
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mol. Neurosci. , 25 June 2019
Sec. Neuroplasticity and Development
Volume 12 - 2019 | https://doi.org/10.3389/fnmol.2019.00152
This article is part of the Research Topic Emerging Mechanisms in Dynamic GABAergic Inhibition View all 16 articles
Behavior and higher cognition rely on the transfer of information between neurons through specialized contact sites termed synapses. Plasticity of neuronal circuits, a prerequisite to respond to environmental changes, is intrinsically coupled with the nerve cell’s ability to form, structurally modulate or remove synapses. Consequently, the synaptic proteome undergoes dynamic alteration on demand in a spatiotemporally restricted manner. Therefore, proper protein localization at synapses is essential for synaptic function. This process is regulated by: (i) protein transport and recruitment; (ii) local protein synthesis; and (iii) synaptic protein degradation. These processes shape the transmission efficiency of excitatory synapses. Whether and how these processes influence synaptic inhibition is, however, widely unknown. Here, we summarize findings on fundamental regulatory processes that can be extrapolated to inhibitory synapses. In particular, we focus on known aspects of posttranscriptional regulation and protein dynamics of the GABA receptor (GABAR). Finally, we propose that local (co)-translational control mechanism might control transmission of inhibitory synapses.
The enormous capacity of the brain to store information and respond to different environmental conditions and challenges crucially rely on underlying mechanisms like synaptic plasticity. This depends on the ability to modulate the strength of transmission between two nerve cells as well as the growth and removal of synapses. Synapses consist of (at least) hundreds of proteins that need to be organized and correctly assembled to ensure proper synaptic function. Changes in synaptic transmission and structure are accompanied and conveyed by local alterations in protein levels. Understanding the regulation of synaptic protein composition is, therefore, crucial to gain insight into complex neurological processes such as learning and memory and, eventually, into neuropsychiatric diseases such as autism spectrum disorders, schizophrenia and bipolar disorders.
In order to remodel the synaptic proteome, neurons exploit different mechanisms that allow spatial and temporal control of protein levels. Protein synthesis was one of the first molecular mechanisms that were discovered to be indispensable for memory formation (Hershkowitz et al., 1975; Shashoua, 1976). Pioneer experiments showed that inhibiting translation blocked the ability of an animal to remember after training (Flexner et al., 1963). In line with this observation, several experiments have shown that strengthening and weakening of synaptic transmission, so called long-term potentiation (LTP) and depression (LTD), respectively, need active translation in a time-dependent manner (Krug et al., 1984; Linden, 1996). The spatial selectivity of synapses to undergo changes upon stimulation raised the question of how a cell knows, which synapse is destined for functional and structural remodeling. This inspired Frey and Morris (1997) to the idea of “synaptic tagging.” Repetitive activation of synapses, therefore, equips such a synapse with a labile molecular “tag.” Eventually, the synaptic tag allows the synapse to recruit newly synthesized proteins. The concept of “synaptic tagging” is a very elegant model to explain processes such as LTP and LTD at excitatory synapses (Frey and Morris, 1997). The precise identity of the tag(s) is still lacking. Furthermore, synaptic plasticity depends on additional processes such as mRNA localization, which is mainly independent of translation activity (Steward et al., 1998). mRNA transport and localization are important determinants of synaptic function (Jung et al., 2014). To date, it is generally believed that mRNAs are assembled into ribonucleoprotein particles (RNPs) consisting of mRNAs and RNA-binding proteins (RBPs). The protein and mRNA composition of these particles differ substantially (Kanai et al., 2004; Fritzsche et al., 2013) giving raise to the idea that different subtypes of particles or granules co-exist in a nerve cell. The function of these RNA granules is: (i) to transport mRNA—in a translationally dormant stage—along cytoskeletal elements such as microtubules to their destination at the synapse; and (ii) to regulate the translation of their target mRNAs. Activity-dependent disassembly of these RNA granules then allows the release of mRNAs and subsequent induction of translation. How neuronal stimulation, recruitment of mRNAs and unpacking of RNPs are synchronized is largely unknown. A pioneer study identified the kinase mechanistic target of rapamycin (mTOR) as a central hub to recruit RNAs. The authors suggest that mTOR might be the tag that controls mRNA recruitment at the synapse (Sosanya et al., 2015). mTOR is essential for proper neuronal function (Costa-Mattioli and Monteggia, 2013; Pernice et al., 2016). It needs to be experimentally verified though whether it might represent an universal synaptic tag or whether it might be specific for a subset of mRNAs.
Local protein expression control comprising mRNA transport, local protein synthesis and recruitment of newly synthesized protein remodel the synaptic proteome. Consequently, protein degradation is compulsive to complete synaptic remodeling. Synaptic protein degradation is induced in an activity-dependent manner (Bingol and Schuman, 2006). Moreover, it is tightly linked to translation to balance the protein need (Klein et al., 2015). In line with this finding, the translation repressor poly(A)-binding protein interacting protein 2A (PAIP2A) is degraded by calpain in neurons upon stimulation (Khoutorsky et al., 2013). Interestingly, calpain also degrades gephyrin (Gphn), a major scaffold protein at inhibitory synapses (Tyagarajan and Fritschy, 2014). This finding indicates that translational activation at excitatory synapses may modulate inhibitory synapses to alter transmission.
In this review article, we provide insight into posttranscriptional regulatory mechanisms that control synaptic protein expression. Since most of these studies investigated these processes at excitatory synapses, we aim to expand these fundamental aspects to inhibitory synapses. We speculate that local expression control also regulates inhibitory transmission to balance neuronal excitation.
With the emergence of the individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) technology (Huppertz et al., 2014), transcriptome-wide identification of RBP mRNA targets and binding site became experimentally addressable. iCLIP has now been performed for a series of RBPs (Tables 1, 2). Interestingly, most of the RBP binding occurs within the 3′-untranslated region (3′-UTR) of transcripts (Andreassi and Riccio, 2009). In addition, it was shown that the median of the 3′-UTR length of mRNAs bound to the RBP Staufen2 that is necessary for RNA transport (Heraud-Farlow and Kiebler, 2014) is longer than the median of the transcriptome (Heraud-Farlow et al., 2013). This finding indicates that a certain 3′-UTR length is needed to allow association with RBPs and, consequently, mRNA transport and/or expression control (Heraud-Farlow and Kiebler, 2014). To test whether mouse GABA receptor (GABAR) subunits show a similar tendency towards longer 3′-UTR length, we analyzed the nucleotide length of their 3′-ends of all GABAA and GABAB receptor subunit isoforms (see “Methods” section). Strikingly, GABAR subunits reveal a significant increase in their 3′-UTR compared to the total mouse 3′-UTRome (Figure 1A). Moreover, the 3′-UTR length was significantly extended when comparing the GABAR subunits with the 3′-UTRome of the somatic and neuropil layer of the hippocampal CA1 region (Cajigas et al., 2012; Figure 1A). An increase in 3′-UTR length is linked with decreased translational activity in HEK cells and human neurons (Floor and Doudna, 2016; Blair et al., 2017) probably due to a higher number of miRNA and RBP binding sites. In addition, 3′-UTR length is extended during neuronal development indicating increased translation regulation in mature neurons compared to developing nerve cells (Blair et al., 2017). Of note, GABAR subunits exhibited a trend towards longer 3′-ends when compared with ionotropic glutamate receptor subunits (Figure 1B). Together, these results suggest that GABAR subunit 3′-UTRs have a high(er) potential to be bound by RBPs. Supportive for this hypothesis is the fact that GABAR subunit mRNAs are enriched in the dendrite containing neuropil layer of CA1 neurons in the hippocampus (Cajigas et al., 2012) suggesting that these mRNAs are localized there. The recognition of mRNA targets by RBPs relies on binding sites within their 3′-UTRs and that each mRNA might have its own specific RNA signature. In detail, these binding sequences consist of both sequence and structural elements (Kiebler and Bassell, 2006; Doyle and Kiebler, 2011; Jung et al., 2014; Sugimoto et al., 2015). Interestingly, GABAR subunits exhibited a lower GC content compared to the total, somatic CA1 and neuropil 3′-UTRome (Figure 1C). Concomitantly, we observed a higher AT content (Figure 1C). Moreover, the same statistically significant effects were detected when comparing ionotropic GluR and GABAR subunit mRNAs (Figure 1D). A lower GC content accounts for less stable secondary structures in the 3′-UTRs of GABAR compared to the total, somatic CA1 and neuropil 3′-UTRome as well as to GluR 3′-ends. Interestingly, the cytoplasmic polyadenylation binding element binding protein (CPEB) binds a short, AT-rich sequence within the 3′-UTR of target mRNAs to control translation and to induce the elongation of polyA tails (Mendez and Richter, 2001). By using RNA immunoprecipitation (RIP), it was shown that CPEB1 and 4 bind different GABAR subunits as well as mRNAs coding for scaffold protein such as Gphn (Parras et al., 2018; see also Tables 1, 2). Moreover, ELAV proteins, among others, bind so-called AU-rich elements (ARE) to stabilize its target mRNAs (Fan and Steitz, 1998; Peng et al., 1998). Therefore, it is tempting to speculate that ELAV proteins also bind mRNAs coding for GABAR subunits to regulate their abundance. Supportive for this idea is an iCLIP-based ELAV target screen from human brain, which detected selective mRNAs encoding GABAR subunits, GABAB receptor auxiliary proteins and GABAR transport proteins (Scheckel et al., 2016; see also Tables 1, 2).
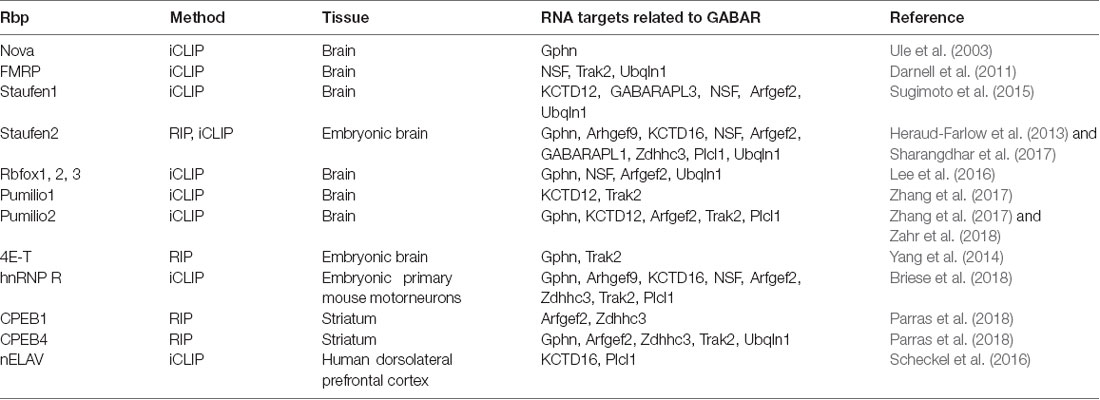
Table 2. Hand-selected list of RBPs with RNAs related to scaffold protein, GABAR auxiliary and transport proteins as targets.
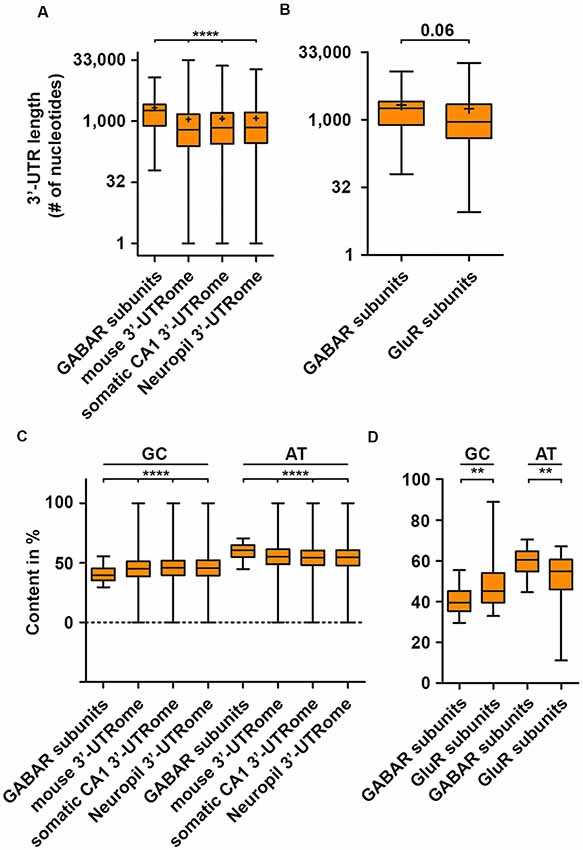
Figure 1. GABA receptor (GABAR) subunits exhibit extended 3′-untranslated region (3′-UTR) length. 3′-UTR lengths of GABAR (GABAA and GABAB receptor) subunits compared to the global mouse, hippocampal CA1, neuropil 3′-UTRome (A) and the 3′-UTR lengths of ionotropic GluR subunits (B). GC and AT content of GABAR subunits 3′-UTRs compared to the global mouse, hippocampal CA1 and neuropil 3′-UTRome (C) as well as ionotropic GluR subunits (D). Abbreviation: +represents the mean. P-values were calculated using the Mann-Whitney U-test, **p < 0.01, ****p < 0.0001.
To date, several GABAR subunits, scaffold, auxiliary and GABAR transport proteins have been detected as targets for RBPs by iCLIP or RIP (Tables 1, 2). Among those, known translation regulators such as fragile X mental retardation protein (FMRP), Pumilio1, 2, 4E-T as well as CPEB1 and 4 all bind GABAR subunit mRNAs. However, how these RBPs act together to locally control the expression of GABAR subunits in dendrites is still unknown. Future studies are clearly needed to unravel the role of RBP mediated protein expression control.
Translation is a multistep process that is regulated by versatile proteins (Jackson et al., 2010). Different sequence features of the mRNA that influence translation activity and association with ribosomal polysomes have been characterized in human cell lines (Floor and Doudna, 2016). In detail, the length and structural stability of the 3′-UTR, the number of miRNA binding sites as well as AU elements in the 3′-UTR are main drivers of translation activity located at the 3′-end of the untranslated region. An increase in these features is associated with decreased translation activity in non-neuronal cells (Floor and Doudna, 2016) as well as nerve cells (Blair et al., 2017). For GABAR subunit 3′-UTRs, we observed an increase in 3′-UTR length and AT content (Figures 1A,C,D). These results suggest that translation of these subunits is strongly regulated. Supportive for this idea is the finding that GABAR subunit mRNAs are recognized and subsequently bound by different RBPs (Table 1). In the last decade, several studies revealed that RBPs control translation of their target mRNAs (Hentze et al., 2018). One extensively studied example is the FMRP. FMRP mediated translational control is crucial for neuronal homeostasis and function since loss-of-function leads to severe neurological impairments in synaptic plasticity which cause intellectual disability and social deficits hallmarked for autism spectrum disorders (Bassell and Warren, 2008; Darnell and Klann, 2013). Furthermore, recent studies showed that FMRP is needed for proper differentiation of neuronal stem cells (Castrén et al., 2005; Gao et al., 2018). FMRP has been shown to co-migrate with translationally active ribosomal polysomes (Stefani et al., 2004). However, this finding was challenged by the same study showing that polysomal co-migration is detergent sensitive (Stefani et al., 2004). A mechanistic study combining in vitro assays and cryoelectron microscopy reported that FMRP inhibits translation through binding to the ribosomal intersubunit space thereby precluding binding of tRNAs and translation elongation factors (Chen et al., 2014). A transcriptome-wide screen for FMRP targets associated with polysomes identified mRNAs coding for subunits of the GABAB receptor complex (Darnell et al., 2011; see Table 1). Moreover, a recent study showed that the GABAA receptor subunit δ was downregulated in an FMRP knock-out mouse model (Gantois et al., 2006). These findings suggest that FMRP may regulate selected subunits of the GABAB and/or GABAA receptor, most likely at the translational level. Another known translation regulator is Pumilio2 (Pum2). For Pum2, it was shown that it represses translation by competing with the eukaryotic initiation factor (eIF4E) for mRNA 5′-cap binding (Cao et al., 2010), an essential step to start translation initiation (Jackson et al., 2010). Moreover, Pum2 is able to form a complex with the miRNA binding protein Argonaute (Ago) and the eukaryotic translation elongation factor 1A to repress translation elongation (Friend et al., 2014). Next to its role as translation regulator, Pum2 regulates transcript stability through recruitment of the polyA deadenylase complex CCR4-NOT (Van Etten et al., 2012), which is the major protein complex to induce RNA degradation (Collart, 2016). Based on a published iCLIP dataset, Pum2 is able to bind subunits of the GABAA and GABAB receptor (Table 1). Interestingly, double knockdown of Pumilio1 and 2 lead to a decrease in the mRNA levels of certain GABAR subunits (Zhang et al., 2017) indicating that they may be regulated posttranscriptionally by Pumilio proteins. Another RBP that impacts the expression of GABAA receptor subunits, is the non-octamer, POU-domain DNA-binding protein (NONO, also known as p54NRB). NONO belongs to the family of polypyrimidine tract-binding protein-associated splicing factors that are known to regulate various aspects of the RNA lifecycle including transcription regulation, splicing, RNA processing and RNA transport (Yarosh et al., 2015). Interestingly, mutations in the NONO locus causes intellectual disability in humans (Mircsof et al., 2015). Moreover, the authors found that the GABAA receptor-mediated inhibition is mainly affected when NONO is depleted (Mircsof et al., 2015) suggesting that this RBP regulates directly or indirectly the expression of the GABAA receptor. Nonetheless, it is widely unknown which GABAR subunits are translationally regulated. However, the binding of RBPs that are known to control RNA metabolism and translation, clearly suggests the existence of posttranscriptional gene regulation mechanisms for GABARs.
It is commonly accepted that the 3′-UTR allows for translational regulation of mRNAs. Research in the last years, however, has shown that the coding sequence (CDS) can also regulate protein synthesis rate, protein folding and protein complex assembly (Hanson and Coller, 2018). Dynamic translation regulation mediated by the CDS became experimentally accessible with the emergence of deep sequencing technologies and ribosome profiling protocols (Ingolia et al., 2009). Studies in cell lines and cultured neurons revealed that longer CDS are associated with translationally active “heavy” polyribosomes; most likely because a longer CDS can accumulate more ribosomes (Floor and Doudna, 2016; Blair et al., 2017). Interestingly, subunits of the GABAAR receptor complex display a shorter CDS compared to ionotropic GluR subunits (Figure 2A) suggestive for differences in translation activity. Another exciting possibility to regulate protein synthesis rate and output is the usage of synonymous codons. Twenty-one amino acids are encoded by 64 codons including three stop codons in the eukaryotic genome (Alberts et al., 2014). This degeneration of the genetic code leads to a codon bias, the preferred usage of certain codons over others to encode the same amino acid. Research in the last decades has shown that the usage bias is not random, but in contrast is driven and influenced by certain features such as translation activity, mRNA stability, protein folding, protein assembly and transcription factor binding (Grantham et al., 1980; Stergachis et al., 2013; Hanson and Coller, 2018). Codons can influence translation speed (Sørensen and Pedersen, 1991) most likely through the levels of cognate and near-cognate tRNAs (Anderson, 1969; Zhang and Ignatova, 2011; Fedyunin et al., 2012; Yu et al., 2015; Hanson and Coller, 2018). Since the nascent chain initiates folding already in the ribosomal exit tunnel (Lu and Deutsch, 2005), the elongation rate can also influence protein folding and, thereby, the protein conformation as it has been shown for the Cystic Fibrosis Transmembrane Regulator (CFTR) in mammalian cells (Kirchner et al., 2017). In line with this finding, Yu et al. (2015) showed using an in vitro translation system that codon usage determines co-translational folding through variation in the elongation rate. In particular for a multi-domain protein, it has been suggested that cluster of rare codons flank the parts of the mRNA that code for protein domains. Thus, ribosomes attenuate at these sites allowing the nascent domains to fold first to prevent misfolding (Schieweck et al., 2016; Hanson and Coller, 2018). Protein domains, that are encoded by the downstream mRNA, can then interact with already folded protein substructures to form a functional complex. Moreover, codon usage dependent protein folding can also influence protein specificity, which was reported for the Multi-Drug Resistance 1 protein (MDR1). A silent mutation in a rare codon changes the specificity of MDR1 (Kimchi-sarfaty et al., 2007). Together, these results strongly indicate that dynamics in the translation elongation rate determine trajectories of (co-)translational folding. Based on these results, an intriguing question raises: can codon usage influence protein folding of transmembrane proteins such as subunits of the GABAA receptor? Interestingly, GABAAR subunits contain more transmembrane helices compared to ionotropic GluR subunits (Figure 2B). This suggests that GABAAR subunits may need more variation in translation speed to allow co-translational folding than ionotropic GluR subunits. Furthermore, GABAAR subunits differ in their codon usage compared to GluR subunits (Figure 2C). Overall, the codon usage profiles between the two receptor groups are similar. For some codons, however, we detected significant differences in their frequency (Figures 2D,E). Interestingly, impaired translation of AGA codons leads to neurodegeneration in a mouse model (Ishimura et al., 2014). Moreover, GABAAR and GluR subunits exploit different stop codons. While GABAAR subunit mRNAs display an almost 1:1:1 ratio, GluR subunits prefer the TGA stop codon that yields the highest readthrough potential in mammalian cell lines (Howard et al., 2000; Bidou et al., 2004; Loughran et al., 2014; Manuvakhova et al., 2014). In addition to co-translational folding, the assembly of large protein complexes can also occur co-translationally (Balchin et al., 2016). It has been shown that this process is crucial for the complex formation in eukaryotic cells (Shiber et al., 2018). It is tempting to speculate that for large neuronal protein complexes such as GABAA receptors, a similar mechanism exists to ensure proper protein-protein interaction. Of note, codon usage and optimality differ dramatically in their impact on RNA stability comparing neurons and non-neuronal cells (Burow et al., 2018). Therefore, a thorough analysis of the neuronal translatome and tRNAome is needed to understand the impact of codon usage on GABAA receptor functioning.
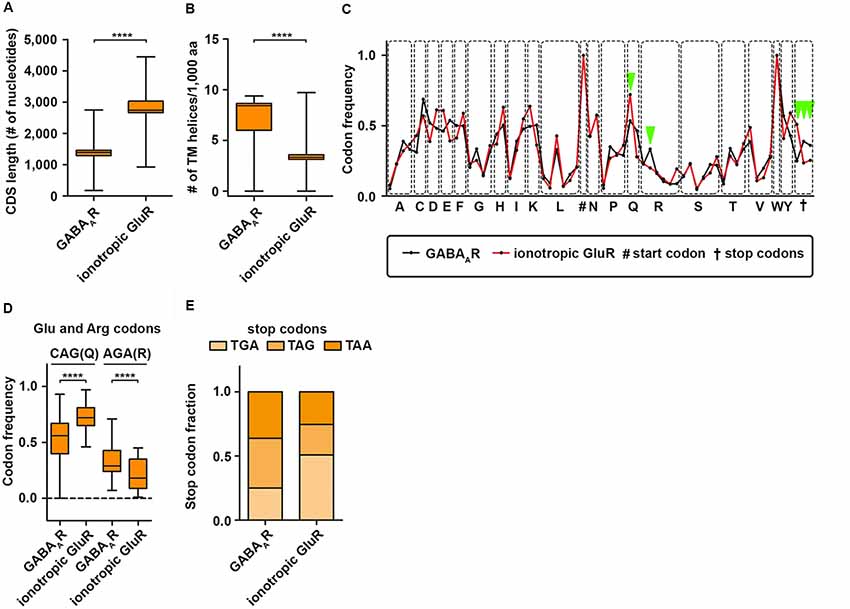
Figure 2. GABAA receptor codon usage differ from ionotropic glutamate receptors. CDS length (A) and the number of transmembrane (TM) helices (B) in GABAAR and ionotropic GluR subunits. (C) Codon usage frequency of GABAAR and GluR for 20 amino acids and stop codons. Dots represent synonymous codons. (D) Codon frequency for CAG (Q) and AGA (R). (E) Relative fraction of stop codon usage between GABAAR and GluR subunits. Abbreviations: CDS, coding sequence; aa, amino acid. P-values were calculated using the Mann-Whitney U-test, ****p < 0.0001.
To sum up, findings from different model organisms and cells demonstrate that translation is a highly dynamic process necessary for many aspects of the protein life cycle. For GABAA receptors, it is widely unknown: (i) whether and how they are translationally regulated; and (ii) whether co-translational folding/assembly is necessary for proper GABAR function. However, our bioinformatic predictions suggest that for some aspects, GABAR are prone to be subject to posttranscriptional regulation. Future studies will be clearly needed to unravel the dynamics and regulatory factors of their translation.
Since the discovery of LTP by Bliss and Lomo (1973), numerous studies have unraveled the plasticity of excitatory synapses in the brain aiming to explain the mechanism of learning and memory formation (Kandel et al., 2014). However, how inhibitory synapses undergo structural and molecular plasticity has been widely overlooked for some time (Gaiarsa and Ben-Ari, 2006). One of the first examples that inhibitory synapses show long-term plasticity was a study on Purkinje cells in the cerebellum published in 1998 (Aizenman et al., 1998). Since that time, various studies have addressed the mechanisms of how inhibitory LTP is conveyed (Castillo et al., 2011). Interestingly, in some aspects, inhibitory and excitatory LTP share similar mechanisms including the exchange of synaptic receptors (de Luca et al., 2017) as well as the importance of scaffold proteins for LTP (Petrini et al., 2014). In this context, it was shown that clustering of Gephyrin (Gphn), the major scaffold protein for inhibitory synapses (Tyagarajan and Fritschy, 2014), is essential for GABAA receptor surface dynamics and iLTP (Petrini et al., 2014). In line with its importance for iLTP, Gphn is posttranslationally modified in response to neuronal activity (Flores et al., 2015; Ghosh et al., 2016), which may represent a molecular hub to control inhibitory transmission. Arguably, one of the most impressive examples showing the dynamics of inhibitory synapse formation is the study by Oh et al. (2016). Upon GABA stimulation, newly formed Gphn cluster appear that are the structural basis for inhibitory synapse formation (Tyagarajan and Fritschy, 2014). Based on our bioinformatic predictions (Figures 1, 2) and RBP target screens (Tables 1, 2), it is tempting to speculate that the appearance of Gphn clusters upon GABA stimulation requires mRNA transport and, subsequently, translation. We propose that these mechanisms are necessary for inhibitory synapse formation (Figure 3). In general, future studies are clearly necessary to address the importance of posttranscriptional gene regulation for GABAergic synaptic transmission. Therefore, it needs to be investigated: (i) which GABAR component is regulated by RBPs; (ii) whether their expression is regulated at the translation, splicing and/or stability level; and (iii) whether their posttranscriptional regulation occurs locally at the synapse. Unraveling the role of RBPs in neuronal inhibition will clearly improve our understanding how neuronal networks are coordinated to find the balance between excitation and inhibition.
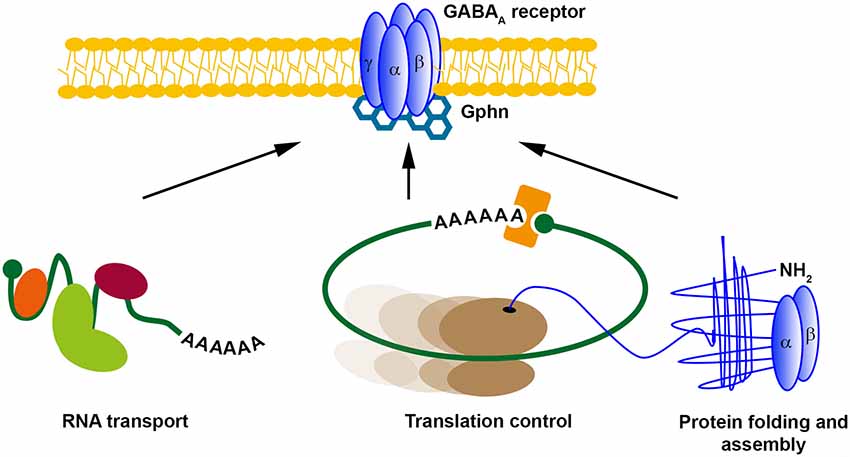
Figure 3. Possible posttranscriptional regulation mechanisms for GABAA receptors. Different posttranscriptional regulatory mechanisms exist. RNA transport, translational control and (co-translational) protein folding and assembly control local protein expression. We propose that GABARs might be regulated at inhibitory synapses in a similar manner. Abbreviation: Gphn, Gephyrin.
For analysis, 3′-UTR sequences and length of transmembrane domains were extracted from the EMSEMBL database (genome assembly GRCm38.p6) using the Gene Ontology ID “GO:0016917” for GABARs, “GO:0008066” for glutamate receptors and “GO:0004970” for ionotropic glutamate receptors. Only annotated mRNA isoforms were analyzed. Statistics were calculated using GraphPad Prism (version 5; GraphPad, San Diego, CA, USA).
All datasets generated for this study are included in the manuscript.
RS and MK conceived, executed and discussed the research that is presented in this article. RS generated the figures, the table and wrote the manuscript. RS and MK edited together.
This work was supported by the DFG (FOR2333, SPP1738 to MK) and the Boehringer Ingelheim Fonds (to RS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We apologize to all colleagues whose work could not be cited or discussed owing to space constraints. We thank members of the Kiebler lab for critical comments.
Aizenman, C. D., Manis, P. B., and Linden, D. J. (1998). Polarity of long-term synaptic gain change is related to postsynaptic spike firing at a cerebellar inhibitory synapse. Neuron 21, 827–835. doi: 10.1016/s0896-6273(00)80598-x
Alberts, B., Johnson, A., Lewis, J., Morgan, D., Raff, M., Roberts, K., et al. (2014). Molecular Biology of the Cell, 6th Edn. New York and Abingdon: Garland Science.
Anderson, W. F. (1969). The effect of tRNA concentration on the rate of protein synthesis. Proc. Natl. Acad. Sci. U S A 62, 566–573. doi: 10.1073/pnas.62.2.566
Andreassi, C., and Riccio, A. (2009). To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 19, 465–474. doi: 10.1016/j.tcb.2009.06.001
Balchin, D., Hayer-Hartl, M., and Hartl, F. U. (2016). In vivo aspects of protein folding and quality control. Science 353:aac4354. doi: 10.1126/science.aac4354
Bassell, G. J., and Warren, S. T. (2008). Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214. doi: 10.1016/j.neuron.2008.10.004
Bidou, L., Hatin, I., Perez, N., Allamand, V., Panthier, J. J., and Rousset, J. P. (2004). Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 11, 619–627. doi: 10.1038/sj.gt.3302211
Bingol, B., and Schuman, E. M. (2006). Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441, 1144–1148. doi: 10.1038/nature04769
Blair, J. D., Hockemeyer, D., Doudna, J. A., Bateup, H. S., and Floor, S. N. (2017). Widespread translational remodeling during human neuronal differentiation. Cell Rep. 21, 2005–2016. doi: 10.1016/j.celrep.2017.10.095
Bliss, T. V. P., and Lomo, T. (1973). Long-lasting potentiation of synaptic transmission in the dentate are of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 331–356. doi: 10.1113/jphysiol.1973.sp010273
Briese, M., Saal-Bauernschubert, L., Ji, C., Moradi, M., Ghanawi, H., Uhl, M., et al. (2018). hnRNP R and its main interactor, the noncoding RNA 7SK, coregulate the axonal transcriptome of motoneurons. Proc. Natl. Acad. Sci. U S A 115, E2859–E2868. doi: 10.1073/pnas.1721670115
Burow, D. A., Martin, S., Quail, J. F., Alhusaini, N., Coller, J., and Cleary, M. D. (2018). Attenuated codon optimality contributes to neural-specific mRNA decay in Drosophila. Cell Rep. 24, 1704–1712. doi: 10.1016/j.celrep.2018.07.039
Cajigas, I. J., Tushev, G., Will, T. J., tom Dieck, S., Fuerst, N., and Schuman, E. M. (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466. doi: 10.1016/j.neuron.2012.02.036
Cao, Q., Padmanabhan, K., and Richter, J. D. (2010). Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 16, 221–227. doi: 10.1261/rna.1884610
Castillo, P., C.Q, C., and Carroll, R. C. (2011). Long-term synaptic plasticity at inhibitory synapses. Curr. Opin. Neurobiol. 21, 328–338. doi: 10.1016/j.conb.2011.01.006
Castrén, M., Tervonen, T., Kärkkäinen, V., Heinonen, S., Castrén, E., Larsson, K., et al. (2005). Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. U S A 102, 17834–17839. doi: 10.1073/pnas.0508995102
Chen, E., Sharma, M. R., Shi, X., Agrawal, R. K., and Joseph, S. (2014). Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417. doi: 10.1016/j.molcel.2014.03.023
Collart, M. A. (2016). The Ccr4-not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 7, 438–454. doi: 10.1002/wrna.1332
Costa-Mattioli, M., and Monteggia, L. M. (2013). mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat. Neurosci. 16, 1537–1543. doi: 10.1038/nn.3546
Darnell, J. C., and Klann, E. (2013). The translation of translational control by FMRP: therapeutic targets for FXS. Nat. Neurosci. 16, 1530–1536. doi: 10.1038/nn.3379
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y. S., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi: 10.1016/j.cell.2011.06.013
de Luca, E., Ravasenga, T., Petrini, E. M., Polenghi, A., Nieus, T., Guazzi, S., et al. (2017). Inter-synaptic lateral diffusion of GABAA receptors shapes inhibitory synaptic currents report inter-synaptic lateral diffusion of GABAA receptors shapes inhibitory synaptic currents. Neuron 95, 63–69.e5. doi: 10.1016/j.neuron.2017.06.022
Doyle, M., and Kiebler, M. A. (2011). Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30, 3540–3552. doi: 10.1038/emboj.2011.278
Fan, X. C., and Steitz, J. A. (1998). Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO 17, 3448–3460. doi: 10.1093/emboj/17.12.3448
Fedyunin, I., Lehnhardt, L., Böhmer, N., Kaufmann, P., Zhang, G., and Ignatova, Z. (2012). TRNA concentration fine tunes protein solubility. FEBS Lett. 586, 3336–3340. doi: 10.1016/j.febslet.2012.07.012
Flexner, J., Flexner, L., and Stellar, E. (1963). Memory in mice as affected by intracerebral puromycin. Science 141, 57–59. doi: 10.1126/science.141.3575.57
Floor, S. N., and Doudna, J. A. (2016). Tunable protein synthesis by transcript isoforms in human cells. Elife 5:e10921. doi: 10.7554/eLife.10921
Flores, C. E., Nikonenko, I., Mendez, P., Fritschy, J., and Tyagarajan, S. K. (2015). Activity-dependent inhibitory synapse remodeling through gephyrin phosphorylation. Proc. Natl. Acad. Sci. U S A 112, 65–72. doi: 10.1073/pnas.1411170112
Frey, U., and Morris, R. G. M. (1997). Synaptic tagging and long-term potentiation. Nature 385, 533–536. doi: 10.1038/385533a0
Friend, K., Campbell, Z. T., Cooke, A., Kroll-Conner, P., Wickens, M. P., and Kimble, J. (2014). A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19, 176–183. doi: 10.1038/nsmb.2214
Fritzsche, R., Karra, D., Bennett, K. L., Ang, F., Heraud-Farlow, J. E., Tolino, M., et al. (2013). Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 5, 1749–1762. doi: 10.1016/j.celrep.2013.11.023
Gaiarsa, J., and Ben-Ari, Y. (2006). “Long-term plasticity at inhibitory synapses: a phenomenon that has been overlooked,” in The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology, eds J. T. Kittler and S. J. Moss (Boca Raton, FL: CRC Press/Taylor & Francis).
Gantois, I., Vandesompele, J., Speleman, F., Reyniers, E., D’Hooge, R., Severijnen, L. A., et al. (2006). Expression profiling suggests underexpression of the GABAA receptor subunit δ in the fragile X knockout mouse model. Neurobiol. Dis. 21, 346–357. doi: 10.1016/j.nbd.2005.07.017
Gao, Y., Li, Y., Stackpole, E. E., Zhao, X., Richter, J. D., Liu, B., et al. (2018). Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. U S A 115, E11397–E11405. doi: 10.1073/pnas.1809588115
Ghosh, H., Auguadri, L., Battaglia, S., Thirouin, Z. S., Zemoura, K., Dieter, A., et al. (2016). Several posttranslational modifications act in concert to regulate gephyrin scaffolding and GABAergic transmission. Nat. Commun. 7:13365. doi: 10.1038/ncomms13365
Grantham, R., Gautier, C., Gouy, M., Mercier, R., and Pavé, A. (1980). Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 8, r49–r62. doi: 10.1093/nar/8.1.197-c
Hanson, G., and Coller, J. (2018). Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30. doi: 10.1038/nrm.2017.91
Hentze, M. W., Castello, A., Schwarzl, T., and Preiss, T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341. doi: 10.1038/nrm.2017.130
Heraud-Farlow, J. E., and Kiebler, M. A. (2014). The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 37, 470–479. doi: 10.1016/j.tins.2014.05.009
Heraud-Farlow, J. E., Sharangdhar, T., Li, X., Pfeifer, P., Tauber, S., Orozco, D., et al. (2013). Staufen2 regulates neuronal target RNAs. Cell Rep. 5, 1511–1518. doi: 10.1016/j.celrep.2013.11.039
Hershkowitz, M., Wilson, J. E., and Glassman, E. (1975). The incorporation of radioactive amino acids into brain subcellular proteins during training. J. Neurochem. 25, 687–694. doi: 10.1111/j.1471-4159.1975.tb04389.x
Howard, M. T., Shirts, B. H., Petros, L. M., Flanigan, K. M., Gesteland, R. F., and Atkins, J. F. (2000). Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 48, 164–169. doi: 10.1002/1531-8249(200008)48:2%3C164::AID-ANA5%3E3.0.CO;2-B
Huppertz, I., Attig, J., D’Ambrogio, A., Easton, L. E., Sibley, C. R., Sugimoto, Y., et al. (2014). iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287. doi: 10.1016/j.ymeth.2013.10.011
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–323. doi: 10.1126/science.1168978
Ishimura, R., Nagy, G., Dotu, I., Zhou, H., Yang, X.-L., Schimmel, P., et al. (2014). RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459. doi: 10.1126/science.1249749
Jackson, R. J., Hellen, C. U. T., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi: 10.1038/nrm2838
Jung, H., Gkogkas, C. G., Sonenberg, N., and Holt, C. E. (2014). Remote control of gene function by local translation. Cell 157, 26–40. doi: 10.1016/j.cell.2014.03.005
Kanai, Y., Dohmae, N., and Hirokawa, N. (2004). Kinesin transports RNA. Neuron 43, 513–525. doi: 10.1016/j.neuron.2004.07.022
Kandel, E. R., Dudai, Y., and Mayford, M. R. (2014). The molecular and systems biology of memory. Cell 157, 163–186. doi: 10.1016/j.cell.2014.03.001
Khoutorsky, A., Yanagiya, A., Gkogkas, C. G., Fabian, M., Prager-Khoutorsky, M., Cao, R., et al. (2013). Control of synaptic plasticity and memory via suppression of poly(A)-Binding protein. Neuron 78, 298–311. doi: 10.1016/j.neuron.2013.02.025
Kiebler, M. A., and Bassell, G. J. (2006). Neuronal RNA granules: movers and makers. Neuron 51, 685–690. doi: 10.1016/j.neuron.2006.08.021
Kimchi-sarfaty, C., Oh, J. M., Kim, I., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., et al. (2007). A silent polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528. doi: 10.1126/science.1135308
Kirchner, S., Cai, Z., Rauscher, R., Kastelic, N., Anding, M., Czech, A., et al. (2017). Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 15:e2000779. doi: 10.1371/journal.pbio.2000779
Klein, M. E., Castillo, P. E., and Jordan, B. A. (2015). Coordination between translation and degradation regulates inducibility of mGluR-LTD. Cell Rep. 10, 1459–1466. doi: 10.1016/j.celrep.2015.02.020
Krug, M., Lössner, B., and Ott, T. (1984). Anisomycin blocks the late phase of long-term potentiation in the dentate gyrus of freely moving rats. Brain Res. Bull. 13, 39–42. doi: 10.1016/0361-9230(84)90005-4
Lee, J. A., Damianov, A., Lin, C. H., Fontes, M., Parikshak, N. N., Anderson, E. S., et al. (2016). Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron 89, 113–128. doi: 10.1016/j.neuron.2015.11.025
Linden, D. J. (1996). A protein synthesis-dependent late phase of cerebellar long-term depression. Neuron 17, 483–490. doi: 10.1016/s0896-6273(00)80180-4
Loughran, G., Chou, M. Y., Ivanov, I. P., Jungreis, I., Kellis, M., Kiran, A. M., et al. (2014). Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938. doi: 10.1093/nar/gku608
Lu, J., and Deutsch, C. (2005). Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 12, 1123–1129. doi: 10.1038/nsmb1021
Manuvakhova, M., Keeling, K., Bedwell, D. M., Manuvakhova, M., Keeling, K., and Bedwell, D. M. (2014). Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. Cold Spring Harb. Lab. Press Oct. 1, 1044–1055. Available at: https://rnajournal.cshlp.org/content/6/7/1044.long.
Mendez, R., and Richter, J. D. (2001). Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529. doi: 10.1038/35080081
Mircsof, D., Langouët, M., Rio, M., Moutton, S., Siquier-pernet, K., Bole-Feysot, C., et al. (2015). Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat. Neurosci. 18, 1731–1736. doi: 10.1038/nn.4169
Murn, J., Zarnack, K., Yang, Y. J., Durak, O., Murphy, E. A., Cheloufi, S., et al. (2015). Control of a neuronal morphology program by an RNA-binding zinc finger protein, Unkempt. Genes Dev. 29, 501–512. doi: 10.1101/gad.258483.115
Oh, W. C., Lutzu, S., Castillo, P. E., and Kwon, H. B. (2016). De novo synaptogenesis induced by GABA in the developing mouse cortex. Science 353, 1037–1040. doi: 10.1126/science.aaf5206
Parras, A., Anta, H., Santos-Galindo, M., Swarup, V., Elorza, A., Nieto-González, J. L., et al. (2018). Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing. Nature 560, 441–446. doi: 10.1038/s41586-018-0423-5
Peng, S. S.-Y., Chen, C. A., Xu, N., and Shyu, A. (1998). RNA stabilization by the AU-rich element binding, HuR, an ELAV protein. EMBO J. 17, 3461–3470. doi: 10.1093/emboj/17.12.3461
Pernice, H. F., Schieweck, R., Kiebler, M. A., and Popper, B. (2016). mTOR and MAPK: from localized translation control to epilepsy. BMC Neurosci. 17:73. doi: 10.1186/s12868-016-0308-1
Petrini, E. M., Ravasenga, T., Hausrat, T. J., Iurilli, G., Olcese, U., Racine, V., et al. (2014). Synaptic recruitment of gephyrin regulates surface GABAA receptor dynamics for the expression of inhibitory LTP. Nat. Commun. 5:3921. doi: 10.1038/ncomms4921
Scheckel, C., Drapeau, E., Frias, M. A., Park, C. Y., Fak, J., Zucker-Scharff, I., et al. (2016). Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife 5:e10421. doi: 10.7554/eLife.10421
Schieweck, R., Popper, B., and Kiebler, M. A. (2016). Co-translational folding: a novel modulator of local protein expression in mammalian neurons? Trends Genet. 32, 788–800. doi: 10.1016/j.tig.2016.10.004
Sharangdhar, T., Sugimoto, Y., Heraud-Farlow, J., Fernández-Moya, S. M., Ehses, J., Ruiz de los Mozos, I., et al. (2017). A retained intron in the 3′-UTR of Calm3 mRNA mediates its Staufen- and activity-dependent localization to neuronal dendrites. EMBO Rep. 18, 1762–1774. doi: 10.15252/embr.201744334
Shashoua, V. E. (1976). Identification of specific changes in the pattern of brain protein synthesis after training. Science 193, 1264–1266. doi: 10.1126/science.959837
Shiber, A., Döring, K., Friedrich, U., Klann, K., Merker, D., Zedan, M., et al. (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272. doi: 10.1038/s41586-018-0462-y
Sørensen, M. A., and Pedersen, S. (1991). Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222, 265–280. doi: 10.1016/0022-2836(91)90211-n
Sosanya, N. M., Cacheaux, L. P., Workman, E. R., Niere, F., Perrone-Bizzozero, N. I., and Raab-Graham, K. F. (2015). Mammalian Target of Rapamycin (mTOR) tagging promotes dendritic branch variability through the capture of Ca2+/Calmodulin-dependent protein kinase II α (CaMKIIα) mRNAs by the RNA-binding Protein HuD. J. Biol. Chem. 290, 16357–16371. doi: 10.1074/jbc.M114.599399
Stefani, G., Fraser, C. E., Darnell, J. C., and Darnell, R. B. (2004). Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004
Stergachis, A. B., Haugen, E., Shafer, A., Fu, W., Vernot, B., Reynolds, A., et al. (2013). Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342, 1367–1372. doi: 10.1126/science.1243490
Steward, O., Wallace, C. S., Lyford, G. L., and Worley, P. F. (1998). Synaptic activation causes the mRNA for the leg Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21, 741–751. doi: 10.1016/s0896-6273(00)80591-7
Sugimoto, Y., Vigilante, A., Darbo, E., Zirra, A., Militti, C., D’Ambrogio, A., et al. (2015). hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519, 491–494. doi: 10.1038/nature14280
Tyagarajan, S. K., and Fritschy, J. M. (2014). Gephyrin: a master regulator of neuronal function? Nat. Rev. Neurosci. 15, 141–156. doi: 10.1038/nrn3670
Ule, J., Jensen, K. B., Ruggiu, M., Mele, A., Ule, A., and Darnell, R. B. (2003). Identifies nova-regutated RNA networks in the brain. Science 302, 1212–1215. doi: 10.1126/science.1090095
Van Etten, J., Schagat, T. L., Hrit, J., Weidmann, C. A., Brumbaugh, J., Coon, J. J., et al. (2012). Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 287, 36370–36383. doi: 10.1074/jbc.M112.373522
Wagnon, J. L., Briese, M., Sun, W., Mahaffey, C. L., Curk, T., Rot, G., et al. (2012). CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS Genet. 8:e1003067. doi: 10.1371/journal.pgen.1003067
Yang, G., Smibert, C. A., Kaplan, D. R., and Miller, F. D. (2014). An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron 84, 723–739. doi: 10.1016/j.neuron.2014.10.022
Yarosh, C. A., Iacona, J. R., Lutz, C. S., and Lynch, K. W. (2015). PSF: nuclear busy-body or nuclear facilitator? Wiley Interdiscip. Rev. RNA 6, 351–367. doi: 10.1002/wrna.1280
Yu, C. H., Dang, Y., Zhou, Z., Wu, C., Zhao, F., Sachs, M. S., et al. (2015). Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59, 744–754. doi: 10.1016/j.molcel.2015.07.018
Zahr, S. K., Yang, G., Kazan, H., Borrett, M. J., Yuzwa, S. A., Voronova, A., et al. (2018). A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520.e6–537.e6. doi: 10.1016/j.neuron.2017.12.045
Zhang, M., Chen, D., Xia, J., Han, W., Cui, X., Neuenkirchen, N., et al. (2017). Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev. 31, 1354–1369. doi: 10.1101/gad.298752.117
Keywords: posttranscriptional regulation, GABA receptors, synapse, co-translational folding/assembly
Citation: Schieweck R and Kiebler MA (2019) Posttranscriptional Gene Regulation of the GABA Receptor to Control Neuronal Inhibition. Front. Mol. Neurosci. 12:152. doi: 10.3389/fnmol.2019.00152
Received: 25 March 2019; Accepted: 29 May 2019;
Published: 25 June 2019.
Edited by:
Andrea Barberis, Istituto Italiano di Tecnologia, ItalyReviewed by:
Carlos B. Duarte, University of Coimbra, PortugalCopyright © 2019 Schieweck and Kiebler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rico Schieweck, cmljby5zY2hpZXdlY2tAbWVkLnVuaS1tdWVuY2hlbi5kZQ==; Michael A. Kiebler, bWtpZWJsZXJAbG11LmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.