- 1Department of Ophthalmology and Visual Sciences, Washington University in St. Louis, St. Louis, MO, United States
- 2Department of Developmental Biology, Washington University in St. Louis, St. Louis, MO, United States
Photoreceptor development of the vertebrate visual system is controlled by a complex transcription regulatory network. OTX2 is expressed in the mitotic retinal progenitor cells (RPCs) and controls photoreceptor genesis. CRX that is activated by OTX2 is expressed in photoreceptor precursors after cell cycle exit. NEUROD1 is also present in photoreceptor precursors that are ready to specify into rod and cone photoreceptor subtypes. NRL is required for the rod fate and regulates downstream rod-specific genes including the orphan nuclear receptor NR2E3 which further activates rod-specific genes and simultaneously represses cone-specific genes. Cone subtype specification is also regulated by the interplay of several transcription factors such as THRB and RXRG. Mutations in these key transcription factors are responsible for ocular defects at birth such as microphthalmia and inherited photoreceptor diseases such as Leber congenital amaurosis (LCA), retinitis pigmentosa (RP) and allied dystrophies. In particular, many mutations are inherited in an autosomal dominant fashion, including the majority of missense mutations in CRX and NRL. In this review, we describe the spectrum of photoreceptor defects that are associated with mutations in the above-mentioned transcription factors, and summarize the current knowledge of molecular mechanisms underlying the pathogenic mutations. At last, we deliberate the outstanding gaps in our understanding of the genotype–phenotype correlations and outline avenues for future research of the treatment strategies.
Introduction
Transcription factors regulate the cell-type specification and differentiation in the retina (Livesey and Cepko, 2001; Harada et al., 2007; Hennig et al., 2008; Byerly and Blackshaw, 2009; Swaroop et al., 2010; Heavner and Pevny, 2012; Brzezinski and Reh, 2015; Stenkamp, 2015; Miesfeld and Brown, 2019; Seritrakul and Gross, 2019; Raeisossadati et al., 2021). Retinal development is highly conserved among vertebrates (Stenkamp, 2015). This review firstly summarizes the up-to-date knowledge of functions of selected transcription factors involved in early stages of photoreceptor development. These transcription factors include OTX2, CRX, NEUROD1, NRL, NR2E3, THRB, and RXRG (Table 1). Secondly, this review describes the congenital disorders that result when these transcription factors are disrupted. Lastly, this review introduces ocular diseases that are associated with distinct forms of mutations in transcription factor genes such as PRDM13 and RAX2.
OTX2 expression is enriched in a large population of retinal progenitor cells, which determines photoreceptor genesis (Ghinia Tegla et al., 2020). CRX and NEUROD1 are expressed in the photoreceptor precursors (Morrow et al., 1999; Hennig et al., 2008; Swaroop et al., 2010). Subsequently, these precursors are fated into rod and cone photoreceptors. Rod lineage is governed by rod-specific transcription factors such as NRL and NR2E3 (Mears et al., 2001; Milam et al., 2002); cone lineage is regulated by transcription factors such as THRB and RXRG (Ng et al., 2001; Deeb, 2006). A precise regulation on the expression of these transcription factors is essential for neurogenesis, cell survival, and homeostasis of photoreceptors. Targetome analysis also helps to determine the overall transcription factor networking involved in photoreceptor development. Therefore, the aberrant or ablated expression of each transcription factor or the networking always results in photoreceptor underdevelopment and degeneration. Mutations in the coding regions of these transcription factors may induce misregulation in target gene expression, thus produce blindness-causing retinopathies, including microphthalmia, Leber congenital amaurosis, retinitis pigmentosa, and cone-rod dystrophy. This review attempts to unveil the relationship between mutations, protein functions and disease phenotypes, and classify (or ‘re-classify) noteworthy mutations of each transcription factor based on mutant protein functions and resulted ocular phenotypes. Interestingly, many cases of missense mutations within the DNA-binding domains, including some CRX and NRL mutations show reduced DNA-binding capabilities and altered binding motif preference or affinity at specific sites. On the other hand, mutations within the coding regions of activation domains or domains that carry regulatory activity often downregulate the expression of target genes, with some exceptional cases. Moreover, a significant number of disease-causing mutations, regardless of the locations in the coding regions, belong to the autosomal dominant class. This review describes several examples to illustrate the potential pathogenic mechanisms.
Selected transcription factors involved in early stage of photoreceptor development and diseases
OTX2
Photoreceptor development starts from the fate specification of the progenitor pool. OTX2, a homeobox gene located on human chromosome 14, encodes a key transcription factor for the development of nervous systems, including brain and retina specification (Acampora et al., 1995; Matsuo et al., 1995; Ang et al., 1996; Cantos et al., 2000; Henderson et al., 2009; Béby et al., 2010; Bernard et al., 2014). OTX2 function in retinal development is briefly introduced in Table 1. OTX2 may function as an oncogene during development. OTX2 overexpression is detected in retinoblastoma (Glubrecht et al., 2009; Li et al., 2015). Pharmacologic inhibition by all-trans retinoic acid (ATRA) reduces OTX2 expression, therefore decreases cell proliferation and tumor growth (Li et al., 2015). OTX2 overexpression is also found in some cases of medulloblastoma, repressing transcription of differentiation markers (Bunt et al., 2012; Lu et al., 2017). OTX2 can directly activate c-MYC expression in medulloblastoma via cis-regulatory elements in MYC promoter (Adamson et al., 2010; Bunt et al., 2011). Notably, concurrent trilateral retinoblastoma and medulloblastoma has been reported (Elias et al., 2001; Jurkiewicz et al., 2010), and aberrant OTX2 expression is a common characteristic.
In a mature retina, cell identity no longer requires Otx2 expression. Otx2 is weakly expressed in rod and cone photoreceptors (Koike et al., 2007), strongly in bipolar cells (Fossat et al., 2007; Kim et al., 2008; Aavani et al., 2017), and in some Müller glia (Brzezinski et al., 2010), regulating their functions by cell-autonomous or non-autonomous actions (Housset et al., 2013; Torero Ibad et al., 2020). Otx2 expression is required for the long-term survival of rod and cone photoreceptors, bipolar cells, and horizontal cells (Béby et al., 2010; Housset et al., 2013). Photoreceptor-specific Otx2 conditional knockout after photoreceptor differentiation induces impaired translocation of arrestin-1 as well as downregulation of ECM components including versican and decorin in the retina (Pensieri et al., 2021). Similarly, in the visual cortex, OTX2 binds to regulate chondroitin sulfate proteoglycans of perineuronal nets (Beurdeley et al., 2012; Bernard et al., 2016), supporting the association of OTX2 with ECMs and cytoskeletons (Boncinelli and Morgan, 2001).
Photoreceptor-specific Otx2 conditional knockout after photoreceptor differentiation does not alter the short-term retinal structure and phototransduction activity (Pensieri et al., 2021), which is thought to be compensated by Crx expression. Another piece of evidence is that loss of OTX2 in Crx−/− photoreceptors worsens the degenerative phenotypes (Hsiau et al., 2007). A possible explanation is that the optimal OTX2-binding site contains the 5′-TAAT-3′ sequence which is recognized by many other homeobox transcription factors such as CRX (Chen et al., 1997; Chatelain et al., 2006; Samuel et al., 2014). Tissue-specificity of transcription regulation is determined by unique sequences flanking this tetranucleotide (Berger et al., 2008; Jolma et al., 2015), not by the bound transcription factors. Such compensatory regulation between OTX2 and CRX is subjected to further investigation.
Interestingly, OTX2 can be transferred to cells that do not express it (Lee et al., 2019; Di Nardo et al., 2020). Exogenous OTX2 promotes the neuroplasticity of the visual cortex (Sugiyama et al., 2008) and survival of retinal ganglion cells and bipolar cells (Torero Ibad et al., 2011; Kim et al., 2015) by transcription regulation (Apulei et al., 2019) or mitochondrial energy complex stabilization (Kim et al., 2015). A proteomic analysis confirms the association of OTX2 with proteins of the mitochondrial energy complex as well as with the neurotransmitter machinery in the retina (Fant et al., 2015). Notably, this type of OTX2 transfer appears to be directional: OTX2 found in type2-off bipolar cells is transferred from photoreceptors (Kim et al., 2015); OTX2 found in ganglion cells is probably transferred from bipolar cells or photoreceptors (Sugiyama et al., 2008); OTX2 found in outer segments of photoreceptors is transferred from RPEs (Pensieri et al., 2021). This phenomenon reflects that OTX2 transfer between retinal cells probably contributes to the non-autonomous action of OTX2 regulating the retinal physiology.
The OTX2 protein has four major domains, namely, a N-terminal domain, a homeodomain, a C-terminal domain, and conserved OTX tail (Figure 1). The homeodomain (location: aa38–97) is a conserved 60-amino acid domain that binds to specific genomic targets (Di Nardo et al., 2018). C-terminal domain is also known as transactivation domain, consisting of nuclear localization signal and transcription regulatory region. In general as shown in reporter assays, OTX2 proteins lacking homeodomain are inactive in DNA-binding, and those lacking the C-terminal domain lose most of the transactivation capacity (Chatelain et al., 2006). In addition, the post-translational modifications of the OTX2 protein are largely unclear. A piece of corroborative evidence presents that interaction between OTX2 and TLE1 is governed by OTX2 phosphorylation during eye formation in xenopus (Satou et al., 2018).
Haploinsufficiency for OTX2 with only a single copy of a coding allele causes microphthalmia in mouse models (Matsuo et al., 1995; Kim et al., 2015) and rare human cases (Wyatt et al., 2008; Tajima et al., 2009). Heterozygous OTX2 mutations in human patients result in severe ocular malformations which are usually associated with abnormal development in brain and pituitary dysfunction (Beby and Lamonerie, 2013). The clinical manifestations include unilateral and bilateral forms of anophthalmia/microphthalmia, optic nerve hypoplasia and coloboma (Ragge et al., 2005; Gorbenko Del Blanco et al., 2012). Notably, OTX2 mutations are linked to the etiology of 2–3% of anophthalmia/microphthalmia cases (Wyatt et al., 2008; Tajima et al., 2009; Jones et al., 2016).
There is no clear genotype–phenotype correlation for OTX2 mutations. Associations can however be proposed between disease phenotypes and domains of the mutant proteins. Firstly, it is worth noting that a large majority of mutations within the coding region for homeodomain including OTX2R40GfsX47, OTX2T46NfsX42, OTX2R68P, OTX2A72HfsX15, OTX2E79X, OTX2R89G, OTX2R89X, OTX2R90S, and OTX2Q97X causes bilateral microphthalmia (Ragge et al., 2005; Ashkenazi-Hoffnung et al., 2010; Gonzalez-Rodriguez et al., 2010; Schilter et al., 2011; Gregory et al., 2021). The functional assays show absent or nearly lost transactivation activity. These mutations generally cause frameshifts or premature stop codons producing mutant proteins with largely truncated or missing transactivation domain. In addition, many of these mutations carry no dominant-negative effect based on the functional analysis of the mutation proteins in cultured cells. Thus, the neuronal disorders are predicted as a result of OTX2 haploinsufficiency. Only few missense mutations have been reported so far. Seven patients of two families carrying the same missense mutation OTX2E79K show pattern dystrophy of RPEs at macula with normal or moderately reduced rod-driven or cone-driven electroretinogram (ERG) responses (Vincent et al., 2014). It is unclear if the dominant OTX2E79K (c.235G > A) carries any DNA-binding specificity and transactivation capacity in the retina and why the manifestations in OTX2E79K patients (Vincent et al., 2014) are different from those with OTX2E79X (c.235G > T) (Gregory et al., 2021).
Secondly, mutations within the coding region for transactivation domain produce variable disease phenotypes. Notably, OTX2P133T (bilateral microphthalmia), OTX2P134A (unilateral anophthalmia), OTX2P134R (unilateral optic nerve aplasia) are missense mutations affecting nuclear localization of mutant proteins (Ragge et al., 2005; Gorbenko Del Blanco et al., 2012). Functional assays indicate that OTX2P134R mutation is dominant and produces the mutant protein with reduced transactivation activity. Mutation proteins produced by OTX2P133T and OTX2P134A have normal transactivation activity (Chatelain et al., 2006). It is unclear if mutant proteins still function in the nucleus and how the dominant-negative effect of OTX2P134R mutation contributes to the disease phenotypes. A large majority of nonsense or frameshift mutations within the coding region for transactivation domain cause reduced or loss-of-function transactivation, including OTX2S135LfsX2 (bilateral optic nerve aplasia), OTX2S136LfsX43 (bilateral optic nerve aplasia), OTX2S138X [Leber congenital amaurosis (LCA) or retinal dystrophy], OTX2S167X (bilateral microphthalmia), OTX2C170X (retinal dystrophy), OTX2Y179X (bilateral microphthalmia) and OTX2G188X (bilateral microphthalmia) (Ragge et al., 2005; Henderson et al., 2009; Tajima et al., 2009; Ashkenazi-Hoffnung et al., 2010; Gregory et al., 2021). These mutations are thought of having intact DNA-binding specificity and showing no dominant-negative effect. OTX2Y179X causes nearly lost transactivation, while OTX2G188X (only 8 aa apart) has 50% reduction. Both causes microphthalmia with many similar manifestations, suggesting high-level (>50%) OTX2 expression is essential to eye development. Detailed functional assays with graded OTX2 expression may help to address this hypothesis. Despite OTX2S138X, OTX2C170X and OTX2Y179X producing no transactivation activity, the disease phenotypes are associated with retinal defects instead of microphthalmia or anophthalmia. It is worth noting that various cases of incomplete penetrance have been reported in patient families (Ragge et al., 2005; Wyatt et al., 2008; Ashkenazi-Hoffnung et al., 2010; Schilter et al., 2011), including patients with OTX2Y179X. Regardless of possible phenotypic variations by incomplete penetrance, further studies need to determine how these mutations specifically affect the retina.
Lastly, sporadic, de novo and familiar OTX2 mutations with complete penetrance account for 37, 42, 16% of reported cases, respectively (Fang et al., 2016). Patients with OTX2 mutations usually develop pituitary hormone deficiency. The frequency of co-existence of pituitary hormone deficiency with ocular defects is however unclear. There are few cases with pituitary dysfunction without an ocular phenotype: OTX2R127W and OTX2N233S (Diaczok et al., 2008; Matsumoto et al., 2020). Furthermore, non-coding regions such as DHS-4 are required to initiate Otx2 expression (Emerson and Cepko, 2011; Muranishi et al., 2011; Wilken et al., 2015; Chan et al., 2020), mutation within these regions has not yet been reported in humans. Due to the complex pathogenetic mechanisms, treatment to OTX2 mutations is currently unavailable.
CRX
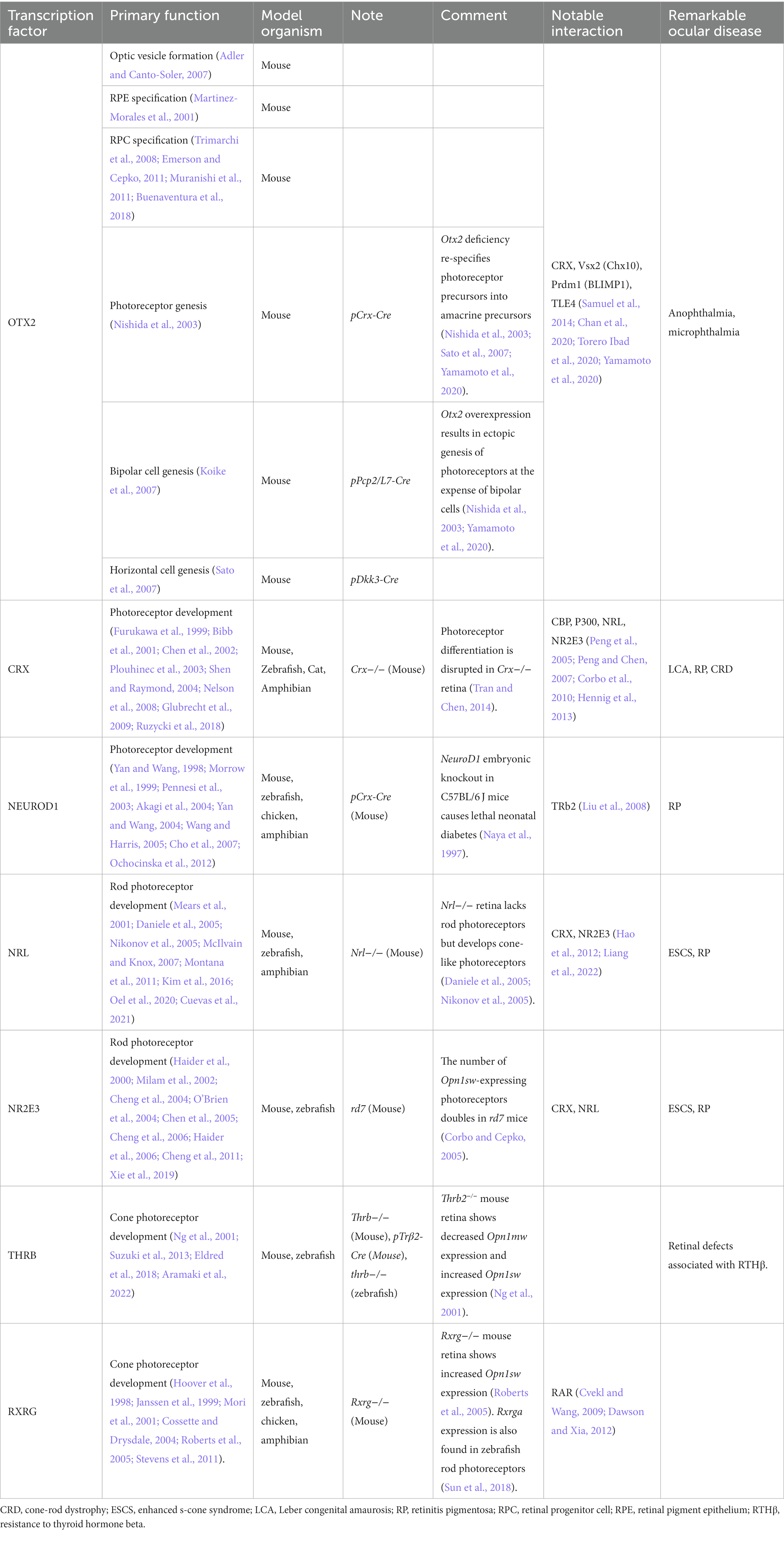
CRX is another homeobox gene that is located on human chromosome 19 and expressed in vertebrate photoreceptors and some bipolar cells as well as in pineal gland (Chen et al., 1997; Furukawa et al., 1997, 1999; Rovsing et al., 2011). CRX function in photoreceptor development is briefly introduced in Table 1. The CRX protein consists of three major domains: the homeodomain at residues 39–99 facilitates the DNA binding; the transactivation domain at residues 113–284, including a WSP motif at residues 158–170, contains binding sites for other transcription coregulators; conserved OTX tail is found at resides 284–295 (Figure 2; Freund et al., 1997; Tran et al., 2014).
Pathogenic CRX mutations are associated with macular dystrophy (Hull et al., 2014), cone-rod dystrophy (CRD) (Freund et al., 1997), retinitis pigmentosa (RP) (Sohocki et al., 2001), and LCA (Freund et al., 1998; Rivolta et al., 2001). CRX mutations are known to occur de novo or to be inherited mostly in an autosomal dominant pattern, consisting of nonsense, missense, and frameshift mutations (Stenson et al., 2014). CRX mutations could cause dominant disorders by two possible mechanisms, namely, the CRX haploinsufficiency, and/or dominant negative or gain-of-function effects of the mutant proteins. Unlike OTX2 mutations, CRX haploinsufficiency may not cause severe phenotypes. The study on Crx+/− mice do not develop any detectable functional defects up to 6 months (Tran and Chen, 2014). Human patients with CRX heterozygosity do not develop LCA either (patients with CRX nullizygosity develop LCA) (Ibrahim et al., 2018). Therefore, the dominant-negative effects are ascribed to the functions of mutant proteins. However, it remains unknown if the mutant CRX allele could partially abrogate the production of a functional CRX from the normal allele. Further studies are needed to address this question in detail. Alternatively, dominant negative activities of mutant proteins have been demonstrated in animal models (Tran and Chen, 2014; Ruzycki et al., 2017). The reported dominant-negative mutations that arise in the homeodomain are mostly missense mutations, and those identified in the transactivation domain are largely frameshifts (Rivolta et al., 2001; Tran and Chen, 2014). Various knockin mouse models harboring mutations identified in human patients have been generated for the pathogenic analysis.
CRXR90W presents a hypomorphic missense mutation located in the homeodomain (Swaroop et al., 1999; Tran et al., 2014), and is associated with a dominant late-onset mild CRD and recessive LCA. The mutant protein has abolished DNA binding activity, and thus cannot transactivate target genes (Swaroop et al., 1999; Tran et al., 2014). CRXE80A and CRXK88N mutations represent distinct antimorphic missense mutations located in the homeodomain (Chen et al., 2002; Nichols II et al., 2010; Terrell et al., 2012), which manifest early-onset dominant CRD and dominant LCA in human patients, respectively (Freund et al., 1997; Nichols II et al., 2010). These mutant proteins are predicted to bind discrete DNA sequences and show different transactivation activities from the wildtype control. Future animal model studies will provide insights into the pathogenesis of these dominant mutations.
CRXE168d2 presents an antimorphic frameshift mutation located in the transactivation domain (Tran et al., 2014), and is associated with dominant LCA in human patients (Freund et al., 1998; Jacobson et al., 1998). This mutation results in the early truncation of the transactivation domain, producing a protein that retains the ability of DNA binding but fails to transactivate target genes (Tran et al., 2014). In addition, CRXE168d2 allele overproduces the mutant protein at about four times more than the wildtype protein in heterozygous mice, which exacerbates the dominant-negative effect on the binding competition (Tran et al., 2014). Cone photoreceptor degeneration occurs prior to rod photoreceptor degeneration in the heterozygous mice, whereas rod photoreceptor appears functional with shorter outer segments at 1 month-old but undergoes progressive cell death till complete loss at 6 month-old (Tran et al., 2014). Interestingly, the ratio of mutant to wildtype CRX proteins directly correlates with the disease phenotype severity (Tran et al., 2014). In addition, truncation at the last exon by frameshift results in premature terminations of transcription (Rivolta et al., 2001; Stenson et al., 2014), producing shortened but stable mutant mRNA that may avoid nonsense-mediated decay (Lejeune and Maquat, 2005). CrxRip presents a unique mouse model with the c.763del1 mutation located in the last exon, causing a skipping of the OTX tail and a non-homologous extension of 133 residues (Roger et al., 2014). The mutant protein does not bind or transactivate target genes (Roger et al., 2014). CrxRip/+ mice show LCA-like phenotypes (Roger et al., 2014). Photoreceptors in CrxRip/+ mice do not form outer segments, due to impaired photoreceptor gene expression and incomplete differentiation at early development (Roger et al., 2014). The dominant-negative effect of CrxRip mutation does not signify a competition between the mutant and WT proteins, but likely arises from the disruption of the photoreceptor gene expression network.
AAV-based CRX gene augmentation can partially rescue the photoreceptor phenotypes and restore expression of phototransduction-related genes in CRXK88N or CRXI138fs48 human retinal organoids (Kruczek et al., 2021). On the other hand, knockout of CRX mutant alleles by CRISPR/Cas9-based gene editing can achieve moderate rescue of photoreceptor phenotypes in CRXK88Q/+ or CRXT155ins4/+ retinal organoids (Chirco et al., 2021). Thus, both gene augmentation and gene-editing-based therapies have translational potential to treat early-onset CRX-associated retinopathies.
NEUROD1
NEUROD1 is a basic helix–loop–helix (bHLH) transcription factor regulating the development of the cerebellum, hippocampal dentate gyrus, olfactory system, inner ear and auditory system, retina, and endocrine pancreas; it forms heterodimers with other bHLH transcription factors and binds to E box-containing promoter sequences to regulate gene expression of target genes (Naya et al., 1997; Poulin et al., 1997; Miyata et al., 1999; Liu et al., 2000; Breslin et al., 2003; Bernardo et al., 2008; Pan et al., 2009; Boutin et al., 2010; Evsen et al., 2013; Mastracci et al., 2013). NEUROD1 is located on human chromosome 2 and well-known of regulating β-cell development, insulin synthesis and secretion, as well as glucose homeostasis (Huang et al., 2002; Petersen et al., 2002; Andrali et al., 2007; Romer et al., 2019). NEUROD1 inactivation during the differentiation of human embryonic stem cells causes neonatal diabetes mellitus and defective β-cell function (Romer et al., 2019). Early-onset diabetes due to homozygous or heterozygous NEUROD1 mutations have also been reported in human patients (Kristinsson et al., 2001; Liu et al., 2007; Gonsorčíková et al., 2008; Rubio-Cabezas et al., 2010; Chapla et al., 2015; Bouillet et al., 2020; Brodosi et al., 2021), thus NEUROD1 is associated with maturity-onset diabetes of the young (MODY), i.e., MODY6 (Horikawa and Enya, 2019). Heterozygous NEUROD1 mutations are also linked to autosomal dominant type 2 diabetes (Malecki et al., 1999, 2003).
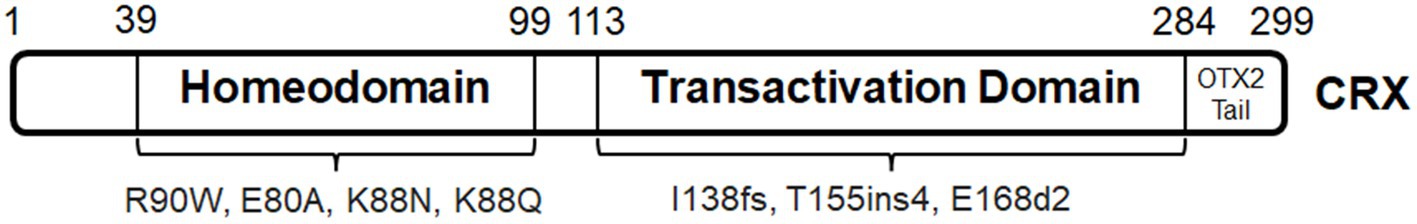
NEUROD1 function in photoreceptor development is briefly introduced in Table 1. Ophthalmological records of patients with NEUROD1 mutations are limited (Figure 3). Patients with homozygous frameshift mutations (NEUROD1D122GfsX12 and NEUROD1L143AfsX55) develop permanent neonatal diabetes and neurological abnormalities including retinal disorders (Rubio-Cabezas et al., 2010; Orosz et al., 2015). The truncated mutant proteins are considered of lacking the transactivation domain for transcription regulatory functions. Patients with NEUROD1L143AfsX55 develop nyctalopia, blurry vision, and visual field constriction from early childhood, and show absent rod- and cone-driven ERG responses (Orosz et al., 2015). These manifestations are similar to those caused by RP and rod–cone dystrophy (RCD). Interestingly, homozygous missense mutation NEUROD1V242I is associated with non-syndromic autosomal recessive RP (Wang et al., 2014). This mutation happens within the coding region for the transactivation domain. Since patients do not develop early-onset defects, this mutation might only affect the functional maintenance of photoreceptors in adulthood. These findings suggest differences in the functional roles of human NEUROD1 and mouse counterpart. Human NEUROD1 transactivation domain, at least a subdomain, is essential for photoreceptor development, while mouse NEUROD1 is required for functional maintenance. In addition, a bioinformatic analysis shows that NEUROD1 is differentially expressed at optic nerve head of patients with primary open-angle glaucoma; histological evidence and patient cases have not been documented (Wang et al., 2017).
The conserved bHLH domain is located at aa101–153. Mutations within the region coding bHLH, including NEUROD1R103P, NEUROD1E111K and NEUROD1M114L, probably abolish the binding of the mutant proteins to the promoters of target genes (Kristinsson et al., 2001; Szopa et al., 2016; Brodosi et al., 2021). These mutations are associated to MODY. However, ophthalmological records of patients with these mutations are unavailable. Furthermore, considering NEUROD1’s important functions in glucose homeostasis, its role in the pathogenesis of diabetic retinopathy has not been reported.
AAV-based NeuroD1-mediated gene therapies can reprogram brain astrocytes into neurons that are able to re-establish synapses and integrate with the survived neurons after ischemic injury in mice (Chen et al., 2020; Wu et al., 2020; Tang et al., 2021). In particular, the reprogrammed neurons form specific projections and functional connectivity in the mouse primary visual cortex, promoting the recovery of visual responses and orientation discrimination (Tang et al., 2021). However, NEUROD1-mediated gene therapy has not been proposed in the retina.
NRL and NR2E3
NRL is a basic-motif leucine zipper transcription factor that is encoded by the gene on human chromosome 14 and expressed in developing lens, developing and mature rod photoreceptors and pineal gland (Swaroop et al., 1992; Liu et al., 1996; Farjo et al., 1997; Swain et al., 2001; Kanda et al., 2007). NRL function in photoreceptor development is briefly introduced in Table 1.
In general, night blindness from early childhood is a common symptom for patients with pathogenic NRL mutations, followed by variable onsets of reduced visual acuity. Mutations can be classified by the protein domains, namely, bZIP domain and minimal transactivation domain (MTD) (Figure 4A). NRL bZIP domain is located at aa159-222. Mutations within the coding region for bZIP domain, such as heterozygous missense mutation NRLL160P (compound with NRLA76GfsX18), homozygous missense mutation NRLR170S, homozygous nonsense mutation NRLQ182X, and homozygous frameshift mutations NRLR218fs and NRLC219fs affect DNA binding and transcription activation of target genes (Nishiguchi et al., 2004; Kanda et al., 2007; Collin et al., 2011; Neveling et al., 2012; Littink et al., 2018; El-Asrag et al., 2022). These mutations cause autosomal recessive RP, some of which are specified as clumped pigment retinal degeneration that is manifested by clusters of pigmented deposits at the peripheral retina, chorioretinal atrophy and attenuated arterioles (Newman et al., 2016; Littink et al., 2018). Autosomal dominant mutation within bZIP-coding region has not been reported. NRL MTD is located at aa30-93. Homozygous mutations within MTD-coding region, including NRLR31X, NRLL75fs, and NRLQ80X (Kanda et al., 2007; Newman et al., 2016; El-Asrag et al., 2022), are considered of lacking bZIP domain. In particular, NRLR31X results in an early truncation of the mutant protein at MTD and causes enhanced S-cone syndrome (ESCS) with no detectable rod-driven ERG response, which matches retinal phenotypes in Nrl−/− mice (Newman et al., 2016). However, the screening analysis in cohorts of ESCS patients indicates that NRL mutation is a rare cause (Acar et al., 2003; Nishiguchi et al., 2004; Wright et al., 2004a; Collin et al., 2011; Neveling et al., 2012). The rest of pathogenic mutations within MTD-coding region belong to the autosomal dominant class, including NRLP49L and mutations at hot spots S50 (NRLS50T, NRLS50P, NRLS50L, and NRLS50del) and P51 (NRLP51L and NRLP51S) (Martinez-Gimeno et al., 2001; DeAngelis et al., 2002; Kanda et al., 2007; Gao et al., 2016; Qin et al., 2017; Mizobuchi et al., 2022). Interestingly, functional assays indicate that these missense mutations produce mutant proteins that have reduced level of phosphorylation but are able to enhance transcription activation at Rho promoter (Bessant et al., 1999; DeAngelis et al., 2002; Kanda et al., 2007). In humans, these mutations cause autosomal dominant RP with the signature phenotype of bone spicule–shaped pigment deposits. A significant number of patients with autosomal dominant mutations in NRL develop RP at adult ages, although only less than 30 cases have been reported so far.

Figure 4. (A) NRL protein domains and associated mutations. (B) NR2E3 protein domains and associated mutations.
Nrl knockout after the completion of photoreceptor fate determination may favor photoreceptor survival in mouse models of Rho−/−, rd10 and RHOP347S (Montana et al., 2013; Yu and Wu, 2018). It is worth noting that Rho and Pde6β are direct target genes of NRL. Only a small population of differentially expressed genes between rod and cone photoreceptors significantly change their expression by Nrl knockout, including Nr2e3 (Yu and Wu, 2018). More importantly, Nrl knockout in young adult mice does not produce retinal rosettes, Müller glia dysfunction and vascular defects which can be found in Nrl−/− retina (Roger et al., 2012; Yu and Wu, 2018). Therefore, NRL knockout can potentially serve as a neuroprotective method to preserve rod photoreceptors from ongoing degeneration without any significant disruption in transcription, structural and functional homeostasis.
A notable downstream target gene of NRL is NR2E3 that is located on human chromosome 15. NR2E3 function in photoreceptor development is briefly introduced in Table 1. ESCS is exclusively associated with autosomal recessive mutations in NR2E3, for example, NR2E3R76Q, NR2E3R104Q, NR2E3L371W, NR2E3L373P, NR2E3L374F, NR2E3R385P, and NR2E3M407K, explaining over 90% of reported cases (Figure 4B; Wright et al., 2004a; Audo et al., 2008; Tsang and Sharma, 2018; de Carvalho et al., 2021). A large number of ESCS-associated mutations are located within the region coding the ligand-binding domain, especially the α-helix (Pachydaki et al., 2009; Tan et al., 2013). ESCS is often diagnosed through the typical features on ERG responses: loss of rod-driven response and increased S-cone-driven response (Vincent et al., 2013; Tsang and Sharma, 2018; de Carvalho et al., 2021). ESCS patients always suffer from nyctalopia at the first decade. They also develop clumped pigment deposits at RPE, dot-like lesions at ONL, and variable loss of visual acuity (Jacobson et al., 1990, 1991; Audo et al., 2008; Garafalo et al., 2018; Tsang and Sharma, 2018). Notably, hyper-sensitivity of S-cone photoreceptors at early onset concentrates at the central field and extends into the peripheral field.
In-depth analysis of ERG and phenotypic findings with ESCS patients at various disease stages suggests a parallel pattern between disease manifestations and observations in rd7 mice (Wright et al., 2004b; Iannaccone et al., 2021). However, differences between human patients and rd7 mice are noteworthy. Firstly, rod-driven ERG response is still detectable in young rd7 mice (Akhmedov et al., 2000; Ueno et al., 2005). ESCS patients show loss of rod-driven ERG response at early childhood. Secondly, ESCS patients only have dysplastic photoreceptors or pseudo-rosettes at ONL, as compared to the more deleterious structure of whorls and rosettes at ONL in rd7 mice (Wang et al., 2009). Lastly, such parallel pattern between ESCS patients and rd7 mice is limited to functional and histological measurements; comparative gene expression profiles have not been documented.
Pathogenic NR2E3 mutations are also associated with autosomal recessive (Gerber et al., 2000; Tan et al., 2013; Al-khuzaei et al., 2020) and autosomal dominant RP (NR2E3G65R) (Coppieters et al., 2007), although only a few cases have been reported. In terms of treatment strategies to NR2E3-associated retinopathies, fate-switch to developmentally altered photoreceptors might be unrealistic. Thus, practical approaches aim to slow down the progression of retinal degeneration. An in vitro study proposes a treatment strategy of knocking down a NR2E3 pathogenic variant by antisense oligonucleotides (Naessens et al., 2019). In addition, in vivo treatment by photoregulin-3 (PR3), a NR2E3 inhibitor, can slow down the photoreceptor degeneration in RhoP23H mice (Nakamura et al., 2017). Therefore, these findings suggest NR2E3 antagonism helps to reduce susceptibility of rod photoreceptors to genetic insults possibly by conferring cone photoreceptor properties. Interestingly, NR2E3 as a genetic modifier directly can serve as a therapeutic target to treat inherited retinal diseases including NR2E3-associated retinopathies. Nr2e3 overexpression yields promising rescue results in mouse models of rd1, rd7, rd16, Rho−/−, and RhoP23H (Li et al., 2021): AAV8-Nr2e3 helps to preserve photoreceptor density, promote cell survival at ONL, and enhance ERG responses. The therapeutic mechanisms of NR2E3 antagonism and overexpression are subjected to further investigation.
THRB and RXRG
Cones with different wavelength sensitivities develop from RPCs and subsequently differentiate for distinct color perceptions, which is reliant on specific transcription factors. THRB and RXRG are two notable transcription factors for this process.
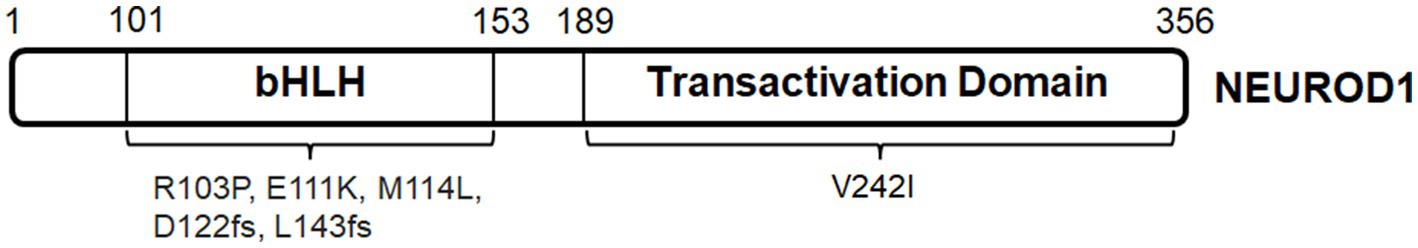
Thyroid hormone receptors are a family of ligand-dependent nuclear receptors, characterized by the conserved protein structure of an N-terminus, a DNA binding domain that binds to the thyroid hormone response elements (TREs), and a ligand binding domain for triiodothyronine (T3) across many vertebrate species including zebrafish, chicken, mouse, and human (Sjöberg and Vennström, 1995; Deeb, 2006; Darras et al., 2011; Ng et al., 2011). T3 is important for many body functions including metabolism, heart rate, and tissue development (Li et al., 2014; Mullur et al., 2014; Bassett and Williams, 2016; Chattergoon, 2019; Vale et al., 2019; Bernal et al., 2022). THRA and THRB are two members of this family (Forrest et al., 2002). THRA is located on human chromosome 17, while THRB is located on human chromosome 14. THRB function in photoreceptor development is briefly introduced in Table 1. In particular, THRB isoform 2, THRB2 (also known as TRβ2) is expressed in cone photoreceptors (Sjöberg et al., 1992; Applebury et al., 2007; Ng et al., 2009; Suzuki et al., 2013; Marelli et al., 2016). Zebrafish is a useful model for understanding the roles of thyroid hormone signaling and trβ2 in cone photoreceptor development. Firstly, Trβ2 binds to activate its own trβ2 promoter, suggesting that trβ2 expression is self-regulating (Suzuki et al., 2013). Secondly, Trβ2 determines the fate and proper L-cone differentiation and regulates the expression of opsins (opn1lw1 and opn1lw2) (Suzuki et al., 2013; Volkov et al., 2020). Samples with ablated thyroid glands maintain a similar level of opn1lw2 expression as the WT controls during development, suggesting that Trβ2 regulates L-cone differentiation independent of thyroid hormones (Mackin et al., 2019). Thirdly, Trβ2 may not be involved in the establishment of cone density ratio during development (Deveau et al., 2020). Lastly, Trβ2 regulates Cyp27c1 expression in zebrafish RPE for the production of vitamin A2-based retinoids, implying that Trβ2 signaling may interact with other signaling pathways to promote retinal development (Volkov et al., 2020). Fate switch of L-cone precursors to UV cones in thrb−/− zebrafish retina generally agrees with the selective changes in Thrb2−/− mouse retina, i.e., decrease in Opn1mw expression and increase in Opn1sw expression, supporting a conserved developmental role (Ng et al., 2001; Volkov et al., 2020).
In general, heterozygous THRB mutations are associated with a metabolic syndrome called resistance to thyroid hormone beta (RTHβ) (Onigata and Szinnai, 2014; Concolino et al., 2019; Pappa, 2021). More than 200 different THRB mutations have already been identified in RTHβ patients. A large number of these mutations happen at the coding region for ligand-binding domain and hinge region (Pappa and Refetoff, 2018), inhibiting TRβ2 binding as homodimers to TREs (Figure 5A). Detailed ophthalmological records of RTHβ patients are infrequent. Of particular interest, a study of clinical observations with 31 RTHβ patients concludes functional defects in RTHβ photoreceptors and deficits in color vision (Campi et al., 2017). Another case report shows that a child with a compound missense THRβR338W/R429W mutation at the coding region for ligand-binding domain has severely reduced M- and L-cone-driven ERG responses and increased S-cone-driven ERG responses (Weiss et al., 2012). Unfortunately, treatment strategies targeting THRB mutations have not been developed for retinal defects.
Retinoid X receptor (RXR) belongs to the nuclear hormone superfamily that comprises three isoforms, namely, α, β, and γ. RXRs share a common protein structure: the N-terminal, a DNA-binding domain and a ligand-binding domain (Rowe, 1997; Dawson and Xia, 2012). RXRs form both homo- and hetero-dimers with a number of nuclear receptors, including thyroid hormone receptors, retinoic acid receptors, and peroxisome-proliferator-activated receptors (Mangelsdorf and Evans, 1995; Chawla et al., 2001), and bind to repeats of the consensus sequence AGGTCA with a 1 base pair spacer (Rowe, 1997).
RXRG is located on human chromosome 1 and expressed in the retina of several species, including human, mouse, chick, zebrafish, xenopus (Hoover et al., 1998; Janssen et al., 1999; Mori et al., 2001; Cossette and Drysdale, 2004; Roberts et al., 2005; Stevens et al., 2011). RXRG function in photoreceptor development is briefly introduced in Table 1. RXRG deficiency can cause metabolic disorders, including type 2 diabetes (Brown et al., 2000; Davies et al., 2001; Wang et al., 2002; Haugen et al., 2004). A mutation in the coding region for helix–helix interface could impair various cellular processes (Zhang et al., 2004; Figure 5B); unfortunately, retina-specific RXRG mutation has not been reported yet. Interestingly, RXRG can serve as a therapeutic target for retinopathies. RXR agonist PA024 can selectively upregulate Rxrg expression and decrease photoreceptor cell death in mixed neuro-glial cultures from rd1 retinas (Volonté et al., 2021).
Other transcription factors
Inherited retinal diseased can be caused by other transcription factors, such as AHR (Zhou et al., 2018), ATF6 (Xu et al., 2015; Chiang et al., 2019), RORB (Sadleir et al., 2020; Morea et al., 2021). It is worth mentioning that disease-associated mutations do not always produce loss-of-function variants, two examples as follows.
PRDM13
North Carolina macular dystrophy (NCMD) is an inheritable abnormality affecting the macula, which usually occurs at birth but progresses little with aging. NCMD is inherited as an autosomal dominant manner and completely penetrant with phenotypic variability. Intragenic mutations in PRDM13 gene have not been reported for NCMD. However, a number of NCMD patients carry missense mutations in the MCDR1 locus upstream of PRDM13 gene in human chromosome 6 (Small et al., 2016, 2019a,b; Namburi et al., 2020). These mutations may alter the spatio-temporal pattern of PRMD13 expression. In the eye, PRDM13 is expressed in the fetal (Small et al., 2016) and adult retina (Green et al., 2021), predominantly in amacrine cells. In particular, PRDM13 regulates the development and subtype specification of amacrine cells in xenopus and mouse retinas (Watanabe et al., 2015; Bessodes et al., 2017). Interestingly, the sequencing analysis on a family of NCMD patients shows a tandem duplication of PRDM13 gene and a partial copy of CCNC gene in MCDR1 locus, suggesting PRDM13 overexpression responsible for NCMD pathogenesis (Bowne et al., 2016). A similar case of PRDM13 duplication also reports NCMD phenotype (Small et al., 2021). Indeed, CG13296 (PRDM13 orthologue) overexpression severely affects the development of eye-antennal imaginal disks in Drosophila melanogaster (Manes et al., 2017). In addition, a single nucleotide variant located 7.8 kb upstream of PRDM13 gene (within the MCDR1 locus) is associated with autosomal dominant progressive bifocal chorioretinal atrophy that is presumably related to NCMD (Silva et al., 2019). The regulatory function of the MCDR1 locus remains to be determined.
RAX2
RAX2 interacts and synergistically functions with CRX (Wang et al., 2004), and is required for photoreceptor differentiation in vertebrate retina (Chen and Cepko, 2002; Nelson et al., 2009; Wu et al., 2009; Irie et al., 2015). Pathogenic variants in RAX2 (human chromosome 19) cause autosomal dominant retinal dystrophies, including CRD, RP and age-related macular degeneration (Wang et al., 2004; Yang et al., 2015; Van de Sompele et al., 2019). Surprisingly, increased transactivation activity has been observed in in vitro functional analysis on RAX2 mutations (Wang et al., 2004), such as RAX2R87Q and RAX2P140_G141dup. RAX2R87Q occurs in the coding region for homeodomain (aa 25–89), while RAX2P140_G141dup is found in the coding region for the transactivation domain. Other reported mutations including RAX2S49P (homozygous), RAX2P52R (heterozygous), RAX2A113Gfs*178 (homozygous), RAX2A156Rfs*131 (heterozygous), RAX2G137R (heterozygous) show reduced transactivation activity. Further disease modeling analysis will inform insights into the roles of RAX2 in transcriptional coactivation with other transcription factors, as well as functions of RAX2 in retinal development and pathogenesis.
Conclusion
All in all, photoreceptor development is regulated by a specific network of transcription factors. Genetic variations in these genes result in autosomal recessive or dominant mutations. This review provides a mechanistic enlightenment of the genotype–phenotype relationship between above-mentioned mutations and ocular disease manifestations. In general, in vitro or in vivo functional analysis of the mutant proteins helps to determine their conformational changes, regulatory capacity, and interference with the action of wildtype proteins, which can be further correlated to the functional roles of specific protein domains. Thus, missense, nonsense and frameshift mutations that happen to the same coding region may produce mutant proteins with different regulatory functions. When the animal model is unavailable for a specific mutation, such as cases of OTX2 mutations, genotype–phenotype relationship would solely reply on in vitro molecular analysis. The study of animal models is conducive to understanding the pathogenic mechanisms of blindness-causing mutations, as well as testing therapeutic approaches. The use of animal models also helps to dissect the disease progression for cell-type specificity, expanding the scope of genotype–phenotype relationship; such examples can be found in CRX-associated retinopathies. Hence, understanding genotype–phenotype relationship benefits two horizons: (1) predictions on the disease onset/progression of an unknown mutation; (2) management of treatment windows. Gene therapy holds a promise in treating early-onset inherited retinal diseases, although significant challenges and unanswered knowledge gaps remain. A long-overlooked issue is how effective a strategy of gene therapy such as gene augmentation can treat an unknown mutation. In order to tackle this issue, genotype–phenotype relationship needs to fulfill excellent predictive power. In-depth analysis of domain-based transcription factor interactome as well as mutant/wildtype protein binding motifs can collectively help to achieve this goal.
Author contributions
CS conceived the contents, drafted the manuscript, and prepared the figures. SC edited the manuscript and figures. All authors contributed to the article and approved the submitted version.
Funding
NIH grants R01 EY012543 and R01 EY032136 (to SC), and Research to Prevent Blindness (to DOVS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aavani, T., Tachibana, N., Wallace, V., Biernaskie, J., and Schuurmans, C. (2017). Temporal profiling of photoreceptor lineage gene expression during murine retinal development. Gene Expr. Patterns 23-24, 32–44. doi: 10.1016/j.gep.2017.03.001
Acampora, D., Mazan, S., Lallemand, Y., Avantaggiato, V., Maury, M., Simeone, A., et al. (1995). Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279–3290. doi: 10.1242/dev.121.10.3279
Acar, C., Mears, A. J., Yashar, B. M., Maheshwary, A. S., Andreasson, S., Baldi, A., et al. (2003). Mutation screening of patients with Leber congenital Amaurosis or the enhanced S-cone syndrome reveals a lack of sequence variations in the NRL gene. Mol. Vis. 9, 14–17.
Adamson, D. C., Shi, Q., Wortham, M., Northcott, P. A., di, C., Duncan, C. G., et al. (2010). OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 70, 181–191. doi: 10.1158/0008-5472.CAN-09-2331
Adler, R., and Canto-Soler, M. V. (2007). Molecular mechanisms of optic vesicle development: complexities, ambiguities and controversies. Dev. Biol. 305, 1–13.
Akagi, T., Inoue, T., Miyoshi, G., Bessho, Y., Takahashi, M., Lee, J. E., et al. (2004). Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 279, 28492–28498. doi: 10.1074/jbc.M400871200
Akhmedov, N. B., Piriev, N. I., Chang, B., Rapoport, A. L., Hawes, N. L., Nishina, P. M., et al. (2000). A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. U. S. A. 97, 5551–5556. doi: 10.1073/pnas.97.10.5551
Al-khuzaei, S., Broadgate, S., Halford, S., Jolly, J. K., Shanks, M., Clouston, P., et al. (2020). Novel pathogenic sequence variants in NR2E3 and clinical findings in three patients. Genes 11:1288. doi: 10.3390/genes11111288
Andrali, S. S., Qian, Q., and Ozcan, S. (2007). Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 282, 15589–15596.
Ang, S. L., Jin, O., Rhinn, M., Daigle, N., Stevenson, L., and Rossant, J. (1996). A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122, 243–252. doi: 10.1242/dev.122.1.243
Applebury, M. L., Farhangfar, F., Glösmann, M., Hashimoto, K., Kage, K., Robbins, J. T., et al. (2007). Transient expression of thyroid hormone nuclear receptor TRbeta2 sets S opsin patterning during cone photoreceptor genesis. Dev. Dyn. 236, 1203–1212. doi: 10.1002/dvdy.21155
Apulei, J., Kim, N., Testa, D., Ribot, J., Morizet, D., Bernard, C., et al. (2019). Non-cell autonomous OTX2 Homeoprotein regulates visual cortex plasticity through Gadd45b/g. Cereb. Cortex 29, 2384–2395. doi: 10.1093/cercor/bhy108
Aramaki, M., Wu, X., Liu, H., Liu, Y., Cho, Y. W., Song, M., et al. (2022). Transcriptional control of cone photoreceptor diversity by a thyroid hormone receptor. Proc. Natl. Acad. Sci. U. S. A. 119:e2209884119. doi: 10.1073/pnas.2209884119
Ashkenazi-Hoffnung, L., Lebenthal, Y., Wyatt, A. W., Ragge, N. K., Dateki, S., Fukami, M., et al. (2010). A novel loss-of-function mutation in OTX2 in a patient with anophthalmia and isolated growth hormone deficiency. Hum. Genet. 127, 721–729. doi: 10.1007/s00439-010-0820-9
Audo, I., Michaelides, M., Robson, A. G., Hawlina, M., Vaclavik, V., Sandbach, J. M., et al. (2008). Phenotypic variation in enhanced S-cone syndrome. Invest. Ophthalmol. Vis. Sci. 49, 2082–2093. doi: 10.1167/iovs.05-1629
Bassett, J. H., and Williams, G. R. (2016). Role of thyroid hormones in skeletal development and bone maintenance. Endocr. Rev. 37, 135–187. doi: 10.1210/er.2015-1106
Beby, F., and Lamonerie, T. (2013). The homeobox gene Otx2 in development and disease. Exp. Eye Res. 111, 9–16. doi: 10.1016/j.exer.2013.03.007
Béby, F., Housset, M., Fossat, N., le Greneur, C., Flamant, F., Godement, P., et al. (2010). Otx2 gene deletion in adult mouse retina induces rapid RPE dystrophy and slow photoreceptor degeneration. PLoS One 5:e11673. doi: 10.1371/journal.pone.0011673
Berger, M. F., Badis, G., Gehrke, A. R., Talukder, S., Philippakis, A. A., Peña-Castillo, L., et al. (2008). Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cells 133, 1266–1276. doi: 10.1016/j.cell.2008.05.024
Bernal, J., Morte, B., and Diez, D. (2022). Thyroid hormone regulators in human cerebral cortex development. J. Endocrinol. 255, R27–r36.
Bernard, C., Kim, H. T., Torero Ibad, R., Lee, E. J., Simonutti, M., Picaud, S., et al. (2014). Graded Otx2 activities demonstrate dose-sensitive eye and retina phenotypes. Hum. Mol. Genet. 23, 1742–1753. doi: 10.1093/hmg/ddt562
Bernard, C., Vincent, C., Testa, D., Bertini, E., Ribot, J., di Nardo, A. A., et al. (2016). A mouse model for conditional secretion of specific single-chain antibodies provides genetic evidence for regulation of cortical plasticity by a non-cell autonomous Homeoprotein transcription factor. PLoS Genet. 12:e1006035. doi: 10.1371/journal.pgen.1006035
Bernardo, A. S., Hay, C. W., and Docherty, K. (2008). Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol. Cell. Endocrinol. 294, 1–9. doi: 10.1016/j.mce.2008.07.006
Bessant, D. A., Payne, A. M., Mitton, K. P., Wang, Q. L., Swain, P. K., Plant, C., et al. (1999). A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat. Genet. 21, 355–356. doi: 10.1038/7678
Bessodes, N., Parain, K., Bronchain, O., Bellefroid, E. J., and Perron, M. (2017). PRDM13 forms a feedback loop with Ptf1a and is required for glycinergic amacrine cell genesis in the Xenopus Retina. Neural Dev. 12:16. doi: 10.1186/s13064-017-0093-2
Beurdeley, M., Spatazza, J., Lee, H. H. C., Sugiyama, S., Bernard, C., di Nardo, A. A., et al. (2012). Otx2 binding to perineuronal nets persistently regulates plasticity in the mature visual cortex. J. Neurosci. 32, 9429–9437. doi: 10.1523/JNEUROSCI.0394-12.2012
Bibb, L. C., Holt, J. K., Tarttelin, E. E., Hodges, M. D., Gregory-Evans, K., Rutherford, A., et al. (2001). Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum. Mol. Genet. 10, 1571–1579. doi: 10.1093/hmg/10.15.1571
Boncinelli, E., and Morgan, R. (2001). Downstream of Otx2, or how to get a head. Trends Genet. 17, 633–636. doi: 10.1016/s0168-9525(01)02418-0
Bouillet, B., Crevisy, E., Baillot-Rudoni, S., Gallegarine, D., Jouan, T., Duffourd, Y., et al. (2020). Whole-exome sequencing identifies the first French MODY 6 family with a new mutation in the NEUROD1 gene. Diabetes Metab. 46, 400–402. doi: 10.1016/j.diabet.2020.03.001
Boutin, C., Hardt, O., de Chevigny, A., Coré, N., Goebbels, S., Seidenfaden, R., et al. (2010). NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 107, 1201–1206. doi: 10.1073/pnas.0909015107
Bowne, S. J., Sullivan, L. S., Wheaton, D. K., Locke, K. G., Jones, K. D., Koboldt, D. C., et al. (2016). North Carolina macular dystrophy (MCDR1) caused by a novel tandem duplication of the PRDM13 gene. Mol. Vis. 22, 1239–1247.
Breslin, M. B., Zhu, M., and Lan, M. S. (2003). NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J. Biol. Chem. 278, 38991–38997. doi: 10.1074/jbc.M306795200
Brodosi, L., Baracco, B., Mantovani, V., and Pironi, L. (2021). NEUROD1 mutation in an Italian patient with maturity onset diabetes of the young 6: a case report. BMC Endocr. Disord. 21:202. doi: 10.1186/s12902-021-00864-w
Brown, N. S., Smart, A., Sharma, V., Brinkmeier, M. L., Greenlee, L., Camper, S. A., et al. (2000). Thyroid hormone resistance and increased metabolic rate in the RXR-gamma-deficient mouse. J. Clin. Invest. 106, 73–79. doi: 10.1172/JCI9422
Brzezinski, J. A. T., Lamba, D. A., and Reh, T. A. (2010). Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629.
Brzezinski, J. A., and Reh, T. A. (2015). Photoreceptor cell fate specification in vertebrates. Development 142, 3263–3273.
Buenaventura, D. F., Ghinia-Tegla, M. G., and Emerson, M. M. (2018). Fate-restricted retinal progenitor cells adopt a molecular profile and spatial position distinct from multipotent progenitor cells. Dev. Biol. 443, 35–49. doi: 10.1016/j.ydbio.2018.06.023
Bunt, J., Hasselt, N. E., Zwijnenburg, D. A., Koster, J., Versteeg, R., and Kool, M. (2011). Joint binding of OTX2 and MYC in promotor regions is associated with high gene expression in medulloblastoma. PLoS One 6:e26058. doi: 10.1371/journal.pone.0026058
Bunt, J., Hasselt, N. E., Zwijnenburg, D. A., Hamdi, M., Koster, J., Versteeg, R., et al. (2012). OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int. J. Cancer 131, E21–E32. doi: 10.1002/ijc.26474
Byerly, M. S., and Blackshaw, S. (2009). Vertebrate retina and hypothalamus development. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 380–389. doi: 10.1002/wsbm.22
Campi, I., Cammarata, G., Bianchi Marzoli, S., Beck-Peccoz, P., Santarsiero, D., Dazzi, D., et al. (2017). Retinal photoreceptor functions are compromised in patients with resistance to thyroid hormone syndrome (RTHβ). J. Clin. Endocrinol. Metab. 102, 2620–2627. doi: 10.1210/jc.2016-3671
Cantos, R., Cole, L. K., Acampora, D., Simeone, A., and Wu, D. K. (2000). Patterning of the mammalian cochlea. Proc. Natl. Acad. Sci. 97, 11707–11713. doi: 10.1073/pnas.97.22.11707
Chan, C. S. Y., Lonfat, N., Zhao, R., Davis, A. E., Li, L., Wu, M. R., et al. (2020). Cell type- and stage-specific expression of Otx2 is regulated by multiple transcription factors and cis-regulatory modules in the retina. Development 147:dev187922. doi: 10.1242/dev.187922
Chapla, A., Mruthyunjaya, M. D., Asha, H. S., Varghese, D., Varshney, M., Vasan, S. K., et al. (2015). Maturity onset diabetes of the young in India - a distinctive mutation pattern identified through targeted next-generation sequencing. Clin. Endocrinol. 82, 533–542. doi: 10.1111/cen.12541
Chatelain, G., Fossat, N., Brun, G., and Lamonerie, T. (2006). Molecular dissection reveals decreased activity and not dominant negative effect in human OTX2 mutants. J. Mol. Med. (Berl) 84, 604–615. doi: 10.1007/s00109-006-0048-2
Chattergoon, N. N. (2019). Thyroid hormone signaling and consequences for cardiac development. J. Endocrinol. 242, T145–t160. doi: 10.1530/JOE-18-0704
Chawla, A., Repa, J. J., Evans, R. M., and Mangelsdorf, D. J. (2001). Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870. doi: 10.1126/science.294.5548.1866
Chen, C. M., and Cepko, C. L. (2002). The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development 129, 5363–5375. doi: 10.1242/dev.00114
Chen, J., Rattner, A., and Nathans, J. (2005). The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 25, 118–129.
Chen, S., Wang, Q. L., Nie, Z., Sun, H., Lennon, G., Copeland, N. G., et al. (1997). Crx, a novel Otx-like paired-homeodomain protein, binds to and Transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030. doi: 10.1016/S0896-6273(00)80394-3
Chen, S., Wang, Q. L., Xu, S., Liu, I., Li, L. Y., Wang, Y., et al. (2002). Functional analysis of cone–rod homeobox (CRX) mutations associated with retinal dystrophy. Hum. Mol. Genet. 11, 873–884. doi: 10.1093/hmg/11.8.873
Chen, Y. C., Ma, N. X., Pei, Z. F., Wu, Z., do-Monte, F. H., Keefe, S., et al. (2020). A NeuroD1 AAV-based gene therapy for functional brain repair after ischemic injury through in vivo astrocyte-to-neuron conversion. Mol. Ther. 28, 217–234. doi: 10.1016/j.ymthe.2019.09.003
Cheng, H., Khanna, H., Oh, E. C., Hicks, D., Mitton, K. P., and Swaroop, A. (2004). Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 13, 1563–1575. doi: 10.1093/hmg/ddh173
Cheng, H., Aleman, T. S., Cideciyan, A. V., Khanna, R., Jacobson, S. G., and Swaroop, A. (2006). In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 15, 2588–2602. doi: 10.1093/hmg/ddl185
Cheng, H., Khan, N. W., Roger, J. E., and Swaroop, A. (2011). Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum. Mol. Genet. 20, 4102–4115. doi: 10.1093/hmg/ddr334
Chiang, W. J., Kroeger, H., Chea, L., and Lin, J. H. (2019). Pathomechanisms of ATF6-associated cone photoreceptor diseases. Adv. Exp. Med. Biol. 1185, 305–310. doi: 10.1007/978-3-030-27378-1_50
Chirco, K. R., Chew, S., Moore, A. T., Duncan, J. L., and Lamba, D. A. (2021). Allele-specific gene editing to rescue dominant CRX-associated LCA7 phenotypes in a retinal organoid model. Stem Cell Reports 16, 2690–2702. doi: 10.1016/j.stemcr.2021.09.007
Cho, J. H., Klein, W. H., and Tsai, M. J. (2007). Compensational regulation of bHLH transcription factors in the postnatal development of BETA2/NeuroD1-null retina. Mech. Dev. 124, 543–550. doi: 10.1016/j.mod.2007.06.001
Collin, R. W. J., van den Born, L. I., Klevering, B. J., de Castro-Miró, M., Littink, K. W., Arimadyo, K., et al. (2011). High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the Dutch population. Invest. Ophthalmol. Vis. Sci. 52, 2227–2239. doi: 10.1167/iovs.10-6185
Concolino, P., Costella, A., and Paragliola, R. M. (2019). Mutational landscape of resistance to thyroid hormone Beta (RTHβ). Mol. Diagn. Ther. 23, 353–368. doi: 10.1007/s40291-019-00399-w
Coppieters, F., Leroy, B. P., Beysen, D., Hellemans, J., de Bosscher, K., Haegeman, G., et al. (2007). Recurrent mutation in the first zinc finger of the orphan nuclear receptor NR2E3 causes autosomal dominant retinitis pigmentosa. Am. J. Hum. Genet. 81, 147–157. doi: 10.1086/518426
Corbo, J. C., and Cepko, C. L. (2005). A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 1:e11. doi: 10.1371/journal.pgen.0010011
Corbo, J. C., Lawrence, K. A., Karlstetter, M., Myers, C. A., Abdelaziz, M., Dirkes, W., et al. (2010). CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525. doi: 10.1101/gr.109405.110
Cossette, S. M., and Drysdale, T. A. (2004). Early expression of thyroid hormone receptor beta and retinoid X receptor gamma in the Xenopus embryo. Differentiation 72, 239–249.
Cuevas, E., Holder, D. L., Alshehri, A. H., Tréguier, J., Lakowski, J., and Sowden, J. C. (2021). NRL(−/−) gene edited human embryonic stem cells generate rod-deficient retinal organoids enriched in S-cone-like photoreceptors. Stem Cells 39, 414–428. doi: 10.1002/stem.3325
Cvekl, A., and Wang, W. L. (2009). Retinoic acid signaling in mammalian eye development. Exp. Eye Res. 89, 280–291. doi: 10.1016/j.exer.2009.04.012
Daniele, L. L., Lillo, C., Lyubarsky, A. L., Nikonov, S. S., Philp, N., Mears, A. J., et al. (2005). Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 46, 2156–2167. doi: 10.1167/iovs.04-1427
Darras, V. M., van Herck, S. L. J., Heijlen, M., and de Groef, B. (2011). Thyroid hormone receptors in two model species for vertebrate embryonic development: chicken and zebrafish. J. Thyroid. Res. 2011:402320. doi: 10.4061/2011/402320
Davies, P. J., Berry, S. A., Shipley, G. L., Eckel, R. H., Hennuyer, N., Crombie, D. L., et al. (2001). Metabolic effects of rexinoids: tissue-specific regulation of lipoprotein lipase activity. Mol. Pharmacol. 59, 170–176. doi: 10.1124/mol.59.2.170
Dawson, M. I., and Xia, Z. (2012). The retinoid X receptors and their ligands. Biochim. Biophys. Acta 1821, 21–56. doi: 10.1016/j.bbalip.2011.09.014
de Carvalho, E. R., Robson, A. G., Arno, G., Boon, C. J. F., Webster, A. A., and Michaelides, M. (2021). Enhanced S-cone syndrome: Spectrum of clinical, imaging, Electrophysiologic, and genetic findings in a retrospective case series of 56 patients. Ophthalmol. Retina 5, 195–214. doi: 10.1016/j.oret.2020.07.008
DeAngelis, M. M., Grimsby, J. L., Sandberg, M. A., Berson, E. L., and Dryja, T. P. (2002). Novel mutations in the NRL gene and associated clinical findings in patients with dominant retinitis pigmentosa. Arch. Ophthalmol. 120, 369–375. doi: 10.1001/archopht.120.3.369
Deeb, S. S. (2006). Genetics of variation in human color vision and the retinal cone mosaic. Curr. Opin. Genet. Dev. 16, 301–307. doi: 10.1016/j.gde.2006.04.002
Deveau, C., Jiao, X., Suzuki, S. C., Krishnakumar, A., Yoshimatsu, T., Hejtmancik, J. F., et al. (2020). Thyroid hormone receptor beta mutations alter photoreceptor development and function in Danio rerio (zebrafish). PLoS Genet. 16:e1008869. doi: 10.1371/journal.pgen.1008869
Di Nardo, A. A., Joliot, A., and Prochiantz, A. (2020). Homeoprotein transduction in neurodevelopment and physiopathology. Sci. Adv. 6:eabc6374. doi: 10.1126/sciadv.abc6374
di Nardo, A. A., Fuchs, J., Joshi, R. L., Moya, K. L., and Prochiantz, A. (2018). The physiology of Homeoprotein transduction. Physiol. Rev. 98, 1943–1982. doi: 10.1152/physrev.00018.2017
Diaczok, D., Fuchs, J., Joshi, R. L., Moya, K. L., and Prochiantz, A. (2008). A novel dominant negative mutation of OTX2 associated with combined pituitary hormone deficiency. J. Clin. Endocrinol. Metab. 93, 4351–4359. doi: 10.1210/jc.2008-1189
El-Asrag, M. E., Corton, M., McKibbin, M., Avila-Fernandez, A., Mohamed, M. D., Blanco-Kelly, F., et al. (2022). Novel homozygous mutations in the transcription factor NRL cause non-syndromic retinitis pigmentosa. Mol. Vis. 28, 48–56.
Eldred, K. C., Hadyniak, S. E., Hussey, K. A., Brenerman, B., Zhang, P.-W., Chamling, X., et al. (2018). Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362:p.eaau6348. doi: 10.1126/science.aau6348
Elias, W. J., Lopes, M. B., Golden, W. L., Jane, J. A. Sr., and Gonzalez-Fernandez, F. (2001). Trilateral retinoblastoma variant indicative of the relevance of the retinoblastoma tumor-suppressor pathway to medulloblastomas in humans. J. Neurosurg. 95, 871–878. doi: 10.3171/jns.2001.95.5.0871
Emerson, M. M., and Cepko, C. L. (2011). Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Dev. Biol. 360, 241–255. doi: 10.1016/j.ydbio.2011.09.012
Evsen, L., Sugahara, S., Uchikawa, M., Kondoh, H., and Wu, D. K. (2013). Progression of neurogenesis in the inner ear requires inhibition of Sox2 transcription by neurogenin1 and neurod1. J. Neurosci. 33, 3879–3890. doi: 10.1523/JNEUROSCI.4030-12.2013
Fang, Q., George, A. S., Brinkmeier, M. L., Mortensen, A. H., Gergics, P., Cheung, L. Y. M., et al. (2016). Genetics of combined pituitary hormone deficiency: roadmap into the genome era. Endocr. Rev. 37, 636–675. doi: 10.1210/er.2016-1101
Fant, B., Samuel, A., Audebert, S., Couzon, A., el Nagar, S., Billon, N., et al. (2015). Comprehensive interactome of Otx2 in the adult mouse neural retina. Genesis 53, 685–694. doi: 10.1002/dvg.22903
Farjo, Q., Jackson, A., Pieke-Dahl, S., Scott, K., Kimberling, W. J., Sieving, P. A., et al. (1997). Human bZIP transcription factor gene NRL: structure, genomic sequence, and fine linkage mapping at 14q11.2 and negative mutation analysis in patients with retinal degeneration. Genomics 45, 395–401. doi: 10.1006/geno.1997.4964
Forrest, D., Reh, T. A., and Rüsch, A. (2002). Neurodevelopmental control by thyroid hormone receptors. Curr. Opin. Neurobiol. 12, 49–56.
Fossat, N., le Greneur, C., Béby, F., Vincent, S., Godement, P., Chatelain, G., et al. (2007). A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors. BMC Dev. Biol. 7:122. doi: 10.1186/1471-213X-7-122
Freund, C. L., Gregory-Evans, C. Y., Furukawa, T., Papaioannou, M., Looser, J., Ploder, L., et al. (1997). Cone-rod dystrophy due to mutations in a novel photoreceptor-specific Homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell(Cambridge,Mass) 91, 543–553. doi: 10.1016/S0092-8674(00)80440-7
Freund, C. L., Wang, Q. L., Chen, S., Muskat, B. L., Wiles, C. D., Sheffield, V. C., et al. (1998). De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat. Genet. 18, 311–312. doi: 10.1038/ng0498-311
Furukawa, T., Morrow, E. M., and Cepko, C. L. (1997). Crx, a novel otx-like Homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cells 91, 531–541. doi: 10.1016/S0092-8674(00)80439-0
Furukawa, T., Morrow, E. M., Li, T., Davis, F. C., and Cepko, C. L. (1999). Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 23, 466–470. doi: 10.1038/70591
Gao, M., Zhang, S., Liu, C., Qin, Y., Archacki, S., Jin, L., et al. (2016). Whole exome sequencing identifies a novel NRL mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol. Vis. 22, 234–242.
Garafalo, A. V., Calzetti, G., Cideciyan, A. V., Roman, A. J., Saxena, S., Sumaroka, A., et al. (2018). Cone vision changes in the enhanced S-cone syndrome caused by NR2E3 gene mutations. Invest. Ophthalmol. Vis. Sci. 59, 3209–3219. doi: 10.1167/iovs.18-24518
Gerber, S., Rozet, J. M., Takezawa, S. I., Coutinho dos Santos, L., Lopes, L., Gribouval, O., et al. (2000). The photoreceptor cell-specific nuclear receptor gene (PNR) accounts for retinitis pigmentosa in the crypto-Jews from Portugal (Marranos), survivors from the Spanish inquisition. Hum. Genet. 107, 276–284. doi: 10.1007/s004390000350
Ghinia Tegla, M. G., Buenaventura, D. F., Kim, D. Y., Thakurdin, C., Gonzalez, K. C., and Emerson, M. M. (2020). OTX2 represses sister cell fate choices in the developing retina to promote photoreceptor specification. eLife 9:e54279. doi: 10.7554/eLife.54279
Glubrecht, D. D., Kim, J. H., Russell, L., Bamforth, J. S., and Godbout, R. (2009). Differential CRX and OTX2 expression in human retina and retinoblastoma. J. Neurochem. 111, 250–263. doi: 10.1111/j.1471-4159.2009.06322.x
Gonsorčíková, L., Průhová, Š., Cinek, O., Ek, J., Pelikánová, T., Jørgensen, T., et al. (2008). Autosomal inheritance of diabetes in two families characterized by obesity and a novel H241Q mutation in NEUROD1. Pediatr. Diabetes 9, 367–372. doi: 10.1111/j.1399-5448.2008.00379.x
Gonzalez-Rodriguez, J., Pelcastre, E. L., Tovilla-Canales, J. L., Garcia-Ortiz, J. E., Amato-Almanza, M., Villanueva-Mendoza, C., et al. (2010). Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia-anophthalmia-coloboma (MAC) spectrum cases. Br. J. Ophthalmol. 94, 1100–1104. doi: 10.1136/bjo.2009.173500
Gorbenko del Blanco, D., Romero, C. J., Diaczok, D., de Graaff, L. C. G., Radovick, S., and Hokken-Koelega, A. C. S. (2012). A novel OTX2 mutation in a patient with combined pituitary hormone deficiency, pituitary malformation, and an underdeveloped left optic nerve. Eur. J. Endocrinol. 167, 441–452. doi: 10.1530/EJE-12-0333
Green, D. J., Lenassi, E., Manning, C. S., McGaughey, D., Sharma, V., Black, G. C., et al. (2021). North Carolina macular dystrophy: phenotypic variability and computational analysis of disease-associated noncoding variants. Invest. Ophthalmol. Vis. Sci. 62:16. doi: 10.1167/iovs.62.7.16
Gregory, L. C., Gergics, P., Nakaguma, M., Bando, H., Patti, G., McCabe, M. J., et al. (2021). The phenotypic spectrum associated with OTX2 mutations in humans. Eur. J. Endocrinol. 185, 121–135. doi: 10.1530/EJE-20-1453
Haider, N. B., Jacobson, S. G., Cideciyan, A. V., Swiderski, R., Streb, L. M., Searby, C., et al. (2000). Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 24, 127–131. doi: 10.1038/72777
Haider, N. B., Demarco, P., Nystuen, A. M., Huang, X., Smith, R. S., Mccall, M. A., et al. (2006). The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis. Neurosci. 23, 917–929. doi: 10.1017/S095252380623027X
Hao, H., Kim, D. S., Klocke, B., Johnson, K. R., Cui, K., Gotoh, N., et al. (2012). Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 8, –e1002649. doi: 10.1371/journal.pgen.1002649
Harada, T., Harada, C., and Parada, L. F. (2007). Molecular regulation of visual system development: more than meets the eye. Genes Dev. 21, 367–378. doi: 10.1101/gad.1504307
Haugen, B. R., Jensen, D. R., Sharma, V., Pulawa, L. K., Hays, W. R., Krezel, W., et al. (2004). Retinoid X receptor γ-deficient mice have increased skeletal muscle lipoprotein lipase activity and less weight gain when fed a high-fat diet. Endocrinology 145, 3679–3685. doi: 10.1210/en.2003-1401
Heavner, W., and Pevny, L. (2012). Eye development and retinogenesis. Cold Spring Harb Perspect Biol 4:a008391. doi: 10.1101/cshperspect.a008391
Henderson, R. H., Williamson, K. A., Kennedy, J. S., Webster, A. R., Holder, G. E., Robson, A. G., et al. (2009). A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Mol. Vis. 15, 2442–2447.
Hennig, A. K., Peng, G.-H., and Chen, S. (2008). Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 1192, 114–133. doi: 10.1016/j.brainres.2007.06.036
Hennig, A. K., Peng, G.-H., and Chen, S. (2013). Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. PLoS One 8:e69721
Hoover, F., Seleiro, E. A. P., Kielland, A., Brickell, P. M., and Glover, J. C. (1998). Retinoid X receptor gamma gene transcripts are expressed by a subset of early generated retinal cells and eventually restricted to photoreceptors. J. Comp. Neurol. 391, 204–213. doi: 10.1002/(SICI)1096-9861(19980209)391:2<204::AID-CNE4>3.0.CO;2-6
Horikawa, Y., and Enya, M. (2019). Genetic dissection and clinical features of MODY6 (NEUROD1-MODY). Curr. Diab. Rep. 19:12. doi: 10.1007/s11892-019-1130-9
Housset, M., Samuel, A., Ettaiche, M., Bemelmans, A., Beby, F., Billon, N., et al. (2013). Loss of Otx2 in the adult retina disrupts retinal pigment epithelium function, causing photoreceptor degeneration. J. Neurosci. 33, 9890–9904. doi: 10.1523/JNEUROSCI.1099-13.2013
Hsiau, T. H. C., Diaconu, C., Myers, C. A., Lee, J., Cepko, C. L., and Corbo, J. C. (2007). The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One 2:e643. doi: 10.1371/journal.pone.0000643
Huang, H. P., Chu, K., Nemoz-Gaillard, E., Elberg, D., and Tsai, M. J. (2002). Neogenesis of BETA-cells in adult BETA2/NeuroD-deficient mice. Mol. Endocrinol. 16, 541–551. doi: 10.1210/me.16.3.541
Hull, S., Arno, G., Plagnol, V., Chamney, S., Russell-Eggitt, I., Thompson, D., et al. (2014). The phenotypic variability of retinal dystrophies associated with mutations in CRX, with report of a novel macular dystrophy phenotype. Invest. Ophthalmol. Vis. Sci. 55, 6934–6944. doi: 10.1167/iovs.14-14715
Iannaccone, A., Brabbit, E., Lopez-Miro, C., Love, Z., Griffiths, V., Kedrov, M., et al. (2021). Interspecies correlations between human and mouse NR2E3-associated recessive disease. J. Clin. Med. 10:475. doi: 10.3390/jcm10030475
Ibad, R. T., Rheey, J., Mrejen, S., Forster, V., Picaud, S., Prochiantz, A., et al. (2011). Otx2 promotes the survival of damaged adult retinal ganglion cells and protects against excitotoxic loss of visual acuity in vivo. J. Neurosci. 31, 5495–5503. doi: 10.1523/JNEUROSCI.0187-11.2011
Ibrahim, M. T., Alarcon-Martinez, T., Lopez, I., Fajardo, N., Chiang, J., and Koenekoop, R. K. (2018). A complete, homozygous CRX deletion causing nullizygosity is a new genetic mechanism for Leber congenital amaurosis. Sci. Rep. 8:5034. doi: 10.1038/s41598-018-22704-z
Irie, S., Sanuki, R., Muranishi, Y., Kato, K., Chaya, T., and Furukawa, T. (2015). Rax Homeoprotein regulates photoreceptor cell maturation and survival in association with Crx in the postnatal mouse retina. Mol. Cell. Biol. 35, 2583–2596. doi: 10.1128/MCB.00048-15
Jacobson, S. G., Marmor, M. F., Kemp, C. M., and Knighton, R. W. (1990). SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. Invest. Ophthalmol. Vis. Sci. 31, 827–838.
Jacobson, S. G., Román, A. J., Román, M. I., Gass, J. D. M., and Parker, J. A. (1991). Relatively enhanced S cone function in the Goldmann-Favre syndrome. Am J. Ophthalmol. 111, 446–453. doi: 10.1016/S0002-9394(14)72379-7
Jacobson, S. G., Cideciyan, A. V., Huang, Y., Hanna, D. B., Freund, C. L., Affatigato, L. M., et al. (1998). Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest. Ophthalmol. Vis. Sci. 39, 2417–2426.
Janssen, J. J., Kuhlmann, E. D., van Vugt, A. H. M., Winkens, H. J., Janssen, B. P. M., Deutman, A. F., et al. (1999). Retinoic acid receptors and retinoid X receptors in the mature retina: subtype determination and cellular distribution. Curr. Eye Res. 19, 338–347. doi: 10.1076/ceyr.19.4.338.5307
Jolma, A., Yin, Y., Nitta, K. R., Dave, K., Popov, A., Taipale, M., et al. (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388. doi: 10.1038/nature15518
Jones, G. E., Robertson, L., Warman, P., Craft, E. V., Cresswell, L., and Vasudevan, P. C. (2016). 14q22.3 microdeletion encompassing OTX2 in a five-generation family with microphthalmia, pituitary abnormalities, and intellectual disability. Ophthalmic Genet. 37, 352–353. doi: 10.3109/13816810.2015.1059463
Jurkiewicz, E., Pakuła-Kościesza, I., Rutynowska, O., and Nowak, K. (2010). Trilateral retinoblastoma: an institutional experience and review of the literature. Childs Nerv. Syst. 26, 129–132. doi: 10.1007/s00381-009-0958-8
Kanda, A., Friedman, J. S., Nishiguchi, K. M., and Swaroop, A. (2007). Retinopathy mutations in the bZIP protein NRL alter phosphorylation and transcriptional activity. Hum. Mutat. 28, 589–598. doi: 10.1002/humu.20488
Kim, D. S., Ross, S. E., Trimarchi, J. M., Aach, J., Greenberg, M. E., and Cepko, C. L. (2008). Identification of molecular markers of bipolar cells in the murine retina. J. Comp. Neurol. 507, 1795–1810. doi: 10.1002/cne.21639
Kim, H. T., Kim, S. J., Sohn, Y. I., Paik, S. S., Caplette, R., Simonutti, M., et al. (2015). Mitochondrial protection by exogenous Otx2 in mouse retinal neurons. Cell Rep. 13, 990–1002. doi: 10.1016/j.celrep.2015.09.075
Kim, J.-W., Yang, H. J., Brooks, M. J., Zelinger, L., Karakülah, G., Gotoh, N., et al. (2016). NRL-regulated transcriptome dynamics of developing rod photoreceptors. Cell Rep. 17, 2460–2473. doi: 10.1016/j.celrep.2016.10.074
Koike, C., Nishida, A., Ueno, S., Saito, H., Sanuki, R., Sato, S., et al. (2007). Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol. Cell. Biol. 27, 8318–8329. doi: 10.1128/MCB.01209-07
Kristinsson, S. Y., Talseth, B., Steingrimsson, E., Thorsson, A. V., Helgason, T., Hreidarsson, A. B., et al. (2001). MODY in Iceland is associated with mutations in HNF-1α and a novel mutation in NeuroD1. Diabetologia 44, 2098–2103. doi: 10.1007/s001250100016
Kruczek, K., Qu, Z., Gentry, J., Fadl, B. R., Gieser, L., Hiriyanna, S., et al. (2021). Gene therapy of dominant CRX-Leber congenital Amaurosis using patient stem cell-derived retinal organoids. Stem Cell Reports 16, 252–263. doi: 10.1016/j.stemcr.2020.12.018
Lee, E. J., Kim, N., Park, J. W., Kang, K. H., Kim, W. I., Sim, N. S., et al. (2019). Global analysis of intercellular homeodomain protein transfer. Cell Rep. 28, 712–722.e3. doi: 10.1016/j.celrep.2019.06.056
Lejeune, F., and Maquat, L. E. (2005). Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17, 309–315.
Li, M., Iismaa, S. E., Naqvi, N., Nicks, A., Husain, A., and Graham, R. M. (2014). Thyroid hormone action in postnatal heart development. Stem Cell Res. 13, 582–591. doi: 10.1016/j.scr.2014.07.001
LI, J., di, C. H. U. N. H. U. I., JING, J., di, Q. U. N., NAKHLA, J., and ADAMSON, D. C. (2015). OTX2 is a therapeutic target for retinoblastoma and may function as a common factor between C-MYC, CRX, and phosphorylated RB pathways. Int. J. Oncol. 47, 1703–1710. doi: 10.3892/ijo.2015.3179
Li, S., Datta, S., Brabbit, E., Love, Z., Woytowicz, V., Flattery, K., et al. (2021). Nr2e3 is a genetic modifier that rescues retinal degeneration and promotes homeostasis in multiple models of retinitis pigmentosa. Gene Ther. 28, 223–241. doi: 10.1038/s41434-020-0134-z
Liang, X., Brooks, M. J., and Swaroop, A. (2022). Developmental genome-wide occupancy analysis of bZIP transcription factor NRL uncovers the role of c-Jun in early differentiation of rod photoreceptors in the mammalian retina. Hum. Mol. Genet. 31, 3914–3933. doi: 10.1093/hmg/ddac143
Littink, K. W., Stappers, P., Riemslag, F., Talsma, H., van Genderen, M., Cremers, F., et al. (2018). Autosomal recessive NRL mutations in patients with enhanced S-cone syndrome. Genes (Basel) 9:68. doi: 10.3390/genes9020068
Liu, Q., Ji, X., Breitman, M. L., Hitchcock, P. F., and Swaroop, A. (1996). Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene 12, 207–211.
Liu, M., Pereira, F. A., Price, S. D., Chu, M. J., Shope, C., Himes, D., et al. (2000). Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 14, 2839–2854. doi: 10.1101/gad.840500
Liu, L., Furuta, H., Minami, A., Zheng, T., Jia, W., Nanjo, K., et al. (2007). A novel mutation, Ser159Pro in the NeuroD1/BETA2 gene contributes to the development of diabetes in a Chinese potential MODY family. Mol. Cell. Biochem. 303, 115–120. doi: 10.1007/s11010-007-9463-0
Liu, H., Etter, P., Hayes, S., Jones, I., Nelson, B., Hartman, B., et al. (2008). NeuroD1 regulates expression of thyroid hormone receptor β2 and cone opsins in the developing mouse retina. J. Neurosci. 28, 749–756. doi: 10.1523/JNEUROSCI.4832-07.2008
Livesey, R., and Cepko, C. (2001). Neurobiology. Developing order. Nature 413, 471–473. doi: 10.1038/35097186
Lu, Y., Labak, C. M., Jain, N., Purvis, I. J., Guda, M. R., Bach, S. E., et al. (2017). OTX2 expression contributes to proliferation and progression in Myc-amplified medulloblastoma. Am. J. Cancer Res. 7, 647–656.
Mackin, R. D., Frey, R. A., Gutierrez, C., Farre, A. A., Kawamura, S., Mitchell, D. M., et al. (2019). Endocrine regulation of multichromatic color vision. Proc. Natl. Acad. Sci. U. S. A. 116, 16882–16891. doi: 10.1073/pnas.1904783116
Malecki, M. T., Jhala, U. S., Antonellis, A., Fields, L., Doria, A., Orban, T., et al. (1999). Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 23, 323–328. doi: 10.1038/15500
Malecki, M. T., Cyganek, K., Klupa, T., and Sieradzki, J. (2003). The Ala45Thr polymorphism of BETA2/NeuroD1 gene and susceptibility to type 2 diabetes mellitus in a polish population. Acta Diabetol. 40, 109–111. doi: 10.1007/s005920300015
Manes, G., Joly, W., Guignard, T., Smirnov, V., Berthemy, S., Bocquet, B., et al. (2017). A novel duplication of PRMD13 causes North Carolina macular dystrophy: overexpression of PRDM13 orthologue in drosophila eye reproduces the human phenotype. Hum. Mol. Genet. 26, 4367–4374. doi: 10.1093/hmg/ddx322
Mangelsdorf, D. J., and Evans, R. M. (1995). The RXR heterodimers and orphan receptors. Cells 83, 841–850.
Marelli, F., Carra, S., Agostini, M., Cotelli, F., Peeters, R., Chatterjee, K., et al. (2016). Patterns of thyroid hormone receptor expression in zebrafish and generation of a novel model of resistance to thyroid hormone action. Mol. Cell. Endocrinol. 424, 102–117. doi: 10.1016/j.mce.2016.01.020
Martinez-Gimeno, M., Maseras, M., Baiget, M., Beneito, M., Antiñolo, G., Ayuso, C., et al. (2001). Mutations P51U and G122E in retinal transcription factor NRL associated with autosomal dominant and sporadic retinitis pigmentosa. Hum. Mutat. 17:520. doi: 10.1002/humu.1135
Martinez-Morales, J. R., Signore, M., Acampora, D., Simeone, A., and Bovolenta, P. (2001). Otx genes are required for tissue specification in the developing eye. Development 128, 2019–2030. doi: 10.1242/dev.128.11.2019
Mastracci, T. L., Anderson, K. R., Papizan, J. B., and Sussel, L. (2013). Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS Genet. 9:e1003278. doi: 10.1371/journal.pgen.1003278
Matsumoto, R., Suga, H., Aoi, T., Bando, H., Fukuoka, H., Iguchi, G., et al. (2020). Congenital pituitary hypoplasia model demonstrates hypothalamic OTX2 regulation of pituitary progenitor cells. J. Clin. Invest. 130, 641–654. doi: 10.1172/JCI127378
Matsuo, I., Kuratani, S., Kimura, C., Takeda, N., and Aizawa, S. (1995). Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 9, 2646–2658. doi: 10.1101/gad.9.21.2646
McIlvain, V. A., and Knox, B. E. (2007). Nr2e3 and Nrl can reprogram retinal precursors to the rod fate in Xenopus retina. Dev. Dyn. 236, 1970–1979.
Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., et al. (2001). Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447–452. doi: 10.1038/ng774
Miesfeld, J. B., and Brown, N. L. (2019). Eye organogenesis: a hierarchical view of ocular development. Curr. Top. Dev. Biol. 132, 351–393.
Milam, A. H., Rose, L., Cideciyan, A. V., Barakat, M. R., Tang, W. X., Gupta, N., et al. (2002). The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc. Natl. Acad. Sci. U. S. A. 99, 473–478. doi: 10.1073/pnas.022533099
Miyata, T., Maeda, T., and Lee, J. E. (1999). NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13, 1647–1652.
Mizobuchi, K., Hayashi, T., Matsuura, T., and Nakano, T. (2022). Clinical characterization of autosomal dominant retinitis pigmentosa with NRL mutation in a three-generation Japanese family. Doc. Ophthalmol. 144, 227–235. doi: 10.1007/s10633-022-09874-y
Montana, C. L., Lawrence, K. A., Williams, N. L., Tran, N. M., Peng, G. H., Chen, S., et al. (2011). Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J. Biol. Chem. 286, 36921–36931. doi: 10.1074/jbc.M111.279026
Montana, C. L., Kolesnikov, A. V., Shen, S. Q., Myers, C. A., Kefalov, V. J., and Corbo, J. C. (2013). Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc. Natl. Acad. Sci. 110, 1732–1737. doi: 10.1073/pnas.1214387110
Morea, A., Boero, G., Demaio, V., Francavilla, T., and la Neve, A. (2021). Eyelid myoclonia with absences, intellectual disability and attention deficit hyperactivity disorder: a clinical phenotype of the RORB gene mutation. Neurol. Sci. 42, 2059–2062. doi: 10.1007/s10072-020-05031-y
Mori, M., Ghyselinck, N. B., Chambon, P., and Mark, M. (2001). Systematic immunolocalization of retinoid receptors in developing and adult mouse eyes. Invest. Ophthalmol. Vis. Sci. 42, 1312–1318.
Morrow, E. M., Furukawa, T., Lee, J. E., and Cepko, C. L. (1999). NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23–36. doi: 10.1242/dev.126.1.23
Mullur, R., Liu, Y. Y., and Brent, G. A. (2014). Thyroid hormone regulation of metabolism. Physiol. Rev. 94, 355–382. doi: 10.1152/physrev.00030.2013
Muranishi, Y., Terada, K., Inoue, T., Katoh, K., Tsujii, T., Sanuki, R., et al. (2011). An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J. Neurosci. 31, 16792–16807. doi: 10.1523/JNEUROSCI.3109-11.2011
Naessens, S., Ruysschaert, L., Lefever, S., Coppieters, F., and de Baere, E. (2019). Antisense oligonucleotide-based downregulation of the G56R pathogenic variant causing NR2E3-associated autosomal dominant retinitis Pigmentosa. Genes (Basel) 10:363. doi: 10.3390/genes10050363
Nakamura, P. A., Shimchuk, A. A., Tang, S., Wang, Z., DeGolier, K., Ding, S., and Reh, T. A. (2017). Small molecule Photoregulin3 prevents retinal degeneration in the rho(P23H) mouse model of retinitis pigmentosa. eLife :6. doi: 10.7554/eLife.30577
Namburi, P., Khateb, S., Meyer, S., Bentovim, T., Ratnapriya, R., Khramushin, A., et al. (2020). A unique PRDM13-associated variant in a Georgian Jewish family with probable North Carolina macular dystrophy and the possible contribution of a unique CFH variant. Mol. Vis. 26, 299–310.
Naya, F. J., Huang, H. P., Qiu, Y., Mutoh, H., DeMayo, F. J., Leiter, A. B., et al. (1997). Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11, 2323–2334. doi: 10.1101/gad.11.18.2323
Nelson, S. M., Park, L., and Stenkamp, D. L. (2009). Retinal homeobox 1 is required for retinal neurogenesis and photoreceptor differentiation in embryonic zebrafish. Dev. Biol. 328, 24–39. doi: 10.1016/j.ydbio.2008.12.040
Nelson, S. M., Frey, R. A., Wardwell, S. L., and Stenkamp, D. L. (2008). The developmental sequence of gene expression within the rod photoreceptor lineage in embryonic zebrafish. Dev. Dyn. 237, 2903–2917. doi: 10.1002/dvdy.21721
Neveling, K., Collin, R. W. J., Gilissen, C., van Huet, R. A. C., Visser, L., Kwint, M. P., et al. (2012). Next-generation genetic testing for retinitis pigmentosa. Hum. Mutat. 33, 963–972. doi: 10.1002/humu.22045
Newman, H., Blumen, S. C., Braverman, I., Hanna, R., Tiosano, B., Perlman, I., et al. (2016). Homozygosity for a recessive loss-of-function mutation of the NRL gene is associated with a variant of enhanced S-cone syndrome. Invest. Ophthalmol. Vis. Sci. 57, 5361–5371. doi: 10.1167/iovs.16-19505
Ng, L., Hurley, J. B., Dierks, B., Srinivas, M., Saltó, C., Vennström, B., et al. (2001). A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98. doi: 10.1038/83829
Ng, L., Ma, M., Curran, T., and Forrest, D. (2009). Developmental expression of thyroid hormone receptor beta2 protein in cone photoreceptors in the mouse. Neuroreport 20, 627–631. doi: 10.1097/WNR.0b013e32832a2c63
Ng, L., Lu, A., Swaroop, A., Sharlin, D. S., Swaroop, A., and Forrest, D. (2011). Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J. Neurosci. 31, 11118–11125. doi: 10.1523/JNEUROSCI.1709-11.2011
Nichols, L. L., Alur, R. P., Boobalan, E., Sergeev, Y. V., Caruso, R. C., Stone, E. M., et al. (2010). Two novel CRX mutant proteins causing autosomal dominant Leber congenital amaurosis interact differently with NRL. Hum. Mutat. 31, E1472–E1483. doi: 10.1002/humu.21268
Nikonov, S. S., Daniele, L. L., Zhu, X., Craft, C. M., Swaroop, A., and Pugh, E. N. Jr. (2005). Photoreceptors of Nrl −/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J. Gen. Physiol. 125, 287–304. doi: 10.1085/jgp.200409208
Nishida, A., Furukawa, A., Koike, C., Tano, Y., Aizawa, S., Matsuo, I., et al. (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6, 1255–1263. doi: 10.1038/nn1155
Nishiguchi, K. M., Friedman, J. S., Sandberg, M. A., Swaroop, A., Berson, E. L., and Dryja, T. P. (2004). Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc. Natl. Acad. Sci. U. S. A. 101, 17819–17824. doi: 10.1073/pnas.0408183101
O’Brien, K. M. B., Cheng, H., Jiang, Y., Schulte, D., Swaroop, A., and Hendrickson, A. E. (2004). Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Invest. Ophthalmol. Vis. Sci. 45, 2807–2812. doi: 10.1167/iovs.03-1317
Ochocinska, M. J., Muñoz, E. M., Veleri, S., Weller, J. L., Coon, S. L., Pozdeyev, N., et al. (2012). NeuroD1 is required for survival of photoreceptors but not pinealocytes: results from targeted gene deletion studies. J. Neurochem. 123, 44–59. doi: 10.1111/j.1471-4159.2012.07870.x
Oel, A. P., Neil, G. J., Dong, E. M., Balay, S. D., Collett, K., and Allison, W. T. (2020). Nrl is dispensable for specification of rod photoreceptors in adult zebrafish despite its deeply conserved requirement earlier in ontogeny. iScience 23:101805. doi: 10.1016/j.isci.2020.101805
Onigata, K., and Szinnai, G. (2014). Resistance to thyroid hormone. Endocr. Dev. 26, 118–129. doi: 10.1159/000363159
Orosz, O., Czeglédi, M., Kántor, I., Balogh, I., Vajas, A., Takács, L., et al. (2015). Ophthalmological phenotype associated with homozygous null mutation in the NEUROD1 gene. Mol. Vis. 21, 124–130.
Pachydaki, S. I., Klaver, C. C., Barbazetto, I. A., Roy, M. S., Gouras, P., Allikmets, R., et al. (2009). Phenotypic features of patients with NR2E3 mutations. Arch. Ophthalmol. 127, 71–75. doi: 10.1001/archophthalmol.2008.534
Pan, N., Jahan, I., Lee, J. E., and Fritzsch, B. (2009). Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 337, 407–428. doi: 10.1007/s00441-009-0826-6
Pappa, T., and Refetoff, S. (2021). Resistance to thyroid hormone beta: a focused review. Front. Endocrinol. :12. doi: 10.3389/fendo.2021.656551
Pappa, T., and Refetoff, S. (2018). Human genetics of thyroid hormone receptor beta: resistance to thyroid hormone beta (RTHβ). Methods Mol. Biol. 1801, 225–240. doi: 10.1007/978-1-4939-7902-8_18
Peng, G.-H., and Chen, S. (2007). Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum. Mol. Genet. 16, 2433–2452. doi: 10.1093/hmg/ddm200
Peng, G. H., Ahmad, O., Ahmad, F., Liu, J., and Chen, S. (2005). The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14, 747–764. doi: 10.1093/hmg/ddi070
Pennesi, M. E., Cho, J. H., Yang, Z., Wu, S. H., Zhang, J., Wu, S. M., et al. (2003). BETA2/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J. Neurosci. 23, 453–461. doi: 10.1523/JNEUROSCI.23-02-00453.2003
Pensieri, P., Mantilleri, A., Plassard, D., Furukawa, T., Moya, K. L., Prochiantz, A., et al. (2021). Photoreceptor cKO of OTX2 enhances OTX2 intercellular transfer in the retina and causes photophobia. eNeuro 8:ENEURO.0229-21.2021, ENEURO.0229–ENEU21.2021. doi: 10.1523/ENEURO.0229-21.2021
Petersen, H. V., Jensen, J. N., Stein, R., and Serup, P. (2002). Glucose induced MAPK signalling influences NeuroD1-mediated activation and nuclear localization. FEBS Lett. 528, 241–245. doi: 10.1016/S0014-5793(02)03318-5
Plouhinec, J.-L., Sauka-Spengler, T., Germot, A., le Mentec, C., Cabana, T., Harrison, G., et al. (2003). The mammalian Crx genes are highly divergent representatives of the Otx5 gene family, a gnathostome orthology class of orthodenticle-related homeogenes involved in the differentiation of retinal photoreceptors and circadian entrainment. Mol. Biol. Evol. 20, 513–521. doi: 10.1093/molbev/msg085
Poulin, G., Turgeon, B., and Drouin, J. (1997). NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17, 6673–6682.
Qin, Y., Liu, F., Yu, S., Yang, L., Gao, M., Tang, Z., et al. (2017). Identification of a novel NRL mutation in a Chinese family with retinitis pigmentosa by whole-exome sequencing. Eye 31, 815–817. doi: 10.1038/eye.2016.327
Raeisossadati, R., Ferrari, M. F. R., Kihara, A. H., AlDiri, I., and Gross, J. M. (2021). Epigenetic regulation of retinal development. Epigenetics Chromatin 14:11. doi: 10.1186/s13072-021-00384-w
Ragge, N. K., Brown, A. G., Poloschek, C. M., Lorenz, B., Henderson, R. A., Clarke, M. P., et al. (2005). Heterozygous mutations of OTX2 cause severe ocular malformations. Am. J. Hum. Genet. 76, 1008–1022. doi: 10.1086/430721
Rivolta, C., Peck, N. E., Fulton, A. B., Fishman, G. A., Berson, E. L., and Dryja, T. P. (2001). Novel frameshift mutations in CRX associated with Leber congenital amaurosis. Hum. Mutat. 18, 550–551. doi: 10.1002/humu.1243
Roberts, M. R., Hendrickson, A., McGuire, C. R., and Reh, T. A. (2005). Retinoid X receptor γ is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 46, 2897–2904. doi: 10.1167/iovs.05-0093
Roger, J. E., Ranganath, K., Zhao, L., Cojocaru, R. I., Brooks, M., Gotoh, N., et al. (2012). Preservation of cone photoreceptors after a rapid yet transient degeneration and remodeling in cone-only Nrl−/− mouse retina. J. Neurosci. 32, 528–541. doi: 10.1523/JNEUROSCI.3591-11.2012
Roger, J. E., Hiriyanna, A., Gotoh, N., Hao, H., Cheng, D. F., Ratnapriya, R., et al. (2014). OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest. 124, 631–643. doi: 10.1172/JCI72722
Romer, A. I., Singer, R. A., Sui, L., Egli, D., and Sussel, L. (2019). Murine perinatal β-cell proliferation and the differentiation of human stem cell-derived insulin-expressing cells require NEUROD1. Diabetes 68, 2259–2271. doi: 10.2337/db19-0117
Rovsing, L., Clokie, S., Bustos, D. M., Rohde, K., Coon, S. L., Litman, T., et al. (2011). Crx broadly modulates the pineal transcriptome. J. Neurochem. 119, 262–274. doi: 10.1111/j.1471-4159.2011.07405.x
Rubio-Cabezas, O., Minton, J. A. L., Kantor, I., Williams, D., Ellard, S., and Hattersley, A. T. (2010). Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes 59, 2326–2331. doi: 10.2337/db10-0011
Ruzycki, P. A., Zhang, X., and Chen, S. (2018). CRX directs photoreceptor differentiation by accelerating chromatin remodeling at specific target sites. Epigenetics Chromatin 11:42.
Ruzycki, P. A., Linne, C. D., Hennig, A. K., and Chen, S. (2017). Crx-L253X mutation produces dominant photoreceptor defects in TVRM65 mice. Invest. Ophthalmol. Vis. Sci. 58, 4644–4653. doi: 10.1167/iovs.17-22075
Sadleir, L. G., de Valles-Ibáñez, G., King, C., Coleman, M., Mossman, S., Paterson, S., et al. (2020). Inherited RORB pathogenic variants: overlap of photosensitive genetic generalized and occipital lobe epilepsy. Epilepsia 61, e23–e29. doi: 10.1111/epi.16475
Samuel, A., Housset, M., Fant, B., and Lamonerie, T. (2014). Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PLoS One 9:e89110. doi: 10.1371/journal.pone.0089110
Sato, S., Inoue, T., Terada, K., Matsuo, I., Aizawa, S., Tano, Y., et al. (2007). Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis 45, 502–507. doi: 10.1002/dvg.20318
Satou, Y., Minami, K., Hosono, E., Okada, H., Yasuoka, Y., Shibano, T., et al. (2018). Phosphorylation states change Otx2 activity for cell proliferation and patterning in the Xenopus embryo. Development 145:dev159640. doi: 10.1242/dev.159640
Schilter, K. F., Schneider, A., Bardakjian, T., Soucy, J. F., Tyler, R. C., Reis, L. M., et al. (2011). OTX2 microphthalmia syndrome: four novel mutations and delineation of a phenotype. Clin. Genet. 79, 158–168. doi: 10.1111/j.1399-0004.2010.01450.x
Seritrakul, P., and Gross, J. M. (2019). Genetic and epigenetic control of retinal development in zebrafish. Curr. Opin. Neurobiol. 59, 120–127. doi: 10.1016/j.conb.2019.05.008
Shen, Y. C., and Raymond, P. A. (2004). Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev. Biol. 269, 237–251. doi: 10.1016/j.ydbio.2004.01.037
Silva, R. S., Arno, G., Cipriani, V., Pontikos, N., Defoort-Dhellemmes, S., Kalhoro, A., et al. (2019). Unique noncoding variants upstream of PRDM13 are associated with a spectrum of developmental retinal dystrophies including progressive bifocal chorioretinal atrophy. Hum. Mutat. 40, 578–587. doi: 10.1002/humu.23715
Sjöberg, M., and Vennström, B. (1995). Ligand-dependent and-independent transactivation by thyroid hormone receptor beta 2 is determined by the structure of the hormone response element. Mol. Cell. Biol. 15, 4718–4726. doi: 10.1128/MCB.15.9.4718
Sjöberg, M., Vennström, B., and Forrest, D. (1992). Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for alpha and N-terminal variant beta receptors. Development 114, 39–47. doi: 10.1242/dev.114.1.39
Small, K. W., DeLuca, A. P., Whitmore, S. S., Rosenberg, T., Silva-Garcia, R., Udar, N., et al. (2016). North Carolina macular dystrophy is caused by dysregulation of the retinal transcription factor PRDM13. Ophthalmology 123, 9–18. doi: 10.1016/j.ophtha.2015.10.006
Small, K. W., Tran, E. M., Small, L., Rao, R. C., and Shaya, F. (2019a). Multimodal imaging and functional testing in a North Carolina macular disease family: toxoplasmosis, fovea Plana, and torpedo maculopathy are Phenocopies. Ophthalmol Retina 3, 607–614. doi: 10.1016/j.oret.2019.03.002
Small, K. W., Vincent, A. L., Knapper, C. L., and Shaya, F. S. (2019b). Congenital toxoplasmosis as one phenocopy of North Carolina macular dystrophy (NCMD/MCDR1). Am J Ophthalmol Case Rep 15:100521. doi: 10.1016/j.ajoc.2019.100521
Small, K. W., van de Sompele, S., Nuytemans, K., Vincent, A., Yuregir, O. O., Ciloglu, E., et al. (2021). A novel duplication involving PRDM13 in a Turkish family supports its role in North Carolina macular dystrophy (NCMD/MCDR1). Mol. Vis. 27, 518–527.
Sohocki, M. M., Daiger, S. P., Bowne, S. J., Rodriquez, J. A., Northrup, H., Heckenlively, J. R., et al. (2001). Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mutat. 17, 42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K
Stenkamp, D. L. (2015). Development of the vertebrate eye and retina. Prog. Mol. Biol. Transl. Sci. 134, 397–414. doi: 10.1016/bs.pmbts.2015.06.006
Stenson, P. D., Mort, M., Ball, E. V., Shaw, K., Phillips, A. D., and Cooper, D. N. (2014). The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9. doi: 10.1007/s00439-013-1358-4
Stevens, C. B., Cameron, D. A., and Stenkamp, D. L. (2011). Plasticity of photoreceptor-generating retinal progenitors revealed by prolonged retinoic acid exposure. BMC Dev. Biol. 11:51.
Sugiyama, S., di Nardo, A. A., Aizawa, S., Matsuo, I., Volovitch, M., Prochiantz, A., et al. (2008). Experience-dependent transfer of Otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell(Cambridge,Mass) 134, 508–520. doi: 10.1016/j.cell.2008.05.054
Sun, C., Galicia, C., and Stenkamp, D. L. (2018). Transcripts within rod photoreceptors of the zebrafish retina. BMC Genomics 19:127.
Suzuki, S. C., Bleckert, A., Williams, P. R., Takechi, M., Kawamura, S., and Wong, R. O. L. (2013). Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. U. S. A. 110, 15109–15114. doi: 10.1073/pnas.1303551110
Swain, P. K., Hicks, D., Mears, A. J., Apel, I. J., Smith, J. E., John, S. K., et al. (2001). Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J. Biol. Chem. 276, 36824–36830. doi: 10.1074/jbc.M105855200
Swaroop, A., Kim, D., and Forrest, D. (2010). Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11:563. doi: 10.1371/journal.pgen.1002649
Swaroop, A., Wang, Q. L., Wu, W., Cook, J., Coats, C., Xu, S., et al. (1999). Leber congenital Amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum. Mol. Genet. 8, 299–305. doi: 10.1093/hmg/8.2.299
Swaroop, A., Xu, J. Z., Pawar, H., Jackson, A., Skolnick, C., and Agarwal, N. (1992). A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl. Acad. Sci. U. S. A. 89, 266–270. doi: 10.1073/pnas.89.1.266
Szopa, M., Ludwig-Galezowska, A. H., Radkowski, P., Skupien, J., Machlowska, J., Klupa, T., et al. (2016). A family with the Arg103Pro mutation in the NEUROD1 gene detected by next-generation sequencing - clinical characteristics of mutation carriers. Eur. J. Med. Genet. 59, 75–79. doi: 10.1016/j.ejmg.2016.01.002
Tajima, T., Ohtake, A., Hoshino, M., Amemiya, S., Sasaki, N., Ishizu, K., et al. (2009). OTX2 loss of function mutation causes anophthalmia and combined pituitary hormone deficiency with a small anterior and ectopic posterior pituitary. J. Clin. Endocrinol. Metab. 94, 314–319. doi: 10.1210/jc.2008-1219
Tan, M. H. E., Zhou, X. E., Soon, F. F., Li, X., Li, J., Yong, E. L., et al. (2013). The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS One 8:e74359. doi: 10.1371/journal.pone.0074359
Tang, Y., Wu, Q., Gao, M., Ryu, E., Pei, Z., Kissinger, S. T., et al. (2021). Restoration of visual function and cortical connectivity after ischemic injury through NeuroD1-mediated gene therapy. Front. Cell Dev. Biol. 9:9. doi: 10.3389/fcell.2021.720078
Terrell, D., Xie, B., Workman, M., Mahato, S., Zelhof, A., Gebelein, B., et al. (2012). OTX2 and CRX rescue overlapping and photoreceptor-specific functions in the drosophila eye. Dev. Dyn. 241, 215–228. doi: 10.1002/dvdy.22782
Torero Ibad, R., Mazhar, B., Vincent, C., Bernard, C., Dégardin, J., Simonutti, M., et al. (2020). OTX2 non-cell autonomous activity regulates inner retinal function. eNeuro 7:ENEURO.0012. doi: 10.1523/ENEURO.0012-19.2020
Tran, N. M., and Chen, S. (2014). Mechanisms of blindness: animal models provide insight into distinct CRX-associated retinopathies. Dev. Dynam. 243, 1153–1166. doi: 10.1002/dvdy.24151
Tran, N. M., Zhang, A., Zhang, X., Huecker, J. B., Hennig, A. K., and Chen, S. (2014). Mechanistically distinct mouse models for CRX-associated retinopathy. PLoS Genet. 10, –e1004111. doi: 10.1371/journal.pgen.1004111
Trimarchi, J. M., Stadler, M. B., and Cepko, C. L. (2008). Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One 3:e1588. doi: 10.1371/journal.pone.0001588
Tsang, S. H., and Sharma, T. (2018). Enhanced S-cone syndrome (Goldmann-Favre syndrome). Adv. Exp. Med. Biol. 1085, 153–156. doi: 10.1007/978-3-319-95046-4_28
Ueno, S., Kondo, M., Miyata, K., Hirai, T., Miyata, T., Usukura, J., et al. (2005). Physiological function of S-cone system is not enhanced in rd7 mice. Exp. Eye Res. 81, 751–758. doi: 10.1016/j.exer.2005.04.013
Vale, C., Neves, J. S., von Hafe, M., Borges-Canha, M., and Leite-Moreira, A. (2019). The role of thyroid hormones in heart failure. Cardiovasc. Drugs Ther. 33, 179–188. doi: 10.1007/s10557-019-06870-4
van de Sompele, S., Smith, C., Karali, M., Corton, M., van Schil, K., Peelman, F., et al. (2019). Biallelic sequence and structural variants in RAX2 are a novel cause for autosomal recessive inherited retinal disease. Genet. Med. 21, 1319–1329. doi: 10.1038/s41436-018-0345-5
Vincent, A., Robson, A. G., and Holder, G. E. (2013). Pathognomonic (diagnostic) ERGs. Retina 33, 5–12. doi: 10.1097/IAE.0b013e31827e2306
Vincent, A., Forster, N., Maynes, J. T., Paton, T. A., Billingsley, G., Roslin, N. M., et al. (2014). OTX2 mutations cause autosomal dominant pattern dystrophy of the retinal pigment epithelium. J. Med. Genet. 51, 797–805. doi: 10.1136/jmedgenet-2014-102620
Volkov, L. I., Kim-Han, J. S., Saunders, L. M., Poria, D., Hughes, A. E. O., Kefalov, V. J., et al. (2020). Thyroid hormone receptors mediate two distinct mechanisms of long-wavelength vision. Proc. Natl. Acad. Sci. 117, 15262–15269. doi: 10.1073/pnas.1920086117
Volonté, Y. A., Ayala-Peña, V. B., Vallese-Maurizi, H., Garelli, A., Rotstein, N. P., Politi, L. E., et al. (2021). Retinoid X receptor activation promotes photoreceptor survival and modulates the inflammatory response in a mouse model of retinitis pigmentosa. Biochim. Biophys. Acta, Mol. Cell Res. 1868:119098. doi: 10.1016/j.bbamcr.2021.119098
Wang, J. C., and Harris, W. A. (2005). The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev. Biol. 285, 101–115.
Wang, F., Li, H., Xu, M., Li, H., Zhao, L., Yang, L., et al. (2014). A homozygous missense mutation in NEUROD1 is associated with nonsyndromic autosomal recessive retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 56, 150–155. doi: 10.1167/iovs.14-15382
Wang, H., Chu, W., Hemphill, C., Hasstedt, S. J., and Elbein, S. C. (2002). Mutation screening and association of human retinoid X receptor gamma variation with lipid levels in familial type 2 diabetes. Mol. Genet. Metab. 76, 14–22. doi: 10.1016/S1096-7192(02)00016-1
Wang, Q. L., Chen, S., Esumi, N., Swain, P. K., Haines, H. S., Peng, G., et al. (2004). QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum. Mol. Genet. 13, 1025–1040. doi: 10.1093/hmg/ddh117
Wang, N. K., Fine, H. F., Chang, S., Chou, C. L., Cella, W., Tosi, J., et al. (2009). Cellular origin of fundus autofluorescence in patients and mice with a defective NR2E3 gene. Br. J. Ophthalmol. 93, 1234–1240. doi: 10.1136/bjo.2008.153577
Wang, X., Gong, K., Li, H., Wang, C., Qu, C., and Li, H. (2017). Gene expression profiling of the optic nerve head of patients with primary open-angle glaucoma. J. Ophthalmol. 2017, 1–7. doi: 10.1155/2017/6896390
Watanabe, S., Sanuki, R., Sugita, Y., Imai, W., Yamazaki, R., Kozuka, T., et al. (2015). Prdm13 regulates subtype specification of retinal amacrine interneurons and modulates visual sensitivity. J. Neurosci. 35, 8004–8020. doi: 10.1523/JNEUROSCI.0089-15.2015
Weiss, A. H., Kelly, J. P., Bisset, D., and Deeb, S. S. (2012). Reduced L- and M- and increased S-cone functions in an infant with thyroid hormone resistance due to mutations in the THRβ2 gene. Ophthalmic Genet. 33, 187–195. doi: 10.3109/13816810.2012.681096
Wilken, M. S., Brzezinski, J. A., la Torre, A., Siebenthall, K., Thurman, R., Sabo, P., et al. (2015). DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin 8:8. doi: 10.1186/1756-8935-8-8
Wright, A. F., Reddick, A. C., Schwartz, S. B., Ferguson, J. S., Aleman, T. S., Kellner, U., et al. (2004a). Mutation analysis of NR2E3 and NRL genes in enhanced S cone syndrome. Hum. Mutat. 24:439. doi: 10.1002/humu.9285
Wright, A. F., Jacobson, S. G., Cideciyan, A. V., Roman, A. J., Shu, X., Vlachantoni, D., et al. (2004b). Lifespan and mitochondrial control of neurodegeneration. Nat. Genet. 36, 1153–1158. doi: 10.1038/ng1448
Wu, H. Y., Perron, M., and Hollemann, T. (2009). The role of Xenopus Rx-L in photoreceptor cell determination. Dev. Biol. 327, 352–365. doi: 10.1016/j.ydbio.2008.12.017
Wu, Z., Parry, M., Hou, X. Y., Liu, M. H., Wang, H., Cain, R., et al. (2020). Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of Huntington’s disease. Nat. Commun. 11:1105. doi: 10.1038/s41467-020-14855-3
Wyatt, A., Bakrania, P., Bunyan, D. J., Osborne, R. J., Crolla, J. A., Salt, A., et al. (2008). Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum. Mutat. 29, E278–E283. doi: 10.1002/humu.20869
Xie, S., Han, S., Qu, Z., Liu, F., Li, J., Yu, S., et al. (2019). Knockout of Nr2e3 prevents rod photoreceptor differentiation and leads to selective L−/M-cone photoreceptor degeneration in zebrafish. Biochim. Biophys. Acta Mol. basis Dis. 1865, 1273–1283. doi: 10.1016/j.bbadis.2019.01.022
Xu, M., Gelowani, V., Eblimit, A., Wang, F., Young, M. P., Sawyer, B. L., et al. (2015). ATF6 is mutated in early onset photoreceptor degeneration with macular involvement. Invest. Ophthalmol. Vis. Sci. 56, 3889–3895. doi: 10.1167/iovs.15-16778
Yamamoto, H., Kon, T., Omori, Y., and Furukawa, T. (2020). Functional and evolutionary diversification of Otx2 and Crx in vertebrate retinal photoreceptor and bipolar cell development. Cell Rep. 30, 658–671.e5. doi: 10.1016/j.celrep.2019.12.072
Yan, R. T., and Wang, S. Z. (1998). neuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J. Neurobiol. 36, 485–496.
Yan, R. T., and Wang, S. Z. (2004). Requirement of neuroD for photoreceptor formation in the chick retina. Invest. Ophthalmol. Vis. Sci. 45, 48–58.
Yang, P., Chiang, P. W., Weleber, R. G., and Pennesi, M. E. (2015). Autosomal dominant retinal dystrophy with electronegative waveform associated with a novel RAX2 mutation. JAMA Ophthalmol 133, 653–661. doi: 10.1001/jamaophthalmol.2015.0357
Yu, W., and Wu, Z. (2018). In vivo applications of CRISPR-based genome editing in the retina. Front. Cell Dev. Biol. 6:53. doi: 10.3389/fcell.2018.00053
Zhang, Z., Burch, P. E., Cooney, A. J., Lanz, R. B., Pereira, F. A., Wu, J., et al. (2004). Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 14, 580–590. doi: 10.1101/gr.2160004
Keywords: photoreceptor development, transcription factor, mutation, inherited retinal disease, pathogenic mechanism
Citation: Sun C and Chen S (2023) Disease-causing mutations in genes encoding transcription factors critical for photoreceptor development. Front. Mol. Neurosci. 16:1134839. doi: 10.3389/fnmol.2023.1134839
Edited by:
Frans Vinberg, The University of Utah, United StatesReviewed by:
Susana Da Silva, University of Pittsburgh, United StatesJonathan Lin, Stanford University, United States
Copyright © 2023 Sun and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chi Sun, c3VuY2hpQHd1c3RsLmVkdQ==
 Chi Sun
Chi Sun Shiming Chen
Shiming Chen