- 1Department of Biological Sciences, Korea Advanced Institute for Science and Technology (KAIST), Daejeon, South Korea
- 2Center for Synaptic Brain Dysfunctions, Institute for Basic Science (IBS), Daejeon, South Korea
- 3Division of National Supercomputing, Korea Institute of Science and Technology Information (KISTI), Daejeon, South Korea
SLC6A20A is a proline and glycine transporter known to regulate glycine homeostasis and NMDA receptor (NMDAR) function in the brain. A previous study found increases in ambient glycine levels and NMDA receptor-mediated synaptic transmission in the brains of Slc6a20a-haploinsufficient mice, but it remained unknown whether Slc6a20a deficiency leads to disease-related behavioral deficits in mice. Here, we report that Slc6a20a heterozygous and homozygous mutant mice display differential behavioral phenotypes in locomotor, repetitive behavioral, and spatial and fear memory domains. In addition, these mice show differential transcriptomic changes in synapse, ribosome, mitochondria, autism, epilepsy, and neuron-related genes. These results suggest that heterozygous and homozygous Slc6a20a deletions in mice lead to differential changes in behaviors and transcriptomes.
Introduction
NMDA receptors (NMDARs) critically regulate the development and function of the nervous system (Paoletti et al., 2013; Zhu and Paoletti, 2015; Hansen et al., 2018). Glycine homeostasis is a key regulator of NMDAR function in the brain (Johnson and Ascher, 1987; Paoletti et al., 2013; Zhu and Paoletti, 2015; Hansen et al., 2018). Glycine levels in the extracellular spaces of the brain are thought to be regulated by two well-known glycine transporters, GlyT1 and GlyT2, which are encoded by Slc6a9 and Slc6a5, respectively (Smith et al., 1992; Liu et al., 1993).
SLC6A20A, which was originally reported to transport mainly proline (Smith et al., 1995; Nash et al., 1998; Kiss et al., 2002; Kowalczuk et al., 2005; Takanaga et al., 2005; Broer and Gether, 2012), was recently shown to also transport glycine and regulate glycine/proline homeostasis and NMDAR function in the mouse brain (Bae et al., 2021). Thus, SLC6A20A could be a novel target for inhibiting glycine uptake in the brain and thereby increasing ambient glycine levels and NMDAR function; this could be relevant for the treatment of NMDAR-related brain disorders (Bae et al., 2021), similar to the targeting of GlyT1 and GlyT2 for the treatment of schizophrenia, alcohol dependence, and pain (Tsai et al., 2004; Lane et al., 2006; Javitt, 2012; Harvey and Yee, 2013). However, it remains unclear whether deletion of Slc6a20a in mice leads to any disease-related behavioral deficits through specific mechanistic deviations.
In the present study, we show that Slc6a20a heterozygous and homozygous mice (Slc6a20a+/– and Slc6a20a–/– mice, respectively) display differential behavioral deficits in the locomotor, repetitive behavioral, and memory domains as well as transcriptomic changes in genes associated with synapses, mitochondria, ribosomes, autism spectrum disorders (ASD), epilepsy, and neurons.
Materials and Methods
Mice
Mice lacking exon 3 of the Slc6a20a gene were previously described (Bae et al., 2021). All mice were maintained under a 12-hr light/dark cycle (light phase: between 1:00 a.m. and 1:00 p.m.), given food and water ad libitum, and weaned at postnatal day 21. Female heterozygous mice and male heterozygous mice were crossed in order to obtain wild-type, heterozygous, and homozygous progenies. All mice were maintained in the mouse facility of Korea Advanced Institute of Science and Technology (KAIST). The experimental procedures were approved by the Committee of Animal Research at KAIST (KA2020-89).
Behavioral Tests
Adult male mice aged 3-6 months were used for all behavioral tests. All behavioral experiments were performed during the dark phase of the light/dark cycle (1:00 p.m. to 1:00 a.m.) in which mice are usually active. The brightness conditions in behavioral experiments described below refer to the local brightness around the center region of the apparatus rather than that in the behavioral room. Mice were habituated to an empty, dark experimental room for 30 min before the start of each behavioral test except for the Morris water maze test. EthoVision XT (Noldus) was used to analyze behavioral results unless otherwise noted.
Open-Field Test
This experiment was performed in order to analyze the locomotor activity and anxiety-like behavior in mice. A mouse was placed into the center of a white box (40 cm x 40 cm x 40 cm; ∼100 lux in the center). Mouse movements were then recorded for an hour, and the distance moved and time spent in the center region of the apparatus were analyzed.
Elevated Plus-Maze Test
This experiment was performed to investigate height-induced anxiety-like behavior in mice. A heightened, cross-shaped device with two arms closed and two arms open was used for the experiment. A mouse was placed in the center region of the apparatus (∼200 lux) and allowed to freely move around the environment for 10 minutes. Time spent in closed arms, open arms, and center was analyzed.
Light-Dark Box Test
This experiment was performed to measure light-induced anxiety-like behavior in mice. The apparatus contained two different boxes/chambers, with one with the roof (dark box), and another without the roof (light box; ∼250 lux). A mouse was placed in the light box at the beginning and allowed to freely move around the environment.
Three-Chamber Test
This test was performed to measure social approach and social novelty recognition (Crawley, 2004; Nadler et al., 2004). A mouse was placed in the center chamber of the three-chambered apparatus (59 cm x 39.5 cm x 21.5 cm; ∼100 lux), and its activity was recorded for 10 min. Then, a stranger mouse (S1) was positioned in the cage placed at the corner of a side chamber, and an object (O) was placed in the cage at the corner of the other side chamber. 129S1 mice from the Jackson Laboratory were used as strangers. After 10-min recording of mouse activity, the object was replaced with another stranger (S2), followed by 10-min recording.
Morris Water Maze Test
This test was performed to measure spatial learning and memory in mice (Vorhees and Williams, 2006). A round pool was filled with white paint-added water. Water temperature was maintained at ∼20°C. The pool was divided into four quadrants, and a platform was positioned in one of the quadrants. For the first six days, mice were taught to find an invisible platform. On the seventh day, the probe test was performed: a platform was removed from the pool. A mouse was then placed in the center of the pool and allowed to freely swim around for a minute. Time spent in quadrants and number of crossings across the platform were measured. After the probe test, a platform was placed in the opposite quadrant. For additional three days, mice were trained to locate the new position of the platform (reversal phase). On the eleventh day, another probe test (reversal probe test) was performed.
Contextual Fear Conditioning Test
This test was performed to measure fear learning and memory in mice. On day 0, a mouse was placed in the chamber with an electrocuting platform and habituated for 5 min. On day 1, foot shocks (0.8 mA) were provided for 2 s at 2, 3, and 4 min after the start of recording. Activity of a mouse was recorded for 5 min. On days 2, 3, and 8, a mouse was placed into the same chamber without any shock, and its activity was recorded for 5 min. Percent of freezing time was calculated.
LABORAS
LABORAS (Laboratory Animal Behavior Observation Registration and Analysis System) experiments were performed for a long-term (4-day) monitoring of various mouse behaviors such as locomotor activity, repetitive behaviors (climbing, grooming, and rearing), drinking, and eating (Quinn et al., 2003, 2006). Total investigation time was 96 h with 12-h light/dark cycle, during which food and water were provided.
RNA-Seq Analysis
Four mice at ∼P120 were used for each group (wild type, heterozygous, and homozygous). The extracted mouse brains were preserved in RNAlater solution (Ambion) and stored at −20°C. Poly-T oligo-attached magnetic beads were utilized to purify poly-A mRNAs. RNA concentrations were quantified using Quant-IT RiboGreen (Invitrogen, R11490), and RNA integrity was determined using TapeStation RNA screen tape (Agilent Technologies), after which only high-quality RNAs (RIN > 7.0) were selected for cDNA library construction using Illumina TruSeq mRNA Sample Prep kit (Illumina). Indexed libraries were submitted to an Illumina NovaSeq (Illumina), and paired-end (2 × 100 bp) sequencing was performed by Macrogen Inc.
Transcript abundance was estimated with Salmon (v 1.1.0) (Patro et al., 2017) in Quasi-mapping-based mode onto the Mus musculus genome (GRCm38) with GC bias correction (−gcBias). The acquired abundance data was imported to R (v.3.5.3) with tximport (Soneson et al., 2015) package and differential gene expression analysis was performed using R/Bioconductor DEseq2 (v1.30.1) (Love et al., 2014). Principal component analysis (PCA) was performed for the regularized log transform (rlog) of the normalized counts using plotPCA (with default parameter) tools implemented in DEseq2. Normalized read counts were computed by dividing the raw read counts by size factors and fitted to a negative binomial distribution. The p-values were adjusted for multiple testing with the Benjamini-Hochberg correction. Genes with an adjusted p-value of less than 0.05 were considered as differentially expressed. Volcano plots were generated using R ggplot2 (v.3.1.1) package. The Gene Ontology (GO) enrichment analyses were performed using DAVID software (version 6.8) (Huang da et al., 2009). Mouse gene names were converted to human homologs using the Mouse Genome Informatics (MGI) database. Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005, 2007) was performed to determine whether a priori-defined gene sets would show statistically significant differences in expression between WT and Slc6a20a-mutant mice. Enrichment Analysis was performed using GSEAPreranked® (gsea-3.0.jar) module on gene set collections downloaded from Molecular Signature Database (MSigDB) v 7.0. GSEAPreranked was applied using the list of all genes expressed, ranked by the fold change and multiplied by the inverse of the p-value with recommended default settings (1,000 permutations and a classic scoring scheme). The false discovery rate (FDR) was estimated to control the false positive finding of a given normalized enrichment score (NES) by comparing the tails of the observed and null distributions derived from 1,000 gene set permutations. The gene sets with an FDR of less than 0.05 were considered as significantly enriched. Integration and visualization of the GSEA results were performed using the EnrichmentMap Cytoscape App (version 3.8.1) (Merico et al., 2010; Isserlin et al., 2014).
Statistical Analysis
GraphPad Prism (version 9.2.0; GraphPad Software) were used to perform the statistical analyses. Outliers were retained. Statistical details are presented in Supplementary Table 1.
Results
Slc6a20a Deficiency in Mice Induces Moderate Hyperactivity Without Affecting Anxiety-Like Behavior
Because glycine levels and NMDAR functions are elevated in Slc6a20a-mutant mice (Bae et al., 2021), and NMDARs critically regulate brain development and function (Paoletti et al., 2013), we subjected Slc6a20a heterozygous and homozygous mutant mice (Slc6a20a+/– and Slc6a20a–/– mice, respectively) to a battery of behavioral tests.
As the first step, we used immunoblot analysis to determine the temporal pattern of Slc6a20a protein expression during brain development. The immunoblot pattern from whole-brain lysates indicated that protein expression was strong at early developmental stages, including late embryonic and neonatal stages, and thereafter decreased to adult levels over the first few weeks of postnatal life (Figure 1A).
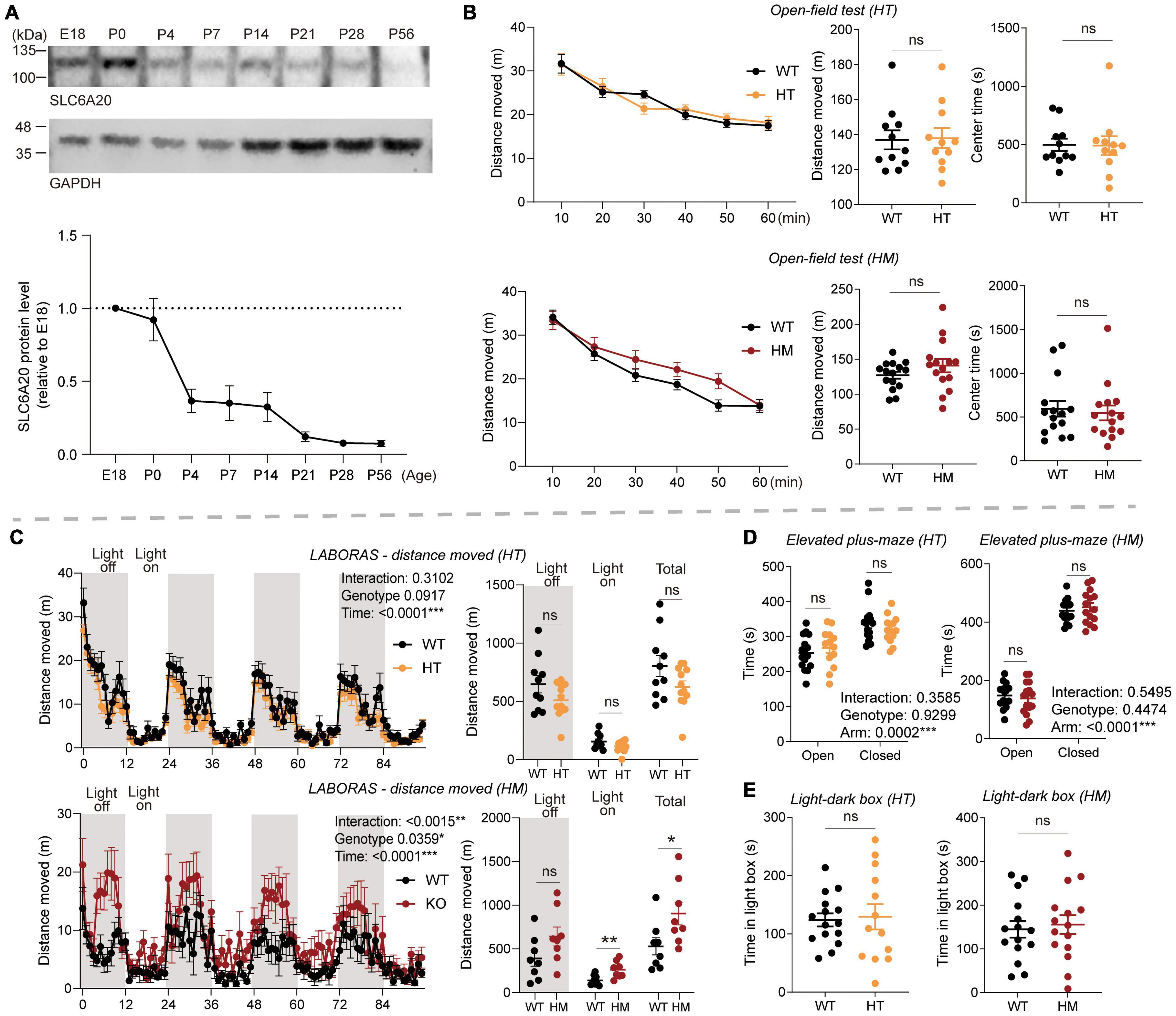
Figure 1. Slc6a20a deficiency in mice induces moderate hyperactivity without affecting anxiety-like behavior. (A) Temporal expression pattern of SLC6A20A proteins in mouse brains at the indicated embryonic (E) and postnatal (P) days. Whole-brain lysates were immunoblotting with SLC6A20 and control (GAPDH) antibodies. SLC6A20 signals were normalized to GAPDH signals for quantification. Note that the utilized pan-SLC6A20 antibody could recognize both SLC6A20A and SLC6A20B proteins in the brain; the expression level of SLC6A20A seems to be about three times greater based on the decrease in the pan-SLC6A20 signal of Slc6a20a homozygous mutant mouse brain (Bae et al., 2021) (n = 4 mice). (B) Locomotor activity in the open-field test is normal in Slc6a20a+/– and Slc6a20a–/– mice (3–6 months; HT and HM, respectively) compared with WT mice, as shown by distance moved. Note that there is no genotype difference in the time spent in the center region of the open-field area, suggestive of normal anxiety-like behavior. (n = 11 mice [WT for HT], 11 [HT], 15 [WT for HM], 15 [HM], two-way repeated-measures/RM-ANOVA [distance moved] and Student’s t-test [total distance moved, time in center]). (C) Locomotor activity is moderately increased in Slc6a20a–/– but not Slc6a20a+/– mice in the Laboras test, where mouse movements were measured for 4 consecutive days. Note that Slc6a20a–/– mice show hyperactivity during the total and light-on periods but not the light-off period, suggestive of disturbed sleep. (n = 10 mice [WT for HT], 12 [HT], 8 [WT for HM], 8 [HM], two-way RM-ANOVA [distance moved across 4 days] and Student’s t-test [total distance moved during total, light-off, and light-on periods]). (D) Anxiety-like behavior is normal in Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) in the elevated plus-maze test, as shown by time spent in the open/closed arms. (n = 16 mice [WT for HT], 13 [HT], 15 [WT for HM], 15 [HM], Student’s t-test). (E) Anxiety-like behavior is normal in Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) in the light-dark test, as shown by time in the light chamber. (n = 15 mice [WT for HT], 13 [HT], 15 [WT for HM], 15 [HM], Student’s t-test). Statistical significance and p values in the graphs; *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
In the open-field test, Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) showed locomotor activities comparable to those of wild-type (WT) mice (Figure 1B), suggestive of normal locomotor activity in a novel environment. In the Laboras test, where mouse movements were monitored for 4 consecutive days (Quinn et al., 2003, 2006), Slc6a20a+/– mice showed normal locomotor activity comparable to that of WT mice (Figure 1C). In contrast, Slc6a20a–/– mice showed moderately increased locomotor activity during the total period and the light-on period, but not during the light-off period (Figure 1C), suggesting that homozygous but not heterozygous Slc6a20a deletion in mice leads to hyperactivity in a familiar environment. The increased activity during the light-on period may also suggest that sleep was disturbed in Slc6a20a–/– mice, as reported previously in mice lacking the receptor tyrosine phosphatase PTPRS and showing disturbed sleep behaviors and rhythms (Park et al., 2020).
Slc6a20a+/– and Slc6a20a–/– mice spent normal amounts of time in the center region of the open-field arena (Figure 1B), suggesting that anxiety-like behavior was not altered. Similarly, Slc6a20a+/– and Slc6a20a–/– mice showed normal levels of anxiety-like behaviors in the elevated plus-maze and light-dark tests (Figures 1D,E).
These results collectively suggest that homozygous but not heterozygous Slc6a20a deletion in mice induces moderate hyperactivity in a familiar environment, without altering anxiety-like behavior.
Slc6a20a Deficiency in Mice Moderately Enhances Repetitive Behavior Without Affecting Social Behavior
We next subjected Slc6a20a-mutant mice to behavioral tests in the social and repetitive behavioral domains. In the three-chamber social-interaction test, Slc6a20a+/– and Slc6a20a–/– mice displayed normal levels of social approach, as shown by time spent exploring social and non-social targets and the social preference index (Figure 2A). In addition, these mice displayed normal levels of social novelty recognition, as shown by time spent exploring novel and familiar social targets (Figure 2A).
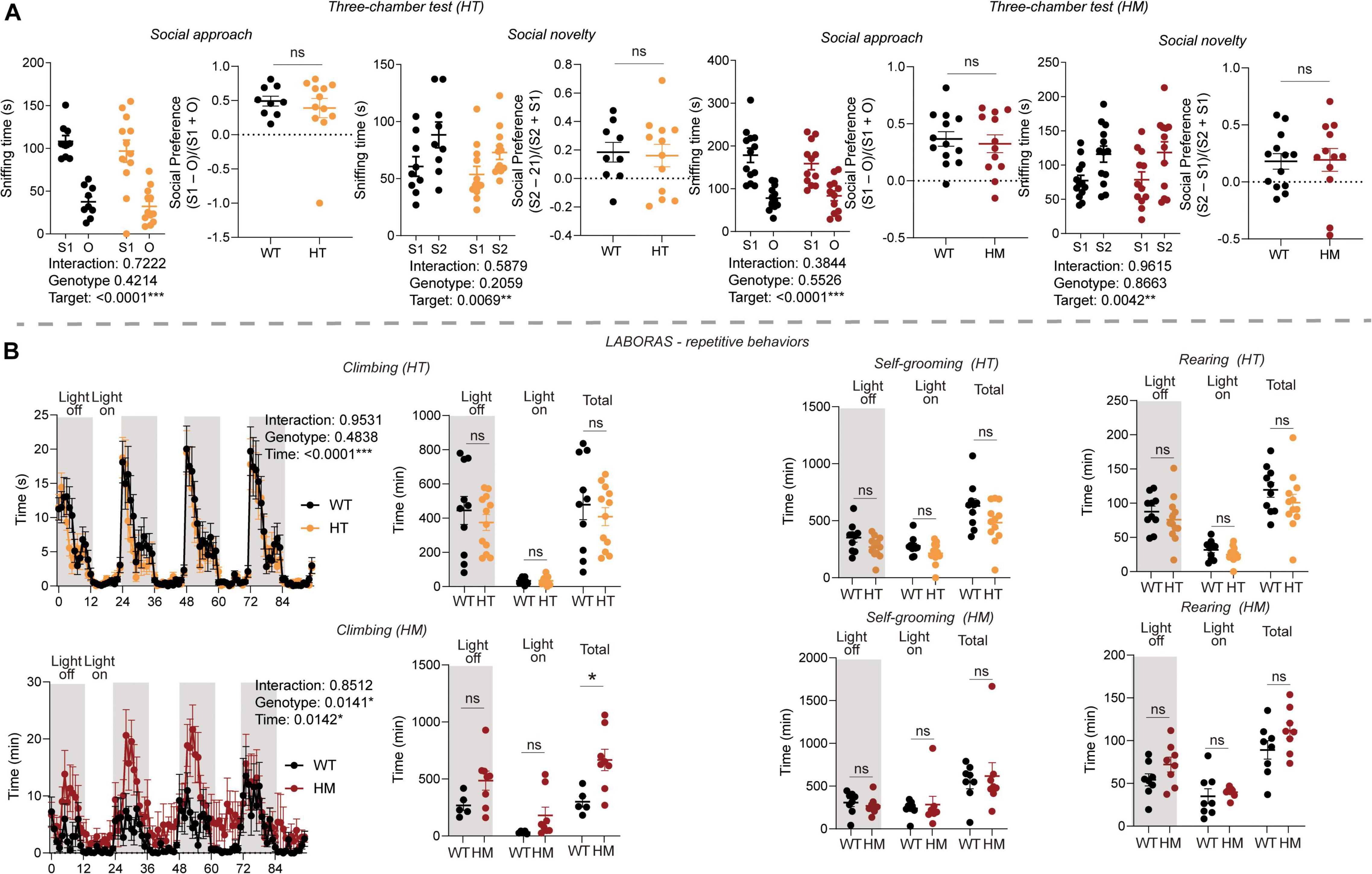
Figure 2. Slc6a20a deficiency in mice moderately enhances repetitive behavior without affecting social behavior. (A) Normal levels of social approach and social novelty recognition are seen in Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) in the three-chamber test, as shown by the time spent exploring/sniffing a social stranger (S1) vs. an object (O) for social approach or a novel stranger (S2) vs. a familiar stranger (S1) for social novelty recognition, and also by the social preference index derived from sniffing time for S1 – O/sniffing time for S1 + O (or sniffing time for S2 – S1/sniffing time for S2 + S1). (n = 9 mice [WT for HT], 12 [HT], 13 [WT for HM], 12 [HM], two-way RM-ANOVA with Bonferroni test [S1 vs. O, or S2 vs. S1], Student’s t-test [preference index]). (B) Increased repetitive climbing is seen in Slc6a20a–/– but not Slc6a20a+/– mice in the Laboras test, where mouse movements were measured for 4 consecutive days. Note that Slc6a20a–/– mice show increased repetitive climbing during the total period but not during the light-off/on period, and that Slc6a20a–/– mice show normal levels of repetitive self-grooming and rearing. (n = 10 mice [WT for HT], 12 [HT], 8 [WT for HM], 8 [HM], two-way RM-ANOVA [climbing across 4 days] and Student’s t-test [climbing/self-grooming/rearing time]). Statistical significance and p values in the graphs; *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
In the Laboras test, Slc6a20a–/– but not Slc6a20a+/– mice showed increased repetitive climbing (Figure 2B), a form of repetitive behavior characterized by overhanging from the wire cage lids (Protais et al., 1976; Riffee et al., 1979; Wilcox et al., 1979; Cabib and Puglisi-Allegra, 1985). However, Slc6a20a+/– and Slc6a20a–/– mice did not show any alteration in other repetitive behaviors, such as self-grooming and rearing (Figure 2B). Other behaviors, such as drinking and eating, were also normal in Slc6a20a–/– and Slc6a20a+/– mice (Supplementary Figure 1).
These results suggest that Slc6a20a deficiency in mice induces a moderate increase in repetitive behavior without affecting social behaviors, with repetitive climbing but not other repetitive behaviors increased in Slc6a20a–/– but not Slc6a20a+/– mice.
Slc6a20a Deficiency in Mice Moderately Enhances Spatial and Fear Learning and Memory
Because NMDARs critically regulate various forms of learning and memory (Collingridge, 1987; Bliss et al., 2014), we next subjected Slc6a20a+/– and Slc6a20a–/– mice to spatial and fear learning and memory paradigms.
In the Morris water maze test, Slc6a20a+/– and Slc6a20a–/– mice showed normal levels of spatial memory acquisition and retrieval during the initial learning and probe phases of the test, respectively, compared with WT mice (Figure 3A). In the reversal test, Slc6a20a+/– but not Slc6a20a–/– mice showed enhancements during the acquisition but not probe phase, compared with WT mice (Figure 3A).
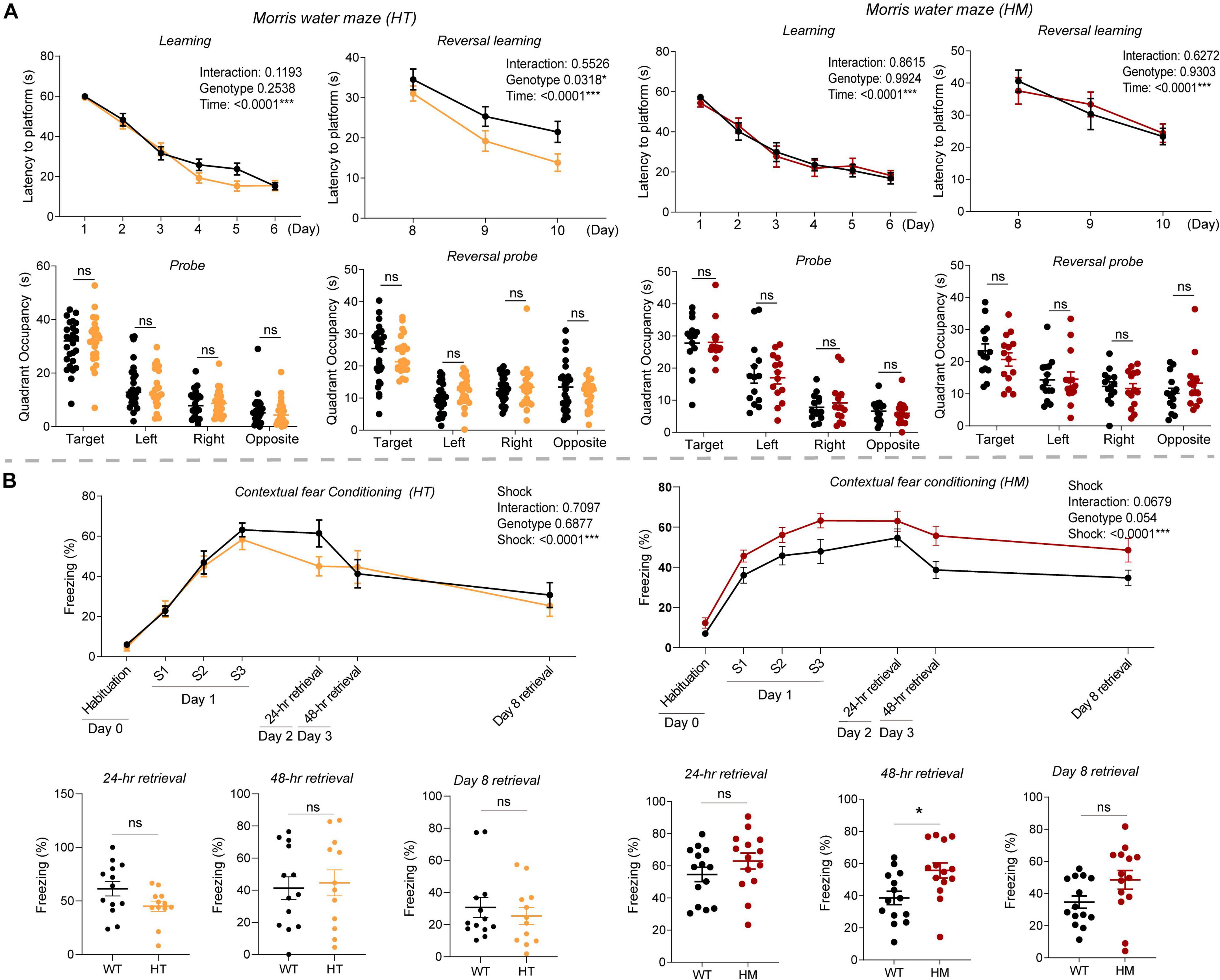
Figure 3. Slc6a20a deficiency in mice moderately enhances spatial and fear memory. (A) Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) show normal levels of spatial memory acquisition and retrieval (probe test) during the initial phase of the Morris water maze test, whereas Slc6a20a+/– but not Slc6a20a–/– mice show enhanced spatial memory acquisition but not retrieval during the reversal phase of the test. (n = 26 mice [WT for HT], 23 [HT], 14 [WT for HM],14 [HM], two-way RM-ANOVA [latency to platform] and Student’s t-test [quadrant occupancy]). (B) Slc6a20a+/– and Slc6a20a–/– mice (3–6 months) show differential levels of fear memory acquisition (day 1) and retrieval (probe tests; days 2, 3, and 8) in the contextual fear conditioning test. (n = 13 mice [WT for HT], 12 [HT], 14 [WT for HM], 14 [HM], two-way RM-ANOVA [acquisition] and Student’s t-test [retrievals]). Statistical significance and p values in the graphs; *p < 0.05, ***p < 0.001, ns, not significant.
In the contextual fear memory conditioning test, Slc6a20a+/– mice showed normal levels of fear memory acquisition on day 1 compared with WT mice (Figure 3B). In memory retrieval tests consecutively performed on day 2 (for 24-h retrieval), day 3 (for 48-h retrieval), and day 8 (7-day retrieval), Slc6a20a+/– mice showed largely normal fear memory retrieval.
Slc6a20a–/– mice showed normal levels of fear memory acquisition on day 1 (Figure 3B), although there was an increasing tendency. In the retrieval tests, Slc6a20a–/– mice showed increased retrieval on day 3 (48-h retrieval); the levels of retrieval on day 2 (24-h retrieval) and day 8 (7-day retrieval) were normal but showed increasing tendencies.
These results collectively suggest that Slc6a20a deficiency in mice induces moderate increases in spatial learning and memory in the Morris water maze test and fear learning and memory in contextual fear conditioning test.
Differentially Expressed Genes (DEGs) in Slc6a20a+/– and Slc6a20a–/– Mice
To investigate the molecular phenotypes of Slc6a20a+/– and Slc6a20a–/– mice, we performed RNA-sequencing (RNA-seq) analyses using whole-brain lysates obtained at ∼P120. Our analysis identified 13 DEGs (7 upregulated and 6 downregulated; cutoff, p value < 0.05) in Slc6a20a+/– mice and 33 DEGs (27 upregulated and 6 downregulated) in Slc6a20a–/– mice, with four DEGs overlapping between the genotypes (Nlgn3/neuroligin-3, Tenm3/teneurin-3, and Cdc73 upregualted and Slc6a20a downregulated) (Figures 4A,B and Supplementary Table 2).
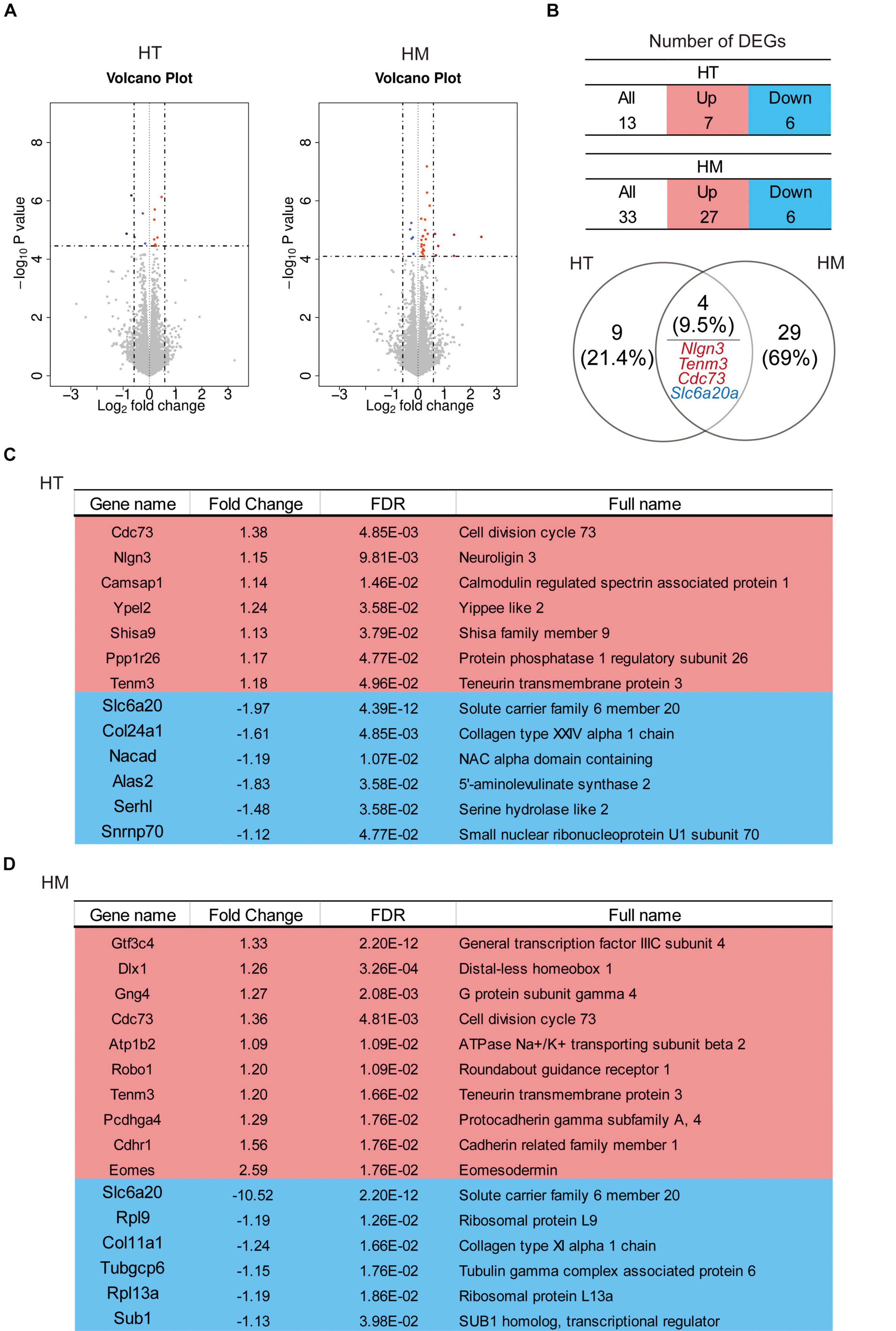
Figure 4. Differentially expressed genes (DEGs) in Slc6a20a+/– and Slc6a20a–/– mice. (A) Volcano plot for DEGs from Slc6a20a+/– and Slc6a20a–/– mice (∼P120). (n = 4 mice for WT-HT, HT, WT-HM, and HM; dotted lines indicate adjusted p-value < 0.05 or | FC| > 1.5; DEGs were defined by adjusted p-values but not fold changes). See Supplementary Table 2 for raw RNA-seq results. (B) Summary tables showing the numbers of up- and downregulated DEGs, and Venn diagrams showing the overlap between DEGs from Slc6a20a+/– and Slc6a20a–/– mice. Upregulated, overlapped DEGs (Nlgn3/neuroligin-3,Tenm3/teneurin-3, and Cdc73) and downregulated, overlapped DEGs (Slc6a20a) are indicated in red and blue colors, respectively. (C,D) Lists of all significantly up- and downregulated DEGs from Slc6a20a+/– and Slc6a20a–/– mice; only the top 10 upregulated DEGs out of 27 are shown for Slc6a20a–/– mice to save space (see Supplementary Table 3 for further details).
In DEGs from Slc6a20a+/– mice, the seven significantly upregulated genes were Cdc73 (cell division cycle 73), Nlgn3 (neuroligin 3), Camsap1 (calmodulin regulated spectrin-associated protein 1), Ypel2 (Yippee like 2), Shisa9 (Shisa family member 9), Ppp1r26 (protein phosphatase 1 regulatory subunit 26), and Tenm3 (teneurin transmembrane protein 3) (Figure 4C and Supplementary Table 3). The products of some of these genes have been associated with synaptic and neuronal functions. Neuroligin-3 is a synaptic adhesion molecule involved in synapse development and regulation and ASD-related brain functions and behaviors (Etherton et al., 2011; Foldy et al., 2013; Jaramillo et al., 2014; Rothwell et al., 2014; Zhang and Sudhof, 2016; Cao et al., 2018; Sudhof, 2018). CAMSAP1 binds to the minus end of microtubules and regulates neuronal polarity/migration and cortical lamination (Meng et al., 2008; Akhmanova and Steinmetz, 2015; Zhou et al., 2020). Shisa9 (also known as CKAMP44) is an AMPA receptor auxiliary protein that regulates the trafficking, subcellular localization, and function of AMPA receptors (von Engelhardt et al., 2010; Jacobi and von Engelhardt, 2018; von Engelhardt, 2019). Teneurin-3 is a transmembrane protein involved in homophilic adhesion as well as heterophilic adhesion with latrophilins that regulates synapse specificity and neural circuit assembly (Mosca et al., 2012; Li et al., 2018; Sando et al., 2019; Pederick and Luo, 2021).
The six significantly downregulated genes in Slc6a20a+/– mice were Slc6a20a (solute carrier family 6 member 20), as expected, followed by Col24a1 (collagen type XXIV alpha 1 chain), Nacad (NAC alpha domain-containing), Alas2 (5′-aminolevulinate synthase 2), and Serhl2 (serine hydrolase-like 2), and Snrnp70 (small nuclear ribonucleoprotein U1 subunit 70) (Figure 4C). Interestingly, Alas2 is an enzyme localized in the mitochondria of erythrocytes; it regulates the heme biosynthetic pathway and is implicated in X-linked sideroblastic anemia (XLSA) (Nzelu et al., 2021).
In Slc6a20a–/– mice, the top ten upregulated genes were Gtf3c4 (general transcription factor IIIC subunit 4), Dlx1 (distal-less homeobox 1), Gng4 (G protein subunit gamma 4), Cdc73 (cell division cycle 73; also upregulated in Slc6a20a+/– mice), Atp1b2 (ATPase Na + /K + transporting subunit beta 2), Robo1 (roundabout guidance receptor 1), Tenm3 (teneurin transmembrane protein 3), Pcdhga4 (protocadherin gamma subfamily A, 4), Cdhr1 (cadherin related family member 1), and Eomes (eomesodermin) (Figure 4D). Dlx1 is a homeobox transcription factor that regulates neuronal differentiation (Petryniak et al., 2007; Lee B. et al., 2018; Pla et al., 2018; Lindtner et al., 2019). Atp1b2 is a sodium-potassium ATPase that regulates neuronal excitability (Larsen et al., 2016). Robo1 is a transmembrane protein that regulates axon guidance and neuronal migration (Seeger et al., 1993; Kidd et al., 1998).
Many DEGs from Slc6a20a–/– mice not mentioned above have also been associated with synaptic and neuronal functions; examples include Mdga1 (MAM domain containing glycosylphosphatidylinositol anchor 1) (Lee et al., 2013; Pettem et al., 2013; Kim et al., 2017; Um and Ko, 2017), Cacng8 (calcium voltage-gated channel auxiliy subunit gamma 8) (Nicoll et al., 2006; Diaz-Alonso and Nicoll, 2021), Sema5a (semaphorin 5A) (Duan et al., 2014), and Nlgn3 (neuroligin 3) (Sudhof, 2018). It is also notable that neuroligin-3 and teneurin-3 are similarly upregulated in Slc6a20a+/– and Slc6a20a–/– mice, suggesting that their upregulations may represent a shared mechanism responding to Slc6a20a deletion. Lastly, the six significantly downregulated genes in Slc6a20a–/– mice were Slc6a20a, Rpl9 (ribosomal protein L9), Col11a1 (collagen type XI alpha 1 chain), Tubgcp6 (tubulin gamma complex associated protein 6), Rpl13a (ribosomal protein L13A), and Sub1 (SUB1 homolog, transcriptional regulator) (Figure 4D).
These results collectively suggest that both Slc6a20a+/– and Slc6a20a–/– mice show upregulation of genes that are associated with synaptic and neuronal functions and downregulation of genes associated with ribosomal and mitochondrial functions. Similar results were obtained from our gene set enrichment analysis (GSEA; see below).
Biological Functions Altered in the Transcriptomes of Slc6a20a+/– and Slc6a20a–/– Mice Revealed by GSEA
Since our DEG analysis yielded small numbers of DEGs, we performed gene set enrichment analysis (GSEA), which can identify altered biological functions using a large number of small but coordinated transcriptomic changes in a less biased manner than analysis of a small number of large transcriptomic changes (Subramanian et al., 2005, 2007).
The transcripts derived from WT and Slc6a20a+/– mice (HT/WT transcripts; ∼P120 whole brain) were positively enriched for gene sets associated with neuronal synapses, as shown by the top five enriched gene sets (Figure 5A and Supplementary Table 4). Clustering of the positively enriched gene sets using CytoScape Enrichment App (Merico et al., 2010; Isserlin et al., 2014) further confirmed that there were positive enrichments for functions associated with neuronal synapses, such as synaptic specialization, presynaptic active zone, ion channels, dendrites, and axons (Figure 5B).
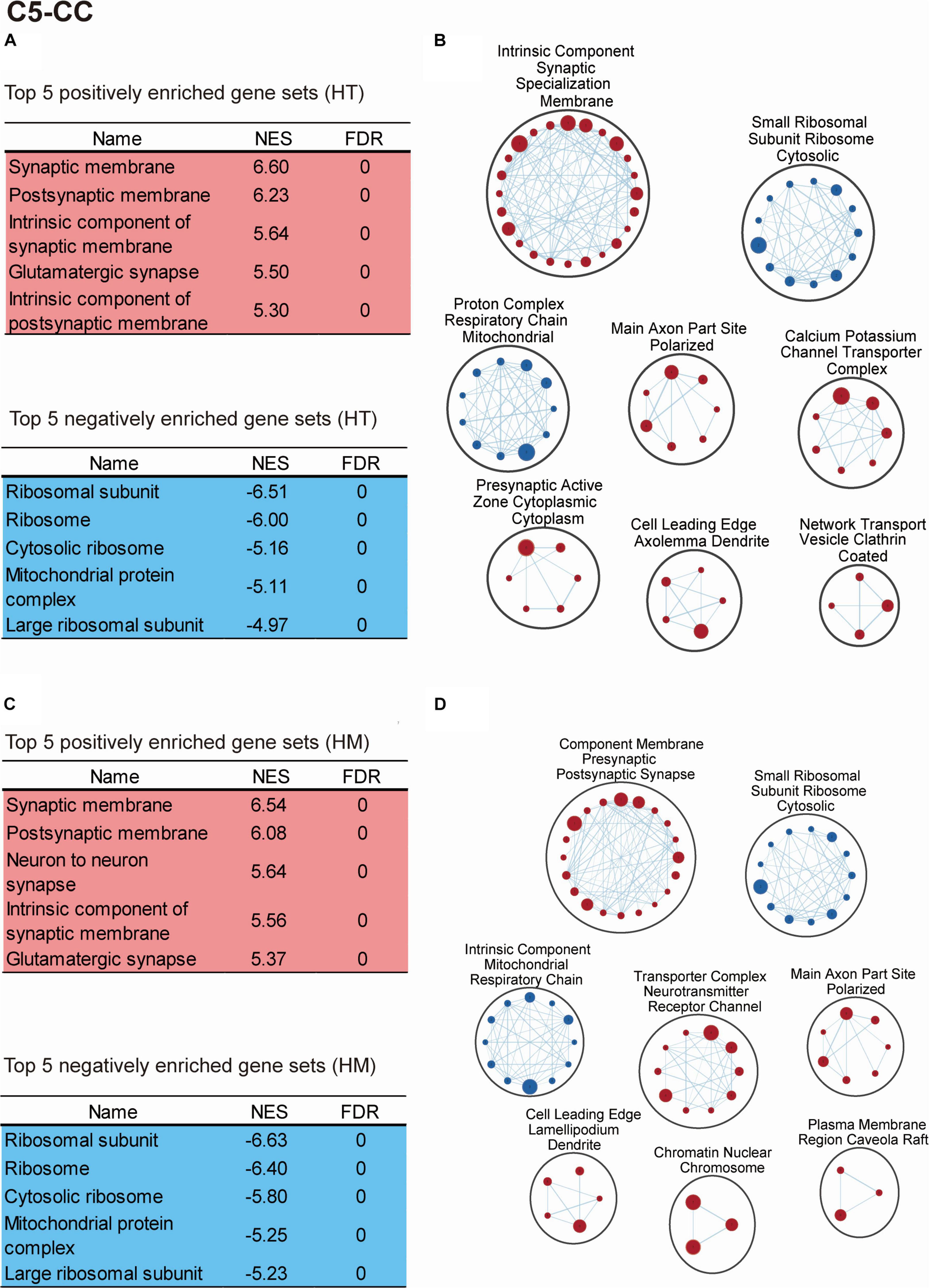
Figure 5. Biological functions altered in the transcriptomes of Slc6a20a+/– and Slc6a20a–/– mice, as revealed by GSEA. (A–D) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts), as shown by the top five most strongly enriched gene sets (A,C) and clustering of the enriched gene sets using CytoScape EnrichmentApp (B,D). See Supplementary Table 4 for enriched gene sets beyond the top five genes shown in the table. Gene set clusters composed of > 3 gene sets are shown. These GSEA results were derived using the gene sets of the cellular component (CC) domain in the C5 database; see Supplementary Figures 2,3 for the GSEA results from the biological process (BP) and molecular function (MF) domains in the C5 database. NES, normalized enrichment score; FDR, false detection rate. [n = 4 mice for WT-HT, HT, WT-HM, and HM; FDR < 0.05 (B,D)].
These results were obtained using the gene sets of the cellular component domain in the C5 database; we also obtained similar results using the gene sets of the biological process and molecular function domains of the C5 database (Supplementary Figures 2A,B, 3A,B and Supplementary Table 4). In addition, the use of gene sets in the KEGG domain indicated positive enrichments for synapse-related gene sets including long-term potentiation and gap junction, known to cooperate with excitatory synapses (Lee et al., 2021), as well as negative enrichments for ribosome/mitochondria-related gene sets (Supplementary Figures 4A,B and Supplementary Table 4).
The HT/WT transcripts were negatively enriched for gene sets associated with ribosomes and mitochondria (Figure 5A and Supplementary Table 4). In addition, CytoScape Enrichment App analysis revealed similar negative enrichments for ribosome/mitochondria-related functions, such as ribosomal subunits and respiratory chain complex (Figure 5B).
The transcripts derived from WT and Slc6a20a–/– mice (HM/WT transcripts) were positively enriched for gene sets associated with neuronal synapses, as supported by the top five gene sets and CytoScape Enrichment App clustering (Figures 5C,D and Supplementary Table 4). In addition, the HM/WT transcripts were negatively enriched for gene sets associated with ribosomes and mitochondria, as supported by the top five gene sets and CytoScape Enrichment App clustering (Figures 5C,D and Supplementary Table 4). Similar results were obtained using the gene sets of the biological process and molecular function domains (Supplementary Figures 2C,D, 3C,D and Supplementary Table 4).
These results collectively suggest that heterozygous and homozygous deletion of Slc6a20a in mice leads to similar increases in synapse-associated genes and similar decreases in ribosome- and mitochondria-related genes.
ASD-Related Transcriptomic Changes in Slc6a20a+/– and Slc6a20a–/– Mice Revealed by GSEA
Previous studies investigated transcriptomic changes associated with ASD (Garbett et al., 2008; Voineagu et al., 2011; Gupta et al., 2014; Parikshak et al., 2016; Velmeshev et al., 2019) and reported gene sets that are up- or downregulated in ASD (termed ASD-related gene sets hereafter), including DEG_Up_Voineagu, Co-Exp_Up_M16_Voineagu, DEG_Down_Voineagu, and Co-Exp_Down_M12_Voineagu (Voineagu et al., 2011; Werling et al., 2016) (details on these gene sets are summarized in Supplementary Table 5).
In addition, a large number of previous studies on ASD led to the compilation of ASD-risk gene sets, including SFARI genes (all genes and high-confidence category 1 genes) (Abrahams et al., 2013)1, FMRP targets (Darnell et al., 2011; Werling et al., 2016), De Novo Missense (protein-disrupting or missense rare de novo variants) (Iossifov et al., 2014; Werling et al., 2016), De Novo Variants (protein-disrupting rare de novo variants) (Iossifov et al., 2014; Werling et al., 2016), and AutismKB (Autism KnowledgeBase) (Xu et al., 2012; Yang et al., 2018; Supplementary Table 5). The genes in these ASD-risk gene sets are thought be generally downregulated in ASD because many of the mutations are missense, nonsense, splice-site, frame-shift, and deletion mutations.
Using these gene sets, we performed GSEA using the HT/WT and HM/WT transcripts from Slc6a20a-mutant mice. The HT/WT and HM/WT transcripts were positively and similarly enriched for ASD-related gene sets that are downregulated in ASD, such as DEG_Down_Voineagu and Co-Exp_Down_M12_Voineagu (Figure 6A and Supplementary Table 4), suggesting that both HT/WT and HM/WT transcripts exhibit patterns opposite those observed in ASD (termed anti-ASD hereafter).
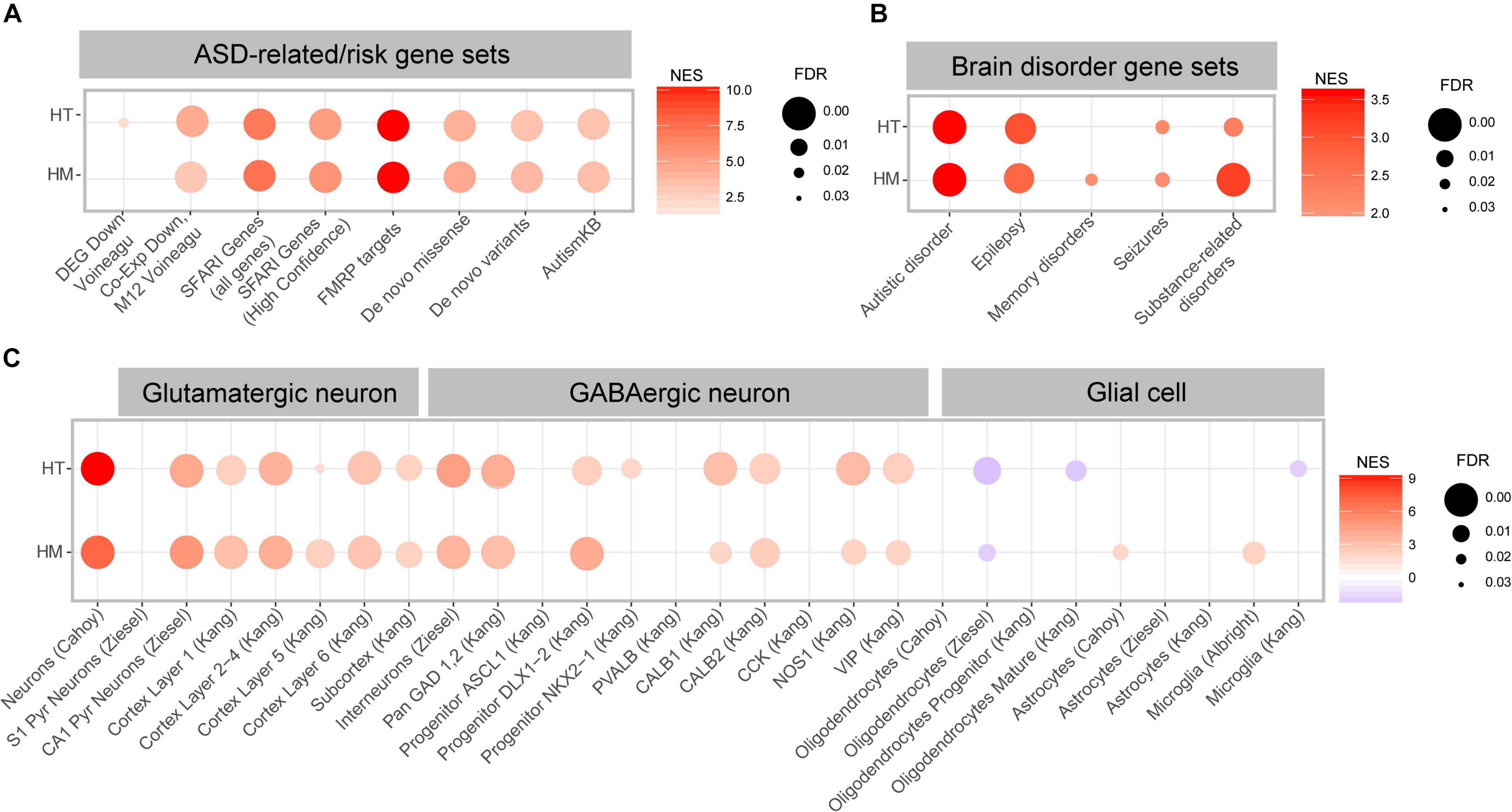
Figure 6. ASD-related transcriptomic changes in Slc6a20a+/– and Slc6a20a–/– mice are revealed by GSEA. (A) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts) for ASD-related gene sets (DEG_Down_Voineagu and Co-Exp_Down_M12_Voineagu) and ASD-risk gene sets (SFARI genes (all genes and high-confidence category 1 genes), FMRP targets, De Novo Missense, De Novo Variants, and AutismKB). See Supplementary Table 4 for further details on the GSEA results. NES, normalized enrichment score; FDR, false detection rate. (n = 4 mice for WT-HT, HT, WT-HM, and HM). (B) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts) for gene sets in the DisGeNet (https://www.disgenet.org/). See Supplementary Table 4 for further details of the GSEA results. NES, normalized enrichment score; FDR, false detection rate. (n = 4 mice for WT-HT, HT, WT-HM, and HM). (C) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts) for cell-type-specific gene sets. See Supplementary Table 4 for further details of the GSEA results. NES, normalized enrichment score; FDR, false detection rate. (n = 4 mice for WT-HT, HT, WT-HM, and HM).
The HT/WT and HM/WT transcripts were also positively and similarly enriched for all of the tested ASD-risk gene sets, including SFARI genes, FMRP targets (most strongly enriched), De Novo Missense, De Novo Variants, and AutismKB (Figure 6A and Supplementary Table 4), and thus again exhibited an anti-ASD pattern.
When tested against gene sets associated with various brain disorders (DisGeNet2 (Pinero et al., 2017, 2020), the HT/WT and HM/WT transcripts were positively and similarly enriched for ASD and epilepsy-related gene sets, and the HM/WT transcripts were positively and strongly enriched for substance use-related gene sets relative to the HT/WT transcripts (Figure 6B and Supplementary Table 4).
Distinct cell-type-specific transcriptomic changes have also been reported in ASD, including downregulation of neuron- and oligodendrocyte-related genes and upregulation of astrocyte- and microglia-related genes (Voineagu et al., 2011; Werling et al., 2016). This led us to test if the HT/WT and HM/WT transcripts are enriched in these previously reported cell type-specific gene sets (Albright and Gonzalez-Scarano, 2004; Cahoy et al., 2008; Kang et al., 2011; Zeisel et al., 2015; Werling et al., 2016; Velmeshev et al., 2019, 2020; Supplementary Table 5).
The HT/WT and HM/WT transcripts were positively enriched for gene sets associated with glutamate and GABA neurons (Figure 6C and Supplementary Table 4), suggestive of anti-ASD transcriptomic patterns. However, the HT/WT and HM/WT transcripts were negatively and moderately enriched for gene sets associated with oligodendrocytes (Figure 6C), suggestive of “ASD-like” or “pro-ASD” transcriptomic patterns. Intriguingly, the HT/WT and HM/WT transcripts displayed weak enrichments for astrocyte and microglia-related gene sets relative to those for glutamate/GABA neurons; thus we observed both positive and negative enrichments for microglia (Figure 6C).
These results collectively suggest that heterozygous and homozygous Slc6a20a deletions in mice lead to largely anti-ASD transcriptomic enrichment patterns, as supported by the enrichment patterns for ASD-related/risk and cell-type-specific gene sets. In addition, the cell-type-specific transcriptomic changes induced by Slc6a20a deletion are stronger in neurons and oligodendrocytes relative to glial cells.
Discussion
In the present study, we analyzed the behavioral and transcriptomic phenotypes of Slc6a20a+/– and Slc6a20a–/– mice. Our results revealed that these mice display differential hyperactivity, repetitive climbing, and moderately enhanced spatial and fear memory, as well as upregulation of synapse-related genes and downregulation of ribosome- and mitochondria-related genes. ASD-related transcriptomic changes are also observed, such as upregulation of ASD-related genes, including FMRP targets, and stronger upregulation of neuron-related genes compared to glia-related genes.
The behavioral phenotypes seem to be stronger in Slc6a20a–/– mice than in Slc6a20a+/– mice in some assays. For instance, Slc6a20a–/– mice are hyperactive in a familiar environment (Laboras results) and show increased self-grooming in Laboras cages and abnormally increased 48-hr fear memory, whereas Slc6a20a+/– mice do not exhibit such changes. However, spatial learning and memory is increased in Slc6a20a+/– but not in Slc6a20a–/– mice during the reversal but not the initial phase of the Morris water maze test. These results collectively suggest that heterozygous and homozygous Slc6a20a deletions in mice lead to differential behavioral phenotypes.
The mechanisms underlying these behavioral deficits are unclear. However, the RNA-seq results obtained from Slc6a20a+/– and Slc6a20a–/– mice indicate that synapse-related genes are upregulated. We note that Slc6a20a+/– mice have been shown to display increased NMDAR function at juvenile stages (Bae et al., 2021). In addition, the increased Slc6a20a expression in Pten-mutant mice has been causally associated with decreased ambient glycine levels and NMDAR function, and increased repetitive self-grooming (Bae et al., 2021). Moreover, neuroligin-3, which is upregulated in both Slc6a20a+/– and Slc6a20a–/– mice, has been associated with the regulation of NMDAR functions (Etherton et al., 2011; Polepalli et al., 2017; Zhang et al., 2017). It is thus tempting to speculate that altered NMDAR function in Slc6a20a+/– and Slc6a20a–/– mice might be associated with the behavioral deficits observed in these animals.
It should be noted that Slc6a20a expression at the protein level is much higher at late embryonic and early postnatal stages than at adult stages, while the behavioral experiments were mainly performed using mutant mice at ages around 3-6 months. Therefore, it is less likely that adult-stage deficiency of Slc6a20a directly causes the behavioral deficits, and the mechanistic deviations occurred at early developmental stages may have long-lasting impacts. Details on such mechanisms remain to be determined. However, given that Slc6a20a deficiency leads to NMDAR hyperfunction at juvenile stages (Bae et al., 2021), as mentioned above, and NMDAR function has profound impacts on the brain development and function across all developmental stages (Paoletti et al., 2013; Zhu and Paoletti, 2015; Hansen et al., 2018), it is probable that the NMDAR function impaired during early development have long-lasting effects on neuronal and synapse/circuit mechanisms that are associated with the observed behavioral deficits.
A notable transcriptomic change associated with the increased synaptic gene expression seen in Slc6a20a+/– and Slc6a20a–/– mice is the decreased expression levels of ribosomal and mitochondrial genes. This leads us to question whether ribosomal and mitochondrial gene downregulation are causally associated with the synaptic gene upregulation and behavioral deficits in the mutant mice. It has been shown that synaptic proteins are reciprocally related to protein synthesis in ASD (Santini and Klann, 2014). In addition, mitochondrial dysfunction has been linked to synaptic deficits (Li et al., 2004; Vos et al., 2010; Sheng and Cai, 2012; Lee A. et al., 2018) and ASD (Hollis et al., 2017; Frye, 2020; Rojas-Charry et al., 2021), and ribosomal dysfunction has been associated with ASD and epilepsy (Lombardo, 2021). These results suggest that ribosomal and mitochondrial gene downregulation observed in our mutant mice may contribute to their synaptic and behavioral deficits.
GSEA using brain disorder-related gene sets shows that the Slc6a20a+/– and Slc6a20a–/– transcriptomes are associated with ASD and epilepsy, and that the Slc6a20a–/– transcriptome is more strongly associated with substance use-related disorders than the Slc6a20a+/– transcriptome. Among the ASD-related gene sets, the Slc6a20a+/– and Slc6a20a–/– transcriptomes are more strongly enriched for FMRP targets associated with the fragile X syndrome. These results suggest that Slc6a20a deletion in mice leads to transcriptomic changes associated with multiple brain disorders. In addition, given that many FMRP targets have been related to synaptic functions (Bagni and Zukin, 2019), these results further support the hypothesis that synaptic deficits may underlie the behavioral deficits in these mutant mice.
GSEA using cell-type-specific gene sets indicates that the Slc6a20a+/– and Slc6a20a–/– transcriptomes are more strongly enriched for neuron (glutamate and GABA)-related gene sets relative to glia (astrocyte/microglia)-related gene sets. Slc6a20a has been shown to be expressed more strongly in glial cells (astrocytes and microglia) than in neurons (glutamate and GABA) (Bae et al., 2021). Our results suggest that the transcriptomic changes observed in synapse/ribosome/mitochondria-related genes in the mutant brain likely represent changes occurring in neurons rather than cell-autonomous changes occurring in glial cells (astrocytes and microglia).
Lastly, it should be pointed out that the differences between Slc6a20a+/– and Slc6a20a–/– mice were greater in behavioral phenotypes than in transcriptomic phenotypes. It is possible that the subtle differences between Slc6a20a+/– and Slc6a20a–/– mice in the expression levels of neuron/glia-related genes (i.e., different cortical layers, GABA subtypes, and glial cell types) may still contribute to the differential behavioral changes. Alternatively, the transcriptomic changes may reflect compensatory changes initiated in an effort to rescue the gene deletion effects, rather than those mediating the pathophysiological changes. Further validation of the mechanistic/functional changes suggested by the transcriptomic changes should be tested at the protein, synaptic, and functional levels.
In summary, we herein show that heterozygous and homozygous Slc6a20a deletions in mice lead to differential behavioral deficits in locomotor, repetitive behavioral, and spatial and fear memory domains and transcriptomic changes in genes associated with synapses, ribosomes, mitochondria, ASD, epilepsy, and neurons.
Data Availability Statement
The RNA-Seq data presented in the study are deposited in the Gene Expression Ombinus (GEO) repository at the National Center for Biotechnology Information (NCBI), accession number GSE193387.
Ethics Statement
The animal study was reviewed and approved by Committee of Animal Research at KAIST (KA2020-89).
Author Contributions
JK, JR, and MB performed behavioral experiments. SK performed immunoblot analysis. HK performed RNA-Seq analyses. MB, HK, and EK wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Korea Institute of Science and Technology Information (K-19-L02-C07-S01 to HK) and the Institute for Basic Science (IBS-R002-D1 to EK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.857820/full#supplementary-material
Supplementary Figure 1 | Normal levels of repetitive drinking and eating and immobility and other parameters in Slc6a20a–/– and Slc6a20a+/– mice in the Laboras test. (A,B) Normal levels of repetitive drinking and eating are seen for Slc6a20a–/– and Slc6a20a+/– mice in the Laboras test, where mouse movements were measured for 4 consecutive days, as shown by total time spent drinking and eating. (n = 10 mice [WT for HT], 12 [HT], 8 [WT for HM], 8 [HM], Student’s t-test). Statistical significance and p values in the graphs; ns, not significant.
Supplementary Figure 2 | Biological functions altered in the transcriptomes of Slc6a20a+/– and Slc6a20a–/– mice, as revealed by GSEA with gene sets in the biological process domain. (A,B) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts), using the gene sets in the biological process domain in the C5 database, as shown by top five most strongly enriched gene sets (A,C) and clustering of the enriched gene sets using CytoScape EnrichmentApp (B,D). See Supplementary Table 4 for enriched gene sets additional to the top five gene sets shown in the table. Gene set clusters composed of > 3 gene sets are shown. NES, normalized enrichment score; FDR, false detection rate. [n = 4 mice for WT-HT, HT, WT-HM, and HM; FDR < 0.05 (B,D)].
Supplementary Figure 3 | Biological functions altered in the transcriptomes of Slc6a20a+/– and Slc6a20a–/– mice, as revealed by GSEA with gene sets in the molecular function domain. (A,B) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts) using the gene sets in the molecular function domain in the C5 database, as shown by top five most strongly enriched gene sets (A,C) and clustering of the enriched gene sets using CytoScape EnrichmentApp (B,D). See Supplementary Table 4 for enriched gene sets additional to the top five gene sets shown in the table. Gene set clusters composed of > 3 gene sets are shown. NES, normalized enrichment score; FDR, false detection rate. [n = 4 mice for WT-HT, HT, WT-HM, and HM; FDR < 0.05 (B,D)].
Supplementary Figure 4 | Biological functions altered in the transcriptomes of Slc6a20a+/– and Slc6a20a–/– mice, as revealed by GSEA with gene sets in the KEGG database. (A,B) GSEA results for transcriptomes from WT and Slc6a20a+/– mice (HT/WT transcripts) and WT and Slc6a20a–/– mice (HM/WT transcripts) using the gene sets in the KEGG database, as shown by top five most strongly enriched gene sets (A,C) and clustering of the enriched gene sets using CytoScape EnrichmentApp (B,D). See Supplementary Table 4 for enriched gene sets additional to the top five gene sets shown in the table. Gene set clusters composed of >3 gene sets are shown. NES, normalized enrichment score; FDR, false detection rate. [n = 4 mice for WT-HT, HT, WT-HM, and HM; FDR < 0.05 (B,D)].
Supplementary Table 1 | Statistical results.
Supplementary Table 2 | Raw RNA-Seq results.
Supplementary Table 3 | DEGs derived from RNA-Seq results.
Supplementary Table 4 | GSEA results.
Supplementary Table 5 | Gene sets used in the GSEA analysis.
Footnotes
References
Abrahams, B. S., Arking, D. E., Campbell, D. B., Mefford, H. C., Morrow, E. M., Weiss, L. A., et al. (2013). SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4:36. doi: 10.1186/2040-2392-4-36
Akhmanova, A., and Steinmetz, M. O. (2015). Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726. doi: 10.1038/nrm4084
Albright, A. V., and Gonzalez-Scarano, F. (2004). Microarray analysis of activated mixed glial (microglia) and monocyte-derived macrophage gene expression. J. Neuroimmunol. 157, 27–38. doi: 10.1016/j.jneuroim.2004.09.007
Bae, M., Roh, J. D., Kim, Y., Kim, S. S., Han, H. M., Yang, E., et al. (2021). SLC6A20 transporter: a novel regulator of brain glycine homeostasis and NMDAR function. EMBO Mol. Med. 13:e12632. doi: 10.15252/emmm.202012632
Bagni, C., and Zukin, R. S. (2019). A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron 101, 1070–1088. doi: 10.1016/j.neuron.2019.02.041
Bliss, T. V., Collingridge, G. L., and Morris, R. G. (2014). Synaptic plasticity in health and disease: introduction and overview. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130129. doi: 10.1098/rstb.2013.0129
Broer, S., and Gether, U. (2012). The solute carrier 6 family of transporters. Br. J. Pharmacol. 167, 256–278. doi: 10.1111/j.1476-5381.2012.01975.x
Cabib, S., and Puglisi-Allegra, S. (1985). Different effects of apomorphine on climbing behavior and locomotor activity in three strains of mice. Pharmacol. Biochem. Behav. 23, 555–557. doi: 10.1016/0091-3057(85)90418-6
Cahoy, J. D., Emery, B., Kaushal, A., Foo, L. C., Zamanian, J. L., Christopherson, K. S., et al. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278. doi: 10.1523/JNEUROSCI.4178-07.2008
Cao, W., Lin, S., Xia, Q. Q., Du, Y. L., Yang, Q., Zhang, M. Y., et al. (2018). Gamma oscillation dysfunction in mPFC leads to social deficits in neuroligin 3 R451C knockin mice. Neuron 97, 1253–1260.e7. doi: 10.1016/j.neuron.2018.02.001
Collingridge, G. (1987). Synaptic plasticity. the role of NMDA receptors in learning and memory. Nature 330, 604–605. doi: 10.1038/330604a0
Crawley, J. N. (2004). Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment. Retard. Dev. Disabil. Res. Rev. 10, 248–258. doi: 10.1002/mrdd.20039
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi: 10.1016/j.cell.2011.06.013
Diaz-Alonso, J., and Nicoll, R. A. (2021). AMPA receptor trafficking and LTP: carboxy-termini, amino-termini and TARPs. Neuropharmacology 197:1087. doi: 10.1016/j.neuropharm.2021.108710
Duan, Y., Wang, S. H., Song, J., Mironova, Y., Ming, G. L., Kolodkin, A. L., et al. (2014). Semaphorin 5A inhibits synaptogenesis in early postnatal- and adult-born hippocampal dentate granule cells. Elife 3:e04390. doi: 10.7554/eLife.04390
Etherton, M., Foldy, C., Sharma, M., Tabuchi, K., Liu, X., Shamloo, M., et al. (2011). Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc. Natl. Acad. Sci. U.S.A. 108, 13764–13769. doi: 10.1073/pnas.1111093108
Foldy, C., Malenka, R. C., and Sudhof, T. C. (2013). Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 78, 498–509. doi: 10.1016/j.neuron.2013.02.036
Frye, R. E. (2020). Mitochondrial dysfunction in autism spectrum disorder: unique abnormalities and targeted treatments. Semin. Pediatr. Neurol. 35:100829. doi: 10.1016/j.spen.2020.100829
Garbett, K., Ebert, P. J., Mitchell, A., Lintas, C., Manzi, B., Mirnics, K., et al. (2008). Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol. Dis. 30, 303–311. doi: 10.1016/j.nbd.2008.01.012
Gupta, S., Ellis, S. E., Ashar, F. N., Moes, A., Bader, J. S., Zhan, J., et al. (2014). Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun. 5:5748. doi: 10.1038/ncomms6748
Hansen, K. B., Yi, F., Perszyk, R. E., Furukawa, H., Wollmuth, L. P., Gibb, A. J., et al. (2018). Structure, function, and allosteric modulation of NMDA receptors. J. Gen. Physiol. 150, 1081–1105. doi: 10.1085/jgp.201812032
Harvey, R. J., and Yee, B. K. (2013). Glycine transporters as novel therapeutic targets in schizophrenia, alcohol dependence and pain. Nat. Rev. Drug Discov. 12, 866–885. doi: 10.1038/nrd3893
Hollis, F., Kanellopoulos, A. K., and Bagni, C. (2017). Mitochondrial dysfunction in autism spectrum disorder: clinical features and perspectives. Curr. Opin. Neurobiol. 45, 178–187. doi: 10.1016/j.conb.2017.05.018
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Iossifov, I., O’Roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D., et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. doi: 10.1038/nature13908
Isserlin, R., Merico, D., Voisin, V., and Bader, G. D. (2014). Enrichment Map – a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res 3:141. doi: 10.12688/f1000research.4536.1
Jacobi, E., and von Engelhardt, J. (2018). AMPA receptor complex constituents: control of receptor assembly, membrane trafficking and subcellular localization. Mol. Cell. Neurosci. 91, 67–75. doi: 10.1016/j.mcn.2018.05.008
Jaramillo, T. C., Liu, S., Pettersen, A., Birnbaum, S. G., and Powell, C. M. (2014). Autism-related neuroligin-3 mutation alters social behavior and spatial learning. Autism Res. 7, 264–272. doi: 10.1002/aur.1362
Javitt, D. C. (2012). “Glycine transport inhibitors in the treatment of schizophrenia,” in Novel Antischizophrenia Treatments. Handbook of Experimental Pharmacology, eds M. Geyer and G. Gross (Berlin: Springer), 367–399. doi: 10.1007/978-3-642-25758-2_12
Johnson, J. W., and Ascher, P. (1987). Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325, 529–531. doi: 10.1038/325529a0
Kang, H. J., Kawasawa, Y. I., Cheng, F., Zhu, Y., Xu, X., Li, M., et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. doi: 10.1038/nature10523
Kidd, T., Brose, K., Mitchell, K. J., Fetter, R. D., Tessier-Lavigne, M., Goodman, C. S., et al. (1998). Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92, 205–215. doi: 10.1016/s0092-8674(00)80915-0
Kim, J. A., Kim, D., Won, S. Y., Han, K. A., Park, D., Cho, E., et al. (2017). Structural insights into modulation of neurexin-neuroligin trans-synaptic adhesion by MDGA1/neuroligin-2 complex. Neuron 94, 1121–1131.e6. doi: 10.1016/j.neuron.2017.05.034
Kiss, H., Darai, E., Kiss, C., Kost-Alimova, M., Klein, G., Dumanski, J. P., et al. (2002). Comparative human/murine sequence analysis of the common eliminated region 1 from human 3p21.3. Mamm. Genome 13, 646–655. doi: 10.1007/s00335-002-3037-y
Kowalczuk, S., Broer, A., Munzinger, M., Tietze, N., Klingel, K., and Broer, S. (2005). Molecular cloning of the mouse IMINO system: an Na+- and Cl–-dependent proline transporter. Biochem. J. 386(Pt 3) 417–422. doi: 10.1042/BJ20050100
Lane, H. Y., Huang, C. L., Wu, P. L., Liu, Y. C., Chang, Y. C., Lin, P. Y., et al. (2006). Glycine transporter I inhibitor, N-methylglycine (sarcosine), added to clozapine for the treatment of schizophrenia. Biol. Psychiatry 60, 645–649. doi: 10.1016/j.biopsych.2006.04.005
Larsen, B. R., Stoica, A., and MacAulay, N. (2016). Managing brain extracellular K+ during neuronal activity: the physiological role of the Na+/K+-ATPase subunit isoforms. Front. Physiol. 7:141. doi: 10.3389/fphys.2016.00141
Lee, A., Hirabayashi, Y., Kwon, S. K., Lewis, T. L. Jr., and Polleux, F. (2018). Emerging roles of mitochondria in synaptic transmission and neurodegeneration. Curr. Opin. Physiol. 3, 82–93. doi: 10.1016/j.cophys.2018.03.009
Lee, B., Kim, J., An, T., Kim, S., Patel, E. M., Raber, J., et al. (2018). Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nat. Commun. 9:2026. doi: 10.1038/s41467-018-04377-4
Lee, E., Lee, S., Shin, J. J., Choi, W., Chung, C., Lee, S., et al. (2021). Excitatory synapses and gap junctions cooperate to improve Pv neuronal burst firing and cortical social cognition in Shank2-mutant mice. Nat. Commun. 12:5116. doi: 10.1038/s41467-021-25356-2
Lee, K., Kim, Y., Lee, S. J., Qiang, Y., Lee, D., Lee, H. W., et al. (2013). MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc. Natl. Acad. Sci. U.S.A. 110, 336–341. doi: 10.1073/pnas.1219987110
Li, J., Shalev-Benami, M., Sando, R., Jiang, X., Kibrom, A., Wang, J., et al. (2018). Structural basis for teneurin function in circuit-wiring: a toxin motif at the synapse. Cell 173, 735–748.e15. doi: 10.1016/j.cell.2018.03.036
Li, Z., Okamoto, K., Hayashi, Y., and Sheng, M. (2004). The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887. doi: 10.1016/j.cell.2004.11.003
Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., et al. (2019). Genomic resolution of DLX-orchestrated transcriptional circuits driving development of forebrain GABAergic neurons. Cell Rep. 28, 2048–2063.e8. doi: 10.1016/j.celrep.2019.07.022
Liu, Q. R., Lopez-Corcuera, B., Mandiyan, S., Nelson, H., and Nelson, N. (1993). Cloning and expression of a spinal cord- and brain-specific glycine transporter with novel structural features. J. Biol. Chem. 268, 22802–22808.
Lombardo, M. V. (2021). Ribosomal protein genes in post-mortem cortical tissue and iPSC-derived neural progenitor cells are commonly upregulated in expression in autism. Mol. Psychiatry 26, 1432–1435. doi: 10.1038/s41380-020-0773-x
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Meng, W., Mushika, Y., Ichii, T., and Takeichi, M. (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959. doi: 10.1016/j.cell.2008.09.040
Merico, D., Isserlin, R., Stueker, O., Emili, A., and Bader, G. D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. doi: 10.1371/journal.pone.0013984
Mosca, T. J., Hong, W., Dani, V. S., Favaloro, V., and Luo, L. (2012). Trans-synaptic teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237–241. doi: 10.1038/nature10923
Nadler, J. J., Moy, S. S., Dold, G., Trang, D., Simmons, N., Perez, A., et al. (2004). Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 3, 303–314. doi: 10.1111/j.1601-183X.2004.00071.x
Nash, S. R., Giros, B., Kingsmore, S. F., Kim, K. M., El-Mestikawy, S., Dong, Q., et al. (1998). Cloning, gene structure and genomic localization of an orphan transporter from mouse kidney with six alternatively-spliced isoforms. Recept. Channels 6, 113–128.
Nicoll, R. A., Tomita, S., and Bredt, D. S. (2006). Auxiliary subunits assist AMPA-type glutamate receptors. Science 311, 1253–1256. doi: 10.1126/science.1123339
Nzelu, D., Shangaris, P., Story, L., Smith, F., Piyasena, C., Alamelu, J., et al. (2021). X-linked sideroblastic anaemia in a female fetus: a case report and a literature review. BMC Med. Genomics 14:296. doi: 10.1186/s12920-021-01146-z
Paoletti, P., Bellone, C., and Zhou, Q. (2013). NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14, 383–400. doi: 10.1038/nrn3504
Parikshak, N. N., Swarup, V., Belgard, T. G., Irimia, M., Ramaswami, G., Gandal, M. J., et al. (2016). Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427. doi: 10.1038/nature20612
Park, H., Choi, Y., Jung, H., Kim, S., Lee, S., Han, H., et al. (2020). Splice-dependent trans-synaptic PTPdelta-IL1RAPL1 interaction regulates synapse formation and non-REM sleep. EMBO J. 39:e104150. doi: 10.15252/embj.2019104150
Patro, R., Duggal, G., Love, M., Irizarry, R., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419.
Pederick, D. T., and Luo, L. (2021). Teneurins. Curr. Biol. 31, R936–R937. doi: 10.1016/j.cub.2021.06.035
Petryniak, M. A., Potter, G. B., Rowitch, D. H., and Rubenstein, J. L. (2007). Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55, 417–433. doi: 10.1016/j.neuron.2007.06.036
Pettem, K. L., Yokomaku, D., Takahashi, H., Ge, Y., and Craig, A. M. (2013). Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J. Cell Biol. 200, 321–336. doi: 10.1083/jcb.201206028
Pinero, J., Bravo, A., Queralt-Rosinach, N., Gutierrez-Sacristan, A., Deu-Pons, J., Centeno, E., et al. (2017). DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839. doi: 10.1093/nar/gkw943
Pinero, J., Ramirez-Anguita, J. M., Sauch-Pitarch, J., Ronzano, F., Centeno, E., Sanz, F., et al. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855. doi: 10.1093/nar/gkz1021
Pla, R., Stanco, A., Howard, M. A., Rubin, A. N., Vogt, D., Mortimer, N., et al. (2018). Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb. Cortex 28, 3797–3815. doi: 10.1093/cercor/bhx241
Polepalli, J. S., Wu, H., Goswami, D., Halpern, C. H., Sudhof, T. C., and Malenka, R. C. (2017). Modulation of excitation on parvalbumin interneurons by neuroligin-3 regulates the hippocampal network. Nat. Neurosci. 20, 219–229. doi: 10.1038/nn.4471
Protais, P., Costentin, J., and Schwartz, J. C. (1976). Climbing behavior induced by apomorphine in mice: a simple test for the study of dopamine receptors in striatum. Psychopharmacology 50, 1–6. doi: 10.1007/BF00634146
Quinn, L. P., Stean, T. O., Chapman, H., Brown, M., Vidgeon-Hart, M., Upton, N., et al. (2006). Further validation of LABORAS using various dopaminergic manipulations in mice including MPTP-induced nigro-striatal degeneration. J. Neurosci. Methods 156, 218–227. doi: 10.1016/j.jneumeth.2006.03.013
Quinn, L. P., Stean, T. O., Trail, B., Duxon, M. S., Stratton, S. C., Billinton, A., et al. (2003). LABORAS: initial pharmacological validation of a system allowing continuous monitoring of laboratory rodent behaviour. J. Neurosci. Methods 130, 83–92. doi: 10.1016/s0165-0270(03)00227-9
Riffee, W. H., Wilcox, R. E., and Smith, R. V. (1979). Stereotypic and hypothermic effects of apomorphine and N-n-propylnorapomorphine in mice. Eur. J. Pharmacol. 54, 273–277. doi: 10.1016/0014-2999(79)90086-4
Rojas-Charry, L., Nardi, L., Methner, A., and Schmeisser, M. J. (2021). Abnormalities of synaptic mitochondria in autism spectrum disorder and related neurodevelopmental disorders. J. Mol. Med. 99, 161–178. doi: 10.1007/s00109-020-02018-2
Rothwell, P. E., Fuccillo, M. V., Maxeiner, S., Hayton, S. J., Gokce, O., Lim, B. K., et al. (2014). Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158, 198–212. doi: 10.1016/j.cell.2014.04.045
Sando, R., Jiang, X., and Sudhof, T. C. (2019). Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363:eaav7969. doi: 10.1126/science.aav7969
Santini, E., and Klann, E. (2014). Reciprocal signaling between translational control pathways and synaptic proteins in autism spectrum disorders. Sci. Signal. 7:re10. doi: 10.1126/scisignal.2005832
Seeger, M., Tear, G., Ferres-Marco, D., and Goodman, C. S. (1993). Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 10, 409–426. doi: 10.1016/0896-6273(93)90330-t
Sheng, Z. H., and Cai, Q. (2012). Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 13, 77–93. doi: 10.1038/nrn3156
Smith, K. E., Borden, L. A., Hartig, P. R., Branchek, T., and Weinshank, R. L. (1992). Cloning and expression of a glycine transporter reveal colocalization with NMDA receptors. Neuron 8, 927–935. doi: 10.1016/0896-6273(92)90207-t
Smith, K. E., Fried, S. G., Durkin, M. M., Gustafson, E. L., Borden, L. A., Branchek, T. A., et al. (1995). Molecular cloning of an orphan transporter. a new member of the neurotransmitter transporter family. FEBS Lett. 357, 86–92. doi: 10.1016/0014-5793(94)01328-x
Soneson, C., Love, M. I., and Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. doi: 10.12688/f1000research.7563.2
Subramanian, A., Kuehn, H., Gould, J., Tamayo, P., and Mesirov, J. P. (2007). GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 23, 3251–3253. doi: 10.1093/bioinformatics/btm369
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sudhof, T. C. (2018). Towards an understanding of synapse formation. Neuron 100, 276–293. doi: 10.1016/j.neuron.2018.09.040
Takanaga, H., Mackenzie, B., Suzuki, Y., and Hediger, M. A. (2005). Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system imino. J. Biol. Chem. 280, 8974–8984. doi: 10.1074/jbc.M413027200
Tsai, G., Lane, H. Y., Yang, P., Chong, M. Y., and Lange, N. (2004). Glycine transporter I inhibitor, N-methylglycine (sarcosine), added to antipsychotics for the treatment of schizophrenia. Biol. Psychiatry 55, 452–456. doi: 10.1016/j.biopsych.2003.09.012
Um, J. W., and Ko, J. (2017). Neural glycosylphosphatidylinositol-anchored proteins in synaptic specification. Trends Cell Biol. 27, 931–945. doi: 10.1016/j.tcb.2017.06.007
Velmeshev, D., Magistri, M., Mazza, E. M. C., Lally, P., Khoury, N., D’Elia, E. R., et al. (2020). Cell-type-specific analysis of molecular pathology in autism identifies common genes and pathways affected across neocortical regions. Mol. Neurobiol. 57, 2279–2289. doi: 10.1007/s12035-020-01879-5
Velmeshev, D., Schirmer, L., Jung, D., Haeussler, M., Perez, Y., Mayer, S., et al. (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. doi: 10.1126/science.aav8130
Voineagu, I., Wang, X., Johnston, P., Lowe, J. K., Tian, Y., Horvath, S., et al. (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384. doi: 10.1038/nature10110
von Engelhardt, J. (2019). AMPA receptor auxiliary proteins of the CKAMP family. Int. J. Mol. Sci. 20:1460. doi: 10.3390/ijms20061460
von Engelhardt, J., Mack, V., Sprengel, R., Kavenstock, N., Li, K. W., Stern-Bach, Y., et al. (2010). CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science 327, 1518–1522. doi: 10.1126/science.1184178
Vorhees, C. V., and Williams, M. T. (2006). Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858. doi: 10.1038/nprot.2006.116
Vos, M., Lauwers, E., and Verstreken, P. (2010). Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front. Synaptic Neurosci. 2:139. doi: 10.3389/fnsyn.2010.00139
Werling, D. M., Parikshak, N. N., and Geschwind, D. H. (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat. Commun. 7:10717. doi: 10.1038/ncomms10717
Wilcox, R. E., Riffee, W. H., and Smith, R. V. (1979). Pharmacological basis for N-n-propylnorapomorphine induced stereotypic cage climbing and behavioral arousal in mice. Pharmacol. Biochem. Behav. 11, 653–659. doi: 10.1016/0091-3057(79)90258-2
Xu, L. M., Li, J. R., Huang, Y., Zhao, M., Tang, X., and Wei, L. (2012). AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 40, D1016–D1022. doi: 10.1093/nar/gkr1145
Yang, C., Li, J., Wu, Q., Yang, X., Huang, A. Y., Zhang, J., et al. (2018). AutismKB 2.0: a knowledgebase for the genetic evidence of autism spectrum disorder. Database 2018:bay106. doi: 10.1093/database/bay106
Zeisel, A., Munoz-Manchado, A. B., Codeluppi, S., Lonnerberg, P., La Manno, G., Jureus, A., et al. (2015). Brain structure. cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. doi: 10.1126/science.aaa1934
Zhang, B., Seigneur, E., Wei, P., Gokce, O., Morgan, J., and Sudhof, T. C. (2017). Developmental plasticity shapes synaptic phenotypes of autism-associated neuroligin-3 mutations in the calyx of Held. Mol. Psychiatry 22, 1483–1491. doi: 10.1038/mp.2016.157
Zhang, B., and Sudhof, T. C. (2016). Neuroligins are selectively essential for NMDAR signaling in cerebellar stellate interneurons. J. Neurosci. 36, 9070–9083. doi: 10.1523/JNEUROSCI.1356-16.2016
Zhou, Z., Xu, H., Li, Y., Yang, M., Zhang, R., Shiraishi, A., et al. (2020). CAMSAP1 breaks the homeostatic microtubule network to instruct neuronal polarity. Proc. Natl. Acad. Sci. U.S.A. 117, 22193–22203. doi: 10.1073/pnas.1913177117
Keywords: glycine and proline transport, NMDA receptors, autism spectrum disorders, epilepsy, learning and memory, synapse, mitochondria, ribosomes
Citation: Kim J, Roh JD, Kim S, Kang H, Bae M and Kim E (2022) Slc6a20a Heterozygous and Homozygous Mutant Mice Display Differential Behavioral and Transcriptomic Changes. Front. Mol. Neurosci. 15:857820. doi: 10.3389/fnmol.2022.857820
Received: 19 January 2022; Accepted: 28 January 2022;
Published: 07 March 2022.
Edited by:
Jaewon Ko, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaReviewed by:
Yi-Ping Hsueh, Institute of Molecular Biology, Academia Sinica, TaiwanSe-Young Choi, Seoul National University, South Korea
Copyright © 2022 Kim, Roh, Kim, Kang, Bae and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eunjoon Kim, a2ltZUBrYWlzdC5hYy5rcg==
 Junhyung Kim
Junhyung Kim Junyeop Daniel Roh
Junyeop Daniel Roh Seongbin Kim
Seongbin Kim Hyojin Kang
Hyojin Kang Mihyun Bae
Mihyun Bae Eunjoon Kim
Eunjoon Kim