- 1Basic Medical Sciences Department, College of Medicine, QU Health, Qatar University, Doha, Qatar
- 2Neurological Disorders Research Center, Qatar Biomedical Research Institute, Hamad Bin Khalifa University, Doha, Qatar
- 3College of Health and Life Sciences, Hamad Bin Khalifa University, Doha, Qatar
- 4School of Medicine, New Giza University, Giza, Egypt
Reprogramming human somatic cells into a pluripotent state, achieved through the activation of well-defined transcriptional factors known as OSKM factors, offers significant potential for regenerative medicine. While OSKM factors are a robust reprogramming method, efficiency remains a challenge, with only a fraction of cells undergoing successful reprogramming. To address this, we explored genes related to genomic integrity and cellular survival, focusing on iPSCs (A53T-PD1) that displayed enhanced colony stability. Our investigation had revealed three candidate genes CCN3, POSTN, and PTHLH that exhibited differential expression levels and potential roles in iPSC stability. Subsequent analyses identified various protein interactions for these candidate genes. POSTN, significantly upregulated in A53T-PD1 iPSC line, showed interactions with extracellular matrix components and potential involvement in Wnt signaling. CCN3, also highly upregulated, demonstrated interactions with TP53, CDKN1A, and factors related to apoptosis and proliferation. PTHLH, while upregulated, exhibited interactions with CDK2 and genes involved in cell cycle regulation. RT-qPCR validation confirmed elevated CCN3 and PTHLH expression in A53T-PD1 iPSCs, aligning with RNA-seq findings. These genes’ roles in preserving pluripotency and cellular stability require further exploration. In conclusion, we identified CCN3, POSTN, and PTHLH as potential contributors to genomic integrity and pluripotency maintenance in iPSCs. Their roles in DNA repair, apoptosis evasion, and signaling pathways could offer valuable insights for enhancing reprogramming efficiency and sustaining pluripotency. Further investigations are essential to unravel the mechanisms underlying their actions.
1 Introduction
The reprogramming of human somatic cells into a pluripotent state has been accomplished by the ectopic expression of well-defined transcriptional factors such as OCT4 (also known as POU5F1), SOX2, KLF4, MYC (also known as c-MYC); these factors are collectively referred to as the OSKM factors (Yamanaka, 2012; Swaidan et al., 2020). Human induced pluripotent stem cells (hiPSCs), which possess embryonic stem cell (ESC)-like characteristics, have increased the possibility of generating patient-specific cells (Dimos et al., 2008; Soldner et al., 2009). These hiPSCs are distinguished by their ability to self-renew and differentiate into any cell type of the body. Consequently, these cells can serve purposes such as comprehending disease progression, developing and testing novel drugs, and conducting autologous replacement therapies for degenerated cells (Dimos et al., 2008). This is particularly relevant to degenerative diseases of the central nervous system (CNS) such as Parkinson’s disease (PD), in which dopaminergic neurons are lost (Soldner et al., 2009).
PD is the second most common chronic progressive neurodegenerative disease after Alzheimer’s disease. It is characterized by the loss of the nigrostriatal dopamine-releasing neurons. Despite the fact that genetic factors are implicated in the pathogenesis of PD, the majority of PD cases are sporadic, not associated with a known genetic mutation (Soldner et al., 2009). The pathological hallmark of PD is the aggregation and deposition of abnormal α-synuclein protein within the neuronal cell bodies (Stefanis, 2012). Numerous mutations have been reported in the gene responsible for encoding this protein. The α-synuclein point mutation (A53T) is believed to be the most frequent and associated with the familial type of PD (Swaidan et al., 2020).
While OSKM factors have demonstrated a robust method for reprogramming somatic cells that has potential for regenerative medicine, the process is still inefficient since only a small fraction of cells undergoes a complete and successful reprogramming (Swaidan et al., 2020). To overcome low reprogramming efficiency, several studies have examined factors that could enhance the activation of the pluripotency pathway and prevent the differentiation of stem cells (M. Wang et al., 2017; Bogliotti et al., 2018; Xu et al., 2016; Theka et al., 2017). In our earlier research, we pinpointed five genes (GBX2, NANONGP8, SP8, PEG3, and ZIC1) exhibiting differential expression in iPSCs generated from fibroblast cells. These genes maintained their differential expression regardless of the presence or absence of the PD A53T mutation, while also showcasing interactions with OSKM factors (Swaidan et al., 2020). Building upon our prior research, this paper focuses on a distinct sample (named A53T-PD1) that displayed enhanced colony integrity and stability in comparison to other samples (A53T-PD2, ID-PD and HC). This observation holds true despite the fact that a smaller number of colonies were formed during the reprogramming process of this sample.
Hence, with the aim of enhancing the regeneration process’s efficiency, our focus lies in investigating genes related to genomic integrity/stability and cellular survival. These genes exhibit differential expression in A53T-PD1 cells and could potentially be genetically modified or suppressed to bolster and sustain iPSC pluripotency.
2 Materials and methods
2.1 Cell lines and cell culture
In this study, four established human iPS cell lines (HC, ID-PD, A53T-PD1, and A53T-PD2) were employed (n = 3 per iPS cell line, total = 12 samples). These stem cells were grown using a feeder-free system with Matrigel (Corning) for cell adherence and maintained in StemFlex media (Termo Fisher Scientifc) under controlled conditions at 37°C and 5% CO2 incubators. Cells were fed every other day, and passage was performed when cell confluency reached approximately 80%, typically within four to 5 days after the previous passage. The passage process involved pre-coating of 35 mm dishes with Matrigel for 30 min, 2 h, or overnight, washing colonies with 1 mL Dulbecco’s Phosphate-Buffered Saline (DPBS), gently dissociating the cells into appropriately sized colonies (∼100 µm) using 500 µL of non-enzymatic reagent (ReLeSR; StemCell Technologies), collecting these colonies with StemFlex media, centrifuging them for 4 min at 800 RPM and 22°C, resuspending the resulting pellet in 1 mL fresh StemFlex media, and finally evenly distributing 70 μL of cells onto the freshly prepared dishes containing 2 mL fresh StemFlex media.
2.2 RNA and RT-qPCR
Total RNA was extracted with a Direct-zol RNA MicroPrep Extraction kit (Zymo Research) according to manufacturer’s instructions. Then, cDNA was synthesized using Superscript IV, First-Strand Synthesis System kit (Invitrogen). The RT-qPCR technique was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems) and TaqMan probes (20×) according to the manufacturer’s protocol. The list of genes: Assay name: POSTN, Assay ID: Hs0166750, PTHLH, Assay ID: Hs00174969, NOV (CCN3), Assay ID: Hs00159631 and housekeeping gene GAPDH, Assay ID: Hs02786624. The total reaction volume was 10 µL per well. RT-qPCR reaction was run in triplicates using Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument under default conditions (95°C for 10 min and then PCR reaction: 40 cycles of 95°C, 15 s and 60°C, 1 min).
2.3 RNA sequencing (RNA-Seq)
The transcriptomic sequencing process encompassed two key steps: the first step involved the preparation of mRNA libraries, followed by RNA sequencing. Initially, the quality of the extracted total RNA samples was assessed using a 2100 Bioanalyzer equipped with the Agilent RNA Nano 6000 chip. Subsequently, we carried out mRNA library preparation using the TruSeq Stranded mRNA Sample Prep LS kit (Illumina). To ensure the quality and quantity of the generated mRNA libraries, a thorough quality control analysis was conducted using the Agilent DNA 1000 chip on a 2100 Bioanalyzer, along with the quantification of DNA library templates using a Qubit assay. For cluster generation, indexed DNA libraries were normalized to 10 nM and placed in a diluted cluster template (DCT) plate, after which they were combined in equal volumes in a pooled DCT plate (PDP). The final step involved RNA sequencing, which was performed utilizing the HiSeq 4000 system (Illumina) at the QBRI genomic core facility.
2.4 RNA-seq analysis
We employed the Bcl2fastq Conversion Software to perform two crucial tasks: firstly, to demultiplex the data, and secondly, to convert the BCL files generated by Illumina sequencing systems into standard FASTQ file formats, which are essential for subsequent RNA-Seq analysis. Quality control checks were conducted using FastQC, a specialized tool designed for high-throughput sequence data analysis, accessible at (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The subsequent analysis followed the standard QIAGEN bioinformatics CLC Genomics Workbench 20.0 pipeline (https://digitalinsights.qiagen.com). This sequential pipeline comprised four key stages: 1) pre-processing of raw sequencing reads, 2) mapping of reads to a reference genome, 3) quantification of genes and transcripts, and 4) differential expression analysis. Data normalization was performed relative to the healthy control iPS cell line, which served as the baseline for comparison.
2.5 Gene ontology molecular analysis
We utilized the Protein Analysis Through Evolutionary Relationships (PANTHER) 14.1 platform, accessible at (http://www.pantherdb.org), as an online classification system. PANTHER is specifically designed for the identification and categorization of proteins and their corresponding genes based on their family/subfamily, molecular function, biological processes, and pathways in which they play integral roles within the cellular context. This software served as a valuable tool for filtering and pinpointing genes relevant to our areas of focus, which encompassed cellular integrity and genomic stability.
2.6 Protein-protein interaction analysis
We employed the Search Tools for the Retrieval of Interacting Genes (STRING) 10.5, an online software dedicated to the exploration of gene and protein interactions, accessible at (https://string-db.org/). Our primary objective with STRING was to establish predicted interactions between our selected genes of interest and protein-encoding genes that are involved in cell cycle checkpoints, DNA repair and replication, TP53 phosphorylation, and apoptosis inhibition. Through the STRING platform, we did not only visualize the nature of interactions between these proteins but also examined the underlying evidence supporting these interactions.
2.7 Databases and literature review
A comprehensive literature review was conducted using medical search engines such as PubMed, Medline, UpToDate, Scopus, Access Medicine, Genetic Home Reference, and Access Genetics. Specific keywords were employed such as “the name of the chosen candidate gene,” “pluripotency/pluripotent,” “reprogramming,” “genomic stability/integrity in Embryonic stem cells/ESCs,” and “induced pluripotent stem cells/iPSCs”. Relevant articles were screened and their findings documented for confirmation of transcriptional factors’ involvement in genomic integrity and cellular survival.
2.8 Statistical analysis
Statistical analyses were conducted using GraphPad Prism 8.4.3 (GraphPad Software, Inc., San Diego, CA). Data were presented as the mean ± standard deviation (SD) for each group (N = 3). Statistical significance was assessed through the Mann–Whitney U test and defined as p-values less than 0.05 (p < 0.05) or 0.01 (p < 0.01).
3 Results
3.1 A set of genes involved in molecular function regulation, including DNA binding, biological adhesion and intracellular signalling
Due to the superior colony stability observed in A53T-PD1 iPSCs, we turned our focus to the identification of differentially expressed candidate genes across distinct iPS cell lines. Utilizing RNA sequencing data, we conducted a comparative analysis of differentially expressed genes between A53T-PD1 and other iPS cell lines: HC, ID-PD, and A53T-PD2. Our selection criteria involved genes exhibiting a false discovery rate (FDR) of less than 0.05 specifically in A53T-PD1 cell line. This screening resulted in the identification of a total of 43 genes, comprising 39 genes with upregulated expression and 4 genes displaying downregulated expression (Figure 1).
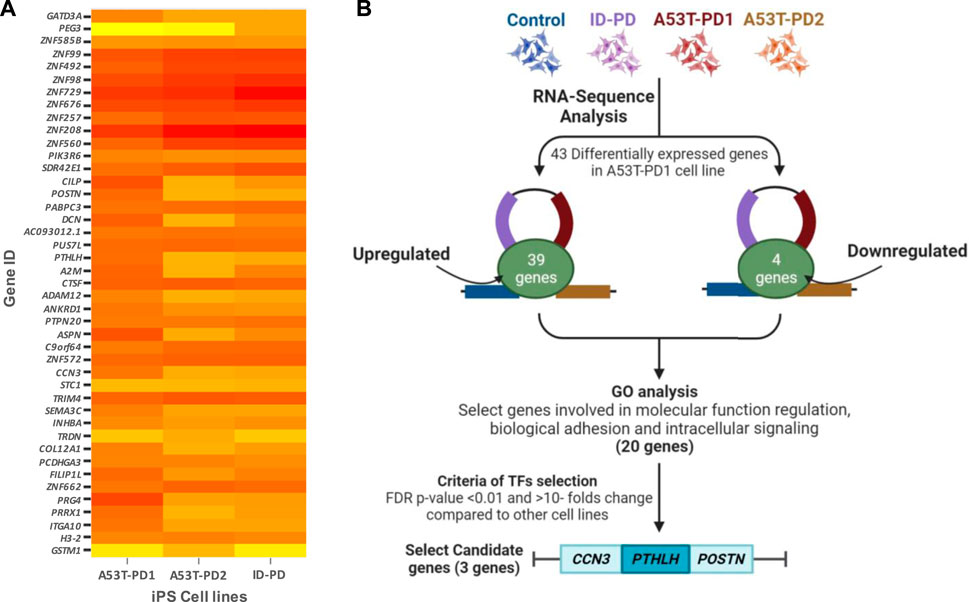
FIGURE 1. Comprehensive visualization of study findings. (A) Heatmap of differentially expressed genes in A53T-PD1 cell line. (B) Illustrative flowchart summarizing the procedural pathway that led to the identification of three candidate genes within the A53T-PD1 iPS cell line as potential contributors to genomic integrity and pluripotency maintenance in iPSCs.
Subsequent to this discovery phase, we proceeded to subject the 43 genes to Gene Ontology analysis (GO), in order to systematically classify them based on their roles and functions within the cellular context (Figure 2). Our selection process was grounded in the prioritization of cellular processes that bear utmost relevance to both cellular integrity and genomic stability. These encompassed processes such as molecular function regulation, DNA binding, biological and cell adhesion, as well as intracellular signaling. Notably, while the statistical significance threshold was not met for DNA binding and cell adhesion molecules (with FDR p-values of 0.42 and 0.25, respectively), the attributes of molecular function regulation, biological adhesion, and intracellular signaling exhibited statistical significance (FDR p-values of 0.003, 0.04, and 0.04, respectively). Guided by these findings, our attention was directed towards refining the gene selection criteria to those involved in molecular functions, biological processes, and protein classes, with a focus on genes that are implicated in molecular function regulation, biological adhesion, and intracellular signaling (as outlined in Figure 2).
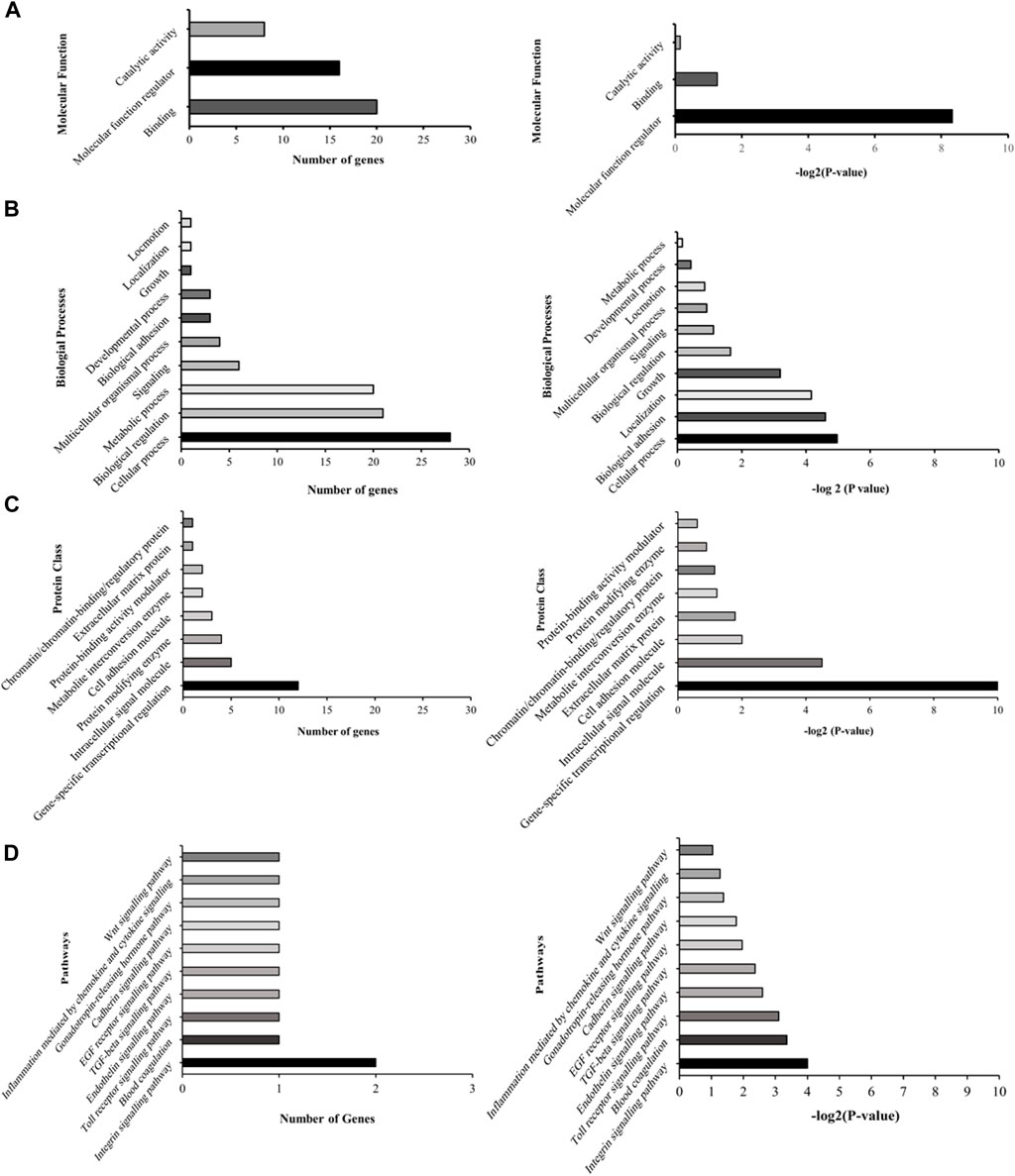
FIGURE 2. Illustrative graphs for gene ontology (GO) analysis. The left panels (A–D) display representative graphs categorizing the 43 differentially expressed genes based on their cellular roles and functions. The right panels (A–D) demonstrate the statistical significance of these categorized genes.
3.2 Selection of candidate genes in A53T-PD1 cell line
We successfully identified a total of 20 genes, effectively categorized into two distinct groups. The initial group comprised 11 zinc finger transcription regulator genes, integral to the orchestration of molecular function regulation. The remaining 9 genes, namely, CCN3, INHBA, PTHLH, PIK3R6, PEG3, H3.2, POSTN, PCDHGA3, and STC1, demonstrated their involvement across various functional domains. Further delving into the expression patterns of these 20 genes within each iPS cell line occurred through an in-depth analysis, utilizing the differential expression data generated in relation to control iPSCs.
For the zinc finger transcription regulator genes, a consistent and statistically significant upregulation was observed across all cell lines (FDR <0.01), compared to control iPSCs (illustrated in Figure 3A). As a result of this uniformity, these genes were subsequently excluded from further gene selection. Conversely, among the remaining genes (Figure 3B), the highest upregulation in A53T-PD1 cell line was notably exhibited by POSTN, soaring 70-fold (FDR <0.001), followed closely by PTHLH with a 59-fold increase (FDR <0.001). In contrast, both POSTN and PTHLH showed downregulation in ID-PD and A53T-PD2 cell lines. Additionally, CCN3, INHBA, and PIK3R6 displayed substantial upregulation in A53T-PD1 cell line, significantly exceeding other cell lines (FDR <0.001 for all).
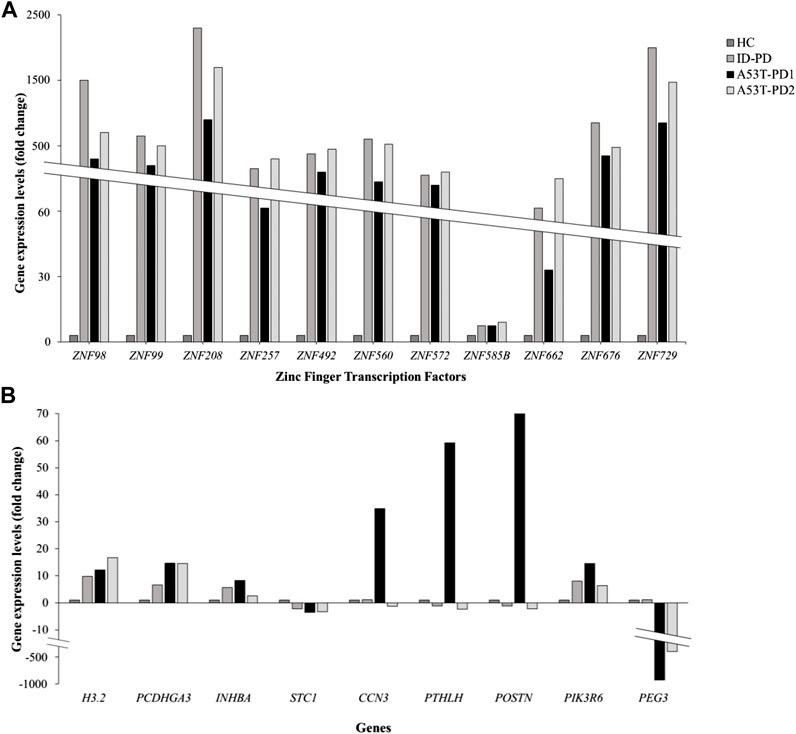
FIGURE 3. Expression profiling of 20 genes involved in genomic integrity and cellular survival. Gene expression levels for a panel of 20 genes associated with genomic integrity/stability and cellular survival were analyzed using RNA-Seq data and represented in two separate graphs. (A) Illustrates the expression levels of 11 zinc finger transcription factors expressed in different iPS cell lines. (B) Illustrates the expression levels of the remaining 9 genes expressed in iPS cell lines.
H3.2. displayed an upregulation of 12-fold in A53-PD1 cell line, in contrast to 16-fold in A53T-PD2 and 9-fold in ID-PD cell lines, all with an FDR <0.001. PCDHGA3, on the other hand, exhibited consistent upregulation across all cell lines, demonstrating a similar 15-fold change in both A53T-PD1 and A53T-PD2 cell lines (FDR <0.001). Unlike all the previously mentioned genes, STC1 and PEG3 displayed downregulation. While STC1 demonstrated downregulation across all cell lines without significant variation among them, PEG3 showcased its most pronounced downregulation in A53T-PD1 cell line at a staggering −990-fold change (FDR <0.001), succeeded by A53T-PD2 cell line at −443-fold change (FDR <0.001) (Figure 3B).
In the quest for a final selection of candidate genes potentially contributing to the generation of the most stable iPSC colonies in A53T-PD1 cell line, we employed two stringent criteria: (a) FDR p-value <0.01 and (b) a significant upregulation or downregulation in A53T-PD1 cell line, displaying a variation of more than 10-fold compared to other cell lines. Consequently, INHBA, PIK3R6, H3.2, STC1, and PCDHGA3 were disregarded. It is important to highlight that PEG3 was excluded from consideration, as it had already been analyzed in our previous work (Swaidan et al., 2020). Therefore, this rigorous process left us with a final selection of three genes: CCN3, POSTN, and PTHLH.
3.3 Protein-protein interaction analysis
Following gene selection, we utilized STRING software to predict potential protein interactions associated with our chosen candidate genes. Individually, each gene underwent scrutiny to anticipate its feasible associations with other proteins. Additionally, each gene underwent evaluation in conjunction with a cluster of genes linked to cell cycle regulation and DNA repair/replication.
An illustrative instance of this predictive analysis is evident in POSTN, which exhibited a remarkable 70-fold upregulation in A53T-PD1 cell line. This gene was predicted to stimulate COL1A1 expression, contributing to the synthesis of a substantial portion of type 1 collagen (as indicated in Figure 4A through the green arrow). Furthermore, POSTN demonstrated unknown interactions with other collagen-related genes, including COL1A2, COL5A2, and COL3A1, as well as the DCN gene, responsible for encoding the decorin protein. Decorin, in turn, contributes to extracellular matrix strength and cell stability. Notably, POSTN also exhibited a binding interaction with TNC, which encodes Tenascin C, a crucial extracellular matrix protein involved in guiding migrating neurons and axons during development, synaptic plasticity, and neuronal regeneration (as indicated in Figure 4A by the blue line).
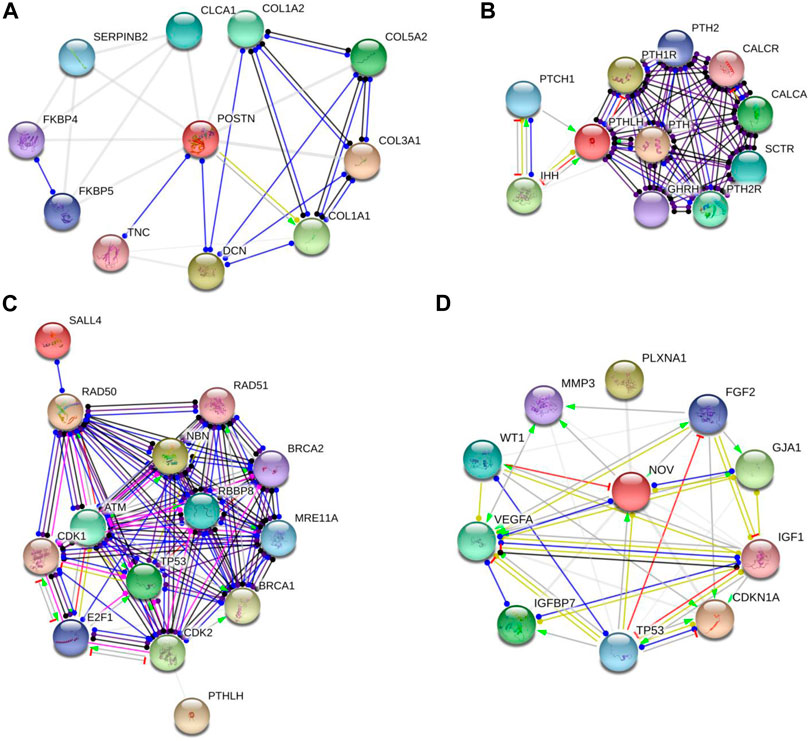
FIGURE 4. Protein-protein interaction networks revealed by String software analysis. (A,B and D) Protein interaction network involving POSTN, PTHLH and CCN3 (also known as NOV). (C) Predicted interaction network of genes responsible for preserving cellular survival and genomic integrity in ESCs. Each network node represents all the proteins generated by a single protein-coding gene locus. Colored nodes denote query proteins and first shell of interactors, while white nodes indicate second shell of interactors. Empty nodes signify proteins with unknown 3D structures, while filled nodes denote proteins with known or predicted 3D structures. It's important to note that these protein associations, while significant, do not necessarily indicate physical binding. The network edges in this figure represent molecular interactions between the associated proteins, with line shape indicating the predicted mode of action. Line colors convey different action types, such as binding (blue line), activation (green line), inhibition (red line), reaction (black line), catalysis (purple line), transcriptional regulation (yellow line), and posttranslational modification (pink line). Unidentified interactions are represented by grey lines. Arrowed lines indicate a positive effect, bar-headed lines signify a negative effect, and circular-ended lines denote an unspecified effect.
In our subsequent exploration, we aimed to determine whether POSTN had any potential interactions with gene sets pivotal to fundamental cellular processes such as cellular stability and genomic integrity in ESCs. These encompassed genes that are responsible for DNA repair and replication, phosphorylation of p53, and other molecules like Rb and ATM. For this analysis, we included genes related to cell cycle checkpoints (CDK1/2), the MRN complex intrinsic to the homologous recombination pathway of DNA repair (MRE11, RAD50, and NBS1), alongside BRCA1/2, RAD51, and SALL4. However, it's important to highlight that POSTN did not display any predicted interactions with the aforementioned genes.
Similarly, PTHLH which experienced a significant upregulation in A53T-PD1 cell line (an impressive 59-fold increase) was predicted to exert an inhibitory influence on the IHH gene (as indicated in Figure 4B by the red line). This particular gene plays a pivotal role in governing the differentiation of progenitor cells into osteoblasts, thus exerting a significant influence over cellular differentiation dynamics. Additionally, PTHLH exhibited interactions with parathyroid hormone-related genes including PTH1R, PTH2, PTH2R, as well as calcitonin-related genes such as CALCR and CALCA. Upon closer examination of PTHLH’s interactions with the aforementioned gene set that is vital to substantial cellular processes, an intriguing and as yet unidentified interaction emerged with cyclin-dependent kinase 2 (CDK2) (Figure 4C). CDK2, a pivotal player in cell cycle control, undertakes the phosphorylation of TP53, thereby impeding apoptosis. Furthermore, CDK2’s involvement extends to the activation of E2F1 gene expression, an essential precursor for the initiation of DNA synthesis. The orchestrated actions of CDK2 and E2F1 contribute to the hyperphosphorylation of the Rb protein-encoding gene, ultimately facilitating ESCs’ efficient transition into the S phase of the cell cycle and thereby shortening the duration of the G1 phase.
Interestingly, further exploration of CCN3 (which exhibited a significant upregulation, 34.9-fold increase in A53T-PD1 cell line) revealed that CCN3’s transcriptional regulation, as an extracellular adhesion-related protein, is directly influenced by the TP53 gene that is recognized as a pivotal tumor suppressor gene. This gene wields substantial influence over cellular survival and apoptotic processes through the p53 signaling pathway (Figure 4D).
Moreover, CCN3 exhibited a mysterious interaction with CDKN1A, a crucial player within the p53 pathway responsible for orchestrating cell apoptosis and survival. Notably, CDKN1A serves as an essential mediator, enabling p53/TP53 to execute its role as a proliferation inhibitor in response to DNA damage. Furthermore, CCN3’s interactions extended to VEGFA, where it contributed to elevated expression levels and binding activity, while concurrently exerting inhibitory effects on WT1, leading to decreased expression levels. This latter gene, WT1, assumes the role of suppressing cellular apoptosis. Additionally, CCN3’s involvement encompassed the transcriptional regulation of the IGF1 gene, known for its capacity to hinder cellular apoptosis by activating the PI3K/AKT pathway and inciting cell proliferation. This effect stands in contrast to the apoptotic influence of p53, thus creating a complex interplay (illustrated in Figure 4D).
3.4 CCN3 and PTHLH are differentially expressed in A53T-PD1 iPS cell line
As a means of corroborating the RNA-seq analysis that identified CCN3, PTHLH, and POSTN as potential players in cellular stability and pluripotency maintenance, we quantified their expression levels across the four distinct iPS cell lines (HC, A53T-PD1, A53T-PD2, and ID-PD) using RT-qPCR. In terms of the genes under examination, the expression levels of POSTN in A53T-PD1 exhibited similarity to those observed in the control (HC) cell line, with insignificant downregulation observed in both the ID-PD and A53T-PD2 iPS cell lines. Conversely, the A53T-PD1 cell line exhibited significantly elevated expression levels of CCN3 (227-fold increase, p < 0.0001) and PTHLH (542-fold increase, p < 0.0001) when contrasted with control iPS cell lines (Figure 5). These findings are consistent with our RNA-seq analysis, providing further support to our observation that additional transcriptional factors may play a role in bolstering the stability of iPS colonies and upholding their pluripotency.
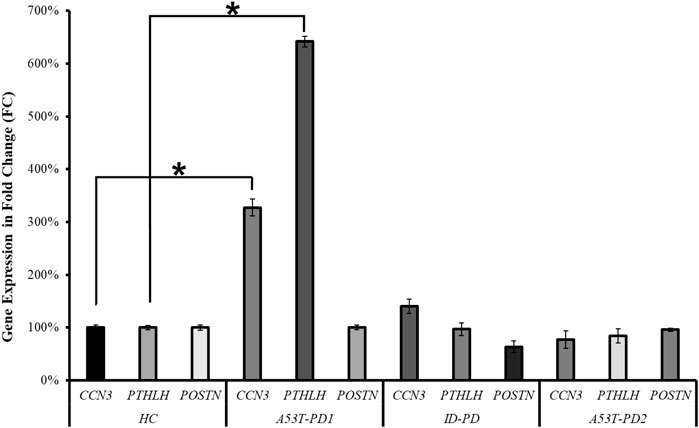
FIGURE 5. RT-qPCR validation of three candidate genes in various iPS cell lines. RT-qPCR analysis was conducted to validate the expression of the three candidate genes (CCN3, PTHLH, POSTN) across different iPS cell lines. The statistical significance was determined using the Mann–Whitney U test, with *p < 0.0001 indicating significance.
It is imperative to note that differences, whether insignificant or significant, may exist between RNA-seq and RT-qPCR analyses of gene expression levels. Such variations can be attributed to several factors, including methodological disparities. RNA-seq provides a comprehensive, unbiased assessment of the entire transcriptome, detecting both known and novel transcripts, whereas RT-qPCR focuses on specific transcripts, offering a more targeted analysis. Additionally, differences in sensitivity and dynamic ranges are evident, with RNA-seq possessing a broader dynamic range capable of detecting low-abundance transcripts and capturing alternative splicing events, while qPCR, though highly sensitive, may have a more limited dynamic range compared to RNA-seq. Furthermore, normalization methods diverge, with RNA-seq utilizing techniques like TPM (transcripts per million) or FPKM (fragments per kilobase of transcript per million mapped reads) to account for library size and transcript length, while qPCR relies on reference genes for normalization, and the choice of reference genes can influence results. Lastly, considerations of gene isoforms and alternative splicing reveal that RNA-seq detects multiple isoforms and alternative splicing events, whereas primers designed for qPCR may target specific isoforms, potentially missing alternative splicing events (Marioni et al., 2008; Mortazavi et al., 2008; Bustin et al., 2009; Wang et al., 2009; Derveaux et al., 2010).
3.5 CCN3 involvement in cell apoptosis and FGFR2 signaling pathways
To further explore CCN3’s role in the regulation of cellular apoptosis and proliferation, we conducted a comprehensive investigation using the Reactome Pathway Database. The data obtained from this database has revealed its participation in cellular apoptosis through an interaction with the POU4F2 domain, known to bind to TP53, thereby modulating the transcription of pro-apoptotic genes (Figure 6A). Inhibition of this pathway has been observed to augment iPSC reprogramming efficiency by fostering cell proliferation. Furthermore, CCN3 has also been found to be a participant in the fibroblast growth factor receptor 2 (FGFR2) signaling pathway, where it interacts with fibroblast growth factor receptor substrate 3 (FSR3), an adapter protein responsible for transmitting signals downstream from FGFR (Figure 6B). Activation of this pathway has been demonstrated to play a crucial role in both the initiation and stabilization phases of reprogramming by promoting cellular proliferation.
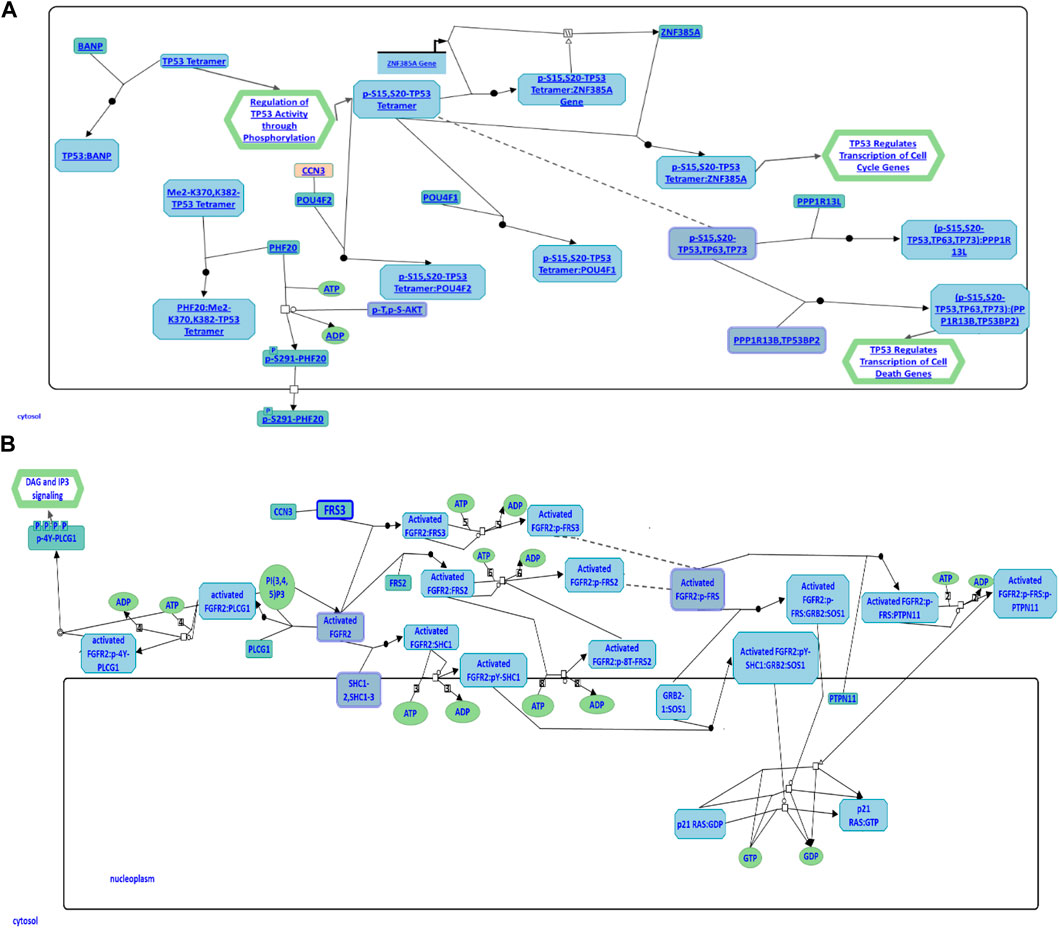
FIGURE 6. CCN3 involvement in cell apoptosis and FGFR2 signaling pathways. (A) CCN3’s role in the p53 apoptosis pathway is illustrated. CCN3 interacts with the POU4F2 domain, which binds to TP53, regulating the transcription of pro-apoptotic genes. (B) CCN3’s involvement in the FGFR2 signaling pathway is represented. CCN3 interacts with FSR3, an adapter protein that transmits signals downstream from FGFR. Both diagrams have been obtained and adapted under the CC-BY 4.0 international license from Reactome.
3.6 PTHLH implication in G protein activation pathway
Similarly, we conducted a comprehensive investigation utilizing the Reactome Pathway Database analysis to elucidate the cellular processes governed by PTHLH, leading to the discovery of its central role in G protein activation (Figure 7). PTHLH’s interaction with a G-protein-coupled receptor (GPCR) forms a liganded-GPCR complex, initiating the exchange of GDP for GTP within the G-protein alpha subunit (GNAS), ultimately resulting in G-protein activation. Once activated, the GNAS directly interacts with the proto-oncogene tyrosine kinase Src (SRC), leading to SRC autophosphorylation and subsequent activation. Functionally, the SRC kinase family plays a pivotal role in cellular growth and cancer. Additionally, the G-protein family, particularly the GNAS, is involved in the regulation of cellular apoptosis and the maintenance of pluripotency in ESCs.
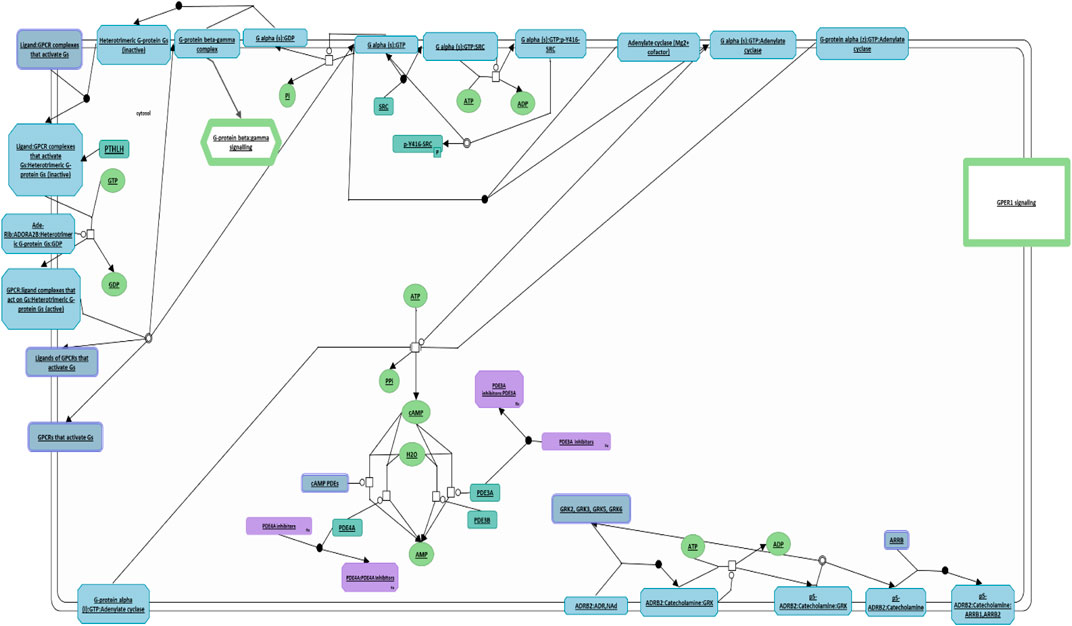
FIGURE 7. PTHLH-mediated G protein activation pathway. This figure illustrates the pivotal role of PTHLH in G protein activation where it binds to G-protein-coupled receptor (GPCR), forming a liganded-GPCR complex. This diagram has been obtained and modified under the CC-BY 4.0 international license from Reactome.
4 Discussion
ESCs and iPSCs offer immense potential, not only in the field of regenerative medicine but also as valuable tools for investigating developmental processes, pathologies, and the assessment of novel drug interventions. However, the maintenance of genomic integrity and cellular stability presents a formidable challenge to sustaining the self-renewal and pluripotency of PSCs. Genomic instability can impair self-renewal pathways, activate the p53 apoptotic pathway, and promote PSC differentiation (Su et al., 2019). It is therefore imperative to safeguard the genomic integrity of PSCs and explore the genes that may be involved in this process, all while promoting robust growth to preserve their vital functions and self-renewal capability. Several factors, including culture conditions, can contribute to genomic instability in ESCs, and exposure to endogenous or exogenous genotoxic stresses can lead to cell apoptosis due to DNA damage (Martins-Taylor and Xu, 2012). It has been noted that ESCs have shown distinct mechanisms to shield themselves from DNA double-strand breaks, setting them apart from somatic cells (Nagaria et al., 2013).
In our previous study (Swaidan et al., 2020), iPS cell lines were successfully generated from a PD patient carrying the A53T mutation (A53T-PD1). Although the generated colonies number was limited, these colonies exhibited remarkable genomic resilience and prolonged viability compared to other iPS cell lines that were generated in parallel. This intriguing finding prompted us to consider additional factors influencing genomic integrity beyond age and disease mutation. Our hypothesis regarding the potential roles of certain molecules specific to A53T-PD1 cell lines, at both nuclear and cellular levels, in maintaining genomic and cellular stability were substantiated by transcriptomic analysis. This analysis revealed three differentially expressed genes in the A53T-PD1 cell line (CCN3, POSTN, and PTHLH), all of which are implicated in cellular stability, DNA repair mechanisms, and apoptosis inhibition.
Among these genes, CCN3, also known as NOV, exhibited significant upregulation in the A53T-PD1 iPS cell line compared to other cell lines (34.9-fold, FDR <0.001). CCN3 belongs to the CCN family of matricellular proteins, known for its pivotal role in extracellular signaling and the maintenance of cellular viability (Kubota and Takigawa, 2013). Previous studies have highlighted the importance of CCN3, particularly its role in cellular adhesion and the regulation of integrin expression (Lafont et al., 2005; Lin et al., 2005; Vallacchi et al., 2008). These findings support our hypothesis that CCN3 plays a crucial role in pluripotency circuits and their maintenance, particularly in cell adhesion receptor systems and integrin expression (Vitillo and Kimber, 2017; Yu et al., 2018). Moreover, the involvement of CCN3 in regulating integrin expression not only underscores its significance in initiating and sustaining pluripotency but also implies its role in maintaining the compact morphology of pluripotent colonies, dependent on integrin activity (Yu et al., 2018). In addition, increased cellular proliferation, a hallmark of cells undergoing reprogramming, can be achieved by inhibiting p53, which is responsible for programmed cell death, and promoting LIN28 expression, which regulates cell cycle genes like CDK genes. This mechanism contributes to the efficiency of reprogramming processes and the stability of hiPSCs (Hong et al., 2009; Kawamura et al., 2009; Hawkins et al., 2014).
Through STRING analysis, we observed that CCN3 is involved in promoting cell survival. Notably, it directly interacts with key players in the p53 pathway, including TP53 and CDKN1A, which regulate cell apoptosis and survival. Reactome analysis further elucidated these interactions, revealing CCN3’s direct interaction with POU domain transcription factors that modulate p53 and regulate the transcription of pro-apoptotic genes (Figure 6A). When the P53 pathway is activated in stem cells, it initiates a cascade of events leading to cell cycle arrest, commitment to a differentiation pathway, and the formation of progenitor cells. P53 serves as a guardian of genomic stability, and its activation is pivotal for maintaining cellular integrity (Levine et al., 2016). Furthermore, CCN3’s interaction with FGF2, a key factor in the FGF2 pathway governing reprogramming initiation and stabilization, provides additional evidence of its role in improving reprogramming efficiency. FGF2 accelerates cell proliferation during the initial phases of reprogramming, and it has been noted that hiPSCs generated in the presence of Activin/Nodal and FGF2 ligands tend to stabilize in the primed state (Jiao et al., 2013; Hawkins et al., 2014). In addition, FGF2 is known to influence the self-renewal and clonogenic capacity of pluripotent stem cells (Aprile et al., 2023). These findings highlight CCN3’s significance in orchestrating key signaling pathways that govern cellular fate and function. Its active participation in both the P53 and FGF2 pathways positions CCN3 as a central player in the intricate network of molecular events that determine cell behavior, stability, and responsiveness to environmental cues. Our results were further substantiated by pluripotency pathways extracted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, which highlighted the significance of the FGF2 pathway in governing reprogramming process in primed stem cells (Figure 8). Interestingly, the CCN3 gene has been documented to collaborate with FGF2 as a substrate, leading to enhanced cell proliferation and improved cell survival when they are combined (Lafont et al., 2005; van Roeyen et al., 2008). Reactome analysis also revealed CCN3’s direct interaction with FSR3, an adapter protein linking FGF receptor to downstream signaling pathways (Figure 6B).
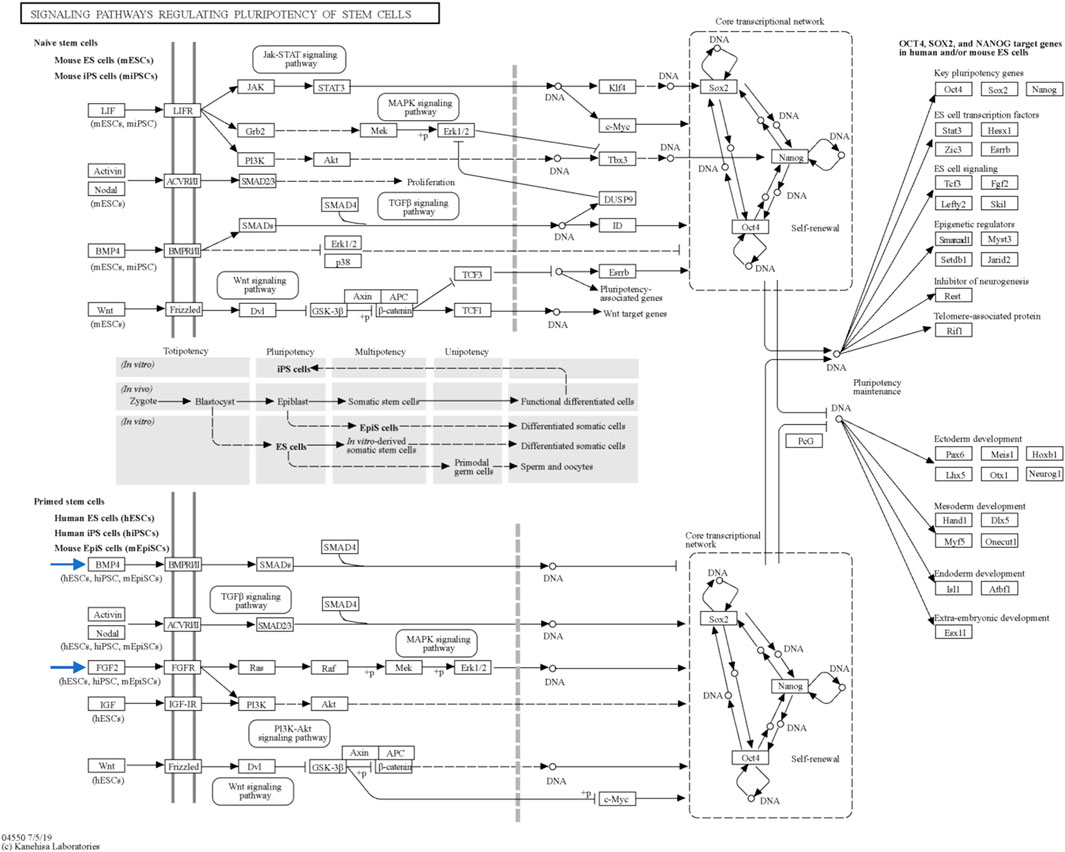
FIGURE 8. Key signaling pathways regulating stem cell pluripotency. This figure, sourced from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, provides an overview of the primary signaling pathways that govern the pluripotency of stem cells. The figure is interactive and can be explored in detail with links to pathway representations available at https://www.genome.jp/pathway/map04550. Of particular interest are the BMP4 and FGF2 pathways within primed stem cells (indicated by the blue arrows), which play significant roles in pluripotency regulation.
Moreover, CCN3 was found to enhance the expression of BMP-4, a protein critical for pluripotency initiation and maintenance through various mechanisms (Qi et al., 2004; Tan et al., 2012; Hawkins et al., 2014; Yu et al., 2020). KEGG database results highlighted the BMP-4 pathway’s involvement in governing reprogramming process in primed stem cells (Figure 8). Collectively, these findings strengthen our hypothesis regarding CCN3’s role in initiating and maintaining the reprogramming process, as well as its contribution to stabilizing and preserving reprogrammed cells from apoptosis.
The role of CCN3 in cellular stability and integrity emerges as a complex and multifaceted process, as evidenced by the findings from various studies employing overexpression and knockdown approaches. A study focused on CCN3 overexpression in bone marrow-derived stroma cell line (ST-2) cells revealed its inhibitory influence on BMP signaling, particularly preventing BMP-2 from phosphorylating Smad1/5/8, ultimately leading to the inhibition of osteoblastogenesis. This implies that CCN3 may play a crucial role in regulating bone homeostasis by modulating BMP signaling, a pathway fundamental to osteoblastic differentiation (Zuo et al., 2010), which in turn indicates the stability of ST-2 cells in its undifferentiated form. Moreover, CCN3’s involvement in the regulation of primitive hematopoietic stem cells adds another layer to its significance in cellular homeostasis. Studies employing CCN3 knockdown or forced expression in CD34+ cells have shown the necessity of CCN3 for the functional self-renewal of hematopoietic stem cells (Zuo et al., 2010).In a different investigation, CCN3 levels were modified by employing adenoviruses that either overexpressed CCN3 (Ad-CCN3) or interfered with CCN3 (Ad-siCCN3) in mouse embryonic fibroblasts (MEFs). This study aimed to examine the impacts and mechanisms of CCN3 on the process of osteoblastic differentiation. The findings indicated a notable hindrance of osteogenic differentiation in MEFs with higher CCN3 expression, primarily attributed to the suppression of BMP signaling and the reciprocal inhibition with DLL1(Su et al., 2018). Mechanistic analyses further uncovered CCN3’s suppressive effects on BMP/Smad and BMP/MAPK signaling pathways, along with its mutual inhibition with DLL1, a prominent membrane protein ligand associated with the Notch signaling pathway. Significantly, Hey1, a target gene shared by both BMP and Notch signaling pathways, emerged as a pivotal factor, partially counteracting the inhibitory impact of CCN3 on osteoblastic differentiation (Su et al., 2018). These regulatory roles of CCN3 in both bone formation and hematopoiesis underscores its vitality in maintaining cellular stability within distinct cellular contexts.
Another gene exclusively upregulated in the A53T-PD1 iPS cell line compared to other cell lines was PTHLH (59-fold, FDR <0.001). STRING analysis unveiled an unknown interaction between PTHLH and CDK2, which plays a role in the cell cycle and TP53 inhibition. Previous research has highlighted PTHLH as a crucial molecular regulator of cellular development and survival (Guo et al., 2012). Additionally, in Pthlh-depleted embryos at the morula stage, the expression of pluripotency-related genes NANOG and POU5F1 was markedly diminished (Min et al., 2022). While there is no direct evidence suggesting PTHLH’s involvement in histone acetylation during early embryonic development before implantation, previous research has indicated its direct influence on histone deacetylase 4 (HDAC4) (Kozhemyakina et al., 2009; Correa et al., 2010). Moreover, PTHLH exerts its influence through a complex network of signaling pathways involving ERK/MAPK pathways (Datta et al., 2005). ERK depletion has been associated with dysregulated pluripotency gene expression, decreased proliferation rates, G1 cell cycle arrest, increased apoptosis, faster telomere shortening, and impaired genomic stability (Chen et al., 2015). This data correlates well our data that PTHLH may play a role in increasing the cell survival and stability.
Reactome analysis confirmed PTHLH’s role in intracellular signaling, through its binding to GPCR leading to the activation of GNAS (Figure 7). G-proteins are essential regulators of cellular apoptotic cascades, with GNAS being linked to both proapoptotic and anti-apoptotic pathways (Melien, 2007; Yanamadala et al., 2009). GNAS overexpression has been shown to inhibit hydrogen peroxide-induced apoptosis by downregulating Bcl-xl (Zhao et al., 2006). Furthermore, endogenous GNAS activation through agonists has inhibited apoptosis mediated by cAMP production and PKA activation, leading to the inhibition of pro-apoptotic transcription factors such as AP-1, NF-κB, and NFAT (Choi et al., 2009). Interestingly, the Gs-alpha pathway has been proposed to regulate pluripotency in ESCs. Activation of Gs-alpha and amplification of cAMP levels have been shown to promote Oct4 expression, stimulating ESC self-renewal and pluripotency (Faherty et al., 2007; Layden et al., 2010). Therefore, we hypothesize that PTHLH may exert indirect influences on the expression of pluripotency-related genes, leading to more cellular stability, although the precise underlying mechanism warrants further investigation.
PTHLH has shown to be an upstream activator of GNAS which directly interacts with SRC, leading to SRC autophosphorylation and subsequent activation. SRC plays a diverse role in iPSCs, influencing key cellular processes such as survival, proliferation, and cell fate decisions. Its signaling pathways support iPSC self-renewal, prevent apoptosis, maintain stability, and impact commitment to specific lineages (Lu et al., 2007; Shoni et al., 2014; Zhang et al., 2014; Chaudhari et al., 2019). SRC also modulates pluripotency-associated pathways, including Wnt/β-catenin (Min et al., 2022), which is crucial for maintaining stem cell identity. During cellular reprogramming, SRC activation facilitates the transition to a pluripotent state, contributing to the erasure of cell identity (Ma et al., 2013; González and Huangfu, 2016). Additionally, SRC is associated with cellular senescence, potentially regulating senescence during reprogramming (Anerillas et al., 2022). The context-dependent nature of SRC in iPSCs, influenced by cellular context, reprogramming stage, and interactions with other pathways, underscores the dynamic complexity of its functions. Altogether, emphasize the important role of PTHLH in maintaining iPSC stability and integrity. Indeed, the downregulation of PTHLH emerged as a critical factor influencing cellular stability and integrity, particularly evident in a MYCN-amplified, TP53-mutated neuroblastoma cell line. The reduction of PTHLH levels led to a consequential decrease in MYCN expression. This event triggered a cascade of effects, including cell cycle arrest, induction of senescence, and impairment of migration and invasion capabilities within the neuroblastoma cell line (García et al., 2019). These findings highlight the intricate role of PTHLH in orchestrating key cellular processes and maintaining the equilibrium necessary for stability and integrity.
POSTN exhibited the highest upregulation, increasing by 70-fold (FDR <0.001), in A53T-PD1 iPS cell line compared to other iPS cell lines (Figure 3B). STRING analysis predicted that POSTN interacts with extracellular matrix components, including collagen-related proteins, Decorin and Tenascin C. Our findings align with previous research indicating that POSTN is highly expressed in a mouse ES cell line treated with Mek inhibitor PD0325901, in contrast to other untreated cell lines (Chen et al., 2015). PD is a pharmacological agent that inhibits the Mek/Erk signaling pathway, promoting self-renewal and pluripotency in mESCs (Chen et al., 2015). Additionally, POSTN has been associated with the augmentation of the Wnt signaling pathway in mouse breast cancer stem cells (Wang et al., 2013). This suggests a potential role for POSTN in maintaining self-renewal and pluripotency through the activation of the Wnt signaling pathway, which is essential for preserving naïve pluripotency and epigenetic stability in mESCs (de Jaime-Soguero et al., 2018; Theka et al., 2019).
Previous studies elucidate the intricate and context-dependent role of POSTN in cellular stability and integrity across various biological systems. In renal cell carcinoma (RCC), POSTN emerges as a key regulator, where its knockdown significantly suppresses epithelial-mesenchymal transition (EMT) through the IKL/AKT/mTOR pathway. Conversely, overexpression of POSTN facilitates EMT (Jia et al., 2021), indicating the crucial role of POSTN in cell stability and integrity. Moreover, overexpression of POSTN in the context of osteoblasts acts as a protective factor by impeding melatonin-induced apoptosis; hence, increase cell stability and integrity. This protective effect is achieved by inhibiting the eIF2α-ATF4 pathway through the suppression of the protein kinase R-like endoplasmic reticulum kinase (PERK) pathway (Zhu et al., 2021). These roles of POSTN highlights its versatile functions in maintaining cellular homeostasis. Furthermore, using mesenchymal stem cells (MSCs), POSTN plays a crucial role in the mineralization of the extracellular matrix (ECM) and supports tendon formation when overexpressed. This emphasizes POSTN’s involvement in ECM dynamics, which is essential for tissue development and integrity (Zhu et al., 2021).
In conclusion, our study sheds light on the potential roles of CCN3, PTHLH, and POSTN in maintaining genomic and cellular stability while preserving pluripotency in iPSCs. These findings warrant further investigations to unravel the precise mechanisms through which these genes contribute to these processes.
5 Conclusion
In summary, we identified three genes that were highly expressed in A53T-PD1 iPS cell line, from which robust and stable colonies were generated. The implications of these identified genes extend to potential selective interactions, which are vital for upholding the genomic integrity and maintaining pluripotency in both ESCs and iPSCs. Their functions may encompass activating DNA repair mechanisms, evading apoptosis through TP53 phosphorylation, and triggering signaling pathways that are crucial for preserving the pluripotent state of ESCs. To substantiate this conceptual framework and gain a deeper understanding of the underlying mechanisms, it becomes imperative to conduct thorough functional investigations. These endeavors hold the promise of unraveling crucial insights into the precise roles of these genes in ensuring genomic stability while upholding the self-renewal and pluripotency of the generated iPSCs.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies involving humans because these are iPS cell lines established in our lab, hence no need for ethical approval. In addition, ethical approval was obtained for the original research work (Swaidan et al., 2020; Scientific Reports) were we established these cell lines. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
NTS: Conceptualization, Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing. NHS: Conceptualization, Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing. ATA: Formal Analysis, Methodology, Validation, Writing–original draft. TD: Formal Analysis, Writing–original draft. RTA: Formal Analysis, Validation, Writing–original draft. AAA: Formal Analysis, Validation, Writing–original draft. RT: Validation, Writing–original draft. RA: Visualization, Writing–original draft. AH: Visualization, Writing–original draft. SS-A: Validation, Writing–original draft. SA: Writing–review and editing, Resources. AMA: Supervision, Writing–review and editing. ME: Project administration, Supervision, Writing–review and editing, Conceptualization, Data curation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Qatar University funded the publication of this article. This research work was supported by QRDI [grant number: UREP27-156-3-044].
Acknowledgments
We extend our gratitude to Qatar University which funded the publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anerillas, C., Herman, A. B., Rossi, M., Munk, R., Lehrmann, E., Martindale, J. L., et al. (2022). Early SRC activation skews cell fate from apoptosis to senescence. Sci. Adv. 8 (14), eabm0756. doi:10.1126/sciadv.abm0756
Aprile, D., Alessio, N., Squillaro, T., Di Bernardo, G., Peluso, G., and Galderisi, U. (2023). Role of glycosphingolipid SSEA-3 and FGF2 in the stemness and lineage commitment of multilineage differentiating stress enduring (MUSE) cells. Cell Prolif. 56 (1), e13345. doi:10.1111/cpr.13345
Bogliotti, Y. S., Wu, J., Vilarino, M., Okamura, D., Alba Soto, D., Zhong, C., et al. (2018). Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. 115 (9), 2090–2095. doi:10.1073/pnas.1716161115
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
Chaudhari, P., Tian, L., Kim, A., Zhu, Q., Anders, R., Schwarz, K. B., et al. (2019). Transient c-src suppression during endodermal commitment of human induced pluripotent stem cells results in abnormal profibrotic cholangiocyte-like cells. Stem cells Dayt. Ohio) 37 (3), 306–317. doi:10.1002/stem.2950
Chen, H., Guo, R., Zhang, Q., Guo, H., Yang, M., Wu, Z., et al. (2015). Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. 112 (44), E5936–E5943. doi:10.1073/pnas.1516319112
Choi, Y. J., Kim, S. Y., Oh, J. M., and Juhnn, Y. S. (2009). Stimulatory heterotrimeric G protein augments gamma ray-induced apoptosis by up-regulation of Bak expression via CREB and AP-1 in H1299 human lung cancer cells. Exp. Mol. Med. 41 (8), 592–600. doi:10.3858/emm.2009.41.8.065
Correa, D., Hesse, E., Seriwatanachai, D., Kiviranta, R., Saito, H., Yamana, K., et al. (2010). Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev. Cell 19 (4), 533–546. doi:10.1016/j.devcel.2010.09.008
Datta, N. S., Chen, C., Berry, J. E., and McCauley, L. K. (2005). PTHrP signaling targets cyclin D1 and induces osteoblastic cell growth arrest. J. Bone Min. Res. 20 (6), 1051–1064. doi:10.1359/jbmr.050106
de Jaime-Soguero, A., Abreu de Oliveira, W. A., and Lluis, F. (2018). The pleiotropic effects of the canonical Wnt pathway in early development and pluripotency. Genes (Basel) 9 (2), 93. doi:10.3390/genes9020093
Derveaux, S., Vandesompele, J., and Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR. Methods (San Diego, Calif.) 50 (4), 227–230. doi:10.1016/j.ymeth.2009.11.001
Dimos, J. T., Rodolfa, K. T., Niakan, K. K., Weisenthal, L. M., Mitsumoto, H., Chung, W., et al. (2008). Induced pluripotent stem cells generated from patients with ALS can Be differentiated into motor neurons. Science 321 (5893), 1218–1221. doi:10.1126/science.1158799
Faherty, S., Fitzgerald, A., Keohan, M., and Quinlan, L. R. (2007). Self-renewal and differentiation of mouse embryonic stem cells as measured by Oct4 expression: the role of the cAMP/PKA pathway. Vitro Cell Dev. Biol. Anim. 43 (1), 37–47. doi:10.1007/s11626-006-9001-5
García, M., Rodríguez-Hernández, C. J., Mateo-Lozano, S., Pérez-Jaume, S., Gonçalves-Alves, E., Lavarino, C., et al. (2019). Parathyroid hormone-like hormone plays a dual role in neuroblastoma depending on PTH1R expression. Mol. Oncol. 13 (9), 1959–1975. doi:10.1002/1878-0261.12542
González, F., and Huangfu, D. (2016). Mechanisms underlying the formation of induced pluripotent stem cells. Wiley Interdiscip. Rev. Dev. Biol. 5 (1), 39–65. doi:10.1002/wdev.206
Guo, L., Qi, S. T., Miao, D. Q., Liang, X. W., Li, H., Ou, X. H., et al. (2012). The roles of parathyroid hormone-like hormone during mouse preimplantation embryonic development. PLoS One 7 (7), e40528. doi:10.1371/journal.pone.0040528
Hawkins, K., Joy, S., and McKay, T. (2014). Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J. Stem Cells 6 (5), 620–628. doi:10.4252/wjsc.v6.i5.620
Hong, H., Takahashi, K., Ichisaka, T., Aoi, T., Kanagawa, O., Nakagawa, M., et al. (2009). Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460 (7259), 1132–1135. doi:10.1038/nature08235
Jia, Y. Y., Yu, Y., and Li, H. J. (2021). POSTN promotes proliferation and epithelial-mesenchymal transition in renal cell carcinoma through ILK/AKT/mTOR pathway. J. Cancer 12 (14), 4183–4195. doi:10.7150/jca.51253
Jiao, J., Dang, Y., Yang, Y., Gao, R., Zhang, Y., Kou, Z., et al. (2013). Promoting reprogramming by FGF2 reveals that the extracellular matrix is a barrier for reprogramming fibroblasts to pluripotency. Stem Cells 31 (4), 729–740. doi:10.1002/stem.1318
Kawamura, T., Suzuki, J., Wang, Y. V., Menendez, S., Morera, L. B., Raya, A., et al. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460 (7259), 1140–1144. doi:10.1038/nature08311
Kozhemyakina, E., Cohen, T., Yao, T. P., and Lassar, A. B. (2009). Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol. Cell Biol. 29 (21), 5751–5762. doi:10.1128/mcb.00415-09
Kubota, S., and Takigawa, M. (2013). The CCN family acting throughout the body: recent research developments. Biomol. Concepts 4 (5), 477–494. doi:10.1515/bmc-2013-0018
Lafont, J., Thibout, H., Dubois, C., Laurent, M., and Martinerie, C. (2005). NOV/CCN3 induces adhesion of muscle skeletal cells and cooperates with FGF2 and IGF-1 to promote proliferation and survival. Cell Commun. Adhesion 12 (1-2), 41–57. doi:10.1080/15419060500383069
Layden, B. T., Newman, M., Chen, F., Fisher, A., and Lowe, W. L. (2010). G protein coupled receptors in embryonic stem cells: a role for Gs-alpha signaling. PLoS One 5 (2), e9105. doi:10.1371/journal.pone.0009105
Levine, A. J., Puzio-Kuter, A. M., Chan, C. S., and Hainaut, P. (2016). The role of the p53 protein in stem-cell biology and epigenetic regulation. Cold Spring Harb. Perspect. Med. 6 (9), a026153. doi:10.1101/cshperspect.a026153
Lin, C. G., Chen, C. C., Leu, S. J., Grzeszkiewicz, T. M., and Lau, L. F. (2005). Integrin-dependent functions of the angiogenic inducer NOV (CCN3): implication in wound healing. J. Biol. Chem. 280 (9), 8229–8237. doi:10.1074/jbc.M404903200
Lu, M., Glover, C. H., Tien, A. H., Humphries, R. K., Piret, J. M., and Helgason, C. D. (2007). Involvement of tyrosine kinase signaling in maintaining murine embryonic stem cell functionality. Exp. Hematol. 35 (8), 1293–1302. doi:10.1016/j.exphem.2007.04.010
Ma, T., Xie, M., Laurent, T., and Ding, S. (2013). Progress in the reprogramming of somatic cells. Circulation Res. 112 (3), 562–574. doi:10.1161/CIRCRESAHA.111.249235
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M., and Gilad, Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18 (9), 1509–1517. doi:10.1101/gr.079558.108
Martins-Taylor, K., and Xu, R. H. (2012). Concise review: genomic stability of human induced pluripotent stem cells. Stem Cells 30 (1), 22–27. doi:10.1002/stem.705
Melien, O. (2007). Heterotrimeric G proteins and disease. Methods Mol. Biol. 361, 119–144. doi:10.1385/1-59745-208-4:119
Min, J. K., Park, H. S., Lee, Y. B., Kim, J. G., Kim, J. I., and Park, J. B. (2022). Cross-talk between Wnt signaling and Src tyrosine kinase. Biomedicines 10 (5), 1112. doi:10.3390/biomedicines10051112
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. methods 5 (7), 621–628. doi:10.1038/nmeth.1226
Nagaria, P., Robert, C., and Rassool, F. V. (2013). DNA double-strand break response in stem cells: mechanisms to maintain genomic integrity. Biochim. Biophys. Acta 1830 (2), 2345–2353. doi:10.1016/j.bbagen.2012.09.001
Qi, X., Li, T. G., Hao, J., Hu, J., Wang, J., Simmons, H., et al. (2004). BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U. S. A. 101 (16), 6027–6032. doi:10.1073/pnas.0401367101
Shoni, M., Lui, K. O., Vavvas, D. G., Muto, M. G., Berkowitz, R. S., Vlahos, N., et al. (2014). Protein kinases and associated pathways in pluripotent state and lineage differentiation. Curr. stem Cell Res. Ther. 9 (5), 366–387. doi:10.2174/1574888x09666140616130217
Soldner, F., Hockemeyer, D., Beard, C., Gao, Q., Bell, G. W., Cook, E. G., et al. (2009). Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136 (5), 964–977. doi:10.1016/j.cell.2009.02.013
Stefanis, L. (2012). α-Synuclein in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2 (2), a009399. doi:10.1101/cshperspect.a009399
Su, J., Zhu, D., Huo, Z., Gingold, J. A., Ang, Y. S., Tu, J., et al. (2019). Genomic integrity safeguards self-renewal in embryonic stem cells. Cell Rep. 28 (6), 1400–1409. doi:10.1016/j.celrep.2019.07.011
Su, X., Wei, Y., Cao, J., Wu, X., Mou, D., Luo, J., et al. (2018). CCN3 and DLL1 co-regulate osteogenic differentiation of mouse embryonic fibroblasts in a Hey1-dependent manner. Cell Death Dis. 9 (12), 1188. doi:10.1038/s41419-018-1234-1
Swaidan, N. T., Salloum-Asfar, S., Palangi, F., Errafii, K., Soliman, N. H., Aboughalia, A. T., et al. (2020). Identification of potential transcription factors that enhance human iPSC generation. Sci. Rep. 10 (1), 21950. doi:10.1038/s41598-020-78932-9
Tan, T. W., Huang, Y. L., Chang, J. T., Lin, J. J., Fong, Y. C., Kuo, C. C., et al. (2012). CCN3 increases BMP-4 expression and bone mineralization in osteoblasts. J. Cell Physiol. 227 (6), 2531–2541. doi:10.1002/jcp.22991
Theka, I., Sottile, F., Aulicino, F., Garcia, A. C., and Cosma, M. P. (2017). Reduced expression of Paternally Expressed Gene-3 enhances somatic cell reprogramming through mitochondrial activity perturbation. Sci. Rep. 7 (1), 9705. doi:10.1038/s41598-017-10016-7
Theka, I., Sottile, F., Cammisa, M., Bonnin, S., Sanchez-Delgado, M., Di Vicino, U., et al. (2019). Wnt/β-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells. Sci. Rep. 9 (1), 948. doi:10.1038/s41598-018-37442-5
Vallacchi, V., Daniotti, M., Ratti, F., Di Stasi, D., Deho, P., De Filippo, A., et al. (2008). CCN3/Nephroblastoma overexpressed matricellular protein regulates integrin expression, adhesion, and dissemination in melanoma. Cancer Res. 68 (3), 715–723. doi:10.1158/0008-5472.can-07-2103
van Roeyen, C. R. C., Eitner, F., Scholl, T., Boor, P., Kunter, U., Planque, N., et al. (2008). CCN3 is a novel endogenous PDGF-regulated inhibitor of glomerular cell proliferation. Kidney Int. 73 (1), 86–94. doi:10.1038/sj.ki.5002584
Vitillo, L., and Kimber, S. J. (2017). Integrin and FAK regulation of human pluripotent stem cells. Curr. Stem Cell Rep. 3 (4), 358–365. doi:10.1007/s40778-017-0100-x
Wang, M., Tang, L., Liu, D., Ying, Q. L., and Ye, S. (2017). The transcription factor Gbx2 induces expression of Kruppel-like factor 4 to maintain and induce naive pluripotency of embryonic stem cells. J. Biol. Chem. 292 (41), 17121–17128. doi:10.1074/jbc.M117.803254
Wang, X., Jia, L., Wang, Z., Huang, Y., Liu, W., Zhu, X., et al. (2013). Periostin contributes to the acquisition of multipotent stem cell-like properties in human mammary epithelial cells and breast cancer cells. PLOS ONE 8 (8), e72962. doi:10.1371/journal.pone.0072962
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10 (1), 57–63. doi:10.1038/nrg2484
Xu, Z., Robitaille, A. M., Berndt, J. D., Davidson, K. C., Fischer, K. A., Mathieu, J., et al. (2016). Wnt/β-catenin signaling promotes self-renewal and inhibits the primed state transition in naïve human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 113 (42), E6382–e6390. doi:10.1073/pnas.1613849113
Yamanaka, S. (2012). Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10 (6), 678–684. doi:10.1016/j.stem.2012.05.005
Yanamadala, V., Negoro, H., and Denker, B. M. (2009). Heterotrimeric G proteins and apoptosis: intersecting signaling pathways leading to context dependent phenotypes. Curr. Mol. Med. 9 (5), 527–545. doi:10.2174/156652409788488784
Yu, L., Li, J., Hong, J., Takashima, Y., Fujimoto, N., Nakajima, M., et al. (2018). Low cell-matrix adhesion reveals two subtypes of human pluripotent stem cells. Stem Cell Rep. 11 (1), 142–156. doi:10.1016/j.stemcr.2018.06.003
Yu, S., Zhou, C., Cao, S., He, J., Cai, B., Wu, K., et al. (2020). BMP4 resets mouse epiblast stem cells to naive pluripotency through ZBTB7A/B-mediated chromatin remodelling. Nat. Cell Biol. 22 (6), 651–662. doi:10.1038/s41556-020-0516-x
Zhang, X., Simerly, C., Hartnett, C., Schatten, G., and Smithgall, T. E. (2014). Src-family tyrosine kinase activities are essential for differentiation of human embryonic stem cells. Stem Cell Res. 13 (3), 379–389. doi:10.1016/j.scr.2014.09.007
Zhao, C., Lai, J. S., Warsh, J. J., and Li, P. P. (2006). Galpha(s) sensitizes human SH-SY5Y cells to apoptosis independently of the protein kinase A pathway. J. Neurosci. Res. 84 (2), 389–397. doi:10.1002/jnr.20875
Zhu, D., Zhou, W., Wang, Z., Wang, Y., Liu, M., Zhang, G., et al. (2021). Periostin: an emerging molecule with a potential role in spinal degenerative diseases. Front. Med. 8, 694800. doi:10.3389/fmed.2021.694800
Keywords: iPSCs, ESCs, transcription factors, genomic integrity, cellular survival
Citation: Swaidan NT, Soliman NH, Aboughalia AT, Darwish T, Almeshal RO, Al-Khulaifi AA, Taha RZ, Alanany R, Hussein AY, Salloum-Asfar S, Abdulla SA, Abdallah AM and Emara MM (2024) CCN3, POSTN, and PTHLH as potential key regulators of genomic integrity and cellular survival in iPSCs. Front. Mol. Biosci. 11:1342011. doi: 10.3389/fmolb.2024.1342011
Received: 21 November 2023; Accepted: 11 January 2024;
Published: 05 February 2024.
Edited by:
Xiaojun Ren, University of Colorado Denver, United StatesReviewed by:
Shilpi Minocha, Indian Institute of Technology Delhi, IndiaDomenico Aprile, University of Campania Luigi Vanvitelli, Italy
Copyright © 2024 Swaidan, Soliman, Aboughalia, Darwish, Almeshal, Al-Khulaifi, Taha, Alanany, Hussein, Salloum-Asfar, Abdulla, Abdallah and Emara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed M. Emara, bWVtYXJhQHF1LmVkdS5xYQ==
†These authors have contributed equally to this work and share first authorship
 Nuha T. Swaidan
Nuha T. Swaidan Nada H. Soliman
Nada H. Soliman Ahmed T. Aboughalia
Ahmed T. Aboughalia Toqa Darwish1
Toqa Darwish1 Azhar A. Al-Khulaifi
Azhar A. Al-Khulaifi Rowaida Z. Taha
Rowaida Z. Taha Rania Alanany
Rania Alanany Ahmed Y. Hussein
Ahmed Y. Hussein Salam Salloum-Asfar
Salam Salloum-Asfar Sara A. Abdulla
Sara A. Abdulla Abdallah M. Abdallah
Abdallah M. Abdallah Mohamed M. Emara
Mohamed M. Emara