- 1Department of General Surgery, Comprehensive Breast Health Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Laboratory of Molecule Synthesis and Function Discovery (Fujian Province University), Department of Chemistry, Fuzhou University, Fuzhou, China
Eleven two-carbon tethered artemisinin–isatin hybrids (4a–k) were designed, synthesized, and evaluated for their antiproliferative activity against MCF-7, MDA-MB-231, and MDA-MB-231/ADR breast cancer cell lines, as well as cytotoxicity toward MCF-10A cells in this paper. Among them, the representative hybrid 4a (IC50: 2.49–12.6 µM) was superior to artemisinin (IC50: 72.4->100 µM), dihydroartemisinin (IC50: 69.6–89.8 µM), and Adriamycin (IC50: 4.46–>100 µM) against the three tested breast cancer cell lines. The structure–activity relationship revealed that the length of the alkyl linker between artemisinin and isatin was critical for the activity, so further structural modification could focus on evaluation of the linker. The in silico studies were used to investigate the mechanism of the most promising hybrid 4a. Target prediction, bioinformatics, molecular docking, and molecular dynamics revealed that the most promising hybrid 4a may exert anti-breast cancer activity by acting on multiple targets such as EGFR, PIK3CA, and MAPK8 and thus participating in multiple tumor-related signaling pathways.
1 Introduction
Breast cancer consists of a group of biologically and molecularly heterogeneous diseases that originate from the breast and can be divided into four major subtypes: luminal-A, luminal-B, human epidermal growth factor receptor 2 (HER2) positive, and triple-negative breast cancer (Feng et al., 2018; Wang et al., 2020a; Tsang and Tse, 2020). At present, breast cancer is the most prevalent malignancy in women (Akr et al., 2017; Lau et al., 2022). In 2020, breast cancer has overtaken lung cancer as the most common malignancy, and an estimated 2.3 million new cases of breast cancer were diagnosed, accounting for nearly one-fourth of women cancer patients (Jaiswal et al., 2021; Sung et al., 2021). Moreover, 685,000 women died due to breast cancer, leading to one-sixth of cancer-related deaths in women (Arnold et al., 2022). Unfortunately, the burden of breast cancer is expected to increase further in the coming years (Wang et al., 2020b; Schröder et al., 2022; Sher et al., 2022).
Advancements in the treatment of breast cancer have resulted in an increasing population of patients living with this disease, and chemotherapy occupies an important position in breast cancer therapy. However, the chemotherapy treatment suffers from the generation of drug resistance (Shaikh and Emens, 2022; Tufail et al., 2022). Hence, development of innovative chemotherapeutics is a promising strategy to improve therapeutic outcomes of breast cancer.
Hybrid molecules, generated by combining two or more molecule entities, could affect multiple targets to fight against various diseases including breast cancers (Bérubé, 2016; Waseem and Ahmad, 2022; Wang et al., 2023a). Artemisinin (ART) and its derivatives (dihydroartemisinin/DHA, artesunate, and artemether), from the traditional Chinese medicine drug, possess a unique peroxy bridge structure, and they could exert the anticancer effects through diverse mechanisms, inclusive of cell cycle inhibition, inhibition of tumor angiogenesis, promotion of DNA damage, and promotion of ferroptosis (Zhang S. et al., 2022a; Hu et al., 2022). Hence, ART derivatives may have significant therapeutic effects on cancers (Gao et al., 2020; Mancuso et al., 2021). Isatin is an exceptionally useful template for developing new anticancer scaffolds; on account of this finding, more and more isatin-based molecules are either in clinical use or in trials (Gupta et al., 2019; Ding et al., 2020). Accordingly, a combination of ART with isatin is a promising strategy to discover novel anti-breast cancer candidates.
In recent years, several series of ART–isatin hybrids have been screened for their potential against various cancer cell lines, and some of them demonstrated promising antiproliferative activity against breast cancer cells (Hou et al., 2021; Zhang Z. et al., 2022b; Dong et al., 2022; Wang et al., 2022). The structure–activity relationship (SAR) revealed that the linker between ART and isatin moieties influenced the antiproliferative activity significantly, and as a linker, alkyl was more favorable than 1,2,3-triazole. Hence, a series of two-carbon tethered ART–isatin hybrids were designed, synthesized, and assessed for their antiproliferative activity against both drug-sensitive (MCF-7 and MDA-MB-231) and Adriamycin-resistant (MDA-MB-231/ADR) breast cancer cell lines in this study. Moreover, the cytotoxicity of the synthesized ART–isatin hybrids toward normal MCF-10A breast cells was also tested. Finally, possible mechanisms were investigated by in silico studies. The purpose of this paper was to find the candidates with promising antiproliferative potential against both drug-sensitive and drug-resistant breast cancer cell lines and high selectivity.
2 Results and discussion
2.1 Experimental studies
2.1.1 Synthesis
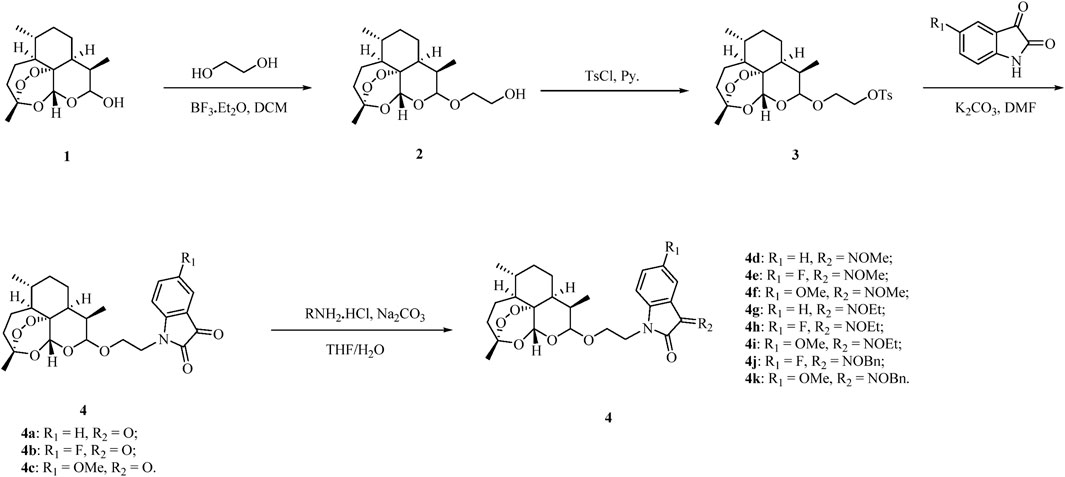
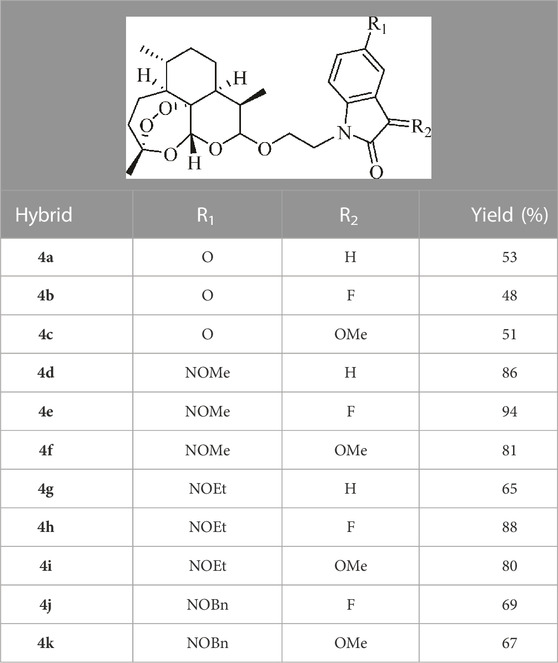
The ART–isatin hybrids 4a–k were prepared according to our method reported previously, and the detailed synthetic route is shown in Scheme 1 (Wang et al., 2023b). Etherification of dihydroartemisinin 1 with ethylene glycol in the presence of boron trifluoride diethyl etherate (BF3•OEt2) generated 2-hydroxyethyl dihydroartemisinin intermediate 2. The conversion of the hydroxy group in intermediate 2 to tosylate (OTs) with pyridine as the base yielded intermediate 3. Alkylation of (5-substituted)isatins with tosylate 3 provided desired ART–isatin hybrids 4a–c. Finally, ART–isatin hybrids 4a–c reacted with methoxylamine/ethoxylamine/benzyloxyamine hydrochlorides using sodium carbonate (Na2CO3) as the base yielded hybrids 4d–k. The structures and yields are listed in Table 1.
2.1.2 Characterization
The desired two-carbon tethered ART–isatin hybrids 4a–k were characterized by high-resolution mass spectrometry (HRMS), proton nuclear magnetic resonance (1H NMR), and carbon-13 nuclear magnetic resonance spectroscopy (13C NMR) (Wang et al., 2023b). The corresponding analytical data and the analytical spectra are included in our previous study and Supplementary Figures S1–S33[27].
2.1.3 Anti-breast cancer potential
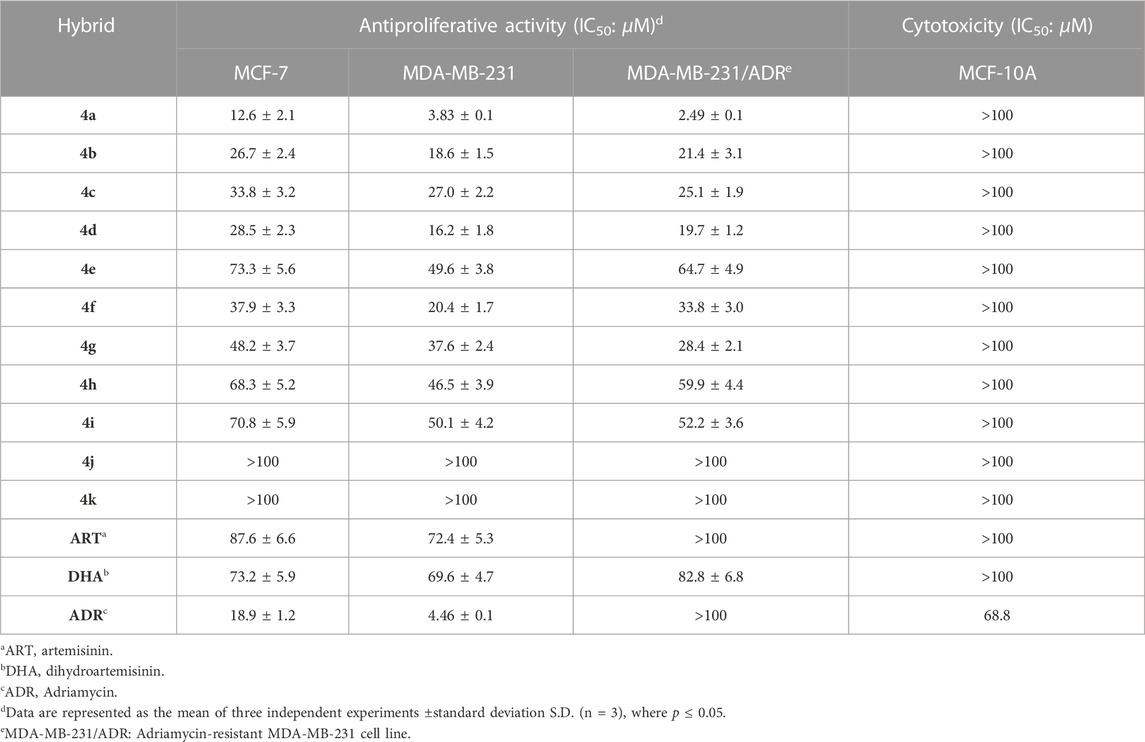
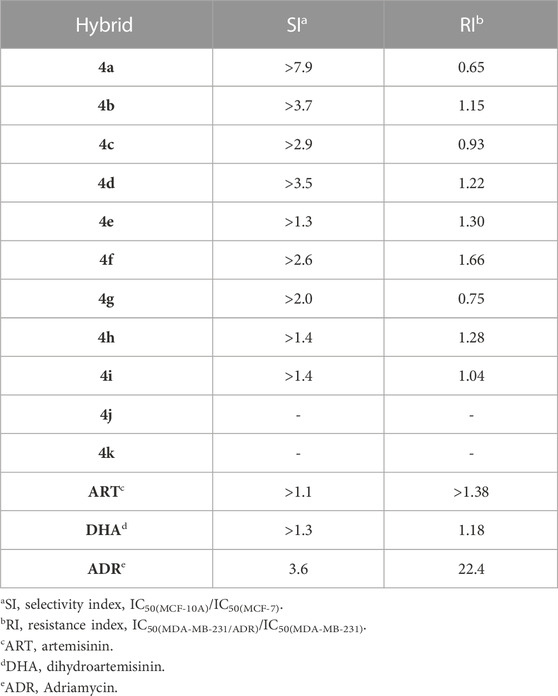
The antiproliferative activity and cytotoxicity of ART–isatin hybrids 4a–k against MCF-7 (CL-0149, purchased from Procell), MDA-MB-231 (CL-0150A, purchased from Procell), and MDA-MB-231/ADR (AC337895, purchased from CASMART) breast cancer cell lines, as well as MCF-10A (CL-0525, purchased from Procell), were assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. ART, DHA, and Adriamycin (ADR) were used as positive controls. The half-maximal inhibitory concentration (IC50) values are presented in Table 2. The selectivity index (SI: IC50(MCF-10A)/IC50(MCF-7)) and resistance index (RI: IC50(MDA-MB-231/ADR)/IC50(MDA-MB-231)) are presented in Table 3.
Table 2 demonstrates that a significant part of the synthesized hybrids (IC50: 2.49–73.3 µM) showed considerable activity against MCF-7, MDA-MB-231, and MDA-MB-231/ADR breast cancer cell lines, and the activity was superior to that of ART (IC50: 72.4->100 µM) and DHA (IC50: 69.6–89.8 µM). The SAR illustrated that the introduction of the (methoxy/ethoxy/benzyloxy)imino group into the C-3 position and fluoro or methoxy group into the C-5 position of the isatin moiety decreased the activity. In particular, incorporation of the benzyloxyimino group into the C-3 position of isatin skeleton led to a loss of activity. Moreover, the length of the alkyl linker between ART and isatin seems to have a significant influence on the activity, as evidenced by that the two-carbon tethered ART–isatin hybrids were more potent than the three-carbon analogs reported in reference 17.
All the synthesized two-carbon tethered hybrids (IC50: >100 µM) were non-toxic toward normal MCF-10A breast cancer cells as ART (IC50: >100 µM) and DHA (IC50: >100 µM), and the cytotoxicity was lower than that of Adriamycin (IC50: 68.8 µM).
Table 3 shows that the selectivity index (SI: IC50(MCF-10A)/IC50(MCF-7)) values of six hybrids were >2.0, which were similar to the previously reported ART–isatin hybrids and controls ART (SI: >1.1), DHA (SI: >1.3), and ADR (SI: 3.6) (Hou et al., 2021; Zhang Z. et al., 2022b; Dong et al., 2022; Wang et al., 2022). It was worth noting that the resistance index (RI: IC50(MDA-MB-231/ADR)/IC50(MDA-MB-231)) values of the active hybrids 4a–k were in a range of 0.65–1.66, which were comparable to the previously reported ART–isatin hybrids and controls ART (RI: >1.38) and DHA (RI: 1.18), and lower than ADR (RI: 22.4), indicating that these hybrids had no or low cross resistance with the current available anti-breast cancer agents (Hou et al., 2021; Zhang Z. et al., 2022b; Dong et al., 2022; Wang et al., 2022).
Amongst the synthesized hybrids, the most active hybrid 4a (IC50: 2.49–12.6 µM) not only was far more potent than the parents ART (IC50: 72.4->100 µM) and DHA (IC50: 69.6–89.8 µM) but also possessed higher activity than Adriamycin (IC50: 4.46->100 µM) against all the three breast cancer lines. The RI value of hybrid 4a was 0.65, implying its potential to overcome drug resistance. In addition, hybrid 4a: >100 µM) was non-toxic toward normal MCF-10A breast cells, and the SI value was >7.9. Accordingly, hybrid 4a could serve as a promising candidate for further preclinical evaluations.
2.2 In silico studies
2.2.1 Target prediction of 4a
SwissTargetPrediction, a tool for predicting the targets of compounds based on the similarity of two-dimensional and three-dimensional structures of known compounds, has become a common tool for predicting the targets of compounds currently due to its better sensitivity and specificity (Daina et al., 2019). We selected the promising candidate 4a and performed target prediction by SwissTargetPrediction. The results showed that 4a may have 109 potential targets. Supplementary Table S1 displays the full list in detail.
2.2.2 Construction of cross-over genes, protein–protein interaction networks, and enrichment analysis
To better explore the cross-over genes, we selected breast cancer-related gene (BC-related gene) in GeneCard to intersect with our predicted targets of 4a (potential target) (Safran et al., 2010). Finally, a total of 51 cross-over genes were selected (Figure 1A). These cross-over genes are the potential gene targets of 4a in the activities against breast cancer. Supplementary Table S2 displays the full list in detail.
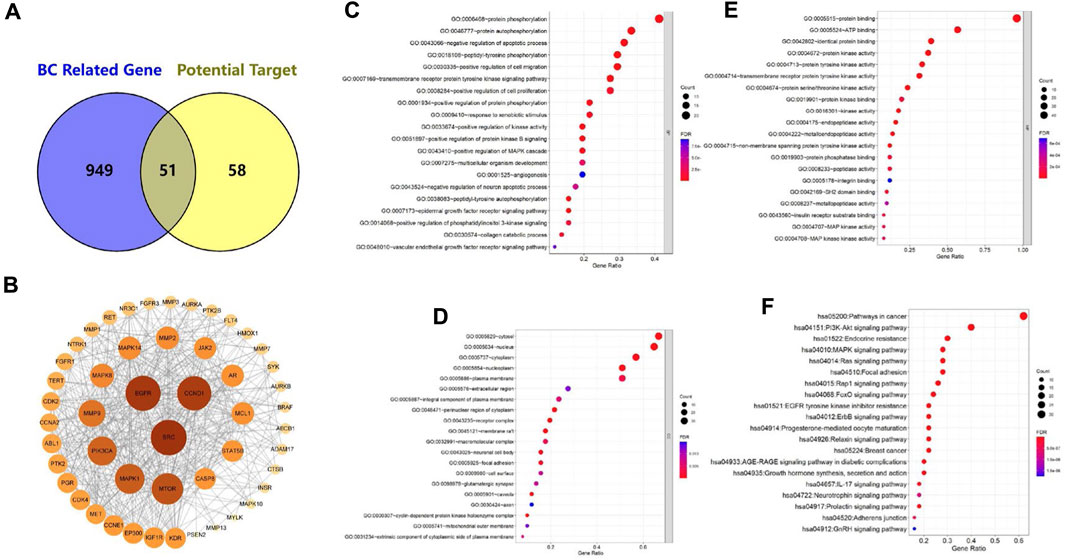
FIGURE 1. (A) Intersection of BC-related gene and potential target (Venn diagram). (B) Protein–protein interaction (PPI) network diagram of candidate targets. (C–E) GO enrichment analysis of target genes: (C) biological processes, (D) cellular components, and (E) molecular functions. (F) KEGG enrichment analysis of target genes.
To obtain the protein–protein interaction (PPI) networks, we imported 51 cross-over genes into the STRING database and selected Homo sapiens as the organism. The results showed 462 interactions. Subsequently, the PPI networks were obtained using Cytoscape for visualization and calculation (Figure 1B). The average closeness centrality (CC) was 0.62, the average betweenness centrality (BC) was 31.5, and the average degree value (DV) was 18.5. Among them, SRC, EGFR, CCND1, MTOR, MAPK1, PIK3CA, MMP9, MAPK8, MMP2, and MAPK14 had higher DV, BC, and CC values than the norm. This suggests that they are the central targets of 4a. Supplementary Table S3 displays the specific node properties in detail.
We performed Gene Ontology (GO) enrichment analysis on selected target genes to further examine them. Based on the GO enrichment analysis, most of the target genes were identified to be involved in biological processes (BPs), cellular components (CCs), and molecular functions (MFs). BP enrichment was mainly involved in the following processes (Figure 1C): protein phosphorylation (21/51), protein autophosphorylation (17/51), negative regulation of the apoptotic process (16/51), and peptidyl-tyrosine phosphorylation (15/51). Positive CC enrichment was observed in the following compartments (Figure 1D): cytosol (34/51), nucleus (33/51), cytoplasm (29/51), nucleoplasm (26/51), and plasma membrane (26/51). MF enrichment includes (Figure 1E) protein binding (49/51), ATP binding (29/51), identical protein binding (20/51), and protein kinase activity (19/51).
We also performed Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on these target genes, and the KEGG pathway enrichment analysis showed that the target genes were mainly involved in pathways in cancer (31/51), the PI3K-Akt signaling pathway (20/51), endocrine resistance (15/51), MAPK signaling pathway (14/50), Ras signaling pathway (14/51), and other pathways (Figure 1F). These suggest that 4a may exert anti-breast cancer effects by acting on multiple tumor signaling pathways.
2.2.3 Molecular docking
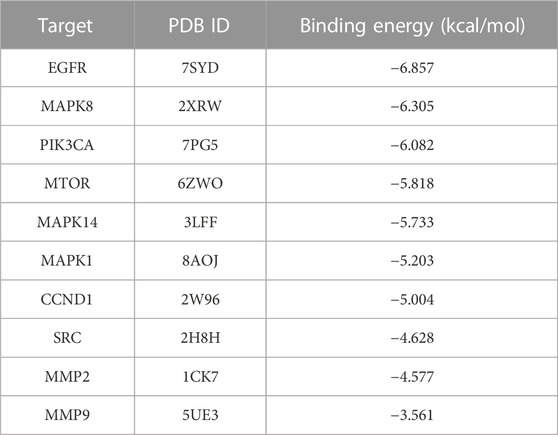
Molecular docking can be used to assess 4a-central target protein interactions. Generally, lower binding energy suggests better interactions. For now, there is no single criterion for screening the binding energy. The screening is usually based on a binding energy ≤ −5.0 kcal/mol (Zhang J. et al., 2022c). The results of molecular docking showed that 7 of the 10 selected central targets were below −5.0 kcal/mol. These results suggest that 4a has good interactions with the seven central targets. All the results are shown in Table 4.
In addition, the visualization analysis allowed us to better understand the spatial structure and interactions between 4a and the central targets. The intermolecular force between 4a and the central targets include van der Waals, hydrogen bond (conventional hydrogen bond, etc.), and hydrophobic (pi–alkyl, alkyl, etc.) forces. Notably, 4a also generates interesting intermolecular forces with central targets, which include pi–sulfur with EGFR, pi–anion with MAPK1 and MAPK14, pi–pi T-shaped with MMP9, pi–cation with MAPK8, amide–pi stacked with MAPK8, and pi–pi stacked with MMP2. These interesting interactions may have a specific effect on the affinity of the 4a-central marker and deserve further investigation. All the results are shown in Supplementary Figures S34–S43.
2.2.4 Molecular dynamics
In order to better understand the kinetics of the interaction between 4a and target proteins, we selected three proteins with the best binding energy—EGFR, MAPK8, and PIK3CA—as key target proteins for molecular dynamics simulation studies.
Root-mean-squared deviation (RMSD), which measures the coordinate deviation of a specific atom with respect to a reference structure, is often used to assess whether a ligand–receptor system has reached stability (Sargsyan et al., 2017). A stable RMSD means that the atoms in the corresponding system become stable, whereas a fluctuating RMSD implies fluctuations. This simulation process is 50 ns (Figure 2A). The simulations show that 4a-MAPK8 and 4a-PIK3CA reach equilibrium soon after the start of the simulation process and that the RMSD values are below 0.3 nm throughout the simulation time. This suggests that the protein–ligand system fluctuates less, which indicates that the docking level is reasonable in the dynamics study (Rastelli et al., 2010; Dai et al., 2023). 4a-EGFR fluctuated more in the initial stage and reached equilibrium after 32 ns, and the RMSD value was approximately 0.5 nm throughout the simulation time, suggesting poor stability in the dynamics study (Li et al., 2010).
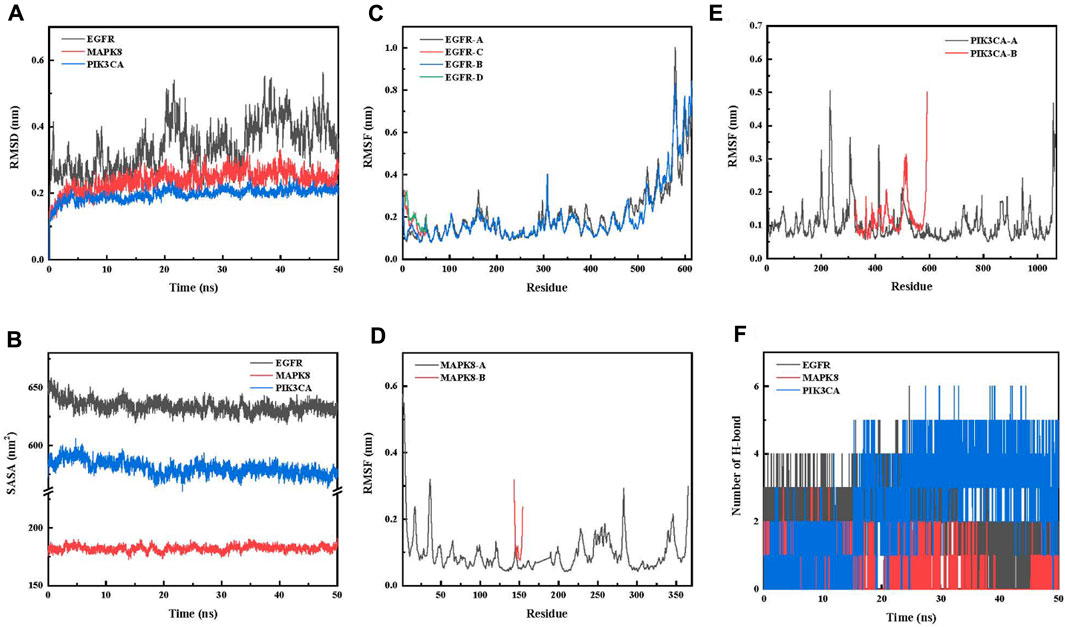
FIGURE 2. (A) Variation curve of RMSD of the protein with time during the simulation of the 4a-key target protein. (B) Variation curve of SASA of the protein with time during the simulation of the 4a-key target protein. (C) RMSF analysis of 4a-EGFR. (D) RMSF analysis of 4a-MAPK8. (E) RMSF analysis of 4a-PIK3CA. (F) Variation in the number of hydrogen bonds with time during the simulation of the 4a-key target protein.
The solvent-accessible surface area (SASA) is calculated by the van der Waals forces interacting with solvent molecules to calculate the solute area (Akoonjee et al., 2022). The lower SASA can be explained by stronger hydrophobic interactions and less inter-complex solvent water, that is, the more compact binding between the 4a-key target protein complexes (VAN DAN BURG et al., 1994). The 50-ns simulation results (Figure 2B) show that all 4a-key target proteins show an acceptable SASA value of the protein–ligand complex during the complex simulation. In addition, the results indicate that the 4a-PIK3CA complex exhibits the most compact structure, followed by the 4a-MAPK8 complex, while the 4a-EGFR complex is the the loosest structure in the dynamics study (Zhou et al., 2023).
Root-mean-squared fluctuation (RMSF) calculates the increase and decrease of each atom relative to its average position, characterizing the change in structure averaged over time, i.e., giving a characterization of the flexibility of each region of the protein (Işık et al., 2022; Xu et al., 2022). All results are shown in Figures 2C–E. First, the residues with small RMSF fluctuations in the protein complex system are consistent with the active residues, which may be related to the interactions such as hydrogen bonds and hydrophobic interactions generated between 4a-key target proteins and forming stable compounds. The regions with larger fluctuations are located in the inactive regions at the edges of the protein, which may be related to interactions such as water and chloride ions. The RMSF values of the 4a-key target protein showed little fluctuation during the 50-ns simulation, indicating that the protein ligands can bind stably in the dynamics study (Radwan et al., 2022).
To explore the interaction of 4a with key target proteins, first, we performed hydrogen bonding analysis (Figure 2F). The average numbers of hydrogen bonds of 4a-EGFR, 4a-MAPK8, and 4a-PIK3CA were 1.97, 0.58, and 2.63, respectively, suggesting that the binding stability of 4a-PIK3CA may be higher than that of the 4a-EGFR and 4a-MAPK8 systems in the dynamics study.
Molecular mechanics-Poisson–Boltzmann surface area (MM-PBSA) is a method for post-processing molecular dynamics trajectories to estimate binding free energies (Homeyer and Gohlke, 2012). To better explain the interaction energy between the ligand and the receptor, we determined the binding energy of all protein–ligand complexes in the equilibrium phase using the gmx_mmpbsa method (Valdés-Tresanco et al., 2021). In the application of the MM-PBSA method, the total binding energy was decomposed into four independent components (electrostatic interactions, van der Waals interactions, and polar and nonpolar solvation interactions). The results of the binding energy of 4a-key target protein are shown in Supplementary Table S4. In the 4a-key target protein complex system, the total binding energies were all negative, indicating their contribution to the binding of the complex system. Among them, the total binding free energies of 4a-EGFR, 4a-MAPK8, and 4a-PIK3CA were −72.310 kJ/mol, −109.794 kJ/mol, and −110.833 kJ/mol, respectively. These results indicate that the total binding free energies of the 4a-key target protein complex system supported the strong binding of 4a-MAPK8 and 4a-PIK3CA complexes, while the 4a-EGFR complex is relatively weaker in the dynamic system (Fu et al., 2017; Kushwaha et al., 2021).
To further understand the 4a-key target protein interaction, we assessed the interaction of each residue with 4a by decomposing the total binding energy into the per-residue contribution energy (Supplementary Figures S44A–C). The important contributing amino acid residues of the 4a-EGFR complex are mainly HIS209 (−8.794 kJ/mol), CYS207 (−5.581 kJ/mol), and ASN210 (−4.140 kJ/mol) in the B-chain of EGFR and HIS209 (−4.043 kJ/mol) in the A-chain of EGFR. The 4a-MAPK8 complex mainly receives contributions from amino acid residues VAL40 (−9.077 kJ/mol), LEU168 (-5.324 kJ/mol), and VAL158 (−4.171 kJ/mol) in the A-chain of MAPK8. LYS678 (−6.364 kJ/mol) and THR679 (−4.450 kJ/mol) in the A-chain of PIK3CA and GLN375 (−4.731 kJ/mol) in the B-chain of PIK3CA are the major amino acid residues of PIK3CA interacting with 4a.
3 Conclusion
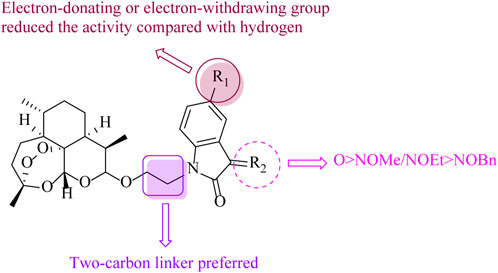
In summary, a series of two-carbon tethered ART–isatin hybrids were synthesized and assessed for their antiproliferative activity against MCF-7, MDA-MB-231, and MDA-MB-231/ADR breast cancer cell lines, as well as cytotoxicity toward MCF-10A cells. SAR (Figure 3) revealed that 1) the length of the alkyl linker between ART and isatin influenced the activity remarkably, and the two-carbon linker was more favorable than the three-carbon linker; 2) the relative contribution order of the functional group at the C-3 position of isatin moiety was carbonyl > methoxy/ethoxyimino > benzyloxyimino; 3) compared with hydrogen, regardless of the electron-donating or electron-withdrawing group at the C-5 position of isatin skeleton, the activity was reduced.
In particular, the representative hybrid 4a (IC50: 2.49–12.6 µM) not only was superior to Adriamycin (IC50: 4.46->100 µM) against all the three breast cancer lines but also exhibited an excellent safety profile and had the potential to overcome drug resistance. Meanwhile, the in silico study provided a preliminary mechanistic study of 4a. The PPI network suggested that 4a may target related proteins expressed by SRC, EGFR, CCND1, MTOR, MAPK1, PIK3CA, MMP9, MAPK8, MMP2, MAPK14, and other genes, which may be central targets for 4a to exert its anti-breast cancer effects. KEGG enrichment analysis suggested that 4a may exert anti-breast cancer effects by participating in the PI3K-Akt signaling pathway, endocrine resistance, MAPK signaling pathway, and other breast cancer-related pathways. Subsequently, molecular docking revealed the binding energy of 4a to the central target and detailed intermolecular force information. Finally, molecular dynamics studies provided dynamic information on the binding pattern of 4a to the three highest interacting key targets (EGFR, MAPK8, and PIK3CA). Therefore, hybrid 4a was a promising anti-breast cancer candidate and merited further preclinical evaluations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
RW: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, validation, visualization, writing–original draft, and writing–review and editing. RH: conceptualization, investigation, methodology, resources, and writing–review and editing. YY: project administration, supervision, and writing–review and editing. ZW: funding acquisition, investigation, project administration, resources, supervision, and writing–review and editing. KS: funding acquisition, project administration, resources, and writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (82002773 and 82072897).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1293763/full#supplementary-material
References
Akoonjee, A., Rampadarath, A., Aruwa, C. E., Ajiboye, T. A., Ajao, A. A. N., and Sabiu, S. (2022). Network pharmacology-and molecular dynamics simulation-based bioprospection of aspalathus linearis for type-2 diabetes care. Metabolites 12 (11), 1013. doi:10.3390/metabo12111013
Akram, M., Iqbal, M., Daniyal, M., and Khan, A. U. (2017). Awareness and current knowledge of breast cancer. Biol. Res. 50, 33–23. doi:10.1186/s40659-017-0140-9
Arnold, M., Morgan, E., Rumgay, H., Mafra, A., Singh, D., Laversanne, M., et al. (2022). Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66, 15–23. doi:10.1016/j.breast.2022.08.010
Bérubé, G. (2016). An overview of molecular hybrids in drug discovery. Expert Opin. drug Discov. 11 (3), 281–305. doi:10.1517/17460441.2016.1135125
Dai, W., Yang, J., Liu, X., Mei, Q., Peng, W., and Hu, X. (2023). Anti-colorectal cancer of Ardisia gigantifolia Stapf. and targets prediction via network pharmacology and molecular docking study. BMC Complementary Med. Ther. 23 (1), 4. doi:10.1186/s12906-022-03822-8
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic acids Res. 47 (W1), W357–W364. doi:10.1093/nar/gkz382
Ding, Z., Zhou, M., and Zeng, C. (2020). Recent advances in isatin hybrids as potential anticancer agents. Arch. Pharm. 353 (3), 1900367. doi:10.1002/ardp.201900367
Dong, M., Zheng, G., Gao, F., Li, M., and Zhong, C. (2022). Three-carbon linked dihydroartemisinin-isatin hybrids: design, synthesis and their antiproliferative anticancer activity. Front. Pharmacol. 13, 834317. doi:10.3389/fphar.2022.834317
Feng, Y., Spezia, M., Huang, S., Yuan, C., Zeng, Z., Zhang, L., et al. (2018). Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes & Dis. 5 (2), 77–106. doi:10.1016/j.gendis.2018.05.001
Fu, M., Chen, L., Zhang, L., Yu, X., and Yang, Q. (2017). Cyclocurcumin, a curcumin derivative, exhibits immune-modulating ability and is a potential compound for the treatment of rheumatoid arthritis as predicted by the MM-PBSA method. Int. J. Mol. Med. 39 (5), 1164–1172. doi:10.3892/ijmm.2017.2926
Gao, F., Sun, Z., Kong, F., and Xiao, J. (2020). Artemisinin-derived hybrids and their anticancer activity. Eur. J. Med. Chem. 188, 112044. doi:10.1016/j.ejmech.2020.112044
Gupta, A. K., Tulsyan, S., Bharadwaj, M., and Mehrotra, R. (2019). Systematic review on cytotoxic and anticancer potential of n-substituted isatins as novel class of compounds useful in multidrug-resistant cancer therapy: in silico and in vitro analysis. Top. Curr. Chem. 377, 15–21. doi:10.1007/s41061-019-0240-9
Homeyer, N., and Gohlke, H. (2012). Free energy calculations by the molecular mechanics Poisson− Boltzmann surface area method. Mol. Inf. 31 (2), 114–122. doi:10.1002/minf.201100135
Hou, H., Qu, B., Su, C., Hou, G., and Gao, F. (2021). Design, synthesis and anti-lung cancer evaluation of 1, 2, 3-Triazole tethered dihydroartemisinin-isatin hybrids. Front. Pharmacol. 12, 3606. doi:10.3389/fphar.2021.801580
Hu, Y., Guo, N., Yang, T., et al. (2022). The potential mechanisms by which artemisinin and its derivatives induce ferroptosis in the treatment of cancer. Oxidative Med. Cell. Longev., 2022. doi:10.1155/2022/1458143
Işık, A., Çevik, U. A., Celik, I., Erçetin, T., Koçak, A., Özkay, Y., et al. (2022). Synthesis, characterization, molecular docking, dynamics simulations, and in silico absorption, distribution, metabolism, and excretion (ADME) studies of new thiazolylhydrazone derivatives as butyrylcholinesterase inhibitors. Z. für Naturforsch. C 77 (11-12), 447–457. doi:10.1515/znc-2021-0316
Jaiswal, P., Tripathi, V., Nayak, A., Kataria, S., Lukashevich, V., Das, A. K., et al. (2021). A Molecular link between diabetes and breast cancer: therapeutic potential of repurposing incretin-based therapies for breast cancer. Curr. Cancer Drug Targets 21 (10), 829–848. doi:10.2174/1568009621666210901101851
Kushwaha, P. P., Singh, A. K., Bansal, T., Yadav, A., Prajapati, K. S., Shuaib, M., et al. (2021). Identification of natural inhibitors against SARS-CoV-2 drugable targets using molecular docking, molecular dynamics simulation, and MM-PBSA approach. Front. Cell. Infect. Microbiol. 11, 728. doi:10.3389/fcimb.2021.730288
Lau, K. H., Tan, A. M., and Shi, Y. (2022). New and emerging targeted therapies for advanced breast cancer. Int. J. Mol. Sci. 23 (4), 2288. doi:10.3390/ijms23042288
Li, Y., Rata, I., Chiu, S.-w., and Jakobsson, E. (2010). Improving predicted protein loop structure ranking using a Pareto-optimality consensus method. BMC Struct. Biol. 10, 22–14. doi:10.1186/1472-6807-10-22
Mancuso, R., Foglio, M., and Olalla Saad, S. (2021). Artemisinin-type drugs for the treatment of hematological malignancies. Cancer Chemother. Pharmacol. 87 (1), 1–22. doi:10.1007/s00280-020-04170-5
Radwan, H. A., Ahmad, I., Othman, I. M., Gad-Elkareem, M. A., Patel, H., Aouadi, K., et al. (2022). Design, synthesis, in vitro anticancer and antimicrobial evaluation, SAR analysis, molecular docking and dynamic simulation of new pyrazoles, triazoles and pyridazines based isoxazole. J. Mol. Struct. 1264, 133312. doi:10.1016/j.molstruc.2022.133312
Rastelli, G., Rio, A. D., Degliesposti, G., and Sgobba, M. (2010). Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. 31 (4), 797–810. doi:10.1002/jcc.21372
Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., et al. (2010). GeneCards Version 3: the human gene integrator. Database 2010, baq020. doi:10.1093/database/baq020
Sargsyan, K., Grauffel, C., and Lim, C. (2017). How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. theory Comput. 13 (4), 1518–1524. doi:10.1021/acs.jctc.7b00028
Schröder, R., Illert, A.-L., Erbes, T., Flotho, C., Lübbert, M., and Duque-Afonso, J. (2022). The epigenetics of breast cancer–Opportunities for diagnostics, risk stratification and therapy. Epigenetics 17 (6), 612–624. doi:10.1080/15592294.2021.1940644
Shaikh, S. S., and Emens, L. A. (2022). Current and emerging biologic therapies for triple negative breast cancer. Expert Opin. Biol. Ther. 22 (5), 591–602. doi:10.1080/14712598.2020.1801627
Sher, G., Salman, N. A., Khan, A. Q., Prabhu, K. S., Raza, A., Kulinski, M., et al. (2022). Epigenetic and breast cancer therapy: promising diagnostic and therapeutic applications. Seminars cancer Biol. 83, 152–165. Elsevier. doi:10.1016/j.semcancer.2020.08.009
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tsang, J., and Tse, G. M. (2020). Molecular classification of breast cancer. Adv. anatomic pathology 27 (1), 27–35. doi:10.1097/PAP.0000000000000232
Tufail, M., Cui, J., and Wu, C. (2022). Breast cancer: molecular mechanisms of underlying resistance and therapeutic approaches. Am. J. Cancer Res. 12 (7), 2920–2949.
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A., and Moreno, E. (2021). gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. theory Comput. 17 (10), 6281–6291. doi:10.1021/acs.jctc.1c00645
Van Dan Burg, B., Dijkstra, B. W., Vriend, G., Van der Vinne, B., Venema, G., and Eijsink, V. G. (1994). Protein stabilization by hydrophobic interactions at the surface. Eur. J. Biochem. 220 (3), 981–985. doi:10.1111/j.1432-1033.1994.tb18702.x
Wang, R., Huang, R., Yuan, Y., Wang, Z., and Shen, K. (2023a). The anti-breast cancer potential of indole/isatin hybrids. Arch. Pharm., e2300402. doi:10.1002/ardp.202300402
Wang, R., Zhang, Q., and Chen, M. (2023b). Artemisinin-isatin hybrids tethered via ethylene linker and their anti-lung cancer activity. Arch. Pharm. 356 (4), 2200563. doi:10.1002/ardp.202200563
Wang, Y., Ding, R., Tai, Z., Hou, H., Gao, F., and Sun, X. (2022). Artemisinin-isatin hybrids with potential antiproliferative activity against breast cancer. Arabian J. Chem. 15 (3), 103639. doi:10.1016/j.arabjc.2021.103639
Wang, Z., Wang, H., Ding, X., Chen, X., and Shen, K. (2020b). A large-cohort retrospective study of metastatic patterns and prognostic outcomes between inflammatory and non-inflammatory breast cancer. Ther. Adv. Med. Oncol. 12, 1758835920932674. doi:10.1177/1758835920932674
Wang, Z., Wang, H., Sun, X., Fang, Y., Lu, S. S., Ding, S. N., et al. (2020a). A risk stratification model for predicting overall survival and surgical benefit in triple-negative breast cancer patients with de novo distant metastasis. Front. Oncol. 10, 14. doi:10.3389/fonc.2020.00014
Waseem, S., and Ahmad, I. (2022). Therapeutic significance of molecular hybrids for breast cancer research and treatment. RSC Med. Chem. doi:10.1039/D2MD00356B
Xu, J., Zhang, S., Wu, T., Fang, X., and Zhao, L. (2022). Discovery of TGFBR1 (ALK5) as a potential drug target of quercetin glycoside derivatives (QGDs) by reverse molecular docking and molecular dynamics simulation. Biophys. Chem. 281, 106731. doi:10.1016/j.bpc.2021.106731
Zhang, J., Wang, R., Qin, Y., and Feng, C. (2022c). Defining the potential targets for biological activity of isoegomaketone based on network pharmacology and molecular docking methods. Life 12 (12), 2115. doi:10.3390/life12122115
Zhang, S., Yi, C., Li, W. W., Luo, Y., Wu, Y. Z., and Ling, H. B. (2022a). The current scenario on anticancer activity of artemisinin metal complexes, hybrids, and dimers. Arch. Pharm. 355 (8), 2200086. doi:10.1002/ardp.202200086
Zhang, Z., Zhang, D., Zhou, Y., Wang, F., Xin, A., Gao, F., et al. (2022b). The anti-lung cancer activity of propylene tethered dihydroartemisinin-isatin hybrids. Arabian J. Chem. 15 (4), 103721. doi:10.1016/j.arabjc.2022.103721
Zhou, H., Bie, S., Li, J., Yuan, L., and Zhou, L. (2023). Comparison on inhibitory effect and mechanism of inhibitors on sPPO and mPPO purified from ‘Lijiang snow’peach by combining multispectroscopic analysis, molecular docking and molecular dynamics simulation. Food Chem. 400, 134048. doi:10.1016/j.foodchem.2022.134048
Keywords: breast cancer, artemisinin, isatin, hybrid molecules, drug resistance, in silico study
Citation: Wang R, Huang R, Yuan Y, Wang Z and Shen K (2023) Two-carbon tethered artemisinin–isatin hybrids: design, synthesis, anti-breast cancer potential, and in silico study. Front. Mol. Biosci. 10:1293763. doi: 10.3389/fmolb.2023.1293763
Received: 13 September 2023; Accepted: 04 October 2023;
Published: 19 October 2023.
Edited by:
Wenhong Deng, Renmin Hospital of Wuhan University, ChinaReviewed by:
Bharathikumar Vellalore Maruthachalam, Janssen Research and Development, United StatesLian-Shun Feng, Peking University, China
Jingjing Wu, Shanghai University of Traditional Chinese Medicine, China
Copyright © 2023 Wang, Huang, Yuan, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunwei Shen, a3dzaGVuQG1lZG1haWwuY29tLmNu; Zheng Wang, emhlbmd3YW5nd2lsc29uQDE2My5jb20=; Yaofeng Yuan, eWFvZmVuZ195dWFuQGZ6dS5lZHUuY24=
†These authors have contributed equally to this work
 Ruo Wang
Ruo Wang Renhong Huang
Renhong Huang Yaofeng Yuan2*
Yaofeng Yuan2* Zheng Wang
Zheng Wang Kunwei Shen
Kunwei Shen