
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 07 June 2024
Sec. Cellular Biochemistry
Volume 11 - 2024 | https://doi.org/10.3389/fmolb.2024.1420365
Protein methylation, similar to DNA methylation, primarily involves post-translational modification (PTM) targeting residues of nitrogen-containing side-chains and other residues. Protein arginine methylation, occurred on arginine residue, is mainly mediated by protein arginine methyltransferases (PRMTs), which are ubiquitously present in a multitude of organisms and are intricately involved in the regulation of numerous biological processes. Specifically, PRMTs are pivotal in the process of gene transcription regulation, and protein function modulation. Abnormal arginine methylation, particularly in histones, can induce dysregulation of gene expression, thereby leading to the development of cancer. The recent advancements in modification mediated by PRMTs and cancer research have had a profound impact on our understanding of the abnormal modification involved in carcinogenesis and progression. This review will provide a defined overview of these recent progression, with the aim of augmenting our knowledge on the role of PRMTs in progression and their potential application in cancer therapy.
Cancer, also known as malignant tumor, is caused by malignant cell proliferation and is aggressive and metastatic (Valastyan and Weinberg, 2011). Arginine is a positively charged amino acid, and as early as 1967, Paik’s team demonstrated the presence of methyl groups in arginine. Arginine methylation is a PTM with a more stable structure than other epigenetic regulators (Bedford and Clarke, 2009). Protein methylation majorly includes lysine methylation and arginine methylation. Both modifications play important role in cellular processes. Arginine methylation, a crucial PTM of histones and various non-histone proteins, plays a crucial role in a wide range of biological activities, including the regulation of gene expression, signal transduction, cell development, and carcinogenesis (Gao et al., 2024). Histones are modified post-translationally through the action of histone modifying enzymes. One of these modifications is methylation, which is carried out by lysine methyltransferases and/or arginine methyltransferases. These enzymes specifically methylate lysine and arginine residues on histones, respectively. Histones H2A and H2B have been less studied than histones H3 and H4 (Zhang et al., 2003), despite being important components of chromatin structure and regulation.
PRMTs play an increasingly important role in physiological processes and development of diseases (including cancer) by mediating the methylation of arginine residues in histone and non-histone proteins (Shishkova et al., 2017) As researchers discovered the link between protein arginine methylation and disease in the early 2000s, they began to extensively explore the possibility of PRMTs as therapeutic targets (Kim D. et al., 2014).
Within the broad category of mammals, there exists a family of nine separate sequence-related protein arginine methyltransferases, also known as PRMT1-9 (Morales et al., 2016), see Table 1. These PRMTs can be divided into three classes based on their catalytic activity: Type I, which includes PRMT1, PRMT2, PRMT3, CARM1 (Coactivator-associated arginine methyltransferase 1, also known as PRMT4), PRMT6, and PRMT8; Type II, encompassing PRMT5 and PRMT9; and Type III is solely represented by PRMT7 (Bedford, 2007). Type I PRMTs have the capacity to methylate 3-methylhistidine-5-methylarginine (MMA) and asymmetric dimethylarginine (ADMA). PRMT1 is capable of methylating H4R3 (Shia et al., 2012) and H2AR11 (Waldmann et al., 2011) in vitro, and PRMT2 can methylate histones H4 (Rowley et al., 2023) and/or H3R8 (Li et al., 2023). PRMT4 methylates H3R2 (Mann et al., 2013), R17 (Hu et al., 2020), and R26 (Mann et al., 2013). PRMT6 has the capacity to methylate not only H4 (Hyllus et al., 2007) and H2AR29 (Waldmann et al., 2011) in vitro, but also H3R2 (Wang J. et al., 2023) in vivo. Type II PRMTs are responsible for the methylation of MMA and symmetric dimethylarginine (SDMA). PRMT5 methylates H4R3 (Sipos et al., 2017), H2AR3 (Yang et al., 2021), H3R2 (Chen et al., 2017) and H3R8 (Yang et al., 2021). Lastly, Type III PRMTs are unique in that they can only catalyze MMA production. PRMT7 catalyzes the mono-methylation of histones, but the specific target of arginine has not been identified (Lee et al., 2005).
Recently, an expansion in the PRMT family has been uncovered by researchers, revealing the presence of two new members, PRMT10 and PRMT11 (Xu et al., 2014; Harada et al., 2015). In this review, we will explore the emerging field of arginine methylation and its associations with tumorigenesis, providing an in-depth understanding of the functional significance and potential molecular pathways of this modification in tumors. The primary objective is to provide the research community with a comprehensive overview of the latest advancements in this specific field of research.
The presence of PRMTs variants could influence the progression of cancer cell phenotypes through their arginine methyltransferase activity. The transcript of PRMT1 gene undergoes mRNA alternative splicing to generate three splicing variants, collectively referred to as v.1, v.2, and v.3 (Scott et al., 1998). PRMT1 v.1 and v.2 expression is significantly reduced in breast cancer. Notably, PRMT1 v.2 is also demonstrated to augment the survival and invasive properties of breast cancer cells (Adamopoulos et al., 2019). Three novel PRMT2 splicing variants, PRMT2α, PRmt2β, and PRMT2γ, were isolated in breast cancer cells (Zhong et al., 2017). These variants lack distinct motifs and therefore produce distinct C-terminal domains, which directly affect the subcellular localization of PRMT2. Furthermore, the PRMT2 variant is able to bind estrogen receptor α both in vivo and in vitro, thereby playing a role in the progression of breast cancer (Zhong et al., 2012).
PRMT8 v. 1 and v. 2 were identified in glioblastoma cell (GBM) lines and three breast cell lines. The PRMT8 v.2 exhibited a pronounced ability to localize to the nucleus in U87MG glioblastoma cells (Hernandez and Dominko, 2016). PRMT8 v.1 was observed to be expressed in both the U-2 OS human osteosarcoma and CRL-2073 human teratoid cancer cell lines (Hernandez et al., 2017). Moreover, PRMT9 genetic mutation has been identified as a significant risk factor for familial lung adenocarcinoma (Zhang et al., 2023). A recent study has highlighted the increased activity of PRMT9 in the cells and leukemia stem cells of individuals diagnosed with acute myeloid leukemia (Dong et al., 2024).
The modification of histone arginine methylation is a sophisticated PTM that has a significant impact on various cellular processes. Specifically, the histone modifications of H4R3me2a and H3R2me2s are associated with transcriptional activation, whereas the modifications of H4R3me2s, H3R2me2a, and H3R8me2s exhibit inhibitory effects on transcription (Di Lorenzo and Bedford, 2011) (Figure 1).
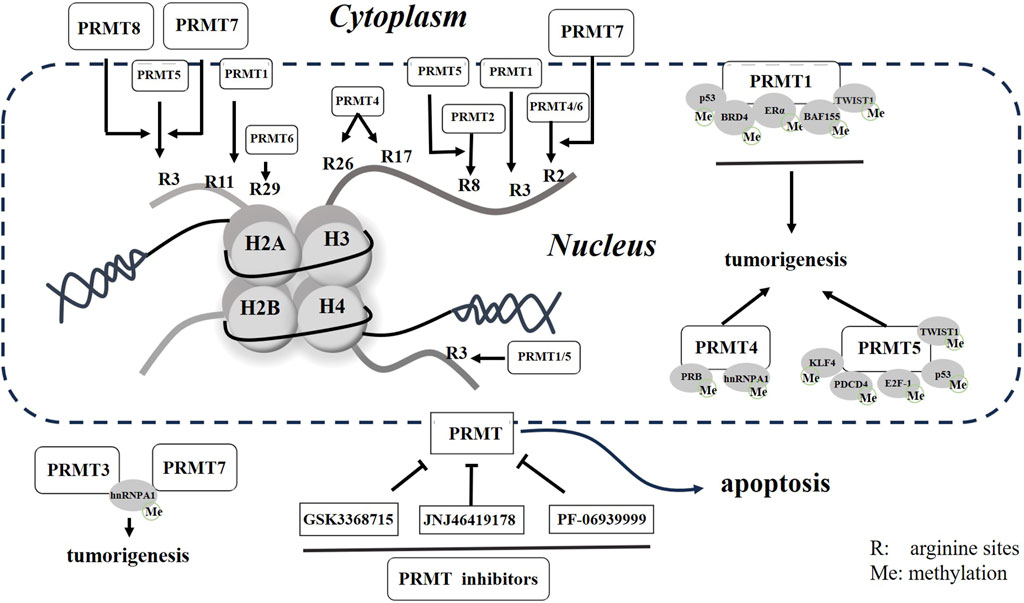
Figure 1. Schematic representation of PRMTs and small molecular inhibitors in cells. Potential arginine residues methylated by PRMTs are presented with numbered R in histones. Both methylated histones to non-histone proteins will promote tumorigenesis, but these processes can be reversed by small molecular inhibitors of PRMTs in cancer.
Promoters are key regions in genes that regulate transcription and are affected by a variety of histone modifications. The presence or absence of histone methylation can significantly affect the activation or inhibition of transcription of related genes. PRMT is recruited to the promoter region of the target gene as a costimulator to promote gene expression, and if arginine methylation is absent, it is associated with gene silencing (Wang et al., 2019).
Methylation of histone H4R3 by PRMT1 is associated with transcriptional activation. The splicing isomer of fusion genes in acute myeloid leukemia, known as AE9a, is capable of recruiting the PRMT1 complex to the promoter region of the AE9a-activated gene. This interaction leads to the methylation of the H4R3 residue, thereby promoting the progression of leukemia cells (Shia et al., 2012). PRMT7-mediated histone arginine methylation is a key regulatory mechanism in the development of B-cell lymphoma, which is able to recruit H4R3me1 and H4R3me2 to the B-cell lymphoma 6 promoter and subsequently modify the H4R3 site on the promoter by methylation (Ying et al., 2015).
PRMT2 enhances the Wnt signaling pathway by asymmetric dimethylation of WNT5a promoter H3R8, which in turn stimulates the growth of renal cancer cells (Li et al., 2023). Liu et al. found that knockdown of PRMT5 in colorectal cancer cells can reduce the accumulation of H4R3me2s and H3R8me2s markers, and reduce the CpG methylation level of cyclin dependent kinase inhibitor 2B promoter (Yang et al., 2021).
PRMT1 can induce the asymmetric dimethylation of histone H4R3, which subsequently promotes the transcription of downstream genes. This process contributes to the maintenance of the tumorigenic properties of esophageal squamous cell carcinoma (Zhao et al., 2019). PRMT5 interacts with specific protein 1 in the transcriptional inhibition complex and preferentially silences miR-29b through dimethylation of H4R3, thereby reducing its anti-leukemia activity (Tarighat et al., 2016). The N-terminal of plant homologous domain PHD finger protein 1 has the ability to recognize PRMT7-mediated H4R3 symmetric dimethylation catalyzed by WD repeat domain 77, and this specific recognition promotes tumorigenesis (Liu et al., 2018).
The expression of PRMT4 induced by high glucose increased the apoptosis of retinal pigment epithelial cells through asymmetric dimethylation of H3R17 (Kim D. I. et al., 2014). Mann et al. found that the interaction of Aminophile, glutamate and leucine-rich protein 1 and PRMT4 promotes the histone H3 arginine dimethylation, which in turn regulates epigenetic changes during breast cancer progression (Mann et al., 2013).
PRMT9 specifically recognizes and targets the R41 and R43 proteins as catalysts for arginine methylation, thereby reducing the activity of the mitochondrial antiviral signaling protein gene (Bai et al., 2022). In Hepatocellular carcinoma (HCC), PRMT9 increases its arginine methylation at R76 and R100 by targeting member 8 of the human heat shock homologous protein family A, accelerates the progression of HCC in vivo (Deng et al., 2023).
The expression level of PRMT5 in hepatocellular carcinoma is significantly high, and an increase in its overexpression can lead to an elevation in the level of H2A/H4 symmetric dimethylation (Sipos et al., 2017). H2AR29 is specifically enriched in PRMT6-inhibited genes, suggesting that H2AR29me2 may be involved in transcriptional repression (Waldmann et al., 2011). In GBM, PRMT6 promotes transcription through histone methylation marker H3R2me2a, which in turn promotes the development of GBM (Wang J. et al., 2023). PRMT7 specifically methylates histone H2AR3 (Karkhanis et al., 2012) and H3R2 (Morita et al., 2021), thus playing a role in regulating cellular responses to DNA damage and the global transcription process of genome activation.
PRMT3 may also play a role in carcinogenesis by regulating specific gene expression within cancer cells. Other scientists have recently discovered that the PRMT3 protein plays a vital role in chemotherapy resistance in cancer. Shi et al. found that PRMT3-mediated insulin-like growth factor 2 mRNA binding protein 1 arginine methylation at the R452 site promoted oxaliplatin resistance in HCC (Shi et al., 2023). Wang et al. also confirmed that PRMT3 interacts with methyltransferase 14 and participates in its arginine methylation, promoting malignant progression and treatment resistance of endometrial cancer (Wang Y. et al., 2023). In the study of drug resistance in pancreatic cancer, PRMT3 enhances chemotherapy resistance in pancreatic cancer by methylating the RNA recognition motif of heterogeneous nuclear ribonucleic acid protein 1, thereby increasing the expression of G-member 2 of the ATP-binding cassette subfamily (Hsu et al., 2018).
PRMT4 specifically modifies additional sex combs like 2 at residue 639/641 through methylation, resulting in a diminished tumor inhibitory function of the mixed lineage leukemia protein 3 (Zhao et al., 2022). Several independent research efforts have uncovered that PRMT5 plays a pivotal role in the regulation of genes that either stimulate or suppress tumor growth. At R361, PRMT5 mechanically triggers Deleted in Pancreatic Cancer 4 methylation, promoting colorectal cancer metastasis (Liu A. et al., 2023). In breast, colorectal, and prostate cancer cells, PRMT4, PRMT5, and PRMT7, as well as their mediated heterogeneous nuclear ribonucleoprotein A1 methylation and splicing isoforms, can effectively promote the growth of cancer cells (Li et al., 2021).
Liu et al. (2020) were the first to find elevated levels of PRMT7 expression in renal cancer tissues. PRMT7 regulates the expression of the proto-oncogene c-MYC and has been shown to promote renal cancer cell growth and accelerate tumor development in a c-MYC dependent manner. PRMT9 is also referred to as F-box unique protein 11 (FBXO11). In contrast to the previously observed high expression, PRMT9 levels were found to be significantly decreased in osteosarcoma and prostate cancer (Grypari et al., 2023). PRMT11, also known as FBXO10, is less studied now, but previous studies have shown that human breast cancer susceptibility-related SNP rs7042509 is present in PRMT11. Xu et al. further found that cellular stress can upregulate PRMT11, and suggested that lens epithelial-derived growth factors may play an important role in this process (Xu et al., 2014). However, in mice, loss of function of PRMT11 did not have a significant effect on B-cell leukemia-lymphoma 2 or B lymphocyte accumulation (Masle-Farquhar et al., 2021).
PRMT1-mediated methylation and activation of phosphoglycerate dehydrogenase enhanced serine synthesis and ameliorated oxidative stress, thereby stimulating the growth and proliferation of HCC cells both in vivo and in vitro (Wang K. et al., 2023). As demonstrated by Liu J. et al. (2023), PRMT1 is crucial in directly methylating cGAS and blocking cGAS/STING sensing signals, thereby facilitating tumor immune evasion. PRMT1 also has the ability to suppress P53-induced transcriptional activity through methylation at multiple regions, thus protecting breast cancer cells from undergoing apoptosis or senescence (Liu et al., 2021).
Studies have shown that high expression of PRMT3 is associated with more aggressive brain cancers. PRMT3 can significantly promote the development and progression of GBM by enhancing glycolysis and expression of hypoxia-inducible factor 1α (Liao et al., 2022). In terms of the mechanism of colorectal cancer tumorigenesis, a specific enzyme, hypoxia-inducible factor 1α R282 methylation, is catalyzed by PRMT3. This process reduces the level of polyubiquitination of hypoxia-inducible factor 1α, leading to a potential stabilization of tumorigenesis in colorectal cancer (Zhang et al., 2021). Moreover, PRMT3 may play a critical role in the regulation of C-creatine, which may contribute to the promotion of tumorigenesis (Hu et al., 2021). Analysis showed that the role of PRMT3 in tumorigenesis is highly dependent on the availability of methyl groups. Du et al. performed research that demonstrated PRMT4 promotes the progression of HCC cells by activating the AKT/mTOR signaling pathway (Du et al., 2021).
Jiang et al. also discovered that activation of the AKT/mT0R pathway caused a significant increase in PRMT6 expression in endometrial cancer, thereby exhibiting a carcinogenic effect (Jiang et al., 2020). PRMT6 acts as a regulator of glucose metabolism through PTM, contributing to the progression of lung cancer (Sun et al., 2023). PRMT7 hinders the proliferation and migration of gastric cancer cells by directly affecting the missing phosphatase and tensin homologues(PTEN) on chromosome 10 (Wang X. et al., 2023).
Research has demonstrated that PRMT9 contributes significantly to the progression of liver cancer and lung cancer through upregulation of the PI3K/Akt/GSK-3β/Snail signaling pathway, leading to enhanced invasion and metastasis (Jiang et al., 2018). PRMT10 is highly expressed in the prostate and has a regulatory effect on the growth of prostate cancer cells by controlling androgen receptor signaling (Harada et al., 2015).
Numerous non-histone proteins have been identified as containing methylated arginine, which are affected by PRMTs through modification of arginine residues on these proteins and play a role in cellular development and tumorigenesis.
In ovarian cancer tissues, PRMT1 overexpression may be involved in tumorigenesis by mediating asymmetric methylation of bromine-containing domain protein 4 (Liu Y. et al., 2023). Arginine methylation plays a crucial regulatory role in E2F transcription factor 1(E2F-1) stability and target gene expression. PRMT4-mediated arginine methylation of retinoblastoma protein is responsible for the dissociation of E2F-1, which subsequently stimulates the transcriptional activation of E2F-1 in dividing cells. This specific methylation negatively regulates the tumor suppressive function of retinoblastoma protein and influences the cell cycle (Kim et al., 2015). However, the reduced arginine methylation of E2F-1 by PRMT5 in tumor cells stabilizes E2F-1, subsequently leading to apoptosis and tumorigenesis (Cho et al., 2012). PRMT5 can modify p53 through arginine methylation, altering its nuclear localization and activity, thereby promoting the occurrence of lymphoma. In addition, PRMT5 can also methylate the zinc finger transcription factor arginine to promote the occurrence of breast cancer tumors. As a common substrate of PRMT1 and PRMT5, Twist-related protein 1 arginine methylation plays a key role in the carcinogenesis and metastasis of head and neck squamous cell carcinoma (Fan et al., 2020). Programmed cell death 4 co-expression with PRMT5 accelerates the growth of breast cancer cells (Kong et al., 2023). Arginine methylation of Heterogeneous nuclear ribonucleic acid protein K can reduce the apoptosis of U-2 OS osteosarcoma cells (Chen et al., 2021).
Despite this, the lack of reliable antibodies and clear mechanisms of demethylases and recognition proteins has, to some extent, hindered the full understanding of the molecular mechanisms and biological functions of arginine methylation.
Due to reports showing that abnormal levels of PRMT are associated with a variety of pathological conditions, researchers are currently working on selective PRMT inhibitors.
Hu et al. (2015) identified a set of cyanine compounds that are effective at suppressing PRMT1, a critical protein involved in cancer development. Through cell proliferation tests, they found that these compounds can significantly inhibit the growth of three leukemia cell lines. Importantly, the PRMT1 inhibitor GSK3368715 has entered phase I clinical trials, and its main tumor types include pancreatic cancer, non-small cell lung cancer, bladder tumors, and blood tumors of diffuse large B-cell lymphoma. In addition, PRMT5 inhibitors JNJ46419178, GSK3326595 and PF-06939999 are also under study. Among them, GSK3326595 has entered clinical phase II research, which has good brain permeability and anti-tumor effects.
The modulation of various biological processes for cell maintenance are mediated by most PRMTs methylation substrates, underscoring the potential implications of arginine methylation dysregulation in the genesis and progression of cancer. PRMTs possesses the capacity to regulate gene expression and protein activity, and has been linked to the pathological procession involved in a multitude of human cancers and inflammation in numerous research studies. Epigenetic regulation is an integral mechanism influencing cell function, predominantly through the induction or repression of histone modifications via PRMTs deposition, thereby enhancing or suppressing expression of key oncogenes, respectively. In summary, it was well known that the expression levels of PRMT1-6 are frequently elevated in tumor tissues and their overexpression is often associated with a poor prognosis. In addition, it was also noted that PRMT7-9 is highly expressed in a majority of tumor specimens, although a minor proportion demonstrates low levels of its expression. As such, PRMTs inhibitors have emerged as a critical role in the global research endeavor to identify efficacious cancer treatment strategies.
In summary, extensive scientific evidence supports the role of arginine methylation in critical biological processes such as transcription, mRNA splicing, DNA damage signaling, and immune signaling. Elevated levels of PRMTs and its variants have been identified in a variety of cancer types, and these associations are frequently linked to increased tumorigenesis and poor clinical outcomes.
XL: Conceptualization, Data curation, Investigation, Methodology, Writing–original draft. YS: Conceptualization, Data curation, Formal Analysis, Resources, Writing–original draft. WM: Conceptualization, Data curation, Resources, Software, Writing–review and editing. XH: Conceptualization, Formal Analysis, Methodology, Resources, Writing–original draft. TB: Conceptualization, Data curation, Resources, Writing–review and editing. SJ: Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Validation, Writing–review and editing, Writing–original draft.
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the National Natural Science Foundation of China (No. 31371386 SJ) and the Natural Science Foundation of Shaanxi Province (No. 2021JZ-27). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ADMA, asymmetric dimethylarginine; SDMA, symmetric dimethylarginine; MMA, mono methylarginine; PRMTs, protein arginine methyltransferases; PTM, post-translational modification; GBM, glioblastoma cell; HCC, hepatocellular carcinoma; E2F-1, early 2 factor transcription factor 1.
Adamopoulos, P. G., Mavrogiannis, A. V., Kontos, C. K., and Scorilas, A. (2019). Novel alternative splice variants of the human protein arginine methyltransferase 1 (PRMT1) gene, discovered using next-generation sequencing. Gene 699, 135–144. doi:10.1016/j.gene.2019.02.072
Bai, X., Sui, C., Liu, F., Chen, T., Zhang, L., Zheng, Y., et al. (2022). The protein arginine methyltransferase PRMT9 attenuates MAVS activation through arginine methylation. Nat. Commun. 13 (1), 5016. doi:10.1038/s41467-022-32628-y
Bedford, M. T. (2007). Arginine methylation at a glance. J. Cell Sci. 120 (Pt 24), 4243–4246. doi:10.1242/jcs.019885
Bedford, M. T., and Clarke, S. G. (2009). Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33 (1), 1–13. doi:10.1016/j.molcel.2008.12.013
Chen, C. C., Yang, J. H., Fu, S. L., Lin, W. J., and Lin, C. H. (2021). Arginine methylation of hnRNPK inhibits the DDX3-hnRNPK interaction to play an anti-apoptosis role in osteosarcoma cells. Int. J. Mol. Sci. 22 (18), 9764. doi:10.3390/ijms22189764
Chen, H., Lorton, B., Gupta, V., and Shechter, D. (2017). A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36 (3), 373–386. doi:10.1038/onc.2016.205
Cho, E. C., Zheng, S., Munro, S., Liu, G., Carr, S. M., Moehlenbrink, J., et al. (2012). Arginine methylation controls growth regulation by E2F-1. Embo J. 31 (7), 1785–1797. doi:10.1038/emboj.2012.17
Deng, W., Ai, J., Zhang, W., Zhou, Z., Li, M., Yan, L., et al. (2023). Arginine methylation of HSPA8 by PRMT9 inhibits ferroptosis to accelerate hepatitis B virus-associated hepatocellular carcinoma progression. J. Transl. Med. 21 (1), 625. doi:10.1186/s12967-023-04408-9
Di Lorenzo, A., and Bedford, M. T. (2011). Histone arginine methylation. FEBS Lett. 585 (13), 2024–2031. doi:10.1016/j.febslet.2010.11.010
Dong, H., He, X., Zhang, L., Chen, W., Lin, Y. C., Liu, S. B., et al. (2024). Targeting PRMT9-mediated arginine methylation suppresses cancer stem cell maintenance and elicits cGAS-mediated anticancer immunity. Nat. Cancer 5, 601–624. doi:10.1038/s43018-024-00736-x
Du, P., Luo, K., Li, R., Zhu, J., Xiao, Q., Li, Y., et al. (2021). PRMT4 promotes hepatocellular carcinoma progression by activating AKT/mTOR signaling and indicates poor prognosis. Int. J. Med. Sci. 18 (15), 3588–3598. doi:10.7150/ijms.62467
Fan, Z., He, L., Li, M., Cao, R., Deng, M., Ping, F., et al. (2020). Targeting methyltransferase PRMT5 retards the carcinogenesis and metastasis of HNSCC via epigenetically inhibiting Twist1 transcription. Neoplasia 22 (11), 617–629. doi:10.1016/j.neo.2020.09.004
Gao, Y., Feng, C., Ma, J., and Yan, Q. (2024). Protein arginine methyltransferases (PRMTs): orchestrators of cancer pathogenesis, immunotherapy dynamics, and drug resistance. Biochem. Pharmacol. 221, 116048. doi:10.1016/j.bcp.2024.116048
Grypari, I. M., Pappa, I., Papastergiou, T., Zolota, V., Bravou, V., Melachrinou, M., et al. (2023). Elucidating the role of PRMTs in prostate cancer using open access databases and a patient cohort dataset. Histol. Histopathol. 38 (3), 287–302. doi:10.14670/HH-18-513
Harada, N., Takagi, T., Nakano, Y., Yamaji, R., and Inui, H. (2015). Protein arginine methyltransferase 10 is required for androgen-dependent proliferation of LNCaP prostate cancer cells. Biosci. Biotechnol. Biochem. 79 (9), 1430–1437. doi:10.1080/09168451.2015.1025035
Hernandez, S., and Dominko, T. (2016). Novel protein arginine methyltransferase 8 isoform is essential for cell proliferation. J. Cell Biochem. 117 (9), 2056–2066. doi:10.1002/jcb.25508
Hernandez, S. J., Dolivo, D. M., and Dominko, T. (2017). PRMT8 demonstrates variant-specific expression in cancer cells and correlates with patient survival in breast, ovarian and gastric cancer. Oncol. Lett. 13 (3), 1983–1989. doi:10.3892/ol.2017.5671
Hsu, M. C., Pan, M. R., Chu, P. Y., Tsai, Y. L., Tsai, C. H., Shan, Y. S., et al. (2018). Protein arginine methyltransferase 3 enhances chemoresistance in pancreatic cancer by methylating hnRNPA1 to increase ABCG2 expression. Cancers (Basel) 11 (1), 8. doi:10.3390/cancers11010008
Hu, B., Li, X., Chen, L., and Liu, Z. (2020). High expression of CARM1 inhibits lung cancer progression by targeting TP53 by regulating CTNNB1. Lung 198 (2), 415–422. doi:10.1007/s00408-020-00324-7
Hu, H., Owens, E. A., Su, H., Yan, L., Levitz, A., Zhao, X., et al. (2015). Exploration of cyanine compounds as selective inhibitors of protein arginine methyltransferases: synthesis and biological evaluation. J. Med. Chem. 58 (3), 1228–1243. doi:10.1021/jm501452j
Hu, Y., Su, Y., He, Y., Liu, W., and Xiao, B. (2021). Arginine methyltransferase PRMT3 promote tumorigenesis through regulating c-MYC stabilization in colorectal cancer. Gene 791, 145718. doi:10.1016/j.gene.2021.145718
Hyllus, D., Stein, C., Schnabel, K., Schiltz, E., Imhof, A., Dou, Y., et al. (2007). PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21 (24), 3369–3380. doi:10.1101/gad.447007
Jiang, H., Zhou, Z., Jin, S., Xu, K., Zhang, H., Xu, J., et al. (2018). PRMT9 promotes hepatocellular carcinoma invasion and metastasis via activating PI3K/Akt/GSK-3β/Snail signaling. Cancer Sci. 109 (5), 1414–1427. doi:10.1111/cas.13598
Jiang, N., Li, Q. L., Pan, W., Li, J., Zhang, M. F., Cao, T., et al. (2020). PRMT6 promotes endometrial cancer via AKT/mTOR signaling and indicates poor prognosis. Int. J. Biochem. Cell Biol. 120, 105681. doi:10.1016/j.biocel.2019.105681
Karkhanis, V., Wang, L., Tae, S., Hu, Y. J., Imbalzano, A. N., and Sif, S. (2012). Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. J. Biol. Chem. 287 (35), 29801–29814. doi:10.1074/jbc.M112.378281
Kim, D., Lim, S., Park, M., Choi, J., Kim, J., Han, H., et al. (2014a). Ubiquitination-dependent CARM1 degradation facilitates Notch1-mediated podocyte apoptosis in diabetic nephropathy. Cell Signal 26 (9), 1774–1782. doi:10.1016/j.cellsig.2014.04.008
Kim, D. I., Park, M. j., Lim, S. k., Choi, J. h., Kim, J. c., Han, H. j., et al. (2014b). High-glucose-induced CARM1 expression regulates apoptosis of human retinal pigment epithelial cells via histone 3 arginine 17 dimethylation: role in diabetic retinopathy. Arch. Biochem. Biophys. 560, 36–43. doi:10.1016/j.abb.2014.07.021
Kim, K. Y., Wang, D. H., Campbell, M., Huerta, S. B., Shevchenko, B., Izumiya, C., et al. (2015). PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation. Mol. Cell Biol. 35 (1), 238–248. doi:10.1128/MCB.00945-14
Kong, J., Wang, Z., Zhang, Y., and Wang, T. (2023). Protein arginine methyltransferases 5 (PRMT5) affect multiple stages of autophagy and modulate autophagy-related genes in controlling breast cancer tumorigenesis. Curr. Cancer Drug Targets 23 (3), 242–250. doi:10.2174/1568009622666220922093059
Lee, J. H., Cook, J. R., Yang, Z. H., Mirochnitchenko, O., Gunderson, S. I., Felix, A. M., et al. (2005). PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280 (5), 3656–3664. doi:10.1074/jbc.M405295200
Lee, W. C., Lin, W. L., Matsui, T., Chen, E. S. W., Wei, T. Y. W., Lin, W. H., et al. (2015). Protein arginine methyltransferase 8: tetrameric structure and protein substrate specificity. Biochemistry 54 (51), 7514–7523. doi:10.1021/acs.biochem.5b00995
Li, W. J., He, Y. H., Yang, J. J., Hu, G. S., Lin, Y. A., Ran, T., et al. (2021). Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 12 (1), 1946. doi:10.1038/s41467-021-21963-1
Li, Z., Chen, C., Yong, H., Jiang, L., Wang, P., Meng, S., et al. (2023). PRMT2 promotes RCC tumorigenesis and metastasis via enhancing WNT5A transcriptional expression. Cell Death Dis. 14 (5), 322. doi:10.1038/s41419-023-05837-6
Liao, Y., Luo, Z., Lin, Y., Chen, H., Chen, T., Xu, L., et al. (2022). PRMT3 drives glioblastoma progression by enhancing HIF1A and glycolytic metabolism. Cell Death Dis. 13 (11), 943. doi:10.1038/s41419-022-05389-1
Liu, A., Yu, C., Qiu, C., Wu, Q., Huang, C., Li, X., et al. (2023a). PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis. Oncogene 42 (19), 1572–1584. doi:10.1038/s41388-023-02674-x
Liu, F., Wan, L., Zou, H., Pan, Z., Zhou, W., and Lu, X. (2020). PRMT7 promotes the growth of renal cell carcinoma through modulating the β-catenin/C-MYC axis. Int. J. Biochem. Cell Biol. 120, 105686. doi:10.1016/j.biocel.2020.105686
Liu, J., Bu, X., Chu, C., Dai, X., Asara, J. M., Sicinski, P., et al. (2023b). PRMT1 mediated methylation of cGAS suppresses anti-tumor immunity. Nat. Commun. 14 (1), 2806. doi:10.1038/s41467-023-38443-3
Liu, L. M., Tang, Q., Hu, X., Zhao, J. J., Zhang, Y., Ying, G. G., et al. (2021). Arginine methyltransferase PRMT1 regulates p53 activity in breast cancer. Life (Basel) 11 (8), 789. doi:10.3390/life11080789
Liu, R., Gao, J., Yang, Y., Qiu, R., Zheng, Y., Huang, W., et al. (2018). PHD finger protein 1 (PHF1) is a novel reader for histone H4R3 symmetric dimethylation and coordinates with PRMT5-WDR77/CRL4B complex to promote tumorigenesis. Nucleic Acids Res. 46 (13), 6608–6626. doi:10.1093/nar/gky461
Liu, Y., Liu, H., Ye, M., Jiang, M., Chen, X., Song, G., et al. (2023c). Methylation of BRD4 by PRMT1 regulates BRD4 phosphorylation and promotes ovarian cancer invasion. Cell Death Dis. 14 (9), 624. doi:10.1038/s41419-023-06149-5
Mann, M., Cortez, V., and Vadlamudi, R. (2013). PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis 34 (7), 1468–1475. doi:10.1093/carcin/bgt091
Masle-Farquhar, E., Russell, A., Li, Y., Zhu, F., Rui, L., Brink, R., et al. (2021). Loss-of-function of Fbxo10, encoding a post-translational regulator of BCL2 in lymphomas, has no discernible effect on BCL2 or B lymphocyte accumulation in mice. PLoS One 16 (4), e0237830. doi:10.1371/journal.pone.0237830
Morales, Y., Cáceres, T., May, K., and Hevel, J. M. (2016). Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 590, 138–152. doi:10.1016/j.abb.2015.11.030
Morita, K., Hatanaka, Y., Ihashi, S., Asano, M., Miyamoto, K., and Matsumoto, K. (2021). Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei. Sci. Rep. 11 (1), 10146. doi:10.1038/s41598-021-89334-w
Rowley, M. J., Prout-Holm, R. A., Liu, R. W., Hendrickson-Rebizant, T., Ige, O. O., Lakowski, T. M., et al. (2023). Protein arginine N-methyltransferase 2 plays a noncatalytic role in the histone methylation activity of PRMT1. J. Biol. Chem. 299 (12), 105360. doi:10.1016/j.jbc.2023.105360
Sayegh, J., Webb, K., Cheng, D., Bedford, M. T., and Clarke, S. G. (2007). Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282 (50), 36444–36453. doi:10.1074/jbc.M704650200
Scott, H. S., Antonarakis, S. E., Lalioti, M. D., Rossier, C., Silver, P. A., and Henry, M. F. (1998). Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2). Genomics 48 (3), 330–340. doi:10.1006/geno.1997.5190
Shi, Y., Niu, Y., Yuan, Y., Li, K., Zhong, C., Qiu, Z., et al. (2023). PRMT3-mediated arginine methylation of IGF2BP1 promotes oxaliplatin resistance in liver cancer. Nat. Commun. 14 (1), 1932. doi:10.1038/s41467-023-37542-5
Shia, W. J., Okumura, A. J., Yan, M., Sarkeshik, A., Lo, M. C., Matsuura, S., et al. (2012). PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119 (21), 4953–4962. doi:10.1182/blood-2011-04-347476
Shishkova, E., Zeng, H., Liu, F., Kwiecien, N. W., Hebert, A. S., Coon, J. J., et al. (2017). Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat. Commun. 8, 15571. doi:10.1038/ncomms15571
Sipos, A., Iván, J., Bécsi, B., Darula, Z., Tamás, I., Horváth, D., et al. (2017). Myosin phosphatase and RhoA-activated kinase modulate arginine methylation by the regulation of protein arginine methyltransferase 5 in hepatocellular carcinoma cells. Sci. Rep. 7, 40590. doi:10.1038/srep40590
Strahan, R. C., McDowell-Sargent, M., Uppal, T., Purushothaman, P., and Verma, S. C. (2017). KSHV encoded ORF59 modulates histone arginine methylation of the viral genome to promote viral reactivation. PLoS Pathog. 13 (7), e1006482. doi:10.1371/journal.ppat.1006482
Sun, M., Li, L., Niu, Y., Wang, Y., Yan, Q., Xie, F., et al. (2023). PRMT6 promotes tumorigenicity and cisplatin response of lung cancer through triggering 6PGD/ENO1 mediated cell metabolism. Acta Pharm. Sin. B 13 (1), 157–173. doi:10.1016/j.apsb.2022.05.019
Tarighat, S. S., Santhanam, R., Frankhouser, D., Radomska, H. S., Lai, H., Anghelina, M., et al. (2016). The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 30 (4), 789–799. doi:10.1038/leu.2015.308
Valastyan, S., and Weinberg, R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147 (2), 275–292. doi:10.1016/j.cell.2011.09.024
Waldmann, T., Izzo, A., Kamieniarz, K., Richter, F., Vogler, C., Sarg, B., et al. (2011). Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11. doi:10.1186/1756-8935-4-11
Wang, J., Xiao, Z., Li, P., Wu, C., Li, Y., Wang, Q., et al. (2023a). PRMT6-CDC20 facilitates glioblastoma progression via the degradation of CDKN1B. Oncogene 42 (14), 1088–1100. doi:10.1038/s41388-023-02624-7
Wang, K., Luo, L., Fu, S., Wang, M., Wang, Z., Dong, L., et al. (2023c). PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma. Nat. Commun. 14 (1), 1011. doi:10.1038/s41467-023-36708-5
Wang, S. M., Dowhan, D. H., and Muscat, G. E. O. (2019). Epigenetic arginine methylation in breast cancer: emerging therapeutic strategies. J. Mol. Endocrinol. 62 (3), R223–r237. doi:10.1530/JME-18-0224
Wang, X., Xu, W., Zhu, C., Cheng, Y., and Qi, J. (2023d). PRMT7 inhibits the proliferation and migration of gastric cancer cells by suppressing the PI3K/AKT pathway via PTEN. J. Cancer 14 (15), 2833–2844. doi:10.7150/jca.88102
Wang, Y., Wang, C., Guan, X., Ma, Y., Zhang, S., Li, F., et al. (2023b). PRMT3-Mediated arginine methylation of METTL14 promotes malignant progression and treatment resistance in endometrial carcinoma. Adv. Sci. (Weinh) 10 (36), e2303812. doi:10.1002/advs.202303812
Xu, X., Powell, D. W., Lambring, C. J., Puckett, A. H., Deschenes, L., Prough, R. A., et al. (2014). Human MCS5A1 candidate breast cancer susceptibility gene FBXO10 is induced by cellular stress and correlated with lens epithelium-derived growth factor (LEDGF). Mol. Carcinog. 53 (4), 300–313. doi:10.1002/mc.21977
Yang, L., Ma, D. W., Cao, Y. P., Li, D. Z., Zhou, X., Feng, J. F., et al. (2021). PRMT5 functionally associates with EZH2 to promote colorectal cancer progression through epigenetically repressing CDKN2B expression. Theranostics 11 (8), 3742–3759. doi:10.7150/thno.53023
Ying, Z., Mei, M., Zhang, P., Liu, C., He, H., Gao, F., et al. (2015). Histone arginine methylation by PRMT7 controls germinal center formation via regulating Bcl6 transcription. J. Immunol. 195 (4), 1538–1547. doi:10.4049/jimmunol.1500224
Zhang, L., Eugeni, E. E., Parthun, M. R., and Freitas, M. A. (2003). Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112 (2), 77–86. doi:10.1007/s00412-003-0244-6
Zhang, Q., Zhao, Y., Song, Z., Zhang, Q., Tian, C., Li, R., et al. (2023). Identification of THSD7B and PRMT9 mutations as risk factors for familial lung adenocarcinoma: a case report. Med. Baltim. 102 (6), e32872. doi:10.1097/MD.0000000000032872
Zhang, X., Wang, K., Feng, X., Wang, J., Chu, Y., Jia, C., et al. (2021). PRMT3 promotes tumorigenesis by methylating and stabilizing HIF1α in colorectal cancer. Cell Death Dis. 12 (11), 1066. doi:10.1038/s41419-021-04352-w
Zhao, Y., Lu, Q., Li, C., Wang, X., Jiang, L., Huang, L., et al. (2019). PRMT1 regulates the tumour-initiating properties of esophageal squamous cell carcinoma through histone H4 arginine methylation coupled with transcriptional activation. Cell Death Dis. 10 (5), 359. doi:10.1038/s41419-019-1595-0
Zhao, Z., Rendleman, E. J., Szczepanski, A. P., Morgan, M. A., Wang, L., and Shilatifard, A. (2022). CARM1-mediated methylation of ASXL2 impairs tumor-suppressive function of MLL3/COMPASS. Sci. Adv. 8 (40), eadd3339. doi:10.1126/sciadv.add3339
Zhong, J., Cao, R. X., Zu, X. Y., Hong, T., Yang, J., Liu, L., et al. (2012). Identification and characterization of novel spliced variants of PRMT2 in breast carcinoma. Febs J. 279 (2), 316–335. doi:10.1111/j.1742-4658.2011.08426.x
Keywords: histone, arginine methylation, arginine methyltransferases, dysregulation, cancer
Citation: Li X, Song Y, Mu W, Hou X, Ba T and Ji S (2024) Dysregulation of arginine methylation in tumorigenesis. Front. Mol. Biosci. 11:1420365. doi: 10.3389/fmolb.2024.1420365
Received: 20 April 2024; Accepted: 22 May 2024;
Published: 07 June 2024.
Edited by:
Vijay Karkal Hegde, Texas Tech University, United StatesReviewed by:
Dhivya Sridaran, Washington University in St. Louis, United StatesCopyright © 2024 Li, Song, Mu, Hou, Ba and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Te Ba, YmF0ZUBzeHRjbS5lZHUuY24=; Shaoping Ji, c2hhb3BpbmdqaUBoZW51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.