
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mol. Neurosci., 20 September 2022
Sec. Methods and Model Organisms
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.956582
This article is part of the Research TopicUsing Zebrafish to Investigate Normal Development and Function of the Nervous System: How nervous system disorders occur and testing of potential therapeutic approachesView all 8 articles
The zebrafish (Danio rerio) is a vertebrate species offering multitude of advantages for the study of conserved biological systems in human and has considerably enriched our knowledge in developmental biology and physiology. Being equally important in medical research, the zebrafish has become a critical tool in the fields of diagnosis, gene discovery, disease modeling, and pharmacology-based therapy. Studies on the zebrafish neuromuscular system allowed for deciphering key molecular pathways in this tissue, and established it as a model of choice to study numerous motor neurons, neuromuscular junctions, and muscle diseases. Starting with the similarities of the zebrafish neuromuscular system with the human system, we review disease models associated with the neuromuscular system to focus on current methodologies employed to study them and outline their caveats. In particular, we put in perspective the necessity to develop standardized and high-resolution methodologies that are necessary to deepen our understanding of not only fundamental signaling pathways in a healthy tissue but also the changes leading to disease phenotype outbreaks, and offer templates for high-content screening strategies. While the development of high-throughput methodologies is underway for motility assays, there is no automated approach to quantify the key molecular cues of the neuromuscular junction. Here, we provide a novel high-throughput imaging methodology in the zebrafish that is standardized, highly resolutive, quantitative, and fit for drug screening. By providing a proof of concept for its robustness in identifying novel molecular players and therapeutic drugs in giant axonal neuropathy (GAN) disease, we foresee that this new tool could be useful for both fundamental and biomedical research.
Over the last 30 years, the zebrafish (Danio rerio) has become the model of choice for many fundamental and translational research studies. With about 70% ortholog genes shared with the human genome (Barbazuk, 2000; Howe et al., 2013), the zebrafish is largely used in the fields of embryology, pharmacology, and toxicology to expand our knowledge on complex biological processes and accelerate the emergence of precision medicine (MacRae and Peterson, 2015). It has also become a preferred species for the modeling of various diseases, including cancer, and cardiovascular, metabolic, and neuromuscular diseases (NMDs) (Seth et al., 2013; Patton et al., 2021).
The success of this animal model largely relies on the numerous advantages it offers to the research community. Regarding practical aspects, the husbandry of zebrafish is relatively simple and inexpensive. Its high fertility rate (∼200 embryos per clutch), rapid and external development, and transparent skin allow direct access to all developmental stages from embryo (one-cell zygote at 0 h post-fertilization, hpf, to hatching at 2–3 days post-fertilization, dpf), to larva (from 3 to 29 dpf) and juvenile (until 6 weeks). As a consequence, zebrafish presents many advantages: (i) genetic manipulation can be achieved at an early stage of embryonic development (injection of antisense oligonucleotides/mRNA for transient effects, or CRISPR agents for the creation of stable transgenic lines), (ii) transparency of embryos allows for the visualization of internal structures and networks at the level of the whole organism, (iii) small-size (0.9–3.5 mm) zebrafish embryos can be kept in 96- or 384-well plates, and (iv) chemical compounds can be administered by simply adding to water (Zon and Peterson, 2005). In practice, the study of zebrafish is cost-effective and compatible with large-scale/high-throughput pharmacological screening (Rennekamp and Peterson, 2015), explaining why it has become the leading model organism for chemical screening and drug discovery.
The zebrafish model has particularly proven to be valuable in vertebrate neuromuscular system studies, which have deepened our knowledge in normal axonal/muscle synapse development and degeneration (Guyon et al., 2007; McLean and Fetcho, 2008; Li et al., 2017) and in identifying the deficits underlying neuromuscular pathologies (Panzer et al., 2005, 2006). The locomotion-sustaining human neuronal circuitry, which is highly conserved in zebrafish, was characterized, thanks to this species (Kimmel et al., 1995; Beattie, 2000). In particular, studies using this model contributed to the identification of molecular signals required for the establishment of motor networks (Berg et al., 2018), neuromuscular junction (NMJ) development, NMJ maintenance, synaptogenesis (Jing et al., 2009, 2010; Banerjee et al., 2011), and identification of muscle precursor types (Devoto et al., 1996).
With the ease of genetic manipulation, various transgenic zebrafish lines represent powerful models to study human diseases, in an era where whole-genome/-exon sequencing generates big data of genetic variants in patients (Pipis et al., 2019; Westra et al., 2019; Herman et al., 2021). In this regard, the zebrafish can be extremely valuable in (1) discovering the disease gene among potential candidates, (2) identifying pathological variants of a known gene, and (3) deciphering pathological mechanisms (McCammon and Sive, 2015; Vaz et al., 2019).
Here, we first describe the key events in zebrafish neuromuscular system development and its resemblance to human counterpart. We then briefly present how the creation of disease models of the neuromuscular system contributes to the understanding of this specific synapse in health, as well as the underlying mechanisms in disease. We then focus on the current behavioral and imaging-based techniques used in the zebrafish, putting in perspective the need to generate tools with greater precision to enable a deeper exploration of the neuromuscular system and to apprehend specificity in disease, an essential aspect for personalized medicine. Finally, we present a novel quantitative and standardized imaging methodology with a proof of concept for its effectiveness in identifying both novel regulators of the neuromuscular system and therapeutic drugs using high-throughput screening.
At the cellular level, the NMJ is made up of the same components across species (Figure 1A). It is formed by two entities: the presynaptic motor neuron (MN) of the nervous system and the postsynaptic muscle fiber, which are separated by a synaptic cleft. The transmission of electrical input along the MN axon is achieved by a spread of action potentials. Upon activation, the fusion of vesicles in MN terminals results in a release of acetylcholine (ACh) into the synaptic cleft, which activates postsynaptic nicotinic ACh receptors (AChRs) in the postsynaptic muscle fibers (Kummer et al., 2006). This event depolarizes the muscle cell and triggers calcium release from the sarcoplasmic reticulum to initiate muscle contraction. Many molecular, histological, and ultrastructural features of NMJ development and integrity are well conserved between mammals and zebrafish (Swenarchuk, 2019). Among other examples, the key role of AChR pre-patterning in guiding MN terminals to the muscle fibers was discovered in the zebrafish model (Panzer et al., 2005), as well as the role of the Wnt pathway in this process (Jing et al., 2010).
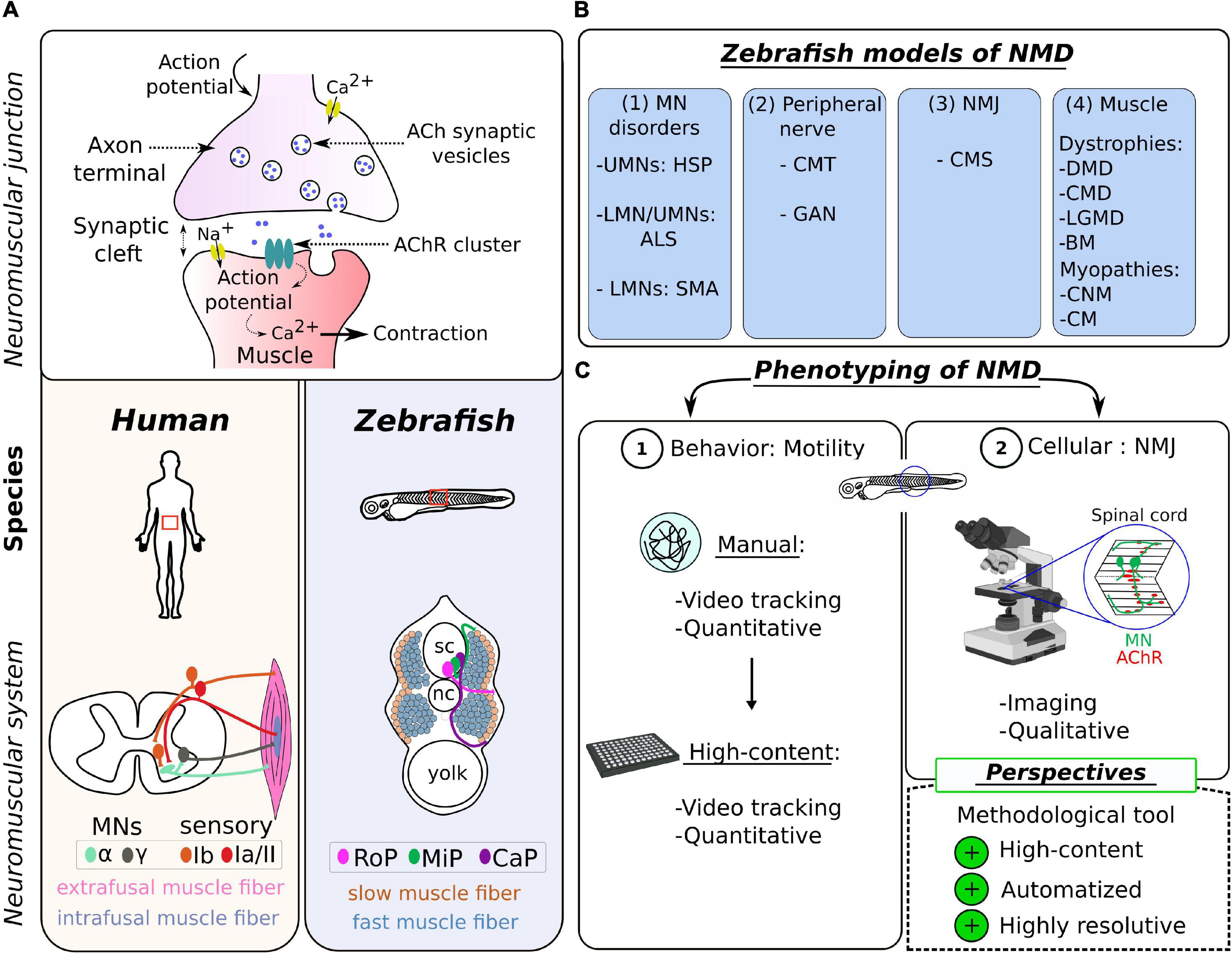
Figure 1. Zebrafish as a model of choice to study the neuromuscular system in human. (A) Schematic representation of the neuromuscular synapse (top) and the topographical differences (bottom) between the zebrafish and the human species. (B) Table summarizing the different NMD pathologies modeled in zebrafish, categorized into four groups according to the first disease target (see Table 1 for details). UMN, upper motor neuron; LMN, lower motor neuron; HSP, hereditary spastic paraplegia; ALS, amyotrophic lateral sclerosis; SMA, spinal muscular atrophy; CMT, Charcot–Marie–Tooth; GAN, giant axonal neuropathy; NMJ, neuromuscular junction; CMS, congenital myasthenic syndrome; DMD, Duchenne muscular dystrophy; CMD, congenital muscular dystrophy; LGMD, limb–girdle muscular dystrophy; BM, Bethlem myopathy; CNM, centronuclear myopathies; CM, congenital myopathy. (C) Scheme summarizing the common tools for NMD phenotyping in the zebrafish, divided into two levels of investigation: (1) the locomotion at the behavioral level and (2) the visualization of the NMJ at the cellular level; and highlighting the methodological challenge presented as perspectives in this review.
The skeletal muscle, which is the only muscle tissue voluntary controlled by the central nervous system, is closely related in zebrafish and humans, at histological, biochemical, and ultrastructural levels (Luna et al., 2015). These similarities include the preservation of two sets of MNs, two main types of muscle fibers (slow and fast), the dystrophin-associated protein (DAPC) complex, and the excitation–contraction coupling system (Berger and Currie, 2012). Differences are restricted to morphological variables such as MN axon diameters and average area of AChR clusters (Jones et al., 2017; Boehm et al., 2020), and the specificity/subclassification of muscle fibers, which have their source in an adaptation of locomotor behavior. In humans, there are two major types of spinal MNs located in the anterior horn of the spinal cord, differentiated according to the size of the soma and their innervation (Figure 1A): α-MNs and γ-MNs (Kanning et al., 2010; Manuel and Zytnicki, 2011). The α-MNs are the most abundant type of MNs in mammals. They innervate extrafusal muscle fibers classified into different subtypes, whose contractile properties differ: slow-twitch, fatigue-resistant (S), and fast-twitch fibers, which are further subdivided into fatigue-resistant (FR) and fatigable (FF) fibers (Brooke and Kaiser, 1970; Burke et al., 1971). The γ-MNs innervate intrafusal muscle fibers responsible for muscle tone and proprioception. Zebrafish spinal MNs are also divided into two groups, differentiated in two distinct waves during development: primary motor neurons (pMNs) and secondary motor neurons (sMNs) innervating specific musculature (Myers, 1985; Eisen et al., 1986). There is also a subclassification of primary motor neurons (pMNs) in each hemisegment, originally designated according to their specific location as caudal primary motor neuron (CaP); middle primary motor neuron (MiP), and rostral primary motor neuron (RoP) (Figure 1A), innervating different muscle fiber territories. Motor neurons from the second wave (sMNs) are considered to be equivalent to α-MNs in humans (Beattie et al., 1997). They have small-size somas, are more numerous, and innervate both deeper fast-twitch muscle and superficial slow-twitch muscle fibers (Menelaou and McLean, 2012; Bello-Rojas et al., 2019). It is worth noting that the proportion of fast muscles is much higher in zebrafish than in mammals, an adaptative response to the need to produce bursting and rapid powerful movements.
The genome is highly conserved between the human and zebrafish species, with a homology for over 80% of disease-causing genes (Howe et al., 2013). This evolutionary characteristic fostered the generation of zebrafish mutants to model pathologies of the human neuromuscular system (Pappalardo et al., 2013). Strategies of transient genetic manipulations using microinjection (Eisen and Smith, 2008) (gene inactivation to model recessive diseases or mRNA overexpression for dominant pathologies), followed by genome editing (Gaj et al., 2013; Schmid and Haass, 2013), have been successfully used to assess the significance of pathological mutations associated with neurodevelopmental and neurodegenerative conditions, including NMDs.
Here, NMDs are defined as pathologies inducing muscle weakness and atrophy (Efthymiou et al., 2016). Thus, NMDs cover a large group of diseases of heterogeneous etiology, affecting the muscles and/or the nerves and comprising (1) motor neuron disorders [e.g., amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), and hereditary spastic paraplegia (HSP)], (2) disorders with a component in the peripheral nerves [e.g., Charcot–Marie–Tooth disease (CMT) and giant axonal neuropathy (GAN)], (3) disorders directly affecting the NMJ [e.g., congenital myasthenic syndrome, (CMS) and Lambert–Eaton syndrome], and (4) primary disorders of the muscle (myopathies and muscular dystrophies) (Figure 1B).
While we present numerous zebrafish models generated to model NMDs (Table 1), we do not intend to describe them all. Instead, we focus on highlighting some features as evidence for great relevance and advantage of the zebrafish research model in these types of studies:
- Zebrafish models of the same disease group, for example, of the MNs (ALS, SMA, and HSP) induce similar deficits, including loss of MNs, aberrant axonal outgrowth, loss of neuromuscular connectivity, muscle denervation, and defective motor performance. The study of these parameters, as easily performed in the zebrafish, enriches our knowledge of the molecular players in the settings and maintenance of the neuromuscular system.
- In a context of diseases with multiple genetic origins (most of the clinical groups described in Table 1), the zebrafish model provides the versatility to validate gene discovery and pathogenic variants using fast and penetrant functional characterization (see Patten et al., 2014; Naef et al., 2019 for review).
- For several diseases, such as GAN (Arribat et al., 2019) and HSP (als2, Gros-Louis et al., 2008), zebrafish models are the first to reproduce the severity of motor and neuromuscular symptoms, where mouse models have so far failed (Cai et al., 2008; Dequen et al., 2008; Ganay et al., 2011). This reinforces the notion of the conservation of the neuromuscular system between the zebrafish and human species.
- The zebrafish represents a highly relevant model for therapeutic development as its physiology is well conserved with human. For example, the zebrafish is an excellent model for muscle pathologies (Jagla et al., 2017), more adapted than mouse (Berger and Currie, 2012). Models with mutations in the zebrafish dystrophin gene (dmd and sapje models) of DMD have been developed (Bassett and Currie, 2004; Guyon et al., 2009), whose robustness of phenotypes (muscle lesions and birefringence, almost 100% penetrant) has led to the development of a therapeutic strategy for human patients (Johnson et al., 2013; Farr et al., 2020), which is now in phase 3 in clinical trial.
The phenotyping of zebrafish models of NMDs and, more generally, the study on the regulation(s) of the neuromuscular system rely on two levels of analysis: the locomotion at the behavioral level (Granato et al., 1996) and the visualization of the NMJ at the cellular level (Fetcho, 2007). We present the most common tools used in zebrafish, highlighting their advantages and their limitations (Figure 1C).
The zebrafish larvae serve as a powerful model to dissect locomotor patterns as their swimming behavior is defined by sequences of stereotyped movements (Wolman and Granato, 2012). At 5 dpf, when the neuromuscular system is established, the zebrafish is capable of fully autonomous and spontaneous movement. At this stage, motility assays have the advantage to be purely motor, contrary to other locomotor tests performed on younger zebrafish embryos, which include a sensory component (e.g., evoked response to stimuli at 48 hpf). The analysis of locomotor activity of larval zebrafish has emerged as a potent tool for phenotypic assessment in the neurosciences and toxicology fields (Basnet et al., 2019). Some commercial tools are available to the community (e.g., Noldus, Viewpoint, or Loligo® Systems): they provide a means to measure different properties of the swimming activity, including quantification of kinetic parameters such as frequency, duration, speed, and total distance traveled within a given time (Noldus et al., 2001). Few academic laboratories developed locomotion-based tracking systems with deeper analysis of the movement patterns of larvae that are suitable for high-content analysis (Colwill and Creton, 2011; Zhou et al., 2014). They were instrumental in providing a better analysis of the sequencing of swimming bouts (Reddy et al., 2022) and in discriminating the great variability in the escape behaviors (Kohashi and Oda, 2008), by measuring the delay, amplitude, duration, frequency, angle, and number of the tail-bending oscillations (Mirat et al., 2013). Overall, the latter methodologies are extremely attractive, translationally relevant, and easily adaptable to high-throughput pharmacological screening strategies but unfortunately still too rarely used by the general academic community.
While important for the comprehension of the locomotor network (Berg et al., 2018), behavioral tests are not systematically used in disease models, as shown in Table 1. Still, when performed, they are valuable to quantify the degree of locomotor deficits: less frequent turning and swimming bouts in the SMA zebrafish model (Hao et al., 2012), shorter traveled distance in the ALS zebrafish model (Armstrong and Drapeau, 2013; Robinson et al., 2019), and severe reduction in both total distance and net velocity (Zhao et al., 2019) in CMT and GAN models (Arribat et al., 2019). Nevertheless, these tools present limitations when it comes to grading the severity of dysfunction between NMD types, or for distinct genes within the same group of diseases. This challenge is mainly due to high variability in the settings applied to the motility tests (type, alternation of light/dark stimulation, duration of the assay, etc.). For example, one can note that the great disparity in the locomotor tests used to describe motor deficits in ALS models, specifically in the SOD1 gene (see Table 1). This aspect has some importance when choice needs to be made to select strong candidates for preclinical studies or discuss the relative contribution of molecular players. Most importantly, current behavioral tests have limited outcomes in defining the functional origin (neuron, NMJ, or muscle) and the cellular signature of individual gene or NMD.
Due to their transparency, zebrafish embryos are particularly suited for cellular imaging (see Pappalardo et al., 2013; Babin et al., 2014; Patten et al., 2014 for review). For the motor system, classic methods are the labeling of the sub-compartments of the NMJ (Panzer et al., 2005) and providing qualitative measures of the integrity and stability of the NMJ. Some pieces of off-line, free, and open-source image processing and analysis software have been used as quantitative tools for the analysis of the NMJ such as axonal length, AChR clustering, and the coefficient of co-localization of NMJ markers (pre- and postsynaptic). Thus, from confocal images, the authors use these methods to (1) measure axonal length [e.g., NeuronJ plugin in ImageJ software (Boon et al., 2009; Robinson et al., 2019; Campanari et al., 2021; Oprişoreanu et al., 2021) or Lucia software (Lemmens et al., 2007)] or (2) define the score for normal versus abnormal (McWhorter et al., 2003; Boyd et al., 2017; Swinnen et al., 2018). Rare laboratories have developed specific metrics for the quantification of motor axon development, as in Smn-deficient zebrafish embryos (Gassman et al., 2013). Overall, the existing analysis tools permit the extraction of quantifiable data of only a limited number (about six or seven) of axons per zebrafish sample. Thus, one important limitation of the current cellular analysis is the difficulty in extracting measurable data for the whole NMJ, especially due to the overlap between pre- and postsynaptic staining. Indeed, unlike other mammalian species such as the mouse, the fish exhibits a spreaded distribution of the motor nerve terminals and a great extent of postsynaptic folding (Slater, 2017), thus particularly challenging to visualize in detail and quantify.
Currently, most cellular studies in zebrafish are conducted manually, using low-throughput imaging techniques. Still, there is a need for standardized, quantitative, and high-resolution techniques to gain in-depth knowledge of the neuromuscular unit and to define specificity of alterations of NMJ (and therefore mechanisms) across pathologies. Moreover, similar to behavioral assays, developing automated tools, compatible with high-throughput screening assays, would be essential to identify and determine the effectiveness of potential drugs in modulating or restoring the neuromuscular system in health and disease. Recent advancements in imaging-based high-content screening technologies for drug discovery and toxicology have mostly found applications in 3D tissue culture or 3D organoid models (Li et al., 2016). Although the zebrafish has earned its place as an efficient model for high-throughput drug screening, the current tools lack a standard pipeline for imaging small organisms. Therefore, designing a quantifiable and standardized methodology with robust assay metrics to quantify parameters of the neuromuscular unit would be critical for both fundamental biology and clinical research.
To address the challenge of standardization and reproducibility of imaging tools in zebrafish, we developed an automated, high-throughput, imaging pipeline in zebrafish, integrating a quantitative analysis of the key parameters of the neuromuscular system: axonal length, AChR clustering, and NMJ synapse. Here, we mainly describe this novel quantitative methodology and briefly illustrate its potency in delineating the specific phenotype and identifying therapeutic drugs in GAN disease.
Our method relies on automated high-resolution image acquisition using multi-well plates (Opera Phenix™ high-content imaging systems, Perkin Elmer) and involves the development of an image processing sequence using Harmony software (v4.9, Perkin Elmer) to prefilter whole-embryo images and exclude regions with artifacts and high background. This protocol allows to define the regions of interest (ROIs) within the spinal cord and make them automatically detectable and accessible to downstream analysis (Figure 2).
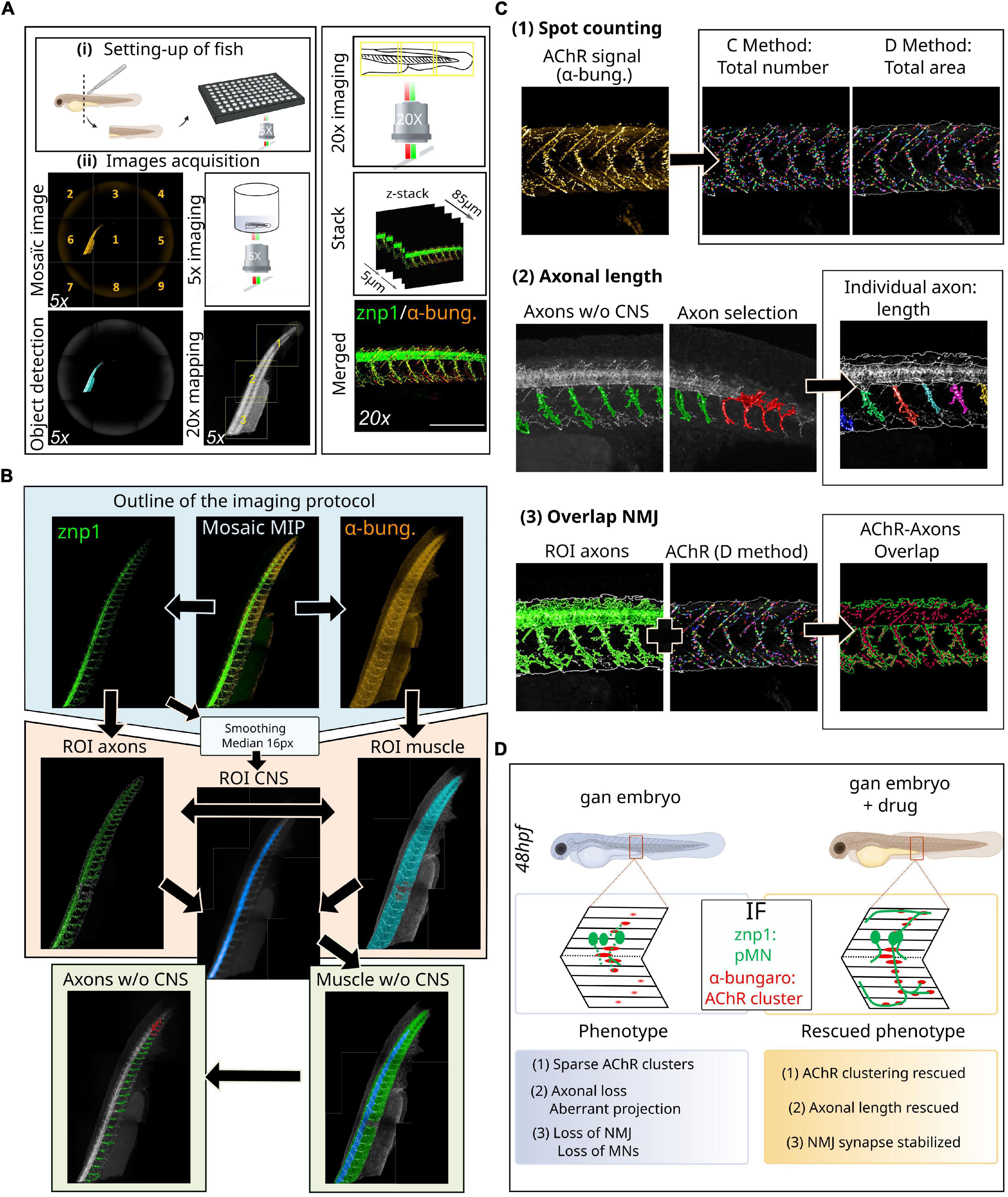
Figure 2. Novel methodology for high-throughput imaging of the neuromuscular system in the zebrafish. (A) Schematic overview of the setup of 48hpf larvae in 96-well plates (i) for automated detection and image acquisition (ii), from object detection (5x) to stacked images (20x). (B) Outline of the imaging and analytic protocol: representative images of NMJ staining (znp1 in green, α-bungarotoxin in red) within the spinal cord of control larvae, from which several steps of ROI segmentation allow to define the different components of the NMJ. (C) Representative images of the different filters resulting in the segmentation of AChR clusters from α-bungarotoxin staining and individual axon from the znp1 staining, enabling the quantitative assessment of spot counting (1), axonal length (2), and NMJ overlapping compounds within the spinal cord (3). See the Supplementary Figure 1 to assess for the quality of the processing tools for each filter: raw data compared to post-analysis pictures at high magnification. (D) Validation of this methodology in the zebrafish model of giant axonal neuropathy (gan). Detailed coverage of NMJ defects in the gan model and identification of hits restoring three parameters of the neuromuscular system, following a high-throughput screening. α-bung, α-bungarotoxin; pMN, primary motor neuron; MN, motor neuron; NMJ, neuromuscular junction; AChR, acetylcholine receptors; MIP, maximum intensity projections; ROI, region of interest; w/o, without; CNS, central nervous system.
• Prerequisite: The first prerequisite concerns the establishment of a model of the neuromuscular system or pathology in the zebrafish, whose phenotype is at least partially described in larval stages. It can include a motor component, with a defect in locomotion with a severity ideally similar to that observed in human patients and/or a cellular defect of varying severity in the neuromuscular system.
• Possible applications: The methodology presented here can have a fundamental and/or therapeutic vocation. Indeed, it allows to scrutinize the neuromuscular phenotypes in greater detail and to identify therapeutic molecules able to restore them. In the latter, it is recommended to carefully select a chemical library of interest (see Peterson and Fishman, 2011 for review).
• Procedure/Methodology: In the next paragraph, we provide a detailed description of the high-throughput imaging approach, which offers high-resolution images of 3D zebrafish larvae mounted in individual wells in 96-well plates. Then, we detail the development of an image analysis sequence, allowing first to define the different ROIs by using maximum intensity projections (MIP) and then to submit them to different analysis filters to extract measurable neuromuscular parameters. Noteworthy, our methodology on high-resolution images allows the quantification of the NMJ, a challenge in the zebrafish model due to dispersed sites of muscle innervation.
Zebrafish embryos of desired aged (see “Advantages of the method” section) are placed in 96-well plates and eventually exposed to small-molecule libraries of compounds that can elicit phenotypic alterations (Figure 2Ai). While three larvae represent the most common settings (Rennekamp and Peterson, 2015), we determined that each well can house up to four zebrafish larvae without compromising proper development. On another note, it is recommended to use plates with a thin and flat plastic or glass bottom. Since the interest here is to image a lateral view of the spinal cord, we recommend cutting the embryo at the anterior part of the yolk extension to enable lateral positioning.
Neuromuscular junctions can be visualized by light microscopy following labeling of both the presynaptic nerve terminal and the postsynaptic acetylcholine receptors (Figure 2A). The most commonly used presynaptic antigens, as in other vertebrate models, include anti-neurofilament (e.g., 3A10), anti-synaptotagmin (e.g., znp-1), anti-neurolin (e.g., zn8), and anti-synaptic vesicle glycoprotein 2 (SV2) to fully label both the pre-synaptic axon and nerve terminal. The fluorescently tagged α-bungarotoxin is the most commonly used protein to label acetylcholine receptors in the postsynaptic membranes of the NMJ. Here, we opted to use the znp-1/α-bungarotoxin combination, which ensures specific marking of the axons and NMJ with little background. Primary antibodies are from the following sources: mouse IgG2a anti-synaptotagmin (1:100, znp-1, DSHB) and anti-α-bungarotoxin (1:50, B35451, Invitrogen). We believe the same methodology can be applied to other NMJ markers mentioned earlier. Indeed, the analysis sequences permit the collection of a large number of measurable parameters, as long as the immunostaining is of required quality.
1. Pre-scan is performed at 5X magnification on the full well surface (nine fields per well with 6% overlap) and red channel only (561 nm) with the widefield mode (Figure 2Aii).
2. Use the “In-the-fly image analysis” tool to create a mosaic image and “Find Image Region” module to detect the whole zebrafish.
3. Use the “Determine Well Layout” module to define a 20X re-scan magnification (with 6% overlap) covering the entire object.
4. Re-scan at 20X magnification on whole fish larvae in the confocal mode. Green (488 nm/znp-1) and red (561 nm/α-bungarotoxin) channels are imaged, with a z-stack of 85 μm (5 μm interval).
5. First, acquire mosaic images (full fish larvae) with MIP for each channel (Figure 2B).
6. To automatically identify the two ROI named axons and muscle, we used the “Find Image Region” module in green (488 nm/znp-1) and red (561 nm/α-bungarotoxin) channels, respectively. The final ROI muscle was obtained by subtracting seven pixels around the initial region to restrict the analysis to the region of interest and to reduce detection of artifacts.
7. The main challenge is to separate each axon into a different entity. To do this, it is imperative to differentiate the axons from the spinal cord. From the same mosaic images with MIP, we applied smoothing with a median filter of 20 px using the “Filter Image” module in the green (488 nm/znp-1) channel. On this new image, we used the “Find Image Region” module to find a new ROI: central nervous system (CNS, spinal cord). Then, we used “Calculate Morphology Properties” to measure the size and the position of the CNS ROI.
8. Using the “Find Surrounding Region” module, the CNS ROI was subtracted from the fish body area (ROI muscle), to create a new surrounding region: body without CNS.
9. Acetylcholine receptors localization/quantification was performed in the ROI muscle using the “Find Spots” module on the α-bungarotoxin channel [Figure 2C(1)]. A total of two distinct methods were used to extract the properties of the AChR spots: method C, more sensitive to the detection of the exact number, and method D, more sensitive to the detection of the area. It is also possible to extract raw data on the number of AChR clusters (method C) and their area (method D) in px2 or μm2, as well as their average labeling intensity (see Supplementary Figure 1).
10. Using the “Find Image Region” module only in the surrounding region (body without the CNS), we created a new ROI: axons without the CNS, corresponding to the axonal region without the spinal cord [Figure 2C(2)]. This is particularly important because the bright spinal cord signal hampers the detection of weaker axonal staining. With this restricted region, we applied a “Modify Population” module with “Cluster by Distance” (2 px distance and area > 300 px2) to individualize axons. An axon selection step can be added to eliminate detection problems with the “Selection Population” module (in case some axons have not been properly individualized).
11. Finally, we used the “Calculate Morphology Properties” module to measure individual axonal length. The data are presented as length units (μm) for length and width and μm2 for axonal area (see Supplementary Figure 1).
12. To measure AChR/axon overlap, we used the “Calculate Position Properties - Cross Population” module to obtain the overlap percentage between the AChR total area and axon ROI [Figure 2C(3)]. This calculation corresponds to the percentage of the AChR cluster area included in the axon ROI area compared to the total AChR area. It is built on the raw data of the axon ROI area (μm2) and the postsynaptic AChR area (μm2) calculated using the method D (see Supplementary Figure 1). To enable the reproducibility of data and use of the analysis sequences obtained by the Harmony software (Perkin Elmer), all the pre-prints are added in HTML format as Supplementary Datasheet 1.
• Validation of this methodology (Figure 2D): We validated this novel methodology with the zebrafish model of giant axonal neuropathy. GAN is a fatal disease, presenting a widespread phenotype which starts with a peripheral neuropathy in infancy and extends to the brain in young adults (Lescouzères and Bomont, 2020) (Figure 1B and Table 1). Since we identified gigaxonin as the defective protein in this disease (Bomont et al., 2000), we have generated a robust zebrafish model of the pathology by (1) transiently repressing gigaxonin expression using morpholino antisense oligonucleotides and (2) creating a knockout line. Both zebrafish models reproduce the loss of motility described in GAN patients (Arribat et al., 2019). Using our high-resolution and quantitative imaging methodology, we refined the role of gigaxonin in controlling (1) AChR clustering, (2) axonal outgrowth/projection, and (3) stability of the NMJ. In addition, the combination of this novel high-content image analysis pipeline with high-throughput screening allowed us to pinpoint drugs capable of restoring all three parameters and develop a therapeutic scheme for preclinical studies (Lescouzères et al., Submitted).
• Advantages of the method: One of the main advantages of this approach is the generation of standardized and quantifiable data for parameters that are usually assessed with either qualitative measures or manual and tedious quantitative tools. In particular, the intention is to address the challenge of quantitative analysis of the NMJ with high throughput. Here, the analysis is automated and allows for robust characterization of a novel phenotype and/or the testing of a large number of compounds in drug screening. Moreover, since the data are obtained from a large number of larvae, it is possible to collect more robust statistics in different populations (diseased/treated larvae, between different genes, etc.). For diseased embryos with modest or less penetrant phenotype, the statistical power of such analysis is extremely high. In addition, our methodology enables the establishment of thresholds from which a specific NMJ-associated parameter is considered affected and/or restored in the treated diseased embryos. Thus, this novel imaging methodology designed for the neuromuscular system presents various advantages that only require a precise positioning of the zebrafish embryos in wells. Noteworthy, the imaging methodology we propose has no time limit and may, in principle, allow the observation of early events of the neuromuscular contacts. To study MN differentiation, (1) the first wave of primary motor neurons at the end of gastrulation (9–10 hpf) or (2) the second wave of secondary motor neurons (14–15 hpf) can be imaged using islet staining (Eisen et al., 1986; Westerfield et al., 1986). Prior to the establishment of motor nerve terminal contacts, postsynaptic muscle fibers form primitive, dynamic, and non-synaptic AChR clusters that are distributed on the membrane of the adaxial cells. This step, called muscle pre-patterning (Panzer et al., 2005; Wu et al., 2010), can be specifically investigated with the α-bungarotoxin staining between 14 and 16 hpf. In addition, axonal branching and outgrowth are studied at more advanced stages, that is, 26–30 hpf for the pMNs (CaP, MiP, and RoP) and 30–50 hpf for sMNs. (Myers, 1985) using znp1 and zn8 staining, respectively. Finally, this workflow for high-throughput image analysis could be easily adapted to study other tissues, such as the retina in the zebrafish. This adaptation would only require the selection of sufficiently specific antibodies, to ensure the reproducible detection thresholds from one embryo to another.
The zebrafish species shares great genetic similarities with humans and is a model of choice for studying the physiology of the neuromuscular system. On the one hand, in the fundamental field, the access to all embryological stages and the ease of use of the model (imaging and housing) allow for the integration of the zebrafish in many comparative studies and the identification of key molecular players in the neuromuscular system. In the field of human genetics, the zebrafish has proven its strength for diagnosis purpose, gene identification, and modeling diseases of the motor neuron, NMJ, and muscle. Taking advantage of the small size and optic transparency of the embryo/larva, behavioral and cellular methodologies uncovered fundamental and disease mechanisms sustaining motor functions. Still, limitations lie in the strength of the assays as one could argue that all NMDs or molecular players look similar, with altered locomotion and decreased axonal outgrowth. Increasing precision is not only necessary to fully decipher the molecular pathways controlling the neuromuscular unit but also mandatory to differentiate diseases and offer the specificity required for personalized medicine. While behavioral assays have been enriched to quantify various parameters of motility in an automated manner, current imaging protocols do not reach this level of development. Here, we present a novel, standardized, automated, and quantitative imaging pipeline for the key parameters of the neuromuscular unit, including the so far challenging quantification of the NMJ. As validated in our GAN disease model, we expect our imaging and analysis tools to be broadly useful to researchers interested in scrutinizing the motor system in detail and executing high-throughput screening in zebrafish models of NMDs.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
This animal study was reviewed and approved by the French ministry (reference N°036) for the creation of the gan zebrafish line.
PB contributed to the conception and design of the work, drafting the manuscript and figures, and critical revisions of the manuscript. LL and BB generated and designed the figures. LL executed the drawing of figures. All authors performed the bibliography search and wrote and reviewed the manuscript.
This work was funded by l’Institut National de la Santé et de la Recherche Médicale (INSERM) to PB and the Fondation pour la Recherche Médicale to LL (FRM, fellowship PLP20170939065). This work was supported by grants from the Association Française contre les Myopathies (AFM grant 17654) and the Fondation Maladies Rares (FMR). BB was supported by the French National Research Agency (ANR-10-INBS-04, « Investments for the future »).
We acknowledge the MRI facility of the France-BioImagingnational infrastructure supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future») for the provision of the Opera Phenix high-content imaging systems and Harmony software. We acknowledge Perkin Elmer for the authorization to share the pre-prints of the analysis sequences. We acknowledge the Aquatic Model Platform of the CRBM Institute (Univ. Montpellier, CNRS, Montpellier) and thank B. Delaval for the coordination of housing and support, and P. Richard and M. Plays for zebrafish husbandry. We are very grateful to H. Barzilai-Tutsch (INMG-MeLiS) for critical comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.956582/full#supplementary-material
Armstrong, G. A. B., and Drapeau, P. (2013). Loss and gain of FUS function impair neuromuscular synaptic transmission in a genetic model of ALS. Hum. Mol. Genet. 22, 4282–4292. doi: 10.1093/hmg/ddt278
Arribat, Y., Mysiak, K. S., Lescouzères, L., Boizot, A., Ruiz, M., Rossel, M., et al. (2019). Sonic hedgehog repression underlies gigaxonin mutation-induced motor deficits in giant axonal neuropathy. J. Clin. Invest. 129, 5312–5326. doi: 10.1172/JCI129788
Babin, P. J., Goizet, C., and Raldúa, D. (2014). Zebrafish models of human motor neuron diseases: Advantages and limitations. Progr. Neurobiol. 118, 36–58. doi: 10.1016/j.pneurobio.2014.03.001
Bailey, E. C., Alrowaished, S. S., Kilroy, E. A., Crooks, E. S., Drinkert, D. M., Karunasiri, C. M., et al. (2019). NAD+ improves neuromuscular development in a zebrafish model of FKRP-associated dystroglycanopathy. Skelet. Muscle 9:21. doi: 10.1186/s13395-019-0206-1
Banerjee, S., Gordon, L., Donn, T. M., Berti, C., Moens, C. B., Burden, S. J., et al. (2011). A novel role for MuSK and non-canonical Wnt signaling during segmental neural crest cell migration. Development 138, 3287–3296. doi: 10.1242/dev.067306
Barbazuk, W. B. (2000). The syntenic relationship of the zebrafish and human genomes. Genome Res. 10, 1351–1358. doi: 10.1101/gr.144700
Basnet, R. M., Zizioli, D., Taweedet, S., Finazzi, D., and Memo, M. (2019). Zebrafish larvae as a behavioral model in neuropharmacology. Biomedicines 7:E23. doi: 10.3390/biomedicines7010023
Bassett, D. I., Bryson-Richardson, R. J., Daggett, D. F., Gautier, P., Keenan, D. G., and Currie, P. D. (2003). Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 130, 5851–5860. doi: 10.1242/dev.00799
Bassett, D., and Currie, P. D. (2004). Identification of a zebrafish model of muscular dystrophy. Clin. Exp. Pharmacol. Physiol. 31, 537–540. doi: 10.1111/j.1440-1681.2004.04030.x
Beattie, C. E. (2000). Control of motor axon guidance in the zebrafish embryo. Brain Res. Bull. 53, 489–500. doi: 10.1016/S0361-9230(00)00382-8
Beattie, C. E., Hatta, K., Halpern, M. E., Liu, H., Eisen, J. S., and Kimmel, C. B. (1997). Temporal separation in the specification of primary and secondary motoneurons in zebrafish. Dev. Biol. 187, 171–182. doi: 10.1006/dbio.1997.8604
Bello-Rojas, S., Istrate, A. E., Kishore, S., and McLean, D. L. (2019). Central and peripheral innervation patterns of defined axial motor units in larval zebrafish. J. Comp. Neurol. 527, 2557–2572. doi: 10.1002/cne.24689
Benedetti, L., Ghilardi, A., Rottoli, E., De Maglie, M., Prosperi, L., Perego, C., et al. (2016). INaP selective inhibition reverts precocious inter- and motorneurons hyperexcitability in the Sod1-G93R zebrafish ALS model. Sci. Rep. 6:24515. doi: 10.1038/srep24515
Berg, E. M., Björnfors, E. R., Pallucchi, I., Picton, L. D., and El Manira, A. (2018). Principles governing locomotion in vertebrates: Lessons from zebrafish. Front. Neural Circuits 12:73. doi: 10.3389/fncir.2018.00073
Berger, J., and Currie, P. D. (2012). Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis. Model Mech. 5, 726–732. doi: 10.1242/dmm.010082
Berger, J., Berger, S., Jacoby, A. S., Wilton, S. D., and Currie, P. D. (2011). Evaluation of exon-skipping strategies for Duchenne muscular dystrophy utilizing dystrophin-deficient zebrafish. J. Cell Mol. Med. 15, 2643–2651. doi: 10.1111/j.1582-4934.2011.01260.x
Boehm, I., Alhindi, A., Leite, A. S., Logie, C., Gibbs, A., Murray, O., et al. (2020). Comparative anatomy of the mammalian neuromuscular junction. J. Anat. 237, 827–836. doi: 10.1111/joa.13260
Bomont, P., Cavalier, L., Blondeau, F., Hamida, C. B., Belal, S., Tazir, M., et al. (2000). The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 26, 370–374. doi: 10.1038/81701
Boon, K.-L., Xiao, S., McWhorter, M. L., Donn, T., Wolf-Saxon, E., Bohnsack, M. T., et al. (2009). Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects. Hum. Mol. Genet. 18, 3615–3625. doi: 10.1093/hmg/ddp310
Boyd, P. J., Tu, W.-Y., Shorrock, H. K., Groen, E. J. N., Carter, R. N., Powis, R. A., et al. (2017). Bioenergetic status modulates motor neuron vulnerability and pathogenesis in a zebrafish model of spinal muscular atrophy. PLoS Genet. 13:e1006744. doi: 10.1371/journal.pgen.1006744
Bragato, C., Gaudenzi, G., Blasevich, F., Pavesi, G., Maggi, L., Giunta, M., et al. (2016). Zebrafish as a model to investigate dynamin 2-related diseases. Sci. Rep. 6:20466. doi: 10.1038/srep20466
Brooke, M. H., and Kaiser, K. K. (1970). Muscle fiber types: How many and what kind? Arch. Neurol. 23, 369–379. doi: 10.1001/archneur.1970.00480280083010
Burke, R. E., Levine, D. N., and Zajac, F. E. (1971). Mammalian motor units: Physiological-histochemical correlation in three types in cat gastrocnemius. Science 174, 709–712. doi: 10.1126/science.174.4010.709
Butti, Z., Pan, Y. E., Giacomotto, J., and Patten, S. A. (2021). Reduced C9orf72 function leads to defective synaptic vesicle release and neuromuscular dysfunction in zebrafish. Commun. Biol. 4:792. doi: 10.1038/s42003-021-02302-y
Buysse, K., Riemersma, M., Powell, G., van Reeuwijk, J., Chitayat, D., Roscioli, T., et al. (2013). Missense mutations in β-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause walker-warburg syndrome. Hum. Mol. Genet. 22, 1746–1754. doi: 10.1093/hmg/ddt021
Cai, H., Shim, H., Lai, C., Xie, C., Lin, X., Yang, W. J., et al. (2008). ALS2/alsin knockout mice and motor neuron diseases. Neurodegener. Dis. 5, 359–366. doi: 10.1159/000151295
Campanari, M.-L., Marian, A., Ciura, S., and Kabashi, E. (2021). TDP-43 regulation of AChE expression can mediate ALS-Like phenotype in zebrafish. Cells 10:221. doi: 10.3390/cells10020221
Chaouch, A., Porcelli, V., Cox, D., Edvardson, S., Scarcia, P., De Grassi, A., et al. (2014). Mutations in the mitochondrial citrate carrier SLC25A1 are associated with impaired neuromuscular transmission. J. Neuromuscul. Dis. 1, 75–90. doi: 10.3233/JND-140021
Chapman, A. L., Bennett, E. J., Ramesh, T. M., De Vos, K. J., and Grierson, A. J. (2013). Axonal transport defects in a mitofusin 2 loss of function model of charcot-marie-tooth disease in zebrafish. PLoS One 8:e67276. doi: 10.1371/journal.pone.0067276
Cheng, L., Guo, X., Yang, X., Chong, M., Cheng, J., Li, G., et al. (2006). Delta-sarcoglycan is necessary for early heart and muscle development in zebrafish. Biochem. Biophys. Res. Commun. 344, 1290–1299. doi: 10.1016/j.bbrc.2006.03.234
Ciura, S., Lattante, S., Le Ber, I., Latouche, M., Tostivint, H., Brice, A., et al. (2013). Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol. 74, 180–187. doi: 10.1002/ana.23946
Colwill, R. M., and Creton, R. (2011). Locomotor behaviors in zebrafish (Danio rerio) larvae. Behav. Process. 86, 222–229. doi: 10.1016/j.beproc.2010.12.003
Da Costa, M. M. J., Allen, C. E., Higginbottom, A., Ramesh, T., Shaw, P. J., and McDermott, C. J. (2014). A new zebrafish model produced by TILLING of SOD1-related amyotrophic lateral sclerosis replicates key features of the disease and represents a tool for in vivo therapeutic screening. Dis. Model Mech. 7, 73–81. doi: 10.1242/dmm.012013
Dequen, F., Bomont, P., Gowing, G., Cleveland, D. W., and Julien, J.-P. (2008). Modest loss of peripheral axons, muscle atrophy and formation of brain inclusions in mice with targeted deletion of gigaxonin exon 1. J. Neurochem. 107, 253–264. doi: 10.1111/j.1471-4159.2008.05601.x
Devoto, S. H., Melançon, E., Eisen, J. S., and Westerfield, M. (1996). Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371–3380. doi: 10.1242/dev.122.11.3371
Dowling, J. J., Low, S. E., Busta, A. S., and Feldman, E. L. (2010). Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum. Mol. Genet. 19, 2668–2681. doi: 10.1093/hmg/ddq153
Dowling, J. J., Vreede, A. P., Low, S. E., Gibbs, E. M., Kuwada, J. Y., Bonnemann, C. G., et al. (2009). Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 5:e1000372. doi: 10.1371/journal.pgen.1000372
Efthymiou, S., Manole, A., and Houlden, H. (2016). Next-generation sequencing in neuromuscular diseases. Curr. Opin. Neurol. 29, 527–536. doi: 10.1097/WCO.0000000000000374
Eisen, J. S., and Smith, J. C. (2008). Controlling morpholino experiments: Don’t stop making antisense. Development 135, 1735–1743. doi: 10.1242/dev.001115
Eisen, J. S., Myers, P. Z., and Westerfield, M. (1986). Pathway selection by growth cones of identified motoneurones in live zebra fish embryos. Nature 320, 269–271. doi: 10.1038/320269a0
Farazi Fard, M. A., Rebelo, A. P., Buglo, E., Nemati, H., Dastsooz, H., Gehweiler, I., et al. (2019). Truncating mutations in UBAP1 cause hereditary spastic paraplegia. Am. J. Hum. Genet. 104, 767–773. doi: 10.1016/j.ajhg.2019.03.001
Farr, G. H., Morris, M., Gomez, A., Pham, T., Kilroy, E., Parker, E. U., et al. (2020). A novel chemical-combination screen in zebrafish identifies epigenetic small molecule candidates for the treatment of Duchenne muscular dystrophy. Skelet. Muscle 10:29. doi: 10.1186/s13395-020-00251-4
Fassier, C., Hutt, J. A., Scholpp, S., Lumsden, A., Giros, B., Nothias, F., et al. (2010). Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat. Neurosci. 13, 1380–1387. doi: 10.1038/nn.2662
Fetcho, J. R. (2007). The utility of zebrafish for studies of the comparative biology of motor systems. J. Exp. Zool. B Mol. Dev. Evol. 308, 550–562. doi: 10.1002/jez.b.21127
Gaj, T., Gersbach, C. A., and Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Ganay, T., Boizot, A., Burrer, R., Chauvin, J., and Bomont, P. (2011). Sensory-motor deficits and neurofilament disorganization in gigaxonin-null mice. Mol. Neurodegener. 6:25. doi: 10.1186/1750-1326-6-25
Gan-Or, Z., Bouslam, N., Birouk, N., Lissouba, A., Chambers, D. B., Vérièpe, J., et al. (2016). Mutations in CAPN1 cause autosomal-recessive hereditary spastic paraplegia. Am. J. Hum. Genet. 98, 1038–1046. doi: 10.1016/j.ajhg.2016.04.002
Gassman, A., Hao, L. T., Bhoite, L., Bradford, C. L., Chien, C.-B., Beattie, C. E., et al. (2013). Small molecule suppressors of Drosophila kinesin deficiency rescue motor axon development in a zebrafish model of spinal muscular atrophy. PLoS One 8:e74325. doi: 10.1371/journal.pone.0074325
Giacomotto, J., Brouilly, N., Walter, L., Mariol, M.-C., Berger, J., Ségalat, L., et al. (2013). Chemical genetics unveils a key role of mitochondrial dynamics, cytochrome c release and IP3R activity in muscular dystrophy. Hum. Mol. Genet. 22, 4562–4578. doi: 10.1093/hmg/ddt302
Gibbs, E. M., Clarke, N. F., Rose, K., Oates, E. C., Webster, R., Feldman, E. L., et al. (2013). Neuromuscular junction abnormalities in DNM2-related centronuclear myopathy. J. Mol. Med. 91, 727–737. doi: 10.1007/s00109-013-0994-4
Goody, M. F., Kelly, M. W., Reynolds, C. J., Khalil, A., Crawford, B. D., and Henry, C. A. (2012). NAD+ biosynthesis ameliorates a zebrafish model of muscular dystrophy. PLoS Biol. 10:e1001409. doi: 10.1371/journal.pbio.1001409
Granato, M., van Eeden, F. J., Schach, U., Trowe, T., Brand, M., Furutani-Seiki, M., et al. (1996). Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123, 399–413.
Gros-Louis, F., Kriz, J., Kabashi, E., McDearmid, J., Millecamps, S., Urushitani, M., et al. (2008). Als2 mRNA splicing variants detected in KO mice rescue severe motor dysfunction phenotype in Als2 knock-down zebrafish. Hum. Mol. Genet. 17, 2691–2702. doi: 10.1093/hmg/ddn171
Gupta, V., Kawahara, G., Gundry, S. R., Chen, A. T., Lencer, W. I., Zhou, Y., et al. (2011). The zebrafish dag1 mutant: A novel genetic model for dystroglycanopathies. Hum. Mol. Genet. 20, 1712–1725. doi: 10.1093/hmg/ddr047
Gupta, V. A., Kawahara, G., Myers, J. A., Chen, A. T., Hall, T. E., Manzini, M. C., et al. (2012). A splice site mutation in laminin-α2 results in a severe muscular dystrophy and growth abnormalities in zebrafish. PLoS One 7:e43794. doi: 10.1371/journal.pone.0043794
Guyon, J. R., Goswami, J., Jun, S. J., Thorne, M., Howell, M., Pusack, T., et al. (2009). Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum. Mol. Genet. 18, 202–211. doi: 10.1093/hmg/ddn337
Guyon, J. R., Mosley, A. N., Zhou, Y., O’Brien, K. F., Sheng, X., Chiang, K., et al. (2003). The dystrophin associated protein complex in zebrafish. Hum. Mol. Genet. 12, 601–615.
Guyon, J. R., Steffen, L. S., Howell, M. H., Pusack, T. J., Lawrence, C., and Kunkel, L. M. (2007). Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta 1772, 205–215. doi: 10.1016/j.bbadis.2006.07.003
Guyon, J., Mosley, A., Jun, S., Montanaro, F., Steffen, L., Zhou, Y., et al. (2005). δ-Sarcoglycan is required for early zebrafish muscle organization. Exp. Cell Res. 304, 105–115. doi: 10.1016/j.yexcr.2004.10.032
Hall, T. E., Bryson-Richardson, R. J., Berger, S., Jacoby, A. S., Cole, N. J., Hollway, G. E., et al. (2007). The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. U.S.A. 104, 7092–7097. doi: 10.1073/pnas.0700942104
Hao, L. T., Burghes, A. H., and Beattie, C. E. (2011). Generation and characterization of a genetic zebrafish model of SMA carrying the human SMN2 gene. Mol. Neurodegener. 6:24. doi: 10.1186/1750-1326-6-24
Hao, L. T., Wolman, M., Granato, M., and Beattie, C. E. (2012). Survival motor neuron affects plastin 3 protein levels leading to motor defects. J. Neurosci. 32, 5074–5084. doi: 10.1523/JNEUROSCI.5808-11.2012
Herman, I., Lopez, M. A., Marafi, D., Pehlivan, D., Calame, D. G., Abid, F., et al. (2021). Clinical exome sequencing in the diagnosis of pediatric neuromuscular disease. Muscle Nerve 63, 304–310. doi: 10.1002/mus.27112
Hewamadduma, C. A. A., Grierson, A. J., Ma, T. P., Pan, L., Moens, C. B., Ingham, P. W., et al. (2013). Tardbpl splicing rescues motor neuron and axonal development in a mutant tardbp zebrafish. Hum. Mol. Genet. 22, 2376–2386. doi: 10.1093/hmg/ddt082
Hirata, H., Watanabe, T., Hatakeyama, J., Sprague, S. M., Saint-Amant, L., Nagashima, A., et al. (2007). Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 134, 2771–2781. doi: 10.1242/dev.004531
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Jagla, K., Kalman, B., Boudou, T., Hénon, S., and Batonnet-Pichon, S. (2017). Beyond mice: Emerging and transdisciplinary models for the study of early-onset myopathies. Semin. Cell Dev. Biol. 64, 171–180. doi: 10.1016/j.semcdb.2016.09.012
Jing, L., Gordon, L. R., Shtibin, E., and Granato, M. (2010). Temporal and spatial requirements of unplugged/MuSK function during zebrafish neuromuscular development. PLoS One 5:e8843. doi: 10.1371/journal.pone.0008843
Jing, L., Lefebvre, J. L., Gordon, L. R., and Granato, M. (2009). Wnt signals organize synaptic prepattern and axon guidance through the zebrafish unplugged/MuSK receptor. Neuron 61, 721–733. doi: 10.1016/j.neuron.2008.12.025
Johnson, N. M., Farr, G. H., and Maves, L. (2013). The HDAC inhibitor TSA ameliorates a zebrafish model of duchenne muscular dystrophy. PLoS Curr. 5:ecurrents.md.8273cf41db10e2d15dd3ab827cb4b027. doi: 10.1371/currents.md.8273cf41db10e2d15dd3ab827cb4b027
Jones, R. A., Harrison, C., Eaton, S. L., Llavero Hurtado, M., Graham, L. C., Alkhammash, L., et al. (2017). Cellular and molecular anatomy of the human neuromuscular junction. Cell Rep. 21, 2348–2356. doi: 10.1016/j.celrep.2017.11.008
Joshi, S., Virdi, S., Etard, C., Geisler, R., and Strähle, U. (2018). Mutation of a serine near the catalytic site of the choline acetyltransferase a gene almost completely abolishes motility of the zebrafish embryo. PLoS One 13:e0207747. doi: 10.1371/journal.pone.0207747
Kabashi, E., Bercier, V., Lissouba, A., Liao, M., Brustein, E., Rouleau, G. A., et al. (2011). FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 7:e1002214. doi: 10.1371/journal.pgen.1002214
Kabashi, E., Lin, L., Tradewell, M. L., Dion, P. A., Bercier, V., Bourgouin, P., et al. (2010). Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 19, 671–683. doi: 10.1093/hmg/ddp534
Kanning, K. C., Kaplan, A., and Henderson, C. E. (2010). Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440. doi: 10.1146/annurev.neuro.051508.135722
Kawahara, G., Guyon, J. R., Nakamura, Y., and Kunkel, L. M. (2010). Zebrafish models for human FKRP muscular dystrophies. Hum. Mol. Genet. 19, 623–633. doi: 10.1093/hmg/ddp528
Kawahara, G., Serafini, P. R., Myers, J. A., Alexander, M. S., and Kunkel, L. M. (2011). Characterization of zebrafish dysferlin by morpholino knockdown. Biochem. Biophys. Res. Commun. 413, 358–363. doi: 10.1016/j.bbrc.2011.08.105
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kohashi, T., and Oda, Y. (2008). Initiation of Mauthner- or non-Mauthner-mediated fast escape evoked by different modes of sensory input. J. Neurosci. 28, 10641–10653. doi: 10.1523/JNEUROSCI.1435-08.2008
Kummer, T. T., Misgeld, T., and Sanes, J. R. (2006). Assembly of the postsynaptic membrane at the neuromuscular junction: Paradigm lost. Curr. Opin. Neurobiol. 16, 74–82. doi: 10.1016/j.conb.2005.12.003
Laird, A. S., Van Hoecke, A., De Muynck, L., Timmers, M., Van den Bosch, L., Van Damme, P., et al. (2010). Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One 5:e13368. doi: 10.1371/journal.pone.0013368
Lattante, S., de Calbiac, H., Le Ber, I., Brice, A., Ciura, S., and Kabashi, E. (2015). Sqstm1 knock-down causes a locomotor phenotype ameliorated by rapamycin in a zebrafish model of ALS/FTLD. Hum. Mol. Genet. 24, 1682–1690. doi: 10.1093/hmg/ddu580
Lemmens, R., Van Hoecke, A., Hersmus, N., Geelen, V., D’Hollander, I., Thijs, V., et al. (2007). Overexpression of mutant superoxide dismutase 1 causes a motor axonopathy in the zebrafish. Hum. Mol. Genet. 16, 2359–2365. doi: 10.1093/hmg/ddm193
Lescouzères, L., and Bomont, P. (2020). E3 ubiquitin ligases in neurological diseases: Focus on gigaxonin and autophagy. Front. Physiol. 11:1022. doi: 10.3389/fphys.2020.01022
Lescouzères, L., Hassen-Khodja, C., Baudot, A., Bordignon, B., and Bomont, P. (Submitted). A Multi-Disciplinary Screening Pipeline in Zebrafish Identifies Therapeutic Drugs for GAN.
Li, L., Zhou, Q., Voss, T. C., Quick, K. L., and LaBarbera, D. V. (2016). High-throughput imaging: Focusing in on drug discovery in 3D. Methods 96, 97–102. doi: 10.1016/j.ymeth.2015.11.013
Li, M., Andersson-Lendahl, M., Sejersen, T., and Arner, A. (2013). Knockdown of desmin in zebrafish larvae affects interfilament spacing and mechanical properties of skeletal muscle. J. Gen. Physiol. 141, 335–345. doi: 10.1085/jgp.201210915
Li, M., Hromowyk, K. J., Amacher, S. L., and Currie, P. D. (2017). Muscular dystrophy modeling in zebrafish. Methods Cell Biol. 138, 347–380. doi: 10.1016/bs.mcb.2016.11.004
Lin, P., Li, J., Liu, Q., Mao, F., Li, J., Qiu, R., et al. (2008). A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42). Am. J. Hum. Genet. 83, 752–759. doi: 10.1016/j.ajhg.2008.11.003
Lin, Y.-Y., White, R. J., Torelli, S., Cirak, S., Muntoni, F., and Stemple, D. L. (2011). Zebrafish Fukutin family proteins link the unfolded protein response with dystroglycanopathies. Hum. Mol. Genet. 20, 1763–1775. doi: 10.1093/hmg/ddr059
Luna, V. M., Daikoku, E., and Ono, F. (2015). “Slow” skeletal muscles across vertebrate species. Cell Biosci. 5:62. doi: 10.1186/s13578-015-0054-6
Lv, X., Zhang, R., Xu, L., Wang, G., Yan, C., and Lin, P. (2022). Tcap deficiency in zebrafish leads to ROS production and mitophagy, and idebenone improves its phenotypes. Front. Cell Dev. Biol. 10:836464. doi: 10.3389/fcell.2022.836464
MacRae, C. A., and Peterson, R. T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14, 721–731. doi: 10.1038/nrd4627
Manuel, M., and Zytnicki, D. (2011). Alpha, beta and gamma motoneurons: Functional diversity in the motor system’s final pathway. J. Integr. Neurosci. 10, 243–276. doi: 10.1142/S0219635211002786
Manzini, M. C., Tambunan, D. E., Hill, R. S., Yu, T. W., Maynard, T. M., Heinzen, E. L., et al. (2012). Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am. J. Hum. Genet. 91, 541–547. doi: 10.1016/j.ajhg.2012.07.009
Mao, F., Li, Z., Zhao, B., Lin, P., Liu, P., Zhai, M., et al. (2015). Identification and functional analysis of a SLC33A1: c.339T>G (p.Ser113Arg) variant in the original SPG42 family. Hum. Mutat. 36, 240–249. doi: 10.1002/humu.22732
Martin, E., Schüle, R., Smets, K., Rastetter, A., Boukhris, A., Loureiro, J. L., et al. (2013). Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am. J. Hum. Genet. 92, 238–244. doi: 10.1016/j.ajhg.2012.11.021
Martin, E., Yanicostas, C., Rastetter, A., Alavi Naini, S. M., Maouedj, A., Kabashi, E., et al. (2012). Spatacsin and spastizin act in the same pathway required for proper spinal motor neuron axon outgrowth in zebrafish. Neurobiol. Dis. 48, 299–308. doi: 10.1016/j.nbd.2012.07.003
McCammon, J. M., and Sive, H. (2015). Challenges in understanding psychiatric disorders and developing therapeutics: A role for zebrafish. Dis. Model Mech. 8, 647–656. doi: 10.1242/dmm.019620
McLean, D. L., and Fetcho, J. R. (2008). Using imaging and genetics in zebrafish to study developing spinal circuits in vivo. Dev. Neurobiol. 68, 817–834. doi: 10.1002/dneu.20617
McMacken, G., Cox, D., Roos, A., Müller, J., Whittaker, R., and Lochmüller, H. (2018). The beta-adrenergic agonist salbutamol modulates neuromuscular junction formation in zebrafish models of human myasthenic syndromes. Hum. Mol. Genet. 27, 1556–1564. doi: 10.1093/hmg/ddy062
McWhorter, M. L., Monani, U. R., Burghes, A. H. M., and Beattie, C. E. (2003). Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 162, 919–931. doi: 10.1083/jcb.200303168
Menelaou, E., and McLean, D. L. (2012). A gradient in endogenous rhythmicity and oscillatory drive matches recruitment order in an axial motor pool. J. Neurosci. 32, 10925–10939. doi: 10.1523/JNEUROSCI.1809-12.2012
Mirat, O., Sternberg, J. R., Severi, K. E., and Wyart, C. (2013). ZebraZoom: An automated program for high-throughput behavioral analysis and categorization. Front. Neural Circuits 7:107. doi: 10.3389/fncir.2013.00107
Müller, J. S., Jepson, C. D., Laval, S. H., Bushby, K., Straub, V., and Lochmüller, H. (2010). Dok-7 promotes slow muscle integrity as well as neuromuscular junction formation in a zebrafish model of congenital myasthenic syndromes. Hum. Mol. Genet. 19, 1726–1740. doi: 10.1093/hmg/ddq049
Myers, P. Z. (1985). Spinal motoneurons of the larval zebrafish. J. Comp. Neurol. 236, 555–561. doi: 10.1002/cne.902360411
Naef, V., Mero, S., Fichi, G., D’Amore, A., Ogi, A., Gemignani, F., et al. (2019). Swimming in deep water: Zebrafish modeling of complicated forms of hereditary spastic paraplegia and spastic ataxia. Front. Neurosci. 13:1311. doi: 10.3389/fnins.2019.01311
Noldus, L. P., Spink, A. J., and Tegelenbosch, R. A. (2001). EthoVision: A versatile video tracking system for automation of behavioral experiments. Behav. Res. Methods Instrum. Comput. 33, 398–414. doi: 10.3758/bf03195394
O’Connor, E., Phan, V., Cordts, I., Cairns, G., Hettwer, S., Cox, D., et al. (2018). MYO9A deficiency in motor neurons is associated with reduced neuromuscular agrin secretion. Hum. Mol. Genet. 27, 1434–1446. doi: 10.1093/hmg/ddy054
O’Connor, E., Töpf, A., Müller, J. S., Cox, D., Evangelista, T., Colomer, J., et al. (2016). Identification of mutations in the MYO9A gene in patients with congenital myasthenic syndrome. Brain 139, 2143–2153. doi: 10.1093/brain/aww130
Oprişoreanu, A.-M., Smith, H. L., Arya, S., Webster, R., Zhong, Z., Eaton-Hart, C., et al. (2019). Interaction of axonal chondrolectin with collagen XIXa1 is necessary for precise neuromuscular junction formation. Cell Rep. 29, 1082–1098.e10. doi: 10.1016/j.celrep.2019.09.033
Oprişoreanu, A.-M., Smith, H. L., Krix, S., Chaytow, H., Carragher, N. O., Gillingwater, T. H., et al. (2021). Automated in vivo drug screen in zebrafish identifies synapse-stabilising drugs with relevance to spinal muscular atrophy. Dis. Model Mech. 14:dmm047761. doi: 10.1242/dmm.047761
Osborn, D. P. S., Pond, H. L., Mazaheri, N., Dejardin, J., Munn, C. J., Mushref, K., et al. (2017). Mutations in INPP5K cause a form of congenital muscular dystrophy overlapping marinesco-sjögren syndrome and dystroglycanopathy. Am. J. Hum. Genet. 100, 537–545. doi: 10.1016/j.ajhg.2017.01.019
Panzer, J. A., Gibbs, S. M., Dosch, R., Wagner, D., Mullins, M. C., Granato, M., et al. (2005). Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Dev. Biol. 285, 340–357. doi: 10.1016/j.ydbio.2005.06.027
Panzer, J. A., Song, Y., and Balice-Gordon, R. J. (2006). In vivo imaging of preferential motor axon outgrowth to and synaptogenesis at prepatterned acetylcholine receptor clusters in embryonic zebrafish skeletal muscle. J. Neurosci. 26, 934–947. doi: 10.1523/JNEUROSCI.3656-05.2006
Pappalardo, A., Pitto, L., Fiorillo, C., Alice Donati, M., Bruno, C., and Santorelli, F. M. (2013). Neuromuscular disorders in zebrafish: State of the art and future perspectives. Neuromol. Med. 15, 405–419. doi: 10.1007/s12017-013-8228-z
Parsons, M. J., Campos, I., Hirst, E. M. A., and Stemple, D. L. (2002). Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development 129, 3505–3512. doi: 10.1242/dev.129.14.3505
Patten, S. A., Armstrong, G. A. B., Lissouba, A., Kabashi, E., Parker, J. A., and Drapeau, P. (2014). Fishing for causes and cures of motor neuron disorders. Dis. Model Mech. 7, 799–809. doi: 10.1242/dmm.015719
Patton, E. E., Zon, L. I., and Langenau, D. M. (2021). Zebrafish disease models in drug discovery: From preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 20, 611–628. doi: 10.1038/s41573-021-00210-8
Peterson, R. T., and Fishman, M. C. (2011). Designing zebrafish chemical screens. Methods Cell Biol. 105, 525–541. doi: 10.1016/B978-0-12-381320-6.00023-0
Pipis, M., Rossor, A. M., Laura, M., and Reilly, M. M. (2019). Next-generation sequencing in Charcot-Marie-Tooth disease: Opportunities and challenges. Nat. Rev. Neurol. 15, 644–656. doi: 10.1038/s41582-019-0254-5
Postel, R., Vakeel, P., Topczewski, J., Knöll, R., and Bakkers, J. (2008). Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev. Biol. 318, 92–101. doi: 10.1016/j.ydbio.2008.03.024
Radev, Z., Hermel, J.-M., Elipot, Y., Bretaud, S., Arnould, S., Duchateau, P., et al. (2015). A TALEN-Exon skipping design for a bethlem myopathy model in zebrafish. PLoS One 10:e0133986. doi: 10.1371/journal.pone.0133986
Ramesh, T., Lyon, A. N., Pineda, R. H., Wang, C., Janssen, P. M. L., Canan, B. D., et al. (2010). A genetic model of amyotrophic lateral sclerosis in zebrafish displays phenotypic hallmarks of motoneuron disease. Dis. Model Mech. 3, 652–662. doi: 10.1242/dmm.005538
Reddy, G., Desban, L., Tanaka, H., Roussel, J., Mirat, O., and Wyart, C. (2022). A lexical approach for identifying behavioural action sequences. PLoS Comput. Biol. 18:e1009672. doi: 10.1371/journal.pcbi.1009672
Rennekamp, A. J., and Peterson, R. T. (2015). 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 24, 58–70. doi: 10.1016/j.cbpa.2014.10.025
Robinson, K. J., Yuan, K. C., Don, E. K., Hogan, A. L., Winnick, C. G., Tym, M. C., et al. (2019). Motor neuron abnormalities correlate with impaired movement in zebrafish that express mutant superoxide dismutase 1. Zebrafish 16, 8–14. doi: 10.1089/zeb.2018.1588
Roscioli, T., Kamsteeg, E.-J., Buysse, K., Maystadt, I., van Reeuwijk, J., van den Elzen, C., et al. (2012). Mutations in ISPD cause walker-warburg syndrome and defective glycosylation of α-dystroglycan. Nat Genet 44, 581–585. doi: 10.1038/ng.2253
Ruparelia, A. A., Oorschot, V., Ramm, G., and Bryson-Richardson, R. J. (2016). FLNC myofibrillar myopathy results from impaired autophagy and protein insufficiency. Hum. Mol. Genet. 25, 2131–2142. doi: 10.1093/hmg/ddw080
Ruparelia, A. A., Zhao, M., Currie, P. D., and Bryson-Richardson, R. J. (2012). Characterization and investigation of zebrafish models of filamin-related myofibrillar myopathy. Hum. Mol. Genet. 21, 4073–4083. doi: 10.1093/hmg/dds231
Sakowski, S. A., Lunn, J. S., Busta, A. S., Oh, S. S., Zamora-Berridi, G., Palmer, M., et al. (2012). Neuromuscular effects of G93A-SOD1 expression in zebrafish. Mol. Neurodegener. 7:44. doi: 10.1186/1750-1326-7-44
Sarparanta, J., Jonson, P. H., Golzio, C., Sandell, S., Luque, H., Screen, M., et al. (2012). Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 44, S1–S2. doi: 10.1038/ng.1103
Schmid, B., and Haass, C. (2013). Genomic editing opens new avenues for zebrafish as a model for neurodegeneration. J. Neurochem. 127, 461–470. doi: 10.1111/jnc.12460
Schmid, B., Hruscha, A., Hogl, S., Banzhaf-Strathmann, J., Strecker, K., van der Zee, J., et al. (2013). Loss of ALS-associated TDP-43 in zebrafish causes muscle degeneration, vascular dysfunction, and reduced motor neuron axon outgrowth. Proc. Natl. Acad. Sci. U.S.A. 110, 4986–4991. doi: 10.1073/pnas.1218311110
See, K., Yadav, P., Giegerich, M., Cheong, P. S., Graf, M., Vyas, H., et al. (2014). SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum. Mol. Genet. 23, 1754–1770. doi: 10.1093/hmg/ddt567
Senderek, J., Müller, J. S., Dusl, M., Strom, T. M., Guergueltcheva, V., Diepolder, I., et al. (2011). Hexosamine biosynthetic pathway mutations cause neuromuscular transmission defect. Am. J. Hum. Genet. 88, 162–172. doi: 10.1016/j.ajhg.2011.01.008
Serafini, P. R., Feyder, M. J., Hightower, R. M., Garcia-Perez, D., Vieira, N. M., Lek, A., et al. (2018). A limb-girdle muscular dystrophy 2I model of muscular dystrophy identifies corrective drug compounds for dystroglycanopathies. JCI Insight 3:e120493. doi: 10.1172/jci.insight.120493
Seth, A., Stemple, D. L., and Barroso, I. (2013). The emerging use of zebrafish to model metabolic disease. Dis. Model Mech. 6, 1080–1088. doi: 10.1242/dmm.011346
Shaw, M. P., Higginbottom, A., McGown, A., Castelli, L. M., James, E., Hautbergue, G. M., et al. (2018). Stable transgenic C9orf72 zebrafish model key aspects of the ALS/FTD phenotype and reveal novel pathological features. Acta Neuropathol. Commun. 6:125. doi: 10.1186/s40478-018-0629-7
Slater, C. R. (2017). The structure of human neuromuscular junctions: Some unanswered molecular questions. Int. J. Mol. Sci. 18:E2183. doi: 10.3390/ijms18102183
Song, Y., Wang, M., Mao, F., Shao, M., Zhao, B., Song, Z., et al. (2013). Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis. Model Mech. 6, 404–413. doi: 10.1242/dmm.009688
Swaminathan, A., Bouffard, M., Liao, M., Ryan, S., Callister, J. B., Pickering-Brown, S. M., et al. (2018). Expression of C9orf72-related dipeptides impairs motor function in a vertebrate model. Hum. Mol. Genet. 27, 1754–1762. doi: 10.1093/hmg/ddy083
Swenarchuk, L. E. (2019). Nerve, muscle, and synaptogenesis. Cells 8:E1448. doi: 10.3390/cells8111448
Swinnen, B., Bento-Abreu, A., Gendron, T. F., Boeynaems, S., Bogaert, E., Nuyts, R., et al. (2018). A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. 135, 427–443. doi: 10.1007/s00401-017-1796-5
Telfer, W. R., Busta, A. S., Bonnemann, C. G., Feldman, E. L., and Dowling, J. J. (2010). Zebrafish models of collagen VI-related myopathies. Hum. Mol. Genet. 19, 2433–2444. doi: 10.1093/hmg/ddq126
Thornhill, P., Bassett, D., Lochmüller, H., Bushby, K., and Straub, V. (2008). Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP). Brain 131, 1551–1561. doi: 10.1093/brain/awn078
Valdmanis, P. N., Meijer, I. A., Reynolds, A., Lei, A., MacLeod, P., Schlesinger, D., et al. (2007). Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152–161. doi: 10.1086/510782
van der Meer, D. L. M., Marques, I. J., Leito, J. T. D., Besser, J., Bakkers, J., Schoonheere, E., et al. (2006). Zebrafish cypher is important for somite formation and heart development. Dev. Biol. 299, 356–372. doi: 10.1016/j.ydbio.2006.07.032
Vaz, R., Hofmeister, W., and Lindstrand, A. (2019). zebrafish models of neurodevelopmental disorders: Limitations and benefits of current tools and techniques. Int. J. Mol. Sci. 20:E1296. doi: 10.3390/ijms20061296
Vettori, A., Bergamin, G., Moro, E., Vazza, G., Polo, G., Tiso, N., et al. (2011). Developmental defects and neuromuscular alterations due to mitofusin 2 gene (MFN2) silencing in zebrafish: A new model for Charcot-Marie-Tooth type 2A neuropathy. Neuromuscul. Disord. 21, 58–67. doi: 10.1016/j.nmd.2010.09.002
Vissing, J., Johnson, K., Töpf, A., Nafissi, S., Díaz-Manera, J., French, V. M., et al. (2019). POPDC3 gene variants associate with a new form of limb girdle muscular dystrophy. Ann. Neurol. 86, 832–843. doi: 10.1002/ana.25620
Westerfield, M., McMurray, J. V., and Eisen, J. S. (1986). Identified motoneurons and their innervation of axial muscles in the zebrafish. J. Neurosci. 6, 2267–2277.
Westra, D., Schouten, M. I., Stunnenberg, B. C., Kusters, B., Saris, C. G. J., Erasmus, C. E., et al. (2019). Panel-based exome sequencing for neuromuscular disorders as a diagnostic service. J. Neuromuscul. Dis. 6, 241–258. doi: 10.3233/JND-180376
Weterman, M. A. J., Sorrentino, V., Kasher, P. R., Jakobs, M. E., van Engelen, B. G. M., Fluiter, K., et al. (2012). A frameshift mutation in LRSAM1 is responsible for a dominant hereditary polyneuropathy. Hum. Mol. Genet. 21, 358–370. doi: 10.1093/hmg/ddr471
Widrick, J. J., Alexander, M. S., Sanchez, B., Gibbs, D. E., Kawahara, G., Beggs, A. H., et al. (2016). Muscle dysfunction in a zebrafish model of Duchenne muscular dystrophy. Physiol. Genomics 48, 850–860. doi: 10.1152/physiolgenomics.00088.2016
Winkler, C., Eggert, C., Gradl, D., Meister, G., Giegerich, M., Wedlich, D., et al. (2005). Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 19, 2320–2330. doi: 10.1101/gad.342005
Wolman, M., and Granato, M. (2012). Behavioral genetics in larval zebrafish: Learning from the young. Dev. Neurobiol. 72, 366–372. doi: 10.1002/dneu.20872
Wood, J. D., Landers, J. A., Bingley, M., McDermott, C. J., Thomas-McArthur, V., Gleadall, L. J., et al. (2006). The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum. Mol. Genet. 15, 2763–2771. doi: 10.1093/hmg/ddl212
Wu, H., Xiong, W. C., and Mei, L. (2010). To build a synapse: Signaling pathways in neuromuscular junction assembly. Development 137, 1017–1033. doi: 10.1242/dev.038711
Xu, L., Geng, H., Lv, X., Wang, G., Yan, C., Zhang, D., et al. (2022). A female carrier of spinal and bulbar muscular atrophy diagnosed with DNAJB6-related distal myopathy. J. Hum. Genet. 67, 441–444. doi: 10.1038/s10038-022-01022-3
Yeh, T.-H., Liu, H.-F., Li, Y.-W., Lu, C.-S., Shih, H.-Y., Chiu, C.-C., et al. (2018). C9orf72 is essential for neurodevelopment and motility mediated by Cyclin G1. Exp. Neurol. 304, 114–124. doi: 10.1016/j.expneurol.2018.03.002
Zhang, R., Yang, J., Zhu, J., and Xu, X. (2009). Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere–membrane interaction, not sarcomere assembly. Hum. Mol. Genet. 18, 4130–4140. doi: 10.1093/hmg/ddp362
Zhao, M., Smith, L., Volpatti, J., Fabian, L., and Dowling, J. J. (2019). Insights into wild-type dynamin 2 and the consequences of DNM2 mutations from transgenic zebrafish. Hum. Mol. Genet. 28, 4186–4196. doi: 10.1093/hmg/ddz260
Zhong, Z., Ohnmacht, J., Reimer, M. M., Bach, I., Becker, T., and Becker, C. G. (2012). Chondrolectin mediates growth cone interactions of motor axons with an intermediate target. J. Neurosci. 32, 4426–4439. doi: 10.1523/JNEUROSCI.5179-11.2012
Zhou, Y., Cattley, R. T., Cario, C. L., Bai, Q., and Burton, E. A. (2014). Quantification of larval zebrafish motor function in multiwell plates using open-source MATLAB applications. Nat. Protoc. 9, 1533–1548. doi: 10.1038/nprot.2014.094
Zivony-Elboum, Y., Westbroek, W., Kfir, N., Savitzki, D., Shoval, Y., Bloom, A., et al. (2012). A founder mutation in Vps37A causes autosomal recessive complex hereditary spastic paraparesis. J. Med. Genet. 49, 462–472. doi: 10.1136/jmedgenet-2012-100742
Zon, L. I., and Peterson, R. T. (2005). In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 4, 35–44. doi: 10.1038/nrd1606
Keywords: zebrafish model, imaging methodology, neuromuscular system, disease mechanisms, drug screening
Citation: Lescouzères L, Bordignon B and Bomont P (2022) Development of a high-throughput tailored imaging method in zebrafish to understand and treat neuromuscular diseases. Front. Mol. Neurosci. 15:956582. doi: 10.3389/fnmol.2022.956582
Received: 30 May 2022; Accepted: 16 August 2022;
Published: 20 September 2022.
Edited by:
Kinga Gawel, Medical University of Lublin, PolandReviewed by:
Catherina Becker, Technical University Dresden, GermanyCopyright © 2022 Lescouzères, Bordignon and Bomont. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pascale Bomont, cGFzY2FsZS5ib21vbnRAaW5zZXJtLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.