Supplementary Figures S6A,B; p < 0.0001); therefore, although we cannot rule out GC bias effects, these sex differences in accessibility appear to be biological in nature. Importantly, male-biased promoter accessibility did not appear to result in increased transcription at those genes, as none of the autosomal genes with male-biased accessibility at promoters showed sex-biased expression. Therefore, the male and female hippocampus may compensate for sex differences in promoter accessibility through different mechanisms to regulate expression.
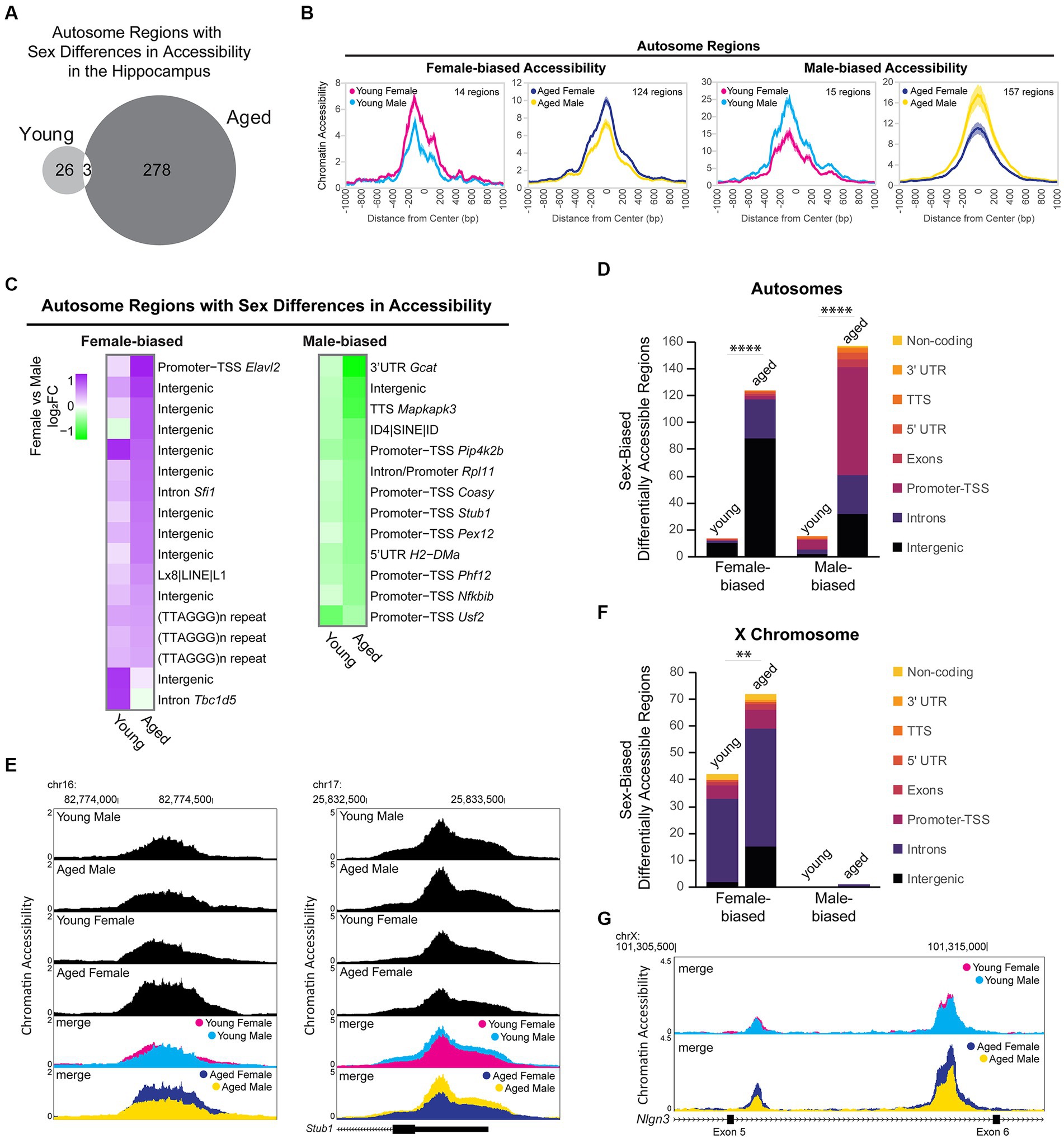
Figure 8. Sex differences in chromatin accessibility. (A) Venn diagram of the number of autosome sex-biased DARs in young adult and aged hippocampus (FDR < 0.05). (B) ATAC-seq profiles for sex-biased DARs on autosomes. Solid lines indicate the average of each condition’s normalized histogram (n = 6 young adult male, 7 aged male, 8 young adult female, 8 aged female) with shading indicating s.e.m. (C) Fold change heat map of sex-biased differentially accessibility for the top 30 autosome regions (top 30 by FDR, clustered by ATAC-seq signal; purple indicates higher accessibility in females and green indicates higher accessibility in males). Each row represents a chromatin region with detailed annotation labeled on the right. (D) Genome annotation of autosome regions that were differently accessible between female and male hippocampus. Chi-square test, young vs. aged X2 = 87.7 more open female, X2 = 117.3 more open male, p < 0.00001 (indicated with ****). (E) Browser track examples of regions showing sex differences in chromatin accessibility. Panel on the left shows the average normalized ATAC-seq signal from young adult and aged male and female hippocampus in the intergenic region chr16: 82773954-82774686 (1.5 Mb from nearest gene, Ncam2) that was significantly more accessible in aged females than males (FDR = 0.003, female vs. male log2FC = 0.79; one of two regions annotated to Ncam2 showing female bias in accessibility). Panel on the right shows the average normalized ATAC-seq signal in the promoter-TSS region chr17: 25832563-25833564 of Stub1 that was significantly more accessible in aged males than females (FDR = 0.002, female vs. male log2FC = −0.61). (F) Genome annotation of chromosome X regions that were differentially accessible between female and male hippocampus. Chi-square test, young vs. aged X2 = 8.1 more open female, p < 0.01 (indicated with **). (G) Browser track example of X chromosome region showing sex differences in chromatin accessibility. The average normalized ATAC-seq signal is shown from young adult and aged male and female hippocampus in the intronic region chrX: 101308500-101316000 between exons 5 (ENSMUSE00001248461) and 6 (ENSMUSE00001264564) of Nlgn3. Two regions were significantly more accessible in aged females than males, with the second ATAC-seq peak also showing female-specific opening with aging (region chrX: 101309331-101309581 aged female vs. male log2FC = 0.88, FDR = 0.001; region chrX: 101313685-101314599 aged female vs. male log2FC = 0.85, FDR =1.19E-11; female aged vs. young log2FC = 0.70, FDR =1.11E-7).
We evaluated sex differences in chromatin accessibility of the X chromosome in young adult and aged hippocampus. Of the 2,595 ATAC-seq regions on the X chromosome, 74 regions (3%) showed sex differences in accessibility, with all but one region showing female-biased accessibility. We found 41 DARs on the X chromosome that showed a sex bias in both young and aged animals, all of which showed female-biased accessibility. Most of these regions were located within the Dxz4 microsatellite and Firre locus important for X-chromosome 3D structure (29 female-biased regions in introns of 4933407K13Rik, Firre, and Gm35612/CrossFirre). Many other female-biased regions were located in the gene body or promoter of female-biased genes (Kdm6a, Ddx3x, Eif2s3x, Kdm5c, and Xist) and/or genes located in the X inactivation center (Xist, Ftx, Mir421, Jpx, and Gm9159) (Yin et al., 2021). The number of regions with female-biased accessibility increased with aging (Figure 8F; chi-square test, young vs. aged X2 = 8.1, p < 0.01), including aging-dependent female bias in accessibility in regions annotated to female-biased genes (Kdm6a, Eif2s3x, and 5530601H04Rik) as well as in the gene bodies of the synaptic regulators Nlgn3 and Srpx2. For example, two regions in an intron of Nlgn3, a gene that encodes the synaptic protein neuroligin-3 (Uchigashima et al., 2021) and whose expression was downregulated with aging in males, showed no sex bias in chromatin accessibility in young animals. However, a female-specific opening with aging resulted in female-biased accessibility in the aged hippocampus (Figure 8G). In the young hippocampus, all X chromosome regions showed a female bias in accessibility. Similarly, in the aged hippocampus, all X chromosome regions were more accessible in females than males, except for one region that showed a male bias in accessibility. This region spans the promoter, first exon, and intron of Gm35612/CrossFirre, a lncRNA that is repressed by the expression of Firre, which is a lncRNA involved in the maintenance of X chromosome inactivation (Fang et al., 2020). Altogether, these findings suggest that CpG-rich regions, including promoters, are more accessible in young and aged male compared to female hippocampus and that there is more divergence in chromatin accessibility in the hippocampus between males and females with aging.
Transcription is a dynamic process involving the controlled coordination of regulatory elements such as promoters and enhancers, and DNA accessibility can indicate active regulatory elements bound by or permissive to chromatin binding factors (Cramer, 2019; Andersson and Sandelin, 2020). Therefore, we wanted to assess the relationship between aging- or sex-related changes in chromatin accessibility and gene expression. For example, many of the genes in the Pcdhb gene cluster were upregulated with aging, and a corresponding increase in chromatin accessibility can be observed in the ATAC-seq signal in this region (Figure 9A). Similarly, Lin28b, a gene encoding an RNA-binding protein that regulates miRNA maturation was upregulated with aging in the hippocampus of males and females, and a region upstream of Lin28b showed an aging-related increase in accessibility in the hippocampus of both sexes (RNA FDR < 0.001 and ATAC peak on far right, upstream/alternative promoter of Lin28b, FDR < 0.01 for both sexes; Figure 9B).
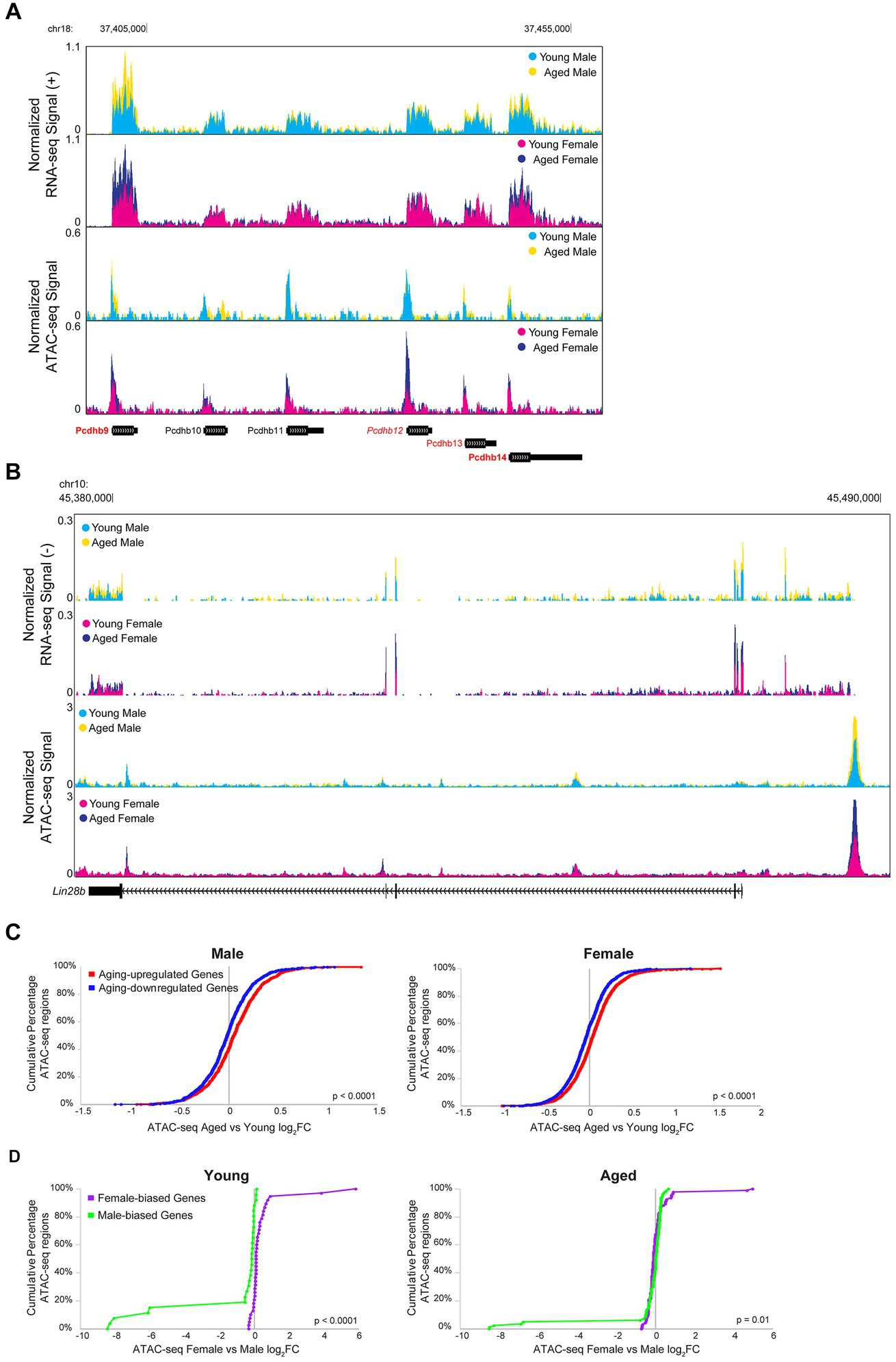
Figure 9. Aging and sex-biased differential expression is correlated with ATAC-seq differential accessibility. (A) Browser track example showing RNA-seq and ATAC-seq signal for a portion of the Pcdhb gene cluster (region chr18: 37397808-37458667), containing four genes that were significantly upregulated with aging. The top two tracks show the normalized plus-strand RNA-seq signal for the male (first row) and female (second row) hippocampus. The bottom two tracks show the normalized ATAC-seq signal for the male (third row) and female (fourth row) hippocampus. Genes that were upregulated with aging in both sexes are indicated with bold red (Pcdhb9: male aged vs. young adult log2FC = 0.82, FDR-adjusted p = 7.21E-33, female aged vs. young adult log2FC = 0.96, FDR-adjusted p = 1.68E-53; Pcdhb14: male aged vs. young adult log2FC = 0.46, FDR-adjusted p = 3.64E-6, female aged vs. young adult log2FC = 0.68, FDR-adjusted p = 6.29E-17). One gene was upregulated with aging in the female hippocampus only, indicated with italic red (Pcdhb12: male aged vs. young log2FC = 0.23, FDR-adjusted p = 0.24, female aged vs. young log2FC = 0.30, FDR-adjusted p = 0.02). One gene was upregulated with aging in the male hippocampus only, indicated with regular font red (Pcdhb13: male aged vs. young log2FC = 0.43, FDR-adjusted p = 0.006, female aged vs. young log2FC = 0.28, FDR-adjusted p = 0.07). Genes that were not DE with aging are indicated with black font. (B) Browser track example as in A, showing RNA-seq and ATAC-seq signal for the Lin28b locus (chr10: 45375000-45491000). RNA signal suggests a long Lin28b isoform, including additional exons present in the ENSMUST00000214555.1/Lin28b-202 transcript from an alternative promoter located at ATAC-seq peak. RNA upregulated with aging female: log2FC = 0.86, p = 4.9E-7; male: log2FC 0.82, p = 8.3E-5; ATAC opened with aging female: log2FC = 0.60, p = 2.2E-3; male: log2FC = 0.75, p = 2.2E-4. (C) Cumulative distributions of aging-related chromatin accessibility fold changes for male (left panel) and female (right panel) hippocampus for ATAC-seq regions annotated to DE genes that were either upregulated (red) or downregulated (blue) with aging (male: Kolmogorov–Smirnov D = 0.14, p < 0.0001; female: Kolmogorov–Smirnov D = 0.15, p < 0.0001). (D) Cumulative distributions of sex-biased chromatin accessibility fold changes for young (left panel) and aged (right panel) hippocampus for ATAC-seq regions annotated to DE genes that were either higher expressed in female (purple) or higher expressed in male (green) hippocampus (young: Kolmogorov–Smirnov D = 0.68, p < 0.0001; aged: Kolmogorov–Smirnov D = 0.24, p = 0.01).
To compare RNA-seq and ATAC-seq results across all DE genes, we first assessed the relationship between aging or sex bias fold changes in expression with the fold changes in chromatin accessibility for ATAC-seq regions closest to the TSS of those genes. We found that aging-related changes in gene expression were significantly correlated with aging-related changes in chromatin accessibility in the male and female hippocampus (male: of 370 DE genes, 316 genes were associated with 2,044 ATAC-seq regions, Spearman rs = 0.12, p < 0.0001; female: of 707 DE genes, 596 were associated with 3,600 regions, Spearman rs = 0.15, p < 0.0001), and a stronger correlation was observed when including only ATAC-seq regions that were within 10 kb of the TSS (male Spearman rs = 0.26, p < 0.0001, female Spearman rs = 0.27, p < 0.0001). Similarly, ATAC-seq regions closest to the TSS of genes that were upregulated with aging in either male or female hippocampus showed more aging-related opening compared to ATAC-seq regions closest to the TSS of genes that were downregulated with aging (Figure 9C; male: Kolmogorov–Smirnov D = 0.14, p < 0.0001; female: Kolmogorov–Smirnov D = 0.15, p < 0.0001).
We also found a correlation between sex bias in expression and chromatin accessibility in the young hippocampus (young: of 17 DE genes, 16 were associated with 66 ATAC-seq regions, Spearman rs = 0.74, p < 0.0001; aged: of 42 genes, 34 were associated with 178 ATAC-seq regions, Spearman rs = 0.00, p = 0.96), and we found significant correlations in both young and aged hippocampus for regions within 10 kb of the TSS (young Spearman rs = 0.88, p < 0.0001, aged Spearman rs = 0.42, p < 0.01). We specifically examined the ATAC-seq regions annotated to the female-biased myelin-related genes from Figure 3D, and found that although some regions showed a trend of female-biased accessibility in the aged hippocampus, many did not, and none showed a significant sex bias (Supplementary Figure S7A). However, overall, ATAC-seq regions closest to the TSS of female- versus male-biased genes showed higher chromatin accessibility in female versus male hippocampus in both young and aged animals (Figure 9D; young: Kolmogorov–Smirnov D = 0.68, p < 0.0001; aged: Kolmogorov–Smirnov D = 0.24, p = 0.01).
To evaluate the relationship of aging and sex bias DE with chromatin accessibility independent of consensus peaks, we analyzed the ATAC-seq signal surrounding DE genes’ TSSs. We found that overall chromatin accessibility increased in the region surrounding the TSS of genes that were upregulated with aging in the female hippocampus (Supplementary Figure S7B; female: Kolmogorov–Smirnov p = 0.03, male: p = 0.11). Furthermore, chromatin accessibility was higher in the female hippocampus in regions surrounding the TSS of female-biased genes, and chromatin accessibility was higher in males in regions surrounding the TSS of male-biased genes (Supplementary Figure S7C; female-biased expression, young: Kolmogorov–Smirnov p = 0.003; aged: p < 0.0001; male-biased, young: p < 0.0001; aged: p = 0.0001). In summary, we found that changes in expression tended to correlate with changes in chromatin accessibility, although we note that these correlations may be underestimated, as enhancers often do not regulate the gene with the nearest TSS (Andersson et al., 2014; Furlong and Levine, 2018).
Memory impairments are a hallmark of aging. In both humans and rodents, the hippocampus is important for long-term memory formation (Bird and Burgess, 2008; Morrison and Baxter, 2012; Allen and Fortin, 2013), and like humans, mice show memory impairments and changes in hippocampal function with aging (Benice et al., 2006; von Bohlen und Halbach et al., 2006; Peleg et al., 2010; Wimmer et al., 2012; Stilling et al., 2014; Pereda et al., 2019). While cell type abundance in the brain, including the hippocampus, does not appear to change with aging (Long et al., 1998; Stilling et al., 2014; Ximerakis et al., 2019; Hahn et al., 2023), a number of cellular processes, including chronic inflammation, dysregulated proteostasis, synaptic function, retrotransposon activation, and genomic instability, are altered with aging and can impact cognitive function and memory formation (Morrison and Baxter, 2012; Pal and Tyler, 2016; Gorbunova et al., 2021; Simpson and Chandra, 2021; López-Otín et al., 2023). Although there are sex differences in aging-related memory deficits and Alzheimer’s disease (Gao et al., 1998; Herlitz and Rehnman, 2008; Jack et al., 2015; McCarrey et al., 2016; Beam et al., 2018; Anstey et al., 2021; Davis et al., 2021), this is the first study, to the best of our knowledge, that examines how aging-related changes in alternative splicing and chromatin accessibility in the hippocampus differ between males and females.
Using a genome-wide approach to understand how aging and sex affect the transcriptome and chromatin accessibility in the mouse hippocampus, our results provide a framework of the molecular mechanisms that may contribute to aging-related memory impairment. Our findings support a number of themes that are consistent with existing hypotheses of aging, including aging-related inflammation, changes in myelination, synaptic alterations, calcium dysregulation, loss of heterochromatin, and increased retrotransposon activity (Morrison and Baxter, 2012; Nikoletopoulou and Tavernarakis, 2012; Mosher and Wyss-Coray, 2014; Pal and Tyler, 2016; Gorbunova et al., 2021; Simpson and Chandra, 2021; Franklin and Simons, 2022; López-Otín et al., 2023). Our results also, however, reveal significant sex differences in the transcriptome and chromatin environment in the hippocampus and further show that aging amplifies sex bias in expression and chromatin accessibility.
Immune response genes were upregulated with aging in both male and female hippocampus, in line with numerous reports of chronic inflammation, upregulation of immune genes, and activation of microglia with aging in the hippocampus and other brain regions (Sheng et al., 1998; Berchtold et al., 2008; Cribbs et al., 2012; Mosher and Wyss-Coray, 2014; Stilling et al., 2014; Mangold et al., 2017; Spittau, 2017; Allen et al., 2023; Hahn et al., 2023). We found more changes in gene expression overall with aging in the female hippocampus than in the male hippocampus (Figure 1D), a result that is consistent with previous microarray studies in both human and mouse hippocampus (Berchtold et al., 2008; Cribbs et al., 2012; Mangold et al., 2017), but that differs from other brain regions (Berchtold et al., 2008). In agreement with the fact that females mount stronger immune responses than males (Klein and Flanagan, 2016; Gal-Oz et al., 2019), we found that female hippocampus showed larger aging-related changes in expression of immune-response genes (e.g., C4b, Figure 2D). Given that the hippocampus has been reported to be more immune-alert than other brain regions (Grabert et al., 2016), the greater aging-related changes in gene expression in female hippocampus may result in part from higher female immune response.
We detected significant aging-dependent changes in the expression and alternative splicing of myelin sheath genes. Produced by oligodendrocytes, myelin is important for axon integrity and action potential conductance (Nave and Werner, 2014) and has been found to be critical for memory retention (Xin and Chan, 2020). During aging, impaired myelin debris clearance and diminished OPC differentiation reduce the efficiency of remyelination (Sim et al., 2002; Franklin and Goldman, 2015; Franklin and Simons, 2022; Chapman et al., 2023). A previous study showed that enhancing OPC proliferation and myelination in the aged hippocampus increased memory retention in mice (Iram et al., 2022). We detected aging-dependent female bias in the expression of genes encoding myelin sheath components, including Mag, Mbp, Mog, and Mal (Figure 3D). The compact myelin gene, Mbp, was upregulated with aging in female but not male hippocampus by RNA-seq and qPCR, although MBP protein did not show sex bias in overall abundance (Figure 4C). There were, however, sex differences in the abundance of specific MBP isoforms in the aged hippocampus (Figure 4C; Supplementary Figures S2D,E), indicating that male and female hippocampus show differential regulation of aging-related Mbp transcript and protein isoform expression. Altogether, the female-biased expression of myelin sheath-related genes with aging could suggest that female hippocampus undergoes less myelin degeneration or more remyelination than male hippocampus during aging, consistent with studies in aged rats showing a female bias in remyelination and white matter volume (Li et al., 2006; Yang et al., 2008). Indeed, the female-biased aging-related upregulation of Bcas1, a marker of early actively myelinating oligodendrocytes (Fard et al., 2017), could reflect more active remyelination in aged female versus male hippocampus. Because an immune response is required for myelin debris clearance and OPC recruitment (Peters, 2002; Franklin and Simons, 2022), one possibility is that a higher immune response in the female hippocampus may lead to more efficient myelin debris clearance and OPC recruitment necessary for remyelination.
We found aging-related splicing differences not only in Mbp but also in myelin sheath genes Mag and Bcas1 in male and female hippocampus (Figure 4B). The alternative splicing of Mag, which encodes a myelin-membrane protein, into large (L-MAG), exon 12-skipped, and small (S-MAG), exon 12-included, isoforms is tightly regulated during development, with L-MAG predominating during early myelinogenesis with potentially larger myelination capacity and S-MAG expression increasing during maturation to maintain myelination (Tropak et al., 1988; Quarles, 2007). Our finding that aging resulted in an increase in S-MAG and a decrease in L-MAG isoforms could point to aging-related reduced myelin formation or capacity. Similarly, the alternative splicing of exon 2 in Mbp is also known to be regulated during development (Harauz and Boggs, 2013; Müller et al., 2013). MBP isoforms containing exon 2 are enriched during active myelin formation, whereas isoforms excluding exon 2 are predominately expressed in adulthood (Woodruff and Franklin, 1998; Li et al., 2000). These isoforms differ in their subcellular location and function, with exon 2-containing isoforms localized to the cytosol and nucleus, where they regulate myelin development and differentiation, whereas exon 2-excluding isoforms are localized to the oligodendrocyte myelin membrane, where they are essential for the formation and function of compact myelin (Harauz and Boggs, 2013). In rats, loss of exon 2-included isoforms (but not other MBP isoforms) with aging was associated with split and retracted myelin sheaths, especially in paranodal regions (Sugiyama et al., 2002). Our finding that exon 2-containing MBP isoform expression decreased with aging suggests that this decrease may result in the loss of molecular signaling pathways regulating remyelination and maintenance of the axo-glial junction. Less is known about alternative splicing of the myelination gene, Bcas1; however, aging-related splicing of Bcas1 has been previously reported in the mouse hippocampus (sex not specified) (Stilling et al., 2014). Notably, in oligodendrocytes, the alternative splicing of Mag, Mbp, and Bcas1 is regulated by the RNA-binding protein quaking (Wu et al., 2002; Zhao et al., 2010; Darbelli et al., 2017). Although we did not detect aging-related differential expression or alternative splicing of the quaking gene (Qk/Qki), phosphorylation of quaking can regulate its binding to and stabilization of Mbp RNA (Zhang et al., 2003). Altogether, it is possible that altered quaking function during aging in the hippocampus may lead to aging-related changes in alternative splicing of Mag, Mbp, and Bcas1, contributing to the decrease in myelination capacity observed with aging.
Previous studies have shown that in the hippocampus, aging is associated with alternations in synaptic function, including deficits in synaptic plasticity induction, reduction in postsynaptic density size, and dysregulated calcium currents (Shankar et al., 1998; Kumar et al., 2009; Morrison and Baxter, 2012). In alignment with these studies, we detected changes in the expression and alternative splicing of genes regulating synaptic function with aging in the hippocampus of male and female mice (Tables 1, 2). We found aging-related changes in the expression of genes encoding both inhibitory and excitatory synapse-related proteins, as well as changes in calcium buffer and calcium channel genes. We found several L- and T-type calcium channel subunit genes that were either downregulated with aging or showed aging-related changes in alternative splicing. Calcium is required for long-lasting synaptic plasticity in the hippocampus and is tightly controlled in neurons through the regulation of calcium buffers and calcium release from internal stores, as well as calcium influx through NMDA receptors and voltage-dependent calcium channels (Nikoletopoulou and Tavernarakis, 2012). Dysregulated calcium homeostasis not only suppresses synaptic plasticity but also causes neurons to be more vulnerable to damage from oxidative stress, and previous studies have shown that aging results in increased L-type voltage-dependent calcium currents that disrupt long-lasting synaptic plasticity and memory in the hippocampus of rodents (Kumar et al., 2009; Nikoletopoulou and Tavernarakis, 2012; Oh et al., 2013; Pereda et al., 2019). Additional genes important for synaptic plasticity and memory, including Camk2a (Elgersma et al., 2002), Ablim3 (Guo et al., 2018), and Cplx2, were downregulated with aging in both sexes. Furthermore, male and female hippocampus showed different aging-related changes in expression and alternative splicing, including male-biased expression of the cysteine/glutamate antiporter gene Slc7a11/xCT, whose deletion in male mice has been shown to improve aging-related memory deficits (Verbruggen et al., 2022).
Sex-biased expression and splicing were observed in both young and aged hippocampus. Sex differences in the brain can result from the influence of gonadal sex hormones and sex chromosome-expressed genes. In both humans and rodents, circulating and brain-derived estrogens as well as circulating androgens diminish with aging, and their loss is linked to memory impairment (Khosla et al., 1998; Edinger and Frye, 2007; Cai and Li, 2020; Low et al., 2020; Taxier et al., 2020). We found that relatively few genes showed sex-biased expression in the young adult hippocampus (Figure 3A), and most were sex chromosome-expressed genes. Two notable exceptions were the male-biased expression of the synaptic vesicle gene, complexin 2 (Cplx2), and the female-biased expression of the neuropeptide gene, Npy. Npy has been shown to be upregulated by estradiol in the hippocampus (Ledoux et al., 2009), and correspondingly, we found female-biased expression in the young adult hippocampus but aging-related downregulation in the female hippocampus. Sex chromosome-expressed genes themselves can also contribute to downstream sex-biased expression. For example, the X chromosome-expressed genes Kdm6a/Utx and Kdm5c are histone demethylases that show female-biased expression, regulate transcription in the brain, and show incomplete functional overlap with their Y-chromosome paralogs Uty and Kdm5d (Iwase et al., 2016; Scandaglia et al., 2017; Tang et al., 2017; Davis et al., 2020; Cabrera Zapata et al., 2022). Therefore, sex hormones and sex chromosome-expressed genes both likely influence the sex-biased expression of hippocampal genes we observe.
Sex-biased alternative splicing can also lead to downstream sex bias in the transcriptome of the hippocampus. We found sex-biased alternative splicing of Hmga1 with male-biased inclusion of the splice isoform corresponding to long exon-containing Hmga1a over the short exon-containing Hmga1b (Figure 5B). HMGA1 is a chromatin modifier and splicing protein whose binding results in more open chromatin (Benecke and Eilebrecht, 2015; Sumter et al., 2016). While the function of the different splice isoforms in chromatin accessibility is not understood, a previous study in female human cell lines has shown that Hmga1a, but not Hmga1b, regulates the splicing pattern of presenilin-2 seen in sporadic Alzheimer’s disease (Manabe et al., 2003). HMGA1a has also been shown to regulate the splicing and DNA binding of estrogen receptor α in breast cancer cells (Massaad-Massade et al., 2002; Ohe et al., 2018). Another gene known to regulate splicing, Miat/Gomafu, also exhibited sex-biased alternative splicing in the young adult hippocampus (Figure 5B). Gomafu expression has been shown to be regulated by estrogen in breast cancer cells (Li et al., 2018) and by synaptic activity in neurons (Barry et al., 2014; Zakutansky and Feng, 2022), and although many alternatively spliced Gomafu isoforms have been identified, little is known about their distinct functions (Zakutansky and Feng, 2022). Therefore, the interplay of sex hormones and sex-biased expression and alternative splicing of regulatory factors may contribute to downstream sex-biased patterns in the hippocampus.
This is the first study, to the best of our knowledge, that examines sex differences in chromatin accessibility with aging in the hippocampus. Our finding that chromatin structure is more open with aging in both sexes (Figures 6C,D) is consistent with previous findings in male hippocampal neurons (Zhang et al., 2022) and with the “heterochromatin loss” model of aging (Villeponteau, 1997; Tsurumi and Li, 2012; Pal and Tyler, 2016). This model suggests that there is a loss of heterochromatin during aging, leading to alterations in the expression of genes residing in these regions (Villeponteau, 1997). Chromatin accessibility is associated with active regulatory elements and is regulated by chromatin and histone modifiers. Histone variants and histone post-translational modifications are associated with open or closed chromatin, and, in general, CpG island methylation is correlated with decreased chromatin accessibility (Collings and Anderson, 2017; Cramer, 2019; Andersson and Sandelin, 2020). Changes in the expression of histone proteins, as well as altered histone modifications and DNA hypo- or hyper-methylation, are widely documented to occur during aging; indeed, aging-related changes in DNA methylation serve as the basis of epigenetic clocks (Horvath et al., 2016; Pal and Tyler, 2016; Simpson and Chandra, 2021; Lu et al., 2023). Therefore, loss of histone proteins, loss of repressive histone marks, and changes in DNA methylation likely contribute to the aging-related opening in chromatin accessibility we detected in the male and female hippocampus.
Our results show that most of the aging-related increases in chromatin accessibility occurred in intergenic and intronic regions, many of which contained retrotransposon LINE-1-derived sequences (Figure 7C). Furthermore, we found increased expression of LINE-1 transcripts with aging in the hippocampus (Figure 7E). Increased retrotransposon activity has been reported to accompany aging in both humans and rodents, and the brain has been reported to exhibit more retrotransposon activity than other tissues (Barter and Foster, 2018; Zhou and Smith, 2019; Gorbunova et al., 2021; Zhang et al., 2022). In young mammalian cells, the histone deacetylase SIRT6 binds to LINE-1 retrotransposon sequences and represses their activity by modulating the heterochromatin status of nearby regions. However, during aging, SIRT6 is depleted from LINE-1 regions, and histone repressive marks and CpG methylation are lost in transposable element regions, leading to the expression of LINE-1 transcripts and the accompanying DNA damage and cellular immune activation. Furthermore, repression of aging-related retrotransposon activity through overexpression of SIRT6 has been shown to increase lifespan (Van Meter et al., 2014; Pal and Tyler, 2016; Gorbunova et al., 2021). We found that full-length intact LINE-1 regions in both male and female hippocampus remained robustly inaccessible with aging (Supplementary Figure S4C), suggesting that transposition-capable sequences as a whole remain suppressed, although individual instances of full-length LINE-1 element activity may occur. LINE-1 derived sequences that opened with aging were truncated sequences incapable of retrotransposition; however, the expression of truncated LINE-1 transcripts has been reported to cause cellular inflammation and DNA damage (Kines et al., 2014; De Cecco et al., 2019; Simon et al., 2019). Thus, loss of repressive histone marks and CpG methylation at LINE-1 regions likely contributed to the aging-related opening of chromatin at those regions and to the increased LINE-1 expression we observed in the male and female hippocampus.
We found that male and female hippocampus showed sex differences in chromatin accessibility on autosomes, and the number of regions with sex-biased accessibility increased with aging (Figure 8D). These differences could be a result of sex-biased expression or activity of histone and/or DNA methylation modifiers. Sex hormones have been shown to affect DNA methylation in human blood (Harbs et al., 2023) and chromatin accessibility in the female ventral hippocampus of young adult mice (Jaric et al., 2019). We detected few sex differences in chromatin accessibility in the young adult hippocampus (Figure 8A); however, differences in aging-related DNA methylation between sexes could contribute to the sex-biased accessibility observed in the aged hippocampus. Indeed, it has been reported that aging-related DNA methylation changes differed between male and female hippocampus in mice (Masser et al., 2017). Furthermore, studies in human blood cells have shown aging-related sex differences in DNA methylation (Li X. et al., 2020; Yusipov et al., 2020; Vershinina et al., 2021). We found male-biased chromatin accessibility in a subset of promoters and CpG-rich regions (Figures 8C,D; Supplementary Figure S6), raising the possibility that CpGs in these regions could be hypermethylated in female compared to male hippocampus. We did not detect sex bias in the expression of DNA methylation-related proteins; therefore, DNA methylation patterns established in development, or other mechanisms regulating the activity of DNA methylation-related proteins, may contribute to these differences in accessibility.
Alternatively, or in conjunction with sex-biased DNA methylation, sex differences in histone modifications may contribute to the male-biased accessibility in promoter regions we observed. As mentioned previously, we detected female-biased expression of the X chromosome-linked histone modifiers Kdm6a and Kdm5c. Kdm5c is a histone demethylase that removes methyl groups from H3K4me2/3 (Brookes et al., 2015; Scandaglia et al., 2017; Ugur et al., 2023). H3K4me3 is a histone mark associated with active promoters, and Kdm5c has been found to reduce H3K4 trimethylation at promoters and suppress transcription (Iwase et al., 2016; Scandaglia et al., 2017; Link et al., 2020). Kdm5c escapes X-inactivation and has been shown to exhibit female-biased expression in the brain (Xu et al., 2008). We found that Kdm5c showed female-biased expression in both young adult and aged hippocampus. In young adult and aged males, the Y-chromosome paralog, Kdm5d, was expressed at slightly lower levels than Kdm5c. Although we found the summed Kdm5c/d expression in males is higher than Kdm5c expression in the female hippocampus, Kdm5d is unable to compensate for Kdm5c function in the brain (Samanta et al., 2022). Indeed, loss-of-function mutations in Kdm5c reduce H3K4me2/3 demethylation in a subset of promoters and enhancers and have been shown to lead to a form of X-linked intellectual disability in males (Jensen et al., 2005; Brookes et al., 2015). Kdm5c shows preferential binding to promoters, especially those containing CpG islands (Iwase et al., 2016), and we found male-biased accessibility at promoters and CpG-rich regions. Therefore, one possibility is that female-biased expression of Kdm5c may result in greater demethylation of H3K4me3 at promoters in the female hippocampus and contribute to lower chromatin accessibility in those regions. A previous study found promoter CpG sites with reduced DNA methylation in the blood of male patients with X-linked intellectual disability and mutations in Kdm5c (Grafodatskaya et al., 2013). Furthermore, the authors showed that these promoter regions also showed female-biased DNA hypermethylation in the brain of controls, as well as DNA hypermethylation that correlated with Kdm5c/Kdm5d dosage (Grafodatskaya et al., 2013). Therefore, female-biased Kdm5c expression may lead to greater histone demethylation and DNA methylation at promoters in females, thereby contributing to the male bias in promoter accessibility we observed. Notably, the male bias in promoter accessibility is not sufficient to drive sex-biased expression of the associated genes. Therefore, other mechanisms may compensate for sex differences in promoter accessibility to regulate expression.
In conclusion, this study provides a comprehensive map of gene expression, splicing, and chromatin accessibility changes in the hippocampus during aging in both male and female mice. Our findings of sex-biased alternative splicing and aging-dependent sex bias in expression and chromatin accessibility illustrate the importance of including female animals in studies of brain aging. These findings also provide novel starting points for future research on chromatin structure and transcriptomics in the hippocampus that can guide treatments for aging-related memory decline.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was approved by UCLA Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
JA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. YT: Conceptualization, Funding acquisition, Investigation, Methodology, Validation, Writing – original draft. FG: Formal analysis, Visualization, Writing – review & editing. C-HL: Formal analysis, Writing – review & editing. MW: Investigation, Validation, Visualization, Writing – review & editing. SN: Methodology, Writing – review & editing. GC: Funding acquisition, Supervision, Writing – review & editing. DB: Funding acquisition, Supervision, Writing – review & editing. KM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by funding from the David Geffen Foundation. YT was supported by the China Scholarship Council CSC-UCLA Scholarship, Whitcome Pre-doctoral Fellowship, and UCLA Dissertation Year Fellowship.
The authors would like to thank the UCLA Broad Stem Cell Research Center Sequencing Core and the UCLA Neuroscience Genomic Core for data collection and Yue Qin for data analysis support. We also thank Patrick B. Chen for comments on the manuscript. The contents of this manuscript have been made available as a preprint (Achiro et al., 2023).
DB has equity and serves on the board of directors for Panorama Medicine. GC is currently employed by Regeneron Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2024.1334862/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE244506
2. ^https://github.com/vsbuffalo/scythe/
4. ^https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE244506
Achiro, J. M., Tao, Y., Gao, F., Lin, C.-H., Watanabe, M., Neumann, S., et al. (2023). Aging differentially alters the transcriptome and landscape of chromatin accessibility in the male and female mouse Hippocampus. bioRxiv. doi: 10.1101/2023.10.17.562606
Allen, W. E., Blosser, T. R., Sullivan, Z. A., Dulac, C., and Zhuang, X. (2023). Molecular and spatial signatures of mouse brain aging at single-cell resolution. Cell 186, 194–208.e18. doi: 10.1016/j.cell.2022.12.010
Allen, T. A., and Fortin, N. J. (2013). The evolution of episodic memory. Proc. Natl. Acad. Sci. U. S. A. 110, 10379–10386. doi: 10.1073/pnas.1301199110
Amemiya, H. M., Kundaje, A., and Boyle, A. P. (2019). The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9:9354. doi: 10.1038/s41598-019-45839-z
Andersson, R., Gebhard, C., Miguel-Escalada, I., Hoof, I., Bornholdt, J., Boyd, M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. doi: 10.1038/nature12787
Andersson, R., and Sandelin, A. (2020). Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87. doi: 10.1038/s41576-019-0173-8
Anstey, K. J., Peters, R., Mortby, M. E., Kiely, K. M., Eramudugolla, R., Cherbuin, N., et al. (2021). Association of sex differences in dementia risk factors with sex differences in memory decline in a population-based cohort spanning 20–76 years. Sci. Rep. 11:7710. doi: 10.1038/s41598-021-86397-7
Asih, P. R., Prikas, E., Stefanoska, K., Tan, A. R. P., Ahel, H. I., and Ittner, A. (2020). Functions of p38 MAP kinases in the central nervous system. Front. Mol. Neurosci. 13:570586. doi: 10.3389/fnmol.2020.570586
Babaev, O., Cruces-Solis, H., Piletti Chatain, C., Hammer, M., Wenger, S., Ali, H., et al. (2018). IgSF9b regulates anxiety behaviors through effects on centromedial amygdala inhibitory synapses. Nat. Commun. 9:5400. doi: 10.1038/s41467-018-07762-1
Bambah-Mukku, D., Travaglia, A., Chen, D. Y., Pollonini, G., and Alberini, C. M. (2014). A positive autoregulatory BDNF feedback loop via C/EBPβ mediates hippocampal memory consolidation. J. Neurosci. 34, 12547–12559. doi: 10.1523/JNEUROSCI.0324-14.2014
Barrientos, R. M., Kitt, M. M., Watkins, L. R., and Maier, S. F. (2015). Neuroinflammation in the normal aging hippocampus. Neuroscience 309, 84–99. doi: 10.1016/j.neuroscience.2015.03.007
Barry, G., Briggs, J. A., Vanichkina, D. P., Poth, E. M., Beveridge, N. J., Ratnu, V. S., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494. doi: 10.1038/mp.2013.45
Barter, J. D., and Foster, T. C. (2018). Aging in the brain: new roles of epigenetics in cognitive decline. Neuroscientist 24, 516–525. doi: 10.1177/1073858418780971
Beam, C. R., Kaneshiro, C., Jang, J. Y., Reynolds, C. A., Pedersen, N. L., and Gatz, M. (2018). Differences between women and men in incidence rates of dementia and Alzheimer’s disease. J. Alzheimers Dis. 64, 1077–1083. doi: 10.3233/JAD-180141
Benecke, A. G., and Eilebrecht, S. (2015). RNA-mediated regulation of HMGA1 function. Biomol. Ther. 5, 943–957. doi: 10.3390/biom5020943
Benice, T. S., Rizk, A., Kohama, S., Pfankuch, T., and Raber, J. (2006). Sex-differences in age-related cognitive decline in C57BL/6J mice associated with increased brain microtubule-associated protein 2 and synaptophysin immunoreactivity. Neuroscience 137, 413–423. doi: 10.1016/j.neuroscience.2005.08.029
Berchtold, N. C., Cribbs, D. H., Coleman, P. D., Rogers, J., Head, E., Kim, R., et al. (2008). Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. U. S. A. 105, 15605–15610. doi: 10.1073/pnas.0806883105
Bhadra, M., Howell, P., Dutta, S., Heintz, C., and Mair, W. B. (2020). Alternative splicing in aging and longevity. Hum. Genet. 139, 357–369. doi: 10.1007/s00439-019-02094-6
Bieri, G., Schroer, A. B., and Villeda, S. A. (2023). Blood-to-brain communication in aging and rejuvenation. Nat. Neurosci. 26, 379–393. doi: 10.1038/s41593-022-01238-8
Bird, C. M., and Burgess, N. (2008). The hippocampus and memory: insights from spatial processing. Nat. Rev. Neurosci. 9, 182–194. doi: 10.1038/nrn2335
Brookes, E., Laurent, B., Õunap, K., Carroll, R., Moeschler, J. B., Field, M., et al. (2015). Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 24, 2861–2872. doi: 10.1093/hmg/ddv046
Brose, N. (2008). For better or for worse: complexins regulate SNARE function and vesicle fusion. Traffic 9, 1403–1413. doi: 10.1111/j.1600-0854.2008.00758.x
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. doi: 10.1038/nmeth.2688
Buenrostro, J. D., Wu, B., Chang, H. Y., and Greenleaf, W. J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:21.29.21-21.29.29. doi: 10.1002/0471142727.mb2129s109
Buraei, Z., and Yang, J. (2010). The ß subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506. doi: 10.1152/physrev.00057.2009
Cabrera Zapata, L. E., Cambiasso, M. J., and Arevalo, M. A. (2022). Epigenetic modifier Kdm6a/Utx controls the specification of hypothalamic neuronal subtypes in a sex-dependent manner. Front. Cell Dev. Biol. 10. doi: 10.3389/fcell.2022.937875
Cai, Z., and Li, H. (2020). An updated review: androgens and cognitive impairment in older men. Front. Endocrinol. 11. doi: 10.3389/fendo.2020.586909
Carlock, C., Wu, J., Shim, J., Moreno-Gonzalez, I., Pitcher, M. R., Hicks, J., et al. (2017). Interleukin33 deficiency causes tau abnormality and neurodegeneration with Alzheimer-like symptoms in aged mice. Transl. Psychiatry 7:e1164. doi: 10.1038/tp.2017.142
Chapman, T. W., Olveda, G. E., Bame, X., Pereira, E., and Hill, R. A. (2023). Oligodendrocyte death initiates synchronous remyelination to restore cortical myelin patterns in mice. Nat. Neurosci. 26, 555–569. doi: 10.1038/s41593-023-01271-1
Collings, C. K., and Anderson, J. N. (2017). Links between DNA methylation and nucleosome occupancy in the human genome. Epigenetics Chromatin 10:18. doi: 10.1186/s13072-017-0125-5
Cramer, P. (2019). Organization and regulation of gene transcription. Nature 573, 45–54. doi: 10.1038/s41586-019-1517-4
Cribbs, D. H., Berchtold, N. C., Perreau, V., Coleman, P. D., Rogers, J., Tenner, A. J., et al. (2012). Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J. Neuroinflammation 9:179. doi: 10.1186/1742-2094-9-179
Cunningham, F., Allen, J. E., Allen, J., Alvarez-Jarreta, J., Amode, M. R., Armean, I. M., et al. (2021). Ensembl 2022. Nucleic Acids Res. 50, D988–D995.
Darbelli, L., Choquet, K., Richard, S., and Kleinman, C. L. (2017). Transcriptome profiling of mouse brains with qkI-deficient oligodendrocytes reveals major alternative splicing defects including self-splicing. Sci. Rep. 7:7554. doi: 10.1038/s41598-017-06211-1
Davis, E. J., Broestl, L., Abdulai-Saiku, S., Worden, K., Bonham, L. W., Miñones-Moyano, E., et al. (2020). A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci. Transl. Med. 12. doi: 10.1126/scitranslmed.aaz5677
Davis, E. J., Solsberg, C. W., White, C. C., Miñones-Moyano, E., Sirota, M., Chibnik, L., et al. (2021). Sex-specific Association of the X chromosome with cognitive change and tau pathology in aging and Alzheimer disease. JAMA Neurol. 78, 1249–1254. doi: 10.1001/jamaneurol.2021.2806
De Cecco, M., Criscione, S. W., Peterson, A. L., Neretti, N., Sedivy, J. M., and Kreiling, J. A. (2013). Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 5, 867–883. doi: 10.18632/aging.100621
De Cecco, M., Ito, T., Petrashen, A. P., Elias, A. E., Skvir, N. J., Criscione, S. W., et al. (2019). L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78. doi: 10.1038/s41586-018-0784-9
Deguchi, M., Hata, Y., Takeuchi, M., Ide, N., Hirao, K., Yao, I., et al. (1998). BEGAIN (brain-enriched guanylate kinase-associated protein), a novel neuronal PSD-95/SAP90-binding protein. J. Biol. Chem. 273, 26269–26272. doi: 10.1074/jbc.273.41.26269
Deng, X., Berletch, J. B., Nguyen, D. K., and Disteche, C. M. (2014). X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378. doi: 10.1038/nrg3687
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dunham, I., Kundaje, A., Aldred, S. F., Collins, P. J., Davis, C. A., Doyle, F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Eagle, A. L., Gajewski, P. A., and Robison, A. J. (2016). Role of hippocampal activity-induced transcription in memory consolidation. Rev. Neurosci. 27, 559–573. doi: 10.1515/revneuro-2016-0010
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Edinger, K. L., and Frye, C. A. (2007). Androgens’ effects to enhance learning may be mediated in part through actions at estrogen receptor-β in the hippocampus. Neurobiol. Learn. Mem. 87, 78–85. doi: 10.1016/j.nlm.2006.07.001
Elgersma, Y., Fedorov, N. B., Ikonen, S., Choi, E. S., Elgersma, M., Carvalho, O. M., et al. (2002). Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron 36, 493–505. doi: 10.1016/S0896-6273(02)01007-3
Ernst, W. L., and Noebels, J. L. (2009). Expanded alternative splice isoform profiling of the mouse Cav3.1/α1G T-type calcium channel. BMC Mol. Biol. 10:53. doi: 10.1186/1471-2199-10-53
Fang, H., Bonora, G., Lewandowski, J. P., Thakur, J., Filippova, G. N., Henikoff, S., et al. (2020). Trans- and cis-acting effects of Firre on epigenetic features of the inactive X chromosome. Nat. Commun. 11:6053. doi: 10.1038/s41467-020-19879-3
Fard, M. K., van der Meer, F., Sánchez, P., Cantuti-Castelvetri, L., Mandad, S., Jäkel, S., et al. (2017). BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med. 9:eaam7816. doi: 10.1126/scitranslmed.aam7816
Forner, S., Kawauchi, S., Balderrama-Gutierrez, G., Kramár, E. A., Matheos, D. P., Phan, J., et al. (2021). Systematic phenotyping and characterization of the 5xFAD mouse model of Alzheimer’s disease. Sci. Data 8:270. doi: 10.1038/s41597-021-01054-y
Franklin, R. J., and Goldman, S. A. (2015). Glia disease and repair-remyelination. Cold Spring Harb. Perspect. Biol. 7:a020594. doi: 10.1101/cshperspect.a020594
Franklin, R. J. M., and Simons, M. (2022). CNS remyelination and inflammation: from basic mechanisms to therapeutic opportunities. Neuron 110, 3549–3565. doi: 10.1016/j.neuron.2022.09.023
Fujita, N., Kemper, A., Dupree, J., Nakayasu, H., Bartsch, U., Schachner, M., et al. (1998). The cytoplasmic domain of the large myelin-associated glycoprotein isoform is needed for proper CNS but not peripheral nervous system myelination. J. Neurosci. 18, 1970–1978. doi: 10.1523/JNEUROSCI.18-06-01970.1998
Furlong, E. E. M., and Levine, M. (2018). Developmental enhancers and chromosome topology. Science 361, 1341–1345. doi: 10.1126/science.aau0320
Gal-Oz, S. T., Maier, B., Yoshida, H., Seddu, K., Elbaz, N., Czysz, C., et al. (2019). ImmGen report: sexual dimorphism in the immune system transcriptome. Nat. Commun. 10:4295. doi: 10.1038/s41467-019-12348-6
Galupa, R., and Heard, E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. doi: 10.1146/annurev-genet-120116-024611
Gao, S., Hendrie, H. C., Hall, K. S., and Hui, S. (1998). The relationships between age, sex, and the incidence of dementia and Alzheimer disease: a meta-analysis. Arch. Gen. Psychiatry 55, 809–815. doi: 10.1001/archpsyc.55.9.809
Gorbunova, V., Seluanov, A., Mita, P., McKerrow, W., Fenyö, D., Boeke, J. D., et al. (2021). The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53. doi: 10.1038/s41586-021-03542-y
Grabert, K., Michoel, T., Karavolos, M. H., Clohisey, S., Baillie, J. K., Stevens, M. P., et al. (2016). Microglial brain region−dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19, 504–516. doi: 10.1038/nn.4222
Grafodatskaya, D., Chung, B. H., Butcher, D. T., Turinsky, A. L., Goodman, S. J., Choufani, S., et al. (2013). Multilocus loss of DNA methylation in individuals with mutations in the histone H3 lysine 4 demethylase KDM5C. BMC Med. Genet. 6:1. doi: 10.1186/1755-8794-6-1
Guo, N., Soden, M. E., Herber, C., Kim, M. T., Besnard, A., Lin, P., et al. (2018). Dentate granule cell recruitment of feedforward inhibition governs engram maintenance and remote memory generalization. Nat. Med. 24, 438–449. doi: 10.1038/nm.4491
Hadad, N., Masser, D. R., Blanco-Berdugo, L., Stanford, D. R., and Freeman, W. M. (2019). Early-life DNA methylation profiles are indicative of age-related transcriptome changes. Epigenetics Chromatin 12:58. doi: 10.1186/s13072-019-0306-5
Hahn, O., Foltz, A. G., Atkins, M., Kedir, B., Moran-Losada, P., Guldner, I. H., et al. (2023). Atlas of the aging mouse brain reveals white matter as vulnerable foci. Cell 186, 4117–4133.e22. doi: 10.1016/j.cell.2023.07.027
Hajdarovic, K. H., Yu, D., Hassell, L.-A., Evans, S. A., Packer, S., Neretti, N., et al. (2022). Single-cell analysis of the aging female mouse hypothalamus. Nat. Aging 2, 662–678. doi: 10.1038/s43587-022-00246-4
Harauz, G., and Boggs, J. M. (2013). Myelin management by the 18.5-kDa and 21.5-kDa classic myelin basic protein isoforms. J. Neurochem. 125, 334–361. doi: 10.1111/jnc.12195
Harbs, J., Rinaldi, S., Keski-Rahkonen, P., Liu, X., Palmqvist, R., Van Guelpen, B., et al. (2023). An epigenome-wide analysis of sex hormone levels and DNA methylation in male blood samples. Epigenetics 18:2196759. doi: 10.1080/15592294.2023.2196759
Hass, J., Walton, E., Kirsten, H., Turner, J., Wolthusen, R., Roessner, V., et al. (2015). Complexin2 modulates working memory-related neural activity in patients with schizophrenia. Eur. Arch. Psychiatry Clin. Neurosci. 265, 137–145. doi: 10.1007/s00406-014-0550-4
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. doi: 10.1016/j.molcel.2010.05.004
Herlitz, A., and Rehnman, J. (2008). Sex differences in episodic memory. Curr. Dir. Psychol. Sci. 17, 52–56. doi: 10.1111/j.1467-8721.2008.00547.x
Hicks, S. C., Okrah, K., Paulson, J. N., Quackenbush, J., Irizarry, R. A., and Bravo, H. C. (2017). Smooth quantile normalization. Biostatistics 19, 185–198. doi: 10.1093/biostatistics/kxx028
Hofer, N. T., Pinggera, A., Nikonishyna, Y. V., Tuluc, P., Fritz, E. M., Obermair, G. J., et al. (2021). Stabilization of negative activation voltages of Cav1.3 L-type Ca2+−channels by alternative splicing. Channels 15, 38–52. doi: 10.1080/19336950.2020.1859260
Horvath, S., Gurven, M., Levine, M. E., Trumble, B. C., Kaplan, H., Allayee, H., et al. (2016). An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 17:171. doi: 10.1186/s13059-016-1030-0
Hu, Z., Liang, M. C., and Soong, T. W. (2017). Alternative splicing of L-type ca(V)1.2 calcium channels: implications in cardiovascular diseases. Genes (Basel) 8:344. doi: 10.3390/genes8120344
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Iram, T., Kern, F., Kaur, A., Myneni, S., Morningstar, A. R., Shin, H., et al. (2022). Young CSF restores oligodendrogenesis and memory in aged mice via Fgf17. Nature 605, 509–515. doi: 10.1038/s41586-022-04722-0
Ishimoto, T., Ninomiya, K., Inoue, R., Koike, M., Uchiyama, Y., and Mori, H. (2017). Mice lacking BCAS1, a novel myelin-associated protein, display hypomyelination, schizophrenia-like abnormal behaviors, and upregulation of inflammatory genes in the brain. Glia 65, 727–739. doi: 10.1002/glia.23129
Ittner, A., Chua, S. W., Bertz, J., Volkerling, A., van der Hoven, J., Gladbach, A., et al. (2016). Site-specific phosphorylation of tau inhibits amyloid-β toxicity in Alzheimer’s mice. Science 354, 904–908. doi: 10.1126/science.aah6205
Iwase, S., Brookes, E., Agarwal, S., Badeaux, A. I., Ito, H., Vallianatos, C. N., et al. (2016). A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep. 14, 1000–1009. doi: 10.1016/j.celrep.2015.12.091
Jack, C. R. Jr., Wiste, H. J., Weigand, S. D., Knopman, D. S., Vemuri, P., Mielke, M. M., et al. (2015). Age, sex, and APOE ε4 effects on memory, brain structure, and β-amyloid across the adult life span. JAMA Neurol. 72, 511–519. doi: 10.1001/jamaneurol.2014.4821
Jaric, I., Rocks, D., Greally, J. M., Suzuki, M., and Kundakovic, M. (2019). Chromatin organization in the female mouse brain fluctuates across the oestrous cycle. Nat. Commun. 10:2851. doi: 10.1038/s41467-019-10704-0
Jensen, L. R., Amende, M., Gurok, U., Moser, B., Gimmel, V., Tzschach, A., et al. (2005). Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236. doi: 10.1086/427563
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. doi: 10.1101/gr.229102
Khosla, S., Melton, L. J. III, Atkinson, E. J., O’Fallon, W. M., Klee, G. G., and Riggs, B. L. (1998). Relationship of serum sex steroid levels and bone turnover markers with bone mineral density in men and women: a key role for bioavailable Estrogen1. J. Clin. Endocrinol. Metabol. 83, 2266–2274.
Kines, K. J., Sokolowski, M., deHaro, D. L., Christian, C. M., and Belancio, V. P. (2014). Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 42, 10488–10502. doi: 10.1093/nar/gku687
Klein, S. L., and Flanagan, K. L. (2016). Sex differences in immune responses. Nat. Rev. Immunol. 16, 626–638. doi: 10.1038/nri.2016.90
Kouser, M., Speed, H. E., Dewey, C. M., Reimers, J. M., Widman, A. J., Gupta, N., et al. (2013). Loss of predominant Shank3 isoforms results in Hippocampus-dependent impairments in behavior and synaptic transmission. J. Neurosci. 33, 18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013
Kumar, A., Bodhinathan, K., and Foster, T. (2009). Susceptibility to calcium dysregulation during brain aging. Frontiers in aging. Neuroscience 1. doi: 10.3389/neuro.24.002.2009
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lanke, V., Moolamalla, S. T. R., Roy, D., and Vinod, P. K. (2018). Integrative analysis of Hippocampus gene expression profiles identifies network alterations in aging and Alzheimer’s disease. Front. Aging Neurosci. 10. doi: 10.3389/fnagi.2018.00153
Ledoux, V. A., Smejkalova, T., May, R. M., Cooke, B. M., and Woolley, C. S. (2009). Estradiol facilitates the release of neuropeptide Y to suppress hippocampus-dependent seizures. J. Neurosci. 29, 1457–1468. doi: 10.1523/JNEUROSCI.4688-08.2009
Li, Y., Jiang, B., Wu, X., Huang, Q., Chen, W., Zhu, H., et al. (2018). Long non-coding RNA MIAT is estrogen-responsive and promotes estrogen-induced proliferation in ER-positive breast cancer cells. Biochem. Biophys. Res. Commun. 503, 45–50. doi: 10.1016/j.bbrc.2018.05.146
Li, W. W., Penderis, J., Zhao, C., Schumacher, M., and Franklin, R. J. (2006). Females remyelinate more efficiently than males following demyelination in the aged but not young adult CNS. Exp. Neurol. 202, 250–254. doi: 10.1016/j.expneurol.2006.05.012
Li, X., Ploner, A., Wang, Y., Magnusson, P. K. E., Reynolds, C., Finkel, D., et al. (2020). Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. elife 9:e51507. doi: 10.7554/eLife.51507
Li, M., Su, S., Cai, W., Cao, J., Miao, X., Zang, W., et al. (2020). Differentially expressed genes in the brain of aging mice with cognitive alteration and depression- and anxiety-like behaviors. Front. Cell Dev. Biol. 8. doi: 10.3389/fcell.2020.00814
Li, Z., Zhang, Y., Li, D., and Feng, Y. (2000). Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 20, 4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Link, J. C., Wiese, C. B., Chen, X., Avetisyan, R., Ronquillo, E., Ma, F., et al. (2020). X chromosome dosage of histone demethylase KDM5C determines sex differences in adiposity. J. Clin. Invest. 130, 5688–5702. doi: 10.1172/JCI140223
Lipscombe, D., Andrade, A., and Allen, S. E. (2013). Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim. Biophys. Acta 1828, 1522–1529. doi: 10.1016/j.bbamem.2012.09.018
Long, J. M., Kalehua, A. N., Muth, N. J., Calhoun, M. E., Jucker, M., Hengemihle, J. M., et al. (1998). Stereological analysis of astrocyte and microglia in aging mouse hippocampus. Neurobiol. Aging 19, 497–503. doi: 10.1016/S0197-4580(98)00088-8
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2023). Hallmarks of aging: an expanding universe. Cell 186, 243–278. doi: 10.1016/j.cell.2022.11.001
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Low, K. L., Tomm, R. J., Ma, C., Tobiansky, D. J., Floresco, S. B., and Soma, K. K. (2020). Effects of aging on testosterone and androgen receptors in the mesocorticolimbic system of male rats. Horm. Behav. 120:104689. doi: 10.1016/j.yhbeh.2020.104689
Lu, A. T., Fei, Z., Haghani, A., Robeck, T. R., Zoller, J. A., Li, C. Z., et al. (2023). Universal DNA methylation age across mammalian tissues. Nat. Aging 3, 1144–1166. doi: 10.1038/s43587-023-00462-6
Manabe, T., Katayama, T., Sato, N., Gomi, F., Hitomi, J., Yanagita, T., et al. (2003). Induced HMGA1a expression causes aberrant splicing of Presenilin-2 pre-mRNA in sporadic Alzheimer’s disease. Cell Death Differ. 10, 698–708. doi: 10.1038/sj.cdd.4401221
Mangold, C. A., Wronowski, B., Du, M., Masser, D. R., Hadad, N., Bixler, G. V., et al. (2017). Sexually divergent induction of microglial-associated neuroinflammation with hippocampal aging. J. Neuroinflammation 14:141. doi: 10.1186/s12974-017-0920-8
Marco, A., Meharena, H. S., Dileep, V., Raju, R. M., Davila-Velderrain, J., Zhang, A. L., et al. (2020). Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 23, 1606–1617. doi: 10.1038/s41593-020-00717-0
Massaad-Massade, L., Navarro, S., Krummrei, U., Reeves, R., Beaune, P., and Barouki, R. (2002). HMGA1 enhances the transcriptional activity and binding of the estrogen receptor to its responsive element. Biochemistry 41, 2760–2768. doi: 10.1021/bi011455j
Masser, D. R., Hadad, N., Porter, H. L., Mangold, C. A., Unnikrishnan, A., Ford, M. M., et al. (2017). Sexually divergent DNA methylation patterns with hippocampal aging. Aging Cell 16, 1342–1352. doi: 10.1111/acel.12681
Massie, A., Boillée, S., Hewett, S., Knackstedt, L., and Lewerenz, J. (2015). Main path and byways: non-vesicular glutamate release by system xc(−) as an important modifier of glutamatergic neurotransmission. J. Neurochem. 135, 1062–1079. doi: 10.1111/jnc.13348
McCarrey, A. C., An, Y., Kitner-Triolo, M. H., Ferrucci, L., and Resnick, S. M. (2016). Sex differences in cognitive trajectories in clinically normal older adults. Psychol. Aging 31, 166–175. doi: 10.1037/pag0000070
McQuail, J. A., Frazier, C. J., and Bizon, J. L. (2015). Molecular aspects of age-related cognitive decline: the role of GABA signaling. Trends Mol. Med. 21, 450–460. doi: 10.1016/j.molmed.2015.05.002
Moortgat, S., Berland, S., Aukrust, I., Maystadt, I., Baker, L., Benoit, V., et al. (2018). HUWE1 variants cause dominant X-linked intellectual disability: a clinical study of 21 patients. Eur. J. Hum. Genet. 26, 64–74. doi: 10.1038/s41431-017-0038-6
Morrison, J. H., and Baxter, M. G. (2012). The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat. Rev. Neurosci. 13, 240–250. doi: 10.1038/nrn3200
Mosher, K. I., and Wyss-Coray, T. (2014). Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem. Pharmacol. 88, 594–604. doi: 10.1016/j.bcp.2014.01.008
Müller, C., Bauer, N. M., Schäfer, I., and White, R. (2013). Making myelin basic protein -from mRNA transport to localized translation. Front. Cell. Neurosci. 7:169. doi: 10.3389/fncel.2013.00169
Nabavi, S., Fox, R., Proulx, C. D., Lin, J. Y., Tsien, R. Y., and Malinow, R. (2014). Engineering a memory with LTD and LTP. Nature 511, 348–352. doi: 10.1038/nature13294
Nahvi, R. J., and Sabban, E. L. (2020). Sex differences in the neuropeptide Y system and implications for stress related disorders. Biomol. Ther. 10. doi: 10.3390/biom10091248
Nave, K. A., and Werner, H. B. (2014). Myelination of the nervous system: mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533. doi: 10.1146/annurev-cellbio-100913-013101
Nikoletopoulou, V., and Tavernarakis, N. (2012). Calcium homeostasis in aging neurons. Front. Genet. 3. doi: 10.3389/fgene.2012.00200
Nyberg, L., Lövdén, M., Riklund, K., Lindenberger, U., and Bäckman, L. (2012). Memory aging and brain maintenance. Trends Cogn. Sci. 16, 292–305. doi: 10.1016/j.tics.2012.04.005
Oh, M. M., Oliveira, F. A., Waters, J., and Disterhoft, J. F. (2013). Altered calcium metabolism in aging CA1 hippocampal pyramidal neurons. J. Neurosci. 33, 7905–7911. doi: 10.1523/JNEUROSCI.5457-12.2013
Ohe, K., Miyajima, S., Abe, I., Tanaka, T., Hamaguchi, Y., Harada, Y., et al. (2018). HMGA1a induces alternative splicing of estrogen receptor alpha in MCF-7 human breast cancer cells. J. Steroid Biochem. Mol. Biol. 182, 21–26. doi: 10.1016/j.jsbmb.2018.04.007
Ou, J., Liu, H., Yu, J., Kelliher, M. A., Castilla, L. H., Lawson, N. D., et al. (2018). ATACseqQC: a bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 19:169. doi: 10.1186/s12864-018-4559-3
Pal, S., and Tyler, J. K. (2016). Epigenetics and aging. Sci. Adv. 2:e1600584. doi: 10.1126/sciadv.1600584
Pandya-Jones, A., and Plath, K. (2016). The “lnc” between 3D chromatin structure and X chromosome inactivation. Semin. Cell Dev. Biol. 56, 35–47. doi: 10.1016/j.semcdb.2016.04.002
Peleg, S., Sananbenesi, F., Zovoilis, A., Burkhardt, S., Bahari-Javan, S., Agis-Balboa, R. C., et al. (2010). Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756. doi: 10.1126/science.1186088
Penzkofer, T., Jäger, M., Figlerowicz, M., Badge, R., Mundlos, S., Robinson, P. N., et al. (2017). L1Base 2: more retrotransposition-active LINE-1s, more mammalian genomes. Nucleic Acids Res. 45, D68–d73. doi: 10.1093/nar/gkw925
Pereda, D., Al-Osta, I., Okorocha, A. E., Easton, A., and Hartell, N. A. (2019). Changes in presynaptic calcium signalling accompany age-related deficits in hippocampal LTP and cognitive impairment. Aging Cell 18:e13008. doi: 10.1111/acel.13008
Peters, A. (2002). The effects of normal aging on myelin and nerve fibers: a review. J. Neurocytol. 31, 581–593. doi: 10.1023/A:1025731309829
Pinggera, A., Lieb, A., Benedetti, B., Lampert, M., Monteleone, S., Liedl, K. R., et al. (2015). CACNA1D de novo mutations in autism spectrum disorders activate Cav1.3 L-type calcium channels. Biol. Psychiatry 77, 816–822. doi: 10.1016/j.biopsych.2014.11.020
Pinggera, A., Mackenroth, L., Rump, A., Schallner, J., Beleggia, F., Wollnik, B., et al. (2017). New gain-of-function mutation shows CACNA1D as recurrently mutated gene in autism spectrum disorders and epilepsy. Hum. Mol. Genet. 26, 2923–2932. doi: 10.1093/hmg/ddx175
Pinggera, A., and Striessnig, J. (2016). Ca(v) 1.3 (CACNA1D) L-type ca(2+) channel dysfunction in CNS disorders. J. Physiol. 594, 5839–5849. doi: 10.1113/JP270672
Quarles, R. H. (2007). Myelin-associated glycoprotein (MAG): past, present and beyond. J. Neurochem. 100, 1431–1448. doi: 10.1111/j.1471-4159.2006.04319.x
Rangwala, S. H., Zhang, L., and Kazazian, H. H. Jr. (2009). Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 10:R100. doi: 10.1186/gb-2009-10-9-r100
Reilly, M. T., Faulkner, G. J., Dubnau, J., Ponomarev, I., and Gage, F. H. (2013). The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 33, 17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Rivera, A. D., Pieropan, F., Chacon-De-La-Rocha, I., Lecca, D., Abbracchio, M. P., Azim, K., et al. (2021). Functional genomic analyses highlight a shift in Gpr17-regulated cellular processes in oligodendrocyte progenitor cells and underlying myelin dysregulation in the aged mouse cerebrum. Aging Cell 20:e13335. doi: 10.1111/acel.13335
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Rozycka, A., and Liguz-Lecznar, M. (2017). The space where aging acts: focus on the GABAergic synapse. Aging Cell 16, 634–643. doi: 10.1111/acel.12605
Samanta, M. K., Gayen, S., Harris, C., Maclary, E., Murata-Nakamura, Y., Malcore, R. M., et al. (2022). Activation of Xist by an evolutionarily conserved function of KDM5C demethylase. Nat. Commun. 13:2602. doi: 10.1038/s41467-022-30352-1
Scandaglia, M., Lopez-Atalaya, J. P., Medrano-Fernandez, A., Lopez-Cascales, M. T., Del Blanco, B., Lipinski, M., et al. (2017). Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep. 21, 47–59. doi: 10.1016/j.celrep.2017.09.014
Schlüter, O. M., Schmitz, F., Jahn, R., Rosenmund, C., and Südhof, T. C. (2004). A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004
Shankar, S., Teyler, T. J., and Robbins, N. (1998). Aging differentially alters forms of long-term potentiation in rat hippocampal area CA1. J. Neurophysiol. 79, 334–341. doi: 10.1152/jn.1998.79.1.334
Shen, S., Park, J. W., Lu, Z. X., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 111, E5593–E5601. doi: 10.1073/pnas.1419161111
Sheng, J. G., Mrak, R. E., and Griffin, W. S. (1998). Enlarged and phagocytic, but not primed, interleukin-1 alpha-immunoreactive microglia increase with age in normal human brain. Acta Neuropathol. 95, 229–234. doi: 10.1007/s004010050792
Sim, F. J., Zhao, C., Penderis, J., and Franklin, R. J. M. (2002). The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J. Neurosci. 22, 2451–2459. doi: 10.1523/JNEUROSCI.22-07-02451.2002
Simon, M., Van Meter, M., Ablaeva, J., Ke, Z., Gonzalez, R. S., Taguchi, T., et al. (2019). LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885.e5. doi: 10.1016/j.cmet.2019.02.014
Simpson, D. J., and Chandra, T. (2021). Epigenetic age prediction. Aging Cell 20:e13452. doi: 10.1111/acel.13452
Spittau, B. (2017). Aging microglia-phenotypes, functions and implications for age-related neurodegenerative diseases. Front. Aging Neurosci. 9:194. doi: 10.3389/fnagi.2017.00194
Stilling, R. M., Benito, E., Gertig, M., Barth, J., Capece, V., Burkhardt, S., et al. (2014). De-regulation of gene expression and alternative splicing affects distinct cellular pathways in the aging hippocampus. Front. Cell. Neurosci. 8:373. doi: 10.3389/fncel.2014.00373
Su, Y., Shin, J., Zhong, C., Wang, S., Roychowdhury, P., Lim, J., et al. (2017). Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 20, 476–483. doi: 10.1038/nn.4494
Sugiyama, I., Tanaka, K., Akita, M., Yoshida, K., Kawase, T., and Asou, H. (2002). Ultrastructural analysis of the paranodal junction of myelinated fibers in 31-month-old-rats. J. Neurosci. Res. 70, 309–317. doi: 10.1002/jnr.10386
Sumter, T. F., Xian, L., Huso, T., Koo, M., Chang, Y. T., Almasri, T. N., et al. (2016). The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr. Mol. Med. 16, 353–393. doi: 10.2174/1566524016666160316152147
Tanabe, Y., Naito, Y., Vasuta, C., Lee, A. K., Soumounou, Y., Linhoff, M. W., et al. (2017). IgSF21 promotes differentiation of inhibitory synapses via binding to neurexin2α. Nat. Commun. 8:408. doi: 10.1038/s41467-017-00333-w
Tang, G.-B., Zeng, Y.-Q., Liu, P.-P., Mi, T.-W., Zhang, S.-F., Dai, S.-K., et al. (2017). The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front. Mol. Neurosci. 10. doi: 10.3389/fnmol.2017.00267
Taxier, L. R., Gross, K. S., and Frick, K. M. (2020). Oestradiol as a neuromodulator of learning and memory. Nat. Rev. Neurosci. 21, 535–550. doi: 10.1038/s41583-020-0362-7
Tereshko, L., Gao, Y., Cary, B. A., Turrigiano, G. G., and Sengupta, P. (2021). Ciliary neuropeptidergic signaling dynamically regulates excitatory synapses in postnatal neocortical pyramidal neurons. elife 10:e65427. doi: 10.7554/eLife.65427
Trabzuni, D., Ramasamy, A., Imran, S., Walker, R., Smith, C., Weale, M. E., et al. (2013). Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 4:2771. doi: 10.1038/ncomms3771
Tropak, M. B., Johnson, P. W., Dunn, R. J., and Roder, J. C. (1988). Differential splicing of MAG transcripts during CNS and PNS development. Brain Res. 464, 143–155. doi: 10.1016/0169-328X(88)90006-X
Tsurumi, A., and Li, W. X. (2012). Global heterochromatin loss: a unifying theory of aging? Epigenetics 7, 680–688. doi: 10.4161/epi.20540
Uchigashima, M., Cheung, A., and Futai, K. (2021). Neuroligin-3: a circuit-specific synapse organizer that shapes normal function and autism Spectrum disorder-associated dysfunction. Front. Mol. Neurosci. 14. doi: 10.3389/fnmol.2021.749164
Uddin, R. K., and Singh, S. M. (2013). Hippocampal gene expression meta-analysis identifies aging and age-associated spatial learning impairment (ASLI) genes and pathways. PLoS One 8:e69768. doi: 10.1371/journal.pone.0069768
Ugur, F. S., Kelly, M. J. S., and Fujimori, D. G. (2023). Chromatin sensing by the auxiliary domains of KDM5C regulates its demethylase activity and is disrupted by X-linked intellectual disability mutations. J. Mol. Biol. 435:167913. doi: 10.1016/j.jmb.2022.167913
Van den Berge, K., Chou, H. J., Roux de Bézieux, H., Street, K., Risso, D., Ngai, J., et al. (2022). Normalization benchmark of ATAC-seq datasets shows the importance of accounting for GC-content effects. Cell Rep. Methods 2:100321. doi: 10.1016/j.crmeth.2022.100321
Van Meter, M., Kashyap, M., Rezazadeh, S., Geneva, A. J., Morello, T. D., Seluanov, A., et al. (2014). SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5:5011. doi: 10.1038/ncomms6011
Verbruggen, L., Ates, G., Lara, O., De Munck, J., Villers, A., De Pauw, L., et al. (2022). Lifespan extension with preservation of hippocampal function in aged system xc−−deficient male mice. Mol. Psychiatry 27, 2355–2368. doi: 10.1038/s41380-022-01470-5
Vershinina, O., Bacalini, M. G., Zaikin, A., Franceschi, C., and Ivanchenko, M. (2021). Disentangling age-dependent DNA methylation: deterministic, stochastic, and nonlinear. Sci. Rep. 11:9201. doi: 10.1038/s41598-021-88504-0
Villeponteau, B. (1997). The heterochromatin loss model of aging. Exp. Gerontol. 32, 383–394. doi: 10.1016/S0531-5565(96)00155-6
Von Bohlen Und Halbach, O., Zacher, C., Gass, P., and Unsicker, K. (2006). Age-related alterations in hippocampal spines and deficiencies in spatial memory in mice. J. Neurosci. Res. 83, 525–531. doi: 10.1002/jnr.20759
Walter, J., Keiner, S., Witte, O. W., and Redecker, C. (2011). Age-related effects on hippocampal precursor cell subpopulations and neurogenesis. Neurobiol. Aging 32, 1906–1914. doi: 10.1016/j.neurobiolaging.2009.11.011
Wimmer, M. E., Hernandez, P. J., Blackwell, J., and Abel, T. (2012). Aging impairs hippocampus-dependent long-term memory for object location in mice. Neurobiol. Aging 33, 2220–2224. doi: 10.1016/j.neurobiolaging.2011.07.007
Woodruff, R. H., and Franklin, R. J. (1998). The expression of myelin basic protein exon 1 and exon 2 containing transcripts during myelination of the neonatal rat spinal cord--an in situ hybridization study. J. Neurocytol. 27, 683–693. doi: 10.1023/A:1006972316697
Wu, J. I., Reed, R. B., Grabowski, P. J., and Artzt, K. (2002). Function of quaking in myelination: regulation of alternative splicing. Proc. Natl. Acad. Sci. U. S. A. 99, 4233–4238. doi: 10.1073/pnas.072090399
Ximerakis, M., Lipnick, S. L., Innes, B. T., Simmons, S. K., Adiconis, X., Dionne, D., et al. (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708. doi: 10.1038/s41593-019-0491-3
Xin, W., and Chan, J. R. (2020). Myelin plasticity: sculpting circuits in learning and memory. Nat. Rev. Neurosci. 21, 682–694. doi: 10.1038/s41583-020-00379-8
Xu, J., Deng, X., and Disteche, C. M. (2008). Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3:e2553. doi: 10.1371/journal.pone.0002553
Yang, C., Bolotin, E., Jiang, T., Sladek, F. M., and Martinez, E. (2007). Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389, 52–65. doi: 10.1016/j.gene.2006.09.029
Yang, S., Li, C., Zhang, W., Wang, W., and Tang, Y. (2008). Sex differences in the white matter and myelinated nerve fibers of Long-Evans rats. Brain Res. 1216, 16–23. doi: 10.1016/j.brainres.2008.03.052
Yin, H., Wei, C., and Lee, J. T. (2021). Revisiting the consequences of deleting the X inactivation center. Proc. Natl. Acad. Sci. U. S. A. 118. doi: 10.1073/pnas.2102683118
Yusipov, I., Bacalini, M. G., Kalyakulina, A., Krivonosov, M., Pirazzini, C., Gensous, N., et al. (2020). Age-related DNA methylation changes are sex-specific: a comprehensive assessment. Aging (Albany NY) 12, 24057–24080. doi: 10.18632/aging.202251
Zakutansky, P. M., and Feng, Y. (2022). The long non-coding RNA GOMAFU in schizophrenia: function, disease risk, and beyond. Cell 11. doi: 10.3390/cells11121949
Zhang, Y., Amaral, M. L., Zhu, C., Grieco, S. F., Hou, X., Lin, L., et al. (2022). Single-cell epigenome analysis reveals age-associated decay of heterochromatin domains in excitatory neurons in the mouse brain. Cell Res. 32, 1008–1021. doi: 10.1038/s41422-022-00719-6
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. doi: 10.1186/gb-2008-9-9-r137
Zhang, Y., Lu, Z., Ku, L., Chen, Y., Wang, H., and Feng, Y. (2003). Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. EMBO J. 22, 1801–1810. doi: 10.1093/emboj/cdg171
Zhao, L., Mandler, M. D., Yi, H., and Feng, Y. (2010). Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl. Acad. Sci. 107, 19061–19066. doi: 10.1073/pnas.1007487107
Zhao, B., Wu, Q., Ye, A. Y., Guo, J., Zheng, X., Yang, X., et al. (2019). Somatic LINE-1 retrotransposition in cortical neurons and non-brain tissues of Rett patients and healthy individuals. PLoS Genet. 15:e1008043. doi: 10.1371/journal.pgen.1008043
Keywords: aging, sex bias, hippocampus, gene expression, alternative splicing, ATAC-seq, chromatin accessibility, LINE-1
Citation: Achiro JM, Tao Y, Gao F, Lin C-H, Watanabe M, Neumann S, Coppola G, Black DL and Martin KC (2024) Aging differentially alters the transcriptome and landscape of chromatin accessibility in the male and female mouse hippocampus. Front. Mol. Neurosci. 17:1334862. doi: 10.3389/fnmol.2024.1334862
Received: 07 November 2023; Accepted: 02 January 2024;
Published: 22 January 2024.
Edited by:
Jason D. Shepherd, The University of Utah, United StatesReviewed by:
Amrutha Swaminathan, Indian Institute of Science Education and Research, Thiruvananthapuram, IndiaCopyright © 2024 Achiro, Tao, Gao, Lin, Watanabe, Neumann, Coppola, Black and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer M. Achiro, amFjaGlyb0BtZWRuZXQudWNsYS5lZHU=; Kesley C. Martin, a2NtYXJ0aW5AbWVkbmV0LnVjbGEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.