
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 07 July 2022
Sec. Pain Mechanisms and Modulators
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.926596
This article is part of the Research Topic Molecular Mechanisms of Nociception View all 14 articles
 Wenting Ma1†
Wenting Ma1† Matthew R. Sapio1†
Matthew R. Sapio1† Allison P. Manalo1
Allison P. Manalo1 Dragan Maric2
Dragan Maric2 Mary Kate Dougherty1
Mary Kate Dougherty1 Taichi Goto1,3
Taichi Goto1,3 Andrew J. Mannes1
Andrew J. Mannes1 Michael J. Iadarola1*
Michael J. Iadarola1*Primary afferent neurons of the dorsal root ganglia (DRG) transduce peripheral nociceptive signals and transmit them to the spinal cord. These neurons also mediate analgesic control of the nociceptive inputs, particularly through the μ-opioid receptor (encoded by Oprm1). While opioid receptors are found throughout the neuraxis and in the spinal cord tissue itself, intrathecal administration of μ-opioid agonists also acts directly on nociceptive nerve terminals in the dorsal spinal cord resulting in marked analgesia. Additionally, selective chemoaxotomy of cells expressing the TRPV1 channel, a nonselective calcium-permeable ion channel that transduces thermal and inflammatory pain, yields profound pain relief in rats, canines, and humans. However, the relationship between Oprm1 and Trpv1 expressing DRG neurons has not been precisely determined. The present study examines rat DRG neurons using high resolution multiplex fluorescent in situ hybridization to visualize molecular co-expression. Neurons positive for Trpv1 exhibited varying levels of expression for Trpv1 and co-expression of other excitatory and inhibitory ion channels or receptors. A subpopulation of densely labeled Trpv1+ neurons did not co-express Oprm1. In contrast, a population of less densely labeled Trpv1+ neurons did co-express Oprm1. This finding suggests that the medium/low Trpv1 expressing neurons represent a specific set of DRG neurons subserving the opponent processes of both transducing and inhibiting nociceptive inputs. Additionally, the medium/low Trpv1 expressing neurons co-expressed other markers implicated in pathological pain states, such as Trpa1 and Trpm8, which are involved in chemical nociception and cold allodynia, respectively, as well as Scn11a, whose mutations are implicated in familial episodic pain. Conversely, none of the Trpv1+ neurons co-expressed Spp1, which codes for osteopontin, a marker for large diameter proprioceptive neurons, validating that nociception and proprioception are governed by separate neuronal populations. Our findings support the hypothesis that the population of Trpv1 and Oprm1 coexpressing neurons may explain the remarkable efficacy of opioid drugs administered at the level of the DRG-spinal synapse, and that this subpopulation of Trpv1+ neurons is responsible for registering tissue damage.
The nociceptive system is a major target of interest for the design of analgesic drugs. When administered at this level of the pain pathway, analgesic agents such as opioids and local anesthetics ultimately act on primary afferent endings by inhibiting presynaptic release of nociceptive signaling molecules, such as substance P and calcitonin gene-related peptide (CGRP) (Julius and Basbaum, 2001; Kondo et al., 2005). A better understanding of which afferents are required for nociception and what constellation of receptors are expressed by these neurons is an important component of designing alternative strategies to selectively inhibit nociception while preserving other somatosensory functions.
Transient receptor potential vanilloid subfamily, member 1 (TRPV1) is expressed by primary afferent nociceptive neurons (Caterina et al., 1997; Szallasi and Blumberg, 1999; Caterina, 2007), and transduces thermal and inflammatory tissue damage stimuli (Karai et al., 2004a; Mishra and Hoon, 2010; Mitchell et al., 2010, 2014). In the search for novel non-opioid analgesics, much attention has been centered on the TRPV1 molecule as an analgesic target (Jancso et al., 1977; Yaksh et al., 1979; Bevan et al., 1992; Urban et al., 2000; Wong and Gavva, 2009; Iadarola and Gonnella, 2013). Genetic ablation or chemical silencing of TRPV1+ afferents has been reported to cause loss of nociceptive responses (Jancso et al., 1977; Karai et al., 2004a; Brown et al., 2005, 2015; Mishra and Hoon, 2010; Brown and Iadarola, 2015; Sapio et al., 2018). However, direct antagonism of the TRPV1 receptor has not led to clinically useful antinociceptive agents, despite promising preclinical rodent studies and demonstrable target engagement in humans (Menéndez et al., 2006; Szallasi et al., 2007; Rowbotham et al., 2011; Quiding et al., 2013). These drugs did not advance in part due to induction of varying degrees of hyperthermia (Wong and Gavva, 2009; Garami et al., 2018; Park et al., 2021), and lack of sustained efficacy (Quiding et al., 2013), as TRPV1 is not the only nociceptive transducing molecule expressed by primary afferents (Goswami et al., 2014). In the complex milieu of tissue damage and inflammation, multiple other algesic mediators remain capable of discharging the primary afferents, even with a high degree of TRPV1 inhibition, which may explain why TRPV1 antagonists are much less analgesic than agonist-mediated chemoaxotomy approaches (Karai et al., 2004a; Iadarola et al., 2018; Sapio et al., 2018). Additionally, patients treated with high concentrations of TRPV1 antagonists become insensitive to noxious hot thermal stimuli, posing a burn risk as they rated temperatures up to 49°C as safe (Rowbotham et al., 2011). Thus, TRPV1+ afferents are the critical transducers of pain associated with tissue damage as well as most forms of warm and hot thermosensation.
Untangling desirable and undesirable pharmacologic effects of TRPV1-based therapies requires clarification of the underlying neurobiology. An interesting observation is that selective lesioning of TRPV1+ nerve endings using the highly potent TRPV1 agonist, resiniferatoxin (RTX), leads to loss of thermal nociceptive sensitivity and with high subcutaneous doses (250 ng intraplantar) can produce near-total insensitivity to tissue-damaging stimuli in the denervated area (Mitchell et al., 2010, 2014). While it is known that TRPV1+ nociceptors comprise several discrete populations of sensory afferent neurons (Sapio et al., 2018), detailed studies of nociceptive cellular neurobiology are needed to elucidate the precise molecular markers of each population. The objective of the present investigations is to delineate the molecular complement of receptors and ion channels in neurons responsible for nociception and analgesic action. Specifically, we hypothesize that we can identify a population of cells using multiplex labeling that contains several nociceptive and analgesic markers, that maybe useful for future studies of pain and analgesia. Delineating these neurons in a quantitative, molecularly-defined fashion enables future studies to directly target these neurons using selective pharmacological analgesic strategies.
Experiments in the present study were approved by the Institutional Animal Care and Use Committee of the Clinical Center, National Institutes of Health (Bethesda, Maryland). Male Sprague-Dawley rats 7–8 weeks old weighing 175–200 g (Charles River Laboratories, Wilmington, MA) were used for all experiments. Rats were housed in pairs with a plastic tube for enrichment/housing. At the time of tissue collection, rats were euthanized under deep isoflurane anesthesia (≥4%) and perfused with cold intracardiac PBS followed by 10% formalin. Bilateral lumbar DRGs were harvested at the level of L3–L6. DRGs were fixed in 10% formalin for 24 h and subsequently transferred to 2.5% formalin before embedding (within 3–4 days). DRG tissues were embedded in paraffin blocks and cut to 6 μm sections and mounted on slides by Histoserv, Inc. (Germantown, MD). For these experiments DRGs from N = 6 rats were collected; with N = 3 stained, analyzed, and scanned for most analyses. Cell types identified were found in approximately the same proportions in each individual animal. Multiple windows on the sections of DRG were selected to capture representative fields for quantification. Sample sizes were estimated from previous experiments (Sapio et al., 2020b).
Fluorescent RNA in situ hybridization was performed using RNAScope® Multiplex Fluorescent V2 Assay (Advanced Cell Diagnostics, Newark, CA) in accordance with the manufacturer's instructions for treating formalin-fixed paraffin-embedded tissue. Target retrieval was performed for 15 mins at 100°C. The catalog numbers of the probes used in these experiments are listed in Table 1. Note that color balancing was performed based on sFPKM data (Sapio et al., 2016) to put weaker signals in channels with the least background autofluorescence (higher wavelengths) to avoid signal bleed. To avoid cross reaction between opioid receptors, the kappa opioid receptor (Oprk1) probe was custom designed to be specific when used together with the other opioid receptor detection probes. This was accomplished by targeting base pairs 1,300–2,293 of NM_017167.3. To validate a neuronal population exhibiting a low signal for Trpm8 hybridization, a second probe was used against a different region of the transcript. This was targeted to base pairs 2,671–3,590 of NM_134371.3.
Hybridized slides were imaged using an Axio Imager.Z2 scanning fluorescence microscope (Zeiss, Oberkochen, Germany), fitted with an ORCA-Flash4.0s CMOS sensor high resolution digital camera (Hamamatsu, Shizuoka, Japan), 20X/0.8 Plan-Apochromat (Phase-2) non-immersion objective (Zeiss), and 200W X-Cite 200DC broad band lamp source (Excelitas Technologies, Waltham MA). Filter sets (Semrock, Rochester NY) for detecting DAPI, Opal520, Opal570, Opal620, and Opal690 fluorescent dyes (Opal Reagent Systems; PerkinElmer, Waltham MA) were custom furnished as described previously (Maric et al., 2021) (Supplementary Table 1). Image tiles sized 600 × 600 μm were captured and seamlessly stitched using ZEN2 imaging software (Zeiss) at 3 pixel/μm spatial resolution. For each emission wavelength, fluorescent microscopy images were captured and displayed as individual layers to generate multi-colored composites. Representative images are enhanced for visibility.
The images were processed with Adobe Photoshop and Fiji (Image J v2.1.0/1.53c) in order to analyze the co-localization patterns of genes that are detected in the multiplex in situ hybridization experiments. For quantification of signal intensity inside individual DRG neurons, polygons were drawn based on the bright field image in a multichannel overlay using Fiji, and fluorescence intensity of this polygonal area was measured. The neuronal diameter was extrapolated from the polygonal area using the formula for the diameter of a circle [diameter = 2√ (area/π)]. For cell counting, cells were counted manually in Photoshop, and tabulated counts were used to plot visualizations of intersecting sets in R (v4.05) using the UpSet package. In these plots, the number of cells positive for each of the four markers is plotted as “set size” and the number of cells co-positive for each given combination of markers is shown as “intersection size.” Cells were identified using a combination of DAPI-labeling and diffusion image contrast imaging which revealed cellular outline. A single 6 μm section was counted for each stain per rat to avoid counting the same cell twice in serial sections. Statistical testing of diameter measurements was conducted using Prism GraphPad (Version 9.3.0, Kruskal-Wallis test with Dunn's multiple comparison test). In these experiments, data from each individual rat was examined before pooling to ensure homogeneity, and the individual values are shown in Supplementary Figure 1. Proportions of cells positive for Oprm1, Oprk1, Oprd1, and Trpv1 markers were determined in a pairwise manner and represented as percentages relative to the total number of cells counted.
To distinguish cell populations by levels of Trpv1, Oprm1, Oprd1, and Trpm8 expression, a surface plot was generated in Fiji to visualize fluorescence intensity per pixel. Surface plots have three axes, with the x and y axes corresponding to the physical dimension of the slide section, and the z-dimension, or height of the surface representing intensity. Intensity in these plots is also represented by color in a flame scale.
For quantitative graphs, each channel was checked for non-specific signal coming from neighboring channels and subtracted if signal bleed was detected to quantify only specific fluorescent signal.
Semiquantitative analysis was performed to estimate the number of cells present or absent for the mRNA encoding the thermosensory nociceptive ion channel Trpv1 and each of the three opioid receptors (Oprm1, Oprk1, and Oprd1). This analysis subdivided the sensory afferents into 12 populations, though not all were prevalent (Figure 1A). In order to concisely depict the overlap between Trpv1 and opioid receptor transcripts, a pairwise matrix of percentages was constructed (Figure 1B). The most prevalent binary combination of markers was Trpv1 and Oprm1, comprising 60.4% of all neurons (ignoring categorization by the other markers). The κ-opioid receptor (encoded by Oprk1) was identified with ~15% of neurons in combination with either Trpv1 or Oprm1. The δ-opioid receptor (Oprd1) was less prevalent, and the rarest combination were neurons co-positive for δ- and κ-opioid receptors (5.4%). These results are based on the aggregated quantification of 755 cells from N = 3 rats. The bottom 15 panels show representative images of the distinct co-expressing populations of Trpv1+/Oprm1+ neurons (Figure 1C), Trpv1+/Oprm1+/Oprk1+ neurons (Figure 1D), and Oprm1+/Oprd1+ neurons (Figure 1E).
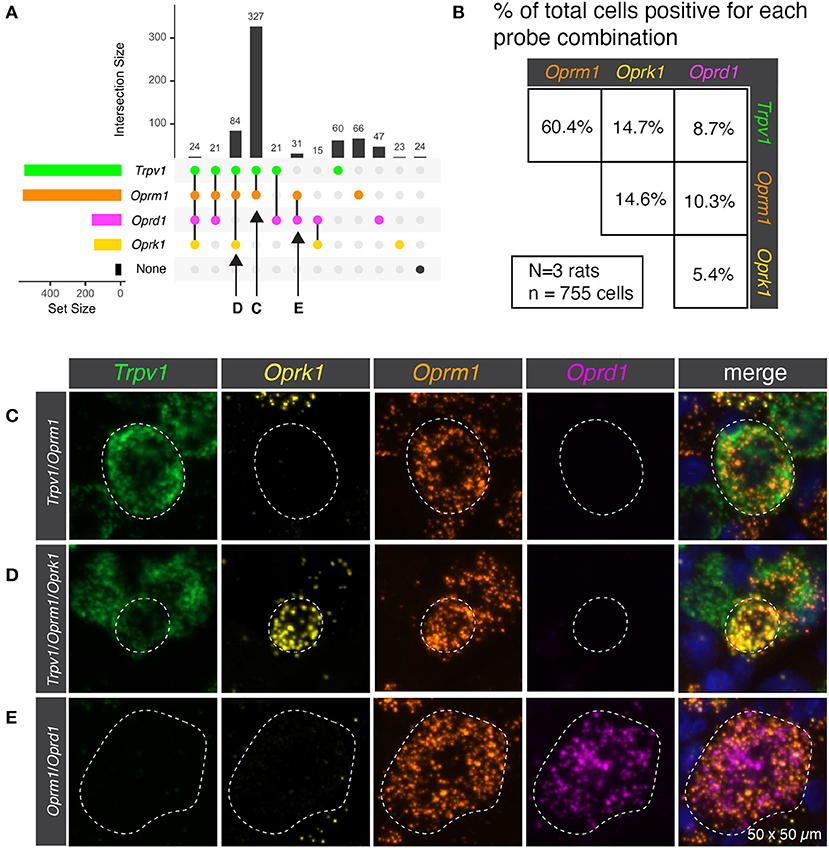
Figure 1. Co-expression of transient receptor potential vanilloid 1 (Trpv1) and the three opioid receptors (Oprm1, Oprk1, Oprd1). (A) Semiquantitative analysis was performed to count the number of cells present or absent for each of the four marker genes. This analysis subdivided the sensory afferents into 12 populations, although not all of these populations were prevalent. (B) In order to simplify the overlap between Trpv1 and Opioid receptor transcripts, a pairwise matrix of percentages is shown. This shows the total number of neurons positive for each marker (set size) as well as the number of cells positive for combinations of markers (intersection size; see Methods section). This display clarifies that Trpv1+/Oprm1+ cells were very prevalent in the DRG, comprising 60.4% of all neurons (ignoring categorization by the other markers.). While all possible combinations were identified, some were quite rare, with the population of cells positive for both Oprk1 and Oprd1 (the rarest combination) comprising ~5% of neurons. (C) Representative multi-channel microscopy images are shown for three of the populations identified in panel A, the first of which being Trpv1+/Oprm1+ neurons. These neurons were the most abundant population at 327 of 755 total cells (or 43.3%). (D) The Trpv1+/Oprm1+/Oprk1+ population is shown, which is the second most abundant subpopulation at 84/755 cells (11.1%). (E) Finally, a representative cell from the Oprm1+/Oprd1+ population is shown. This population is rare, but is large and expresses very high levels of both Oprm1 and Oprd1. Outlines of neurons are shown to enhance visibility (dotted lines).
While examining binary expression patterns (presence vs. absence), we noted substantial variation in expression of Trpv1 transcript. Previously, it has been reported that Trpv1 expression level is high in small diameter thermosensitive C-fibers, consistent with our observation that the highest intensity signals were observed within sparsely distributed small diameter neurons (Figure 2A, arrows). Based on this variation in Trpv1 expression levels we quantified the fluorescence intensity for Trpv1 to further characterize if quantitative expression levels of this ion channel gene indicated cell type differences. A lower intensity of Trpv1 fluorescent signal was evident in many other cells (Figures 2B,C, arrowheads), consistent with the observation of multiple Trpv1+ neuronal populations containing a range of expression (Figure 2D) (Cavanaugh et al., 2011a; Goswami et al., 2014; Mitchell et al., 2014). Quantification of fluorescence intensity revealed three loosely separated populations (inflection points or “knees,” in the plot of Figure 2D; N = 3 rats, n = 146 cells) corresponding to high, medium, and low expressing Trpv1+ neurons, consistent with previous findings from single-cell RNA-Seq (Usoskin et al., 2015; Li et al., 2016; Sapio et al., 2018). A surface plot was generated to visualize the range of expression profiles in the DRG neurons. Note that the high expressors appear as isolated peaks in this view, indicating that they are quantitatively separate from other Trpv1+ neurons (Figure 2E, arrows). Additionally, high intensity Trpv1+ neurons were smaller in diameter than the lower intensity Trpv1+ neurons (Figure 2F). In imaging Trpv1+ neurons, the exposure was set to minimize oversaturation of the signal at the highest level of expression, leading to apparent low signal in some cells. This is visualized showing the raw data (Figure 2G, upper row) and an enhanced image in which brightness is adjusted (Figure 2G, lower row), which shows the wide dynamic range of the signal. Note that even lower expressing Trpv1+ neurons contain a high enough density of puncta to distinguish their signal from background.
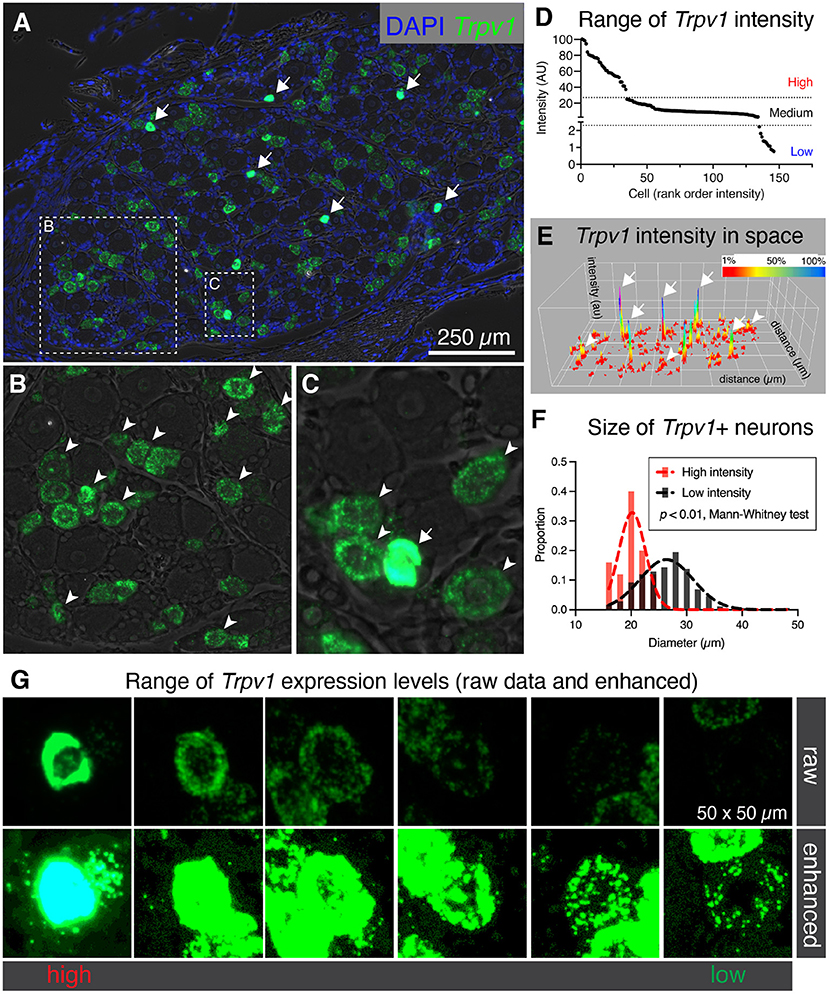
Figure 2. Heterogeneous labeling of Trpv1 mRNA in rat dorsal root ganglion (DRG). (A) Fluorescent in situ hybridization was performed and imaged for whole rat DRG sections (N = 3). Very high levels of expression were observed in a small number of small diameter neurons (white arrows). (B) An enlargement of a neuron-rich region is shown with Trpv1+ neurons (arrowheads). (C) A second enlargement includes one high-expressing Trpv1+ neuron (arrow) and several other Trpv1+ neurons with moderate staining intensity (arrowheads). (D) Intensity values for Trpv1 were plotted by rank order. Based on these values, Trpv1 expression was divided into high, medium and low expression. (E) A surface plot was generated for Trpv1 intensity (arbitrary units, Z-axis) across the entire DRG section shown in (A). This plot shows the relationship between the high-expressing Trpv1 DRG neurons and other cells in the ganglion. Note the high peaks indicating a quantitatively separate population (white arrows). (F) Diameter of the high intensity Trpv1+ neurons was examined by measuring the area in Fiji. High intensity Trpv1+ neurons had a stereotyped small diameter as represented by the narrow Gaussian (red) relative to the broader distribution of medium/low Trpv1+ neurons (black Gaussian). This difference was significant based on a Mann-Whitney test (p < 0.01). (G) Representative fields of 6 Trpv1+ DRG neurons are shown, spanning a range of expression levels. Scanning parameters are tuned so as not to saturate the brightest cells. Note that the range of expression values is such that when the lower expressing Trpv1+ neurons are visible, the highest cells are saturated.
The finding that Trpv1 was unevenly distributed in rat DRG prompted an investigation of Oprm1 and Oprd1. We also examined the distribution of expression intensity for two of the analgesic opioid receptors, the μ-opioid receptor transcript (Oprm1), and the delta opioid receptor transcript (Oprd1), which are responsible for opioid analgesic responses in vivo (reviewed in Valentino and Volkow, 2018). In a wide field we show an enhanced zoomed out image (brightened for clarity; Figures 3A,B). From this zoomed out field we did not observe a similar evident difference in expression level analogous to what was observed for Trpv1 in either Oprm1 or Oprd1. Note that Oprm1 is expressed in a much greater number of neurons than Oprd1. In order to quantify this further, we used a surface plot to represent intensity over area for Oprm1 (Figures 3C,D) and Oprd1 (Figures 3E,F). In the angled view, the peaks represent intensity for staining with these labels, and generally fit the overall expression pattern of these receptors with evident peaks corresponding to the majority of positive cells. This is in contrast to what was observed for Trpv1, which had a small number of highly evident peaks on a linear axis (Figure 2E). Rank order plots were created to quantify expression, and test whether coefficient of variation differed between measured intensity values for Trpv1, Oprm1 and Oprd1. These analyses showed a greater coefficient of variation in Trpv1 intensity as compared with Oprm1 or Oprd1 (Supplementary Figure 2).
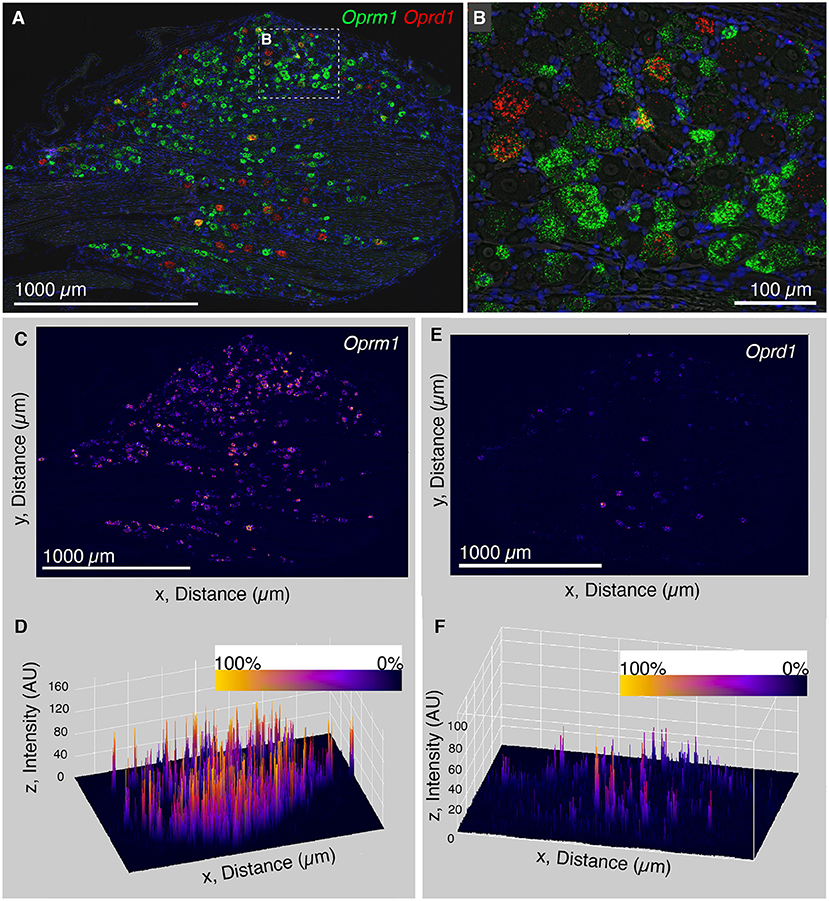
Figure 3. Analysis of intensity distribution for the opioid receptor genes Oprm1 and Oprd1. The finding that Trpv1 was unevenly distributed in rat DRG prompted an investigation of Oprm1 and Oprd1 to answer whether these analgesic opioid receptors were also distributed unevenly across cell populations. (A) In a wide field view of a DRG stained for Oprm1 and Oprd1, we did not observe marked differences in expression across the field. The gene encoding the mu-opioid receptor (Oprm1) is much more widely expressed than the delta receptor gene (Opd1), and both show a range of expression values. (B) However, this range appeared qualitative smaller than that observed for Trpv1. (C) In order to quantify this further, we used a surface plot to represent intensity over area for Oprm1. (D) When viewed from an angle, this analysis shows many peaks of similar height across the DRG, consistent with the wide expression pattern of Oprm1. This is in contrast to Trpv1, which showed several very high peaks (Figure 2E). (E) The same analysis was also performed for Oprd1, which is expressed in many fewer cells than either Trpv1 or Oprm1. (F) Viewed at an angle, most of these Oprd1 peaks are evident in the same linear axis. In Supplementary Figure 2, we quantified differences in distribution and coefficient of variation in these measurements, showing that Trpv1 had significantly higher coefficient of variation than either Oprm1 or Oprd1.
RNA-Seq of rat dorsal root ganglia showed modest expression of Oprm1 and comparatively higher expression of the opioid-related nociceptin receptor 1 (Oprl1) gene (7.9 and 25.8 sFPKM respectively) (Sapio et al., 2016). This receptor binds the opioid-like heptadecapeptide nociceptin, derived from the pronociceptin precursor and has sequence similarity (Phe-Gly-Gly-Phe-Thr-Gly-Ala-Arg-Lys-Ser-Ala-Arg-Lys-Leu-Ala-Asn-Gln) to dynorphin A1-17 (Tyr-Gly-Gly-Phe-Leu-Arg-Arg-Ile-Arg-Pro-Lys-Leu-Lys-Trp-Asp-Asn-Gln). To assess the overlap between expression of Oprm1 and Oprl1 and the nociceptive transducing ion channel genes Trpv1 and Trpa1, multiplex fluorescent in situ hybridization was performed for these four transcripts together. The plot in Figure 4A is derived from 805 cells, counted in 3 sections from 3 animals. Co-expression was assessed for each of the 4 genes. The most prevalent neuronal type identified by these four labels was Oprl1 alone neurons, which accounted for 272 cells (33.8%). When coexpression was taken into account, Oprl1 was observed to be expressed in a majority of DRG neurons (71.6%). Cells positive for all four labels (133 cells, 16.5%) were also among the most prevalent co-expression combinations, indicating a strong overlap between the nociceptive ion channels Trpv1 and Trpa1 with opioid receptor genes Oprm1 and Oprl1. Populations containing the nociceptive marker gene Trpv1 and Oprm1 with or without Oprl1 (59 cells with all three and 147 cells with Trpv1/Oprm1 only) were also prevalent, reinforcing the overlap between this thermal nociceptive ion channel and the main opioid analgesia-producing receptor. A representative panel of neurons is shown in Figure 4B, with neurons positive for all 4 labels indicated with arrows, and Trpv1+/Oprm1+ neurons indicated with arrowheads. These arrows/arrowheads are maintained in all four channels to show individual signal. The overlap between the two nociceptive ion channel genes (Trpv1 and Trpa1) is visualized in Figure 4C. Within these populations, a wide range of signal intensity was detected for Oprm1 indicating variable expression of the μ-opioid receptor gene in the populations described with this 4-plex labeling scheme. In the visualization of Oprl1 alone, note the broad labeling of many primary afferent neurons (Figure 4E). Analogous to the visualization in Figure 4C, the two opioid receptors (Oprm1 and Oprl1) detected in this staining panel were visualized together in Figure 4F. Note the incomplete overlap between these two labels.

Figure 4. Co-expression patterns of nociceptive ion channels and opioid receptors. To assess the overlap between nociceptive transducing ion channels and opioid receptors, multiplex fluorescent hybridization was performed for Trpv1, Trpa1, Oprm1 and Oprl1. This analysis was performed in N = 3 rats total, and 805 cells. (A) Counts were tabulated for co-expression of these four markers in each cell, and the intersections of these are plotted. The most prevalent count group was Oprl1 alone, due to the near-ubiquitous nature of this marker (272 cells). Cells positive for all four labels (133 cells) were also prevalent, indicating overlap between nociceptive ion channels (Trpv1 and Trpa1) and the mu-opioid receptor Oprm1 as well as Oprl1. Populations containing Trpv1 and Oprm1 with or without Oprl1 (59 cells with all three and 147 cells with Trpv1/Oprm1 only) were also prevalent, reinforcing the overlap between these two markers. (B) A representative panel showing a diverse field of cells is shown, with quad-positive neurons highlighted (arrows). Additionally, Trpv1+/Oprm1+ cells are indicated (arrowheads). (C–F) To increase the visibility of the highly multiplexed image, individual channels and channel pairs are shown for the image in (B), with the channel identity found in the bottom right of each panel. (C) A panel showing only Trpv1 and Trpa1 was selected to show the relationship between these two nociceptive ion channels. (D) A range of intensities was observed for Oprm1. Note the presence of less prominent puncta in Trpv1+/Oprm1+ cells (arrowheads). (E) Oprl1 alone. (F) Oprm1 and Oprl1. Scale bar applies to all images.
Individual neuronal perikarya of three distinct populations were examined by multiplex fluorescent in situ hybridization (Figure 5A). The high Trpv1 expressing neurons were selected out for separate analysis based on their unique signature of Trpv1 expression (see Figure 2). The major populations colocalizing Trpv1+/Oprm1+ were low and medium expressing Trpv1 neurons (middle row, Figure 5A Trpv1 medium). Similarly, while not all low Trpv1 expressors are positive for all four labels, we focused on such quad+ neurons for visualization (bottom row, Figure 5A Trpv1 low). This is the population that coexpressed mRNA encoding the chemo-nociceptive ion channel Trpa1. As we observed that the cells labeled with all four markers were generally low for Trpv1, we examined the relationship between Trpv1 and Trpa1 in greater detail. In the representative field (Figure 5B), we observed that Trpv1 and Trpa1 were generally not strongly expressed in the same cells, although some co-positive cells were observed with low levels of both markers. Note the cells strongly expressing Trpv1 (arrowheads) or Trpa1 (arrows), as well as cells co-expressing both markers (asterisks). Cell counts by marker positivity were performed to evaluate the findings of our peripheral ganglionic comparison study between the nodose, DRG and superior cervical ganglia (Sapio et al., 2020b) (Figure 5C). We found that the most prominent populations of cells were Trpv1+/Trpa1- (Trpa1-; 233 cells, 53.8%) followed closely by the faintly labeled Trpv1+/Trpa1+ (190 cells, 43.9%) population. The Trpv1-/Trpa1+ cells (10 cells, 2.3%) comprised the rarest subgroup. In a quantitative analysis of expression levels, Trpv1 and Trpa1 intensity measurements tended toward the axes (Figure 5D, shaded areas), indicating an inverse correlation with expression levels. Note that no cells are identified in the center of the graph, which would indicate bright co-expression of these two markers. Pairwise coincidence is shown for each of the four labels in this experiment (Figure 5E). All of these combinations are common (with the rarest being approximately 19.1% of all cells) for Trpa1 and Oprm1. In these analyses, we tried to identify differences among the high Trpv1+ population, and noticed that they seemed less likely to express high levels of Oprm1. This was formally quantified, and is displayed in pie charts (Figure 5F) showing that the subgroup of Trpv1+ high-expressing neurons (N = 33) expressed Oprm1 only 18.1% of the time, whereas Trpv1+ neurons overall expressed Oprm1 85.7% of the time. We also noted that these cells expressed a low level of Oprm1 on average (Supplementary Figure 3).

Figure 5. Quantification of nociceptive ion channel and opioid receptors. We further examined the subpopulations of DRG neurons shown in Figure 3. (A) Enlarged and enhanced fields were shown for each of three representative cell populations corresponding to high, medium and low Trpv1 expression. Note that several populations of cells have medium Trpv1 expression, but that one of the most common subtypes (Trpv1 and Oprm1 alone), was selected. Similarly, a quad-positive neuron was selected as representative of low Trpv1 expression. (B) To examine the relationship between Trpv1 and Trpa1 in particular, expression levels in perikarya of cells expressing either marker were examined in further detail. In this representative field, neurons strongly enriched for Trpv1 (arrowheads) or Trpa1 (arrow) are indicated, as well as two neurons (asterisks) that co-express these two ion channel genes. (C) Using cell counting, we identified the coincidence of these two markers. More cells express Trpv1 than Trpa1 with the majority of Trpa1+ cells co-expressing Trpv1, and only 10 Trpa1+ cells expressing no detectable Trpv1. (D) However, cells expressing high levels of either Trpv1 or Trpa1 express very low levels of the other transcript. Therefore, the expression of these two markers while coincident, appears to be anticorrelated (note that points fall on the axes rather than in the middle of the plot. (E) Pairwise coincidence is shown for each of the four labels in this experiment. All of these combinations are common (with the rarest being approximately 19.1% of all cells). High Trpv1 neurons, in particular, seemed to differ from other Trpv1 neurons in terms of Oprm1 expression. (F) We addressed this by counting high Trpv1 neurons separately from other Trpv1 neurons (shown in pie charts). Whereas average Trpv1 neurons expressed Oprm1 the majority of the time (85.7%), the high Trpv1 neurons expressed Oprm1 only 18.1% of the time. On average, the expression in these cells was also lower (Supplementary Figure 3). Outlines (dotted lines) of neuronal perikarya are shown to enhance visbility.
In addition to Trpv1, several other temperature sensitive ion channels are implicated in noxious stimuli transduction and nociceptive signaling in baseline and disease states (reviewed in Koivisto et al., 2022). These ion channel genes are useful markers of nociceptive cell populations and have been targeted for therapeutic interventions. While Trpv1 responds to temperatures in the warm to hot temperature range, another ion channel, Trpm8 is activated by cold temperatures and participates in cold allodynia (Bautista et al., 2007; Knowlton et al., 2010). Multiplex fluorescent in situ hybridization revealed heterogeneous expression of the Trpm8 mRNA, with most of the signal concentrated within sparsely distributed, small diameter neurons (Figure 6A, arrows). The results for Trpm8 expression were similar to what was observed for Trpv1 in Figure 2, showing a range of expression consisting of a small subpopulation of brightly labeled neurons and a second set of neurons with weaker signal. An enlarged visualization of Trpm8 alone (Figures 6B,C) showed the densely labeled small to medium sized neurons (arrows), as well as additional neurons showing less intense labeling (arrowheads). The convergence of transcripts was examined using multiplex fluorescent in situ hybridization with four labels (Trpv1, Trpm8, and Oprm1). This staining combination revealed that virtually all Trpm8+ neurons co-expressed Oprm1, albeit at a range of expression values as seen in the representative field (Figure 6D). A surface plot was generated to visualize the range of expression profiles in the DRG neurons. Note that the high expressors appear as isolated peaks in this view, indicating that they are quantitatively separable from other Trpm8+ neurons (Figure 6E). Also note that the lower expressing Trpm8+ neurons required enhancement for visualization, indicating they express Trpm8 at a much lower level than the high-expressing neurons. This lower expression was validated by using a second Trpm8 probe to rule out origination from nonspecific signal (Supplementary Figure 3).
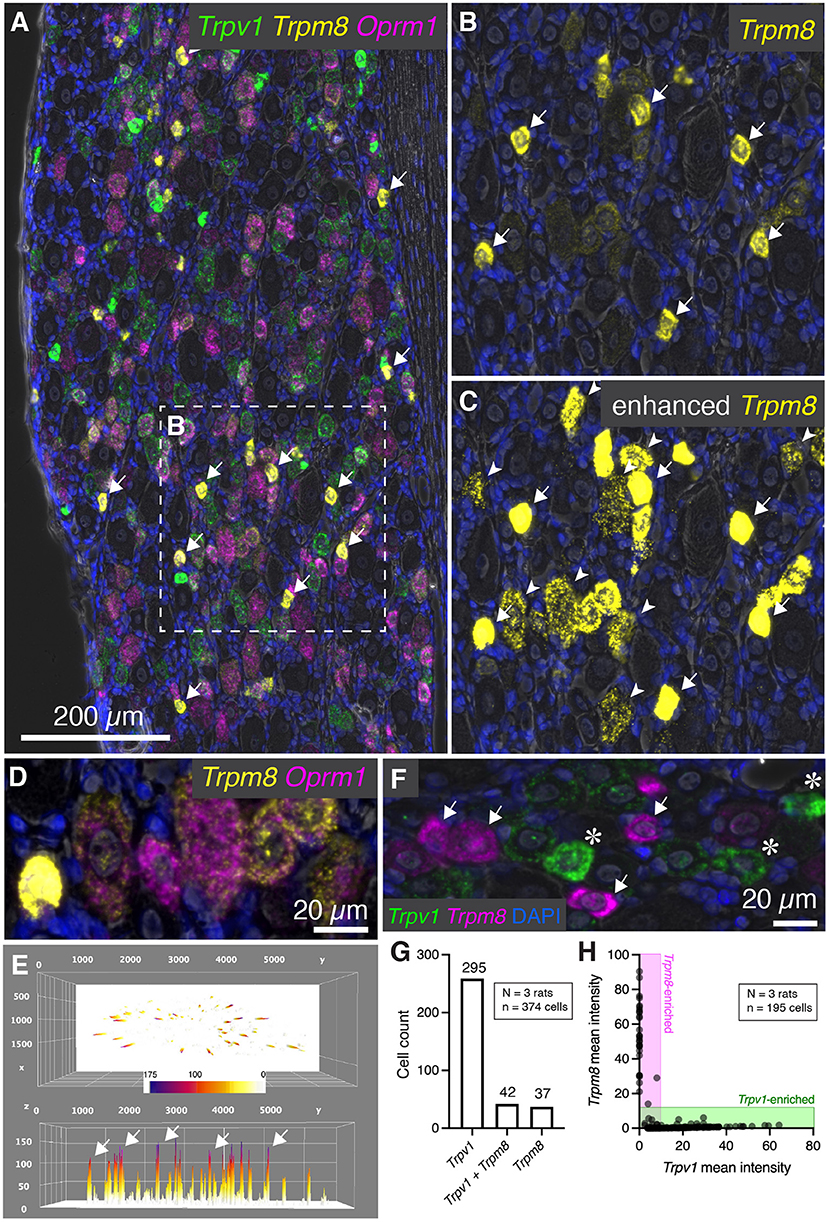
Figure 6. Characterization of Trpm8 expression levels in rat DRG. To assess the overlap between hot and cold thermal nociceptive transducing ion channels, multiplex fluorescent hybridization was performed for Trpv1, and Trpm8 alongside Oprm1. (A) Panoramic view of triple-labeled rat DRG. (B) Cells positive for Trpm8 were brightly labeled small diameter neurons (white arrows). However, a number of other neurons were also visible with lower levels of Trpm8. (C) Similar to what was performed for Trpv1, enhancement of the image saturates the high-expressing Trpm8 neurons, but allows for visualization of the much lower intensity staining observed in other neurons. Note that these neurons still have clear punctate staining pattern and that this staining pattern was reproduced with a different probe for confirmation (Supplementary Figure 4). (D) Another characteristic of Trpm8 neurons is that 100% of them co-express Oprm1 to some extent. This relationship is visualized in the representative field. (E) A surface plot of Trpm8 expression levels is shown with arrows pointing to the highest expressing Trpm8+ neurons. Note the lower peaks span a range of expression levels. (F) The relationship between Trpv1 and Trpm8 was further explored by quantifying levels of expression of these two thermal transducing ion channels shown in the representative field. (G) The Trpv1+ population was much larger than the Trpm8+ population, and approximately half of Trpm8+ neurons co-expressed Trpv1. (H) However, with quantification we saw that no cell co-expressed these markers strongly, and that their expression was anticorrelated (note that the points fall on the axes rather than in the center of the plot).
We further examined the co-expression pattern of the hot and cold thermosensory ion channel transcripts Trpv1 and Trpm8, respectively. In the representative image (Figure 6F), densely labeled Trpm8+ (arrows) and Trpv1+ neurons (asterisks) are indicated. Co-expression analysis showed that the most prominent population was the Trpv1+/Trpm8- population (295 cells, 76.0%), as Trpv1 is more broadly expressed than Trpm8 (Figure 5G). The Trpv1+/Trpm8+ co-positive cells (42 cells, 10.8%) and Trpm8+ populations (37 cells, 9.5%) were less abundant, with approximately half of Trpm8+ cells coexpressing Trpv1. Similar to what was shown for Trpv1 and Trpa1, fluorescence intensities of Trpm8 vs. Trpv1 signals tended to plot along the axes (shaded areas, Figure 6H) indicating that strong expression of either marker gene was observed mainly in separate sensory neurons (cell quantification from N=3 rats). This indicates that strongly Trpv1+ neurons never strongly expressed Trpm8, and strongly Trpm8+ neurons never strongly express Trpv1.
To further assess the overlap between nociceptive transducing ion channels and opioid receptors, multiplex fluorescent in situ hybridization was performed for the thermally responsive ion channels Trpv1 and Trpm8, in conjunction with the μ-opioid receptor gene Oprm1. An additional marker of nociceptive neurons, the voltage-gated sodium channel subunit (NaV1.9) gene, Scn11a, was also included. Human mutations in SCN11A cause pain insensitivity or episodic pain syndromes depending on the nature of the mutation (Leipold et al., 2013; Zhang et al., 2013). The total number of cells counted was 570 from N = 3 rats. Upon quantification of co-expression of these four markers, the most prevalent subgroup was composed of neurons co-positive for Trpv1, Oprm1 and Scn11a (197 cells 34.6%, Figure 7A). Notably, many neurons were also unlabeled (167 cells 29.2%). In a panoramic visualization (Figure 7B) of these cell populations, a representative field was chosen to highlight the staining patterns. Examples of the most prevalent cell type (Trpv1+/Oprm1+/Scn11a+) are indicated with arrowheads (Figures 7C–G). Large diameter cells with no reactivity for these markers are also indicated (asterisks). A putative thermosensory C-fiber with very high levels of Trpv1 is indicated (arrow). This type of cell does not show reactivity for the other three markers, indicating minimum co-localization of either nociceptive or analgesic markers in the highest expressing Trpv1+ neurons. Note the varying levels of Oprm1 in Trpv1+/Oprm1+/Scn11a+ neurons (arrowheads), which could indicate heterogeneity in this labeled class. In an overlay of Trpv1 and Trpm8 (Figure 7E), note that cells were generally high in one or the other but not both markers indicating the specialized role of Trpm8 in cold thermonociception and cold pain, and its localization within a small, discrete subclass of sensory afferents in the rat. Consistent with its role in nociception, the sodium channel subunit Scn11a was prevalent in Trpv1+/Oprm1+/Scn11a+ neurons (arrowheads), as well as strongly expressed in many cells throughout the ganglion. We also considered the Trpm8+ cell populations quantitatively (Supplementary Figure 4), showing that high/medium Trm8+ cells were negative for Trpv1 and Scn11a, whereas low expressing Trpm8+ cells have low levels of these markers.
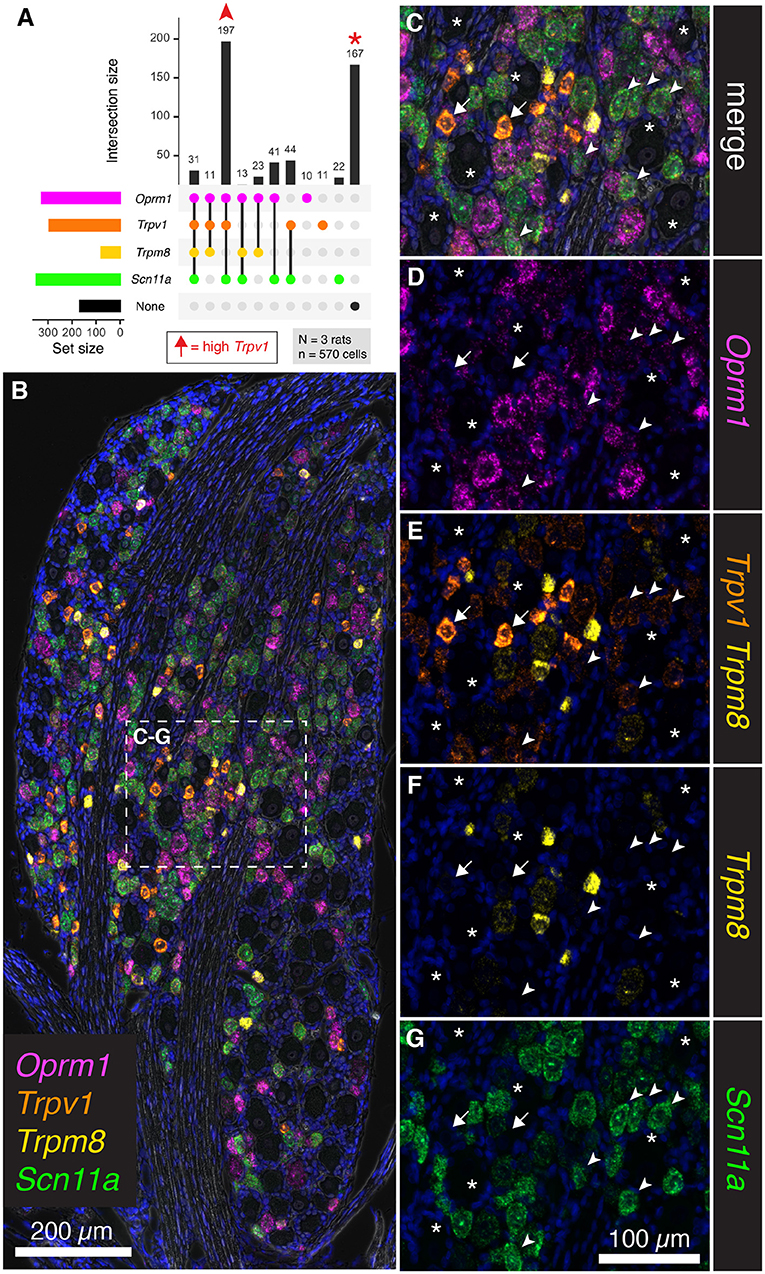
Figure 7. Co-expression analysis for Trpv1, Trpm8, Oprm1, and Scn11a. To further assess the overlap between nociceptive transducing ion channels and opioid receptors, multiplex fluorescent hybridization was performed for Trpv1, Trpm8, Oprm1 and Scn11a in N = 3 rats (n = 570 cells). In this experiment, we investigated the mRNA coding for the cold-pain transducing nociceptive ion channel Trpm8 as well as the mRNA encoding the voltage-gated ion channel subunit Scn11a. (A) Counts were tabulated for co-expression of these four markers as before. The most prevalent group were neurons co-positive for Trpv1, Oprm1 and Scn11a (197 cells). Notably, a large number of neurons was also unlabeled (167 cells). Arrows in the plot in (A) are used to identify cells in the representative fields in subsequent panels. High Trpv1 neurons (arrows) are not in the plot in (A), but were highlighted in the representative fields. (B) A panoramic stitched image is shown to demonstrate the anatomical location of these markers within the greater structure. In the center of the field, a representative area was enlarged for subsequent panels. (C) Overlay of the 4-plex label with DAPI and brightfield is shown. Note that examples of the most prevalent cell type, Trpv1+/Oprm1+/Scn11a+ neurons are indicated (arrowheads). Larger diameter cells with no reactivity for these markers are indicated (asterisks). A singular putative thermosensing c-fiber is indicated with very high levels of Trpv1 (arrow). (D). Note the varying levels of Oprm1 in Trpv1+/Oprm1+/Scn11a+ neurons (arrowheads), and absence of Oprm1 in the high-expressing Trpv1+ neuron (arrow). (E) In an overlay of Trpv1 and Trpm8, note that cells were generally high in one or the other but not both markers. (F) Also note the sparse labeling of Trpm8, indicative of its specialized function. (G) The sodium channel subunit Scn11a was prevalent in Trpv1+/Oprm1+/Scn11a+ neurons (arrowheads), as well as strongly expressed in many cells throughout the ganglion. Scale bar applies to all images. Scale bar applies to (C–G).
To extend the multiplex fluorescent in situ hybridization staining experiments to include a non-nociceptive marker, we examined co-expression of algesic and analgesic markers with osteopontin (Spp1), which labels large diameter neurons (with an average diameter of approximately 38 μm). These neurons are thought to be chiefly implicated in proprioception (Ichikawa et al., 2000; Usoskin et al., 2015; Saito-Diaz et al., 2021). In the co-expression plot in Figure 8A, all of the Spp1+ neurons co-expressed the nociceptin receptor Oprl1, while none showed expression of Trpv1 or Oprm1 (160 Spp1+/Oprl1+ neurons, red arrow). In a panoramic view of whole rat DRG, Spp1+ neurons constitute a distinct population (Figure 8B). Note also that these neurons tended to be spatially arranged in a particular area of the DRG section (Figures 8C,D).
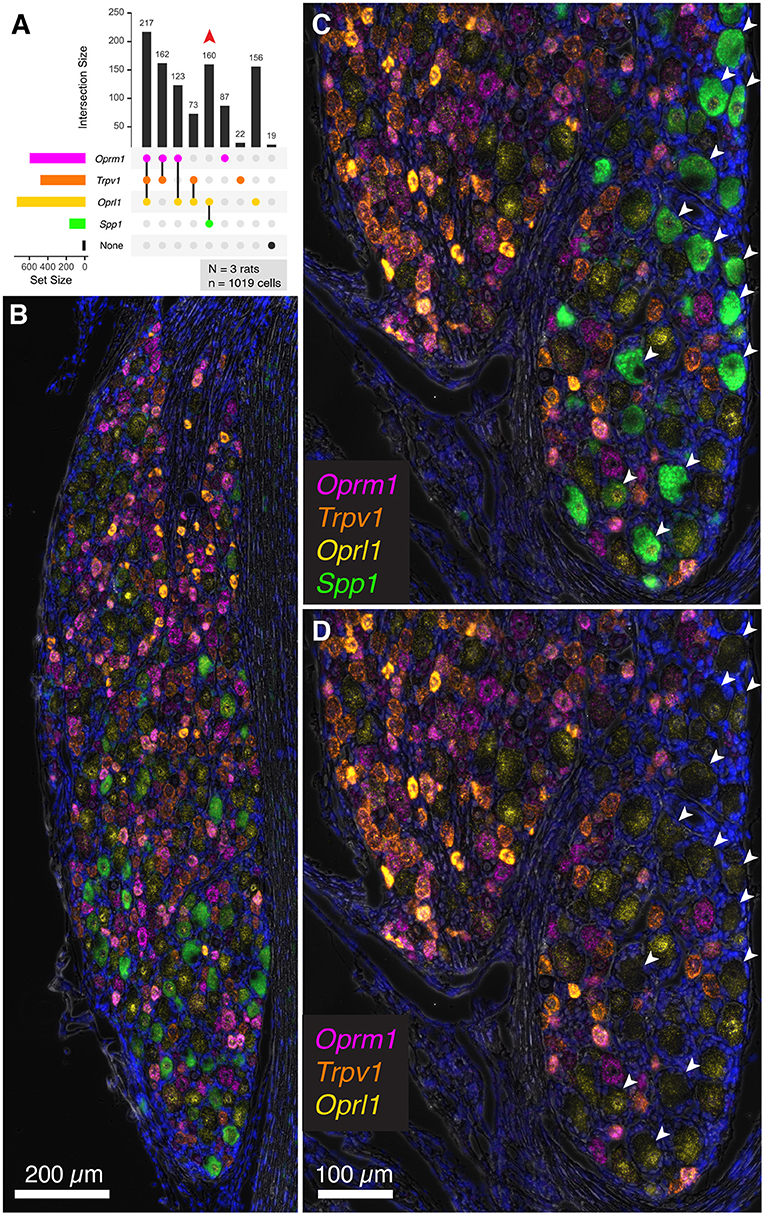
Figure 8. Co-expression of the nociceptive and analgesic markers Trpv1, Oprm1, and Oprl1 with Secreted Phosphoprotein 1/Osteopontin (Spp1) transcript. Co-expression of Trpv1, Oprm1, and Oprl1 with Spp1, a marker of DRG neurons thought to be chiefly implicated in proprioception. (A) In a co-expression plot, all of the Spp1+ neurons co-expressed Oprl1, while none showed expression of Trpv1 or Oprm1 (83 Spp1+/Oprl1+ neurons, red arrow). (B) A panoramic view of a whole rat DRG stained for this combination of markers. Note the distinct signal for Spp1. (C) An enlargement of a different DRG neuron shows spatially distinct areas with and without Spp1 neurons. (D) Note that Spp1+ neurons (arrows) do not contain any Trpv1 or Oprm1 signal.
While numerous categories of co-expression are documented, five readily distinguishable neuronal groups are shown in Figure 9 with frequency distributions of their cell diameters. The high Trpv1-expressing neurons were the smallest in size with a 20 μm mean diameter (n = 25 cells). The Trpv1/Trpa1 cell population (n = 120 cells; often “quad positive,” as in Figure 4; ~26.8 μm average diameter) contains two algesic-sensing ion channels and the μ-opioid receptor that can confer analgesia. This analysis was performed using the double label as a selection criteria, but note that a subpopulation of these cells (such as that in the representative image) has additional labels. The diameter of the high expressing Trpm8 cells was on average 33.0 μm (n = 68 cells), indicating a larger diameter than high Trpv1 neurons (21 μm) or Trpv1/Trpa1 cells (26 μm). The largest neuron identified was the co-positive for the μ-δ opioid receptor (Oprm1+/Ord1+) neurons (44 μm average diameter). These were even larger than the osteopontin (Spp1) expressing proprioceptive neurons (38 μm average diameter; n = 102 cells). In mouse, the osteopontin immunoreactive neurons give rise to the spiral endings innervating muscle spindles (Ichikawa et al., 2000; Chiu et al., 2014).
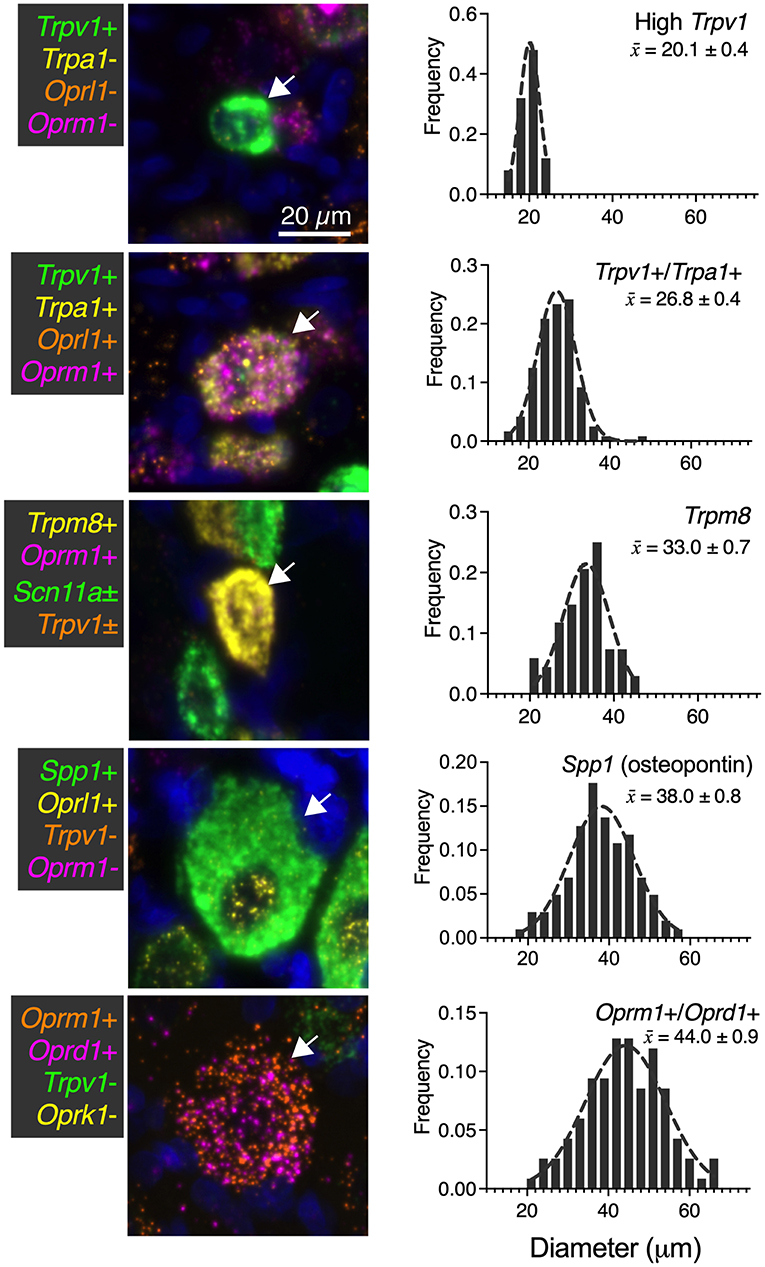
Figure 9. Summary of identified cell types with size analysis. For each of the major identified cell types in the present study, a representative image (66 μm square) is shown. Additionally, histogram information of the cell size for each population is identified in ascending order of mean size ± SEM. High Trpv1 neurons were the smallest cells (20.1 μm ± 0.4). Oprm1+/Oprd1+ neurons were the largest cells identified, with an average size of 44.0 μm ± 0.9. Scale bar applies to all images. These analyses were performed with at least N = 3 rats.
We identified a significant difference in diameter between each of the groups (p < 0.05). Significance testing was performed between the five groups (all comparisons) using a Kruskal-Wallis test with Dunn's correction for multiple comparisons (Prism 9, GraphPad). These findings were calculated from the same sections as other N = 3 analyses in other figures, although sampling was expanded (n = 117 cells in Figure 9) for this analysis due to the rarity of the Oprm1+/Oprd1+ population.
This study examined rat primary afferent neurons using multiplex fluorescent mRNA in situ hybridization to understand nociceptive and analgesia mechanisms in the peripheral nervous system. We consider identifying critical neuronal populations essential knowledge for developing potential peripherally acting non-opioid analgesics that do not exhibit central nervous system side effects. The investigation focused on determining co-expression patterns of nociceptive-, and analgesia-transducing molecular markers in DRG neurons. We hypothesized the conjuncture of these two types of molecules in one or more neuronal subpopulations would provide a colocalization matrix that could be used to generate a molecularly informed approach to peripheral analgesia and analgesic drug development.
The populations of nociceptive Trpv1+ neurons were heterogeneous, expressing varying levels of Trpv1 and the μ-opioid receptor Oprm1. Co-expression analysis indicated differing degrees of μ-opioid expression among the two populations of Trpv1+ neurons that we hypothesize subserve thermosensory and tissue-injury nociception. We identified a population of very high expressors of Trpv1 that are absent of Oprm1. In contrast, most low and medium expressors of Trpv1 do express Oprm1, and are the evident candidates for the neuronal population inhibited by μ-opioid agonist analgesics, especially when given intrathecally or epidurally, administration paradigms where the injectate bathes the central projection of dorsal root ganglion axons entering the spinal cord. The co-expression of Trpv1 and Oprm1 receptors within the same cell types provides both cellular and molecular level explanations for the efficacy of opioid agonists. That is, opioid drugs act by inhibiting the very cells responsible for transmitting certain types of nociceptive stimuli from the periphery to the central nervous system. Available evidence suggests that opioid analgesia occurs, in part, through presynaptic inhibition of transmitter release from Trpv1+ DRG afferent terminals in spinal cord and potentially spinal cord μ-expressing neurons (Kondo et al., 2005; Chen and Pan, 2006; Xanthos and Sandkühler, 2014; Che et al., 2021). Co-expression of Trpv1 was never observed with Spp1, a marker for large diameter proprioceptive neurons, a finding that validates that nociception and proprioception are mediated by distinct neuronal populations. Additionally, the lower expressors of Trpv1 co-expressed other markers implicated in pathological pain states. For example, Trpm8, a nociceptive ion channel implicated in cold thermosensation (Bautista et al., 2007) and cold allodynia (Xing et al., 2007) was identified in some lower-expressing Trpv1 sensory neurons. Additionally, the ion channel NaV 1.9 (encoded by SCN11A), whose gain-of-function mutation leads to familial episodic pain (Leipold et al., 2013), was also identified in the low to medium-expressing Trpv1 population. In contrast, the small diameter high Trpv1 expressing neurons were negative for all of these markers, most likely indicating an exclusive role as a dedicated labeled line for thermonociception and/or non-noxious thermosensation (Kobayashi et al., 2005; Craig, 2018). Coexpression analysis is also a critical step for gaining insight into the algesic and analgesic receptors on DRG neurons, and could be important for understanding indirect interactions between painful and analgesic target receptors. In particular, there are established interactions between pain transducing ion channels and opioid receptors (Chew et al., 2019).
The μ-opioid receptor is one of the most clinically important targets for analgesia, with the majority of highly impactful orally and intravenously available analgesic agents acting through this receptor. While there are three canonical opioid receptors (μ, δ, and κ), and a fourth opioid-like paralog (Oprl1), only μ-receptor agonists are used clinically at this time, although extensive medicinal chemical and preclinical development on the other receptors has occurred or is in progress (Calo et al., 2000; Floyd et al., 2009; Bardoni et al., 2014; Viscusi et al., 2021). Additionally, some controversy exists over detection of these receptors, as they are frequently expressed to a low degree and difficult to quantify accurately (Imlach and Christie, 2014; Iadarola et al., 2015; Sapio et al., 2016). In this regard, the present study fills a critical knowledge gap by delineating the co-expression matrix of these receptors in detail. We characterize the expression pattern of all four receptors, and, in particular, the canonical three together. One point of contention is the pharmacological relevance of opioid receptor heterodimers. In order for the heterodimer to form (Jordan and Devi, 1999; Erbs et al., 2015; Gaborit and Massotte, 2021), both receptors would, at a minimum, have to be expressed in the same cell. Our data confirm that co-expression of the μ-, δ- and κ-opioid receptors does indeed occur in some combinations, albeit in a very small population of neurons. This heterogeneity is confirmed by recording and staining from rat DRG neurons, although some differences in proportions of cells between the present study and the in vitro recording investigation are seen and are likely due to methodological differences (Rau et al., 2005). Interestingly, some large cells were observed with high concentrations of μ- and δ-opioid receptors (Figure 1E), consistent with putative mechanosensory nociceptors, based on genetic anatomical tracing studies done in the mouse (Bardoni et al., 2014). These anatomical findings showing colocalization of Oprm1 and Oprd1 may be related to the observed analgesic synergy between μ- and δ-opioid agonists in the periphery (Bruce et al., 2019). Another study using genetic labeling in mice showed that the κ-opioid receptor was frequently expressed in myelinated lanceolate or circumferential nerve endings around hair follicles, and demonstrated that κ-opioid receptor function at these afferents modulated pain and itch (Snyder et al., 2018). In our rat study the majority, although not all κ-opioid receptor expressing neurons, also expressed the μ-opioid receptor indicating that this pair of receptors likely functions in the same cell to modulate some modalities of nociception.
Another opioid-like receptor examined in this study is the nociceptin/orphanin FQ receptor (NOP receptor, encoded by Oprl1). This is the least studied of the four opioid-like receptors, and although it has been proposed to have multiple avenues toward clinical potential (Lambert, 2008), none have been realized to date. One finding that is particularly relevant to pain and analgesia, is the observation that intrathecal administration of nociceptin, the endogenous agonist at the NOP receptor was analgesic, and this analgesia was not naltrexone reversible, suggesting NOP-dependent analgesic actions are distinct from analgesia mediated at other opioid receptors (Ko et al., 2006). In our staining, we identified signal for Oprl1 expression in almost all DRG neurons, which does not exclude its utility as an antinociceptive agent, but suggests very broad function(s), and may result in off target side effects on non-nociceptive fibers. Notably, broad action, or expression of an analgesic target outside of the nociceptive neurons is not necessarily indicative of reduced analgesic potential, but the ubiquity of Oprl1 signal does contradict the supposition that this receptor is involved in endogenous analgesia in a specific fashion.
One of the key nociceptive markers examined in the present study is Trpv1. This receptor is the endogenous receptor for capsaicin, the pungent ingredient in hot peppers and resiniferatoxin, a new analgesic currently undergoing clinical trials (NCT03542838, NCT02522611, NCT00804154, NCT04885972, and others) (Caterina et al., 1997). It has long been appreciated that Trpv1 is expressed in several subclasses of neurons, and more recently these populations have been described to a greater extent in several species (Goswami et al., 2014; Isensee et al., 2014; Usoskin et al., 2015; Sapio et al., 2018; Tavares-Ferreira et al., 2022). The overall pattern of expression of Trpv1 is similar to previous studies, although it appears that the RNAScope procedure, in particular, labels more neurons than most antibody-based approaches (Mitchell et al., 2010, 2014; Goswami et al., 2014; Poulson et al., 2020; Sapio et al., 2020b; Shiers et al., 2020; Hall et al., 2022; Tavares-Ferreira et al., 2022). It is also notable that the percentage of Trpv1+ neurons as assessed by a technique as sensitive as RNAScope is not necessarily identical to the number of capsaicin-responsive neurons. For example, the percentage of neurons killed by capsaicin incubation in culture is estimated to be approximately 37% in one study (Wood et al., 1988), which is also similar to the number of cultured mouse lumbar DRG neurons responding to 300 nM capsaicin (39%) (Teichert Russell et al., 2012). Therefore, it appears that at a certain level of expression is required to produce robust enough calcium influx to compromise cellular integrity (Karai et al., 2004b). That is, approximately half of the neurons detected as Trpv1+ in our study (presumably the most strongly labeled neurons) are likely to be capsaicin responsive.
Generally, it has been observed that there is a small number of densely immunoreactive small diameter Trpv1+ neurons, as well as a larger number of larger diameter and less densely stained neurons (Tominaga et al., 1998; Cavanaugh et al., 2011b; Goswami et al., 2014). Presumably, this encompasses at least two functionally defined TRPV1+ subclasses: the Trpv1+ C-fibers and A-δ fibers (Mitchell et al., 2010, 2014). The molecular identity of the TRPV1+ A-δ fibers is still being determined (Raithel et al., 2018), in part due to the small percentage of A-δ neurons in DRG (Lawson et al., 2019). Our study suggests that the small diameter high expressing Trpv1+ fibers in the rat contain a narrower repertoire of noci-responsive TRP channels, potentially implicating a more exclusive thermal nociceptive role which, at least in the rat, does not appear to be sensitive to opioid-induced analgesia. Differential thermal activation of A-δ and C-fibers suggest that, based on behavioral withdrawal, the C-fiber population is the one that is less sensitive to μ-opioid receptor agonists (Mitchell et al., 2014) and the A-δ neurons are a subpopulation of the medium to low TRPV1 expressing neurons. To some extent, a similar phenomenon has been examined in humans in a study using high dose transdermal capsaicin patch. In this study, sensitivity to noxious (55°C) thermal stimulation was confined to the capsaicin-treated region, while intensity ratings for less noxious (44°C) or non-noxious (38°C) thermal stimuli were reduced distal to the patch-treated skin (Van Neerven and Mouraux, 2020). This is potentially consistent with the idea that higher expressing Trpv1+ fibers are more capsaicin sensitive and primarily involved in thermonociceptive sensation, while the more broadly, polymodal nociceptive fibers may have less Trpv1 but co-express multiple noci-responsive channels. However, notably, this type of experiment may also be influenced by other variables such as receptive field size and central modulatory effects. Further studies will be needed in humans to tease out the function of these Trpv1+ populations.
Another TRP channel Trpa1 was examined in regard to both Trpv1 and analgesic-related opioid receptors. Trpa1 is the receptor for chemical nociception and is responsible for transducing stimuli such as allyl isothiocyanate, the active pungent ingredient in foods such as wasabi (Jordt et al., 2004; Mcnamara et al., 2007). In our previous study using transcriptomics and multiplex fluorescent in situ hybridization, we examined the expression pattern of Trpv1 and Trpa1 in DRG and nodose ganglion sensory afferents (Sapio et al., 2020b). In that study, we found that Trpv1 and Trpa1 were more highly co-expressed in nodose, whereas in the DRG, neurons that were high in either Trpv1 or Trpa1 tended to be low for the other, suggesting a separate, although not exclusive set of sensory pathways for thermal and chemical nociception in the peripheral nervous system. The present study confirms this result (Figure 6). The largely differential expression is also consistent with previous studies showing partial depletion of Trpa1 with Trpv1 agonists (Pecze et al., 2009) or coexpression of these receptors. However, TRPA1 is apparently expressed in low expressing TRPV1 population, which could explain why these neurons are resistant to TRPV1 agonist actions and could also explain the failure to deplete TRPA1-expressing neurons with TRPV1 agonists in some experiments (Isensee et al., 2014; Sapio et al., 2018, 2020b). The functional implication of co-expression of these receptors is being investigated. For example, heteromers have been reported between these two ion channels with different pharmacologic and biophysical properties and have been proposed to be involved in nociceptive sensitization (Fischer et al., 2014; Patil et al., 2020). Cells with both Trpv1 and Trpa1 also tended to have moderate to high amounts of Oprm1 expression (see Figure 3, Quad+ neurons), consistent with their proposed dual nociceptive and analgesia-conferring properties (Akopian, 2011).
Trpm8 is the primary receptor for cold and cold pain (Bautista et al., 2007) and serves as a marker for cold-sensing sensory afferents (Renthal, 2020). We describe populations of high and low Trpm8 expression, where presumably the high Trpm8 expressing cells are those that have primarily been described in previous studies as cold thermosensory afferents (Le Pichon and Chesler, 2014; Jankowski et al., 2017), and the lower levels of Trpm8 in other cells are currently of unknown significance. However, these could be polymodal neurons, or cells that become recruited at colder temperatures and/or during cold allodynia. Notably, while Trpm8+ afferents have been shown to be involved in responding to environmental cooling, a small population of Trpm8+ fibers has been reported to respond to noxious mechanical stimulation (Jankowski et al., 2017), and it has been suggested that in injury states a subpopulation of Trpm8+ fibers may become responsive to additional noxious modalities. Additionally, the processes underlying thermal encoding in the DRG can be complex, with populations of cells responding in a graded fashion to various temperatures (Wang et al., 2018). Furthermore, electrical recordings have shown that cooling-responsive cells comprise only about 2.3% of all DRG neurons (Lawson et al., 2019), whereas the total number of Trpm8+ cells we quantified was much larger than this estimate. While examining the full range of thermal-encoding cells is beyond the scope of this study, the coincidence of expression of Trpv1 and Trpm8, which encode two major encoding ion channels of temperature, may be useful information for future investigations. This is also an indication that we are examining multiple classes of DRG neurons among the Trpm8+ population. This is one area where additional electrical characterization of these subtypes would be informative, as this can elucidate functional response characteristics to further segregate the DRG afferents (Petruska et al., 2000; Rau et al., 2005; Lawson et al., 2019). This is also interesting given that Trpm8 expression level may be related to tuning thermal responsiveness, which is notable given the finding that various channels and/or channel combinations may be involved in thermal coding based on mouse studies (Paricio-Montesinos et al., 2020).
The overlap between Trpm8 and Oprm1 expression in the rat makes sense given reduction in clinical responses to cold sensation upon morphine (a mu-opioid receptor agonist) administration (Cleeland et al., 1996). This is also corroborated by the finding that morphine alleviated cold allodynia in rat models of chronic pain (Erichsen et al., 2005). Both cases support the idea that Trpm8+ cells are inhibited by μ-opioid receptor agonists. The Trpm8+ neurons also appear to be largely Trpv1-, indicating discrete types of peripheral neurons for the detection of hot and cold. This lack of overlap between Trpv1 and Trpm8, is consistent with previous evidence demonstrating that Trpm8+ neurons are (in general) a small-diameter, relatively rare Trpv1- population of sensory afferents (Dhaka et al., 2008; Pecze et al., 2009; Le Pichon and Chesler, 2014; Jankowski et al., 2017). Although we did detect Trpm8+/Trpv1+ neurons, the expression was largely anticorrelated (Figure 6H) indicating that these two receptors are never strongly co-expressed. While developmentally, Trpm8 is co-expressed in a subset of Trpv1-lineage neurons in the mouse (Mishra et al., 2011) pharmacologic ablation of Trpv1+ fibers in adult rodents suggests a further differentiation of a distinct subpopulation of Trpm8+/Trpv1- neurons (Cavanaugh et al., 2009) consistent with our anatomical findings. This is also consistent with ISH and single-cell RNA-Seq in DRG and trigeminal ganglion showing very low levels of Trpv1 in Trpm8+ neurons (Kobayashi et al., 2005; Von Buchholtz et al., 2020). The idea of multiple Trpm8+ populations (Xing et al., 2006), where the Trpv1-coexpressing population may be more nociceptive in nature would potentially explain our results in context of the broader literature. We also cannot rule out an interaction between these input pathways at the level of the spinal cord, where Trpv1-specific and Trpm8-specific input pathways may be processed together in spinal circuits. For example, it is known that there is some interaction between Trpv1-specific and Trpm8-specific behavioral outcomes (Anderson et al., 2014).
The tetrodotoxin-resistant sodium channel subunit NaV1.9 (encoded by Scn11a) has come into focus as a potential nociceptor-specific target for the development of new non-opioid analgesics. Alongside two other sodium channel subunits in this family (Scn9a and Scn10a), Scn11a has been suggested as an interesting and understudied mediator of pain signaling. For example, the related ion channel subunit gene, Scn9a has been extensively studied in rodents and humans where it is strongly linked to pain and nociceptive signaling (Hisama et al., 1993; Dib-Hajj et al., 2005, 2007, 2013; Yang et al., 2018). Similarly, Scn10a has been used extensively as a nociceptive specific marker (Akopian et al., 1996; Dib-Hajj et al., 1999; Stirling et al., 2005; Shiers et al., 2020). The current study focuses on Scn11a largely due to the fact that it has been studied less intensively than Scn9a and Scn10a, and because mutations in this gene are associated with pain insensitivity or episodic pain syndromes in humans (Leipold et al., 2013; Zhang et al., 2013). For example, NaV1.9 has been found to be enriched in cells in the Trpv1 lineage (Goswami et al., 2014) or depleted with TRPV1 agonist treatment (Isensee et al., 2014; Sapio et al., 2018), suggesting specificity for the nociceptive population. This channel has also been implicated in nociception and hyperalgesia in animal models, suggesting potential utility as an analgesic pharmacological target (Priest et al., 2005). NaV1.9 is required for cold-triggered nociception in the mouse (Lolignier et al., 2015), consistent with our observation that Scn11a is found in high-expressing Trpm8+ neurons. However, our staining shows a broad expression pattern of Scn11a consistent with involvement in other nociceptive functions. For example, Scn11a is frequently co-expressed with Trpv1, which is consistent with human mutation studies of NaV1.9 in nociception in which loss or gain of function SCN11A mutations can cause pain insensitivity or episodic pain syndromes (Leipold et al., 2013; Zhang et al., 2013). The phenotype depends on the location of mutation in the channel. Importantly, the finding that gain of function mutations that inhibit transmitter release leads to pain insensitivity suggests that NaV1.9 is expressed in enough nociceptive afferents to be sufficient to support a pain insensitive phenotype. These findings are also corroborated by other studies using single cell RNA-Seq showing that Scn11a is expressed in Trpv1+ cells, but also in several other classes of sensory afferents (Li et al., 2016; Sapio et al., 2018).
In our staining analysis, we examined secreted phosphoprotein 1 (Spp1) positive neurons, which are non-nociceptive large diameter primary afferents that innervate muscle spindles. The Spp1 gene encodes osteopontin (Ichikawa et al., 2000). While the function of osteopontin in neurons has not been determined, it has been suggested as a regulator of myelination, and hypothesized that its expression is related to the maintenance or formation of axons with high conduction velocities (Higo et al., 2010). These neurons are thought to be chiefly implicated in proprioception (Ichikawa et al., 2000; Usoskin et al., 2015; Saito-Diaz et al., 2021). Anatomically, osteopontin immunoreactivity localizes to spiral axon terminals in muscle spindle fibers. In our staining, we found that Spp1+ cells were negative for Trpv1 and Oprm1, but co-expressed Oprl1. These neurons were also generally large, and their staining pattern, showing distinction from Trpv1 expressors, further suggests their non-nociceptive nature in the DRG.
The current study has several limitations that must be addressed in future studies. For the present investigation, experiments are conducted in male rats, and as such may not be fully reflective of the neuroanatomy and neurochemistry in the mouse or across both sexes (Sadler et al., 2022). For behavioral studies, the rat has many advantages compared to the mouse, however, at this point in time, a large set of molecular-genetic rat manipulations are not available for investigation as they are for the mouse nervous system (Ellenbroek and Youn, 2016; Homberg et al., 2017). Thus, the use of multiplex fluorescence represents a concerted effort to assemble the combinatorial molecular-cellular expression patterns in rat that mediate algesia and analgesia in sensory ganglia. The ultimate goal of these investigations is to gain insights into conserved molecular biological phenomena in both sexes and in humans. Future studies will concentrate on direct human investigation in male and female organ donor tissue to corroborate and extend these findings into human health, disease, and pharmacology (Iadarola et al., 2022), as such studies are more likely to yield information that may be more clinically relevant and applicable. As an additional area for future directions, much attention has been paid to the use of single cell and single nucleus RNA-Seq for the determination and characterization of neuronal cell types in the DRG in mouse, monkey and human (Usoskin et al., 2015; Kupari et al., 2021; Tavares-Ferreira et al., 2022). These studies are useful in the determination of cell types but often lack the precision to determine the expression profile of an individual gene. In the existing databases, the opioid receptors are weakly detected, and probably encountered a limit of detection. Additionally, for genes such as Trpv1 and Trpm8, the mouse and monkey databases have shown correlation between Trpv1 and Trpm8, where the cell population with the highest level of one also has the highest level of the other, which is in direct contrast to our findings (Usoskin et al., 2015; Kupari et al., 2021). However, this could be due to multiple technical issues or species differences and is beyond the scope of the current investigation. Finally, another limitation is that this study does not assess the impact of pain and nerve injury on expression and co-expression of these markers. Nerve injury in particular induces strong transcriptional changes and may alter the baseline expression patterns of nociceptive and analgesic targets (Ray et al., 2018; Sapio et al., 2020a). Future studies can expand upon these foundational data to understand pain conditions.
While the field of nociceptive neuroscience has expended substantial effort over many years toward understanding key receptors involved in nociceptive circuits, there is a need for exact delineation of neuronal populations that contribute to clinical pain and pain control as well as identification of molecular signatures which can define these distinct populations. Our study utilized multiplex high sensitivity mRNA in situ hybridization, which allowed us to obtain precise answers, leading to enhanced clarity of these issues. Importantly, our findings suggest that the medium to low expressing Trpv1+ neurons indeed represent the population that transmit nociceptive signaling associated with tissue-damage and this population is coincident with opioid-induced anti-nociception. Deeper analysis of the coexpression matrix of these nociceptive and analgesic target genes provides additional rationale for identifying susceptible neuronal populations for early-stage novel therapeutics development, with either pharmacological agents or other methods such as in vivo gene transfer, by better defining the neuronal populations directly responsible for clinically relevant pain and pain control.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by NIH Clinical Center.
The project was conceptualized by MI, WM, and MS. Experiments were performed by WM, with technical involvement from MS, DM, and TG. Staining and slide scanning was performed by WM with assistance from AM and supervision from DM in fluorescence microscopy and image analysis. Stained sections were analyzed in Adobe Photoshop by WM with assistance from AM and MS. Visualizations, final figures, and formal analysis were generated primarily by MS, with assistance from WM, AM, and MD. Initial drafts of the manuscript were prepared by MS, WM, and MI with editing and suggestions from all of the authors. Funding was obtained by WM, DM, MI, and AM. The project was supervised by MS, AM, and MI. All authors revised and approved the final manuscript.
Funding for this work was supported by the Intramural Research Program of the NIH Clinical Center (1ZIACL090033-09, 1ZIACL090034-09, and 1ZIACL090035-08 to AM), and the NINDS. This work was also supported by a funds from the National Center for Complementary and Integrative Health (1ZIAAT000017-03), and the Office of Behavioral and Social Sciences Research. TG was supported by the Japan Society for the Promotion of Science Overseas Research Fellowship. This research was made possible through the National Institutes of Health (NIH) Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, Genentech, the American Association for Dental Research, the Colgate-Palmolive Company, Elsevier, alumni of student research programs, and other individual supporters via contributions to the Foundation for the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Ms. Pranavi Nara for technical assistance with several of the in situ hybridization studies.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.926596/full#supplementary-material
Akopian, A. N., Sivilotti, L., and Wood, J. N. (1996). A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 379, 257–262. doi: 10.1038/379257a0
Akopian, A. (2011). Regulation of nociceptive transmission at the periphery via TRPA1-TRPV1 interactions. Curr. Pharm. Biotechnol. 12, 89–94. doi: 10.2174/138920111793937952
Anderson, E. M., Jenkins, A. C., Caudle, R. M., and Neubert, J. K. (2014). The effects of a co-application of menthol and capsaicin on nociceptive behaviors of the rat on the operant orofacial pain assessment device. PLoS ONE 9, e89137. doi: 10.1371/journal.pone.0089137
Bardoni, R., Tawfik, V. L., Wang, D., Francois, A., Solorzano, C., Shuster, S. A., et al. (2014). Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron 81, 1312–1327. doi: 10.1016/j.neuron.2014.01.044
Bautista, D. M., Siemens, J., Glazer, J. M., Tsuruda, P. R., Basbaum, A. I., Stucky, C. L., et al. (2007). The menthol receptor TRPM8 is the principal detector of environmental cold. Nature 448, 204–208. doi: 10.1038/nature05910
Bevan, S., Hothi, S., Hughes, G., James, I. F., Rang, H. P., Shah, K., et al. (1992). Capsazepine: a competitive antagonist of the sensory neurone excitant capsaicin. Br. J. Pharmacol. 107, 544–552. doi: 10.1111/j.1476-5381.1992.tb12781.x
Brown, D. C., Agnello, K., and Iadarola, M. J. (2015). Intrathecal resiniferatoxin in a dog model: efficacy in bone cancer pain. Pain 156, 1018–1024. doi: 10.1097/j.pain.0000000000000115
Brown, D. C., and Iadarola, M. J. (2015). “TRPV1 agonist cytotoxicity for chronic pain relief,” in: TRP Channels as Therapeutic Targets, eds A. Szallasi (Boston: Academic Press).
Brown, D. C., Iadarola, M. J., Perkowski, S. Z., Erin, H., Shofer, F., Laszlo, K. J., et al. (2005). Physiologic and antinociceptive effects of intrathecal resiniferatoxin in a canine bone cancer model. Anesthesiology 103, 1052–1059. doi: 10.1097/00000542-200511000-00020
Bruce, D. J., Peterson, C. D., Kitto, K. F., Akgün, E., Lazzaroni, S., Portoghese, P. S., et al. (2019). Combination of a δ-opioid receptor agonist and loperamide produces peripherally-mediated analgesic synergy in mice. Anesthesiology 131, 649–663. doi: 10.1097/ALN.0000000000002840
Calo, G., Guerrini, R., Rizzi, A., Salvadori, S., and Regoli, D. (2000). Pharmacology of nociceptin and its receptor: a novel therapeutic target. Br. J. Pharmacol. 129, 1261–1283. doi: 10.1038/sj.bjp.0703219
Caterina, M. J. (2007). Transient receptor potential ion channels as participants in thermosensation and thermoregulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R64–76. doi: 10.1152/ajpregu.00446.2006
Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D., and Julius, D. (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824. doi: 10.1038/39807
Cavanaugh, D. J., Chesler, A. T., Braz, J. M., Shah, N. M., Julius, D., and Basbaum, A. I. (2011a). Restriction of transient receptor potential vanilloid-1 to the peptidergic subset of primary afferent neurons follows its developmental downregulation in nonpeptidergic neurons. J. Neurosci. 31, 10119–10127. doi: 10.1523/JNEUROSCI.1299-11.2011
Cavanaugh, D. J., Chesler, A. T., Jackson, A. C., Sigal, Y. M., Yamanaka, H., Grant, R., et al. (2011b). Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J. Neurosci. 31, 5067–5077. doi: 10.1523/JNEUROSCI.6451-10.2011
Cavanaugh, D. J., Lee, H., Lo, L., Shields, S. D., Zylka, M. J., Basbaum, A. I., et al. (2009). Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc. Natl. Acad. Sci. 106, 9075–9080. doi: 10.1073/pnas.0901507106
Che, T., Dwivedi-Agnihotri, H., Shukla, A. K., and Roth, B. L. (2021). Biased ligands at opioid receptors: Current status and future directions. Sci. Signal. 14:aav0320. doi: 10.1126/scisignal.aav0320
Chen, S.-R., and Pan, H.-L. (2006). Blocking μ opioid receptors in the spinal cord prevents the analgesic action by subsequent systemic opioids. Brain Res. 1081, 119–125. doi: 10.1016/j.brainres.2006.01.053
Chew, L. A., Bellampalli, S. S., Dustrude, E. T., and Khanna, R. (2019). Mining the Nav1.7 interactome: opportunities for chronic pain therapeutics. Biochem. Pharmacol. 163, 9–20. doi: 10.1016/j.bcp.2019.01.018
Chiu, I. M., Barrett, L. B., Williams, E. K., Strochlic, D. E., Lee, S., Weyer, A. D., et al. (2014). Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife 3:e34. doi: 10.7554/eLife.04660.034
Cleeland, C. S., Nakamura, Y., Howland, E. W., Morgan, N. R., Edwards, K. R., and Backonja, M. (1996). Effects of oral morphine on cold pressor tolerance time and neuropsychological performance. Neuropsychopharmacology 15, 252–262. doi: 10.1016/0893-133X(95)00205-R
Craig, A. D. (2018). “Chapter 19 - Central neural substrates involved in temperature discrimination, thermal pain, thermal comfort, and thermoregulatory behavior,” in Handbook of Clinical Neurology, ed A. A. Romanovsky (Amsterdam: Elsevier).
Dhaka, A., Earley, T. J., Watson, J., and Patapoutian, A. (2008). Visualizing cold spots: TRPM8-expressing sensory neurons and their projections. J. Neurosci. 28, 566–575. doi: 10.1523/JNEUROSCI.3976-07.2008
Dib-Hajj, S. D., Cummins, T. R., Black, J. A., and Waxman, S. G. (2007). From genes to pain: Na v 1.7 and human pain disorders. Trends Neurosci. 30, 555–563. doi: 10.1016/j.tins.2007.08.004
Dib-Hajj, S. D., Rush, A. M., Cummins, T. R., Hisama, F. M., Novella, S., Tyrrell, L., et al. (2005). Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 128, 1847–1854. doi: 10.1093/brain/awh514
Dib-Hajj, S. D., Tyrrell, L., Cummins, T. R., Black, J. A., Wood, P. M., and Waxman, S. G. (1999). Two tetrodotoxin-resistant sodium channels in human dorsal root ganglion neurons. FEBS Lett. 462, 117–120. doi: 10.1016/S0014-5793(99)01519-7
Dib-Hajj, S. D., Yang, Y., Black, J. A., and Waxman, S. G. (2013). The Na(V)1.7 sodium channel: from molecule to man. Nat. Rev. Neurosci. 14, 49–62. doi: 10.1038/nrn3404
Ellenbroek, B., and Youn, J. (2016). Rodent models in neuroscience research: is it a rat race? Dis. Model. Mech. 9, 1079–1087. doi: 10.1242/dmm.026120
Erbs, E., Faget, L., Scherrer, G., Matifas, A., Filliol, D., Vonesch, J. L., et al. (2015). A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct. Funct. 220, 677–702. doi: 10.1007/s00429-014-0717-9
Erichsen, H. K., Hao, J. X., Xu, X. J., and Blackburn-Munro, G. (2005). Comparative actions of the opioid analgesics morphine, methadone and codeine in rat models of peripheral and central neuropathic pain. Pain 116, 347–358. doi: 10.1016/j.pain.2005.05.004
Fischer, M. J. M., Balasuriya, D., Jeggle, P., Goetze, T. A., Mcnaughton, P. A., Reeh, P. W., et al. (2014). Direct evidence for functional TRPV1/TRPA1 heteromers. Pflügers Archiv. Eur. J. Physiol. 466, 2229–2241. doi: 10.1007/s00424-014-1497-z
Floyd, B. N., Camilleri, M., Busciglio, I., Sweetser, S., Burton, D., Wong, G. Y., et al. (2009). Effect of a kappa-opioid agonist, i.v. JNJ-38488502, on sensation of colonic distensions in healthy male volunteers. Neurogastroenterol. Motil. 21, 281–290. doi: 10.1111/j.1365-2982.2008.01202.x
Gaborit, M., and Massotte, D. (2021). Therapeutic potential of opioid receptor heteromers in chronic pain and associated comorbidities. Br. J. Pharmacol. doi: 10.1111/bph.15772. [Epub ahead of print].
Garami, A., Pakai, E., Mcdonald, H. A., Reilly, R. M., Gomtsyan, A., Corrigan, J. J., et al. (2018). TRPV1 antagonists that cause hypothermia, instead of hyperthermia, in rodents: Compounds' pharmacological profiles, in vivo targets, thermoeffectors recruited and implications for drug development. Acta Physiol. 223, e13038. doi: 10.1111/apha.13038
Goswami, S. C., Mishra, S. K., Maric, D., Kaszas, K., Gonnella, G. L., Clokie, S. J., et al. (2014). Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J. Pain 15, 1338–1359. doi: 10.1016/j.jpain.2014.09.010
Hall, B. E., Macdonald, E., Cassidy, M., Yun, S., Sapio, M. R., Ray, P., et al. (2022). Transcriptomic analysis of human sensory neurons in painful diabetic neuropathy reveals inflammation and neuronal loss. Sci. Rep. 12, 4729. doi: 10.1038/s41598-022-08100-8
Higo, N., Sato, A., Yamamoto, T., Nishimura, Y., Oishi, T., Murata, Y., et al. (2010). SPP1 is expressed in corticospinal neurons of the macaque sensorimotor cortex. J. Comp. Neurol 518, 2633–2644. doi: 10.1002/cne.22356
Hisama, F. M., Dib-Hajj, S. D., and Waxman, S. G. (1993). “SCN9A-Related Inherited Erythromelalgia,” in: GeneReviews(R), eds R. A. Pagon, M. P. Adam, H. H. Ardinger, S. E. Wallace, A. Amemiya, L. J. H. Bean, et al. (Seattle, WA).
Homberg, J. R., Wöhr, M., and Alenina, N. (2017). Comeback of the rat in biomedical research. ACS Chem. Neurosci. 8, 900–903. doi: 10.1021/acschemneuro.6b00415
Iadarola, M. J., and Gonnella, G. L. (2013). Resiniferatoxin for pain treatment: an interventional approach to personalized pain medicine. Open Pain J. 6, 95–107. doi: 10.2174/1876386301306010095
Iadarola, M. J., Sapio, M. R., and Mannes, A. J. (2015). A new splice of life for the mu-opioid receptor. J. Clin. Invest. 125, 2558–2561. doi: 10.1172/JCI82060
Iadarola, M. J., Sapio, M. R., and Mannes, A. J. (2022). Be in it for the long haul: a commentary on human tissue recovery initiatives. J. Pain. doi: 10.1016/j.jpain.2022.04.009
Iadarola, M. J., Sapio, M. R., Raithel, S. J., Mannes, A. J., and Brown, D. C. (2018). Long-term pain relief in canine osteoarthritis by a single intra-articular injection of resiniferatoxin, a potent TRPV1 agonist. Pain 159, 2105–2114. doi: 10.1097/j.pain.0000000000001314
Ichikawa, H., Itota, T., Nishitani, Y., Torii, Y., Inoue, K., and Sugimoto, T. (2000). Osteopontin-immunoreactive primary sensory neurons in the rat spinal and trigeminal nervous systems. Brain Res. 863, 276–281. doi: 10.1016/S0006-8993(00)02126-0
Imlach, W., and Christie Macdonald, J. (2014). The light touch of delta opioid receptors. Neuron 81, 1220–1222. doi: 10.1016/j.neuron.2014.03.004
Isensee, J., Wenzel, C., Buschow, R., Weissmann, R., Kuss, A. W., and Hucho, T. (2014). Subgroup-elimination transcriptomics identifies signaling proteins that define subclasses of TRPV1-positive neurons and a novel paracrine circuit. PLoS ONE 9, e115731. doi: 10.1371/journal.pone.0115731
Jancso, G., Kiraly, E., and Jancso-Gabor, A. (1977). Pharmacologically induced selective degeneration of chemosensitive primary sensory neurones. Nature 270, 741–743. doi: 10.1038/270741a0
Jankowski, M. P., Rau, K. K., and Koerber, H. R. (2017). Cutaneous TRPM8-expressing sensory afferents are a small population of neurons with unique firing properties. Physiol. Rep. 5, e13234. doi: 10.14814/phy2.13234
Jordan, B. A., and Devi, L. A. (1999). G-protein-coupled receptor heterodimerization modulates receptor function. Nature 399, 697–700. doi: 10.1038/21441
Jordt, S. E., Bautista, D. M., Chuang, H. H., Mckemy, D. D., Zygmunt, P. M., Högestätt, E. D., et al. (2004). Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature 427, 260–265. doi: 10.1038/nature02282
Julius, D., and Basbaum, A. I. (2001). Molecular mechanisms of nociception. Nature 413, 203–210. doi: 10.1038/35093019
Karai, L., Brown, D. C., Mannes, A. J., Connelly, S. T., Brown, J., Gandal, M., et al. (2004a). Deletion of vanilloid receptor 1-expressing primary afferent neurons for pain control. J. Clin. Invest. 113, 1344–1352. doi: 10.1172/JCI20449
Karai, L. J., Russell, J. T., Iadarola, M. J., and Olah, Z. (2004b). Vanilloid receptor 1 regulates multiple calcium compartments and contributes to Ca2+-induced Ca2+ release in sensory neurons. J. Biol. Chem. 279, 16377–16387. doi: 10.1074/jbc.M310891200
Knowlton, W. M., Bifolck-Fisher, A., Bautista, D. M., and Mckemy, D. D. (2010). TRPM8, but not TRPA1, is required for neural and behavioral responses to acute noxious cold temperatures and cold-mimetics in vivo. Pain 150, 340–350. doi: 10.1016/j.pain.2010.05.021
Ko, M. C. H., Wei, H., Woods, J. H., and Kennedy, R. T. (2006). Effects of intrathecally administered nociceptin/orphanin fq in monkeys: behavioral and mass spectrometric studies. J. Pharmacol. Exp. Therap. 318, 1257–1264. doi: 10.1124/jpet.106.106120
Kobayashi, K., Fukuoka, T., Obata, K., Yamanaka, H., Dai, Y., Tokunaga, A., et al. (2005). Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with aδ/c-fibers and colocalization with trk receptors. J. Comp. Neurol. 493, 596–606. doi: 10.1002/cne.20794
Koivisto, A.-P., Belvisi, M. G., Gaudet, R., and Szallasi, A. (2022). Advances in Trp channel drug discovery: from target validation to clinical studies. Nat. Rev. Drug Discov. 21, 41–59. doi: 10.1038/s41573-021-00268-4
Kondo, I., Marvizon, J. C., Song, B., Salgado, F., Codeluppi, S., Hua, X. Y., et al. (2005). Inhibition by spinal mu- and delta-opioid agonists of afferent-evoked substance P release. J. Neurosci. 25, 3651–3660. doi: 10.1523/JNEUROSCI.0252-05.2005
Kupari, J., Usoskin, D., Parisien, M., Lou, D., HU, Y., Fatt, M., et al. (2021). Single cell transcriptomics of primate sensory neurons identifies cell types associated with chronic pain. Nat. Commun. 12, 1510. doi: 10.1038/s41467-021-21725-z
Lambert, D. G. (2008). The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat. Rev. Drug Discov. 7, 694–710. doi: 10.1038/nrd2572
Lawson, S. N., Fang, X., and Djouhri, L. (2019). Nociceptor subtypes and their incidence in rat lumbar dorsal root ganglia (DRGs): focussing on C-polymodal nociceptors, Aβ-nociceptors, moderate pressure receptors and their receptive field depths. Curr. Opin. Physiol. 11, 125–146. doi: 10.1016/j.cophys.2019.10.005
Le Pichon, C. E., and Chesler, A. T. (2014). The functional and anatomical dissection of somatosensory subpopulations using mouse genetics. Front. Neuroanat. 8, 21. doi: 10.3389/fnana.2014.00021
Leipold, E., Liebmann, L., Korenke, G. C., Heinrich, T., Giesselmann, S., Baets, J., et al. (2013). A de novo gain-of-function mutation in SCN11A causes loss of pain perception. Nat. Genet. 45, 1399–1404. doi: 10.1038/ng.2767
Li, C. L., Li, K. C., Wu, D., Chen, Y., Luo, H., Zhao, J. R., et al. (2016). Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 26, 967. doi: 10.1038/cr.2016.90
Lolignier, S., Bonnet, C., Gaudioso, C., Noël, J., Ruel, J., Amsalem, M., et al. (2015). The Nav1.9 channel is a key determinant of cold pain sensation and cold allodynia. Cell Rep. 11, 1067–1078. doi: 10.1016/j.celrep.2015.04.027
Maric, D., Jahanipour, J., LI, X. R., Singh, A., Mobiny, A., Van Nguyen, H., et al. (2021). Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat. Commun. 12, 1550. doi: 10.1038/s41467-021-21735-x
Mcnamara, C. R., Mandel-Brehm, J., Bautista, D. M., Siemens, J., Deranian, K. L., Zhao, M., et al. (2007). TRPA1 mediates formalin-induced pain. Proc. Natl. Acad. Sci. USA. 104, 13525–13530. doi: 10.1073/pnas.0705924104
Menéndez, L., Juárez, L., García, E., García-Suárez, O., Hidalgo, A., and Baamonde, A. (2006). Analgesic effects of capsazepine and resiniferatoxin on bone cancer pain in mice. Neurosci. Lett. 393, 70–73. doi: 10.1016/j.neulet.2005.09.046
Mishra, S. K., and Hoon, M. A. (2010). Ablation of TrpV1 neurons reveals their selective role in thermal pain sensation. Mol. Cell. Neurosci. 43, 157–163. doi: 10.1016/j.mcn.2009.10.006
Mishra, S. K., Tisel, S. M., Orestes, P., Bhangoo, S. K., and Hoon, M. A. (2011). TRPV1-lineage neurons are required for thermal sensation. EMBO J. 30, 582–593. doi: 10.1038/emboj.2010.325
Mitchell, K., Bates, B. D., Keller, J. M., Lopez, M., Scholl, L., Navarro, J., et al. (2010). Ablation of rat TRPV1-expressing Adelta/C-fibers with resiniferatoxin: analysis of withdrawal behaviors, recovery of function and molecular correlates. Mol. Pain 6, 94. doi: 10.1186/1744-8069-6-94
Mitchell, K., Lebovitz, E. E., Keller, J. M., Mannes, A. J., Nemenov, M. I., and Iadarola, M. J. (2014). Nociception and inflammatory hyperalgesia evaluated in rodents using infrared laser stimulation after Trpv1 gene knockout or resiniferatoxin lesion. Pain 155, 733–745. doi: 10.1016/j.pain.2014.01.007
Paricio-Montesinos, R., Schwaller, F., Udhayachandran, A., Rau, F., Walcher, J., Evangelista, R., et al. (2020). The sensory coding of warm perception. Neuron 106, 830-841.e3. doi: 10.1016/j.neuron.2020.02.035
Park, C. W., Kim, B. J., Lee, Y. W., Won, C., Park, C. O., Chung, B. Y., et al. (2021). Asivatrep, a TRPV1 antagonist, for the topical treatment of atopic dermatitis: a phase III, randomized, vehicle-controlled study (CAPTAIN-AD). J. Allergy Clin. Immunol. doi: 10.1016/j.jaci.2021.09.024
Patil, M. J., Salas, M., Bialuhin, S., Boyd, J. T., Jeske, N. A., and Akopian, A. N. (2020). Sensitization of small-diameter sensory neurons is controlled by TRPV1 and TRPA1 association. FASEB J. 34, 287–302. doi: 10.1096/fj.201902026R
Pecze, L., Pelsoczi, P., Kecskes, M., Winter, Z., Papp, A., Kaszas, K., et al. (2009). Resiniferatoxin mediated ablation of TRPV1+ neurons removes TRPA1 as well. Can. J. Neurol. Sci. 36, 234–241. doi: 10.1017/S0317167100006600
Petruska, J. C., Napaporn, J., Johnson, R. D., GU, J. G., and Cooper, B. Y. (2000). Subclassified acutely dissociated cells of rat DRG: histochemistry and patterns of capsaicin-, proton-, and ATP-activated currents. J. Neurophysiol. 84, 2365–2379. doi: 10.1152/jn.2000.84.5.2365
Poulson, S. J., Aldarraji, A., Arain, I. I., Dziekonski, N., Motlana, K., Riley, R., et al. (2020). Naked mole-rats lack cold sensitivity before and after nerve injury. Mol. Pain 16, 1744806920955103. doi: 10.1177/1744806920955103
Priest, B. T., Murphy, B. A., Lindia, J. A., Diaz, C., Abbadie, C., Ritter, A. M., et al. (2005). Contribution of the tetrodotoxin-resistant voltage-gated sodium channel NaV1.9 to sensory transmission and nociceptive behavior. Proc. Natl. Acad. Sci. USA. 102, 9382–9387. doi: 10.1073/pnas.0501549102
Quiding, H., Jonzon, B., Svensson, O., Webster, L., Reimfelt, A., Karin, A., et al. (2013). TRPV1 antagonistic analgesic effect: a randomized study of AZD1386 in pain after third molar extraction. Pain 154, 808–812. doi: 10.1016/j.pain.2013.02.004
Raithel, S. J., Sapio, M. R., Iadarola, M. J., and Mannes, A. J. (2018). Thermal A-delta nociceptors, identified by transcriptomics, express higher levels of anesthesia-sensitive receptors than thermal c-fibers and are more suppressible by low-dose isoflurane. Anesth. Analg. 127, 263–266. doi: 10.1213/ANE.0000000000002505
Rau, K. K., Caudle, R. M., Cooper, B. Y., and Johnson, R. D. (2005). Diverse immunocytochemical expression of opioid receptors in electrophysiologically defined cells of rat dorsal root ganglia. J. Chem. Neuroanat. 29, 255–264. doi: 10.1016/j.jchemneu.2005.02.002
Ray, P., Torck, A., Quigley, L., Wangzhou, A., Neiman, M., Rao, C., et al. (2018). Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159, 1325–1345. doi: 10.1097/j.pain.0000000000001217
Renthal, W. (2020). “Chapter 23—pain genetics,” in Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease (Sixth Edition), eds R. N. Rosenberg and J. M. Pascual (New York: Academic Press). doi: 10.1016/B978-0-12-813866-3.00023-0
Rowbotham, M. C., Nothaft, W., Duan, R. W., Wang, Y., Faltynek, C., Mcgaraughty, S., et al. (2011). Oral and cutaneous thermosensory profile of selective TRPV1 inhibition by ABT-102 in a randomized healthy volunteer trial. Pain 152, 1192–1200. doi: 10.1016/j.pain.2011.01.051
Sadler, K. E., Mogil, J. S., and Stucky, C. L. (2022). Innovations and advances in modelling and measuring pain in animals. Nat. Rev. Neurosci. 23, 70–85. doi: 10.1038/s41583-021-00536-7
Saito-Diaz, K., Street, J. R., Ulrichs, H., and Zeltner, N. (2021). Derivation of peripheral nociceptive, mechanoreceptive, and proprioceptive sensory neurons from the same culture of human pluripotent stem cells. Stem Cell Rep. 16, 446–457. doi: 10.1016/j.stemcr.2021.01.001
Sapio, M. R., Goswami, S. C., Gross, J. R., Mannes, A. J., and Iadarola, M. J. (2016). Transcriptomic analyses of genes and tissues in inherited sensory neuropathies. Exp. Neurol. 283, 375–395. doi: 10.1016/j.expneurol.2016.06.023
Sapio, M. R., Iadarola, M. J., Loydpierson, A. J., Kim, J. J., Thierry-Mieg, D., Thierry-Mieg, J., et al. (2020a). Dynorphin and enkephalin opioid peptides and transcripts in spinal cord and dorsal root ganglion during peripheral inflammatory hyperalgesia and allodynia. J. Pain 21, 988–1004. doi: 10.1016/j.jpain.2020.01.001
Sapio, M. R., Neubert, J. K., Lapaglia, D. M., Maric, D., Keller, J. M., Raithel, S. J., et al. (2018). Pain control through selective chemo-axotomy of centrally projecting TRPV1+ sensory neurons. J. Clin. Invest. 128, 1657–1670. doi: 10.1172/JCI94331
Sapio, M. R., Vazquez, F. A., Loydpierson, A. J., Maric, D., Kim, J. J., Lapaglia, D. M., et al. (2020b). Comparative analysis of dorsal root, nodose and sympathetic ganglia for the development of new analgesics. Front. Neurosci. 14, 615362. doi: 10.3389/fnins.2020.615362
Shiers, S., Klein, R. M., and Price, T. J. (2020). Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain 161, 2410–2424. doi: 10.1101/2020.03.06.981597
Snyder, L. M., Chiang, M. C., Loeza-Alcocer, E., Omori, Y., Hachisuka, J., Sheahan, T. D., et al. (2018). Kappa opioid receptor distribution and function in primary afferents. Neuron 99, 1274-1288.e6. doi: 10.1016/j.neuron.2018.08.044
Stirling, L. C., Forlani, G., Baker, M. D., Wood, J. N., Matthews, E. A., Dickenson, A. H., et al. (2005). Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain 113, 27–36. doi: 10.1016/j.pain.2004.08.015
Szallasi, A., and Blumberg, P. M. (1999). Vanilloid (Capsaicin) receptors and mechanisms. Pharmacol. Rev. 51, 159–212.
Szallasi, A., Cortright, D. N., Blum, C. A., and Eid, S. R. (2007). The vanilloid receptor TRPV1: 10 years from channel cloning to antagonist proof-of-concept. Nat. Rev. Drug Discov. 6, 357–372. doi: 10.1038/nrd2280
Tavares-Ferreira, D., Shiers, S., Ray, P. R., Wangzhou, A., Jeevakumar, V., Sankaranarayanan, I., et al. (2022). Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci. Transl. Med. 14, eabj8186. doi: 10.1126/scitranslmed.abj8186
Teichert Russell, W., Smith Nathan, J., Raghuraman, S., Yoshikami, D., Light Alan, R., and Olivera Baldomero, M. (2012). Functional profiling of neurons through cellular neuropharmacology. Proc. Nat. Acad. Sci. 109, 1388–1395. doi: 10.1073/pnas.1118833109
Tominaga, M., Caterina, M. J., Malmberg, A. B., Rosen, T. A., Gilbert, H., Skinner, K., et al. (1998). The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543. doi: 10.1016/S0896-6273(00)80564-4
Urban, L., Campbell, E. A., Panesar, M., Patel, S., Chaudhry, N., Kane, S., et al. (2000). In vivo pharmacology of SDZ 249-665, a novel, non-pungent capsaicin analogue. Pain 89, 65–74. doi: 10.1016/S0304-3959(00)00349-3
Usoskin, D., Furlan, A., Islam, S., Abdo, H., Lonnerberg, P., Lou, D., et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153. doi: 10.1038/nn.3881
Valentino, R. J., and Volkow, N. D. (2018). Untangling the complexity of opioid receptor function. Neuropsychopharmacology 43, 2514–2520. doi: 10.1038/s41386-018-0225-3
Van Neerven, S. G. A., and Mouraux, A. (2020). Capsaicin-induced skin desensitization differentially affects a-delta and c-fiber-mediated heat sensitivity. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.00615
Viscusi, E. R., Torjman, M. C., Munera, C. L., Stauffer, J. W., Setnik, B. S., and Bagal, S. N. (2021). Effect of difelikefalin, a selective kappa opioid receptor agonist, on respiratory depression: a randomized, double-blind, placebo-controlled trial. Clin. Transl. Sci. 14, 1886–1893. doi: 10.1111/cts.13042
Von Buchholtz, L. J., Lam, R. M., Emrick, J. J., Chesler, A. T., and Ryba, N. J. P. (2020). Assigning transcriptomic class in the trigeminal ganglion using multiplex in situ hybridization and machine learning. Pain 161, 2212–2224. doi: 10.1097/j.pain.0000000000001911
Wang, F., Bélanger, E., Côté, S. L., Desrosiers, P., Prescott, S. A., Côt,é, D. C., et al. (2018). Sensory afferents use different coding strategies for heat and cold. Cell Rep. 23, 2001–2013. doi: 10.1016/j.celrep.2018.04.065
Wong, G. Y., and Gavva, N. R. (2009). Therapeutic potential of vanilloid receptor TRPV1 agonists and antagonists as analgesics: recent advances and setbacks. Brain Res. Rev. 60, 267–277. doi: 10.1016/j.brainresrev.2008.12.006
Wood, J. N., Winter, J., James, I. F., Rang, H. P., Yeats, J., and Bevan, S. (1988). Capsaicin-induced ion fluxes in dorsal root ganglion cells in culture. J. Neurosci. 8, 3208–3220. doi: 10.1523/JNEUROSCI.08-09-03208.1988
Xanthos, D. N., and Sandkühler, J. (2014). Neurogenic neuroinflammation: inflammatory CNS reactions in response to neuronal activity. Nat. Rev. Neurosci. 15, 43–53. doi: 10.1038/nrn3617
Xing, H., Chen, M., Ling, J., Tan, W., and Gu, J. G. (2007). TRPM8 mechanism of cold allodynia after chronic nerve injury. J. Neurosci. 27, 13680–13690. doi: 10.1523/JNEUROSCI.2203-07.2007
Xing, H., Ling, J., Chen, M., and Gu, J. G. (2006). Chemical and cold sensitivity of two distinct populations of TRPM8-expressing somatosensory neurons. J. Neurophysiol. 95, 1221–1230. doi: 10.1152/jn.01035.2005
Yaksh, T. L., Farb, D. H., Leeman, S. E., and Jessell, T. M. (1979). Intrathecal capsaicin depletes substance P in the rat spinal cord and produces prolonged thermal analgesia. Science 206, 481–483. doi: 10.1126/science.228392
Yang, Y., Mis, M. A., Estacion, M., Dib-Hajj, S. D., and Waxman, S. G. (2018). NaV1.7 as a pharmacogenomic target for pain: moving toward precision medicine. Trends Pharmacol. Sci. 39, 258–275. doi: 10.1016/j.tips.2017.11.010
Keywords: dorsal root ganglia (DRG), opioid receptor, transient receptor potential vanilloid-1 (TRPV1), nociception, transient receptor potential A1 (TRPA1), mu opioid (MOP) receptor, Kappa opioid receptor (KOR), TRPM8
Citation: Ma W, Sapio MR, Manalo AP, Maric D, Dougherty MK, Goto T, Mannes AJ and Iadarola MJ (2022) Anatomical Analysis of Transient Potential Vanilloid Receptor 1 (Trpv1+) and Mu-Opioid Receptor (Oprm1+) Co-expression in Rat Dorsal Root Ganglion Neurons. Front. Mol. Neurosci. 15:926596. doi: 10.3389/fnmol.2022.926596
Received: 22 April 2022; Accepted: 09 June 2022;
Published: 07 July 2022.
Edited by:
Fabien Marchand, INSERM U1107 Douleur et Biophysique Neurosensorielle (Neuro-Dol), FranceReviewed by:
Braulio Alfredo Muñoz Ramirez, Indiana University Bloomington, United StatesCopyright © 2022 Ma, Sapio, Manalo, Maric, Dougherty, Goto, Mannes and Iadarola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Iadarola, bWljaGFlbC5pYWRhcm9sYUBuaWguZ292
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.