- 1School of Biosciences, Division of Natural Sciences, University of Kent, Canterbury, United Kingdom
- 2MRC Toxicology Unit, Gleeson Building, University of Cambridge, Cambridge, United Kingdom
- 3Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom
In vitro transcribed, modified messenger RNAs (IVTmRNAs) have been used to vaccinate billions of individuals against the SARS-CoV-2 virus, and are currently being developed for many additional therapeutic applications. IVTmRNAs must be translated into proteins with therapeutic activity by the same cellular machinery that also translates native endogenous transcripts. However, different genesis pathways and routes of entry into target cells as well as the presence of modified nucleotides mean that the way in which IVTmRNAs engage with the translational machinery, and the efficiency with which they are being translated, differs from native mRNAs. This review summarises our current knowledge of commonalities and differences in translation between IVTmRNAs and cellular mRNAs, which is key for the development of future design strategies that can generate IVTmRNAs with improved activity in therapeutic applications.
1 Introduction
The notion that in vitro transcribed modified mRNAs (IVTmRNAs) could be used for therapeutic purposes originates from experiments conducted in the 1990s. These early experiments demonstrated that the injection of pure, in vitro transcribed mRNA into mouse muscle resulted in the detectable production of the encoded protein (Wolff et al., 1990). Early delivery mechanisms utilised liposomes (Zhang et al., 2022) which were subsequently superseded by the development of lipid nanoparticle (LNP) formulations which enhanced tissue uptake of the RNAs while at the same time protecting them efficiently from nuclease digest (Hou et al., 2021). Naked (non-protein bound) IVTmRNAs are prone to elicit immune responses, which could be controlled through the development of chemical modifications that suppress the immunogenicity of the RNA molecule (Karikó et al., 2005). These were key developments in the journey into applications as vaccines, which ultimately led to the rapid development of the COVID-19 vaccines, many other vaccines currently in clinical development, and a growing array of additional applications beyond vaccines (Damase et al., 2021; Zhang et al., 2022).
IVTmRNA therapeutics are translated into proteins by the cellular protein synthesis machinery. It may be assumed that the translation of such RNAs uses the same or similar pathways as natural endogenous in vivo transcribed RNAs. While this is likely true in the most general terms, there is considerable scope for variation in how the cell translates such RNAs. This topic is little discussed, and variations in the process of protein synthesis between natural endogenous transcripts and IVTmRNAs are in general not well understood. Patents on the existing COVID-19 vaccines as well as the documented thought processes that led the to the approval of IVTmRNAs for use in the clinic1 strongly focus on formulation and on the immune responses elicited by both the RNA and the encoded proteins, whereas little attention is paid to the actual processes by which the IVTmRNAs are translated.
At the time of writing, billions of IVTmRNA doses have been administered. Given the widespread use, a better understanding of the mechanisms of action of coding therapeutic RNAs, including the process of translation into protein, is key. Moreover, current sequence design approaches are based on those commonly used for recombinant DNA constructs (see below), and better understanding of IVTmRNA-specific translation mechanisms is required to generate more efficient sequence design approaches. With these aims in mind, this review summarises the current literature and understanding of the mechanism of protein synthesis from IVTmRNAs and its relation to protein synthesis on natural transcripts.
2 Cytoplasmic entry and engagement with the translational machinery of mRNAs
All eukaryotic mRNAs, whether mature endogenous transcripts or IVTmRNAs, consist of a number of non-coding elements alongside the protein coding open reading frame (ORF, Figure 1). The non-coding regions usually include a 5′ 7-methyl-GTP cap and a 5′ untranslated region (UTR) of variable length in front of the ORF, and a 3’ UTR (again of variable length) following the ORF. Transcripts end in a poly(A) tail structure, which have typical initial lengths of 200–250 nucleotides in humans (Eckmann et al., 2011) but are typically 130 nucleotides in IVTmRNAs if they are encoded on the in vitro transcription template. All these UTR elements play key roles in recruiting ribosomes and translation factors to the RNA.
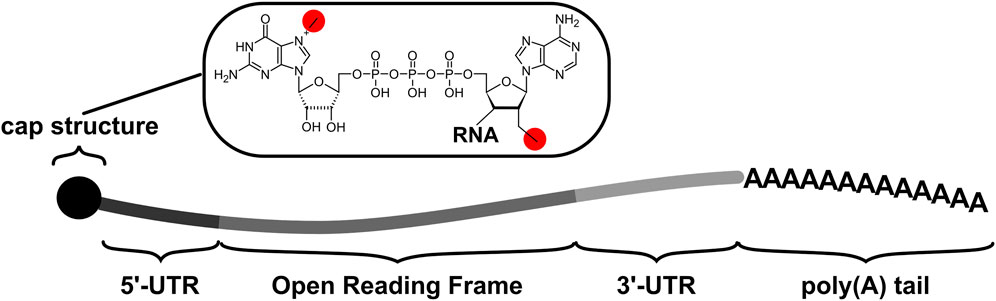
FIGURE 1. The principal components of native transcripts as well as IVTmRNAs comprise untranslated regions (UTRs) which flank the protein-coding Open Reading Frame or ORF. Cap structures are introduced co-transcriptionally in vivo and may be introduced co- or post-transcriptionally in vitro. The poly(A) tail is added post-transcriptionally in vivo, and may be added as part of the transcribed sequence or post-transcriptionally in vitro. The chemical detail illustrates the cap1 structure which is characterized by the two methyl group highlighted in red (alternative cap structures differ in the number and locations of the methyl groups).
Whilst such elements are common between IVTmRNAs and endogenous mRNAs, their effect on translation can differ between natural transcripts and IVTmRNAs due to their differing routes of entry into the cytoplasm and differing modes of engagement with the translational machinery (Figure 2). A particular hallmark of the natural pathway is that the processes of mRNA maturation and export deposit combinations of protein-based markers and RNA nucleotide modifications that record the transcripts’ genesis pathway. Such protein and RNA “marks” have the potential to affect translation in multiple ways. These marks are entirely absent from IVTmRNAs when they first engage with the translational apparatus, which can thus be described as a “naked” mRNA.
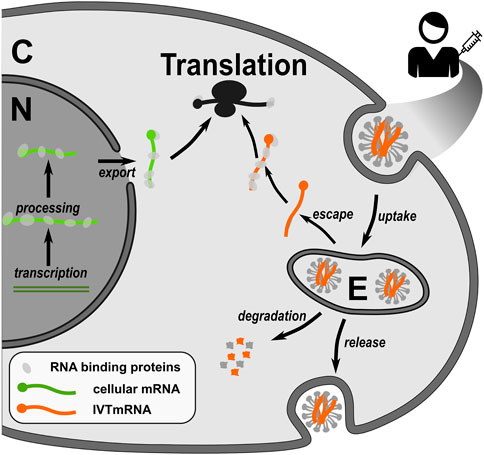
FIGURE 2. Native endogenous transcripts and IVTmRNAs have different genesis pathways which determine differences in translational efficiency. Native transcripts engage with the translational machinery following nuclear processing and export, in a protein-bound mRNP state where mRNP composition is controlled by the preceding processing steps. IVTmRNAs enter cells through the endosomal pathway, from which they escape inefficiently. In how far IVTmRNAs are able to form functional mRNPs is unknown. C, cytoplasm, N, nucleus, E, endosome.
2.1 The native RNA pathway
Native mRNAs engage the translational machinery following nuclear transcription, processing and export to the cytoplasm. During this journey, transcripts do not exist as naked RNAs, instead, they associate with RNA binding proteins to form messenger Ribonucleoproteins (mRNPs). According to the current view in the field, mRNPs change composition as they progress through the different steps of a transcript’s life cycle, in a manner that involves coordinated handover events between the different protein complexes (Pichon et al., 2012). An important aspect of this concept is that the distinct protein components of the mRNP record the life history of a transcript, allowing events early on in a transcript’s history to affect stability, localisation, and translational activity later on. Relevant concepts have been reviewed in depth (Singh et al., 2015; Björk and Wieslander, 2017; Gehring et al., 2017; Khong and Parker, 2020; Zarnack et al., 2020). Here, we focus on steps leading to the first engagement with the translational machinery which are most distinct for endogenous native mRNAs compared to IVTmRNAs.
Hallmarks of nuclear mRNPs following transcription, splicing and export include a dimeric nuclear cap-binding complex (CBC) on the mRNA cap structure. On spliced mRNAs, an additional hallmark is the presence of exon junction complexes (EJCs) which are deposited over exon:exon junctions during splicing and which signal the location of such sites to the translational machinery and the cellular mRNA surveillance pathways. The first or “pioneer” round of translation is thought to occur on mRNPs in this state (Ishigaki et al., 2001). During the pioneer round EJCs are removed from the transcript by the translating ribosome. Because on native mRNAs introns are usually restricted to 5′-UTRs and the open reading frame, but are generally excluded from 3′-UTRs, ribosomes normally terminate on transcripts where all EJCs have been removed. Termination upstream of a remaining EJC thus flags the presence of a premature termination codon and elicits Non-sense Mediated Decay (NMD), one of the cellular mRNA surveillance pathways. Connections between the pioneer round of translation and other quality control pathways have been demonstrated but are less well understood than for NMD. For example, the CBC was recently reported to interact with the RNA component of the signal recognition particle, thereby supporting appropriate targeting of signal sequence-containing proteins to the ER (Park et al., 2021). CBC-dependent translation was also reported to support some forms of antigen presentation, for example on MHC-I (Weinstein-Marom et al., 2019) and on class I HLA (Uchihara et al., 2022).
Over the lifetime of natural transcripts, association between the cap-structure and different cap-binding complexes and enzymes is one of the important signals that accompany entry into and exit from the translationally active state (Figure 3). Following the pioneer round of translation, CBC is replaced by the cytoplasmic cap-binding protein or eukaryotic initiation factor (eIF) 4E, which is part of the eIF4F complex that additionally contains eIF4A, eIF4G, and the poly(A) tail binding protein PAB. This complex forms a central hub for active translation and ribosome recruitment, via interactions with additional translation initiation factors. The active translational state can be interrupted by several eIF4E binding proteins which disrupt eIF4E:eIF4G contacts (Lin et al., 1994; Clemens, 2001), and translational activity can be modulated by the formation of complexes containing different eIF4E and eIF4G isoforms (Landon et al., 2014; Ho et al., 2016). Following a period of translation which coincides with gradual shortening of the poly(A) tail, eIF4E is eventually replaced by decapping enzyme-containing complexes which initiate the mRNA decay process (Fenger- Grøn et al.,. 2005). The CBC-eIF4E handover has been reported to be catalysed by components of the nuclear export machinery (Sato and Maquat, 2009). Moreover, there is an emerging role for RGG-motif containing proteins, which also begin to interact with mRNPs around the time of mRNA export (Poornima et al., 2021), in both CBC-eIF4E and eIF4E-decapping complex handover events (Chowdhury and Jin, 2022).
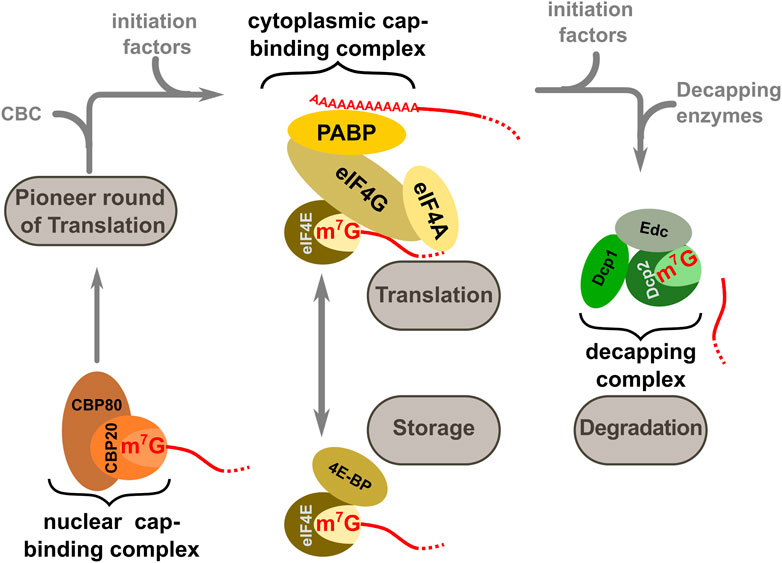
FIGURE 3. Different cap-binding complexes mediate entry into and exit from the translational state of mRNAs in the cell. The nuclear cap binding complex CBC (consisting of CBP20 and CBP80) is present during nuclear processing events. Following export, this complex is exchanged against a cytoplasmic cap-binding complex comprising members of the eIF4 group of translation factors and the poly(A) binding protein PAB (this complex is collectively termed eIF4F). The productive eIF4F complex can be temporarily disrupted by the 4E binding proteins or 4E-BPs and in this inactive form transcripts may be stored in specific cellular structures such a P-bodies. At the end of the translationally active phase the eIF4F complex is disassembled and replaced by the decapping factors.
2.2 The IVTmRNA pathway
All RNA therapeutics currently used in the clinic are transcribed in vitro from tilizing DNA templates in which the 5′-UTR, ORF, 3′-UTR and poly(A) tail are placed downstream of a T7 promoter sequence (Whitley et al., 2022). A number of strategies are available for introducing the 5′-cap structure and poly(A) tail. For capping, post-transcriptional enzymatic strategies are available (usually based on the capping enzymes from vaccinia virus), as well as non-enzymatic co-transcriptional strategies (Muttach et al., 2017). Current vaccines predominantly use the co-transcriptional route to introduce double methylated “cap1” structures (Figure 1), and achieve capping efficiencies of 90%–95%. For polyadenylation, a poly(A) tail can be encoded on the transcription template but poly(A) tail length is then restricted to around 130 nucleotides due to DNA synthesis constraints. A strategy based on “segmented” poly A tails has been proposed in order to avoid problems with synthesis and stability of template-encoded pure poly(A) tails (Trepotec et al., 2019). Alternatively, poly(A) tails can be introduced enzymatically following in vitro transcription which allows generation of longer tails, although this introduces tail length heterogeneity.
Transcribed and capped RNA is purified away from the DNA template and reaction impurities, before being encapsulated in lipid nanoparticles (LNPs, Mukai et al., 2022). LNPs typically consist of mixtures of ionizable lipids, modified polyethylene glycol moieties, and varying helper lipids that form micellar structures around a mixture of the nucleic acid cargo and cholesterol. The different lipids each have defined roles, and LNP composition changes in controlled ways upon injection into patient tissues: for example, the PEG derivatives stabilize LNPs in blood, but need to be shed with controlled rates to enable efficient cellular uptake, whereas the ionizable lipids attract varying surface charges during passage through the endosome thereby promoting endosomal escape at specific times following endocytosis. It has been noted that significant amounts of impurities in the form of truncated IVTmRNAs can still be present in these purified formulations1, the impact of which is currently poorly understood.
The fate of individual lipid nanoparticles and their RNA cargo has been determined using a mixture of approaches, relying primarily on cultured cells and animal models. Transfection efficiency varies between cultured cells and intact tissues (Paunovska et al., 2018) and between the same tissue in different animal species (Hatit et al., 2022). Results obtained with cultured cells may thus not be directly transferable to events in tissues of vaccine recipients. However, data from these studies have revealed the principal mechanisms by which IVTmRNAs enter cells and engage with the translational machinery.
The local LNP concentration around the injection site stabilises within the first few hours post-injection (Hassett et al., 2019), during which time the LNPs diffuse into the local muscle and drain into adjacent lymph nodes (Lindsay et al., 2019). A small proportion of LNPs also spreads systemically and reaches organs such as the liver and spleen, where the LNP concentration peaks after about 8 h (Hassett et al., 2019). Individual cells within the injected tissues take up LNPs through clathrin- and alveolae-mediated endocytosis (Rejman et al., 2005). Although most relevant work has been done on muscle tissue, the same pathways also mediate non-intramuscular delivery such as aerosol delivery to the lungs (Li et al., 2020) and delivery to the eye (Patel et al., 2019).
The efficiency by which individual cells take up LNPs can differ with cell type as well as transcriptional state of the cells (Dobrowolski et al., 2022). The primary destination of injected nanoparticles appear to be muscle cells, various types of immune cells which infiltrate the injection site, and monocytes and other antigen presenting cells in the draining lymph nodes (Liang et al., 2017; Lindsay et al., 2019). Not all cells that take up LNPs also efficiently translate their mRNA cargo, for example, monocytes and dendritic cells produce protein encoded on LNP-delivered transcripts more efficiently than neutrophils despite similar uptake rates (Liang et al., 2017).
Following endocytosis, LNPs and their RNA cargo are routed through the various stages of the endosomal machinery. IVTmRNAs need to escape from the endosome into the cytoplasm to be translated, and this is thought to be one of the most limiting steps of IVTmRNA vaccination strategies (Jiang et al., 2020). In a detailed study of endosomal escape of siRNA-loaded LNPs, escape of endocytosed LNPs was observed within a narrow window around the time of the Rab5-Rab7 conversion step of endosome maturation, 5–15 min after the initiation of endocytic uptake (Wittrup et al., 2015). Endosomal escape strategies are engineered by manipulating LNP composition, for example by including lipids with pKas that interact with the changing pH observed during maturation of endosomal compartments (Hajj et al., 2019; Delehedde et al., 2021). Despite these engineering strategies, typical endosomal escape rates are very low (Deprey et al., 2020; Delehedde et al., 2021), with the great majority of LNPs and their cargo either being degraded, or routed into vesicles which may lead to their re-release into the extracellular medium and subsequent uptake by other cells (Maugeri et al., 2019). Cytoplasmic entry is clearly a limitation in utilising endosomal escape as a mechanism for delivery of IVTmRNAs and almost certainly means much higher amounts of IVTmRNA are required to be administered than if escape were more efficient or alternative routes could be harnessed.
How IVTmRNAs engage with the translational machinery post-endosomal escape is not known in detail, although indirect evidence suggests some possible scenarios. Upon endosomal escape into the cytosol, IVTmRNAs are initially non-protein bound or “naked,” although they may remain associated with some of the LNP components. Cells contain hundreds of unspecific RNA binding proteins (Baltz et al., 2012; Castello et al., 2012), and the crowded intracellular environment (Zimmerman and Minton, 1993) likely drives rapid association of the IVTmRNA with such proteins.
The efficient translation of transfected IVTmRNAs depends on cis-acting sequences, as is the case for cellular transcripts, including 5′- and 3′-UTRs, cap-structure and poly(A) tail (Malone et al., 1989; Jani and Fuchs, 2012). In current IVTmRNA-based therapeutics, the protein-coding ORFs are typically combined with untranslated regions (UTRs) from efficiently translated cellular mRNAs, or with non-natural UTR sequences that have been selected for high translational efficiency (Cao et al., 2021; Fang et al., 2022). The sensitivity of unmodified transcripts to Protein Kinase R (PKR, which is activated by double and single stranded RNA) further suggests that requirements for eIF2 activity (which is the target of PKR) are similar to natural transcripts (Anderson et al., 2010; Cesaro and Michiels, 2021), and the overall time between the introduction of IVTmRNAs into the cell and the emergence of their translation products is similar to transcription-translation delays observed in vivo (Reiser et al., 2019). Despite their different route of entry into the cytoplasm, IVTmRNAs are thus most likely translated via similar mechanisms as natural cellular transcripts, albeit less efficiently as outlined below.
2.3 Consequences of differing entry routes
One of the fundamental differences between natural transcripts and IVTmRNAs is the fact that the latter do not pass through the natural mRNP states that precede engagement of the translational machinery. Any of the processes that have been identified as functionally dependent on the pioneer round of translation, including secretion and antigen presentation, may thus be less efficient for IVTmRNAs.
The synergistic effect from the presence of both a cap structure and the poly(A) tail in IVTmRNAs (Mockey et al., 2006) is similar to that observed in other contexts, where the synergy is known to depend on assembly of a functional cap-binding complex (Borman, 2000; von der Haar et al., 2004). This indicates that IVTmRNAs also require full assembly of this complex. However, unlike for natural transcripts where association with the cytoplasmic cap-binding protein eIF4E has been reported to be catalysed by components of the perinuclear mRNP, IVTmRNAs must somehow associate with non-cap-bound eIF4E encountered in the cytoplasm. Estimates for abundance of eIF4E in human cells vary, but estimates at the lower end (Duncan et al., 1987) are similar to estimates for the total capped transcript content (Marinov et al., 2014), which indicates that available eIF4E may be limiting in actively translating cells. Association between IVTmRNAs and eIF4E may thus limit translational efficiency of the latter. Interestingly, IVTmRNAs were observed to be most efficiently translated when localised near the nucleus (Sayers et al., 2019), the same site that is also the location of first engagement with the translational machinery for most natural transcripts. However, it remains unclear how far perinuclear IVTmRNAs engage with mRNP components that precede the translationally active state.
Multiple lines of evidence indicate that introduction of IVTmRNA-containing LNPs into cells is perceived as a stress that activates various translational control mechanisms. The interaction of nanoparticles with cells can be a cause of stress in itself (Cameron et al., 2022), although the LNPs used for IVTmRNA delivery have been designed to avoid toxic effects (Delehedde et al., 2021). Another cause of cell stress associated with LNP-mediated IVTmRNA delivery are the endosomal rupture events that release LNP cargo into the cytosol, which can result in inflammasome activation, induction of cell death (pyroptosis) of the affected cell, and inflammation in the surrounding tissue (Forster et al., 2022).
In addition to LNP-induced cell stress, the IVTmRNA cargo itself can induce stress pathways. Naked RNA can act as seed material that directly promotes stress granule formation (Bounedjah et al., 2014). Cells transfected with IVTmRNAs frequently show strong activation of PKR (Anderson et al., 2010) which phosphorylates eIF2, thereby ablating global translational activity in transfected cells (Lee et al., 1993). The PKR-dependent loss of translational activity in transfected cells upon introduction of IVTmRNAs can be partially controlled by co-application of inhibitors of the integrated stress response like ISRIB (Ohto et al., 2019) or by using modified uridines in the in vitro transcription process (Anderson et al., 2010; Kirschman et al., 2017). The use of modified nucleotides is discussed in more detail in the following section.
3 Nucleotide modifications and translational efficiency
Transfection of IVTmRNA frequently results in activation of PKR, which reduces the overall translational efficiency of the transfected cell (see above). Moreover, in vitro transcribed RNA stimulates various innate immune receptors, particularly endosomal and cytosolic pattern recognition receptors (PRRs) such as toll-like receptors 7, 8, and 3 that sense single and double stranded RNA, thereby generating its own immune response (Mu and Hur, 2021). Both are undesired activities in the context of RNA therapeutics, and both have been attributed to a self-priming activity associated with the T7 polymerase commonly used for in vitro transcription which results in the aberrant production of double stranded RNA (Mu et al., 2018). Although the dsRNA-dependent stimulation of immune receptors appears to be the main component of immune responses against IVTmRNAs, single stranded RNA has also been reported to stimulate immune responses in a RIG-I dependent manner (Pichlmair et al., 2006).
The activation of both PKR and innate immune response pathways can be suppressed by introducing various modified nucleotides into the in vitro transcribed RNA (Karikó et al., 2005; Anderson et al., 2010; Durbin et al., 2016). Most current studies agree that the incorporation of such modified nucleotides is necessary to avoid undesired PKR stimulation and immunogenicity, although it is worth noting that some studies reported differing results for particular sequences (Thess et al., 2015; Kauffman et al., 2016). Despite ongoing debate concerning these results, the most commonly administered current IVTmRNA vaccines are transcribed using N1-methyl-pseudouridine instead of uridine (Morais et al., 2021).
Pseudouridine (Ψ) is the most common natural base modification in RNAs, including mRNAs (Schwartz et al., 2014). Although in Ψ the base moiety is rotated relative to the ribose when compared to normal uridine, the orientation of the main hydrogen bond donor and acceptor atoms on the Watson-Crick face are identical in the two bases (Figure 4). Consistently, Ψ can be found base-pairing with adenine in a fashion similar to unmodified uridine in experimental RNA structures (Chawla et al., 2015), as well as forming wobble base pairs with uridine and guanine, again similar to unmodified uridine. Thus, from the point of view of codon decoding and the interaction between Ψ containing codons and their base-pairing anticodons, Ψ is expected to behave similar to uridine.
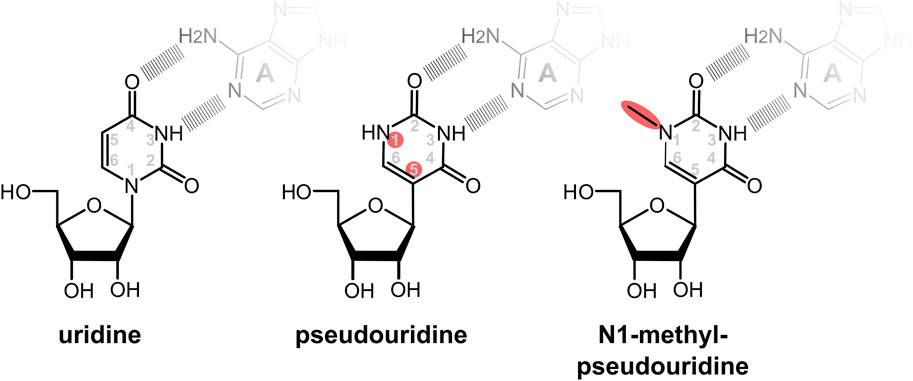
FIGURE 4. Modified nucleotides used to ablate stimulation of immune responses and PKR activity by IVTmRNAs. In pseudouridine the uracil base is rotated so that the glycosidic bond between the sugar and base is transferred from the N1 to the C5 atom of the base. In the commonly used base N1-methyl-pseudouridine, the N1 atom is attached to an additional methyl group. Despite the rotation of the base, the potential to form base pairing patterns via the Watson-Crick face of the uridine is maintained in the modified bases. Sites of changes relative to native uridine are highlighted in red.
Despite the similar geometry of their base-pairing hydrogen bonds, experimental studies have shown that U and Ψ form base pairs with other bases with different affinities. In addition to stabilising the canonical base-pair with adenine, Ψ also stabilises wobble-base pairs with guanine and uridine, but not with cytidine (Kierzek et al., 2014). These effects were attributed to changes in the ability of the base-pairs to stack with neighbouring bases, and in the case of the Ψ:A base pair also to a potential additional hydrogen bond involving the N1 atom which is not available for hydrogen bonding in unmodified uridine (Hudson et al., 2013; Kierzek et al., 2014).
Following initial work with pseudouridine, it was found that methylation of its N1 atom (Figure 3) further enhanced the ability of Ψ to suppress activation of immune receptors and the global translation inhibition through activation of PKR (Andries et al., 2015). Presumably for these reasons the predominant current COVID-19 vaccines contain N1-methyl-pseudouridine (m1Ψ) in place of uridine.
It is difficult to compare the translational activity of uridine-, Ψ- and m1Ψ-containing IVTmRNAs directly because effects of the modifications on the transcript itself need to be disentangled from the PKR-dependent global regulation of translation. Using a massively parallel survey of 5′-UTRs, Sample et al. (2019) showed that secondary structures within the UTRs of Ψ and m1Ψ-modified transcripts more strongly inhibit ribosome recruitment compared to natural uridine-containing ones. Svitkin et al. (2017) showed that modified IVTmRNAs also show evidence of stronger ribosomal pausing and decreased translation elongation rates. Thus, on both counts the local effect of the modifications on the transcript is actually inhibitory, and the increased protein yields observed with modified transcripts are most likely due to the overriding effect of alleviating the global inhibition of translation observed with unmodified transcripts.
The inhibitory effect of Ψ and m1Ψ in both UTRs and ORFs may be in part attributable to the effect of the modifications on the stability of secondary structure elements, although increased stability could also result in a prolonged lifetime and therefore prolonged expression of the target antigen. A direct inhibitory effect of Ψ on codon decoding was confirmed in vitro with bacterial ribosomes (Eyler et al., 2019). In this study isolated Phe-tRNAPheGAA was observed to show two-fold slower GTPase activation on individual UUU codons in which one or more positions were replaced with Ψ. In crystal structures of E. coli ribosomes containing this tRNA and modified codons, placement of the tRNA acceptor stem in the peptidyl transferase centre (PTC) was impaired, indicating that communication between the decoding centre and the PTC is less efficient on Ψ containing codons.
In decoding systems in vivo, the overall time required to decode a codon is dependent on the number and type of non-cognate tRNAs that need to be rejected before a cognate tRNA is accepted (Chu et al., 2011; Tarrant and von der Haar, 2014). A particular problem in this respect are near-cognate tRNAs, which can form partial mini-helices between the codon and anti-codon (Plant et al., 2007). Near-cognate tRNAs take much longer to reject than other non-cognates, and carry a non-negligible risk of misincorporation (Plant et al., 2007; Tarrant and von der Haar, 2014). Since Ψ:G and Ψ:U interactions appear to be stabilised compared to U:G and U:U (Kierzek et al., 2014) and both types of interaction occur in some near-cognate tRNAs, Ψ-containing codons can be expected to be more prone to interference from near- and non-cognate tRNAs. Consistent with this expectation, translation elongation rates on fully Ψ-modified transcripts in reconstituted bacterial translation systems were reduced 3-fold and thus greater than expected if the reduced GTPase activation had been the sole contributing factor. Moreover, increased amino acid misincorporation was observed specifically on Ψ-containing codons in bacterial and cultured human cells (Eyler et al., 2019), Ψ incorporation into stop codons was observed to cause increased non-sense suppression (Karijolich and Yu, 2011), and a recent study provided direct evidence for changes in near-cognate tRNA interactions on m1Ψ-modified codons (Monroe et al., 2022). Together these results suggest that modification of transcripts with Ψ or m1Ψ alters competition from near-cognate tRNAs, thereby leading to general ribosome slow-down as well as reduced translational accuracy.
The effects resulting from intended uridine modifications may overlap with effects caused by inadvertent modifications that can occur in IVTmRNAs, notably those arising from the oxidation of nucleotides in RNA. Oxidatively damaged nucleotides in RNA can lead to amino acid misincorporation (Tanaka et al., 2007) and ribosome stalling (Shan et al., 2007). To our knowledge no studies have yet directly assessed levels of oxidative nucleotide damage in RNA vaccines, but oxidation of LNP components such as cholesterol has been shown to occur and may be exacerbated by the presence of impurities in the PEG lipid used in the production of LNP formulations (Uddin et al., 2021).
One consequence of reduced ribosome speed can be ribosome collisions, which cells can perceive as signalling events that elicit a variety of cellular responses. Ribosome collisions can activate Gcn2, p38 and JNK kinases and reduce global translational activity in the affected cell (Wu et al., 2020; Snieckute et al., 2022). At the same time, colliding ribosomes can activate the E3 ubiquitin ligases of the Ribosomal Quality Control pathway (Juszkiewicz et al., 2020) and the ribosomal RNAse activity which is part of the No-Go decay pathway (Doma and Parker, 2006). These surveillance activities jointly promote the degradation of the stalled ribosome:RNA:nascent peptide complex. Svitkin et al. observed that membrane association could relieve m1Ψ-dependent ribosome collisions in vitro (Svitkin et al., 2022), potentially as a consequence of the altered translational dynamics of membrane associated ribosomes. In sum, whether ribosome collisions occur on modified or unmodified therapeutic mRNAs in vivo is unknown, although if collisions do occur they are likely to reduce the efficiency with which such transcripts are translated.
How much protein an mRNA can produce can be limited either by the translation initiation rate or by the translation elongation rate, depending on the ratio of the two activities (Chu et al., 2014; Tarrant and von der Haar, 2014). While the majority of cellular mRNAs is likely limited by initiation rates, regulatory regions of many recombinant protein encoding constructs are derived from highly translated natural transcripts and such constructs are therefore frequently limited by their elongation rates unless the codon usage of the transcript has been specifically optimised (Chu et al., 2014). If IVTmRNAs containing modified uridines are decoded more slowly than natural transcripts, elongation rates are likely more limiting, potentially increasing the importance of codon usage optimisation for efficient protein expression. The RNA vaccines currently in clinical use show clear signs of codon optimisation as they avoid the use of codons that are infrequently used in humans (Figure 4). Although some analyses of the codon usage in these vaccines have been published (Xia 2021) the exact optimisation algorithms used to generate these sequences have not been disclosed.
Different studies have emphasized various design principles for nucleic acid-based vaccines. These include copying codon usage from highly expressed mammalian genes, which improves translational dynamics and also increases the GC content of the ORF (André et al., 1998); more fine-grained design considerations that allow controlling protein folding and posttranslational modifications which may be adversely affected by “over-optimising” codon usage (Katneni et al., 2022), strategies that avoid PKR activation and immune-stimulation without the use of modified nucleotides (Thess et al., 2015), and strategies that emphasize the control of a transcript’s half-life by controlling secondary structure motifs (Geisberg et al., 2014). While the currently used vaccines were clearly designed with some reference to human codon usage frequencies this has not been implemented rigorously, with both the Pfizer and Moderna vaccines containing some sub-optimal codons for most amino acids (Figure 5). These non-optimal codon choices may have been introduced due to functional considerations such as those outlined above, although to or knowledge information on what such considerations may have entailed has not been made public.

FIGURE 5. Codon usage for sense codons in two clinically used SARS-CoV-2 vaccines, compared to the average codon usage in open reading frames in the human genome (top) and to codon usage in a representative native spike protein encoding sequence from SARS-CoV-2 (bottom, from Genbank ID MN908947).
4 Summary
Although IVTmRNAs are translated by the cellular gene expression machinery, their genesis and route of entry into the cell are fundamentally different from native transcripts. In consequence the location of their first engagement with the translational machinery, their arrival as a non-protein bound, “naked” RNA molecule rather than a pre-formed mRNP, and the presence of modified nucleotides in high densities all have consequences for the efficiency with which IVTmRNAs are translated. Understanding these differences in better detail is required if protein synthesis rates of therapeutic mRNAs are to be improved.
Although therapeutics currently in clinical use are clearly efficient enough to form effective vaccines, the expression levels required to stimulate immune responses are relatively low. The various modes for non-vaccine therapeutics that are currently being explored will require higher expression levels, and for such applications removing the translational limitations of current constructs is of key importance.
Author contributions
All authors contributed to writing of the manuscript and proofread the final version. TvdH and TM designed figures.
Funding
The authors acknowledge funding from the Wellcome Leap R3 program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1See for example the European Medicines Agency assessment reports for Comirnaty (https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-public-assessment-report_en.pdf) and for Spikevax (https://www.ema.europa.eu/en/documents/assessment-report/spikevax-previously-covid-19-vaccine-moderna-epar-public-assessment-report_en.pdf).
References
Anderson, B. R., Muramatsu, H., Nallagatla, S. R., Bevilacqua, P. C., Sansing, L. H., Weissman, D., et al. (2010). Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892. doi:10.1093/nar/gkq347
André, S., Seed, B., Eberle, J., Schraut, W., Bültmann, A., and Haas, J. (1998). Increased immune response elicited by DNA vaccination with a synthetic Gp120 Sequence with optimized codon Usage. Journal of Virology 72, 1497–1503. doi:10.1128/JVI.72.2.1497-1503.1998
Andries, O., Mc Cafferty, S., De Smedt, S. C., Weiss, R., Sanders, N. N., and Kitada, T. (2015). N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344. doi:10.1016/j.jconrel.2015.08.051
Baltz, A. G., Munschauer, M., Schwanhäusser, B., Vasile, A., Murakawa, A., Schueler, M., et al. (2012). The MRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690. doi:10.1016/j.molcel.2012.05.021
Björk, P., and Wieslander, L. (2017). Integration of mRNP formation and export. Cell. Mol. Life Sci. 74, 2875–2897. doi:10.1007/s00018-017-2503-3
Borman, A. M., Michel, Y. M., and Kean, K. M. (2000). Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: The eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5’-end. Nucleic Acids Res. 28, 4068–4075. doi:10.1093/nar/28.21.4068
Bounedjah, O., Desforges, B., Wu, T.-D., Pioche-Durieu, C., Marco, S., Hamon, L., et al. (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691. doi:10.1093/nar/gku582
Cameron, S. J., Sheng, J., Hosseinian, F., and Willmore, W. G. (2022). Nanoparticle effects on stress response pathways and nanoparticle–protein interactions. IJMS 23, 7962. doi:10.3390/ijms23147962
Cao, J., Novoa, E. M., Zhang, Z., Chen, W. C. W., Liu, D., Choi, G. C. G., et al. (2021). High-throughput 5′ UTR engineering for enhanced protein production in non-viral gene therapies. Nat. Commun. 12, 4138. doi:10.1038/s41467-021-24436-7
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., et al. (2012). Insights into RNA biology from an atlas of mammalian MRNA-binding proteins. Cell 149, 1393–1406. doi:10.1016/j.cell.2012.04.031
Cesaro, T., and Michiels, T. (2021). Inhibition of PKR by viruses. Front. Microbiol. 12, 757238. doi:10.3389/fmicb.2021.757238
Chawla, M., Oliva, R., Bujnicki, J. M., and Cavallo, L. (2015). An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic Acids Res. 43, 6714–6729. doi:10.1093/nar/gkv606
Chowdhury, M. N., and Jin, H. (2022). The RGG motif proteins: Interactions, functions, and regulations. WIREs RNA 14, e1748. doi:10.1002/wrna.1748
Chu, D., Barnes, D. J., and von der Haar, T. (2011). The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 39, 6705–6714. doi:10.1093/nar/gkr300
Chu, D., Kazana, E., Bellanger, N., Singh, T., Tuite, M. F., and von der Haar, T. (2014). Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34. doi:10.1002/embj.201385651
Clemens, M. J. (2001). Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J. Cell. Mol. Med. 5, 221–239. doi:10.1111/j.1582-4934.2001.tb00157.x
Damase, T. R., Sukhovershin, R., Boada, C., Taraballi, F., Pettigrew, R. I., and Cooke, J. P. (2021). The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 9, 628137. doi:10.3389/fbioe.2021.628137
Delehedde, C., Even, L., Midoux, P., Pichon, C., and Perche, F. (2021). Intracellular routing and recognition of lipid-based mRNA nanoparticles. Pharmaceutics 13, 945. doi:10.3390/pharmaceutics13070945
Deprey, K., Batistatou, N., and Kritzer, J. A. (2020). A critical analysis of methods used to investigate the cellular uptake and subcellular localization of RNA therapeutics. Nucleic Acids Res. 48, 7623–7639. doi:10.1093/nar/gkaa576
Dobrowolski, C., Paunovska, K., Schrader Echeverri, E., Loughrey, D., Da Silva Sanchez, A. J., Ni, H., et al. (2022). Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery. Nat. Nanotechnol. 17, 871–879. doi:10.1038/s41565-022-01146-9
Doma, M. K., and Parker, R. (2006). Endonucleolytic cleavage of eukaryotic MRNAs with stalls in translation elongation. Nature 440, 561–564. doi:10.1038/nature04530
Duncan, R., Milburn, S. C., and Hershey, J. W. (1987). Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 262, 380–388. doi:10.1016/S0021-9258(19)75938-9
Durbin, A. F., Wang, C., Marcotrigiano, J., and Gehrke, L. (2016). RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7, 008333–e916. doi:10.1128/mBio.00833-16
Eckmann, C. R., Rammelt, C., and Wahle, E. (2011). Control of poly(A) tail length: Control of poly(A) tail length. WIREs RNA 2, 348–361. doi:10.1002/wrna.56
Eyler, D. E., Franco, M. K., Batool, Z., Wu, M. Z., Dubuke, M. L., Dobosz-Bartoszek, M., et al. (2019). Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. U.S.A. 116, 23068–23074. doi:10.1073/pnas.1821754116
Fang, E., Liu, X., Li, M., Zhang, Z., Song, L., Zhu, B., et al. (2022). Advances in COVID-19 mRNA vaccine development. Sig Transduct. Target Ther. 7, 94. doi:10.1038/s41392-022-00950-y
Fenger-Grøn, M., Fillman, C., Norrild, B., and Lykke-Andersen, J. (2005). Multiple processing body factors and the ARE binding protein TTP activate MRNA decapping. Mol. Cell 20, 905–915. doi:10.1016/j.molcel.2005.10.031
Forster, J., Nandi, D., and Kulkarni, A. (2022). mRNA-carrying lipid nanoparticles that induce lysosomal rupture activate NLRP3 inflammasome and reduce mRNA transfection efficiency. Biomater. Sci. 10, 5566–5582. doi:10.1039/D2BM00883A
Gehring, N. H., Wahle, E., and Fischer, U. (2017). Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem. Sci. 42, 369–382. doi:10.1016/j.tibs.2017.02.004
Geisberg, J. V., Moqtaderi, Z., Fan, X., Ozsolak, F., and Struhl, K. (2014). Global analysis of MRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156, 812–824. doi:10.1016/j.cell.2013.12.026
Hajj, K. A., Ball, R. L., Deluty, S. B., Singh, S. R., Strelkova, D., Knapp, C. M., et al. (2019). Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small 15, 1805097. doi:10.1002/smll.201805097
Hassett, K. J., Benenato, K. E., Jacquinet, E., Lee, A., Woods, A., Yuzhakov, O., et al. (2019). Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. - Nucleic Acids 15, 1–11. doi:10.1016/j.omtn.2019.01.013
Hatit, M. Z. C., Lokugamage, M. P., Dobrowolski, C. N., Paunovska, K., Ni, H., Zhao, K., et al. (2022). Species-dependent in vivo mRNA delivery and cellular responses to nanoparticles. Nat. Nanotechnol. 17, 310–318. doi:10.1038/s41565-021-01030-y
Ho, J. J. D., Wang, M., Audas, T. E., Kwon, D., Carlsson, S. K., Timpano, S., et al. (2016). Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 14, 1293–1300. doi:10.1016/j.celrep.2016.01.036
Hou, X., Zaks, T., Langer, R., and Dong, Y. (2021). Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater 6, 1078–1094. doi:10.1038/s41578-021-00358-0
Hudson, G. A., Bloomingdale, R. J., and Znosko, B. M. (2013). Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 19, 1474–1482. doi:10.1261/rna.039610.113
Ishigaki, Y., Li, X., Serin, G., and Maquat, L. E. (2001). Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617. doi:10.1016/S0092-8674(01)00475-5
Jani, B., and Fuchs, R. (2012). <em>in vitro</em> Transcription and Capping of <em>Gaussia</em> Luciferase mRNA Followed by HeLa Cell Transfection. JoVE 26, 3702. doi:10.3791/3702
Jiang, Y., Lu, Q., Wang, Y., Xu, E., Ho, A., Singh, P., et al. (2020). Quantitating endosomal escape of a library of polymers for mRNA delivery. Nano Lett. 20, 1117–1123. doi:10.1021/acs.nanolett.9b04426
Juszkiewicz, S., Slodkowicz, G., Lin, Z., Freire-Pritchett, P., Peak-Chew, S.-Y., and Hegde, R. S. (2020). Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 9, e60038. doi:10.7554/eLife.60038
Karijolich, J., and Yu, Y.-T. (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398. doi:10.1038/nature10165
Karikó, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. doi:10.1016/j.immuni.2005.06.008
Katneni, U. K., Alexaki, A., Hunt, R. C., Hamasaki-Katagiri, N., Hettiarachchi, G. K., Kames, J. M., et al. (2022). Structural, functional, and immunogenicity implications of F9 gene recoding. Blood Adv. 6, 3932–3944. doi:10.1182/bloodadvances.2022007094
Kauffman, K. J., Mir, F. F., Jhunjhunwala, S., Kaczmarek, J. C., Hurtado, J. E., Yang, J. H., et al. (2016). Efficacy and immunogenicity of unmodified and pseudouridine-modified MRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78–87. doi:10.1016/j.biomaterials.2016.09.006
Khong, A., and Parker, R. (2020). The landscape of eukaryotic mRNPs. RNA 26, 229–239. doi:10.1261/rna.073601.119
Kierzek, E., Malgowska, M., Lisowiec, J., Turner, D. H., Gdaniec, Z., and Kierzek, R. (2014). The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42, 3492–3501. doi:10.1093/nar/gkt1330
Kirschman, J. L., Bhosle, S., Vanover, D., Blanchard, E. L., Loomis, K. H., Zurla, C., et al. (2017). Characterizing exogenous mRNA delivery, trafficking, cytoplasmic release and RNA–protein correlations at the level of single cells. Nucleic Acids Res. 45, e113. doi:10.1093/nar/gkx290
Landon, A. L., Muniandy, P. A., Shetty, A. C., Lehrmann, E., Volpon, L., Houng, S., et al. (2014). MNKs act as a regulatory switch for EIF4E1 and EIF4E3 driven MRNA translation in DLBCL. Nat. Commun. 5, 5413. doi:10.1038/ncomms6413
Lee, S. B., Melkova, Z., Yan, W., Williams, B. R. G., Hovanessian, A. G., and Esteban, M. (1993). The interferon-induced double-stranded RNA-activated human P68 protein kinase potently inhibits protein synthesis in cultured cells. Virology 192, 380–385. doi:10.1006/viro.1993.1048
Li, Q., Chan, C., Peterson, N., Hanna, R. N., Alfaro, A., Allen, K. L., et al. (2020). Engineering caveolae-targeted lipid nanoparticles to deliver mRNA to the lungs. ACS Chem. Biol. 15, 830–836. doi:10.1021/acschembio.0c00003
Liang, F., Lindgren, G., Lin, A., Thompson, E. A., Ols, S., Röhss, J., et al. (2017). Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 25, 2635–2647. doi:10.1016/j.ymthe.2017.08.006
Lin, T. A., Kong, X., Haystead, T. A. J., Pause, A., Belsham, G., Sonenberg, N., et al. (1994). PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266, 653–656. doi:10.1126/science.7939721
Lindsay, K. E., Bhosle, S. M., Zurla, C., Beyersdorf, J., Rogers, K. A., Vanover, D., et al. (2019). Visualization of early events in mRNA vaccine delivery in non-human primates via PET–CT and near-infrared imaging. Nat. Biomed. Eng. 3, 371–380. doi:10.1038/s41551-019-0378-3
Malone, R. W., Felgner, P. L., and Verma, I. M. (1989). Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. U.S.A. 86, 6077–6081. doi:10.1073/pnas.86.16.6077
Marinov, G. K., Williams, B. A., McCue, K., Schroth, G. P., Gertz, J., Myers, R. M., et al. (2014). From single-cell to cell-pool transcriptomes: Stochasticity in gene expression and RNA splicing. Genome Res. 24, 496–510. doi:10.1101/gr.161034.113
Maugeri, M., Nawaz, M., Papadimitriou, A., Angerfors, A., Camponeschi, A., Na, M., et al. (2019). Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 10, 4333. doi:10.1038/s41467-019-12275-6
Mockey, M., Gonçalves, C., Dupuy, F. P., Lemoine, F. M., Pichon, C., and Midoux, P. (2006). mRNA transfection of dendritic cells: Synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophysical Res. Commun. 340, 1062–1068. doi:10.1016/j.bbrc.2005.12.105
Monroe, J. G., Mitchell, L., Deb, I., Roy, B., Frank, A. T., and Koutmou, K. (2022). N1-Methylpseudouridine and pseudouridine modifications modulate MRNA decoding during translation. bioRxiv. doi:10.1101/2022.06.13.495988
Morais, P., Adachi, H., and Yu, Y.-T. (2021). The critical contribution of pseudouridine to mRNA COVID-19 vaccines. Front. Cell Dev. Biol. 9, 789427. doi:10.3389/fcell.2021.789427
Mu, X., Greenwald, E., Ahmad, S., and Hur, S. (2018). An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 46, 5239–5249. doi:10.1093/nar/gky177
Mu, X., and Hur, S. (2021). Immunogenicity of in vitro -transcribed RNA. Acc. Chem. Res. 54, 4012–4023. doi:10.1021/acs.accounts.1c00521
Mukai, H., Ogawa, K., Kato, N., and Kawakami, S. (2022). Recent advances in lipid nanoparticles for delivery of nucleic acid, mRNA, and gene editing-based therapeutics. Drug Metabolism Pharmacokinet. 44, 100450. doi:10.1016/j.dmpk.2022.100450
Muttach, F., Muthmann, N., and Rentmeister, A. (2017). Synthetic MRNA capping. Beilstein J. Org. Chem. 13, 2819–2832. doi:10.3762/bjoc.13.274
Ohto, T., Konishi, M., Tanaka, H., Onomoto, K., Yoneyama, M., Nakai, Y., et al. (2019). Inhibition of the inflammatory pathway enhances both the in vitro and in vivo transfection activity of exogenous in vitro-transcribed mRNAs delivered by lipid nanoparticles. Biol. Pharm. Bull. 42, 299–302. doi:10.1248/bpb.b18-00783
Park, J., Chang, J., Hwang, H. J., Jeong, K., Lee, H.-J., Ha, H., et al. (2021). The pioneer round of translation ensures proper targeting of ER and mitochondrial proteins. Nucleic Acids Res. 49, 12517–12534. doi:10.1093/nar/gkab1098
Patel, S., Ryals, R. C., Weller, K. K., Pennesi, M. E., and Sahay, G. (2019). Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release 303, 91–100. doi:10.1016/j.jconrel.2019.04.015
Paunovska, K., Sago, C. D., Monaco, C. M., Hudson, W. H., Castro, M. G., Rudoltz, T. G., et al. (2018). A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 18, 2148–2157. doi:10.1021/acs.nanolett.8b00432
Pichlmair, A., Schulz, O., Tan, C. P., Näslund, T. I., Liljeström, P., Weber, F., et al. (2006). RIG-I-Mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314, 997–1001. doi:10.1126/science.1132998
Pichon, X., Wilson, A. L., Stoneley, M., Bastide, A., A King, H., Somers, J., et al. (2012). RNA binding protein/RNA element interactions and the control of translation. CPPS 13, 294–304. doi:10.2174/138920312801619475
Plant, E. P., Nguyen, P., Russ, J. R., Pittman, Y. R., Nguyen, T., Quesinberry, J. T., et al. (2007). Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS ONE 2, e517. doi:10.1371/journal.pone.0000517
Poornima, G., Srivastava, G., Roy, B., Kuttanda, I. A., Kurbah, I., and Rajyaguru, P. I. (2021). RGG-motif containing mRNA export factor Gbp2 acts as a translation repressor. RNA Biol. 18, 2342–2353. doi:10.1080/15476286.2021.1910403
Reiser, A., Woschée, D., Mehrotra, N., Krzysztoń, R., Strey, H. H., and Rädler, J. O. (2019). Correlation of mRNA delivery timing and protein expression in lipid-based transfection. Integr. Biol. 11, 362–371. doi:10.1093/intbio/zyz030
Rejman, J., Bragonzi, A., and Conese, M. (2005). Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 12, 468–474. doi:10.1016/j.ymthe.2005.03.038
Sample, P. J., Wang, B., Reid, D. W., Presnyak, V., McFadyen, I. J., Morris, D. R., et al. (2019). Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 37, 803–809. doi:10.1038/s41587-019-0164-5
Sato, H., and Maquat, L. E. (2009). Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 23, 2537–2550. doi:10.1101/gad.1817109
Sayers, E. J., Peel, S. E., Schantz, A., England, R. M., Beano, M., Bates, S. M., et al. (2019). Endocytic profiling of cancer cell models reveals critical factors influencing LNP-mediated mRNA delivery and protein expression. Mol. Ther. 27, 1950–1962. doi:10.1016/j.ymthe.2019.07.018
Schwartz, S., Bernstein, D. A., Mumbach, M. R., Jovanovic, M., Herbst, R. H., León-Ricardo, B. X., et al. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of NcRNA and MRNA. Cell 159, 148–162. doi:10.1016/j.cell.2014.08.028
Shan, X., Chang, Y., and Glenn Lin, C. (2007). Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 21, 2753–2764. doi:10.1096/fj.07-8200com
Singh, G., Pratt, G., Yeo, G. W., and Moore, M. J. (2015). The clothes make the mRNA: Past and present trends in mRNP fashion. Annu. Rev. Biochem. 84, 325–354. doi:10.1146/annurev-biochem-080111-092106
Snieckute, G., Genzor, A. V., Vind, A. C., Ryder, L., Stoneley, M., Chamois, S., et al. (2022). Ribosome stalling is a signal for metabolic regulation by the ribotoxic stress response. Cell Metab. 34, 2036–2046.e8. doi:10.1016/j.cmet.2022.10.011
Svitkin, Y. V., Cheng, Y. M., Chakraborty, T., Presnyak, V., John, M., and Sonenberg, N. (2017). N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036. doi:10.1093/nar/gkx135
Svitkin, Y. V., Gingras, A. C., and Sonenberg, N. (2022). Membrane-dependent relief of translation elongation arrest on pseudouridine- and N 1-methyl-pseudouridine-modified MRNAs. Nucleic Acids Res. 50, 7202–7215. doi:10.1093/nar/gkab1241
Tanaka, M., Chock, P. B., and Stadtman, E. R. (2007). Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U.S.A. 104, 66–71. doi:10.1073/pnas.0609737104
Tarrant, D., and von der Haar, T. (2014). Synonymous codons, ribosome speed, and eukaryotic gene expression regulation. Cell. Mol. Life Sci. 71, 4195–4206. doi:10.1007/s00018-014-1684-2
Thess, A., Grund, S., Mui, B. L., Hope, M. J., Baumhof, P., Fotin-Mleczek, M., et al. (2015). Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464. doi:10.1038/mt.2015.103
Trepotec, Z., Geiger, J., Plank, C., Aneja, M. K., and Rudolph, C. (2019). Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting MRNA translation efficiency or half-life. RNA 25, 507–518. doi:10.1261/rna.069286.118
Uchihara, Y., Permata, T. B. M., Sato, H., Kawabata-Iwakawa, R., Katada, S., Gu, W., et al. (2022). DNA damage promotes HLA class I presentation by stimulating a pioneer round of translation-associated antigen production. Mol. Cell 82, 2557–2570.e7. doi:10.1016/j.molcel.2022.04.030
Uddin, M. N., and Roni, M. A. (2021). Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines 9, 1033. doi:10.3390/vaccines9091033
von der Haar, T., Gross, J. D., Wagner, G., and McCarthy, J. E. G. (2004). The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11, 503–511. doi:10.1038/nsmb779
Weinstein-Marom, H., Hendel, L., Laron, E. A., Sharabi-Nov, A., Margalit, A., and Gross, G. (2019). MHC-I presentation of peptides derived from intact protein products of the pioneer round of translation. FASEB J. 33, 11458–11468. doi:10.1096/fj.201802717RRR
Whitley, J., Zwolinski, C., Denis, C., Maughan, M., Hayles, L., Clarke, D., et al. (2022). Development of mRNA manufacturing for vaccines and therapeutics: mRNA platform requirements and development of a scalable production process to support early phase clinical trials. Transl. Res. 242, 38–55. doi:10.1016/j.trsl.2021.11.009
Wittrup, A., Ai, A., Liu, X., Hamar, P., Trifonova, R., Charisse, K., et al. (2015). Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870–876. doi:10.1038/nbt.3298
Wolff, J. A., Malone, R. W., Williams, P., Chong, W., Acsadi, G., Jani, A., et al. (1990). Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468. doi:10.1126/science.1690918
Wu, C. C.-C., Peterson, A., Zinshteyn, B., Regot, S., and Green, R. (2020). Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182, 404–416. doi:10.1016/j.cell.2020.06.006
Xia, X. (2021). Detailed dissection and critical evaluation of the pfizer/BioNTech and Moderna MRNA vaccines. Vaccines 9, 734. doi:10.3390/vaccines9070734
Zarnack, K., Balasubramanian, S., Gantier, M. P., Kunetsky, V., Kracht, M., Schmitz, M. L., et al. (2020). Dynamic mRNP remodeling in response to internal and external stimuli. Biomolecules 10, 1310. doi:10.3390/biom10091310
Zhang, M., Hussain, A., Yang, H., Zhang, J., Liang, X.-J., and Huang, Y. (2022). mRNA-based modalities for infectious disease management. Nano Res. 16, 672–691. doi:10.1007/s12274-022-4627-5
Keywords: RNA therapeutic, translation, protein synthesis, translational control, RNA vaccines
Citation: von der Haar T, Mulroney TE, Hedayioglu F, Kurusamy S, Rust M, Lilley KS, Thaventhiran JE, Willis AE and Smales CM (2023) Translation of in vitro-transcribed RNA therapeutics. Front. Mol. Biosci. 10:1128067. doi: 10.3389/fmolb.2023.1128067
Received: 20 December 2022; Accepted: 30 January 2023;
Published: 08 February 2023.
Edited by:
Susan Gerarda Campbell, Sheffield Hallam University, United KingdomReviewed by:
Daniel L. Kiss, Houston Methodist Research Institute, United StatesAnton A. Komar, Cleveland State University, United States
Tracy Nissan, Stockholm University, Sweden
Copyright © 2023 von der Haar, Mulroney, Hedayioglu, Kurusamy, Rust, Lilley, Thaventhiran, Willis and Smales. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tobias von der Haar, VC52b24tZGVyLUhhYXJAa2VudC5hYy51aw==
 Tobias von der Haar
Tobias von der Haar Thomas E. Mulroney2
Thomas E. Mulroney2 Maria Rust
Maria Rust Kathryn S. Lilley
Kathryn S. Lilley James E. Thaventhiran
James E. Thaventhiran C. Mark Smales
C. Mark Smales