
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Neurosci. , 12 April 2012
Sec. Molecular Signalling and Pathways
Volume 5 - 2012 | https://doi.org/10.3389/fnmol.2012.00038
This article is part of the Research Topic The neuronal functions of EF-hand Ca(2+)-binding proteins View all 22 articles
It is well-known that the opening of L-type voltage-gated calcium channels can be regulated by calmodulin (CaM). One of the main regulatory mechanisms is calcium-dependent inactivation (CDI), where binding of apo-CaM to the cytoplasmic C-terminal domain of the channel can effectively sense an increase in the local calcium ion concentration. Calcium-bound CaM can bind to the IQ-motif region of the C-terminal region and block the calcium channel, thereby providing a negative feedback mechanism that prevents the rise of cellular calcium concentrations over physiological limits. Recently, an additional Ca2+/CaM-binding motif (NSCaTE, N-terminal spatial Ca2+ transforming element) was identified in the amino terminal cytoplasmic region of Cav1.2 and Cav1.3. This motif exists only in Cav1.2 and Cav1.3 channels, and a pronounced N-lobe (Ca2+/CaM) CDI effect was found for Cav1.3. To understand the molecular basis of this interaction, the complexes of Ca2+/CaM with the biosynthetically produced N-terminal region (residues 1–68) and NSCaTE peptide (residues 48–68) were investigated. We discovered that the NSCaTE motif in the N-terminal cytoplasmic region adopts an α-helical conformation, most likely due to its high alanine content. Additionally, the complex exhibits an unusual 1:2 protein:peptide stoichiometry when bound to Ca2+-CaM, and the N-lobe of CaM has a much stronger affinity for the peptide than the C-lobe. The complex structures of the isolated N- and C-lobe of Ca2+/CaM and the NSCaTE peptide were determined by nuclear magnetic resonance spectroscopy and data-driven protein-docking methods. Moreover, we also demonstrated that calcium binding protein 1, which competes with CaM for binding to the C-terminal cytoplasmic domain, binds only weakly to the NSCaTE region. The structures provide insights into the possible roles of this motif in the calcium regulatory network. Our study provides structural evidence for the CaM-bridge model proposed in previous studies.
It has long been known that calmodulin (CaM) can regulate the activity of the voltage-gated calcium channels (VGCCs) (Qin et al., 1999; Zuhlke et al., 1999, 2000; Kim et al., 2004). The interactions between CaM and the IQ-motif in the C-terminal cytoplasmic tail of VGCC occupy the central position in the calcium-dependent feedback regulation loop (Fallon et al., 2005; Van Petegem et al., 2005; Dunlap, 2007). CaM seems to be mainly involved in two opposing mechanisms to control the calcium influx: calcium-dependent inactivation (CDI) and calcium-dependent facilitation (CDF). Depending on the type of channel, CaM can bind to the IQ-motif with either a parallel (L-type) or an anti-parallel (P/Q type) orientation (Fallon et al., 2005; Van Petegem et al., 2005; Kim et al., 2008; Mori et al., 2008). Apo- and Ca2+-CaM both have a flexible dumbbell structure in solution (Yamniuk and Vogel, 2004; Ishida and Vogel, 2006). The N- and C-lobe of CaM play different roles in CDI and CDF (Zuhlke and Reuter, 1998; Peterson et al., 1999; Zuhlke et al., 1999, 2000; DeMaria et al., 2001; Pitt et al., 2001; Dick et al., 2008). Recently, complexes of Ca2+-CaM with the IQ-motifs from both L-type and P/Q-type channels have been crystallized. These high-resolution structures have resolved many ambiguities from previous studies but have also raised some new questions (Fallon et al., 2005; Van Petegem et al., 2005; Houdusse et al., 2006; Mori et al., 2008).
In addition to CaM, the main channel-pore forming α1c subunit of the channel is also regulated by Cavβ subunits, CaMKII and the CaBP-family proteins (Yuan and Bers, 1994; Hudmon et al., 2005; Grueter et al., 2006; Buraei and Yang, 2010). Recently, it has been found that a sequence of ∼10 amino acid residues in the N-terminal cytoplasmic domains of Cav1.2 and Cav1.3, known as the N-terminal spatial Ca2+ transforming element (NSCaTE) motif (Dick et al., 2008), is also involved in CDI regulation. The functional significance of this particular region has been clearly demonstrated for Cav1.2 and Cav1.3 in vivo (Dick et al., 2008). Interestingly, P/Q-type VGCCs lack this motif in their N-terminal region. However, upon the artificial introduction of this motif into the Cav2.2 N-terminus, local calcium selectivity could also be demonstrated. The results of these experiments illustrate an important function for the NSCaTE region in the regulation of specific L-type VGCCs.
To better understand the function of this motif at the molecular level, the complex of Ca2+/CaM and the NSCaTE peptide was investigated in this work. First, two constructs [long (1–68) and short (48–68)] of the Cav1.2 N-terminus, both including the NSCaTE sequence, were made biosynthetically. The interactions between these peptides and CaM were characterized by isothermal titration calorimetry (ITC), steady state fluorescence and nuclear magnetic resonance (NMR) spectroscopy. Although the majority of the N-terminal domain is unstructured, surprisingly, a helical conformation was found for the NSCaTE Ca2+/CaM-binding motif even in aqueous solution. Moreover, an unusual 1:2 stoichiometry of CaM to peptide binding was observed. It was also found that the N-lobe of CaM could bind to Cav1.2NT more tightly than the C-lobe. We also demonstrate that the NSCaTE region binds poorly to the related calcium binding protein 1 (CaBP1) protein (Oz et al., 2011). The relatively low affinity between CaM and the NSCaTE peptide prevented the determination of a complex structure of the intact protein by NMR spectroscopy. Instead, the structure of each lobe of CaM in complex with the NSCaTE was determined separately, making use of the fact that the two isolated lobes of CaM are known to retain their native structure (Thulin et al., 1984). The N-lobe Ca2+-CaM/NSCaTE complex was solved by routine NMR methods, and a comparison between the NMR determined complex structure and a high ambiguity-driven biomolecular docking (HADDOCK)-generated complex structure was made (Dominguez et al., 2003). The C-lobe Ca2+-CaM/NSCaTE complex structure was generated by a combined NMR and data-driven docking strategy. In this paper, we present a model for this complicated regulatory network. The cytoplasmic N- and C-termini of the calcium channel, CaM and CaBP1 are all involved in this network, and the possible roles for these interacting partners are discussed below.
Both CaM (and its N- and C-lobes) and CaBP1 were expressed using the pET30b vector (Novagen). The N-lobe of CaM contains residues 1–77, and the C-lobe contains residues 78–148. The gene for human CaBP1 was synthesized by BlueHeron, Ltd. (Rockville, MD, USA) and was optimized for Escheichia coli expression. Compared with the wild-type S-CaBP1, the first 15 AAs of this CaBP1 variant are missing, similar to the CaBP1(Δ2–15) construct used in the recent CaBP1 structure determined by X-ray crystallography (Findeisen and Minor, 2010). The calmodulin protein and its isolated lobes were purified using a phenyl-Sepharose hydrophobic column (Pharmacia). A previously described protocol for the purification of most CaBPs was used with minor modifications (Queiroz et al., 2001). The frozen pellet was resuspended in 50 mM Tris-HCl, 2 mM ethylenedimainetetraacetic acid (EDTA), 1 mM DTT, 150 mM KCl, 1.5 mg/ml phenylmethanesulfonylfluoride (PMSF), pH 7.5. The sample was always incubated on ice, and all operations were performed in the cold room (∼4°C). The sample was homogenized, and a French press was used to lyse the cells. The soluble protein and debris were separated by centrifugation at 18,000 rpm for 1 h. The supernatant was loaded slowly onto the first hydrophobic column, which had been pre-equilibrated with 50 mM Tris-HCl, 2 mM EDTA, 1 mM DTT, 150 mM KCl, pH 7.5 (buffer A). A total of 200–250 mL of the flow-through solution was collected. CaCl2 and MgCl2 were then added to give final concentrations of 10 mM Ca2+ and 5 mM Mg2+ prior to loading onto the second column, which was pre-equilibrated with 50 mM Tris-HCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM DTT, 100 mM KCl, pH 7.5 (buffer B). After loading, the column was washed with approximately 100 mL of buffer B, after which the proteins were immediately eluted with buffer C (50 mM Tris-HCl, 50 mM KCl, 2 mM EDTA, and 1 mM DTT; pH 7.5) because of the relatively low affinity of the N- and C-lobes of CaM and CaBP1 for the hydrophobic column. The fractions were then pooled together and were further purified using anion exchange. An AKTA purifier system with a Resource Q anion-exchange column (GE Healthcare) was used. Following the anion-exchange purification, all fractions containing the desired protein were collected and dialyzed against 1 g/L ammonium bicarbonate. After five or six buffer exchanges, the sample was flash frozen and lyophilized. The protein powder was stored at −20°C for further studies.
The DNA sequence of the full-length (124 AAs) N-terminal cytoplasmic domain of Cav1.2α was ordered from GENEART, Life Technologies. The codons of this construct were optimized for expression in E. coli. The expression constructs were made in a modified pET15b vector that included a 6× His tag and a small ubiquitin-like modifier protein (SUMO) tag before a tobacco etch virus (TEV) protease-cleavage site (Panavas et al., 2009). Two DNA sequences were successfully cloned between the KpnI and BamHI sites. In initial expression experiments, multiple bands were observed for the 1–124 construct; we, therefore, analyzed shorter constructs and found that a 1–68 construct gave rise to one band. A shorter construct (residues 48–68), which is slightly longer than the NSCaTE motif (residues 52–60) was also made. The longer construct started at the N-terminal end of Cav1.2α and ended in the same region as the short peptide (residues 1–68). The proteins were purified on a Ni-NTA column. After the cleavage of the SUMO tag with TEV protease, the cleaved protein was reloaded onto the Ni column. The flow-through solution containing the peptides was concentrated, and the concentration was determined by OD280 using extinction coefficients obtained from ProtParam (ExPASy), 8480 M−1 cm−1 for the long peptide and 5500 M−1 cm−1 for the short peptide. Since the peptides contain Trp residues, their concentration can be accurately determined. Also the concentration of Ca2+-CaM can be readily determined from the absorbance spectra of the two Tyr residues. The peptide purity was verified by SDS-PAGE and mass spectrometry.
Circular dichroism (CD) spectra were recorded on a Jasco J-810 spectrometer (Jasco, Inc., Easton, MD, USA). Samples were prepared in 10 mM Tris (pH 7.0) with a concentration of 20 uM. Trifluoroethanol (TFE) was added as a co-solvent in solution. A 1 mm pathlength cuvette was used for all spectra.
All ITC experiments were performed on a Microcal VP-ITC instrument. The ITC buffer consisted of 20 mM HEPES, 100 mM KCl, and 5 mM CaCl2, pH 7.0. For experiments with apo-CaM, 5 mM EDTA was used instead of 1 mM CaCl2. The ITC cell was filled with 50 μM intact CaM, N- or C-lobe CaM, or CaBP1. A highly concentrated solution of the NSCaTE peptide was then injected (5 μL each) into the cell. The concentration of the C-lobe of CaM was determined by the absorbance at OD280. However, the N-lobe of CaM has no Trp or Tyr residues. The N-lobe concentration, therefore, was estimated by weight and verified by the Bio-Rad protein assay. The concentration of CaBP1 was measured by OD280 with an extinction coefficient of 2980 M−1 cm−1. All experiments were run at 25°C. The data were fit to either a one-site binding model (N-/C-lobe of CaM and CaBP1) or a two-site binding model (intact CaM). Control experiments were performed to adjust the baseline of each experiment.
All experiments were performed on a Varian Cary Eclipse spectrofluorimeter. The NSCaTE peptide contains one Trp residue and was selectively excited at 295 nm with a 5 nm excitation slit width. The emission spectra were recorded from 300 to 450 nm with a 10 nm emission slit width. All samples were dissolved in a solution of 20 mM HEPES and 100 mM KCl at pH 7.0. A 50 μM peptide concentration was used in the cell. Concentrated CaM or N-/C-lobe CaM (0.7–1.0 mM) was titrated into 1 mL of the peptide in 5 μL aliquots. The changes in the fluorescence intensity for each titration at Δmax were recorded and used to calculate the binding constant, Ka:
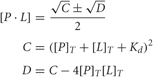
where [P]T and [L]T are the total concentrations of the protein and the peptide. The Caligator 1.05 software, developed by the group of S. Linse, was used to calculate the Ka (Andre and Linse, 2002).
All NMR samples were prepared in buffer containing 20 mM Bis-Tris, 100 mM KCl, pH 7.0. For all complexes, the saturation of the labeled component was monitored with 1H–15N HSQC experiments. The titrations were complete when no further peak shifts were observed.
All 1H–15N HSQC experiments were performed at 25°C on 500 MHz and 700 MHz NMR instruments. For the 1H(F2) and 15N(F1) dimensions, 1024 × 256 real data points were used.
The backbone assignments were completed for the following peptides/proteins in the free or bound state: the 13C/15N-labeled free-state long NSCaTE peptide, the 13C/15N-labeled short NSCaTE peptide saturated with the unlabeled N-/C-lobe CaM and the 13C/15N-labeled CaM N-/C-lobe bound to the unlabeled short NSCaTE peptide. Routine 3D NMR backbone assignment experiments (CBCACONH, HNCACB, HNCO, and HNCACO) were used. The side chain assignments were made using CCCONH, HCCCONH, and HCCH-TOCSY experiments. All of these experiments are the same as those used in the structure determination of the soybean calmodulin isoform 4 (SCaM4)/MAPK-phosphatase peptide complex (Ishida et al., 2009).
A sample containing the 13C/15N-labeled Ca2+/CaM N-lobe saturated with the unlabeled NSCaTE peptide was used to acquire the 13C/15N double-filtered 2D-NOESY spectrum, and the intramolecular NOEs between the CaM N-lobe and peptide were determined. To obtain the structures of the short peptide in complex with the N-lobe of CaM, another sample containing the 13C/15N-labeled NSCaTE peptide saturated with the deuterated Ca2+/CaM N-lobe was examined. The side chain and the 15N-edited 3D-NOESY experiments were performed to obtain unambiguous chemical shift assignments for the NSCaTE peptide and intrapeptide NOEs.
Heteronuclear 1H–15N NOE experiments were performed for the long Cav1.2NT peptide. A 5 sec pre-saturation time was used. The experiment was performed on a 500 MHz NMR instrument.
The structures of the complex of the Ca2+/CaM-NT with the NSCaTE peptide and of Ca2+/CaM-CT saturated with the NSCaTE peptide were determined using the NOE-based proton–proton distances derived from the 15N-edited 3D-NOESY. The dihedral angles along the sequence of each protein were calculated by TALOS (Cornilescu et al., 1999). These restraints were used in the calculation of the final structure by CYANA 2.0 (Guntert, 2004).
The NSCaTE motif only exists in the Cav1.2 and Cav1.3 L-type calcium channels (Figure 1). This sequence does not conform to any other known CaM-binding motif. Most linear peptides do not adopt stable secondary structures in aqueous solution. However, our experimental results show that part of the long NSCaTE (1–68) peptide can adopt a relatively stable helical conformation in aqueous solution from residue S51 to residue A62. The 1H–15N HSQC peak dispersion of the isotope-labeled peptide is only around approximately 1 ppm (7.7–8.7 ppm; see Figure 2A), which is normal for a peptide in a random coil conformation. However, the NSCaTE region, which extends from S51 to A62, is helical according to the secondary structure prediction based on the chemical shift index (CSI) of the backbone heavy atoms (Figure 2B). The remainder of the N-terminal domain of the channel is unstructured, which is consistent with the outcome of protein disorder predictions (see Appendix Figure A1) NMR relaxation experiments were also performed with the long peptide (Figure 2C). The {1H}–15N NOE experiments only showed positive values for the structured helix portion, while all other residues had negative values. The average NOE values for the structured region were also quite small (0.251 ± 0.033), indicating the relative flexibility of this region. The CD spectrum of this peptide indicated the presence of a mixture of random coil and helical conformations. We also showed that the percentage of the helical conformation with increasing amounts of TFE as a co-solvent in solution (see Appendix Figure A2). As expected, the TFE cosolvent appeared to further stabilize the small helix (Zhang et al., 1993). The intensities of the 1H–15N HSQC peaks were not evenly distributed along the sequence; the peaks for the helical portion had the strongest intensity. The backbone sequence assignment could be completed for almost the entire peptide sequence. We noticed that the NSCaTE region had a high percentage of alanine (4/11, residues L50–R60). Of all amino acids, alanine has the highest helix-forming propensity (Pace and Scholtz, 1998), which could be a likely reason for the spontaneous helix formation in this region. However, the severe overlap of the alanine signals hindered a complete backbone assignment. Several small, unassigned peaks in the spectrum may have arisen from a minor conformation of the peptide, possibly arising from a cis-trans conformational exchange from several proline residues in the random coil region.
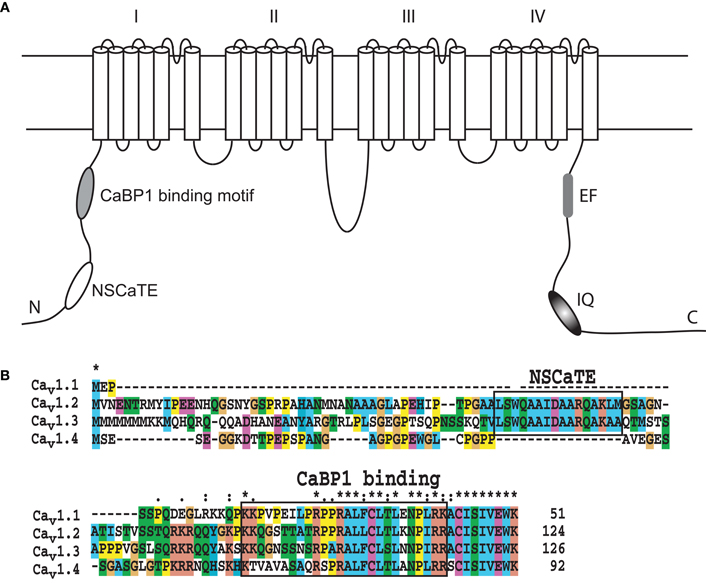
Figure 1. (A) Schematic of the pore-forming α-subunit of the L-type voltage-gated calcium channels Cav1.2 and Cav1.3 indicating the position of the various regulatory elements. (B). Sequence alignment of the N-terminal sequences of the human L-type voltage-gated calcium channels. The alignment was performed with ClustalW, and the default color settings were used. The Ca2+/CaM and CaBP1 binding motifs are highlighted with black boxes.
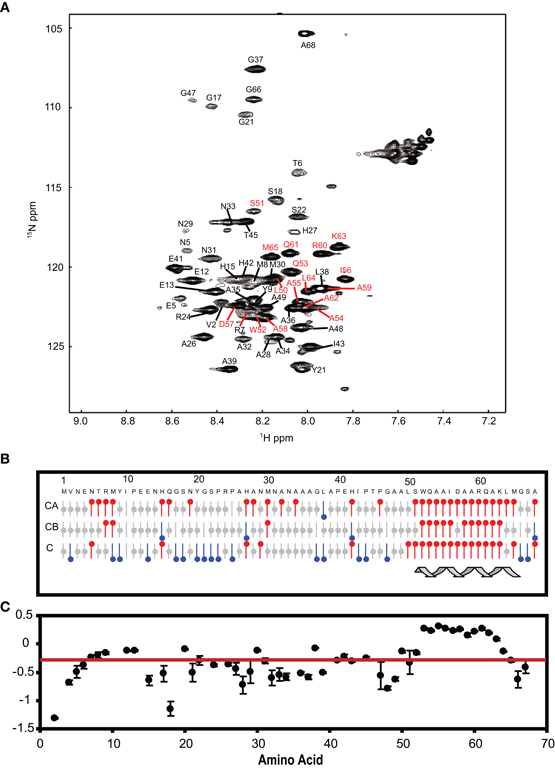
Figure 2. (A) 1H–15N HSQC spectrum of the long NSCaTE (AA 1–68) peptide. (B) Secondary structure prediction based on the 13C chemical shifts of the long NSCaTE construct (Cav1.2NT1–68). (C) Heteronuclear {1H}–15N NOE data for the long NSCaTE construct (Cav1.2NT1–68).
The ITC experiments demonstrated that the binding of the NSCaTE peptide to wild-type CaM is exothermic (Figures 3A,B). An unusual 2:1 stoichiometry (peptide:protein) was observed, which is similar to the binding between the GAD peptide and CaM (Yuan and Vogel, 1998) and the binding of MAPK-phosphatase to CaM (Rainaldi et al., 2007). The NMR titration experiments also confirmed this result, as the HSQC peaks only ceased to shift after more than two equivalents of peptide were added (data not shown). It should be noted that this stoichiometry is different from the 1:1 binding reported by Benmocha et al. (2009). The two-site binding model showed that the two sites had different binding affinities, which was confirmed by studying the binding of each isolated lobe to the peptide (Figures 3C,D). This sequential mode of binding resembles that reported for the MAPK-phosphatase, where both lobes of CaM can bind with different affinities (Rainaldi et al., 2007). A 1:1 stoichiometry was found for each lobe, and the N-lobe had a stronger affinity than the C-lobe (Table 1). However, the binding affinities of the NSCaTE peptide (Kd ∼ μM) to both domains were relatively weaker than other CaM-binding peptides, such as the IQ peptide from the C-terminal tail of the same channel (Kd ∼ nM) (Van Petegem et al., 2005). Compared with the IQ peptide, which has a total of 21 AAs (K1617–E1637) involved in binding with residue Ile1624 in the middle, the effective binding region of the NSCaTE peptide was much shorter (10 AAs, S51–R60). This short region could not span both lobes of Ca2+/CaM in the classical 1:1 mode. The same was observed for the MAPK-phosphatase/Ca2+-CaM interaction, where the CaM-binding domain is also only 10 residues long (Ishida et al., 2009).
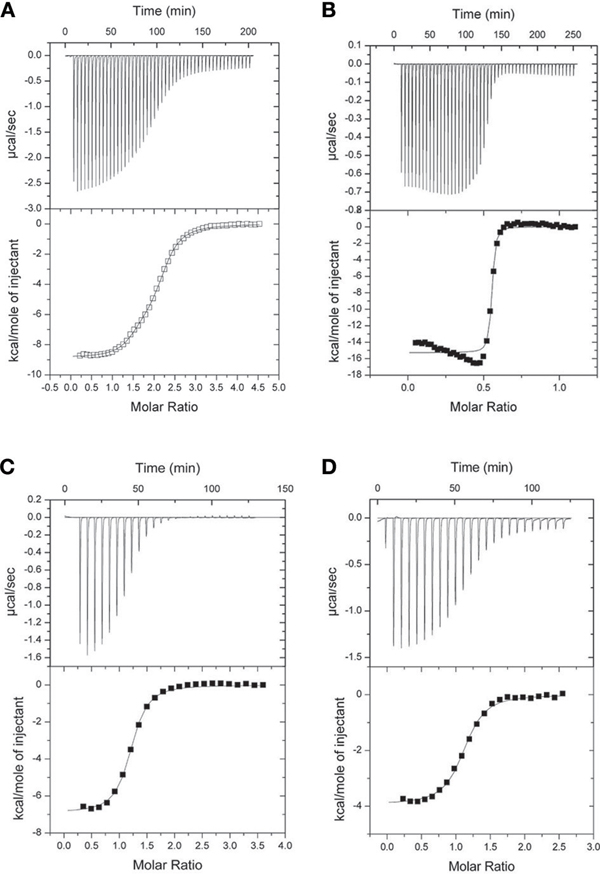
Figure 3. ITC characterization of the interactions of Ca2+/CaM with the long NSCaTE peptide. (A) NSCaTE peptide titrated into Ca2+/CaM. (B) Ca2+/CaM titrated into NSCaTE peptide. (C) NSCaTE peptide titrated into Ca2+/CaM-NT. (D) NSCaTE peptide titrated into Ca2+/CaM-CT.
From previous in vivo studies, we know that three residues (W52, I56, and R60) are critical for the binding of Ca2+/CaM to the NSCaTE peptide in causing the CDI effect (Dick et al., 2008). W52 is especially important, as the W52A mutation completely abolishes the NSCaTE CDI function in vivo. Using site-directed mutagenesis, we also made W52A and I56A mutations in our long construct as well as an L64A mutation. Whereas the L64A mutation had no effect on the binding to CaM (as determined by ITC), the W52A mutation completely abolished and the I56A mutation decreased the Ca2+-CaM affinity (data not shown), thereby confirming the importance of these two residues for productive binding. As CaM has no Trp residues, the chemical environment of the indole-ring of the W52 residue of the NSCaTE peptide in the complex could be directly investigated by steady state fluorescence spectroscopy (Figure A3). Similar to other unstructured CaM-binding peptides containing tryptophan residues, the highest intensity of the broad fluorescence emission of the long peptide in aqueous solution occurred at approximately 355 nm, indicating a fully solvent-exposed Trp. As the peptide was saturated with the intact CaM or with the separate N- and C-lobes of CaM, a blue shift was observed for the highest emission peak at approximately 325 nm with an increase in the fluorescence intensity. This result is similar to those obtained upon the binding of CaM-binding domain peptides from MLCK, CaMKI, and CaMKII (Van Lierop et al., 2002; Yamniuk and Vogel, 2004). The 1:2 stoichiometry was also confirmed by fluorescence spectroscopy titration experiments. The maximum fluorescence emission intensity did not increase after one peptide was saturated with the N-lobe of CaM. However, the intensity slowly increased even after a 2.5-fold excess of C-lobe CaM bound to the peptide. The binding constants determined by this method were 1.39 × 106 M−1 for the N-lobe and 2.32 × 105 M−1 for the C-lobe, which were similar to the values determined by the ITC experiments.
Severe peak broadening in the NMR spectra prevented us from determination of the structure of the complex formed between the intact CaM and the NSCaTE peptide. We investigated different strategies in an attempt to overcome this problem. First, we tried to use the fast structure determination method that relies on the use of CaM's methionine-methyl NOEs (Gifford et al., 2011). However, because of the intermediate binding affinity, the methionine-CH3 group peaks involved in the binding interface was too broad to be confidently assigned. A similar situation has been encountered for the CaM-binding peptide of the N-terminal β-subunit of the olfactory rod CNG channel (Orsale et al., 2003). Because each lobe of CaM can bind one peptide, we superimposed the 1H–15N HSQC from the N- and C-lobe complexes onto the intact CaM complex (Figure 4B). The high degree of overlap indicated that the truncation induced only minor perturbations in the complex structure of each lobe. We first determined the solution structures of the protein lobes in the complex using regular NMR methods. To obtain unambiguous chemical shifts of the peptide in each lobe complex, the backbone and side-chain experiments were also run for the 13C/15N-labeled peptides in the complex. Compared with the peptide in aqueous solution (Figure 2A), the bound peptides had better dispersed peaks (Figures 4C,D). Secondary structure predictions were performed based on the CSI, and the helical conformation we observed for the unbound state of NSCaTE was maintained in both peptides in the complex. The peaks from the C-lobe CaM-bound peptide were broader than those for the N-lobe, which is consistent with the lower affinity of the C-lobe complex. The 13C/F3-filtered and 13C/F1-edited NOESY–HSQC exhibited only a few intermolecular NOEs for the C-lobe complex, much fewer than those obtained for the N-lobe complex. These differences prevented the direct determination of the C-lobe complex structure based on isotope-filtered NMR methods. On the other hand, the N-lobe complex could be defined in this fashion.
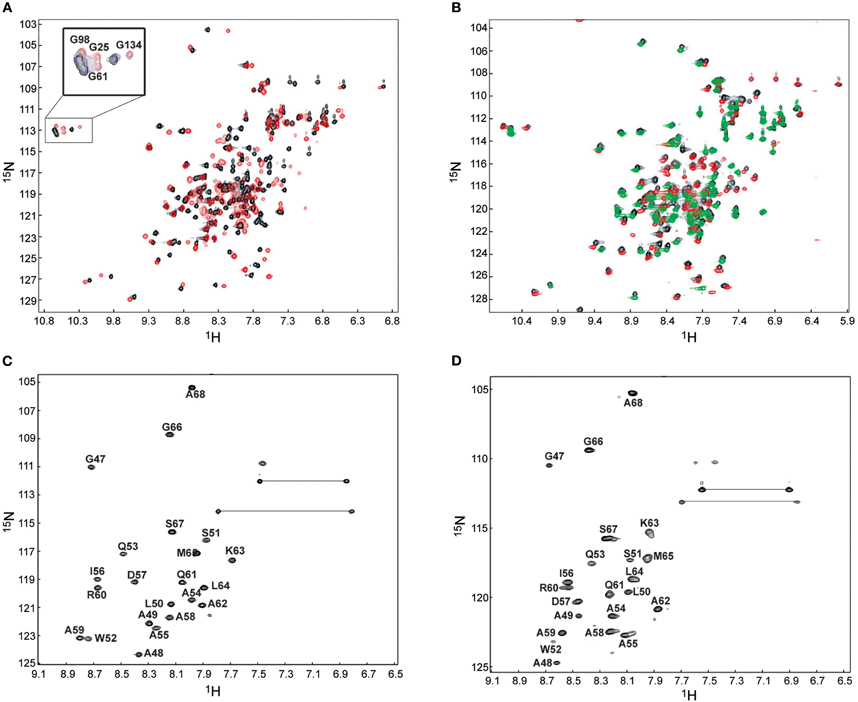
Figure 4. 1H–15N HSQC for the binding of Ca2+/CaM to the short NSCaTE peptide. (A) Ca2+/CaM is shown in black, and fully peptide saturated Ca2+/CaM is red. The conformational change as indicated by the peaks shifts of the four EF-hand Gly residue is highlighted in the top left corner. (B) Superposed HSQC spectra of intact Ca2+/CaM, Ca2+/CaM-NT (red) and Ca2+/CaM-CT (green) in the complex with the peptide. (C)1H–15N HSQC spectrum of the 13C/15N-labeled NSCaTE peptide saturated with Ca2+/CaM-NT. (D)1H–15N HSQC spectrum of the 13C/15N-labeled NSCaTE peptide saturated with Ca2+/CaM-CT.
We also made several comparisons between the isolated lobe and the intact CaM protein-peptide complex. From the backbone assignments for the intact Ca2+-CaM/NSCaTE complex, we calculated the weighted average secondary shifts (WASS) along the sequence (Figure A4A). Combined with the heteronuclear 1H–15N NOE data (Figure A4B), we confirmed through the WASS, as expected from the isolated structures, that the regular secondary structures of Ca2+/CaM are maintained in this complex, including the flexible N-/C-termini and the central linker. The residues of the loop region between the two EF-hands for each lobe also had lower NOE values, which indicated their higher flexibility. In the sequence, eight continuous segments had positive WASS values. These segments corresponded to the eight helices of the four EF-hands that are normally observed in CaM complexes.
The Ca2+/CaM N-lobe complex structure was determined with a routine NMR method. The backbone and side chain chemical shifts of both the CaM N-lobe and the NSCaTE peptide were assigned with various triple-resonance NMR experiments, and the intermolecular NOEs were acquired from the half-filtered NOESY–HSQC experiments. In total, more than 95% of the backbone and side chain heavy atoms and protons were assigned. Many of the intermolecular NOEs came from the N-lobe methionine-methyl or aromatic ring protons to the peptide protons. About half of the intermolecular NOEs in the half-filtered spectrum were confirmed and used in the final complex structure determination. To validate our structure calculation, we also generated a complex model with the protein–protein docking program HADDOCK 2.0. The structures determined for the Ca2+/CaM N-lobe and the peptide in the complex were used as the initial models. The hydrophobic pocket of the N-lobe (A15, L18, F19, L32, V35, M36, L39, M51, V55, F65, M71, and M72) was used as the N-lobe binding interface. The three residues of the peptide (W52, I56, and R60) that have been confirmed to be critical for CaM-binding were defined as the active peptide residues, and the remainder of the peptide residues was defined as passive residues. All of the residues involved in the binding surface were semi-flexible, which allows for the rotation of the side chain atoms. In total, 1000 structures were calculated with the HADDOCK web server (http://haddock.chem.uu.nl)(Dominguez et al., 2003), and the first 20 structures in the cluster with the lowest Haddock score were chosen as the representative complex ensemble. Because the intermolecular NOEs were not available for the C-lobe complex, this data-driven docking method was the only way to generate the complex structure. The Ca2+/CaM-CT structure in this complex was calculated first, and the hydrophobic surface was defined as the following residues: A88, V91, F92, L105, V108, M109, L112, M124, A128, F141, M144, and M145. The peptide structure from the N-lobe complex was used as the initial peptide structure, and the same three residues (W52, I56, and R60) were used as the active residues for the HADDOCK calculation. Both Ca2+-CaM NT/CT in the complex were further refined with the addition of backbone H–N RDC restraints [XPLOR-NIH, version 2.21 (Schwieters et al., 2003)].
Both complexes of the separate N- and C-lobes of CaM bound to the NSCaTE peptide are shown in Figure 5. For the N-lobe complex, the 20 lowest energy solution structures based on the NMR restraints are shown as an ensemble on the top left (Figure 5A). The backbone root mean-square deviation (RMSD) was 0.54 Å (For a summary of the NMR data see Table 2). As indicated in the 1H–15N heteronuclear NOE experiment, the loop region connecting the two EF-hands had a larger RMSD. Similar to most Ca2+/CaM-target peptide complexes, the N-lobe of CaM is composed of two EF-hands. Each EF-hand contains two helices connected by a calcium binding loop, and the two EF-hands were bridged by a short pair of anti-parallel β-strands from the two calcium binding loops (Gifford et al., 2007). The two helices in each EF-hand were perpendicular to each other and exposed hydrophobic residues, forming a continuous hydrophobic surface which can interact with the NSCaTE peptide. As shown using site-directed mutagenesis and fluorescence spectroscopy, W52 of the peptide is the most important residue for CaM binding. In Figure 6, we list the contact residues that are within 4 Å for the side chains of CaM and the peptide. As expected, W52 contacted the most residues of CaM. The total W52 side chain contact surface was 208.5 Å2, which was much larger than the 95.4 Å2 side chain contact surface of the other hydrophobic residue, I56. The total interacting interface between the NSCaTE peptide and CaM was 1520.7 Å2. The peptide and CaM-NT adopted more of an anti-parallel orientation in which the N-terminus of the peptide was close to the C-terminus of CaM-NT. As not all intermolecular NOEs could be confidently assigned (shown in Figure 6C), we compared the HADDOCK-generated structures to the NOE-based complex structures. When we overlaid the CaM region of the 20 Haddock complex structures, the peptide backbone structures were not as convergent (RMSD = 2.34 ± 1.08 Å) as we found for the NOE-based structure due to the relatively wide hydrophobic interacting interface on Ca2+-CaM N-lobe. Additionally, in the ribbon representation (Figure 6B), we observed a different arrangement of the W52 aromatic ring. However, the overall orientation of the peptide backbone relative to CaM-NT was not significantly different in the two methods. This consistency validated the NOE-based structure. Interestingly, the peptide complexed with the C-lobe adopted a parallel orientation. Compared with the CaM/GAD complex, which also exhibits 2:1 stoichiometry, the orientation between the C-lobe of CaM and the peptide was reversed. This difference may be a result of the position of the tryptophan residue in the peptide sequence.
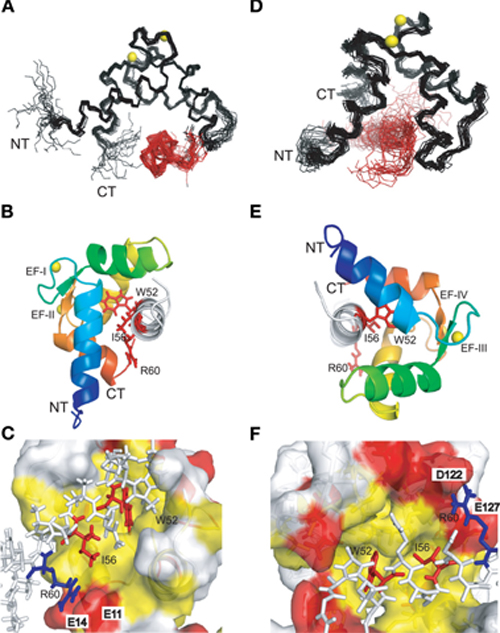
Figure 5. NMR solution structures of the complexes of the N-lobe (A, B, C) and the C-lobe (D, E, F) of Ca2+-CaM with the short NSCaTE peptide. (A, D) Backbone representation of the ensemble of the 20 lowest energy structures of the Ca2+/CaM-NT-NSCaTE complex (A, calculated by CYANA2.1) or the Ca2+/CaM-CT-NSCaTE complex (D, generated by HADDOCK). The CaM lobes are shown in black, and the peptide bundles are red. The calcium ions in the EF-hand loop are yellow. The peptide is shown in white, and the three critical residues (W52, I56, and R60) are highlighted in red. (B, E) Ribbon representations of the lowest energy structures for the Ca2+/CaM NT (B) and CT (E) complexes. The four helices in each lobe follow the default PyMOL rainbow color setting (blue–green–yellow–orange) from the N-terminus to the C-terminus. (C, F) Binding surface representations of the Ca2+/CaM NT (C) and CT (F) complexes. The binding pocket of each lobe is highlighted in yellow (hydrophobic residues) and red (negatively charged residues). The NSCaTE peptide is shown in a stick representation (white), and the two critical hydrophobic residues (W52 and I56, red) and the positively charged residue (R60, blue) are highlighted.
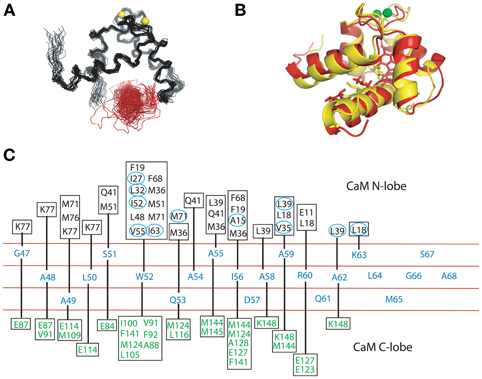
Figure 6. (A). Structure ensemble of the complex of the Ca2+/CaM NT and the NSCaTE motif (generated by HADDOCK 2.0). The colors used here are similar to those in Figure 4A. (B) Ribbon representation of the overlap of the calculated (red) and HADDOCK-generated (yellow) structures. (C) Contact residues between the CaM lobe (NT, calculated by CYANA 2.1, or CT, generated by HADDOCK 2.0) and the NSCaTE peptide (<4 Å). The peptide is shown in the middle, and each residue is listed along the sequence. The contact residues from the N-lobe of CaM are listed above the peptide diagram. The residues with the actual NOE restraints used in the calculations are highlighted with blue circles. The contact residues from the C-lobe of CaM are listed below the peptide diagram.
The structure of the C-lobe complex structure generated by HADDOCK had a much higher peptide backbone RMSD (Figure 5D) than the N-lobe structure (Figure 5A). While the backbone of the Ca2+-CaM C-lobe in the complex is well defined, the position of the peptide is not well defined, which gives rise to the relatively high numbers. In addition, the area near the anchoring residue W52 had low RMSDs, while the C-terminal part of the peptide had diverse orientations. This finding was consistent with the lower peptide affinity observed for the CaM C-lobe. To better understand the preferential peptide binding of the CaM NT, we compared the two complex structures. The average total binding interface area for the C-lobe complex was 1440.0 Å, which is smaller than the interface area we found for the N-lobe. The buried surface for W52 in this complex was 194.4 Å, which was also somewhat smaller than the corresponding value in the N-lobe. However, the contribution from the sidechain of the I56 residue was similar to that in the N-lobe. In both complexes, the tip of the sidechain of the R60 was in close proximity to negatively charged residues (E11 of the N-lobe and E123/127 of the C-lobe). Possible salt bridges between these residues could provide extra anchoring contacts.
It is always interesting to compare different CaM-binding peptides, as many different complex structures have been found (Ishida and Vogel, 2006). The NSCaTE is the first peptide known to bind preferentially to the Ca2+/CaM N-lobe. The Nicotianatabacum MAPK phosphatase (NtMKP1) peptide from N. tabacum MAPK-phosphatase is one of the few naturally occurring CaM-binding peptides that can bind only to the Ca2+/CaM C-lobe (Ishida et al., 2009). However, although both the NSCaTE and NtMKP1 peptides could bind to the CaM C-lobe, the NtMKP1 peptide bound much more tightly and in an opposite orientation than the NSCaTE peptide (Figure 7E). By comparing the sequences of both peptides, we detected many more positively charged residues at the N-terminus of the NtMKP1 peptide, along with two hydrophobic “double-anchoring” residues (W3620 and L3623) (Ishida et al., 2009). There were no positive charges close to the main anchoring residue, W52, of the NSCaTE peptide. The positively charged residue R60 of NSCaTE was close to the negatively charged residue E123 on the third helix of the CaM C-lobe (Figure 7C). Similar to the 2:1 stoichiometry of the NSCaTE peptide to Ca2+/CaM that we found in this study, two carboxy-terminal peptides of plant glutamate decarboxylase (GAD) could interact with the two lobes of Ca2+/CaM simultaneously (Yap et al., 2003). These two GAD peptides were anti-parallel to each other and formed an X shape in the complex, which could not be identified in our complex. As noted in the paper by Yap et al., the high percentage of negative charges in the GAD peptide could pair with the basic residues, and the electrostatic interactions between charged residues were critical for creating the dimer interface. This result was confirmed by examining the potential surface of the GAD peptide dimer in the complex (Figure 7D). The N/C terminal positively charged residues of one peptide could approach the C/N terminal negatively charged residues of the second GAD peptide in the bound butterfly-shaped peptide dimer. A continuous positively charged patch was formed on each side of the peptide dimer and facilitated the binding with each CaM lobe. However, we found only one positively charged residue (R60) and one negatively charged residue (D57) in the NSCaTE peptide, which would not provide enough electrostatic interactions for a peptide dimer similar to GAD in the complex of CaM with NSCaTE. The lack of multiple anchoring hydrophobic and positively charged residues combined with the short length is the main reasons for the unique binding character of the NSCaTE peptide.
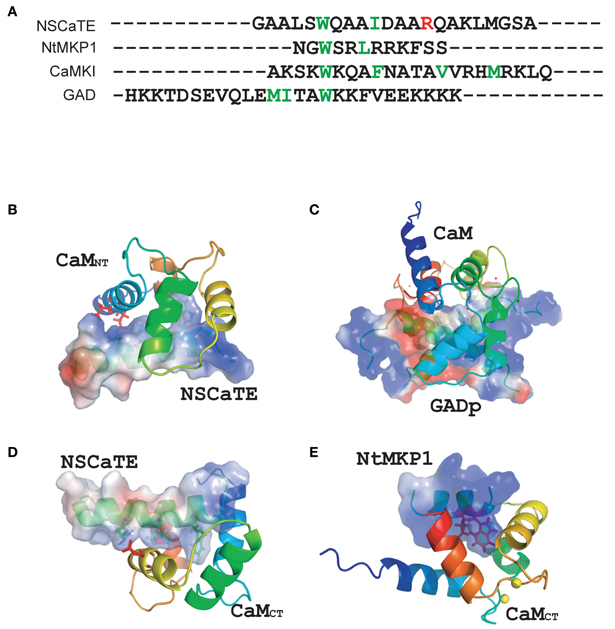
Figure 7. (A) Sequence comparisons of different calmodulin-binding motifs (NSCaTE, NtMKP1, CaMKI, and GAD). (B) Complex structure representations of the NSCaTE peptide and Ca2+-CaM NT. (C) Complex structure representations of the NSCaTE peptide and Ca2+-CaM CT. (D) Complex structure representations of the GAD peptide and Ca2+-CaM (PDB ID: 1NWD). (E) Complex structure representations of the NtMKP1 peptide and Ca2+-CaM CT (PDB ID: 2KN2).
As one of the main sources for calcium influx in a cell, voltage-gated calcium channels play a variety of important roles in neuron transduction, in smooth and cardiac muscle contraction and in gene transcription. Each type of calcium channel has its own build-in control mechanisms, depending on the presence of various additional channel subunits (Cavβ) (McGee et al., 2004; Takahashi et al., 2005; Grueter et al., 2006), regulatory calcium binding proteins and other molecules (Peterson et al., 1999; DeMaria et al., 2001; Fallon et al., 2005). Among the four types of voltage-gated calcium channel (L-, P/Q-, N-, and R-types), the NSCaTE motif was found only in two subtypes (Cav1.2 and Cav1.3) of the L-type VGCCs, which is clearly evident from the alignment of the N-terminal sequences of the four L-type channels (Figure 1B). The C-terminal portion of the N-terminal cytoplasmic domains of the L-type channels has nearly identical sequences, but their N-terminal ends are very different. Each of these four types of channels has a unique and preferred expression pattern. Cav1.1 is mainly expressed in skeletal muscle, and its full function relies on ryanodine receptors. Cav1.4 is mainly located in neurons, especially in the retina. Both Cav1.1 and 1.4 have a relatively weak CDI and play important functions in the dark-adapted light sense (Zhou et al., 2004). Compared with Cav1.1/1.4, Cav1.2 and 1.3 exhibit much stronger CDI, consistent with their functions in cardiac muscle-related EC coupling, which requires rapid and regular muscle contraction and relaxation. Recently, the importance of Cav1 L-type channels in the nervous system has been summarized by Calin-Jageman and Lee (2008). Cav1 channels are widely expressed in pre- and post-synaptic region of neuron in the cerebral cortex, hippocampus and retina. Through the regulation of the neuronal Ca2+ signal, Cav1 channels can trigger electrical and signaling cascades of various activities, such as synaptic plasticity, gene expression, and long-term memory. CDI and CDF are the main mechanisms of the VGCCs-related regulation. It has been shown that the binding of the C-terminal IQ-motif to CaM provides the main basis for the self-control of the VGCC through the mechanisms of CDI and CDF. The CDI and CDF effects, however, do not only depend on the sequences and CaM-binding patterns of the IQ-motif in the C-terminal cytoplasmic region of the channels. These processes are also finely tuned by other interaction partners, including the amino terminal cytoplasmic region of the channel. The whole N-terminal sequence was re-examined by the Lee group (Oz et al., 2011), and they found that the initial sequence is critical for the voltage-dependent inactivation (VDI) kinetics of both long and short isoforms. Interestingly, they also identified a new motif at the C-terminal end of N-terminal cytoplasmic domain of the channel. This new motif cannot bind CaM, but it can bind to CaBP1 in either the calcium- or apo-form in vivo. Considering the high conservation of this motif in L-type channels, a consistent function for this region can be expected.
In our in vitro experiments with NSCaTE, a 1:2 stoichiometric ratio was identified with various techniques. However, CaM is considered as an associated channel subunit currently in most research, with a stoichiometry of 1:1. Although channel dimerization has also been observed, more evidence is still needed to support such a model. If the model presented in this study is correct, this unique 1:2 CaM to NSCaTE peptide ratio could affect channel dimerization in the presence of exogenousCaM. In addition, we found that the NSCaTE motif spontaneously adopted a helical conformation in aqueous solution, which was observed by both NMR and CD spectroscopy. This conformation is different from the conformation observed in most CaM-binding domain peptides, where a transition from the random coil of the unbound state to the helical conformation of the bound state is required for most peptides. The high alanine content of NSCaTE is a likely reason for the naturally occurring helical conformation. Considering the relative weak-binding affinity of this motif toCaM, the presence of the preformed helical conformation is consistent with its role as a minor CDI regulator in special channels, as smaller entropy–enthalpy changes would occur due to the lack of a drastic conformational change.
Another interesting property of the NSCaTE motif is that the motif bound more tightly to the Ca2+-CaM N-lobe than to the C-terminal lobe of Ca2+-CaM. This binding preference is different from that of most CaM-binding peptides, especially when compared with the IQ-motif from the C-terminal cytoplasmic domain of the same calcium channel. From the crystal structure of the CaM/Cav1.2 IQ-motif complex (Fallon et al., 2005; Van Petegem et al., 2005), we already have a deeper understanding of the fundamental channel self-control mechanism in L-type channels, as apo-CaM can also bind to the IQ region, and thereby it can provide a quick response to the elevation of the local calcium ion concentrations close to the channel pore. Once bound to calcium, CaM binds to the IQ-motif in an unique parallel mode due to the higher positive charge in the C-terminal half of the IQ-peptide. This special binding mode allows the Ca2+-CaM C-lobe to dominate the regulation of CDI. A single mutation (IQ/AQ) or double mutation (IQ/AA) will abolish CDI and alleviate an apparent CDF that is severely blocked in the wild-type protein. Van Petegem et al. (2005) proposed a possible in vivo model where the Ca2+-CaM C-lobe is anchored to the C-terminal IQ-motif, but other regions of the channel can compete for the N-lobe of Ca2+-CaM and finely tune the CDI dominated by the C-terminal IQ-motif. This model provides a reasonable explanation for the real function of NSCaTE. Although a trial pull-down assay failed to provide in vitro evidence for this model, in vivo FRET experiments revealed a FRET ratio (FR) larger than one and a meaningful dissociation constant of approximately 20–30 uM in both chimeric and wild-type constructs (Dick et al., 2008). The removal of the critical tryptophan residues of NSCaTE will abolish any interactions between the N-/C-terminal regions bridged by CaM and the corresponding CDI. The mutations of the other two important residues within the motif (Ile and Arg) did modulate its regulatory action by weaker binding and attenuated CDI. The correlation between the NSCaTE motif affinity to CaM and the CDI effect has been systematically investigated by alanine scanning along the motif sequence, and the concept of the local Ca2+ selectivity for this motif has been strengthened in the work done by Tadross et al. (2008).
Other calcium binding proteins, known as CaBPs, are often coexpressed with VGCCs in addition to CaM in neurons, especially those in the presynaptic density. One isoform, CaBP1, can alleviate the CDI caused by CaM and induce CDF. Zhou et al. demonstrated that the N-terminus of Cav1.2 is critical for CaBP1 function and that the removal of the N-terminus will completely abolish the effect of CaBP1 (Zhou et al., 2004, 2005). In addition, another sequence, which does not overlap with the N-terminus of α1c, has been identified as a CaBP1-specific binding motif. However, this identification cannot completely exclude the possibility of an interaction between CaBP1 and the NSCaTE region, due to the high amino acid sequence similarity between CaM and the C-terminal lobe of CaBP1. We, therefore, also investigated the binding of CaBP1 to the NSCaTE region (See Figure A5). Our data indicate that the calcium-bound CaBP1 interacts only very weakly with the NSCaTE motif through its C-lobe. A recently solved CaBP1 X-ray structure (Findeisen and Minor, 2010) can help us understand the possible role of NSCaTE in the complicated CDI regulatory network involving CaM, CaBP1, IQ, and the N-terminus (containing NTI, NSCaTE, and the CaBP1 binding motif) of Cav1.2 and Cav1.3. Extensive mutation and domain swap studies have shown that the N-lobe and residue E94 in the central linker are critical for CaBP1-inhibited CDI and induced CDF. However, we also notice that Ca2+-CaBP1 can bind to the IQ-motif, and unlike CaM, the binding site is the same for the N- and C-lobes of CaBP1. In addition, the C-lobe of Ca2+-CaBP1 cannot compete with the C-lobe of Ca2+-CaM for the IQ motif binding, and mutating either or both EF-hands of C-lobe CaBP1 will partially recover inhibited CDI (Findeisen and Minor, 2010).
In conclusion, although the interactions between the IQ-motif in the C-terminal cytoplasmic region of the channels and CaM seem to dominate the regulation of the voltage-gated calcium channel, other parts of the channels and channel-binding proteins can further fine tune each channel, to allow them to carry out their unique function in various cells and tissues. A simplified model is presented in Figure 8 to describe this complicated regulatory network for Cav1.2 and Cav1.3 at different calcium binding stages. In the resting state, apo-CaM is bound near the IQ-motif region, and apo-CaBP1 interacts with its N-terminal binding motif. Once the channel opens due to a change in the membrane potential, the calcium flows into the cytoplasm and initially saturates CaM and CaBP1 that are located near the channel pore. Ca2+-CaM then binds to the IQ-motif with a parallel domain arrangement and induces CDI/CDF. The N-lobe of Ca2+-CaM can also bind to the NSCaTE motif and accelerate the closing of the channel. In doing so, Ca2+-CaM acts almost like an “adaptor” protein bridging between the two cytoplasmic domains of the channel [for discussions see Yamniuk et al. (2007)]. Simultaneously, the N-lobe of CaBP1 continues to interact with its motif in the N-terminal, while its C-lobe adopts an open conformation. The C-lobe of Ca2+-CaBP1 can also interact with NSCaTE and compete weakly with the N-lobe of Ca2+-CaM, which in turn may inhibit the acceleration of CDI. However, this simplified model does not include other regulatory partners, such as the I–II loop of the α1c subunit of the channel, the Cavβ-subunit and regulation through CaMKII. More binding studies and structural details, therefore, are needed to deconstruct this complicated regulatory network.
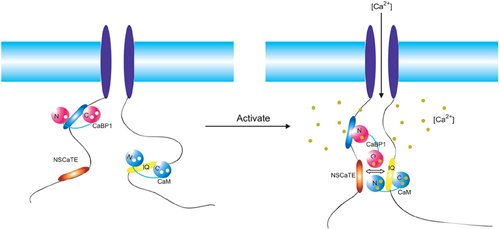
Figure 8. Overview of the regulatory network of the Cav1.2 (α1c). Two motifs (NSCaTE, Ca2+/CaM specific, red; CaBP1 binding motif, blue) in the N-terminus and the IQ motif (yellow) in the C-terminus are represented by oval bars. The regulatory calcium binding proteins CaBP1 and CaM are shown in red and blue, respectively. These proteins are bound to the N-terminus of the channel and the IQ motif in the resting state. Upon membrane depolarization and channel opening, both CaM and CaBP1 bind calcium ions and interact with different motifs. The cross-interference among these binding partners is critical for the distinct regulatory functions of each type of channel.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This project was funded by an operating grant from the Canadian Institutes of Health Research. Hans J. Vogel is the holder of a Scientist award from the Alberta Heritage Foundation for Medical Research.
CaM, calmodulin; CaBP1, calcium binding protein 1; NSCaTE, N-terminal spatial Ca2+ transforming element; VGCCs, voltage-gated calcium channels; SCaM4, soybean calmodulin isoform 4; SCaM4-CT, the C-lobe of soybean calmodulin isoform 4; MAPK, mitogen-activated protein kinase; NtMKP1, N. tabacum MAPK phosphatase; GAD, plant glutamate decarboxylase; SUMO, small ubiquitin-like modifier protein; RMSD, root mean-square deviation; ITC, isothermal titration calorimetry; NMR, nuclear magnetic resonance; CD, dircular dichroism; CDI, calcium-dependent inactivation; CDF, calcium-dependent facilitation; VDI, voltage-dependent inactivation; HADDOCK, high ambiguity-driven biomolecular docking.
Andre, I., and Linse, S. (2002). Measurement of Ca2+-binding constants of proteins and presentation of the Ca Ligator software. Anal. Biochem. 305, 195–205.
Benmocha, A., Almagor, L., Oz, S., Hirsch, J. A., and Dascal, N. (2009). Characterization of the calmodulin-binding site in the N terminus of Cav1.2. Channels (Austin) 3, 337–342.
Buraei, Z., and Yang, J. (2010). The β subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506.
Calin-Jageman, I., and Lee, A. (2008). Cav1 L-type Ca2+channel signaling complexes in neurons. J. Neurochem. 105, 573–583.
Cornilescu, G., Delaglio, F., and Bax, A. (1999). Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302.
DeMaria, C. D., Soong, T. W., Alseikhan, B. A., Alvania, R. S., and Yue, D. T. (2001). Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature 411, 484–489.
Dick, I. E., Tadross, M. R., Liang, H., Tay, L. H., Yang, W., and Yue, D. T. (2008). A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of Cav channels. Nature 451, 830–834.
Dominguez, C., Boelens, R., and Bonvin, A. M. J. J. (2003). HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737.
Fallon, J. L., Halling, D. B., Hamilton, S. L., and Quiocho, F. A. (2005). Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Cav1.2 calcium channel. Structure 13, 1881–1886.
Findeisen, F., and Minor, D. L. Jr. (2010). Structural basis for the differential effects of CaBP1 and calmodulin on Cav1.2 calcium-dependent inactivation. Structure 18, 1617–1631.
Gifford, J. L., Ishida, H., and Vogel, H. J. (2011). Fast methionine-based solution structure determination of calcium calmodulin complexes. J. Biomol. NMR 50, 71–81.
Gifford, J. L., Walsh, M. P., and Vogel, H. J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405, 199–221.
Grueter, C. E., Abiria, S. A., Dzhura, I., Wu, Y., Ham, A. J., Mohler, P. J., Anderson, M. E., and Colbran, R. J. (2006). L-type Ca2+ channel facilitation mediated by phosphorylation of the beta subunit by CaMKII. Mol. Cell 23, 641–650.
Guntert, P. (2004). Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378.
Houdusse, A., Gaucher, J. F., Krementsova, E., Mui, S., Trybus, K. M., and Cohen, C. (2006). Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci. U.S.A. 103, 19326–19331.
Hudmon, A., Schulman, H., Kim, J., Maltez, J. M., Tsien, R. W., and Pitt, G. S. (2005). CaMKII tethers to L-type Ca2+ channels, establishing a local and dedicated integrator of Ca2+ signals for facilitation. J. Cell Biol. 171, 537–547.
Ishida, H., Rainaldi, M., and Vogel, H. J. (2009). Structural studies of soybean calmodulin isoform 4 bound to the calmodulin-binding domain of tobacco mitogen-activated protein kinase phosphatase-1 provide insights into a sequential target binding mode. J. Biol. Chem. 284, 28292–28305.
Ishida, H., and Vogel, H. J. (2006). Protein-peptide interaction studies demonstrate the versatility of calmodulin target protein binding. Protein Pept. Lett. 13, 455–465.
Kim, E. Y., Rumpf, C. H., Fujiwara, Y., Cooley, E. S., Van Petegem, F., and Minor, D. L. Jr. (2008). Structures of Cav2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure 16, 1455–1467.
Kim, J., Ghosh, S., Nunziato, D. A., and Pitt, G. S. (2004). Identification of the components controlling inactivation of voltage-gated Ca2+ channels. Neuron 41, 745–754.
McGee, A. W., Nunziato, D. A., Maltez, J. M., Prehoda, K. E., Pitt, G. S., and Bredt, D. S. (2004). Calcium channel function regulated by the SH3-GK module in beta subunits. Neuron 42, 89–99.
Mori, M. X., Vander Kooi, C. W., Leahy, D. J., and Yue, D. T. (2008). Crystal structure of the Cav2 IQ domain in complex with Ca2+/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2+. Structure 16, 607–620.
Orsale, M., Melino, S., Contessa, G. M., Torre, V., Andreotti, G., Motta, A., Paci, M., Desideri, A., and Cicero, D. O. (2003). Two distinct calcium-calmodulin interactions with N-terminal regions of the olfactory and rod cyclic nucleotide-gated channels characterized by NMR spectroscopy. FEBS Lett. 548, 11–16.
Oz, S., Tsemakhovich, V., Christel, C. J., Lee, A., and Dascal, N. (2011). CaBP1 regulates voltage-dependent inactivation and activation of Cav1.2 (L-type) calcium channels. J. Biol. Chem. 286, 13945–13953.
Pace, C. N., and Scholtz, J. M. (1998). A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75, 422–427.
Panavas, T., Sanders, C., and Butt, T. R. (2009). SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 497, 303–317.
Peterson, B. Z., DeMaria, C. D., Adelman, J. P., and Yue, D. T. (1999). Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron 22, 549–558.
Pitt, G. S., Zuhlke, R. D., Hudmon, A., Schulman, H., Reuter, H., and Tsien, R. W. (2001). Molecular basis of calmodulin tethering and Ca2+-dependent inactivation of L-type Ca2+ channels. J. Biol. Chem. 276, 30794–30802.
Qin, N., Olcese, R., Bransby, M., Lin, T., and Birnbaumer, L. (1999). Ca2+-induced inhibition of the cardiac Ca2+ channel depends on calmodulin. Proc. Natl. Acad. Sci. U.S.A. 96, 2435–2438.
Queiroz, J. A., Tomaz, C. T., and Cabral, J. M. (2001). Hydrophobic interaction chromatography of proteins. J. Biotechnol. 87, 143–159.
Rainaldi, M., Yamniuk, A. P., Murase, T., and Vogel, H. J. (2007). Calcium-dependent and -independent binding of soybean calmodulin isoforms to the calmodulin binding domain of tobacco MAPK phosphatase-1. J. Biol. Chem. 282, 6031–6042.
Schwieters, C. D., Kuszewski, J. J., Tjandra, N., and Clore, G. M. (2003). The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73.
Tadross, M. R., Dick, I. E., and Yue, D. T. (2008). Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell 133, 1228–1240.
Takahashi, S. X., Miriyala, J., Tay, L. H., Yue, D. T., and Colecraft, H. M. (2005). A Cavβ SH3/guanylate kinase domain interaction regulates multiple properties of voltage-gated Ca2+ channels. J. Gen. Physiol. 126, 365–377.
Thulin, E., Andersson, A., Drakenberg, T., Forsen, S., and Vogel, H. J. (1984). Metal ion and drug binding to proteolytic fragments of calmodulin: proteolytic, cadmium-113, and proton nuclear magnetic resonance studies. Biochemistry 23, 1862–1870.
Van Lierop, J. E., Wilson, D. P., Davis, J. P., Tikunova, S., Sutherland, C., Walsh, M. P., and Johnson, J. D. (2002). Activation of smooth muscle myosin light chain kinase by calmodulin. Role of LYS30 and GLY40. J. Biol. Chem. 277, 6550–6558.
Van Petegem, F., Chatelain, F. C., and Minor, D. L. Jr. (2005). Insights into voltage-gated calcium channel regulation from the structure of the Cav1.2 IQ domain-Ca2+/calmodulin complex. Nat. Struct. Mol. Biol. 12, 1108–1115.
Yamniuk, A. P., Rainaldi, M., and Vogel, H. J. (2007). Calmodulin has the potential to function as a Ca-dependent adaptor protein. Plant Signal. Behav. 2, 354–357.
Yamniuk, A. P., and Vogel, H. J. (2004). Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33–57.
Yap, K. L., Yuan, T., Mal, T. K., Vogel, H. J., and Ikura, M. (2003). Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J. Mol. Biol. 328, 193–204.
Yuan, W., and Bers, D. M. (1994). Ca-dependent facilitation of cardiac Ca current is due to Ca-calmodulin-dependent protein kinase. Am. J. Physiol. 267, H982–H993.
Yuan, T., and Vogel, H. J. (1998). Calcium-calmodulin-induced dimerization of the carboxyl-terminal domain from petunia glutamate decarboxylase: a novel calmodulin-peptide interaction motif. J. Biol. Chem. 273, 30328–30335.
Zhang, M., Yuan, T., and Vogel, H. J. (1993). A peptide analog of the calmodulin-binding domain of myosin light chain kinase adopts an α-helical structure in aqueous trifluoroethanol. Protein Sci. 2, 1931–1937.
Zhou, H., Kim, S. A., Kirk, E. A., Tippens, A. L., Sun, H., Haeseleer, F., and Lee, A. (2004). Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Cav1.2 (L-type) Ca2+ channels. J. Neurosci. 24, 4698–4708.
Zhou, H., Yu, K., McCoy, K. L., and Lee, A. (2005). Molecular mechanism for divergent regulation of Cav1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J. Biol. Chem. 280, 29612–29619.
Zuhlke, R. D., Pitt, G. S., Deisseroth, K., Tsien, R. W., and Reuter, H. (1999). Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature 399, 159–162.
Zuhlke, R. D., Pitt, G. S., Tsien, R. W., and Reuter, H. (2000). Ca2+-sensitive inactivation and facilitation of L-type Ca2+ channels both depend on specific amino acid residues in a consensus calmodulin-binding motif in the α1c subunit. J. Biol. Chem. 275, 21121–21129.
Zuhlke, R. D., and Reuter, H. (1998). Ca2+-sensitive inactivation of L-type Ca2+ channels depends on multiple cytoplasmic amino acid sequences of the α1c subunit. Proc. Natl. Acad. Sci. U.S.A. 95, 3287–3294.
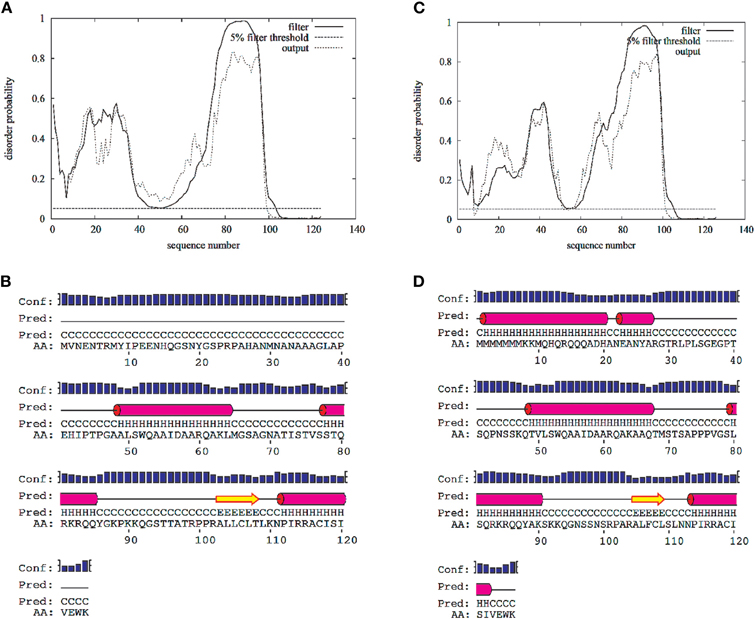
Figure A1. Disorder and secondary structure predictions for the N-terminal cytoplasmic domain of the L-type voltage-gated calcium channel Cav1.2 (1–124 AAs)(A,B) and Cav1.3 (1–126) (C,D). Predictions were calculated with the web-server DISOPRED2 (http://bioinf.cs.ucl.ac.uk/disopred/) and PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred).
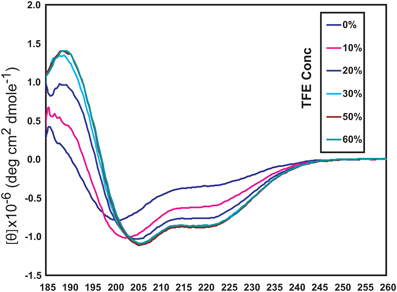
Figure A2. Circular dichroism (CD) spectroscopy of the long NSCaTE motif in aqueous solution with various concentrations of TFE (trifluoroethanol).
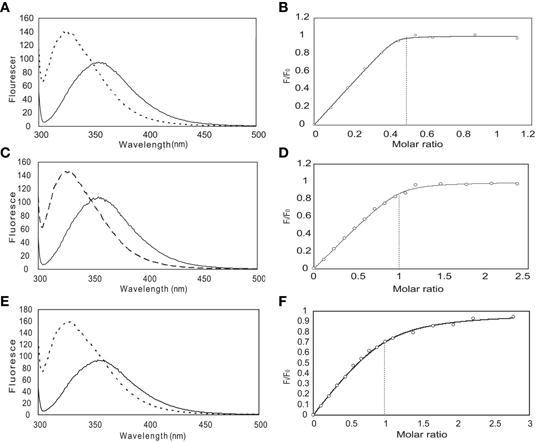
Figure A3. Steady-state fluorescence spectroscopy (left) and binding curves (right) obtained for the long NSCaTE peptide with CaM. (In panels A, C and E, the solid line is recorded without protein and the dotted line is after addition of protein). (A, B) Intact CaM, (C, D) N-lobe of CaM, (E, F) C-lobe of CaM.
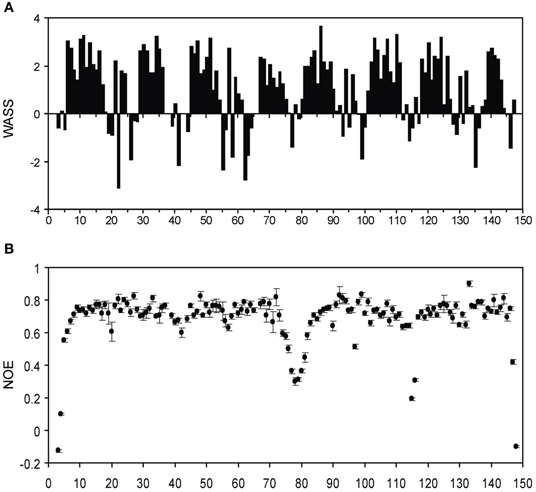
Figure A4. (A) Weighted average secondary shifts (WASS) for the short NSCaTE bound state of intact Ca2+/CaM. A total of 2.5 equivalents of unlabeled peptide was added to saturate the protein. (B) {1H}−15N NOE data for the short NSCaTE bound state of Ca2+/CaM.
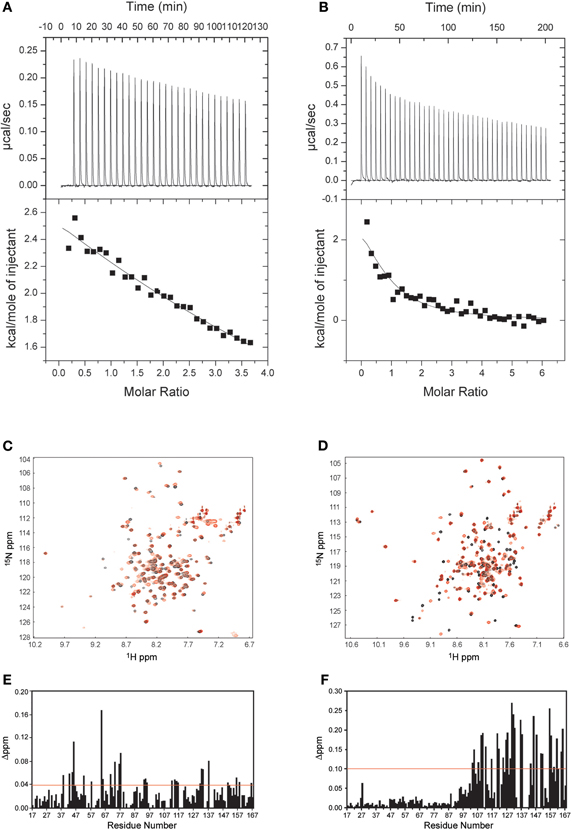
Figure A5. Interactions between CaBP1 and the long NSCaTE motif. (A,C,E) Apo-CaBP1 with NSCaTE peptide: ITC (A), 1H–15N HSQC (C) and chemical shift perturbation (E). (B,D,F) Ca2+/CaBP1 with NSCaTE peptide: ITC (B), 1H–15N HSQC (D) and chemical shift perturbation (F). Please note that compared to Ca2+-CaBP1, the binding of NSCaTE to apo-CaBP1 was extremely weak as indicated by the ITC and NMR data. The ITC data show that both binding events are exothermic.
Keywords: calmodulin, NSCaTE, L-type voltage-gated calcium channel, calcium-dependent inactivation, NMR
Citation: Liu Z and Vogel HJ (2012) Structural basis for the regulation of L-type voltage-gated calcium channels: interactions between the N-terminal cytoplasmic domain and Ca2+-calmodulin. Front. Mol. Neurosci. 5:38. doi: 10.3389/fnmol.2012.00038
Received: 01 February 2012; Paper pending published: 19 February 2012;
Accepted: 13 March 2012; Published online: 12 April 2012.
Edited by:
Michael R. Kreutz, Leibniz-Institute for Neurobiology, GermanyReviewed by:
Karl-Wilhelm Koch, Carl von Ossietzky University Oldenburg, GermanyCopyright: © 2012 Liu and Vogel. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hans J. Vogel, Biochemistry Research Group, Department of Biological Sciences, University of Calgary, 2500 University Drive N.W., Calgary, AB T2N 1N4, Canada. e-mail:dm9nZWxAdWNhbGdhcnkuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.