
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 22 November 2021
Sec. Molecular Signalling and Pathways
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.786226
This article is part of the Research Topic Bidirectional Communication between Synapses and Nucleus in Brain Physiology and Disease View all 7 articles
Long-lasting changes of synaptic efficacy are largely mediated by activity-induced gene transcription and are essential for neuronal plasticity and memory. In this scenario, transcription factors have emerged as pivotal players underlying synaptic plasticity and the modification of neural networks required for memory formation and consolidation. Hippocampal synaptic dysfunction is widely accepted to underlie the cognitive decline observed in some neurodegenerative disorders including Alzheimer’s disease. Therefore, understanding the molecular pathways regulating gene expression profiles may help to identify new synaptic therapeutic targets. The nuclear receptor 4A subfamily (Nr4a) of transcription factors has been involved in a variety of physiological processes within the hippocampus, ranging from inflammation to neuroprotection. Recent studies have also pointed out a role for the activity-dependent nuclear receptor subfamily 4, group A, member 2 (Nr4a2/Nurr1) in hippocampal synaptic plasticity and cognitive functions, although the underlying molecular mechanisms are still poorly understood. In this review, we highlight the specific effects of Nr4a2 in hippocampal synaptic plasticity and memory formation and we discuss whether the dysregulation of this transcription factor could contribute to hippocampal synaptic dysfunction, altogether suggesting the possibility that Nr4a2 may emerge as a novel synaptic therapeutic target in brain pathologies associated to cognitive dysfunctions.
Nuclear receptor subfamily 4, group A (Nr4a), which belongs to the nuclear receptor superfamily, consists in a family of three close-related immediate early genes that encode three orphan nuclear receptors (Nr4a1/NGFI-B, Nr4a2/Nurr1, and Nr4a3/NOR-1). They are expressed in a wide variety of metabolically demanding and energy dependent tissues, such as skeletal muscle, adipose tissue, heart, kidney, T-cells, liver and distinct but overlapping regions of the brain (Zetterström et al., 1996). The three members exhibit tissue-specific expression and hence their roles are context as well as tissue-specific.
Nr4a proteins function as transcription factors that recognize DNA response elements to regulate the expression of a variety of genes involved in multiple biological processes including proliferation, metabolism, immunity, cellular stress, apoptosis, DNA repair, and angiogenesis (Safe et al., 2016). Nr4a activity depends not only on its transcriptional levels (including miRNA targeting) but also on alternative splicing, post-translational modifications, subcellular localization and interaction with other nuclear receptors (Maxwell and Muscat, 2006). These transcriptional regulators may be stably expressed or induced as immediate early genes in response to a wide range of physiological signals, including synaptic activity (Pegoraro et al., 2010). The ability to sense and rapidly respond to changes in the cellular environment appears to be a hallmark of this subfamily of orphan nuclear receptors.
In peripheral tissues, these receptors have been involved in the onset and progression of various types of cancer, inflammation, atherosclerosis and obesity (Ranhotra, 2015). Notably, Nr4a transcription factors have also emerged as essential mediators in several physio-pathological situations in the central nervous system (CNS). The Nr4a family of transcriptional regulators has been reported to modulate hippocampal synaptic plasticity and hippocampal-dependent memory (Hawk and Abel, 2011; Bridi and Abel, 2013) and the alteration of expression or function of these factors has been associated to neurodegenerative diseases (Skerrett et al., 2014), intellectual disability such as autism (Chuang et al., 2015), schizophrenia (Rojas et al., 2007; Ruiz-Sánchez et al., 2021) and epilepsy (Zhang et al., 2016).
This review will primarily focus on the current knowledge about Nr4a2 contribution to the active-dependent processes that underlie synaptic plasticity, learning and memory and the evidence supporting a critical role of this factor in several brain pathologies associated to cognitive dysfunction.
Nuclear receptor subfamily 4, group A, member 2 (Nr4a2), which belongs to the Nr4a subfamily of transcription factors, is primarily expressed in neurons of diverse areas of the CNS, particularly in the substantia nigra pars compacta, ventral tegmental area and limbic area (Zetterström et al., 1996). Moreover, it is also expressed in the hippocampus, subiculum, temporal cortex, olfactory bulb, cerebellum, posterior hypothalamus, and habenuclear nuclei (Saucedo-Cardenas and Conneely, 1996; Quina et al., 2009). Nevertheless, Nr4a2 is expressed not only in neurons, but also in non-neuronal cells such as microglia, astrocytes or endothelial cells (Fan et al., 2009; Saijo et al., 2009) and it is found not only in the CNS but also in other tissues, including the bone, synovial tissues, adrenal gland or the intestine.
Nr4a2 structure (Figure 1A) is composed by a modulator domain, referred to as the activation function (AF-1) or the N-terminal domain (NTD), a conserve DNA-binding domain (DBD) and a ligand-binding domain (LBD) containing its transactivation-dependent AF-2 in the C-terminal domain (CTD) (Ichinose et al., 1999). Classically, nuclear receptor activation is accomplished by the binding of a lipophilic ligand in a hydrophobic pocket within the LBD. By contrast, the crystal structure of Nr4a2 reveals that the Nr4a2-LBD lacks a classical biding pocket for coactivators and/or ligands because of the tight packing of side chains from several bulky hydrophobic side-chain residues, adopting a canonical protein fold resembling that of an agonist-bound, transcriptionally active LBD (Wang et al., 2003). Thus, it is believed that a major difference between Nr4a2 and classical nuclear receptors is the absence of endogenous ligand regulation. Therefore, Nr4a2 activity will be regulated at the level of gene expression and protein stability (Maxwell and Muscat, 2006). Nevertheless, during the last years, several studies have identified synthetic ligands, known as Nr4a2 agonists, which can modulate Nr4a2 transcriptional function (Hammond et al., 2015; Kim et al., 2015; Chatterjee et al., 2020). Identification of such Nr4a2 agonists triggered the possibility of a pharmacological therapeutic approach for diseases related to Nr4a2 (Skerrett et al., 2014; Kim et al., 2015; Moon et al., 2019; Chatterjee et al., 2020; Jeon et al., 2020). The canonical view of Nr4a2 as an orphan nuclear receptor has been recently challenged by some studies showing that the Nr4a2-LBD is highly dynamic with solvent accessibility and the capability to expand (de Vera et al., 2019) allowing the binding of putative endogenous ligands (Bruning et al., 2019; Rajan et al., 2020).
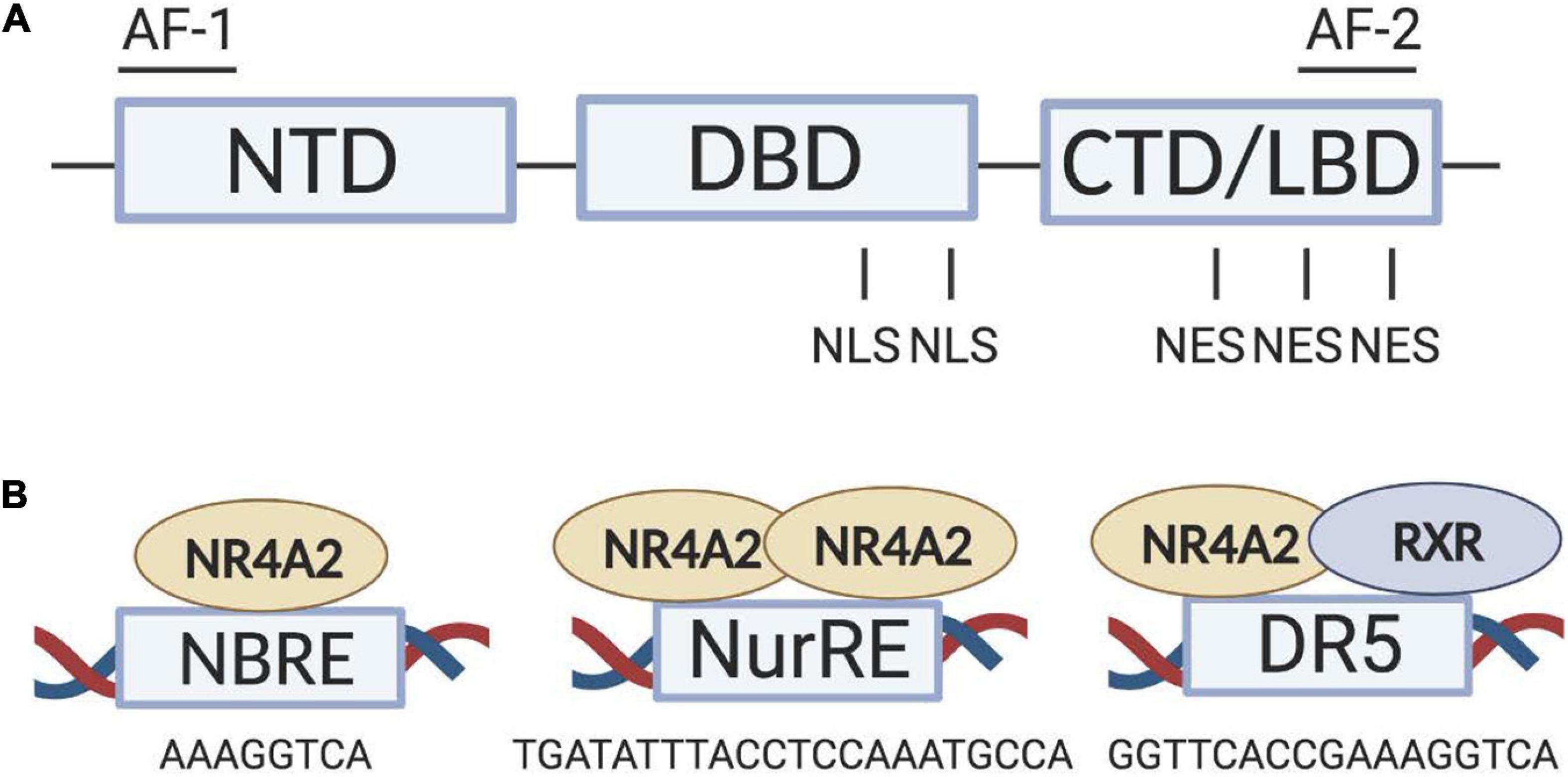
Figure 1. Nr4a2 structure and binding to target genes. (A) Structural domains of Nr4a2, which include NTD, DBD and LBD. Modulator domains (AF-1 and AF-2) and localization signals (NLS and NES) are also depicted. (B) Nr4a2 DNA-binding sites as monomers at NBRE sites, as dimers (homodimers or heterodimers, not shown) at NurRE sites and as heterodimers with RXR at DR5 sites. AF, activation function; CTD, C-terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain, NBRE, nerve growth factor-inducible-β-binding response element; NES, nuclear export signal; NLS, nuclear localization signal, NTD, N-terminal domain; NurRE, nur-response element; RXR, retinoic X receptor.
Nr4a2 protein regulates both positively and negatively the transcription of its target genes by directly binding to response elements in their promoters. Two zinc fingers of the highly conserved Nr4a2-DBD as a monomer or homodimer can bind the nerve growth factor-inducible-β-binding response element (NBRE; 5′-AAAGGTCA-3′) or as homodimer or heterodimer with Nr4a1 can attach to the Nur response element (NurRE; 5′-TGACCTTT-N6-AAAGGTCA-3′). Moreover, as monomer, homodimer or heterodimer, Nr4a2 can dimerize with the retinoic X receptor (RXR) and bind to a motif referred to as DR5 (Figure 1B). This union is permissive; meaning that ligand binding of RXR typically causes full activation of the entire heterodimer (Jiang et al., 2019).
Nr4a2 is known to be a key regulator of the midbrain dopaminergic neurons differentiation, maintenance and survival (Saucedo-Cardenas et al., 1998; Sousa et al., 2007; Jo et al., 2009), being responsible for the transcription of several genes involved in the dopaminergic neuronal phenotype, ranging from genes regulating dopaminergic metabolism, differentiation and neurotransmission (tyrosine hydroxylase –TH–, vesicular amine transporter 2 –VMAT–, dopamine transporter –DAT–), mitochondrial function (sodium oxide dismutase 1 –SOD1–, Ts translation elongation factor mitochondrial –TSFM–, cyclo-oxigenase 5β –COX5β–) and dopaminergic neuronal survival (Ret) (Kadkhodaei et al., 2009; Decressac et al., 2013). In dopaminergic neurons, Nr4a2 also has an essential role in the neuroprotection and anti-inflammatory responses after exposure to neuropathological stress or insults. It has been reported that Nr4a2-null heterozygous dopaminergic neurons exhibit greater vulnerability to neurotoxins (Le et al., 1999). Importantly, Nr4a2 protects dopaminergic neurons from neuroinflammation insults not only through its function in neurons, but also glial cells (Bensinger and Tontonoz, 2009), since it is able to suppress inflammatory gene expression in microglia and astrocytes through transcriptional repression of the NFκB transcription factor, essential to induce inflammatory responses (Saijo et al., 2009).
Nr4a2 contains a “half-CRE” site in its promoter, being controlled by the CREB signaling pathway in many systems, including the hippocampus, which is a cascade critical for the transcription of diverse plasticity- and memory-related genes (Volakakis et al., 2010; Barneda-Zahonero et al., 2012). Although CREB binds with higher affinity to the full 8-base consensus CRE site, it also binds to several variations of this sequence, including the so-called “half-CRE” site (5′CGTCA-3′ and 5′-TGACG-3′), which may be found in many promoters of CREB target genes including Nr4a2 (Zhang et al., 2005). CREB has been extensively studied in the brain since it is involved in a variety of relevant processes such as learning and memory or neuronal survival, among others (Benito and Barco, 2015). For example, it is known that CREB activity is crucial for the maintenance of long-term plasticity and memory formation (Bourtchuladze et al., 1994; Bartsch et al., 1998; Kaldun and Sprecher, 2019). Moreover, the genetic program downstream CREB is complex and different transcriptional programs could be elicited depending on the triggering stimulus, cellular context, and upstream signaling events (Nonaka et al., 2014). Furthermore, selective gene transcription by CREB is highly modulated by the recruitment of specific coactivators such as the CREB binding protein (CBP/p300) or CREB regulated Transcription Coactivator (CRTC1 and CRTC2) (Altarejos and Montminy, 2011). Reports showing that CREB modulates transcription of Nr4a2 (Volakakis et al., 2010; Barneda-Zahonero et al., 2012) has fostered the interest for studying the putative role of Nr4a2 in synaptic plasticity and learning and memory processes.
Several reports have highlighted a role for the Nr4a family of transcription factors in hippocampal synaptic plasticity and cognitive functions and there is an overwhelming amount of evidence showing that synaptic plasticity is a fundamental mechanism contributing to learning and memory storage (Martin et al., 2000; Abraham et al., 2019). Studies revealing the implication of the Nr4a family of transcription factors in hippocampal synaptic plasticity have supported a putative role of these factors in learning and memory formation (Hawk and Abel, 2011). Early studies showed an increase in Nr4a1 mRNA in CA1 after contextual fear conditioning during consolidation (von Hertzen and Giese, 2005; Keeley et al., 2006). Later, it was found that expression of a dominant-negative form of Nr4a in the mouse forebrain, including the hippocampus, impaired long-term hippocampus-dependent contextual fear memory without affecting short-term contextual fear memory or hippocampus-independent cued fear memory (Hawk et al., 2012). Furthermore, it was also observed that histone deacetylase (HDAC) inhibitors-mediated memory enhancement was accompanied by an increase in the expression of Nr4a family members meanwhile blocking Nr4a family expression in the hippocampus prevented HDAC inhibitors-mediated improvement of long-term memory (Hawk et al., 2012). Supporting the role of Nr4a transcription factors in hippocampal synaptic plasticity associated to learning and memory, a strongly impaired transcription-dependent hippocampal long-term potentiation (LTP) maintenance of Schaffer collateral to CA1 synapses was observed in hippocampal slices from Nr4a dominant negative mutant mice whereas no deficits in basal synaptic transmission were observed (Bridi and Abel, 2013).
Both hippocampal LTP and long-term memory are enhanced by the pharmacological inhibition or genetic ablation of HDAC enzymes, which increase histone acetylation in a CREB-dependent manner (Vecsey et al., 2007). Importantly, HDAC inhibition failed to enhance LTP in hippocampal slices from Nr4a dominant negative mutant mice, indicating that Nr4a transcription factors are necessary for mediating the effects of HDAC inhibition on synaptic plasticity (Bridi and Abel, 2013).
Although these results supported the importance of the Nr4a family members for hippocampal memory formation, it is still uncertain which are the specific role for each member: Nr4a1, Nr4a2, and Nr4a3. A differential role in learning for Nr4a1 and Nr4a2 was reported by McNulty and co-workers (McNulty et al., 2012). Using small interfering RNA (siRNA), they found that downregulation of Nr4a2 disrupted long-term memory in the object location and object recognition tasks whereas Nr4a1 seemed to be only required for object location memory. Bridi and co-workers reported an enhancement of LTP when hippocampal slices were incubated with Nr4a1 activators, para-phenyl substituted di-indolylmethane compounds or DIM-C (Bridi et al., 2017). DIM-C-pPhOCH3 or DIM-C-pPhBr enhanced LTP at the CA3–CA1 hippocampal synapses. This enhancement was absent when LTP was triggered in slices from dominant-negative Nr4a mice. Several studies have evaluated the role of Nr4a2 in memory acquisition and consolidation, reporting an increase in the hippocampal expression of Nr4a family members after learning of hippocampus-dependent tasks. A specific increase of Nr4a2 in the hippocampus was observed after different memory-inducing activities. Acquisition of a spatial food search task in adult rats produced an increase in the levels of Nr4a2 mRNA in CA1 and CA3 subregions of the dorsal hippocampus (Peña de Ortiz et al., 2000) and the injection of antisense oligodeoxynucleotides targeting Nr4a2 in the CA3 hippocampal area impaired long-term memory and reversal learning in an appetitive spatial learning task (Colón-Cesario et al., 2006). Relatedly, contextual fear conditioning increased Nr4a2 mRNA and promoter acetylation in a CREB/CBP-dependent manner (Vecsey et al., 2007; Bridi et al., 2017; Oliveira et al., 2018). The role of Nr4a2 in other hippocampal-dependent behavioral tasks like passive avoidance was also reported when Nr4a2 heterozygous mice showed reduced ability to form long-term emotional memories (Rojas et al., 2007). Also, deep brain stimulation in rats by intracranial self-stimulation treatment immediately after the acquisition session of a two-way active avoidance conditioning showed both increased Nr4a2 protein levels in the hippocampus and improved retention (Aldavert-Vera et al., 2013). Accordingly with the above-commented results observed with HDAC inhibitors, it was also reported that a Nr4a2-dependent enhancement of novel object recognition long-term memory was observed when HDAC3 expression was deleted in the CA1 area of the mice dorsal hippocampus (McQuown et al., 2011).
Cognitive decline is also associated with aging and Nr4a2 expression gradually decreases in the gerbil hippocampus with increasing age (Ahn et al., 2018). Furthermore, Nr4a2 has also been recently related to long-term memory impairments in aged animals (Kwapis et al., 2019). Whereas adult or aged rats with intact cognitive functions showed an increase in Nr4a2 expression in the dorsal hippocampus after the object recognition memory training, Nr4a2 failed to be induced in the cognitively impaired aged rats. Learning-induced Nr4a2 was also impaired in the hippocampus of aging mice. Thus, a cross-species impairment of Nr4a2 hippocampal expression is observed in mice and rats with cognitive deficits. Interestingly, failed induction of Nr4a2 expression is reversed when HDAC3 is deleted in the dorsal hippocampus. Deletion of HDAC3 or hippocampal overexpression of Nr4a2 were able to ameliorate object location memory impairment in aged mice (Kwapis et al., 2019).
Indeed, Nr4a2 has also been described to play a role in hippocampal neurogenesis and increasing evidence exists supporting that neurogenesis in the sub granular zone of the hippocampus could play a key role in long-term spatial memory. Interestingly, Nr4a2 is abundantly expressed in adult hippocampal neural precursor cells and it stimulates their proliferation and differentiation both in vitro and in vivo (Kim et al., 2016). Thus, Nr4a2 could have an important role in hippocampal learning and memory not only by a functional modulation of synaptic plasticity but also by acting through hippocampal neurogenesis.
Although a great deal of evidence has highlighted a role for Nr4a2 in hippocampal-dependent cognitive functions (Table 1), the underlying molecular mechanisms are still poorly understood. Nr4a2 is induced by neuronal activity and its expression is mainly dependent on CREB/CRTC1 and calcium channels activation (España et al., 2010; Parra-Damas et al., 2014, 2017a,b; Tokuoka et al., 2014). However, knowledge of the molecular pathways involved in Nr4a2 function in synaptic plasticity in the hippocampus is still incomplete. It is known that Nr4a2 transcription factor regulates several genes implicated in hippocampal synaptic plasticity and memory formation (Volpicelli et al., 2007; Hawk et al., 2012; Kim et al., 2020). One example is Fosl2, a member of the AP-1 family of transcription factors, a family that is known to be important for memory storage (Fleischmann et al., 2003). Other intriguing Nr4a2 target genes include two receptor protein tyrosine phosphatases, a class of molecules implicated in excitatory synapse formation (Dunah et al., 2005) and the myristoylated alanine-rich C kinase substrate (MARCKS), which can modulate memory formation (McNamara et al., 2005). Of particular interest as a candidate target gene by which Nr4a2 influences synaptic plasticity and memory is BDNF, a neurotrophic factor that regulates synaptic plasticity (Zagrebelsky and Korte, 2014) and contributes to the formation and long-term persistence of hippocampus-dependent memories (Bekinschtein et al., 2014; Miranda et al., 2019). Nr4a2 transcriptional activity regulates BDNF levels in different systems. Volpicelli et al. (2007) have shown that Nr4a2 regulates Bdnf gene expression in rat midbrain neurons and similar results were also reported in cerebellar granule cells (Barneda-Zahonero et al., 2012) and in activated rat microglia overexpressing Nr4a2 (Chen et al., 2018). Moreover, BDNF was early on implicated in modulating neuronal activity, and its own production and release has been shown to be activity-dependent (Kovács et al., 2007; Parra-Damas et al., 2014). However, relatively little is known about the mechanisms underlying the activity-dependent regulation of BDNF expression. Future studies are needed for a fully comprehensive knowledge of the cellular and molecular mechanisms involved in the role of Nr4a2 in hippocampal synaptic plasticity, learning, and memory.

Table 1. Overview of the overall effects of Nr4a2 in hippocampal synaptic plasticity, learning and memory.
Long-term memory involves persistent changes in synaptic proteome and structure that require the regulation of gene transcription (Alberini, 2009; Heo et al., 2018). Transcription factors involved in learning and memory will translocate in and out the nucleus in response to the signaling cascades activated at synapses (Ch’ng and Martin, 2011). Several pathways, typically requiring calcium signaling, have been described to connect synaptic inputs to gene transcription (Hagenston and Bading, 2011). Activation of different protein kinases regulates the phosphorylated state of transcription factors and their cytosol-nuclear localization (Klann and Dever, 2004). However, the view that activity-dependent transcriptional regulation is based on post-translational modification of transcription factors located in the cytosol or nucleus is changing. Several transcription modulators involved in learning and memory, such as CRTCs, NFκB or HDAC4, have been reported to be located at synapses and to be transported to nucleus upon synapse stimulation (Uchida and Shumyatsky, 2018). CRTC1 has been found in the postsynaptic density (PSD) subfraction obtained from adult mouse brain and in excitatory synapses from hippocampal cultures. It translocates to nucleus in response to localized synaptic activity or induction of LTP (Ch’ng Toh et al., 2012). Histone deacetylase 4 (HDAC4) shuttles between the cytosol and the nucleus in response to neuronal activity. Although HDAC4 is mainly located in the neuronal cytoplasm with a more variable nuclear presence, immunohistochemistry studies have shown a strong PSD immunoreactivity in dendritic spines (Darcy et al., 2010). Another transcriptional modulator with a synaptic localization is NFκB (Kaltschmidt et al., 1993; Meffert et al., 2003), which is activated upon depolarization (Meffert et al., 2003). Other transcriptional modulators are known to be present at synapses, such as Arc and ATF2 (Lai et al., 2008; Shepherd and Bear, 2011) and it is likely that the list of transcriptional regulators that participate in synapse to nucleus signaling will increase in the future.
Nr4a2 contains a bipartite nuclear localization signal (NLS) within its DBD and three leucine-rich nuclear export signals (NES) in its LBD. Together, these signals regulate Nr4a2 shuttling in and out of the nucleus (García-Yagüe et al., 2013). Notably, data from our laboratory support the presence of Nr4a2 in the postsynaptic density fraction of mature hippocampal neuronal cultures treated with the GABA-A receptor antagonist bicuculline (Figure 2A). Moreover, Nr4a2 was also detected in the postsynaptic density fraction of synaptoneurosomes obtained from adult mouse hippocampus (Figure 2B). These data open the possibility that Nr4a2 could also be considered as a synaptically localized transcriptional regulator that will be transported, by an unknown mechanism, upon synaptic stimulation (Figure 2C). Further future work is needed to confirm the presence of Nr4a2 in the synapse and the importance of this subcellular localization in the already described role of Nr4a2 in hippocampal synaptic plasticity and memory formation.
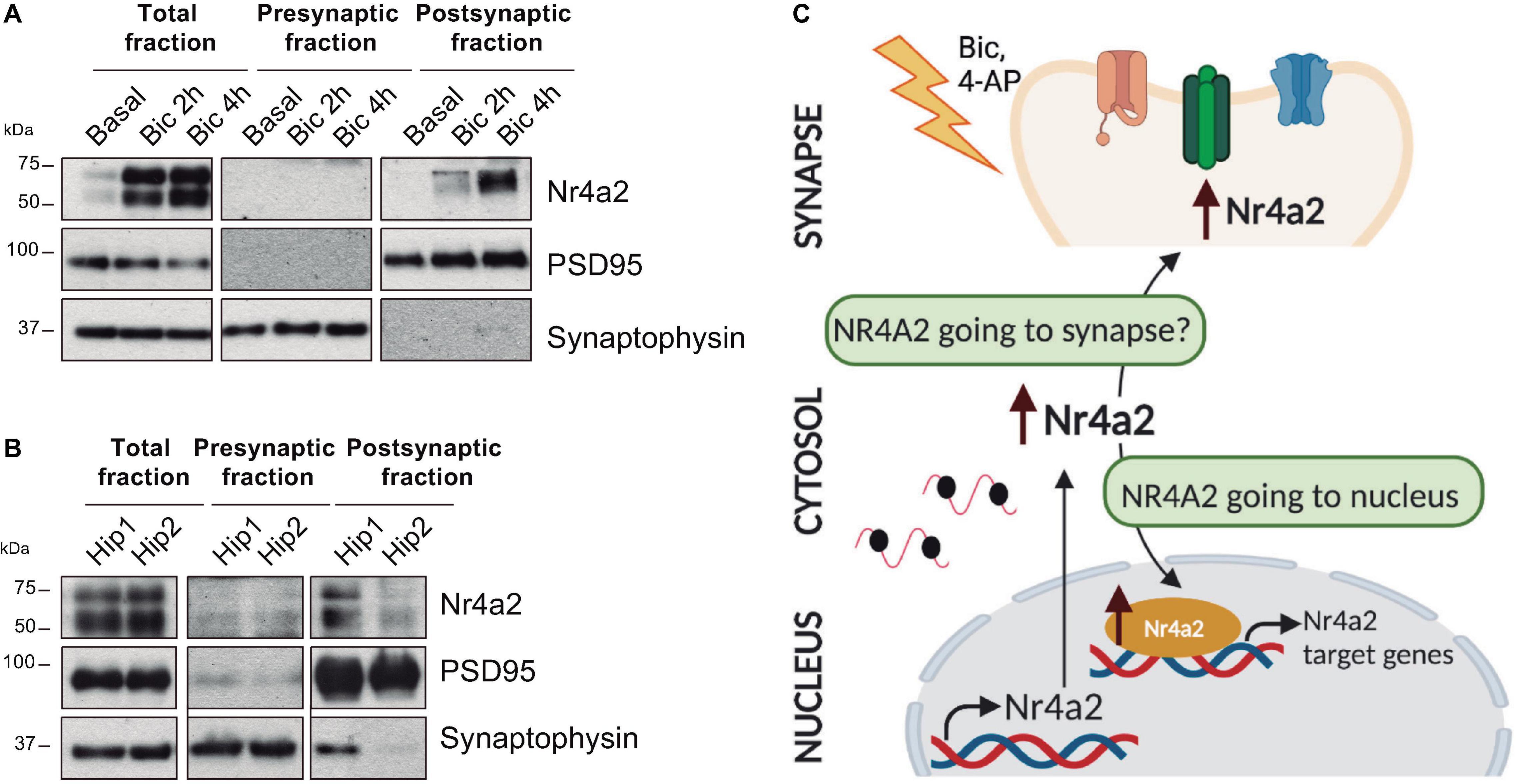
Figure 2. Nr4a2 transcription factor is present at the postsynaptic density both in mature hippocampal-cultured neurons and the adult mouse hippocampus. (A) Synaptoneurosomes from mature hippocampal-cultured neurons after bicuculline (bic; 50 μM) treatment. (B) Synaptoneurosomes of adult mouse hippocampus (hip). All figures correspond to representative images from Western blotting analysis. n ≥ 3. (C) Schematic model of Nr4a2 mobilization to nucleus and postsynaptic density fractions after neuronal activity.
Parkinson’s disease (PD), which results from the degeneration of midbrain dopaminergic neurons, was the first brain pathology that was related to Nr4a2. Early reports showed that Nr4a2 expression was diminished in both aged and PD brains (Chu et al., 2002). Later, Nr4a2 gene expression was found reduced not only in the postmortem brain tissue, but also in the peripheral blood of PD patients (Le et al., 2008; Montarolo et al., 2016). Moreover, Nr4a2 mutations or polymorphisms were detected in sporadic and familial forms of PD (Xu et al., 2002; Le et al., 2003; Zheng et al., 2003; Grimes et al., 2006; Sleiman et al., 2009). Besides PD, evidence supports a role for Nr4a2 in the pathogenesis of different CNS disorders (Figure 3). In recent years, much attention has been directed to Nr4a2 function in hippocampal synaptic plasticity, learning, and memory, addressing much interest to the role of Nr4a2 in brain pathologies that course with cognitive or intellectual disabilities. One of the first brain diseases, besides PD, that was related to Nr4a2 was Alzheimer’s disease (AD).

Figure 3. Nr4a2 involvement in different cognitive and neuropsychiatric brain disorders. Schematic representation of Nr4a2 functions and/or alterations in neurodegenerative disorders (Parkinson’s disease, Alzheimer’s disease), schizophrenia and autism spectrum disorder.
The potential of some nuclear receptors to serve as therapeutic targets in disorders of cognition including AD has been reported (Mandrekar-Colucci and Landreth, 2011; Skerrett et al., 2014). CREB is one of the transcription factors most extensively studied in AD, being disrupted by oligomeric forms of amyloid-β (oAβ) and leading to synaptic plasticity and memory deficits in AD (Vitolo et al., 2002; Saura and Valero, 2011). Moreover, CREB transcription factor represents the crossroad of different synapse-to-nucleus pathways associated with changes in gene expression that underlie memory decline in AD (Marcello et al., 2018). Pioneer genome-wide transcriptome profile analyses of AD transgenic mice hippocampus revealed deregulation of a transcriptional program dependent on CREB/CRTC1. Specifically, APPSw,Ind mice at 6 months of age showed down-regulation of diverse genes compared with wild type mice after spatial training. These genes include Nr4a2 (España et al., 2010; Parra-Damas et al., 2014). Indeed, a specific decrease of Nr4a2 levels in Alzheimer’s pathology has been observed. Nr4a2 mRNA levels were found diminished in Aβ1–42-treated neuronal cells (Terzioglu-Usak et al., 2017) and in AD mouse models. A reduction in Nr4a2 mRNA was also seen in APPSw,Ind mice when initial hippocampal-dependent spatial memory deficits appeared (España et al., 2010; Parra-Damas et al., 2014). A similar decrease was also reported in postmortem brains of human AD patients, specifically in the frontal cortex, and the hippocampal formation (Parra-Damas et al., 2014; Moon et al., 2019). A role of Nr4a2 in AD pathology was further supported by the observation that Nr4a2 protein was prominently expressed in brain areas with Aβ accumulation in the 5XFAD mouse model of AD and, notably, it was highly co-expressed with Aβ at mice ages mimicking early stages of the disease (Oakley et al., 2006). In addition, the number of Nr4a2-expressing cells significantly declined in the 5XFAD mouse in an age-dependent manner, accompanied by increased plaque deposition, suggesting a possible causal-effect relation between Nr4a2 levels and AD progression (Moon et al., 2015). Moreover, in a recent study performed by the same laboratory (Moon et al., 2019), they found that knockdown of Nr4a2 significantly aggravated AD pathology while its overexpression alleviated it, including a decrease in Aβ accumulation and neurodegeneration. Similar observations were obtained in 5XFAD mice treated with Nr4a2 activators.
It is noteworthy to mention that the functional roles of Nr4a2 in AD could not be limited to its implication in synaptic function, but also be extended to its involvement in the inflammatory and neuronal death processes associated to AD. For example, it was described that inflammatory stimulus up-regulated Nr4a2 expression in microglia (Fan et al., 2009). This up-regulation could trigger a compensatory response through modulation of the NFκB action. Nr4a2 binds to target inflammatory gene promoters thought association with NFκB and the CoREST repressor complex down-regulating the expression of several pro-inflammatory genes in microglia and astrocytes (Saijo et al., 2009). Furthermore, Nr4a2 neuroprotective effects are not limited to dopaminergic neurons. In glutamatergic neurons, Nr4a2 also mediates CREB-induced neuroprotection in response to stress by increasing BDNF levels in cerebellar granule cells (Barneda-Zahonero et al., 2012) or up-regulating an anti-apoptotic gene program in hippocampal neurons (Volakakis et al., 2010). Altogether, this anti-inflammatory and pro-survival role in brain cells could also be involved in the reduction of neuroinflammation and neuronal death seen in AD mice models (Moon et al., 2019).
Some reports have also associated Nr4a2 with schizophrenia. This psychiatric disease is believed to result from subtle disturbances during brain development that lead to improper function of synaptic transmission and plasticity of several neurotransmitter systems (Birnbaum and Weinberger, 2017). A consistent neuropathological observation in postmortem brains of schizophrenic patients is the reduction in the density of small dendritic spines (MacDonald et al., 2017), which are more plastic than bigger spines and, therefore, they are related to learning and memory processes (Matsuzaki et al., 2001; Trachtenberg et al., 2002). Missense mutations in the third exon of Nr4a2 (Buervenich et al., 2000) or altered expression of Nr4a2 in postmortem prefrontal cortex were detected in schizophrenic patients (Guillozet-Bongaarts et al., 2014; Corley et al., 2016). Moreover, schizophrenic patients homozygous for variants of the Nr4a2 promoter showed impairments in short-term memory assessed by the Backward Digit Span task (Ruiz-Sánchez et al., 2021). A putative relationship between Nr4a2 and schizophrenia was further supported by studies with rodents. Nr4a2 heterozygous mice displayed a pleiad of symptoms and behaviors related to schizophrenia (Rojas et al., 2007; Vuillermot et al., 2011). These mice showed hyperactivity in novel environments and were deficient in the passive avoidance learning task. Moreover, a depression-like profile was also noticed in the forced-swimming test (Rojas et al., 2007). The behavioral profile of Nr4a2 heterozygous mice regarding schizophrenia-relevant phenotypes was further extended by Vuillermot et al. (2011). They reported that Nr4a2-deficient male mice show sensorimotor gating deficits consistent with an alteration in the glutamatergic system. This deficit in sensorimotor gating has been consistently reported in schizophrenia patients (Swerdlow and Geyer, 1998; Braff et al., 2001). Altogether, these findings indicate that Nr4a2-deficient mice display some phenotypic characteristics of schizophrenia-like dysfunction.
The presence of Nr4a2 deletions in patients with autism spectrum disorder has also related Nr4a2 with intellectual disability. In some reports, the deletion affected Nr4a2 and GPD2 genes and caused intellectual disability and language impairment (Barge-Schaapveld et al., 2013; Leppa Virpi et al., 2016). However, other studies reported an individual with mild intellectual disability and prominent speech and language impairment that presented a deletion encompassing only Nr4a2 (Reuter et al., 2017; Lévy et al., 2018). These studies provide further evidence for Nr4a2 haploinsufficiency being associated with intellectual disability and autism spectrum disorder.
All these studies clearly show that the involvement of Nr4a2 in brain diseases expands beyond PD and point out this transcription factor as a promising candidate for the development of novel therapeutic approaches. Future work should provide solid grounds to determine the precise contribution of Nr4a2 to each brain disorder.
As discussed above, until recently, it was considered that Nr4a family members were orphan nuclear receptors since their LBD seemed to avoid the binding of ligands due to the tight packing of bulky hydrophobic side chain residues (Wang et al., 2003). However, during the last decade, several nuclear magnetic resonance (NMR) studies of the isolated Nr4a2-LBD have suggested that several synthetic small molecules could bind Nr4a2 in canonical and non-canonical binding pockets (Kim et al., 2015; de Vera et al., 2019). Moreover, recent studies have shown that the Nr4a2-LBD could be highly dynamic (de Vera et al., 2019) allowing the binding of putative endogenous ligands such as docosahexaenoic acid, prostaglandin E1 and the dopamine metabolite 5,6-dihydroxyndole (de Vera et al., 2016; Bruning et al., 2019; Rajan et al., 2020). At present, it is still unknown the physiological importance of these putative endogenous ligands on Nr4a2-mediated role in the CNS.
The identification of synthetic small molecules that could modulate Nr4a2 transcriptional activity opens the possibility to provide target-based therapies for brain diseases related to Nr4a2. Among the panoply of Nr4a2 modulators identified so far (Table 2), of special interest are the antimalarial drugs amodiaquine (AQ) and chloroquine (CQ), which increase the transcriptional function of Nr4a2 interacting with its LBD through direct physical binding. These drugs were identified as Nr4a2 agonists after screening a chemical library composed by Food and Drug Administration (FDA)-approved drugs in a cell-based luciferase assay system (Kim et al., 2015). NMR spectroscopy suggested the binding of both drugs to the Nr4a2-LBD. Accordingly, AQ and CQ were able to increase the transcriptional activity of Nr4a2 in human neuroblastoma cell lines (Kim et al., 2015). In addition, in preclinical studies, these compounds meaningfully improved behavioral deficits in rat models of PD without any noticeable sign of dyskinesia-like behavior (Kim et al., 2015). Autophagic-lysosomal blockade was also reported as one of their functions (Qiao et al., 2013). Recently, AQ has also been found to enhance cognitive functions by increasing adult hippocampal neurogenesis (Kim et al., 2016). Importantly, 5XFAD mouse model of AD treated with the Nr4a2 agonist AQ showed robust reduction in typical AD features including deposition of Aβ plaques, neuronal loss, microgliosis and impairment of adult hippocampal neurogenesis, leading to significant improvement of cognitive functions in the Y-maze, which is a behavioral paradigm widely accepted to evaluate spatial working memory. The same study also showed for the first time that AQ treatment significantly inhibited γ-secretase activity and enhanced degradation of Aβ via up-regulation of insulin-degrading enzyme, an Aβ-degrading protease (Moon et al., 2019).
Mercaptopurine was the first Nr4a2 agonist to be identified (Ordentlich et al., 2003). However, although it has been reported that produces anti-inflammatory responses in microglia by attenuation of TNF-α production (Huang et al., 2016), it is not considered a specific Nr4a2 agonist (Wansa et al., 2003; Huang et al., 2016). Other bicyclic compounds described to enhance Nr4a2 transcriptional activity as homodimer or heterodimer are isoxazolopyridinone-based compounds developed by Novartis AG (Hintermann et al., 2007). Among them, isoxazolo-pyridinone 7e (IP7e) has an excellent oral bioavailability and a fast and extensive brain uptake and has been reported to ameliorate neuroinflammation and neurodegeneration in an experimental model of autoimmune encephalomyelitis (Montarolo et al., 2014). However, a recent study suggests that IP7e affects general transcription via Nr4a2-independent mechanisms (Munoz-Tello et al., 2020) seeding doubts about its role as a specific Nr4a2 agonist. Another bicyclic compound, SA00025, a 2-aryl-6-phenylimidazol[1,2-α]pyridine derivative produced by Sanofi Aventis, was reported to exhibit partial neuroprotective effect in PD mouse models (Smith et al., 2015). Although ligand screening shows an EC50 on the low nanomolar range (Jang et al., 2021), no data have shown its direct binding to Nr4a2.
Another group of small molecules that have been identified to specifically activate members of the Nr4a family of transcription factors are the paraphenyl substituted diindolylmethane analogs or C-DIM (Li et al., 2012). Some of the C-DIM compounds, as C-DIM5 and C-DIM8, have a greater specificity for Nr4a1 than for Nr4a2. By contrast, C-DIM12 has higher affinity for Nr4a2 and it is able to exert a potent anti-inflammatory and neuroprotective role in a mouse model of PD (De Miranda et al., 2015). Moreover, it enhances long-term spatial memory in young mice and rescues memory deficits in aged mice (Chatterjee et al., 2020). In summary, the possibility that Nr4a2 could be drug-targeted with several small synthetic molecules opens an attractive and promising future for developing therapeutic strategies that include Nr4a2 as a druggable target.
During the last decade, many reports have supported a major role for the Nr4a family of nuclear receptors in hippocampal synaptic plasticity and memory processes. The role of Nr4a2, a member of this family, has received special attention. Several data suggest that Nr4a2 activation is necessary for hippocampal LTP and hippocampal-mediated learning tasks. However, many questions remain unanswered. It is still unknown which are the signaling pathways that link synaptic activity to Nr4a2 induction. We don’t know yet how Nr4a2 activation is enhancing hippocampal plasticity and learning. Which are the genes activated or repressed by Nr4a2? Does Nr4a2 act on excitatory and/or inhibitory synapses? Is Nr4a2 modulating the pre and/or postsynaptic function? Which impact has glial Nr4a2 in synaptic plasticity? Work during the following years will confirm whether Nr4a2 is present at the synapse and could act as a synaptonuclear messenger and how its subcellular compartmentalization is modulated.
Future studies should improve our knowledge of the role of Nr4a2 in the cognitive impairment associated to several brain pathologies. More pre-clinical studies are needed to confirm the importance of Nr4a2 activation in the hippocampus to overcome the learning and memory deficits observed in animal models of AD. It is necessary to extend the studies on the role of Nr4a2 in AD beyond the 5XFAD mouse model. Also, an extended behavioral characterization of learning and memory in these animals is needed. It would be also relevant to know whether Nr4a2 activation has any impact on Aβ plaques and neurofibrillary tangles, which are the neurohistological hallmarks of AD. Moreover, the relationship between Nr4a2 and some psychiatric diseases has been already established. Thus, it would be also important to address the impact of Nr4a2 on behavioral and psychological symptoms with new studies in animals’ models for these diseases such as schizophrenia or autism spectrum disorder.
All these studies will benefit from the already known Nr4a2 ligands that allow us to consider Nr4a2 as a druggable target. However, to provide future target-based therapeutic interventions, more work should be done to modify existing chemical structures to increase the specificity of Nr4a2 agonists, increase their blood-brain barrier permeability and improve their pharmacokinetic characteristics.
Finally, another important question needs to be answered during the next decade. Is Nr4a2 an orphan nuclear receptor? Although this question has been considered affirmative until now, recent studies suggest that endogenous ligands could indeed bind to Nr4a2-LBD. Future confirmation of the existence of such endogenous ligands will open an important scenario, since additional mechanisms, besides protein levels, could regulate Nr4a2 activity.
JC-S, AM-M, and JR-Á conceived the idea for this review and reviewed the literature. JC-S wrote the first draft of the manuscript and created the figures. JR-Á wrote the final draft. JC-S and AM-M revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the grants from Ministerio de Ciencia, Innovación y Universidades (SAF2017-89271-R and PID2020-117510RB-I00), CIBERNED (CB06/05/0042) and Generalitat de Catalunya (SGR2017-0749) to JR-Á. JC-S was a recipient of a Fellowship from the Ministerio de Ciencia, Innovación y Universidades (FPU-14/05392).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abraham, W. C., Jones, O. D., and Glanzman, D. L. (2019). Is plasticity of synapses the mechanism of long-term memory storage? NPJ Sci. Learn. 4:9.
Ahn, J. H., Lee, J. S., Cho, J. H., Park, J. H., Lee, T. K., Song, M., et al. (2018). Age-dependent decrease of Nurr1 protein expression in the gerbil hippocampus. Biomed. Rep. 8, 517–522. doi: 10.3892/br.2018.1094
Alberini, C. M. (2009). Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 89, 121–145. doi: 10.1152/physrev.00017.2008
Aldavert-Vera, L., Huguet, G., Costa-Miserachs, D., Ortiz, S. P. D., Kádár, E., Morgado-Bernal, I., et al. (2013). Intracranial self-stimulation facilitates active-avoidance retention and induces expression of c-Fos and Nurr1 in rat brain memory systems. Behav. Brain Res. 250, 46–57. doi: 10.1016/j.bbr.2013.04.025
Altarejos, J. Y., and Montminy, M. (2011). CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151. doi: 10.1038/nrm3072
Barge-Schaapveld, D. Q. C. M., Ofman, R., Knegt, A. C., Alders, M., Höhne, W., Kemp, S., et al. (2013). Intellectual disability and hemizygous GPD2 mutation. Am. J. Med. Genet. Part A 161, 1044–1050. doi: 10.1002/ajmg.a.35873
Barneda-Zahonero, B., Servitja, J.-M., Badiola, N., Miñano-Molina, A. J., Fadó, R., Saura, C. A., et al. (2012). Nurr1 protein is required for N-Methyl-d-Aspartic Acid (n.d.) receptor-mediated neuronal survival. J. Biol. Chem. 287, 11351–11362. doi: 10.1074/jbc.M111.272427
Bartsch, D., Casadio, A., Karl, K. A., Serodio, P., and Kandel, E. R. (1998). CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 95, 211–223. doi: 10.1016/s0092-8674(00)81752-3
Bekinschtein, P., Cammarota, M., and Medina, J. H. (2014). BDNF and memory processing. Neuropharmacology 76, 677–683. doi: 10.1016/j.neuropharm.2013.04.024
Benito, E., and Barco, A. (2015). The neuronal activity-driven transcriptome. Mol. Neurobiol. 51, 1071–1088.
Bensinger, S. J., and Tontonoz, P. (2009). A Nurr1 Pathway for Neuroprotection. Cell 137, 26–28. doi: 10.1016/j.cell.2009.03.024
Birnbaum, R., and Weinberger, D. R. (2017). Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 18, 727–740. doi: 10.1038/nrn.2017.125
Bourtchuladze, R., Frenguelli, B., Blendy, J., Cioffi, D., Schutz, G., and Silva, A. J. (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. doi: 10.1016/0092-8674(94)90400-6
Braff, D. L., Geyer, M. A., and Swerdlow, N. R. (2001). Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology 156, 234–258. doi: 10.1007/s002130100810
Bridi, M. S., and Abel, T. (2013). The NR4A orphan nuclear receptors mediate transcription-dependent hippocampal synaptic plasticity. Neurobiol. Learn. Mem. 105, 151–158. doi: 10.1016/j.nlm.2013.06.020
Bridi, M. S., Hawk, J. D., Chatterjee, S., Safe, S., and Abel, T. (2017). Pharmacological activators of the NR4A nuclear receptors enhance LTP in a CREB/CBP-dependent manner. Neuropsychopharmacology 42, 1243–1253. doi: 10.1038/npp.2016.253
Bruning, J. M., Wang, Y., Oltrabella, F., Tian, B., Kholodar, S. A., Liu, H., et al. (2019). Covalent modification and regulation of the nuclear receptor Nurr1 by a dopamine metabolite. Cell Chem. Biol. 26, 674.e676–685.e676. doi: 10.1016/j.chembiol.2019.02.002
Buervenich, S., Carmine, A., Arvidsson, M., Xiang, F., Zhang, Z., Sydow, O., et al. (2000). NURR1 Mutations in cases of schizophrenia and manic-depressive disorder. Am. J. Med. Genet. 96, 808–813. doi: 10.1002/1096-8628(20001204)96:6<808::aid-ajmg23>3.0.co;2-e
Chatterjee, S., Walsh, E. N., Yan, A. L., Giese, K. P., Safe, S., and Abel, T. (2020). Pharmacological activation of Nr4a rescues age-associated memory decline. Neurobiol. Aging 85, 140–144. doi: 10.1016/j.neurobiolaging.2019.10.001
Chen, X.-X., Qian, Y., Wang, X.-P., Tang, Z.-W., Xu, J.-T., Lin, H., et al. (2018). Nurr1 promotes neurogenesis of dopaminergic neuron and represses inflammatory factors in the transwell coculture system of neural stem cells and microglia. CNS Neurosci. Therapeutics 24, 790–800. doi: 10.1111/cns.12825
Ch’ng, T. H., and Martin, K. C. (2011). Synapse-to-nucleus signaling. Curr. Opin. Neurobiol. 21, 345–352.
Ch’ng Toh, H., Uzgil, B., Lin, P., Avliyakulov Nuraly, K., O’Dell Thomas, J., and Martin Kelsey, C. (2012). Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell 150, 207–221.
Chu, Y., Kompoliti, K., Cochran, E. J., Mufson, E. J., and Kordower, J. H. (2002). Age-related decreases in Nurr1 immunoreactivity in the human substantia nigra. J. Comparat. Neurol. 450, 203–214. doi: 10.1002/cne.10261
Chuang, H.-C., Huang, T.-N., and Hsueh, Y.-P. (2015). T-Brain-1 – A potential master regulator in autism spectrum disorders. Autism Res. 8, 412–426. doi: 10.1002/aur.1456
Colón-Cesario, W. I., Martínez-Montemayor, M. M., Morales, S., Félix, J., Cruz, J., Adorno, M., et al. (2006). Knockdown of Nurr1 in the rat hippocampus: Implications to spatial discrimination learning and memory. Learn. Mem. 13, 734–744. doi: 10.1101/lm.407706
Corley, S. M., Tsai, S.-Y., Wilkins, M. R., and Shannon Weickert, C. (2016). Transcriptomic analysis shows decreased cortical expression of NR4A1, NR4A2 and RXRB in schizophrenia and provides evidence for nuclear receptor dysregulation. PLoS One 11:e0166944. doi: 10.1371/journal.pone.0166944
Darcy, M. J., Calvin, K., Cavnar, K., and Ouimet, C. C. (2010). Regional and subcellular distribution of HDAC4 in mouse brain. J. Comparat. Neurol. 518, 722–740. doi: 10.1002/cne.22241
De Miranda, B. R., Popichak, K. A., Hammond, S. L., Jorgensen, B. A., Phillips, A. T., Safe, S., et al. (2015). The Nurr1 activator 1,1-Bis(3′-Indolyl)-1-(p-Chlorophenyl)methane blocks inflammatory gene expression in BV-2 microglial cells by inhibiting nuclear factor κB. Mol. Pharmacol. 87, 1021–1034. doi: 10.1124/mol.114.095398
de Vera, I. M. S., Giri, P. K., Munoz-Tello, P., Brust, R., Fuhrmann, J., Matta-Camacho, E., et al. (2016). Identification of a binding site for unsaturated fatty acids in the orphan nuclear receptor Nurr1. ACS Chem. Biol. 11, 1795–1799. doi: 10.1021/acschembio.6b00037
de Vera, I. M. S., Munoz-Tello, P., Zheng, J., Dharmarajan, V., Marciano, D. P., Matta-Camacho, E., et al. (2019). Defining a canonical ligand-binding pocket in the orphan nuclear receptor Nurr1. Structure 27, 66.e65–77.e65.
Decressac, M., Volakakis, N., Björklund, A., and Perlmann, T. (2013). NURR1 in Parkinson disease—from pathogenesis to therapeutic potential. Nat. Rev. Neurol. 9, 629–636. doi: 10.1038/nrneurol.2013.209
Dunah, A. W., Hueske, E., Wyszynski, M., Hoogenraad, C. C., Jaworski, J., Pak, D. T., et al. (2005). LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat. Neurosci. 8, 458–467. doi: 10.1038/nn1416
España, J., Valero, J., Miñano-Molina, A. J., Masgrau, R., Martín, E., Guardia-Laguarta, C., et al. (2010). β-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 30, 9402–9410. doi: 10.1523/JNEUROSCI.2154-10.2010
Fan, X., Luo, G., Ming, M., Pu, P., Li, L., Yang, D., et al. (2009). Nurr1 expression and its modulation in Microglia. Neuroimmunomodulation 16, 162–170. doi: 10.1159/000204229
Fleischmann, A., Hvalby, O., Jensen, V., Strekalova, T., Zacher, C., Layer, L. E., et al. (2003). Impaired long-term memory and NR2A-Type NMDA receptor-dependent synaptic plasticity in mice lacking c-fos in the CNS. J. Neurosci. 23:9116. doi: 10.1523/JNEUROSCI.23-27-09116.2003
García-Yagüe, Á. J., Rada, P., Rojo, A. I., Lastres-Becker, I., and Cuadrado, A. (2013). Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J. Biol. Chem. 288, 5506–5517. doi: 10.1074/jbc.M112.439190
Grimes, D. A., Han, F., Panisset, M., Racacho, L., Xiao, F., Zou, R., et al. (2006). Translated mutation in the Nurr1 gene as a cause for Parkinson’s disease. Mov. Disord. 21, 906–909.
Guillozet-Bongaarts, A. L., Hyde, T. M., Dalley, R. A., Hawrylycz, M. J., Henry, A., Hof, P. R., et al. (2014). Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry 19, 478–485. doi: 10.1038/mp.2013.30
Hagenston, A. M., and Bading, H. (2011). Calcium signaling in synapse-to-nucleus communication. Cold Spring Harb. Perspect. Biol. 3:a004564.
Hammond, S. L., Safe, S., and Tjalkens, R. B. (2015). A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neurosci. Lett. 607, 83–89. doi: 10.1016/j.neulet.2015.09.015
Hawk, J. D., and Abel, T. (2011). The role of NR4A transcription factors in memory formation. Brain Res. Bull. 85, 21–29. doi: 10.1016/j.brainresbull.2011.02.001
Hawk, J. D., Bookout, A. L., Poplawski, S. G., Bridi, M., Rao, A. J., Sulewski, M. E., et al. (2012). NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J. Clin. Investigat. 122, 3593–3602.
Heo, S., Diering, G. H., Na, C. H., Nirujogi, R. S., Bachman, J. L., Pandey, A., et al. (2018). Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc. Natl. Acad. Sci. U.S.A. 115:E3827. doi: 10.1073/pnas.1720956115
Hintermann, S., Chiesi, M., von Krosigk, U., Mathé, D., Felber, R., and Hengerer, B. (2007). Identification of a series of highly potent activators of the Nurr1 signaling pathway. Bioorganic Med. Chem. Lett. 17, 193–196. doi: 10.1016/j.bmcl.2006.09.062
Huang, H.-Y., Chang, H.-F., Tsai, M.-J., Chen, J.-S., and Wang, M.-J. (2016). 6-Mercaptopurine attenuates tumor necrosis factor-α production in microglia through Nur77-mediated transrepression and PI3K/Akt/mTOR signaling-mediated translational regulation. J. Neuroinflamm. 13:78. doi: 10.1186/s12974-016-0543-5
Ichinose, H., Ohye, T., Suzuki, T., Sumi-Ichinose, C., Nomura, T., Hagino, Y., et al. (1999). Molecular cloning of the human Nurr1 gene: characterization of the human gene and cDNAs. Gene 230, 233–239. doi: 10.1016/s0378-1119(99)00065-7
Jang, Y., Kim, W., Leblanc, P., Kim, C.-H., and Kim, K.-S. (2021). Potent synthetic and endogenous ligands for the adopted orphan nuclear receptor Nurr1. Exp. Mol. Med. 53, 19–29. doi: 10.1038/s12276-021-00555-5
Jeon, S. G., Yoo, A., Chun, D. W., Hong, S. B., Chung, H., Kim, J.-I., et al. (2020). The critical role of Nurr1 as a mediator and therapeutic target in Alzheimer’s disease-related pathogenesis. Aging Dis. 11, 705–724. doi: 10.14336/AD.2019.0718
Jiang, L., Dai, S., Li, J., Liang, X., Qu, L., Chen, X., et al. (2019). Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements. J. Biol. Chem. 294, 19795–19803. doi: 10.1074/jbc.RA119.010730
Jo, A. Y., Kim, M.-Y., Lee, H.-S., Rhee, Y.-H., Lee, J.-E., Baek, K.-H., et al. (2009). Generation of dopamine neurons with improved cell survival and phenotype maintenance using a degradation-resistant Nurr1 mutant. Stem Cells 27, 2238–2246. doi: 10.1002/stem.146
Kadkhodaei, B., Ito, T., Joodmardi, E., Mattsson, B., Rouillard, C., Carta, M., et al. (2009). Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J. Neurosci. 29:15923.
Kaldun, J. C., and Sprecher, S. G. (2019). Initiated by CREB: resolving gene regulatory programs in learning and memory. BioEssays 41:1900045. doi: 10.1002/bies.201900045
Kaltschmidt, C., Kaltschmidt, B., and Baeuerle, P. A. (1993). Brain synapses contain inducible forms of the transcription factor NF-κB. Mechan. Dev. 43, 135–147. doi: 10.1016/0925-4773(93)90031-r
Keeley, M. B., Wood, M. A., Isiegas, C., Stein, J., Hellman, K., Hannenhalli, S., et al. (2006). Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn. Mem. 13, 135–142. doi: 10.1101/lm.86906
Kim, C.-H., Han, B.-S., Moon, J., Kim, D.-J., Shin, J., Rajan, S., et al. (2015). Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 112, 8756–8761. doi: 10.1073/pnas.1509742112
Kim, J.-I., Jeon, S. G., Kim, K. A., Kim, Y. J., Song, E. J., Choi, J., et al. (2016). The pharmacological stimulation of Nurr1 improves cognitive functions via enhancement of adult hippocampal neurogenesis. Stem Cell Res. 17, 534–543. doi: 10.1016/j.scr.2016.09.027
Kim, S. M., Cho, S. Y., Kim, M. W., Roh, S. R., Shin, H. S., Suh, Y. H., et al. (2020). Genome-Wide analysis identifies NURR1-controlled network of new synapse formation and cell cycle arrest in human neural stem cells. Mol. Cells 43, 551–571. doi: 10.14348/molcells.2020.0071
Klann, E., and Dever, T. E. (2004). Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 5, 931–942.
Kovács, K. A., Steullet, P., Steinmann, M., Do, K. Q., Magistretti, P. J., Halfon, O., et al. (2007). TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 104:4700. doi: 10.1073/pnas.0607524104
Kwapis, J. L., Alaghband, Y., López, A. J., Long, J. M., Li, X., Shu, G., et al. (2019). HDAC3-mediated repression of the &It;em>Nr4a&It;/em> family contributes to age-related impairments in long-term memory. J. Neurosci. 39:4999.
Lai, K.-O., Zhao, Y., Ch’ng, T. H., and Martin, K. C. (2008). Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl. Acad. Sci. U.S.A. 105:17175. doi: 10.1073/pnas.0803906105
Le, W., Pan, T., Huang, M., Xu, P., Xie, W., Zhu, W., et al. (2008). Decreased NURR1 gene expression in patients with Parkinson’s disease. J. Neurol. Sci. 273, 29–33.
Le, W.-D., Conneely, O. M., Zou, L., He, Y., Saucedo-Cardenas, O., Jankovic, J., et al. (1999). Selective agenesis of mesencephalic dopaminergic neurons in Nurr1-Deficient mice. Exp. Neurol. 159, 451–458. doi: 10.1006/exnr.1999.7191
Le, W.-D., Xu, P., Jankovic, J., Jiang, H., Appel, S. H., Smith, R. G., et al. (2003). Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 33, 85–89.
Leppa Virpi, M., Kravitz Stephanie, N., Martin Christa, L., Andrieux, J., Le Caignec, C., Martin-Coignard, D., et al. (2016). Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am. J. Hum. Genet. 99, 540–554. doi: 10.1016/j.ajhg.2016.06.036
Lévy, J., Grotto, S., Mignot, C., Maruani, A., Delahaye-Duriez, A., Benzacken, B., et al. (2018). NR4A2 haploinsufficiency is associated with intellectual disability and autism spectrum disorder. Clin. Genet. 94, 264–268. doi: 10.1111/cge.13383
Li, X., Lee, S.-O., and Safe, S. (2012). Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem. Pharmacol. 83, 1445–1455. doi: 10.1016/j.bcp.2012.02.021
MacDonald, M. L., Alhassan, J., Newman, J. T., Richard, M., Gu, H., Kelly, R. M., et al. (2017). Selective loss of smaller spines in schizophrenia. Am. J. Psychiatry 174, 586–594. doi: 10.1176/appi.ajp.2017.16070814
Mandrekar-Colucci, S., and Landreth, G. E. (2011). Nuclear receptors as therapeutic targets for Alzheimer’s disease. Expert Opin. Therapeutic Targets 15, 1085–1097. doi: 10.1517/14728222.2011.594043
Marcello, E., Di Luca, M., and Gardoni, F. (2018). Synapse-to-nucleus communication: from developmental disorders to Alzheimer’s disease. Curr. Opin. Neurobiol. 48, 160–166. doi: 10.1016/j.conb.2017.12.017
Martin, S. J., Grimwood, P. D., and Morris, R. G. M. (2000). Synaptic plasticity and memory: an evaluation of the hypothesis. Annu. Rev. Neurosci. 23, 649–711.
Matsuzaki, M., Ellis-Davies, G. C. R., Nemoto, T., Miyashita, Y., Iino, M., and Kasai, H. (2001). Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4, 1086–1092. doi: 10.1038/nn736
Maxwell, M. A., and Muscat, G. E. (2006). The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 4:e002. doi: 10.1621/nrs.04002
McNamara, R. K., Hussain, R. J., Simon, E. J., Stumpo, D. J., Blackshear, P. J., Abel, T., et al. (2005). Effect of myristoylated alanine-rich C kinase substrate (MARCKS) overexpression on hippocampus-dependent learning and hippocampal synaptic plasticity in MARCKS transgenic mice. Hippocampus 15, 675–683. doi: 10.1002/hipo.20089
McNulty, S. E., Barrett, R. M., Vogel-Ciernia, A., Malvaez, M., Hernandez, N., Davatolhagh, M. F., et al. (2012). Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn. Mem. 19, 588–592. doi: 10.1101/lm.026385.112
McQuown, S. C., Barrett, R. M., Matheos, D. P., Post, R. J., Rogge, G. A., Alenghat, T., et al. (2011). HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 31:764.
Meffert, M. K., Chang, J. M., Wiltgen, B. J., Fanselow, M. S., and Baltimore, D. (2003). NF-κB functions in synaptic signaling and behavior. Nat. Neurosci. 6, 1072–1078. doi: 10.1038/nn1110
Miranda, M., Morici, J. F., Zanoni, M. B., and Bekinschtein, P. (2019). Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain. Front. Cell. Neurosci. 13:363. doi: 10.3389/fncel.2019.00363
Montarolo, F., Perga, S., Martire, S., Navone, D. N., Marchet, A., Leotta, D., et al. (2016). Altered NR4A subfamily gene expression level in peripheral blood of Parkinson’s and Alzheimer’s disease patients. Neurotoxicity Res. 30, 338–344. doi: 10.1007/s12640-016-9626-4
Montarolo, F., Raffaele, C., Perga, S., Martire, S., Finardi, A., Furlan, R., et al. (2014). Effects of isoxazolo-pyridinone 7e, a potent activator of the Nurr1 signaling pathway, on experimental autoimmune encephalomyelitis in mice. PLoS One 9:e108791. doi: 10.1371/journal.pone.0108791
Moon, M., Jeong, I., Kim, C.-H., Kim, J., Lee, P. K. J., Mook-Jung, I., et al. (2015). Correlation between orphan nuclear receptor Nurr1 expression and amyloid deposition in 5XFAD mice, an animal model of Alzheimer’s disease. J. Neurochem. 132, 254–262. doi: 10.1111/jnc.12935
Moon, M., Jung, E. S., Jeon, S. G., Cha, M.-Y., Jang, Y., Kim, W., et al. (2019). Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell 18:e12866. doi: 10.1111/acel.12866
Munoz-Tello, P., Lin, H., Khan, P., de Vera, I. M. S., Kamenecka, T. M., and Kojetin, D. J. (2020). Assessment of NR4A ligands that directly bind and modulate the orphan nuclear receptor Nurr1. J. Med. Chem. 63, 15639–15654. doi: 10.1021/acs.jmedchem.0c00894
Nonaka, M., Kim, R., Fukushima, H., Sasaki, K., Suzuki, K., Okamura, M., et al. (2014). Region-Specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron 84, 92–106. doi: 10.1016/j.neuron.2014.08.049
Oakley, H., Cole, S. L., Logan, S., Maus, E., Shao, P., Craft, J., et al. (2006). Intraneuronal β-Amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129. doi: 10.1523/JNEUROSCI.1202-06.2006
Oliveira, A., Martinho, R., Serrão, P., and Moreira-Rodrigues, M. (2018). Epinephrine released during traumatic events may strengthen contextual fear memory through increased hippocampus mRNA expression of Nr4a transcription factors. Front. Mol. Neurosci. 11:334. doi: 10.3389/fnmol.2018.00334
Ordentlich, P., Yan, Y., Zhou, S., and Heyman, R. A. (2003). Identification of the antineoplastic agent 6-Mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1*. J. Biol. Chem. 278, 24791–24799. doi: 10.1074/jbc.M302167200
Parra-Damas, A., Chen, M., Enriquez-Barreto, L., Ortega, L., Acosta, S., Perna, J. C., et al. (2017a). CRTC1 function during memory encoding is disrupted in neurodegeneration. Biol. Psychiatry 81, 111–123. doi: 10.1016/j.biopsych.2016.06.025
Parra-Damas, A., Rubió-Ferrarons, L., Shen, J., and Saura, C. A. (2017b). CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters. Sci. Rep. 7:18004. doi: 10.1038/s41598-017-18215-y
Parra-Damas, A., Valero, J., Chen, M., España, J., Martín, E., Ferrer, I., et al. (2014). Crtc1 activates a transcriptional program deregulated at early Alzheimer's disease-related stages. J. Neurosci. 34:5776. doi: 10.1523/JNEUROSCI.5288-13.2014
Pegoraro, S., Broccard, F. D., Ruaro, M. E., Bianchini, D., Avossa, D., Pastore, G., et al. (2010). Sequential steps underlying neuronal plasticity induced by a transient exposure to gabazine. J. Cell. Physiol. 222, 713–728. doi: 10.1002/jcp.21998
Peña de Ortiz, S., Maldonado-Vlaar, C. S., and Carrasquillo, Y. (2000). Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol. Learn. Mem. 74, 161–178. doi: 10.1006/nlme.1999.3952
Qiao, S., Tao, S., Rojo de la Vega, M., Park, S. L., Vonderfecht, A. A., Jacobs, S. L., et al. (2013). The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation- and chemotherapy-induced cell death. Autophagy 9, 2087–2102. doi: 10.4161/auto.26506
Quina, L. A., Wang, S., Ng, L., and Turner, E. E. (2009). Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J. Neurosci. 29:14309. doi: 10.1523/JNEUROSCI.2430-09.2009
Rajan, S., Jang, Y., Kim, C. H., Kim, W., Toh, H. T., Jeon, J., et al. (2020). PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat. Chem. Biol. 16, 876–886. doi: 10.1038/s41589-020-0553-6
Ranhotra, H. S. (2015). The NR4A orphan nuclear receptors: mediators in metabolism and diseases. J. Receptors Signal Transduct. 35, 184–188. doi: 10.3109/10799893.2014.948555
Reuter, M. S., Krumbiegel, M., Schlüter, G., Ekici, A. B., Reis, A., and Zweier, C. (2017). Haploinsufficiency of NR4A2 is associated with a neurodevelopmental phenotype with prominent language impairment. Am. J. Med. Genet. Part A 173, 2231–2234. doi: 10.1002/ajmg.a.38288
Rojas, P., Joodmardi, E., Hong, Y., Perlmann, T., and Ögren, S. O. (2007). Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol. Psychiatry 12, 756–766. doi: 10.1038/sj.mp.4001993
Ruiz-Sánchez, E., Jiménez-Genchi, J., Alcántara-Flores, Y. M., Castañeda-González, C. J., Aviña-Cervantes, C. L., Yescas, P., et al. (2021). Working memory deficits in schizophrenia are associated with the rs34884856 variant and expression levels of the NR4A2 gene in a sample Mexican population: a case control study. BMC Psychiatry 21:86. doi: 10.1186/s12888-021-03081-w
Safe, S., Jin, U.-H., Morpurgo, B., Abudayyeh, A., Singh, M., and Tjalkens, R. B. (2016). Nuclear receptor 4A (NR4A) family – orphans no more. J. Steroid Biochem. Mol. Biol. 157, 48–60. doi: 10.1016/j.jsbmb.2015.04.016
Saijo, K., Winner, B., Carson, C. T., Collier, J. G., Boyer, L., Rosenfeld, M. G., et al. (2009). A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137, 47–59. doi: 10.1016/j.cell.2009.01.038
Saucedo-Cardenas, O., and Conneely, O. M. (1996). Comparative distribution of NURR1 and NUR77 nuclear receptors in the mouse central nervous system. J. Mol. Neurosci. 7, 51–63. doi: 10.1007/BF02736848
Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W.-D., Smidt, M. P., Cox, J. J., De Mayo, F., et al. (1998). Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. U.S.A. 95, 4013–4018. doi: 10.1073/pnas.95.7.4013
Saura, C. A., and Valero, J. (2011). The role of CREB signaling in Alzheimer’s disease and other cognitive disorders. Rev. Neurosci. 22, 153–169.
Shepherd, J. D., and Bear, M. F. (2011). New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 14, 279–284. doi: 10.1038/nn.2708
Skerrett, R., Malm, T., and Landreth, G. (2014). Nuclear receptors in neurodegenerative diseases. Neurobiol. Dis. 72, 104–116. doi: 10.1016/j.nbd.2014.05.019
Sleiman, P. M. A., Healy, D. G., Muqit, M. M. K., Yang, Y. X., Van Der Brug, M., Holton, J. L., et al. (2009). Characterisation of a novel NR4A2 mutation in Parkinson’s disease brain. Neurosci. Lett. 457, 75–79. doi: 10.1016/j.neulet.2009.03.021
Smith, G. A., Rocha, E. M., Rooney, T., Barneoud, P., McLean, J. R., Beagan, J., et al. (2015). A Nurr1 agonist causes neuroprotection in a Parkinson’s disease lesion model primed with the toll-like receptor 3 dsRNA inflammatory stimulant Poly(I:C). PLoS One 10:e0121072. doi: 10.1371/journal.pone.0121072
Sousa, K. M., Mira, H., Hall, A. C., Jansson-Sjöstrand, L., Kusakabe, M., and Arenas, E. (2007). Microarray analyses support a role for Nurr1 in resistance to oxidative stress and neuronal differentiation in neural stem cells. Stem Cells 25, 511–519. doi: 10.1634/stemcells.2006-0238
Swerdlow, N. R., and Geyer, M. A. (1998). Using an animal model of deficient sensorimotor gating to study the pathophysiology and new treatments of schizophrenia. Schizophrenia Bull. 24, 285–301. doi: 10.1093/oxfordjournals.schbul.a033326
Terzioglu-Usak, S., Negis, Y., Karabulut, D. S., Zaim, M., and Isik, S. (2017). Cellular model of Alzheimer’s disease: Aβ1-42 peptide induces amyloid deposition and a decrease in topo isomerase IIβ and Nurr1 expression. Curr. Alzheimer Res. 14, 636–644. doi: 10.2174/1567205014666170117103217
Tokuoka, H., Hatanaka, T., Metzger, D., and Ichinose, H. (2014). Nurr1 expression is regulated by voltage-dependent calcium channels and calcineurin in cultured hippocampal neurons. Neurosci. Lett. 559, 50–55. doi: 10.1016/j.neulet.2013.11.033
Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E., et al. (2002). Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420, 788–794.
Uchida, S., and Shumyatsky, G. P. (2018). Synaptically localized transcriptional regulators in memory formation. Neuroscience 370, 4–13.
Vecsey, C. G., Hawk, J. D., Lattal, K. M., Stein, J. M., Fabian, S. A., Attner, M. A., et al. (2007). Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. J. Neurosci. 27:6128. doi: 10.1523/JNEUROSCI.0296-07.2007
Vitolo, O. V., Sant’Angelo, A., Costanzo, V., Battaglia, F., Arancio, O., and Shelanski, M. (2002). Amyloid β-peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13217–13221. doi: 10.1073/pnas.172504199
Volakakis, N., Kadkhodaei, B., Joodmardi, E., Wallis, K., Panman, L., Silvaggi, J., et al. (2010). NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. U.S.A. 107:12317. doi: 10.1073/pnas.1007088107
Volpicelli, F., Caiazzo, M., Greco, D., Consales, C., Leone, L., Perrone-Capano, C., et al. (2007). Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J. Neurochem. 102, 441–453. doi: 10.1111/j.1471-4159.2007.04494.x
von Hertzen, L. S. J., and Giese, K. P. (2005). Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J. Neurosci. 25:1935. doi: 10.1523/JNEUROSCI.4707-04.2005
Vuillermot, S., Joodmardi, E., Perlmann, T., Ove Ögren, S., Feldon, J., and Meyer, U. (2011). Schizophrenia-relevant behaviors in a genetic mouse model of constitutive Nurr1 deficiency. Genes Brain Behav. 10, 589–603. doi: 10.1111/j.1601-183X.2011.00698.x
Wang, Z., Benoit, G., Liu, J., Prasad, S., Aarnisalo, P., Liu, X., et al. (2003). Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423, 555–560.
Wansa, K., Harris, J., Yan, G., Ordentlich, P., and Muscat, G. (2003). The AF-1 Domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-Mercaptopurine. J. Biol. Chem. 278, 24776–24790. doi: 10.1074/jbc.M300088200
Xu, P. Y., Liang, R., Jankovic, J., Hunter, C., Zeng, Y. X., Ashizawa, T., et al. (2002). Association of homozygous 7048G7049 variant in the intron six of Nurr1 gene with Parkinson’s disease. Neurology 58:881. doi: 10.1212/wnl.58.6.881
Zagrebelsky, M., and Korte, M. (2014). Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology 76, 628–638. doi: 10.1016/j.neuropharm.2013.05.029
Zetterström, R. H., Williams, R., Perlmann, T., and Olson, L. (1996). Cellular expression of the immediate early transcription factors Nurr1 and NGFI-B suggests a gene regulatory role in several brain regions including the nigrostriatal dopamine system. Mol. Brain Res. 41, 111–120. doi: 10.1016/0169-328x(96)00074-5
Zhang, X., Odom, D. T., Koo, S.-H., Conkright, M. D., Canettieri, G., Best, J., et al. (2005). Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102:4459. doi: 10.1073/pnas.0501076102
Zhang, Y., Chen, G., Gao, B., Li, Y., Liang, S., Wang, X., et al. (2016). NR4A1 knockdown suppresses seizure activity by regulating surface expression of NR2B. Sci. Rep. 6:37713.
Keywords: Nr4a2, transcription factor, hippocampus, synaptic plasticity, brain, learning, memory
Citation: Català-Solsona J, Miñano-Molina AJ and Rodríguez-Álvarez J (2021) Nr4a2 Transcription Factor in Hippocampal Synaptic Plasticity, Memory and Cognitive Dysfunction: A Perspective Review. Front. Mol. Neurosci. 14:786226. doi: 10.3389/fnmol.2021.786226
Received: 29 September 2021; Accepted: 27 October 2021;
Published: 22 November 2021.
Edited by:
Toh Hean Ch’ng, Nanyang Technological University, SingaporeReviewed by:
Timothy J. Jarome, Virginia Tech, United StatesCopyright © 2021 Català-Solsona, Miñano-Molina and Rodríguez-Álvarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: José Rodríguez-Álvarez, am9zZS5yb2RyaWd1ZXpAdWFiLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.