
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 25 October 2023
Sec. Brain Disease Mechanisms
Volume 16 - 2023 | https://doi.org/10.3389/fnmol.2023.1253040
This article is part of the Research TopicMethods and Models of Retina Regeneration and Degeneration - Volume IIView all 4 articles
 Johanna L. Heinz1,2
Johanna L. Heinz1,2 Sigrid M. A. Swagemakers3
Sigrid M. A. Swagemakers3 Joanna von Hofsten4,5
Joanna von Hofsten4,5 Marie Helleberg6,7
Marie Helleberg6,7 Michelle M. Thomsen1,2
Michelle M. Thomsen1,2 Kerstin De Keukeleere1,2
Kerstin De Keukeleere1,2 Joke H. de Boer8
Joke H. de Boer8 Tomas Ilginis9
Tomas Ilginis9 Georges M. G. M. Verjans10
Georges M. G. M. Verjans10 Peter M. van Hagen11
Peter M. van Hagen11 Peter J. van der Spek3
Peter J. van der Spek3 Trine H. Mogensen1,2*
Trine H. Mogensen1,2*Purpose: Herpes simplex virus (HSV) and varicella-zoster virus (VZV) are neurotropic human alphaherpesviruses endemic worldwide. Upon primary infection, both viruses establish lifelong latency in neurons and reactivate intermittently to cause a variety of mild to severe diseases. Acute retinal necrosis (ARN) is a rare, sight-threatening eye disease induced by ocular VZV or HSV infection. The virus and host factors involved in ARN pathogenesis remain incompletely described. We hypothesize an underlying genetic defect in at least part of ARN cases.
Methods: We collected blood from 17 patients with HSV-or VZV-induced ARN, isolated DNA and performed Whole Exome Sequencing by Illumina followed by analysis in Varseq with criteria of CADD score > 15 and frequency in GnomAD < 0.1% combined with biological filters. Gene modifications relative to healthy control genomes were filtered according to high quality and read-depth, low frequency, high deleteriousness predictions and biological relevance.
Results: We identified a total of 50 potentially disease-causing genetic variants, including missense, frameshift and splice site variants and on in-frame deletion in 16 of the 17 patients. The vast majority of these genes are involved in innate immunity, followed by adaptive immunity, autophagy, and apoptosis; in several instances variants within a given gene or pathway was identified in several patients.
Discussion: We propose that the identified variants may contribute to insufficient viral control and increased necrosis ocular disease presentation in the patients and serve as a knowledge base and starting point for the development of improved diagnostic, prophylactic, and therapeutic applications.
Varicella-zoster virus (VZV) and herpes simplex virus (HSV)-1 and -2 are double-stranded (ds) DNA alphaherpesviruses which infect most individuals in a native community (Fields et al., 2013d), usually, leading to rather mild disease. VZV is most commonly acquired in early childhood during which primary infection gives rise to varicella (chickenpox). Subsequently, VZV becomes latent in dorsal root sensory neurons and cranial nerves and often reactivates later in life, then causing herpes zoster (shingles) (Fields et al., 2013c). Similarly, HSV becomes latent after primary infection in sensory ganglia; HSV-1 typically resides within trigeminal ganglia, HSV-2 within lumbar-and sacral ganglia. Classically, both primary infection and reactivation is followed by cold sores or genital herpes (Fields et al., 2013b). In rare cases, infection with both viruses may spread to the central nervous system (CNS), causing severe neurological disease, such as meningitis or encephalitis (Kleinschmidt-DeMasters and Gilden, 2001; Tyler, 2004; Alvarez et al., 2020).
Involvement of the ocular sensory nerves during alphaherpesvirus infection may lead to acute retinal necrosis (ARN), a rare and sight-threatening inflammation of the posterior part of the eye affecting various layers of the uvea, blood vessels, and full thickness necrosis of the retinal neuroepithelium, as described by the American Uveitis Society in 1994 (Holland, 1994). The exact molecular mechanism by which viral reactivation might lead to ARN onset and whether this pathway is the same for the different alphaherpesviruses remains incompletely understood. The primary site of latency most likely is the trigeminal ganglion with the virus spreading through the 1. branch of the trigeminal nerve, although reactivation from other sites may also be envisaged. ARN is a rare disease manifestation, the prevalence only amounting to 1 in 2 million individuals per year (Muthiah et al., 2007). The necrosis of affected tissues leads to various degrees of visual symptoms, including potentially permanent loss of central vision or the visual field due to retinal detachment, optic nerve or macula damage. In the absence of treatment the condition progresses rapidly and often with disadvantageous visual outcome in one or even both eyes. Unfortunately, even with systemic antiviral treatment the prognosis remains poor, with detachment of the retina as the most common complication (Lau et al., 2007; Hillenkamp et al., 2009; Iwahashi-Shima et al., 2013). VZV infection is most commonly associated with ARN, followed by HSV-1 and HSV-2 (Lau et al., 2007; Grahn and Studahl, 2015; Cox and Miller, 2022). Since many patients have reached adulthood before disease onset, it is reasonable to assume that ARN is most often a complication of reactivation rather than primary infection. Importantly, herpes simplex encephalitis (HSE) appears to be a risk factor for developing ARN later in life, highlighting the connections between the two phenotypes (Ahmadieh et al., 1991; Levinson et al., 1999; Ganatra et al., 2000; Maertzdorf et al., 2001; von Hofsten et al., 2019) and the origin of the retina as a neuroepithelium.
CNS infection by alphaherpesviruses preferentially affects individuals of the extremes of age, children and the elderly, as well as the immunocompromised (Whitley and Gnann, 2002; Wiegering et al., 2011; Fields et al., 2013a). However, occurrence in young, otherwise healthy individuals suggests a role for specific, so far undescribed inborn errors of immunity (IEI) (Kennedy and Mogensen, 2020; Herlin et al., 2021). Differences in virulence between different viral strains have also been suggested as a determinant of disease severity (Richards et al., 1981; Kolb et al., 2016; Pandey et al., 2017; Shipley et al., 2018; Pandey and Szpara, 2019). However, these observations cannot explain all interindividual variability in disease severity, thus implicating a major role of the host immune response in at least part of the cases (Brandt, 2005; Thompson et al., 2014; Kennedy and Mogensen, 2020). Clinical VZV isolates exhibit only minor sequence variation, which so far has not been conclusively connected to disease outcome in humans (Breuer, 2010). In a case of VZV ARN only minor sequence diversity in the viral genome was described (von Hofsten et al., 2020).
Numerous IEI have been described to cause severe disease after infection with VZV, HSV or other viral pathogens. Most strikingly, individuals with severe combined immunodeficiency (SCID) or natural killer (NK) deficiency are especially vulnerable to potentially fatal infection with a wide range of pathogens (Biron et al., 1989; Brooks et al., 1990). In addition, a number of single gene IEI have been demonstrated to underly increased susceptibility to severe viral infections, including mutations in GATA2 (Mace et al., 2013), DOCK2 (Dobbs et al., 2015), DOCK8 (Zhang et al., 2009), IFNGR1 (Roesler et al., 1999), TYK2 (Kreins et al., 2015), STAT1 (Dupuis et al., 2003), CORA1A (Yee et al., 2016), MCM4 (Gineau et al., 2012), and STK4 (Abdollahpour et al., 2012; Jørgensen et al., 2021). Moreover, IEI may also lead to a more narrow disease phenotypes, as in the case of HSE with disease causing mutations identified in TLR3 (Zhang et al., 2007; Mørk et al., 2015) and genes encoding downstream signaling molecules UNC93B (Casrouge et al., 2006), TRIF (Sancho-Shimizu et al., 2011), TRAF3 (Pérez de Diego et al., 2010), TBK1 (Herman et al., 2012), IRF3 (Andersen et al., 2015), and IFNAR1 (Bastard et al., 2021). Of note, a mutation in TLR3 has also been described in a patient with recurrent herpes zoster ophthalmicus (Liang et al., 2020). In addition to these interferon (IFN)-related genes, IEI affecting genes encoding for the RNA debranching molecule DBR1 (Zhang et al., 2018) as well as for SNORA31 (Lafaille et al., 2019) have been identified. More recently, mutations in genes involved in the autophagy pathway, MAP1LC3B2 and ATG4, have been shown to predispose to recurrent HSV-2 lymphocytic Mollaret meningitis (Hait et al., 2020). Finally, single gene IEI in different subunits of RNA polymerase III have been identified first in children with severe acute VZV CNS or lung infection upon primary infection (Ogunjimi et al., 2017) and subsequently in otherwise healthy adults upon VZV reactivation (Carter-Timofte et al., 2018a, 2019). This collection of single gene IEIs in severe disease after viral infection suggest potentially similar role in ARN pathogenesis.
Despite the current literature and findings on IEI and associated alpha-herpesvirus infections of the CNS, the underlying genetic etiology underlying of a large fraction of the severely affected patients remains unexplained. Here, we describe potentially disease-causing variants in genes encoding molecules regulating innate and adaptive immune responses, apoptosis and autophagy, in a cohort of 17 adult ARN patients. With the identification of rare, potentially damaging genetic variants in patients suffering from this rare ocular disease, we hope to shed light on pathways involved in sensing and responding to alphaherpesviruses and to promote understanding of the pathogenesis of viral infection and associated complications in the CNS.
The study enrolled adult patients from medical Ophthalmology clinics at Rigshospitalet, Copenhagen, University Medical Centre Utrecht, and Halland Hospital Halmstad in the time period from January 2017 until June 2023. The enrolled patients had a clinical diagnosis of acute retinal necrosis in at least one eye fulfilling the criteria described by the American Uveitis Society in 1994 (Holland, 1994). There was no predefined limitation of time interval between the diagnosis of ARN and blood collection for genetic testing. Inclusion criteria involved the presence of necrotizing retinitis affecting the peripheral retina and positive diagnostic tests for either HSV or VZV DNA or antibodies by polymerase chain reaction (PCR) or Goldmann-Witmer Coefficient (GWC) in aqueous or vitreous specimens. In case of the Dutch patients, all individuals have positive results for both tests. The clinical characteristics of all patients meeting the inclusion criteria are summarized in Table 1. Exclusion criteria encompassed the inability to provide informed consent, known primary or secondary immunodeficiency, pregnancy or malignancy, or positive tests for toxoplasmosis and syphilis.
Whole blood from P1-P8 was collected aseptically into EDTA containing tubes from each patient. Genomic DNA was isolated from the whole blood with help of the DNeasy Blood and Tissue Kit (Qiagen) according to manufacturer’s instructions. P9-P17 were included at the University Medical Center Utrecht, the Netherlands, genomic DNA purified from blood samples. The samples were selected from the Uveitis Biobank and all patients signed informed consent.
Genomic DNA purified from whole blood samples from P1–P8 was subjected to Whole Exome Sequencing (WES) at the Department of Molecular Medicine (MOMA), Aarhus University Hospital, using KAPA HTP library preparation and Nimblegen SeqCap EZ MedExome Plus kits, followed by analysis with Nextseq version2 chemistry [2 × 150 base pairs] from Illumina. Single nucleotide polymorphisms (SNPs) were called relative to hg19. Variant call files were further filtered using VarSeq 2.3.0 (Golden Helix). Variants were filtered according to (a) quality and read-depth, (b) frequency, only keeping undescribed or rare variants, (c) predicted variant deleteriousness, and (d) biological relevance.
Genomic DNA purified from whole blood samples from P9-P17 was subjected to Whole Genome Sequencing (WGS) at BGI (BGI Europe A/S, Copenhagen, Denmark),1 using DNBSEQ™ Sequencing Technology and BGISEQ-500 platform. All clean reads were aligned to the human reference genome (GRCh37/HG19) using Burrows-Wheeler Aligner (BWA V0.7.12). Variant calling was performed using best Practices for variant analysis with the Genome Analysis Toolkit [HaplotypeCaller of GATK(v3.3.0)].2 VCF files were annotated using ANNOVAR (Wang et al., 2010).
Variants were filtered using Tibco Spotfire Analyst according to (a) quality and read-depth, (b) frequency, only keeping undescribed or rare variants, (c) predicted variant deleteriousness and (d) biological relevance. Detailed Description for all Filters (see Figure 1 for overview of filtering of variants):
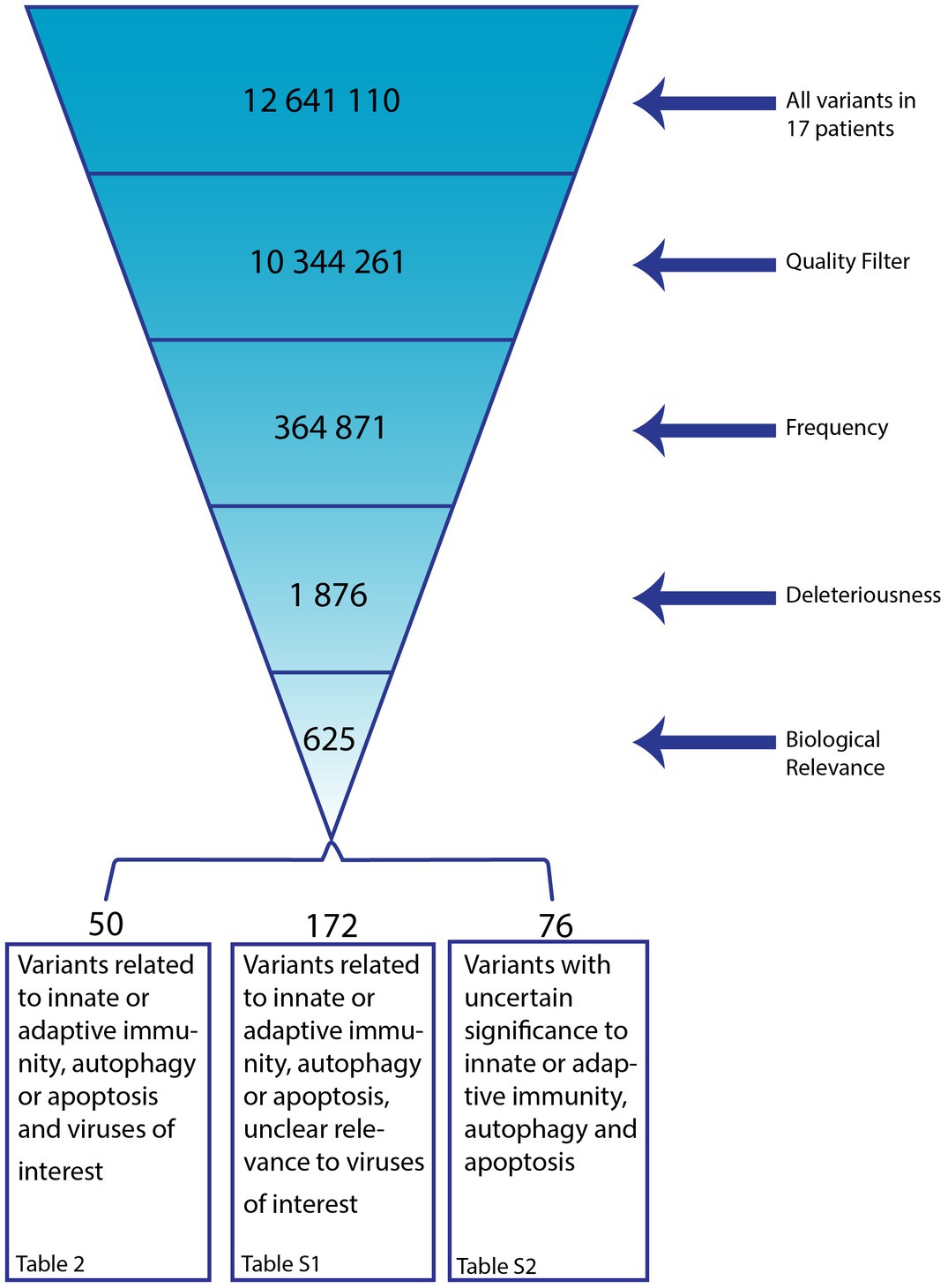
Figure 1. Overview on Filtering Process following Whole Exome Sequencing (WES). From the total of variants identified in 17 patients with alphaherpesvirus-induced ARN we included those of high quality, frequencies of <0.1%, predicted to be Loss of Function (LoF), missense, or splice-site and predicted to be deleterious by at least one out of six damage prediction tools (SIFT, PolyPhen-2, MutationTaster, MutationAssessor, FATHMM, FATHMM MKL). In addition, variants were selected with a CADD score < 15, classified to be Likely Pathogenic, Pathogenic, VUS, VUS/Conflicting, VUS/Weak Pathogenic, VUS/Weak Benign by the ACMG Sample Classifier AND with a REVEL score < 0.25. Variants of biological relevance were subsequently selected. The remaining variants were manually evaluated and listed in Table 2 and Supplementary Tables S1, S2.
(a) Quality: only kept variants that pass the Variant Quality Score Recalibration (VQSR) filtering (or missing) AND have a frequency in the sample fitting a hemi-, hetero-, or homozygous phenotype (or missing) AND have a technical read-depth of 30 or more AND achieve a Genotype Quality Score of more than 20
(b) Frequency: exclude variants with reported frequencies >0.1% AND exclude variants in 1% most exonically variable genes in human public database
(c) Deleteriousness: Only keep variants that are LoF, missense OR splice site variants AND only keep variants predicted to be damaging by at least one out of six damage prediction tools [SIFT (Kumar et al., 2009), PolyPhen-2 (Adzhubei et al., 2010), MutationTaster (Schwarz et al., 2014), MutationAssessor (Reva et al., 2011), FATHMM, FATHMM MKL (Shihab et al., 2014)]AND have a CADD score > 15 (Kircher et al., 2014) AND are classified to be Likely Pathogenic, Pathogenic, VUS, VUS/Conflicting, VUS/Weak Pathogenic, VUS/Weak Benign by the ACMG Sample Classifier (Richards et al., 2015) AND have a REVEL score < 0.25 (Ioannidis et al., 2016)
(d) Biological Relevance: using gene lists related to VZV, HSV, immune response, immunodeficiency and autophagy
P9-P17 were included at the University Medical Center Utrecht, the Netherlands, genomic DNA purified from blood samples. Following, the remaining variants were examined manually and sorted in three categories. Variants with roles in innate or adaptive immunity, autophagy or apoptosis and connection to the relevant pathogen are considered to be most likely disease contributing and are displayed in Table 2. Variants in genes with roles in immunity, autophagy or apoptosis but no connection to the virus infection in the patient are collected in Supplementary Table S1. Variants in genes with no connection to the relevant pathogen and non-substantial biological relevance are in Supplementary Table S2. Variants with a MSC score (Itan et al., 2016) higher than CADD score were excluded, as were variants in genes described to be causative for diseases not present in the regarding patient.
Additionally, variants were filtered using Tibco Spotfire Analyst according to different MOI patterns without filtering for biological relevance:
1. Dominant (patients 0/1, PAV (protein affecting variants), control cohorts (GnomAD, ExAC) <0.0001, CADD (version 1.6) score > 20).
2. Compound heterozygous (control cohorts (GnomAD, ExAC) < 0.03 AND per patient 0/1). Select genes with two different variants per patient.
3. Homozygous (control cohorts (GnomAD, ExAC) < 0.03 AND per patient 1/1).
QIAGEN IPA (QIAGEN Inc.)3 was used to get neighborhood genes linked to known candidates (TLR3, UNC93B1, TRAF3, TICAM1, TBK1, IRF3 and DBR1).
Neighborhood genes were added as a column to the variants file in Tibco Spotfire Analyst. Results of this additional filtering can be found in Table 3.
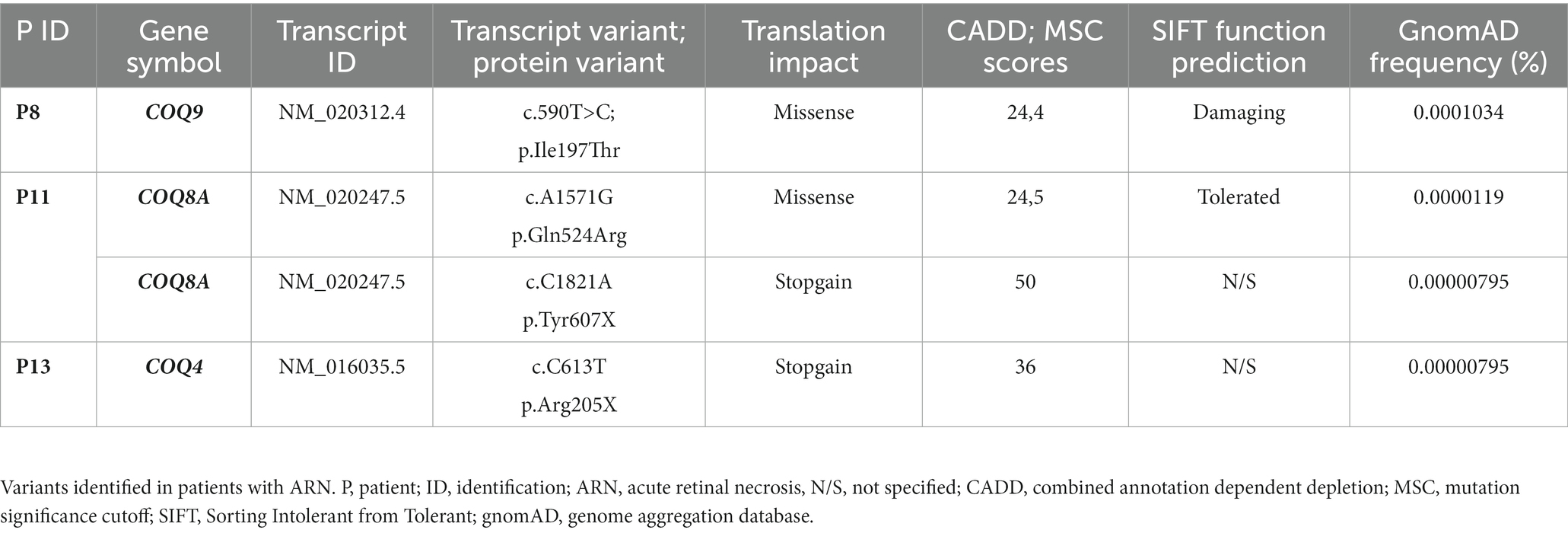
Table 3. Selected rare genetic variants in patients with ARN identified through unbiased analysis without biological filters.
Swedish patients P1-3 were included in accordance with The Helsinki Declaration and with approval from the Swedish Ethical board (no. 2020-04850).
Danish patients P4–P8 were included in accordance with The Helsinki Declaration and Danish ethics guidelines after approval from the Danish National Committee on Health Ethics (no. 1-10-72-275-15) and the Data Protection Agency.
Dutch patients P5-P17 were included after approval form The Biobank Research Ethics Committee (TCBio) UMC Utrecht (TCBio-protocolnr. 21-264).
Oral and written consent from each patient was obtained.
Variant-containing genes after the last evaluation step (50 genes; Table 2) were subjected to STRING analysis for known and predicted interactions of gene products. Based on (Szklarczyk et al., 2015) Version 11.5, STRING Consortium 2023.
For this study, 17 immunocompetent ARN patients from Sweden, Denmark and the Netherlands were included. The patients had clinical signs of ARN and clinical diagnosis was confirmed and viral infection determined by detection of viral DNA and/or intra-ocular production of virus-specific antibodies in aqueous and vitreous humor by real-time quantitative PCR and/or Goldman-Witmer-Coefficient (GWF) (Fekkar et al., 2008). As reported by others, Varicella zoster virus was the most common viral etiology detected in the ARN cohort, namely in 12 patients (70.6%), HSV-1 in three (17.6%) and HSV-2 in two (11.8%). Our cohort thereby follows the distribution previously described and is representative for ARN (Lau et al., 2007; Cox and Miller, 2022). Age of disease onset was between 18 and 72 years, with five patients between 18 and 30 years old (29.4%), six patients between 31 and 60 years (35.3%) and six above 60 years (35.3%). In the youngest group HSV-2 or VZV were identified in two cases each, and HSV-1 in one. In the middle age and oldest group VZV was detected in five cases and HSV-1 for one each. Notably, HSV-2 only occurred in the youngest age group in our cohort, which is in concordance with this virus being described as more common among young ARN patients (Cox and Miller, 2022). Only P14 suffered from a previous episode of encephalitis, which is described as risk factor for ARN (Ahmadieh et al., 1991; Levinson et al., 1999; Ganatra et al., 2000; Maertzdorf et al., 2001; von Hofsten et al., 2019). More detailed demographic and clinical information of the ARN patients is provided in Table 1.
Despite the advanced age of some of the patients, the development of ARN by common human alphaherpesviruses reactivating from latency is a rare event, given the rarity of the disease suggesting an underlying IEI. To identify potential genetic causes for the infectious phenotype we subjected DNA from all patients to WES analysis and analyzed genetic variants relative the human reference genome (GRCh37/HG19). Subsequently, these identified variants were filtered by quality and read-depth, frequency predicted variant deleteriousness and biological relevance. Figure 1 shows a schematic of the filtering process. In total, 625 variants were left after automatic filtering. Of these, 327 variants were excluded based on manual quality control, if the combined annotation dependent (CADD) score < mutation significance cut-off (MSC), if present in a low complexity region of the genes that is known to be difficult to sequence or if the gene was described as causative for a disease the regarding patient did not have and that would have developed by age of ARN diagnosis. The remaining 298 variants were sorted in three categories. First, 50 variants in genes involved in immune signaling, autophagy or apoptosis, having been described to play a role in infection with the associated virus. These have the highest probability of being disease contributing/causing (Table 2). Second, 172 variants were found in genes with a role in immune signaling, autophagy or apoptosis, but without any known direct involvement in VZV or HSV antiviral pathways (Supplementary Table S1). Lastly, 76 variants in genes without evidence of a substantial role in immune signaling, autophagy, apoptosis or virus interaction remained (Supplementary Table S2). In 16 of 17 patients, the search for potentially disease contributing variants was successful (Table 2). The number of variants per patient ranges from one to eight. Among the 50 variants presented in Table 2, 39 are missense, three are frameshift, seven splice site variants, and one an in-frame deletion.
Remarkably, despite the small size of the cohort, variants in a specific gene occurred in more than one patient. For instance, P2 and P3 display unique variants in ASB2. Additionally, P3 harbors a variant in ASB3 from the same gene family. This accumulation of variants might indicate a prominent role for ASB proteins in preventing ARN. The same holds true for TRRAP with variants in P6 and P8, NCSTN with variants in P6 and P17 and APC2 with variants in P8 and P17. Similarly, P1 has a variant in ATG9A and P5 in its paralog ATG9B, suggesting a possible important role for these genes within the autophagy pathway.
Among the identified variant containing genes, similar functions and common signaling pathways are shared, strengthening the hypothesis of an underlying functional defect. We used STRING analysis to check for protein–protein interaction within the 45 genes from Table 2. The analysis reveals five partly overlapping main clusters of different gene functions based on known and predicted protein interactions (Figure 2). Further, the analysis of the identified genes shows significantly more interactions than expected from a random group of 45 genes (39 interactions instead of 18, protein–protein interaction enrichment value of p = 1.02 × 105). This enrichment further indicates functional biological connectivity and involvement of dysfunction of the indicated pathways in the development of ARN.
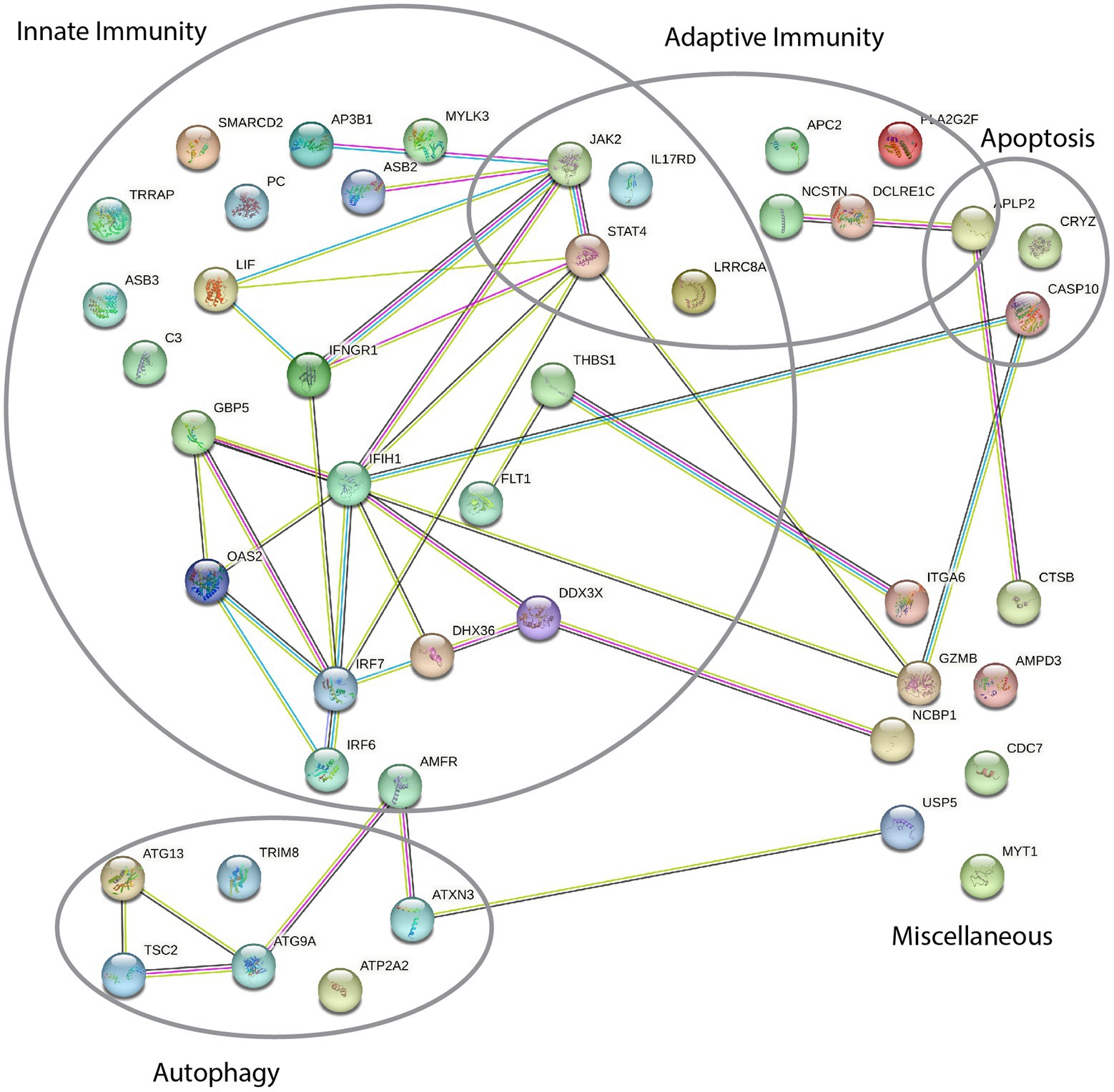
Figure 2. STRING protein–protein interaction network of proteins implicated in the acute retinal necrosis (ARN) patient cohort. Each node depicted represents the gene-product of genes with variants in at least one patient in our ARN cohort. Each line indicates known or predicted interactions between the proteins: Light blue, known interaction from curated database; magenta, known interaction experimentally determined; dark green, predicted interaction based on gene neighborhood; red, predicted interaction based on gene fusion; dark blue, predicted interaction based on gene co-occurrence; light green, text mining; brown co-expression, purple protein homology.
The largest group of variants lies within genes with a function in innate immunity. Here, a number of genes directly involved in the IFN response were identified, including IRF7 (P9), IFNGR1 (P11) and IRF6 (P17). Analogously, DDX3X (P7) leads to IFN-β induction mediated by IRF3 and TBK1 and IKBKE. Also, IFIH1 (MDA5; P17) senses viral nucleic acids directly in the cytoplasm, leading to induction of type I IFN and IFN-induced genes (ISG) (Gu et al., 2013; Lamborn et al., 2017; Wu et al., 2020; Liu et al., 2021). Importantly, transcription of IFIH1 has been shown to be highly elevated in VZV-infected cells (Oh et al., 2019). OAS2 (P4) is an IFN-inducible gene activated by dsRNA and regulating antiviral activity via RNAseL, which has been ascribed a role in the context of HSV-2 infection (Sarkar et al., 1999a,b; Nixon et al., 2014). Two of the variants are in genes involved in sensing of dsRNA, PC (P9) and DHX36 (P12). PC acts as an activator of the dsRNA sensor RIG-I (Vastag et al., 2011), while DHX36 is a part of a TICAM1 complex, which also senses dsRNA (Kim et al., 2010). Production of significant amounts of dsRNA is a common feature in viruses with a DNA genome (Weber et al., 2006). Several genes are also involved in the JAK–STAT signaling pathway induced by type I IFN, a pathway known to be counteracted by herpesviruses (Yokota et al., 2004; Gerada et al., 2020). Among these are LiF (P8), STAT4 (P14), ASB2 (P2 & P3) and ASB3 (P3), and importantly JAK2 (P9). STAT4 also plays a role in adaptive immunity as a key factor for differentiation of T-helper cells (Morinobu et al., 2002; Zhang et al., 2022).
Within innate immunity, several other well represented pathways among variants are the family of MAPK and, more specifically, JNK signaling. Notably, these pathways have been demonstrated to be upregulated in VZV infection, highlighting their relevance in this context (Hargett et al., 2005; Kurapati et al., 2017; Hu et al., 2020). As an example, TRRAP, mutated in P6 and P8, interacts with ELK1, a substrate of JNKs (Zeke et al., 2016). Interestingly, replication efficiency of VZV has been shown to be increased upon inhibition of the JNK pathway (Rahaus et al., 2004). Similarly, IL17RD (P10) also regulates JNK signaling and TLR signaling (Girondel and Meloche, 2021). Additionally the IL17 receptor has been ascribed a role in HSV-1 infection (Hirose et al., 2020). FLT1 (P14) activates the MAPK signaling pathway and its expression is increased in HSV-infected sensory neurons (Kramer et al., 2003). The Wnt/β-catenin signaling circuit is another pathway known to play a role in innate and adaptive immunity and to be manipulated by herpesviruses (Zwezdaryk et al., 2016; Ljungberg et al., 2019). Variants were identified in Wnt signaling regulators APC2 (P8 & P17), NCSTN (P6 & P17) and AMFR (P4). AMFR further acts as a ubiquitin-protein ligase (Shimizu et al., 1999).
Moreover, variants were found in SMARCD2 (P3), involved in differentiation of neutrophils (Witzel et al., 2017), THBS1 (P3), involved in regulation of dendritic cell activation (Doyen et al., 2003), the activator of inflammasome assembly GPB5 (P14) (Shenoy et al., 2012), the complement factor C3 (C17), MYLK3 (P16), involved in IL1β and NF-κB signaling, AP3B1 (P3), affecting natural killer (NK) and NKT cell granule-associated protein release (Shin J. et al., 2021) and TRIM8 (P4). TRIM8 has broad roles in innate immunity within IFN-γ, TNF-α and IL1β signaling (Toniato et al., 2002; Yan et al., 2017; Ye et al., 2017). Interestingly, TRIM8 also regulates immune responses by mediating autophagic degradation of cytoplasmic regulators of innate immunity, thus acting at the interface between innate immunity and autophagy in human cells (Kimura et al., 2015). Similarly, LRRC8A (P7) has a role as 2′-3’-cGAMP transporter and is involved in B-cell development, bridging between innate and adaptive immunity. Finally, PLA2G2F (P3) and DCLRE1C (P8) only have roles in adaptive immunity, within V(D)J recombination (Ma et al., 2002; Murakami et al., 2018). PLA2G2F expression is altered in context of VZV infection (Wu et al., 2021). Deficiency of the DCLRE1C gene product ARTEMIS leads to severe combined immunodeficiency deficiency, while partially impaired function has been described with persistent VZV infection (Volk et al., 2015). VZV is the ARN causing pathogen in both P3 and P8.
Autophagy is another main functional group with identified variants in the ARN cohort. As mentioned above, we identified variants both in ATG9A (P1) and ATG9B (P5), phospholipid scramblases with key roles in autophagosomal membrane expansion (Orsi et al., 2012). ATP2A (P15) is involved in formation of autophagic membranes as well (Zhao et al., 2017). Furthermore, we present variants in ATG13 (P16), part of the autophagy initiation complex (Jung et al., 2009), as well as in TSC2 (P2) and ATXN3 (P17) playing roles in initiating autophagy (Ashkenazi et al., 2017; Rehbein et al., 2021). Some of these, like ATG13 and ATXN3, are upregulated in VZV-infected cells (Grose et al., 2016), whereas, e.g., ATP2A2 is downregulated upon infection (Jones and Arvin, 2003). Upregulation of these autophagy proteins upon infection may suggest a role in anti-viral defense whereas downregulation indicates their expression being counteracted by the virus, both leading to the theory that abolished gene function might be deleterious for a functional antiviral immune response. All individuals in which we identified variants in the above mentioned genes are VZV ARN patients.
Three of the identified variant containing genes encode proteins with a main role in apoptosis: CRYZ (P3) stabilizes BCL2, with anti-apoptotic function in VZV infected cells (Kennedy et al., 2020) and APLP2 (P8) interferes with apoptosis. Importantly, APLP2 also reduces MHC class I expression, providing it with a double role in apoptosis and adaptive immunity (Pandey et al., 2016). Meanwhile, CASP10 (P14) activates the apoptotic cascade (Wang et al., 2001). Interestingly, variants in genes with anti-apoptotic function are found in patients with ARN secondary to VZV infection, whereas the variant in a pro-apoptotic gene is found in an HSV ARN patient, possibly indicating different functions of apoptosis in VZV versus HSV infection.
Several other cellular and immunological pathways and molecules were implicated through the presence of gene variants in this analysis. A variant was identified in the gene encoding for the deubiquitinating enzyme USP5 (P3), that modulates neuropathic and inflammatory pain by interacting with calcium channels also implicated in postherpetic neuralgia (García-Caballero et al., 2014; Li et al., 2021). CTSB (P6) is a lysosomal protease that is upregulated in VZV infected fibroblasts (Tommasi et al., 2020), and GZMB (P11), a cytotoxic molecule secreted by cytotoxic T lymphocytes and NK cells to induce apoptosis and able to cleave viral proteins, combining several different pathways (Tibbs and Cao, 2022). Importantly, among GZMB’s target viruses are both HSV and VZV (Gerada et al., 2019). ITGA6 (P12) is a cell surface receptor downregulated by VZV (Bowles et al., 2012). MYT1 (P16) phosphorylates CDC2, which in turn phosphorylates VZV glycoprotein I. AMPD3 (P17) is a metabolic enzyme alternatively expressed in MeWo-IE63 cells compared with control cells (Habran et al., 2007), indicating possible relevance in context of VZV infection. The Patients 3, 6, 11, 12, 16 and 17 were diagnosed with ARN secondary to VZV infection. CDC7 (P7) is a kinase involved in the cell division cycle and HSV-1 infection (Hou et al., 2017). Lastly, NCBP1 (P10) is involved in splicing and translation regulation and decay and nuclear mRNA export out with particular importance in cellular stress situations, such as viral infection (Gebhardt et al., 2015). Furthermore, NCBP1 is differentially expressed after HSV infection (Dembowski and DeLuca, 2015). P7 and P10 developed ARN secondary to HSV-2 and HSV-1, respectively.
Finally, a different filtering approach was added. In short, pre-determined gene lists for filtering of biological relevance were left out while other quality and damage prediction filters were even more strict (more details described in the method section). The main findings of this filtering method are variants in different subunits of Coenzyme Q which are shown in Table 3. A variant in COQ4 was found in P13, and additional variants in COQ9 and COQ8A were identified in P8 and P11, respectively. All variants are of very low frequency and were not present in a cohort of 1,354 healthy individuals above the age of 80 years used as a control cohort (Erikson et al., 2016). Coenzyme Q is an essential component of the mitochondrial respiratory chain, localized to the inner mitochondrial membrane and linked to mitochondrial diseases in general (Desbats et al., 2015). Its dual role as key electron transfer factor in the mitochondria, and as the main antioxidant in the prevention of oxidative damage in cell membranes makes it an essential molecule in cellular survival. LoF mutations in COQ genes have been previously described causative for primary COQ10 deficiency (Blumkin et al., 2014; Desbats et al., 2015; Traschütz et al., 2020). Neurological abnormalities that can occur in primary coenzyme Q10 deficiency include seizures, intellectual disability, poor muscle tone (hypotonia), involuntary muscle contractions (dystonia), progressive muscle stiffness (spasticity), abnormal eye movements (nystagmus), and of note, vision loss caused by retinal degeneration (Blumkin et al., 2014; Desbats et al., 2015; Traschütz et al., 2020). While not traditionally regarded as part of the immune system, COQ8A is present in the list of neighborhood genes for TLR signaling regulator UNC93B1 (Hubel et al., 2019) and preclinical and clinical studies have shown that COQ10 has anti-inflammatory and antioxidant effects, as well as effects on mitochondrial function, which have been linked to the inflammatory response (Sifuentes-Franco et al., 2022), thereby opening up the possibility of a functional role of COQ10 gene variants in ARN pathogenesis.
In this study, we present the WES analysis of a cohort consisting of 17 ARN patients secondary to VZV (n = 12), HSV-1 (n = 3) and HSV-2 (n = 2) infection. Thereby, we are aiming to unravel presently unknown host-virus interactions and restriction factors and signaling pathways essential for antiviral defenses against herpesviruses in the ocular tissue and the CNS. Remarkably, in 16 out of 17 patients we were able to identify rare genetic variants predicted to be damaging. In total, we here describe 50 rare variants in 45 separate genes predicted to be deleterious in gene function by different bioinformatic damage prediction tools. The gene products have functions within viral sensing, innate and adaptive immunity, autophagy or apoptosis and have previously been described in context of alphaherpesvirus infection. This strict selection process ensures that the described variants are prime candidates for causing infectious susceptibility and pathology, thereby representing possible novel candidates for IEI in humans.
STRING analysis of all variants shows significantly more interactions than expected for a random group of genes (39 instead of 18, Figure 2). This can partly be explained by the use of pre-selected gene lists during the analysis, but nevertheless indicates an enrichment of related pathways in variants of the cohort. Further, the analysis indicates that most variant containing genes can be classified into one or more biologically functional groups: innate immunity, adaptive immunity, autophagy and apoptosis.
The innate immune response plays a critical role in herpesvirus pathogenesis (Moraru et al., 2012; Caselli et al., 2017; Gerada et al., 2020); it is therefore not surprising that many IEI previously described by us and others are located within the innate immune system (Roesler et al., 1999; Dupuis et al., 2003; Zhang et al., 2007; Andersen et al., 2015; Kreins et al., 2015; Thomsen et al., 2021). In the present study, the largest functional group of variants lies within innate immune response genes (see Figure 2), indicating functional impairment of innate immune sensing and signaling as one of the leading causes of severe disease in ARN patients. With IRF7, IFNGR1, IRF6, DDX3X, IFIH1, and OAS2 we identified several variants in genes directly involved in the IFN response, implicating that diminished function of any of these could impair the IFN response upon viral reactivation, potentially facilitating viral spread to the CNS and the ocular nerve. As stated in the introduction, single gene IEIs in genes involved in the IFN response have been found to be causative for both HSV and VZV CNS infection. For example, DDX3X mutations in patients with Natural killer/T-cell lymphoma have been found to underly disturbances of NK cell function (Jiang et al., 2015), ASB2 is involved in NK cell migration (Shin J. H. et al., 2021) and Lif has been described to alter receptor expression on NK cells (Abdoli et al., 2021). In addition, STAT4 appears to be imperative for IL-12 mediated cytotoxicity and IFN-γ production in NK cells (Wang et al., 2000). NK cell function is found to be of particular importance in herpesvirus virulence (Moraru et al., 2012; Caselli et al., 2017), and NK deficiency was among the first described predisposing factors for severe VZV infection (Biron et al., 1989; Gineau et al., 2012).
Most remarkably, despite the relatively small size of the cohort and the rareness of the disease in general, we found not only variants in related pathways, but in the same gene in different patients. This serves as a strong indicator for the importance of proper gene function for prevention of ARN in VZV/HSV infection. ASB2 and ASB3 with variants identified in several patients, play a role in suppression and degradation of cytokines, such as JAK2 (Kohroki et al., 2005; Nie et al., 2011). Given that ARN is characterized by excessive inflammation in the eye, it is possible that uncontrolled activation of JAK–STAT signaling will lead to pathological inflammation, thereby contributing to the observed necrosis. ASB2 is strongly upregulated in VZV-infected cells (Kim et al., 2015). Similar considerations may be made for the variants present in the histone acetyltransferase TRRAP, a protein that alters gene transcription in response to binding with phosphorylated Elk1, a JNK substrate (Zeke et al., 2016). TRRAP plays a role in repressing IFN stimulated genes, pointing toward excessive inflammation in the absence of impaired TRRAP function (Detilleux et al., 2022).
Intriguingly, two of the identified variants, DHX36 and PC, are within genes related to sensing of viral RNA, in accordance with a failure to detect dsRNA intermediates of both viruses would be detrimental to the host. Previously, a number of patients with severe VZV infection were demonstrated to harbor gene variants in different subunits of RNA polymerase III, a cytosolic DNA sensor converting AT-rich DNA into 5′ 3-phosphorylated RNA (Ogunjimi et al., 2017; Carter-Timofte et al., 2018a,b, 2019; Thomsen et al., 2021). Therefore it may appear somewhat surprising that in this cohort with 12 patients developing ARN secondary to VZV infection we did not observe any potentially disease causing variants in POL III genes under the defined selection criteria for deleteriousness and frequency. Among the previously identified patients with defective POL III sensing were children with encephalitis, cerebellitis and pneumonia as well as adults diagnosed with meningoencephalitis and CNS vasculitis (Ogunjimi et al., 2017; Carter-Timofte et al., 2018a, 2019). Our main explanation for why POL III variants were not identified in the present ARN cohort is either the cohort is relatively small and therefore not given to identify POL III variants which are only present in a minority of patients with VZV infection, or that ARN is mainly a reactivation event, whereas most, but not all, POL III variants were originally identified in children with VZV encephalitis during primary infection (varicella) – immune mechanism may differ and involve different molecules and pathways in primary infection as opposed to reactivation in the eye to cause ARN. Intriguingly, however, two of the identified variants, DHX36 and PC, are within genes related to sensing of viral RNA, in accordance with a failure to detect dsRNA intermediates of both viruses would be detrimental to the host.
A particular interesting finding of the present study was several variants in autophagy-related genes. ATG9A and its paralogue ATG9B encode autophagy proteins with are key roles in autophagic membrane elongation (Orsi et al., 2012). Additional variants in autophagy related genes were present within this cohort, mostly involving initiation of autophagy. For a long time, autophagy has been known as an anti-viral defense mechanism (Lee and Iwasaki, 2008). With regards to HSV-1 infection, there is good evidence that autophagy plays a protective role, especially in the CNS (Wilcox et al., 2015; Yakoub and Shukla, 2015). We have previously described two patients with variants in autophagy-related genes contributing to development of recurrent HSV2 lymphocytic Mollaret meningitis (Hait et al., 2020) and have described more patients harboring autophagy related gene variants (Hait et al., 2021). Whereas it has been demonstrated that autophagy is upregulated in VZV infection (Eisfeld et al., 2006; Takahashi et al., 2009; Girsch et al., 2020); the importance of autophagy in antiviral immunity against VZV is more controversial and unresolved. (Extensively reviewed in Heinz et al., 2021).
Previous work from our group on a VZV encephalitis cohort also showed a significant portion of variants within the autophagy pathway (Thomsen et al., 2021), implicating variants in ATG2A and ATG2B, ULK1, TGM2 and LAMP2, alongside yet another variant in ATG9A and one in TSC1. Accumulation of variants in autophagy-related genes in VZV CNS infection (Thomsen et al., 2021) and the present ARN cohort may suggest autophagy defects to be unfavorable for these patients. For this reason, we believe the description of novel variants in relation to VZV infection provides especially valuable information about the relevance of this pathway in VZV antiviral defenses.
Genetic variants in apoptosis-related genes were previously identified in poliomyelitis and a Mollaret meningitis cohort (Andersen et al., 2019; Hait et al., 2021); however, the present study represents the first description of major variants related to apoptosis in patients with VZV infection. Considering how herpesviruses establish latency in their hosts it seems plausible that a careful regulation of cell death pathways is of central importance. Here we identified two variants in anti-apoptotic genes, the impaired functionality of which might lead to excessive cell death thereby possibly contributing to necrosis of ocular tissue. On the other hand, we report one variant in a pro-apoptotic gene, indicating that impaired apoptosis might interfere with viral control earlier in the re-activation phase and thereby enable increased production of viral particles, which in turn increases the risk for viral dissemination to the ocular nerve. Functional studies are needed to further elucidate this fascinating potential role for apoptosis dysregulation in severe infection.
The canonical Wnt signaling pathway, of which NCSTN belongs, is established to provide anti-viral activity against different herpesviruses (Zwezdaryk et al., 2016). Intriguingly, Wnt signaling is described to be upregulated in VZV infected neurons, but not in fibroblasts (Markus et al., 2014), indicating a particular importance of this pathway in the context of CNS infection. Moreover, not only Wnt signaling, but also NCSTN is significantly upregulated in VZV infection (Jones and Arvin, 2003). However, the exact mechanism by which reduced function of NCSTN may benefit VZV needs further experimental validation.
Finally, we found potentially damaging variants in COQ4, COQ8A and COQ9 in three unrelated patients suffering from VZV-induced ARN. The co-enzyme Q10 complex plays a role in inhibition of ROS production by reducing oxidative damage and inflammation and; importantly, is expressed in the retina (Qu et al., 2009). A normal functioning COQ10 complex reduces cell death of retinal cells and delays progression of retinal diseases such as age-related macular degeneration, retinitis pigmentosa, diabetic retinopathy, glaucoma and last but not least ARN (Zhang et al., 2017; Verbakel et al., 2018; Jurkute et al., 2022; Fuller et al., 2023). Of note, VZV, the diagnosed pathogen in all three patients with COQ variants, is known to disrupt proper mitochondrial function and production of reactive oxygen species (Keller et al., 2016; Andrieux et al., 2021). Evolutionary conservation of this pathway has been described in Drosophila melanogaster (Hura et al., 2022), further underlining the essential role of this biological pathway, possibly also in antiviral defense against VZV infection of ocular tissue. Of note, Q10-related variants have not previously been identified in any other WES studies (Andersen et al., 2019; Hait et al., 2021; Thomsen et al., 2021), underlining a potential specificity of COQ10 biology for ARN development.
The present analysis represents the largest cohort of ARN patients investigated genetically, particularly given the rareness of the disease. Our findings can be seen as a starting point for furter studies on the pathogenesis of ARN, a knowledge that will in the long term be of advantage for prognosis, prevention and treatment methods. However, confirmation of these findings in larger cohorts is needed. Perhaps most importantly, a tremendous amount of work is left to functionally evaluate the importance of each of the identified genes as well as the mechanism by which defective function of that gene promotes development of ARN. Each of the variants must be functionally validated through knock-out and Knock-in approaches in patient cells and experimental cell models, and the patient cells must be reconstituted to normal by overexpression of the wild-type protein of interest. The incompleteness of the method represents another limitation of the study: since WES analysis does not cover intronic and non-coding regions of the genome some disease causing genetic alterations may be missed. Although most disease causing variants presently known are localized within the coding regions, there is a risk of missing potentially disease causing variants in introns and regulatory regions of the genome. Another interesting, so far unanswered, question is, whether the identified gene defects lead to more frequent viral reactivation, or do not change the frequency of reactivation per se, but rather increase the risk of developing complications in response to these. Indeed, genetic defects leading to more frequent reactivation have been described previously (Estefanía et al., 2007; Rizzo et al., 2012; Caselli et al., 2014), although for other IEI underlying severe viral disease, the pathogenesis rather involves unrestricted replication once the virus is reactivated.
Here, we identify several genes and pathways that appear to be important for disease pathogenesis. Further functional studies on each of these gene variants are necessary in the future. While in some cases disease phenotype will not be explained by a single-gene IEI, overall, we believe that our findings contribute to explaining the development of severe retinal disease in at least some of these patients and provide new insight into the pathomechanisms of severe alphaherpesvirus infection of the brain and related tissues. Collectively the present manuscript represents a descriptive genetic study with the aim of providing a map of some genes/molecular targets and pathways in immune circuits and restriction factors within cells that may be disturbed or defective in patients with insufficient immune responses or inability to maintain latency of VZV and HSV1 and 2. These results may then inspire further experimental studies and comparison of identified variants by whole exome/whole genome sequencing in patients with VZV, HSV1 or similar severe viral infections. Ultimately, we expect this knowledge to be valuable for future development of improved patient diagnosis, prophylaxis, and treatment of this rare, yet sight-threatening ocular disease and to elucidate so far unknown aspects of neuropathogenesis of alphaherpesvirus infection in humans.
The datasets presented in this article are not readily available because Danish GDPR rules do not allow sharing entire genetic sequences from humans in a repository. Requests to access the datasets should be directed to TM.
The studies involving humans were approved by Swedish patients P1-3 were included in accordance with The Helsinki Declaration and with approval from the Swedish Ethical board (no. 2020-04850). Danish patients P4–P8 were included in accordance with The Helsinki Declaration and Danish ethics guidelines after approval from the Danish National Committee on Health Ethics (no. 1-10-72-275-15) and the Data Protection Agency. Dutch patients P5-P17 were included after approval form The Biobank Research Ethics Committee (TCBio) UMC Utrecht (TCBio-protocolnr. 21-264). Oral and written consent from each patient was obtained. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
TM and GV conceived the idea. TM, JHo, TI, MH, and JB cared for patients. JHe, MT, JHo, TI, MH, and JB collected and isolated patient material. JHe analyzed data together with MT, SS, PS, PH, KK, and TM. TM and JHe wrote the first draft of the manuscript with contributions from GV, JHo, TI, MH, JB, MT, KK, SS, PS, and PH. TM and JHe revised the manuscript. All authors contributed, read, commented, and approved the final version of the manuscript.
TM was funded by the Independent Research Fund Denmark (4004-00047B), Aarhus University Research Fund (AUFF-E-2015-FLS-66), and The Lundbeck Foundation (R268-2016-3927).
We wish to thank all patients participating in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1253040/full#supplementary-material
Abdoli, Z., Assarehzadegan, M.-A., Pipelzadeh, M. H., Iranparast, S., Dashti Gerdabi, N., Parsanahad, M., et al. (2021). Leukemia inhibitory factor suppresses NKG2D mRNA expression and presentation on human natural killer cells. Iran. J. Allergy Asthma Immunol. 20, 98–105. doi: 10.18502/ijaai.v20i1.5416
Abdollahpour, H., Appaswamy, G., Kotlarz, D., Diestelhorst, J., Beier, R., Schäffer, A. A., et al. (2012). The phenotype of human STK4 deficiency. Blood 119, 3450–3457. doi: 10.1182/blood-2011-09-378158
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. doi: 10.1038/nmeth0410-248
Ahmadieh, H., Sajjadi, S. H., Azarmina, M., and Kalani, H. (1991). Association of herpetic encephalitis with acute retinal necrosis syndrome. Ann. Ophthalmol. 23, 215–219.
Alvarez, J. C., Alvarez, J., Tinoco, J., Mellado, P., Miranda, H., Ferrés, M., et al. (2020). Varicella-zoster virus meningitis and encephalitis: an understated cause of central nervous system infections. Cureus 12:e11583. doi: 10.7759/cureus.11583
Andersen, N.-S. B., Larsen, S. M., Nissen, S. K., Jørgensen, S. E., Mardahl, M., Christiansen, M., et al. (2019). Host genetics, innate immune responses, and cellular death pathways in poliomyelitis patients. Front. Microbiol. 10:1495. doi: 10.3389/fmicb.2019.01495
Andersen, L. L., Mørk, N., Reinert, L. S., Kofod-Olsen, E., Narita, R., Jørgensen, S. E., et al. (2015). Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 212, 1371–1379. doi: 10.1084/jem.20142274
Andrieux, P., Chevillard, C., Cunha-Neto, E., and Nunes, J. P. S. (2021). Mitochondria as a cellular hub in infection and inflammation. Int. J. Mol. Sci. 22:11338. doi: 10.3390/ijms222111338
Ashkenazi, A., Bento, C. F., Ricketts, T., Vicinanza, M., Siddiqi, F., Pavel, M., et al. (2017). Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111. doi: 10.1038/nature22078
Bastard, P., Manry, J., Chen, J., Rosain, J., Seeleuthner, Y., AbuZaitun, O., et al. (2021). Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency. J. Clin. Invest. 131:e139980. doi: 10.1172/JCI139980
Biron, C. A., Byron, K. S., and Sullivan, J. L. (1989). Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320, 1731–1735. doi: 10.1056/NEJM198906293202605
Blumkin, L., Leshinsky-Silver, E., Zerem, A., Yosovich, K., Lerman-Sagie, T., and Lev, D. (2014). Heterozygous mutations in the ADCK3 gene in siblings with cerebellar atrophy and extreme phenotypic variability. JIMD Reports 12, 103–107. doi: 10.1007/8904_2013_251
Bowles, J. B., Steain, M., Slobedman, B., and Abendroth, A. (2012). Inhibition of integrin α6 expression by cell-free varicella-zoster virus. J. Gen. Virol. 93, 1725–1730. doi: 10.1099/vir.0.039917-0
Brandt, C. R. (2005). The role of viral and host genes in corneal infection with herpes simplex virus type 1. Exp. Eye Res. 80, 607–621. doi: 10.1016/j.exer.2004.09.007
Breuer, J. (2010). VZV molecular epidemiology. Curr. Top. Microbiol. Immunol. 342, 15–42. doi: 10.1007/82_2010_9
Brooks, E. G., Schmalstieg, F. C., Wirt, D. P., Rosenblatt, H. M., Adkins, L. T., Lookingbill, D. P., et al. (1990). A novel X-linked combined immunodeficiency disease. J. Clin. Invest. 86, 1623–1631. doi: 10.1172/JCI114884
Carter-Timofte, M. E., Hansen, A. F., Christiansen, M., Paludan, S. R., and Mogensen, T. H. (2019). Mutations in RNA polymerase III genes and defective DNA sensing in adults with varicella-zoster virus CNS infection. Genes Immun. 20, 214–223. doi: 10.1038/s41435-018-0027-y
Carter-Timofte, M. E., Hansen, A. F., Mardahl, M., Fribourg, S., Rapaport, F., Zhang, S.-Y., et al. (2018a). Varicella-zoster virus CNS vasculitis and RNA polymerase III gene mutation in identical twins. Neurol. Neuroimmunol. Neuroinflamm. 5:e500. doi: 10.1212/NXI.0000000000000500
Carter-Timofte, M. E., Paludan, S. R., and Mogensen, T. H. (2018b). RNA polymerase III as a gatekeeper to prevent severe VZV infections. Trends Mol. Med. 24, 904–915. doi: 10.1016/j.molmed.2018.07.009
Caselli, E., Bortolotti, D., Marci, R., Rotola, A., Gentili, V., Soffritti, I., et al. (2017). HHV-6A infection of endometrial epithelial cells induces increased endometrial NK cell-mediated cytotoxicity. Front. Microbiol. 8:2525. doi: 10.3389/fmicb.2017.02525
Caselli, E., Rizzo, R., Ingianni, A., Contini, P., Pompei, R., and Di Luca, D. (2014). High prevalence of HHV8 infection and specific killer cell immunoglobulin-like receptors allotypes in Sardinian patients with type 2 diabetes mellitus. J. Med. Virol. 86, 1745–1751. doi: 10.1002/jmv.23771
Casrouge, A., Zhang, S.-Y., Eidenschenk, C., Jouanguy, E., Puel, A., Yang, K., et al. (2006). Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312. doi: 10.1126/science.1128346
Cox, J. T., and Miller, J. B. (2022). Retinal detachment associated with acute retinal necrosis. Int. Ophthalmol. Clin. 62, 157–172. doi: 10.1097/IIO.0000000000000414
Dembowski, J. A., and DeLuca, N. A. (2015). Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 11:e1004939. doi: 10.1371/journal.ppat.1004939
Desbats, M. A., Lunardi, G., Doimo, M., Trevisson, E., and Salviati, L. (2015). Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J. Inherit. Metab. Dis. 38, 145–156. doi: 10.1007/s10545-014-9749-9
Detilleux, D., Raynaud, P., Pradet-Balade, B., and Helmlinger, D. (2022). The TRRAP transcription cofactor represses interferon-stimulated genes in colorectal cancer cells. elife 11:e69705. doi: 10.7554/eLife.69705
Dobbs, K., Domínguez Conde, C., Zhang, S.-Y., Parolini, S., Audry, M., Chou, J., et al. (2015). Inherited DOCK2 deficiency in patients with early-onset invasive infections. N. Engl. J. Med. 372, 2409–2422. doi: 10.1056/NEJMoa1413462
Doyen, V., Rubio, M., Braun, D., Nakajima, T., Abe, J., Saito, H., et al. (2003). Thrombospondin 1 is an autocrine negative regulator of human dendritic cell activation. J. Exp. Med. 198, 1277–1283. doi: 10.1084/jem.20030705
Dupuis, S., Jouanguy, E., Al-Hajjar, S., Fieschi, C., Al-Mohsen, I. Z., Al-Jumaah, S., et al. (2003). Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33, 388–391. doi: 10.1038/ng1097
Eisfeld, A. J., Turse, S. E., Jackson, S. A., Lerner, E. C., and Kinchington, P. R. (2006). Phosphorylation of the varicella-zoster virus (VZV) major transcriptional regulatory protein IE62 by the VZV open reading frame 66 protein kinase. J. Virol. 80, 1710–1723. doi: 10.1128/JVI.80.4.1710-1723.2006
Erikson, G. A., Bodian, D. L., Rueda, M., Molparia, B., Scott, E. R., Scott-Van Zeeland, A. A., et al. (2016). Whole-genome sequencing of a healthy aging cohort. Cells 165, 1002–1011. doi: 10.1016/j.cell.2016.03.022
Estefanía, E., Gómez-Lozano, N., Portero, F., de Pablo, R., Solís, R., Sepúlveda, S., et al. (2007). Influence of KIR gene diversity on the course of HSV-1 infection: resistance to the disease is associated with the absence of KIR2DL2 and KIR2DS2. Tissue Antigens 70, 34–41. doi: 10.1111/j.1399-0039.2007.00844.x
Fekkar, A., Bodaghi, B., Touafek, F., Le Hoang, P., Mazier, D., and Paris, L. (2008). Comparison of immunoblotting, calculation of the Goldmann-Witmer coefficient, and real-time PCR using aqueous humor samples for diagnosis of ocular toxoplasmosis. J. Clin. Microbiol. 46, 1965–1967. doi: 10.1128/JCM.01900-07
Fields, B.N., and Knipe, D.M. & and Howley, P.M. (2013a) Fields virology: Chapter 12: Epidemiologz, 6th. Wolters Kluwer Health/Lippincott WIlliams & Wilkins: Philadelphia.
Fields, B.N., and Knipe, D.M. & and Howley, P.M. (2013b) Fields virology: Chapter 60: pathogenesis of human infection, 6th. Wolters Kluwer Health/Lippincott WIlliams & Wilkins: Philadelphia.
Fields, B.N., and Knipe, D.M. & and Howley, P.M. (2013c) Fields virology: Chapter 63: Pathogenesis & pathology, 6th. Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia.
Fields, B.N., and Knipe, D.M. & and Howley, P.M. (2013d) Fields virology: Chapter 63: Varicella-zoster virus, 6th. Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia.
Fuller, J. T., Barnes, S., Sadun, L. A., Ajmera, P., Alexandrova, A. N., and Sadun, A. A. (2023). Coenzyme Q10 trapping in mitochondrial complex I underlies Leber's hereditary optic neuropathy. Proc. Natl. Acad. Sci. U. S. A. 120:e2304884120. doi: 10.1073/pnas.2304884120
Ganatra, J. B., Chandler, D., Santos, C., Kuppermann, B., and Margolis, T. P. (2000). Viral causes of the acute retinal necrosis syndrome. Am J. Ophthalmol. 129, 166–172. doi: 10.1016/s0002-9394(99)00316-5
García-Caballero, A., Gadotti, V. M., Stemkowski, P., Weiss, N., Souza, I. A., Hodgkinson, V., et al. (2014). The deubiquitinating enzyme USP5 modulates neuropathic and inflammatory pain by enhancing Cav3.2 channel activity. Neuron 83, 1144–1158. doi: 10.1016/j.neuron.2014.07.036
Gebhardt, A., Habjan, M., Benda, C., Meiler, A., Haas, D. A., Hein, M. Y., et al. (2015). mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nature. Communications 6:8192. doi: 10.1038/ncomms9192
Gerada, C., Campbell, T. M., Kennedy, J. J., McSharry, B. P., Steain, M., Slobedman, B., et al. (2020). Manipulation of the innate immune response by varicella zoster virus. Front. Immunol. 11:1. doi: 10.3389/fimmu.2020.00001
Gerada, C., Steain, M., Campbell, T. M., McSharry, B., Slobedman, B., and Abendroth, A. (2019). Granzyme B cleaves multiple herpes simplex virus 1 and varicella-zoster virus (VZV) gene products, and VZV ORF4 inhibits natural killer cell cytotoxicity. J. Virol. 93, 10–1128. doi: 10.1128/JVI.01140-19
Gineau, L., Cognet, C., Kara, N., Lach, F. P., Dunne, J., Veturi, U., et al. (2012). Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J. Clin. Invest. 122, 821–832. doi: 10.1172/JCI61014
Girondel, C., and Meloche, S. (2021). Interleukin-17 receptor D in physiology, inflammation and cancer. Front. Oncol. 11:656004. doi: 10.3389/fonc.2021.656004
Girsch, J. H., Jackson, W., Carpenter, J. E., Moninger, T. O., Jarosinski, K. W., and Grose, C. (2020). Exocytosis of progeny infectious varicella-zoster virus particles via a Mannose-6-phosphate receptor pathway without Xenophagy following secondary envelopment. J. Virol. 94, 10–1128. doi: 10.1128/JVI.00800-20
Grahn, A., and Studahl, M. (2015). Varicella-zoster virus infections of the central nervous system – prognosis, diagnostics and treatment. J. Infect. 71, 281–293. doi: 10.1016/j.jinf.2015.06.004
Grose, C., Buckingham, E. M., Carpenter, J. E., and Kunkel, J. P. (2016). Varicella-zoster virus infectious cycle: ER stress, autophagic flux, and amphisome-mediated trafficking. Pathogens 5:67. doi: 10.3390/pathogens5040067
Gu, L., Fullam, A., Brennan, R., and Schröder, M. (2013). Human DEAD box helicase 3 couples IκB kinase ε to interferon regulatory factor 3 activation. Mol. Cell. Biol. 33, 2004–2015. doi: 10.1128/MCB.01603-12
Habran, L., El Mjiyad, N., Di Valentin, E., Sadzot-Delvaux, C., Bontems, S., and Piette, J. (2007). The varicella-zoster virus immediate-early 63 protein affects chromatin-controlled gene transcription in a cell-type dependent manner. BMC Mol. Biol. 8:99. doi: 10.1186/1471-2199-8-99
Hait, A. S., Olagnier, D., Sancho-Shimizu, V., Skipper, K. A., Helleberg, M., Larsen, S. M., et al. (2020). Defects in LC3B2 and ATG4A underlie HSV2 meningitis and reveal a critical role for autophagy in antiviral defense in humans. Sci. Immunol. 5:eabc2691. doi: 10.1126/sciimmunol.abc2691
Hait, A. S., Thomsen, M. M., Larsen, S. M., Helleberg, M., Mardahl, M., Barfod, T. S., et al. (2021). Whole-exome sequencing of patients with recurrent HSV-2 lymphocytic mollaret meningitis. J. Infect. Dis. 223, 1776–1786. doi: 10.1093/infdis/jiaa589
Hargett, D., McLean, T., and Bachenheimer, S. L. (2005). Herpes simplex virus ICP27 activation of stress kinases JNK and p38. J. Virol. 79, 8348–8360. doi: 10.1128/JVI.79.13.8348-8360.2005
Heinz, J., Kennedy, P. G. E., and Mogensen, T. H. (2021). The role of autophagy in varicella zoster virus infection. Viruses 13:1053. doi: 10.3390/v13061053
Herlin, L. K., Hansen, K. S., Bodilsen, J., Larsen, L., Brandt, C., Andersen, C. Ø., et al. (2021). Varicella zoster virus encephalitis in Denmark from 2015 to 2019-A nationwide prospective cohort study. Clin. Infect. Dis. 72, 1192–1199. doi: 10.1093/cid/ciaa185
Herman, M., Ciancanelli, M., Ou, Y.-H., Lorenzo, L., Klaudel-Dreszler, M., Pauwels, E., et al. (2012). Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209, 1567–1582. doi: 10.1084/jem.20111316
Hillenkamp, J., Nölle, B., Bruns, C., Rautenberg, P., Fickenscher, H., and Roider, J. (2009). Acute retinal necrosis: clinical features, early vitrectomy, and outcomes. Ophthalmology 116, 1971–5.e2. doi: 10.1016/j.ophtha.2009.03.029
Hirose, S., Jaggi, U., Wang, S., Tormanen, K., Nagaoka, Y., Katsumata, M., et al. (2020). Role of TH17 responses in increasing herpetic keratitis in the eyes of mice infected with HSV-1. Invest. Ophthalmol. Vis. Sci. 61:20. doi: 10.1167/iovs.61.6.20
Holland, G. N. (1994). Standard diagnostic criteria for the acute retinal necrosis syndrome. Executive Committee of the American Uveitis Society. Am J. Ophthalmol. 117, 663–667. doi: 10.1016/s0002-9394(14)70075-3
Hou, J., Zhang, Z., Huang, Q., Yan, J., Zhang, X., Yu, X., et al. (2017). Antiviral activity of PHA767491 against human herpes simplex virus in vitro and in vivo. BMC Infect. Dis. 17:217. doi: 10.1186/s12879-017-2305-0
Hu, K., Fu, M., Wang, J., Luo, S., Barreto, M., Singh, R., et al. (2020). HSV-2 infection of human genital epithelial cells upregulates TLR9 expression through the SP1/JNK signaling pathway. Front. Immunol. 11:356. doi: 10.3389/fimmu.2020.00356
Hubel, P., Urban, C., Bergant, V., Schneider, W. M., Knauer, B., Stukalov, A., et al. (2019). A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 20, 493–502. doi: 10.1038/s41590-019-0323-3
Hura, A. J., Hawley, H. R., Tan, W. J., Penny, R. J., Jacobsen, J. C., and Fitzsimons, H. L. (2022). Loss of Drosophila Coq8 results in impaired survival, locomotor deficits and photoreceptor degeneration. Mol. Brain 15:15. doi: 10.1186/s13041-022-00900-3
Ioannidis, N. M., Rothstein, J. H., Pejaver, V., Middha, S., McDonnell, S. K., Baheti, S., et al. (2016). REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885. doi: 10.1016/j.ajhg.2016.08.016
Itan, Y., Shang, L., Boisson, B., Ciancanelli, M. J., Markle, J. G., Martinez-Barricarte, R., et al. (2016). The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods 13, 109–110. doi: 10.1038/nmeth.3739
Iwahashi-Shima, C., Azumi, A., Ohguro, N., Okada, A. A., Kaburaki, T., Goto, H., et al. (2013). Acute retinal necrosis: factors associated with anatomic and visual outcomes. Jpn. J. Ophthalmol. 57, 98–103. doi: 10.1007/s10384-012-0211-y
Jiang, L., Gu, Z.-H., Yan, Z.-X., Zhao, X., Xie, Y.-Y., Zhang, Z.-G., et al. (2015). Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 47, 1061–1066. doi: 10.1038/ng.3358
Jones, J. O., and Arvin, A. M. (2003). Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77, 1268–1280. doi: 10.1128/jvi.77.2.1268-1280.2003
Jørgensen, S. E., Al-Mousawi, A., Assing, K., Hartling, U., Grosen, D., Fisker, N., et al. (2021). STK4 deficiency impairs innate immunity and interferon production through negative regulation of TBK1-IRF3 signaling. J. Clin. Immunol. 41, 109–124. doi: 10.1007/s10875-020-00891-7
Jung, C. H., Jun, C. B., Ro, S.-H., Kim, Y.-M., Otto, N. M., Cao, J., et al. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003. doi: 10.1091/mbc.e08-12-1249
Jurkute, N., Cancellieri, F., Pohl, L., Li, C. H. Z., Heaton, R. A., Reurink, J., et al. (2022). Biallelic variants in coenzyme Q10 biosynthesis pathway genes cause a retinitis pigmentosa phenotype. NPJ. Genomic Med. 7:60. doi: 10.1038/s41525-022-00330-z
Keller, A. C., Badani, H., McClatchey, P. M., Baird, N. L., Bowlin, J. L., Bouchard, R., et al. (2016). Varicella zoster virus infection of human fetal lung cells alters mitochondrial morphology. J. Neurovirol. 22, 674–682. doi: 10.1007/s13365-016-0457-0
Kennedy, P. G., Graner, M. W., Gunaydin, D., Bowlin, J., Pointon, T., and Yu, X. (2020). Varicella-Zoster Virus infected human neurons are resistant to apoptosis. J. Neurovirol. 26, 330–337. doi: 10.1007/s13365-020-00831-6
Kennedy, P. G., and Mogensen, T. H. (2020). Determinants of neurological syndromes caused by varicella zoster virus (VZV). J. Neurovirol. 26, 482–495. doi: 10.1007/s13365-020-00857-w
Kim, J.-A., Park, S.-K., Kumar, M., Lee, C.-H., and Shin, O. S. (2015). Insights into the role of immunosenescence during varicella zoster virus infection (shingles) in the aging cell model. Oncotarget 6, 35324–35343. doi: 10.18632/oncotarget.6117
Kim, T., Pazhoor, S., Bao, M., Zhang, Z., Hanabuchi, S., Facchinetti, V., et al. (2010). Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase a helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 107, 15181–15186. doi: 10.1073/pnas.1006539107
Kimura, T., Jain, A., Choi, S. W., Mandell, M. A., Schroder, K., Johansen, T., et al. (2015). TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 210, 973–989. doi: 10.1083/jcb.201503023
Kircher, M., Witten, D. M., Jain, P., O'Roak, B. J., Cooper, G. M., and Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315. doi: 10.1038/ng.2892
Kleinschmidt-DeMasters, B. K., and Gilden, D. H. (2001). Varicella-Zoster virus infections of the nervous system: clinical and pathologic correlates. Arch. Pathol. Lab. Med. 125, 770–780. doi: 10.5858/2001-125-0770-VZVIOT
Kohroki, J., Nishiyama, T., Nakamura, T., and Masuho, Y. (2005). ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 579, 6796–6802. doi: 10.1016/j.febslet.2005.11.016
Kolb, A. W., Lee, K., Larsen, I., Craven, M., and Brandt, C. R. (2016). Quantitative trait locus based virulence determinant mapping of the HSV-1 genome in murine ocular infection: genes involved in viral regulatory and innate immune networks contribute to virulence. PLoS Pathog. 12:e1005499. doi: 10.1371/journal.ppat.1005499
Kramer, M. F., Cook, W. J., Roth, F. P., Zhu, J., Holman, H., Knipe, D. M., et al. (2003). Latent herpes simplex virus infection of sensory neurons alters neuronal gene expression. J. Virol. 77, 9533–9541. doi: 10.1128/jvi.77.17.9533-9541.2003
Kreins, A. Y., Ciancanelli, M. J., Okada, S., Kong, X.-F., Ramírez-Alejo, N., Kilic, S. S., et al. (2015). Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212, 1641–1662. doi: 10.1084/jem.20140280
Kumar, P., Henikoff, S., and Ng, P. C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081. doi: 10.1038/nprot.2009.86
Kurapati, S., Sadaoka, T., Rajbhandari, L., Jagdish, B., Shukla, P., Ali, M. A., et al. (2017). Role of the JNK pathway in varicella-zoster virus lytic infection and reactivation. J. Virol. 91, 10–1128. doi: 10.1128/JVI.00640-17
Lafaille, F. G., Harschnitz, O., Lee, Y. S., Zhang, P., Hasek, M. L., Kerner, G., et al. (2019). Human SNORA31 variations impair cortical neuron-intrinsic immunity to HSV-1 and underlie herpes simplex encephalitis. Nat. Med. 25, 1873–1884. doi: 10.1038/s41591-019-0672-3
Lamborn, I. T., Jing, H., Zhang, Y., Drutman, S. B., Abbott, J. K., Munir, S., et al. (2017). Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 214, 1949–1972. doi: 10.1084/jem.20161759
Lau, C. H., Missotten, T., Salzmann, J., and Lightman, S. L. (2007). Acute retinal necrosis features, management, and outcomes. Ophthalmology 114, 756–762. doi: 10.1016/j.ophtha.2006.08.037
Lee, H. K., and Iwasaki, A. (2008). Autophagy and antiviral immunity. Curr. Opin. Immunol. 20, 23–29. doi: 10.1016/j.coi.2008.01.001
Levinson, R. D., Reidy, R., and Chiu, M. T. (1999). Acute retinal necrosis after neonatal herpes encephalitis. Br. J. Ophthalmol. 83, 123–124. doi: 10.1136/bjo.83.1.123a
Li, R., Ou, M., Yang, S., Huang, J., Chen, J., Xiong, D., et al. (2021). Change in Cav3.2 T-type calcium channel induced by varicella-zoster virus participates in the maintenance of herpetic neuralgia. Front. Neurol. 12:741054. doi: 10.3389/fneur.2021.741054
Liang, F., Glans, H., Enoksson, S. L., Kolios, A. G. A., Loré, K., and Nilsson, J. (2020). Recurrent herpes zoster ophthalmicus in a patient with a novel toll-like receptor 3 variant linked to compromised activation capacity in fibroblasts. J. Infect. Dis. 221, 1295–1303. doi: 10.1093/infdis/jiz229
Liu, G., Lee, J.-H., Parker, Z. M., Acharya, D., Chiang, J. J., van Gent, M., et al. (2021). ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 6, 467–478. doi: 10.1038/s41564-021-00884-1
Ljungberg, J. K., Kling, J. C., Tran, T. T., and Blumenthal, A. (2019). Functions of the WNT signaling network in shaping host responses to infection. Front. Immunol. 10:2521. doi: 10.3389/fimmu.2019.02521
Ma, Y., Pannicke, U., Schwarz, K., and Lieber, M. R. (2002). Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cells 108, 781–794. doi: 10.1016/s0092-8674(02)00671-2
Mace, E. M., Hsu, A. P., Monaco-Shawver, L., Makedonas, G., Rosen, J. B., Dropulic, L., et al. (2013). Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood 121, 2669–2677. doi: 10.1182/blood-2012-09-453969
Maertzdorf, J., van der Lelij, A., Baarsma, G. S., Osterhaus, A. D., and Verjans, G. M. (2001). Herpes simplex virus type 1 (HSV-1)--induced retinitis following herpes simplex encephalitis: indications for brain-to-eye transmission of HSV-1. Ann. Neurol. 49, 104–106. doi: 10.1002/1531-8249(200101)49:1<104::aid-ana15>3.0.co;2-y
Markus, A., Waldman Ben-Asher, H., Kinchington, P. R., and Goldstein, R. S. (2014). Cellular transcriptome analysis reveals differential expression of pro-and antiapoptosis genes by varicella-zoster virus-infected neurons and fibroblasts. J. Virol. 88, 7674–7677. doi: 10.1128/JVI.00500-14
Moraru, M., Cisneros, E., Gómez-Lozano, N., de Pablo, R., Portero, F., Cañizares, M., et al. (2012). Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 188, 4412–4420. doi: 10.4049/jimmunol.1103434
Morinobu, A., Gadina, M., Strober, W., Visconti, R., Fornace, A., Montagna, C., et al. (2002). STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 99, 12281–12286. doi: 10.1073/pnas.182618999
Mørk, N., Kofod-Olsen, E., Sørensen, K. B., Bach, E., Ørntoft, T. F., Østergaard, L., et al. (2015). Mutations in the TLR3 signaling pathway and beyond in adult patients with herpes simplex encephalitis. Genes Immun. 16, 552–566. doi: 10.1038/gene.2015.46
Murakami, M., Yamamoto, K., and Taketomi, Y. (2018). Phospholipase A2 in skin biology: new insights from gene-manipulated mice and lipidomics. Inflammat. Regenerat. 38:31. doi: 10.1186/s41232-018-0089-2
Muthiah, M. N., Michaelides, M., Child, C. S., and Mitchell, S. M. (2007). Acute retinal necrosis: a national population-based study to assess the incidence, methods of diagnosis, treatment strategies and outcomes in the UK. Br. J. Ophthalmol. 91, 1452–1455. doi: 10.1136/bjo.2007.114884
Nie, L., Zhao, Y., Wu, W., Yang, Y.-Z., Wang, H.-C., and Sun, X.-H. (2011). Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell Res. 21, 754–769. doi: 10.1038/cr.2010.165
Nixon, B., Fakioglu, E., Stefanidou, M., Wang, Y., Dutta, M., Goldstein, H., et al. (2014). Genital herpes simplex virus type 2 infection in humanized HIV-transgenic mice triggers HIV shedding and is associated with greater neurological disease. J. Infect. Dis. 209, 510–522. doi: 10.1093/infdis/jit472
Ogunjimi, B., Zhang, S.-Y., Sørensen, K. B., Skipper, K. A., Carter-Timofte, M., Kerner, G., et al. (2017). Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J. Clin. Invest. 127, 3543–3556. doi: 10.1172/JCI92280
Oh, S.-J., Lim, S., Song, M. J., Ahn, J. H., Lee, C. H., and Shin, O. S. (2019). Whole transcriptome analyses reveal differential mRNA and microRNA expression profiles in primary human dermal fibroblasts infected with clinical or vaccine strains of varicella zoster virus. Pathogens 8:183. doi: 10.3390/pathogens8040183
Orsi, A., Razi, M., Dooley, H. C., Robinson, D., Weston, A. E., Collinson, L. M., et al. (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873. doi: 10.1091/mbc.E11-09-0746
Pandey, U., Renner, D. W., Thompson, R. L., Szpara, M. L., and Sawtell, N. M. (2017). Inferred father-to-son transmission of herpes simplex virus results in near-perfect preservation of viral genome identity and in vivo phenotypes. Sci. Rep. 7:13666. doi: 10.1038/s41598-017-13936-6
Pandey, P., Sliker, B., Peters, H. L., Tuli, A., Herskovitz, J., Smits, K., et al. (2016). Amyloid precursor protein and amyloid precursor-like protein 2 in cancer. Oncotarget 7, 19430–19444. doi: 10.18632/oncotarget.7103
Pandey, U., and Szpara, M. L. (2019). Herpes simplex virus disease management and diagnostics in the era of high-throughput sequencing. Clin. Microbiol. Newsl. 41, 41–48. doi: 10.1016/j.clinmicnews.2019.02.003
Pérez de Diego, R., Sancho-Shimizu, V., Lorenzo, L., Puel, A., Plancoulaine, S., Picard, C., et al. (2010). Human TRAF3 adaptor molecule deficiency leads to impaired toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33, 400–411. doi: 10.1016/j.immuni.2010.08.014
Qu, J., Kaufman, Y., and Washington, I. (2009). Coenzyme Q10 in the human retina. Invest. Ophthalmol. Vis. Sci. 50, 1814–1818. doi: 10.1167/iovs.08-2656
Rahaus, M., Desloges, N., and Wolff, M. H. (2004). Replication of varicella-zoster virus is influenced by the levels of JNK/SAPK and p38/MAPK activation. J. Gen. Virol. 85, 3529–3540. doi: 10.1099/vir.0.80347-0
Rehbein, U., Prentzell, M. T., Cadena Sandoval, M., Heberle, A. M., Henske, E. P., Opitz, C. A., et al. (2021). The TSC complex-mTORC1 Axis: from lysosomes to stress granules and Back. Front. Cell Development. Biol. 9:751892. doi: 10.3389/fcell.2021.751892
Reva, B., Antipin, Y., and Sander, C. (2011). Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39:e118. doi: 10.1093/nar/gkr407
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424. doi: 10.1038/gim.2015.30
Richards, J. T., Kern, E. R., Overall, J. C., and Glasgow, L. A. (1981). Differences in neurovirulence among isolates of Herpes simplex virus types 1 and 2 in mice using four routes of infection. J. Infect. Dis. 144, 464–471. doi: 10.1093/infdis/144.5.464
Rizzo, R., Gentili, V., Casetta, I., Caselli, E., de Gennaro, R., Granieri, E., et al. (2012). Altered natural killer cells' response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J. Neuroimmunol. 251, 55–64. doi: 10.1016/j.jneuroim.2012.07.004
Roesler, J., Kofink, B., Wendisch, J., Heyden, S., Paul, D., Friedrich, W., et al. (1999). Listeria monocytogenes and recurrent mycobacterial infections in a child with complete interferon-gamma-receptor (IFNgammaR1) deficiency: mutational analysis and evaluation of therapeutic options. Exp. Hematol. 27, 1368–1374. doi: 10.1016/s0301-472x(99)00077-6
Sancho-Shimizu, V., Pérez de Diego, R., Lorenzo, L., Halwani, R., Alangari, A., Israelsson, E., et al. (2011). Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121, 4889–4902. doi: 10.1172/JCI59259
Sarkar, S. N., Bandyopadhyay, S., Ghosh, A., and Sen, G. C. (1999a). Enzymatic characteristics of recombinant medium isozyme of 2′-5′ oligoadenylate synthetase. J. Biol. Chem. 274, 1848–1855. doi: 10.1074/jbc.274.3.1848
Sarkar, S. N., Ghosh, A., Wang, H. W., Sung, S. S., and Sen, G. C. (1999b). The nature of the catalytic domain of 2′-5′-oligoadenylate synthetases. J. Biol. Chem. 274, 25535–25542. doi: 10.1074/jbc.274.36.25535
Schwarz, J. M., Cooper, D. N., Schuelke, M., and Seelow, D. (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362. doi: 10.1038/nmeth.2890
Shenoy, A. R., Wellington, D. A., Kumar, P., Kassa, H., Booth, C. J., Cresswell, P., et al. (2012). GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485. doi: 10.1126/science.1217141
Shihab, H. A., Gough, J., Mort, M., Cooper, D. N., Day, I. N. M., and Gaunt, T. R. (2014). Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genomics 8:11. doi: 10.1186/1479-7364-8-11
Shimizu, K., Tani, M., Watanabe, H., Nagamachi, Y., Niinaka, Y., Shiroishi, T., et al. (1999). The autocrine motility factor receptor gene encodes a novel type of seven transmembrane protein. FEBS Lett. 456, 295–300. doi: 10.1016/s0014-5793(99)00966-7
Shin, J. H., Moreno-Nieves, U. Y., Zhang, L. H., Chen, C., Dixon, A. L., Linde, M. H., et al. (2021). AHR regulates NK cell migration via ASB2-mediated ubiquitination of Filamin A. Front. Immunol. 12:624284. doi: 10.3389/fimmu.2021.624284
Shin, J., Nile, A., and Oh, J.-W. (2021). Role of adaptin protein complexes in intracellular trafficking and their impact on diseases. Bioengineered 12, 8259–8278. doi: 10.1080/21655979.2021.1982846
Shipley, M. M., Renner, D. W., Ott, M., Bloom, D. C., Koelle, D. M., Johnston, C., et al. (2018). Genome-wide surveillance of genital herpes simplex virus type 1 from multiple anatomic sites over time. J. Infect. Dis. 218, 595–605. doi: 10.1093/infdis/jiy216
Sifuentes-Franco, S., Sánchez-Macías, D. C., Carrillo-Ibarra, S., Rivera-Valdés, J. J., Zuñiga, L. Y., and Sánchez-López, V. A. (2022). Antioxidant and anti-inflammatory effects of coenzyme Q10 supplementation on infectious diseases. Healthcare 10:487. doi: 10.3390/healthcare10030487
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43:D447-52. doi: 10.1093/nar/gku1003
Takahashi, M.-N., Jackson, W., Laird, D. T., Culp, T. D., Grose, C., Haynes, J. I., et al. (2009). Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. J. Virol. 83, 5466–5476. doi: 10.1128/JVI.02670-08
Thompson, R. L., Williams, R. W., Kotb, M., and Sawtell, N. M. (2014). A forward phenotypically driven unbiased genetic analysis of host genes that moderate herpes simplex virus virulence and stromal keratitis in mice. PLoS One 9:e92342. doi: 10.1371/journal.pone.0092342
Thomsen, M. M., Tyrberg, T., Skaalum, K., Carter-Timofte, M., Freytag, M. R., Norberg, P., et al. (2021). Genetic variants and immune responses in a cohort of patients with varicella zoster virus encephalitis. J. Infect. Dis. 224, 2122–2132. doi: 10.1093/infdis/jiab254
Tibbs, E., and Cao, X. (2022). Emerging canonical and non-canonical roles of Granzyme B in health and disease. Cancers 14:1436. doi: 10.3390/cancers14061436
Tommasi, C., Rogerson, C., Depledge, D. P., Jones, M., Naeem, A. S., Venturini, C., et al. (2020). Kallikrein-mediated cytokeratin 10 degradation is required for varicella zoster virus propagation in skin. J. Invest. Dermatol. 140, 774–784.e11. doi: 10.1016/j.jid.2019.08.448
Toniato, E., Chen, X. P., Losman, J., Flati, V., Donahue, L., and Rothman, P. (2002). TRIM8/GERP RING finger protein interacts with SOCS-1. J. Biol. Chem. 277, 37315–37322. doi: 10.1074/jbc.M205900200
Traschütz, A., Schirinzi, T., Laugwitz, L., Murray, N. H., Bingman, C. A., Reich, S., et al. (2020). Clinico-genetic, imaging and molecular delineation of COQ8A-Ataxia: a multicenter study of 59 patients. Ann. Neurol. 88, 251–263. doi: 10.1002/ana.25751
Tyler, K. L. (2004). Herpes simplex virus infections of the central nervous system: encephalitis and meningitis, including Mollaret's. Herpes 11, 57A–64A.
Vastag, L., Koyuncu, E., Grady, S. L., Shenk, T. E., and Rabinowitz, J. D. (2011). Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 7:e1002124. doi: 10.1371/journal.ppat.1002124
Verbakel, S. K., van Huet, R. A. C., Boon, C. J. F., den Hollander, A. I., Collin, R. W. J., Klaver, C. C. W., et al. (2018). Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 66, 157–186. doi: 10.1016/j.preteyeres.2018.03.005
Volk, T., Pannicke, U., Reisli, I., Bulashevska, A., Ritter, J., Björkman, A., et al. (2015). DCLRE1C (ARTEMIS) mutations causing phenotypes ranging from atypical severe combined immunodeficiency to mere antibody deficiency. Hum. Mol. Genet. 24, 7361–7372. doi: 10.1093/hmg/ddv437
von Hofsten, J., Bergström, T., and Zetterberg, M. (2019). Alpha herpes virus type and viral load in intraocular fluids in patients with acute retinal necrosis. BMJ Open Ophthalmol. 4:e000247. doi: 10.1136/bmjophth-2018-000247
von Hofsten, J., Ringlander, J., Norberg, P., Zetterberg, M., Andersson, M., Lindh, M., et al. (2020). Deep sequencing of varicella-zoster virus in aqueous humor from a patient with acute retinal necrosis presenting with acute glaucoma. Open Forum Infect. Dis. 7:ofaa198. doi: 10.1093/ofid/ofaa198
Wang, J., Chun, H. J., Wong, W., Spencer, D. M., and Lenardo, M. J. (2001). Caspase-10 is an initiator caspase in death receptor signalling. Proc. Natl. Acad. Sci. U. S. A. 98, 13884–13888. doi: 10.1073/pnas.241358198
Wang, K. S., Frank, D. A., and Ritz, J. (2000). Interleukin-2 enhances the response of natural killer cells to interleukin-12 through up-regulation of the interleukin-12 receptor and STAT4. Blood 95, 3183–3190. doi: 10.1182/blood.V95.10.3183
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603
Weber, F., Wagner, V., Rasmussen, S. B., Hartmann, R., and Paludan, S. R. (2006). Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80, 5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006
Whitley, R. J., and Gnann, J. W. (2002). Viral encephalitis: familiar infections and emerging pathogens. Lancet 359, 507–513. doi: 10.1016/S0140-6736(02)07681-X
Wiegering, V., Schick, J., Beer, M., Weissbrich, B., Gattenlöhner, S., Girschick, H. J., et al. (2011). Varicella-zoster virus infections in immunocompromised patients - a single Centre 6-years analysis. BMC Pediatr. 11:31. doi: 10.1186/1471-2431-11-31
Wilcox, D. R., Wadhwani, N. R., Longnecker, R., and Muller, W. J. (2015). Differential reliance on autophagy for protection from HSV encephalitis between newborns and adults. PLoS Pathog. 11:e1004580. doi: 10.1371/journal.ppat.1004580
Witzel, M., Petersheim, D., Fan, Y., Bahrami, E., Racek, T., Rohlfs, M., et al. (2017). Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet. 49, 742–752. doi: 10.1038/ng.3833
Wu, S., Yang, S., Ou, M., Chen, J., Huang, J., Xiong, D., et al. (2021). Transcriptome analysis reveals the role of cellular calcium disorder in varicella zoster virus-induced post-herpetic neuralgia. Front. Mol. Neurosci. 14:665931. doi: 10.3389/fnmol.2021.665931
Wu, X. M., Zhang, J., Li, P. W., Hu, Y. W., Cao, L., Ouyang, S., et al. (2020). NOD1 promotes antiviral signaling by binding viral RNA and regulating the interaction of MDA5 and MAVS. J. Immunol. 204, 2216–2231. doi: 10.4049/jimmunol.1900667
Yakoub, A. M., and Shukla, D. (2015). Autophagy stimulation abrogates herpes simplex virus-1 infection. Sci. Rep. 5:9730. doi: 10.1038/srep09730
Yan, F.-J., Zhang, X.-J., Wang, W.-X., Ji, Y.-X., Wang, P.-X., Yang, Y., et al. (2017). The E3 ligase tripartite motif 8 targets TAK1 to promote insulin resistance and steatohepatitis. Hepatology 65, 1492–1511. doi: 10.1002/hep.28971
Ye, W., Hu, M.-M., Lei, C.-Q., Zhou, Q., Lin, H., Sun, M.-S., et al. (2017). TRIM8 negatively regulates TLR3/4-mediated innate immune response by blocking TRIF-TBK1 interaction. J. Immunol. 199, 1856–1864. doi: 10.4049/jimmunol.1601647
Yee, C. S., Massaad, M. J., Bainter, W., Ohsumi, T. K., Föger, N., Chan, A. C., et al. (2016). Recurrent viral infections associated with a homozygous CORO1A mutation that disrupts oligomerization and cytoskeletal association. J. Allergy Clin. Immunol. 137, 879–88.e2. doi: 10.1016/j.jaci.2015.08.020
Yokota, S., Yokosawa, N., Okabayashi, T., Suzutani, T., Miura, S., Jimbow, K., et al. (2004). Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J. Virol. 78, 6282–6286. doi: 10.1128/JVI.78.12.6282-6286.2004
Zeke, A., Misheva, M., Reményi, A., and Bogoyevitch, M. A. (2016). JNK signaling: regulation and functions based on complex protein-protein partnerships. Microbiol Mol Biol Rev 80, 793–835. doi: 10.1128/MMBR.00043-14
Zhang, S.-Y., Clark, N. E., Freije, C. A., Pauwels, E., Taggart, A. J., Okada, S., et al. (2018). Inborn errors of RNA lariat metabolism in humans with brainstem viral infection. Cells 172, 952–965.e18. doi: 10.1016/j.cell.2018.02.019
Zhang, Q., Davis, J. C., Lamborn, I. T., Freeman, A. F., Jing, H., Favreau, A. J., et al. (2009). Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 361, 2046–2055. doi: 10.1056/NEJMoa0905506
Zhang, S.-Y., Jouanguy, E., Ugolini, S., Smahi, A., Elain, G., Romero, P., et al. (2007). TLR3 deficiency in patients with herpes simplex encephalitis. Science 317, 1522–1527. doi: 10.1126/science.1139522
Zhang, X., Tohari, A. M., Marcheggiani, F., Zhou, X., Reilly, J., Tiano, L., et al. (2017). Therapeutic potential of co-enzyme Q10 in retinal diseases. Curr. Med. Chem. 24, 4329–4339. doi: 10.2174/0929867324666170801100516
Zhang, Y. S., Xin, D. E., Wang, Z., Peng, W., Zeng, Y., Liang, J., et al. (2022). Acetylation licenses Th1 cell polarization to constrain Listeria monocytogenes infection. Cell Death Differ. 29, 2303–2315. doi: 10.1038/s41418-022-01017-9
Zhao, Y. G., Chen, Y., Miao, G., Zhao, H., Qu, W., Li, D., et al. (2017). The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell 67, 974–989.e6. doi: 10.1016/j.molcel.2017.08.005
Zwezdaryk, K. J., Combs, J. A., Morris, C. A., and Sullivan, D. E. (2016). Regulation of Wnt/β-catenin signaling by herpesviruses. World J. Virol. 5, 144–154. doi: 10.5501/wjv.v5.i4.144
Keywords: acute retinal necrosis (ARN), interferon, autophagy, apoptosis, whole exome sequencing
Citation: Heinz JL, Swagemakers SMA, von Hofsten J, Helleberg M, Thomsen MM, De Keukeleere K, de Boer JH, Ilginis T, Verjans GMGM, van Hagen PM, van der Spek PJ and Mogensen TH (2023) Whole exome sequencing of patients with varicella-zoster virus and herpes simplex virus induced acute retinal necrosis reveals rare disease-associated genetic variants. Front. Mol. Neurosci. 16:1253040. doi: 10.3389/fnmol.2023.1253040
Received: 04 July 2023; Accepted: 09 October 2023;
Published: 25 October 2023.
Edited by:
Francesca Caccuri, University of Brescia, ItalyReviewed by:
Elisabetta Caselli, University of Ferrara, ItalyCopyright © 2023 Heinz, Swagemakers, von Hofsten, Helleberg, Thomsen, De Keukeleere, de Boer, Ilginis, Verjans, van Hagen, van der Spek and Mogensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Trine H. Mogensen, dHJpbm1vZ2VAcm0uZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.