- 1Department of Endocrinology, Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing, China
- 2School of Biomedical Sciences, University of Queensland, Brisbane, QLD, Australia
Organisms have developed common behavioral and physiological adaptations to the influence of the day/night cycle. The CLOCK system forms an internal circadian rhythm in the suprachiasmatic nucleus (SCN) during light/dark input. The SCN may synchronize the growth hormone (GH) secretion rhythm with the dimming cycle through somatostatin neurons, and the change of the clock system may be related to the pulsatile release of GH. The GH—insulin-like growth factor 1 (IGF-1) axis and clock system may interact further on the metabolism through regulatory pathways in peripheral organs. We have summarized the current clinical and animal evidence on the interaction of clock systems with the GH—IGF-1 axis and discussed their effects on metabolism.
Introduction
All animals live under the influence of the 24-h cycle of the earth’s rotation. Organisms sense these regular external changes and synchronize their physical activities, such as behavior, food intake, energy metabolism, sleep, reproductive activity, and immune function, to increase their survival abilities (Takahashi et al., 2008). Organisms have developed a highly conservative and complex molecular clock system, which creates an internal circadian rhythm during light/dark input (Buhr and Takahashi, 2013). The output of this regulatory system is linked to numerous organs and tissues, relaying different signals released by the central circadian system (Hastings et al., 2007). The mammalian brain’s central clock system consists of pairs of SCN which locate at the base of the hypothalamus. These clusters of about 10,000 GABA-enabled neurons including ventricular cores that receive direct neural control from the retina and brainstem region (Reppert and Weaver, 2002). SCN controls the endocrine cycle and metabolic rhythm in two ways. First, SCN determines the timing of sleep-dependent events, such as nocturnal secretions of prolactin and growth hormone, by dissecting the centers that control sleep and wakefulness. In addition, the central clock system can regulate the rhythmic release of hormones such as melatonin and cortisol by linking to the neuroendocrine and autonomic nervous systems independently of sleep-driving hormones and other rhythms (Hastings et al., 2007). The peripheral clock system plays a role in almost all organs and tissues. The activity of the peripheral clock system is synchronized with the central master clock system through body fluids and neural connections. The central and peripheral clocks use the same set of transcription factors, including CLOCK and BMAL1, to generate circadian pattern of gene expression (Kalsbeek et al., 2006; Nicolaides et al., 2014).
SCN may act on somatostatinergic neurons and GH-releasing hormones (GHRH) to synchronize the GH rhythm with the light-dark cycle (Willoughby and Martin, 1978; Vaccarino et al., 1995; Davies et al., 2004). At the same time, as the aging process progresses, the decrease in the clock rhythm (Hastings et al., 2003) is also consistent with the decrease in the pulsatile release of GH (Kuwahara et al., 2004). Therefore, there may be a connection between the clock system and the rhythmic release of hormones. A large number of experiments and reviews have confirmed the interaction between circadian rhythm regulation and the hypothalamus-pituitary-adrenal cortisol (HPA) axis (Nader et al., 2010; Nicolaides et al., 2014). There is currently a lack of relevant review to summarize the interaction between circadian rhythm regulation and the GH/IGF-1 axis. Our previous experiments have confirmed that the circadian rhythm disorder caused by changes in lighting interferes with the pulsatile release pattern of growth hormone in male mice and is accompanied by changes in the expression of peripheral clock genes in the liver (Wang et al., 2021). Therefore, this review will focus on the interaction between the circadian rhythm and the GH-IGF-1 axis and its effect on metabolism.
Circadian Clock System
The CLOCK transcript forms a heterodimer with brain and muscle arnt-like protein 1 (BMAL1). Under the control of the biological clock system, the heterodimer CLOCK/BMAL1 and a series of other transcription factors are responsible for the circadian oscillation of gene expression (Hastings et al., 2007; Takahashi et al., 2008). The molecular mechanism of the circadian oscillation of gene expression is mediated by the transcription/translation feedback loop (Nader et al., 2010). The CLOCK/BMAL1 heterodimer combines with the E-box response element which located in the promoter region to stimulate the expression of other target genes, the core of which is the transcriptional expression of other clock genes, such as Periods (PER1, PER2, and PER3), and Cryptochromes (CRY1 and CRY2). The activated Pers and Crys stimulate the activities of casein kinases 1ε/δ and inhibit the transcriptional activity of the CLOCK/BMAL1 by inhibiting the binding to the E-box response element. A negative feedback transcription cycle is eventually formed to maintain the oscillation of the gene expression (Kiyohara et al., 2006; Kondratov et al., 2006b). In addition to the regulation of this main transcription loop, CLOCK/BMAL1 stimulates the expression of other clock-related transcriptions, such as REV-ERBα, retinoic acid-borne orphaned binders a (RORα), DEC1, DEC2, and albumin D-binding (DBP), which form an auxiliary loop that stabilizes the main regulatory loop (Ripperger and Schibler, 2006; Padmanabhan et al., 2012). Importantly, transcription factors in the main regulatory and auxiliary loops control many downstream circadian clock-related genes and affect a variety of biological activities such as sleep/wake cycle, eating pattern, energy consumption, and glucose metabolism (Takahashi et al., 2008; Figure 1). In addition to neural connections, the central clock system also synchronizes the circadian rhythms of the peripheral clock system through hormones or factors, such as arginine vasopressin (AVP) and tumor necrosis factor (TNF)α (Kraves and Weitz, 2006; Hastings et al., 2007).
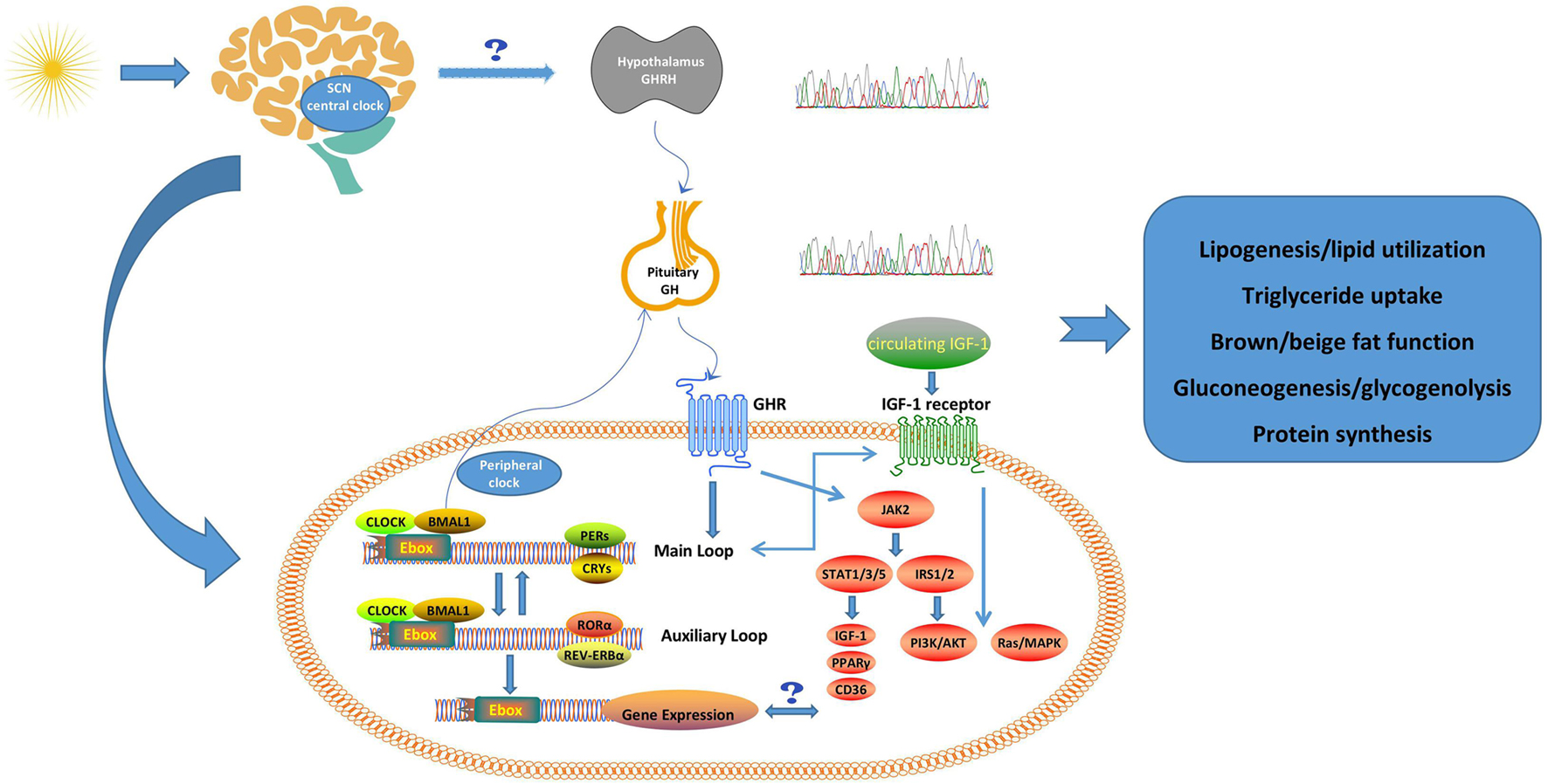
Figure 1. Possible crosstalk between clock system and GH/IGF-1 axis. The CLOCK system forms an internal circadian rhythm in the SCN during light/dark cycles. The SCN may synchronize the GH secretion rhythm with the dimming cycle through somatostatin neurons, and the change of the clock system may be related to the pulsatile release of GH. The GH/IGF-1 axis and clock system may interact further on the metabolism through several pathways in peripheral tissues (e.g., liver, fat). The physiological roles of GH and IGF-1 are also summarized in this figure (Huang et al., 2020b). SCN, suprachiasmatic nucleus; GHRH, Growth hormone-releasing hormone; GH, growth hormone; GHR, growth hormone receptor; IGF-1, insulin-like growth factor 1; BMAL1, brain-muscle-arnt-like protein 1; PERs, periods; CRYS, Cryptochromes; RORα, retinoic acid-related orphan receptor α; Ebox, enhancer motif; JAK2, Janus kinase 2; STAT, signal transducer and activator of transcription; IRS, insulin receptor substrate; PPARγ, peroxisome proliferator-activated receptor γ; PI3K/Akt, phosphatidylinositol 3-kinase/protein kinase B.
The clock system mainly regulates metabolism in the following three ways. The first one is to control nuclear receptors. The turnover of carbohydrates, proteins, and lipids, and the production/storage of energy are necessary for survival. Approximately 10% of energy-controlling gene expression, including those encoding nuclear hormone receptors and glucose and lipid metabolism enzymes, are regulated by circadian rhythm in a tissue-specific manner (Panda et al., 2002; Storch et al., 2002; Yang et al., 2006). Nuclear receptors constitute a superfamily of ligand-activated transcription factors, which regulate critical physiological processes including growth, development, hormonal signals, reproduction, and energy metabolism (Sonoda et al., 2008). Special nuclear receptors are used as sensors for metabolites such as hormones, vitamins, and lipids. The expression of some nuclear receptors is regulated by CLOCK and BMAL1. These receptors include retinoic acid-related orphan receptor α (RORα), REV-ERBα, and peroxisome proliferator-activated receptor (PPAR)α (Oishi et al., 2005). One of the nuclear receptors, REV-ERBα, is also a negative regulator of the rhythmic CLOCK transcription circuit. It may inhibit glucogenesis, lipid metabolism, adipocyte differentiation, and the transcriptional activities of several other nuclear receptors, including PPARγ and RORα (Yin et al., 2007; Duez and Staels, 2008; Figure 1). CLOCK−/− and BMAL1−/− mice exhibit disorders of glucose metabolism and circadian changes in circulating glucose and triglycerides, which lead to obesity, hyperlipidemia, and diabetes (Rudic et al., 2004; Turek et al., 2005; Sahar and Sassone-Corsi, 2012; Albrecht, 2017). The mice have increased expression of plasminogen activator inhibitor-1 (PAI-1), which is a known risk factor for obesity, diabetes, and cardiovascular disease (Oishi et al., 2006; Oishi, 2009). Another circadian clock protein PER2 inhibits the expression of PAI-1 in a CLOCK/BMAL1-dependent manner as an important factor in the development of these metabolic diseases after the circadian clock system is dysregulated (Oishi et al., 2009). In addition, the mRNA expression of BMAL1, PER2, and CRY1 in visceral fat is closely related to the increase in waist circumference which is an indicator of metabolic syndrome (Gómez-Abellán et al., 2008).
The second is that the circadian clock system may control the rate-limiting steps of the metabolic process (Panda et al., 2002). For example, the activation of the rate-limiting enzyme HMG-CoA reductase (HMGCR) in cholesterol biosynthesis shows circadian rhythm (Demierre et al., 2005), and the activity is the highest during the night. In addition, the circadian clock system may control the expression of nicotinamide phosphosarcosyltransferase (NAMPT), which is a key rate-limiting enzyme in the salvage pathway of NAD+ biosynthesis (Ramsey et al., 2009). The rhythm of the enzyme expression drives the oscillation of NAD + levels, and the synthesis of NAD+ is involved in the process of aging and lipid metabolism (Belenky et al., 2007). NAD+ regulates the circadian clock system through SIRT1. SIRT1 is a histone deacetylase, which may regulate the transcriptional activity of BMAL1/CLOCK, forming a metabolic feedback loop again between the circadian clock system and metabolism (Nakahata et al., 2009; Ramsey et al., 2009).
The last pathway is that the circadian clock system controls cell metabolism by regulating the nutrient sensors Sirt1 and AMP-activated protease (AMPK) (Sahar and Sassone-Corsi, 2012). Sirt1 regulates gene expression through histone deacetylation. Circadian gene expression and BMAL1 acetylation are disturbed in liver-specific SIRT1 mutant mice (Nakahata et al., 2008). AMPK is a key factor in energy regulation. The activity of AMPK is found to be rhythmic in mouse liver, hypothalamus, and fibroblasts (Lamia et al., 2009). AMPK may regulate the circadian rhythm by phosphorylating CRY1 (Lamia et al., 2009) and casein kinase 1 (CK1)ε (Yang et al., 2017). CK1ε plays an important role in regulating the circadian rhythm by phosphorylating PER protein and controlling its degradation (Shirogane et al., 2005). Interestingly, the activation of AMPK also leads to an increase in NAD+ levels (Cantó et al., 2009). Therefore, AMPK may indirectly regulate the expression of circadian genes through the activation of SIRT1.
The circadian rhythm system, CLOCK gene mechanism and metabolic pathways are intertwined by neural circuits which transmit the environmental signals to peripheral organs through hormones, chemokines and neuropeptides (Mazzoccoli et al., 2012). The desynchronization among central and peripheral clock system, metabolic pathways and regulators may impair the metabolic homeostasis which could contribute to the progress of obesity, metabolic syndrome, and diabetes.
GH-IGF-1 Axis
One major function of GH is to stimulate tissue growth. Lack of GH may lead to dwarfism, while excessive GH may lead to giantism. A variety of neurotransmitter pathways, as well as various peripheral feedback signals, regulate the secretion of GH by acting directly on the anterior pituitary gland and/or by regulating the release of GHRH or somatostatin in the hypothalamus (Giustina and Veldhuis, 1998). GH secreted from the pituitary gland acts on the peripheral organs and stimulates the production of IGF-1 (Cuttler, 1996). Growth hormone and IGF-1 play a variety of regulatory roles in the body. One of the major functions of GH and IGF-1 is to promote linear growth. However, GH and IGF-1 have different effects on glucose and lipid metabolism. GH may antagonize some actions of insulin, to promote lipolysis and hinder adipogenesis, while IGF-1 has the opposite effect (Moøller and Joørgensen, 2009). In the feeding state, GH secretion decreases while insulin secretion increases, leading to increased glucose uptake by skeletal muscle and fat accumulation. In the fasting state, GH increases lipolysis and hepatic glucose output while insulin concentration decreases (Rabinowitz et al., 1965). However, a meta-analysis revealed that fasting and energy restricting diets did not generate a significant effect on circulating IGF-1 (Rahmani et al., 2019). A concept of insulin-growth hormone balance has been proposed that the ratio of two hormones is closely related to the glucose and lipid metabolism and energy metabolism of obese patients (Huang et al., 2020b). After adulthood, the secretion of GH and IGF-1 continues to decrease, and the secretion of elderly people over 60 years old is significantly diminished (Zadik et al., 1985). Studies have revealed that the GH/IGF-1 axis plays a key role in the aging process of humans and animals (Junnila et al., 2013). At the same time, the GH/IGF-1 axis is also involved in the pathogenesis of obesity (Berryman et al., 2013), cardiovascular disease (Colao, 2008), and tumor (Chhabra et al., 2011).
GH receptor is a member of the class I cytokine receptor family and exists in almost all cell types in the human body (Waters et al., 1999). GH activates Janus kinase 2 (JAK2)/STAT and Src/MAPK pathways after binding to GH receptor (Brooks and Waters, 2010). The former mainly regulates metabolism, while the latter regulates mitotic function. JAK2 controls the metabolic effects by activating STAT1, 3, and 5, of which STAT5 is the most prominent one. It may also promote the production of IGF-1 to accelerate linear growth (Woelfle et al., 2003). Studies have shown that JAK2 may also phosphorylate insulin receptor substrate 1/2 (IRS1/2) and activate the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) pathway (Ridderstrale et al., 1995; Argetsinger et al., 1996; Yamauchi et al., 1998; Figure 1). However, these studies either used super-physiological GH doses (Ridderstrale et al., 1995; Yamauchi et al., 1998) or did not evaluate the physiological effects of GH administration (Argetsinger et al., 1996), and the conclusions may not apply to physiological situations. Another study has shown that GH increases lipolysis by activating MEK/ERK and inhibiting PPARγ and fat-specific protein 27 (FSP27) (Sharma et al., 2019). GH also stimulates muscle lipid uptake by increasing muscle lipoprotein lipase (LPL) activity (LeRoith and Yakar, 2007). The free fatty acids released from white adipose tissue (WAT) are absorbed and oxidized in other tissues, so the net effect of elevated GH promotes the reduction of body fat accumulation. Recent studies have also found that the GHR-JAK2-STAT5 signal inhibits lipid uptake and neo-adipogenesis in the liver, partly by inhibiting PPARγ and downstream CD36 (Sos et al., 2011; Liu et al., 2016; Chhabra et al., 2019). This shows that GH has a direct effect on lipid metabolism in the liver and may reduce the occurrence of non-alcoholic fatty liver.
Previous studies have revealed that GH controls the generation of IGF-1 by targeting the gene transcription of IGF-1 via STAT5 in diverse physiological situations (Chia et al., 2010; Rotwein, 2012). The liver is the major organ of the synthesis of endocrine factors including IGF-1 and IGF-2, as well as the binding proteins (IGFBPs) (Adamek and Kasprzak, 2018). The secretion of IGF-1 and IGFBPs is not only under the control of endocrine and nutritional factors, but also autocrine and paracrine factors (Voci et al., 1999). The main signaling pathways downstream of the IGF-1 receptor are Ras/MAPK and PI3K/Akt, while these two pathways are responsible for the glucose and lipid metabolism in the liver (Huang et al., 2020b).
Crosstalk Between Circadian Clock System and GH/IGF-1 Axis
Regulatory Effect of the GH/IGF-1 Axis on the Circadian Clock System
There are very few reports to identify the regulatory effect of the GH/IGF-1 axis on the circadian clock system. One study raised an interesting point: circadian clock-related gene expression may exist in the pituitary, and this expression is related to the expression of GH gene (Kim et al., 2015). Fast-growing transgenic coho salmon including the OnMTGH1 gene construct was used as the target model (Kim et al., 2015). Vital clock genes in this research revealed various responses to the overexpression of GH. In this study, the correlation between the CLOCK gene and the BMAL1 gene was not high, which was consistent with the view that different parts of the circadian clock system linked to different functions as circadian oscillators (Guillaumond et al., 2005). Most of the core clock genes (CLOCK, PER1, PER2, CRY3, NRLD2) showed the difference of expression oscillation in the pituitary of GH transgenic and wild type coho salmon, which indicated that the GH gene might have a regulatory effect on the expression of pituitary clock genes. There are several possible explanations for the different expression patterns (amplitude and phase) of core clock genes between transgenic and wild type coho salmon. GH regulates its own production in the pituitary through negative feedback control. This interference process may integrate metabolic processes in other parts of the body, such as the production of IGF-1 in the liver, to respond to changes in seasonal and nutritional challenges by affecting normal regulatory processes (Beckman, 2011). In addition, GH transgenic Coho salmon exhibited pathophysiological effects of increased GH expression, such as changes in reproduction, metabolism, stress, and disease resistance (Pitkänen et al., 1999; Bessey et al., 2004; Kim et al., 2013), which were all affected by the action of pituitary hormones, and might even be affected by changes in pituitary structure. It is known that overexpression of GH may affect the molting of coho salmon, which is a complex physiological process, and this process may affect the expression level, pattern, and/or effect of clock gene (Devlin et al., 2000). Although most studies have been carried out in vertebrates to determine the interaction between metabolism and clock genes (Delezie and Challet, 2011), transgenic GH over-expressing Coho salmon may be a useful model to improve the understanding of the interaction between circadian molecular clocks and nutritional status.
There are a few related human studies. Six patients with hypopituitarism were studied to determine the relationship between peripheral clock gene expression and GH (Sjögren et al., 2007). After 2 weeks of GH treatment (0.5 mg/day), systemic metabolic testing and skeletal muscle biopsy were performed in these patients. It was found that the plasma IGF-1 levels increased after GH treatment, accompanied by an increased expression of the CLOCK gene and decreased expression of the PER1 gene in muscle tissues. This study suggested for the first time that GH might regulate the peripheral clock system by affecting the expression of CLOCK and PERIOD genes in humans. The most interesting observation was that there was an opposite effect on expression between the CLOCK gene and the PER1 gene by GH. GH may be a potential medium for SCN to regulate the peripheral clock system. The rhythmic release of GH secreted by the pituitary gland is controlled by growth hormone-releasing hormone and somatostatin in the hypothalamus (Anderson et al., 2004). It may act as an endocrine factor to transmit signals from SCN to peripheral tissues.
Regulation of Circadian Clock System on the GH/IGF-1 Axis
Human GH (hGH) transgenic mice were used to express the hGH gene in mouse pituitary somatotrophs (Vakili et al., 2016). This change of cell/tissue-specific expression was a result of a transgene containing the whole hGH gene and locus control region (LCR) in a fragment of human chromosome (Jin et al., 2009; Vakili et al., 2011, 2016). LCR was used as tissue or cell-specific enhancer and provided a suitable site for gene integration and independent expression. All mice presented normal growth patterns and specifically express (but not overexpress) of hGH gene in somatotrophs of the pituitary gland. Sequences of the hGH gene promoter revealed that the enhancer motif (Ebox) element could bind the circadian transcriptional regulators (CLOCK and BMAL1). In addition, CLOCK/BMAL1 was responsible for the transactivation of the hGH gene promotor. The article proves that the synthesis of hGH, especially the expression of the hGH gene, is under the control of circadian rhythm, and it is also the target of CLOCK/BMAL1 signals.
Previous studies reported that mice with defective clock genes [BMAL1−/− and CRY−/− (Cry1−/− and Cry2−/−)] had significantly slower growth and more weight loss (Masuki et al., 2005; Kondratov et al., 2006a; Sun et al., 2006; Bur et al., 2009). This growth defect became obvious 2–3 weeks after birth, which coincided with the maturation time of the somatotroph axis (Clark et al., 1985). It is well known that pulsatile release of GH may increase the synthesis of major urine proteins (MUP) in the liver of male mice and subsequent accumulation in the urine (Norstedt and Palmiter, 1984; Low et al., 2001). The study found that the main urine protein in the urine of CRY−/− male mice was reduced compared with that of wild-type mice, which was related to the downregulation of MUP1 gene expression in the liver. This phenomenon was not observed in female CRY−/− mice, and the deletion of the CRY gene did not change the distribution of GH values in female mice. On the contrary, the random GH level of CRY gene-deficient male mice was significantly increased, and only 20% of the random GH values were lower than 1 ng/ml, indicating that the duration of the GH trough was shorter than that in the control group. These data revealed that the GH secretion profile of CRY−/− male mice were changed with reduced GH secretion. It was suggested that the expression of clock genes (such as CRY) was related to the pulsatile release of GH in male mice. Such change in male mice may contribute to metabolic sex dimorphism (Bur et al., 2009). In addition, the total content of GH in the pituitary of wild-type male mice is scattered in a wide range of values, which may indicate the ever-changing process of GH peaks and troughs. The secretion pattern with a low-amplitude irregular pulsatile profile in CRY−/− male mice was similar to that of female mice. This study also found that the MUP1 mRNA levels were restored in CRY−/− male mice with the injection of bovine GH and octreotide (to inhibit endogenous GH secretion). The injection also reversed the feminization expression pattern of CYP2B9, CYP2D9, CYP4A12, CYP7B1, ELOVL3 (dominant expression genes in female mice) in the liver to that of wild-type male mice. The results further confirmed the role of the GH axis in the sex dimorphism of CRY−/− male mice (Bur et al., 2009). A similar conclusion was reached in another study using genetically modified male rats (expressing human GH) as an experimental model (Hirao et al., 2010). Another hypothesis is that the circadian rhythm in the pituitary may synchronize the unitary ultradian activities of GH-secreting cells through a long-distance homotypic cell network (Bonnefont et al., 2000; Bonnefont and Mollard, 2003), although this possibility remains to be studied.
In addition, studies have confirmed that the disruption of the circadian clock system affects the release of GH and GH-mediated signal pathways. The BMAL1 knockout mouse (BMAL1−/−) model was used to explore the effects of circadian clock dysfunction on the GH circulating levels and downstream pathways (Lyu et al., 2020). In BMAL1−/− mice, the GH receptor (GHR) signaling was decreased, including reduced phosphorylation of GHR, JAK2, and STAT1/3/5. Such reduction might be due to the increased expression of the negative regulators, such as SOCS. Interestingly, the study tested the 24-h serum GH concentration of male and female mice (blood collection every 1 h) and found that there was no significant difference in serum GH concentration between BMAL1−/− mice and control mice. The level of IGF-1 in serum, however, was reduced. According to our previous extensive research of GH profiles (Steyn et al., 2011), the GH secretion profile in male mice is characterized by a high-amplitude pulse pattern for less than 30 min every 3–3.5 h with some variability. Therefore, the sampling time with a 1-h interval may not be enough to reflect the GH release pattern of mice compared with a 10-min interval routinely used in this laboratory (Steyn et al., 2011; Huang et al., 2020a).
There were also some indirect evidences to demonstrate the relationship between circadian clock and pulsatile GH secretion. In night workers, the sleep-related GH pulse was lowered, but the reduction was compensated for by the large individual GH pulses occurring during waking periods (Brandenberger and Weibel, 2004). The total GH secretion during the 24 h was constant. Our previous results also found that rotating light disturbed the GH secreted model with more GH pulse numbers and lower GH pulse altitude (Wang et al., 2021). The interaction between pulsatile GH secretion and circadian clock system warrants further research.
Along the aging, circadian rhythm is progressively perturbed and circulating IGF-1 level is reduced, resulting in defects in multiple systematic physiological functions (Tevy et al., 2013). There are also potential interactive pathways between circadian rhythm and IGF-1 levels. Recently, it was revealed that circulating/hepatic IGF-1 levels presented a circadian rhythm in mouse fed ad libitum. The level of IGF-1 was higher in the serum during daytime and in the liver during night-time, but lower in the serum during night-time and in the liver during daytime (Patel et al., 2016). Therefore, expression of circadian clock genes may influence the IGF-1 levels. It was reported that BMAL1 deficient mice had altered circadian rhythm of circulating IGF-1 levels (Patel et al., 2016), and CRY1 and 2 deficient mice had reduced IGF-1 production (Chaudhari et al., 2017). IGF-1 levels may reset the liver circadian clock (Ikeda et al., 2018).
Due to the difficulty in measuring pulsatile GH secretion, the interaction between the GH-IGF-1 axis and circadian clock warrants further detailed research. Changes of pulsatile GH secretion profile need to be investigated, probably using the animal models with selected circadian clock gene knock-out. Rhythmic changes of circadian clock-related genes by GH demand further investigation in GH-knock-out or mutant animal models. Evidence in observational human studies is requested in future to link animal study to human physiology. The rhythmic changes of circadian clock-related genes in patients with acromegaly or GH deficiency could be helpful. In addition, the pulsatile secretion of GH profiles in patients with sleep deprivation would provide some indirect evidence of circadian change on GH secretion. In summary, the relationship between the GH/IGF-1 axis and the circadian clock system needs to be carefully investigated in the future.
Conclusion
Both the central circadian clock system and the control center of the GH/IGF-1 axis are located in the hypothalamus and related genes/hormones are expressed/released in circadian rhythm, which regulate metabolism through multi-level interactions. Many people engage in nightshift work with sleep rhythm disorders. The subsequent weight gain, metabolic disorders, and related cardiovascular and cerebrovascular diseases turn into an important public health problem (Scheer et al., 2009; Leproult et al., 2014; Morris et al., 2016). At present, there is no definitive evidence of the interaction between the circadian clock system and the GH/IGF-1 axis and the pathophysiological mechanism causing metabolic disorders. It is utterly necessary to further confirm whether the GH/IGF-1 axis plays a major regulatory role in circadian regulation.
Author Contributions
LG and CC made substantial contributions to conception and design and revised it critically for important intellectual content. WW, XD, ZH, and QP involved in drafting the manuscript. WW and XD made the figure. All authors contributed to thearticle and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81670763 and 81471050) and the Australian NHMRC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamek, A., and Kasprzak, A. (2018). Insulin-like growth factor (IGF) system in liver diseases. Int. J. Mol. Sci. 19, 1–24. doi: 10.3390/ijms19051308
Albrecht, U. (2017). The circadian clock, metabolism and obesity. Obes. Rev. 18, 25–33. doi: 10.1111/obr.12502
Anderson, L. L., Jeftinija, S., and Scanes, C. G. (2004). Growth hormone secretion: molecular and cellular mechanisms and in vivo approaches. Exp. Biol. Med. 229, 291–302. doi: 10.1177/153537020422900403
Argetsinger, L. S., Norstedt, G., Billestrup, N., White, M. F., and Carter-Su, C. (1996). Growth hormone, interferon-γ, and leukemia inhibitory factor utilize insulin receptor substrate-2 in intracellular signaling. J. Biol. Chem. 271, 29415–29421. doi: 10.1074/jbc.271.46.29415
Beckman, B. R. (2011). Perspectives on concordant and discordant relations between insulin-like growth factor 1 (IGF1) and growth in fishes. Gen. Comp. Endocrinol. 170, 233–252. doi: 10.1016/j.ygcen.2010.08.009
Belenky, P., Bogan, K. L., and Brenner, C. (2007). NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19. doi: 10.1016/j.tibs.2006.11.006
Berryman, D. E., Glad, C. A. M., List, E. O., and Johannsson, G. (2013). The GH/IGF-1 axis in obesity: pathophysiology and therapeutic considerations. Nat. Rev. Endocrinol. 9, 346–356. doi: 10.1038/nrendo.2013.64
Bessey, C., Devlin, R. H., Liley, N. R., and Biagi, C. A. (2004). Reproductive performance of growth-enhanced transgenic coho salmon. Trans. Am. Fish. Soc. 133, 1205–1220. doi: 10.1577/T04-010.1
Bonnefont, X., and Mollard, P. (2003). Electrical activity in endocrine pituitary cells in situ: a support for a multiple-function coding. FEBS Lett. 548, 49–52. doi: 10.1016/S0014-5793(03)00727-0
Bonnefont, X., Fiekers, J., Creff, A., and Mollard, P. (2000). Rhythmic bursts of calcium transients in acute anterior pituitary slices. Endocrinology 141, 868–875. doi: 10.1210/endo.141.3.7363
Brandenberger, G., and Weibel, L. (2004). The 24-h growth hormone rhythm in men: sleep and circadian influences questioned. J. Sleep Res. 13, 251–255. doi: 10.1111/j.1365-2869.2004.00415.x
Brooks, A. J., and Waters, M. J. (2010). The growth hormone receptor: mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 6, 515–525. doi: 10.1038/nrendo.2010.123
Buhr, E. D., and Takahashi, J. S. (2013). Molecular components of the mammalian circadian clock. Handb. Exp. Pharmacol. 217, 3–27. doi: 10.1007/978-3-642-25950-0_1
Bur, I. M., Cohen-Solal, A., Carmignac, D., Abecassis, P., Chauvet, N., Martin, A. O., et al. (2009). The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J. Biol. Chem. 284, 9066–9073. doi: 10.1074/jbc.M808360200
Cantó, C., Gerhart-Hines, Z., Feige, J. N., Lagouge, M., Noriega, L., Milne, J. C., et al. (2009). AMPK regulates energy expenditure by modulating NAD + metabolism and SIRT1 activity. Nature 458, 1056–1060. doi: 10.1038/nature07813
Chaudhari, A., Gupta, R., Patel, S., Velingkaar, N., and Kondratov, R. (2017). Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol. Biol. Cell 28, 834–842. doi: 10.1091/mbc.e16-08-0624
Chhabra, Y., Nelson, C. N., Plescher, M., Barclay, J. L., Smith, A. G., Andrikopoulos, S., et al. (2019). Loss of growth hormone–mediated signal transducer and activator of transcription 5 (STAT5) signaling in mice results in insulin sensitivity with obesity. FASEB J. 33, 6412–6430. doi: 10.1096/fj.201802328R
Chhabra, Y., Waters, M. J., and Brooks, A. J. (2011). Role of the growth hormone-IGF-1 axis in cancer. Expert Rev. Endocrinol. Metab. 6, 71–84. doi: 10.1586/eem.10.73
Chia, D. J., Varco-Merth, B., and Rotwein, P. (2010). Dispersed chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J. Biol. Chem. 285, 17636–17647. doi: 10.1074/jbc.M110.117697
Clark, R. G., Jansson, J. O., Isaksson, O., and Robinson, I. C. A. F. (1985). Intravenous growth hormone: growth responses to patterned infusions in hypophysectomized rats. J. Endocrinol. 104, 53–61. doi: 10.1677/joe.0.1040053
Colao, A. (2008). The GH-IGF-I axis and the cardiovascular system: clinical implications. Clin. Endocrinol. (Oxf.) 69, 347–358. doi: 10.1111/j.1365-2265.2008.03292.x
Cuttler, L. (1996). The regulation of growth hormone secretion. Endocrinol. Metab. Clin. North Am. 25, 541–571. doi: 10.1016/S0889-8529(05)70340-6
Davies, J. S., Carter, D. A., and Wells, T. (2004). Photic stimulation inhibits growth hormone secretion in rats: a hypothalamic mechanism for transient entrainment. Endocrinology 145, 2950–2958. doi: 10.1210/en.2003-1236
Delezie, J., and Challet, E. (2011). Interactions between metabolism and circadian clocks: reciprocal disturbances. Ann. N. Y. Acad. Sci. 1243, 30–46. doi: 10.1111/j.1749-6632.2011.06246.x
Demierre, M. F., Higgins, P. D. R., Gruber, S. B., Hawk, E., and Lippman, S. M. (2005). Statins and cancer prevention. Nat. Rev. Cancer 5, 930–942. doi: 10.1038/nrc1751
Devlin, R. H., Swanson, P., Clarke, W., Plisetskaya, E., Dickhoff, W., Moriyama, S., et al. (2000). Seawater adaptability and hormone levels in growth-enhanced transgenic coho salmon, Oncorhynchus kisutch. Aquaculture 191, 367–385. doi: 10.1016/S0044-8486(00)00484-1
Duez, H., and Staels, B. (2008). Rev-erbα gives a time cue to metabolism. FEBS Lett. 582, 19–25. doi: 10.1016/j.febslet.2007.08.032
Giustina, A., and Veldhuis, J. D. (1998). Pathophysiology of the neuroregulation of growth hormone secretion in experimental animals and the human. Endocr. Rev. 19, 717–797. doi: 10.1210/er.19.6.717
Gómez-Abellán, P., Hernández-Morante, J. J., Luján, J. A., Madrid, J. A., and Garaulet, M. (2008). Clock genes are implicated in the human metabolic syndrome. Int. J. Obes. 32, 121–128. doi: 10.1038/sj.ijo.0803689
Guillaumond, F., Dardente, H., Giguère, V., and Cermakian, N. (2005). Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403. doi: 10.1177/0748730405277232
Hastings, M. H., Reddy, A. B., and Maywood, E. S. A. (2003). clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661. doi: 10.1038/nrn1177
Hastings, M., O’Neill, J. S., and Maywood, E. S. (2007). Circadian clocks: regulators of endocrine and metabolic rhythms. J. Endocrinol. 195, 187–198. doi: 10.1677/JOE-07-0378
Hirao, J., Niino, N., Arakawa, S., Shibata, S., Mori, K., Ando, Y., et al. (2010). Circadian modulation of hepatic transcriptome in transgenic rats expressing human growth hormone. J. Toxicol. Sci. 35, 673–685. doi: 10.2131/jts.35.673
Huang, Z., Huang, L., Wang, C., Zhu, S., Qi, X., Chen, Y., et al. (2020a). Dapagliflozin restores insulin and growth hormone secretion in obese mice. J. Endocrinol. 245, 1–12. doi: 10.1530/JOE-19-0385
Huang, Z., Huang, L., Waters, M. J., and Chen, C. (2020b). Insulin and growth hormone balance: implications for obesity. Trends Endocrinol. Metab. 31, 642–654. doi: 10.1016/j.tem.2020.04.005
Ikeda, Y., Kamagata, M., Hirao, M., Yasuda, S., Iwami, S., Sasaki, H., et al. (2018). Glucagon and/or IGF-1 production regulates resetting of the liver circadian clock in response to a protein or amino acid-only diet. EBioMedicine 28, 210–224. doi: 10.1016/j.ebiom.2018.01.012
Jin, Y., Lu, S. Y., Fresnoza, A., Detillieux, K., Duckworth, M. L., and Cattini, P. (2009). Differential placental hormone gene expression during pregnancy in a transgenic mouse containing the human growth hormone/chorionic Somatomammotropin Locus. Placenta 30, 226–235. doi: 10.1016/j.placenta.2008.12.011
Junnila, R. K., List, E. O., Berryman, D. E., Murrey, J. W., and Kopchick, J. J. (2013). The GH/IGF-1 axis in ageing and longevity. Nat. Rev. Endocrinol. 9, 366–376. doi: 10.1038/nrendo.2013.67
Kalsbeek, A., Palm, I. F., Fleur, S. L., Scheer, F., Perreau-Lenz, S., Ruiter, M., et al. (2006). SCN outputs and the hypothalamic balance of life. J. Biol. Rhythms 21, 458–469. doi: 10.1177/0748730406293854
Kim, J. H., Balfry, S., and Devlin, R. H. (2013). Disease resistance and health parameters of growth-hormone transgenic and wild-type coho salmon, Oncorhynchus kisutch. Fish Shellfish Immunol. 34, 1553–1559. doi: 10.1016/j.fsi.2013.03.365
Kim, J. H., White, S. L., and Devlin, R. H. (2015). Interaction of growth hormone overexpression and nutritional status on pituitary gland clock gene expression in coho salmon, Oncorhynchus kisutch. Chronobiol. Int. 32, 113–127. doi: 10.3109/07420528.2014.958160
Kiyohara, Y. B., Tagao, S., Tamanini, F., Morita, A., Sugisawa, Y., Yasuda, M., et al. (2006). The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. U.S.A. 103, 10074–10079. doi: 10.1073/pnas.0601416103
Kondratov, R. V., Kondratova, A. A., Gorbacheva, V. Y., Vykhovanets, O. V., and Antoch, M. P. (2006a). Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873. doi: 10.1101/gad.1432206
Kondratov, R. V., Kondratova, A. A., Lee, C., Gorbacheva, V., Chernov, M., and Antoch, M. (2006b). Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle 5, 890–895. doi: 10.4161/cc.5.8.2684
Kraves, S., and Weitz, C. J. (2006). A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat. Neurosci. 9, 212–219. doi: 10.1038/nn1633
Kuwahara, S., Sari, D. K., Tsukamoto, Y., Tanaka, S., and Sasaki, F. (2004). Age-related changes in growth hormone (GH) cells in the pituitary gland of male mice are mediated by GH-releasing hormone but not by somatostatin in the hypothalamus. Brain Res. 998, 164–173. doi: 10.1016/j.brainres.2003.10.060
Lamia, K. A., Sachdeva, U. M., DiTacchio, L., Williams, E. C., Alvarez, J. G., Egan, D. F., et al. (2009). AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440. doi: 10.1126/science.1172156
Leproult, R., Holmbäck, U., and Van Cauter, E. (2014). Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes 63, 1860–1869. doi: 10.2337/db13-1546
LeRoith, D., and Yakar, S. (2007). Mechanisms of disease: metabolic effects of growth hormone and insulin-like growth factor 1. Nat. Clin. Pract. Endocrinol. Metab. 3, 302–310. doi: 10.1038/ncpendmet0427
Liu, Z., Cordoba-Chacon, J., Kineman, R., Cronstein, B., Muzumdar, R., Gong, Z., et al. (2016). Growth hormone control of hepatic lipid metabolism. Diabetes 65, 3598–3609. doi: 10.2337/db16-0649
Low, M. J., Otero-Corchón, V., Parlow, A., Ramírez, J. L., Kumar, U., Patel, Y., et al. (2001). Somatostatin is required for masculinization of growth hormone-regulated hepatic gene expression but not of somatic growth. J. Clin. Invest. 107, 1571–1580. doi: 10.1172/JCI11941
Lyu, X., Wang, G., Pi, Z., and Wu, L. (2020). Circadian clock disruption attenuated growth hormone(GH)-mediated signalling. Gen. Comp. Endocrinol. 302:113670. doi: 10.1016/j.ygcen.2020.113670
Masuki, S., Todo, T., Nakano, Y., Okamura, H., and Nose, H. (2005). Reduced α-adrenoceptor responsiveness and enhanced baroreflex sensitivity in Cry-deficient mice lacking a biological clock. J. Physiol. 566, 213–224. doi: 10.1113/jphysiol.2005.086728
Mazzoccoli, G., Pazienza, V., and Vinciguerra, M. (2012). Clock genes and clock-controlled genes in the regulation of metabolic rhythms. Chronobiol. Int. 29, 227–251. doi: 10.3109/07420528.2012.658127
Moøller, N., and Joørgensen, J. O. L. (2009). Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr. Rev. 30, 152–177. doi: 10.1210/er.2008-0027
Morris, C. J., Purvis, T. E., Hu, K., and Scheer, F. A. J. L. (2016). Circadian misalignment increases cardiovascular disease risk factors in humans. Proc. Natl. Acad. Sci. U.S.A. 113, E1402–E1411. doi: 10.1073/pnas.1516953113
Nader, N., Chrousos, G. P., and Kino, T. (2010). Interactions of the circadian CLOCK system and the HPA axis. Trends Endocrinol. Metab. 21, 277–286. doi: 10.1016/j.tem.2009.12.011
Nakahata, Y., Kaluzová, M., Grimaldi, B., Sahar, S., Hirayama, J., Chen, D., et al. (2008). The NAD+-Dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. doi: 10.1016/j.cell.2008.07.002
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M., and Sassone-Corsi, P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657. doi: 10.1126/science.1170803
Nicolaides, N. C., Charmandari, E., Chrousos, G. P., and Kino, T. (2014). Circadian endocrine rhythms: the hypothalamic–pituitary– adrenal axis and its actions. Ann. N. Y. Acad. Sci. 1318, 71–80. doi: 10.1038/jid.2014.371
Norstedt, G., and Palmiter, R. (1984). Secretory rhythm of growth hormone regulates sexual differentiation of mouse liver. Cell 36, 805–812. doi: 10.1016/0092-8674(84)90030-8
Oishi, K. (2009). Plasminogen activator inhibitor-1 and the circadian clock in metabolic disorders. Clin. Exp. Hypertens. 31, 208–219. doi: 10.1080/10641960902822468
Oishi, K., Miyazaki, K., Uchida, D., Ohkura, N., Wakabayashi, M., Doi, R., et al. (2009). PERIOD2 is a circadian negative regulator of PAI-1 gene expression in mice. J. Mol. Cell. Cardiol. 46, 545–552. doi: 10.1016/j.yjmcc.2009.01.001
Oishi, K., Ohkura, N., Wakabayashi, M., Shirai, H., Sato, K., Matsuda, J., et al. (2006). CLOCK is involved in obesity-induced disordered fibrinolysis in ob/ob mice by regulating PAI-1 gene expression. J. Thromb. Haemost. 4, 1774–1780. doi: 10.1111/j.1538-7836.2006.02032.x
Oishi, K., Shirai, H., and Ishida, N. (2005). CLOCK is involved in the circadian transactivation of peroxisome- proliferator-activated receptor α (PPARα) in mice. Biochem. J. 386, 575–581. doi: 10.1042/BJ20041150
Padmanabhan, K., Robles, M. S., Westerling, T., and Weitz, C. (2012). Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337, 599–602. doi: 10.1126/science.1221592
Panda, S., Antoch, M., Miller, B. H., Su, A., Schook, A. B., Straume, M., et al. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. doi: 10.1016/S0092-8674(02)00722-5
Patel, S. A., Chaudhari, A., Gupta, R., Velingkaar, N., and Kondratov, R. V. (2016). Circadian clocks govern calorie restriction-mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. FASEB J. 30, 1634–1642. doi: 10.1096/fj.15-282475
Pitkänen, T. I., Krasnov, A., Teerijoki, H., and Mölsä, H. (1999). Transfer of growth hormone (GH) transgenes into Arctic charr (Salvelinus alpinus L.). I. Growth response to various GH constructs. Genet. Anal. Biomol. Eng. 15, 91–98. doi: 10.1016/S1050-3862(99)00011-X
Rabinowitz, D., Klassen, G. A., and Zierler, K. L. (1965). Effect of human growth hormone on muscle and adipose tissue metabolism. J. Clin. Invest. 44, 51–61. doi: 10.1172/JCI105126
Rahmani, J., Varkaneh, H., Clark, C. C., Zand, H., Bawadi, H., Ryan, P., et al. (2019). The influence of fasting and energy restricting diets on IGF-1 levels in humans: a systematic review and meta-analysis. Ageing Res. Rev. 53:100910. doi: 10.1016/j.arr.2019.100910
Ramsey, K. M., Yoshino, J., Brace, C. S., Abrassart, D., Kobayashi, Y., Marcheva, B., et al. (2009). Circadian clock feedback cycle through NAMPT-Mediated NAD+ biosynthesis. Science 324, 651–654. doi: 10.1126/science.1171641
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian clocks in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Ridderstrale, M., Degerman, E., and Tornqvist, H. (1995). Growth hormone stimulates the tyrosine phosphorylation of the insulin receptor substrate-1 and its association with phosphatidylinositol 3-kinase in primary adipocytes. J. Biol. Chem. 270, 3471–3474. doi: 10.1074/jbc.270.8.3471
Ripperger, J. A., and Schibler, U. (2006). Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374. doi: 10.1038/ng1738
Rotwein, P. (2012). Mapping the growth hormone-Stat5b-IGF-I transcriptional circuit. Trends Endocrinol. Metab. 23, 186–193. doi: 10.1016/j.tem.2012.01.001
Rudic, R. D., McNamara, P., Curtis, A., Boston, R., Panda, S., Hogenesch, J., et al. (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2:e377. doi: 10.1371/journal.pbio.0020377
Sahar, S., and Sassone-Corsi, P. (2012). Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol. Metab. 23, 1–8. doi: 10.1016/j.tem.2011.10.005
Scheer, F. A. J. L., Hilton, M. F., Mantzoros, C. S., and Shea, S. A. (2009). Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. U.S.A. 106, 4453–4458. doi: 10.1073/pnas.0808180106
Sharma, V. M., Vestergaard, E., Jessen, N., Kolind-Thomsen, P., Nellemann, B., Nielsen, T., et al. (2019). Growth hormone acts along the PPARγ-FSP27 axis to stimulate lipolysis in human adipocytes. Am. J. Physiol. Endocrinol. Metab. 316, E34–E42. doi: 10.1152/ajpendo.00129.2018
Shirogane, T., Jin, J., Ang, X. L., and Harper, J. W. (2005). SCFβ-TRCP controls Clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–26872. doi: 10.1074/jbc.M502862200
Sjögren, K., Leung, K., Kaplan, W., Gardiner-Garden, M., Gibney, J., and Ho, K. (2007). Growth hormone regulation of metabolic gene expression in muscle: a microarray study in hypopituitary men. Am. J. Physiol. Endocrinol. Metab. 293, 364–371. doi: 10.1152/ajpendo.00054.2007
Sonoda, J., Pei, L., and Evans, R. M. (2008). Nuclear receptors: decoding metabolic disease. FEBS Lett. 582, 2–9. doi: 10.1016/j.febslet.2007.11.016
Sos, B. C., Harris, C., Nordstrom, S. M., Tran, J. L., Balázs, M., Caplazi, P., et al. (2011). Abrogation of growth hormone secretion rescues fatty liver in mice with hepatocytespecific deletion of JAK2. J. Clin. Invest. 121, 1412–1423. doi: 10.1172/JCI42894
Steyn, F. J., Huang, L., Ngo, S., Leong, J. W., Tan, H. Y., Xie, T. Y., et al. (2011). Development of a method for the determination of pulsatile growth hormone secretion in mice. Endocrinology 152, 3165–3171. doi: 10.1210/en.2011-0253
Storch, K. F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F. C., Wong, W. H., et al. (2002). Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. doi: 10.1038/nature744
Sun, Y., Yang, Z., Niu, Z., Wang, W., Peng, J., Li, Q., et al. (2006). The mortality of MOP3 deficient mice with a systemic functional failure. J. Biomed. Sci. 13, 845–851. doi: 10.1007/s11373-006-9108-4
Takahashi, J. S., Hong, H. K., Ko, C. H., and McDearmon, E. L. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. doi: 10.1038/nrg2430
Tevy, M. F., Giebultowicz, J., Pincus, Z., Mazzoccoli, G., and Vinciguerra, M. (2013). Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol. Metab. 24, 229–237. doi: 10.1016/j.tem.2012.12.002
Turek, F. W., Joshu, C., Kohsaka, A., Lin, E., Ivanova, G., McDearmon, E., et al. (2005). Obesity and metabolic syndrome in circadian Clock mutant nice. Science 308, 1043–1045. doi: 10.1126/science.1108750
Vaccarino, F. J., Sovran, P., Baird, J. P., and Ralph, M. R. (1995). Growth hormone-releasing hormone mediates feeding-specific feedback to the suprachiasmatic circadian clock. Peptides 16, 595–598. doi: 10.1016/0196-9781(95)00018-F
Vakili, H., Jin, Y., and Cattini, P. A. (2016). Evidence for a circadian effect on the reduction of human growth hormone gene expression in response to excess caloric intake. J. Biol. Chem. 291, 13823–13833. doi: 10.1074/jbc.M116.722744
Vakili, H., Jin, Y., Nagy, J. I., and Cattini, P. A. (2011). Transgenic mice expressing the human growth hormone gene provide a model system to study human growth hormone synthesis and secretion in non-tumor-derived pituitary cells: differential effects of dexamethasone and thyroid hormone. Mol. Cell. Endocrinol. 345, 48–57. doi: 10.1016/j.mce.2011.07.010
Voci, A., Arvigo, M., Massajoli, M., Garrone, S., Bottazzi, C., Demori, I., et al. (1999). IGF-I production by adult rat hepatocytes is stimulated by transforming growth factor-a and transforming growth factor-b1. Eur. J. Endocrinol. 140, 577–582. doi: 10.1530/eje.0.1400577
Wang, W., Huang, Z., Huang, L., Tan, C., Chen, W., Roelfsema, F., et al. (2021). Rotating day and night disturb growth hormone secretion profiles, body energy metabolism and insulin levels in mice. Neuroendocrinology1–12. doi: 10.1159/000518338
Waters, M. J., Shang, C. A., Behncken, S. N., Tam, S. P., Li, H., Shen, B., et al. (1999). Growth hormone as a cytokine. Clin. Exp. Pharmacol. Physiol. 26, 760–764. doi: 10.1046/j.1440-1681.1999.03129.x
Willoughby, J. O., and Martin, J. B. (1978). The suprachiasmatic nucleus synchronizes growth hormone secretory rhythms with the light-dark cycle. Brain Res. 151, 413–417. doi: 10.1016/0006-8993(78)90899-5
Woelfle, J., Billiard, J., and Rotwein, P. (2003). Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J. Biol. Chem. 278, 22696–22702. doi: 10.1074/jbc.M301362200
Yamauchi, T., Kaburagi, Y., Ueki, K., Tsuji, Y., Stark, G., Kerr, I., et al. (1998). Growth hormone and prolactin stimulate tyrosine phosphorylation of insulin receptor substrate-1, -2, and -3, their association with p85 phosphatidylinositol 3-kinase (PI3-kinase), and concomitantly PI3-kinase activation via JAK2 kinase. J. Biol. Chem. 273, 15719–15726. doi: 10.1074/jbc.273.25.15719
Yang, X., Downes, M., Yu, R. T., Bookout, A. L., He, W., Straume, M., et al. (2006). Nuclear receptor expression links the circadian clock to metabolism. Cell 126, 801–810. doi: 10.1016/j.cell.2006.06.050
Yang, Y., Xu, T., Zhang, Y., and Qin, X. (2017). Molecular basis for the regulation of the circadian clock kinases CK1δ and CK1ε. Cell. Signal. 31, 58–65. doi: 10.1016/j.cellsig.2016.12.010
Yin, L., Wu, N., Curtin, J. C., Qatanani, M., Szwergold, N. R., Reid, R. A., et al. (2007). Rev-erba, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789. doi: 10.1126/science.1150179
Keywords: GH, IGF-1, metabolism, clock, circadian rhythm
Citation: Wang W, Duan X, Huang Z, Pan Q, Chen C and Guo L (2021) The GH-IGF-1 Axis in Circadian Rhythm. Front. Mol. Neurosci. 14:742294. doi: 10.3389/fnmol.2021.742294
Received: 16 July 2021; Accepted: 23 August 2021;
Published: 09 September 2021.
Edited by:
Jun Hirayama, Komatsu University, JapanReviewed by:
Sooyoung Chung, Ewha Womans University, South KoreaYu Tahara, Waseda University, Japan
Copyright © 2021 Wang, Duan, Huang, Pan, Chen and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Chen, Y2hlbi5jaGVuQHVxLmVkdS5hdQ==; Lixin Guo, Z2x4MTIxOEAxNjMuY29t
†These authors have contributed equally to this work
 Weihao Wang
Weihao Wang Xiaoye Duan
Xiaoye Duan Zhengxiang Huang
Zhengxiang Huang Qi Pan
Qi Pan Chen Chen
Chen Chen Lixin Guo
Lixin Guo