- 1 Division of Medical Pharmacology, Leiden/Amsterdam Center for Drug Research, Leiden, Netherlands
- 2 Department of Human Genetics, Leiden University Medical Center, Leiden, Netherlands
A limiting factor in brain research still is the difficulty to evaluate in vivo the role of the increasing number of proteins implicated in neuronal processes. We discuss here the potential of antisense-mediated RNA targeting approaches. We mainly focus on those that manipulate splicing (exon skipping and exon inclusion), but will also briefly discuss mRNA targeting. Classic knockdown of expression by mRNA targeting is only one possible application of antisense oligonucleotides (AON) in the control of gene function. Exon skipping and inclusion are based on the interference of AONs with splicing of pre-mRNAs. These are powerful, specific and particularly versatile techniques, which can be used to circumvent pathogenic mutations, shift splice variant expression, knock down proteins, or to create molecular models using in-frame deletions. Pre-mRNA targeting is currently used both as a research tool, e.g., in models for motor neuron disease, and in clinical trials for Duchenne muscular dystrophy and amyotrophic lateral sclerosis. AONs are particularly promising in relation to brain research, as the modified AONs are taken up extremely fast in neurons and glial cells with a long residence, and without the need for viral vectors or other delivery tools, once inside the blood brain barrier. In this review we cover (1). The principles of antisense-mediated techniques, chemistry, and efficacy. (2) The pros and cons of AON approaches in the brain compared to other techniques of interfering with gene function, such as transgenesis and short hairpin RNAs, in terms of specificity of the manipulation, spatial, and temporal control over gene expression, toxicity, and delivery issues. (3) The potential applications for Neuroscience. We conclude that there is good evidence from animal studies that the central nervous system can be successfully targeted, but the potential of the diverse AON-based approaches appears to be under-recognized.
Introduction
In mammals the brain orchestrates a variety of different processes ranging from maintaining homeostasis to complex behavioral outputs. Abnormalities in brain functioning may immediately affect survival or have major consequences for health and general functioning of the individual. A limiting factor in brain research still is the difficulty to evaluate the role of the increasing number of proteins implicated in neuronal processes in vivo. More often than not, specific receptor ligands or inhibitors are lacking, targets are “un-druggable,” and transgenic approaches costly, time-consuming, and/or dependent on viral delivery. Here, we will review the potential of antisense-mediated approaches targeting RNA, an area that has gone through considerable development over the last years. We will mainly focus on those applications aimed at manipulation of splicing (exon skipping and exon inclusion), but will also discuss mRNA targeting or classical knockdown. Most applications of antisense-mediated manipulation have been directly inspired by splice events that are relevant for a number of human genetic diseases, but the approach may well be incorporated in the general molecular toolkit we have available for tackling current neuroscientific questions in vivo.
AON Mechanisms
Antisense oligonucleotides (AONs) are small pieces of modified RNA or DNA that can hybridize to RNA. In this manner they can generate different effects depending on the AON chemistry and target site (see Figure 1; Table 1). Initially, AONs were used to induce gene knockdown (Dias and Stein, 2002). This can be achieved through RNase H, a ubiquitous enzyme that cleaves RNA:RNA or RNA:DNA hybrids (Figure 1A). The AONs used for this application are generally modified with a phosphorothioate backbone, which increases AON stability and enhances uptake of the AON over cell membranes. Gene knockdown can also be achieved using AONs targeting the translation start site (translation block, Figure 1B). Here, AONs can be modified further to render them RNase H-resistant by addition of a methyl or methoxy-ethyl group to the 2′O sugar ribose, which is the target cleavage site of the RNase H enzyme. Alternatively, nucleotides have been modified even further, e.g., using phosphorodiamidate morpholino oligomers (PMOs), peptide nucleic acids or locked nucleic acids. PMOs have been used for developmental studies in zebrafish embryos (Nasevicius and Ekker, 2000; Nasevicius et al., 2000). Multiple RNase H-dependent AONs are in clinical trials including one against high-grade glioma in phase IIb (trabedersen (Bogdahn et al., 2011), and one has even been registered as a drug for cytomegalovirus induced retinitis (vitravene; Marwick, 1998).
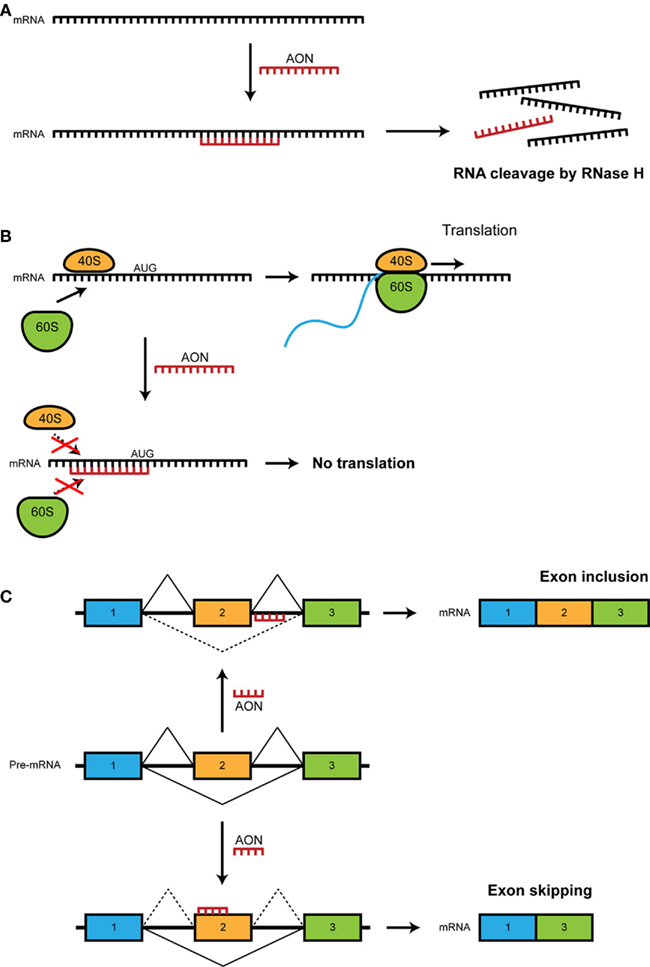
Figure 1. Schematic representation of different modes of action of antisense oligonucleotides. (A) RNase H-dependent pathway. Binding of antisense oligodeoxyribonucleotides (AONs) with a phosphorothioate backbone results in a RNA:DNA hybrid, which activates RNase H. RNase H will cleave the mRNA and prevents the translation into a protein. (B) RNase-independent translational block. 2′OH modified RNase H-resistant oligomers targeting the translation start site prevent translation and elongation. AONs binding to the AUG initiation site or downstream prevents binding of the ribosomal units or results in steric blockage. (C) Alternative splicing. 2′OH modified RNase H-resistant or alternatively modified AONs complementary to the target pre-mRNA can result in: (1) inclusion of an exon by binding to the exonic splicing silencers (ESEs) or intronic splicing silencers (ISSs). (2) exclusion of an exon by binding to the 3′ or 5′ slice sites or exon-internal sequences, resulting in an in-frame transcript and translation of a shorter partly functional protein. Full lines indicate possible splicing events while dashed lines indicate non-possible events.
However, with the availability of shRNA and siRNA, which generally gives a more robust gene knockdown (or complete knockout when cre-recombinase systems are used), the use of AONs is often not the method of first choice to achieve knockdown (in spite of advantages related to cellular uptake – see below). Meanwhile, other AON applications that use different mechanisms of action are gaining more interest. The best-known application is the manipulation of splicing. Using AONs that target splice sites or exonic/intronic inclusion signals located within exons or introns, exons can be hidden from the splicing machinery, resulting in the skipping of the target exon (Figure 1C). This can have multiple applications, e.g., switching from one isoform to another, skipping an aberrantly introduced exon to restore the normal transcript, or introducing an out-of-frame deletion to knock down expression of a gene. The latter approach may also be considered as a complementary method to AON-induced knockout through RNAse H-dependent cleavage of RNA:DNA hybrids (Aartsma-Rus et al., 2009). Exon skipping resulting in the expression of truncated, non-functional proteins may be of particular interest in relation with genes or gene pathways which are considered “un-druggable.” Since specific ligands or antagonists cannot always target molecules of interest, AON-mediated RNA targeting can be a good alternative to achieve partial and/or reversible knockdown of such proteins.
Finally, another application of exon skipping is to reframe transcripts allowing the production of an internally deleted, partially functional protein rather than a prematurely truncated non-functional protein (Figure 1C). This has been extensively studied as a therapeutic approach for Duchenne muscular dystrophy (DMD). Protein restoration has been shown in patient-derived cell cultures and in animal models this led to a rescued phenotype (Aartsma-Rus et al., 2006; Heemskerk et al., 2009, 2010). After encouraging results in phase I and I/II clinical trials (Lu et al., 2003; Alter et al., 2006; van Deutekom et al., 2007; Kinali et al., 2009; Goemans et al., 2011), this approach is currently tested in phase III clinical trials. As will be detailed below, this strategy to generate deletion mutants bears much promise for experimental neuroscience too. In other cases, intron splicing silencers may be targeted, resulting in exon inclusion and therefore increase of the expression of a gene or isoform. Here, the most prominent application is rescue of spinal muscular atrophy (SMA) by AON-mediated stimulation of the expression of a functional homolog (see below; Williams et al., 2009; Burghes and McGovern, 2010; Hua et al., 2010; Nlend Nlend et al., 2010).
Specificity
A very important aspect of all splicing-modulation or gene-silencing operations is specificity to the selected target. siRNAs exert their actions in the cytoplasm via interactions with the RNA-induced silencing complex (RISC; Krebs and Alsberg, 2011). Off-target effects appear when siRNA strands interact with partially complementary regions of mRNAs other than the fully complementary target mRNAs (Ma et al., 2006; Vickers et al., 2009; Petri et al., 2011). AON development has faced the same issues in the past (Gaglione and Messere, 2010) and the solutions included modifications of the backbone to reduce base-pair affinity, thus reducing off-target effects (Yoo et al., 2004; Guterstam et al., 2008). Luckily, these modifications can be applied to siRNAs as well (Gaglione and Messere, 2010). A problem that might arise is cell death due to oversaturation of cellular RNA pathways by siRNAs (Grimm et al., 2006) that are necessary for normal cellular function. However, this problem does not exist with AONs since they exert their activity in the nucleus without the need for anything equivalent to the RISC complex (Smith et al., 2006).
Cellular Delivery and Associated Safety
In all instances of RNA or DNA interference in the brain, delivery is an issue. In vivo manipulation of gene expression with shRNA very often depends on the use of viral vectors (Di Benedetto et al., 2009; Ehlert et al., 2010; Kubo et al., 2010), as do cre-recombinase mediated gene excision (Kolber et al., 2008) or gene overexpression models (Ulusoy et al., 2010; Woldbye et al., 2010). However, AONs after reaching the brain, are readily taken up by neurons, and are therefore independent of viral transduction of neurons (Figure 2A). The ease of delivery of the present day modified AONs seems to be linked with a lack of any major adverse side effects.
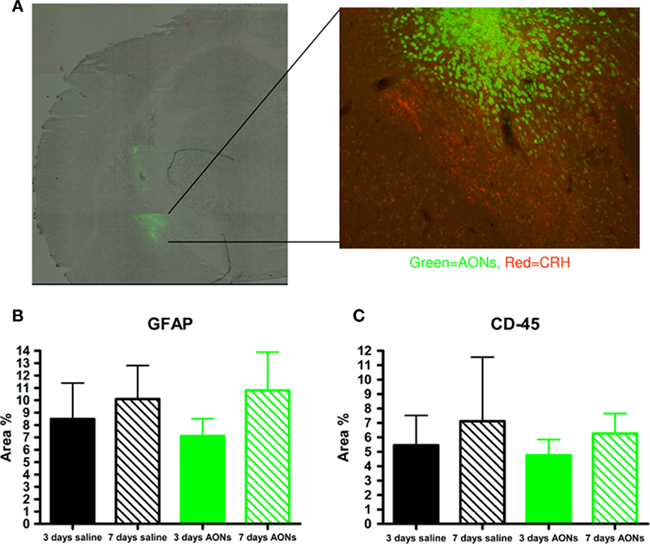
Figure 2. Efficient uptake and low toxicity after local AON injections in the central amygdala (CeA) of the mouse brain. (A) Green fluorescent signal in the mouse CeA (magnification 50×). In the right panel, colocalization of AONs (green) and CRH (red) in the CeA (10×). (B) The area of GFAP (marker for astrocytes) positive cells is not significantly different between animals injected with saline and animals injected with AONs 3 and 7 days after a single local injection in the CeA. (C) Area of CD-45 (marker for activated microglia) positive cells is not significantly different between animals injected with saline and animals injected with AONs 3 and 7 days after a single local injection in the CeA (n = 4–7 per group). Graphs (B) and (C) show that a single injection of AONs induces a similar immune response to a single injection of saline (vehicle).
Delivery of viral vectors has been associated with various levels of toxicity in the brain, mainly depending on viral type used. For example, AAV vectors have been shown to induce neurotoxicity when delivered to the central nervous system (CNS; Oshiro et al., 1995; Driesse et al., 2000; Ehlert et al., 2010; Jayandharan et al., 2011), although serotypes may differ in that aspect (Sanchez et al., 2011). Other viral types, such as retrovirus, show milder toxicity, but they are not suitable for investigation of long-term effects and have limits in the cellular types they can infect (Kaplitt et al., 1998). Lentivirus causes less inflammatory and immune response, but it still shares the disadvantage that preexisting immunity to the parental wild-type virus may cause an accentuated immune response. In contrast, for 2′-O-modified-phosphorothioate AONs only very mild toxicity has been reported, which did not interfere with their desirable effects (Liebsch et al., 1999; Hua et al., 2010) after delivery in the brain via the ventricles, or in cultured neuronal cells (Muller et al., 2000). Although it has been shown that phosphorothioate AONs and siRNAs can have an immunostimulatory effect via toll-like receptors (TLRs; Sioud et al., 2007; Okun et al., 2009), appropriate 2′-O modifications, such as 2′-O-methylation can suppress these effects (Robbins et al., 2007; Sioud et al., 2007; Hamm et al., 2010). The toxic effects that have been reported in some studies after AON delivery in the brain may be due to the vehicle used (Chiasson et al., 1994) or lack of appropriate modifications. Importantly, one can only use AONs bearing 2′-O modifications for modulation of splicing, as an AON without such modification will result in RNAse H activation. Preliminary results from our group have confirmed lack of immune responses to 2′-O-methyl-phosphorothioate AONs compared with saline treatment after a single local injection in the central nucleus of the amygdala (CeA) of the mouse brain (Figure 2B,C). However, it is important to mention that possible toxic and immunostimulatory effects may also be a function of dosage, concentration or duration of treatment (Hua et al., 2010).
Compared to viral delivery methods, AONs have a very rapid uptake and initiation of the effect (Pitts et al., 2009; Ma et al., 2011; within minutes to hours), which allows for administration between different stages of the same experiment (Pitts et al., 2009; Pitts and Takahashi, 2010). Secondly, AONs administration allows better dosage control that can give the optimal effect while reducing potential toxic effects due to, e.g., complete or too high levels of knockdown (Smith et al., 2006; Heemskerk et al., 2010; Hua et al., 2010). In contrast, virally mediated methods tend to produce an all-or-nothing effect, particularly when cre-recombinase systems are used (Pfeifer et al., 2001; Kolber et al., 2008). Another characteristic of AON targeting is the possibility to discontinue treatment (Smith et al., 2006). Although AONs have a longer half-life than, for instance, siRNAs (Smith et al., 2006), eventually they are degraded allowing gene expression to return to basal levels. Viral vectors, however, have a virtually permanent action, although long-term effects may depend on viral type (Kaplitt et al., 1998). Obviously, in instances where long-term manipulation is the goal, a single treatment with a long-term effect may be desirable (Hua et al., 2010). Finally other advantages include rapid production and lack of GMO safety related issues, since no genetically engineered viruses are involved and there is no risk of recombination or reversion to wild-type virus (Naldini et al., 1996; Kaplitt et al., 1998)
Even when methods of virus-independent, direct delivery of siRNA are considered, for example based on conjugations (Iorns et al., 2007) several other issues become apparent. These methods are characterized by various inherent challenges, such as high degradation rate of the siRNA, low cellular uptake and efficiency (Shim and Kwon, 2010), and induction of interferon responses (Sledz et al., 2003; Grimm et al., 2006; Pan et al., 2011). In comparison, AONs have a lower turnover rate (Smith et al., 2006), more prolonged action (Vickers et al., 2009) and, as they are single stranded rather than double stranded, better cellular uptake (our results), which can be achieved even in the absence of auxiliary transfectants (Stein et al., 2010).
In conclusion, AON treatments appear as an attractive approach not only in cases where they restore protein function (such as DMD) but in many other cases where modulation of gene expression is required. Moreover, they offer advantages over other approaches such as siRNA interference that may be very advantageous in certain contexts. In terms of non-specific reactions, we have to emphasize that no definitive lack of any adverse reactions has been proven, but from the available evidence we conclude that side effects in terms of astrogliosis and microgliosis are very limited.
Brain Delivery of AONs
A major challenge of both AON and shRNA applications in neuroscience and in particular for possible clinical use in neurodegenerative disorders is the actual delivery to the brain. The blood brain barrier (BBB) is a physiological obstruction for molecules to enter the brain and molecules can only enter the brain interstitial fluid by transport through the brain capillary endothelial cells (Pardridge, 2002). Intravenous or intraperitoneal administration of phosphorothioate oligonucleotides in rodents showed a very low uptake in brain (Agrawal et al., 1991; Cossum et al., 1993). Increased brain uptake of AON after peripheral delivery can be achieved by increasing the permeability of the BBB (Riley et al., 1998) or through encapsulating the AON in liposomes conjugated to monoclonal antibodies (Zhang et al., 2002; Brignole et al., 2003). Another way to solve this problem is by local injections in the desired brain region if spatial specificity is important or by injection in the cerebrospinal fluid if broad distribution in the brain is deemed more important.
Direct injection in specific brain regions is a method that has been widely used both in rodent studies and in human patients (Olbricht et al., 2010). Experimentally, they offer insight in local effects of widespread factors (Ambroggi et al., 2009), and can have the advantage of contralateral controls in the same animal. Moreover, it provides the options of single injections or repeated/continuous delivery via cannulation. Importantly, it also offers the possibility of reducing the injected dose, thus decreasing potential toxic or immunogenic effects. In human patients intracranial delivery is used in the context of glioblastoma treatment with AONs (Bogdahn et al., 2011).
The alternative of intraventricular (or intrathecal) delivery into the cerebrospinal fluid has also proven successful. Continuous infusion into the ventricle of rodent and non-human primate brains showed significant concentrations of AON throughout the brain, brain stem, and spinal cord. Significant reduction of targeted mRNA indicated that the AON is readily taken up by cells (Smith et al., 2006). The advantage of ventricular infusion through a surgically implanted pump is that there is constant delivery where the dosage can be accurately regulated (Dash and Cudworth, II. 1998). Furthermore, the disadvantage of the AONs’ restricted ability to cross the BBB also is a clear advantage, since after ventricular infusion the AONs will remain in the brain (Hua et al., 2010) thereby reducing side effects on peripheral organs like liver and kidney that readily take up AONs.
In conclusion, while AONs for use in the CNS cannot be administered systemically, they have excellent entry into cells once they passed the BBB. For several backbone chemistries, it has been shown that local injection and distribution via the CSF seem to be devoid of any major toxicity.
Current Disease Models
There are several neurodegenerative disorders where AONs are a promising therapeutic strategy and we will show some examples where AON treatment resulted in therapeutic benefit in animal models and/or clinical trials. First we will discuss neurodegenerative diseases where the aim of AON treatment is to reduce transcript levels of disease-causing proteins such as in multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS), and Alzheimer disease (AD). Next, we will discuss SMA where the AON therapeutic strategy is aimed at modulating pre-mRNA splicing events. Other possible applications of AONs may include Huntington’s disease where allele-specific lowering strategies are explored (Sah and Aronin, 2011).
Multiple sclerosis is an autoimmune disease of the CNS where multifocal infiltration of autoreactive T lymphocytes across the BBB takes place. Lymphocytes in MS patients display high levels of α-4 integrin on their surface (Cannella and Raine, 1995) and this plays an important role in lymphocyte migration to sites of inflammation (Rose et al., 2007). Decreasing leukocyte trafficking into various organs has been successful using monoclonal antibodies against α-4 integrin (Lobb and Hemler, 1994). In a commonly used mouse model for MS, the experimental autoimmune encephalomyelitis model, AON-induced blocking of α-4 integrin expression reduced the incidence and severity of paralytic symptoms (Myers et al., 2005). The 20-mer AONs with 2′-methoxy-ethyl modification and a phosphorothioate backbone were designed to target a sequence just 3′ of the translation start site of the murine α-4 integrin mRNA to block its translation (Figure 1B). Subcutaneous daily injections reduced α-4 integrin surface expression. Although the site of actions of this particular AON is unknown, it is thought that α-4 integrin levels are reduced in peripheral lymphoid tissue and this prevents trafficking of activated mononuclear cells into brain and spinal cord (Myers et al., 2005).
Alzheimer’s disease is the most common form of dementia, in which AONs are considered in yet another mode. Cleavage of β-amyloid precursor protein (APP) at the β-secretase and γ-secretase site causes elevated levels of β-amyloid peptide (Aβ). This is considered a key event in the pathogenesis of AD (Van Broeck et al., 2007). Point mutations near the β-secretase site in the human gene for APP lead to a dominantly inherited form of AD (Selkoe and Kopan, 2003). In a transgenic mouse model of AD containing this mutated β-secretase site, translation of the APP–mRNA was blocked by AONs that bind specifically to the mutated β-secretase site (Figure 1B). The AONs used in this study had a methoxy-ethyl group attached to the 2′O sugar ribose capped at 5′- and 3′-ends with a phosphorothioate (PS) backbone. Repeated injections into the third ventricle (once a week for 4 weeks) reduced the levels of toxic Aβ and increased the levels of soluble α-cleaved β-APP, indicating that this could be a possible strategy to treat familial AD (Chauhan and Siegel, 2007).
Amyotrophic lateral sclerosis is a progressive neurodegenerative disorder caused by degeneration of motor neurons in the brain and spinal cord. This eventually leads to muscle weakening, twitching, and an inability to move the arms, legs, and body (Al-Chalabi and Leigh, 2000). Only 5% of ALS cases are familial and about 20% of all familial cases result from a specific genetic defect that leads to mutation of the enzyme known as superoxide dismutase 1 (SOD1) rendering the protein toxic and prone to aggregation (Bossy-Wetzel et al., 2004). The AONs that have been used in ALS were designed to lower mRNA levels of the SOD1 transcripts and were phosphorothioate modified chimeric nucleotides with 5 2′-O methoxy-ethyl modifications on both the 5′ and 3′ ends and 10 deoxynucleotides in the center to support RNase H activity (Figure 1A). Continuous ventricular infusion reduced levels of mutant SOD1 in a rodent model of ALS and significantly slowed disease progression (Smith et al., 2006). A phase I study to test the safety of this AON in subjects with familial ALS with a SOD1 mutation began in 2009 and the first results are expected in 2011.
Another motor neuron disease where the use of antisense oligonucleotides is under investigation is SMA. SMA is an autosomal recessive neuromuscular disorder caused by dysfunction and loss of motor neurons in the anterior horn of the spinal cord and lower brain stem. The underlying cause of SMA is a homozygous deletion of survival motor neuron 1 (SMN1). SMN1 depletion is viable because of the presence of the almost identical SMN2 gene. However, the majority of SMN2 mRNA transcripts are lacking exon 7, due to a silent mutation within this exon that hampers exon inclusion which results in a truncated protein and reduced expression of functional SMN protein (Lorson et al., 2010). Current therapeutic strategies are aimed at modulation of the splicing of SMN2 by blocking exonic splicing silencers (ESEs) or intronic splicing silencers (ISSs). Transfecting fibroblasts with an AON (termed ISS-N1) blocking an ISS in intron 7 of SMN2 was found to result in inclusion of SMN2 exon 7 (Singh et al., 2006). Improved efficacy of the AON was achieved by incorporation of a uniform 2′-O-2-methoxy-ethyl (MOE) chemistry into the ribose sugars and a single injection of this AONs into the cerebral ventricles in a severe mouse model of SMA showed increased exon 7 inclusion and SMN protein levels in the spinal cord resulting in increased muscle size and strength (Passini et al., 2011). An increased exon 7 inclusion could also be achieved by targeting the 3′SS region of exon 8 with 2′-O-methyl and phosphorothioate backbone modified AONs (Lim and Hertel, 2001). 2′-O-methyl modified AONs with a phosphorothioate backbone were found to result in exon 7 inclusion and elevated SMN protein expression levels in vivo (Williams et al., 2009; Hua et al., 2010)(Figure 1C). Another strategy is to add a functional moiety to the AON to replace the missing splicing enhancer protein to enhance the recruitment of exon 7 by the splicing machinery (Cartegni and Krainer, 2003; Skordis et al., 2003). Several studies applying this strategy in animal models have since been published. Dickson et al. (2008) targeted the 3′SS region of exon 8 with an AON that included a functional sequence to attract hnRNP A1. Baughan et al. (2009) used an AON targeting the 1 ISS element in intron 6 of SMN2 with an ESE tail to recruit positive slicing factors. Both showed an increase in brain SMN protein levels after intraventricular injection in SMA mouse models. A transgenic mouse model expressing a modified snRNP gene that induced exon 7 inclusion also showed that adding a functional moiety to AONs has the potential to revert the phenotype and increase survival (Voigt et al., 2010).
AONs as Experimental Tools
Knockdown
The most widely used application of AON-mediated RNA targeting in the CNS has been the downregulation of gene expression through intranuclear RNase H-mediated cleavage of DNA:RNA hybrids (Chiasson et al., 1994; Dias and Stein, 2002; Figure 1A). Thus, the AON in this case is targeted against an mRNA sequence of interest (Chiasson et al., 1994). This approach offers an alternative, with certain advantages, to knockdown induced by viral vectors and siRNAs which are mediated by the RISC complex. We present a few recent examples from which the advantages of “classical” knockdown use of AONs is apparent.
Ma et al. (2011) used AONs to knock down BDNF expression in various brain areas and studied its involvement in conditioned taste aversion memory formation (Ma et al., 2011). They showed that BDNF synthesis in the CeA is necessary for long-term memory formation of conditioned taste aversion and especially for conditioned taste aversion consolidation. Likewise, AONs have been also used to knock down temporally the expression of CRH in the CeA, (Pitts et al., 2009; Pitts and Takahashi, 2010). In a series of experiments targeting CRH mRNA it was shown that CRH plays an important role in contextual fear conditioning consolidation in the CeA (Pitts et al., 2009). Furthermore, it was shown that CRH involvement in this context may be important up to 24 h after training for successful consolidation of contextual fear (Pitts and Takahashi, 2010). These studies illustrate the advantage of infusing AONs at different time points (Ma et al., 2011).
Antisense oligonucleotide-mediated knockdown has been combined with other gene-silencing techniques to serve specific experimental purposes, or even to delineate the mechanisms underlying, for example, RNA interference. Hemmings-Mieszczak et al. (2003) used mixtures of siRNAs and AONs to achieve a higher degree of reduction of the expression of the pain receptor P2X3, in vitro, and a more pronounced functional outcome. The effect was stronger when the siRNA and the AON targeted mRNA sequences distant from each other, because sequences close to each other may lead to masking of the complementary either the siRNA or the AON (Hemmings-Mieszczak et al., 2003).
Antisense oligonucleotides were recently used in an elegant way to inhibit the expression of proteins associated with the RISC complex. AON-mediated downregulation of Argonaute proteins Ago1 and Ago2, combined with modified cleavage deficient siRNAs, showed that off-target effects of siRNAs are independent from Ago2 cleavage, but they require interaction with Ago proteins and the RISC complex (Vickers et al., 2009). A similar approach was used to investigate the involvement of the RISC complex in pre-rRNA processing. Targeting of Dicer, Drosha, or Ago2 lead to impairments in pre-rRNA processing, indicating that these proteins are involved in the biogenesis of rRNA (Liang and Crooke, 2011). The great advantage of AON-mediated knockdown here is that its action depends on an entirely different mechanism from siRNA allowing interference with one without affecting the other.
Thus, RNase H-mediated cleavage of DNA:RNA hybrids still is broadly used in basal and clinical research. In addition, exon skipping and inclusion offer a number of possibilities that are unique for AONs.
In – and Out-of-Frame Deletions
The possibilities for in and out-of-frame exon deletions (Figure 1C) that can be achieved with AONs are virtually endless. Out-of-frame deletions may be used as an alternative to gene-silencing, or to generate truncated proteins. In particular, in-frame deletions can be used to create mutations that otherwise would require a costly and time-consuming knock-in approach. Here we elaborate on the use of the domain structure of steroid receptors such as the glucocorticoid receptor (GR) gene.
The GR has been shown to be involved in various functions including stress responses, inhibition of inflammatory responses, and metabolic effects. The relationship between the structure and the function of the GR has been extensively studied (Giguere et al., 1986; Mittelstadt and Ashwell, 2003). In short, the GR protein contains domains that arise from eight exons (2–9, exon 1 of the mRNA is not translated): exon 2 codes for the N-terminal half of the protein which contains the major transcriptional activation domain τ1, exons 3 and 4 code for the central part of the protein which contains two zinc fingers involved in DNA-binding and homodimerization. The C-terminal region of the protein, encoded by exons 5–9, include among others, the domains responsible for transcriptional activation (τ2) and ligand binding (Figure 3A; Giguere et al., 1986; Danielian et al., 1992; Mittelstadt and Ashwell, 2003).
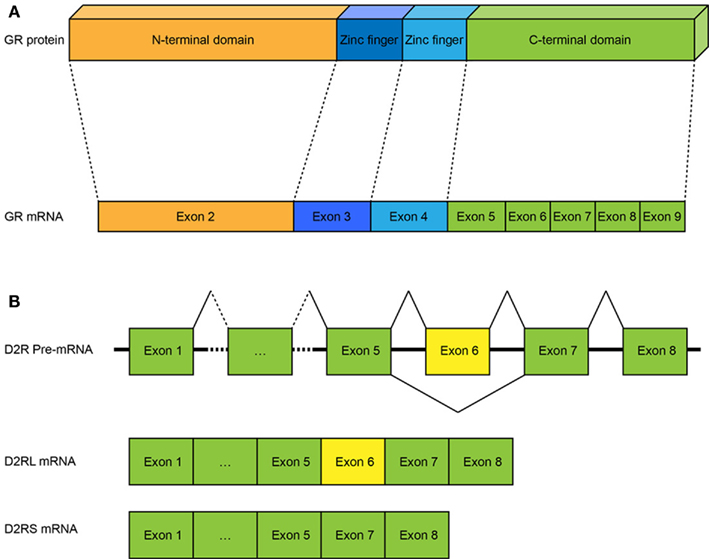
Figure 3. Relation between GR mRNA and protein and splicing events of the D2R gene. (A) The 8 coding exons of the GR gene and the protein domains they code for. Exon 2 codes for the N-terminal domain of the protein which contains the major transcriptional activation domain τ1. Exons 3 and 4 code for two zinc-finger domains that are involved in DNA-binding and homodimerization. Finally exons 5–9 code for the C-terminal end of the protein which contains the domains for transcriptional activation and ligand binding. (B) Splicing events of the D2R RNA. The D2R gene generate two isoforms, D2R long and short. The expression of the isoforms depends on the inclusion or exclusion of exon 6. Inclusion of exon 6 leads to expression of the long isoform (D2RL), whereas exclusion of exon 6 results in the expression of the short isoform (D2Rs).
A good example of the potential of exon skipping is provided by exon 4 of the GR mRNA, which codes for a zinc-finger domain that is involved in DNA-binding. The GR can act in a DNA-binding dependent mode, but many of its effects are mediated by interactions with other transcription factors, rather than with DNA (Reichardt et al., 1998; Oitzl et al., 2001). Considerable effort has been made to generate the GRdim/dim knock-in mouse. As a consequence of a point mutation in exon 4, this mouse has impaired GR binding to the DNA, but GR can still transrepress transcription factors like AP1 and NF-κB. Even though this has proven to be a very valuable model, the separation between DNA-binding and transcription factor binding is incomplete (Adams et al., 2003). Skipping of exon 4 would result in the loss of the second zinc-finger domain of the protein, which is expected to abrogate DNA-binding completely. This would be a cheaper and faster solution than the knock-in approach that would be otherwise used to generate a similar phenotype. In addition, local injection in the brain can be used to address steroid signaling in specific brain areas, for which no specific cre-driving promoters exist.
The GR is just an example of a potential target of in-frame deletions via AON-mediated RNA targeting. Other nuclear receptors, such as the mineralocorticoid receptor and the estrogen receptor, share similar features with the GR, such as the 117-nucleotide long exon that codes for a zinc-finger domain. One can appreciate that the potential of this approach may have virtually no limits: theoretically any exon divisible by three can be targeted for skipping. Combined with the possibilities for out-of-frame deletions, that can be extended, practically, to every gene, we conclude that AON-mediated RNA targeting is a very powerful and versatile research tool.
Natural Splice Variants
For a vast number of genes, splice variants occur. Especially in the CNS alternative splicing events are very common and potentially important for numerous pathways (Fagnani et al., 2007). In relation to the aforementioned brain glucocorticoid signaling, we have observed that splice variants of the GR partner steroid receptor coactivator-1 (SRC-1) are major determinants in the effectiveness of GR, with one variant potentiating repression, and the other variants potentiating stimulation of the target gene corticotropin releasing hormone (CRH) in different brain regions (van der Laan et al., 2008; Lachize et al., 2009). Splice variant diversity has been observed for CRH receptor genes (Markovic et al., 2008; Zmijewski and Slominski, 2009), Cannabinoid receptor genes (Ryberg et al., 2005; Liu et al., 2009), and many others (Fagnani et al., 2007; Trifonov et al., 2010). Below we elaborate on yet one more of these examples, that of dopamine receptor D2 splice variants.
D2 Receptor Splice Variants (Isoform Switching)
The dopaminergic system plays an essential role in the brain, controlling a wide range of physiological processes, varying from motor and cognitive functions to responses to natural rewards and drug addiction (Centonze et al., 2004; Fetsko et al., 2005; De Mei et al., 2009). These effects are mediated by five different receptor subtypes which can be divided in two groups: the D1-like receptors (D1 and D5 receptors) and the D2-like receptors (D2, D3, and D4 receptors; van Ham et al., 2007; De Mei et al., 2009). These two groups of receptors differ in their pharmacological properties and the endocellular pathways in which they are involved (Tan et al., 2003; van Ham et al., 2007).
The D2R gene consists of eight exons, seven of which are coding (Vallone et al., 2000). Alternative splicing of the D2 gene generates two isoforms. The expression of each isoform depends on the in- or exclusion of exon 6 in the mRNA (Figure 3B). Inclusion of exon 6 results in the expression of the long isoform (D2L) whereas its exclusion results in the expression of the short isoform (D2S). Presynaptic neurons show a higher expression of D2S while postsynaptic neurons show a higher expression of D2L. The functional importance of the D2L/D2S ratio has been extensively studied in both humans and animal models. In humans, regulation of the D2L/D2S ratio has been shown to be relevant for cognitive and motor performance, working memory, emotion processing, schizophrenia, and drug addiction (Bertolino et al., 2009; Blasi et al., 2009; De Mei et al., 2009; Glatt et al., 2009; Fazio et al., 2011; Moyer et al., 2011). The importance of the D2L/D2S ratio has been shown in rodent studies, where a higher D2L/D2S strain-specific ratio may account for differences in drug-induced stereotypic behavior, responses to pharmacological agents, and susceptibility to drug abuse (Colelli et al., 2010). Moreover, mice deficient only for D2L show decreased levels of rearing and locomotion, reduced home cage activity and are less sensitive to haloperidol-induced catalepsy (Wang et al., 2000) and raclopride treatment (Xu et al., 2002). Also, they show learning impairments when compared with their wild-type counterparts (Fetsko et al., 2005).
In this context, AON-mediated RNA targeting can offer further refinements of existing models as well as new opportunities. First, exon skipping or exon inclusion can be used to reverse the naturally occurring D2L/D2S ratio in different animal strains, instead of comparing different strains in the same experiment. Secondly, new possibilities for developing D2S-knock-out animals arise. Since it is practically impossible to generate a real D2S-knock-out animal with conventional methods, an exon inclusion approach can be utilized to achieve a similar result. Here, new possibilities of targeted modulation in favor of D2L can be explored, leading to animals exhibiting very limited expression of D2S, thus opening new avenues for the exploration of the in vivo effects of the D2R isoforms. Thirdly, both exon skipping and exon inclusion approaches, combined with the transient effects of AONs, can be incorporated in classical experiments, but this time in a repeated-measures setup. In this manner, the same animals can be used to look into before and after conditions regarding the D2L/D2S ratio or each isoform individually. Lastly, taking into consideration the vast extent of the D2R isoforms in human brain functions, a potential therapeutic approach that could reset and normalize the D2L/D2S ratio, could be of medical importance in relation to Parkinson’s disease, schizophrenia, and drug abuse.
In conclusion, to date, classical methods have been used to study the differential contribution of the two isoforms in vivo. These include SNP analysis and fMRI in humans and D2 and D2L knock-out mice. The broad involvement of the D2R isoforms in pathways in the brains, the implication for health and disease and the limited tools available for the study of the two isoforms qualify it as an excellent example of potential applications of antisense-mediated RNA targeting both as a research tool and as potential therapeutic intervention.
Concluding Remarks
Antisense oligonucleotide-mediated gene-silencing and exclusion/inclusion of exons offer many options for clinical and basal neuroscience alike. The good uptake in neurons once the BBB is passed is a major strength, in combination with good efficacy of interference with translation and splicing events. Interference with splicing allows shifting ratios of endogenous splice variants, knockdown, as well as the generation of internal deletions and truncations. Temporal and spatial control of the manipulations is considerable, and AON-mediated gene manipulation will often be more rapid that alternative transgenic approaches. We therefore consider such approaches as a highly promising part of the general neuroscientist’s toolkit.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aartsma-Rus, A., Janson, A. A., Heemskerk, J. A., De Winter, C. L., Van Ommen, G. J., and Van Deutekom, J. C. (2006). Therapeutic modulation of DMD splicing by blocking exonic splicing enhancer sites with antisense oligonucleotides. Ann. N. Y. Acad. Sci. 1082, 74–76.
Aartsma-Rus, A., van Vliet, L., Hirschi, M., Janson, A. A., Heemskerk, H., de Winter, C. L., de Kimpe, S., van Deutekom, J. C., t Hoen, P. A., and van Ommen, G. J. (2009). Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol. Ther. 17, 548–553.
Adams, M., Meijer, O. C., Wang, J., Bhargava, A., and Pearce, D. (2003). Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol. Endocrinol. 17, 2583–2592.
Agrawal, S., Temsamani, J., and Tang, J. Y. (1991). Pharmacokinetics, biodistribution, and stability of oligodeoxynucleotide phosphorothioates in mice. Proc. Natl. Acad. Sci. U.S.A. 88, 7595–7599.
Al-Chalabi, A., and Leigh, P. N. (2000). Recent advances in amyotrophic lateral sclerosis. Curr. Opin. Neurol. 13, 397–405.
Alter, J., Lou, F., Rabinowitz, A., Yin, H., Rosenfeld, J., Wilton, S. D., Partridge, T. A., and Lu, Q. L. (2006). Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med. 12, 175–177.
Ambroggi, F., Turiault, M., Milet, A., Deroche-Gamonet, V., Parnaudeau, S., Balado, E., Barik, J., van der Veen, R., Maroteaux, G., Lemberger, T., Schutz, G., Lazar, M., Marinelli, M., Piazza, P. V., and Tronche, F. (2009). Stress and addiction: glucocorticoid receptor in dopaminoceptive neurons facilitates cocaine seeking. Nat. Neurosci. 12, 247–249.
Baughan, T. D., Dickson, A., Osman, E. Y., and Lorson, C. L. (2009). Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum. Mol. Genet. 18, 1600–1611.
Bertolino, A., Fazio, L., Caforio, G., Blasi, G., Rampino, A., Romano, R., Di Giorgio, A., Taurisano, P., Papp, A., Pinsonneault, J., Wang, D., Nardini, M., Popolizio, T., and Sadee, W. (2009). Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain 132, 417–425.
Blasi, G., Lo Bianco, L., Taurisano, P., Gelao, B., Romano, R., Fazio, L., Papazacharias, A., Di Giorgio, A., Caforio, G., Rampino, A., Masellis, R., Papp, A., Ursini, G., Sinibaldi, L., Popolizio, T., Sadee, W., and Bertolino, A. (2009). Functional variation of the dopamine D2 receptor gene is associated with emotional control as well as brain activity and connectivity during emotion processing in humans. J. Neurosci. 29, 14812–14819.
Bogdahn, U., Hau, P., Stockhammer, G., Venkataramana, N. K., Mahapatra, A. K., Suri, A., Balasubramaniam, A., Nair, S., Oliushine, V., Parfenov, V., Poverennova, I., Zaaroor, M., Jachimczak, P., Ludwig, S., Schmaus, S., Heinrichs, H., and Schlingensiepen, K. H. (2011). Targeted therapy for high-grade glioma with the TGF-beta2 inhibitor trabedersen: results of a randomized and controlled phase IIb study. Neurooncology 13, 132–142.
Bossy-Wetzel, E., Schwarzenbacher, R., and Lipton, S. A. (2004). Molecular pathways to neurodegeneration. Nat. Med. 10(Suppl.), S2–S9.
Brignole, C., Pagnan, G., Marimpietri, D., Cosimo, E., Allen, T. M., Ponzoni, M., and Pastorino, F. (2003). Targeted delivery system for antisense oligonucleotides: a novel experimental strategy for neuroblastoma treatment. Cancer Lett. 197, 231–235.
Burghes, A. H., and McGovern, V. L. (2010). Antisense oligonucleotides and spinal muscular atrophy: skipping along. Genes Dev. 24, 1574–1579.
Cannella, B., and Raine, C. S. (1995). The adhesion molecule and cytokine profile of multiple sclerosis lesions. Ann. Neurol. 37, 424–435.
Cartegni, L., and Krainer, A. R. (2003). Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Mol. Biol. 10, 120–125.
Centonze, D., Gubellini, P., Usiello, A., Rossi, S., Tscherter, A., Bracci, E., Erbs, E., Tognazzi, N., Bernardi, G., Pisani, A., Calabresi, P., and Borrelli, E. (2004). Differential contribution of dopamine D2S and D2L receptors in the modulation of glutamate and GABA transmission in the striatum. Neuroscience 129, 157–166.
Chauhan, N. B., and Siegel, G. J. (2007). Antisense inhibition at the [beta]-secretase-site of [beta]-amyloid precursor protein reduces cerebral amyloid and acetyl cholinesterase activity in Tg2576. Neuroscience 146, 143–151.
Chiasson, B. J., Armstrong, J. N., Hooper, M. L., Murphy, P. R., and Robertson, H. A. (1994). The application of antisense oligonucleotide technology to the brain: some pitfalls. Cell. Mol. Neurobiol. 14, 507–521.
Colelli, V., Fiorenza, M. T., Conversi, D., Orsini, C., and Cabib, S. (2010). Strain-specific proportion of the two isoforms of the dopamine D2 receptor in the mouse striatum: associated neural and behavioral phenotypes. Genes Brain Behav. 9, 703–711.
Cossum, P. A., Sasmor, H., Dellinger, D., Truong, L., Cummins, L., Owens, S. R., Markham, P. M., Shea, J. P., and Crooke, S. (1993). Disposition of the 14C-labeled phosphorothioate oligonucleotide ISIS 2105 after intravenous administration to rats. J. Pharmacol. Exp. Ther. 267, 1181–1190.
Danielian, P. S., White, R., Lees, J. A., and Parker, M. G. (1992). Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 11, 1025–1033.
Dash, A. K., and Cudworth, G. C. II. (1998). Therapeutic applications of implantable drug delivery systems. J. Pharmacol. Toxicol. Methods 40, 1–12.
De Mei, C., Ramos, M., Iitaka, C., and Borrelli, E. (2009). Getting specialized: presynaptic and postsynaptic dopamine D2 receptors. Curr. Opin. Pharmacol. 9, 53–58.
Di Benedetto, B., Wefers, B., Wurst, W., and Kuhn, R. (2009). Local knockdown of ERK2 in the adult mouse brain via adeno-associated virus-mediated RNA interference. Mol. Biotechnol. 41, 263–269.
Dias, N., and Stein, C. A. (2002). Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1, 347–355.
Dickson, A., Osman, E., and Lorson, C. L. (2008). A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum. Gene Ther. 19, 1307–1315.
Driesse, M. J., Esandi, M. C., Kros, J. M., Avezaat, C. J., Vecht, C., Zurcher, C., van der Velde, I., Valerio, D., Bout, A., and Sillevis Smitt, P. A. (2000). Intra-CSF administered recombinant adenovirus causes an immune response-mediated toxicity. Gene Ther. 7, 1401–1409.
Ehlert, E. M., Eggers, R., Niclou, S. P., and Verhaagen, J. (2010). Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 11, 20.
Fagnani, M., Barash, Y., Ip, J. Y., Misquitta, C., Pan, Q., Saltzman, A. L., Shai, O., Lee, L., Rozenhek, A., Mohammad, N., Willaime-Morawek, S., Babak, T., Zhang, W., Hughes, T. R., van der Kooy, D., Frey, B. J., and Blencowe, B. J. (2007). Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 8, R108.
Fazio, L., Blasi, G., Taurisano, P., Papazacharias, A., Romano, R., Gelao, B., Ursini, G., Quarto, T., Lo Bianco, L., Di Giorgio, A., Mancini, M., Popolizio, T., Rubini, G., and Bertolino, A. (2011). D2 receptor genotype and striatal dopamine signaling predict motor cortical activity and behavior in humans. Neuroimage 54, 2915–2921.
Fetsko, L. A., Xu, R., and Wang, Y. (2005). Effects of age and dopamine D2L receptor-deficiency on motor and learning functions. Neurobiol. Aging 26, 521–530.
Gaglione, M., and Messere, A. (2010). Recent progress in chemically modified siRNAs. Mini Rev. Med. Chem. 10, 578–595.
Giguere, V., Hollenberg, S. M., Rosenfeld, M. G., and Evans, R. M. (1986). Functional domains of the human glucocorticoid receptor. Cell 46, 645–652.
Glatt, S. J., Faraone, S. V., Lasky-Su, J. A., Kanazawa, T., Hwu, H. G., and Tsuang, M. T. (2009). Family-based association testing strongly implicates DRD2 as a risk gene for schizophrenia in Han Chinese from Taiwan. Mol. Psychiatry 14, 885–893.
Goemans, N. M., Tulinius, M., van den Akker, J. T., Burm, B. E., Ekhart, P. F., Heuvelmans, N., Holling, T., Janson, A. A., Platenburg, G. J., Sipkens, J. A., Sitsen, J. M., Aartsma-Rus, A., van Ommen, G. J., Buyse, G., Darin, N., Verschuuren, J. J., Campion, G. V., de Kimpe, S. J., and van Deutekom, J. C. (2011). Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 364, 1513–1522.
Grimm, D., Streetz, K. L., Jopling, C. L., Storm, T. A., Pandey, K., Davis, C. R., Marion, P., Salazar, F., and Kay, M. A. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541.
Guterstam, P., Lindgren, M., Johansson, H., Tedebark, U., Wengel, J., El Andaloussi, S., and Langel, Ü. (2008). Splice-switching efficiency and specificity for oligonucleotides with locked nucleic acid monomers. Biochem. J. 412, 307–313.
Hamm, S., Latz, E., Hangel, D., Müller, T., Yu, P., Golenbock, D., Sparwasser, T., Wagner, H., and Bauer, S. (2010). Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology 215, 559–569.
Heemskerk, H., de Winter, C., van Kuik, P., Heuvelmans, N., Sabatelli, P., Rimessi, P., Braghetta, P., van Ommen, G. J., de Kimpe, S., Ferlini, A., Aartsma-Rus, A., and van Deutekom, J. C. (2010). Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol. Ther. 18, 1210–1217.
Heemskerk, H. A., de Winter, C. L., de Kimpe, S. J., van Kuik-Romeijn, P., Heuvelmans, N., Platenburg, G. J., van Ommen, G. J., van Deutekom, J. C., and Aartsma-Rus, A. (2009). In vivo comparison of 2′-O-methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. J. Gene Med 11, 257–66.
Hemmings-Mieszczak, M., Dorn, G., Natt, F. J., Hall, J., and Wishart, W. L. (2003). Independent combinatorial effect of antisense oligonucleotides and RNAi-mediated specific inhibition of the recombinant Rat P2X3 receptor. Nucleic Acids Res. 31, 2117–2126.
Hua, Y., Sahashi, K., Hung, G., Rigo, F., Passini, M. A., Bennett, C. F., and Krainer, A. R. (2010). Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644.
Iorns, E., Lord, C. J., Turner, N., and Ashworth, A. (2007). Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 6, 556–568.
Jayandharan, G. R., Aslanidi, G., Martino, A. T., Jahn, S. C., Perrin, G. Q., Herzog, R. W., and Srivastava, A. (2011). Activation of the NF-κB pathway by adeno-associated virus (AAV) vectors and its implications in immune response and gene therapy. Proc. Natl. Acad. Sci. U.S.A. 108, 3743–3748.
Kaplitt, M. G., Darakchiev, B., and During, M. J. (1998). Prospects for gene therapy in pediatric neurosurgery. Pediatr. Neurosurg. 28, 3–14.
Kinali, M., Arechavala-Gomeza, V., Feng, L., Cirak, S., Hunt, D., Adkin, C., Guglieri, M., Ashton, E., Abbs, S., Nihoyannopoulos, P., Elena Garralda, M., Rutherford, M., McCulley, C., Popplewell, L., Graham, I. R., Dickson, G., Wood, M. J. A., Wells, D. J., Wilton, S. D., Kole, R., Straub, V., Bushby, K., Sewry, C., Morgan, J. E., and Muntoni, F. (2009). Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 8, 918–928.
Kolber, B. J., Roberts, M. S., Howell, M. P., Wozniak, D. F., Sands, M. S., and Muglia, L. J. (2008). Central amygdala glucocorticoid receptor action promotes fear-associated CRH activation and conditioning. Proc. Natl. Acad. Sci. U.S.A. 105, 12004–12009.
Krebs, M. D., and Alsberg, E. (2011). Localized, targeted, and sustained siRNA delivery. Chem. Eur. J. 17, 3054–3062.
Kubo, K., Tomita, K., Uto, A., Kuroda, K., Seshadri, S., Cohen, J., Kaibuchi, K., Kamiya, A., and Nakajima, K. (2010). Migration defects by DISC1 knockdown in C57BL/6, 129X1/SvJ, and ICR strains via in utero gene transfer and virus-mediated RNAi. Biochem. Biophys. Res. Commun. 400, 631–637.
Lachize, S., Apostolakis, E. M., van der Laan, S., Tijssen, A. M., Xu, J., de Kloet, E. R., and Meijer, O. C. (2009). Steroid receptor coactivator-1 is necessary for regulation of corticotropin-releasing hormone by chronic stress and glucocorticoids. Proc. Natl. Acad. Sci. U.S.A. 106, 8038–8042.
Liang, X. H., and Crooke, S. T. (2011). Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 39, 4875–4889.
Liebsch, G., Landgraf, R., Engelmann, M., Lorscher, P., and Holsboer, F. (1999). Differential behavioural effects of chronic infusion of CRH1 and CRH2 receptor antisense oligonucleotides into the rat brain. J. Psychiatr. Res. 33, 153–163.
Lim, S. R., and Hertel, K. J. (2001). Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 276, 45476–45483.
Liu, Q. R., Pan, C. H., Hishimoto, A., Li, C. Y., Xi, Z. X., Llorente-Berzal, A., Viveros, M. P., Ishiguro, H., Arinami, T., Onaivi, E. S., and Uhl, G. R. (2009). Species differences in cannabinoid receptor 2 (CNR2 gene): identification of novel human and rodent CB2 isoforms, differential tissue expression and regulation by cannabinoid receptor ligands. Genes Brain Behav. 8, 519–530.
Lobb, R. R., and Hemler, M. E. (1994). The pathophysiologic role of alpha 4 integrins in vivo. J. Clin. Invest. 94, 1722–1728.
Lorson, C. L., Rindt, H., and Shababi, M. (2010). Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum. Mol. Genet. 19, R111–R118.
Lu, Q. L., Mann, C. J., Lou, F., Bou-Gharios, G., Morris, G. E., Xue, S. A., Fletcher, S., Partridge, T. A., and Wilton, S. D. (2003). Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat. Med. 9, 1009–1014.
Ma, L., Wang, D. D., Zhang, T. Y., Yu, H., Wang, Y., Huang, S. H., Lee, F. S., and Chen, Z. Y. (2011). Region-specific involvement of BDNF secretion and synthesis in conditioned taste aversion memory formation. J. Neurosci. 31, 2079–2090.
Ma, Y., Creanga, A., Lum, L., and Beachy, P. A. (2006). Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359–363.
Markovic, D., Lehnert, H., Levine, M. A., and Grammatopoulos, D. K. (2008). Structural determinants critical for localization and signaling within the seventh transmembrane domain of the type 1 corticotropin releasing hormone receptor: lessons from the receptor variant R1d. Mol. Endocrinol. 22, 2505–2519.
Mittelstadt, P. R., and Ashwell, J. D. (2003). Disruption of glucocorticoid receptor exon 2 yields a ligand-responsive C-terminal fragment that regulates gene expression. Mol. Endocrinol. 17, 1534–1542.
Moyer, R. A., Wang, D., Papp, A. C., Smith, R. M., Duque, L., Mash, D. C., and Sadee, W. (2011). Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology 36, 753–762.
Muller, Y. L., Reitstetter, R., and Yool, A. J. (2000). Antisense knockdown of calcium-dependent K+ channels in developing cerebellar Purkinje neurons. Dev. Brain Res. 120, 135–140.
Myers, K. J., Witchell, D. R., Graham, M. J., Koo, S., Butler, M., and Condon, T. P. (2005). Antisense oligonucleotide blockade of alpha 4 integrin prevents and reverses clinical symptoms in murine experimental autoimmune encephalomyelitis. J. Neuroimmunol. 160, 12–24.
Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M., and Trono, D. (1996). In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267.
Nasevicius, A., and Ekker, S. C. (2000). Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26, 216–220.
Nasevicius, A., Larson, J., and Ekker, S. C. (2000). Distinct requirements for zebrafish angiogenesis revealed by a VEGF-A morphant. Yeast 17, 294–301.
Nlend Nlend, R., Meyer, K., and Schumperli, D. (2010). Repair of pre-mRNA splicing: prospects for a therapy for spinal muscular atrophy. RNA Biol. 7, 430–440.
Oitzl, M. S., Reichardt, H. M., Joels, M., and de Kloet, E. R. (2001). Point mutation in the mouse glucocorticoid receptor preventing DNA binding impairs spatial memory. Proc. Natl. Acad. Sci. U.S.A. 98, 12790–12795.
Okun, E., Lathia, J. D., and Mattson, M. P. (2009). Adhesion- and migration-related side effects of phosphothioated CpG oligodeoxynucleotides. Cell Adh. Migr. 3, 272–274.
Olbricht, W. L., Neeves, K. B., and Foley, C. P. (2010). Microfluidic probes in the treatment of brain-related diseases. Drug News Perspect. 23, 491–497.
Oshiro, E. M., Viola, J. J., Oldfield, E. H., Walbridge, S., Bacher, J., Frank, J. A., Blaese, R. M., and Ram, Z. (1995). Toxicity studies and distribution dynamics of retroviral vectors following intrathecal administration of retroviral vector-producer cells. Cancer Gene Ther. 2, 87–95.
Pan, Q., de Ruiter, P. E., von Eije, K. J., Smits, R., Kwekkeboom, J., Tilanus, H. W., Berkhout, B., Janssen, H. L., and van der Laan, L. J. (2011). Disturbance of the microRNA pathway by commonly used lentiviral shRNA libraries limits the application for screening host factors involved in hepatitis C virus infection. FEBS Lett. 585, 1025–1030.
Pardridge, W. M. (2002). Drug and gene targeting to the brain with molecular trojan horses. Nat. Rev. Drug Discov. 1, 131–139.
Passini, M. A., Bu, J., Richards, A. M., Kinnecom, C., Sardi, S. P., Stanek, L. M., Hua, Y., Rigo, F., Matson, J., Hung, G., Kaye, E. M., Shihabuddin, L. S., Krainer, A. R., Bennett, C. F., and Cheng, S. H. (2011). Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 3, 72ra18.
Petri, S., Dueck, A., Lehmann, G., Putz, N., Rüdel, S., Kremmer, E., and Meister, G. (2011). Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA 17, 737–749.
Pfeifer, A., Brandon, E. P., Kootstra, N., F. Gage, H., and Verma, I. M. (2001). Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc. Natl. Acad. Sci. U.S.A. 98, 11450–11455.
Pitts, M. W., and Takahashi, L. K. (2010). The central amygdala nucleus via corticotropin-releasing factor is necessary for time-limited consolidation processing but not storage of contextual fear memory. Neurobiol. Learn. Mem. 95, 86–91.
Pitts, M. W., Todorovic, C., Blank, T., and Takahashi, L. K. (2009). The central nucleus of the amygdala and corticotropin-releasing factor: insights into contextual fear memory. J. Neurosci. 29, 7379–7388.
Reichardt, H. M., Kaestner, K. H., Tuckermann, J., Kretz, O., Wessely, O., Bock, R., Gass, P., Schmid, W., Herrlich, P., Angel, P., and Schutz, G. (1998). DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93, 531–541.
Riley, M., Gary, I., Norman Kim, N., Vance Watson, E., Pierre Gobin, Y., LeBel, C. P., Black, K. L., and Bartus, R. T. (1998). Intra-arterial administration of carboplatin and the blood brain barrier permeabilizing agent, RMP-7: a toxicologic evaluation in swine. J. Neurooncol. 36, 167–178.
Robbins, M., Judge, A., Liang, L., McClintock, K., Yaworski, E., and MacLachlan, I. (2007). 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15, 1663–1669.
Rose, D. M., Alon, R., and Ginsberg, M. H. (2007). Integrin modulation and signaling in leukocyte adhesion and migration. Immunol. Rev. 218, 126–134.
Ryberg, E., Vu, H. K., Larsson, N., Groblewski, T., Hjorth, S., Elebring, T., Sjögren, S., and Greasley, P. J. (2005). Identification and characterisation of a novel splice variant of the human CB1 receptor. FEBS Lett. 579, 259–264.
Sah, D. W. Y., and Aronin, N. (2011). Oligonucleotide therapeutic approaches for Huntington disease. J. Clin. Invest. 121, 500–507.
Sanchez, C. E., Tierney, T. S., Gale, J. T., Alavian, K. N., Sahin, A., Lee, J. S., Mulligan, R. C., and Carter, B. S. (2011). Recombinant adeno-associated virus type 2 pseudotypes: comparing safety, specificity, and transduction efficiency in the primate striatum. J. Neurosurg. 114, 672–680.
Selkoe, D., and Kopan, R. (2003). Notch and presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26, 565–597.
Shim, M. S., and Kwon, Y. J. (2010). Efficient and targeted delivery of siRNA in vivo. FEBS J. 277, 4814–4827.
Singh, N. K., Singh, N. N., Androphy, E. J., and Singh, R. N. (2006). Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 26, 1333–1346.
Sioud, M., Furset, G., and Cekaite, L. (2007). Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 361, 122–126.
Skordis, L. A., Dunckley, M. G., Yue, B., Eperon, I. C., and Muntoni, F. (2003). Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 100, 4114–4119.
Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H., and Williams, B. R. G. (2003). Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5, 834–839.
Smith, R. A., Miller, T. M., Yamanaka, K., Monia, B. P., Condon, T. P., Hung, G., Lobsiger, C. S., Ward, C. M., McAlonis-Downes, M., Wei, H., Wancewicz, E. V., Bennett, C. F., and Cleveland, D. W. (2006). Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Investig. 116, 2290–2296.
Stein, C. A., Bo Hansen, J., Lai, J., Wu, S., Voskresenskiy, A., Høg, A., Worm, J., Hedtjärn, M., Souleimanian, N., Miller, P., Soifer, H. S., Castanotto, D., Benimetskaya, L., Ørum, H., and Koch, T. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 38, e3.
Tan, S., Hermann, B., and Borrelli, E. (2003). Dopaminergic mouse mutants: Investigating the roles of the different dopamine receptor subtypes and the dopamine transporter. Int. Rev. Neurobiol. 54, 145–197.
Trifonov, S., Houtani, T., Shimizu, J., Hamada, S., Kase, M., Maruyama, M., and Sugimoto, T. (2010). GPR155: gene organization, multiple mRNA splice variants and expression in mouse central nervous system. Biochem. Biophys. Res. Commun. 398, 19–25.
Ulusoy, A., Decressac, M., Kirik, D., and Bjorklund, A. (2010). Viral vector-mediated overexpression of alpha-synuclein as a progressive model of Parkinson’s disease. Prog. Brain Res. 184, 89–111.
Vallone, D., Picetti, R., and Borrelli, E. (2000). Structure and function of dopamine receptors. Neurosci. Biobehav. Rev. 24, 125–132.
Van Broeck, B., Van Broeckhoven, C., and Kumar-Singh, S. (2007). Current insights into molecular mechanisms of alzheimer disease and their implications for therapeutic approaches. Neurodegener. Dis. 4, 349–365.
van der Laan, S., Lachize, S. B., Vreugdenhil, E., de Kloet, E. R., and Meijer, O. C. (2008). Nuclear receptor coregulators differentially modulate induction and glucocorticoid receptor-mediated repression of the corticotropin-releasing hormone gene. Endocrinology 149, 725–732.
van Deutekom, J. C., Janson, A. A., Ginjaar, I. B., Frankhuizen, W. S., Aartsma-Rus, A., Bremmer-Bout, M., den Dunnen, J. T., Koop, K., van der Kooi, A. J., Goemans, N. M., de Kimpe, S. J., Ekhart, P. F., Venneker, E. H., Platenburg, G. J., Verschuuren, J. J., and van Ommen, G. J. B. (2007). Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 357, 2677–2686.
van Ham, II Banihashemi, B., Wilson, A. M., Jacobsen, K. X., Czesak, M., and Albert, P. R. (2007). Differential signaling of dopamine-D2S and -D2L receptors to inhibit ERK1/2 phosphorylation. J. Neurochem. 102, 1796–1804.
Vickers, T. A., Lima, W. F., Wu, H., Nichols, J. G., Linsley, P. S., and Crooke, S. T. (2009). Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 “slicer” independent and can be mediated by Ago1. Nucleic Acids Res. 37, 6927–6941.
Voigt, T., Meyer, K., Baum, O., and Schümperli, D. (2010). Ultrastructural changes in diaphragm neuromuscular junctions in a severe mouse model for spinal muscular atrophy and their prevention by bifunctional U7 snRNA correcting SMN2 splicing. Neuromuscul. Disord. 20, 744–752.
Wang, Y. Y., Xu, R., Sasaoka, T., Tonegawa, S., Kung, M. P., and Sankoorikal, E. B. (2000). Dopamine D2 long receptor-deficient mice display alterations in striatum-dependent functions. J. Neurosci. 20, 8305–8314.
Williams, J. H., Schray, R. C., Patterson, C. A., Ayitey, S. O., Tallent, M. K., and Lutz, G. J. (2009). Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J. Neurosci. 29, 7633–7638.
Woldbye, D. P., Angehagen, M., Gotzsche, C. R., Elbrond-Bek, H. A., Sorensen, T., Christiansen, S. H., Olesen, M. V., Nikitidou, L., Hansen, T. V., Kanter-Schlifke, I., and Kokaia, M. (2010). Adeno-associated viral vector-induced overexpression of neuropeptide Y Y2 receptors in the hippocampus suppresses seizures. Brain 133, 2778–2788.
Xu, R., Hranilovic, D., Fetsko, L. A., Bucan, M., and Wang, Y. (2002). Dopamine D2S and D2L receptors may differentially contribute to the actions of antipsychotic and psychotic agents in mice. Mol. Psychiatry 7, 1075–1082.
Yoo, B. H., Bochkareva, E., Bochkarev, A., Mou, T. C., and Gray, D. M. (2004). 2′-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 32, 2008–2016.
Zhang, Y., Jeong Lee, H., Boado, R. J., and Pardridge, W. M. (2002). Receptor-mediated delivery of an antisense gene to human brain cancer cells. J. Gene Med. 4, 183–194.
Keywords: antisense oligonucleotides, exon skipping, central nervous system, receptors, splicing, gene therapy
Citation: Zalachoras I, Evers MM, van Roon-Mom WMC, Aartsma-Rus AM, and Meijer OC (2011) Antisense-mediated RNA targeting: versatile and expedient genetic manipulation in the brain. Front. Mol. Neurosci. 4:10. doi: 10.3389/fnmol.2011.00010
Received: 11 April 2011;
Paper pending published: 11 May 2011;
Accepted: 08 July 2011;
Published online: 19 July 2011.
Edited by:
Alistair N. Garratt, Max Delbrück Center for Molecular Medicine, GermanyReviewed by:
Alistair N. Garratt, Max Delbrück Center for Molecular Medicine, GermanyF. Gregory Wulczyn, Charité – Universitätsmedizin Berlin, Germany
Copyright: © 2011 Zalachoras, Evers, van Roon-Mom, Aartsma-Rus and Meijer. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Onno C. Meijer, Faculty of Science, Gorlaeus Laboratories, Division of Medical Pharmacology, Leiden/Amsterdam Center for Drug Research, Leiden University, Einsteinweg 55, Leiden 2333 CC, Netherlands. e-mail:by5tZWlqZXJAbGFjZHIubGVpZGVudW5pdi5ubA==
 Ioannis Zalachoras1
Ioannis Zalachoras1