- 1Qatar Biomedical Research Institute (QBRI), Qatar Foundation, Hamad Bin Khalifa University, Doha, Qatar
- 2Department of Biochemistry, Jamia Hamdard, New Delhi, India
Multiple drug resistance is the main obstacle in the treatment of bacterial diseases. Resistance against antibiotics demands the exploration of new antimicrobial drug targets. A variety of in silico and genetic approaches show that the enzymes of the riboflavin biosynthetic pathway are crucial for the survival of bacteria. This pathway is absent in humans thus enzymes of the riboflavin biosynthetic pathway are emerging drug targets for resistant pathogenic bacterial strains. Exploring the structural details, their mechanism of action, intermediate elucidation, and interaction analysis would help in designing suitable inhibitors of these enzymes. The riboflavin biosynthetic pathway consists of seven distinct enzymes, namely, 3,4-dihydroxy-2-butanone 4-phosphate synthase, GTP cyclohydrolase II, pyrimidine deaminase/reductase, phosphatase, lumazine synthase, and riboflavin synthase. The present review summarizes the research work that has been carried out on these enzymes in terms of their structures, active site architectures, and molecular mechanism of catalysis. This review also walks through small molecule inhibitors that have been developed against several of these enzymes.
Introduction
Riboflavin (also known as vitamin B2) is the originator for flavocoenzymes, flavin mononucleotide (FMN), and flavin adenine dinucleotide (FAD), two necessary cofactors that play key roles in various crucial cellular functions (Muller, 2018). They are involved in a wide variety of redox reactions (single as well as two electron transfer reactions) that are vital to energy metabolism essential for all living organisms (Mansoorabadi et al., 2007; Muller, 2018). Apart from their involvement in redox reactions, they also take part in a remarkable number of non-redox processes including DNA damage repair, circadian clock regulation, signal transduction, nitrogen fixation, light sensing, and bioluminescence (Salomon et al., 2001; Bornemann, 2002; Thompson and Sancar, 2002; Wijnen and Young, 2006; Macheroux et al., 2011). Lumazine binding proteins, which can bind riboflavin, FMN, 6, 7-dimethyl-8-ribityllumazine non-covalently as a fluorophore, take part in the bioluminescence process as an optical transponders in several bacterial species (Lee, 1993; Petushkov and Lee, 1997). This astonishing photochemical reaction reveals the remarkable flexibility of the isoalloxazine chromophore, in that the flavin cofactors side chains assist mainly as anchors that locked the binding of the fluorophore to the cognate proteins. Flavin cofactors also participate to form complex catalytic centers, sometimes including more than one coenzyme, various flavin adducts and/or other cofactors such as iron-sulfur clusters (Roberts et al., 2003; Hudson et al., 2005; Cheng et al., 2006).
Riboflavin is biosynthesized de novo in microorganisms and plants, whereas in animals they depend on dietary supplements. The riboflavin biosynthetic pathway was extensively studied and consists of seven distinct enzymes catalyzing the reaction, starting with one guanosine triphosphate (GTP) and two molecules of ribulose 5-phosphate (Ru5P) as the initial precursors (Figure 1). GTP is transformed into 2,5-diamino-6-ribosylamino-4(3H)- pyrimidinedione 5′-phosphate (DARPP) by GTP cyclohydrolase II. The bifunctional enzyme deaminase/reductase catalyses the deamination and subsequent reduction to convert DARPP to 5-amino-6-ribitylamino-2.4(1H, 3H)-pyrimidinedione 5′-phosphate (ArPP). ARPP is dephosphorylated by yet unknown phosphatase, resulting in 5-amino-6-ribitylamino-2.4(1H, 3H)-pyrimidinedione (ArP). Along the parallel line, 3,4-dihydroxy-2-butanone-4-phosphate synthase (DHBPS) catalyzes the conversion of ribulose-5-phosphate (Ru5P) to 3,4-dihydroxy-2-butanone 4-phosphate (DHBP). The resulting products ArPP and DHBP are condensed by lumazine synthase (LS) to form 6,7-dimethyl-8-ribityllumazine (DRL) with the release of inorganic phosphate. The final step involves an unusual dismutation reaction of two DRL, converting them into one riboflavin and one molecule of ArP. This reaction encompasses the transfer of a four-carbon unit between two identical substrates and the reaction is catalyzed by riboflavin synthase (RS). ArP formed in the final reaction is recycled back and used as a substrate by LS (Foor and Brown, 1975; Burrows and Brown, 1978; Volk and Bacher, 1991; Kis et al., 1995; Fischer and Bacher, 2008; 2011; Ladenstein et al., 2013).
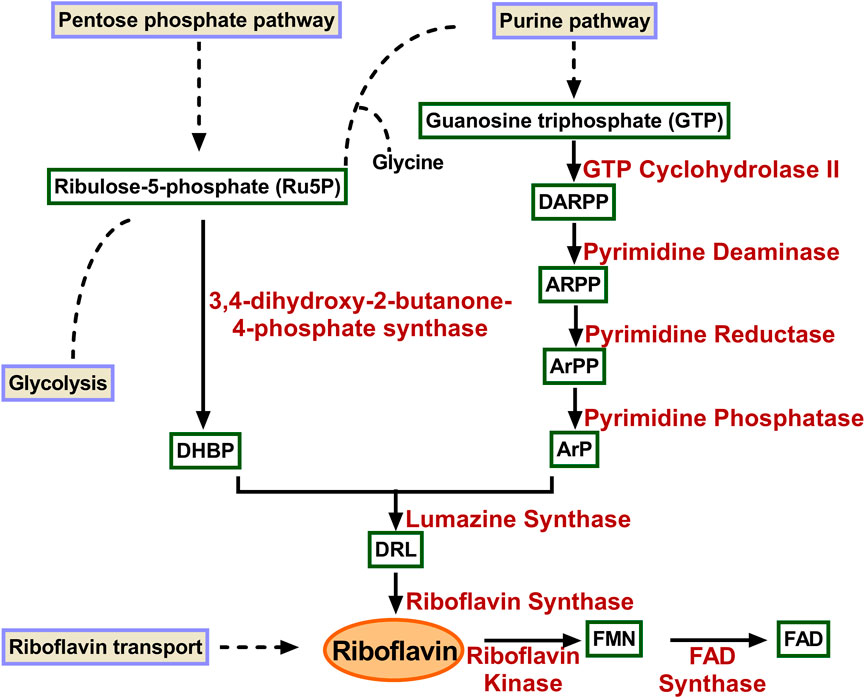
FIGURE 1. Enzymes involved in riboflavin biosynthetic pathway. GTP is converted to ArP through four reactions catalysed consecutively by GTP cyclohydrolase II, pyrimidine deaminase, pyrimidine reductas, and pyrimidine phosphatase. Similarly, DHBP is formed Ru5P catalyzed by 3,4-dihydroxy-2-butanone-4-phosphate synthase. The two products condensed to form DRL in a reaction catalyzed by lumazine synthase. Riboflavin synthase converts two molecules of DRL to riboflavin and ArP. DARPP, 2,5-diamino-6-ribosyl-amino-4(3H)pyrimidinedione 5′-phosphate; ARPP, 5-amino-6-ribosyl-amino-2.4(1H, 3H)pyrimidinedione 5′-phosphate; ArPP 5-amino-6-ribitylamino-2.4-(1H, 3H)-pyrimidinedione-5′-phosphate; ArP 5-amino-6-ribitylamino-2.4-(1H, 3H)-pyrimidinedione; DHBP 3,4-dihydroxy-2-butanone-4-phosphate; DRL 6,7-dimethyl-8-ribityl-lumazine; FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide.
Identification and evaluation of novel drug targets drive the advancement of new inhibitors against pathogenic microbial strains resistant to common antibiotics. Several in silico approaches were engaged to investigate genomic data to identify potential drug targets that are critical for the survival and virulence of bacterial pathogens and with no counterpart in humans or other animal proteins (Gupta et al., 2019). Several approaches including a whole-genome transposon random approach to ascertain the critical gene set found in various branches of life and were applied to the genomes of Haemophilus influenza, Mycobacterium tuberculosis, Pseudomonas aeruginosa, E. coli and Mycoplasma genitalium (Akerley et al., 2002; Gerdes et al., 2002; Lamichhane et al., 2003; Sassetti et al., 2003; Glass et al., 2006; Liberati et al., 2006). Gene deletion techniques were also used for defining important genes in Bacillus subtilis and Escherichia coli (Kobayashi et al., 2003; Baba et al., 2006). These studies suggested that the enzymes of the riboflavin biosynthesis pathway are vital and can be used as drug targets.
Plants and many microorganisms including bacterial pathogens can manufacture riboflavin de novo, but animals do not have this ability, which they fulfill through their diet. Bacterial pathogens especially the Gram-negative bacteria like M. tuberculosis and Salmonella typhimurium stringently need endogenous riboflavin due to the absence of a riboflavin uptake system from the environment (Wang, 1992; Dahl et al., 2004; Long et al., 2010). Hence, the enzymes of the riboflavin biosynthetic pathway could be measured as selective therapeutic targets. Various research works reinforced the significance of de novo riboflavin biosynthesis in several pathogenic bacterial antibiotics-resistant microbes and enzymes of this pathway as therapeutic targets. In fact, it has also been established that riboflavin biosynthetic genes and enzymes are virulence factors in Salmonella enterica (Becker et al., 2006; Rollenhagen and Bumann, 2006). Hence, the development of potential drugs against the enzymes involved in the riboflavin (vitamin B2) biosynthetic pathway opens a fresh line of attack for the treatment of bacterial infections. Designing novel antimicrobial drugs that selectively target pathogens are immediately required to tackle the multidrug resistance that arises in various pathogens.
3,4-Dihydroxy-2-butanone 4-phosphate synthase (DHBPS)
DHBPS is one of the first enzymes involved in the riboflavin biosynthetic pathway and converts Ru5P to DHBP and formate (Figure 1). DHBPS can exist as a single enzyme or it can form a bi-functional enzyme. It combined with GTP cyclohydrolase II (GCH II) to form a bi-functional enzyme, although DHBPS and GCH II can exist as two independent enzymes. The catalytic mechanism proposed for DHBPS involves several intermediate steps including skeletal rearrangement and fragmentation. These intermediate stages culminate with the release of formate and stable endiol intermediate that undergoes protonation with the formation of DHBP (Volk and Bacher, 1990; Richter et al., 1992). The chemical transformation includes a skeletal rearrangement and proposes a central role for acid/base catalysis with substrate/intermediates interaction requires divalent metal ion Mg2+ (Volk and Bacher, 1990). DHBPS enzyme class from various microorganisms has a primary sequence length of 204–233 amino acid residues and with 25%–60% sequence homology.
The three-dimensional structures of DHBPS were solved from E. coli, M. grisea, M. jannaschii, Candida albicans, S. typhimurium, M. tuberculosis, and V. cholera (Liao et al., 2001a; Liao et al., 2002; Steinbacher et al., 2003; Echt et al., 2004; Kumar et al., 2010; Singh et al., 2011; Islam et al., 2015). All of the structures solved to date indicate that DHBPS exists as a homodimer and present a characteristic alpha + beta fold containing mainly beta strands with a complex linkage. The homodimeric nature of DHBPS creates two active surfaces of the enzyme and is located at the interface between two subunits in the vicinity of the residues conserved among species. The surface of each active site is primarily shaped by one monomeric unit with an additional surface being created by neighboring monomeric unit residues (Figure 2). The active pocket of DHBPS was identified by crystallographic studies from the archaeon M. jannaschii and C. albicans in a complex with ribulose-5-phosphate and metal ions (Steinbacher et al., 2003; Echt et al., 2004) and are highlighted in Figure 2. Structures available till date suggest a degree of flexibility in the active cavity, especially at the loop regions. The DHBPS structure shows two different conformations of loop present near the active site. It can be either open or closed depending on the substrate availability. An open conformation exists in the absence of substrate where these loops points away from the active site cavity, while it closes up in the presence of substrate or substrate along with metals, which are required for the completion of catalysis, as a number of residues from these loops interact with substrate and metals (Liao et al., 2002; Steinbacher et al., 2003; Kumar et al., 2010; Islam et al., 2015). The substrate (Ru5P) present in the active site cavity also shows flexibility in its conformation. Moreover, the divalent metal ions (“dimetal center”) present at the active site (Figure 2) are crucial for the catalytic activity of DHBPS and are proposed to be involved in substrate and intermediates stabilization (Liao et al., 2002; Steinbacher et al., 2003; Islam et al., 2015). The detailed investigation of DHBPS structures in complexes with several metals highlighted the breathing of metal or shift in the position of two metals during the course of the reaction (Liao et al., 2002; Islam et al., 2015). Recently, two transient intermediates were identified using time-dependent structural studies and helps in defining the mechanism of DHBPS (Kenjić et al., 2022).
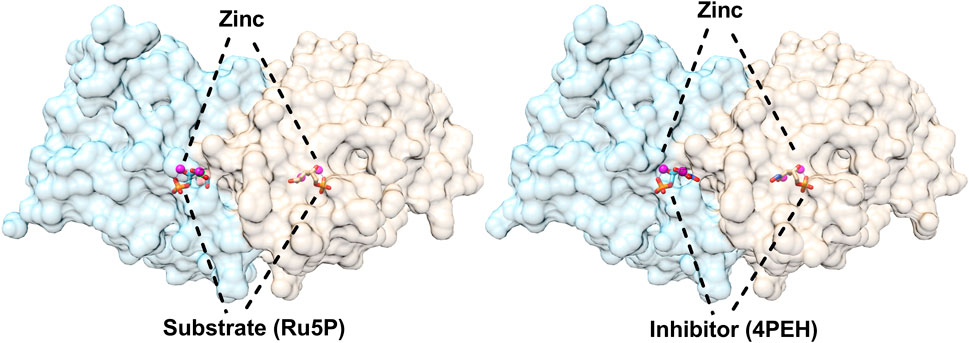
FIGURE 2. Structures of 3,4-dihydroxy-2-butanone 4-phosphate synthase (DHBPS) from V. cholera. Crystal structure of DHBPS in complexed with substrate (Ru5P) and metal ions (PDBID: 4P8E) showing the active site architecture (left). Similarly, crystal structure of DHBPS in complexed with inhibitor (4PEH) and zinc ions (PDBID: 4P6P) highlighting the binding of inhibitor and metal (right).
GTP cyclohydrolase II (GCH II)
GCH II converts GTP into DARPP (2,5-diamino-6-ribosylamino-4(3H)-pyrimidinedione 5′-phosphate), formate, and inorganic pyrophosphate with the aid of divalent metal ion (Foor and Brown, 1975; Foor and Brown, 1980; Ritz et al., 2001). Formate is released from the imidazole ring while pyrophosphate is released from the nucleotide precursor affording the formation of DARPP (Foor and Brown, 1980; Ren et al., 2005). Biochemical studies suggested the role of zinc ion, which plays a crucial part in guanine ring opening although it is not needed for inorganic pyrophosphate release (Kaiser et al., 2002). The GCH II has no significant amino acid sequence similarity with GCH I and differs in functional state. The two GTP cyclohydrolase have completely different oligomeric states, while GCH II exists as a homodimer, GCH I forms as a homodecamer (Nar et al., 1995).
Structures of GCHII in apo as well as in complex form have been solved from E. coli, and Helicobacter pylori (Ren et al., 2005; Yadav and Karthikeyan, 2015). It has an alpha/beta fold which mainly consists of a central core of antiparallel β-sheets connected by loops and helices (Figure 3). The three dimensional structure of E. coli GCH II was also solved with GMPCPP (substrate analogue), which showed the main residues that form the active site cavity and participate in substrate interaction. No major conformational changes were observed between the apo and holo forms of the enzymes with only a small deviation of the side chains of the residues involved in the binding of the substrate or substrate analogue (Ren et al., 2005). A zinc ion was bound intrinsically to the GCHII and is coordinated through three cysteines (Figure 3). Loss of any one of the three cysteines is enough to bring to an end the binding as well as its enzymatic function (Kaiser et al., 2002). The active region of GCH II contains substrate, GMP and zinc ion where the metal coordinated by three cysteines of a CX2GX7CXC motif. The metal was spaning the α and β phosphate moieties of the triphosphate motif and interacted with the amino acid residue of the protein (Figure 3). The complex structure also highlighted Arg128 interaction with the α-phosphonate for pyrophosphate release and formation of the proposed covalent guanylyl-GTP cyclohydrolase II intermediate (Ren et al., 2005). The structure of GCHII from H. pylori is similar to E. coli, although it is without any intrinsically bound zinc ion, probably exhibiting the inactive state of GCHII (Ren et al., 2005; Yadav and Karthikeyan, 2015).
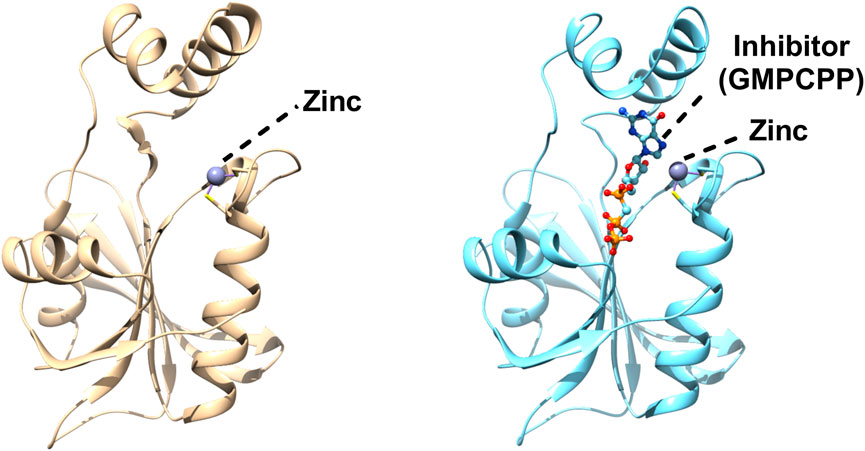
FIGURE 3. Structures of Escherichia coli GTP cyclohydrolase II (GCH II). Cartoon representation of Escherichia coli GCH II in apo form (PDBID: 2BZ1) revealing the intrinsic metal binding (left). Crystal Structure of Escherichia coli GCH II in complex with GMPCPP and Zinc (PDBID: 2BZ0) enlightening the inhibitor binding (right). Zinc is shown as grey ball.
Pyrimidine deaminase/reductase
DARPP formed by GCHII is transformed into ArPP (5-amino-6-ribitylamino-2.4(1H, 3H)-pyrimidinedione 5′-phosphate) in two steps comprising the deamination of the pyrimidine ring and reduction of the ribosyl side chain. These two reactions are catalyzed by bifunctional pyrimidine deaminase/reductase enzyme (Richter et al., 1997). The order of the two processes is different in various organisms studied. In bacteria and plants, deamination precedes reduction; however, reduction precedes deamination in yeast and archaea (Burrows and Brown, 1978; Nielsen and Bacher, 1981; Fischer et al., 2004). The bifunctional enzyme has a deaminase domain at the N-terminal, which contains a zinc ion, and an NADPH-binding reductase domain at the C-terminal. The reductive reaction requires the NADPH in reduced form and deaminase activity abolition does not disturb the reductase activity (Magalhães et al., 2008).
The x-ray crystal structure of bifunctional deaminase/reductase from E. coli, B. subtilis, M. jannaschii and Acinetobacter baumannii has been solved with several complexes (Chatwell et al., 2006; Chen et al., 2006; 2009; Stenmark et al., 2007; Dawson et al., 2013). It forms as a homodimer in E. coli, while exists in a homotetrameric form in B. subtilis (Chen et al., 2006; Stenmark et al., 2007). Each monomer is made up of two separate functional domains, comprises of an N-terminal deaminase domain and a C-terminal reductase domain with a short linker region dividing these domains (Figure 4). The N-terminal deaminase domain contains beta-sheet surrounded by alpha-helices with central portion dominated by mixed beta-strands. Similarly, the reductase domain mainly consists of beta-sheet with seven parallel strands and a β-hairpin. The deaminase domain occupies the far end and the homodimeric interface is primarily constructed by two consecutive segments of the reductase domain. The deaminase domain binds zinc with tetrahedral coordination by a zinc-binding motif having two cysteines, one histidine, and a water molecule (Dawson et al., 2013). The binding of NADP occurred at the surface of the reductase domain in a very extended conformation (Figure 4) with slight variations at the nicotinamide ring (Stenmark et al., 2007). The reductase active pocket was covered by the loop, which interacts with the cofactor and substrate analogue in the binary complexes (Ren et al., 2005). The substrate/substrate analogue ribose 5-phosphate binds to subunit A of the enzyme but does not bind to subunit B (Figure 4). A ternary complex of the reductase domain was created by a combination of the substrate with the ribitylimino intermediate and cofactor NADP binary structures (Chen et al., 2009).
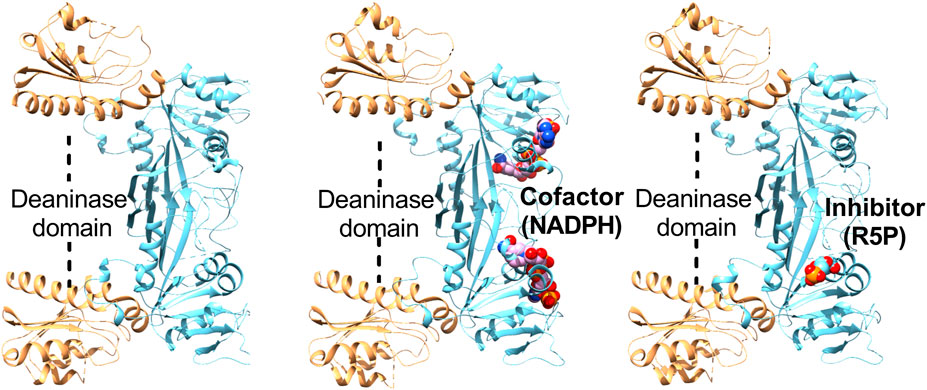
FIGURE 4. Structures of Escherichia coli bifunctional deaminase/reductase. Cartoon representation of the crystal structure of Escherichia coli deaminase/reductase in apo form (PDBID: 2G6V) underlining the domain organization and their relative orientation (left). The crystal structure of Escherichia coli bifunctional deaminase/reductase in complex with cofactor, NADPH (PDBID: 2O7P) binding in the active site of the reductase domain (middle). Structure of E. coli deaminase/reductase in complex with a substrate analogue, ribose 5-phosphate (R5P), bound to the active site of the reductase domain (right).
Lumazine synthase (LS)
LS facilitates the condensation of two substrates, namely, DHBP and ArPP, leading to the biosynthesis of DRL. The subsequent dismutation reaction of two DRL converts it into one riboflavin and one ArPP, catalyzed by riboflavin synthase (RS). Numerous structural, biophysical and mechanistic studies were conducted to understand the mechanism of the reaction carried out by LS. The enzyme-catalyzed reaction was anticipated to have several intermediary steps and start with the substrate binding, nucleophilic attack, formation of Schiff base intermediate, proton abstraction, and phosphate elimination with subsequent ring closure. Finally, the release of water would terminate the reaction, resulting in product creation in the form of DRL (Kis et al., 1995; Bacher et al., 1996; Zhang et al., 2003).
The crystal structures of LS from different organisms were reported. All the structural and oligomeric studies performed suggest a pentameric assembly as a basic unit, although, various degrees of the quaternary higher oligomeric states were observed for different organisms. The LS proteins were reported in the pentameric form in Saccharomyces cerevisiae, Schizosaccharomyces pombe, M. grisea, and M. tuberculosis (Persson et al., 1999; Meining et al., 2000; Gerhardt et al., 2002a; Morgunova et al., 2005), as dimers of pentamers in Brucella abortus (Braden et al., 2000; Zylberman et al., 2004) and as icosahedral capsids consisting of 60 identical subunits, which can be designated as dodecamers of pentamers in B. subtilis, Aquifex aeolicus, S. oleracea and S. typhimurium (Schott et al., 1990; Ladenstein et al., 1994; Persson et al., 1999; Zhang et al., 2001; Kumar et al., 2011). Moreover, in B. subtilis a new arrangement of LS and RS was seen, they exist as a 1 MDa protein complex having LS forming the outer capsid, while 3 RS subunits occupied the core (Ladenstein et al., 1988; Schott et al., 1990). The mega-dalton protein complex of LS and RS proposed to improve the catalytic productivity of the two processes carried by these two enzymes by substrate channeling especially at low substrate concentrations (Ritsert et al., 1995). The topology of the higher oligomeric form (icosahedral and decameric forms of protein) looks like that of the homopentameric LSs (Figure 5). Comparative analysis of several crystal structures of LS from various species displayed a flavodoxin-like fold irrespective of the oligomeric status, either pentameric or higher quaternary structures of the protein. The overall architecture of the single subunit contains a four-stranded beta-sheet with alpha helices covering the central beta strands. Structural studies of LS structures reveal the topologically equivalent active sites and were located between adjacent subunits creating a cavity to accommodate the substrate (Figure 5). LS binds two substrates, namely, DHBP and ArPP and their location were identified through several structural studies of the protein-ligand complexes formed between the protein and metabolically stable substrate, intermediate, and product analogues (Ritsert et al., 1995; Persson et al., 1999; Meining et al., 2000; Morgunova et al., 2005).
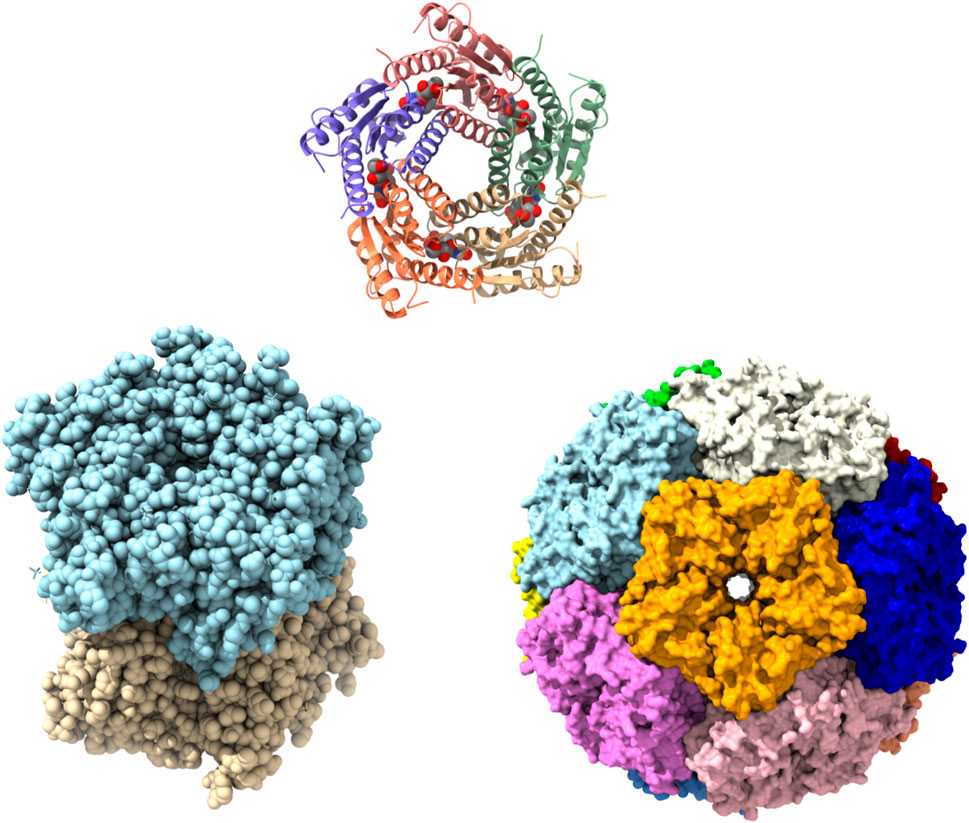
FIGURE 5. Structures of lumazine synthase (LS) in several oligomeric states. Pentamer assembly of LS from M. tuberculosis bound to inhibitor TS50 (5-(1,3,7-trihydro-9-day-ribityl-2,4,8-purinetrione-7-yl)pentane 1-phosphate) at active sites (PDBID: 2C94). Each subunit is represented in different colour (upper). Crystal Structure of LS from B. abortus (PDBID: 1XN1) showing as dimer of pentamer (lower left). Surface representation of icosahedral assembly of S. typhimurium LS showing each pentamer in different colour (PDBID: 3MK3) (lower right).
Riboflavin synthase (RS)
RS is an ultimate protein in the biosynthetic pathway of riboflavin, that catalyzes the dismutation of two DRL to yield one riboflavin and one molecule of ArPP (PLAUT, 1963; Harvey and Plaut, 1966). In essence, it performs the transfer of a butane unit (4-carbon unit) from one molecule of DRL to another DRL molecule and in the process creating riboflavin and ArPP, hence outlining the terminal reaction of the riboflavin biosynthetic pathway. The second product of the reaction, ArPP, can be recycled and serve as a substrate for LS, the penultimate enzyme in the biosynthetic pathway of riboflavin. The enzyme-catalyzed and the uncatalyzed reactions proceed with identical regiochemistry involving a head-to-tail arrangement of the two 4-carbon moieties from which the xylene ring of riboflavin is formed (Paterson and Wood, 1969; Sedlmaier et al., 1987; Ladenstein et al., 2013). The enzyme-catalyzed reaction necessitates the simultaneous occurrence of two substrates in an anti-parallel orientation at the active site of the enzyme. A proposed reaction mechanism of RS proceeds through a pentacyclic intermediate structure, which is formed by the dimerization of DRL and the sequence of two elimination reaction that converts the pentacyclic intermediate into equimolar amounts of riboflavin and ArPP (Illarionov et al., 2001; 2005).
RS from bacteria, yeast, and plants are shown to form homotrimers (Bacher et al., 1980; Liao et al., 2001b; Gerhardt et al., 2002b), while, in archaea, it forms homopentamer with completely different protein sequences (Ramsperger et al., 2006). In all the RS studied so far, the homotrimeric RS subunits contain intramolecular sequence similarity possibly due to two molecules of similar substrate. The comparison of proximal and distal domains of homotrimeric RS showed a high structural similarity, suggesting that both domains have grown from a common ancestor probably through gene duplication. The three-dimensional structure of RS from E. coli, B. abortus, and S. pombe was determined and showed that the protein was a homotrimer. Each subunit consists of two repeating β-barrel domains that share high sequence and structural similarity. The only difference between the two β-barrels is the presence of a C-terminal helix at the C-terminal β-barrel which is suggested to play a role in the trimerization of RS (Liao et al., 2001b; Gerhardt et al., 2002b; Serer et al., 2014). The structural studies on RS also revealed that the active site is present between the N-terminal β-barrel of one subunit and the C-terminal β-barrel of another subunit with its substrate binding in anti-parallel orientation. The interactions between the subunits of the trimer are primarily mediated by the arrangement of the C-termini (which do not participate in the intra-subunit sequence similarity) in a triple helical motif (Figure 6). The trimeric RS from E. coli was solved in the apo form, while RS from S. pombe was solved complex with substrate analogue (Liao et al., 2001b; Gerhardt et al., 2002b). The homotrimeric structure of RS from B. abortus was solved in the presence of riboflavin and in complex with two product analogues, namely, roseoflavin and 5-nitro-6-ribitylamino-2.4(1H, 3H)-pyrimidinedione (Serer et al., 2014) (Figure 6). Additionally, the crystal structure of N-terminal domain of E. coli RS in the presence of riboflavin was solved and showed that it can bind riboflavin and exists as a homodimer which could be superposed with a single subunit of full-length enzyme. This model was used to find out the differences in substrate and product binding at the C and N-terminal domains and validated the proposed reaction mechanism (Meining et al., 2003).
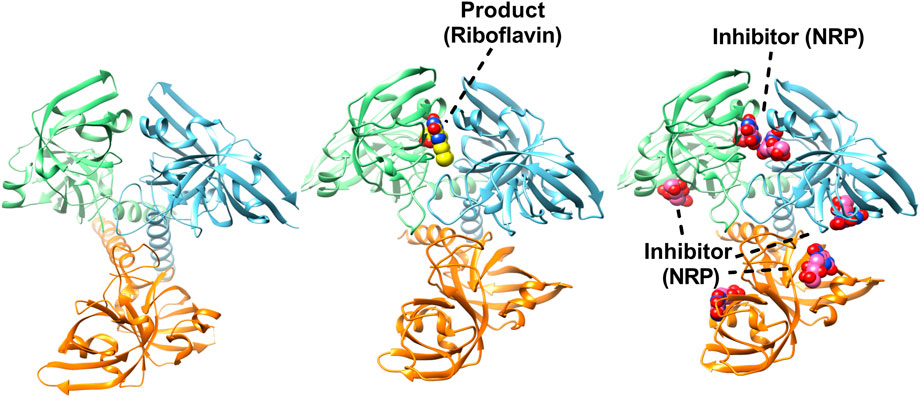
FIGURE 6. Structures of trimeric riboflavin synthase (RS). Crystal structure of RS from Escherichia coli (PDBID: 1I8D). Each subunit is shown in different colour (left). Structure of trimeric RS from B. abortus in complex with one riboflavin (PDBID: 4E0F) (middle). Crystal structure of RS from B. abortus in complex with 5-Nitro-6-(D-Ribitylamino)-2.4(1H, 3H) Pyrimidinedione (PDBID: 4GQN) highlighting the inhibitor binding in active sites (right).
Inhibitors of riboflavin pathway enzymes
The compound 4-phosphoerythronohydroxamic acid (4PEH) was studied for its inhibitory ability due to structurally mimicking the substrate Ru5P of DHBPS. The compound was found to inhibit DHBPS activity with a Ki value of around 100 μM (Islam et al., 2015). Apart from DHBPS, 4PEH was shown to act as a competitive inhibitor for M. tuberculosis ribose-5-phosphate isomerase B (Roos et al., 2005). Structure determination with inhibitor confirms the competitive mechanism where it occupies the active site and was unable to accommodate an intermediate that was critical for DHBPS activity (Islam et al., 2015).
GCH II, being the rate-limiting step in the biosynthesis of riboflavin, makes it a potential novel selective antimicrobial drug target. Phosphomethylphosphonic acid guanyl ester, GMPCPP was known to inhibit the GCH II and act as a GTP substrate analogue. The bound inhibitor structure of E. coli GCH II with GMPCPP revealed the detailed stereochemistry of the enzyme active site and helped in the structure-based design of inhibitors of GCH II (Ren et al., 2005). E. coli bifunctional deaminase/reductase structure complexed with the substrate analogue ribose-5-phosphate (RP5) was helpful in defining the binding pocket useful for inhibitor design (Stenmark et al., 2007). To gain structural insights and inhibitor design, the structure of B. subtilis bifunctional deaminase/reductase in complex with 5-diamino-6-ribosylamino-2.4(1H, 3H)-pyrimidinedione 5′-phosphate (AROPP) was solved at 2.56-Å resolution (Chen et al., 2009). GMP molecule, a substrate analogue for the deaminase reaction was observed in the NADPH-binding site of the reductase domain, occupying the adenine-binding pocket (Dawson et al., 2013). These initial molecules may be exploited to provide starting points for a structure-based approach to antibacterial drug discovery.
Most of the inhibitors were designed or studied centered around LS and RS due to their involvement in the last step of the pathway. The development of LS and RS inhibitors was focused on the modifications of the central pyrimidinedione core, side chain extension from the core and various stable substrates, intermediate and product analogues. Based on the crystal structures complexed with substrate and product of the reaction catalyzed by LS, several organic compounds have been synthesized and characterized by various docking, simulation and spectrophotometrically as inhibitors of LS from M. tuberculosis, B. abortus and C. albicans (Cushman et al., 1998; Braden et al., 2000; Cushman et al., 2001; Cushman et al., 2002a; Cushman et al., 2004; Chen et al., 2005; Cushman et al., 2005; Zhang et al., 2008). The crystal structure of icosahedral B. subtilis LS capsids with 5-nitro-6-ribitylamino-2.4-(1H, 3H) pyrimidinedione, a substrate analogue inhibitor, was solved allowing a detailed description of the substrate analogue binding site (Ritsert et al., 1995). Based on the structure-based drug designing, several substrate analogous inhibitors of LS from C. albicans have been synthesized, namely, 1,3,7-Trihydro-9-day-ribityl-2,4,8-purinetrione-7-yl (TS13); 3-(1,3-dihydro-9-day-ribityl-2,4,8-purinetrione-7-yl)propane 1-phosphate (TS44); 4-(6.7(5H, 8H)-dioxo-8-day-ribityllumazine-5-yl)butane 1-phosphate (GJ43); and [4-(6-chloro-2,4-dioxo-1,2,3,4 tetrahydropyrimidin-5-yl)butyl] phosphate (JC33). The binding affinities of these inhibitors were measured using isothermal titration calorimetry and were found to be in the micromolar range (Morgunova et al., 2007). Several amide derivatives of 5-phosphonoalkyl-6-D-ribitylaminopyrimidinedione were synthesized and their selectivity for inhibition for LS as well as RS was evaluated. These amide derivatives showed better inhibitory potency to RS than LS and this was further validated by molecular docking demonstrating that the active site of the RS protein could gladly harbor two substrate/substrate analogue/inhibitor molecules (Cushman et al., 2002b).
Several phosphonate inhibitors of LS were manufactured in that the phosphorus atom was separated from the pyrimidinedione ring by a carbon linker. These inhibitors were metabolically stable analogues of intermediate structure designed using enzyme-inhibitor complex. These phosphonates show remarkable LS inhibitory potency with Ki in the range of 100–500 μM (Cushman et al., 1999c; 1999b). Three bis(6,7-dimethyl-8-D-ribityllumazines) containing two lumazine moieties and (ribitylamino) uracils bearing fluorosulfonyl, sulfonic acid, and carboxylic acid were synthesized. These compounds showed very weak inhibitors of RS, although more potent for LS (Cushman et al., 1997; 1999a).
The inhibitors created by the replacement of the pyrimidinedione core with a purine showed inhibition for B. subtilis LS and E. coli RS in mid micromolar range which further improved to lower micromolar range upon the addition of a ketone group (Cushman et al., 1998; Cushman et al., 2001). A series of inhibitors based on 8-aza derivative with alkyl and phosphate moiety were synthesized. All the synthesized compounds were exceptionally potent inhibitors of LS of M. tuberculosis, M. grisea, C. albicans, and S. pombe with inhibition potency in the low nanomolar to sub nanomolar range (Zhang et al., 2007). The designs of dual inhibitors were rapidly progressed because of their effectiveness against LS and RS and the fact it would be rare to mutate both enzymes simultaneously to create a resistant pathogen. Several dual inhibitors were synthesized and found to be effective in inhibiting the LS and RS of various organisms (Cushman et al., 2002a; Cushman et al., 2005; Talukdar et al., 2012).
A robust high-throughput screening (HTS) platform was developed for screening the inhibitors against DHBPS, LS, and RS (Kaiser et al., 2007; Talukdar et al., 2009; Zhao et al., 2009). Covalent hydrates of trifluoromethylated Pyrazoles were recognized as inhibitors of RS, and showed potent antimicrobial activity against M. tuberculosis (Zhao et al., 2009). Recently, HTS approach has identified ten molecules from initial 44,000 low molecular weight compounds against RS from Brucella spp. With inhibition in the low micromolar range. Several of the most effective inhibitors were subsequently optimized and represent a promising and effective antimicrobial for brucellosis (Serer et al., 2019). Table 1 summarizes all the inhibitors of riboflavin biosynthetic enzymes mentioned in this section.
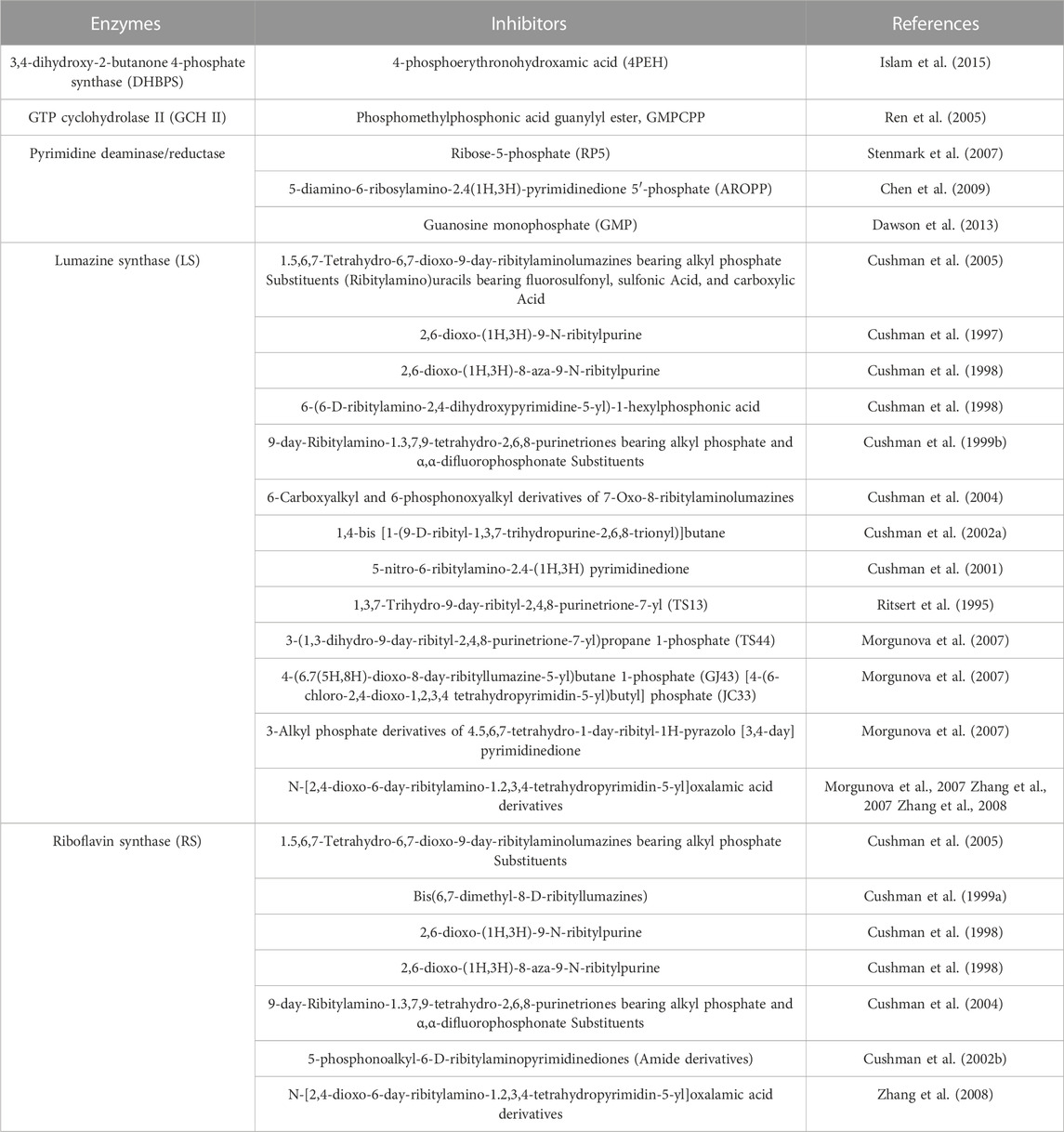
TABLE 1. List of inhibitors (substrate, intermediate and product analogues) for riboflavin biosynthetic enzymes.
Conclusion
Structures of all the enzymes of the riboflavin biosynthetic pathway were solved from several bacterial pathogens. High-resolution structures along with enzyme kinetics were helpful in finalizing the reaction mechanism with the formation of various possible intermediates. Structure-based drug designing has started due to the abundant complex structures of these enzymes with substrate, substrate analogs, intermediates, product and product analogs. Several stable compounds were synthesized based on modification of the pyrimidine core, phosphate, and alkyl chain especially for LS and RS. The inhibitors were tested on various microorganisms and found to be effective which provides a template to generate new lead compounds in the development of therapeutics.
Author contributions
ZI and PK contributed to conception and design of the review. ZI wrote the first draft of the manuscript. PK wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1228763/full#supplementary-material
References
Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N., and Mekalanos, J. J. (2002). A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 99, 966–971. doi:10.1073/pnas.012602299
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The keio collection. Mol. Syst. Biol. 2, 0008. doi:10.1038/msb4100050
Bacher, A., Baur, R., Eggers, U., Harders, H. D., Otto, M. K., and Schnepple, H. (1980). Riboflavin synthases of Bacillus subtilis. Purification and properties. J. Biol. Chem. 255, 632–637. doi:10.1016/s0021-9258(19)86223-3
Bacher, A., Fischer, M., Kis, K., Kugelbrey, K., Mörtl, S., Scheuring, J., et al. (1996). Biosynthesis of riboflavin: Structure and mechanism of lumazine synthase. Biochem. Soc. Trans. 24, 89–94. doi:10.1042/bst0240089
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., et al. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307. doi:10.1038/nature04616
Bornemann, S. (2002). Flavoenzymes that catalyse reactions with no net redox change. Nat. Prod. Rep. 19, 761–772. doi:10.1039/b108916c
Braden, B. C., Velikovsky, C. A., Cauerhff, A. A., Polikarpov, I., and Goldbaum, F. A. (2000). Divergence in macromolecular assembly: X-Ray crystallographic structure analysis of lumazine synthase from Brucella abortus. J. Mol. Biol. 297, 1031–1036. doi:10.1006/jmbi.2000.3640
Burrows, R. B., and Brown, G. M. (1978). Presence of Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J. Bacteriol. 136, 657–667. doi:10.1128/jb.136.2.657-667.1978
Chatwell, L., Krojer, T., Fidler, A., Römisch, W., Eisenreich, W., Bacher, A., et al. (2006). Biosynthesis of riboflavin: Structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5’-phosphate reductase of Methanocaldococcus jannaschii. J. Mol. Biol. 359, 1334–1351. doi:10.1016/j.jmb.2006.04.045
Chen, J., Illarionov, B., Bacher, A., Fischer, M., Haase, I., Georg, G., et al. (2005). A high-throughput screen utilizing the fluorescence of riboflavin for identification of lumazine synthase inhibitors. Anal. Biochem. 338, 124–130. doi:10.1016/j.ab.2004.11.033
Chen, S.-C., Chang, Y.-C., Lin, C.-H., Lin, C.-H., and Liaw, S.-H. (2006). Crystal structure of a bifunctional deaminase and reductase from Bacillus subtilis involved in riboflavin biosynthesis. J. Biol. Chem. 281, 7605–7613. doi:10.1074/jbc.M510254200
Chen, S.-C., Lin, Y.-H., Yu, H.-C., and Liaw, S.-H. (2009). Complex structure of Bacillus subtilis RibG: The reduction mechanism during riboflavin biosynthesis. J. Biol. Chem. 284, 1725–1731. doi:10.1074/jbc.M805820200
Cheng, V. W. T., Ma, E., Zhao, Z., Rothery, R. A., and Weiner, J. H. (2006). The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J. Biol. Chem. 281, 27662–27668. doi:10.1074/jbc.M604900200
Cushman, M., Jin, G., Sambaiah, T., Illarionov, B., Fischer, M., Ladenstein, R., et al. (2005). Design, synthesis, and biochemical evaluation of 1,5,6,7-tetrahydro-6,7-dioxo-9-D-ribitylaminolumazines bearing alkyl phosphate substituents as inhibitors of lumazine synthase and riboflavin synthase. J. Org. Chem. 70, 8162–8170. doi:10.1021/jo051332v
Cushman, M., Mavandadi, F., Kugelbrey, K., and Bacher, A. (1997). Design and synthesis of (Ribitylamino)uracils bearing fluorosulfonyl, sulfonic acid, and carboxylic acid functionality as inhibitors of lumazine synthase. J. Org. Chem. 62, 8944–8947. doi:10.1021/jo9712963
Cushman, M., Mavandadi, F., Kugelbrey, K., and Bacher, A. (1998). Synthesis of 2,6-dioxo-(1H,3H)-9-N-ribitylpurine and 2,6-dioxo-(1H,3H)-8-aza-9-N-ribitylpurine as inhibitors of lumazine synthase and riboflavin synthase. Bioorg. Med. Chem. 6, 409–415. doi:10.1016/s0968-0896(98)00013-3
Cushman, M., Mavandadi, F., Yang, D., Kugelbrey, K., Kis, K., and Bacher, A. (1999a). Synthesis and biochemical evaluation of bis(6,7-dimethyl-8-D-ribityllumazines) as potential bisubstrate analogue inhibitors of riboflavin synthase. J. Org. Chem. 64, 4635–4642. doi:10.1021/jo9821731
Cushman, M., Mihalic, J. T., Kis, K., and Bacher, A. (1999c). Design and synthesis of 6-(6-D-ribitylamino-2,4-dihydroxypyrimidin-5-yl)-1-hexyl phosphonic acid, a potent inhibitor of lumazine synthase. Bioorg. Med. Chem. Lett. 9, 39–42. doi:10.1016/s0960-894x(98)00687-8
Cushman, M., Mihalic, J. T., Kis, K., and Bacher, A. (1999b). Design, synthesis, and biological evaluation of homologous phosphonic acids and sulfonic acids as inhibitors of lumazine synthase. J. Org. Chem. 64, 3838–3845. doi:10.1021/jo9821729
Cushman, M., Sambaiah, T., Jin, G., Illarionov, B., Fischer, M., and Bacher, A. (2004). Design, synthesis, and evaluation of 9-D-ribitylamino-1,3,7,9-tetrahydro-2,6,8-purinetriones bearing alkyl phosphate and alpha,alpha-difluorophosphonate substituents as inhibitors of tiboflavin synthase and lumazine synthase. J. Org. Chem. 69, 601–612. doi:10.1021/jo030278k
Cushman, M., Yang, D., Gerhardt, S., Huber, R., Fischer, M., Kis, K., et al. (2002a). Design, synthesis, and evaluation of 6-carboxyalkyl and 6-phosphonoxyalkyl derivatives of 7-oxo-8-ribitylaminolumazines as inhibitors of riboflavin synthase and lumazine synthase. J. Org. Chem. 67, 5807–5816. doi:10.1021/jo0201631
Cushman, M., Yang, D., Kis, K., and Bacher, A. (2001). Design, synthesis, and evaluation of 9-D-ribityl-1,3,7-trihydro-2,6,8-purinetrione, a potent inhibitor of riboflavin synthase and lumazine synthase. J. Org. Chem. 66, 8320–8327. doi:10.1021/jo010706r
Cushman, M., Yang, D., Mihalic, J. T., Chen, J., Gerhardt, S., Huber, R., et al. (2002b). Incorporation of an amide into 5-phosphonoalkyl-6-D-ribitylaminopyrimidinedione lumazine synthase inhibitors results in an unexpected reversal of selectivity for riboflavin synthase vs lumazine synthase. J. Org. Chem. 67, 6871–6877. doi:10.1021/jo020144r
Dahl, S. G., Sylte, I., and Ravna, A. W. (2004). Structures and models of transporter proteins. J. Pharmacol. Exp. Ther. 309, 853–860. doi:10.1124/jpet.103.059972
Dawson, A., Trumper, P., Chrysostomou, G., and Hunter, W. N. (2013). Structure of diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase from Acinetobacter baumannii. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 69, 611–617. doi:10.1107/S174430911301292X
Echt, S., Bauer, S., Steinbacher, S., Huber, R., Bacher, A., and Fischer, M. (2004). Potential anti-infective targets in pathogenic yeasts: Structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans. J. Mol. Biol. 341, 1085–1096. doi:10.1016/j.jmb.2004.06.053
Fischer, M., and Bacher, A. (2011). Biosynthesis of vitamin B2: A unique way to assemble a xylene ring. Chembiochem 12, 670–680. doi:10.1002/cbic.201000681
Fischer, M., and Bacher, A. (2008). Biosynthesis of vitamin B2: Structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 474, 252–265. doi:10.1016/j.abb.2008.02.008
Fischer, M., Römisch, W., Saller, S., Illarionov, B., Richter, G., Rohdich, F., et al. (2004). Evolution of vitamin B2 biosynthesis: Structural and functional similarity between pyrimidine deaminases of eubacterial and plant origin. J. Biol. Chem. 279, 36299–36308. doi:10.1074/jbc.M404406200
Foor, F., and Brown, G. M. (1980). GTP cyclohydrolase II from Escherichia coli. Methods Enzymol. 66, 303–307. doi:10.1016/0076-6879(80)66473-8
Foor, F., and Brown, G. M. (1975). Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J. Biol. Chem. 250, 3545–3551. doi:10.1016/s0021-9258(19)41549-4
Gerdes, S. Y., Scholle, M. D., D’Souza, M., Bernal, A., Baev, M. V., Farrell, M., et al. (2002). From genetic footprinting to antimicrobial drug targets: Examples in cofactor biosynthetic pathways. J. Bacteriol. 184, 4555–4572. doi:10.1128/JB.184.16.4555-4572.2002
Gerhardt, S., Haase, I., Steinbacher, S., Kaiser, J. T., Cushman, M., Bacher, A., et al. (2002a). The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase. J. Mol. Biol. 318, 1317–1329. doi:10.1016/s0022-2836(02)00116-x
Gerhardt, S., Schott, A.-K., Kairies, N., Cushman, M., Illarionov, B., Eisenreich, W., et al. (2002b). Studies on the reaction mechanism of riboflavin synthase: X-Ray crystal structure of a complex with 6-carboxyethyl-7-oxo-8-ribityllumazine. Structure 10, 1371–1381. –81. doi:10.1016/s0969-2126(02)00864-x
Glass, J. I., Assad-Garcia, N., Alperovich, N., Yooseph, S., Lewis, M. R., Maruf, M., et al. (2006). Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103, 425–430. doi:10.1073/pnas.0510013103
Gupta, R., Verma, R., Pradhan, D., Jain, A. K., Umamaheswari, A., and Rai, C. S. (2019). An in silico approach towards identification of novel drug targets in pathogenic species of Leptospira. PLoS One 14, e0221446. doi:10.1371/journal.pone.0221446
Harvey, R. A., and Plaut, G. W. (1966). Riboflavin synthetase from yeast. J. Biol. Chem. 241, 2120–2136. doi:10.1016/s0021-9258(18)96675-5
Hudson, J. M., Heffron, K., Kotlyar, V., Sher, Y., Maklashina, E., Cecchini, G., et al. (2005). Electron transfer and catalytic control by the iron-sulfur clusters in a respiratory enzyme, E. coli fumarate reductase. J. Am. Chem. Soc. 127, 6977–6989. doi:10.1021/ja043404q
Illarionov, B., Eisenreich, W., and Bacher, A. (2001). A pentacyclic reaction intermediate of riboflavin synthase. Proc. Natl. Acad. Sci. U. S. A. 98, 7224–7229. doi:10.1073/pnas.131610698
Illarionov, B., Haase, I., Fischer, M., Bacher, A., and Schramek, N. (2005). Pre-steady-state kinetic analysis of riboflavin synthase using a pentacyclic reaction intermediate as substrate. Biol. Chem. 386, 127–136. doi:10.1515/BC.2005.016
Islam, Z., Kumar, A., Singh, S., Salmon, L., and Karthikeyan, S. (2015). Structural basis for competitive inhibition of 3,4-dihydroxy-2-butanone-4-phosphate synthase from Vibrio cholerae. J. Biol. Chem. 290, 11293–11308. doi:10.1074/jbc.M114.611830
Kaiser, J., Illarionov, B., Rohdich, F., Eisenreich, W., Saller, S., den Brulle, J. Van, et al. (2007). A high-throughput screening platform for inhibitors of the riboflavin biosynthesis pathway. Anal. Biochem. 365, 52–61. doi:10.1016/j.ab.2007.02.033
Kaiser, J., Schramek, N., Eberhardt, S., Püttmer, S., Schuster, M., and Bacher, A. (2002). Biosynthesis of vitamin B2. Eur. J. Biochem. 269, 5264–5270. doi:10.1046/j.1432-1033.2002.03239.x
Kenjić, N., Meneely, K. M., Wherritt, D. J., Denler, M. C., Jackson, T. A., Moran, G. R., et al. (2022). Evidence for the chemical mechanism of RibB (3,4-dihydroxy-2-butanone 4-phosphate synthase) of riboflavin biosynthesis. J. Am. Chem. Soc. 144, 12769–12780. doi:10.1021/jacs.2c03376
Kis, K., Volk, R., and Bacher, A. (1995). Biosynthesis of riboflavin. Studies on the reaction mechanism of 6,7-dimethyl-8-ribityllumazine synthase. Biochemistry 34, 2883–2892. doi:10.1021/bi00009a019
Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., et al. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100, 4678–4683. doi:10.1073/pnas.0730515100
Kumar, P., Singh, M., Gautam, R., and Karthikeyan, S. (2010). Potential anti-bacterial drug target: Structural characterization of 3,4-dihydroxy-2-butanone-4-phosphate synthase from Salmonella typhimurium LT2. Proteins 78, 3292–3303. doi:10.1002/prot.22837
Kumar, P., Singh, M., and Karthikeyan, S. (2011). Crystal structure analysis of icosahedral lumazine synthase from Salmonella typhimurium, an antibacterial drug target. Acta Crystallogr. D. Biol. Crystallogr. 67, 131–139. doi:10.1107/S0907444910053370
Ladenstein, R., Fischer, M., and Bacher, A. (2013). The lumazine synthase/riboflavin synthase complex: Shapes and functions of a highly variable enzyme system. FEBS J. 280, 2537–2563. doi:10.1111/febs.12255
Ladenstein, R., Ritsert, K., Huber, R., Richter, G., and Bacher, A. (1994). The lumazine synthase/riboflavin synthase complex of Bacillus subtilis. X-ray structure analysis of hollow reconstituted beta-subunit capsids. Eur. J. Biochem. 223, 1007–1017. doi:10.1111/j.1432-1033.1994.tb19079.x
Ladenstein, R., Schneider, M., Huber, R., Bartunik, H. D., Wilson, K., Schott, K., et al. (1988). Heavy riboflavin synthase from Bacillus subtilis. Crystal structure analysis of the icosahedral beta 60 capsid at 3.3 A resolution. J. Mol. Biol. 203, 1045–1070. doi:10.1016/0022-2836(88)90128-3
Lamichhane, G., Zignol, M., Blades, N. J., Geiman, D. E., Dougherty, A., Grosset, J., et al. (2003). A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: Application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100, 7213–7218. doi:10.1073/pnas.1231432100
Lee, J. (1993). Lumazine protein and the excitation mechanism in bacterial bioluminescence. Biophys. Chem. 48, 149–158. doi:10.1016/0301-4622(93)85006-4
Liao, D.-I., Zheng, Y.-J., Viitanen, P. V., and Jordan, D. B. (2002). Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase. Biochemistry 41, 1795–1806. doi:10.1021/bi015652u
Liao, D. I., Calabrese, J. C., Wawrzak, Z., Viitanen, P. V., and Jordan, D. B. (2001a). Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure 9, 11–18. doi:10.1016/s0969-2126(00)00550-5
Liao, D. I., Wawrzak, Z., Calabrese, J. C., Viitanen, P. V., and Jordan, D. B. (2001b). Crystal structure of riboflavin synthase. Structure 9, 399–408. doi:10.1016/s0969-2126(01)00600-1
Liberati, N. T., Urbach, J. M., Miyata, S., Lee, D. G., Drenkard, E., Wu, G., et al. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103, 2833–2838. doi:10.1073/pnas.0511100103
Long, Q., Ji, L., Wang, H., and Xie, J. (2010). Riboflavin biosynthetic and regulatory factors as potential novel anti-infective drug targets. Chem. Biol. Drug Des. 75, 339–347. doi:10.1111/j.1747-0285.2010.00946.x
Macheroux, P., Kappes, B., and Ealick, S. E. (2011). Flavogenomics-a genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634. doi:10.1111/j.1742-4658.2011.08202.x
Magalhães, M. L. B., Argyrou, A., Cahill, S. M., and Blanchard, J. S. (2008). Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase involved in riboflavin biosynthesis. Biochemistry 47, 6499–6507. doi:10.1021/bi800264g
Mansoorabadi, S. O., Thibodeaux, C. J., and Liu, H. (2007). The diverse roles of flavin coenzymes-nature’s most versatile thespians. J. Org. Chem. 72, 6329–6342. doi:10.1021/jo0703092
Meining, W., Eberhardt, S., Bacher, A., and Ladenstein, R. (2003). The structure of the N-terminal domain of riboflavin synthase in complex with riboflavin at 2.6A resolution. J. Mol. Biol. 331, 1053–1063. doi:10.1016/s0022-2836(03)00844-1
Meining, W., Mörtl, S., Fischer, M., Cushman, M., Bacher, A., and Ladenstein, R. (2000). The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue. J. Mol. Biol. 299, 181–197. doi:10.1006/jmbi.2000.3742
Morgunova, E., Meining, W., Illarionov, B., Haase, I., Jin, G., Bacher, A., et al. (2005). Crystal structure of lumazine synthase from Mycobacterium tuberculosis as a target for rational drug design: Binding mode of a new class of purinetrione inhibitors. Biochemistry 44, 2746–2758. doi:10.1021/bi047848a
Morgunova, E., Saller, S., Haase, I., Cushman, M., Bacher, A., Fischer, M., et al. (2007). Lumazine synthase from Candida albicans as an anti-fungal target enzyme: Structural and biochemical basis for drug design. J. Biol. Chem. 282, 17231–17241. doi:10.1074/jbc.M701724200
Muller, F. (2018). in Chemistry and biochemistry of flavoenzymes. Editor F. Müller (CRC Press). doi:10.1201/9781351070577
Nar, H., Huber, R., Auerbach, G., Fischer, M., Hösl, C., Ritz, H., et al. (1995). Active site topology and reaction mechanism of GTP cyclohydrolase I. Proc. Natl. Acad. Sci. U. S. A. 92, 12120–12125. doi:10.1073/pnas.92.26.12120
Nielsen, P., and Bacher, A. (1981). Biosynthesis of riboflavin. Characterization of the product of the deaminase. Biochim. Biophys. Acta 662, 312–317. doi:10.1016/0005-2744(81)90044-9
Paterson, T., and Wood, H. C. S. (1969). Deuterium exchange of C-methyl protons in 6,7-dimethyl-8-D-ribityl-lumazine, and studies of the mechanism of riboflavin biosynthesis. J. Chem. Soc. D. Chem. Commun. 290, 290. doi:10.1039/c29690000290
Persson, K., Schneider, G., Jordan, D. B., Viitanen, P. V., and Sandalova, T. (1999). Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly. Protein Sci. 8, 2355–2365. doi:10.1110/ps.8.11.2355
Petushkov, V. N., and Lee, J. (1997). Purification and characterization of flavoproteins and cytochromes from the yellow bioluminescence marine bacterium Vibrio fischeri strain Y1. Eur. J. Biochem. 245, 790–796. doi:10.1111/j.1432-1033.1997.00790.x
Plaut, G. W. (1963). Studies on the nature of the enzymic conversion of 6,7-dimethyl-8-ribityllumazine to riboflavin. J. Biol. Chem. 238, 2225–2243. doi:10.1016/s0021-9258(18)67964-5
Ramsperger, A., Augustin, M., Schott, A.-K., Gerhardt, S., Krojer, T., Eisenreich, W., et al. (2006). Crystal structure of an archaeal pentameric riboflavin synthase in complex with a substrate analog inhibitor: Stereochemical implications. J. Biol. Chem. 281, 1224–1232. doi:10.1074/jbc.M509440200
Ren, J., Kotaka, M., Lockyer, M., Lamb, H. K., Hawkins, A. R., and Stammers, D. K. (2005). GTP cyclohydrolase II structure and mechanism. J. Biol. Chem. 280, 36912–36919. doi:10.1074/jbc.M507725200
Richter, G., Fischer, M., Krieger, C., Eberhardt, S., Lüttgen, H., Gerstenschläger, I., et al. (1997). Biosynthesis of riboflavin: Characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179, 2022–2028. doi:10.1128/jb.179.6.2022-2028.1997
Richter, G., Volk, R., Krieger, C., Lahm, H. W., Röthlisberger, U., and Bacher, A. (1992). Biosynthesis of riboflavin: Cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174, 4050–4056. doi:10.1128/jb.174.12.4050-4056.1992
Ritsert, K., Huber, R., Turk, D., Ladenstein, R., Schmidt-Bäse, K., and Bacher, A. (1995). Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: Crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution. J. Mol. Biol. 253, 151–167. doi:10.1006/jmbi.1995.0542
Ritz, H., Schramek, N., Bracher, A., Herz, S., Eisenreich, W., Richter, G., et al. (2001). Biosynthesis of riboflavin: Studies on the mechanism of GTP cyclohydrolase II. J. Biol. Chem. 276, 22273–22277. doi:10.1074/jbc.M100752200
Roberts, G. A., Celik, A., Hunter, D. J. B., Ost, T. W. B., White, J. H., Chapman, S. K., et al. (2003). A self-sufficient cytochrome p450 with a primary structural organization that includes a flavin domain and a [2Fe-2S] redox center. J. Biol. Chem. 278, 48914–48920. doi:10.1074/jbc.M309630200
Rollenhagen, C., and Bumann, D. (2006). Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74, 1649–1660. doi:10.1128/IAI.74.3.1649-1660.2006
Roos, A. K., Burgos, E., Ericsson, D. J., Salmon, L., and Mowbray, S. L. (2005). Competitive inhibitors of Mycobacterium tuberculosis ribose-5-phosphate isomerase B reveal new information about the reaction mechanism. J. Biol. Chem. 280, 6416–6422. doi:10.1074/jbc.M412018200
Salomon, M., Eisenreich, W., Dürr, H., Schleicher, E., Knieb, E., Massey, V., et al. (2001). An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. U. S. A. 98, 12357–12361. doi:10.1073/pnas.221455298
Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi:10.1046/j.1365-2958.2003.03425.x
Schott, K., Ladenstein, R., König, A., and Bacher, A. (1990). The lumazine synthase-riboflavin synthase complex of Bacillus subtilis. Crystallization of reconstituted icosahedral beta-subunit capsids. J. Biol. Chem. 265, 12686–12689. doi:10.1016/s0021-9258(19)38398-x
Sedlmaier, H., Müller, F., Keller, P. J., and Bacher, A. (1987). Enzymatic synthesis of riboflavin and FMN specifically labeled with 13C in the xylene ring. Z. Naturforsch. C 42, 425–429. doi:10.1515/znc-1987-0416
Serer, M. I., Bonomi, H. R., Guimarães, B. G., Rossi, R. C., Goldbaum, F. A., and Klinke, S. (2014). Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility. Acta Crystallogr. D. Biol. Crystallogr. 70, 1419–1434. doi:10.1107/S1399004714005161
Serer, M. I., Carrica, M. D. C., Trappe, J., López Romero, S., Bonomi, H. R., Klinke, S., et al. (2019). A high-throughput screening for inhibitors of riboflavin synthase identifies novel antimicrobial compounds to treat brucellosis. FEBS J. 286, 2522–2535. doi:10.1111/febs.14829
Singh, M., Kumar, P., and Karthikeyan, S. (2011). Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis. J. Struct. Biol. 174, 374–384. doi:10.1016/j.jsb.2011.01.013
Steinbacher, S., Schiffmann, S., Richter, G., Huber, R., Bacher, A., and Fischer, M. (2003). Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: Implications for the catalytic mechanism. J. Biol. Chem. 278, 42256–42265. doi:10.1074/jbc.M307301200
Stenmark, P., Moche, M., Gurmu, D., and Nordlund, P. (2007). The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: Implications for the reductive mechanism. J. Mol. Biol. 373, 48–64. doi:10.1016/j.jmb.2006.12.009
Talukdar, A., Breen, M., Bacher, A., Illarionov, B., Fischer, M., Georg, G., et al. (2009). Discovery and development of a small molecule library with lumazine synthase inhibitory activity. J. Org. Chem. 74, 5123–5134. doi:10.1021/jo900238q
Talukdar, A., Zhao, Y., Lv, W., Bacher, A., Illarionov, B., Fischer, M., et al. (2012). O-Nucleoside, S-nucleoside, and N-nucleoside probes of lumazine synthase and riboflavin synthase. J. Org. Chem. 77, 6239–6261. doi:10.1021/jo3010364
Thompson, C. L., and Sancar, A. (2002). Photolyase/cryptochrome blue-light photoreceptors use photon energy to repair DNA and reset the circadian clock. Oncogene 21, 9043–9056. doi:10.1038/sj.onc.1205958
Volk, R., and Bacher, A. (1991). Biosynthesis of riboflavin. Studies on the mechanism of L-3,4-dihydroxy-2-butanone 4-phosphate synthase. J. Biol. Chem. 266, 20610–20618. doi:10.1016/s0021-9258(18)54753-0
Volk, R., and Bacher, A. (1990). Studies on the 4-carbon precursor in the biosynthesis of riboflavin. Purification and properties of L-3,4-dihydroxy-2-butanone-4-phosphate synthase. J. Biol. Chem. 265, 19479–19485. doi:10.1016/s0021-9258(17)45398-1
Wang, A. (1992). Isolation of vitamin B2 auxotroph and preliminary genetic mapping in Salmonella typhimurium. Yi Chuan Xue Bao 19, 362–368. Available at: http://www.ncbi.nlm.nih.gov/pubmed/1466913.
Wijnen, H., and Young, M. W. (2006). Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40, 409–448. doi:10.1146/annurev.genet.40.110405.090603
Yadav, S., and Karthikeyan, S. (2015). Structural and biochemical characterization of GTP cyclohydrolase II from Helicobacter pylori reveals its redox dependent catalytic activity. J. Struct. Biol. 192, 100–115. doi:10.1016/j.jsb.2015.08.004
Zhang, X., Meining, W., Cushman, M., Haase, I., Fischer, M., Bacher, A., et al. (2003). A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus. J. Mol. Biol. 328, 167–182. doi:10.1016/s0022-2836(03)00186-4
Zhang, X., Meining, W., Fischer, M., Bacher, A., and Ladenstein, R. (2001). X-Ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile aquifex aeolicus at 1.6 A resolution: Determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306, 1099–1114. doi:10.1006/jmbi.2000.4435
Zhang, Y., Illarionov, B., Morgunova, E., Jin, G., Bacher, A., Fischer, M., et al. (2008). A new series of N-[2,4-dioxo-6-d-ribitylamino-1,2,3,4-tetrahydropyrimidin-5-yl]oxalamic acid derivatives as inhibitors of lumazine synthase and riboflavin synthase: Design, synthesis, biochemical evaluation, crystallography, and mechanistic implications. J. Org. Chem. 73, 2715–2724. doi:10.1021/jo702631a
Zhang, Y., Jin, G., Illarionov, B., Bacher, A., Fischer, M., and Cushman, M. (2007). A new series of 3-alkyl phosphate derivatives of 4,5,6,7-tetrahydro-1-D-ribityl-1H-pyrazolo[3,4-d]pyrimidinedione as inhibitors of lumazine synthase: Design, synthesis, and evaluation. J. Org. Chem. 72, 7176–7184. doi:10.1021/jo070982r
Zhao, Y., Bacher, A., Illarionov, B., Fischer, M., Georg, G., Ye, Q.-Z., et al. (2009). Discovery and development of the covalent hydrates of trifluoromethylated Pyrazoles as riboflavin synthase inhibitors with antibiotic activity against Mycobacterium tuberculosis. J. Org. Chem. 74, 5297–5303. doi:10.1021/jo900768c
Keywords: riboflavin biosynthetic pathway, inhibitors, 3,4-dihydroxy-2-butanone 4-phosphate synthase, lumazine synthase, riboflavin synthase
Citation: Islam Z and Kumar P (2023) Inhibitors of riboflavin biosynthetic pathway enzymes as potential antibacterial drugs. Front. Mol. Biosci. 10:1228763. doi: 10.3389/fmolb.2023.1228763
Received: 25 May 2023; Accepted: 03 July 2023;
Published: 11 July 2023.
Edited by:
Ghazala Muteeb, King Faisal University, Saudi ArabiaReviewed by:
Shazi Shakil, King Abdulaziz University, Saudi ArabiaGautam Srivastava, UCONN Health, United States
Gulam M. Rather, The State University of New Jersey, United States.
Copyright © 2023 Islam and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeyaul Islam, emlzbGFtQGhia3UuZWR1LnFh Pankaj Kumar, cGFua2Fqa3VtYXJAamFtaWFoYW1kYXJkLmFjLmlu
 Zeyaul Islam
Zeyaul Islam Pankaj Kumar
Pankaj Kumar