- Department of Thyroid and Breast Surgery, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai, China
Introduction: Circular RNAs (circRNAs) regulatory network is important in human cancer. We, therefore, mapped the regulatory networks driven by circRNA in luminal-subtype breast cancer.
Methods: Breast cancer-related microarray datasets from GEO database were analyzed for the differentially expressed circRNAs, miRNAs, and mRNAs. The potential downstream RNAs were collected using Circular RNA Interactome or Targetscan database. Protein-protein interaction (PPI) analysis was performed for the filtered genes to identify hub genes. The functions were annotated by the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. CircRNA-miRNA-mRNA networks were mapped using Cytoscape software. Hsa_circ_0086735-miR-1296-5p-STAT1 axis was used for verification. The expression levels of hsa_circ_0086735, miR-1296-5p, and STAT1 mRNA were confirmed by qRT-PCR in luminal-subtype tissues and cell lines. The interactions among them were verified by Luciferase reporter assay and RNA pull-down assay. Cell proliferation and apoptosis were assayed. Overall and distant metastasis-free survival was analyzed.
Results: A total of 70 genes were finally targeted and enriched in multi-process and multi-pathway. Networks containing 96 circRNA-miRNA-mRNA axes were constructed. Hsa_circ_0086735 and STAT1 mRNA was upregulated in luminal breast cancer, while miR-1296-5p was downregulated. Hsa_circ_0086735-miR-1296-5p-STAT1 axis promotes breast cancer progression and contributes to tamoxifen resistance. High hsa_circ_0086735 was associated with poor overall and distant metastasis-free survival.
Discussion: This study identified the hsa_circ_0086735-miR-1296-5p-STAT1 as an important regulatory axis in luminal-subtype breast cancer, aiding to determine potential therapeutic targets.
Introduction
Breast cancer is a global female malignancy and the leading cause of female cancer deaths (Loibl et al., 2021). It is a molecularly heterogeneous disease, identified as at least five molecularly distinct subtypes based on gene expression (Tsang and Tse, 2020). These molecular subtypes have distinct prognostic values across multiple treatment settings (Cheang et al., 2009). Among these subtypes, luminal breast cancers are termed hormone-receptor-positive breast cancer, including estrogen receptor (ER)-positive-luminal A (luminal A) and ER-positive-luminal B (luminal B) (Pellegrino et al., 2021). Luminal breast cancer is the most common type in women diagnosed with breast cancer, characterized by a high risk of relapse (Adachi et al., 2016). Currently, ER status is one predictive marker with an associated targeted therapy. The therapy choice for two-thirds of luminal breast cancer is endocrine therapy with tamoxifen or aromatase inhibitors (Katsura et al., 2022). Endocrine therapy has contributed to a recent decrease in the risk of the composite outcome of recurrence or death in breast cancer (Kerr et al., 2022). However, the emergence of acquired tamoxifen resistance, in approximately 40% of patients, is the major problem (Mishra et al., 2021). Although endocrine therapy would benefit many lower-risk patients, it may fail to prevent recurrence in some patients with high-risk resistance. Endocrine resistance can emerge from impaired estrogen signaling, MYC overexpression, amplification of cyclin D1 or MDM2 gene, lower activity of CYP2D6, or aberrant promoter methylation that activates Akt/mTOR pathway or other estrogen-independent growth-promoting pathways (Treeck et al., 2023). Thus, there is a need to unveil the underly mechanism of subtype breast cancer, the novel therapy targets overcoming the tamoxifen resistance, and the biomarkers to circumvent poor disease prognosis.
Though the traditional notion of gene regulation is centered around DNA and mRNAs, the protein-coding region only accounts for 2% of the human whole-genome transcriptome (Kong et al., 2022). A large proportion of the sequence, more than 90%, is found transcription for non-coding RNAs (ncRNAs) (Cohen and Jia, 2014). Though the ncRNAs do not control protein synthesis, they demonstrate their importance in tumorigenesis and tumor progression (Yan and Bu, 2021). micro-RNAs (miRNAs), a set of small ncRNAs of −20–23 nucleotides in length, is a type of well-studied ncRNA now (Ashekyan et al., 2022). Circular RNAs (circRNAs) are a recently understood significant class of ncRNAs, whose 5′ and 3′ ends are covalently attached via “back-splicing” (Yu and Kuo, 2019). CircRNAs demonstrate a more stable potential and are resistant to degradation by exonucleases due to the absence of 5′ or 3′ end (Jeck et al., 2013). Whereas miRNAs silence mRNA and regulate gene expression, circRNAs function through several mechanisms at transcriptional/translational levels (Kristensen et al., 2019). Both miRNAs and circRNAs have various roles in biological process regulation. A pan-cancer analysis has reported circRNA CDR1as acts as a mediator in the alteration of the tumor microenvironment (Zou et al., 2019). Moreover, circRNAs harboring miRNA response elements can sponge miRNAs and interfere with the splicing of other RNAs as endogenous RNAs (ceRNAs) (Kristensen et al., 2019). The binding and inhibition of miRNA by circRNA are one of the most influential and deregulated mechanisms in cancer (Zhang et al., 2021). The possible interaction between circRNAs and miRNAs makes circRNA possible to cross paths with various significant pathways related to cancers, acting as oncogenes or tumor suppressors depending on the function of the miRNA-downstream genes (Ashekyan et al., 2022). As circRNA could sponge to tens of miRNAs and miRNAs have thousands of targeting genes, even a minor dysregulation in the circRNA/miRNA/mRNA pathway could be ruinous (Khan et al., 2021). The circRNA-miRNA-mRNA axis could interfere with many drugs and relate to the progression of many tumors, including breast cancer (Ghazimoradi and Babashah, 2022). Therefore, this impactful axis, including the network resulting from it, needs to be discovered.
In a search for a circRNA-miRNA-mRNA network that might influence luminal subtype and tamoxifen resistance, the related GEO datasets were downloaded and analyzed. The differentially expressed circRNAs, miRNAs, and mRNAs were used to map the network. To elucidate the regulatory and molecular mechanism of the circRNA-miRNA-mRNA axis, cell functional assays, including cell proliferation and cell apoptosis, were performed.
Materials and methods
Associated circRNA expression dataset extraction
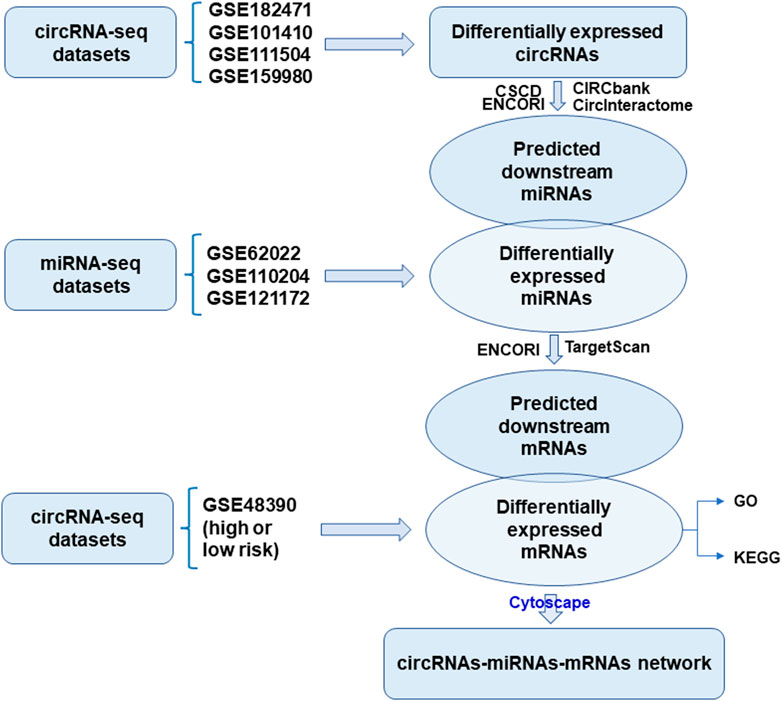
The GEO database was exhaustively searched for breast-cancer-related microarray datasets. Four GEO datasets, with accession numbers GSE182471, GSE101410, GSE111504, and GSE159980, were selected for the screen of circRNAs dysregulated in tamoxifen-resistant and metastatic luminal breast cancer. Dataset GSE182471 summarizes circRNA expression profiles from five pairs of breast cancer tissue samples and adjacent non-tumor tissue samples by microarray and RT-qPCR. GSE101410 comprises RNA-seq Luminal subtype-specific circRNAs in breast cancer cells. GSE111504 identified the circRNA expression signatures for breast cancer metastasis. GSE159980 is composed of microarray analysis of tamoxifen-sensitive and -resistant cells. Raw expression data were scored and extracted/normalized using the GEO-online tool GEO2R after applying log2 transformation. The overlapped circRNAs among these four datasets were available by VENNY diagram.
Associated miRNA expression dataset collection
GSE62022, GSE110204, and GSE121172 were analyzed using GEO2R for the luminal-specific miRNAs. The dysregulated miRNAs were subjected to a volcano-diagram presentation. The potential miRNAs for the obtained circRNAs were collected using Circular RNA Interactome (https://circinteractome.nia.nih.gov/index.html). The dysregulated miRNAs from GSE datasets were integrated with the circRNA-targeting miRNAs. GSE48390 was normalized and log2 transformed using GEO2R for differentially expressed genes between the high- and low-risk groups. Genes with absolute logFC values more than 2 and adjusted p-value less than 0.05 were screened as significantly differentially expressed.
Construction of protein-protein interaction networks
Significantly differentially expressed genes were subjected to STRING for the prediction of protein interactions. Networks were constructed using Cytoscape software and the hub genes in the network were identified using cyto-Hubba, a plug-in Cytoscape.
Functional and pathway enrichment analysis
Gene symbols were transformed for the analysis of gene-centric functional and canonical pathway enrichment analysis using OmicShare Tools.
Construction of the circRNA-miRNA-mRNA network
Combining RNA interactions and expression levels, the circRNA-miRNA-mRNA networks were established as two distinct sets using Cytoscape software. The circRNA-miRNA-mRNA axes containing hub genes were used for the visualization of final biological networks.
Patients and sample study
To be eligible for this study, women had to be at their first diagnosis of luminal-subtype breast cancer (Engstrøm et al., 2013). The candidates received no cancer-related therapy before surgery and immediately underwent breast surgery, followed by tamoxifen treatments. The tumor core biopsies, along with the adjacent normal tissues, were obtained from all patients and preserved in RNAlater for circRNA, miRNA, and mRNA expression evaluation. All women included in this study provided written informed consent. The study was approved by the Research Ethics Committee at Shanghai Tenth People’s Hospital, Tongji University School of Medicine. Information on survival time and distant metastasis was obtained from electronic or paper medical records in our institution.
Cell culture
The cell lines used in the analysis include MCF7, ZR-75-1, MCF7 Tam1, and ZR-75-1 Tam1, purchased from American Type Culture Collection (Rockville, MD, United States). Whereas ZR-75-1 and MCF7 were respectively cultured in 10% FBS-supplemented RPMI-1640 and EMEM, culture media for MCF7 was additionally contained and 10 μg/mL human insulin (Sigma. St. Louis, MO, United States). The tamoxifen-resistant cell lines, MCF-7 Tam1 and ZR-75-1 Tam1, used the same culture media as the parental cell lines, additionally containing 1 µM 4-hydroxytamoxifen (Sigma. St. Louis, MO, United States).
Transient transfections
Hsa_circ_0086735 siRNA (si-circ), STAT1 siRNA (si-STAT1), and non-silencing control plasmids (si-NC and ctr-STAT1) were purchased from Guangzhou RiboBio. Mi-1296-5p inhibitor (in-miR) and non-silencing control (in-NC) were purchased from Guangzhou Genecopoeia. The above vectors were transfected into cells by transient transfection using Lipofectamine 3,000 transfection reagents from Invitrogen.
RNA isolation and quantitative real-time PCR (qRT-PCR)
First, total RNA was extracted from tissue homogenate and cells using TRIzol (Life Technologies, Carlsbad, CA, United States). Then, total RNA was purified using the miRNeasy Mini kit (QIAGEN, Hilden, Germany) and assessed for purity using a NanoVue spectrophotometer (Ge BioSciences, United States). Total RNA, combined with anchored oligo (dT)18 primer and reverse transcriptase from the Transcriptor High FidelitycDNA Synthesis Kit (Roche, Germany), was subjected to the first strand synthesis for cDNA. The cDNA samples were examined by qRT-PCR using FastStart™ universal SYBR® Green (Roche) for determination of miR-1296-5p and STAT1 mRNA, using U6 and beta-actin as the endogenous control. For hsa_circ_0086735, total RNA was prepared to the linear RNA using RNase R digestion (Epicentre Technologies, Madison, WI, United States). RNase R-treated RNA was reversely transcribed to cDNA using SuperScript III First Strand Synthesis System (Invitrogen). qRT-PCR was performed using Power Up Sybr Green Master Mix (Thermo Fisher Scientific). Each sample was assayed in triplicate determinations at least.
Luciferase reporter assay
Luciferase reporter plasmids, including WT-circ, MUT-circ, WT-STAT1, and MUT-STAT1, were constructed by LandM Biotechnology (Guangzhou, China). These reporter plasmids were singly transfected into MCF7 and ZR-75-1 cells with miR-1296-5p mimic or inhibitor. After incubation, each cells lysate was prepared using two kinds of buffers, Firefly luciferase substrate and Firefly luciferase inhibitor/Renilla luciferase substrate, corresponding to the first measure of the Firefly luciferase activity and second measure of the Renilla luciferase activity. Finally, the normalized luciferase activity was calculated by Firefly luciferase activity/Renilla luciferase activity. Data were collected from at least three independent experiments for each subgroup.
RNA pull-down assay
The combination of miR-1296-5p and hsa_circ_0086735, as well as miR-1296-5p and STAT1, was determined by RNA pull-down assay. The miR-1296-5p probe (biotin-miR) and its negative control (biotin-NC) were constructed by Ribobio. Harvested MCF7 and ZR-75-1 were lysed using lysis buffer and then incubated with a biotin-labeled miT-1296-5p probe and streptavidin agarose magnetic beads. Hsa_circ_0086735 and STAT1 mRNA in miR-1296-5p probe pull-down complex were detected by qRT-PCR. Data were collected from at least three independent experiments for each subgroup.
Proliferation assays
An appropriate number of cells (1 × 105 of MCF7 and 8 × 104 of ZR-75-1 or ZR-75-1 Tam1) were seeded into a 96-well plate. Different periods after plating, 0, 12, 24, 48, and 72 h for MCF7 and ZR-75-1 and 0, 2, 4, 6, 9 days for ZR-75-1 Tam1, 10 μL of CCK-8 solution (Millipore-Sigma) was added to each well, followed by a 2-h incubation. The optical density of each well was measured at 450 nm using a BioTek spectrophotometer.
For tamoxifen-sensitive studies with the ZR-75-1 tamoxifen-resistant cell line, cells were seeded with increasing tamoxifen concentrations (0–120 μg/mL). After 6 days, cell viability was evaluated by CCK-8 solution (Millipore-Sigma). Data were collected in biological duplicates run in triplicate for each time point.
Annexin V-FITC/PI double staining for apoptosis
To quantify apoptosis, transfected cells cultured for 48 h were harvested, digested, and pipetted to achieve a uniform single-cell suspension. Then cells were doubly stained with the Annexin V-FITC Apoptosis Detection Kit (Sigma-Aldrich). To score Annexin V and/or PI-positive cells, cells were subjected to analysis on a BD Accuri C6 Plus Flow Cytometer (Franklin Lakes, NJ, United States). All data were obtained from at least three independent culture experiments.
Statistical analysis
Hsa_circ_0086735, miR-1296-5p, and STAT1 mRNA expression levels were calculated from qRT-PCR CT values using 2−ΔΔCT. The paired t-test was conducted for comparison of the expression differences between the tumor and normal tissues. Based on the mean value of hsa_circ_0086735 in the enrolled patients’ tissues, patients were classified into a low-expression group and a high-expression group. Chi-square test or Spearman coefficient of correlation was used to compare tumor characteristics across categories of hsa_circ_0086735 expression levels. The prognostic significance of relevant variables was assessed by Cox’s proportional-hazards regression from multivariate analysis. Overall and distant metastasis-free survival was assessed by Kaplan-Meier analysis with the log-rank test. Whereas normally distributed variables were compared using two-sided t-tests, non-normally distributed variables were compared using Mann-Whitney U tests. A p-value less than 0.05 was the cutpoint to be considered significant.
Results
Differential expression of circRNAs in tamoxifen-resistant luminal breast cancer
The flowchart of construction for the circRNA-miRNA-mRNA network was shown in Figure 1. To filter differentially expressed circRNAs, GSE18247 was analyzed by GEO2R (Figure 2A). To determine differentially expressed circRNAs in tamoxifen-resistant luminal breast cancer, the circRNAs obtained from GSE182471 were crossed and overlapped with those from GSE101410, GSE111504, and GSE159980. There were eight circRNAs significantly dysregulated in tamoxifen-resistant luminal breast cancer (Figure 2B). The differential expression of these eight circRNAs, hsa_circ_0086735, hsa_circ_0008267, hsa_circ_0001921, hsa_circ_0000458, hsa_circ_0008817, hsa_circ_0022601, hsa_circ_0004036, and hsa_circ_0006314, was presented as a heat map in Figure 2C.
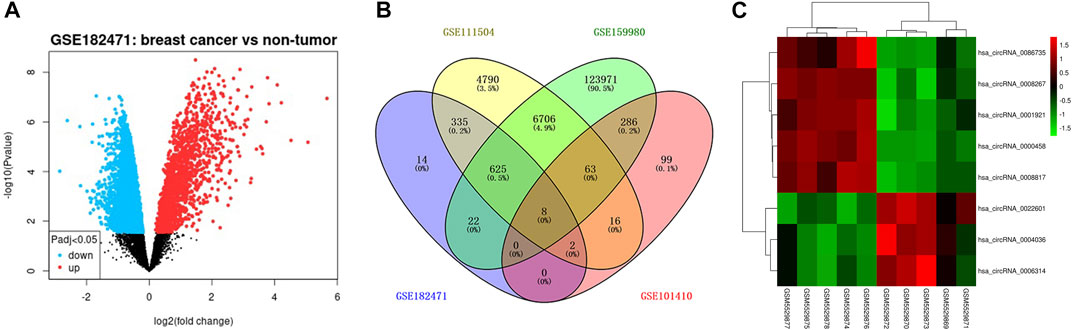
FIGURE 2. The differentially expressed circRNAs in tamoxifen-resistant and luminal-subtype breast cancer. (A) Volcano map of up/downregulated circRNAs in GSE182471 analyzed by GEOR2. The red dots indicated upregulated circRNAs and the blue dots indicated downregulated circRNAs. (B) Venn diagrams of the shared differentially expressed circRNAs among GSE182471, GSE111504, GSE159980, and 101,410. (C) Heatmap of eight up/downregulated circRNAs according to the expression in GSE182471.
Differentially expressed miRNAs targeted by the dysregulated circRNAs
The luminal-related miRNAs were obtained from GSE32022, GSE110204, and GSE121172, and presented as a Volcano diagram (Figure 3A). The downstream miRNAs of the eight dysregulated circRNAs were collected and overlapped with the luminal-subtype-related differentially expressed miRNAs (DE-miRNAs) (Figures 3B,C). In total, 26 miRNAs were classified as present across the panel of eight dysregulated circRNAs. These target miRNAs were used for further differential analysis.
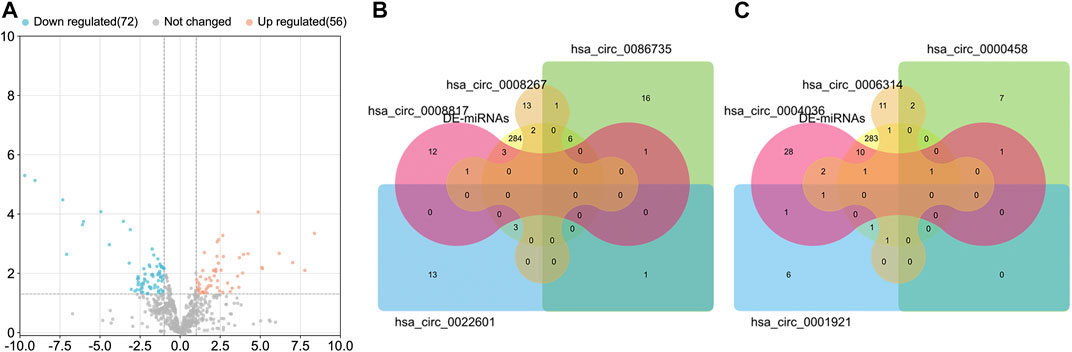
FIGURE 3. The differentially expressed miRNAs (DE-miRNAs) in luminal-subtype breast cancer. (A) Volcano map of the shared miRNAs from GSE32022, GSE110204, and GSE121172. The orange dots indicated upregulated micRNAs and the blue dots indicated downregulated miRNAs. (B) and (C) Venn diagrams of the overlapped miRNAs among DE-miRNAs and the circRNA-targeting miRNAs.
The prognostic gene corresponding to differential expressed circRNA-miRNA axes
The prognostic gene signature from GSE48390 for Han Chinese breast cancers was analyzed by GEO2R. A total of 508 differentially expressed and prognosis-related genes were identified (Figure 4A). Subsequently, 70 genes were screened to cover the downstream genes of the above 26 miRNAs, and their PPI was shown in Figure 4B with orphaned nodes deleted. The top 10 hub nodes in the PPI network were identified as PTPRC, IL7R, STAT1, LCK, SLAMF1, CCR5, ITK, CD38, CD3D, and CD24. We further conducted the GO and KEGG pathway enrichment analysis of 70 genes. The GO-enriched results showed that the genes mainly enriched in the main immune system process, developmental process, response to stimulus, and multicellular organismal process (Figure 4C). KEGG enrichment analysis showed that they were enriched for 124 pathways (Figure 4D). In KEGG, the top 20 enriched pathways included PD-L1 expression and PD-1 checkpoint pathway in cancer, cAMP signaling pathway, and JAK-STAT signaling pathway (Figure 4E).

FIGURE 4. Bioinformatics of the differentially expressed mRNAs. (A) Volcano map of differentially expressed mRNAs in GSE48390 analyzed by GEOR2. (B) Protein-protein interaction (PPI) networks of the shared mRNA between differentially expressed mRNAs and miRNA-targeting mRNAs. (C) The enriched GO terms by the shared mRNAs. (D) The enriched KEGG pathways of the shared mRNAs. (E) The top 20 pathways enriched by the shared mRNAs.
Maps of the ceRNA (circRNA-miRNA-mRNA) network
A total of 96 circRNA-miRNA-mRNA axis, including 47 correctly sequenced ones (Figure 5A) and 49 invalid-ordered ones (Figure 5B), were obtained with the above-obtained RNAs. As for the hub genes (Supplementary Table S1), the circRNA-miRNA-mRNA-pathway network was mapped (Figure 5C). Given STAT1 was one of the hub genes that ranked second (Score = 10) in the interaction network by Degree method (Supplementary Table S1), we chose the hsa_circ_0086735-miR-1296-5p-STAT1 axis for further validation.
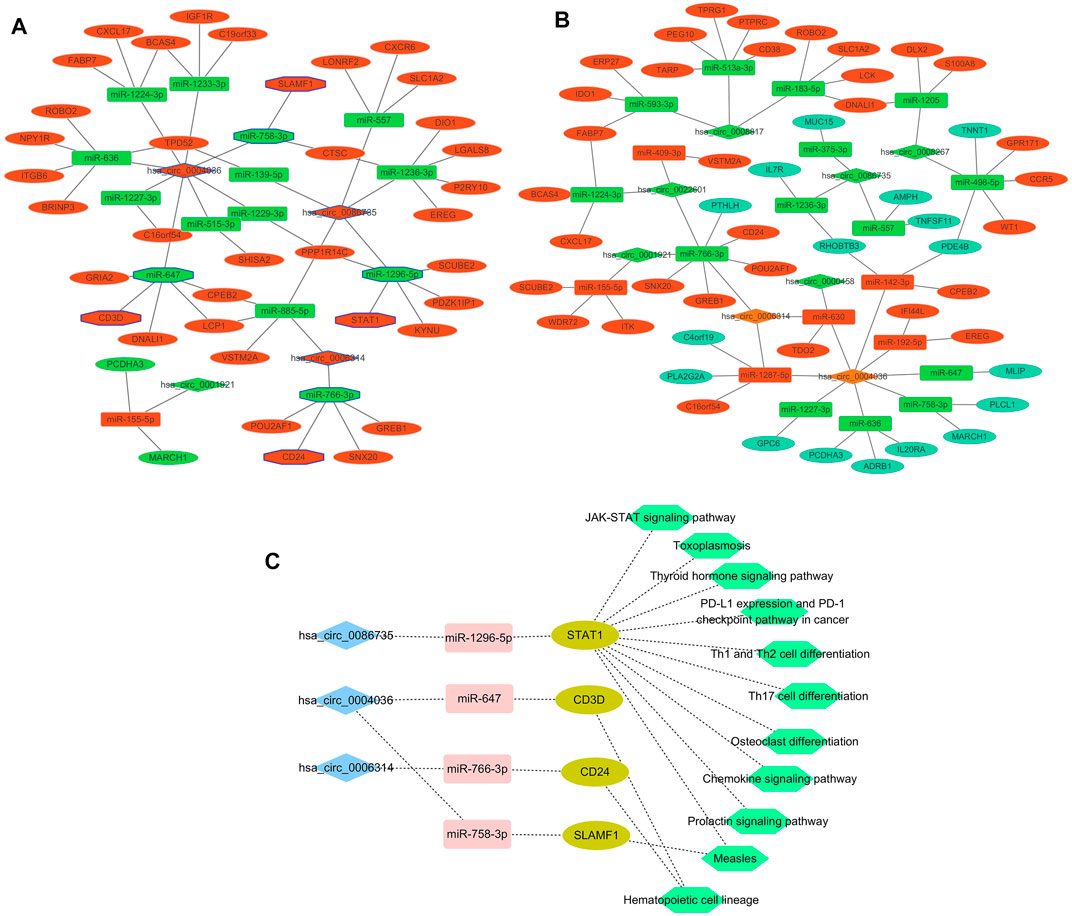
FIGURE 5. Networks. (A) and (B) Differentially expressed circRNA-miRNA-mRNA networks. The diamond represented circRNAs, the rectangles miRNAs, and ellipses mRNAs. Red and yellow color represent up- and downregulation in luminal-B tumor tissues, respectively. The black border indicated the pathway derived from the top 10 hub genes. (C) Network of the main circRNA-miRNA-mRNA-pathway.
Validation of hsa_circ_0086735, miR-1296-5p, and STAT1 expression
qRT-PCR was used to validate the expression levels of hsa_circ_0086735, miR-1296-5p, and STAT1 in luminal-subtype breast cancer. Hsa_circ_0086735 was significantly upregulated in luminal breast tumor tissues (p < 0.001, Figure 6A), which corresponded to the array data from GSE182471. In contrast, miR-1296-5p was significantly downregulated in the tumor tissues (p < 0.001, Figure 6B). STAT1 mRNA was an upregulated mRNA in luminal cancerous tissues (p < 0.001, Figure 6C). Luminal-subtype cell lines showed the same altered tends correspondingly (p < 0. 01, Figures 6D–F).
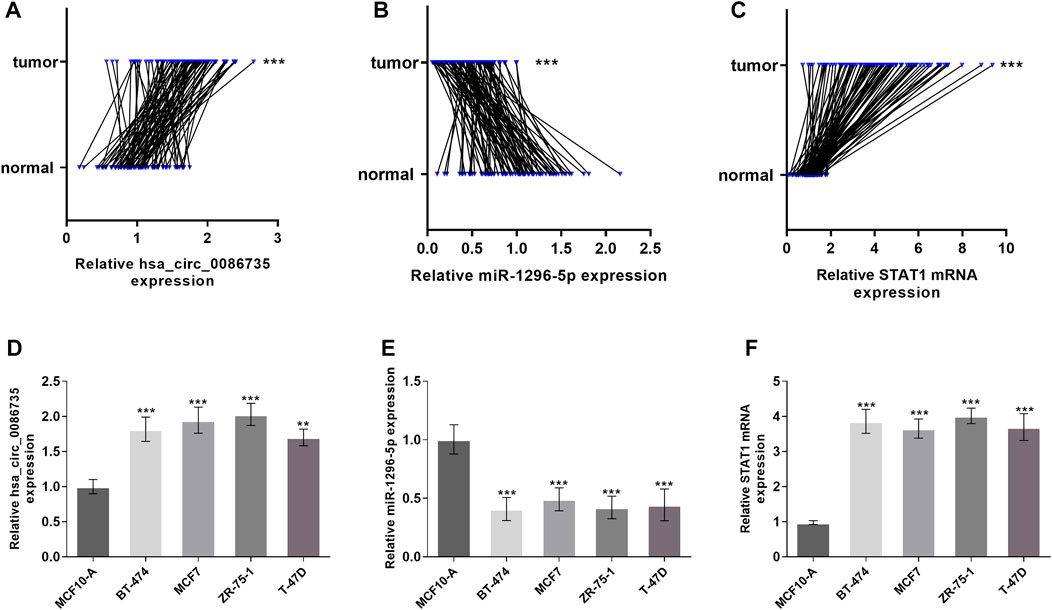
FIGURE 6. Expression of hsa_circ_0086735 in luminal-subtype breast cancer. Luminal-subtype breast cancer tissues expressed higher hsa_circ_0086735 (A), lower miR-1296-5p (B), and higher STAT1 mRNA (C) compared to those in adjacent normal tissues. Expression of hsa_circ_0086735 (D), miR-1296-5p (E), and STAT1 mRNA (F) in luminal-subtype cell lines compared to those in MCF10-A. ***p < 0.001.
Validation of hsa_circ_0086735-miR-1296-5p-STAT1 axis
To confirm the interaction between hsa_circ_0086735 and miR-1296-5p, as well as miR-1296-5p and STAT1, Luciferase reporter assay and RNA pull-down assay, were introduced. The binding sites between hsa_circ_0086735 and miR-1296-5p were shown in Figure 7A, to gain the specific sequence of the combination of the two and thus to construct Luciferase report plasmids and sequence used in RNA pull-down assay. The expression of hsa_circ_0086735 was inversely correlated to that of miR-1296-5p (Figure 7B). Then the plasmids containing mutant- or wild-type hsa_circ_0086735 were used to study the influence of miR-1296-5p dysregulation on their luciferase activity. As expected, the reporter that carried binding sites was significantly influenced by miR-1296-5p mimics or inhibitors in MCF7 cells, whereas a luciferase reporter without binding was negligible (p < 0.001, Figure 7C). Then, biotin-labeled RNA pull-down assays demonstrated that biotin-labeled specific miR-1296-5p probes could pull down abundant hsa_circ_0086735 in ZR-75-1 cells (p < 0.001, Figure 7D). We simultaneously tested the binding of miR-1296-5p to STAT1 (Figure 7E). MiR-1296-5p was inversely expressed with STAT1 mRNA (Figure 7F). With Luciferase reporter assay, we assessed the binding of the wild-STAT1-containing reporter to miR-1296-5p and found that wild-type STAT1 resulted in repression of fluorescence intensity by miR-1296-5p mimics or induction by a miR-1296-5p inhibitor (p < 0.001, Figure 7G). RNA pull-down assay confirmed their binding (p < 0.001, Figure 7H).
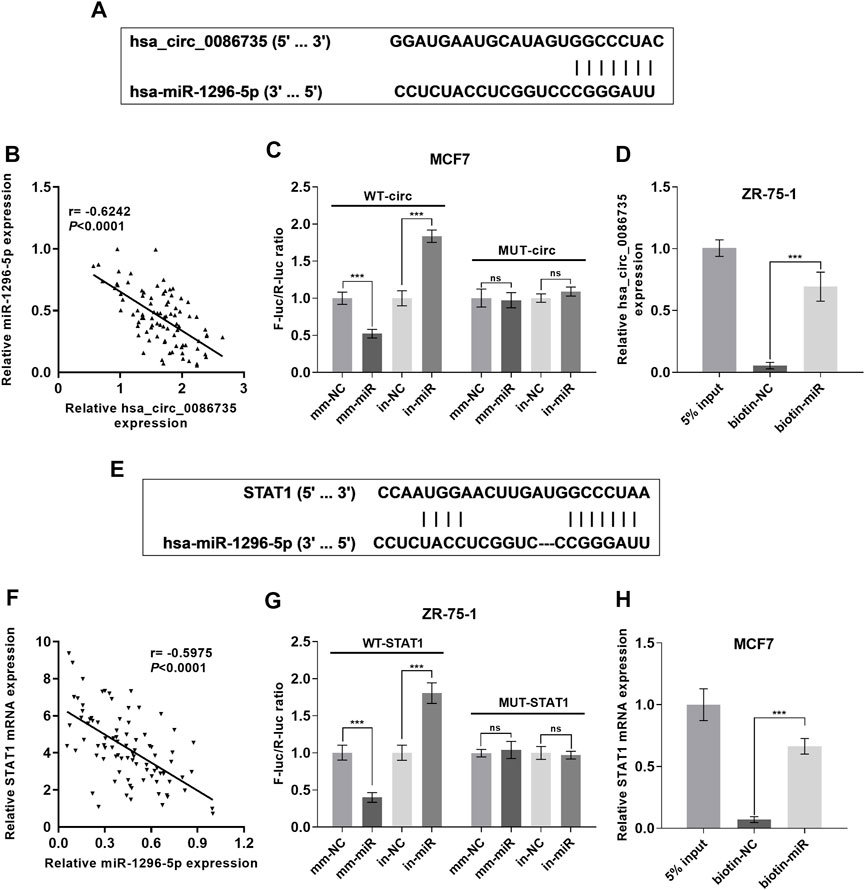
FIGURE 7. Hsa_circ_0086735 sponged to miR-1296-5p, and miR-1296-5p targeted to STAT1. (A) Predicted complementary binding sites of hsa_circ_0086735 against miR-1296-5p. (B) The expression level of hsa_circ_0086735 was reversely related to that of miR-1296-5p. (C) Luciferase reporter assays confirm the binding between hsa_circ_0086735 and miR-1296-5p. The pmirGL3 reporter vectors were cloned with the wild-type hsa_circ_0086735 with the potential miR-1296-5p binding sites to construct WT-circ plasmids or mutant of these sites to construct MUT-circ plasmids, and then transfected into MCF-7 cells with miR-1296-5p mimic (mm-miR) or inhibitor (in-miR) or corresponding negative controls (mm-NC or in-NC). (D) In vitro pull-down of hsa_circ_0086735 with biotin-labeled miR-1296-5p (biotin-miR) or negative control (biotin-NC) in ZR-75-1 cells. (E) Predicted complementary binding sites of miR-1296-5p against STAT1. (F) Luciferase reporter assays confirm the binding between miR-1296-5p and STAT1. The pmirGL3 reporter vectors were cloned with the wild-type STAT1 with the potential miR-1296-5p binding sites to construct WT-STAT1 plasmids or mutant of these sites to construct MUT-STATA1 plasmids, and then transfected into ZR-75-1 cells with miR-1296-5p mimic (mm-miR) or inhibitor (in-miR) or corresponding negative controls (mm-NC or in-NC). (G) In vitro pull-down of STAT1 mRNA with biotin-labeled miR-1296-5p (biotin-miR) or negative control (biotin-NC) in MCF7 cells. (H) The expression level of miR-1296-5p was reversely related to that of STAT1. ***p < 0.001.
Hsa_circ_0086735-miR-1296-5p-STAT1 axis promoted luminal breast cancer progression
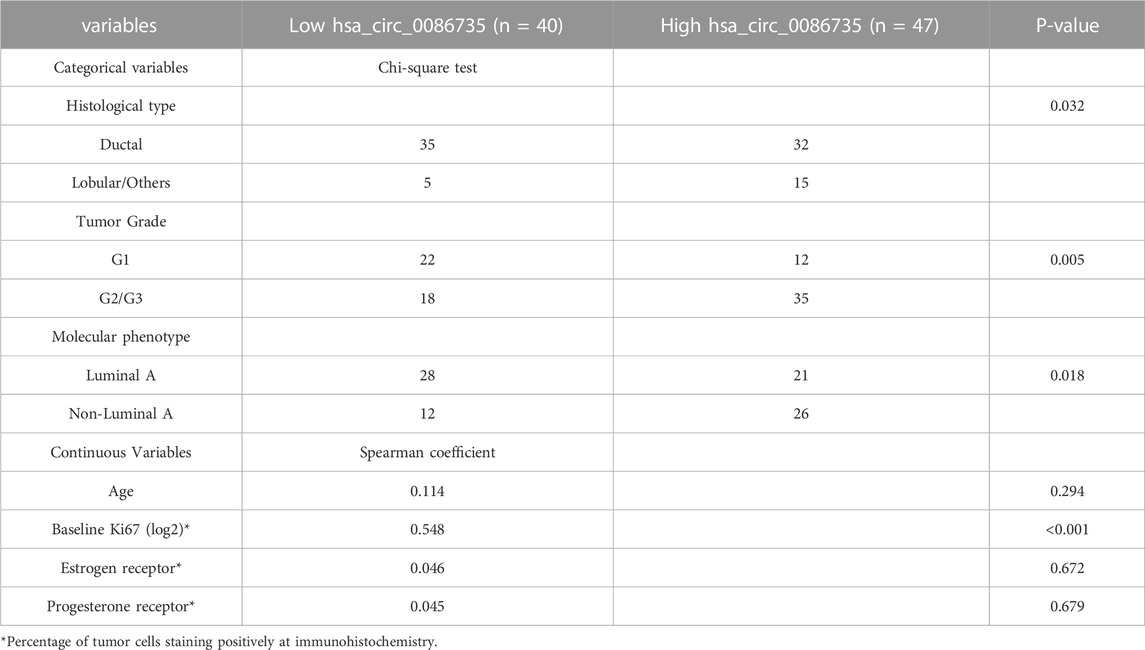
To determine whether hsa_circ_0086735-miR-1296-5p-STAT1 axis was associated with luminal breast cancer cell function or patients’ survival, we assessed cell proliferation, cell apoptosis, and patients’ overall survival. Cell transfection was verified by the expression of STAT1 mRNA (Figures 8A,B). We observed a robust reduction in the proliferation of luminal breast cancer cells if hsa_circ_0086735 was individually inhibited, but increased again if both hsa_circ_0086735 and miR-1296-5p were inhibited until there was another reversal if hsa_circ_0086735, miR-1296-5p, and STAT1 were all inhibited (Figures 8C,D). The opposite trend occurs in cell apoptosis suggesting that hsa_circ_0086735 and STAT1 deficiency induced the apoptotic capacity, while miR-1296-5p inhibition reduced the apoptotic capacity (Figures 8E,F). The mean level of hsa_circ_0086735 (1.66) was defined as the cutoff for grouping of high expression and low expression. Next, high hsa_circ_0086735 was significantly correlated with patients’ histological type, tumor grade, molecular phenotype, and baseline Ki67 (Table 1). Additionally, high hsa_circ_0086735 was associated with poor overall survival in luminal breast cancer patients by Kaplan-Meier analysis (Figure 8G) and Multivariate Cox proportional hazards analysis (Table 2; HR 11.889, p = 0.007).
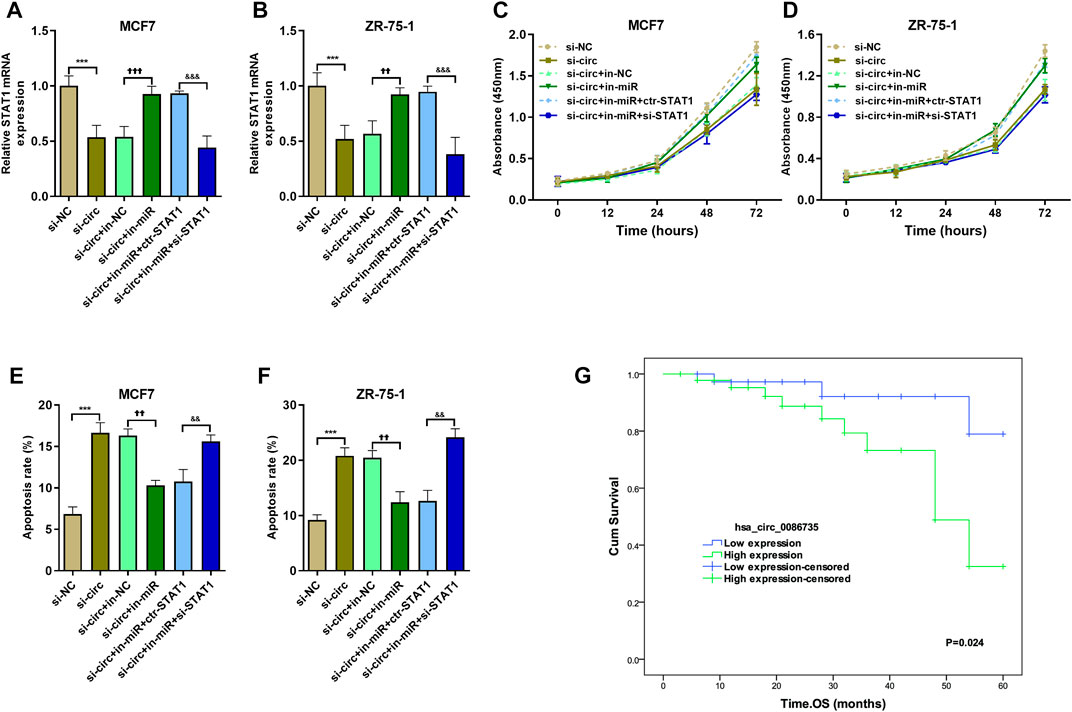
FIGURE 8. The effect of hsa_circ_0086735-miR-1296-5p-STAT1 axis on proliferation and apoptosis of luminal-subtype cell lines. (A) and (B) The transfection efficiency was confirmed by qRT-PCR analysis. CCK8 was performed on MCF7 (C) and ZR-75-1 (D) cells. Apoptosis assay by AnnexinV-FITC/PI staining for MCF7 (E) and ZR-75-1 (F) cells. (G) Overall survival of 87 breast cancer patients whose tumors had high or low hsa_circ_0086735 expression. ***p < 0.001, compared with si-NC group. ††p < 0.01, †††p < 0.001, compared with si-circ+in-NC group. &&p < 0.01, &&&p < 0.001, compared with si-circ+in-miR + ctr-STAT1 group. si-NC: negative siRNA control for hsa_circ_0086735; si-circ: siRNA for hsa_circ_0086735; in-NC: inhibitor negative control; in-miR: miR-1296-5p inhibitor; ctr-STAT1: negative siRNA control for STAT1; si-STAT1: siRNA for STAT1.
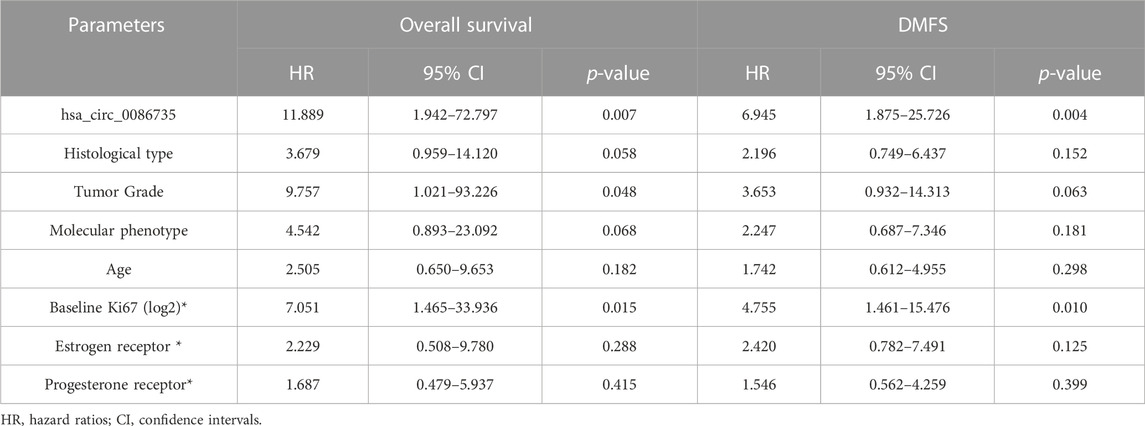
TABLE 2. Multivariable analysis of predictors for the prognosis of overall survival and distant metastasis-free survival (DMFS).
Hsa_circ_0086735-miR-1296-5p-STAT1 axis affects tamoxifen sensitivity
To determine whether hsa_circ_0086735-miR-1296-5p-STAT1 axis contributes to tamoxifen resistance, the area under the ROC curve (AUC) for STAT1, along with the other three genes in circRNA-miRNA-mRNA-pathway network, discriminating the non-response and response groups, was obtained from CTR-DB datasets (Supplementary Figure S1). Then, the expression of hsa_circ_0086735 was determined as an upregulated status in tamoxifen-resistant cell lines compared to that in the parental cell lines (Figure 9A). ZR-75-1 Tam1 cell line with a higher hsa_circ_0086735 expression was transfected for the subsequent experiments (Figure 9B). ZR-75-1 Tam1 developed a vulnerability to tamoxifen after hsa_circ_0086735 knockdown or hsa_circ_0086735/miR-1296-5p/STAT1 inhibition but become resistant to tamoxifen when hsa_circ_0086735 and miR-1296-5p were simultaneously inhibited, presented as the change in IC50 values (Figure 9C). Subsequent functional assays suggested that inhibited hsa_circ_0086735 reduced the cell proliferation potential, while miR-1296-5p can weaken this effect but STAT1 can consolidate it (Figure 9D). The induction of hsa_circ_0086735 on cell apoptosis rate can be abolished by miR-1296-5p and reproduced with the inhibition of STAT1 (Figure 9E). In patients treated with tamoxifen, high hsa_circ_0086735 was associated with poor distant metastasis-free survival (DMFS) (Figure 9F; Table 2, HR 6.945, p = 0.004).
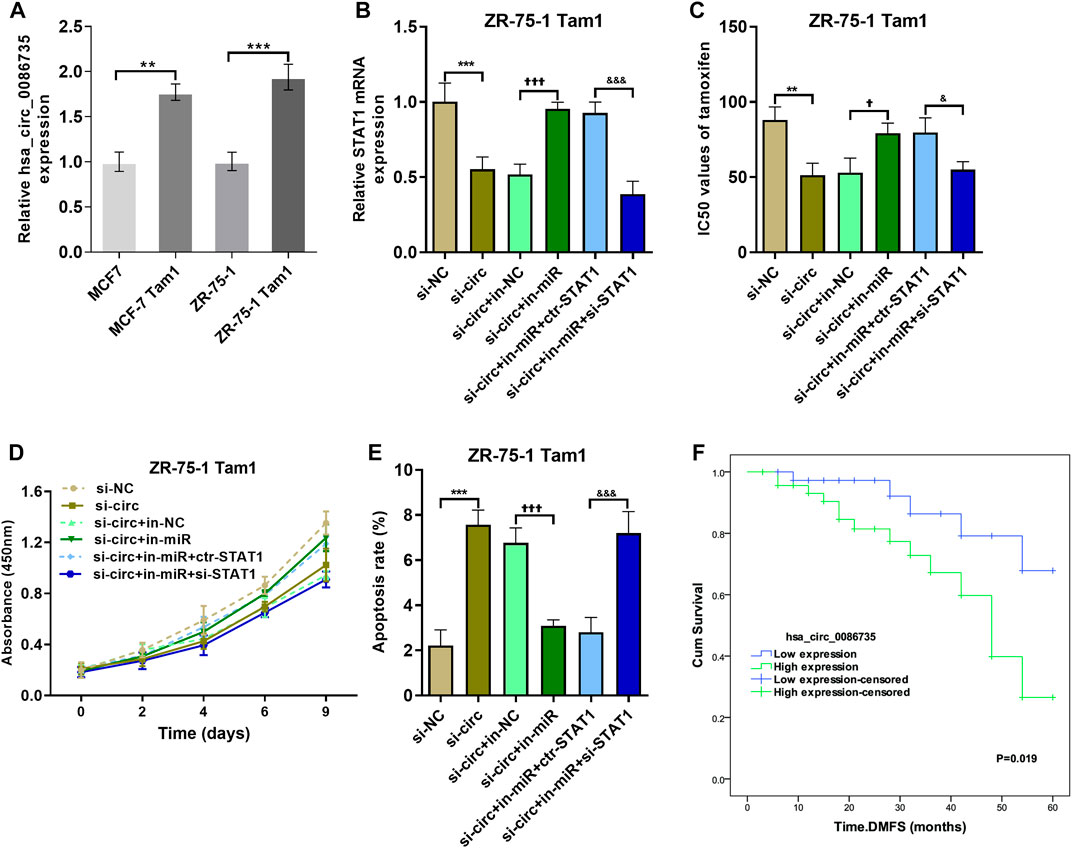
FIGURE 9. The effect of hsa_circ_0086735-miR-1296-5p-STAT1 axis on the proliferation of tamoxifen-resistant cells. (A) Hsa_circ_0086735 was upregulated in tamoxifen-resistant cells. (B) The transfection efficiency was confirmed by qRT-PCR analysis. (C) The IC50 values of different cell lines. (D) The cell proliferation by CCK-8 assay. (E) Apoptosis assay by AnnexinV-FITC/PI staining. (F) Distant metastasis-free survival of 87 breast cancer patients whose tumors had high or low hsa_circ_0086735 expression. ***p < 0.001, compared with si-NC group. ††p < 0.01, †††p < 0.001, compared with si-circ+in-NC group. &&p < 0.01, &&&p < 0.001, compared with si-circ+in-miR + ctr-STAT1 group. si-NC: negative siRNA control for hsa_circ_0086735; si-circ: siRNA for hsa_circ_0086735; in-NC: inhibitor negative control; in-miR: miR-1296-5p inhibitor; ctr-STAT1: negative siRNA control for STAT1; si-STAT1: siRNA for STAT1.
Discussion
The discovery of circRNA subsets has added a new layer of complexity to the alterations of gene expression associated with epigenetic changes, primarily via the ceRNA network comprised of various circRNA-miRNA-mRNA axes (Shen et al., 2015; Fontemaggi et al., 2021). The dysfunction of circRNA-miRNA-mRNA regulatory signal represents another important epigenetic-controlled mechanism of the pathogenic gene expression in human cancer, which are important nodes in cancer development (Tang et al., 2021; Mo et al., 2022). Mapping the circRNA-miRNA-mRNA network is an important way to elucidate the regulatory role of circRNA being investigated (Rengganaten et al., 2020; Pang et al., 2022). In this work, we screened luminal-specific and tamoxifen-resistant-related circRNAs, as well as differentially expressed miRNAs and mRNAs, from GEO breast cancer datasets. The differentially expressed circRNAs, miRNAs, and mRNA were subsequently used in the mapping of a novel circRNA-miRNA-mRNA network. Finally, the obtained hsa_circ_0086735-miR-1296-5p-STAT1 was proved to promote the progression of luminal breast cancer and tamoxifen resistance.
Growing evidence has validated the ceRNA function of circRNAs in cancers (Becatti et al., 2023). For instance, circularLRRC7 can exert as a potential glioblastoma suppressor by functioning as ceRNA for miR-1281 (Kong et al., 2021). In the current study, microarray datasets from the GEO database were downloaded and analyzed to identify robust differentially expressed circRNAs, miRNAs, and mRNAs that are specific to luminal-subtype breast cancer. Using the GEO2R method and other online analysis plates, our study systematically integrated multiple microarray datasets on breast cancer in GEO. The PPI network was constructed and used to screen hub genes. GO and KEGG enrichment analyses were conducted to annotate the functions of differentially expressed genes. The network was constructed, and the pathway network composed of hub genes was mapped. Based on the circRNA-miRNA-mRNA-pathway network, hsa_circ_0086735-miR-1296-5p-STAT1 axis was used for verification. The expression levels of hsa_circ_0086735, miR-1296-5p, and STAT1 mRNA were confirmed by qRT-PCR in luminal-subtype tissues and cell lines. The interactions among them were verified by Luciferase reporter assay and RNA pull-down assay.
A proven research result is that circRNA can influence cancer cell function. For example, the upregulation of circ_0008812 and circ_0001583 can contribute to the proliferation of breast cancer cells (Lin et al., 2022). Though hsa_circ_0086735 has few reports, hsa_circUBAP2 has been reported as an upregulated circRNA in breast cancer (Ma et al., 2021), just like the upregulated expression in luminal-subtype in this study. We also revealed the promoting role of hsa_circ_0086735 in breast cancer cell growth, which was in line with its role in glioma (Wang et al., 2021). The prognosis significance of hsa_circ_0086735 was verified in our 87 patients, which was mentioned in a previous meta-analysis (Ma et al., 2021). MiR-1296-5p has been reported to decrease ERBB2-positive breast cancer and exerts a tumor-suppressive function in breast cancer (Chen et al., 2017). The other known circRNAs that can target miR-1296-5p include hsa_circs_0000517, hsa_circ_0017639, and circ_0048764 (Zang et al., 2020; Chang et al., 2022). STAT1 was verified to accelerate breast cancer via deregulating homeostasis of the tumor microenvironment (Zellmer et al., 2017; Hou et al., 2018). Many cancers, including cancer, demonstrate. Inhibiting STAT1 activation, along with facilitating STAT3 activation, can aid the removal of breast malignant cells via EGFR moderation (Concha-Benavente et al., 2013). Therefore, the hsa_circ_0086735-miR-1296-5p-STAT1 axis can promote the cell proliferation and inhibited cell apoptosis of luminal-subtype breast cancer (Supplementary Figure S2).
CircRNAs can influence breast cancer cell response to endocrine therapies by mechanisms involving different estrogen signaling via modulating the expression of proteins acting as coregulators of estrogen signaling (Molibeli et al., 2021; Bahramy et al., 2023). Possible involvements of hsa_circ_0086735-miR-1296-5p-STAT1 axis in resistance of tamoxifen were investigated in this work. We revealed that hsa_circ_0086735 contributed to the resistance of tamoxifen. miR-1296-5p inhibitor has been discovered to restore the decreased cisplatin resistance in cisplatin-resistant non-small cell lung cancer cells (Chang et al., 2022). In this study, miR-1296-5p inhibitor restored the tamoxifen resistance in ZR-75-1 Tam1 cells. Depletion of STAT1 can decrease levels of ERα protein and cell proliferation in tamoxifen-resistant cell line LCC2 (Hou et al., 2018). A previous study has identified that suggest that increased STAT1 signaling is important in endocrine resistance and that STAT inhibitors may represent potential therapies in endocrine-resistant breast cancer (Huang et al., 2014; Chen et al., 2023). STAT1/c-Myc pathway is involved in POR-induced TAM-resistant breast cancer (Chen et al., 2023). The present study reveals a possible tamoxifen-resistant mechanism by which hsa_circ_0086735 may sponge miR-1296-5p, indirectly moderating STAT1 and contributing to tamoxifen resistance. Targeting of hsa_circ_0086735 may be a potential treatment strategy for tamoxifen-resistant luminal breast cancers.
A circRNA-miRNA-mRNA network was mapped in luminal breast cancer in this work. Hsa_circ_0086735-miR-1296-5p-STAT1 axis in the network can promote cancer progression and resistance to tamoxifen. Hsa_circ_0086735-miR-1296-5p-STAT1 axis may serve as therapeutic considerations to eradicate tamoxifen -resistance in breast cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the All women included in this study provided written informed consent. The study was approved by the Research Ethics Committee in Shanghai Tenth People's Hospital, Tongji University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author contributions
YH and MQ made substantial contributions to conception and design, performed all the experiments, and was a major contributor in writing the manuscript. JC, LC, and WJ contributed to acquisition of patients and tissues specimens, analysis and interpretation of data. GW has been involved in drafting the manuscript and revising it critically for important intellectual content.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1162259/full#supplementary-material
References
Adachi, Y., Ishiguro, J., Kotani, H., Hisada, T., Ichikawa, M., Gondo, N., et al. (2016). Comparison of clinical outcomes between luminal invasive ductal carcinoma and luminal invasive lobular carcinoma. BMC Cancer 16, 248. doi:10.1186/s12885-016-2275-4
Ashekyan, O., Abdallah, S., Shoukari, A. A., Chamandi, G., Choubassy, H., Itani, A. R. S., et al. (2022). Spotlight on exosomal non-coding RNAs in breast cancer: An in silico analysis to identify potential lncRNA/circRNA-miRNA-Target Axis. Int. J. Mol. Sci. 23, 8351. doi:10.3390/ijms23158351
Bahramy, A., Zafari, N., Rajabi, F., Aghakhani, A., Jayedi, A., Khaboushan, A. S., et al. (2023). Prognostic and diagnostic values of non-coding RNAs as biomarkers for breast cancer: An umbrella review and pan-cancer analysis. Front. Mol. Biosci. 10, 1096524. doi:10.3389/fmolb.2023.1096524
Becatti, M., Dmello, C., and Kumar, B. (2023). Editorial: Exploring circular RNAs and their applications within health and disease. Front. Mol. Biosci. 10, 1145738. doi:10.3389/fmolb.2023.1145738
Chang, F., Li, J., Sun, Q., Wei, S., and Song, Y. (2022). Hsa_circ_0017639 regulates cisplatin resistance and tumor growth via acting as a miR-1296-5p molecular sponge and modulating sine oculis homeobox 1 expression in non-small cell lung cancer. Bioengineered 13, 8806–8822. doi:10.1080/21655979.2022.2053810
Cheang, M. C., Chia, S. K., Voduc, D., Gao, D., Leung, S., Snider, J., et al. (2009). Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 101, 736–750. doi:10.1093/jnci/djp082
Chen, G., He, M., Yin, Y., Yan, T., Cheng, W., Huang, Z., et al. (2017). miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell. Int. 17, 95. doi:10.1186/s12935-017-0466-y
Chen, S., Wu, D., Liu, Q., Jin, F., Yao, F., and Fang, Y. (2023). POR overexpression induces tamoxifen-resistance in breast cancer through the STAT1/c-Myc pathway. Mol. Carcinog. 62, 249–260. doi:10.1002/mc.23481
Cohen, A. L., and Jia, S. (2014). Noncoding RNAs and the borders of heterochromatin. Wiley Interdiscip. Rev. RNA 5, 835–847. doi:10.1002/wrna.1249
Concha-Benavente, F., Srivastava, R. M., Ferrone, S., and Ferris, R. L. (2013). EGFR-mediated tumor immunoescape: The imbalance between phosphorylated STAT1 and phosphorylated STAT3. Oncoimmunology 2, e27215. doi:10.4161/onci.27215
Engstrøm, M. J., Opdahl, S., Hagen, A. I., Romundstad, P. R., Akslen, L. A., Haugen, O. A., et al. (2013). Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res. Treat. 140, 463–473. doi:10.1007/s10549-013-2647-2
Fontemaggi, G., Turco, C., Esposito, G., and Di Agostino, S. (2021). New molecular mechanisms and clinical impact of circRNAs in human cancer. Cancers (Basel) 13, 3154. doi:10.3390/cancers13133154
Ghazimoradi, M. H., and Babashah, S. (2022). The role of CircRNA/miRNA/mRNA axis in breast cancer drug resistance. Front. Oncol. 12, 966083. doi:10.3389/fonc.2022.966083
Hou, Y., Li, X., Li, Q., Xu, J., Yang, H., Xue, M., et al. (2018). STAT1 facilitates oestrogen receptor α transcription and stimulates breast cancer cell proliferation. J. Cell. Mol. Med. 22, 6077–6086. doi:10.1111/jcmm.13882
Huang, R., Faratian, D., Sims, A. H., Wilson, D., Thomas, J. S., Harrison, D. J., et al. (2014). Increased STAT1 signaling in endocrine-resistant breast cancer. PLoS One 9, e94226. doi:10.1371/journal.pone.0094226
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 19, 141–157. doi:10.1261/rna.035667.112
Katsura, C., Ogunmwonyi, I., Kankam, H. K., and Saha, S. (2022). Breast cancer: Presentation, investigation and management. Br. J. Hosp. Med. 83, 1–7. doi:10.12968/hmed.2021.0459
Kerr, A. J., Dodwell, D., Mcgale, P., Holt, F., Duane, F., Mannu, G., et al. (2022). Adjuvant and neoadjuvant breast cancer treatments: A systematic review of their effects on mortality. Cancer Treat. Rev. 105, 102375. doi:10.1016/j.ctrv.2022.102375
Khan, S., Jha, A., Panda, A. C., and Dixit, A. (2021). Cancer-associated circRNA-miRNA-mRNA regulatory networks: A meta-analysis. Front. Mol. Biosci. 8, 671309. doi:10.3389/fmolb.2021.671309
Kong, D., Yu, H., Sim, X., White, K., Tai, E. S., Wenk, M., et al. (2022). Multidisciplinary effort to drive precision-medicine for the future. Front. Digit. Health 4, 845405. doi:10.3389/fdgth.2022.845405
Kong, X., Xu, R., Wang, W., Zeng, M., Li, Y., Lin, M., et al. (2021). CircularLRRC7 is a potential tumor suppressor associated with miR-1281 and PDXP expression in glioblastoma. Front. Mol. Biosci. 8, 743417. doi:10.3389/fmolb.2021.743417
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Lin, H., Long, F., Zhang, X., Wang, P., and Wang, T. (2022). Upregulation of circ_0008812 and circ_0001583 predicts poor prognosis and promotes breast cancer proliferation. Front. Mol. Biosci. 9, 1017036. doi:10.3389/fmolb.2022.1017036
Loibl, S., Poortmans, P., Morrow, M., Denkert, C., and Curigliano, G. (2021). Breast cancer. Lancet 397, 1750–1769. doi:10.1016/S0140-6736(20)32381-3
Ma, Y., Niu, X., Yan, S., Liu, Y., Dong, R., and Li, Y. (2021). Circular RNA profiling facilitates the diagnosis and prognostic monitoring of breast cancer: A pair-wise meta-analysis. J. Clin. Lab. Anal. 35, e23575. doi:10.1002/jcla.23575
Mishra, A., Srivastava, A., Pateriya, A., Tomar, M. S., Mishra, A. K., and Shrivastava, A. (2021). Metabolic reprograming confers tamoxifen resistance in breast cancer. Chem. Biol. Interact. 347, 109602. doi:10.1016/j.cbi.2021.109602
Mo, M., Liu, B., Luo, Y., Tan, J. H. J., Zeng, X., Zeng, X., et al. (2022). Construction and comprehensive analysis of a circRNA-miRNA-mRNA regulatory network to reveal the pathogenesis of hepatocellular carcinoma. Front. Mol. Biosci. 9, 801478. doi:10.3389/fmolb.2022.801478
Molibeli, K. M., Hu, R., Liu, Y., Xiong, D., and Tang, L. (2021). Potential clinical applications of exosomal circular RNAs: More than diagnosis. Front. Mol. Biosci. 8, 769832. doi:10.3389/fmolb.2021.769832
Pang, Q., Lin, X., Sun, J., Hu, J., Dai, S., Shen, Y., et al. (2022). Comprehensive analysis of circular RNA expression in ceRNA networks and identification of the effects of hsa_circ_0006867 in keloid dermal fibroblasts. Front. Mol. Biosci. 9, 800122. doi:10.3389/fmolb.2022.800122
Pellegrino, B., Hlavata, Z., Migali, C., De Silva, P., Aiello, M., Willard-Gallo, K., et al. (2021). Luminal breast cancer: Risk of recurrence and tumor-associated immune suppression. Mol. Diagn Ther. 25, 409–424. doi:10.1007/s40291-021-00525-7
Rengganaten, V., Huang, C. J., Tsai, P. H., Wang, M. L., Yang, Y. P., Lan, Y. T., et al. (2020). Mapping a circular RNA-microRNA-mRNA-signaling regulatory Axis that modulates stemness properties of cancer stem cell populations in colorectal cancer spheroid cells. Int. J. Mol. Sci. 21, 7864. doi:10.3390/ijms21217864
Shen, T., Han, M., Wei, G., and Ni, T. (2015). An intriguing RNA species--perspectives of circularized RNA. Protein Cell. 6, 871–880. doi:10.1007/s13238-015-0202-0
Tang, X., Ren, H., Guo, M., Qian, J., Yang, Y., and Gu, C. (2021). Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 19, 910–928. doi:10.1016/j.csbj.2021.01.018
Treeck, O., Haerteis, S., and Ortmann, O. (2023). Non-coding RNAs modulating estrogen signaling and response to endocrine therapy in breast cancer. Cancers (Basel) 15, 1632. doi:10.3390/cancers15061632
Tsang, J. Y. S., and Tse, G. M. (2020). Molecular classification of breast cancer. Adv. Anat. Pathol. 27, 27–35. doi:10.1097/PAP.0000000000000232
Wang, J., Li, T., and Wang, B. (2021). Circ-UBAP2 functions as sponges of miR-1205 and miR-382 to promote glioma progression by modulating STC1 expression. Cancer Med. 10, 1815–1828. doi:10.1002/cam4.3759
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65, 625–639. doi:10.1042/EBC20200032
Yu, C. Y., and Kuo, H. C. (2019). The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 26, 29. doi:10.1186/s12929-019-0523-z
Zang, H., Li, Y., Zhang, X., and Huang, G. (2020). Circ_0000517 contributes to hepatocellular carcinoma progression by upregulating TXNDC5 via sponging miR-1296-5p. Cancer Manag. Res. 12, 3457–3468. doi:10.2147/CMAR.S244024
Zellmer, V. R., Schnepp, P. M., Fracci, S. L., Tan, X., Howe, E. N., and Zhang, S. (2017). Tumor-induced stromal STAT1 accelerates breast cancer via deregulating tissue homeostasis. Mol. Cancer Res. 15, 585–597. doi:10.1158/1541-7786.MCR-16-0312
Zhang, M., Bai, X., Zeng, X., Liu, J., Liu, F., and Zhang, Z. (2021). Diffuse painful erythema on right thigh. Clin. Chim. Acta 523, 120–123. doi:10.1016/j.jdcr.2021.06.032
Keywords: network, luminal breast cancer, circRNA, tamoxifen resistance, hsa_circ_0086735-miR-1296-5p-STAT1
Citation: Huang Y, Qian M, Chu J, Chen L, Jian W and Wang G (2023) Identification of circRNA-miRNA-mRNA network in luminal breast cancers by integrated analysis of microarray datasets. Front. Mol. Biosci. 10:1162259. doi: 10.3389/fmolb.2023.1162259
Received: 09 February 2023; Accepted: 17 April 2023;
Published: 28 April 2023.
Edited by:
Arun Malhotra, University of Miami, United StatesReviewed by:
Snehal Dinkar Nirgude, Children’s Hospital of Philadelphia, United StatesYutian Zou, Sun Yat-sen University Cancer Center (SYSUCC), China
Copyright © 2023 Huang, Qian, Chu, Chen, Jian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Wang, d2FuZ2dhbmcxOTgzY25AMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yixiang Huang
Yixiang Huang Mingping Qian†
Mingping Qian† Gang Wang
Gang Wang