- Department of Biology, Ashoka University, Sonipat, Haryana, India
Entamoeba histolytica is the causative agent of amoebiasis. DNA replication studies in E. histolytica first started with the ribosomal RNA genes located on episomal circles. Unlike most plasmids, Entamoeba histolytica rDNA circles lacked a fixed origin. Replication initiated from multiple sites on the episome, and these were preferentially used under different growth conditions. In synchronized cells the early origins mapped within the rDNA transcription unit, while at later times an origin in the promoter-proximal upstream intergenic spacer was activated. This is reminiscent of eukaryotic chromosomal replication where multiple potential origins are used. Biochemical studies on replication and recombination proteins in Entamoeba histolytica picked up momentum once the genome sequence was available. Sequence search revealed homologs of DNA replication and recombination proteins, including meiotic genes. The replicative DNA polymerases identified included the α, δ, ε of polymerase family B; lesion repair polymerases Rev1 and Rev3; a translesion repair polymerase of family A, and five families of polymerases related to family B2. Biochemical analysis of EhDNApolA confirmed its polymerase activity with expected kinetic constants. It could perform strand displacement, and translesion synthesis. The purified EhDNApolB2 had polymerase and exonuclease activities, and could efficiently bypass some types of DNA lesions. The single DNA ligase (EhDNAligI) was similar to eukaryotic DNA ligase I. It was a high-fidelity DNA ligase, likely involved in both replication and repair. Its interaction with EhPCNA was also demonstrated. The recombination-related proteins biochemically characterized were EhRad51 and EhDmc1. Both shared the canonical properties of a recombinase and could catalyse strand exchange over long DNA stretches. Presence of Dmc1 indicates the likelihood of meiosis in this parasite. Direct evidence of recombination in Entamoeba histolytica was provided by use of inverted repeat sequences located on plasmids or chromosomes. In response to a variety of stress conditions, and during encystation in Entamoeba invadens, recombination-related genes were upregulated and homologous recombination was enhanced. These data suggest that homologous recombination could have critical roles in trophozoite growth and stage conversion. Availability of biochemically characterized replication and recombination proteins is an important resource for exploration of novel anti-amoebic drug targets.
1 Introduction
Entamoeba histolytica is a parasitic protist that causes amoebic dysentery and liver abscess, which if not timely treated can be fatal (Stanley, 2003). The parasite spreads by the faecal-oral route and is ingested through food and water contaminated with E. histolytica cysts. These migrate to the colon and get converted to trophozoites that rapidly multiply in the lumen. In asymptomatic infections trophozoites get re-converted to cysts which are excreted to complete the life cycle. In a minority of individuals, the trophozoites become invasive and invade the colonic epithelium causing intestinal ulcers, and may invade other organs, mainly the liver to form liver abscesses.
Entamoeba histolytica is an early-branching eukaryote with interesting unique features. It lacks typical mitochondria that are replaced by mitosomes which lack DNA, and may perform functions like sulfate activation and oxygen detoxification (van der Giezen and Tovar, 2005; Mi-ichi et al., 2009; Maralikova et al., 2010). Its ∼20 Mb genome is exclusively nuclear-localized (Lorenzi et al., 2010) and is composed of a number of repetitive elements. The non-long terminal repeat (LTR) retrotransposons, E. histolytica long and short interspersed nuclear elements (EhLINEs and EhSINEs respectively), are present on all chromosomes, and together occupy about 11% of the genome (Sharma et al., 2001; Lorenzi et al., 2008; Kaur et al., 2021). Another class of repeated DNAs are the ribosomal RNA genes, located exclusively on extrachromosomal circular molecules (Bhattacharya et al., 1998) rather than on linear chromosomes (Bagchi et al., 1999). The rDNA circles account for ∼10% of the genome. Due to their abundance, and the availability of their complete nucleotide sequence, early studies on E. histolytica DNA replication focused on the rDNA episome (Sehgal et al., 1994).
2 Replication of the ribosomal DNA episome of E. histolytica
In E. histolytica strain HM-1:IMSS the rRNA genes are located in the nucleus on extrachromosomal circles of size 24.5 kb. Each circle, present in about 200 copies, contains two rDNA transcription units of 5.8 kb each arranged as inverted repeats. They are separated by upstream and downstream intergenic spacers (IGS) (Bhattacharya et al., 1998). The rDNA circles analysed in different E. histolytica strains are very similar to the strain HM-1:IMSS. However, some circles contain one rDNA unit in place of two. These circular DNAs afford a convenient system for DNA replication studies.
Amongst unicellular eukaryotes the best studied nuclear plasmids are the Saccharomyces cerevisiae 2 μm plasmid and Dictyostelium discoideum plasmids. Much like bacterial plasmids where DNA replication initiates at a relatively fixed site, called the origin of replication (ori) (Couturier et al., 1988) here too, DNA replication initiates at fixed sites in the replicons. The yeast 2 μm plasmid initiates DNA replication at a unique site called the autonomously replicating sequence (ARS) which contains an 11-bp core and an AT-rich region to the 3′- side of the core (Volkert et al., 1989). The D. discoideum plasmids also initiate replication at a specific site that contains a 49-bp element present in three copies (Hughes et al., 1988).
The replication origin of the E. histolytica rDNA circles was mapped by using neutral/neutral two-dimensional gel electrophoresis. In this method (Brewer and Fangman, 1987) the replicon to be studied is digested with restriction enzymes. Electrophoretic migration of the restriction fragments is determined by gel electrophoresis in the first dimension based on size, and in the second dimension based on conformation. A restriction fragment that contains the ori where a replication fork starts and proceeds bidirectionally would have the shape of a bubble, while a fragment in which replication forks originating from the outside enter the fragment would contain Y arcs in place of bubbles. A fragment in which forks moving in opposite directions meet and terminate would have an X shape. These molecules can be readily distinguished by two-dimensional gel electrophoresis as their migration is retarded by varying degrees in the second dimension (Brewer and Fangman, 1987). The analysis of E. histolytica rDNA circles with this method showed that all fragments contained both simple Ys and bubbles, indicating the presence of multiple oris dispersed throughout the molecule, rather than a single fixed ori (Dhar et al., 1996). This unexpected observation was further confirmed by electron microscopy. The rDNA circle was linearized with restriction enzymes and the location of replication bubbles from the cut ends was measured (Dhar et al., 1996). These data corroborated the gel electrophoresis study and showed that replication bubbles were located throughout the molecule and appeared with greater frequency within the rDNA transcription units.
Although multiple oris are unusual for plasmid replication, they are the norm for eukaryotic chromosomes which contain many potential oris. These are activated to initiate replication in a time and context-dependent manner (Bogan et al., 2000; Bell and Dutta, 2002; Kumar and Remus, 2016). Further analysis of the E. histolytica rDNA oris showed that here too, all potential origins are not used equally at all times (Ghosh et al., 2003). In exponentially growing cells the most commonly used ori mapped upstream of the rDNA transcription unit, close to the promoter of rRNA genes (Panigrahi et al., 2009), and terminated in the downstream intergenic spacer. However, when cells were starved of serum for 24 h, after which serum was replenished to resume replication in a synchronous manner, the early oris mapped within the rDNA transcription unit. At later times the ori in the promoter-proximal upstream IGS was activated, while the early oris in the transcription unit were silenced. These data suggest that while the upstream IGS is the preferred ori in exponentially growing cells, other potential oris exist. The latter are probably recruited when the upstream ori is not in an active state (e.g., in cells recovering from stress), but become silenced when the upstream ori is activated. Another significant observation was that the upstream IGS fragment of the rDNA episome did not show ori activity when it was located ectopically in an artificial plasmid. Other fragments of the rDNA episome could, however, function as oris in the ectopic location (Ghosh et al., 2003).
3 DNA replication enzymes
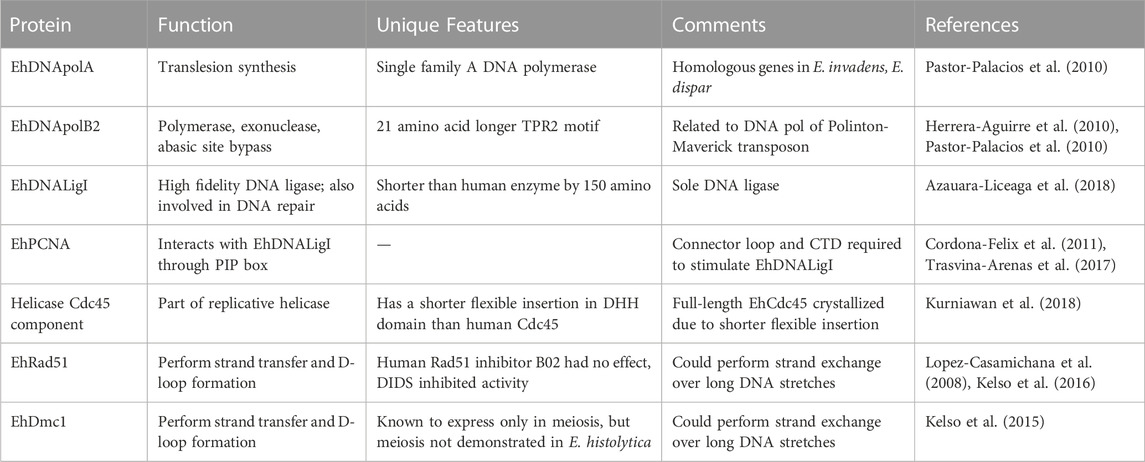
The availability of E. histolytica genome sequence in 2005 (Loftus et al., 2005), gave a major boost to biochemical studies on enzymes involved in major metabolic pathways (Table 1). Entamoeba histolytica genome sequence was searched for homologs of the canonical DNA polymerase enzymes. Entamoeba histolytica contains homologs of the replicative DNA polymerases α, δ, ε of the polymerase family B; lesion repair polymerases Rev1 and Rev3; and another translesion repair polymerase belonging to family A. In addition, there are five families of polymerases related to the family B2 encoded by DNA transposons and bacteriophage φ29 (Loftus et al., 2005; Lorenzi et al., 2010; Pastor-Palacios et al., 2010).
3.1 Family A DNA polymerase in E. histolytica
Family A DNA polymerases in eukaryotes are generally of three types. DNA polymerase γ of this family is the mitochondrial replicative DNA polymerase, while DNA Pol ν (Marini et al., 2003) and DNA Pol θ (Takata et al., 2006) are nuclear localized and are involved in translesion synthesis. A single putative DNA polymerase A member has been identified in E. histolytica by sequence homology with Klenow fragment. This EhDNApolA has been biochemically characterized and shown to be nuclear-localized and engaged in translesion synthesis (Pastor-Palacios et al., 2010). Homologous genes have also been identified in Entamoeba invadens (the reptilian parasite that is used as a model system for encystation studies) and Entamoeba dispar (the non-pathogenic species found in humans). A shared feature of the Entamoeba DNApolA members is the absence of a 3′-5′ exonuclease active site, while the polymerization site is well-conserved. As seen by homology modelling, EhDNApolA adopts a structure similar to the Klenow fragment bound to DNA, with the fingers, palm and thumb subdomains forming a DNA-binding cleft. EhDNApolA could incorporate dNTPs into an annealed primer-template, confirming its polymerase activity with kinetic constants similar to those of known family A DNA polymerases (Pastor-Palacios et al., 2010). It also had moderate strand displacement activity, but lacked 3′-5′ exonuclease activity. EhDNApolA had efficient translesion synthesis activity against a non-blocking lesion like that due to 8-oxoguanosine, and was also active against a blocking lesion like thymine glycol.
3.2 Family B2 DNA polymerase in E. histolytica
The archetypical Family B2 DNA polymerase is the bacteriophage φ29 enzyme (Rodriguez et al., 2005). In addition to the polymerization and 3′-5′ exonuclease domains, family B2 DNA polymerases contain two extra domains called terminal protein regions (TPR) 1 and 2 which confer processivity and strand displacement activities. The E. histolytica genome encodes four family B2 DNA polymerases (Herrera-Aguirre et al., 2010; Pastor-Palacios et al., 2010; Vicente et al., 2012). The closest orthologs of the E. histolytica enzymes are the DNA polymerases encoded by the DNA transposon class called Polinton-Maverick found in E. invadens (Kapitonov and Jurka, 2006; Pastor-Palacios et al., 2012). The amino acid sequences required for polymerization and exonuclease activities are well conserved in all 4 E. histolytica sequences (Pastor-Palacios et al., 2012). One of the 4 copies showing best match with φ29 enzyme was cloned and expressed. Amino acid sequence alignment of this copy (EhDNApolB2) with the φ29 enzyme showed 38% identity in the polymerase and exonuclease domains. The TPR2 motif was 21 amino acids longer in E. histolytica. In a structural model these 21 amino acids were shown to lie adjacent to the finger sub domain. The recombinant protein was shown to have polymerase and exonuclease activities. Enzyme activity was inhibited by aphidicolin, an inhibitor specific to family B DNA pols. EhDNApolB2 could also efficiently bypass lesions due to abasic sites or 8-oxoguanosine, although it could not bypass the strong block due to thymine glycol, which is expected from family B DNA polymerases. EhDNApolB2 achieves lesion bypass by incorporating dATP opposite the lesion. From deletion studies it was shown that the TPR2 motif was involved in efficient bypass of abasic sites, in addition to its known role in processivity and strand displacement.
3.3 The DNA ligase of E. histolytica
In contrast to vertebrates, E. histolytica encodes a single DNA ligase (EhDNAligI) which is similar to eukaryotic DNA ligase I (Azuara-Liceaga et al., 2018). It is an ATP-dependent DNA ligase of 685 amino acids with 35% identity to human DNA ligase I (Cardona-Felix et al., 2010). It is shorter than the human enzyme by 150 amino acids at the N-terminal. Being the sole DNA ligase, it is likely involved in both DNA replication in the maturation of Okazaki fragments, and in DNA repair processes. Biochemical analysis of recombinant EhDNAligI showed that it could carry out the essential steps in DNA ligation, namely, adenylation, binding to 5′-phosphorylated nicked DNA and sealing the nick. As expected of a family I DNA ligase it could ligate a RNA strand upstream of the nick, but not downstream of it. It was a high-fidelity DNA ligase and could discriminate 11 of the 12 possible mismatches on either side of the nick on the two DNA strands. The only mismatch efficiently ligated was the T:G mismatch at the 5′-phosphate strand (Azauara-Liceaga et al., 2018). This wobble base-pair mismatch is seen in ligases of Escherichia coli and Thermus thermophilus as well (Chauleau and Shuman, 2016). The possible involvement of EhDNAligI in DNA repair was suggested by various observations. In cells subjected to oxidative DNA damage EhDNAligI was shown to translocate to the nuclear periphery and co-localize with PCNA and the 8-oxoguanosine adduct. Following recovery from stress and elimination of 8-oxoguanosine, EhDNAligI was no longer concentrated at the nuclear periphery and was seen diffusely in the cytoplasm.
3.4 EhPCNA
PCNA is a highly conserved, essential accessory protein in DNA replication and repair. It serves as a DNA clamp; forms a homo trimeric ring around the DNA and slides along the DNA length. PCNA recruits a variety of proteins involved in DNA replication, repair and chromatin structure (González-Magaña and Blanco, 2020). It interacts with its protein partners via specific motifs, one of which is the PCNA-interacting peptide (PIP) box. The E. histolytica homolog of PCNA was identified by sequence search. Biochemical and structural studies with the recombinant EhPCNA showed that it formed a homotrimer and had a PIP box through which it interacted with EhDNAligI (Cordona-Felix et al., 2011). The interaction of EhDNAligI and EhPCNA was studied in more detail (Trasvina-Arenas et al., 2017). The N-terminal region of EhDNAligI is only 39 amino acids (compared with 262 amino acids in the human enzyme HsDNAligI), and is thought to be involved in protein-protein interactions. The PIP box is located at residues 2 to 9. Two deletion mutants of EhDNAligI were generated-one lacking the first 39 amino acids (∆PIP) and another lacking the first 58 amino acids in which, along with PIP, two putative alpha helices of the DNA binding domain (∆DBD) were deleted. Both deletion mutants could perform the first step of ligation reaction, that is self-adenylation, although the ∆DBD had reduced activity compared with full-length. ∆PIP could carry out the second step of ligation reaction, that is binding of the ligase to 5′- phosphate of the DNA nick with kinetics comparable to the full-length, and the increase in binding affinity in the presence of EhPCNA was also comparable; showing that PIP box of EhDNAligI was not needed for interaction with EhPCNA. Although ∆DBD did not bind to nicked DNA substrate, addition of EhPCNA could restore this capability. These data suggest that the initial interaction between EhDNAligI and EhPCNA occurs through the PIP box, and a second interaction takes place between the two proteins through a specific interface. Using affinity chromatography it was shown that EhPCNA could bind with both deletion mutants, showing that the two proteins interacted independent of the PIP box. Further it was shown that the nick-sealing activity of ∆DBD was restored by EhPCNA. The interdomain connector loop and C-terminal domain of EhPCNA was required for stimulating the activity of EhDNAligI. The C-terminal of EhPCNA probably allows allosteric transition of EhDNAligI from a spread-shape to a ring structure.
3.5 The Cdc45 component of E. histolytica replicative helicase
Successful DNA replication requires unwinding of the DNA duplex which is achieved by specialized replicative helicases. These are molecular motors that unwind DNA at the front of the replication fork, and translocate along single-stranded DNA using energy from nucleotide hydrolysis. In eukaryotes, replisome assembly at the ori starts with loading of the hexameric minichromosome maintenance complex (Mcm) 2–7. This is activated through a series of events, leading to the recruitment of cell division cycle 45 (Cdc45) and Go-Ichi-Ni-San (GINS) proteins, which together form the active replicative helicase Cdc45/Mcm2-7/GINS (CMG) holoenzyme. In model organisms, the molecular mechanism by which the replicative helicase operates is being revealed through single molecule studies (Spinks et al., 2021) and cryo electron microscopy (EM) (Kurniawan et al., 2018). It has been shown that Cdc45 and GINS bind to the outer surface of the N-tier ring of the hexameric Mcm2-7 complex to stabilize its closed conformation (Petojevic et al., 2015). This allows the ATPase motor of Mcm2-7 to catalyze translocation of CMG complex along the DNA strand. Orthologs of the replicative helicase proteins exist in E. histolytica but their biochemical properties have not been studied. However, the E. histolytica Cdc45 protein has been studied by cryo EM (Kurniawan et al., 2018). This study was motivated by the fact that it was difficult to crytallize full-length human Cdc45 due to the presence of a flexible insertion within the N-terminal DHH domain of this protein. Crystal structure could be obtained only after deleting an11-amino acid stretch from this region. The 543-residue, E. histolytica Cdc45 protein has a shorter flexible insertion in this region, making it possible to crystallize the intact protein. The E. histolytica Cdc45 consists of the conserved N-terminal DHH and C-terminal DHHA1 domains connected through an α-helical connector segment. Its structure could be solved to 1.66 Å resolution. There was overall similarity to human Cdc45, but the E. histolytica Cdc45 had several unique features. There was a distinct orientation of the C-terminal DHHA1 domain due to which there was disordering of the adjacent protruding α-helical segment implicated in DNA polymerase ɛ interactions. The authors suggest that Cdc45 could adopt multiple conformations and thereby regulate protein-protein interactions important in DNA replication.
4 DNA recombination in E. histolytica
Parasitic protists are continuously exposed to environmental toxins and host immune pressure, which can affect the stability of their genome. Homologous recombination (HR) is a conserved mechanism that can repair DNA damage due to double strand breaks (DSBs). These could occur due to genotoxic agents or during normal processes like meiotic division, or during DNA synthesis to enable bypass of lesions that block the replicative polymerase (Brandsma and Gent, 2012). HR is important to generate genetic diversity which parasites use to evade the host immune response (Deitsch et al., 1999; Conway et al., 2002). DSBs, caused by DNA damage or by the Spo11 endonuclease during meiosis, trigger HR (Keeney et al., 1997; San Filippo et al., 2008). DSBs are processed by the Mrn/Mrx complex, which includes Mre11p, to generate single strand tails with 3′-OH ends. The ssDNA forms a nucleoprotein filament in association with Rad51p and Dmc1p (Masson and West, 2001) which is stimulated by the Hop2-Mnd1 proteins to invade a homologous sequence and form a D-loop intermediate (Tsubouchi and Roeder, 2002). The Msh2 protein (which interacts with Mlh1p) is part of a complex involved in strand invasion. Subsequent steps lead to the capture of the second DSB end and the formation and resolution of Holliday junctions, promoted by the Msh4, Msh5 and Mer3 proteins (Hunter and Kleckner, 2001).
A search of the E. histolytica genome showed the presence of many HR-related genes. Some meiotic genes like Spo11, Dmc1, Mnd1, and many HR specific genes like Mlh1, Msh2, Rad21, and Rad51 are present in both E. histolytica and E. invadens (Bhattacharya et al., 2000; Ramesh et al., 2005; Malik et al., 2008; Singh et al., 2013; Kelso et al., 2016). HR-related genes were upregulated when DSBs were induced by UV-C irradiation (López-Casamichana et al., 2008) or when cells were subjected to growth stress (Singh et al., 2013). Further, HR could be directly demonstrated, and its efficiency increased during growth stress in E. histolytica and during encystation in E. invadens (Singh et al., 2013). These data suggest that HR operates in Entamoeba and could have critical roles in trophozoite growth and stage conversion.
4.1 Direct demonstration of HR in E. histolytica
The presence of a large number of HR-related genes in E. histolytica suggests that this conserved process should occur in this organism. However, the absence of markers for genetic selection, lack of any obvious sexual stages, and the inability to knock out genes by homologous recombination made it difficult to directly demonstrate HR in E. histolytica cells. Using an alternative approach based on recombination between inverted repeats it was possible to directly demonstrate the process of HR (Singh et al., 2013). Entamoeba histolytica and E. invadens cells were stably transfected with a plasmid containing two identical inverted repeats of 730 bp each, separated by a 1.6 kb fragment of luciferase (LUC) sequence. Recombination between the inverted repeats would cause flipping of the LUC sequence. This was measured by PCR using one primer from the LUC sequence and another from flanking sequence in the plasmid backbone. In the parental construct the two primers would be in the same orientation and would fail to give an amplicon. If recombination occurred between the inverted repeats the two primers would now be facing each other due to flipping of sequence, and a PCR product of expected size would be obtained. As an internal control a primer pair was designed from the LUC sequence which would give an amplicon in both the parental plasmid and that obtained post-transfection. This strategy showed that HR indeed takes place in E. histolytica cells, as an amplicon was obtained from the plasmid DNA of transfected cells but not from the original plasmid. The amplicons were sequenced to further demonstrate flipping of the sequence exactly at the expected recombination junction. Apart from cells grown under normal growth conditions HR was also checked in cells under different stress conditions, namely, serum starvation, heat stress, oxygen stress and UV-C irradiation. HR was activated in all stresses, by 1.4- to 2.7-fold. The same experimental protocol was used to check HR during cyst formation in E. invadens as well (Singh et al., 2013). Within 8 h of transfer of trophozoites to encystation medium recombination efficiency went up and remained high (3–4 fold) throughout encystation. Active recombination was also observed during excystation when the cysts obtained were transferred to fresh growth medium.
HR events were also demonstrated at chromosomal loci using the short interspersed nuclear element (SINE) sequences which are abundant in the E. histolytica genome. From the genome sequence database two SINE pairs were selected which were organized as inverted repeats in relatively close proximity on the same scaffold (Huntley et al., 2010). As in the plasmid study, recombined molecules could be detected here as well by using PCR primers that would give an amplicon only when recombination occurred between the inverted SINE repeats. HR occurred in exponentially growing cells and was enhanced under conditions of growth stress, concomitant with the upregulation of HR genes. Thus, inverted repeat sequences, either located on a plasmid or chromosomally, are capable of recombining in E. histolytica.
4.2 Transcriptional changes in HR genes during genotoxic stress in E. histolytica
Cells undergoing stress can suffer DNA damage, which is a trigger for HR. Several studies have looked at transcriptional changes in HR-related genes of E. histolytica in a variety of stresses.
4.2.1 UV-irradiation-induced stress
HR was induced in E. histolytica trophozoites irradiated with 254 nm UV-C (López-Casamichana et al., 2008; Charcas-Lopez et al., 2014). The appearance of DSBs was shown by TUNEL and comet assays, and was also suggested by histone phosphorylation. It is known that DSBs induce phosphorylation of human histone H2AX at the serine residue located in the SQ motif at the C-terminus (Chen et al., 2000). The same SQ motif is present in E. histolytica histone H2A as well, since the canonical histone H2A sequence of E. histolytica has been replaced with H2AX (Sullivan et al., 2006). Two gene sequences homologous to H2AX have been identified in E. histolytica and both have the conserved SQ motif (Lopez-Casamichana et al., 2008). The amount of phosphorylated H2AX, measured by Western blot analysis increased 5-fold in E. histolytica cells after UV-C treatment for 10 min, indicating the presence of DNA DSBs. This system was used to look at transcriptional changes (measured by semi-quantitative RT-PCR) in E. histolytica cells responding to DNA DSBs induced by UV-C treatment. It was found that EhMre11, EhRad51, EhRad51c and EhRad52 genes were transcribed at a very low level in non-irradiated trophozoites, and their expression levels went up after UV-C treatment. EhRad51 levels went up 16-fold, 30 min after UV-C treatment. On the other hand, the expression levels of EhNbs1, EhRad54 and EhRad59 genes which were abundantly transcribed in untreated trophozoites, dropped after UV-C treatment (Lopez-Casamachina et al., 2008). Possible post-transcriptional regulation of these genes in UV-treated cells was not studied. This system was further studied to look at global transcriptional changes in UV-C treated cells, using cDNA microarrays (Weber et al., 2009). Most genes showed less than 2-fold change in transcription levels. Interestingly, the genes coding for Fe-S clusters-containing proteins were over-expressed, showing that these could be involved in adaptation to DNA damage. Several genes coding for cytoskeleton proteins were downregulated, suggesting that actin dynamics could be impaired after UV irradiation.
4.2.2 Serum starvation-induced growth stress
Serum starvation induces a similar set of proteins as found during UV irradiation in HeLa cells (Glazer et al., 1989). Starvation is also known to trigger trophozoite to cyst conversion in Entamoeba, a process which could depend on meiotic recombination (Sanchez et al., 1994). To check for changes in expression of HR genes during serum starvation, exponentially growing E. histolytica cells were transferred to serum-deprived medium and starved for 24 h. Samples were removed at various time points and RNA levels of candidate genes were measured by qRT-PCR and northern hybridization (Singh et al., 2013). The analysed genes represented major steps of the recombination pathway, e.g., formation of DSBs (Spo11), sister chromatid pairing (Rad21), resection of ends following DSBs (Mre11), formation of nucleoprotein filament (Rad51, Dmc1), recombinase enhancement by stabilizing the presynaptic filament (Mnd1), mismatch repair during strand invasion (Msh2), and resolution of Holliday junctions (Mlh1). The expression levels of all genes went up in starved cells, and by 18 h of starvation the increase was between 1.2- to 5.5- fold. The relative expression of these genes was compared by Northern blot analysis. EhSpo11, EhMnd1, EhRad21 and EhMre11 were expressed at low levels in normal proliferating cells whereas EhDmc1 was expressed at moderate levels. These data showed that not only are homologues of HR genes present in E. histolytica, they are transcribed into mRNAs of the predicted size, and are upregulated during growth stress.
4.3 Transcriptional changes in HR and meiosis-specific genes during trophozoite to cyst conversion in Entamoeba invadens
Entamoeba invadens has been the model organism used to study encystation in Entamoeba, as it was not possible to obtain E. histolytica cysts in lab cultures. Exponentially growing E. histolytica trophozoites were transferred to encystation medium in which it typically took 72 h to complete the process of encystation. To monitor transcriptional changes in HR genes, cell samples were removed at various time points, and the RNA was analysed by qRT-PCR and Northern blotting (Singh et al., 2013). The HR genes studied were EiRad51, EiRad21, EiMlh1, EiMre11, and EiMsh2; whereas the meiosis-specific genes studied were EiSpo11A, EiSpo11B, EiDmc1 and EiMnd1. All the tested genes (except EiRad51) were upregulated during encystation. Expression levels went up 2- to 7.8-fold after 16–24 h of induction after which they began to decline. By 72 h, when encystation was complete, the expression levels of all genes had dropped. Northern analysis showed that EiSPO11a and EiSPO11b were poorly expressed in E. invadens trophozoites whereas EiDMC1 was expressed at moderate levels.
4.4 Biochemical characterization of the HR-related proteins EhRad51 and EhDmc1
4.4.1 EhRad51
Rad51 and Dmc1 are the eukaryotic orthologs of E. coli RecA recombinase that is responsible for homology search and strand exchange (Sung and Robberson, 1995). It forms the nucleoprotein filament which invades the homologous duplex and results in a displacement loop (D-loop). DNA synthesis is primed from the invading strand to restore damaged DNA. Rad51 is ubiquitously expressed whereas Dmc1 is specific to meiotic cells (Bishop et al., 1992). Sequences homologous to both Rad51 and Dmc1 are present in the E. histolytica genome. EhRad51 has been studied in detail. It encodes a protein of predicted molecular weight 40.3 kDa. The recombinant protein expressed in E. coli was biochemically characterized (Lopez-Casamichana et al., 2008). It could bind both ss- and ds-DNA. When incubated with a circular dsDNA and homologous ssDNA it could perform strand transfer and D-loop formation, thus demonstrating the canonical properties of Rad51 recombinase. Another study further characterized EhRad51 using a highly purified recombinant protein expressed in E. coli (Kelso et al., 2016). As seen with Rad51 from other organisms (Chi et al., 2006), EhRad51 could hydrolyse ATP, and the activity was stimulated in the presence of DNA, especially ssDNA. In keeping with this observation, EhRad51 was shown to bind DNA, with a strong preference for ssDNA. A nuclease protection assay was used to demonstrate the ability of EhRad51 to form a presynaptic filament in the presence of ATP. The non-hydrolyzable analogs of ATP, and surprisingly, even ADP could support filament formation, showing that nucleotide binding is required for this reaction but hydrolysis is not. Further, the ability of EhRad51 to perform strand exchange was demonstrated by using φX174 circular ssDNA along with φX174 linear dsDNA which is 5.4 kb. Strand exchange reaction was measured by the appearance of ds nicked circular DNA. This is formed by extension of the initial joint molecule by EhRad51 through its strand displacement activity (Kelso et al., 2016). Thus, EhRad51 could perform in vitro strand exchange over at least 5.4 kb EhRad51 could weakly catalyze the formation of D-loops, and this activity was stimulated 4-fold in the presence of calcium, as previously observed for human Rad51 (Bugreev and Mazin, 2004). D-loop formation was greatly enhanced by the addition of murine Hop2-Mnd1 which is a known accessory factor for Rad51 (Petukhova et al., 2005). This observation also shows the strong evolutionary conservation of Hop2-Mnd1 function.
4.4.2 EhDmc1
Dmc1 has the same properties as Rad51, with the exception that it is expressed only in meiotic cells. A sequence homologous to Dmc1 has been identified by sequence search. The recombinant EhDmc1 protein has been purified and biochemically characterized (Kelso et al., 2015). Much like EhRad51, EhDmc1 was also found to catalyze strand exchange over long DNA stretches. This activity was stimulated by calcium and the accessory factor Hop2-Mnd1. Although meiosis has not been demonstrated in E. histolytica, EhDmc1 was found to express in proliferating cells of E. histolytica. This raises the question whether meiosis, indeed, occurs in E. histolytica trophozoites, or whether EhDmc1 has another unidentified role, for instance in maintaining the polyploid state of E. histolytica (Willhoeft and Tannich, 1999).
4.5 HR as a therapeutic drug target
DNA repair and recombination are essential for parasite growth, and for it to cope with environmental stresses and evade the host immune response. Thus, this process could be a potential target for new therapies.
Two of the known inhibitors of human Rad51, DIDS (4,4′-diisothiocyanostilbene-2,2′-disulfonic acid) and B02 were tested with EhRad51 (Kelso et al., 2016). While DIDS inhibited the strand exchange activity of EhRad51, B02 did not have any significant effect. The same was also seen with EhDmc1. DIDS inhibited EhDmc1, and, importantly, it could strongly inhibit encystation in E. invadens whereas it had no effect on cell proliferation. Although it is possible that DIDS could inhibit encystation through an independent mechanism, these data do support the view that HR may be essential during encystation in Entamoeba. In addition, independent of the mechanism of action, DIDS could be one of a promising class of molecules of therapeutic potential to reduce infectivity by inhibiting cyst formation. Although DIDS had an inhibitory effect on EhRad51 and EhDmc1, it cannot itself be used directly as a drug due to its high toxicity in humans (Ishida et al., 2009).
5 Discussion
DNA replication studies in E. histolytica started with the rDNA episome which, due to its small size and high copy number, was an attractive system. Unlike most other prokaryotic and eukaryotic plasmids, this episome did not initiate replication from a fixed origin. Rather it shared some hallmarks of eukaryotic chromosomal DNA replication where there are many potential origins, of which some are fired at a given time and context (Bell and Dutta, 2002). The E. histolytica rDNA episome also contained many potential origins, which were not used equally at all times (Dhar et al., 1996). In exponentially growing cells the most commonly used ori mapped upstream of the rDNA transcription unit, close to the promoter of rRNA genes. However, when cells were starved of serum and replication was resumed in a synchronous manner by serum replenishment, the early oris mapped within the rDNA transcription unit, while at later times the ori in the promoter-proximal upstream IGS was activated (Ghosh et al., 2003). It is interesting that the upstream ori mapped very close to the rDNA promoter. A similar pattern is observed in other eukaryotes and it has been suggested that binding of transcription factors could influence the initiation of DNA replication. In Xenopus oocytes when cells divide rapidly without transcription during early embryogenesis, replication initiates from multiple oris in the rDNA locus. At mid-blastula stage when rDNA transcription is activated, the ori shifts to the IGS (Hyrien et al., 1995).
Context-dependent ori usage has also been reported in the β-globin locus. Here the deletion of an upstream region called the locus control region (LCR) represses transcription of the locus and also the ori (Aladjem et al., 1998). However, when the β-globin ori is placed in an ectopic location, it can function without the LCR. Hence, in addition to the ori sequence, factors like chromatin organisation which could change in different genomic contexts may influence initiation. These data suggest that the pattern of replication initiation and ori usage seen in the E. histolytica rDNA episome has strong similarities with eukaryotic chromosomes rather than plasmids. This is possibly because episomal rRNA genes in various protozoa (Ravel-Chapuis et al., 1985; Clark and Cross, 1987) probably originate from chromosomal loci, and have thus retained the same regulatory signals. Rather than being arranged in hundreds to thousands of tandemly-repeated copies on linear chromosomes as in most eukaryotes, these protozoa maintain their rRNA genes on multi-copy plasmids that were probably ‘pinched off’ from chromosomal locations.
Homologs of most of the DNA replication and recombination proteins could be found in E. histolytica by genome sequence analysis. Many of these genes were cloned and expressed, and the purified proteins were analysed biochemically. Some of them, like the family B2 DNA polymerases, show interesting evolutionary origins. The closest orthologs of the E. histolytica Family B2 DNA polymerase enzymes are those encoded by the DNA transposon class called Polinton-Maverick found in E. invadens (Kapitonov and Jurka, 2006; Pastor-Palacios et al., 2012). The archetypical Family B2 DNA polymerase is the bacteriophage φ29 enzyme (Rodriguez et al., 2005). Compared with this enzyme the TPR2 motif (found in family B2 DNA polymerases) was 21 amino acids longer in E. histolytica. From deletion studies it was shown that the TPR2 motif was involved in efficient bypass of abasic sites, in addition to its known role in processivity and strand displacement. Thus, the E. histolytica enzyme seems to have acquired additional specificities through the extra 21 amino acid motif. It was possibly derived from a DNA transposon which is still found in the E. invadens genome, although very few copies of it exist in extant E. histolytica.
The presence of most of the conserved recombination-related genes in E. histolytica indicates that the process operates in this parasite. However, the absence of markers for genetic selection, lack of any obvious sexual stages, and the inability to knock out genes by homologous recombination made it difficult to experimentally demonstrate HR in E. histolytica cells. Several lines of evidence now point to the existence of HR in Entamoeba. These include differential expression of HR-related genes in response to DSBs induced by UV-C irradiation (López-Casamichana et al., 2008) or when cells were subjected to growth stress (Singh et al., 2013). Importantly, HR could be directly demonstrated during growth stress in E. histolytica and during encystation in E. invadens (Singh et al., 2013) using inverted repeat sequences. Detailed analysis of E. invadens cells undergoing encystation showed the upregulation of HR and meiotic genes at 16–24 h after transfer to encystation medium. This indicates that HR and meiotic recombination is likely to be important in conversion of the uninucleate trophozoite to tetra nucleate cyst. Although there is no direct evidence of DNA exchange in E. invadens, studies with Giardia intestinalis indicate that DNA exchange occurs following nuclear fusion (karyogamy) during encystation in this organism (Poxleitner et al., 2008). It has been suggested that the presence of homologous gene sequences corresponding to the core meiotic machinery in Giardia and other protists (Entamoeba, Plasmodium, Trypanosoma and Leishmania) is indicative of an ancestral origin of meiosis and there is a central role of this process in all extant eukaryotes (Ramesh et al., 2005).
Although HR does operate in E. histolytica, its efficiency during normal growth may be low. That could be a possible reason for difficulty in obtaining gene knockouts in E. histolytica. The observation that HR is activated in E. histolytica cells immediately after exposure to a variety of stress conditions could be exploited to obtain gene knock-out in this organism (Singh et al., 2013). Efficiency of gene knock-out could be enhanced if the cells transfected with the suitable DNA construct were exposed to a brief duration of stress. HR is an essential process for parasite growth and differentiation, making it an attractive drug target. However, since HR is a highly conserved process, and is also essential for the host, the challenge would be to find species-specific inhibitors of HR proteins that selectively inhibit E. histolytica HR (Kelso et al., 2017). The fact that the human HR inhibitor B02 had no effect on E. histolytica HR shows that it should be possible to find E. histolytica -specific inhibitors as well (Kelso et al., 2016). High throughput screens could be set up using recombinant EhRad51 to look for small molecule inhibitors of the E. histolytica enzyme which do not inhibit the human enzyme. Future studies may provide structural information about E. histolytica recombinases, which could be used to design Entamoeba-specific inhibitor molecules.
Author contributions
SB has conceived and prepared the manuscript.
Funding
This study was supported by a fellowship from the Indian National Science Academy.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aladjem, M. I., Groudine, M., Brody, L. L., Dieken, E. S., Fournier, R. E., and Wahl, G. M. (1998). Genetic dissection of a mammalian replicator in the human beta-globin locus. Sci. (New York, N.Y.) 281 (5379), 1005–1009. doi:10.1126/science.281.5379.1005
Azuara-Liceaga, E., Betanzos, A., Cardona-Felix, C. S., Castañeda-Ortiz, E. J., Cárdenas, H., Cárdenas-Guerra, R. E., et al. (2018). The sole DNA ligase in Entamoeba histolytica is a high-fidelity DNA ligase involved in DNA damage repair. Front. Cell. Infect. Microbiol. 8, 214. doi:10.3389/fcimb.2018.00214
Bagchi, A., Bhattacharya, A., and Bhattacharya, S. (1999). Lack of a chromosomal copy of the circular rDNA plasmid of Entamoeba histolytica. Int. J. Parasitol. 29 (9), 1775–1783. doi:10.1016/S0020-7519(99)00125-3
Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. doi:10.1146/annurev.biochem.71.110601.135425
Bhattacharya, A., Satish, S., Bagchi, A., and Bhattacharya, S. (2000). The genome of Entamoeba histolytica. Int. J. Parasitol. 30 (4), 401–410. doi:10.1016/s0020-7519(99)00189-7
Bhattacharya, S., Som, I., and Bhattacharya, A. (1998). The ribosomal DNA plasmids of Entamoeba. Parasitol. Today 14 (5), 181–185. doi:10.1016/S0169-4758(98)01222-8
Bishop, D. K., Park, D., Xu, L., and Kleckner, N. (1992). DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456. doi:10.1016/0092-8674(92)90446-j
Bogan, J. A., Natale, D. A., and Depamphilis, M. L. (2000). Initiation of eukaryotic DNA replication: Conservative or liberal? J. Cell. Physiology, 184(2), 139–150. doi:10.1002/1097-4652(200008)184:2<139:AID-JCP1>3.0.CO;2-8
Brandsma, I., and Gent, D. C. (2012). Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 3 (1), 9. doi:10.1186/2041-9414-3-9
Brewer, B. J., and Fangman, W. L. (1987). The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51 (3), 463–471. doi:10.1016/0092-8674(87)90642-8
Bugreev, D. V., and Mazin, A. V. (2004). Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U. S. A. 101, 9988–9993. doi:10.1073/pnas.0402105101
Cardona-Felix, C. S., Lara-Gonzalez, S., and Brieba, L. G. (2011). Structure and biochemical characterization of proliferating cellular nuclear antigen from a parasitic protozoon. Acta Crystallogr. Sect. D. Biol. Crystallogr. 67, 497–505. doi:10.1107/S0907444911010547
Cardona-Felix, C. S., Pastor-Palacios, G., Cardenas, H., Azuara-Liceaga, E., and Brieba, L. G. (2010). Biochemical characterization of the DNA ligase I from Entamoeba histolytica. Mol. Biochem. Parasitol. 174 (1), 26–35. doi:10.1016/j.molbiopara.2010.06.010
Charcas-Lopez, M., Garcia-Morales, L., Pezet-Valdez, M., Lopez-Camarillo, C., Zamorano-Carrillo, A., and Marchat, L. A. (2014). Expression of EhRAD54, EhRAD51, and EhBLM proteins during DNA repair by homologous recombination in Entamoeba histolytica. Parasite (Paris, Fr. 21, 7. doi:10.1051/parasite/2014006
Chauleau, M., and Shuman, S. (2016). Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA). Nucleic Acids Res. 44 (5), 2298–2309. doi:10.1093/nar/gkw049
Chen, H. T., Bhandoola, A., Difilippantonio, M. J., Zhu, J., Brown, M. J., Tai, X., et al. (2000). Response to RAG-mediated VDJ cleavage by NBS1 and γ-H2AX. Sci. (New York, N.Y.) 290 (5498), 1962–1965. doi:10.1126/science.290.5498.1962
Chi, P., Van Komen, S., Sehorn, M. G., Sigurdsson, S., and Sung, P. (2006). Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair 5, 381–391. doi:10.1016/j.dnarep.2005.11.005
Clark, C. G., and Cross, G. A. (1987). rRNA genes of Naegleria gruberi are carried exclusively on a 14-kilobase-pair plasmid. Mol. Cell. Biol. 7 (9), 3027–3031. doi:10.1128/mcb.7.9.3027
Conway, C., Proudfoot, C., Burton, P., Barry, J. D., and McCulloch, R. (2002). Two pathways of homologous recombination in Trypanosoma brucei. Mol. Microbiol. 45 (6), 1687–1700. doi:10.1046/j.1365-2958.2002.03122.x
Couturier, M., Bex, F., Bergquist, P. L., and Maas, W. K. (1988). Identification and classification of bacterial plasmids. Microbiol. Rev. 52 (3), 375–395. doi:10.1128/mr.52.3.375-395.1988
Deitsch, K. W., Calderwood, M. S., Wellems, T. E., and Scherf, A. (1999). Intra-cluster recombination and var transcription switches in the antigenic variation of Plasmodium falciparum. Mol. Biochem. Parasitol. 101 (1-2), 107–116. doi:10.1016/s0166-6851(99)00062-6
Dhar, S. K., Roy Choudhury, N., Mittal, V., Bhattacharya, A., and Bhattacharya, S., (1996). Replication initiates at multiple dispersed sites in the ribosomal DNA plasmid of the protozoan parasite Entamoeba histolytica. Mol. Cell. Biol., 16(5), 2314–2324. doi:10.1128/MCB.16.5.2314
Ghosh, S., Satish, S., Tyagi, S., Bhattacharya, A., and Bhattacharya, S. (2003). Differential use of multiple replication origins in the ribosomal DNA episome of the protozoan parasite Entamoeba histolytica. Nucleic Acids Res. 31 (8), 2035–2044. doi:10.1093/nar/gkg320
Glazer, P., Greggio, N., Metherall, J., and Summers, W. (1989). UV-induced DNA-binding proteins in human cells. Proc. Natl. Acad. Sci. U. S. A. 86 (4), 1163–1167. doi:10.1073/pnas.86.4.1163
González-Magaña, A., and Blanco, F. J. (2020). Human PCNA structure, function and interactions. Biomolecules 10 (4), 570. doi:10.3390/biom10040570
Herrera-Aguirre, M. E., Luna-Arias, J. P., Labra-Barrios, M. L., and Orozco, E., (2010). Identification of four Entamoeba histolytica organellar DNA polymerases of the family B and cellular localization of the Ehodp1 gene and EhODP1 protein.J. Biomed. Biotechnol., 2010, 734898. doi:10.1155/2010/734898
Hughes, J. E., Dworkin, M., and Sussman, M. (1988). Nuclear plasmids in the Dictyostelium slime molds. Dev. Genet. 9 (4-5), 495–504. doi:10.1002/dvg.1020090426
Hunter, N., and Kleckner, N. (2001). The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106 (1), 59–70. doi:10.1016/s0092-8674(01)00430-5
Huntley, D. M., Pandis, I., Butcher, S. A., and Ackers, J. P. (2010). Bioinformatic analysis of Entamoeba histolytica SINE1 elements. BMC Genomics 11, 321. doi:10.1186/1471-2164-11-321
Hyrien, O., Maria, C., and Méchali, M. (1995). Transition in specification of embryonic metazoan DNA replication origins. Sci. (New York, N.Y.) 270 (5238), 994–997. doi:10.1126/science.270.5238.994
Ishida, T., Takizawa, Y., Kainuma, T., Inoue, J., Mikawa, T., Shibata, T., et al. (2009). DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Res. 37 (10), 3367–3376. doi:10.1093/nar/gkp200
Kapitonov, V. V., and Jurka, J. (2006). Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 103 (12), 4540–4545. doi:10.1073/pnas.0600833103
Kaur, D., Agrahari, M., Singh, S. S., Mandal, P. K., Bhattacharya, A., and Bhattacharya, S. (2021). Transcriptomic analysis of Entamoeba histolytica reveals domain-specific sense strand expression of LINE-encoded ORFs with massive antisense expression of RT domain. Plasmid 114, 102560. doi:10.1016/j.plasmid.2021.102560
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 (3), 375–384. doi:10.1016/s0092-8674(00)81876-0
Kelso, A. A., Say, A. F., Sharma, D., Ledford, L. L., Turchik, A., Saski, C. A., et al. (2015). Entamoeba histolytica Dmc1 catalyzes homologous DNA pairing and strand exchange that is stimulated by calcium and Hop2-Mnd1. PloS one 10 (9), e0139399. doi:10.1371/journal.pone.0139399
Kelso, A. A., Waldvogel, S. M., Luthman, A. J., and Sehorn, M. G. (2017). Homologous recombination in protozoan parasites and recombinase inhibitors. Front. Microbiol. 8, 1716. doi:10.3389/fmicb.2017.01716
Kelso, A. A., Waldvogel, S. M., Luthman, A. J., Sehorn, M. G., Turchick, A., Sharma, D., et al. (2016). Characterization of the recombination activities of the Entamoeba histolytica Rad51 recombinase. Mol. Biochem. Parasitol. 210 (1-2), 71–84. doi:10.1016/j.molbiopara.2016.09.001
Kumar, C., and Remus, D. (2016). Eukaryotic replication origins: Strength in flexibility. Nucl. (Austin, Tex.) 7 (3), 292–300. doi:10.1080/19491034.2016.1187353
Kurniawan, F., Shi, K., Kurahashi, K., Bielinsky, A., and Aihara, H. (2018). Crystal structure ofEntamoeba histolytica Cdc45 suggests a conformational switch that may regulate DNA replication. iScience 3, 102–109. doi:10.1016/j.isci.2018.04.011
Loftus, B., Anderson, I., Davies, R., Alsmark, U. C. M., Samuelson, J., Amedeo, P., et al. (2005). The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868. doi:10.1038/nature03291
López-Casamichana, M., Orozco, E., Marchat, L. A., García-Rivera, G., Guillen, N., Weber, C., et al. (2008). Transcriptional profile of the homologous recombination machinery and characterization of the EhRAD51 recombinase in response to DNA damage in Entamoeba histolytica. BMC Mol. Biol. 9, 35. doi:10.1186/1471-2199-9-35
Lorenzi, H. A., Puiu, D., Miller, J. R., Brinkac, L. M., Amedeo, P., Hall, N., et al. (2010). New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Neglected Trop. Dis. 4 (6), e716. doi:10.1371/journal.pntd.0000716
Lorenzi, H., Thiagarajan, M., Haas, B., Wortman, J., Hall, N., and Caler, E. (2008). Genome-wide survey, discovery and evolution of repetitive elements in three Entamoeba species. BMC Genomics 9, 595. doi:10.1186/1471-2164-9-595
Malik, S.-B., Pightling, A. W., Stefaniak, L. M., Schurko, A. M., and Logsdon, J. M. (2008). An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PloS One 3 (8), e2879. doi:10.1371/journal.pone.0002879
Maralikova, B., Ali, V., Nakada-Tsukui, K., Nozaki, T., van der Giezen, M., Henze, K., et al. (2010). Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 12 (3), 331–342. doi:10.1111/j.1462-5822.2009.01397.x
Marini, F., Kim, N., Schuffert, A., and Wood, R. D. (2003). POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 278 (34), 32014–32019. doi:10.1074/jbc.M305646200
Masson, J. Y., and West, S. C. (2001). The Rad51 and Dmc1 recombinases: A non-identical twin relationship. Trends Biochem. Sci. 26 (2), 131–136. doi:10.1016/s0968-0004(00)01742-4
Mi-ichi, F., Abu Yousuf, M., Nakada-Tsukui, K., and Nozaki, T. (2009). Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. U. S. A. 106 (51), 21731–21736. doi:10.1073/pnas.0907106106
Panigrahi, S. K., Jhingan, G. D., Som, I., Bhattacharya, A., Petri, W. A., and Bhattacharya, S. (2009). Promoter analysis of palindromic transcription units in the ribosomal DNA circle of Entamoeba histolytica. Eukaryot. cell 8 (1), 69–76. doi:10.1128/EC.00254-08
Pastor-Palacios, G., Azuara-Liceaga, E., and Brieba, L. G. (2010). A nuclear family A DNA polymerase from Entamoeba histolytica bypasses thymine glycol. PLoS neglected Trop. Dis. 4 (8), e786. doi:10.1371/journal.pntd.0000786
Pastor-Palacios, G., López-Ramírez, V., Cardona-Felix, C. S., and Brieba, L. G. (2012). A transposon-derived DNA polymerase from Entamoeba histolytica displays intrinsic strand displacement, processivity and lesion bypass. PloS one 7 (11), e49964. doi:10.1371/journal.pone.0049964
Petojevic, T., Pesavento, J. J., Costa, A., Liang, J., Wang, Z., Berger, J. M., et al. (2015). Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc. Natl. Acad. Sci. USA. 112, E249–E258. doi:10.1073/pnas.1422003112
Petukhova, G. V., Pezza, R. J., Vanevski, F., Ploquin, M., Masson, J. Y., and Camerini-Otero, R. D. (2005). The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 12, 449–453. doi:10.1038/nsmb923
Poxleitner, M. K., Carpenter, M. L., Mancuso, J. J., Wang, C. J., Dawson, S. C., and Cande, W. Z. (2008). Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science 319 (5869), 1530–1533. doi:10.1126/science.1153752
Ramesh, M. A., Malik, S. B., and Logsdon, J. M. (2005). A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 15 (2), 185–191. doi:10.1016/j.cub.2005.01.003
Ravel-Chapuis, P., Michot, B., Nigon, V., Neyret, O., and Freyssinet, G. (1985). Extrachromosomal circular nuclear rDNA in Euglena gracilis. Nucleic Acids Res. 13 (20), 7529–7537. doi:10.1093/nar/13.20.7529
Rodriguez, I., Lazaro, J. M., Blanco, M., Kamtekar, S., Berman, A. J., Wang, J., et al. (2005). A specific subdomain in phi29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. 102 (18), 6407–6412. doi:10.1073/pnas.0500597102
San Filippo, J., Sung, P., and Klein, H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257. doi:10.1146/annurev.biochem.77.061306.125255
Sánchez, L., Enea, V., and Eichinger, D. (1994). Identification of a developmentally regulated transcript expressed during encystation of Entamoeba invadens. Mol. Biochem. Parasitol. 67 (1), 125–135. doi:10.1016/0166-6851(94)90102-3
Sehgal, D., Mittal, V., Ramachandran, S., Dhar, S. K., Bhattacharya, A., and Bhattacharya, S. (1994). Nucleotide sequence organisation and analysis of the nuclear ribosomal DNA circle of the protozoan parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 67 (2), 205–214. doi:10.1016/0166-6851(94)00129-4
Sharma, R., Bagchi, A., Bhattacharya, A., and Bhattacharya, S. (2001). Characterization of a retrotransposon-like element from Entamoeba histolytica. Mol. Biochem. Parasitol. 115 (2), 45–53. doi:10.1016/S0166-6851(01)00300-0
Singh, N., Bhattacharya, A., and Bhattacharya, S. (2013). Homologous recombination occurs in Entamoeba and is enhanced during growth stress and stage conversion. PloS one 8 (9), e74465. doi:10.1371/journal.pone.0074465
Spinks, R. R., Spenkelink, L. M., Dixon, N. E., and van Oijen, A. M. (2021). Single-molecule insights into the dynamics of replicative helicases. Front. Mol. Biosci.; 8: 741718. doi:10.3389/fmolb.2021.741718
Sullivan, W. J., Naguleswaran, A., and Angel, S. O. (2006). Histones and histone modifications in protozoan parasites. Cell. Microbiol. 8 (12), 1850–1861. doi:10.1111/j.1462-5822.2006.00818.x
Sung, P., and Robberson, D. L. (1995). DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82 (3), 453–461. doi:10.1016/0092-8674(95)90434-4
Takata, K., Shimizu, T., Iwai, S., and Wood, R. D. (2006). Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 281 (33), 23445–23455. doi:10.1074/jbc.M604317200
Trasviña-Arenas, C. H., Cardona-Felix, C. S., Azuara-Liceaga, E., Diaz-Quezada, C., and Brieba, L. G. (2017). Proliferating cell nuclear antigen restores the enzymatic activity of a DNA ligase I deficient in DNA binding. FEBS open bio 7 (5), 659–674. doi:10.1002/2211-5463.12209
Tsubouchi, H., and Roeder, G. S. (2002). The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 22 (9), 3078–3088. doi:10.1128/MCB.22.9.3078-3088.2002
van der Giezen, M., and Tovar, J. (2005). Degenerate mitochondria. EMBO Rep. 6 (6), 525–530. doi:10.1038/sj.embor.7400440
Vicente, J. B., Tran, V., Pinto, L., Teixeira, M., and Singh, U. (2012). A detoxifying oxygen reductase in the anaerobic protozoan Entamoeba histolytica. Eukaryot. cell 11 (9), 1112–1118. doi:10.1128/EC.00149-12
Volkert, F. C., Wilson, D. W., and Broach, J. R. (1989). Deoxyribonucleic acid plasmids in yeasts. Microbiol. Rev. 53 (3), 299–317. doi:10.1128/mr.53.3.299-317.1989
Weber, C., Marchat, L. A., Guillen, N., and Lopez-Camarillo, C. (2009). Effects of DNA damage induced by UV irradiation on gene expression in the protozoan parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 164 (2), 165–169. doi:10.1016/j.molbiopara.2008.12.005
Keywords: entamoeba histolytica, replication, homologous recombination, meiotic genes, ribosomal DNA episome, Entamoeba invadens, encystation
Citation: Bhattacharya S (2023) Episomal and chromosomal DNA replication and recombination in Entamoeba histolytica. Front. Mol. Biosci. 10:1212082. doi: 10.3389/fmolb.2023.1212082
Received: 25 April 2023; Accepted: 26 May 2023;
Published: 08 June 2023.
Edited by:
Anand Srivastava, National Institute of Animal Biotechnology (NIAB), IndiaReviewed by:
Victor M. Ayala-Garcia, Juárez University of the State of Durango, MexicoLuis G. Brieba, National Polytechnic Institute of Mexico (CINVESTAV), Mexico
Sudip K. Ghosh, Indian Institute of Technology Kharagpur, India
Copyright © 2023 Bhattacharya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudha Bhattacharya, c2JqbnUxMTBAZ21haWwuY29t
 Sudha Bhattacharya
Sudha Bhattacharya