
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 09 December 2022
Sec. Brain Disease Mechanisms
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.1010101
This article is part of the Research Topic Biomarkers, Mechanisms, and Treatment Strategies for Epilepsy View all 8 articles
 Shitao Wang1,2
Shitao Wang1,2 Xuemei Cai3
Xuemei Cai3 Shiyong Liu4
Shiyong Liu4 Qixin Zhou5
Qixin Zhou5 Ting Wang1
Ting Wang1 Sunbing Du1
Sunbing Du1 Dan Wang1
Dan Wang1 Fei Yang1
Fei Yang1 Qian Wu1
Qian Wu1 Yanbing Han1*
Yanbing Han1*Background: Genetic factors have been found to be associated with the efficacy and adverse reactions of antiseizure medications. BCL11A is an important regulator of the development of neuronal networks. However, the role of BCL11A in epilepsy remains unclear. This study aimed to evaluate the genetic association of BCL11A with the susceptibility to develop epileptic seizures and therapeutic response of patients with epilepsy in Han Chinese.
Methods: We matched 450 epilepsy cases with 550 healthy controls and 131 drug-resistant epilepsy patients with 319 drug-responsive epilepsy patients from two different centers. Genetic association analysis, genetic interaction analysis, expression quantitative trait loci analysis and protein–protein interaction analysis were conducted.
Results: Our results showed that rs2556375 not only increases susceptibility to develop epileptic seizures (OR = 2.700, 95% = 1.366–5.338, p = 0.004 and OR = 2.984, 95% = 1.401–6.356, p = 0.005, respectively), but also increases the risk of drug resistance(OR = 21.336, 95%CI =2.489–183.402, p = 0.005). The interaction between rs2556375 and rs12477097 results in increased risk for pharma coresistant. In addition, rs2556375 regulated BCL11A expression in human brain tissues (p = 0.0096 and p = 0.033, respectively). Furthermore, the protein encoded by BCL11A interacted with targets of approved antiepileptic drugs.
Conclusion: BCL11A may be a potential therapeutic target for epilepsy. Rs2556375 may increase the risks of epilepsy and drug resistance by regulating BCL11A expression in human brain tissues. Moreover, the interaction between rs2556375 and rs12477097 results in increased risk for drug resistance.
Epilepsy is a neurological disease with genetic factors in its pathogenesis (International League Against Epilepsy Consortium on Complex Epilepsies, 2014). The heritability of epilepsy is estimated to be more than 50% of all epilepsies (Pal et al., 2010). So far, a large number of genes associated with epilepsy has been identified with the help of advances in genetic technologies (Wang et al., 2017). However, evidence for the associations of some of these genes with epilepsy is controversial partly due to study power and scope (Cavalleri et al., 2007; Kasperaviciūte et al., 2010; Guo et al., 2012; Leu et al., 2012; Steffens et al., 2012; Franco and Perucca, 2015). Genetic factors have been found to be associated with the efficacy and adverse reactions of antiepileptic drugs (Franco and Perucca, 2015; Balestrini and Sisodiya, 2018). Currently, approximately one-third of epileptic seizures cannot be effectively controlled with available antiepileptic drugs, so the discovery of new drug targets has become increasingly important.
The BCL11A gene is expressed at high levels in hematopoietic tissues and the brain (Nakamura et al., 2000; Funnell et al., 2015). Functional deficits in BCL11A can result in persistence of HbF (Satterwhite et al., 2001; Sankaran et al., 2008). BCL11A-L is an important protein product of BCL11A gene. In hippocampal neurons, downregulation of BCL11A-L expression upregulates axon branching and dendrite outgrowth and increases the arbor complexity of axons and dendrites, while overexpression of BCL11A-L reduces neurite arborization (Kuo et al., 2009). BCL11A-L may regulate deleted in colorectal cancer and microtubule-associated protein 1b expression through an interaction with calmodulin-dependent serine protein kinase, thereby limiting neuritogenesis (Kuo et al., 2010). Changes in the expression of BCL11A will lead to abnormal development of neurons, which can cause abnormal discharge of neurons, resulting in seizures. To date, BCL11A mutations have been shown to be associated with intellectual disability syndrome (Dias et al., 2016; Cai et al., 2017; Soblet et al., 2018). Two de novo BCL11A heterozygous variants were reported to be associated with patients with epileptic encephalopathy (Yoshida et al., 2018), and it also indicated that a de novo mutation in the BCL11A gene result in epilepsy (Korenke et al., 2020). However, the role of common BCL11A single nucleotide polymorphisms(SNPs) in therapeutic response of patients with epilepsy has not been evaluated, and the role of common BCL11A SNPs in epilepsy in Han Chinese remains unknown.
In this study, we analyzed BCL11A SNPs to explore their potential associations with epilepsy risk and therapeutic response of patients with epilepsy, and then performed expression quantitative trait loci (eQTL) analysis to investigate effects of identified SNPs on BCL11A expression. Furthermore, protein–protein interaction(PPI) analysis was performed to further evaluate the role of BCL11A in epilepsy treatment.
We recruited 450 epilepsy cases diagnosed at the First Affiliated Hospital of Kunming Medical University and Xinqiao Hospital. In parallel, we recruited 550 healthy controls without a personal history of epilepsy or a family history of epilepsy for routine health checkups from the same hospitals. All samples were stored in the biobanks of the First Affiliated Hospital of Kunming Medical University and Kunming Medical University. The diagnoses of epilepsy and drug-resistant epilepsy are based on 2017 and 2010 International League Against Epilepsy criteria, respectively. Carbamazepine, valproic acid, levetiracetam and Lamotrigine were prescribed to the epilepsy patients. Symptomatic epilepsy was ruled out through blood tests, disease history review, imaging examination, etc. All participants were required to sign the informed consent form.
BCL11A Tagging SNPs with a minor allele frequency greater than 0.05 in Han Chinese were selected from the 1,000 Genomes database1 using Haploview software. The Tissue DNA Kit (OMEGA, United States) and the Blood DNA Mini Kit (OMEGA, United States) were used to extract genomic DNA from brain tissues and peripheral blood, respectively. Brain tissue samples was derived from abnormal discharge lesions in the temporal lobe of patients with drug-resistant epilepsy patients undergoing epilepsy surgery. Genotyping was conducted using the Bio-Rad CFX96 (BioRad, United States) platform, and the primers were designed with Primer Premier V6.0 (Premier Biosoft Inc., United States). Details on PCR primers are provided in Supplementary Table 1.
We performed eQTL analysis using 16 brain tissues from patients with drug-resistant epilepsy to investigate whether identified SNPs regulate BCL11A expression in human brain tissues. Tissue RNA Kit (Omega, United States) and the FastQuant cDNA kit (Tiangen, China) are used for genomic RNA extraction and cDNA reverse transcription, respectively, and the SYBR®Green I (Vazyme, China) and the ABI QuantStudio 6 Flex™ (ABI, United States) analyzer are used for Quantitative PCR. We designed the primers with Primer Premier V6.0 (Premier Biosoft Inc., United States). Details on Quantitative PCR premiers are provided in Supplementary Table 2. Functional effect of identified SNPs on BCL11A expression was further verified by data from the Genotype-Tissue Expression project (GTEx).2
To further evaluate the role of BCL11A in epilepsy treatment, we obtained approved antiepileptic drug targets from DrugBank5.0 (Wishart et al., 2018) and the Therapeutic Target Database 2020 (Wang et al., 2020). The interactions between the protein encoded by BCL11A and approved antiepileptic drug targets were evaluated with Cytoscape V.3.7.2 (Su et al., 2014).
Associations of BCL11A SNPs with epilepsy and therapeutic response were analyzed with binary logistic regression. Hardy–Weinberg equilibrium and difference in gender were assessed using the Chi-square (χ2) test and Fisher’s exact test. The eQTL analysis and difference in mean age were analyzed by independent samples t-test. A p value <0.05 (two-tailed) was considered statistically significant. For Bonferroni correction, A p value <0.0056 (0.05/9) was considered statistically significant. All analyses were conducted with SPSS23.0(IBM Corp, Armonk, NY) and Graphpad Prism9.0.3
The baseline characteristics are summarized in Table 1.
We selected nine tagging SNPs with a minor allele frequency greater than 0.05 from the 1,000 Genomes database (see Footnote 1) using Haploview software, which complied with the Hardy–Weinberg equilibrium in the control group(p > 0.05 for all; Tables 2, 3).

Table 3. Associations between BCL11A SNP genotypes and therapeutic response of patients with epilepsy.
Of the nine tagging SNPs, rs2556375 is associated with increased risk of epilepsy (OR = 2.700, 95% = 1.366–5.338, p = 0.004 and OR = 2.984, 95% = 1.401–6.356, p = 0.005, respectively) after adjusting for gender and age and Bonferroni correction at genotypic level (Table 2). In addition, the rs6747099 genotype distribution was also different between the epilepsy and control groups after adjusting for gender and age(OR = 0.275, 95%CI = 0.077–0.980, p = 0.046; Table 2), but did not reach significance after Bonferroni correction(p > 0.0056).
Similarly, rs2556375 is associated with increased risk of drug resistance (OR = 21.336, 95%CI = 2.489–183.402, p = 0.005) after adjusting for gender and age at genotypic level (Table 3). At the same time, we also found that rs356977, rs10189857, rs12477097 and rs13018474 are associated with drug resistance after adjusting for gender and age(OR = 0.306, 95%CI = 0.097–0.968, p = 0.044; OR = 3.285, 95%CI = 1.200–8.989, p = 0.021; OR = 2.849, 95%CI = 1.051–7.727, p = 0.040 and OR = 0.464, 95%CI = 0.256–0.842, p = 0.012, respectively) at genotypic level (Table 3). However, only rs2556375 reached significance after Bonferroni correction (p < 0.0056).
The interactions among nine SNPs were detected by multifactor dimensionality reduction(MDR). For the association with epilepsy risk, no significant model was detected by MDR. For the association with therapeutic response, Table 4 provided the results testing by MDR. Two-to three-locus models were significant (p = 0.0107). An interaction dendrogram showed the strongest interaction between rs2556375 and rs12477097 (Figure 1).
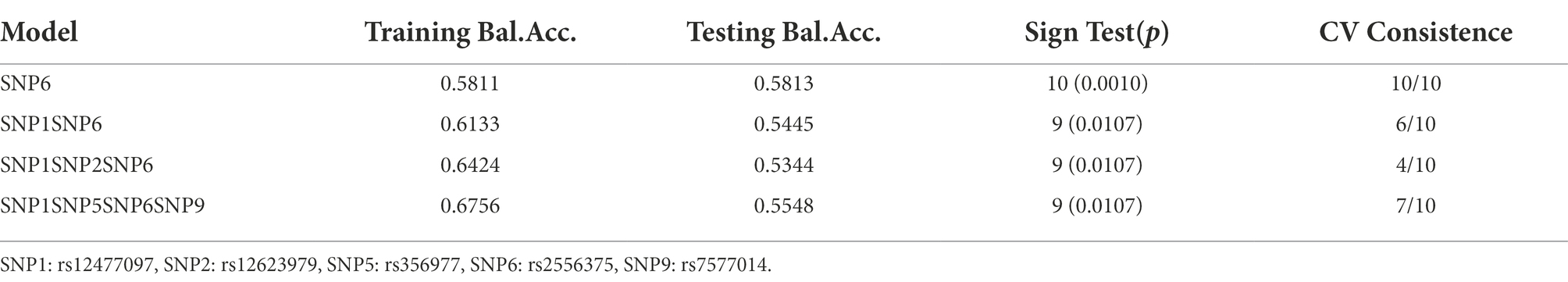
Table 4. MDR analysis of BCL11A SNPs showed different interactions for therapeutic response of patients with epilepsy.

Figure 1. Different types of interaction dendrogram among nine SNPs. SNP1: rs12477097, SNP2: rs12623979, SNP5: rs356977, SNP6: rs2556375, SNP9: rs7577014.
To obtain the OR and 95%CI for a joint effect of rs2556375 and rs12477097, we conducted genetic interaction analysis by using logistic regression analyses. As shown in Table 5, the participants with rs12477097 AC genotype and rs2556375 GG genotype had higher risk of drug resistance than the participants with rs12477097 CC genotype and rs2556375TT genotype.
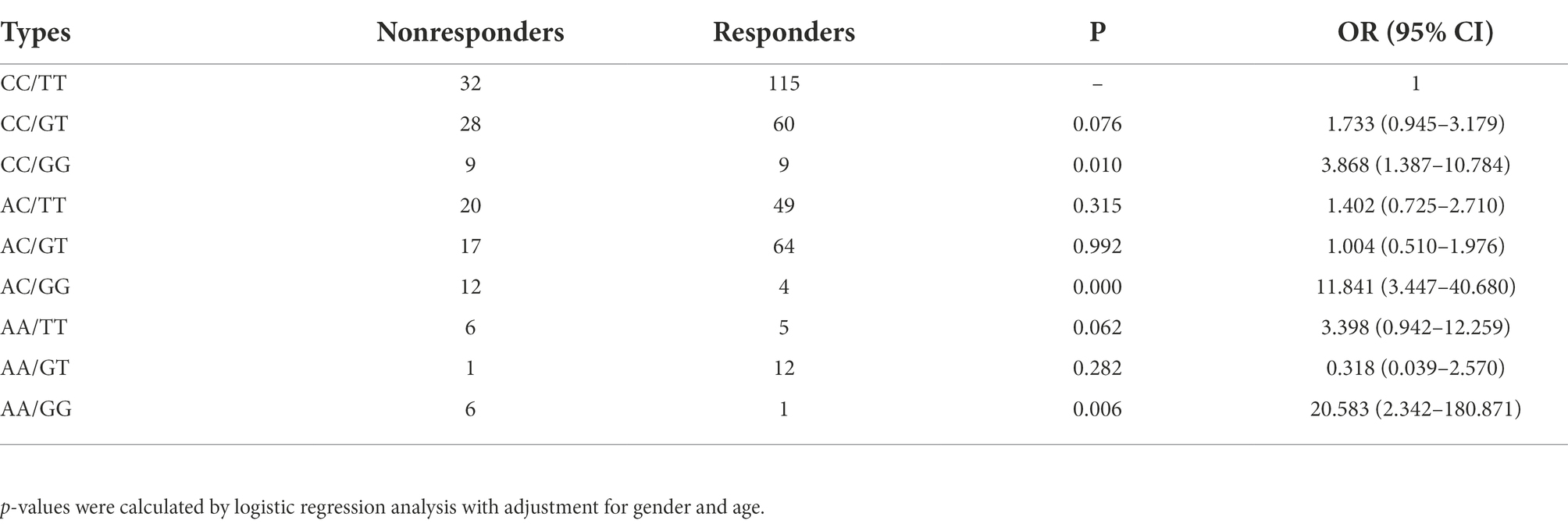
Table 5. Joint effects analysis of rs2556375 and rs12477097 for therapeutic response of patients with epilepsy.
The result of functional analysis showed that rs2556375 is an eQTL in drug-resistant epileptic brain tissue, and the carriers of the G allele exhibited downregulated STX1B gene expression (Figure 2). Furthermore, this finding was confirmed by data from the brain tissue database GTEx (Figure 3).5
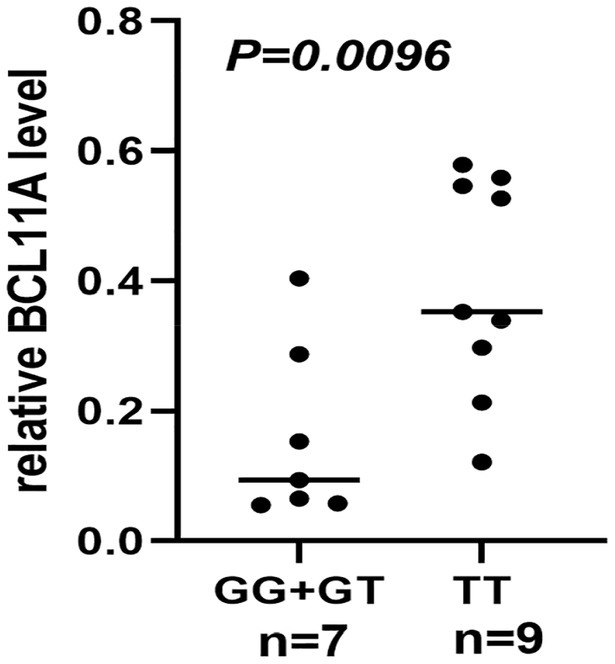
Figure 2. The rs2556375 is an eQTL in brain tissue of drug-resistant epilepsy patients. The carriers of the G allele exhibited downregulated BCL11A gene expression.
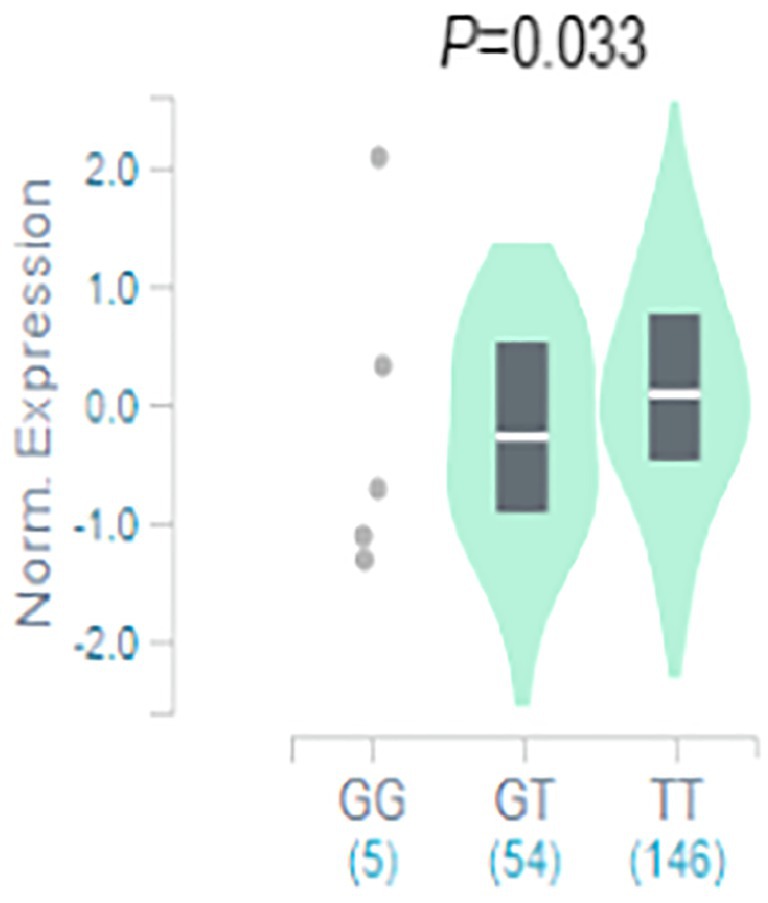
Figure 3. The rs2556375 is an eQTL in cortex. Data were retrieved from the brain tissue database GTEx (https://gtexportal.org/).
115 genes targeted by approved antiepileptic drugs were obtained from DrugBank5.0 (Wishart et al., 2018) and the Therapeutic Target Database 2020 (Wang et al., 2020). Details on 115 genes are provided in Supplementary Table 3. PPI analysis showed that the protein encoded by BCL11A interacts with approved antiepileptic drug targets (Figure 4).
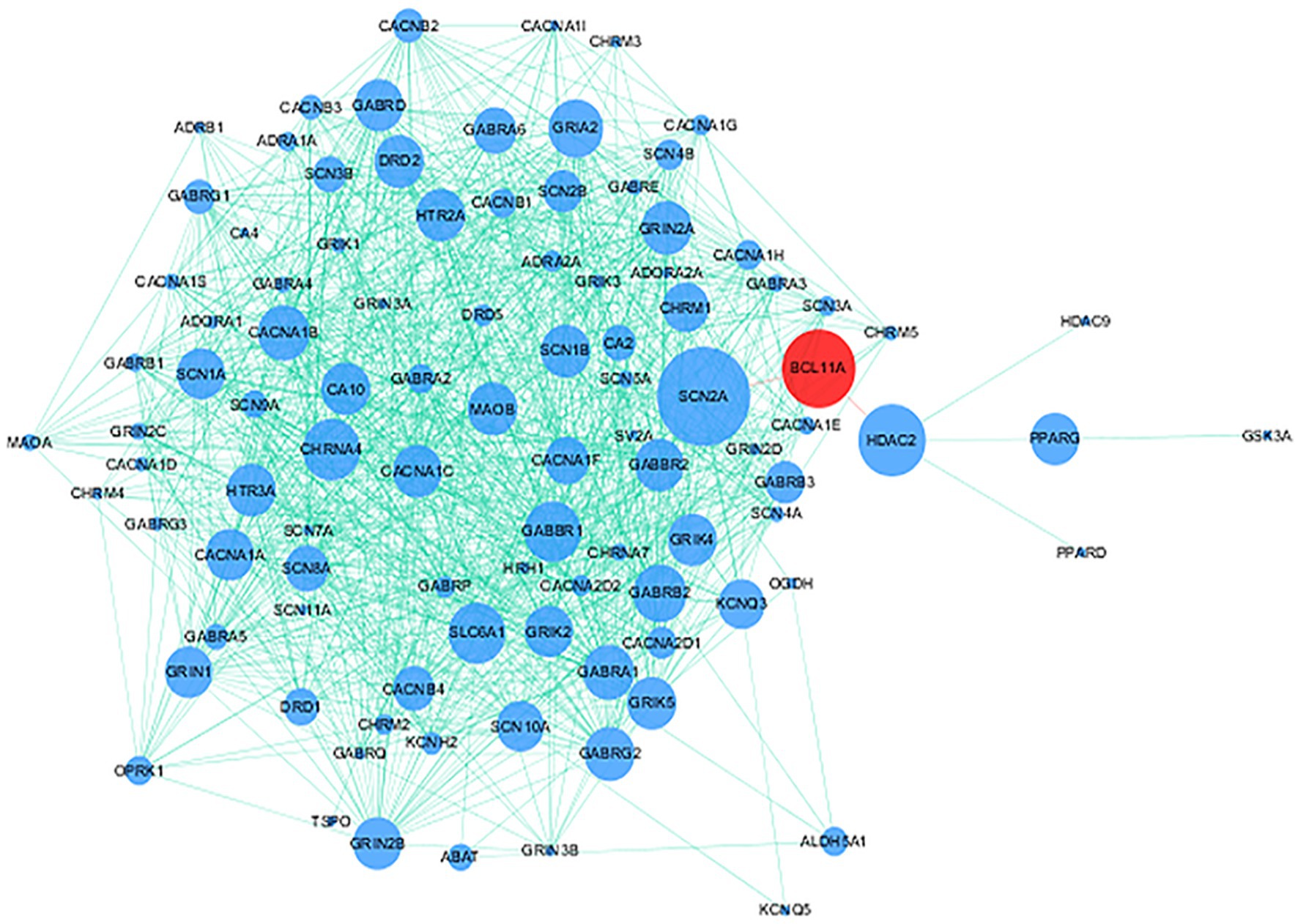
Figure 4. Protein–protein interaction (PPI) network of BCL11A gene and genes targeted by approved antiepileptic drugs. Red node and blue nodes represent BCL11A gene and genes targeted by approved antiepileptic drugs, respectively. The proteins connected by the red line mean that these proteins have direct interaction.
BCL11A is an important regulator of the development of neuronal networks, and its proper expression is very important for axon branching and dendrite growth (Kuo et al., 2009, 2010). Based on the above results, we speculate that BCL11A may affect the risks of epilepsy and drug resistance by regulating the development of neuronal networks.
Gene SNPs have been found to be associated with the risk of diseases and the efficacy of drug treatment (Tawfeek and Alhassanin, 2018; Ebid et al., 2020; Pozhidaev et al., 2020; de Assis et al., 2021). However, the association between the common BCL11A SNPs and epilepsy and the association between the common BCL11A SNPs and therapeutic response of patients with epilepsy in Han Chinese remain unclear. In this study, by conducting genetic association analysis, genetic interaction analysis, eQTL analysis and PPI analysis, we have identified a novel SNP (rs2556375) associated with epilepsy and therapeutic response of patients with epilepsy. To our knowledge, this is the first study that evaluate the associations of common BCL11A SNPs with epilepsy risk and therapeutic response of patients with epilepsy in Han Chinese.
We performed a two-stage case–control study to evaluate the associations between BCL11A SNPs and epilepsy. In the Stage I study, we found that rs2556375 increases epilepsy risk (Table 2). In the Stage II study, further analyses suggested that rs2556375 increases the risk of drug resistance (Table 3). Furthermore, we also found that the participants with rs12477097 AC genotype and rs2556375GG genotype had higher risk of drug resistance than the participants with rs12477097 CC genotype and rs2556375TT genotype (Table 5), suggesting an important pathogenic mechanism that rs2556375 affects the risk of drug resistance by interacting with rs12477097. Combined with the above findings, we can reasonably speculate that rs2556375 may play an important role in the risk and therapeutic response of patients with epilepsy.
Among all the tagging SNPs, rs2556375, rs6747099, rs356977, rs10189857, rs12477097and rs13018474 in BCL11A also showed associations with epilepsy or therapeutic response of patients with epilepsy, but the association did not exist after Bonferroni correction. However, the role of these SNPs in the risk and therapeutic response of patients with epilepsy could not be simply ignored, as further verification of our findings is necessary. Although we did not find that rs12477097 alone affected therapeutic response of patients with epilepsy, we found that the interaction between rs2556375 and rs12477097 could affect therapeutic response of patients with epilepsy.
Previous studies have found that gene SNPs often affect the risk of diseases by regulating gene expression (Maurano et al., 2012; Wang et al., 2021a,b). To clarify how rs2556375 affects the risks of epilepsy and drug resistance, we included 16 brain tissues from patients with drug-resistant epilepsy, and further used data from GTEx database for eQTL analysis to evaluate whether rs2556375 regulates the expression level of BCL11A in human brain tissue. Our results showed that rs2556375 regulates the expression of BCL11A in human brain tissue, especially in brain tissue of patients with drug-resistant epilepsy (Figure 2), which further demonstrates what we found in genetic association studies. Furthermore, the results of PPI analysis verify the role of BCL11A in epilepsy treatment. These above results will contribute to the discovery of new therapeutic targets for epilepsy.
Although we conducted genetic association analysis, genetic interaction analysis, eQTL analysis and PPI analysis and found that the BCL11A may be associated with the risk of epilepsy and therapeutic response of patients with epilepsy in Han Chinese. However, the sample size included in this study is relatively small, and we only performed functional analysis in human brain tissue. Therefore, it is necessary to performed further research with large samples and functional analysis to verify our conclusions.
BCL11A may be a potential therapeutic target for epilepsy. Rs2556375 may increase the risks of epilepsy and drug resistance by regulating BCL11A expression in human brain tissues. Furthermore, the interaction between rs2556375 and rs12477097 results in increased risk for drug resistance. These findings will facilitate translation of genetic findings to clinical treatment in the future.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Ethics Committee of the First Affiliated Hospital of Kunming Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
YH and SW contributed to study design. SD, TW, and DW performed the experiments and contributed to the data collection. QW, XC, and FY performed the statistical analysis. YH, SW, QZ, and SL were involved in the writing of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81660228 and 81601134), Yunnan Province Talent Training Program (2017HB048), Yunnan Health Training Project of High Level Talents (L-2019019 and H-2018056), and Yunnan Science and Research Funding Program (2016NS029).
We thank all the participants for providing blood and brain tissue samples. We also appreciate Zhenrong Xie for sample storage and management.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2022.1010101/full#supplementary-material
Balestrini, S., and Sisodiya, S. M. (2018). Pharmacogenomics in epilepsy. Neurosci. Lett. 667, 27–39. doi: 10.1016/j.neulet.2017.01.014
Cai, T., Chen, X., Li, J., Xiang, B., Yang, L., Liu, Y., et al. (2017). Identification of novel mutations in the HbF repressor gene BCL11A in patients with autism and intelligence disabilities. Am. J. Hematol. 92, E653–E656. doi: 10.1002/ajh.24902
Cavalleri, G. L., Weale, M. E., Shianna, K. V., Singh, R., Lynch, J. M., Grinton, B., et al. (2007). Multicentre search for genetic susceptibility loci in sporadic epilepsy syndrome and seizure types: a case-control study. Lancet Neurol. 6, 970–980. doi: 10.1016/S1474-4422(07)70247-8
de Assis, G. G., Hoffman, J. R., Bojakowski, J., Murawska-Ciałowicz, E., Cieszczyk, P., and Gasanov, E. V. (2021). The Val66 and Met66 alleles-Specifific expression of BDNF in human muscle and their metabolic responsivity. Front. Mol. Neurosci. 14:638176. doi: 10.3389/fnmol.2021.638176
Dias, C., Estruch, S. B., Graham, S. A., McRae, J., Sawiak, S. J., Hurst, J. A., et al. (2016). BCL11A Haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am. J. Hum. Genet. 99, 253–274. doi: 10.1016/j.ajhg.2016.05.030
Ebid, A. I. M., Ahmed, O. A., Agwa, S. H., Abdel-Motaleb, S. M., and Hagag, R. S. (2020). Impact of IL28B gene polymorphism on efficacy and safety of direct acting antivirals in hepatitis C Egyptian patients. Int. J. Clin. Pharm. 42, 1207–1216. doi: 10.1007/s11096-020-01085-2
Franco, V., and Perucca, E. (2015). The pharmacogenomics of epilepsy. Expert. Rev. Neurother. 15, 1161–1170. doi: 10.1586/14737175.2015.1083424
Funnell, A. P., Prontera, P., Ottaviani, V., Piccione, M., Giambona, A., Maggio, A., et al. (2015). 2p15-p16.1 microdeletions encompassing and proximal to BCL11A are associated with elevated HbF in addition to neurologic impairment. Blood 126, 89–93. doi: 10.1182/blood-2015-04-638528
Guo, Y., Baum, L. W., Sham, P. C., Wong, V., Ng, P. W., Lui, C. H., et al. (2012). Two-stage genome-wide association study identifies variants in CAMSAP1L1 as susceptibility loci for epilepsy in Chinese. Hum. Mol. Genet. 21, 1184–1189. doi: 10.1093/hmg/ddr550
International League Against Epilepsy Consortium on Complex Epilepsies (2014). Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 13, 893–903. doi: 10.1016/S1474-4422(14)70171-1
Kasperaviciūte, D., Catarino, C. B., Heinzen, E. L., Depondt, C., Cavalleri, G. L., Caboclo, L. O., et al. (2010). Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain 133, 2136–2147. doi: 10.1093/brain/awq130
Korenke, G. C., Schulte, B., Biskup, S., Neidhardt, J., and Owczarek-Lipska, M. (2020). A novel de novo frameshift mutation in the BCL11A gene in a patient with intellectual disability syndrome and epilepsy. Mol. Syndromol. 11, 135–140. doi: 10.1159/000508566
Kuo, T. Y., Chen, C. Y., and Hsueh, Y. P. (2010). Bcl11A/CTIP1 mediates the effect of the glutamate receptor on axon branching and dendrite outgrowth. J. Neurochem. 114, 1381–1392. doi: 10.1111/j.1471-4159.2010.06852.x
Kuo, T. Y., Hong, C. J., and Hsueh, Y. P. (2009). Bcl11A/CTIP1 regulates expression of DCC and MAP1b in control of axon branching and dendrite outgrowth. Mol. Cell. Neurosci. 42, 195–207. doi: 10.1016/j.mcn.2009.07.006
Leu, C., de Kovel, C. G., Zara, F., Striano, P., Pezzella, M., Robbiano, A., et al. (2012). Genome-wide linkage meta-analysis identifies susceptibility loci at 2q34 and 13q31.3 for genetic generalized epilepsies. Epilepsia 53, 308–318. doi: 10.1111/j.1528-1167.2011.03379.x
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic localization of common diseaseassociated variation in regulatory DNA. Science 337, 1190–1195. doi: 10.1126/science.1222794
Nakamura, T., Yamazaki, Y., Saiki, Y., Moriyama, M., Largaespada, D. A., Jenkins, N. A., et al. (2000). Evi9 encodes a novel zinc finger protein that physically interacts with BCL6, a known human B-cell proto-oncogene product. Mol. Cell. Biol. 20, 3178–3186. doi: 10.1128/mcb.20.9.3178-3186.2000
Pal, D. K., Pong, A. W., and Chung, W. K. (2010). Genetic evaluation and counseling for epilepsy. Nat. Rev. Neurol. 6, 445–453. doi: 10.1038/nrneurol.2010.92
Pozhidaev, I. V., Paderina, D. Z., Fedorenko, O. Y., Kornetova, E. G., Semke, A. V., Loonen, A. J. M., et al. (2020). 5-Hydroxytryptamine receptors and tardive dyskinesia in schizophrenia. Front. Mol. Neurosci. 13:63. doi: 10.3389/fnmol.2020.00063
Sankaran, V. G., Menne, T. F., Xu, J., Akie, T. E., Lettre, G., Van Handel, B., et al. (2008). Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–1842. doi: 10.1126/science.1165409
Satterwhite, E., Sonoki, T., Willis, T. G., Harder, L., Nowak, R., Arriola, E. L., et al. (2001). The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood 98, 3413–3420. doi: 10.1182/blood.V98.12.3413
Soblet, J., Dimov, I., Graf von Kalckreuth, C., Cano-Chervel, J., Baijot, S., Pelc, K., et al. (2018). BCL11A frameshift mutation associated with dyspraxia and hypotonia affecting the fine, gross, oral, and speech motor systems. Am. J. Med. Genet. A 176, 201–208. doi: 10.1002/ajmg.a.38479
Steffens, M., Leu, C., Ruppert, A. K., Zara, F., Striano, P., Robbiano, A., et al. (2012). Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum. Mol. Genet. 21, 5359–5372. doi: 10.1093/hmg/dds373
Su, G., Morris, J. H., Demchak, B., and Bader, G. D. (2014). Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics 47, 1–24. doi: 10.1002/0471250953.bi0813s47
Tawfeek, G. A., and Alhassanin, S. (2018). HLA-G Gene Polymorphism in Egyptian Patients with Non-Hodgkin Lymphoma and its Clinical Outcome. Immunol. Invest. 47, 315–325. doi: 10.1080/08820139.2018.1430826
Wang, J., Lin, Z. J., Liu, L., Xu, H. Q., Shi, Y. W., Yi, Y. H., et al. (2017). Epilepsy-associated genes. Seizure 44, 11–20. doi: 10.1016/j.seizure.2016.11.030
Wang, Y., Zhang, S., Li, F., Zhou, Y., Zhang, Y., Wang, Z., et al. (2020). Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 48, D1031–D1041. doi: 10.1093/nar/gkz981
Wang, S., Zhang, X., Zhou, L., Wu, Q., and Han, Y. (2021a). Analysis of GABRG2 C588T polymorphism in genetic epilepsy and evaluation of GABRG2 in drug treatment. Clin. Transl. Sci. 1–9. doi: 10.1111/cts.12997
Wang, S., Zhou, L., He, C., Wang, D., Cai, X., Yu, Y., et al. (2021b). The association between STX1B polymorphisms and treatment response in patients with epilepsy. Front. Pharmacol. 12:701575. doi: 10.3389/fphar.2021.701575
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. doi: 10.1093/nar/gkx1037
Keywords: BCL11A, polymorphism, epilepsy, gene, drug resistance
Citation: Wang S, Cai X, Liu S, Zhou Q, Wang T, Du S, Wang D, Yang F, Wu Q and Han Y (2022) A novel BCL11A polymorphism influences gene expression, therapeutic response and epilepsy risk: A multicenter study. Front. Mol. Neurosci. 15:1010101. doi: 10.3389/fnmol.2022.1010101
Received: 02 August 2022; Accepted: 18 November 2022;
Published: 09 December 2022.
Edited by:
Shaunik Sharma, The University of Iowa, United StatesReviewed by:
Ana Fortuna, University of Coimbra, PortugalCopyright © 2022 Wang, Cai, Liu, Zhou, Wang, Du, Wang, Yang, Wu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbing Han, eW5oeWJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.