
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 03 September 2021
Sec. Molecular Signalling and Pathways
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.718387
This article is part of the Research Topic Transcription Regulation - Brain Development and Homeostasis - A Finely Tuned and Orchestrated Scenario in Physiology and Pathology View all 12 articles
 Gennaro Ruggiero1,2
Gennaro Ruggiero1,2 Zohar Ben-Moshe Livne3
Zohar Ben-Moshe Livne3 Yair Wexler3
Yair Wexler3 Nathalie Geyer1
Nathalie Geyer1 Daniela Vallone1
Daniela Vallone1 Yoav Gothilf3,4
Yoav Gothilf3,4 Nicholas S. Foulkes1*
Nicholas S. Foulkes1*The zebrafish represents a powerful model for exploring how light regulates the circadian clock due to the direct light sensitivity of its peripheral clocks, a property that is retained even in organ cultures as well as zebrafish-derived cell lines. Light-inducible expression of the per2 clock gene has been predicted to play a vital function in relaying light information to the core circadian clock mechanism in many organisms, including zebrafish. To directly test the contribution of per2 to circadian clock function in zebrafish, we have generated a loss-of-function per2 gene mutation. Our results reveal a tissue-specific role for the per2 gene in maintaining rhythmic expression of circadian clock genes, as well as clock-controlled genes, and an impact on the rhythmic behavior of intact zebrafish larvae. Furthermore, we demonstrate that disruption of the per2 gene impacts on the circadian regulation of the cell cycle in vivo. Based on these results, we hypothesize that in addition to serving as a central element of the light input pathway to the circadian clock, per2 acts as circadian regulator of tissue-specific physiological functions in zebrafish.
The circadian clock is an endogenous and self-sustaining timing mechanism present in most organisms, which evolved to anticipate daily environmental changes and thereby to coordinate physiological and behavioral adaptations (Pittendrigh, 1993). Consistent with its central coordinating role within physiology, disruption of the circadian timing system is associated with many pathological conditions (Toh et al., 2001; Turek et al., 2005; Savvidis and Koutsilieris, 2012). A vital feature of the internal clock is that external environmental signals (zeitgebers, primarily light, but also food and temperature changes) can regularly adjust the phase of the circadian system to ensure synchronization with the environmental day-night cycle.
In vertebrates, at the core of the molecular mechanism of the circadian clock is a series of interlocking transcription-translation feedback loops. The positive limb of these regulatory loops is constituted by the transcription factors CLOCK and BMAL, which heterodimerize, bind to E-box enhancer promoter elements and thereby activate the transcription of downstream clock target genes. These include genes which constitute the negative limb of the clock mechanism, the period (Per) and cryptochrome (Cry) genes. Following translation, the PER and CRY proteins heterodimerize, translocate back to the nucleus, and inhibit transcriptional activation directed by CLOCK/BMAL (Partch et al., 2014). CLOCK and BMAL also regulate the expression of other, clock-controlled genes (CCGs) including the transcription factors Rev-erbα and Rorα which form a stabilizing regulatory loop within the core clock mechanism.
Over the course of vertebrate evolution, the regulatory mechanisms, as well as the anatomical organization which underlies the circadian timing system, have undergone several changes (Menaker et al., 1997). At the anatomical level, in mammals the circadian timing system is characterized by a “master” clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus with multiple independent “peripheral” clocks distributed in most tissues, organs and cells. This SCN clock receives light input indirectly from the retina and is thereby synchronized with the external solar day. It subsequently relays this timing information to the peripheral clocks via a variety of endocrine and systemic cues (Schibler and Sassone-Corsi, 2002; Schibler et al., 2015). In contrast, in non-mammalian vertebrates, a directly light-entrainable circadian oscillator is distributed in multiple tissues including the pineal gland, retina, and various brain nuclei, predicting the widespread expression of photoreceptors and elements of the clock light input pathway (Fukada and Okano, 2002). An extreme independence of central clock regulation can be seen in teleost fish where all peripheral clocks can be directly light-entrained (Whitmore et al., 1998; Sassone-Corsi et al., 2000; Foulkes, 2016). At the genomic level, several clock genes have undergone duplications and probably species-specific and tissue-specific sub-functionalization. The per gene has three homologs in mammals and four homologs in fish, per1a, per1b, per2, and per3.
The per2 gene has been predicted to play an important role in the photic entrainment mechanism of the circadian clock in the vertebrate circadian timing mechanism. Indeed, levels of per1 and per2 mRNA expression are transiently induced in response to light exposure, in the mouse SCN (Albrecht et al., 1997; Bae et al., 2001). Furthermore, in zebrafish the expression of the per2 gene is induced robustly following direct exposure of intact larvae, isolated tissues, cells and even cell lines to light via the effect of a D-box enhancer promoter element located in the per2 gene promoter region (Vatine et al., 2009; Mracek et al., 2012). In addition to light-entrainment, PER2 is linked with the clock mechanism itself, as the S752G PER2 mutation in humans leads to hypo-phosphorylation, PER2 stabilization, and a familial advanced sleep-phase syndrome (FASPS) phenotype (Toh et al., 2001). Via its ability to downregulate transactivation driven by the CLOCK/BMAL complex within the core clock mechanism, the PER2 protein appears to contribute to the circadian regulation of a wide range of cellular functions (Albrecht et al., 2007), including metabolism and cell cycle (Fu et al., 2002; Grimaldi et al., 2010; Gu et al., 2012).
Several studies have also pinpointed a direct role for the PER2 protein, independent of its function within the core circadian clock. Thus, in zebrafish, the PER2 protein has been implicated in the direct transcriptional regulation of the bmal1 gene via the retinoic acid—related orphan receptor response element (RORE) binding sites in zebrafish (Wang et al., 2015). Furthermore, we have shown that light-induced expression of per2 during early embryonic development is a prerequisite for the development of a functional circadian clock system (Ziv and Gothilf, 2006). In addition, in mouse, it has been implicated as a tumor suppressor gene (Fu et al., 2002). Therefore, current evidence points to this clock protein playing a diverse role in the dynamic control of physiological systems, including the cell cycle.
In this report, we have explored the function of the per2 clock gene in zebrafish by generating a new per2 knockout (KO) zebrafish line. Specifically, using TALEN technology, we introduced a truncation mutation into the zebrafish per2 locus, and then characterized the resulting phenotype of the per2 KO zebrafish line. We show that loss of per2 gene function results in an abnormal pattern of rhythmic locomotor activity in per2 KO larvae under different lighting conditions and conclude that per2 plays an essential role in the regulation of circadian phase and amplitude of behavioral rhythms and their entrainment by light. Moreover, we demonstrate a tissue-specific function for the per2 gene in the maintenance of rhythmic expression of core circadian clock genes and CCGs. Finally, we reveal that disruption of per2 gene function impacts on circadian regulation of the cell cycle in vivo. Therefore, these results point to a pleiotropic function for the per2 gene in circadian regulation of tissue specific function.
Wild type (WT) and corresponding per2 KO sibling AB strain zebrafish lines were raised at 28°C under a 14 h:10 h light/dark cycle from the hatching stage. Lights were turned on at 8:00 and turned off at 22:00 and the fish were fed twice daily. To generate embryos, male and female zebrafish were paired in the evening, and spawning occurred the next day within 1 h after lights on. For locomotor activity analysis, embryos were transferred into 48-well plates (one larva per well) during the 4th or 5th day of development and placed into the DanioVision observation chamber (Noldus Information Technology). All zebrafish procedures were approved by the Tel-Aviv University Animal Care Committee (04-18-051) and conducted in accordance with the National Council for Animal Experimentation, Ministry of Health, Israel. At the Karlsruhe Institute of Technology, all husbandry and experimental procedures were performed in accordance with European Legislation for the Protection of Animals used for Scientific Purposes (Directive 2010/63/EU), the German Animal Protection Law [May 18th, 2006 (BGBl. I S. 1206, 1313), last changed March 29th, 2017 (BGBl. I S. 626)]. Research was also approved by the Local Government of Baden-Württemberg, Karlsruhe, Germany (35-9185.81/G-131/16 and 35-9185.82/A-9/18). General license for fish maintenance and breeding: Az.: 35-9185.64.
Genome editing with the transcription activator-like effector nucleases (TALEN) system was used to generate per2 KO fish, registered in the Zebrafish Model Organism Database (ZFIN) as per2tlv02. Specific TALENs designed to target the 2nd exon of per2 (TALE F target sequence 5′-tcagcactactggtgtca-3′, TALE R target sequence 5′-tgaaaatcacaaattacc-3′) were obtained from Addgene (TAL3138 and TAL3139, Addgene plasmids #41312 and #41313). The TALE nuclease expression vectors were linearized with PmeI and transcribed using mMESSAGE mMACHINE T7 kit (Ambion) followed by the Poly-A tailing kit (Ambion) according to the manufacturer’s protocol. Approximately 2 nl of the TALENs mRNA at concentration of 100 ng/μl each were microinjected into one-cell stage embryos (F0). The injected embryos were raised, and their progeny (F1) were fin-clipped and screened by PCR (using the primers: per2-E2-F 5′-gccagtttcgcagaaggcactg-3′, per2-I2-R 5′-agccatcaggtctcaactgtttgtca-3′) followed by T7E1 assay for identifying mutations in per2 coding sequence. A male and female F1 fish carrying the same 8 bp deletion mutation in per2 exon 2 (Figure 1) were identified by sequencing and crossed to produce homozygous KO fish (F2) and their WT siblings. The progeny of the F2 mutated homozygotes and of their WT siblings were used for behavioral analysis, raising a possibility of observing maternal effects of the mutation. However, the lack of any conclusive evidence for maternal inheritance of circadian clock function in zebrafish makes such a maternal effect unlikely (Whitmore et al., 1998).
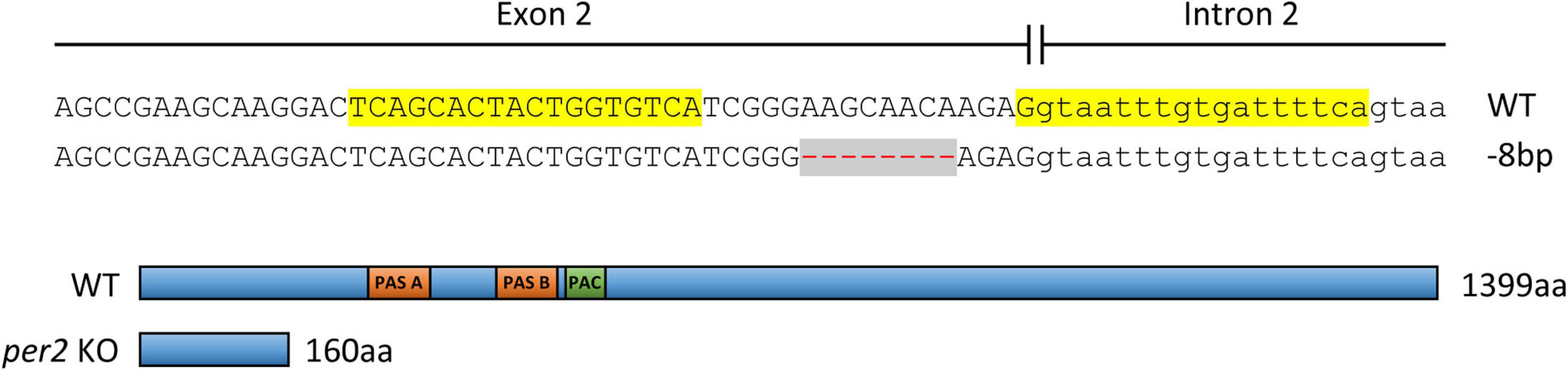
Figure 1. Above: Establishment of a TALEN-mediated per2 KO zebrafish line. A pair of TALENs was used to target the 2nd exon of the zebrafish per2 gene (TALEN left and right target sites are highlighted in yellow). A deletion of 8 bp (red dashes against a gray background) resulted in a frameshift mutation. Below: The consequent introduction of a premature stop codon and a predicted truncated PER2 protein of 160 aa compared with the 1399 aa WT protein. PER-ARNT-SIM domains (PAS, orange) and C-terminal of PAS domain (PAC, green) in the WT protein are indicated.
Homozygous per2 KO embryos and control embryos (progeny of WT siblings) were raised in a light- and temperature-controlled incubator under 12-h:12-h LD cycles or constant darkness at 28°C. On the 4th or 5th day of development, larvae were placed in 48-well plates in the observation chamber of the DanioVision tracking system (Noldus Information Technology) for acclimation under controlled temperature (28°C) and lighting conditions (LED; intensity of “light” and “dim light” were 1.8 W/m2 and 0.013 W/m2, respectively) according to the desired protocol. Starting from the 6th day of development, movement was tracked and analyzed by the EthoVision XT 11 software (Noldus Information Technology). Locomotor activity was measured across three daily cycles by the total distance moved (cm) by each larva per 10 min time-bins. All experiments were repeated independently two to four times, the results shown in Figure 2 are of one representative experiment.
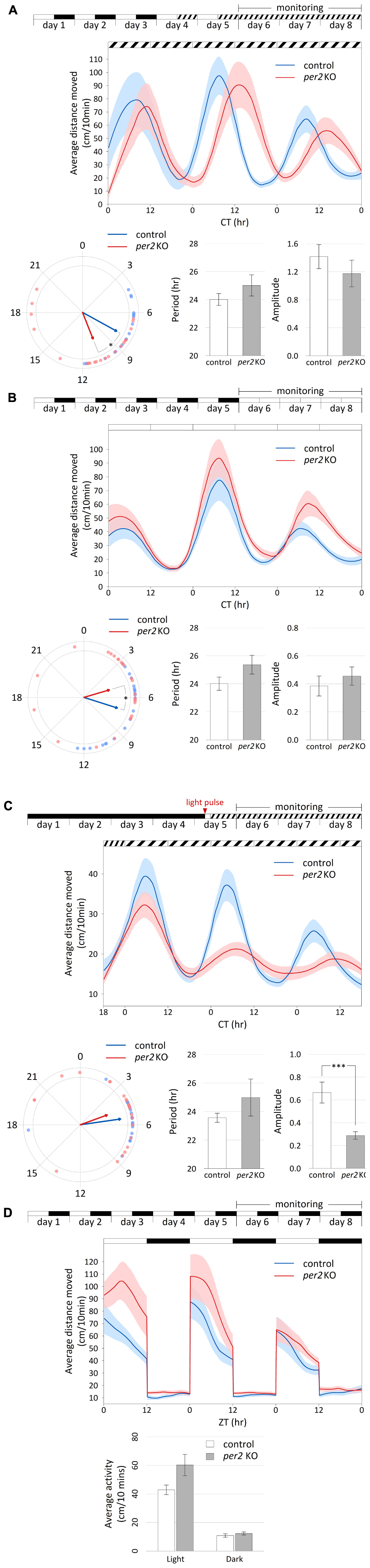
Figure 2. Per2 KO affects the phase of circadian rhythms of locomotor activity and their entrainment by light. Analysis of locomotor activity of 6–8 dpf per2 KO and control larvae under various lighting conditions. (A–D) Top, experimental design of the photic treatment prior to and throughout activity monitoring. White boxes represent light, black boxes represent dark, and diagonally lined boxes represent dim light. Middle, the average distance moved (cm/10 min) is plotted on the y-axis and circadian time (CT) for panels (A–C) or zeitgeber time (ZT) for panel (D) is plotted on the x-axis; error bars indicate SE. (A–C) Bottom from left to right, comparison of average phase, period (±SE) and amplitude (±SE) between genotypes. In the circular plot of circadian phase, arrow direction represents the average phase for each genotype and the length represents the variance (longer arrow stands for low variance and vice versa). (D) Bottom, comparison of average activity (±SE) between genotypes throughout the light and dark segments. (A) Circadian rhythms of locomotor activity under DimDim, after entrainment by 3 LD cycles and 2 light-dim light (LDim) cycles; per2 KO larvae (n = 21) display a phase delay of 2.7 h compared to control larvae (n = 20; p < 0.05 (denoted by *), Watson–Williams test). (B) Circadian rhythms of locomotor activity under LL, after entrainment by 5 LD cycles; per2 KO larvae (n = 24) display a phase advance of 2.3 h compared to control larvae (n = 22; p < 0.05 (denoted by *), Watson–Williams test). (C) Circadian rhythms of locomotor activity under DimDim, after exposure to a 3-h light pulse (indicated by red arrowhead); per2 KO larvae (n = 23) display a decreased amplitude of activity rhythmicity compared to control larvae (n = 23; p < 0.001 (denoted by ***), t-test). (D) Locomotor activity under LD cycles is not affected by per2 KO; no significant difference in average activity was observed between genotypes during both the light and the dark segments (n = 24 per2 KO; n = 23 controls).
For the analysis of circadian activity under constant conditions (Figures 2A–C), normalization of the data was obtained by dividing each activity value by the mean activity value. Short-term trends were removed by a LOESS-smoothed 75th-percentile function (Benjamini et al., 2011) with half window widths of 3.33 h (20 sliding points) for the moving percentiles and 8.33 h (50 sliding points) for the LOESS curve. Peaks (local maxima) and troughs (local minima) in the normalized smoothed curves were used to compute the period, phase and amplitude of the circadian locomotor activity rhythms. Period was estimated as the weighted mean time difference (in hr) between each pair of consecutive peaks. Phase was estimated as the weighted mean direction [mean of circular quantities; (Jammalamadaka and SenGupta, 2001)] of peak time relative to the estimated period. Since higher amplitudes are less sensitive to noise, weight was assigned to each peak in proportion to its amplitude for estimating period and phase. The reported amplitude was defined as half the difference in normalized activity between the peak of the 2nd day of tracking and the preceding trough. Values are reported as mean ± standard error (SE). Statistical differences in period and amplitude between groups were determined by t-test, and statistical differences in phase were determined by Watson–Williams test for the homogeneity of means.
In an experiment with LD cycles (Figure 2D), smoothing was performed separately on each cycle with half window widths of 2.5 h (15 sliding points) for the moving percentiles and 4.17 h (25 sliding points) for the LOESS curve. In order to assess the difference in activity between groups under the light and dark conditions, the average distance moved (cm/10 min) was calculated for each larva separately for the light and for the dark segments. The activity values (log-transformed) of the two genotypes under the light and dark segments were compared by mixed model ANOVA.
Data plotted in Figures 2A–D (middle chart) is the average across larvae ± SE of the non-normalized LOESS-smoothed percentile functions, with each group consisting of 20–24 larvae.
Total RNA of zebrafish tissues was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The concentrations of RNA samples were assessed with a NanoDrop ND-1000 spectrometer (PeqLab). The quality of the RNA was determined after electrophoresis on an agarose gel to visualize the integrity of the ribosomal 28S, 18S, and 5S RNA bands. The first strand cDNA synthesis of total RNA was performed according to the manufacturer’s protocol (Promega). Quantitative PCR was performed using the Step One Plus Real-Time PCR System (Applied Biosystems) and SYBR Green (Promega) master mix according to the manufacturers’ recommendations. Primer sequences are shown in Supplementary Table 1. The relative expression levels for each gene were calculated by the 2–ΔΔCT method and normalized using the relative expression of β-actin.
Protein extracts were prepared by homogenizing sample tissues in 1X Laemmli (6% SDS, 20% glycerol, 125 mM Tris pH 6.8, 0.01% bromophenol blue, 100 mM DTT) containing a 1X cocktail of protease inhibitors (Sigma-Aldrich) buffer. The samples were electrophoresed on an SDS polyacrylamide gel and transferred to an Immun-Blot [polyvinylidene difluoride (PVDF)] membrane (Millipore) by electroblotting. Antibody incubation and washing was performed following the manufacturers’ recommendations and visualization was performed by using the ECL detection system (Bio-Rad). Images were acquired and analyzed by using the Image Lab Software (Bio-Rad).
Significance in the difference in gene expression dynamics between mutants and controls was assessed via a two-way ANOVA, fitted independently for each gene in each tissue. The ANOVA model consisted of three fixed effects: genotype (per2 KO or WT siblings), time, and their interaction genotype × time (see detailed results in Table 1). A significant interaction indicates an alteration in the gene’s expression dynamics in the mutants. A significant genotype effect indicates a total increase or reduction in expression level in the mutants across all time points. A significant time effect indicates non-constant expression over time as would be expected in the rhythmic genes chosen in this work. Post hoc analysis was performed in instances of significant interaction (p < 0.05), comparing the two genotypes at each time point individually. P-values were corrected using Sidak’s method. Analysis was carried out using GraphPad Prism 7.0. All estimates are expressed as means ± SD of biological or technical replicates.
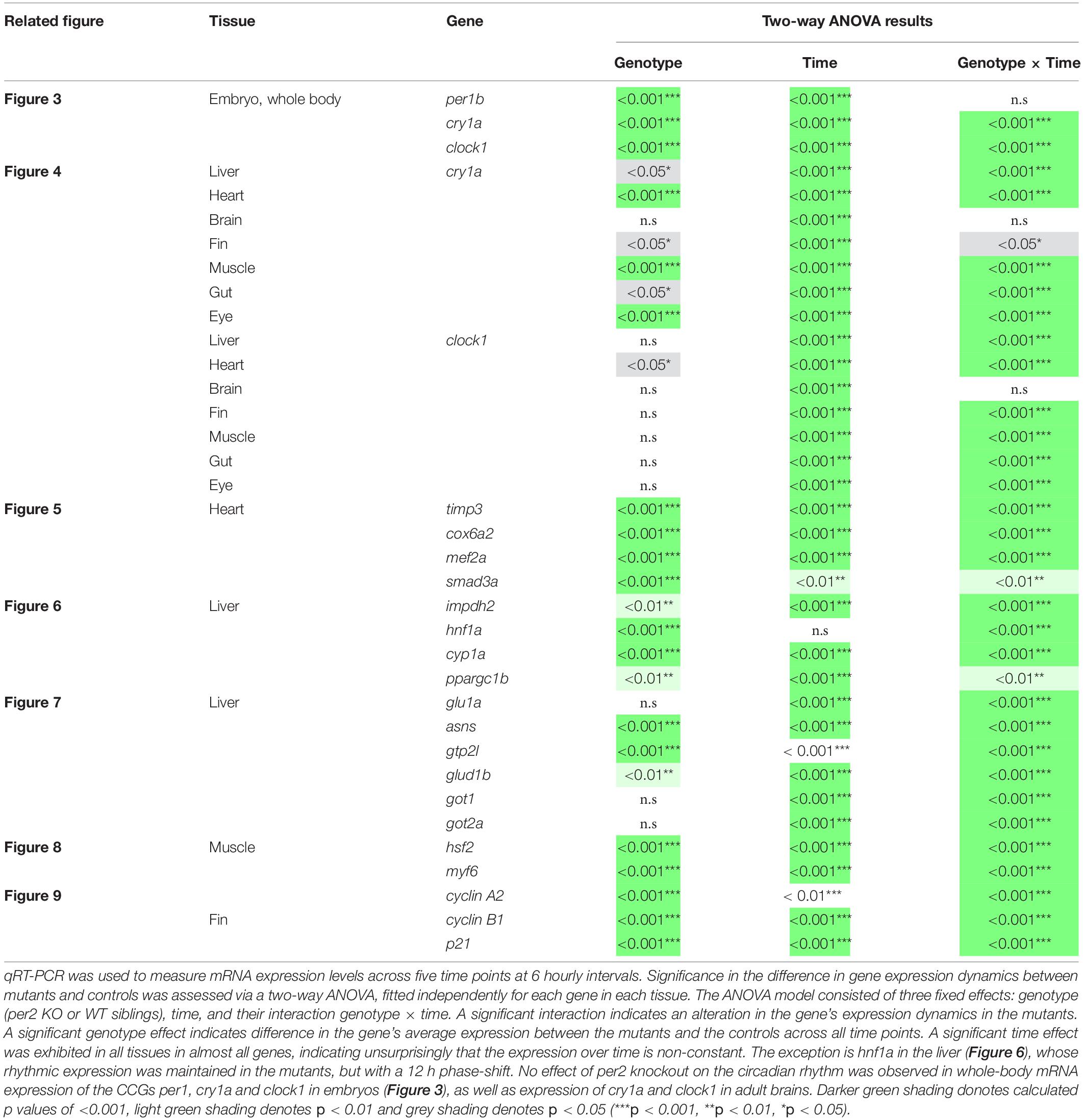
Table 1. Under LD conditions, the per2 knockout alters circadian rhythms of mRNA expression in (i) CCGs in the liver, heart, fins, muscles, gut and eyes (Figure 4), (ii) clock-controlled genes in the heart (Figure 5), (iii) regulators of key physiological hepatic processes in the liver (Figure 6), (iv) genes encoding enzymes involved in biosynthesis of non-essential amino acids in the liver (Figure 7), (v) regulators of skeletal muscle myogenesis and regeneration in the muscles (Figure 8), and in (vi) cell-cycle regulators in the fin (Figure 9).
In order to directly address the function of per2 in zebrafish, we established a TALEN-based per2 KO fish line. An 8 bp deletion was generated in the 2nd exon of per2, resulting in a frameshift which is predicted to encode a truncated protein of 160 amino acids (aa) instead of the 1399 aa WT PER2 protein (Figure 1). Without the crucial PAS protein-protein interaction domains, this small, truncated protein is predicted to lack the normal function that involves direct interaction with other transcriptional regulatory factors. The presence of this mutation was subsequently validated by PCR and DNA sequencing in each subsequent generation of the per2 KO line.
In order to assess the impact of this TALEN-generated mutation on clock-regulated behavioral rhythms and their entrainment by light, we initially analyzed the rhythmic locomotor activity of per2 KO and control larvae under different photic regimes. Following entrainment by LD cycles (Cahill et al., 1998) or by exposure to a single pulse of light (Ben-Moshe et al., 2014), WT larval zebrafish display daily rhythms of locomotor activity under constant conditions with higher activity levels during the subjective daytime, a pattern that is highly reproducible amongst independently raised families.
Locomotor activity was measured in LD-entrained per2 KO and WT control larvae that were placed under constant dim light (DimDim; Figure 2A) or under constant light (LL; Figure 2B). Under both conditions, rhythms of locomotor activity were maintained with no significant difference in period or amplitude (DimDim, periods of 24.0 ± 0.4 and 25.0 ± 0.8 and amplitudes of 1.42 ± 0.17 and 1.17 ± 0.19 for control and per2 KO larvae, respectively; LL, periods of 24.0 ± 0.5 h and 25.4 ± 0.7 h and amplitudes of 0.38 ± 0.07 and 0.46 ± 0.07 for control and per2 KO larvae, respectively). However, substantial phase differences between the genotypes were observed under both conditions. Interestingly, the two lighting conditions induced an opposite effect. While under DimDim, per2 KO larvae exhibited a phase delay of 2.7 h compared to the controls, under LL, per2 KO larvae exhibited a phase advance of 2.3 h (DimDim, phases of 7.9 ± 0.5 and 10.6 ± 0.8 for control and per2 KO larvae, respectively; p < 0.05, Watson–Williams test; LL, phases of 7.2 ± 0.6 and 4.9 ± 0.8 for control and per2 KO larvae, respectively; p < 0.05, Watson–Williams test). Thus, loss of per2 function induces a differential effect on the phase of locomotor activity rhythms which depends on the lighting conditions. The results under DimDim (Figure 2A) agree with a previous report with a different per2 mutant, in which per2 mutant larvae presented an approximately 2-h phase delay and a ∼1.1-h lengthened period under constant darkness (Wang et al., 2015).
When larvae raised in constant darkness were exposed to a single 3-h light pulse on the 5th day post-fertilization (dpf) and then monitored under DimDim (Figure 2C), a procedure which is sufficient to trigger and set the phase of high amplitude rhythms of locomotor activity (Ben-Moshe et al., 2014), per2 KO larvae displayed a significantly lower amplitude of activity (0.66 ± 0.09 and 0.29 ± 0.03 for control and per2 KO larvae, respectively, p < 0.001, t-test). However, the period and phase did not change significantly (periods of 23.6 ± 0.3 h and 25.0 ± 1.3 h and phases of 5.5 ± 0.4 and 4.7 ± 0.8 for control and per2 KO larvae, respectively). These observations reflect the predicted role of PER2 in light-entrainment and are consistent with previous findings obtained by morpholino-mediated per2 knock-down using a similar experimental setup (Ben-Moshe et al., 2014), where circadian locomotor activity rhythms were similarly disrupted by this manipulation.
Under LD cycles (Figure 2D), the locomotor activity patterns of per2 KO larvae were unaltered compared to control larvae. Both groups exhibited significantly higher activity during the light phase compared to the dark phase (p < 0.0001, ANOVA) due to a masking effect, in which activity is mainly determined by the lighting conditions and not by the clock. No significant differences were observed between the activity of the two groups during both the light and the dark segments, an observation that does not correspond to a previously reported experiment with another per2 mutant, where per2 mutant larvae displayed reduced overall locomotor activity under LD conditions compared to control larvae (Wang et al., 2015). This dissimilar outcome from two per2 mutant lines may reflect different experimental conditions or genetic backgrounds. Overall, our analysis supports an essential role for PER2 in the regulation of circadian phase and amplitude of behavioral rhythms and their entrainment by light.
In order to explore the consequences of loss of per2 function at the gene expression level, we examined the pattern of rhythmic core clock gene expression in the per2 mutants under normal LD (12 h light-12 h dark) cycle conditions. We initially compared the dynamic mRNA expression pattern of a subset of circadian clock genes (clock1, cry1a, and per1b) in whole body RNA extracts of WT sibling and per2 mutant zebrafish larvae raised under LD cycle conditions (Figure 3). While our results revealed a small reduction in expression levels in the mutants in the case of all 3 clock genes, the overall expression pattern for these clock genes was comparable in the WT and mutant larvae.
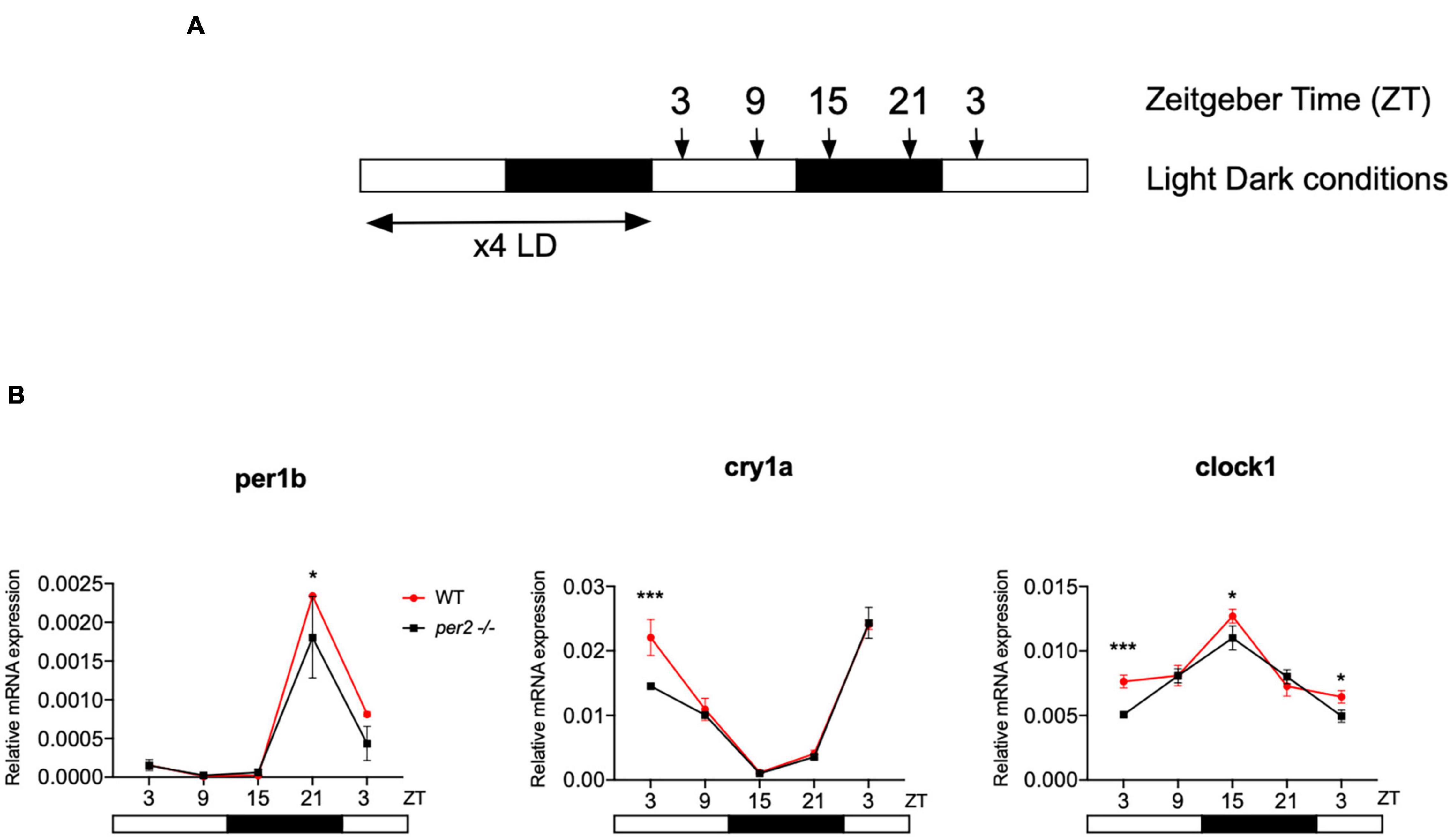
Figure 3. Circadian clock gene expression analysis in per2 KO and WT larvae. (A) Schematic representation of the experimental design. The horizontal bars represent the lighting conditions before and during sampling; white and black boxes indicate light and dark periods, respectively; the arrows show the sampling times. WT and per2 mutant larvae were kept 4 dpf in LD cycle (12 h light-12 h dark) conditions. On the 5th dpf, total RNA was extracted from pools of larvae collected at 6-h intervals during a sampling window of 24 h. (B) qRT-PCR analysis of mRNA expression levels of the circadian clock genes (clock1, cry1a, and per1b) in WT and per2 mutant larvae. Mean mRNA relative expression (n = 2–3) ± SD is plotted on the y-axis, while zeitgeber time (ZT) is plotted on the x-axis. ZT0 corresponds to lights-on, ZT12 to lights-off. A significant, small decrease in expression level was observed in mutants in all three genes (***p < 0.001, ANOVA genotype effect, Table 1). Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, *p < 0.05).
Since the whole-body expression pattern of clock genes did not differ greatly between per2 mutants and WT fish, we next tested for tissue-specific differences in the expression of clock genes. We examined the rhythmic expression of the cry1a and clock1 clock genes in liver, heart, brain, fin, muscle, gut, and eyes of WT and per2 mutant zebrafish adults maintained under LD cycle conditions (Figure 4). Interestingly, a change in the rhythmic profile of clock1 and cry1a expression was detected in the heart, liver, gut and muscle of the per2 mutants relative to the WT fish.
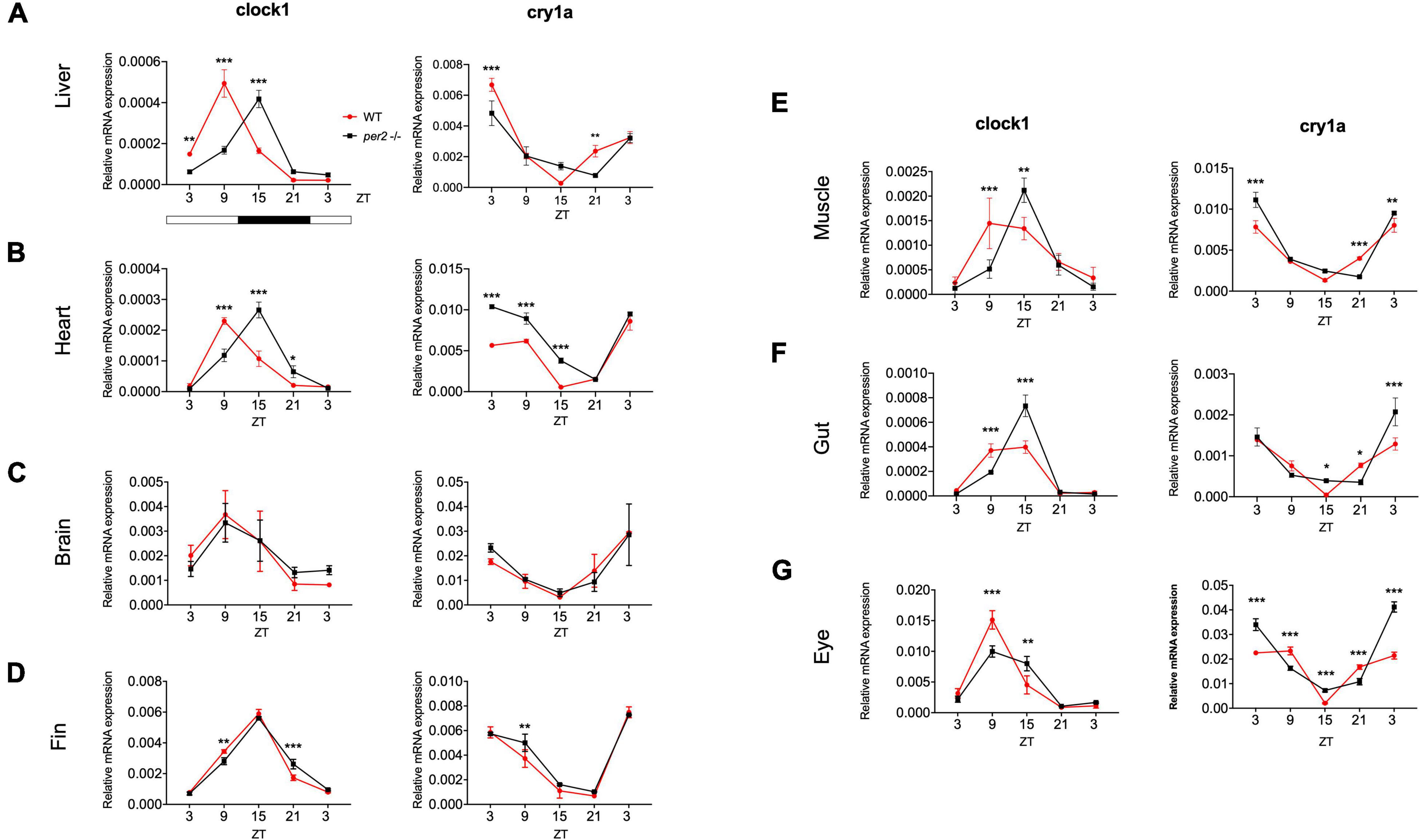
Figure 4. Per2 knockout alters rhythmic mRNA expression of the cry1a and clock1 clock genes in adult zebrafish tissues. qRT-PCR analysis of expression levels of the circadian clock genes clock1 and cry1a in the (A) liver, (B) heart, (C) brain, (D) fin, (E) muscle, (F) gut and (G) eyes of WT and per2 mutant adult fish. Sets (n = 3) of 4 WT and per2 KO fishes (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, tissues were dissected and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for both genes in liver, heart, fin, muscle, gut, and eyes, as determined by a significant ANOVA Genotype × Time interaction effect (p < 0.001, see Table 1). For both genes, no effect was observed in the brain. Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, **p < 0.01, *p < 0.05).
Given the observation of tissue-specific changes in the rhythmic expression of certain clock genes in our zebrafish mutant, we next performed a gene expression analysis of CCGs in per2 KO zebrafish heart, liver, and muscle under normal LD cycle conditions, as a first step toward evaluating the possible impact of the per2 mutation on zebrafish cardiac, hepatic and skeletal muscle physiology.
For the heart, we examined the expression of the following CCGs: timp3, encoding a tissue inhibitor of matrix metalloproteinases that was identified as being circadian clock regulated in the mouse heart (Durgan and Young, 2010), mef2a a clock controlled transcription factor involved in heart development and myofibril assembly (Wang et al., 2005, 2007), cox6a2 a nuclear-encoded cytochrome oxidase subunit involved in mitochondrial electron transport, that shows circadian modulation in the mouse heart (Martino et al., 2004) and smad3a, a TGF-β signaling gene exhibiting a circadian expression pattern throughout the brain of zebrafish larvae (Sato et al., 2012; Sloin et al., 2018; Finger et al., 2021). The circadian expression of each of these CCGs in per2 KO heart tissue was affected, with a significant alteration of the circadian expression pattern, thus suggesting a potential involvement of per2 in the circadian regulation of heart physiology in zebrafish (Figure 5). We then analyzed the expression of the impdh2 [IMP dehydrogenase, a rate limiting enzyme in de novo purine synthesis (Li et al., 2013)], cyp1a [cytochrome P4501A, involved in detoxification (Carmona-Antoñanzas et al., 2017)], ppargc1b [Peroxisome proliferator-activated receptor gamma coactivator 1-beta, a transcriptional coactivator involved in multiple aspects of cellular energy metabolism (Lin et al., 2003)] and hnf1a [hepatocyte nuclear factor 4, alpha, involved in regulating liver-specific gene expression (Courtois et al., 1987)] genes which all encode regulators of key physiological hepatic processes and have been reported to show circadian rhythms of expression in zebrafish larvae (Li et al., 2013). The cyp1a, ppargc1b, and hnf1a genes all exhibited a significantly altered circadian expression pattern in the per2 KO liver (Figure 6). Specifically, cyp1a showed a general reduction in expression levels and disrupted rhythmic expression. Furthermore, ppargc1b and hnf1a exhibited a 6 and 12 h phase delay of the rhythmic pattern, respectively. Interestingly, however, the phase of rhythmic expression of impdh2 resembled that observed in WT liver controls, suggesting that the precise pattern of disrupted rhythmic gene expression upon loss of per2 function differs between CCGs. Given the central role played by the circadian clock in regulating key metabolic pathways including amino acid biosynthesis (Krishnaiah et al., 2017), we next chose to test if the circadian expression of a set of CCGs encoding key or rate-limiting enzymes involved in the biosynthetic pathways for non-essential amino acids (Li et al., 2013), was affected in the per2 KO liver. We specifically tested expression of got1 and got2a (glutamic-oxaloacetic transaminase 1, and 2a) which are linked with the aspartate biosynthesis pathway), asparagine synthetase (asns) mediating asparagine production, glutamate dehydrogenase 1b (glud1b), mediating glutamate synthesis, glutamine synthetase 1a (glu1a), catalyzing glutamine formation and glutamic pyruvate transaminase 2-like (gpt2l), involved in alanine biosynthesis. Significant changes in the pattern of rhythmic expression of all six CCGs were observed in per2 KO liver (Figure 7). Specifically, gtp2l, asns, and got1 all displayed a reduced amplitude of rhythmic expression. Furthermore, glud1b, asns, got1, and got2a exhibited a phase shift of approximately 6 h, as previously observed for clock gene expression.
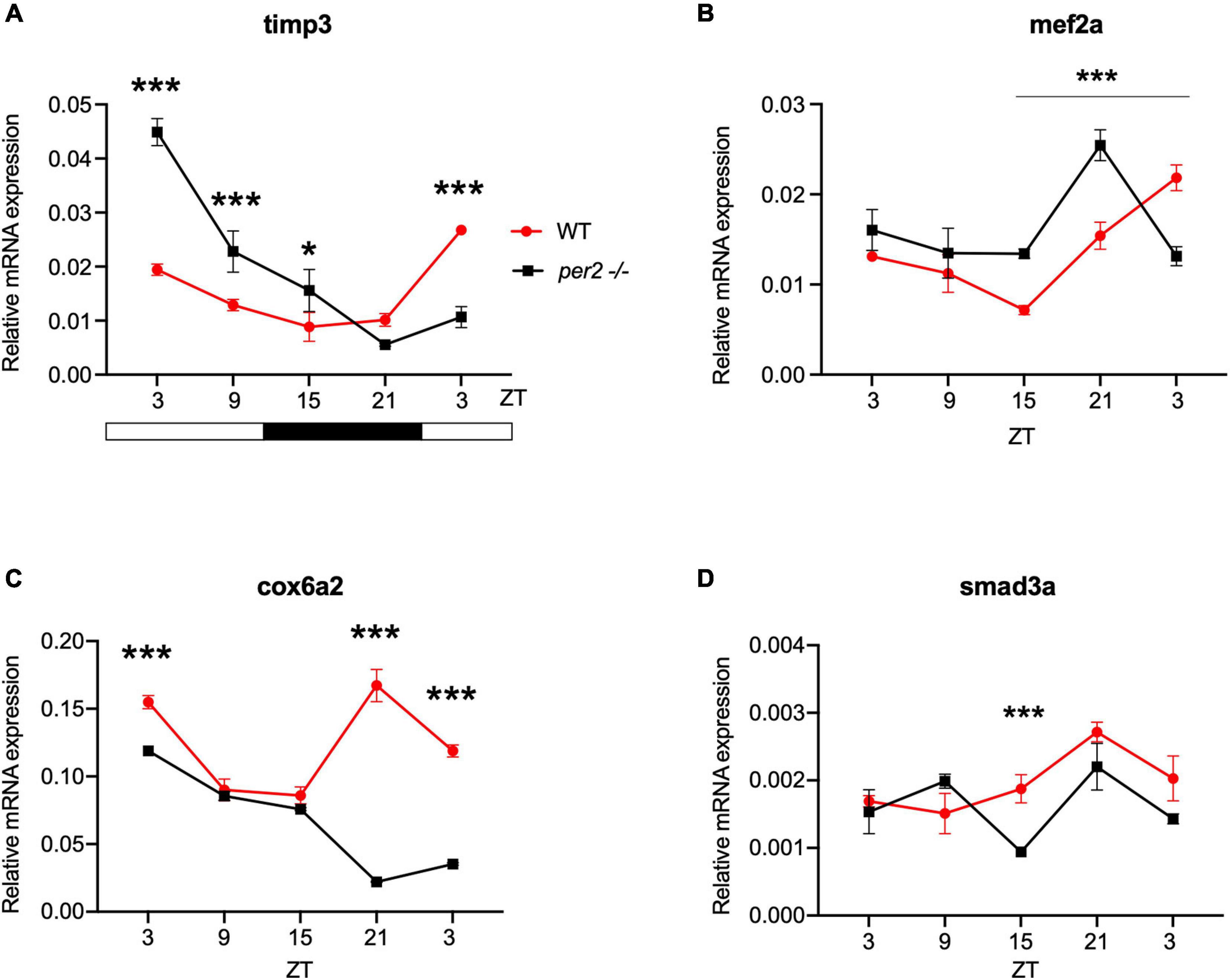
Figure 5. Per2 knockout alters the rhythmic mRNA expression of clock-controlled genes in the adult zebrafish heart. qRT-PCR analysis of expression levels of four putative CCGs (A) timp3, (B) mef2a, (C) cox6a2, and (D) smad3a in WT and per2 KO heart tissues. Sets (n = 3) of 4 WT and per2 KO fish (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, hearts were dissected and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for timp3, cox6a2, mef2a, and smad3a, as determined by a significant ANOVA Genotype × Time interaction effect for all genes (p < 0.01, see Table 1). Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, *p < 0.05).
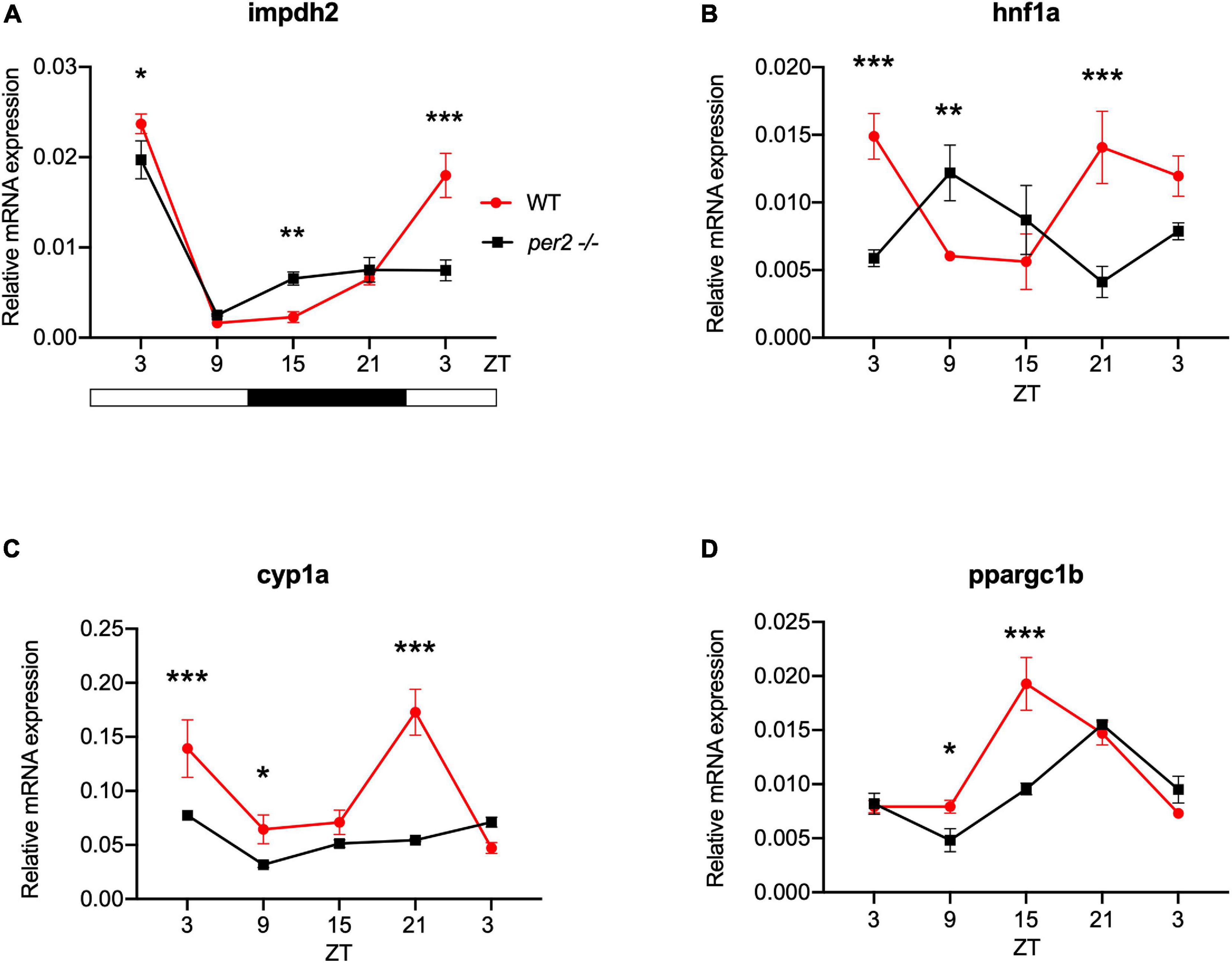
Figure 6. Per2 knockout alters the rhythmic mRNA expression of regulators of key physiological hepatic processes in the adult zebrafish liver. qRT-PCR analysis of expression levels of four CCGs (A) impdh2, (B) hnf1a, (C) cyp1a, and (D) ppargc1b in WT and per2 KO liver. Sets of 4 WT and per2 KO fishes (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, livers were dissected and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression (n = 2–3) ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for impdh2, hnf1a, cyp1a and ppargc1b, as determined by a significant ANOVA Genotype × Time interaction effect for all genes (p < 0.01, see Table 1). Furthermore, hnf1a exhibited a 12 h phase delay, and ppargc1b exhibited a 6 h phase delay. Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, **p < 0.01, *p < 0.05).
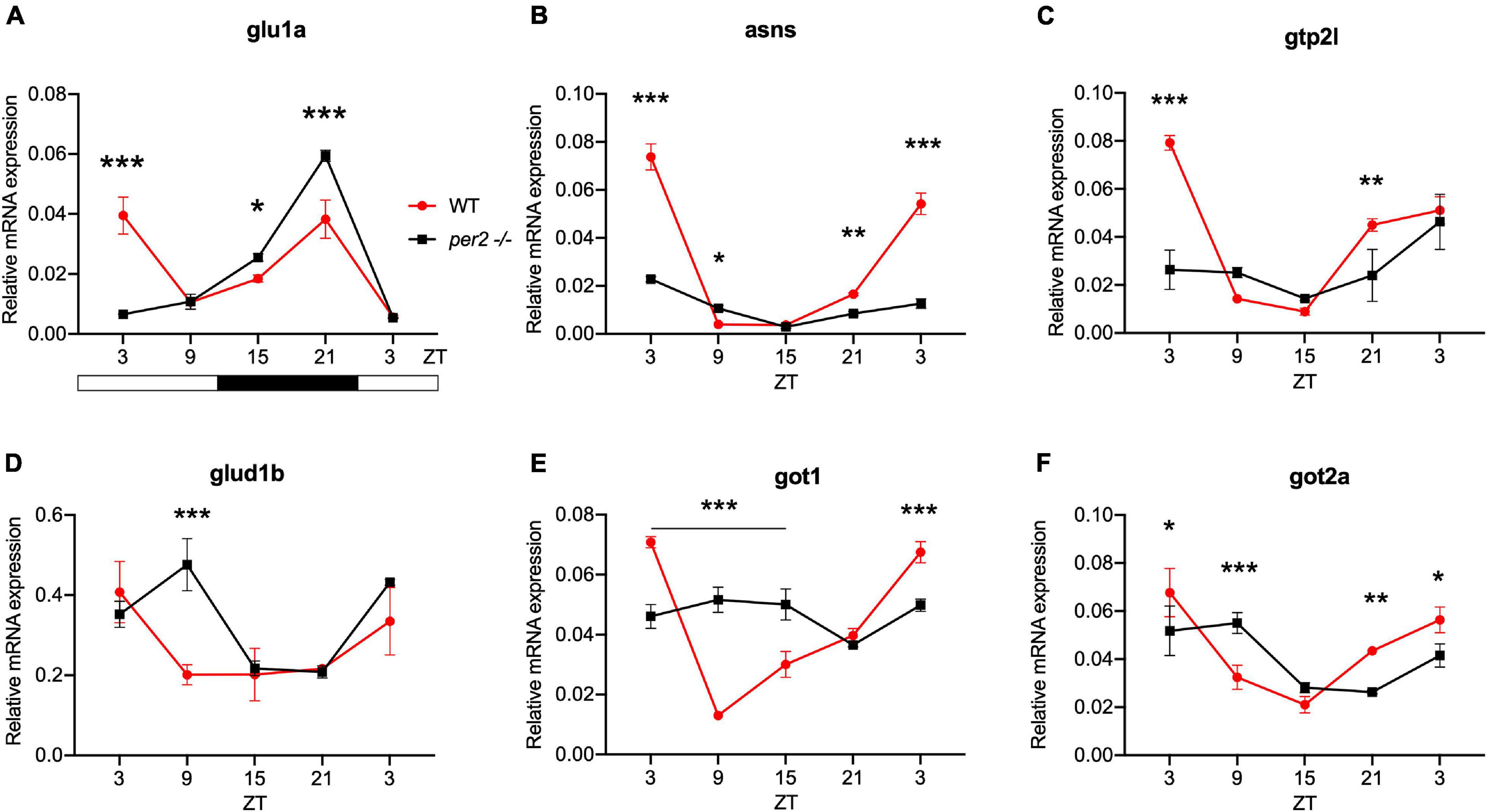
Figure 7. Per2 knockout alters the rhythmic mRNA expression of CCGs encoding key enzymes in non-essential amino acid synthesis in the adult zebrafish liver. qRT-PCR analysis of expression levels of six CCGs (A) glu1a, (B) asns, (C) gpt2l (D) glud1b (E) got1, and (F) got2a in WT and per2 KO liver tissues as described in the legend for Figure 6. A change in rhythm was observed for all genes, as determined by a significant ANOVA Genotype × Time interaction effect for all genes (p < 0.001, see Table 1). Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, **p < 0.01, *p < 0.05).
Expression of CCGs in the per2 KO skeletal muscle was also tested. In particular, based on a previous study that identified putative CCGs in skeletal muscle of the zebrafish (Amaral and Johnston, 2012), we tested the expression of myf6 and hsf2 (Figure 8), that play an important role during skeletal muscle myogenesis and regeneration (McArdle et al., 2006; Hinits et al., 2007; Wang et al., 2008; Chong et al., 2009; Amaral and Johnston, 2012). We revealed that both hsf2 and myf6 exhibited a significant alteration in their rhythmic expression pattern in per2 KO skeletal muscle. Rhythmic Myf6 expression showed a phase delay of 6 h that matches the phase shift observed in the expression pattern of the clock genes cry1a and clock1 while hsf2 exhibited a robust reduction of rhythm amplitude. Thus, taken together, our findings (Figures 4–8) implicate the per2 gene in playing a role in circadian clock regulation in a tissue- and gene-specific manner.
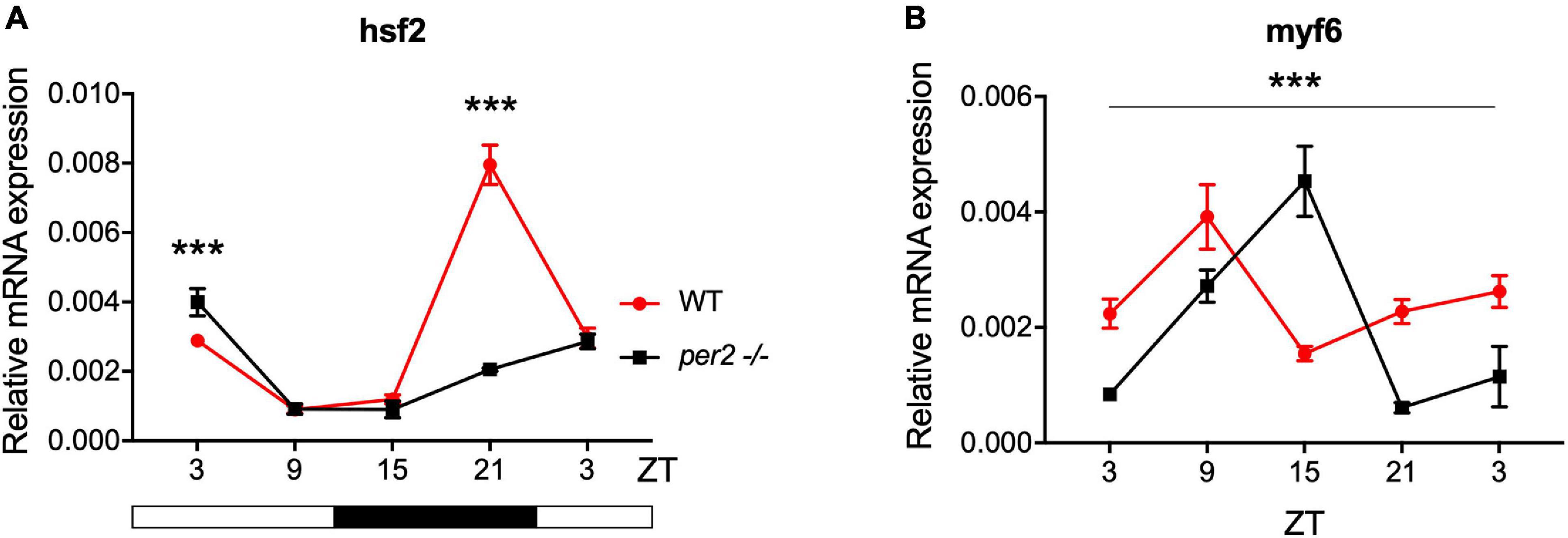
Figure 8. Per2 knockout alters the rhythmic mRNA expression of CCGs in adult zebrafish skeletal muscle. qRT-PCR analysis of expression levels of the CCGs (A) hsf2 and (B) myf6 in WT and per2 KO skeletal muscle. Sets of 4 WT and per2 KO fish (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, muscle tissue was dissected and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression (n = 2–3) ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for both CCGs, as determined by a significant ANOVA Genotype × Time interaction effect for all genes (p < 0.001, see Table 1). myf6 exhibited a 6 h phase delay. Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001).
Given the proposed role of per2 as a tumor suppressor gene we next investigated the contribution of the per2 gene to the circadian regulation of the cell cycle in zebrafish. We tested the gene expression of the clock-controlled cell cycle checkpoint regulators cyclin A2, cyclin B1 and p21 in WT control and per2 KO fins sampled in vivo. p21 is a potent cyclin-dependent kinase inhibitor (CKI) which functions as a regulator of cell cycle progression from G1 to S phase, while cyclin B1 and cyclin A2 serve as regulators of the entry into M and S phase, respectively. In all cases, in the mutant samples we observed significant changes in the rhythmic profile of gene expression with a phase delay compared to WT fin controls (Figure 9).
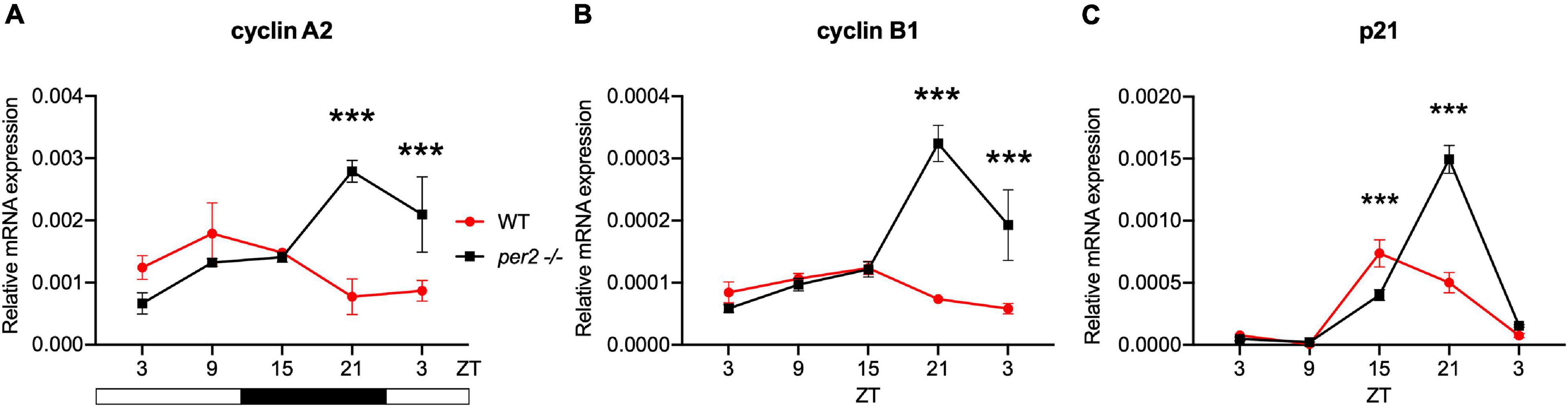
Figure 9. Per2 knockout alters the rhythmic mRNA expression of cell cycle regulator genes in adult zebrafish fin tissues. qRT-PCR analysis of expression levels of some cell cycle CCGs (A) cyclin A2, (B) cyclin B1, and (C) p21 in WT and per2 KO fin tissue. Sets of 4 WT and per2 KO fishes (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, a portion of caudal fin tissue was amputated and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression (n = 2–3) ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for cyclin A2, cyclin B1, and p21, as determined by a significant ANOVA Genotype × Time interaction effect for all genes (p < 0.001, see Table 1). Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001).
In our previous studies, we have shown that M phase progression in zebrafish adult fin tissues, is gated to occur preferentially during the dark phase as a result of circadian clock regulation (Idda et al., 2012). Therefore, we tested whether dynamic changes in the levels of M phase were affected in the per2 KO zebrafish adult fin tissues. We used a western blot assay to quantify levels of the phospho-H3 protein, a marker of chromatin compaction associated with mitosis, in the whole fin protein extracts of the WT and per2 KO zebrafish lines. In agreement with our previous results (Idda et al., 2012), WT fin tissues exhibited a peak of phospho-H3 protein levels around ZT16 (Figure 10). Instead, in the per2 KO fin tissues this peak was significantly reduced, consistent with abnormal circadian clock regulation of cell cycle progression in the per2 KO proliferative fin tissues.
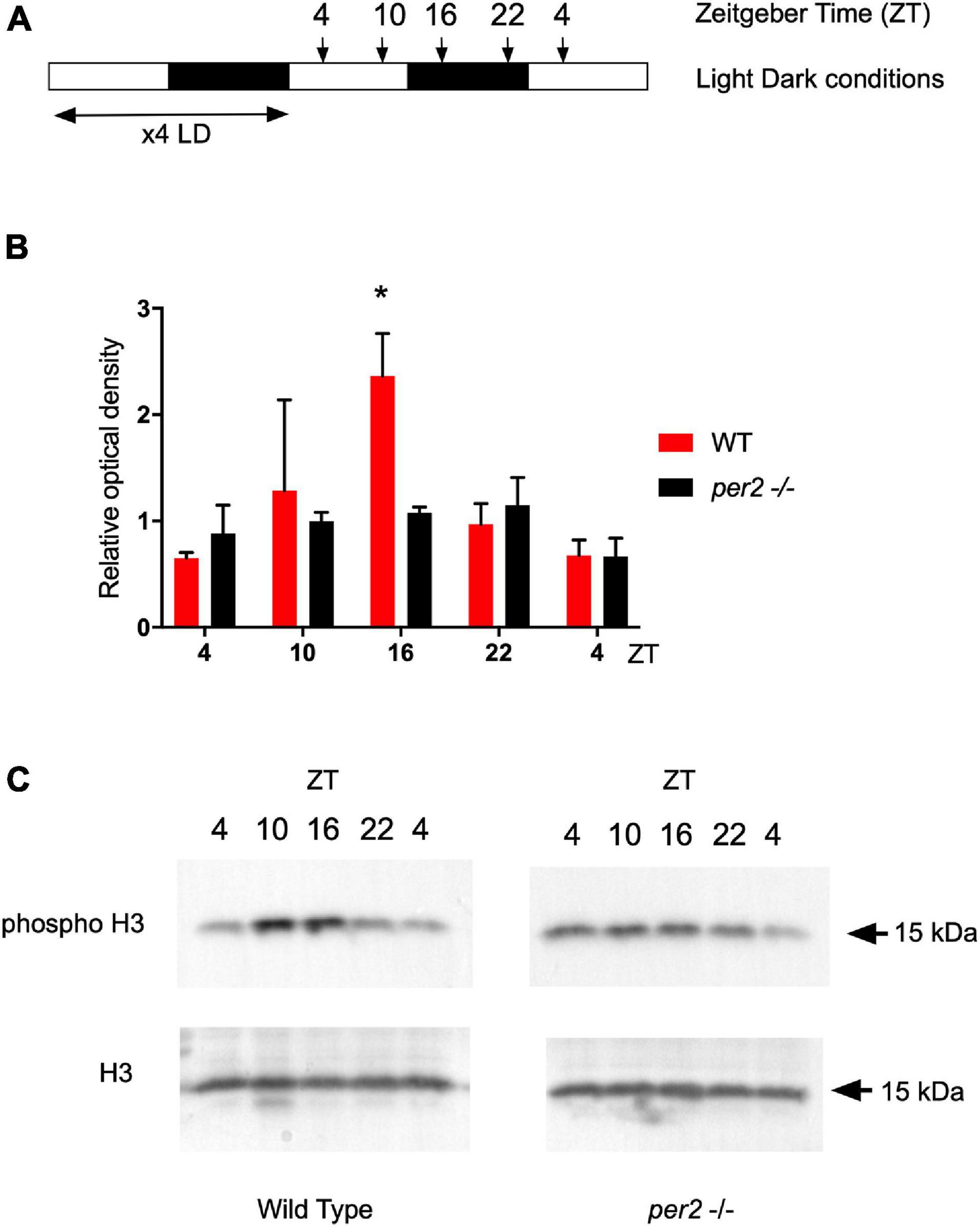
Figure 10. Western blot analysis and quantification of the phospho-H3 protein in fins of the WT and per2 KO zebrafish line. (A) Schematic representation of the experimental design. (B) Quantification of Western blot analysis of Phospho-Histone H3 (p-H3) ser10 expression level normalized to using Histone H3 (H3) as loading control. Relative optical density values for pH3-specific bands were calculated from measurements of ECL images and plotted on the y-axis. (C) Representative images of western blots obtained with pH3 and H3 specific antibodies used for the quantification in panel (B). Asterisks represent levels of significance in comparing the two genotypes at each timepoint individually, corrected by Sidak’s method (*p < 0.05).
The data presented in this study point to a complex role for the per2 gene in the circadian timing system in fish. In particular, we have demonstrated that the per2 gene loss-of-function mutation affects rhythmic behavior of zebrafish larvae, the phase of rhythmic core clock gene expression, as well as the expression of certain CCGs in a tissue-specific manner. Finally, we reveal that loss of per2 gene function is associated with abnormalities in the circadian regulation of the cell cycle in vivo.
The light inducible expression of the per2 gene in the mammalian SCN as well as in the pineal gland, brain and peripheral tissues in fish point to an important role for the per2 gene in the entrainment of the circadian clock by light. This prediction is supported here by the difference between WT and per2 KO larvae in the amplitude and robustness of their locomotor activity rhythms in response to a 3-h light pulse. Nevertheless, the fact that the clock of per2 KO fish is still somewhat entrained by light indicates the existence of mechanisms which compensate for the loss of per2 gene function. It is tempting to speculate that the clock gene cry1a may be an element of such a mechanism. Cry1a is another robustly light-induced clock gene which is able to interact with other core clock proteins, such as CLOCK and BMAL (Tamai et al., 2007). In addition, we have previously shown that 1-h exposure to light is sufficient to induce the expression of several core clock genes or accessory clock genes (Ben-Moshe et al., 2014) suggesting the existence of additional compensatory factors.
In addition to light-entrainment, per2 loss-of-function also strongly affects the phase of the locomotor activity rhythms, suggesting a role for PER2 in the clock mechanism itself. Interestingly, the effect of per2 KO on the circadian phase is context-dependent – under constant dim-light per2 KO led to a phase delay of 2.7 h while under constant light per2 KO led to a phase advance of 2.3 h. Since zebrafish per2 is a light-responsive gene, the light-dependence of per2 KO effects may be predicted. It is tempting to speculate that the different phase shifts observed under different light intensities in the mutant might reflect accompanying differences in light-induced signal transduction in the larvae which subsequently differentially target PER2 function. Furthermore, the phase of the locomotor activity rhythms in the mutants shows a higher variability relative to WT controls pointing to a disruption of the entire clock system by the per2 mutation. Thus, using rhythmic locomotor activity as a clock output we have demonstrated that per2 is involved in both the core clock mechanism and its entrainment by light.
Period proteins together with the cryptochromes are classically regarded as negative elements in the transcription translation feedback loop mechanism at the core of the circadian clock (Partch et al., 2014). However, the lack of a major effect on cycling clock gene expression in per2 KO larvae would tend to argue against such a global role for PER2 in zebrafish. We did observe significant tissue-specific differences in the dynamic expression pattern of core clock genes in our per2 KO fish. For example, rhythmic expression of clock1 mRNA was affected in the liver, heart, muscle and gut of the per2 KO, but not in other tissues. PER2 shares amino acid sequence motifs with both coactivators and corepressors of hormone receptors. For example, the mouse PER2 protein is characterized by two LXXLL motifs in both of its predicted protein–protein interaction domains (Albrecht et al., 2007). This motif is present in different coactivators which interact with nuclear receptors such as the steroid hormone receptor coactivator-1 (SRC-1) (Oñate et al., 1995). Moreover, it has been demonstrated that the PER2 protein upregulates bmal1b gene expression by directly binding to the Rorα nuclear receptor in zebrafish (Wang et al., 2015). Interestingly, a ROR/REV-ERB response element (RORE) has also been identified in the zebrafish clock1 promoter, an observation that would potentially account for the dysregulation of clock1 gene expression in certain tissues of per2 KO adults. Thus, light-induced per2 expression might serve to adjust the phase and amplitude of rhythmic expression of the clock gene. An unresolved issue remains how the observed changes in the profile and timing of clock1 and cry1a mRNA expression in certain tissues does not lead to corresponding alterations in the rhythmic expression of other core clock component genes, such as per1b as well as other CCGs. It is tempting to speculate that the effects of PER2 are manifest in a gene-specific fashion and may reflect the different constellations of clock-regulated enhancer elements present in the promoters of various CCGs and core clock genes.
The contribution of per2 to shaping the profile of tissue specific rhythmic core clock gene expression raises the question of the extent to which the per2 gene may also play a role in regulating tissue-specific physiology. In order to address this question, we also analyzed CCG expression in the liver, heart and muscle of the per2 KO line. In particular, we focused attention on the expression of genes that are involved in the regulation of molecular mechanisms which underlie important physiological processes ranging from metabolism, development and maintenance of homeostasis to basic cellular processes, including cell growth and division (proliferation), cell movement (migration), controlled cell death (apoptosis) and cell differentiation. In the per2 KO heart we observed a significant impact on the circadian expression of the timp3, mef2a, cox6a2, and smad3a genes (Wang et al., 2005, 2007; Sloin et al., 2018). Our results therefore implicate per2 in the circadian clock-mediated regulation of various cardiac functions. This is of potential medical importance because there is a well-documented time of day-dependent increase in the sensitivity to myocardial infarction (Peckova et al., 1998) and disruption of circadian rhythms is a major contributor to heart pathophysiology (Crnko et al., 2019).
We also investigated the involvement of the per2 gene in the circadian clock regulation of liver-specific CCGs, such as impdh2, cyp1a, ppargc1b, and hnf1a as well as genes encoding rate-limiting enzymes involved in the biosynthetic pathways for non-essential amino acids, namely got1, got2a, asns, glud1b, glu1a, and gpt2l. The results reveal an impact of the per2 mutation on the rhythmic expression of many of these key metabolic regulatory-specific genes. Interestingly, the per2 gene has already been associated with the regulation of liver-specific metabolic pathways in mammals. In particular, REV-ERBα and PPARα interact with the PER2 protein in the liver to regulate the transcription of their target genes (Schmutz et al., 2010). Moreover, using per2 KO mice, it has been shown that PER2 directly represses the nuclear receptor PPARγ, critical for adipogenesis, and hepatic insulin sensitivity (Grimaldi et al., 2010). Therefore, these previous findings together with our own CCG expression analysis support the notion that the per2 gene plays an important role in liver physiology. We also reveal disruption of rhythmic expression of the CCGs myf6 and hsf2 in the per2 KO skeletal muscle. Thus, PER2 may play an important role in the temporal coordination of the mechanisms which direct the repair of muscle damage generated during daytime-elevated locomotor activity.
The involvement of per2 gene function in the circadian regulation of the cell cycle has been widely demonstrated in mammals (Fu et al., 2002; Gu et al., 2012; Yang et al., 2012; Tan et al., 2015). Consistently, we observed a robust effect of the loss of per2 gene function on the phase of rhythmic expression pattern of important cell cycle regulators, p21, cyclin A2, and cyclin B1 in adult fin tissue. It is important to note that in the same tissue, rhythmic expression of clock genes is apparently normal indicating that the per2-regulated expression of clock-controlled cell cycle regulators in peripheral tissues may be regulated by a distinct mechanism from the transcriptional control circuits within the core circadian clock mechanism itself. Namely, the cell cycle control represents a specific, clock output function for per2. In the case of the p21 gene in zebrafish, transcriptional regulation by the core clock mechanism via E-box enhancers has been shown to direct rhythmic expression (Laranjeiro et al., 2013). Furthermore, at a mechanistic level, it has already been demonstrated that the PER2 protein modulates p53 stability and transcriptional activity in normal human cells, thus affecting the gene expression of cell cycle regulators, including p21, in response to DNA damage in mammals (Gotoh et al., 2014). Therefore, the phase-setting effects on p21 rhythmic expression may rely on the loss of protein-protein interaction between PER2 and p53 in the per2 KO zebrafish line. Previously, it has been shown that M phase progression in zebrafish adult fin tissue, shows a light-entrained, circadian clock regulation (Idda et al., 2012). Our quantification of the levels of mitosis throughout the LD cycle, revealed a dampened M-phase rhythm in the per2 KO fin tissue. The dysregulation of rhythmic M phase progression, together with the abnormal gene expression profile of p21, cyclin A2, and cyclin B1, indicates that per2 may play a role in timing of the G1/S or G2/M cell cycle checkpoints. Given the impact of per2 gene function on the regulation of the expression levels of these important cell cycle regulators, it could be anticipated that PER2 may play a key role during the early stages of zebrafish embryonic development. However, the normal early embryonic development observed in per2 KO mutants would tend to argue against this. Instead, this observation would tend to support a hypothesis that the contribution of PER2 to cell cycle regulation is cell type-or developmental stage-specific.
In conclusion, our results suggest that the per2 gene plays a crucial role in the circadian regulation of multiple tissue-specific cellular and physiological processes in zebrafish.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the National Council for Animal Experimentation, Ministry of Health, Israel Local Government of Baden-Württemberg, Karlsruhe, Germany.
DV, NF, and YG designed the experiments. GR, ZB-ML, NG, and YW performed and interpreted the experiments. GR, ZB-ML, DV, YW, YG, and NF prepared the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the German-Israeli Foundation for Scientific Research and Development (GIF grant number I-1320-203.13/2015 to NF and YG) as well as the Helmholtz funding program, BIFTM (to NF, DV, and NG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank B. Rapp and O. Kassel for support and discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.718387/full#supplementary-material
Albrecht, U., Bordon, A., Schmutz, I., and Ripperger, J. (2007). The multiple facets of Per2. Cold Spring Harb. Sym. 72, 95–104. doi: 10.1101/sqb.2007.72.001
Albrecht, U., Sun, Z. S., Eichele, G., and Lee, C. C. (1997). A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055–1064. doi: 10.1016/s0092-8674(00)80495-x
Amaral, I. P. G., and Johnston, I. A. (2012). Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302, R193–R206. doi: 10.1152/ajpregu.00367.2011
Bae, K., Jin, X., Maywood, E. S., Hastings, M. H., Reppert, S. M., and Weaver, D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN Circadian Clock. Neuron 30, 525–536. doi: 10.1016/s0896-6273(01)00302-6
Benjamini, Y., Fonio, E., Galili, T., Havkin, G. Z., and Golani, I. (2011). Quantifying the buildup in extent and complexity of free exploration in mice. Proc. Natl. Acad. Sci. 108, 15580–15587. doi: 10.1073/pnas.1014837108
Ben-Moshe, Z., Alon, S., Mracek, P., Faigenbloom, L., Tovin, A., Vatine, G. D., et al. (2014). The light-induced transcriptome of the zebrafish pineal gland reveals complex regulation of the circadian clockwork by light. Nucleic Acids Res. 42, 3750–3767. doi: 10.1093/nar/gkt1359
Cahill, G. M., Hurd, M. W., and Batchelor, M. M. (1998). Circadian rhythmicity in the locomotor activity of larval zebrafish. Neuroreport 9, 3445–3449. doi: 10.1097/00001756-199810260-00020
Carmona-Antoñanzas, G., Santi, M., Migaud, H., and Vera, L. M. (2017). Light- and clock-control of genes involved in detoxification. Chronobiol. Int. 34, 1–16. doi: 10.1080/07420528.2017.1336172
Chong, S. W., Korzh, V., and Jiang, Y. J. (2009). Myogenesis and molecules—insights from zebrafish Danio rerio. J. Fish Biol. 74, 1693–1755. doi: 10.1111/j.1095-8649.2009.02174.x
Courtois, G., Morgan, J., Campbell, L., Fourel, G., and Crabtree, G. (1987). Interaction of a liver-specific nuclear factor with the fibrinogen and alpha 1-antitrypsin promoters. Science 238, 688–692. doi: 10.1126/science.3499668
Crnko, S., Pré, B. C. D., Sluijter, J. P. G., and Laake, L. W. V. (2019). Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat. Rev. Cardiol. 16, 437–447. doi: 10.1038/s41569-019-0167-4
Durgan, D. J., and Young, M. E. (2010). The cardiomyocyte circadian clock. Circ. Res. 106, 647–658. doi: 10.1161/circresaha.109.209957
Finger, A.-M., Jäschke, S., del Olmo, M., Hurwitz, R., Granada, A. E., Herzel, H., et al. (2021). Intercellular coupling between peripheral circadian oscillators by TGF-β signaling. Sci. Adv. 7:eabg5174. doi: 10.1126/sciadv.abg5174
Fu, L., Pelicano, H., Liu, J., Huang, P., and Lee, C. C. (2002). The circadian gene period2 Plays an important role in tumor suppression and DNA Damage Response In Vivo. Cell 111, 41–50. doi: 10.1016/s0092-8674(02)00961-3
Fukada, Y., and Okano, T. (2002). Circadian clock system in the pineal gland. Mol. Neurobiol. 25, 19–30. doi: 10.1385/mn:25:1:019
Gotoh, T., Vila-Caballer, M., Santos, C. S., Liu, J., Yang, J., and Finkielstein, C. V. (2014). The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol. Biol. Cell. 25, 3081–3093. doi: 10.1091/mbc.e14-05-0993
Grimaldi, B., Bellet, M. M., Katada, S., Astarita, G., Hirayama, J., Amin, R. H., et al. (2010). PER2 controls lipid metabolism by direct regulation of PPARγ. Cell. Metab. 12, 509–520. doi: 10.1016/j.cmet.2010.10.005
Gu, X., Xing, L., Shi, G., Liu, Z., Wang, X., Qu, Z., et al. (2012). The circadian mutation PER2S662G is linked to cell cycle progression and tumorigenesis. Cell. Death Differ. 19, 397–405. doi: 10.1038/cdd.2011.103
Hinits, Y., Osborn, D. P. S., Carvajal, J. J., Rigby, P. W. J., and Hughes, S. M. (2007). Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene. Expr. Patterns 7, 738–745. doi: 10.1016/j.modgep.2007.06.003
Idda, M. L., Kage, E., Lopez-Olmeda, J. F., Mracek, P., Foulkes, N. S., and Vallone, D. (2012). Circadian timing of injury-induced cell proliferation in Zebrafish. PLoS One 7:e34203. doi: 10.1371/journal.pone.0034203
Jammalamadaka, S. R., and SenGupta, A. (2001). Topics in Circular Statistics. Singapore: World Scientific Publishing Co Pte Ltd.
Krishnaiah, S. Y., Wu, G., Altman, B. J., Growe, J., Rhoades, S. D., Coldren, F., et al. (2017). Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell. Metab. 25, 961–974. doi: 10.1016/j.cmet.2017.03.019
Laranjeiro, R., Laranjeiro, R., Tamai, T. K., Peyric, E., Peyric, E., Krusche, P., et al. (2013). Cyclin-dependent kinase inhibitor p20 controls circadian cell-cycle timing. Proc. Natl. Acad. Sci. U.S.A. 110, 6835–6840. doi: 10.1073/pnas.1217912110
Li, Y., Li, G., Wang, H., Du, J., and Yan, J. (2013). Analysis of a gene regulatory cascade mediating circadian rhythm in zebrafish. Plos Comput .Biol. 9:e1002940. doi: 10.1371/journal.pcbi.1002940
Lin, J., Tarr, P. T., Yang, R., Rhee, J., Puigserver, P., Newgard, C. B., et al. (2003). PGC-1β in the regulation of hepatic glucose and energy metabolism∗. J. Biol. Chem. 278, 30843–30848. doi: 10.1074/jbc.m303643200
Martino, T., Arab, S., Straume, M., Belsham, D. D., Tata, N., Cai, F., et al. (2004). Day/night rhythms in gene expression of the normal murine heart. J. Mol. Med. 82, 256–264. doi: 10.1007/s00109-003-0520-1
McArdle, A., Broome, C. S., Kayani, A. C., Tully, M. D., Close, G. L., Vasilaki, A., et al. (2006). HSF expression in skeletal muscle during myogenesis: Implications for failed regeneration in old mice. Exp. Gerontol. 41, 497–500. doi: 10.1016/j.exger.2006.02.002
Menaker, M., Moreira, L. F., and Tosini, G. (1997). Evolution of circadian organization in vertebrates. Braz. J. Med. Biol. Res. 30, 305–313. doi: 10.1590/s0100-879x1997000300003
Mracek, P., Santoriello, C., Idda, M. L., Pagano, C., Ben-Moshe, Z., Gothilf, Y., et al. (2012). Regulation of per and cry genes reveals a central role for the D-Box enhancer in light-dependent gene expression. PLoS One 7:e51278. doi: 10.1371/journal.pone.0051278
Oñate, S. A., Tsai, S. Y., Tsai, M.-J., and O’Malley, B. W. (1995). Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270, 1354–1357. doi: 10.1126/science.270.5240.1354
Partch, C. L., Green, C. B., and Takahashi, J. S. (2014). Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24, 90–99. doi: 10.1016/j.tcb.2013.07.002
Peckova, M., Fahrenbruch, C. E., Cobb, L. A., and Hallstrom, A. P. (1998). Circadian variations in the occurrence of cardiac arrests. Circulation 98, 31–39. doi: 10.1161/01.cir.98.1.31
Pittendrigh, C. S. (1993). Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54. doi: 10.1146/annurev.ph.55.030193.000313
Sassone-Corsi, P., Foulkes, N. S., and Whitmore, D. (2000). Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404, 87–91. doi: 10.1038/35003589
Sato, F., Sato, H., Jin, D., Bhawal, U. K., Wu, Y., Noshiro, M., et al. (2012). Smad3 and Snail show circadian expression in human gingival fibroblasts, human mesenchymal stem cell, and in mouse liver. Biochem. Bioph. Res. Co. 419, 441–446. doi: 10.1016/j.bbrc.2012.02.076
Savvidis, C., and Koutsilieris, M. (2012). Circadian rhythm disruption in cancer biology. Mol. Med. 18, 1249–1260. doi: 10.2119/molmed.2012.00077
Schibler, U., Gotic, I., Saini, C., Gos, P., Curie, T., Emmenegger, Y., et al. (2015). Clock-Talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 80, 223–232. doi: 10.1101/sqb.2015.80.027490
Schibler, U., and Sassone-Corsi, P. (2002). A web of circadian pacemakers. Cell 111, 919–922. doi: 10.1016/s0092-8674(02)01225-4
Schmutz, I., Ripperger, J. A., Baeriswyl-Aebischer, S., and Albrecht, U. (2010). The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Gene Dev. 24, 345–357. doi: 10.1101/gad.564110
Sloin, H. E., Ruggiero, G., Rubinstein, A., Storz, S. S., Foulkes, N. S., and Gothilf, Y. (2018). Interactions between the circadian clock and TGF-β signaling pathway in zebrafish. PloS One 13:e0199777. doi: 10.1371/journal.pone.0199777
Tamai, T. K., Young, L. C., and Whitmore, D. (2007). Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc. Natl. Acad. Sci. 104, 14712–14717. doi: 10.1073/pnas.0704588104
Tan, X.-M., Ye, H., Yang, K., Chen, D., Wang, Q.-Q., Tang, H., et al. (2015). Circadian variations of clock gene Per2 and cell cycle genes in different stages of carcinogenesis in golden hamster buccal mucosa. Sci. Rep. UK 5:9997. doi: 10.1038/srep09997
Toh, K. L., Jones, C. R., He, Y., Eide, E. J., Hinz, W. A., Virshup, D. M., et al. (2001). An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043. doi: 10.1126/science.1057499
Turek, F. W., Joshu, C., Kohsaka, A., Lin, E., Ivanova, G., McDearmon, E., et al. (2005). Obesity and metabolic syndrome in circadian clock mutant mice. Science 308, 1043–1045. doi: 10.1126/science.1108750
Vatine, G., Vallone, D., Appelbaum, L., Mracek, P., Ben-Moshe, Z., Lahiri, K., et al. (2009). Light directs zebrafish period2 expression via Conserved D and E Boxes. PLoS Biol. 7:e1000223. doi: 10.1371/journal.pbio.1000223
Wang, M., Zhong, Z., Zhong, Y., Zhang, W., and Wang, H. (2015). The zebrafish period2 protein positively regulates the circadian clock through mediation of retinoic acid receptor (RAR)-related orphan receptor α (Rorα). J. Biol. Chem. 290, 4367–4382. doi: 10.1074/jbc.m114.605022
Wang, Y., Li, C., Lee, G., Tsay, H., Tsai, H., and Chen, Y. (2008). Inactivation of zebrafish mrf4 leads to myofibril misalignment and motor axon growth disorganization. Dev. Dynam. 237, 1043–1050. doi: 10.1002/dvdy.21478
Wang, Y.-X., Qian, L.-X., Liu, D., Yao, L.-L., Jiang, Q., Yu, Z., et al. (2007). Bone morphogenetic protein-2 acts upstream of myocyte-specific enhancer factor 2a to control embryonic cardiac contractility. Cardiovasc. Res. 74, 290–303. doi: 10.1016/j.cardiores.2007.02.007
Wang, Y.-X., Qian, L.-X., Yu, Z., Jiang, Q., Dong, Y.-X., Liu, X.-F., et al. (2005). Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 579, 4843–4850. doi: 10.1016/j.febslet.2005.07.068
Whitmore, D., Foulkes, N. S., Strähle, U., and Sassone-Corsi, P. (1998). Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat. Neurosci. 1, 701–707. doi: 10.1038/3703
Yang, X., He, X., Yang, Z., and Jabbari, E. (2012). Mammalian PER2 regulates AKT activation and DNA damage response. Biochem. Cell. Biol. 90, 675–682. doi: 10.1139/o2012-025
Keywords: circadian clock, zebrafish, period, cell cycle, behavior, metabolism
Citation: Ruggiero G, Ben-Moshe Livne Z, Wexler Y, Geyer N, Vallone D, Gothilf Y and Foulkes NS (2021) Period 2: A Regulator of Multiple Tissue-Specific Circadian Functions. Front. Mol. Neurosci. 14:718387. doi: 10.3389/fnmol.2021.718387
Received: 31 May 2021; Accepted: 03 August 2021;
Published: 03 September 2021.
Edited by:
Estela Maris Muñoz, CONICET Dr. Mario H. Burgos Institute of Histology and Embryology (IHEM), ArgentinaReviewed by:
Elise Cau, CNRS Délégation Occitanie, FranceCopyright © 2021 Ruggiero, Ben-Moshe Livne, Wexler, Geyer, Vallone, Gothilf and Foulkes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas S. Foulkes, bmljaG9sYXMuZm91bGtlc0BraXQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.