
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 24 January 2024
Sec. Molecular Diagnostics and Therapeutics
Volume 11 - 2024 | https://doi.org/10.3389/fmolb.2024.1327014
This article is part of the Research Topic 14-3-3 Proteins: Possible importance in Neurodegenerative Diseases View all 5 articles
Cell signaling regulates several physiological processes by receiving, processing, and transmitting signals between the extracellular and intracellular environments. In signal transduction, phosphorylation is a crucial effector as the most common posttranslational modification. Selectively recognizing specific phosphorylated motifs of target proteins and modulating their functions through binding interactions, the yeast 14-3-3 proteins Bmh1 and Bmh2 are involved in catabolite repression, carbon metabolism, endocytosis, and mitochondrial retrograde signaling, among other key cellular processes. These conserved scaffolding molecules also mediate crosstalk between ubiquitination and phosphorylation, the spatiotemporal control of meiosis, and the activity of ion transporters Trk1 and Nha1. In humans, deregulation of analogous processes triggers the development of serious diseases, such as diabetes, cancer, viral infections, microbial conditions and neuronal and age-related diseases. Accordingly, the aim of this review article is to provide a brief overview of the latest findings on the functions of yeast 14-3-3 proteins, focusing on their role in modulating the aforementioned processes.
The 14-3-3 protein family includes highly conserved acidic proteins of approximately 30 kDa in size. These proteins are abundantly expressed in all eukaryotes and often in multiple isoforms. The yeast Saccharomyces cerevisiae expresses only two isoforms, Bmh1 and Bmh2. Encoded by the BMH1 and BMH2 genes, these isoforms are essential in most laboratory strains (van Heusden et al., 1992). Disrupting one of the BMH genes has a negligible effect on yeast cells, whereas simultaneous deleting both genes is lethal, clearly indicating that these isoforms have redundant but nonetheless crucial functions (van Heusden et al., 1995).
Bmh proteins commonly function as adaptors in signal transduction pathways by binding phosphorylated proteins, thereby activating, inactivating or sequestering them (van Heusden and Yde Steensma, 2006; van Heusden, 2009). Both Bmh proteins form stable dimers, similar to the 14-3-3 protein isoforms in higher eukaryotes (Yaffe et al., 1997), with a clear preference for heterodimer formation (Chaudhri et al., 2003). A thorough proteomic analysis using multistep immunoaffinity purification and mass spectrometry has identified 271 yeast proteins that specifically interact with Bmh proteins in a phosphorylation-dependent manner (Kakiuchi et al., 2007). Phosphorylation-dependent interactions with Bmh proteins underlie a wide range of physiological roles (Obsilova and Obsil, 2022). Since protein phosphorylation regulates both cell cycle and metabolism, the analysis of the budding yeast cell cycle phosphoproteome has unsurprisingly revealed more than 200 sites on metabolic enzymes and transporters regulated by phosphorylation (Zhang et al., 2019). Among the thousands of sites whose phosphorylation increased during the cell cycle, both Cdk-phosphorylated and R-R-X-S motifs were highly enriched. Moreover, such motifs are phosphorylated by PKA and recognized by 14-3-3 proteins (Zhang et al., 2019). These data suggest that binding interactions with Bmh proteins participate in a number of key cellular processes.
The aim of this review is to provide an overview of the latest findings on the functions of yeast 14-3-3 proteins. Notwithstanding our efforts to update readers on the state of the art on the role of Bmh proteins in key yeast processes, they should also refer to excellent reviews on Bmh proteins previously published since the turn of the century (van Hemert et al., 2001; van Heusden, 2005; van Heusden and Yde Steensma, 2006; van Heusden, 2009; Conrad et al., 2014; Kumar, 2017).
Yeasts, unlike higher eukaryotes, usually express only one or two 14-3-3 protein isoforms. In the yeast S. cerevisiae, these isoforms are encoded by the BMH1 and BMH2 genes. In 1991, the gene for the Bmh1 protein was discovered by chance when the neighboring PDA1 gene was cloned, and this newly discovered protein was named brain modulosignaling homologue (Bmh) for its similarity to 14-3-3 proteins previously characterized in brain tissue and known to specifically modulate other proteins (van Heusden et al., 1992). Three years later, the second gene, BMH2, was identified by sequence homology (van Heusden et al., 1995; van Heusden, 2005).
Both Bmh proteins are interchangeable because deleting either gene has little effect on yeast cell viability, growth, sporulation efficiency, or even spore viability. However, simultaneous disrupting both BMH genes is lethal in most strains, suggesting that 14-3-3 proteins play key roles in yeast cellular processes (van Heusden et al., 1992; van Heusden and Yde Steensma, 2006). Further demonstrating that Bmh proteins have crucial functions in yeast, BMH1 is not required for yeast growth in rich medium, but deleting both BMH genes causes severe growth defects and increases sensitivity to various stressors (Kumar and Srivastava, 2016; Kumar, 2017).
As for other yeast 14-3-3 proteins, Schizosaccharomyces pombe also express two genes encoding 14-3-3 proteins. These proteins were named rad24 and rad25 because they were isolated for their association with radiation sensitivity (Ford et al., 1994). Similarly, the dimorphic yeast Yarrowia lipolytica also contains two 14-3-3 genes, YlBMH1 and YlBMH2 (Hurtado and Rachubinski, 2002). Conversely, the pathogenic yeast Candida albicans thrives despite expressing only one 14-3-3 gene (Cognetti et al., 2002). All yeast 14-3-3 proteins have nevertheless remained highly conserved during evolution, as did 14-3-3 isoforms of higher eukaryotes, albeit with some variability. This variability is persistently found in loops connecting individual α-helices and especially in the C-terminal stretch.
All 14-3-3 isoforms, including Bmh1/2 proteins, recognize three canonical phosphorylated motifs, namely R[S/Φ][+]pSXP (mode I) (Yaffe et al., 1997), RX[Φ/S][+]pSXP (mode II) (Rittinger et al., 1999) and pS/pT-X1–2-COOH (C-terminal mode III) (Ganguly et al., 2005), where pS is phosphoserine, Φ is an aromatic residue, + is a basic residue, and X is any type of residue (usually Leu, Glu, Ala, and Met). In addition to these canonical motifs, 14-3-3 proteins can also bind to motifs with considerably divergent features, including non-phosphorylated moieties (reviewed in (Obsilova and Obsil, 2022)). As shown by structural studies, 14-3-3 proteins form highly helical dimers, with each protomer consisting of nine antiparallel α-helices (H1-H9) (Eisenreichova et al., 2016; Alblova et al., 2017; Smidova et al., 2019). The shape of the 14-3-3 dimer molecule resembles that of a cup with approximate dimensions of 35 Å × 35 Å × 20 Å (width, length and depth) (Figure 1A). The 14-3-3 dimer can simultaneously bind to two phosphorylated motifs located either within the same target protein or in two different proteins thanks to two amphiphilic grooves formed by the α-helices H3, H5, H7 and H9 (Liu et al., 1995; Xiao et al., 1995; Obsil et al., 2001; Obsil et al., 2003; Veisova et al., 2012).
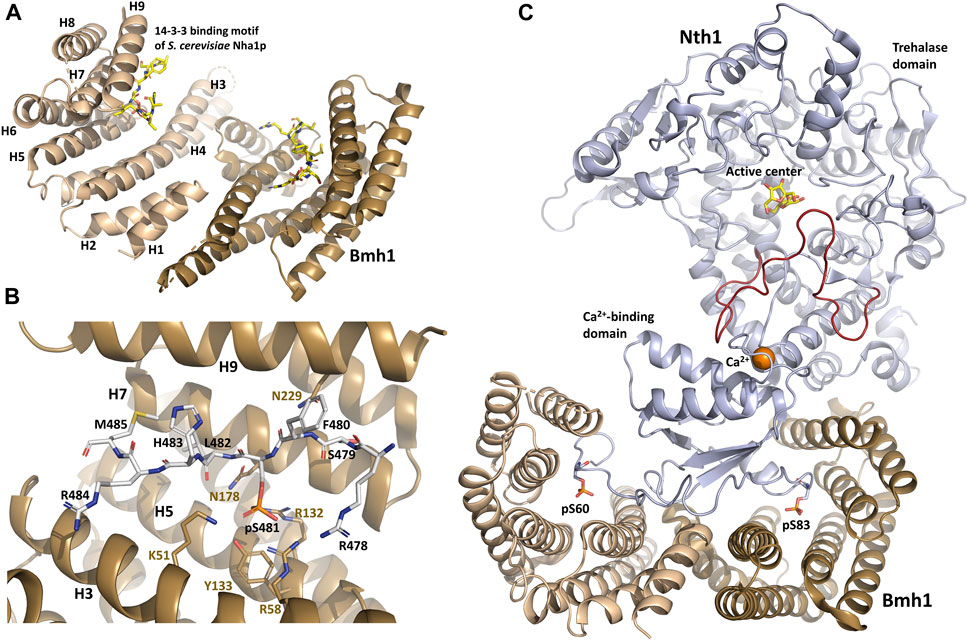
FIGURE 1. Structure of the yeast Bmh1 protein. (A) Crystal structure of the Saccharomyces cerevisiae Nha1p (Na+/H+ antiporter) 14-3-3 binding motif Ser481 bound to Bmh1 dimer (PDB ID: 6QK8) (Smidova et al., 2019). (B) Detailed view of interactions between the ligand binding groove of Bmh1 and the 14-3-3 binding motif Ser481 of Saccharomyces cerevisiae Nha1p (RSFpS481LHRM). (C) Crystal structure of the Saccharomyces cerevisiae Nth1:Bmh1 complex (PDB ID: 5N6N) (Alblova et al., 2017). The flexible part of the catalytic trehalase domain that is stabilized by the formation of the complex is shown in red. The figure was prepared with PyMOL (https://pymol.org/2/).
When bound to various 14-3-3 isoforms, short synthetic phosphopeptides adopt an extended conformation with a fixed orientation in the ligand binding groove of 14-3-3, without inducing substantial structural changes in the 14–3-3 dimer (Yaffe et al., 1997; Rittinger et al., 1999; Yang et al., 2006; Ottmann et al., 2007; Sluchanko and Gusev, 2017; Psenakova et al., 2018; Kalabova et al., 2020; Horvath et al., 2021; Pohl et al., 2021). The phosphate group of pSer/pThr binds to a positively charged pocket within the amphipatic ligand-binding groove of 14-3-3 formed by conserved residues Lys51, Arg58, Arg132 and Tyr133 (Bmh1 numbering) located in helices H3 and H5 (Figure 1B). In addition, the Pro residue often found at position +2 with respect to pSer/pThr in 14-3-3 ligand-binding motifs creates a kink in the polypeptide chain. This abrupt change in the direction of the polypeptide chain enables its exit from the binding groove, thereby enhancing binding affinity and stabilizing 14-3-3 protein-ligand interactions (Yaffe et al., 1997; Rittinger et al., 1999).
Although Bmh1 and Bmh2 show a high degree of homology to 14-3-3 proteins of other organisms, they differ from the isoforms of higher eukaryotes in the sequence and length of their C-terminal segment (van Heusden and Yde Steensma, 2006; van Heusden, 2009). The C-terminal segment of Bmh proteins is considerably longer and contains a polyglutamine repeat with an unknown function. Bmh1 contains a stretch of 10 consecutive glutamines, whereas Bmh2 contains as many as 17 consecutive glutamines. Regardless of their length, these polyQ stretches have prevailed during evolution and may be related to the ability to interact with prion-like domains, as previously shown for other polyQ-containing proteins (Ripaud et al., 2014; Gerbich and Gladfelter, 2021). These features prevent the C-terminal segment from interacting with the ligand binding groove and functioning as an auto-inhibitor, as described for some mammalian isoforms (Truong et al., 2002; Obsilova et al., 2004; Silhan et al., 2004; Veisova et al., 2010).
14-3-3 proteins regulate their binding partners through several mechanisms, including conformational modulation of the ligand. When the protein bound to 14-3-3 is an enzyme, this interaction can modulate its enzymatic activity. Such modulation is exemplified by the activation of the yeast neutral trehalase Nth1, as discussed in Section 3.2. 14-3-3 binding can also mask the binding site(s) of other proteins or binding partners, such as an RNA-binding site, in regulating Rim4 during meiosis (Zhang et al., 2023), as discussed in Section 3.3. And in yet another mechanism, 14-3-3 proteins regulate the subcellular localization of their binding partners, as in the regulation of the mitochondrial retrograde (RTG) signaling pathway (Rios-Anjos et al., 2017) and the stress-responsive transcriptional activators Msn2 and Msn4 (Beck and Hall, 1999), which are discussed below, in Sections 3.6 and 3.7, respectively.
Glucose repression (also known as catabolite repression) is an essential mechanism for budding yeast whereby high levels of glucose and fructose, the preferred carbon sources, repress the expression of enzymes required for the catabolism of other carbon sources (Gancedo, 1998). The key component of the glucose repression pathway is the protein kinase Snf1, the yeast homolog of AMP-activated protein kinase (AMPK) (Carlson, 1999). Snf1 is inhibited by glucose and activated in its absence by phosphorylation of Thr210 in the activation loop (Rubenstein et al., 2008). This phosphorylation is promoted by the activity of three partly redundant upstream kinases, namely Sak1, Tos3 and Elm1, and counteracted by protein phosphatase 1 (PP1) comprising the regulatory subunit Reg1 and the catalytic subunit Glc7 (Conrad et al., 2014). Activated Snf1 stimulates transcription of genes needed for growth on nonglucose carbon sources by phosphorylating Mig1 (a zinc finger and DNA binding protein that mediates glucose repression), thereby promoting its transport from the nucleus to the cytoplasm (De Vit et al., 1997). However, more recent studies have shown that Mig1 constantly shuttles between the nucleus and cytoplasm regardless of the glucose presence and Snf1 activation (Bendrioua et al., 2014; Wollman et al., 2017). Snf1 also controls the transcription of glucose-repressed genes, such as transcription factors Cat8 and Adr1, which subsequently control the expression of hundreds of glucose-repressed genes through direct binding and transcription activation (Bruckmann et al., 2004; Ichimura et al., 2004; Braun et al., 2014). Adr1 activity is regulated at multiple levels and affected by Bmh binding (Young et al., 2012).
The involvement of Bmh proteins in glucose repression was first revealed by their interaction with Reg1 (Mayordomo et al., 2003; Dombek et al., 2004). More specifically, Bmh proteins bind to the conserved 187-232 region within the N-terminal part of phosphorylated Reg1. This interaction is required to maintain complete repression, but Bmh proteins also function independently of Reg1 during glucose repression. As a case in point, simultaneous deletion of both genes encoding these proteins causes synergistic release from suppression, at least partly due to direct Bmh binding to the phosphorylated regulatory domain of promoter-bound transcription factor Adr1 (Figure 2A) (Braun et al., 2013).
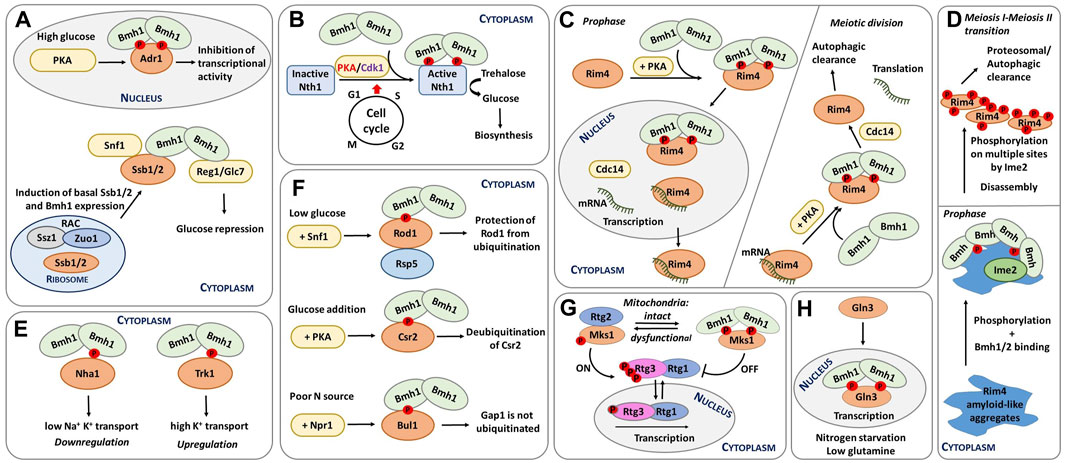
FIGURE 2. Schematic representation of selected functions of yeast 14-3-3 proteins. (A) Transcriptional regulation of glucose-repressed genes. Under high-glucose conditions, Adr1 is phosphorylated by PKA, which leads to Bmh binding. The formation of this complex inactivates Adr1 (Braun et al., 2013). In addition, Ssb1/2 and Bmh1 regulate Snf1, with Ssb directly interacting with Bmh. The resulting complex functions as a chaperone module of Snf1 (Hubscher et al., 2016; Hubscher et al., 2017). Furthermore, RAC is also regulates Snf1 activity by increasing Ssb1/2 and Bmh1/2 mRNA levels during growth on glucose (Hubscher et al., 2016; Yamada et al., 2023). (B) Regulation of neutral trehalase Nth1. Nth1 activity is regulated by the coordinated action of PKA, Cdk1 and Bmh during the cell cycle transition from G1 to S phase. Phosphorylation and Bmh binding activate Nth1, which subsequently converts trehalose into glucose for further biosynthetic events (Veisova et al., 2012; Dengler et al., 2021). (C) Spatiotemporal control of meiosis by 14-3-3 proteins. Left: before meiotic divisions, Cdc14 is upregulated; as a result, the Rim4-mRNA complex is transferred to the nucleus, and the Rim4-Bmh1/2 complex breaks down. Right: During meiotic anaphase, increased PKA activity releases Rim4 from mRNA, thus forming the Rim4-Bmh1/2 complex. In the cytoplasm, Cdc14 induces the disassembly of the Rim4-Bmh1/2 complex, Rim4 degradation, and translation of Rim4-sequestered mRNAs. Modified from (Herod et al., 2022; Zhang et al., 2023). (D) Bmh participate in the phosphorylation-mediated clearance of Rim4 aggregates during meiosis, thereby contributing to global protein aggregate homeostasis. Bmh proteins bind to Rim4 amyloid-like aggregates and facilitate Rim4 phosphorylation by the meiosis-specific kinase Ime2. This phosphorylation subsequently leads to the disassembly of Rim4 aggregates. Modified from (Herod et al., 2022). (E) Different mechanisms of regulation of yeast transporters by 14-3-3 proteins. 14-3-3 binding downregulates the cation efflux activity of Nha1 but upregulates the activity of the potassium transporter Trk1 (Smidova et al., 2019; Masaryk et al., 2023). (F) Endocytosis regulation. Under low glucose conditions, Rod1 is phosphorylated by Snf1 and inactivated by the formation of a complex with 14-3-3 proteins. This complex protects Rod1 from ubiquitination (Becuwe et al., 2012b). Adding glucose induces Csr2 phosphorylation by PKA and subsequent 14-3-3 protein binding, keeping Csr2 deubiquitylated (Hovsepian et al., 2017). Under low nitrogen levels, the Npr1 kinase phosphorylates the Bul adaptors, which are inhibited by 14-3-3 protein binding. 14-3-3 protein binding, in turn, prevents Gap1 ubiquitylation, maintaining this general amino acid permease active at the plasma membrane (Merhi and Andre, 2012). (G) Dynamic regulation of mitochondrial retrograde signaling. When cells have functional mitochondria, the RTG pathway is inactive, phosphorylated Rtg3 is bound to Rtg1 in the cytoplasm, and phosphorylated Mks1 is bound to 14-3-3. Dysfunctional mitochondria trigger the activation of the RTG pathway. Under these conditions, Mks1 is dephosphorylated, binding to Rtg2 and, thus, prompting the translocation of the Rtg1-Rtg3 complex into the nucleus and the transcription of genes involved in the retrograde response. Adapted from (Bui and Labedzka-Dmoch, 2023). (H) Regulation of rapamycin-mediated cell signaling and nitrogen catabolite repression. Upon rapamycin treatment, Gat1 and Gln3 translocate into the nucleus and activate the transcription of target genes. Upon nitrogen starvation and low glutamine, Gln3 remains in the nucleus bound to Bmh and mediates NCR-sensitive transcription (Tate et al., 2017).
Adr1 is a nutrient-regulated transcription factor that coactivates genes needed for growth in the absence of a fermentable carbon source. Under high-glucose conditions, Adr1 is phosphorylated by PKA, which enables Bmh binding to Adr1, preventing transcriptional activation by Adr1. Under low glucose conditions, Adr1 is activated by dephosphorylation by the protein phosphatase complex Glc7-Reg1 in a Snf1-dependent manner. In short, Bmh inhibits Adr1 at high glucose concentrations, and the Glc7-Reg1 complex overrides Adr1 repression at low glucose concentrations. Snf1 may phosphorylate and consequently activate Adr1 by phosphorylating a site or sites in the DNA-binding domain of Adr1 (Young et al., 2008; Ratnakumar et al., 2009), whereas PKA inactivates Adr1 by phosphorylating sites C-terminal to the DNA-binding domain, particularly Ser230, thus allowing Bmh binding. So, Bmh controls the transcriptional activation by Adr1 by directly binding to PKA-dependent phosphorylation sites, most likely modulating the ability of Adr1 to activate transcription (Parua et al., 2010; Parua and Young, 2014).
Another adaptor protein crucial for glucose repression through its interaction with Bmh is the yeast Hsp70 homolog Ssb1 (Dombek et al., 2004). As a chaperone, Ssb1 directly interacts with unfolded polypeptide substrates while residing on the ribosome, but half of the Ssb molecules remain in the cytoplasm (Peisker et al., 2010). In turn, cytosolic Ssb1/2 is associated with the Snf/Glc7 signaling pathway as a member of the Snf1 complex, which also contains Reg1 (Elbing et al., 2006). Ssb1/2 and Bmh1 post-translationally regulate Snf1, with Ssb directly interacting with Bmh and the resulting complex functioning as a novel chaperone module that modulates Snf1 (Hubscher et al., 2016; Hubscher et al., 2017).
The conserved yeast ribosome-associated complex (RAC) also regulates Snf1 activity (Hubscher et al., 2016; Zhang et al., 2017). This multiprotein complex is formed by Ssz1 (non-canonical Hsp70) and Zuo1 (J-protein). Zuo1 is also involved in glucose repression, possibly by increasing Ssb1/2 and Bmh1/2 mRNA levels during growth on glucose (Yamada et al., 2023). However, the exact mechanism whereby RAC and Ssb collaborate with Bmh to modulate Snf1 under various physiological conditions remains unknown. Further structural research must be conducted to elucidate the details of these interactions and their functional significance.
Yeast metabolism, growth and division must adapt to available nutrients. This adaption mechanism is mediated by cross-talk between nutrient signaling, metabolism, growth and the cell cycle (Ewald, 2018). In the G1 phase, excess nutrients can be stored as trehalose and glycogen. These storage carbohydrates are then used for biosynthetic processes in the S, G2 and M phases of the cell cycle (Figure 2B) (Crowe et al., 1984). Intracellular trehalose levels are therefore under strong cell cycle control.
This tight control is enabled by the cell cycle kinase Cdk1 and the metabolic regulator cAMP-dependent protein kinase (PKA). These two kinases phosphorylate and thus activate the neutral trehalase Nth1, the enzyme responsible for hydrolyzing trehalose into two glucose molecules (Ewald et al., 2016; Zhao et al., 2016; Solaki and Ewald, 2018). In contrast to trehalases from prokaryotic and higher eukaryotic organisms, yeast neutral trehalase Nth1 is regulated through a unique mechanism involving phosphorylation, calcium binding and association with Bmh proteins (Wera et al., 1999; Panni et al., 2008; Schepers et al., 2012; Veisova et al., 2012).
The N-terminal segment of Nth1 contains five sites, of which four are phosphorylated by PKA (Ser20, Ser21, Ser60 and Ser83) and one by Cdk1 (Ser66). Ser20 and Ser83 are constitutively phosphorylated, whereas Ser21 and Ser60 are phosphorylated to a lesser extent. Ser60 and Ser83 phosphorylation creates Bmh-binding motifs. However, under nutrient-poor conditions, with low PKA activity, Ser60 is not sufficiently phosphorylated; instead Nth1 activity is also regulated by Ser66 phosphorylation by Cdk1. This modification likely creates an alternative Bmh-binding motif (Dengler et al., 2021). Therefore, Cdk1 phosphorylates and increases Nth1 activity only when PKA activity is low.
Structural analysis of yeast Nth1 alone and in a complex with Bmh1 revealed that the formation of this complex establishes a suitable spatial arrangement of the catalytic and calcium-binding domains of Nth1by stabilizing the flexible portion of the catalytic domain (Figure 1C) and triggering Nth1 activity (Macakova et al., 2013; Kopecka et al., 2014; Alblova et al., 2017). The calcium ion bound to the calcium-binding domain also stabilizes the interaction between this domain and the flexible portion of the catalytic domain, thus further increasing trehalase activity.
In the plant pathogen Fusarium graminearum, the interaction between Nth1 and Bmh1 is enhanced by validamycin A, albeit through an unclear mechanism (Ren et al., 2022). Nevertheless, Nth1 activation by Bmh proteins is a classic example of a mode of action whereby 14-3-3 proteins modulate the enzymatic activity of the bound protein by changing the structure of its active site (Obsilova et al., 2014). In this case, though, Bmh1 binds to Nth1 far from the active center, so this allosteric regulation results from an induced conformational change.
Trehalose also enhances autophagy, thereby mediating neuroprotection in various animal models of Alzheimer´s and Parkinson´s disease (Sarkar et al., 2007; Castillo et al., 2013). When yeast cells face stress, one of the yeast enzymes that catalyze trehalose synthesis is trehalose-6-phosphate phosphatase (Tps2), which dephosphorylates trehalose-6-phosphate to trehalose (De Virgilio et al., 1993). Tps2 positively regulates autophagy in S. cerevisiae through processes involving Bmh proteins (Kim et al., 2021). Under nutrient-rich conditions, the Tor protein kinase complex 1 (TORC1) phosphorylates Rim15 kinase, thus inducing its interaction with Bmh1/2 and inactivation. Under nitrogen starvation, Rim15 is dephosphorylated by Tps2, leading to its dissociation from Bmh1/2 and translocation to the nucleus (Pedruzzi et al., 2003).
In meiosis, diploid cells divide into four daughter cells, each of which with half the number of chromosomes of the parent cell. As such, meiosis is a key process that produces haploid gametes for sexual reproduction through two rounds of cell division involving DNA replication (meiosis I) and subsequent segregation of the two chromosomes (meiosis II). Despite the importance of this process, only a few genes involved in meiosis have been identified so far (Tsuchiya et al., 2014; Ballew and Lacefield, 2019), including six novel regulators of meiosis commitment in budding yeast (Gavade et al., 2022), namely nutrient sensing regulator Bcy1, CDK-like kinase Ime2, Polo kinase Cdc5, RNA-binding protein Pes4 and Bmh1/2 (Gavade et al., 2022). Even more recently, two other genes have been implicated in this process–PKA and phosphatase Cdc14 (Zhang et al., 2023). Time will tell whether the limited number of genes associated with yeast meiosis reflects functional redundancy, species-specific differences or lack of knowledge on mechanisms of post-transcriptional regulation.
In S. cerevisiae, meiotic gene expression is tightly controlled by RNA-binding proteins, such as Rim4, a major suppressor of the translation of meiotic transcripts (Figure 2C) (Enyenihi and Saunders, 2003; Carpenter et al., 2018). During meiosis, the spatiotemporal regulation of the Rim4 ribonucleoprotein complex is controlled by PKA phosphorylation, yeast 14-3-3 protein binding and dephosphorylation by Cdc14 (Zhang et al., 2023). Accordingly, phosphorylated Rim4 forms a complex with the Bmh1/Bmh2 heterodimer, thereby preventing interactions between Rim4 and mRNA. Within the RNA-binding region of Rim4, two sites (Thr216 and Ser525) were identified as Bmh-binding motifs after phosphorylation by PKA. These are the primary motifs for robust Rim4-Bmh1/2 interactions during prophase I, when PKA-mediated phosphorylation of Rim4 peaks.
The conserved phosphatase Cdc14 upregulates meiosis-specific autophagy (Feng et al., 2022). Prior to meiotic division, Cdc14 mainly promotes the formation of the Rim4-mRNA complex to its nuclear localization. During meiotic anaphase, increased PKA activity facilitates Rim4 release from bound mRNA, which enables the formation of the Rim4-Bmh1/2 complex and subsequent Cdc14 upregulation and translocation from the nucleus to the cytoplasm. As a result, the Rim4-Bmh1/2 complex breaks down during anaphase.
In the cytoplasm, Cdc14 functions in opposition to PKA, removing Bmh1/2 from Rim4. This dissociation determines the timing of Rim4 degradation by autophagy due to the loss of protection by Bmh1/2 binding and of the translation of Rim4-sequestered mRNAs (Zhang et al., 2023). By regulating the interaction between Rim4 and Bmh1/2, and thus the formation of the Rim4-mRNA complex, spatiotemporally controlled Rim4 phosphorylation by PKA and dephosphorylation by Cdc14 phosphatase determine the distribution, function and stability of Rim4.
Some human and yeast RNA-binding proteins contain prion-like domains rich in Asn and Glu residues that form amyloid aggregates (King et al., 2012). For example, Rim4 has a prion domain and forms amyloid-like aggregates (Berchowitz et al., 2015) to which yeast 14-3-3 proteins bind during meiosis, facilitating Rim4 phosphorylation by the meiosis-specific kinase Ime2 (Herod et al., 2022). This phosphorylation subsequently mediates the disassembly of Rim4 aggregates and contributes to overall protein homeostasis and cellular protection against pathological protein aggregates (Figure 2D). In addition to potentially protecting cells against pathological protein aggregation and age-related diseases, 14-3-3 proteins may also play a key role in Rim4 biology because Bmh proteins contain polyQ stretches and the prion domains of Rim4 are also highly enriched in Glu and Asn residues. Therefore, Bmh proteins might have co-evolved with Rim4 to regulate aggregate disassembly.
Another protein involved in yeast meiosis is the RNA-binding protein Pes4, which regulates the timing and translation of several mRNAs during meiosis II progression after activation by Polo kinase (Esposito and Esposito, 1974; Tsuchiya et al., 2014; Jin et al., 2017). In this meiotic commitment model, Ime2 kinase phosphorylates and activates the middle meiosis transcription factor Ndt80 at the end of prophase I (Gavade et al., 2022). Ndt80 subsequently induces Cdc5, Pas4 and Ime2 gene expression. Bmh1 then directly interacts with Ndt80, Cdc5 and Pes4, stabilizing and maintaining Ndt80 levels, increasing Cdc5 kinase activity and modulating Pes4 to maintain meiotic commitment. In short, Bmh proteins are required to maintain normal Ndt80 levels, to activate Polo kinase and to interact with Pes4, which in turn regulates the timing of translation of several mRNAs important for meiosis II (Gavade et al., 2022). Moreover, these findings support a model in which Bmh proteins, alongside other proteins, control meiotic commitment by protecting cells from meiosis upon nutrient addition.
Another yeast process involving interactions with Bmh proteins is ion transport across the plasma membrane. In S. cerevisiae, for instance, intracellular cation and pH homeostasis are mediated by plasma membrane transporters, such as H+-ATPase, K+ uptake uniporter, K+ efflux channel, Na+(K+)/H+ antiporter and Na+(K+)-ATPase (Arino et al., 2010, 2019). Potassium storage is mediated by two independent transporters, Trk1 and Trk2 (Madrid et al., 1998). K+ is also actively exported from cells by the antiporter Nha1 and the ATPase Ena1, which concurrently export toxic sodium ions. Both Ena1 and Nha1 export four alkali metal cations (Na+, K+ and their analogs Rb+ and Li+), but only Ena1 uses ATP hydrolysis as an energy source. Nha1 uses, instead, the input H+ gradient generated by the H+-ATPase Pma1 (Kinclova et al., 2001; Ruiz and Arino, 2007).
14-3-3 proteins regulate several plasma-membrane ion transporters, including the Na+/H+ exchanger and the Arabidopsis thaliana potassium channel Tpk1 (Lehoux et al., 2001; Latz et al., 2007). In S. cerevisiae, the interaction between Bmh1 and Nha1 was validated at the protein level by bimolecular fluorescence complementation, which showed that this interaction enhances cell survival under salt stress (Zahradka et al., 2012). In subsequent in vivo and in vitro experiments, the phenotypes resulting from disruption of the interaction between Nha1 and Bmh1 were described, whereas biophysical characterization of the C-terminal portion of Nha1 revealed that Bmh binds to phosphorylated Ser481 and, to a lesser extent, to Ser479 motifs. Upon Bmh binding, the C-terminus of Nha1 transitions from a disordered to an ordered state. Because mutating Ser481 to Ala increases cation efflux activity, Bmh binding is likely necessary for Nha1 inhibition, which should be low under standard growth conditions when yeast need to accumulate high levels of K+ (Figure 2E) (Smidova et al., 2019).
Another transporter fully or partly regulated by phosphorylation through Bmh binding is the potassium transporter Trk1. However, unlike the aforementioned negative regulation of Nha1, Bmh binding activates Trk1. At low K+ levels, Trk1 activity depends on two residues of the second intracellular loop (Ser882 and Thr900), but Thr900 phosphorylation alone is responsible for Bmh binding, in line with the finding that the Thr900-to-Ala mutation markedly slows growth under low K+ (Masaryk et al., 2023). Yet again, though, the precise molecular mechanisms whereby Bmh proteins modulate the function of Nha1 and Trk1 channels remain unknown.
Without endocytosis, yeast cells cannot adapt to environmental changes by inhibiting intracellular signaling, thus downregulating plasma membrane receptors (Horak, 2013). In S. cerevisiae, the endocytosis of transporters is mediated by their ubiquitylation by the ubiquitin ligase Rsp5 and its arrestin-related trafficking adaptor (Art) proteins (Polo and Di Fiore, 2008; Becuwe et al., 2012a; MacGurn et al., 2012; Merhi and Andre, 2012). The endocytic activity of Rsp5 adaptor proteins is modulated by phosphorylation and subsequent Bmh binding, as shown in several studies.
The arrestin Rod1 (Art4), for example, is part of the glucose signaling pathway involving the kinase Snf1 and the phosphatase Reg1-Glc7/PP1. In the presence of glucose, Rod1 is activated, but when yeast cells are grown in lactate medium, Rod1 is phosphorylated at Ser447 by Snf1 and kept in an inhibited state, sequestered by Bmh proteins (Figure 2F) (Shinoda and Kikuchi, 2007; Alvaro et al., 2016). In other words, phosphorylation prevents Rod1 ubiquitination and hence its activation.
In addition to activating Rod1, glucose also induces endocytosis of the lactate transporter Jen1, associated with its dephosphorylation by Glc7/PP1, and subsequent Jen1 ubiquitination by Rsp5 (Paiva et al., 2002; Becuwe et al., 2012b). Upon endocytosis or glucose depletion, Rod1 is again phosphorylated and bound to Bmh, leading to Rod1 dissociation from the trans-Golgi network, its relocalization to the cytosol and Jen1 recycling to the cell membrane (Becuwe and Leon, 2014). Thus, nutrient availability intrinsincally modulates the plasma membrane expression of nutrient transporters primarily by controlling their ubiquitination and subsequent endocytosis.
Rod1 also regulates the endocytosis of the glucose transporters Hxt1, Hxt3 and Hxt6 through a similar mechanism (via Snf1/Bmh). In addition to Rod1, other Art proteins that regulate Rsp5-dependent transporter downregulation, such as Rog3 (Art7), also bind to Bmh. These interactions are modulated by the carbon source (Andoh et al., 2002; Llopis-Torregrosa et al., 2016).
Another study has shown that the Art protein Csr2 (Art8) is regulated similarly to Rod1 and triggers high-affinity glucose endocytosis (Hovsepian et al., 2017). Csr2 is activated by ubiquitination, whereas glucose addition transcriptionally represses Csr2 to its inactive, deubiquitinated form. Moreover, glucose-induced Csr2 deubiquitination correlates with its association with Bmh in a PKA phosphorylation-dependent manner (Figure 2F).
Bul1 and Bul2 are two other Rsp5 adaptor proteins that interact with Bmh after their phosphorylation by the kinase Npr1 (Belgareh-Touze et al., 2008). Their interaction with Bmh protects the amino acid permease Gap1 from downregulation under low nitrogen conditions. Under high nitrogen conditions, conversely, Sit4 phosphatase dephosphorylates Bul1 and Bul2, which dissociate from Bmh proteins and are subsequently ubiquitylated, ultimately downregulating Gap1 (MacGurn et al., 2011; Merhi and Andre, 2012).
Mitochondrial retrograde (RTG) signaling is known in all eukaryotes, from yeast to humans, but its molecular mechanisms are highly diverse (Butow and Avadhani, 2004). S. cerevisiae was the first organism in which the RTG pathway was investigated because mitochondrial DNA is not required for its growth. The retrograde reaction is an adaptation of cellular metabolism to disrupted mitochondrial function (Trendeleva and Zvyagilskaya, 2018). Cells with dysfunctional mitochondria experience three main problems, namely glutamate deficiency, energy and adenosine triphosphate (ATP) deficiency, and increased levels of reactive oxygen species (Bui and Labedzka-Dmoch, 2023).
In yeast, RTG signaling is mediated by three transcription factors (Rtg1-3) (Liu and Butow, 2006). As long as mitochondria are functional, the RTG pathway remains switched off, phosphorylated Rtg3 stays bound to Rtg1, and their complex resides in the cytoplasm (Figure 2G). But when mitochondria are damaged, Rtg2 activates the Rtg1/3 complex, either directly through the histone acetyltransferase complex or indirectly through interaction with Mks1, a transcription regulator and a key negative modulator of the RTG pathway (Chiu et al., 2022). So under conditions of lower ATP levels, the RTG pathway becomes active due to Mks1 sequestration by Rtg2, which functions as a cytoplasmic sensor of this pathway (Zhang et al., 2013).
This Rtg2-Mks1 interaction also protects Mks1 from hyperphosphorylation and binding to Bmh1/2 proteins. In turn, Bmh1/2 binding prevents Mks1 degradation by the Skp, Cullin, F-box-containing (SCF) E3 ubiquitin ligase complex. When the RTG pathway becomes inactive again, Mks1 detaches from Rtg2 and binds to Bmh1/2, thereby inhibiting the transfer of the Rtg1/3 complex to the nucleus. Free Mks1 is then degraded by glucose repression resistant protein 1 (Grr1) of the SCF E3 ubiquitin ligase complex (Liu et al., 2005). Mks1 degradation ensures an efficient transition between Rtg2-Mks1 and Bmh1/2-Mks1 complexes. The RTG pathway is also inhibited by Lst8, a seven WD40-repeat protein required for targeting amino acid permeases to the plasma membrane in response to nutrient sensing through TOR signaling (Liu et al., 2001; Rios-Anjos et al., 2017; Bui and Labedzka-Dmoch, 2023).
Rapamycin is a small molecule that forms a complex with Fpr1 and Tor (target of rapamycin), thereby blocking their activity in TOR signaling pathway. In S. cerevisiae, TOR signaling programmed cell growth in response to nutrients, such as nitrogen and carbon (Beck and Hall, 1999). Treating yeast with rapamycin triggers a rapid and robust starvation response, including changes in the expression of many genes.
In addition to various other roles in cells, yeast 14-3-3 proteins are also involved in rapamycin-mediated cell signaling by binding to the transcription factors Msn2, Msn4 and Rtg3 and their sequestration in the cytoplasm (Bertram et al., 1998). Upon rapamycin treatment, these transcription factors are released and transferred to the nucleus (Beck and Hall, 1999; van Heusden and Steensma, 2001). Therefore, by controlling the subcellular localization of aforementioned transcription factors in the cytoplasm with Bmh, the TOR signaling pathway regulates nutrient metabolism. Moreover, overexpressing both BMH genes represses the inhibitory effects of rapamycin, whereas deleting BMH genes increases the sensitivity of cells to rapamycin (Bertram et al., 1998).
Bmh1 and Bmh2 regulate rapamycin-mediated transcription differently. Both Bmh proteins are required for rapamycin-induced regulation of different but overlapping sets of genes as they associate with the promoters of at least some of these genes. However, only BMH2 suppresses genes implicated in ribosome biogenesis and blocks the activation of genes sensitive to nitrogen catabolite repression (NCR) (Trembley et al., 2014). The ability of S. cerevisiae to prioritize high-quality nitrogen sources over poor sources is based on the regulation of the nitrogen-responsive transcriptional activators Gln3 and Gat1 of the GATA family (Kulkarni et al., 2006).
Under high-nitrogen conditions, GATA factors are found in the cytoplasm and NCR-sensitive transcription is minimal. Conversely, when nitrogen levels are low, Gln3 is translocated into the nucleus, markedly increasing GATA factor-mediated transcription (Figure 2H). This regulation is based on TORC1-mediate control of Gln3 as rapamycin treatment induces Gat1 and Gln3 translocation into the nucleus, activating their transcriptional activity (Trembley et al., 2014).
Both Gat1 and Gln3 interact with Bmh1/2 proteins in vivo, suggesting their role in NCR regulation (Kakiuchi et al., 2007). But the subcellular localization of Gln3 is also regulated by the general amino acid control (GAAC) pathway mediated by the protein kinase Gcn2 (Tate et al., 2017). Together with the transcription factor Gcn4 and Bmh1/2, the GAAC pathway is required for the NCR-sensitive nuclear localization of Gln3 (Hinnebusch, 2005; Tate et al., 2017). Gcn2, Gcn4 or Bmh1/2 loss decreases Gln3 phosphorylation and therefore lowers NCR transcription although exactly how Bmh1/2 proteins control Gln3 localization and function is still unclear.
Bmh proteins may also be involved in regulating lifespan, apoptosis and heavy metal resistance. Nutrient-sensitive caloric restriction, TOR and PKA contribute to lifespan extension by enhancing the stress response, protecting cells from the accumulation of age-dependent oxidative damage. As a case in point, Bmh1 phosphorylation at Ser238 by PKA increases during chronological aging (Wang et al., 2009). Bmh1 removal then extends lifespan by activating the stress response, presumably by preventing inhibitory Bmh1 effects on these longevity factors, thereby extending lifespan.
Disrupted in many diseases, programmed cell death (PCD) is an important stress-induced process in which 14-3-3 proteins play various regulatory roles (Zhou et al., 2019). For instance, 14-3-3 proteins prevent the human proapoptotic protein Bax from inducing PCD in yeast (Clapp et al., 2012a; Clapp et al., 2012b). In addition, 14-3-3 proteins protect yeast cells against apoptosis induced by rapamycin, leucine deficiency and various stresses, such as cadmium or cycloheximide (Clapp et al., 2012b). Yet, when subjecting normal and PCD-resistant yeast cells to various stresses, cells expressing pro-survival 14-3-3 proteins are more resistant to rapamycin and copper-induced stress, but less so to other stresses such as iron exposure (Eid et al., 2017), suggesting the need for functional vacuoles, which mediate iron and 14-3-3 effects on PCD in yeast cells. In summary, PCD in yeast cells involves a complex interplay between processes that activate and inhibit responses to various stresses via 14-3-3 proteins.
The involvement of Bmh proteins in various signaling pathways has also been demonstrated by recent, large-scale screenings involving yeast collections with one or two deletions. One such study screened targets for improved cadmium tolerance in S. cerevisiae, identifying seven target genes, including BMH1 (Chen et al., 2021). However, cadmium treatment did not induce any significant reduction in BMH1 mRNA levels. Another study used transposon mutagenesis to investigate mutational tolerance and genome-wide genetic interactions in copy number variant (CNV) strains. These strains not only contribute to evolutionary adaptation but can also have harmful effects and cause CNV-associated diseases such as cancer. In three CNV strains, namely Aneu, ComQuad and Trip1A, a positive genetic interaction was identified between BMH1 and GAP1, and evidence suggests that some CNV strains have altered mitochondrial activity or function, enhanced mitochondrial mutational tolerance, and altered patterns of interactions with BMH1 (Avecilla et al., 2023). Bmh is a negative regulator of retrograde signaling; therefore, if mitochondria are dysfunctional and retrograde signaling is constitutively activated, mutations in BMH1 can be tolerated. Genetic approaches have also been used in another recent study on the MAPK signaling pathway, which controls fiber growth (Gonzalez et al., 2023). This study proposed the existence of a Ste20p-independent branch (Ste20p is a yeast p21-activated kinase) of this pathway involving adaptor protein Bem4p, whereas the Ste20p-dependent branch requires Bmh1/2 proteins. Moreover, a recent genome-scale screening in S. cerevisiae identified BMH1 among multicopy suppressors of the growth defect caused by glutathione stress (Yasukawa et al., 2023).
Thanks to recent advances in biochemical, structural, and bioinformatic methods of analysis, we can now better understand the various functions of 14-3-3 proteins, including how 14-3-3 proteins recognize their targets and control their localization and function. 14-3-3 proteins mediate various regulatory functions and are pivotal players in many signaling processes. Nevertheless, several aspects related to 14-3-3 function remain unresolved, particularly structural and functional details about the complexity and modulation of bound targets and the underlying mechanisms of their self-regulation. The main problem remains the lack of available structural data on yeast 14-3-3 protein complexes with full length binding partners. We can only hope that recent advances in cryo-EM methodology will increase the number of available structures of yeast 14-3-3 protein complexes with their full-length binding partners. Since yeast 14-3-3 proteins regulate carbon metabolism, glucose repression, and surface localization of sugar transporters, global protein homeostasis, they may be involved in metabolic diseases associated with sugar metabolism, including diabetes, age-related diseases, including Alzheimer´s disease, Parkinson´s disease and transthyretin amyloidosis as suggested by several studies (Thandavarayan et al., 2008; Watanabe et al., 2008; Wang et al., 2009; Herod et al., 2022) and reviewed in (Kleppe et al., 2011; Knowles et al., 2014; Rial et al., 2023). Further research on the yeast 14-3-3 protein complexes, including structural studies, may therefore help us to gain insights into the role of these key regulators of cellular pathways towards developing effective strategies for targeting and modulating their function.
VO: Writing–original draft, Writing–review and editing. TO: Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Research on 14-3-3 proteins is funded in our laboratory by the Czech Science Foundation (Grant No. 23-04686S) and the Czech Academy of Sciences (RVO:67985823 of the Institute of Physiology).
The authors thank Carlos V. Melo for editing the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alblova, M., Smidova, A., Docekal, V., Vesely, J., Herman, P., Obsilova, V., et al. (2017). Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1. Proc. Natl. Acad. Sci. U. S. A. 114 (46), E9811–E9820. doi:10.1073/pnas.1714491114
Alvaro, C. G., Aindow, A., and Thorner, J. (2016). Differential phosphorylation provides a switch to control how α-arrestin Rod1 down-regulates mating pheromone response in Saccharomyces cerevisiae. Genetics 203 (1), 299–317. doi:10.1534/genetics.115.186122
Andoh, T., Hirata, Y., and Kikuchi, A. (2002). PY motifs of Rod1 are required for binding to Rsp5 and for drug resistance. FEBS Lett. 525 (1-3), 131–134. doi:10.1016/s0014-5793(02)03104-6
Arino, J., Ramos, J., and Sychrova, H. (2010). Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74(1), 95–120. doi:10.1128/Mmbr.00042-09
Arino, J., Ramos, J., and Sychrova, H. (2019). Monovalent cation transporters at the plasma membrane in yeasts. Yeast 36 (4), 177–193. doi:10.1002/yea.3355
Avecilla, G., Spealman, P., Matthews, J., Caudal, E., Schacherer, J., and Gresham, D. (2023). Copy number variation alters local and global mutational tolerance. Genome Res. 33 (8), 1340–1353. doi:10.1101/gr.277625.122
Ballew, O., and Lacefield, S. (2019). The DNA damage checkpoint and the spindle position checkpoint maintain meiotic commitment in Saccharomyces cerevisiae. Curr. Biol. 29 (3), 449–460. doi:10.1016/j.cub.2018.12.043
Beck, T., and Hall, M. N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402 (6762), 689–692. doi:10.1038/45287
Becuwe, M., Herrador, A., Haguenauer-Tsapis, R., Vincent, O., and Leon, S. (2012a). Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem. Res. Int. 2012, 242764. doi:10.1155/2012/242764
Becuwe, M., and Leon, S. (2014). Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. Elife 3, e03307. doi:10.7554/eLife.03307
Becuwe, M., Vieira, N., Lara, D., Gomes-Rezende, J., Soares-Cunha, C., Casal, M., et al. (2012b). A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell. Biol. 196 (2), 247–259. doi:10.1083/jcb.201109113
Belgareh-Touze, N., Leon, S., Erpapazoglou, Z., Stawiecka-Mirota, M., Urban-Grimal, D., and Haguenauer-Tsapis, R. (2008). Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36 (Pt 5), 791–796. doi:10.1042/BST0360791
Bendrioua, L., Smedh, M., Almquist, J., Cvijovic, M., Jirstrand, M., Goksor, M., et al. (2014). Yeast AMP-activated protein kinase monitors glucose concentration changes and absolute glucose levels. J. Biol. Chem. 289 (18), 12863–12875. doi:10.1074/jbc.M114.547976
Berchowitz, L. E., Kabachinski, G., Walker, M. R., Carlile, T. M., Gilbert, W. V., Schwartz, T. U., et al. (2015). Regulated Formation of an amyloid-like translational repressor governs gametogenesis. Cell. 163 (2), 406–418. doi:10.1016/j.cell.2015.08.060
Bertram, P. G., Zeng, C., Thorson, J., Shaw, A. S., and Zheng, X. F. (1998). The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr. Biol. 8 (23), 1259–1267. doi:10.1016/s0960-9822(07)00535-0
Braun, K. A., Parua, P. K., Dombek, K. M., Miner, G. E., and Young, E. T. (2013). 14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 33 (4), 712–724. doi:10.1128/MCB.01226-12
Braun, K. A., Vaga, S., Dombek, K. M., Fang, F., Palmisano, S., Aebersold, R., et al. (2014). Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci. Signal 7 (333), ra64. doi:10.1126/scisignal.2005000
Bruckmann, A., Steensma, H. Y., Teixeira De Mattos, M. J., and Van Heusden, G. P. (2004). Regulation of transcription by Saccharomyces cerevisiae 14-3-3 proteins. Biochem. J. 382 (Pt 3), 867–875. doi:10.1042/BJ20031885
Bui, T. H. D., and Labedzka-Dmoch, K. (2023). RetroGREAT signaling: the lessons we learn from yeast. IUBMB Life 76, 26–37. doi:10.1002/iub.2775
Butow, R. A., and Avadhani, N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell. 14 (1), 1–15. doi:10.1016/s1097-2765(04)00179-0
Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2 (2), 202–207. doi:10.1016/S1369-5274(99)80035-6
Carpenter, K., Bell, R. B., Yunus, J., Amon, A., and Berchowitz, L. E. (2018). Phosphorylation-mediated clearance of amyloid-like assemblies in meiosis. Dev. Cell. 45 (3), 392–405. doi:10.1016/j.devcel.2018.04.001
Castillo, K., Nassif, M., Valenzuela, V., Rojas, F., Matus, S., Mercado, G., et al. (2013). Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9 (9), 1308–1320. doi:10.4161/auto.25188
Chaudhri, M., Scarabel, M., and Aitken, A. (2003). Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 300 (3), 679–685. doi:10.1016/s0006-291x(02)02902-9
Chen, Y., Liang, J., Chen, Z., Wang, B., and Si, T. (2021). Genome-scale screening and combinatorial optimization of gene overexpression targets to improve cadmium tolerance in Saccharomyces cerevisiae. Front. Microbiol. 12, 662512. doi:10.3389/fmicb.2021.662512
Chiu, S. T., Tseng, W. W., and Wei, A. C. (2022). Mathematical modeling and analysis of mitochondrial retrograde signaling dynamics. iScience 25 (12), 105502. doi:10.1016/j.isci.2022.105502
Clapp, C., Portt, L., Khoury, C., Sheibani, S., Eid, R., Greenwood, M., et al. (2012a). Untangling the roles of anti-apoptosis in regulating programmed cell death using humanized yeast cells. Front. Oncol. 2, 59. doi:10.3389/fonc.2012.00059
Clapp, C., Portt, L., Khoury, C., Sheibani, S., Norman, G., Ebner, P., et al. (2012b). 14-3-3 protects against stress-induced apoptosis. Cell. Death Dis. 3 (7), e348. doi:10.1038/cddis.2012.90
Cognetti, D., Davis, D., and Sturtevant, J. (2002). The Candida albicans 14-3-3 gene, BMH1, is essential for growth. Yeast 19 (1), 55–67. doi:10.1002/yea.804
Conrad, M., Schothorst, J., Kankipati, H. N., Van Zeebroeck, G., Rubio-Texeira, M., and Thevelein, J. M. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38 (2), 254–299. doi:10.1111/1574-6976.12065
Crowe, J. H., Crowe, L. M., and Chapman, D. (1984). Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science 223 (4637), 701–703. doi:10.1126/science.223.4637.701
Dengler, L., Ord, M., Schwab, L. M., Loog, M., and Ewald, J. C. (2021). Regulation of trehalase activity by multi-site phosphorylation and 14-3-3 interaction. Sci. Rep. 11 (1), 962. doi:10.1038/s41598-020-80357-3
De Virgilio, C., Burckert, N., Bell, W., Jeno, P., Boller, T., and Wiemken, A. (1993). Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur. J. Biochem. 212 (2), 315–323. doi:10.1111/j.1432-1033.1993.tb17664.x
De Vit, M. J., Waddle, J. A., and Johnston, M. (1997). Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 8 (8), 1603–1618. doi:10.1091/mbc.8.8.1603
Dombek, K. M., Kacherovsky, N., and Young, E. T. (2004). The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279 (37), 39165–39174. doi:10.1074/jbc.M400433200
Eid, R., Zhou, D. R., Arab, N. T. T., Boucher, E., Young, P. G., Mandato, C. A., et al. (2017). Heterologous expression of anti-apoptotic human 14-3-3β/α enhances iron-mediated programmed cell death in yeast. PLoS One 12 (8), e0184151. doi:10.1371/journal.pone.0184151
Eisenreichova, A., Klima, M., and Boura, E. (2016). Crystal structures of a yeast 14-3-3 protein from Lachancea thermotolerans in the unliganded form and bound to a human lipid kinase PI4KB-derived peptide reveal high evolutionary conservation. Acta Crystallogr. Sect. F. Struct. Biol. Commun. 72 (Pt 11), 799–803. doi:10.1107/S2053230X16015053
Elbing, K., McCartney, R. R., and Schmidt, M. C. (2006). Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 393 (Pt 3), 797–805. doi:10.1042/BJ20051213
Enyenihi, A. H., and Saunders, W. S. (2003). Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163 (1), 47–54. doi:10.1093/genetics/163.1.47
Esposito, R. E., and Esposito, M. S. (1974). Genetic recombination and commitment to meiosis in Saccharomyces. Proc. Natl. Acad. Sci. U. S. A. 71 (8), 3172–3176. doi:10.1073/pnas.71.8.3172
Ewald, J. C. (2018). How yeast coordinates metabolism, growth and division. Curr. Opin. Microbiol. 45, 1–7. doi:10.1016/j.mib.2017.12.012
Ewald, J. C., Kuehne, A., Zamboni, N., and Skotheim, J. M. (2016). The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol. Cell. 62 (4), 532–545. doi:10.1016/j.molcel.2016.02.017
Feng, W., Arguello-Miranda, O., Qian, S., and Wang, F. (2022). Cdc14 spatiotemporally dephosphorylates Atg13 to activate autophagy during meiotic divisions. J. Cell. Biol. 221 (5), e202107151. doi:10.1083/jcb.202107151
Ford, J. C., al-Khodairy, F., Fotou, E., Sheldrick, K. S., Griffiths, D. J., and Carr, A. M. (1994). 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265 (5171), 533–535. doi:10.1126/science.8036497
Gancedo, J. M. (1998). Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62 (2), 334–361. doi:10.1128/MMBR.62.2.334-361.1998
Ganguly, S., Weller, J. L., Ho, A., Chemineau, P., Malpaux, B., and Klein, D. C. (2005). Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. U. S. A. 102 (4), 1222–1227. doi:10.1073/pnas.0406871102
Gavade, J. N., Puccia, C. M., Herod, S. G., Trinidad, J. C., Berchowitz, L. E., and Lacefield, S. (2022). Identification of 14-3-3 proteins, Polo kinase, and RNA-binding protein Pes4 as key regulators of meiotic commitment in budding yeast. Curr. Biol. 32 (7), 1534–1547.e9. doi:10.1016/j.cub.2022.02.022
Gerbich, T. M., and Gladfelter, A. S. (2021). Moving beyond disease to function: physiological roles for polyglutamine-rich sequences in cell decisions. Curr. Opin. Cell. Biol. 69, 120–126. doi:10.1016/j.ceb.2021.01.003
Gonzalez, B., Mirzaei, M., Basu, S., Pujari, A. N., Vandermeulen, M. D., Prabhakar, A., et al. (2023). Turnover and bypass of p21-activated kinase during Cdc42-dependent MAPK signaling in yeast. J. Biol. Chem. 299 (11), 105297. doi:10.1016/j.jbc.2023.105297
Herod, S. G., Dyatel, A., Hodapp, S., Jovanovic, M., and Berchowitz, L. E. (2022). Clearance of an amyloid-like translational repressor is governed by 14-3-3 proteins. Cell. Rep. 39 (5), 110753. doi:10.1016/j.celrep.2022.110753
Hinnebusch, A. G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450. doi:10.1146/annurev.micro.59.031805.133833
Horak, J. (2013). Regulations of sugar transporters: insights from yeast. Curr. Genet. 59 (1-2), 1–31. doi:10.1007/s00294-013-0388-8
Horvath, M., Petrvalska, O., Herman, P., Obsilova, V., and Obsil, T. (2021). 14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites. Commun. Biol. 4 (1), 986. doi:10.1038/s42003-021-02518-y
Hovsepian, J., Defenouillere, Q., Albanese, V., Vachova, L., Garcia, C., Palkova, Z., et al. (2017). Multilevel regulation of an α-arrestin by glucose depletion controls hexose transporter endocytosis. J. Cell. Biol. 216 (6), 1811–1831. doi:10.1083/jcb.201610094
Hubscher, V., Mudholkar, K., Chiabudini, M., Fitzke, E., Wolfle, T., Pfeifer, D., et al. (2016). The Hsp70 homolog Ssb and the 14-3-3 protein Bmh1 jointly regulate transcription of glucose repressed genes in Saccharomyces cerevisiae. Nucleic Acids Res. 44 (12), 5629–5645. doi:10.1093/nar/gkw168
Hubscher, V., Mudholkar, K., and Rospert, S. (2017). The yeast Hsp70 homolog Ssb: a chaperone for general de novo protein folding and a nanny for specific intrinsically disordered protein domains. Curr. Genet. 63 (1), 9–13. doi:10.1007/s00294-016-0610-6
Hurtado, C. A. R., and Rachubinski, R. A. (2002). YlBMH1 encodes a 14-3-3 protein that promotes filamentous growth in the dimorphic yeast Yarrowia lipolytica. Microbiol. Read. 148 (Pt 11), 3725–3735. doi:10.1099/00221287-148-11-3725
Ichimura, T., Kubota, H., Goma, T., Mizushima, N., Ohsumi, Y., Iwago, M., et al. (2004). Transcriptomic and proteomic analysis of a 14-3-3 gene-deficient yeast. Biochemistry 43 (20), 6149–6158. doi:10.1021/bi035421i
Jin, L., Zhang, K., Sternglanz, R., and Neiman, A. M. (2017). Predicted RNA binding proteins Pes4 and Mip6 regulate mRNA levels, translation, and localization during sporulation in budding yeast. Mol. Cell. Biol. 37 (9), e00408-16. doi:10.1128/MCB.00408-16
Kakiuchi, K., Yamauchi, Y., Taoka, M., Iwago, M., Fujita, T., Ito, T., et al. (2007). Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46 (26), 7781–7792. doi:10.1021/bi700501t
Kalabova, D., Filandr, F., Alblova, M., Petrvalska, O., Horvath, M., Man, P., et al. (2020). 14-3-3 protein binding blocks the dimerization interface of caspase-2. FEBS J. 287 (16), 3494–3510. doi:10.1111/febs.15215
Kim, B., Lee, Y., Choi, H., and Huh, W. K. (2021). The trehalose-6-phosphate phosphatase Tps2 regulates ATG8 transcription and autophagy in Saccharomyces cerevisiae. Autophagy 17 (4), 1013–1027. doi:10.1080/15548627.2020.1746592
Kinclova, O., Ramos, J., Potier, S., and Sychrova, H. (2001). Functional study of the Saccharomyces cerevisiae Nha1p C-terminus. Mol. Microbiol. 40 (3), 656–668. doi:10.1046/j.1365-2958.2001.02412.x
King, O. D., Gitler, A. D., and Shorter, J. (2012). The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80. doi:10.1016/j.brainres.2012.01.016
Kleppe, R., Martinez, A., Doskeland, S. O., and Haavik, J. (2011). The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell. Dev. Biol. 22 (7), 713–719. doi:10.1016/j.semcdb.2011.08.008
Knowles, T. P., Vendruscolo, M., and Dobson, C. M. (2014). The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell. Biol. 15 (6), 384–396. doi:10.1038/nrm3810
Kopecka, M., Kosek, D., Kukacka, Z., Rezabkova, L., Man, P., Novak, P., et al. (2014). Role of the EF-hand-like motif in the 14-3-3 protein-mediated activation of yeast neutral trehalase Nth1. J. Biol. Chem. 289 (20), 13948–13961. doi:10.1074/jbc.M113.544551
Kulkarni, A., Buford, T. D., Rai, R., and Cooper, T. G. (2006). Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res. 6 (2), 218–229. doi:10.1111/j.1567-1364.2006.00031.x
Kumar, R. (2017). An account of fungal 14-3-3 proteins. Eur. J. Cell. Biol. 96 (2), 206–217. doi:10.1016/j.ejcb.2017.02.006
Kumar, R., and Srivastava, S. (2016). Quantitative proteomic comparison of stationary/G0 phase cells and tetrads in budding yeast. Sci. Rep. 6, 32031. doi:10.1038/srep32031
Latz, A., Becker, D., Hekman, M., Mueller, T., Beyhl, D., Marten, I., et al. (2007). TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J. 52 (3), 449–459. doi:10.1111/j.1365-313X.2007.03255.x
Lehoux, S., Abe, J., Florian, J. A., and Berk, B. C. (2001). 14-3-3 Binding to Na+/H+ exchanger isoform-1 is associated with serum-dependent activation of Na+/H+ exchange. J. Biol. Chem. 276 (19), 15794–15800. doi:10.1074/jbc.M100410200
Liu, D., Bienkowska, J., Petosa, C., Collier, R. J., Fu, H., and Liddington, R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376 (6536), 191–194. doi:10.1038/376191a0
Liu, Z., and Butow, R. A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185. doi:10.1146/annurev.genet.40.110405.090613
Liu, Z., Sekito, T., Epstein, C. B., and Butow, R. A. (2001). RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 20 (24), 7209–7219. doi:10.1093/emboj/20.24.7209
Liu, Z., Spirek, M., Thornton, J., and Butow, R. A. (2005). A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol. Biol. Cell. 16 (10), 4893–4904. doi:10.1091/mbc.e05-06-0516
Llopis-Torregrosa, V., Ferri-Blazquez, A., Adam-Artigues, A., Deffontaines, E., van Heusden, G. P., and Yenush, L. (2016). Regulation of the yeast Hxt6 hexose transporter by the Rod1 α-arrestin, the Snf1 protein kinase, and the Bmh2 14-3-3 protein. J. Biol. Chem. 291 (29), 14973–14985. doi:10.1074/jbc.M116.733923
Macakova, E., Kopecka, M., Kukacka, Z., Veisova, D., Novak, P., Man, P., et al. (2013). Structural basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1. Biochim. Biophys. Acta 1830 (10), 4491–4499. doi:10.1016/j.bbagen.2013.05.025
MacGurn, J. A., Hsu, P. C., and Emr, S. D. (2012). Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259. doi:10.1146/annurev-biochem-060210-093619
MacGurn, J. A., Hsu, P. C., Smolka, M. B., and Emr, S. D. (2011). TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 147 (5), 1104–1117. doi:10.1016/j.cell.2011.09.054
Madrid, R., Gomez, M. J., Ramos, J., and Rodriguez-Navarro, A. (1998). Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273 (24), 14838–14844. doi:10.1074/jbc.273.24.14838
Masaryk, J., Kale, D., Pohl, P., Ruiz-Castilla, F. J., Zimmermannova, O., Obsilova, V., et al. (2023). The second intracellular loop of the yeast Trk1 potassium transporter is involved in regulation of activity, and interaction with 14-3-3 proteins. Comput. Struct. Biotechnol. J. 21, 2705–2716. doi:10.1016/j.csbj.2023.04.019
Mayordomo, I., Regelmann, J., Horak, J., and Sanz, P. (2003). Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 544 (1-3), 160–164. doi:10.1016/s0014-5793(03)00498-8
Merhi, A., and Andre, B. (2012). Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32 (22), 4510–4522. doi:10.1128/MCB.00463-12
Obsil, T., Ghirlando, R., Anderson, D. E., Hickman, A. B., and Dyda, F. (2003). Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry 42 (51), 15264–15272. doi:10.1021/bi0352724
Obsil, T., Ghirlando, R., Klein, D. C., Ganguly, S., and Dyda, F. (2001). Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 105 (2), 257–267. doi:10.1016/s0092-8674(01)00316-6
Obsilova, V., Herman, P., Vecer, J., Sulc, M., Teisinger, J., and Obsil, T. (2004). 14-3-3zeta C-terminal stretch changes its conformation upon ligand binding and phosphorylation at Thr232. J. Biol. Chem. 279 (6), 4531–4540. doi:10.1074/jbc.M306939200
Obsilova, V., Kopecka, M., Kosek, D., Kacirova, M., Kylarova, S., Rezabkova, L., et al. (2014). Mechanisms of the 14-3-3 protein function: regulation of protein function through conformational modulation. Physiol. Res. 63 (Suppl. 1), S155–S164. doi:10.33549/physiolres.932659
Obsilova, V., and Obsil, T. (2022). Structural insights into the functional roles of 14-3-3 proteins. Front. Mol. Biosci. 9, 1016071. doi:10.3389/fmolb.2022.1016071
Ottmann, C., Marco, S., Jaspert, N., Marcon, C., Schauer, N., Weyand, M., et al. (2007). Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell. 25 (3), 427–440. doi:10.1016/j.molcel.2006.12.017
Paiva, S., Kruckeberg, A. L., and Casal, M. (2002). Utilization of green fluorescent protein as a marker for studying the expression and turnover of the monocarboxylate permease Jen1p of Saccharomyces cerevisiae. Biochem. J. 363 (Pt 3), 737–744. doi:10.1042/0264-6021:3630737
Panni, S., Landgraf, C., Volkmer-Engert, R., Cesareni, G., and Castagnoli, L. (2008). Role of 14-3-3 proteins in the regulation of neutral trehalase in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 8 (1), 53–63. doi:10.1111/j.1567-1364.2007.00312.x
Parua, P. K., Ratnakumar, S., Braun, K. A., Dombek, K. M., Arms, E., Ryan, P. M., et al. (2010). 14-3-3 (Bmh) proteins inhibit transcription activation by Adr1 through direct binding to its regulatory domain. Mol. Cell. Biol. 30 (22), 5273–5283. doi:10.1128/MCB.00715-10
Parua, P. K., and Young, E. T. (2014). Binding and transcriptional regulation by 14-3-3 (Bmh) proteins requires residues outside of the canonical motif. Eukaryot. Cell. 13 (1), 21–30. doi:10.1128/EC.00240-13
Pedruzzi, I., Dubouloz, F., Cameroni, E., Wanke, V., Roosen, J., Winderickx, J., et al. (2003). TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. 12 (6), 1607–1613. doi:10.1016/s1097-2765(03)00485-4
Peisker, K., Chiabudini, M., and Rospert, S. (2010). The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803 (6), 662–672. doi:10.1016/j.bbamcr.2010.03.005
Pohl, P., Joshi, R., Petrvalska, O., Obsil, T., and Obsilova, V. (2021). 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains. Commun. Biol. 4 (1), 899. doi:10.1038/s42003-021-02419-0
Polo, S., and Di Fiore, P. P. (2008). Finding the right partner: science or ART? Cell. 135 (4), 590–592. doi:10.1016/j.cell.2008.10.032
Psenakova, K., Petrvalska, O., Kylarova, S., Lentini Santo, D., Kalabova, D., Herman, P., et al. (2018). 14-3-3 protein directly interacts with the kinase domain of calcium/calmodulin-dependent protein kinase kinase (CaMKK2). Biochim. Biophys. Acta 1862 (7), 1612–1625. doi:10.1016/j.bbagen.2018.04.006
Ratnakumar, S., Kacherovsky, N., Arms, E., and Young, E. T. (2009). Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 182 (3), 735–745. doi:10.1534/genetics.109.103432
Ren, L., Hou, Y. P., Zhu, Y. Y., Zhao, F. F., Duan, Y. B., Wu, L. Y., et al. (2022). Validamycin A enhances the interaction between neutral trehalase and 14-3-3 protein Bmh1 in Fusarium graminearum. Phytopathology 112 (2), 290–298. doi:10.1094/PHYTO-05-21-0214-R
Rial, S. A., Shishani, R., Cummings, B. P., and Lim, G. E. (2023). Is 14-3-3 the combination to unlock new pathways to improve metabolic homeostasis and beta-cell function? Diabetes 72 (8), 1045–1054. doi:10.2337/db23-0094
Rios-Anjos, R. M., Camandona, V. L., Bleicher, L., and Ferreira-Junior, J. R. (2017). Structural and functional mapping of Rtg2p determinants involved in retrograde signaling and aging of Saccharomyces cerevisiae. PLoS One 12 (5), e0177090. doi:10.1371/journal.pone.0177090
Ripaud, L., Chumakova, V., Antonin, M., Hastie, A. R., Pinkert, S., Korner, R., et al. (2014). Overexpression of Q-rich prion-like proteins suppresses polyQ cytotoxicity and alters the polyQ interactome. Proc. Natl. Acad. Sci. U. S. A. 111 (51), 18219–18224. doi:10.1073/pnas.1421313111
Rittinger, K., Budman, J., Xu, J., Volinia, S., Cantley, L. C., Smerdon, S. J., et al. (1999). Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell. 4 (2), 153–166. doi:10.1016/s1097-2765(00)80363-9
Rubenstein, E. M., McCartney, R. R., Zhang, C., Shokat, K. M., Shirra, M. K., Arndt, K. M., et al. (2008). Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 283 (1), 222–230. doi:10.1074/jbc.M707957200
Ruiz, A., and Arino, J. (2007). Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot. Cell. 6 (12), 2175–2183. doi:10.1128/EC.00337-07
Sarkar, S., Davies, J. E., Huang, Z., Tunnacliffe, A., and Rubinsztein, D. C. (2007). Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 282 (8), 5641–5652. doi:10.1074/jbc.M609532200
Schepers, W., Van Zeebroeck, G., Pinkse, M., Verhaert, P., and Thevelein, J. M. (2012). In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J. Biol. Chem. 287, 44130–44142. doi:10.1074/jbc.M112.421503
Shinoda, J., and Kikuchi, Y. (2007). Rod1, an arrestin-related protein, is phosphorylated by Snf1-kinase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 364 (2), 258–263. doi:10.1016/j.bbrc.2007.09.134
Silhan, J., Obsilova, V., Vecer, J., Herman, P., Sulc, M., Teisinger, J., et al. (2004). 14-3-3 protein C-terminal stretch occupies ligand binding groove and is displaced by phosphopeptide binding. J. Biol. Chem. 279 (47), 49113–49119. doi:10.1074/jbc.M408671200
Sluchanko, N. N., and Gusev, N. B. (2017). Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J. 284 (9), 1279–1295. doi:10.1111/febs.13986
Smidova, A., Stankova, K., Petrvalska, O., Lazar, J., Sychrova, H., Obsil, T., et al. (2019). The activity of Saccharomyces cerevisiae Na(+), K(+)/H(+) antiporter Nha1 is negatively regulated by 14-3-3 protein binding at serine 481. Biochim. Biophys. Acta Mol. Cell. Res. 1866 (12), 118534. doi:10.1016/j.bbamcr.2019.118534
Solaki, M., and Ewald, J. C. (2018). Fueling the cycle: CDKs in carbon and energy metabolism. Front. Cell. Dev. Biol. 6, 93. doi:10.3389/fcell.2018.00093
Tate, J. J., Buford, D., Rai, R., and Cooper, T. G. (2017). General amino acid control and 14-3-3 proteins bmh1/2 are required for nitrogen catabolite repression-sensitive regulation of Gln3 and Gat1 localization. Genetics 205 (2), 633–655. doi:10.1534/genetics.116.195800
Thandavarayan, R. A., Watanabe, K., Ma, M., Veeyaveedu, P. T., Gurusamy, N., Palaniyandi, S. S., et al. (2008). 14-3-3 protein regulates Ask1 signaling and protects against diabetic cardiomyopathy. Biochem. Pharmacol. 75 (9), 1797–1806. doi:10.1016/j.bcp.2008.02.003
Trembley, M. A., Berrus, H. L., Whicher, J. R., and Humphrey-Dixon, E. L. (2014). The yeast 14-3-3 proteins BMH1 and BMH2 differentially regulate rapamycin-mediated transcription. Biosci. Rep. 34 (2), e00099. doi:10.1042/BSR20130096
Trendeleva, T. A., and Zvyagilskaya, R. A. (2018). Retrograde signaling as a mechanism of yeast adaptation to unfavorable factors. Biochem. (Mosc) 83 (2), 98–106. doi:10.1134/S0006297918020025
Truong, A. B., Masters, S. C., Yang, H., and Fu, H. (2002). Role of the 14-3-3 C-terminal loop in ligand interaction. Proteins 49 (3), 321–325. doi:10.1002/prot.10210
Tsuchiya, D., Yang, Y., and Lacefield, S. (2014). Positive feedback of NDT80 expression ensures irreversible meiotic commitment in budding yeast. PLoS Genet. 10 (6), e1004398. doi:10.1371/journal.pgen.1004398
van Hemert, M. J., van Heusden, G. P., and Steensma, H. Y. (2001). Yeast 14-3-3 proteins. Yeast 18 (10), 889–895. doi:10.1002/yea.739
van Heusden, G. P. (2005). 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life 57 (9), 623–629. doi:10.1080/15216540500252666
van Heusden, G. P. (2009). 14-3-3 Proteins: insights from genome-wide studies in yeast. Genomics 94 (5), 287–293. doi:10.1016/j.ygeno.2009.07.004
van Heusden, G. P., Griffiths, D. J., Ford, J. C., Chin, A. W. T. F., Schrader, P. A., Carr, A. M., et al. (1995). The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229 (1), 45–53. doi:10.1111/j.1432-1033.1995.tb20435.x
van Heusden, G. P., and Steensma, H. Y. (2001). 14-3-3 Proteins are essential for regulation of RTG3-dependent transcription in Saccharomyces cerevisiae. Yeast 18 (16), 1479–1491. doi:10.1002/yea.765
van Heusden, G. P., Wenzel, T. J., Lagendijk, E. L., de Steensma, H. Y., and van den Berg, J. A. (1992). Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 302 (2), 145–150. doi:10.1016/0014-5793(92)80426-h
van Heusden, G. P., and Yde Steensma, H. (2006). Yeast 14-3-3 proteins. Yeast 23 (3), 159–171. doi:10.1002/yea.1338
Veisova, D., Macakova, E., Rezabkova, L., Sulc, M., Vacha, P., Sychrova, H., et al. (2012). Role of individual phosphorylation sites for the 14-3-3-protein-dependent activation of yeast neutral trehalase Nth1. Biochem. J. 443 (3), 663–670. doi:10.1042/BJ20111615
Veisova, D., Rezabkova, L., Stepanek, M., Novotna, P., Herman, P., Vecer, J., et al. (2010). The C-terminal segment of yeast BMH proteins exhibits different structure compared to other 14-3-3 protein isoforms. Biochemistry 49 (18), 3853–3861. doi:10.1021/bi100273k
Wang, C., Skinner, C., Easlon, E., and Lin, S. J. (2009). Deleting the 14-3-3 protein Bmh1 extends life span in Saccharomyces cerevisiae by increasing stress response. Genetics 183 (4), 1373–1384. doi:10.1534/genetics.109.107797
Watanabe, K., Ma, M., Narasimman, G., Suresh, P. S., Juan, W., and Punniyakoti, V. T. (2008). P361 14-3-3 Protein prevents development of diabetic cardiomyopathy. Int. J. Cardiol. 125, S69. doi:10.1016/s0167-5273(08)70272-9
Wera, S., De Schrijver, E., Geyskens, I., Nwaka, S., and Thevelein, J. M. (1999). Opposite roles of trehalase activity in heat-shock recovery and heat-shock survival in Saccharomyces cerevisiae. Biochem. J. 343 Pt 3, 621–626. doi:10.1042/0264-6021:3430621
Wollman, A. J., Shashkova, S., Hedlund, E. G., Friemann, R., Hohmann, S., and Leake, M. C. (2017). Transcription factor clusters regulate genes in eukaryotic cells. Elife 6, e27451. doi:10.7554/eLife.27451
Xiao, B., Smerdon, S. J., Jones, D. H., Dodson, G. G., Soneji, Y., Aitken, A., et al. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376 (6536), 188–191. doi:10.1038/376188a0
Yaffe, M. B., Rittinger, K., Volinia, S., Caron, P. R., Aitken, A., Leffers, H., et al. (1997). The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 91 (7), 961–971. doi:10.1016/s0092-8674(00)80487-0
Yamada, Y., Shiroma, A., Hirai, S., and Iwasaki, J. (2023). Zuo1, a ribosome-associated J protein, is involved in glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 23, foad038. doi:10.1093/femsyr/foad038
Yang, X., Lee, W. H., Sobott, F., Papagrigoriou, E., Robinson, C. V., Grossmann, J. G., et al. (2006). Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U. S. A. 103 (46), 17237–17242. doi:10.1073/pnas.0605779103
Yasukawa, T., Iwama, R., Yamasaki, Y., Masuo, N., and Noda, Y. (2023). Yeast Rim11 kinase responds to glutathione-induced stress by regulating the transcription of phospholipid biosynthetic genes. Mol. Biol. Cell. 35, ar8. doi:10.1091/mbc.E23-03-0116
Young, E. T., Tachibana, C., Chang, H. W., Dombek, K. M., Arms, E. M., and Biddick, R. (2008). Artificial recruitment of mediator by the DNA-binding domain of Adr1 overcomes glucose repression of ADH2 expression. Mol. Cell. Biol. 28 (8), 2509–2516. doi:10.1128/MCB.00658-07
Young, E. T., Zhang, C., Shokat, K. M., Parua, P. K., and Braun, K. A. (2012). The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae. J. Biol. Chem. 287 (34), 29021–29034. doi:10.1074/jbc.M112.380147
Zahradka, J., van Heusden, G. P., and Sychrova, H. (2012). Yeast 14-3-3 proteins participate in the regulation of cell cation homeostasis via interaction with Nha1 alkali-metal-cation/proton antiporter. Biochimica Biophysica Acta 1820 (7), 849–858. doi:10.1016/j.bbagen.2012.03.013
Zhang, F., Pracheil, T., Thornton, J., and Liu, Z. (2013). Adenosine triphosphate (ATP) is a candidate signaling molecule in the mitochondria-to-nucleus retrograde response pathway. Genes. (Basel) 4 (1), 86–100. doi:10.3390/genes4010086
Zhang, L., Winkler, S., Schlottmann, F. P., Kohlbacher, O., Elias, J. E., Skotheim, J. M., et al. (2019). Multiple layers of phospho-regulation coordinate metabolism and the cell cycle in budding yeast. Front. Cell. Dev. Biol. 7, 338. doi:10.3389/fcell.2019.00338
Zhang, R., Feng, W., Qian, S., Li, S., and Wang, F. (2023). Regulation of Rim4 distribution, function, and stability during meiosis by PKA, Cdc14, and 14-3-3 proteins. Cell. Rep. 42 (9), 113052. doi:10.1016/j.celrep.2023.113052
Zhang, Y., Sinning, I., and Rospert, S. (2017). Two chaperones locked in an embrace: structure and function of the ribosome-associated complex RAC. Nat. Struct. Mol. Biol. 24 (8), 611–619. doi:10.1038/nsmb.3435
Zhao, G., Chen, Y., Carey, L., and Futcher, B. (2016). Cyclin-dependent kinase Co-ordinates carbohydrate metabolism and cell cycle in S. cerevisiae. Mol. Cell. 62 (4), 546–557. doi:10.1016/j.molcel.2016.04.026
Keywords: 14-3-3 proteins, yeast, protein-protein interaction, phosphorylation, molecular mechanism, scaffolding, adaptor protein
Citation: Obsilova V and Obsil T (2024) The yeast 14-3-3 proteins Bmh1 and Bmh2 regulate key signaling pathways. Front. Mol. Biosci. 11:1327014. doi: 10.3389/fmolb.2024.1327014
Received: 24 October 2023; Accepted: 15 January 2024;
Published: 24 January 2024.
Edited by:
Alan Tartakoff, Case Western Reserve University, United StatesReviewed by:
Elton Young, American Association of Retired Persons, United StatesCopyright © 2024 Obsilova and Obsil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veronika Obsilova, dmVyb25pa2Eub2JzaWxvdmFAZmd1LmNhcy5jeg==; Tomas Obsil, b2JzaWxAbmF0dXIuY3VuaS5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.