
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 28 September 2023
Sec. Brain Disease Mechanisms
Volume 16 - 2023 | https://doi.org/10.3389/fnmol.2023.1246842
This study explores the implications of a novel germline missense mutation (R38C) in the succinate dehydrogenase (SDH) subunit B, which has been linked to neurodegenerative diseases. The mutation was identified from the SDH mutation database and corresponds to the SDH2R32C allele, mirroring the human SDHBR38C mutation. By subjecting the mutant yeast model to hydrogen peroxide (H2O2) stress, simulating oxidative stress, we observed heightened sensitivity to oxidative conditions. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis revealed significant regulation (p < 0.05) of genes associated with antioxidant systems and energy metabolism. Through gas chromatography-mass spectrometry (GC-MS) analysis, we examined yeast cell metabolites under oxidative stress, uncovering insights into the potential protective role of o-vanillin. This study elucidates the biological mechanisms underlying cellular oxidative stress responses, offering valuable insights into its repercussions. These findings shed light on innovative avenues for addressing neurodegenerative diseases, potentially revolutionizing therapeutic strategies.
The only enzyme that is shared by both the tricarboxylic acid (TCA) cycle and the electron transport chain is succinate dehydrogenase (SDH), also known as succinate-coenzyme Q reductase (SQR) or Complex II (Yamaguchi et al., 2022). The complex is located within the inner mitochondrial membrane. It has a mitochondrial matrix-oriented and a mitochondrial membrane gap integration domain, with SDHA and SDHB hydrophilic subunits implanted in the inner mitochondrial membrane and SDHC and SDHD hydrophobic subunits positioned on the mitochondrial matrix side (Moosavi et al., 2020).
According to Schweizer et al. (2020), SDHB deficiency, or inadequate succinate dehydrogenase B subunit, is frequently present in paragangliomas (PCPGs). In SDHB mutant PCPGs, the levels of reactive oxygen species (ROS) are elevated, the tricarboxylic cycle is metabolically reprogrammed, and the electron transport chain is altered (Goffrini et al., 2009). Cancer is characterized by oxidative stress, which speeds up the growth of tumors and the spread of the disease (Quinlan et al., 2012; Bezawork-Geleta et al., 2018; Sun et al., 2020). Examples include PCPGs with SDHB mutations and malignancies with IDH1 mutations. Cellular metabolism and redox balance were found to be altered by the functional disruption of mitochondrial complex II brought on by SDHB mutations in cancer cells (Mauer et al., 2020).
Diabetes, cardiovascular disease, and many other illnesses are all linked to oxidative stress (Rhee and Bae, 2015). Numerous studies have shown over time that oxidative stress plays a significant role in the development of illnesses (Zhang et al., 2022). Given that oxidative stress has a role in paragangliomatosis, antioxidant therapy may be employed to treat the disorder (Henderson et al., 2009). However, more fundamental research is required to completely comprehend the mechanisms through which oxidative stress contributes to the development of pheochromocytoma/paraganglioma (PGLs), as these treatments have not yet shown evidence of therapeutic benefit.
Recently, researchers have used yeast models to examine the functional effects of gene mutations on the enzymatic activity and respiratory activity of SDH (Szeto et al., 2007; Beimers et al., 2022). Yeast is a good model for studying gene functions related to human illnesses because it can survive without a working mitochondrial respiratory chain (Baruffini and Lodi, 2010; Invernizzi et al., 2013; Abugable et al., 2017; Prevost et al., 2018; Basu et al., 2020; Gilea et al., 2021). The ability of yeast cells to quickly delete, modify, and reinsert S. cerevisiae genes, in contrast to human cells, provides a wealth of important information for understanding the molecular causes of disease (Kumar and Reichert, 2021). This study uses the SDH2 gene, which is the yeast equivalent of the mammalian SDHB gene, as a model for a typical family of PGL mutations.
The TCA Circulating Gene Mutation Database1 provides an overview of complex II mutations (Hoekstra and Bayley, 2013). Around 15% of paragangliomas are thought to be caused by all Complex II mutations, which are thought to have tumor-suppressive roles (Pęczkowska et al., 2013; Richter et al., 2019; Wurth et al., 2021). The reported number of mutations for each component gene varies significantly for unknown reasons. Five categories of gene mutations exist: harmful, potentially pathogenic, unknown clinical significance (VUS), potentially benign, and benign. Some diseases are brought on by a gene mutation that can be seen in the clinical manifestation. The newly identified mutation, on the other hand, is typically not supported by any conclusive literature and is indicated in the disease mutation database as having questionable clinical relevance. Since the mutation with unknown clinical implications may be pathogenic, numerous functional investigations can be done to demonstrate whether the mutation is deleterious at this time (Makhnoon et al., 2019).
In the study presented here, the R38C missense mutation in the B subunit of a new kind of succinate dehydrogenase (SDH) linked to neurodegenerative illnesses is examined for its functional effects. To our knowledge, this particular mutation has not been previously described, and it is yet unclear how it affects cellular function and disease pathogenesis. Our research intends to close this information gap and illuminate the molecular significance of this mutation in relation to neurodegenerative disorders.
In this work, phenotypic changes of yeast cell development were evaluated first. The internal ultrastructure of yeast cells was then described at the organelle level using scanning TEM. The molecular expression of genes related to energy metabolism and antioxidant enzyme systems was examined using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The metabolites of yeast cells under oxidative stress were then analyzed using GC-MS. This study seeks to provide a novel perspective on the molecular mechanisms affecting metabolites and yeast cell resistance to oxidative stress caused by SDH2 mutation (Figure 1).
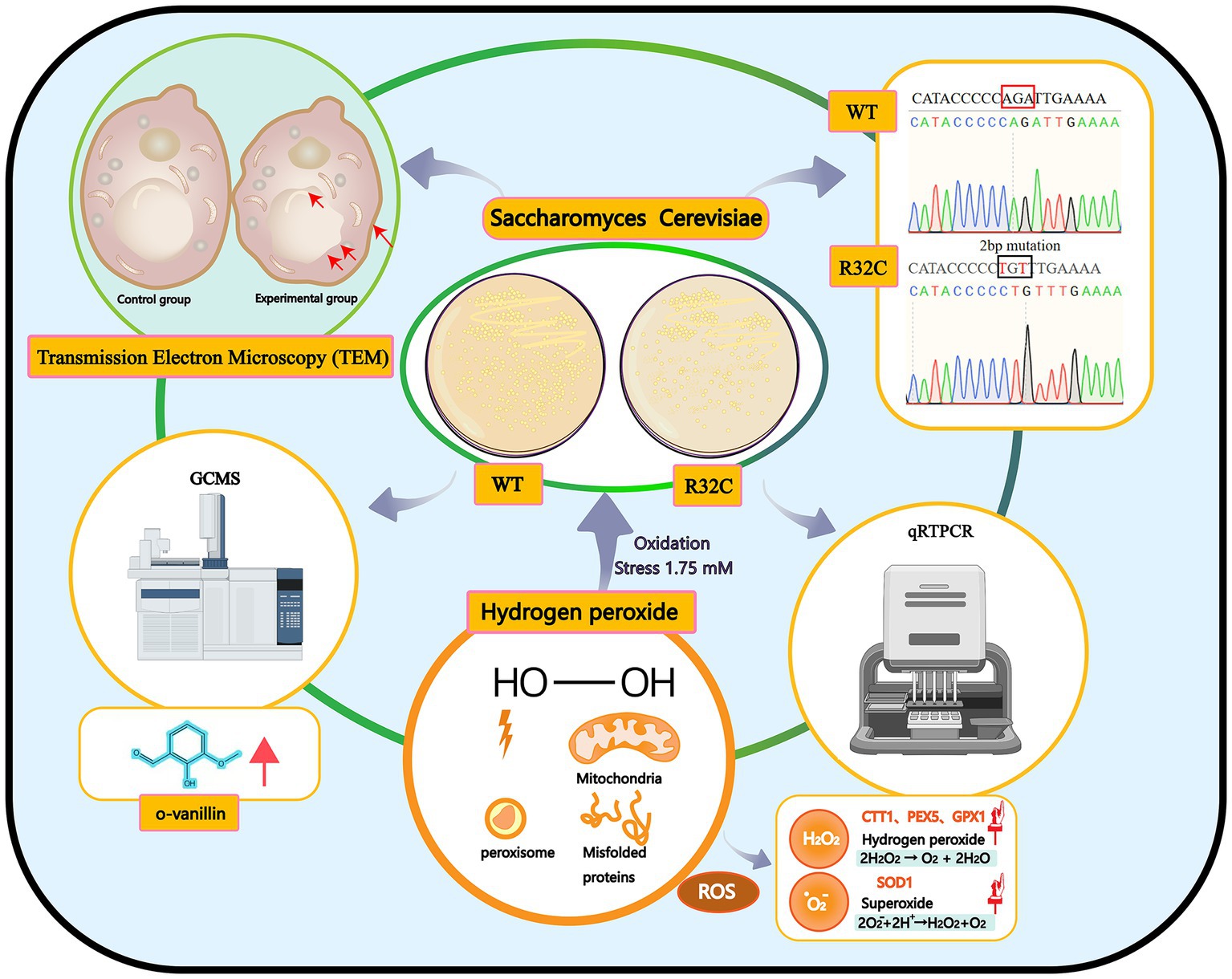
Figure 1. Graphical summary of the methods and results used in this study. Schematic representation of the interactions of the effects of hydrogen peroxide stress on Saccharomyces cerevisiae. Production of peroxisomes and misfolded proteins under oxidative stress.
The strain used in this study was Saccharomyces cerevisiae wild type BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). The YPD medium (1% yeast extract, 2% peptone, and 2% glucose) was used to culture the cells. To check for SDH2 transformants, cells were cultured in a YPD medium with 200 μg/mL genomycin as an addition. To test the sensitivity under oxidative conditions, different concentrations of hydrogen peroxide were applied to YPD medium. Cells were incubated in a liquid YPD medium while being agitated at 220 rpm and 29°C for other tests.
As previously stated (Lopez-Berges et al., 2010), our work used the plasmid pUG6 and the primers specified in Table 1 to replace a particular gene by PCR-mediated homologous integration. Geneticin resistance (KanMX) was used as a screenable marker. Using knockout primers, a 1.8 kb DNA fragment from the pUG6 genome was amplified by PCR to produce a knockout cassette for SDH2. A genomic analysis of the transformed strain was conducted using PCR to verify the insertion of outside genes into the structure.
Table 2 contains a list of the investigation’s primers, and PYX212 was the plasmid used in this study. Using the BY4741 genome as a template, SDH2-F and SDH2-R were used as primers for PCR amplification of the SDH2 fragment with the enzymatic cut site (Tao et al., 2020; Liang et al., 2021).
To determine S. cerevisiae cells’ receptivity to oxidative stress, they were grown for a whole day at 29°C in a YPD liquid medium. The cells were suspended in fresh YPD liquid the following day with an OD600 value set to 1. Before being spotted in 3 μL onto the control and 1, 1.5, and 1.75 mM H2O2 YPD solid medium and incubated upside down at 30°C for 2–3 d, the samples were diluted to 10−1, 10−2, 10−3, 10−4, and 10−5. The growth was observed, noted, and visually documented. To investigate the growth inhibition of BY4741 and mutant strains under oxidative stress, cells were grown in a liquid medium containing YPD, and 1, 1.5, and 1.75 mM H2O2 and OD600 values were checked every 3 h. Data were acquired in order to plot growth curves (Edwards et al., 2020).
In order to perform the MTT experiment, yeast cells were grown to the logarithmic growth stage, treated with hydrogen peroxide at doses of 0 and 1.75 mM, respectively. Each well received a total of 20 μL of a 5 mg/mL MTT solution (Biyun Tian, China). The supernatant was removed by centrifugation after 4 h of incubation, and 200 μL of dimethyl sulfoxide was then added. The culture plate was placed on an enzyme marker (Thermo, United States) and the optical density (OD) value was determined at 490 nm after shaking for 10 min.
2′,7′-Dichlorofluorescein diacetate (DCFH-DA, Molecular Probes, United States) was used to quantify ROS levels (Serpa et al., 2021). Overnight, strains were grown in YPD medium. After being collected, the cells were diluted in fresh YPD to an OD600 of 0.5, and then incubated for 6 h at 30°C. One hundred and eighty revolutions per minute, then 1.75 mM of hydrogen peroxide was added. The cells were then exposed to hydrogen peroxide for one additional hour. Control and treated cells were stained with 20 mg/mL DCFH-DA and then incubated at 30°C for 30 min after being washed with phosphate free saline (PBS). Following a second PBS buffer wash, cells were examined with a fluorescent enzyme marker (Abdullah et al., 2022).
The level of lipid peroxidation and cellular activity in the cells were determined using the malondialdehyde (MDA) assay kit, superoxide dismutase (SOD), glutathione GPx, and succinate dehydrogenase (SDH) assay kits (Nanjing Jiancheng Bioengineering Institute), respectively (V et al., 2022).
Using pESC-URA plasmid, SDH2-R32C and SNCA genes (synthesized by Kingsley Biotechnology Co., Ltd., China) were exogenously inserted and introduced into yeast cells for expression (Khulape et al., 2015).
Regarding the aforementioned technique, cells were given a hydrogen peroxide treatment. Cells under oxidative stress and under control were collected, split using liquid nitrogen, and extracted with RNAiso Plus (TaKaRa) for RNA extraction. With the help of the Taq Pro Universal SYBR qPCR Master Mix from Vazyme Biotechnology in China, real-time fluorescence quantitative PCR was carried out (Qian et al., 2021; Wang et al., 2023). Table 3 provides a list of the primers employed in qRT-PCR tests.
After being treated with hydrogen peroxide, yeast was examined using transmission electron microscopy, as previously mentioned (Bayley et al., 2005). In a nutshell, cells were isolated by centrifugation, twice-washed with PBS, fixed with 2.5% glutaraldehyde at 4°C overnight, followed by 1% osmium acid solution for 1–2 h. Using an ethanol gradient, cells were dehydrated, and TEM analysis was performed (Ubhe, 2023).
The metabolites in S. cerevisiae were identified via gas chromatography–mass spectrometry (GC-MS). For metabolomics analysis, samples were quenched, wall-breaking was eliminated, and they were derivatized. The sample was separated at a controlled flow rate of 1 mL/min using helium with a purity of 99.999% as the carrier gas and a temperature program of 60°C for 1 min, followed by a continuous temperature increase of 12°C/min to 240°C and a target temperature increase of 40°C/min to 320°C for 5 min. The sample was injected at a 4:1 ratio of 1 L with a solvent delay time of 5 min at an input temperature of 250°C and an electron impact (EI) off-source temperature of 230°C. 70 V was utilized as the EI voltage. In full scan mode, data were collected for mass-to-charge ratios (m/z) between 50 and 600 (Liu et al., 2020; Roberts et al., 2023; Tufariello et al., 2023).
Survival data under oxidative stress conditions were determined using the MTT assay in the presence and absence of treatment with 50 μM o-vanillin, respectively.
The experiments were conducted in triplicate, and the results were analyzed using one-way and two-way ANOVA (Analysis of Variance) to determine their significance. The data is presented in bar graphs, showing the mean and standard deviation of the results, which demonstrate that the experiments were statistically significant.
The missense mutation c.112C>T, in which the base C at position 112 is transformed to base T, correlating to the amino acid alteration of arginine to cysteine at position 38, was found to be a novel mutation in the SDHB gene. This mutation was categorized as having a clinically unknown clinical significance (VUS) mutation. According to the results of the sequence comparison and the preferred codons, the yeast SDH2 gene differs from the human SDHB gene in that it contains base mutations at positions 32 and A, as shown in Figure 2A. Figure 2B illustrates that this residue is conserved. Figures 2C,D show the sequencing profile before and after the base mutation.
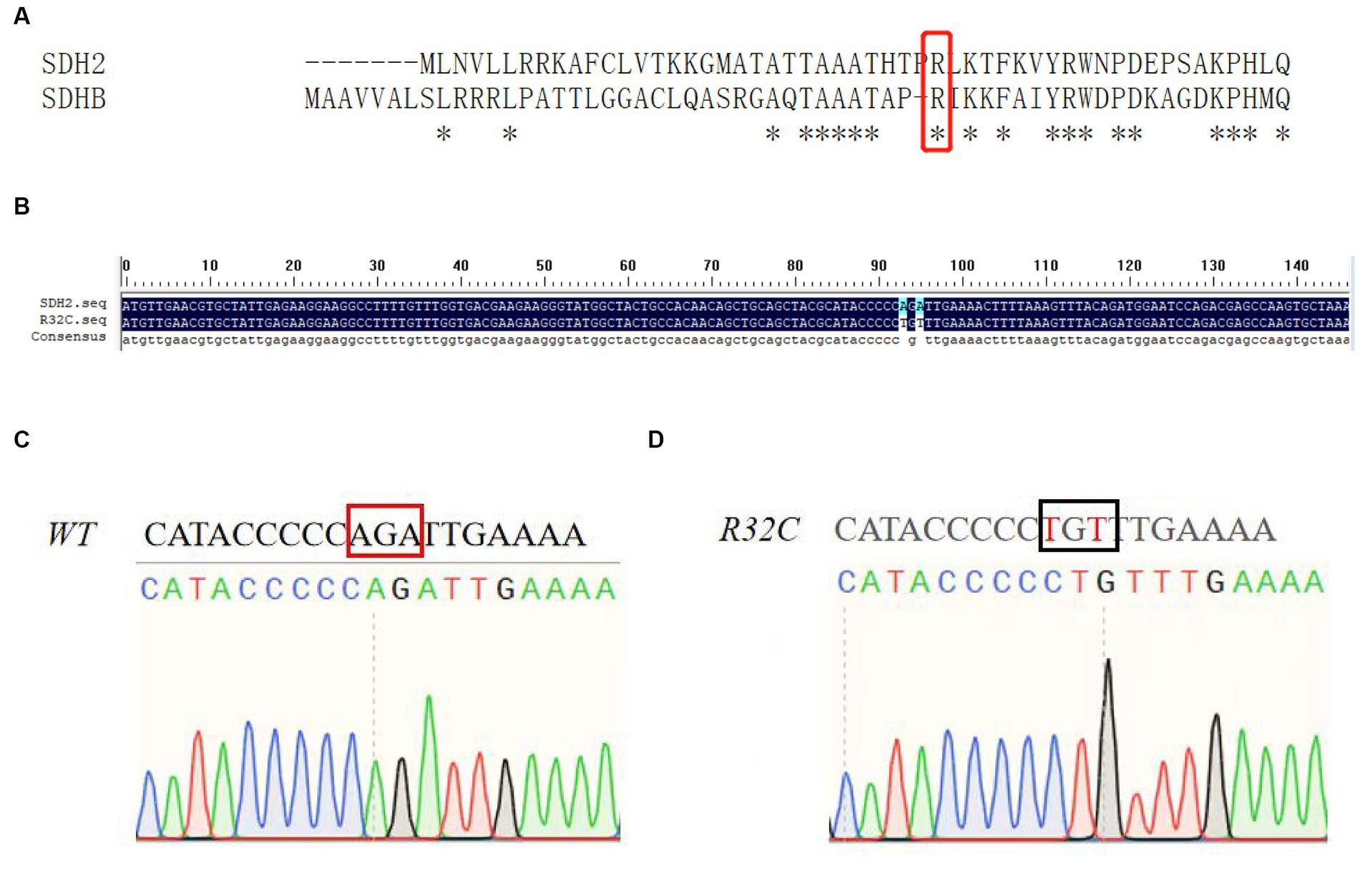
Figure 2. Mutation analysis. (A) Sequence alignment of yeast SDH2 gene with human SDHB gene; (B) Base sequence alignment of yeast SDH2 gene with R32C mutant bacteria. This indicates that the residue is conserved; (C, D) Sequencing map of yeast SDH2 wild type gene with R32C mutant bacteria.
The R32C mutation results in changes in the structure or function of the active site, which in turn affects SDH2 activity. The effect of the R32C mutant on the active site was assessed using a succinate dehydrogenase activity assay. The results show that the R32C SDH enzyme activity is lower than that of WT (Figure 3A).
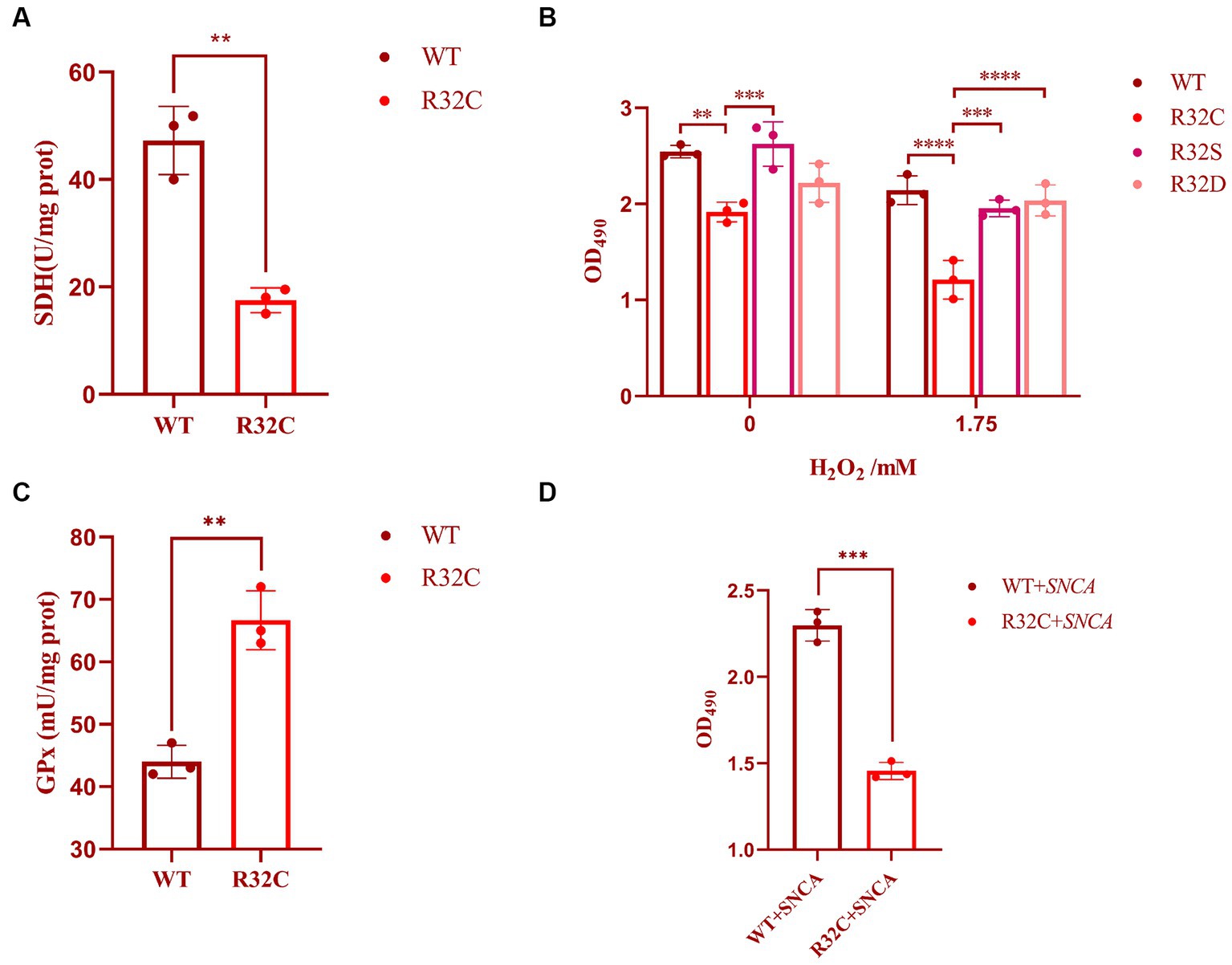
Figure 3. Cellular characterization of SDH2 and R32C. (A) Determination of succinate dehydrogenase (SDH) activity by Saccharomyces cerevisiae WT and R32C. (B) Determination of survival data of WT, R32C, R32S, and R32D by treatment with different concentrations of hydrogen peroxide. (C) Determination of WT and R32C glutathione peroxidase (GPx). (D) Determination of survival of WT and R32C after introduction of SNCA, respectively (mean ± sd). 0.01 < p < 0.05 (**); p < 0.01 (***); p < 0.0001 (****).
The R32C mutation changes an amino acid residue from arginine (R) to cysteine (C). Construction of a three-dimensional structural model of the mutant SDH2 subunit protein suggests that this amino acid sequence change alters the normal interactions between SDH2 and its complex partners, resulting in altered function or impaired protein–protein interactions (Figure 4).
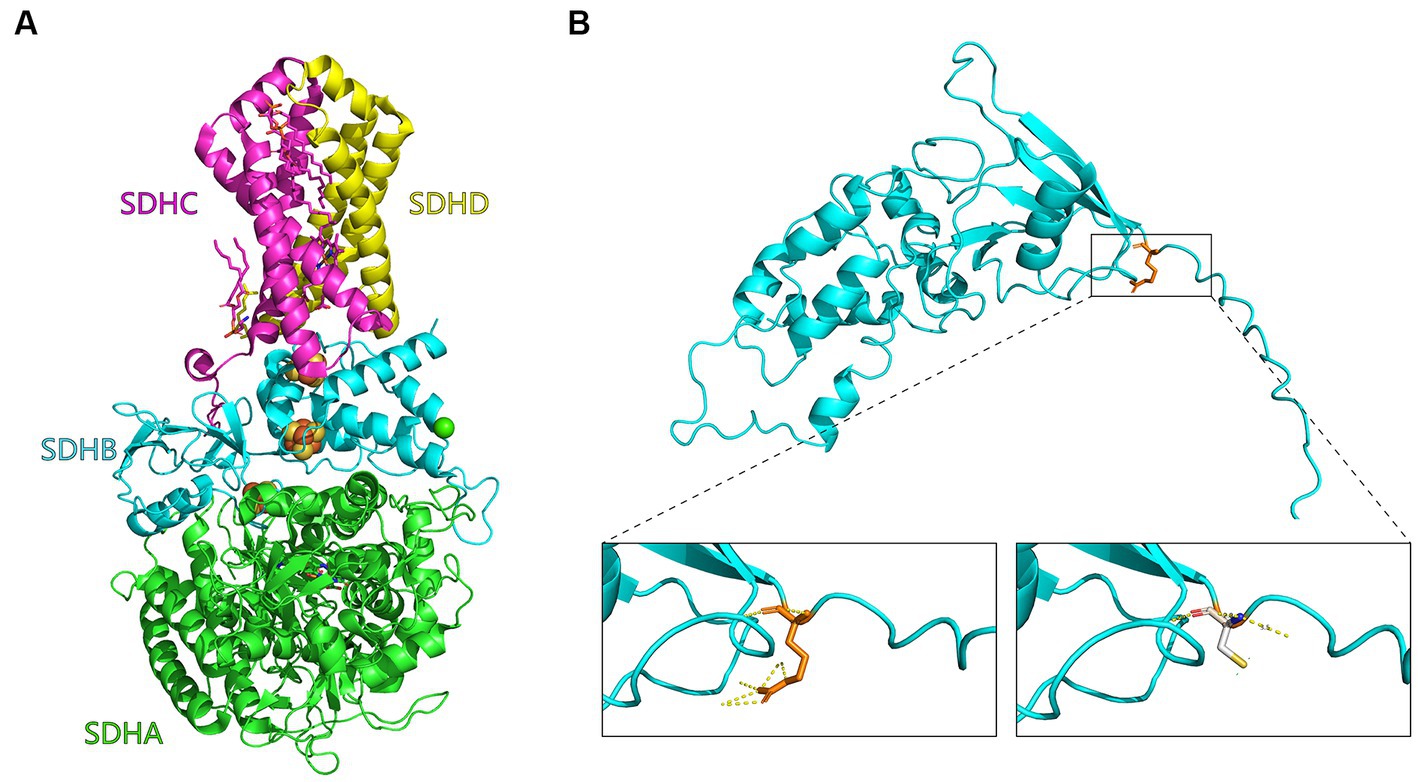
Figure 4. Mutant SDH2 subunit model. (A) Three-dimensional structural diagram of succinate dehydrogenase (1NEK.pdb) constructed with PyMOL. the SDHA, SDHB, SDHC, and SDHD subunits are indicated in green, cyan, purple, and yellow, respectively. (B) Enlarged view of the Saccharomyces cerevisiae SDH2 gene before and after the R32C mutation. On the left is the arginine (R32, orange) of the yeast SDH2 subunit before mutation, and interactions with other amino acids are indicated by yellow dashed lines. On the right is modeling of the mutation of arginine to cysteine on the yeast SDH2 subunit (C32, white).
The R32C mutation leads to an additional cysteine residue in the SDH2 protein. Cysteine residues can be oxidized and are known as redox switches. Other SDHB mutations recapitulated in yeast were not all reported to be sensitive to oxidative stress (Panizza et al., 2013), so this is something more specific to the mutation.
In this study, the effect of substitution on oxidative stress sensitivity was assessed by replacing cysteine with serine (S) and aspartic (D) acids, and cell survival was measured using MTT. The experimental results showed that the survival rate of R32C was significantly lower than that of WT and R32S and R32D under both 0 and 1.75 mM hydrogen peroxide stress (p < 0.05), while the cell survival of R32S and R32D was not significantly different from that of WT (p > 0.05, Figure 3B).
The effect of R32C on intracellular oxidative stress is interesting, so we investigated the intrinsic effects of R32C without added hydrogen peroxide. The effect of H2O2 scavenging can be assessed indirectly by measuring glutathione peroxidase (GPx). As shown in Figure 3C, the enzyme activity of GPx was significantly higher in R32C compared to WT. Therefore, R32C has an enhanced ability to scavenge H2O2 and maintained cellular homeostasis by increasing GPx enzyme activity.
To study the link between neurodegeneration and R32C mutants, alpha-synuclein (encoded by the SNCA gene) was used in yeast to screen R32C mutants for increased sensitivity to the protein. The results of the experiments are shown in Figure 3D. After introducing the SNCA gene in WT and R32C, respectively, the MTT method was used to compare the growth survival rates of the two strains, and it was found that the R32C mutant showed higher sensitivity compared to the WT strain, which may indicate a potential link between the mutant and neurodegeneration.
According to the dot plate experiment results, BY4741 was less impacted. At the same time, R32C growth was considerably hindered at 1.75 mM H2O2 stress (Figure 5A). Based on the results of growth curve experiments, R32C’s growth was found to be inhibited at 1.5 mM H2O2 stress, although BY4741’s growth was less impacted. Both strains’ growth was inhibited at 1.75 mM H2O2 stress (Figure 5B). The normal cells exhibited an intact cell wall structure, a clear cell membrane, an evident nucleus and vacuole, and mitochondria were visible in the cell pulp in the absence of further hydrogen peroxide stress (Figure 6A). However, in the hydrogen peroxide-treated cells, the cell morphology appeared depressed and atrophied, the typical cell wall and cell membrane structure vanished, the gap between the cell wall and cytoplasm expanded, the cytoplasm atrophied, electron-dense particles appeared in the middle electron-sparing layer, the contents of the cytoplasm were disorganized, and some of the contents were seen to leak out (Figure 6B). It is shown that oxidative stress causes cellular damage in yeast and its mutant strains, which may be related to the development of neurodegenerative diseases.
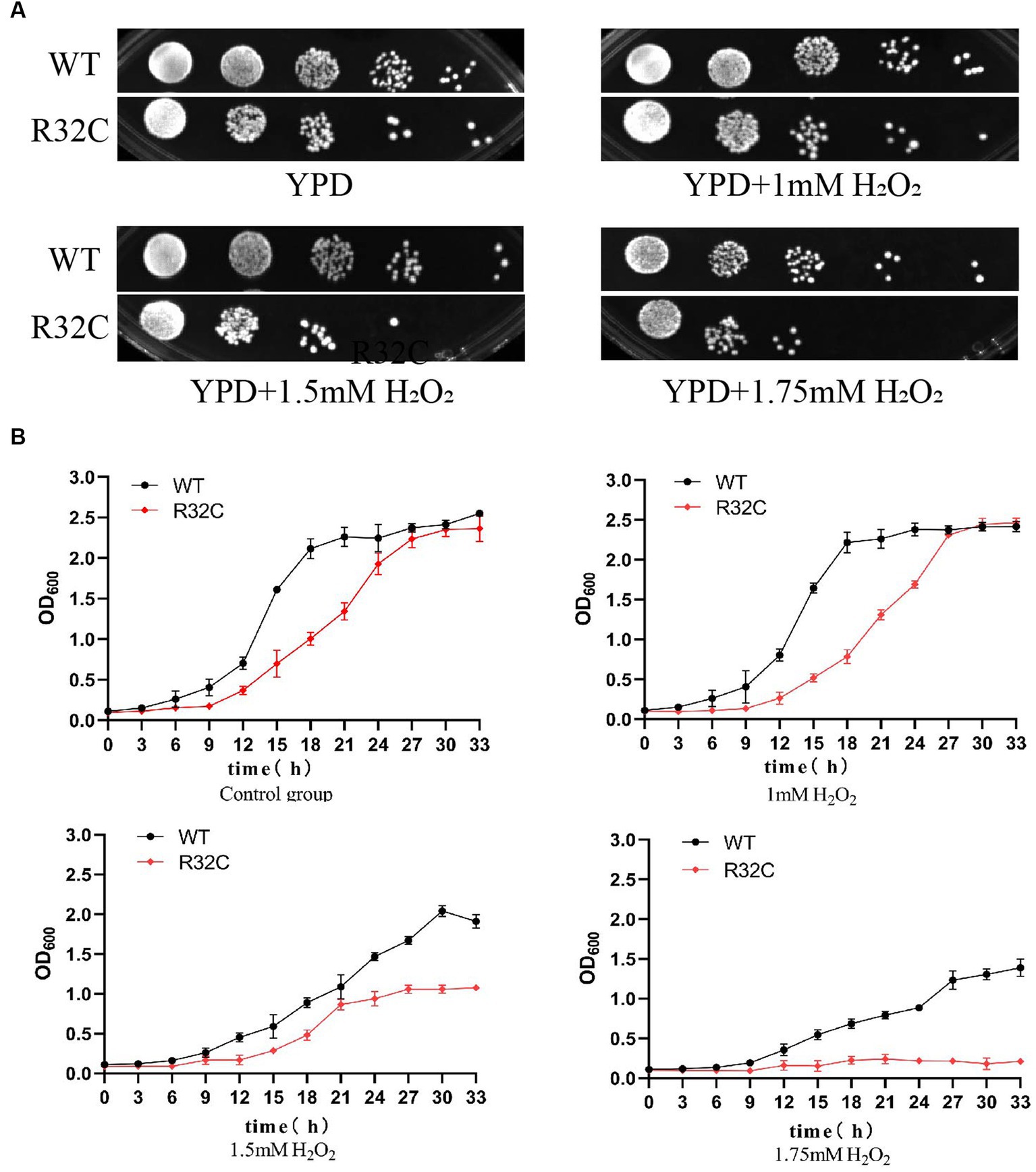
Figure 5. The R32C mutant strain resulted in increased susceptibility to oxidative stress. (A) Wild-type strain BY4741 and mutant strain R32C were cultured on solid YPD medium containing 0 mM, 1 mM, 1.5 mM, and 1.75 mM H2O2. (B) OD600 was measured at different H2O2 concentrations to compare the growth inhibition of BY4741 and R32C.
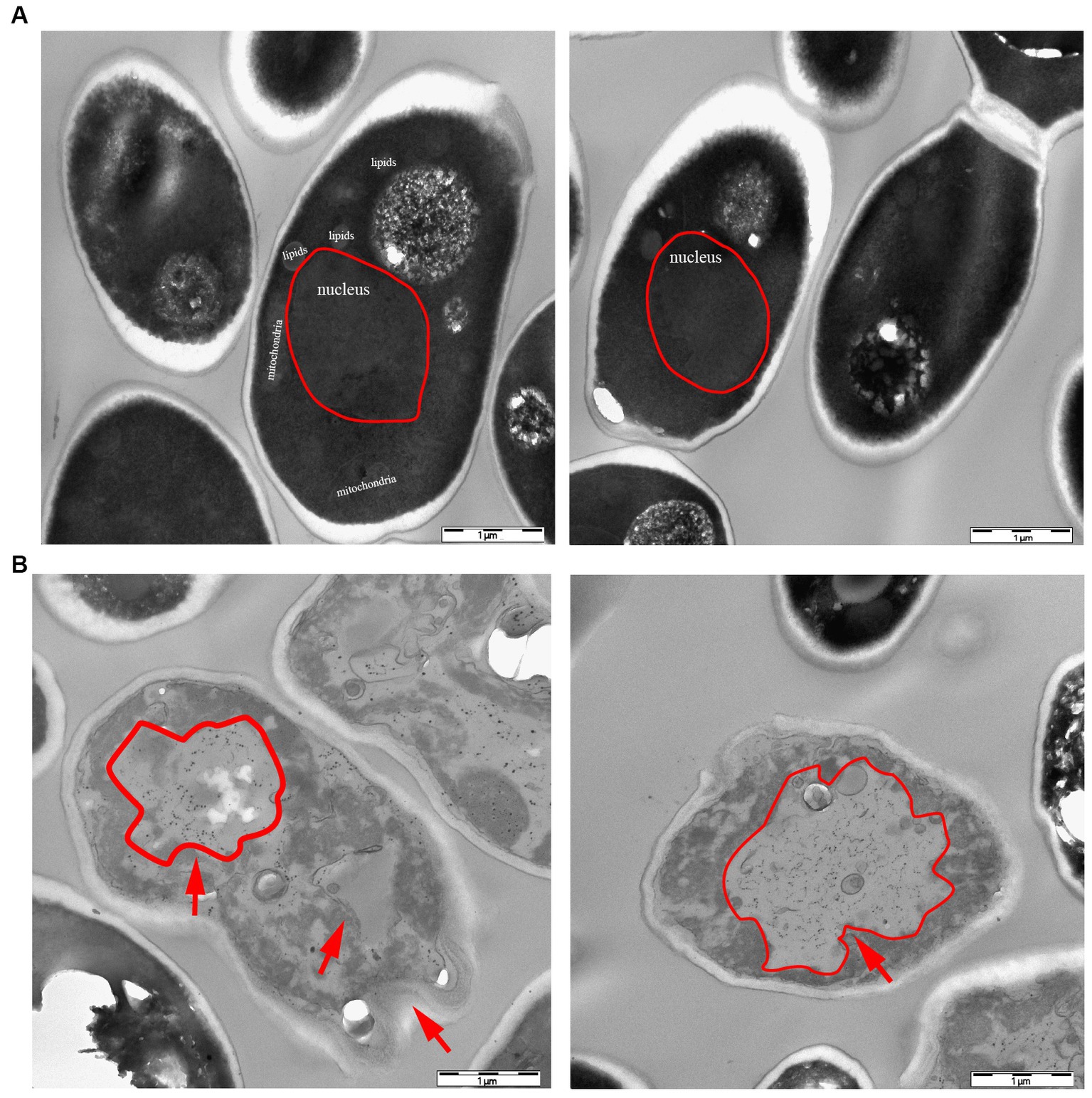
Figure 6. Transmission electron microscopy (TEM) characterization of yeast morphology. (A) WT control group without hydrogen peroxide stress. (B) Phenotypic characteristics of strain R32C under 1.75 mM hydrogen peroxide stress. As shown by the red arrows, in the hydrogen peroxide-treated cells, the cell morphology appears to be concave and atrophied.
An essential indicator of oxidative stress in living things is ROS. MDA is a significant marker in the lipid peroxidation response, which can indirectly represent the degree of free radical damage that cells have been exposed to Wu et al. (2023) and impact the structure of cell membranes (Tsikas, 2017). Hydrogen peroxide stress up-regulated the intracellular ROS content of S. cerevisiae and its mutant strains (p < 0.05) (Figure 7A) and increased the MDA content at a later stage (p < 0.05) (Figure 7B). These preliminary results indicated that lipid peroxidation of yeast cell membranes was exacerbated under oxidative stress.
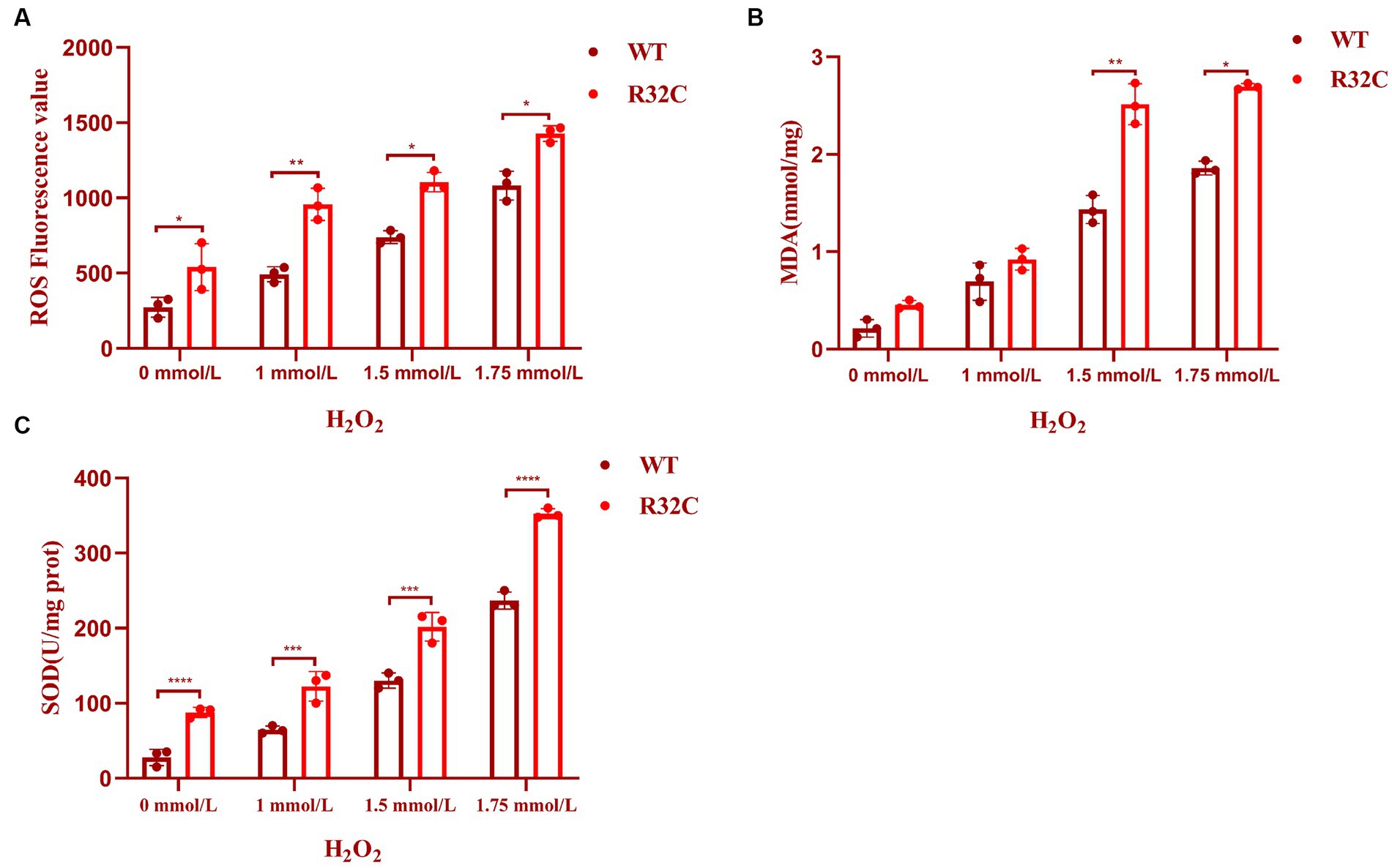
Figure 7. R32C mutation causes oxidative damage to the oxidative system. (A) Measurement of intracellular ROS content in Saccharomyces cerevisiae under different concentrations of H2O2 treatment; (B) Measurement of intracellular MDA content in Saccharomyces cerevisiae under different concentrations of H2O2 treatment. (C) Determination of intracellular SOD content of Saccharomyces cerevisiae under different concentrations of H2O2 treatment (mean ± sd). p < 0.05(*); 0.01 < p < 0.05(**); p < 0.01(***); p < 0.0001 (****).
A typical family of biological enzymes called superoxide dismutase plays a critical function in the oxidative and antioxidant balance of the body and can protect cells by scavenging superoxide anion free radicals. As seen in Figure 7C, the SOD enzyme activity increased in wild-type BY4741 and R32C as the concentration of hydrogen peroxide stress rose, demonstrating that hydrogen peroxide stress would enhance the intracellular oxidative stress response and harm the antioxidant system. Additionally, at 1.75 mM H2O2, the R32C mutant strain exhibited the maximum enzyme activity.
A few genes with antioxidant activity must be expressed in the organism to prevent oxidative damage because cells exposed to high amounts of hydrogen peroxide produce more ROS. To ascertain the relative expression levels of genes linked to oxidative stress and antioxidant, we performed culture in wild-type BY4741 and R32C mutant strains with or without the addition of hydrogen peroxide. The findings demonstrated that the hydrogen peroxide-treated strains had greater levels of SOD1 gene expression than the untreated strains (Figure 8A). Since hydrogen peroxide stress increases intracellular ROS, it is known that the transcript levels of SOD1 were elevated in the hydrogen peroxide-treated medium, indicating that intracellular oxidative stress is more significant and that more SOD is needed in the organism to remove oxidative stress.
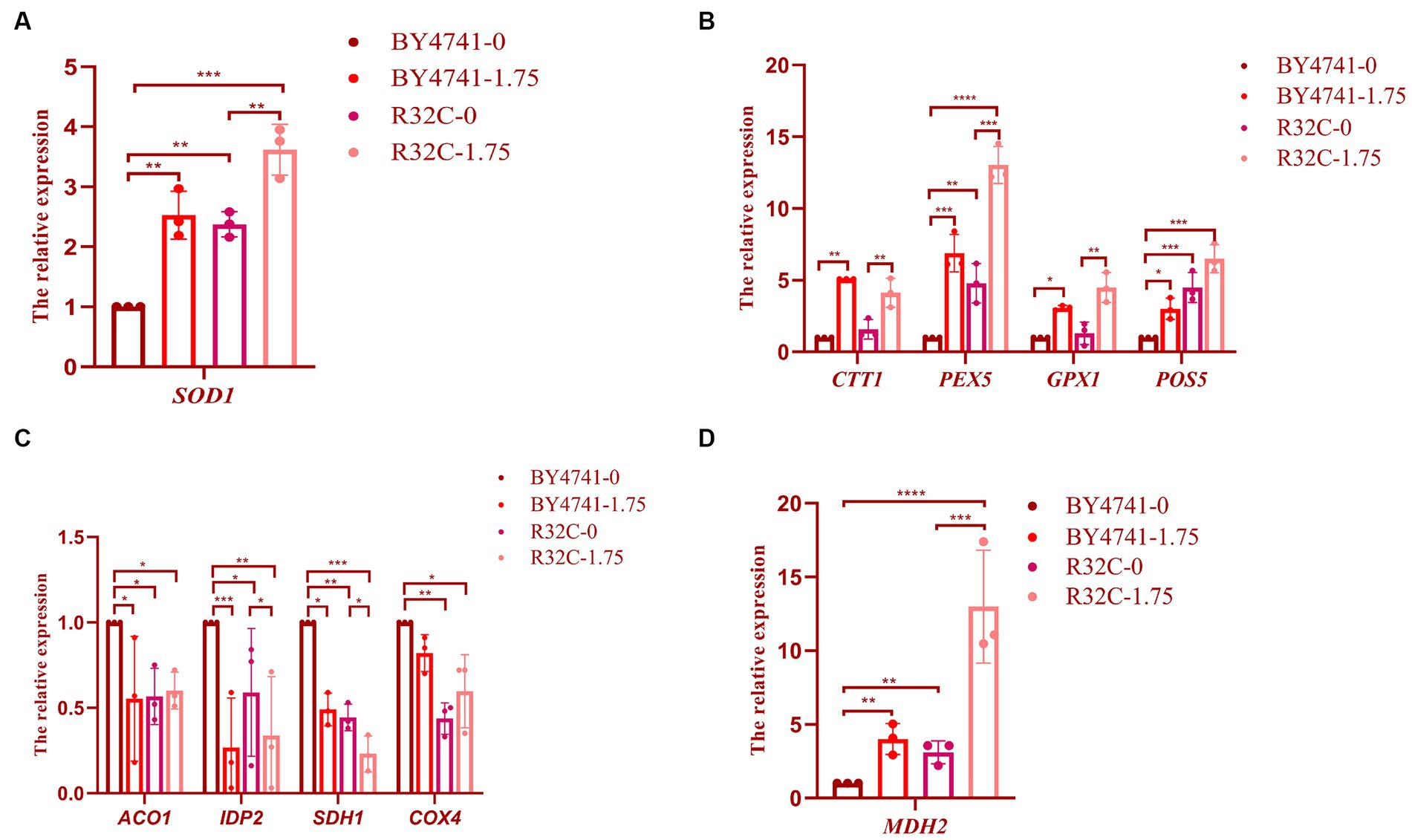
Figure 8. Susceptibility of R32C mutant strain to oxidative stress associated with antioxidant system and energy metabolism. Real-time fluorescence quantitative PCR assay (A): SOD1 gene transcript levels (Note: BY4741-0, i.e., BY4741 normal culture; BY4741-1.75, i.e., BY4741 cultured at 1.75 mM H2O2, as described below), (B) CTT1, PEX5, GPX1, POS5 gene transcript levels. (C) ACO1, IDP2, SDH1, COX4 gene transcription levels. (D) MDH2 gene transcription levels. Data presented is the results of three independent experiments (mean ± sd). p < 0.05(*); 0.01 < p < 0.05(**); p < 0.01(***); p < 0.0001(****).
When the expression levels of the CTT1, PEX5, GPX1, and POS5 genes were compared to those without H2O2 treatment, the transcript levels of CTT1 were considerably higher, as shown in Figure 8B. The CTT1 gene may be able to withstand oxidative damage due to its high expression. While the expression level of the GPX1 gene did not change significantly, the expression levels of the PEX5 and POS5 genes were significantly higher in the R32C mutant compared to BY4741 under normal circumstances, indicating that the level of oxidative stress was significantly increased in the mutant cells. After exposure to H2O2, the transcript levels of the PEX5, GPX1, and POS5 genes rose. The transcript level of the PEX5 gene increased 14.5-fold when compared to WT, demonstrating that PEX5 is a crucial gene in the antioxidant pathway.
During oxidative stress, cells undergo various changes to mitigate the damaging effects. For instance, previous studies have reported that the peroxisomal protein PEX5 plays a crucial role in maintaining peroxisome functionality and protecting against oxidative stress (Wang and Subramani, 2017). PEX5 is involved in the import of matrix proteins into peroxisomes and has been shown to be upregulated in response to oxidative stress conditions (Walton et al., 2017).
We used real-time fluorescence quantitative PCR analysis on the expression of important essential genes to clarify the resistance mechanism of hydrogen peroxide stress on the respiratory energy metabolism of yeast. After H2O2 therapy, the relative expression of the genes ACO1, IDP2, SDH1, and COX4 was much lower than it was in the group that received no treatment (Figure 8C). Malate dehydrogenase, which is encoded by the MDH2 gene, showed the most substantial up-regulation of expression (Figure 8D), demonstrating that H2O2 may efficiently control energy metabolism in S. cerevisiae (Supplementary Figure S1).
S. cerevisiae that had been subjected to various hydrogen peroxide treatments underwent GC-MS analysis. The peak area normalization method was used to compute the relative amounts of each chemical component in the samples, and the retention durations of the metabolites were compared to the mass spectrometry data. The yeast metabolites under stress from hydrogen peroxide were o-vanillin, as shown in Figure 9A. The R32C mutant strain had a higher o-vanillin content than wild-type BY4741 but less than that treated with H2O2, and the R32C mutant strain treated with H2O2 had the highest content of o-vanillin (Figure 9B). O-vanillin is a compound that can be used to study oxidative stress and antioxidant responses. In some studies, o-vanillin has been used as an antioxidant or a compound that increases the levels of endogenous antioxidants such as glutathione (Paul et al., 2021).
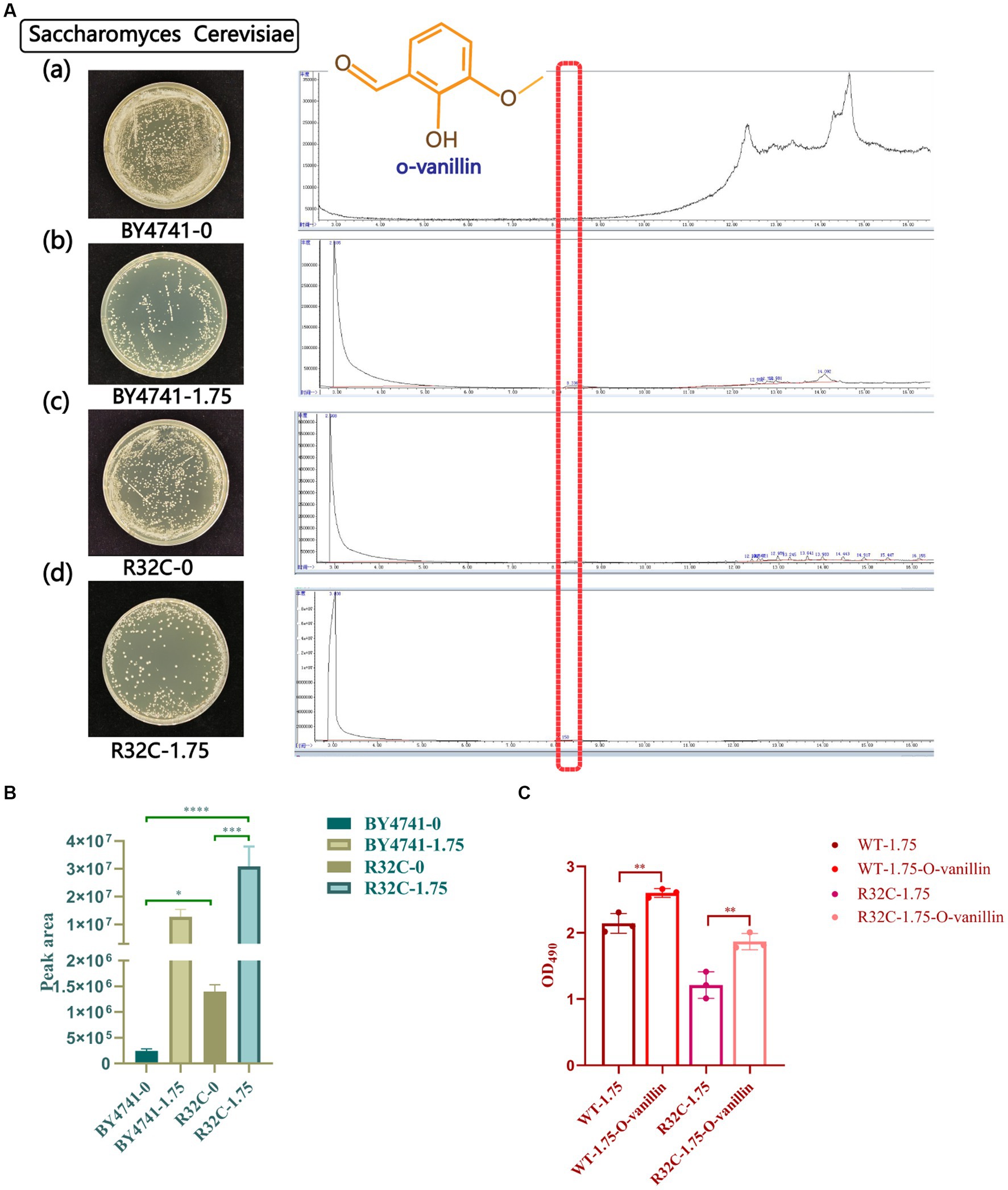
Figure 9. Gas chromatography–mass spectrometry (GC-MS) scans of metabolite profiles of wild-type and mutant yeast strains under hydrogen peroxide stress. (A) (a-d) represent the strains with different concentrations of hydrogen peroxide. Red highlights in the scan plot indicate specific peaks of o-vanillin. (B) Under different hydrogen peroxide concentration stresses area map of o-vanillin peak. (C) Survival data of cells treated with o-vanillin under oxidative stress. p < 0.05(*); 0.01 < p < 0.05(**); p < 0.01(***); p < 0.0001 (****).
To clarify that increased o-vanillin levels may be protective for yeast under oxidative stress, cell survival data after treatment with o-vanillin were measured. The results are shown in Figure 9C, where the survival of both WT and R32C cells treated with o-vanillin under 1.75 mM H2O2 was higher than that of the untreated ones, suggesting that o-vanillin has a protective effect on the cells under oxidative stress conditions.
In this study a combination of real-time fluorescence quantitative PCR (polymerase chain reaction) and GC-MS (gas chromatography–mass spectrometry) analytical methods were used.
The transcript levels were quantified using real-time fluorescence quantitative PCR analysis. Total RNA was isolated from the cells and then reverse transcribed into complementary DNA (cDNA). The resulting cDNA samples were subjected to real-time PCR using specific primers for the target transcripts of interest. Furthermore, to analyze the metabolite profiles, the cells were subjected to GC-MS analysis. This technique separates and identifies the metabolites present in the sample based on their retention times and mass spectra.
To better understand the connections between genes and metabolites, we carried out experimental assessments between the transcript levels of genes involved in antioxidant and energy metabolism and metabolites in S. cerevisiae. The antioxidant system is essential for protecting cells from harmful outside influences and scavenging oxygen free radicals. As shown in Figure 10, oxidative stress can lead to an increase in the amount of reactive oxygen species in S. cerevisiae, which in turn causes an increase in the amount of superoxide anion radicals and H2O2 in the organism and an increase in the expression levels of antioxidant-related genes SOD1, CTT1, PEX5, and GPX1. Conversely, the transcriptional expression levels of genes related to energy metabolism ACO1, IPD2, SDH1, and COX4 decreased. MDH2 gene transcript expression levels, however, were markedly up-regulated. The level of o-vanillin rose under oxidative stress, according to the examination of S. cerevisiae metabolites by GC-MS, and o-vanillin is crucial for the body’s antioxidation process. This encouraged the cells to produce more o-vanillin to combat the adverse reaction and prevent damage to the organism.
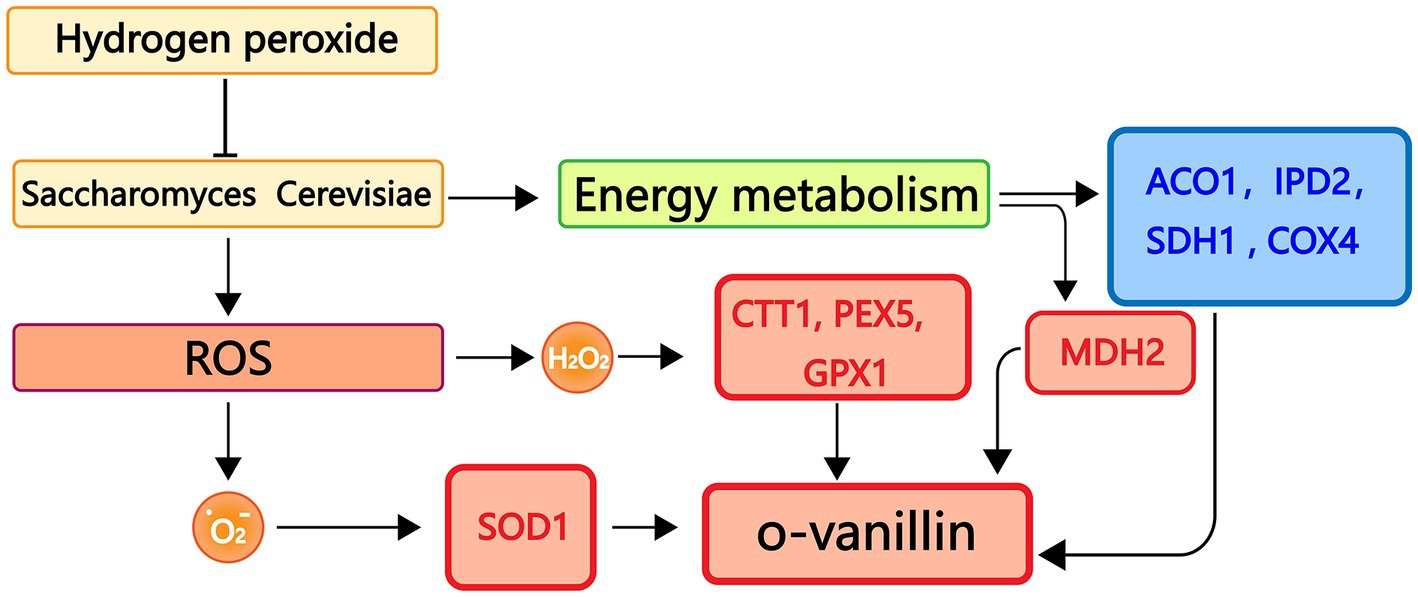
Figure 10. Real-time fluorescence quantitative PCR, GC-MS binding analysis graph. Red represents up-regulated genes, blue represents down-regulated genes.
Numerous illnesses such as neurodegenerative diseases are brought on by oxidative stress. We can offer theoretical direction for comprehending the development of oxidative stress-related disorders and creating treatment approaches by investigating the molecular regulatory systems and metabolic alterations of cells under oxidative stress.
There is growing evidence that succinate dehydrogenase (SDH) is a source and regulator of reactive oxygen species (ROS). Both losses of the function of SDH can lead to intracellular production of ROS with associated effects on the development of pathophysiological conditions, cancer and neurodegenerative diseases (Hadrava Vanova et al., 2020). The addition of ROS concentrations can initiate different cellular actions. Physically, they support signaling pathways involved in cell growth and protection, while their high levels lead to cell damage and death. The balance between ROS production and ROS clearance needs to be strictly regulated (Dröge, 2002).
SDH mutations both lead to increased ROS production in cancer and neurodegenerative diseases (Yankovskaya et al., 2003; Bardella et al., 2011; Iverson et al., 2012). Oxidative stress is a key factor in the emergence of disease and many diseases are associated with oxidative stress. In this study, Exogenous addition of H2O2 mimics the oxidative stress environment in humans. Compare the response of R32C mutant and wild-type cells to oxidative stress sensitivity. A yeast model was constructed based on the R32C mutant strain of the succinate dehydrogenase SDH2 gene, in order to confirm the mutation’s pathogenic significance—whether it causes succinate dehydrogenase SDHB-related neurodegenerative diseases in humans associated neurodegenerative diseases.
Recently, Mayr et al. (2010) discovered a patient with a homozygote (Y12C) mutation in the nuclear gene ATP5E, which codes for the ATP synthase subunit. This mutation has been demonstrated to have a considerable impact on the assembly of the human ATP synthase complex, in contrast to its negligible impact on ATP synthase activity in yeast, indicating that the biogenesis of the complex is significantly different. A novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) found in individuals with glomerular tumors has been the subject of functional studies in a yeast model. These studies suggest the C191Y SDHB mutation suppresses SDH enzyme activity, increasing ROS formation and mtDNA mutability in our yeast model (Goffrini et al., 2009; Panizza et al., 2013) results of study demonstrate that the yeast is a good functional model to validate the pathogenic significance of SDHB missense mutations.
The reduced SDH enzyme activity of the R32C mutant strain suggests that mutations in the gene encoding the SDH2 complex reduce its catalytic activity, leading to reduced respiratory rate and cellular damage, which in turn affects cellular activity. The R32C mutation may have an effect not only on the cellular localization of SDH2, but also on the protein abundance of SDH2, which is normally located in mitochondria and functions in the mitochondrial respiratory chain complex II. Mutations may result in aberrant localization of SDH2, preventing it from correctly localizing to mitochondria or interacting abnormally with other organelles (Garcia Seisdedos et al., 2022). The mutated protein abundance may be different from that of the wild-type (WT) protein (Pastor et al., 2009). Based on a three-dimensional structural model of succinate dehydrogenase, we know that the arginine residue establishes interactions with the other amino acids of the SDH2 subunit, and that replacing the residue with cysteine alters these interactions. So we hypothesized that the changes in these interactions may be related to the development of neurodegenerative diseases.
While our study provides insights into the ultrastructural changes observed in the cells, it is important to consider potential confounding factors that could contribute to these alterations. The treatment of cells with H2O2, known to induce oxidative stress, could potentially trigger secondary effects that contribute to the observed ultrastructural changes. For example, H2O2 treatment might lead to osmotic stress or alterations in pH, which can independently impact cellular morphology and organelle integrity (Erkmen, 2021).
In fact, although the mutation itself induces oxidative stress, according to experimental results, R32C mutants have a diminished ability to combat oxidative stress when exposed to H2O2 stress. Here are some possible reasons: Dysregulation of redox signaling pathways: R32C mutations may disrupt redox signaling pathways, which play a key role in coordinating cellular responses to oxidative stress. Increased ROS production: Although the R32C mutation induces oxidative stress, the mutation may also result in increased ROS production under basal conditions. Higher ROS levels may have burdened the antioxidant defense system, making the mutant cells more susceptible to additional oxidative stress induced by H2O2. Impaired Repair Mechanisms: R32C mutations may affect repair mechanisms for oxidative damage, such as DNA repair or protein quality control systems. Impairment of these repair processes may lead to the accumulation of damaged molecules, further reducing the ability to cope with exogenous oxidative stress.
The primary method for supplying energy is the tricarboxylic acid cycle. Mutations in TCA cycling enzymes such as SDH predispose carriers to Neurodegenerative diseases. Our findings suggest that R32C mutations lead to significant dysregulation of key stress-responsive genes, including up-regulation of genes involved in antioxidant defense and down-regulation of genes related to energy metabolism. This dysregulation suggests that the normal transcriptional control of stress-responsive genes is disturbed, which may lead to cellular dysfunction and susceptibility to oxidative damage. MDH2 was significantly upregulated in this study. The following are some potential causes: (1) Hydrogen peroxide can impair the integrity and function of S. cerevisiae cell membranes, which indirectly inhibits energy metabolism; (2) hydrogen peroxide triggers a burst of reactive oxygen species in S. cerevisiae, which damages DNA and impairs mitochondrial function among other things, inhibiting energy metabolic pathways; and (3) (Carere et al., 2014) indicated that the decrease in the amount of ATP in Clostridium thermocellum resulted in a deficiency of NADPH and NADP+ and as a compensation, the cells increased the activity of MDH, resulting in more NADPH production. After hydrogen peroxide treatment, the tricarboxylic acid cycle was broken.
O-vanillin is a potent ROS scavenger in several antioxidant assays. It implies that more o-vanillin is promoted under oxidative stress, improving cellular resistance to negative effects. According to Li et al. (2022) O-vanillin disturbed the ultrastructure of mitochondria, and qRT-PCR dramatically changed the relative expression levels of enzyme genes related to the TCA cycle. As a result, o-vanillin is linked to energy metabolism. Since SDHB is an important component of mitochondria, its function is crucial for cell growth and development. However, SDHB mutations alter metabolites (Figure 9A), which may modulate the innate immune response and determine the pro-tumor immune response in vivo (Macias-Ceja et al., 2019). For example, lactate and succinate have been reported as pro-tumor metabolites that regulate tumor-associated macrophage polarization, a common feature of malignant solid tumors. Indeed, in SDHB mutant cells, altered metabolites may lead to different anti-tumor vs. pro-tumor responses, which overcomes the defect of SDHB dysfunction in cell growth and development.
Our research demonstrates that yeast is an appropriate and functional model for testing the pathogenic potential of new missense SDHB mutations. Furthermore, conservation analysis throughout evolution could provide additional information for functional testing to determine the pathogenicity of mutations.
Our results indicate that the R32C SDH2 mutation inhibits cell development and growth, resulting in significant accumulation of ROS formation, increased antioxidant activity and secondary metabolites in the yeast model. While other yeast models for SDH mutations exist, we establish a novel specific model of the VUS R38C and interpret the biological mechanism of cellular oxidative stress. These findings contribute to our understanding of the mechanisms behind the onset of neurodegenerative diseases and highlight the yeast model as an effective method for investigating the potential pathogenic significance of novel mutations and uncommon polymorphisms in SDH.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
JZ, DW, and HC: investigation. HC: project administration. DW: resources. HZ: supervision. JZ: writing—original draft. SS, SL and LL: writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the 973 Program of China (No. 2013CB733904), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX18_1116).
We thank Prof. Liqin Du of Guangxi University for the gift of plasmid pUG6, and Prof. Hongwei Yu of Zhejiang University for the gift of plasmid pESC-URA. We thank Tete Daniel Koudama for linguistic guidance and color correction.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2023.1246842/full#supplementary-material
Abdullah, M. F., Cinkilic, N., Vatan, O., Inci, D., and Aydin, R. (2022). Antiproliferative effects of novel copper (II) complexes on lung Cancer cell line. BioRxiv. doi: 10.1101/2022.08.12.503805
Abugable, A. A., Awwad, D. A., Fleifel, D., Ali, M. M., El-Khamisy, S., and Elserafy, M. (2017). Personalised medicine: genome maintenance lessons learned from studies in yeast as a model organism. Adv. Exp. Med. Biol. 1007, 157–178. doi: 10.1007/978-3-319-60733-7_9
Bardella, C., Pollard, P. J., and Tomlinson, I. (2011). SDH mutations in cancer. Biochim. Biophys. Acta 1807, 1432–1443. doi: 10.1016/j.bbabio.2011.07.003
Baruffini, E., and Lodi, T. (2010). Construction and validation of a yeast model system for studying in vivo the susceptibility to nucleoside analogues of DNA polymerase gamma allelic variants. Mitochondrion 10, 183–187. doi: 10.1016/j.mito.2009.10.002
Basu, U., Bostwick, A. M., Das, K., Dittenhafer-Reed, K. E., and Patel, S. S. (2020). Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J. Biol. Chem. 295, 18406–18425. doi: 10.1074/jbc.REV120.011202
Bayley, J. P., Devilee, P., and Taschner, P. E. M. (2005). The SDH mutation database: an online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Med. Genet. 6:39. doi: 10.1186/1471-2350-6-39
Beimers, W., Braun, M., Schwinefus, K., Pearson, K., Wilbanks, B., and Maher, L. J. (2022). A suppressor of dioxygenase inhibition in a yeast model of SDH deficiency. Endocr. Relat. Cancer 29, 345–358. doi: 10.1530/ERC-21-0349
Bezawork-Geleta, A., Wen, H., Dong, L. F., Yan, B., Vider, J., Boukalova, S., et al. (2018). Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat. Commun. 9:2221. doi: 10.1038/s41467-018-04603-z
Carere, C. R., Rydzak, T., Cicek, N., Levin, D. B., and Sparling, R. (2014). Role of transcription and enzyme activities in redistribution of carbon and electron flux in response to N₂ and H₂ sparging of open-batch cultures of Clostridium thermocellum ATCC 27405. Appl. Microbiol. Biotechnol. 98, 2829–2840. doi: 10.1007/s00253-013-5500-y
Dröge, W. (2002). Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95. doi: 10.1152/physrev.00018.2001
Edwards, H., Yang, Z., and Xu, P. (2020). Characterization of met 25 as a color associated genetic marker in Yarrowia lipolytica. Metab. Eng. Commun. 11:e00147. doi: 10.1016/j.mec.2020.e00147
Erkmen, O. (2021). “Osmotic pressure and pH effects on microorganisms” in Laboratory Practices in Microbiology, 199–204.
Garcia Seisdedos, H., Levin, T., Shapira, G., Freud, S., and Levy, E. D. (2022). Mutant libraries reveal negative design shielding proteins from supramolecular self-assembly and relocalization in cells. Proc. Natl. Acad. Sci. 119:e2101117119. doi: 10.1073/pnas.2101117119
Gilea, A. I., Ceccatelli Berti, C., Magistrati, M., di Punzio, G., Goffrini, P., Baruffini, E., et al. (2021). Saccharomyces cerevisiae as a tool for studying mutations in nuclear genes involved in diseases caused by mitochondrial DNA instability. GENES 12:1866. doi: 10.3390/genes12121866
Goffrini, P., Ercolino, T., Panizza, E., Giache, V., Cavone, L., Chiarugi, A., et al. (2009). Functional study in a yeast model of a novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) diagnosed in a patient affected by a glomus tumor. Hum. Mol. Genet. 18, 1860–1868. doi: 10.1093/hmg/ddp102
Hadrava Vanova, K., Kraus, M., Neuzil, J., and Rohlena, J. (2020). Mitochondrial complex II and reactive oxygen species in disease and therapy. Redox Rep. 25, 26–32. doi: 10.1080/13510002.2020.1752002
Henderson, A., Douglas, F., Perros, P., Morgan, C., and Maher, E. R. (2009). SDHB-associated renal oncocytoma suggests a broadening of the renal phenotype in hereditary paragangliomatosis. Familial Cancer 8, 257–260. doi: 10.1007/s10689-009-9234-z
Hoekstra, A. S., and Bayley, J. P. (2013). The role of complex II in disease. BBA-Bioenergetics 1827, 543–551. doi: 10.1016/j.bbabio.2012.11.005
Invernizzi, F., Tigano, M., Dallabona, C., Donnini, C., Ferrero, I., Cremonte, M., et al. (2013). A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum. Mutat. 34, 1619–1622. doi: 10.1002/humu.22441
Iverson, T. M., Maklashina, E., and Cecchini, G. (2012). Structural basis for malfunction in complex II. J. Biol. Chem. 287, 35430–35438. doi: 10.1074/jbc.R112.408419
Khulape, S. A., Maity, H. K., Pathak, D. C., Mohan, C. M., and Dey, S. (2015). Antigenic validation of recombinant hemagglutinin-neuraminidase protein of Newcastle disease virus expressed in Saccharomyces cerevisiae. Acta Virol. 59, 240–246. doi: 10.4149/av_2015_03_240
Kumar, R., and Reichert, A. S. (2021). Common principles and specific mechanisms of Mitophagy from yeast to humans. Int. J. Mol. Sci. 22:4363. doi: 10.3390/ijms22094363
Liang, Z. C., Deng, M. L., Zhang, Z., Li, M. R., Zhou, S. J., Zhao, Z. G., et al. (2021). One-step construction of a food-grade expression system based on the URA3 gene in Kluyveromyces lactis. Plasmid 116:102577. doi: 10.1016/j.plasmid.2021.102577
Li, Q., Zhao, Y., Zhu, X., and Xie, Y. (2022). Antifungal effect of o -vanillin on mitochondria of aspergillus flavus: ultrastructure and TCA cycle are destroyed. Int. J. Food Sci. Technol. 57, 3142–3149. doi: 10.1111/ijfs.15647
Liu, B., Wang, X. F., Zhao, J. H., Qin, L. H., Shi, L., Zhou, T., et al. (2020). Effects of salinity on the synthesis of 3-methylthiopropanol, 2-phenylethanol, and isoamyl acetate in Zygosaccharomyces rouxii and Z. rouxii 3-2. Bioprocess Biosyst. Eng. 43, 831–838. doi: 10.1007/s00449-019-02279-3
Lopez-Berges, M. S., Rispail, N., Prados-Rosales, R. C., and Di Pietro, A. (2010). A nitrogen response pathway regulates virulence in plant pathogenic fungi: role of TOR and the bZIP protein mea B. Plant Signal. Behav. 5, 1623–1625. doi: 10.4161/psb.5.12.13729
Macias-Ceja, D. C., Ortiz-Masiá, D., Salvador, P., Gisbert-Ferrándiz, L., Hernández, C., Hausmann, M., et al. (2019). Succinate receptor mediates intestinal inflammation and fibrosis. Mucosal Immunol. 12, 178–187. doi: 10.1038/s41385-018-0087-3
Makhnoon, S., Garrett, L. T., Burke, W., Bowen, D. J., and Shirts, B. H. (2019). Experiences of patients seeking to participate in variant of uncertain significance reclassification research. J Community Genet. 10, 189–196. doi: 10.1007/s12687-018-0375-3
Mauer, C. B., Reys, B., and Wickiser, J. (2020). De novo SDHB gene mutation in a family with extra-adrenal paraganglioma. Familial Cancer 19, 269–271. doi: 10.1007/s10689-020-00174-5
Mayr, J. A., Havlíčková, V., Zimmermann, F., Magler, I., Kaplanová, V., Ješina, P., et al. (2010). Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 ε subunit. Hum. Mol. Genet. 19, 3430–3439. doi: 10.1093/hmg/ddq254
Moosavi, B., Zhu, X.-L., Yang, W.-C., and Yang, G.-F. (2020). Molecular pathogenesis of tumorigenesis caused by succinate dehydrogenase defect. Eur. J. Cell Biol. 99:151057. doi: 10.1016/j.ejcb.2019.151057
Panizza, E., Ercolino, T., Mori, L., Rapizzi, E., Castellano, M., Opocher, G., et al. (2013). Yeast model for evaluating the pathogenic significance of SDHB, SDHC and SDHD mutations in PHEO-PGL syndrome. Hum. Mol. Genet. 22, 804–815. doi: 10.1093/hmg/dds487
Pastor, M. M., Proft, M., and Pascual-Ahuir, A. (2009). Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast*. J. Biol. Chem. 284, 30307–30317. doi: 10.1074/jbc.M109.050682
Paul, V., Rai, D. C., Lakshmi, T. S. R., Srivastava, S. K., and Tripathi, A. D. (2021). A comprehensive review on vanillin: its microbial synthesis, isolation and recovery. Food Biotechnol. 35, 22–49. doi: 10.1080/08905436.2020.1869039
Pęczkowska, M., Kowalska, A., Sygut, J., Waligórski, D., Malinoc, A., Janaszek-Sitkowska, H., et al. (2013). Testing new susceptibility genes in the cohort of apparently sporadic phaeochromocytoma/paraganglioma patients with clinical characteristics of hereditary syndromes. Clin. Endocrinol. 79, 817–823. doi: 10.1111/cen.12218
Prevost, C. T., Peris, N., Seger, C., Pedeville, D. R., Wershing, K., Sia, E. A., et al. (2018). The influence of mitochondrial dynamics on mitochondrial genome stability. Curr. Genet. 64, 199–214. doi: 10.1007/s00294-017-0717-4
Qian, X., Jin, H., Chen, Z. J., Dai, Q. Q., Sarsaiya, S., Qin, Y. T., et al. (2021). Comparative transcriptome analysis of genes involved in Sesquiterpene alkaloid biosynthesis in Trichoderma longibrachiatum MD33 and UN32. Front. Microbiol. 12:800125. doi: 10.3389/fmicb.2021.800125
Quinlan, C. L., Orr, A. L., Perevoshchikova, I. V., Treberg, J. R., Ackrell, B. A., and Brand, M. D. (2012). Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 287, 27255–27264. doi: 10.1074/jbc.M112.374629
Rhee, S. G., and Bae, S. H. (2015). The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap 1 and Nrf 2 activation and by inhibition of mTORC1. Free Radic. Biol. Med. 88, 205–211. doi: 10.1016/j.freeradbiomed.2015.06.007
Richter, S., Gieldon, L., Pang, Y., Peitzsch, M., Huynh, T., Leton, R., et al. (2019). Metabolome-guided genomics to identify pathogenic variants in isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase genes in pheochromocytoma and paraganglioma. Genet. Med. 21, 705–717. doi: 10.1038/s41436-018-0106-5
Roberts, R., Khomenko, I., Eyres, G. T., Bremer, P., Silcock, P., Betta, E., et al. (2023). Investigation of geraniol biotransformation by commercial Saccharomyces yeast strains by two headspace techniques: solid-phase microextraction gas chromatography/mass spectrometry (SPME-GC/MS) and proton transfer reaction-time of flight-mass spectrometry (PTR-ToF-MS). Fermentation-Basel 9:294. doi: 10.3390/fermentation9030294
Schweizer, L., Thierfelder, F., Thomas, C., Soschinski, P., Suwala, A., Stichel, D., et al. (2020). Molecular characterization of CNS paragangliomas identifies cauda equina paragangliomas as a distinct tumor entity. Acta Neuropathol. 140, 893–906. doi: 10.1007/s00401-020-02218-7
Serpa, P. B. S., Woolcock, A., Taylor, S. D., and dos Santos, A. P. (2021). Validation of a flow cytometric assay to detect intraerythrocytic reactive oxygen species in horses. Vet. Clin. Pathol. 50, 20–27. doi: 10.1111/vcp.12976
Sun, L. L., Zhou, F. Y., Shao, Y. N., Lv, Z. M., and Li, C. H. (2020). The iron-sulfur protein subunit of succinate dehydrogenase is critical in driving mitochondrial reactive oxygen species generation in Apostichopus japonicus. Fish Shellfish Immunol. 102, 350–360. doi: 10.1016/j.fsi.2020.04.060
Szeto, S. S. W., Reinke, S. N., Sykes, B. D., and Lemire, B. D. (2007). Ubiquinone-binding site mutations in the Saccharomyces cerevisiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. J. Biol. Chem. 282, 27518–27526. doi: 10.1074/jbc.M700601200
Tao, C. C., Yang, Y., Li, F., Qiao, L., Wu, Y., Sun, X. D., et al. (2020). Cloning short DNAinto plasmids byone-step PCR. Thoracic Cancer 11, 3409–3415. doi: 10.1111/1759-7714.13660
Tsikas, D. (2017). Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal. Biochem. 524, 13–30. doi: 10.1016/j.ab.2016.10.021
Tufariello, M., Palombi, L., Rizzuti, A., Musio, B., Capozzi, V., Gallo, V., et al. (2023). Volatile and chemical profiles of Bombino sparkling wines produced with autochthonous yeast strains. Food Control 145:109462. doi: 10.1016/j.foodcont.2022.109462
Ubhe, A. S. (2023). Imaging of liposomes by negative staining transmission Electron microscopy and cryogenic transmission Electron microscopy. Methods Mol. Biol. (Clifton, N.J.) 2622, 245–251. doi: 10.1007/978-1-0716-2954-3_22
V, S. V. H., Pudi, S., Gade, R. R., Vudi, S., Bn, V. K. D. K., and Thota, S. S. B. (2022). Assessment of salivary malondialdehyde and superoxide dismutase levels in completely edentulous patients: an in vivo study. Cureus 14, –e27949. doi: 10.7759/cureus.27949
Walton, P. A., Brees, C., Lismont, C., Apanasets, O., and Fransen, M. (2017). The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1833–1843. doi: 10.1016/j.bbamcr.2017.07.013
Wang, D., Sarsaiya, S., Qian, X., Jin, L., Shu, F., Zhang, C., et al. (2023). Analysis of the response mechanisms of Pinellia ternata to terahertz wave stresses using transcriptome and metabolic data. Front. Plant Sci. 14:1227507. doi: 10.3389/fpls.2023.1227507
Wang, W., and Subramani, S. (2017). Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis. Cell Cycle 16, 2037–2045. doi: 10.1080/15384101.2017.1376149
Wurth, R., Jha, A., Kamilaris, C., Gill, A. J., Poplawski, N., Xekouki, P., et al. (2021). A case of carney triad complicated by renal cell carcinoma and a germline SDHA pathogenic variant. Endocrinol. Diabetes Metab. Case Rep. 2021, 20–0170. doi: 10.1530/EDM-20-0170
Wu, Z.-F., Liu, X.-Y., Deng, N.-H., Ren, Z., and Jiang, Z.-S. (2023). Outlook of Ferroptosis-targeted lipid peroxidation in cardiovascular disease. Curr. Med. Chem. 30, 3550–3561. doi: 10.2174/0929867330666221111162905
Yamaguchi, Y., Yokoyama, M., Takemoto, A., Nakamura, Y., Fukuda, S., Uehara, S., et al. (2022). Succinate dehydrogenase-deficient malignant paraganglioma complicated by succinate dehydrogenase-deficient renal cell carcinoma. IJU Case Rep 5, 480–483. doi: 10.1002/iju5.12520
Yankovskaya, V., Horsefield, R., Törnroth, S., Luna-Chavez, C., Miyoshi, H., Léger, C., et al. (2003). Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704. doi: 10.1126/science.1079605
Keywords: Saccharomyces cerevisiae , succinate dehydrogenase, R32C, qRT-PCR, GC-MS, hydrogen peroxide stress
Citation: Zheng J, Liu S, Wang D, Li L, Sarsaiya S, Zhou H and Cai H (2023) Unraveling the functional consequences of a novel germline missense mutation (R38C) in the yeast model of succinate dehydrogenase subunit B: insights into neurodegenerative disorders. Front. Mol. Neurosci. 16:1246842. doi: 10.3389/fnmol.2023.1246842
Received: 27 June 2023; Accepted: 13 September 2023;
Published: 28 September 2023.
Edited by:
Jonasz Jeremiasz Weber, Ruhr University Bochum, GermanyReviewed by:
Agustin Krisna Wardani, University of Brawijaya, IndonesiaCopyright © 2023 Zheng, Liu, Wang, Li, Sarsaiya, Zhou and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heng Cai, Y2hlbmdAbmp0ZWNoLmVkdS5jbg==
† These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.