- 1CNR Institute of Cell Biology and Neurobiology, Monterotondo, Italy
- 2School of Medicine, Institute of Otolaryngology, Catholic University, Rome, Italy
- 3Department of Science, Roma Tre University, Rome, Italy
- 4Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy
- 5Department of Physics and Astronomy G. Galilei, University of Padua, Padua, Italy
- 6CNR Institute of Neuroscience, Padua Section, Padua, Italy
- 7Department of Clinical Neurobiology, Deutches Krebforschungzentrum, University of Heidelberg, Heidelberg, Germany
- 8Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai, China
Panx1 forms plasma membrane channels in brain and several other organs, including the inner ear. Biophysical properties, activation mechanisms and modulators of Panx1 channels have been characterized in detail, however the impact of Panx1 on auditory function is unclear due to conflicts in published results. To address this issue, hearing performance and cochlear function of the Panx1−/− mouse strain, the first with a reported global ablation of Panx1, were scrutinized. Male and female homozygous (Panx1−/−), hemizygous (Panx1+/−) and their wild type (WT) siblings (Panx1+/+) were used for this study. Successful ablation of Panx1 was confirmed by RT-PCR and Western immunoblotting in the cochlea and brain of Panx1−/− mice. Furthermore, a previously validated Panx1-selective antibody revealed strong immunoreactivity in WT but not in Panx1−/− cochleae. Hearing sensitivity, outer hair cell-based “cochlear amplifier” and cochlear nerve function, analyzed by auditory brainstem response (ABR) and distortion product otoacoustic emission (DPOAE) recordings, were normal in Panx1+/− and Panx1−/− mice. In addition, we determined that global deletion of Panx1 impacts neither on connexin expression, nor on gap-junction coupling in the developing organ of Corti. Finally, spontaneous intercellular Ca2+ signal (ICS) activity in organotypic cochlear cultures, which is key to postnatal development of the organ of Corti and essential for hearing acquisition, was not affected by Panx1 ablation. Therefore, our results provide strong evidence that, in mice, Panx1 is dispensable for hearing acquisition and auditory function.
Introduction
Panx1 is the most thoroughly characterized member of the pannexin gene family (Panchin et al., 2000; Bruzzone et al., 2003; Baranova et al., 2004) encoding Panx1, Panx2 and Panx3 proteins1 that form plasma membrane channels known as pannexons (Sosinsky et al., 2011). Biophysical properties, activation mechanisms and modulators of Panx1 channels have been extensively reviewed (Dahl et al., 2013; Penuela et al., 2013; Dahl and Muller, 2014; Patel et al., 2014; Esseltine and Laird, 2016).
Quantification of Panx1 mRNA levels by quantitative real-time polymerase chain reaction (QPCR) in mouse central and peripheral nervous system, and various organs, revealed highest values in trigeminal ganglia > bladder > spleen, followed at distance by hippocampus > cortex ~ calvaria > heart > cerebellum, with lowest levels in kidney and spleen (Hanstein et al., 2013). Furthermore, Panx1 has been localized in eye (Ray et al., 2005; Kurtenbach et al., 2014), taste buds (Huang et al., 2007), olfactory system (Zhang et al., 2012) and inner ear (Tang et al., 2008; Wang et al., 2009; Zhao, 2016). As for the latter, the impact of Panx1 on auditory function is unclear due to recent publication of conflicting results.
The first Panx1 knockout mice (Panx1tm1Mony, International strain designation B6;129-Panx1tm1.1Fama/Cnrm; EMMA ID:11476)2, hereafter referred to briefly as Panx1−/−, were reported in Anselmi et al. (2008). A subsequent study used in situ hybridization and western blot in brain extracts to confirm successful Panx1 deletion in these mice (Bargiotas et al., 2011). The same study determined that the following antibodies were not specific: goat anti-Panx1, sc-49695 (Santa Cruz); chicken anti-Panx1, no. ANT0027 (Diatheva); chicken anti-human Panx1, #4515, provided by G. Dahl (University of Miami School of Medicine); rabbit anti-Panx1, provided by G. Zoidl (Bochum, Germany). Panx1−/− mice displayed altered electroretinograms in response to light flashes (Kranz et al., 2013), whereas no auditory phenotype was detected by auditory brainstem response (ABR) analysis (Anselmi et al., 20083).
Other researchers used mice with loxP sites flanking exon 2 of Panx1, obtained from Genentech. Deletion of exon 2 (which introduces a frameshit and premature stop codon), was accomplished by breeding to C57BL/6-Gt(ROSA)26Sortm16(Cre) Arte (TaconicArtemis), a ubiquitously active Cre deleter line (Otto et al., 2009). Offspring were backcrossed several times to C57BL/6 mice, to breed out the Cre recombinase, resulting in a knockout strain henceforth referred to as Genentech-Panx1−/−. Successful ablation of Panx1 was confirmed in several organs on these mice (muscle, lung, liver, kidney, tail, brain, heart, thymus, spleen and skin tissues; Qu et al., 2011; Cone et al., 2013; Penuela et al., 2014) as well as in the cochlea (Abitbol et al., 2016). Genentech-Panx1−/− mice exhibited neither reduced hearing sensitivity nor cochlear nerve defects; furthermore, susceptibility to noise-induced hearing loss in these mice was indistinguishable from that of their wild type (WT) controls (Abitbol et al., 2016).
A strain carrying the Panx1tm1a(KOMP)Wtsi allele was generated by the Knock Out Mouse Project (KOMP4) using the multipurpose tm1a knockout-first promoter-driven selection cassette, which has been adopted also by other major mouse knockout programs such as EUCOMM (EUropean Conditional Mouse Mutagenesis Program5; Collins et al., 2007). The versatile tm1a allele contains an IRES:lacZ trapping cassette and a floxed promoter-driven neo cassette inserted into the intron of a gene, disrupting gene function6. However, mice homozygous for the Panx1tm1a(KOMP)Wtsi knockout-first promoter-driven allele were found to express about 30% residual Panx1 mRNA in all organs examined, leading to the conclusion that Panx1tm1a(KOMP)Wtsi is a hypomorphic allele (Hanstein et al., 2013). Hypomorphism has been reported also for other knockout-first alleles (Shpargel et al., 2012; Ryder et al., 2014).
Aiming to achieve Panx1 conditional deletion in the cochlea, Chen et al. (2015) crossed the hypomorphic Panx1tm1a(KOMP)Wtsi mice with Foxg1-Cre mice (Hébert and McConnell, 2000; Bredenkamp et al., 2007), and identified homozygous Foxg1Cre:Panx1f/f mice as conditional Panx1 knockout mice (henceforth referred to as Foxg1-cPanx1−/−). Intense immunofluorescence labeling with the non-specific chicken anti-human Panx1 antibody #4515 was detected in the spiral limbus and organ of Corti, but not in the lateral wall of Foxg1-cPanx1−/− mice, which had significantly elevated hearing thresholds at all frequencies, reduced endocochlear potential and reduced cochlear microphonics (Chen et al., 2015).
Zhao et al. (2015) crossed Panx1tm1a(KOMP)Wtsi mice with Paired box 2 Cre (Pax2-Cre) mice (Tian et al., 2006; Ohyama, 2009) to generate yet another strain of Panx1 conditional knockout in the cochlea (henceforth referred to as Pax2-cPanx1−/−). ABR thresholds of Pax2-cPanx1−/− mice were 40 dB sound pressure level (SPL) greater than in WT littermates at P80 (where P0 is day of birth). Likewise, DPOAEs were significantly decreased in Pax2-cPanx1−/− mice compared to WT (Zhao et al., 2015) at P50.
In the light of these contrasting results, the goal of the present study was to clarify the role of Panx1 in hearing. To this end, we re-evaluated hearing performance and cochlear function of Panx1−/− mice using in vivo electrophysiology, plus a variety of biochemical and biophysical assays.
Materials and Methods
Ethics Statement
Animal work was performed in accordance with a protocol approved by the Italian Ministry of Health (Authorization n.1005/2016-PR, date 21/10/2016, DGSAF Prot. No. 002451-P-25/10/2016 and No. 0001276-P-19/01/2016).
Animals and Genotyping
Animals were bread and genotyped in the CNR Monterotondo node of the European Mouse Mutant Archive (EMMA; del Hierro et al., 2016), an ESFRI/INFRAFRONTIER Distributed Research Infrastructure7.
Mice tested were P5 pups and adults (1–3 months of age). Male and female homozygous (Panx1−/−), hemizygous (Panx1+/−) and their WT siblings (Panx1+/+) were used for this study. The background strains of these mice was C57BL/6N.
Panx1 mice were genotyped according to published protocols by standard PCR on extracted mouse tail tips (Anselmi et al., 2008; Bargiotas et al., 2011) using the following primers:
• Panx1 f: 5′-GGAAAGTCAACAGAGGTACCC-3′
• Panx1 r: 5′-CTTGGCCACGGAGTATGTGTT-3′
• LacZ: 5′-GTCCCTCTCACCACTTTTCTTACC-3′
The Panx1+/+ allele was targeted by the above f and r primers and identified by a 330 bp band, whereas Panx1−/− was targeted by primers Panx1 f and LacZ, and was identified by a 630 bp band. Panx1+/− was identified by the simultaneous presence of a 330 bp and a 630 bp band.
For some internal control experiments, mice with ubiquitous deletion of connexin 30 (Cx30−/−; International strain name B6.129P2-Gjb6tm1Kwi/Cnrm; EMMA ID: 00323; MGI ID: 2447863; Teubner et al., 2003; Cohen-Salmon et al., 2007; Schütz et al., 2010; Johnson et al., 2017) were also used. The background strains of these mice was C57BL/6J. Cx30−/− mice were genotyped as indicated in the protocol provided by INFRAFRONTIER:
• Cx30 f: 5′-GGTACCTTCTACTAATTAGCTTGG-3′
• Cx30 r: 5′-AGGTGGTACCCATTGTAGAGGAAG-3′
• LacZ: 5′-AGCGAGTAACAACCCGTCGGATTC-3′
Cx30+/+ mice were identified by a 544 bp band, whereas Cx30−/− mice were identified by a 460 bp band.
RNA Extraction and RT-PCR
Following euthanasia, cochleae and brains were dissected from adult and P5 Panx1+/+ and Panx1−/− mice and flash-frozen in liquid nitrogen. Total RNA was extracted with RNeasy mini kit (Qiagen, Cat. No. 74106) and retrotranscribed with oligo–dT 12–18 primers (ThermoFisher, Cat. No. 18418012) using Omniscript RT kit (Qiagen, Cat. No. 205111). Subsequently RT-PCR was performed using PCR BIO HS Taq Mix Red (PB10.23–02). The PCR profile was: 95°C for 5 min, 94°C for 15 s, 62°C for 30 s, 72°C for 15 s and 72°C for 2 min for 34 cycles. Primers used were:
• Panx1 ex3 f: 5′-ACACCTCTGCTCAGACCTGAA-3′
• Panx1 ex4 r: 5′-TGCACAGAAACTCGCGTCCGAGA-3′
• GAPDH f: 5′-ATGTGTCCGTCGTGGATCTGAC-3′
• GAPDH r: 5′-AGACAACCTGGTCCTCAGTGTAG-3′
The amplified products were run on a 1, 5% agarose gel with SYBR Safe (ThermoFisher, Cat. No. S33102) for visualization of the Panx1 band (336 bp) and the GAPDH band (132 bp) (Figure 1).
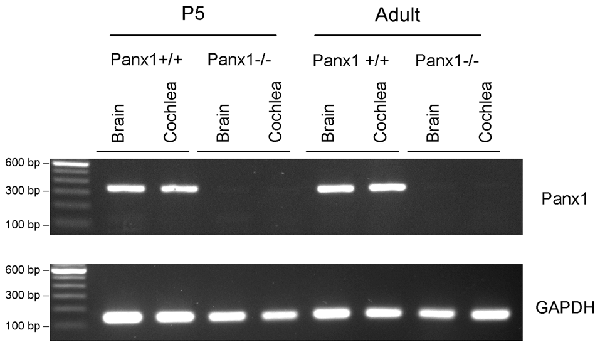
Figure 1. RT-PCR analysis of brain and cochlea Panx1 and GAPDH mRNA transcription in wild type (Panx1+/+) and Panx1−/− mice.
QPCR
QPCR was performed on cDNA to amplify Cx26 and Cx30 and was normalized to GAPDH. Gene expression relative to GAPDH was estimated according to a published method (Pfaffl, 2001). Amplification was carried out using Power SYBR Green (Applied Biosystems, Cat. No. 4367659) on the ABI 7700 sequence detection system equipped with ABI Prism 7700 SDS software (Applied Biosystems) through the following amplification cycles: 50°C for 2 min, 95°C for 10 min, 95°C for 15 min, 60°C for 1 min (40 cycles). Primers used are as follows:
• Cx26 f: 5′-TCACAGAGCTGTGCTATTTG-3′
• Cx26 r: 5′-ACTGGTCTTTTGGACTTTCC-3′
• Cx30 f: 5′-GGCCGAGTTGTGTTACCTGCT-3′
• Cx30 r: 5′-TCTCTTTCAGGGCATGGTTGG-3′
• GAPDH f: 5′-ATGTGTCCGTCGTGGATCTGAC-3′
• GAPDH r: 5′-AGACAACCTGGTCCTCAGTGTAG-3′
Western Immunoblotting
Total proteins were extracted from brains and cochleae of P5 and adult Panx1+/+ (n = 4) and Panx1−/− mice (n = 4). Tissues were dissected, collected on liquid nitrogen, stored at −80°C and homogenized by using ice cold RIPA buffer (Pierce; 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% DOC, 1% Triton X-100, 0.1% SDS, and 1× protease, phosphatase-1, and phosphatase-2 inhibitor cocktails [Sigma]). The lysate was sonicated three times at 10 Hz (Hielscher, Ultrasound Technology UP50H/UP100H), centrifuged (13,000 rpm, 15 min, 4°C), and a 5 μl aliquot of the supernatant was assayed to determine the protein concentration (microBCA kit, Pierce). SDS-PAGE reducing sample buffer was added to the supernatant, and samples were heated to 95°C for 5 min. Protein lysates (70 μg) were loaded onto 12% Tris-glycine polyacrylamide gels for electrophoretic separation. ColorburstTM Electrophoresis markers (Sigma) were used as molecular mass standards. Proteins were then transferred onto nitrocellulose membranes at 100 V for 2 h at 4°C in transfer buffer containing 25 mM Tris, 192 mM glycine, 0.1% SDS and 20% methanol. Membranes were incubated for 1 h with blocking buffer (5% skim milk in TBST), and then incubated overnight at 4°C with primary antibodies directed against Panx1 (1 mg/ml, ThermoFisher, Cat. No. 487900), Cx26 (1 mg/ml, ThermoFisher, Cat. No. 512800), Cx30 (1 mg/ml, ThermoFisher, Cat. No. 712200) and GAPDH (1:2500, Abcam). After three 10 min rinses in TBST, membranes were incubated for 1 h at room temperature (22–25°C) with HRP-conjugated secondary antibodies (Cell Signaling, 1:2500). The membranes were then washed, and the bands were visualized with an enhanced chemiluminescence detection kit (GE Healthcare, UK). Protein expression was evaluated and documented by using a UVItec Cambridge Alliance system.
Immunohistochemistry and Confocal Imaging
Cochleae were extracted from P5 and adult mice and processed as previously described (Crispino et al., 2011, 2017). Briefly, samples were fixed in 4% paraformaldehyde and decalcified in ethylenediaminetetraacetic acid (EDTA, 0.3 M). Specimens were included in 3% agarose dissolved in PBS and cut in 100 μm thickness steps using a vibratome (VT 1000 S, Leica). Tissue slices were permeabilized with 0.1% Triton X-100, dissolved in bovine serum albumin 2% solution. Panx1 was immunolabeled by overnight incubation at 4°C with a chicken anti-Panx1 specific antibody (1:500; extracellular loop epitope: VQQKSSLQSES, AvesLab #6358; Hanstein et al., 2013) provided by Prof. Eliana Scemes. Cx30 and Cx26 were immunolabeled by overnight incubation at 4°C respectively with a rabbit polyclonal Cx30 and a mouse monoclonal Cx26 selective antibody (10 μg/ml, ThermoFisher, Cat. No. 712200 for Cx30; Cat. No. 335800 for Cx26). Secondary antibodies (10 μg/ml) were: Alexa Fluor® 488 goat anti-chicken IgY (H+L), ThermoFisher, Cat. No. A11039; Alexa Fluor® 488 goat anti-rabbit IgG, ThermoFisher, Cat. No. A11008; Alexa Fluor® 488 goat anti-mouse IgG, ThermoFisher, Cat. No. A11029), applied at room temperature (22–25°C). F–Actin was stained by incubation with AlexaFluor 568 phalloidin (1 U/ml, ThermoFisher, Cat. No. A12380), and nuclei were stained with 4′,6′diamidino–2′phenylindole (DAPI, ThermoFisher, Cat. No. D1306; 1:200). The same immunostaining procedure was used also for organotypic cultures from P5 pups. All samples were mounted onto glass slides with a mounting medium (FluorSaveTM Reagent, Merk, Cat. No. 345789) and analyzed using a confocal microscope (TCS SP5, Leica) equipped with an oil–immersion objective (40× HCX PL APO 1.25 N.A., Leica).
ABR and DPOAE Measurement
Auditory function was assessed in a sound-attenuating enclosure (ETS-Lindgren SD Test Enclosure, MDL Technologies Limited, Hitchin, UK) using an ABR Workstation (Tucker-Davis Technologies, Inc., Alachua, FL, USA) comprising: Z-Series 3-DSP Bioacoustic System w/Attenuators and Optic fiber; Medusa 4-Channel Pre-Amp/Digitizer; Medusa 4-Channel Low Imped. Headstage; MF1-M Multi Field Magnetic Speakers—Mono; AEP/OAE Software for RZ6; Experiment Control Workstation. Sound levels were calibrated using a inch Free Field Measure Calibration Microphone Kit (Model 480C02; PCB).
Mice were anesthetized with intraperitoneal injections of ketamine (70 mg/g for males, 100 mg/g for females) and medetomidine (1 mg/g). The depth of anesthesia was periodically verified by the lack of foot-pinch response. Body temperature was maintained at 37°C using a heating pad under feedback control. Corneal drying was prevented by application of ophthalmic gel to the eyes of the animals.
For ABR recordings (Scimemi et al., 2014), acoustic stimuli consisted of clicks (100 μs duration) and tone bursts (1 ms rise–fall time with 3 ms plateau) of 4, 8, 16, 24 and 32 kHz, and were delivered in the free field using a MF1-M speaker. Bioelectrical potentials were collected with gauge 27, 13 mm needle electrodes (Cat. No. S83018-R9, Rochester) inserted subdermally at the vertex (active), ventrolateral to the left ear (reference) and above the tail (ground). Potentials were amplified, filtered (0.3–3 kHz) and averaged over 512 presentations of the same stimulus. Hearing threshold levels were determined offline as the SPL at which a Wave II peak, could be visually identified above the noise floor (0.1 μV).
Otoacoustic emissions (Kemp, 1978) were evoked using a pair of equal intensity primary tones, f1 = 14,544 Hz, and f2 = 17,440 kHz delivered at intensities ranging from 20 to 80 dB SPL in 10 dB SPL increments. Each primary tone (20.97 ms duration, 47/s) was emitted by a separate MF1-M speaker, configured for closed field stimulation, and delivered to the mouse ear via a small tube as prescribed by the manufacturer. The cubic distortion product 2f1 – f2 = 11,648 kHz, was detected using a small microphone (ER10B+ Low Noise Probe and Microphone, Etymotic Research, IL, USA) coupled to the ear canal.
Preparation of Cochlea Organotypic Cultures
Cochleae from P5 mouse pups were quickly dissected in ice-cold Hepes buffered (pH 7.2) HBSS (ThermoFisher, Cat. No. 14025050), placed onto 12 mm glass coverslips coated with Cell-Tak (Biocoat, Cat. No. 354240) and incubated overnight at 37°C in DMEM/F12 (ThermoFisher, Cat. No. 11320-074) supplemented with 5% FBS (ThermoFisher, Cat. No. 10270-106) and 100 μg/ml ampicillin (Sigma-Aldrich, Cat. No. A0166).
Dye Transfer Assays in Cochlear Organotypic Cultures
To visualize gap junction coupling among non-sensory cells of the lesser epithelial ridge (LER), we performed dye-transfer assays using the fluorescent tracer Lucifer Yellow (LY, CH Lithium Salt, Thermofisher, #L12926) for microinjection. Cochlear cultures were transferred on the stage of a spinning disk confocal microscope (DSU, Olympus) and perfused for 5 min at 1 ml/min with EXM, an extracellular solution containing (in mM): NaCl 135, KCl 5.8, CaCl2 1.3, NaH2PO4 0.7, MgCl2 0.9, Hepes−NaOH 10, d−glucose 6, pyruvate 2, amino acids and vitamins (pH 7.48, 307 mOsm). For dye delivery, patch pipettes were fabricated from glass capillaries (G85150T-4, Harvard Apparatus, Edenbridge, UK) using a double stage vertical puller (PP-830, Narishige) and were filled with LY dissolved at 220 μM (final concentration) in a 320 mOsm intracellular solution containing (in mM): KCl 134, NaCl 4, MgCl2 1, HEPES 20, EGTA 10 (adjusted to pH 7.3 with KOH) and filtered through 0.22 μm pores (Millipore). Pipette resistances were 3–4 MOhm when immersed in the bath. One cell (donor) was patch clamped and maintained in the cell-attach configuration for a few seconds to establish a baseline. The patch of membrane under the pipette sealed to the donor cell was subsequently ruptured, allowing the LY to fill the cell, while leaving the seal intact (whole cell recording conditions). The cell was held at its zero current level using the current clamp configuration of the patch clamp amplifier (Axopatch 200B, Molecular devices). LY diffusion among (first order) cells adjacent to the injected cell was monitored over time by acquiring fluorescence images at a rate of 1 frame per second with a typical exposure time of 70 ms. Fluorescence images were displayed as (F − Fbck)/(Fmax − Fbck), where Fmax is the maximal value reached in the injected cell at the end of the recordin interval and Fbck is autofluorescence. Experiments were performed at room temperature (22–25°C).
Multiphoton Microscopy and Ca2+ Imaging in Cochlear Organotypic Cultures
To record spontaneous intercellular Ca2+ signal (ICS) activity in nonsensory cells of the mouse cochlea, organotypic cultures of sensory epithelium were incubated for 45 min at 37°C in DMEM/F12 supplemented with the acetoxymethyl (AM) ester of the selective Ca2+ sensor Fluo-Forte (16 μM, Enzo Life Science, #ENZ-52014). The incubation medium contains also pluronic F-127 (0.1% w/v, Sigma-Aldrich, #P2443) and sulfinpyrazone (250 μM, Sigma-Aldrich, #S9509) to prevent dye sequestration and secretion. Cultures were then transferred to an upright microscope stage (see below) and continually perfused with EXM (see above) for 15 min at 1 ml/min in a dark environment at 25°C to allow for dye de-esterification. All subsequent imaging experiments were also performed at 25°C.
To record Ca2+ signals, we used a two-photon microscope (Bergamo II, Thorlabs) equipped with a resonant scanner and coupled with a mode-locked Ti:Sapphire pulsed laser (Chameleon-Ultra II, Coherent). Fluo-Forte was excited at 940 nm by focusing the Ti:Sapphire beam onto the sample through a water-immersion objective (XLPlan N, 25× 1.05 NA, Olympus). Average power at the sample was ~20 mW. The fluorescence signal, collected by the same objective, was reflected towards the detection arm of the microscope by the 705 nm primary dichroic mirror of the microscope (Semrock, FF705-Di01), placed at 45° above the objective. After traversing a 680 nm short pass filter (FF01-680/SP-25, Semrock) and 495 nm dichroic beam-splitter (T495lpxru, Chroma Technology), the Fluo-Forte signal was selected in the range 435–485 nm by a single band-pass filter (ET460/50m-2p, Chroma Technology) placed in front of a non-descanned GaAsP detector (H7422–50, Hamamatsu). Mechanical ultra-fast shutters were used to limit light exposure to the bear minimum required for image acquisition.
Sequences of 512 × 512 pixels frames were acquired, averaged in lots of nine and presented at a final rate of 5 per second. Illumination intensity, frame average, frame rate and the number of pixels in each frame were adjusted so as to minimize photobleaching and phototoxicity, while achieving sufficient signal to noise ratio and temporal resolution. Image sequences were acquired using ThorImage LS 3.1 software (Thorlabs).
Ca2+ signals were quantified as pixel-by-pixel relative changes of fluorescence emission intensity, i.e., ΔF(t)/F0 where t is time, F(t) is fluorescence at time t, F0 is the fluorescence at the onset of the recording and ΔF(t) = F(t) − F0. All data were processed off-line and presented using Vimmaging (F. Mammano and C. Ciubutaru, VIMM, Padova, Italy), a custom-made software routine developed under MATLAB™ environment (The MathWorks Inc., Natick, MA, USA).
Results
Panx1 Is Absent in the Cochlea of Panx1−/− Mice
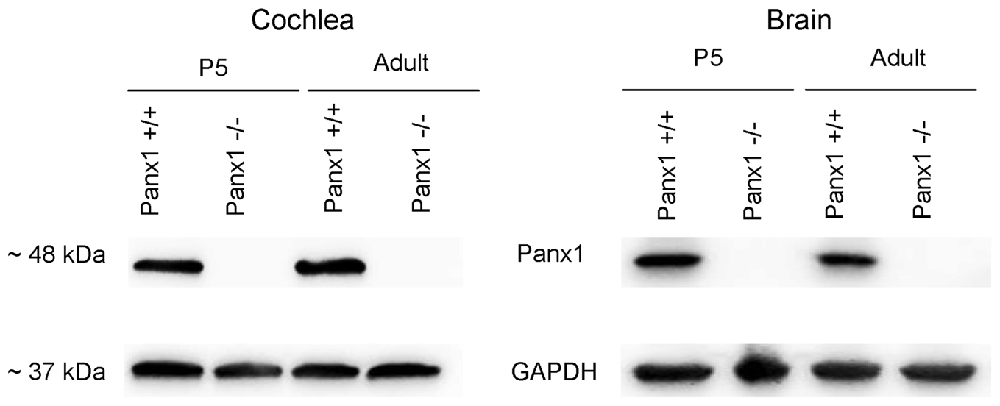
Generation and genotyping of Panx1−/− mice were previously described (Anselmi et al., 2008; Bargiotas et al., 2011). Here we report an additional data set based on Panx1 expression analyses by RT-PCT and Western immunoblotting. Panx1 mRNA transcript expression was detected in the cochlea and brain of WT mice at both P5 and adult stage, but not in Panx1−/− mice (Figure 1). Consistent with these results, Western blots failed to reveal Panx1 expression in the cochlea of Panx1−/− mice at both P5 and in the adult stage, whereas bands with the correct molecular weight (~48 kDa) were present in Panx1+/+ (i.e., WT) extracts (Figure 2).
Using a previously validated antibody that targets an extracellular epitope of the Panx1 protein (AvesLab #6358; Hanstein et al., 2013), we detected strong immunoreactivity in epithelial cells lining the endolymphatic surface of the sensory epithelium (inner sulcus, outer sulcus), supporting and epithelial cells of the organ of Corti, Reissner’s membrane, and spiral ganglion neurons of WT mice. Weak immunostaining was also detected in the spiral limbus and spiral ligament of these mice, whereas the AvesLab #6358 antibody failed to label cochlear tissue from Panx1−/− mice (Figure 3). Altogether these results confirm successful ablation of Panx1 in the (brain and) cochlea of Panx1−/− mice.
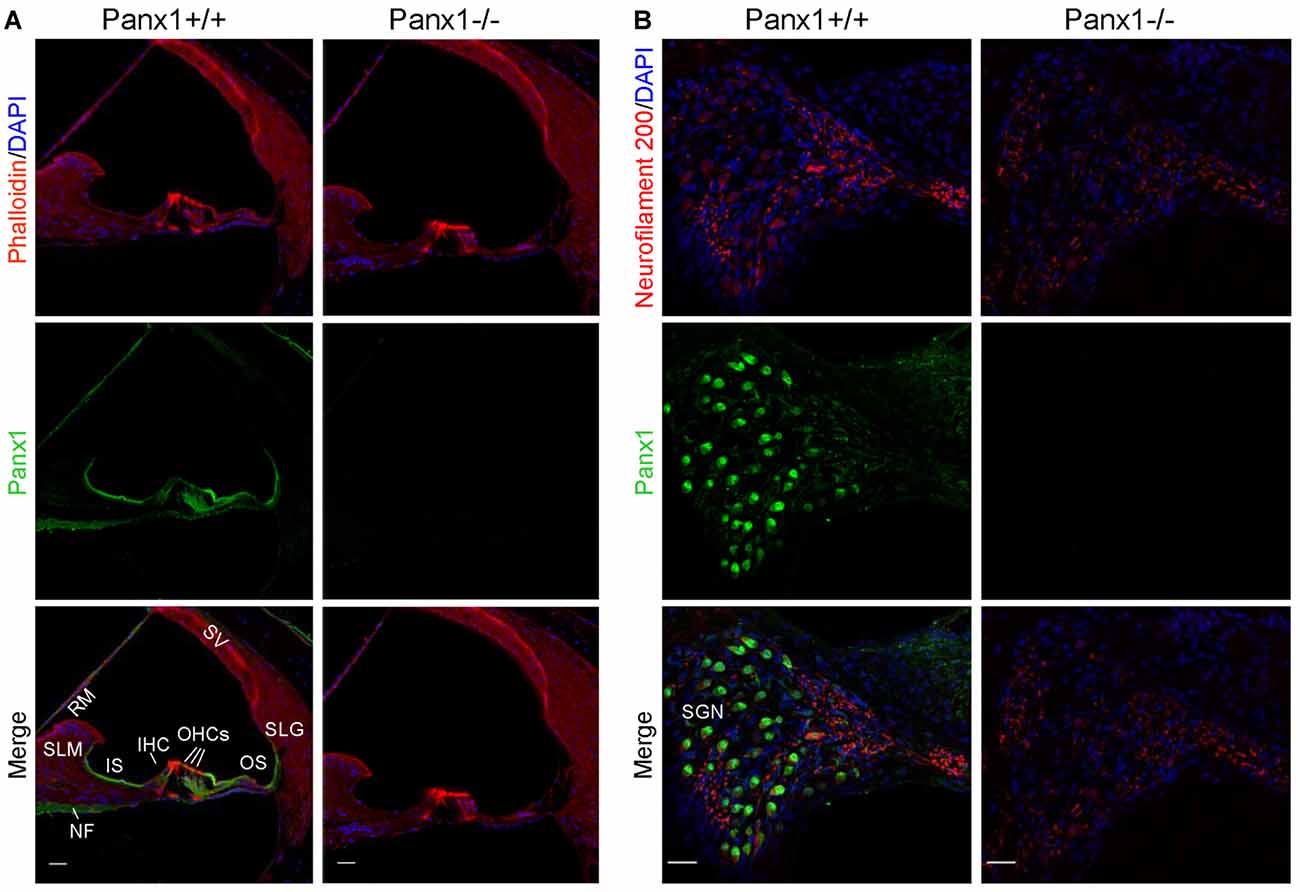
Figure 3. Immunofluorescence with a validated Panx1-selective antibody in cochlear midmodiolar sections. (A) Representative transversal sections of scala media. Shown are maximal projection rendering of three consecutive confocal optical sections taken at 0.5 μm intervals in the apical cochlear turn; actin filaments were stained with phalloidin (red) and nuclei with DAPI (blue); Panx1 expression was detected with the AvesLab #6358 selective antibody (green). IHC, inner hair cells; OHCs, outer hair cells; RM, Reissner’s membrane; SV, stria vascularis; SLM, spiral limbus; SLG, spiral ligament; IS, inner sulcus; OS, outer sulcus; NF, nerve fibers. Gamma filters were applied to both red (γ = 0.40) and green (γ = 0.55) channels. Scale bars: 25 μm. (B) Representative transversal sections of spiral ganglion. Maximal projection rendering of 10 consecutive confocal optical sections taken at 1.5 μm intervals in the medial cochlear turn; nerve fibers were stained with a neurofilament 200 selective antibody (red). SGN, spiral ganglion neurons. Gamma filters were applied to both red (γ = 0.55) and green (γ = 0.9) channels. Scale bars: 25 μm.
ABRs and DPOAEs in Panx1−/− Mice Are Indistinguishable from WT Controls
Next, we sought to corroborate and extend the results obtained by Anselmi et al. (2008) by analyzing in greater detail the hearing performance of Panx1−/− mice. In humans and mice alike, sound-evoked ABR potentials appear as a series of consecutive relative maxima (peaks), termed Waves and labeled with Roman numerals, which arise from the synchronous short-latency synaptic activity of successive nuclei along the peripheral afferent auditory neural pathway (Zheng et al., 1999; Legatt, 2002; Zhou et al., 2006). The first peak (Wave I) arises from the cochlea and/or compound action potential of the auditory nerve ~1 ms after the stimulus onset (latency). Waves from II to V originate from cochlear nuclei, contralateral superior olivary complex, lateral lemniscus and contralateral lateral inferior colliculus. For reference, Table I of Scimemi et al. (2014) presents means and standard deviations (SD) of latency and amplitude values of ABR peaks I–V for C57BL/6 mice.
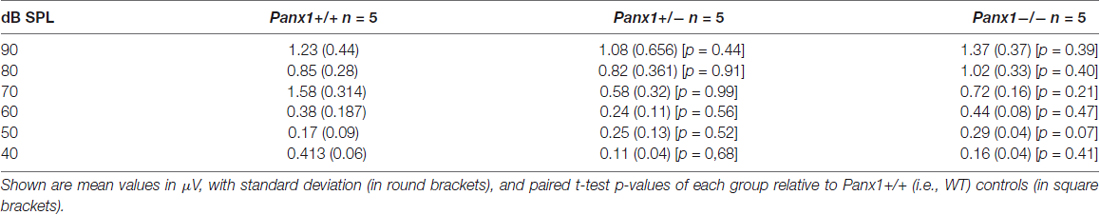
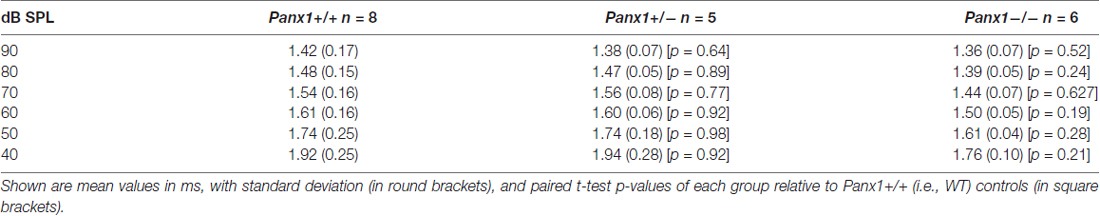
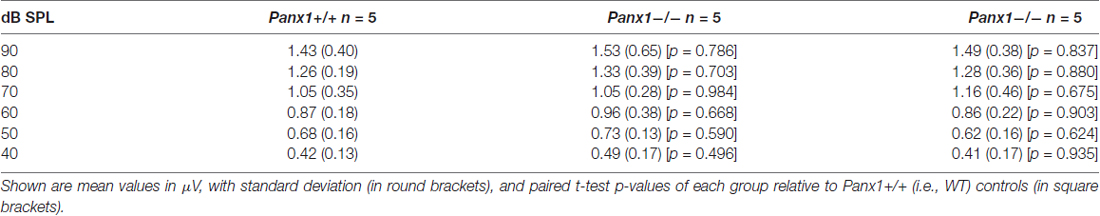
Hearing threshold estimates from click and pure-tone ABR analysis, as well as latency and amplitude of Wave I and Wave II in Panx1−/− mice aged between P30 and P90, were indistinguishable from those of age-matched Panx1+/− and WT control mice (Figures 4, 5, Tables 1–5).
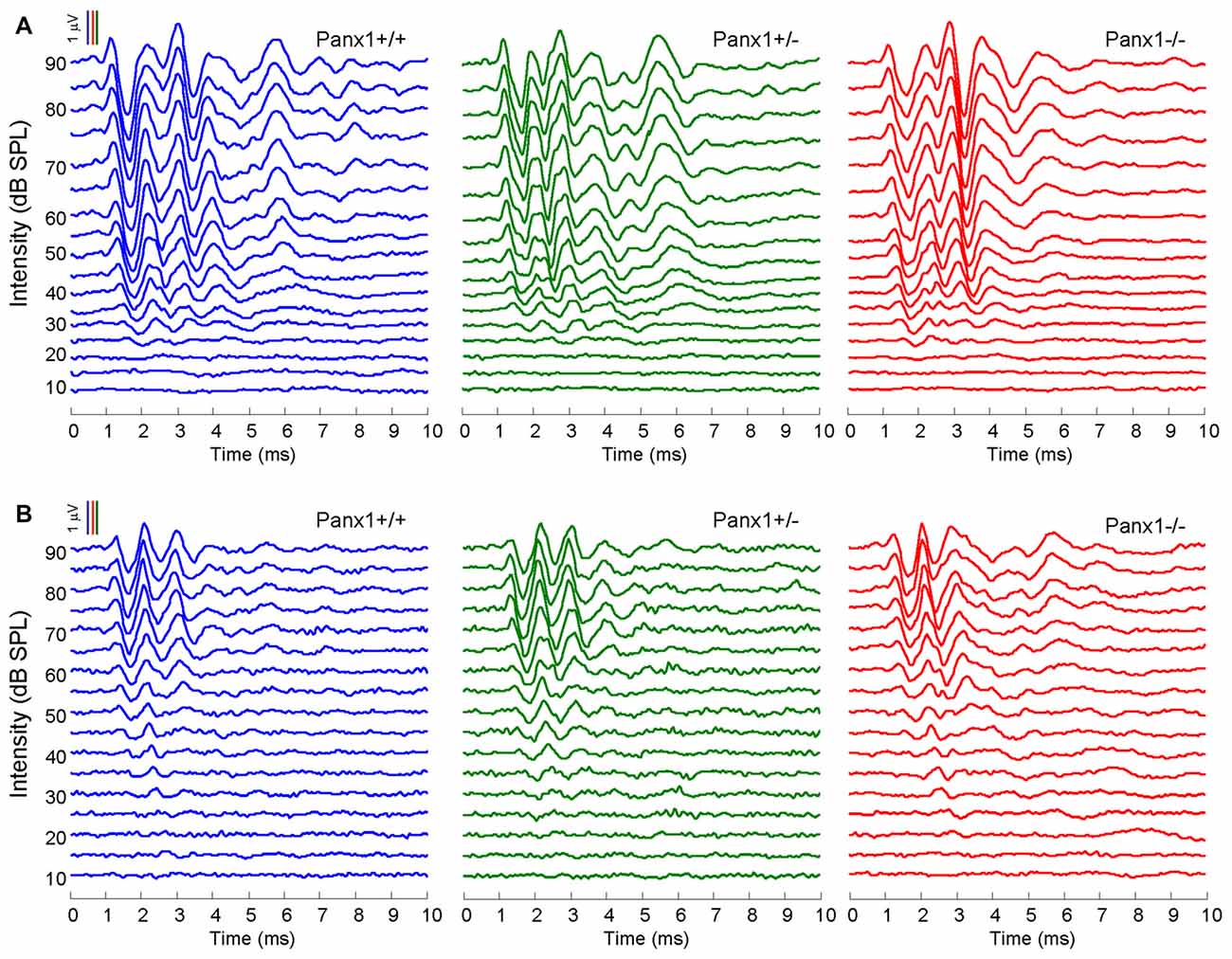
Figure 4. In vivo electrophysiological recordings. (A,B) Representative recordings of auditory brainstem responses (ABRs) evoked by clicks (A) and 24 kHz tone burst stimuli (B).
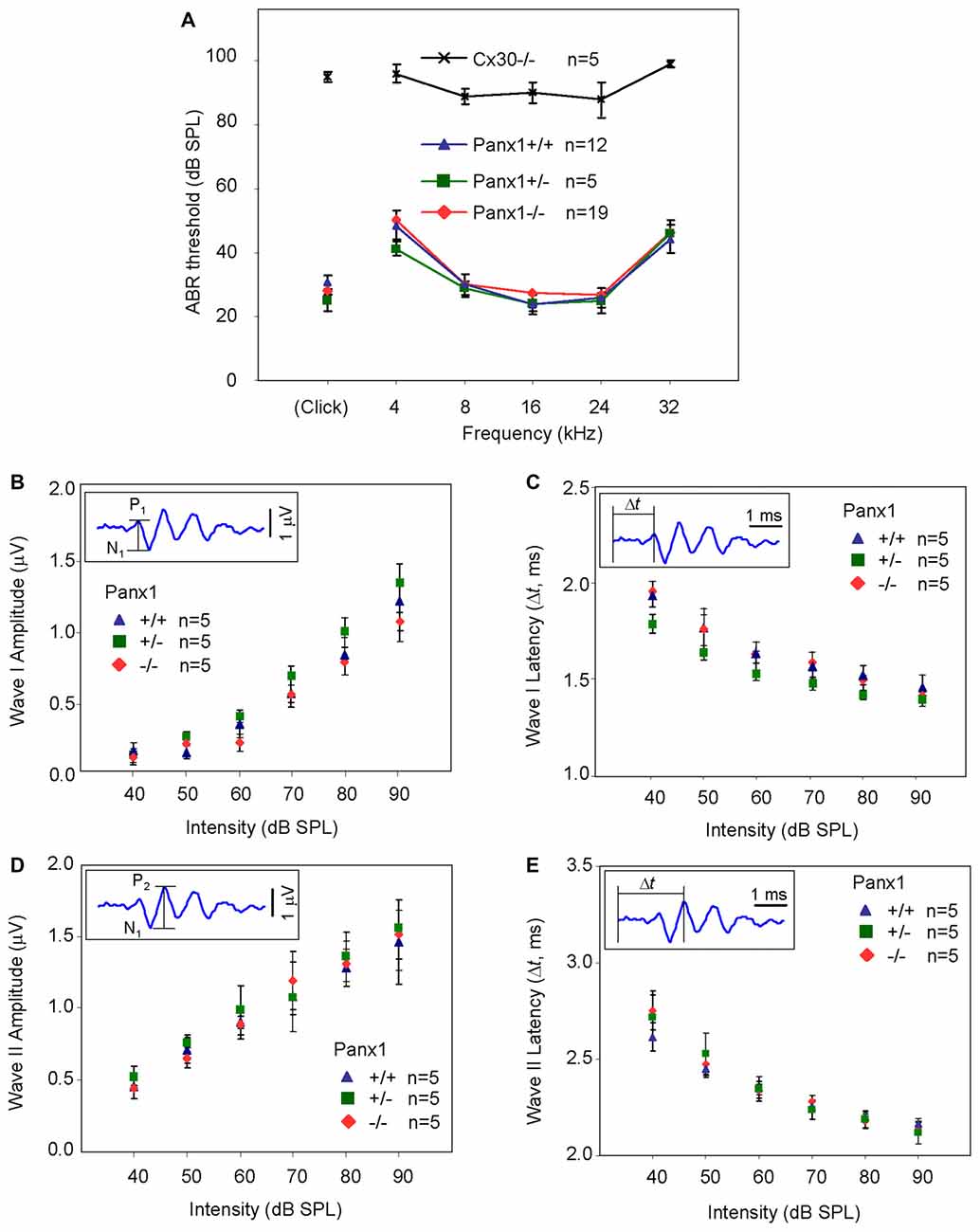
Figure 5. ABR analysis. (A) Hearing threshold estimated from Wave II for clicks and tone bursts at 4, 8, 16, 24, 32 kHz; Cx30−/− mice were used as an internal control. (B) Amplitude of Wave I (see inset) evoked by 24 kHz tone vs. stimulus intensity. (C) Latency of Wave I (see inset) evoked by 24 kHz tone pips vs. stimulus intensity. (D) Amplitude of Wave II (see inset) evoked by 24 kHz tone vs. stimulus intensity. (E) Latency of Wave II (see inset) evoked by 24 kHz tone pips vs. stimulus intensity. Error bars represent SEM. See also Tables 1–5.
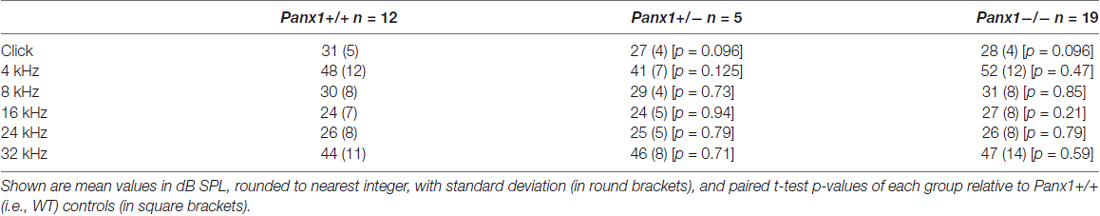
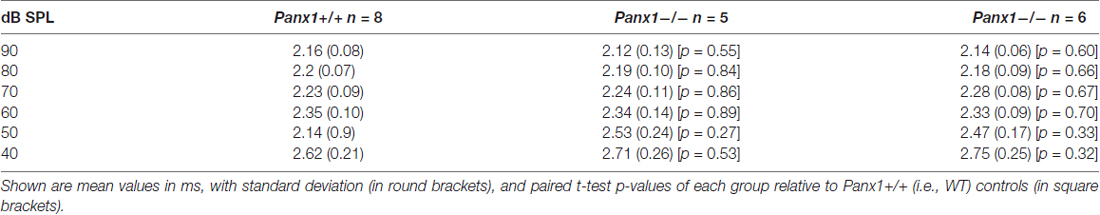
Table 1. Auditory brainstem response (ABR) thresholds for click stimuli and for tone bursts at 4, 8, 16, 24, 32 kHz obtained from Panx1+/+, Panx1+/− and Panx1−/− mice.
Sound generated within the mammalian inner ear as a reflection of outer hair cell (OHC) mechanical activity (Nobili and Mammano, 1996; Nobili et al., 2003) can be detected with a sensitive microphone placed in the auditory meatus (Kemp, 1978, 2002). Therefore, as a further non-invasive indicator of cochlear function, we measured the cubic (2f1 − f2) DPOAE (see “Materials and Methods” section) and found no significant differences in the DPOAE growth function of Panx1−/− mice and age-matched WT controls (Figure 6).
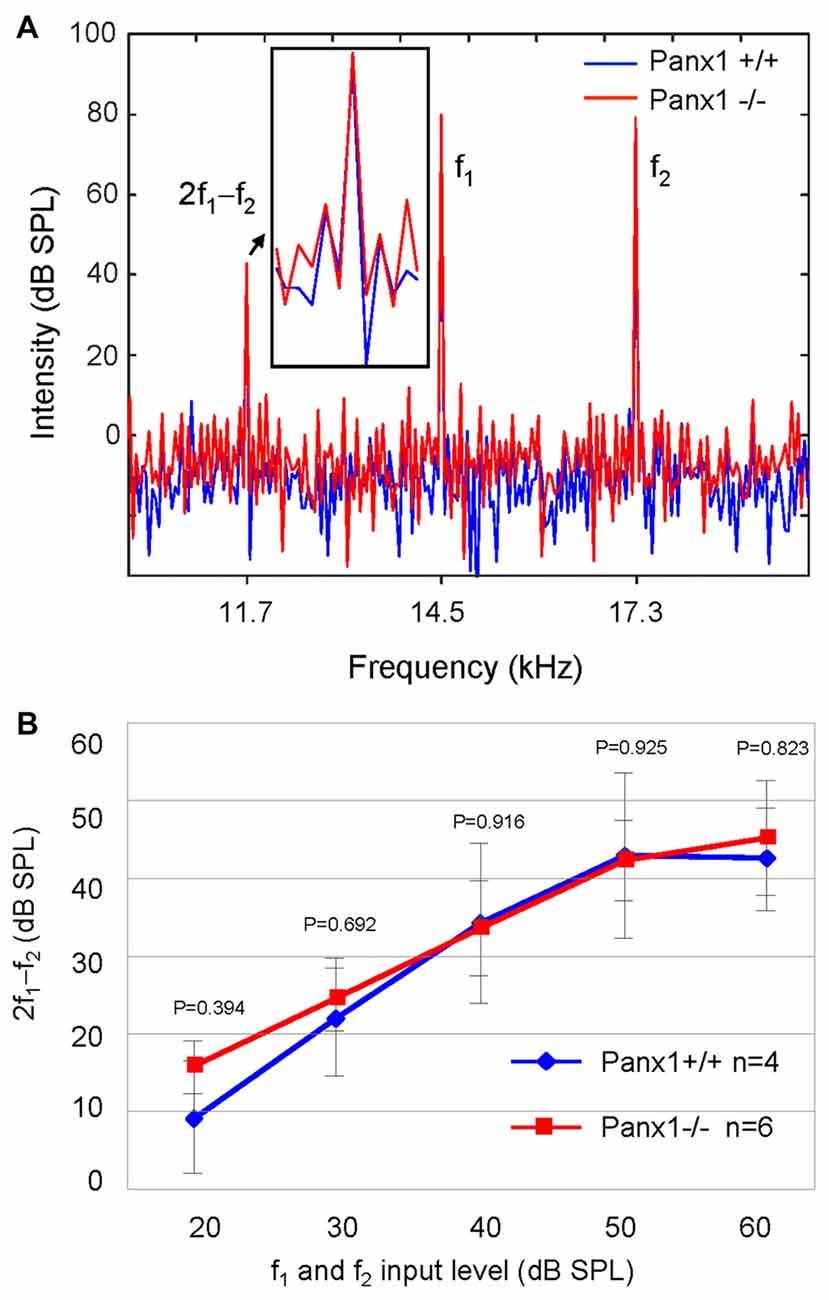
Figure 6. Analysis of the 2f1 − f2 cubic distortion product. (A) Representative spectra from WT (Panx1+/+, blue) and Panx1−/− mice (red) for a pair of 80 dB sound pressure level (SPL) primary frequencies (f1 and f2); the inset shows a magnified view of the spectrum in the region of the 2f1 − f2 cubic distortion product. (B) Growth function of the 2f1 − f2 cubic distortion product; error bars represent SEM; p-values were determined by two-tail t-test.
Altogether, these results indicate absence of detectable defects in auditory function of Panx1−/− mice. This conclusion is based on their normal hearing sensitivity, normal function of the outer hair cell-based “cochlear amplifier” (Frolenkov et al., 1998; Nobili et al., 1998; Ashmore, 2008) and absence of cochlear nerve defects.
Connexin Expression and Function Is Normal in Panx1−/− Mice
Pannexins bear significant sequence homology with the invertebrate gap junction proteins, innexins, and more distant similarities in their membrane topologies and pharmacological sensitivities with the gap junction proteins, connexins (Sosinsky et al., 2011).
Non-sensory cells of the mammalian cochlea express two closely related gap junction proteins, connexin 26 (Cx26) and connexin 30 (Cx30; Lautermann et al., 1998; Ahmad et al., 2003; Forge et al., 2003; Zhao et al., 2006), the expression of which is coordinately regulated (Ortolano et al., 2008). Mouse models indicate that altered expression levels of these connexins in the early postnatal days impacts on organ of Corti development and hair cell maturation (Johnson et al., 2017), preventing normal hearing acquisition (Cohen-Salmon et al., 2002; Teubner et al., 2003; Ahmad et al., 2007; Sun et al., 2009; Crispino et al., 2011, 2017; Qu et al., 2012; Zhu et al., 2013). As regulatory mechanism may potentially be shared between connexins and pannexins, we examined the expression of Cx26 and Cx30 by Western blot analysis and QPCR, and found no significant differences between Panx1−/− mice and age-matched WT controls (Figure 7). The spatial distribution of Cx26 at P5 was investigated also by immunofluorescence and, again, no differences between Panx1−/− mice and age-matched WT controls were detected (Figure 8).
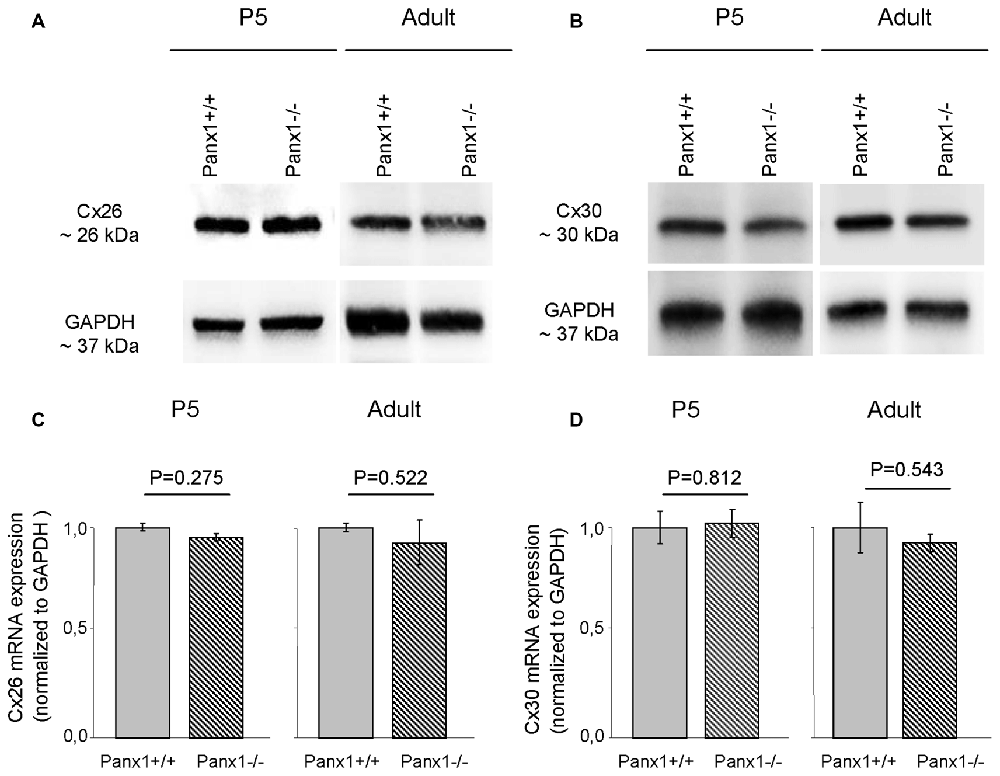
Figure 7. Level of Cx26 and Cx30 expression in Panx1−/− and Panx1+/+ (i.e., WT) cochleae. (A,B) Western blots antibodies selective for Cx26 (A) and Cx30 (B). (C,D) QPCR results for mRNA transcription in P5 and adult cochleae mice from WT and Panx1−/− mice (n = 3) normalized to WT, for Cx26 (C) and Cx30 (D). Error bars represent SEM; p-values were determined by two-tail t-test.
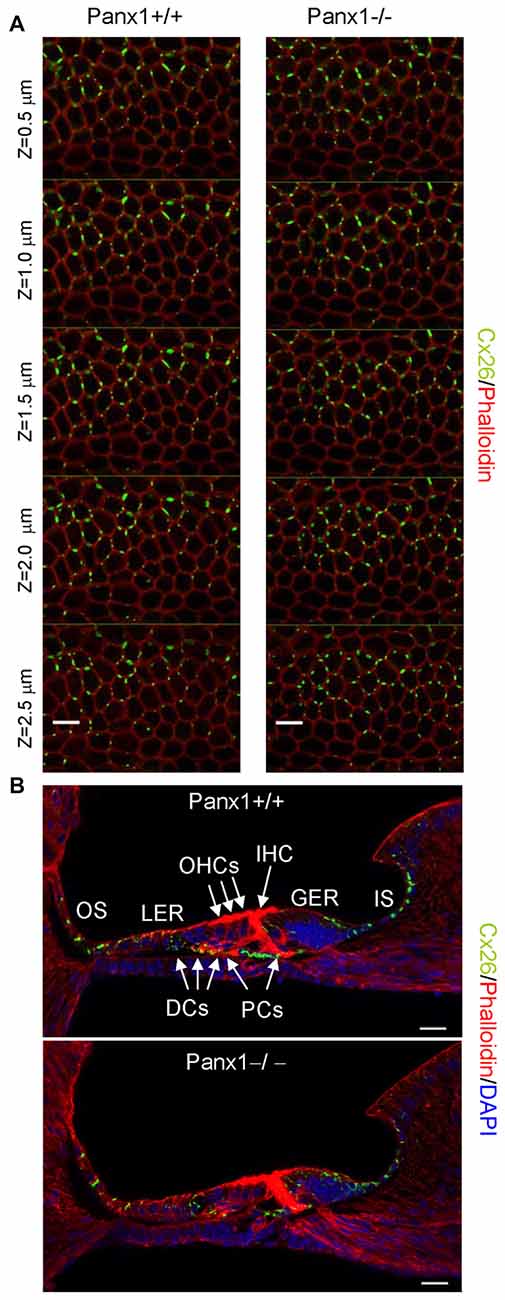
Figure 8. Cx26 expression pattern in the sensory epithelium of Panx1+/+ (i.e., WT) and Panx1−/− cochleae from P5 mice. (A) Confocal through-focus sequence (z-stack) acquired at 0.5 μm increments showing Cx26 expression detected with a selective antibody (green) in epithelial cells of the LER counterstained with phalloidin (red); scale bars: 10 μm. (B) Representative transversal sections of the organ of Corti. Shown are maximal projection rendering of three consecutive midmodiolar confocal optical sections taken at 0.5 μm intervals in the medial cochlear turn; nuclei were stained with DAPI (blue). IHC, inner hair cells; OHCs, outer hair cells; DCs, Deiters’ cells; LER, lesser epithelial ridge; greater epithelial ridge (GER) greater epithelial ridge; PCs, Pillar cells; scale bars: 20 μm.
To assess whether the expressed connexins confer cell-to-cell connectivity, we quantified dye transfer in the LER of organotypic cochlear cultures from P5 mice (see “Materials and Methods” section). To gauge transfer rate, we measured the slope of the LY fluorescence growth function at the onset of dye delivery in the donor cell (m1) and in its nearest neighbors (m2), and found no significant differences in the m2/m1 ratio of Panx1−/− mice and WT controls (Figure 9).
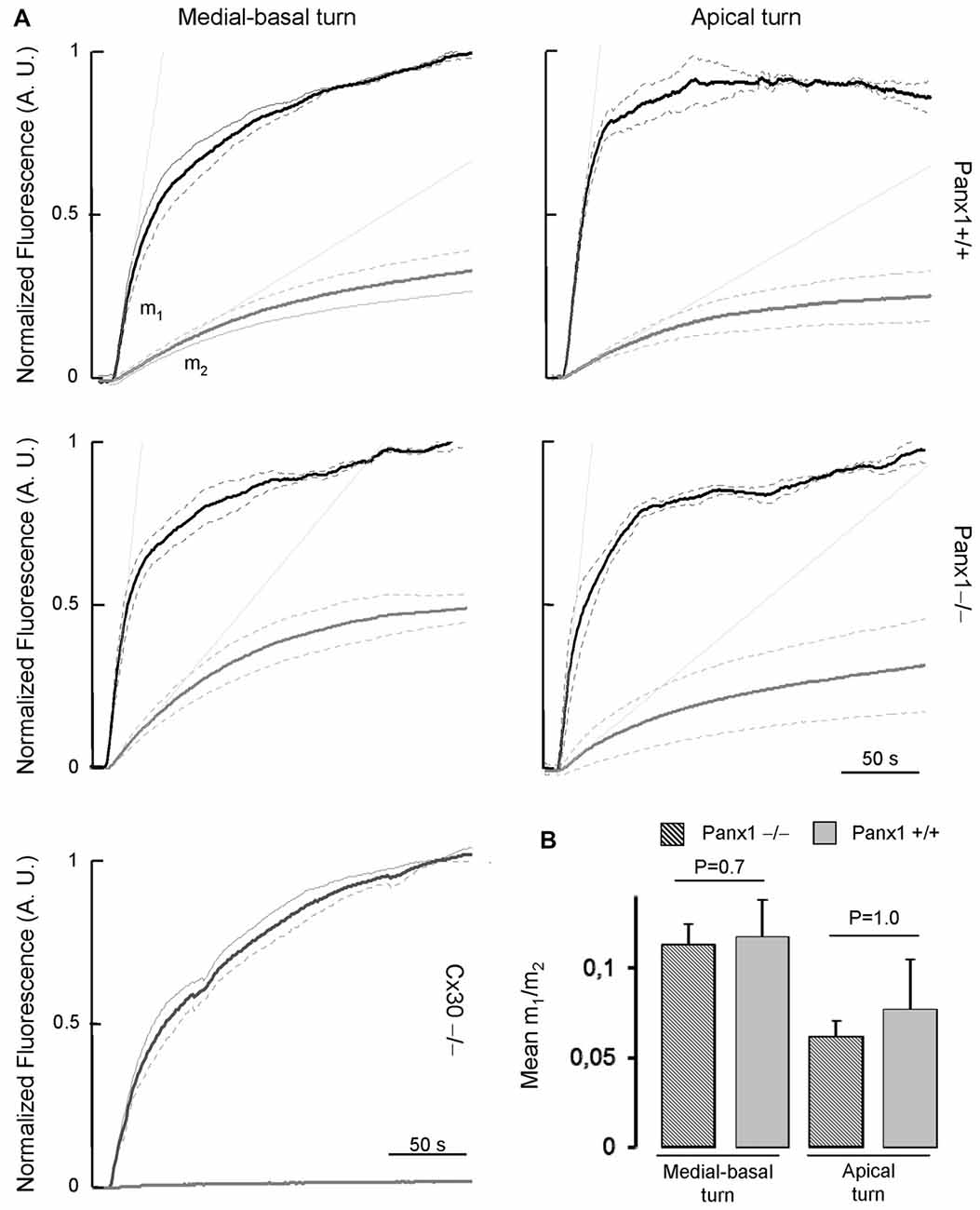
Figure 9. (A) Lucifer yellow fluorescence emission averaged over the cell body of (first order) cells (gray solid lines, n = 5) adjacent to the injected cell and normalized to the maximal fluorescence emission detected in the injected cell (black solid lines); data are mean values ± SEM (dot lines) for n = 8 cells in each condition. (B) For each experiment, the interpolating line of the curve related to first order cells (with computed slope m2) and to the injected cell (with computed slope m1) was computed over the first 10 s of recording. Histograms show mean values of the ratio between m2 and m1 for WT (Panx+/+, dashed bars) and Panx1−/− mice (filled bars). Error bars represent SEM.
Altogether the results presented in Figures 7–9 indicate that lack of Panx1 in Panx1−/− mice impacts neither on connexins expression, nor on gap-junction coupling in the developing organ of Corti.
Spontaneous Ca2+ Signaling Activity Is Normal in the Developing Cochlea of Panx1−/− Mice
Connexins play a crucial development role in the postnatal cochlea (as mentioned above), also by supporting ICS activity both in the LER (Beltramello et al., 2005; Piazza et al., 2007; Anselmi et al., 2008; Majumder et al., 2010; Ceriani et al., 2016) and in the greater epithelial ridge (GER; Tritsch et al., 2007; Schütz et al., 2010; Rodriguez et al., 2012; Wang and Bergles, 2015; Johnson et al., 2017; Mammano and Bortolozzi, 2017). Using focal ATP delivery or photostimulation with caged IP3, Anselmi et al. (2008) showed ICS failure in cultures with deficient expression of Cx26 and Cx30, whereas ICS in organotypic cultures from Panx1−/− mice was indistinguishable from those of WT controls.
Here, we used multiphoton microscopy to monitor spontaneous ICS activity (Tritsch et al., 2007; Schütz et al., 2010; Rodriguez et al., 2012; Wang and Bergles, 2015; Johnson et al., 2017; Mammano and Bortolozzi, 2017) in organotypic cochlear cultures from P5 mice loaded with the Ca2+ indicator Fluo Forte AM (Mammano and Bortolozzi, 2017). Specifically, we examined the frequency of occurrence of spontaneous Ca2+ transients (events) in the apical cochlear turn by counting all occurrences within the portion of the GER in the field view from P5 Panx1−/− mice and age–matched WT siblings (Figure 10). We found a similar mean frequencies of occurrence (15.5 ± 6.0 events/min in Panx1+/+ vs. 15.0 ± 3.4 events/min in Panx1−/− cultures). Likewise, amplitude and inter-peak interval distributions of spontaneous Ca2+ transients of Panx1−/− cultures overlapped with those of Panx1+/+ cultures. Altogether, these results indicate that spontaneous ICS activity in the GER of the postnatal cochlea is not affected by Panx1 ablation.
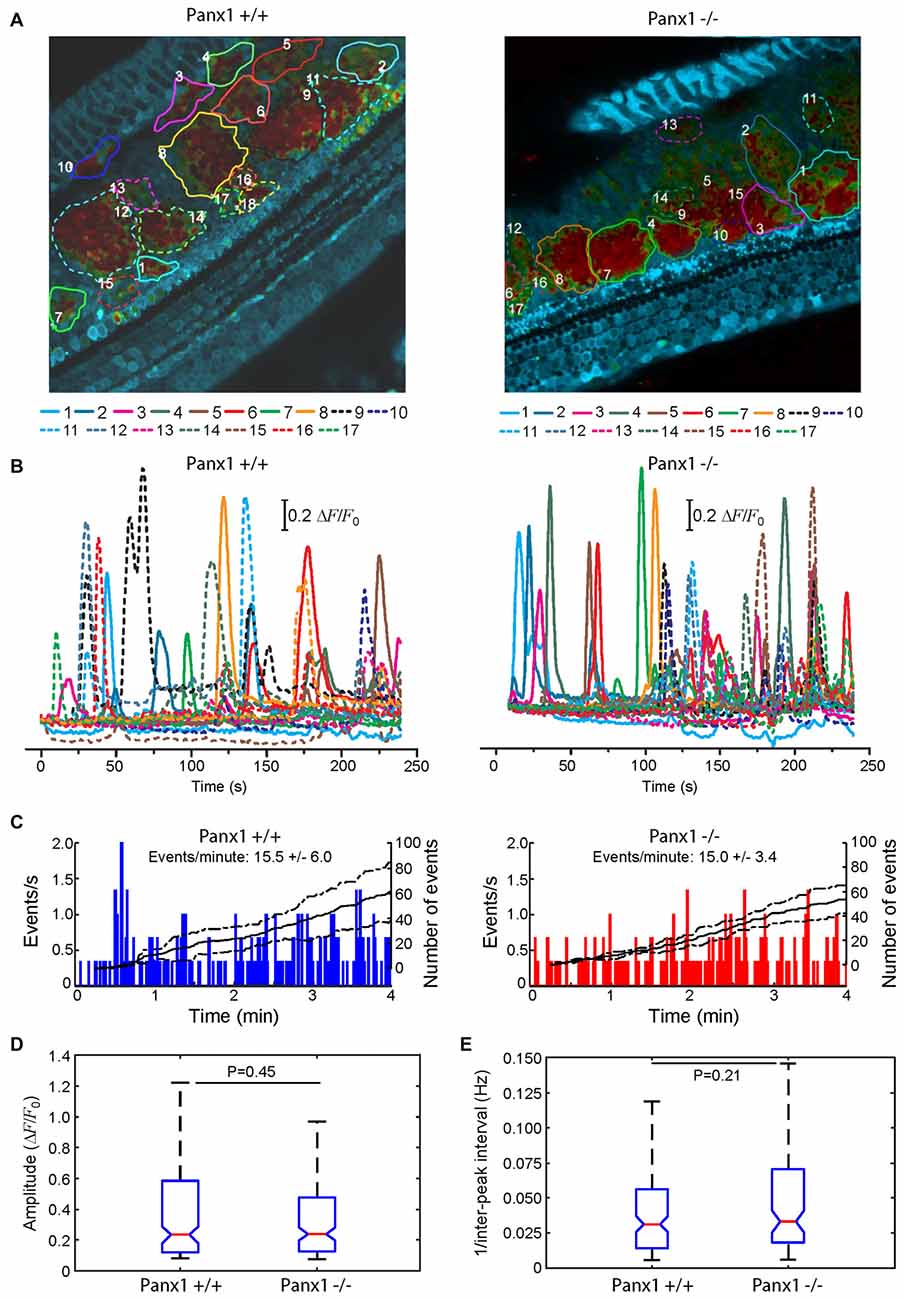
Figure 10. Spontaneous cytosolic Ca2+ transients in the GER of WT (Panx+/+) and Panx1−/− postnatal cochlear cultures. (A) Representative false-color images of Fluo-Forte fluorescence changes (ΔF/F0), encoded as shown by the color scale bar, obtained as maximal projection rendering of all frames recorded in an apical-middle turn culture from a P5 mouse imaged for 4 min at 5 frames per second; regions of interests are shown superimposed on different GER areas invaded by spontaneous Ca2+ waves. Scale bar, 50 μm. (B) Fluo-forte traces generated as pixel averages from the color-matched regions of interests shown in (A). (C) Frequency histograms of spontaneous cytosolic Ca2+ transients (events) in cultures from P5 Panx+/+ and Panx1−/− mice (pooled data). (D) Distributions of events amplitude (ΔFmax/F0), from data in (C). The values of the median and inter-quartile range (IQR) are, respectively: 0.2340 and 0.4641 (Panx+/+); 0.2398 and 0.3501 (Panx1−/−). (E) Distributions of the frequency (i.e., the reciprocal of the inter-peak interval) between consecutive events from data in (C). The values of the median and IQR are, respectively (in Hz): 0.0312 and 0.0420 (Panx+/+); 0.0331 and 0.0520 (Panx1−/−). P-values in (C,D) were computed with the Mann-Whitney U test.
Discussion
The Panx1−/− strain we have analyzed was the first with a reported global ablation of Panx1 (Anselmi et al., 2008; Bargiotas et al., 2011). The present results confirm successful ablation of Panx1 in Panx1−/− mice, while our ABR and DPOAE data indicate normal hearing sensitivity, normal function of the outer hair cell-based “cochlear amplifier” (Frolenkov et al., 1998; Nobili et al., 1998; Ashmore, 2008) and absence of cochlear nerve defects, in agreement with the initial observation that Panx1−/− mice do not exhibit a detectable hearing phenotype (Anselmi et al., 20088).
We also confirmed that lack of Panx1 affects neither the expression of inner ear connexins nor gap junction communication in the organ of Corti. Furthermore, our experiments with cochlear organotypic cultures indicate that the ATP-release mechanism underlying the spontaneous ICS activity of cells in the GER is intact. Preservation of this mechanism is essential for hearing acquisition (Schütz et al., 2010; Rodriguez et al., 2012; Mammano and Bortolozzi, 2017) and maturation of sensory hair cells (Johnson et al., 2017). Recent results with a monoclonal antibody that inhibits Cx26 hemichannels substantiate the notion that, in the cochlear sensory epithelium, ATP is released from such hemichannels (Xu et al., 2017).
Ensuring that an allele derived from the tm1a cassette is a full null, rather that a hypomorph such as the Panx1tm1a(KOMP)Wtsi strain, and alleviating potential off-target gene mis-regulation, requires modification of tm1a, which can be performed in embryonic stem (ES) cells or in crosses with transgenic Flp and Cre mice. Flp deletion converts tm1a to a conditional allele (tm1c), restoring gene activity, whereas the promoter-driven selection cassette and floxed exon of the tm1a allele can be deleted by Cre to generate a lacZ-tagged allele (tm1b; Skarnes et al., 2011). This is usually accomplished by breeding the mice to a source of Cre expressed in the germline, followed by outcrossing and selection of knockout offspring that fail to carry the Cre driver (Skarnes et al., 2011). This was also the strategy followed by the International Mouse Phenotyping Consortium (IMPC9 Brown and Moore, 2012) to convert the tm1a allele to tm1b for subsequent phenotyping (Ryder et al., 2014). The Panx1tm1b(KOMP)Wtsi strain generated in this way was analyzed by the IMPC and reported to have no significant hearing/vestibular phenotype10.
Thus, altogether, our results and conclusions are consistent with hearing assessments both in the Genentech-Panx1−/− strain (Abitbol et al., 2016) and in the Panx1tm1b(KOMP)Wtsi strain generated and analyzed by the IMPC.
The lack of any measurable auditory phenotype in three strains, which are all global knockouts of Panx1, is in stark contrast with the phenotype reported for Pax2-cPanx1−/− (Zhao et al., 2015), and even more so for Foxg1-cPanx1−/− mice that, despite descending from the hypomorphic Panx1tm1a(KOMP)Wtsi strain, retained strong immunoreactivity for the Panx1 #4515 chicken anti-human antibody (which is not specific: Bargiotas et al., 2011; see “Introduction” section) in the organ of Corti and the spiral limbus (Chen et al., 2015). However, it should be considered that both the Pax2-cPanx1−/− and the Foxg1-cPanx1−/− strain must necessarily express a Cre recombinase (Friedel et al., 2011), whereas the three global knockout strains mentioned above do not express Cre. It is well known that Cre expression in mammalian cells can induce chromosomal aberrations and toxicity that is dependent on the level of Cre activity (Loonstra et al., 2001). Indeed, Caspase-3 activation and cell degeneration are reported hallmarks of the organ of Corti in Pax2-cPanx1−/− mice (Zhao et al., 2015). Furthermore, Foxg1Cre mice that are homozygous for the targeted mutation die perinatally (Tian et al., 201211), whereas heterozygous Gfi1Cre mice, one of the many driver lines used for conditional cell-specific gene deletion/reporter gene activation in the inner ear (Cox et al., 2012), were recently reported with an early onset progressive hearing loss, which was absent in their wild-type littermates (Matern et al., 2017).
Another risk factor that is worth considering is related to the structure of the tm1a selection cassette (Testa et al., 2004; Skarnes et al., 2011) used to generate the hypomorphic Panx1tm1a(KOMP)Wtsi mice. As the latter were not crossed with a Flp deleter line before being used to create Pax2-cPanx1−/− and Foxg1-cPanx1−/− mice, it is not clear from the data provided if the Cre-mediated deletion removed only exon 2 of Panx1 or also the neo cassette, nor from which tissue the sample tested was obtained.
It is well known that retention of a neo cassette can cause unexpected phenotypes in “knockout” mice due to neighborhood effects (Pham et al., 1996; Scacheri et al., 2001; Ren et al., 2002; Meier et al., 2010). Indeed, removal of the neo cassette and critical exon from the tm1a allele is regarded as an essential procedure that alleviates potential off-target gene mis-regulation caused by the neo promoter, and to ensure that the allele is a full null rather then a hypomorph (Skarnes et al., 2011).
In this vein, it was recently argued that the hearing loss phenotype exhibited by Cx30−/− mice (Teubner et al., 2003) depends on the cumulative effect of deletion of Cx30 and 3’ insertion of a lacZ and neo cassette. Indeed, in a strictly related knockout mouse model (Cx30Δ/Δ) in which Cx30 was removed without perturbing the surrounding sequences, hearing thresholds determined by ABR analysis are normal (Boulay et al., 2013; Crispino et al., 2017).
In conclusion, our extended characterization of Panx1−/− mice provides strong evidence that Panx1 is dispensable for hearing acquisition and auditory function.
Data and Code Availability
Data and computer code used to analyze data are available from the authors upon request.
Author Contributions
FaM designed the studies, provided resources to conduct the studies and wrote the manuscript; HM provided Panx1−/− mice and genotyping protocols; VZ, MP and FC performed animal genotyping; MR and FS were in charge of animal welfare and performed quality controls; VZ and FP performed in vivo electrophysiology; AC wrote software code to filter ABR waveforms; CDC wrote image acquisition and analysis software; VZ, FP and CN analyzed ABR and DPOAE data; VZ performed immunofluorescence studies; FP performed Western blot analyses; VZ and GZ generated organotypic cochlear cultures; GZ performed patch clamp, dye transfer in cochlear organotypic cultures and analyzed data; CP and FlM performed multiphoton microscopy and Ca2+ imaging in cochlear organotypic cultures, and analyzed data; AMS, ARF and FaM supervised the work of junior colleagues; VZ, GC, FC, FS, ARF and AMS edited the text.
Funding
This work was supported by Consiglio Nazionale delle Ricerche (CNR) Progetto di Interesse Invecchiamento (grant DSB.AD009.001.004/INVECCHIAMENTO-IBCN) and Fondazione Telethon (grant GGP13114) to FaM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The AvesLab #6358 antibody used in this study was a gift of Prof. Eliana Scemes (Dominick P. Purpura Department of Neuroscience, Kennedy Center, Albert Einstein College of Medicine, Bronx, NY, USA). The authors thank E. Perlas of EMBL—Rome Histology Facility and G. Bolasco of EMBL—Rome Microscopy Facility for assistance with histology and microscopy; I. Losso of EMMA–INFRAFRONTIER Monterotondo for technical assistance.
Footnotes
- ^ http://www.genenames.org/cgi-bin/genefamilies/set/288
- ^ http://www.informatics.jax.org/allele/MGI:5310802
- ^ http://www.informatics.jax.org/marker/phenotypes/MGI:1860055
- ^ http://www.knockoutmouse.org
- ^ http://www.eucomm.org
- ^ http://www.komp.org/alleles.php#conditional-promoter-csd
- ^ https://www.infrafrontier.eu/
- ^ http://www.informatics.jax.org/marker/phenotypes/MGI:1860055
- ^ http://www.mousephenotype.org/
- ^ http://www.mousephenotype.org/data/genes/MGI:1860055#section-associations
- ^ http://jaxmice.jax.org/strain/004337.html
References
Abitbol, J. M., Kelly, J. J., Barr, K., Schormans, A. L., Laird, D. W., and Allman, B. L. (2016). Differential effects of pannexins on noise-induced hearing loss. Biochem. J. 473, 4665–4680. doi: 10.1042/bcj20160668
Ahmad, S., Chen, S., Sun, J., and Lin, X. (2003). Connexins 26 and 30 are co-assembled to form gap junctions in the cochlea of mice. Biochem. Biophys. Res. Commun. 307, 362–368. doi: 10.1016/s0006-291x(03)01166-5
Ahmad, S., Tang, W., Chang, Q., Qu, Y., Hibshman, J., Li, Y., et al. (2007). Restoration of connexin26 protein level in the cochlea completely rescues hearing in a mouse model of human connexin30-linked deafness. Proc. Natl. Acad. Sci. U S A 104, 1337–1341. doi: 10.1073/pnas.0606855104
Anselmi, F., Hernandez, V. H., Crispino, G., Seydel, A., Ortolano, S., Roper, S. D., et al. (2008). ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc. Natl. Acad. Sci. U S A 105, 18770–18775. doi: 10.1073/pnas.0800793105
Ashmore, J. (2008). Cochlear outer hair cell motility. Physiol. Rev. 88, 173–210. doi: 10.1152/physrev.00044.2006
Baranova, A., Ivanov, D., Petrash, N., Pestova, A., Skoblov, M., Kelmanson, I., et al. (2004). The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 83, 706–716. doi: 10.1016/j.ygeno.2003.09.025
Bargiotas, P., Krenz, A., Hormuzdi, S. G., Ridder, D. A., Herb, A., Barakat, W., et al. (2011). Pannexins in ischemia-induced neurodegeneration. Proc. Natl. Acad. Sci. U S A 108, 20772–20777. doi: 10.1073/pnas.1018262108
Beltramello, M., Piazza, V., Bukauskas, F. F., Pozzan, T., and Mammano, F. (2005). Impaired permeability to Ins(1,4,5)P3 in a mutant connexin underlies recessive hereditary deafness. Nat. Cell Biol. 7, 63–69. doi: 10.1038/ncb1205
Boulay, A., del Castillo, F. J., Giraudet, F., Hamard, G., Giaume, C., Petit, C., et al. (2013). Hearing is normal without connexin30. J. Neurosci. 33, 430–434. doi: 10.1523/jneurosci.4240-12.2013
Bredenkamp, N., Seoighe, C., and Illing, N. (2007). Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev. Genes Evol. 217, 227–233. doi: 10.1007/s00427-006-0128-x
Brown, S. D., and Moore, M. W. (2012). The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–640. doi: 10.1007/s00335-012-9427-x
Bruzzone, R., Hormuzdi, S. G., Barbe, M. T., Herb, A., and Monyer, H. (2003). Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. U S A 100, 13644–13649. doi: 10.1073/pnas.2233464100
Ceriani, F., Pozzan, T., and Mammano, F. (2016). Critical role of ATP-induced ATP release for Ca2+ signaling in nonsensory cell networks of the developing cochlea. Proc. Natl. Acad. Sci. U S A 113, E7194–E7201. doi: 10.1073/pnas.1616061113
Chen, J., Zhu, Y., Liang, C., Chen, J., and Zhao, H. B. (2015). Pannexin1 channels dominate ATP release in the cochlea ensuring endocochlear potential and auditory receptor potential generation and hearing. Sci. Rep. 5:10762. doi: 10.1038/srep10762
Cohen-Salmon, M., Ott, T., Michel, V., Hardelin, J. P., Perfettini, I., Eybalin, M., et al. (2002). Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr. Biol. 12, 1106–1111. doi: 10.1016/s0960-9822(02)00904-1
Cohen-Salmon, M., Regnault, B., Cayet, N., Caille, D., Demuth, K., Hardelin, J. P., et al. (2007). Connexin30 deficiency causes instrastrial fluid-blood barrier disruption within the cochlear stria vascularis. Proc. Natl. Acad. Sci. U S A 104, 6229–6234. doi: 10.1073/pnas.0605108104
Cone, A. C., Ambrosi, C., Scemes, E., Martone, M. E., and Sosinsky, G. E. (2013). A comparative antibody analysis of pannexin1 expression in four rat brain regions reveals varying subcellular localizations. Front. Pharmacol. 4:6. doi: 10.3389/fphar.2013.00006
Cox, B. C., Liu, Z., Lagarde, M. M., and Zuo, J. (2012). Conditional gene expression in the mouse inner ear using Cre-loxP. J. Assoc. Res. Otolaryngol. 13, 295–322. doi: 10.1007/s10162-012-0324-5
Crispino, G., Di Pasquale, G., Scimemi, P., Rodriguez, L., Galindo Ramirez, F., De Siati, R. D., et al. (2011). BAAV mediated GJB2 gene transfer restores gap junction coupling in cochlear organotypic cultures from deaf Cx26Sox10Cre mice. PLoS One 6:e23279. doi: 10.1371/journal.pone.0023279
Crispino, G., Galindo Ramirez, F., Campioni, M., Zorzi, V., Praetorius, M., Di Pasquale, G., et al. (2017). In vivo genetic manipulation of inner ear connexin expression by bovine adeno-associated viral vectors. Sci. Rep. 7:6567. doi: 10.1038/s41598-017-06759-y
Dahl, G., Qiu, F., and Wang, J. (2013). The bizarre pharmacology of the ATP release channel pannexin1. Neuropharmacology 75, 583–593. doi: 10.1016/j.neuropharm.2013.02.019
Dahl, G., and Muller, K. J. (2014). Innexin and pannexin channels and their signaling. FEBS Lett. 588, 1396–1402. doi: 10.1016/j.febslet.2014.03.007
del Hierro, M. J., Fernandez, J., Castrillo, M., Martin-Dorado, I., Montoliu, L., Hagn, M., et al. (2016). EMMA: the european mouse mutant archive. Transgenic Res. 25, 228–228. Accession Number: WOS:000371155900078
Esseltine, J. L., and Laird, D. W. (2016). Next-generation connexin and pannexin cell biology. Trends Cell Biol. 26, 944–955. doi: 10.1016/j.tcb.2016.06.003
Forge, A., Becker, D., Casalotti, S., Edwards, J., Marziano, N., and Nevill, G. (2003). Gap junctions in the inner ear: comparison of distribution patterns in different vertebrates and assessement of connexin composition in mammals. J. Comp. Neurol. 467, 207–231. doi: 10.1002/cne.10916
Friedel, R. H., Wurst, W., Wefers, B., and Kuhn, R. (2011). Generating conditional knockout mice. Methods Mol. Biol. 693, 205–231. doi: 10.1007/978-1-60761-974-1_12
Frolenkov, G. I., Atzori, M., Kalinec, F., Mammano, F., and Kachar, B. (1998). The membrane-based mechanism of cell motility in cochlear outer hair cells. Mol. Biol. Cell 9, 1961–1968. doi: 10.1091/mbc.9.8.1961
Hanstein, R., Negoro, H., Patel, N. K., Charollais, A., Meda, P., Spray, D. C., et al. (2013). Promises and pitfalls of a Pannexin1 transgenic mouse line. Front. Pharmacol. 4:61. doi: 10.3389/fphar.2013.00061
Hébert, J. M., and McConnell, S. K. (2000). Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222, 296–306. doi: 10.1006/dbio.2000.9732
Huang, Y. J., Maruyama, Y., Dvoryanchikov, G., Pereira, E., Chaudhari, N., and Roper, S. D. (2007). The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proc. Natl. Acad. Sci. U S A 104, 6436–6441. doi: 10.1073/pnas.0611280104
International Mouse Knockout Consortium, Collins, F. S., Rossant, J., and Wurst, W. (2007). A mouse for all reasons. Cell 128, 9–13. doi: 10.1016/j.cell.2006.12.018
Johnson, S. L., Ceriani, F., Houston, O., Polishchuk, R., Polishchuk, E., Crispino, G., et al. (2017). Connexin-mediated signaling in nonsensory cells is crucial for the development of sensory inner hair cells in the mouse cochlea. J. Neurosci. 37, 258–268. doi: 10.1523/jneurosci.2251-16.2017
Kemp, D. T. (1978). Stimulated acoustic emissions from within the human auditory system. J. Acoust. Soc. Am. 64, 1386–1391. doi: 10.1121/1.382104
Kemp, D. T. (2002). Otoacoustic emissions, their origin in cochlear function, and use. Br. Med. Bull. 63, 223–241. doi: 10.1093/bmb/63.1.223
Kranz, K., Dorgau, B., Pottek, M., Herrling, R., Schultz, K., Bolte, P., et al. (2013). Expression of Pannexin1 in the outer plexiform layer of the mouse retina and physiological impact of its knockout. J. Comp. Neurol. 521, 1119–1135. doi: 10.1002/cne.23223
Kurtenbach, S., Kurtenbach, S., and Zoidl, G. (2014). Emerging functions of pannexin 1 in the eye. Front. Cell. Neurosci. 8:263. doi: 10.3389/fncel.2014.00263
Lautermann, J., ten Cate, W. J., Altenhoff, P., Grümmer, R., Traub, O., Frank, H., et al. (1998). Expression of the gap-junction connexins 26 and 30 in the rat cochlea. Cell Tissue Res. 294, 415–420. doi: 10.1007/s004410051192
Legatt, A. D. (2002). Mechanisms of intraoperative brainstem auditory evoked potential changes. J. Clin. Neurophysiol. 19, 396–408. doi: 10.1097/00004691-200210000-00003
Loonstra, A., Vooijs, M., Beverloo, H. B., Allak, B. A., van Drunen, E., Kanaar, R., et al. (2001). Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U S A 98, 9209–9214. doi: 10.1073/pnas.161269798
Majumder, P., Crispino, G., Rodriguez, L., Ciubotaru, C. D., Anselmi, F., Piazza, V., et al. (2010). ATP-mediated cell-cell signaling in the organ of Corti: the role of connexin channels. Purinergic Signal. 6, 167–187. doi: 10.1007/s11302-010-9192-9
Mammano, F., and Bortolozzi, M. (2017). Ca2+ signaling, apoptosis and autophagy in the developing cochlea: milestones to hearing acquisition. Cell Calcium doi: 10.1016/j.ceca.2017.05.006 [Epub ahead of print].
Matern, M., Vijayakumar, S., Margulies, Z., Milon, B., Song, Y., Elkon, R., et al. (2017). Gfi1Cre mice have early onset progressive hearing loss and induce recombination in numerous inner ear non-hair cells. Sci. Rep. 7:42079. doi: 10.1038/srep42079
Meier, I. D., Bernreuther, C., Tilling, T., Neidhardt, J., Wong, Y. W., Schulze, C., et al. (2010). Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 24, 1714–1724. doi: 10.1096/fj.09-140749
Nobili, R., and Mammano, F. (1996). Biophysics of the cochlea. II: stationary nonlinear phenomenology. J. Acoust. Soc. Am. 99, 2244–2255. doi: 10.1121/1.415412
Nobili, R., Mammano, F., and Ashmore, J. (1998). How well do we understand the cochlea? Trends Neurosci. 21, 159–167. doi: 10.1016/S0166-2236(97)01192-2
Nobili, R., Vetesnik, A., Turicchia, L., and Mammano, F. (2003). Otoacoustic emissions from residual oscillations of the cochlear basilar membrane in a human ear model. J. Assoc. Res. Otolaryngol. 4, 478–494. doi: 10.1007/s10162-002-3055-1
Ohyama, T. (2009). Unraveling inner ear induction by gene manipulation using Pax2-Cre BAC transgenic mice. Brain Res. 1277, 84–89. doi: 10.1016/j.brainres.2009.02.036
Ortolano, S., Di Pasquale, G., Crispino, G., Anselmi, F., Mammano, F., and Chiorini, J. A. (2008). Coordinated control of connexin 26 and connexin 30 at the regulatory and functional level in the inner ear. Proc. Natl. Acad. Sci. U S A 105, 18776–18781. doi: 10.1073/pnas.0800831105
Otto, C., Fuchs, I., Kauselmann, G., Kern, H., Zevnik, B., Andreasen, P., et al. (2009). GPR30 does not mediate estrogenic responses in reproductive organs in mice. Biol. Reprod. 80, 34–41. doi: 10.1095/biolreprod.108.071175
Panchin, Y., Kelmanson, I., Matz, M., Lukyanov, K., Usman, N., and Lukyanov, S. (2000). A ubiquitous family of putative gap junction molecules. Curr. Biol. 10, R473–R474. doi: 10.1016/s0960-9822(00)00576-5
Patel, D., Zhang, X., and Veenstra, R. D. (2014). Connexin hemichannel and pannexin channel electrophysiology: how do they differ? FEBS Lett. 588, 1372–1378. doi: 10.1016/j.febslet.2013.12.023
Penuela, S., Gehi, R., and Laird, D. W. (2013). The biochemistry and function of pannexin channels. Biochim. Biophys. Acta 1828, 15–22. doi: 10.1016/j.bbamem.2012.01.017
Penuela, S., Kelly, J. J., Churko, J. M., Barr, K. J., Berger, A. C., and Laird, D. W. (2014). Panx1 regulates cellular properties of keratinocytes and dermal fibroblasts in skin development and wound healing. J. Invest. Dermatol. 134, 2026–2035. doi: 10.1038/jid.2014.86
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pham, C. T., MacIvor, D. M., Hug, B. A., Heusel, J. W., and Ley, T. J. (1996). Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. U S A 93, 13090–13095. doi: 10.1073/pnas.93.23.13090
Piazza, V., Ciubotaru, C. D., Gale, J. E., and Mammano, F. (2007). Purinergic signalling and intercellular Ca2+ wave propagation in the organ of Corti. Cell Calcium 41, 77–86. doi: 10.1016/j.ceca.2006.05.005
Qu, Y., Misaghi, S., Newton, K., Gilmour, L. L., Louie, S., Cupp, J. E., et al. (2011). Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J. Immunol. 186, 6553–6561. doi: 10.4049/jimmunol.1100478
Qu, Y., Tang, W., Zhou, B., Ahmad, S., Chang, Q., Li, X., et al. (2012). Early developmental expression of connexin26 in the cochlea contributes to its dominate functional role in the cochlear gap junctions. Biochem. Biophys. Res. Commun. 417, 245–250. doi: 10.1016/j.bbrc.2011.11.093
Ray, A., Zoidl, G., Weickert, S., Wahle, P., and Dermietzel, R. (2005). Site-specific and developmental expression of pannexin1 in the mouse nervous system. Eur. J. Neurosci. 21, 3277–3290. doi: 10.1111/j.1460-9568.2005.04139.x
Ren, S. Y., Angrand, P. O., and Rijli, F. M. (2002). Targeted insertion results in a rhombomere 2-specific Hoxa2 knockdown and ectopic activation of Hoxa1 expression. Dev. Dyn. 225, 305–315. doi: 10.1002/dvdy.10171
Rodriguez, L., Simeonato, E., Scimemi, P., Anselmi, F., Cali, B., Crispino, G., et al. (2012). Reduced phosphatidylinositol 4,5-bisphosphate synthesis impairs inner ear Ca2+ signaling and high-frequency hearing acquisition. Proc. Natl. Acad. Sci. U S A 109, 14013–14018. doi: 10.1073/pnas.1211869109
Ryder, E., Doe, B., Gleeson, D., Houghton, R., Dalvi, P., Grau, E., et al. (2014). Rapid conversion of EUCOMM/KOMP-CSD alleles in mouse embryos using a cell-permeable Cre recombinase. Transgenic Res. 23, 177–185. doi: 10.1007/s11248-013-9764-x
Scacheri, P. C., Crabtree, J. S., Novotny, E. A., Garrett-Beal, L., Chen, A., Edgemon, K. A., et al. (2001). Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis 30, 259–263. doi: 10.1002/gene.1072
Schütz, M., Scimemi, P., Majumder, P., De Siati, R. D., Crispino, G., Rodriguez, L., et al. (2010). The human deafness-associated connexin 30 T5M mutation causes mild hearing loss and reduces biochemical coupling among cochlear non-sensory cells in knock-in mice. Hum. Mol. Genet. 19, 4759–4773. doi: 10.1093/hmg/ddq402
Scimemi, P., Santarelli, R., Selmo, A., and Mammano, F. (2014). Auditory brainstem responses to clicks and tone bursts in C57 BL/6J mice. Acta Otorhinolaryngol. Ital. 34, 264–271.
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8:e1002964. doi: 10.1371/journal.pgen.1002964
Skarnes, W. C., Rosen, B., West, A. P., Koutsourakis, M., Bushell, W., Iyer, V., et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342. doi: 10.1038/nature10163
Sosinsky, G. E., Boassa, D., Dermietzel, R., Duffy, H. S., Laird, D. W., MacVicar, B., et al. (2011). Pannexin channels are not gap junction hemichannels. Channels (Austin) 5, 193–197. doi: 10.4161/chan.5.3.15765
Sun, Y., Tang, W., Chang, Q., Wang, Y., Kong, W., and Lin, X. (2009). Connexin30 null and conditional connexin26 null mice display distinct pattern and time course of cellular degeneration in the cochlea. J. Comp. Neurol. 516, 569–579. doi: 10.1002/cne.22117
Tang, W., Ahmad, S., Shestopalov, V. I., and Lin, X. (2008). Pannexins are new molecular candidates for assembling gap junctions in the cochlea. Neuroreport 19, 1253–1257. doi: 10.1097/WNR.0b013e32830891f5
Testa, G., Schaft, J., van der Hoeven, F., Glaser, S., Anastassiadis, K., Zhang, Y., et al. (2004). A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38, 151–158. doi: 10.1002/gene.20012
Teubner, B., Michel, V., Pesch, J., Lautermann, J., Cohen-Salmon, M., Sohl, G., et al. (2003). Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Hum. Mol. Genet. 12, 13–21. doi: 10.1093/hmg/12.1.13
Tian, C., Gong, Y., Yang, Y., Shen, W., Wang, K., Liu, J., et al. (2012). Foxg1 has an essential role in postnatal development of the dentate gyrus. J. Neurosci. 32, 2931–2949. doi: 10.1523/JNEUROSCI.5240-11.2012
Tian, Y., James, S., Zuo, J., Fritzsch, B., and Beisel, K. W. (2006). Conditional and inducible gene recombineering in the mouse inner ear. Brain Res. 1091, 243–254. doi: 10.1016/j.brainres.2006.01.040
Tritsch, N. X., Yi, E., Gale, J. E., Glowatzki, E., and Bergles, D. E. (2007). The origin of spontaneous activity in the developing auditory system. Nature 450, 50–55. doi: 10.1038/nature06233
Wang, H. C., and Bergles, D. E. (2015). Spontaneous activity in the developing auditory system. Cell Tissue Res. 361, 65–75. doi: 10.1007/s00441-014-2007-5
Wang, X. H., Streeter, M., Liu, Y. P., and Zhao, H. B. (2009). Identification and characterization of pannexin expression in the mammalian cochlea. J. Comp. Neurol. 512, 336–346. doi: 10.1002/cne.21898
Xu, L., Carrer, A., Zonta, F., Qu, Z., Ma, P., Li, S., et al. (2017). Design and characterization of a human monoclonal antibody that modulates mutant connexin 26 hemichannels implicated in deafness and skin disorders. Front. Mol. Neurosci. 10:298. doi: 10.3389/fnmol.2017.00298
Zhang, H., Chen, Y., and Zhang, C. (2012). Patterns of heterogeneous expression of pannexin 1 and pannexin 2 transcripts in the olfactory epithelium and olfactory bulb. J. Mol. Histol. 43, 651–660. doi: 10.1007/s10735-012-9443-x
Zhao, H. B. (2016). Expression and function of pannexins in the inner ear and hearing. BMC Cell Biol. 17:16. doi: 10.1186/s12860-016-0095-7
Zhao, H. B., Kikuchi, T., Ngezahayo, A., and White, T. W. (2006). Gap junctions and cochlear homeostasis. J. Membr. Biol. 209, 177–186. doi: 10.1007/s00232-005-0832-x
Zhao, H. B., Zhu, Y., Liang, C., and Chen, J. (2015). Pannexin 1 deficiency can induce hearing loss. Biochem. Biophys. Res. Commun. 463, 143–147. doi: 10.1016/j.bbrc.2015.05.049
Zheng, Q. Y., Johnson, K. R., and Erway, L. C. (1999). Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear. Res. 130, 94–107. doi: 10.1016/s0378-5955(99)00003-9
Zhou, X., Jen, P. H., Seburn, K. L., Frankel, W. N., and Zheng, Q. Y. (2006). Auditory brainstem responses in 10 inbred strains of mice. Brain Res. 1091, 16–26. doi: 10.1016/j.brainres.2006.01.107
Keywords: cochlea, pannexins, connexins, hair cells, non-sensory cells, auditory brainstem responses, distortion product otoacoustic emissions
Citation: Zorzi V, Paciello F, Ziraldo G, Peres C, Mazzarda F, Nardin C, Pasquini M, Chiani F, Raspa M, Scavizzi F, Carrer A, Crispino G, Ciubotaru CD, Monyer H, Fetoni AR, Salvatore AM and Mammano F (2017) Mouse Panx1 Is Dispensable for Hearing Acquisition and Auditory Function. Front. Mol. Neurosci. 10:379. doi: 10.3389/fnmol.2017.00379
Received: 02 October 2017; Accepted: 30 October 2017;
Published: 28 November 2017.
Edited by:
Isabel Varela-Nieto, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Maria E. Rubio, University of Pittsburgh, United StatesMiguel A. Merchán, Universidad de Salamanca, Spain
Copyright © 2017 Zorzi, Paciello, Ziraldo, Peres, Mazzarda, Nardin, Pasquini, Chiani, Raspa, Scavizzi, Carrer, Crispino, Ciubotaru, Monyer, Fetoni, Salvatore and Mammano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabio Mammano, ZmFiaW8ubWFtbWFub0B1bmlwZC5pdA==
 Veronica Zorzi
Veronica Zorzi Fabiola Paciello
Fabiola Paciello Gaia Ziraldo
Gaia Ziraldo Chiara Peres
Chiara Peres Flavia Mazzarda
Flavia Mazzarda Chiara Nardin
Chiara Nardin Miriam Pasquini
Miriam Pasquini Francesco Chiani
Francesco Chiani Marcello Raspa
Marcello Raspa Ferdinando Scavizzi
Ferdinando Scavizzi Andrea Carrer
Andrea Carrer Giulia Crispino
Giulia Crispino Catalin D. Ciubotaru
Catalin D. Ciubotaru Hannah Monyer
Hannah Monyer Anna R. Fetoni
Anna R. Fetoni Anna M. Salvatore
Anna M. Salvatore Fabio Mammano
Fabio Mammano