- 1Department of Pathology, Bishan Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Gastroenterology, Bishan Hospital of Chongqing Medical University, Chongqing, China
The consistent notion holds that hepatocellular carcinoma (HCC) initiation, progression, and clinical treatment failure treatment failure are affected by the accumulation of various genetic and epigenetic alterations. MicroRNAs (miRNAs) play an irreplaceable role in a variety of physiological and pathological states. meanwhile, epithelial-mesenchymal transition (EMT) is a crucial biological process that controls the development of HCC. miRNAs regulate the intermediation state of EMTor mesenchymal-epithelial transition (MTE)thereby regulating HCC progression. Notably, miRNAs regulate key HCC-related molecular pathways, including the Wnt/β-catenin pathway, PTEN/PI3K/AKT pathway, TGF-β pathway, and RAS/MAPK pathway. Therefore, we comprehensively reviewed how miRNAs produce EMT effects by multiple signaling pathways and their potential significance in the pathogenesis and treatment response of HCC. emphasizing their molecular pathways and progression in HCC initiation. Additionally, we also pay attention to regulatory mechanisms that are partially independent of signaling pathways. Finally, we summarize and propose miRNA-targeted therapy and diagnosis and defense strategies forHCC. The identification of the mechanism leading to the activation of EMT programs during HCC disease processes also provides a new protocol for the plasticity of distinct cellular phenotypes and possible therapeutic interventions. Consequently, we summarize the latest progress in this direction, with a promising path for further insight into this fast-moving field.
1 Introduction
Hepatocellular carcinoma is a malignant tumor derived from hepatocytes and hepatobiliary cells. The latest data show that liver cancer ranks sixth in new cases and third in mortality worldwide (Sung et al., 2021). In recent years, although great progress has been made on HCC molecular mechanism and therapy techniques, the prognosis of HCC patients is generally poor due to high metastasis and recurrence, which makes us need to examine this urgent problem from a broader perspective (Avila et al., 2006). Studies have shown chronic alcohol consumption or aflatoxin intoxication, chronic hepatitis B or C virus (HBV, HCV, etc.) infection or a high-fat diet that leads to obesity are clear etiological factors that contribute to the development of HCC for decades (Forner et al., 2012). The formation of HCC is a heterogeneous formation of hepatocytes and progenitor cells, both of which are epithelial cell types, and the plastic changes of the latter lead to the generation of EMT. At the same time, the production of EMT in hepatoma cells is systematically regulated by miRNAs (Gao et al., 2014). MicroRNA (miRNA) is a class of endogenous small non-coding RNA of 18–25 nucleotides long. It is well-recognized that miRNAs play a crucial role in the initiation and proliferation of various types of cancer through the regulation of dissimilar signaling pathways (Doghish et al., 2022). EMT endows cells with migration and invasion properties, induces stem cell properties, effectively prevents apoptosis and senescence, and contributes to immunosuppression. The mesenchymal state of cells is closely related to the ability of cells to migrate to distant organs. At the same time, the induced stemness of cells allows their subsequent differentiation into multiple cell types during development and the onset of metastasis (Thiery et al., 2009). Meanwhile, EMT is a key intermediate process that drives tumor metastasis. Accumulating evidence suggests that dysregulation of some microRNAs is involved in this process. more clearly speaking, ample evidence exists for the important regulatory potential of other miRNAs in conditions associated with TMT processes in HCC (Szabo and Bala, 2013). Notably, the precise regulation of HCC progression by EMT-mediated miRNAs needs to be better summarized and considered despite numerous efforts by researchers (Chen et al., 2016).
2 Classical miRNA production pathways
As integral parts of small regulatory RNAs, miRNAs do not encode proteins but play a critical role in post-transcriptional regulation by targeting the 3′-UTR of target gene transcripts to reduce the expression of homologous proteins and regulate cellular biological processes (Feng et al., 2022). The formation of miRNA is accompanied by four processes. When nuclear RNA polymerase II transcribes pri-miRNA with a hairpin structure (Lee et al., 2004), it is cleaved by the microprocessor to form pre-miRNAand then exported from the nucleus to the cytoplasm by Exportin five and RAN-GTP (Monica Ballarino et al., 2009). In the cytoplasm, pre-miRNA is further converted into miRNAs* by Dicer, and with the help of the chaperones HSC70/HSP90, the formed miRNA* is loaded into the Argonaute protein to form a silencing complex, whereby targeting and regulating the target gene (Iwasaki et al., 2010). Mechanistically, the silencing complex forms 3′-end extensive pairing on the one hand and then cleaves mRNA in animal cells through endonuclease properties (Bartel, 2018). On the other hand, the eukaryotic translation initiation factor 4E (eIF4E) transporter (4E-T) forms a translation barrier by recruiting DDX6 to bind the CCR4-NOT complex (Kamenska et al., 2014; Kamenska et al., 2016), preventing miRNA translation initiation and recruiting TNRC6 and tail-related regions through the Argonaute protein (PABPC) binding accelerates the rapid degradation of miRNAs (Jonas and Izaurralde, 2015). The schematic diagram of the canonical miRNA production and action mechanism is shown in Figure 1.
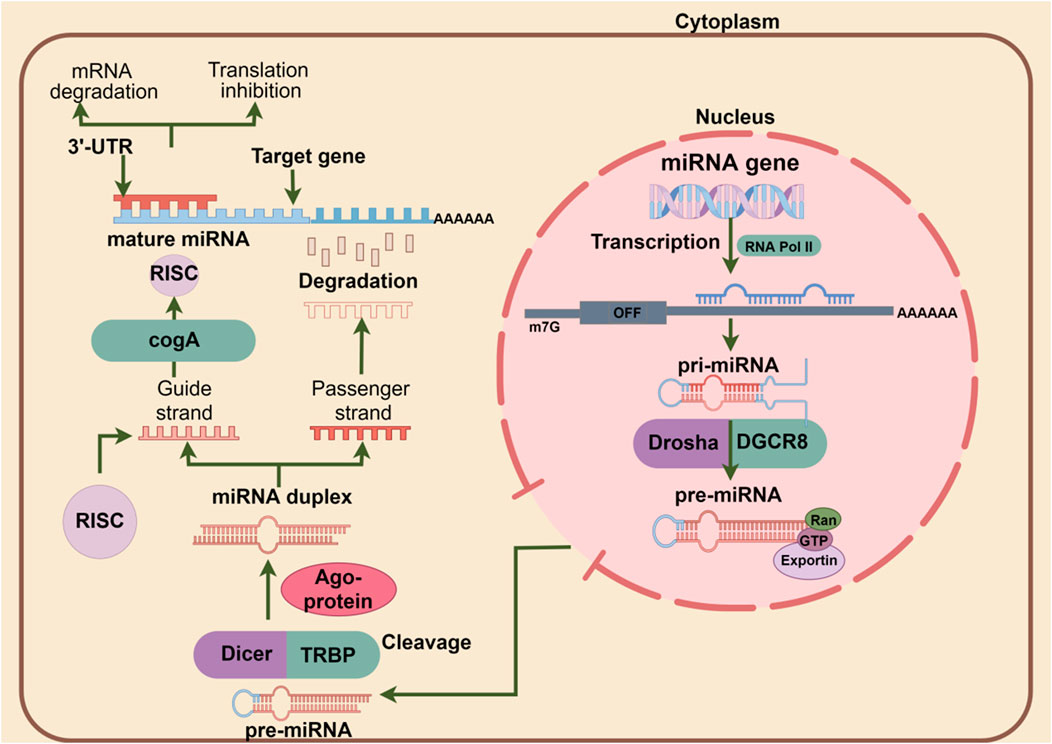
Figure 1. The pri-miRNAs in the nucleus are synthesized by RNA polymerase II. After processing by the microprocessor, it is transported to the cytoplasm and then treated by DICER. Finally, mature miRNAs are loaded onto AGO proteins and bind to target genes to exert their biological functions.
3 EMT
The concept of EMT was first proposed by Betty Hay in 1963 and described its pivotal role in embryonic development (Trelstad et al., 1967). EMT is mainly divided into three types according to the biological background of occurrence. Type I EMT occurs during embryonic development, and type II and type III EMT occurs during wound healing, tissue regeneration, and progression of cancer respectively (Lamouille et al., 2014). Notably, type III EMT is always accompanied by systemic metastasis of liver cancer cells, including lymph nodes, lungs, kidneys, and spleens. The characteristics of EMT during cancer metastasis is the suppression of the expression of the epithelial gene E-cadherin, resulting in the typical polygonal, cobblestone-like shape of epithelial cells with intercellular adhesions such as cell junctions, including tight junctions, adherens junctions, desmosomes, and gaps the junction gradually disintegrates and is accompanied by a loss of cell polarity. In addition, cells acquire a spindle-shaped mesenchymal morphology and express markers associated with the mesenchymal cell state, in particular N-cadherin, matrix metallopeptidase family proteins (MMPs), vimentin, and fibronectin (Trelstad et al., 1967; Lamouille et al., 2014; Dongre and Weinberg, 2018).
4 Multiple signaling pathways involved in EMT-mediated miRNA regulation of HCC
To shed light on a portion of the molecular underpinnings of miRNA regulating the EMT in cells, we, at least in part, introduce the interplay of multiple signaling pathways during HCC metastasis. Correspondingly, Studies have shown that EMT progression is regulated by a variety of signaling pathways. In addition to the common signaling pathway that promotes transforming growth factor-β (TGFβ) fibrosis through SMAD proteins, it can also activate PI3K-AKT, RAS/MAPK, and p38 MAPK and other key signaling pathways in HCC (Derynck, 2005; Xu et al., 2009). Phosphorylation of TβRI creates a site between growth factor receptor binding protein 2 (GRB2) and Sevenless (SOS) and initiates the RAS-RAF-MEK-ERK-MAPK pathway, where TGFβ-induced p38 MAPK (Lee et al., 2007). Some growth factors induce EMT through receptor tyrosine kinases (RTKs), including epidermal growth factor (EGF), fibroblast growth factor (FGF), hepatocyte growth factor (HGF), and vascular endothelial growth factor (VEGF). Furthermore, activated ERK1 and ERK2, which initiate MAPK, promote EMT by increasing the expression of EMT transcription factors and regulators of cell motility and invasion (Stoker, 1985; Rivera-Feliciano et al., 2006; Wanami et al., 2008; Shirakihara et al., 2011). Meanwhile, WNT/β-catenin signaling promotes EMT by inhibiting glycogen synthase. Glycogen synthase kinase-3 (GSK3β) is found to stabilize β-catenin, which translocates to the nucleus to engage the transcription factors lymphoid enhancer-binding factor 1 (LEF) and T cell factor (TCF) and promote EMT-friendly Gene Expression Program (Yang et al., 2006; Nawshad et al., 2007). Hypoxia in the tumor environment can also promote EMT through hypoxia-inducible factor 1α (HIF1α), thereby activating the expression of TWIST (Yang et al., 2008). Notably, the intervention of exosomes makes the interaction between various signaling pathways more effective, and the remote regulation of HCC by miRNAs through exosomes has also received further attention.
4.1 miRNAs regulate the EMT process of hepatoma cells via the Wnt/β-catenin signaling pathway
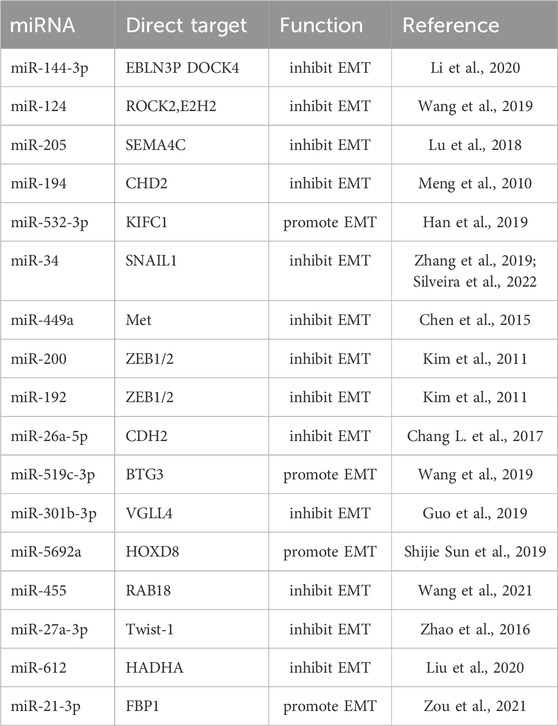
High expression of miR-5188 in HCC patients predicts poor prognosis. Existing evidence indicates that miR-5188 directly targets FOXO1, which interacts with β-catenin in the cytoplasm to reduce nuclear translocation of β-catenin and promote Wnt signaling and downstream tumor activation EMT. The highly expressed miR-5188 inhibited the expression of FOXO1, promoted the accumulation of β-catenin in the nucleus, and activated the expression of interstitial genes to promote cell proliferation and invasion (Lin et al., 2019). High levels of miR-197 expression promote EMT and invasion of HCC cells in vitro and in vivo, and mechanistically, miR-197 directly targets Wnt/β-catenin signaling negative regulators Axin-2, Naked cuticle 1 (NKD1) and Dickkopf-related protein 2 (DKK2) thereby increasing the activity of TCF/LEF and accelerating the nuclear accumulation of β-catenin (Hu et al., 2018). Available data show that in clinical liver cancer patient samples and liver cancer cells, miR-498 is significantly lower than that in normal patients and liver cells. The highly expressed miR-498 can inhibit the proliferation, migration, and invasion of hepatoma cells by inhibiting ZEB2 (Zhang T. et al., 2018). Frizzled Class Receptor 7(FZD7), a co-receptor of Wnt signaling, is a key factor in the regulation of Wnt. Studies have shown that miR-542-3p has been demonstrated as a tumor suppressor in many cancers, inhibiting the proliferation and invasion of HCC by inhibiting the activation of the Wnt signaling pathway through targeted regulation FZD7 in liver cancer cells (Wu et al., 2017). Hepatic stellate cells trigger miR-1246, which targets binding to RORα and impairs the interaction of RORα protein with β-catenin, resulting in increased nuclear levels of β-catenin and decreased cytoplasmic β-catenin levels in vitro and in vivo (Huang et al., 2020). Similarly, miR-504 targets FZD7 and β-catenin, and restoration of FZD7 expression significantly reversed the inhibitory effects of miR-504 on HCC cell proliferation, invasion, and Wnt/β-catenin signaling (Quan et al., 2018). miR-298 directly targets catenin delta 1 (CTNND1), and the low expression of miR-298 may be a powerful assistant for the proliferation and invasion of liver cancer cells. Conversely, miR-298 overexpression and CTNND1 knockdown inhibited Wnt/β-catenin signaling and resulted in decreased expression of β-catenin, WNT11, Cyclin D1, and MMP7 in HCCLM3 cells, while inhibiting EMT (Cao N. et al., 2018). miR-708 directly targets ZEB1 in vitro, which is involved in HSCs by regulating the Wnt/β-catenin signaling pathway, and highly expressed miR-708 reverses EMT (Yang B. et al., 2020). miR-122 expression is closely related to tumor size, vascular invasion, and patient survival in HCC patients. Mechanistic studies reveal that miR-122 overexpression inhibits the EMT process by targeting Snail1 and Snail2, and regulates their expression levels in HCC cells, while the upregulated expression of miR-122 inhibited the Wnt/β-catenin signaling pathway and slowed the EMT process of tumors (Jin et al., 2017). CircRNA mitochondrial tRNA translation optimization 1 (circMTO1) acts as a cavernous body of miR-541-5p to co-regulate HCC cell proliferation, migration, and invasion. The study found that upregulation of miR-541-5p inhibited the expression of ZIC1, promoted HCC cell proliferation, migration, and invasion, and inhibited cell apoptosis. In addition, the latter activated Wnt/β-catenin signaling to promote EMT. Silencing CircMTO1 could upregulate Wnt/β-catenin pathway markers, β-catenin, cyclin D1, c-myc and mesenchymal markers N-cadherin, Vimentin, and MMP2 downstream ZIC1 regulator expression, while the epithelial marker E-cadherin expression was downregulated (Li D.et al., 2021). Drug-induced, dependent on the Wnt/βcatenin signaling pathway, miR-219-5p targeted inhibition of glypican-3 (GPC3) inhibited HCC cell proliferation, migration, and invasion (Gong et al., 2019). miR-194 targets the protein regulator of cytokinesis 1 (PRC1) to inhibit EMT, growth, proliferation, invasion, and migration in HCC cells in vitro, and mouse xenograft growth in vivo, which is Wnt/-dependent by biosignature analysis inactivation of catenin signaling (Zhang et al., 2019). As shown in Figure 2, miRNAs regulate Wnt/β-catenin by targeting multiple genes.
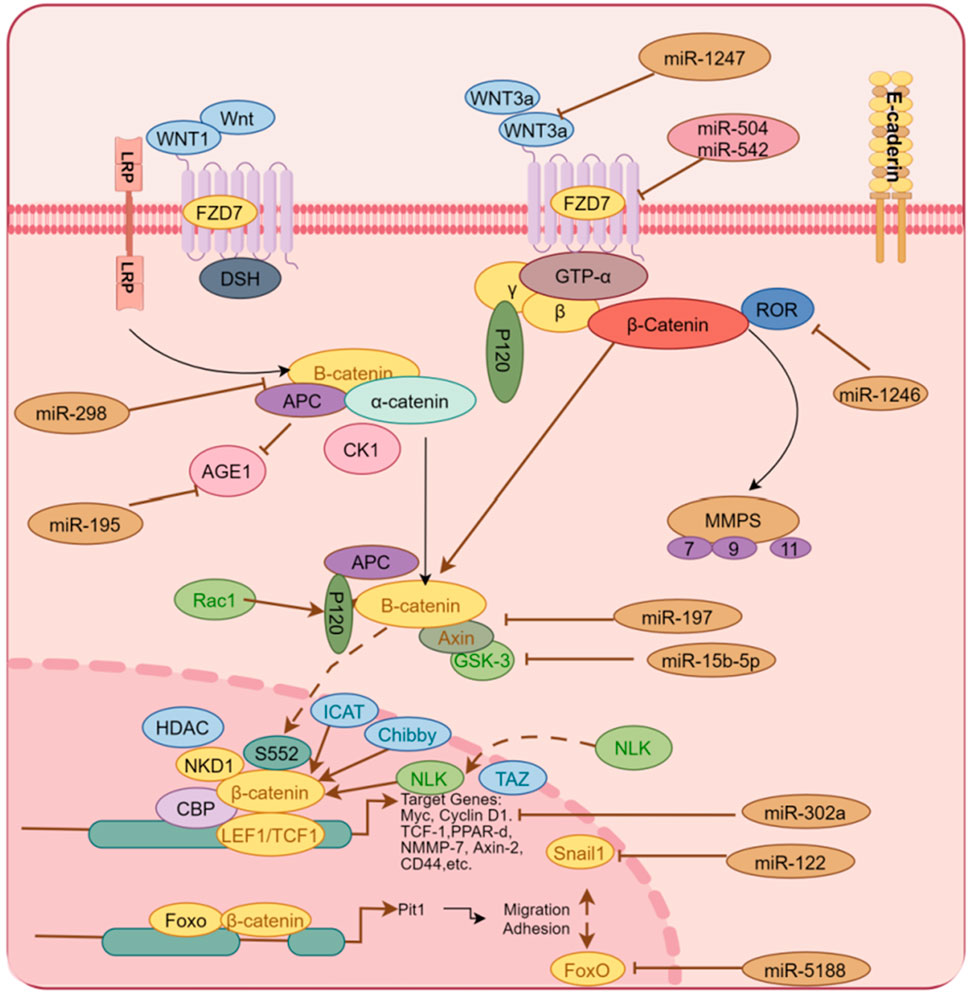
Figure 2. Multiple miRNAs regulate the activity of the Wnt/β-catenin signaling pathway by targeting some genes in the Wnt/β-catenin signaling pathway, including Wnt3 a, FDZ7, Axin, β-catenin and other proteins, thereby activating or blocking the EMT process of liver cancer cells.
4.2 miRNA-dependent PTEN/PI3K/AKT signaling pathway regulates EMT process in HCC
Previous experimental data showed that miR-25-3p, miR-130b-3p, and miR-425-5p were transferred to macrophages via exosomes to induce M2 polarization in macrophages through CXCL12/CXCR4-mediated activation of the PI3K/Akt signaling pathway to regulate PTEN. The latter enhances EMT and secretes vascular endothelial growth factor (VEGF) to promote liver cancer cell metastasis (Wang et al., 2020). It has been reported that the vast majority of HCC is caused by liver fibrosis or cirrhosis (Bruix et al., 2014), miRNAs have attracted much attention in regulating hepatic fibrosis and promoting the metastasis of hepatocytes to cancer. We found that miRNA-21 is associated with greater activation of cancer-associated fibroblasts (CAFs) and higher blood vessel density in HCC patients, as HCC cells secrete exosomes miRNA-21 that directly targets PTEN, resulting in hepatic stellate cells (HSCs) Activation of PDK1/AKT signaling pathway. Activated CAFs further promote cancer progression by secreting angiogenic cytokines, including VEGF, MMP2, MMP9, b-FGF, and TGF-β, and enhance EMT progression in hepatoma cells (Zhou et al., 2018). miR-425-5p targets both SCAI and PTEN and promotes HCC cell invasion and metastasis through SCAI-mediated dysregulation of integrin β1-Fak/Src-RhoA/CDC42, PTEN-AKT, and TIMP2-MMP2/MMP9 signaling, clinical experimental results also showed that high expression of miR-425-5p also predicted poor prognosis and low long-term survival after surgery (Fang et al., 2017). Previous studies suggest that miR-345 indirectly regulates transcription factors Slug, Snail, and Twist by targeting interferon regulatory factor 1 (IRF1), and activates the mTOR/STAT3/Akt signaling pathway, thereby slowing the EMT process of liver cancer (Yu Q. et al., 2017). Targeting tyrosine kinase receptor factor (Met) by miR-148a reduces Snail accumulation in the nucleus, promotes hepatocyte growth, activates downstream Akt Ser473 phosphorylation, and inhibits GSK-3β-Ser9 phosphorylation, thereby alleviating EMT cancer in the liver (Zhang et al., 2014).
Numerous pieces of evidence show that the malignant potential of HCC is regulated by the tumor microenvironment (TME) (Berindan-Neagoe and Calin, 2014; Steinbichler et al., 2017). This is partly owing to cancer-associated fibroblasts (CAFs) helping regulate tumor progression. Thus, understanding how they function in HCC could improve patient outcomes. In addition, another team found that exosomal miR-21 and miR-10b induced by the acidic microenvironment of HCC promoted the proliferation and metastasis of cancer cells, and also triggered the activation of HIF-1α and HIF-2α to exacerbate the metastasis of HCC cells and ectopic colonization. Mechanistically, HIF-1α and HIF-2α can bind to the promoter HRE sites of miR-21 and miR-10b, activate the overexpression of miR-21 and miR-10b to inhibit the tumor suppressor PTEN and activate p-AKT/mTOR signaling pathway, thereby inhibiting epithelial gene expression, promoting mesenchymal gene synthesis, and ultimately accelerating the EMT process (Tian et al., 2019). Studies have shown that miR-150-3p inhibits HCC migration and invasion through exosomes. Although the specific mechanism has not been well explained, however, The notion that low expression of miR-150-3p in HCC tissue is an important factor in the recurrence of HCC patients is well supported by clinical data (Yugawa et al., 2021). In recent years, the regulation of HCC progression by exosomal miR-155-5p has attracted the attention of researchers. miR-155-5p activates the PI3K/Akt pathway in HCC by directly targeting the 3′-UTR of the tumor suppressor PTEN to promote proliferation, Invasion, and migration, inhibiting apoptosis (Fu et al., 2017; Sun et al., 2019). Interestingly, both miRNA-32-5p and miR-92a-3p have the same mechanism of action as miR-155-5p in the hepatocyte EMT process and enhance the resistance of hepatoma cells to multiple drugs (Fu et al., 2018; Yang J. et al., 2020).
In addition, the evidence provides a clear view that has-miR-181a promotes liver metastasis in colorectal cancer. The study found that has-miR-181a promotes cell proliferation and invasion by combining with the tumor suppressor WIF-1. On the contrary, inhibiting the expression of has-miR-181a and increasing the expression of WIF-1 can inhibit tumor growth and metastasis (Ji et al., 2014). The EMT inhibitory effect of miR-451 on hepatoma cells is mainly reflected in that miR-451 targets the oncogene c-Myc to inhibit cell proliferation and reduces the activation of Erk1/2 signaling to control EMT-related marker proteins including E-cadherin and its MMPs family member (Huang JY et al., 2015). miR-101 inhibits liver cancer cell proliferation and tumorigenicity by regulating the expression of SOX9, further research indicates that downregulation of miR-101 promotes liver cell metastasis in vitro and associates poor prognosis of the patients with HCC (Zhang et al., 2012). The highly expressed hematopoietic-substrate-1-associated protein X-1 (HAX1) is targeted by miR-125 in hepatoma cells. Overexpression of miR-125 reduces the expression of mesenchymal phenotype-related genes including VEGF, N-cadherin, and vimentin, which results in inhibiting the proliferation and migration of hepatoma cells (Zha and Li, 2020). In addition, previous study shows that miR-492, miR-519a, miR-21, miR-93, and other regulatory factors are involved in the PTEN/AKT/GSK signaling pathway, and these regulatory factors promote cell proliferation and invasion by promoting cell proliferation and invasion to aggravate the EMT process of hepatoma cells (Bao et al., 2013; Jiang J et al., 2014; Ohta et al., 2015; Tu et al., 2016). Furthermore, the downregulation of miR-204 under hypoxia-inducible conditions contributes to the upregulation of VASP at the post-transcriptional level. Studies have shown that overexpression of miR-204 targets the abundance of regulated proteins under VASP inhibits the AKT/ERK signaling pathway, upregulates the E-cadherin gene, downregulates E-cadherin, Twist, MMPs family proteins, and reverses hypoxia induction. EMT process of liver cancer cells (Liu et al., 2018). Figure 3 shows that different miRNAs synergistically target and regulate the PTEN/PI3K/AKT signaling pathway to induce EMT development.
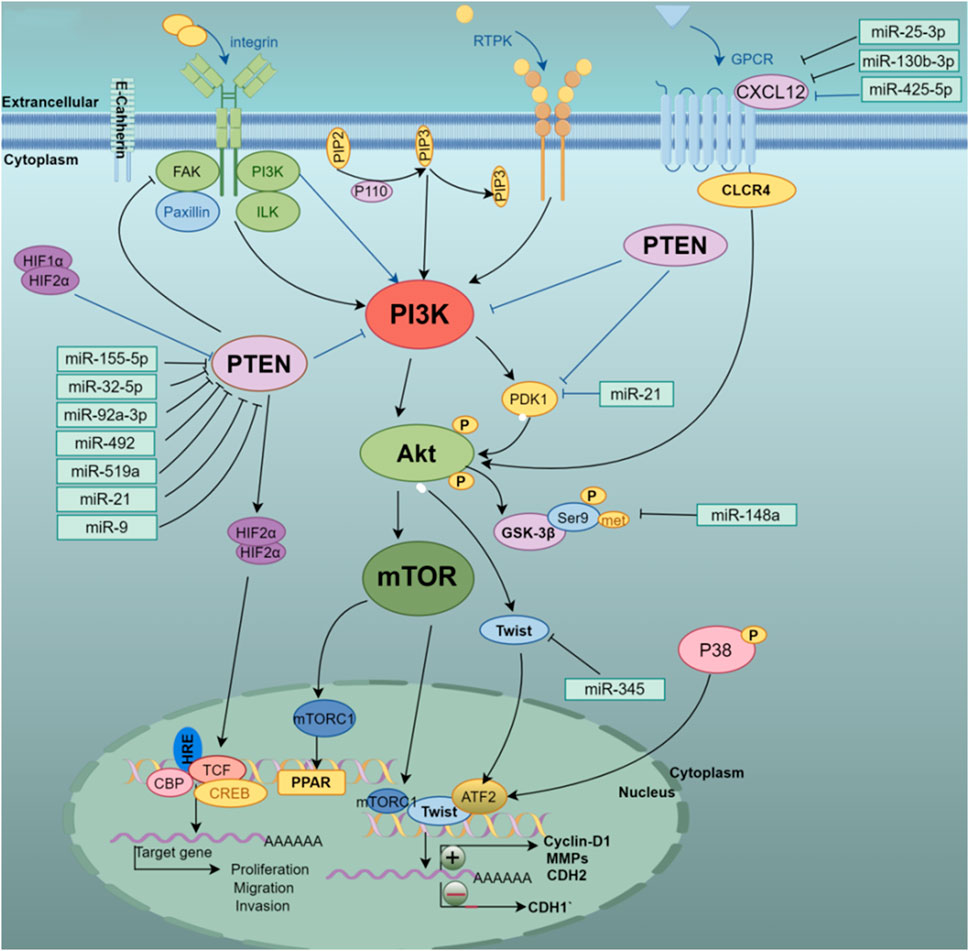
Figure 3. Multi-miRNA targeted regulation of PTEN/PI3K/AKT pathway-related genes, thereby activating or inhibiting the PI3K/Akt/mTOR signaling pathway to affect the occurrence and development of EMT in HCC cells.
4.3 miRNA regulates the EMT process of hepatoma cells by the TGF-β signaling pathway
The transforming growth factor (TGF)-β family is a multifunctional cytokine family that plays an important regulatory role in the development and pathogenesis of various diseases including fibrotic diseases, cardiovascular diseases, and cancer. there is a bidirectional regulatory relationship between miRNA and the TGF-β signaling pathway (Gregory et al., 2011). EMT is an orchestrated process by transforming growth factor (TGF)-β1, which is stored in ECM-rich tissue embedded in HCC cells (Giannelli et al., 2016). Previous studies have found that TGF-β activation regulates miR-105, miR-199a, miR-215, miR-421, miR-529, miR- 21, miR-181, miR-155, miR-192, and conversely, consistent with the diversity of miRNA target genes, computational prediction of miRNA targets suggests that multiple components of the TGF-β signaling pathway are targeted by multiple miRNAs This includes miR-21, miR-17/92 cluster, miR-106b, miR-211, miR-590 (Suzuki, 2018). In particular, the miR-200 family consisting of TGF-β-downregulated miRNAs targets TGF-β, TβR-I, and Smad2 Thus, downregulation of the miR-200 family by TGF-β enhances TGF-β signaling Conduction and induction of EMT (Gregory et al., 2011). In addition, miR-15b-5p, miR-421, miR-1303, miR-221-3p, and miR-486-5p are involved in TGF-β/BMP signaling pathway, angiogenesis, autophagy, and inflammation, promoting HCC malignant development and EMT transformation (Critelli et al., 2017). Hypermethylation is one of the factors of abnormal expression of miRNA, and also one of the signs of malignant transformation of tumors. Experiment data suggest that miR-142 in tumor tissue is hypermethylated, which targets and regulates the expression of TGF-β and inhibits the activation of its signaling pathway. Low expression of miR-142 accelerates the occurrence of EMT in hepatoma cells (Yu M. et al., 2017). miR-200c and miR-34a targeting EMT-related transcription factors Twist and Snail-1 have been reported in many studies to inhibit EMT development and regulate cell invasion and metastasis in hepatoma cells by activating TGF-β (Iwahashi et al., 2016). miR-630 inhibits EMT by targeting Slug. Interestingly, the TGF-β-Erk/SP1 and JNK/c-Jun signaling pathways inhibit miR-630 by occupying transcription factor binding sites and thereby form a loop regulation, while Clinical data indicate that miR-630, as a tumor suppressor factor, is low expressed in patients with higher recurrence rate and shorter overall survival (Chen et al., 2016). Another study also showed that TGF-β activation could lead to miR-374a-5p targeting GADD45A via exosomes to promote EMT development (Lin et al., 2020a). Interestingly, it was mentioned above that miR-542-3p can play a role in the β-catenin signaling pathway, not only that, the existing experimental results show that miR-542-3p can directly target TGF-β1 to affect TGF-β/Smad signaling pathway, thereby inhibiting EMT development, which is consistent with clinical data that low expression of miR-542-3p is associated with poor prognosis (Zhang X. et al., 2018). previous research has established that miR-199b-5p binds to the 3′-UTR of N-cadherin mRNA, thereby reducing N-cadherin expression in HCC cells. Overexpression of miR-199b-5p inhibited TGF-b1-induced Akt phosphorylation, and inhibition of PI3K/Akt pathway blocked TGF-b1-induced N-cadherin expression to promote cell aggregation, inhibiting Cell migration and invasion in HCC cells and inhibition of tumor metastasis in nude mouse (Zhou et al., 2017). miR-744-5p targets TGF-β1 to inhibit this signaling pathway, thereby reversing EMT and suppressing HCC proliferation and invasion (Huang et al., 2021). Figure 4 shows the model diagram of miRNA regulating the TGF-β signaling pathway to affect the development of EMT in liver cancer cells.
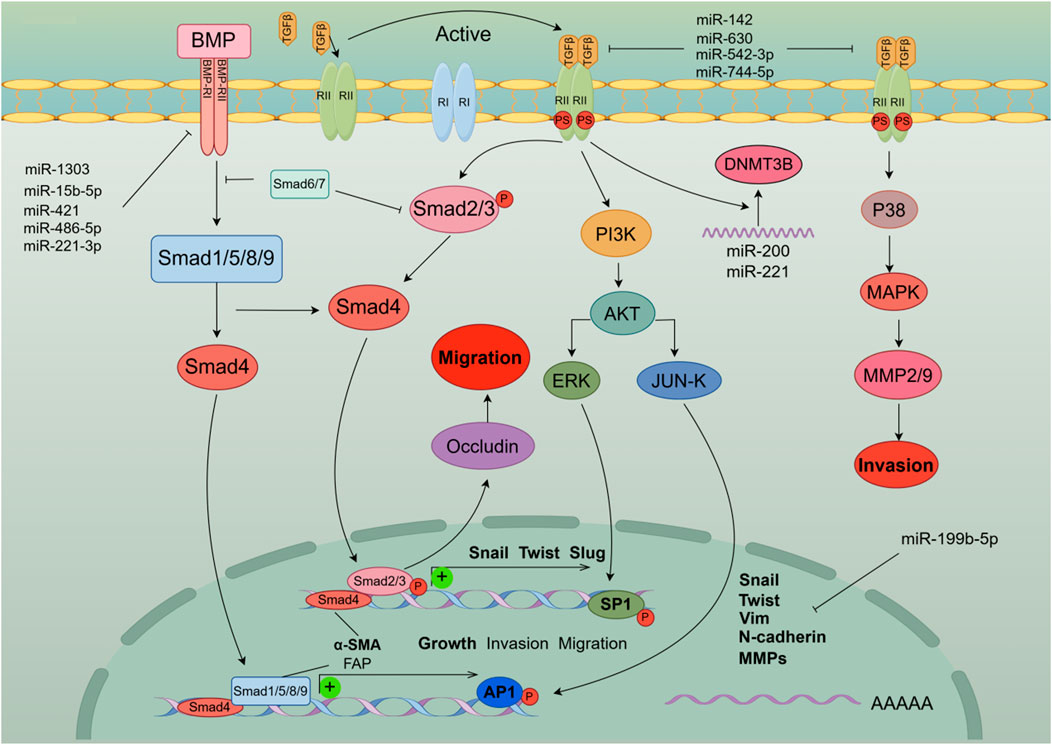
Figure 4. MiRNAs affect the occurrence and development of EMT in liver cancer cells by activating the TGF-β signaling pathway and cross-talking multiple signaling pathways including PI3 K and MAPK. In addition, the activation of the TGF-β signaling pathway initiates the expression of EMT-TFs genes and induces EMT in liver cancer cells.
4.4 miRNA regulates the EMT process of hepatoma cells through the RAS/MAPK signaling pathway
Mitogen-activated protein kinase (MAPK) pathways are characterized by their distinct kinase proteins and link extracellular signals to mechanisms that control physiological cellular processes such as growth, differentiation, proliferation, migration, and apoptosis. Alters in the MAPK cascade play critical roles in cancer development and progression (Dhillon et al., 2007). Different families of miRNA target and regulate the MAPK signaling pathway to affect the development of diseases, especially in liver cancer cells. A large number of studies have shown that miRNAs target this pathway protein to control the EMT process (Vasuri et al., 2018). miR-346 targets the tumor-promoting factor SMYD3, which is highly expressed to activate MAP kinase signaling and promote the formation of Ras-driven cancers, resulting in the methylation of MAP3K2. Therefore, highly expressed miR-346 can effectively inhibit liver cancer cells. proliferation and EMT transformation (Weiyou Zhu et al., 2017). NRG1 is a direct target of miR-296-5p and binds ERBB family members to activate downstream Janus kinase (JAK)/signal transducer and (STAT) transcriptional activators, phosphatidylinositol 3′-kinase (PI3K)-Akt and MAPK, which leads to cell proliferation, invasion, and angiogenesis, and promotes EMT. Highly expressed miR-296 reverses the proliferation and invasion of hepatoma cells (Shi et al., 2018). The hypermethylation of miR-129-3p promoter in metastatic tissues may be one of the inducements for hypermetastatic HCC. Mechanistically, miR-129-3p targeting Aurora-A, as a serine/threonine protein kinase, activates PI3K/Akt and p38-MAPK signaling to promote EMT, invasion in vitro and metastasis in vivo in HCC cells (Shiyun Cui et al., 2016). miR-181 directly regulates MAPK phosphatase-5 (MKP-5) to inhibit the phosphorylation of downstream p38 MAPK to block polycyclic aromatic hydrocarbons (PAH)-induced hepatocellular carcinoma cell invasion and migration (Song et al., 2013). The low expression of miR-143 in liver cancer patients targets the regulation transcription factor GATA-binding factor 6 (GATA6) to inhibit the growth and invasion of liver cancer cells (Xue et al., 2017). miR-125a targets MMP11, SIRT7, and c-Raf to regulate MAPK signaling and inhibit hepatocellular carcinoma cell proliferation (Coppola et al., 2017). As an activator of RAS, GAB1 is highly expressed in HCC patients. Studies have found that miR-200a targets and binds to GAB1 to inhibit RAS/MAPK signaling, thereby reducing the migration and invasion ability of liver cancer cells (Wang et al., 2017). Similarly, high levels of miR-150 reduced GAB1 expression, subsequently downregulated phosphorylated ERK1/2 and inhibited EMT, while low levels of miR-150 were associated with worse outcomes in HCC patients. Clinicopathological features are significantly associated with poor prognosis (Wei Sun et al., 2016). Experimental results demonstrate that upregulated miR-487a correlates with a poor prognosis for HCC patients. miR-487a directly binds the germination-associated EVH1 domain containing 2 (SPRED2) or phosphoinositide-3-Kinase regulatory subunit 1 (PIK3R1) to enhance HCC cell proliferation and metastasis. Importantly, miR-487a promotes MAPK signaling through SPRED2 and promotes proliferation via PIK3R1-mediated AKT signaling (Chang R.M. et al., 2017). miR-188-5p relies on MAPK signaling to inhibit HCC cell proliferation and metastasis by targeting fibroblast growth factor 5 (FGF5) (Fang et al., 2015). Figure 5 shows that miRNAs affect the EMT process of liver cancer cells through multi-target regulation of the RAS/MAPK signaling pathway.
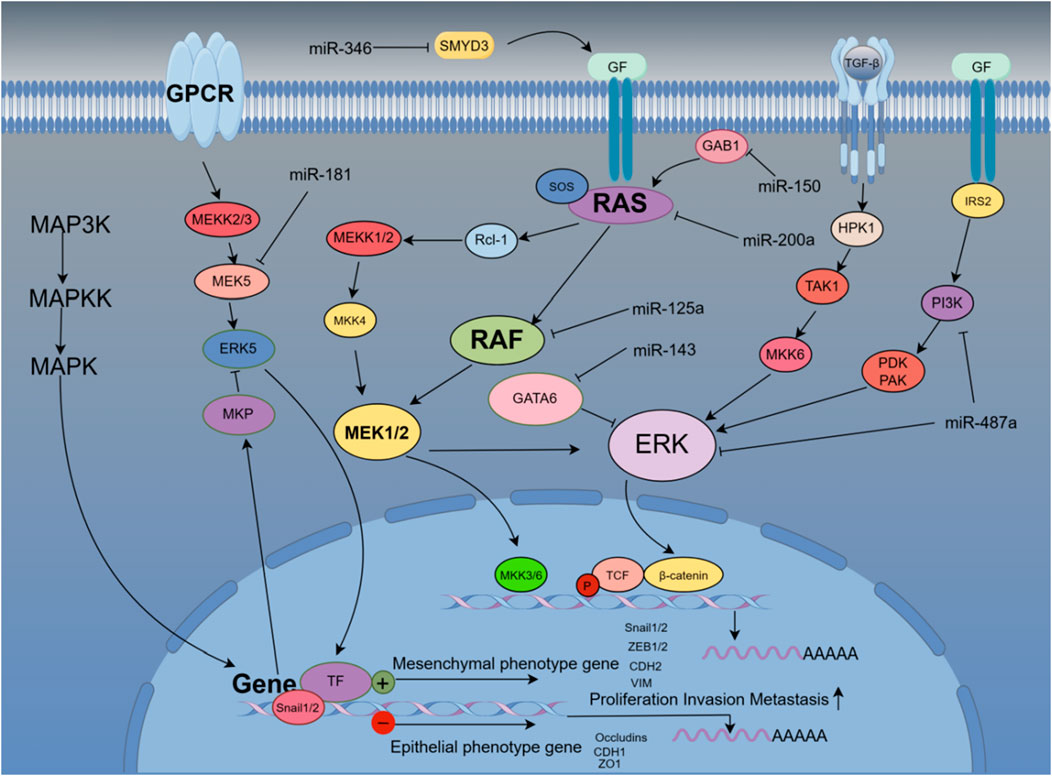
Figure 5. miRNAs accurately regulate the occurrence and development of EMT in liver cancer cells by targeting genes related to the Ras/MAPK pathway.
All the miRNAs involved in this part depend on different signaling pathways to regulate biological functions summarized in Table 1.
5 miRNAs regulate EMT independently of the aforementioned Signaling pathways
Exosomes are important carriers of confidence transmission among T cells, B cells, dendritic cells, epithelial cells, and tumor cells, and their size ranges from 30 to 100 nm in diameter. Multiple studies have demonstrated that abundant cell-free exosome-derived miRNAs can induce miRNA-mediated target gene repression and function effectively as oncogenes or tumor suppressors in various pathological processes in recipient normal or tumor cells (Valadi et al., 2007; Taylor and Gercel-Taylor, 2008). There is emerging evidence indicating that exosomes play a central role in cell-to-cell communication. Existing studies have shown that dysregulation of exosomal microRNAs (miRNAs), including miR-125b-5p, miR-374a-5p, miR-24-3p, miR-200b-3p, and miR-21-5p, play an important role in the progression of hepatocarcinogenesis (Lin et al., 2020b).
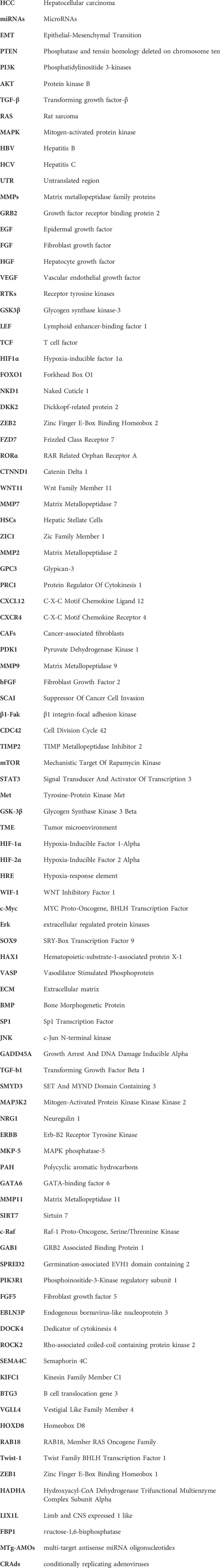
The study has found that the expression of miR-144-3p in patients with advanced liver cancer was significantly reduced, further evidence indicates that miR-144-3p targets the regulation of endogenous bornavirus-like nucleoprotein EBLN3P and dedicator of cytokinesis 4 DOCK4 to inhibit the proliferation and invasion of liver cancer cells. Interestingly, an in-depth study of the mechanism by which EBLN3P competes with miR-144-3p to reduce the latter’s inhibitory effect on DOCK4 promotes EMT transition in HCC cells. Therefore, inhibiting the expression of miR-144-3p and increasing EBLN3P played a similar promoting effect, while overexpression of miR-144-3p and reducing DOCK4 exerted a similar inhibitory effect on EMT (Li et al., 2020). Another research found that low expression of miR-124 promotes cell migration, and overexpression of miR-124 can directly target Rho-associated coiled-coil and (EZH2) genes containing protein kinase 2 (ROCK2) to destabilize its mRNA and inhibit EMT (Zheng et al., 2012). miR-205 targeting axon guidance factor 4C (SEMA4C) promotes apoptosis, inhibits proliferation, and attenuates tumor cell EMT In liver cancer cells (Lu et al., 2018). In contrast, expression of miR-194 in liver stromal cells leads to decreased levels of the mesenchymal phenotype gene N-cadherin, preventing cell migration and invasion (Meng et al., 2010). miR-532-3p targets and regulates kinesin family member C1 (KIFC1) a tumor-promoting factor, which regulates HCC cell metastasis and invasion through Gankyrin-dependent activation of Twist (Han et al., 2019). Low expression of miR-34 leads to poor prognosis and enhanced hepatocyte migration and invasion in vitro in HCC patients (Zhang et al., 2019; Silveira et al., 2022). Similarly, miR-449a can also target the regulation of Met protein abundance, reduce the accumulation of Snail in the nucleus, reduce the level of mesenchymal markers, and increase the expression of epithelial markers, ultimately blocking the EMT of liver cancer cells. Aberrantly expressed miR-449a is often accompanied by high metastasis and enhanced the invasive ability of liver cancer cells (Chen et al., 2015). Activation of p53 promoted the expression of miR-200 and miR-192, which inhibited the expression of ZEB1/2 and blocked the EMT transition of hepatoma cells (Kim et al., 2011). Previous clinical studies clarified that the prognosis of patients with low expression of miR-26a-5p was significantly worse than that of patients with high expression, and a strong downregulation of miR-26a-5p was observed in tumor tissue, research suggests that low expression of miR-26a-5p upregulated vimentin expression and downregulated E-cadherin expression (Chang L. et al., 2017). miR-519c-3p promotes HCC cell proliferation, migration, and invasion in vitro and enhances HCC cell growth and metastasis in vivo. Mechanistically, B cell translocation gene 3 (BTG3) was identified as a direct downstream target of miR-519c-3p, which acts as a tumor promoter in regulating HCC growth and metastasis by targeting BTG3 and may become a new therapeutic target for HCC (Wang et al., 2019). miR-301b-3p directly binds to the 3′UTR of degenerate-like family member 4 (VGLL4) and negatively regulates its expression. Meanwhile, high miR-301b-3p levels predict a significant decrease in overall survival in HCC patients. Knockdown of miR-301b-3p inhibits cell proliferation, causes cell cycle arrest in the G2/M phase, and induces apoptosis in Huh7 and Hep3B cells, inhibiting cell invasion and migration, thereby blocking EMT progression (Guo et al., 2019). miR-5692a has an oncogenic effect in HCC by targeting HOXD8, and reducing miR-5692a can effectively improve the proliferation and invasion of HCC cells and promote cell apoptosis (Shijie Sun et al., 2019). miR-455 normally acts as a tumor suppressor in various cancers, and in liver cancer cells, miR-455 inhibits cell viability and invasion by directly targeting RAB18. Increasing miR-455 expression is expected to improve patient prognosis (Wang et al., 2021). The regulation of EMT by miRNA targeting Twist-1 has been well reported in numerous works of literature (Feng et al., 2022). Interestingly, Twist-1 can also regulate miRNA to form miRNA/Twist-1/miRNA loop regulation, and the upregulation of Twist-1 leads to 18 differential expressions of miRNAs, among which miR-27a-3p downregulation clamps the EMT process of hepatoma cells (Zhao et al., 2016). As an activator of EMT, ZEB1 is a member of the zinc finger family, encoding zinc finger and homeodomain transcriptional cytokines (Li L. Y. et al., 2021). It has been widely reported that miRNAs controlling ZEB1 can reduce the proliferation and invasion of hepatoma cells to block EMT (Xu et al., 2018; Feng et al., 2022).
Of note, an increasing number of studies have shown that miRNAs enable control of cellular EMT processes through cellular metabolism. On the one hand, miR-612 reduces HCC pseudopodia formation and inhibits metastasis and invasion through HADHA-mediated lipid reprogramming, which is consistent with clinical data that patients with low miR-612 levels and high HADHA levels have a poorer prognosis and shorter overall survival (Liu et al., 2020). In terms of glucose metabolism, the Limb and CNS expressed 1-like (LIX1L) regulates miR-21-3p expression to target fructose-1,6-bisphosphatase (FBP1). The study found that LIX1L promotes miR-21-3p expression to inhibit FBP1 protein abundance, resulting in attenuated gluconeogenesis, thereby promoting EMT development. Conversely, knockdown of LIX1L and inhibition of miR-21-3p expression can block the transformation of hepatocytes into carcinogenesis. and reverse the EMT process (Zou et al., 2021). In addition, numerous reports have demonstrated that miRNAs also play critical roles in metabolic homeostasis, and miR-122 plays a central role in maintaining liver homeostasis by affecting various genes involved in hepatic cholesterol and lipid metabolism (Szabo and Bala, 2013). The biological functions of miRNAs independent of signaling pathways are summarized in Table 2.
6 Developing promising therapeutic strategies for miRNA
The application of microRNA in cancer treatment is a new and exciting method to interfere with the molecular mechanism of malignant tumors. There are several major advantages for the clinical application of miRNA, the first is as a biomarker for cancer prediction and diagnosis, easy to detect and sample (Volinia S et al., 2006). Secondly, miRNAs have a wide range of miRNA, according to statistics, more than 30% of the genes can accept the regulation from miRNA (Corsini et al., 2012). In addition, multi-site, multi-target artificial miRNA development strategies can be designed for specific tumors (Wang, 2011). However, clone selection is laborious and targeting miRNAs generated from multiple loci remains a challenge (Lu and Rothenberg, 2018). To provide a more reference and promising research direction, and to better serve clinical practice, multi-treatment strategies have been accurately proposed in the field of non-coding RNA. Firstly, miRNA has been used as a biomarker in various studies to promote the identification and characterization of tumorigenesis, which can rapidly diagnose diseases in clinical diagnosis (Acunzo et al., 2015). Five strategies have been proposed for targeting endogenous oncogenic miRNAs including knocking down in vivo expression, development of antisense oligonucleotide strands for miRNAs, development of inhibitors, development of miRNA sponges such as long non-coding RNAs and circular non-coding RNAs, and depletion-type single-stranded fragments developed for target gene mRNA sequences (Qadir MI, 2017). In particular, miRNA oligomers are subdivided into anti-miRNA oligonucleotides (AMOs) and lock-nucleic acid antisense oligonucleotides (lock-nucleic acid antisense oligonucleotides, LNAs), miRNA sponges, miRNA masks, nanoparticles, antagonists, and multi-target antisense miRNA oligonucleotides (MTg-AMOs). Anti-miRNA-221 developed for miRNA-221 has a significant inhibitory effect on HCC and improved survival, and clinical trials have also obtained encouraging results (Bae et al., 2015). In addition, based on the reintroduction of single or multiple mimetic micro-RNA (non-natural double-stranded micro-RNA homologous RNA fragments) into a group of tumor cells, attempts are made to reconstruct the endogenous normal expression profile and restore or lose function. They include gene therapy, miRNA mimics, siRNAs miRNA, antagonists, and small-molecule inhibitors (Bader et al., 2011). Based on the tolerance of cancer cells to miRNA participation in a special period, the development of interventions targeting multiple miRNA mimics may inhibit multiple signaling pathways, thereby improving the efficacy of miRNA mimics to achieve satisfactory results (Li and Lu, 2014). Previous studies have shown that oncolytic viruses have been shown to interact with miRNA expression in HCC. Such carriers have tumor-specific effects and can cause cytolysis of tumor cells; therefore, healthy tissues have low toxicity in vivo. Therefore, oncogenic viruses can be used as an optimization strategy for cancer treatment (Slaney and Darcy, 2015). Regulatory miRNA sequences are inserted into the genome of conditionally replicating adenoviruses (CRAds) to enable the liver to specifically inhibit viral replication. In addition, oncolytic viruses have also been used in HCC xenotransplantation models to develop let-7-dependent adenoviruses, which can only replicate in cancer cells and not in normal healthy liver tissue. miRNA-199-dependent CRAd has also been developed, which can replicate in liver cancer cells but is less common in normal healthy liver tissue (Ulasov et al., 2013).
Notably, although nucleic acid drugs such as miRNAs have targeted regulatory effects, however, similar to other therapeutic oligonucleotides, the main challenge remains the successful delivery of therapeutic miRNAs to target tissues without compromising miRNA integrity, because naked ribonucleic acid is subject to nuclease-dependent rapid degradation and is therefore unstable in the liquid environment of the organism (Davis et al., 2008; Whitehead et al., 2009; Kaasgaard T 2010). Therefore, many therapeutic applications of RNAi are limited to local administration and limit the exposure of RNAi reagents to potential degradation mechanisms to a minimum (Akhtar and Benter, 2007). However, topical administration is only applicable to a limited local area of the target tissue and is generally not conducive to the exposure of all diseased cells to the drug. Therefore, systemic administration is a better route of administration, and the development and exploration of miRNA drug delivery systems is also an important part of miRNA targeted therapy (Bader et al., 2011). Another advantage of the in-depth development of miRNA targeted therapy is that it will be used as an emerging means of preclinical diagnosis (He et al., 2020). A typical example is the upregulation of miR-221 in liver cancer cells whichhas been confirmed by multiple studies as a potential biomarker for the detection of hepatocellular carcinoma (Karakatsanis et al., 2011; Yasser et al., 2021; Di Martino et al., 2022). More importantly, as an innovative diagnostic indicator, miRNA is usually obtained from gastric juice, urine, saliva, and blood as a non-invasive detection method, which greatly increases its application value (Baraniskin et al., 2011; Vila-Navarro et al., 2017; Cao Y. et al., 2018). The high-sensitivity detection technology of miR-21 in breast cancer developed with the sensing effect of nanomaterials has promoted the early diagnosis of breast cancer (Salahandish et al., 2018). Equally exciting is the use of Nano-String technology, or nCounter technology, which is a quantitative method without digital amplification, to identify the content of MicroRNA-1246 in the dysregulated serum of prostate cancer, to facilitate clinical diagnosis (Bhagirath et al., 2018).
Therapeutic miRNAs constitute a special form of targeted therapy, and tools and diagnoses that have been promoted for personalized medicine and used to accurately characterize cancer genotypes in patients will become valuable resources for the implementation of miRNA-based treatments in diseases (Weidhaas et al., 2007). In addition, miRNA-based therapies have strong anti-cancer activity in animal models, and therapeutic miRNAs may fulfill their promise by potentially safely and unimpeded translation of miRNA replacements into clinical practice. The recent role of miRNAs in cancer stem cells, and their ability to make cancer cells more sensitive to radiation and conventional chemotherapy, will create new opportunities for more effective treatment of cancer (Liu et al., 2011; Lai et al., 2018; Luo J et al., 2019).
Finally, we are also concerned that miRNA therapy can be combined with chemotherapy, radiotherapy, and immunotherapy, to providing new methods for miRNA-based therapy and the use of alternative therapies and miRNA mimics. Meanwhile, miRNA, together with other ncRNAs such as lncRNA and circRNA, has important clinical significance in the pathogenesis, diagnosis, and treatment of human cancer. More relevant studies should be valued by researchers to fully understand the pathogenesis of cancer and improve targeted therapy.
7 Conclusion and perspectives
Based on the complex biological background and elusive regulatory network of HCC, in-depth exploration of its molecular pathogenesis requires a systematic understanding of the body’s system solution strategy between various external stresses and internal mutations. miRNAs, as a crucial part of the complex regulatory network in organisms, play a pivotal role in post-transcriptional regulation. As an essential biological process in cancer metastasis and invasion, EMT is systematically regulated by miRNAs, and this system is reflected in the interactive dialogue of multiple signaling pathways. Distinct pathways have predominant roles in the initiation and progression of EMT and regulate changes in morphology and gene expression that convert epithelial cells into motile mesenchymal cells for hepatocellular carcinoma initiation and progression. Identification of the background in pharmacy, molecular biology, and cell biology will identify the cross-regulation of multiple signaling pathways in vivo. In addition, a comprehensive and objective understanding of the EMT-mediated miRNA regulatory network in the process of hepatocellular carcinogenesis is imminent to provide new insights for further mechanism research and clinical drug development and screening. Further efforts need to focus on clinical development, especially in the areas of miRNA family drug resistance and tumor-targeted therapy, according to these discovered mechanisms. If all these efforts are completed, new treatment strategies for advanced hepatocellular carcinoma will be determined, and better methods for outcome prediction will be provided.
Author contributions
JC: Writing–review and editing, Writing–original draft, Software, Project administration, Funding acquisition. FH: Writing–review and editing, Resources, Project administration. HP: Writing–review and editing, Resources. JG: Writing–review and editing, Supervision, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the molecular pathology research project of atrophic gastritis in chongqing Medical University, project number BYKY-CX2023018; chongqing Medical University Innovative Team Atrophic Gastritis Outcome Mechanism and Standardized Diagnosis and Treatment project funding, project number W0183; Chongqing Municipal Education Commission GAMT (guanidinoacetic acid N-methyltransferase) enhances the stemness of tumor stem cells through metabolic reprogramming to promote the mechanism of colorectal cancer progression funded research project, project number KJQN202200463.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acunzo, M., Romano, G., Wernicke, D., and Croce, C. M. (2015). MicroRNA and cancer – a brief overview. Adv. Biol. Regul. 57, 1–9. doi:10.1016/j.jbior.2014.09.013
Akhtar, S., and Benter, I. F. (2007). Nonviral delivery of synthetic siRNAs in vivo. J. Clin. Investigation 117 (12), 3623–3632. doi:10.1172/JCI33494
Avila, M. A., Berasain, C., Sangro, B., and Prieto, J. (2006). New therapies for hepatocellular carcinoma. Oncogene 25 (27), 3866–3884. doi:10.1038/sj.onc.1209550
Bader, A. G., Brown, D., Stoudemire, J., and Lammers, P. (2011). Developing therapeutic microRNAs for cancer. Gene Ther. 18 (12), 1121–1126. doi:10.1038/gt.2011.79
Bae, H. J., Jung, K. H., Eun, J. W., Shen, Q., Kim, H. S., Park, S. J., et al. (2015). MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J. Hepatology 63 (2), 408–419. doi:10.1016/j.jhep.2015.03.019
Bao, L., Yan, Y., Xu, C., Ji, W., Shen, S., Xu, G., et al. (2013). MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett. 337 (2), 226–236. doi:10.1016/j.canlet.2013.05.007
Baraniskin, A., Kuhnhenn, J., Schlegel, U., Chan, A., Deckert, M., Gold, R., et al. (2011). Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117 (11), 3140–3146. doi:10.1182/blood-2010-09-308684
Berindan-Neagoe, I., and Calin, G. A. (2014). Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 20 (24), 6247–6253. doi:10.1158/1078-0432.CCR-13-2500
Bhagirath, D., Yang, T. L., Bucay, N., Sekhon, K., Majid, S., Shahryari, V., et al. (2018). microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 78 (7), 1833–1844. doi:10.1158/0008-5472.CAN-17-2069
Bruix, J., Gores, G. J., and Mazzaferro, V. (2014). Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 63 (5), 844–855. doi:10.1136/gutjnl-2013-306627
Cao, N., Mu, L., Yang, W., Liu, L., Liang, L., and Zhang, H. (2018). MicroRNA-298 represses hepatocellular carcinoma progression by inhibiting CTNND1-mediated Wnt/β-catenin signaling. Biomed. Pharmacother. 106, 483–490. doi:10.1016/j.biopha.2018.06.135
Cao, Y., Green, K., Quattlebaum, S., Milam, B., Lu, L., Gao, D., et al. (2018). Methylated genomic loci encoding microRNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. Clin. Epigenetics 10 (1), 43. doi:10.1186/s13148-018-0470-7
Chang, L., Li, K., and Guo, T. (2017). miR-26a-5p suppresses tumor metastasis by regulating EMT and is associated with prognosis in HCC. Clin. Transl. Oncol. 19 (6), 695–703. doi:10.1007/s12094-016-1582-1
Chang, R.-M., Xiao, S., Lei, X., Yang, H., Fang, F., and Yang, L.-Y. (2017). miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin. Cancer Res. 23 (10), 2593–2604. doi:10.1158/1078-0432.CCR-16-0851
Chen, S. P., Liu, B. X., Xu, J., Pei, X. F., Liao, Y. J., Yuan, F., et al. (2015). MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer 15, 706. doi:10.1186/s12885-015-1738-3
Chen, W. X., Zhang, Z. G., Ding, Z. Y., Liang, H. F., Song, J., Tan, X. L., et al. (2016). MicroRNA-630 suppresses tumor metastasis through the TGF-β- miR-630-Slug signaling pathway and correlates inversely with poor prognosis in hepatocellular carcinoma. Oncotarget 7 (16), 22674–22686. doi:10.18632/oncotarget.8047
Coppola, N., de Stefano, G., Panella, M., Onorato, L., Iodice, V., Minichini, C., et al. (2017). Lowered expression of microRNA-125a-5p in human hepatocellular carcinoma and up-regulation of its oncogenic targets sirtuin-7, matrix metalloproteinase-11, and c-Raf. Oncotarget 8 (15), 25289–25299. doi:10.18632/oncotarget.15809
Corsini, L. R., Terrasi, M., Amodeo, V., Fanale, D., Fiorentino, E., Cicero, G., et al. (2012). The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin. Ther. Targets 16 (2), S103–S109. doi:10.1517/14728222.2011.650632
Critelli, R., Milosa, F., Faillaci, F., Condello, R., Turola, E., Marzi, L., et al. (2017). Microenvironment inflammatory infiltrate drives growth speed and outcome of hepatocellular carcinoma: a prospective clinical study. Cell Death Dis. 8 (8), e3017. doi:10.1038/cddis.2017.395
Davis, M. E., Chen, Z., and Shin, D. M. (2008). Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug Discov. 7 (9), 771–782. doi:10.1038/nrd2614
Derynck, X.-H. F. a. R., and Derynck, R. (2005). Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693. doi:10.1146/annurev.cellbio.21.022404.142018
Dhillon, A. S., Hagan, S., Rath, O., and Kolch, W. (2007). MAP kinase signalling pathways in cancer. Oncogene 26 (22), 3279–3290. doi:10.1038/sj.onc.1210421
Di Martino, M. T., Arbitrio, M., Caracciolo, D., Cordua, A., Cuomo, O., Grillone, K., et al. (2022). miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: a systematic review. Mol. Ther. - Nucleic Acids 27, 1191–1224. doi:10.1016/j.omtn.2022.02.005
Doghish, A. S., Ismail, A., El-Mahdy, H. A., Elkady, M. A., Elrebehy, M. A., and Sallam, A.-A. M. (2022). A review of the biological role of miRNAs in prostate cancer suppression and progression. Int. J. Biol. Macromol. 197, 141–156. doi:10.1016/j.ijbiomac.2021.12.141
Dongre, A., and Weinberg, R. A. (2018). New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20 (2), 69–84. doi:10.1038/s41580-018-0080-4
Fang, F., Chang, R.-m., Yu, L., Lei, X., Xiao, S., Yang, H., et al. (2015). MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J. Hepatology 63 (4), 874–885. doi:10.1016/j.jhep.2015.05.008
Fang, F., Song, T., Zhang, T., Cui, Y., Zhang, G., and Xiong, Q. (2017). MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. oncotarget 8 (19), 31745–31757. doi:10.18632/oncotarget.15958
Feng, J., Hu, S., Liu, K., Sun, G., and Zhang, Y. (2022). The role of MicroRNA in the regulation of tumor epithelial–mesenchymal transition. Cells 11 (13), 1981. doi:10.3390/cells11131981
Forner, A., Llovet, J. M., and Bruix, J. (2012). Hepatocellular carcinoma. Lancet 379 (9822), 1245–1255. doi:10.1016/S0140-6736(11)61347-0
Fu, X., Liu, M., Qu, S., Ma, J., Zhang, Y., Shi, T., et al. (2018). Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 37 (1), 52. doi:10.1186/s13046-018-0677-7
Fu, X., Wen, H., Jing, L., Yang, Y., Wang, W., Liang, X., et al. (2017). MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 108 (4), 620–631. doi:10.1111/cas.13177
Gao, Q., Wang, X.-Y., Zhou, J., and Fan, J. (2014). Multiple carcinogenesis contributes to the heterogeneity of HCC. Nat. Rev. Gastroenterology Hepatology 12 (1), 13. doi:10.1038/nrgastro.2014.6-c1
Giannelli, G., Koudelkova, P., Dituri, F., and Mikulits, W. (2016). Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatology 65 (4), 798–808. doi:10.1016/j.jhep.2016.05.007
Gong, T., Ning, X., Deng, Z., Liu, M., Zhou, B., Chen, X., et al. (2019). Propofol-induced miR-219-5p inhibits growth and invasion of hepatocellular carcinoma through suppression of GPC3-mediated Wnt/β-catenin signalling activation. J. Cell Biochem. 120 (10), 16934–16945. doi:10.1002/jcb.28952
Gregory, P. A., Bracken, C. P., Smith, E., Bert, A. G., Wright, J. A., Roslan, S., et al. (2011). An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 22 (10), 1686–1698. doi:10.1091/mbc.E11-02-0103
Guo, Y., Yao, B., Zhu, Q., Xiao, Z., Hu, L., Liu, X., et al. (2019). MicroRNA-301b-3p contributes to tumour growth of human hepatocellular carcinoma by repressing vestigial like family member 4. J. Cell Mol. Med. 23 (8), 5037–5047. doi:10.1111/jcmm.14361
Han, J., Wang, F., Lan, Y., Wang, J., Nie, C., Liang, Y., et al. (2019). KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene 38 (3), 406–420. doi:10.1038/s41388-018-0440-8
He, B., Zhao, Z., Cai, Q., Zhang, Y., Zhang, P., Shi, S., et al. (2020). miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 16 (14), 2628–2647. doi:10.7150/ijbs.47203
Hu, Z., Wang, P., Lin, J., Zheng, X., Yang, F., Zhang, G., et al. (2018). MicroRNA-197 promotes metastasis of hepatocellular carcinoma by activating wnt/β-catenin signaling. Cell. Physiology Biochem. 51 (1), 470–486. doi:10.1159/000495242
Huang, J.-L., Fu, Y.-P., Gan, W., Liu, G., Zhou, P.-Y., Zhou, C., et al. (2020). Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 476, 140–151. doi:10.1016/j.canlet.2020.02.012
Huang, W., Chen, Q., Dai, J., Zhang, Y., Yi, Y., Wei, X., et al. (2021). miR-744-5p suppresses tumor proliferation and metastasis by targeting transforming growth factor-beta 1 (TGF-β1) in hepatocellular carcinoma (HCC). J. Gastrointest. Oncol. 12 (4), 1811–1822. doi:10.21037/jgo-21-319
Huang Jy, Z. K., Chen, D. Q., Chen, J., Feng, B., Song, H., Chen, Y., et al. (2015). MicroRNA-451: epithelial-mesenchymal transition inhibitor and prognostic biomarker of hepatocelluar carcinoma. Oncotarget 6 (21), 18613–18630. doi:10.18632/oncotarget.4317
Iwahashi, S., Shimada, M., Utsunomiya, T., Imura, S., Morine, Y., Ikemoto, T., et al. (2016). Epithelial–mesenchymal transition-related genes are linked to aggressive local recurrence of hepatocellular carcinoma after radiofrequency ablation. Cancer Lett. 375 (1), 47–50. doi:10.1016/j.canlet.2016.02.041
Iwasaki, S., Kobayashi, M., Yoda, M., Sakaguchi, Y., Katsuma, S., Suzuki, T., et al. (2010). Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39 (2), 292–299. doi:10.1016/j.molcel.2010.05.015
Ji, D., Li, M., Zhan, T., Yao, Y., Zhang, Z., Xi, J., et al. (2014). MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol. Cancer 13, 86. doi:10.1186/1476-4598-13-86
Jiang J, Z. Y., Yu, C., Li, Z., Pan, Y., and Sun, C. (2014). MicroRNA-492 expression promotes the__progression of hepatic cancer by targeting PTEN. Cancer Cell Int. 24 (1), 95.
Jin, Y., Wang, J., Han, J., Luo, D., and Sun, Z. (2017). MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/β-cadherin signaling pathway. Exp. Cell Res. 360 (2), 210–217. doi:10.1016/j.yexcr.2017.09.010
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16 (7), 421–433. doi:10.1038/nrg3965
Kaasgaard T, A. T., and Andresen, T. L. (2010). Liposomal cancer therapy: exploiting tumor characteristics. Expert Opin. Drug Deliv. 7 (2), 225–243. doi:10.1517/17425240903427940
Kamenska, A., Lu, W. T., Kubacka, D., Broomhead, H., Minshall, N., Bushell, M., et al. (2014). Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing. Nucleic Acids Res. 42 (5), 3298–3313. doi:10.1093/nar/gkt1265
Kamenska, A., Simpson, C., Vindry, C., Broomhead, H., Bénard, M., Ernoult-Lange, M., et al. (2016). The DDX6–4E-T interaction mediates translational repression and P-body assembly. Nucleic Acids Res. 44 (13), 6318–6334. doi:10.1093/nar/gkw565
Karakatsanis, A., Papaconstantinou, I., Gazouli, M., Lyberopoulou, A., Polymeneas, G., and Voros, D. (2011). Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol. Carcinog. 52 (4), 297–303. doi:10.1002/mc.21864
Kim, T., Veronese, A., Pichiorri, F., Lee, T. J., Jeon, Y. J., Volinia, S., et al. (2011). p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 208 (5), 875–883. doi:10.1084/jem.20110235
Lai, X., Gupta, S. K., Schmitz, U., Marquardt, S., Knoll, S., Spitschak, A., et al. (2018). MiR-205-5p and miR-342-3p cooperate in the repression of the E2F1 transcription factor in the context of anticancer chemotherapy resistance. Theranostics 8 (4), 1106–1120. doi:10.7150/thno.19904
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15 (3), 178–196. doi:10.1038/nrm3758
Lee, M. K., Pardoux, C., Hall, M. C., Lee, P. S., Warburton, D., Qing, J., et al. (2007). TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 26, 3957–3967. doi:10.1038/sj.emboj.7601818
Lee, Y., Kim, M., Han, J., Yeom, K.-H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23 (20), 4051–4060. doi:10.1038/sj.emboj.7600385
Li, D., Zhang, J., Yang, J., Wang, J., Zhang, R., Li, J., et al. (2021). CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating Wnt/β-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 13 (1), 12. doi:10.1038/s41419-021-04464-3
Li, L.-Y., Yang, J.-f., Rong, F., Luo, Z.-p., Hu, S., Fang, H., et al. (2021). ZEB1 serves an oncogenic role in the tumourigenesis of HCC by promoting cell proliferation, migration, and inhibiting apoptosis via Wnt/β-catenin signaling pathway. Acta Pharmacol. Sin. 42 (10), 1676–1689. doi:10.1038/s41401-020-00575-3
Li, H., Wang, M., Zhou, H., Lu, S., and Zhang, B. (2020). Long noncoding RNA EBLN3P promotes the progression of liver cancer via alteration of microRNA-144-3p/DOCK4 signal. Cancer Manag. Res. 12, 9339–9349. doi:10.2147/CMAR.S261976
Li, Y., and Lu, H. (2014). Noncoding RNAs: 'our turn' to join the p53 network. J. Mol. Cell Biol. 6 (3), 179–180. doi:10.1093/jmcb/mju022
Lin, Q., Bai, M. J., Zhu, D., Chen, J. W., Wang, H. F., Li, M. A., et al. (2020a). Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 12 (3), 1080–1095.
Lin, Q., Zhou, C. R., Bai, M.-J., Zhu, D., Chen, J.-W., Wang, H.-F., Li, M.-A, et al. (2020b). Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 12 (3), 1080–1095.
Lin, X., Zuo, S., Luo, R., Li, Y., Yu, G., Zou, Y., et al. (2019). HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics 9 (25), 7583–7598. doi:10.7150/thno.37717
Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17 (2), 211–215. doi:10.1038/nm.2284
Liu, Y., Lu, L. L., Wen, D., Liu, D. L., Dong, L. L., Gao, D. M., et al. (2020). MiR-612 regulates invadopodia of hepatocellular carcinoma by HADHA-mediated lipid reprogramming. J. Hematol. Oncol. 13 (1), 12. doi:10.1186/s13045-019-0841-3
Liu, Z., Wang, Y., Dou, C., Xu, M., Sun, L., Wang, L., et al. (2018). Hypoxia-induced up-regulation of VASP promotes invasiveness and metastasis of hepatocellular carcinoma. Theranostics 8 (17), 4649–4663. doi:10.7150/thno.26789
Lu, J., Lin, Y., Li, F., Ye, H., Zhou, R., Jin, Y., et al. (2018). MiR-205 suppresses tumor growth, invasion, and epithelial-mesenchymal transition by targeting SEMA4C in hepatocellular carcinoma. FASEB J. 32, 6123–6134. doi:10.1096/fj.201800113R
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141 (4), 1202–1207. doi:10.1016/j.jaci.2017.08.034
Luo J, S. Z., Li, T., Li, J. Q., Zhang, Z. Q., Chen, G. S., Qi, H. Z., et al. (2019). MicroRNA-146a-5p enhances radiosensitivity in hepatocellular carcinoma through replication protein A3-induced activation of the DNA repair pathway. Am. J. Physiol. Cell Physiol. 16 (3), C299–C311. doi:10.1152/ajpcell.00189.2018
Meng, Z., Fu, X., Chen, X., Zeng, S., Tian, Y., Jove, R., et al. (2010). miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 52 (6), 2148–2157. doi:10.1002/hep.23915
Monica Ballarino, F. P., Girardi, E., Morlando, M., Cacchiarelli, D., Marchioni, M., Proudfoot, N. J., et al. (2009). Coupled RNA processing and transcription of intergenic primary microRNAs. M OLECULAR C ELLULAR B IOLOGY 29 (20), 5632–5638. doi:10.1128/MCB.00664-09
Nawshad, A., Medici, D., Liu, C.-C., and Hay, E. D. (2007). TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J. Cell Sci. 120 (9), 1646–1653. doi:10.1242/jcs.003129
Ohta, K., Wang, J., Ono, S., Iida, Y., Hata, K., Huang, S. K., et al. (2015). MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget 6 (5), 3211–3224. doi:10.18632/oncotarget.3085
Qadir Mi, R. S., and Rizvi, S. Z. (2017). miRNA in hepatocellular carcinoma: pathogenesis and therapeutic approaches. Crit. Rev. Eukaryot. Gene Expr. 27 (4), 355–361. doi:10.1615/CritRevEukaryotGeneExpr.2017019539
Quan, H., Li, B., and Yang, J. (2018). MicroRNA-504 functions as a tumor suppressor in hepatocellular carcinoma through inhibiting Frizzled-7-mediated-Wnt/β-catenin signaling. Biomed. Pharmacother. 107, 754–762. doi:10.1016/j.biopha.2018.07.150
Rivera-Feliciano, J., Lee, K.-H., Kong, S. W., Rajagopal, S., Ma, Q., Springer, Z., et al. (2006). Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 133 (18), 3607–3618. doi:10.1242/dev.02519
Salahandish, R., Ghaffarinejad, A., Omidinia, E., Zargartalebi, H., Majidzadeh-A, K., Naghib, S. M., et al. (2018). Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 120, 129–136. doi:10.1016/j.bios.2018.08.025
Shi, D.-M., Li, L.-X., Bian, X.-Y., Shi, X.-J., Lu, L.-L., Zhou, H.-X., et al. (2018). miR-296-5p suppresses EMT of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3 signaling. J. Exp. Clin. Cancer Res. 37 (1), 294. doi:10.1186/s13046-018-0957-2
Shijie Sun, N. W., Sun, Z., Wang, X., Cui, H., and Cui, H. (2019). MiR-5692a promotes proliferation and inhibits apoptosis by targeting HOXD8 in hepatocellular carcinoma. J. BUON 24 (1), 178–186.
Shirakihara, T., Horiguchi, K., Miyazawa, K., Ehata, S., Shibata, T., Morita, I., et al. (2011). TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 30 (4), 783–795. doi:10.1038/emboj.2010.351
Shiyun Cui, K. Z., Chen, Li, Chen, J., Pan, Y., Feng, B., Lu, L., et al. (2016). Methylation-associated silencing of microRNA-129-3p promotes epithelial-mesenchymal transition, invasion and metastasis of hepatocelluar cancer by targeting Aurora-A. Oncotarget 7 (47), 78009–78028. doi:10.18632/oncotarget.12870
Silveira, D. A., Gupta, S., Sinigaglia, M., and Mombach, J. C. M. (2022). The Wnt pathway can stabilize hybrid phenotypes in the epithelial-mesenchymal transition: a logical modeling approach. Comput. Biol. Chem. 99, 107714. doi:10.1016/j.compbiolchem.2022.107714
Slaney, C. Y., and Darcy, P. K. (2015). Releasing the brake on oncolytic viral therapy. Clin. Cancer Res. 21 (24), 5417–5419. doi:10.1158/1078-0432.CCR-15-1769
Song, M.-K., Park, Y.-K., and Ryu, J.-C. (2013). Polycyclic aromatic hydrocarbon (PAH)-mediated upregulation of hepatic microRNA-181 family promotes cancer cell migration by targeting MAPK phosphatase-5, regulating the activation of p38 MAPK. Toxicol. Appl. Pharmacol. 273 (1), 130–139. doi:10.1016/j.taap.2013.08.016
Steinbichler, T. B., Dudas, J., Riechelmann, H., and Skvortsova, (2017). The role of exosomes in cancer metastasis. Semin. Cancer Biol. 44, 170–181. doi:10.1016/j.semcancer.2017.02.006
Stoker, M. P. M., and Perryman, M. (1985). An epithelial scatter factor released by embryo fibroblasts. J. Cell Sci. 77, 209–223. doi:10.1242/jcs.77.1.209
Sun, J. F., Zhang, D., Gao, C. J., Zhang, Y. W., and Dai, Q. S. (2019). Exosome-mediated MiR-155 transfer contributes to hepatocellular carcinoma cell proliferation by targeting PTEN. Med. Sci. Monit. Basic Res. 25, 218–228. doi:10.12659/MSMBR.918134
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Suzuki, H. (2018). MicroRNA control of TGF-β signaling. Int. J. Mol. Sci. 19 (7), 1901. doi:10.3390/ijms19071901
Szabo, G., and Bala, S. (2013). MicroRNAs in liver disease. Nat. Rev. Gastroenterology Hepatology 10 (9), 542–552. doi:10.1038/nrgastro.2013.87
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110 (1), 13–21. doi:10.1016/j.ygyno.2008.04.033
Thiery, J. P., Acloque, H., Huang, R. Y. J., and Nieto, M. A. (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139 (5), 871–890. doi:10.1016/j.cell.2009.11.007
Tian, X. P., Wang, C. Y., Jin, X. H., Li, M., Wang, F. W., Huang, W. J., et al. (2019). Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics 9 (7), 1965–1979. doi:10.7150/thno.30958
Trelstad, R. L., Hay, E. D., and Revel, J. D. (1967). Cell contact during early morphogenesis in the chick embryo. Dev. Biol. 16 (1), 78–106. doi:10.1016/0012-1606(67)90018-8
Tu, K., Liu, Z., Yao, B., Han, S., and Yang, W. (2016). MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int. J. Oncol. 48 (3), 965–974. doi:10.3892/ijo.2015.3309
Ulasov, I., Callegari, E., Elamin, B. K., D’Abundo, L., Falzoni, S., Donvito, G., et al. (2013). Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PLoS ONE 8 (9), e73964. doi:10.1371/journal.pone.0073964
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9 (6), 654–659. doi:10.1038/ncb1596
Vasuri, F., Visani, M., Acquaviva, G., Brand, T., Fiorentino, M., Pession, A., et al. (2018). Role of microRNAs in the main molecular pathways of hepatocellular carcinoma. World J. Gastroenterology 24 (25), 2647–2660. doi:10.3748/wjg.v24.i25.2647
Vila-Navarro, E., Vila-Casadesús, M., Moreira, L., Duran-Sanchon, S., Sinha, R., Ginés, À., et al. (2017). MicroRNAs for detection of pancreatic neoplasia: biomarker discovery by next-generation sequencing and validation in 2 independent cohorts. Ann. Surg. 265 (6), 1226–1234. doi:10.1097/SLA.0000000000001809
Volinia S, C. G., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103 (7), 2257–2261. doi:10.1073/pnas.0510565103
Wanami, L., Chen, H., Peiro, S., Garciadeherreros, A., and Bachelder, R. (2008). Vascular endothelial growth factor-A stimulates Snail expression in breast tumor cells: implications for tumor progression. Exp. Cell Res. 314 (13), 2448–2453. doi:10.1016/j.yexcr.2008.05.004
Wang, C., Zhu, G., Yu, M., Mi, X., and Qu, H. (2021). miR-455 inhibits the viability and invasion by targeting RAB18 in hepatocellular carcinoma. J. Oncol. 2021, 9923454. doi:10.1155/2021/9923454
Wang, D., Wang, X., Si, M., Yang, J., Sun, S., Wu, H., et al. (2020). Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 474, 36–52. doi:10.1016/j.canlet.2020.01.005
Wang, J., Song, W., Shen, W., Yang, X., Sun, W., Qu, S., et al. (2017). MicroRNA-200a suppresses cell invasion and migration by directly targeting GAB1 in hepatocellular carcinoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 25 (1), 1–10. doi:10.3727/096504016X14685034103798
Wang, L., Mo, H., Jiang, Y., Wang, Y., Sun, L., Yao, B., et al. (2019). MicroRNA-519c-3p promotes tumor growth and metastasis of hepatocellular carcinoma by targeting BTG3. Biomed. Pharmacother. 118, 109267. doi:10.1016/j.biopha.2019.109267
Wang, Z. (2011). The guideline of the design and validation of MiRNA mimics. MicroRNA Cancer 676, 211–223. doi:10.1007/978-1-60761-863-8_15
Weidhaas, J. B., Babar, I., Nallur, S. M., Trang, P., Roush, S., Boehm, M., et al. (2007). MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 67 (23), 11111–11116. doi:10.1158/0008-5472.CAN-07-2858
Wei Sun, Z. Z., Wang, J., Shang, R., Zhou, L., Wang, X., Duan, J., et al. (2016). MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget 7 (10), 11595–11608. doi:10.18632/oncotarget.7292
Weiyou Zhu, J. Q., Ma, L., Ma, P., Yang, F., Shu, Y., and Shu, Y. (2017). MiR-346 suppresses cell proliferation through SMYD3 dependent approach in hepatocellular carcinoma. oncotarget 8 (26), 65218–65229. doi:10.18632/oncotarget.18060
Whitehead, K. A., Langer, R., and Anderson, D. G. (2009). Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8 (2), 129–138. doi:10.1038/nrd2742
Wu, W., Dang, S., Feng, Q., Liang, J., Wang, Y., and Fan, N. (2017). MicroRNA-542-3p inhibits the growth of hepatocellular carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem. Biophysical Res. Commun. 482 (1), 100–105. doi:10.1016/j.bbrc.2016.10.136
Xu, J., Lamouille, S., and Derynck, R. (2009). TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19 (2), 156–172. doi:10.1038/cr.2009.5
Xu, X., Tao, Y., Shan, L., Chen, R., Jiang, H., Qian, Z., et al. (2018). The role of MicroRNAs in hepatocellular carcinoma. J. Cancer 9 (19), 3557–3569. doi:10.7150/jca.26350
Xue, F., Yin, J., Xu, L., and Wang, B. (2017). MicroRNA-143 inhibits tumorigenesis in hepatocellular carcinoma by downregulating GATA6. Exp. Ther. Med. 13 (6), 2667–2674. doi:10.3892/etm.2017.4348
Yang, B., Feng, X., Liu, H., Tong, R., Wu, J., Li, C., et al. (2020). High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene 39 (42), 6529–6543. doi:10.1038/s41388-020-01450-5
Yang, J., Tao, Q., Zhou, Y., Chen, Q., Li, L., Hu, S., et al. (2020). MicroRNA-708 represses hepatic stellate cells activation and proliferation by targeting ZEB1 through Wnt/β-catenin pathway. Eur. J. Pharmacol. 871, 172927. doi:10.1016/j.ejphar.2020.172927
Yang, L., Lin, C., and Liu, Z.-R. (2006). P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell 127 (1), 139–155. doi:10.1016/j.cell.2006.08.036
Yang, M. H., Wu, M. Z., Chiou, S. H., Chen, P. M., Chang, S. Y., Liu, C. J., et al. (2008). Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 10 (3), 295–305. doi:10.1038/ncb1691
Yasser, M. B., Abdellatif, M., Emad, E., Jafer, A., Ahmed, S., Nageb, L., et al. (2021). Circulatory miR-221 and miR-542 expression profiles as potential molecular biomarkers in Hepatitis C Virus mediated liver cirrhosis and hepatocellular carcinoma. Virus Res. 296, 198341. doi:10.1016/j.virusres.2021.198341
Yu, M., Xue, H., Wang, Y., Shen, Q., Jiang, Q., Zhang, X., et al. (2017). miR-345 inhibits tumor metastasis and EMT by targeting IRF1-mediated mTOR/STAT3/AKT pathway in hepatocellular carcinoma. Int. J. Oncol. 50 (3), 975–983. doi:10.3892/ijo.2017.3852
Yu, Q., Xiang, L., Yin, L., Liu, X., Yang, D., and Zhou, J. (2017). Loss-of-function of miR-142 by hypermethylation promotes TGF-β-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 50 (6), e12384. doi:10.1111/cpr.12384
Yugawa, K., Yoshizumi, T., Mano, Y., Itoh, S., Harada, N., Ikegami, T., et al. (2021). Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur. J. Surg. Oncol. 47 (2), 384–393. doi:10.1016/j.ejso.2020.08.002
Zha, Z., and Li, J. (2020). MicroRNA125a5p regulates liver cancer cell growth, migration and invasion and EMT by targeting HAX1. Int. J. Mol. Med. 46 (5), 1849–1861. doi:10.3892/ijmm.2020.4729
Zhang, J. P., Zeng, C., Xu, L., Gong, J., Fang, J. H., and Zhuang, S. M. (2014). MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene 33 (31), 4069–4076. doi:10.1038/onc.2013.369
Zhang, L., Liao, Y., and Tang, L. (2019). MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 38 (1), 53. doi:10.1186/s13046-019-1059-5
Zhang, T., Liu, W., Meng, W., Zhao, H., Yang, Q., Gu, S.-j., et al. (2018). Downregulation of miR-542-3p promotes cancer metastasis through activating TGF-β/Smad signaling in hepatocellular carcinoma. OncoTargets Ther. 11, 1929–1939. doi:10.2147/OTT.S154416
Zhang, X., Xu, X., Ge, G., Zang, X., Shao, M., Zou, S., et al. (2018). miR-498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol. Rep. 41, 1638-1648. doi:10.3892/or.2018.6948
Zhang, Y., Guo, X., Xiong, L., Kong, X., Xu, Y., Liu, C., et al. (2012). MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 586 (24), 4362–4370. doi:10.1016/j.febslet.2012.10.053
Zhao, N., Sun, H., Sun, B., Zhu, D., Zhao, X., Wang, Y., et al. (2016). miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: an essential role for Twist-1 in HCC. Sci. Rep. 6 (1), 23091. doi:10.1038/srep23091
Zheng, F., Liao, Y. J., Cai, M. Y., Liu, Y. H., Liu, T. H., Chen, S. P., et al. (2012). The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61 (2), 278–289. doi:10.1136/gut.2011.239145
Zhou, S.-j., Liu, F.-y., Zhang, A.-h., Liang, H.-f., Wang, Y., Ma, R., et al. (2017). MicroRNA-199b-5p attenuates TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br. J. Cancer 117 (2), 233–244. doi:10.1038/bjc.2017.164
Zhou, Y., Ren, H., Dai, B., Li, J., Shang, L., Huang, J., et al. (2018). Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 37 (1), 324. doi:10.1186/s13046-018-0965-2
Zou, J., Zhu, X., Xiang, D., Zhang, Y., Li, J., Su, Z., et al. (2021). LIX1-like protein promotes liver cancer progression via miR-21-3p-mediated inhibition of fructose-1,6-bisphosphatase. Acta Pharm. Sin. B 11 (6), 1578–1591. doi:10.1016/j.apsb.2021.02.005
Glossary
Keywords: HCC, miRNA, EMT, signaling pathway, targeted therapy
Citation: Chen J, He F, Peng H and Guo J (2024) The underlying mechanism and targeted therapy strategy of miRNAs cross-regulating EMT process through multiple signaling pathways in hepatocellular carcinoma. Front. Mol. Biosci. 11:1378386. doi: 10.3389/fmolb.2024.1378386
Received: 29 January 2024; Accepted: 04 March 2024;
Published: 22 March 2024.
Edited by:
Chengliang Zhu, Zhejiang University, ChinaReviewed by:
Dhivya Sridaran, Washington University in St. Louis, United StatesNahid Arghiani, Stockholm University, Sweden
Copyright © 2024 Chen, He, Peng and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinjun Guo, MTQyMDM3QGhvc3BpdGFsLmNxbXUuZWR1LmNu
 Juan Chen1
Juan Chen1 Jinjun Guo
Jinjun Guo