
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Neurosci., 27 May 2022
Sec. Brain Disease Mechanisms
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.918664
This article is part of the Research TopicThe Legacy of Dr. Candance Pert: Recent Advances in the Neuropeptides and Neuroreceptors ResearchView all 11 articles
Vesicle transport through interaction with t-SNAREs 1A (Vti1a), a member of the N-ethylmaleimide-sensitive factor attachment protein receptor protein family, is involved in cell signaling as a vesicular protein and mediates vesicle trafficking. Vti1a appears to have specific roles in neurons, primarily by regulating upstream neurosecretory events that mediate exocytotic proteins and the availability of secretory organelles, as well as regulating spontaneous synaptic transmission and postsynaptic efficacy to control neurosecretion. Vti1a also has essential roles in neural development, autophagy, and unconventional extracellular transport of neurons. Studies have shown that Vti1a dysfunction plays critical roles in pathological mechanisms of Hepatic encephalopathy by influencing spontaneous neurotransmission. It also may have an unknown role in amyotrophic lateral sclerosis. A VTI1A variant is associated with the risk of glioma, and the fusion product of the VTI1A gene and the adjacent TCF7L2 gene is involved in glioma development. This review summarizes Vti1a functions in neurons and highlights the role of Vti1a in the several nervous system disorders.
Vesicle trafficking is an essential process in neuronal information transmission and is regulated by various regulators, such as the N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein family and Munc18-1 (Tang et al., 2021; Koike and Jahn, 2022). Membrane fusion mediated by the SNARE protein family is critical for vesicle trafficking (Mima, 2019). Based on their different functions in membrane fusion, SNARE proteins are divided into transport vesicle-associated SNAREs [v-SNAREs, such as Synaptobrevin (Syb)/vesicle-associated membrane protein (VAMP)] and SNAREs associated with target membrane localization [t-SNAREs, such as SNAP25 and Syntaxin (Stx)]; they spontaneously form complexes to carry out their functions (Katz and Brennwald, 2000). Furthermore, SNAREs are differentiated into Q-SNAREs and R-SNAREs based on the polar side chains associated with each SNARE central layer that contain highly conserved glutamine (Q) or arginine (R) residues (Kloepper et al., 2007). There are three Q-SNARE subfamilies, Qa, Qb, and Qc, and each SNARE complex contains one R-SNARE and three Q-SNAREs (Sutton et al., 1998). Currently, more than 30 SNARE proteins can be combined into unique complexes that drive specific membrane fusions (Bock et al., 2001).
The SNARE complex catalyzes the fusion of synaptic vesicles with the presynaptic membrane (Chen et al., 2021; Sauvola and Littleton, 2021). The canonical neuronal SNARE complex that mediates this process consists of the vesicular protein VAMP2 and the plasma membrane-associated proteins Syntaxin-1 and SNAP25 (Sudhof, 2004). These proteins are necessary for normal synaptic transmission, but some types of transmission are less dependent on these proteins. For example, deletion of the VAMP2 or SNAP25 genes in mice results in severely impaired stimulation-evoked neurotransmitter release (Liu et al., 2019). However, among the forms of spontaneous neurotransmitter release, a deficiency of VAMP2 or SNAP25 is less consequential (Ramirez and Kavalali, 2012; Liu et al., 2019). These results indicate the existence of other non-canonical SNARE proteins that are involved in synaptic vesicle fusion and could preferentially support spontaneous synaptic transmission (Kononenko and Haucke, 2012). VAMP7 and vesicle transport via interaction with t-SNARE homolog 1A (Vti1a) are synaptic vesicle proteins that cause spontaneous neurotransmitter release and regulate neuronal activity by mediating spontaneous neurotransmission (Ramirez and Kavalali, 2012; Crawford et al., 2017).
The Vti protein was originally discovered in yeast (Fischer von Mollard and Stevens, 1998). In mammals, the Vti protein has two orthologs, Vti1a and Vti1b, which are widely expressed in tissues (Kreykenbohm et al., 2002). Vti1a is primarily localized to the trans-Golgi network (TGN) in cells, while Vti1b is localized to late endosomes (Kreykenbohm et al., 2002). In neurons, Vti1a is also localized to the cell body and presynaptic terminals, and splice variants of this protein are enriched in purified synaptic vesicles (Antonin et al., 2000b; Takamori et al., 2006). Vti1a and Vti1b appear to have different functions. For example, Vti1a plays an essential role in insulin-stimulated glucose transport (Bose et al., 2005); and regulates exocytosis in adrenal chromaffin cells (Walter et al., 2014). The loss of Vti1a impairs exocytosis, while cells lacking Vti1b do not exhibit any secretory defects (Walter et al., 2014). Vti1a and vti1b may have key overlaps in certain functions. For instance, deletion of either vti1a or vti1b alone is tolerable in mice, whereas deletion of both results in extensive neurodegeneration and perinatal lethality (Kunwar et al., 2011). Additionally, in Vti1a/b double-deficient neurons, impaired synaptic density, decreased secretion efficiency, and Golgi cargo accumulation have been observed, which could be rescued by the expression of Vti1a or Vti1b (Emperador-Melero et al., 2018).
VTI genes are highly conserved, and in the published genomics data, only six loss-of-function mutations have been reported, of which three are VTI1A (Lek et al., 2016). The VTI1A gene is located on chromosome 10q25.2 (Zhang et al., 2018). Although 14 heterozygous deletions or duplications in VTI1A and two duplications in VTI1B have been reported in patients diagnosed with neurodevelopmental delay and intellectual disability, these numbers are lower compared to other SNARE genes (Firth et al., 2009). However, studies have reported that the fusion product between the human VTI1A gene and adjacent genes plays an important role in the occurrence of cancer (Zhang et al., 2018). Furthermore, VTI1A variants are associated with cancer risk (Su et al., 2015). For example, in lung cancer caused by smoking, the hypomethylation of VTI1A variants is involved in carcinogenesis (Gao et al., 2016). Interestingly, hypomethylation of VTI1A variants also is involved in the development of hypertriglyceridemia (Guardiola et al., 2022).
It exists a low sequence homology between Vti1a and Vti1b (31–33% homology) (Emperador-Melero et al., 2019). All Vti proteins contain a C-terminal type II transmembrane domain, a Qb-SNARE motif, and an N-terminal triple helix Habc domain (Figure 1; Antonin et al., 2002). The Habc domain of Vti proteins has multiple functions, including the recruitment of tethered proteins and regulators and the correct classification of SNARE proteins (Gossing et al., 2013). Its abnormality can affect the stability of protein structure and impair the function. In carboxypeptidase Y (CPY) trafficking in yeast, the folded Habc domain is critical for proper CPY trafficking and late endosomal SNARE complex assembly (Conibear and Stevens, 1998). Two temperature-sensitive mutants (Q29R and W79R) in the Habc domain of Vti have been found to cause CPY sorting defects (Gossing et al., 2013). Vti1a is a vesicle-transporting V-SNARE protein considered a Qb-SNARE due to the central layer of aspartate residues which acts similar to glutamine (Zwilling et al., 2007). It regulates cellular secretion and has important physiological functions in the nervous system (Tang, 2020).
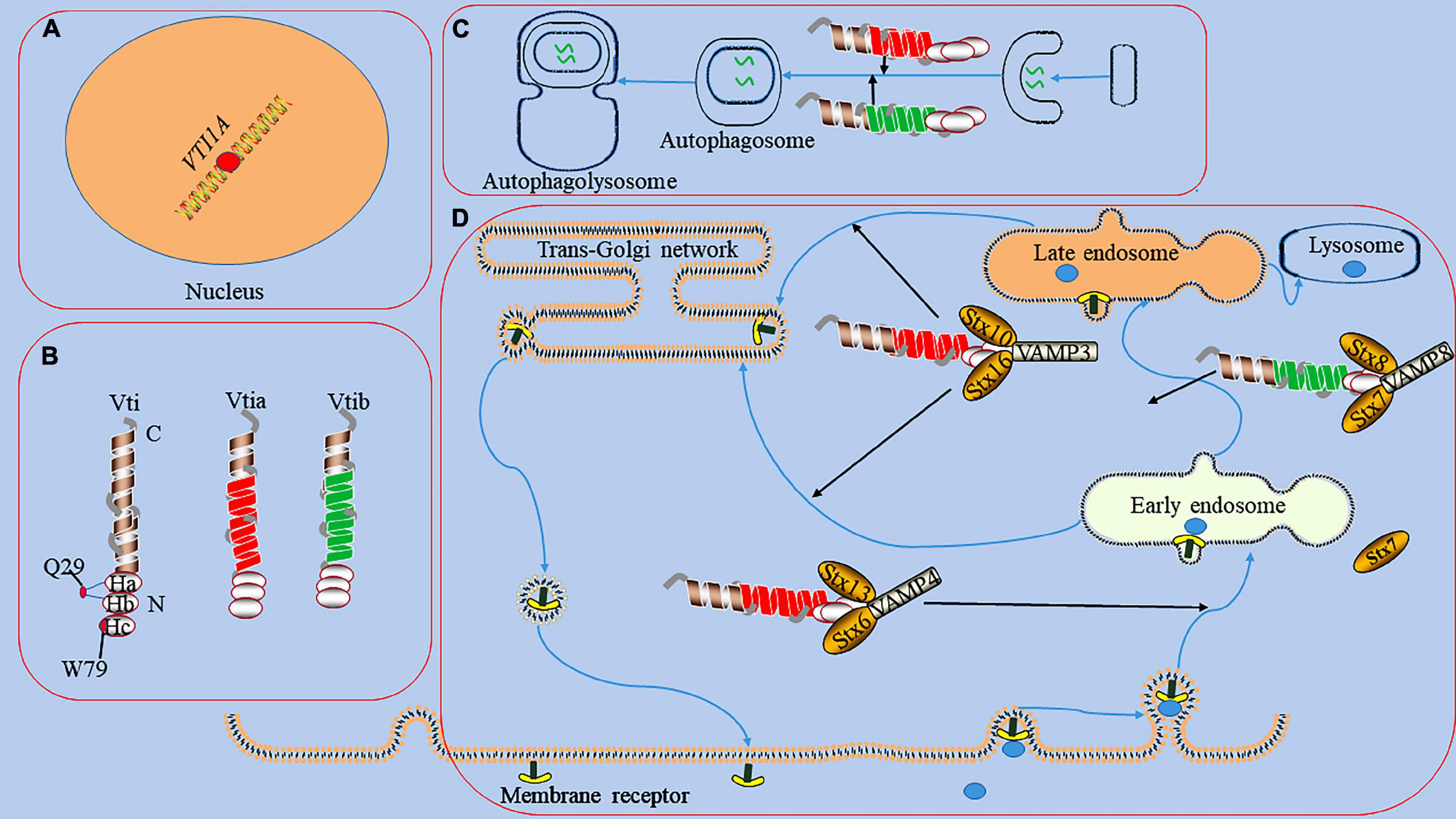
Figure 1. The functions of Vti1a in endosomal circulation and the formation of autophagosome. panel (A), it represents the dot of VTI1A on Mutations/variations; the mutations/variations and gene fusions of VTI1A (red dots) in glial cells may be oncogenic. Panel (B), it shows the domains and structures of Vti1a and Vti1b, a Vti protein contains a C-terminal transmembrane domain, a Qb-SNARE motif, and an N-terminal Habc domain; the Habc domain exists 2 mutants (Q29R localized in the loop between Ha and Hb and W79R localized on Hc); there has a low sequence homology between Vti1a and Vti1b. Panel (C), it displays the process of the formation of autophagosome; Vti1a and Vti1b mediates autophagosome maturation. Panel (D), it reveals endosomal circulation; Vti1a forms a SNARE complex with Syntaxin (Stx)10, Stx16, and VAMP3 to mediate the transport of related substances from the endosome to the Golgi apparatus. Vti1a co-immunoprecipitates with VAMP4, Stx6, and Stx16 in early circulating endosomal transport. Vti1b forms a complex with Stx7, Stx8, and VAMP8 and plays a role in late endosomal fusion.
Glial cells and neurons are the main secretory cells in the nervous system, and SNAREs are the important regulators (Verkhratsky et al., 2016; Karim and Brett, 2018; Vilcaes et al., 2021). Lysosome exocytosis in astrocytes is mainly dependent on VAMP7, and the downregulation of VAMP7 expression inhibits the fusion of ATP storage vesicles and the propagation of ATP-mediated intercellular Ca2+ wave (Verderio et al., 2012). A study had found that Vti1a is closely related to glioma (Wang et al., 2017). Although there is an evidence that Vti1a plays a pivotal role in intracellular signaling by regulating the secretion of intracellular organelles such as Golgi apparatus, lysosomes, and endosomes (Emperador-Melero et al., 2019). But there is no direct evidence for how Vti1a works in glial cells. Vti1a appears to carry out actions in neurons, including regulation of neurosecretion by affecting upstream neurosecretory events, spontaneous synaptic transmission, and postsynaptic efficacy, as well as regulation of neural development, autophagic activity, and unconventional extracellular transport of neurons (Ganley et al., 2008; Flowerdew and Burgoyne, 2009; Kunwar et al., 2011; Lu et al., 2013; Crawford et al., 2017; Emperador-Melero et al., 2018).
The Vti1a protein regulates neuronal secretion by controlling exocytosis proteins and availability of secretory organelles (Ganley et al., 2008; Emperador-Melero et al., 2018). Neuronal communication primarily depends on the secretion of signaling molecules carried by synaptic vesicles (SVs) and dense core vesicles (DCVs) (Emperador Melero et al., 2017). SVs store neurotransmitters and are locally recovered after exocytosis (Haucke et al., 2011; Jahn and Fasshauer, 2012). DCVs are continuously produced in the TGN and mainly stores neuropeptides and neurotrophic factors (Wong et al., 2012; Moro et al., 2021). Studies have indicated that a single deletion of Vti1a or combined deletion of Vti1a and Vti1b decreases SV and DCV release related to the Ca2+-dependent exocytosis of SV and DCV regulated by Vti1a (Figure 2; Walter et al., 2014; Crawford et al., 2017). In Vti1a/b double knockout neurons, SV and DCV secretion was reduced, and the exocytotic proteins SNAP25 and Munc13-1 were decreased to a similar extent; overexpression of Vti1a or Vti1b rescued this change (Emperador-Melero et al., 2018).
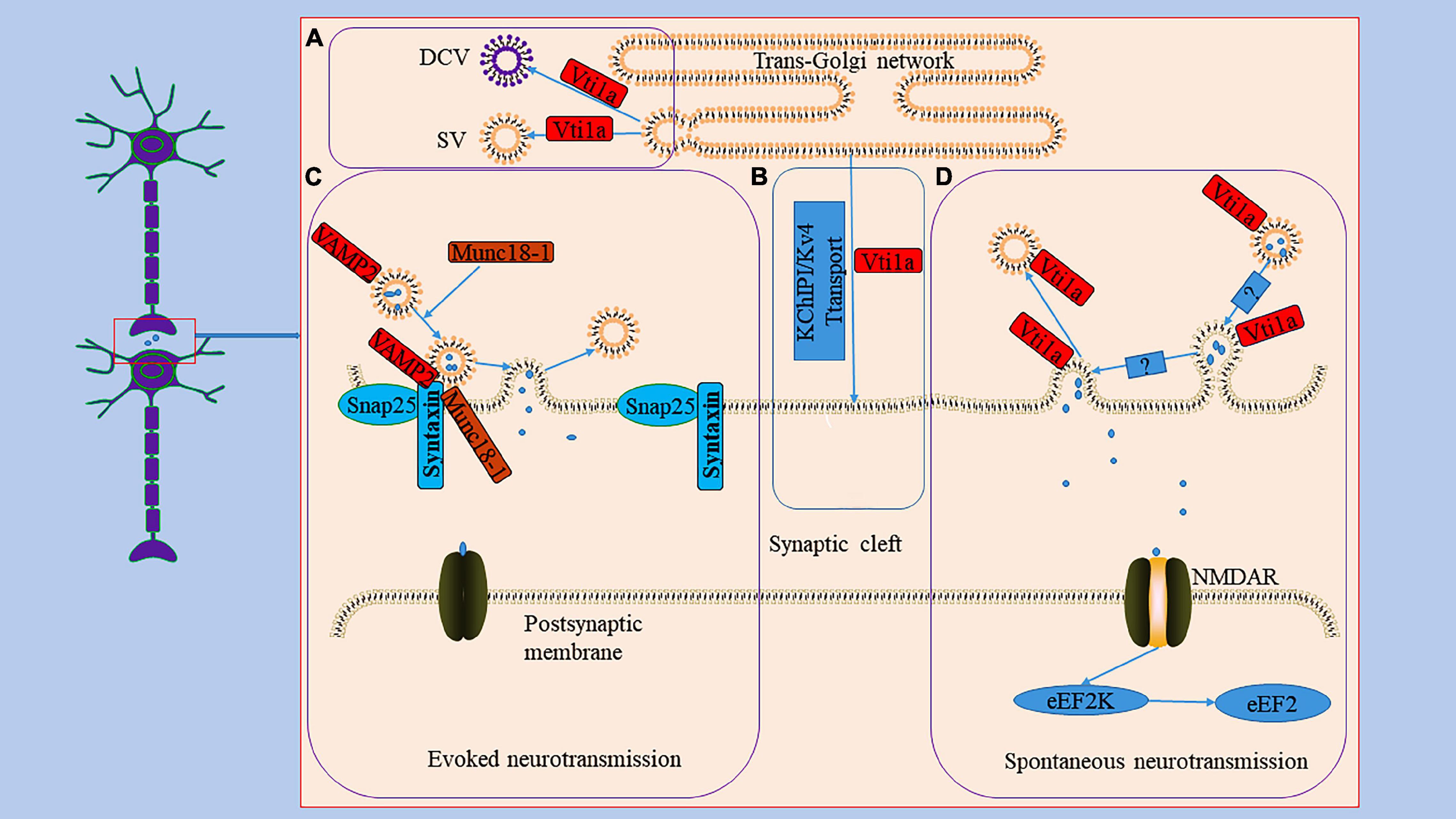
Figure 2. The role Vti1a in spontaneous transmission. Panel (A), it reveals the generation of DCV and SV in TGN; Vti1a is mainly localized to TGN to regulate the secretion of SV and DCV. Panel (B), it shows an unconventional exocrine pathway; Vti1a transports KChIP1/Kv4 through an unconventional exocrine pathway. Evoked neurotransmission and spontaneous neurotransmission are the two main ways of synaptic transmission. Panel (C), it represents the process of the evoked neurotransmission; the synaptic vesicle protein VAMP2 forms a SNARE complex with SNP25 and Syntaxin and mediates vesicle trafficking in evoked neurotransmission under the regulation of Munc18-1. Panel (D), it displays the process of the spontaneous transmission; Vti1a drives synaptic vesicle activity to mediate spontaneous transmission with an unclear mechanism to activate NMDAR to activate eEF2K to dephosphorylate eEF2.
Vti1a also cooperates with several SNAREs associated with the TGN and endosomes to mediate vesicular trafficking processes (Figure 1; Simonsen et al., 1998; Antonin et al., 2000a; Wendler and Tooze, 2001; Ganley et al., 2008). For example, Vti1a co-immunoprecipitates with VAMP4, Stx6, and Stx16 and assembles into a structurally conserved SNARE complex that mediates early, circulating endosomal transport of Shiga toxin and TGN46 (Simonsen et al., 1998; Wendler and Tooze, 2001). In addition, Vti1a forms a SNARE complex with Stx10, Stx16, and VAMP3 to mediate mannose 6-phosphate receptor (MPR) transport from endosomes to the Golgi (Ganley et al., 2008). Vti1b also plays a role in late endosomal fusion involving Stx7, Stx8, and VAMP8 (Antonin et al., 2000a). Vti1a/b-deficient neurons exhibit impaired synaptic density, secretion efficiency, Golgi size, and Golgi cargo accumulation (Emperador-Melero et al., 2018). Vti1a also may indirectly affect neurotransmitter transmission by influencing vesicle formation at the TGN as well as DCV generation (Walter et al., 2014).
It is known that action potential-evoked neurotransmitter release is a central process in the synaptic transmission of information (Rama et al., 2018). Furthermore, studies have demonstrated that spontaneous vesicle fusion plays a vital role in this process and is autonomous and heterogeneous in interneuron communication (Figure 2; Kavalali, 2015; Astacio et al., 2022). Spontaneous neurotransmitter release is not related to neuronal electrical activity but is equally involved in regulating synaptic stability and plasticity (Frank et al., 2006). Currently, it is debated whether the spontaneous neurotransmitter release has a dedicated pool of vesicles. For example, the same vesicles were involved in active and spontaneous release in cultured neurons from mouse (Wilhelm et al., 2010). However, in another study, both evoked and spontaneous vesicle pools were generated in rat hippocampal neurons (Fredj and Burrone, 2009). Vti1a is thought to regulate spontaneous fusion vesicles, driving synaptic vesicle activity to mediate spontaneous neurotransmitter transmission (Hoopmann et al., 2010; Kononenko and Haucke, 2012; Ramirez and Kavalali, 2012). An evidence suggests that endosome sorting is required for the production of exocytosing synaptic vesicles (Hoopmann et al., 2010). And Vti1a regulates the endosome sorting process (Emperador-Melero et al., 2019). In neurons from Vti1a/Vti1b double-deficient mice, whole-cell patch-clamp recordings demonstrated an 80–90% reduction in post-stimulation-evoked neurotransmitter release and a 65% reduction in the frequency of spontaneous fusion events (Emperador-Melero et al., 2018). Interestingly, the levels of exocytotic proteins SNAP25 and Munc13-1, Synaptobrevin-2, Stx1, and Munc18-1 were reduced; and the delivery of SNAP25 and DCV cargoes to the axon was reduced, and these molecules accumulate in the Golgi apparatus (Emperador-Melero et al., 2018). These secretion defects can be almost completely rescued by exogenous expression of Vti1a (Emperador-Melero et al., 2018). Thus, these evidences seem to suggest that Vti1a may mediate vesicle fusion by affecting horizontal synapses of associated exocytotic proteins as well as upstream events of apical vesicle secretion. However, whether Vti1a is directly involved in the fusion of synaptic vesicle membranes remains unknown. Deletion of Vti1a impairs spontaneous high-frequency glutamate release, which exerts pathogenic effects (Crawford et al., 2017). Vti1a knockdown results in reduced spontaneous SV fusion (Ramirez et al., 2012; Crawford et al., 2017). Vti1a exhibits robust trafficking under neuronal resting conditions, which selectively maintains spontaneous neurotransmitter release (Ramirez et al., 2012). In addition, studies have shown that synaptotagmin-11 inhibits spontaneous release primarily by inhibiting Vti1a-containing vesicles (Li et al., 2021). The evidences suggest that spontaneous synaptic transmission is independent of evoked synaptic transmission and that Vti1a is a joint regulator of this process. However, the downstream pathways of Vti1a-mediated regulation of vesicle activity and the corresponding molecular mechanisms remain unclear.
Vti1a also acts as a key regulator of the postsynaptic efficacy of spontaneous synaptic transmission (Figure 2; Crawford et al., 2017). Eukaryotic elongation factor-2 kinase (eEF2K) is a Ca2+/calmodulin-dependent serine/threonine kinase that is essential for regulating the elongation in protein translation (Heise et al., 2014). Its inactivation leads to the dephosphorylation of postsynaptic eEF2, which inhibits brain-hippocampus-derived neurotrophic factor (BDNF) protein synthesis, affecting neuronal activity (Verpelli et al., 2010; Suzuki and Monteggia, 2020). Loss of Vti1a impairs eEF2K signaling, resulting in decreased eEF2 phosphorylation (Crawford et al., 2017).
Normal neural development involves various stages of synaptic development and various links of neuronal connections, and this process is regulated by a variety of proteins, such as synaptic cell adhesion molecules and SNARE (Hepp and Langley, 2001; Ko, 2012). Major neurodevelopmental events associated with SNAREs include progenitor cell proliferation, neurite outgrowth, neuronal migration, synapse formation and transmission, and neurodegeneration (Hepp and Langley, 2001; Kimura et al., 2003; Wang and Tang, 2006). Double deletion of the VTI1A/1B gene impairs neuronal process outgrowth and projection leading to abnormal neuronal differentiation or maturation (Sokpor et al., 2021). For example, axonal loss and neurodegeneration in peripheral ganglia were observed in VTI1A/1B double-deficient mouse embryos, resulting in severely impaired neuronal development (Kunwar et al., 2011). In addition, a VTI1A/1B null mutation in mice resulted in a dysplastic cortex in late embryos (Sokpor et al., 2021). However, individual deletions of Vti1a or Vti1b are tolerated in mice, whereas simultaneous deletions cause extensive neurodegeneration and lethal effects (Atlashkin et al., 2003; Kunwar et al., 2011). Although the Vti1a deficiency is not fatal, it adversely affects neuronal development and metabolism (Kunwar et al., 2011).
Autophagy is a cellular degradation pathway which autophagosomes fuse with lysosomes to form autophagolysosomes and degrade their contents (Iriondo et al., 2021; Gubas and Dikic, 2022). The autophagy pathway involves a series of membrane fusion events and requires the participation of SNARE proteins, such as Syn17, VAMP3, VAMP7, YKT6, Vti1 (Tian et al., 2021). For example, mutation of Vti1p in yeast disrupts autophagosome-vacuole fusion (Ishihara et al., 2001). In mammals, the abnormality of Vti1a and Vti1b affects the formation of autophagosomes (Figure 1; Lu et al., 2013; Chou et al., 2021). Mutations in the CHMP2B gene can cause frontotemporal dementia, the pathogenicity of which is primarily thought to be the result of autophagy-endolysosomal dysfunction (Deng et al., 2022; Roos et al., 2022). Stx13, as a strong genetic modifier of CHMP2B, is involved in phagocyte maturation and affects autophagosome formation (Lu et al., 2013). Knockdown of Stx13 or its binding partner Vti1a leads to accumulation of the autophagy marker LC3, affects autophagosome maturation, and blocks autophagic flux (Lu et al., 2013). Although studies have shown that regulation of Vti1a and Vti1b can inhibit phagocytosis in phagocytes and may play a regulatory role in the process of autophagy (Cai et al., 2011). However, the specific mechanism by which Vti1a regulates the autophagy process remains unclear.
The K channel interacting proteins (KChIPs) belong to the neuronal calcium receptor family and assemble into a natural complex with the α subunit of the voltage-gated Kv4 potassium channel, encoding A-type K+ current to regulate neuronal excitability (Bähring, 2018; Kise et al., 2021). The specific assembly contributes to forming and stabilizing voltage-gated potassium channel tetramers and increases channel transport to the cell membrane surface (Alfaro-Ruíz et al., 2020). For example, siRNA-mediated knockdown of Vti1a or VAMP7 inhibited Kv4/KChIP1 transport to the Neuro2A cell membrane (Flowerdew and Burgoyne, 2009). The VAMP7/Vti1a SNARE complex controls an unconventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels (Figure 2; Flowerdew and Burgoyne, 2009). These evidences suggest that neurons depend on an unconventional exocrine pathway of Vti1a and VAMP7 for Kv4/KChIP1 trafficking. However, the nature of this pathway and the membrane transport mechanism still need to be explored further.
Studies have shown that Vti1a plays a substantial role in some neurological disorders (Table 1); Vti1a gene variants and fusion transcripts with adjacent genes are involved in gliomas (Wang et al., 2017); Vti1a plays a key role hepatic encephalopathy (Popek et al., 2018) by regulating spontaneous neurotransmitter transmission. Furthermore, Vti1a is involved in ALS, but its specific role is unclear (Nagao and Hayashi, 2010).
Vti1a forms fusion products with adjacent genes in human cancer tissues, and Vti1a variants are associated with the development of various cancers (Table 2; Gao et al., 2016; Wang et al., 2016, 2017; Zhang et al., 2018; Tsuge et al., 2019). A previous study identified VTI1A as one of the susceptibility genes for glioma in European populations (Kinnersley et al., 2015). Interestingly, in 473 Chinese glioma, the VTI1A single nucleotide polymorphism (SNP) variant rs11196067 was significantly associated with its risk, suggesting that VTI1A variants might increase the susceptibility of individuals to glioma (Wang et al., 2017). In addition, the results of a meta-analysis of four genome-wide association studies on glioma revealed that the VTI1A SNP variant rs11196067 was a susceptibility gene in glioma (Kinnersley et al., 2015).
In GBM of glioma, genomic and transcriptome sequencing revealed gene fusions in approximately 30–50% of patients (Shah et al., 2013). The transcription factor 7-like 2 (TCF7L2) gene is located on chromosome 10q25.2, about 131 kb downstream of the VTI1A gene (Zhang et al., 2018). Several variants were identified in the VTI1A-TCF7L2 fusion gene region that were associated with tumor risk, including glioma (Zhang et al., 2018). Surprisingly, the VTI1A-TCF7L2 gene fusion transcripts were detected in the tumor tissue and plasma of GBM (Wang et al., 2020). The TCF7L2 gene product is a transcription factor containing a high mobility group cassette that plays an essential role in the Wnt signaling pathway (Su et al., 2015; Torres-Aguila et al., 2022). The Wnt signaling pathway has been shown to play an important role in the progression of glioma by affecting the differentiation, proliferation, migration, and apoptosis of neural cells (Tompa et al., 2019; Pei et al., 2022). Recent studies have shown the presence of glioma stem cells in GBM (Ma et al., 2020). Wnt/β-catenin is a major signaling pathway in brain development, regulating the self-renewal and differentiation of neural stem and progenitor cells (Tompa et al., 2018). Studies have shown that fusion of the VTI1A and TCF7L2 genes, encoding a VTI1A-TCF4 fusion protein containing truncated TCF4, regulates the Wnt signaling pathway and participates in colorectal cancer development (Davidsen et al., 2018). The Vti1a-TCF4 fusion product may interfere with oncogenic signaling in the brain to promote gliomagenesis through a similar mechanism. In addition, the VTI1A gene encodes the v-SNARE protein, which mediates the transport of vesicles from the endosome to the TGN. A sorting molecule, the Wnt-interacting receptor, was found to circulate from the plasma membrane to the Golgi through early endosomes (Harterink et al., 2011). This process may be regulated by Vti1a (Harterink et al., 2011). In short, abnormal Vti1a may affect Wnt-interacting receptor signaling or promote tumor cell migration in an as yet undetermined manner, but the exact mechanism is unclear.
Hepatic encephalopathy is a complex neuropsychiatric syndrome, usually resulting from acute or chronic liver failure, and is associated with decreased excitatory neurotransmission (Albrecht et al., 2010; Palomero-Gallagher and Zilles, 2013). Impaired release of synaptic neurotransmitter transmission with concomitant reduction of kinesin Vti1a was observed in mice with azomethane-induced hepatic encephalopathy (Popek et al., 2018). This alteration may be related to inefficient Vti1a protein recruitment and impaired SV transport to neurotransmitter release sites (Popek et al., 2018). Another study on the expression of glycogen synthase kinase 3beta in the spinal cord of amyotrophic lateral sclerosis (ALS) reported decreased TGN GSK3beta expression in motor neurons, as well as Vti1a (Nagao and Hayashi, 2010). However, the exact mechanism of action is still unknown.
In conclusion, Vti1a is a critical vesicular protein that primarily regulates neurosecretory. It has essential roles in spontaneous neurotransmitter transmission, neuronal autophagy, neural development, and unconventional extracellular transport. Furthermore, Vti1a forms transcriptional complexes with adjacent genes, and Vti1a variants play critical roles in glioma. Therefore, Vti1a is highly relevant in neurological diseases and presents substantial research potential. For example, one study found that the Vti1a gene transcript could be detected in tumor tissue and serum of GBM (Wang et al., 2020). This evidence suggests that Vti1a has the potential to be a candidate biomarker for GBM. However, the sample size of that particular study was small, and further research is needed to confirm the results.
Nevertheless, several issues still need to be resolved. For example, how do VTI1A variants or mutants combine with gene fusion products or drive tumorigenesis on their own? As a protein involved in spontaneous neurotransmission, what is the relationship between VAMP7 and Vti1a? How do these proteins function in complex neurotransmission? Increased understanding of the function and regulatory mechanisms of Vti1a might provide potential therapeutic targets for related diseases.
FT and DX: conceptualization. FT, DX, XZ, and XL: software. FT, DX, ZZ, and XL: resources. FT and JF: writing–original draft preparation. FT, DX, XZ, ZZ, and XL: writing-review and editing. XZ and XL: visualization. DX and XL: supervision. All authors read and approved the final manuscript.
This study was supported by the National Key R&D Program of China (grant nos. 2017YFA 0104201 and 2017YFA 0104200), the National Science Foundation of China (grant nos. 82071353, 82001593, 81330016, 81630038, and 81771634), and the Science and Technology Bureau of Chengdu City (grant no. 2015-HM01-00424-SF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank EditSprings (https://www.editsprings.com) for editing English language of our manuscript.
Vti1a, Vesicle transport through interaction with t-SNAREs 1A; SNARE, N-ethylmaleimide -sensitive factor attachment protein receptor; VAMP, vesicle-associated membrane protein; Syb, Synaptobrevin; Stx, Syntaxin; TGN, trans-Golgi network; SV, synaptic vesicle; DCV, dense core vesicle; eEF2K, eukaryotic elongation factor-2 kinase; BDNF, brain-hippocampus-derived neurotrophic factor; KChIP, K channel interacting proteins; TCF7L2, The transcription factor 7-like 2; SNP, single nucleotide polymorphism; GBM, glioblastoma; NMDAR, N-methyl -D-aspartate receptor; AMPA, amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid.
Albrecht, J., Zielińska, M., and Norenberg, M. (2010). Glutamine as a mediator of ammonia neurotoxicity: a critical appraisal. Biochem. Pharmacol. 80, 1303–1308. doi: 10.1016/j.bcp.2010.07.024
Alfaro-Ruíz, R., Aguado, C., Martín-Belmonte, A., Moreno-Martínez, A., and Luján, R. (2020). Cellular and subcellular localisation of Kv4-associated KChIP proteins in the rat cerebellum. Int. J. Mol. Sci. 21:6403. doi: 10.3390/ijms21176403
Antonin, W., Dulubova, I., Arac, D., Pabst, S., Plitzner, J., Rizo, J., et al. (2002). The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J. Biol. Chem. 277, 36449–36456. doi: 10.1074/jbc.M204369200
Antonin, W., Riedel, D., and von Mollard, G. (2000b). The SNARE Vti1a-beta is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci. 20, 5724–5732. doi: 10.1523/jneurosci.20-15-05724.2000
Antonin, W., Holroyd, C., Fasshauer, D., Pabst, S., Von Mollard, G., and Jahn, R. (2000a). A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19, 6453–6464. doi: 10.1093/emboj/19.23.6453
Astacio, H., Vasin, A., and Bykhovskaia, M. (2022). Stochastic properties of spontaneous synaptic transmission at individual active zones. J. Neurosci. 42, 1001–1019. doi: 10.1523/jneurosci.1162-21.2021
Atlashkin, V., Kreykenbohm, V., Eskelinen, E., Wenzel, D., Fayyazi, A., and Fischer von Mollard, G. (2003). Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol. Cell. Biol. 23, 5198–5207. doi: 10.1128/mcb.23.15.5198-5207.2003
Bähring, R. (2018). Kv channel-interacting proteins as neuronal and non-neuronal calcium sensors. Channels 12, 187–200. doi: 10.1080/19336950.2018.1491243
Bock, J., Matern, H., Peden, A., and Scheller, R. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. doi: 10.1038/35057024
Bose, A., Guilherme, A., Huang, S., Hubbard, A., Lane, C., Soriano, N., et al. (2005). The v-SNARE Vti1a regulates insulin-stimulated glucose transport and Acrp30 secretion in 3T3-L1 adipocytes. J. Biol. Chem. 280, 36946–36951. doi: 10.1074/jbc.M508317200
Cai, D., Ho, Y., Chiow, K., Wee, S., Han, Y., Peh, M., et al. (2011). Aspirin regulates SNARE protein expression and phagocytosis in dendritic cells. Mol. Membr. Biol. 28, 90–102. doi: 10.3109/09687688.2010.525756
Chen, F., Chen, H., Chen, Y., Wei, W., Sun, Y., Zhang, L., et al. (2021). Dysfunction of the SNARE complex in neurological and psychiatric disorders. Pharmacol. Res. 165, 105469. doi: 10.1016/j.phrs.2021.105469
Chou, H., Lee, Y., Lin, Y., Wen, J., Peng, W., Hsieh, P., et al. (2021). PTPN9-mediated dephosphorylation of VTI1B promotes ATG16L1 precursor fusion and autophagosome formation. Autophagy 17, 2750–2765. doi: 10.1080/15548627.2020.1838117
Conibear, E., and Stevens, T. H. (1998). Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta 1404, 211–230. doi: 10.1016/S0167-4889(98)00058-5
Crawford, D., Ramirez, D., Trauterman, B., Monteggia, L., and Kavalali, E. (2017). Selective molecular impairment of spontaneous neurotransmission modulates synaptic efficacy. Nat. Commun. 8:14436. doi: 10.1038/ncomms14436
Davidsen, J., Larsen, S., Coskun, M., Gögenur, I., Dahlgaard, K., Bennett, E., et al. (2018). The VTI1A-TCF4 colon cancer fusion protein is a dominant negative regulator of Wnt signaling and is transcriptionally regulated by intestinal homeodomain factor CDX2. PLoS One 13:e0200215. doi: 10.1371/journal.pone.0200215
Deng, X., Sun, X., Yue, W., Duan, Y., Hu, R., Zhang, K., et al. (2022). CHMP2B regulates TDP-43 phosphorylation and cytotoxicity independent of autophagy via CK1. J. Cell Biol. 221:e202103033. doi: 10.1083/jcb.202103033
Emperador Melero, J., Nadadhur, A., Schut, D., Weering, J., Heine, V., Toonen, R., et al. (2017). Differential maturation of the two regulated secretory pathways in human iPSC-derived neurons. Stem Cell Rep. 8, 659–672. doi: 10.1016/j.stemcr.2017.01.019
Emperador-Melero, J., Huson, V., van Weering, J., Bollmann, C., Fischer von Mollard, G., Toonen, R., et al. (2018). Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat. Commun. 9:3421. doi: 10.1038/s41467-018-05699-z
Emperador-Melero, J., Toonen, R. F., and Verhage, M. (2019). Vti proteins: beyond endolysosomal trafficking. Neuroscience 420, 32–40. doi: 10.1016/j.neuroscience.2018.11.014
Firth, H., Richards, S., Bevan, A., Clayton, S., Corpas, M., Rajan, D., et al. (2009). DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533. doi: 10.1016/j.ajhg.2009.03.010
Fischer von Mollard, G., and Stevens, T. (1998). A human homolog can functionally replace the yeast vesicle-associated SNARE Vti1p in two vesicle transport pathways. J. Biol. Chem. 273, 2624–2630. doi: 10.1074/jbc.273.5.2624
Flowerdew, S., and Burgoyne, R. (2009). A VAMP7/Vti1a SNARE complex distinguishes a non-conventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels. Biochem. J. 418, 529–540. doi: 10.1042/bj20081736
Frank, C., Kennedy, M., Goold, C., Marek, K., and Davis, G. (2006). Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron 52, 663–677. doi: 10.1016/j.neuron.2006.09.029
Fredj, N., and Burrone, J. (2009). A resting pool of vesicles is responsible for spontaneous vesicle fusion at the synapse. Nat. Neurosci. 12, 751–758. doi: 10.1038/nn.2317
Ganley, I., Espinosa, E., and Pfeffer, S. (2008). A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 180, 159–172. doi: 10.1083/jcb.200707136
Gao, X., Zhang, Y., Breitling, L., and Brenner, H. (2016). Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget 7, 59017–59028. doi: 10.18632/oncotarget.10007
Gossing, M., Chidambaram, S., and Fischer von Mollard, G. (2013). Importance of the N-terminal domain of the Qb-SNARE Vti1p for different membrane transport steps in the yeast endosomal system. PLoS One 8:e66304. doi: 10.1371/journal.pone.0066304
Guardiola, M., Ibarretxe, D., Plana, N., Masana, L., and Ribalta, J. (2022). DNA methylation pattern of hypertriglyceridemic subjects. Clin. Investig. Arterioscler. 34, 27–32. doi: 10.1016/j.arteri.2021.09.002
Gubas, A., and Dikic, I. (2022). A guide to the regulation of selective autophagy receptors. FEBS J. 289, 75–89. doi: 10.1111/febs.15824
Harterink, M., Port, F., Lorenowicz, M., McGough, I., Silhankova, M., Betist, M., et al. (2011). A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923. doi: 10.1038/ncb2281
Haucke, V., Neher, E., and Sigrist, S. (2011). Protein scaffolds in the coupling of synaptic exocytosis and endocytosis. Nat. Rev. Neurosci. 12, 127–138. doi: 10.1038/nrn2948
Heise, C., Gardoni, F., Culotta, L., di Luca, M., Verpelli, C., and Sala, C. (2014). Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. Front. Cell Neurosci. 8:35. doi: 10.3389/fncel.2014.00035
Hepp, R., and Langley, K. (2001). SNAREs during development. Cell Tissue Res. 305, 247–253. doi: 10.1007/s004410100359
Hoopmann, P., Punge, A., Barysch, S., Westphal, V., Bückers, J., Opazo, F., et al. (2010). Endosomal sorting of readily releasable synaptic vesicles. Proc. Natl. Acad. Sci. U.S.A. 107, 19055–19060. doi: 10.1073/pnas.1007037107
Iriondo, M., Etxaniz, A., Antón, Z., Montes, L., and Alonso, A. (2021). Molecular and mesoscopic geometries in autophagosome generation. A review. Biochim. Biophys. Acta Biomembr. 1863:183731. doi: 10.1016/j.bbamem.2021.183731
Ishihara, N., Hamasaki, M., Yokota, S., Suzuki, K., Kamada, Y., Kihara, A., et al. (2001). Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12, 3690–3702. doi: 10.1091/mbc.12.11.3690
Jahn, R., and Fasshauer, D. (2012). Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207. doi: 10.1038/nature11320
Karim, M. A., and Brett, C. L. (2018). The Na(+)(K(+))/H(+) exchanger Nhx1 controls multivesicular body-vacuolar lysosome fusion. Mol. Biol Cell 29, 317–325. doi: 10.1091/mbc.E17-08-0496
Katz, L., and Brennwald, P. (2000). Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell 11, 3849–3858. doi: 10.1091/mbc.11.11.3849
Kavalali, E. (2015). The mechanisms and functions of spontaneous neurotransmitter release. Nat. Rev. Neurosci. 16, 5–16. doi: 10.1038/nrn3875
Kimura, K., Mizoguchi, A., and Ide, C. (2003). Regulation of growth cone extension by SNARE proteins. J. Histochem. Cytochem. 51, 429–433. doi: 10.1177/002215540305100404
Kinnersley, B., Labussière, M., Holroyd, A., Di Stefano, A., Broderick, P., Vijayakrishnan, J., et al. (2015). Genome-wide association study identifies multiple susceptibility loci for glioma. Nat. Commun. 6:8559. doi: 10.1038/ncomms9559
Kise, Y., Kasuya, G., Okamoto, H., Yamanouchi, D., Kobayashi, K., Kusakizako, T., et al. (2021). Structural basis of gating modulation of Kv4 channel complexes. Nature 599, 158–164. doi: 10.1038/s41586-021-03935-z
Kloepper, T., Kienle, C., and Fasshauer, D. (2007). An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18, 3463–3471. doi: 10.1091/mbc.e07-03-0193
Ko, J. (2012). The leucine-rich repeat superfamily of synaptic adhesion molecules: LRRTMs and Slitrks. Mol. Cells. 34, 335–340. doi: 10.1007/s10059-012-0113-3
Koike, S., and Jahn, R. (2022). SNARE proteins: zip codes in vesicle targeting? Biochem. J. 479, 273–288. doi: 10.1042/bcj20210719
Kononenko, N., and Haucke, V. (2012). Spontaneous neurotransmission: a SNARE for the rest. Neuron 73, 3–5. doi: 10.1016/j.neuron.2011.12.015
Kreykenbohm, V., Wenzel, D., Antonin, W., Atlachkine, V., and von Mollard, G. (2002). The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81, 273–280. doi: 10.1078/0171-9335-00247
Kunwar, A., Rickmann, M., Backofen, B., Browski, S., Rosenbusch, J., Schöning, S., et al. (2011). Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc. Natl. Acad. Sci. U.S.A. 108, 2575–2580. doi: 10.1073/pnas.1013891108
Lek, M., Karczewski, K., Minikel, E., Samocha, K., Banks, E., Fennell, T., et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. doi: 10.1038/nature19057
Li, W., Wang, Y., Li, C., Gao, P., Zhang, F., Hu, M., et al. (2021). Synaptotagmin-11 inhibits spontaneous neurotransmission through vti1a. J. Neurochem. 159, 729–741. doi: 10.1111/jnc.15523
Liu, Y., Sugiura, Y., Südhof, T., and Lin, W. (2019). Ablation of all synaptobrevin vSNAREs blocks evoked but not spontaneous neurotransmitter release at neuromuscular synapses. J. Neurosci. 39, 6049–6066. doi: 10.1523/jneurosci.0403-19.2019
Lu, Y., Zhang, Z., Sun, D., Sweeney, S., and Gao, F. (2013). Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell 52, 264–271. doi: 10.1016/j.molcel.2013.08.041
Ma, D., Hou, L., Xia, H., Li, H., Fan, H., Jia, X., et al. (2020). PER2 inhibits proliferation and stemness of glioma stem cells via the Wnt/β-catenin signaling pathway. Oncol. Rep. 44, 533–542. doi: 10.3892/or.2020.7624
Mima, J. (2019). Reconstituted proteoliposome fusion mediated by yeast SNARE-family proteins. Methods Mol. Biol. 1860, 303–322. doi: 10.1007/978-1-4939-8760-3_20
Moro, A., Hoogstraaten, R., Persoon, C., Verhage, M., and Toonen, R. (2021). Quantitative analysis of dense-core vesicle fusion in rodent CNS neurons. STAR Protoc. 2:100325. doi: 10.1016/j.xpro.2021.100325
Nagao, M., and Hayashi, H. (2010). Expression of glycogen synthase kinase-3beta in motor neurons of Amyotrophic lateral sclerosis [Conference Abstract]. Neuropathology. 30:315. doi: 10.1111/j.1440-1789.2010.01036.x
Palomero-Gallagher, N., and Zilles, K. (2013). Neurotransmitter receptor alterations in hepatic encephalopathy: a review. Arch Biochem. Biophys. 536, 109–121. doi: 10.1016/j.abb.2013.02.010
Pei, Y., Zhang, H., Lu, K., Tang, X., Li, J., Zhang, E., et al. (2022). Circular RNA circRNA_0067934 promotes glioma development by modulating the microRNA miR-7/Wnt/β-catenin axis. Bioengineered 13, 5792–5802. doi: 10.1080/21655979.2022.2033382
Popek, M., Bobula, B., Sowa, J., Hess, G., Polowy, R., Filipkowski, R., et al. (2018). Cortical synaptic transmission and plasticity in acute liver failure are decreased by presynaptic events. Mol. Neurobiol. 55, 1244–1258. doi: 10.1007/s12035-016-0367-4
Rama, S., Zbili, M., and Debanne, D. (2018). Signal propagation along the axon. Curr. Opin. Neurobiol. 51, 37–44. doi: 10.1016/j.conb.2018.02.017
Ramirez, D., and Kavalali, E. (2012). The role of non-canonical SNAREs in synaptic vesicle recycling. Cell. Logist. 2, 20–27. doi: 10.4161/cl.20114
Ramirez, D., Khvotchev, M., Trauterman, B., and Kavalali, E. (2012). Vti1a identifies a vesicle pool that preferentially recycles at rest and maintains spontaneous neurotransmission. Neuron 73, 121–134. doi: 10.1016/j.neuron.2011.10.034
Roos, P., Johannsen, P., Lindquist, S., Brown, J., Waldemar, G., Duno, M., et al. (2022). Six generations of CHMP2B-mediated frontotemporal dementia: clinical features, predictive testing, progression, and survival. Acta Neurol. Scand. 145, 529–540. doi: 10.1111/ane.13578
Sauvola, C., and Littleton, J. (2021). SNARE regulatory proteins in synaptic vesicle fusion and recycling. Front. Mol. Neurosci. 14:733138. doi: 10.3389/fnmol.2021.733138
Shah, N., Lankerovich, M., Lee, H., Yoon, J., Schroeder, B., and Foltz, G. (2013). Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 14:818. doi: 10.1186/1471-2164-14-818
Simonsen, A., Bremnes, B., Rønning, E., Aasland, R., and Stenmark, H. (1998). Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol. 75, 223–231. doi: 10.1016/s0171-9335(98)80116-7
Sokpor, G., Rosenbusch, J., Kunwar, A., Rickmann, M., Tuoc, T., Rizzoli, S., et al. (2021). Ablation of Vti1a/1b triggers neural progenitor pool depletion and cortical layer 5 malformation in late-embryonic mouse cortex. Neuroscience 463, 303–316. doi: 10.1016/j.neuroscience.2021.03.021
Su, W., Chen, Z., Zhang, X., Su, J., Xie, Z., Yan, H., et al. (2015). Single nucleotide polymorphisms in VTI1A gene contribute to the susceptibility of Chinese population to non-small cell lung cancer. Int. J. Biol. Markers 30, e286–e293. doi: 10.5301/jbm.5000140
Sudhof, T. (2004). The synaptic vesicle cycle. Annu. Rev. Neurosci. 27, 509–547. doi: 10.1146/annurev.neuro.26.041002.131412
Sutton, R., Fasshauer, D., Jahn, R., and Brunger, A. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. doi: 10.1038/26412
Suzuki, K., and Monteggia, L. (2020). The role of eEF2 kinase in the rapid antidepressant actions of ketamine. Adv. Pharmacol. 89, 79–99. doi: 10.1016/bs.apha.2020.04.005
Takamori, S., Holt, M., Stenius, K., Lemke, E., Grønborg, M., Riedel, D., et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. doi: 10.1016/j.cell.2006.10.030
Tang, B. (2020). Vesicle transport through interaction with t-SNAREs 1a (Vti1a)’s roles in neurons. Heliyon 6:e04600. doi: 10.1016/j.heliyon.2020.e04600
Tang, F., Xiao, D., Chen, L., Gao, H., and Li, X. (2021). Role of Munc18-1 in the biological functions and pathogenesis of neurological disorders (Review). Mol. Med. Rep. 23:198. doi: 10.3892/mmr.2021.11837
Tian, X., Teng, J., and Chen, J. (2021). New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy 17, 2680–2688. doi: 10.1080/15548627.2020.1823124
Tompa, M., Kalovits, F., Nagy, A., and Kalman, B. (2018). Contribution of the Wnt pathway to defining biology of glioblastoma. Neuromol. Med. 20, 437–451. doi: 10.1007/s12017-018-8514-x
Tompa, M., Nagy, A., Komoly, S., and Kalman, B. (2019). Wnt pathway markers in molecular subgroups of glioblastoma. Brain Res. 1718, 114–125. doi: 10.1016/j.brainres.2019.05.008
Torres-Aguila, N., Salonna, M., Hoppler, S., and Ferrier, D. (2022). Evolutionary diversification of the canonical Wnt signaling effector TCF/LEF in chordates. Dev. Growth Differ. 64, 120–137. doi: 10.1111/dgd.12771
Tsuge, S., Saberi, B., Cheng, Y., Wang, Z., Kim, A., Luu, H., et al. (2019). Detection of novel fusion transcript VTI1A-CFAP46 in hepatocellular carcinoma. Gastrointest. Tumors 6, 11–27. doi: 10.1159/000496795
Verderio, C., Cagnoli, C., Bergami, M., Francolini, M., Schenk, U., Colombo, A., et al. (2012). TI-VAMP/VAMP7 is the SNARE of secretory lysosomes contributing to ATP secretion from astrocytes. Biol. Cell 104, 213–228. doi: 10.1111/boc.201100070
Verkhratsky, A., Matteoli, M., Parpura, V., Mothet, J.-P., and Zorec, R. (2016). Astrocytes as secretory cells of the central nervous system: idiosyncrasies of vesicular secretion. EMBO J. 35, 239–257. doi: 10.15252/embj.201592705
Verpelli, C., Piccoli, G., Zibetti, C., Zanchi, A., Gardoni, F., Huang, K., et al. (2010). Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci. 30, 5830–5842. doi: 10.1523/jneurosci.0119-10.2010
Vilcaes, A. A., Chanaday, N. L., and Kavalali, E. T. (2021). Interneuronal exchange and functional integration of synaptobrevin via extracellular vesicles. Neuron 109, 971.e5–983.e5. doi: 10.1016/j.neuron.2021.01.007
Walter, A., Kurps, J., de Wit, H., Schöning, S., Toft-Bertelsen, T., Lauks, J., et al. (2014). The SNARE protein vti1a functions in dense-core vesicle biogenesis. EMBO J. 33, 1681–1697. doi: 10.15252/embj.201387549
Wang, L., Yekula, A., Muralidharan, K., Small, J., Rosh, Z., Kang, K., et al. (2020). Novel gene fusions in glioblastoma tumor tissue and matched patient plasma. Cancers 12:1219. doi: 10.3390/cancers12051219
Wang, N., Deng, Z., Wang, M., Li, R., Xu, G., and Bao, G. (2017). Additional evidence supports association of common genetic variants in VTI1A and ETFA with increased risk of glioma susceptibility. J. Neurol. Sci. 375, 282–288. doi: 10.1016/j.jns.2017.02.013
Wang, Y., and Tang, B. (2006). SNAREs in neurons–beyond synaptic vesicle exocytosis (Review). Mol. Membr. Biol. 23, 377–384. doi: 10.1080/09687860600776734
Wang, Z., Seow, W., Shiraishi, K., Hsiung, C., Matsuo, K., Liu, J., et al. (2016). Meta-analysis of genome-wide association studies identifies multiple lung cancer susceptibility loci in never-smoking Asian women. Hum. Mol. Genet. 25, 620–629. doi: 10.1093/hmg/ddv494
Wendler, F., and Tooze, S. (2001). Syntaxin 6: the promiscuous behaviour of a SNARE protein. Traffic 2, 606–611. doi: 10.1034/j.1600-0854.2001.20903.x
Wilhelm, B., Groemer, T., and Rizzoli, S. (2010). The same synaptic vesicles drive active and spontaneous release. Nat. Neurosci. 13, 1454–1456. doi: 10.1038/nn.2690
Wong, M., Zhou, C., Shakiryanova, D., Lloyd, T., Deitcher, D., and Levitan, E. (2012). Neuropeptide delivery to synapses by long-range vesicle circulation and sporadic capture. Cell 148, 1029–1038. doi: 10.1016/j.cell.2011.12.036
Zhang, M., Tang, M., Fang, Y., Cui, H., Chen, S., Li, J., et al. (2018). Cumulative evidence for relationships between multiple variants in the VTI1A and TCF7L2 genes and cancer incidence. Int. J. Cancer 142, 498–513. doi: 10.1002/ijc.31074
Keywords: Vti1a, N-ethylmaleimide-sensitive factor attachment protein receptor protein, neurosecretion, spontaneous synaptic transmission, gene fusion
Citation: Tang F, Fan J, Zhang X, Zou Z, Xiao D and Li X (2022) The Role of Vti1a in Biological Functions and Its Possible Role in Nervous System Disorders. Front. Mol. Neurosci. 15:918664. doi: 10.3389/fnmol.2022.918664
Received: 12 April 2022; Accepted: 09 May 2022;
Published: 27 May 2022.
Edited by:
Jaewon Ko, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaReviewed by:
Sangyong Jung, Institute of Molecular and Cell Biology (A*STAR), SingaporeCopyright © 2022 Tang, Fan, Zhang, Zou, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xihong Li, bGl4aWhvbmdoeGV5QDE2My5jb20=; Dongqiong Xiao, bTEzODgxNzQ5NDk0QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.