
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci. , 03 July 2024
Sec. Molecular Diagnostics and Therapeutics
Volume 11 - 2024 | https://doi.org/10.3389/fmolb.2024.1417306
This article is part of the Research Topic Advances in Diagnostics, Prevention, and Therapeutics by Drug-Delivery System View all 3 articles
Lung cancer is the leading cause of death among malignant tumors in the world. High lung cancer mortality rate is due to most of patients diagnosed at advanced stage. The Liquid biopsy of lung cancer have received recent interest for early diagnosis. One of the components of liquid biopsy is the exosome. The exosome cargos non-coding-RNAs, especially long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs). The lung cancer derived exosomal non-coding RNAs play the pivotal roles of lung cancer in carcinogenesis, diagnosis, therapy, drug resistance and prognosis of lung cancer. Given ceRNA (competitive endogenous RNA) mechanism, lncRNA or circRNA can act as ceRNA to compete to bind miRNAs and alter the expression of the targeted mRNA, contributing to the development and progression of lung cancer. The current research progress of the roles of the exosomal non-coding-RNAs and the interplay of ceRNAs and miRNAs in mediated lung cancer is illustrated in this article. Hence, we presented an experimentally validated lung cancer derived exosomal non-coding RNAs-regulated target gene axis from already existed evidence in lung cancer. Then LncRNA/circRNA-miRNA-mRNA axis may be a potential target for lung cancer treatment and has great potential in the diagnosis and prognosis of lung cancer.
Lung cancer stands as the most prevalent malignancies globally in 2022, responsible for more than 1.8 million death (Bray et al., 2024). The mortality rate associated with lung cancer is notably high, primarily due to delayed detection and diagnosis, resulting in reduced overall survival rates. Patients with early-stage lung cancer have significantly better prognosis than those with advanced disease (Li et al., 2023). The lung cancer can be classified into two main types: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), representing 15% and 85% of cases respectively (Zhang Y. et al., 2023). The 5-year survival rate for early-stage NSCLC (1A1) can reach 90% (Memon et al., 2024), and even early-stage SCLC patients can achieve a 5-year survival rate of 30% (Liu Q. et al., 2024). However, for patients with advanced-stage lung cancer, the 5-year survival rate dramatically drops to 10%. The absence of clear early symptoms and reliable biomarkers often leads to the late-stage diagnosis of lung cancer, resulting in poorer prognosis. Despite notable advancements in the diagnosis and treatment of lung cancer, the 5-year survival rate remains only 17.9%. This rate further declines to only 4% for patients with distant metastasis (Liao et al., 2023). Therefore, it is crucial to identify new biomarkers to enhance diagnostic accuracy and discover effective therapeutic targets that can facilitate precision treatment.
Tumor biomarkers play a crucial role in the early diagnosis, treatment, therapeutic monitoring and prognosis prediction of lung cancer. Traditional diagnostic methods for early detection of lung cancer include CT and tissue biopsy. However, the CT have limitations such as false positives and radiation exposure, while tissue biopsy are invasive and restricted by tumor heterogeneity (Ettinger et al., 2022). In contrast, liquid biopsy involves monitoring tumors through blood or bronchoalveolar lavage fluid. It offers the advantages of non-invasiveness and bypassing tumor heterogeneity, making it a potentially effective method for early lung cancer diagnosis (Chabon et al., 2020). Currently, the most commonly detected biomarkers in liquid biopsy are circulating tumor cells (CTCs), circulating tumor DNAs (ctDNAs) and exosomes (Casagrande et al., 2023).
Exosomes are small extracellular vesicles with diameters ranging from 30 to 150 nm that are secreted by most cells including the tumor cells through the endoplasmic reticulum pathway (Kalluri and LeBleu, 2020). Exosomes are released through exocytosis once multivesicular bodies fuse with the cell membrane. Exosomes can be isolated from various body fluids, including saliva, blood, pleural effusion, bronchoalveolar lavage fluid, and sputum (Ren et al., 2024). Common methods for exosome isolation comprise ultracentrifugation, ultrafiltration (UF), immune affinity capture and commercial kits like the exoEasy Maxi kit (Qiagen) (Ma et al., 2024a). Exosomes play a role in removing surplus or unnecessary components from cells to maintain cellular homeostasis. Furthermore, they act as vehicles for intercellular substance and information exchange, facilitating communication between tumor cells and nearby or distant cells, as well as stromal cells (Jacobsen Skanderup et al., 2023). Exosomes derived from lung cancer can be obtained from plasma, serum, pleural effusion or bronchoalveolar lavage fluid. Notably, Exosomes secreted by lung cancer cells are more abundant than those secreted by non-tumor cells, making them potential biomarkers for liquid biopsy in lung cancer diagnosis (Choi et al., 2019).
In recent years, an increasing number of studies have indicated that exosome carrying non-coding RNAs (ncRNAs) hold potential as therapeutic targets for lung cancer and serve as new biomarkers for its diagnosis and prognosis (Lai et al., 2023). The dysregulated expression of these ncRNAs within exosome play an important role in driving the development and progression of lung cancer. These vesicles transport essential information, including proteins, lipids and nucleic acids (www.exocarta.org), with a particular focus on ncRNAs (miRNAs, lncRNAs and circRNAs), which elevates their significance (Clancy and D'Souz-Schorey, 2023). Exosome not only serves vital roles in intercellular material and information transfer but also exhibit close associations with tumor development and progression, making them promising early diagnostic markers for various cancers. Sequencing of exosomal RNA enables rapid and efficient acquisition of comprehensive information, rendering it an ideal approach for disease diagnosis and prognosis (Gupta, 2022). Current research on ncRNAs in exosome predominantly centers on the significant regulatory roles of miRNA, lncRNA and circRNA in transcription, post-transcription and translation, positioning them as potential biological targets for the prevention, diagnosis and treatment of lung cancer (Zhao et al., 2023).
The miRNA refers to endogenous ncRNA molecules approximately 18–22 nucleotides in size, capable of binding to complementary sequences in the 3′untranslated region (UTR) of target gene’s mRNA through miRNA response elements (MRE). This binding inhibits the expression of protein-coding genes and leads to the renewal, conversion or degradation of mRNA transcripts (Kilikevicius et al., 2022). In 2011, Salmena et al. proposed the hypothesis of a competitive endogenous RNA (ceRNA) interaction mechanism between non-coding RNA and mRNA (Salmena et al., 2011). According to this hypothesis, the lncRNA and circRNA containing MREs can alleviate the inhibitory effect of miRNA on target genes by binding to miRNA (Figure 1A). Consequently, the ceRNA can regulate the translation of protein encoded by specific mRNA by modify their binding with miRNAs through the sponge effect. In recent years, numerous studies have focused on the investigation of ncRNAs in relation to lung cancer. Specifically, the intricate crosstalk between various CeRNAs and miRNA has emerged as a promising avenue for both therapeutic targeting and biomarker development in lung cancer (Wu X. et al., 2020). This review aims to provide an overview and discussion of the potential roles of three types of exosomal ncRNAs in liquid biopsy, with specific emphasis on their applications in the diagnosis, therapy and prognosis of lung cancer. Moreover, highlight the hope of the lncRNA/circRNA-miRNA-mRNA axis for reliable identification, prediction and therapy of lung cancer.
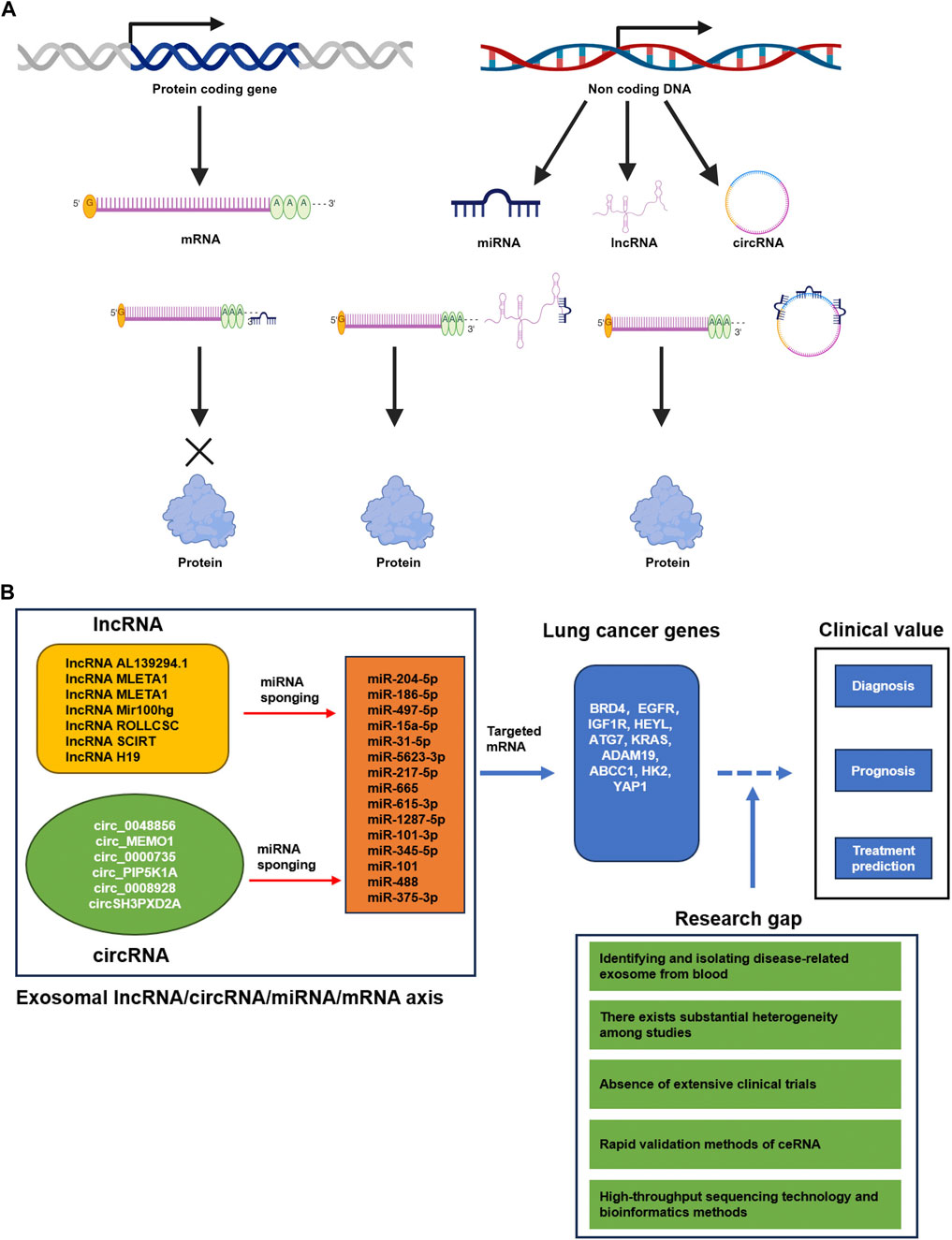
Figure 1. (A) The CeRNA mechanism (Created with BioRender.com). Protein-coding genes undergo transcription, resulting in the production of mRNA that is translated to proteins. Non-coding DNA is transcribed into various types of ncRNAs, including miRNA, lncRNA, and circRNA. These ncRNAs play an important role in regulating mRNA translation. Specifically, miRNAs bind to partially complementary sequences located in either the 3′UTR or ORF regions of target mRNAs, thereby impeding their translation. These partially complementary sequences are denoted as microRNA response elements (MREs). Likewise, lncRNAs and circRNAs also harbor numerous MREs, and they competitively bind to miRNAs, acting as sponges, ultimately enhancing mRNA translation into proteins. These ncRNAs that engage in competitive interactions are referred to as competing endogenous RNAs (ceRNAs). (B) Summary of the current potential lncRNA/circRNA-miRNA-mRNA axis derived from lung cancer exosomes in diagnosis, prognosis and treatment prediction of lung cancer. The GAP of basic research and clinical application in lung cancer.
Exosomes, along with the enclosed miRNA, are taken up by neighboring or distant cells, regulating the translation of target mRNA and thereby modulating processes related to tumor immunity and the tumor microenvironment. This modulation has the potential to facilitate tumor growth, invasion, metastasis, angiogenesis and drug resistance. Consequently, the role of exosomal miRNA in the regulation of cancer progression is substantial, making them promising candidates as diagnostic and therapeutic biomarkers (Sun et al., 2018). Additionally, besides their involvement in tumors, evidence indicates that exosome miRNA is implicated in corneal diseases (Verma et al., 2023; Arora and Verma, 2024), complications of diabetes (Xu et al., 2022), and autoimmune skin diseases (Zhang R. et al., 2023). Hence, they possess the potential to serve as biomarkers and therapeutic targets in these conditions. Recent studies have shown that the exosomal secretion of miRNA has potential as an ideal diagnostic biomarker for the clinical application of lung cancer. Certain exosomal miRNAs found in plasma or serum exhibit high expression levels in NSCLC, thus serving as specific biomarkers for its diagnosis and prognosis (Table 1). Notably, the exosomal miR-146a-5p/miR-486-5p (Wu Q. et al., 2020), miR-619-5p/miR-4454 (Feng et al., 2023), miR-21/miR-25/miR-155/miR-210/miR-486 (Liu et al., 2020) and EV-miR-486-5p/miR-21-5p (Tong et al., 2024) were upregulated in NSCLC patients than in healthy people. The high expression of these three panels EV-miRNAs is positive correlation with NSCLC diagnosis. Cazzoli et al. observed higher expression levels of four miRNAs (miR-378A/miR-379/miR-139-5P/miR−200B-5P) in exosomes extracted from the blood of patients with lung adenocarcinoma compared to healthy smokers in 2013 (Cazzoli et al., 2013). Additionally, mi1290/miR-29C-3p (Zhang Q. et al., 2023) and miR-7977 (Chen et al., 2020) showed elevated expression levels in exosomes and served as effective diagnostic biomarkers for lung adenocarcinoma. These miRNAs present promising candidates for the development of highly sensitive and non-invasive early diagnostic biomarkers for NSCLC.
Exosomal miRNAs not only served as diagnostic markers but also function as valuable prognostic indicators for lung cancer patients. The level of miR-4497 (Zheng et al., 2023), miR-1290 (Wu Y. et al., 2020), and miR-133a-3p (Yang et al., 2023) are all significantly and inversely associated with patients’ progression-free survival (PFS). Moreover, miR-451a (Kanaoka et al., 2018) also reliably predicts OS in NSCLC. These exosomal miRNAs play a crucial role in lung cancer prognosis.
LncRNAs are typically characterized as non-coding transcripts exceeding 200 nucleotides in length, which regulate protein expression at various levels (Herman et al., 2022). Increasing evidences confirms the distinct expression patterns of lncRNAs in tumor tissues and tumor-derived exosomes. Significantly, exosomal lncRNAs can serve as potential biomarkers for identification the lung cancer (Table 1). Notably, exosomal lncRNAs TBILA/AGAP2-AS1 (Tao et al., 2020) and DLX6-AS1 (Zhang et al., 2019) were found to be upregulated in NSCLC. They exhibited significant positive correlations with the tumor size, lymph node metastasis and TNM stage of the NSCLC, thus serving as diagnostic markers for NSCLC. Furthermore, exosomal lncRNA SLC9A3-AS1/PCAT6 showed elevated expression in lung cancer patients and can potentially be employed for identifying lung cancer (Bai et al., 2019). Additionally, the lncRNA GAS5 (Li et al., 2019) and Lnc-RNA-RP11-510M2 (Mohamed Gamal El-Din et al., 2022) were observed to be expressed at lower levels in NSCLC and lung cancer compared to healthy controls respectively, also making them potential biomarkers for distinguishing lung cancer. Furthermore, lncCRLA exhibited specific expression in adenocarcinoma cells, mouse adenocarcinoma tissues and patient plasma exosomes, rendering it valuable for predicting the occurrence of lung adenocarcinoma (Fan et al., 2020). Notably, in lung SCC, the exosomal lncRNA SOX2-OT displayed significant upregulation when compared to exosomes from healthy individuals. The findings demonstrated a positive correlation between exosomal SOX2-OT levels and tumor size, TNM staging and lymph node metastasis, thereby enabling the detection of SCC (Teng et al., 2019). Additionally, exosomal lncRNA RP5-977B1 exhibited higher expression levels in NSCLC compared to the healthy control group. This aberrant expression was associated with the diagnosis of NSCLC and a shorter OS (Min et al., 2022). Subsequent research revealed that lnc00917 (Xiong et al., 2021), lnc01125 (Xian et al., 2021) and HAGLR (Le et al., 2019) were all highly expressed in NSCLC patients and demonstrated positive correlations with tumor stage and lymph node metastasis. Consequently, they have emerged as potential candidate biomarkers for diagnosing and predicting the prognosis of NSCLC.
CircRNAs are a rapidly growing subgroup of non-coding RNA molecules, which have captured considerable attention because of their implicated roles in oncogenesis. The growing researches have suggested that exosomal circRNAs can serve as vital diagnostic and prognostic biomarkers for different types of lung cancer (Table 1) (Zhang F. et al., 2023). Notably the synergistic utilization of circ_0047921/circ_0056285/circ_0007761 has demonstrated a substantial improvement in the diagnostic accuracy for NSCLC (Xian et al., 2020). Current investigations have also shown that the levels of serum exosomal circ_0001492/circ_0001439/circ_0000896 are significantly elevated in individuals with lung adenocarcinoma compared to healthy subjects, and their expression levels is experienced a pronounced decrease after surgical intervention. Moreover, the combination of these three specific serum exosomal circRNAs has exhibited superior efficacy, heightened sensitivity and specificity in the detection of lung adenocarcinoma (Kang et al., 2022). Based on these evidences, exosomal circRNAs are emerging as exceptionally promising and reliable biomarkers for the detection and prognosis of lung cancer.
These exosomal ncRNAs (miRNAs, lncRNA and circRNA) play an important role in regulating mRNA translation. Specifically, miRNAs bind to partially complementary sequences located in either the 3′UTR or ORF regions of target mRNAs, thereby impeding their translation. These partially complementary sequences are denoted as microRNA response elements (MREs). Likewise, lncRNAs and circRNAs also harbor numerous MREs, and they competitively bind to miRNAs, acting as sponges, ultimately enhancing mRNA translation into proteins. These ncRNAs that engage in competitive interactions are referred to as competing endogenous RNAs (ceRNAs). Exosomes secreted by lung cancer cells can transport the cancer cell secreted lncRNAs or ciricRNAs to nearby or distant cells, as well as stromal cells. And then the lncRNAs or ciricRNAs can upregulated the targeted gene expression by the ceRNA mechnisms in these cells. Finaly, the ceRNA can influence tumor progression and tumor microenvironment. Exosomal lncRNA/circRNA-miRNA-mRNA regulatory networks have the ability to upregulate target gene expression and effectively regulate the progression of lung cancer (Salmena et al., 2011). Thus, the exosomal lncRNA/circRNA–miRNA–mRNA axis holds significant potential as a diagnostic and identification tool for lung cancer. For instance, recent studies have demonstrated that lncRNA AL139294.1 was transported to recipient cells through exosome. LncRNA AL139294.1 promotes lung cancer metastasis in vitro and in vivo by competitively binding with miR-204-5p to regulate BRD4 and activate the Wnt and NF-κB2 pathways. Moreover, the lncRNA AL139294.1 is upregulated in the exosomes of NSCLC patients’ serum, while miR-204-5p is downregulated. The increased level of lncRNA AL139294.1 and decreased level of miR-204-5p are positively correlated with advanced pathological stage, lymph node metastasis and distant metastasis in NSCLC patients (Ma et al., 2024b). Thus, the AL139294.1-miR-204-5p-BRD4 axis has the potential to serve as a diagnostic biomarker for non-small cell lung cancer. Similarly, exosomal lncRNA MLETA1miR-186-5p-EGFR and MLETA1-miR-497-5p-IGF1R axes have been found to promote lung cancer metastasis. And then their expression within exosomes negatively correlates with the non-metastatic survival of patients, making them as prognostic biomarkers and potential therapeutic targets for lung cancer (Hsu et al., 2023). Shi Q. et al. (2023) demonstrated that the exosomal transport of lncRNA Mir100 hg enhances glycolysis and promotes lung adenocarcinoma metastasis through interaction with miRNA-15a-5p/31-5p (Shi L. et al., 2023). Zhang et al. (2024) found that exosomal lncRNA ROLLCSC stimulates lipid metabolism and promotes lung cancer metastasis through its ceRNA function with miR-5623-3p and miR-217-5p (Zhang et al., 2024). Additionally, these exosomal lncRNA ceRNA regulatory networks may serve as biomarkers for DFS. Furthermore, the transfer of ncRNA H19 through exosomes has been found to promote resistance to Erlotinib in non-small cell lung cancer via the miR-615-3p-ATG7 axis, and it can potentially predict the efficacy of Erlotinib treatment (Pan and Zhou, 2020).
Exosomal circRNAs have been implicated in ceRNA regulatory networks associated with the progression of lung cancer. Specifically, Circ-MEMO1 (Ding et al., 2020), circ_0048856 (He et al., 2022), and circ_0000735 (Liu S. et al., 2024) derived from exosomes exhibit increased levels in the serum and cell lines of NSCLC. Exosomal circ_0048856-miR-1287-5p, circ-MEMO1-miR-101-3p-KRAS axis, and circ_0000735by-miR-345-5p-ADAM19 pathway promotes NSCLC development. These regulatory networks encompassing circ_0048856-miR-1287-5p, circ-MEMO1-miR-101-3p-KRAS, and circ_00007355-miR-345-5p-ADAM19 play a role of detection of non-small cell lung cancer. Additionally, silencing exosomal circ_PIP5K1A suppresses NSCLC progression and enhances cisplatin sensitivity through modulation of the miR-101-ABCC1 axis (Shao et al., 2021), whereas knockdown of exosomal circ_0008928 inhibits NSCLC progression and augments cisplatin sensitivity via regulation of the miR-488-HK2 axis (Shi Q. et al., 2023). Moreover, exosomal circSH3PXD2A mitigates DDP chemotherapy resistance in SCLC by modulating the miR-375-3p-YAP1 axis (Chao et al., 2023). These discoveries introduce novel potential biomarkers for predicting the efficacy of drug resistance treatment in both non-small cell lung cancer and SCLC.
Studying non-invasive and effective methods for the detection of lung cancer has significant importance in reducing the risk of death in patients with this disease. The research on exosome ncRNA has made continuous progress, and its functional role as stable and non-invasive biomarkers for the diagnosis, prognosis, and treatment response of various cancers, especially lung cancer, has garnered much attention. In this review, we primarily summarize the clinical value of exosome ncRNA, including miRNA, lncRNA, circRNA and the complex crosslink of ceRNA, in the diagnosis, prognosis, and treatment prediction of lung cancers (Figure 1B). The study of exosome ceRNA complex crosslink in cancer research is an emerging field, and advancements in understanding the role of exosomal lncRNA/circRNA-miRNA-mRNA axis in lung cancer have been made through basic research. There is mounting evidence to suggests that exosomal ncRNA biomarkers have prospective applications in the diagnosis and prognosis of lung cancer. However, the clinical use of exosomal noncoding RNA biomarkers in lung cancer patients is still in the preclinical stage, and there are certain obstacles in clinical translation. The present challenge in the field revolves around the efficient isolation and identification of disease-related exosomes from blood. Additionally, there is a need to identify specific ncRNAs, such as miRNA, lncRNA, circRNA and ceRNA, as potential early diagnostic markers derived from disease-related exosomes. Furthermore, precise cut-off values are required to identify specific ncRNAs, including miRNA, lncRNA, circRNA and ceRNA, as potential early diagnostic markers derived from disease-related Exosomes. Moreover, there exists substantial heterogeneity among studies. Additionally, the absence of extensive clinical trials hinders the confirmation of their potential value (Li et al., 2021). The development of high-throughput sequencing technology and bioinformatics methods such as whole exosome sequencing (WES) will enable future studies to compare and analyze the exosomal ncRNA and lncRNA/circRNA-miRNA-mRNA of lung cancer tissues under various physiological and pathological conditions, providing a global understanding of exosomal lncRNA/circRNA-miRNA-mRNA axis (George et al., 2024). This comprehensive understanding will contribute to the discovery of novel exosomal lncRNA/circRNA-miRNA-mRNA axis. Furthermore, it is crucial to explore rapid validation methods and decipher the mechanisms of lncRNA/circRNA-miRNA-mRNA axis in lung cancer development. Currently, there is limited research on the clinical application of lncRNA/circRNA-miRNA-mRNA, and further studies are needed to demonstrate their clinical value (Figure 1B). Hopefully, in the near future, non-invasive detection of exosomal ncRNA can be implemented for the screening of lung cancer patients.
RW: Conceptualization, Writing–original draft. YX: Writing–original draft, Writing–review and editing. LT: Writing–review and editing. XZ: Supervision, Writing–original draft. SZ: Conceptualization, Writing–original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported Guangzhou Municipal Science and Technology Bureau- 202201011011, Guang Dong Basic and Applied Basic Research Foundation 2022A1515111041, the State Key Laboratory of Respiratory Disease (SKLRD) Open Project SKLRD-Z-202115.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arora, S., and Verma, N. (2024). Exosomal microRNAs as potential biomarkers and therapeutic targets in corneal diseases. Mol. Vis. 30, 92–106.
Bai, Y., Qu, Y., Wu, Z., Ren, Y., Cheng, Z., Lu, Y., et al. (2019). Absolute quantification and analysis of extracellular vesicle lncRNAs from the peripheral blood of patients with lung cancer based on multi-colour fluorescence chip-based digital PCR. Biosens. Bioelectron. 142, 111523. doi:10.1016/j.bios.2019.111523
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263. doi:10.3322/caac.21834
Casagrande, G. M. S., Silva, M. O., Reis, R. M., and Leal, L. F. (2023). Liquid biopsy for lung cancer: up-to-date and perspectives for screening programs. Int. J. Mol. Sci. 24, 2505. doi:10.3390/ijms24032505
Cazzoli, R., Buttitta, F., Di Nicola, M., Malatesta, S., Marchetti, A., Rom, W. N., et al. (2013). microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 8, 1156–1162. doi:10.1097/JTO.0b013e318299ac32
Chabon, J. J., Hamilton, E. G., Kurtz, D. M., Esfahani, M. S., Moding, E. J., Stehr, H., et al. (2020). Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251. doi:10.1038/s41586-020-2140-0
Chao, F., Zhang, Y., Lv, L., Wei, Y., Dou, X., Chang, N., et al. (2023). Extracellular vesicles derived circSH3PXD2A inhibits chemoresistance of small cell lung cancer by miR-375-3p/YAP1. Int. J. Nanomedicine 18, 2989–3006. doi:10.2147/IJN.S407116
Chen, L., Cao, P., Huang, C., Wu, Q., Chen, S., and Chen, F. (2020). Serum exosomal miR-7977 as a novel biomarker for lung adenocarcinoma. J. Cell. Biochem. 121, 3382–3391. doi:10.1002/jcb.29612
Choi, D., Montermini, L., Jeong, H., Sharma, S., Meehan, B., and Rak, J. (2019). Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano 13, 10499–10511. doi:10.1021/acsnano.9b04480
Clancy, J. W., and D'Souza-Schorey, C. (2023). Tumor-derived extracellular vesicles: multifunctional entities in the tumor microenvironment. Annu. Rev. Pathol. 18, 205–229. doi:10.1146/annurev-pathmechdis-031521-022116
Ding, C., Xi, G., Wang, G., Cui, D., Zhang, B., Wang, H., et al. (2020). Exosomal circ-MEMO1 promotes the progression and Aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS Axis. Front. Genet. 11, 962. doi:10.3389/fgene.2020.00962
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J. R., Bharat, A., et al. (2022). Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 20, 497–530. doi:10.6004/jnccn.2022.0025
Fan, T., Sun, N., and He, J. (2020). Exosome-derived LncRNAs in lung cancer. Front. Oncol. 10, 1728. doi:10.3389/fonc.2020.01728
Feng, L., Feng, Z., Hu, J., Gao, J., Li, A., He, X., et al. (2023). Identification of hsa-miR-619-5p and hsa-miR-4454 in plasma-derived exosomes as a potential biomarker for lung adenocarcinoma. Front. Genet. 14, 1138230. doi:10.3389/fgene.2023.1138230
George, J., Maas, L., Abedpour, N., Cartolano, M., Kaiser, L., Fischer, R. N., et al. (2024). Evolutionary trajectories of small cell lung cancer under therapy. Nature 627, 880–889. doi:10.1038/s41586-024-07177-7
Gupta, D. K. (2022). Editorial: exosomal biomarkers: roles in diagnostics and therapeutics. Front. Mol. Biosci. 9, 1127540. doi:10.3389/fmolb.2022.1127540
He, Y., Liu, Y., Cha, N., Gao, Y., Li, F., Zhang, M., et al. (2022). Exosomal circ_0048856 derived from non-small cell lung cancer contributes to aggressive cancer progression through downregulation of miR-1287-5p. Pathol. Res. Pract. 232, 153659. doi:10.1016/j.prp.2021.153659
Herman, A. B., Tsitsipatis, D., and Gorospe, M. (2022). Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 82, 2252–2266. doi:10.1016/j.molcel.2022.05.027
Hsu, X.-R., Wu, J.-E., Wu, Y.-Y., Hsiao, S.-Y., Liang, J.-L., Wu, Y.-J., et al. (2023). Exosomal long noncoding RNA MLETA1 promotes tumor progression and metastasis by regulating the miR-186-5p/EGFR and miR-497-5p/IGF1R axes in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 42, 283. doi:10.1186/s13046-023-02859-y
Jacobsen Skanderup, A., Ang, S. F., and Tan, D. S. W. (2023). Liquid biopsies for cancer genotyping: coming of age? Clin. Cancer Res. 29, 1381–1383. doi:10.1158/1078-0432.CCR-22-3087
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367, eaau6977. doi:10.1126/science.aau6977
Kanaoka, R., Iinuma, H., Dejima, H., Sakai, T., Uehara, H., Matsutani, N., et al. (2018). Usefulness of plasma exosomal MicroRNA-451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncology 94, 311–323. doi:10.1159/000487006
Kang, Y., You, J., Gan, Y., Chen, Q., Huang, C., Chen, F., et al. (2022). Serum and serum exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as diagnostic biomarkers for lung adenocarcinoma. Front. Oncol. 12, 912246. doi:10.3389/fonc.2022.912246
Kilikevicius, A., Meister, G., and Corey, D. R. (2022). Reexamining assumptions about miRNA-guided gene silencing. Nucleic Acids Res. 50, 617–634. doi:10.1093/nar/gkab1256
Lai, X., Zhong, J., Zhang, B., Zhu, T., and Liao, R. (2023). Exosomal non-coding RNAs: novel regulators of macrophage-linked intercellular communication in lung cancer and inflammatory lung diseases. Biomolecules 13, 536. doi:10.3390/biom13030536
Le, R., Luo, L., Luo, L., Chen, S., Ran, R., and Liu, X. (2019). Identification of plasma exosomes long non-coding RNA HAGLR and circulating tumor cells as potential prognosis biomarkers in non-small cell lung cancer. Transl. Cancer Res. 8, 2264–2273. doi:10.21037/tcr.2019.09.43
Li, C., Lv, Y., Shao, C., Chen, C., Zhang, T., Wei, Y., et al. (2019). Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell Physiol. 234, 20721–20727. doi:10.1002/jcp.28678
Li, M.-Y., Liu, L.-Z., and Dong, M. (2021). Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer 20, 22. doi:10.1186/s12943-021-01312-y
Li, Y., Yan, B., and He, S. (2023). Advances and challenges in the treatment of lung cancer. Biomed. Pharmacother. 169, 115891. doi:10.1016/j.biopha.2023.115891
Liao, W., Coupland, C. A. C., Burchardt, J., Baldwin, D. R., Gleeson, F. V., Hippisley-Cox, J., et al. (2023). Predicting the future risk of lung cancer: development, and internal and external validation of the CanPredict (lung) model in 19·67 million people and evaluation of model performance against seven other risk prediction models. Lancet Respir. Med. 11, 685–697. doi:10.1016/S2213-2600(23)00050-4
Liu, C., Kannisto, E., Yu, G., Yang, Y., Reid, M. E., Patnaik, S. K., et al. (2020). Non-invasive detection of exosomal MicroRNAs via tethered cationic lipoplex nanoparticles (tCLN) biochip for lung cancer early detection. Front. Genet. 11, 258. doi:10.3389/fgene.2020.00258
Liu, Q., Zhang, J., Guo, C., Wang, M., Wang, C., Yan, Y., et al. (2024). Proteogenomic characterization of small cell lung cancer identifies biological insights and subtype-specific therapeutic strategies. Cell 187, 184–203.e28. doi:10.1016/j.cell.2023.12.004
Liu S., S., Wu, X., Wang, Y., and Chen, Y. (2024). Exosomal circ_0000735 contributes to non-small lung cancer malignant progression. J. Biochem. Mol. Toxicol. 38, e23700. doi:10.1002/jbt.23700
Ma, X., Chen, Z., Chen, W., Chen, Z., and Meng, X. (2024a). Exosome subpopulations: the isolation and the functions in diseases. Gene 893, 147905. doi:10.1016/j.gene.2023.147905
Ma, X., Chen, Z., Chen, W., Chen, Z., Shang, Y., Zhao, Y., et al. (2024b). LncRNA AL139294.1 can be transported by extracellular vesicles to promote the oncogenic behaviour of recipient cells through activation of the Wnt and NF-κB2 pathways in non-small-cell lung cancer. J. Exp. Clin. Cancer Res. 43, 20. doi:10.1186/s13046-023-02939-z
Memon, D., Schoenfeld, A. J., Ye, D., Fromm, G., Rizvi, H., Zhang, X., et al. (2024). Clinical and molecular features of acquired resistance to immunotherapy in non-small cell lung cancer. Cancer Cell 42, 209–224.e9. doi:10.1016/j.ccell.2023.12.013
Min, L., Zhu, T., Lv, B., An, T., Zhang, Q., Shang, Y., et al. (2022). Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int. J. Clin. Oncol. 27, 1013–1024. doi:10.1007/s10147-022-02129-5
Mohamed Gamal El-Din, G., Ibrahim, F. K., Shehata, H. H., Osman, N. M., Abdel-Rahman, O. M., and Ali, M. (2022). Exosomal expression of RAB27A and its related lncRNA Lnc-RNA-RP11-510M2 in lung cancer. Arch. Physiol. Biochem. 128, 1479–1485. doi:10.1080/13813455.2020.1778036
Pan, R., and Zhou, H. (2020). Exosomal transfer of lncRNA H19 promotes Erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 Axis. Cancer Manag. Res. 12, 4283–4297. doi:10.2147/CMAR.S241095
Ren, F., Fei, Q., Qiu, K., Zhang, Y., Zhang, H., and Sun, L. (2024). Liquid biopsy techniques and lung cancer: diagnosis, monitoring and evaluation. J. Exp. Clin. Cancer Res. 43, 96. doi:10.1186/s13046-024-03026-7
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. doi:10.1016/j.cell.2011.07.014
Shao, N., Song, L., and Sun, X. (2021). Exosomal circ_PIP5K1A regulates the progression of non-small cell lung cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol. Cell Biochem. 476, 2253–2267. doi:10.1007/s11010-021-04083-8
Shi, L., Li, B., Zhang, Y., Chen, Y., Tan, J., Chen, Y., et al. (2023). Exosomal lncRNA Mir100hg derived from cancer stem cells enhance glycolysis and promote metastasis of lung adenocarcinoma through mircroRNA-15a-5p/31-5p. Cell Commun. Signal 21, 248. doi:10.1186/s12964-023-01281-3
Shi, Q., Ji, T., Ma, Z., Tan, Q., and Liang, J. (2023). Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 Axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother Radiopharm. 38, 558–571. doi:10.1089/cbr.2020.4490
Sun, Z., Shi, K., Yang, S., Liu, J., Zhou, Q., Wang, G., et al. (2018). Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 17, 147. doi:10.1186/s12943-018-0897-7
Tao, Y., Tang, Y., Yang, Z., Wu, F., Wang, L., Yang, L., et al. (2020). Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int. J. Biol. Sci. 16, 471–482. doi:10.7150/ijbs.39123
Teng, Y., Kang, H., and Chu, Y. (2019). Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet. Test. Mol. Biomarkers 23, 235–240. doi:10.1089/gtmb.2018.0103
Tong, Z., Yang, D., Shen, C., Li, C., Xu, X., Li, Q., et al. (2024). Rapid automated extracellular vesicle isolation and miRNA preparation on a cost-effective digital microfluidic platform. Anal. Chim. Acta 1296, 342337. doi:10.1016/j.aca.2024.342337
Verma, N., Khare, D., Poe, A. J., Amador, C., Ghiam, S., Fealy, A., et al. (2023). MicroRNA and protein cargos of human limbal epithelial cell-derived exosomes and their regulatory roles in limbal stromal cells of diabetic and non-diabetic corneas. Cells 12, 2524. doi:10.3390/cells12212524
Wu, Q., Yu, L., Lin, X., Zheng, Q., Zhang, S., Chen, D., et al. (2020). Combination of serum miRNAs with serum exosomal miRNAs in early diagnosis for non-small-cell lung cancer. Cancer Manag. Res. 12, 485–495. doi:10.2147/CMAR.S232383
Wu, X., Sui, Z., Zhang, H., Wang, Y., and Yu, Z. (2020). Integrated analysis of lncRNA-mediated ceRNA network in lung adenocarcinoma. Front. Oncol. 10, 554759. doi:10.3389/fonc.2020.554759
Wu, Y., Wei, J., Zhang, W., Xie, M., Wang, X., and Xu, J. (2020). Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther. 13, 7809–7818. doi:10.2147/OTT.S263934
Xian, J., Su, W., Liu, L., Rao, B., Lin, M., Feng, Y., et al. (2020). Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the Chinese population. J. Mol. Diagn 22, 1096–1108. doi:10.1016/j.jmoldx.2020.05.011
Xian, J., Zeng, Y., Chen, S., Lu, L., Liu, L., Chen, J., et al. (2021). Discovery of a novel linc01125 isoform in serum exosomes as a promising biomarker for NSCLC diagnosis and survival assessment. Carcinogenesis 42, 831–841. doi:10.1093/carcin/bgab034
Xiong, D., Wang, C., Yang, Z., Han, F., and Zhan, H. (2021). Clinical significance of serum-derived exosomal LINC00917 in patients with non-small cell lung cancer. Front. Genet. 12, 728763. doi:10.3389/fgene.2021.728763
Xu, Y.-X., Pu, S.-D., Li, X., Yu, Z.-W., Zhang, Y.-T., Tong, X.-W., et al. (2022). Exosomal ncRNAs: novel therapeutic target and biomarker for diabetic complications. Pharmacol. Res. 178, 106135. doi:10.1016/j.phrs.2022.106135
Yang, X., Yu, F., Huang, G., Ni, Y., Zhang, T., Zou, Z., et al. (2023). Exosomal miR-133a-3p promotes the growth and metastasis of lung cancer cells following incomplete microwave ablation. Int. J. Hyperth. 40, 2190065. doi:10.1080/02656736.2023.2190065
Zhang, F., Jiang, J., Qian, H., Yan, Y., and Xu, W. (2023). Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 16, 67. doi:10.1186/s13045-023-01452-2
Zhang, Q., Zheng, K., Gao, Y., Zhao, S., Zhao, Y., Li, W., et al. (2023). Plasma exosomal miR-1290 and miR-29c-3p as diagnostic biomarkers for lung cancer. Heliyon 9, e21059. doi:10.1016/j.heliyon.2023.e21059
Zhang, R., Wei, Y., Wang, T., Nie, X., Shi, Z., Deng, Y., et al. (2023). Exosomal miRNAs in autoimmune skin diseases. Front. Immunol. 14, 1307455. doi:10.3389/fimmu.2023.1307455
Zhang, X., Guo, H., Bao, Y., Yu, H., Xie, D., and Wang, X. (2019). Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol. Lett. 18, 5197–5204. doi:10.3892/ol.2019.10892
Zhang, Y., Vaccarella, S., Morgan, E., Li, M., Etxeberria, J., Chokunonga, E., et al. (2023). Global variations in lung cancer incidence by histological subtype in 2020: a population-based study. Lancet Oncol. 24, 1206–1218. doi:10.1016/S1470-2045(23)00444-8
Zhang, Y.-H., Chen, Y., Shi, L., Han, X., Xie, J.-C., Chen, Y., et al. (2024). A novel lung cancer stem cell extracellular vesicles lncRNA ROLLCSC modulate non-stemness cancer cell plasticity through miR-5623-3p and miR-217-5p targeting lipid metabolism. Int. J. Biol. Macromol. 256, 128412. doi:10.1016/j.ijbiomac.2023.128412
Zhao, J., Li, X., Liu, L., Zhu, Z., and He, C. (2023). Exosomes in lung cancer metastasis, diagnosis, and immunologically relevant advances. Front. Immunol. 14, 1326667. doi:10.3389/fimmu.2023.1326667
Keywords: exosome, non-coding RNAs, lncRNA/circRNA-miRNA-mRNA axis, lung cancer, clinical application
Citation: Wang R, Xu Y, Tong L, Zhang X and Zhang S (2024) Recent progress of exosomal lncRNA/circRNA–miRNA–mRNA axis in lung cancer: implication for clinical application. Front. Mol. Biosci. 11:1417306. doi: 10.3389/fmolb.2024.1417306
Received: 14 April 2024; Accepted: 11 June 2024;
Published: 03 July 2024.
Edited by:
Yasunari Matsuzaka, The University of Tokyo, JapanReviewed by:
Seyyed Hossein Khatami, Shahid Beheshti University of Medical Sciences, IranCopyright © 2024 Wang, Xu, Tong, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Zhang, emhhbmdfeGlhb0BnaWJoLmFjLmNu; Sheng Zhang, emhhbmdfc2hlbmdAZ2liaC5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.