
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci., 03 January 2022
Sec. Molecular Signalling and Pathways
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.801882
This article is part of the Research TopicMolecular Mechanisms of Glutamatergic Synapse Function and DysfunctionView all 14 articles
Tetraspanins (Tspans) comprise a membrane protein family structurally defined by four transmembrane domains and intracellular N and C termini that is found in almost all cell types and tissues of eukaryotes. Moreover, they are involved in a bewildering multitude of diverse biological processes such as cell adhesion, motility, protein trafficking, signaling, proliferation, and regulation of the immune system. Beside their physiological roles, they are linked to many pathophysiological phenomena, including tumor progression regulation, HIV-1 replication, diabetes, and hepatitis. Tetraspanins are involved in the formation of extensive protein networks, through interactions not only with themselves but also with numerous other specific proteins, including regulatory proteins in the central nervous system (CNS). Interestingly, recent studies showed that Tspan7 impacts dendritic spine formation, glutamatergic synaptic transmission and plasticity, and that Tspan6 is correlated with epilepsy and intellectual disability (formerly known as mental retardation), highlighting the importance of particular tetraspanins and their involvement in critical processes in the CNS. In this review, we summarize the current knowledge of tetraspanin functions in the brain, with a particular focus on their impact on glutamatergic neurotransmission. In addition, we compare available resolved structures of tetraspanin family members to those of auxiliary proteins of glutamate receptors that are known for their modulatory effects.
A large family of abundantly expressed transmembrane proteins, found in all multicellular eukaryotes and comprising 33 members in humans, create an important protein network involved in a wide range of cellular processes such as cell proliferation, adhesion, signaling, fusion, and migration (Boucheix and Rubinstein, 2001). Back in 1988, a melanoma-associated antigen (ME491) was identified to consist of four transmembrane domains (TMs) (Hotta et al., 1988). Two years later its sequence homology to the Sm23 antigen found in the parasitic trematode Schistosoma mansoni was recognized (Wright et al., 1990). Further investigations revealed that the proteins CD37 (Classon et al., 1989) and TAPA-1 (target of the antiproliferative antibody 1, later denoted as CD81) (Oren et al., 1990) also share structural similarity. For classification purposes, the term “tetraspanins” was proposed for all members of the transmembrane four superfamily (TM4SF) (Maecker et al., 1997) (see Table 1 for overview of tetraspanin nomenclature). These proteins are believed to function through their shared exceptional ability to interact with each other and numerous partner proteins to create a dynamic network of interactions known as the “tetraspanin
web” or “tetraspanin-enriched microdomains” (TEMs) on the cell surface (Hemler, 2003; Charrin et al., 2009; Dornier et al., 2012). Consequently, tetraspanins are also regarded as molecular organizers (Boucheix and Rubinstein, 2001). For example, the interaction of tetraspanins with integrins has been well documented (Schmidt et al., 1996; Bassani and Cingolani, 2012). Integrins are cell adhesion molecules consisting of an α- and a β-subunit and can mediate both cell–cell and cell–extracellular matrix interactions (Hynes, 2002; Bassani and Cingolani, 2012). In this contect, tetraspanins, in the form of TEMs, are believed to exert a scaffolding function and organize proteins spatially and temporally in biological membranes (Perot and Ménager, 2020). For example, TEMs may be enriched in integrins and thus act as functional units involved in cell adhesion (Termini and Gillette, 2017; Perot and Ménager, 2020). With respect to the role of TEMs in the brain, interactions between Tspan28 (CD81) and Tspan29 (CD9) with integrins α3β1 and α6β1 have been shown to influence neurite growth (Schmidt et al., 1996; Stipp and Hemler, 2000; Bassani and Cingolani, 2012). Thus, these distinct units organized by tetraspanins play diverse roles in a variety of biological processes such as in viral infections, in cell–cell adhesion through interplay with the aforementioned integrins, and in cancer development and metastasis (Perot and Ménager, 2020). Viruses have been reported to preferentially enter cells via TEMs, which presumably can happen with or without direct binding to tetraspanins as receptors for virus entry (Pileri et al., 1998; Martin et al., 2005; Charrin et al., 2009; He et al., 2013; Hantak et al., 2019).
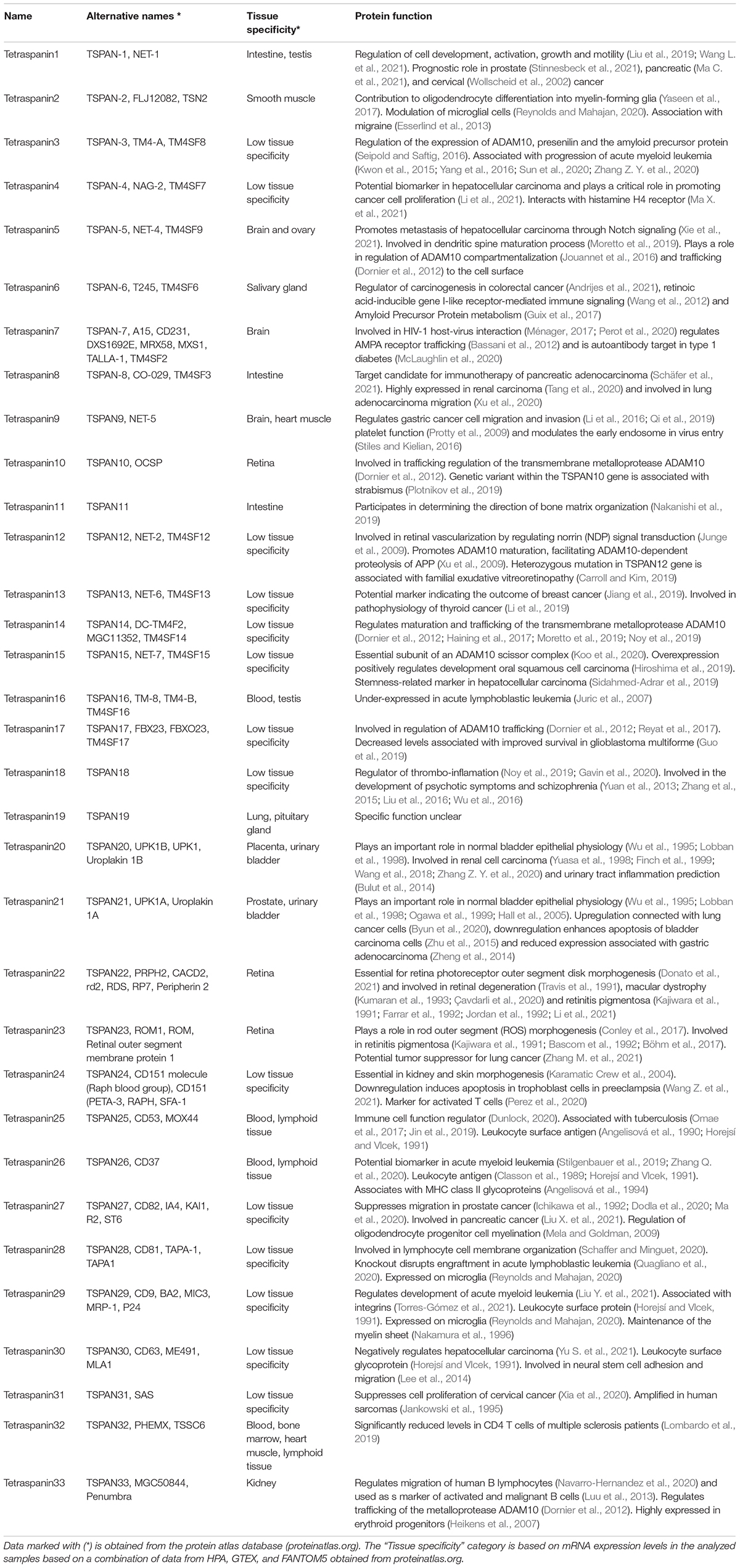
Table 1. Summary of human tetraspanins with their alternative names, tissue specificity and the functions.
Tetraspanins are small integral membrane proteins that range in size from 204 (Tspan13) to 355 (Tspan10) amino acids (Charrin et al., 2009) and have a molecular weight of 20–30 kDa (Bassani and Cingolani, 2012). Topologically, tetraspanins possess four TMs, where the first two (TM1 and TM2) are linked by a small extracellular loop (EC1) of 20–28 amino acids and the last two (TM3 and TM4) are connected by a large extracellular loop (EC2) of 76–131 amino acids (Berditchevski, 2001; Huang et al., 2005; Lazo, 2007). TM2 and TM3 are joined by a very short intracellular loop (IL) (Figure 1). In addition, tetraspanins comprise short cytosolic N- and C-termini (Huang et al., 2005; Seigneuret, 2006). Tetraspanins are distinguished from other proteins with four TMs by several distinct features. These include the unequal size of the two extracellular loops, the presence of two to four cysteine pairs, and a CCG motif (Cys-Cys-Gly sequence) in EC2 (Figure 1). In addition, over 50% of tetraspanins carry a PxxCC (Pro-x-x-Cys-Cys) motif, where x can be any amino acid (Berditchevski, 2001; Huang et al., 2005; Seigneuret, 2006). The cysteine pairs in EC2 play an essential role for the correct folding of the domain by forming disulfide bridges (Berditchevski, 2001). Furthermore, a highly conserved subfamily of tetraspanins known as TspanC8, comprising Tspan5, Tspan10, Tspan14, Tspan15, Tspan17, and Tspan33, is characterized by their unique property of possessing eight cysteines within their EC2 (Boucheix and Rubinstein, 2001; Huang et al., 2005). Most tetraspanins are potentially N-glycosylated in EC2 (Maecker et al., 1997). Tetraspanins known to be un-glycosylated are Tspan28 (CD81) and Tspan12 (Net-2), while Tspan29 (CD9) contains an N-glycosylation site in EC1 (Boucheix and Rubinstein, 2001). Presumably, most tetraspanins are also S-acylated. For example, palmitoylation has been demonstrated in three independent studies for each tetraspanin tested (Seehafer et al., 1988; Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002; Stipp et al., 2003). In addition, ubiquitination has already been demonstrated for Tspan6 (Wang et al., 2012), Tspan24 (CD151), Tspan27 (CD82) (Tsai et al., 2007), and Tspan28 (CD81) (Lineberry et al., 2008; Charrin et al., 2009; Termini and Gillette, 2017).
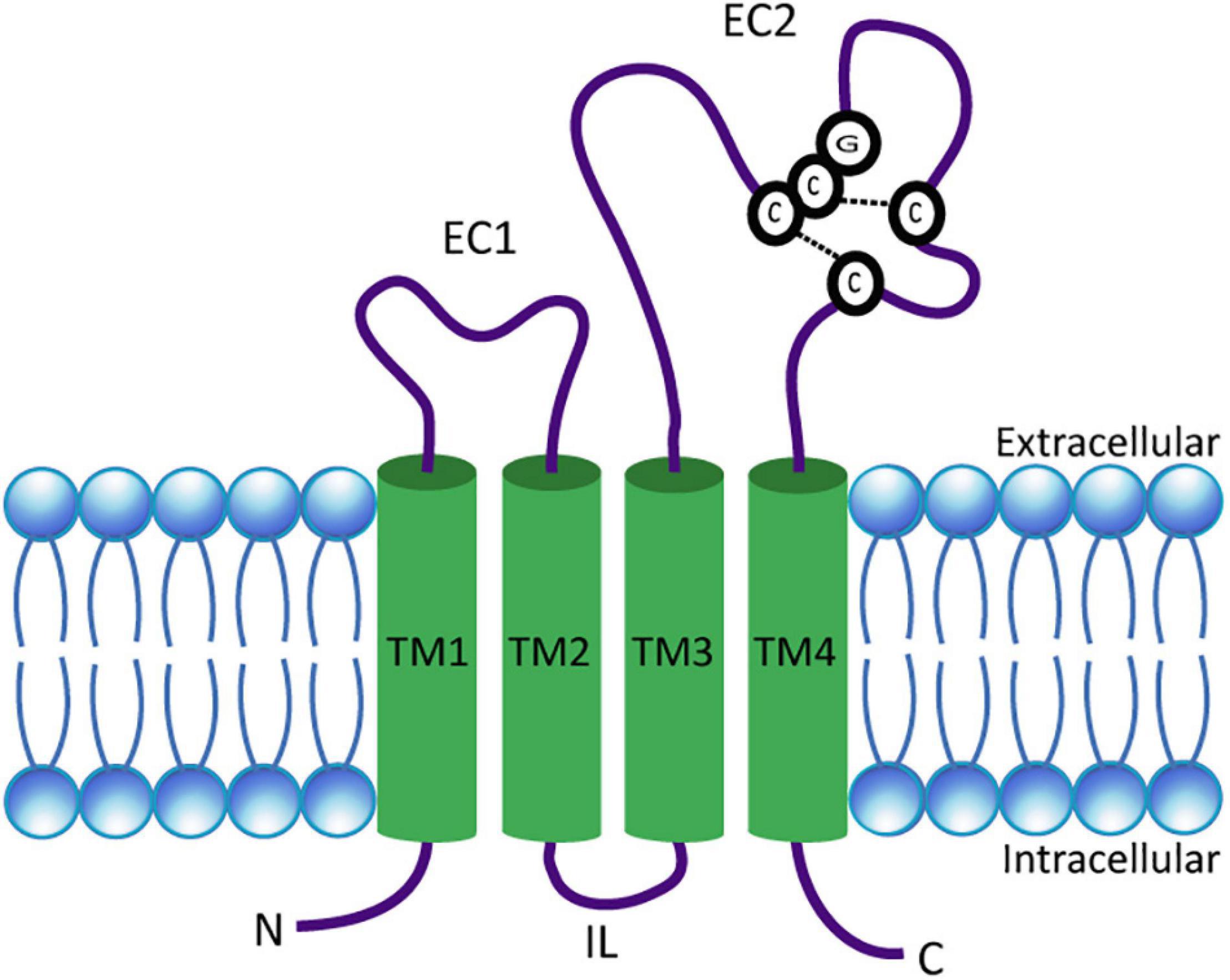
Figure 1. Schematic presentation of a tetraspanin. The model depicts the conserved structural features of tetraspanins. EC1 and EC2 represent the short and large extracellular domains, respectively. Transmembrane domains TM1–TM4 and the CCG motif along with disulfide bonds between two pairs of cysteine residues in EC2 are shown. N- and C-termini as well as the intracellular loop (IL) are located in the cytosol.
In addition to the research dedicated to uncovering all of the structural peculiarities of this protein family, a major effort has been made over the last 20 years to shed more light on the involvement of these proteins in various physiological functions in the body. Many tetraspanins are involved in the process of cell development, activation, growth regulation, and motility. It has been shown that Tspan1 significantly reduces cell migration, tissue invasion, and increases apoptosis of human pancreatic cancer cells (Tian et al., 2018). Furthermore, the expression of Tspan3, Tspan4, and Tspan7 in cells with high migratory potential suggests a role in the regulation of migration processes (Kashef et al., 2013). Down-regulation of Tspan8 inhibits the proliferation and migration of colorectal cancer cells, while over-expression promotes the opposite effects (Zhang et al., 2019). Overexpression of Tspan9 significantly inhibits the proliferation, migration, and invasion of human gastric cancer cells (Li et al., 2016). Direction of bone matrix organization orthogonal to osteoblast alignment is controlled by Tspan11-mediated focal adhesion assembly (Nakanishi et al., 2019).
Another context in which tetraspanins are involved, is the immune system and regulation of inflammatory pathways. Tspan18 knockout mice have reduced thrombus size in a deep vein thrombosis model, and reduced platelet deposition in the microcirculation following myocardial ischemia-reperfusion injury (Noy et al., 2019). Tspan26 is a B-cell surface antigen widely expressed on mature B cells, and it is involved in immune regulation and tumor suppression (Xu-Monette et al., 2016). Tspan28 (CD81) knockout cells show impaired viral DNA replication and produce greatly diminished viral titers in Herpes simplex virus 1 infection in a neuroblastoma cell model (Benayas et al., 2020).
Since Tspan28, Tspan29, and Tspan30 are abundantly expressed on extracellular vesicles categorized as exosomes, they are also used as exosomal markers (Lai et al., 2010; Oksvold et al., 2015). Tspan31 is a critical regulator of transduction of survival and apoptotic signals in hepatocellular carcinoma cells (Wang et al., 2018). Two tetraspanins (Tspan20 and Tspan21), better known as uroplakins (UPKs), are involved in normal bladder epithelial physiology. Tspan20, also known as uroplakin 1B (UPK1B), promotes the invasion and metastasis of bladder cancer via regulation of the Wnt/β-catenin pathway (Wang et al., 2018; Zhang Z. Y. et al., 2020), while the reduced expression of Tspan21 (UPK1A) might play a role in the progression of gastric cancer (Zheng et al., 2014).
The standardized tetraspanin nomenclature (Charrin et al., 2009) utilizes a simple numbering system from Tspan1 to Tspan33. However, in certain research fields other names for some of the tetraspanin proteins are more frequently used. A summary of all human tetraspanins including their alternative names, tissue-specific RNA expression, and their reported involvement in different physiological or pathophysiological processes is listed in Table 1.
One of the first evidences of tetraspanins playing an important role in the CNS came from research on the Drosophila melanogaster tetraspanin late bloomer (lbl) in the middle of the 1990s. It was observed that in fly mutants lacking this protein, adjacent motoneurons showed increased ectopic sprouting, and synapse formation was delayed. These results, which indicate that lbl promotes the formation of motor neuron synapses in the fruit fly, suggest that other tetraspanins may also be involved in this process and that a similar mechanism may also be found in mammals (Kopczynski et al., 1996). Furthermore, RNA in situ analyses of all tetraspanin family genes found in Drosophila revealed that most tetraspanins are either specifically expressed in the nervous system or intestines or have ubiquitously low levels of expression. In addition, other studies suggested that the tetraspanins are not critical for neurite growth but that their absence delays the formation of the final synaptic contact points of motor neuron axons. Moreover, the tetraspanins expressed by motor neurons showed partial compensation for the role of deleted lbl (Fradkin et al., 2002).
Following these studies in Drosophila, numerous research efforts in the following years focused on demonstrating the importance and involvement of tetraspanins in various processes in the CNS. Since the tetraspanins are referred to as “molecular organizers,” it became clear that certain tetraspanins playing an important role in regulating ADAM10 (A disintegrin and metalloproteinase 10), are involved in the Notch signaling pathway, and are essential for photoreceptor function.
Tetraspanins have been reported to regulate metalloproteases. Particularly important for the nervous system is ADAM10 also known as Kuzbanian in Drosophila, which works as “molecular scissors” and is responsible for cleaving the extracellular portion of dozens of transmembrane proteins including amyloid precursor protein (APP), notch, and cadherin (Hartmann et al., 2002; Reiss et al., 2005; Kuhn et al., 2010; Dornier et al., 2012; Groot et al., 2014; Seipold and Saftig, 2016). It has been shown that Tspan3 acts as a stabilizing factor of active ADAM10, APP, and the γ-secretase complex at the plasma membrane and Tspan12 and Tspan15 may be involved in Alzheimer’s disease physiopathology (Seipold et al., 2017). It was found that all members of the TspanC8 subfamily are directly involved in the regulation of ADAM10 exit from the ER and play an important role in its enzymatic maturation by promoting its trafficking to the cell surface (Dornier et al., 2012). TspanC8s, indeed, differentially regulate ADAM10 membrane compartmentalization through atypical association. Four TspanC8 members, in particular Tspan5, Tspan14, Tspan15, and Tspan33, increase ADAM10 expression at the cell surface (Dornier et al., 2012). Mass spectrometric analysis suggested a reduced interaction of ADAM10 with other components of the tetraspanin web in Tspan15-expressing cells. By contrast, ADAM10 association with other members of the C8 tetraspanin subfamily was increased in Tspan5-transfected cells (Jouannet et al., 2016). Recently, with the help of the first monoclonal antibodies generated against Tspan15, it was shown that ADAM10 and Tspan15 exist together at the cell surface and that ADAM10 is required for Tspan15 expression in cell lines (Koo et al., 2020). This suggests that the direct interaction of Tspan15 with ADAM10 makes it crucial for the ADAM10 scissor complex (Koo et al., 2020). In an independent study, mass spectrometric analysis revealed Tspan12 as a partner for ADAM10 that is involved in its maturation and increases ADAM10-mediated cleavage of the APP (Xu et al., 2009). Tspan15 was identified as the only member of the TspanC8 family involved in the promotion of ADAM10-dependent N-cadherin cleavage when overexpressed in human embryonic kidney cells (Noy et al., 2016). Some integrin–tetraspanin interactions facilitate maturation and cell surface expression of the integrin receptors. Since many tetraspanins contain PDZ domain-binding motifs, it is possible that some of them serve as linkers between integrins and intracellular PDZ scaffolding proteins or signal transduction molecules (Bassani and Cingolani, 2012).
Notch has been established as one of the major substrates of the ADAM10 protease (Groot et al., 2014). Several studies have suggested that Notch has a crucial role in the synaptic plasticity of the mammalian brain (Costa et al., 2003, 2005; Wang et al., 2004). Depending on the level of receptor activation, Notch might decrease or increase long-term potentiation (LTP) (Wang et al., 2004; Dahlhaus et al., 2008). Human Tspan5 and Tspan33, the orthologs of Caenorhabditis elegans TSP-12, appear to facilitate γ-secretase-mediated cleavage and thus promote Notch activity (Dunn et al., 2010). Further investigation revealed that other members of TspanC8, Tspan5, and Tspan14, also appear to be promoters of ligand-induced Notch activity in U2OS and HeLa cells, while Tspan15 has the opposite effect (Dornier et al., 2012; Jouannet et al., 2016). The latter observation explains why the knockdown of Tspan15 enhances Notch activation (Jouannet et al., 2016).
Human Tspan22, also known as peripherin 2 (PRPH2) or retinal degeneration slow (RDS), is located in the retina and abundantly expressed in the outer segment (OS) of photoreceptors (Molday and Molday, 1987; Milstein et al., 2020). PRPH2 is thought to be essential for the vision and morphogenesis of the OS, which consists of hundreds of disc membranes mainly responsible for the conversion of light signals into electrical signals. Membranous discs in OS undergo constant renewal (Young, 1967). In this regard, PRPH2 in a complex with rom-1 (rod outer segment membrane protein 1, which is another related tetraspanin) is required for the regulation of morphogenesis and structural integrity of the OS (Stuck et al., 2016; Murru et al., 2018). It was found that any gene-level mutation or defect in PRPH2 leads to a broad range of progressive retinal degeneration events in humans (Kohl et al., 1998), affecting mainly either rod or cone cell photoreceptors (Stuck et al., 2016). PRPH2-deficient mice show disrupted photoreceptor morphogenesis (Sanyal et al., 1980). In addition, PRPH2 serves as a molecular bridge between rhodopsin and the rod cyclic nucleotide-gated (CNG) channel complex in rod OS (Becirovic et al., 2014).
To point out other tetraspanins that may play important roles in the CNS, we extracted RNA expression level data in different brain regions from “The Human Protein Atlas” database (Uhlén et al., 2015) in Figure 2.
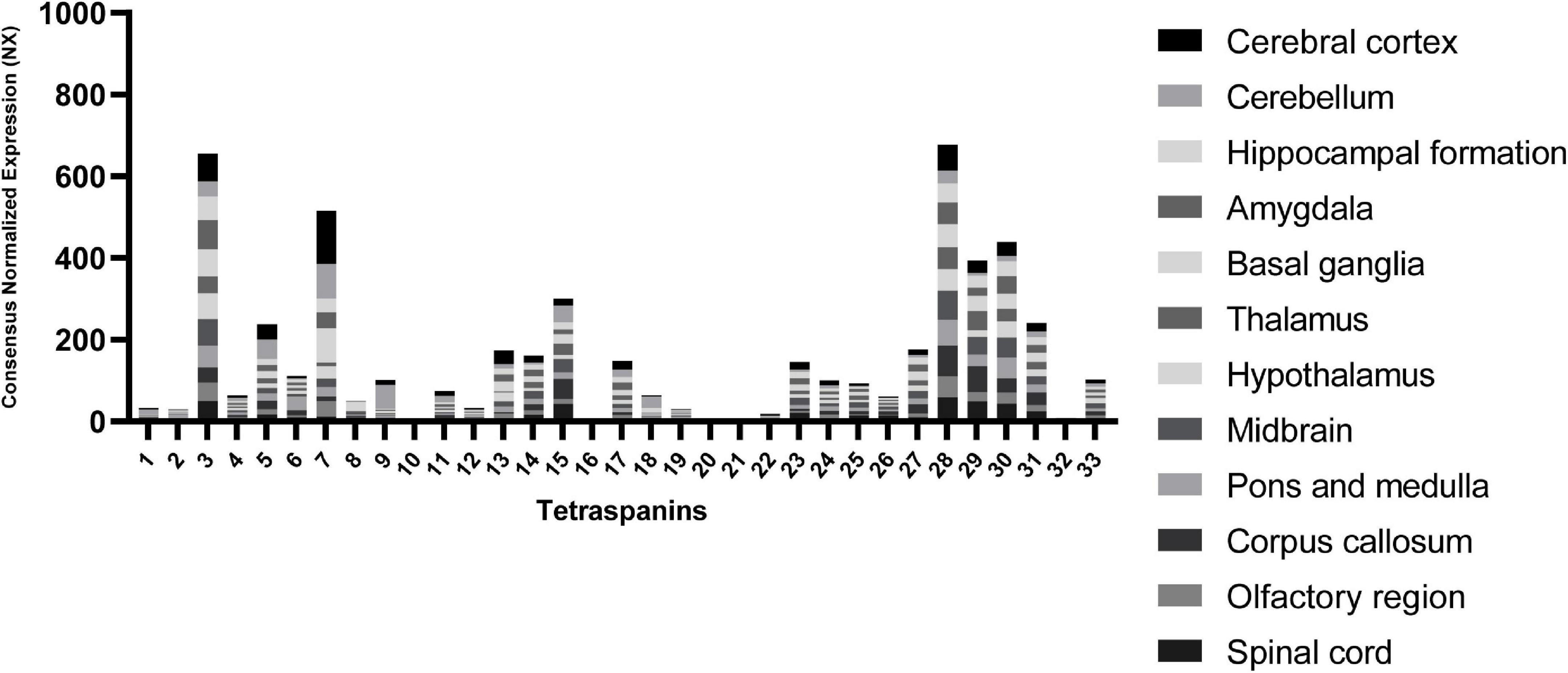
Figure 2. RNA expression levels of tetraspanins in different regions of the brain. Consensus Normalized eXpression (NX) levels shown are created by combining the data from three transcriptomics datasets (HPA, GTEx, and FANTOM5) using normalization pipeline. Each bar contains combined data of all brain regions indicated. Data used to compose the graph obtained from the Human Protein Atlas database (proteinatlas.org).
The tetraspanin with the strongest expression in the brain tissue is Tspan7 (Table 1), which also shows the strongest localization in cerebral cortex in comparison to other regions of the brain (Figure 2). Tspan7 was characterized as a key player in the morphological and functional maturation of glutamatergic synapses (Bassani and Passafaro, 2012). Tspan3 stands out as having one of the highest RNA expression levels in the brain among all tetraspanins (Figure 2).
Interestingly, the tetraspanins Tspan28, Tspan29, and Tspan30 exhibit overall high levels of RNA expression. Tspan29 and Tspan28 are shown to be expressed on microglial cells (Reynolds and Mahajan, 2020). Tspan29, more commonly called CD9, is involved in neurite outgrowth and cell migration, and it is associated with the α6β1 integrin and the neural adhesion molecule L1 in mouse brain (Schmidt et al., 1996). Tspan29 was also detected in human adult nervous tissue, and its expression is correlated with myelination where it may interact with the extracellular matrix and participate in the maintenance of the myelin sheath (Nakamura et al., 1996). It also enhances myelin membrane adhesion to extracellular matrices at very late stages of development (Kagawa et al., 1997). Furthermore, Tspan29 plays a role in glial cells in pathological conditions such as the devastating transmissible spongiform encephalopathy (TSE) (Doh-Ura et al., 2000).
Involvement of Tspan13 in modulating the coupling between the voltage sensor activation and pore opening of voltage-gated Ca2+ channels (encoded by the caveolin genes, abbreviated as CAV) was confirmed where Tspan13 has been identified as an interaction partner of the α1 subunit of N-type CaV2.2 (Mallmann et al., 2013).
Tspan27 (CD82) is a critical molecule in the regulation of oligodendrocyte progenitor cell migration and myelination (Mela and Goldman, 2009). Downregulation of this molecule in oligodendrocytes inhibits differentiation, reduces myelin protein accumulation, and leads to regression to less mature stages (Mela and Goldman, 2013). In addition, there is evidence that the CD82-TRPM7-Numb pathway is associated with age-related synapse/memory impairments (Zhao et al., 2020). Further, TIMP-1 (tissue inhibitor of metalloproteinase-1) has been identified as a potent key regulator of Tspan30 (CD63) and β1-integrin-mediated signaling that regulates human neural stem cell adhesion and migration (Lee et al., 2014).
Ionotropic glutamate receptors (iGluRs) are ligand-gated cation selective ion channels that can be activated by the brain’s major excitatory neurotransmitter L-glutamate. The family of iGluRs can be further subdivided into kainate (KA), N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) and Delta receptors (Hollmann and Heinemann, 1994; Sobolevsky et al., 2009; Traynelis et al., 2010). AMPARs are typically located at the post-synapse and mediate the majority of rapid excitatory neurotransmission (Gasic and Hollmann, 1992). They play a critical role in mechanisms such as synaptic plasticity and are associated with various neurodegenerative and neuropsychiatric disorders (Palmer et al., 2005). The AMPA receptor (AMPAR) subfamily consists of a total of four members (GluA1–GluA4) and they are functional as homomeric and heteromeric tetramers (Sager et al., 2009). Many different auxiliary subunits have been discovered, which influence AMPAR function in the CNS with respect to trafficking and gating. The most prominent and intensively studied auxiliary proteins are the so-called transmembrane AMPAR modulatory proteins (TARPs), of which there are six members in total (γ2, γ3, γ4, γ5, γ7, and γ8) (Tomita, 2010), with γ2 being the most extensively member studied to date. For example, γ2 promotes trafficking of AMPARs to the plasma membrane and influences their biophysical functions (Tomita, 2010). Thus, γ2 is able to slow receptor deactivation and at the same time strongly reduces receptor desensitization accompanied by a faster recovery from desensitization (Priel et al., 2005; Tomita, 2010; Ben-Yaacov et al., 2017). In addition, γ2 increases agonist affinity and influences receptor gating by increasing channel conductance and opening probability (Tomita, 2010; Ben-Yaacov et al., 2017).
Research over the last 20 years has revealed many interacting proteins that play a key role in the regulation of glutamate receptor trafficking and functions, which in turn leads to stabilization and strengthening of synaptic plasticity. Major directly interacting proteins of AMPARs include GRIP1 (glutamate receptor-interacting proteins 1) (Osten et al., 2000) and PICK1 (protein interacting with C kinase 1) (Xia et al., 1999), required for either insertion of AMPARs at the synaptic surface (Osten et al., 2000) or removal from the synaptic membrane (Braithwaite et al., 2002) as well as for the regulation of spine morphology.
Tspan6, associated with the neurological condition EFMR (epilepsy female-restricted with mental retardation, now referred to as intellectual disability), has been identified as a novel regulator of hippocampal synaptic transmission and LTP, with a key role in synapse development and AMPAR trafficking (Depienne et al., 2011; Vincent et al., 2012; Salas et al., 2017). It was speculated that the increased synaptic transmission observed by some authors was caused by an increased response to glutamate (Salas et al., 2017). However, no altered expression of synaptic proteins was detected (Salas et al., 2017). In fact, the expression of postsynaptic density protein 95 (PSD-95), AMPARs, NMDARs, and metabotropic glutamate receptors (mGluRs) was unchanged in the postsynaptic membrane (Salas et al., 2017). This observation applies equally to intracellular receptors, which are receptors that are not located in the plasma membrane but in intracellular compartments of the neuron, such as the ER or endosomes, and cell-surface receptors as well as to synaptically and non-synaptically localized GluA1-containing AMPARs (Salas et al., 2017). Moreover, no change in phosphorylation could be detected for the GluA1 subunit (Salas et al., 2017). The authors combine their seemingly contradictory results of an observed increase in basal synaptic transmission in combination with reduced LTP in the hypothesis of “occluded” LTP (Salas et al., 2017). This hypothesis posits that sustained, increased basal synaptic transmission inhibits the induction of LTP because further excitatory stimulation is not possible anymore. It has also been speculated that Tspan6 alters the biophysical properties of AMPARs, possibly in a TARP-like manner (Salas et al., 2017), so a direct interaction between Tspan6 and AMPARs may be possible.
Tspan7, which is highly homologous to Tspan6, is associated with the neurological disease X-linked intellectual disability (Zemni et al., 2000; Abidi et al., 2002; Bassani et al., 2012). Mutation of the TM4SF2 gene (=Tspan7) is a cause of a severe intellectual disability and cognitive impairment. The mutation is connected with alterations in AMPAR expression levels, which cause changes in excitatory synapse structure and function (Murru et al., 2017). The ampakine CX516 has been shown to have positive effects on Tspan7 knockout mice, rescuing the intellectual disability phenotype, suggesting pharmacological modulation of AMPARs as a potential therapeutic target (Murru et al., 2017).
Tspan7 has also been found to modulate glutamatergic synaptic transmission as well as synaptic plasticity (Bassani et al., 2012; Salas et al., 2017). In particular, it was shown to promote dendritic spine formation (Bassani et al., 2012). Effectively, Tspan7 influences neuronal morphogenesis by regulating filopodia density and dendritic spine morphology (Bassani et al., 2012). This is presumably mediated by interactions with integrin β1 and/or phosphatidylinositol 4-kinase (PI4K), and ultimately results in remodeling of the actin cytoskeleton, as speculated by the authors (Bassani et al., 2012). The authors furthermore demonstrated that Tspan7 interacts with the PDZ domain of PICK1 to regulate AMPAR trafficking and hippocampal spine development in vitro (Bassani et al., 2012). It was shown that the interaction of Tspan7 with PICK1 attenuates the internalization of AMPARs, which is typically mediated by the interaction of the latter with PICK1 (Perez et al., 2001). This subsequently leads to an increased availability of AMPARs at the postsynaptic membrane and thus to an enhanced excitability of the postsynaptic neuron (Bassani et al., 2012). Because co-immunoprecipitation experiments have shown that the C-terminus of Tspan7 knocks down PICK1 and β1 integrin as well as the two AMPAR subunits GluA2 and GluA3, a macromolecular complex between the aforementioned proteins, in which Tspan7 serves as an organizer, has been hypothesized (Bassani et al., 2012). However, a direct modulation of AMPARs by Tspan7, similar to what has been proposed for Tspan6, appears equally plausible, as it could also explain the findings described in Bassani et al. (2012). In fact, like the prototypical TARP γ2, Tspan7 may preferentially interact with the GluA2 subunit, as it colocalizes with it most predominantly (Bassani et al., 2012), and hence could be responsible for the maintenance of normal synaptic plasticity. The fact that in the co-immunoprecipitation experiments Tspan7 pulled down two AMPAR subunits, namely GluA2 and GluA3, in addition to other proteins, may also indicate a direct interaction between the aforementioned proteins, although none of the proteomics studies published to date have found tetraspanins among the AMPAR-associated proteins discovered with these methods (Schwenk et al., 2012, 2014; Li et al., 2013; Chen et al., 2014; Bettler and Fakler, 2017). However, this does not necessarily mean that tetraspanins do not interact with AMPARs, as these studies rely on the specific properties of certain mild detergents, which may not, however, be suitable for maintaining the specific interactions of tetraspanins. An overview of the functions of Tspan7 in neurons described here and found by Bassani et al. (2012) is shown in Figure 3.
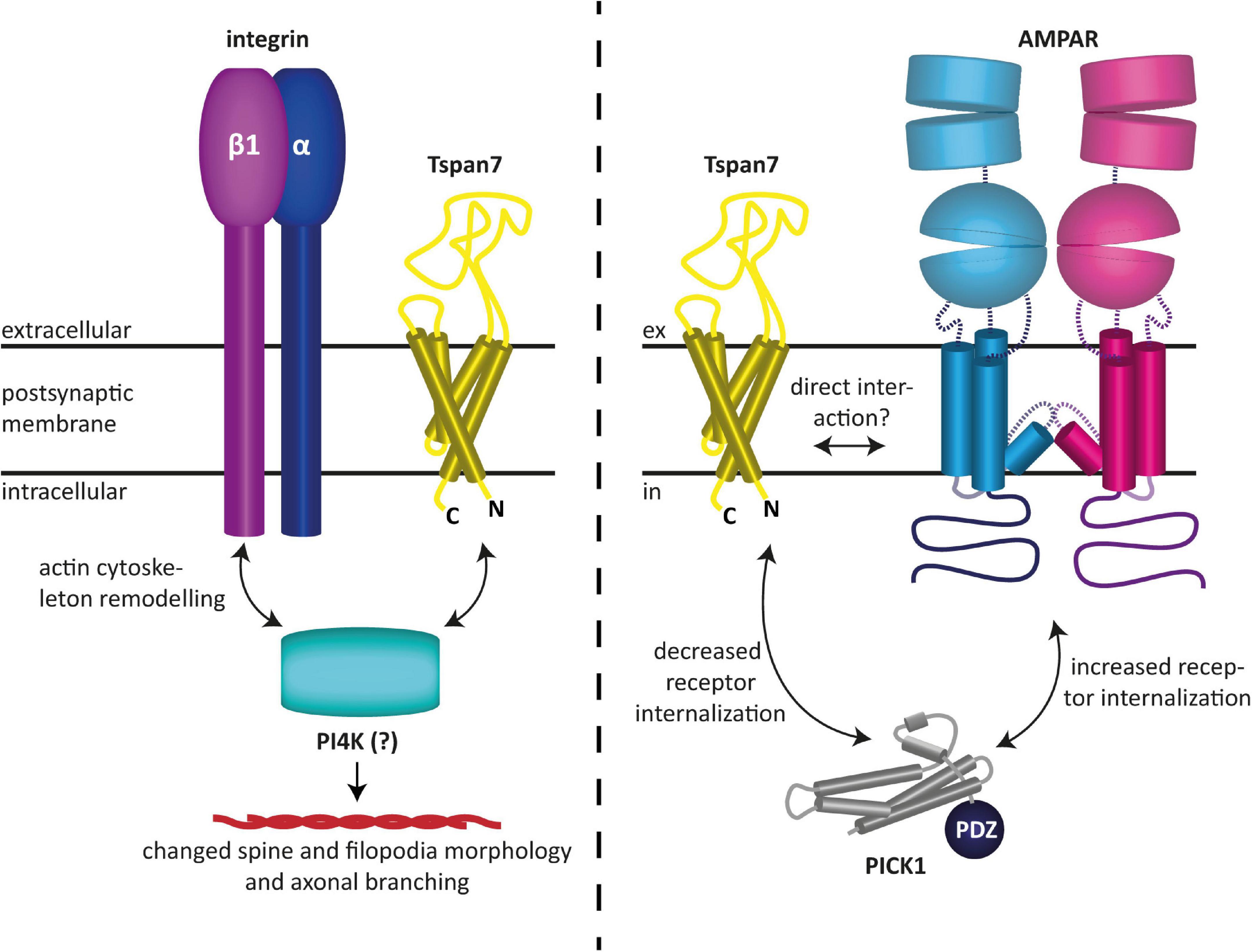
Figure 3. Reported functions of Tspan7 in neurons. (Left) Tspan7 influences neuronal morphogenesis by regulating filopodia density and dendritic spine morphology. This is presumably mediated by interactions with integrin β1 and/or PI4K and ultimately results in remodeling of the actin cytoskeleton (Bassani et al., 2012). (Right) Tspan7 regulates synaptic function at glutamatergic synapses. Interaction of Tspan7 with PICK1 attenuates the internalization of AMPARs, which is typically mediated by the interaction of the latter with PICK1 (Perez et al., 2001). This subsequently leads to an increased availability of AMPARs at the postsynaptic membrane and thus to an enhanced excitability of the postsynaptic neuron. In this process, Tspan7 interacts with PICK1 via its C-terminus (Bassani et al., 2012). The figures are based on the illustrations of Bassani and Passafaro (2012) and Perot and Ménager (2020).
From the entire tetraspanin family, only the full-length structures of Tspan28 (CD81) (Zimmerman et al., 2016; Yang et al., 2020; Susa et al., 2021), Tspan29 (CD9) (Umeda et al., 2020), and Tspan25 (CD53) (Yang et al., 2020) have been resolved to date, and the structures were found to be highly similar. Thus, the four TMs of Tspan25, Tspan28, and Tspan29 each form an ice cream cone-shaped structure with two TM pairs forming the sides of this structure (Figure 4B; Zimmerman et al., 2016; Umeda et al., 2020; Yang et al., 2020). The TM domains of all solved tetraspanins structures are virtually superimposable (Figure 4B), with Tspan25 in the open conformation, without bound lipid, adopting the same structure as Tspan28 and Tspan29 in the closed form with bound cholesterol (Yang et al., 2020). The EC2 domains of the beforementioned tetraspanins can be generally described as mushroom-like structures that have a total of five helices A to E. Here, helices A and E form the fungal stem, while helix B forms the fungal head together with the variable C–D region (Figure 4A) (Zimmerman et al., 2016; Umeda et al., 2020; Yang et al., 2020). The general mushroom shape is visible in all resolved structures, whereas slightly larger structural variations are apparent in the C–D region (Figure 4B).
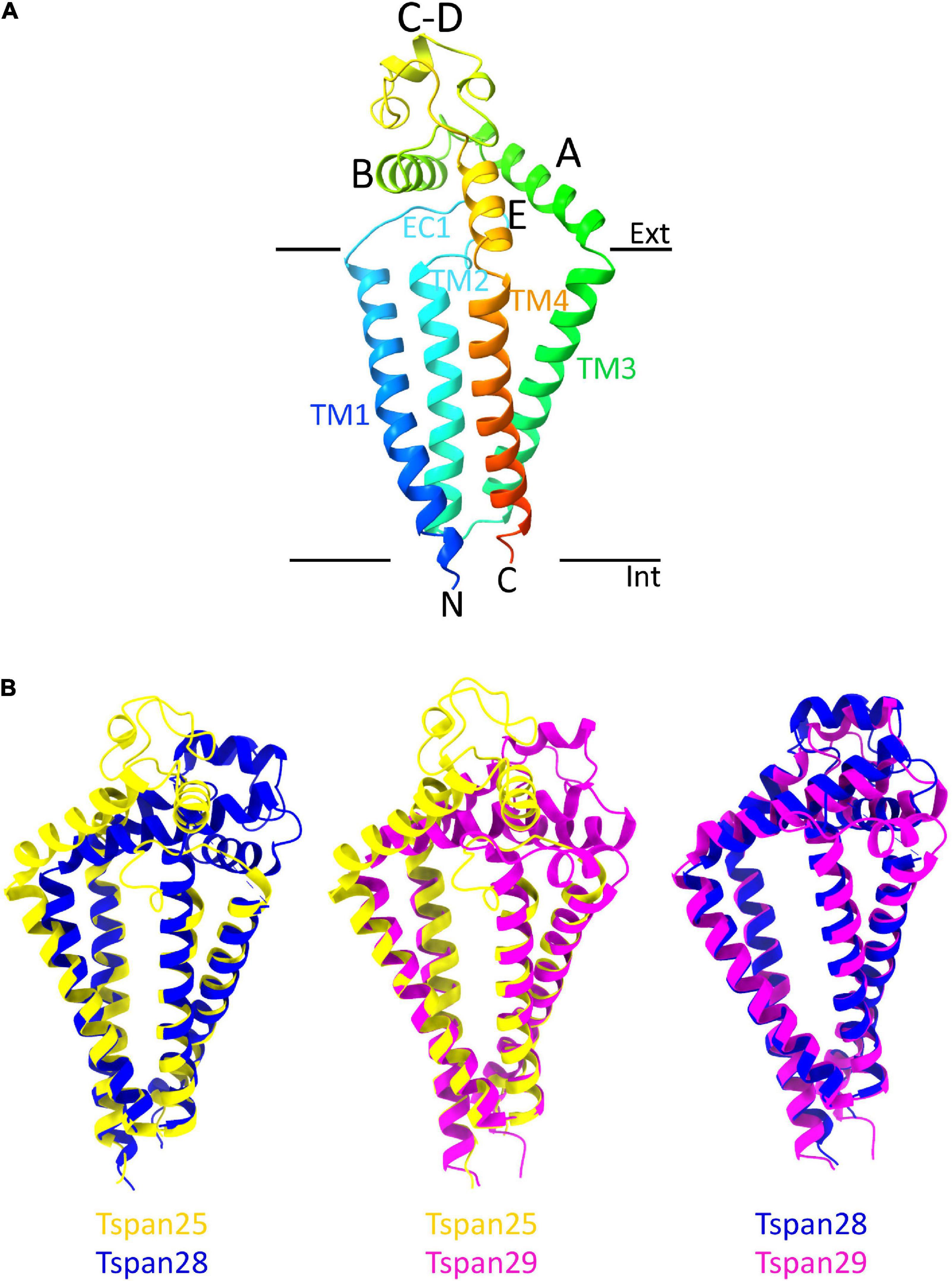
Figure 4. Overall structure of a tetraspanin (A). Helices A–E are indicated in the EC2 domain between TM3 and TM4. Helices A, B, and E represent conserved regions, while helices C–D are variable among tetraspanins. Superposition of all previously resolved structures of tetraspanins (B). Shown is the overlay of Tspan25 (pdb model 6WVG in yellow) with Tspan28 (pdb model 5TCX in blue) and of Tspan25 (yellow) with Tspan29 (pdb model 6K4J in magenta) as well as of Tspan28 (blue) with Tspan29 (magenta).
The EC2 domain is thought to be responsible for the partnering of the respective tetraspanins with other proteins, a property facilitated by its sequence and structural variability (Zimmerman et al., 2016; Umeda et al., 2020; Yang et al., 2020). The shorter EC1 domain appears disordered in most structural resolutions representing the closed state of the respective molecule, suggesting a high flexibility of this domain (Yang et al., 2020; Susa et al., 2021). Based on the recently resolved structures of the open state, in which the EC1 domain is well visible, it can be assumed that EC1 stabilizes the open conformation through interaction with EC2 (Yang et al., 2020; Susa et al., 2021).
Looking at the membrane topology, the close similarity between TARPs and tetraspanins is obvious (Figure 5A). Both proteins have four TM domains, although these adopt a more conical form in the previously resolved structures of tetraspanins compared with those of TARPs (Figure 5B). In the extracellular domain, both protein families are topologically very similar. Both feature a large extracellular loop (Ex1 in TARPs and EC2 in tetraspanins) and a small loop (Ex2 in TARPs and EC1 in tetraspanins). These loops are folded differently in the structures resolved so far. The small Ex2 domain in TARPs contains a β-sheet and a loop according to the available resolved structures, which unfortunately have been fully resolved so far only for TARP γ5 (pdb model 7RZ5) (Klykov et al., 2021). In tetraspanins, the small EC1 loop either does not contain a secondary structure element, i.e., comprises exclusively a loop, as in Tspan25 (pdb model 6WVG) (Yang et al., 2020), or EC1 contains an α-helix along with a loop, as in Tspan28 (pdb model 7JIC) (Susa et al., 2021) and Tspan29 (pdb model 6K4J) (Umeda et al., 2020). The small loops are overall shorter in tetraspanins than in TARPs and presumably serve to stabilize the larger EC2 loop (Yang et al., 2020). For TARPs, both loops are thought to have a receptor-modulating function, although there is no consensus so far, as different observations have been reported (Twomey et al., 2016; Riva et al., 2017; Herguedas et al., 2019). Remarkable differences exist in the formation of secondary structural elements. The Ex1 loop forms one α-helix and four β-strands in TARPs, whereas the large loop in the tetraspanins exclusively forms α-helical elements (Figures 5B,C). The formation of disulfide bridges in the large EC2 domain of tetraspanins is likely important for proper folding. However, TARPs also have several cysteine residues in their large Ex1 loop that form disulfide bridges (Figure 5C).
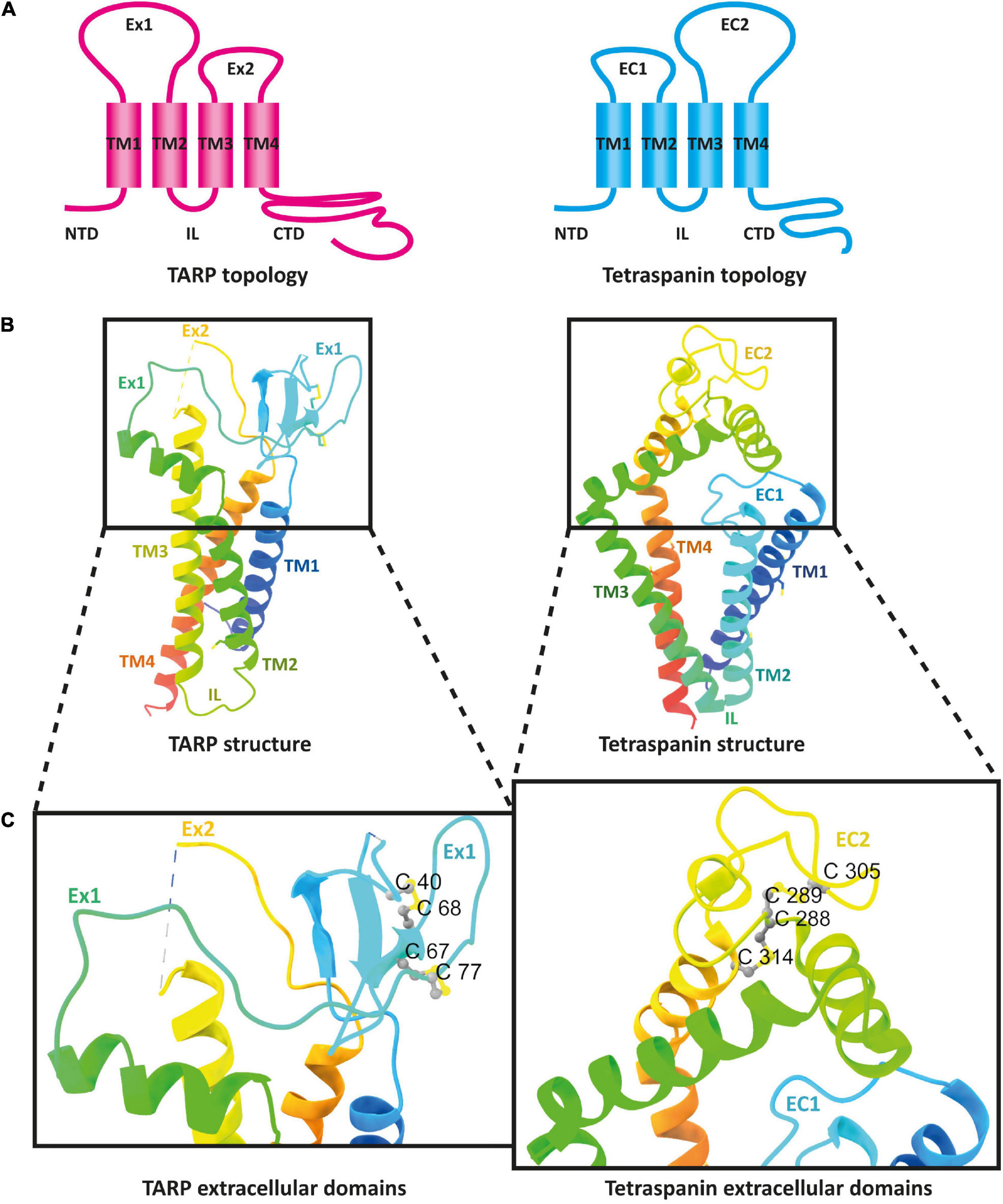
Figure 5. The architecture of transmembrane AMPAR regulatory proteins (TARPs) and tetraspanins (TSPANs). (A) Overall topology of TARPs (left) in comparison to tetraspanins (right). (B) Ribbon diagrams, rainbow-colored (from N-terminus in blue to C-terminus in red), representing the structures of TARPs (left) based on the structure of human TARP γ2, originally named stargazin and resolved by cryo-EM (pdb model 6DLZ, published by Twomey et al., 2018), and human Tspan25 (right), also known as CD53, resolved by crystallization and X-ray diffraction (pdb model 6WVG, published by Yang et al., 2020). Cysteine residues and disulfide bridges are presented as yellow heteroatoms and sticks, respectively. Distinct domains are labeled. Black boxes indicate the areas of TARPs and tetraspanins enlarged in panel (C). Cysteine residues forming disulfide bridges in the big loops are labeled (one-letter amino acid code with number of each residue) and presented in ball and stick style with sulfur and carbon shown in yellow and gray, respectively.
Unfortunately, the structures resolved to date do not include tetraspanins associated with CNS functions such as Tspan6 or Tspan7. So far, the structure of many different proteins, identified as auxiliary subunits of AMPARs, has been elucidated. These include structures of the TARPs γ2 (Twomey et al., 2016), γ5 (Klykov et al., 2021), and γ8 (Herguedas et al., 2019), GSG1L (germ cell-specific gene 1-like protein) (Chen and Gouaux, 2019), and CNIH3 (cornichon family AMPA receptor auxiliary protein 3) (Nakagawa, 2019) as well as cornichon family AMPA receptor auxiliary protein 2 (CNIH2) (Zhang D. et al., 2021). If some tetraspanins do indeed interact with or modulate the function of AMPARs, as has been postulated, it may be assumed that they are structurally similar to already known AMPAR auxiliary subunits. In Figure 6, already known structures of selected AMPAR auxiliary subunits are shown in comparison to the previously resolved structures of Tspan25, Tspan28, and Tspan29. In the absence of solved structures for Tspan6 and Tspan7, which serve essential roles in the CNS (Bassani et al., 2012; Salas et al., 2017), predicted structures of Tspan6 and Tspan7 were used which have been mapped using the predictions of DeepMind’s new artificial intelligence called AlphaFold (Jumper et al., 2021; Tunyasuvunakool et al., 2021).
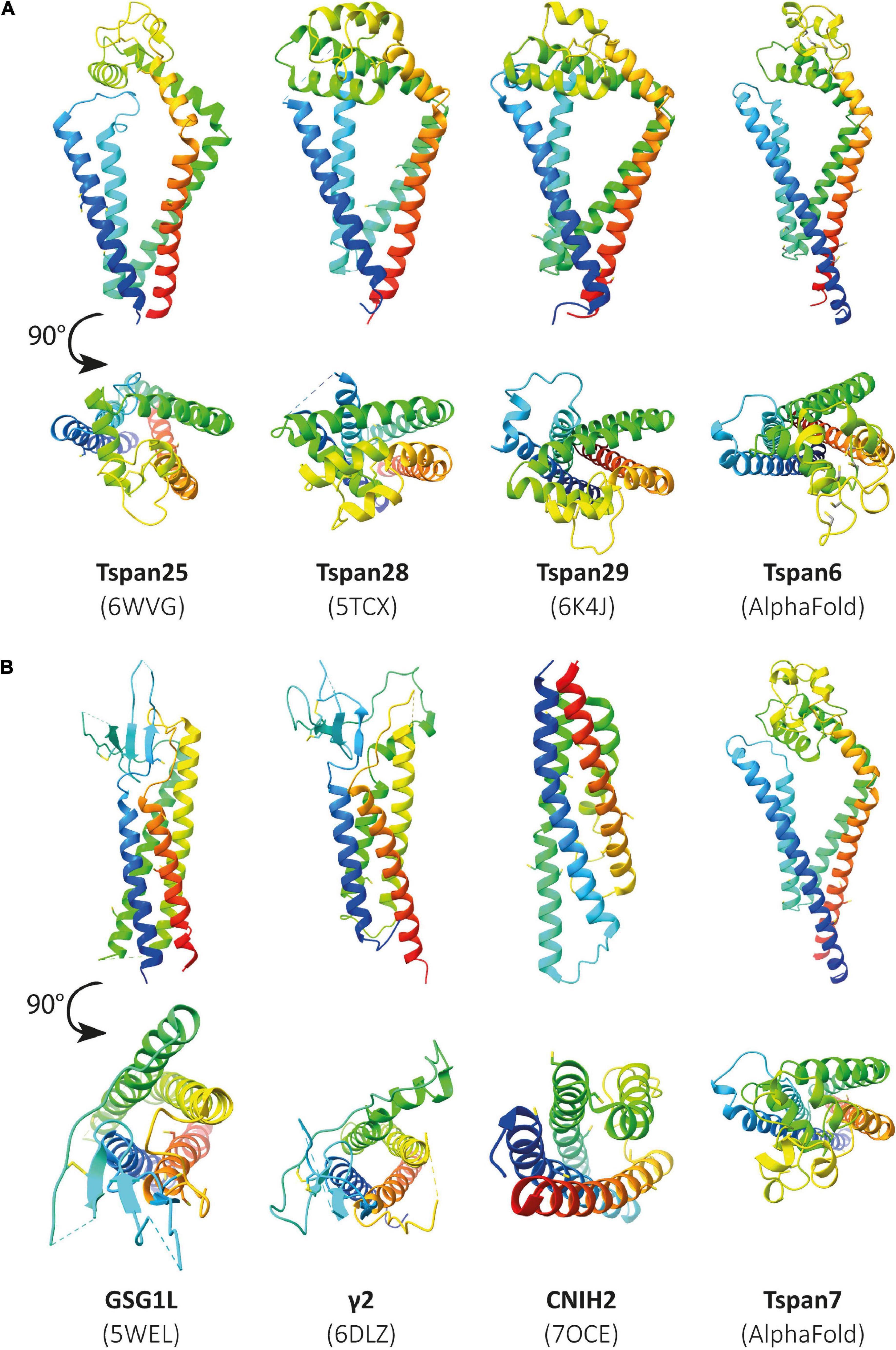
Figure 6. Illustration of previously resolved and predicted tetraspanin structures in comparison to resolved structures of known AMPAR auxiliary subunits. Ribbon diagrams rainbow-colored (from N-terminus in blue to C-terminus in red) showing the structures of the chosen proteins from Homo sapiens, displayed with different views, one parallel to the membrane (upper) and another from the extracellular side rotated by a 90° angle (lower). In panel (A) the following structures are depicted (from left to right): Tspan25 (pdb model 6WVG), Tspan28 (pdb model 5TCX), Tspan29 (pdb model 6K4J), and Tspan6 (AlphaFold database). Panel (B) shows the structures of GSG1L (pdb model 5WEL), γ2 (pdb model 6DLZ), CNIH2 (pdb model 7OCE), and Tspan7 (AlphaFold database) from left to right.
Figure 6A (three structures on the left) shows all previously known tetraspanin structures (Tspan25 or CD53, Tspan28 or CD81, and Tspan29 or CD9) in two views: parallel to the membrane, and in a birds-eye view from above the membrane. Both views are compared to three selected AMPAR auxiliary subunits (Figure 6B): GSG1L, TARP γ2, and CNIH2. GSG1L and γ2 are structurally strikingly similar, whereas CNIH2 adopts a different fold. The structures predicted by AlphaFold for Tspan6 and Tspan7 (Figures 6A,B, two structures on the right), for each of which an essential function in the CNS has already been demonstrated (Bassani et al., 2012; Murru et al., 2017; Salas et al., 2017), are shown in comparison to γ2 (Figure 6B, center). Both are predicted to have a similar conformation, which differs markedly from that of the previously resolved tetraspanin structures (see Figure 7). Tspan6 (blue) and Tspan7 (gray) are generally less conical, their first as well as last TM domains are much longer, and the large extracellular loop, while also forming α-helices, has a significantly different spatial structure by comparison. Of all the tetraspanin structures resolved so far, Tspan25 (yellow) has the most similar structure to Tspan6, whereas Tspan28 (pink) and Tspan29 (orange) show stronger structural deviations from Tspan6.
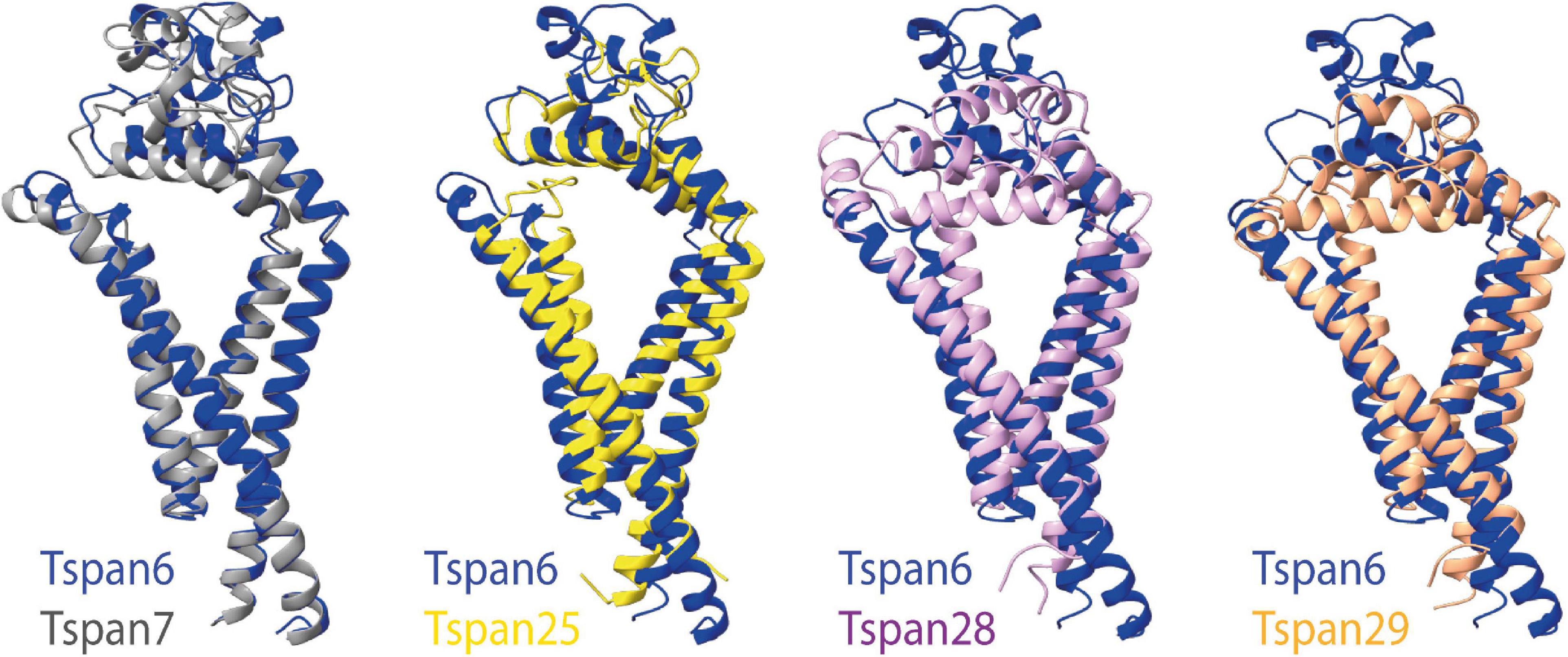
Figure 7. Superposition of selected tetraspanin structures with the predicted structure of Tspan6. The left side shows the superposition of the structures of Tspan6 (blue) and Tspan7 (gray) predicted by AlphaFold; both tetraspanins are known to influence glutamatergic synapses. For comparison, the overlays of the structure of Tspan6 with already known resolved structures of tetraspanins are shown on the right (from left to right): overlay of Tspan6 with Tspan25 (yellow) (PDB model 6WVG) as well as with Tspan28 (pale pink) (PDB model 5TCX), and with Tspan29 (orange) (PDB model 6K4J).
To compare the prediction of AlphaFold with experimentally resolved protein structures, the prediction for γ2 was superimposed on a structure of γ2 elucidated by cryo-EM (Figure 8, left). Overall, the two structures from Homo sapiens match well. As expected, the motile parts of the loops diverge slightly as they represent flexible elements. Somewhat unexpectedly, the TM4 of γ2 is predicted by AlphaFold to be much longer than was determined by experimental analysis. The predicted structure for Tspan28 (AlphaFold), on the other hand, overlaps almost perfectly with the resolved structure (Figure 8, right), confirming AlphaFold as a trustworthy, powerful tool for in silico analysis of the proteins studied here.
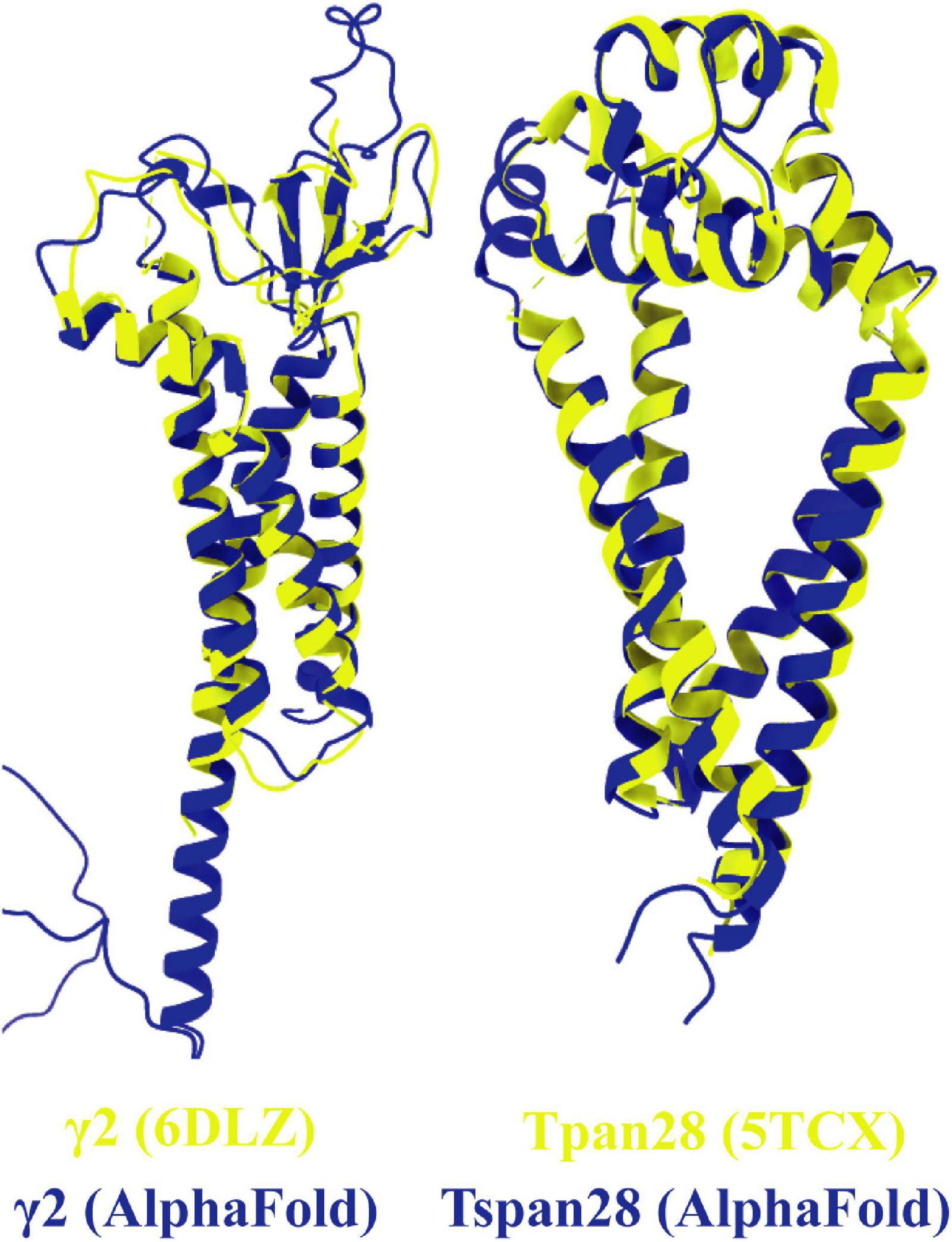
Figure 8. Overlays of predicted and resolved structures. To obtain an impression of the accuracy and reliability of the structures predicted by AlphaFold, the corresponding pdb files of the solved structures of γ2 [(left) pdb model 6DLZ, colored in yellow] and Tspan28 [(right) pdb model 5TCX, colored in yellow] were superimposed on the respective AlphaFold predictions (colored in blue).
To predict which other tetraspanins might play an important role in the glutamatergic synapse, we superimposed structures of all other tetraspanins (predicted by AlphaFold) with Tspan7 using the ChimeraX program. Out of all aligned structures, four tetraspanins (Tspan3, Tspan6, Tspan13, and Tspan31) stand out (Figure 9) because of their small Root mean square distance (RMSD) value in comparison to other tetraspanins when aligned with Tspan7 (Table 2). On the basis of the small RMSD values which indicate high structural similarity of aligned proteins (Carugo, 2007), we hypothesize that these particular tetraspanins (Tspan3, Tspan13, and Tspan31) may share functional characteristics with Tspan7, that have already been demonstrated for Tspan6 (Salas et al., 2017) and thus may play an important role in the CNS. Both direct and indirect interactions with CNS proteins are conceivable. Figure 10 shows a hypothetical direct interaction of Tspan3 and Tspan31 with the AMPAR subunit GluA2 at a glutamatergic synapse. Next to it, TARP γ8 is shown, which functions in a similar manner as γ2 with respect to its AMPAR-modulating effects. Further investigation is required to characterize their functional properties in relevance to the CNS expression pattern of these tetraspanins.
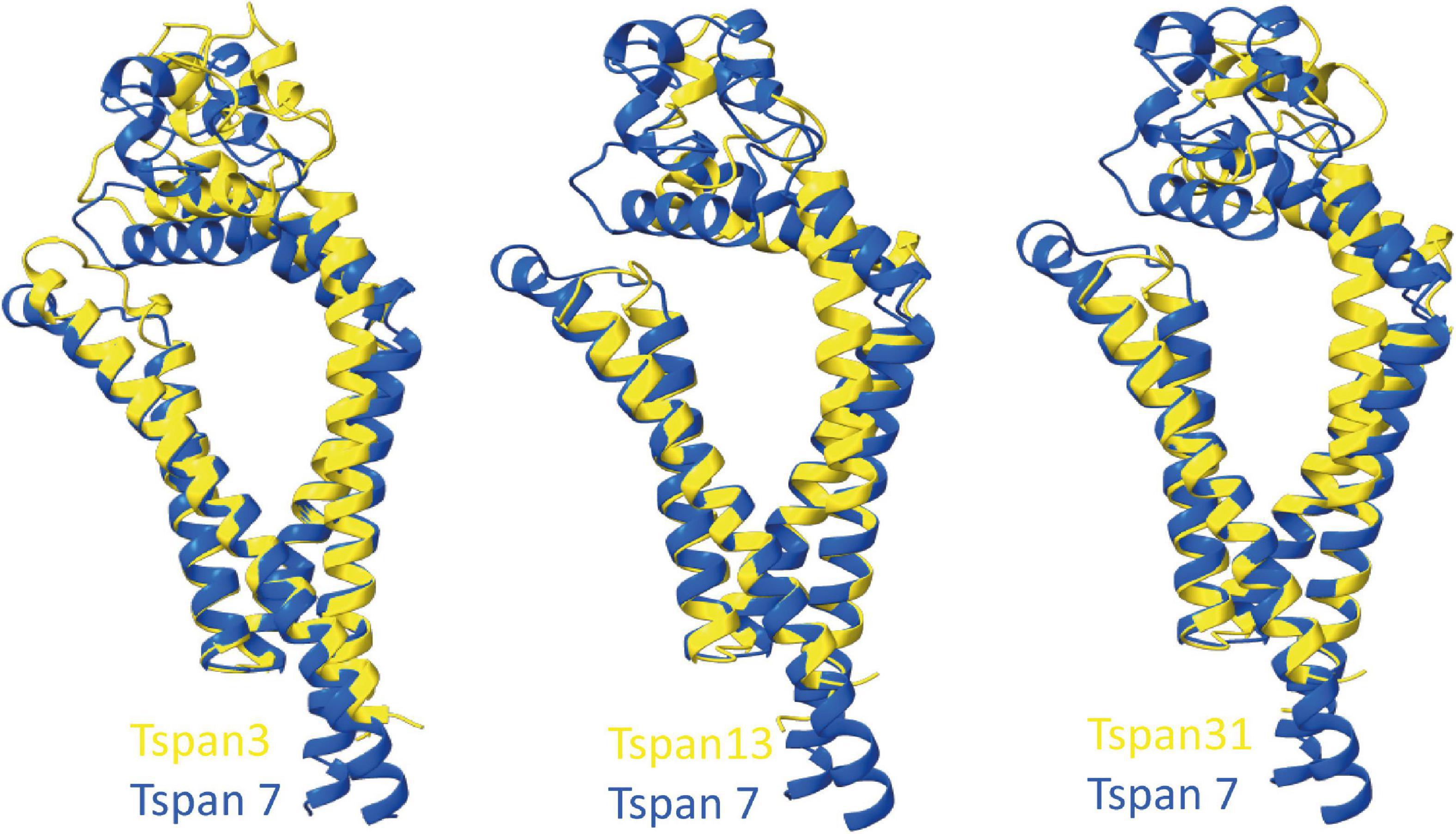
Figure 9. Superimposition of AlphaFold-predicted structures of Tspan3, Tspan13, and Tspan31 with Tspan7 using ChimeraX. Aligned structures of Tspan3, Tspan13, and Tspan31 (yellow) and Tspan7 (blue). RMSD values for Tspan3/7, Tspan13/7, and Tspan31/7 are 0.9, 0.829, and 0.854 Å, respectively.
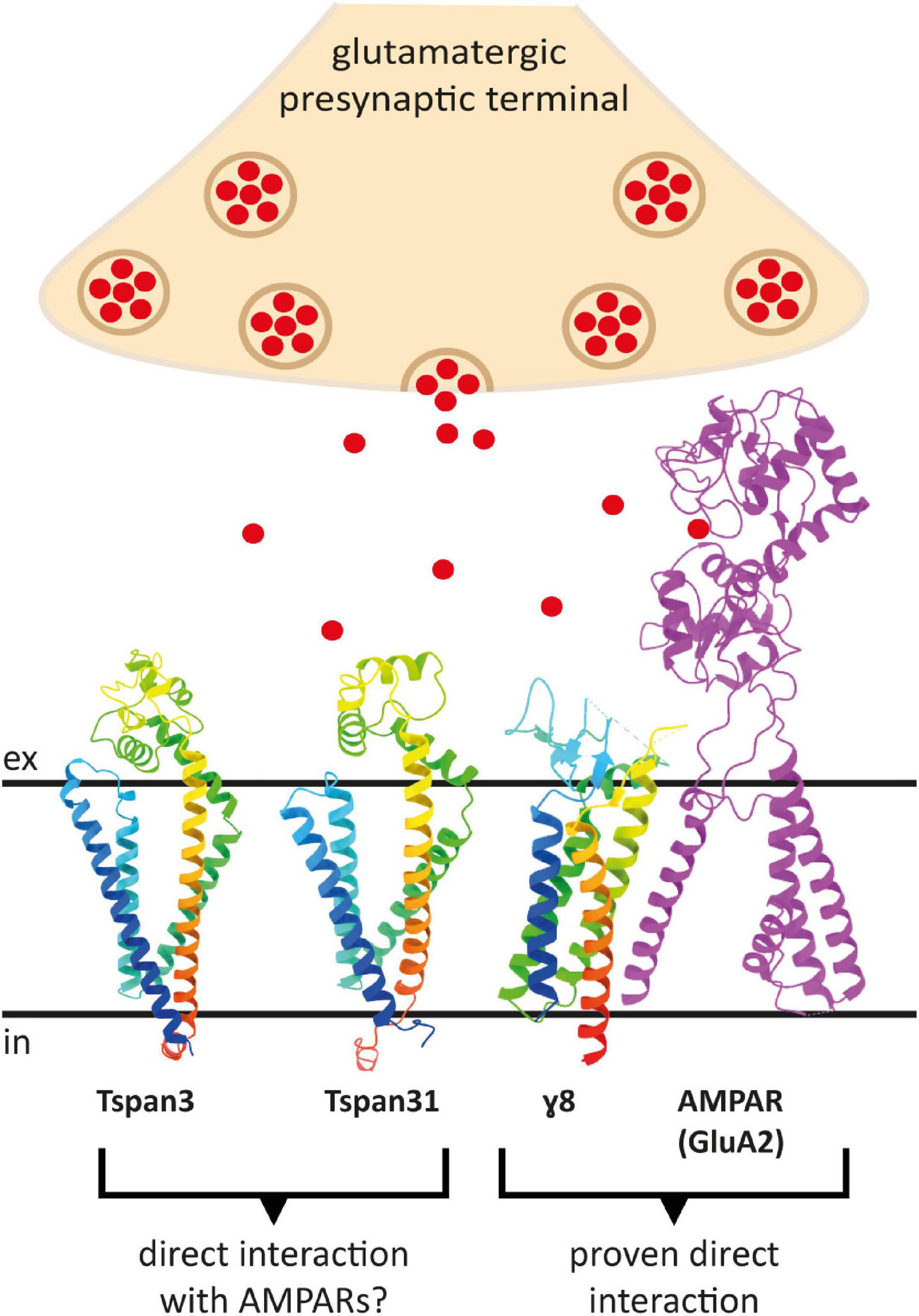
Figure 10. Putative direct interactions of tetraspanins with AMPARs at the postsynaptic membrane of a glutamatergic synapse. Shown is a GluA2 subunit in the D position in magenta, which has been proven to interact with the TARP γ8 (in rainbow colors) (right). To the left are the AlphaFold prediction models of Tspan3 and Tspan31, also colored in rainbow colors, which may also affect AMPAR function in a TARP-like manner. Red dots represent glutamate molecules. The proteins are displayed in ribbon style. The AMPAR subunit GluA2 lacks the C-terminus as well as the amino terminal domain (ATD) (PDB model: 7LEP) (Yu J. et al., 2021).
Studies in the past two decades have deepened our knowledge about the role of tetraspanins in diverse cellular processes. Although there has been considerable progress in understanding the relation between tetraspanins and its partner molecules involved in the regulation of numerous cellular functions, not much is known about structure-related functions, interaction, expression, and localization of tetraspanins in association with glutamate receptors.
Data from “The Human Protein Atlas” database indicated that Tspan3, Tspan5, Tspan6, Tspan7, Tspan15, Tspan28, Tspan29, and Tspan30 RNA is detected in functionally relevant quantities in different brain regions. According to the literature, all these tetraspanins have significant role in CNS, whereas only for Tspan6 and Tspan7 it has been proven to interact with glutamate receptors. However, since only three tetraspanin structures have been resolved so far, therefore we suggest that the structural similarity of predicted tetraspanin structures with the Tspan7 might indicate other unexplored members of tetraspanin family with the potential to contribute to the organization of the glutamatergic synapse. Moreover, beside the reported role of Tspan6 and its closest paralog Tspan7 at AMPAR-mediated synaptic transmission, three additional tetraspanins, Tspan3, Tspan13, and Tspan31 also point toward their potential involvement in important processes at the glutamatergic synapse based on their smaller RMSD value in comparison to other tetraspanins.
In addition, Tspan7 mutation and Tspan6 mRNA upregulation has been linked to intellectual disability and Alzheimer’s disease, respectively, therefore, understanding the regulation of glutamatergic synapses by tetraspanins may help to develop new targets for therapeutic interventions of several CNS diseases. Considering the abundant expression and broad spectrum of functions of tetraspanins in CNS, further investigation is required into the synaptic role of structurally related members of the tetraspanin family as well as understanding the mechanism of their actions as potential auxiliary subunits of AMPARs.
All authors contributed to the article and approved the submitted version.
This work was supported by in-house funding of the Faculty of Chemistry and Biochemistry, Ruhr University Bochum (AB, JL, and JS). We acknowledge support by the Open Access Publication Funds of the Ruhr University Bochum.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abidi, F. E., Holinski-Feder, E., Rittinger, O., Kooy, F., Lubs, H. A., Stevenson, R. E., et al. (2002). A novel 2 bp deletion in the TM4SF2 gene is associated with MRX58. J. Med. Genet. 39, 430–433. doi: 10.1136/jmg.39.6.430
Andrijes, R., Hejmadi, R. K., Pugh, M., Rajesh, S., Novitskaya, V., Ibrahim, M., et al. (2021). Tetraspanin 6 is a regulator of carcinogenesis in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 118:e2011411118. doi: 10.1073/pnas.2011411118
Angelisová, P., Hilgert, I., and Horejsí, V. (1994). Association of four antigens of the tetraspans family (CD37, CD53, TAPA-1, and R2/C33) with MHC class II glycoproteins. Immunogenetics 39, 249–256. doi: 10.1007/bf00188787
Angelisová, P., Vlcek, C., Stefanová, I., Lipoldová, M., and Horejsí, V. (1990). The human leucocyte surface antigen CD53 is a protein structurally similar to the CD37 and MRC OX-44 antigens. Immunogenetics 32, 281–285. doi: 10.1007/bf00187099
Bascom, R. A., Manara, S., Collins, L., Molday, R. S., Kalnins, V. I., and Mcinnes, R. R. (1992). Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron 8, 1171–1184. doi: 10.1016/0896-6273(92)90137-3
Bassani, S., and Cingolani, L. A. (2012). Tetraspanins: interactions and interplay with integrins. Int. J. Biochem. Cell Biol. 44, 703–708. doi: 10.1016/j.biocel.2012.01.020
Bassani, S., Cingolani, L. A., Valnegri, P., Folci, A., Zapata, J., Gianfelice, A., et al. (2012). The X-linked intellectual disability protein TSPAN7 regulates excitatory synapse development and AMPAR trafficking. Neuron 73, 1143–1158. doi: 10.1016/j.neuron.2012.01.021
Bassani, S., and Passafaro, M. (2012). TSPAN7: a new player in excitatory synapse maturation and function. Bioarchitecture 2, 95–97. doi: 10.4161/bioa.20829
Becirovic, E., Nguyen, O. N., Paparizos, C., Butz, E. S., Stern-Schneider, G., Wolfrum, U., et al. (2014). Peripherin-2 couples rhodopsin to the CNG channel in outer segments of rod photoreceptors. Hum. Mol. Genet. 23, 5989–5997. doi: 10.1093/hmg/ddu323
Benayas, B., Sastre, I., López-Martín, S., Oo, A., Kim, B., Bullido, M. J., et al. (2020). Tetraspanin CD81 regulates HSV-1 infection. Med. Microbiol. Immunol. 209, 489–498. doi: 10.1007/s00430-020-00684-0
Ben-Yaacov, A., Gillor, M., Haham, T., Parsai, A., Qneibi, M., and Stern-Bach, Y. (2017). Molecular mechanism of AMPA receptor modulation by TARP/Stargazin. Neuron 93, 1126–1137.e4. doi: 10.1016/j.neuron.2017.01.032
Berditchevski, F. (2001). Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114, 4143–4151.
Berditchevski, F., Odintsova, E., Sawada, S., and Gilbert, E. (2002). Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 277, 36991–37000. doi: 10.1074/jbc.M205265200
Bettler, B., and Fakler, B. (2017). Ionotropic AMPA-type glutamate and metabotropic GABA(B) receptors: determining cellular physiology by proteomes. Curr. Opin. Neurobiol. 45, 16–23. doi: 10.1016/j.conb.2017.02.011
Böhm, S., Riedmayr, L. M., Nguyen, O. N. P., Gießl, A., Liebscher, T., Butz, E. S., et al. (2017). Peripherin-2 and Rom-1 have opposing effects on rod outer segment targeting of retinitis pigmentosa-linked peripherin-2 mutants. Sci. Rep. 7:2321. doi: 10.1038/s41598-017-02514-5
Boucheix, C., and Rubinstein, E. (2001). Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205. doi: 10.1007/pl00000933
Braithwaite, S. P., Xia, H., and Malenka, R. C. (2002). Differential roles for NSF and GRIP/ABP in AMPA receptor cycling. Proc. Natl. Acad. Sci. U.S.A. 99, 7096–7101. doi: 10.1073/pnas.102156099
Bulut, I. K., Mir, S., Berdeli, A., and Sozeri, B. (2014). The predictive value of urinary UPIb mRNA levels in VUR and recurrent urinary tract infections. Clin. Nephrol. 81, 152–158. doi: 10.5414/cn107778
Byun, Y., Choi, Y. C., Jeong, Y., Yoon, J., and Baek, K. (2020). Long Noncoding RNA Expression Profiling Reveals Upregulation of Uroplakin 1A and Uroplakin 1A Antisense RNA 1 under Hypoxic Conditions in Lung Cancer Cells. Mol. Cells 43, 975–988. doi: 10.14348/molcells.2020.0126
Carroll, R. M., and Kim, B. J. (2019). Asymptomatic adults in a single family with familial exudative vitreoretinopathy and TSPAN12 variant. Ophthalmic Genet. 40, 474–479. doi: 10.1080/13816810.2019.1686157
Carugo, O. (2007). Statistical validation of the root-mean-square-distance, a measure of protein structural proximity. Protein Eng. Des. Sel. 20, 33–37. doi: 10.1093/protein/gzl051
Çavdarli, C., Çavdarlı, B., and Alp, M. N. (2020). Investigating the role of BEST1 and PRPH2 variants in the molecular aetiology of adult-onset vitelliform macular dystrophies. Ophthalmic Genet. 41, 585–590. doi: 10.1080/13816810.2020.1821385
Charrin, S., Le Naour, F., Silvie, O., Milhiet, P. E., Boucheix, C., and Rubinstein, E. (2009). Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154. doi: 10.1042/bj20082422
Charrin, S., Manié, S., Oualid, M., Billard, M., Boucheix, C., and Rubinstein, E. (2002). Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516, 139–144. doi: 10.1016/s0014-5793(02)02522-x
Chen, N., Pandya, N. J., Koopmans, F., Castelo-Székelv, V., Van Der Schors, R. C., Smit, A. B., et al. (2014). Interaction proteomics reveals brain region-specific AMPA receptor complexes. J. Proteome Res. 13, 5695–5706. doi: 10.1021/pr500697b
Chen, S., and Gouaux, E. (2019). Structure and mechanism of AMPA receptor - auxiliary protein complexes. Curr. Opin. Struct. Biol. 54, 104–111. doi: 10.1016/j.sbi.2019.01.011
Classon, B. J., Williams, A. F., Willis, A. C., Seed, B., and Stamenkovic, I. (1989). The primary structure of the human leukocyte antigen CD37, a species homologue of the rat MRC OX-44 antigen. J. Exp. Med. 169, 1497–1502. doi: 10.1084/jem.169.4.1497
Conley, S. M., Stuck, M. W., Watson, J. N., and Naash, M. I. (2017). Rom1 converts Y141C-Prph2-associated pattern dystrophy to retinitis pigmentosa. Hum. Mol. Genet. 26, 509–518. doi: 10.1093/hmg/ddw408
Costa, R. M., Drew, C., and Silva, A. J. (2005). Notch to remember. Trends Neurosci. 28, 429–435. doi: 10.1016/j.tins.2005.05.003
Costa, R. M., Honjo, T., and Silva, A. J. (2003). Learning and memory deficits in Notch mutant mice. Curr. Biol. 13, 1348–1354. doi: 10.1016/s0960-9822(03)00492-5
Dahlhaus, M., Hermans, J. M., Van Woerden, L. H., Saiepour, M. H., Nakazawa, K., Mansvelder, H. D., et al. (2008). Notch1 signaling in pyramidal neurons regulates synaptic connectivity and experience-dependent modifications of acuity in the visual cortex. J. Neurosci. 28, 10794–10802. doi: 10.1523/jneurosci.1348-08.2008
Depienne, C., Trouillard, O., Bouteiller, D., Gourfinkel-An, I., Poirier, K., Rivier, F., et al. (2011). Mutations and deletions in PCDH19 account for various familial or isolated epilepsies in females. Hum. Mutat. 32, E1959–E1975. doi: 10.1002/humu.21373
Dodla, P., Bhoopalan, V., Khoo, S. K., Miranti, C., and Sridhar, S. (2020). Gene expression analysis of human prostate cell lines with and without tumor metastasis suppressor CD82. BMC Cancer 20:1211. doi: 10.1186/s12885-020-07675-7
Doh-Ura, K., Mekada, E., Ogomori, K., and Iwaki, T. (2000). Enhanced CD9 expression in the mouse and human brains infected with transmissible spongiform encephalopathies. J. Neuropathol. Exp. Neurol. 59, 774–785. doi: 10.1093/jnen/59.9.774
Donato, L., Abdalla, E. M., Scimone, C., Alibrandi, S., Rinaldi, C., Nabil, K. M., et al. (2021). Impairments of photoreceptor outer segments renewal and phototransduction due to a peripherin rare haplotype variant: insights from molecular modeling. Int. J. Mol. Sci. 22:3484. doi: 10.3390/ijms22073484
Dornier, E., Coumailleau, F., Ottavi, J. F., Moretti, J., Boucheix, C., Mauduit, P., et al. (2012). TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 199, 481–496. doi: 10.1083/jcb.201201133
Dunlock, V. E. (2020). Tetraspanin CD53: an overlooked regulator of immune cell function. Med. Microbiol. Immunol. 209, 545–552. doi: 10.1007/s00430-020-00677-z
Dunn, C. D., Sulis, M. L., Ferrando, A. A., and Greenwald, I. (2010). A conserved tetraspanin subfamily promotes Notch signaling in Caenorhabditis elegans and in human cells. Proc. Natl. Acad. Sci. U.S.A. 107, 5907–5912. doi: 10.1073/pnas.1001647107
Esserlind, A. L., Christensen, A. F., Le, H., Kirchmann, M., Hauge, A. W., Toyserkani, N. M., et al. (2013). Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur. J. Neurol. 20, 765–772. doi: 10.1111/ene.12055
Farrar, G. J., Kenna, P., Jordan, S. A., Kumar-Singh, R., Humphries, M. M., Sharp, E. M., et al. (1992). Autosomal dominant retinitis pigmentosa: a novel mutation at the peripherin/RDS locus in the original 6p-linked pedigree. Genomics 14, 805–807. doi: 10.1016/s0888-7543(05)80193-4
Finch, J. L., Miller, J., Aspinall, J. O., and Cowled, P. A. (1999). Cloning of the human uroplakin 1B cDNA and analysis of its expression in urothelial-tumor cell lines and bladder-carcinoma tissue. Int. J. Cancer 80, 533–538. doi: 10.1002/(sici)1097-0215(19990209)80:4<533::aid-ijc9>3.0.co;2-5
Fradkin, L. G., Kamphorst, J. T., Diantonio, A., Goodman, C. S., and Noordermeer, J. N. (2002). Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc. Natl. Acad. Sci. U.S.A. 99, 13663–13668. doi: 10.1073/pnas.212511099
Gasic, G. P., and Hollmann, M. (1992). Molecular neurobiology of glutamate receptors. Annu. Rev. Physiol. 54, 507–536. doi: 10.1146/annurev.ph.54.030192.002451
Gavin, R. L., Koo, C. Z., and Tomlinson, M. G. (2020). Tspan18 is a novel regulator of thrombo-inflammation. Med. Microbiol. Immunol. 209, 553–564. doi: 10.1007/s00430-020-00678-y
Groot, A. J., Habets, R., Yahyanejad, S., Hodin, C. M., Reiss, K., Saftig, P., et al. (2014). Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol. Cell. Biol. 34, 2822–2832. doi: 10.1128/mcb.00206-14
Guix, F. X., Sannerud, R., Berditchevski, F., Arranz, A. M., Horré, K., Snellinx, A., et al. (2017). Tetraspanin 6: a pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol. Neurodegener. 12:25. doi: 10.1186/s13024-017-0165-0
Guo, X. B., Zhang, X. C., Chen, P., Ma, L. M., and Shen, Z. Q. (2019). miR-378a-3p inhibits cellular proliferation and migration in glioblastoma multiforme by targeting tetraspanin 17. Oncol. Rep. 42, 1957–1971. doi: 10.3892/or.2019.7283
Haining, E. J., Matthews, A. L., Noy, P. J., Romanska, H. M., Harris, H. J., Pike, J., et al. (2017). Tetraspanin Tspan9 regulates platelet collagen receptor GPVI lateral diffusion and activation. Platelets 28, 629–642. doi: 10.1080/09537104.2016.1254175
Hall, G. D., Weeks, R. J., Olsburgh, J., Southgate, J., Knowles, M. A., Selby, P. J., et al. (2005). Transcriptional control of the human urothelial-specific gene, uroplakin Ia. Biochim. Biophys. Acta 1729, 126–134. doi: 10.1016/j.bbaexp.2005.04.004
Hantak, M. P., Qing, E., Earnest, J. T., and Gallagher, T. (2019). Tetraspanins: architects of viral entry and exit platforms. J. Virol. 93:e01429-17. doi: 10.1128/jvi.01429-17
Hartmann, D., De Strooper, B., Serneels, L., Craessaerts, K., Herreman, A., Annaert, W., et al. (2002). The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624. doi: 10.1093/hmg/11.21.2615
He, J., Sun, E., Bujny, M. V., Kim, D., Davidson, M. W., and Zhuang, X. (2013). Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog. 9:e1003701. doi: 10.1371/journal.ppat.1003701
Heikens, M. J., Cao, T. M., Morita, C., Dehart, S. L., and Tsai, S. (2007). Penumbra encodes a novel tetraspanin that is highly expressed in erythroid progenitors and promotes effective erythropoiesis. Blood 109, 3244–3252. doi: 10.1182/blood-2006-09-046672
Hemler, M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. doi: 10.1146/annurev.cellbio.19.111301.153609
Herguedas, B., Watson, J. F., Ho, H., Cais, O., García-Nafría, J., and Greger, I. H. (2019). Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Science 364:eaav9011. doi: 10.1126/science.aav9011
Hiroshima, K., Shiiba, M., Oka, N., Hayashi, F., Ishida, S., Fukushima, R., et al. (2019). Tspan15 plays a crucial role in metastasis in oral squamous cell carcinoma. Exp. Cell Res. 384:111622. doi: 10.1016/j.yexcr.2019.111622
Hollmann, M., and Heinemann, S. (1994). Cloned glutamate receptors. Annu. Rev. Neurosci. 17, 31–108. doi: 10.1146/annurev.ne.17.030194.000335
Horejsí, V., and Vlcek, C. (1991). Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. 288, 1–4. doi: 10.1016/0014-5793(91)80988-f
Hotta, H., Ross, A. H., Huebner, K., Isobe, M., Wendeborn, S., Chao, M. V., et al. (1988). Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res. 48, 2955–2962.
Huang, S., Yuan, S., Dong, M., Su, J., Yu, C., Shen, Y., et al. (2005). The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86, 674–684. doi: 10.1016/j.ygeno.2005.08.004
Hynes, R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687. doi: 10.1016/s0092-8674(02)00971-6
Ichikawa, T., Ichikawa, Y., Dong, J., Hawkins, A. L., Griffin, C. A., Isaacs, W. B., et al. (1992). Localization of metastasis suppressor gene(s) for prostatic cancer to the short arm of human chromosome 11. Cancer Res. 52, 3486–3490.
Jankowski, S. A., De Jong, P., and Meltzer, P. S. (1995). Genomic structure of SAS, a member of the transmembrane 4 superfamily amplified in human sarcomas. Genomics 25, 501–506. doi: 10.1016/0888-7543(95)80051-m
Jiang, L., Zhang, X., Geradts, J., Wei, Q., Hochwald, S., Xu, H., et al. (2019). Expression of tetraspanins NET-6 and CD151 in breast cancer as a potential tumor biomarker. Clin. Exp. Med. 19, 377–384. doi: 10.1007/s10238-019-00554-x
Jin, H. S., Cho, J. E., and Park, S. (2019). Association between CD53 genetic polymorphisms and tuberculosis cases. Genes Genomics 41, 389–395. doi: 10.1007/s13258-018-0764-3
Jordan, S. A., Farrar, G. J., Kumar-Singh, R., Kenna, P., Humphries, M. M., Allamand, V., et al. (1992). Autosomal dominant retinitis pigmentosa (adRP; RP6): cosegregation of RP6 and the peripherin-RDS locus in a late-onset family of Irish origin. Am. J. Hum. Genet. 50, 634–639.
Jouannet, S., Saint-Pol, J., Fernandez, L., Nguyen, V., Charrin, S., Boucheix, C., et al. (2016). TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci. 73, 1895–1915. doi: 10.1007/s00018-015-2111-z
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Junge, H. J., Yang, S., Burton, J. B., Paes, K., Shu, X., French, D. M., et al. (2009). TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell 139, 299–311. doi: 10.1016/j.cell.2009.07.048
Juric, D., Lacayo, N. J., Ramsey, M. C., Racevskis, J., Wiernik, P. H., Rowe, J. M., et al. (2007). Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 25, 1341–1349. doi: 10.1200/jco.2006.09.3534
Kagawa, T., Mekada, E., Shishido, Y., and Ikenaka, K. (1997). Immune system-related CD9 is expressed in mouse central nervous system myelin at a very late stage of myelination. J. Neurosci. Res. 50, 312–320. doi: 10.1002/(SICI)1097-4547(19971015)50:2<312::AID-JNR19>3.0.CO;2-9
Kajiwara, K., Hahn, L. B., Mukai, S., Travis, G. H., Berson, E. L., and Dryja, T. P. (1991). Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature 354, 480–483. doi: 10.1038/354480a0
Karamatic Crew, V., Burton, N., Kagan, A., Green, C. A., Levene, C., Flinter, F., et al. (2004). CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood 104, 2217–2223. doi: 10.1182/blood-2004-04-1512
Kashef, J., Diana, T., Oelgeschläger, M., and Nazarenko, I. (2013). Expression of the tetraspanin family members Tspan3, Tspan4, Tspan5 and Tspan7 during Xenopus laevis embryonic development. Gene Expr. Patterns 13, 1–11. doi: 10.1016/j.gep.2012.08.001
Klykov, O., Gangwar, S. P., Yelshanskaya, M. V., Yen, L., and Sobolevsky, A. I. (2021). Structure and desensitization of AMPA receptor complexes with type II TARP γ5 and GSG1L. Mol. Cell. [Epub ahead of print]. doi: 10.1016/j.molcel.2021.09.030
Kohl, S., Giddings, I., Besch, D., Apfelstedt-Sylla, E., Zrenner, E., and Wissinger, B. (1998). The role of the peripherin/RDS gene in retinal dystrophies. Acta Anat. 162, 75–84. doi: 10.1159/000046471
Koo, C. Z., Harrison, N., Noy, P. J., Szyroka, J., Matthews, A. L., Hsia, H. E., et al. (2020). The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex. J. Biol. Chem. 295, 12822–12839. doi: 10.1074/jbc.RA120.012601
Kopczynski, C. C., Davis, G. W., and Goodman, C. S. (1996). A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science 271, 1867–1870. doi: 10.1126/science.271.5257.1867
Kuhn, P. H., Wang, H., Dislich, B., Colombo, A., Zeitschel, U., Ellwart, J. W., et al. (2010). ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 29, 3020–3032. doi: 10.1038/emboj.2010.167
Kumaran, N., Pennesi, M. E., Yang, P., Trzupek, K. M., Schlechter, C., Moore, A. T., et al. (1993). “Leber congenital amaurosis/early-onset severe retinal dystrophy overview,” in GeneReviews(®), eds M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, G. Mirzaa, et al. (Seattle, WA: University of Washington).
Kwon, H. Y., Bajaj, J., Ito, T., Blevins, A., Konuma, T., Weeks, J., et al. (2015). Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell 17, 152–164. doi: 10.1016/j.stem.2015.06.006
Lai, R. C., Arslan, F., Lee, M. M., Sze, N. S., Choo, A., Chen, T. S., et al. (2010). Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 4, 214–222. doi: 10.1016/j.scr.2009.12.003
Lazo, P. A. (2007). Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 98, 1666–1677. doi: 10.1111/j.1349-7006.2007.00584.x
Lee, S. Y., Kim, J. M., Cho, S. Y., Kim, H. S., Shin, H. S., Jeon, J. Y., et al. (2014). TIMP-1 modulates chemotaxis of human neural stem cells through CD63 and integrin signalling. Biochem. J. 459, 565–576. doi: 10.1042/bj20131119
Li, K. W., Chen, N., and Smit, A. B. (2013). Interaction proteomics of the AMPA receptor: towards identification of receptor sub-complexes. Amino Acids 44, 1247–1251. doi: 10.1007/s00726-013-1461-9
Li, P., Dong, M., and Wang, Z. (2019). Downregulation of TSPAN13 by miR-369-3p inhibits cell proliferation in papillary thyroid cancer (PTC). Bosn. J. Basic Med. Sci. 19, 146–154. doi: 10.17305/bjbms.2018.2865
Li, P. Y., Lv, J., Qi, W. W., Zhao, S. F., Sun, L. B., Liu, N., et al. (2016). Tspan9 inhibits the proliferation, migration and invasion of human gastric cancer SGC7901 cells via the ERK1/2 pathway. Oncol. Rep. 36, 448–454. doi: 10.3892/or.2016.4805
Li, W. N., Du, X. J., Zhang, Y. T., Wang, L. Y., and Zhu, J. (2021). A novel mutation in the PRPH2 gene in a Chinese pedigree with retinitis pigmentosa and angle-closure glaucoma. BMC Ophthalmol. 21:302. doi: 10.1186/s12886-021-02064-5
Lineberry, N., Su, L., Soares, L., and Fathman, C. G. (2008). The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. J. Biol. Chem. 283, 28497–28505. doi: 10.1074/jbc.M805092200
Liu, G., Wang, Y., Yang, L., Zou, B., Gao, S., Song, Z., et al. (2019). Tetraspanin 1 as a mediator of fibrosis inhibits EMT process and Smad2/3 and beta-catenin pathway in human pulmonary fibrosis. J. Cell Mol. Med. 23, 3583–3596. doi: 10.1111/jcmm.14258
Liu, J., Li, M., and Su, B. (2016). GWAS-identified schizophrenia risk SNPs at TSPAN18 are highly diverged between Europeans and East Asians. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 1032–1040. doi: 10.1002/ajmg.b.32471
Liu, X., Guo, X., Li, H., Chen, J., and Han, H. (2021). Effects of KAI gene expression on ferroptosis in pancreatic cancer cells. Mol. Med. Rep. 23:163. doi: 10.3892/mmr.2020.11802
Liu, Y., Wang, G., Zhang, J., Chen, X., Xu, H., Heng, G., et al. (2021). CD9, a potential leukemia stem cell marker, regulates drug resistance and leukemia development in acute myeloid leukemia. Stem Cell Res. Ther. 12:86. doi: 10.1186/s13287-021-02155-6
Lobban, E. D., Smith, B. A., Hall, G. D., Harnden, P., Roberts, P., Selby, P. J., et al. (1998). Uroplakin gene expression by normal and neoplastic human urothelium. Am. J. Pathol. 153, 1957–1967. doi: 10.1016/s0002-9440(10)65709-4
Lombardo, S. D., Mazzon, E., Basile, M. S., Campo, G., Corsico, F., Presti, M., et al. (2019). Modulation of Tetraspanin 32 (TSPAN32) expression in T cell-mediated immune responses and in multiple sclerosis. Int. J. Mol. Sci. 20:4323. doi: 10.3390/ijms20184323
Luu, V. P., Hevezi, P., Vences-Catalan, F., Maravillas-Montero, J. L., White, C. A., Casali, P., et al. (2013). TSPAN33 is a novel marker of activated and malignant B cells. Clin. Immunol. 149, 388–399. doi: 10.1016/j.clim.2013.08.005
Ma, C., Cui, Z., Wang, Y., Zhang, L., Wen, J., Guo, H., et al. (2021). Bioinformatics analysis reveals TSPAN1 as a candidate biomarker of progression and prognosis in pancreatic cancer. Bosn. J. Basic Med. Sci. 21, 47–60. doi: 10.17305/bjbms.2020.5096
Ma, X., Verweij, E. W. E., Siderius, M., Leurs, R., and Vischer, H. F. (2021). Identification of TSPAN4 as Novel Histamine H(4) Receptor Interactor. Biomolecules 11:1127. doi: 10.3390/biom11081127
Ma, Z., Gao, Y., Liu, W., Zheng, L., Jin, B., Duan, B., et al. (2020). CD82 Suppresses ADAM17-Dependent E-cadherin cleavage and cell migration in prostate cancer. Dis. Markers 2020:8899924. doi: 10.1155/2020/8899924
Maecker, H. T., Todd, S. C., and Levy, S. (1997). The tetraspanin superfamily: molecular facilitators. FASEB J. 11, 428–442. doi: 10.1096/fasebj.11.6.9194523
Mallmann, R. T., Wilmes, T., Lichvarova, L., Bührer, A., Lohmüller, B., Castonguay, J., et al. (2013). Tetraspanin-13 modulates voltage-gated CaV2.2 Ca2+ channels. Sci. Rep. 3:1777. doi: 10.1038/srep01777
Martin, F., Roth, D. M., Jans, D. A., Pouton, C. W., Partridge, L. J., Monk, P. N., et al. (2005). Tetraspanins in viral infections: a fundamental role in viral biology? J. Virol. 79, 10839–10851. doi: 10.1128/jvi.79.17.10839-10851.2005
McLaughlin, K. A., Tombs, M. A., and Christie, M. R. (2020). Autoimmunity to tetraspanin-7 in type 1 diabetes. Med. Microbiol. Immunol. 209, 437–445. doi: 10.1007/s00430-020-00674-2
Mela, A., and Goldman, J. E. (2009). The tetraspanin KAI1/CD82 is expressed by late-lineage oligodendrocyte precursors and may function to restrict precursor migration and promote oligodendrocyte differentiation and myelination. J. Neurosci. 29, 11172–11181. doi: 10.1523/jneurosci.3075-09.2009
Mela, A., and Goldman, J. E. (2013). CD82 blocks cMet activation and overcomes hepatocyte growth factor effects on oligodendrocyte precursor differentiation. J. Neurosci. 33, 7952–7960. doi: 10.1523/jneurosci.5836-12.2013
Ménager, M. M. (2017). TSPAN7, effector of actin nucleation required for dendritic cell-mediated transfer of HIV-1 to T cells. Biochem. Soc. Trans. 45, 703–708. doi: 10.1042/bst20160439
Milstein, M. L., Cavanaugh, B. L., Roussey, N. M., Volland, S., Williams, D. S., and Goldberg, A. F. X. (2020). Multistep peripherin-2/rds self-assembly drives membrane curvature for outer segment disk architecture and photoreceptor viability. Proc. Natl. Acad. Sci. U.S.A. 117, 4400–4410. doi: 10.1073/pnas.1912513117
Molday, L. L., and Molday, R. S. (1987). Glycoproteins specific for the retinal rod outer segment plasma membrane. Biochim. Biophys. Acta 897, 335–340. doi: 10.1016/0005-2736(87)90430-5
Moretto, E., Longatti, A., Murru, L., Chamma, I., Sessa, A., Zapata, J., et al. (2019). TSPAN5 enriched microdomains provide a platform for dendritic spine maturation through neuroligin-1 clustering. Cell Rep. 29, 1130–1146.e8. doi: 10.1016/j.celrep.2019.09.051
Murru, L., Moretto, E., Martano, G., and Passafaro, M. (2018). Tetraspanins shape the synapse. Mol. Cell. Neurosci. 91, 76–81. doi: 10.1016/j.mcn.2018.04.001
Murru, L., Vezzoli, E., Longatti, A., Ponzoni, L., Falqui, A., Folci, A., et al. (2017). Pharmacological Modulation of AMPAR rescues intellectual disability-like phenotype in Tm4sf2-/y Mice. Cereb. Cortex 27, 5369–5384. doi: 10.1093/cercor/bhx221
Nakagawa, T. (2019). Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366, 1259–1263. doi: 10.1126/science.aay2783
Nakamura, Y., Iwamoto, R., and Mekada, E. (1996). Expression and distribution of CD9 in myelin of the central and peripheral nervous systems. Am. J. Pathol. 149, 575–583.
Nakanishi, Y., Matsugaki, A., Kawahara, K., Ninomiya, T., Sawada, H., and Nakano, T. (2019). Unique arrangement of bone matrix orthogonal to osteoblast alignment controlled by Tspan11-mediated focal adhesion assembly. Biomaterials 209, 103–110. doi: 10.1016/j.biomaterials.2019.04.016
Navarro-Hernandez, I. C., López-Ortega, O., Acevedo-Ochoa, E., Cervantes-Díaz, R., Romero-Ramírez, S., Sosa-Hernández, V. A., et al. (2020). Tetraspanin 33 (TSPAN33) regulates endocytosis and migration of human B lymphocytes by affecting the tension of the plasma membrane. FEBS J. 287, 3449–3471. doi: 10.1111/febs.15216
Noy, P. J., Gavin, R. L., Colombo, D., Haining, E. J., Reyat, J. S., Payne, H., et al. (2019). Tspan18 is a novel regulator of the Ca(2+) channel Orai1 and von Willebrand factor release in endothelial cells. Haematologica 104, 1892–1905. doi: 10.3324/haematol.2018.194241
Noy, P. J., Yang, J., Reyat, J. S., Matthews, A. L., Charlton, A. E., Furmston, J., et al. (2016). TspanC8 Tetraspanins and A Disintegrin and Metalloprotease 10 (ADAM10) Interact via Their Extracellular Regions: EVIDENCE FOR DISTINCT BINDING MECHANISMS FOR DIFFERENT TspanC8 PROTEINS. J. Biol. Chem. 291, 3145–3157. doi: 10.1074/jbc.M115.703058
Ogawa, K., Johansson, S. L., and Cohen, S. M. (1999). Immunohistochemical analysis of uroplakins, urothelial specific proteins, in ovarian Brenner tumors, normal tissues, and benign and neoplastic lesions of the female genital tract. Am. J. Pathol. 155, 1047–1050. doi: 10.1016/s0002-9440(10)65206-6
Oksvold, M. P., Neurauter, A., and Pedersen, K. W. (2015). Magnetic bead-based isolation of exosomes. Methods Mol. Biol. 1218, 465–481. doi: 10.1007/978-1-4939-1538-5_27
Omae, Y., Toyo-Oka, L., Yanai, H., Nedsuwan, S., Wattanapokayakit, S., Satproedprai, N., et al. (2017). Pathogen lineage-based genome-wide association study identified CD53 as susceptible locus in tuberculosis. J. Hum. Genet. 62, 1015–1022. doi: 10.1038/jhg.2017.82
Oren, R., Takahashi, S., Doss, C., Levy, R., and Levy, S. (1990). TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 10, 4007–4015. doi: 10.1128/mcb.10.8.4007-4015.1990
Osten, P., Khatri, L., Perez, J. L., Köhr, G., Giese, G., Daly, C., et al. (2000). Mutagenesis reveals a role for ABP/GRIP binding to GluR2 in synaptic surface accumulation of the AMPA receptor. Neuron 27, 313–325. doi: 10.1016/s0896-6273(00)00039-8
Palmer, C. L., Cotton, L., and Henley, J. M. (2005). The molecular pharmacology and cell biology of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol. Rev. 57, 253–277. doi: 10.1124/pr.57.2.7
Perez, J. L., Khatri, L., Chang, C., Srivastava, S., Osten, P., and Ziff, E. B. (2001). PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 21, 5417–5428. doi: 10.1523/jneurosci.21-15-05417.2001
Perez, M. D., Seu, L., Lowman, K. E., Moylan, D. C., Tidwell, C., Samuel, S., et al. (2020). The tetraspanin CD151 marks a unique population of activated human T cells. Sci. Rep. 10:15748. doi: 10.1038/s41598-020-72719-8
Perot, B. P., García-Paredes, V., Luka, M., and Ménager, M. M. (2020). Dendritic cell maturation regulates TSPAN7 function in HIV-1 transfer to CD4(+) T lymphocytes. Front. Cell. Infect. Microbiol. 10:70. doi: 10.3389/fcimb.2020.00070
Perot, B. P., and Ménager, M. M. (2020). Tetraspanin 7 and its closest paralog tetraspanin 6: membrane organizers with key functions in brain development, viral infection, innate immunity, diabetes and cancer. Med. Microbiol. Immunol. 209, 427–436. doi: 10.1007/s00430-020-00681-3
Pileri, P., Uematsu, Y., Campagnoli, S., Galli, G., Falugi, F., Petracca, R., et al. (1998). Binding of hepatitis C virus to CD81. Science 282, 938–941. doi: 10.1126/science.282.5390.938
Plotnikov, D., Shah, R. L., Rodrigues, J. N., Cumberland, P. M., Rahi, J. S., Hysi, P. G., et al. (2019). A commonly occurring genetic variant within the NPLOC4-TSPAN10-PDE6G gene cluster is associated with the risk of strabismus. Hum. Genet. 138, 723–737. doi: 10.1007/s00439-019-02022-8
Priel, A., Kolleker, A., Ayalon, G., Gillor, M., Osten, P., and Stern-Bach, Y. (2005). Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J. Neurosci. 25, 2682–2686. doi: 10.1523/jneurosci.4834-04.2005
Protty, M. B., Watkins, N. A., Colombo, D., Thomas, S. G., Heath, V. L., Herbert, J. M., et al. (2009). Identification of Tspan9 as a novel platelet tetraspanin and the collagen receptor GPVI as a component of tetraspanin microdomains. Biochem. J. 417, 391–400. doi: 10.1042/bj20081126
Qi, Y., Lv, J., Liu, S., Sun, L., Wang, Y., Li, H., et al. (2019). TSPAN9 and EMILIN1 synergistically inhibit the migration and invasion of gastric cancer cells by increasing TSPAN9 expression. BMC Cancer 19:630. doi: 10.1186/s12885-019-5810-2
Quagliano, A., Gopalakrishnapillai, A., Kolb, E. A., and Barwe, S. P. (2020). CD81 knockout promotes chemosensitivity and disrupts in vivo homing and engraftment in acute lymphoblastic leukemia. Blood Adv. 4, 4393–4405. doi: 10.1182/bloodadvances.2020001592
Reiss, K., Maretzky, T., Ludwig, A., Tousseyn, T., De Strooper, B., Hartmann, D., et al. (2005). ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 24, 742–752. doi: 10.1038/sj.emboj.7600548
Reyat, J. S., Chimen, M., Noy, P. J., Szyroka, J., Rainger, G. E., and Tomlinson, M. G. (2017). ADAM10-Interacting Tetraspanins Tspan5 and Tspan17 Regulate VE-Cadherin Expression and Promote T Lymphocyte Transmigration. J. Immunol. 199, 666–676. doi: 10.4049/jimmunol.1600713
Reynolds, J. L., and Mahajan, S. D. (2020). Transmigration of Tetraspanin 2 (Tspan2) siRNA Via Microglia Derived Exosomes across the Blood Brain Barrier Modifies the Production of Immune Mediators by Microglia Cells. J. Neuroimmune Pharmacol. 15, 554–563. doi: 10.1007/s11481-019-09895-6
Riva, I., Eibl, C., Volkmer, R., Carbone, A. L., and Plested, A. J. (2017). Control of AMPA receptor activity by the extracellular loops of auxiliary proteins. eLife 6:e28680. doi: 10.7554/eLife.28680
Sager, C., Tapken, D., Kott, S., and Hollmann, M. (2009). Functional modulation of AMPA receptors by transmembrane AMPA receptor regulatory proteins. Neuroscience 158, 45–54. doi: 10.1016/j.neuroscience.2007.12.046
Salas, I. H., Callaerts-Vegh, Z., Arranz, A. M., Guix, F. X., D’hooge, R., Esteban, J. A., et al. (2017). Tetraspanin 6: a novel regulator of hippocampal synaptic transmission and long term plasticity. PLoS One 12:e0171968. doi: 10.1371/journal.pone.0171968
Sanyal, S., De Ruiter, A., and Hawkins, R. K. (1980). Development and degeneration of retina in rds mutant mice: light microscopy. J. Comp. Neurol. 194, 193–207. doi: 10.1002/cne.901940110
Schäfer, D., Tomiuk, S., Küster, L. N., Rawashdeh, W. A., Henze, J., Tischler-Höhle, G., et al. (2021). Identification of CD318, TSPAN8 and CD66c as target candidates for CAR T cell based immunotherapy of pancreatic adenocarcinoma. Nat. Commun. 12:1453. doi: 10.1038/s41467-021-21774-4
Schaffer, A. M., and Minguet, S. (2020). Caveolin-1, tetraspanin CD81 and flotillins in lymphocyte cell membrane organization, signaling and immunopathology. Biochem. Soc. Trans. 48, 2387–2397. doi: 10.1042/bst20190387
Schmidt, C., Künemund, V., Wintergerst, E. S., Schmitz, B., and Schachner, M. (1996). CD9 of mouse brain is implicated in neurite outgrowth and cell migration in vitro and is associated with the alpha 6/beta 1 integrin and the neural adhesion molecule L1. J. Neurosci. Res. 43, 12–31. doi: 10.1002/jnr.490430103
Schwenk, J., Baehrens, D., Haupt, A., Bildl, W., Boudkkazi, S., Roeper, J., et al. (2014). Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84, 41–54. doi: 10.1016/j.neuron.2014.08.044
Schwenk, J., Harmel, N., Brechet, A., Zolles, G., Berkefeld, H., Müller, C. S., et al. (2012). High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621–633. doi: 10.1016/j.neuron.2012.03.034
Seehafer, J. G., Tang, S. C., Slupsky, J. R., and Shaw, A. R. (1988). The functional glycoprotein CD9 is variably acylated: localization of the variably acylated region to a membrane-associated peptide containing the binding site for the agonistic monoclonal antibody 50H.19. Biochim. Biophys. Acta 957, 399–410. doi: 10.1016/0167-4838(88)90231-2
Seigneuret, M. (2006). Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 90, 212–227. doi: 10.1529/biophysj.105.069666
Seipold, L., Damme, M., Prox, J., Rabe, B., Kasparek, P., Sedlacek, R., et al. (2017). Tetraspanin 3: a central endocytic membrane component regulating the expression of ADAM10, presenilin and the amyloid precursor protein. Biochim. Biophys. Acta 1864, 217–230. doi: 10.1016/j.bbamcr.2016.11.003
Seipold, L., and Saftig, P. (2016). The emerging role of tetraspanins in the proteolytic processing of the amyloid precursor protein. Front. Mol. Neurosci. 9:149. doi: 10.3389/fnmol.2016.00149
Sidahmed-Adrar, N., Ottavi, J. F., Benzoubir, N., Ait Saadi, T., Bou Saleh, M., Mauduit, P., et al. (2019). Tspan15 is a new stemness-related marker in hepatocellular carcinoma. Proteomics 19:e1900025. doi: 10.1002/pmic.201900025
Sobolevsky, A. I., Rosconi, M. P., and Gouaux, E. (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. doi: 10.1038/nature08624
Stiles, K. M., and Kielian, M. (2016). Role of TSPAN9 in Alphavirus Entry and Early Endosomes. J. Virol. 90, 4289–4297. doi: 10.1128/jvi.00018-16
Stilgenbauer, S., Aurran Schleinitz, T., Eichhorst, B., Lang, F., Offner, F., Rossi, J. F., et al. (2019). Phase 1 first-in-human trial of the anti-CD37 antibody BI 836826 in relapsed/refractory chronic lymphocytic leukemia. Leukemia 33, 2531–2535. doi: 10.1038/s41375-019-0475-z
Stinnesbeck, M., Kristiansen, A., Ellinger, J., Hauser, S., Egevad, L., Tolkach, Y., et al. (2021). Prognostic role of TSPAN1, KIAA1324 and ESRP1 in prostate cancer. APMIS 129, 204–212. doi: 10.1111/apm.13117
Stipp, C. S., and Hemler, M. E. (2000). Transmembrane-4-superfamily proteins CD151 and CD81 associate with alpha 3 beta 1 integrin, and selectively contribute to alpha 3 beta 1-dependent neurite outgrowth. J. Cell Sci. 113(Pt 11), 1871–1882.
Stipp, C. S., Kolesnikova, T. V., and Hemler, M. E. (2003). Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28, 106–112. doi: 10.1016/s0968-0004(02)00014-2
Stuck, M. W., Conley, S. M., and Naash, M. I. (2016). PRPH2/RDS and ROM-1: historical context, current views and future considerations. Prog. Retin. Eye Res. 52, 47–63. doi: 10.1016/j.preteyeres.2015.12.002
Sun, H., Sun, Y., Chen, Q., and Xu, Z. (2020). LncRNA KCNQ1OT1 contributes to the progression and chemoresistance in acute myeloid leukemia by modulating Tspan3 through suppressing miR-193a-3p. Life Sci. 241:117161. doi: 10.1016/j.lfs.2019.117161
Susa, K. J., Rawson, S., Kruse, A. C., and Blacklow, S. C. (2021). Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81. Science 371, 300–305. doi: 10.1126/science.abd9836
Tang, Y., Xie, J., Chen, Y., Zhang, T., Zhuo, H., Chen, J., et al. (2020). Tspan8 is highly expressed in clear cell renal cell carcinoma and indicates poor prognosis. Ann. Clin. Lab. Sci. 50, 638–644.
Termini, C. M., and Gillette, J. M. (2017). Tetraspanins function as regulators of cellular signaling. Front. Cell. Dev. Biol. 5:34. doi: 10.3389/fcell.2017.00034
Tian, J., Zhang, R., Piao, H., Li, X., Sheng, W., Zhou, J., et al. (2018). Silencing Tspan1 inhibits migration and invasion, and induces the apoptosis of human pancreatic cancer cells. Mol. Med. Rep. 18, 3280–3288. doi: 10.3892/mmr.2018.9331
Tomita, S. (2010). Regulation of ionotropic glutamate receptors by their auxiliary subunits. Physiology 25, 41–49. doi: 10.1152/physiol.00033.2009
Torres-Gómez, Á, Cardeñes, B., Díez-Sainz, E., Lafuente, E. M., and Cabañas, C. (2021). Functional integrin regulation through interactions with tetraspanin CD9. Methods Mol. Biol. 2217, 47–56. doi: 10.1007/978-1-0716-0962-0_5
Travis, G. H., Christerson, L., Danielson, P. E., Klisak, I., Sparkes, R. S., Hahn, L. B., et al. (1991). The human retinal degeneration slow (RDS) gene: chromosome assignment and structure of the mRNA. Genomics 10, 733–739. doi: 10.1016/0888-7543(91)90457-p
Traynelis, S. F., Wollmuth, L. P., Mcbain, C. J., Menniti, F. S., Vance, K. M., Ogden, K. K., et al. (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496. doi: 10.1124/pr.109.002451
Tsai, Y. C., Mendoza, A., Mariano, J. M., Zhou, M., Kostova, Z., Chen, B., et al. (2007). The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 13, 1504–1509. doi: 10.1038/nm1686
Tunyasuvunakool, K., Adler, J., Wu, Z., Green, T., Zielinski, M., Žídek, A., et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596. doi: 10.1038/s41586-021-03828-1
Twomey, E. C., Yelshanskaya, M. V., Grassucci, R. A., Frank, J., and Sobolevsky, A. I. (2016). Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science 353, 83–86. doi: 10.1126/science.aaf8411
Twomey, E. C., Yelshanskaya, M. V., Vassilevski, A. A., and Sobolevsky, A. I. (2018). Mechanisms of channel block in calcium-permeable AMPA receptors. Neuron 99, 956–968.e4. doi: 10.1016/j.neuron.2018.07.027
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419
Umeda, R., Satouh, Y., Takemoto, M., Nakada-Nakura, Y., Liu, K., Yokoyama, T., et al. (2020). Structural insights into tetraspanin CD9 function. Nat. Commun. 11:1606. doi: 10.1038/s41467-020-15459-7
Vincent, A. K., Noor, A., Janson, A., Minassian, B. A., Ayub, M., Vincent, J. B., et al. (2012). Identification of genomic deletions spanning the PCDH19 gene in two unrelated girls with intellectual disability and seizures. Clin. Genet. 82, 540–545. doi: 10.1111/j.1399-0004.2011.01812.x
Wang, F. H., Ma, X. J., Xu, D., and Luo, J. (2018). UPK1B promotes the invasion and metastasis of bladder cancer via regulating the Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 5471–5480. doi: 10.26355/eurrev_201809_15807
Wang, L., Gao, P., Yuan, P., Zhou, P., Fan, H., Lin, X., et al. (2021). miR-573 suppresses pancreatic cancer cell proliferation, migration, and invasion through targeting TSPAN1. Strahlenther. Onkol. 197, 438–448. doi: 10.1007/s00066-020-01728-3
Wang, Z., Cai, B., Cao, C., Lv, H., Dai, Y., Zheng, M., et al. (2021). Downregulation of CD151 induces oxidative stress and apoptosis in trophoblast cells via inhibiting ERK/Nrf2 signaling pathway in preeclampsia. Free Radic. Biol. Med. 164, 249–257. doi: 10.1016/j.freeradbiomed.2020.12.441
Wang, Y., Chan, S. L., Miele, L., Yao, P. J., Mackes, J., Ingram, D. K., et al. (2004). Involvement of Notch signaling in hippocampal synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 101, 9458–9462. doi: 10.1073/pnas.0308126101
Wang, Y., Tong, X., Omoregie, E. S., Liu, W., Meng, S., and Ye, X. (2012). Tetraspanin 6 (TSPAN6) negatively regulates retinoic acid-inducible gene I-like receptor-mediated immune signaling in a ubiquitination-dependent manner. J. Biol. Chem. 287, 34626–34634. doi: 10.1074/jbc.M112.390401
Wollscheid, V., Kühne-Heid, R., Stein, I., Jansen, L., Köllner, S., Schneider, A., et al. (2002). Identification of a new proliferation-associated protein NET-1/C4.8 characteristic for a subset of high-grade cervical intraepithelial neoplasia and cervical carcinomas. Int. J. Cancer 99, 771–775. doi: 10.1002/ijc.10442
Wright, M. D., Henkle, K. J., and Mitchell, G. F. (1990). An immunogenic Mr 23,000 integral membrane protein of Schistosoma mansoni worms that closely resembles a human tumor-associated antigen. J. Immunol. 144, 3195–3200.
Wu, L., Chen, F., Wei, J., Shen, Y., and Xu, Q. (2016). Study of the tetraspanin 18 association with schizophrenia in a Han Chinese population. Psychiatry Res. 241, 263–266. doi: 10.1016/j.psychres.2016.03.057
Wu, X. R., Medina, J. J., and Sun, T. T. (1995). Selective interactions of UPIa and UPIb, two members of the transmembrane 4 superfamily, with distinct single transmembrane-domained proteins in differentiated urothelial cells. J. Biol. Chem. 270, 29752–29759. doi: 10.1074/jbc.270.50.29752
Xia, J., Zhang, X., Staudinger, J., and Huganir, R. L. (1999). Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron 22, 179–187. doi: 10.1016/s0896-6273(00)80689-3
Xia, Y., Deng, Y., Zhou, Y., Li, D., Sun, X., Gu, L., et al. (2020). TSPAN31 suppresses cell proliferation in human cervical cancer through down-regulation of its antisense pairing with CDK4. Cell Biochem. Funct. 38, 660–668. doi: 10.1002/cbf.3526
Xie, Q., Guo, H., He, P., Deng, H., Gao, Y., Dong, N., et al. (2021). Tspan5 promotes epithelial-mesenchymal transition and tumour metastasis of hepatocellular carcinoma by activating Notch signalling. Mol. Oncol. 15, 3184–3202. doi: 10.1002/1878-0261.12980
Xu, D., Sharma, C., and Hemler, M. E. (2009). Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 23, 3674–3681. doi: 10.1096/fj.09-133462
Xu, Y., Wu, H., Wu, L., Xu, L., Li, J., Wang, Q., et al. (2020). Silencing of long non-coding RNA SOX21-AS1 inhibits lung adenocarcinoma invasion and migration by impairing TSPAN8 via transcription factor GATA6. Int. J. Biol. Macromol. 164, 1294–1303. doi: 10.1016/j.ijbiomac.2020.07.172
Xu-Monette, Z. Y., Li, L., Byrd, J. C., Jabbar, K. J., Manyam, G. C., Maria De Winde, C., et al. (2016). Assessment of CD37 B-cell antigen and cell of origin significantly improves risk prediction in diffuse large B-cell lymphoma. Blood 128, 3083–3100. doi: 10.1182/blood-2016-05-715094
Yang, X., Claas, C., Kraeft, S. K., Chen, L. B., Wang, Z., Kreidberg, J. A., et al. (2002). Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell 13, 767–781. doi: 10.1091/mbc.01-05-0275
Yang, Y., Liu, X. R., Greenberg, Z. J., Zhou, F., He, P., Fan, L., et al. (2020). Open conformation of tetraspanins shapes interaction partner networks on cell membranes. EMBO J. 39:e105246. doi: 10.15252/embj.2020105246
Yang, Y. G., Sari, I. N., Zia, M. F., Lee, S. R., Song, S. J., and Kwon, H. Y. (2016). Tetraspanins: spanning from solid tumors to hematologic malignancies. Exp. Hematol. 44, 322–328. doi: 10.1016/j.exphem.2016.02.006
Yaseen, I. H., Monk, P. N., and Partridge, L. J. (2017). Tspan2: a tetraspanin protein involved in oligodendrogenesis and cancer metastasis. Biochem. Soc. Trans. 45, 465–475. doi: 10.1042/bst20160022
Young, R. W. (1967). The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72. doi: 10.1083/jcb.33.1.61
Yu, J., Rao, P., Clark, S., Mitra, J., Ha, T., and Gouaux, E. (2021). Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition. Nature 594, 448–453. doi: 10.1038/s41586-021-03540-0
Yu, S., Chen, J., Quan, M., Li, L., Li, Y., and Gao, Y. (2021). CD63 negatively regulates hepatocellular carcinoma development through suppression of inflammatory cytokine-induced STAT3 activation. J. Cell. Mol. Med. 25, 1024–1034. doi: 10.1111/jcmm.16167
Yuan, J., Jin, C., Qin, H. D., Wang, J., Sha, W., Wang, M., et al. (2013). Replication study confirms link between TSPAN18 mutation and schizophrenia in Han Chinese. PLoS One 8:e58785. doi: 10.1371/journal.pone.0058785
Yuasa, T., Yoshiki, T., Tanaka, T., Kim, C. J., Isono, T., and Okada, Y. (1998). Expression of uroplakin Ib and uroplakin III genes in tissues and peripheral blood of patients with transitional cell carcinoma. Jpn. J. Cancer Res. 89, 879–882. doi: 10.1111/j.1349-7006.1998.tb00643.x
Zemni, R., Bienvenu, T., Vinet, M. C., Sefiani, A., Carrié, A., Billuart, P., et al. (2000). A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat. Genet. 24, 167–170. doi: 10.1038/72829
Zhang, B., Li, D. X., Lu, N., Fan, Q. R., Li, W. H., and Feng, Z. F. (2015). Lack of Association between the TSPAN18 Gene and Schizophrenia Based on New Data from Han Chinese and a Meta-Analysis. Int. J. Mol. Sci. 16, 11864–11872. doi: 10.3390/ijms160611864
Zhang, D., Watson, J. F., Matthews, P. M., Cais, O., and Greger, I. H. (2021). Gating and modulation of a hetero-octameric AMPA glutamate receptor. Nature 594, 454–458. doi: 10.1038/s41586-021-03613-0
Zhang, M., Jiang, J., and Ma, C. (2021). The Outer Retinal Membrane Protein 1 Could Inhibit Lung Cancer Progression as a Tumor Suppressor. Comput. Math. Methods Med. 2021:6651764. doi: 10.1155/2021/6651764
Zhang, Q., Han, Q., Zi, J., Song, C., and Ge, Z. (2020). CD37 high expression as a potential biomarker and association with poor outcome in acute myeloid leukemia. Biosci. Rep. 40:BSR20200008. doi: 10.1042/bsr20200008
Zhang, Z. Y., Zhang, J. L., Zhao, L. X., Yang, Y., Guo, R., Zhou, N., et al. (2020). NAA10 promotes proliferation of renal cell carcinoma by upregulating UPK1B. Eur. Rev. Med. Pharmacol. Sci. 24, 11553–11560. doi: 10.26355/eurrev_202011_23796
Zhang, T., Sun, X., Han, J., and Han, M. (2019). Genetic variants of TSPAN12 gene in patients with retinopathy of prematurity. J. Cell. Biochem. 120, 14544–14551. doi: 10.1002/jcb.28715
Zhao, Y., Kiss, T., Delfavero, J., Li, L., Li, X., Zheng, L., et al. (2020). CD82-TRPM7-Numb signaling mediates age-related cognitive impairment. Geroscience 42, 595–611. doi: 10.1007/s11357-020-00166-4
Zheng, Y., Wang, D. D., Wang, W., Pan, K., Huang, C. Y., Li, Y. F., et al. (2014). Reduced expression of uroplakin 1A is associated with the poor prognosis of gastric adenocarcinoma patients. PLoS One 9:e93073. doi: 10.1371/journal.pone.0093073
Zhu, H., Tang, Y., Zhang, X., Jiang, X., Wang, Y., Gan, Y., et al. (2015). Downregulation of UPK1A suppresses proliferation and enhances apoptosis of bladder transitional cell carcinoma cells. Med. Oncol. 32:84. doi: 10.1007/s12032-015-0541-y
Keywords: tetraspanins, CNS, synaptic function, glutamate receptor, modulator structures
Citation: Becic A, Leifeld J, Shaukat J and Hollmann M (2022) Tetraspanins as Potential Modulators of Glutamatergic Synaptic Function. Front. Mol. Neurosci. 14:801882. doi: 10.3389/fnmol.2021.801882
Received: 25 October 2021; Accepted: 07 December 2021;
Published: 03 January 2022.
Edited by:
Dhrubajyoti Chowdhury, Yale University, United StatesReviewed by:
Janosch P. Heller, Dublin City University, IrelandCopyright © 2022 Becic, Leifeld, Shaukat and Hollmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Hollmann, bWljaGFlbC5ob2xsbWFubkBydWIuZGU=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.