- 1Structural Genomics Consortium, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2Department of Chemistry, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 3The Neuro’s Early Drug Discovery Unit (EDDU), McGill University, Montreal, QC, Canada
- 4UNC Lineberger Comprehensive Cancer Center, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 5Structural Genomics Consortium, University of Toronto, Toronto, ON, Canada
Cilia are cellular signaling hubs. Given that human kinases are central regulators of signaling, it is not surprising that kinases are key players in cilia biology. In fact, many kinases modulate ciliogenesis, which is the generation of cilia, and distinct ciliary pathways. Several of these kinases are understudied with few publications dedicated to the interrogation of their function. Recent efforts to develop chemical probes for members of the cyclin-dependent kinase like (CDKL), never in mitosis gene A (NIMA) related kinase (NEK), and tau tubulin kinase (TTBK) families either have delivered or are working toward delivery of high-quality chemical tools to characterize the roles that specific kinases play in ciliary processes. A better understanding of ciliary kinases may shed light on whether modulation of these targets will slow or halt disease onset or progression. For example, both understudied human kinases and some that are more well-studied play important ciliary roles in neurons and have been implicated in neurodevelopmental, neurodegenerative, and other neurological diseases. Similarly, subsets of human ciliary kinases are associated with cancer and oncological pathways. Finally, a group of genetic disorders characterized by defects in cilia called ciliopathies have associated gene mutations that impact kinase activity and function. This review highlights both progress related to the understanding of ciliary kinases as well as in chemical inhibitor development for a subset of these kinases. We emphasize known roles of ciliary kinases in diseases of the brain and malignancies and focus on a subset of poorly characterized kinases that regulate ciliary biology.
1 Introduction
Kinases are a class of proteins that regulate a diversity of pathways via phosphorylation. Mutations, loss, or overexpression of kinases have each been implicated in various pathologies. Their involvement in essential disease-driving processes makes kinases an attractive protein class for pharmacological manipulation in cancer, neurological disorders, viruses, and other diseases. Another very attractive feature of kinases is that they bind small molecules in their ATP-binding site and thus are highly tractable. The more than 80 FDA-approved small molecule drugs (Roskoski, 2023) support their tractability and provide tangible evidence of the significant success that has been realized when targeting these proteins, especially for cancer. Cancer is just one disease area in which kinases play key roles. It has been suggested that vast therapeutic potential can be realized through targeting this protein class for other disorders in need of more effective therapeutic options (Krahn et al., 2020; Axtman, 2021).
Ciliogenesis is a highly regulated process involving the assembly of cilia extruding from the plasma membrane. There has been a growing pool of knowledge which implicates kinases as regulators of ciliogenesis (Goto et al., 2017). Kinases are also responsive to changes in cilia (Christensen et al., 2012). Some kinases are localized to cilia, while others are nuclear or located within different organelles, but still impact ciliogenesis (Wirschell et al., 2011; Christensen et al., 2012; Abraham et al., 2022). The many facets of cilia biology, including their evolution, architecture, composition, generation, localization, regulation, and subtype characterization have been extensively described by others (Hildebrandt et al., 2011; Lee and Gleeson, 2011; Christensen et al., 2012; Christensen et al., 2017; Goto et al., 2017; Reiter and Leroux, 2017; Modarage et al., 2022; Arora et al., 2023) and thus will not be the focus of this review. Dysfunctional ciliogenesis and/or aberration of other ciliary signaling pathways can result in human ciliopathies, an emerging class of more than 30 disparate, single-gene, developmental and degenerative disorders characterized by defects in ciliary structure and function (Hildebrandt et al., 2011; Lee and Gleeson, 2011; Goto et al., 2017). These typically result from absent or disrupted function of primary cilia. Primary cilia play a role in sensory detection rather than motility, represent signaling nodes, exist on the surface of almost every quiescent, differentiated cell in the human body, and are essential for the development and homeostasis of human tissues (Lee and Gleeson, 2011; Christensen et al., 2012; Goto et al., 2017; Reiter and Leroux, 2017; Loukil et al., 2021). Primary cilia coordinate signaling pathways in cell cycle control, differentiation, migration, neurotransmission, and other key cellular processes, making them critical organelles (Christensen et al., 2012).
While ciliopathies are rare disorders, they often present with shared clinical features. These include cystic kidneys, situs inversus, retinal issues, brain malformation and/or intellectual disability, heterotaxy, hydrocephaly, craniofacial and skeletal abnormalities, liver disease, anosmia, congenital heart diseases and cardiac fibrosis, infertility, improper circulation of cerebral spinal fluid, hypoplasia, obesity, retinal degeneration, blindness, diabetes, tumorigenesis, and polydactyly (Lee and Gleeson, 2011; Wirschell et al., 2011; Christensen et al., 2012; Goto et al., 2017; Villalobos et al., 2019; Arora et al., 2023; Benmerah et al., 2023). From this list it is clear that ciliopathies impact several vital organs including the brain, heart, kidneys, liver, eyes, respiratory tract, and reproductive system as well as digits. Ciliopathies can be quite organ-specific with examples such as polycystic kidney disease, retinitis pigmentosa, and nephronophthisis. They can also be pleiotropic disorders, like cerebello-oculo-renal syndrome, Primary Ciliary Dyskinesia (PCD), Bardet-Biedl syndrome, Meckel-Gruber syndrome, orofaciodigital syndrome 1, Joubert syndrome, STAR syndrome, and Jeune asphyxiating thoracic dystrophy (Zaghloul and Katsanis, 2009; Hildebrandt et al., 2011; Lee and Gleeson, 2011; Wirschell et al., 2011; Guen et al., 2016; Goto et al., 2017; Smith et al., 2022; Benmerah et al., 2023). More comprehensive reviews of ciliopathies provide additional details on their inheritance, genetics, clinical symptoms, and other features (Hildebrandt et al., 2011; Lee and Gleeson, 2011; Christensen et al., 2012; Abraham et al., 2022; Modarage et al., 2022). Advances in genomics, proteomics, next-generation sequencing, and transcriptomics has led to the identification of disease-causative ciliary gene mutations and allowed for a better understanding of the pathogenesis of these disorders (Lee and Gleeson, 2011; Wirschell et al., 2011; Modarage et al., 2022). Despite these advances, difficulties persist in diagnosing ciliopathies that present with similar phenotypes (Modarage et al., 2022).
Primary cilia also play important roles in brain development and are involved in patterning, maintenance, and proliferation of the progenitor pool (Youn and Han, 2018; Rocha and Prinos, 2022). These organelles influence neuronal differentiation, connectivity, and activity (Lepanto et al., 2016; Bowie and Goetz, 2020; Rocha and Prinos, 2022). Mutations in some ciliary genes are associated with brain abnormalities and may result in neurological manifestations. Examples of neuronal developmental disorders that result, in part, from ciliary dysfunction include autisms and Joubert syndrome, while Parkinson’s disease (PD) and amyotrophic lateral sclerosis (ALS) are examples of neurodegenerative diseases in which aberrant cilia are involved (Doherty, 2009; Trulioff et al., 2017; Youn and Han, 2018; Ma et al., 2022; Rocha and Prinos, 2022).
Various cancer cells have been shown to lack cilia expression (Menzl et al., 2014; Cao and Zhong, 2016; Higgins et al., 2019). Changes in primary cilia have been noted in renal, prostate, cholangiocarcinoma, pancreatic, skin, brain and breast cancers (Seeley et al., 2009; Basten et al., 2013; Basten and Giles, 2013; Hassounah et al., 2013; Seeger-Nukpezah et al., 2013; Menzl et al., 2014; Snedecor et al., 2015; Higgins et al., 2019). Renal epithelial and human primary melanoma cells demonstrate significant losses of cilia in response to carcinogens (Basten et al., 2013; Snedecor et al., 2015; Higgins et al., 2019). Pancreatic cancer cells and intraepithelial neoplasia lesions from human pancreatic ductal adenocarcinoma demonstrate significantly suppressed ciliogenesis (Seeley et al., 2009; Emoto et al., 2014; Higgins et al., 2019). Many of the examples above link loss of cilia to cancer, but this is not always the case. There is evidence suggesting the existence of primary cilia could be a hallmark of aggressive pancreatic ductal adenocarcinoma (Emoto et al., 2014). Moreover, medulloblastomas with activation in Hedgehog (Shh) or Wnt signaling were found to have primary cilia, but primary cilia were not found in medulloblastomas in other molecular subgroups (Han et al., 2009).
Changes in cilia, while not fully understood, have long been associated with cancer development and progression. Using agents that inhibit or promote ciliogenesis has been considered as a therapeutic approach for different cancers (Liu et al., 2018; Lee et al., 2021). An established interaction between cilia and autophagy, a cellular clearance mechanism, has provided a hypothesis for at least one essential process that is disrupted when cilia are lost in cancer (Cao and Zhong, 2016). The role of cilia in suppressing abnormal cell proliferation through regulating cell cycle entry/exit is another key pathway disrupted when cilia are absent (Cao and Zhong, 2016). Links between primary cilia and important signaling pathways with cancer implications, such as Hedgehog, Wnt, and Notch, have been extensively reviewed (Goetz et al., 2009; Egeberg et al., 2012; Seeger-Nukpezah et al., 2013; Liu et al., 2018; Higgins et al., 2019; Mill et al., 2023) and will not be the focus herein.
2 Ciliary kinases
Embedded in the disease-causative ciliary networks are many regulatory kinases that represent druggable nodes. While some of these kinases have been the focus of drug discovery campaigns, others have not been investigated. These “understudied” kinases have had significantly less effort devoted to their study and thus their functions remain poorly annotated in the literature. Still, there are described links between understudied kinases and ciliary function. We provide herein a discussion of three subfamilies of understudied kinases that play a role in ciliary function and/or in pathways that result in ciliopathies. Furthermore, we provide a summary of tool molecules that can be used in efforts to study these ciliary kinases.
Since many human kinases, both well-studied and understudied, have described roles in ciliary pathways, a subset has been explored as potential targets for pharmacological intervention in the pursuit of therapies for patients that suffer from ciliopathies or other conditions, such as cancer or neurodegeneration. We provide a list of human kinases with reported links to ciliary pathways and dysfunction in Table 1. This table specifies the described roles of each kinase in cilia, which includes the following categories: ciliary signaling, ciliary resorption, ciliary function, ciliary motility, ciliogenesis, cilia length, ciliary trafficking, ciliary stability, ciliary structure, ciliary localization, and ciliary dynamics. These categories better define the ciliary function(s) of each of the kinases in Table 1. While our list is extensive, it is not a compendium of all human kinases that regulate cilia. The distribution of these kinases around the human kinome tree (Figure 1) highlights the involvement of disparate kinases in ciliary function(s) and demonstrates that these kinases are not clustered in a single family. Both Table 1; Figure 1 reinforce the idea that kinases are ubiquitous in ciliary pathways and play an indispensable role in their generation, structure, stability, resorption, and function.
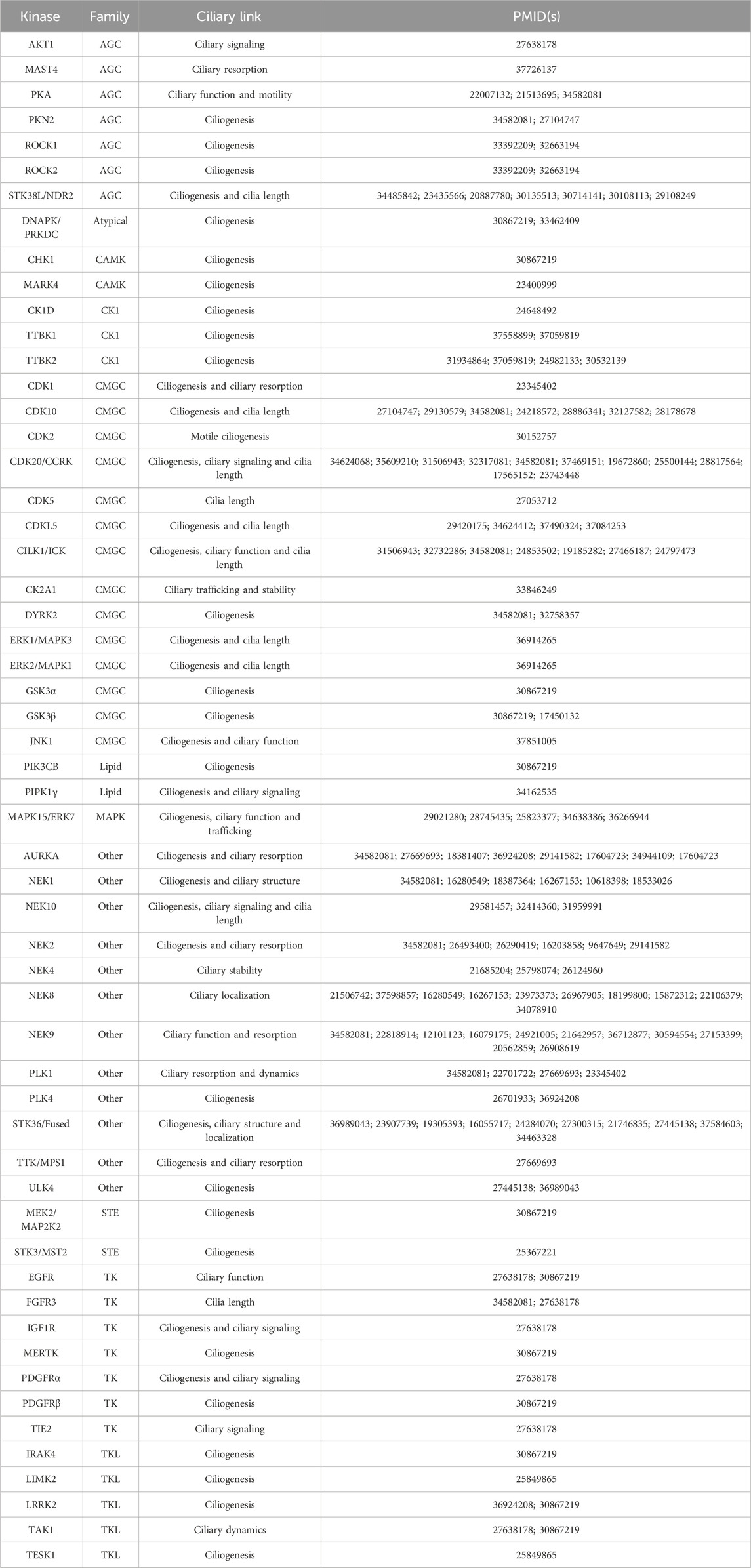
TABLE 1. Selected kinases and their reported links to cilia formation and dysfunction. References are provided for further reading on these kinases. Additional details are provided in the following sections for CDKL, NEK, and TTBK family members as well as LRRK2, PLK1, CDK20/CCRK, CILK1/ICK, CDK10, PKN2, MAPK15, STK38L, STK36, and ULK4.
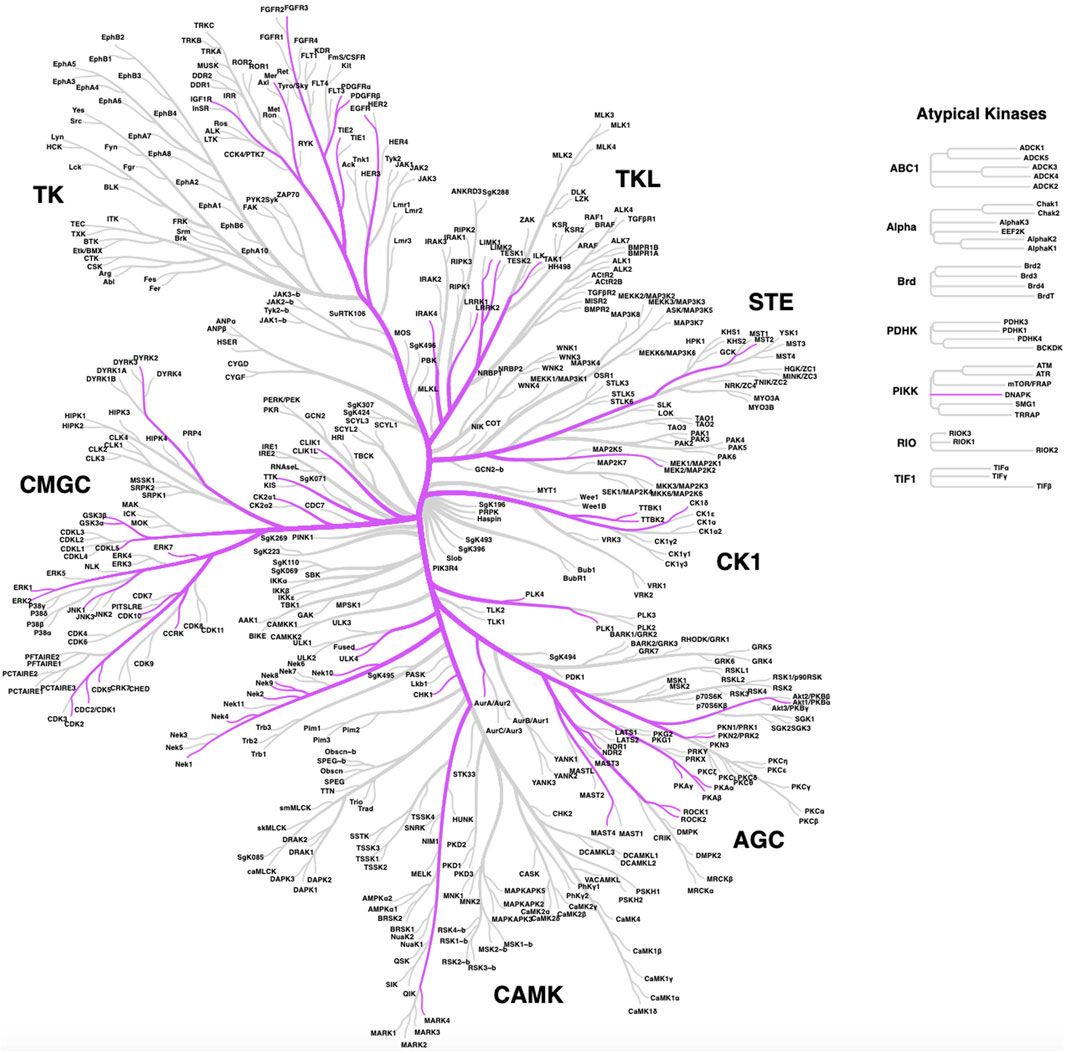
FIGURE 1. Distribution of ciliary kinases on the phylogenic tree of the human kinome. The same kinases included in Table 1 are shown except for lipid kinases PIPK1γ and PIK3CB, which are not pictured here.
Kinase activity is essential for ciliary activity and, thus, kinase dysfunctions are responsible for a plethora of pathological conditions, including neurological disorders and cancer. In the next subsection examples of important ciliary kinases associated with neuronal tissue in health and disease will first be reviewed. We then provide a summary of how ciliary kinases are hijacked by cancers to drive oncogenic processes.
2.1 Ciliary kinases with roles in neurodevelopment and neurological diseases
Several more studied and a few lesser studied kinases have connections to neuronal development and disorders. While we will introduce the roles of members of the CDKL, NEK, and TTBK families in these processes in the next section, examples of other kinases from Table 1 with to that modulate ciliary function in the brain are included below. Table 2 serves as a quick summary of the ciliary kinases from Table 1 that have been linked to neurological issues and which are discussed in some detail herein.
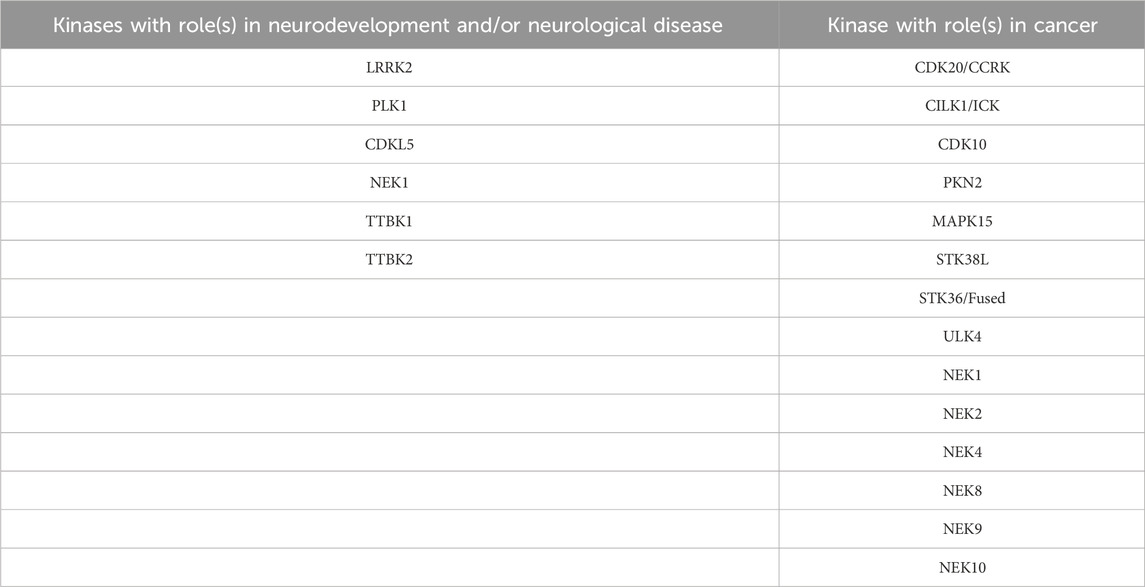
TABLE 2. Subset of ciliary kinases from Table 1 with links to neurodevelopment, neurological disorders, or cancer. Further reading on each of these kinases is provided in various subsections.
2.1.1 LRRK2
Cilia alterations are implicated in neurodegenerative disorders such as Parkinson’s disease (PD). Leucine-rich repeat kinase 2 (LRRK2) encodes a kinase that is involved in vesicular membrane trafficking and is one the genes mutated in familial cases of Parkinson’s disease (Alessi and Sammler, 2018). Primary cilia status has been assessed in mouse and human studies of PD. Cilia loss was detected in neurons and astrocytes from mice carrying the G2019S familial LRRK2 mutation and was associated with dysregulation of Hedgehog signaling (Khan et al., 2021). The G2019S LRRK2 mutation was also found to result in primary cilia loss in iPSC (induced pluripotent stem cell) derived neurons from PD patients (Dhekne et al., 2018). Many small molecule inhibitors have been developed for LRRK2 (Azeggagh and Berwick, 2022), several of which have advanced into clinical trials for PD (Kingwell, 2023).
2.1.2 PLK1
Polo-like kinase (PLK1) localizes to the primary cilium transitional zone and is activated during cilia disassembly (Seeger-Nukpezah et al., 2012). In NIH3T3 cells, PLK1 is recruited to the pericentriolar matrix by PCM1 (pericentriolar material 1), leading to cilia disassembly by activating HDAC6 prior to mitosis entry (Wang et al., 2013). The interaction between PCM1 and PLK1 is dependent on the phosphorylation of PCM1 by CDK1 (cyclin-dependent kinase 1) (Wang et al., 2013). PLK1 interacts with Treacle (encoded by TCOF1 gene), which is a centrosome and kinetochore associated-protein important for mitotic progression and proper neurogenesis (Sakai et al., 2012). Treacle interacts with Plk1 and promotes proliferation in the developing cortex of mice, demonstrating that Plk1 is necessary for neural progenitor mitotic progression (Sakai et al., 2012).
2.2 Ciliary kinases hijacked to propagate cancer
Ciliary kinases regulate signaling pathways such as Hedgehog, Wnt, Hippo, and other pathways that are functionally linked to ciliogenesis and cancer. Many of the ciliary kinases discussed below are deregulated in tumors. Herein we will focus on a subset of ciliary kinases from Table 1 that are linked to cancer with the aim to promote efforts to understand their function. Table 2 provides a quick reference of the ciliary kinases from Table 1 with identified roles in cancer propagation and/or progression, all of which are highlighted in more detail in the following sections.
2.2.1 CDK20/CCRK
Cyclin-dependent kinase 20 (CDK20), or more commonly referred to as cell cycle-related kinase (CCRK), is a member of the CDK family. CDK20/CCRK has been functionally linked to cell cycle checkpoint control, is essential for cell proliferation, and is centrally involved in the development of many malignancies (Chivukula and Malkhed, 2023). Several studies have reported the overexpression of CDK20 in cancers from the brain, colon, liver, lung, and ovary (Chivukula and Malkhed, 2023). CDK20 upregulation in some of these cancers is clinically significant as it correlates with tumor staging, shorter patient survival, and poor prognosis (Wu et al., 2009; Feng et al., 2015). Downstream of Smo and upstream of Gli, CDK20 regulates ciliogenesis and Hedgehog signaling across organisms from Chlamydomonas to C. elegans to humans (Snouffer et al., 2017). CDK20 has been functionally linked to glioblastoma where it behaves as an oncogene and contributes to increased proliferation (Ng et al., 2007). Glioblastoma cells display deregulated, high levels of CDK20, and its depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner (Yang et al., 2013). In this context, the effects of CDK20 on ciliogenesis were found to be mediated by its substrate, intestinal cell kinase (ICK) (Yang et al., 2013). Depletion of CDK20 leads to accumulation of ICK at ciliary tips, altered ciliary transport, and inhibition of cell cycle re-entry in NIH3T3 fibroblasts (Yang et al., 2013). All of this evidence makes CDK20 a promising drug target. The CDK20 protein structure was recently predicted using AlphaFold. This structure was used in AI-accelerated hit discovery for CDK20 to produce a novel small molecule inhibitor of CDK20 (Ren et al., 2023).
2.2.2 CILK1/ICK
Ciliogenesis associated kinase 1 (CILK1), previously known as intestinal cell kinase (ICK), is now recognized as a ubiquitously expressed member of the RCK family of serine/threonine kinases (Fu et al., 2019). Inactivating loss-of-function mutations in the human CILK1 gene produce lethal developmental ciliopathies, namely, the endocrine-cerebro-osteodysplasia (ECO) syndrome (MIM 612651) (Lahiry et al., 2009) and short-rib polydactyly syndrome (SRPS) type II (MIM 263520) (Paige Taylor et al., 2016). In mice, both Cilk1 knock-out and Cilk1 knock-in mutations have recapitulated human ciliopathies. CILK1 has a fundamental role in the function of cilia, and it is required for ciliogenesis (Chaya et al., 2014) by controlling ciliary length (Moon et al., 2014). CILK1 regulates the ciliary localization of Shh pathway components and the localization of intraflagellar transport (IFT) components at ciliary tips (Chaya et al., 2014). CILK1 is activated by phosphorylation at Thr157 by its upstream kinase CDK20, which triggers its autophosphorylation at Tyr159. CILK1 phosphorylates Raptor, Scythe, GSK3β, and KIF3A, linking CILK1 function to cellular metabolism, ciliogenesis, and Hedgehog signaling (Fu et al., 2019). KIF3A, a kinesin motor protein, controls IFT anterograde transport thus raising the possibility that effects of CILK1 on ciliogenesis are mediated through its phosphorylation of KIF3A. Through phosphorylation of these substrates, CILK1 is involved in the regulation of mTOR, Wnt, Hedgehog, and FGFR signaling pathways. Interestingly, CILK1/ICK was found to mediate the effect of CDK20 on ciliogenesis in glioblastoma cells (Yang et al., 2013) and it also regulates the inhibitory effect of fibroblast growth factor on cilia by interacting with FGFR3 (Kunova Bosakova et al., 2019). Thus, ICK seems to be a signaling hub that integrates multiple signaling pathways in ciliary control (Fu et al., 2019).
2.2.3 CDK10 and PKN2
CDK10 is a kinase of the CDK family that forms a heterodimer with Cyclin M (Guen et al., 2013; Guen et al., 2017). The heterodimer promotes cell proliferation via phosphorylating the oncogene ETS2 and is an important regulator of triple negative breast cancer response to endocrine therapy (Iorns et al., 2008). CDK10/cyclin M regulates ciliogenesis through phosphorylation of PKN2 as well as regulating RhoA and the actin cytoskeleton. Mutations in CDK10/Cyclin M cause STAR syndrome, a developmental disorder affecting the skeleton and limbs (Guen et al., 2013). Homozygous mutations in CDK10 cause Al Kaissi syndrome, a neurodevelopmental disorder displaying growth retardation and spine and craniofacial malformations (Windpassinger et al., 2017). Cells from Cdk10/Cyclin M deficient mice or STAR mutants were reported to present with elongated cilia (Guen et al., 2016; Windpassinger et al., 2017). CDK10 phosphorylates PKN2, a kinase that is also critical for ciliogenesis. This suggests that the effects of CDK10 on cilia are mediated, in part, by PKN2 (Guen et al., 2016). PKN2 also regulates RhoA cytoskeleton stress fiber dynamics and cell migration (Guen et al., 2016). Because PKN2 has been linked to several types of cancer including breast, colorectal, renal, head and neck and prostate cancers, it is considered an emerging target (Patel et al., 2020). Dihydropyrrolopyridinone-based PKN2 chemical tools that could enable studies around this kinase were reported in the last few years (Scott et al., 2020; Scott et al., 2022). More recently, computational docking approaches identified promising chemical leads for PKN2 (Al-Sha’er et al., 2023). Although there are not yet specific CDK10 inhibitors, recent efforts toward identifying small molecules targeting CDK10 are underway (Robert et al., 2020).
2.2.4 MAPK15 (aka ERK7/ERK8)
MAPK15, originally known as ERK7/ERK8, is an atypical member of the MAPK kinase family. It is an understudied MAP kinase as its functions have only recently started to be elucidated (Deniz et al., 2023). MAPK15 is functionally implicated in a variety of cellular activities such as cell proliferation, apoptosis, autophagy, and maintenance of genomic integrity (Deniz et al., 2023). This kinase also plays an evolutionarily conserved, essential role in ciliogenesis (Kazatskaya et al., 2017). Knockdown of MAPK15 diminishes the number and the length of cilia in X. laevis, C. elegans, and human neurons (Miyatake et al., 2015). In addition, MAPK15 regulates the localization of ciliary proteins involved in cilium structure, transport, and signaling and regulates apical body migration by phosphorylating CAPZIP (Miyatake et al., 2015; Kazatskaya et al., 2017). MAPK15 has been functionally linked to the Shh subgroup of medulloblastoma where it regulates Hedgehog signalling and tumorigenesis in a cilia-dependent fashion (Pietrobono et al., 2021).
2.2.5 STK38L
STK38L, also known as NDR2 kinase, is a member of the nuclear Dbf2-related (NDR) serine/threonine kinase family. The NDR protein kinases play crucial roles in the control of cell proliferation, apoptosis, and morphogenesis (Santos et al., 2023). These kinases are also important regulators of the Hippo signaling pathway through phosphorylating YAP/TAZ (Hergovich, 2016). STK38L/NDR2 is involved in primary cilium formation (Chiba et al., 2013) and is mutated in a naturally occurring canine ciliopathy, termed early retinal degeneration (Goldstein et al., 2010). Stk38l deletion in mice caused decreased proliferation of retinal amacrine cells by increasing the expression of neuronal stress genes while decreasing the expression of synaptic genes (Léger et al., 2018). STK38L is crucial for ciliogenesis via phosphorylating Rabin8 and impairing pre-ciliary membrane biogenesis at the pericentrosome (Chiba et al., 2013). Genetic variants in STK38L have been associated with increased glioma risk (Chen et al., 2019) and coding mutations were reported in microsatellite-unstable colorectal cancer (Kondelin et al., 2018). STK38L was reported to be overexpressed in KRAS-dependent pancreatic cancer cell lines where it was found to be essential for cell proliferation (Grant et al., 2017).
2.2.6 STK36/Fused
STK36/Fused is a member of the serine/threonine protein kinase (STK) family. This kinase is similar to a Drosophila protein, Fused, that plays a key role in the Hedgehog signaling pathway by regulating the activity of Gli transcription factors by promoting their nuclear localization and opposing the effect of Suppressor of Fused (SUFU) (Murone et al., 2000). STK36 is required for postnatal development through regulating the homeostasis of cerebrospinal fluid (CSF) and ciliary function. STK36 was shown to be essential for the construction of the central pair apparatus of motile cilia (Wilson et al., 2009). Mutation of STK36 leads to primary ciliary dyskinesia with a central pair defect (Edelbusch et al., 2017). Knockout of the homologous mouse gene leads to severe growth retardation and congenital hydrocephalus due to a functional defect in motile cilia (Merchant et al., 2005). The STK36 effects on ciliogenesis and CSF flow are functionally linked to its binding to ULK4, which is another kinase indispensable for motile ciliogenesis (Zhang et al., 2023).
2.2.7 ULK4
Unc51-like kinase 4 (ULK4) belongs to the Unc-51-like serine/threonine kinase (STK) family and encodes a pseudokinase with unclear function (Luo et al., 2022). It is a paralogue of STK36, which is also involved in ciliogenesis. ULK4 has known roles in the remodeling of cytoskeletal components, and it regulates neurite branching and elongation as well as neuron cell motility (Lang et al., 2014). Accumulating evidence indicates that ULK4 participates in corticogenesis, cilia maintenance, myelination, and white matter integrity (Lang et al., 2014). Ulk4 deletion in mice causes decreased intermediate neural progenitors and increased apoptosis, thus disrupting normal cortical development (Liu et al., 2016a). Likewise, Ulk4 null knockout mice present disturbed motile cilia development and disorganized ciliary beating, which impairs CSF flow eventually leading to congenital hydrocephalus (Vogel et al., 2012; Liu et al., 2016b). These phenotypes are identical to STK36 hypomorphic mutants, supporting that ULK4 and STK36 interact as part of a complex (Zhang et al., 2023). The ULK4 protein contains a pseudokinase domain at the N-terminus and is predicted to be catalytically inactive. Its pseudokinase domain interacts with STK36, indicating that ULK4 can directly regulate active kinases despite being catalytically inactive itself (Zeqiraj and van Aalten, 2010). The structure of ULK4 has been resolved, enabling virtual and experimental screens that have identified promising chemical scaffolds for efforts to design specific ULK4 inhibitors (Khamrui et al., 2020). Functional genomic analysis identified the master transcription factor Foxj1 and Foxj1 pathway in cilia as well as an array of other ciliogenesis factors are specifically regulated by ULK4 (Liu et al., 2016a). Furthermore, ULK4 was recently reported to be a component of primary cilia in the neuroepithelium where it acted as a positive regulator of Shh signaling (Mecklenburg et al., 2021). Altogether, these studies demonstrate that ULK4 plays a vital role in ciliogenesis and that deficiency of ULK4 causes hydrocephalus and other ciliopathy-related phenotypes prevalent in neurodevelopmental and neuropsychiatric disorders (Luo et al., 2022).
3 Understudied ciliary kinase families
The primary literature provides insights into the roles of the kinases in Table 1, including those related to cilia and in the pathology of ciliopathies as well as other diseases. In general, fewer papers have been dedicated to certain families of kinases. In the sections that follow, we provide an in-depth discussion of the roles of understudied members of the cyclin-dependent kinase like (CDKL), never in mitosis gene A (NIMA) related kinase (NEK), and tau tubulin kinase (TTBK) families as they relate to cilia. To spur research on the ciliary pathway functions and regulation by these understudied kinases, we provide more details about what has been published that connects kinases within these families to cilia. Furthermore, we provide structures of and associated references for the best available chemical probes or high-quality inhibitors of these understudied ciliary kinases that can be used in follow-up studies. Figure 2 illustrates the ciliary pathways driven by members of these understudied kinase families and highlights the expected consequences of kinase inhibition on cilia.
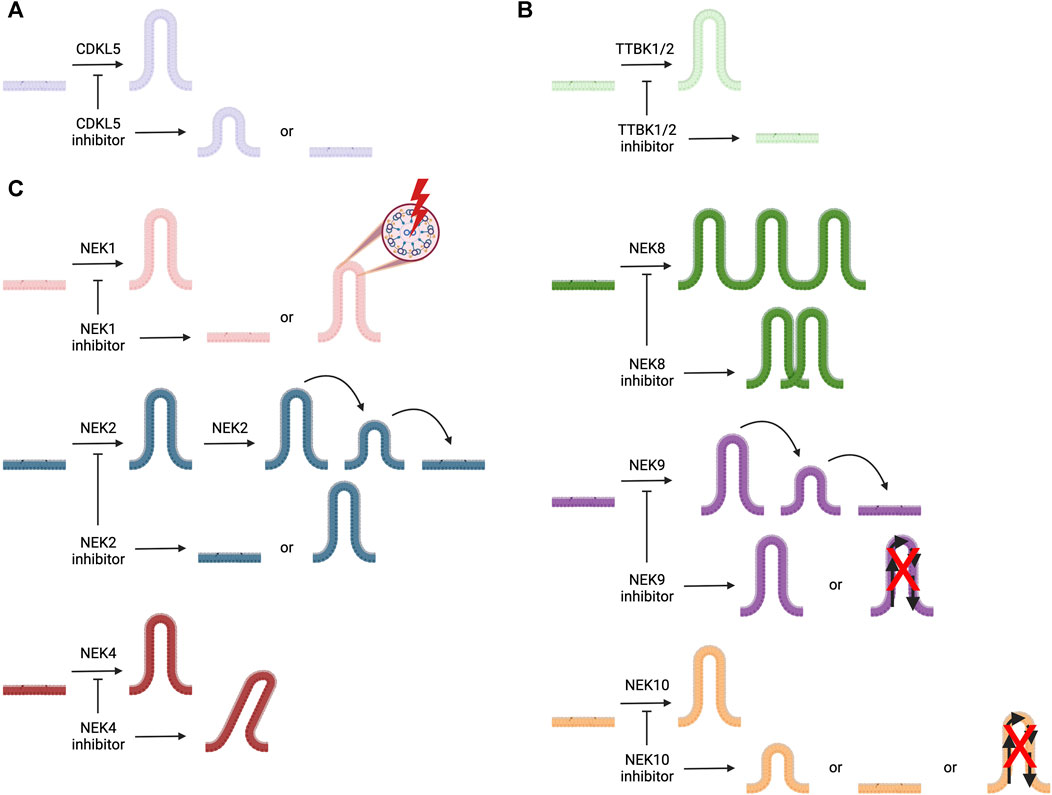
FIGURE 2. Illustration of cilia, represented in different colors for each kinase, the roles of ciliary kinases CDKL5, NEK1, NEK2, NEK4, NEK8, NEK9, NEK10, TTBK1, and TTBK2, and how inhibition of these kinases is expected to impact ciliary morphology and/or function(s). (A) CDKL5 is involved in ciliogenesis and regulating cilia length. (B) TTBK1 and TTBK2 both play a role in ciliogenesis. (C) NEK1 regulates ciliogenesis and ciliary structure; NEK2 plays a role in ciliogenesis and ciliary resorption; NEK4 helps to stabilize cilia; NEK8 is involved with ciliary localization; NEK9 regulates ciliary function and its resorption; and NEK10 modulates ciliogenesis, ciliary signaling, and cilia length.
3.1 CDKL family
CDKL kinases are a family of five relatively underexplored serine/threonine human kinases: CDKL1, CDKL2, CDKL3, CDKL4, and CDKL5. This family has the highest sequence similarity to cyclin-dependent kinases (CDKs) (Canning et al., 2018; Ong et al., 2023). CDKLs contain a cyclin binding domain, although no cyclin-dependent functions have been ascribed. While little is known about CDKL1–4 regarding their function and role(s) in human biology, CDKL5 has been identified as a regulator of ciliogenesis and cilia length (Canning et al., 2018). Structural characterization of all CDKLs, except for CDKL4, has confirmed them to contain a conserved N-terminal kinase domain with variable C-termini (Canning et al., 2018). The C-termini of CDKL2 and CDKL3 were found to have an atypical αJ helix necessary for their catalytic functions, which is absent in CDKL1 and CDKL5 (Canning et al., 2018). Notably, for CDKL5, the C-terminus is involved in trafficking CDKL5 to its subcellular compartments during various developmental stages (Rusconi et al., 2008; Canning et al., 2018; Ong et al., 2023).
CDKL5, also known as serine/threonine kinase 9 (STK9), is the only member of the CDKL family with strong connections to ciliogenesis in humans. CDKL5 is ubiquitously expressed in human tissues, but is observed at higher levels in the hippocampus, cerebellar, striatum, and cortex regions of the CNS in humans and mice, consistent with its roles in dendritic spine growth, brain development, and excitatory synapse composition (Rusconi et al., 2008; Fagerberg et al., 2014; Canning et al., 2018; Ong et al., 2023). A study using RNAi targeting CDKL5, which caused reduced neurite growth and dendritic arborization in rat cortical neurons, confirmed that this kinase regulates neuronal morphogenesis (Chen et al., 2010).
CDKL5 localizes to the basal body of primary cilia, and when aberrantly overexpressed, can lead to jeopardized ciliogenesis (Canning et al., 2018). Hippocampal neurons from Cdkl5 deficient mice presented elongated primary cilia, however no changes in the levels of Wnt and Shh proteins were visualized by Western blot (Di Nardo et al., 2022). Missense mutations in the CDKL5 gene leads to a syndrome known as CDKL5-deficiency disorder (CDD). CDD is characterized by severe encephalopathy, noncanonical developmental physiology, and intellectual disability with intractable epilepsy onset from early age, reflective of a significant impact on brain function (Olson et al., 2019; Di Nardo et al., 2022; Castano et al., 2023; Ong et al., 2023).
To date, there have only been two high-quality chemical probes published by members of the Structural Genomics Consortium (SGC) for CDKL5 (Figure 3). SGC-CAF382-1 (B1 in the original publication) demonstrates a cell-free CDKL5 enzymatic IC50 = 6.7 nM and engages with CDKL5 in cells with an IC50 = 11 nM (Castano et al., 2023). SGC-CDKL5/GSK3-1 is a CDKL5 and GSK3 (GSK3⍺ and GSK3β) probe with a cell-free CDKL5 enzymatic IC50 = 6.5 nM and a nearly equal in-cell CDKL5 target engagement IC50 = 4.6 nM (Ong et al., 2023).
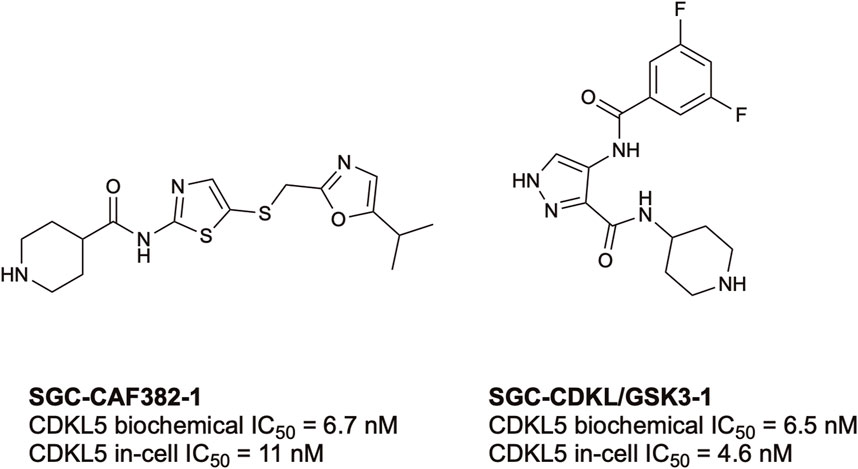
FIGURE 3. Structures and available potency data for the most advanced CDKL5 chemical probes, SGC-CAF382-1 and SGC-CDKL5/GSK3-1.
CDKL1, the only other CDKL kinase with connections to ciliogenesis, has only been studied in model systems (zebrafish and C. elegans). The human and C. elegans CDKL1 proteins share 77% identity, and both have similar phosphorylation regulatory sites: Thr14, Tyr15, and Thr161 in humans and Ser14, Tyr15, and Thr159 in zebrafish (Hsu et al., 2011). CDKL1, however, has been shown to influence cilia length and Hedgehog signaling in zebrafish, a pathway that is also linked to primary cilia (Hsu et al., 2011; Bangs and Anderson, 2017; Park et al., 2021). In lieu of a high-quality chemical tool for CDKL1, genomics, siRNA, and other techniques have been used to begin to uncover the functions of this kinase. In C. elegans, CDKL1 localizes to the transition zone at the base of the cilium, in a CEP-290-dependent manner, and regulates the length of the growing cilia by interacting with IFT anterograde kinesin motor proteins involved in axoneme formation of the cilia (Li et al., 2016; Park et al., 2021). Due to the high sequence identity, known roles of orthologs, and strong connection between Hedgehog signaling and primary cilia, it is proposed that CDKL1 might have an underlying role in human ciliogenesis (Bangs and Anderson, 2017).
3.2 NEK family
The NIMA-related kinase family comprises 11 relatively understudied kinases (NEK1–NEK11) that play roles in many important biological processes and have been linked to several diseases. Recent reviews discuss their biological and disease relevance and provide information about inhibitors (Fry et al., 2012; Meirelles et al., 2014; Wells et al., 2018; Pavan et al., 2021; Panchal and Evan Prince, 2023b; Nguyen et al., 2023). These and other reviews highlight that mutations in all NEK family members have been identified in different cancers and the functions of each NEK in cancer progression delineated (Moniz et al., 2011; Panchal and Evan Prince, 2023a). NEK1 mutations have also been identified as a genetic cause of amyotrophic lateral sclerosis (Kenna et al., 2016). Members of the NEK family play various roles in ciliogenesis and are implicated in distinct ciliopathies (Table 1). Statistical analysis has linked the evolution of the NEKs and centrioles, both of which are responsible for organization of microtubule development as well as the basal bodies of cilia (Quarmby and Mahjoub, 2005).
NEK1 is localized to the basal body region and centrosomes (Shalom et al., 2008). Nek1 mutant mice exhibited a range of defects including progressing PKD (Upadhya et al., 2000), which has a well-established tie to ciliary dysfunction (White and Quarmby, 2008). NEK1 serves as a coordinator between ciliogenesis and cell cycle progression, a role that is speculated to involve signaling between the nucleus and the primary cilium (White and Quarmby, 2008). Remarkably, both overexpression of NEK1 and the complete removal of NEK1 severely decreased the percentage of cells bearing a primary cilium, suggesting that ciliogenesis depends on tightly regulated NEK1 expression (Shalom et al., 2008). The authors proposed that this observation could be due to either NEK1 blocking the upregulation of ciliary proteins or due to it physically interfering with the formation of complexes at the centrosome (White and Quarmby, 2008). NEK1 has also been shown to bind KIF3A, a kinesin motor protein crucial for ciliogenesis, further underscoring its role in directly impacting IFT (Quarmby and Mahjoub, 2005; White and Quarmby, 2008).
While fewer papers have been written about its role in ciliogenesis, NEK2 has a confirmed function in cilia homeostasis. NEK2 phosphorylates C-Nap1, Cep68, and Rootletin at the beginning of mitosis, allowing for the formation of the mitotic spindle (Fry et al., 1998; Bahe et al., 2005). This kinase is required for centriole splitting and is involved in the HDAC6 pathway, which promotes tubulin deacetylation (Endicott et al., 2015; DeVaul et al., 2017). NEK2 has been specifically shown to phosphorylate KIF24, ensuring that cilia resorption occurs prior to mitosis, and increased levels of NEK2 facilitate cilia depolymerization (Mirvis et al., 2018). Conversely, reduction of NEK2 leads to centriole defects and deficiencies in cilia biogenesis (Endicott et al., 2015).
NEK4 is localized to the base of primary cilium in RPE cells, suggesting that its role may include cilia stabilization (Basei et al., 2015). This kinase interacts with RPGRIP1 and RPGRIP1L, two proteins associated with ciliopathies (Patnaik et al., 2015). In a study from Coene et al., NEK4 knockdown resulted in a decrease in ciliated cells. The authors hypothesized that the interaction between RPGRIP1/RPGRIP1L and NEK4 forms a scaffold for the assembly of cilium-related kinases and substrates (Coene et al., 2011).
NEK8 is involved in the regulation of ciliary physiology and associated human ciliopathies (Choi et al., 2013; Grampa et al., 2016; Claus et al., 2022). In a mouse model of juvenile cystic kidney disease, mice bearing mutations in Nek8 display defects in ciliary localization that were potentially causal in the emergence of nephronophthisis (Otto et al., 2008). In the same mouse model, mice were observed to have an abnormal interaction between Nek8 and the polycystin complex (Valkova et al., 2005). During the transition from the cell cycle stage to ciliogenesis, NEK8 is activated and then ultimately degraded (Zalli et al., 2012). Mutations in NEK8, including missense and loss-of-function mutations, affected the regulation of signaling in the Hippo pathway through its main effector, YAP (Grampa et al., 2016).
NEK9 has roles in cell division as studies link phospho-NEK9 with microtubule and centrosomal organization, cytokinesis, and centrosome maturation (Roig et al., 2002; Roig et al., 2005; Sdelci et al., 2012; Meirelles et al., 2014). NEK9 has been implicated in the pathologies of Perthes disease, upward gaze palsy, arthrogryposis, congenital contracture syndrome, fetal akinesia, skeletal ciliopathies, meningiomas, and nevus comedonicus (Casey et al., 2016; Levinsohn et al., 2016; Dunn et al., 2019; Abraham et al., 2022; Liu et al., 2022). Ciliogenesis is bidirectionally regulated by selective autophagic cellular processes, whereby ciliogenesis promotes autophagy and vice-versa. NEK9 has been shown to have a role in upregulating autophagy via its binding to ATG8 proteins, which regulate autophagy (Behrends et al., 2010). ATG8 proteins are regulated via a domain on NEK9 called LC3 interacting region (LIR). Negative regulators of autophagy in the ATG8 family include GABARAP and GABARAPL1. Finally, autophagy of NEK9 is required for cilia formation, as NEK9 regulates ciliogenesis by interacting with autophagy adaptor MYH9 and myosin IIA, a suppressor of ciliogenesis (Yamamoto et al., 2021).
NEK10 has a role in promoting optimal cilia length during post-mitotic cilia assembly via interactions with pericentriolar matrix protein 1 (PCM1). NEK10 also stimulates ciliary transport as well as ciliary number and structure (Chivukula et al., 2020). Removal of NEK10 led to a decline in ciliated cells and a decrease in NEK10-promoted cilia resorption (Al Mutairi et al., 2020). Loss-of-function mutations in NEK10 resulted in PCD in humans (Al Mutairi et al., 2020). NEK10 forms a trimeric complex with PCM1 and RIIβ. A protein complex that includes NEK10 is found at centriolar satellites, and the role of NEK10 in this complex makes it essential for ciliogenesis (Porpora et al., 2018).
According to the chemical probes portal and SGC databases there are no high-quality chemical probes targeting any member(s) of the NEK family. To date, there are several lead compounds, sourced from the literature, that represent candidates for optimization. Examples of potential tool molecules are summarized in Figure 4. BAY 61-3606 was originally reported as a SYK inhibitor (Yamamoto et al., 2003), but interesting biological activity led to a deeper dive into its kinase target profile, revealing that NEK1 was also a target, with a IC50 of 159 nM (Lau et al., 2012). NEK2 is the most well-studied member of the NEK family, and, as such, there are several inhibitors available, including CRUK ICR compound (R)-21 with a NEK2 IC50 = 22 nM (Innocenti et al., 2012; Wells et al., 2018). This compound shows good selectivity over PLK1, an often seen off-target of NEK2 inhibitors. Some additional NEK2 inhibitors can be found in a NEK-family review (Wells et al., 2018). Abbott compound 17 was originally identified as a MAP3K8 inhibitor (George et al., 2008), and subsequently was found to bind to NEK4 with an IC50 = 50 nM (Metz et al., 2011). No further structure–activity relationship (SAR) studies on this scaffold for NEK4 have been reported. The BRAF inhibitor dabrafenib has been approved by the FDA for melanoma. Because dabrafenib showed activity against some cancer lines that differentiated it from other BRAF inhibitors, further kinase profiling was performed. These experiments demonstrated useful and consequential inhibition of CDK16 and NEK9 (Phadke et al., 2018). While some of these candidates bind to their associated NEK with high affinity, the modest kinome-wide selectivity of these compounds has thus far precluded them from being considered chemical probes (Arrowsmith et al., 2015). Through informed SAR campaigns, it should be possible to refine these leads into chemical probes, which may prove to be valuable tools in understanding the role of the NEKs in ciliary biology.
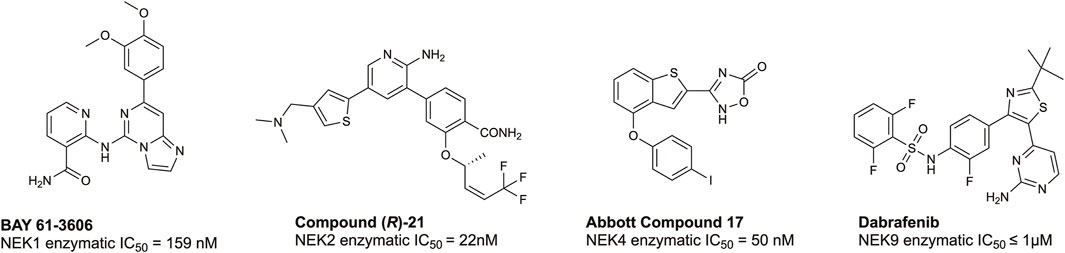
FIGURE 4. Structures and available potency data for promising chemical leads for NEK1, NEK2, NEK4, and NEK9.
3.3 TTBK family
Tau tubulin kinases are a family of serine/threonine/tyrosine kinases that belong to the larger casein kinase superfamily (CMGC) (Sato et al., 2006; Nozal and Martinez, 2019; Bashore et al., 2023). There are two isoforms of TTBK, tau tubulin kinase 1 and tau tubulin kinase 2, and their kinase domains share 88% identity and 96% similarity. Their catalytic residues, K63 and D164 for TTBK1 and K50 and D141 for TTBK2, are similar; however, their non-catalytic domains are distinct from one another (whole sequence similarity: 63% and 35% identity) (Nozal and Martinez, 2019; Bashore et al., 2023). TTBK2 regulates the initiation of ciliogenesis by acting at the distal end of the mother centriole to remove capping protein CP110 from the mother centriole (Čajánek and Nigg, 2014) and facilitate the recruitment of IFT proteins needed for the subsequent assembly of the ciliary axonemal microtubules (Čajánek and Nigg, 2014; Bashore et al., 2023; Binó and Čajánek, 2023). TTBK2 is not only highly expressed in the granular cell layer, cerebellum Purkinje cells, hippocampus, midbrain, and substantia nigra regions of the brain, but also ubiquitously expressed in most human tissues (Houlden et al., 2007; Bashore et al., 2023). As TTBK2 is essential for initiating cilia assembly, it is not surprising that a pathogenic mutation of TTBK2 leads to a neurological disorder known as spinocerebellar ataxia 11 (SCA11), characterized by atrophy of the Purkinje cells of the cerebellum (Bowie et al., 2018; Bashore et al., 2023). This truncated gene product is lacking the C-terminus, which is involved in trafficking TTBK2 to the basal body of primary cilium and also disrupts its interaction with CEP164, a key substrate that leads to initiation of ciliogenesis (Bowie et al., 2018; Bashore et al., 2023). Defects in ciliary assembly, stability, and function are observed in mice carrying the mutated alleles (Bowie et al., 2018). Another study showed cerebellar degeneration, altered intracellular levels of calcium, and loss of VGLUT2+ synapses in Ttbk2 mutated mice (Bowie and Goetz, 2020).
TTBK1 expression is confined to hippocampal, cortical, and entorhinal cortex neurons (Nozal and Martinez, 2019; Bashore et al., 2023). In human pluripotent TTBK2 knockout stem cells, TTBK1 was able to compensate for the loss of TTBK2 and regulate the assembly of primary cilia during neural rosette formation (Binó and Čajánek, 2023). Binó et al. further showed that in a TTBK2 rescue experiment, TTBK1 activity and expression levels increase enough to compensate for the loss of TTBK2 during neural rosette formation (Binó and Čajánek, 2023). Although TTBK1 lacks the C-terminal CEP164-binding domain that directs TTBK2 to the mother centriole, TTBK1 can still phosphorylate the key substrates of ciliogenesis outside of the mother centriole (Binó and Čajánek, 2023). This study brings to light the first indication that TTBK1 is a regulatory kinase of ciliogenesis. Furthermore, TTBK1 is known to act on neuropathic proteins like tau at pathogenically relevant sites and is overexpressed in Alzheimer’s disease (AD) with single nucleotide polymorphisms associated with late-onset AD (Ikezu and Ikezu, 2014; Bashore et al., 2023). TTBK1 also co-localizes with other neuropathic proteins and neurofibrillary tangles associated with amyotrophic lateral sclerosis and frontotemporal dementia (Ikezu and Ikezu, 2014; Bashore et al., 2023).
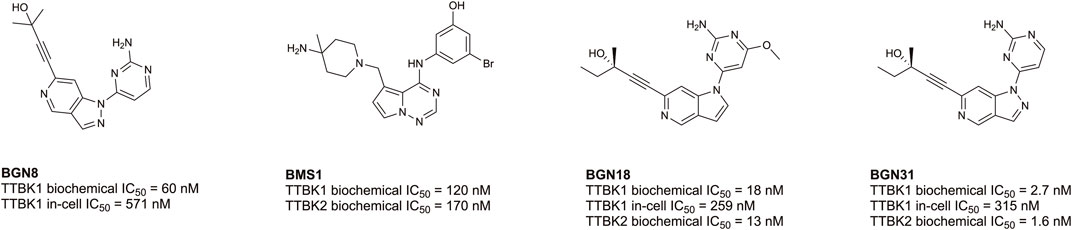
Despite being understudied, recent efforts have been made towards developing inhibitors of TTBK1/2. Biogen, Bristol-Myers Squibb, and AstraZeneca have published co-crystal structures of TTBK1 bound inhibitors (AZ1, AZ2, BMS1, and BGN18, Figure 5) (Xue et al., 2013; Kiefer et al., 2014; Halkina et al., 2021; Bashore et al., 2023). In 2013, AstraZeneca reported TTBK1 inhibitors AZ1 and AZ2, with Kd values of 0.24 µM and 4.1 µM, respectively (Xue et al., 2013). Later, a group optimized this same scaffold to yield compound 29, with selectivity for TTBK1 over TTBK2 (TTBK1 IC50 = 0.24 µM, TTBK2 IC50 = 4.22 µM) (Figure 5) (Nozal et al., 2022). Compound 29 was brain penetrant and lowered levels of phosphorylated TDP-43 in vitro and in vivo in a TDP-43 transgenic mouse model (Nozal et al., 2022). In 2014, Bristol-Myers Squibb published BMS1 and co-crystallized it in complex with TTBK1 (Kiefer et al., 2014). This compound has cell-free IC50 values for TTBK1 of 120 nM and for TTBK2 of 170 nM (Kiefer et al., 2014). In 2021, Biogen published a series of brain penetrant, azaindazole-based TTBK1/2 inhibitors: BGN8, BGN18, and BGN31 (Figure 5) (Halkina et al., 2021). Azaindazole BGN8 had a TTBK1 biochemical IC50 = 60 nM and in-cell TTBK1 IC50 = 571 nM (Halkina et al., 2021). This compound was further optimized to yield BGN18, which demonstrated TTBK1/2 biochemical IC50 values of 13–18 nM and TTBK1 in-cell potency of 259 nM (Halkina et al., 2021). Finally, the campaign yielded an azaindole-based analog, BGN31, which is the most advanced inhibitor of the series (Halkina et al., 2021). This compound has single-digit nM TTBK1/2 biochemical IC50 values and displays TTBK1 in-cell potency of 315 nM (Halkina et al., 2021). BGN31 was advanced to in vivo studies of tau phosphorylation, utilizing a mouse model of hypothermia and a developmental rat model, and was shown to reduce phosphorylation of tau at disease relevant sites (Halkina et al., 2021; Bashore et al., 2023).
The results of an academic effort by the SGC at the University of North Carolina to produce TTBK1/2 inhibitors was published in 2023 (Bashore et al., 2023). The indole scaffold of these inhibitors was discovered from an investigation of the off-target activity of a published Amgen NF-κB inhibitor, AMG28, which included inhibition of TTBK1/2 (Bashore et al., 2023). Compounds 9 and 10 were the most advanced leads from that campaign and these analogs demonstrated TTBK1/2 enzymatic IC50 values of 384 nM and 175 nM, respectively, for compound 9 and TTBK1/2 enzymatic IC50 values 579 nM and 258 nM, respectively, for compound 10 (Figure 5) (Bashore et al., 2023). Both compounds have been shown to inhibit ciliogenesis in human pluripotent stem cell-based models and phenocopy what is observed due to genetic editing of TTBK2 (Bashore et al., 2023; Binó and Čajánek, 2023). The available TTBK1/2 inhibitors in Figure 5 are mostly devoid of isoform selectivity, and many of the inhibitors lack overall kinome-wide selectivity data. The high sequence homology between the kinase domains of TTBK1 and TTBK2 suggests that selectivity between these kinases may be very difficult to achieve. To date, no potent and selective chemical probe for either TTBK1 or TTBK2 exists, but delivery of such a compound will facilitate deconvolution of the importance and roles of these kinases in ciliogenesis, canonical signaling pathways, and disease.
4 Conclusion
The importance of cilia and the kinases that aid in their regulation is clear. Many of these kinases fall into the understudied category, including members of the CDKL, NEK, and TTBK families, suggesting that we still may uncover new kinases and uncharacterized roles of kinases in ciliary biology. Continued research dedicated to these essential enzymes and the pathways into which they fit will further our understanding of normal and aberrant ciliary functions in human biology and disease. Since many ciliopathies, cancers, and nervous system disorders are caused in part by dysfunctional ciliary pathways, additional knowledge will move the field closer to a more complete understanding of ciliary function and potential treatments for related diseases. The small molecule tools discussed herein are being developed to better understand the roles of cilia in biology and disease. These pre-clinical chemical tools will aid in the dissection of ciliary networks to pinpoint those nodes that, when inhibited, could result in therapeutic benefit for patients.
Author contributions
RF: Writing–original draft, Writing–review and editing. PR: Writing–original draft, Writing–review and editing. CR: Writing–original draft, Writing–review and editing. BA: Writing–original draft, Writing–review and editing. JC: Writing–original draft. TD: Funding acquisition, Supervision, Writing–original draft. DD: Funding acquisition, Supervision, Writing–original draft, Writing–review and editing. PP: Funding acquisition, Writing–original draft, Writing–review and editing. AA: Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Structural Genomics Consortium (SGC) is a registered charity (number 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Eshelman Institute for Innovation, Genentech, Genome Canada through Ontario Genomics Institute, EU/EFPIA/OICR/McGill/KTH/Diamond, Innovative Medicines Initiative 2 Joint Undertaking, Janssen, Merck KGaA (aka EMD in Canada and United States), Pfizer, the São Paulo Research Foundation-FAPESP, and Takeda. Research reported in this publication was supported in part by NIH R01CA273095, 1R21NS112770-01A1, and U24DK116204 as well as Department of Defense ALSRP award AL220105.
Acknowledgments
Coral was used to make the kinome illustration in Figure 1. Coral was developed in the Phanstiel Lab at UNC; http://phanstiel-lab.med.unc.edu/CORAL (Metz et al., 2018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is responsibility of the authors and does not necessarily represent the official views of the funders.
References
Abraham, S. P., Nita, A., Krejci, P., and Bosakova, M. (2022). Cilia kinases in skeletal development and homeostasis. Dev. Dyn. 251, 577–608. doi:10.1002/dvdy.426
Alessi, D. R., and Sammler, E. (2018). LRRK2 kinase in Parkinson's disease. Science 360, 36–37. doi:10.1126/science.aar5683
Al Mutairi, F., Alkhalaf, R., Alkhorayyef, A., Alroqi, F., Yusra, A., Umair, M., et al. (2020). Homozygous truncating NEK10 mutation, associated with primary ciliary dyskinesia: a case report. BMC Pulm. Med. 20, 141. doi:10.1186/s12890-020-1175-1
Al-Sha'er, M. A., Basheer, H. A., and Taha, M. O. (2023). Discovery of new PKN2 inhibitory chemotypes via QSAR-guided selection of docking-based pharmacophores. Mol. Divers 27, 443–462. doi:10.1007/s11030-022-10434-4
Arora, S., Rana, M., Sachdev, A., and D’souza, J. S. (2023). Appearing and disappearing acts of cilia. J. Biosci. 48, 8. doi:10.1007/s12038-023-00326-6
Arrowsmith, C. H., Audia, J. E., Austin, C., Baell, J., Bennett, J., Blagg, J., et al. (2015). The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541. doi:10.1038/nchembio.1867
Axtman, A. D. (2021). Characterizing the role of the dark kinome in neurodegenerative disease – a mini review. Biochimica Biophysica Acta (BBA) - General Subj. 1865, 130014. doi:10.1016/j.bbagen.2021.130014
Azeggagh, S., and Berwick, D. C. (2022). The development of inhibitors of leucine-rich repeat kinase 2 (LRRK2) as a therapeutic strategy for Parkinson's disease: the current state of play. Br. J. Pharmacol. 179, 1478–1495. doi:10.1111/bph.15575
Bahe, S., Stierhof, Y. D., Wilkinson, C. J., Leiss, F., and Nigg, E. A. (2005). Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell. Biol. 171, 27–33. doi:10.1083/jcb.200504107
Bangs, F., and Anderson, K. V. (2017). Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175. doi:10.1101/cshperspect.a028175
Basei, F. L., Meirelles, G. V., Righetto, G. L., Dos Santos Migueleti, D. L., Smetana, J. H. C., and Kobarg, J. (2015). New interaction partners for Nek4.1 and Nek4.2 isoforms: from the DNA damage response to RNA splicing. Proteome Sci. 13, 11. doi:10.1186/s12953-015-0065-6
Bashore, F. M., Marquez, A. B., Chaikuad, A., Howell, S., Dunn, A. S., Beltran, A. A., et al. (2023). Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis. Sci. Rep. 13, 6118. doi:10.1038/s41598-023-32854-4
Basten, S. G., and Giles, R. H. (2013). Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia 2, 6. doi:10.1186/2046-2530-2-6
Basten, S. G., Willekers, S., Vermaat, J. S., Slaats, G. G., Voest, E. E., Van Diest, P. J., et al. (2013). Reduced cilia frequencies in human renal cell carcinomas versus neighboring parenchymal tissue. Cilia 2, 2. doi:10.1186/2046-2530-2-2
Behrends, C., Sowa, M. E., Gygi, S. P., and Harper, J. W. (2010). Network organization of the human autophagy system. Nature 466, 68–76. doi:10.1038/nature09204
Benmerah, A., Briseño-Roa, L., Annereau, J.-P., and Saunier, S. (2023). Repurposing small molecules for nephronophthisis and related renal ciliopathies. Kidney Int. 104, 245–253. doi:10.1016/j.kint.2023.04.027
Binó, L., and Čajánek, L. (2023). Tau tubulin kinase 1 and 2 regulate ciliogenesis and human pluripotent stem cells–derived neural rosettes. Sci. Rep. 13, 12884. doi:10.1038/s41598-023-39887-9
Bowie, E., and Goetz, S. C. (2020). TTBK2 and primary cilia are essential for the connectivity and survival of cerebellar Purkinje neurons. Elife 9, e51166. doi:10.7554/eLife.51166
Bowie, E., Norris, R., Anderson, K. V., and Goetz, S. C. (2018). Spinocerebellar ataxia type 11-associated alleles of Ttbk2 dominantly interfere with ciliogenesis and cilium stability. PLoS Genet. 14, e1007844. doi:10.1371/journal.pgen.1007844
Čajánek, L., and Nigg, E. A. (2014). Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. U. S. A. 111, E2841–E2850. doi:10.1073/pnas.1401777111
Canning, P., Park, K., Gonçalves, J., Li, C., Howard, C. J., Sharpe, T. D., et al. (2018). CDKL family kinases have evolved distinct structural features and ciliary function. Cell. Rep. 22, 885–894. doi:10.1016/j.celrep.2017.12.083
Cao, M., and Zhong, Q. (2016). Cilia in autophagy and cancer. Cilia 5, 4. doi:10.1186/s13630-016-0027-3
Casey, J. P., Brennan, K., Scheidel, N., Mcgettigan, P., Lavin, P. T., Carter, S., et al. (2016). Recessive NEK9 mutation causes a lethal skeletal dysplasia with evidence of cell cycle and ciliary defects. Hum. Mol. Genet. 25, 1824–1835. doi:10.1093/hmg/ddw054
Castano, A., Silvestre, M., Wells, C. I., Sanderson, J. L., Ferrer, C. A., Ong, H. W., et al. (2023). Discovery and characterization of a specific inhibitor of serine-threonine kinase cyclin-dependent kinase-like 5 (CDKL5) demonstrates role in hippocampal CA1 physiology. eLife 12, e88206. doi:10.7554/eLife.88206
Chaya, T., Omori, Y., Kuwahara, R., and Furukawa, T. (2014). ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. Embo J. 33, 1227–1242. doi:10.1002/embj.201488175
Chen, H., Chen, G., Li, G., Zhang, S., Chen, H., Chen, Y., et al. (2019). Two novel genetic variants in the STK38L and RAB27A genes are associated with glioma susceptibility. Int. J. Cancer 145, 2372–2382. doi:10.1002/ijc.32179
Chen, Q., Zhu, Y. C., Yu, J., Miao, S., Zheng, J., Xu, L., et al. (2010). CDKL5, a protein associated with rett syndrome, regulates neuronal morphogenesis via Rac1 signaling. J. Neurosci. 30, 12777–12786. doi:10.1523/JNEUROSCI.1102-10.2010
Chiba, S., Amagai, Y., Homma, Y., Fukuda, M., and Mizuno, K. (2013). NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. Embo J. 32, 874–885. doi:10.1038/emboj.2013.32
Chivukula, R. R., Montoro, D. T., Leung, H. M., Yang, J., Shamseldin, H. E., Taylor, M. S., et al. (2020). A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance. Nat. Med. 26, 244–251. doi:10.1038/s41591-019-0730-x
Chivukula, S., and Malkhed, V. (2023). The role of CDK20 protein in carcinogenesis. Curr. Drug Targets 24, 790–796. doi:10.2174/1389450124666230719102112
Choi, H. J., Lin, J. R., Vannier, J. B., Slaats, G. G., Kile, A. C., Paulsen, R. D., et al. (2013). NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell. 51, 423–439. doi:10.1016/j.molcel.2013.08.006
Christensen, S. T., Clement, C. A., Satir, P., and Pedersen, L. B. (2012). Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J. Pathol. 226, 172–184. doi:10.1002/path.3004
Christensen, S. T., Morthorst, S. K., Mogensen, J. B., and Pedersen, L. B. (2017). Primary cilia and coordination of receptor tyrosine kinase (RTK) and transforming growth factor β (TGF-β) signaling. Cold Spring Harb. Perspect. Biol. 9, a028167. doi:10.1101/cshperspect.a028167
Claus, L., Stallworth, J., Van Jaarsveld, R., Turner, J., Hawks, A., May, M., et al. (2022). FC044: heterozygous variants in kinase domain of NEK8 cause an autosomal-dominant ciliopathy. Nephrol. Dial. Transplant. 37. doi:10.1093/ndt/gfac104.004
Coene, K. L. M., Mans, D. A., Boldt, K., Gloeckner, C. J., Van Reeuwijk, J., Bolat, E., et al. (2011). The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum. Mol. Genet. 20, 3592–3605. doi:10.1093/hmg/ddr280
Deniz, O., Hasygar, K., and Hietakangas, V. (2023). Cellular and physiological roles of the conserved atypical MAP kinase ERK7. FEBS Lett. 597, 601–607. doi:10.1002/1873-3468.14521
DeVaul, N., Koloustroubis, K., Wang, R., and Sperry, A. O. (2017). A novel interaction between kinase activities in regulation of cilia formation. BMC Cell. Biol. 18, 33. doi:10.1186/s12860-017-0149-5
Dhekne, H. S., Yanatori, I., Gomez, R. C., Tonelli, F., Diez, F., Schüle, B., et al. (2018). A pathway for Parkinson's Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. Elife 7, e40202. doi:10.7554/eLife.40202
Di Nardo, A., Rühmkorf, A., Award, P., Brennecke, A., Fagiolini, M., and Sahin, M. (2022). Phenotypic characterization of Cdkl5-knockdown neurons establishes elongated cilia as a functional assay for CDKL5 Deficiency Disorder. Neurosci. Res. 176, 73–78. doi:10.1016/j.neures.2021.10.001
Doherty, D. (2009). Joubert syndrome: insights into brain development, cilium biology, and complex disease. Semin. Pediatr. Neurol. 16, 143–154. doi:10.1016/j.spen.2009.06.002
Dunn, J., Ferluga, S., Sharma, V., Futschik, M., Hilton, D. A., Adams, C. L., et al. (2019). Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 40, 77–91. doi:10.1016/j.ebiom.2018.12.048
Edelbusch, C., Cindrić, S., Dougherty, G. W., Loges, N. T., Olbrich, H., Rivlin, J., et al. (2017). Mutation of serine/threonine protein kinase 36 (STK36) causes primary ciliary dyskinesia with a central pair defect. Hum. Mutat. 38, 964–969. doi:10.1002/humu.23261
Egeberg, D. L., Lethan, M., Manguso, R., Schneider, L., Awan, A., Jørgensen, T. S., et al. (2012). Primary cilia and aberrant cell signaling in epithelial ovarian cancer. Cilia 1, 15. doi:10.1186/2046-2530-1-15
Emoto, K., Masugi, Y., Yamazaki, K., Effendi, K., Tsujikawa, H., Tanabe, M., et al. (2014). Presence of primary cilia in cancer cells correlates with prognosis of pancreatic ductal adenocarcinoma. Hum. Pathol. 45, 817–825. doi:10.1016/j.humpath.2013.11.017
Endicott, S. J., Basu, B., Khokha, M., and Brueckner, M. (2015). The NIMA-like kinase Nek2 is a key switch balancing cilia biogenesis and resorption in the development of left-right asymmetry. Development 142, 4068–4079. doi:10.1242/dev.126953
Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. doi:10.1074/mcp.M113.035600
Feng, H., Yu, Z., Tian, Y., Lee, Y. Y., Li, M. S., Go, M. Y., et al. (2015). A CCRK-EZH2 epigenetic circuitry drives hepatocarcinogenesis and associates with tumor recurrence and poor survival of patients. J. Hepatol. 62, 1100–1111. doi:10.1016/j.jhep.2014.11.040
Fry, A. M., Mayor, T., Meraldi, P., Stierhof, Y. D., Tanaka, K., and Nigg, E. A. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell. Biol. 141, 1563–1574. doi:10.1083/jcb.141.7.1563
Fry, A. M., O'regan, L., Sabir, S. R., and Bayliss, R. (2012). Cell cycle regulation by the NEK family of protein kinases. J. Cell. Sci. 125, 4423–4433. doi:10.1242/jcs.111195
Fu, Z., Gailey, C. D., Wang, E. J., and Brautigan, D. L. (2019). Ciliogenesis associated kinase 1: targets and functions in various organ systems. FEBS Lett. 593, 2990–3002. doi:10.1002/1873-3468.13600
George, D., Friedman, M., Allen, H., Argiriadi, M., Barberis, C., Bischoff, A., et al. (2008). Discovery of thieno[2,3-c]pyridines as potent COT inhibitors. Bioorg Med. Chem. Lett. 18, 4952–4955. doi:10.1016/j.bmcl.2008.08.037
Goetz, S. C., Ocbina, P. J., and Anderson, K. V. (2009). The primary cilium as a Hedgehog signal transduction machine. Methods Cell. Biol. 94, 199–222. doi:10.1016/S0091-679X(08)94010-3
Goldstein, O., Kukekova, A. V., Aguirre, G. D., and Acland, G. M. (2010). Exonic SINE insertion in STK38L causes canine early retinal degeneration (erd). Genomics 96, 362–368. doi:10.1016/j.ygeno.2010.09.003
Goto, H., Inaba, H., and Inagaki, M. (2017). Mechanisms of ciliogenesis suppression in dividing cells. Cell. Mol. Life Sci. 74, 881–890. doi:10.1007/s00018-016-2369-9
Grampa, V., Delous, M., Zaidan, M., Odye, G., Thomas, S., Elkhartoufi, N., et al. (2016). Novel NEK8 mutations cause severe syndromic renal cystic dysplasia through YAP dysregulation. PLoS Genet. 12, e1005894. doi:10.1371/journal.pgen.1005894
Grant, T. J., Mehta, A. K., Gupta, A., Sharif, A. a.D., Arora, K. S., Deshpande, V., et al. (2017). STK38L kinase ablation promotes loss of cell viability in a subset of KRAS-dependent pancreatic cancer cell lines. Oncotarget 8, 78556–78572. doi:10.18632/oncotarget.20833
Guen, V. J., Gamble, C., Flajolet, M., Unger, S., Thollet, A., Ferandin, Y., et al. (2013). CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proc. Natl. Acad. Sci. U. S. A. 110, 19525–19530. doi:10.1073/pnas.1306814110
Guen, V. J., Gamble, C., Lees, J. A., and Colas, P. (2017). The awakening of the CDK10/Cyclin M protein kinase. Oncotarget 8, 50174–50186. doi:10.18632/oncotarget.15024
Guen, V. J., Gamble, C., Perez, D. E., Bourassa, S., Zappel, H., Gärtner, J., et al. (2016). STAR syndrome-associated CDK10/Cyclin M regulates actin network architecture and ciliogenesis. Cell. Cycle 15, 678–688. doi:10.1080/15384101.2016.1147632
Halkina, T., Henderson, J. L., Lin, E. Y., Himmelbauer, M. K., Jones, J. H., Nevalainen, M., et al. (2021). Discovery of potent and brain-penetrant tau tubulin kinase 1 (TTBK1) inhibitors that lower tau phosphorylation in vivo. J. Med. Chem. 64, 6358–6380. doi:10.1021/acs.jmedchem.1c00382
Han, Y. G., Kim, H. J., Dlugosz, A. A., Ellison, D. W., Gilbertson, R. J., and Alvarez-Buylla, A. (2009). Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 15, 1062–1065. doi:10.1038/nm.2020
Hassounah, N. B., Nagle, R., Saboda, K., Roe, D. J., Dalkin, B. L., and Mcdermott, K. M. (2013). Primary cilia are lost in preinvasive and invasive prostate cancer. PLoS One 8, e68521. doi:10.1371/journal.pone.0068521
Hergovich, A. (2016). The roles of NDR protein kinases in Hippo signalling. Genes. (Basel) 7, 21. doi:10.3390/genes7050021
Higgins, M., Obaidi, I., and Mcmorrow, T. (2019). Primary cilia and their role in cancer. Oncol. Lett. 17, 3041–3047. doi:10.3892/ol.2019.9942
Hildebrandt, F., Benzing, T., and Katsanis, N. (2011). Ciliopathies. N. Engl. J. Med. 364, 1533–1543. doi:10.1056/NEJMra1010172
Houlden, H., Johnson, J., Gardner-Thorpe, C., Lashley, T., Hernandez, D., Worth, P., et al. (2007). Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat. Genet. 39, 1434–1436. doi:10.1038/ng.2007.43
Hsu, L. S., Liang, C. J., Tseng, C. Y., Yeh, C. W., and Tsai, J. N. (2011). Zebrafish cyclin-dependent protein kinase-like 1 (zcdkl1): identification and functional characterization. Int. J. Mol. Sci. 12, 3606–3617. doi:10.3390/ijms12063606
Ikezu, S., and Ikezu, T. (2014). Tau-tubulin kinase. Front. Mol. Neurosci. 7, 33. doi:10.3389/fnmol.2014.00033
Innocenti, P., Cheung, K. M., Solanki, S., Mas-Droux, C., Rowan, F., Yeoh, S., et al. (2012). Design of potent and selective hybrid inhibitors of the mitotic kinase Nek2: structure-activity relationship, structural biology, and cellular activity. J. Med. Chem. 55, 3228–3241. doi:10.1021/jm201683b
Iorns, E., Turner, N. C., Elliott, R., Syed, N., Garrone, O., Gasco, M., et al. (2008). Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 13, 91–104. doi:10.1016/j.ccr.2008.01.001
Kazatskaya, A., Kuhns, S., Lambacher, N. J., Kennedy, J. E., Brear, A. G., Mcmanus, G. J., et al. (2017). Primary cilium formation and ciliary protein trafficking is regulated by the atypical MAP kinase MAPK15 in Caenorhabditis elegans and human cells. Genetics 207, 1423–1440. doi:10.1534/genetics.117.300383
Kenna, K. P., Van Doormaal, P. T., Dekker, A. M., Ticozzi, N., Kenna, B. J., Diekstra, F. P., et al. (2016). NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 48, 1037–1042. doi:10.1038/ng.3626
Khamrui, S., Ung, P. M. U., Secor, C., Schlessinger, A., and Lazarus, M. B. (2020). High-resolution structure and inhibition of the schizophrenia-linked pseudokinase ULK4. J. Am. Chem. Soc. 142, 33–37. doi:10.1021/jacs.9b10458
Khan, S. S., Sobu, Y., Dhekne, H. S., Tonelli, F., Berndsen, K., Alessi, D. R., et al. (2021). Pathogenic LRRK2 control of primary cilia and Hedgehog signaling in neurons and astrocytes of mouse brain. Elife 10, e67900. doi:10.7554/eLife.67900
Kiefer, S. E., Chang, C. J., Kimura, S. R., Gao, M., Xie, D., Zhang, Y., et al. (2014). The structure of human tau-tubulin kinase 1 both in the apo form and in complex with an inhibitor. Acta Crystallogr. F. Struct. Biol. Commun. 70, 173–181. doi:10.1107/S2053230X14000144
Kingwell, K. (2023). LRRK2-targeted Parkinson disease drug advances into phase III. Nat. Rev. Drug Discov. 22, 3–5. doi:10.1038/d41573-022-00212-0
Kondelin, J., Salokas, K., Saarinen, L., Ovaska, K., Rauanheimo, H., Plaketti, R. M., et al. (2018). Comprehensive evaluation of coding region point mutations in microsatellite-unstable colorectal cancer. EMBO Mol. Med. 10, e8552. doi:10.15252/emmm.201708552
Krahn, A. I., Wells, C., Drewry, D. H., Beitel, L. K., Durcan, T. M., and Axtman, A. D. (2020). Defining the neural kinome: strategies and opportunities for small molecule drug discovery to target neurodegenerative diseases. ACS Chem. Neurosci. 11, 1871–1886. doi:10.1021/acschemneuro.0c00176
Kunova Bosakova, M., Nita, A., Gregor, T., Varecha, M., Gudernova, I., Fafilek, B., et al. (2019). Fibroblast growth factor receptor influences primary cilium length through an interaction with intestinal cell kinase. Proc. Natl. Acad. Sci. U. S. A. 116, 4316–4325. doi:10.1073/pnas.1800338116
Lahiry, P., Wang, J., Robinson, J. F., Turowec, J. P., Litchfield, D. W., Lanktree, M. B., et al. (2009). A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am. J. Hum. Genet. 84, 134–147. doi:10.1016/j.ajhg.2008.12.017
Lang, B., Pu, J., Hunter, I., Liu, M., Martin-Granados, C., Reilly, T. J., et al. (2014). Recurrent deletions of ULK4 in schizophrenia: a gene crucial for neuritogenesis and neuronal motility. J. Cell. Sci. 127, 630–640. doi:10.1242/jcs.137604
Lau, K. S., Zhang, T., Kendall, K. R., Lauffenburger, D., Gray, N. S., and Haigis, K. M. (2012). BAY61-3606 affects the viability of colon cancer cells in a genotype-directed manner. PLoS One 7, e41343. doi:10.1371/journal.pone.0041343
Lee, J., Park, K. C., Sul, H. J., Hong, H. J., Kim, K.-H., Kero, J., et al. (2021). Loss of primary cilia promotes mitochondria-dependent apoptosis in thyroid cancer. Sci. Rep. 11, 4181. doi:10.1038/s41598-021-83418-3
Lee, J. E., and Gleeson, J. G. (2011). A systems-biology approach to understanding the ciliopathy disorders. Genome Med. 3, 59. doi:10.1186/gm275
Léger, H., Santana, E., Leu, N. A., Smith, E. T., Beltran, W. A., Aguirre, G. D., et al. (2018). Ndr kinases regulate retinal interneuron proliferation and homeostasis. Sci. Rep. 8, 12544. doi:10.1038/s41598-018-30492-9
Lepanto, P., Badano, J. L., and Zolessi, F. R. (2016). Neuron's little helper: the role of primary cilia in neurogenesis. Neurogenes. (Austin) 3, e1253363. doi:10.1080/23262133.2016.1253363
Levinsohn, J. L., Sugarman, J. L., Mcniff, J. M., Antaya, R. J., and Choate, K. A. (2016). Somatic mutations in NEK9 cause nevus comedonicus. Am. J. Hum. Genet. 98, 1030–1037. doi:10.1016/j.ajhg.2016.03.019
Li, C., Jensen, V. L., Park, K., Kennedy, J., Garcia-Gonzalo, F. R., Romani, M., et al. (2016). MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol. 14, e1002416. doi:10.1371/journal.pbio.1002416
Liu, F., Dai, L., Li, Z., and Yin's, X. (2022). Novel variants of NEK9 associated with neonatal arthrogryposis: two case reports and a literature review. Front. Genet. 13, 989215. doi:10.3389/fgene.2022.989215
Liu, H., Kiseleva, A. A., and Golemis, E. A. (2018). Ciliary signalling in cancer. Nat. Rev. Cancer 18, 511–524. doi:10.1038/s41568-018-0023-6
Liu, M., Guan, Z., Shen, Q., Flinter, F., Domínguez, L., Ahn, J. W., et al. (2016a). Ulk4 regulates neural stem cell pool. Stem Cells 34, 2318–2331. doi:10.1002/stem.2423
Liu, M., Guan, Z., Shen, Q., Lalor, P., Fitzgerald, U., O'brien, T., et al. (2016b). Ulk4 is essential for ciliogenesis and CSF flow. J. Neurosci. 36, 7589–7600. doi:10.1523/JNEUROSCI.0621-16.2016
Loukil, A., Barrington, C., and Goetz, S. C. (2021). A complex of distal appendage-associated kinases linked to human disease regulates ciliary trafficking and stability. Proc. Natl. Acad. Sci. U. S. A. 118, e2018740118. doi:10.1073/pnas.2018740118
Luo, S., Zheng, N., and Lang, B. (2022). ULK4 in neurodevelopmental and neuropsychiatric disorders. Front. Cell. Dev. Biol. 10, 873706. doi:10.3389/fcell.2022.873706
Ma, R., Kutchy, N. A., Chen, L., Meigs, D. D., and Hu, G. (2022). Primary cilia and ciliary signaling pathways in aging and age-related brain disorders. Neurobiol. Dis. 163, 105607. doi:10.1016/j.nbd.2021.105607
Mecklenburg, N., Kowalczyk, I., Witte, F., Görne, J., Laier, A., Mamo, T. M., et al. (2021). Identification of disease-relevant modulators of the SHH pathway in the developing brain. Development 148, dev199307. doi:10.1242/dev.199307
Meirelles, G. V., Perez, A. M., De Souza, E. E., Basei, F. L., Papa, P. F., Melo Hanchuk, T. D., et al. (2014). Stop Ne(c)king around": how interactomics contributes to functionally characterize Nek family kinases. World J. Biol. Chem. 5, 141–160. doi:10.4331/wjbc.v5.i2.141
Menzl, I., Lebeau, L., Pandey, R., Hassounah, N. B., Li, F. W., Nagle, R., et al. (2014). Loss of primary cilia occurs early in breast cancer development. Cilia 3, 7. doi:10.1186/2046-2530-3-7
Merchant, M., Evangelista, M., Luoh, S. M., Frantz, G. D., Chalasani, S., Carano, R. A., et al. (2005). Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol. Cell. Biol. 25, 7054–7068. doi:10.1128/MCB.25.16.7054-7068.2005
Metz, J. T., Johnson, E. F., Soni, N. B., Merta, P. J., Kifle, L., and Hajduk, P. J. (2011). Navigating the kinome. Nat. Chem. Biol. 7, 200–202. doi:10.1038/nchembio.530
Metz, K. S., Deoudes, E. M., Berginski, M. E., Jimenez-Ruiz, I., Aksoy, B. A., Hammerbacher, J., et al. (2018). Coral: clear and customizable visualization of human kinome data. Cell. Syst. 7, 347–350. doi:10.1016/j.cels.2018.07.001
Mill, P., Christensen, S. T., and Pedersen, L. B. (2023). Primary cilia as dynamic and diverse signalling hubs in development and disease. Nat. Rev. Genet. 24, 421–441. doi:10.1038/s41576-023-00587-9
Mirvis, M., Stearns, T., and James Nelson, W. (2018). Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 475, 2329–2353. doi:10.1042/BCJ20170453
Miyatake, K., Kusakabe, M., Takahashi, C., and Nishida, E. (2015). ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat. Commun. 6, 6666. doi:10.1038/ncomms7666
Modarage, K., Malik, S. A., and Goggolidou, P. (2022). Molecular Diagnostics of ciliopathies and insights into novel developments in diagnosing rare diseases. Br. J. Biomed. Sci. 79, 10221. doi:10.3389/bjbs.2021.10221
Moniz, L., Dutt, P., Haider, N., and Stambolic, V. (2011). Nek family of kinases in cell cycle, checkpoint control and cancer. Cell. Div. 6, 18. doi:10.1186/1747-1028-6-18
Moon, H., Song, J., Shin, J. O., Lee, H., Kim, H. K., Eggenschwiller, J. T., et al. (2014). Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc. Natl. Acad. Sci. U. S. A. 111, 8541–8546. doi:10.1073/pnas.1323161111
Murone, M., Luoh, S. M., Stone, D., Li, W., Gurney, A., Armanini, M., et al. (2000). Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell. Biol. 2, 310–312. doi:10.1038/35010610
Ng, S. S., Cheung, Y. T., An, X. M., Chen, Y. C., Li, M., Li, G. H., et al. (2007). Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma. J. Natl. Cancer Inst. 99, 936–948. doi:10.1093/jnci/djm011
Nguyen, K., Boehling, J., Tran, M. N., Cheng, T., Rivera, A., Collins-Burow, B. M., et al. (2023). NEK family review and correlations with patient survival outcomes in various cancer types. Cancers (Basel) 15, 2067. doi:10.3390/cancers15072067
Nozal, V., and Martinez, A. (2019). Tau Tubulin Kinase 1 (TTBK1), a new player in the fight against neurodegenerative diseases. Eur. J. Med. Chem. 161, 39–47. doi:10.1016/j.ejmech.2018.10.030
Nozal, V., Martínez-González, L., Gomez-Almeria, M., Gonzalo-Consuegra, C., Santana, P., Chaikuad, A., et al. (2022). TDP-43 modulation by tau-tubulin kinase 1 inhibitors: a new avenue for future amyotrophic lateral sclerosis therapy. J. Med. Chem. 65, 1585–1607. doi:10.1021/acs.jmedchem.1c01942
Olson, H. E., Demarest, S. T., Pestana-Knight, E. M., Swanson, L. C., Iqbal, S., Lal, D., et al. (2019). Cyclin-dependent kinase-like 5 deficiency disorder: clinical review. Pediatr. Neurol. 97, 18–25. doi:10.1016/j.pediatrneurol.2019.02.015
Ong, H. W., Liang, Y., Richardson, W., Lowry, E. R., Wells, C. I., Chen, X., et al. (2023). A potent and selective CDKL5/GSK3 chemical probe is neuroprotective. USA: ACS Chem Neurosci In Press.
Otto, E. A., Trapp, M. L., Schultheiss, U. T., Helou, J., Quarmby, L. M., and Hildebrandt, F. (2008). NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J. Am. Soc. Nephrol. 19, 587–592. doi:10.1681/ASN.2007040490
Paige Taylor, S., Kunova Bosakova, M., Varecha, M., Balek, L., Barta, T., Trantirek, L., et al. (2016). An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum. Mol. Genet. 25, 3998–4011. doi:10.1093/hmg/ddw240
Panchal, N. K., and Evan Prince, S. (2023a). The NEK family of serine/threonine kinases as a biomarker for cancer. Clin. Exp. Med. 23, 17–30. doi:10.1007/s10238-021-00782-0
Panchal, N. K., and Evan Prince, S. (2023b). The NEK family of serine/threonine kinases as a biomarker for cancer. Clin. Exp. Med. 23, 17–30. doi:10.1007/s10238-021-00782-0
Park, K., Li, C., Tsiropoulou, S., Gonçalves, J., Kondratev, C., Pelletier, L., et al. (2021). CDKL kinase regulates the length of the ciliary proximal segment. Curr. Biol. 31, 2359–2373.e7. doi:10.1016/j.cub.2021.03.068
Patel, H., Li, J., Herrero, A., Kroboth, J., Byron, A., Kriegsheim, A. V., et al. (2020). Novel roles of PRK1 and PRK2 in cilia and cancer biology. Sci. Rep. 10, 3902. doi:10.1038/s41598-020-60604-3
Patnaik, S. R., Raghupathy, R. K., Zhang, X., Mansfield, D., and Shu, X. (2015). The role of RPGR and its interacting proteins in ciliopathies. J. Ophthalmol. 2015, 414781. doi:10.1155/2015/414781
Pavan, I. C. B., Peres De Oliveira, A., Dias, P. R. F., Basei, F. L., Issayama, L. K., Ferezin, C. C., et al. (2021). On broken Ne(c)ks and broken DNA: the role of human NEKs in the DNA damage response. Cells 10, 507. doi:10.3390/cells10030507
Phadke, M., Remsing Rix, L. L., Smalley, I., Bryant, A. T., Luo, Y., Lawrence, H. R., et al. (2018). Dabrafenib inhibits the growth of BRAF-WT cancers through CDK16 and NEK9 inhibition. Mol. Oncol. 12, 74–88. doi:10.1002/1878-0261.12152
Pietrobono, S., Franci, L., Imperatore, F., Zanini, C., Stecca, B., and Chiariello, M. (2021). MAPK15 controls hedgehog signaling in medulloblastoma cells by regulating primary ciliogenesis. Cancers (Basel) 13, 4903. doi:10.3390/cancers13194903
Porpora, M., Sauchella, S., Rinaldi, L., Delle Donne, R., Sepe, M., Torres-Quesada, O., et al. (2018). Counterregulation of cAMP-directed kinase activities controls ciliogenesis. Nat. Commun. 9, 1224. doi:10.1038/s41467-018-03643-9
Quarmby, L. M., and Mahjoub, M. R. (2005). Caught Nek-ing: cilia and centrioles. J. Cell. Sci. 118, 5161–5169. doi:10.1242/jcs.02681
Reiter, J. F., and Leroux, M. R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell. Biol. 18, 533–547. doi:10.1038/nrm.2017.60
Ren, F., Ding, X., Zheng, M., Korzinkin, M., Cai, X., Zhu, W., et al. (2023). AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 14, 1443–1452. doi:10.1039/d2sc05709c
Robert, T., Johnson, J. L., Guichaoua, R., Yaron, T. M., Bach, S., Cantley, L. C., et al. (2020). Development of a CDK10/CycM in vitro kinase screening assay and identification of first small-molecule inhibitors. Front. Chem. 8, 147. doi:10.3389/fchem.2020.00147
Rocha, C., and Prinos, P. (2022). Post-transcriptional and post-translational modifications of primary cilia: how to fine tune your neuronal antenna. Front. Cell. Neurosci. 16, 809917. doi:10.3389/fncel.2022.809917
Roig, J., Groen, A., Caldwell, J., and Avruch, J. (2005). Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell. 16, 4827–4840. doi:10.1091/mbc.e05-04-0315
Roig, J., Mikhailov, A., Belham, C., and Avruch, J. (2002). Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes. Dev. 16, 1640–1658. doi:10.1101/gad.972202
Roskoski, R. (2023). Properties of FDA-approved small molecule protein kinase inhibitors: a 2023 update. Pharmacol. Res. 187, 106552. doi:10.1016/j.phrs.2022.106552
Rusconi, L., Salvatoni, L., Giudici, L., Bertani, I., Kilstrup-Nielsen, C., Broccoli, V., et al. (2008). CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J. Biol. Chem. 283, 30101–30111. doi:10.1074/jbc.M804613200
Sakai, D., Dixon, J., Dixon, M. J., and Trainor, P. A. (2012). Mammalian neurogenesis requires Treacle-Plk1 for precise control of spindle orientation, mitotic progression, and maintenance of neural progenitor cells. PLoS Genet. 8, e1002566. doi:10.1371/journal.pgen.1002566
Santos, P. F., Fazendeiro, B., Luca, F. C., Ambrósio, A. F., and Léger, H. (2023). The NDR/LATS protein kinases in neurobiology: key regulators of cell proliferation, differentiation and migration in the ocular and central nervous system. Eur. J. Cell. Biol. 102, 151333. doi:10.1016/j.ejcb.2023.151333
Sato, S., Cerny, R. L., Buescher, J. L., and Ikezu, T. (2006). Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 98, 1573–1584. doi:10.1111/j.1471-4159.2006.04059.x
Scott, F., Fala, A. M., Pennicott, L. E., Reuillon, T. D., Massirer, K. B., Elkins, J. M., et al. (2020). Development of 2-(4-pyridyl)-benzimidazoles as PKN2 chemical tools to probe cancer. Bioorg Med. Chem. Lett. 30, 127040. doi:10.1016/j.bmcl.2020.127040
Scott, F., Fala, A. M., Takarada, J. E., Ficu, M. P., Pennicott, L. E., Reuillon, T. D., et al. (2022). Development of dihydropyrrolopyridinone-based PKN2/PRK2 chemical tools to enable drug discovery. Bioorg Med. Chem. Lett. 60, 128588. doi:10.1016/j.bmcl.2022.128588
Sdelci, S., Schütz, M., Pinyol, R., Bertran, M. T., Regué, L., Caelles, C., et al. (2012). Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 22, 1516–1523. doi:10.1016/j.cub.2012.06.027
Seeger-Nukpezah, T., Liebau, M. C., Höpker, K., Lamkemeyer, T., Benzing, T., Golemis, E. A., et al. (2012). The centrosomal kinase Plk1 localizes to the transition zone of primary cilia and induces phosphorylation of nephrocystin-1. PLoS One 7, e38838. doi:10.1371/journal.pone.0038838
Seeger-Nukpezah, T., Little, J. L., Serzhanova, V., and Golemis, E. A. (2013). Cilia and cilia-associated proteins in cancer. Drug Discov. Today Dis. Mech. 10, e135–e142. doi:10.1016/j.ddmec.2013.03.004
Seeley, E. S., Carrière, C., Goetze, T., Longnecker, D. S., and Korc, M. (2009). Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 69, 422–430. doi:10.1158/0008-5472.CAN-08-1290
Shalom, O., Shalva, N., Altschuler, Y., and Motro, B. (2008). The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 582, 1465–1470. doi:10.1016/j.febslet.2008.03.036
Smith, A. O., Jonassen, J. A., Preval, K. M., Davis, R. J., and Pazour, G. J. (2022). c-JUN n-terminal kinase (JNK) signaling in autosomal dominant polycystic kidney disease. J. Cell. Signal 3, 62–78. doi:10.33696/Signaling.3.068
Snedecor, E. R., Sung, C. C., Moncayo, A., Rothstein, B. E., Mockler, D. C., Tonnesen, M. G., et al. (2015). Loss of primary cilia in melanoma cells is likely independent of proliferation and cell cycle progression. J. Investig. Dermatol 135, 1456–1458. doi:10.1038/jid.2015.22
Snouffer, A., Brown, D., Lee, H., Walsh, J., Lupu, F., Norman, R., et al. (2017). Cell Cycle-Related Kinase (CCRK) regulates ciliogenesis and Hedgehog signaling in mice. PLoS Genet. 13, e1006912. doi:10.1371/journal.pgen.1006912
Trulioff, A., Ermakov, A., and Malashichev, Y. (2017). Primary cilia as a possible link between left-right asymmetry and neurodevelopmental diseases. Genes. (Basel) 8, 48. doi:10.3390/genes8020048
Upadhya, P., Birkenmeier, E. H., Birkenmeier, C. S., and Barker, J. E. (2000). Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. U. S. A. 97, 217–221. doi:10.1073/pnas.97.1.217
Valkova, N., Yunis, R., Mak, S. K., Kang, K., and Kültz, D. (2005). Nek8 mutation causes overexpression of galectin-1, sorcin, and vimentin and accumulation of the major urinary protein in renal cysts of jck mice. Mol. Cell. Proteomics 4, 1009–1018. doi:10.1074/mcp.M500091-MCP200
Villalobos, E., Criollo, A., Schiattarella, G. G., Altamirano, F., French, K. M., May, H. I., et al. (2019). Fibroblast primary cilia are required for cardiac fibrosis. Circulation 139, 2342–2357. doi:10.1161/CIRCULATIONAHA.117.028752
Vogel, P., Read, R. W., Hansen, G. M., Payne, B. J., Small, D., Sands, A. T., et al. (2012). Congenital hydrocephalus in genetically engineered mice. Vet. Pathol. 49, 166–181. doi:10.1177/0300985811415708
Wang, G., Chen, Q., Zhang, X., Zhang, B., Zhuo, X., Liu, J., et al. (2013). PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J. Cell. Sci. 126, 1355–1365. doi:10.1242/jcs.114918
Wells, C. I., Kapadia, N. R., Couñago, R. M., and Drewry, D. H. (2018). In depth analysis of kinase cross screening data to identify chemical starting points for inhibition of the Nek family of kinases. Medchemcomm 9, 44–66. doi:10.1039/c7md00510e
White, M. C., and Quarmby, L. M. (2008). The NIMA-family kinase, Nek1 affects the stability of centrosomes and ciliogenesis. BMC Cell. Biol. 9, 29. doi:10.1186/1471-2121-9-29
Wilson, C. W., Nguyen, C. T., Chen, M. H., Yang, J. H., Gacayan, R., Huang, J., et al. (2009). Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature 459, 98–102. doi:10.1038/nature07883
Windpassinger, C., Piard, J., Bonnard, C., Alfadhel, M., Lim, S., Bisteau, X., et al. (2017). CDK10 mutations in humans and mice cause severe growth retardation, spine malformations, and developmental delays. Am. J. Hum. Genet. 101, 391–403. doi:10.1016/j.ajhg.2017.08.003
Wirschell, M., Yamamoto, R., Alford, L., Gokhale, A., Gaillard, A., and Sale, W. S. (2011). Regulation of ciliary motility: conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme. Archives Biochem. Biophysics 510, 93–100. doi:10.1016/j.abb.2011.04.003
Wu, G. Q., Xie, D., Yang, G. F., Liao, Y. J., Mai, S. J., Deng, H. X., et al. (2009). Cell cycle-related kinase supports ovarian carcinoma cell proliferation via regulation of cyclin D1 and is a predictor of outcome in patients with ovarian carcinoma. Int. J. Cancer 125, 2631–2642. doi:10.1002/ijc.24630
Xue, Y., Wan, P. T., Hillertz, P., Schweikart, F., Zhao, Y., Wissler, L., et al. (2013). X-ray structural analysis of tau-tubulin kinase 1 and its interactions with small molecular inhibitors. ChemMedChem 8, 1846–1854. doi:10.1002/cmdc.201300274
Yamamoto, N., Takeshita, K., Shichijo, M., Kokubo, T., Sato, M., Nakashima, K., et al. (2003). The orally available spleen tyrosine kinase inhibitor 2-[7-(3,4-dimethoxyphenyl)-imidazo[1,2-c]pyrimidin-5-ylamino]nicotinamide dihydrochloride (BAY 61-3606) blocks antigen-induced airway inflammation in rodents. J. Pharmacol. Exp. Ther. 306, 1174–1181. doi:10.1124/jpet.103.052316
Yamamoto, Y., Chino, H., Tsukamoto, S., Ode, K. L., Ueda, H. R., and Mizushima, N. (2021). NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/myosin IIA. Nat. Commun. 12, 3292. doi:10.1038/s41467-021-23599-7
Yang, Y., Roine, N., and Mäkelä, T. P. (2013). CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner. EMBO Rep. 14, 741–747. doi:10.1038/embor.2013.80
Youn, Y. H., and Han, Y. G. (2018). Primary cilia in brain development and diseases. Am. J. Pathol. 188, 11–22. doi:10.1016/j.ajpath.2017.08.031
Zaghloul, N. A., and Katsanis, N. (2009). Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Investigation 119, 428–437. doi:10.1172/JCI37041
Zalli, D., Bayliss, R., and Fry, A. M. (2012). The Nek8 protein kinase, mutated in the human cystic kidney disease nephronophthisis, is both activated and degraded during ciliogenesis. Hum. Mol. Genet. 21, 1155–1171. doi:10.1093/hmg/ddr544
Zeqiraj, E., and Van Aalten, D. M. (2010). Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 20, 772–781. doi:10.1016/j.sbi.2010.10.001
Keywords: kinase, cilia, ciliogenesis, ciliopathy, understudied, chemical probe, cancer, neurological disorder
Citation: Flax RG, Rosston P, Rocha C, Anderson B, Capener JL, Durcan TM, Drewry DH, Prinos P and Axtman AD (2024) Illumination of understudied ciliary kinases. Front. Mol. Biosci. 11:1352781. doi: 10.3389/fmolb.2024.1352781
Received: 08 December 2023; Accepted: 29 January 2024;
Published: 08 March 2024.
Edited by:
Joao Goncalves, Deep Genomics Inc., CanadaCopyright © 2024 Flax, Rosston, Rocha, Anderson, Capener, Durcan, Drewry, Prinos and Axtman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison D. Axtman, YWxpc29uLmF4dG1hbkB1bmMuZWR1
†These authors have contributed equally to this work
 Raymond G. Flax
Raymond G. Flax Peter Rosston1,2†
Peter Rosston1,2† Cecilia Rocha
Cecilia Rocha Thomas M. Durcan
Thomas M. Durcan David H. Drewry
David H. Drewry Panagiotis Prinos
Panagiotis Prinos Alison D. Axtman
Alison D. Axtman