
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 30 November 2023
Sec. Structural Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmolb.2023.1306483
This article is part of the Research Topic Allosteric Functions and Inhibitions: Structural Insights View all 7 articles
Recruitment of enzymes to intracellular membranes often modulates their catalytic activity, which can be important in cell signaling and membrane trafficking. Thus, re-localization is not only important for these enzymes to gain access to their substrates, but membrane interactions often allosterically regulate enzyme function by inducing conformational changes across different time and amplitude scales. Recent structural, biophysical and computational studies have revealed how key enzymes interact with lipid membrane surfaces, and how this membrane binding regulates protein structure and function. This review summarizes the recent progress in understanding regulatory mechanisms involved in enzyme-membrane interactions.
Protein-membrane interactions play a crucial role in cell signaling and membrane trafficking (Cho and Stahelin, 2005; Niggli, 2005; Leonard and Hurley, 2011). Membranes serve as sites for signaling activity and as dynamic scaffolds for the recruitment of signaling molecules. These interactions can re-localize enzymes to substrates on membranes and regulate enzyme function through allosterically-induced conformational changes. Allosteric regulation upon membrane binding allows the enzyme to respond to changes in membrane lipid composition and allows for different activities in different subcellular compartments. Allosteric sites may also serve as targets for small molecule inhibitors for drug development, which may act to block appropriate protein-membrane interactions and/or conformational changes necessary for protein function. In this Review, we focus on peripheral membrane binding enzymes that leverage various strategies for reversible membrane interaction, often leading to changes in enzyme function. Two major methods to facilitate membrane interactions are modular membrane-targeting domains that recognize specific lipids in the membrane (Lemmon, 2008; Stahelin, 2009) and amphipathic secondary structures (Drin and Antonny, 2010; Madsen et al., 2010). There are also post-translational modifications (e.g., palmitoylation) that add a hydrophobic anchor that can act together with protein components to relocalize proteins to membranes and regulate enzyme function.
Here, we emphasize recent biophysical studies that help to describe the mechanisms of allosteric regulation upon enzyme-membrane interactions, help to identify common themes amongst these proteins, and provide a future road map for further understanding this critical class of enzymes. We divide these enzymes into two major groups, those that primarily use an amphipathic helix for membrane engagement and those that use specialized membrane-interacting domains, including C1/C2, PH, PX and FERM domains. The examples are meant to be illustrative, not exhaustive.
Many membrane-associated proteins contain an amphipathic α-helix that is often unfolded in solution but becomes folded upon membrane interaction. Such a protein segment may initially be attracted to the membrane through electrostatic interaction, but then folds into an α-helix with its nonpolar residues inserted into the membrane while polar residues face the lipid head groups. It has been proposed that these amphipathic helices especially bind to membranes with high curvature owing to their lipid packing defects (Drin and Antonny, 2010).
Two well-studied enzymes known to interact with membranes through their amphipathic helices are phosphocholine cytidylyltransferase (CCT) and phospholipase A2 (PLA2). CCT is a rate-limiting and regulatory enzyme of phosphatidylcholine (PC) metabolism (Vance and Choy, 1979). Chemical structures of PC and other important lipids are illustrated in Figure 1. CCTα/CCT1 and CCTβ/CCT2 are two major isoforms in eukaryotic organisms, which have similar structures (Lykidis et al., 1999) and are regulated similarly by membrane lipids. CCT undergoes allosteric activation when binding to membranes, which turns the soluble form (CCTsol) to a membrane-bound form (CCTmem) and boosts the catalytic efficiency by over 200-fold. The work by the Cornell lab (Cornell, 2016; Cornell, 2020) has especially been instrumental in our understanding of how the CCT-membrane binding activates CCT.
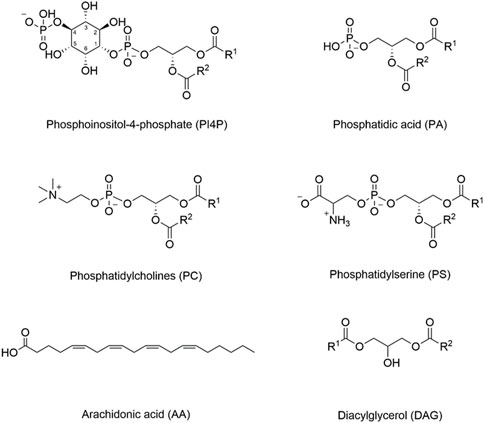
FIGURE 1. Structures of important membrane lipids. The inositol ring is numbered in phosphoinositol-4-phosphate (PI4P), as different hydroxyl groups can be phosphorylated to generate different phosphoinositide lipids (PIPs). PIPs can also be phosphorylated at more than one position. For example, PI(4,5)P2 represents phosphorylation at both the 4- and 5-positions. Different internal membranes are enriched in different PIP lipids, and some PIP-interacting domains may have increased affinity for PIPs phosphorylated at specific positions.
Upon membrane binding, the autoinhibitory (AI) segment of the membrane–lipid sensor domain (domain M) dissociates from the four-helix complex (2 αE + 2 AI helices) at the base of the catalytic domain (see Figure 2), as suggested by photo-crosslinking and deuterium exchange analysis (Huang et al., 2013). Molecular dynamics (MD) simulations suggest that this dissociation destabilizes the αE helices and enables it to sample new conformations (Ramezanpour et al., 2018). Following the change in the αE helices, membrane insertion and folding of the “leash” of domain M leads to a conformational change in the linker region (Taneva et al., 2019). The allosteric linker, composed of the αE hinge, the αEC, and the J segment, can mediate the communication between domain M and the active site. Fluorescence resonance energy transfer (FRET) experiments suggest that folding of the allosteric linker can pull the active site close to the membrane (Knowles et al., 2019).
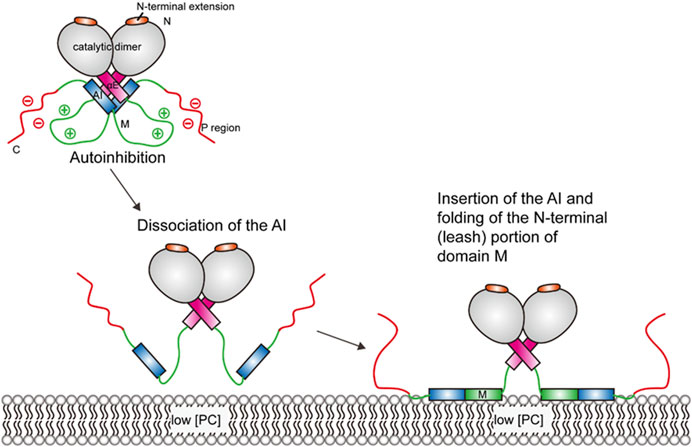
FIGURE 2. Schematic illustration of CCT activation. The CCT enzyme is kept in an autoinhibited state until interactions with the membrane lead to a series of conformational changes, which enables the amphipathic M domain to interact with the membrane.
Membrane lipid composition and the phosphorylation status of the C-terminal region are two regulators of CCT membrane binding (Cornell, 2016). High negative charge density and lipid packing defects on the membrane caused by low PC content trigger the membrane binding and the conversion of CCT, leading to PC synthesis. In the absence of high negative charge density and lipid packing defects, the rate of PC synthesis is lower. This feedback regulation helps to maintain cellular PC homeostasis (Cornell, 2016).
Perhaps the most interesting aspect is the dual function of the amphipathic helix domain M: it acts as both an autoinhibitory device in CCTsol and an activating device in CCTmem. Membrane binding of domain M disrupts its autoinhibitory contact with the active site. The deletion of domain M or specifically the AI segment only leads to partial activation. For full activation, domain M must be present (Ding et al., 2012). Amphipathic helices contribute to protein–membrane binding since they can overcome the water–lipid energy barrier (Drin and Antonny, 2010). The amphipathic helix domain M can essentially sense the features of low PC content membrane, enabling membrane binding (Cornell, 2016). Mechanistic studies suggest that the amphipathic helix binds to negatively charged membranes through electrostatic interactions and the hydrophobic effect. With no electrostatic effect, it then relies on the insertion of its hydrophobic residues into lipid packing defects (Drin and Antonny, 2010). This mechanism may be applicable to other membrane binding proteins with similar amphipathic helix. Although there is strong evidence showing that the linker is important for allosteric control, a high-resolution CCT-membrane binding structure in its active form is still not solved (Cornell, 2020), which would provide much needed insight.
PLA2 catalyzes the hydrolysis of acyl chains at the sn-2 position of membrane phospholipids to produce fatty acids that are important for downstream signaling. The activity of PLA2 is higher in the presence of lipid aggregates (vesicles, micelles, etc.) than monomeric substrates (Singer et al., 2002; Gelb et al., 2003). The PLA2 superfamily consists of 16 groups and many subgroups, and can be described as six types. Each type has a distinctive structure with an active site binds to specific phospholipid substrates and an interfacial surface that can bind to the membranes (Mouchlis et al., 2015; Mouchlis et al., 2018).
The Dennis lab (Dennis, 2022; Mouchlis and Dennis, 2022) has made many important insights into the allosteric interaction between lipid bilayer membranes and PLA2, especially through their judicial use of hydrogen–deuterium exchange mass spectrometry (HDX-MS) combined with MD simulations to understand the association mechanism of the recombinant human form of group IVA cytosolic (cPLA2), group V secreted (sPLA2), group VIA calcium-independent (iPLA2), and group VIIA lipoprotein-associated phospholipase A2 (GVIIA Lp-PLA2); they propose that PLA2 exists in at least three forms: the “closed” form in the cytosol, the “open” membrane-associated “unbound” form, and the “open” membrane-associated “bound” form when binding to a substrate (Mouchlis and Dennis, 2022).
HDX-MS studies on the iPLA2 with phospholipid vesicles and MD simulations suggest that an amphipathic helix (residues 708–730) close to the active site penetrates the membrane. The hydrophilic residues of the amphipathic helix interact with the headgroups of the lipids and the hydrophobic residues interacts with the fatty acid chains (Hsu et al., 2009; Mouchlis et al., 2015). Studies on sPLA2 also show an amphipathic helix working in a similar way (Burke et al., 2008a). HDX-MS indicates that an amphipathic loop (residues 640–648) and the amphipathic helix (residues 708–730) are important for regulating the volume of the binding pocket of iPLA2. Indeed, MD simulation studies suggest that the binding pocket volume of iPLA2 is larger in the presence of a membrane.
cPLA2 contains a protein kinase C (PKC) conserved region 2 (C2) domain and a catalytic domain. HDX-MS showed lower deuteration levels in some peptide regions derived from the C2 and catalytic domains in the presence of phospholipid vesicles (Burke et al., 2008b). According to their model, part of the C2 domain (residues 35–39 and 96–98) penetrates the membrane and “pulls” the catalytic domain to the membrane. Two amphipathic helixes of the catalytic domain (residues 268–279 and 466–470) also interact with the membrane (Burke et al., 2008b; Mouchlis et al., 2015).
Based on the HDX-MS data, two amphipathic helices of Lp-PLA2 (residues 114–120 and 360–368) are involved in membrane binding. MD simulations show that the volume of the active site is increased upon membrane binding and the amphipathic helical region (residues 100–130) is responsible for this conformational change (Mouchlis et al., 2022). A site-directed tryptophan fluorescence experiment also indicated that this peptide region is in a more polar environment because of the conformational change in the presence of vesicles.
Altogether, these data support the hypothesis that the membrane binding sites of the PLA2s serve as allosteric sites, and that membrane binding changes the enzyme conformation from a closed, inactive form to an open, active form on the membrane surface.
PKC conserved region 1 (C1) domains were first identified in PKCs and most of them bind to diacylglycerol (DAG) and phorbol ester (Colón-González and Kazanietz, 2006). They are zinc finger-like domains composed of ∼50 amino acids. The interaction between C1 and DAG is the key part of PKC activation. Structures of PKC membrane binding domains are shown in Figure 3.
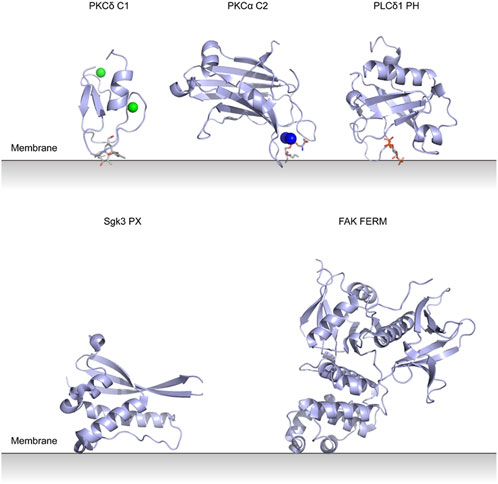
FIGURE 3. Lipid binding domains on the membrane with their putative membrane-binding pose. PKCδ C1 domain with Zn2+ and phorbol-1,3-acetate (PDB entry 1PTR). PKCα C2 domain with Ca2+ and phosphatidylserine (PDB entry 1DSY). PLCδ1 PH domain with inositol trisphosphate (PDB entry 1MAI). Sgk3 PX domain (PDB entry 6EDX). FAK FERM domain (PDB entry 2AL6).
PKC is a Ser/Thr kinase and is one of the best-studied paradigms of enzymes that are re-localized to membranes and then activated by lipids. All PKC isoenzymes consist of the N-terminal regulatory domain, C-terminal catalytic domain and a hinge between them. Conventional PKC isoenzymes (cPKCs; α, βI/II, and γ) are activated by two second messengers Ca2+ and DAG. Novel PKC isoenzymes (nPKCs; ε, δ, θ, and η) are activated by DAG alone. Atypical PKCs (aPKCs; ζ and ι/λ) are regulated by protein-protein interactions.
The activation of cPKCs requires a conformational transition from an inactive cytosolic form to an active membrane -bound form. The inactive form is autoinhibited by trapping the pseudosubstrate (PS) region into the active site of kinase domain. The autoinhibitory interaction is released upon interaction of the regulatory domain with lipid membrane.
One model (see Figure 4) (Antal et al., 2014) proposes that PKCβII is originally in an open conformation and all the domains are unmasked (Dutil and Newton, 2000). Phosphorylation of PKCβII at three sites (T500, T641, S660) then leads to an autoinhibited conformation (Behn-Krappa and Newton, 1999; Feng et al., 2000; Antal et al., 2015). The Ca2+-sensing C2 domain clamps the autoinhibitory PS region in the active site of the kinase domain, and the DAG-sensing C1 domains is masked. Hydrolysis of phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) generates second messengers, DAG and inositol-1,4,5-trisphosphate (IP3). IP3 stimulates the release of Ca2+ from the endoplasmic reticulum. There are two major steps leading to the activation. First, mutagenesis and single-molecule studies show that Ca2+ binds to C2 domain and mediates the engagement of PKC on the plasma membrane through bridging the C2 domain to phosphatidylserine and PI(4,5)P2 (Nalefski and Newton, 2001; Corbalán-García et al., 2003; Evans et al., 2006). Thus, the C2 domain is removed from the kinase domain and the regulatory and catalytic domain are separated. Second, PKC binds its membrane-embedded activator DAG via the C1 domain (Giorgione et al., 2003; Dries et al., 2007; Ziemba et al., 2014), which results in an allosteric conformational change that releases the PS from the substrate-binding cavity and the full activation of PKC (Orr and Newtont, 1994). It should be noted that the number of C1 domains involved in the membrane binding is different among PKC isoenzymes (Ziemba et al., 2014; Antal et al., 2015).
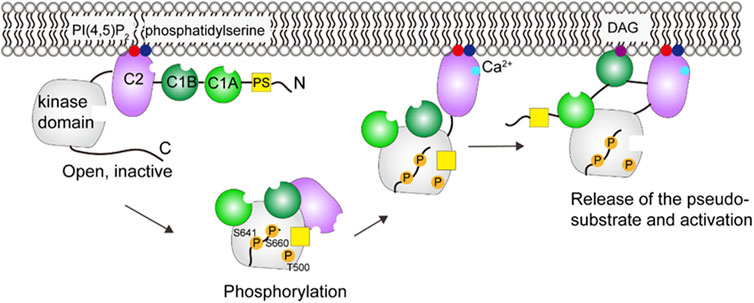
FIGURE 4. Schematic illustration of PKCβII activation. In the open, inactive form, the C2 domain binds PI(4,5)P2 and phosphatidylserine, but the DAG site in C1B is blocked. Phosphorylation and Ca2+ binding induces a series of conformational changes that allows the C1B to engage with DAG. These events lead to the activation of PKC. This figure was adapted in part from ref. Antal et al. (2015).
Studies of energetics suggest that membrane binding of C1 and C2 provides the energy to release the PS domain from the active site (Newton and Johnson, 1998). However, the atomic-level structure of the full-length active/inactive PKC and the spatiotemporal activation sequence has remained elusive. The details of how the intra-molecular rearrangement enables the PS release are still unknown. The nature of all domain-domain interactions also remains to be understood.
C2 domains have a common fold of an antiparallel β-sheet sandwich (Sutton et al., 1995). The majority of C2 domains interact with lipids in a Ca2+-dependent manner. Some C2 domains bind to anionic lipids like phosphatidylserine and phosphoinositides (PIPs) and some bind to zwitterionic lipids like PC (Nalefski et al., 1998; Perisic et al., 1998; Verdaguer et al., 1999; Sánchez-Bautista et al., 2006).
Phosphatase and tensin homolog (PTEN) desphosphorylates PI(3,4,5)P3 to PI(4,5)P2 in the PTEN/PI3Kα/Akt signaling pathway. The product PI(4,5)P2, also the allosteric activator, facilitates PTEN hydrolysis of PI(3,4,5)P3, which creates a positive feedback loop (Campbell et al., 2003). The catalytic activity of PTEN is stimulated by 5–8 fold in the presence of vesicles containing PI(4,5)P2 (McConnachie et al., 2003; Walker et al., 2004). Molecular modeling and NMR studies identified the allosteric site, the N-terminal PI(4,5)P2 binding domain (PBD), near the active site (Wei et al., 2015). Mutagenesis studies have shown that PTEN binds to the plasma membrane with three major anchor points (Walker et al., 2004; Shenoy et al., 2012; Wei et al., 2015; Masson et al., 2016; Irvine et al., 2019; Jang et al., 2021): the PBD, the arginine loop in the catalytic phosphatase domain, and the lysine-rich, phosphatidylserine-binding motif CBR3 loop in the Ca2+-independent C2 domain.
X-ray scattering, HDX-MS and cross-linking studies show that the phosphorylated C-terminal tail (CTT) (S380, T382, T383, S385) covers the CBR3 loop and Cα2 in C2 domain and the phosphatase domain, which interferes with the membrane-binding interface and results in a closed, inactive state of PTEN in the cytosol (see Figure 5) (Odriozola et al., 2007; Rahdar et al., 2009; Bolduc et al., 2013; Chen et al., 2016; Masson et al., 2016). PBD binds to the phosphatase domain at the same time. Dephosphorylation of the CTT and the following release of autoinhibition expose the active site (Bolduc et al., 2013). PTEN then binds to the membrane using the arginine loop and CBR3 loop. MD simulations suggest that the coordination of PI(3,4,5)P3 to the P loop, one of the core loops of the active site, may allosterically promote unfolding of the α-helix PBD (Jang et al., 2021). The unfolded PBD is subsequently released from the phosphatase domain in the presence of PI(4,5)P2, translocated onto the membrane, which stabilizes the protein through salt bridges between lipids and a polybasic patch of the PBD, leading to the coordination of two PI(4,5)P2 and full activation of PTEN (Jang et al., 2021).
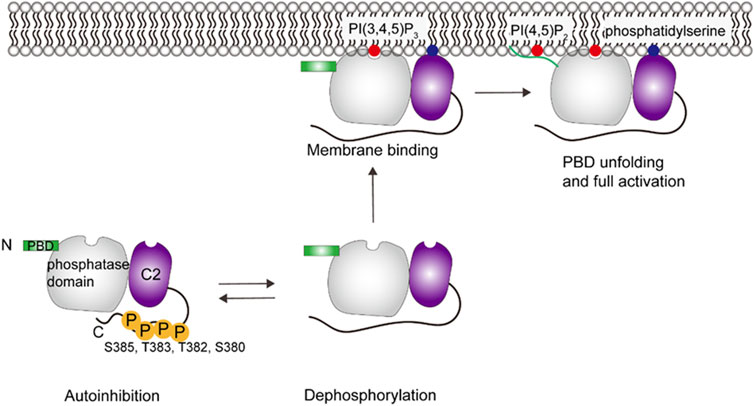
FIGURE 5. Schematic illustration of PTEN activation. PTEN remains in an autoinhibited state until removal of phosphate groups in the C-terminal region, which then allows membrane interaction. Specific interactions include C2 domain interacting with phosphatidylserine and the PBD domain interacting with PI(4,5)P2, which then allows the phosphatase domain to gain access to its PI(3,4,5)P3 substrate. This figure was adapted in part from ref. Jang et al. (2021).
There are some questions about the PTEN regulation that remain to be studied. First, since CTT is an intrinsically disordered region (IDR), it is difficult to obtain a picture of the proposed closed state (Maccario et al., 2007; Malaney et al., 2013; Dempsey et al., 2021; Smith et al., 2022). Second, previous MD simulations showed that membrane binding of PTEN leads to an orientation change between the phosphatase and C2 domain (Kalli et al., 2014), which requires experimental evidence. In addition, it has been proposed that the full activation of PTEN requires the formation of PTEN dimer (Papa et al., 2014; Heinrich et al., 2015). PTEN dimers seem to have a more compact conformation, which allows for better cooperativity between the phosphatase and C2 domains that may increase of the catalytic activity (Heinrich et al., 2015). How PTEN dimers engage in the PTEN-membrane interaction remains to be understood.
The Src homology 2 domain-containing inositol 5-phosphatase (SHIP) dephosphorylates PI(3,4,5)P3 to generate PI(3,4)P2. The SHIP family, consisting of SHIP1 and SHIP2, share a high level of sequence and structural homology, but have different tissue expression patterns. A previous study (Ong et al., 2007) showed that adding PI(3,4)P2 to the enzyme reaction activates full-length SHIP1, but not SHIP1 without the C2 domain. Protein lipid overlay (PLO) assays suggested that purified C2 domain bound to PI(3,4)P2, although there are some concerns about PLO assays being artifact prone. These data indicate that the C2 domain is needed for the allosteric activation of SHIP1 and that allosteric activators such as PI(3,4)P2 might bind to the C2 domain.
A recent study (Waddell et al., 2023) used single molecule total internal reflection fluorescence (TIRF) microscopy to visualize membrane association and dissociation dynamics of fluorescently labeled SHIP1 on supported lipid bilayers (SLBs). They found that the SHIP1 truncation containing the central phosphatase domain flanked by C2 domain binds to PI(3,4)P2 and pleckstrin homology-related (PHR) domain (PHR-PP-C2) binds to PI(3,4,5)P3. Molecular dissection showed that the full-length SHIP1 was autoinhibited by the N-terminal Src homology 2 (SH2) domain and the disordered C-terminus compared to the central PHR-PP-C2. They also found that phosphatidylserine enhanced the activity of full-length SHIP1, but the overall activity was lower than that of PHR-PP-C2 lacking autoinhibition. They suggest that the autoinhibition of SHIP1 is regulated by a mechanism that is partially resistant to phosphatidylserine-mediated activation, and phosphatidylserine may bind to the C2 domain, leading to allosteric activation.
Studies by the Lietha lab established that the C2 domain stabilizes the phosphatase (Ptase) domain and promotes membrane binding (Le Coq et al., 2017). With surface plasmon resonance (SPR) experiments, they found that both the Ptase and Ptase-C2 bind phosphatidylserine, and the C2 domain enhances the binding. The C2 domain affects the catalytic activity of the Ptase domain, which may indicate allosteric communication. MD simulations and X-ray crystal structure analysis suggested that communication between the C2 domain and the active site in the Ptase is modulated by the conformational dynamics of α5–7 helices and L4 loop.
More recent MD simulations (John et al., 2023) with SHIP2 bound to a lipid membrane suggest that C2 causes conformational changes in L4 and α5–7, but there is no change in flexibility in these regions, which may be because that the presence of a lipid membrane changes protein conformational dynamics. Another study (Le Coq et al., 2021) focusing on the function of the PHR domain of SHIP2 showed that the reaction product PI(3,4)P2 induces higher activity of PHR-Ptase-C2 compared to Ptase-C2 and Ptase. The direct binding of the PHR domain and PI(3,4)P2 was indicated by a fluorescence polarization assay. They proposed a model where both the PHR and C2 domains interacted with the membrane, resulting in optimal positioning toward the substrate and allosteric activation of SHIP2. They also proposed allosteric communication via the PHR-Ptase linker interacting with the Ptase-C2 linker.
A simulation and calculation work also compared the role of C2 domains in PTEN and SHIP2 (John et al., 2023). This study showed that the C2 domain of PTEN plays an important role in membrane binding and adjusts the posture of PTEN for substrate binding which is similar to the role of other C2 domains. The C2 domain in SHIP2 binds relatively weakly to phospholipid membranes but contributes to allosteric interdomain changes essential for catalytic activity.
Pleckstrin homology (PH) domains consist of a 7-stranded β sandwich structure together with a C-terminal α helix (see Figure 3) (Lemmon et al., 1995). They are found in many types of proteins and are thought to bind PIPs and target proteins to specific membranes. The specific interactions with PIPs are still unclear for most PH domain-containing proteins (Singh et al., 2021).
Ser and Thr kinase Akt, also known as protein kinase B (PKB), is one of the key proteins in the phosphoinositide 3-kinase signaling pathways. There are three Akt isoforms, Akt1, Akt2 and Akt3, that all have an N-terminal PH domain, an unstructured linker sequence, a kinase domain, and a C-terminal regulatory domain. PI(3,4,5)P3 or PI(3,4)P2 binding to the PH domain allosterically activates Akt by unmasking the substrate binding site (Ebner et al., 2017).
The conformational changes of Akt upon PI(3,4,5)P3 binding have been elucidated by small-angle X-ray scattering (SAXS) and HDX-MS experiments (Lučic et al., 2018). In the cytosol, the interaction between the PH domain and the kinase domain inhibits substrate binding and keeps Akt in a closed and inactive state, with the activation loop and the hydrophobic motif sequestered in the autoinhibited conformation. The generation of PI(3,4,5)P3 or PI(3,4)P2 in the plasma membrane leads to the binding of Akt and release of autoinhibition, the PH domain is displaced from the catalytic cleft, the activation loop and the hydrophobic motif are exposed for phosphorylation(see Figure 6) (Ebner et al., 2017). The phosphorylation of T308 by 3-phosphoinositide-dependent kinase 1 (PDK1) orders the activation loop and helps with substrate binding (Stokoe et al., 1977; Alessi et al., 1997), and phosphorylation on S473 by mTOR complex 2 (mTORC2) in the hydrophobic motif orders the C helix and organizes the catalytic residues (Sarbassov et al., 2005; Lučic et al., 2018). The phosphorylated residues are protected by ATP from dephosphorylation (Yang et al., 2002a; Yang et al., 2002b). Fluorescence imaging techniques and live cell spectroscopy show that dissociation from PI(3,4,5)P3 is the rate-limiting step in Akt dephosphorylation and the presence of PI(3,4,5)P3 is required for sustained Akt phosphorylation (Ebner et al., 2017). Turnover of PI(3,4,5)P3 and PI(3,4)P2 results in Akt inactivation by returning it to the autoinhibited form. The phosphorylated activation loop and hydrophobic motif are released and then accessible for dephosphorylation (Lučic et al., 2018). In summary, both PI(3,4,5)P3/PI(3,4)P2 binding and phosphorylation are required for Akt activation. The significance of activation in the presence of PI(3,4,5)P3 or PI(3,4)P2 is thought to enhance the substrate specificity and reduce the potential crosstalk between signaling pathways (Ebner et al., 2017).
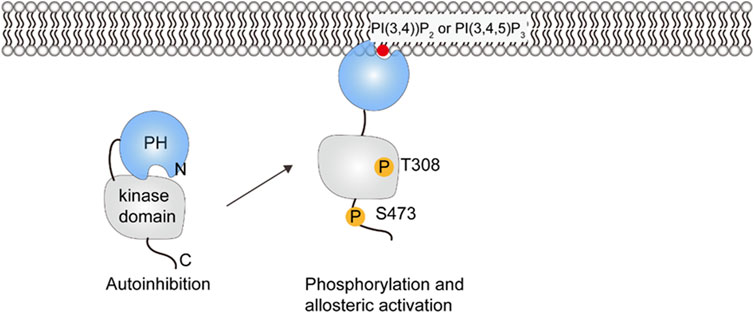
FIGURE 6. Schematic illustration of PKB/Akt activation. Phosphorylation at positions 308 and 473 releases the PH domain so that it can interact with PIP lipids in the membrane.
Bruton’s tyrosine kinase (Btk) is a protein kinase that is critical in activation of B cells. Btk consists of a PH domain fused with a Tec homology (PH-TH) domain, a proline-rich region (PRR), a Src homology 3 (SH3) domain, a SH2 domain, and a C-terminal kinase domain. The activation of Btk depends on the recruitment to the plasma membrane. The PH-TH domain binds two PI(3, 4, 5)P3, which allosterically mediate Btk dimer formation and activation (Chung et al., 2019; Wang et al., 2019).
NMR and HDX-MS reveal the autoinhibitory interactions in cytosolic Btk between the PH-TH and the kinase domain, and between SH3 and the SH2-kinase linker (see Figure 7) (Devkota et al., 2017; Joseph et al., 2017; Amatya et al., 2019). Displacement of the SH3 domain from the kinase domain by PRR transiently opens the autoinhibited structure of Btk (Joseph et al., 2017), which may promote PI(3, 4, 5)P3 binding. After Btk is recruited to the membrane containing PI(3, 4, 5)P3, a domain rearrangement can occur and likely leads to a second autoinhibitory structure with the PH-TH domain adopting the Saraste dimer (Hyvönen and Saraste, 1997), a structure associated with membrane binding. The second autoinhibitory structure based on crystallography (Wang et al., 2015)may suggest an intermediate between the fully autoinhibited and active state (Amatya et al., 2019; Kueffer et al., 2021). When there is sufficient amount of PI(3, 4, 5)P3 in the membrane, Btk binds to PI(3, 4, 5)P3 through both the canonical and peripheral sites on the PH domain to stabilize membrane interactions (Chung et al., 2019). The binding of the second PI(3, 4, 5)P3 may also be involved in an allosteric structural change or electrostatic interaction necessary for dimerization (Chung et al., 2019; Wang et al., 2019). This further structural change allows the SH2 domain to contact the kinase domain N-lobe and helps stabilize the kinase domain in its active state. Btk then undergoes trans-autophosphorylation and achieves activation (Rawlings et al., 1996).
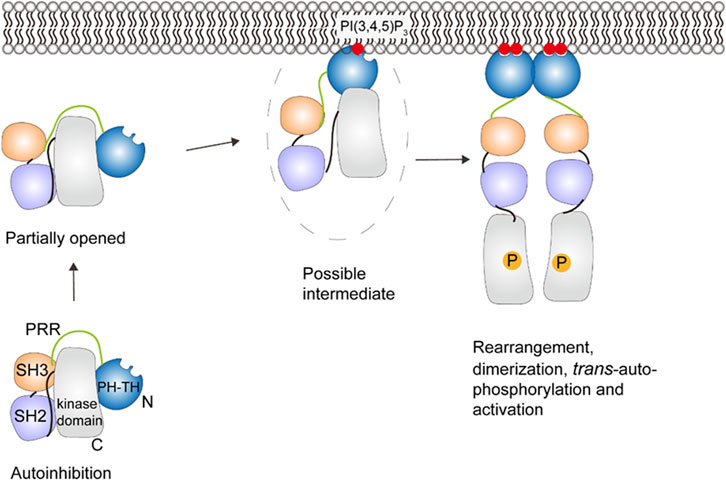
FIGURE 7. Schematic illustration of Btk activation. A series of conformational changes, including changes in domain-domain interactions, must occur to allow the PH-TH domain to interact with PI(3,4,5)P3. There is a proposed intermediate state that interacts with the membrane, but further conformational changes, dimerization and trans-autophosphorphorylation are required for full activation. This figure was adapted in part from ref. Kueffer et al. (2021).
Fluorescence correlation spectroscopy (FCS) shows that by binding to multiple PI(3, 4, 5)P3, the PH-TH module is more sensitive to the PI(3, 4, 5)P3 concentration on the surface thus leading to better regulation of Btk (Chung et al., 2019). However, the role and structural basis for the PI(3, 4, 5)P3-induced dimerization in Btk activation is not clear and awaits further study.
Myotubularins (MTM) are phosphatidylinositol phosphatases, which dephosphorylate the D-3 position of PI3P and PI(3,5)P2 and generate phosphatidylinositol and PI5P, respectively (Begley and Dixon, 2005). The product PI5P is an allosteric activator that promotes oligomerization of MTM1 into a heptamer, which enhances the activity of MTM1 (Schaletzky et al., 2003). The PH domain overlapping with the Glucosyl transferases, Rab-like GTPase activators and myotubularins (GRAM) motif has been reported to bind PIPs and mediate the allosteric activation (Lorenzo et al., 2005). The structural details about the activation and the oligomerization require further studies.
Phospholipase D (PLD) is a transphosphatidylase that hydrolyzes PC to generate phosphatidic acid (PA) (Mohn et al., 1992). Two canonical mammalian isoforms PLD1 and PLD2 have similar structures (Sung et al., 1999a; Sung et al., 1999b). The tandem phox homology-pleckstrin homology (PX-PH) domains in PLDs are thought to activate the enzyme by association with PI(4,5)P2, and they may play different roles in membrane associations and activation in different PLD isoforms (Yao et al., 2021). A recent study based on crystallography and mutagenesis demonstrated that a polybasic pocket in the catalytic domain likely binds PI(4,5)P2 (Bowling et al., 2020). Structural determination of full-length PLD is also needed to identify the PIP binding sites and activation mechanisms.
PX domains are membrane binding domains that bind to phosphatidylinositol and PIPs and consist of three anti-parallel β-strands followed by three α-helices (see Figure 3) (Ellson et al., 2002). Besides PLD, PX domain also exists in serum- and glucocorticoid-regulated kinase 3 (Sgk3). Sgk3 is a serine/threonine protein kinase allosterically activated by PI3P. The allosteric activation process of Sgk3 is similar to that of Akt. HDX-MS provides evidence for conformational changes in Sgk3 in the presence of vesicles (Pokorny et al., 2021). Cytosolic Sgk3 is autoinhibited by its PX domain, PI3P binding relieves the autoinhibition and renders Sgk3 a substrate for PDK1. Then PDK1 phosphorylates the Sgk3 activation loop, which completes the activation of Sgk3 (Kobayashi and Cohen, 1999).
Four-point-one, ezrin, radixin, moesin (FERM) domains have three compact lobes (see Figure 3) and are found in a variety of cytoskeletal-associated proteins that link the plasma membrane with cytoskeleton at specific cellular locations (Chishti et al., 1998; Frame et al., 2010).
Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase and is composed of a N-terminal FERM domain, a central kinase domain, an unstructured proline-rich region, and a C-terminal focal adhesion targeting (FAT) domain. FAK binding to PI(4,5)P2-rich membranes in focal adhesion induces a multistep activation sequence, which causes FAK oligomerization and an conformational change for autophosphorylation and activation (Goñi et al., 2014; Acebrón et al., 2020).
In the autoinhibited form, the FERM domain binds to the kinase domain, blocks the active site and sequesters phosphorylation sites (see Figure 8) (Lietha et al., 2007). FAT domain targets FAK into focal adhesions (Arold et al., 2002; Hayashi et al., 2002; Gao et al., 2004). Locally increased concentration of FAK close to the adhesome induces FAK dimerization through FERM-FERM interactions (Brami-Cherrier et al., 2014). Vesicle pull-down experiments show that a basic patch on the FERM domain binds to PI(4,5)P2 in the membrane through electrostatic interactions (Goñi et al., 2014). Cryo-electron microscopy (cryo-EM) of FAK bound to a PI(4,5)P2 -containing membrane shows that binding induces FAK oligomerization between FAK dimers and release of the autoinhibition: the kinase domain is released, and it undergoes a conformational change, binds to PI(4,5)P2 in the membrane and places the active site towards the membrane (Hall and Schaller, 2017; Acebrón et al., 2020). The conformational change upon PI(4,5)P2 binding also exposes Y397 in the linker between the FERM and kinase domains to trans -autophosphorylation by another FAK dimer. The Src kinase is then recruited to the autophosphorylated linker and phosphorylates Y576 and Y577 in the activation loop of FAK, which activates FAK (Calalb et al., 1995; Lietha et al., 2007). The active site of FAK faces toward the membrane in the cryo-EM structure, making it difficult for Src to access the activation loop and for substrates to access FAK catalytic site. An additional level of regulation, likely the tension force, is thought to expose the active site and trigger the activation of FAK in the presence of PI(4,5)P2 (Acebrón et al., 2020). The understanding of tension force within the focal adhesion requires further experiments. In addition, the proximity of FAK may promote FAT dimerization (Le Coq et al., 2022) through a helix-swapped structure observed in crystallography (Arold et al., 2002), which has not been proven since FAT domain is missing from the cryo-EM structure (Goñi et al., 2014).
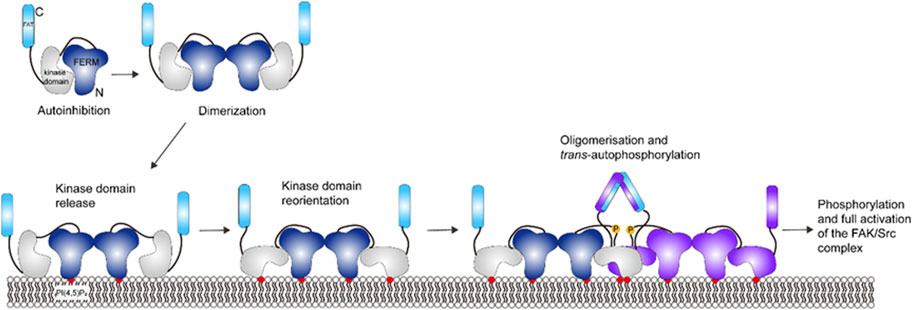
FIGURE 8. Schematic illustration of FAK activation. Activation of FAK requires dimerization to release the kinase domain and allow for domain reorientation. FAK can further oligomerize upon trans-phosphorylation. This figure was adapted in part from ref. Acebrón et al. (2020).
There are other enzymes that are allosterically regulated by membranes, but most mechanisms are unclear. PI(4,5)P2 binds to a basic region of regulatory domain in P21 activated kinase 1 (Pak1) to promote its activation synergistically with Rac1/Cdc42 (Strochlic et al., 2010; Malecka et al., 2013). Arachidonic acid was shown to activate protein phosphatase 5 (PP5) by binding to its regulatory tetratricopeptide repeat (TPR) domain (Chen and Cohen, 1997). Another example is Sac1, where its activity is stimulated by binding of anionic phospholipids like phosphatidylinositol and phosphatidylserine. Lipid binding in the cationic groove induces a conformational switch of the catalytic P-loop and activates Sac1 (Zhong et al., 2012).
Here, we have described the mechanism of membrane binding and subsequent conformational changes for key peripheral membrane binding enzymes. These actions not only re-localize these enzymes to these membranes, but also modulate their catalytic activity. We have categorized enzymes into where an amphipathic α -helix or a specialized lipid-binding domain is most important for membrane engagement. The amphipathic α-helices in CCT and PLA2 bind to the core of the protein in the unbound, inactive form. Rotation of these helices exposes the hydrophobic side for binding to the membrane. Amphipathic helices enable the protein to sense changes in physical properties of the bilayer. Thus, proteins containing amphipathic helices not only bind to specific lipids but respond more generally to negative surface charge and lipid packing voids of the membrane.
In contrast, lipid-binding domains consist of a binding pocket for a specific lipid, especially different phosphoinositide lipids, and enable the protein to be regulated by the concentration of a specific lipid in the membrane. In addition, the presence of more than one lipid-binding domain allows cooperation between lipids to increase the binding affinity and enzyme activity, diversifying enzyme regulation. For example, regulation of PKC’s function by two lipid-binding domains provides higher sensitivity in the activation process.
The general regulation strategy used by many peripheral proteins is to displace an autoinhibitory domain or release the intramolecular interaction upon membrane binding. Some peripheral proteins undergo oligomerization or dimer dissociation during membrane binding, where the changes of intramolecular and intermolecular interactions both contribute to the allosteric regulation. Moreover, positive feedback via allosteric activation by reaction products is observed in some phosphoinositide phosphatases, which is an important part in lipid metabolism. These processes are also often induced by changes in the phosphorylation state of these proteins. Other post-translational modifications (PTMs), especially palmitoylation and acylation, are also important for membrane recruitment and are responsible for fine-tuning enzyme function. Cysteine palmitoylation attaches a long chain fatty acid to the protein, increases its hydrophobicity and membrane-binding affinity (Salaun et al., 2010). Palmitoylation regulates function and cellular localization of Btk-C isoform and Akt. The phosphorylation levels of Btk-C and Akt are decreased if they are unpalmitoylated (Blaustein et al., 2021; Kokabee et al., 2022). Palmitoylation of PKC facilitates its interaction with membrane (Ford et al., 1998). Lysine acetylation neutralizes the positive charge of the protein, resulting in weaker binding with negatively charged membranes (Okada et al., 2021). Acylation at the PH domain of Akt blocks membrane-binding and reduces Akt phosphorylation, while deacylation promotes membrane-binding and activation (Sundaresan et al., 2011). Acylation of PTEN in the catalytic cleft attenuates its activity (Okumura et al., 2006).
While much has been revealed about membrane binding and consequent allosteric regulation, high-resolution protein-membrane binding structures from X-ray crystallography or cryo-EM are still required to map the spatiotemporal activation sequence for most enzymes mentioned above. Imaging studies to reveal the co-localization of membrane-bound proteins with specific lipids in vivo may also help us to understand how membrane localization and allosteric activation happen inside cells. These studies will continue to reveal important concepts in membrane binding and allosteric regulation, and provide novel mechanisms by which to modulate these functions through small molecules, serving both as chemical probes and potential starting points for pharmaceutical development.
JY: Visualization, Writing–original draft, Writing–review and editing. DB: Conceptualization, Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grant AI104878.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acebrón, I., Righetto, R. D., Schoenherr, C., Buhr, S. de, Redondo, P., Culley, J., et al. (2020). Structural basis of focal adhesion kinase activation on lipid membranes. EMBO J. 39 (19), e104743. doi:10.15252/EMBJ.2020104743
Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R. J., Reese, C. B., et al. (1997). Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 7 (4), 261–269. doi:10.1016/S0960-9822(06)00122-9
Amatya, N., Wales, T. E., Kwon, A., Yeung, W., Joseph, R. E., Bruce Fulton, D., et al. (2019). Lipid-targeting pleckstrin homology domain turns its autoinhibitory face toward the TEC kinases. Proc. Natl. Acad. Sci. U. S. A. 116 (43), 21539–21544. doi:10.1073/pnas.1907566116
Antal, C. E., Callender, J. A., Kornev, A. P., Taylor, S. S., and Newton, A. C. (2015). Intramolecular C2 domain-mediated autoinhibition of protein kinase C βii. Cell Rep. 12 (8), 1252–1260. doi:10.1016/J.CELREP.2015.07.039
Antal, C. E., Violin, J. D., Kunkel, M. T., Skovsø, S., and Newton, A. C. (2014). Intramolecular conformational changes optimize protein kinase C signaling. Chem. Biol. 21 (4), 459–469. doi:10.1016/J.CHEMBIOL.2014.02.008
Arold, S. T., Hoellerer, M. K., and Noble, M. E. M. (2002). The structural basis of localization and signaling by the focal adhesion targeting domain. Structure 10 (3), 319–327. doi:10.1016/S0969-2126(02)00717-7
Begley, M. J., and Dixon, J. E. (2005). The structure and regulation of myotubularin phosphatases. Curr. Opin. Struct. Biol. 15 (6), 614–620. doi:10.1016/J.SBI.2005.10.016
Behn-Krappa, A., and Newton, A. C. (1999). The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr. Biol. 9 (14), 728–737. doi:10.1016/S0960-9822(99)80332-7
Blaustein, M., Piegari, E., Martínez Calejman, C., Vila, A., Amante, A., Manese, M. V., et al. (2021). Akt is S-palmitoylated: a new layer of regulation for Akt. Front. Cell Dev. Biol. 9, 626404. doi:10.3389/fcell.2021.626404
Bolduc, D., Rahdar, M., Tu-Sekine, B., Sivakumaren, S. C., Raben, D., Amzel, L. M., et al. (2013). Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. Elife 2013 (2), e00691. doi:10.7554/ELIFE.00691
Bowling, F. Z., Salazar, C. M., Bell, J. A., Huq, T. S., Frohman, M. A., and Airola, M. V. (2020). Crystal structure of human PLD1 provides insight into activation by PI(4,5)P2 and RhoA. Nat. Chem. Biol. 16 (4), 400–407. doi:10.1038/s41589-020-0499-8
Brami-Cherrier, K., Gervasi, N., Arsenieva, D., Walkiewicz, K., Boutterin, M. C., Ortega, A., et al. (2014). FAK dimerization controls its kinase-dependent functions at focal adhesions. EMBO J. 33 (4), 356–370. doi:10.1002/EMBJ.201386399
Burke, J. E., Hsu, Y. H., Deems, R. A., Li, S., Woods, V. L., and Dennis, E. A. (2008b). A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2. J. Biol. Chem. 283 (45), 31227–31236. doi:10.1074/jbc.M804492200
Burke, J. E., Karbarz, M. J., Deems, R. A., Li, S., Woods, V. L., and Dennis, E. A. (2008a). Interaction of group IA phospholipase A2 with metal ions and phospholipid vesicles probed with deuterium exchange mass spectrometry. Biochemistry 47 (24), 6451–6459. doi:10.1021/BI8000962
Calalb, M. B., Polte, T. R., and Hanks, S. K. (1995). Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell Biol. 15 (2), 954–963. doi:10.1128/MCB.15.2.954
Campbell, R. B., Liu, F., and Ross, A. H. (2003). Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 278 (36), 33617–33620. doi:10.1074/JBC.C300296200
Chen, M. X., and Cohen, P. T. W. (1997). Activation of protein phosphatase 5 by limited proteolysis or the binding of polyunsaturated fatty acids to the TPR domain. FEBS Lett. 400 (1), 136–140. doi:10.1016/S0014-5793(96)01427-5
Chen, Z., Dempsey, D. R., Thomas, S. N., Hayward, D., Bolduc, D. M., and Cole, P. A. (2016). Molecular features of phosphatase and tensin homolog (PTEN) regulation by C-terminal phosphorylation. J. Biol. Chem. 291 (27), 14160–14169. doi:10.1074/jbc.M116.728980
Chishti, A. H., Kim, A. C., Marfatia, S. M., Lutchman, M., Hanspal, M., Jindal, H., et al. (1998). The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 23 (8), 281–282. doi:10.1016/S0968-0004(98)01237-7
Cho, W., and Stahelin, R. V. (2005). Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34, 119–151. doi:10.1146/ANNUREV.BIOPHYS.33.110502.133337
Chung, J. K., Nocka, L. M., Decker, A., Wang, Q., Kadlecek, T. A., Weiss, A., et al. (2019). Switch-like activation of bruton’s tyrosine kinase by membrane-mediated dimerization. Proc. Natl. Acad. Sci. U. S. A. 166 (22), 10798–10803. doi:10.1073/pnas.1819309116
Colón-González, F., and Kazanietz, M. G. (2006). C1 domains exposed: from diacylglycerol binding to protein–protein interactions. Biochimica Biophysica Acta (BBA) - Mol. Cell Biol. Lipids 1761 (8), 827–837. doi:10.1016/J.BBALIP.2006.05.001
Corbalán-García, S., García-García, J., Rodríguez-Alfaro, J. A., and Gómez-Fernández, J. C. (2003). A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase Calpha. J. Biol. Chem. 278 (7), 4972–4980. doi:10.1074/JBC.M209385200
Cornell, R. B. (2016). Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochimica Biophysica Acta (BBA) - Mol. Cell Biol. Lipids 1861 (8), 847–861. doi:10.1016/J.BBALIP.2015.12.022
Cornell, R. B. (2020). Membrane lipids assist catalysis by CTP: phosphocholine cytidylyltransferase. J. Mol. Biol. 432 (18), 5023–5042. doi:10.1016/j.jmb.2020.03.024
Dempsey, D. R., Viennet, T., Iwase, R., Park, E., Henriquez, S., Chen, Z., et al. (2021). The structural basis of PTEN regulation by multi-site phosphorylation. Nat. Struct. Mol. Biol. 28 (10), 858–868. doi:10.1038/s41594-021-00668-5
Dennis, E. A. (2022). Allosteric regulation by membranes and hydrophobic subsites in phospholipase A2 enzymes determine their substrate specificity. J. Biol. Chem. 298 (5), 101873. doi:10.1016/j.jbc.2022.101873
Devkota, S., Joseph, R. E., Boyken, S. E., Fulton, D. B., and Andreotti, A. H. (2017). An autoinhibitory role for the pleckstrin homology domain of interleukin-2-inducible tyrosine kinase and its interplay with canonical phospholipid recognition. Biochemistry 56 (23), 2938–2949. doi:10.1021/acs.biochem.6b01182
Ding, Z., Taneva, S. G., Huang, H. K. H., Campbell, S. A., Semenec, L., Chen, N., et al. (2012). A 22-mer segment in the structurally pliable regulatory domain of metazoan CTP: phosphocholine cytidylyltransferase facilitates both silencing and activating functions. J. Biol. Chem. 287 (46), 38980–38991. doi:10.1074/jbc.M112.402081
Dries, D. R., Gallegos, L. L., and Newton, A. C. (2007). A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J. Biol. Chem. 282 (2), 826–830. doi:10.1074/JBC.C600268200
Drin, G., and Antonny, B. (2010). Amphipathic helices and membrane curvature. FEBS Lett. 584 (9), 1840–1847. doi:10.1016/J.FEBSLET.2009.10.022
Dutil, E. M., and Newton, A. C. (2000). Dual role of pseudosubstrate in the coordinated regulation of protein kinase C by phosphorylation and diacylglycerol. J. Biol. Chem. 275 (14), 10697–10701. doi:10.1074/jbc.275.14.10697
Ebner, M., Lučić, I., Leonard, T. A., and Yudushkin, I. (2017). PI(3,4,5)P3 engagement restricts Akt activity to cellular membranes. Mol. Cell 65 (3), 416–431.e6. doi:10.1016/J.MOLCEL.2016.12.028
Ellson, C. D., Andrews, S., Stephens, L. R., and Hawkins, P. T. (2002). The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 115 (6), 1099–1105. doi:10.1242/JCS.115.6.1099
Evans, J. H., Murray, D., Leslie, C. C., and Falke, J. J. (2006). Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell 17 (1), 56–66. doi:10.1091/mbc.e05-06-0499
Feng, X., Becker, K. P., Stribling, S. D., Peters, K. G., and Hannun, Y. A. (2000). Regulation of receptor-mediated protein kinase C membrane trafficking by autophosphorylation. J. Biol. Chem. 275 (22), 17024–17034. doi:10.1074/JBC.275.22.17024
Ford, D. A., Horner, C. C., and Gross, R. W. (1998). Protein kinase C acylation by palmitoyl coenzyme A facilitates its translocation to membranes. Biochemistry 37 (34), 11953–11961. doi:10.1021/bi980565w
Frame, M. C., Patel, H., Serrels, B., Lietha, D., and Eck, M. J. (2010). The FERM domain: organizing the structure and function of FAK. Nat. Rev. Mol. Cell Biol. 11 (11), 802–814. doi:10.1038/nrm2996
Gao, G., Prutzman, K. C., King, M. L., Scheswohl, D. M., DeRose, E. F., London, R. E., et al. (2004). NMR solution structure of the focal adhesion targeting domain of focal adhesion kinase in complex with a paxillin LD peptide: evidence for a two-site binding model. J. Biol. Chem. 279 (9), 8441–8451. doi:10.1074/jbc.M309808200
Gelb, M. H., Jain, M. K., Hanel, A. M., and Berg, O. G. (2003). INTERFACIAL enzymology of glycerolipid hydrolases: lessons from secreted phospholipases A2. Annu. Rev. Biochem. 64, 653–688. doi:10.1146/ANNUREV.BI.64.070195.003253
Giorgione, J., Hysell, M., Harvey, D. F., and Newton, A. C. (2003). Contribution of the C1A and C1B domains to the membrane interaction of protein kinase C. Biochemistry 42 (38), 11194–11202. doi:10.1021/bi0350046
Goñi, G. M., Epifano, C., Boskovic, J., Camacho-Artacho, M., Zhou, J., Bronowska, A., et al. (2014). Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. U. S. A. 111 (31), E3177–E3186. doi:10.1073/pnas.1317022111
Hall, J. E., and Schaller, M. D. (2017). Phospholipid binding to the FAK catalytic domain impacts function. PLoS One 12 (2), e0172136. doi:10.1371/JOURNAL.PONE.0172136
Hayashi, I., Vuori, K., and Liddington, R. C. (2002). The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin. Nat. Struct. Biol. 9 (2), 101–106. doi:10.1038/nsb755
Heinrich, F., Chakravarthy, S., Nanda, H., Papa, A., Pandolfi, P. P., Ross, A. H., et al. (2015). The PTEN tumor suppressor forms homodimers in solution. Structure 23 (10), 1952–1957. doi:10.1016/j.str.2015.07.012
Hsu, Y. H., Burke, J. E., Li, S., Woods, V. L., and Dennis, E. A. (2009). Localizing the membrane binding region of group VIA Ca2+-independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 284 (35), 23652–23661. doi:10.1074/jbc.M109.021857
Huang, H. K. H., Taneva, S. G., Lee, J., Silva, L. P., Schriemer, D. C., and Cornell, R. B. (2013). The membrane-binding domain of an amphitropic enzyme suppresses catalysis by contact with an amphipathic helix flanking its active site. J. Mol. Biol. 425 (9), 1546–1564. doi:10.1016/J.JMB.2012.12.003
Hyvönen, M., and Saraste, M. (1997). Structure of the PH domain and Btk motif from bruton’s tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16 (12), 3396–3404. doi:10.1093/EMBOJ/16.12.3396
Irvine, W. A., Flanagan, J. U., and Allison, J. R. (2019). Computational prediction of amino acids governing protein-membrane interaction for the PIP3 cell signaling system. Structure 27 (2), 371–380. doi:10.1016/J.STR.2018.10.014
Jang, H., Smith, I. N., Eng, C., and Nussinov, R. (2021). The mechanism of full activation of tumor suppressor PTEN at the phosphoinositide-enriched membrane. iScience 24 (5), 102438. doi:10.1016/J.ISCI.2021.102438
John, L. H., Naughton, F. B., Sansom, M. S. P., and Larsen, A. H. (2023). The role of C2 domains in two different phosphatases: PTEN and SHIP2. Membranes 13 (4), 408. doi:10.3390/MEMBRANES13040408
Joseph, R. E., Wales, T. E., Fulton, D. B., Engen, J. R., and Andreotti, A. H. (2017). Achieving a graded immune response: BTK adopts a range of active/inactive conformations dictated by multiple interdomain contacts. Structure 25 (10), 1481–1494. doi:10.1016/J.STR.2017.07.014
Kalli, A. C., Devaney, I., and Sansom, M. S. P. (2014). Interactions of phosphatase and tensin homologue (PTEN) proteins with phosphatidylinositol phosphates: insights from molecular dynamics simulations of PTEN and voltage sensitive phosphatase. Biochemistry 53 (11), 1724–1732. doi:10.1021/bi5000299
Knowles, D. G., Lee, J., Taneva, S. G., and Cornell, R. B. (2019). Remodeling of the interdomain allosteric linker upon membrane binding of CCTα pulls its active site close to the membrane surface. J. Biol. Chem. 294 (42), 15531–15543. doi:10.1074/JBC.RA119.009850
Kobayashi, T., and Cohen, P. (1999). Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 339 (2), 319–328. doi:10.1042/BJ3390319
Kokabee, M., Wang, X., Voorand, E., Alin, E., Kokabee, L., Khan, F., et al. (2022). Palmitoylation of the alternative amino terminus of the BTK-C isoform controls subcellular distribution and signaling. Cancer Genomics Proteomics 19 (4), 415–427. doi:10.21873/CGP.20329
Kueffer, L. E., Joseph, R. E., and Andreotti, A. H. (2021). Reining in BTK: interdomain interactions and their importance in the regulatory control of BTK. Front. Cell Dev. Biol. 9, 655489. doi:10.3389/fcell.2021.655489
Le Coq, J., Acebrón, I., Martin, B. R., Navajas, P. L., and Lietha, D. (2022). New insights into FAK structure and function in focal adhesions. J. Cell Sci. 135 (20), jcs259089. doi:10.1242/jcs.259089
Le Coq, J., Camacho-Artacho, M., Velázquez, J., Santiveri, C. M., Gallego, L. H., Campos-Olivas, R., et al. (2017). Structural basis for interdomain communication in SHIP2 providing high phosphatase activity. Elife 6, e26640. doi:10.7554/ELIFE.26640
Le Coq, J., López Navajas, P., Rodrigo Martin, B., Alfonso, C., and Lietha, D. (2021). A new layer of phosphoinositide-mediated allosteric regulation uncovered for SHIP2. FASEB J. 35 (8), e21815. doi:10.1096/fj.202100561R
Lemmon, M. A. (2008). Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9 (2), 99–111. doi:10.1038/nrm2328
Lemmon, M. A., Ferguson, K. M., O’Brien, R., Sigler, P. B., and Schlessinger, J. (1995). Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. 92 (23), 10472–10476. doi:10.1073/PNAS.92.23.10472
Leonard, T. A., and Hurley, J. H. (2011). Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 21 (6), 785–791. doi:10.1016/J.SBI.2011.07.006
Lietha, D., Cai, X., Ceccarelli, D. F. J., Li, Y., Schaller, M. D., and Eck, M. J. (2007). Structural basis for the autoinhibition of focal adhesion kinase. Cell 129 (6), 1177–1187. doi:10.1016/J.CELL.2007.05.041
Lorenzo, Ó., Urbé, S., and Clague, M. J. (2005). Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J. Cell Sci. 118 (9), 2005–2012. doi:10.1242/JCS.02325
Lučic, I., Rathinaswamy, M. K., Truebestein, L., Hamelin, D. J., Burke, J. E., and Leonard, T. A. (2018). Conformational sampling of membranes by Akt controls its activation and inactivation. Proc. Natl. Acad. Sci. U. S. A. 115 (17), E3940–E3949. doi:10.1073/pnas.1716109115
Lykidis, A., Baburina, I., and Jackowski, S. (1999). Distribution of CTP:phosphocholine cytidylyltransferase (CCT) isoforms. Identification of a new CCTbeta splice variant. J. Biol. Chem. 274 (38), 26992–27001. doi:10.1074/jbc.274.38.26992
Maccario, H., Perera, N. M., Davidson, L., Downes, C. P., and Leslie, N. R. (2007). PTEN is destabilized by phosphorylation on Thr366. Biochem. J. 405 (3), 439–444. doi:10.1042/BJ20061837
Madsen, K. L., Bhatia, V. K., Gether, U., and Stamou, D. (2010). BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 584 (9), 1848–1855. doi:10.1016/J.FEBSLET.2010.01.053
Malaney, P., Uversky, V. N., and Davé, V. (2013). The PTEN long N-tail is intrinsically disordered: increased viability for PTEN therapy. Mol. Biosyst. 9 (11), 2877–2888. doi:10.1039/C3MB70267G
Malecka, K. A., Szentpetery, Z., and Peterson, J. R. (2013). Synergistic activation of P21-activated kinase 1 by phosphatidylinositol 4,5-bisphosphate and rho GTPases. J. Biol. Chem. 288 (13), 8887–8897. doi:10.1074/jbc.M112.428904
Masson, G. R., Perisic, O., Burke, J. E., and Williams, R. L. (2016). The intrinsically disordered tails of PTEN and PTEN-L have distinct roles in regulating substrate specificity and membrane activity. Biochem. J. 473 (2), 135–144. doi:10.1042/BJ20150931
McConnachie, G., Pass, I., Walker, S. M., and Downes, C. P. (2003). Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 371 (3), 947–955. doi:10.1042/BJ20021848
Mohn, H., Chalifa, V., and Liscovitch, M. (1992). Substrate specificity of neutral phospholipase D from rat brain studied by selective labeling of endogenous synaptic membrane phospholipids in vitro. J. Biol. Chem. 267 (16), 11131–11136. doi:10.1016/S0021-9258(19)49885-2
Mouchlis, V. D., Bucher, D., McCammon, J. A., and Dennis, E. A. (2015). Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. U. S. A. 112 (6), E516–E525. doi:10.1073/pnas.1424651112
Mouchlis, V. D., Chen, Y., Andrew McCammon, J., and Dennis, E. A. (2018). Membrane allostery and unique hydrophobic sites promote enzyme substrate specificity. J. Am. Chem. Soc. 140 (9), 3285–3291. doi:10.1021/jacs.7b12045
Mouchlis, V. D., and Dennis, E. A. (2022). Membrane association allosterically regulates phospholipase A 2 enzymes and their specificity. Acc. Chem. Res. 2022, 34. doi:10.1021/ACS.ACCOUNTS.2C00497/ASSET/IMAGES/LARGE/AR2C00497_0004.JPEG
Mouchlis, V. D., Hayashi, D., Vasquez, A. M., Cao, J., Andrew McCammon, J., and Dennis, E. A. (2022). Lipoprotein-associated phospholipase A2: a paradigm for allosteric regulation by membranes. Proc. Natl. Acad. Sci. U. S. A. 119 (2), e2102953118. doi:10.1073/pnas.2102953118
Nalefski, E. A., McDonagh, T., Somers, W., Seehra, J., Falke, J. J., and Clark, J. D. (1998). Independent folding and ligand specificity of the C2 calcium-dependent lipid binding domain of cytosolic phospholipase A2. J. Biol. Chem. 273 (3), 1365–1372. doi:10.1074/jbc.273.3.1365
Nalefski, E. A., and Newton, A. C. (2001). Membrane binding kinetics of protein kinase C betaII mediated by the C2 domain. Biochemistry 40 (44), 13216–13229. doi:10.1021/bi010761u
Newton, A. C., and Johnson, J. E. (1998). Protein kinase C: a paradigm for regulation of protein function by two membrane-targeting modules. Biochimica Biophysica Acta (BBA) - Rev. Biomembr. 1376 (2), 155–172. doi:10.1016/S0304-4157(98)00003-3
Niggli, V. (2005). REGULATION OF PROTEIN ACTIVITIES BY PHOSPHOINOSITIDE PHOSPHATES. Annu. Rev. Cell Dev. Biol. 21, 57–79. doi:10.1146/ANNUREV.CELLBIO.21.021704.102317
Odriozola, L., Singh, G., Hoang, T., and Chan, A. M. (2007). Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 282 (32), 23306–23315. doi:10.1074/jbc.M611240200
Okada, A. K., Teranishi, K., Ambroso, M. R., Isas, J. M., Vazquez-Sarandeses, E., Lee, J. Y., et al. (2021). Lysine acetylation regulates the interaction between proteins and membranes. Nat. Commun. 12 (1), 6466–6512. doi:10.1038/s41467-021-26657-2
Okumura, K., Mendoza, M., Bachoo, R. M., DePinho, R. A., Cavenee, W. K., and Furnari, F. B. (2006). PCAF modulates PTEN activity. J. Biol. Chem. 281 (36), 26562–26568. doi:10.1074/jbc.M605391200
Ong, C. J., Ming-Lum, A., Nodwell, M., Ghanipour, A., Yang, L., Williams, D. E., et al. (2007). Small-molecule agonists of SHIP1 inhibit the phosphoinositide 3-kinase pathway in hematopoietic cells. Blood 110 (6), 1942–1949. doi:10.1182/BLOOD-2007-03-079699
Orr, J. W., and Newtont, A. C. (1994). Intrapeptide regulation of protein kinase C. J. Biol. Chem. 269 (11), 8383–8387. doi:10.1016/S0021-9258(17)37205-8
Papa, A., Wan, L., Bonora, M., Salmena, L., Song, M. S., Hobbs, R. M., et al. (2014). Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell 157 (3), 595–610. doi:10.1016/j.cell.2014.03.027
Perisic, O., Fong, S., Lynch, D. E., Bycroft, M., and Williams, R. L. (1998). Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J. Biol. Chem. 273 (3), 1596–1604. doi:10.1074/jbc.273.3.1596
Pokorny, D., Truebestein, L., Fleming, K. D., Burke, J. E., and Leonard, T. A. (2021). In vitro reconstitution of Sgk3 activation by phosphatidylinositol 3-phosphate. J. Biol. Chem. 297 (2), 100919. doi:10.1016/j.jbc.2021.100919
Rahdar, M., Inoue, T., Meyer, T., Zhang, J., Vazquez, F., and Devreotes, P. N. (2009). A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. U. S. A. 106 (2), 480–485. doi:10.1073/pnas.0811212106
Ramezanpour, M., Lee, J., Taneva, S. G., Peter Tieleman, D., and Cornell, R. B. (2018). An auto-inhibitory helix in CTP:phosphocholine cytidylyltransferase hijacks the catalytic residue and constrains a pliable, domain-bridging helix pair. J. Biol. Chem. 293 (18), 7070–7084. doi:10.1074/JBC.RA118.002053
Rawlings, D. J., Scharenberg, A. M., Park, H., Wahl, M. I., Lin, S., Kato, R. M., et al. (1996). Activation of BTK by a phosphorylation mechanism initiated by SRC family kinases. Science 271 (5250), 822–825. doi:10.1126/SCIENCE.271.5250.822
Salaun, C., Greaves, J., and Chamberlain, L. H. (2010). The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191 (7), 1229–1238. doi:10.1083/JCB.201008160
Sánchez-Bautista, S., Marín-Vicente, C., Gómez-Fernández, J. C., and Corbalán-García, S. (2006). The C2 domain of PKCalpha is a Ca2+ -dependent PtdIns(4,5)P2 sensing domain: a new insight into an old pathway. J. Mol. Biol. 362 (5), 901–914. doi:10.1016/J.JMB.2006.07.093
Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-MTOR complex. Science 307 (5712), 1098–1101. doi:10.1126/SCIENCE.1106148
Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J., and Barr, F. A. (2003). Phosphatidylinositol-5-Phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr. Biol. 13 (6), 504–509. doi:10.1016/S0960-9822(03)00132-5
Shenoy, S. S., Nanda, H., and Lösche, M. (2012). Membrane association of the PTEN tumor suppressor: electrostatic interaction with phosphatidylserine-containing bilayers and regulatory role of the C-terminal tail. J. Struct. Biol. 180 (3), 394–408. doi:10.1016/J.JSB.2012.10.003
Singer, A. G., Ghomashchi, F., Le Calvez, C., Bollinger, J., Bezzine, S., Rouault, M., et al. (2002). Interfacial kinetic and binding properties of the complete set of human and mouse groups I, II, V, X, and XII secreted phospholipases A2. J. Biol. Chem. 277 (50), 48535–48549. doi:10.1074/jbc.M205855200
Singh, N., Reyes-Ordoñez, A., Compagnone, M. A., Moreno, J. F., Leslie, B. J., Ha, T., et al. (2021). Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 12 (1), 4339–4413. doi:10.1038/s41467-021-24639-y
Smith, I. N., Dawson, J. E., Krieger, J., Thacker, S., Bahar, I., and Eng, C. (2022). Structural and dynamic effects of PTEN C-terminal tail phosphorylation. J. Chem. Inf. Model 62 (17), 4175–4190. doi:10.1021/acs.jcim.2c00441
Stahelin, R. V. (2009). Lipid binding domains: more than simple lipid effectors. J. Lipid Res. 50 (Suppl. L), S299–S304. doi:10.1194/JLR.R800078-JLR200
Stokoe, D., Stephens, L. R., Copeland, T., Gaffney, P. R. J., Reese, C. B., Painter, G. F., et al. (1977). Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277 (5325), 567–570. doi:10.1126/science.277.5325.567
Strochlic, T. I., Viaud, J., Rennefahrt, U. E. E., Anastassiadis, T., and Peterson, J. R. (2010). Phosphoinositides are essential coactivators for P21-activated kinase 1. Mol. Cell 40 (3), 493–500. doi:10.1016/J.MOLCEL.2010.10.015
Sundaresan, N. R., Pillai, V. B., Wolfgeher, D., Samant, S., Vasudevan, P., Parekh, V., et al. (2011). The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal 4 (182), ra46. doi:10.1126/scisignal.2001465
Sung, T. C., Altshuller, Y. M., Morris, A. J., and Frohman, M. A. (1999a). Molecular analysis of mammalian phospholipase D2. J. Biol. Chem. 274 (1), 494–502. doi:10.1074/jbc.274.1.494
Sung, T. C., Zhang, Y., Morris, A. J., and Frohman, M. A. (1999b). Structural analysis of human phospholipase D1. J. Biol. Chem. 274 (6), 3659–3666. doi:10.1074/jbc.274.6.3659
Sutton, R. B., Davletov, B. A., Berghuis, A. M., Sudhof, T. C., and Sprang, S. R. (1995). Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell 80 (6), 929–938. doi:10.1016/0092-8674(95)90296-1
Taneva, S. G., Lee, J., Knowles, D. G., Tishyadhigama, C., Chen, H., and Cornell, R. B. (2019). Interdomain communication in the phosphatidylcholine regulatory enzyme, CCTα, relies on a modular αE helix. J. Biol. Chem. 294 (42), 15517–15530. doi:10.1074/JBC.RA119.009849
Vance, D. E., and Choy, P. C. (1979). How is phosphatidylcholine biosynthesis regulated? Trends Biochem. Sci. 4 (7), 145–148. doi:10.1016/0968-0004(79)90001-X
Verdaguer, N., Corbalan-Garcia, S., Ochoa, W. F., Fita, I., and Gómez-Fernández, J. C. (1999). Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 18 (22), 6329–6338. doi:10.1093/EMBOJ/18.22.6329
Waddell, G. L., Drew, E. E., Rupp, H. P., and Hansen, S. D. (2023). Mechanisms controlling membrane recruitment and activation of the autoinhibited SHIP1 inositol 5-phosphatase. J. Biol. Chem. 299 (8), 105022. doi:10.1016/J.JBC.2023.105022
Walker, S. M., Leslie, N. R., Perera, N. M., Batty, I. H., and Downes, C. P. (2004). The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem. J. 379 (2), 301–307. doi:10.1042/BJ20031839
Wang, Q., Pechersky, Y., Sagawa, S., Pan, A. C., and Shaw, D. E. (2019). Structural mechanism for bruton’s tyrosine kinase activation at the cell membrane. Proc. Natl. Acad. Sci. U. S. A. 116 (19), 9390–9399. doi:10.1073/pnas.1819301116
Wang, Q., Vogan, E. M., Nocka, L. M., Rosen, C. E., Zorn, J. A., Harrison, S. C., et al. (2015). Autoinhibition of bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. Elife 2015 (4), e06074. doi:10.7554/ELIFE.06074
Wei, Y., Stec, B., Redfield, A. G., Weerapana, E., and Roberts, M. F. (2015). Phospholipid-binding sites of phosphatase and tensin homolog (PTEN): EXPLORING the mechanism of phosphatidylinositol 4,5-BISPHOSPHATE activation. J. Biol. Chem. 290 (3), 1592–1606. doi:10.1074/JBC.M114.588590
Yang, J., Cron, P., Good, V. M., Thompson, V., Hemmings, B. A., and Barford, D. (2002a). Crystal structure of an activated akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9 (12), 940–944. doi:10.1038/nsb870
Yang, J., Cron, P., Thompson, V., Good, V. M., Hess, D., Hemmings, B. A., et al. (2002b). Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell 9 (6), 1227–1240. doi:10.1016/S1097-2765(02)00550-6
Yao, Y., Li, J., Lin, Y., Zhou, J., Zhang, P., and Xu, Y. (2021). Structural insights into phospholipase D function. Prog. Lipid Res. 81, 101070. doi:10.1016/J.PLIPRES.2020.101070
Zhong, S., Hsu, F., Stefan, C. J., Wu, X., Patel, A., Cosgrove, M. S., et al. (2012). Allosteric activation of the phosphoinositide phosphatase sac1 by anionic phospholipids. Biochemistry 51 (15), 3170–3177. doi:10.1021/bi300086c
Keywords: protein-membrane interactions, allosteric regulation, conformational change, signaling, lipid metabolism, lipid kinase, phosphatidylinositol phosphate lipids
Citation: Yu J and Boehr DD (2023) Regulatory mechanisms triggered by enzyme interactions with lipid membrane surfaces. Front. Mol. Biosci. 10:1306483. doi: 10.3389/fmolb.2023.1306483
Received: 03 October 2023; Accepted: 17 November 2023;
Published: 30 November 2023.
Edited by:
Nicolas Doucet, Université du Québec, CanadaCopyright © 2023 Yu and Boehr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David D. Boehr, ZGRiMTJAcHN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.