- 1EdinOmics, RRID:SCR_021838, University of Edinburgh, Max Born Crescent, Edinburgh, United Kingdom
- 2Institute of Quantitative Biology, Biochemistry and Biotechnology, School of Biological Sciences, University of Edinburgh, Edinburgh, United Kingdom
This mini review focuses on the opportunities provided by current and emerging separation techniques for mass spectrometry metabolomics. The purpose of separation technologies in metabolomics is primarily to reduce complexity of the heterogeneous systems studied, and to provide concentration enrichment by increasing sensitivity towards the quantification of low abundance metabolites. For this reason, a wide variety of separation systems, from column chemistries to solvent compositions and multidimensional separations, have been applied in the field. Multidimensional separations are a common method in both proteomics applications and gas chromatography mass spectrometry, allowing orthogonal separations to further reduce analytical complexity and expand peak capacity. These applications contribute to exponential increases in run times concomitant with first dimension fractionation followed by second dimension separations. Multidimensional liquid chromatography to increase peak capacity in metabolomics, when compared to the potential of running additional samples or replicates and increasing statistical confidence, mean that uptake of these methods has been minimal. In contrast, in the last 15 years there have been significant advances in the resolution and sensitivity of ion mobility spectrometry, to the point where high-resolution separation of analytes based on their collision cross section approaches chromatographic separation, with minimal loss in sensitivity. Additionally, ion mobility separations can be performed on a chromatographic timescale with little reduction in instrument duty cycle. In this review, we compare ion mobility separation to liquid chromatographic separation, highlight the history of the use of ion mobility separations in metabolomics, outline the current state-of-the-art in the field, and discuss the future outlook of the technology. “Where there is one, you’re bound to divide it. Right in two”, James Maynard Keenan.
1 Introduction
Global untargeted metabolomics aims at facilitating our understanding of the dynamics of the chemical composition of biological systems. Metabolites in heterogenous systems are chemically diverse and vary in their abundances, with many endogenous metabolites existing at very low concentrations. The primary goal of metabolomics is the unbiased relative quantification of each metabolite in a biological system. For comprehensive metabolite coverage of biological systems to be successful, multiple separation techniques are needed.
In this review, we outline the principles of separation systems as applied to mass spectrometry based metabolomics. We focus on the capabilities of the increasingly commonly used technique of ion mobility, hyphenated (or not) to liquid chromatography. We evaluate the current state-of-the-art of ion mobility metabolomics in its various guises and provide a forward-looking summary of where we can expect the technology to move in future.
2 Liquid chromatography
The term liquid chromatography was originally described by Michael Tswett in 1906 (Tswett, 1968). The method initially used to separate plant pigments (hence the name) has now become a powerful and ubiquitous technique routinely applied to separate and purify complex mixtures of molecules. Briefly, analytes are dissolved in a solvent and then passed through a column filled with a stationary phase. The stationary phase can be made of various materials, such as silica or polymer beads, which are packed uniformly into the column. As the analytes pass through the column, different molecules interact with the stationary phase, depending on their physicochemical properties. This interaction causes the molecules to separate and travel through the column at different rates, creating peaks.
There are many different stationary phases, allowing separations to be tailored depending on the particular characteristics of the analytes in question. A detailed analysis of stationary phases is beyond the scope of this article, and there are already many excellent reviews on the subject (Zou et al., 2002; Zhang, 2008; West et al., 2010; Sobańska, 2021), but the most commonly applicable phases in metabolomics in general are reversed phase (RP, which separates on the basis of hydrophobicity) and hydrophilic interaction liquid chromatography (HILIC, which separates on the basis of hydrophilicity).
2.1 Advantages of liquid chromatography separation
While there have been many excellent studies in the metabolomics discipline conducted using direct infusion mass spectrometry (Maleki et al., 2018; Dou et al., 2023; Sun et al., 2023; Wolthuis et al., 2023) and flow injection mass spectrometry (Zang et al., 2018), chromatographic separations are commonly hyphenated to mass spectrometry. This coupling adds a further dimension of separation, beyond mass spectrometry itself, which in essence is a separation.
The main aim of applying chromatographic separations up-front of a mass spectral analysis is to reduce the complexity of highly complex biological samples, in an effort to ameliorate the effects of dynamic range (Want et al., 2005) and ion suppression (Furey et al., 2013). Furthermore, chromatography provides a very valuable concentration of each analyte as it leaves the column as a peak, substantially improving its signal-to-noise ratio and making it more likely to be detected and quantified, as opposed to direct infusion mass spectrometry.
2.2 Limitations of liquid chromatography for global metabolomics
One of the main limitations of chromatography is its lack of reproducibility and its heavy reliance on a few common stationary phases. Reversed-phase chromatography, for example, is now the most well-developed and ubiquitous separation medium in analytical chromatography, and rather than use a different column, it is common to use ion-pairing reagents to adapt the chemistry of an analyte to a reversed-phase system (Gong, 2015). Unfortunately, the vast majority of ion-pairing reagents are not mass spectrometry compatible, comprising (as many do) inorganic salts or highly ionizable organic compounds in millimolar concentrations that overwhelm the detection of less abundant endogenous compounds in the system being studied. Various powerful metabolomics methods exploit these properties to perform more targeted analyses, but for broad-based untargeted metabolomics, analysts are typically limited to RP or HILIC with volatile organic acids, bases and salts used as separation systems. Thus, a major limitation of liquid chromatography, as applied to mass spectrometry is that the separation gradient is responsible for much of the selectivity of the system, which can lead to situations where global separations are poorly suited to the separation of closely related compounds.
3 Ion mobility spectrometry
Ion mobility spectrometry (IMS), also known as gaseous electrophoresis, plasma chromatography (Revercomb and Mason, 1975), or ion chromatography (Helden et al., 1995), is an electrophoretic technique that separates ions based on their mobility in gas phase when subjected to an electric field. Traditional IMS measurements determine the drift velocities of gaseous ions in a weak electric field at a constant temperature. The applied electrical field accelerates ions through the drift region, which is counteracted by the drift gas that impedes ion progress. The speed of ion movement or drift velocity (νd) is therefore proportional to the strength of the applied electric field (E), with mobility (K) of the ion being the constant of proportionality. The mobility of an ion is determined by its shape, size, and charge in the given drift gas. As a result, ions traverse the drift region at a velocity that is proportional to the inverse of their collision cross section (CCS), a physical property that reflects the geometric shape of the ion in the specific gas (Borsdorf and Eiceman, 2006). More compact structural conformations undergo fewer collisions with the drift gas and therefore have smaller CCS values, than extended planar structures. Thus, the CCS feature provides insight into the overall shape of the molecule, while ion mobility allows rapid separation of mixtures, including isobaric ions, typically within milliseconds.
3.1 Types of ion mobility spectrometry
There are four major types of ion mobility spectrometry (Figure 1). The most common and oldest ion mobility analysers consist of a drift tube containing an inert gas, typically dubbed ‘drift tube ion mobility spectrometry or DTIMS’ (Kirk et al., 2019), as described above. Travelling wave ion mobility spectrometry (TWIMS) is a more recent modification of the traditional drift tube on similar hardware, with the addition of an electrostatic pulse waveform that propagates along the tube, allowing nonlinear resolution of ions at the expense of ion heating (Shvartsburg and Smith, 2008). Ion heating is one of the more significant limitations of TWIMS as ion temperatures can reach 551–774 K (Merenbloom et al., 2012) and can lead to dissociation in small molecules (Morsa et al., 2011). More recent developments in this technology include structures for lossless ion manipulation (SLIM) and cyclic ion mobility (Giles et al., 2019). The SLIM architecture consists of a sandwich structure, with parallel, mirror image electrode arrays that can be configured for either conventional or travelling wave ion mobility (Ibrahim et al., 2017). Currently, available commercial instruments based on the technology configure the system in a serpentine array, significantly extending the ion mobility path length to a total of 13 m utilizing 44 U-turns. Resolving power varies depending on the configuration of the ion optics and the voltages applied, but resolutions of around 300 are available (May et al., 2021). The cyclic IMS instrument incorporates a doughnut-shaped cyclic ion mobility (cIM) device that allows ions to be separated via a user-designated number of repetitive 98 cm paths. The drawback to the instrument is that faster, lighter ions eventually end up ‘lapping’ slower ones, and to this end the instrument brackets the cIM with ion traps, allowing isolation and selection of pre-and post-separation ions. This allows some unprecedented and complex ion mobility experiments, such as the one described by (Sisley et al., 2020), to be performed.
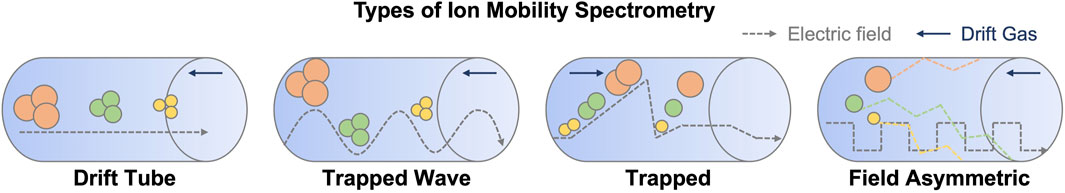
FIGURE 1. Schematic representation of the different types of ion mobility and the principles of separation. The direction of drift gas flow and electric field are indicated with dark blue and grey arrows, respectively.
Another type of ion mobility analyser is high-field asymmetric ion mobility spectrometry (FAIMS). This relies on the use of a changing compensation voltage as a filter across a gas flow counter to the direction of the ions. FAIMS was originally published by Buryakov et al. (1993), and has since found its way into several commercial instruments. It is generally employed as a filter (Canterbury et al., 2008), rather than a separation device and has therefore been sparsely used in liquid chromatography coupled to ion mobility multidimensional separations.
Recently, the Park group introduced the trapped ion mobility spectrometry (TIMS) device (Ridgeway et al., 2016). Unlike the previous systems, in TIMS the ions are held in a trapping device and exposed to a moving column of gas, based typically on a modification of ion funnel technology. This trap and release substantially reduce the dimensions of the ion mobility device. Ions are separated based on their physicochemical properties as in conventional drift tube ion mobility, however, the movement of the gas and ions are reversed. In TIMS, ramping the electric field gradient (E) releases ions in descending order of their mobility (K). For an excellent review of TIMS, see (Ridgeway et al., 2018). Parallel accumulation, serial fragmentation (PASEF) is an experimental methodology available on TIMS instruments that uses the unique capabilities of the ion mobility trapping device to accumulate ions, and then sequentially pulse them into a quadrupole for isolation and fragmentation (Meier et al., 2018). Thus, in a single drift spectrum, fragment patterns for each MSMS spectrum are resolved via ion mobility. This dramatically increases the acquisition rate of data dependent acquisition experiments (Meier et al., 2015). It has recently been modified to support data-independent acquisition (dia-PASEF) for proteomics applications (Demichev et al., 2022), which provides an intriguing possibility for improving metabolomics analysis.
3.2 Advantages of ion mobility as a separation technology
Regardless of the specific type of ion mobility device, there are two main advantages of ion mobility spectrometry. Firstly, it allows the separation of isomeric analytes. As discussed in Section 2.2, one of the major limitations of chromatography is that while an optimal liquid chromatography gradient can separate closely related ions, the generally applicable rapid gradients used with untargeted analysis often preclude the separation of isomeric compounds with closely related structures. While this is an issue for chromatography, modern ion mobility instruments with resolutions greater than 100 can now separate many isomeric ions within milliseconds. By coupling ion mobility to chromatography, multidimensional separation can be performed, allowing unprecedented resolution of complex mixtures without extending separation times or employing complex fractionation strategies.
Similar to the first, the second advantage of ion mobility comes from the modern high-resolution IM instruments. Collisional cross section is a fundamental property of an ion, and it can therefore be calculated with greater accuracy than predicting retention times on columns. While ion mobility devices measure ‘drift times’ rather than CCS directly, all modern instruments can calculate CCS from drift times, and this directly addresses one of the biggest challenges in metabolomics—the unambiguous identification of metabolites.
This latter poses an important challenge for the application of IMS to metabolomics. CCSs are able to be determined from experimental parameters via the Mason-Schamp equation (Siems et al., 2012) in DTIMS instruments, while for other types of IMS, calibration standards with known CCS values (derived from DTIMS experiments) must be used (Dodds and Baker, 2019). In practice, even on DTIMS instruments, a typical use for metabolomics, e.g., single field multiplexed analysis, require calibration standards derived from slower, more complex stepped-field methodologies (Dodds and Baker, 2019). Due to the development of large-scale, experimentally determined libraries (Zheng et al., 2017; Picache et al., 2019), as well as appropriate calibration, CCS features provide an additional criterion for metabolite annotation.
3.3 Limitations of ion mobility
Sensitivity has been the perennial issue with ion mobility mass spectrometry. In standard ion mobility analysis, analytes are introduced to the ion mobility device as a packet, followed by ion separation and concomitant detection in the mass spectrometer. Before the next packet of ions can be introduced to the ion mobility device, the slowest ion in the current packet must pass through and exit the IM device. This means that during ion mobility separation, all ions that would otherwise be analysed are lost. Furthermore, due to imperfect electronics, ions of interest can also be lost during the ion mobility separation itself. There have been many attempts to improve sensitivity, such as improving overall instrument design as outlined in 3.1 (Deng et al., 2016), advances in the construction of the instrument with improved lens designs and electric field generation, and the use of multiplexing. Multiplexing is a technique used to decrease ion packet losses in ion mobility mass spectrometry. Multiple ion packets are pulsed into a single ion mobility device in a pseudo-random order. This creates a patterning effect in the ion mobility analysis that can be deconvoluted by the use of a sliding window algorithm during data analysis. This technique has been reported to reduce sensitivity losses from 99% to 50% (Reinecke et al., 2019).
Current, commercially available ion mobility instruments possess resolving power in the hundreds, compared to the tens of thousands of theoretical plates typical in chromatography (for a fascinating discussion on the relationship between resolution parameters for chromatography and spectrometry see (Rokushika et al., 1985)). Consequently, reproducibility is a key parameter to confidently assign collisional cross sections to analytes. Interlaboratory studies have been performed, demonstrating RSDs of 0.14% and ∼1.5% in TWIMS-based devices (Hernández-Mesa et al., 2020; Righetti et al., 2020) and 0.29% for DTIMS-based devices (Stow et al., 2017), which compares favourably to chromatography retention time RSD of 5%–20% depending on analyte (Madji Hounoum et al., 2015).
A further limitation to ion mobility is that it does not get around the problem of matrix effect, a somewhat complex term in ‘omics disciplines, particularly metabolomics. Typically, the “matrix” corresponds to all of the components of a sample one does not want to analyse, while the “analytes” are the components one wants to detect. While all sample preparation methodologies are biased, a good ‘targeted’ analysis aims to strip out the vast majority of the “matrix” while leaving the “analytes” in place. In untargeted metabolomics one does not have this luxury, and the “matrix” and the analytes are (apart from contaminants, buffers, salts, etc.) mostly the same thing. The interaction between these contaminants, as well as the more abundant analytes within the ion mobility device can result in ion suppression (or ion enhancement).
Ion suppression is commonly associated with electrospray ionization, although matrix effects and suppression of ionization are also found in electron ionization used with gas chromatography (Yarita et al., 2015) and nanoelectrospray ionization (Kourtchev et al., 2020). Ion suppression occurs in the ion source, but matrix effects such as ion-ion interactions and space charging can occur in trapping instruments (Hohenester et al., 2020). Thus, ion mobility separations, occurring as they do, after ionization, provide no reduction on in-source ion suppression. Consequently, suppression resulting from, flow injection or direct infusion mass spectrometry will not be ameliorated by the use of IMS, and could result in additional artifacts in measurement (Levin et al., 2014).
4 Hyphenating ion mobility to mass spectrometry reduces sample complexity
As previously stated, the primary purpose of separation in mass spectrometry studies is the reduction in sample complexity and reduction of matrix effects. The human metabolome database currently contains 217,920 compounds (Wishart et al., 2022), demonstrating the scale of metabolomics researchers’ analytical challenge. Of these, 8,369 metabolites are currently listed as ‘detected’ in human tissues, with the remainder either predicted or derived from other sources, such as the microbiome and the diet. With the impact of ion suppression (see 3.3) and the limited dynamic range of mass spectrometers (Want et al., 2005), it is very common to perform some separation of analytes before they are presented to the mass spectrometer.
An exception to this is mass spectrometry imaging (MSI), a technique where mass spectrometry is used to raster across a tissue or other spatially resolved sample, providing a spectrum per pixel. Individual ions can be collated to generate images of the distribution of a molecular species to, for example, a tissue structure or feature. Of course, a typical matrix-assisted laser desorption ionization (MALDI) (Tanaka et al., 1988) or desorption electrospray ionization (DESI) (Takáts et al., 2004) process is extremely challenging to hyphenate to chromatography as extraction of analytes and ionization are performed directly from the sample. Therefore, MSI sees considerable benefits from the incorporation of ion mobility separation prior to mass spectrometry (Spraggins et al., 2019), regardless of any impact of matrix effects, as high-resolution IMS can separate isobaric compounds without prior chromatographic separation, while also maintaining acquisition speed.
Outside the world of MSI, hyphenation of chromatography to mass spectrometry has been used to ameliorate ion suppression and the effects of limited dynamic range for decades, and the recent advances in ion mobility peak capacities have made this a viable addition, or alternative, to chromatography separations.
5 The use of multidimensional separation and best practices
Typical multidimensional separation workflows consist of coupling multiple liquid chromatography-based separation chemistries and hyphenating to mass spectrometry. A full overview of metabolomics applications with liquid chromatography is beyond the scope of this article. For a comprehensive review of recent methods please read (Lv et al., 2019).
Many examples of ion mobility used as a separation method for metabolomics studies appear in the literature (see Table 1). Of the 28 tabulated articles, nine perform no prior separation to analysis via ion mobility-mass spectrometry. Of these, four are single-cell studies using a variety of methods (laser ablation electrospray ionization, matrix-assisted laser desorption ionization, microsampling), where the addition of chromatography would be challenging, if not impossible. For the remainder, a variety of chromatography methods are hyphenated. The most common remains the use of RP chromatography employing C18 columns, but a growing number of HILIC chromatography methods are beginning to be applied. A single method by Causon et al. incorporates heart-cutting two-dimensional liquid chromatography into a metabolomics workflow, while an additional two methods incorporate both HILIC and RP chromatography, performed separately, into their analyses.
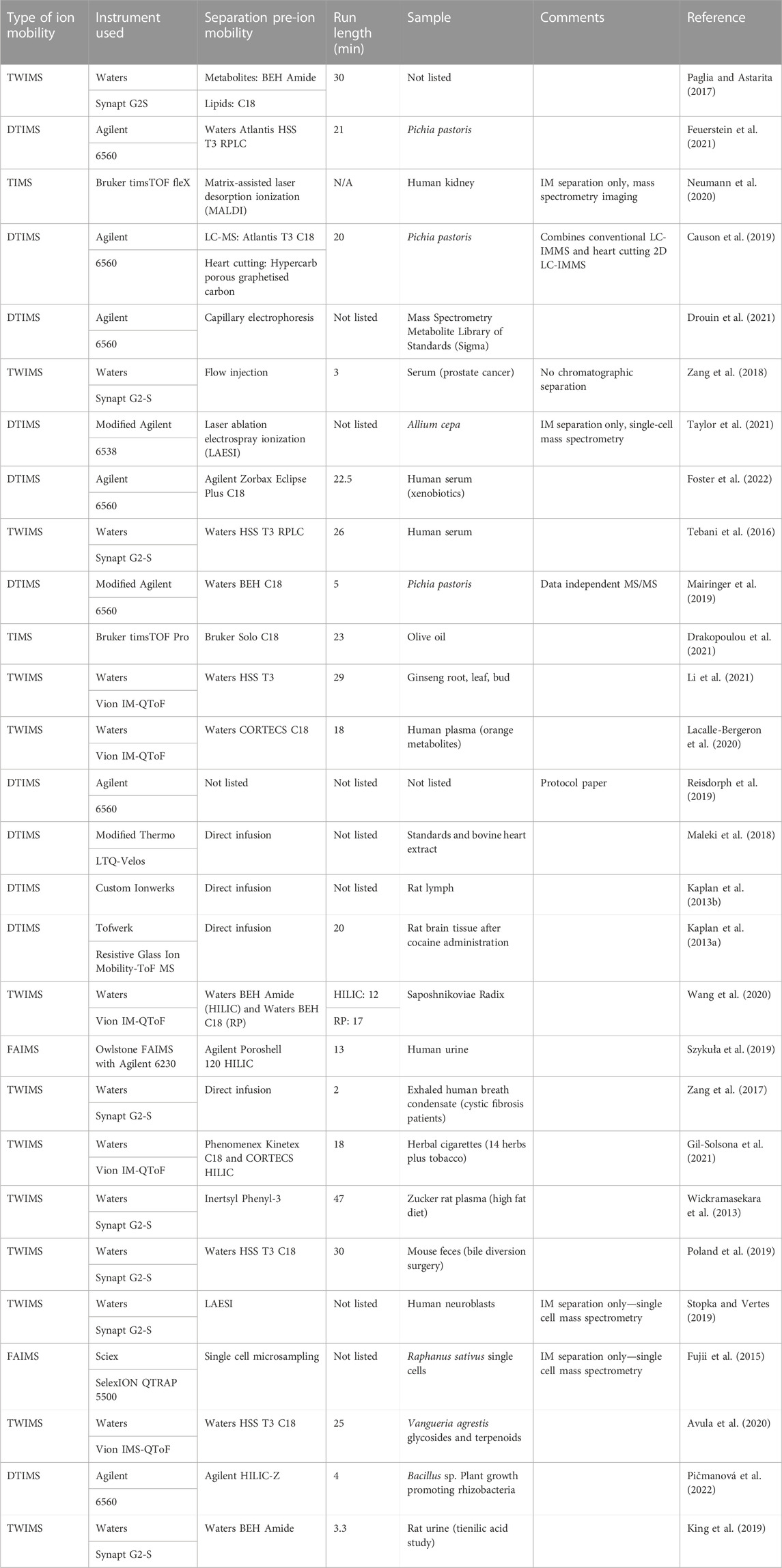
TABLE 1. Typical workflows in liquid chromatography ion mobility mass spectrometry utilizing multidimensional separation principles.
Liquid chromatography hyphenated with ion mobility mass spectrometry has become a powerful technique for reducing complexity in untargeted metabolomics. Publications should, of course, adhere to the most appropriate rigorous standards for reporting, the most recent of which is currently (Kirwan et al., 2022), but ion mobility mass spectrometry is a relatively recent development and standards are still evolving. At minimum, the parameters for the ion mobility device should be recorded along with those for the mass spectrometer it is hyphenated to. This is relatively straightforward for dedicated hybrid instruments, but can be more complex for aftermarket or less intrinsically-linked hardware. Additionally, calibration standards should be noted and the parameters of the calibration should be recorded. Typically these are performed before every batch, but IM calibration samples at both the beginning and the end of a run would provide good evidence that the system was working to appropriate parameters during sample acquisition. CCS libraries, both predicted (Zhou et al., 2020) and experimentally determined (Nichols et al., 2018) are now commonly used in ion mobility mass spectrometry analyses. Both are useful but which library and what criteria (typically a 1%–2% window is used to match CCS) were used is critically important information in the quality assessment of identifications, and should be rigorously reported.
Of course, libraries are not valuable without software to support their use, and academic software that supports the diversity of ion mobility datasets has been relatively slow in development in comparison to the advances in instrumentation. The three most commonly used packages in the field are MS-Dial (Tsugawa et al., 2015), Skyline (MacLean et al., 2010) and MzMine (Schmid et al., 2023). All three are graphical applications that provide peak picking, metabolite annotation and statistics, with Skyline being more closely aimed at targeted applications and MS-Dial having the benefit of integrated large-scale libraries. Ideally, the development of modular tools accessible via R and/or Python would support rapid advancement in terms of new features, that could then be backported into more attractive GUI applications while allowing the possibility of building pipelines into interfaces such as Workflow4Metabolomics (Giacomoni et al., 2015).
6 Conclusion and prospects
Ion mobility spectrometry hyphenated to mass spectrometry has seen rapid technological advancement in the previous 10 years. Initially confined to a few specialists, it is finding itself now routinely applied in proteomics and metabolomics studies for the valuable additional information it provides. Given its capacity for extremely rapid separations, we anticipate a great future for the technology and expect to see further developments to improvements in resolution for the technology.
Author contributions
TM and KB: writing original draft, preparation of figures and tables, conceptualisation and revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Biotechnology and Biological Sciences Research Council grant BB/W020270/1 and the Engineering and Physical Sciences Research Council grant EP/V038095/1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Avula, B., Bae, J. Y., Wang, Y. H., Wang, M., Osman, A. G., Smith, K., et al. (2020). Chemical profiling and characterization of phenolic acids, flavonoids, terpene glycosides from Vangueria agrestis using ultra-high-performance liquid chromatography/ion mobility quadrupole time-of-flight mass spectrometry and metabolomics approach. Biomed. Chromatogr. 34, e4840. doi:10.1002/bmc.4840
Borsdorf, H., and Eiceman, G. A. (2006). Ion mobility spectrometry: Principles and applications. Appl. Spectrosc. Rev. 41, 323–375. doi:10.1080/05704920600663469
Buryakov, I. A., Krylov, E. V., Nazarov, E. G., and Rasulev, U. Kh. (1993). A new method of separation of multi-atomic ions by mobility at atmospheric pressure using a high-frequency amplitude-asymmetric strong electric field. Int. J. Mass Spectrom. Ion. Process 128, 143–148. doi:10.1016/0168-1176(93)87062-w
Canterbury, J. D., Yi, X., Hoopmann, M. R., and MacCoss, M. J. (2008). Assessing the dynamic range and peak capacity of nanoflow LC-FAIMS-MS on an ion trap mass spectrometer for proteomics. Anal. Chem. 80, 6888–6897. doi:10.1021/ac8004988
Causon, T. J., Si-Hung, L., Newton, K., Kurulugama, R. T., Fjeldsted, J., and Hann, S. (2019). Fundamental study of ion trapping and multiplexing using drift tube-ion mobility time-of-flight mass spectrometry for non-targeted metabolomics. Anal. Bioanal. Chem. 411, 6265–6274. doi:10.1007/s00216-019-02021-8
Demichev, V., Szyrwiel, L., Yu, F., Teo, G. C., Rosenberger, G., Niewienda, A., et al. (2022). dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts. Nat. Commun. 13, 3944. doi:10.1038/s41467-022-31492-0
Deng, L., Ibrahim, Y. M., Baker, E. S., Aly, N. A., Hamid, A. M., Zhang, X., et al. (2016). Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. Anal. Chem. 1, 2396–2399. doi:10.1002/slct.201600460
Dodds, J. N., and Baker, E. S. (2019). Ion mobility spectrometry: Fundamental concepts, instrumentation, applications, and the road ahead. J. Am. Soc. Mass Spectrom. 30, 2185–2195. doi:10.1007/s13361-019-02288-2
Dou, Y., Mäkinen, M., and Jänis, J. (2023). Analysis of volatile and nonvolatile constituents in gin by direct-infusion ultrahigh-resolution ESI/APPI FT-ICR mass spectrometry. J. Agric. Food Chem. 71, 7082–7089. doi:10.1021/acs.jafc.3c00707
Drakopoulou, S. K., Damalas, D. E., Baessmann, C., and Thomaidis, N. S. (2021). Trapped ion mobility incorporated in LC-HRMS workflows as an integral analytical platform of high sensitivity: Targeted and untargeted 4D-metabolomics in extra virgin olive oil. J. Agric. Food Chem. 69, 15728–15737. doi:10.1021/acs.jafc.1c04789
Drouin, N., Mielcarek, A., Wenz, C., and Rudaz, S. (2021). Evaluation of ion mobility in capillary electrophoresis coupled to mass spectrometry for the identification in metabolomics. Electrophoresis 42, 342–349. doi:10.1002/elps.202000120
Feuerstein, M. L., Kurulugama, R. T., Hann, S., and Causon, T. (2021). Novel acquisition strategies for metabolomics using drift tube ion mobility-quadrupole resolved all ions time-of-flight mass spectrometry (IM-QRAI-TOFMS). Anal. Chim. Acta 1163, 338508. doi:10.1016/j.aca.2021.338508
Foster, M., Rainey, M., Watson, C., Dodds, J. N., Kirkwood, K. I., Fernández, F. M., et al. (2022). Uncovering PFAS and other xenobiotics in the dark metabolome using ion mobility spectrometry, mass defect analysis, and machine learning. Environ. Sci. Technol. 56, 9133–9143. doi:10.1021/acs.est.2c00201
Fujii, T., Matsuda, S., Tejedor, M. L., Esaki, T., Sakane, I., Mizuno, H., et al. (2015). Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat. Protoc. 10, 1445–1456. doi:10.1038/nprot.2015.084
Furey, A., Moriarty, M., Bane, V., Kinsella, B., and Lehane, M. (2013). Ion suppression; A critical review on causes, evaluation, prevention and applications. Talanta 115, 104–122. doi:10.1016/j.talanta.2013.03.048
Giacomoni, F., Le Corguillé, G., Monsoor, M., Landi, M., Pericard, P., Pétéra, M., et al. (2015). Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 31, 1493–1495. doi:10.1093/bioinformatics/btu813
Gil-Solsona, R., Sancho, J. V., Gassner, A. L., Weyermann, C., Hernández, F., Delémont, O., et al. (2021). Use of ion mobility-high resolution mass spectrometry in metabolomics studies to provide near MS/MS quality data in a single injection. J. Mass Spectrom. 56, e4718. doi:10.1002/jms.4718
Giles, K., Ujma, J., Wildgoose, J., Pringle, S., Richardson, K., Langridge, D., et al. (2019). A cyclic ion mobility-mass spectrometry system. Anal. Chem. 91, 8564–8573. doi:10.1021/acs.analchem.9b01838
Gong, L. (2015). Comparing ion-pairing reagents and counter anions for ion-pair reversed-phase liquid chromatography/electrospray ionization mass spectrometry analysis of synthetic oligonucleotides. Rapid Commun. Mass Spectrom. 29, 2402–2410. doi:10.1002/rcm.7409
Helden, G. Von, Bowers, M. T., and Wyttenbach, T. (1995). Inclusion of a MALDI ion source in the ion chromatography technique: Conformational information on polymer and biomolecular ions. Int. J. Mass Spectrom. Ion. Process 147, 349–364. doi:10.1016/0168-1176(95)04211-3
Hernández-Mesa, M., D’Atri, V., Barknowitz, G., Fanuel, M., Pezzatti, J., Dreolin, N., et al. (2020). Interlaboratory and interplatform study of steroids collision cross section by traveling wave ion mobility spectrometry. Anal. Chem. 92, 5013–5022. doi:10.1021/acs.analchem.9b05247
Hohenester, U. M., Barbier Saint-Hilaire, P., Fenaille, F., and Cole, R. B. (2020). Investigation of space charge effects and ion trapping capacity on direct introduction ultra-high-resolution mass spectrometry workflows for metabolomics. J. Mass Spectrom. 55, e4613. doi:10.1002/jms.4613
Ibrahim, Y. M., Hamid, A. M., Deng, L., Garimella, S. V. B., Webb, I. K., Baker, E. S., et al. (2017). New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst 142, 1010–1021. doi:10.1039/c7an00031f
Kaplan, K. A., Chiu, V. M., Lukus, P. A., Zhang, X., Siems, W. F., Schenk, J. O., et al. (2013a). Neuronal metabolomics by ion mobility mass spectrometry: Cocaine effects on glucose and selected biogenic amine metabolites in the frontal cortex, striatum, and thalamus of the rat. Anal. Bioanal. Chem. 405, 1959–1968. doi:10.1007/s00216-012-6638-7
Kaplan, K., Jackson, S., Dwivedi, P., Davidson, W. S., Yang, Q., Tso, P., et al. (2013b). Monitoring dynamic changes in lymph metabolome of fasting and fed rats by matrix-assisted laser desorption/ionization-ion mobility mass spectrometry (MALDI-IMMS). Int. J. Ion Mobil. Spectrom. 16, 177–184. doi:10.1007/s12127-012-0102-4
King, A. M., Mullin, L. G., Wilson, I. D., Coen, M., Rainville, P. D., Plumb, R. S., et al. (2019). Development of a rapid profiling method for the analysis of polar analytes in urine using HILIC–MS and ion mobility enabled HILIC–MS. Metabolomics 15, 17–11. doi:10.1007/s11306-019-1474-9
Kirk, A. T., Bohnhorst, A., Raddatz, C. R., Allers, M., and Zimmermann, S. (2019). Ultra-high-resolution ion mobility spectrometry — Current instrumentation, limitations, and future developments. Anal. Bioanal. Chem. 411, 6229–6246. doi:10.1007/s00216-019-01807-0
Kirwan, J. A., Gika, H., Beger, R. D., Bearden, D., Dunn, W. B., Goodacre, R., et al. (2022). Quality assurance and quality control reporting in untargeted metabolic phenotyping: mQACC recommendations for analytical quality management. Metabolomics 18, 70–16. doi:10.1007/s11306-022-01926-3
Kourtchev, I., Szeto, P., O’Connor, I., Popoola, O. A. M., Maenhaut, W., Wenger, J., et al. (2020). Comparison of heated electrospray ionization and nanoelectrospray ionization sources coupled to ultra-high-resolution mass spectrometry for analysis of highly complex atmospheric aerosol samples. Anal. Chem. 92, 8396–8403. doi:10.1021/acs.analchem.0c00971
Lacalle-Bergeron, L., Portolés, T., López, F. J., Sancho, J. V., Ortega-Azorín, C., Asensio, E. M., et al. (2020). Ultra-Performance liquid chromatography-ion mobility separation-quadruple time-of-flight ms (UHPLC-IMS-QTOF ms) metabolomics for short-term biomarker discovery of orange intake: A randomized, controlled crossover study. Nutrients 12, 1916. doi:10.3390/nu12071916
Levin, M., Krisilov, A., Zon, B., and Eiceman, G. (2014). The effect of space charge in ion mobility spectrometry. Int. J. Ion Mobil. Spectrom. 17, 73–77. doi:10.1007/s12127-014-0151-y
Li, W., Yang, X., Chen, B., Zhao, D., Wang, H., Sun, M., et al. (2021). Ultra-high performance liquid chromatography/ion mobility time-of-flight mass spectrometry-based untargeted metabolomics combined with quantitative assay unveiled the metabolic difference among the root, leaf, and flower bud of Panax notoginseng. Arabian J. Chem. 14, 103409. doi:10.1016/j.arabjc.2021.103409
Lv, W., Shi, X., Wang, S., and Xu, G. (2019). Multidimensional liquid chromatography-mass spectrometry for metabolomic and lipidomic analyses. Trends Anal. Chem. 120, 115302. doi:10.1016/j.trac.2018.11.001
MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., et al. (2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. doi:10.1093/bioinformatics/btq054
Madji Hounoum, B., Blasco, H., Nadal-Desbarats, L., Diémé, B., Montigny, F., Andres, C. R., et al. (2015). Analytical methodology for metabolomics study of adherent mammalian cells using NMR, GC-MS and LC-HRMS. Anal. Bioanal. Chem. 407, 8861–8872. doi:10.1007/s00216-015-9047-x
Mairinger, T., Kurulugama, R., Causon, T. J., Stafford, G., Fjeldsted, J., and Hann, S. (2019). Rapid screening methods for yeast sub-metabolome analysis with a high-resolution ion mobility quadrupole time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 33, 66–74. doi:10.1002/rcm.8420
Maleki, H., Karanji, A. K., Majuta, S., Maurer, M. M., and Valentine, S. J. (2018). Ion mobility spectrometry-mass spectrometry coupled with gas-phase hydrogen/deuterium exchange for metabolomics analyses. J. Am. Soc. Mass Spectrom. 29, 230–241. doi:10.1007/s13361-017-1798-5
May, J. C., Leaptrot, K. L., Rose, B. S., Moser, K. L. W., Deng, L., Maxon, L., et al. (2021). Resolving power and collision cross section measurement accuracy of a prototype high-resolution ion mobility platform incorporating structures for lossless ion manipulation. J. Am. Soc. Mass Spectrom. 32, 1126–1137. doi:10.1021/jasms.1c00056
Meier, F., Beck, S., Grassl, N., Lubeck, M., Park, M. A., Raether, O., et al. (2015). Parallel accumulation-serial fragmentation (PASEF): Multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. J. Proteome Res. 14, 5378–5387. doi:10.1021/acs.jproteome.5b00932
Meier, F., Brunner, A. D., Koch, S., Koch, H., Lubeck, M., Krause, M., et al. (2018). Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545. doi:10.1074/mcp.TIR118.000900
Merenbloom, S. I., Flick, T. G., and Williams, E. R. (2012). How hot are your ions in TWAVE ion mobility spectrometry? J. Am. Soc. Mass Spectrom. 23, 553–562. doi:10.1007/s13361-011-0313-7
Morsa, D., Gabelica, V., and De Pauw, E. (2011). Effective temperature of ions in traveling wave ion mobility spectrometry. Anal. Chem. 83, 5775–5782. doi:10.1021/ac201509p
Neumann, E. K., Migas, L. G., Allen, J. L., Caprioli, R. M., Van De Plas, R., and Spraggins, J. M. (2020). Spatial metabolomics of the human kidney using MALDI trapped ion mobility imaging mass spectrometry. Anal. Chem. 92, 13084–13091. doi:10.1021/acs.analchem.0c02051
Nichols, C. M., Dodds, J. N., Rose, B. S., Picache, J. A., Morris, C. B., Codreanu, S. G., et al. (2018). Untargeted molecular discovery in primary metabolism: Collision cross section as a molecular descriptor in ion mobility-mass spectrometry. Anal. Chem. 90, 14484–14492. doi:10.1021/acs.analchem.8b04322
Paglia, G., and Astarita, G. (2017). Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat. Protoc. 12, 797–813. doi:10.1038/nprot.2017.013
Picache, J. A., Rose, B. S., Balinski, A., Leaptrot, K. L., Sherrod, S. D., May, J. C., et al. (2019). Collision cross section compendium to annotate and predict multi-omic compound identities. Chem. Sci. 10, 983–993. doi:10.1039/c8sc04396e
Pičmanová, M., Moses, T., Cortada-Garcia, J., Barrett, G., Florance, H., Pandor, S., et al. (2022). Rapid HILIC-Z ion mobility mass spectrometry (RHIMMS) method for untargeted metabolomics of complex biological samples. Metabolomics 18, 16–12. doi:10.1007/s11306-022-01871-1
Poland, J. C., Schrimpe-Rutledge, A. C., Sherrod, S. D., Flynn, C. R., and McLean, J. A. (2019). Utilizing untargeted ion mobility-mass spectrometry to profile changes in the gut metabolome following biliary diversion Surgery. Anal. Chem. 91, 14417–14423. doi:10.1021/acs.analchem.9b02924
Reinecke, T., Naylor, C. N., and Clowers, B. H. (2019). Ion multiplexing: Maximizing throughput and signal to noise ratio for ion mobility spectrometry. Trends Anal. Chem. 116, 340–345. doi:10.1016/j.trac.2019.03.014
Reisdorph, R., Michel, C., Quinn, K., Doenges, K., and Reisdorph, N. (2019). Untargeted differential metabolomics analysis using drift tube ion mobility-mass spectrometry. Methods Mol. Biol. 2084, 55–78. doi:10.1007/978-1-0716-0030-6_3
Revercomb, H. E., and Mason, E. A. (1975). Theory of plasma chromatography/gaseous electrophoresis. Review. Anal. Chem. 47, 970–983. doi:10.1021/ac60357a043
Ridgeway, M. E., Lubeck, M., Jordens, J., Mann, M., and Park, M. A. (2018). Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom. 425, 22–35. doi:10.1016/j.ijms.2018.01.006
Ridgeway, M. E., Wolff, J. J., Silveira, J. A., Lin, C., Costello, C. E., and Park, M. A. (2016). Gated trapped ion mobility spectrometry coupled to fourier transform ion cyclotron resonance mass spectrometry. Int. J. Ion Mobil. Spectrom. 19, 77–85. doi:10.1007/s12127-016-0197-0
Righetti, L., Dreolin, N., Celma, A., McCullagh, M., Barknowitz, G., Sancho, J. V., et al. (2020). Travelling wave ion mobility-derived collision cross section for mycotoxins: Investigating interlaboratory and interplatform reproducibility. J. Agric. Food Chem. 68, 10937–10943. doi:10.1021/acs.jafc.0c04498
Rokushika, S., Hatano, H., Bairn, M. A., and Hill, H. H. (1985). Resolution measurement for ion mobility spectrometry. Anal. Chem. 57, 1902–1907. doi:10.1021/ac00286a023
Schmid, R., Heuckeroth, S., Korf, A., Smirnov, A., Myers, O., Dyrlund, T. S., et al. (2023). Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 41, 447–449. doi:10.1038/s41587-023-01690-2
Shvartsburg, A. A., and Smith, R. D. (2008). Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 80, 9689–9699. doi:10.1021/ac8016295
Siems, W. F., Viehland, L. A., and Hill, H. H. (2012). Improved momentum-transfer theory for ion mobility. 1. Derivation of the fundamental equation. Anal. Chem. 84, 9782–9791. doi:10.1021/ac301779s
Sisley, E. K., Ujma, J., Palmer, M., Giles, K., Fernandez-Lima, F. A., and Cooper, H. J. (2020). LESA cyclic ion mobility mass spectrometry of intact proteins from thin tissue sections. Anal. Chem. 92, 6321–6326. doi:10.1021/acs.analchem.9b05169
Sobańska, A. W. (2021). Emerging or underestimated silica-based stationary phases in liquid chromatography. Crit. Rev. Anal. Chem. 51, 631–725. doi:10.1080/10408347.2020.1760782
Spraggins, J. M., Djambazova, K. V., Rivera, E. S., Migas, L. G., Neumann, E. K., Fuetterer, A., et al. (2019). High-Performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem. 91, 14552–14560. doi:10.1021/acs.analchem.9b03612
Stopka, S. A., and Vertes, A. (2019). Metabolomic profiling of adherent mammalian cells in situ by LAESI-MS with ion mobility separation. Methods Mol. Biol. 2084, 235–244. doi:10.1007/978-1-0716-0030-6_15
Stow, S. M., Causon, T. J., Zheng, X., Kurulugama, R. T., Mairinger, T., May, J. C., et al. (2017). An interlaboratory evaluation of drift tube ion mobility-mass spectrometry collision cross section measurements. Anal. Chem. 89, 9048–9055. doi:10.1021/acs.analchem.7b01729
Sun, X., Jia, Z., Zhang, Y., Zhao, X., Zhao, C., Lu, X., et al. (2023). A strategy for uncovering the serum metabolome by direct-infusion high-resolution mass spectrometry. Metabolites 13, 460. doi:10.3390/metabo13030460
Szykuła, K. M., Meurs, J., Turner, M. A., Creaser, C. S., and Reynolds, J. C. (2019). Combined hydrophilic interaction liquid chromatography-scanning field asymmetric waveform ion mobility spectrometry-time-of-flight mass spectrometry for untargeted metabolomics. Anal. Bioanal. Chem. 411, 6309–6317. doi:10.1007/s00216-019-01790-6
Takáts, Z., Wiseman, J. M., Gologan, B., and Cooks, R. G. (2004). Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Sci. (1979) 306, 471–473. doi:10.1126/science.1104404
Tanaka, K., Waki, H., Ido, Y., Akita, S., Yoshida, Y., Yoshida, T., et al. (1988). Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2, 151–153. doi:10.1002/rcm.1290020802
Taylor, M. J., Mattson, S., Liyu, A., Stopka, S. A., Ibrahim, Y. M., Vertes, A., et al. (2021). Optical microscopy-guided laser ablation electrospray ionization ion mobility mass spectrometry: Ambient single cell metabolomics with increased confidence in molecular identification. Metabolites 11, 200. doi:10.3390/metabo11040200
Tebani, A., Schmitz-Afonso, I., Rutledge, D. N., Gonzalez, B. J., Bekri, S., and Afonso, C. (2016). Optimization of a liquid chromatography ion mobility-mass spectrometry method for untargeted metabolomics using experimental design and multivariate data analysis. Anal. Chim. Acta 913, 55–62. doi:10.1016/j.aca.2016.02.011
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., et al. (2015). MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526. doi:10.1038/nmeth.3393
Tswett, M. (1968). Physical chemical studies on chlorophyll adsorptions. A Source Book Chem. 1900-1950, 23–28.
Wang, S., Qian, Y., Sun, M., Jia, L., Hu, Y., Li, X., et al. (2020). Holistic quality evaluation of Saposhnikoviae Radix (Saposhnikovia divaricata) by reversed-phase ultra-high performance liquid chromatography and hydrophilic interaction chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry-based untargeted metabolomics. Arabian J. Chem. 13, 8835–8847. doi:10.1016/j.arabjc.2020.10.013
Want, E. J., Cravatt, B. F., and Siuzdak, G. (2005). The expanding role of mass spectrometry in metabolite profiling and characterization. ChemBioChem 6, 1941–1951. doi:10.1002/cbic.200500151
West, C., Elfakir, C., and Lafosse, M. (2010). Porous graphitic carbon: A versatile stationary phase for liquid chromatography. J. Chromatogr. A 1217, 3201–3216. doi:10.1016/j.chroma.2009.09.052
Wickramasekara, S., Zandkarimi, F., Morré, J., Kirkwood, J., Legette, L., Jiang, Y., et al. (2013). Electrospray quadrupole travelling wave ion mobility time-of-flight mass spectrometry for the detection of plasma metabolome changes caused by xanthohumol in obese zucker (fa/fa) rats. Metabolites 3, 701–717. doi:10.3390/metabo3030701
Wishart, D. S., Guo, A. C., Oler, E., Wang, F., Anjum, A., Peters, H., et al. (2022). Hmdb 5.0: The human metabolome database for 2022. Nucleic Acids Res. 50, D622–D631. doi:10.1093/nar/gkab1062
Wolthuis, J. C., Magnúsdóttir, S., Stigter, E., Tang, Y. F., Jans, J., Gilbert, M., et al. (2023). Multi-country metabolic signature discovery for chicken health classification. Metabolomics 19, 9–14. doi:10.1007/s11306-023-01973-4
Yarita, T., Aoyagi, Y., and Otake, T. (2015). Evaluation of the impact of matrix effect on quantification of pesticides in foods by gas chromatography-mass spectrometry using isotope-labeled internal standards. J. Chromatogr. A 1396, 109–116. doi:10.1016/j.chroma.2015.03.075
Zang, X., Monge, M. E., Gaul, D. A., and Fernández, F. M. (2018). Flow injection-traveling-wave ion mobility-mass spectrometry for prostate-cancer metabolomics. Anal. Chem. 90, 13767–13774. doi:10.1021/acs.analchem.8b04259
Zang, X., Pérez, J. J., Jones, C. M., Monge, M. E., McCarty, N. A., Stecenko, A. A., et al. (2017). Comparison of ambient and atmospheric pressure ion sources for cystic fibrosis exhaled breath condensate ion mobility-mass spectrometry metabolomics. J. Am. Soc. Mass Spectrom. 28, 1489–1496. doi:10.1007/s13361-017-1660-9
Zhang, W. (2008). Fluorocarbon stationary phases for liquid chromatography applications. J. Fluor Chem. 129, 910–919. doi:10.1016/j.jfluchem.2008.07.001
Zheng, X., Aly, N. A., Zhou, Y., Dupuis, K. T., Bilbao, A., Paurus, V. L., et al. (2017). A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem. Sci. 8, 7724–7736. doi:10.1039/c7sc03464d
Zhou, Z., Luo, M., Chen, X., Yin, Y., Xiong, X., Wang, R., et al. (2020). Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 11, 4334–4413. doi:10.1038/s41467-020-18171-8
Keywords: chromatography, separation, drift time, drift tube, travelling wave, trapped, high-field asymmetric
Citation: Moses T and Burgess K (2023) Right in two: capabilities of ion mobility spectrometry for untargeted metabolomics. Front. Mol. Biosci. 10:1230282. doi: 10.3389/fmolb.2023.1230282
Received: 28 May 2023; Accepted: 27 July 2023;
Published: 04 August 2023.
Edited by:
Michael Anton Witting, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Maria Monge, Consejo Nacional de Investigaciones Científicas y Técnicas, ArgentinaGiuseppe Paglia, University of Milano Bicocca, Italy
Copyright © 2023 Moses and Burgess. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karl Burgess, a2FybC5idXJnZXNzQGVkLmFjLnVr
 Tessa Moses
Tessa Moses Karl Burgess
Karl Burgess