- 1Division of Pulmonary and Critical Care Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 2Institute of Respiratory Diseases, Sun Yat-sen University, Guangzhou, China
- 3Department of Emergency Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
Background: The idiopathic pulmonary fibrosis (IPF) is a progressive and lethal interstitial lung disease with high morbidity and mortality. IPF is characterized by excessive extracellular matrix accumulation (ECM) and epithelial-mesenchymal transformation (EMT). To date, few anti-fibrotic therapeutics are available to reverse the progression of pulmonary fibrosis, and it is important to explore new profibrotic molecular regulators mediating EMT and pulmonary fibrosis.
Methods: Based on our model of TGF-β1-induced EMT in BEAS-2B cells, we performed the genome-wide CRISPR/Cas9 knockout (GeCKO) screening technique, pathway and functional enrichment analysis, loss-of-function experiment, as well as other experimental techniques to comprehensively investigate profibrotic regulators contributing to EMT and the pathogenesis of pulmonary fibrosis.
Results: Utilizing the GeCKO library screening, we identified 76 top molecular regulators. Ten candidate genes were subsequently confirmed by integrating the high-throughput data with findings from pathway and functional enrichment analysis. Among the candidate genes, knockout of COL20A1 and COL27A1 led to decreased mRNA expression of ECM components (Fibronectin and Collagen-I), as well as an increased rate of cell apoptosis. The mRNA expression of Collagen-I, together with the cell viability and migration, were inhibited when knocking out the WNT11. In addition, a decrease in the protein deposition of ECM components was observed by suppressing the expression of COL20A1, COL27A1, and WNT11.
Conclusion: Our study demonstrates that the COL20A1, COL27A1, and WNT11 serve as key profibrotic regulators of EMT. Gaining understanding and insights into these key profibrotic regulators of EMT paves the way for the discovery of new therapeutic targets against the onset and progression of IPF.
1 Introduction
Idiopathic pulmonary fibrosis (IPF) is an irreversible and progressive interstitial lung disease characterized by dyspnea and decline in pulmonary function (Onishchenko et al., 2022; Raghu et al., 2022). The annual incidence of acute exacerbation of IPF is high, varying from 4.8% to 43% (Collard et al., 2016). Patients with IPF experience poor prognosis, with survival rates of 88% at 1–2 years and only 31% at over 5 years (Khor et al., 2020). Of note, the median survival time for IPF patients ranges from only 2–3 years after diagnosis (Ley et al., 2011). Although the use of a few drugs (e.g., nintedanib) slowed the annual rate of decline in pulmonary function in patients with IPF (Antoniou et al., 2020), currently there are no effective therapeutics available to reverse the disease progression.
It has been demonstrated that differentiation of fibroblasts into myofibroblasts and excessive accumulation of extracellular matrix (ECM) in lung play a key role in the pathogenesis of IPF (Upagupta et al., 2018; Alvarez-Palomo et al., 2020). Importantly, prior studies have provided evidence that alveolar epithelial cells are progenitors of fibroblasts and myofibroblasts (Kim et al., 2006; Willis et al., 2006), and this conversion of alveolar epithelial cells into the mesenchymal phenotype is termed the epithelial-mesenchymal transformation (EMT). The EMT is closely related to the development of IPF. For example, there is a profound association between the EMT and transforming growth factor-beta 1 (TGF-β1) signaling pathway (Di Gregorio et al., 2020),which has been demonstrated to play a central role in pathogenesis of IPF (Ye and Hu, 2021). Although the TGF-β1 signaling has been proven as a predictive marker and pharmacological target for IPF, few anti-fibrotic drugs targeting this signaling are put into clinical use due to side effects and the need for large doses (Shi et al., 2022). Thus, the identification of new molecules that regulate the EMT and development of pulmonary fibrosis using novel high-throughput screening techniques may offer promising strategies for discovering new therapeutic targets against IPF.
In the past decade, high-throughput screening techniques, such as small interfering RNA screening (Oh et al., 2018) and microarray (Hu et al., 2018) have been applied to identify new molecular regulators contributing to the development of IPF. Despite that some genes and signaling pathways associated with IPF have been discovered, these findings vary due to specific limitations of the technology employed, as well as differences in experimental conditions and the phenotypic readout chosen for analyses. Among the high-throughput screening techniques, the genome-wide CRISPR/Cas9 screening is an advanced and powerful tool for the screening and discovery of unknown genes that contribute to specific biological phenotypes and diseases (Shalem et al., 2015). This technique has been applied to various diseases (Xu et al., 2023; Pfeifer et al., 2024) and also used for screening biological factors regulating disease susceptibility (Zhu et al., 2021). In this study, to systematically investigate molecular regulators that are closely involved in EMT and the pathogenesis of pulmonary fibrosis, we applied the genome-wide CRISPR/Cas9 knockout (GeCKO) screening technique in TGF-β1-treated human bronchial epithelial cells (BEAS-2B). Our results demonstrate that the COL20A1, COL27A1, and WNT11 are key profibrotic regulators of IPF, and the knockout of these genes attenuated the TGF-β1-induced EMT and pulmonary fibrosis in vitro. Our findings show that these profibrotic regulators may serve as specific therapeutic targets, paving the way for the discovery of new medications against the onset and progression of IPF.
2 Materials and methods
2.1 Cell culture
Human bronchial epithelial cell line (BEAS-2B, CRL-3588) was purchased from the American Type Culture Collection, and 293 TN cells (LV900A-1) were obtained from the System Bioscience. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, United States) supplemented with 10% fetal bovine serum (Gibco, United States). All cells were routinely tested and free from microbial contamination.
2.2 EMT induction by TGF-β1
Cells were seeded at a density of 1 × 105 per well onto six-well culture plates and maintained in the DMEM in a humidified atmosphere of 5% CO2 at 37°C. On the following day, BEAS-2B cells were treated with human recombinant TGF-β1 (2 μg/mL, rTGF-β1, PHG9204, Gibco, United States) at the concentration of 5 ng/mL, 10 ng/mL, 100 ng/mL, and 1,000 ng/mL for 48 h, respectively. Cells cultured in the absence of rTGF-β1 served as the control. After the treatment, cell extracts were collected for further analyses.
2.3 Lentivirus production
A human genome-wide CRISPR knockout pooled library (GeCKO v2) targeting 19,050 genes and 1,864 microRNAs (miRNAs) was purchased from the Addgene (#1000000048, United States). To produce the lentivirus for the GeCKO v2 sgRNA library, 3 × 106 293 TN cells were transduced with 9 μg of GeCKO v2 plasmids, 3 μg of pCMV-VSV-G, 3 μg of pMDLg, 3 μg of pRRE, and 3 μg of pRSV-Rev in a 150 mm dish containing 90 μL polyethylenimine. After 6 h, cells were washed with PBS, and fresh complete DMEM was added. The lentiviral supernatant was harvested after 48 h of incubation at 37°C and 5% CO2. The supernatant was then centrifuged at 3,000 rpm for 15 min at 4°C and stored for further use.
2.4 Genome-wide CRISPR/Cas9 knockout library screening
Genome-Cas9-edited BEAS-2B (BEAS-2B-GeCKO) cells were generated as previously described (Wei et al., 2019). Briefly, BEAS-2B cells were transduced with GeCKO v2 sgRNA library at a low MOI (∼0.3). After incubating with puromycin for 14 days to eliminate untransduced cells, viable cells were collected to evaluate the transduction efficiency. (With regard to the knockout of individual candidate genes, corresponding gRNAs were introduced into the lentiCRISPR v2.1 vector (Supplementary Table S1), followed by the cell transduction.) Thereafter, 2 × 107 BEAS-2B-GeCKO cells were treated with rTGF-β1 (10 ng/mL). We performed four consecutive rounds of TGF-β1 selection according to the EMT readout assay. After four rounds of rTGF-β1 treatment, viable cells were collected for subsequent analyses. Genomic DNA was extracted from TGF-β1-resistant BEAS-2B-GeCKO cells using the HiPure Tissue DNA Kit (Magen, China). The single guide RNA (sgRNA) sequences were amplified by polymerase chain reaction (PCR). The sequence of forward primer is 5′-TCTTGTGGAAAGGACGAAACACCG-3’; The sequence of reverse primer is 5′-ACCTTCTCTAGGCACCGGAT-3’. The gRNA sequences were amplified using 2 × PrimStar MasterMix (Takara, Japan) through the PCR. The conditions of PCR were as follows: 98°C for 30 s; 35 cycles (98°C for 10 s; 60°C for 10 s; 72°C for 50 s); 72°C for 1 min; 4°C. The distribution of sgRNA was assessed by Illumina next-generation sequencing (NGS).
2.5 Screening enrichment analysis
Following the NGS, the top-scoring gene hits were identified by applying a threshold of total reads of sgRNAs>300 and the sgRNA diversity (the counts of sgRNAs targeting each gene) > 1. In addition, a dataset (GSE104908) was downloaded from the Gene Expression Omnibus database and differentially expressed genes (LogFC>2, q < 0.05) were chosen for analyses. A Venn diagram was constructed using the R software v4.3.3 to compare the overlap between the top-scoring genes and GSE104908. To investigate the expression levels of the identified key profibrotic regulators in patients with pulmonary fibrosis, the GSE40839 and GSE24206 datasets were also downloaded from the Gene Expression Omnibus database. Prior to the statistical analysis, the gene chip data were subjected to deduplication, log2 transformation, and quality control. The Mann-Whitney U Test was applied to analyze the expression levels of key profibrotic regulators between the pulmonary fibrosis patients and control groups. Data were visualized in violin plots using the R software v4.3.3. To identify significantly enriched signaling pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were conducted using the R software v4.3.3. To characterize the biological process, cellular component, and molecular function, the Gene ontology (GO) analysis was performed using the WEB-based Gene Set Analysis Toolkit (https://www.webgestalt.org/). The STRING was used to visualize the protein-protein interaction (PPI) network.
2.6 RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
First, the 1 mL of Trizol was added to cells (5 × 106) and vortexed briefly. Then 200 μL of chloroform was added to the cell lysate, followed by incubation (5 min at room temperature) and centrifugation (10,000 rpm, 5 min at room temperature). After centrifugation, 500 μL isopropanol was added to the aqueous phase and mixed gently. Samples were then centrifuged, and the supernatant was discarded. The pellet was subsequently washed with 70% ethanol, and the supernatant was removed after centrifugation. The pellet was allowed to air dry and resuspended in 20 μL of RNase-free water. The purity and concentration of RNA was measured using the NanoDrop 2000 (Thermo Fisher Scientific; United States). The cDNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific; United States) according to the manufacturer’s instructions. The RT-qPCR was carried out using the PowerUp SYBR Green master mix (Applied Biosystems; United States) and target-specific primers (shown below) on a CFX96 Touch Real-Time PCR Detection System (BIO-RAD; United States). The β-actin was used as the reference gene for normalization. Relative mRNA expression of the targets was calculated as the fold change relative to PBS-treated cells using the delta-delta Ct method (2−ΔΔCT). Sequences of primers for target genes are listed below:
β-actin FW: TGGCACCAGCACAATGAA.
β-actin RV: CTAAGTCATAGTCCGCCTAGAAGC.
Fibronectin FW: AGCAAGCCCGGTTGTTATGA.
Fibronectin RV: CCCACTCGGTAAGTGTTCCC.
Collagen-I FW: ATTCCAGTTCGAGTATGGCGG.
Collagen-I RV: CTTGAGGTTGCCAGTCTGCT.
2.7 Cell transduction and Western blotting
The pLKO.1-shRNA constructs targeting COL20A1, COL27A1, and WNT11 were purchased from IGEbio (China). The stable knockdown of these three genes was conducted through the lentiviral transduction of shRNA expressing virus, followed by puromycin selection. The pLKO.1-scramble shRNA was used as the control. The sequences of shRNAs are the following:
shRNA-COL20A1: GCTACACCTTGCAGATCTTCG.
shRNA-COL27A1: GCAGTGGCTATTCGATCTTCC.
shRNA-WNT11: GCCTGTGAAGGACTCGGAACT.
shRNA-scrambled: CCTAAGGTTAAGTCGCCCTCG.
After the rTGF-β1 treatment, cells were washed twice with ice-cold PBS. Cells were then lysed by adding radioimmunoprecipitation assay buffer (Beyotime, China) with the protease inhibitor (Beyotime, China), followed by centrifugation (12,000 rpm, 15 min at 4°C). After discarding the supernatant, the loading buffer (EpiZyme, United States) was added, and cells were heated at 95°C for 10 min. Samples were electrophoresed in the 10% Sodium Dodecyl Sulfate polyacrylamide gels, and proteins were subsequently transferred to polyvinylidene difluoride membranes. Membranes were blocked with skimmed milk for 2 h at room temperature and incubated with primary antibodies overnight at 4°C. On the next day, polyvinylidene difluoride membranes were washed with TBST (100 mM NaCl, 10 mM Tris, 0.1% Tween 20), and blots were incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. After washing the TBST, membranes were developed with ECL chemiluminescent substrate (Tanon, China). Data quantification was conducted using the Fiji software (ImageJ, United States). Relative protein levels were calculated after normalization to GAPDH.
Antibodies used are as follows: rabbit anti-Fibronectin (proteintech #15613-1-AP, 1:2000), mouse anti-Collagen Type I (proteintech #67288-1-Ig, 1:5000), and mouse anti-GAPDH (proteintech #60004-1-Ig, 1:10,000). HRP-conjugated secondary antibodies include goat anti-mouse (proteintech #RGAM001, 1:5000) and goat anti-rabbit (proteintech # RGAR001, 1:5000).
2.8 CCK-8 assay
The cell viability was assessed using the cell counting kit-8 (CCK-8, Beyotime, China). Cells (3 × 103 per well) were seeded onto 96-well plates in triplicate and incubated at 37% with 5% CO2 for 24 h. Cells were then treated with 10 ng/mL of rTGF-β1 for 48 h. Subsequently, 10 μL of CCK-8 solution was added, followed by an additional 2 h of incubation. The absorbance at 450 nm was detected using a microplate reader (BioTeke, United States).
2.9 Cell migration assay
Cells (1 × 104) were seeded onto the transwell inserts (Corning, United States) containing 200 μL of serum-free DMEM. The DMEM containing 20% fetal bovine serum (500 μL) was added into the lower chamber. After 48 h of rTGF-β1 treatment (10 ng/mL), cells that migrated/invaded into the lower surface were washed with sterile PBS, fixed with 4% paraformaldehyde (20 min), and stained with 0.1% crystal violet (15 min). Excess crystal violet and unfixed cells were then washed away with PBS, followed by the observation under a bright-field light microscopy (Olympus, Japan) at ×100 magnification. Relative cell migration was calculated as the ratio of migrated cells in experimental groups to the background migration in the control group.
2.10 Cell apoptosis analysis
Cells (1 × 105 per well) were seeded onto six-well culture plates and cultured overnight. Cells were then treated with rTGF-β1 (10 ng/mL) for 48 h. Subsequently, cells were stained with 5 μL of Annexin V-APC (BioLegend, United States) and 10 μL of 7-amino-actinomycin D (eBioscience, United States) for 15 min at room temperature. The apoptotic cells were determined and quantified by flow cytometry, and data were analyzed using the FlowJo software (Tree Star, United States).
2.11 Statistical analysis
All data were expressed as mean ± SEM. Data analysis was performed using the GraphPad Prism 9.30 (GraphPad Software, United States). Data were analyzed using unpaired two-tailed t-test or Mann-Whitney U Test for comparison between two groups. The one-way analysis of variance (ANOVA) with Tukey’s post-test was applied for multiple comparisons. The p value < 0.05 was considered statistically significant.
3 Results
3.1 Identification of EMT and ECM-generating phenotype in BEAS-2B and BEAS-2B-GeCKO cells
To explore the successful induction of EMT in human bronchial epithelial cells and its potential association with ECM synthesis and deposition, we aimed to establish a TGF-β1-induced EMT in BEAS-2B cells and measured the level of Collagen-I and Fibronectin, both of which are essential components of ECM (Zhang et al., 2018). To identify the optimal concentration of rTGF-β1, BEAS-2B cells were treated with different doses of rTGF-β1 (0, 5, 10, 100, and 1,000 ng/mL) for 48 h, followed by detecting the mRNA expression of Collagen-I and Fibronectin. As shown in Figure 1A, the mRNA expression levels of Collagen-I and Fibronectin gradually increased with the rise of rTGF-β1 concentration, peaking at 10 ng/mL. This result demonstrates the presence of an ECM-generating phenotype in BEAS-2B cells and the successful induction of EMT. However, a significant decrease in the expression of Collagen-I and Fibronectin was observed at 100 ng/mL and 1,000 ng/mL (Figure 1A), possibly due to the detrimental effect of higher TGF-β1 concentrations on cell viability. We therefore confirmed the use of 10 ng/mL of rTGF-β1 for subsequent experiments. After four consecutive rounds of rTGF-β1 treatment, higher levels of Collagen-I and Fibronectin mRNA expression were found in BEAS-2B-GeCKO cells compared with control. This result confirms the successful induction of EMT in BEAS-2B-GeCKO cells, paving the way for the establishment of the GeCKO screening (Figure 1B).
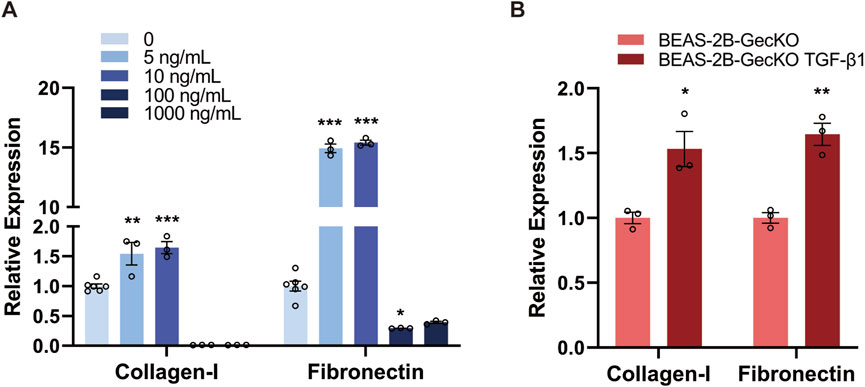
Figure 1. TGF-β1 treatment induces the EMT and ECM-generating phenotype in BEAS-2B and BEAS-2B-GeCKO cells. (A) The mRNA expression of Collagen-I and Fibronectin in BEAS-2B cells was determined by RT-qPCR following treatment with different doses of rTGF-β1. The optimized concentration of rTGF-β1 (10 ng/mL) was confirmed for subsequent experiments. (B) The mRNA expression of Collagen-I and Fibronectin in BEAS-2B-GeCKO cells was determined by RT-qPCR following rTGF-β1 treatment. Data are expressed as mean ± SEM, and analyzed using one-way ANOVA with Tukey’s multiple comparisons test (A) or unpaired two-tailed t-test (B). In (A), n = 6 for the control group; n = 3 for rTGF-β1 treated groups; In (B), n = 3 per group. * indicates significant difference compared with the control. *P < 0.05, **P < 0.01, ***P < 0.001. GeCKO, Genome-wide CRISPR/Cas9 knockout; TGF-β1, transforming growth factor-beta 1.
3.2 Conditional screening of BEAS-2B-GeCKO cells and identification of top molecular regulators mediating EMT and pulmonary fibrosis
To uncover new molecular regulators of the TGF-β1-induced EMT, BEAS-2B cells were transduced with GeCKO v2 sgRNA library to generate the BEAS-2B-GeCKO cells. The Figure 2A illustrates the schematic flow of EMT induction by treating BEAS-2B-GeCKO cells with rTGF-β1 and subsequent high-throughput sequencing. We analyzed two samples in the testing set using the NGS. For each sample, a minimum of 5.1 GB of data were generated. A total of 19,050 genes and 1,864 miRNAs were detected through the NGS, in line with the number of genes and miRNAs targeted by the GeCKO pooled library. Amplified sgRNA fragments were able to be detected in BEAS-2B-NC cells and BEAS-2B-GeCKO cells as compared to BEAS-2B cells, proving that the GeCKO v2 sgRNA library was successfully transduced into BEAS-2B cells (Figure 2B). By analyzing 14M Illumina NGS reads from 3.6×107 BEAS-2B-GeCKO cells post the rTGF-β1 selection, we observed the enrichment of 70,255 sgRNAs, representing 57% of total 123,411 sgRNAs in the GeCKO library. In addition, we detected an enrichment of 20,368 genes, including miRNAs, with 828 or 886 genes represented by six independent sgRNAs in the library, 3,247 or 3,132 genes represented by five independent sgRNAs, 5,553 or 5,607 genes represented by four independent sgRNAs, 5,755 or 5,861 genes represented by three independent sgRNAs, and 3,546 or 3,436 genes represented by two independent sgRNAs (Figures 2C, D).
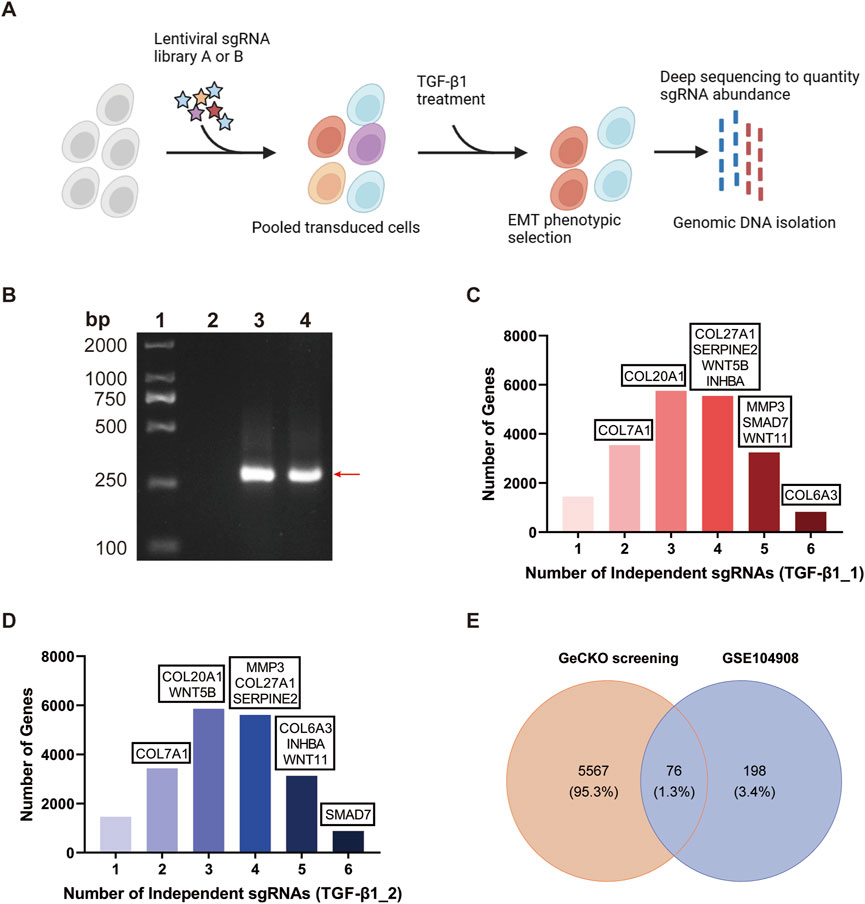
Figure 2. Conditional screening of BEAS-2B-GeCKO cells identifies 76 top molecular regulators mediating EMT and pulmonary fibrosis. (A) Schematic flow of generating BEAS-2B-GeCKO cells and subsequent TGF-β1 treatment and NGS. (B) A representative image demonstrating the successful transduction of GeCKO v2 sgRNA library into BEAS-2B cells. From the left to right, strips represent the markers, and the detection of GeCKO v2 sgRNA library in BEAS-2B cells, BEAS-2B-NC cells, and BEAS-2B-GeCKO cells, respectively. (C, D) The number of enriched genes represented by 1, 2, 3, 4, 5, or six independent sgRNAs. Genes were identified by total reads of sgRNAs and sgRNA diversity. (E) The Venn diagram analysis of data from our GeCKO screening and GSE104908 confirmed 76 top molecular regulators. EMT, epithelial mesenchymal transformation; GeCKO, Genome-wide CRISPR/Cas9 knockout; sgRNA, single guide RNA; TGF-β1, transforming growth factor-beta 1.
Among the enriched genes, we identified a total of 5,643 top-scoring genes by applying the threshold of total reads of sgRNAs>300 and sgRNA diversity>1. This result suggests the potential profibrotic properties of these top-scoring genes and demonstrates that our GeCKO screening can accurately screen essential and functional molecular regulators involved in the TGF-β1-induced EMT. To identify crucial molecular regulators for further study, we compared our GeCKO screening data with the differentially expressed genes from GSE104908 (Figure 2E). Through the analysis of Venn diagram, 76 top molecular regulators were confirmed (Figure 2E).
3.3 Pathway and functional enrichment analysis
To explore the PPI network among the identified top molecular regulators, we conducted the STRING analysis. As illustrated in Figure 3A, the PPI network comprising 76 nodes and 53 edges was constructed by applying a threshold of p < 0.05 and interaction score>0.4 (Disconnected nodes were hidden). Moreover, we performed KEGG pathway analysis for the top molecular regulators, and results exhibited enrichment in the TGF-β signaling pathway as well as other pathways associated with ECM dysregulation and EMT (e.g., focal adhesion and ECM-receptor interaction) (Figure 3B). Notably, we observed the enrichment of INHBA and found that the INHBA is involved in the TGF-β signaling pathway (Figure 3B and Supplementary Table S2). This result is in line with prior studies demonstrating the crosstalk between INHBA, EMT initiation and TGF-β pathway activation (Yu et al., 2021). In addition, the enrichment of COL6A3 was found, and it participates in ECM-receptor interaction, focal adhesion, and the PI3K-Akt signaling pathway (Figure 3B and Supplementary Table S2), all of which are associated with the pathogenesis of pulmonary fibrosis. The WNT11 and Wnt signaling pathway were also found to be enriched (Supplementary Table S2). Furthermore, the top-ranked biological process, cellular component, and molecular function of those top molecular regulators were analyzed through the GO analysis. As shown in Figure 3C and Supplementary Table S3, other molecular regulators (e.g., COL20A1, COL27A1, COL6A3, SMAD7, and WNT11) are involved in ECM structural constituent, collagen trimer, ECM component, adherens junction, collagen-containing ECM, as well as ECM organization. (Detailed information of KEGG and GO analyses is provided in Supplementary Tables S2, S3) Altogether, as these enriched genes are closely associated with ECM accumulation and TGF-β pathway activation, we therefore deduced that they may play a central role in regulating the TGF-β1-induced EMT and pulmonary fibrosis.
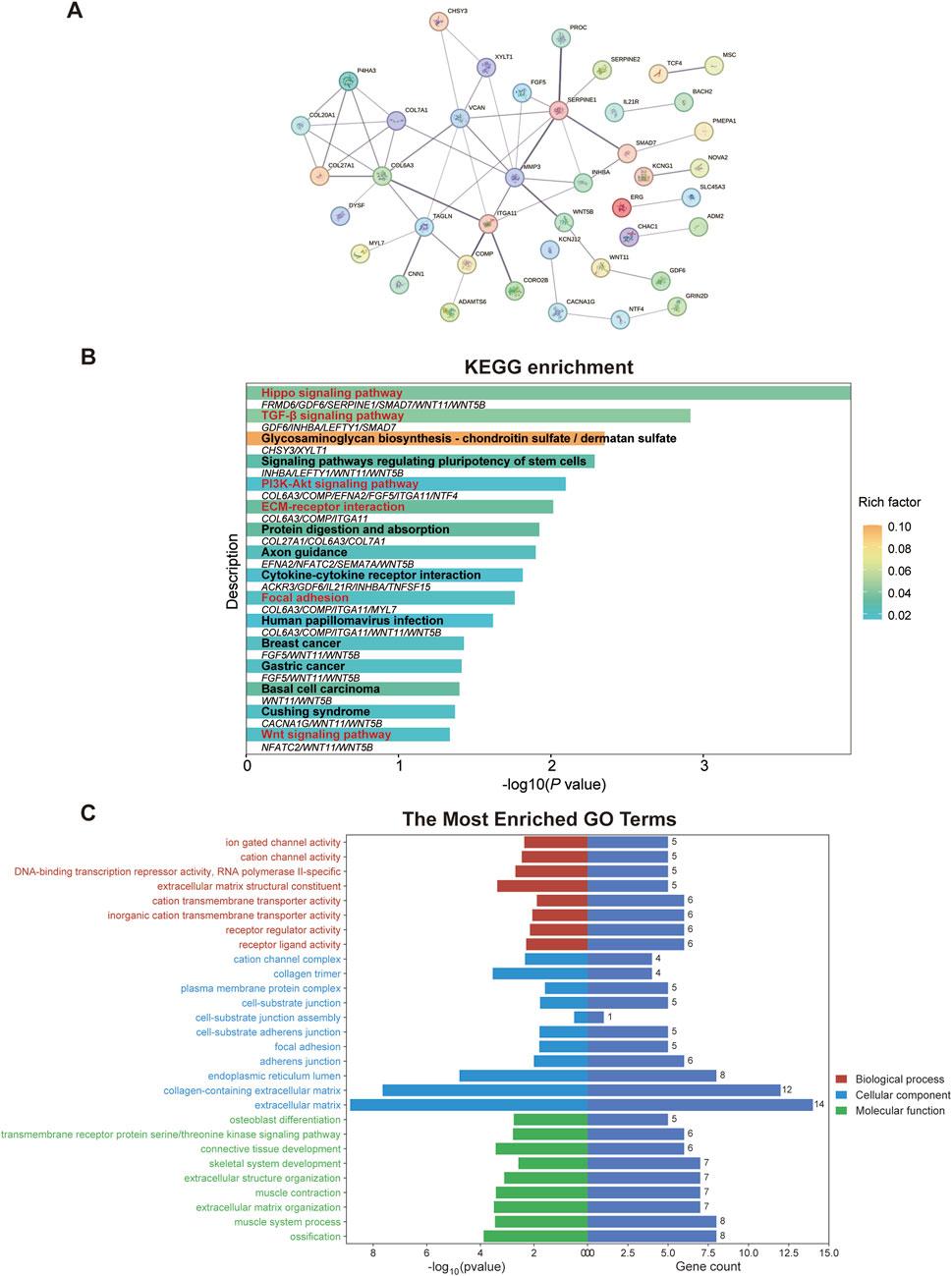
Figure 3. Pathway and functional enrichment analysis of top molecular regulators reveals a strong association with EMT and ECM dysregulation. (A) Protein-protein interaction network among the top molecular regulators was constructed in the STRING database. (B) KEGG pathway analysis of the top molecular regulators. Signaling pathways that are closely associated with ECM dysregulation, EMT, and pulmonary fibrosis are highlighted in red. Genes enriched in corresponding pathways are shown below the bars. (C) Through the GO analysis of top molecular regulators, the enriched GO terms indicate the close correlation with ECM. ECM, extracellular matrix; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PI3K-Akt, phosphatidylinositol 3-kinase-protein kinase B; TGF-β, transforming growth factor-beta.
3.4 Confirmation of the candidate genes and the effect of their knockout on TGF-β1-induced EMT
By integrating results of the Venn diagram with findings from pathway and functional enrichment analysis, the candidate genes (COL6A3, MMP3, WNT11, SMAD7, COL27A1, WNT5B, SERPINE2, INHBA, COL20A1, and COL7A1) were finally confirmed. The sgRNA counts for each candidate gene are shown in Figure 4A.
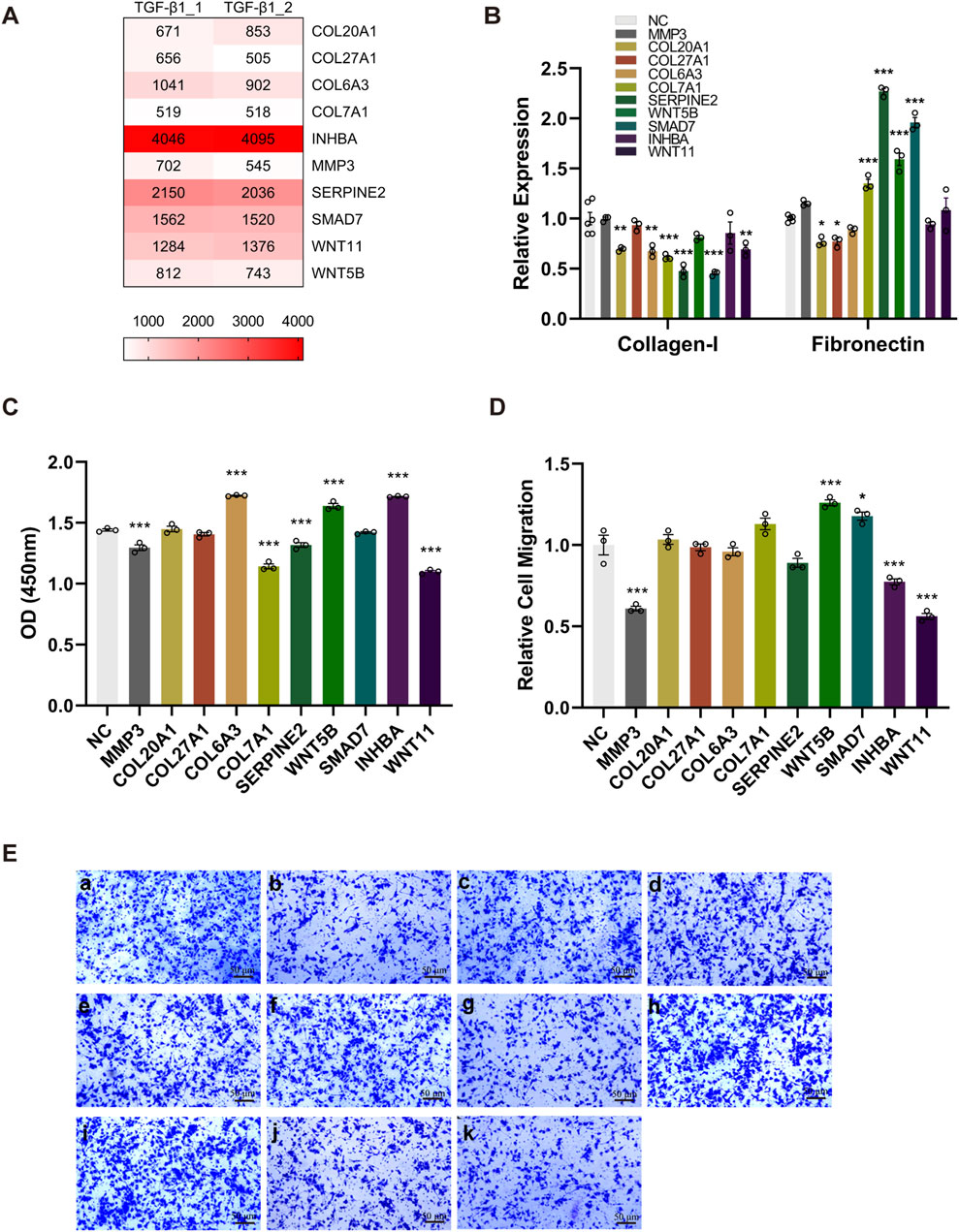
Figure 4. The knockout of candidate genes alters the phenotype of TGF-β1-induced EMT. Ten BEAS-2B cell lines were generated by specifically knocking out each candidate gene, followed by the rTGF-β1 treatment. PBS-treated cell lines served as the control. (A) A heatmap displaying the sgRNA counts of the candidate genes. (B) Upon the knockout of candidate genes, alterations in the mRNA expression of Collagen-I and Fibronectin was determined via RT-qPCR. (C) The effect of knocking out each candidate gene on cell viability was evaluated by CCK-8 assay. (D, E) Compared with control group (a), the effect of knocking out MMP3 (b), COL20A1 (c), COL27A1 (d), COL6A3 (e), COL7A1 (f), SERPINE2 (g), WNT5B (h), SMAD7 (i), INHBA (j), and WNT11 (k) on relative cell migration was assessed using the transwell system (magnification: ×100). Relative cell migration was calculated as the ratio of migrated cells in experimental groups to the background migration in the control group. Data are expressed as mean ± SEM, and analyzed using one-way ANOVA with Tukey’s multiple comparisons test. In (B), n = 6 for the control group; n = 3 for other groups (candidate gene knockout); In (C) and (D), n = 3 per group. * indicates significant difference compared with the control. *P < 0.05, **P < 0.01, ***P < 0.001. NGS, next-generation sequencing; OD, optical density; sgRNA, single guide RNA; TGF-β1, transforming growth factor-beta 1.
To assess whether these candidate genes play a key role in regulating the TGF-β1- induced EMT, we generated 10 BEAS-2B cell lines through the specific knockout of every single candidate gene, followed by the rTGF-β1 treatment. As presented in Figure 4B, the knockout of candidate genes (COL20A1, COL6A3, COL7A1, SERPINE2, SMAD7, and WNT11) greatly decreased the mRNA expression levels of Collagen-I. Moreover, although higher mRNA levels of Fibronectin were detected after knocking out the COL7A1, SERPINE2, WNT5B, and SMAD7, the substantial reduction in Fibronectin mRNA was observed upon the knockout of COL20A1 and COL27A1 (Figure 4B). In addition, post the rTGF-β1 treatment, a significant reduction in cell viability was observed when knocking out the candidate genes (MMP3, COL7A1, SERPINE2, and WNT11) compared with the control (Figure 4C). In contrast, the knockout of candidate genes (COL6A3, WNT5B, and INHBA) in BEAS-2B cells promoted cell proliferation (Figure 4C). Furthermore, Figures 4D, E show that the relative migration/invasion abilities of BEAS-2B cells following rTGF-β1 treatment were inhibited upon the knockout of MMP3, INHBA, and WNT11, whereas the knockout of WNT5B and SMAD7 facilitated the migration/invasion of BEAS-2B cells.
3.5 The effect of knocking out key profibrotic regulators on cell apoptosis and ECM components deposition
Based on the ten BEAS-2B cell lines with specific knockout of every single candidate gene, we performed the flow cytometry to assess whether the loss-of-function of candidate genes affected the cell apoptosis. As shown in Figures 5A, B, only the knockout of COL20A1, COL27A1, and COL7A1 greatly enhanced the rate of apoptosis in BEAS-2B cells after rTGF-β1 treatment, whereas knocking out the COL6A3 led to a decrease in the rate of apoptosis. Moreover, after suppressing the expression of COL20A1, COL27A1, and WNT11, the relative protein levels of Fibronectin and Collagen-I (key fibrosis-regulating proteins) were diminished (Figure 5C). These results prove that the COL20A1, COL27A1, and WNT11 may serve as key profibrotic regulators and contribute to the ECM deposition and TGF-β1-induced EMT.
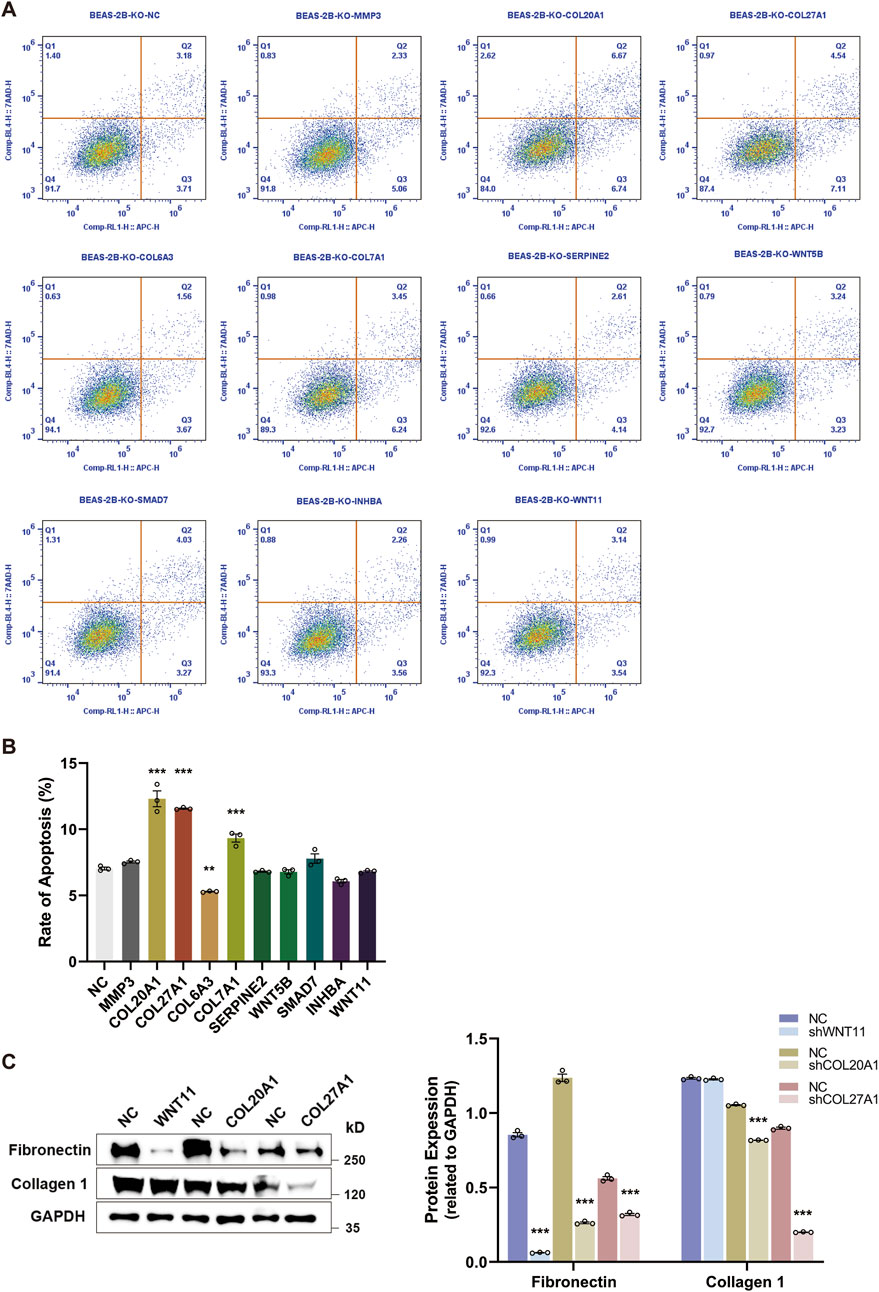
Figure 5. The knockout of key profibrotic regulators promotes cell apoptosis and reduces ECM components deposition. Ten BEAS-2B cell lines were generated by specifically knocking out each candidate gene, followed by the rTGF-β1 treatment. PBS-treated cell lines served as the control. (A) Representative plots of cell apoptosis for BEAS-2B cell lines. (B) Upon the knockout of candidate genes, the rate of apoptosis for BEAS-2B cell lines was analyzed by flow cytometry. (C) After suppressing the expression of key profibrotic regulators (COL20A1, COL27A1, and WNT11), alterations in the protein expression levels of Fibronectin and Collagen-I were analyzed and quantified by Western blot. Analysis of protein levels in relation to GAPDH. Data are expressed as mean ± SEM, and analyzed using one-way ANOVA with Tukey’s multiple comparisons test (B) or unpaired two-tailed t-test between each condition (C). In (B) and (C), n = 3 per group. * indicates significant difference compared with the control. **P < 0.01, ***P < 0.001. KO, knockout; NC, negative control.
3.6 The expression of COL20A1, COL27A1, and WNT11 in patients with pulmonary fibrosis
To further consolidate the findings of our study, we analyzed the expression levels of COL20A1, COL27A1, and WNT11 in patients with pulmonary fibrosis. As presented in Figures 6A, B, although there were no differences in the expression of COL20A1 between IPF patients and the control in GSE24206 dataset, a substantial increase in COL27A1 expression was observed in IPF patients. In addition, the WNT11 expression was significantly higher in patients with pulmonary fibrosis as compared with the control group in GSE40839 dataset (Figure 6C).
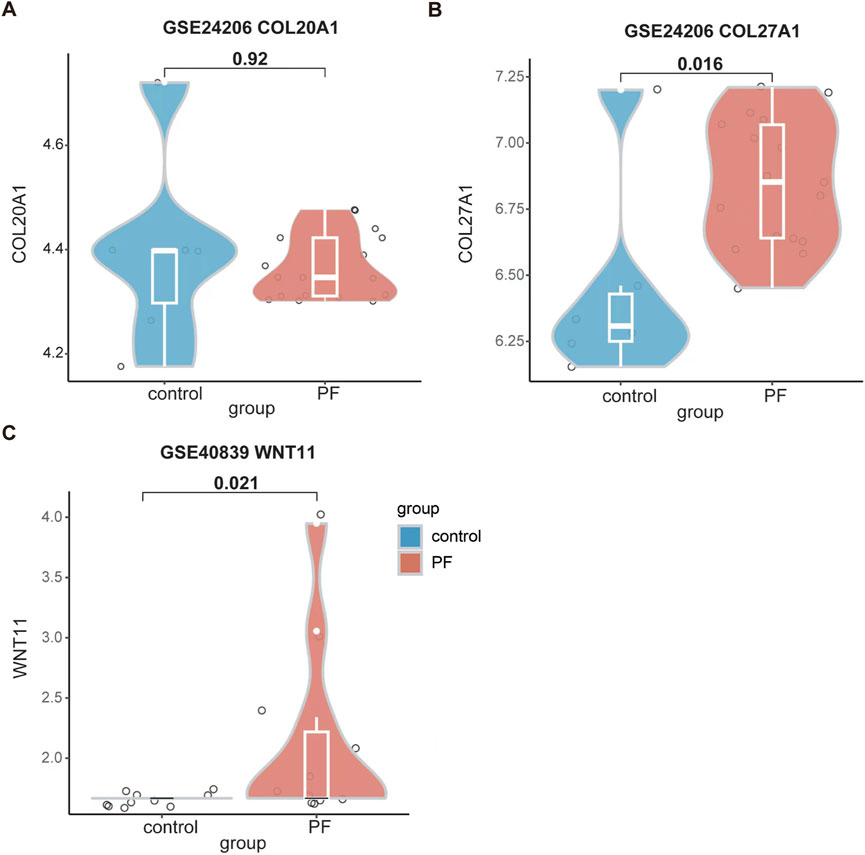
Figure 6. Enhanced expression levels of COL27A1 and WNT11 were found in patients with pulmonary fibrosis. (A) The expression level of COL20A1 in GSE24206. (B) The expression level of COL27A1 in GSE24206. (C) The expression level of WNT11 in GSE40839. Data are shown in violin plots, and analyzed using Mann-Whitney U Test. In (A) and (B), n = 6 for the control group; n = 17 for the PF group. In (C), n = 10 for the control group; n = 11 for the PF group. All the violin plots were generated using R software v4.3.3. PF, pulmonary fibrosis.
Altogether, all of our findings demonstrate that the COL20A1, COL27A1, and WNT11 serve as key profibrotic regulators and may play a crucial role in regulating the TGF-β1-induced EMT and the pathogenesis of pulmonary fibrosis.
4 Discussion
IPF is characterized by aberrant remodeling of lung parenchyma (Li et al., 2023) that results from the chronic, continuous tissue repair process with excessive formation and deposition of collagen-rich ECM (Gauldie et al., 2002). Given that the ECM is mainly produced by fibroblasts and myofibroblasts (D'Urso and Kurniawan, 2020; Mei et al., 2021), most scholars believe that these cells are derived from resident tissue fibroblasts (Phan, 2008) and, importantly, from the epithelial cells undergoing EMT (Willis et al., 2006). Therefore, elucidating EMT-related signaling pathways and studying medications that target EMT may provide potential therapeutic strategies against the onset and progression of IPF. In order to explore the key molecular regulators of EMT and pulmonary fibrosis, the genome-wide CRISPR/Cas9 knockout technique was utilized in this research. The CRISPR/Cas9 based screening technique has proved high reagent consistency, strong phenotypic effects, and high validation rates (Shalem et al., 2015). It is a reliable and powerful method to realize genome-level analyses and to identify important genes and signaling pathways associated with the phenotype of interest, which has been used in a variety of diseases, including the organ fibrosis (Turner et al., 2020).
In the present study, we investigated the key molecular regulators of EMT by performing the GeCKO screening in TGF-β1-treated BEAS-2B cells to evaluate their phenotypic readouts, such as the abilities of cell migration/invasion and deposition of ECM components (Collagen-I and Fibronectin). By integrating the GeCKO screening results, pathway and functional enrichment analysis, and data from GSE104908, we confirmed 10 candidate genes. Among the identified candidate genes, the COL6A3 (Nance et al., 2014), COL7A1 (Horimasu et al., 2017), WNT5B (Malizia et al., 2009), and MMP3 (DePianto et al., 2015) have been demonstrated to be associated with pulmonary fibrosis. Importantly, these aforementioned genes are involved in ECM-receptor interaction, TGF-β signaling pathway, focal adhesion, adherens junction, and Hippo signaling pathway (Figure 3). Besides the ECM-receptor interaction and TGF-β signaling pathway, the focal adhesion (Zhao et al., 2016), adherens junction (Lappi-Blanco et al., 2013), and Hippo signaling pathway (Liu et al., 2015) have also been proven to contribute to the EMT and pathogenesis of pulmonary fibrosis. Therefore, these results provide a comprehensive understanding of the critical role of these genes in the EMT and pulmonary fibrosis, paving the way for the subsequent loss-of-function experiment.
Among the confirmed candidate genes, the specific knockout of COL20A1, COL27A1, and WNT11 in TGF-β1-treated BEAS-2B cells led to increased rate of cell apoptosis and decreased protein deposition of ECM components (Figure 5), particularly Collagen-I. As the Collagen-I plays a crucial role in fibrogenesis (Yang et al., 2013) and the pulmonary fibrosis is characterized by its accumulation (Ding et al., 2021), the reduction in Collagen-I deposition caused by knocking out the COL20A1, COL27A1, and WNT11 may indicate a potential direction for pulmonary fibrosis treatment. Furthermore, the cell viability and migration were also inhibited when knocking out the WNT11 (Figure 4). Importantly, significantly higher expression of COL27A1 and WNT11 was found in patients with pulmonary fibrosis (Figure 6). These results provide strong evidence that these three candidate genes play a key role in EMT and the pathogenesis of pulmonary fibrosis. Interestingly, enhanced expression of the collagen protein coding gene-COL20A1 was detected in experimental models of pulmonary fibrosis (Nguyen et al., 2021; Lamichhane et al., 2022). It is noteworthy that, in contrast to these studies, we performed the loss-of-function experiment on COL20A1 to further verify its property of promoting ECM deposition in our model of TGF-β1-treated human bronchial epithelial cells, demonstrating the crucial role of COL20A1 in regulating the EMT and pulmonary fibrosis. Additionally, in line with our findings, a strong upregulation of COL27A1 was also found in IPF lungs (Selman et al., 2006), indicating its potential role in mediating the onset and progression of pulmonary fibrosis. Indeed, our loss-of-function experiment further confirmed the key role of COL27A1 in facilitating ECM deposition and TGF-β1-induced EMT because the specific knockout of COL27A1 attenuated the deposition of ECM components. Of note, apart from its role in ECM deposition, the expression of COL27A1 has been reported to be associated with apoptosis to maintain the tissue homeostasis (Gonzaga-Jauregui et al., 2020). Since elevated levels of cell apoptosis were observed upon the knockout of COL27A1, we assume that the enhanced expression of COL27A1 may reduce cell apoptosis, thereby promoting the survival and proliferation of cells with ECM-generating phenotype and then resulting in the development of pulmonary fibrosis. Consistent with our assumptions, impaired apoptosis in myofibroblasts (Chang et al., 2010) and apoptosis-resistant fibroblasts (Ajayi et al., 2013) are found in the IPF lung. Surprisingly, blocking the inhibitor of apoptosis proteins abrogated bleomycin-induced pulmonary fibrosis (Ashley et al., 2016), suggesting that induction of apoptosis in fibrosis effector cells may serve as an alternative therapeutic strategy for the treatment of IPF. With regard to the WNT11, a previously published study reported a remarkable increase of WNT11 in renal epithelial cells following TGF-β1 treatment, and the WNT11 interacted with TGF-β for the development of renal fibrosis (Zhang et al., 2012). Despite that prior studies have investigated the role of WNT11 and Wnt signaling in both in vivo and in vitro models of pulmonary fibrosis (Henderson et al., 2010; Liu et al., 2021), the direct molecular targets or downstream signaling of WNT11 and the mechanism by which WNT11 interacts with other profibrotic molecules to regulate the pathogenesis of pulmonary fibrosis still remain unclear. This requires to be further explored by future studies. Moreover, although our results show that significantly elevated expression of COL27A1 and WNT11 was found in pulmonary fibrosis patients, we did not find any diagnostic or prognostic information related to these genes, indicating that further in vivo experiments and preclinical studies require to be carried out to explore their potential diagnostic and prognostic value in pulmonary fibrosis. Notably, our results present that silencing WNT5B promoted the TGF-β1-induced EMT by enhancing cell viability and mRNA expression of Fibronectin, as well as the cell migration/invasion abilities. Nevertheless, the suppression of WNT5B mitigated the EMT in EBV-treated epithelial cells (Malizia et al., 2009). This suggests that the regulatory mechanism of some candidate genes needs to be further verified using in vivo models of pulmonary fibrosis. Collectively, by combining the GeCKO screening with loss-of-function experiment, our results provide a more profound and reliable validation on the profibrotic properties of the candidate genes identified in this study, and the COL20A1, COL27A1, and WNT11 were confirmed as key profibrotic regulators.
Our study has several limitations. First, due to limited resources, we focused on screening and investigating key profibrotic regulators of pulmonary fibrosis using the GeCKO screening and loss-of-function experiment in the present study. In our follow-up experiments, the overexpression of key profibrotic regulators and multiple experimental techniques will be applied to explore potential molecular targets and downstream profibrotic pathways that these key profibrotic regulators may interact with for the pathogenesis of IPF. Second, we performed the GeCKO screening and subsequent loss-of-function experiment in vitro, whereas the impact of systemic and organ milieu and the potential crosstalk between different effector cells cannot be overlooked in the onset and development of IPF. Therefore, we already established the mouse model of bleomycin-induced pulmonary fibrosis, and we are planning to further investigate the profibrotic properties of the key profibrotic regulators using the in vivo GeCKO library screening technique in our future research.
5 Conclusion
Through the combination of GeCKO library screening, pathway and functional enrichment analysis, and other experimental techniques, we identified 10 candidate genes for TGF-β1-induced EMT. By applying the loss-of-function experiment, the COL20A1, COL27A1, and WNT11 were confirmed as key profibrotic regulators of EMT and development of pulmonary fibrosis. Gaining understanding and insights into these profibrotic regulators may promote the identification of novel therapeutic targets for the prevention and treatment of IPF.
Data availability statement
The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA1221872.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
CT: Conceptualization, Methodology, Validation, Writing–original draft. JW: Data curation, Investigation, Methodology, Writing–original draft. XY: Investigation, Methodology, Writing–original draft. KK: Data curation, Software, Writing–review and editing. YeL: Validation, Visualization, Writing–review and editing. FL: Formal Analysis, Visualization, Writing–review and editing. HY: Formal Analysis, Project administration, Supervision, Writing–review and editing. YiL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Science and Technology Planning Project of Guangdong Province, grant number 2014A020212152.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2025.1507163/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Sequences and total reads of sgRNAs targeting the candidate genes.
SUPPLEMENTARY TABLE S2 | Results of KEGG pathway analysis for the 76 top molecular regulators.
SUPPLEMENTARY TABLE S3 | Results of enriched GO terms for the 76 top molecular regulators, categorized by biological process, cellular component, and molecular function.
Abbreviations
ANOVA, analysis of variance; CCK-8, cell counting kit-8; DMEM, Dulbecco’s Modified Eagle Medium; ECM, extracellular matrix; EMT, epithelial-mesenchymal transformation; GeCKO, genome-wide CRISPR/Cas9 knockout; GO, Gene ontology; IPF, idiopathic pulmonary fibrosis; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNAs, microRNAs; NGS, next-generation sequencing; PBS, phosphate buffered saline; PCR, polymerase chain reaction; PPI, protein-protein interaction; RT-qPCR, Reverse transcription-quantitative polymerase chain reaction; sgRNA, single guide RNA; TGF-β, transforming growth factor-beta.
References
Ajayi, I. O., Sisson, T. H., Higgins, P. D., Booth, A. J., Sagana, R. L., Huang, S. K., et al. (2013). X-linked inhibitor of apoptosis regulates lung fibroblast resistance to Fas-mediated apoptosis. Am. J. Respir. Cell Mol. Biol. 49 (1), 86–95. doi:10.1165/rcmb.2012-0224OC
Alvarez-Palomo, B., Sanchez-Lopez, L. I., Moodley, Y., Edel, M. J., and Serrano-Mollar, A. (2020). Induced pluripotent stem cell-derived lung alveolar epithelial type II cells reduce damage in bleomycin-induced lung fibrosis. Stem Cell Res. Ther. 11 (1), 213. doi:10.1186/s13287-020-01726-3
Antoniou, K., Markopoulou, K., Tzouvelekis, A., Trachalaki, A., Vasarmidi, E., Organtzis, J., et al. (2020). Efficacy and safety of nintedanib in a Greek multicentre idiopathic pulmonary fibrosis registry: a retrospective, observational, cohort study. ERJ Open Res. 6 (1). doi:10.1183/23120541.00172-2019
Ashley, S. L., Sisson, T. H., Wheaton, A. K., Kim, K. K., Wilke, C. A., Ajayi, I. O., et al. (2016). Targeting inhibitor of apoptosis proteins protects from bleomycin-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol. 54 (4), 482–492. doi:10.1165/rcmb.2015-0148OC
Chang, W., Wei, K., Jacobs, S. S., Upadhyay, D., Weill, D., and Rosen, G. D. (2010). SPARC suppresses apoptosis of idiopathic pulmonary fibrosis fibroblasts through constitutive activation of beta-catenin. J. Biol. Chem. 285 (11), 8196–8206. doi:10.1074/jbc.M109.025684
Collard, H. R., Ryerson, C. J., Corte, T. J., Jenkins, G., Kondoh, Y., Lederer, D. J., et al. (2016). Acute exacerbation of idiopathic pulmonary fibrosis. An international working group report. Am. J. Respir. Crit. Care Med. 194 (3), 265–275. doi:10.1164/rccm.201604-0801CI
DePianto, D. J., Chandriani, S., Abbas, A. R., Jia, G., N'Diaye, E. N., Caplazi, P., et al. (2015). Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax 70 (1), 48–56. doi:10.1136/thoraxjnl-2013-204596
Di Gregorio, J., Robuffo, I., Spalletta, S., Giambuzzi, G., De Iuliis, V., Toniato, E., et al. (2020). The epithelial-to-mesenchymal transition as a possible therapeutic target in fibrotic disorders. Front. Cell Dev. Biol. 8, 607483. doi:10.3389/fcell.2020.607483
Ding, H., Chen, J., Qin, J., Chen, R., and Yi, Z. (2021). TGF-β-induced α-SMA expression is mediated by C/EBPβ acetylation in human alveolar epithelial cells. Mol. Med. 27 (1), 22. doi:10.1186/s10020-021-00283-6
D'Urso, M., and Kurniawan, N. A. (2020). Mechanical and physical regulation of fibroblast-myofibroblast transition: from cellular mechanoresponse to tissue pathology. Front. Bioeng. Biotechnol. 8, 609653. doi:10.3389/fbioe.2020.609653
Gauldie, J., Kolb, M., and Sime, P. J. (2002). A new direction in the pathogenesis of idiopathic pulmonary fibrosis? Respir. Res. 3 (1), 1. doi:10.1186/rr158
Gonzaga-Jauregui, C., Yesil, G., Nistala, H., Gezdirici, A., Bayram, Y., Nannuru, K. C., et al. (2020). Functional biology of the Steel syndrome founder allele and evidence for clan genomics derivation of COL27A1 pathogenic alleles worldwide. Eur. J. Hum. Genet. 28 (9), 1243–1264. doi:10.1038/s41431-020-0632-x
Henderson, W. R., Chi, E. Y., Ye, X., Nguyen, C., Tien, Y. T., Zhou, B., et al. (2010). Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc. Natl. Acad. Sci. U. S. A. 107 (32), 14309–14314. doi:10.1073/pnas.1001520107
Horimasu, Y., Ishikawa, N., Taniwaki, M., Yamaguchi, K., Hamai, K., Iwamoto, H., et al. (2017). Gene expression profiling of idiopathic interstitial pneumonias (IIPs): identification of potential diagnostic markers and therapeutic targets. BMC Med. Genet. 18 (1), 88. doi:10.1186/s12881-017-0449-9
Hu, W., Pei, W., Zhu, L., Nie, J., Pei, H., Zhang, J., et al. (2018). Microarray profiling of TGF-β1-induced long non-coding RNA expression patterns in human lung bronchial epithelial BEAS-2B cells. Cell Physiol. Biochem. 50 (6), 2071–2085. doi:10.1159/000495052
Khor, Y. H., Ng, Y., Barnes, H., Goh, N. S. L., McDonald, C. F., and Holland, A. E. (2020). Prognosis of idiopathic pulmonary fibrosis without anti-fibrotic therapy: a systematic review. Eur. Respir. Rev. 29 (157), 190158. doi:10.1183/16000617.0158-2019
Kim, K. K., Kugler, M. C., Wolters, P. J., Robillard, L., Galvez, M. G., Brumwell, A. N., et al. (2006). Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl. Acad. Sci. U. S. A. 103 (35), 13180–13185. doi:10.1073/pnas.0605669103
Lamichhane, R., Patial, S., and Saini, Y. (2022). Higher susceptibility of males to bleomycin-induced pulmonary inflammation is associated with sex-specific transcriptomic differences in myeloid cells. Toxicol. Appl. Pharmacol. 454, 116228. doi:10.1016/j.taap.2022.116228
Lappi-Blanco, E., Lehtonen, S. T., Sormunen, R., Merikallio, H. M., Soini, Y., and Kaarteenaho, R. L. (2013). Divergence of tight and adherens junction factors in alveolar epithelium in pulmonary fibrosis. Hum. Pathol. 44 (5), 895–907. doi:10.1016/j.humpath.2012.08.016
Ley, B., Collard, H. R., and King, T. E. (2011). Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 183 (4), 431–440. doi:10.1164/rccm.201006-0894CI
Li, Z., Wang, S., Zhao, H., Yan, P., Yuan, H., Zhao, M., et al. (2023). Artificial neural network identified the significant genes to distinguish Idiopathic pulmonary fibrosis. Sci. Rep. 13 (1), 1225. doi:10.1038/s41598-023-28536-w
Liu, F., Lagares, D., Choi, K. M., Stopfer, L., Marinkovic, A., Vrbanac, V., et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 308 (4), L344–L357. doi:10.1152/ajplung.00300.2014
Liu, T., Gonzalez De Los Santos, F., Hirsch, M., Wu, Z., and Phan, S. H. (2021). Noncanonical Wnt signaling promotes myofibroblast differentiation in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 65 (5), 489–499. doi:10.1165/rcmb.2020-0499OC
Malizia, A. P., Lacey, N., Walls, D., Egan, J. J., and Doran, P. P. (2009). CUX1/Wnt signaling regulates epithelial mesenchymal transition in EBV infected epithelial cells. Exp. Cell Res. 315 (11), 1819–1831. doi:10.1016/j.yexcr.2009.04.001
Mei, Q., Liu, Z., Zuo, H., Yang, Z., and Qu, J. (2021). Idiopathic pulmonary fibrosis: an update on pathogenesis. Front. Pharmacol. 12, 797292. doi:10.3389/fphar.2021.797292
Nance, T., Smith, K. S., Anaya, V., Richardson, R., Ho, L., Pala, M., et al. (2014). Transcriptome analysis reveals differential splicing events in IPF lung tissue. PLoS One 9 (3), e92111. doi:10.1371/journal.pone.0092111
Nguyen, X. X., Renaud, L., and Feghali-Bostwick, C. (2021). Identification of impacted pathways and transcriptomic markers as potential mediators of pulmonary fibrosis in transgenic mice expressing human IGFBP5. Int. J. Mol. Sci. 22 (22), 12609. doi:10.3390/ijms222212609
Oh, R. S., Haak, A. J., Smith, K. M. J., Ligresti, G., Choi, K. M., Xie, T., et al. (2018). RNAi screening identifies a mechanosensitive ROCK-JAK2-STAT3 network central to myofibroblast activation. J. Cell Sci. 131 (10), jcs209932. doi:10.1242/jcs.209932
Onishchenko, D., Marlowe, R. J., Ngufor, C. G., Faust, L. J., Limper, A. H., Hunninghake, G. M., et al. (2022). Screening for idiopathic pulmonary fibrosis using comorbidity signatures in electronic health records. Nat. Med. 28 (10), 2107–2116. doi:10.1038/s41591-022-02010-y
Pfeifer, M., Brammeld, J. S., Price, S., Pilling, J., Bhavsar, D., Farcas, A., et al. (2024). Genome-wide CRISPR screens identify the YAP/TEAD axis as a driver of persister cells in EGFR mutant lung cancer. Commun. Biol. 7 (1), 497. doi:10.1038/s42003-024-06190-w
Phan, S. H. (2008). Biology of fibroblasts and myofibroblasts. Proc. Am. Thorac. Soc. 5 (3), 334–337. doi:10.1513/pats.200708-146DR
Raghu, G., Remy-Jardin, M., Richeldi, L., Thomson, C. C., Inoue, Y., Johkoh, T., et al. (2022). Idiopathic pulmonary fibrosis (an update) and progressive pulmonary fibrosis in adults: an official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 205 (9), e18–e47. doi:10.1164/rccm.202202-0399ST
Selman, M., Pardo, A., Barrera, L., Estrada, A., Watson, S. R., Wilson, K., et al. (2006). Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am. J. Respir. Crit. Care Med. 173 (2), 188–198. doi:10.1164/rccm.200504-644OC
Shalem, O., Sanjana, N. E., and Zhang, F. (2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16 (5), 299–311. doi:10.1038/nrg3899
Shi, N., Wang, Z., Zhu, H., Liu, W., Zhao, M., Jiang, X., et al. (2022). Research progress on drugs targeting the TGF-β signaling pathway in fibrotic diseases. Immunol. Res. 70 (3), 276–288. doi:10.1007/s12026-022-09267-y
Turner, R. J., Golz, S., Wollnik, C., Burkhardt, N., Sternberger, I., Andag, U., et al. (2020). A whole genome-wide arrayed CRISPR screen in primary organ fibroblasts to identify regulators of kidney fibrosis. SLAS Discov. 25 (6), 591–604. doi:10.1177/2472555220915851
Upagupta, C., Shimbori, C., Alsilmi, R., and Kolb, M. (2018). Matrix abnormalities in pulmonary fibrosis. Eur. Respir. Rev. 27 (148), 180033. doi:10.1183/16000617.0033-2018
Wei, L., Lee, D., Law, C. T., Zhang, M. S., Shen, J., Chin, D. W., et al. (2019). Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat. Commun. 10 (1), 4681. doi:10.1038/s41467-019-12606-7
Willis, B. C., duBois, R. M., and Borok, Z. (2006). Epithelial origin of myofibroblasts during fibrosis in the lung. Proc. Am. Thorac. Soc. 3 (4), 377–382. doi:10.1513/pats.200601-004TK
Xu, W. W., Liao, L., Dai, W., Zheng, C. C., Tan, X. P., He, Y., et al. (2023). Genome-wide CRISPR/Cas9 screening identifies a targetable MEST-PURA interaction in cancer metastasis. EBioMedicine 92, 104587. doi:10.1016/j.ebiom.2023.104587
Yang, J., Wheeler, S. E., Velikoff, M., Kleaveland, K. R., LaFemina, M. J., Frank, J. A., et al. (2013). Activated alveolar epithelial cells initiate fibrosis through secretion of mesenchymal proteins. Am. J. Pathol. 183 (5), 1559–1570. doi:10.1016/j.ajpath.2013.07.016
Ye, Z., and Hu, Y. (2021). TGF‑β1: gentlemanly orchestrator in idiopathic pulmonary fibrosis (Review). Int. J. Mol. Med. 48 (1), 132. doi:10.3892/ijmm.2021.4965
Yu, Y., Wang, W., Lu, W., Chen, W., and Shang, A. (2021). Inhibin β-A (INHBA) induces epithelial-mesenchymal transition and accelerates the motility of breast cancer cells by activating the TGF-β signaling pathway. Bioengineered 12 (1), 4681–4696. doi:10.1080/21655979.2021.1957754
Zhang, P., Cai, Y., Soofi, A., and Dressler, G. R. (2012). Activation of Wnt11 by transforming growth factor-β drives mesenchymal gene expression through non-canonical Wnt protein signaling in renal epithelial cells. J. Biol. Chem. 287 (25), 21290–21302. doi:10.1074/jbc.M112.357202
Zhang, R., Wu, Y., Xie, F., Zhong, Y., Wang, Y., Xu, M., et al. (2018). RGMa mediates reactive astrogliosis and glial scar formation through TGFβ1/Smad2/3 signaling after stroke. Cell Death Differ. 25 (8), 1503–1516. doi:10.1038/s41418-018-0058-y
Zhao, X. K., Cheng, Y., Liang Cheng, M., Yu, L., Mu, M., Li, H., et al. (2016). Focal adhesion kinase regulates fibroblast migration via integrin beta-1 and plays a central role in fibrosis. Sci. Rep. 6, 19276. doi:10.1038/srep19276
Keywords: idiopathic pulmonary fibrosis, epithelial-mesenchymal transformation, extracellular matrix, genome-wide CRISPR/Cas9 knockout screening, TGF-β1, COL20A1, COL27A1, WNT11
Citation: Tan C, Wang J, Ye X, Kasimu K, Li Y, Luo F, Yi H and Luo Y (2025) Genome-wide CRISPR/Cas9 screening identifies key profibrotic regulators of TGF-β1-induced epithelial-mesenchymal transformation and pulmonary fibrosis. Front. Mol. Biosci. 12:1507163. doi: 10.3389/fmolb.2025.1507163
Received: 07 October 2024; Accepted: 09 January 2025;
Published: 17 February 2025.
Edited by:
Rohit Upadhyay, Tulane University, United StatesReviewed by:
Raffaella Gallo, Magna Græcia University, ItalyAbhishek Mishra, University of Arkansas for Medical Sciences, United States
Copyright © 2025 Tan, Wang, Ye, Kasimu, Li, Luo, Yi and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Yi, eWlodWkyQG1haWwuc3lzdS5lZHUuY24=; Yifeng Luo, bHlpZkBtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Chunjiang Tan
Chunjiang Tan Juan Wang1,2†
Juan Wang1,2† Yifeng Luo
Yifeng Luo