
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 03 December 2021
Sec. Molecular Signalling and Pathways
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.742790
This article is part of the Research TopicProtein Misfolding, Altered Mechanisms and NeurodegenerationView all 9 articles
 Mauro Montalbano1,2
Mauro Montalbano1,2 Elizabeth Jaworski3
Elizabeth Jaworski3 Stephanie Garcia1,2
Stephanie Garcia1,2 Anna Ellsworth1,2
Anna Ellsworth1,2 Salome McAllen1,2
Salome McAllen1,2 Andrew Routh3,4
Andrew Routh3,4 Rakez Kayed1,2*
Rakez Kayed1,2*Tau protein is a known contributor in several neurodegenerative diseases, including Alzheimer’s disease (AD) and frontotemporal dementia (FTD). It is well-established that tau forms pathological aggregates and fibrils in these diseases. Tau has been observed within the nuclei of neurons, but there is a gap in understanding regarding the mechanism by which tau modulates transcription. We are interested in the P301L mutation of tau, which has been associated with FTD and increased tau aggregation. Our study utilized tau-inducible HEK (iHEK) cells to reveal that WT and P301L tau distinctively alter the transcription and alternative polyadenylation (APA) profiles of numerous nuclear precursors mRNAs, which then translate to form proteins involved in chromatin remodeling and splicing. We isolated total mRNA before and after over-expressing tau and then performed Poly(A)-ClickSeq (PAC-Seq) to characterize mRNA expression and APA profiles. We characterized changes in Gene Ontology (GO) pathways using EnrichR and Gene Set Enrichment Analysis (GSEA). We observed that P301L tau up-regulates genes associated with reactive oxygen species responsiveness as well as genes involved in dendrite, microtubule, and nuclear body/speckle formation. The number of genes regulated by WT tau is greater than the mutant form, which indicates that the P301L mutation causes loss-of-function at the transcriptional level. WT tau up-regulates genes contributing to cytoskeleton-dependent intracellular transport, microglial activation, microtubule and nuclear chromatin organization, formation of nuclear bodies and speckles. Interestingly, both WT and P301L tau commonly down-regulate genes responsible for ubiquitin-proteosome system. In addition, WT tau significantly down-regulates several genes implicated in chromatin remodeling and nucleosome organization. Although there are limitations inherent to the model systems used, this study will improve understanding regarding the nuclear impact of tau at the transcriptional and post-transcriptional level. This study also illustrates the potential impact of P301L tau on the human brain genome during early phases of pathogenesis.
Tau is a neuronal protein found both inside and outside of the nucleus that contributes to the pathology of neurodegenerative diseases such as frontotemporal dementia (FTD) and Alzheimer’s disease (AD) (Sultan et al., 2011). It is primarily described as a microtubule-associated protein (Violet et al., 2014). Nuclear tau has been found to “protect” DNA (Hua and He, 2003; Sultan et al., 2011; Violet et al., 2014) during reactive oxygen species (ROS)-induced heat stress. However, nuclear and cytosolic tau interact with RNA to form droplets (Zhang et al., 2017) and aggregates (Kampers et al., 1996). Tau has also been observed altering nuclear structure (Monroy-Ramírez et al., 2013; Montalbano et al., 2019) in the human nuclei of neuroblastoma (Loomis et al., 1990; Shea and Cressman, 1998) and in HEK-293 cells. More specifically, phosphorylation of nuclear tau negatively regulates its nuclear function in pluripotent neuronal cells and neuroblastoma cells (Ulrich et al., 2018). Previous studies have revealed that nuclear tau plays a role in the DNA damage response (DDR) through deadenylation, which triggers major mRNA decay pathways (Baquero et al., 2019; Farmer et al., 2020). Most recently, we found that oligomeric assemblies of tau containing RNA-binding proteins impair chromatin remodeling and nuclear lamina formation through associations with histones and chromatin components in the nuclear compartment (Montalbano et al., 2020).
Despite the well-established importance of tau in the cytoskeleton of neurons (Venkatramani and Panda, 2019), there is growing evidence that tau is notably involved in nucleolar transcription and cellular stress responses (Maina et al., 2018a,b). Recently, it was shown that mutations and/or the phosphorylation of tau results in the deformation of the neuronal nuclear membrane and can disrupt nucleocytoplasmic transport (Lester and Parker, 2018) in FTD (Montalbano et al., 2019; Paonessa et al., 2019) and AD (Eftekharzadeh et al., 2018; Tripathi et al., 2019). Related studies analyzed the direct impact in transcriptional activity due to tau and found that nuclear tau regulates the expression of VGluT1, a gene that controls glutamatergic synaptic transmission, and that tau displacement from microtubules (MTs) increases nuclear accumulation of tau (Siano et al., 2019). Furthermore, tau modifies histone acetylation and was shown to have a broad epigenomic impact in the aging and pathology of AD human brains (Klein et al., 2019). It has also been observed that tau interacts with neuronal pericentromeric DNA regions, particularly in association with HP1 and H3K9me3 (Mansuroglu et al., 2016), this observation spots tau protein as potential chromatin remodeling factor. Lastly, tau exhibits binding interactions with genic and intergenic DNA sequences of primary cultured neurons, especially in positions ± 5,000 bp away from the start site of transcription (Benhelli-Mokrani et al., 2018).
In eukaryotic cells, the maturation of 3′ ends in mRNA involves endonucleolytic cleavage of the nascent RNA followed by the synthesis of a poly(A) tail on the 3′ terminus of the cleaved product by a poly(A) polymerase (PAP) (Stewart, 2019). This reaction is called polyadenylation and is fundamentally linked to transcription termination. The sequences for the mRNA precursors and the proteins required for polyadenylation are well understood. It has been clearly elucidated that a single gene can give rise to many possible transcripts, each with different polyadenylation sites [poly(A)-sites, or PASs], and that differential usage of these sites can lead to the formation of mRNA isoforms. This phenomenon is called alternative polyadenylation (APA) (Gruber and Zavolan, 2019) and is a common event in eukaryotic cells. In fact, researchers have determined that 50% of mammalian mRNA-encoding genes express APA isoforms (Tian et al., 2005; Shepard et al., 2011). Considering this information, we used tau-inducible HEK (iHEK) cell lines to obtain and analyze transcriptomic and APA profiles in the presence of WT and P301L tau. To characterize transcriptional and post-transcriptional profiles modified by WT and P301L, we utilized Poly(A)-ClickSeq (PAC-Seq) to measure changes in the expression of the host mRNA transcript whilst simultaneously characterizing changes in the PAS usage or creation of mRNA isoforms. In addition, we employed Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO) to study the main gene domains modulated by tau.
In this study we used two different versions of tau-inducible HEK (iHEK) cells: iHEK overexpressing WT tau and iHEK overexpressing mutated P301L tau. They were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. To induce WT and mutant tau overexpression, iHEK cells were treated with 1 μg/ml of Tetracycline (Tet) for 24 h in FBS-depleted DMEM (Gibco™ LS11965118, Thermo Fisher Scientific). iHEK cells not treated with Tet were named control (Ctr). After 24 h, two washes with medium were done to remove excess Tet. Immediately after the washes, the cells were stained and collected. Detachment of cells was completed with Trypsin (GibcoTM Trypsin-EDTA, 0.25% Phenol red, LS25200114 Thermo Fisher Scientific), and the cells warmed for 3 min in the incubator following the addition of Trypsin. The cells were then centrifuged at 1,000 rpm for 5 min. Lastly, cell pellets were harvested and used for protein fractionation, and mRNA extraction.
Total mRNA was collected by using TRIzol extraction reagent according to established protocol (Rio et al., 2010). RNA samples for Real Time Analysis (RT-PCR) were quantified using a NanoDrop Spectrophotometer (NanoDrop Technologies), followed by analysis on an RNA Nano chip using the Agilent 2100 Bioanalyzer (Agilent Technologies). Only samples with high quality total RNA were used (RIN: 7.5–10.0) for the study. Synthesis of cDNA was performed with either 0.5 or 1 μg of total RNA in a 20 μl reaction using the reagents available within the Taqman Reverse Transcription Reagents Kit from Life Technologies (#N8080234). Q-PCR amplifications (performed in duplicate or triplicate) were done using 1 μl of cDNA in a total volume of 20 μl using the iTaq Universal SYBR Green Supermix (Bio-Rad #1725125). The final concentrations of the primers were 300 nM. Relative RT-QPCR assays are performed with either 18S RNA gene as a normalizer. Absolute RNA quantification analysis was performed using known amounts of a synthetic transcript created from the gene of interest.
Protocols for Poly(A)-ClickSeq (PAC-Seq) have been described in detail by Jaworski and Routh (2018) and Elrod et al. (2019). Approximately 1 μg of total cellular RNA per sample was used as a template in reverse-transcription reactions supplemented with 40 μM Azido-VTPs and primed using an oligo-dT primer containing a partial Illumina i7 indexing adaptor. Azido-terminated cDNA fragments were “click-ligated” to hexynyl-functionalized click-adaptors containing the Illumina i5 universal sequencing adaptor. Single-stranded cDNA libraries were indexed in a final PCR reaction for 15–18 PCR cycles. Final libraries were size extracted by gel-electrophoresis and submitted for sequencing using an Illumina NextSeq550 to prepare 1 × 150 SE reads. RNAseq datasets is uploaded to NCBI SRA, reference number: PRJNA744518.
PAC-Seq data were analyzed using the Differential Poly-A Clustering (DPAC) program, which ran with default settings as previously described (Routh, 2019). DPAC trims and quality-filters raw FASTQ data and therefore requires each read to have at least 25 “As” at the 3′ end of the read. These reads are then trimmed using cutadapt. Trimmed reads are mapped to the reference human genome (hg19) using HISAT2 (Kim et al., 2019). The 3′end of mapped reads are thus used to annotate poly(A)-sites and annotated based upon overlaps with gene annotations obtained from UCSC genome browser. Gene counts were extracted and DESeq2 was used to calculated changes in gene expression as well as relative changes in expression in individual poly(A)-sites found within single genes. Differential gene expression was assigned when a gene had a fold change greater than ± 1.5-fold with a p-adj value less than 0.1. Alternative polyadenylation is assigned when a single gene has two or more clustered poly(A)-sites wherein at least one of these sites has a differential usage greater than a ± 1.5-fold, a p-adj value less than 0.1, and a change of the relative usage of a poly(A)-cluster within the gene of greater than 10%.
Immunoblot (IB) analyses were performed with iHEK cell fraction samples as previously described (Montalbano et al., 2020). Approximately 10 μg of protein preparations were loaded onto precast NuPAGE 4–12% Bis-Tris gels (NP0335BOX, Invitrogen) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses. Gels were subsequently transferred onto nitrocellulose membranes and blocked overnight at 4°C with 10% non-fat dry milk. Membranes were then probed for 1 h at room temperature with Pan-Tau (Tau13, 1:10,000, MMS-520R Covance), (GAPDH, 1:1,000, ab9485 Abcam), Histone3 (1:1,000, ab201456 Abcam), RCC1 (1:100, Clone E-6 sc-55559 Santa Cruz Tech.), DNAJC2 (1:5,000, ab134572 Abcam), Histone 1.2 (1:500, ab4086 Abcam), HMGB1 (1:500, ab18256 Abcam), SMARCA5 (1:10,000, #PA5-78253, Invitrogen), SMARCC1 (0.4 μg/ml, #PA5-55058, Invitrogen), and β-Actin (1:5,000, #A1978, Sigma-Aldrich). Antibodies were diluted in 5% non-fat dry milk. Immunoreactivity was detected using a horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG, 1:10,000, NA934 GE Healthcare). Tau13 and Tau5 immunoreactivity were detected using an anti-mouse IgG (1:10,000, NA931 GE Healthcare) diluted in 5% milk. ECL Plus (K-12045-D50, GE Healthcare) was used to visualize protein bands. LaminB1/Histone3 and GAPDH were used to normalize and quantify nuclear and cytoplasmic proteins, respectively. The compartment extraction was conducted with Qproteome Cell Compartment Kits (Qiagen, #37502); nuclear, cell membrane, and cytoplasmic proteins were isolated and preserved for IB analysis.
Cells on a 24-well coverslip were fixed with 0.5 ml of 4% PFA/PBS for 15 min. The cells were then washed 3 times in phosphate buffered saline (PBS), for 5 min for each wash. The cells were permeabilized in 0.5 ml PBS and 0.2% Triton X-100 in phosphate buffered saline containing 0.5% Tween (PBST) for 5 min. Blocking was done in 0.5 ml of 5% normal goat serum (NGS) in PBST for 1 h. Primary antibody was diluted in 5% NGS/PBST overnight at 4°C for incubation, and then washed 3 times in PBST, for 10 min each. Secondary antibody diluted in 5% NGS/PBST was incubated for 2 h at room temperature. All the secondary antibodies were purchased from Thermo Fisher Scientific and used at a 1:800 dilution for staining. After applying secondary antibodies, cells were incubated in DAPI (nuclei staining) diluted 1:10,000 in PBST (5 mg/ml stock solution) for 5 min after the first wash. The cells were then washed 2 times with PBST, and once with PBS (10 min each) prior to mounting coverslips. Coverslips were mounted on glass microscope slides using 8–10 μl of Prolong Gold Antifade mounting media with DAPI (Invitrogen, P36941) per coverslip. Slides were air-dried in fume hood or stored at 4°C until ready to be dried in the fume hood. The primary antibodies used in this study for immunocytochemistry (ICC) are as follows: Histone 1.2 (Abcam ab4086—1 μg/ml), Ki-67 (Abcam ab92742—1 μg/ml), SMARCC1 (Invitrogen PA5-55058—0.25 μg/ml, SMARCA5 (Invitrogen PA5-78253—1 μg/ml, MCM2 (Abcam ab108935—1/1,000), RCC1 (Santa Cruz, Inc. sc-55559—1:50), and Tau13 (Bio Legend MMS-520R—1/200). After three washes with PBS, cells were probed with mouse and rabbit-specific fluorescent-labeled secondary antibodies (1:200, Alexa Fluor 488 and 633, Life Technologies). Single frame images were collected using the Keyence BZ-X 710 Microscope. Images for quantification of area and integrated density were taken in nuclear target areas guided by the DAPI fluorescence. We then performed single extraction analysis using BZ-X Analyzer software (Keyence). We used 200 nuclei per target area and used the Nikon 20X objective for imaging and quantification analysis.
All in vitro experiments were performed in at least three biological replicates. All data are presented as means ± SD and were analyzed using GraphPad Prism Software 6.0. Statistical analyses included the Student’s t-test or one-way ANOVA followed by Tukey’s Multiple Comparisons Test. Column means were compared using one-way ANOVA with treatment as the independent variable. In addition, group means were compared using two-way ANOVA considering factors for each treatment, respectively. When ANOVA showed a significant difference, pair-wise comparisons between group means were examined by the Tukey and Dunnett Multiple Comparison Test.
Firstly, we evaluated changes in gene expression profiles upon expression of WT and P301L tau in iHEK cells that were induced with tetracycline (Tet). After 24 h of Tet induction, we confirmed tau expression in the cytoplasm and nuclei of iHEK cells (Supplementary Figure 1A). Total cellular RNA from WT and P301L tau [untreated (Control) and treated (+ Tet)] study groups was extracted using TRIzol reagent and by following established protocol (Montalbano et al., 2019, 2020). RNA was sequenced using Poly(A)-ClickSeq (PAC-Seq) to measure changes in gene expression and poly(A)-site usage (Elrod et al., 2019). A schematic of the experimental design is provided in Figure 1A. Volcano scatterplots from WT and P301L tau iHEK (Figures 1B,C, respectively) demonstrate a substantial difference in the number of genes regulated by WT tau and P301L tau. After Tet induction in the WT tau iHEK cell system, we observed up-regulation of 88 genes and down-regulation of 30 genes (gene names listed in Figure 1D). In the P301L tau iHEK cell system, these numbers dropped to 10 up-regulated genes and only 1 down-regulated (gene names listed in Figure 1E).
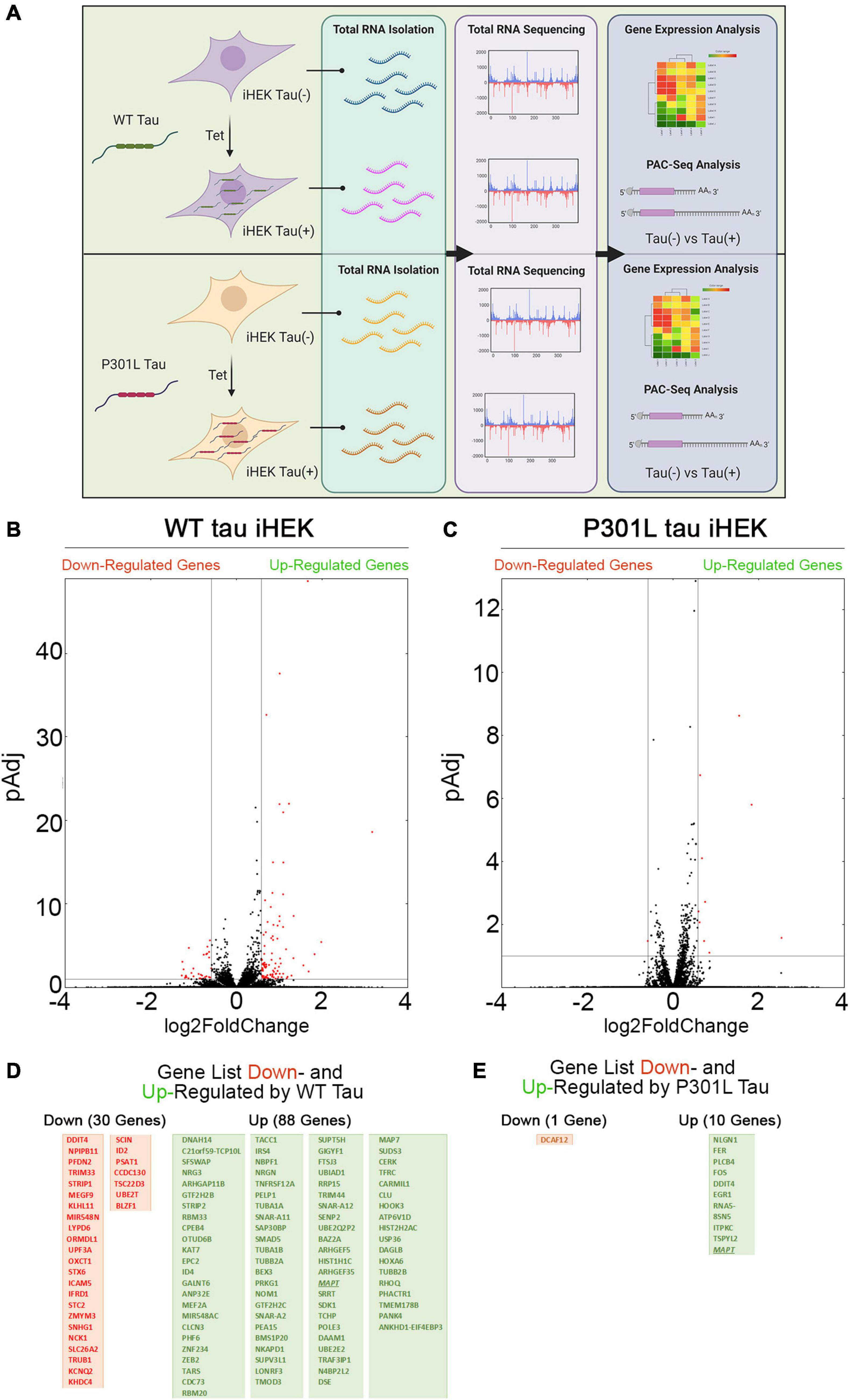
Figure 1. Tau-dependent Gene expression. (A) Schematic representation of experimental plan, from Tet induction in WT and P301L Tau iHEK to RNA isolation, sequencing to gene expression analysis. (B) Volcano Plot for Down- and Up-regulated gene in WT tau iHEK. (C) Volcano Plot for Down- and Up-regulated gene in P301L tau iHEK. (D) Gene Lists of Down-Regulated (Red Boxes) and Up-Regulated (Green Boxes) Genes in WT tau iHEK. (E) Gene Lists of Down-Regulated (Red Boxes) and Up-Regulated (Green Boxes) Genes in P301L tau iHEK.
Supplementary Figure 1B displays the scatterplots of WT and P301L tau gene expression, while Supplementary Figure 1C reports the Principal Component Analysis (PCA). PCA demonstrates significant variation among the study groups. More specifically, the analysis suggests a significant difference in transcriptional activity of WT tau due to the higher number of genes modulated in comparison to the mutant P301L tau form. Using EnrichR (Kuleshov et al., 2016), we established Gene Ontology (GO) of the biological processes, molecular functions, and cellular components altered by both the up-regulated and the down-regulated sets of genes. WT tau GO is summarized in Figure 2. WT tau up-regulated genes belonging mainly to classes of cytoskeleton-dependent intracellular transport genes (GO: 0030705, TUBA1A, TUBB2B TUBA1B, TUBB2A, and HOOK3) and genes responsible for the regulation of cytoskeleton organization (GO: 0051493). Imbalanced expression of tubulin and tau induces neuronal dysfunction in C. elegans (Miyasaka et al., 2018), indicating that tau itself can disturb tubulin gene expression. The reason behind this pronounced involvement of TUBB genes could be due to the fact that TUBB1B, TUBB2B, TUBA1A, and TUBB2A are clustered together within the genome (Bittermann et al., 2019).
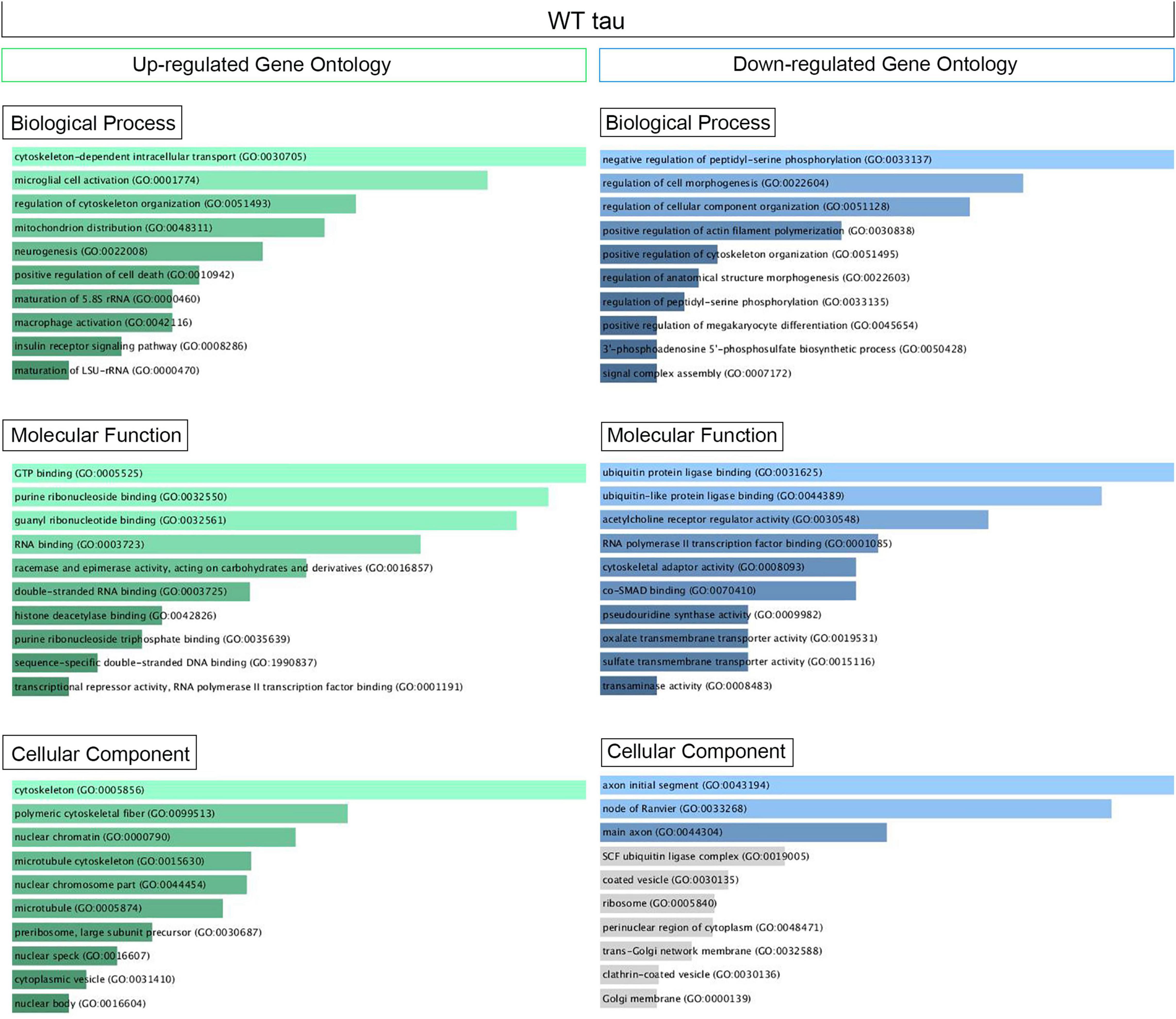
Figure 2. Up- and Down regulated genes in WT Tau iHEK Gene Ontology. Left Column (Green) Up-regulated genes analyzed by Enrich GO and divided by Biological Process, Molecular Function and Cellular Component. Right Column (Blue) Down-regulated genes analyzed by Enrich GO and divided by Biological Process, Molecular Function and Cellular Component. Gray bars represent not significant correlation.
It is important to note that biological process such as microglial cell activation (GO: 0001774) and macrophage activation (GO: 0042116) were also observed as being up-regulated, which confirms known effects of tau on the neuro-inflammatory response commonly observed in neurodegenerative diseases (Kwon and Koh, 2020). Neuro-immunomodulation can also effect cytoskeleton reorganization (Morales et al., 2013). Our GO analysis revealed up-regulated genes involved in mitochondrion distribution (GO: 0048311, MAPT, and MEF2A genes), morphogenesis (GO: 0070584, SUPV3L1, p = 0.03892), neurogenesis (GO 0022008, NOM1, MAPT, and DAGLB genes), and positive regulation of cell death (GO 0010942, SAP30BP, MAPT, and CLU genes). The increase of CLU expression was a particularly interesting observation. Clusterin is a multifunctional, secreted chaperone involved in several basic biological events, including cell death, tumor progression and neurodegeneration. The CLU gene is notably associated with an increased AD risk (Karch and Goate, 2015). In terms of molecular functions, the up-regulated genes we observed have several enriched pathways, including RNA binding (GO: 0003723, USP36, NOM1, TFRC, BAZ2A, SUPTSH, PHF6, FTSJ3, SUPV3L1, TUBA1B, RBM20, MAPT, RBM33, PELP1, HIST1H1C, CPEB4) and several nuclear functions, such as histone deacetylase binding (GO: 0042826, MEF2A, SUDS3 and PHF6) and sequence-specific double stranded DNA binding (GO: 1990837, MEF2A, KAT7, and MAPT).
Transcriptional products of up-regulated genes are mostly localized in the cytoplasm and nuclear compartments. We detected transcripts associated with nuclear chromatin (GO: 0000790), such as MEF2A, ZEB2, ANP32E, SUDS3, HIST2H2AC, and HIST1H1C. We also examined nuclear speck transcripts (GO: 0016607), such as CARMIL1, USP36, GTF2H2C, BAZ2A, and MAPT genes, which are also included in nuclear body components (GO: 0016604), along with SUDS3 and SENP2. The other cell compartment well represented in our GO analysis is the cytoplasm. In particular, the microtubule cytoskeleton (GO: 0015630) contained the following up-regulated genes: TUBB2B, SAP30BP, TUBA1B, TUBA1A, TMOD3, MAP7, TARS, TACC1, MAPT, CLU, and RHOQ. A complete Enrich-GO list of significant up-regulated genes observed in WT tau is presented in Supplementary Table 1.
Overall, thirty genes were significantly downregulated by WT tau protein. The main biological process affected was the regulation of cellular component organization (GO: 0051128) as it relates to cytoskeleton organization and structure morphogenesis. Molecular functions associated with the aforementioned genes are closely related to ubiquitin protein ligase binding (GO: 0031625) and ubiquitin-like protein ligase binding (GO: 0044389). Genes important to neuronal components included genes essential to the structure of initial axonal segments, nodes of Ranvier, and main axons. These three groups typically involve the gene KCNQ2. This gene encodes for Potassium voltage-gated channel subfamily KQT member 2, which plays a critical role in determining the subthreshold electrical excitability of neurons as well as the responsiveness of neurons to synaptic inputs. Therefore, KCNQ2 is important in the regulation of neuronal excitability and the loss-of-function or gain-of function of this gene can lead to various forms of neonatal epilepsy (Niday et al., 2017).
Furthermore, Cullin-RING E3 ubiquitin-ligase complex component KLHL11 is down-regulated, as well as the STX6 gene. STX6 encodes for Syntaxin-6, which is involved in intracellular vesicle trafficking and is integrally associated with the Golgi apparatus. Another Golgi protein that is down-regulated is Golgin-45 (BLZF1). It is required for normal Golgi structure and for protein transport from the Endoplasmic Reticulum (ER) through the Golgi apparatus to the cell surface (Short et al., 2001). Lastly, the ER gene STC2 is downregulated and encodes for Stanniocalcin-2. This glycoprotein has an anti-hypocalcemic action on calcium and phosphate homeostasis (Ito et al., 2004).
We also detected two nucleolus-localized genes among the down-regulated group: UBE2T (ubiquitin-conjugating enzyme with E2 T) and UPF3A, (a regulator of non-sense transcript 3A). The mitochondrial genes that were down-regulated included OXCT1 (Succinyl-CoA: 3-ketoacid coenzyme A transferase 1, mitochondrial enzyme), TRUB1 and PFDN2 (Prefoldin subunit 2). An Enrich-GO list of downregulated genes present in WT tau is depicted in Supplementary Table 2.
Although there are limitations inherent with the model used, these data suggest that WT tau intrinsically and significantly impacts the cell at a transcriptional level. More specifically, a higher number of genes are up-regulated and down-regulated by WT tau when compared to P301L tau. This suggests that the P301L mutation of tau has not impact at the transcriptional level. This sort of loss could have detrimental effects on cell structure and organization.
The GO pathways and cellular compartments upregulated and downregulated by P301L tau are listed In Supplementary Tables 3, 4, respectively. As observed in WT tau iHEK cells, the MAPT gene is on the upregulated gene list for P301L tau, as expected after Tet induction of the iHEK cells. Within the group of axonal and cytoskeleton genes, we noticed up-regulation of NLGN1, a gene that encodes for Neuroligin-1. Neuroligin is a postsynaptic neuronal surface protein involved in cell-to-cell interactions via its interactions with neurexin family members (Bemben et al., 2015). It has been established that the NLGN1 gene is associated with amyloid-β oligomers (AβOs) in AD-causing synaptic impairment (Brito-Moreira et al., 2017). In addition, NLGN1 is typically altered in AD hippocampi and also modulates amyloid-beta oligomer toxicity (Dufort-Gervais et al., 2020). Neuroligin-1 plays an influential role in synaptic function and synaptic signal transmission, most likely through its ability to recruit and cluster together other synaptic proteins (Bemben et al., 2015). For instance, neuroligin-1 may promote the initial formation of synapses (Craig and Kang, 2007), but is not essential for the complete formation of synapsyes. In vitro, Neuroligin-1 triggers the de novo formation of presynaptic structures. NLGN1 may also be involved in specification of excitatory synapses (Bemben et al., 2015). For example, NLGN1 functions to maintain wakefulness quality and normal synchrony of cerebral cortex activity during wakefulness and sleep (El Helou et al., 2013). Neuroligin-1 is predominantly located in synaptic cleff of the cell membrane (Wu et al., 2019).
When we analyzed upregulated genes, we detected a considerable number of genes related to nuclear body (GO: 0016604) and nuclear speck (GO: 0016607) domains including the genes ITPKC and MAPT. Interestingly, it has been observed that the FER gene participates in several different cytoplasmic and nuclear functions. For example, FER is associated with nuclear chromatin (GO: 0000790) and the microtubule skeleton (GO: 0015630). The FER gene also encodes for a tyrosine-protein kinase that plays a role in synapse organization, trafficking of synaptic vesicles, the generation of excitatory post-synaptic currents, and neuron-to-neuron synaptic transmission (Lee et al., 2008). Lastly, FER plays a role in neuronal cell death after brain damage (Lee et al., 2008). The only gene down-regulated by P301L tau is DCAF12, which is a component of the Cullin-RING ubiquitin ligase complex (Patrón et al., 2019). This gene is also down-regulated by WT tau and belongs to genes associated with ubiquitinization processes. The failure of ubiquitinization pathways is known to have a strong connection to neurodegenrative diseases (Zheng et al., 2014). Supplementary Figure 2 summerizes upregulated and downregulated genes in P301L tau, subcatagorized by biological process, molecular function and cellular component.
In summary, the P301L mutation upregulates genes involved in positive regulation of neuronal death and responsiveness to reactive oxygen species (ROS) production. This is in contrast to the genes altered by WT tau that have a greater effect on cell structural processes. The most important molecular function altered by such genes would be sequence-specific double-stranded DNA binding, transcriptional expression, and chromatin remodeling. Overall, our GO data suggests no impact on transcription in mutated P301L tau that may relate to pathology only through the protein level resulting in toxic aggregation. Modulating genes known to be associated with neurodegenerative disease suggests that mutated tau engenders harmful transcription patterns that contribute to the well-established effects of tau proteinaceous-aggregation toxicity.
Gene Set Enrichment Analysis (GSEA) offers an opportunity to evaluate and identify classes of genes or proteins that are over-represented in a large set of genes or proteins and may have an association with disease phenotypes. Due to the differences in gene numbers modulated by WT tau vs. P301L tau, we performed GSEA. This analysis compared models with and without WT tau. We observed that WT tau down-regulates the expression of numerous genes linked to chromatin organization (Figure 3A) and chromatin remodeling (Figure 3B) domains. By looking at the chromatin organization and remodeling gene clusters, we identified that several high-mobility group box proteins (HMG) HMGN5, HMGB2, and HMGA1 are up-regulated while HMGB1 and HMGN1 are down-regulated. It is important to note that HMGB1 is an activator of neuro-inflammatory responses and has been implicated in AD (Paudel et al., 2020). In addition, several components of the SWI/SFN chromatin remodeling complex are downregulated. The identification of genes SMARCE1, SMARCA5, and SMARCC1, imply that tau has a substantial impact on chromatin remodeling in the cells. The heterogeneous nuclear ribonucleoproteins (hnRNPs), HNRNPU and HNRNPC, were also found to be downregulated in WT tau. Down-regulation of several factors implicated in DNA replication and repair processes, indicates that WT tau also significantly affects the nuclear compartment of cells in terms of structure and content. Several of these genes are clustered as covalent chromatin modification in GO (Figure 3C).
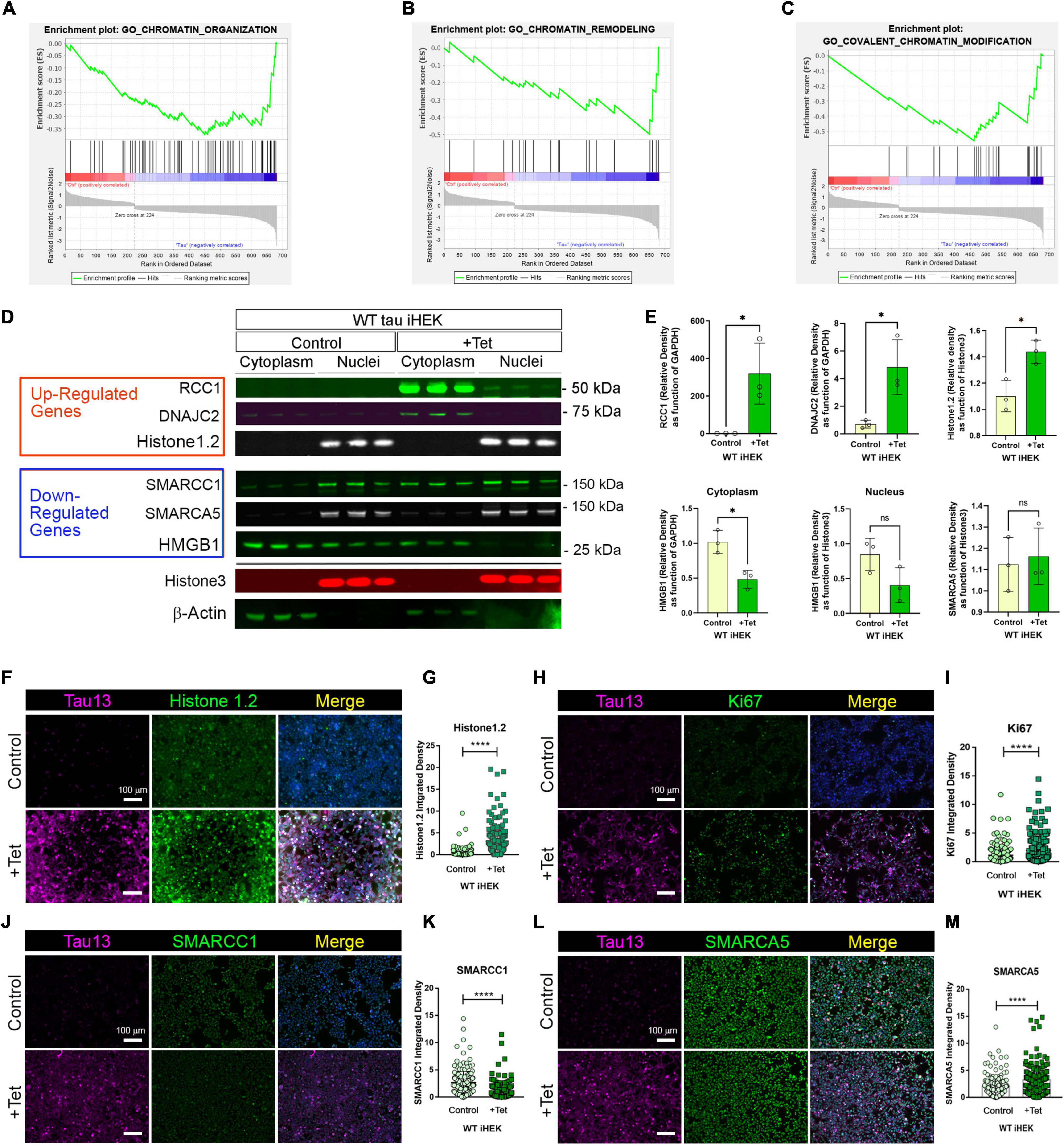
Figure 3. WT tau modulates gene expression of chromatin organization and remodeling factors. (A) Enrichment plot for GO Chromatin organization. (B) Enrichment plot for GO Chromatin remodeling. (C) Enrichment plot for GO-Covalent Chromatin modification. (D) IB of Up-regulated genes: RCC1, DNAJC2 and Histone 1.2 (red box) and Down-regulated genes: SMARCC1, SMARCA5 and HMGB1 (blue box) in cytoplasm and nuclear fractions from WT and P301L tau iHEK. Histone 3 and β-Actin has been used as loading control for nuclear and cytoplasmatic fractions, respectively. (E) Immunoblot quantification of RCC1 (*p < 0.01), DNAJC2 (*p < 0.01), Histone1.2 (*p < 0.01), cytoplasmic HMGB1 (*p < 0.01), nuclear HMGB1 and SMARCA5 proteins in cytoplasmatic and nuclear fractions of WT tau iHEK. (F) Representative Tau 13 (magenta) and Histone 1.2 (green) Co-IF of control (-Tet) and treated WT tau iHEK. (G) Histone 1.2 integrated density quantification in control and + Tet cells (Unpaired t-test, ****p < 0.0001). (H) Representative Tau 13 (magenta) and Ki67 (green) Co-IF of control (-Tet) and treated WT tau iHEK. (I) Ki67 integrated density quantification in control and + Tet cells (Unpaired t-test, ****p < 0.0001). (J) Representative Tau 13 (magenta) and SMARCC1 (green) Co-IF of control (-Tet) and treated WT tau iHEK. (K) SMARCC1 integrated density quantification in control and + Tet cells (Unpaired t-test, ****p < 0.0001). (L) Representative Tau 13 (magenta) and SMARCA5 (green) Co-IF of control (-Tet) and treated WT tau iHEK. (M) SMARCA5 integrated density quantification in control and + Tet cells (Unpaired t-test, ****p < 0.0001).
To validate gene expression changes observed in GSEA analysis, we verified multiple proteins via western blot by using the up-regulated and down-regulated lists generated from Histone Binding GO. We verified up-regulation of RCC1, DnaJC2 and Histone1.2 proteins in the cytoplasm and in nuclear fractions of WT tau iHEK cells (Figure 3D). We also confirmed RCC1 expression and noticed its accumulation in the cytoplasm for both cell lines. Interestingly, we discerned that RCC1 is not imported into the nuclei where it should function as a regulator of chromatin condensation. Instead, DnaJC2 in P301L tau iHEK cells appear to be downregulated (Supplementary Figure 3). However, Histone 1.2 is upregulated in both cell lines. We did not observe down-regulation of the chromatin remodeling complex factors SMARCC1 and SMARCA5. Instead, we detected their accumulation in the cytoplasmic fractions while in the presence of tau, which suggests a deficit in these factors in the nuclei, as observed in our western blots. Lastly, HMGB1 and β-Actin are down-regulated, but HMGB1 is not detected in the nuclei when in the presence of tau. Histone 3 was used as a nuclear loading control. We performed quantification of bands to compare protein levels in WT tau iHEK of RCC1, DNAJC2, Histone1.2, HMGB1 (Cytoplasm/Nuclei) and SMARCA5 (Figure 3E).
To verify gene expression results, alongside western blots, we performed co-immunofluorescence in WT tau iHEK cells. We evaluated integrated density of Histone 1.2 (Figures 3F,G), Ki67 (Figures 3H,I), SMARCC1 (Figures 3J,K), and SMARCA5 (Figures 3L,M). Analysis was performed by considering nuclear integrated density of “–” and “+” tau WT iHEK proteins. To detect and confirm tau expression, we used the Tau13 antibody. MCM2 and RCC1 images and their relative integrated density quantifications are presented in Supplementary Figure 4.
GSEA analysis for WT tau revealed significant down-regulation in the pathways for histone-binding (Figure 4A) and nucleosome organization clusters (Figure 4B). Several genes were detected in the histone and nucleosome domains, which were recurring and can be viewed in the chromatin gene list showed in Figure 3. In addition, we observed an up-regulation of RCC1 (a regulator of chromosome condensation), CTSL (Cathepsin L), MCM2 (Minichromosome maintenance complex component 2), and DNAJC2 (DnaJ heat shock protein member C2). In Nucleosome GO, we observed up-regulated HMGB2 and HMGA1 (high mobility group box B2 and A1). On the contrary, several Lysine acetylation regulators were downregulated: BRD3 and BRD9 (from BRD family), HDAC2, KDM5B, KAT7, and SFTD2.
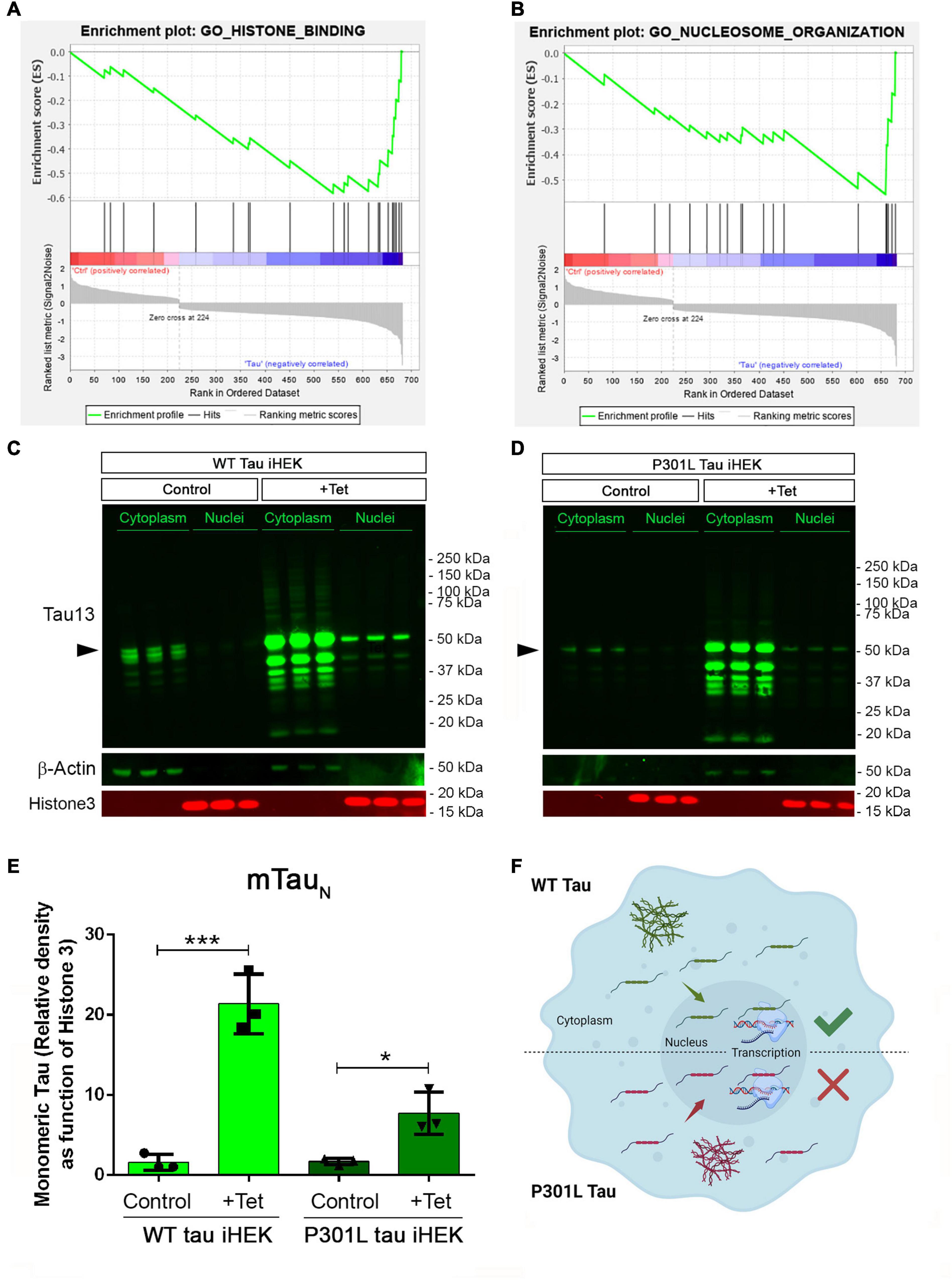
Figure 4. Tau nuclear shuttling. (A) GSEA GO-Histone Binding heat map in WT tau. (B) GSEA GO-Nucleosome organization heat map in WT tau. (C) Immunoblot (IB) with Tau13 (1:1,000) and β-Actin (1:1,000) of cytoplasm and nuclear fraction from WT (left panel) and (D) P301L (right panel) tau induced with Tet. Monomeric tau is indicated by rightwards filled arrows (E) Relative density of nuclear monomeric tau (mTauN, normalized with Histone3). Unpaired t-test has been performed to compare column means [(–) WT tau vs. WT tau ***p = 0.0009, (–) P301l tau vs. P301L tau *p = 0.0169]. (F) Schematic model on tau nuclear import in the two iHEK cell lines.
We also used western blotting to verify tau levels in cytoplasm and nuclear fractions of WT and P301L tau iHEK cells (Figures 4C,D, respectively). We found that upon Tet induction in both compartments, tau was detected, which was previously observed (Montalbano et al., 2019) and expected. Western blot analysis demonstrated that tau is represented mainly in its monomeric form (mTauN) when probing the nucleus. We compared the level of mTauN in both cell lines and we determined that mTauN increased in both cell lines after Tet induction. However, the WT mTauN was present in a significantly higher level when compared to the P301L mTauN (Figure 4E). This difference is due to the higher MAPT transgene expression efficiency in WT tau iHEK cell lines as was confirmed by RT-qPCR in a previous study (Montalbano et al., 2019). These observations suggest that the monomeric form of tau protein predominantly carries out transcriptional activity and that the P301L mutation did not affect the nuclear import of tau, but instead modulated transcriptional activity. Cytoplasmic mTau was quantified as well (Supplementary Figure 4). In general, we propose that WT and P301L tau both shuttle into the nuclei but then modulate transcription differently. The schematic model for this idea is represented in Figure 4F. In summary, many nuclear factor genes involved in several nuclear activities, including chromatin condensation, are downregulated in WT tau, which indicates a potential role of WT and P301L tau in the control of chromatin factors, expression and subsequent cellular localization.
From PAC-seq analysis, we identified 110 genes with shortened 3′UTRs. The majority of these shortened genes belong to significant pathways associated with mRNA processing (GO: 0006397), RNA Splicing (GO: 0000377, GO: 0000398), and RNA metabolic processes (GO: 0016070) (Figure 5A). These domains share several genes: HNRNPA3, SRRT, PRPF4B, CCAR1, LSM8; SNRNP40, HNRNPK, ZMAT2, ZC3H11A, HNRNPF, PCBP2, SNRPE, and HNRNPC. The regulation of responses to DNA damage (GO: 2001020) comprise the following genes: BCLAF1, FMR1, USP1, and HMGA2 (others listed in Figure 5B).
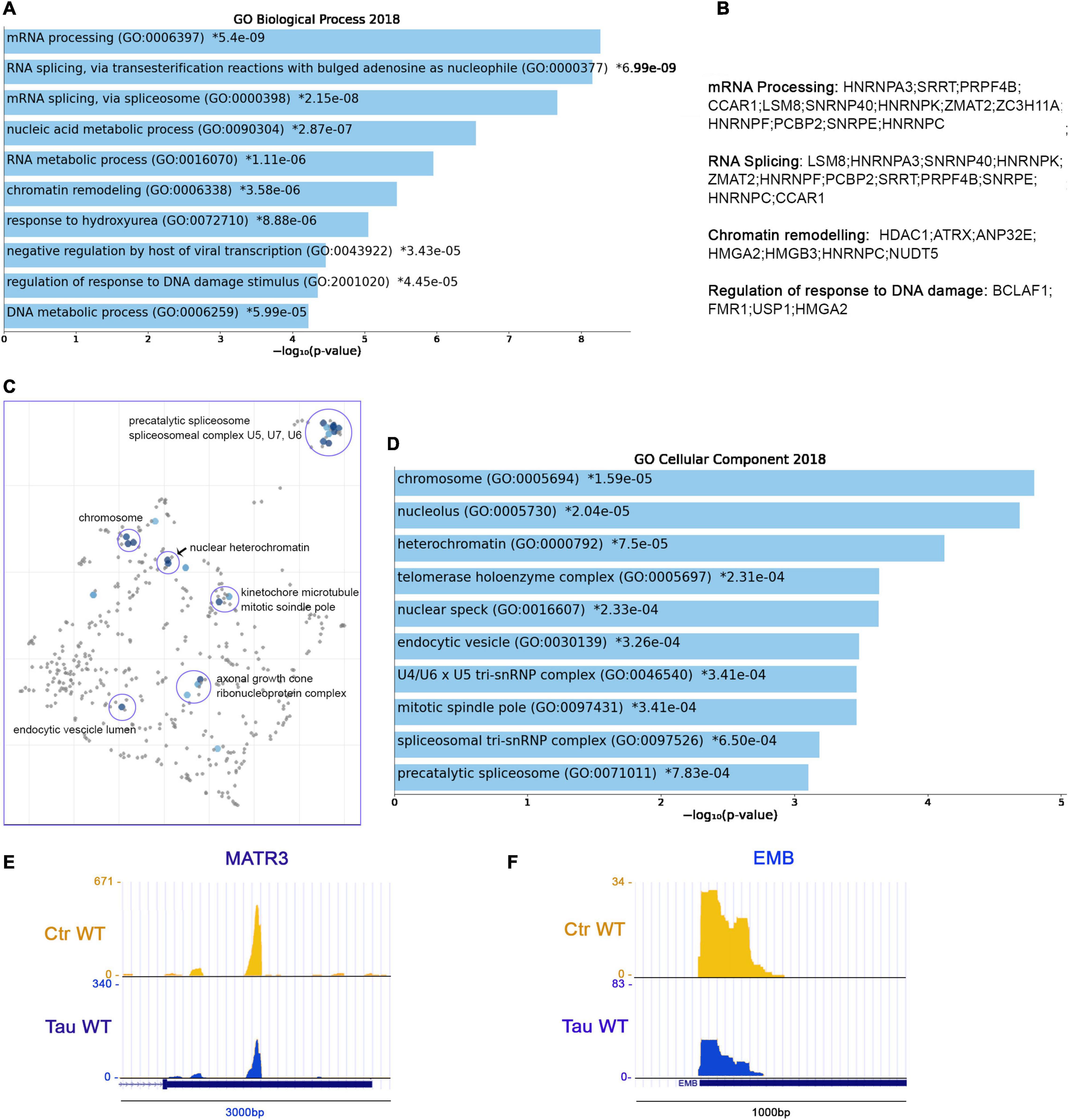
Figure 5. RNA metabolism, chromatin organization and HNRNPs precursor’s display shortened APAs in the presence of WT tau. (A) Enrich-GO Biological Process of WT tau shortened APAs (p-value reported). (B) Partial list of biological process genes with shortened APAs upon presence of WT tau (mRNA processing, RNA splicing, chromatin remodeling and regulation of response to DNA damage). (C) The Cellular Components scatterplot is organized so that similar gene sets are clustered together. The larger blue points represent significantly enriched terms—the darker the blue, the more significant the term and the smaller the p-value. The gray points are not significant. Plots has been generated and downloaded using scatter plot visualization Appyter. (D) Enrich-GO Cellular Component of WT tau shortened APAs (p-value reported). Representative poly-A site usage for shortened poly-A tails from PAC-Seq of MATR3 (E) and EMB (F) transcripts. An asterisk (*) next to a p-value indicates the term also has a significant adjusted p-value (<0.05).
Within the shortened APA precursors, various genes are related to nuclear function, such as the chromosome related genes (GO: 0005694) IK, FMR1, HMGA2, SMC4, SMC3, SMC2, and SMC6. Structural maintenance of chromosome (SMC) proteins are ATPases that are essential to chromosomal condensation, sister-chromatid cohesion, recombination, DNA repair, and epigenetic silencing of gene expression (Yatskevich et al., 2019). Eukaryotes have at least six genes encoding SMCs (SMC1-SMC6) (Aragon et al., 2013). They inherently work as heterodimers: SMC1/SMC3 (Cohesion Complex), SMC2/SMC4 (Condensin Complex) and SMC5/SMC6 (Aragon et al., 2013).
Several nucleolar (GO: 0005730) genes have altered poly(A) site usage by WT tau including: PARP1, FMR1, CHD7, DDX21, PWP1, PPM1E, SMC2, RSL1D1, ILF3, NCL, S100A13, KIF20B, RAN, and GET4. As we saw in the shortened 3′UTRs, the most affected genes for lengthened 3′ UTRs lie within the RNA binding function domain (GO: 0003723). MRNA processing, RNA splicing, and nucleic acid metabolic processes received the top scores, indicating a strong impact of WT tau in the regulation of mRNA isoforms at different levels. All significant enrichment terms are clustered and represented in a scatterplot in Figure 5C and Enrich-GO Cellular Component of WT Tau shortened APAs is represented in Figure 5D. In the mRNA processing domain (GO: 0006397) we identified several heterogeneous nuclear ribonucleoproteins (hnRNPs) genes (HNRNPA3, HNRNPK, HNRNPF, HNRNPC, HNRNPDL). HnRNPs are involved in alternative splicing, transcriptional and translational regulation, stress granules formation, cell cycle regulation, and axonal transport (Geuens et al., 2016). Their dysfunction has been shown have neurological implications, but their roles have not been comprehensively investigated. Several neurodegenerative diseases, including AD, FTD, and amyotrophic lateral sclerosis (ALS) have been associated with hnRNPs when it comes to the progression of these pathologies (Lee et al., 2013). More specifically, hnRNPK has been linked to the transcripts of several cytoskeletal genes, including MAPT, which is needed for axonogenesis (Liu and Szaro, 2011).
In Alzheimer’s disease, hnRNPC promotes APP translation (Lee et al., 2010) and stabilizes the APP precursors mRNA, which could suggest that increasing hnRNPC levels may promote Aβ secretion (Rajagopalan et al., 1998). Within the hnRNPs group, hnRNPA3, hnRNPF and hnRNPDL are all detected in pathological inclusions of ALS and FTD brains (Lee et al., 2013; Mori et al., 2013; Gami-Patel et al., 2016). Moreover, hnRNPK is a regulator of p53 (Low et al., 2021), which we and others recently discovered was present in elevated amounts in AD cortices (Baquero et al., 2019; Farmer et al., 2020). It has been also determined that hnRNPK sumoylation mediates p53 activity (Pelisch et al., 2012). All this evidence places hnRNPs in a central position for further experimental analysis in human brain tissues to elucidate more valuable information about the localization and function of this large family of ribonucleoproteins.
HnRNPA3 has been identified in neuronal cytoplasmic and intranuclear inclusions in patients with GGGGCC expansion repeats (Mori et al., 2013) and hnRNP F were also found to co-localize with GGGGCC expansion foci in immunoprecipitation studies (Mandler et al., 2014). In addition, western blot analyses imply that hnRNP may be in part responsible for the toxicity incurring by C9orf72 mutations, considering important RNA processes such as splicing are compromised. hnRNP A3 and K have been found associated with TDP-43 (Moujalled et al., 2017). Implications of tau-mediated APAs in hnRNPs open new venues for investigators to study new mechanistic insights of these proteins in several proteinophaties. Within RBPs group, we also observed the MATR3 gene. This gene encodes for Matrin3, a DNA/RNA-binding protein. Mutations in this gene cause familial ALS/FTD, and MATR3 pathology is a feature of sporadic disease, suggesting that its dysfunction is inherently linked to ALS pathogenesis (Malik et al., 2018).
Shorter 3′UTR are generally associated with enhanced translation of the mRNA APA in the presence of WT tau, which supports the finding that high-levels of hnRNPs sustain dysfunction of stress granules in ALS and FTD. Recent proteomic analysis in AD human Neurofibrillary Tangles (NFTs) showed that phospho-tau in NFTs is associated with more than 500 proteins (Drummond et al., 2020). We observed several of these proteins in the APAs shortened WT tau, such as HNRNPK, ILF3, AP2B1, RAN, RAB11A, HSP90B1, PARP1, PPIA, NCL, HNRNPA3, HSP90AA1, HNRNPC, MATR3 (Figure 5E) and EMB (Figure 5F). It is intriguing that the presence of chaperone Hsp90, a tau-regulated gene, plays a crucial role in neurodegenerative pathologies and has been studied in AD or a long time (Campanella et al., 2018).
These observations suggest that tau has early effects on gene expression that results in later stages of toxic associations commonly found in neurodegeneration. Enrich-GO (Cellular Function) of shortened-APAs genes by WT tau is provided in “Supplemental Information” section. GO-Cellular Process, Molecular Process and Cellular Components bar charts of shortened APAs are shown in Supplementary Figure 4.
Further analysis revealed 173 genes with lengthened APAs. The complete list of the 173 genes with lengthened APAs is reported in “Supplemental Information” section. Among these genes, we found that many of them are related to three major biological process: chromatin remodeling (GO: 0006338), negative regulation of gene expression (GO: 0010629), and mRNA processing (GO: 0006397) (Figure 6A). To be more specific, we noticed several genes belonging to the ATP-dependent chromatin remodeling complex npBAF (mammalian SWI/SFN, GO: 0071564): SMARCC2, ARID1A, SMARCA2, and SMARCA4. This complex is found in neuronal progenitor’s cells and post-mitotic neurons, and it is essential for the maturation of the post-mitotic neuronal phenotype as well as long-term memory formation (Sokpor et al., 2017). Along with the chromatin remodeling complex, other genes contained altered APAs, including pericentric chromatin components (GO: 0005721, HELLS, and CBX3), and nuclear chromatin factors (GO: 0000790, SMARCC2, CBX3, H3F3A, NUCKS1, ARID1A, SMARCA2, SMARCA4, HIST2H2AC, RAD50, NASP, MYC, NSMF, TCF3) (Figure 6B).
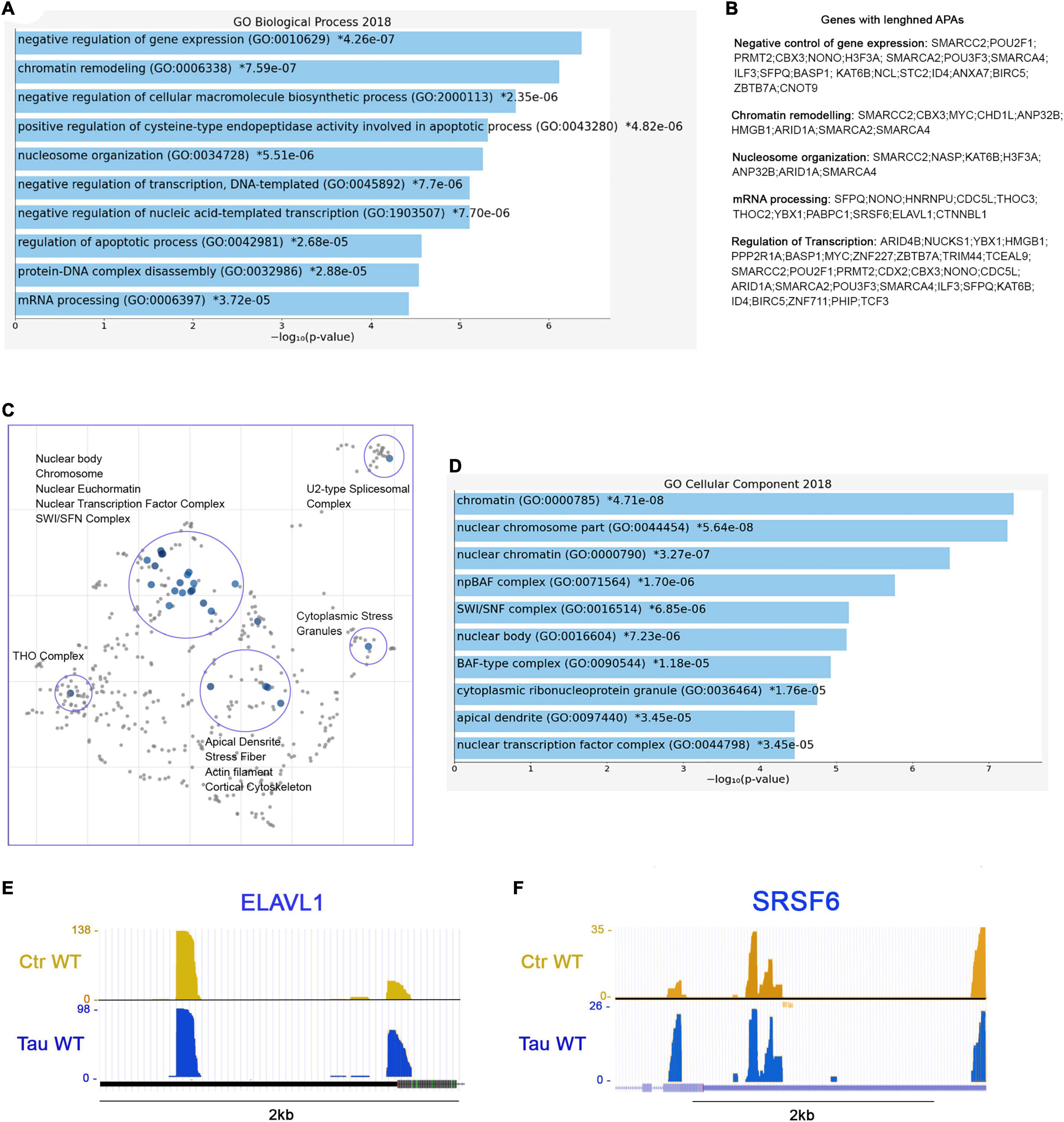
Figure 6. RNA processing and splicing precursor’s display lengthened APAs in presence of WT tau. (A) Enrich-GO Biological Process of WT tau lengthened APAs (p-value reported). (B) Partial list of biological process genes with lengthened APAs upon presence of WT tau negative control of gene expression, chromatin remodeling, nucleosome organization, mRNA processing and regulation of transcription). (C) The Cellular Components scatterplot of lengthened APAs in WT tau is organized so that similar gene sets are clustered together. The larger blue points represent significantly enriched terms—the darker the blue, the more significant the term and the smaller the p-value. The gray points are not significant. Plots has been generated and downloaded using scatter plot visualization Appyter. (D) Enrich-GO Cellular Component of WT tau lengthened APAs (p-value reported). Representative poly-A site usage for lengthened poly-A tails from PAC-Seq of ELAVL (E) and SRSF6 (F) transcripts. An asterisk (*) next to a p-value indicates the term also has a significant adjusted p-value (<0.05).
Several nuclear speck (GO: 0016607) genes were also identified: BASP1, POM,; ERBI, YLPM1, HNRNPU, LUC7L3, CDC5L, TCF3, SRSF6 (Figure 6F), and KIF20B. Cytoplasmic ribonucleoprotein granule (GO: 0036464) and cytoplasmic stress granules (GO: 0010494) genes were delineated as MBNL1, CARHSP1, NCL, HNRNPU, IQGAP1, YBX1, RAC1, PABPC1, CNOT9. Within the domain of RNA processing, two genes THOC2 and THOC3 were also identified. They are components of the THO complex (GO: 0000445) involved in efficient export of poly-adenylated RNA and spliced RNAs (Stewart, 2019).
The THO complex appears to coordinate transcripts for synapses development and dopamine neuron survival (Maeder et al., 2018). Recently, it has been found to interact with ZC3H14, which regulates the processing of neuronal transcripts (Morris and Corbett, 2018), so it is not surprising to find in our dataset another polyadenosine RNA-binding protein ZC3H15 on the list of lengthened APAs. These observations indicate that export complex RNA precursors are meaningfully affected by WT tau.
Not surprisingly, many translation initiation factors (GO: 0003743) were also discovered in our analysis including EIF2S3, EIF3E, EIF3A, EIF1, and EIF4G1. It is important to note that many APA-lengthened proteins in our study are RNA-Binding Proteins (RBPs). In fact, 46/173, or ∼27% of the total were. RBPs are implicated in the pathogenesis and progression of numerous neurodegenerative diseases, and they are linked to toxic interactions and aggregations in amyloidogenic proteins such Amyloid-beta and tau. The subsequent dysfunction of RBPs is closely related to distinct pathways that are altered in proteinophaties (Maziuk et al., 2017).
Considering the above, we also studied the presence of lengthened APAs of ELAVL1 (Figure 6E). This gene encodes for HuR (RBPs), which is a neuroprotective protein. This protein has been demonstrated in the regulation of oxidative metabolism in neurons as a way to protect from neurodegeneration (Skliris et al., 2015).
Apical dendrites (GO: 0097440) (MAP1B, NSMF and CLU) and other cytoskeletal genes (ACTR2, LIMA1, TPM4, PPP2R1A, BASP1, TARS, PHIP, NSMF, IQGAP1, RAC1, CLU, and SMARCA2) display lengthened poly-A tails as well. Enrich-GO (Cellular Function) of lengthened-APAs genes in WT tau is provided in “Supplemental Information” section. GO-Cellular Process, Molecular Process and Cellular Components bar charts of lengthened-APAs are shown in Supplementary Figure 4.
In P301L tau precursor APAs, we detected 23 lengthened genes in total. More specifically, the THOC2 gene, which is a component of the THO complex (GO: 0000445) was lengthened in WT tau. Another gene of the small nuclear ribonucleoprotein complex (SNRPE) was detected. SNRPE is also a gene for the spliceosome complex (GO: 0005681) (Figure 7A). Lastly, the nuclear replication fork (GO: 0043596) gene BAZ1B was also observed.

Figure 7. Mutant P301L tau modulates APAs associated with spliceosome and nuclear chromatin. (A) Scatterplot of gene clusters from lengthened mRNA precursors upon P301L Tau expression. (B) Scatterplot of gene clusters from shortened mRNA precursors upon P301L Tau expression. (C) Model for nuclear tau activity to transcriptional and post-transcriptional levels.
In contrast to WT tau, P301L tau induces lengthening of the HNRNPF gene. HnRNPs represent a large RNA-Binding protein family that contributes to many aspects of nucleic acid metabolism, including alternative splicing, mRNA stabilization, transcriptional, and translational regulation (Geuens et al., 2016). Dysregulation of RNA metabolism is crucial in the pathogenesis of several neurodegenerative diseases as Parkinson’s (Lu et al., 2014), FTD and overlaps with aspects of ALS. Some studies revealed possible involvement of hnRNPs in the pathogenesis and progression of these diseases (Bampton et al., 2020). Furthermore, hnRNP F has been uncovered in RNA foci in human brain tissue of FTD-ALS patients (Lee et al., 2013). Affinity pull-down assays and genome-wide analysis also revealed a hnRNP F-bound splicing complex that regulates neuronal and oligodendroglial differentiation pathways in the developing brain (Mandler et al., 2014). As observed for WT tau, the mutant P301L form also modulates several RNA-Binding Proteins (GO: 0003723): SLFN11, HNRNPF, FASN, HUWE1, PRRC2C, THOC2, HMGN2, SRSF7, and GIGYF7. We found 34 genes in total with evidence of APA and shortened 3′UTRs (Figure 7B). The three top-scored cellular components were nuclear speck (GO: 0016607), nuclear body (GO: 0016604) with RBM39 (ALS associated gene; Couthouis et al., 2011) and Nuclear heterochromatin genes (GO: 0005720). Nuclear speak and body genes consisted of LUC7L3, SRSF4, NSRP1, and SRSF11. Nuclear heterochromatin genes detected were H2AFY and HIST1H1E. H2AFY encodes for a variant of the H2A histone that is present in a subset of nucleosomes where its role is to represses transcription (Doyen et al., 2006).
The Cellular Components scatterplot of lengthened APAs in WT Tau is presented in Figure 6C and GO Cellular component bar charts in Figure 6D.
These data suggest that the mutant P301L form of tau reduces activity in transcription and alternative poly(A) tails processes due to loss-of-function. However, P301L tau does generate different mRNA isoforms of transcripts mainly translated in splicing factors, nuclear speckle/body structures and chromatin remodeling proteins. Enrich-GO (Cellular Function) of shortened and lengthened-APAs by P301L tau is provided in “Supplemental Information” section. GO-Cellular Process, Molecular Process and Cellular Components bar charts of shortened and lengthened-APAs are shown in Supplementary Figure 5.
In this study, we revealed new mechanistic insights into non-canonical tau functions. In particular, we showed novel tau activities in transcription and alternative poly-adenylation (APA) pathways. APA is a widespread mechanism of gene regulation that generates 3′ ends in transcripts made by RNA polymerase II (Tian and Manley, 2013). APA is regulated in cell proliferation, differentiation and extracellular cues. It occurs in the 3′UTR and leads to the production of mRNA isoforms, followed by splicing which leads to the production of distinct protein isoforms (Tian and Manley, 2013). Tau is typically described as an abundant neuronal microtubule-binding protein. Recently, we observed its presence within non-neuronal human cell lines and neuronal nuclei in AD brains (Montalbano et al., 2019, 2020) alongside other study (Violet et al., 2014). We were particularly interested in the possibility of non-canonical tau functions. We hypothesized that nuclear tau acts as a transcriptional regulator. To test our hypothesis, we used the tau inducible HEK system, which is a well-established cell line capable of studying mechanisms related to the tau aggregation process within a controlled system of MAPT gene expression (Koren et al., 2019). Our study employed new technologies such as Poly(A)-ClickSeq to resolve whether genes were upregulated or downregulated by WT and P301L tau in an in vitro model. Furthermore, we analyzed alternative polyadenylation (APA) profiles under the presence of WT and P301L tau (Couthouis et al., 2011).
Our results suggest that both WT and P301L tau are able to shuttle into the nuclei (Figure 4). This observation confirmed our previous observations (Montalbano et al., 2019). We did not investigate the effect of the P301L mutation on nuclei-cytoplasm shuttling in this report. The decreased number of genes expressed in P301L cells suggests that this particular mutation of tau impairs transcriptional activity. We did not investigate the consequences of P301L tau in great detail, but our observations suggest new mechanistic insights linked to alternative nuclear tau function.
One APA transcript of significance is the SFPQ gene, which we identified in WT tau expression as having a lengthened 3′UTR. SFPQ has been associated with tau as a critical factor for rapid progression of AD, and it has been observed as downregulated in post-mortem brain tissue of rapidly progressive AD patients (Younas et al., 2020). Therefore, the lengthened APAs in this gene could explain the down-regulation in the presence of a high level of tau, which mimics late-stage AD. In vitro data of SFPQ down-regulation due to human tau suggest a causal role of tau, possibly through the alternative poly-adenylation of SFPQ transcripts.
Further analysis comparing 3′UTRs lengthened between WT and P301L tau revealed that a significant number of RBPs showed lengthened 3′UTRs in P301L compared to WT tau. For example, we detected 72 RBPs including FUS (found in Supplemental Information section). These data suggest a significant difference in RNA isoforms based on genetic tau background, which then subsequently modulates different aspects of RNA metabolism in neurons.
Using the same cellular models, we determined that the prominent form of nuclear tau is monomeric, but Tet induction causes tau oligomerization within the nuclei (Montalbano et al., 2019). The formation of large and nuclear oligomeric forms is another possible explanation observed as a consequence of mutated tau. Mutant P301L tau shows a distinct aggregation mechanism compared to WT (Strang et al., 2018) and aggregates faster than WT (Barghorn et al., 2000; Aoyagi et al., 2007). For example, monomeric tau in the cytoplasm of cells producing (WT or P301L) tau aggregate and subsequently avoid nuclear translocation. In addition, aggregation in the cytoplasm and within the nuclei of tau reduces the pool of monomeric nuclear tau. This pathological mechanism can compete with functional monomeric and oligomeric tau, which then alters tau transcriptional activity. This phenomenon should be investigated in the near future using neuronal models. Another function of tau is binding DNA in vitro. Overall, the multifunctional nature of nuclear tau should be thoroughly scrutinized in order to identify unrevealed functions connected to DNA expression and RNA processing. We suggest that the nature of nuclear tau as a transcriptional factor, chromatin remodeler and/or transcriptional co-factor must be elucidated using proper models such as induced pluripotent stem cells or mouse primary neurons carrying mutation on P301 site. At this stage, we can only hypothesize the direct and indirect effects of tau during transcription.
This study utilized PAC-ClickSeq technology to identify the APA modulated by P301L and WT tau. Alternative Poly-A (APA) sites in human genome have been identify mainly in 3′UTRs (UTR-APA) sites, which harbor diverse regulatory sequences. This type of APA can change the length and composition of 3′UTR, which subsequently affects the binding of miRNAs and/or RBPs. This post-transcriptional modification leads to differences in mRNA stability, export, localization, translational efficiency (Gruber and Zavolan, 2019). Although the currently accepted theory is that genes with longer 3′UTR tend to show decreased expression levels, this does not necessarily mean that every single gene with a longer 3′UTR is less stable those with a shorter one.
We plan to investigate these findings using primary neurons and in vivo models in the near future. We are choosing these alternative models because the iHEK cell model have inherent limitations in terms of reliability as a neuronal system. However, the iHEK cells used in this study are an established model used by many researchers to study the mechanistic insights of tau aggregation and toxicity. The results presented in this study support non-canonical functions of tau. Therefore, we report broad tau-driven, post-transcriptional regulation in APAs by both WT and P301L tau considering both cell lines produced high levels of monomeric and aggregated tau. In this study, we did not investigate which tau isoform regulates APA in cells and by what method tau regulates APAs, but we established a new category of interest in post-translational modification. We hope further studies of nuclear tau and its relation to DNA and RNA processing will identify new targets in tauopathies and eventually find new therapeutic targets.
As mentioned in the discussion, the main limitation of this study is the nature of tau inducible HEK cells. We are aware that further study on neuronal cells is necessary. However, iHEK models are commonly used to study mechanisms that are tau-dependent and several of them have been translated into neurons models. All relevant datasets used and/or analyzed in this current study are available upon request from the corresponding author.
The source data underlying all main and Supplementary Figures are provided as a Source Data file. RNAseq datasets is uploaded to NCBI SRA, reference number: PRJNA744518. Figures 1A, 4F, 7C were generated using BioRender Software1.
While tau biology has been extensively studied and closely linked to several neurodegenerative diseases, our current understanding of tau’s functions in the nucleus is limited. Given the role of tau in disease progression and pathogenesis, elucidating the function of tau activity in transcription and its nuclear accumulation may reveal novel therapeutic targets; therefore, helping identify new upstream pathways that have yet to be investigated. In this study, we used tau-inducible cell lines to uncover new molecular mechanisms by which tau functions in the nucleus. This study systematically investigates the changes in transcriptomic and alternative polyadenylation profiles modulated by WT and mutant P301L tau protein. In this manuscript, we report following new findings (i) tau modulates gene expression of transcripts associated with chromatin remodeling and splicing complexes; (ii) WT and mutant P301L tau regulate, differentially, transcription and alternative polyadenylation (APA) profiles; and (iii) P301L mutation affects the transcription mediated by tau protein. The potential role of tau in mediating transcription and alternative polyadenylation processes is not well studied, representing a novelty in the field. Therefore, this research establishes a new direction for investigating tau nuclear function in both human and mouse brains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MM, AR, and RK: conceptualization and methodology. MM, EJ, SM, AE, and SG: investigation. AR and EJ: transcriptomic analysis. MM: writing—original draft. RK: funding acquisition and resources. MM and RK: supervision. All authors: writing—review and editing.
This work was supported by the Mitchell Center for Neurodegenerative Diseases, the Gillson Longenbaugh Foundation and National Institute of Health grants: R01AG054025, R01NS094557, R01AG055771, R01AG060718, and the American Heart Association collaborative grant 17CSA33620007 (RK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the members of the RK and AR labs for their support and help. We thank Bergman Isabelle B. and Leiana Fung for editing and proofreading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.742790/full#supplementary-material
AD, Alzheimer’s disease; APA, alternative polyadenylation; DDR, DNA damage response; FTD, frontal temporal dementia; GO, Gene Ontology; GSEA, gene set enrichment analysis; MTs, microtubules; PAC, poly(A) cluster; PAC-Seq; Poly(A)-ClickSeq, PAP, poly-A polymerase; PAS, poly(A) site; RNA, ribonucleotide acid; Tet, Tetracycline; 3′ UTR, 3′ untranslated region; iHEK, inducible human embryonic kidney cells; ER, endoplasmic reticulum.
Aoyagi, H., Hasegawa, M., and Tamaoka, A. (2007). Fibrillogenic nuclei composed of P301L mutant tau induce elongation of P301L tau but not wild-type Tau. J. Biol. Chem. 282, 20309–20318. doi: 10.1074/jbc.M611876200
Aragon, L., Martinez-Perez, E., and Merkenschlager, M. (2013). Condensin, cohesin and the control of chromatin states. Curr. Opin. Genet. Dev. 23, 204–211. doi: 10.1016/j.gde.2012.11.004
Bampton, A., Gittings, L. M., Fratta, P., Lashley, T., and Gatt, A. (2020). The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 140, 599–623. doi: 10.1007/s00401-020-02203-0
Baquero, J., Varriano, S., Ordonez, M., Kuczaj, P., Murphy, M. R., Aruggoda, G., et al. (2019). Nuclear Tau, p53 and Pin1 regulate PARN-mediated deadenylation and gene expression. Front. Mol. Neurosci. 12:242. doi: 10.3389/fnmol.2019.00242
Barghorn, S., Zheng-Fischhöfer, Q., Ackmann, M., Biernat, J., von Bergen, M., Mandelkow, E. M., et al. (2000). Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39, 11714–11721. doi: 10.1021/bi000850r
Bemben, M. A., Shipman, S. L., Nicoll, R. A., and Roche, K. W. (2015). The cellular and molecular landscape of neuroligins. Trends Neurosci. 38, 496–505. doi: 10.1016/j.tins.2015.06.004
Benhelli-Mokrani, H., Mansuroglu, Z., Chauderlier, A., Albaud, B., Gentien, D., Sommer, S., et al. (2018). Genome-wide identification of genic and intergenic neuronal DNA regions bound by Tau protein under physiological and stress conditions. Nucleic Acids Res. 46, 11405–11422. doi: 10.1093/nar/gky929
Bittermann, E., Abdelhamed, Z., Liegel, R. P., Menke, C., Timms, A., Beier, D. R., et al. (2019). Differential requirements of tubulin genes in mammalian forebrain development. PLoS Genet. 15:e1008243. doi: 10.1371/journal.pgen.1008243
Brito-Moreira, J., Lourenco, M. V., Oliveira, M. M., Ribeiro, F. C., Ledo, J. H., Diniz, L. P., et al. (2017). Interaction of amyloid-β (Aβ) oligomers with neurexin 2α and neuroligin 1 mediates synapse damage and memory loss in mice. J. Biol. Chem. 292, 7327–7337. doi: 10.1074/jbc.M116.761189
Campanella, C., Pace, A., Caruso Bavisotto, C., Marzullo, P., Marino Gammazza, A., Buscemi, S., et al. (2018). Heat shock proteins in Alzheimer’s disease: role and targeting. Int. J. Mol. Sci. 19:2603. doi: 10.3390/ijms19092603
Couthouis, J., Hart, M. P., Shorter, J., DeJesus-Hernandez, M., Erion, R., Oristano, R., et al. (2011). A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. U.S.A. 108, 20881–20890. doi: 10.1073/pnas.1109434108
Craig, A. M., and Kang, Y. (2007). Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17, 43–52. doi: 10.1016/j.conb.2007.01.011
Doyen, C.-M., An, W., Angelov, D., Bondarenko, V., Mietton, F., Studitsky, V. M., et al. (2006). Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 26, 1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006
Drummond, E., Pires, G., MacMurray, C., Askenazi, M., Nayak, S., Bourdon, M., et al. (2020). Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain 143, 2803–2817. doi: 10.1093/brain/awaa223
Dufort-Gervais, J., Provost, C., Charbonneau, L., Norris, C. M., Calon, F., Mongrain, V., et al. (2020). Neuroligin-1 is altered in the hippocampus of Alzheimer’s disease patients and mouse models, and modulates the toxicity of amyloid-beta oligomers. Sci. Rep. 10:6956. doi: 10.1038/s41598-020-63255-6
Eftekharzadeh, B., Daigle, J. G., Kapinos, L. E., Coyne, A., Schiantarelli, J., Carlomagno, Y., et al. (2018). Tau protein disrupts nucleocytoplasmic transport in alzheimer’s disease. Neuron 99, 925.e7–940.e7. doi: 10.1016/j.neuron.2018.07.039
El Helou, J., Bélanger-Nelson, E., Freyburger, M., Dorsaz, S., Curie, T., La Spada, F., et al. (2013). Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 9974–9979. doi: 10.1073/pnas.1221381110
Elrod, N. D., Jaworski, E. A., Ji, P., Wagner, E. J., and Routh, A. (2019). Development of Poly(A)-ClickSeq as a tool enabling simultaneous genome-wide poly(A)-site identification and differential expression analysis. Methods 155, 20–29. doi: 10.1016/j.ymeth.2019.01.002
Farmer, K. M., Ghag, G., Puangmalai, N., Montalbano, M., Bhatt, N., and Kayed, R. (2020). P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer’s disease. Acta Neuropathol. Commun. 8:132. doi: 10.1186/s40478-020-01012-6
Gami-Patel, P., Bandopadhyay, R., Brelstaff, J., Revesz, T., and Lashley, T. (2016). The presence of heterogeneous nuclear ribonucleoproteins in frontotemporal lobar degeneration with FUS-positive inclusions. Neurobiol. Aging 46, 192–203. doi: 10.1016/j.neurobiolaging.2016.07.004
Geuens, T., Bouhy, D., and Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. doi: 10.1007/s00439-016-1683-5
Gruber, A. J., and Zavolan, M. (2019). Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 20, 599–614. doi: 10.1038/s41576-019-0145-z
Hua, Q., and He, R. (2003). Tau could protect DNA double helix structure. Biochim. Biophys. Acta 1645, 205–211. doi: 10.1016/s1570-9639(02)00538-1
Ito, D., Walker, J. R., Thompson, C. S., Moroz, I., Lin, W., Veselits, M. L., et al. (2004). Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol. Cell. Biol. 24, 9456–9469. doi: 10.1128/MCB.24.21.9456-9469.2004
Jaworski, E., and Routh, A. (2018). ClickSeq: replacing fragmentation and enzymatic ligation with click-chemistry to prevent sequence chimeras. Methods Mol. Biol. 1712, 71–85. doi: 10.1007/978-1-4939-7514-3_6
Kampers, T., Friedhoff, P., Biernat, J., Mandelkow, E. M., and Mandelkow, E. (1996). RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 399, 344–349. doi: 10.1016/s0014-5793(96)01386-5
Karch, C. M., and Goate, A. M. (2015). Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51. doi: 10.1016/j.biopsych.2014.05.006
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Klein, H.-U., McCabe, C., Gjoneska, E., Sullivan, S. E., Kaskow, B. J., Tang, A., et al. (2019). Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 22, 37–46. doi: 10.1038/s41593-018-0291-1
Koren, S. A., Hamm, M. J., Meier, S. E., Weiss, B. E., Nation, G. K., Chishti, E. A., et al. (2019). Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 137, 571–583. doi: 10.1007/s00401-019-01970-9
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377
Kwon, H. S., and Koh, S.-H. (2020). Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl. Neurodegen. 9:42. doi: 10.1186/s40035-020-00221-2
Lee, E. K., Kim, H. H., Kuwano, Y., Abdelmohsen, K., Srikantan, S., Subaran, S. S., et al. (2010). hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 17, 732–739. doi: 10.1038/nsmb.1815
Lee, S.-H., Peng, I.-F., Ng, Y. G., Yanagisawa, M., Bamji, S. X., Elia, L. P., et al. (2008). Synapses are regulated by the cytoplasmic tyrosine kinase Fer in a pathway mediated by p120catenin, Fer, SHP-2, and beta-catenin. J. Cell Biol. 183, 893–908. doi: 10.1083/jcb.200807188
Lee, Y.-B., Chen, H.-J., Peres, J. N., Gomez-Deza, J., Attig, J., Stalekar, M., et al. (2013). Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186. doi: 10.1016/j.celrep.2013.10.049
Lester, E., and Parker, R. (2018). The Tau of nuclear-cytoplasmic transport. Neuron 99, 869–871. doi: 10.1016/j.neuron.2018.08.026
Liu, Y., and Szaro, B. G. (2011). hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development 138, 3079–3090. doi: 10.1242/dev.066993
Loomis, P. A., Howard, T. H., Castleberry, R. P., and Binder, L. I. (1990). Identification of nuclear tau isoforms in human neuroblastoma cells. Proc. Natl. Acad. Sci. U.S.A. 87, 8422–8426. doi: 10.1073/pnas.87.21.8422
Low, Y.-H., Asi, Y., Foti, S. C., and Lashley, T. (2021). Heterogeneous nuclear ribonucleoproteins: implications in neurological diseases. Mol. Neurobiol. 58, 631–646. doi: 10.1007/s12035-020-02137-4
Lu, B., Gehrke, S., and Wu, Z. (2014). RNA metabolism in the pathogenesis of Parkinson×s disease. Brain Res. 1584, 105–115. doi: 10.1016/j.brainres.2014.03.003
Maeder, C. I., Kim, J.-I., Liang, X., Kaganovsky, K., Shen, A., Li, Q., et al. (2018). The THO complex coordinates transcripts for synapse development and dopamine neuron survival. Cell 174, 1436.e20–1449.e20. doi: 10.1016/j.cell.2018.07.046
Maina, M. B., Bailey, L. J., Doherty, A. J., and Serpell, L. C. (2018a). The involvement of Abeta42 and Tau in nucleolar and protein synthesis machinery dysfunction. Front. Cell. Neurosci. 12:220. doi: 10.3389/fncel.2018.00220
Maina, M. B., Bailey, L. J., Wagih, S., Biasetti, L., Pollack, S., Quinn, J., et al. (2018b). The involvement of tau in nucleolar transcription and the stress response. Acta Neuropathol. Commun. 6:70. doi: 10.1186/s40478-018-0565-6
Malik, A. M., Miguez, R. A., Li, X., Ho, Y.-S., Feldman, E. L., and Barmada, S. J. (2018). Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization. eLife 7:e35977. doi: 10.7554/eLife.35977
Mandler, M. D., Ku, L., and Feng, Y. (2014). A cytoplasmic quaking I isoform regulates the hnRNP F/H-dependent alternative splicing pathway in myelinating glia. Nucleic Acids Res. 42, 7319–7329. doi: 10.1093/nar/gku353
Mansuroglu, Z., Benhelli-Mokrani, H., Marcato, V., Sultan, A., Violet, M., Chauderlier, A., et al. (2016). Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep. 6:33047. doi: 10.1038/srep33047
Maziuk, B., Ballance, H. I., and Wolozin, B. (2017). Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front. Mol. Neurosci. 10:89. doi: 10.3389/fnmol.2017.00089
Miyasaka, T., Shinzaki, Y., Yoshimura, S., Yoshina, S., Kage-Nakadai, E., Mitani, S., et al. (2018). Imbalanced expression of Tau and tubulin induces neuronal dysfunction in C. elegans models of tauopathy. Front. Neurosci. 12:415. doi: 10.3389/fnins.2018.00415
Monroy-Ramírez, H. C., Basurto-Islas, G., Mena, R., Cisneros, B., Binder, L. I., Avila, J., et al. (2013). Alterations in the nuclear architecture produced by the overexpression of tau protein in neuroblastoma cells. J. Alzheimers Dis. 36, 503–520. doi: 10.3233/JAD-122401
Montalbano, M., McAllen, S., Puangmalai, N., Sengupta, U., Bhatt, N., Johnson, O. D., et al. (2020). RNA-binding proteins Musashi and tau soluble aggregates initiate nuclear dysfunction. Nat. Commun. 11:4305. doi: 10.1038/s41467-020-18022-6
Montalbano, M., McAllen, S., Sengupta, U., Puangmalai, N., Bhatt, N., Ellsworth, A., et al. (2019). Tau oligomers mediate aggregation of RNA-binding proteins Musashi1 and Musashi2 inducing Lamin alteration. Aging Cell 18:e13035. doi: 10.1111/acel.13035
Morales, I., Jiménez, J. M., Mancilla, M., and Maccioni, R. B. (2013). Tau oligomers and fibrils induce activation of microglial cells. J. Alzheimers Dis. 37, 849–856. doi: 10.3233/JAD-131843
Mori, K., Lammich, S., Mackenzie, I. R. A., Forné, I., Zilow, S., Kretzschmar, H., et al. (2013). hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 125, 413–423. doi: 10.1007/s00401-013-1088-7
Morris, K. J., and Corbett, A. H. (2018). The polyadenosine RNA-binding protein ZC3H14 interacts with the THO complex and coordinately regulates the processing of neuronal transcripts. Nucleic Acids Res. 46, 6561–6575. doi: 10.1093/nar/gky446
Moujalled, D., Grubman, A., Acevedo, K., Yang, S., Ke, Y. D., Moujalled, D. M., et al. (2017). TDP-43 mutations causing amyotrophic lateral sclerosis are associated with altered expression of RNA-binding protein hnRNP K and affect the Nrf2 antioxidant pathway. Hum. Mol. Genet. 26, 1732–1746. doi: 10.1093/hmg/ddx093
Niday, Z., Hawkins, V. E., Soh, H., Mulkey, D. K., and Tzingounis, A. V. (2017). Epilepsy-associated KCNQ2 channels regulate multiple intrinsic properties of layer 2/3 pyramidal neurons. J. Neurosci. 37, 576–586. doi: 10.1523/JNEUROSCI.1425-16.2016
Paonessa, F., Evans, L. D., Solanki, R., Larrieu, D., Wray, S., Hardy, J., et al. (2019). Microtubules deform the nuclear membrane and disrupt nucleocytoplasmic transport in Tau-mediated frontotemporal dementia. Cell Rep. 26, 582.e5–593.e5. doi: 10.1016/j.celrep.2018.12.085
Patrón, L. A., Nagatomo, K., Eves, D. T., Imad, M., Young, K., Torvund, M., et al. (2019). Cul4 ubiquitin ligase cofactor DCAF12 promotes neurotransmitter release and homeostatic plasticity. J. Cell Biol. 218, 993–1010. doi: 10.1083/jcb.201805099
Paudel, Y. N., Angelopoulou, E., Piperi, C., Othman, I., Aamir, K., and Shaikh, M. F. (2020). Impact of HMGB1, RAGE, and TLR4 in Alzheimer’s disease (AD): from risk factors to therapeutic targeting. Cells 9:383. doi: 10.3390/cells9020383
Pelisch, F., Pozzi, B., Risso, G., Muñoz, M. J., and Srebrow, A. (2012). DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J. Biol. Chem. 287, 30789–30799. doi: 10.1074/jbc.M112.390120
Rajagopalan, L. E., Westmark, C. J., Jarzembowski, J. A., and Malter, J. S. (1998). hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 26, 3418–3423. doi: 10.1093/nar/26.14.3418
Rio, D. C., Ares, M. J., Hannon, G. J., and Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010:db.rot5439. doi: 10.1101/pdb.prot5439
Routh, A. (2019). DPAC: a tool for differential poly(A)-cluster usage from poly(A)-targeted RNAseq data. G3 9, 1825–1830. doi: 10.1534/g3.119.400273
Shea, T. B., and Cressman, C. M. (1998). A 26-30 kDa developmentally-regulated tau isoform localized within nuclei of mitotic human neuroblastoma cells. Int. J. Dev. Neurosci. 16, 41–48. doi: 10.1016/s0736-5748(97)00044-0
Shepard, P. J., Choi, E.-A., Lu, J., Flanagan, L. A., Hertel, K. J., and Shi, Y. (2011). Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772. doi: 10.1261/rna.2581711
Short, B., Preisinger, C., Körner, R., Kopajtich, R., Byron, O., and Barr, F. A. (2001). A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–884. doi: 10.1083/jcb.200108079
Siano, G., Varisco, M., Caiazza, M. C., Quercioli, V., Mainardi, M., Ippolito, C., et al. (2019). Tau modulates VGluT1 expression. J. Mol. Biol. 431, 873–884. doi: 10.1016/j.jmb.2019.01.023
Skliris, A., Papadaki, O., Kafasla, P., Karakasiliotis, I., Hazapis, O., Reczko, M., et al. (2015). Neuroprotection requires the functions of the RNA-binding protein HuR. Cell Death Diff. 22, 703–718. doi: 10.1038/cdd.2014.158
Sokpor, G., Xie, Y., Rosenbusch, J., and Tuoc, T. (2017). Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 10:243. doi: 10.3389/fnmol.2017.00243
Stewart, M. (2019). Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 294, 2977–2987. doi: 10.1074/jbc.REV118.005594
Strang, K. H., Croft, C. L., Sorrentino, Z. A., Chakrabarty, P., Golde, T. E., and Giasson, B. I. (2018). Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 293, 2408–2421. doi: 10.1074/jbc.M117.815357
Sultan, A., Nesslany, F., Violet, M., Bégard, S., Loyens, A., Talahari, S., et al. (2011). Nuclear tau, a key player in neuronal DNA protection. J. Biol. Chem. 286, 4566–4575. doi: 10.1074/jbc.M110.199976
Tian, B., Hu, J., Zhang, H., and Lutz, C. S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212. doi: 10.1093/nar/gki158
Tian, B., and Manley, J. L. (2013). Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 38, 312–320. doi: 10.1016/j.tibs.2013.03.005
Tripathi, T., Prakash, J., and Shav-Tal, Y. (2019). Phospho-Tau impairs nuclear-cytoplasmic transport. ACS Chem. Neurosci. 10, 36–38. doi: 10.1021/acschemneuro.8b00632
Ulrich, G., Salvade, A., Boersema, P., Calì, T., Foglieni, C., Sola, M., et al. (2018). Phosphorylation of nuclear Tau is modulated by distinct cellular pathways. Sci. Rep. 8:17702. doi: 10.1038/s41598-018-36374-4
Venkatramani, A., and Panda, D. (2019). Regulation of neuronal microtubule dynamics by tau: implications for tauopathies. Int. J. Biol. Macromol. 133, 473–483. doi: 10.1016/j.ijbiomac.2019.04.120
Violet, M., Delattre, L., Tardivel, M., Sultan, A., Chauderlier, A., Caillierez, R., et al. (2014). A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front. Cell. Neurosci. 8:84. doi: 10.3389/fncel.2014.00084
Wu, X., Morishita, W. K., Riley, A. M., Hale, W. D., Südhof, T. C., and Malenka, R. C. (2019). Neuroligin-1 signaling controls LTP and NMDA receptors by distinct molecular pathways. Neuron 102, 621.e3–635.e3. doi: 10.1016/j.neuron.2019.02.013
Yatskevich, S., Rhodes, J., and Nasmyth, K. (2019). Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482. doi: 10.1146/annurev-genet-112618-043633
Younas, N., Zafar, S., Shafiq, M., Noor, A., Siegert, A., Arora, A. S., et al. (2020). SFPQ and Tau: critical factors contributing to rapid progression of Alzheimer’s disease. Acta Neuropathol. 140, 317–339. doi: 10.1007/s00401-020-02178-y
Zhang, X., Lin, Y., Eschmann, N. A., Zhou, H., Rauch, J. N., Hernandez, I., et al. (2017). RNA stores tau reversibly in complex coacervates. PLoS Biol. 15:e2002183. doi: 10.1371/journal.pbio.2002183
Keywords: alternative polyadenylation, frontal temporal dementia, gene ontology, gene set enrichment analysis, tau, neurodegenaration
Citation: Montalbano M, Jaworski E, Garcia S, Ellsworth A, McAllen S, Routh A and Kayed R (2021) Tau Modulates mRNA Transcription, Alternative Polyadenylation Profiles of hnRNPs, Chromatin Remodeling and Spliceosome Complexes. Front. Mol. Neurosci. 14:742790. doi: 10.3389/fnmol.2021.742790
Received: 16 July 2021; Accepted: 09 November 2021;
Published: 03 December 2021.
Edited by:
Neha Gogia, Yale University, United StatesReviewed by:
Victoria Campos-Peña, Manuel Velasco Suárez Instituto Nacional de Neurología y Neurocirugía, MexicoCopyright © 2021 Montalbano, Jaworski, Garcia, Ellsworth, McAllen, Routh and Kayed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakez Kayed, cmFrYXllZEB1dG1iLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.