- 1Infection and Epigenetics Laboratory, School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT), Bhubaneswar, Odisha, India
- 2Environmental Science Laboratory, School of Applied Sciences, Kalinga Institute of Industrial Technology (KIIT), Bhubaneswar, Odisha, India
- 3School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT), Bhubaneswar, Odisha, India
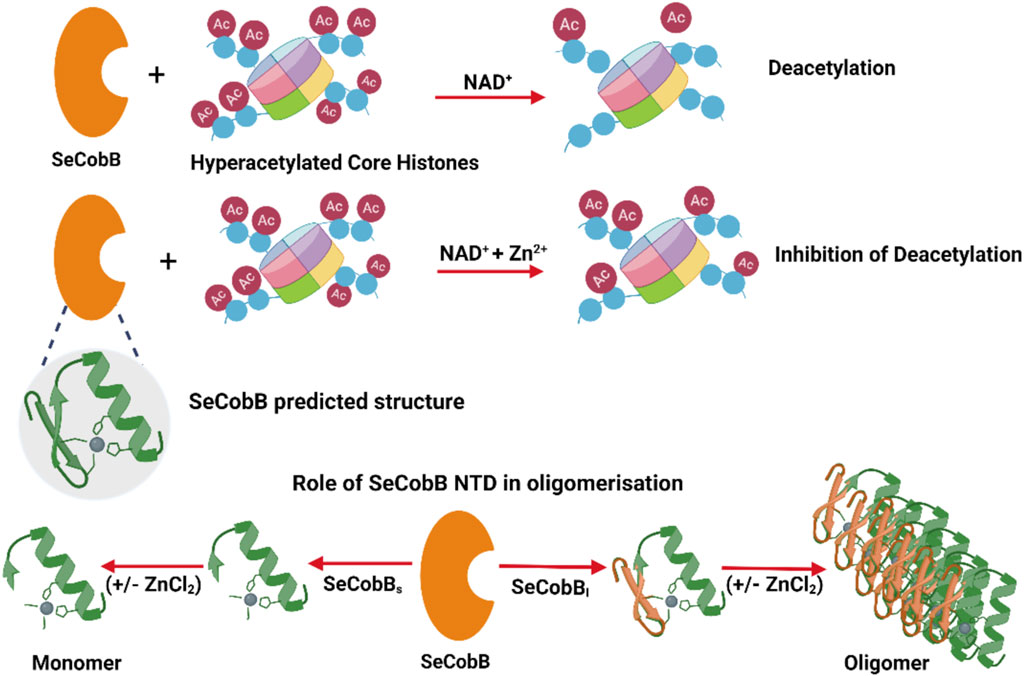
Prokaryotic deacetylases are classified into nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins and Zn2+-dependent deacetylases. NAD+ is a coenzyme for redox reactions, thus serving as an essential component for energy metabolism. The NAD+-dependent deacetylase domain is quite conserved and well characterized across bacterial species like CobB in Escherichia coli and Salmonella, Rv1151c in Mycobacterium, and SirtN in Bacillus subtilis. E. coli CobB is the only bacterial deacetylase with a known crystal structure (PDB ID: 1S5P), which has 91% sequence similarity with Salmonella CobB (SeCobB). Salmonella encodes two CobB isoforms, SeCobBS and SeCobBL, with a difference of 37 amino acids in its N-terminal domain (NTD). The hydrophobic nature of NTD leads to the stable oligomerization of SeCobBL. The homology modeling-based predicted structure of SeCobB showed the presence of a zinc-binding motif of unknown function. Tryptophan fluorescence quenching induced by ZnCl2 showed that Zn2+ has a weak interaction with SeCobBS but higher binding affinity toward SeCobBL, which clearly demonstrated the crucial role of NTD in Zn2+ binding. In the presence of Zn2+, both isoforms had significantly reduced thermal stability, and a greater effect was observed on SeCobBL. Dynamic light scattering (DLS) studies reflected a ninefold increase in the scattering intensity of SeCobBL upon ZnCl2 addition in contrast to an ∼onefold change in the case of SeCobBS, indicating that the Zn2+ interaction leads to the formation of large particles of SeCobBL. An in vitro lysine deacetylase assay showed that SeCobB deacetylated mammalian histones, which can be inhibited in the presence of 0.25–1.00 mM ZnCl2. Taken together, our data conclusively showed that Zn2+ strongly binds to SeCobBL through the NTD that drastically alters its stability, oligomeric status, and enzymatic activity in vitro.
1 Introduction
The sirtuin-dependent protein deacylation system in prokaryotes is known to have a significant influence on bacterial physiology by the regulation of gene expression (Lima et al., 2011), maintenance of energy homeostasis (Chan et al., 2011), and modulation of acetate–glucose metabolism by restoring the activity of acetyl-coenzyme A synthetase (Acs) (Castano-Cerezo et al., 2011; Castano-Cerezo et al., 2015). Acetylation can be either enzyme-mediated or non-enzymatic, whereas deacetylation is always an enzyme-mediated process (AbouElfetouh et al., 2015). Prokaryotic deacetylases can be classified into two groups: nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins and Zn2+-dependent deacetylases (Frye, 2000; Gregoretti et al., 2004). NAD+-dependent deacetylases are well studied in both Gram-negative and Gram-positive bacteria like CobB in E. coli (de Diego Puente et al., 2015), Salmonella (Tucker and Escalante-Semerena, 2010), Vibrio cholerae (Liimatta et al., 2018), and Yersinia pestis (Liu et al., 2018), Rv1151c in Mycobacterium (Liu et al., 2014), SirtA in Streptomyces (VanDrisse and Escalante-Semerena, 2018), and SirtN in B. subtilis (Gardner and Escalante-Semerena, 2009). Multiple sequence alignments of CobB homologs in selective gastrointestinal bacteria demonstrate 70%–80% sequence homology and a conserved NAD+-binding domain [(Mishra et al., 2022), Supplementary Figure S1]. They play diverse roles in various bacterial systems such as regulation of the TacT–TacA toxin–antitoxin system (VanDrisse et al., 2017) and PhoP–PhoQ two-component system in Salmonella (Ren et al., 2016), biofilm formation in Mycobacterium tuberculosis, resistance to a first-line drug (isoniazid) in M. smegmatis (Gu et al., 2015), chemotaxis in Y. pestis, and growth homeostasis in B. subtilis (Gardner and Escalante-Semerena, 2009). Salmonella and other members of the Enterobacteriaceae family encode two CobB isoforms, CobBs (236 aa) and CobBL (273 aa), with a difference of 37 amino acids in its N-terminal. Both the isoforms (SeCobBS and SeCobBL) are functional deacetylases, with SeCobBS being enzymatically more active (Tucker and Escalante-Semerena, 2010). E. coli CobB (EcCobB, PDB ID: 1S5P) is the only bacterial deacetylase whose crystal structure is solved so far, which displays 91% sequence similarity with Salmonella CobB (SeCobB). Our homology modeling and the AlphaFold structure database showed that like EcCobB, Salmonella CobB also contains a zinc-binding motif with unknown function.
Zinc acquisition and homeostasis contribute significantly toward bacterial physiology and pathogenesis (Nairz et al., 2010; Ammendola et al., 2016). This homeostasis is crucial not only for the expression of metallozymes (Hood and Skaar, 2012) and other proteins related to bacterial metabolism but also for the adequate expression of virulent factors to cause infection (Waldron and Robinson, 2009; Vickers, 2017). Among micronutrients like copper, zinc, manganese, and iron, maintaining the intracellular and extracellular levels of zinc is of utmost importance for the structural and catalytic regulation of various bacterial proteins involved in processes like DNA replication and oxidative stress response (Cerasi et al., 2013). This regulation is achieved by zinc efflux and influx transporters like P1B-type ATPase ZntA and ZnuABC in E. coli, respectively (Hazan et al., 2001; Wei and Fu, 2006; Romiguier and Roux, 2017). Zinc serves as a crucial bridge between bacterial metabolism and defense mechanisms against the host in Salmonella by ZnuABC, a zinc uptake transporter. Zn metal has a provident impact on the regulation of the protein structure and function due to its strong affinity toward amino acid residues, especially cysteine (Tainer et al., 1991; Giles et al., 2003). Zn metal–cysteine complexes have multifaceted functions like the inhibition of enzymatic activity in dimethylarginine dimethylaminohydrolase (DDAH-1) (Maret, 2013a), function as a redox switch in betaine–homocysteine methyltransferase (BHMT) (Maret, 2005), and act as a stabilizing bridge between protein complexes like in endothelial NOS isoform (NOS3) (Fischmann et al., 1999).
Here, we report that SeCobBL is an oligomeric protein, whereas SeCobBS is a monomer in solution, which clearly indicates that oligomerization is mediated through a 37-amino acid N-terminal domain (NTD). Zn2+ binds to both isoforms, SeCobBL and SeCobBS, albeit with different affinities. SeCobBS has weak binding affinity for Zn2+, which indicates a very weak interaction with the predicted Zn-binding motif. SeCobBL strongly binds to Zn2+, which is presumably mediated through the NTD. The SeCobB–Zn2+ interaction greatly enhances the kinetic and thermal stability of both the proteins in the solution. Dynamic light scattering (DLS) showed that ZnCl2 induces a ninefold increase in the SeCobBL scattering intensity in the solution compared to the ∼ onefold change in the case of SeCobBS, which indicated that Zn2+ induced the formation of larger particles of SeCobBL. Both SeCobBS and SeCobBL deacetylase activities are inhibited at a higher concentration of Zn2+. Taken together, our study is the first report to demonstrate the function of NTD of SeCobBL and effect of Zn2+ on the stability and activity of SeCobB. We also show that the predicted Zn-binding domain plays a limited role in Zn2+ binding in vitro, and we predict that similar effects will be observed in SeCobB homologs.
2 Materials and methods
2.1 Bacterial strains, plasmids, and culture conditions
The bacterial strains used in this study are E. coli DH5α and E. coli BL21 codon plus (DE3). These strains were grown in LB broth at 37°C, 150 rpm. Antibiotics were added as required for the culture at the indicated concentration—chloramphenicol (20 μg ml−1), tetracycline (20 μg ml−1), and kanamycin (50 μg ml−1). Both the larger and shorter isoforms of the SeCobB gene were cloned using genomic DNA of Salmonella enterica subspecies I serovar Enteritidis str. P125109. The pET 28a (+) plasmid was used for both cloning and recombinant protein expression. The detailed protocol for cloning is given in Supplementary Material.
2.2 Purification and characterization of SeCobB
Both shorter and full-length isoforms of the SeCobB protein (SeCobBS and SeCobBL, respectively) were purified using Ni2+–NTA affinity chromatography (Supplementary Figure S2). The oligomeric status of SeCobB isoforms was determined by size exclusion chromatography (SEC) using a Sephacryl S-200 16/60 GPC column attached to AKTA Pure (GE Healthcare). The column was pre-equilibrated with a protein elution buffer (25 mM Tris pH 8, 200 mM NaCl, 2 mM β-mercaptoethanol, and 5% glycerol) at a flow rate of 0.5 ml/min. It was calibrated using SEC standard protein markers (Gel Filtration Markers Kit, Sigma-Aldrich: MWGF200), and the void volume (Vo) was determined by passing blue dextran under the same conditions. Each of the eluted peaks were analyzed by SDS-PAGE to identify the presence of SeCobBS and SeCobBL. The apparent molecular weight of SeCobBS and SeCobBL was determined by interpolating the peak elution volume (Gupta et al., 2000) on the SEC standard plot. The SEC standard plot is a linear calibration curve produced by plotting the logarithms of the known molecular masses (log MW) of protein standards versus their respective Ve/Vo values, where Ve is the elution volume and Vo is the void volume.
The secondary structure of recombinant SeCobBS and SeCobBL was determined using circular dichroism (McDevitt et al., 2011) spectra using a Chirascan CD spectrometer (Applied Photophysics) at the Central Research Facility of Institute of Life Sciences, Bhubaneswar, India. A graph was plotted between wavelength (λ) and molar ellipticity (ϴ) after baseline correction. All the data are represented as an average of three scans, with the protein purified in three independent batches.
2.3 Solution-state structures of SeCobBS and SeCobBL obtained using DLS
To gain insights into how the solution-state structure of the protein changes in the presence of a divalent metal, DLS experiments were conducted using a multi-angle particle size analyzer from Photocore Ltd. (Russia). DLS was utilized to determine the size distribution of the protein in both the absence and presence of ZnCl2.
Ni–NTA-purified recombinant SeCobB in the protein elution buffer (25 mM Tris, pH 8, 200 mM NaCl, 2 mM β-mercaptoethanol, and 5% glycerol) was subjected to high-speed centrifugation at 12,000 rpm, 10 min at 4°C to avoid any possible air bubbles. The supernatant was used to prepare suitable dilutions using the protein elution buffer as the solvent. They were transferred to a clean and dry cylindrical glass vial of 10 mm diameter for carrying out DLS. The outer surface of the cuvette was gently wiped with lint-free tissue before placing into the instrument to remove dust or dirt to avoid unnecessary scattering from the glass wall.
In the DLS study, the protein suspension at a very dilute concentration is illuminated with a laser light of wavelength (λ) 654 nm, and the scattering intensity is collected at a scattering angle θ (=90o) using a photon detector. The intensity auto-correlation is calculated as (Lima et al., 2011)
In Eq. 2, I(t) is the correlation function at time t and
The correlation functions are analyzed using a CONTIN-based method, and the corresponding size distributions are obtained by fitting the correlation function with Eq. 2 (see Figure 1).
where Γ is the characteristic decay rate, which relates the translational free diffusion coefficient Do as
Here, Q is the scattering vector and is related to the scattering angle θ by
Here, kB is the Boltzmann constant, T is the absolute temperature, η is the viscosity of the solvent, and Rh is the hydrodynamic radius.
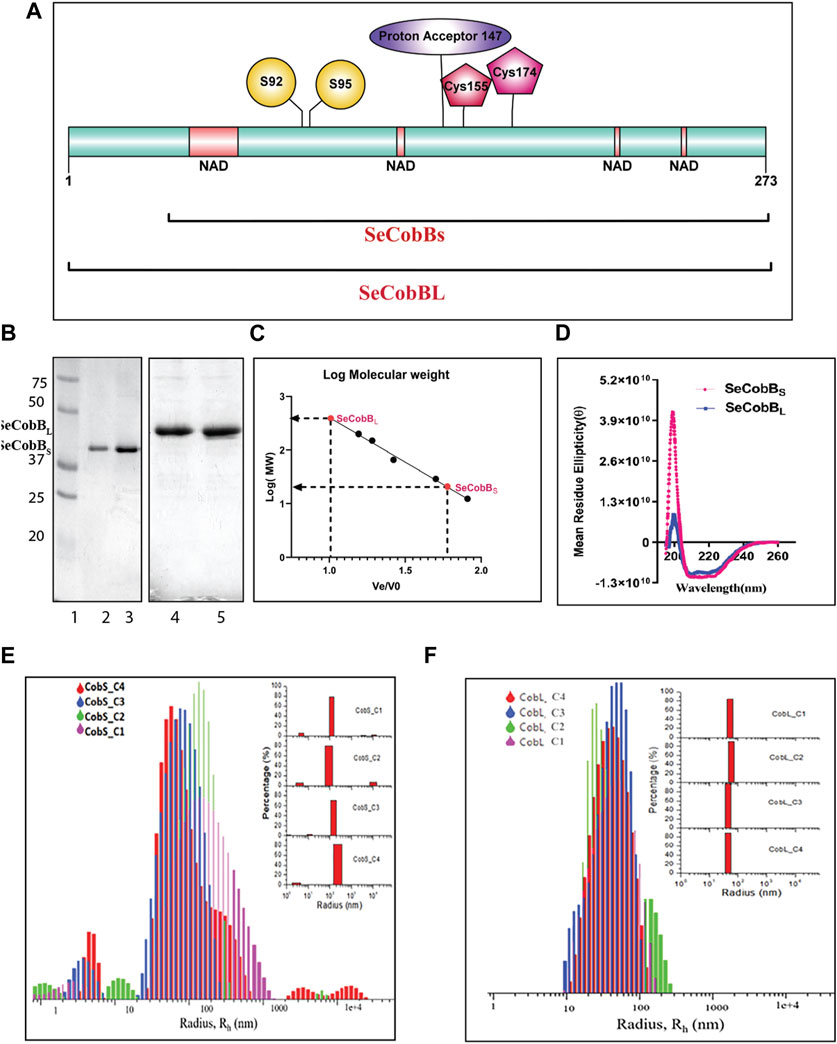
FIGURE 1. Characterization of the S. enterica deacetylase protein isoforms, SeCobBS and SeCobBL. (A) Domain organization of the S. enterica CobB protein. SeCobBS lacks the initial 37 amino acid residues present in SeCobBL. The predicted domains are represented in different colors: NAD+-binding domain in peach, Zn2+-binding sites in maroon (Cys 155 and Cys 174), substrate-binding site in yellow (S92 and S95), and active site in purple (147). The full-length protein is represented in cyan. (1–273). (B) SeCobB protein purification profile. Lane 1—Bio-Rad protein ladder; lanes 2 and 3—purified SeCobBS at 30 kDa; lanes 4 and 5—purified SeCobBL at 34 KDa. (C) Size exclusion chromatography profile of SeCobBS and SeCobBL using a HiPrep 16/60 Sephacryl S-200 column. The standard curve was generated by plotting Ve/Vo vs. log of molecular weight of the SEC standards (Ve—elution volume of each protein; Vo—void volume). The solution molecular weight of SeCobBS and SeCobBL was determined by interpolating the Ve/Vo value on the standard curve (dotted line). (D) Determination of secondary structures of SeCobBS and SeCobBL using circular dichroism spectroscopy. Molar residue ellipticity (θ) values plotted against the wavelength (λ) showed two peaks at 208 and 222 nm. Size distribution plots of hydrodynamic radius Rh (in nm) of recombinant SeCobBS (E) and SeCobBL (F) at different concentrations. The concentrations CobS_C1–CobS_C4 correspond to 93 μg/ml, 187 μg/ml, 375 μg/ml, and 750 μg/ml of the SeCobBS protein, respectively. The concentrations CobL_C1–CobL_C4 correspond to 100 μg/ml, 200 μg/ml, 500 μg/ml, and 800 μg/ml, respectively. The inset bar diagram represents the % size distribution for SeCobBS (E) and SeCobBL (F) against the radius (nm) of different particle sizes.
2.4 Prediction and optimization of the predicted 3D-modeled structure
The primary sequence of the NAD+-dependent deacylase protein of Salmonella Enteritidis PT4 (strain P125109) was retrieved from the UniProtKB database (accession ID: A0A6C7HR52); the protein had a length of 273 aa (SeCobBL) (Consortium, 2015). However, we deleted the first 37-amino acid (aa) sequence to check their binding affinities against Zn2+ in a comparative way, where the 236-aa sequence was referred to as the shorter isoform (SeCobBS) and the 273-aa sequence was termed the longer isoform (SeCobBL). The three-dimensional (3D) structure of both SeCobBL and SeCobBS was determined using online servers like I-TASSER (Roy et al., 2010). The best I-TASSER-generated structure was selected for refinement based on its C-score value. Qualitative analysis of the best modeled structure of both protein isoforms was done through different online servers.
Servers like PROCHECK (Roman et al., 1993), Verify3D (Lüthy et al., 1992), and ERRAT (Colovos and Yeates, 1993) were used for the quality factor analysis of the model protein from the SAVES meta-server. The ProSA webserver (Wiederstein and Sippl, 2007) was used for the analysis of the Z-score of the target protein. However, the best modeled structure was obtained from the I-TASSER server, with 2.1% residues for SeCobBL and 1.5% residues for SeCobBS in the outliers and 72% in the case of SeCobBL and 74% in the case of SeCobBS in the favorable region. These structures were then selected for refinement based on their respective C-score values in the GalaxyRefine webserver (Ko et al., 2012). This course of action was repeated until the quality of the structural conformation failed to increase any further.
2.5 In vitro histone deacetylase assay
The in vitro histone deacetylase assay was standardized in the laboratory (Mishra et al., 2022). In brief, HCT-116 cells were treated with sodium butyrate (NaBU) for 24 h to hyperacetylate all the proteins including histones in the cells. Mammalian hyperacetylated histones were enriched by TCA precipitation and used as a substrate for HDAC assays. Then, 50–200 ng of recombinant SeCobB was incubated with 10 μg hyperacetylated, acid-extracted core histones in the presence of NAD+ (5 mM) as a cofactor in the HDAC assay buffer (50 mM Tris-Cl, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, and 0.2 mM PMSF) for 60 min at 37°C.
We also performed an in vitro histone deacetylase assay in the presence of zinc (ZnCl2). A measure of 0.5 M ZnCl2 stock was prepared by dissolving 681.45 mg ZnCl2 (SRL-87288) in a minimum of 2 N HCl and then increasing the volume up to 10 ml with distilled water. A measure of 100 ng recombinant SeCobB was incubated with 10 μg hyperacetylated, acid-extracted core histones in the presence of NAD+ (5 mM), along with varying concentrations of ZnCl2 (0.25–2.5 mM) under the same conditions as mentioned above. The immunoblots were probed with anti-acetyl lysine and anti-H3 antibodies. The in vitro deacetylase assay for both SeCobBS and SeCobBL was performed separately.
2.6 Molecular docking studies
To check the binding mode interaction between Zn2+ and SeCobBS/SeCobBL, docking studies were performed using AutoDock v4.2.6 software (Olson and Olson, 2010). It was initialized with protein and ion preparations. The grid scale was positioned at 40 × 40 × 40 xyz points with a grid spacing of 0.375 Å, and the grid core was chosen at dimensions (x, y, and z) 54.535, 58.898, and 50.521, respectively. The docking analysis was carried out using a rigid protein and genetic algorithm with the following default parameters: the maximum number of generations = 2700, maximum number of seeds = 2,500,000 runs, population size of 150, and 100 GA runs. Following that, docking was done by setting the parameters to default values, followed by using the command-line interface for autogrid and autodock applications.
We performed a specific docking approach on both sites and a blind docking approach to check the binding affinity of Zn2+ toward SeCobBS using AutoDock Vina software and the online server Metal Ion-Binding site prediction and modeling server (MIB2), respectively. A webserver that was used to create the expected metal ion-bound 3D structure and prediction of metal ion-binding residues was defined. To create a binding template, areas that bind 12 different types of metal ion-binding residues were taken into consideration. The query protein and the template were compared structurally using the fragment transformation approach without any data training. The template contained residues that are within 3.5 Å of the metal ions by modifying the scoring algorithms based on structural and binding residue similarity. The prediction of binding residues for 18 different types of metal ions, namely, Ca2+, Cu2+, Fe3+, Mg2+, Mn2+, Zn2+, Cd2+, Fe2+, Ni2+, Hg2+, Co2+, Cu+, Au+, Ba2+, Pb2+, Pt2+, Sm3+, and Sr2+, is supported. The metal ion docking after prediction is also provided using MIB (Lu et al., 2012; Lin et al., 2016).
2.7 Fluorescence quenching studies to determine the SeCobB–Zn2+ interaction
SeCobBS and SeCobBL (5 µM in the protein elution buffer, pH 8.0) were titrated with ZnCl2 (0–100 μM, at a rate of 5 µM per addition). Tryptophan fluorescence intensity of this bivalent metal ion-bound SeCobBS and SeCobBL was recorded with an excitation wavelength of 295 nm using a spectrofluorometer (FLS1000, Edinburgh Instruments, United Kingdom). The temperature of the samples was maintained at 25°C by using Peltier attached to the spectrofluorometer. Stern–Volmer plots and Scatchard analysis were done using corrected fluorescence data considering the effect of dilution. The linear fit of the data was obtained using the Stern–Volmer equation,
and the Scatchard equation,
where F0 and F are the emission intensities of SeCobBS and SeCobBL in the absence and presence of the zinc ions, respectively, provided the Stern–Volmer quenching constant (KSV), the binding constant (Kb), and the number of binding sites (n). Here, [Q] stands for [zinc ions].
2.8 Effect of Zn2+ on temperature-induced unfolding of SeCobB
The thermal stability of SeCobBS and SeCobBL was determined using a thermal-induced denaturation experiment. In brief, both protein isoforms (5 µM in the protein elution buffer, pH 8.0) were incubated in the absence and presence of ZnCl2 (0–100 µM). Intrinsic tryptophan fluorescence spectra of all the samples were recorded in the 310–400-nm region using an excitation wavelength of 295 nm. The change in tryptophan fluorescence at 334 nm was recorded stepwise between 25°C and 90°C. As mentioned previously (Nandi et al., 2013), Vant Hoff enthalpy (ΔHVH) and entropy (ΔS) were calculated from the thermal melting data.
3 Results
3.1 Characterization of the S. enterica deacetylase protein SeCobB
The schematic domain organization of SeCobB, illustrating both shorter and larger isoforms, the predicted NAD+-binding domain, Zn2+-binding site, substrate-binding site, and active site, is shown in Figure 1A. To further characterize Salmonella CobB (SeCobB), we cloned it in E. coli using a pET-28a (+) vector, and the N-terminal 6×His-tagged protein was extracted and purified by Ni–NTA agarose metal affinity chromatography (Figure 1B, lanes 2–3 and 4–5). Size exclusion chromatography showed that SeCobBS exists as a monomer, whereas SeCobBL exists as an oligomer in solution (Figure 1C). Circular dichroism spectroscopy revealed that both isoforms have a similar secondary structure that predominantly harbors alpha helices, illustrated by peaks at 208 and 222 nm, respectively (Figure 1D).
We determined the oligomeric status of SeCobB isoforms by multi-angle dynamic light scattering. The concentration-dependent correlation function for SeCobBS (denoted as CobS_C1-CobS_C4) and SeCobBL (denoted as CobL_C1-CobL_C4) is shown in Supplementary Figure S3. The data are fitted with the distribution of decay rates G (Γ) using Eq. 2. The experimental function (Eq. 1) agreed well with the theoritical fit function, and the obtained distribution of the hydrodynamic radius (Eq. 5) was plotted as shown in Figures 1E, F for SeCobBS and SeCobBL, respectively. It is worth noting that for all concentrations ranging from 93 to 750 μg/ml of SeCobBS, the size distribution curve has multiple peaks that vary from 5 nm to several microns, which clearly demonstrates the heterogeneous nature of protein particles with a large variation in their sizes coexisting in the solution state (Figure 1E). SeCobBL gives a single distribution peak that overlaps with each other for the entire range of concentrations (Figure 1F), demonstrating a homogeneous particle size distribution within the protein solution.
The bar diagram representation corresponding to the hydrodynamic radius plot is demonstrated as a function of the percentage (%) of size distribution for SeCobBS and SeCobBL in the inset of Figures 1E, F, respectively. The major contribution (∼75%) in CobS_C1 to CobS_C4 comes from the main peak that corresponds to a mean radius of 147 nm ± 60 nm, whereas the radius of the small peak varies from 2 to 11 nm with a population of ˂ 5% of total proteins in the solution coexisting with a small % of micrometer-sized aggregates. These proteins, which are monomers with a radius range of 2–11 nm, can be aligned to their molecular weight estimated in GPC. In addition, we also observe large aggregates of the protein at a high concentration (CobS_C3 and CoBS_C4) amounting to a small population (<5%). SeCobBL, which is an oligomer in solution, displays a single peak corresponding to a mean radius of 49.99 nm ± 15 nm that coincides with the molecular weight estimated in GPC.
3.2 Salmonella CobB is a NAD+-dependent functional protein lysine deacetylase
A deacetylase protein of Archaeoglobus fulgidus, Sir2-Af2 deacetylating C-terminal of p53 peptide, and yeast Hst2 deacetylating acetylated histone H4 peptide have been reported (Tanny et al., 2004). Thus, we investigated whether SeCobB can deacetylate core histones. To establish the same, we performed in vitro histone deacetylase assay on acid-extracted, hyperacetylated core histones with SeCobBs and SeCobBL separately in the presence and absence of NAD+, followed by Western blotting with anti-acetyl lysine and anti-H3 antibodies. The acetyl lysine antibody detected the concentration-dependent deacetylation of core histones by both isoforms in the presence of NAD+ (Figures 2A, B, lanes 2–4), whereas no deacetylation occurred in the absence of NAD+ (lane 5, top panel, Figures 2A, B). A gradual decrease in the band intensity was representative of concentration-dependent deacetylation. An equal amount of substrate was used in each reaction, as indicated by the uniform band intensity of H3 using the anti-H3 antibody (Figures 2A, B, bottom panel).
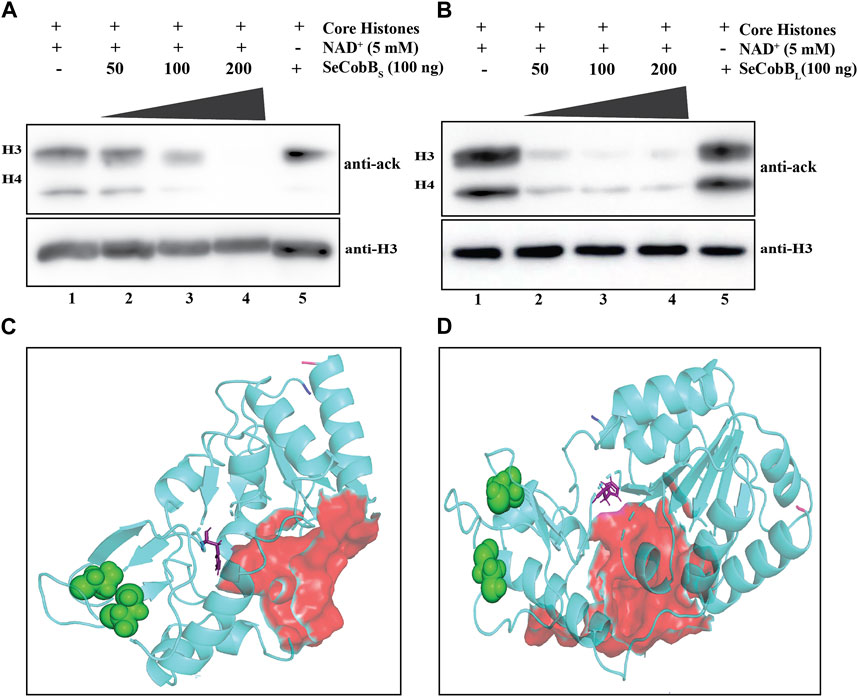
FIGURE 2. (A, B) In vitro deacetylase assay to show that SeCobB is a NAD+-dependent deacetylase. In vitro histone lysine deacetylase assay with SeCobBS (A) and SeCobBL (B) using hyperacetylated mammalian core histones as the substrate. The upper panel shows Western blotting with the anti-acetyl lysine antibody. Lane 1—acid-extracted core histones (substrate); lanes 2–4—core histones incubated with purified SeCobBS (A) and SeCobBL (B) at different concentrations (50, 100, and 200 ng), along with NAD+ as the cofactor; lane 5—core histones incubated with 200 ng purified enzyme without NAD+. Immunoblots were re-probed with the anti-H3 antibody to confirm equal sample loading (bottom panel). The assay was performed in three biological replicates. (C, D) Homology modeling to illustrate the predicted structure of SeCobB. Cartoon representation of the modeled structure of SeCobBS (C) and SeCobBL (D) using the I-TASSER server. The N-terminal and C-terminal ends are shown in blue and hot pink, respectively. The NAD+-binding sites are shown as surface-shaped (red), zinc-binding sites (sphere-shaped in green), and the active sites are shown as stick-shaped in purple.
3.3 Three-dimensional structure prediction of SeCobB
No structural information is available for NAD+-dependent protein deacylase (CobB) of Salmonella Enteritidis PT4 (strain P125109). Due to a lack of validated structural information about the target protein, Zn2+-binding site and NAD+-binding site residues were predicted using UniProt (Figure 1A) and I-TASSER servers. The I-TASSER-modeled structure of SeCobBL was compared with the AlphaFold database-predicted structure to check their structural similarity. The structural alignment showed that the RMSD between these two structures was 1.549 Å (Supplementary Figure S5), which is less than the default cut-off of RMSD of 2 Å, and that the extent of dissimilarity between two structures were not greater, as per their structural comparison. Likewise, the predicted NAD+-binding sites and zinc ion-binding sites are shown in Figure 1. The protein 3D structure generated through the I-TASSER server represents the good quality after validation through Ramachandran plot analysis, Verify3D, ERRAT, and the ProSA server (Tables 3, 4). The Ramachandran plot analysis of the SeCobBS model structure before refinement revealed that 74.0% of residues were in the allowed region and 1.5% in the disallowed region. However, after protein structure refinement using the GalaxyRefine webserver, the number of amino acid residues in the allowed region increased to 92%, and only 1% of residues were in the disallowed area. Similarly, the number of residues in the SeCobBL modeled structure was shifted from 72.0% to 89.8% in the allowed region, whereas it was shifted from 2.1% to 1.7% in the disallowed region. The validation of our selected model through the Verify3D program (checks for the compatibility of a three-dimensional atomic model with its own one-dimensional amino acid sequence) revealed that there were 74.15 residues for SeCobBS and 72.53 residues for SeCobBL with an average 3D–1D score >0.2 (Supplementary Figures S4E, F). The server “ERRAT” provided the overall quality factor (expressed as the percentage of the protein for which the calculated error values fall within the 95% rejection limit) of the model as 93.1818 for SeCobBS and 94.024 for SeCobBL (Supplementary Figures S4C, D). We studied the quality of the target protein using the ProSA webserver, and it reported a Z-score value of −7.05 (SeCobBS) and −7.92 (SeCobBL), which is well within the range of the native conformation of the crystal structure. The ProSA analysis of the model structure revealed an improvement in the Z-score after structural refinement.
3.4 Zn2+ has an inhibitory effect on the NAD+-dependent in vitro deacetylase activity of SeCobB
The predicted 3D model structure of Salmonella CobB (Figure 3C) shows that it contains a zinc-binding motif. Most of the Sirt2 family proteins contain a Cys-X-X-Cys-(X)15–20-Cys-X-X-Cys sequence as a characteristic feature of the Zn2+-binding motif within their conserved domain. The predicted zinc-binding sites in SeCobB according to “UniProt” software are cysteine at positions 155 and 174 (Figure 1A). To explore the effect of the Zn2+–cysteine interaction on the activity of Salmonella CobB, we performed an in vitro deacetylase assay at different concentrations of ZnCl2 with SeCobBS and SeCobBL separately (Figures 3A, B). The anti-acetyl lysine antibody detected a gradual decrease in the band intensity with decreasing ZnCl2 concentration, representative of the increase in the deacetylase activity (Figures 3A, B, lanes 2–6). The band intensity was the lowest in the absence of ZnCl2, indicative of maximum deacetylation by restoring the deacetylase activity (Figures 3A, B, lane 7). An equal loading of the substrate was confirmed by re-probing the blots with the anti-H3 antibody (Figures 3A, B, bottom panel).
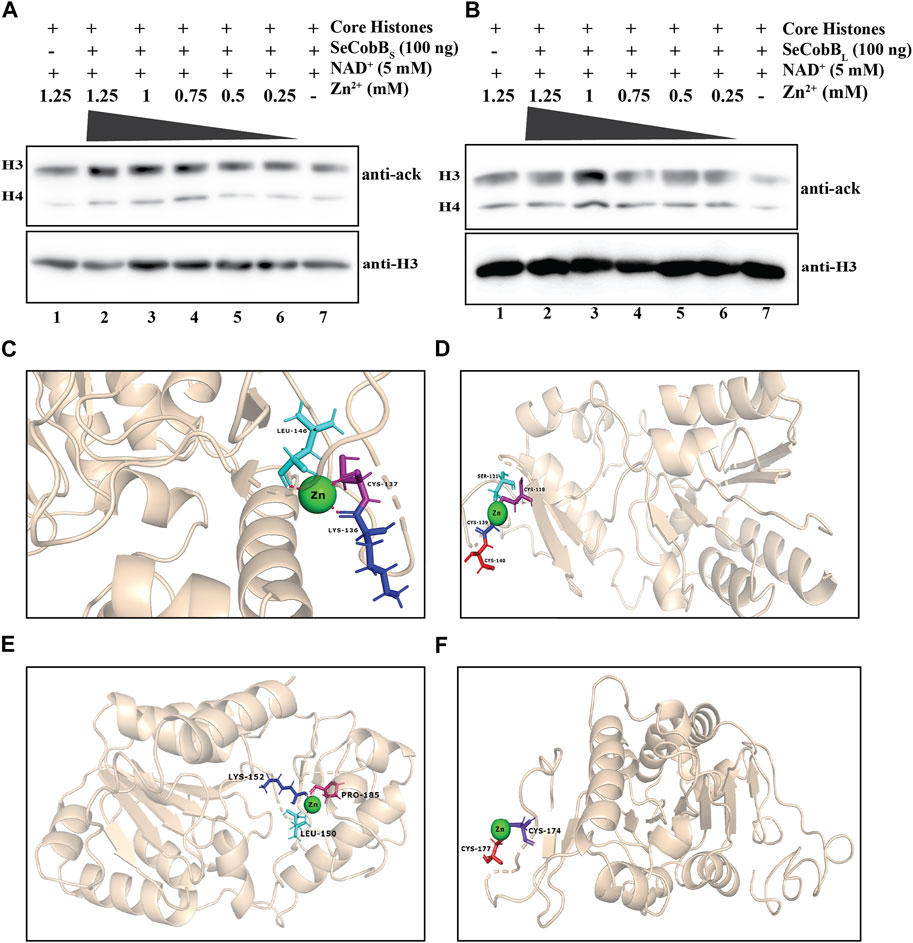
FIGURE 3. (A, B) Effect of Zn2+ on SeCobB activity. In vitro histone lysine deacetylase assay with 100 ng of purified SeCobBS (A), SeCobBL (B), and ZnCl2 using hyperacetylated HCT-116 core histones as the substrate. Immunoblots were probed with pan-acetyl lysine (upper panel) and anti-H3 (bottom panel) antibodies. Lane 1 (A, B)—acid-extracted core histones (substrate); lanes 2–7 (A, B)—core histones incubated with ZnCl2 at different concentrations (1.25, 1, 0.75, 0.5, and 0.25 mM), along with NAD+(5 mM) as the cofactor. Immunoblots were re-probed with the anti-H3 antibody to confirm equal loading of the sample. The assay was performed with three independent batches of purified protein. (C, D) Molecular docking study to predict the amino acid residues involved in the Zn2+–SeCobBS interaction. Cartoon representation of the docked complex of the SeCobBS modeled structure against Zn2+ through a site-specific approach using AutoDock Vina (C) and (D) blind docking approach using the MIB2 webserver. (E, F) Molecular docking study to predict the amino acid residues involved in the Zn2+–SeCobBL interaction. Cartoon representation of the docked complex of the SeCobBL modeled structure against Zn2+ through a site-specific approach using AutoDock Vina (E) and (F) blind docking approach using the MIB2 webserver. The target protein is shown in wheat, whereas Zn2+ (sphere-shaped) (green) and the H-bond residues are stick-shaped.
Since a predicted zinc-binding motif exists within SeCobB, whose function is unknown, and an in vitro HDAC assay illustrated that zinc mediated inhibition in the deacetylase activity, we performed a docking analysis of Zn2+ ions with the predicted 3D model structure of SeCobBS (Figures 3C, D). The predicted Zn2+-binding sites in the case of SeCobBL are Cys-155 and Cys-174, while it is Cys-118 and Cys-137 in the case of SeCobBS. The site-specific docking study of SeCobBL–Zn2+ revealed that Zn2+ showed three H-bond interactions (Leu-150, Lys-152, and Pro-185) (Figure 3E), none of which is present within the predicted Zn2+-binding site. In the case of SeCobBS, the H-bond interactions are Lys-136, Cys-137, and Leu-146 (Figure 3C), among which only CYS-137 is present within the predicted Zn2+-binding sites. These H-bond interactions showed a binding affinity of −1.2 kcal/mol toward Zn2+ for both isoforms. The blind docking study revealed that Zn2+ showed two H-bond interactions (Cys-174 and Cys-177) for SeCobBL and four H-bond interactions (Cys-118, Ser-121, Cys-139, and Cys-140) for SeCobBS (Figures 3D, F), of which Cys-174 for SeCobBL and Cys-118 in the case of SeCobBS are the only residues present within the predicted Zn2+-binding sites. The hydrogen bond interactions showed a binding score of 3.222 for the longer isoform and 3.272 for the shorter isoform with zinc-binding sites.
3.5 Solution structure of SeCobB in the presence of ZnCl2
Divalent cations are known to induce protein aggregation in solution, leading to a change in particle size, which can be studied by DLS and other techniques. This, coupled with fluorescence spectroscopy and other techniques, helps further elucidate the complex interplay of factors that influence a protein’s size and aggregation behavior in the presence of metal ions and salt. In the DLS experiment, the protein solution was treated with 250 μM ZnCl2, and we monitored changes in the size and evolution of size aggregates over 600 s (Figure 4). Upon the introduction of the salt, both SeCobBS and SeCobBL exhibited two types of size distributions in the solution state (Eq. 5), designated as A and B in Figure 4. For both SeCobBS and SeCobBL, within distribution 1, the hydrodynamic radius remained relatively stable at approximately 30 nm throughout the experiment. In contrast, within distribution 2, the hydrodynamic radius showed a remarkable increase from 90 to 300 nm over the observed time frame for SeCobBL. Conversely, for SeCobBS, within distribution 2, the variation in the hydrodynamic radius was much narrower, spanning from 35 to 50 nm. This substantial change in the hydrodynamic radius for SeCobBL within distribution 2 suggests the progressive formation of larger protein complexes. This phenomenon is likely attributed to the strong interaction between the protein molecules and ZnCl2. Importantly, this observation strongly supports the findings of the experimental studies in fluorescence spectroscopy, which indicated the presence of more available zinc-binding sites within SeCobBL than within SeCobBS. The average scattering intensity is known to be proportional to the size distribution of proteins in the solution. Therefore, we investigated its time-dependent variation immediately after the induction with ZnCl2. Figure 4C shows the average intensity as a function of time, denoted as It (normalized with the intensity at time zero), for SeCobBS and SeCobBL following ZnCl2 induction. It is evident from the plot that the average value of It/Io for SeCobBS is lower than that for SeCobBL and remains relatively constant throughout the observed time period. In contrast, for SeCobBL, the average intensity experiences a rapid increase immediately after induction, peaking at 50 s, after which the rate of increase in intensity slows down with time. The higher average scattering intensity and the rapid initial increase observed in SeCobBL, compared to SeCobBS, are consistent with the presence of a larger size variation within SeCobBL.
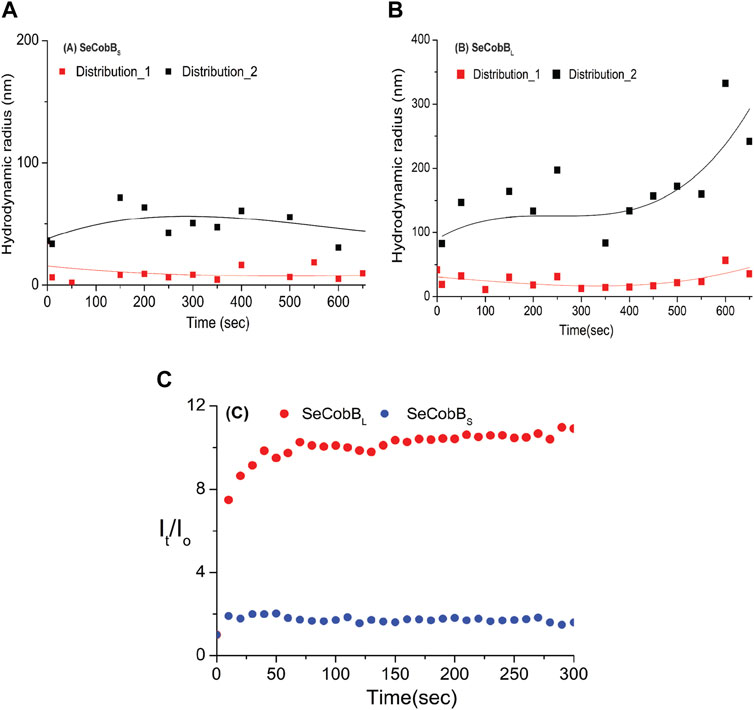
FIGURE 4. DLS studies (at T = 20°C) were conducted to investigate the kinetics of the solution-state structure of SeCobBS compared to SeCobBL after induction with 250 μM ZnCl2 solution. Two distinct types of size distributions, based on hydrodynamic radius, were observed for SeCobBS (A) and SeCobBL (B). The time-dependent evolution of the hydrodynamic radius is plotted AutoDock Vina (A,B). The line drawn on the data points in (A) and (B) is guided to the eye. Additionally, the corresponding average intensity, denoted as It and normalized to the intensity at t = 0, is plotted as a function of time (C). The experiment was performed in three individual batches.
3.6 SeCobB–Zn2+ interaction studies by fluorescence spectroscopy
The proteins containing an intrinsic fluorophore tryptophan can be used to monitor the binding of metal ions (Mattocks et al., 2021). The binding of Zn2+ ion with SeCobBS and SeCobBL was monitored by the quenching of tryptophan fluorescence upon the addition of zinc ions. A measure of 5 µM of SeCobBS and SeCobBL was titrated with zinc ions from 0 to 100 µM. Quenching of fluorescence intensity was observed upon the addition of Zn2+, which is suggestive of the interaction between Zn2+ and both protein isoforms (Figures 5A, 6A). It is observed from Figures 5A, 6A that the quenching in SeCobBS is ∼6%, while that in SeCobBL is ∼14%. This shows that zinc interacts strongly with SeCobBL compared to SeCobBS.
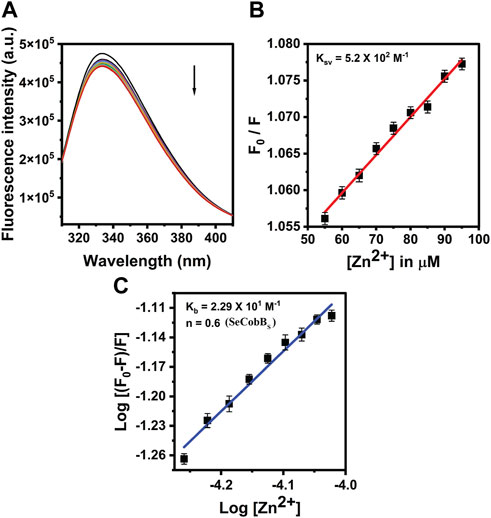
FIGURE 5. Fluorescence quenching studies of SeCobBS. (A) Intrinsic tryptophan fluorescence spectra of SeCobBS (5 µM) in the presence of zinc ions (0–100 μM, at the rate of change of 5 µM per addition). Tryptophan fluorescence spectra were recorded in the range of 310–400 nm at 25°C. The excitation wavelength was 295 nm. The arrow indicates the effect of increasing concentration of Zn2+ on the tryptophan fluorescence emission of SeCobBS. (B) The linear fit of F0/F vs. [Zn2+] and Stern–Volmer quenching constant (KSV) was calculated using Eq. 3. (C) The plot represents the linear fit of log [(F0−F)/F] vs. log [Zn2+] for zinc ions, and the binding constant (Kb) was estimated using Eq. 4. The experiment was performed in three biological replicates.
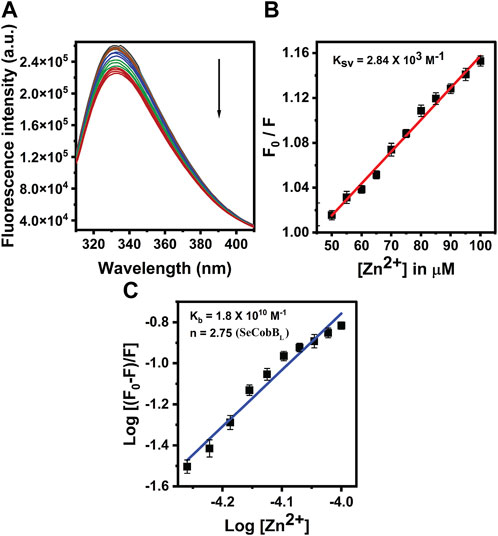
FIGURE 6. Fluorescence quenching studies of Zn2+ with SeCobBL. (A) Intrinsic tryptophan fluorescence spectra of SeCobBL (5 µM) in the presence of zinc ions (0–100 μM, at the rate of change of 5 µM per addition). Tryptophan fluorescence spectra were recorded in the range of 310–400 nm at 25°C. The excitation wavelength was 295 nm. The arrow indicates the effect of increasing concentration of Zn2+ on the tryptophan fluorescence emission of SeCobBL. (B) The linear fit of F0/F vs. [Zn2+] and Stern–Volmer quenching constant (KSV) was calculated using Eq. 3. (C) The plot represents the linear fit of log [(F0−F)/F] vs. log [Zn2+] for zinc ions, and the binding constant (Kb) was estimated using Eq. 4. The experiment was performed in three biological replicates.
The intrinsic fluorescence intensity of both protein isoforms decreased gradually upon increasing the concentration of zinc ions, but the saturation due to zinc quenching was observed earlier in SeCobBS than that in SeCobBL (Figures 5A, 6A). Stern–Volmer quenching (Eq. 6) and Scatchard analysis (Eq. 7) revealed that the binding affinity of Zn2+ with SeCobBL is more than that with SeCobBS (Table 1). The Stern–Volmer quenching constant for Zn2+ with SeCobBS and SeCobBL is 5.20 × 102 M−1 and 2.84 × 103 M−1, respectively (Figure 5B, 6B and Table 1). The extent of fluorescence quenching signifies the association of zinc ions with both protein isoforms. The state of equilibrium between free and bound proteins upon binding with small molecules is defined by the Scatchard equation.
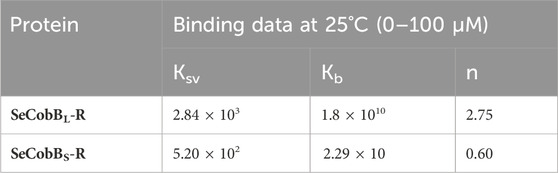
TABLE 1. Stern–Volmer (Ksv) and binding constant (Kb) values of quenching using intrinsic tryptophan fluorescence between Zn2+ and SeCobB.
The association binding constant (Kb) for SeCobBS and SeCobBL with zinc is 2.29 × 10 M−1 and 1.8 × 1010 M−1, with the binding stoichiometry/site (n) 0.60 and 2.75, respectively (Figures 5C, 6C and Table 1). This shows that the tendency of Zn2+ to bind with SeCobBL is much higher than that with SeCobBS, which is also reflected in the number of binding sites. The in vitro deacetylase assay demonstrates that there is maximum inhibition in the deacetylase activity of SeCobBS and SeCobBL at 0.75 and 1 mM ZnCl2, respectively. Beyond this concentration, saturation is attained in the band intensity. According to the relative intensity graph, 1.98% and 2.79% residual deacetylase activity of SeCobBS and SeCobBL was observed in the presence of 0.75 mM ZnCl2 compared to the activity in the absence of ZnCl2 (Figures 3A, B and Supplementary Figure S4). This result also corroborates with the fact that the number of zinc-binding sites in SeCobBS is less than that in SeCobBL.
The structural stability and integrity of a protein are essential to exhibit its function. Therefore, to assess the stability of SeCobBS and SeCobBL, thermal denaturation experiments were conducted. The thermal denaturation of SeCobBS and SeCobBL carried out in the absence of zinc ions showed a bi-dose response model fitting, which revealed two thermal melting (Tm) values for both protein isoforms (Figure 7). The Tm values for SeCobBL are 31.20°C and 71.32°C, and those for SeCobBS are 28.67°C and 67.94°C (Table 2) in the absence of zinc ions (Figure 7) (Table 2 and Figures 7A, B).
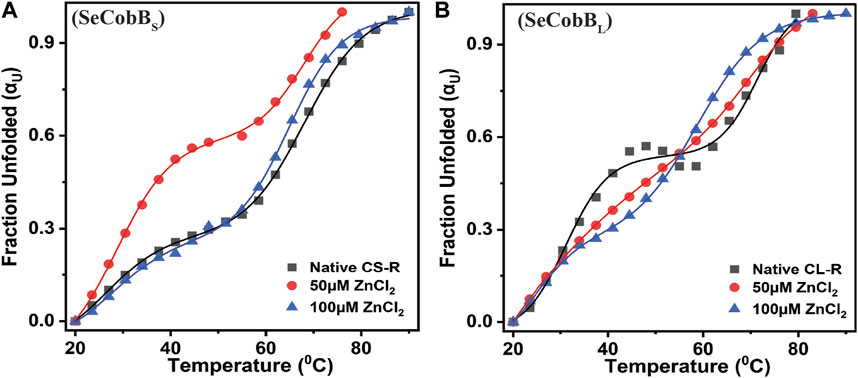
FIGURE 7. Effect of Zn2+ on the thermal stability of SeCobBS (A) and SeCobBL (B). Thermal unfolding profiles for 5 µM SeCobBS and SeCobBL in the absence or presence of zinc ions (0–100 µM) in protein elution buffer (pH 8.0). Temperature-induced changes in the fraction of the unfolded state (αU) for SeCobBS and SeCobBL proteins. The profile has been normalized to a scale of 0–1. Symbols represent the experimental data points, and the solid lines represent the best fit according to the bi-dose curve fitting. The experiment was performed in three biological replicates.
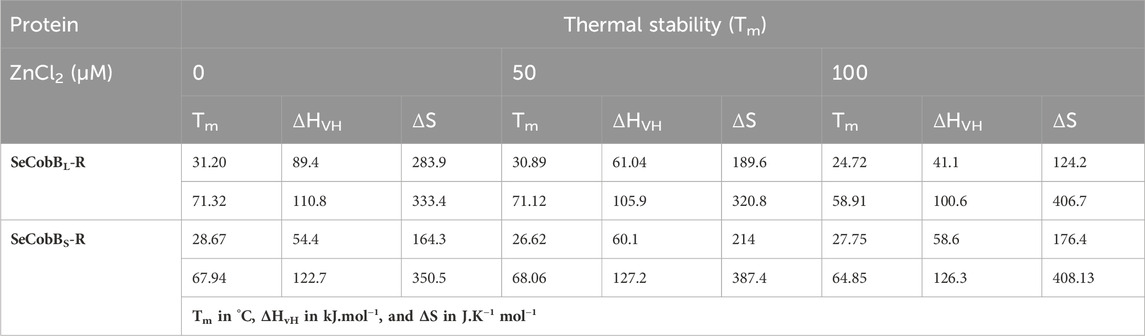
TABLE 2. Mid-point transition or Tm, van’t Hoff enthalpy (ΔHvH), and entropy (ΔS) values associated with the thermal denaturation of SeCobBL and SeCobBS at different zinc ion concentrations.
A similar bi-dose response fitting was also reported by Anand et al. (2019) for the Eis protein from M. tuberculosis and M. smegmatis. This shows that both protein isoforms have a sequential thermal denaturation mechanism. The thermal denaturation data clearly show that SeCobBL is thermally more stable than SeCobBS at ∼ 3°C–5°C. In the presence of Zn2+, the thermal stability of both SeCobBS and SeCobBL decreases but more significantly for SeCobBL. The addition of 100 µM Zn2+ reduces the Tm1 value of SeCobBS from 28.67°C to 27.75°C (ΔTm1 = 0.92°C) and the Tm2 value of SeCobBS from 67.94°C to 64.85°C (ΔTm2 = 3.09°C). In SeCobBL, an increase in the zinc ion concentration decreases the Tm1 value from 31.20°C to 24.72°C (ΔTm1 = 6.48°C) and the Tm2 value from 71.32°C to 58.91°C (ΔTm2 = 12.41°C), respectively. The ΔTm1 (0.92°C and 6.48°C for SeCobBS and SeCobBL, respectively) and ΔTm2 values (3.09°C and 12.41°C for SeCobBS and SeCobBL, respectively) in the presence of 100 µM ZnCl2 revealed that the interaction or binding of zinc ions decreased the thermal stability of SeCobBL to a greater extent than that of SeCobBS.
The higher decrease in the thermal stability of the former in the presence of zinc ions may be due to the greater binding of the zinc ions with SeCobBL than with SeCobBS. The calculation of the van’t Hoff enthalpy of the thermal denaturation curves revealed that the van’t Hoff enthalpy for SeCobBL for the first thermal melting transition decreased significantly from 89.4 to 41.1 kJ mol−1 (Table 2), while for the second thermal melting transition, it decreased from 110.8 to 100.6 kJ mol−1 (Table 2 and Figure 7) upon the addition of zinc ions. The greater decrease in the van’t Hoff enthalpy for the first denaturation transition may be due to the perturbation in the oligomeric assembly of SeCobBL upon the addition of zinc ions. Since SeCobBS is monomeric in nature, the change in van’t Hoff enthalpy upon the addition of zinc ions is not very significant.
4 Discussion
4.1 SeCobB is an NAD+-dependent lysine histone deacetylase
Prokaryotic NAD+-dependent deacetylases, predominantly known as CobB, the mammalian SIRT5 isoforms, are fairly conserved in Gram-negative bacteria and have been extensively investigated in S. enterica and E. coli (Mishra et al., 2022). Gram-negative species like Vibrio, Mycobacterium, Shigella, and Klebsiella are of clinical importance due to their ability to cause infection in the host. The sirtuin core domain in SeCobB is evolutionarily more conserved in archaea than in Eukarya (Buck et al., 2004). Numerous biological processes including cellular metabolism, transcriptional repression, and epigenetic alterations are influenced by the sirtuin-mediated regulation of acetylation. The deacetylase protein, frequently mentioned as CobB in the majority of the bacterial operations, has profound relevance in Salmonella physiology, like survival under stress conditions and regulation of virulence (Li et al., 2010; Liu et al., 2018). Escalante-Semerena (2010) reported predominant deacetylase activity of SeCobBS over SeCobBL in removing the lysine residue (K609) from acetylated acetyl CoA synthase (AcsAc). Our data are the first report to biochemically prove SeCobB to be a NAD+-dependent histone lysine deacetylase (Figure 2), justifying them to be a class III deacetylase. We found the complete absence of acetylated H3 and H4 bands at 200 ng of SeCobBS (Figure 2A), which are still visible in the case of SeCobBL (Figure 2B). The higher deacetylase activity of SeCobBS than that of SeCobBL is consistent with an earlier report. Only two such previous reports demonstrated eukaryotic histones as a substrate for a bacterial deacetylase (Zhao et al., 2004; Mishra et al., 2022). In vitro deacetylation of core histones by SeCobB clearly indicates the ability of a bacterial deacetylase to alter host proteins during an infection.
4.2 Structure prediction and molecular docking studies
Zinc metalloenzymes are therapeutic targets in cancer, cardiac disease, bacterial infection, and Alzheimer’s disease. The majority of these enzymes are targeted by a potential drug candidate by investigating the interaction with the Zn2+ bound to them. As a result, the precise prediction of the protein–Zn2+ interaction is a key part of computational docking and virtual screening against Zn2+-binding proteins.
The Ramachandran plot was used to define the best modeled structure of SeCobBS based on the C-score, and software applications like PROCHECK, ERRAT, and Verify3D were used for further refinement (Supplementary Figure S4). The polished ERRAT server output of the SeCobB structure is shown as a function of error values versus amino acids in Supplementary Figure S4. With a resolution of 2–3 Å, the protein structure received a score >90%. The red and yellow sections of the ERRAT graph reflect uncertain parts of the structure, while the white portions show the definite parts. This plot analysis identifies residues with error values >95% and 99% in very less time (Tables 3, 4).
Predictability of an anticipated model is determined using the ProSA webserver. It reveals the accuracy of the modeled protein with respect to the experimentally crystallized structure. It also generates a Z-score that represents the overall model quality. Its value is reflected in a plot of all experimentally determined protein chains in the current PDB. Different colors differentiate groupings of structures from different sources (X-ray and NMR) in this figure.
The software application used here to determine the amino acid residues involved in the SeCobB–Zn2+ interaction follows different algorithms and force fields issued for docking purposes. The residues that most often bind to metal ions are CYS, HIS, GLU, and ASP (Auld, 2001; Golovin et al., 2005) because the atoms of their charged side chains can coordinate with metal ions. We performed site-specific docking using AutoDock, in which Cys 118 and Cys 137 were defined in a grid box to facilitate the specific interaction of Zn2+ toward these two predicted sites for SeCobBS. The same exercise was also performed for SeCobBL. Cys 118 and Cys 137 are annotated as Cys 155 and Cys 174 in the larger isoform. However, no such specific Zn2+-binding sites were defined in the case of blind docking; instead, the designed server had specified the grid box based on their template structure. Since the parameters used for both the techniques are different, Zn2+ might have more binding affinity toward one cysteine than the other. Further experiments are required to ascertain the binding affinity of Zn2+ toward either of the cysteine residues.
4.3 Zn2+ inhibits the in vitro deacetylase activity of SeCobB
NAD+-dependent protein deacetylase activity is critical to the physiological function of proteins belonging to the SIR2 family (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000). Zinc has two major functions; either it can serve as a cofactor of enzyme catalysis, or it can act as an inhibitor of enzyme function. Zinc-mediated inhibition of enzyme activity can occur through three different mechanisms, namely, direct binding to the enzyme active site, allosteric mode of inhibition, and inhibition induced by binding to Zn2+ consecutive to catalytic zinc. The binding affinity of Zn2+ toward a protein, which is necessary to establish the physiological significance of zinc-mediated inhibition, varies from micromolar to picomolar concentration. Zinc-mediated inhibition of the enzyme phosphoglucomutase has been reported, which uses magnesium as its cofactor. The concentration of Zn2+ required for its inhibition is > 32 pmol/L (Magneson et al., 1987). The Zn2+ concentration beyond 1 mM is cytotoxic to the cells (McDevitt et al., 2011), hence determining the pathophysiological concentration. We performed in vitro histone deacetylase assay using 100 ng of purified SeCobBS and SeCobBL separately with various concentrations of ZnCl2 (0.25–1.25 mM). With the increasing concentration of ZnCl2, there was a sequential inhibition in the catalytic activity of both isoforms, which was restored in the absence of ZnCl2 (Figures 3A, B). The inhibition and restoration of deacetylase activity were ascertained by the enhanced and diminished band intensity captured from chemiluminescence data. This manifested the role of Zn2+ ions in inhibiting the catalytic activity of both isoforms. However, the extent of inhibition in the biochemical activity was different for both. The maximum inhibition of SeCobBS activity was achieved with 1.25 mM ZnCl2, whereas for SeCobBL, it was at 1 mM (Supplementary Figure S6).
4.4 Fluorescence spectroscopy studies
Interesting insights into the importance of Zn2+ in the field of nutrition, enzyme catalysis, protein biochemistry, and cellular biology have been obtained with the development of analytical techniques, like fluorescence spectroscopy, with a high sensitive level of detection (Maret et al., 2001; Maret, 2013b). This has shed light on the zinc-mediated alteration in the structure–function regulation of enzymes involved in various cellular processes. Since SeCobB contains three tryptophan residues, we used fluorescence spectroscopy to record any changes in the thermal and structural stability of the protein due to the Zn2+–SeCobB interaction. A quenching study was done to define the binding affinity of Zn2+ with both SeCobBS and SeCobBL. Data obtained from fluorescence quenching studies displayed a reasonable interaction between Zn2+ and SeCobB. The Ksv value for SeCobBS is 10 times lower than that of SeCobBL (Table 1), suggesting the requirement of a comparatively higher ZnCl2 concentration for interaction equivalent to SeCobBL. The binding constant (Kb) for SeCobBS is 109 times less than that for SeCobBL, also supported by the number of Zn2+-binding sites (Table 1). The binding affinity of Zn2+ toward SeCobBL is higher than that toward SeCobBS. SeCobBL is an oligomer in solution, whereas SeCobBS is a monomer (Figure 1C). Thus, being an oligomer, SeCobBL confers a greater number of available zinc-binding sites, resulting in adequate binding and relevant interactions.
The thermal denaturation graph of both isoforms illustrates the biphasic melting curve both in the absence and presence of Zn2+ (Figure 7). This may be because of the independent melting of the NAD+-binding Rossmann fold domain and Zn2+-binding domain, which are the characteristic domains of the SeCobB predicted structure (Gao et al., 2020).
The initial 37-amino acid stretch in the N-terminus of SeCobBL is 37% hydrophobic, which is a potential reason for its oligomerization. Moreover, DLS results demonstrate the narrow size distribution in SeCobBL (Figure 1F) that facilitates more Zn2+-binding sites than in SeCobBS (Figure 1E). However, we conducted more detailed DLS studies to understand the effect of Zn2+ on the structural properties of SeCobB and their dependence on temperature (Figure 4) in order to correlate with fluorescence spectroscopy studies. Induction with 250 µM ZnCl2 at 20°C led to an increase in the scattering intensity of both SeCobBS and SeCobBL within 50 s of salt addition. However, the time-dependent leap in the scattering intensity (It) upon ZnCl2 addition was ∼9 times more in SeCobBL than that in SeCobBS, implying that zinc mediated a profound impact on the larger isoform than on the shorter isoform.
5 Conclusion
This is the first study to experimentally report that the predicted zinc-binding motif of S. enterica CobB, present within the shorter isoform, SeCobBS (38–273 amino acid position), has a low affinity for Zn2+, and we hypothesize that it may be true for other bacterial CobB proteins. The NTD of SeCobBL helps in the formation of a stable oligomer, and it is the main site for the Zn2+–CobB interaction. This is supported by a ninefold increase in the scattering intensity of recombinant SeCobBL on the addition of ZnCl2, whereas there was hardly any change in the scattering intensity of SeCobBS (≤1.5-fold). Higher affinity toward SeCobBL is also validated by the association constant (KSV) and binding constant (Kb) values, which are 10 times and 109 times more than that for SeCobBS, respectively. The definite number of zinc-binding sites in the larger isoform also corroborates to negligible binding with its truncated form, i.e., SeCobBS. Thermal stability of SeCobBL was significantly reduced compared to that of SeCobBS in the presence of Zn2+. Zn2+ inhibited histone deacetylase activity of SeCobB at a higher concentration. Taken together, Zn2+ induced structural changes, and inhibition of deacetylase activity of SeCobB delineates the function of the predicted zinc-binding motif of bacterial CobB. We further predict that the zinc-binding domain is not a suitable target for future drug development.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
SB: conceptualization, data curation, formal analysis, investigation, methodology, software, visualization, and writing–original draft. PP: data curation, formal analysis, methodology, and writing–original draft. AD: software, visualization, and writing–original draft. AJ: data curation and writing–original draft. AD: data curation and writing–original draft. PM: data curation, resources, software, supervision, and writing–original draft. AP: data curation, methodology, supervision, validation, writing–original draft, and writing–review and editing. RM: conceptualization, funding acquisition, investigation, methodology, project administration, resources, supervision, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research of the RM laboratory was funded by the Department of Biotechnology (DBT) (BT/PR15263/MED/29/995/2015), Government of India. RM also thanks the DBT for partial funding (BT/MED/30/SP19662/2018-reg). AD thanks DST INSPIRE, Government of India, for Ph.D. fellowship (ref. no. IF140066). SB was supported by ICMR-SRF fellowship (Fellowship ID 2020-9157). SB was supported by ICMR-SRF fellowship (Fellowship ID 2020-9157, Reference Id: 45/07/2020/BIO/BMS).
Acknowledgments
The authors acknowledge support from the Central Research Facility, KIIT Deemed to be University, for using the spectrofluorometer. The authors thank Dileep Vasudevan T., Institute of Life Sciences, Bhubaneswar, India, for providing the circular dichroism facility and KIIT-TBI Bioprocess facility.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1345158/full#supplementary-material
References
AbouElfetouh, A., Kuhn, M. L., Hu, L. I., Scholle, M. D., Sorensen, D. J., Sahu, A. K., et al. (2015). The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen 4 (1), 66–83. doi:10.1002/mbo3.223
Ammendola, S., D'Amico, Y., Chirullo, B., Drumo, R., Ciavardelli, D., Pasquali, P., et al. (2016). Zinc is required to ensure the expression of flagella and the ability to form biofilms in Salmonella enterica sv Typhimurium. Metallomics 8 (10), 1131–1140. doi:10.1039/c6mt00108d
Anand, S., Ganaie, A. A., and Sharma, C. (2019). Differential thermal stability, conformational stability and unfolding behavior of Eis proteins from Mycobacterium smegmatis and Mycobacterium tuberculosis. PLoS One 14 (3), e0213933. doi:10.1371/journal.pone.0213933
Auld, D. S. (2001). “Zinc coordination sphere in biochemical zinc sites,” in Zinc biochemistry, physiology, and homeostasis: recent insights and current trends (Berlin, Germany: Springer), 85–127.
Buck, S. W., Gallo, C. M., and Smith, J. S. (2004). Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 75 (6), 939–950. doi:10.1189/jlb.0903424
Castano-Cerezo, S., Bernal, V., Blanco-Catalá, J., Iborra, J. L., and Cánovas, M. (2011). cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82 (5), 1110–1128. doi:10.1111/j.1365-2958.2011.07873.x
Castano-Cerezo, S., Bernal, V., Röhrig, T., Termeer, S., and Cánovas, M. (2015). Regulation of acetate metabolism in Escherichia coli BL21 by protein N(ε)-lysine acetylation. Appl. Microbiol. Biotechnol. 99 (8), 3533–3545. doi:10.1007/s00253-014-6280-8
Cerasi, M., Ammendola, S., and Battistoni, A. (2013). Competition for zinc binding in the host-pathogen interaction. Front. Cell Infect. Microbiol. 3, 108. doi:10.3389/fcimb.2013.00108
Chan, C. H., Garrity, J., Crosby, H. A., and Escalante-Semerena, J. C. (2011). In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Mol. Microbiol. 80 (1), 168–183. doi:10.1111/j.1365-2958.2011.07566.x
Colovos, C., and Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2 (9), 1511–1519. doi:10.1002/pro.5560020916
Consortium, U. (2015). UniProt: a hub for protein information. Nucleic acids Res. 43 (D1), D204–D212. doi:10.1093/nar/gku989
de Diego Puente, T., Gallego-Jara, J., Castaño-Cerezo, S., Bernal Sánchez, V., Fernández Espín, V., García de la Torre, J., et al. (2015). The protein acetyltransferase PatZ from Escherichia coli is regulated by autoacetylation-induced oligomerization. J. Biol. Chem. 290 (38), 23077–23093. doi:10.1074/jbc.M115.649806
Fischmann, T. O., Hruza, A., Niu, X. D., Fossetta, J. D., Lunn, C. A., Dolphin, E., et al. (1999). Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat. Struct. Biol. 6 (3), 233–242. doi:10.1038/6675
Frye, R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273 (2), 793–798. doi:10.1006/bbrc.2000.3000
Gao, K., Oerlemans, R., and Groves, M. R. (2020). Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 12 (1), 85–104. doi:10.1007/s12551-020-00619-2
Gardner, J. G., and Escalante-Semerena, J. C. (2009). In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol. 191 (6), 1749–1755. doi:10.1128/JB.01674-08
Giles, N. M., Watts, A. B., Giles, G. I., Fry, F. H., Littlechild, J. A., and Jacob, C. (2003). Metal and redox modulation of cysteine protein function. Chem. Biol. 10 (8), 677–693. doi:10.1016/s1074-5521(03)00174-1
Golovin, A., Dimitropoulos, D., Oldfield, T., Rachedi, A., and Henrick, K. (2005). MSDsite: a database search and retrieval system for the analysis and viewing of bound ligands and active sites. Proteins Struct. Funct. Bioinforma. 58 (1), 190–199. doi:10.1002/prot.20288
Gregoretti, I. V., Lee, Y. M., and Goodson, H. V. (2004). Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338 (1), 17–31. doi:10.1016/j.jmb.2004.02.006
Gu, L., Chen, Y., Wang, Q., Li, X., Mi, K., and Deng, H. (2015). Functional characterization of sirtuin-like protein in Mycobacterium smegmatis. J. Proteome Res. 14 (11), 4441–4449. doi:10.1021/acs.jproteome.5b00359
Gupta, R. P., Verma, P. C., and Garg, S. R. (2000). Effect of experimental zinc deficiency on immunological responses in Salmonella-infected Guinea-pigs. J. Comp. Pathol. 123 (1), 1–6. doi:10.1053/jcpa.2000.0376
Hazan, R., Sat, B., Reches, M., and Engelberg-Kulka, H. (2001). Postsegregational killing mediated by the P1 phage "addiction module" phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183 (6), 2046–2050. doi:10.1128/JB.183.6.2046-2050.2001
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 (8), 525–537. doi:10.1038/nrmicro2836
Imai, S., Johnson, F. B., Marciniak, R. A., McVey, M., Park, P. U., and Guarente, L. (2000). Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol. 65, 297–302. doi:10.1101/sqb.2000.65.297
Ko, J., Park, H., Heo, L., and Seok, C. (2012). GalaxyWEB server for protein structure prediction and refinement. Nucleic acids Res. 40 (W1), W294–W297. doi:10.1093/nar/gks493
Landry, J., Sutton, A., Tafrov, S. T., Heller, R. C., Stebbins, J., Pillus, L., et al. (2000). The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U. S. A. 97 (11), 5807–5811. doi:10.1073/pnas.110148297
Li, R., Gu, J., Chen, Y. Y., Xiao, C. L., Wang, L. W., Zhang, Z. P., et al. (2010). CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 76 (5), 1162–1174. doi:10.1111/j.1365-2958.2010.07125.x
Liimatta, K., Flaherty, E., Ro, G., Nguyen, D. K., Prado, C., and Purdy, A. E. (2018). A putative acetylation system in Vibrio cholerae modulates virulence in arthropod hosts. Appl. Environ. Microbiol. 84 (21), e01113. doi:10.1128/AEM.01113-18
Lima, B. P., Antelmann, H., Gronau, K., Chi, B. K., Becher, D., Brinsmade, S. R., et al. (2011). Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter. Mol. Microbiol. 81 (5), 1190–1204. doi:10.1111/j.1365-2958.2011.07742.x
Lin, Y. F., Cheng, C. W., Shih, C. S., Hwang, J. K., Yu, C. S., and Lu, C. H. (2016). MIB: metal ion-binding site prediction and docking server. J. Chem. Inf. Model 56 (12), 2287–2291. doi:10.1021/acs.jcim.6b00407
Liu, F., Yang, M., Wang, X., Yang, S., Gu, J., Zhou, J., et al. (2014). Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell Proteomics 13 (12), 3352–3366. doi:10.1074/mcp.M114.041962
Liu, W., Tan, Y., Cao, S., Zhao, H., Fang, H., Yang, X., et al. (2018). Protein acetylation mediated by YfiQ and CobB is involved in the virulence and stress response of Yersinia pestis. Infect. Immun. 86 (6), e00224. doi:10.1128/iai.00224-18
Lu, C. H., Lin, Y. F., Lin, J. J., and Yu, C. S. (2012). Prediction of metal ion-binding sites in proteins using the fragment transformation method. PLoS One 7 (6), e39252. doi:10.1371/journal.pone.0039252
Lüthy, R., Bowie, J. U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356 (6364), 83–85. doi:10.1038/356083a0
Magneson, G. R., Puvathingal, J. M., and Ray, W. J. (1987). The concentrations of free Mg2+ and free Zn2+ in equine blood plasma. J. Biol. Chem. 262 (23), 11140–11148. doi:10.1016/s0021-9258(18)60936-6
Maret, W. (2005). Zinc coordination environments in proteins determine zinc functions. J. Trace Elem. Med. Biol. 19 (1), 7–12. doi:10.1016/j.jtemb.2005.02.003
Maret, W. (2013a). Inhibitory zinc sites in enzymes. Biometals 26 (2), 197–204. doi:10.1007/s10534-013-9613-7
Maret, W. (2013b). Zinc biochemistry: from a single zinc enzyme to a key element of life. Adv. Nutr. 4 (1), 82–91. doi:10.3945/an.112.003038
Maret, W., Yetman, C. A., and Jiang, L.-J. (2001). Enzyme regulation by reversible zinc inhibition: glycerol phosphate dehydrogenase as an example. Chemico-Biological Interact. 130, 891–901. doi:10.1016/s0009-2797(00)00243-x
Mattocks, J. A., Tirsch, J. L., and Cotruvo, J. A. (2021). “Determination of affinities of lanthanide-binding proteins using chelator-buffered titrations,” in Rare-earth element biochemistry: characterization and applications of lanthanide-binding biomolecules (Amsterdam, Netherlands: Elsevier Science), 23–61.
McDevitt, C. A., Ogunniyi, A. D., Valkov, E., Lawrence, M. C., Kobe, B., McEwan, A. G., et al. (2011). A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 7 (11), e1002357. doi:10.1371/journal.ppat.1002357
Mishra, P., Beura, S., Sikder, S., Dhal, A. K., Vasudevan, M., Roy, M., et al. (2022). vp1524, a Vibrio parahaemolyticus NAD +-dependent deacetylase, regulates host response during infection by induction of host histone deacetylation. J. Biochem. 171 (6), 673–693. doi:10.1093/jb/mvac027
Nairz, M., Schroll, A., Sonnweber, T., and Weiss, G. (2010). The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 12 (12), 1691–1702. doi:10.1111/j.1462-5822.2010.01529.x
Nandi, S. K., Rehna, E. A. A., Panda, A. K., Shiburaj, S., Dharmalingam, K., and Biswas, A. (2013). A S52P mutation in the 'α-crystallin domain' of Mycobacterium leprae HSP18 reduces its oligomeric size and chaperone function. FEBS J. 280 (23), 5994–6009. doi:10.1111/febs.12519
Olson, O. T. a.A. J., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Ren, J., Sang, Y., Tan, Y., Tao, J., Ni, J., Liu, S., et al. (2016). Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 12 (3), e1005458. doi:10.1371/journal.ppat.1005458
Roman, A., Laskowski, D. S. M., and Janet, M. (1993). Thornton Main Chain Bond lengths and bond angles in protein structures. J. Mol. Biol. 231 (231), 1049–1067. doi:10.1006/jmbi.1993.1351
Romiguier, J., and Roux, C. (2017). Analytical biases associated with GC-content in molecular evolution. Front. Genet. 8, 16. doi:10.3389/fgene.2017.00016
Roy, A., Kucukural, A., and Yang, Z. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi:10.1038/nprot.2010.5
Smith, J. S., Brachmann, C. B., Celic, I., Kenna, M. A., Muhammad, S., Starai, V. J., et al. (2000). A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 97 (12), 6658–6663. doi:10.1073/pnas.97.12.6658
Tainer, J. A., Roberts, V. A., and Getzoff, E. D. (1991). Metal-binding sites in proteins. Curr. Opin. Biotechnol. 2 (4), 582–591. doi:10.1016/0958-1669(91)90084-i
Tanny, J. C., Kirkpatrick, D. S., Gerber, S. A., Gygi, S. P., and Moazed, D. (2004). Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell Biol. 24 (16), 6931–6946. doi:10.1128/MCB.24.16.6931-6946.2004
Tucker, A. C., and Escalante-Semerena, J. C. (2010). Biologically active isoforms of CobB sirtuin deacetylase in Salmonella enterica and Erwinia amylovora. J. Bacteriol. 192 (23), 6200–6208. doi:10.1128/JB.00874-10
VanDrisse, C. M., and Escalante-Semerena, J. C. (2018). In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and Nɛ -lysine acetylation. Mol. Microbiol. 107 (4), 577–594. doi:10.1111/mmi.13901
VanDrisse, C. M., Parks, A. R., and Escalante-Semerena, J. C. (2017). A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. MBio 8 (3), e00708. doi:10.1128/mbio.00708-17
Vickers, N. J. (2017). Animal communication: when I'm calling you, will you answer too? Curr. Biol. 27 (14), R713–R715. doi:10.1016/j.cub.2017.05.064
Waldron, K. J., and Robinson, N. J. (2009). How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7 (1), 25–35. doi:10.1038/nrmicro2057
Wei, Y., and Fu, D. (2006). Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 281 (33), 23492–23502. doi:10.1074/jbc.M602254200
Wiederstein, M., and Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids Res. 35 (Suppl. l_2), W407–W410. doi:10.1093/nar/gkm290
Keywords: Salmonella nicotinamide adenine dinucleotide-dependent deacetylase (CobB), Zn2+–CobB interaction, CobB oligomerization, CobB homology modeling, inhibition of CobB deacetylase activity, CobB thermal stability
Citation: Beura S, Pritam P, Dhal AK, Jana A, Dash A, Mohanty P, Panda AK and Modak R (2024) An insight into the role of the N-terminal domain of Salmonella CobB in oligomerization and Zn2+ mediated inhibition of the deacetylase activity. Front. Mol. Biosci. 11:1345158. doi: 10.3389/fmolb.2024.1345158
Received: 27 November 2023; Accepted: 24 January 2024;
Published: 13 March 2024.
Edited by:
Diletta Ami, University of Milano-Bicocca, ItalyReviewed by:
Ninganagouda R. Patil, B V B College of Engg. and Tech., IndiaManuela Leri, University of Florence, Italy
Copyright © 2024 Beura, Pritam, Dhal, Jana, Dash, Mohanty, Panda and Modak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rahul Modak, cmFodWwubW9kYWtAa2lpdGJpb3RlY2guYWMuaW4=; Alok Kumar Panda, YWxvay5wYW5kYWZjaEBraWl0LmFjLmlu
 Shibangini Beura
Shibangini Beura Pulak Pritam
Pulak Pritam Ajit Kumar Dhal1
Ajit Kumar Dhal1 Arindam Jana
Arindam Jana Alok Kumar Panda
Alok Kumar Panda Rahul Modak
Rahul Modak