
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci., 27 September 2021
Sec. Pain Mechanisms and Modulators
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.720973
This article is part of the Research TopicMolecular Mechanisms of NociceptionView all 14 articles
 Tomislav Kokotović1,2,3
Tomislav Kokotović1,2,3 Michiel Langeslag4,5,6
Michiel Langeslag4,5,6 Ewelina M. Lenartowicz1,3
Ewelina M. Lenartowicz1,3 John Manion7
John Manion7 Christopher W. Fell1,2,3
Christopher W. Fell1,2,3 Elham Alehabib8
Elham Alehabib8 Abbas Tafakhori9
Abbas Tafakhori9 Hossein Darvish10
Hossein Darvish10 Eric J. Bellefroid11
Eric J. Bellefroid11 G. Gregory Neely7
G. Gregory Neely7 Michaela Kress4
Michaela Kress4 Josef M. Penninger12,13
Josef M. Penninger12,13 Vanja Nagy1,2,3*
Vanja Nagy1,2,3*PR domain-containing member 12 (PRDM12) is a key developmental transcription factor in sensory neuronal specification and survival. Patients with rare deleterious variants in PRDM12 are born with congenital insensitivity to pain (CIP) due to the complete absence of a subtype of peripheral neurons that detect pain. In this paper, we report two additional CIP cases with a novel homozygous PRDM12 variant. To elucidate the function of PRDM12 during mammalian development and adulthood, we generated temporal and spatial conditional mouse models. We find that PRDM12 is expressed throughout the adult nervous system. We observed that loss of PRDM12 during mid-sensory neurogenesis but not in the adult leads to reduced survival. Comparing cellular biophysical nociceptive properties in developmental and adult-onset PRDM12 deletion mouse models, we find that PRDM12 is necessary for proper nociceptive responses throughout life. However, we find that PRDM12 regulates distinct age-dependent transcriptional programs. Together, our results implicate PRDM12 as a viable therapeutic target for specific pain therapies even in adults.
Congenital insensitivity to pain (CIP), a subtype of hereditary sensory and autonomic neuropathies (HSAN), primarily affects the peripheral nervous system (PNS). Recently, mutations in methyltransferase PR domain-containing member 12 (PRDM12) have been demonstrated to cause CIP, which based on clinical symptoms was subtyped as HSAN type VIII (OMIM 616488) (Chen et al., 2015). PRDM12 belongs to a large family of transcription factors responsible for cell fate decisions (Hohenauer and Moore, 2012). At present, there are 24 reported patients with PRDM12-CIP, including the complete absence of all modalities of acute and chronic pain perception, frequently accidental or self-inflicted injuries, frequent bone fractures, the absence of corneal reflex and impaired tear production, and skin ulcerations and recurrent bacterial infections often requiring amputations. Unlike the association of CIP syndromes with other genes, such as nerve growth factor beta (NGFβ), a very few PRDM12-CIP patients present with central nervous system (CNS) symptoms, such as intellectual disability (ID) (Chen et al., 2015; Imhof et al., 2020). Skin biopsies of patients revealed the absence of protein gene product 9.5- (PGP9.5) reactive nerve fiber endings in the epidermis, suggesting the participation of PRDM12 in nociceptor development or the survival in humans (Chen et al., 2015).
Nociceptors are pseudounipolar primary afferent sensory neurons in the PNS that detect noxious, potentially tissue-damaging, and painful stimuli. Their somata reside in the dorsal root ganglia (DRG) and trigeminal ganglion, with one axonal process in the periphery and one that synapses in the spinal cord (SC) or brain stem. Nociceptors express a variety of specialized noxious stimuli receptors for their function. These include transient receptor potential (TRP) cation channels, including TRPV1 and TRPA1 activated by capsaicin (Caterina et al., 1997) or noxious cold or electrophilic chemicals such as allyl isothiocyanate and cinnamaldehyde (Bandell et al., 2004; Julius, 2013). Additionally, potentially tissue-damaging temperatures are detected by a variety of voltage-gated Na+ channels, including Nav1.7, Nav1.8, and Nav1.9. Indeed, deleterious variants in genes encoding for Nav1.7 (SCN9A) and Nav1.9 (SCN11A) were also identified to be causative for a type of CIP (OMIM 243000 and OMIM 615548, respectively). Strong noxious stimulation of nociceptors will trigger afferent action potentials (APs) that send nociceptive signals to the CNS to induce the conscious sensation of pain.
PR domain-containing member 12 had no intrinsic enzymatic activity and was reported to recruit euchromatic histone-lysine N-methyltransferase 2 (EHMT2), also known as G9a, for the trimethylation of histone 3 lysine 9 (H3K9me3) resulting in subsequent regulation of target genes (Yang and Shinkai, 2013; Matsukawa et al., 2015). Detailed studies on Danio rerio, Xenopus laevis, and Drosophila melanogaster demonstrated that PRDM12 regulates a transcriptional program critical in cell fate decisions during early sensory neuronal development required for the specification and function of nociceptors (Matsukawa et al., 2015; Nagy et al., 2015; Thelie et al., 2015). Reduction of Hamlet, an ortholog of PRDM12, in D. melanogaster sensory neurons caused reduced nociceptive responses to noxious heat in fly embryos, a phenotype that could not be rescued by reconstitution with human PRDM12 disease-causing variants, providing evidence of the contribution of variants to pathology (Nagy et al., 2015).
In mammals, embryonic mouse studies demonstrated that PRDM12 is expressed in DRGs and is necessary for the initiation and maintenance of an NGF-β receptor, neurotrophic receptor tyrosine kinase 1 (TrkA), by modulating neurogenins 1 and 2 (Ngn1 and Ngn2) activity, thereby promoting nociceptive lineage from neural crest cell progenitors (Kinameri et al., 2008; Bartesaghi et al., 2019; Desiderio et al., 2019). Correspondingly, the analysis of DRG from constitutive knock-out (KO, Prdm12−/−) mice revealed a significant reduction in DRG size, decreased density of cutaneous myelinated sensory branches and the absence of nociceptor-specific markers, all suggesting the absence of nociceptors (Bartesaghi et al., 2019; Desiderio et al., 2019). This specific reduction in nociceptor lineage seems to be a result of a proliferation defect during neurogenesis (Bartesaghi et al., 2019; Landy et al., 2021), however, an increase in apoptotic cell death with no change in proliferation was also reported (Bartesaghi et al., 2019; Desiderio et al., 2019). Constitutive embryonic PRDM12 deficiency resulted in early neonatal death suggesting that in mice, unlike humans, PRDM12 function is essential for survival (Bartesaghi et al., 2019; Desiderio et al., 2019).
Here, we aim to understand the functional role of PRDM12 in nociception in the mammal system. We first present two patients from a large consanguineous family with typical CIP symptoms, including the complete absence of pain perception, confirmed to have a previously unreported homozygous variant in PRDM12. To clarify the role played by PRDM12 in pain perception, we developed several different mouse models. To model human patient variants, we generated a mouse model with the previously reported CIP W160C PRDM12 single base-pair mutation (Prdm12W160C) and S159AfsTer2 (Prdm12S159AFsTer2) using clustered regularly interspaced short palindromic repeats- (CRISPR-) associated system 9 (Cas9) genome editing (Chen et al., 2015; Nagy et al., 2015). We additionally generated conditional mouse mutants, and therefore crossed Prdm12fl/fl to Sox10-Cre (Prdm12fl/fl; Sox10-Cre) to ablate PRDM12 expression in the neural crest cell population during embryonic development, to Avil-Cre (Prdm12fl/fl; Avil-Cre) to ablate PRDM12 expression in the developing DRG and to tamoxifen- (TAM-) inducible Rosa26-CreERT2 mice (Prdm12fl/fl; Rosa26-CreERT2) to be able to induce whole-body PRDM12 KOs in adults. To combine genetic, cellular, electrophysiological, and molecular approaches, we were able to ascertain that PRDM12 is critical for survival and functional nociception in mice. Additionally, we were able to delineate the transcriptional program regulated by PRDM12, in developing and adult mammalian systems containing both core-PRDM12 regulated genes and those that are age-specific.
Following informed consent and approval from patients and participating relatives, all procedures were performed. The procedures are in accordance with the ethical standards and approval of the Shahid Beheshti University of Medical Sciences ethics board and with the current update of the Declaration of Helsinki.
Whole-exome sequencing (WES) was performed at the Biomedical Sequencing Facility (BSF) in CeMM Center for Molecular Medicine of the Austrian Academy of Sciences (CeMM). Briefly, genomic DNA was extracted from the whole blood of patients, parents, and participating siblings using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The quantity and quality of DNA from index patient 1 were assessed by the Qubit 2.0 Fluorometric Quantitation system (Life Technologies, Carlsbad, CA, USA). Libraries were prepared using the Nextera DNA Flex Exome Library Prep Kit (Illumina, San Diego, CA, USA). Briefly, genomic DNA was tagmented, size-selected, and amplified followed by the two rounds of hybridization with biotinilated baits and capturing with streptavidin-conjugated magnetic beads. After enrichment, library fragments represented in the total 45 Mb coding region were amplified and size-selected. Final library pools were quality controlled and sequenced on a HiSeq 3000 instrument (Illumina, San Diego, CA, USA) using 75-bp paired-end chemistry. DNA sequences were mapped to GRCh37 (hg19) version of a human reference genome using a Burrows–Wheeler Aligner with default parameters. Single-nucleotide variants (SNVs) and indels were annotated with SNPEff (Cingolani et al., 2012), Combined Annotation Dependent Depletion (CADD; Kircher et al., 2014), the single-nucleotide polymorphism database (dbSNP; Sherry et al., 2001), the genome aggregation database (gnomAD; https://gnomad.broadinstitute.org/) (Karczewski et al., 2020), and ClinVar (Landrum et al., 2018) data. Subsequent filtering of the remaining variants of interest was based on a homozygous inheritance pattern, a variant type, the population frequency, and the gene lists of interest in relation to the symptoms of the patient. Further, the segregation analysis was performed in patient 2, as well as in both sets of parents and participating siblings by the conventional Sanger sequencing.
Prdm12W160 experiments were approved by the University of Sydney Animal Ethics Committee under the animal ethics protocol (938), and the reporting complies with the animals in research: reporting in vivo experiments (ARRIVE) guidelines. Experimentation protocols comply with National Health and Medical Research Council (NHMRC) guidelines. Mice were housed in specific pathogen-free facilities at Australian BioResources, The Charles Perkins Centre and the Medical Foundation Building at the University of Sydney, Australia. Prdm12W160 mice were generated by CRISPR Cas9-mediated genome editing and were confirmed by the Sanger sequencing using the following primers: forward primer 5′-CTGCTTGGGAGTCTCTTAGAGAG-3′; reverse primer 5′-ATACAGCTGAACGAGGGTGA-3′; and sequencing primer (forward) 5′- TGACTCTCTATTGATCTTTTGCTTC-3′.
Prdm12−/− or Prdm12fl/fl mice were generated on C57BL/6 background using the promoter-driven knockout-first gene trap technique with En2SA-IRES-LacZ cassette in first intron and loxP sites around the second exon as described previously (Desiderio et al., 2019). Female mice carrying an conditional allele were crossed to Advillin-Cre or Rosa26-Cre-ERT2 male mice to generate corresponding developmental or TAM-inducible Prdm12 conditional knockout offspring. Both Prdm12fl/fl; Rosa26-CreERT2+ and their Prdm12fl/fl; Rosa26-CreERT2− littermates received 80 mg/kg intraperitoneal injections of TAM (Sigma-Aldrich, St. Louis, MO, USA) once per day for 4 consecutive days. TAM-injections were carried out at the age of 8 weeks, and animals were then tested 3 weeks following injections. TAM-injected Prdm12fl/fl; Rosa26-CreERT2− animals are referred to as “controls.” These mice were housed at the Animal Facility of the Medical University of Vienna, and Comparative Medicine Mousehouse Vienna Biocenter, Vienna, Austria. All mice were maintained at a 12-h light/dark cycle and provided with food and water ad libitum. Experiments were approved by the Bundesministerium fur Wissenschaft, Forschung und Wirtschaft (BMWFW-66.015/0011-WF/V/3b/2017) and were carried out according to EU-directive 2010/63/EU. Genotyping was determined using the primers as follows: for Cre, forward primer 5′-GCTCGACCAGTTTAGTTACCC-3′, reverse primer 5′-TCGCGATTATCTTCTATATCTTCAG-3′; for Prdm12 forward primer 5′-GCTGATCGAGTCCAGGAGAC-3′, reverse primer 5′-CCAAACATCCACAACCTTCA-3′. Select animals in each cohort were confirmed by qRT-PCR of DRG or whole brain (WB) tissue.
Tissue samples were collected in an RIPA (Sigma-Aldrich, St. Louis, MO, USA) buffer with Proteinase and Phosphatase inhibitor cocktail (Thermo Fischer Scientific, Waltham, MA USA) and homogenized using a Precellys 24 Tissue Homogenizer (Bertin Instruments, Frankfurt, Germany, 3 s × 20 s, 5,000 rpm). Protein concentration was measured using the Bradford assay (Bio-Rad, Hercules, CA, USA). Laemmli buffer- (Sigma-Aldrich, St. Louis, MO, USA) treated samples with standardized concentration of 1 μg/μl were incubated at 95°C for 5 min. About 25 μg of protein per lane was loaded on a 10% polyacrylic acid (PAA) gel, followed by protein transfer to a methanol-activated poly vinylidene fluoride (PVDF) membrane (GE Healthcare Life Sciences, Marlborough, MA, USA) for 10 h at 4°C with a constant current of 0.12 A. The membrane was then blocked with 5% bovine serum albumin (BSA) in TBS-T (0.01% Tween 20) for 1 h at room temperature (RT). Anti-PRDM12 (1:500, Santa Cruz, sc-130242) and loading control anti-HSP90α/β (1:1000, Santa Cruz, sc-13119) primary antibodies were incubated for 12 h at 4°C, followed by 1 h of incubation at RT with anti-mouse horseradish peroxidase- (HRP-) conjugated secondary antibody (1:30000, Sigma-Aldrich, St. Louis, MO, USA; GENA931). To control e specificity of the anti-PRDM12 antibody, human embryonic kidney (HEK) cell lysates overexpressing complementary DNA (cDNA) of human wild type (WT) PRDM12 (PRDM12WT) as a positive control or CIP-causing PRDM12 variant PRDM12S58fs previously determined (Nagy et al., 2015) not to have a protein product, as a negative control were run alongside tissue lysates as labeled. The signal was visualized using a chemiluminescent reagent (ECL, GE Healthcare Life Sciences, Marlborough, MA, USA) and imaged using the ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA).
Lumbar DRG from WT mice were dissected into ice cold phosphate-buffered saline (PBS), fixed in 4% formaldehyde for 20 min, and washed with PBS. Following cryopreservation in 30% sucrose, 20-μm sections were cut on a Microm Cryostat (HM560, Microm) at the Histopathology Service Facility in the Vienna Biocenter Core Facilities (VBCF), the member of the Vienna Biocenter (VBC), Austria, and stored in −80°C until needed. PBS hydrated tissue was permeabilized with 0.5% TritonX-PBS for 15 min and blocked for 1 h at RT with blocking buffer 10% normal goat serum in 0.25% TritonX-PBS. Following 2 × 3 min washes in 0.1% Tween20-PBS, the sections were incubated for 1 h at RT with goat F(ab) anti-mouse IgG (1:2000, Abcam, ab6668, Cambridge, UK) in 0.1% Tween20-PBS. Primary antibodies raised against IB4 (1:1000, Thermo Fisher, I21412, Waltham, MA, USA), Nav1.8 (1:200, Sigma-Aldrich Handels GmbH, Vienna, Austria, S2071), TrkA (1:200, GeneTex GTX54856, Irvine, CA, USA), or CGRP (1:500, Millipore, Burlington, MA, USA), with Guinea pig polyclonal anti-PRDM12 (1:10,000, clone 910) (Desiderio et al., 2019), diluted in a blocking buffer were incubated O/N at 4°C. Following 3 × 3 min washes in 0.01% TritonX-PBS, appropriate secondary antibodies (1:500 goat anti-mouse or anti-rabbit Alexa Fluor 488 or 546, Invitrogen, Waltham, MA, USA) diluted in 0.01% TritonX-PBS were incubated for 1 h at RT. Finally, following 3 × 3 min washes in 0.01% TritonX-PBS, the sections were mounted and imaged on the Zeiss LSM780 confocal microscope.
Skin biopsies were collected from hind paws, and flattened and fixed in 4% formaldehyde in PBS O/N at 4°C. Following cryopreservation in 30% sucrose, 20-μm sections were prepared as described above and stored at −80°C. Tissue was hydrated in PBS, incubated in 50 mM glycine/PBS for 1 h at RT, and blocked with 10% normal goat serum and 1% BSA in 0.1% TritonX-PBS. The sections were then incubated at 4°C O/N with a primary antibody raised against PGP9.5 (1:200, Zytomed 516-3344, Berlin, Germany) in blocking buffer. The sections were washed 3 × 3 min in 0.01% TritonX/PBS, followed by appropriate secondary antibody and nuclear label DAPI (1:2000, Carl Roth, Karlsruhe, Germany) in 0.01% TritonX-PBS and incubated for 1 h at RT. Following 3 × 3 min washes with 0.01% TritonX-PBS, the sections were mounted and imaged as mentioned earlier.
Behavioral assays were performed on both male and female mice at 8–15 weeks of age separated by at least 24 h between the tests to reduce handling stress. Experimenter was blind to the genotype of the test subjects.
Intraplantar capsaicin injections: about 1 μg of capsaicin (Sigma, St. Louis, MO, USA) diluted in 15 μl of normal saline was injected intraplantar in the hind paw of the animal. The animal was then observed for a total of 5 min, which is the time for the duration of reaction to the injection. Counted reactions included licking, shaking, or lifting of the injected paw. At the completion of 5 min, the animal was returned to its home cage. Prdm12fl/fl; Avil-Cre+ N = 33 was compared to the corresponding control littermates; and N = 26 of Prdm12fl/fl; Rosa26-CreERT2+ to N = 35 of their corresponding control littermates.
Von frey test: the measurements were performed according to Lau et al. (2019). Briefly, mice were habituated on 3 separate days and then tested three times with filaments ranging from 0.04 to 2 g with 10 applications per filament.
The Hargreaves test: mice were habituated on 3 separate days and then tested on 3 consecutive days with a low and a high input resistance (IR) intensity. The number of test animals in both von Frey and Hargraves assays was N = 16, compared to N = 18 of littermate controls.
Open-field test (OFT) was performed by the Preclinical Phenotyping Facility at the VBCF, the member of the Vienna Biocenter (pcPHENO, VBCF), Austria. Briefly, mice were placed in a 28 × 28 cm gray, plexi-glass arena and allowed to freely explore for 5 min. Time spent in the center or along the walls of the arena, as well as the distance traveled during exploration, were measured using an automated activity system (TSE-Systems, Bad Homburg, Germany). Reluctance to enter the center of the arena as compared to controls was scored and interpreted as the levels of anxiety. Distance traveled during the test was compared to controls and used as a readout for locomotion deficits.
Morris water maze: mice were trained in the Morris water maze (pcPHENO, VBCF) as described previously (Nagy et al., 2019). Briefly, mice were trained for two sessions a day, with four trials per session, using alternating entry points in different quadrants for each trial. Mice were video tracked using the software Topscan 3.0 (Cleversys, Inc., Reston, VA, USA). On day 1, the visual capacity of the mice was checked by making the platform visible with a black flag and letting the mice explore the pool for 1 min or until they reached the platform (data not shown). Short-term memory test was performed after the last trial, on days 8 and 11 to test for long-term memory. The time spent on searching the target quadrant and target zone (exact location of the platform) was recorded and used as a readout for memory. For both, OFT and Morris water maze N = 8 of Prdm12fl/fl; Rosa26-CreERT2+ were compared to N = 12 of their corresponding control littermates.
For microscopic x-ray imaging, samples were contrasted with IKI (Lugol's iodine solution) (Metscher, 2009) and mounted in an aqueous medium (Metscher, 2011). MicroCT images were acquired using the Xradia MicroXCT system (www.zeiss.com/microscopy/int/x-ray.html) in the Theoretical Biology Department at the University of Vienna. Scans were made with the source settings of 60 kVp at 133 μA and source-sample-detector distances chosen to optimize the field of view for the sample. Tomographic slices were reconstructed using the Xradia XMReconstructor software and saved as TIFF image stacks with an isotropic voxel size of 7.5 μm. The segmentation and processing of images were performed manually using Amira Software 6.4.
Sensory neuron culture: lumbar DRG were harvested from adult mice (age > 8 weeks) as discussed in previous publications (Langeslag et al., 2014; Namer et al., 2017). The sensory neurons were plated on glass coverslips coated with poly-L-lysine/laminin-1 (Sigma-Aldrich, Merck, Burlington, MA, USA) and cultivated in supplemented neruronal culture media (TNB) (Biochrom) containing 25 ng/ml mNGF 2.5S (Alomone Labs, Jerusalem, Israel) at 37°C and 5% CO2 in a humidified incubator for 16–24 h.
Cultured sensory neurons were used for electrophysiological experiments 16–24 h after seeding. Only DRG neurons with size smaller than <35 μm were selected, representing the small- to medium-sized neurons. Glass coverslips were mounted in a recording chamber and placed on a Zeiss Axiovert 200 microscope. All measurements were recorded with an EPC 10 and the Patchmaster v2.73 software (HEKA) at RT. For more details on the methodology for single-cell recordings, please see Supplementary Materials.
From the isolated sensory neurons, cellular voltage and current recordings were performed in a whole-cell patch-clamp configuration. The DRG neurons were kept in extracellular solution (ECS) containing (in mM); NaCl (150), KCl (5), CaCl2 (2), MgCl2 (1), HEPES (10), glucose (10), and the pH was set to 7.3 with NaOH. Borosilicate glass pipettes (Science Products) were pulled with a horizontal puller (P-97, Sutter Instrument Company, Novato, CA, USA) and filled with intracellular solution (ICS) containing (in mM); K-gluconate (98), KCl (50), CaCl2 (0.5), MgCl2 (2), EGTA (5), HEPES (10), MgATP (2), NaGTP (0.2), and the pH was adjusted to 7.3 with KOH.
In current-clamp recordings, the recorded neurons were kept at the holding current of 0 pA. The IR of sensory neurons was determined by four increasing hyperpolarizing current injections (Δ-5 pA from a holding current of 0 pA, 5 kHz).
The minimal current to evoke a single AP within 50 ms (IAP) for each neuron was obtained by 5 pA stepwise increasing, depolarizing instanteous, block pulses, and APs were recorded at 20 kHz. The amplitudes of 5 s ramp-shaped depolarizations were set at 1x, 2x, and 3x IAP and sampled at 5 kHz. The amplitude of the 20 s depolarizing pulse was set to 2x IAP for the corresponding neuron, and the voltage changes were recorded at 5 kHz.
A seven-barrel system with the common outlet was used for the heat stimulation of single neurons (Dittert et al., 1998). In voltage clamp recordings, the neurons were clamped at a holding potential of −60 mV. Heat-activated inward currents (IHeat) were elicited by applying ramp-shaped heat stimuli at 60 s intervals (linear temperature increases from RT to 50°C within 5 s). Neurons were assigned as heat responsive when they showed a non-linear inward current with a threshold below −0.2 nA from the baseline current.
Action potential analysis: the average IR was calculated according to Ohm's law. From the AP evoked by injecting depolarizing current pulses (50 ms, sampled at 20 kHz), the resting membrane potential (Vmem), afterhyperpolarization (AHP), and overshoot (OS) of Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Avil-Cre− DRG neurons were determined. From the first derivative of the evoked APs, the maximal speeds of depolarization (S1) and of the biphasic repolarization (S2 and S3) were derived. Further, the time between S1, S2, and/or S3 (t1-t2; t2-t3; and t1-t3) was determined to compare AP width. The corresponding membrane voltage was deducted at the point where the falling slope of the 1st derivative reversed into a rising slope, and the AP threshold was determined (Langeslag et al., 2014; Namer et al., 2017).
RNA isolation: lumbar DRG tissues were dissected from 10 to 14-week-old animals, immediately placed in RNALater solution (Ambion, Austin, TX, USA), and snap frozen in liquid nitrogen until further isolation. Tissues were lysed using Precellys 24 Tissue Homogenizer (Bertin Instruments, Paris, France) 3×30 s at 5000 rpm, by QIAshredder (Qiagen, Valencia, CA, USA) homogenization. RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) with on-column DNease treatment with RNase-Free Dnase Set (Qiagen, Valencia, CA, USA). A minimum of three biological replicates per genotype were used.
Real-time q-PCR: reverse transcription was performed using the qScript cDNA Synthesis Kit (Quantabio, Beverly, MA, USA) with an input amount of 200 ng of RNA, as per the instructions of the manufacturer. Real-time quantitative PCRs (RT-qPCRs) were done using an iTaq Universal SYBR Green Supermix (Bio Rad, Los Angeles, CA, USA) and run on a StepOnePlus Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). The comparative Ct method (ΔΔCt) was used to determine relative gene expression, normalizing to GAPDH. The primers used for qPCR are Gapdh forward: 5'-GGTCGTATTGGGCGCCTGGTCACC-3'; reverse: 5'-CACACCCATGACGAACATGGGGGC-3'; Prdm12 (exon 3) forward: 5'-CTACATCAAGTGTGCCCGGA−3'; reverse: 5'- TGGCCTTGTAGAAGATGCTCG-3'.
RNA sequencing: the amount of RNA was quantified using a Qubit 2.0 Fluorometric Quantitation System (Life Technologies, Carlsbad, CA, USA), and an Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA, USA) was used to calculate the RNA integrity score. RNA-sequencing (RNA-seq) libraries were prepared using the TruSeq Stranded messenger RNA (mRNA) LT sample preparation kit Illumina, San Diego, CA, USA) using Sciclone and Zephyr liquid handling robotics (PerkinElmer, Spokane, WA, USA) at pre- and post-PCR steps. Library concentrations were determined using a Qubit 2.0 Fluorometric Quantitation system (Life Technologies, Carlsbad, CA, USA), and the distribution of sizes was determined using an Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA, USA). For sequencing, the 14 libraries were pooled, diluted to equimolar amounts, and sequenced on an Illumina HiSeq 3000/4000 using 50-bp single-end chemistry. Base calls provided by the real-time analysis (RTA) software (Illumina, San Diego, CA, USA) were converted into a multiplexed, unaligned BAM format before demultiplexing into sample-specific unaligned BAM files.
Next generation sequencing (NGS) samples (passing quality filtering) were aligned to the UCSC Genome Browser (Mus musculus genome assembly mm10, Mus musculus transcriptome mm10_e100) of the genome reference consortium (GrCh38/GrCm38) using the STAR alignment tool (Dobin et al., 2013). The Bioconductor DESeq2 package (Love et al., 2014) was used for testing differentially expressed genes (DEGs) using the negative binomial distribution.
Overrepresentation analysis of the unranked list of significantly (FDR < 0.05) DEGs common to both animal models was performed. The analysis utilized g:profiler package (version e103_eg50_p1) (Reimand et al., 2007), specifically g:GOSt Functional profiling tool with the Benjamini-Hochberg FDR threshold cutoff of 0.05. Gene sets utilized in analysis included gene ontology molecular function (GO:MF) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway sets. The 10 most enriched terms for both GO:MF and KEGG pathways were represented in a decreasing order using –log10(FDR) value.
Gene set enrichment analysis (GSEA) was performed using the GSEA software. As GSEA input expression data set values, DeSeq2 normalized counts were used. The analysis performed for MSigDB gene sets (Subramanian et al., 2005; Liberzon et al., 2011) from C2: KEGG pathways, C5: GO:MF, and cutoffs for the value of p < 0.05 and FDR < 0.25. GO:MF gene sets were further imported into Cytoscape (v.3.8.2) (Shannon et al., 2003), and the pathways were clustered into networks using EnrichmentMap (v.3.3.1) (Merico et al., 2010). For network generation (Reimand et al., 2019), a similarity cutoff of 0.375 for combined Overlap+Jaccard score and combined score of 0.5 was used.
All electrophysiological data were analyzed using the Origin software (Originlab, Origin Pro 8, North Hampton, NH, USA). Statistical analysis (Graphpad Prism 7.0, GraphPad Software Inc., San Diego, CA) was performed by either the Mann–Whitney test in case of non-normal-distributed data or t-test for normal-distributed data (Shapiro–Wilk). The distribution of heat-responsive sensory neurons was tested using a Fisher's exact test. Statistical analysis of behavioral data and qPCR was performed by unpaired Student's t-test (capsaicin injections, Von Frey assay, Hargreaves test, and OFT, qRT-PCR). For Morris water maze, unpaired Student's t-test was used to analyze the short- and long-term memory data and two-way ANOVA with Sidak's multiple comparisons test for the latency to reach the platform.
Currently, there are 24 known PRDM12-CIP patients from different ethnic backgrounds (Figure 1A). To contribute a better understanding of prevalence and the contribution of a PRDM12 variant to nociceptive dysfunction, we present two cases harboring an unreported PRDM12 variant. Two affected first-order male cousins from an Iranian Farsi consanguineous family presented to the clinics with the absence of acute and chronic pain perception starting from birth (Figure 1B, Table 1). There is no history of CIP in the family. Index patient [patient 1 (P1)] was first presented with a tongue ulceration, and was later confirmed to have disturbed temperature and vibration sensation, sensorineural deafness, corneal abrasions, frequent skin and bone infections, and scoliosis. Patient 2 (P2) exhibited very similar symptoms with a few minor differences (Table 1). While both patients were determined to exhibit autism spectrum disorder (ASD), ID, and muscular weakness, patient 1 also presented with mild cortical malformation as evaluated by MRI (data not shown). Due to clinical similarity to other PRDM12-CIP cases, the targeted Sanger sequencing was performed on the DNA sample from patient 1 that revealed a homozygous splice acceptor site variant within intron 1 of PRDM12 NM_021619:c.224-2 A>T (chr9: 133541993) (Figure 1B). Considering that ASD and ID are not commonly associated with PRDM12-CIP, and to rule out any other known CIP-causing gene contribution, we performed WES on the DNA sample from patient 1. WES analysis confirmed the presence of the PRDM12 variant with a CADD score (Kircher et al., 2014) of 29.4. Subsequently, we confirmed that the variant segregated with the disease in a recessive fashion: in a homozygous state in patients 1 and 2 and in a heterozygous state in each of the four parents (Figure 1B). A different homozygous variant in the same position (c.224-2A>G) has previously been published in a patient suffering from CIP accompanied with global developmental delay, similarly to the patients we describe (Saini et al., 2017). Subsequent filtering of the WES failed to detect any additional rare, high CADD-PHRED scoring variants within the other known CIP genes, including NGF, NTRK1, SCN9A, SCN11A, ZFHX2, FAAHP1, and CLTCL1. To identify potential variants contributing to the autism and ID phenotype, we performed a search for rare variants in the genes associated with ASDs and ID as per PanelApp corresponding panels (Martin et al., 2019). The analysis identified rare variants within nine genes confirmed or predicted to be causative for ADD and/or ID in patient 1, however, subsequent Sanger sequencing of these loci in patients 1 and 2 revealed that none were shared (Table 2).
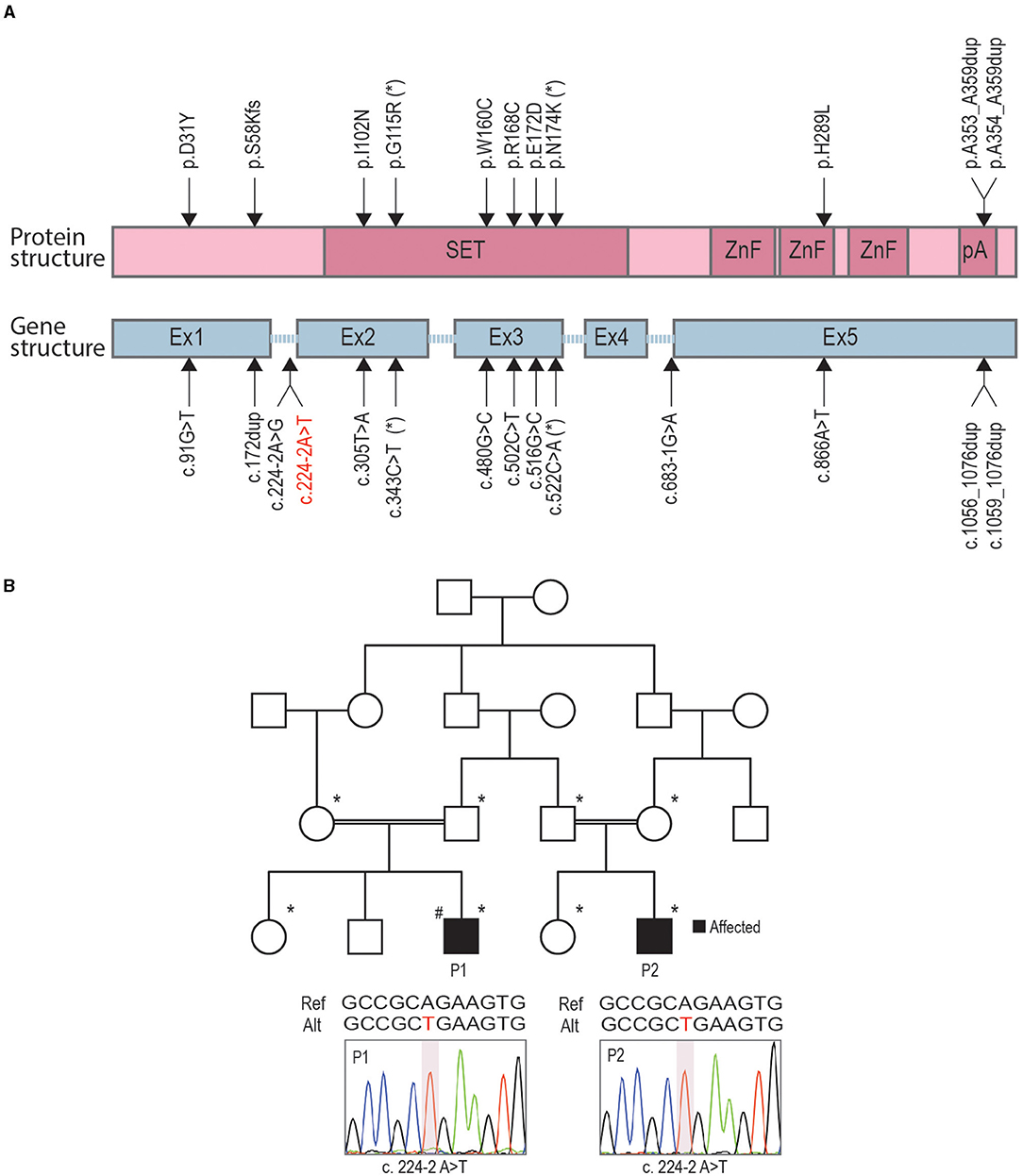
Figure 1. Identification of novel variants in two related congenital insensitivity to pain (CIP) cases. (A) A schematic representation of PR-domain containing member 12 (PRDM12) protein domain structure (pink) and gene structure (blue) with marked locations of previously published causative CIP variants and a novel splice-site variant reported here (red letters). (B) Family pedigree, with affected individuals shaded in black; family members included in segregation analysis indicated with asterisks; hashtag denotes index patient (patient 1, P1) for which whole-exome sequencing (WES) was performed. The position of the missense mutation is depicted below the family pedigree for patients 1 (P1) and 2 (P2) with an illustrative Sanger chromatogram of genomic PCR amplicons.
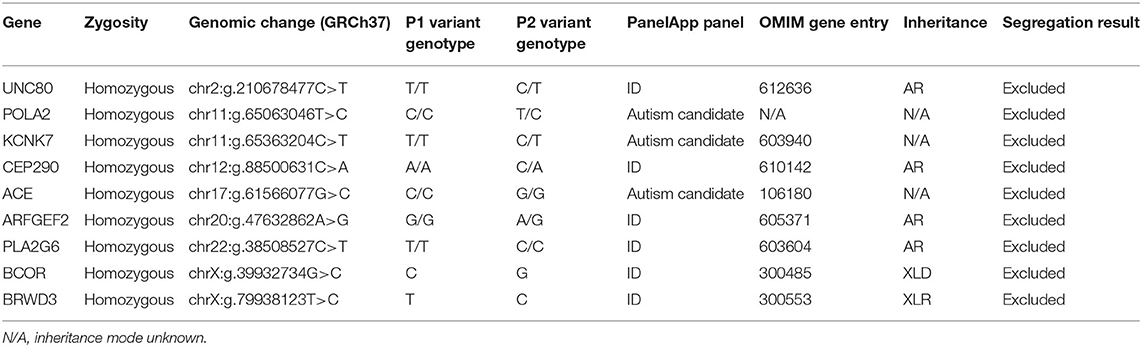
Table 2. Rare homozygous variants in autism and/or intellectual disability- (ID-) associated genes identified in patient 1 whole-exome sequencing (WES).
To understand PRDM12 expression pattern in the mouse, we performed the western blotting analysis of various tissues harvested from adult WT mice. HEK293 cell lysates overexpressing either WT PRDM12 (PRDM12WT) or previously published frameshift mutation Prdm12S58fs (PRDM12S58fs) that leads to no protein product (Nagy et al., 2015) were blotted to ensure antibody specificity (Figure 2A). In addition, low expression in the heart, we did not detect PRDM12 immunoreactivity in any other non-nervous tissue we examined (Figure 2A, upper panel). We detected PRDM12 immunoreactivity in both peripheral and CNS tissue samples (Figure 2A, lower panel). Interestingly, PRDM12 expression was noted in all brain regions examined. As previously reported in embryonic tissue, immunohistochemical double labeling of adult WT DRG sections revealed PRDM12 to remain co-expressed with various polymodal nociceptive markers, including IB4, Nav1.8, CGRP, and TRKA (Figures 2B–E) (Desiderio et al., 2019).
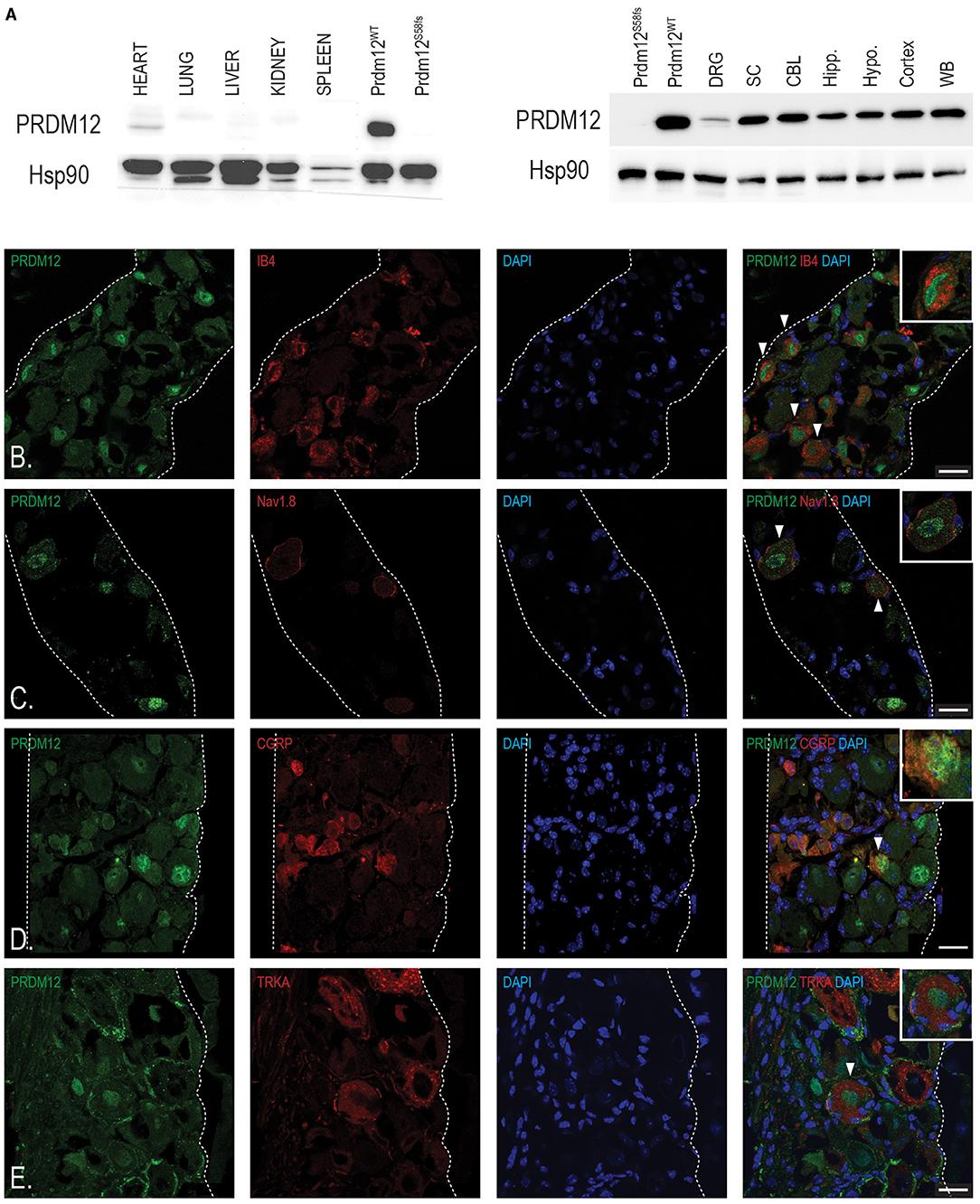
Figure 2. Tissue analyses reveal PRDM12 expression in adult mouse peripheral and central nervous tissue. (A) Upper panel, western blot analysis shows a slight PRDM12 protein expression in adult heart tissue, but no detectable expression in the lungs, livers, kidneys, or spleen. Lower panel, western blot indicates PRDM12 protein level expression in the central nervous system (CNS): whole brain (WB), cortex, hypothalamus (Hypo.), hippocampus (Hipp.), cerebellum (CBL), and spinal cord (SC), as well as in peripheral nervous system (PNS), and dorsal root ganglia (DRG). Antibody positive control (PRDM12WT) is HEK293 cell lysate overexpressing wild type (WT) PRDM12 complementary DNA (cDNA) and negative control (PRDM12S58fs) is HEK203 cell lysate overexpressing PRDM12S58fs cDNA. HSP90 was used as loading control. (B–E) Representative immunohistochemical images of adult mouse lumbar DRG sections indicating PRDM12 co-expression with classical nociceptive markers: IB4, Nav1.8, CGRP, and tyrosine kinase 1 (TRKA), as labeled. White arrows indicate cells with co-labeling, with digitally enlarged insets of select co-labeled cells. Scale bar in all images is 20 μm.
Constitutive, “classic” KO (Figure 3A) of Prdm12 (Prdm12−/−) results in a significant reduction of DRG volume (Figure 3B) and early neonatal death of unknown cause (Desiderio et al., 2019). We were, therefore, motivated to generate a mouse model that more closely resembles human disease as most CIP-causing variants of PRDM12 do not affect its protein expression (Nagy et al., 2015). Therefore, using the Cas9 methodology, we generated a frameshift at position c.537A_del (p.S159AfsTer2; Prdm12S159AfsTer2) and point mutation at c.542G>C (p.W160C; Prdm12W160C) in murine Prdm12 gene (NM_001123362, Supplementary Figure 1A) to reflect the previously reported PRDM12-CIP mutation W160C (Nagy et al., 2015). To improve viability, mice were maintained on a mixed FVB/NJ/C57/BL6 background. Prdm12S159AfsTer2 mutation is predicted to cause an early stop codon and likely mimic the constitutive Prdm12−/− absence of PRDM12 protein expression. Both Prdm12S159AfsTer2 and Prdm12W160C mice, similarly to the constitutive Prdm12−/−, showed complete penetrance of prenatal lethality in homozygous mutants. Heterozygous Prdm12S159AfsTer2+/− mice had normal pain perception as compared to WT littermates when tested in acute pain assays (Supplementary Figure 1B).
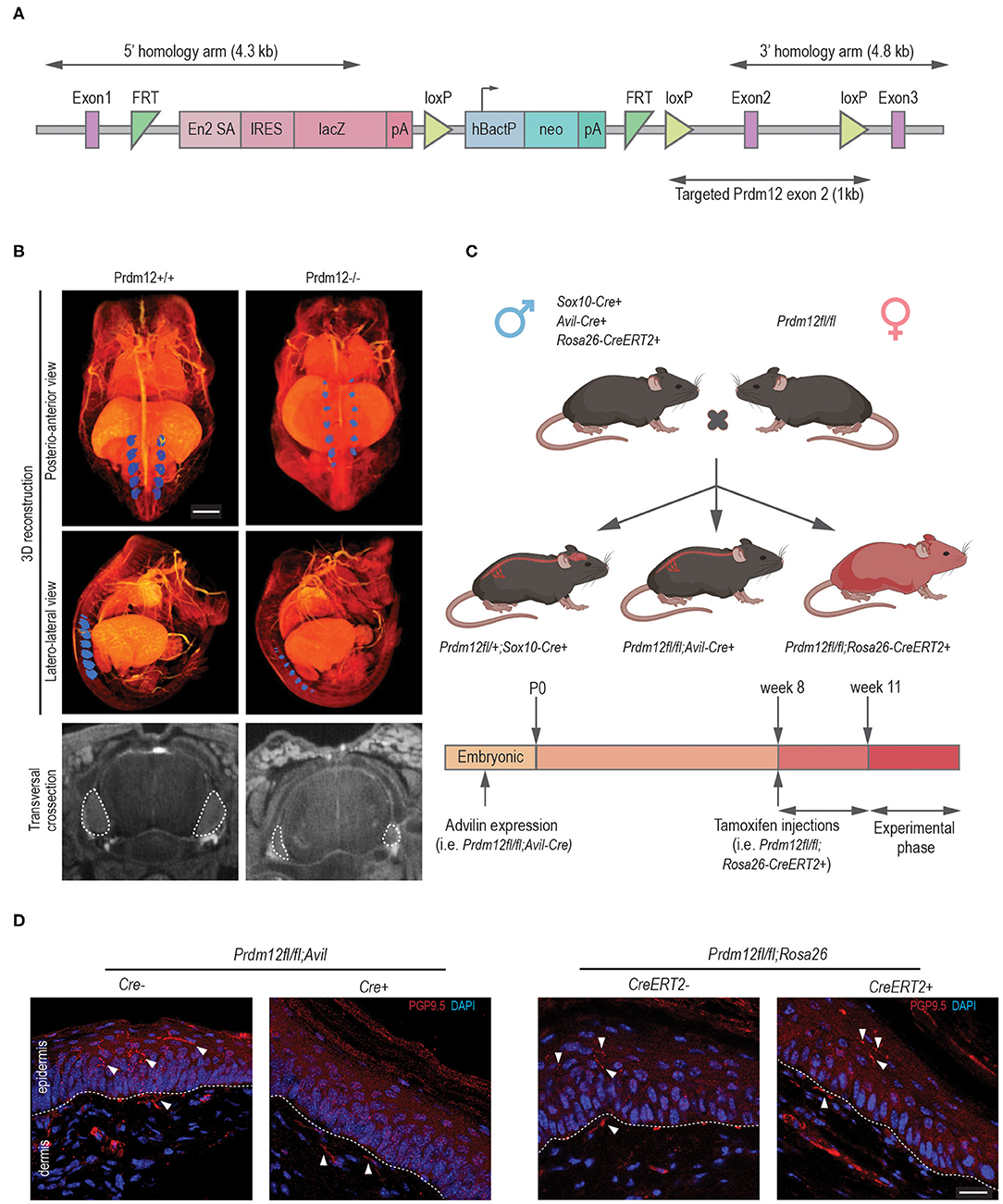
Figure 3. Validation of animal models generated for the study. (A) A schematic representation of the lethal classical Prdm12 knock-out (KO) allele. (B) Lower panels, representative lumbar DRG cross-section micro-CT images of WT and Prdm12−/− E15.5 embryos depicting a reduction of DRG volume in the Prdm12−/−. Middle and upper panels—pseudocolored (blue) 3D reconstructed micro-CT images of lumbar DRG in WT and Prdm12−/− 15.5 embryos, as indicated—posteroanterior (upper panels) and latero-lateral view (middle panels). Scale bar is 1,000 μm. Transversal cross-section of the SC with DRG outlined with a white dotted line (bottom panel). (C) Schematic of the genetic crosses performed to generate developmental constitutive Prdm12fl/fl; Avil-Cre and Prdm12fl/fl; Sox10-Cre, and inducible Prdm12fl/fl; Rosa26-CreERT2, with the timeline of tamoxifen (TAM) injection, analysis or tissue harvest below. (D) Left two panels: representative immunohistochemical images of hind paw skin biopsy from PRDM12fl/fl; Avil-Cre+ show the complete absence of PGP9.5-reactive nerve fibers in the epidermal layer as compared to the Cre− controls. In contrast, PGP9.5-reactive nerve fibers are clearly visible in the epidermal layers in representative images of TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ hind paw skin biopsy (right two panels) as compared to the Cre− controls. Dotted line delineates epidermis/dermis border. Scale bar is 20 μm.
To understand the functional role of PRDM12, we generated several conditional Prdm12 mutant mouse models. We aimed to determine if PRDM12 function was necessary for pain perception when ablated during early development and/or in adults. We first crossed Prdm12fl/fl to Sox10-Cre (Prdm12fl/fl; Sox10-Cre), thereby eliminating PRDM12 expression in the entire neural crest cell population during embryonic development (Figure 3C). Notably, we did not detect any viable Prdm12fl/fl; Sox10-Cre+ offspring. Thermal and chemical acute pain tests (hot plate assay and capsaicin intraplantar injection, respectively) on heterozygous Prdm12fl/+; Sox10-Cre+ did not reveal any reduction in pain perception when compared to control littermates (data not shown).
We next sought to ablate PRDM12 expression specifically in the DRG, thus we crossed Prdm12fl/fl to Avil-Cre (Prdm12fl/fl; Avil-Cre) to remove PRDM12 expression approximately at embryonic day 12.5 (Figure 3C) (Hasegawa et al., 2007; Desiderio et al., 2019). RT-qPCR and the western blotting of lumbar DRG tissue confirmed a significant reduction of Prdm12 transcript and protein expression, respectively (Supplementary Figure 2A). We noted a reduction of the survival of Prdm12fl/fl; Avil-Cre+ animals (Supplementary Figure 2B). The animals that died did so before weaning for reasons that have yet to be determined. In Prdm12fl/fl; Avil-Cre+ survivors, we noted corneal abrasions and facial scarring similar to PRDM12-CIP patients (Supplementary Figure 2C) (Desiderio et al., 2019; Chen et al., 2020).
To delete PRDM12 in adults, we crossed Prdm12fl/fl to TAM-inducible Rosa26-CreERT2 mice (Prdm12fl/fl; Rosa26-CreERT2). Following TAM injections at 8 weeks of age, we generated whole-body PRDM12 KOs in adults 3 weeks following injection as confirmed by RT-qPCR in lumbar DRG and WB tissue (Figure 3C, Supplementary Figure 3A). Unlike the constitutive (Prdm12−/−) or neuronal crest cell progenitor PRDM12 KOs (Prdm12fl/fl; Sox10-Cre+), there was no noted reduction in the survival of adult TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ as compared to control littermates. As PRDM12 is expressed in the mouse brain, we aimed to identify any confounding CNS effects that may have resulted by the reduction of PRDM12 expression in the CNS. OFT of TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ revealed no deficiencies in the distance traveled or increased anxiety as compared to control littermates (Supplementary Figure 3B). Furthermore, the Morris water maze test and the gold standard for hippocampal-dependent spatial learning, revealed no deficiencies in swimming ability, learning acquisition, or short- or long-term memory retention (Supplementary Figure 3C).
To determine the effect of nociceptor development in our conditional mouse models and the extent to which they phenocopy the symptoms of patients, we performed the immunohistochemical analysis of hind paw skin biopsies from both Prdm12fl/fl; Avil-Cre and TAM-injected Prdm12fl/fl; Rosa26-CreERT2 adult mice with anti-PGP9.5 antibody, a marker for small caliber, intraepidermal nerve fibers. PGP9.5 immunostaining is considered as a diagnostic tool in the study of peripheral neuropathies, which, in PRDM12-CIP case, revealed the absence of PGP9.5 reactive nerve endings in patient epidermis (Chen et al., 2015; Van Acker et al., 2016). As it was noted for PRDM12-CIP patients, Prdm12fl/fl; Avil-Cre+ biopsies had no detectable PGP9.5 reactive fibers in the epidermis, in contrast to control littermates (Figure 3D). TAM-injected Prdm12fl/fl; Rosa26-CreERT2+, on the other hand, had normal epidermal PGP9.5+ innervation (Figure 3D).
To investigate the functional nociceptive role of PRDM12 in the two mouse models at a cellular level, we performed single-cell electrophysiological studies on cultured DRG. We isolated DRG from Prdm12fl/fl; Avil-Cre+ and TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and their respective controls and cultured them for ~16 h. The resting membrane potential (Vmem) of Prdm12fl/fl; Avil-Cre+ DRG neurons was similar to Prdm12fl/fl; Avil-Cre− DRG neurons in whole-cell current-clamp recordings (Table 3). However, the IR in the Prdm12fl/fl; Avil-Cre+ DRG neurons was significantly low. Consequently, the minimal current to evoke an AP within 50 ms (IAP) was increased to more than two times in Prdm12fl/fl; Avil-Cre+ neurons. The ablation of the Prdm12 gene after TAM-injection in Prdm12fl/fl; Rosa26-CreERT2 DRG neurons, however, showed no differences in the resting membrane potential, IR and IAP compared to DRG neurons were derived from control, TAM-injected Prdm12fl/fl; Rosa26-CreERT2−, mice (Table 3).
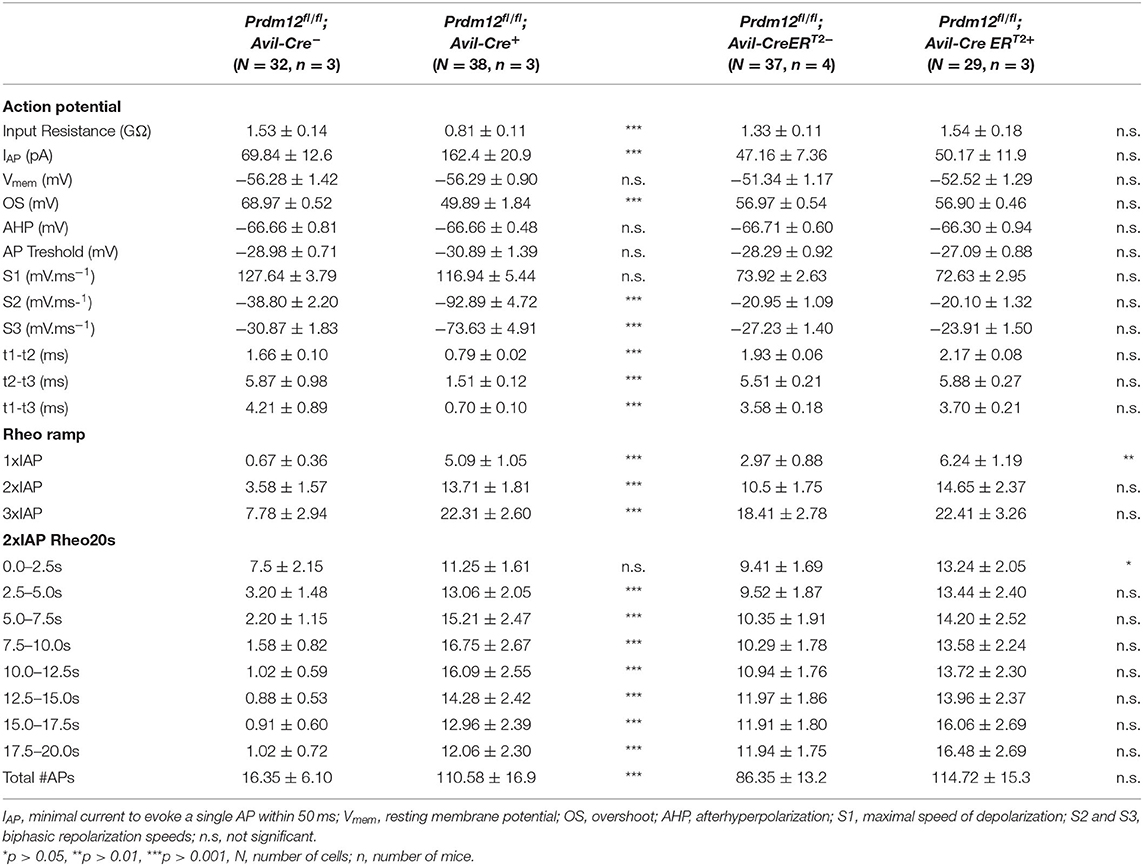
Table 3. Action potential (AP) characteristics of Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Rosa26-CreERT2+ nociceptive neurons and corresponding controls.
To investigate the properties of APs between the corresponding genotypes, we injected the currents at IAP + 10 pA of the recorded neurons. Remarkably, APs evoked from Prdm12fl/fl; Avil-Cre+ were significantly different from those derived from Prdm12fl/fl; Avil-Cre−. The AP width recorded from cultured Prdm12fl/fl; Avil-Cre+ DRG neurons was significantly smaller than those from Prdm12fl/fl; Avil-Cre− neurons and seem to lack the typical biphasic repolarization phase (also known as “hump” or “shoulder”), which is a characteristic of nociceptive DRG (Figure 4A) (Ritter and Mendell, 1992). This reduction in AP width (t1–t3, Table 3) in Prdm12fl/fl; Avil-Cre+ is correlated with increased repolarization speeds (dV/dt min1 and dV/dt min2, Table 3), whereas the depolarization speed (dV/dt max) is unchanged. Additionally, the AP peak (OS) was significantly lower in Prdm12fl/fl; Avil-Cre+ DRG neurons, whereas the AHP was unchanged (Figure 4A, Table 3). The AP threshold did not differ between Prdm12fl/fl; Avil-Cre− and Prdm12fl/fl; Avil-Cre+ DRG neurons. The AP characteristics Vmem, IR, and IAP recorded from Prdm12fl/fl; Rosa26-CreERT2+ and Prdm12fl/fl; Rosa26-CreERT2−-derived neurons have not shown any differences (Figure 4B, Table 3).
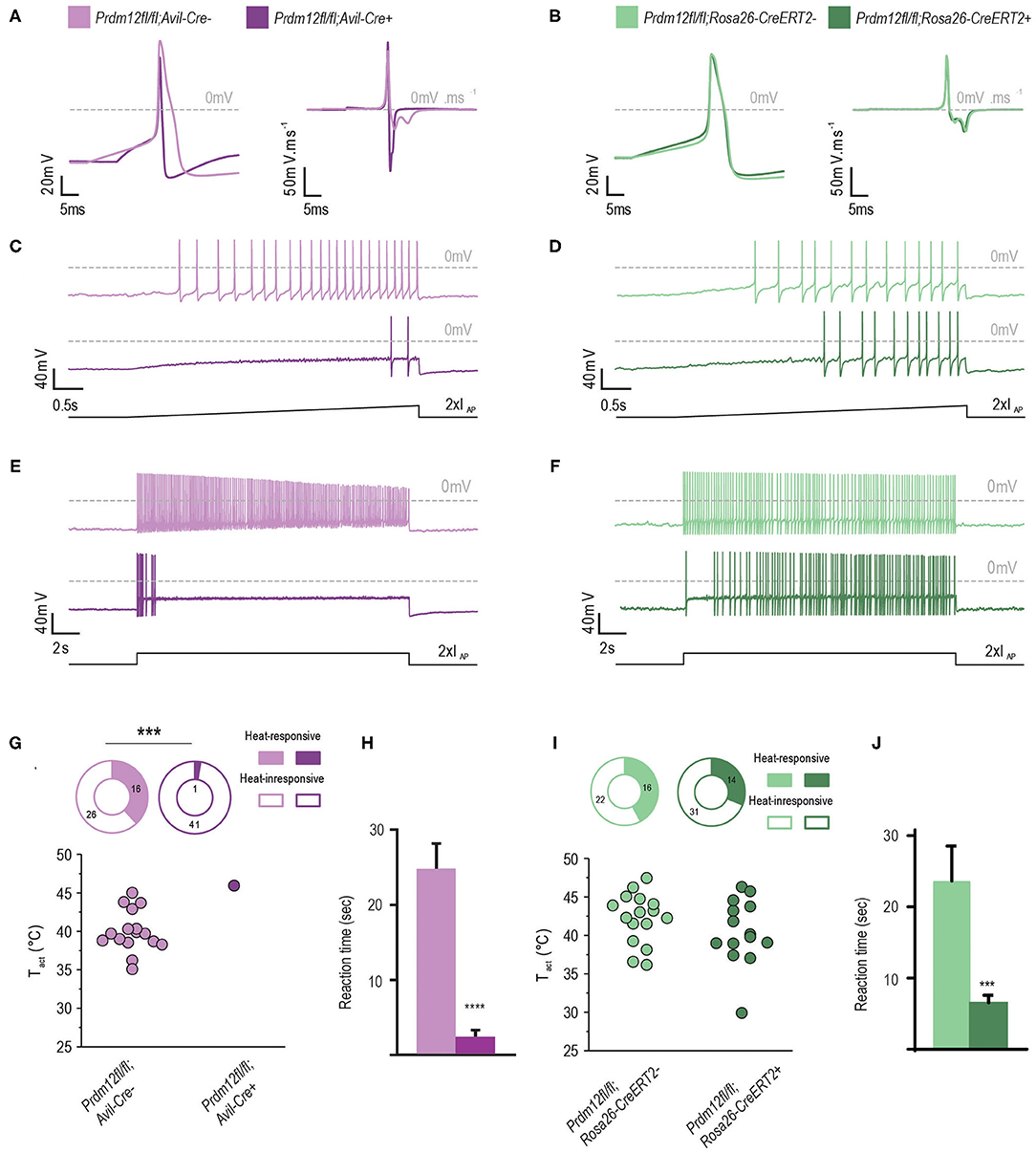
Figure 4. PRDM12 expression is required for proper biophysical properties of cultured DRG. (A) and (B) Representative action potentials (APs) and their first derivatives from Prdm12fl/fl; Avil-Cre+ [(A), dark purple trace], Prdm12fl/fl; Avil-Cre− [(A), light purple trace], Prdm12fl/fl; Rosa26-CreERT2+ [(B), dark green trace], and Prdm12fl/fl; Rosa26-CreERT2− [(B), light green trace] cultured DRG neurons. Note the reduced AP width and lack of biphasic repolarization in Prdm12fl/fl; Avil-Cre+ (A). (C,D) Typical examples of AP generation upon 2 x IAP ramp-shaped depolarization showing the inability of Prdm12fl/fl; Avil-Cre+ DRG neurons to generate similar number of APs compared to the Prdm12fl/fl; Avil-Cre− DRG neurons (C). Note the delay of AP firing for initial 2 s in the ramp-shaped depolarization in Prdm12fl/fl; Avil-CreERT2+, as compared to littermate controls (D). (E) Prdm12fl/fl; Avil-Cre+ neurons dot not generate APs over the full time course of 2 x IAP current injection compared to Prdm12fl/fl; Avil-Cre− neurons. (F) The TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and Prdm12fl/fl; Rosa26-CreERT2− generate APs over the full length of 2 x IAP current injection although the TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ show a reduced number of APs in the beginning of the 2 x IAP current injection. (G,I) Fraction of neurons that showed temperature-evoked currents (top panels) and temperature activation thresholds (Tact) of the recorded neurons (bottom panels). Prdm12fl/fl; Avil-Cre+ neurons do not respond to the applied ramp-shaped temperature increase (G), whereas in both fractions of TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and Prdm12fl/fl; Rosa26-CreERT2− neurons their respective Tact were similar (I). (H) Prdm12fl/fl; Avil-Cre+ exhibited reduced behavioral reactions to intraplantar capsaicin injections as compared to their respective littermate controls (N = 33, mean values 12.38 ± 1.702 SEM vs. N = 18 1.216 ± 0.4454 SEM). (J) TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ exhibited reduced behavioral reactions to intraplantar capsaicin injections as compared to their respective littermate controls (N = 26, mean values 23.66 ± 4.887 SEM vs. N = 35 6.555 ± 1.04 SEM). Values of ***p ≤ 0.005, ****p ≤ 0.0001, ns: not significant. Lower black traces are examples of the applied stimulus protocol.
We applied two different depolarization protocols to evaluate the excitability of the DRG neurons. During a ramp-shaped depolarization, the Prdm12fl/fl; Avil-Cre+ neurons generated fewer APs (Figures 4C,D, Table 3). Additionally, the number of APs generated in a 20-s long, block-shaped depolarization by Prdm12fl/fl; Avil-Cre+ neurons were significantly low (Figure 4E, Table 3) and the Prdm12fl/fl; Avil-Cre+ neurons predominantly fired within the seconds of the depolarization (Table 3). In contrast to Prdm12fl/fl; Avil-Cre+, the Prdm12fl/fl;Avil-Cre− neurons generated more APs during ramp-shaped depolarizations and constantly during the long block-shaped depolarization. Meanwhile, there were no differences observed in the AP characteristics between TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and Prdm12fl/fl; Rosa26-CreERT2− neurons, both excitability protocols showed a minor difference between the two genotypes. In the 1 × IAP depolarizing ramp, the Prdm12fl/fl; Rosa26-CreERT2+ showed a reduced number of Aps, and additionally, in the prolonged 2× IAP depolarization, they showed a reduced number of APs within the first few seconds (Figure 4F, Table 3).
Non-specific activation of Na+ channels by prepulse at −120 mV revealed larger currents in Prdm12fl/fl; Avil-Cre+ DRG neurons compared to controls (Supplementary Figure 4A). The difference was not observed between the TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and littermate controls. To determine the contribution of tetrodotoxin- (TTX-) sensitive and resistant inward currents observed in Prdm12fl/fl; Avil-Cre+, subsequent inhibition of TTX-sensitive inward currents was performed by 250 ms prepulse at −40 mV showing a reduction of inward currents only in Prdm12fl/fl; Avil-Cre+, and not in TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ neurons (Supplementary Figure 4B), as compared to their respective littermate controls. These results imply a larger fraction of TTX-S Na+ channels in Prdm12fl/fl; Avil-Cre+ (Supplementary Figures 4C–E).
PR domain-containing member 12-deficient and control neurons were also tested for their thermal responsiveness by an increase in a ramp-shaped temperature. Strikingly, only a single Prdm12fl/fl;Avil-Cre+ neuron (of the 42 recorded neurons) responded to the thermal stimulus with a relatively high temperature threshold (>45.0°C, Figure 4G). From the Prdm12fl/fl; Avil-Cre,, ~38% of the neurons responded with an averaged threshold temperature of 40.0°C. The percentage of thermal-responsive neurons was similar between TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ and Prdm12fl/fl; Rosa26-CreERT2− neurons and the temperature threshold of the thermal-activated currents (Figures 4H,I).
To determine the behavioral responses of our mouse models to noxious chemical stimuli, we injected intraplantar capsaicin, a pungent ingredient in chili peppers, and recorded the behavioral response. Capsaicin activates its receptor TrpV1 on nociceptive C-fibers, generating a painful sensation completely absent in CIP patients. We found that both conditional mouse models Prdm12fl/fl; Avil-Cre+ and TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ had significantly reduced responses to capsaicin as compared to their respective control littermates (Figures 4H,J).
To identify the effect of Prdm12 deletion on the key genes responsible for acute pain perception between our two mouse models, we performed mRNA sequencing on bulk lumbar DRG samples from at least three animals of both models and corresponding controls. We sought to clarify the cellular phenotypic difference of the development and adult mouse model. Prdm12fl/fl; Avil-Cre+ had 5,851 significantly (false discovery rate, FDR < 0.05) DEGs as compared to corresponding controls, including markers of nociceptors, some of which including Trpv1, Ntrk1, Calca, and Scn10a are additionally validated by RT-qPCR (Supplementary Figure 5A). Out of 5,851 DEGs, 868 genes had an expression level fold change larger than 2 for upregulated DEGs, or smaller than −2 for downregulated DEGs (Figure 5A, top panel). In comparison, TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ had 67 significantly (FDR < 0.05) DEGs (Figure 5A, bottom panel).
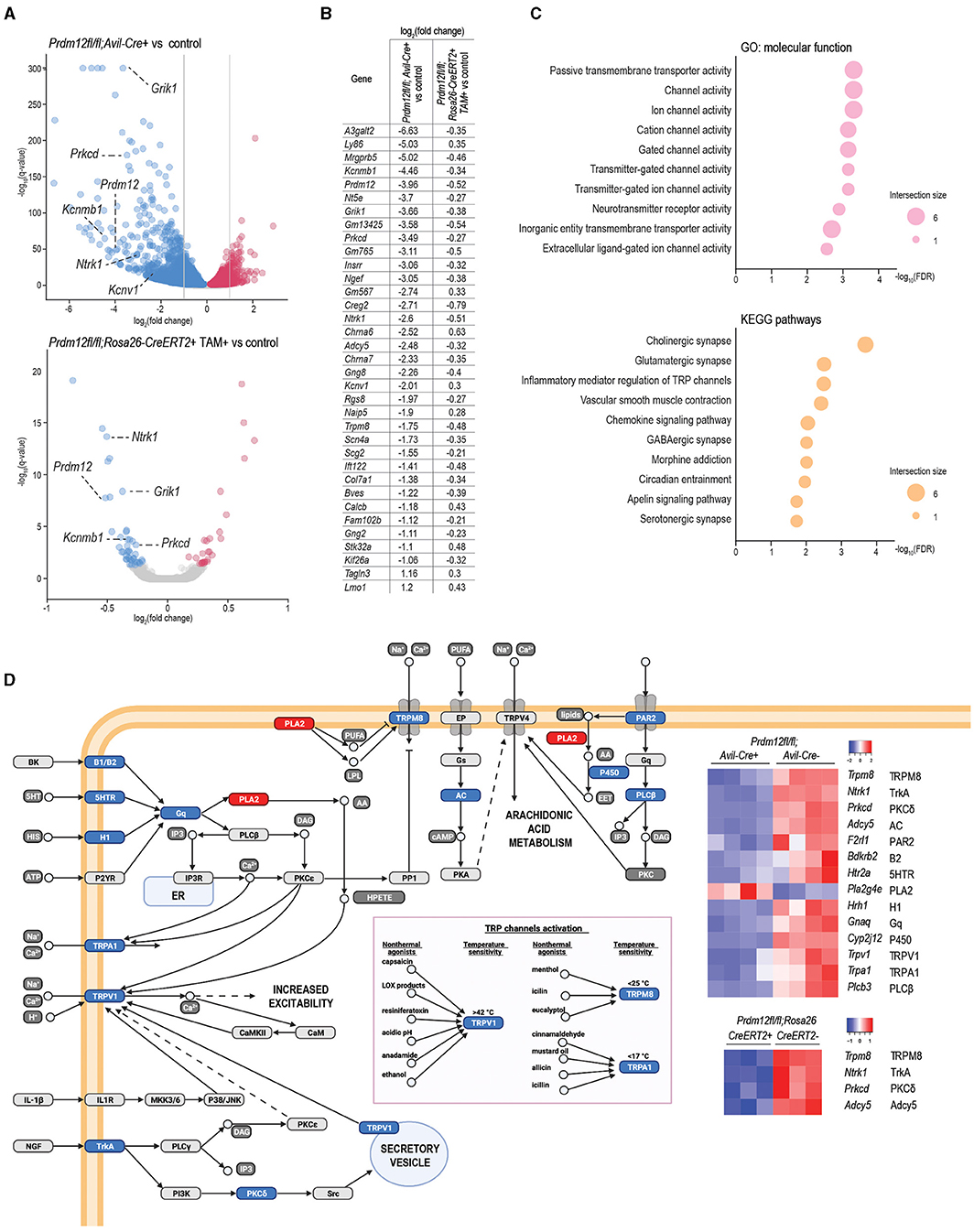
Figure 5. Transcriptomics analysis of DRG of Prdm12-deficient animal models. (A) Volcano plot of differentially expressed genes (DEGs) depicts significantly upregulated and downregulated genes in pooled lumbar DRG isolated from adult Prdm12fl/fl; Avil-Cre+ (upper panel, four biological replicates) and Prdm12fl/fl; Rosa26-ERT2+ (lower panel, three biological replicates) as compared to their respective control littermates. X-axis represents log2 fold change and the y-axis represents –log10(q-values). Genes with the value of q (FDR) < 0.05 were considered differentially regulated, downregulated genes in blue and upregulated genes in red. Gray dotted lines in the top panel demark the cutoff for the most DEGs with at least 2-fold upregulation or downregulation. (B) The list of core DEGs (value of q < 0.05) common to both Prdm12fl/fl; Rosa26-ERT2+ and Prdm12fl/fl; Avil-Cre+ model, and the most DEGs (value of q < 0.05, |log2(fold change)|>1) of the Prdm12fl/fl; Avil-Cre+ model as compared to their corresponding controls. (C) About 10 most enriched terms in the overrepresentation analysis of the core DEGs for the GO:MF database (upper panel) and kyoto encyclopedia of genes and genomes (KEGG) pathways (lower panel). Values on the x-axis present –log10 of the Benjamini–Hochenberg FDR score for a particular pathway enrichment significance. (D) Transient receptor potential (TRP) channels pathway with heatplot of Z-scores for DEG members of the TRP pathway in both developmental and adult Prdm-12 deficiency model. Protein symbols corresponding to the dysregulated transcripts are denoted next to the transcript symbol, and colors within the pathway illustration correspond to the downregulation (blue) or upregulation (red) of the transcripts represented in the heatmap.
To gain insights into the possible contribution of DEGs to the PRDM12 core transcriptional program crucial for the function of nociceptors, we considered all 67 Prdm12fl/fl; Rosa26-CreERT2+ DEGs and 868 most significantly dysregulated genes of the Prdm12fl/fl; Avil-Cre+ model and identified 35 transcripts common to both models, now referred to as PRDM12 core DEGs (Figure 5B). In addition to Prdm12, the core DEGs include the genes encoding for TRP channels and TRP channel-associated signaling including Ntrk1, Trpm8, Adcy5, Prkcd, and ion channels like Kcnv1, Kcnb1, Scn4a, and neurotransmitter receptor activity like Grik1 and Chrna7. Overrepresentation analysis of MF and KEGG pathway gene sets with a g:profiler toolbox revealed that PRDM12 core DEGs are mainly involved in the function of ion channel activity, either passive or gated (Figure 5C). Among KEGG pathways, top enriched pathways are associated with synaptic signaling and pathways regulating TRP-mediated signaling (Figure 5C, bottom panel). DEG protein products included in TRP channel signaling are illustrated in Figure 5D, with the associated heat map representing DEGs from both mouse models.
To examine if dysregulated transcriptomes of Prdm12fl/fl; Avil-Cre+ and TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ models vs. corresponding controls reflect transcriptome footprints of nociceptors or some other sensory cell types in the DRG as benchmark gene sets, we used the top 50 genes marking each of the 14 different distinct sensory cellular subtypes described by Sharma et al. (2020) and performed GSEA. Prdm12fl/fl; Avil-Cre+ model is predominantly enriched for the cell types associated with proprioception and mechanoreception (Figure 6A, top). Moreover, in both Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Rosa26-CreERT2+ experimental set-up, the subtypes of nociceptors were enriched in the corresponding controls as compared to the loss-of-function models (Figure 6A, Supplementary Figures 5B–D). This finding shows that the deletion of Prdm12 depletes DRG of nociceptor-specific genes, and in particular the subtypes of somatostatin-positive and CGRP-positive nociceptors. Intriguingly, this observation is common to both the developmental Prdm12fl/fl; Avil-Cre+ as well as adult TAM-inducible Prdm12fl/fl; Rosa26-CreERT2+models. Recently, Landy et al. (2021) have reported a transcriptomic analysis of bulk RNA in DRG harvested from a spatially restricted adult-inducible Prdm12 KO (Prdm12AvilERT2CKO). Between 150 genes that were dysregulated in TAM-inducible Prdm12AvilERT2CKO described by Landy et al. (2021) and 67 DEGs that were found in our TAM-inducible Prdm12fl/fl; Rosa26-CreERT2 model, we find an overlap in 2 genes—Prdm12 and Chrna6. However, between DEGs in Landy et al. (2021) model and our Prdm12fl/fl; Avil-Cre model, we identified 40 overlapping genes, 21 of which are expressed in the DRG, including Prdm12, Chrna6, Trpm3, and are important for the development of nervous system, such as Nnat. Mapping of the possible functional consequences of the DEGs in both models was performed by GSEA of GO MF (GO:MF) gene sets, followed by generating the clustered network of the enriched terms (Figure 6B). Developmental model Prdm12fl/fl; Avil-Cre+ vs. controls exhibited an enrichment in the control samples (conversely the depletion in Prdm12fl/fl; Avil-Cre+) for gene sets that cluster into networks including “ion channel activity,” “neurotransmitter associated activity,” and “neuropeptides” (Figure 6B). Clusters in the adult model TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ vs. control maintain the depletion of gene sets associated with “ion channel activity” (Figure 6A).
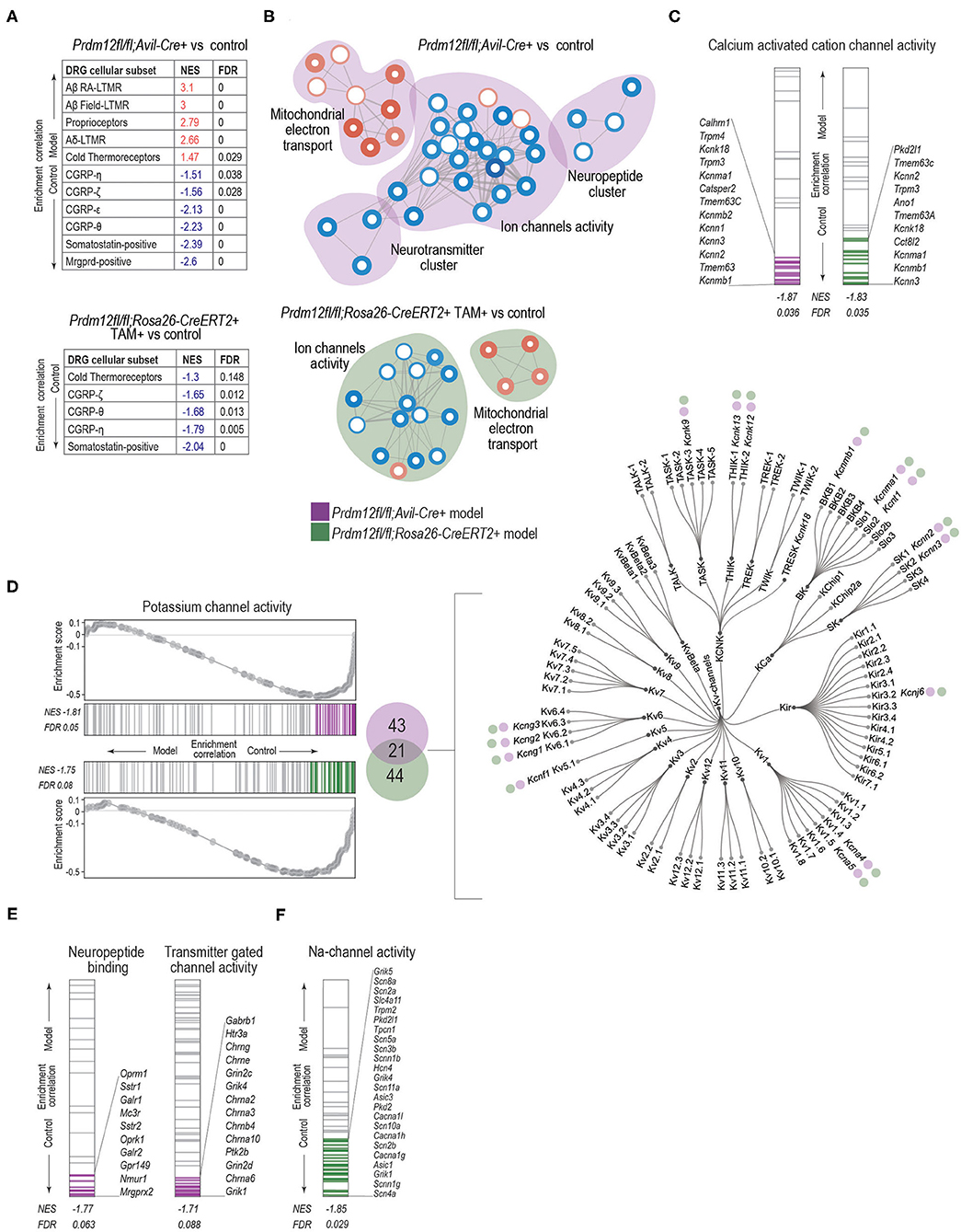
Figure 6. GSEA analysis with enrichment mapping shows distinct clusters of dysregulated MF:GO terms. (A) Dysregulation of bulk RNA DRG transcriptomes as demarked by gene sets most significant by sensory neuron subtypes detected in the DRG, for both Prdm12fl/fl; Avil-Cre+ (top table) or TAM-injected Prdm12fl/fl; Rosa26-ERT2+ (bottom table) as detected by GSEA with the cutoff value of p of 0.05 and FDR cutoff of 0.25. (B) Clusters of GSEA enriched-term nodes in the GO:MF terms. Nodes in red either in Prdm12fl/fl; Avil-Cre+ (pink cloud, left) or in Prdm12fl/fl; Rosa26-ERT2+ (blue cloud, right) are enriched in DRG, while nodes in blue represent the depletion of MF terms, as labeled. Thickness of the node border corresponds to the –log10(FDR), while depth of the color corresponds to normalized enrichment score (NES) of a particular node as compared to the whole gene set. Thickness of the edge lines corresponds to the strength of the used similarity coefficient. Clusters of highly connected nodes are grouped together and annotated with the closest common functional term. (C) GSEA enrichment graphs showing the distribution of calcium-activated cation channel activity as an example of a significantly enriched GO:MF term common to both in Prdm12fl/fl; Avil-Cre+ (left) or TAM-injected Prdm12fl/fl; Rosa26-ERT2+ (right) showing genes contributing to the leading edge in purple and green for Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Rosa26-ERT2+, respectively. NES and FDR values are denotated in the figure. (D) Enrichment graphs depicting the distribution of GOM:MF potassium channel activity genes enrichment in the controls of both Prdm12fl/fl; Avil-Cre+ (left panel, top) and Prdm12fl/fl; Rosa26-ERT2+ (left panel, bottom). Graphs for each model show running enrichment scores, as well as NES and FDR. Genes within the leading edge are colored purple and green for Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Rosa26-ERT2+ models, respectively. In the middle panel, Venn diagram depicts the number of genes within the leading edge of Prdm12fl/fl; Avil-Cre+ (43 genes, purple) and Prdm12fl/fl; Rosa26-ERT2+ (44 genes, green) and 21 overlapping genes. Dendogram of voltage-gated potassium channel families (right) denotes the type of Kv-channel transcripts shared between the two models, note the purple and green circles. (E) Enrichment graphs showing the distribution of terms specific for Prdm12fl/fl; Avil-Cre+ model, showing the depletion of GO:MF neuropeptide binding genes and transmitter-gated channel activity genes in Prdm12fl/fl; Avil-Cre+ model as compared to the corresponding controls. Positions of genes contributing to the significant enrichment are colored in purple. (F) Enrichment graphs showing the distribution of GO:MF sodium channel activity genes in the transcriptome of Prdm12fl/fl; Rosa26-ERT2+ model, showing an enrichment in the controls as compared to the deficiency model. Positions of genes contributing to the significant enrichment are colored in green. NES and FDR values are denoted in the figures.
To explore the enrichment patterns within ion channel activity clusters between the models, we investigated various gene sets to identify if any specific GO:MF terms are common in the two models. “Calcium activated ion channel activity” (Figure 6C) and “potassium channel activity” terms (Figure 6D, Supplementary Figure 5E) are enriched in controls of both models. Within the genes contributing to the leading edge of both models, 21 genes are common (Figure 6D, middle panel). Most of the common genes belong to the group of voltage-gated potassium channels (Figure 6D, right panel). To identify specific gene sets that differ between Prdm12fl/fl; Avil-Cre+ and Prdm12fl/fl; Rosa26-CreERT2+ models, we analyzed neuropeptide and neurotransmitter clusters in the Prdm12fl/fl; Avil-Cre+. Within a neuropeptide cluster, GO terms associated with neuropeptide binding were depleted in the Prdm12fl/fl; Avil-Cre+ (and enriched in the controls), with transcripts associated with opioid receptors Oprm1 and Oprk, as well as galanin receptors Galr1 and Galr2 within the leading edge (Figure 6E, left panel). In the neurotransmitter cluster, genes associated with neurotransmitter-mediated ion channel activity were depleted in Prdm12fl/fl; Avil-Cre+ model as compared to controls, with the subunits of glutamate receptors (Grin2c, Grik4, Grin2d, and Grik1), cholinergic nicotinic receptors contributing the most to the leading edge (Figure 6E, right panel). Furthermore, the analysis of ion channel cluster in the Prdm12fl/fl; Rosa26-CreERT2+ model revealed the depletion of sodium channel activity genes, including markers of nociceptors, voltage-gated sodium channels encoded by Scn11a, and Scn10a, members of TRP pathways like acid sensing channels encoded by Asic1 and Asic3 as well as glutamate receptor subunits Grik1, Grik4, and Grik5 (Figure 6F).
Here, we present two novel cases of CIP in a large, consanguineous family, exhibiting symptoms in line with defective nociception that share a novel homozygous variant in PRDM12. Generating several developmental and adult-onset deletion mouse models of Prdm12, we could determine that early developmental expression of PRDM12 is essential for survival in mice. We show that PRDM12 expression is essential for acute noxious pain detection throughout life. Detailed electrophysiological analyses revealed different but significantly altered biophysical properties of cultured DRG in the developmental and in the adult PRDM12 ablation models. Indeed, different transcriptional programs are regulated by PRDM12 at different maturation stages, accounting for a difference in the corresponding cellular phenotypes. Taken together, these data confirm that developmental PRDM12 is essential in pain perception in mice by regulating key nociceptor-specific genes required for the development and function of nociceptors. While PRDM12 expression in adults is no longer essential for survival or needed for certain types of CNS functions, it is still required for nociceptive responses.
Human PRDM12 expression is exclusive to the PNS making it a promising therapeutic target (Chen et al., 2015). However, considering the restricted PRDM12 expression, it is not clear why a small number of PRDM12-CIP patients, including those in this report, have CNS dysfunctions. The analysis of WES of patient 1 identified eight rare variants in genes associated with autism and/or ID, but these are not shared with patient 2 and are unlikely to explain the CNS symptoms in this family. Interestingly, a splice-site acceptor variant reported here is in the same position as an already reported PRDM12-CIP case of different ethnic background, presenting with typical CIP symptoms including ID (Saini et al., 2017). Why should variants at this position but not in others have an impact on CNS function is not clear. Therefore, the molecular mechanisms underlying CNS symptoms that are associated with PRDM12-CIP in a small number of cases remain to be elucidated.
We report PRDM12 expression in all adult mouse brain regions examined, suggesting a role for PRDM12 beyond nociception. Generating the Prdm12W160C mouse model that proved to be neonatally lethal allowed us to conclude that fully functional PRDM12 is required for the survival in mice. While the cause of death is still not clear, lethality of the neuronal progenitor KO model Prdm12fl/fl; Sox10-Cre leads us to posit that the PRDM12 essentiality may be intrinsic to the cells derived from the Sox10+ lineage, not necessarily of neuronal origin. The heart or brain, for example, expresses PRDM12 but also contain Sox10+ cardiomyocytes and glia, respectively (Kuhlbrodt et al., 1998; Montero et al., 2002; Sande-Melón et al., 2019; Tang et al., 2019). The role for PRDM12 in mouse CNS has recently gained attention. Hypothalamic PRDM12 regulates Pomc expression, food intake, adiposity, and weight (Chen et al., 2020; Woo et al., 2020). Here, we report no effect of ablating PRDM12 expression in adults on survival, locomotion, anxiety, or hippocampal-dependent learning. These results suggest that whatever the role for developmental PRDM12 is in the mouse CNS, it is no longer required for the behaviors tested here in the adult. It would be of great interest to fully characterize the role for PRDM12 in the mouse CNS, and in light of a growing number of PRDM12-CIP patients with CNS symptoms, to explore the possibility for its function in the human brain.
Analyses of PRDM12 developmental and adult-ablation models show a dependence of proper nociceptive responses on the presence of Prdm12. Contrary to our results, recent publication indicates that, while the adult murine model of Prdm12 deficiency has an effect on transcriptional regulation, it does not translate to the impairment of pain sensation behaviorally (Landy et al., 2021). We posit that these differences could be explained by the different experimental mouse models used in the two studies. TAM-inducible Prdm12fl/fl; Rosa26-CreERT2+ adult model used here targets Prdm12 exon 2, mimicking the splice acceptor mutation described in our patients, under the ubiquitous Rosa26 promoter, while Landy et al. (2021) describe a model that targets exon 5, and is under the TAM-inducible DRG-specific Avil promoter targeting a more specific cellular subtype only in the periphery. The reduction in behavioral responses to capsaicin injections together with the significant dysfunction of cultured nociceptor electrophysiological responses in our Prdm12fl/fl; Rosa26-CreERT2+ mouse model confirm PRDM12 nociceptive regulation in the PNS even in adults. However, a variety of behavioral pain responses were not observed in the DRG-specific adult-ablation model in Landy et al. (2021), implying a possible contribution of PRDM12 action in CNS pain responses that would warrant further study.
Electrophysiological studies of developmental homozygous mouse model clearly determined that it is the peripherally expressed PRDM12 during the embryonic/developmental stage that is required for the detection and signaling of nociceptive stimuli. We find that, while resting membrane potential is unaffected in the developmental PRDM12-ablation model, AP morphology is impaired, as well as the number of APs generated upon different stimulation protocols. Moreover, based on the prepulse inhibition experiments, we find that the ratio of currents generated by TTX-sensitive Na+ channels is increased in Prdm12fl/fl; Avil-Cre+ neurons as compared to control littermates. A significant reduction of pain-related genes encoding for Nav1.8, Nav1.9, TrpV1, TrkA, and CGRP, in the Prdm12fl/fl; Avil-Cre+ strongly corroborates the electrophysiological nociceptive dysfunctions are due to the lack of nociceptors in developmental models as described previously (Desiderio et al., 2019).
Transcriptome analysis of the bulk DRG shows Prdm12fl/fl; Avil-Cre+ dysregulation of more than 5,000 genes, 868 of which are dysregulated by at least a 2-fold increase or decrease. Downregulated expression of nociceptor signature genes including Ntrk1, Scn10a and Scn11a, Calca in Prdm12fl/fl; Avil-Cre+ compared to controls is a consequence of the lack of nociceptor development as described previously (Desiderio et al., 2019), and confirmed by the lack of skin PGP9.5 positive innervation in Prdm12fl/fl; Avil-Cre+ animals. Nevertheless, as Prdm12fl/fl; Rosa26-CreERT2+ TAM-inducible model overlaps with Prdm12fl/fl; Avil-Cre+ in 35 DEGs, which in the overrepresentation analysis is associated with ion channels, and more specifically TRP channels pathways, indicate that Prdm12 maintains a role in regulating ion channels essential for the function of nociceptors even in the adulthood, supported by the electrophysiological and behavioral data. Indeed, the comparison of our Prdm12fl/fl; Avil-Cre model transcriptomic data to adult TAM-inducible model Prdm12AvilERT2CKO proposed by Landy et al. (2021) shows an overlap in 40 DEGs, out of which 21 are DRG genes.
Gene set enrichment analysis highlights that both developmental and adult models share the depletion of the terms associated with an ion channel cluster. Many of these terms in adult model are the members of distinct subfamilies of voltage-gated potassium channels essential for the generation and propagation of APs, corroborating the electrophysiological changes and behavioral responses to capsaicin injections. In contrast, developmental model exhibits specific clusters of dysregulated gene sets including neurotransmitter- (e.g., Grik1, Grik4, Grin2d, and Grin2c) and neuropeptide- (e.g., Oprm1, Oprk1, Galr1, Galr2, and Nmur1) related clusters. Specific gene set that is selectively depleted in Prdm12fl/fl; Rosa26-CreERT2+ but not in Prdm12fl/fl; Avil-Cre+ is associated with sodium channel activity. Interestingly, these include nociceptor-specific genes Scn10a, encoding for Nav1.8 and Scn11a encoding for Nav1.9, possibly contributing to the observed differences in AP firing in TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ as compared to the corresponding controls.
PR domain-containing member 12 ablation in Prdm12fl/fl; Avil-Cre+ animals results in electrophysiological and behavioral deficiencies and the lack of nociceptor skin innervation due to the essential role played by PRDM12 in the specification of nociceptor development. However, TAM-injected Prdm12fl/fl; Rosa26-CreERT2+ adult-deletion model maintains a significant dysregulation of a small subset of genes involved in TRP channel and ion channel activities. Clearly, PRDM12 is an indispensable component of nociceptor specification and function required for pain perception throughout life. Together with its uniquely restricted expression in humans and the requirement for adult pain perception make it an intriguing therapeutic target with immense clinical potential.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA734482.
The studies involving human participants were reviewed and approved by Shahid Beheshti University of Medical Sciences ethics board (with the current update of the Declaration of Helsinki). Written informed consent to participate in this study and for the publication of any potentially identifiable images or data included in this article was provided by the participants' legal guardian/next of kin. The animal study was reviewed and approved by Bundesministerium fur Wissenschaft, Forschung und Wirtschaft (BMWFW-66.015/0011-WF/V/3b/2017), Austria and carried out according to EU-directive 2010/63/EU. Prdm12W160 and Prdm12S159AfsTer2+/- experiments were approved by the University of Sydney Animal Ethics Committee under animal ethics protocol (938) and reporting complies with the ARRIVE guidelines.
TK: curated data, performed the formal analysis, investigation, validation of the data, visualization, and assisted in writing the manuscript. ML: curated data, performed the formal analysis, investigation, and assisted in writing the manuscript. EL, JM, and CF: curated data, performed the formal analysis, and investigation. EA and AT: curated patient data. HD: participated in project administration and supervision. EB: provided resources. GN and MK: provided resources, project administration, supervision, and funding acquisition. JP: conceptualized the project and methodology, provided resources, and funding acquisition. VN: conceptualized the project and methodology, curated data, participated in the investigation, formal analysis, resources, project administration, supervision, funding acquisition, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by Austrian Science Fund (FWF): P32924 and Ludwig Boltzmann Gesellschaft. TK was funded by a DOC-fellowship of the Austrian Academy of Sciences (OeAW): 25408; CF was funded by a DOC-fellowship of the Austrian Academy of Sciences (OeAW): 25525.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank patients and their families for participating in this study. WES was performed at the BSF in CeMM Center for Molecular Medicine of the Austrian Academy of Sciences (CeMM). We would like to thank Brian Metcher, Department of Evolutionary Biology, University of Vienna, for his help with the microCT imaging and Sophie Imhof, Medical University of Vienna, for help with figure design. Figure illustrations were created with BioRender, Adobe Illustrator 2021, and Python (Pandas, Numpy, Seaborn libraries).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.720973/full#supplementary-material
Bandell, M., Story, G. M., Hwang, S. W., Viswanath, V., Eid, S. R., Petrus, M. J., et al. (2004). Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron 41, 849–857. doi: 10.1016/S0896-6273(04)00150-3
Bartesaghi, L., Wang, Y., Fontanet, P., Wanderoy, S., Berger, F., Wu, H., et al. (2019). PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Rep. 26, 3484–3492.e4. doi: 10.1016/j.celrep.2019.02.098
Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D., and Julius, D. (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389, 816–824. doi: 10.1038/39807
Chen, X., Wyler, S. C., Li, L., Arnold, A. G., Wan, R., Jia, L., et al. (2020). Comparative transcriptomic analyses of developing melanocortin neurons reveal new regulators for the anorexigenic neuron identity. J. Neurosci. 40, 3165–3177. doi: 10.1523/JNEUROSCI.0155-20.2020
Chen, Y.-C., Auer-Grumbach, M., Matsukawa, S., Zitzelsberger, M., Themistocleous, A. C., Strom, T. M., et al. (2015). Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 47, 803–808. doi: 10.1038/ng.3308
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff SNPs in the genome of Drosophila melanogaster strain. Fly 6, 80–92. doi: 10.4161/fly.19695
Desiderio, S., Vermeiren, S., Van Campenhout, C., Kricha, S., Malki, E., Richts, S., et al. (2019). Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Rep. 26, 3522–3536.e5. doi: 10.1016/j.celrep.2019.02.097
Dittert, I., Vlachová, V., Knotková, H., Vitásková, Z., Vyklicky, L., Kress, M., et al. (1998). A technique for fast application of heated solutions of different composition to cultured neurones. J. Neurosci. Methods. 82, 195–201. doi: 10.1016/S0165-0270(98)00051-X
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Hasegawa, H., Abbott, S., Han, B. X., Qi, Y., and Wang, F. (2007). Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J. Neurosci. 27, 14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007
Hohenauer, T., and Moore, A. W. (2012). The Prdm family: expanding roles in stem cells and development. Dev. 139, 2267–2282. doi: 10.1242/dev.070110
Imhof, S., Kokotović, T., and Nagy, V. (2020). PRDM12: new opportunity in pain research. Trends Mol. Med. 26, 895–897. doi: 10.1016/j.molmed.2020.07.007
Julius, D. (2013). TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384. doi: 10.1146/annurev-cellbio-101011-155833
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans, Genome Aggregation Database Consortium. Nature 581:19. doi: 10.1038/s41586-020-2308-7
Kinameri, E., Inoue, T., Aruga, J., Imayoshi, I., Kageyama, R., Shimogori, T., et al. (2008). Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE 3, 1–10. doi: 10.1371/journal.pone.0003859
Kircher, M., Witten, D. M., Jain, P., O'roak, B. J., Cooper, G. M., and Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315. doi: 10.1038/ng.2892
Kuhlbrodt, K., Herbarth, B., Sock, E., Hermans-Borgmeyer, I., and Wegner, M. (1998). Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998
Landrum, M. J., Lee, J. M., Benson, M., Brown, G. R., Chao, C., Chitipiralla, S., et al. (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46:gkx1153. doi: 10.1093/nar/gkx1153
Landy, M. A., Goyal, M., Casey, K. M., Liu, C., and Lai, H. C. (2021). Loss of Prdm12 during development, but not in mature nociceptors, causes defects in pain sensation. Cell Rep. 34:108913. doi: 10.1016/j.celrep.2021.108913
Langeslag, M., Malsch, P., Welling, A., and Kress, M. (2014). Reduced excitability of gp130-deficient nociceptors is associated with increased voltage-gated potassium currents and Kcna4 channel upregulation. Pflugers Arch. Eur. J. Physiol. 466, 2153–2165. doi: 10.1007/s00424-014-1443-0
Lau, M. T., Manion, J., Littleboy, J. B., Oyston, L., Khuong, T. M., Wang, Q. P., et al. (2019). Molecular dissection of box jellyfish venom cytotoxicity highlights an effective venom antidote. Nat. Commun. 10:1655. doi: 10.1038/s41467-019-09681-1
Liberzon, A., Subramanian, A., Pinchback, R., Thorvaldsdóttir, H., Tamayo, P., and Mesirov, J. P. (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. doi: 10.1093/bioinformatics/btr260
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:8. doi: 10.1186/s13059-014-0550-8
Martin, A. R., Williams, E., Foulger, R. E., Leigh, S., Daugherty, L. C., Niblock, O., et al. (2019). PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat. Genet. 51, 1560–1565. doi: 10.1038/s41588-019-0528-2
Matsukawa, S., Miwata, K., Asashima, M., and Michiue, T. (2015). The requirement of histone modification by PRDM12 and Kdm4a for the development of pre-placodal ectoderm and neural crest in Xenopus. Dev. Biol. 399, 164–176. doi: 10.1016/j.ydbio.2014.12.028
Merico, D., Isserlin, R., Stueker, O., Emili, A., and Bader, G. D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5:13984. doi: 10.1371/journal.pone.0013984
Metscher, B. D. (2009). MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev. Dyn. 246, 368–380. doi: 10.1002/dvdy.21857
Metscher, B. D. (2011). X-ray microtomographic imaging of intact vertebrate embryos. Cold Spring Harb. Protoc. 2011:prot067033. doi: 10.1101/pdb.prot067033
Montero, J. A., Giron, B., Arrechedera, H., Cheng, Y. C., Scotting, P., Chimal-Monroy, J., et al. (2002). Expression of Sox8, Sox9 and Sox10 in the developing valves and autonomic nerves of the embryonic heart. Mech. Dev. 118, 199–202. doi: 10.1016/S0925-4773(02)00249-6
Nagy, V., Cole, T., Van Campenhout, C., Khoung, T. M., Leung, C., Vermeiren, S., et al. (2015). The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 14, 1799–1808. doi: 10.1080/15384101.2015.1036209
Nagy, V., Hollstein, R., Pai, T. P., Herde, M. K., Buphamalai, P., Moeseneder, P., et al. (2019). HACE1 deficiency leads to structural and functional neurodevelopmental defects. Neurol. Genet. 5:e330. doi: 10.1212/NXG.0000000000000330
Namer, B., Ørstavik, K., Schmidt, R., Mair, N., Kleggetveit, I. P., Zeidler, M., et al. (2017). Changes in ionic conductance signature of nociceptive neurons underlying fabry disease phenotype. Front. Neurol. 8:335. doi: 10.3389/fneur.2017.00335
Reimand, J., Isserlin, R., Voisin, V., Kucera, M., Tannus-Lopes, C., Rostamianfar, A., et al. (2019). Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517. doi: 10.1038/s41596-018-0103-9
Reimand, J., Kull, M., Peterson, H., Hansen, J., and Vilo, J. (2007). G: profiler-a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35:W193. doi: 10.1093/nar/gkm226
Ritter, A. M., and Mendell, L. M. (1992). Somal membrane properties of physiologically identified sensory neurons in the rat: effects of nerve growth factor. J. Neurophysiol. 68, 2033–2041. doi: 10.1152/jn.1992.68.6.2033
Saini, A. G., Padmanabh, H., Sahu, J. K., Kurth, I., Voigt, M., and Singhi, P. (2017). Hereditary sensory polyneuropathy, pain insensitivity and global developmental delay due to novel mutation in PRDM12 gene. Indian J. Pediatr. 84, 332–333. doi: 10.1007/s12098-016-2284-y
Sande-Melón, M., Marques, I. J., Galardi-Castilla, M., Langa, X., Pérez-López, M., Botos, M. A., et al. (2019). Adult sox10+ cardiomyocytes contribute to myocardial regeneration in the Zebrafish. Cell Rep. 29, 1041–1054.e5. doi: 10.1016/j.celrep.2019.09.041
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, N., Flaherty, K., Lezgiyeva, K., Wagner, D. E., Klein, A. M., and Ginty, D. D. (2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. doi: 10.1038/s41586-019-1900-1
Sherry, S. T., Ward, M.-H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucl. Acids Res. 29, 308–11. doi: 10.1093/nar/29.1.308
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Tang, W., Martik, M. L., Li, Y., and Bronner, M. E. (2019). Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish. Elife. 8:e47929. doi: 10.7554/eLife.47929.018
Thelie, A., Desiderio, S., Hanotel, J., Quigley, I., Driessche, B., Van Rodari, A., et al. (2015). Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Development 142, 3416–3428. doi: 10.1242/dev.121871
Van Acker, N., Ragé, M., Sluydts, E., Knaapen, M. W. M., De Bie, M., Timmers, M., et al. (2016). Automated PGP9.5 immunofluorescence staining: a valuable tool in the assessment of small fiber neuropathy? BMC Res. Notes. 9:280. doi: 10.1186/s13104-016-2085-4
Woo, J., Kang, H., Lee, E. Y., Park, S., and Cho, Y. E. (2020). Investigation of PRDM7 and PRDM12 expression pattern during mouse embryonic development by using a modified passive clearing technique. Biochem. Biophys. Res. Commun. 524, 346–353. doi: 10.1016/j.bbrc.2019.12.133
Keywords: Prdm12, TRPV1, capsaicin, pain, nociception, CIP, dorsal root ganglia
Citation: Kokotović T, Langeslag M, Lenartowicz EM, Manion J, Fell CW, Alehabib E, Tafakhori A, Darvish H, Bellefroid EJ, Neely GG, Kress M, Penninger JM and Nagy V (2021) PRDM12 Is Transcriptionally Active and Required for Nociceptor Function Throughout Life. Front. Mol. Neurosci. 14:720973. doi: 10.3389/fnmol.2021.720973
Received: 05 June 2021; Accepted: 20 August 2021;
Published: 27 September 2021.
Edited by:
Cyril Goudet, INSERM U1191 Institut de Génomique Fonctionnelle (IGF), FranceReviewed by:
Fabien Marchand, INSERM U1107 Douleur et Biophysique Neurosensorielle (Neuro-Dol), FranceCopyright © 2021 Kokotović, Langeslag, Lenartowicz, Manion, Fell, Alehabib, Tafakhori, Darvish, Bellefroid, Neely, Kress, Penninger and Nagy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vanja Nagy, dmFuamEubmFneUBydWQubGJnLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.