- 1Central Laboratory and Key Laboratory of Neurophysiology, Linyi People’s Hospital, Shandong University, Linyi, China
- 2Department of Medicinal Chemistry, Key Laboratory of Chemical Biology (Ministry of Education), School of Pharmaceutical Sciences, Shandong University, Jinan, China
- 3Chemistry Department and Center for Diagnostics and Therapeutics, Georgia State University, Atlanta, GA, United States
- 4Department of Neurology, Linyi People’s Hospital, Shandong University, Linyi, China
Glioma is characterized by a high recurrence rate, short survival times, high rates of mortality and treatment difficulties. Surgery, chemotherapy and radiation (RT) are the standard treatments, but outcomes rarely improve even after treatment. With the advancement of molecular pathology, recent studies have found that the development of glioma is closely related to various epigenetic phenomena, including DNA methylation, abnormal microRNA (miRNA), chromatin remodeling and histone modifications. Owing to the reversibility of epigenetic modifications, the proteins and genes that regulate these changes have become new targets in the treatment of glioma. In this review, we present a summary of the potential therapeutic targets of glioma and related effective treating drugs from the four aspects mentioned above. We further illustrate how epigenetic mechanisms dynamically regulate the pathogenesis and discuss the challenges of glioma treatment. Currently, among the epigenetic treatments, DNA methyltransferase (DNMT) inhibitors and histone deacetylase inhibitors (HDACIs) can be used for the treatment of tumors, either individually or in combination. In the treatment of glioma, only HDACIs remain a good option and they provide new directions for the treatment. Due to the complicated pathogenesis of glioma, epigenetic applications to glioma clinical treatment are still limited.
Introduction
Glioma is the most common form of primary malignant brain tumors, accounting for nearly 30% of all brain tumors, and also one of the most lethal (Chien et al., 2016). Some symptoms of glioma may be subtle and gradually worsen, while others may be present as an acute illness. The exact mechanism of occurrence of glioma remains unclear.
Recently, the World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS; Wen and Huse, 2017) categorized glioma into four grades. Grades I and II are considered low-grade glioma, whereas grades III and IV are high-grades, with the degree of malignancy increasing in the higher grades. The types of glioma include low-grade glioma (WHO II: mainly refers to diffuse astrocytoma, oligodendroglioma, and oligodendrocyte astrocytoma), anaplastic glioma (WHO III), glioblastoma (GBM, WHO IV), brain gliomatosis (pathologically astrocytoma-based, can be divided into WHO II, III, IV) and ependymomas (WHO II, III). Each grade has a relatively specific prognosis to guide the clinical treatment, but most of the glioma are WHO III grade anaplastic glioma and WHO grade IV GBM, which is devoid of cure with the current therapeutic options.
Unfortunately, most high-grade gliomas do not have a definitive cure. The current treatment for these high-grade tumors mainly focuses on surgical resection, followed by chemotherapy and radiotherapy. Temozolomide (TMZ) can traverse the blood-brain barrier and it is often used to treat these tumors. However, various side effects and adverse prognosis exist, such as the hematological toxicity (Stupp et al., 2005), and glioma have proven to be particularly resistant to radiotherapy and chemotherapy. The main mechanism of drug resistance is correlated with the O6-methyl guanine-DNA methyltransferase (MGMT; Hegi et al., 2005), which can bind to an alkyl compound on the O6 of DNA guanine and reduce it so that protein complexes cannot recognize the mismatched base pair, resulting in cell tolerance to O6 guanine methylation. As a result, the efficacy of these treatments is not promising and the prognosis of patients remains poor.
Therapies aimed to overcome these limitations have been presented in recent preclinical and clinical studies, including targeted molecular therapy, immunotherapy, gene therapy and stem cell therapy. The success of targeted molecular therapy for some tumor types, such as non-small cell lung cancer (Antonelli et al., 2016), malignant melanoma (Sullivan and Atkins, 2009) and chronic myeloid leukemia (Smirnikhina et al., 2016), has important guiding significance for targeted malignant glioma therapy.
With the advancement of the molecular pathology of malignant glioma, it is now evident that epigenetic abnormalities, including aberrant DNA methylation, abnormal microRNA (miRNA), chromatin remodeling and histone modifications, are closely related to the occurrence of glioma (Kondo et al., 2014; Hashizume, 2017; Figure 1). Multiple enzymes and genes that regulate the epigenetic modifications have become new targets not only for glioma treatments but for the treatment of other cancers as well.
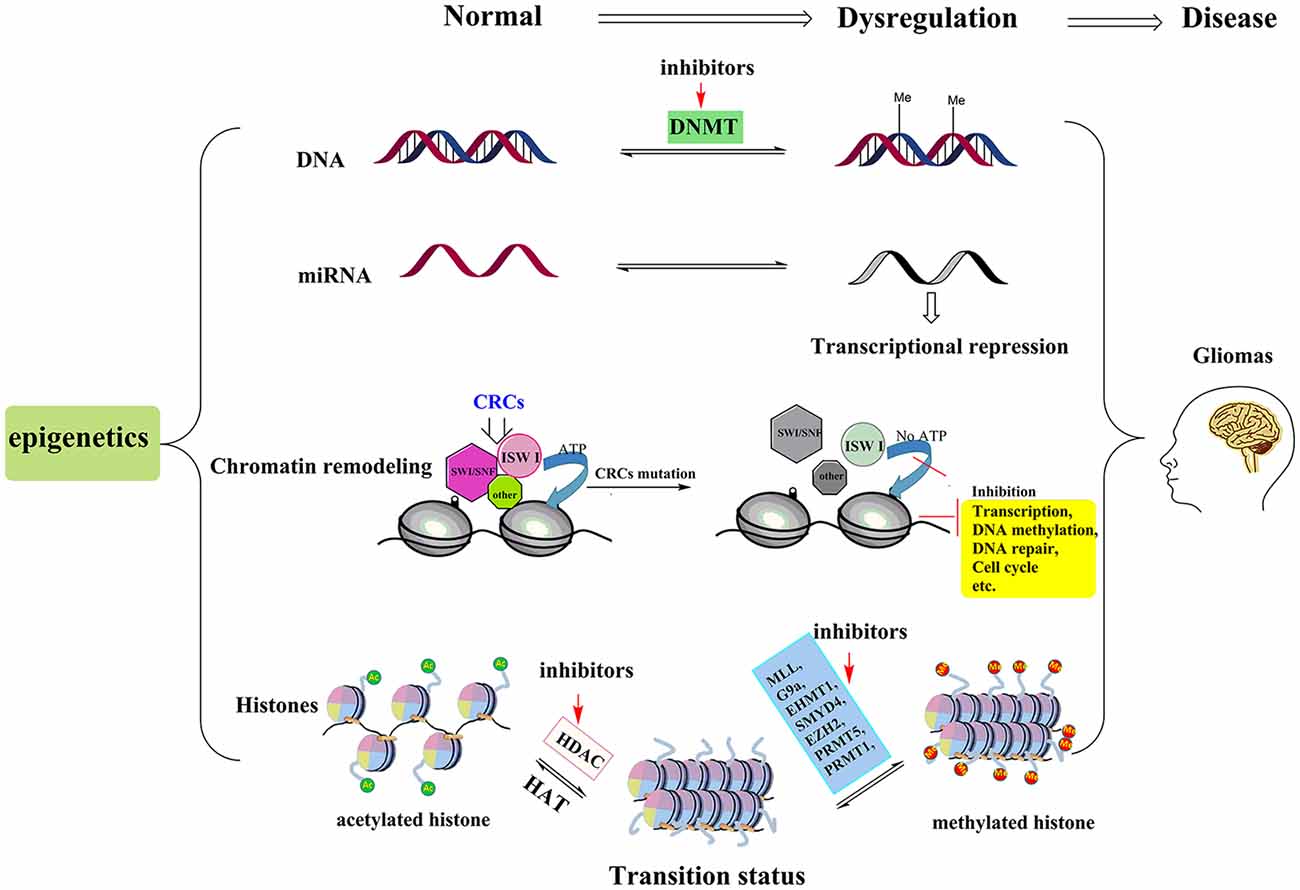
Figure 1. Pathogenesis and treatment options for glioma. The pathogenesis of glioma involves multiple processes. Here we show four epigenetic abnormalities (from top to bottom) linked to the occurrence of glioma: aberrant DNA methylation, abnormal microRNA (miRNA), chromatin remodeling and histone modifications. Among them, chromatin remodeling complexes (CRCs, such as SWI/SNF, ISW I, and other types of complexes) rely on the hydrolysis of ATP to provide energy to complete the chromatin structure changes. When the key proteins of the CSCs are mutated, this leads to abnormalities in the expression of tumor suppressor genes or those genes involved in cell cycle regulation, leading to the occurrence of glioma. In histone modification section, by inhibiting the activities of histone methyltransferases and histone deacetylase (HDAC), more sites in histone tails are free to be acetylated and this process can reverse the aberrant histone modifications, and then further suppress tumor cell proliferation and induce apoptosis. The red arrows represent potential epigenetic-based therapeutic approaches against glioma. For example, in DNA methylation section, DNMT inhibitor, 5-aza-20-deoxycytidine is the representative drug. In histone modifications, the HDACIs (vorinostat, panobinostat, valproic acid, etc.) and relevant histone methyltransferases inhibitors are potential treatment drugs in clinic.
This review summarizes the potential epigenetic-based therapeutic targets and related drugs for glioma. We illustrate how the epigenetic mechanisms dynamically regulate the pathogenesis of the disease. To provide information that will assist clinicians, we discuss preclinical and clinical trials of epigenetic-based treatments of glioma and include the results from these studies. This review also highlights the limitations of the current treatment and suggests future potential areas for research and advancements in the prognosis of the disease.
DNA Methylation
DNA methylation is one of the earliest discovered epigenetic modifications pathways. There are four possible sites for DNA methylation (Jones, 2012; Lovkvist et al., 2016)—the N-6 position of adenine, the N-4 position of cytosine, the N-7 position of guanine and the C-5 position of cytosine. In mammalian cells, DNA methylation occurs predominantly in cytosine of 5’-CpG-3’ to produce 5-methylcytosine (5mC). The methylation reaction is site-specific and it is performed by the enzyme DNMT with the help of methyl donor s-adenosylmethionine (SAM) as a co-factor (Bird, 2002). Currently, according to the differences in their structure and function, these DNMTs are divided into three categories, DNMT1, DNMT2, DNMT3 (DNMT3a and DNMT3b), with DNMT1 and DNMT3 as the representatives. DNMT1 is involved in the maintenance and extension of methylation, a necessity for non-CpG site methylation. DNMT3a and DNMT3b are de novo methyltransferases that methylate CpG. De novo methyltransferases may be involved in the regulation of cell growth and differentiation, in which DNMT3b plays an important role in tumor gene methylation (Chédin, 2011). DNMT2 binds to a specific site on DNA and its main target is tRNA (Kaiser et al., 2017). A number of studies (Hashimshony et al., 2003; Krausz et al., 2012; Shimooka et al., 2013) show that DNA methylation can contribute to changes in chromatin structure, DNA conformation, DNA stability, interactions between DNA and proteins, and can also control gene expression. DNA methylation has additionally become an important study of epigenetics and epigenetic genomics due to the close relationship between DNA methylation, human development and tumor diseases, particularly the transcriptional inactivation of tumor suppressor genes caused by methylation of CpG islands content.
Aberrant DNA Methylation in Glioma
Many studies (de Souza et al., 2018; Liao et al., 2018) have shown that the patterns of DNA methylation in glioma cells are different from those in normal cells. Most notably, the coexistence of extensive hypomethylation and CpG island hypermethylation are characteristic of tumor cells. The DNA methylation status of some relevant genes in glioma is therefore a good biomarker for clinical glioma diagnosis (Figure 1; Qu et al., 2013; Choudhury et al., 2015). Hypermethylation of the gene promoter region is the clearest epigenetic change that occurs in the tumor. The methylation status of the promoter region in the human genome regulates gene expression. Almost all housekeeping genes and roughly half of the tissue-specific genes are under the control of the promoter region. Under normal physiological conditions, most CpG islands are in a hypomethylated state; however, some housekeeping genes, such as DNA repair genes and tumor suppressor genes, are often hypermethylated in tumor tissues (Jin et al., 2017). This aberrant methylation can lead to gene transcription suppression and the loss of their biological function. DNA-5-hydroxymethylcytosine (5hmC), an epigenetic mark resulting from 5mC oxidation, correlates with the progression of glioma (Fernandez et al., 2018). Recent evidence (Kraus et al., 2012) has shown that DNA 5hmC negatively correlates with tumor grade. The CpG island methylator phenotype (G-CIMP) is also used as an indicator of glioma prognosis in infancy, pediatric and adults (Mack et al., 2014; Malta et al., 2018). Importantly, another study (Jha et al., 2014) found that pediatric GBM have a distinct methylome compared with that of adults, suggesting that the G-CIMP indicator of glioma prognosis in adult GBM cannot simply be extrapolated to pediatric GBM and there is a strong need for identification of separate prognostic markers.
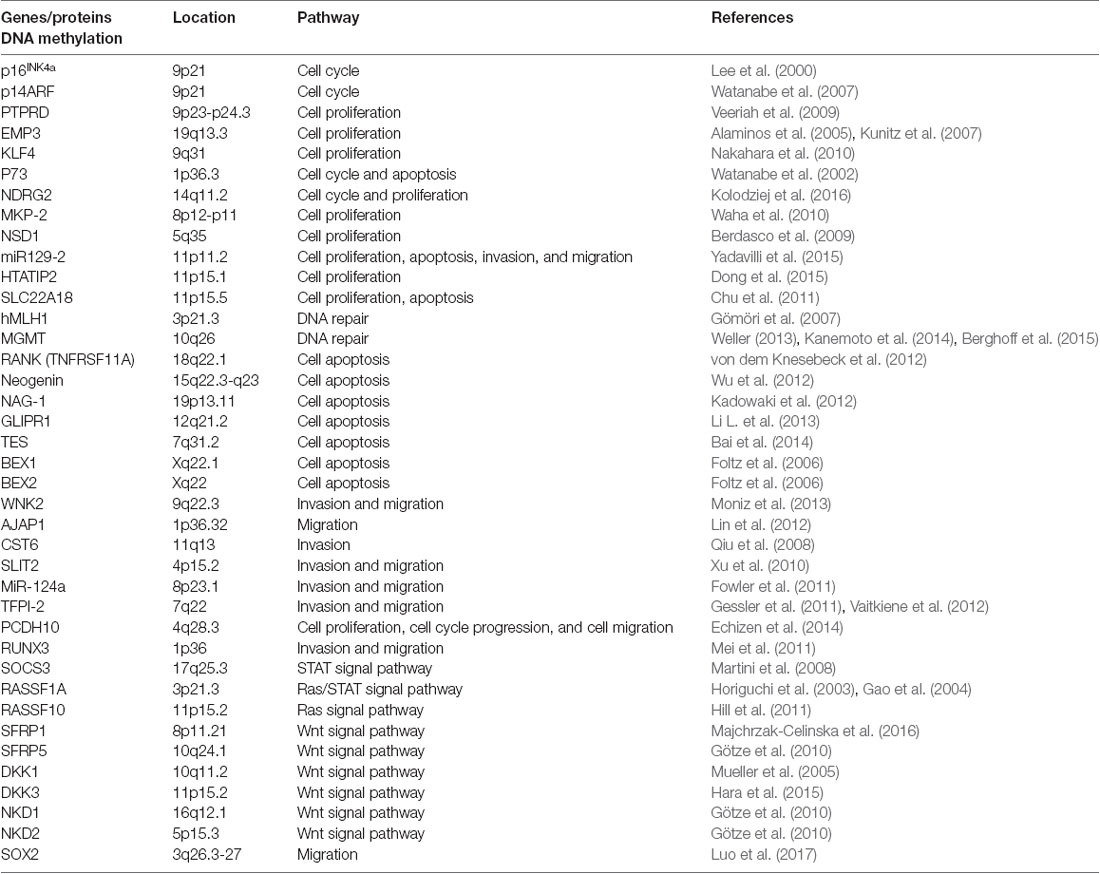
Abnormal DNA methylation is an important indicator of the inactivation of tumor suppressor genes. Many tumor suppressor genes in glioma have also been identified, including p16INK4a (Lee et al., 2000), p14ARF (Watanabe et al., 2007), MLH1 (Gömöri et al., 2007) and NDRG2 (Kolodziej et al., 2016; see Table 1). The p16INK4a gene maintains the dephosphorylated activating state of the retinoblastoma tumor suppressor protein (pRb) in the normal cyclinD-Rb pathway to control cell cycle progression. A high frequency (more than 50%) of homozygous p16INK4a gene deletion has been demonstrated in GBM tissues, and p16INK4a is altered in 80% of glioma cell lines. Therefore, restoration of p16INK4a would suppress cell proliferation and induce cell growth arrest (Lee et al., 2000). MGMT is a crucial DNA damage repair gene that can repair alkyl damage caused by BCNU. Esteller et al. (2000) found that MGMT promoter hypermethylation existed in approximately 40% of glioma tissues. The methylation level is related to the occurrence and prognosis of the tumor, which is more important than the prognosis of age and tumor grade (Mur et al., 2015). Besides, the methylation level of MGMT promoter is the most important indicator to assess TMZ sensitivity in glioma treatment, and down-regulated MGMT can substantially restore TMZ chemosensitivity in vitro and in vivo (Xipell et al., 2016; Yu et al., 2018). Except for the genes mentioned above, the CpG island methylation in gene promoter regions of p73 (Watanabe et al., 2002), LATS1, LATS2 (Jiang et al., 2006), and the genes that are listed in Table 1 are also closely related with the occurrence and development of glioma.
Isocitrate dehydrogenases 1 (IDH1) is not only a major source of NADPH in the human brain (Bleeker et al., 2010) but also in other body tissues (Kim et al., 2009). The mutations of methylation regulatory proteins IDH1/2 can be detected in most low-grade diffuse astrocytoma (75% mutation rate) and anaplastic astrocytoma (66% mutation rate) as well as oligodendroglioma, mixed promyelocytoma and secondary sex polymorphic neuroblastoma (GBM, 76% mutation rate). Noushmehr et al. (2010) found that the IDH1 mutation is highly correlated with the G-CIMP, and its mutation was associated with the prognosis of secondary GBM and positively correlated with the survival rate of patients (Rossetto et al., 2011). Studies have shown that the hypermethylated phenotype of IDH mutations causes insulator proteins to separate from IDH mutant binding sites (Flavahan et al., 2016). These free insulator proteins are then linked to the normally resting platelet-derived growth factor receptor (PDGFRA) oncogene promoter to form a complex that can further stimulate tumor cell growth. By using demethylated drugs to restore normal function in these insulator proteins, PDGFRA can inhibit the growth of IDH mutant astrocytoma cells (Flavahan et al., 2016). In another study, they associated known subtypes with specific alterations in NF1 and PDGFRA/IDH1 in order to provide a framework for the investigation of targeted therapies (Verhaak et al., 2010). As a critical gene of glioma, IDH has vast potential for the diagnosis, treatment and prognosis of glioma.
MicroRNA
MiRNAs are short non-coding endogenous RNAs that involve in post-transcriptional gene expression regulation in animals and plants (Bartel, 2004). miRNAs can target complementary regions of the 3’-UTR of the mRNA, inhibiting post-transcriptional processes or degrading mRNA and ultimately reducing protein levels (Pileggi et al., 2013). There are many miRNAs in the human genome, targeting tens of thousands of mRNAs.
MicroRNAs and Glioma
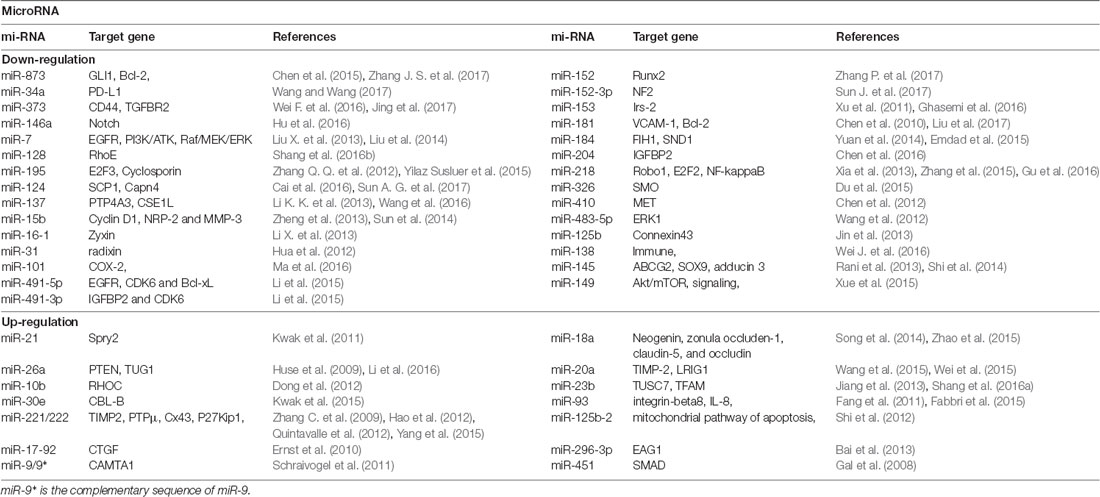
In recent years, some studies have shown that miRNAs play key roles in the transcriptional regulation and growth and proliferation of various tumor genes (Yan et al., 2017). Therefore, miRNA-based individual therapy and gene editing methods may play important roles in the diagnosis and treatment of glioma. It is currently estimated that about half of the miRNA genes are located in glioma cancer genes or their fragile sites (Table 2), and these miRNA genes can regulate 3% of the entire glioma tumor genes and 30% of the coding genes. Also, a single miRNA can simultaneously affect a hundred mRNAs of GBM (Berindan-Neagoe et al., 2014), whereas a single mRNA of glioma can be modulated by one or more miRNAs (Lakomy et al., 2011).
miRNAs play many critical roles in the progression of glioma diseases. In particular, miRNAs regulate the expression of cancer-related genes, participate in the regulation of tumorigenesis and regulatory pathways, regulate the differentiation of glioma stem cells, and are encoded by oncolytic viruses and involved in tumor processes. Zhang C. et al. (2012), for instance, found miR-221/222 positively correlated with the degree of glioma infiltration and cell invasion, whereas knockdown of miR-221/222 decreased cell invasion via modulating the levels of the TIMP3 target. Knockdown of miR-221/222 additionally increased TIMP3 expression and considerably inhibited tumor growth in a xenograft model. Another study indicated that the over-expression of miR-221/222 reduced p27kipl levels (Zhang C. et al., 2009). P27kipl prevented cell cycle from G1 to S phase by binding to CDK2 and cyclin E complexes. Therefore, down-regulated miR-221/222 can up-regulate p27kipl to inhibit tumor proliferation.
Chromatin Remodeling
Chromatin remodeling complexes (CRCs) have ATPase activity and they rely on the hydrolysis of ATP to provide energy to complete the chromatin structure changes (Stanton et al., 2017). Depending on the different subunits that can hydrolyze ATP, the complexes can be divided into SWI/SNF, ISW I and other types of complexes (Figure 1). The SWI/SNF complex and the ISW I complex family were the first to be found in yeast and Drosophila (Biegel et al., 2014). The human SWI/SNF complex is a polymer with many molecules, including BRG1, hBMR and tumor suppressor protein Hsnf5, which mainly activates gene transcription and is also involved in the recombination of immunoglobulin and TCR genes (Pulice and Kadoch, 2016). The ISW I complex family include three complexes—RSF, HuCHRAC and CAF1 (Loyola et al., 2003). RSF is a heterodimer that mainly consists of Hsnf-h, which is involved in transcription initiation (Sheu et al., 2010); HucHRAC contains Hsnf-2 h and chromatin assembly factor Hacf1, which is related to the maintenance of the heterochromatin replication status (Hanai et al., 2008); CAF1 is involved in chromatin assembly, altering the state of chromatin to correlate with DNA function (Endo et al., 2017). These complexes and related proteins are associated with activation and inhibition of transcription, DNA methylation, DNA repair and cell cycle.
Chromatin Remodeling and Glioma
Human diseases caused by abnormal chromatin remodeling are often due to mutations in the key proteins of the remodeling complex. This can lead to the failure of chromatin remodeling in which nucleosomes cannot be correctly positioned, preventing basic transcriptional machinery and the complexes that can repair DNA damage from accessing DNA, which can lead to aberrant gene expression. If these mutations lead to abnormalities of tumor suppressor genes or proteins that regulate cell cycle, they can finally lead to the occurrence of cancer (Marfella et al., 2008; Choi et al., 2015).
Liau et al. (2017) recently indicated that chromatin remodeling regulated GBM drug resistance. GBM stem cells (GSC) can reversibly change to a slow-cycling, long-lasting state when targeted by kinase inhibitors. Under this state, the notch signaling pathway is activated and histone demethylase KDM6A/B is significantly up-regulated. This leads to the removing of trimethylation of H3K27 in genome cis-regulatory region and further leads to the increased levels of H3K27Ac. Chromatin remodeling played a key role in this cellular shift, and this research provided a novel target for the development of effective treatments in the future. By targeting epigenetic and developmental pathways, it is possible to eradicate drug-resistant tumor cells and prevent disease recurrence. Another study (Xiao et al., 2017) revealed evidence demonstrating up-regulated chromatin remodeling factor lymphoid-specific helicase (LSH) promoted the development of glioma. Research (Xiao et al., 2017) indicates that the up-regulated transcription factor E2F1 and glycogen synthase kinase-3β (GSK-3β, an intact complex of E2F1) in astrocytomas and GBM were associated with the progression of glioma and correlated with LSH expression. The depletion of E2F1 decreased LSH expression and cell growth, while inhibition of GSK3β increased the enrichment of E2F1 to the LSH promoter, and increased LSH expression. Lipoprotein receptor-related protein 6 (LRP6), an upstream regulator of GSK3β signaling pathway, was also over expressed in glioma tissue. Knockdown of LRP6 reduced LSH expression level through decreased recruitment of E2F1 to the LSH promoter, finally leading to inhibition of cell growth. Taken together, a mechanistic link between LSH expression and activation of the LPR6/GSK3β/E2F1 axis in glioma illustrates a novel role of LSH in malignant astrocytomas and GBM. Understanding the roles of LSH in glioma progression will not only enrich our knowledge of glioma but also frame LSH as a potential therapeutic target for the treatment of these deadly brain cancers.
Histone Modifications
In the mammalian epigenome, histone modifications can occur in many ways. The basic unit of histone, called nucleosome, is an octamer that consists of two H2A, two H2B, two H3, two H4 and 147 base pairs wound outside of the composition (de Ruijter et al., 2003; Figure 1). The core histone has C-terminal and N-terminal binding regions. Of these, the N-terminal is particularly relevant, since lysine residue of the N-terminal extends out of the nucleosome and is accessible for modifications, including acetylation, methylation, phosphorylation, ubiquitination and ADP ribosylation. These modifications subsequently alter the expression of the gene without altering the base pair (Wang et al., 2014; Mathias et al., 2015). The total process, known as epigenetic regulation, involves a variety of enzymes. Scientists have classified these enzymes according to their functions (Liu et al., 2011): “writers” (enzymes that add groups such as methyl, acetyl and glycans), “erasers” (enzymes that remove post-translational modifications) and “readers” (enzymes that recognize these epigenetic markers and regulate epigenetic effects). The protein complexes that promote the movement of nucleosomes on chromatin are called “movers.” From this enormous pool, many therapeutic targets are derived as a single target or as combinations, apart from the most prominent DNMTs and histone deacetylases (HDACs).
Dysregulation of Histone Modifications in Glioma
Aberrant histone modifications can lead to transcriptional abnormalities in gene expression that eventually lead to the development and progression of glioma. Among the various histone modification proteins, two have attracted more attention than the others—HDACs, which cause histone deacetylation, and histone methyltransferases, which cause methylation at various sites of histone (Zang et al., 2014). Among the different classes of HDAC enzymes, HDAC1 (Wang et al., 2017), HDAC2, HDAC3 (Leng et al., 2016), HDAC5 and HDAC9 (Milde et al., 2010) have undergone significant changes in glioma cells. H3 acetylation levels are elevated in high-grade astrocytoma compared to low-grade medulloblastoma and normal brain tissues, as are the expression of HDAC5 and HDAC9 (Milde et al., 2010) in high-grade medulloblastoma. One survey demonstrated that mRNA levels of class II and IV HDACs were down-regulated in GBM compared to low-grade astrocytomas and normal brain tissues (Lucio-Eterovic et al., 2008). The use of HDAC inhibitors (HDACIs) for the treatment of cancer is an area of active investigation. In glioma treatment, HDACIs have been used for the treatment of GBM in combination with RT therapy and chemotherapy (Shi et al., 2016; Ghiaseddin et al., 2018). The anti-tumor mechanism of HDACIs includes blocking cell cycle and promoting cell differentiation, and inducing apoptosis and inhibiting angiogenesis, which can inhibit proliferation and apoptosis of various tumor cells (Marks and Breslow, 2007).
In glioma cells, histone methyltransferases G9a, EZH2, MLL1 and MLL2 (Chang et al., 2009; Cheung et al., 2012; Liu F. et al., 2013; Kondengaden et al., 2016; Wiese et al., 2016; Banasavadi-Siddegowda et al., 2018) regulated the methylation level of lysine located in histone (Table 3); these modifications were closely related to gene transcription regulation and genome integration (Heddleston et al., 2012; Zhou et al., 2016). Protein arginine methyltransferase 5 (PRMT5) is another candidate gene for the diagnosis and treatment of glioma, its nuclear expression correlates with poor survival in glioma patients. Banasavadi-Siddegowda et al. (2018) revealed that GBM cells treated with PRMT5 inhibitor mirrored the effects of PRMT5 knockdown, wherein it led to apoptosis of differentiated GBM cells and drove undifferentiated primary patient-derived GBM cells into a non-replicative senescent state.
Inhibiting the activities of histone methyltransferases or HDACs can suppress glioma cell proliferation and induce apoptosis (Sharma et al., 2010; Vargas et al., 2014), suggesting that the inhibitors of these proteins could be candidate drugs for the treatment of glioma. Recently, Ghildiyal and Sen (2017) reported that histone methyltransferase G9a that regulated H3K9 dimethylation has also correlated with the development and progression of glioma, and its inhibitors have also been reported as potential agents for the treatment of glioma (Guo et al., 2016).
It is worth noting that no post-translational modifications processes exist in isolation but rather act with mutual influence and coordination, usually referred to as histone crosstalk. In our earlier research (Zang et al., 2017), we found that simultaneously inhibiting the activity of HDAC and G9a would yield better effects than inhibiting single targets. In our anti-proliferation experiment, multiple cancer phenotypes including leukemia, prostatic carcinoma, hepatocellular carcinoma and pulmonary carcinoma and breast carcinoma were used in this study. Compared to the single target suppression effect, simultaneous inhibition of the activities for two protein targets showed a better anti-proliferation effect in parts of the tumor cell lines, such as breast carcinoma. As illustrated in Figure 1, under the cooperation of G9a inhibitor and HDACIs, more sites in histone tails are free to be acetylated. This acetylation status can activate cancer suppression gene transcription and alleviate the disease. Thus, developing high activity HDAC and G9a hybrid inhibitors is another effective route in targeted epigenetic therapy.
The Polycomb group (PcG) protein family is a group of gene regulatory factors that play a role in embryonic development (Zhao et al., 2017). They are divided into two protein complexes based on function, namely PRC1 and PRC2 (Collinson et al., 2016). PRC2 is a multi-protein complex responsible for the methylation of H3 at lysine 27 (H3K27Me). Zeste Gene Enhancer Homolog 2 (EZH2) is a catalytic subunit that constitutes the PRC2 protein complex (Collinson et al., 2016). Current studies (Orzan et al., 2011; Chen et al., 2017) indicated that EZH2 is over expressed in many tumor tissues, including glioma, and is closely related to the malignant progression, invasion and metastasis of the tumors. In the cellular level of research, EZH2 gene silencing technology or using EZH2 inhibitors prevented glioma cell proliferation (Kurmasheva et al., 2017). Therefore, focusing on EZH2 as a new target may pave a new way for the treatment of clinical glioma (Zhang Y. et al., 2017). In pediatric GBM, H3F3A involves two critical single-point mutations in the histone tail at lysine (K) 27 (K27M) and glycine (G) 34 (G34R/V) that are both involved with key regulatory post-transcriptional modifications (Schwartzentruber et al., 2012). The discovery of these molecular markers provides a basis for the diagnosis of this type of glioma and will lay the foundation for further diagnosis and treatment research.
Present Clinical Workflow
DNA methylation by DNMT leads to the gene silencing of tumor suppressor genes (Dammann et al., 2017). The inhibition of DNMTs can achieve reactivation of transcription of these critical genes (Castillo-Aguilera et al., 2017). Thus, the study of DNMT inhibitors has become a new bright spot in the treatment of glioma. Among the DNMT inhibitors that have now entered the clinical trials, 5-aza-2′-deoxycytidine (Chu et al., 2013) is the most representative. In tumor cells, 5-aza-2′-deoxycytidine is blended with DNA in the form of phosphate, and then inhibit DNMT activity, eventually leading to the desired low methylation status to exert antitumor effects (Sun et al., 2011). Currently the clinical research of 5-aza-2′-deoxycytidine is limited in leukemia (Roboz et al., 2018) and part of solid tumors (Garrido-Laguna et al., 2013; Fan et al., 2014; lung cancer, etc., not including glioma). For glioma treatment, most of the studies with 5-aza-2′-deoxycytidine are still in pre-clinical research stage (Oi et al., 2009; Zhang et al., 2014).
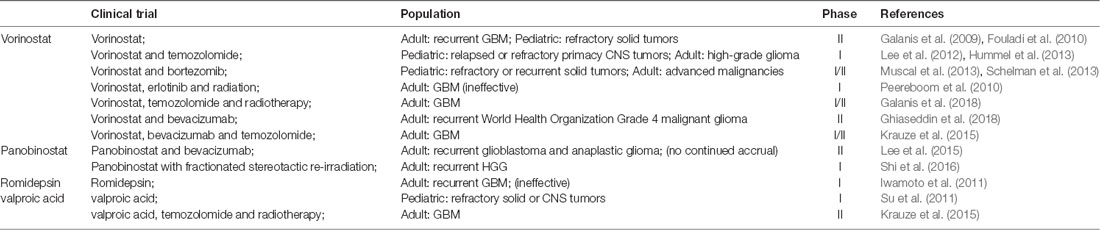
HDACIs can inhibit glioma oncogene transcription and have a variety of effects on cell life cycle. HDACIs can arrest cell division in G1 and G2 phases, induce cell differentiation and apoptosis (Hazane-Puch et al., 2016), destroy the combination of heat shock protein and substrate protein, promote degradation of oncoprotein, and also inhibit the growth and proliferation of glioma by inhibiting tumor angiogenesis (Pei et al., 2016). DNMT inhibitors and HDACIs can be used for the treatment of various tumors, either individually or as a synergistic combination (Xu et al., 2014; Pathania et al., 2016). In glioma, as a new therapeutic drug, HDACIs provide new directions for the treatment of glioma. Already many HDACIs have entered the phase I/II clinical trials (Table 4) alone or in combination with other chemotherapeutic agents such as TMZ and radiotherapy for the treatment of various types of glioma, including diffuse intrinsic pontine glioma (DIPG), progressive, or recurrent GBM (Lee et al., 2012; Krauze et al., 2015; Kim et al., 2017).
One phase I study conducted by the Children’s Oncology Group (COG) investigated vorinostat with TMZ in relapsed or refractory primary CNS tumors (Hummel et al., 2013). Five-day cycles of vorinostat in combination with TMZ were well tolerated in children with recurrent CNS malignancies, with myelosuppression as the dose-limiting toxicities (DLT). Accumulation of acetylated H3 in peripheral blood mononuclear cells (PBMC) was observed after administration of vorinostat. One phase II trial of vorinostat on recurrent GBM was reported by the North Central Cancer Treatment Group (Galanis et al., 2009). In this study, vorinostat monotherapy was well tolerated in patients with recurrent GBM, and there were obvious increases in acetylation levels of H2B, H3 and H4 after treatment. Microarray RNA analysis showed changes in genes regulated by vorinostat, such as up-regulation of E-cadherin.
With regards to panobinostat, one phase II study of panobinostat in combination with bevacizumab (BEV) was attempted in individuals with recurrent GBM and anaplastic glioma, but part of this study did not meet the criteria for continued accrual and was closed (Lee et al., 2015). Prior to closure, the treatment was reasonably well tolerated in both cohorts, but the addition of panobinostat to BEV did not significantly improve 6-month progression-free survival (PFS6) compared to historical controls of BEV monotherapy in either cohort. More preclinical evidences have shown that panobinostat may act as a radiosensitizer. A phase I trial combining panobinostat with stereotactic re-irradiation in patients with recurrent HGG has been reported (Shi et al., 2016). The results were more promising than panobinostat with BEV, with a PFS6 of 83% in the panobinostat and stereotactic re-irradiation therapy group, compared to 30.4% in the panobinostat with BEV group.
Another phase I study of valproic acid (VPA) in pediatric patients with refractory solid or CNS tumors was conducted by COG (Su et al., 2011). Histone hyperacetylation was observed in half of the patients at steady state. Krauze et al. (2015) recently conducted a phase II study of concurrent radiation therapy, TMZ, and VPA for patients with GBM. The results of this study demonstrated that the addition of VPA to concurrent RT/TMZ in patients with newly diagnosed GBM was well tolerated. VPA may result in improved outcomes compared to historical data and merits further study.
Overall, HDACIs as monotherapy or a combination therapy seem promising in improving prognosis in this difficulty to treat malignancy glioma (Table 4). For the possible toxicity that epigenetic drugs may present during treatment, improving the dosage regimen (Issa et al., 2004) or developing new epigenetic therapies or gaining knowledge of how to synchronize them with other treatment modalities are good choices to alleviate this toxic effect. So far, HDAC inhibitors, vorinostat and valproic acid can both combine with TMZ and/or RT to exert good effects in clinical trials to treat children with refractory or recurrent CNS malignancies or adult GBM (Lee et al., 2012; Hummel et al., 2013; Krauze et al., 2015). This is a good trend for our future clinical research. Vorinostat combined with erlotinib or panobinostat with BEV did not show obvious effects (Peereboom et al., 2010; Lee et al., 2015). Though the results are not optimistic, they provide valuable information for future research. In recent years, immunotherapy combined with other drugs has been a hot topic in cancer treatment, but few clinical trials were reported on combining the HDACIs with Gene-Mediated Cytotoxic Immunotherapy (G-MCI). Most G-MCI were preferred to combine with TMZ and standard of care (SOC) radiation after surgery, and survival outcomes were most notably improved in patients with minimal residual disease after gross total resection (Chiocca et al., 2011; Wheeler et al., 2016).
Although there are few clinical trials on the combination of glioma immunotherapy and epigenetics, many basic studies on immunotherapy and epigenetics of glioma were reported recently (Gallagher et al., 2017; Bhat et al., 2018). Studies have shown that tumor cells can use epigenetic mechanisms to alter their autoimmune origin and disrupt the process of recognition between tumor cells and the immune system. By DNA methylation or histone modifications, tumor cells can directly or indirectly down-regulate the expression levels of key molecules in the tumor immunoreaction process, thereby destroying the immune recognition and killing tumor cells (Maio et al., 2015). At present, the immunotherapeutic drugs that can combine with epigenetic drugs mainly include cytokines immunosuppressive agents, polypeptide vaccines, immunological adjuvants and tumor cell vaccine agents. Epigenetic drugs combined with tumor immunotherapy drugs will become an important research direction for the future treatment of tumors, and also provide new ideas for the treatment of glioma. With the development of improved medical standards, we believe more high-quality phase trials in newly diagnosed and recurrent GBM are imperative.
Conclusions and Perspectives
Glioma is a common primary malignant brain tumor with high recurrence rates, short survival times, high rates of mortality and treatment difficulties. For all grades of glioma patients, the largest range of safe resection remains the central step in current comprehensive treatment strategies (Kreth et al., 2013). Prior treatments of these tumors had taught us that conventional surgeries and chemo-radiotherapy protocols can only minimally improve the quality of life and slightly prolong the survival of some patients. The postoperative treatments, including radiation, chemotherapy, the dose and the cycle, should be implemented after comprehensive evaluation based on factors such as patient’s age, operation circumstances, histopathological classification and molecular characteristics (Jiang et al., 2016). Further investigations and reviews of the treatment strategies for malignant glioma are needed.
The genetic instability and heterogeneity of glioma are prominent. The mutual regulation mechanisms of related signal transduction pathways, which are not yet fully understood, are essential for the determination of therapeutic targets and drug development process. In the study of epigenetic therapy, multi-target combined inhibition is a critical concept in targeted drug development. In addition to single drug with multi-target inhibition, the combined use of multiple targeted drugs is also important, including those mentioned in this study (e.g., HDAC and G9a). However, even if the targeted drugs are tested in combination, the number of trials is quite large and pre-clinical pre-screening of drugs is therefore necessary. Whether the targeted drug can act on the expected target site and whether it can effectively inhibit the downstream signaling pathways, the potential toxic and side effects, are related to the safety and efficacy of targeted therapy. These issues need to be studied in depth.
Overall, epigenetic modifications are closely related to glioma proliferation, metastasis, invasion and prognosis. Various epigenetic modifications closely interact to participate in the occurrence and progression of glioma. Breakthroughs in the treatment of glioma require advances in scientific research, improvements in therapeutic technologies and protocols, and the development of diagnosis and treatment around individualized protocols. The epigenetic phenomena of glioma, including DNA methylation, abnormal microRNA, chromatin remodeling and histone modifications, have excellent potential significance and application to the diagnosis, treatment, and prognosis of glioma. Among the four fields, the methylation level of the gene promoter region can be taken as a guide for glioma diagnosis, and also related to the prognosis of the glioma (Mur et al., 2015), miRNAs play many critical roles in the progression of glioma (Zhang C. et al., 2012), the protein levels of enzymes that regulate histone modifications are candidates for the diagnosis of the glioma, and its inhibitors are good candidate drugs for the treatment of glioma (Sharma et al., 2010; Vargas et al., 2014). These phenomena might also help monitor high-risk groups, and assist in tumor risk assessment, judgment of tumor recurrence, prediction of tumor treatment efficacy and prognosis and development of specific new target drugs. It is believed that with the improvement of detection methods and experimental methods, promising results will be achieved in the fields of glioma prevention, diagnosis and treatment. Biotherapy, including gene therapy, immunotherapy and targeted molecular therapy, provides a new hope for the treatment of glioma. Some of these therapies have been shown to be effective in preclinical studies and safe in phase I clinical trials (Maio et al., 2015). Yet, clinical trials in phase II and III have been conducted. Based on our review, except HDACIs, a new therapeutic and epigenetic drug that can be taken alone or combined with other drugs or other treatments, the combination of epigenetic drugs with biotherapy is also a particularly interesting and novel direction for the future treatment of tumors. Due to the complicated pathogenesis of glioma, epigenetic applications to glioma clinical treatment are still limited. Discovering more effective therapeutic targets, developing novel targeted drugs, improving the efficacy of existing drugs in clinical research, and reducing the side effects of existing drugs are the problems that we need to face and solve in clinical treatment. With continued research, epigenetics understanding is certain to improve and, so is the epigenetic-based treatment of glioma.
Author Contributions
XH and LZ conceived and designed the project. Each author has contributed significantly to the submitted work. LZ drafted the manuscript. SK, FC and LW revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation (number 2018M632679); Medical and Health Science and Technology Development Project of Shandong Province (number 2017WS639); Linyi Science and Technology Development Project (number 201717024); Linyi People’s Hospital Doctoral Research Foundation (number 2016LYBS02); Key Research Project program of Shandong Province (number 2016GSF201056); and the Natural Science Foundation of Shandong Province (number ZR2014HM077).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MC and handling editor declared their shared affiliation at the time of the review.
References
Alaminos, M., Dávalos, V., Ropero, S., Setién, F., Paz, M. F., Herranz, M., et al. (2005). EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 65, 2565–2571. doi: 10.1158/0008-5472.can-04-4283
Antonelli, G., Libra, M., Panebianco, V., Russo, A. E., Vitale, F. V., Colina, P., et al. (2016). Molecular-targeted therapy for elderly patients with advanced non-small cell lung cancer. Oncol. Lett. 11, 3–8. doi: 10.3892/ol.2015.3901
Bai, Y., Liao, H., Liu, T., Zeng, X., Xiao, F., Luo, L., et al. (2013). MiR-296–3p regulates cell growth and multi-drug resistance of human glioblastoma by targeting ether-à-go-go (EAG1). Eur. J. Cancer 49, 710–724. doi: 10.1016/j.ejca.2012.08.020
Bai, Y., Zhang, Q.-G., and Wang, X.-H. (2014). Downregulation of TES by hypermethylation in glioblastoma reduces cell apoptosis and predicts poor clinical outcome. Eur. J. Med. Res. 19:66. doi: 10.1186/s40001-014-0066-4
Banasavadi-Siddegowda, Y. K., Welker, A. M., An, M., Yang, X., Zhou, W., Shi, G., et al. (2018). PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol. 20, 753–763. doi: 10.1093/neuonc/nox206
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Berdasco, M., Ropero, S., Setien, F., Fraga, M. F., Lapunzina, P., Losson, R., et al. (2009). Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc. Natl. Acad. Sci. U S A 106, 21830–21835. doi: 10.1073/pnas.0906831106
Berghoff, A. S., Hainfellner, J. A., Marosi, C., and Preusser, M. (2015). Assessing MGMT methylation status and its current impact on treatment in glioblastoma. CNS Oncol. 4, 47–52. doi: 10.2217/cns.14.50
Berindan-Neagoe, I., Monroig Pdel, C., Pasculli, B., and Calin, G. A. (2014). MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 64, 311–336. doi: 10.3322/caac.21244
Bhat, J., Kouakanou, L., Peters, C., Yin, Z., and Kabelitz, D. (2018). Immunotherapy with human gamma delta T cells—synergistic potential of epigenetic drugs? Front. Immunol. 9:512. doi: 10.3389/fimmu.2018.00512
Biegel, J. A., Busse, T. M., and Weissman, B. E. (2014). SWI/SNF chromatin remodeling complexes and cancer. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 350–366. doi: 10.1002/ajmg.c.31410
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. doi: 10.1101/gad.947102
Bleeker, F. E., Atai, N. A., Lamba, S., Jonker, A., Rijkeboer, D., Bosch, K. S., et al. (2010). The prognostic IDH1R132 mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 119, 487–494. doi: 10.1007/s00401-010-0645-6
Cai, J. J., Qi, Z. X., Chen, L. C., Yao, Y., Gong, Y., and Mao, Y. (2016). miR-124 suppresses the migration and invasion of glioma cells in vitro via Capn4. Oncol. Rep. 35, 284–290. doi: 10.3892/or.2015.4355
Castillo-Aguilera, O., Depreux, P., Halby, L., Arimondo, P. B., and Goossens, L. (2017). DNA methylation targeting: the DNMT/HMT crosstalk challenge. Biomolecules 7:E3. doi: 10.3390/biom7010003
Chang, Y., Zhang, X., Horton, J. R., Upadhyay, A. K., Spannhoff, A., Liu, J., et al. (2009). Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 16, 312–317. doi: 10.1038/nsmb.1560
Chédin, F. (2011). The DNMT3 family of mammalian de novo DNA methyltransferases. Prog. Mol. Biol. Transl. Sci. 101, 255–285. doi: 10.1016/b978-0-12-387685-0.00007-x
Chen, P. H., Chang, C. K., Shih, C. M., Cheng, C. H., Lin, C. W., Lee, C. C., et al. (2016). The miR-204–3p-targeted IGFBP2 pathway is involved in xanthohumol-induced glioma cell apoptotic death. Neuropharmacology 110, 362–375. doi: 10.1016/j.neuropharm.2016.07.038
Chen, X., Ma, H., Wang, Z., Zhang, S., Yang, H., and Fang, Z. (2017). EZH2 palmitoylation mediated by ZDHHC5 in p53-mutant glioma drives malignant development and progression. Cancer Res. 77, 4998–5010. doi: 10.1158/0008-5472.can-17-1139
Chen, L., Zhang, J., Feng, Y., Li, R., Sun, X., Du, W., et al. (2012). MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem. Cell Biol. 44, 1711–1717. doi: 10.1016/j.biocel.2012.06.027
Chen, X., Zhang, Y., Shi, Y., Lian, H., Tu, H., Han, S., et al. (2015). MiR-873 acts as a novel sensitizer of glioma cells to cisplatin by targeting Bcl-2. Int. J. Oncol. 47, 1603–1611. doi: 10.3892/ijo.2015.3143
Chen, G., Zhu, W., Shi, D., Lv, L., Zhang, C., Liu, P., et al. (2010). MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol. Rep. 23, 997–1003. doi: 10.3892/or_00000725
Cheung, H. C., Yatsenko, S. A., Kadapakkam, M., Legay, H., Su, J., Lupski, J. R., et al. (2012). Constitutional tandem duplication of 9q34 that truncates EHMT1 in a child with ganglioglioma. Pediatr. Blood Cancer 58, 801–805. doi: 10.1002/pbc.23219
Chien, L.-N., Gittleman, H., Ostrom, Q.-T., Hung, K.-S., Sloan, A.-E., Hsieh, Y.-C., et al. (2016). Comparative brain and central nervous system tumor incidence and survival between the united states and taiwan based on population-based registry. Front. Public Health 4:151. doi: 10.3389/fpubh.2016.00151
Chiocca, E. A., Aguilar, L. K., Bell, S. D., Kaur, B., Hardcastle, J., Cavaliere, R., et al. (2011). Phase IB study of gene-mediated cytotoxic immunotherapy adjuvant to up-front surgery and intensive timing radiation for malignant glioma. J. Clin. Oncol. 29, 3611–3619. doi: 10.1200/JCO.2011.35.5222
Choi, Y. J., Yoo, N. J., and Lee, S. H. (2015). Mutation of HELLS, a chromatin remodeling gene, gastric and colorectal cancers. Pathol. Oncol. Res. 21, 851–852. doi: 10.1007/s12253-014-9862-y
Choudhury, S. R., Cui, Y., Milton, J. R., Li, J., and Irudayaraj, J. (2015). Selective increase in subtelomeric DNA methylation: an epigenetic biomarker for malignant glioma. Clin. Epigenetics 7:107. doi: 10.1186/s13148-015-0140-y
Chu, S. H., Feng, D. F., Ma, Y. B., Zhang, H., Zhu, Z. A., Li, Z. Q., et al. (2011). Promoter methylation and downregulation of SLC22A18 are associated with the development and progression of human glioma. J. Transl. Med. 9:156. doi: 10.1186/1479-5876-9-156
Chu, B. F., Karpenko, M. J., Liu, Z., Aimiuwu, J., Villalona-Calero, M. A., Chan, K. K., et al. (2013). Phase I study of 5-aza-2’-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother. Pharmacol. 71, 115–121. doi: 10.1007/s00280-012-1986-8
Collinson, A., Collier, A. J., Morgan, N. P., Sienerth, A. R., Chandra, T., Andrews, S., et al. (2016). Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 17, 2700–2714. doi: 10.1016/j.celrep.2016.11.032
Dammann, R. H., Richter, A. M., Jiménez, A. P., Woods, M., Kuster, M., and Witharana, C. (2017). Impact of natural compounds on DNA methylation levels of the tumor suppressor gene RASSF1A in cancer. Int. J. Mol. Sci. 18:E2160. doi: 10.3390/ijms18102160
de Ruijter, A. J., van Gennip, A. H., Caron, H. N., Kemp, S., and van Kuilenburg, A. B. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749. doi: 10.1042/bj20021321
de Souza, C. F., Sabedot, T. S., Malta, T. M., Stetson, L., Morozova, O., Sokolov, A., et al. (2018). A distinct DNA methylation shift in a subset of glioma CpG island methylator phenotypes during tumor recurrence. Cell Rep. 23, 637–651. doi: 10.1016/j.celrep.2018.03.107
Dong, X., Deng, Q., Nie, X., Zhang, M., Jia, W., Chen, C., et al. (2015). Downregulation of HTATIP2 expression is associated with promoter methylation and poor prognosis in glioma. Exp. Mol. Pathol. 98, 192–199. doi: 10.1016/j.yexmp.2015.01.013
Dong, C. G., Wu, W. K., Feng, S. Y., Wang, X. J., Shao, J. F., and Qiao, J. (2012). Co-inhibition of microRNA-10b and microRNA-21 exerts synergistic inhibition on the proliferation and invasion of human glioma cells. Int. J. Oncol. 41, 1005–1012. doi: 10.3892/ijo.2012.1542
Du, W., Liu, X., Chen, L., Dou, Z., Lei, X., Chang, L., et al. (2015). Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro Oncol. 17, 243–253. doi: 10.1093/neuonc/nou217
Echizen, K., Nakada, M., Hayashi, T., Sabit, H., Furuta, T., Nakai, M., et al. (2014). PCDH10 is required for the tumorigenicity of glioblastoma cells. Biochem. Biophys. Res. Commun. 444, 13–18. doi: 10.1016/j.bbrc.2013.12.138
Emdad, L., Janjic, A., Alzubi, M. A., Hu, B., Santhekadur, P. K., Menezes, M. E., et al. (2015). Suppression of miR-184 in malignant gliomas upregulates SND1 and promotes tumor aggressiveness. Neuro Oncol. 17, 419–429. doi: 10.1093/neuonc/nou220
Endo, A., Ly, T., Pippa, R., Bensaddek, D., Nicolas, A., and Lamond, A. I. (2017). The chromatin assembly factor complex 1 (CAF1) and 5-azacytidine (5-AzaC) affect cell motility in src-transformed human epithelial cells. J. Biol. Chem. 292, 172–184. doi: 10.1074/jbc.m116.751024
Ernst, A., Campos, B., Meier, J., Devens, F., Liesenberg, F., Wolter, M., et al. (2010). De-repression of CTGF via the miR-17–92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29, 3411–3422. doi: 10.1038/onc.2010.83
Esteller, M., Garcia-Foncillas, J., Andion, E., Goodman, S. N., Hidalgo, O. F., Vanaclocha, V., et al. (2000). Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 343, 1350–1354. doi: 10.1056/NEJM200011093431901
Fabbri, E., Brognara, E., Montagner, G., Ghimenton, C., Eccher, A., Cantu, C., et al. (2015). Regulation of IL-8 gene expression in gliomas by microRNA miR-93. BMC Cancer 15:661. doi: 10.1186/s12885-015-1659-1
Fan, H., Lu, X., Wang, X., Liu, Y., Guo, B., Zhang, Y., et al. (2014). Low-dose decitabine-based chemoimmunotherapy for patients with refractory advanced solid tumors: a phase I/II report. J. Immunol. Res. 2014:371087. doi: 10.1155/2014/371087
Fang, L., Deng, Z., Shatseva, T., Yang, J., Peng, C., Du, W. W., et al. (2011). MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene 30, 806–821. doi: 10.1038/onc.2010.465
Fernandez, A. F., Bayón, G. F., Sierra, M. I., Urdinguio, R. G., Toraño, E. G., García, M., et al. (2018). Loss of 5hmC identifies a new type of aberrant DNA hypermethylation in glioma. Hum. Mol. Genet. 27, 3046–3059. doi: 10.1093/hmg/ddy214
Flavahan, W. A., Drier, Y., Liau, B. B., Gillespie, S. M., Venteicher, A. S., Stemmer-Rachamimov, A. O., et al. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. doi: 10.1038/nature16490
Foltz, G., Ryu, G. Y., Yoon, J. G., Nelson, T., Fahey, J., Frakes, A., et al. (2006). Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumor suppressor genes in malignant glioma. Cancer Res. 66, 6665–6674. doi: 10.1158/0008-5472.can-05-4453
Fouladi, M., Park, J. R., Stewart, C. F., Gilbertson, R. J., Schaiquevich, P., Sun, J., et al. (2010). Pediatric phase I trial and pharmacokinetic study of vorinostat: a Children’s Oncology Group phase I consortium report. J. Clin. Oncol. 28, 3623–3629. doi: 10.1200/JCO.2009.25.9119
Fowler, A., Thomson, D., Giles, K., Maleki, S., Mreich, E., Wheeler, H., et al. (2011). miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur. J. Cancer 47, 953–963. doi: 10.1016/j.ejca.2010.11.026
Gal, H., Pandi, G., Kanner, A. A., Ram, Z., Lithwick-Yanai, G., Amariglio, N., et al. (2008). MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 376, 86–90. doi: 10.1016/j.bbrc.2008.08.107
Galanis, E., Anderson, S. K., Miller, C. R., Sarkaria, J. N., Jaeckle, K., Buckner, J. C., et al. (2018). Phase I/II trial of vorinostat combined with temozolomide and radiation therapy for newly diagnosed glioblastoma: results of Alliance N0874/ABTC 02. Neuro Oncol. 20, 546–556. doi: 10.1093/neuonc/nox161
Galanis, E., Jaeckle, K. A., Maurer, M. J., Reid, J. M., Ames, M. M., Hardwick, J. S., et al. (2009). Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J. Clin. Oncol. 27, 2052–2058. doi: 10.1200/JCO.2008.19.0694
Gallagher, S. J., Shklovskaya, E., and Hersey, P. (2017). Epigenetic modulation in cancer immunotherapy. Curr. Opin. Pharmacol. 35, 48–56. doi: 10.1016/j.coph.2017.05.006
Gao, Y., Guan, M., Su, B., Liu, W., Xu, M., and Lu, Y. (2004). Hypermethylation of the RASSF1A gene in gliomas. Clin. Chim. Acta 349, 173–179. doi: 10.1016/j.cccn.2004.07.006
Garrido-Laguna, I., McGregor, K. A., Wade, M., Weis, J., Gilcrease, W., Burr, L., et al. (2013). A phase I/II study of decitabine in combination with panitumumab in patients with wild-type (wt) KRAS metastatic colorectal cancer. Invest. New Drugs 31, 1257–1264. doi: 10.1007/s10637-013-9947-6
Gessler, F., Voss, V., Seifert, V., Gerlach, R., and Kögel, D. (2011). Knockdown of TFPI-2 promotes migration and invasion of glioma cells. Neurosci. Lett. 497, 49–54. doi: 10.1016/j.neulet.2011.04.027
Ghasemi, A., Fallah, S., and Ansari, M. (2016). MiR-153 as a tumor suppressor in glioblastoma multiforme is downregulated by DNA methylation. Clin. Lab. 62, 573–580. doi: 10.7754/clin.lab.2015.150738
Ghiaseddin, A., Reardon, D., Massey, W., Mannerino, A., Lipp, E. S., Herndon, J. E., et al. (2018). Phase II study of bevacizumab and vorinostat for patients with recurrent world health organization grade 4 malignant glioma. Oncologist 23:157-e21. doi: 10.1634/theoncologist.2017-0501
Ghildiyal, R., and Sen, E. (2017). Concerted action of histone methyltransferases G9a and PRMT-1 regulates PGC-1α-RIG-I axis in IFNγ treated glioma cells. Cytokine 89, 185–193. doi: 10.1016/j.cyto.2015.12.008
Gömöri, E., Pál, J., Mészáros, I., Dóczi, T., and Matolcsy, A. (2007). Epigenetic inactivation of the hMLH1 gene in progression of gliomas. Diagn. Mol. Pathol. 16, 104–107. doi: 10.1097/pdm.0b013e318033f140
Götze, S., Wolter, M., Reifenberger, G., Müller, O., and Sievers, S. (2010). Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int. J. Cancer 126, 2584–2593. doi: 10.1002/ijc.24981
Gu, J. J., Gao, G. Z., and Zhang, S. M. (2016). MiR-218 inhibits the tumorgenesis and proliferation of glioma cells by targeting Robo1. Cancer Biomark. 16, 309–317. doi: 10.3233/cbm-160568
Guo, A. S., Huang, Y. Q., Ma, X. D., and Lin, R. S. (2016). Mechanism of G9a inhibitor BIX01294 acting on U251 glioma cells. Mol. Med. Rep. 14, 4613–4621. doi: 10.3892/mmr.2016.5815
Hanai, K., Furuhashi, H., Yamamoto, T., Akasaka, K., and Hirose, S. (2008). RSF governs silent chromatin formation via histone H2Av replacement. PLoS Genet. 4:e1000011. doi: 10.1371/journal.pgen.1000011
Hao, J., Zhang, C., Zhang, A., Wang, K., Jia, Z., Wang, G., et al. (2012). miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol. Rep. 27, 1504–1510. doi: 10.3892/or.2012.1652
Hara, K., Kageji, T., Mizobuchi, Y., Kitazato, K. T., Okazaki, T., Fujihara, T., et al. (2015). Blocking of the interaction between Wnt proteins and their co-receptors contributes to the anti-tumor effects of adenovirus-mediated DKK3 in glioblastoma. Cancer Lett. 356, 496–505. doi: 10.1016/j.canlet.2014.09.045
Hashimshony, T., Zhang, J., Keshet, I., Bustin, M., and Cedar, H. (2003). The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34, 187–192. doi: 10.1038/ng1158
Hashizume, R. (2017). Epigenetic targeted therapy for diffuse intrinsic pontine glioma. Neurol. Med. Chir. 57, 331–342. doi: 10.2176/nmc.ra.2017-0018
Hazane-Puch, F., Arnaud, J., Trocmé, C., Faure, P., Laporte, F., and Champelovier, P. (2016). Sodium selenite decreased HDAC activity, cell proliferation and induced apoptosis in three human glioblastoma cells. Anticancer Agents Med. Chem. 16, 490–500. doi: 10.2174/1871520615666150819095426
Heddleston, J. M., Wu, Q., Rivera, M., Minhas, S., Lathia, J. D., Sloan, A. E., et al. (2012). Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ. 19, 428–439. doi: 10.1038/cdd.2011.109
Hegi, M. E., Diserens, A. C., Gorlia, T., Hamou, M. F., de Tribolet, N., Weller, M., et al. (2005). MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003. doi: 10.1016/s0513-5117(08)70328-4
Hill, V. K., Underhill-Day, N., Krex, D., Robel, K., Sangan, C. B., Summersgill, H. R., et al. (2011). Epigenetic inactivation of the RASSF10 candidate tumor suppressor gene is a frequent and an early event in gliomagenesis. Oncogene 30, 978–989. doi: 10.1038/onc.2010.471
Horiguchi, K., Tomizawa, Y., Tosaka, M., Ishiuchi, S., Kurihara, H., Mori, M., et al. (2003). Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene 22, 7862–7865. doi: 10.1038/sj.onc.1207082
Hua, D., Ding, D., Han, X., Zhang, W., Zhao, N., Foltz, G., et al. (2012). Human miR-31 targets radixin and inhibits migration and invasion of glioma cells. Oncol. Rep. 27, 700–706. doi: 10.3892/or.2011.1555
Hu, H.-Q., Sun, L.-G., and Guo, W.-J. (2016). Decreased miRNA-146a in glioblastoma multiforme and regulation of cell proliferation and apoptosis by target Notch1. Int. J. Biol. Markers 31, e270–e275. doi: 10.5301/jbm.5000194
Hummel, T. R., Wagner, L., Ahern, C., Fouladi, M., Reid, J. M., McGovern, R. M., et al. (2013). A pediatric phase 1 trial of vorinostat and temozolomide in relapsed or refractory primary brain or spinal cord tumors: a Children’s Oncology Group phase 1 consortium study. Pediatr. Blood Cancer 60, 1452–1457. doi: 10.1002/pbc.24541
Huse, J. T., Brennan, C., Hambardzumyan, D., Wee, B., Pena, J., Rouhanifard, S. H., et al. (2009). The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 23, 1327–1337. doi: 10.1101/gad.1777409
Issa, J. P., Garcia-Manero, G., Giles, F. J., Mannari, R., Thomas, D., Faderl, S., et al. (2004). Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103, 1635–1640. doi: 10.1182/blood-2003-03-0687
Iwamoto, F. M., Lamborn, K. R., Kuhn, J. G., Wen, P. Y., Yung, W. K., Gilbert, M. R., et al. (2011). A phase I/II trial of the histone deacetylase inhibitor romidepsin for adults with recurrent malignant glioma: north american brain tumor consortium study 03–03. Neuro Oncol. 13, 509–516. doi: 10.1093/neuonc/nor017
Jha, P., Pia Patric, I. R., Shukla, S., Pathak, P., Pal, J., Sharma, V., et al. (2014). Genome-wide methylation profiling identifies an essential role of reactive oxygen species in pediatric glioblastoma multiforme and validates a methylome specific for H3 histone family 3A with absence of G-CIMP/isocitrate dehydrogenase 1 mutation. Neuro Oncol. 16, 1607–1617. doi: 10.1093/neuonc/nou113
Jiang, Z., Li, X., Hu, J., Zhou, W., Jiang, Y., Li, G., et al. (2006). Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci. Res. 56, 450–458. doi: 10.1016/j.neures.2006.09.006
Jiang, T., Mao, Y., Ma, W., Mao, Q., You, Y., Yang, X., et al. (2016). CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 375, 263–273. doi: 10.1016/j.canlet.2016.01.024
Jiang, J., Yang, J., Wang, Z., Wu, G., and Liu, F. (2013). TFAM is directly regulated by miR-23b in glioma. Oncol. Rep. 30, 2105–2110. doi: 10.3892/or.2013.2712
Jin, C., Li, M., Ouyang, Y., Tan, Z., and Jiang, Y. (2017). MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J. Neurooncol. 133, 247–255. doi: 10.1007/s11060-017-2438-4
Jin, Z., Xu, S., Yu, H., Yang, B., Zhao, H., and Zhao, G. (2013). miR-125b inhibits Connexin43 and promotes glioma growth. Cell. Mol. Neurobiol. 33, 1143–1148. doi: 10.1007/s10571-013-9980-1
Jing, S.-Y., Jing, S.-Q., Liu, L.-L., Xu, L.-F., Zhang, F., and Gao, J.-L. (2017). Down-expression of miR-373 predicts poor prognosis of glioma and could be a potential therapeutic target. Eur. Rev. Med. Pharmacol. Sci. 21, 2421–2425.
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/nrg3230
Kadowaki, M., Yoshioka, H., Kamitani, H., Watanabe, T., Wade, P. A., and Eling, T. E. (2012). DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. Int. J. Cancer 130, 267–277. doi: 10.1002/ijc.26082
Kaiser, S., Jurkowski, T. P., Kellner, S., Schneider, D., Jeltsch, A., and Helm, M. (2017). The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 14, 1241–1251. doi: 10.1080/15476286.2016.1236170
Kanemoto, M., Shirahata, M., Nakauma, A., Nakanishi, K., Taniguchi, K., Kukita, Y., et al. (2014). Prognostic prediction of glioblastoma by quantitative assessment of the methylation status of the entire MGMT promoter region. BMC Cancer 14:641. doi: 10.1186/1471-2407-14-641
Kim, J., Kim, K. Y., Jang, H.-S., Yoshida, T., Tsuchiya, K., Nitta, K., et al. (2009). Role of cytosolic NADP+-dependent isocitrate dehydrogenase in ischemia-reperfusion injury in mouse kidney. Am. J. Physiol. Renal Physiol. 296, F622–F633. doi: 10.1152/ajprenal.90566.2008
Kim, W. J., Newman, W. C., and Amankulor, N. M. (2017). Phase I/II trial of combination of temozolomide chemotherapy and immunotherapy with fusions of dendritic and glioma cells in patients with glioblastoma. Neurosurgery 81:N11. doi: 10.1093/neuros/nyx263
Kolodziej, M. A., Weischer, C., Reinges, M. H. T., Uhl, E., Weigand, M. A., Schwarm, F. P., et al. (2016). NDRG2 and NDRG4 expression is altered in glioblastoma and influences survival in patients with MGMT-methylated tumors. Anticancer Res. 36, 887–897.
Kondengaden, S. M., Luo, L. F., Huang, K., Zhu, M., Zang, L., Bataba, E., et al. (2016). Discovery of novel small molecule inhibitors of lysine methyltransferase G9a and their mechanism in leukemia cell lines. Eur. J. Med. Chem. 122, 382–393. doi: 10.1016/j.ejmech.2016.06.028
Kondo, Y., Katsushima, K., Ohka, F., Natsume, A., and Shinjo, K. (2014). Epigenetic dysregulation in glioma. Cancer Sci. 105, 363–369. doi: 10.1111/cas.12379
Kraus, T. F., Globisch, D., Wagner, M., Eigenbrod, S., Widmann, D., Münzel, M., et al. (2012). Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int. J. Cancer 131, 1577–1590. doi: 10.1002/ijc.27429
Krausz, C., Sandoval, J., Sayols, S., Chianese, C., Giachini, C., Heyn, H., et al. (2012). Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 7:e44479. doi: 10.1371/journal.pone.0044479
Krauze, A. V., Myrehaug, S. D., Chang, M. G., Holdford, D. J., Smith, S., Shih, J., et al. (2015). A phase 2 study of concurrent radiation therapy, temozolomide and the histone deacetylase inhibitor valproic acid for patients with glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 92, 986–992. doi: 10.1016/j.ijrobp.2015.04.038
Kreth, F. W., Thon, N., Simon, M., Westphal, M., Schackert, G., Nikkhah, G., et al. (2013). Gross total but not incomplete resection of glioblastoma prolongs survival in the era of radiochemotherapy. Ann. Oncol. 24, 3117–3123. doi: 10.1093/annonc/mdt388
Kunitz, A., Wolter, M., van den Boom, J., Felsberg, J., Tews, B., Hahn, M., et al. (2007). DNA hypermethylation and aberrant expression of the EMP3 gene at 19q13.3 in Human Gliomas. Brain Pathol. 17, 363–370. doi: 10.1111/j.1750-3639.2007.00083.x
Kurmasheva, R. T., Sammons, M., Favours, E., Wu, J., Kurmashev, D., Cosmopoulos, K., et al. (2017). Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the pediatric preclinical testing program. Pediatr. Blood Cancer 64:e26218. doi: 10.1002/pbc.26218
Kwak, S. Y., Kim, B. Y., Ahn, H. J., Yoo, J. O., Kim, J., Bae, I. H., et al. (2015). Ionizing radiation-inducible miR-30e promotes glioma cell invasion through EGFR stabilization by directly targeting CBL-B. FEBS J. 282, 1512–1525. doi: 10.1111/febs.13238
Kwak, H. J., Kim, Y. J., Chun, K. R., Woo, Y. M., Park, S. J., Jeong, J. A., et al. (2011). Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 30, 2433–2442. doi: 10.1038/onc.2010.620
Lakomy, R., Sana, J., Hankeova, S., Fadrus, P., Kren, L., Lzicarova, E., et al. (2011). MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 102, 2186–2190. doi: 10.1111/j.1349-7006.2011.02092.x
Lee, S. H., Kim, M. S., Kwon, H. C., Park, I. C., Park, M. J., Lee, C. T., et al. (2000). Growth inhibitory effect on glioma cells of adenovirus-mediated p16/INK4a gene transfer in vitro and in vivo. Int. J. Mol. Med. 6, 559–563. doi: 10.3892/ijmm.6.5.559
Lee, E. Q., Puduvalli, V. K., Reid, J. M., Kuhn, J. G., Lamborn, K. R., Cloughesy, T. F., et al. (2012). Phase I study of vorinostat in combination with temozolomide in patients with high-grade gliomas: north american brain tumor consortium study 04–03. Clin. Cancer Res. 18, 6032–6039. doi: 10.1158/1078-0432.ccr-12-1841
Lee, E. Q., Reardon, D. A., Schiff, D., Drappatz, J., Muzikansky, A., Grimm, S. A., et al. (2015). Phase II study of panobinostat in combination with bevacizumab for recurrent glioblastoma and anaplastic glioma. Neuro Oncol. 17, 862–867. doi: 10.1093/neuonc/nou350
Leng, Y., Wang, J., Wang, Z., Liao, H.-M., Wei, M., Leeds, P., et al. (2016). Valproic acid and other HDAC inhibitors upregulate FGF21 gene expression and promote process elongation in glia by inhibiting HDAC2 and 3. Int. J. Neuropsychopharmacol. 19:pyw035. doi: 10.1093/ijnp/pyw035
Li, J., An, G., Zhang, M., and Ma, Q. (2016). Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem. Biophys. Res. Commun. 477, 743–748. doi: 10.1016/j.bbrc.2016.06.129
Li, X., Ling, N., Bai, Y., Dong, W., Hui, G. Z., Liu, D., et al. (2013). MiR-16–1 plays a role in reducing migration and invasion of glioma cells. Anat. Rec. 296, 427–432. doi: 10.1002/ar.22626
Li, X., Liu, Y., Granberg, K. J., Wang, Q., Moore, L. M., Ji, P., et al. (2015). Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene 34, 1619–1628. doi: 10.1038/onc.2014.98
Li, K. K., Yang, L., Pang, J. C., Chan, A. K., Zhou, L., Mao, Y., et al. (2013). MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol. 23, 426–439. doi: 10.1111/bpa.12015
Li, L., Yang, G., Ren, C., Tanimoto, R., Hirayama, T., Wang, J., et al. (2013). Glioma pathogenesis-related protein 1 induces prostate cancer cell death through Hsc70-mediated suppression of AURKA and TPX2. Mol. Oncol. 7, 484–496. doi: 10.1016/j.molonc.2012.12.005
Liao, P., Ostrom, Q. T., Stetson, L., and Barnholtz-Sloan, J. S. (2018). Models of epigenetic age capture patterns of DNA methylation in glioma associated with molecular subtype, survival and recurrence. Neuro Oncol. 20, 942–953. doi: 10.1093/neuonc/noy003
Liau, B. B., Sievers, C., Donohue, L. K., Gillespie, S. M., Flavahan, W. A., Miller, T. E., et al. (2017). Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 20, 233.e7–246.e7. doi: 10.1016/j.stem.2016.11.003
Lin, N., Di, C., Bortoff, K., Fu, J., Truszkowski, P., Killela, P., et al. (2012). Deletion or epigenetic silencing of AJAP1 on 1p36 in glioblastoma. Mol. Cancer Res. 10, 208–217. doi: 10.1158/1541-7786.mcr-10-0109
Liu, F., Barsyte-Lovejoy, D., Allali-Hassani, A., He, Y., Herold, J. M., Chen, X., et al. (2011). Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J. Med. Chem. 54, 6139–6150. doi: 10.1021/jm200903z
Liu, F., Barsyte-Lovejoy, D., Li, F., Xiong, Y., Korboukh, V., Huang, X. P., et al. (2013). Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J. Med. Chem. 56, 8931–8942. doi: 10.1021/jm401480r
Liu, X., Li, G., Su, Z., Jiang, Z., Chen, L., Wang, J., et al. (2013). Poly(amido amine) is an ideal carrier of miR-7 for enhancing gene silencing effects on the EGFR pathway in U251 glioma cells. Oncol. Rep. 29, 1387–1394. doi: 10.3892/or.2013.2283
Liu, Y. S., Lin, H. Y., Lai, S. W., Huang, C. Y., Huang, B. R., Chen, P. Y., et al. (2017). MiR-181b modulates EGFR-dependent VCAM-1 expression and monocyte adhesion in glioblastoma. Oncogene 36, 5006–5022. doi: 10.1038/onc.2017.129
Liu, Z., Jiang, Z., Huang, J., Huang, S., Li, Y., Yu, S., et al. (2014). miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int. J. Oncol. 44, 1571–1580. doi: 10.3892/ijo.2014.2322
Lovkvist, C., Dodd, I. B., Sneppen, K., and Haerter, J. O. (2016). DNA methylation in human epigenomes depends on local topology of CpG sites. Nucleic Acids Res. 44, 5123–5132. doi: 10.1093/nar/gkw124
Loyola, A., Huang, J. Y., LeRoy, G., Hu, S., Wang, Y. H., Donnelly, R. J., et al. (2003). Functional analysis of the subunits of the chromatin assembly factor RSF. Mol. Cell. Biol. 23, 6759–6768. doi: 10.1128/mcb.23.19.6759-6768.2003
Lucio-Eterovic, A. K., Cortez, M. A., Valera, E. T., Motta, F. J., Queiroz, R. G., Machado, H. R., et al. (2008). Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer 8:243. doi: 10.1186/1471-2407-8-243
Luo, G., Luo, W., Sun, X., Lin, J., Wang, M., Zhang, Y., et al. (2017). MicroRNA21 promotes migration and invasion of glioma cells via activation of Sox2 and βcatenin signaling. Mol. Med. Rep. 15, 187–193. doi: 10.3892/mmr.2016.5971
Ma, C., Zheng, C., Bai, E., and Yang, K. (2016). miR-101 inhibits glioma cell invasion via the downregulation of COX-2. Oncol. Lett. 12, 2538–2544. doi: 10.3892/ol.2016.4939
Mack, S. C., Witt, H., Piro, R. M., Gu, L., Zuyderduyn, S., Stutz, A. M., et al. (2014). Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450. doi: 10.1038/nature13108
Maio, M., Covre, A., Fratta, E., Di Giacomo, A. M., Taverna, P., Natali, P. G., et al. (2015). Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin. Cancer Res. 21, 4040–4047. doi: 10.1158/1078-0432.ccr-14-2914
Majchrzak-Celinska, A., Slocinska, M., Barciszewska, A. M., Nowak, S., and Baer-Dubowska, W. (2016). Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival. J. Appl. Genet. 57, 189–197. doi: 10.1007/s13353-015-0312-7
Malta, T. M., de Souza, C. F., Sabedot, T. S., Silva, T. C., Mosella, M. Q. S., Kalkanis, S. N., et al. (2018). Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 20, 608–620. doi: 10.1093/neuonc/nox183
Marfella, C. G., Henninger, N., LeBlanc, S. E., Krishnan, N., Garlick, D. S., Holzman, L. B., et al. (2008). A mutation in the mouse Chd2 chromatin remodeling enzyme results in a complex renal phenotype. Kidney Blood Press. Res. 31, 421–432. doi: 10.1159/000190788
Marks, P. A., and Breslow, R. (2007). Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90. doi: 10.1038/nbt1272
Martini, M., Pallini, R., Luongo, G., Cenci, T., Lucantoni, C., and Larocca, L. M. (2008). Prognostic relevance of SOCS3 hypermethylation in patients with glioblastoma multiforme. Int. J. Cancer 123, 2955–2960. doi: 10.1002/ijc.23805
Mathias, R. A., Guise, A. J., and Cristea, I. M. (2015). Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell. Proteomics 14, 456–470. doi: 10.1074/mcp.o114.046565
Mei, P. J., Bai, J., Liu, H., Li, C., Wu, Y. P., Yu, Z. Q., et al. (2011). RUNX3 expression is lost in glioma and its restoration causes drastic suppression of tumor invasion and migration. J. Cancer Res. Clin. Oncol. 137, 1823–1830. doi: 10.1007/s00432-011-1063-4
Milde, T., Oehme, I., Korshunov, A., Kopp-Schneider, A., Remke, M., Northcott, P., et al. (2010). HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin. Cancer Res. 16, 3240–3252. doi: 10.1158/1078-0432.ccr-10-0395
Moniz, S., Martinho, O., Pinto, F., Sousa, B., Loureiro, C., Oliveira, M. J., et al. (2013). Loss of WNK2 expression by promoter gene methylation occurs in adult gliomas and triggers Rac1-mediated tumour cell invasiveness. Hum. Mol. Genet. 22, 84–95. doi: 10.1093/hmg/dds405
Mueller, W., Lass, U., Wellmann, S., Kunitz, F., and von Deimling, A. (2005). Mutation analysis of DKK1 and in vivo evidence of predominant p53-independent DKK1 function in gliomas. Acta Neuropathol. 109, 314–320. doi: 10.1007/s00401-004-0969-1
Mur, P., Rodríguez de Lope, Á., Díaz-Crespo, F. J., Hernández-Iglesias, T., Ribalta, T., Fiaño, C., et al. (2015). Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J. Neurooncol. 122, 441–450. doi: 10.1007/s11060-015-1738-9
Muscal, J. A., Thompson, P. A., Horton, T. M., Ingle, A. M., Ahern, C. H., McGovern, R. M., et al. (2013). A phase I trial of vorinostat and bortezomib in children with refractory or recurrent solid tumors: a Children’s Oncology Group phase I consortium study (ADVL0916). Pediatr. Blood Cancer 60, 390–395. doi: 10.1002/pbc.24271
Nakahara, Y., Northcott, P. A., Li, M., Kongkham, P. N., Smith, C., Yan, H., et al. (2010). Genetic and epigenetic inactivation of Kruppel-like factor 4 in medulloblastoma. Neoplasia 12, 20–27. doi: 10.1593/neo.91122
Noushmehr, H., Weisenberger, D. J., Diefes, K., Phillips, H. S., Pujara, K., Berman, B. P., et al. (2010). Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522. doi: 10.1016/j.ccr.2010.03.017
Oi, S., Natsume, A., Ito, M., Kondo, Y., Shimato, S., Maeda, Y., et al. (2009). Synergistic induction of NY-ESO-1 antigen expression by a novel histone deacetylase inhibitor, valproic acid, with 5-aza-2’-deoxycytidine in glioma cells. J. Neurooncol. 92, 15–22. doi: 10.1007/s11060-008-9732-0
Orzan, F., Pellegatta, S., Poliani, P. L., Pisati, F., Caldera, V., Menghi, F., et al. (2011). Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol. Appl. Neurobiol. 37, 381–394. doi: 10.1111/j.1365-2990.2010.01132.x
Pathania, R., Ramachandran, S., Mariappan, G., Thakur, P., Shi, H., Choi, J. H., et al. (2016). Combined inhibition of DNMT and HDAC blocks the tumorigenicity of cancer stem-like cells and attenuates mammary tumor growth. Cancer Res. 76, 3224–3235. doi: 10.1158/0008-5472.can-15-2249
Peereboom, D. M., Shepard, D. R., Ahluwalia, M. S., Brewer, C. J., Agarwal, N., Stevens, G. H., et al. (2010). Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J. Neurooncol. 98, 93–99. doi: 10.1007/s11060-009-0067-2
Pei, Y., Liu, K. W., Wang, J., Garancher, A., Tao, R., Esparza, L. A., et al. (2016). HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell 29, 311–323. doi: 10.1016/j.ccell.2016.02.011
Pileggi, A., Klein, D., Fotino, C., Bravo-Egaña, V., Rosero, S., Doni, M., et al. (2013). MicroRNAs in islet immunobiology and transplantation. Immunol. Res. 57, 185–196. doi: 10.1007/s12026-013-8436-5
Pulice, J. L., and Kadoch, C. (2016). Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb. Symp. Quant. Biol. 81, 53–60. doi: 10.1101/sqb.2016.81.031021
Qiu, J., Ai, L., Ramachandran, C., Yao, B., Gopalakrishnan, S., Fields, C. R., et al. (2008). Invasion suppressor cystatin E/M (CST6): high-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab. Invest. 88, 910–925. doi: 10.1038/labinvest.2008.66
Qu, Y., Dang, S., and Hou, P. (2013). Gene methylation in gastric cancer. Clin. Chim. Acta 424, 53–65. doi: 10.1016/j.cca.2013.05.002
Quintavalle, C., Garofalo, M., Zanca, C., Romano, G., Iaboni, M., del Basso De Caro, M., et al. (2012). miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 31, 858–868. doi: 10.1038/onc.2011.280
Rani, S. B., Rathod, S. S., Karthik, S., Kaur, N., Muzumdar, D., and Shiras, A. S. (2013). MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 15, 1302–1316. doi: 10.1093/neuonc/not090
Roboz, G. J., Ritchie, E. K., Dault, Y., Lam, L., Marshall, D. C., Cruz, N. M., et al. (2018). Phase I trial of plerixafor combined with decitabine in newly diagnosed older patients with acute myeloid leukemia. Haematologica 103, 1308–1316. doi: 10.3324/haematol.2017.183418
Rossetto, M., Ciccarino, P., Boisselier, B., Labussiere, M., and Sanson, M. (2011). Metabolism of glioma and IDH1/IDH2 mutations. Rev. Neurol. Paris. 167, 699–703. doi: 10.1016/j.neurol.2011.08.002
Schelman, W. R., Traynor, A. M., Holen, K. D., Kolesar, J. M., Attia, S., Hoang, T., et al. (2013). A phase I study of vorinostat in combination with bortezomib in patients with advanced malignancies. Invest. New Drugs 31, 1539–1546. doi: 10.1007/s10637-013-0029-6
Schraivogel, D., Weinmann, L., Beier, D., Tabatabai, G., Eichner, A., Zhu, J. Y., et al. (2011). CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 30, 4309–4322. doi: 10.1038/emboj.2011.301
Schwartzentruber, J., Korshunov, A., Liu, X. Y., Jones, D. T., Pfaff, E., Jacob, K., et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231. doi: 10.1038/nature10833
Shang, C., Guo, Y., Hong, Y., and Xue, Y. X. (2016a). Long non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front. Cell. Neurosci. 10:235. doi: 10.3389/fncel.2016.00235
Shang, C., Hong, Y., Guo, Y., Liu, Y. H., and Xue, Y. X. (2016b). miR-128 regulates the apoptosis and proliferation of glioma cells by targeting RhoE. Oncol. Lett. 11, 904–908. doi: 10.3892/ol.2015.3927
Sharma, V., Koul, N., Joseph, C., Dixit, D., Ghosh, S., and Sen, E. (2010). HDAC inhibitor, scriptaid, induces glioma cell apoptosis through JNK activation and inhibits telomerase activity. J. Cell. Mol. Med. 14, 2151–2161. doi: 10.1111/j.1582-4934.2009.00844.x
Sheu, J. J., Guan, B., Choi, J. H., Lin, A., Lee, C. H., Hsiao, Y. T., et al. (2010). Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J. Biol. Chem. 285, 38260–38269. doi: 10.1074/jbc.m110.138735
Shi, W., Palmer, J. D., Werner-Wasik, M., Andrews, D. W., Evans, J. J., Glass, J., et al. (2016). Phase I trial of panobinostat and fractionated stereotactic re-irradiation therapy for recurrent high grade gliomas. J. Neurooncol. 127, 535–539. doi: 10.1007/s11060-016-2059-3
Shi, L., Wang, Z., Sun, G., Wan, Y., Guo, J., and Fu, X. (2014). miR-145 inhibits migration and invasion of glioma stem cells by targeting ABCG2. Neuromolecular Med. 16, 517–528. doi: 10.1007/s12017-014-8305-y
Shi, L., Zhang, S., Feng, K., Wu, F., Wan, Y., Wang, Z., et al. (2012). MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int. J. Oncol. 40, 119–129. doi: 10.3892/ijo.2011.1179
Shimooka, Y., Nishikawa, J., and Ohyama, T. (2013). Most methylation-susceptible DNA sequences in human embryonic stem cells undergo a change in conformation or flexibility upon methylation. Biochemistry 52, 1344–1353. doi: 10.1021/bi301319y
Smirnikhina, S. A., Lavrov, A. V., Chelysheva, E. Y., Adilgereeva, E. P., Shukhov, O. A., Turkina, A., et al. (2016). Whole-exome sequencing reveals potential molecular predictors of relapse after discontinuation of the targeted therapy in chronic myeloid leukemia patients. Leuk. Lymphoma 57, 1669–1676. doi: 10.3109/10428194.2015.1132420
Song, Y., Wang, P., Zhao, W., Yao, Y., Liu, X., Ma, J., et al. (2014). MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp. Cell Res. 324, 54–64. doi: 10.1016/j.yexcr.2014.03.009
Stanton, B. Z., Hodges, C., Crabtree, G. R., and Zhao, K. (2017). A general non-radioactive ATPase assay for chromatin remodeling complexes. Curr. Protoc. Chem. Biol. 9, 1–10. doi: 10.1002/cpch.16
Stupp, R., Mason, W. P., van den Bent, M. J., Weller, M., Fisher, B., Taphoorn, M. J., et al. (2005). Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996. doi: 10.1016/j.canrad.2005.05.001
Su, J. M., Li, X.-N., Thompson, P., Ou, C. N., Ingle, A. M., Russell, H., et al. (2011). Phase 1 study of valproic acid in pediatric patients with refractory solid or CNS tumors: a children’s oncology group report. J. Clin. Oncol. 17, 589–597. doi: 10.1158/1078-0432.ccr-10-0738
Sullivan, R. J., and Atkins, M. B. (2009). Molecular-targeted therapy in malignant melanoma. Expert Rev. Anticancer Ther. 9, 567–581. doi: 10.1586/era.09.20
Sun, G., Shi, L., Yan, S., Wan, Z., Jiang, N., Fu, L., et al. (2014). MiR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed. Res. Int. 2014:687826. doi: 10.1155/2014/687826
Sun, J., Tian, X., Zhang, J., Huang, Y., Lin, X., Chen, L., et al. (2017). Regulation of human glioma cell apoptosis and invasion by miR-152–3p through targeting DNMT1 and regulating NF2: MiR-152-3p regulate glioma cell apoptosis and invasion. J. Exp. Clin. Cancer Res. 36:100. doi: 10.1186/s13046-017-0567-4
Sun, A. G., Wang, M. G., Li, B., and Meng, F. G. (2017). Down-regulation of miR-124 target protein SCP-1 inhibits neuroglioma cell migration. Eur. Rev. Med. Pharmacol. Sci. 21, 723–729. doi: 10.1007/s13577-017-0180-z
Sun, J.-Z., Yang, X.-X., Li, X.-H., Xu, W.-W., Wang, Y., Zhu, W., et al. (2011). Aberrant CpG island hypermethylation and down-regulation of Oct-6 mRNA expression in human hepatocellular carcinoma. Dig. Dis. Sci. 56, 3072–3077. doi: 10.1007/s10620-011-1686-y
Vaitkiene, P., Skiriute, D., Skauminas, K., and Tamasauskas, A. (2012). Associations between TFPI-2 methylation and poor prognosis in glioblastomas. Medicina 48, 345–349. doi: 10.3390/medicina48070051
Vargas, J. E., Filippi-Chiela, E. C., Suhre, T., Kipper, F. C., Bonatto, D., and Lenz, G. (2014). Inhibition of HDAC increases the senescence induced by natural polyphenols in glioma cells. Biochem. Cell Biol. 92, 297–304. doi: 10.1139/bcb-2014-0022
Veeriah, S., Brennan, C., Meng, S., Singh, B., Fagin, J. A., Solit, D. B., et al. (2009). The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. U S A 106, 9435–9440. doi: 10.1073/pnas.0900571106
Verhaak, R. G., Hoadley, K. A., Purdom, E., Wang, V., Qi, Y., Wilkerson, M. D., et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110. doi: 10.1016/j.ccr.2009.12.020
von dem Knesebeck, A., Felsberg, J., Waha, A., Hartmann, W., Scheffler, B., Glas, M., et al. (2012). RANK (TNFRSF11A) is epigenetically inactivated and induces apoptosis in gliomas. Neoplasia 14, 526–534. doi: 10.1596/neo.12360
Waha, A., Felsberg, J., Hartmann, W., von dem Knesebeck, A., Mikeska, T., Joos, S., et al. (2010). Epigenetic downregulation of mitogen-activated protein kinase phosphatase MKP-2 relieves its growth suppressive activity in glioma cells. Cancer Res. 70, 1689–1699. doi: 10.1158/0008-5472.can-09-3218
Wang, X.-Q., Bai, H.-M., Li, S.-T., Sun, H., Min, L.-Z., Tao, B.-B., et al. (2017). Knockdown of HDAC1 expression suppresses invasion and induces apoptosis in glioma cells. Oncotarget 8, 48027–48040. doi: 10.18632/oncotarget.18227
Wang, L., Liu, J., Zhong, Z., Gong, X., Liu, W., Shi, L., et al. (2016). PTP4A3 is a target for inhibition of cell proliferatin, migration and invasion through Akt/mTOR signaling pathway in glioblastoma under the regulation of miR-137. Brain Res. 1646, 441–450. doi: 10.1016/j.brainres.2016.06.026
Wang, C., Schroeder, F. A., and Hooker, J. M. (2014). Visualizing epigenetics: current advances and advantages in HDAC PET imaging techniques. Neuroscience 264, 186–197. doi: 10.1016/j.neuroscience.2013.09.018
Wang, L., Shi, M., Hou, S., Ding, B., Liu, L., Ji, X., et al. (2012). MiR-483–5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett. 586, 1312–1317. doi: 10.1016/j.febslet.2012.03.035
Wang, Y., and Wang, L. (2017). miR-34a attenuates glioma cells progression and chemoresistance via targeting PD-L1. Biotechnol. Lett. 39, 1485–1492. doi: 10.1007/s10529-017-2397-z
Wang, Z., Wang, B., Shi, Y., Xu, C., Xiao, H. L., Ma, L. N., et al. (2015). Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene 34, 1407–1419. doi: 10.1038/onc.2014.75
Watanabe, T., Huang, H., Nakamura, M., Wischhusen, J., Weller, M., Kleihues, P., et al. (2002). Methylation of the p73 gene in gliomas. Acta Neuropathol. 104, 357–362. doi: 10.1007/s00401-002-0549-1
Watanabe, T., Katayama, Y., Yoshino, A., Yachi, K., Ohta, T., Ogino, A., et al. (2007). Aberrant hypermethylation of p14ARF and O6-methylguanine-DNA methyltransferase genes in astrocytoma progression. Brain Pathol. 17, 5–10. doi: 10.1111/j.1750-3639.2006.00030.x
Wei, J., Nduom, E. K., Kong, L. Y., Hashimoto, Y., Xu, S., Gabrusiewicz, K., et al. (2016). MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 18, 639–648. doi: 10.1093/neuonc/nov292
Wei, J., Qi, X., Zhan, Q., Zhou, D., Yan, Q., Wang, Y., et al. (2015). miR-20a mediates temozolomide-resistance in glioblastoma cells via negatively regulating LRIG1 expression. Biomed. Pharmacother. 71, 112–118. doi: 10.1016/j.biopha.2015.01.026
Wei, F., Wang, Q., Su, Q., Huang, H., Luan, J., Xu, X., et al. (2016). miR-373 inhibits glioma cell u251 migration and invasion by down-regulating CD44 and TGFBR2. Cell. Mol. Neurobiol. 36, 1389–1397. doi: 10.1007/s10571-016-0338-3
Weller, M. (2013). Assessing the MGMT status in glioblastoma: one step forward, two steps back? Neuro Oncol. 15, 253–254. doi: 10.1093/neuonc/not014
Wen, P. Y., and Huse, J. T. (2017). 2016 world health organization classification of central nervous system tumors. Continuum 23, 1531–1547. doi: 10.1212/CON.0000000000000536
Wheeler, L. A., Manzanera, A. G., Bell, S. D., Cavaliere, R., McGregor, J. M., Grecula, J. C., et al. (2016). Phase II multicenter study of gene-mediated cytotoxic immunotherapy as adjuvant to surgical resection for newly diagnosed malignant glioma. Neuro Oncol. 18, 1137–1145. doi: 10.1093/neuonc/now002
Wiese, M., Schill, F., Sturm, D., Pfister, S., Hulleman, E., Johnsen, S. A., et al. (2016). No significant cytotoxic effect of the EZH2 inhibitor tazemetostat (EPZ-6438) on pediatric glioma cells with wildtype histone 3 or mutated histone 3.3. Hum. Pathol. 228, 113–117. doi: 10.1055/s-0042-105292
Wu, X., Li, Y., Wan, X., Kayira, T. M., Cao, R., Ju, X., et al. (2012). Down-regulation of neogenin accelerated glioma progression through promoter Methylation and its overexpression in SHG-44 induced apoptosis. PLoS One 7:e38074. doi: 10.1371/journal.pone.0038074
Xia, H., Yan, Y., Hu, M., Wang, Y., Wang, Y., Dai, Y., et al. (2013). MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κB activity. Neuro Oncol. 15, 413–422. doi: 10.1093/neuonc/nos296
Xiao, D., Huang, J., Pan, Y., Li, H., Fu, C., Mao, C., et al. (2017). Chromatin remodeling factor LSH is upregulated by the LRP6-GSK3β-E2F1 axis linking reversely with survival in gliomas. Theranostics 7, 132–143. doi: 10.7150/thno.17032
Xipell, E., Aragón, T., Martínez-Velez, N., Vera, B., Idoate, M. A., Martínez-Irujo, J. J., et al. (2016). Endoplasmic reticulum stress-inducing drugs sensitize glioma cells to temozolomide through downregulation of MGMT, MPG, and Rad51. Neuro Oncol. 18, 1109–1119. doi: 10.1093/neuonc/now022
Xu, J., Liao, X., Lu, N., Liu, W., and Wong, C. W. (2011). Chromatin-modifying drugs induce miRNA-153 expression to suppress Irs-2 in glioblastoma cell lines. Int. J. Cancer 129, 2527–2531. doi: 10.1002/ijc.25917
Xu, Y., Li, W. L., Fu, L., Gu, F., and Ma, Y. J. (2010). Slit2/Robo1 signaling in glioma migration and invasion. Neurosci. Bull. 26, 474–478. doi: 10.1007/s12264-010-0730-9
Xu, S., Ren, J., Chen, H. B., Wang, Y., Liu, Q., Zhang, R., et al. (2014). Cytostatic and apoptotic effects of DNMT and HDAC inhibitors in endometrial cancer cells. Curr. Pharm. Des. 20, 1881–1887. doi: 10.2174/13816128113199990527
Xue, L., Wang, Y., Yue, S., and Zhang, J. (2015). Low MiR-149 expression is associated with unfavorable prognosis and enhanced Akt/mTOR signaling in glioma. Int. J. Clin. Exp. Pathol. 8, 11178–11184.
Yadavilli, S., Scafidi, J., Becher, O. J., Saratsis, A. M., Hiner, R. L., Kambhampati, M., et al. (2015). The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget 6, 12141–12155. doi: 10.18632/oncotarget.3716
Yan, Y., Xu, Z., Li, Z., Sun, L., and Gong, Z. (2017). An insight into the increasing role of LncRNAs in the pathogenesis of gliomas. Front. Mol. Neurosci. 10:53. doi: 10.3389/fnmol.2017.00053
Yang, F., Wang, W., Zhou, C., Xi, W., Yuan, L., Chen, X., et al. (2015). MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 36, 3763–3773. doi: 10.1007/s13277-014-3017-3
Yilaz Susluer, S., Biray Avci, C., Dodurga, Y., Ozlem Dogan Sigva, Z., Oktar, N., and Gunduz, C. (2015). Downregulation of miR-195 via cyclosporin A in human glioblastoma cells. J. BUON. 20, 1337–1340.
Yu, Z., Chen, Y., Wang, S., Li, P., Zhou, G., and Yuan, Y. (2018). Inhibition of NF-kappaB results in anti-glioma activity and reduces temozolomide-induced chemoresistance by down-regulating MGMT gene expression. Cancer Lett. 428, 77–89. doi: 10.1016/j.canlet.2018.04.033
Yuan, Q., Gao, W., Liu, B., and Ye, W. (2014). Upregulation of miR-184 enhances the malignant biological behavior of human glioma cell line A172 by targeting FIH-1. Cell. Physiol. Biochem. 34, 1125–1136. doi: 10.1159/000366326
Zang, L., Kondengaden, S. M., Zhang, Q., Li, X., Sigalapalli, D. K., Kondengadan, S. M., et al. (2017). Structure based design, synthesis and activity studies of small hybrid molecules as HDAC and G9a dual inhibitors. Oncotarget 8, 63187–63207. doi: 10.18632/oncotarget.18730
Zang, L.-L., Wang, X.-J., Li, X.-B., Wang, S.-Q., Xu, W.-R., Xie, X.-B., et al. (2014). SAHA-based novel HDAC inhibitor design by core hopping method. J. Mol. Graph. Model. 54, 10–18. doi: 10.1016/j.jmgm.2014.08.005
Zhang, Y., Dong, W., Zhu, J., Wang, L., Wu, X., and Shan, H. (2017). Combination of EZH2 inhibitor and BET inhibitor for treatment of diffuse intrinsic pontine glioma. Cell Biosci. 7:56. doi: 10.1186/s13578-017-0184-0
Zhang, Y., Han, D., Wei, W., Cao, W., Zhang, R., Dong, Q., et al. (2015). MiR-218 inhibited growth and metabolism of human glioblastoma cells by directly targeting E2F2. Cell. Mol. Neurobiol. 35, 1165–1173. doi: 10.1007/s10571-015-0210-x
Zhang, C., Kang, C., You, Y., Pu, P., Yang, W., Zhao, P., et al. (2009). Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int. J. Oncol. 34, 1653–1660. doi: 10.3892/ijo_00000296
Zhang, Q.-M., Shen, N., Xie, S., Bi, S.-Q., Luo, B., Lin, Y.-D., et al. (2014). MAGED4 expression in glioma and upregulation in glioma cell lines with 5-aza-2′-deoxycytidine treatment. Asian Pac. J. Cancer Prev. 15, 3495–3501. doi: 10.7314/apjcp.2014.15.8.3495
Zhang, P., Sun, H., Yang, B., Luo, W., Liu, Z., Wang, J., et al. (2017). miR-152 regulated glioma cell proliferation and apoptosis via Runx2 mediated by DNMT1. Biomed. Pharmacother. 92, 690–695. doi: 10.1016/j.biopha.2017.05.096
Zhang, Q.-Q., Xu, H., Huang, M.-B., Ma, L.-M., Huang, Q.-J., Yao, Q., et al. (2012). MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion. Neuro Oncol. 14, 278–287. doi: 10.1093/neuonc/nor216
Zhang, C., Zhang, J., Hao, J., Shi, Z., Wang, Y., Han, L., et al. (2012). High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 10:119. doi: 10.1186/1479-5876-10-119
Zhang, J.-S., Zhao, Y., Lv, Y., Liu, P.-Y., Ruan, J.-X., Sun, Y.-L., et al. (2017). miR-873 suppresses H9C2 cardiomyocyte proliferation by targeting GLI1. Gene 626, 426–432. doi: 10.1016/j.gene.2017.05.062
Zhao, W., Tong, H., Huang, Y., Yan, Y., Teng, H., Xia, Y., et al. (2017). Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J. Biol. Chem. 292, 2773–2784. doi: 10.1074/jbc.m116.763961
Zhao, Y.-Y., Zhao, L.-N., Wang, P., Miao, Y.-S., Liu, Y.-H., Wang, Z.-H., et al. (2015). Overexpression of miR-18a negatively regulates myocyte enhancer factor 2D to increase the permeability of the blood-tumor barrier via Krüppel-like factor 4-mediated downregulation of zonula occluden-1, claudin-5, and occludin. J. Neurosci. Res. 93, 1891–1902. doi: 10.1002/jnr.23628
Zheng, X., Chopp, M., Lu, Y., Buller, B., and Jiang, F. (2013). MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 329, 146–154. doi: 10.1016/j.canlet.2012.10.026
Keywords: glioma, epigenetics, DNA methylation, miRNA, chromatin remodeling, histone modifications
Citation: Zang L, Kondengaden SM, Che F, Wang L and Heng X (2018) Potential Epigenetic-Based Therapeutic Targets for Glioma. Front. Mol. Neurosci. 11:408. doi: 10.3389/fnmol.2018.00408
Received: 12 July 2018; Accepted: 16 October 2018;
Published: 15 November 2018.
Edited by:
Alberto Javier Ramos, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Marianela Candolfi, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaTimothy J. Jarome, Virginia Tech, United States
Copyright © 2018 Zang, Kondengaden, Che, Wang and Heng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Wang, d2FuZ2xqNzMwQDE2My5jb20=
Xueyuan Heng, aGVuZ3h1ZXl1YW4xMkAxNjMuY29t
 Lanlan Zang
Lanlan Zang Shukkoor Muhammed Kondengaden
Shukkoor Muhammed Kondengaden Fengyuan Che
Fengyuan Che Lijuan Wang
Lijuan Wang Xueyuan Heng
Xueyuan Heng