
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 05 May 2021
Sec. Molecular Signalling and Pathways
Volume 14 - 2021 | https://doi.org/10.3389/fnmol.2021.638176
This article is part of the Research Topic The Role and Mechanism of Metabolic Dysfunction in the Development of Neurodegenerative Disease View all 5 articles
 Gilmara Gomes de Assis1,2*
Gilmara Gomes de Assis1,2* Jay R. Hoffman3
Jay R. Hoffman3 Jacek Bojakowski4
Jacek Bojakowski4 Eugenia Murawska-Ciałowicz5
Eugenia Murawska-Ciałowicz5 Paweł Cięszczyk1
Paweł Cięszczyk1 Eugene V. Gasanov6*
Eugene V. Gasanov6*Brain-derived neurotrophic factor (BDNF) plays an essential role in nervous system formation and functioning, including metabolism. Present only in humans, the “Val66Met” polymorphism of the BDNF gene (BDNF) is suggested to have a negative influence on the etiology of neurological diseases. However, this polymorphism has only been addressed, at the molecular level, in nonhuman models. Knowledge about Val66- and Met66-variant differences, to date, has been achieved at the protein level using either cell culture or animal models. Thus, the purpose of our study was to analyze the impact of the Val66Met polymorphism on BDNF expression in healthy humans and compare the allele-specific responses to metabolic stress. Muscle biopsies from 13 male recreational athletes (34 ± 9 years, 1.80 ± 0.08 m, 76.4 ± 10.5 kg) were obtained before and immediately following a VO2max test. Allele-specific BDNF mRNA concentrations were quantified by droplet digital PCR (ddPCR) in heterozygous and homozygous subjects. The results indicated that BDNF expression levels were influenced by the genotype according to the presence of the polymorphism. BDNF expression from the Met66-coding alleles, in heterozygotes, was 1.3-fold lower than that from the Val66-coding alleles. Total BDNF mRNA levels in these heterozygotes remained below the whole sample’s mean. A partial dominance was detected for the Val66-coding variant on the Met66-coding’s. BDNF expression levels decreased by an average of 1.8-fold following the VO2max test, independent of the individual’s genotype. The results of this study indicate that metabolic stress downregulates BDNF expression but not plasma BDNF concentrations. No correlation between expression level and plasma BDNF concentrations was found.
Brain-derived neurotrophic factor (BDNF) is a neurotrophin, a small secretory dimeric protein that appears in all vertebrates with a highly conserved structure (Rafieva and Gasanov, 2016). Showing the most pervasive and abundant expression throughout the brain among the neurotrophins, BDNF plays a pivotal role in cell proliferation and differentiation, and supports neural tissue formation, muscle–neuron interaction, and the organization of the central nervous system (CNS) (Friedman, 2010). In addition, BDNF is essential for neuronal outgrowth and continuous regulation of processes such as synaptic transmission, long-term potentiation, and depreciation (Lamb et al., 2015; Sasi et al., 2017). BDNF is largely expressed in neurons and neural tissue and is also produced by muscle (Matthews et al., 2009), although the role of muscle production has yet to be elucidated in humans to date.
In humans, a single nucleotide polymorphism resulting in a substitution of an adenine (A) base by guanine (G) in the BDNF gene (BDNF) has been identified in ∼20% of the population (Maisonpierre et al., 1991; Shimizu et al., 2004). This polymorphism is named “Val66Met” due to a Valine to Methionine substitution in the 66th amino-acid position of the synthesized protein (BDNF) with respect to the A or G genotype (Rafieva and Gasanov, 2016; Hunt et al., 2018). Considering the pivotal function of BDNF, it has been suggested that the BDNF Val66Met may have a role in the etiology of several neurological diseases (Egan et al., 2003; Notaras and van den Buuse, 2018; Shen et al., 2018) and psychiatric disorders (Kishi et al., 2018).
The BDNF protein (mature form, referred from here on as BDNF) is synthesized by cells as a precursor, pro-BDNF, that undergoes a multistage processing, which includes the loss of the pro-domain part containing the polymorphic 66th amino-acid position. Therefore, the Val66Met variance does not appear in the structure of released BDNF. However, a change in the pro-domain part of pro-BDNF, the Val66Met substitution, could influence the secretory pathway direction and release of BDNF (Chen et al., 2004) affecting the efficiency of BDNF synthesis and function (Bath and Lee, 2006; Pruunsild et al., 2007).
In humans, the influence of the Val66Met polymorphism has been observed through the association between Met66 genotypes and pathological changes in the morphology of specific areas of the brain (Egan et al., 2003). Importantly, the polymorphism fails to parcel out as a determinant for the development of neuropathological conditions among other factors such as ethnicity, gender, and other influences (Shimizu et al., 2004; Kishi et al., 2018; Shen et al., 2018). When examining various clinical subpopulations, there are no reliable associations between the genotype and plasma BDNF concentrations (Yoshimura et al., 2011), suggesting that the association of this polymorphism with risk for disease, based on circulating BDNF concentrations, is limited (Shen et al., 2018). Nevertheless, there appears to be a pattern in the acute stress response of individuals carrying the Met66 allele in BDNF that results in an upregulation in their cortisol response to stress, which may provide support for the influence of the Val66Met polymorphism in the development of stress-related disorders including neurological disease (Kishi et al., 2018; de Assis and Gasanov, 2019).
The present understanding of Val66- and Met66-variant differences, to date, has been achieved at the protein level using either cell culture or animal models (Egan et al., 2003; Chen et al., 2004; Lou et al., 2005; Anastasia et al., 2013; Uegaki et al., 2017). However, BDNF production could also be affected at the level of gene function. Serving as a template for the pro-BDNF translation, mRNA initiates BDNF production, reflecting gene activity, and is the first level of BDNF regulation. In regard to gene expression, Val66- and Met66-coding alleles may be transcribed either equally or not at all, being in competition for the mRNA synthesis machinery and potentially interfering with their expression levels. However, to the best of our knowledge, the influence of the Val66Met polymorphism on BDNF functioning (i.e., transcription) has never been previously analyzed.
In consideration of previous observations of BDNF responsivity to aerobic exercise (Dinoff et al., 2016; de Assis et al., 2018), we have developed a method for analyzing the gene expression of human BDNF regarding the Val66Met polymorphism in vivo using a maximal endurance exercise protocol. In order to reveal the relation between the Met66 and Val66 alleles of BDNF, and its impact on BDNF functioning (i.e., transcription), we have analyzed the quantities of BDNF mRNA in the muscle of healthy subjects at rest and following a maximal aerobic capacity (VO2max) test.
In accordance with the European Union regulation 2016/679 of 27 April 2016 and the Ethical Committee of the Central Clinic Hospital of the MSWiA in Warsaw, Poland, 25 physically active healthy volunteers (37.5 ± 9.9 years, among them five women) following an explanation of all procedures, risks, and benefits, provided his or her informed consent to participate in this study. All participants were genotyped for the BDNF Val66Met polymorphism. Thirteen of the participants, all males (age: 34.5 ± 9 years, BMI: 22.85 ± 1.6, and VO2max: 54.3 ± 6.2 ml/kg/min), were included in the experiment (Table 1).
All study participants reported to the Human Performance Laboratory in the Mossakowski Medical Research Centre, Polish Academy of Science, Warsaw, Poland. During the visit, all participants were provided with a brief medical exam that included anthropometric measurements. A blood draw (3–4 ml) was obtained by a nurse from a cubital vein, divided to aliquots, and immediately embedded in dry ice and stored at −80°C until analysis. A 0.5 ml aliquot from a blood sample was used for genomic DNA (gDNA) extraction by a NucleoSpin Tissue kit (Macherey-Nagel GmbH & Co., KG, Germany) according to the manufacturer’s procedures. The resulting DNA concentrations were determined by a NanoDropTM 2000 Spectrophotometer (Thermo Fisher Scientific, United States). The gDNA was used as a template in a one-step amplified refractory mutation system PCR according to Sheikh et al. (2010). In this protocol, two primers (P1 and P2) surrounding the BDNF’s area of interest (polymorphism bearing ∼400 bp) and two primers (P3 and P4) corresponding to Val- and Met-coding sequences, respectively (internal to the first pair and in opposite orientation to each other) were added together with a PCR Mix Plus mixture (A&A Biotechnology, Poland) according to the manufacturer’s procedures. The reaction was performed in a C1000 Touch Thermal Cycler (Bio-Rad, United States). PCR results were analyzed in a 6% polyacrylamide or 2.5% agarose gel-electrophoresis with ethidium bromide staining. Images were generated by Bio-Rad Gel Documentation System. Results showed the full-length BDNF-specific product (∼400 bp) and the corresponding products to the presence of Val (∼250 bp) and Met (∼200 bp) in BDNF variants (see Figure 1).
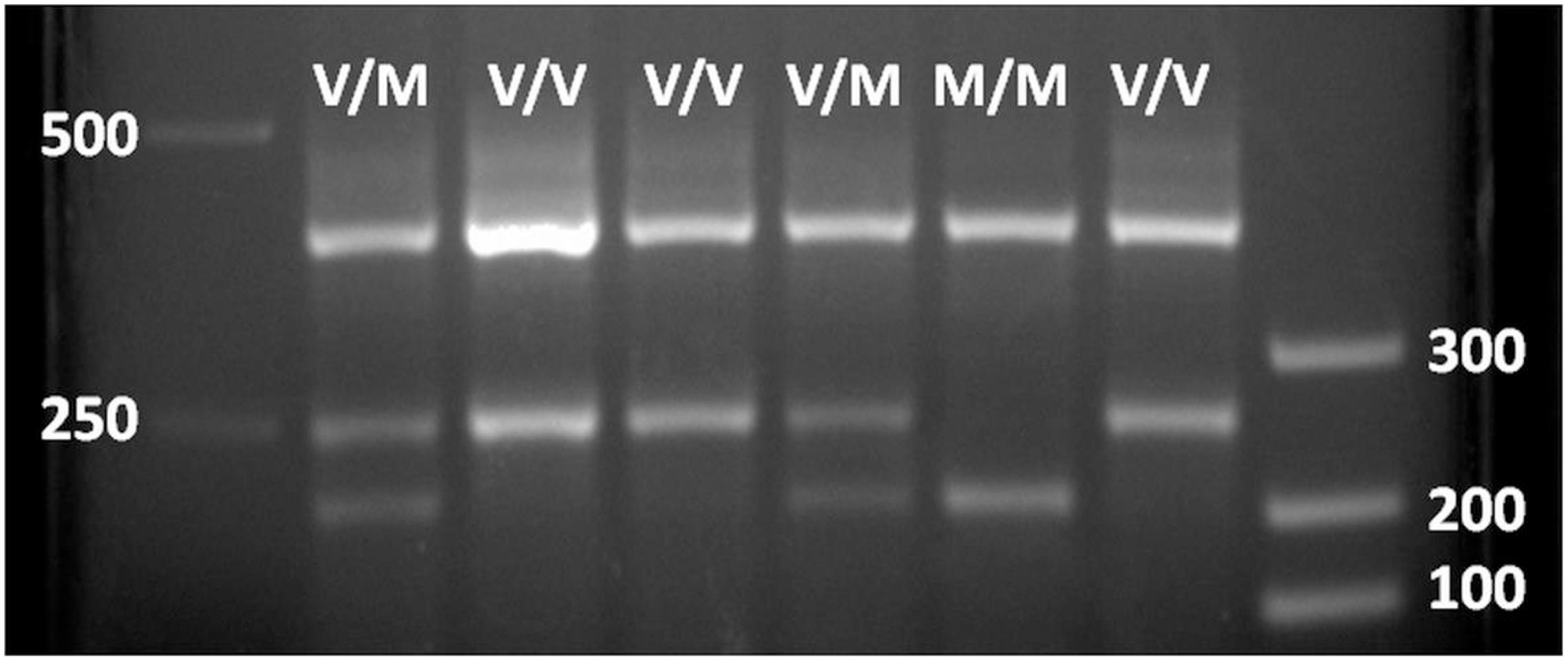
Figure 1. Agarose gel electrophoresis of PCR BDNF polymorphism analysis. 400 bp, BDNF gene full-length fragment (internal positive PCR control); 250 bp, Val66-coding BDNF gene fragment; 200 bp, Met66-coding BDNF gene fragment.
Participants reported to the laboratory and rested comfortably in a supine position for 30 min. Heart rate and blood pressure were obtained prior to the initial muscle biopsy. Following the biopsy (∼15 min), participants performed a maximum aerobic capacity (VO2max) test using the Bruce treadmill protocol (Bruce et al., 2004). Change in both the gradient and the speed of the treadmill occurred every 3 min (seven stages) that began at 2.7 kph/10% gradient and progressed up to 8.9 kph/20% grade, or until the participants’ volitional exhaustion. Immediately (30–60 s) following the conclusion of the VO2max test, a second muscle biopsy was obtained. Blood samples were collected from an antecubital vein at the same time-point of the muscle biopsies.
Approximately 50 mg (wet weight) of skeletal muscle from Vastus lateralis was obtained per sampling by suction using a semiautomatic needle biopsy (14 g, 100 mm). The sample was immediately embedded in dry ice and stored at −80°C until analysis. All procedures occurred between 3 p.m. and 5 p.m. (GMT+2).
Muscle samples were manually homogenized in Eppendorf tubes and mixed with 0.5 ml of a TRIzol reagent (T9424, Sigma-Aldrich). The TRIzol-chloroform RNA extraction was performed according to the manufacturer’s protocol, and the total RNA was dissolved in 0.03 ml of RNAse-free water; RNA concentrations were determined by a NanoDropTM 2000 Spectrophotometer, and the RNA solutions were stored at −80°C. One microgram of RNA was later used for cDNA synthesis in 0.02 ml of the reaction mixture using the iScriptTM Reverse Transcription kit (Bio-Rad) according to the manufacturer’s protocol. Subsequently, 1 μl of the cDNA solution was used as a template for ddPCR analyses.
A housekeeping gene was primarily established as a reference for the patterns of BDNF expression in the ddPCRTM system (de Assis et al., 2020). For this, the cDNA of three subjects’ samples (one woman) both at rest and post VO2max were applied in 20 μl reactions for the Bio-Rad PrimePCR Pathway Plate—Reference Genes H96, Human—containing 14 housekeeping gene candidates recommended for muscle tissue analysis, internal PCR, reverse transcription, RNA quality, and gDNA admixture controls. RT-PCRs were performed in triplicate according to the manufacturer’s protocol (Bio-Rad, United States). The beta-2-microglobulin (B2M) and the ribosomal protein S18 (RPS18) were among the three of the most reliable candidate genes applicable for ddPCRTM analysis [inter- and intraindividual threshold cycle, C(t), difference did not exceed 2.5 cycles]. The 13 cDNA samples were then applied to the ddPCRTM system with an allele-specific BDNF assay containing hydrolyzed probes (TaqMan) with both HEX (hexachlorofluorescein) and FAM (6-carboxyfluorescein) detection of Val- and Met-coding alleles, respectively (ddPCRTM Mutation Assay: rs6265, dHsaMDS320493890), using PrimePCRTM ddPCRTM Supermix for Probes (no dUTP), as well as the PrimePCRTM ddPCRTM Expression Probe assay, B2M, Human (HEX-probe detection, qHsaCPE5053101) in parallel (all designed by Bio-Rad). RNA samples without a reverse transcription step were also used in parallel as a template, in the same quantity according to the RNA template in cDNA samples as a genomic DNA (gDNA) admixture control. The quantities of reaction products in these samples were subtracted from the corresponding samples analyzed for BDNF expression (de Assis et al., 2020). The gDNA of Val66Met heterozygotes was used as a control of the BDNF allele ratio and as a positive control of the reaction. The ratios between Val and Met-alleles of BDNF in gDNA samples were 1 ± 0.03.
To exclude the reference gene influence, the second part of mRNA samples were treated by DNase I (M0303, NEB, United States) according to the manufacturer’s protocol and used for cDNA synthesis as described above. One microliter of cDNA solution was used in ddPCR for the allele-specific BDNF expression analysis with the RPS18 gene as a reference using the PrimePCRTM Probe assay, RPS18, Human (FAM-detection, qHsaCEP0040177, Bio-Rad). B2M and RPS18 expression was also compared in the same reaction using FAM- and HEX-fluorescence detection. All reactions were performed in duplicate.
To measure the BDNF concentration in plasma, blood samples were collected and drawn into tubes containing ethylene-diaminetetraacetic acid (EDTA) and stored at 4°C for 2–3 h. Then samples were centrifuged at 2,500 × g for 15 min at 4°C, and the supernatants were stored at −80°C until analysis. During analysis, thawed samples were centrifuged in Eppendorf tubes at 10,000 × g for 10 min. BDNF concentrations in plasma samples were analyzed based on the manufacturer’s guidance by the sandwich enzyme-linked immunosorbent assay (ELISA) Human BDNF PicoKine kit (Boster Bio, United States). The absorbance at 450 nm was measured with a Spark multimode microplate reader (Tecan, Switzerland) to determine BDNF values using the lyophilized human BDNF (Boster Bio, United States) dilutions as a standard. The ELISA kit sensitivity was <15 pg/ml, the coefficient of variation was <7.5%, and all blood samples were analyzed in duplicate.
Pearson product-moment correlations were applied to the pre-post VO2max values of BDNF/B2M∗103 expression and BDNF plasma concentrations, as well as for their concentration deltas. The association of age, BMI, and VO2max with BDNF expression and plasma BDNF was also verified by Pearson correlation analysis. Paired Student’s t-tests were used to verify pre-post differences in BDNF expression and BDNF blood concentrations. Two-way analysis of variance (ANOVA) was applied to verify for significance in Val66Met genotype influence on pre-post VO2max BDNF expression changes and allele-specific expression difference in heterozygotes. In the event of a significant F ratio, Bonferroni post hoc analysis was used to examine pairwise differences.
To compare the allele-specific expression in the heterozygotes regardless of conditions, the means of single allele mRNA levels were calculated in both pre- and post-VO2max test (pre- and post-means for each allele). Next, the levels of Val66- and Met66-coding alleles’ expression in pre- and post-VO2max conditions were standardized by calculating their ratios with their pre- and post-means, respectively. The standardized numbers of Val66- and Met66-coding alleles’ expression were then analyzed by a paired t-test. An alpha level of p ≤ 0.05 was used to determine statistical significance. All the data are presented as mean ± SD.
Characteristics and genotypes of the study participants including plasma BDNF concentrations are presented in Table 1. Comparison between B2M and RPS18 showed that these genes’ expression patterns were fully coherent, with no difference in pre-post VO2max test results (Supplementary Figures 1, 2). Therefore, all the calculations of BDNF expression quantities were related to B2M mRNA quantities. The results of BDNF mRNA/B2M∗103 expression analyses from skeletal muscle samples showed a high homogeneity at rest (1.78 ± 0.53 BDNF/103 B2M mRNA molecules) and even higher homogeneity after the VO2max test conditions (0.98 ± 0.33 BDNF/103 B2M mRNA molecules) (Table 2). The BDNF mRNA concentrations ranged from 0.528 to 2.982 molecules of BDNF per 103 molecules of B2M. Neither the individual differences nor the changes evoked by the VO2max test exceeded a threefold change in BDNF mRNA levels.

Table 2. Allele-specific quantification of BDNF mRNA in human muscle tissue by ddPCR with B2M mRNA as a reference.
The BDNF mRNA expression was significantly higher at rest compared to post-VO2max test conditions (p < 0.001). A 44 ± 9.7% decrease in BDNF mRNA levels following the VO2max test was observed (Table 3). There was a strong positive correlation between resting- and post-VO2max test values of BDNF mRNA (r = 0.859; p < 0.001). Participants whose BDNF expression levels were above the group average at rest maintained BDNF expression levels above the group mean following the VO2max test, whereas those whose expression levels were below the mean remained below the average BDNF expression levels of the group. A strong negative correlation was detected between resting BDNF expression levels and the pre-post changes in BDNF expression (pre/post Δ) (r = −0.838; p < 0.001), indicating a greater fall in BDNF expression levels for those displaying higher basal levels of BDNF expression. ANOVA performed between BDNF Val66Met homo- and heterozygotes indicated a significant effect of the VO2max test on gene expression [F(1,22) = 12.02, p = 0.002, part. eta sq. = 0.33] with no influence of the genotype (p = 0.1656). Post hoc analysis confirmed the significant effect of the VO2max test on BDNF expression (p < 0.001) and the absence of significant genotype influence (p > 0.05) (Figures 2, 3).

Figure 2. Spaghetti plot of BDNF expression level change. Pre—Total BDNF expression in rest conditions; Post—Total BDNF expression after VO2max test. Val66Val homozygotes are colored dark blue, Val66Met heterozygotes: magenta, Met66Met homozygote: green, Mean: the dotted red line. All BDNF expression values are related to B2M mRNA.
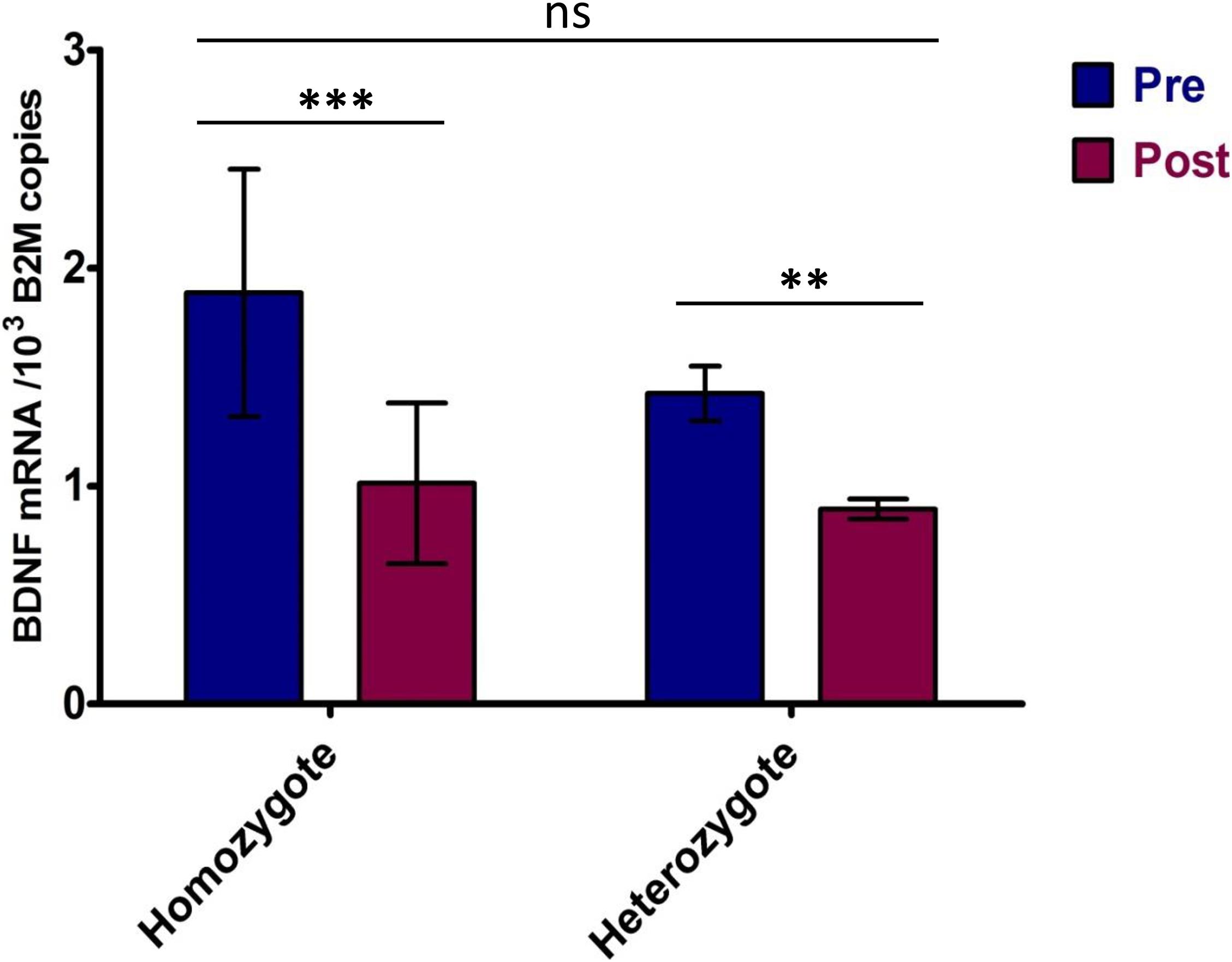
Figure 3. BDNF expression in homo- and heterozygotes in response to maximum effort test. Pre—Total BDNF expression in rest conditions; Post—Total BDNF expression after VO2max test. Significance levels marked by asterisks, where **p < 0.05, ***p < 0.01. Bars are given with ± SD.
Regarding allele specificity, Met66-coding BDNF mRNA levels in heterozygotes quantified 76.1 ± 11.2% of Val66-coding mRNA level values independent of metabolic conditions, which means 1.3-fold lower. This was confirmed by a comparison of the allele-specific expression levels standardized with the condition-specific expression means (p < 0.001) (Table 4).
Two-way ANOVA performed between the Val66- and Met66-coding allele expression levels in BDNF heterozygotes pre- and post-VO2max test confirmed both the metabolic stress effect [F(1,8) = 82.35, p < 0.0001, part. eta sq. = 0.58] on BDNF expression and the BDNF allele expression difference [F(1,8) = 47.05, p = 0.0001, part. eta sq. = 0.33] (see Figure 4). In addition, a significant interaction [F(1,8) = 5.19, p = 0.052, part. eta sq. = 0.04) was noted between Val66 and Met66 allele expression differences in response to the VO2max test.
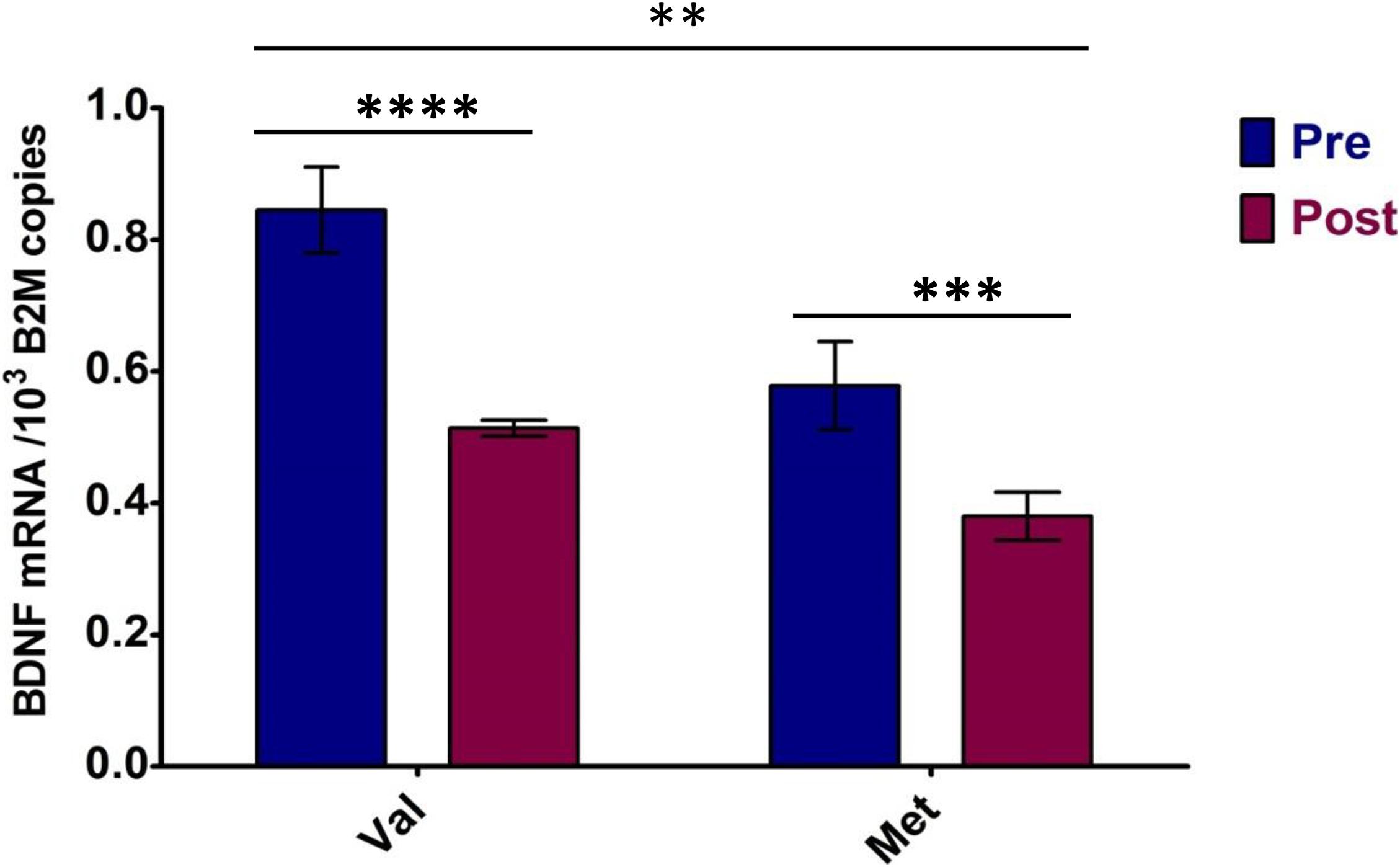
Figure 4. Allele-specific BDNF expression of heterozygotes in response to maximum effort test. Pre—Total BDNF expression in rest conditions; Post—Total BDNF expression after VO2max test. Val, Val66-coding BDNF mRNA; Met, Met66-coding BDNF mRNA. Significance levels marked by asterisks, where **p < 0.05, ***p < 0.01, ****p < 0.001. Bars are given with ± SD.
The pre-/post-VO2max test changes in BDNF expression, as well as the Val66-/Met66-coding allele relations, were confirmed in independent measurements with RPS18 expression as a reference (Table 5). A 38.3 ± 12.5% decrease in BDNF mRNA levels following the VO2max test was observed (Supplementary Figure 2). Met66-coding BDNF mRNA levels in heterozygotes quantified 78.5 ± 9.2% of Val66-coding mRNA level values (1.29 ± 0.14-fold lower) independent of metabolic conditions (Table 5).
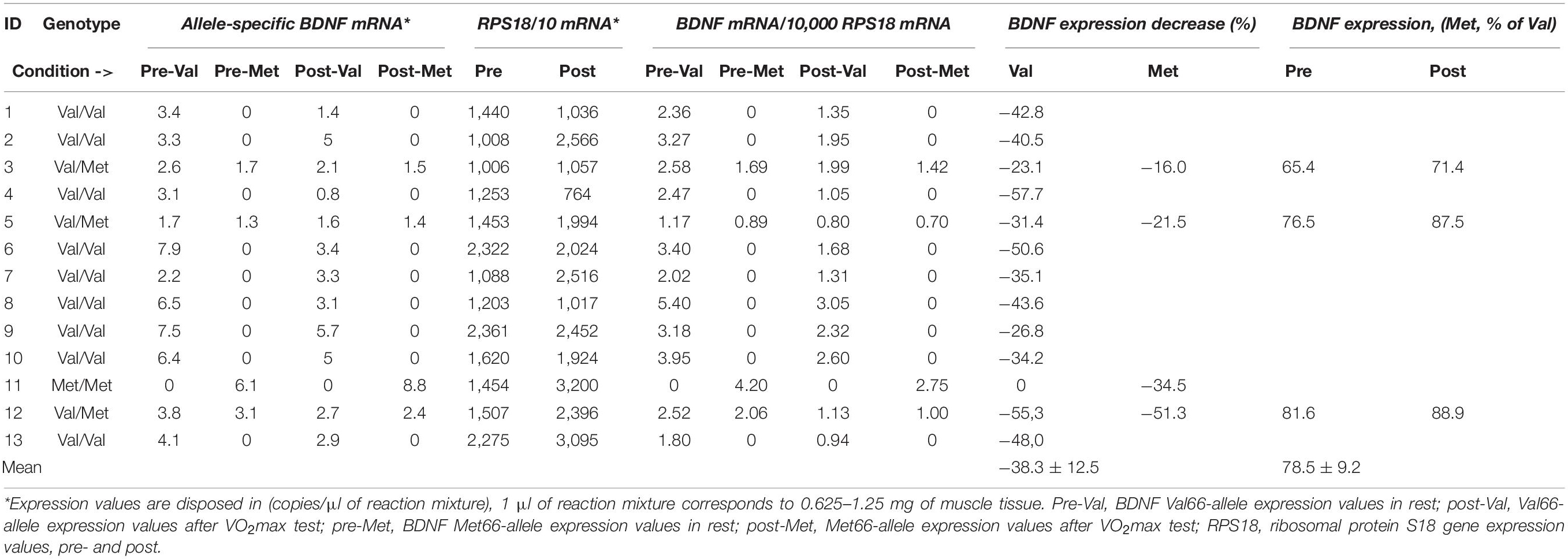
Table 5. Allele-specific quantification of BDNF mRNA in human muscle tissue by ddPCR with RPS18 mRNA as a reference, pre-post VO2 max test BDNF differences, and Val66-Met66 allele BDNF expression ratio in heterozygotes related to RSP18 mRNA.
The differences between resting and post-VO2max test plasma BDNF concentrations were not significantly different. In addition, the genotype did not appear to influence plasma BDNF concentrations. However, a strong negative correlation (r = −0.821; p < 0.001) was noted between plasma BDNF concentrations pre-VO2max test and the changes evoked by the test—the ΔBDNF (plasma post-BDNF – pre-BDNF). This suggests that participants with higher resting plasma BDNF concentrations experienced a decrease in post-test BDNF concentrations, while those with lower plasma BDNF concentrations experienced an increase in post-exercise BDNF concentrations (Figure 5). A moderate negative correlation was noted between the BDNF expression level changes (pre/post Δ) and plasma BDNF ones (ΔBDNF) (r = −0.531, p = 0.062). This means that a greater decrease in the BDNF expression level was associated with an increase in the plasma BDNF concentrations resulting from the VO2max test, while a lighter decrease in the BDNF expression tended to reflect a decrease in plasma BDNF concentrations. No association was noted between BDNF mRNA and plasma concentrations of BDNF at rest (r = −0.15, p = 0.634) nor post-VO2max assessments (r = 0.36, p = 0.221), as well as between any of the participants’ physiological parameters (VO2max, height, weight, and age).
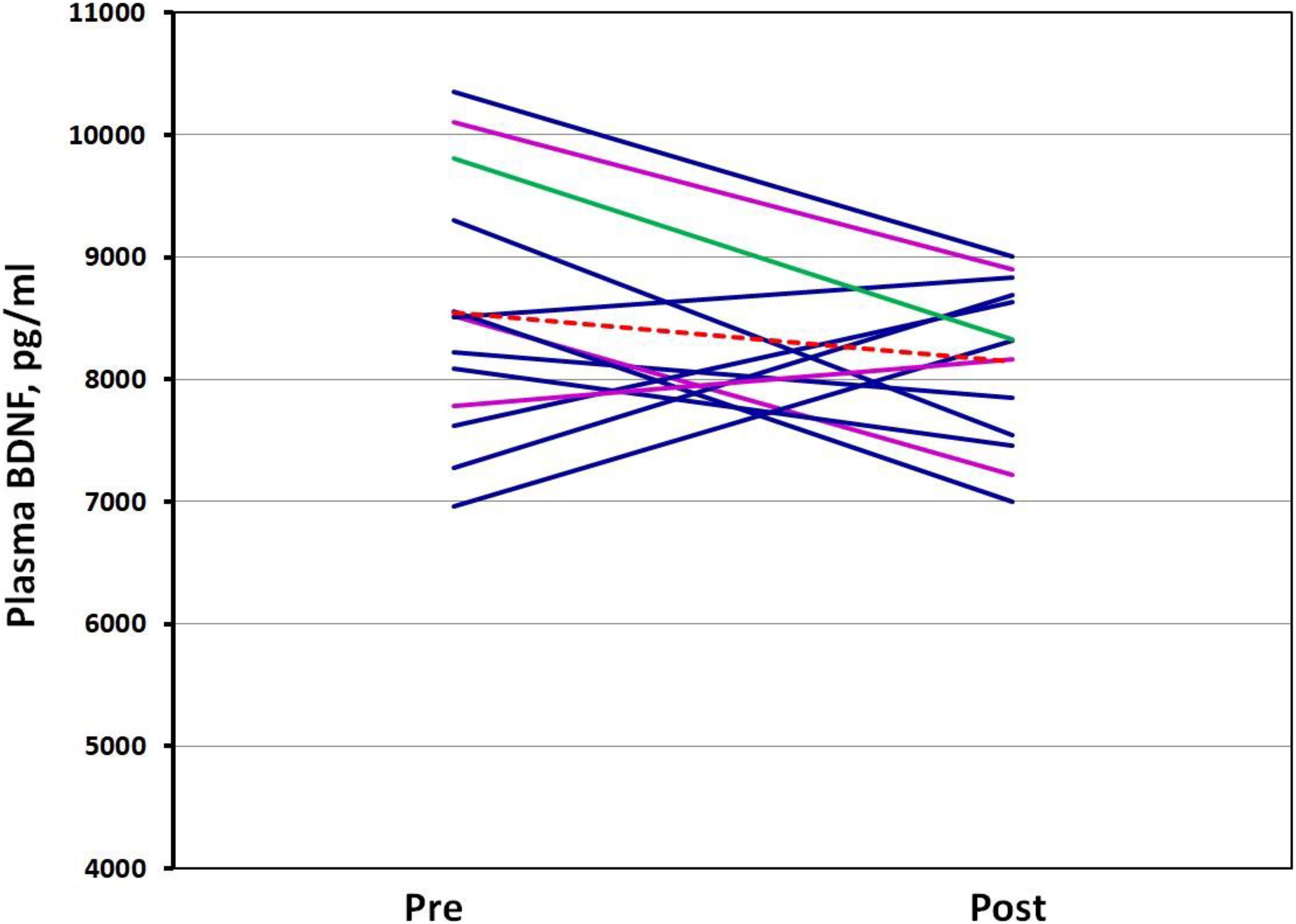
Figure 5. Spaghetti plot of plasma BDNF level change. Pre—plasma BDNF concentrations in rest conditions; Post—plasma BDNF concentrations after VO2max test. Val66Val homozygotes are colored dark blue, Val66Met heterozygotes: magenta, Met66Met homozygote: green, Mean: the dotted red line.
Our results indicated that BDNF expression in muscle was within a narrow range among all participants, regardless of the genotype and conditions (rest and metabolic stress), suggesting that muscle BDNF expression is very stable. The absolute quantification of muscle BDNF expression by ddPCR provided an opportunity to detect relatively small changes resulting from different metabolic conditions and genotype-induced expression variations. The effect of exercise on changes in BDNF expression levels was not dependent on the participants’ age or genotype suggesting a consistent response. This is supported by previous research of Matthews et al. (2009) who reported an increase in BDNF expression from 2 to 24 h following endurance exercise (120 min cycling at 60% of VO2max) using quantitative RT-PCR. However, immediately after the bout of exercise, BDNF expression levels were shown to decrease at the same time-point as our results. It is likely that an initial decay in BDNF expression is seen during an acute metabolic stress, which is followed by a compensatory rise during recovery. The results of the present study indicated that the higher the pre-exercise BDNF expression levels, the more pronounced their decline during exercise. This perhaps is suggestive of a stability feature for BDNF expression, as well as an immediate sensitivity to metabolic changes.
Plasma BDNF concentrations failed to correlate with BDNF expression levels at both rest and post-exercise, with no one-directional changes noted in plasma BDNF following the exercise stress. Instead, an increase in plasma BDNF concentrations was noted in the participants with lower resting BDNF concentrations in response to the VO2max test, while the participants with higher plasma BDNF concentrations at rest experienced a decrease following maximal effort exercise. This bidirectional response, which appears to be related to resting BDNF concentrations, may reflect a stabilizing tendency to maintain circulating BDNF concentrations within a certain biological range during metabolic stress, such as exercise. Another potential explanation is also related to the exhaustive exercise protocol in which participants have a specific switch point that is activated when the metabolic system moves from predominantly aerobic to anaerobic. This is supported by previous studies that reported a difference in the circulating BDNF response to aerobic vs. anaerobic exercise (Dinoff et al., 2016; de Assis et al., 2018).
A weak correlation was observed between changes in both expression and circulating levels of BDNF. Those participants showing greater changes in gene expression experienced smaller changes in plasma BDNF concentrations. This suggests negative regulatory feedback from circulating BDNF concentrations to its gene expression, suggesting an indirect or delayed connection between BDNF expression and blood concentrations. The discrepancy between BDNF expression and secretion may be related to possible differences in the response of BDNF producing blood and muscle cells to a metabolic stress, especially if the latter are not a source of circulating BDNF (Matthews et al., 2009). Furthermore, the appearance of BDNF in the circulation requires a considerable time lag from mRNA translation to dimeric active protein secretion. BDNF released by cells into the circulation involves secretory vesicle trafficking and a complex multistage process that includes the cleavage of pro-BDNF with the pro-domain removal. Increases in BDNF expression leads to an increase in pro-BDNF synthesis with a slow accumulation in secretory vesicles where pro-BDNF conversion to BDNF begins (Chen et al., 2004; Lou et al., 2005; Pruunsild et al., 2007; Rafieva and Gasanov, 2016). Thus, changes in BDNF expression levels are unlikely to immediately affect BDNF production.
The release of BDNF in response to metabolic stress and other types of signals is characteristic of the regulated pathway of neurotrophin secretion (Sasi et al., 2017). It represents a stimulus-dependent liberation by cells of secretory vesicles containing pre-synthesized BDNF that results in increases in circulating BDNF concentrations (Chen et al., 2004; Lou et al., 2005; Rafieva and Gasanov, 2016). The temporal gap between BDNF mRNA synthesis and BDNF release is dependent on the stimulus but not on the current level of BDNF expression. This likely contributes to the lack of an association between BDNF gene expression levels and changes in circulating concentrations.
The discrepancy between BDNF expression and circulating BDNF concentrations should be addressed with respect to the efficiency in the conversion of pro-BDNF to BDNF. Secretory vesicles contain unprocessed pro-BDNF; once secreted, it is subsequently cleaved to BDNF by blood and tissue proteases, increasing the gap between gene expression and mature factor appearance (Friedman, 2010; Rafieva and Gasanov, 2016). The portion of secreted unprocessed pro-BDNF may potentially increase when metabolically stimulated. This has been supported by investigators reporting an increase in the pro-BDNF/BDNF ratio during an acute aerobic stress (Brunelli et al., 2012). Thus, relying solely on blood BDNF concentrations may not provide the full spectrum of BDNF production. Measuring BDNF expression levels, as well as pro-BDNF circulating concentrations, may provide a more complete picture.
To the best of our knowledge, this appears to be the first investigation to demonstrate that the two alleles of human BDNF are expressed simultaneously in both non-polymorphic and polymorphic individuals, carrying Val66- and Met66-coding alleles. By comparing the total BDNF expression in all participants and detecting the allele-specific expression in the heterozygotes, we were able to conclude that both alleles of human BDNF are active. At the same time, our data showed an inequality between Val66 and Met66 BDNF allele activity. Despite the low number of heterozygotes occurring in healthy volunteers (3 out of 25 participants), the difference between the Val66-coding and the Met66-coding BDNF mRNA levels was detected in each one of the heterozygotes, at rest or during a metabolic stress (e.g., maximal exercise).
Gene activity (i.e., effectiveness of transcription) depends on the cis-regulatory elements represented in the gene and its neighboring DNA sequence, as well as on trans-regulatory elements represented by a set of regulatory genes. In the cellular environment of a heterozygous organism, all the trans-acting elements are shared between the two alleles of a gene so that their interaction with the gene’s alleles (in our case, BDNF) is determined by the relative effectiveness of the alleles’ cis-regulatory elements. By this, the heterogeneity in the gene alleles’ expression depends on its sequence only. Analyzing the transcription effectiveness of two different BDNF alleles in one subject, excluding differences in non-genetic and trans-acting factors, we compared the alleles’ relative activity, i.e., dominant/recessive relations (Yan and Zhou, 2004). Our results suggested that the Val66Met polymorphism is a cis-acting element affecting the gene expression in which the Val66-coding allele variant exhibits a partial dominance on the Met66-coding allele variant.
To exclude the metabolic stress factor, a mathematical averaging of BDNF expression at rest and post-exercise in heterozygotes was performed. A difference in Val66 and Met66 BDNF gene alleles was confirmed. The dominant-recessive relation of BDNF Val66 and Met66 BDNF alleles was also supported by the expression pattern of the Met66-homozygote subject, where in the absence of Val66-coding allele suppression, it demonstrated one of the highest levels of BDNF expression.
The total expression of BDNF in Met66-heterozygotes remained below the mean expression level of the whole sample. A trend for a stabilizing shape was also observed for total BDNF expression—the higher the resting expression levels, the greater the decrease following maximal exercise. Interestingly, this effect was also attributed to an allele-specific reaction. The Val66-allele, which exposed higher levels of expression in comparison to the Met66 one, demonstrated the more pronounced exercise-induced decrease, while the Met66-allele’s expression decline after exercise was milder.
There are several limitations to our study. Our observations are based on comparing three heterozygotes to 10 homozygote participants. Although results are consistent, further investigations are needed in larger samples of genotyped populations. As allele interplay is possible in heterozygotes only, this group should be analyzed apart from the homozygotes where no such BDNF expression suppressive effect is possible. Therefore, association studies dividing populations traditionally into groups of Val66-homozygotes and Met66-carriers (with Met66-homozygotes and Val66Met-heterozygotes inside) can be led to a diminishment of significant findings and/or incorrect interpretations. The results of our study support the hypothesis that the Val66Met polymorphism impacts the BDNF gene function (Egan et al., 2003; Chen et al., 2004; Bath and Lee, 2006; Kishi et al., 2018; Notaras and van den Buuse, 2018; Shen et al., 2018; Skibinska et al., 2018; Toh et al., 2018). However, alteration in BDNF expression levels does not appear to directly impact BDNF release/secretion. It appears that the post-translation processes prior to BDNF release can be differentially affected by the presence of Methionine, which might imply the Val66Met polymorphism influence on BDNF release and functions (Egan et al., 2003; Chen et al., 2004; Yoshimura et al., 2011; Uegaki et al., 2017).
Our study showed that BDNF expresses both alleles simultaneously. Results indicated that the expression of BDNF immediately after maximal exercise decreases 1.8 ± 0.4-fold, regardless of the genotype. Resting BDNF expression levels positively correlate with post-exercise levels and negatively correlate with the magnitude of exercise-induced changes. Furthermore, BDNF expression levels do not correlate with plasma BDNF. The levels of BDNF expression appear to be modulated by the Met66 allele presence in heterozygotes. The Val66-allele variant exhibits a partial dominance over the Met66-allele variant. We believe that these findings further our understanding on the mechanisms of BDNF production and function. To elucidate the physiological implications of Val66Met polymorphism on nervous systems-related pathologies, further studies on BDNF expression and protein release must be addressed to specific genotyped populations.
The datasets generated during and/or analyzed during the current study are available in the Zenodo repository, https://zenodo.org/record/3633677#X8EAyc1KiUk.
This study was performed in accordance with the European Union regulation 2016/679 of 27 April 2016 and the 96 Ethical Committee of the Central Clinic Hospital of the MSWiA in Warsaw, Poland.
GA, PC, and EG: conceptualization. GA, JB, and EG: sample collection and analysis, and resources. GA and EG: data curation and project administration. GA, JH, and EG: writing—original draft preparation. EM-C and PC: writing—review and editing, and funding acquisition. PC: supervision. All authors contributed to the article and approved the submitted version.
This research was supported by grant nos. 2016/21/B/NZ3/00354 and UMO-2017/27/B/NZ7/00204 from the National Science Center (Kraków, Poland). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Prof. Jacek Kuznicki, the Head of Laboratory of Neurodegeneration of the International Institute of Molecular and Cell Biology in Warsaw, and his Deputy Dr. Lukasz Majewski for their help and support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.638176/full#supplementary-material
Supplementary Figure 1 | B2M and RPS18 expression levels in samples. cDNA samples numbered; mRNA samples represent the gDNA admixture, and reaction specificity control.
Supplementary Figure 2 | B2M and RPS18 expression level ratio stability in Pre- and Post-VO2max test samples.
Supplementary Figure 3 | Spaghetti plot of BDNF expression level change related to RPS18 mRNA. Pre—Total BDNF expression in rest conditions; Post—Total BDNF expression after VO2max test. Val66Val homozygotes are colored dark blue, Val66Met heterozygotes: magenta, Met66Met homozygote: green, Mean: the dotted red line.
Anastasia, A., Deinhardt, K., Chao, M. V., Will, N. E., Irmady, K., Lee, F. S., et al. (2013). Val66Met polymorphism of BDNF alters prodomain structure to induce neuronal growth cone retraction. Nat. Commun. 4:2490. doi: 10.1038/ncomms3490
Bath, K. G., and Lee, F. S. (2006). Variant BDNF (Val66Met) impact on brain structure and function. Cogn. Affect. Behav. Neurosci. 6, 79–85. doi: 10.3758/CABN.6.1.79
Bruce, R. A., Blackmon, J. R., Jones, J. W., and Strait, G. (2004). Exercising testing in adult normal subjects and cardiac patients. Ann. Noninvasive Electrocardiol. 9, 291–303. doi: 10.1111/j.1542-474X.2004.93003.x
Brunelli, A., Dimauro, I., Sgrò, P., Emerenziani, G. P., Magi, F., Baldari, C., et al. (2012). Acute exercise modulates BDNF and pro-BDNF protein content in immune cells. Med. Sci. Sports Exerc. 44, 1871–1880. doi: 10.1249/MSS.0b013e31825ab69b
Chen, Z. Y., Patel, P. D., Sant, G., Meng, C. X., Teng, K. K., Hempstead, B. L., et al. (2004). Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J. Neurosci. 24, 4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004
de Assis, G. G., and Gasanov, E. V. (2019). BDNF and cortisol integrative system – plasticity vs. degeneration: implications of the Val66Met polymorphism. Front. Neuroendocrinol. 55:100784. doi: 10.1016/j.yfrne.2019.100784
de Assis, G. G., Gasanov, E. V., de Sousa, M. B. C., Kozacz, A., and Murawska-Cialowicz, E. (2018). Brain derived neutrophic factor, a link of aerobic metabolism to neuroplasticity. J. Physiol. Pharmacol. 69, 351–359. doi: 10.26402/jpp.2018.3.12
de Assis, G. G., Hoffman, J. R., and Gasanov, E. V. (2020). BDNF Val66Met polymorphism, the allele-specific analysis by qRT-PCR – a novel protocol. Int. J. Med. Sci. 17, 3058–3064. doi: 10.7150/ijms.50643
Dinoff, A., Herrmann, N., Swardfager, W., Liu, C. S., Sherman, C., Chan, S., et al. (2016). The effect of exercise training on resting concentrations of peripheral brain-derived neurotrophic factor (BDNF): a meta-analysis. PLoS One 11:e0163037. doi: 10.1371/journal.pone.0163037
Egan, M. F., Kojima, M., Callicott, J. H., Goldberg, T. E., Kolachana, B. S., Bertolino, A., et al. (2003). The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112, 257–269. doi: 10.1016/S0092-8674(03)00035-7
Friedman, W. J. (2010). Proneurotrophins, seizures, and neuronal apoptosis. Neuroscientist 16, 244–252. doi: 10.1177/1073858409349903
Hunt, S. E., McLaren, W., Gil, L., Thormann, A., Schuilenburg, H., Sheppard, D., et al. (2018). Ensembl variation resources. Database 2018:bay119. doi: 10.1093/database/bay119
Kishi, T., Yoshimura, R., Ikuta, T., and Iwata, N. (2018). Brain-derived neurotrophic factor and major depressive disorder: evidence from meta-analyses. Front. Psychiatry 8:308. doi: 10.3389/fpsyt.2017.00308
Lamb, Y. N., Mckay, N. S., Thompson, C. S., Hamm, J. P., Waldie, K. E., and Kirk, I. J. (2015). Brain-derived neurotrophic factor Val66Met polymorphism, human memory, and synaptic neuroplasticity. Wiley Interdiscip. Rev. Cogn. Sci. 6, 97–108. doi: 10.1002/wcs.1334
Lou, H., Kim, S. K., Zaitsev, E., Snell, C. R., Lu, B., and Loh, Y. P. (2005). Sorting and activity-dependent secretion of BDNF require interaction of a specific motif with the sorting receptor carboxypeptidase E. Neuron 45, 245–255. doi: 10.1016/j.neuron.2004.12.037
Maisonpierre, P. C., Le Beau, M. M., Espinosa, R. III, Ip, N. Y., Belluscio, L., de la Monte, S. M., et al. (1991). Human and rat brain-derived neurotrophic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics 10, 558–568. doi: 10.1016/0888-7543(91)90436-I
Matthews, V. B., Aström, M. B., Chan, M. H., Bruce, C. R., Krabbe, K. S., Prelovsek, O., et al. (2009). Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMP-activated protein kinase. Diabetologia 52, 1409–1418. doi: 10.1007/s00125-009-1364-1
Notaras, M., and van den Buuse, M. (2018). Brain-derived neurotrophic factor (BDNF): novel insights into regulation and genetic variation. Neuroscientist 25, 434–454. doi: 10.1177/1073858418810142
Pruunsild, P., Kazantseval, A., Aid, T., Palm, K., and Timmusk, T. (2007). Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90, 397–406. doi: 10.1016/j.ygeno.2007.05.004
Rafieva, L. M., and Gasanov, E. V. (2016). Neurotrophin propeptides: biological functions and molecular mechanisms. Curr. Protein Pept. Sci. 17, 298–305. doi: 10.2174/1389203716666150623104145
Sasi, M., Vignoli, B., Canossa, M., and Blum, R. (2017). Neurobiology of local and intercellular BDNF signaling. Pflugers Arch. Eur. J. Physiol. 469, 1–18. doi: 10.1007/s00424-017-1964-4
Sheikh, H. I., Hayden, E. P., Kryski, K. R., Smith, H. J., and Singh, S. M. (2010). Genotyping the BDNF rs6265 (val66met) polymorphism by one-step amplified refractory mutation system PCR. Psychiatr. Genet. 20, 109–112. doi: 10.1097/YPG.0b013e32833a2038
Shen, T., You, Y., Joseph, C., Mirzaei, M., Klistorner, A., Graham, S. L., et al. (2018). BDNF polymorphism: a review of its diagnostic and clinical relevance in neurodegenerative disorders. Aging Dis. 9, 523–536. doi: 10.14336/AD.2017.0717
Shimizu, E., Hashimoto, K., and Iyo, M. (2004). Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. Am. J. Med. Genet. 126B, 122–123. doi: 10.1002/ajmg.b.2011
Skibinska, M., Groszewska, A., Kapelski, P., Rajewska-Rager, A., Pawlak, J., Dmitrzak-Weglarz, M., et al. (2018). Val66Met functional polymorphism and serum protein level of brain-derived neurotrophic factor (BDNF) in acute episode of schizophrenia and depression. Pharmacol. Rep. 70, 55–59. doi: 10.1016/j.pharep.2017.08.002
Toh, Y. L., Ng, T., Tan, M., Tan, A., and Chan, A. (2018). Impact of brain-derived neurotrophic factor genetic polymorphism on cognition: a systematic review. Brain Behav. 8:e01009. doi: 10.1002/brb3.1009
Uegaki, K., Kumanogoh, H., Mizui, T., Hirokawa, T., Ishikawa, Y., and Kojima, M. (2017). BDNF binds its pro-peptide with high affinity and the common Val66Met polymorphism attenuates the interaction. Int. J. Mol. Sci. 18:1042. doi: 10.3390/ijms18051042
Yan, H., and Zhou, W. (2004). Allelic variations in gene expression. Curr. Opin. Oncol. 16, 39–43. doi: 10.1097/00001622-200401000-00008
Yoshimura, R., Kishi, T., Suzuki, A., Umene-Nakano, W., Ikenouchi-Sugita, A., Hori, H., et al. (2011). The brain-derived neurotrophic factor (BDNF) polymorphism Val66Met is associated with neither serum BDNF level nor response to selective serotonin reuptake inhibitors in depressed Japanese patients. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 1022–1025. doi: 10.1016/j.pnpbp.2011.02.009
Keywords: Val66Met polymorphism, BDNF, allele-specific gene expression, mRNA level, metabolism, VO2max test
Citation: de Assis GG, Hoffman JR, Bojakowski J, Murawska-Ciałowicz E, Cięszczyk P and Gasanov EV (2021) The Val66 and Met66 Alleles-Specific Expression of BDNF in Human Muscle and Their Metabolic Responsivity. Front. Mol. Neurosci. 14:638176. doi: 10.3389/fnmol.2021.638176
Received: 05 December 2020; Accepted: 18 March 2021;
Published: 05 May 2021.
Edited by:
Ildikó Rácz, University Hospital Bonn, GermanyReviewed by:
Masami Kojima, National Institute of Advanced Industrial Science and Technology (AIST), JapanCopyright © 2021 de Assis, Hoffman, Bojakowski, Murawska-Ciałowicz, Cięszczyk and Gasanov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilmara Gomes de Assis, Z2lsbWFyYS5nb21lc2RlYXNzaXNAYXdmLmdkYS5wbA==; Eugene V. Gasanov, ZWdhc2Fub3ZAaWltY2IuZ292LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.