- Department of Biological Chemistry and the Center for Bioengineering, The Edmond and Lily Safra Center for Brain Science, The Hebrew University of Jerusalem, Jerusalem, Israel
MicroRNAs (miRNAs) can notably control many targets each and regulate entire cellular pathways, but whether miRNAs can regulate complete neurotransmission processes is largely unknown. Here, we report that miRNAs with complementary sequence motifs to the key genes involved in acetylcholine (ACh) synthesis and/or packaging show massive overlap with those regulating ACh degradation. To address this topic, we first searched for miRNAs that could target the 3′-untranslated regions of the choline acetyltransferase (ChAT) gene that controls ACh synthesis; the vesicular ACh transporter (VAChT), encoded from an intron in the ChAT gene and the ACh hydrolyzing genes acetyl- and/or butyrylcholinesterase (AChE, BChE). Intriguingly, we found that many of the miRNAs targeting these genes are primate-specific, and that changes in their levels associate with inflammation, anxiety, brain damage, cardiac, neurodegenerative, or pain-related syndromes. To validate the in vivo relevance of this dual interaction, we selected the evolutionarily conserved miR-186, which targets both the stress-inducible soluble “readthrough” variant AChE-R and the major peripheral cholinesterase BChE. We exposed mice to predator scent stress and searched for potential associations between consequent changes in their miR-186, AChE-R, and BChE levels. Both intestinal miR-186 as well as BChE and AChE-R activities were conspicuously elevated 1 week post-exposure, highlighting the previously unknown involvement of miR-186 and BChE in psychological stress responses. Overlapping miRNA regulation emerges from our findings as a recently evolved surveillance mechanism over cholinergic neurotransmission in health and disease; and the corresponding miRNA details and disease relevance may serve as a useful resource for studying the molecular mechanisms underlying this surveillance.
Introduction
MicroRNAs (miRNAs) are small, 20–25 nucleotides long, non-coding RNA molecules, each of which can predictably target many protein-coding messenger RNA (mRNA) transcripts to silence them post-transcriptionally. Mammalian miRNAs bind target mRNAs via a short “seed” sequence, such that many miRNAs can target the same mRNAs, and different mRNAs may be targeted by a single miRNA gene (Bartel, 2009). Interestingly, miRNAs often target different mRNAs all involved in a particular biological function (Chen et al., 2004). This has been extensively studied in various cancers (Kefas et al., 2008; Papagiannakopoulos et al., 2008; Levy et al., 2010; Lupini et al., 2013), but the control by miRNAs of specific neurotransmission processes remained largely unexplored. In principle, one would predict that the synthesis, packaging in neuronal vesicles and destruction or re-uptake of a certain neurotransmitter should be co-regulated; this, in turn implies that some miRNAs may co-suppress two or more of the mRNA transcripts involved in regulating the levels of certain neurotransmitters, and that modified expression of such miRNAs might be involved in diseases associated with impaired regulation of this neurotransmission pathway. Based on this working hypothesis, we studied miRNA-mediated regulation of mRNA transcripts involved in the synthesis, vesicle packaging, and destruction of acetylcholine (ACh).
MiRNA-binding sequence motifs are primarily located at the 3′-untranslated region (3′-UTR) of the mRNA transcript (Bartel, 2009). Therefore, we interrogated the 3′-UTR domains in the choline acetyltransferase (ChAT) gene, which is responsible for ACh synthesis and the vesicular acetylcholine transporter (VAChT), encoded from the first intron in the ChAT gene (Erickson et al., 1994; Eiden, 1998). The VAChT transcript has its own 3′-UTR, which might suggest that it can be regulated by distinct miRNAs. Given that released ACh is degraded by the ACh hydrolyzing enzymes acetyl- and butyrylcholinesterase (AChE, BChE) (Meshorer and Soreq, 2006), and since increased AChE synthesis may be linked with decreased ChAT production (Kaufer et al., 1998), we further searched for potential overlaps in the predicted miRNAs between the human ChAT/VAChT 3′-UTRs and the alternative 3′-UTR choices of the major variants of AChE mRNA, the “synaptic” AChE-S and the “read-through” variant AChE-R which shares its 3′-UTR with the “erythrocytic” AChE-E isoform and is known to be induced under stress (Meshorer and Soreq, 2006). Of note, the AChE-S variant possesses a considerably shorter 3′-UTR compared to AChE-R (Hanin and Soreq, 2011) with further likelihood of differential miRNA regulation for the AChE-R and AChE-S targets. The BChE gene, completing the series of regions to be analyzed, has a different 3′-UTR with lower G, C-content (Soreq and Seidman, 2001), demonstrating sequence differences. All of these regions are functionally involved in cholinergic signaling, and we therefore designated the corresponding miRNAs “CholinomiRs.” A schematic overview of the transcripts involved in this pathway is given in Figure 1.
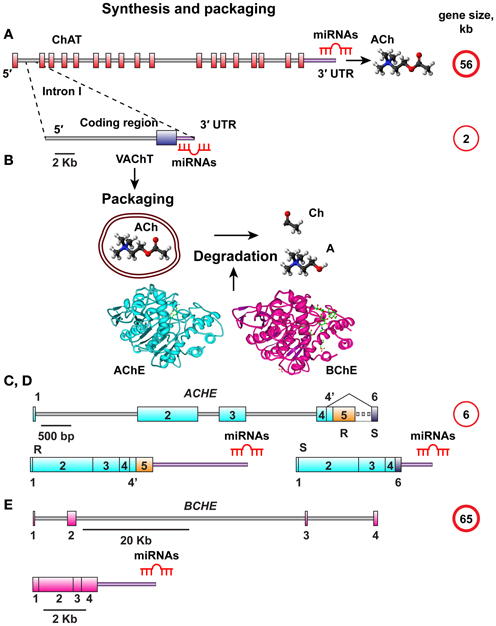
Figure 1. Schematic overview of ACh synthesis, packaging, and destruction. (A) The choline acetyl transferase (ChAT) gene responsible for ACh synthesis is 56 kb in size and all its splice variants have the same 3′UTR. (B) The vesicular acetylcholine transporter (VAChT) controlling ACh packaging into vesicles is encoded from the first intron of the ChAT gene. The VAChT gene is only 2 kb in length, yet it has its own 3′UTR. Once released, ACh should be degraded into Acetate and Choline by one of the splice variants of AChE or BChE, depending on its site of release. (C,D) The AChE gene is 6 kb in size, and its mRNA transcript is spliced to yield the major AChE-S and AChE-R splice variants with distinct 3′-UTR domains. (E) The BChE gene is 65 kb in size and yields only one known splice variant.
There were only a few overlaps between the predicted CholinomiRs regulating the synthesis and destruction of ACh. In contrast, we identified numerous overlaps between those CholinomiRs controlling ACh packaging and its synthesis; suggesting that miRNAs play an important role in selectively co-regulating cholinergic signaling by adapting the rates and efficacy of ACh packaging and destruction. Changes in these CholinomiRs, of which many are primate-specific, were further reported by others in inflammation and anxiety, brain damage, pain, cardiac, and neurodegenerative diseases, all of which are known for cholinergic signaling impairments.
To test the relevance of our predictions for in vivo conditions, we subjected mice to the long-lasting predator scent stress (Zimmerman et al., 2012) and tested, 1 week later, for changes in one miRNA, miR-186 and its predicted targets AChE-R and BChE. Given our previous findings of miRNA regulation of cholinergic-mediated production of intestinal miR-132 (Shaked et al., 2009), we quantified miR-186 levels in intestinal sections and measured cholinesterase activities. We found that predator scent stress induces intestinal increases in both the cholinesterases-targeting miR-186 and in the activities of the targeted cholinesterases. Our findings support the hypothesis that overlapping CholinomiR regulation serves as a recently evolved surveillance mechanism that can balance cholinergic signaling in brain and peripheral systems. The detailed lists of these miRNAs and their potential involvements with different diseases may be a valuable resource for researchers interested in both basic and translational aspects of key neuroinflammation and pain-related disorders.
Materials and Methods
Bioinformatics Approaches
3′-UTR sequences of the human ChAT, VAChT, AChE-S, AChE-R, and BChE transcripts were acquired from the NCBI nucleotide database (Entrez Nucleotide, 2010 http://www.ncbi.nlm.nih.gov/nuccore/). These sequences are 380, 58, 219, 963, and 477 nucleotides long, respectively. MiRNA-mRNA interactions were addressed by using four different algorithms, miRBase (Last update in June 2013, http://www.mirbase.org/), TargetScan (Last update in June 2012, http://www.targetscan.org/vert_50/), microcosm (version 5) (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) and miRanda (Last update in August 2010, http://www.microrna.org/microrna/home.do) which were last updated in June 2013, June 2012 and August 2010, respectively. All predictions ensured a threshold P-value < 0.05, and analysis specifications allowed both evolutionarily conserved and non-conserved miRNAs, which further enabled us to differentiate between primate-specific and evolutionarily conserved miRNAs.
To gain more information on the identified miRNAs and assess their prospects to interact with their targets, we determined the G, C content of all identified miRNAs using G, C content calculator algorithms (http://www.endmemo.com/bio/gc.php). We further focused on the overlapping miRNAs targeting more than one 3′-UTR of the 5 studied transcripts and distinguished primate-specific from evolutionarily conserved miRNAs that appeared in our list using the HomoloGen conservation score1 that reports genomic conservation values between tested genes from different species. For data-mining regarding the relation to specific diseases we utilized PubMed and Google Scholar.
Experimental in vivo Tests
To experimentally test the putative association between changes in the identified miRNAs and their protein targets under stressful conditions, we exposed C57/B6J mice to predator scent and additionally injected them for four consecutive days with 50 μ g kg−1 saline, essentially as in (Zimmerman et al., 2012). 7 days later, we removed intestinal sections from these mice and matched male control mice, as in (Shaked et al., 2009). The mice were housed four per cage, at 21 ± 1°C, in a 12-h light/dark cycle. The RNA extraction procedure followed that of (Maharshak et al., 2013) for human intestinal biopsies. Extracted RNA from the intestinal sections was used to quantify miR-186 and RNU6 levels by qRT-PCR (PerfeCTa® microRNA Assay). RNU6 snRNA levels were used to normalize the levels of miR-186. The following primers were employed: RNU6 (Quanta, Cat. # HS-RNU6), miR-186-5p (Quanta, Cat. # HSMIR-0186-5p), PerfeCTa® Universal PCR Primer (Quanta, Cat. # 95109-500). Protein extraction from the intestinal sections was performed by using Solution D (0.01 M TRIS, 1 M NaCl, 1 mM EGTA, 1% T-X100). Cholinesterase activities in the tissue homogenates were determined by quantifying acetylthiocholine hydrolysis rates (Ellman et al., 1961) in the presence or absence of iso-OMPA to selectively block BChE activities, all as under (Arbel et al., 2014). Animal procedures followed the ethics instructions at The Hebrew University of Jerusalem (Ethics number of research: NS-10205-4).
Statistical Analysis
Collected data was summarized and displayed as mean ± standard deviation (Erickson et al., 1994) using SPSS software (version 19.0, SPSS INC, Chicago, IL, USA). Normally distributed variables were compared using Student's t test. The level of significance used for all of the above analyses was 2-tailed p < 0.05.
Results
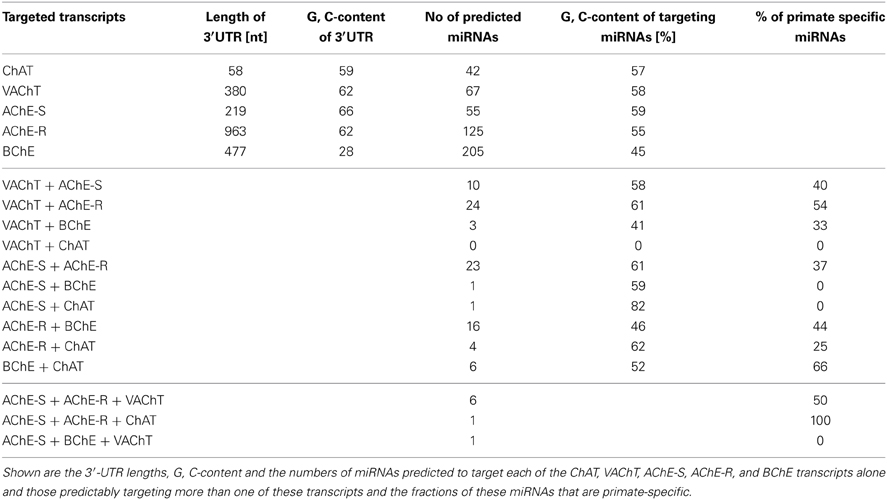
3′-UTR sequences of the human ChAT, VAChT, AChE-S, AChE-R, and BChE transcripts were acquired from the NCBI nucleotide database. These sequences span 380, 58, 219, 963, and 477 nucleotides, respectively and differ in their G, C content. We identified 42, 67, 55, 125, and 205 complementary miRNAs predicted to bind to the interrogated 3′-UTRs, respectively (Table 1, for details see Supplementary Table 1). Thus, the density of miRNA binding sites in these transcripts did not simply reflect their length, and the AChE-R and BChE mRNA transcripts encoding for soluble, secreted proteins emerged as particularly susceptible for miRNA targeting. Interestingly, we found no overlap between the VAChT and ChAT targeting miRNAs, suggesting differential regulation of the ACh synthesis and packaging processes in spite of these transcripts being subject to joint transcriptional control. In contradistinction, 26% of the ChAT targeting miRNAs are also predicted to target cholinesterases, and ACh packaging and degradation seemed to share yet more miRNAs: of 67 VAChT-targeting miRNAs, 55% predictably recognize binding sites in cholinesterases as well, and 5 of them co-target both the alternatively spliced “synaptic” AChE-S and the stress and inflammation-inducible “read-through” AChE-R transcript, enabling more profound suppression of ACh hydrolysis (for details see Supplementary Table 2). Notable examples involve the conserved neurodevelopment-associated hsa-miR-125b (Martino et al., 2009) and the primate-specific hsa-miR-608 and -765 (primate specificity determined by HomoloGen conservation score1).
Our working hypothesis predicted that miRNAs targeting more than one of the five 3′-UTR domains would be more likely than others to be causally involved in regulating this entire pathway. Also, many believe that recent evolutionary processes re-shaped the miRNA landscape in primates, contributing to human higher brain functions (Khaitovich et al., 2006). To find out if this re-shaping process affected the regulation of the cholinergic system, we searched for primate specificity within the group of 76 identified miRNAs that target more than one cholinergic transcript (Supplementary Table 3). About half (49%) of the VAChT and cholinesterases co-targeting miRNAs were found to be primate-specific (Supplementary Table 3), suggesting recently evolved miRNA-mediated mechanisms for co-regulation of ACh packaging and degradation (Table 1).
Of the cholinesterase targeting miRNAs, we found 16 that predictably target both BChE and AChE-R, whereas 23 miRNAs show “seed” complementarity to both AChE-R and AChE-S. However, BChE, and the synaptic AChE-S variant share only one single miRNA (Table 1), hsa-miR-491-5p (further details on shared miRNAs can be found in Supplementary Table 2). BChE has a much lower G, C content than the other four transcripts (Soreq et al., 1990); therefore, we wanted to find out whether this is reflected in its putatively targeting miRNAs. Not surprisingly, we found the average G, C content of BChE-targeting miRNAs to be 45%, approximately 12% lower than the average G, C content of the miRNAs targeting the other four transcripts (Table 1, Supplementary Table 4).
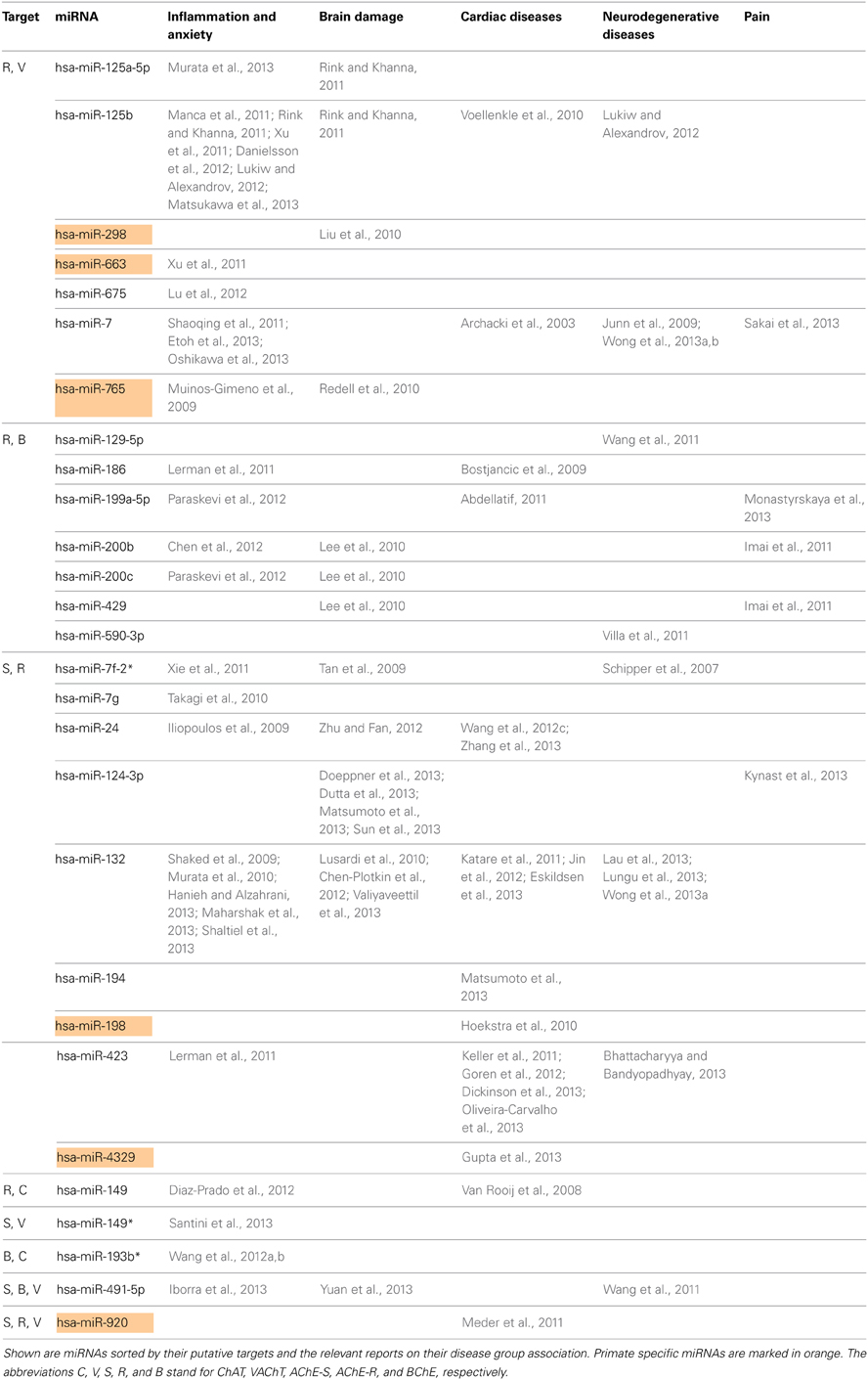
Next, we searched for the relevance of miRNAs targeting more than one transcript involved in ACh metabolism to known disease phenotypes (Supplementary Table 5). Compatible with the cancer-associated bias in the miRNA field, we found numerous studies on cancer-related miRNAs that target more than one transcript involved in ACh metabolism (Supplementary Table 5), and yet more are likely to accumulate in the near future. However, after excluding all of these cancer-related publications, we revealed 28 out of the identified 76 miRNAs that associate with other diseases. These could be classified into five groups: inflammation and anxiety, brain damage, cardiac or neurodegenerative diseases and pain-related syndromes (Supplementary Table 6). Table 2 lists these 28 miRNAs including the transcripts they are targeting as well as their reported disease associations. Interestingly, 67% of these miRNAs play key roles in inflammation-associated diseases and 61% of them target more than one disease group.
Figure 2 presents the miRNAs co-targeting AChE-S and AChE-R, BChE, and AChE-R or VAChT and AChE-R in a pie chart classifying the shared targets and relevant diseases and highlighting the observed interactions between these miRNAs and the disease phenotypes, as well as the primate specificity of part of these miRNAs. Disease group association is shown as surrounding lines of differential thicknesses, reflecting the number of miRNAs in each group. A notable example is the evolutionarily conserved neural-expressed miR-125b, which targets both VAChT and AChE-R (Table 2) and associates with all five groups: inflammation and anxiety (Manca et al., 2011; Xu et al., 2011; Danielsson et al., 2012; Matsukawa et al., 2013), brain damage (Rink and Khanna, 2011), neurodegenerative diseases (Lukiw and Alexandrov, 2012) and cardiac diseases (Voellenkle et al., 2010) and diverse pain syndromes (Imai et al., 2011; Kynast et al., 2013; Monastyrskaya et al., 2013; Sakai et al., 2013). Known association of each of these diseases with impaired cholinergic signaling (Shenhar-Tsarfaty et al., 2013a,b) supports the notion of physiological significance for surveillance by the overlapping miRNAs. A primate-specific example involves miR-765, which co-targets AChE-R and VAChT. Redell (Redell et al., 2010) found that miR-765 is upregulated in the plasma of patients after traumatic brain injury, compatible with changes in serum cholinesterases in post-stroke patients (Ben Assayag et al., 2010). Additionally, miR-765 targets the neurotrophin-3 receptor 3′UTR, and Muiños-Gimeno (Muinos-Gimeno et al., 2009) discovered a single nucleotide change located in the miR-765 binding site of this receptor's mRNA 3′UTR to be associated with panic disorder. Neurotrophin receptors regulate cholinergic signaling (Naumann et al., 2002), which associates with stress reactions (Kaufer et al., 1998; Sklan et al., 2004) as well as with RNA metabolism changes in the Alzheimer's brain (Berson et al., 2012; Lau et al., 2013); predicting that disrupted association of miRNAs with these receptors would impair cholinergic signaling and could increase the risk of anxiety-related diseases.
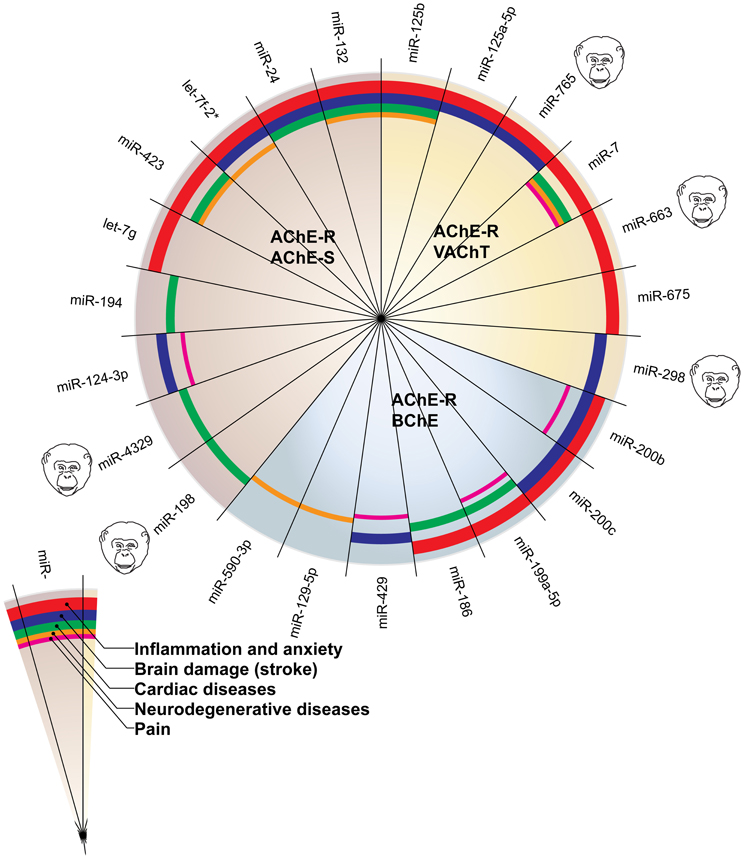
Figure 2. MiRNAs targeting AChE-S and AChE-R, BChE and AChE-R or VAChT and AChE-R show associations to different disease groups related to cholinergic signaling. The pie chart classifies the shared targets, as well as the relevant diseases and highlights the observed interactions between them. Disease group association is shown as surrounding lines of differential thicknesses, reflecting the number of miRNAs in each group. The red, blue, green, orange and purple colored lines symbolize disease associations with the groups of “inflammation and anxiety,” “brain damage (e.g., stroke),” “cardiac diseases,” “neurodegenerative diseases,” and “pain,” respectively. Note that miR-132 and miR-125b associate with all disease groups. The monkey heads next to specific miRNA numbers symbolize primate specificity.
To experimentally test for in vivo association of stress-induced changes in the identified miRNAs and their putative target genes, we selected the evolutionarily conserved miR-186 which predictably targets the two soluble cholinesterases, BChE and AChE-R. Of those, AChE-R increases under psychological stress were well documented (Kaufer et al., 1998; Cohen et al., 2002; Meshorer et al., 2005; Shaltiel et al., 2013), including the serum (Sklan et al., 2004), but BChE's involvement was never approached. In our current study, we quantified miR-186 levels in intestinal biopsies prepared from male C57BJ mice 7 days following 10 min exposure to cat litter (predator scent stress) and injection for four consecutive days with 50 μ g kg−1 saline (Zimmerman et al., 2012) compared to matched controls (n = 5 mice per group). In the intestinal biopsies, miR-186 expression normalized to the house-keeping short RNA RNU6 showed a 1.6-fold increase (p < 0.016) in pre-stressed mice. In parallel, these mice showed a 1.8-fold elevation in total cholinesterase activities (p < 0.003, Student's t test) as well as a less pronounced 1.6-fold increase in AChE levels (p < 0.054). Figure 3 presents these findings, demonstrating directly associated changes in the intestinal levels of the miR-186 and its two cholinergic regulating targets.
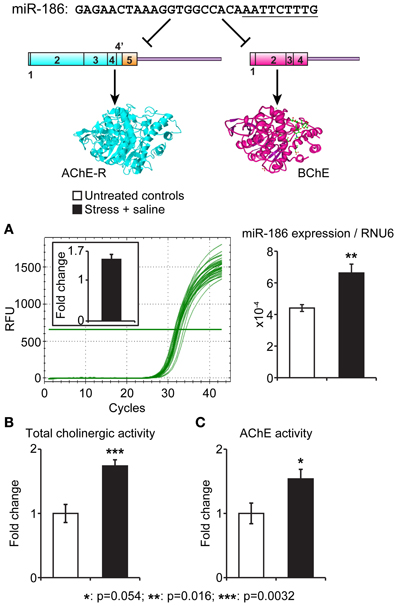
Figure 3. Intestinal miR-186 increases under predator scent stress are accompanied by elevated BChE and AChE activities. Top: The nucleotide sequence of the BChE and AChE-R-targeting miR-186 and schemes of its AChE-R and BChE mRNA targets. The “seed” sequence is underlined. (A) qRT-PCR quantification normalized to RNU6 levels demonstrates reproducible RFU values and 1.6-fold excess of miR-186 in intestinal sections from stressed mice, 7 days post-predator scent exposure (p = 0.016). (B,C) Highly significant intestinal elevation of total acetylthiocholine hydrolytic activity (p = 0.0032) accompanied by less pronounced AChE activity measured in the presence of 10 μM iso-OMPA (p = 0.05). N = 5 mice per group, in all tests.
Discussion
Combined use of four different bioinformatics algorithms identified a large number of miRNAs putatively targeting the 3′UTRs of ChAT, VAChT, AChE-S, AChE-R, and BChE. MiRNAs can notably regulate whole biological pathways; for example, miR-181 controls mouse hematopoiesis (Chen et al., 2004), miR-608 targets two inflammation-related transcripts, CDC42 and IL6 (Jeyapalan et al., 2011; Kang et al., 2011) and miR-221 controls multiple cancer pathways (Lupini et al., 2013). To challenge the possibility that certain miRNAs likewise regulate ACh metabolism and belong to the family of CholinomiRs, we searched for miRNAs targeting more than one of the five transcripts involved in the process of ACh synthesis, packaging and degradation. Intriguingly, packaging more than synthesis of ACh emerged as being co-regulated with its degradation, suggesting a dynamically controlled surveillance of these two processes. Furthermore, these findings highlight the option of differential post-transcriptional regulation of VAChT and ChAT, which share the same promoter but have distinct 3′-UTR domains.
The numbers of miRNAs we identified are likely to be under-estimated due to the exclusion of all the cancer-related miRNAs, which may be linked to cholinergic signaling as well. However, it is noteworthy that many of the miRNAs associated with ACh metabolism are primate-specific. This implies that there are no mouse models to study their function and influence, decreasing the likelihood of experimental animal studies of these miRNAs. Nevertheless, we found many human disease-association studies that demonstrate putative links of these miRNAs to inflammation and possibly reflecting the inflammatory blockade by ACh (Tracey, 2010). In comparison, only a few of the identified miRNAs appear to be largely modified in neurodegenerative diseases such as Alzheimer's disease, perhaps because cholinergic neurons decline early in the Alzheimer's brains (Mcgeer et al., 1984), so that it may be too late to find miRNAs playing a role in ACh metabolism in late stage brain samples from Alzheimer's patients. An exception is the AChE-targeted miR-132, which shows a drastic decline in the Alzheimer's brain (Lau et al., 2013).
The apparent relevance of CholinomiRs to diverse pain syndromes is particularly intriguing (Kress et al., 2013). Thus miR-199a-5p is expressed in the bladder's smooth muscle and urothelium and may play a role in bladder pain syndrome (Monastyrskaya et al., 2013) by suppressing LIN7C, ARHGAP12, PALS1, RND1 and PVRL1. In addition, miR-199a-5p is predicted to target both BChE and AChE-R, suggesting that its increase would up-regulate cholinergic stimulation in the bladder, which could also contribute to pain reactions. Likewise, miR-200b and miR-429 predictably target BChE and AChE-R, and changes in their levels were reported under neuropathic pain following sciatic nerve ligation within the limbic forebrain's nucleus accumbens (Imai et al., 2011). This has been attributed to miR-200b/429 targeting of DNA methyltransferase 3a (DNMT3a), which indeed accumulated in post-synaptic neurons in the nucleus accumbens under a neuropathic pain-like state. Such an increase may lead to long-term silencing of several genes at the transcriptional level, and enhanced cholinergic stimulation might contribute to this effect. An inverse effect has intriguingly been observed for miR-7a, which predictably targets VAChT and AChE-R and may hence reduce the packaging efficiency of ACh and limit cholinergic signals: miR-7a alleviates the maintenance of neuropathic pain through regulating neuronal excitability by targeting the β 2 subunit of the voltage-dependent sodium channel (Sakai et al., 2013). In all of these cases, the cholinergic targets may thus modulate the observed pain-related effects.
Our experimental test of miR-186 relevance for stress-related conditions revealed a direct dual association between elevated miR-186 and parallel increases in BChE levels in intestinal tissues from predator scent-injection-stressed mice. These effects were long-lasting and were only significant for BChE, highlighting for the first time, the changes in this protein as associated with psychological stress. That both miR-186 and its BChE target show intestinal increases under stress may indicate that these miRNA changes reflect a feedback response limiting excessive ACh stimulation; supporting this notion, serum BChE increases in post-stroke patients were associated with better prospects of recovery (Shenhar-Tsarfaty et al., 2013a). Further studies will be required to explore the molecular mechanisms underlying this involvement.
Several limitations need to be taken into account regarding this study. First, the search algorithms for miRNA candidates appear to differ substantially, each yielding different results. In our current study, we studied those miRNAs found in each of these algorithms, to improve the prospects of identifying as many relevant miRNAs as possible. Second, as our study spanned all of the miRNAs that predictably target the 3′-UTRs in all of the transcripts of interest, further studies will be required to functionally validate these miRNAs not only as single targeting but also as dually targeting more than one of these ACh metabolism-related transcripts. Third, we utilized a data-mining approach as before (Hanin and Soreq, 2011), and relied on explorative studies which link the identified miRNAs to disease association, but it remains unclear if such associations reflect the disease outcome or inversely, an effort of the system to protect itself from the disease.
Taken together, our current study highlights the importance of interrogating the extent and dynamics of miRNA regulation at a pathway level as a novel approach for studying the molecular mechanisms underlying specific processes in health and disease. Moreover, the large fraction of primate-specific miRNAs that were identified in our study calls for paying special attention to such cases. Given that miRNAs are considered “druggable” molecules, for example by antisense oligonucleotide manipulations (Shaked et al., 2009; Shaltiel et al., 2013), it is imperative to search for high throughput datasets from human tissue studies of relevant diseases and perhaps engineer new mouse models with knocked-in primate-specific miRNAs, such as miR-608. This can become a novel approach for identifying targets for therapeutic intervention with diseases where primate-specific miRNAs are subjected to major changes. In either case, our current approach represents the peak of an iceberg; however, it provides an initial proof of principle for the concept of joint regulation over different transcripts involved in specific neurotransmission pathways. This study should further be regarded as a first step in a long pathway, since we only focused on five transcripts out of many involved in only one pathway, the cholinergic signaling pathway; but there are many more transcripts, such as neurotrophin or nicotinic and muscarinic ACh receptors playing a role in cholinergic signaling. Our findings thus indicate overlapping miRNA regulation as a new surveillance mechanism that can balance cholinergic neurotransmission and may be of value for both basic and translational aspects of neuroinflammation-related disorders.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Shani Shenhar-Tsarfaty, Mr. Yochai Wolf and Mr. Uriya Bekenstein, Jerusalem for helpful advice and discussions. This study was supported by a European Research Council Advanced Research Award 321501 (to Hermona Soreq) and The European Commission FP-7 Health-2013-Innovation grant number 602133 (to Hermona Soreq). Bettina Nadorp is an incumbent of a PhD fellowship from The Hebrew University's Bioengineering program.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnmol.2014.00009/abstract
Footnotes
References
Abdellatif, M. (2011). Cardioprotective microRNAs. Pediatr. Cardiol. 32, 311–316. doi: 10.1007/s00246-010-9882-7
Arbel, Y., Shenhar-Tsarfaty, S., Waiskopf, N., Finkelstein, A., Halkin, A., Revivo, M., et al. (2014). Decline in serum cholinesterase activities predicts 2 year major adverse cardiac events. Mol. Med. doi: 10.2119/molmed.2013.00139. [Epub ahead of print].
Archacki, S. R., Angheloiu, G., Tian, X. L., Tan, F. L., Dipaola, N., Shen, G. Q., et al. (2003). Identification of new genes differentially expressed in coronary artery disease by expression profiling. Physiol. Genom. 15, 65–74. doi: 10.1152/physiolgenomics.00181.2002
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Ben Assayag, E., Shenhar-Tsarfaty, S., Ofek, K., Soreq, L., Bova, I., Shopin, L., et al. (2010). Serum cholinesterase activities distinguish between stroke patients and controls and predict 12-month mortality. Mol. Med. 16, 278–286. doi: 10.2119/molmed.2010.00015
Berson, A., Barbash, S., Shaltiel, G., Goll, Y., Hanin, G., Greenberg, D. S., et al. (2012). Cholinergic-associated loss of hnRNP-A/B in Alzheimer's disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 4, 730–742. doi: 10.1002/emmm.201100995
Bhattacharyya, M., and Bandyopadhyay, S. (2013). Studying the differential co-expression of microRNAs reveals significant role of white matter in early Alzheimer's progression. Mol. Biosyst. 9, 457–466. doi: 10.1039/c2mb25434d
Bostjancic, E., Zidar, N., and Glavac, D. (2009). MicroRNA microarray expression profiling in human myocardial infarction. Dis. Markers 27, 255–268. doi: 10.1155/2009/641082
Chen, C. Z., Li, L., Lodish, H. F., and Bartel, D. P. (2004). MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86. doi: 10.1126/science.1091903
Chen, Y., Ge, W., Xu, L., Qu, C., Zhu, M., Zhang, W., et al. (2012). miR-200b is involved in intestinal fibrosis of Crohn's disease. Int. J. Mol. Med. 29, 601–606. doi: 10.3892/ijmm.2012.894
Chen-Plotkin, A. S., Unger, T. L., Gallagher, M. D., Bill, E., Kwong, L. K., Volpicelli-Daley, L., et al. (2012). TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 32, 11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012
Cohen, O., Erb, C., Ginzberg, D., Pollak, Y., Seidman, S., Shoham, S., et al. (2002). Neuronal overexpression of ‘readthrough’ acetylcholinesterase is associated with antisense-suppressible behavioral impairments. Mol. Psychiatry 7, 874–885. doi: 10.1038/sj.mp.4001103
Danielsson, K., Wahlin, Y. B., Gu, X., Boldrup, L., and Nylander, K. (2012). Altered expression of miR-21, miR-125b, and miR-203 indicates a role for these microRNAs in oral lichen planus. J. Oral Pathol. Med. 41, 90–95. doi: 10.1111/j.1600-0714.2011.01084.x
Diaz-Prado, S., Cicione, C., Muinos-Lopez, E., Hermida-Gomez, T., Oreiro, N., Fernandez-Lopez, C., et al. (2012). Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet. Disord. 13:144. doi: 10.1186/1471-2474-13-144
Dickinson, B. A., Semus, H. M., Montgomery, R. L., Stack, C., Latimer, P. A., Lewton, S. M., et al. (2013). Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 15, 650–659. doi: 10.1093/eurjhf/hft018
Doeppner, T. R., Doehring, M., Bretschneider, E., Zechariah, A., Kaltwasser, B., Muller, B., et al. (2013). MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. 126, 251–265. doi: 10.1007/s00401-013-1142-5
Dutta, R., Chomyk, A. M., Chang, A., Ribaudo, M. V., Deckard, S. A., Doud, M. K., et al. (2013). Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann. Neurol. 73, 637–645. doi: 10.1002/ana.23860
Eiden, L. E. (1998). The cholinergic gene locus. J. Neurochem. 70, 2227–2240. doi: 10.1046/j.1471-4159.1998.70062227.x
Ellman, G. L., Courtney, K. D., Andres, V. Jr., and Feather-Stone, R. M. (1961). A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95. doi: 10.1016/0006-2952(61)90145-9
Erickson, J. D., Varoqui, H., Schafer, M. K., Modi, W., Diebler, M. F., Weihe, E., et al. (1994). Functional identification of a vesicular acetylcholine transporter and its expression from a “cholinergic” gene locus. J. Biol. Chem. 269, 21929–21932.
Eskildsen, T. V., Jeppesen, P. L., Schneider, M., Nossent, A. Y., Sandberg, M. B., Hansen, P. B., et al. (2013). Angiotensin II Regulates microRNA-132/-212 in Hypertensive Rats and Humans. Int. J. Mol. Sci. 14, 11190–11207. doi: 10.3390/ijms140611190
Etoh, M., Jinnin, M., Makino, K., Yamane, K., Nakayama, W., Aoi, J., et al. (2013). microRNA-7 down-regulation mediates excessive collagen expression in localized scleroderma. Arch. Dermatol. Res. 305, 9–15. doi: 10.1007/s00403-012-1287-4
Goren, Y., Kushnir, M., Zafrir, B., Tabak, S., Lewis, B. S., and Amir, O. (2012). Serum levels of microRNAs in patients with heart failure. Eur. J. Heart Fail. 14, 147–154. doi: 10.1093/eurjhf/hfr155
Gupta, M. K., Halley, C., Duan, Z. H., Lappe, J., Viterna, J., Jana, S., et al. (2013). miRNA-548c: a specific signature in circulating PBMCs from dilated cardiomyopathy patients. J. Mol. Cell. Cardiol. 62, 131–141. doi: 10.1016/j.yjmcc.2013.05.011
Hanieh, H., and Alzahrani, A. (2013). MicroRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur. J. Immunol. 43, 2771–2782. doi: 10.1002/eji.201343486
Hanin, G., and Soreq, H. (2011). Cholinesterase-Targeting microRNAs Identified in silico affect specific biological processes. Front. Mol. Neurosci. 4:28. doi: 10.3389/fnmol.2011.00028
Hoekstra, M., Van Der Lans, C. A., Halvorsen, B., Gullestad, L., Kuiper, J., Aukrust, P., et al. (2010). The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem. Biophys. Res. Commun. 394, 792–797. doi: 10.1016/j.bbrc.2010.03.075
Iborra, M., Bernuzzi, F., Correale, C., Vetrano, S., Fiorino, G., Beltran, B., et al. (2013). Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 173, 250–258. doi: 10.1111/cei.12104
Iliopoulos, D., Hirsch, H. A., and Struhl, K. (2009). An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706. doi: 10.1016/j.cell.2009.10.014
Imai, S., Saeki, M., Yanase, M., Horiuchi, H., Abe, M., Narita, M., et al. (2011). Change in microRNAs associated with neuronal adaptive responses in the nucleus accumbens under neuropathic pain. J. Neurosci. 31, 15294–15299. doi: 10.1523/JNEUROSCI.0921-11.2011
Jeyapalan, Z., Deng, Z., Shatseva, T., Fang, L., He, C., and Yang, B. B. (2011). Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 39, 3026–3041. doi: 10.1093/nar/gkq1003
Jin, W., Reddy, M. A., Chen, Z., Putta, S., Lanting, L., Kato, M., et al. (2012). Small RNA sequencing reveals microRNAs that modulate angiotensin II effects in vascular smooth muscle cells. J. Biol. Chem. 287, 15672–15683. doi: 10.1074/jbc.M111.322669
Junn, E., Lee, K. W., Jeong, B. S., Chan, T. W., Im, J. Y., and Mouradian, M. M. (2009). Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. U.S.A. 106, 13052–13057. doi: 10.1073/pnas.0906277106
Kang, J. G., Majerciak, V., Uldrick, T. S., Wang, X., Kruhlak, M., Yarchoan, R., et al. (2011). Kaposi's sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol. 225, 378–389. doi: 10.1002/path.2962
Katare, R., Riu, F., Mitchell, K., Gubernator, M., Campagnolo, P., Cui, Y., et al. (2011). Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circ. Res. 109, 894–906. doi: 10.1161/CIRCRESAHA.111.251546
Kaufer, D., Friedman, A., Seidman, S., and Soreq, H. (1998). Acute stress facilitates long-lasting changes in cholinergic gene expression. Nature 393, 373–377. doi: 10.1038/30741
Kefas, B., Godlewski, J., Comeau, L., Li, Y., Abounader, R., Hawkinson, M., et al. (2008). microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 68, 3566–3572. doi: 10.1158/0008-5472.CAN-07-6639
Keller, A., Leidinger, P., Bauer, A., Elsharawy, A., Haas, J., Backes, C., et al. (2011). Toward the blood-borne miRNome of human diseases. Nat. Methods 8, 841–843. doi: 10.1038/nmeth.1682
Khaitovich, P., Enard, W., Lachmann, M., and Paabo, S. (2006). Evolution of primate gene expression. Nat. Rev. Genet. 7, 693–702. doi: 10.1038/nrg1940
Kress, M., Huttenhofer, A., Landry, M., Kuner, R., Favereaux, A., Greenberg, D., et al. (2013). microRNAs in nociceptive circuits as predictors of future clinical applications. Front. Mol. Neurosci. 6:33. doi: 10.3389/fnmol.2013.00033
Kynast, K. L., Russe, O. Q., Moser, C. V., Geisslinger, G., and Niederberger, E. (2013). Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain 154, 368–376. doi: 10.1016/j.pain.2012.11.010
Lau, P., Bossers, K., Janky, R., Salta, E., Frigerio, C. S., Barbash, S., et al. (2013). Alteration of the microRNA network during the progression of Alzheimer's disease. EMBO Mol. Med. 5, 1613–1634. doi: 10.1002/emmm.201201974
Lee, S. T., Chu, K., Jung, K. H., Yoon, H. J., Jeon, D., Kang, K. M., et al. (2010). MicroRNAs induced during ischemic preconditioning. Stroke 41, 1646–1651. doi: 10.1161/STROKEAHA.110.579649
Lerman, G., Avivi, C., Mardoukh, C., Barzilai, A., Tessone, A., Gradus, B., et al. (2011). MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE 6:e20916. doi: 10.1371/journal.pone.0020916
Levy, C., Khaled, M., Iliopoulos, D., Janas, M. M., Schubert, S., Pinner, S., et al. (2010). Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 40, 841–849. doi: 10.1016/j.molcel.2010.11.020
Liu, D.-Z., Tian, Y., Ander, B. P., Xu, H., Stamova, B. S., Zhan, X., et al. (2010). Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 30, 92–101. doi: 10.1038/jcbfm.2009.186
Lu, T. X., Sherrill, J. D., Wen, T., Plassard, A. J., Besse, J. A., Abonia, J. P., et al. (2012). MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J. Allergy Clin. Immunol. 129, 1064.e1069–1075.e1069. doi: 10.1016/j.jaci.2012.01.060
Lukiw, W. J., and Alexandrov, P. N. (2012). Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer's disease (AD) brain. Mol. Neurobiol. 46, 11–19. doi: 10.1007/s12035-012-8234-4
Lungu, G., Stoica, G., and Ambrus, A. (2013). MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci. Lett. 553, 153–158. doi: 10.1016/j.neulet.2013.08.001
Lupini, L., Bassi, C., Ferracin, M., Bartonicek, N., D'abundo, L., Zagatti, B., et al. (2013). miR-221 affects multiple cancer pathways by modulating the level of hundreds messenger RNAs. Front. Genet. 4:64. doi: 10.3389/fgene.2013.00064
Lusardi, T. A., Farr, C. D., Faulkner, C. L., Pignataro, G., Yang, T., Lan, J., et al. (2010). Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J. Cereb. Blood Flow Metab. 30, 744–756. doi: 10.1038/jcbfm.2009.253
Maharshak, N., Shenhar-Tsarfaty, S., Aroyo, N., Orpaz, N., Guberman, I., Canaani, J., et al. (2013). MicroRNA-132 modulates cholinergic signaling and inflammation in human inflammatory bowel disease. Inflamm. Bowel Dis. 19, 1346–1353. doi: 10.1097/MIB.0b013e318281f47d
Manca, S., Magrelli, A., Cialfi, S., Lefort, K., Ambra, R., Alimandi, M., et al. (2011). Oxidative stress activation of miR-125b is part of the molecular switch for Hailey-Hailey disease manifestation. Exp. Dermatol. 20, 932–937. doi: 10.1111/j.1600-0625.2011.01359.x
Martino, S., Di Girolamo, I., Orlacchio, A., Datti, A., and Orlacchio, A. (2009). MicroRNA implications across neurodevelopment and neuropathology. J. Biomed. Biotechnol. 2009:654346. doi: 10.1155/2009/654346
Matsukawa, T., Sakai, T., Yonezawa, T., Hiraiwa, H., Hamada, T., Nakashima, M., et al. (2013). MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res. Ther. 15, R28. doi: 10.1186/ar4164
Matsumoto, S., Sakata, Y., Suna, S., Nakatani, D., Usami, M., Hara, M., et al. (2013). Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 113, 322–326. doi: 10.1161/CIRCRESAHA.113.301209
Mcgeer, P. L., Mcgeer, E. G., Suzuki, J., Dolman, C. E., and Nagai, T. (1984). Aging, Alzheimer's disease, and the cholinergic system of the basal forebrain. Neurology 34, 741–745. doi: 10.1212/WNL.34.6.741
Meder, B., Keller, A., Vogel, B., Haas, J., Sedaghat-Hamedani, F., Kayvanpour, E., et al. (2011). MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 106, 13–23. doi: 10.1007/s00395-010-0123-2
Meshorer, E., Bryk, B., Toiber, D., Cohen, J., Podoly, E., Dori, A., et al. (2005). SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Mol. Psychiatry 10, 985–997. doi: 10.1038/sj.mp.4001735
Meshorer, E., and Soreq, H. (2006). Virtues and woes of AChE alternative splicing in stress-related neuropathologies. Trends Neurosci. 29, 216–224. doi: 10.1016/j.tins.2006.02.005
Monastyrskaya, K., Sanchez-Freire, V., Hashemi Gheinani, A., Klumpp, D. J., Babiychuk, E. B., Draeger, A., et al. (2013). miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am. J. Pathol. 182, 431–448. doi: 10.1016/j.ajpath.2012.10.020
Muinos-Gimeno, M., Guidi, M., Kagerbauer, B., Martin-Santos, R., Navines, R., Alonso, P., et al. (2009). Allele variants in functional MicroRNA target sites of the neurotrophin-3 receptor gene (NTRK3) as susceptibility factors for anxiety disorders. Hum. Mutat. 30, 1062–1071. doi: 10.1002/humu.21005
Murata, K., Furu, M., Yoshitomi, H., Ishikawa, M., Shibuya, H., Hashimoto, M., et al. (2013). Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE 8:e69118. doi: 10.1371/journal.pone.0069118
Murata, K., Yoshitomi, H., Tanida, S., Ishikawa, M., Nishitani, K., Ito, H., et al. (2010). Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 12, R86. doi: 10.1186/ar3013
Naumann, T., Casademunt, E., Hollerbach, E., Hofmann, J., Dechant, G., Frotscher, M., et al. (2002). Complete deletion of the neurotrophin receptor p75NTR leads to long-lasting increases in the number of basal forebrain cholinergic neurons. J. Neurosci. 22, 2409–2418.
Oliveira-Carvalho, V., Da Silva, M. M., Guimaraes, G. V., Bacal, F., and Bocchi, E. A. (2013). MicroRNAs: new players in heart failure. Mol. Biol. Rep. 40, 2663–2670. doi: 10.1007/s11033-012-2352-y
Oshikawa, Y., Jinnin, M., Makino, T., Kajihara, I., Makino, K., Honda, N., et al. (2013). Decreased miR-7 expression in the skin and sera of patients with dermatomyositis. Acta Derm. Venereol. 93, 273–276. doi: 10.2340/00015555-1459
Papagiannakopoulos, T., Shapiro, A., and Kosik, K. S. (2008). MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 68, 8164–8172. doi: 10.1158/0008-5472.CAN-08-1305
Paraskevi, A., Theodoropoulos, G., Papaconstantinou, I., Mantzaris, G., Nikiteas, N., and Gazouli, M. (2012). Circulating MicroRNA in inflammatory bowel disease. J. Crohns Colitis 6, 900–904. doi: 10.1016/j.crohns.2012.02.006
Redell, J. B., Moore, A. N., Ward, N. H. 3rd., Hergenroeder, G. W., and Dash, P. K. (2010). Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 27, 2147–2156. doi: 10.1089/neu.2010.1481
Rink, C., and Khanna, S. (2011). MicroRNA in ischemic stroke etiology and pathology. Physiol. Genomics 43, 521–528. doi: 10.1152/physiolgenomics.00158.2010
Sakai, A., Saitow, F., Miyake, N., Miyake, K., Shimada, T., and Suzuki, H. (2013). miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain 136, 2738–2750. doi: 10.1093/brain/awt191
Santini, P., Politi, L., Vedova, P. D., Scandurra, R., and Scotto D'abusco, A. (2013). The inflammatory circuitry of miR-149 as a pathological mechanism in osteoarthritis. Rheumatol. Int. doi: 10.1007/s00296-013-2754-8. [Epub ahead of print].
Schipper, H. M., Maes, O. C., Chertkow, H. M., and Wang, E. (2007). MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Biol. 1, 263–274. Available online at: http://www.la-press.com/microrna-expression-in-alzheimerblood-mononuclear-cells-article-a483
Shaked, I., Meerson, A., Wolf, Y., Avni, R., Greenberg, D., Gilboa-Geffen, A., et al. (2009). MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973. doi: 10.1016/j.immuni.2009.09.019
Shaltiel, G., Hanan, M., Wolf, Y., Barbash, S., Kovalev, E., Shoham, S., et al. (2013). Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct. Funct. 218, 59–72. doi: 10.1007/s00429-011-0376-z
Shaoqing, Y., Ruxin, Z., Guojun, L., Zhiqiang, Y., Hua, H., Shudong, Y., et al. (2011). Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am. J. Rhinol. Allergy 25, e242–e246. doi: 10.2500/ajra.2011.25.3682
Shenhar-Tsarfaty, S., Berliner, S., Bornstein, N. M., and Soreq, H. (2013a). Cholinesterases as biomarkers for parasympathetic dysfunction and inflammation-related disease. J. Mol. Neurosci. doi: 10.1007/s12031-013-0176-4. [Epub ahead of print].
Shenhar-Tsarfaty, S., Waiskopf, N., Ofek, K., Shopin, L., Usher, S., Berliner, S., et al. (2013b). Atherosclerosis and arteriosclerosis parameters in stroke patients associate with paraoxonase polymorphism and esterase activities. Eur. J. Neurol. 20, 891–898. doi: 10.1111/ene.12074
Sklan, E. H., Lowenthal, A., Korner, M., Ritov, Y., Landers, D. M., Rankinen, T., et al. (2004). Acetylcholinesterase/paraoxonase genotype and expression predict anxiety scores in health, risk factors, exercise training, and genetics study. Proc. Natl. Acad. Sci. U.S.A. 101, 5512–5517. doi: 10.1073/pnas.0307659101
Soreq, H., Ben-Aziz, R., Prody, C. A., Seidman, S., Gnatt, A., Neville, L., et al. (1990). Molecular cloning and construction of the coding region for human acetylcholinesterase reveals a G + C-rich attenuating structure. Proc. Natl. Acad. Sci. U.S.A. 87, 9688–9692. doi: 10.1073/pnas.87.24.9688
Soreq, H., and Seidman, S. (2001). Acetylcholinesterase–new roles for an old actor. Nat. Rev. Neurosci. 2, 294–302. doi: 10.1038/35067589
Sun, Y., Gui, H., Li, Q., Luo, Z. M., Zheng, M. J., Duan, J. L., et al. (2013). MicroRNA-124 protects neurons against apoptosis in cerebral ischemic stroke. CNS Neurosci. Ther. 19, 813–819. doi: 10.1111/cns.12142
Takagi, T., Naito, Y., Mizushima, K., Hirata, I., Yagi, N., Tomatsuri, N., et al. (2010). Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J. Gastroenterol. Hepatol. 25(Suppl. 1), S129–S133. doi: 10.1111/j.1440-1746.2009.06216.x
Tan, K. S., Armugam, A., Sepramaniam, S., Lim, K. Y., Setyowati, K. D., Wang, C. W., et al. (2009). Expression profile of MicroRNAs in young stroke patients. PLoS ONE 4:e7689. doi: 10.1371/journal.pone.0007689
Tracey, K. J. (2010). Understanding immunity requires more than immunology. Nat. Immunol. 11, 561–564. doi: 10.1038/ni0710-561
Valiyaveettil, M., Alamneh, Y. A., Miller, S.-A., Hammamieh, R., Arun, P., Wang, Y., et al. (2013). Modulation of cholinergic pathways and inflammatory mediators in blast-induced traumatic brain injury. Chem. Biol. Interact. 203, 371–375. doi: 10.1016/j.cbi.2012.10.022
Van Rooij, E., Sutherland, L. B., Thatcher, J. E., Dimaio, J. M., Naseem, R. H., Marshall, W. S., et al. (2008). Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13027–13032. doi: 10.1073/pnas.0805038105
Villa, C., Fenoglio, C., De Riz, M., Clerici, F., Marcone, A., Benussi, L., et al. (2011). Role of hnRNP-A1 and miR-590-3p in neuronal death: genetics and expression analysis in patients with Alzheimer disease and frontotemporal lobar degeneration. Rejuvenation Res. 14, 275–281. doi: 10.1089/rej.2010.1123
Voellenkle, C., Van Rooij, J., Cappuzzello, C., Greco, S., Arcelli, D., Di Vito, L., et al. (2010). MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genomics 42, 420–426. doi: 10.1152/physiolgenomics.00211.2009
Wang, H.-J., Zhang, P.-J., Chen, W.-J., Feng, D., Jia, Y.-H., and Xie, L.-X. (2012a). Four serum microRNAs identified as diagnostic biomarkers of sepsis. J. Trauma Acute Care Surg. 73, 850–854. doi: 10.1097/TA.0b013e31825a7560
Wang, H., Zhang, P., Chen, W., Feng, D., Jia, Y., and Xie, L. (2012b). Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS ONE 7:e38885. doi: 10.1371/journal.pone.0038885
Wang, J., Huang, W., Xu, R., Nie, Y., Cao, X., Meng, J., et al. (2012c). MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J. Cell. Mol. Med. 16, 2150–2160. doi: 10.1111/j.1582-4934.2012.01523.x.
Wang, W.-X., Huang, Q., Hu, Y., Stromberg, A. J., and Nelson, P. T. (2011). Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121, 193–205. doi: 10.1007/s00401-010-0756-0
Wong, H.-K. A., Veremeyko, T., Patel, N., Lemere, C. A., Walsh, D. M., Esau, C., et al. (2013a). De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer's disease. Hum. Mol. Genet. 22, 3077–3092. doi: 10.1093/hmg/ddt164
Wong, M. S., Chen, L., Foster, C., Kainthla, R., Shay, J. W., and Wright, W. E. (2013b). Regulation of telomerase alternative splicing: a target for chemotherapy. Cell Rep. 3, 1028–1035. doi: 10.1016/j.celrep.2013.03.011
Xie, Y.-F., Shu, R., Jiang, S.-Y., Liu, D.-L., and Zhang, X.-L. (2011). Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int. J. Oral Sci. 3, 125–134. doi: 10.4248/IJOS11046
Xu, N., Brodin, P., Wei, T., Meisgen, F., Eidsmo, L., Nagy, N., et al. (2011). MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J. Invest. Dermatol. 131, 1521–1529. doi: 10.1038/jid.2011.55
Yuan, M., Zhan, Q., Duan, X., Song, B., Zeng, S., Chen, X., et al. (2013). A functional polymorphism at miR-491-5p binding site in the 3′-UTR of MMP-9 gene confers increased risk for atherosclerotic cerebral infarction in a Chinese population. Atherosclerosis 226, 447–452. doi: 10.1016/j.atherosclerosis.2012.11.026
Zhang, H.-B., Li, R.-C., Xu, M., Xu, S.-M., Lai, Y.-S., Wu, H.-D., et al. (2013). Ultrastructural uncoupling between T-tubules and sarcoplasmic reticulum in human heart failure. Cardiovasc. Res. 98, 269–276. doi: 10.1093/cvr/cvt030
Zhu, H., and Fan, G. C. (2012). Role of microRNAs in the reperfused myocardium towards post-infarct remodelling. Cardiovasc. Res. 94, 284–292. doi: 10.1093/cvr/cvr291
Keywords: acetlycholinesterase, butyrylcholinesterase, choline acetyltransferase, vesicular acetylcholine transporter, microRNA-186, cholinergic signaling, primate-specific microRNAs
Citation: Nadorp B and Soreq H (2014) Predicted overlapping microRNA regulators of acetylcholine packaging and degradation in neuroinflammation-related disorders. Front. Mol. Neurosci. 7:9. doi: 10.3389/fnmol.2014.00009
Received: 23 September 2013; Accepted: 21 January 2014;
Published online: 10 February 2014.
Edited by:
Bernard Attali, Tel Aviv University, IsraelReviewed by:
Alessandro Vercelli, Università Degli Studi di Siena, ItalyBjörn Spittau, Albert-Ludwigs-University Freiburg, Germany
Copyright © 2014 Nadorp and Soreq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hermona Soreq, Department of Biological Chemistry and the Center for Bioengineering, The Edmond and Lily Safra Center for Brain Science, The Hebrew University of Jerusalem, The Edmond J. Safra Campus - Givat Ram, Jerusalem 91904, Israel e-mail:aGVybW9uYS5zb3JlcUBtYWlsLmh1amkuYWMuaWw=
 Bettina Nadorp
Bettina Nadorp Hermona Soreq
Hermona Soreq