
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 06 January 2025
Sec. Metabolomics
Volume 11 - 2024 | https://doi.org/10.3389/fmolb.2024.1406055
This article is part of the Research Topic Metabolomics in Personalized Cancer Medicine View all 6 articles
Background: Prostate cancer (PCa), the most prevalent malignant neoplasm in males, involves complex biological mechanisms and risk factors, many of which remain unidentified. By employing a novel two-sample Mendelian randomization (MR) approach, this study aims to elucidate the causal relationships between the circulating metabolome and PCa risk, utilizing comprehensive data on genetically determined plasma metabolites and metabolite ratios.
Methods: For the MR analysis, we utilized data from the GWAS Catalog database to analyze 1,091 plasma metabolites and 309 ratios in relation to PCa outcomes within two independent GWAS datasets. The inverse variance weighted (IVW) method was the primary approach for determining the existence of the causal relationship, supplemented by additional MR methods for heterogeneity, pleiotropy, and cross-validation. The false discovery rate (FDR) and Bonferroni correction were applied to identify the most significant causative associations. Additionally, reverse MR and Steiger filtering were conducted to ascertain whether PCa influenced the observed metabolite levels. Furthermore, metabolic pathway analysis was conducted with MetaboAnalyst 6.0 software.
Results: In the MR analysis, our findings reveal three overlapped metabolite ratios (arginine to glutamate, phosphate to uridine, and glycerol to mannitol/sorbitol) inversely associated with PCa risk. Following FDR correction (FDR < 0.05), cysteinylglycine disulfide was identified as a potential reducer of PCa risk, whereas Uridine and N-acetyl-L-glutamine (NAG) were pinpointed as potential risk factors. Notably, NAG (OR 1.044; 95% CI 1.025–1.063) emerged as a metabolite with significant causal influence, as confirmed by stringent Bonferroni correction (P < 0.05/1400). Steiger’s directionality test (P < 0.001) and reverse MR confirmed the proposed causal direction. Furthermore, metabolic pathway analysis revealed a significant association between the “Glutathione Metabolism” pathway and PCa development.
Conclusion: This study provides novel insights into the potential causal effects of plasma metabolites and metabolite ratios on PCa. The identified metabolites and ratios could serve as candidate biomarkers, contributing to the elucidation of PCa’s biological mechanisms.
Prostate cancer (PCa) is projected to be the most frequently diagnosed cancer (29%) and the second leading cause of cancer-related mortality (11%) among males in the United States in 2024 (Siegel et al., 2024), as well as a significant cause of mortality worldwide (Sung et al., 2021). The effective prevention and treatment of PCa are crucial in reducing morbidity and mortality. Recent epidemiological studies have identified multiple genetic, lifestyle, and environmental factors associated with PCa risk (Maomao et al., 2022; Jiang et al., 2023). Notably, an increasing number of genetic susceptibility variants involved in the biological mechanisms of PCa have been identified through Genome-Wide Association Studies (GWAS) (Chen et al., 2024). However, these factors alone cannot fully explain the etiology and biological mechanisms of PCa. Moreover, the complex interplay among these factors complicates the determination of potential causality underlying their associations with PCa risk.
Circulating plasma metabolites, small molecules originating from cells, tissues, and biological fluids, encompass a variety of compounds such as amino acids, peptides, carbohydrates, lipids, and xenobiotics. It is noteworthy that metabolomics can offer novel insights into the biological mechanisms of diseases by revealing intermediate metabolites and altered metabolic pathways, a technique frequently employed to study physiological and pathophysiological processes in cancer (Chen et al., 2024; Wishart, 2019; Irajizad et al., 2023; Pavlova et al., 2022; Pan et al., 2024). Several studies have suggested that metabolites, as functional intermediates, can elucidate potential biological mechanisms related to the genetics of PCa (Chen et al., 2024; Lin et al., 2022). However, previous studies have predominantly focused on a limited subset of metabolites and were constrained by small sample sizes, potential confounders, and reverse causality. Moreover, many metabolites, including enzymes, transporters, substrates, and products of enzymatic reactions, reflect biological processes that individual plasma metabolites alone cannot reveal. Investigating cancer risks through metabolite ratios, such as substrate-to-product or metabolite pairs sharing an enzyme or transporter, warrants consideration (Chen et al., 2023). A recent mediation Mendelian randomization (MR) study indicated that the succinate-to-acetoacetate ratio mediated the effect of CD62L-monocyte % monocyte on PCa, with a mediation proportion of 16.6% (95% CI: −163%–196%) (Wu et al., 2024).
MR analysis, an epidemiological research strategy, utilizes genetic variants as instrumental variables (IVs) to link exposure with outcome, thereby assessing causal relationships. Compared to other epidemiological research strategies, MR offers unbiased estimates on genotypes determined at conception, which are not commonly susceptible to confounding factors and reverse causality (Hemani et al., 2018). Owing to this significant advantage, MR has been extensively applied over the past decade to infer the causality of risk exposures to diseases, utilizing publicly available GWAS summary statistics (Lin et al., 2024; Dai et al., 2024; Li et al., 2022). Recently, Chen YH et a (Chen et al., 2023) extended the comprehensive genomic atlas of the plasma metabolome, prioritizing metabolites implicated in human diseases, including 1,091 metabolite levels and 309 metabolite ratios. Additionally, Wang A et al. (Wang et al., 2023) reported the most recent GWAS summary statistics on PCa, featuring the largest sample size to date (726,828).
Herein, we hypothesize that this genetically determined 1,400 plasma metabolome atlas could elucidate the causality of the plasma metabolome on PCa. Consequently, we implemented a two-sample MR approach to assess the causal effects of the human plasma metabolome on PCa and identify potential metabolic pathways, which might elucidate the mechanism of PCa. To validate the significant causal associations identified, we conducted a series of complementary analyses to reinforce their reliability and robustness.
This study systematically investigates the causal relationship between the plasma metabolome (exposures) and PCa (outcome) using MR. The methods of this study were in compliance with the STROBE-MR checklist (Skrivankova et al., 2021). Three foundational assumptions necessary for MR analysis were satisfied: (1) genetically determined IVs are strongly associated with the plasma metabolome; (2) the genetic IVs are not associated with any confounding factors related to the plasma metabolome and PCa; and (3) the genetic IVs influence PCa only through their effect on the plasma metabolome. This study utilized publicly available datasets which had already received ethical approval and informed consent; thus, no additional ethical declaration or consent was required. An overview of this study is depicted in Figure 1.
To mitigate non-independence arising from the overlap of exposure and outcome samples, datasets for exposure and outcome were sourced from different databases. In selecting exposure and outcome GWAS summary statistics, criteria encompassed the European origin of the population, the larger size of population samples and SNPs, and the time of data publication. Recently, Chen et al. (Chen et al., 2023) reported a highly comprehensive exploration of the genetic influences on the plasma metabolome, with a focus on metabolites implicated in human diseases. In their study, a total of 8,299 unrelated individuals of European ancestries were recruited from the Canadian Longitudinal Study on Aging (CLSA) cohort (Raina et al., 2019), all of whom had genome-wide genotyping and measurements of circulating plasma metabolites. Following rigorous quality control and data curation, 1,091 metabolite levels were quantified in plasma samples by Metabolon, Inc. (https://www.metabolon.com/) using the Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) platform. Of these, 850 known metabolites were categorized into eight super pathways: lipid, amino acid, nucleotide, xenobiotics, cofactor and vitamins, carbohydrate, peptide, and energy. Furthermore, to elucidate biological processes not apparent from the study of individual plasma metabolites alone, 309 genetic determinants of metabolite ratios were calculated, including substrate to product ratios and metabolite pairs sharing an enzyme or transporter. Notably, the most recent GWAS summary statistics for PCa, with the largest samples to date (122,188 cases; 604,640 controls; 25,146,978 SNPs) (Wang et al., 2023), was selected for the primary MR analysis. To validate the conclusions drawn, GWAS data were obtained from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) Consortium, comprising 79,148 cases, 61,106 controls, and 19,716,640 SNPs (Schumacher et al., 2018). An overview of the GWAS summary statistics utilized in this study is available in the GWAS Catalog (https://www.ebi.ac.uk/gwas/) (Table 1).
To identify the IVs for metabolite levels and ratios, several procedures were undertaken to verify the validity of the first assumption. Initially, genetic variants were identified through association thresholds of p < 1 × 10−5, a standard practice in MR analysis to encompass a broader range of variation when SNPs available for exposure are limited (Liu et al., 2024). Subsequently, SNPs showing independent inheritance and minimal linkage disequilibrium (LD) (r2 < 0.001, kb = 10,000) within the 1,000 Genomes European subset (Genomes Project et al., 2015) were identified through a clumping procedure in R software. SNPs that were ambiguous or palindromic were also excluded. To mitigate the risk of weak instrument bias, the F statistic (F > 10) was employed to ascertain if the identified IVs possessed sufficient power to represent metabolites or metabolite ratios (Burgess et al., 2011). We also utilized the NHGRI-EBI GWAS Catalog (www.ebi.ac.uk/gwas), which adheres to the findable, accessible, interoperable, and reusable (FAIR) data principles, to determine whether selected SNPs (or those in LD with them) were previously associated with other traits or diseases (P < 1 × 10⁻⁵) (Sollis et al., 2023). Using the LDtrait tool, SNPs associated with potential confounders, such as type 2 diabetes mellitus, non-PCa cancer, smoking, alcohol use, stroke, autoimmune diseases, coronary artery disease, and hyperlipidemia, were excluded to minimize confounding between significantly associated metabolites and PCa. To augment the reliability of our findings, an online power calculator (mRnd) was utilized to calculate the statistical power for MR (https://cnsgenomics.shinyapps.io/mRnd/).
In this study, five complementary MR methods were utilized to estimate the causal relationship between exposure and outcome, namely MR-Egger (Bowden et al., 2015), weighted median (Bowden et al., 2016), inverse variance weighted (IVW), simple mode, and weighted mode. Among these, the IVW method, which provides the most accurate causal estimation between exposure and outcome, served as the primary approach (Burgess et al., 2013). Furthermore, several supplementary analyses were performed to validate the robustness of our MR results. The MR-Egger method not only identifies violations of the IVs assumption but also calculates intercepts to assess potential horizontal pleiotropy and bias from ineffective IVs, providing effect estimates unaffected by these violations (Bowden et al., 2015). Cochran’s Q statistic was applied to estimate heterogeneity among the variables (SNPs) (Cohen et al., 2015). Moreover, the MR-PRESSO global test, an additional MR method designed to identify and correct horizontal pleiotropic outliers, was utilized to assess the existence of horizontal pleiotropy (Verbanck et al., 2018). A “leave-one-out (LOO)” sensitivity analysis was conducted to determine whether results were affected by a single SNP (Burgess and Thompson, 2017). Finally, scatter plots and funnel plots were used to visually display the relationships and interplay between each genetic instrument.
Notably, all analyses utilized the “TwoSampleMR” (version 0.4.22) and MR-PRESSO packages in R software (version 3.6.0). A P-value of <0.05 was deemed statistically significant. Moreover, in light of multiple testing concerns, the false discovery rate (FDR) and Bonferroni correction were applied to identify statistically significant results in multiple comparisons. According to prior studies, FDR < 0.05 and Bonferroni correction are regarded as indicative of significant causal relationships, with Bonferroni correction being notably more stringent.
To investigate whether the researched outcomes had an impact on the plasma metabolome, a reverse MR analysis was performed. In this reverse analysis, SNPs (P < 5 × 10−8, r2 < 0.001, kb = 10,000) selected from GWAS data on PCa as IVs, with blood plasma metabolite levels as the outcome, were used to assess the bidirectionality of the previously determined causal relationship.
Metabolic pathway analysis was conducted using the online tool Metaconflict 6.0 (https://www.metaboanalyst.ca/) (Chong et al., 2018). Functional enrichment analyses and the pathway analyses module were utilized to identify metabolite groups or pathways potentially relevant to the biological processes underlying PCa. Importantly, this study focused solely on metabolites that surpassed the recommended association threshold as determined by IVW (PIVW-mre < 0.05).
Gene expression data were obtained from the publicly available microarray dataset GSE46602, hosted in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). This dataset consists of 14 benign prostate gland samples and 36 PCa samples (Mortensen et al., 2015). Subsequently, Gene Set Enrichment Analysis (GSEA) was conducted using the R package “clusterProfiler” to identify significantly enriched biological pathways. Given the focus of our study, we specifically examined gene sets related to glutathione metabolism. The analysis was performed using default parameters, and the resulting p-values were adjusted using the FDR method to control for multiple hypothesis testing.
After a rigorous selection process, MR analysis was conducted to evaluate the causality of 1,091 metabolite levels and 309 metabolite ratios on PCa using independent GWAS summary datasets. The IVs generated for the plasma metabolome ranged from 11 to 93 SNPs, with a minimum F statistic of 19.50, indicating that all IVs were sufficiently robust for MR analysis. Supplementary Tables S1, S2 displays the characteristics of SNPs and their genetic associations with the 1,400 plasma metabolome and PCa.
The IVW method of MR was utilized to establish causality between the 1,400 plasma metabolome components and PCa, using independent GWAS summary data. A total of 152 suggestive and unique causal associations (PIVW-mre < 0.05) were identified, comprising 117 plasma metabolite levels and 35 metabolite ratios, as detailed in Supplementary Tables S3, S4. Of the 117 plasma metabolites, 91 were identified as known metabolites (including 40 lipids, 33 amino acids, 7 xenobiotics, 4 nucleotides, 4 cofactor and vitamins, 2 carbohydrates, and 1 peptide), while 26 were unknown or partially characterized, as classified by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000). Importantly, all overlapping components of the plasma metabolome exhibited consistent causal effects on PCa across various GWAS dataset pairs, as shown in Figure 2. Additionally, the outcomes of heterogeneity analysis employing Cochran’s Q statistic (applied to both IVW and MR-Egger methods) and pleiotropy analysis using MR-Egger intercepts and the MR-PRESSO global test for these suggestive causal associations are detailed in Supplementary Tables S5, S6.
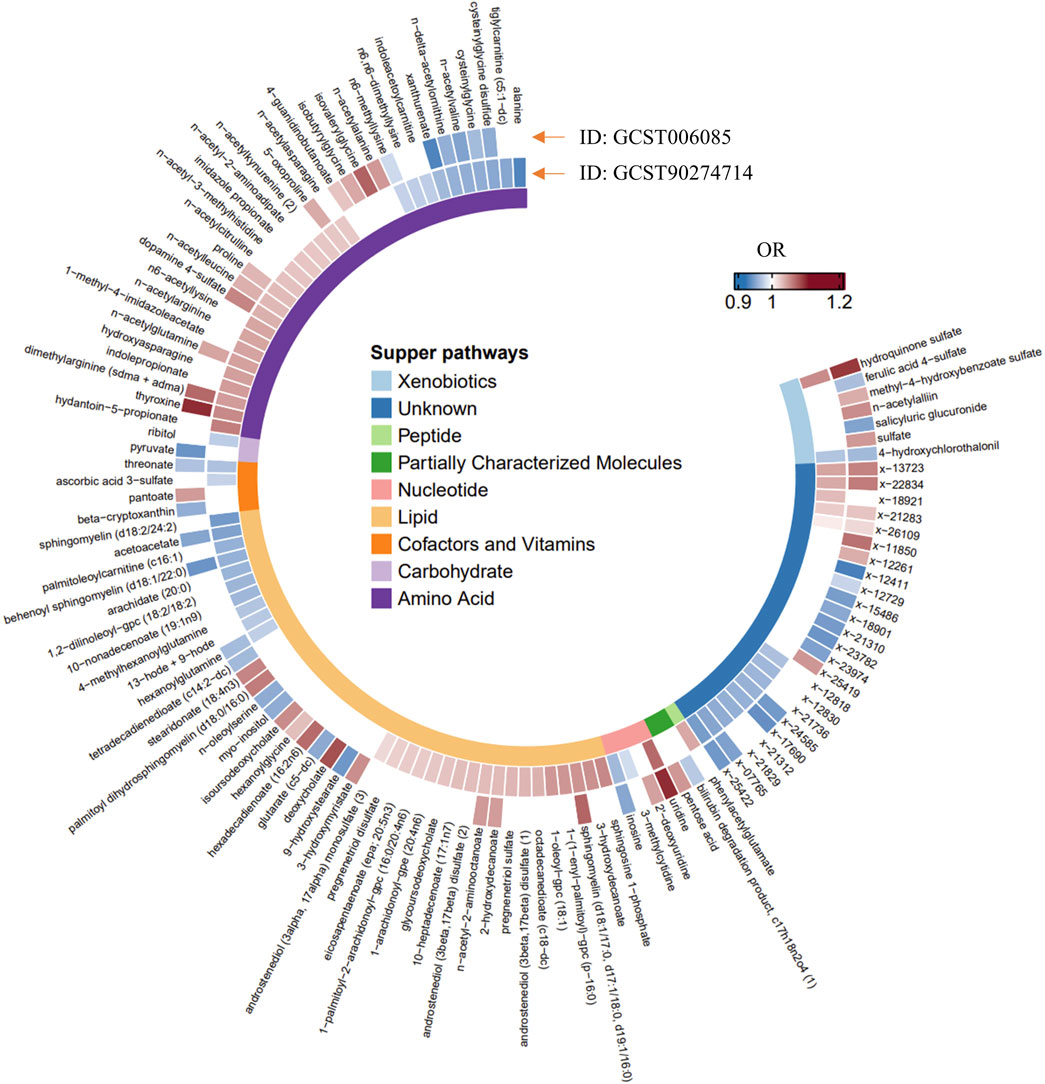
Figure 2. Mendelian randomization associations of 117 plasma metabolite levels on the risk of the two different GWAS datasets of PCa (derived from the random-effect IVW analysis). OR, Odds Ratio.
To confirm the robustness of our findings, further screening was performed on the 152 suggestive associations. Associations within the plasma metabolome consistently identified across all five complementary MR methods (IVW, simple mode, weighted mode, weighted median, and MR-Egger) were included. Figure 3 demonstrates that 20 causal associations were consistent across two distinct GWAS datasets of PCa. All three causative associations among the overlapped metabolite ratios were negatively correlated with PCa risk, specifically: arginine to glutamate ratio, phosphate to uridine ratio, and glycerol to mannitol/sorbitol ratio, with corresponding odds ratios (OR) and confidence intervals detailed in the text. Additionally, no evidence of pleiotropy or heterogeneity was observed among these associations. However, after applying FDR correction to these P-values, no significant confirmative associations were observed, categorizing the results as suggestive causal associations (Figure 3).
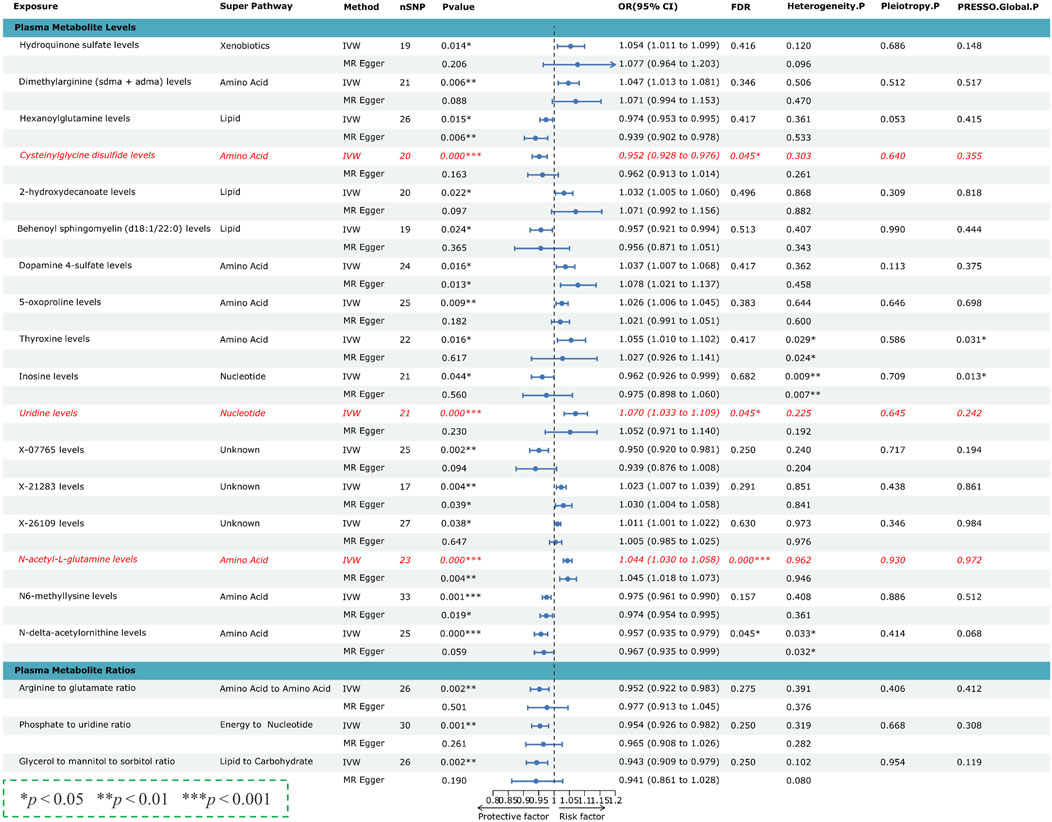
Figure 3. Forest plots presenting the causal effects of 20 identified overlapping plasma metabolites and metabolite ratios on prostate cancer (PCa) risk. Three metabolites, highlighted in red fonts, surpassed the FDR correction threshold (FDR < 0.05), exhibiting no pleiotropy or heterogeneity. SNPs, Single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; FDR, P-value corrected by FDR.
Among the 17 overlapping metabolite levels, 14 causative associations passed all sensitivity analyses without evidence of heterogeneity or horizontal pleiotropy, with three showing significant association with PCa risk (FDR < 0.05). Significant associations were observed with cysteinylglycine disulfide levels negatively correlated with PCa risk, and uridine and N-acetyl-L-glutamine (NAG) levels positively correlated, as detailed with corresponding OR and confidence intervals in Figure 3. Bonferroni correction (P < 0.05/1,400; 3.571 × 10−5) was utilized to identify the most significant causal relationships among the 1,400 plasma metabolome components and PCa. Results indicated that NAG levels are significantly associated with an increased risk of PCa. The statistical power for the association of NAG levels with PCa was 1.0, demonstrating robustness against a type I error rate of 0.05. Fixed-effect IVW estimates confirmed the association of NAG levels with an increased risk of PCa across two datasets. Specifically, additional methods including the weighted median, weighted mode, simple mode, and MR-Egger, indicated consistent results, as shown in Figure 4 and Supplementary Figure S1, S2. Furthermore, based on the “leave-one-out” analysis, none of the associations were driven solely by a single SNP, as evidenced in Supplementary Figures S3–S6. The MR Steiger directionality test results confirmed the accuracy of our causal direction estimate (P < 0.001). Reverse MR analysis showed no causal relationship from PCa to NAG levels, indicating directionality in the observed association, as detailed in Supplementary Table S7. Consequently, it was determined that NAG levels might be causally associated with PCa, underscoring the reliability of the results.
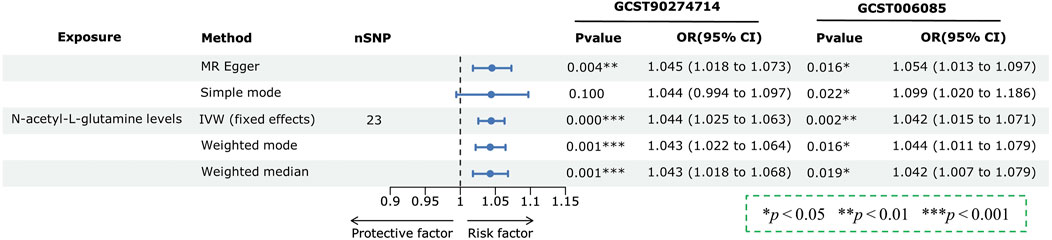
Figure 4. Forest plots depicting the causal effect estimates of N-acetyl-L-glutamine (NAG) levels on prostate cancer (PCa) risk across two independent datasets, utilizing five complementary MR methods.
Metabolic pathway analysis identified five pathways significantly associated with the development of PCa (P < 0.05): Glutathione metabolism, Biosynthesis of unsaturated fatty acids, Arginine and proline metabolism, Tyrosine metabolism, and Pyrimidine metabolism. Notably, the Glutathione metabolism pathway emerged as a recurrent feature across two independent PCa datasets, underscoring its potential role in PCa pathophysiology (Table 2). To further delineate the molecular underpinnings of this finding, we performed GSEA focused on glutathione-related gene sets. This approach enabled a more integrated assessment of dysregulated metabolic signatures in PCa, transcending the metabolite-level observations. The GSEA results revealed a consistent downregulation of multiple glutathione metabolism-associated gene sets in malignant prostate tissues compared to benign controls. Among these, GOMF_Glutathione_Transferase_Activity and WP_Glutathione_Metabolism showed statistically significant enrichment patterns (FDR q < 0.05) (Supplementary Figure S7).
In this comprehensive MR analysis, leveraging large-scale GWAS data, we identified 152 significant associations. These suggest potential causal influences of the plasma metabolome—including 117 metabolite levels and 35 metabolite ratios—on the risk of PCa. Of these, 17 metabolite levels and three metabolite ratios exhibited consistent associations across two independent PCa datasets. Notably, following stringent Bonferroni correction, plasma NAG emerged as the most robust causal factor. Furthermore, integrating metabolite-focused MR findings with GSEA of transcriptomic data implicated glutathione-related metabolic pathways in PCa pathogenesis. Collectively, these results underscore the complex interplay between genetic variation, metabolic profiles, and PCa, and may inform the development of novel prevention and therapeutic strategies.
Effective methods for cancer risk assessment and prevention remain a paramount priority. Advances in metabolomics have heightened interest in circulating metabolites as non-invasive biomarkers that reflect both endogenous metabolic states and environmental exposures (Qiu et al., 2023). Certain metabolites have been implicated in tumor biology; for example, proline—a highly abundant amino acid in collagen—contributes to the formation of extracellular matrices by cancer-associated fibroblasts, thereby fostering a tumor-promoting microenvironment (Kay et al., 2022). Moreover, metabolites involved in nucleotide metabolism have shown potential as therapeutic targets, as their modulation can impede tumor progression in preclinical models (Stine et al., 2022). In the present study, the observed positive association between asymmetric dimethylarginine (ADMA), an endogenous nitric oxide synthase (NOS) inhibitor, and PCa risk is consistent with emerging molecular evidence (Reddy et al., 2018; Asha Parveen et al., 2023). Similarly, dopamine 4-sulfate, a dopamine metabolite implicated in tumor proliferation, was positively associated with PCa risk. Prior studies have indicated its potential as a predictive biomarker for non-muscle invasive bladder cancer (NMIBC) (Cheng et al., 2018), suggesting that it may exert broader oncogenic influences.
Among the 1,400 plasma metabolites and ratios analyzed, plasma NAG demonstrated the most pronounced causal relationship with PCa. Mechanistically, NAG is intimately linked to glutamine metabolism. Glutamine—a central amino acid supporting biosynthesis, bioenergetics, and cell signaling—furnishes cancer cells with essential nitrogen and carbon for the synthesis of nucleotides, amino acids, and lipids, while also maintaining redox balance (Pavlova et al., 2022). By undergoing N-acetylation, glutamine’s stability, transport, and downstream metabolic availability may be enhanced, potentially providing tumor cells with more refined control over glutamine-derived intermediates. This regulatory mechanism could bolster cancer cell growth, survival, and invasive capabilities (Caldovic et al., 2010). Moreover, within the tumor microenvironment, PCa cells frequently display metabolic reprogramming driven by oncogenic signals and genetic alterations, which optimize glutamine uptake and utilization. Studies in irradiation-resistant HepG2 cells have further highlighted elevated NAG levels under ferroptotic stress, suggesting a role for this metabolite in stress response pathways (Yuan et al., 2022). Although our MR analysis strongly implicates NAG in PCa etiology, additional experimental studies are warranted to elucidate the precise molecular mechanisms involved.
Consideration of multiple-testing adjustments strengthens the validity of our findings. Beyond the Bonferroni correction, FDR correction identified cysteinylglycine and uridine as significantly associated with PCa risk, acting as a protective and risk factor, respectively. Previous research has documented that elevated extracellular cysteinylglycine, a metabolite related to glutathione, can suppress ferroptosis in certain cancer contexts, thereby fostering tumor growth (Hayashima and Katoh, 2022). Elevated serum cysteinylglycine levels have also been associated with a heightened risk of breast cancer (Lin et al., 2007), although other studies have reported inverse or null associations with certain malignancies (Houghton et al., 2019; Miranti et al., 2016). Uridine, a pyrimidine nucleoside abundant in plasma, fulfills multiple biological roles, including those related to antioxidant capacity and aging (Lai et al., 2023; Zhang et al., 2022). Functionally, uridine may enhance cancer cell proliferation and survival by serving as an energy source and modifying protein O-GlcNAcylation patterns (Yang et al., 2024). Our MR results, which identify uridine as a risk factor for PCa, align with these mechanistic insights. Taken together, these findings suggest that NAG, cysteinylglycine, and uridine may influence PCa onset and progression, potentially through pathways involving oxidative stress or ferroptosis regulation.
To our knowledge, this study is the first MR investigation to integrate genomic data with plasma metabolite ratios to assess causality in PCa risk. Our analyses identified three metabolite ratios—arginine to glutamate, phosphate to uridine, and glycerol to mannitol/sorbitol—that were significantly and inversely associated with PCa risk across datasets. These ratios integrate multiple metabolic dimensions: for instance, arginine, as a precursor to nitric oxide, may enhance immune surveillance, whereas glutamate supports tumorigenic processes. A higher arginine-to-glutamate ratio could thus reflect metabolic conditions less conducive to cancer cell proliferation (Huang et al., 2019). Likewise, while phosphate contributes to ATP production and phosphorylation signaling, uridine supports RNA and nucleotide biosynthesis. Thus, a metabolic environment favoring phosphate over uridine may be less advantageous for the rapid proliferation of malignant cells (Pavlova et al., 2022; Lai et al., 2023). The glycerol-to-mannitol/sorbitol ratio may similarly indicate restricted use of alternative carbohydrate pathways by cancer cells, potentially limiting their ability to adapt to oxidative stress and altering their energy metabolism (Schwab et al., 2024).
Our pathway analyses indicate that alterations in glutathione metabolism may contribute to the redox imbalances, growth advantages, and oxidative stress resistance commonly observed in PCa. Glutathione, as a key intracellular antioxidant, is integral to maintaining cellular redox homeostasis. Tumors can exploit glutathione-related pathways to mitigate oxidative stress, enhance survival under challenging microenvironmental conditions, and promote therapeutic resistance. By modulating components of glutathione synthesis, recycling, or transport, it may be possible to disrupt this carefully orchestrated balance, rendering cancer cells more susceptible to oxidative damage. Agents designed to inhibit key enzymes, transporters, or regulatory factors within this pathway could diminish the cancer cells’ metabolic flexibility. When combined with standard treatments—such as androgen deprivation therapy, chemotherapy, or emerging targeted therapies—these metabolic interventions may lower the threshold for cancer cell death, delay resistance, and enhance long-term treatment efficacy.
This study is not without limitations. First, the accuracy of MR analyses is contingent upon the selection and interpretation of genetic instruments. The lack of sex-specific instruments and potential genetic heterogeneity may introduce residual confounding. Additionally, while MR offers robust evidence for causal inference, our findings require experimental validation to unravel the underlying biological mechanisms. These initial results, however, provide a foundation for future research. We advocate for enhanced screening protocols in populations with metabolic dysregulation and recommend longitudinal cohort studies to identify metabolite-based biomarkers of cancer recurrence. Such efforts may ultimately improve the clinical prevention, early detection, and prognosis of PCa.
This study reveals novel causal relationships between specific plasma metabolites and PCa risk, notably identifying N-acetyl-L-glutamine (NAG) as a key factor and underscoring glutathione metabolism as a pivotal pathway. By employing a robust two-sample MR framework, these findings illuminate the intricate metabolic underpinnings of PCa, thus offering valuable new insights into its pathophysiological mechanisms. Such revelations not only enrich our understanding of disease etiology but also point toward promising biomarkers for early detection, prevention, and targeted therapeutic strategies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
CF: Conceptualization, Validation, Visualization, Writing–original draft, Writing–review and editing. HL: Writing–original draft, Writing–review and editing. CZ: Writing–original draft, Writing–review and editing. YZ: Writing–original draft, Writing–review and editing. HZ: Writing–original draft, Writing–review and editing. PZ: Writing–original draft, Writing–review and editing. SZ: Writing–original draft, Writing–review and editing. LW: Writing–original draft, Writing–review and editing. JY: Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by Ph.D. Startup Fund of The First People’s Hospital of Lianyungang (BS202206).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1406055/full#supplementary-material
Asha Parveen, S. M., Kami Reddy, K. R., and Ummanni, R. (2023). Dimethylarginine Dimethylaminohydrolase - 1 expression is increased under tBHP-induced oxidative stress regulates nitric oxide production in PCa cells attenuates mitochondrial ROS-mediated apoptosis. Nitric Oxide 138-139, 70–84. doi:10.1016/j.niox.2023.07.002
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44 (2), 512–525. doi:10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40 (4), 304–314. doi:10.1002/gepi.21965
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37 (7), 658–665. doi:10.1002/gepi.21758
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32 (5), 377–389. doi:10.1007/s10654-017-0255-x
Burgess, S., Thompson, S. G., and Collaboration, C. C. G. (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40 (3), 755–764. doi:10.1093/ije/dyr036
Caldovic, L., Ah Mew, N., Shi, D., Morizono, H., Yudkoff, M., and Tuchman, M. (2010). N-acetylglutamate synthase: structure, function and defects. Mol. Genet. Metab. 100 (Suppl. 1), S13–S19. doi:10.1016/j.ymgme.2010.02.018
Chen, Y., Lu, T., Pettersson-Kymmer, U., Stewart, I. D., Butler-Laporte, G., Nakanishi, T., et al. (2023). Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55 (1), 44–53. doi:10.1038/s41588-022-01270-1
Chen, Y., Xie, Y., Ci, H., Cheng, Z., Kuang, Y., Li, S., et al. (2024). Plasma metabolites and risk of seven cancers: a two-sample Mendelian randomization study among European descendants. BMC Med. 22 (1), 90. doi:10.1186/s12916-024-03272-8
Cheng, X., Liu, X., Liu, X., Guo, Z., Sun, H., Zhang, M., et al. (2018). Metabolomics of non-muscle invasive bladder cancer: biomarkers for early detection of bladder cancer. Front. Oncol. 8, 494. doi:10.3389/fonc.2018.00494
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46 (W1), W486–W494. doi:10.1093/nar/gky310
Cohen, J. F., Chalumeau, M., Cohen, R., Korevaar, D. A., Khoshnood, B., and Bossuyt, P. M. (2015). Cochran's Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 68 (3), 299–306. doi:10.1016/j.jclinepi.2014.09.005
Dai, X., Wang, H., Zhong, R., Li, J., and Hou, Y. (2024). Causality of genetically determined metabolites on susceptibility to prevalent urological cancers: a two-sample Mendelian randomization study and meta-analysis. Front. Genet. 15, 1398165. doi:10.3389/fgene.2024.1398165
Genomes Project, C., Auton, A., Brooks, L. D., Durbin, R. M., Garrison, E. P., Kang, H. M., et al. (2015). A global reference for human genetic variation. Nature 526 (7571), 68–74. doi:10.1038/nature15393
Hayashima, K., and Katoh, H. (2022). Expression of gamma-glutamyltransferase 1 in glioblastoma cells confers resistance to cystine deprivation-induced ferroptosis. J. Biol. Chem. 298 (3), 101703. doi:10.1016/j.jbc.2022.101703
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, e34408. doi:10.7554/eLife.34408
Houghton, S. C., Eliassen, A. H., Zhang, S. M., Selhub, J., Rosner, B. A., Willett, W. C., et al. (2019). Plasma B-vitamin and one-carbon metabolites and risk of breast cancer before and after folic acid fortification in the United States. Int. J. Cancer 144 (8), 1929–1940. doi:10.1002/ijc.31934
Huang, J., Mondul, A. M., Weinstein, S. J., Derkach, A., Moore, S. C., Sampson, J. N., et al. (2019). Prospective serum metabolomic profiling of lethal prostate cancer. Int. J. Cancer 145 (12), 3231–3243. doi:10.1002/ijc.32218
Irajizad, E., Kenney, A., Tang, T., Vykoukal, J., Wu, R., Murage, E., et al. (2023). A blood-based metabolomic signature predictive of risk for pancreatic cancer. Cell Rep. Med. 4 (9), 101194. doi:10.1016/j.xcrm.2023.101194
Jiang, J., Yang, J., Chen, B., Li, J., Zhang, T., Tan, D., et al. (2023). The impact of nutrient supply on prostate cancer risk worldwide. Nutrients 15 (24), 5131. doi:10.3390/nu15245131
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kay, E. J., Paterson, K., Riera-Domingo, C., Sumpton, D., Däbritz, J. H. M., Tardito, S., et al. (2022). Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat. Metab. 4 (6), 693–710. doi:10.1038/s42255-022-00582-0
Lai, K., Song, C., Gao, M., Deng, Y., Lu, Z., Li, N., et al. (2023). Uridine alleviates sepsis-induced acute lung injury by inhibiting ferroptosis of macrophage. Int. J. Mol. Sci. 24 (6), 5093. doi:10.3390/ijms24065093
Li, R., Wang, X., Zhang, Y., Xu, X., Wang, L., Wei, C., et al. (2022). Analysis of tryptophan and its main metabolite kynurenine and the risk of multiple cancers based on the bidirectional mendelian randomization analysis. Front. Oncol. 12, 852718. doi:10.3389/fonc.2022.852718
Lin, J., Manson, J. E., Selhub, J., Buring, J. E., and Zhang, S. M. (2007). Plasma cysteinylglycine levels and breast cancer risk in women. Cancer Res. 67 (23), 11123–11127. doi:10.1158/0008-5472.CAN-07-3061
Lin, Y., Yang, Z., Li, J., Sun, Y., Zhang, X., Qu, Z., et al. (2022). Effects of glutamate and aspartate on prostate cancer and breast cancer: a Mendelian randomization study. BMC Genomics 23 (1), 213. doi:10.1186/s12864-022-08442-7
Lin, Y., Zhang, Y., Wang, S., Cao, L., Zhao, R., Ma, X., et al. (2024). Pharmacological targets of SGLT2 inhibition on prostate cancer mediated by circulating metabolites: a drug-target Mendelian randomization study. Front. Pharmacol. 15, 1443045. doi:10.3389/fphar.2024.1443045
Liu, S., Ding, D., Liu, F., Guo, Y., Xie, L., and Han, F. J. (2024). Exploring the causal role of multiple metabolites on ovarian cancer: a two sample Mendelian randomization study. J. Ovarian Res. 17 (1), 22. doi:10.1186/s13048-023-01340-w
Maomao, C., He, L., Dianqin, S., Siyi, H., Xinxin, Y., Fan, Y., et al. (2022). Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol. Med. 19 (8), 1121–1138. doi:10.20892/j.issn.2095-3941.2022.0231
Miranti, E. H., Freedman, N. D., Weinstein, S. J., Abnet, C. C., Selhub, J., Murphy, G., et al. (2016). Prospective study of serum cysteine and cysteinylglycine and cancer of the head and neck, esophagus, and stomach in a cohort of male smokers. Am. J. Clin. Nutr. 104 (3), 686–693. doi:10.3945/ajcn.115.125799
Mortensen, M. M., Hoyer, S., Lynnerup, A. S., Ørntoft, T. F., Sørensen, K. D., Borre, M., et al. (2015). Expression profiling of prostate cancer tissue delineates genes associated with recurrence after prostatectomy. Sci. Rep. 5, 16018. doi:10.1038/srep16018
Pan, R., Liu, J., Xiao, M., Sun, C., Zhu, J., Wan, L., et al. (2024). Causal links of human serum metabolites on the risk of prostate cancer: insights from genome-wide Mendelian randomization, single-cell RNA sequencing, and metabolic pathway analysis. Front. Endocrinol. (Lausanne) 15, 1443330. doi:10.3389/fendo.2024.1443330
Pavlova, N. N., Zhu, J., and Thompson, C. B. (2022). The hallmarks of cancer metabolism: still emerging. Cell Metab. 34 (3), 355–377. doi:10.1016/j.cmet.2022.01.007
Qiu, S., Cai, Y., Yao, H., Lin, C., Xie, Y., Tang, S., et al. (2023). Small molecule metabolites: discovery of biomarkers and therapeutic targets. Signal Transduct. Target Ther. 8 (1), 132. doi:10.1038/s41392-023-01399-3
Raina, P., Wolfson, C., Kirkland, S., Griffith, L. E., Balion, C., Cossette, B., et al. (2019). Cohort profile: the Canadian longitudinal study on aging (CLSA). Int. J. Epidemiol. 48 (6), 1752–1753j. doi:10.1093/ije/dyz173
Reddy, K. R. K., Dasari, C., Duscharla, D., Supriya, B., Ram, N. S., Surekha, M. V., et al. (2018). Dimethylarginine dimethylaminohydrolase-1 (DDAH1) is frequently upregulated in prostate cancer, and its overexpression conveys tumor growth and angiogenesis by metabolizing asymmetric dimethylarginine (ADMA). Angiogenesis 21 (1), 79–94. doi:10.1007/s10456-017-9587-0
Schumacher, F. R., Al Olama, A. A., Berndt, S. I., Benlloch, S., Ahmed, M., Saunders, E. J., et al. (2018). Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50 (7), 928–936. doi:10.1038/s41588-018-0142-8
Schwab, A., Siddiqui, M. A., Ramesh, V., Gollavilli, P. N., Turtos, A. M., Møller, S. S., et al. (2024). Polyol pathway-generated fructose is indispensable for growth and survival of non-small cell lung cancer. Cell Death Differ. doi:10.1038/s41418-024-01415-1
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49. doi:10.3322/caac.21820
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA 326 (16), 1614–1621. doi:10.1001/jama.2021.18236
Sollis, E., Mosaku, A., Abid, A., Buniello, A., Cerezo, M., Gil, L., et al. (2023). The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51 (D1), D977–d985. doi:10.1093/nar/gkac1010
Stine, Z. E., Schug, Z. T., Salvino, J. M., and Dang, C. V. (2022). Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 21 (2), 141–162. doi:10.1038/s41573-021-00339-6
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50 (5), 693–698. doi:10.1038/s41588-018-0099-7
Wang, A., Shen, J., Rodriguez, A. A., Saunders, E. J., Chen, F., Janivara, R., et al. (2023). Characterizing prostate cancer risk through multi-ancestry genome-wide discovery of 187 novel risk variants. Nat. Genet. 55 (12), 2065–2074. doi:10.1038/s41588-023-01534-4
Wishart, D. S. (2019). Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 99 (4), 1819–1875. doi:10.1152/physrev.00035.2018
Wu, X., Zou, W., and Liu, Z. (2024). The relationship between immune cells and prostate cancer, and the mediating role of metabolites: a Mendelian randomization study. Sci. Rep. 14 (1), 26217. doi:10.1038/s41598-024-78085-z
Yang, Y., Ye, Y., Deng, Y., and Gao, L. (2024). Uridine and its role in metabolic diseases, tumors, and neurodegenerative diseases. Front. Physiol. 15, 1360891. doi:10.3389/fphys.2024.1360891
Yuan, Z., Liu, T., Huo, X., Wang, H., Wang, J., and Xue, L. (2022). Glutamine transporter SLC1A5 regulates ionizing radiation-derived oxidative damage and ferroptosis. Oxid. Med. Cell Longev. 2022, 3403009. doi:10.1155/2022/3403009
Keywords: metabolites, metabolites ratios, prostate cancer (PCa), risk, Mendelian randomization (MR)
Citation: Feng C, Li H, Zhang C, Zhou Y, Zhang H, Zheng P, Zhao S, Wang L and Yang J (2025) Exploring the causal role of plasma metabolites and metabolite ratios in prostate cancer: a two-sample Mendelian randomization study. Front. Mol. Biosci. 11:1406055. doi: 10.3389/fmolb.2024.1406055
Received: 24 March 2024; Accepted: 18 December 2024;
Published: 06 January 2025.
Edited by:
Jacopo Troisi, University of Salerno, ItalyReviewed by:
Qingmo Yang, The First Affiliated Hospital of Xiamen University, ChinaCopyright © 2025 Feng, Li, Zhang, Zhou, Zhang, Zheng, Zhao, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Yang, MTg5NjEzMjIzNTVAMTg5LmNu; Lei Wang, d2FuZ2xlaXdlbmR5QDEyNi5jb20=; Shaolin Zhao, emhhb3NoYW9saW4xQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.