- 1Department of Neurology, Xiangya Hospital, Central South University, Changsha, China
- 2National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
- 3Clinical Research Center for Epileptic Disease of Hunan Province, Central South University, Changsha, China
- 4Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, China
- 5Department of Emergency, Xiangya Hospital, Central South University, Changsha, China
Background: An increasing number of observational studies have revealed an association among the gut microbiota, gut metabolites, and epilepsy. However, this association is easily influenced by confounders such as diet, and the causality of this association remains obscure.
Methods: Aiming to explore the causal relationship and ascertain specific gut microbe taxa for epilepsy, we conducted a bi-directional Mendelian randomization (MR) study based on the genome-wide association study (GWAS) data of epilepsy from the International League Against Epilepsy, with the gut microbiota GWAS results from MiBioGen, and summary-level GWAS data of gut microbiota-dependent metabolites trimethylamine N-oxide and its predecessors.
Results: Nine phyla, 15 classes, 19 orders, 30 families, and 96 genera were analyzed. A suggestive association of host-genetic-driven increase in family Veillonellaceae with a higher risk of childhood absence epilepsy (odds ratio [OR]: 1.033, confidential interval [CI]: 1.015–1.051, PIVW = 0.0003), class Melainabacteria with a lower risk of generalized epilepsy with tonic-clonic seizures (OR = 0.986, CI = 0.979–0.994, PIVW = 0.0002), class Betaproteobacteria (OR = 0.958, CI = 0.937–0.979, PIVW = 0.0001), and order Burkholderiales (OR = 0.960, CI = 0.937–0.984, PIVW = 0.0010) with a lower risk of juvenile myoclonic epilepsy were identified after multiple-testing correction. Our sensitivity analysis revealed no evidence of pleiotropy, reverse causality, weak instrument bias, or heterogeneity.
Conclusion: This is the first MR analysis to explore the potential causal relationship among the gut microbiota, metabolites, and epilepsy. Four gut microbiota features (two class levels, one order level, and one family level) were identified as potential interventional targets for patients with childhood absence epilepsy, generalized epilepsy with tonic-clonic seizures, and juvenile myoclonic epilepsy. Previous associations in numerous observational studies may had been interfered by confounders. More rigorous studies were needed to ascertain the relationship among the gut microbiota, metabolites, and epilepsy.
Introduction
Epilepsy is a common neurological disease characterized by recurrent unprovoked seizures, besetting over 50 million patients worldwide, with an incidence rate of 50–70 per 100,000 people (Deuschl et al., 2020). With the main manifestation of epileptic seizures and abnormal electroencephalogram results, epilepsy is a disease with strong heterogeneity, which can be further classified into several subtypes such as focal epilepsy and generalized epilepsy. The etiology of epilepsy is also complex as it involves genetic, structural, infectious, metabolic, immune, and unknown factors (Beghi, 2020).
Recently, the mutual interaction between the gut microbiota and the human body is revealed, and the gut microbiota is considered as a promising therapeutic target through probiotic supplements and fecal microbiota transplantation (FMT) (Gomaa, 2020). Among these microbiome-to-host interactions, the central nervous system (CNS) and gut microbiota communication, termed the microbiota–gut–brain (MGB) axis, is a research hotspot, and its interaction routes are related to metabolites, immune responses, and the enteric nervous system (Cryan et al., 2019). The potential functions of the MGB axis in psychiatric and CNS disorders have been previously summarized (Iannone et al., 2019; Socała et al., 2021). Epilepsy, one of the most common diseases of the CNS, is also related to the gut microbiota and metabolites through the MGB axis at both laboratory and clinical levels (Holmes et al., 2020; Ding et al., 2021). For example, a distinct gut microbiome profile has been detected in patients with epilepsy, especially in those with anti-seizure medication resistance (Peng et al., 2018; Şafak et al., 2020). Furthermore, high-fat ketogenic diet (KD) is recommended for patients with epilepsy, diet-related gut microbiome alterations are observed with an anti-seizure effect (Olson et al., 2018; Ang et al., 2020). Therefore, the gut microbiota is a promising biomarker and therapeutic target for epilepsy (De Caro et al., 2019; Arulsamy and Shaikh, 2022; Russo, 2022). However, the causality among the gut microbiota, metabolites, and epilepsy remains unclear and requires more direct evidence.
Randomized controlled trials (RCTs) are the gold standard for exploring causal relationships. However, an RCT is not only costly but also difficult for investigating the gut microbiome and neurological disorders because of potential confounders. Meanwhile, the Mendelian randomization (MR) study is an alternative tool for exploring the causal relationship between exposure and outcome, utilizing single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) (Smith and Ebrahim, 2003). An MR study has several types such as two-sample MR, two-step MR, and bi-directional MR. The two-sample MR refers to MR based on the exposure and outcome using a genome-wide association study (GWAS) dataset without overlap. The bi-directional MR is utilized to test the effect of exposure on the outcome, and the outcome on exposure by retrieving different IVs from exposure or outcome datasets to ascertain the robustness of direction. With increasing publicly available GWAS data, an MR study is more feasible for conducting epidemiological research. Moreover, abundant GWAS data in the gut microbiome, gut-microbiome metabolites, and epilepsy have been reported recently, thus providing the research foundation of our MR analysis.
In this study, we conducted the first bi-directional MR analysis to examine the causal relationship among the gut microbiome, metabolites, and epilepsy following the “STROBE-MR” guidelines (Skrivankova et al., 2021a,b). The summary statistics of epilepsy, gut microbiota, and metabolites are derived from the International League Against Epilepsy (ILAE) consortium and large-scale GWAS data. This research not only improves our understanding of the mutual interaction among the gut microbiota, metabolites, and epilepsy but also reveals the research direction.
Methods
Study design and data sources
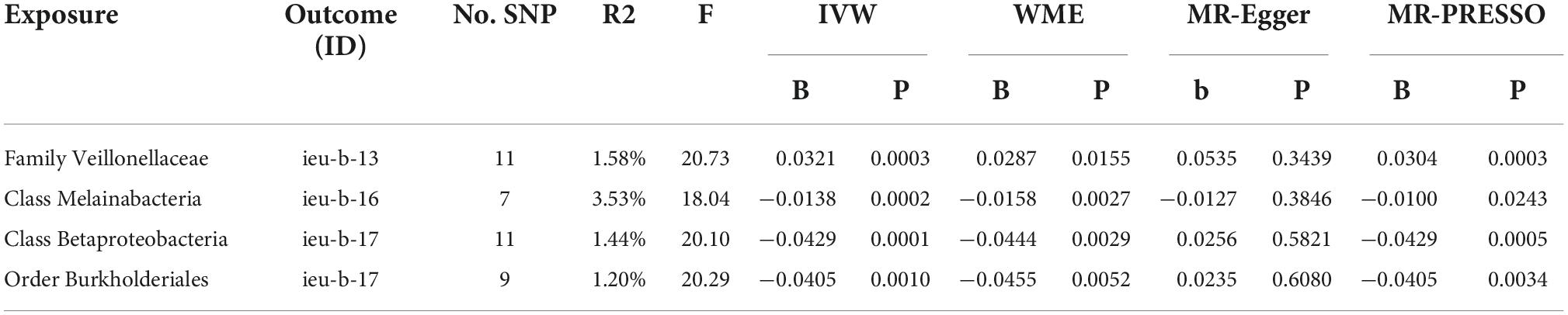
The study flowchart is presented in Figure 1A. We conducted a bidirectional MR study to investigate the causal relationship among the gut microbiota, metabolites, and epilepsy. First, genetic variants from previous GWAS summary-level data are retrieved and used as IVs. Then, a two-sample MR is conducted using the R software (4.1.3) following the guideline of the R package ‘‘two-sample MR’’ (0.5.6)1 including three MR methods. Several sensitivity analyses, such as the pleiotropy test, heterogeneity test, and leave-one-out analysis, are performed sequentially. Finally, we adopted a reverse MR method to explore whether a bidirectional relationship exists among epilepsy, the gut microbiota, and metabolites.
Figure 1. Study design and Mendelian randomization core assumption. (A) Data resource and study design of our bi-directional MR. (B) Three assumptions in the Mendelian randomization study.
Genome-wide association study (GWAS) statistics summary-level data of the gut microbiome have been generated from the largest genome-wide meta-analysis to date, the MiBioGen study (Kurilshikov et al., 2021). This MiBioGen consortium curated and analyzed 16S faucal microbiome data and genome-wide genotypes from 24 cohorts (18,340 individuals) and identified 31 loci affecting the gut microbiome with genome-wide significance (P < 5 × 10–8). As for gut metabolite data, we leveraged GWAS summary statistics of the human metabolome in a community-based cohort containing 2076 participants (Rhee et al., 2013). GWAS summary statistics for epilepsy are retrieved from the OpenGWAS database API2 (Elsworth et al., 2020). The GWAS data in this database included epilepsy (GWAS ID: ieu-b-8), genetic generalized epilepsy (ieu-b-9), focal epilepsy (ieu-b-10), focal epilepsy-documented lesion negative (ieu-b-11), juvenile absence epilepsy (ieu-b-12), childhood absence epilepsy (ieu-b-13), focal epilepsy-documented hippocampal sclerosis (ieu-b-14), focal epilepsy-documented lesions other than hippocampal sclerosis (ieu-b-15), generalized epilepsy with tonic-clonic seizures (ieu-b-16), and juvenile myoclonic epilepsy (ieu-b-17) based on the ILAE classification position paper on epilepsies (Scheffer et al., 2017; Ilae Consortium, 2018). Detailed information on the dataset used, without overlap between the exposure and outcome data, is summarized in Table 1. Additional information, including demographic characteristics, eligibility criteria, and ethics approval can be found in the original article (Ilae Consortium, 2018).
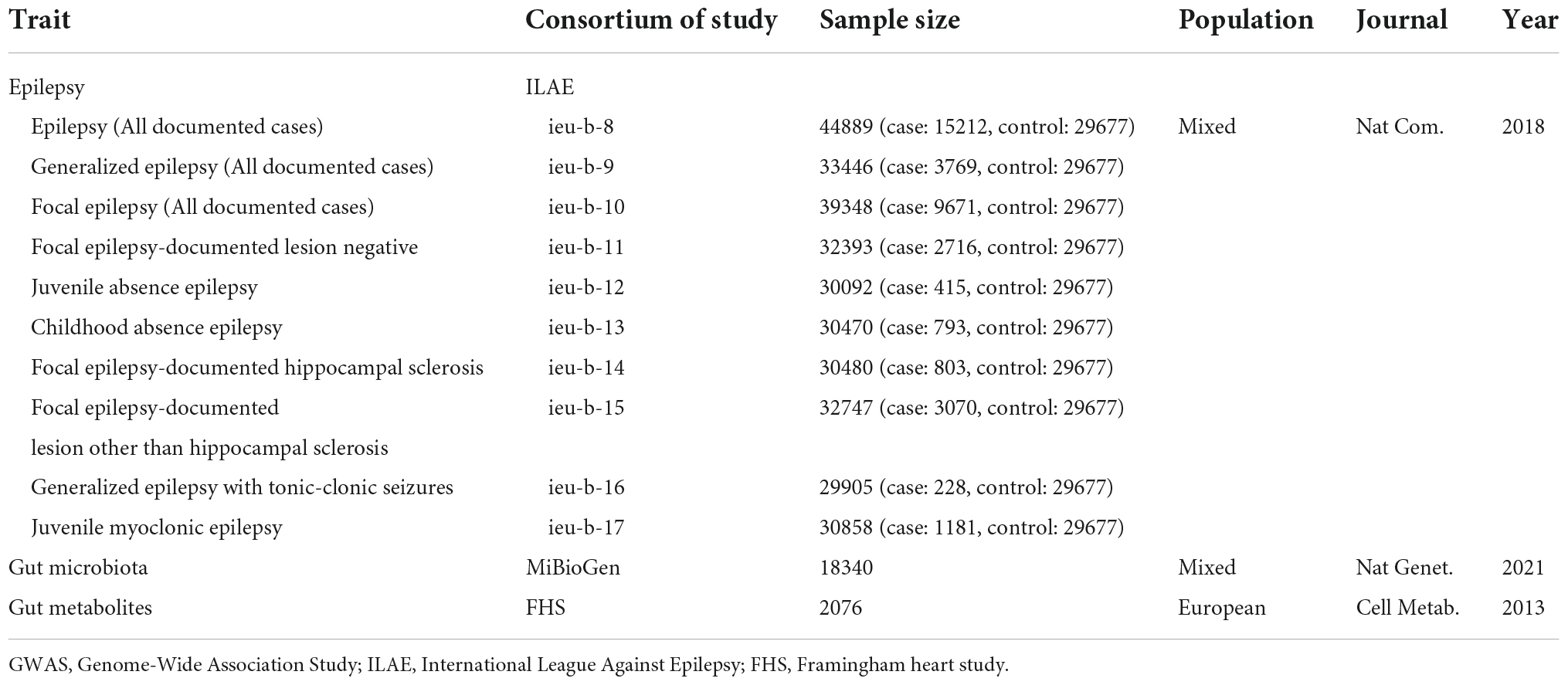
Table 1. Characteristics of included GWAS summary-level data of epilepsy, gut microbiota, and gut metabolites.
Instrumental variables selection
Bacterial taxa were classified and analyzed at six levels (phylum, class, order, family, genus, and species) based on SNPs available in the gut microbiome GWAS summary data. Candidate IVs were identified at a significance level of P < 1.0 × 10–5 according to the previously published studies (Sanna et al., 2019; Ni et al., 2021). The parameters of the clump function in the R package were set at r2< 0.1 and kb = 500 kb, guaranteeing the independence of each IV, which was the same as in a previous publication, to minimize the impact of linkage disequilibrium violating the randomized allele allocation (Ni et al., 2021). Furthermore, as our significance level was set at P < 1.0 × 10–5, the F statistic was used to exclude the weak instrument bias violating the first assumption of MR (Burgess and Thompson, 2011). Lastly, the palindromic SNPs were also excluded from the MR. The IVs adopted in this study are listed in Supplementary Table 1. Similarly, IVs of gut metabolites were extracted under a suggestive significance level of P < 5.0 × 10–5 (Zhuang et al., 2021). The parameters of the clump function in the R package were set at r2< 0.2 and kb = 10,000 kb (Zhuang et al., 2021).
For the outcome data, we collected epilepsy GWAS data from the MR database (see text footnote 1). The gut microbiome, metabolites, and epilepsy data were harmonized for subsequent MR. As the exposure and outcome GWAS datasets were large-scale GWAS research, the threshold of minor allele frequency was set at 0.01.
Mendelian randomization study
As presented in Figure 1B, this MR was conducted following the MR model with selected IVs in the previous step, conforming to three assumptions as follows: (1) SNPs were robustly associated with the gut microbiome/metabolites; (2) SNPs were not associated with confounders; (3) SNPs do not affect the epilepsy outcomes except through the potential effects of the gut microbiome or metabolites.
A two-sample MR analysis was conducted using three primary methods: inverse variance weighted median (IVW), weighted median, and MR Egger to evaluate the causal relationship among the gut microbiome, metabolites, and epilepsy (Bowden et al., 2015, 2016; Burgess et al., 2015). The MR analysis was conducted using two-sample MR packages according to the developers’ guidelines. IVW was a classic method based on the meta-analysis of each SNPs Wald ratio, while the weighted median mode calculated the median effects of SNPs. The MR Egger analysis, albeit with lower statistical power than IVW, can be applied in the presence of horizontal pleiotropy. Additionally, we conducted an MR-PRESSO test using the MR-PRESSO R package (1.0) to evaluate whether a horizontal pleiotropy effect violates the assumption of MR (Verbanck et al., 2018). Based on the MR-PRESSO Global test for overall horizontal pleiotropy and outlier test for each SNP pleiotropy significance evaluation, outlier SNPs were removed until the P-value of the global test remained >0.05. Moreover, the multiple-testing significance threshold at each level (phylum, class, order, family, and genus) was set as 0.05/n, where n was the effective number of independent bacterial taxa at each taxonomic level. However, owing to the sample size and restricted power of the gut microbiota GWAS data, IVs at the species level were insufficient for MR analysis. Therefore, we conducted MR at the phylum, class, order, family, and genus levels.
Sensitivity analysis
The MR-Egger regression and MR-PRESSO tests were conducted to exclude potential pleiotropy. The Q test in the IVW test was performed to evaluate the heterogeneity of results. The leave-one-out analysis excluded SNPs individually and recomputed the effect to test the robustness of the results. The MR Steiger directionality test was adopted to explore the robustness of the causality direction.
Reverse Mendelian randomization analysis
A reverse MR analysis was conducted to explore the reverse causality from epilepsy (as exposures) to gut microbiota and metabolites (as outcomes). The procedure was the same as the abovementioned protocol for the two-sample MR.
This bidirectional MR and sensitivity analysis adhered to the guidelines of the two-sample MR and MR-PRESSO packages.
Results
Two-sample Mendelian randomization of gut microbiota (exposure) on epilepsy (outcome)
Twelve exposure and outcome datasets were acquired for MR analysis with detailed information such as the consortium, sample size, and population (Table 1). Under a suggestive significance level of P < 1 × 10–5, the significant SNPs were selected from the GWAS summary data of gut microbiota in the nine phyla, 16 classes, 20 orders, 35 families, and 96 genera. After clumping and harmonization, an MR analysis was conducted between each pair of exposure and outcome to explore causality. The significant threshold for each level was corrected based on multiple testing as follows: phylum P = 5.56 × 10–3 (0.05/9); class P = 3.13 × 10–3 (0.05/16); order P = 2.50 × 10–3 (0.05/20); family P = 1.43 × 10–3 (0.05/35); genus P = 5.21 × 10–4 (0.05/96); and gut metabolites P = 1.25 × 10–2 (0.05/4).
After data preprocessing, an MR analysis was performed for each pair of exposure (gut microbiota) and outcome (epilepsy) based on three MR methods (IVW, weighted median, and MR Egger). Under the corrected significant threshold, four correlations between gut microbiome features and epilepsy were identified using the IVW method (Table 2) as follows: family Veillonellaceae with a higher risk of childhood absence epilepsy (PIVW = 3.00 × 10–4), class Melainabacteria with a lower risk of generalized epilepsy with tonic-clonic seizures (PIVW = 2.00 × 10–4), and class Betaproteobacteria (PIVW = 1.18 × 10–4) and order Burkholderiales (PIVW = 1.03 × 10–3) with a lower risk of juvenile myoclonic epilepsy. These correlations are depicted in scatter and forest plots in Figures 2, 3, respectively. More information on single SNP is summarized in Supplementary Table 1.
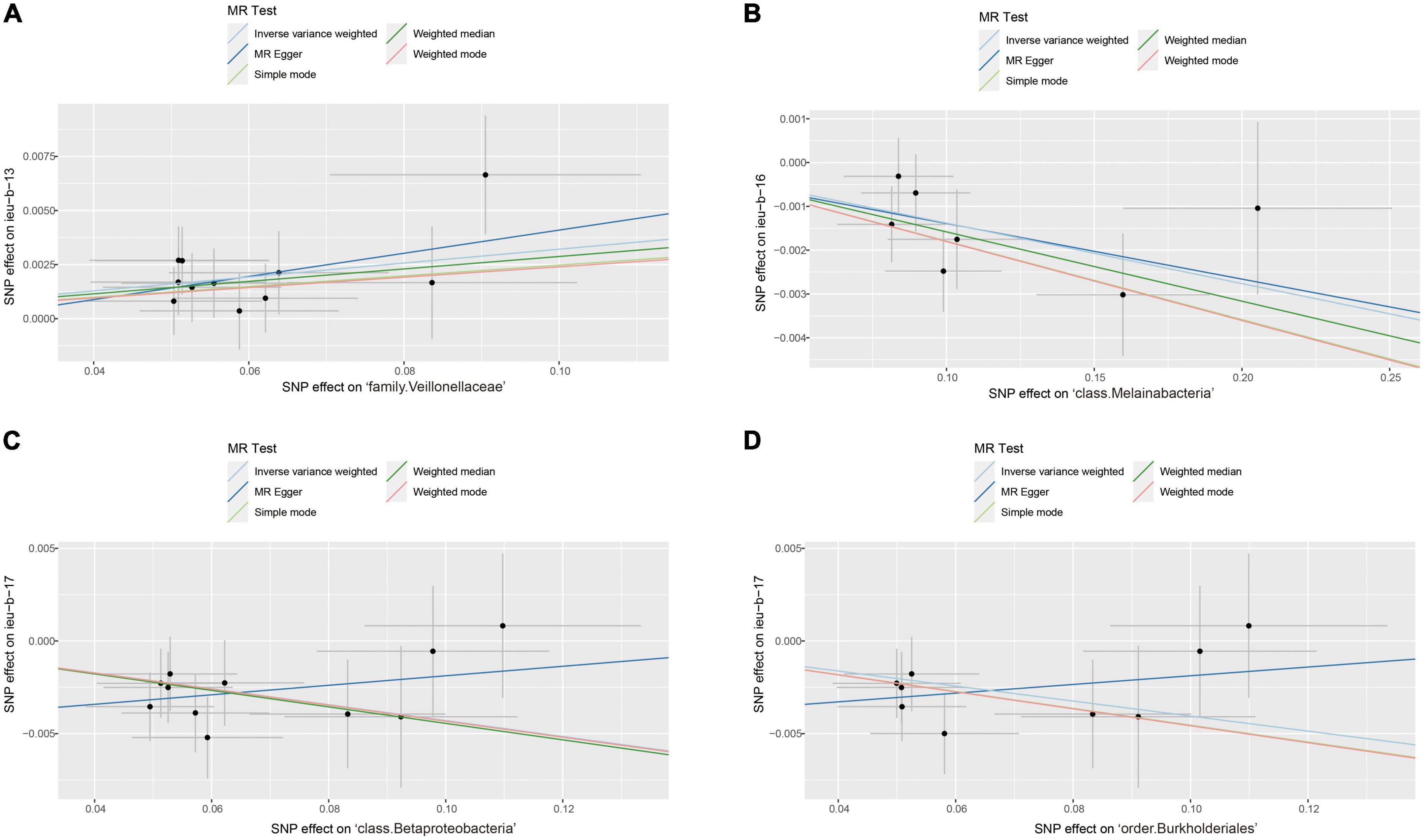
Figure 2. Scatter plots of significant causality of the gut microbiota and epilepsy. (A) Scatter plots of the family Veillonellaceae on childhood absence epilepsy. (B) Scatter plots of the class Melainabacteria on generalized epilepsy with tonic-clonic seizures. (C) Scatter plots of the class Betaproteobacteria and (D) The order Burkholderiales on juvenile myoclonic epilepsy. The lines move obliquely upward from left to right exhibiting a positive correlation between the gut microbiota and epilepsy with horizontal and vertical lines indicating the 95% confidence interval of each association. The lines with a negative correlation are inclined downward from left to right, indicating a protective effect of the gut microbiota on epilepsy.
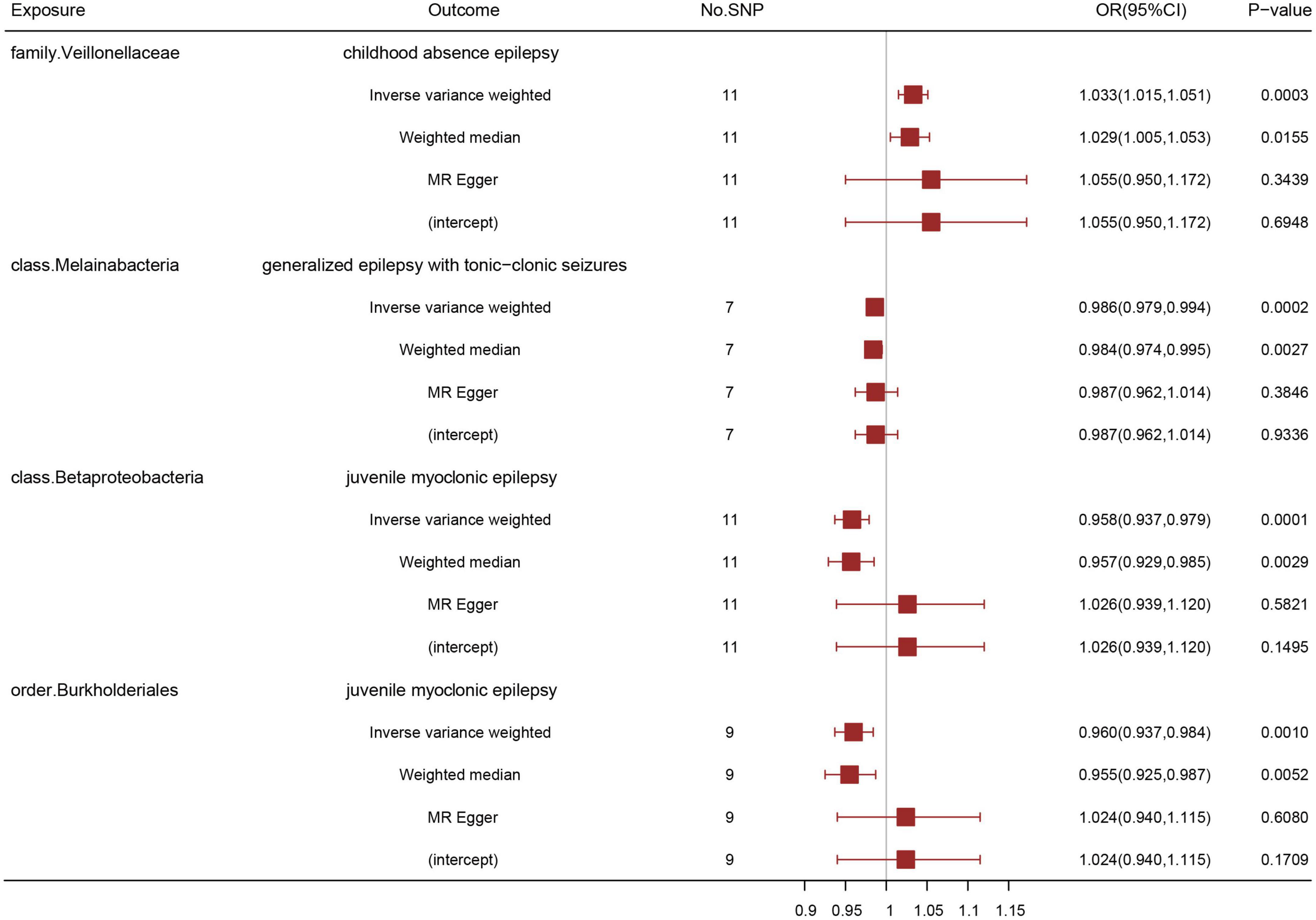
Figure 3. Association of genetically predicted gut microbiota with epilepsy by three different MR methods: inverse variance weighted, weighted median, and MR Egger. A positive correlation between the family Veillonellaceae and childhood absence epilepsy, negative correlations between the class Melainabacteria and generalized epilepsy with tonic-clonic seizures, between the class Betaproteobacteria and juvenile myoclonic epilepsy, and between the order Burkholderials and juvenile myoclonic epilepsy were suggested. OR: odds ratio; CI: confidential interval.
In the heterogeneity (IVW test and MR-Egger regression), pleiotropy (MR-PRESSO test and MR-Egger regression test) and weak instrument bias tests (F statistic), no evidence of heterogeneity, pleiotropy, or weak instrument bias was noted. Additional details are summarized in Supplementary Table 2. Furthermore, the MR Steiger directionality test revealed a robust direction from the gut microbiota to epilepsy in all results. The leave-one-out sensitivity analysis illustrated the robustness of our results, as no single SNP drives causal association (Supplementary Figure 1). Funnel plots of these four significant results excluded a potential bias (Supplementary Figure 2).
Reverse Mendelian randomization analysis of epilepsy (exposure) on gut microbiota (outcome)
With a significant P-value set at 1 × 10–5, IVs were extracted from significant epilepsy GWAS datasets in previous MR analysis of gut microbiota on epilepsy; however, no significant results were identified in this reverse MR analysis. The reverse analysis indicated the absence of causality from epilepsy to the gut microbiota, which was in accordance with our MR steiger results.
Bi-directional Mendelian randomization analysis Mendelian randomization of gut metabolites and neurological disorders
In the gut metabolite and epilepsy MR analysis, the SNPs concerning carnitine, choline, trimethylamine N-oxide (TMAO), and betaine were extracted from the GWAS summary data. With a genome-wide significance level set at P < 5 × 10–5, which was the same as the parameters in previous publications, SNPs were extracted for subsequent MR analysis after clumping (r2 = 0.2, kb = 10,000) and harmonization (Zhuang et al., 2021). Similar to the gut microbiota and epilepsy MR analysis, three MR methods (IVW, weighted median, and MR-Egger) were adopted to explore the potential causality with multiple sensitivity analyses. No significant causality between gut metabolites and epilepsy was found. For reverse MR analysis, IVs extracted from epilepsy under significance levels were not identified in the gut metabolite GWAS dataset.
All results on phylum, class, order, family, genus, or metabolite levels are summarized in Supplementary Tables 3–8.
Discussion
To the best of our knowledge, this is the first MR study to reveal the potential causal relationship among the gut microbiome, metabolites, and epilepsy based on large GWAS summary-level data. The potential contributory or protective effect of the family Veillonellaceae on childhood absence epilepsy, class Melainabacteria on generalized epilepsy with tonic-clonic seizures, class Betaproteobacteria, and order Burkholderials on juvenile myoclonic epilepsy are demonstrated in this MR analysis.
The association between the gut microbiota and epilepsy has been summarized in a systematic review published in 2020 with an increasing number of studies published over the past two years; however, the gut microbiota alteration is inconsistent in all the studies (Arulsamy et al., 2020). Some studies have revealed that the richness of numerous gut microbiomes, such as Proteobacteria and Fusobacterial phyla, as possible biomarkers for epilepsy diagnosis increased in patients with epilepsy (Şafak et al., 2020; Dong et al., 2022). Regarding the gut microbiota characteristics of patients with drug-resistant epilepsy (DRE), the Firmicutes phylum is commonly increased in DRE, and bifidobacteria exhibits a potential protective function against epilepsy (Peng et al., 2018; Lee et al., 2020, 2021). In addition to the altered gut microbiota composition of patients with epilepsy, KD therapy, commonly recommended for patients with DRE, also exerts a therapeutic effect due to its influence on the gut microbiota (Fan et al., 2019; Tang et al., 2021). Decreased Proteobacteria and Firmicutes and increased Bacteroidetes are commonly observed after KD therapy (Xie et al., 2017; Zhang et al., 2018; Gong et al., 2021). In addition to KD therapy, FMT is also a promising strategy for epilepsy, and a case report has demonstrated that epilepsy in a girl with Crohn’s disease was cured after FMT (He et al., 2017). Therefore, we summarized the diverse gut microbiota changes to directly exhibit these observational results of the gut microbiota and epilepsy in Supplementary Table 9.
The abovementioned studies have confirmed the close relationship between the gut microbiota and epilepsy, and KD therapy may alter the bacteria to further control seizures. However, a direct causal relationship is unclear and may be affected by confounding factors. Intriguingly, numerous correlations summarized in Supplementary Table 9 are not identified in our MR analysis after multiple-testing corrections for several reasons. Supplementary Tables 3–8 exhibit that the function of one gut microbiome in epilepsy is diverse in different subtypes of epilepsy. As previous observational studies did not focus on the specific subtype of epilepsy, the association identified in our MR analysis may be more specific and robust. Moreover, although confounders such as antibiotic use have been controlled in most studies, previous observational studies of Proteobacteria and epilepsy may be affected by other influencing factors such as diet or age (Sullivan et al., 2001; Greenhalgh et al., 2016; Rinninella et al., 2019).
Family Veillonellaceae, which belongs to the Firmicutes phylum, is a potential risk factor for childhood absence epilepsy. Similar to previous results, the abundance of Firmicutes is increased in patients with DRE (Xie et al., 2017; Peng et al., 2018; Lee et al., 2021). Furthermore, a relatively increased level of Firmicutes in other neurological disorders, such as Parkinson’s disease, has been reported (Bedarf et al., 2017). Although direct evidence between Veillonellaceae and epilepsy is lacking, the family Veillonellaceae is positively related to non-social fear behavior in infants, which requires further elucidation based on a subsequent longitudinal study of endocrine, metabolites, and immune alteration (Carlson et al., 2021). Family Veillonellaceae is also negatively correlated with orientation and delayed recall scores in anamnestic mild cognitive impairment research (Liu et al., 2021). Moreover, Veillonellaceae can influence normal brain functions and is involved in several neurological disorders. From another perspective, Ruminococcus, another microbiome belonging to the Firmicutes phylum, reportedly has a lower level of N-acetyl aspartic acid (NAA) and serotonin (Bedarf et al., 2017; Mudd et al., 2017). However, NAA is important for neuronal health. As serotonin can inhibit the T-type calcium channel with reduced bursting electrical activity, increased Ruminococcus may sensitize epileptiform discharge because of decreased NAA and serotonin (Petersen et al., 2017). Whether Veillonellaceae influences childhood absence epilepsy by modulating neurotransmitter levels, similarly with Ruminococcus, is also worth exploring.
Additionally, our MR analysis identified the class Betaproteobacteria and order Burkholderia, which belong to the Proteobacteria phylum, are potential protective factors of juvenile myoclonic epilepsy. Previous associations between the microbiome and the phylum Proteobacteria have been widely reported, as discussed above and summarized in Supplementary Table 9. Alterations in the Proteobacteria phylum in patients with epilepsy are not consistent. As a common phylum in the human body, Proteobacteria are related to diseases such as obesity (Shin et al., 2015; Greenhalgh et al., 2016). Although Escherichia particularly E. coli, Salmonella, and Vibrio, which belong to the Gammaproteobacteria class, are notorious pathogens, the Proteobacteria phylum should not be considered a risk factor for all neurological diseases (Singh et al., 2015). A more specific association at the class, order, or family level of the phylum Proteobacteria is absent. Unfortunately, as for mechanisms underlying this association, studies that have identified a direct correlation among juvenile myoclonic epilepsy and class Betaproteobacteria and order Burkholderial are lacking. From the MGB axis perspective, the gut microbiota is associated with neurodevelopment, and a recent observational study has reported that gut microbiota such as Bifidobacterium, Bacteroidetes, and Lachnospiraceae can influence neurodevelopment in infants (Oliphant et al., 2021; Beghetti et al., 2022). The neurodevelopmental modulation capability of the gut microbiota may be correlated with the release of neuroactive substances, short-chain fatty acids (SCFAs), and alterations in intestinal or blood-brain barrier integrity alteration (Louis et al., 2010; Hsiao et al., 2013; Sarkar et al., 2016). As the association between juvenile myoclonic epilepsy and neurodevelopment has been discussed previously, the causality of class Betaproteobacteria or order Burkholderial and juvenile myoclonic epilepsy may be relevant to their neurodevelopment modulation capability (Lin et al., 2014). Additionally, the effect of each gut microbiome on epilepsy may not rely on one specific pathway but an integrative model including the immune system, nervous system, neurotransmitters, SCFAs, and the hypothalamic–pituitary–adrenal axis (Ding et al., 2021). Another interesting result of class Melainabacteria in generalized epilepsy with tonic-clonic seizures is that the phylum Melainabacteria has been identified as an accurate biomarker of zinc status (Chen et al., 2021). Because zinc levels are associated with neurodevelopment, Melainabacteria may influence generalized epilepsy by modulating zinc levels. Additionally, Melainabacteria, which interacts with other gut microbiomes and exerts an influence on the human body, is also essential for gut biodiversity. Lastly, after our MR analysis of the gut microbiota, the causality of gut microbiota and the three subtypes of generalized epilepsy, childhood absence epilepsy, juvenile myoclonic epilepsy, and generalized epilepsy with tonic-clonic seizures, are identified without any focal epilepsy. A possible reason for this result is that the gut microbiota releases factors to the systemic circulation with a potential role in triggering an immune response or impacting the permeability of the blood-brain barrier, functioning in the bilateral brain rather than a focal area (Logsdon et al., 2018).
Regarding gut metabolites, TMAO may be generated from L-carnitine, betaine, choline, and other choline-containing compounds that participate in the gut microbiome and hepatic flavin-dependent monooxygenases (Zeisel and Warrier, 2017). The relationship between TMAO and its predecessors with multiple chronic diseases, such as cardiovascular diseases and cancer, has also been previously studied (Cho and Caudill, 2017). The neuroprotective function of dietary choline and the therapeutic potential of carnitine in multiple neurological disorders, including epilepsy, have been reviewed previously (Blusztajn et al., 2017; Maldonado et al., 2020). Moreover, in a kainite-induced temporal lobe epilepsy model, acetyl-L-carnitine exerts its anticonvulsant effect by ameliorating oxidative stress, pyroptosis, and neuroinflammation (Tashakori-Miyanroudi et al., 2022). However, after multiple testing corrections of our MR analysis, the causality of genetically predicted TMAO, choline, carnitine, and betaine on epilepsy is not determined. A previous MR analysis has also revealed no direct causality of TMAO in Alzheimer’s disease (Zhuang et al., 2021). Hence, the previous association between TMAO and multiple neurological disorders may be interfered with by confounders. The role of TMAO in the CNS of the human body requires further investigation.
This MR study has multiple advantages over previous studies. First, this is the first MR to explore the potential causal relationship between the gut microbiota, metabolites, and epilepsy. Second, our MR study is based on the largest GWAS-summary-level data on the gut microbiome from the MiBioGen study. A bidirectional MR analysis is conducted to ensure the robustness of our results. Several microbiomes have been identified as potential therapeutic targets in patients with epilepsy. As the gut microbiota GWAS dataset is based on three ancestries, the generalizability of our results applies to different populations. The non-significant causalities of our results also provide important information that previous observational studies may had been easily interfered with by confounders. Lastly, whether gut metabolites such as TMAO are potential predisposing factors for epilepsy has also been studied. Our MR analysis indicates that specific gut microbiome functions should be considered in a specific subtype of epilepsy, rather than from a general perspective.
Nevertheless, this study has some limitations. First, as SNPs in the MiBioGen study less than 5 × 10–8 are not sufficient for MR analysis, the significance level of gut microbiota IV selection is set at 1 × 10–5 instead of 5 × 10–8, which is the same as that in a previous publication (Ni et al., 2021). However, the F-statistics are guaranteed to be > 10 to exclude potential weak instrument bias and make the statistical results more robust (Burgess and Thompson, 2011). Second, as the epilepsy ILAE GWAS dataset lacks exposure levels and periods, the effect of gut microbiomes and metabolites on epilepsy exposed to different levels or timings must be further ascertained. Third, although previous observational studies mainly focused on the relationship between the gut microbiota and DRE, MR has not been conducted to explore the causality between gut microbiota, metabolites, and DRE due to the lack of GWAS summary-level data on DRE, which is worth researching in future. Fourth, direct mechanistic research to support our findings is still lacking. To obtain more direct evidence of the relationship between the gut microbiota and epilepsy, more research is required to explore the effects of these bacteria on the immune response, blood-brain barrier permeability, neuronal excitability, and brain development. Lastly, the gut microbiota is easily influenced by environmental factors, such as diet. However, horizontal pleiotropy is excluded from our MR analysis based on the following two sensitivity analyses: MR-Egger and MR-PRESSO results.
Collectively, we comprehensively analyzed the potential causal relationship among the gut microbiota, metabolites, and epilepsy. This bi-directional MR study ascertained the predisposing or protective effects of the family Veillonellaceae on childhood absence epilepsy, class Melainabacteria on generalized epilepsy with tonic-clonic seizures, class Betaproteobacteria, and order Burkholderials on juvenile myoclonic epilepsy, which are promising gut biomarkers and novel therapeutic targets of epilepsy.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
YO, YC, and LL designed the study. YO and YC performed the analysis and drafted the manuscript. GW and YS participated in revising the manuscript. LL, ZY, and BX reviewed the manuscript for its intellectual content and revise the entire work. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2021YFC1005305), National Natural Science Foundation of China (82171454), Key Research and Development Program of Hunan Province (2022SK2042), Natural Science Foundation of Hunan Province Project (2020JJ5914 and 2020JJ5952), and Innovative Construction Foundation of Hunan Province (2021SK4001).
Acknowledgments
The authors thank the members of the International League Against Epilepsy (ILAE), Framingham Heart Study (FHS), MiBioGen consortium for providing summary-level data publicity available.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.994270/full#supplementary-material
Supplementary Figure 1 | Mendelian randomization leave-one-out sensitivity analysis of significant results. (A) Leave-one-out analysis of the family Veillonellaceae on childhood absence epilepsy. (B) Leave-one-out analysis of the class Melainabacteria on generalized epilepsy with tonic-clonic seizures. Leave-one-out analysis of (C) the class Betaproteobacteria and (D) the order Burkholderiales on juvenile myoclonic epilepsy.
Supplementary Figure 2 | Funnel plots demonstrating significant results. (A) Funnel plots of the family Veillonellaceae on childhood absence epilepsy. (B) Funnel plots of the class Melainabacteria on generalized epilepsy with tonic-clonic seizures. Funnel plots of (C) the class Betaproteobacteria and (D) the order Burkholderiales on juvenile myoclonic epilepsy.
Abbreviations
CNS, central nervous system; DRE, drug-resistant epilepsy; FMT, fecal microbiota transplantation; GWAS, genome-wide association study; ILAE, International League Against Epilepsy; IVs, instrumental variables; IVW, inverse variance weighted median; KD, ketogenic diet; MGB, microbiota–gut–brain; MR, Mendelian randomization; NAA, N-acetyl aspartic acid; RCT, randomized controlled trial; SNP, single nucleotide polymorphism; TMAO, trimethylamine N-oxide.
Footnotes
References
Ang, Q. Y., Alexander, M., Newman, J. C., Tian, Y., Cai, J., Upadhyay, V., et al. (2020). Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell 181, 1263–1275.e16. doi: 10.1016/j.cell.2020.04.027
Arulsamy, A., and Shaikh, M. F. (2022). Epilepsy-associated comorbidities among adults: a plausible therapeutic role of gut microbiota. Neurobiol. Dis. 165:105648. doi: 10.1016/j.nbd.2022.105648
Arulsamy, A., Tan, Q. Y., Balasubramaniam, V., O’Brien, T. J., and Shaikh, M. F. (2020). Gut microbiota and epilepsy: a systematic review on their relationship and possible therapeutics. ACS Chem. Neurosci. 11, 3488–3498. doi: 10.1021/acschemneuro.0c00431
Bedarf, J. R., Hildebrand, F., Coelho, L. P., Sunagawa, S., Bahram, M., Goeser, F., et al. (2017). Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 9:39. doi: 10.1186/s13073-017-0428-y
Beghetti, I., Barone, M., Turroni, S., Biagi, E., Sansavini, A., Brigidi, P., et al. (2022). Early-life gut microbiota and neurodevelopment in preterm infants: any role for Bifidobacterium? Eur. J. Pediatr. 181, 1773–1777.
Beghi, E. (2020). The epidemiology of epilepsy. Neuroepidemiology 54, 185–191. doi: 10.1159/000503831
Blusztajn, J. K., Slack, B. E., and Mellott, T. J. (2017). Neuroprotective actions of dietary choline. Nutrients 9:815. doi: 10.3390/nu9080815
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965
Burgess, S., Scott, R. A., Timpson, N. J., Davey Smith, G., and Thompson, S. G. (2015). Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552. doi: 10.1007/s10654-015-0011-z
Burgess, S., and Thompson, S. G. (2011). Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat. Med. 30, 1312–1323. doi: 10.1002/sim.4197
Carlson, A. L., Xia, K., Azcarate-Peril, M. A., Rosin, S. P., Fine, J. P., Mu, W., et al. (2021). Infant gut microbiome composition is associated with non-social fear behavior in a pilot study. Nat. Commun. 12:3294. doi: 10.1038/s41467-021-23281-y
Chen, L., Wang, Z., Wang, P., Yu, X., Ding, H., Wang, Z., et al. (2021). Effect of long-term and short-term imbalanced zn manipulation on gut microbiota and screening for microbial markers sensitive to Zinc status. Microbiol. Spectr. 9:e0048321. doi: 10.1128/Spectrum.00483-21
Cho, C. E., and Caudill, M. A. (2017). Trimethylamine-N-Oxide: friend, foe, or simply caught in the cross-fire? Trends Endocrinol. Metab. 28, 121–130. doi: 10.1016/j.tem.2016.10.005
Cryan, J. F., O’Riordan, K. J., Cowan, C. S. M., Sandhu, K. V., Bastiaanssen, T. F. S., Boehme, M., et al. (2019). The microbiota-gut-brain axis. Physiol. Rev. 99, 1877–2013. doi: 10.1152/physrev.00018.2018
De Caro, C., Iannone, L. F., Citraro, R., Striano, P., De Sarro, G., Constanti, A., et al. (2019). Can we ‘seize’ the gut microbiota to treat epilepsy? Neurosci. Biobehav. Rev. 107, 750–764. doi: 10.1016/j.neubiorev.2019.10.002
Deuschl, G., Beghi, E., Fazekas, F., Varga, T., Christoforidi, K. A., Sipido, E., et al. (2020). The burden of neurological diseases in Europe: an analysis for the Global Burden of Disease Study 2017. Lancet Public Health 5, e551–e567.
Ding, M., Lang, Y., Shu, H., Shao, J., and Cui, L. (2021). Microbiota-Gut-Brain axis and epilepsy: a review on mechanisms and potential therapeutics. Front. Immunol. 12:742449. doi: 10.3389/fimmu.2021.742449
Dong, L., Zheng, Q., Cheng, Y., Zhou, M., Wang, M., Xu, J., et al. (2022). Gut microbial characteristics of adult patients with epilepsy. Front. Neurosci. 16:803538. doi: 10.3389/fnins.2022.803538
Elsworth, B., Lyon, M., Alexander, T., Liu, Y., Matthews, P., Hallett, J., et al. (2020). The MRC IEU OpenGWAS data infrastructure. bioRxiv [preprint] doi: 10.1101/2020.08.10.244293
Fan, Y., Wang, H., Liu, X., Zhang, J., and Liu, G. (2019). Crosstalk between the ketogenic diet and epilepsy: from the perspective of gut microbiota. Med. Inflamm. 2019:8373060. doi: 10.1155/2019/8373060
Gomaa, E. Z. (2020). Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek 113, 2019–2040. doi: 10.1007/s10482-020-01474-7
Gong, X., Cai, Q., Liu, X., An, D., Zhou, D., Luo, R., et al. (2021). Gut flora and metabolism are altered in epilepsy and partially restored after ketogenic diets. Microb. Pathog. 155:104899. doi: 10.1016/j.micpath.2021.104899
Greenhalgh, K., Meyer, K. M., Aagaard, K. M., and Wilmes, P. (2016). The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 18, 2103–2116. doi: 10.1111/1462-2920.13318
He, Z., Cui, B. T., Zhang, T., Li, P., Long, C. Y., Ji, G. Z., et al. (2017). Fecal microbiota transplantation cured epilepsy in a case with Crohn’s disease: the first report. World J. Gastroenterol. 23, 3565–3568. doi: 10.3748/wjg.v23.i19.3565
Holmes, M., Flaminio, Z., Vardhan, M., Xu, F., Li, X., Devinsky, O., et al. (2020). Cross talk between drug-resistant epilepsy and the gut microbiome. Epilepsia 61, 2619–2628. doi: 10.1111/epi.16744
Hsiao, E. Y., McBride, S. W., Hsien, S., Sharon, G., Hyde, E. R., McCue, T., et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. doi: 10.1016/j.cell.2013.11.024
Iannone, L. F., Preda, A., Blottière, H. M., Clarke, G., Albani, D., Belcastro, V., et al. (2019). Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev. Neurother. 19, 1037–1050. doi: 10.1080/14737175.2019.1638763
Ilae Consortium. (2018). Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 9:5269. doi: 10.1038/s41467-018-07524-z
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165.
Lee, H., Lee, S., Lee, D. H., and Kim, D. W. (2021). A comparison of the gut microbiota among adult patients with drug-responsive and drug-resistant epilepsy: an exploratory study. Epilepsy Res. 172:106601. doi: 10.1016/j.eplepsyres.2021.106601
Lee, K., Kim, N., Shim, J. O., and Kim, G. H. (2020). Gut bacterial dysbiosis in children with intractable epilepsy. J. Clin. Med. 10:5. doi: 10.3390/jcm10010005
Lin, J. J., Dabbs, K., Riley, J. D., Jones, J. E., Jackson, D. C., Hsu, D. A., et al. (2014). Neurodevelopment in new-onset juvenile myoclonic epilepsy over the first 2 years. Ann. Neurol. 76, 660–668. doi: 10.1002/ana.24240
Liu, P., Jia, X. Z., Chen, Y., Yu, Y., Zhang, K., Lin, Y. J., et al. (2021). Gut microbiota interacts with intrinsic brain activity of patients with amnestic mild cognitive impairment. CNS Neurosci. Ther. 27, 163–173. doi: 10.1111/cns.13451
Logsdon, A. F., Erickson, M. A., Rhea, E. M., Salameh, T. S., and Banks, W. A. (2018). Gut reactions: how the blood-brain barrier connects the microbiome and the brain. Exp. Biol. Med. 243, 159–165. doi: 10.1177/1535370217743766
Louis, P., Young, P., Holtrop, G., and Flint, H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314. doi: 10.1111/j.1462-2920.2009.02066.x
Maldonado, C., Vázquez, M., and Fagiolino, P. (2020). Potential therapeutic role of carnitine and acetylcarnitine in neurological disorders. Curr. Pharm. Des. 26, 1277–1285.
Mudd, A. T., Berding, K., Wang, M., Donovan, S. M., and Dilger, R. N. (2017). Serum cortisol mediates the relationship between fecal Ruminococcus and brain N-acetylaspartate in the young pig. Gut Microbes 8, 589–600. doi: 10.1080/19490976.2017.1353849
Ni, J. J., Xu, Q., Yan, S. S., Han, B. X., Zhang, H., Wei, X. T., et al. (2021). Gut microbiota and psychiatric disorders: a two-sample mendelian randomization study. Front. Microbiol. 12:737197. doi: 10.3389/fmicb.2021.737197
Oliphant, K., Ali, M., D’Souza, M., Hughes, P. D., Sulakhe, D., Wang, A. Z., et al. (2021). Bacteroidota and Lachnospiraceae integration into the gut microbiome at key time points in early life are linked to infant neurodevelopment. Gut Microbes 13:1997560. doi: 10.1080/19490976.2021.1997560
Olson, C. A., Vuong, H. E., Yano, J. M., Liang, Q. Y., Nusbaum, D. J., and Hsiao, E. Y. (2018). The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173, 1728–1741.e13. doi: 10.1016/j.cell.2018.04.027
Peng, A., Qiu, X., Lai, W., Li, W., Zhang, L., Zhu, X., et al. (2018). Altered composition of the gut microbiome in patients with drug-resistant epilepsy. Epilepsy Res. 147, 102–107. doi: 10.1016/j.eplepsyres.2018.09.013
Petersen, A. V., Jensen, C. S., Crépel, V., Falkerslev, M., and Perrier, J. F. (2017). Serotonin regulates the firing of principal cells of the subiculum by inhibiting a T-type Ca(2+) current. Front. Cell Neurosci. 11:60. doi: 10.3389/fncel.2017.00060
Rhee, E. P., Ho, J. E., Chen, M. H., Shen, D., Cheng, S., Larson, M. G., et al. (2013). A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 18, 130–143. doi: 10.1016/j.cmet.2013.06.013
Rinninella, E., Cintoni, M., Raoul, P., Lopetuso, L. R., Scaldaferri, F., Pulcini, G., et al. (2019). Food components and dietary habits: keys for a healthy gut microbiota composition. Nutrients 11:2393. doi: 10.3390/nu11102393
Russo, E. (2022). The gut microbiota as a biomarker in epilepsy. Neurobiol. Dis. 163:105598. doi: 10.1016/j.nbd.2021.105598
Şafak, B., Altunan, B., Topçu, B., and Eren Topkaya, A. (2020). The gut microbiome in epilepsy. Microb. Pathog. 139:103853. doi: 10.1016/j.micpath.2019.103853
Sanna, S., van Zuydam, N. R., Mahajan, A., Kurilshikov, A., Vich Vila, A., Võsa, U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605. doi: 10.1038/s41588-019-0350-x
Sarkar, A., Lehto, S. M., Harty, S., Dinan, T. G., Cryan, J. F., and Burnet, P. W. J. (2016). Psychobiotics and the manipulation of bacteria-gut-brain signals. Trends Neurosci. 39, 763–781. doi: 10.1016/j.tins.2016.09.002
Scheffer, I. E., Berkovic, S., Capovilla, G., Connolly, M. B., French, J., Guilhoto, L., et al. (2017). ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia 58, 512–521. doi: 10.1111/epi.13709
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Singh, V., Yeoh, B. S., Xiao, X., Kumar, M., Bachman, M., Borregaard, N., et al. (2015). Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat. Commun. 6:7113. doi: 10.1038/ncomms8113
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Davies, N. M., Swanson, S. A., VanderWeele, T. J., et al. (2021a). Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ 375:n2233. doi: 10.1136/bmj.n2233
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021b). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA 326, 1614–1621. doi: 10.1001/jama.2021.18236
Smith, G. D., and Ebrahim, S. (2003). ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J. Epidemiol. 32, 1–22. doi: 10.1093/ije/dyg070
Socała, K., Doboszewska, U., Szopa, A., Serefko, A., Włodarczyk, M., Zielińska, A., et al. (2021). The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol. Res. 172:105840. doi: 10.1016/j.phrs.2021.105840
Sullivan, A., Edlund, C., and Nord, C. E. (2001). Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1, 101–114.
Tang, Y., Wang, Q., and Liu, J. (2021). Microbiota-gut-brain axis: a novel potential target of ketogenic diet for epilepsy. Curr. Opin. Pharmacol. 61, 36–41. doi: 10.1016/j.coph.2021.08.018
Tashakori-Miyanroudi, M., Ramazi, S., Hashemi, P., Nazari-Serenjeh, M., Baluchnejadmojarad, T., and Roghani, M. (2022). Acetyl-L-Carnitine exerts neuroprotective and anticonvulsant effect in kainate murine model of temporal lobe epilepsy. J. Mol. Neurosci. 72, 1224–1233. doi: 10.1007/s12031-022-01999-8
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698.
Xie, G., Zhou, Q., Qiu, C. Z., Dai, W. K., Wang, H. P., Li, Y. H., et al. (2017). Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J. Gastroenterol. 23, 6164–6171. doi: 10.3748/wjg.v23.i33.6164
Zeisel, S. H., and Warrier, M. (2017). Trimethylamine N-Oxide, the microbiome, and heart and kidney disease. Annu. Rev. Nutr. 37, 157–181.
Zhang, Y., Zhou, S., Zhou, Y., Yu, L., Zhang, L., and Wang, Y. (2018). Altered gut microbiome composition in children with refractory epilepsy after ketogenic diet. Epilepsy Res. 145, 163–168. doi: 10.1016/j.eplepsyres.2018.06.015
Keywords: gut microbiota, gut metabolites, epilepsy, bi-directional Mendelian randomization study, causality
Citation: Ouyang Y, Chen Y, Wang G, Song Y, Zhao H, Xiao B, Yang Z and Long L (2022) Genetically proxied gut microbiota, gut metabolites with risk of epilepsy and the subtypes: A bi-directional Mendelian randomization study. Front. Mol. Neurosci. 15:994270. doi: 10.3389/fnmol.2022.994270
Received: 14 July 2022; Accepted: 11 October 2022;
Published: 03 November 2022.
Edited by:
Longbo Zhang, Yale University, United StatesReviewed by:
Xiaoqing Jiang, Peking University, ChinaKonark Mukherjee, Virginia Tech, United States
Copyright © 2022 Ouyang, Chen, Wang, Song, Zhao, Xiao, Yang and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Long, bG9uZ2xpbGkxOTgyQDEyNi5jb20=; Zhuanyi Yang, bmV1cm95enlAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yuzhen Ouyang
Yuzhen Ouyang Yu Chen
Yu Chen Ge Wang
Ge Wang Yanmin Song
Yanmin Song Haiting Zhao
Haiting Zhao Bo Xiao
Bo Xiao Zhuanyi Yang
Zhuanyi Yang Lili Long
Lili Long