- 1Institut Pasteur, Université de Paris Cité, CNRS UMR3528, Biochemistry of Macromolecular Interactions Unit, Paris, France
- 2Molecular Microbiology and Structural Biochemistry Laboratory, CNRS UMR 5086, University of Lyon, IBCP, Lyon, France
- 3Université de Paris Cité, Paris, France
- 4Institut Pasteur, Université de Paris Cité, CNRS UMR3528, Biological NMR and HDX-MS Technological Platform, Paris, France
- 5Institut National de la Santé et de la Recherche Médicale (INSERM), Lyon, France
Introduction
The adenylate cyclase (CyaA) toxin is a major virulence factor produced by Bordetella pertussis, the causative agent of whooping cough and is involved in the early stages of respiratory tract colonization (Guiso, 2017; Novak et al., 2017). This toxin invades eukaryotic cells through a unique but poorly understood mechanism that involves a direct translocation of its N-terminal adenylate cyclase catalytic domain (ACD) across the host plasma membrane (Voegele et al., 2018a). ACD produces supraphysiological levels of cAMP (Karst et al., 2010; Ahmad et al., 2016; O'Brien et al., 2017; Skopova et al., 2017), leading to cell death (Sakamoto et al., 1992; Ladant and Ullmann, 1999; Vojtova et al., 2006; Novak et al., 2017). The study of the translocation mechanism proved to be a formidable challenge, as the protein most likely features different conformations during the different steps of the translocation process, and no high-resolution structure is available yet. In this Opinion paper, we summarize the progress made so far in understanding the translocation mechanism and discuss possible avenues to address open questions in the field.
State of the art
CyaA is a 1706-residue long multi-domain protein (Ladant and Ullmann, 1999), consisting of five domains: (1) the adenylate cyclase domain (ACD, residues 1-364) which is activated by calmodulin (CaM) binding and produces cAMP at the expense of ATP (Guo et al., 2005; Karst et al., 2010; O'Brien et al., 2017); (2) the translocation region (TR, residues 365-527) which is implicated in ACD translocation into target cells (Karst et al., 2012); (3) the hydrophobic region (HR, residues 528-710) which is able to form pores into membranes (Benz, 2016); (4) the acylation region (AR, residues 711-1005) which contains two lysine residues, K860 and K983, that are post-translationally acylated by a dedicated B. pertussis acyltransferase CyaC (Barry et al., 1991; Hackett et al., 1994; Westrop et al., 1996; O'Brien et al., 2019); this modification is essential for proper refolding of CyaA (Karst et al., 2014) and ACD translocation in vivo and in vitro (Barry et al., 1991; Novak et al., 2017); (5) the cell-receptor binding domain (RD, residues 1006-1706), made up of approximately 40 copies of calcium-binding RTX (Repeat-in-Toxin) motifs (Linhartová et al., 2010; Alouf et al., 2015; O'Brien et al., 2015; Benz, 2016). The C-terminal extremity of RD contains a secretion signal recognized by the dedicated type 1 secretion system (T1SS), made of B. pertussis CyaB, CyaD, and CyaE proteins (Masure et al., 1990). The secretion process is a processive mechanism initiated from the C-terminus of the protein as it exits first from the T1SS machinery. Once secreted through the T1SS, the toxin binds calcium in the extracellular medium (Bauche et al., 2006; Sotomayor-Perez et al., 2015; O'Brien et al., 2018). A calcium-induced disorder-to-order transition of RD occurs upon CyaA secretion from the low-calcium concentration of the bacterial cytosol to the calcium-rich extracellular environment (Rose et al., 1995; Bauche et al., 2006; Chenal et al., 2009; Chenal et al., 2010; Sotomayor Perez et al., 2010; Sotomayor-Pérez et al., 2011; Sotomayor-Pérez et al., 2013; O'Brien et al., 2015; Bumba et al., 2016; O'Brien et al., 2018). CyaA folding is an acylation-dependent and calcium-driven sequential process (Karst et al., 2014; Cannella et al., 2017; O'Brien et al., 2019).
Intoxication of target cells occurs via a unique process among known bacterial toxins. CyaA can intoxicate target cells via a high affinity interaction between RD and the CD11b/C18 integrin that is expressed by a subset of leukocytes (Guermonprez et al., 2001; Guermonprez et al., 2002; El-Azami-El-Idrissi et al., 2003; Masin et al., 2005; Goldsmith et al., 2022) (neutrophils, dendritic cells (DC) and macrophages). CyaA can also invades host cells, such as epithelial cells (Angely et al., 2017; Angely et al., 2020), in the absence of CD11b/CD18 receptors by a direct interaction with the plasma membrane of eukaryotic cells; this process can be advantageously analyzed on mammalian erythrocytes as these cells are devoid of any membrane trafficking (Guermonprez et al., 2001; Guermonprez et al., 2002; El-Azami-El-Idrissi et al., 2003; Masin et al., 2005; Fiser et al., 2007; Chenal and Ladant, 2018). CyaA membrane translocation is calmodulin dependent (Voegele et al., 2021), calcium dependent, membrane-potential dependent and requires acylation on the lysine residues K860 and K983 located in AR (Fiser et al., 2007; Bumba et al., 2010; Karst et al., 2012; Uribe et al., 2013; Alouf et al., 2015). We propose that ACD is translocated across the plasma membrane into the cytoplasm through a transient and local membrane destabilization mediated by TR (Subrini et al., 2013; Voegele et al., 2018b; Voegele et al., 2021) (see Figure 1). Once translocated in the cytoplasm, TR interacts with CaM, and this high affinity TR:CaM complex exerts an entropic pulling effect on ACD located in the extracellular environment (Voegele et al., 2021). We hypothesize that the TR:CaM entropic pulling effect induces the unfolding of ACD followed by its translocation across the plasma membrane (Voegele et al., 2021). Calmidazolium, a potent CaM inhibitor, prevents CaM:TR complex formation and subsequently ACD translocation (Voegele et al., 2021; Léger et al., 2022). Once in the cytoplasm, ACD refolds upon CaM binding (Karst et al., 2010; O'Brien et al., 2017), which activates the enzymatic activity of ACD, leading to overproduction of cAMP and cell death.
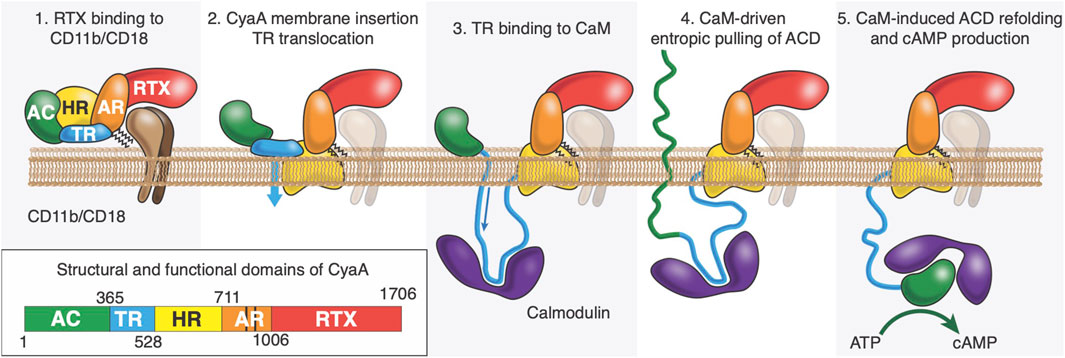
Figure 1. After its synthesis in the bacteria, CyaA is secreted through the Type-1 secretion system (T1SS). Once refolded in the extracellular space, CyaA interacts with CD11b/CD18 receptor via its RTX domain (Panel 1). CyaA may also directly interact with the plasma membrane of the host. It is proposed that the HR would interact with the host membrane (Panel 2) and TR would cross it (flip event) while binding with high affinity to Calmodulin, an abundant protein in eukaryotic cells (Panel 3). We also hypothesize that the complex between P454 and CaM favors the translocation of ACD by exerting an entropic pulling effect that induces the unfolding of ACD (Panel 4). Once in the cytoplasm, ACD refolds upon CaM binding (O’Brien et al., 2017), which activates the enzymatic activity of ACD, leading to overproduction of cAMP.
In an effort to understand the determinants of protein translocation, we identified a peptide, corresponding to residues 454 to 485 of CyaA, coined P454, that exhibits membrane-active properties and interacts with high affinity with calmodulin. P454 forms a helix upon interaction with membranes and exhibits membrane-active properties (Subrini et al., 2013): not only it binds to membranes, but it also permeabilizes lipid vesicles. These experimental observations were supported by molecular dynamics simulations (Voegele et al., 2018b). Most importantly, deletion of the P454 segment abolishes CyaA intoxication of target cells (Karst et al., 2012).
How does P454 contribute to the transport of ACD across the lipid membranes into the cytosol? We hypothesized that in an early step of the translocation process, the P454 segment in CyaA can destabilize the cell membrane and translocate across the lipid bilayer to reach the cytosolic compartment, where it can bind the intracellular CaM; CaM binding would then drag the polypeptide chain inside cell via an entropic pooling force (Voegele et al., 2021). The structure and folding state of ACD before and upon membrane translocation (i.e., in the absence of CaM) is unknown yet, however, we showed that ACD is weakly stable in solution, a state favorable for its translocation across the plasma membrane (Karst et al., 2010; O'Brien et al., 2017; Voegele et al., 2018b).
One powerful method to investigate peptide translocation relies on the use of droplet interface bilayers (DIB) (Bayley et al., 2008). Briefly, two populations of droplets containing a buffer solution are delimited by a phospholipid monolayer, surrounded by a hydrophobic solvent; one population (cis droplets) contains a dye-labelled (fluorescent) peptide, while the other population (trans droplet) contains holo-CaM. When the lipid monolayer of the cis droplet contacts the lipid monolayer of the trans droplet, a lipid bilayer is formed at the interface. The transfer of fluorescence from the cis (fluorescent) droplet to the trans (non-fluorescent) droplet across the lipid bilayer interface can be measured. We performed the experiment with P454, and observed fluorescent rings staining the cis droplets at the beginning of the experiment, indicating that the peptides interact with the cis lipid monolayer. After a few minutes, fluorescence was detected within the trans compartment, indicating translocation of P454 and P454:CaM complex formation (Voegele et al., 2021). In the context of the full length CyaA toxin, we proposed that the CaM/P454 complex provides the energy required to overcome the penalty of ACD translocation across the plasma membrane by an entropic pulling effect (Voegele et al., 2021).
Questions regarding the role of membrane composition in peptide/membrane interaction and peptide translocation are very relevant for a mechanistic understanding of ACD translocation across plasma membrane. Experiments show that P454 binding depends on the lipid headgroup charge, acyl chain fluidity (i.e., acyl chain length and degree of saturation) and lipid polymorphism (Voegele et al., 2017; Voegele et al., 2018b). Unsaturated and negatively charged lipids promote P454 membrane partitioning, while negative membrane curvature and increased membrane rigidity (i.e., saturated acyl chains and cholesterol) do the opposite (Voegele et al., 2018b). The affinity of P454 for negatively charged membranes may be explained by the presence of two arginine residues, R461 and R474. Both arginine residues from P454 contribute to membrane active properties and calmodulin affinity; there is a correlation between the charge, the permeabilization efficiency, the free energy of solution-to-membrane partitioning and CaM binding (Voegele et al., 2018b). Substitutions of arginine by glutamate residues strongly reduce the membrane active properties and CaM affinity for P454 (Voegele et al., 2021). CyaA mutants harboring these mutations (R to E) still binds to the plasma membrane but the translocation of ACD is completely abolished.
Based on available data, we proposed a model of CyaA translocation into eukaryotic cells (Figure 1). We propose that, once TR is translocated into the cytoplasm, P454:CaM binding with high affinity induces unfolding of ACD. The disordered state of ACD would favor its translocation across membrane. The free energy gain due to the formation of the P454:CaM complex would effectively pull the unfolded ACD domain across the membrane. However, this hypothesis requires translocation of TR across the host membrane, which has been verified only for the isolated P454 peptide on model membranes using DIBs. We hypothesize that the translocation of TR across the plasma membrane requires membrane potential and calcium influx induced by CyaA membrane insertion.
Discussion
The mechanism of protein translocation illustrated in Figure 1 is supported by experimental evidence (Voegele et al., 2021), but its details are not established, and some questions remain open. How does the entropic pulling work? What are the sequence determinants for the translocation of the P454 peptide? Does translocation across the host membrane require partial or full unfolding of ACD? Overall, in our view, a better understanding of CyaA translocation requires a quantitative, molecular-level description of the series of events from CyaA secretion to ACD translocation.
First of all, a better knowledge of CyaA structure and dynamics will greatly contribute to understanding protein translocation. We expect that a combination of cryo-EM, SEC-SAXS, bioinformatics, and computer simulations will be necessary to describe the conformational preferences and the dynamics of CyaA. Novel structure prediction algorithms based on machine learning, such as Alphafold (Jumper et al., 2021) (AF2), may contribute to a better understanding of protein structure; however, these algorithms are based on knowledge extracted from databases of existing protein structures, that is rather limited when it comes to membrane proteins. One possible workaround is to use AF2 only for protein regions known not to be inserted in the membrane, and physics-based models or experimental structural information for membrane-interacting fragments. As for experimental information, cryo-EM methods hold great promise, as their success is not limited to soluble proteins: a number of membrane protein structures have been resolved at high resolution in recent years (Renaud et al., 2018). As for computer simulations, we expect them to make a significant contribution to the study of both the structure and the dynamics of the system. Different scales relevant to the translocation process should be considered: simulations at the atomistic scale can be used to explore the conformational flexibility of relatively short peptides (such as P454) in different environments (i.e., the cytosol vs. the membrane interior) but remain prohibitively expensive for large systems (Gupta et al., 2022), such as the one including the entire CyaA protein, embedded in a membrane with complex (plasma membrane-like) lipid composition. They also remain too expensive to enable predictions on the actual dynamics of peptide translocation, that probably takes place on time scales of milliseconds or longer. On the other hand, coarse-grained (CG) simulations can be used to reach millisecond time scales (Gupta et al., 2022; Marrink et al., 2023) and to explore translocation dynamics and thermodynamics of peptides (Kabelka et al., 2021), particularly when combined to enhanced sampling techniques (Barducci et al., 2011), but are generally less reliable in the exploration of protein conformational changes (Monticelli et al., 2008; Periole et al., 2009; Borges-Araújo et al., 2023). This limitation also applies to the latest version of the Martini CG force field (Souza et al., 2021), but we expect that further development will improve the description of conformational flexibility in proteins (Borges-Araújo et al., 2023). The major strength of CG approaches remains speed, and can be harnessed by calculating translocation properties for series of mutant peptides, to clarify the molecular driving forces of the translocation process.
In general, differences between in silico, in vitro, and in vivo systems should be carefully considered, as results obtained in model systems cannot always be directly transferred to living systems. On one hand, in model systems (in silico and in vitro) it is easier to control and change system properties (composition, membrane potential, calcium concentration, etc.). On the other hand, simplifications contained in such model systems sometimes result in failure to reproduce results obtained in cells.
Another important aspect that deserves a more quantitative description is the partitioning of CyaA and its subdomains into membranes. Numerous methods are available for this, and we recently developed a label-free NMR method, B2LiVe, based on selective excitation of amide resonances, to quantify affinity of proteins and peptides for membranes (Anderluh, 2023; Sadi et al., 2023).
Besides protein binding, also protein translocation should be described more quantitatively, for instance by investigating the translocation energetics of TR and ACD, in the presence and in the absence of the CD11b/CD18 receptor. We are also interested to determine the contribution of the factors involved in the translocation process, i.e., the membrane potential, the calcium gradient, the TR:CaM entropic pulling, and the contribution of lipid composition, polymorphism and charge, as well as membrane fluidity, rigidity, thickness and curvature. A number of different approaches are available to study translocation, for example, tethered bilayer membranes (Veneziano et al., 2013; Veneziano et al., 2017). This approach demonstrated that the translocation of the catalytic domain of CyaA is strictly dependent on the presence of CaM, calcium gradient and on the application of a negative trans-membrane potential (Veneziano et al., 2013), as found for entry into eukaryotic cells (Otero et al., 1995). However, this method is laborious and time consuming, and DIBs are emerging as a promising alternative (Bayley et al., 2008; Voegele et al., 2021).
Finally, questions on the molecular determinants of P454 binding to CaM and peptide translocation can be explored by studying mutants, both at the experimental and at the computational level. In perspective, we expect that mutants of P454 (or larger portions of the TR domain) could be optimized to design inactivated CyaA recombinant proteins with improved translocation capabilities. This would be particularly interesting for enhancing the antigen-delivery efficacy of recombinant CyaA-based vaccines (Fayolle et al., 1996; Karimova et al., 1998; Masin et al., 2005).
Author contributions
AA: Writing–original draft, Writing–review and editing. M-HN: Writing–original draft, Writing–review and editing. DL: Writing–original draft, Writing–review and editing. LM: Conceptualization, Writing–original draft, Writing–review and editing. AC: Conceptualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We acknowledge CC-IN2P3 (https://cc.in2p3.fr) for computing services (data storage and backup). LM acknowledges funding by the Institut National de la Santé et de la Recherche Médicale (INSERM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, J. N., Cerny, O., Linhartova, I., Masin, J., Osicka, R., and Sebo, P. (2016). cAMP signalling of Bordetella adenylate cyclase toxin through the SHP-1 phosphatase activates the BimEL-Bax pro-apoptotic cascade in phagocytes. Cell Microbiol. 18, 384–398. doi:10.1111/cmi.12519
Alouf, J., Ladant, D., and Popoff, M. R. (2015). The comprehensive sourcebook of bacterial protein toxins. Elsevier.
Anderluh, G. (2023). A label-free approach for measuring interactions of proteins with lipid membranes. Cell Rep. Methods 3, 100649. doi:10.1016/j.crmeth.2023.100649
Angely, C., Ladant, D., Planus, E., Louis, B., Filoche, M., Chenal, A., et al. (2020). Functional and structural consequences of epithelial cell invasion by Bordetella pertussis adenylate cyclase toxin. PLoS One 15, e0228606. doi:10.1371/journal.pone.0228606
Angely, C., Nguyen, N. M., Andre Dias, S., Planus, E., Pelle, G., Louis, B., et al. (2017). Exposure to Bordetella pertussis adenylate cyclase toxin affects integrin-mediated adhesion and mechanics in alveolar epithelial cells. Biol. Cell 109, 293–311. doi:10.1111/boc.201600082
Barducci, A., Bonomi, M., and Parrinello, M. (2011). Metadynamics. WIREs Comput. Mol. Sci. 1, 826–843. doi:10.1002/wcms.31
Barry, E. M., Weiss, A. A., Ehrmann, I. E., Gray, M. C., Hewlett, E. L., and Goodwin, M. S. (1991). Bordetella pertussis adenylate cyclase toxin and hemolytic activities require a second gene, cyaC, for activation. J. Bacteriol. 173, 720–726. doi:10.1128/jb.173.2.720-726.1991
Bauche, C., Chenal, A., Knapp, O., Bodenreider, C., Benz, R., Chaffotte, A., et al. (2006). Structural and functional characterization of an essential RTX subdomain of bordetella pertussis adenylate cyclase toxin. J. Biol. Chem. 281, 16914–16926. doi:10.1074/jbc.M601594200
Bayley, H., Cronin, B., Heron, A., Holden, M. A., Hwang, W. L., Syeda, R., et al. (2008). Droplet interface bilayers. Mol. Biosyst. 4, 1191–1208. doi:10.1039/b808893d
Benz, R. (2016). Channel formation by RTX-toxins of pathogenic bacteria: basis of their biological activity. Biochim. Biophys. Acta 1858, 526–537. doi:10.1016/j.bbamem.2015.10.025
Borges-Araújo, L., Patmanidis, I., Singh, A. P., Santos, L. H. S., Sieradzan, A. K., Vanni, S., et al. (2023). Pragmatic coarse-graining of proteins: models and applications. J. Chem. Theory Comput. 19, 7112–7135. doi:10.1021/acs.jctc.3c00733
Bumba, L., Masin, J., Fiser, R., and Sebo, P. (2010). Bordetella adenylate cyclase toxin mobilizes its beta2 integrin receptor into lipid rafts to accomplish translocation across target cell membrane in two steps. PLoS Pathog. 6, e1000901. doi:10.1371/journal.ppat.1000901
Bumba, L., Masin, J., Macek, P., Wald, T., Motlova, L., Bibova, I., et al. (2016). Calcium-driven folding of RTX domain β-rolls ratchets translocation of RTX proteins through type I secretion ducts. Mol. Cell 62, 47–62. doi:10.1016/j.molcel.2016.03.018
Cannella, S. E., Ntsogo Enguéné, V. Y., Davi, M., Malosse, C., Sotomayor Pérez, A. C., Chamot-Rooke, J., et al. (2017). Stability, structural and functional properties of a monomeric, calcium–loaded adenylate cyclase toxin, CyaA, from Bordetella pertussis. Sci. Rep. 7, 42065. doi:10.1038/srep42065
Chenal, A., Guijarro, J. I., Raynal, B., Delepierre, M., and Ladant, D. (2009). RTX calcium binding motifs are intrinsically disordered in the absence of calcium: implication for protein secretion. J. Biol. Chem. 284, 1781–1789. doi:10.1074/jbc.M807312200
Chenal, A., Karst, J. C., Pérez, A. C. S., Wozniak, A. K., Baron, B., England, P., et al. (2010). Calcium-induced folding and stabilization of the intrinsically disordered RTX domain of the CyaA toxin. Biophys. J. 99, 3744–3753. doi:10.1016/j.bpj.2010.10.016
Chenal, A., and Ladant, D. (2018). Bioengineering of bordetella pertussis adenylate cyclase toxin for antigen-delivery and immunotherapy. Toxins 10, 302. doi:10.3390/toxins10070302
El-Azami-El-Idrissi, M., Bauche, C., Loucka, J., Osicka, R., Sebo, P., Ladant, D., et al. (2003). Interaction of Bordetella pertussis adenylate cyclase with CD11b/CD18: role of toxin acylation and identification of the main integrin interaction domain. J. Biol. Chem. 278, 38514–38521. doi:10.1074/jbc.M304387200
Fayolle, C., Sebo, P., Ladant, D., Ullmann, A., and Leclerc, C. (1996) In vivo induction of CTL responses by recombinant adenylate cyclase of Bordetella pertussis carrying viral CD8+ T cell epitopes. J. Immunol. Balt. 156, 4697–4706. doi:10.4049/jimmunol.156.12.4697
Fiser, R., Masín, J., Basler, M., Krusek, J., Spuláková, V., Konopásek, I., et al. (2007). Third activity of Bordetella adenylate cyclase (AC) toxin-hemolysin. Membrane translocation of AC domain polypeptide promotes calcium influx into CD11b+ monocytes independently of the catalytic and hemolytic activities. J. Biol. Chem. 282, 2808–2820. doi:10.1074/jbc.M609979200
Goldsmith, J. A., DiVenere, A. M., Maynard, J. A., and McLellan, J. S. (2022). Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin. Cell Rep. 40, 111196. doi:10.1016/j.celrep.2022.111196
Guermonprez, P., Fayolle, C., Rojas, M.-J., Rescigno, M., Ladant, D., and Leclerc, C. (2002). In vivo receptor-mediated delivery of a recombinant invasive bacterial toxoid to CD11c + CD8 alpha -CD11bhigh dendritic cells. Eur. J. Immunol. 32, 3071–3081. doi:10.1002/1521-4141(200211)32:11<3071::AID-IMMU3071>3.0.CO;2-A
Guermonprez, P., Khelef, N., Blouin, E., Rieu, P., Ricciardi-Castagnoli, P., Guiso, N., et al. (2001). The adenylate cyclase toxin of Bordetella pertussis binds to target cells via the alpha(M)beta(2) integrin (CD11b/CD18). J. Exp. Med. 193, 1035–1044. doi:10.1084/jem.193.9.1035
Guiso, N. (2017). Bordetella adenylate cyclase-hemolysin toxins. Toxins 9, 277. doi:10.3390/toxins9090277
Guo, Q., Shen, Y., Lee, Y.-S., Gibbs, C. S., Mrksich, M., and Tang, W.-J. (2005). Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin. EMBO J. 24, 3190–3201. doi:10.1038/sj.emboj.7600800
Gupta, C., Sarkar, D., Tieleman, D. P., and Singharoy, A. (2022). The ugly, bad, and good stories of large-scale biomolecular simulations. Curr. Opin. Struct. Biol. 73, 102338. doi:10.1016/j.sbi.2022.102338
Hackett, M., Guo, L., Shabanowitz, J., Hunt, D. F., and Hewlett, E. L. (1994). Internal lysine palmitoylation in adenylate cyclase toxin from Bordetella pertussis. Science 266, 433–435. doi:10.1126/science.7939682
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kabelka, I., Brožek, R., and Vácha, R. (2021). Selecting collective variables and free-energy methods for peptide translocation across membranes. J. Chem. Inf. Model. 61, 819–830. doi:10.1021/acs.jcim.0c01312
Karimova, G., Fayolle, C., Gmira, S., Ullmann, A., Leclerc, C., and Ladant, D. (1998). Charge-dependent translocation of Bordetella pertussis adenylate cyclase toxin into eukaryotic cells: implication for the in vivo delivery of CD8(+) T cell epitopes into antigen-presenting cells. Proc. Natl. Acad. Sci. U. S. A. 95, 12532–12537. doi:10.1073/pnas.95.21.12532
Karst, J. C., Barker, R., Devi, U., Swann, M. J., Davi, M., Roser, S. J., et al. (2012). Identification of a region that assists membrane insertion and translocation of the catalytic domain of Bordetella pertussis CyaA toxin. J. Biol. Chem. 287, 9200–9212. doi:10.1074/jbc.M111.316166
Karst, J. C., Ntsogo Enguéné, V. Y., Cannella, S. E., Subrini, O., Hessel, A., Debard, S., et al. (2014). Calcium, acylation, and molecular confinement favor folding of Bordetella pertussis adenylate cyclase CyaA toxin into a monomeric and cytotoxic form. J. Biol. Chem. 289, 30702–30716. doi:10.1074/jbc.M114.580852
Karst, J. C., Sotomayor Pérez, A. C., Guijarro, J. I., Raynal, B., Chenal, A., and Ladant, D. (2010). Calmodulin-induced conformational and hydrodynamic changes in the catalytic domain of Bordetella pertussis adenylate cyclase toxin. Biochemistry 49, 318–328. doi:10.1021/bi9016389
Ladant, D., and Ullmann, A. (1999). Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 7, 172–176. doi:10.1016/s0966-842x(99)01468-7
Léger, C., Pitard, I., Sadi, M., Carvalho, N., Brier, S., Mechaly, A., et al. (2022). Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium. BMC Biol. 20, 176. doi:10.1186/s12915-022-01381-5
Linhartová, I., Bumba, L., Mašín, J., Basler, M., Osička, R., Kamanová, J., et al. (2010). RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34, 1076–1112. doi:10.1111/j.1574-6976.2010.00231.x
Marrink, S. J., Monticelli, L., Melo, M. N., Alessandri, R., Tieleman, D. P., and Souza, P. C. T. (2023). Two decades of Martini: better beads, broader scope. WIREs Comput. Mol. Sci. 13, e1620. doi:10.1002/wcms.1620
Masin, J., Basler, M., Knapp, O., El-Azami-El-Idrissi, M., Maier, E., Konopasek, I., et al. (2005). Acylation of lysine 860 allows tight binding and cytotoxicity of Bordetella adenylate cyclase on CD11b-expressing cells. Biochemistry 44, 12759–12766. doi:10.1021/bi050459b
Masure, H. R., Au, D. C., Gross, M. K., Donovan, M. G., and Storm, D. R. (1990). Secretion of the Bordetella pertussis adenylate cyclase from Escherichia coli containing the hemolysin operon. Biochemistry 29, 140–145. doi:10.1021/bi00453a017
Monticelli, L., Kandasamy, S. K., Periole, X., Larson, R. G., Tieleman, D. P., and Marrink, S.-J. (2008). The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4, 819–834. doi:10.1021/ct700324x
Novak, J., Cerny, O., Osickova, A., Linhartova, I., Masin, J., Bumba, L., et al. (2017). Structure-function relationships underlying the capacity of bordetella adenylate cyclase toxin to disarm host phagocytes. Toxins 9, 300. doi:10.3390/toxins9100300
O'Brien, D. P., Cannella, S. E., Voegele, A., Raoux-Barbot, D., Davi, M., Douché, T., et al. (2019). Post-translational acylation controls the folding and functions of the CyaA RTX toxin. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 33, 10065–10076. doi:10.1096/fj.201802442RR
O'Brien, D. P., Durand, D., Voegele, A., Hourdel, V., Davi, M., Chamot-Rooke, J., et al. (2017). Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis. PLoS Biol. 15, e2004486. doi:10.1371/journal.pbio.2004486
O'Brien, D. P., Hernandez, B., Durand, D., Hourdel, V., Sotomayor-Perez, A. C., Vachette, P., et al. (2015). Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion. Sci. Rep. 5, 14223. doi:10.1038/srep14223
O'Brien, D. P., Perez, A. C. S., Karst, J., Cannella, S. E., Enguéné, V. Y. N., Hessel, A., et al. (2018). Calcium-dependent disorder-to-order transitions are central to the secretion and folding of the CyaA toxin of Bordetella pertussis, the causative agent of whooping cough. Toxicon 149, 37–44. doi:10.1016/j.toxicon.2018.01.007
Otero, A. S., Yi, X. B., Gray, M. C., Szabo, G., and Hewlett, E. L. (1995). Membrane depolarization prevents cell invasion by Bordetella pertussis adenylate cyclase toxin. J. Biol. Chem. 270, 9695–9697. doi:10.1074/jbc.270.17.9695
Periole, X., Cavalli, M., Marrink, S.-J., and Ceruso, M. A. (2009). Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5, 2531–2543. doi:10.1021/ct9002114
Renaud, J.-P., Chari, A., Ciferri, C., Liu, W., Rémigy, H.-W., Stark, H., et al. (2018). Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492. doi:10.1038/nrd.2018.77
Rose, T., Sebo, P., Bellalou, J., and Ladant, D. (1995). Interaction of calcium with Bordetella pertussis adenylate cyclase toxin. Characterization of multiple calcium-binding sites and calcium-induced conformational changes. J. Biol. Chem. 270, 26370–26376. doi:10.1074/jbc.270.44.26370
Sadi, M., Carvalho, N., Léger, C., Vitorge, B., Ladant, D., Guijarro, J. I., et al. (2023). B2LiVe, a label-free 1D-NMR method to quantify the binding of amphitropic peptides or proteins to membrane vesicles. Cell Rep. Methods 3, 100624. doi:10.1016/j.crmeth.2023.100624
Sakamoto, H., Bellalou, J., Sebo, P., and Ladant, D. (1992). Bordetella pertussis adenylate cyclase toxin. Structural and functional independence of the catalytic and hemolytic activities. J. Biol. Chem. 267, 13598–13602. doi:10.1016/s0021-9258(18)42254-5
Skopova, K., Tomalova, B., Kanchev, I., Rossmann, P., Svedova, M., Adkins, I., et al. (2017). Cyclic AMP-elevating capacity of adenylate cyclase toxin-hemolysin is sufficient for lung infection but not for full virulence of bordetella pertussis. Infect. Immun. 85, e00937–16. doi:10.1128/IAI.00937-16
Sotomayor Perez, A. C., Karst, J. C., Davi, M., Guijarro, J. I., Ladant, D., and Chenal, A. (2010). Characterization of the regions involved in the calcium-induced folding of the intrinsically disordered RTX motifs from the bordetella pertussis adenylate cyclase toxin. J. Mol. Biol. 397, 534–549. doi:10.1016/j.jmb.2010.01.031
Sotomayor-Pérez, A. C., Ladant, D., and Chenal, A. (2011). Calcium-induced folding of intrinsically disordered repeat-in-toxin (RTX) motifs via changes of protein charges and oligomerization states. J. Biol. Chem. 286, 16997–17004. doi:10.1074/jbc.M110.210393
Sotomayor-Perez, A. C., Ladant, D., and Chenal, A. (2015). Disorder-to-order transition in the CyaA toxin RTX domain: implications for toxin secretion. Toxins Basel 7, 1–20. doi:10.3390/toxins7010001
Sotomayor-Pérez, A.-C., Subrini, O., Hessel, A., Ladant, D., and Chenal, A. (2013). Molecular crowding stabilizes both the intrinsically disordered calcium-free state and the folded calcium-bound state of a repeat in toxin (RTX) protein. J. Am. Chem. Soc. 135, 11929–11934. doi:10.1021/ja404790f
Souza, P. C. T., Alessandri, R., Barnoud, J., Thallmair, S., Faustino, I., Grünewald, F., et al. (2021). Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388. doi:10.1038/s41592-021-01098-3
Subrini, O., Sotomayor-Pérez, A.-C., Hessel, A., Spiaczka-Karst, J., Selwa, E., Sapay, N., et al. (2013). Characterization of a membrane-active peptide from the Bordetella pertussis CyaA toxin. J. Biol. Chem. 288, 32585–32598. doi:10.1074/jbc.M113.508838
Uribe, K. B., Etxebarria, A., Martín, C., and Ostolaza, H. (2013). Calpain-mediated processing of adenylate cyclase toxin generates a cytosolic soluble catalytically active N-terminal domain. PloS One 8, e67648. doi:10.1371/journal.pone.0067648
Veneziano, R., Rossi, C., Chenal, A., Brenner, C., Ladant, D., and Chopineau, J. (2017). Synthesis and characterization of tethered lipid assemblies for membrane protein reconstitution (Review). Biointerphases 12, 04E301. doi:10.1116/1.4994299
Veneziano, R., Rossi, C., Chenal, A., Devoisselle, J. M., Ladant, D., and Chopineau, J. (2013). Bordetella pertussis adenylate cyclase toxin translocation across a tethered lipid bilayer. Proc. Natl. Acad. Sci. U. S. A. 110, 20473–20478. doi:10.1073/pnas.1312975110
Voegele, A., O’Brien, D. P., Subrini, O., Sapay, N., Cannella, S. E., Enguene, V. Y. N., et al. (2018a). Translocation and calmodulin-activation of the adenylate cyclase toxin (CyaA) of Bordetella pertussis. Pathog. Dis. 76. doi:10.1093/femspd/fty085
Voegele, A., O’Brien, D. P., Subrini, O., Sapay, N., Cannella, S. E., Enguéné, V. Y. N., et al. (2018b). Translocation and calmodulin-activation of the adenylate cyclase toxin (CyaA) of Bordetella pertussis. Pathog. Dis. 76. doi:10.1093/femspd/fty085
Voegele, A., Sadi, M., O’Brien, D. P., Gehan, P., Raoux-Barbot, D., Davi, M., et al. (2021). A high-affinity calmodulin-binding site in the CyaA toxin translocation domain is essential for invasion of eukaryotic cells. Adv. Sci. 8, 2003630. doi:10.1002/advs.202003630
Voegele, A., Subrini, O., Sapay, N., Ladant, D., and Chenal, A. (2017). Membrane-active properties of an amphitropic peptide from the CyaA toxin translocation region. Toxins 9, 369. doi:10.3390/toxins9110369
Vojtova, J., Kamanova, J., and Sebo, P. (2006). Bordetella adenylate cyclase toxin: a swift saboteur of host defense. Curr. Opin. Microbiol. 9, 69–75. doi:10.1016/j.mib.2005.12.011
Keywords: membrane protein, bacterial toxin, translocation across membranes, biological membrane, molecular modeling & simulation
Citation: Abettan A, Nguyen M-H, Ladant D, Monticelli L and Chenal A (2024) CyaA translocation across eukaryotic cell membranes. Front. Mol. Biosci. 11:1359408. doi: 10.3389/fmolb.2024.1359408
Received: 21 December 2023; Accepted: 23 January 2024;
Published: 22 March 2024.
Edited by:
Gianvito Grasso, Dalle Molle Institute for Artificial Intelligence Research, SwitzerlandReviewed by:
Víctor G. Almendro Vedia, Complutense University of Madrid, SpainClaudio Canale, University of Genoa, Italy
Copyright © 2024 Abettan, Nguyen, Ladant, Monticelli and Chenal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Monticelli, bHVjYS5tb250aWNlbGxpQGluc2VybS5mcg==; Alexandre Chenal, YWxleGFuZHJlLmNoZW5hbEBwYXN0ZXVyLmY=r
 Amiel Abettan
Amiel Abettan Minh-Ha Nguyen
Minh-Ha Nguyen Daniel Ladant
Daniel Ladant Luca Monticelli
Luca Monticelli Alexandre Chenal
Alexandre Chenal